Abstract
Background
PHOSPHATE1 (PHO1) gene family members have diverse roles in plant growth and development, and they have been studied in Arabidopsis, rice, and Physcomitrella. However, it has yet to be described in other plants. Therefore, we surveyed the evolutionary patterns of genomes within the plant PHO1 gene family, focusing on soybean (Glycine max) due to its economic importance.
Results
Our data show that PHO1 genes could be classified into two major groups (Class I and Class II). Class I genes were only present and expanded in dicotyledonous plants and Selaginella moellendorffii; Class II genes were found in all land plants. Class I sequence losses in other lineages may be attributed to gene loss after duplication events in land plant evolution. Introns varied from 7 to 14, and ancestral state reconstruction analyses revealed that genes with 13 introns were ancestral, thus suggesting that the intron loss was a chief constituent of PHO1 gene evolution. In the soybean genome, only 12 PHO1-like genes (GmaPHO1) were detected at the mRNA level. These genes display tissue-specific or tissue-preferential expression patterns during soybean plant and fruit development. Class I genes were more broadly expressed than Class II. GmaPHO1 genes had altered expression in response to salt, osmotic, and inorganic phosphate stresses.
Conclusions
Our study revealed that PHO1 genes originated from a eukaryotic ancestor and that two major classes formed in land plants. Class I genes are only present in dicots and lycophytes. GmaPHO1genes had diverse expression patterns in soybean, indicating their dramatic functional diversification.
Keywords: Evolution, Gene expression, PHO1 gene family, Phylogeny, Soybean
Background
PHOSPHATE1 (PHO1) genes play diverse roles in plant growth and development; for example, some are important for inorganic phosphate (Pi) sensing and transport. Arabidopsis thaliana AthPHO1 (At3g23430), the first species in which the PHO1 gene family was identified, was reported to be key for long-distance transfer of Pi from the root to the shoot [1,2], as well as signal transduction of the long-distance Pi-deficiency response [3,4]. AthPHO1 is predominantly expressed in root vascular tissues and is involved in root Pi loading into the apoplastic space of the xylem vessels [1]. Gene ectopic expression of AthPHO1 in leaves [5] and in mesophyll protoplasts [6] mediates cellular specific phosphate efflux. The Arabidopsis genome encodes 10 additional genes (identified as AthPHO1; H1 to AthPHO1; H10) which are homologous to AthPHO1[7]. However, only AthPHO1; H1 complements athpho1 mutants when expressed under control of the AthPHO1 promoter, indicating that only these two members are involved in long-distance Pi transport from root to shoots [8]. The rice genome has only three PHO1 homologs (OsaPHO1; H1, H2, H3) clustering with AthPHO1 and AthPHO1; H1, but only OsaPHO1; H2 complements athpho1 mutants, suggesting a role in phosphate homeostasis [9].
Seven PHO1-like genes are present in the non-vascular land plant Physcomitrella patens genome; however these gene functions are unclear [10]. In Arabidopsis, some PHO1 homologues are also expressed in non-vascular tissues such as hydathodes, trichomes, and pollen grains [7]. AthPHO1; H4 plays a role in the response of hypocotyls to blue light [11], seed size and flowering [12-14]. AthPHO1; H10 was reported to be induced by numerous stresses, including local wounding responses [15,16]. Recently, AthPHO1 has been suggested to play an important role in the stomatal response to abscisic acid (ABA); its expression in guard cells is induced by ABA treatment [17]. Therefore, plant PHO1 genes have undergone a functional diversification and acquired diverse roles beyond Pi transport and homeostasis. Furthermore, the PHO1 homolog xenotropic and polytropic retrovirus receptor 1 (XPR1) in mammals functions as a receptor [18]; and the suppressor of yeast Gα protein deletion (SYG1) protein is involved in the mating pheromone signal transduction pathway [19]. Such functions suggest that PHO1 genes have diversified over time.
