Abstract
Background
Increasing evidence accumulates on the central involvement of microRNAs (miRNAs) in disease pathophysiology. We identified distinctly deregulated miRNAs in human renal allograft biopsies from patients undergoing acute cellular rejection, antibody-mediated rejection (ABMR), and delayed graft function (DGF).
Methods
Sixty-five posttransplantation kidney biopsy samples covering 41 cases with acute rejection (15 vascular rejection, 15 interstitial rejection, and 11 ABMR), 14 DGF cases, and 10 protocol biopsies serving as controls were analyzed using the Affymetrix GeneChip miRNA Array. Differentially regulated miRNAs were identified by Student’s t test and Bonferroni correction. Target genes for the set of miRNAs were retrieved from miRTarBase (experimentally verified targets) as well as by applying the target prediction routines DIANAmT, miRanda, and Targetscan.
Results
Patients with acute cellular rejection, ABMR, and DGF discriminate from the control group (protocol biopsies) in unsupervised clustering of miRNA profiles, clearly identifying deregulated miRNAs in rejection and DGF. Angiogenesis, apoptosis, and transforming growth factor-β signaling were identified as relevant pathways in ischemic response following an integrative analysis of miRNA targets and mRNA expression profiles. Inflammation by chemokine and cytokine signaling, T-cell activation, and B-cell activation were identified as relevant in acute rejection accordingly.
Conclusion
These data suggest that distinct miRNA signatures playing a role in specific biological pathways discriminate acute cellular and humoral rejection and DGF. This finding serves as valuable tool for a rational selection of diagnostic, prognostic, and potentially therapeutic molecular targets of posttransplantation events.
Keywords: Acute rejection, Antibody-mediated rejection, Delayed graft function, miRNA, Integrative bioinformatics
Postischemic injury leading to delayed allograft function and early alloimmune responses such as cellular and humoral rejection are among the main contributors to reduced long-term allograft function and survival (1). However, knowledge on the molecular regulation and pathophysiology of these events is still limited. Omics, and in particular transcriptomics, have brought forward deregulated protein coding transcripts allowing an assessment of molecular processes and pathways characterizing delayed graft function (DGF) and rejection (2–10). Transcriptomics studies have been further complemented by proteomics covering tissue (11) as well as urine (12). Next to transcripts of protein coding genes and proteins, microRNAs (miRNAs) were recently identified as powerful regulatory elements specifically determining transcriptional and translational control.
miRNAs are a class of small, noncoding, 18- to 24-nucleotide RNAs with diverse functions, including translational control of mRNAs for a wide variety of proteins (13).
The impact of miRNA regulation involved in allograft function and immunity has not been investigated thoroughly, although it has long been acknowledged that posttranscriptional regulation occurs on the cellular level in the recipients’ immune system and the donor organ (14). miRNAs have the potency to change the abundance of target proteins via silencing their transcripts. As an example, Godwin et al. (15) showed that unique miRNA signatures determined the course of renal injury after warm ischemia in mice. Anglicheau et al. (16) recently identified a set of miRNAs being predictive of acute cellular rejection (AREJ) of human renal allografts. However, antibody-mediated rejection (ABMR) and DGF in human posttransplantation kidney biopsy samples have not been investigated so far on the level of miRNAs.
This study therefore seeks to elucidate the regulation of miRNAs in the development of DGF and acute cellular and humoral rejection episodes in renal grafts after kidney transplantation. We used miRNA expression profiling to identify miRNAs deregulated in postischemic acute renal transplant failure and acute rejection episodes after renal transplantation when compared with protocol biopsy (PBx) samples. Furthermore, we identified downstream regulatory pathways of these clinical entities to uncover potential biomarkers and therapeutic targets.
RESULTS
Patients
miRNA profiles of 65 renal allograft biopsies obtained from 63 adult recipients were investigated in this study. Demographic data on transplant donors and recipients are provided in Table 1.
TABLE 1.