Soybean (Glycine max) is a crop of economic significance and a world-wide source of high quality protein and vegetable oils [20]. Understanding mechanisms of nutrient homeostasis regulation, especially Pi, may increase soybean yield. PHO1 gene family molecular evolution and functional diversification remains understudied, so whether soybean PHO1 genes are involved in Pi sensing and transfer is unclear. To address this gap in the scientific literature, we experimentally characterized PHO1 genes of G. max (GmaPHO1) in Nannong1138-2 cultivars, and investigated their evolutionary patterns on a broad-scale phylogenetic framework. Soybean is a diploidized ancient tetraploid species [21,22], and GmaPHO1 genes might have distinct evolutionary patterns that differ from their orthologs in other plant species. GmaPHO1 genes may have unique expression patterns in response to various stimuli such as Pi stress, contributing to plant diversification. Our detailed gene expression analysis revealed that these genes may be important for plant organ development and responses to various abiotic stimuli.
Results
Phylogenetic analysis of the plant PHO1 genes
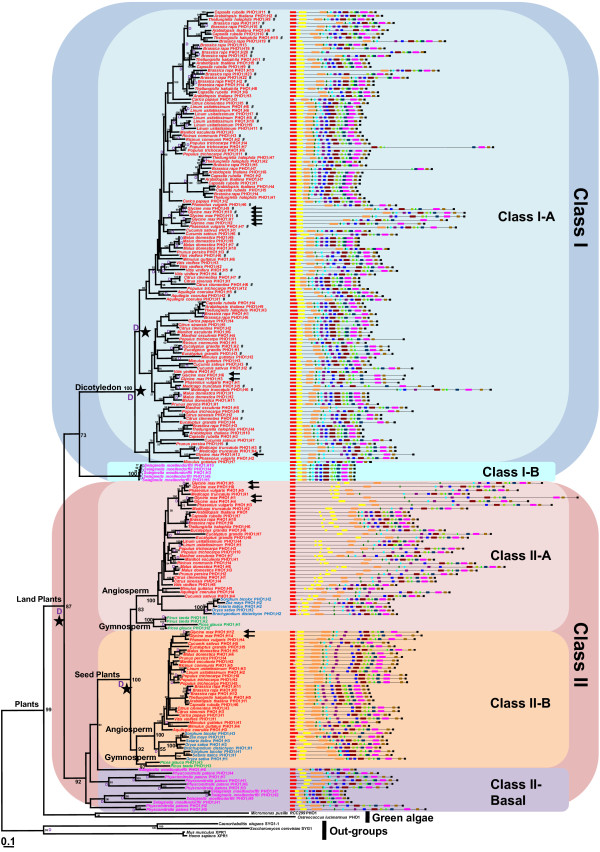
Phylogenetic analyses allow us to identify evolutionary conservation and divergence of genes. To understand the evolution of the plants PHO1 gene family, we used AthPHO1 to query the NCBI and Phytozome databases (see Methods). We obtained 223 of PHO1-like sequences from 32 plant species, including two chlorophyta species, one moss, one lycophyte, two gymnosperms, and 26 angiosperms. Whole-genome sequences of most plant species are available, except for the two gymnosperms (see Additional file 1: Table S1). Currently, 6 chlorophyta genomes were released; however, only two PHO1 genes were found in two species of Ostreococcus lucimarinus and Micromonas pusilla RCC299. We observed that few homologs had evolved in yeast and animals; however, no PHO1-like sequences were found in prokaryotes, suggesting that PHO1-like genes originated in a common eukaryotic ancestor.
A maximum likelihood tree was generated using amino acid sequences of the deduced full-length peptides (733 aligned positions) with the best-fit evolutionary JTT (Jones, Taylor and Thornton) model (Figure 1). The WAG (Whelan and Goldman) and LG (Le and Gascuel) models were also tested (see Methods), and resultant tree topologies were consistent with the JTT model; only a few gene relationships were varied within each Class (see Additional file 2: Figure S1). Proteins SYG1 in Saccharomyces cerevisiae, SYG-1 in Caenorhabditis elegans, and XPR1 in Mus musculus and Homo sapiens were used as out-groups in phylogenetic analyses. PHO1 genes from algae formed the basal lineage, whereas PHO1-like genes from land plants were monophyletic, apparently forming two major groups (Classes I, II) based on the current tree topology, with a well-supported bootstrap value (87%; Figure 1). One hundred twenty-five PHO1-like genes from dicots were specifically assigned to Class IA (with 100% bootstrap value support) with a basal lineage (Class IB) containing 5 sequences from the basal land plants Selaginella moellendorffii. In contrast, 91 members were present in Class II, which could be subdivided into Class IIA (41 genes) and Class IIB (38 genes) with basal grade (Class II-Basal) that was a non-monophyletic group including 5 Selaginella genes and 7 Physcomitrella genes. Class IIA and Class IIB consisted of the sequences from all seed plants with a 100% bootstrap value.