Demographic data of kidney donors and recipients
| Variable | PBx | DGF | P | AREJ | P | ABMR | P |
|---|---|---|---|---|---|---|---|
| Number of biopsies | 10 | 14 | N/A | 30 | N/A | 11 | N/A |
| Number of donors | 10 | 13 | N/A | 29 | N/A | 11 | N/A |
| Donor age, yr | 59.5 (54.0; 67.0) | 51.0 (46.0; 64.0) | 0.42 | 54.0 (48.0; 60.0) | 0.18 | 53.0 (44.0; 64.0) | 0.40 |
| Donor sex, F/M | 6/4 | 5/8 | 0.41 | 12/17 | 0.47 | 4/7 | 0.37 |
| Donor source, living/deceased | 6/4 | 0/13 | <0.01a | 3/26 | <0.01a | 0/11 | 0.01a |
| Donor last creatinine, mg/dL | 0.82 (0.69; 0.92) | 0.95 (0.7; 1.4) | 0.17 | 1.00 (0.82; 1.17) | 0.04 | 1.04 (0.7; 1.4) | 0.14 |
| Number of recipients | 10 | 13 | N/A | 29 | N/A | 11 | N/A |
| Recipient sex, F/M | 3/7 | 2/11 | 0.62 | 7/22 | 0.70 | 5/6 | 0.66 |
| Recipient age, yr | 50.8 (41.3; 59.1) | 53.5 (47.2; 65.1) | 0.34 | 49.1 (36.7; 61.3) | 0.86 | 59.9 (47.0; 65.3) | 0.38 |
| Transplant number, 1/2/3/4/5 | 10/0/0/0/0 | 10/1/1/0/1 | 1.00a | 26/3/0/0/0 | 0.56a | 7/2/2/0/0 | 0.09a |
| Cold ischemic time, hr | 1.0 (0.0; 8) | 11.5 (10; 17.5) | <0.01 | 13 (7; 16) | <0.01 | 20 (15; 22) | <0.01 |
| PRA latest, % | 0.0 (0.0; 0.0) | 0.0 (0.0; 5.0) | 0.02 | 3.0 (0.0; 4.0) | <0.01 | 40.0 (3.0; 60.0) | <0.01 |
| Sum of HLA mismatches, 0/1/2/3/4/5 | 0/1/2/4/2/1 | 3/3/1/4/1/1 | 0.57 | 2/2/6/7/10/2 | 0.87 | 0/0/1/5/5/0 | 0.51 |
| Immunosuppression, CNI/else | 9/1 | 12/1 | 1.00a | 23/5 | 0.31a | 11/0 | 1.00a |
| Induction therapy, none/IL-2/ATG | 6/3/1 | 11/1/1 | 0.26a | 23/4/1 | 0.50a | 6/3/2 | 1.00a |
| Time of biopsy, days after transplantation | 104.5 (102; 109.5) | 7 (2; 8) | 16.5 (7; 44.5) | 14 (12; 55.5) |
Fisher’s exact test.
Continuous data are provided as median and interquartile range, and categorical data are shown as counts. P values refer to the comparison with PBx.
ABMR, antibody-mediated rejection; AREJ, acute cellular rejection; ATG, antithymocyte globulin; CNI, calcineurin inhibitors; DGF, delayed graft function; HLA, human leukocyte antigen; IL, interleukin; N/A, not applicable; PBx, protocol biopsy; PRA, panel reactive antibody.
The control group (PBx) consisted mainly of live donors and served as example of optimal allograft function and morphology. Cold ischemic time and latest determined panel reactive antibody concentrations were significantly lower in the PBx group compared with the DGF, AREJ, and ABMR groups. Donor last creatinine values were lower in the control group compared with the AREJ group. All other demographic values and clinical parameters did not show significant differences between groups.
MicroRNA Profiling
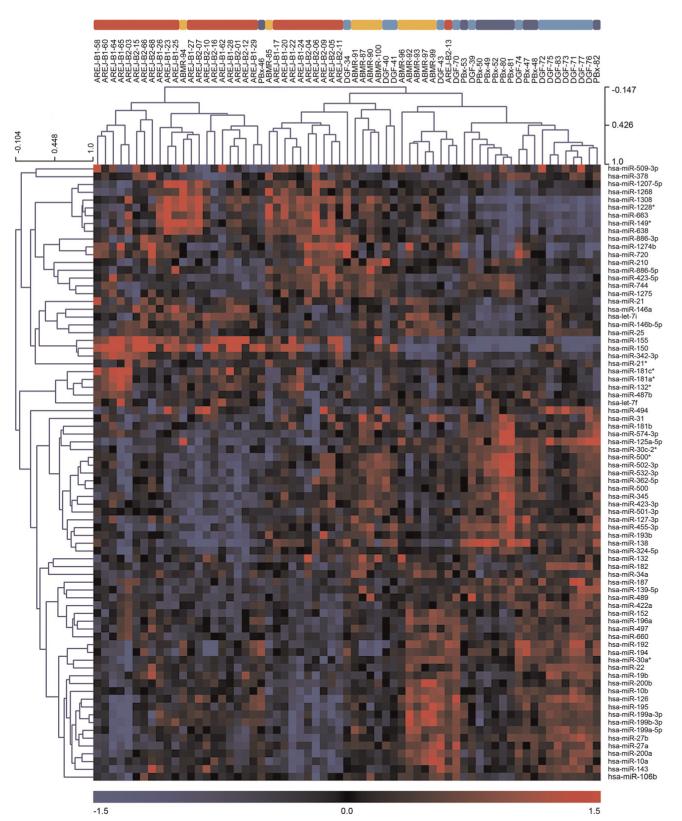
Correlation of the miRNA profiles in the control biopsy group showed no clustering of live versus deceased donors, suggesting no selection bias due to donor status. Figure 1 shows the unsupervised hierarchical clustering of the 65 miRNA profiles. Patients developing rejection (30 samples for cellular rejection and 11 samples for humoral rejection) clearly separated from the control group (10 samples). Accordingly, the DGF kidneys (14 samples) showed a distinctly different miRNA profile compared with the control biopsies.
FIGURE 1.
Unsupervised clustering of miRNAs with high variance (top 20%, 80 miRNAs); AREJ (red), ABMR (orange), DGF (light blue), and PBx (blue). The heat map was produced using clustering of rows (miRNA expression values) and columns (samples) of the data matrix using the complete linkage algorithm and Pearson correlation. (left) miRNA clustering tree; (top) sample clustering tree. The samples are clustering broadly into two groups: rejection and no rejection. (bottom) The color scale illustrates the relative expression level of the indicated miRNA across all samples: (red) expression>0 and (blue) expression<0. ABMR, antibody-mediated rejection; AREJ, acute cellular rejection; DGF, delayed graft function; miRNA, microRNA; PBx, protocol biopsy.
The sets of significantly differentially regulated miRNAs comparing controls and DGF and rejection, respectively, are provided in Table 2. DGF kidneys exhibited an activation of seven miRNAs (miR-182, miR-106b, miR-20a, miR-21*, miR-18a, miR-17, and miR-106a). miR-182 and miR-21* were also identified as up-regulated in ABMR, indicating that processes involved in DGF may also be relevant in the pathogenesis of ABMR. Deregulation of miR-182 and miR-21* were confirmed in DGF and ABMR kidneys by quantitative real-time poly-merase chain reaction (qRT-PCR; see Figure S1 and Figure S3, SDC, http://links.lww.com/TP/A774).
TABLE 2.
Significant differentially regulated miRNAs (t test with Bonferroni correction) ranked by fold change
| miRNA | miRBase accession | Fold change | Adjusted P | |
|---|---|---|---|---|
| DGF vs. PBx | ||||
| Up-regulated | hsa-miR-182 | MIMAT0000259 | 1.9 | 0.005 |
| hsa-miR-106b | MIMAT0000386 | 1.66 | 0.044 | |
| hsa-miR-20a | MIMAT0000075 | 1.57 | 0.017 | |
| hsa-miR-21* | MIMAT0004494 | 1.54 | 0.02 | |
| hsa-miR-18a | MIMAT0000072 | 1.49 | 0.016 | |
| hsa-miR-17 | MIMAT0000070 | 1.32 | 0.03 | |
| hsa-miR-106a | MIMAT0000103 | 1.26 | 0.042 | |
| AREJ vs. PBx | ||||
| Up-regulated | hsa-miR-150 | MIMAT0000451 | 6.46 | 0.009 |
| hsa-miR-155 | MIMAT0000646 | 4.96 | <0.001 | |
| hsa-miR-663a | MIMAT0003326 | 2.27 | 0.001 | |
| hsa-miR-638 | MIMAT0003308 | 2.02 | 0.011 | |
| Down-regulated | hsa-miR-138 | MIMAT0000430 | 0.31 | 0.013 |
| hsa-miR-125a | MIMAT0000443 | 0.38 | <0.001 | |
| hsa-miR-455 | MIMAT0004784 | 0.42 | 0.002 | |
| hsa-miR-30c-2* | MIMAT0004550 | 0.49 | <0.001 | |
| hsa-miR-574-3p | MIMAT0003239 | 0.49 | 0.042 | |
| hsa-miR-502-3p | MIMAT0004775 | 0.51 | 0.031 | |
| hsa-miR-181b | MIMAT0000257 | 0.51 | 0.017 | |
| hsa-miR-99b | MIMAT0000689 | 0.52 | <0.001 | |
| hsa-miR-139-5p | MIMAT0000250 | 0.53 | 0.001 | |
| hsa-miR-27b | MIMAT0000419 | 0.57 | 0.01 | |
| hsa-miR-424* | MIMAT0004749 | 0.59 | 0.004 | |
| hsa-miR-193b | MIMAT0002819 | 0.61 | 0.028 | |
| hsa-miR-99b* | MIMAT0004678 | 0.63 | 0.002 | |
| hsa-let-7b | MIMAT0000063 | 0.64 | 0.002 | |
| hsa-miR-181a | MIMAT0000256 | 0.65 | 0.046 | |
| hsa-miR-23b | MIMAT0000418 | 0.68 | <0.001 | |
| hsa-miR-361-5p | MIMAT0000703 | 0.69 | 0.003 | |
| hsa-miR-125b-2* | MIMAT0004603 | 0.70 | 0.033 | |
| ABMR vs. PBx | ||||
| Up-regulated | hsa-miR-663 | MIMAT0003326 | 2.56 | 0.001 |
| hsa-miR-146b-5p | MIMAT0002809 | 2.35 | <0.001 | |
| hsa-miR-1228 | MIMAT0005582 | 2.10 | 0.006 | |
| hsa-let-7i | MIMAT0000415 | 2.09 | 0.023 | |
| hsa-miR-21* | MIMAT0004494 | 1.59 | 0.008 | |
| hsa-miR-182 | MIMAT0000259 | 1.58 | 0.003 |
ABMR, antibody-mediated rejection; AREJ, acute cellular rejection; DGF, delayed graft function; miRNA, microRNA; PBx, protocol biopsy.