Figure 1.
Phylogenetic tree and gene structure of the PHO1 gene family in plants. The maximum likelihood tree of the land plant PHO1 genes constructed based on the deduced full-length peptide amino acid sequences using the proteins SYG1 in Saccharomyces cerevisiae, SYG-1 in Caenorhabditis elegans, and XPR1in Mus musculus and Homo sapiens as out-groups. Bootstrap values (>50%) for this tree are shown on each branch. The different classes are marked with different colored backgrounds. The genes from the basal land plants are indicated in rose pink; dicot in red, monocots in blue and gymnosperm in green, respectively. The PHO1-like genes in soybean are indicated by arrows. The symbol ‘D’ indicates the detected gene duplication events, and the symbol ‘#’ indicates genes resulted from tandem duplication in their hosts. The black star indicates the 4 important major duplication events during the expansion of PHO1 genes in land plants. Gene structure was predicted based on the sequences from the Phytozome database. Colored boxes represent exons and connecting lines indicate the introns, respectively. The structure of PHO1 genes in the gymnosperm Pinus taeda and Picea glauca are not represented for lack of their genome sequences.
Gene family copy number varied from 2 to 23 among the species, which was not correlated with the genome size (R=−0.15, P=0.57) but slightly correlated with number of whole-genome duplication events (WGD) occurring within Viridiplantae (R=0.51, P=0.04, see Additional file 3: Table S2) [23-25]. Brassica rapa had the largest PHO1 gene family (23 genes) with a 500 Mb genome size and 4 WGD events. Brachypodium distachyon and Zea mays had only 2 genes (355 Mb and 2400 Mb genome sizes) with 2 and 3 WGD events occurring. Class I contained 125 genes from 21 dicots and 5 genes from S. moellendorffii, while Class II had only 91 genes from all 30 land plants studied (Figure 1; see Additional file 3: Table S2), indicating that the PHO1 gene family asymmetrically evolved between Class I and Class II.
To investigate PHO1 gene family expansion and gene diversification, a gene tree was reconciled with the species tree. Analyses revealed 90 duplications and 29 losses, with 164 of the D/L score (duplication events =‘D’; four major duplication events for gene family expansion in land plants are indicated by stars; Figure 1). Class I and II diverged from ancestral duplications, and subclass Class IA arose from at least two major duplication events. Class IIA (close to the well-known AthPHO1) and Class IIB (close to the AthPHO1; H1) arose from one duplication event occurring in seed plant ancestors, and each class could be divided further into angiosperm and gymnosperm lineages. Class I genes in the dicots and S.moellendorffii were preserved and may have resulted from gene loss after the first duplication in other lineages. Inspection of the physical chromosomal location of PHO1 genes suggested that tandem duplication may have contributed to evolution of the Class I PHO1 gene family in the specific host. Fifty-five of the Class IA 125 members appeared on native host chromosomes as 2 to 3 gene tandem repeats (highlighted by ‘#’ in Figure 1; see Additional file 4: Table S3).
Gene structure variation throughout plant PHO1 gene family
Comparisons of 215 cDNA sequences and genomic DNA from 28 species revealed exons and introns number and position for each individual gene in land plants (Figure 1; see Additional file 5: Table S4 and Additional file 6: Figure S2). Six PHO1-like transcripts from gymnosperms were not included; their gene sequences were unavailable. Two PHO1 gene sequences from green algae had no introns, and diverse gene structures in this family resulted from multiple intron losses and rare intron gains in land plants. Intron numbers of these genes varied from 7 to 14, with widely divergent lengths. All genes in the Class II-basal grade had more than 9 introns, a characteristic that seemed to be maintained during seed plant evolution. Around 87.0% of Class I members and 93.0% of the genes in Class IIA and IIB had more than 9 introns; genes from Class IIA had more than 11 introns, and 73.0% of the PHO1 genes within Class IIA had 14 introns.