Biopsies with documented AREJ showed activation of miR-150, miR-155, miR-663, and miR-638 as well as suppression of 18 miRNAs (Table 2).
ABMR separated from cellular rejection and the control group by activation of miR-146-5p, miR-1228, let-7i, miR-21*, and miR-182. miR-155, miR-125a, and miR-146b were validated by qRT-PCR (see Figure S2 and Figure S3, SDC, http://links.lww.com/TP/A774).
Comparative Pathway Analyses
The workflow of comparative pathway analysis is illustrated in Figure 2, and affected pathways in acute rejection and DGF are provided in Table 3. Acute rejection was characterized by enriched pathways of immune response, inflammation mediated by chemokine and cytokine signaling, and T-cell as well as B-cell activation. DGF, on the other hand, was characterized by pathways of angiogenesis, proliferation, and apoptosis.
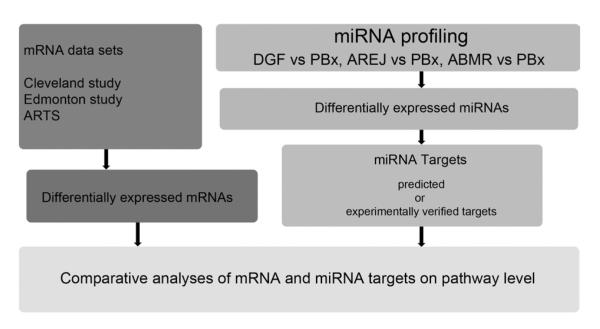
FIGURE 2.
Schematic representation of our combinatorial approach for identifying miRNAs, miRNA targets, genes, and molecular pathways in DGF and acute rejection. Differentially expressed miRNAs were obtained by comparing DGF, AREJ, and ABMR compared with allografts with normal function (PBx). miRNA target genes were predicted by DIANAmT, miRanda, and Targetscan (42–44), and experimentally validated targets were identified by using miRTarBase (45). Posttransplantation gene expression profiles were derived from the Cleveland study (2) and from the Edmonton study (6). ARTS was obtained from analysis of an independent gene expression data set of renal allograft biopsies with acute rejection (8). Comparison of miRNA target lists and gene lists were performed to derive set of pathways core to acute rejection and DGF. ABMR, antibody-mediated rejection; AREJ, acute cellular rejection; ARTS, acute rejection transcript set; DGF, delayed graft function; miRNA, microRNA; PBx, protocol biopsy.
TABLE 3.
Pathway enrichment analysis of predicted and validated miRNA targets and differentially regulated transcripts in DGF and acute rejection
| DGF |
Predicted targets |
Experimentally validated targets |
Cleveland data set |
Cleveland data set (ATN) |
|---|---|---|---|---|
| Pathways | P | P | P | P |
| Angiogenesis | <0.001 | <0.001 | 0.015 | 0.035 |
| Apoptosis signaling pathway | <0.001 | 0.008 | 0.014 | 0.011 |
| TGF-ß signaling pathway | <0.001 | <0.001 | 0.001 | 0.001 |
| Endothelin signaling pathway | 0.001 | 0.038 | 0.002 | 0.002 |
| VEGF signaling pathway | 0.011 | <0.001 | 0.028 | 0.016 |
| PDGF signaling pathway | <0.001 | 0.002 | 0.025 | 0.037 |
| Acute rejection |
Predicted targets |
Experimentally validated targets |
Cleveland data set (acute rejection) |
ARTS |
|---|---|---|---|---|
| Pathways | P | P | P | P |
| EGFR signaling pathway | <0.001 | 0.014 | 0.001 | 0.012 |
| Inflammation mediated by chemokine and cytokine signaling pathway |
<0.001 | <0.001 | 0.039 | <0.001 |
| T-cell activation | 0.003 | 0.044 | <0.001 | <0.001 |
| Cytoskeletal regulation by Rho GTPase | 0.001 | 0.011 | 0.005 | 0.047 |
| B-cell activation | 0.013 | 0.020 | 0.031 | 0.034 |
ARTS, acute rejection transcript set; ATN, acute tubular necrosis; DGF, delayed graft function; EGFR, epidermal growth factor receptor; miRNA, microRNA; PDGF, platelet-derived growth factor; TGF, transforming growth factor; VEGF, vascular endothelial growth factor.