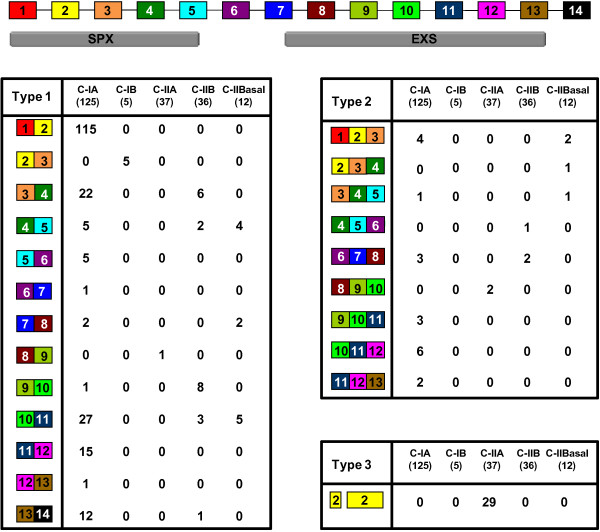
To understand PHO1 gene structures evolution, we reconstructed the ancestral states of exon patterns in 28 land plants excluding gymnosperms (see Additional file 6: Figure S2). Data show that 14-exon structure of PHO1genes could represent ancestral land plants structure. Diverse structures of other members in the PHO1 gene family could be traced back to the 14-exon structure, and their ancestral components could be clearly distinguished. Given that all genes were 14 putative EXONs, exons from 4 to13 were highly conserved with respect to lengths and sequences within this family. These exons were observed to encode part of the SPX (Pfam PF03105) domain and the entirety of the EXS (Pfam PF03124) domain (Figure 2), domains which are characteristic of the PHO1 protein family [26-28]. PHO1 gene structure diversity apparently resulted from intron losses and novel intron gains after the origin of the 14-exon structure (see Additional file 5: Table S4).
Figure 2.
Structural evolutions of plant PHO1 genes. Variation in exon and intron structure of plant PHO1 genes. The ancient gene structure with 14-EXONS is given. Boxes in different colors indicate different exon and the lines represent introns. Different types of intron loss or intron gain processes in PHO1 gene family are listed below the 14-exon structure.
Intron loss, as a major molecular event, happened twice at most in the consequent and adjacent exons in PHO1 land plants genes evolution, whereas novel intron gain was unique to Class IIA. Thus, the divergent patterns of intron number variations could be summarized into three types (Figure 2). Type 1 included the exon-fusion events that happened once in adjacent exons; Type 2 included exon-fusion events that occurred twice in the adjacent exons (involving the three consequent exons), while Type 3 represented a novel intron gain event. Among the 215 genes studied, the exon-fusion events occurred 243 times in pattern Type 1, 28 times in pattern Type 2 and 29 times in pattern Type 3, suggesting that intron loss was main molecular event occurring during evolution of this gene family. In pattern Type 1, we observed 13 exon-fusion events which were location dependent. Such intron loss could occur multiple times (maximally 4 times) on one gene. Pattern Type 2 consisted of 9 events that were location-specific and could occur with Type 1 pattern on one gene. Intron gains (Type 3) occurred 29 times in exon 2 only of Class IIA in 37 genes. We also assessed the intron phase for all PHO1 genes (see Additional file 6: Figure S2). Intron phases of adjacent exons were virtually conserved in these differently classed genes irrespective of how introns were lost or gained within the 14-exon structure.
Genomic identification of the PHO1 gene family in soybean
Release of Glycine max (Williams 82) draft soybean genome [22] facilitated the isolation of GmaPHO1 genes in this investigation. Data-mining using 11 Arabidopsis PHO1 homologs as queries revealed 14 putative PHO1 homologous genes (designated as GmaPHO1; H1 to GmaPHO1; H14) in the soybean genome. GmaPHO1; H2, H3, H6, H7, H9, H10, H11 and H13 belong to Class Ι; GmaPHO1; H1, H4, H5 and H8 are from Class IIA and GmaPHO1; H12 and H14 consist of Class IIB (Figure 1, highlighted by arrows). These genes were found to be distributed on 7 of the 20 chromosomes (Table 1; see Additional file 7: Figure S3), and the gene number on each chromosome varied widely, one gene on each chromosome of 1, 7, 9, and 18; two on chromosome 10, three on chromosome 2, and 5 genes were localized to chromosome 20. The number of introns varied from 12 to 14 (Figure 1; see Additional file 8: Table S5). Encoded proteins were predicted to contain the SPX and EXS domains (see Additional file 9: Figure S4), and were estimated to be located on plasma membranes. Each GmaPHO1 protein contained at least 5 trans-membrane segments (Table 1), suggesting the potential function of these proteins as transporters of phosphate absorbed from the soil. GmaPHO1-like proteins range from 753 to 817 amino acids in length and have 30-94% overall identities (Table 1; see Additional file 10: Table S6). Interestingly, several pairs of GmaPHO1-like genes on different chromosomes showed high identities in protein sequences, for example, there was 94% identity between GmaPHO1; H1 and GmaPHO1; H4, 93% between GmaPHO1; H12 and GmaPHO1; H14 and 91% between GmaPHO1; H2 and GmaPHO1; H7. These data reflected genome duplication during the soybean evolution [29,30]. The open reading frame (ORF) sequence of these genes was confirmed by isolation of the cDNA from soybean Nanong1138-2. Gene-specific primers were designed according to the predicted sequences from soybean Williams 82 (see Additional file 11: Table S7). Ultimately we obtained 12 of the 14 predicted cDNA sequences, revealing 12 expressed functional PHO1-like genes in the soybean genome. No transcription was observed for GmaPHO1; H11 on chromosome 2 and GmaPHO1; H13 on chromosome 20 in Nannong1138-2.
Table 1.
Molecular characterization of GmaPHO1 genes in soybean
| Gene | Locus name | Chra | PLb | SLc | TMSd |
|---|---|---|---|---|---|
|
GmaPHO1; H1 |
Glyma02g00640 |
02 |
763 |
PM, Nuc |
7 |
|
GmaPHO1; H2 |
Glyma07g35520 |
07 |
801 |
PM, Nuc |
8 |
|
GmaPHO1; H3 |
Glyma09g37000 |
09 |
756 |
PM, Nuc |
6 |
|
GmaPHO1; H4 |
Glyma10g00720 |
10 |
764 |
PM, Nuc |
5 |
|
GmaPHO1; H5 |
Glyma10g32670 |
10 |
771 |
PM, Nuc |
7 |
|
GmaPHO1; H6 |
Glyma18g49680 |
18 |
773 |
PM, Nuc |
6 |
|
GmaPHO1; H7 |
Glyma20g03960 |
20 |
784 |
PM, Nuc |
5 |
|
GmaPHO1; H8 |
Glyma20g34930 |
20 |
771 |
PM, Nuc |
7 |
|
GmaPHO1; H9 |
Glyma20g04130 |
20 |
792 |
PM, Nuc, ER |
7 |
|
GmaPHO1; H10 |
Glyma20g04150 |
20 |
804 |
PM, Nuc, ER |
9 |
|
GmaPHO1; H11 |
Glyma20g04160 |
20 |
817 |
PM, Nuc |
8 |
|
GmaPHO1; H12 |
Glyma01g22990 |
01 |
804 |
PM, Nuc, ER |
7 |
|
GmaPHO1; H13 |
Glyma02g12320 |
02 |
753 |
PM, ER |
6 |
| GmaPHO1; H14 | Glyma02g14440 | 02 | 789 | PM, Vac | 6 |
PM, plasma membrane; Nuc, nuclear; Vac, vacuolar membrane; ER, endoplasmic reticulum. Chra, Location of GmaPHO1 genes on chromosomes. PLb Peptide length (aa). SLc, Subcellular location of GmaPHO1 proteins. TMSd, Number of transmembrane segments.
Organ-specific expression of GmaPHO1 genes during soybean development
The expression of GmaPHO1 genes using real-time RT-PCR approach provided clues to their functional divergence. Total RNAs used in this study was taken from roots, leaves, stems, flowers, and developing fruits of soybean cultivar Nannong1138-2.