The numbers of predicted and experimentally verified targets of the differentially regulated miRNAs and mRNAs are provided in Tables S1 and S2, respectively (see SDC, http://links.lww.com/TP/A774).
The comparison of acute cellular and humoral rejection yielded as relevant pathways inflammation, chemokine and cytokine signaling, apoptosis signaling, and interleukin signaling in both rejection types. Nicotinic acetylcholine receptor signaling and cytoskeletal regulation by Rho GTPase were only enriched in AREJ (see Table S3, SDC, http://links.lww.com/TP/A774).
DISCUSSION
The present study provides evidence for specific miRNAs regulation in processes after renal transplantation, including DGF, AREJ, and acute humoral rejection. Similar discrimination was shown previously on the mRNA level of biopsies with acute rejection as example (9), allowing a descriptive analysis of identified features in the context of biology and pathogenesis of renal allograft rejection.
Because hierarchical clustering of miRNA profiles by itself does not allow accounting for the various mRNA targets of each miRNA, we used pathway enrichment analysis for analyzing the impact of differentially regulated miRNAs on the expression level of mRNAs, thereby allowing a functional interpretation on the molecular process and pathway level (17).
We identified seven miRNAs in DGF kidneys, which are all involved in cell death and proliferation, specifically including miR-21*. The major sequence of miR-21 is one of the most discussed miRNAs in the literature and is up-regulated in various pathophysiologic entities including tumors, fibrosis, and ischemic injury (15, 18, 19).
In the present study, miR-182 was found to be differentially regulated in both DGF and ABMR. miR-182 was shown to be up-regulated in ischemic-reperfusion injury (IRI) and in a PT-Dicer−/− IRI mouse model (20). miR-182 is found to be activated by interleukin-2 and STAT5 and inhibits FOXO1, leading to clonal expansion of T helper cells (21). miR-182 was recently identified as key regulator and potential biomarker of heart allograft rejection in mice (22), and miR-182 expression increases early after ischemic preconditioning in brain (23). Further, in a natural model of ischemic tolerance, miR-182 was significantly decreased in brains of ground squirrels during hibernation torpor (24). These analogies indicate that miR-182 may play a critical role in the regulation of both ischemic and immunologic injuries.
In AREJ, four miRNAs were found to be significantly up-regulated. miR-150 plays an important role in B-cell development and controls c-Myb expression in vivo (25–27). miR-155 is a key player in adaptive immunity and T-cell–mediated antibody response, and T-cell receptor stimulation is known to induce miR-155 expression (25, 26). Ni et al. (28) found that a high level of miR-663 stimulated by oscillatory shear stress initiates an inflammatory response of endothelial cells. Both miR-638 and miR-663 were found to be up-regulated in PBMCs as well as in kidney biopsies of patients with lupus nephritis (29, 30), indicating a high inflammatory state in kidney tissue. miR-663, miR-30c, miR-139, and let-7b were also associated with chronic allograft dysfunction as investigated by Scian et al. (31).
In a recent study by Anglicheau et al. (16), 53 significantly differentially regulated miRNAs (P<0.05) were identified in a training set of four normal and three acute rejection biopsies. Six miRNAs identified in the present work (miR-155, miR-125a, miR-30c, miR-27b, miR-193b, and miR-125b) are also quoted in this set.
In ABMR, six up-regulated miRNAs could be identified. miR-21* and miR-182 were also overexpressed in DGF, miR-663 was also up-regulated in AREJ, and miR-146b could also be identified by Anglicheau et al. in biopsies developing acute rejection. The concordance of these miRNAs indicates strongly that processes involved in DGF and AREJ are also important in the pathogenesis of ABMR.
Pathway enrichment analysis of differentially regulated mRNA and miRNA targets revealed biological processes relevant for acute rejection and injury.
Recent studies have shown that angiogenesis, the vascular endothelial growth factor signaling pathway, and the endothelin signaling pathway play an important role in IRI (32, 33). Transforming growth factor-β signaling up-regulates miR-21 expression (15) and is also known to be involved in ischemic injury (34). The platelet-derived growth factor signaling pathway was recently identified as a key pathway affected after ischemic kidney injury. Platelet-derived growth factor expression is increased in tubules, endothelium, and macrophages after injury (35).
Central pathways in the pathophysiology of acute rejection are inflammation by chemokine and cytokine signaling pathway, T-cell activation, and B-cell activation (36), and distinct identification of these pathways demonstrates the value of the combinatorial approach involving miRNA targets and mRNA expression profiles on the level of pathway analysis.
Certainly, there are still challenges and limitations in the interpretation of the given kidney biopsy miRNA profiles, specifically the incomplete human miRNA list and their experimentally validated target lists, only partially compensated by target prediction models (13). For handling the significant false-positive rate on miRNA target predictions, a further interpretation of target lists on the level of processes and pathways, as also applied in this work, appears a promising strategy (37, 38).
In summary, we identified the central role of miRNA regulation of key posttransplantation events such as acute rejection and DGF, and further studies will investigate the utility of miRNA candidates as biomarkers for these clinical entities. Ultimately, modulations of these pathways by miRNA therapeutics such as antagomiRs or mimics might offer a novel treatment option in transplantation.
MATERIALS AND METHODS
Renal Transplant Biopsy Specimen
Biopsy specimens included in this study were derived from the Vienna Renal Biopsy Registry at the Clinical Institute of Pathology of the Medical University of Vienna (H.R.). All biopsies were obtained between 2003 and 2009 and were treated following a homogenous, standardized protocol. In brief, needle biopsies were fixed in buffered 4.5% formalin at pH 7 for a minimum of 24 hr, dehydrated, and paraffin embedded according to standard pathology procedures. The study protocol was approved by the institutional review board (Ethical Committee of the Medical University of Vienna # EK-343/2010, to be found at http://ohrp.cit.nih.gov/search).
Histopathologic diagnoses were completed according to the updated Banff ‘07 criteria by H.R. (39). The AREJ sample set included AREJ, BANFF-1, and BANFF-2, and the ABMR sample set included biopsies of subjects with rapidly increasing creatinine and positive C4d staining and morphologic evidence of humoral tissue injury. The DGF sample set was defined by acute tubular necrosis without rejection and the need of more than one dialysis within the first week after transplantation, and the control group (PBx) consisted of biopsies without any sign of morphological damage (40, 41).
Sample size calculation was based on reference data, indicating a number of 10 biopsies in each group as sufficient to detect a difference in mean expression of one (SD, 0.6) with 0.8 power at an adjusted P<0.05 using Bonferroni correction for multiple testing.
MicroRNA Profiling
RNA from formalin-fixed, paraffin-embedded tissue was shown as suitable source for miRNA expression profiling (15). Total RNA from 10 slides (5 μm thick) was isolated and purified using miRNeasy FFPE Kit (Qiagen, Valencia, CA). Affymetrix GeneChip miRNA Arrays holding 904 unique small RNA sequences based on the Sanger miRBASE version 11 were used for miRNA profiling. Total RNA (1 μg/sample) was labeled using the Flash-Tag RNA Labeling Kit (Genisphere, Hatfield, PA) and hybridized to the arrays as described by the manufacturer (Affymetrix, Santa Clara, CA).
Affymetrix data were preprocessed, normalized, and summarized using the robust multiaverage method with quantile normalization and annotated using the corresponding annotation file (.cdf ). The unpaired Students t-test and Bonferroni correction (adjusted α<0.05) was used to discover differentially regulated miRNAs between groups. Microarray data are available in the Gene Expression Omnibus at National Center for Biotechnology Information with the identification GSE30282 and supplementary information can be found at our homepage at http://www.meduniwien.ac.at/nephrogene.
MicroRNA Target Prediction, Functional Annotation, and Pathway Enrichment Analysis
Three algorithms were used to predict miRNA targets, namely, DIANAmT, miRanda, and Targetscan (42–44). Only miRNA targets identified by all three prediction algorithms were included in further analysis. Additionally, experimentally validated targets were extracted for identified miRNAs from miRTarBase (45).
Target genes were subsequently analyzed with respect to their molecular function, associated biological processes, and subcellular location using gene ontology terms as provided by the Gene Ontology Consortium (46). Functional grouping of genes was based on gene ontology terms and Protein Analysis Through Evolutionary Relationships ontologies (47, 48). Target genes of differentially regulated miRNAs as well as differentially regulated mRNAs were further analyzed via pathway enrichment analysis using the Protein Analysis Through Evolutionary Relationships platform as provided at www.pantherdb.org (47).