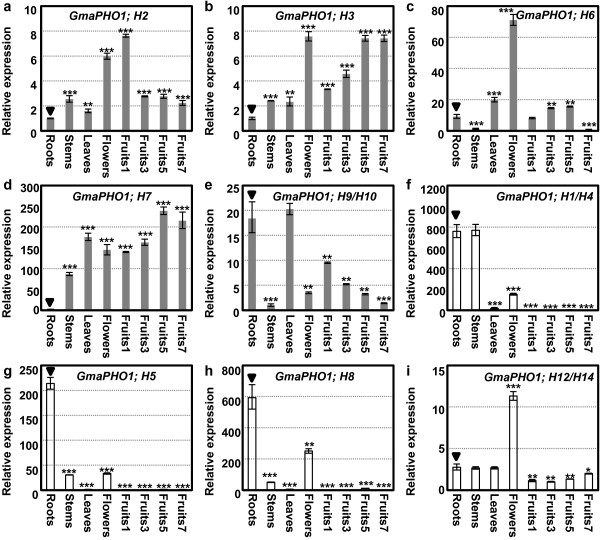
Two-tailed Student’s t-test of gene expression variation among these organs confirmed diverse expression patterns (see Additional file 12: Figure S5). Here, we report difference in gene expression in plant organs compared with that in roots. Under normal condition, most GmaPHO1 genes were constitutively expressed in different tissues, but several genes had tissue-specific or preferential expression patterns (Figure 3). In Class Ι (Figure 3a-e, highlighted in grey column), GmaPHO1; H2 and GmaPHO1; H3 were found to be expressed in all organs examined; however, they were significantly abundant in flowers and fruits (P<0.001). GmaPHO1; H2 was significantly down-regulated while GmaPHO1; H3 was significantly up-regulated during fruit development (Figure 3a and b). GmaPHO1; H6 was predominantly expressed in flowers and leaves (P<0.001) and was moderately expressed in roots and earlier fruit developments with low expression in stems and 7-day fruits after pollination (Figure 3c). The expression level of GmaPHO1; H7 was found to be significantly higher in most organs and maintained a higher level during fruit development, but undetectable in roots (P<0.001) (Figure 3d). GmaPHO1; H9/H10 were expressed in most tissues; however, their expression levels were found to be much higher in roots and leaves (P<0.01) and extremely lower in stems and 7-day fruits (P<0.001). Notably, these genes were observed to be gradually repressed during fruit development (Figure 3e). However, genes in Class II had limited organ expression patterns with clear difference between Class IIA and Class IIB (Figure 3f-i, highlighted in empty column). Genes in Class IIA were predominantly expressed in roots; however, they were expressed to a lesser degree in flowers and were not expressed in leaves and developing fruits (P<0.001). GmaPHO1; H1/H4 had an equivalent expression in stems and roots, while GmaPHO1; H5 and GmaPHO1; H8 were less expressed in stems (Figure 3f-h). GmaPHO1; H12/H14 in Class IIB were constitutively expressed, and predominantly so in mature flowers (P<0.001) (Figure 3i). These findings suggest that GmaPHO1 genes may exert certain roles in soybean development with lineage-specific divergent patterns.
Figure 3.
Organ-specific expressions of GmaPHO1 genes during development. (a-e) Organ-specific expressions of the Class I genes. (f-i) Organ-specific expressions of the Class II genes. The gene is indicated in each graph. Roots, stems, leaves, mature flowers and fruits (1-, 3-, 5- and 7-day fruits after pollination) are involved. Actin was used as an internal control. The experiments were performed based on three independent biological samples. Error bars=standard deviations. The star (*P < 0.05, **P < 0.01 and ***P < 0.001) represented the statistical significance of the gene expression variation in other tissues compared to that in roots indicated by triangles.