The workflow for comparative analysis of miRNA and mRNA data sets and qRT-PCR validation can be found in Figure 2 and in the supplemental digital content (see SDC, http://links.lww.com/TP/A774).
Statistical Analysis of Clinical Data
Statistical assessment of the AREJ, the ABMR, the DGF, and the PBx sample set used for deriving miRNA expression profiles was performed by using SAS for Windows 9.2 (SAS Institute, Cary, NC).
Continuous data were analyzed by analysis of variance or Wilcoxon rank-sum test. Categorical data were evaluated by chi-square tests or Fisher’s exact tests where appropriate. P<0.05 was considered statistically significant.
Supplementary Material
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
Acknowledgments
This study was supported by a grant from the Austrian Science Fund (FWF P-21436 to R.O.).
Footnotes
The authors declare no conflicts of interest.
J.W. wrote the article, performed the microarray experiments and bioinformatical analysis, and collected the clinical data. H.R. evaluated and selected the renal biopsies and the reviewed article. P.P. designed the project and supervised the bioinformatical analysis. A.K. performed the statistical analysis and the reviewed article. A.S. designed the project and reviewed the article. F.M. reviewed the article. B.M. supervised the bioinformatical analysis and reviewed article. R.O. designed and supervised the project and critically reviewed the article.
Additional information for reviewing purposes: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=thyptkoqamgkkrc&acc=GSE30282.
REFERENCES
- 1.Chang SH, Russ GR, Chadban SJ, et al. Trends in kidney transplantation in Australia and New Zealand, 1993–2004. Transplantation. 2007;84:611. doi: 10.1097/01.tp.0000280553.23898.ef. [DOI] [PubMed] [Google Scholar]
- 2.Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauser P, Schwarz C, Mitterbauer C, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Lab Invest. 2004;84:353. doi: 10.1038/labinvest.3700037. [DOI] [PubMed] [Google Scholar]
- 4.Kainz A, Mitterbauer C, Hauser P, et al. Alterations in gene expression in cadaveric vs. live donor kidneys suggest impaired tubular counterbalance of oxidative stress at implantation. Am J Transplant. 2004;4:1595. doi: 10.1111/j.1600-6143.2004.00554.x. [DOI] [PubMed] [Google Scholar]
- 5.Mengel M, Chang J, Kayser D, et al. The molecular phenotype of 6-week protocol biopsies from human renal allografts: reflections of prior injury but not future course. Am J Transplant. 2011;11:708. doi: 10.1111/j.1600-6143.2010.03339.x. [DOI] [PubMed] [Google Scholar]
- 6.Mueller TF, Einecke G, Reeve J, et al. Microarray analysis of rejection in human kidney transplants using pathogenesis-based transcript sets. Am J Transplant. 2007;7:2712. doi: 10.1111/j.1600-6143.2007.02005.x. [DOI] [PubMed] [Google Scholar]
- 7.Perco P, Kainz A, Wilflingseder J, et al. Histogenomics: association of gene expression patterns with histological parameters in kidney biopsies. Transplantation. 2009;87:290. doi: 10.1097/TP.0b013e318191b4c0. [DOI] [PubMed] [Google Scholar]
- 8.Saint-Mezard P, Berthier CC, Zhang H, et al. Analysis of independent microarray datasets of renal biopsies identifies a robust transcript signature of acute allograft rejection. Transpl Int. 2009;22:293. doi: 10.1111/j.1432-2277.2008.00790.x. [DOI] [PubMed] [Google Scholar]
- 9.Sarwal M, Chua MS, Kambham N, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 10.Wilflingseder J, Kainz A, Muhlberger I, et al. Impaired metabolism in donor kidney grafts after steroid pretreatment. Transpl Int. 2010;23:796. doi: 10.1111/j.1432-2277.2010.01053.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen G, Huang JB, Mi J, et al. Characterization of acute renal allograft rejection by proteomic analysis of renal tissue in rat. Mol Biol Rep. 2012;39:1315. doi: 10.1007/s11033-011-0864-5. [DOI] [PubMed] [Google Scholar]
- 12.Srivastava M, Eidelman O, Torosyan Y, et al. Elevated expression levels of ANXA11, integrins beta3 and alpha3, and TNF-alpha contribute. Proteomics Clin Appl. 2011;5:311. doi: 10.1002/prca.201000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fessler BJ, Paliogianni F, Hama N, et al. Glucocorticoids modulate CD28 mediated pathways for interleukin 2 production in human T cells: evidence for posttranscriptional regulation. Transplantation. 1996;62:1113. doi: 10.1097/00007890-199610270-00016. [DOI] [PubMed] [Google Scholar]