Gene expression in response to various stresses in soybean
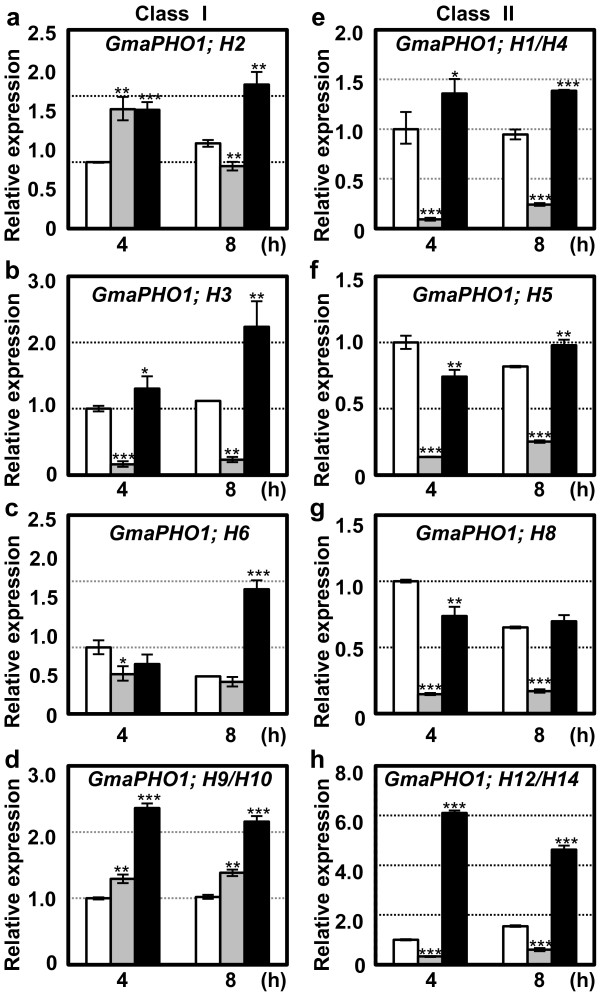
Examination of soybean PHO1 genes transcription in roots challenged with osmotic, salt stress, and Pi stresses in the 3-week old seedlings (see Methods) identified genes that respond to abiotic stresses. Total RNA from roots was subjected to real-time RT-PCR for GmaPHO1genes (except for GmaPHO1; H7 due to its silencing in roots; Figure 3d). Relative to untreated controls (Figure 4, empty column), Class I genes had different responses to osmotic stress (Figure 4a-d, highlighted in grey column), while the genes in Class II responded similarly to osmotic stress (Figure 4e-h, highlighted in grey column). In Class I, GmaPHO1; H3 was rapidly and constantly repressed (P<0.01) (Figure 4b), and GmaPHO1; H9/H10 were induced by osmotic stresses (P<0.01) (Figure 4d). The genes GmaPHO1; H2 or GmaPHO1; H6 were slightly influenced by osmotic treatments (P<0.05) (Figure 4a and c). Class IIA genes were repressed by osmotic stresses (P<0.001) (Figure 4e-h, highlighted in grey column). Under salt stresses, all Class Ι genes were induced to express (P<0.05) (Figure 4a-d, highlighted in black column). In Class IIA, genes were moderately changed under the influence of salt stress (Figure 4e-g). In contrast, GmaPHO1; H12/H14 in Class IIB were strongly induced under salt stress conditions (P<0.001); their expression increased more than 6 times compared with controls (Figure 4h). The two types of Pi stress treatments altered GmPHO1 gene transcription significantly at most treatment time-points (see Additional file 13: Figure S6); however, each gene had complex but similar pattern under the Pi stress treatments, suggesting that GmPHO1 genes have an complex response to Pi alteration around the roots. Thus, these genes have different roles in response to salt, osmotic and Pi stresses.
Figure 4.
Expression of GmaPHO1 genes in response to salt and drought stresses. (a-d) Root expression of the Class I gene in response to PEG-6000 (grey column) or NaCl (black column). (e-h) Root expression of the Class II gene in response to PEG-6000 (grey column) or NaCl (black column). Genes are indicated in each graph. Empty column represents the untreated control. The 3-week old seedlings of ‘Nannong1138-2’ were treated with for 4 h and 8 h. Actin was used as an internal control. The experiments were performed based on three independent biological samples. Error bars=standard deviations. Star indicates the significance of gene expression variation under each treatment in comparison to the untreated control (*P < 0.05, **P < 0.01 and ***P < 0.001).
Discussion
Limited functional characterization of the PHO1 gene family suggests it has important roles in plant developmental and physiological processes [1,3-7,12,13,15-17]. Here we report our investigations into the phylogeny and structural evolution of PHO1-like genes in plants and share data about their functional divergence. Our data were confirmed with comprehensive expression analyses of these genes in soybean.