- 15.Godwin JG, Ge X, Stephan K, et al. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci U S A. 2010;107:14339. doi: 10.1073/pnas.0912701107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A. 2009;106:5330. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 18.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 19.Nair VS, Maeda LS, Ioannidis JP. Clinical outcome prediction by microRNAs in human cancer: a systematic review. J Natl Cancer Inst. 2012;104:528. doi: 10.1093/jnci/djs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Q, Bhatt K, He HZ, et al. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2010;21:756. doi: 10.1681/ASN.2009070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stittrich AB, Haftmann C, Sgouroudis E, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 22.Wei L, Wang M, Qu X, et al. Differential expression of microRNAs during allograft rejection. Am J Transplant. 2012;12:1113. doi: 10.1111/j.1600-6143.2011.03958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee ST, Chu K, Jung KH, et al. MicroRNAs induced during ischemic preconditioning. Stroke. 2010;41:1646. doi: 10.1161/STROKEAHA.110.579649. [DOI] [PubMed] [Google Scholar]
- 24.Lee YJ, Johnson KR, Hallenbeck JM. Global protein conjugation by ubiquitin-like-modifiers during ischemic stress is regulated by microRNAs and confers robust tolerance to ischemia. PLoS One. 2012;7:e47787. doi: 10.1371/journal.pone.0047787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris A, Krams SM, Martinez OM. MicroRNAs as immune regulators: implications for transplantation. Am J Transplant. 2010;10:713. doi: 10.1111/j.1600-6143.2010.03032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 27.Zhou B, Wang S, Mayr C, et al. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni CW, Qiu H, Jo H. MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells. Am J Physiol Heart Circ Physiol. 2011;300:H1762. doi: 10.1152/ajpheart.00829.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai Y, Sui W, Lan H, et al. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int. 2009;29:749. doi: 10.1007/s00296-008-0758-6. [DOI] [PubMed] [Google Scholar]
- 30.Te JL, Dozmorov IM, Guthridge JM, et al. Identification of unique microRNA signature associated with lupus nephritis. PLoS One. 2010;5:e10344. doi: 10.1371/journal.pone.0010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scian MJ, Maluf DG, David KG, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am J Transplant. 2011;11:2110. doi: 10.1111/j.1600-6143.2011.03666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basile DP, Fredrich K, Chelladurai B, et al. Renal ischemia reperfusion inhibits VEGF expression and induces ADAMTS-1, a novel VEGF inhibitor. Am J Physiol Renal Physiol. 2008;294:F928. doi: 10.1152/ajprenal.00596.2007. [DOI] [PubMed] [Google Scholar]
- 33.Siedlecki AM, Jin X, Thomas W, et al. RGS4, a GTPase activator, improves renal function in ischemia-reperfusion injury. Kidney Int. 2011;80:263. doi: 10.1038/ki.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Favreau F, Thuillier R, Cau J, et al. Anti-thrombin therapy during warm ischemia and cold preservation prevents chronic kidney graft fibrosis in a DCD model. Am J Transplant. 2010;10:30. doi: 10.1111/j.1600-6143.2009.02924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen YT, Chang FC, Wu CF, et al. Platelet-derived growth factor receptor signaling activates pericyte-myofibroblast transition in obstructive and post-ischemic kidney fibrosis. Kidney Int. 2011;80:1170. doi: 10.1038/ki.2011.208. [DOI] [PubMed] [Google Scholar]
- 36.Spivey TL, Uccellini L, Ascierto ML, et al. Gene expression profiling in acute allograft rejection: challenging the immunologic constant of rejection hypothesis. J Transl Med. 2011;9:174. doi: 10.1186/1479-5876-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowarsch A, Preusse M, Marr C, et al. miTALOS: analyzing the tissue-specific regulation of signaling pathways by human and mouse microRNAs. RNA. 2011;17:809. doi: 10.1261/rna.2474511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 17:1169. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 39.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8:753. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- 40.Kainz A, Wilflingseder J, Mitterbauer C, et al. Steroid pretreatment of organ donors to prevent postischemic renal allograft failure: a randomized, controlled trial. Ann Intern Med. 2010;153:222. doi: 10.7326/0003-4819-153-4-201008170-00003. [DOI] [PubMed] [Google Scholar]
- 41.Mengel M, Sis B, Haas M, et al. Banff 2011 meeting report: new concepts in antibody-mediated rejection. Am J Transplant. 2012;12:563. doi: 10.1111/j.1600-6143.2011.03926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maragkakis M, Alexiou P, Papadopoulos GL, et al. Accurate micro-RNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 45.Hsu SD, Lin FM, Wu WY, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mi H, Lazareva-Ulitsky B, Loo R, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).