Evolution of the PHO1 gene family during evolution of plants
Our results revealed that PHO1 homologs were prevalent in eukaryotes only. Unlike previous reports [9,10,28], we identified two PHO1-like cDNAs in two of the six released genomes of chlorophyta, indicating that the gene is not required for all unicellular green algae. However, PHO1-like genes were expanded tremendously in multicellular plants (Figure 5). Phylogenetic reconstruction with an 87% bootstrap value revealed that these genes formed two major classes in land plants (Class I, II), indicating that the PHO1-like genes from all land plants share a common ancestor; the two classes are sister groups. The topology of the PHO1 gene family of land plants might have originated through several duplications, followed by gene loss in some descendants. Genes in Class I, occupying 58.8% (130/221) of the total identified genes, were from 21 dicots and one lycophyte, while 91 members from land plants were present in Class II (Figure 5). One duplication event led to the division of Class II into Class IIA and Class IIB of the genes in seed plants, while multiple duplication events including tandem duplications in dicots that specifically occurred in Class I led to its expansion. These data indicated that lineage-specific expansion and divergence events of the PHO1-like genes occurred in seed plants.
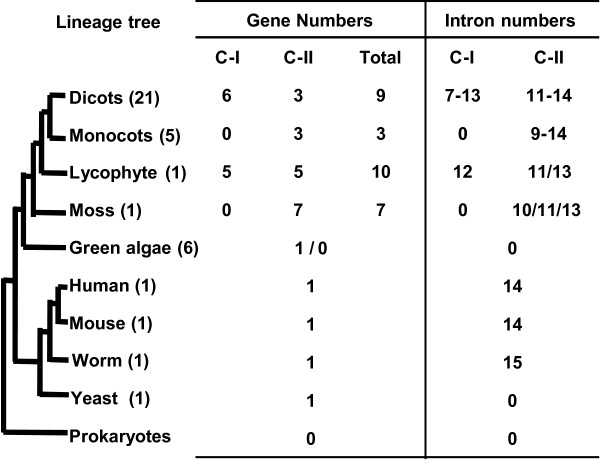
Figure 5.
Evolutionary patterns of the PHO1 gene family. Variations in both gene copy and introns of the PHO1 genes in the major lineages of organisms are summarized. Liverworts and gymnosperms are not included as their e whole genome sequences were not available. The number of the species with whole genome sequenced is given in the parenthesis behind of each lineage. Gene copy number of the lineages with multiple species is given as the mean per species. Class I and Class II, are abbreviated as C-I and C-II respectively.
Gene duplication is a permanent and continual factor contributing to the complexity of genetic material [23,31-33]. Gene copy variation, a result of gene duplication and gene loss, was prominent in the PHO1 gene family. The gene family also evolved in yeast, animals and green algae with a single copy, while copy numbers of these genes in land plants varied between 2 and 23. The variable copy numbers are not consistent with the genome sizes of different species, evidence for which was obtained from an early vascular plant with a small genome (100 Mb). There are 10 copies of PHO1 genes in S.moellendorffii, whereas in Z.mays (2400 Mb) only 2 copies survived. However, gene family expansion slightly correlates to the whole-genome duplication and triplication events which occurred among Viridiplantae [23-25], indicating an important role of gene losses during PHO1 gene family evolution. Monocots were found to possess lesser PHO1-like genes compared to dicots and basal land plants, like P.patens and S.moellendorffii. There are 185 members in 21 dicots compared to 13 members in 5 monocots (Figure 5); thus, gene loss events are key to monocots evolution. These results imply that the expansion of the PHO1 gene family may be associated with the origin of diversification of land plants.
A genome-wide study compared orthologous groups of genes in 4 model genomes (Arabidopsis, poplar, rice, and P. patens) and revealed that genes related to intracellular components, regulation of metabolism, hormone metabolism, transcriptional regulation, cell communication, and responses to hormone stimuli are more likely to have undergone non-tandem duplication. In contrast, genes involved in stress responses had an increased probability of being retained in a single-lineage fashion after tandem duplication, suggesting that tandem duplicates are probably important for adaptations to rapidly changing environments [33]. Interestingly, we found that almost 50% of Class ΙA genes (55/125) underwent tandem duplication. However, found no tandem repeats in Class II. PHO1-like genes in Class II have been functionally characterized in Arabidopsis and rice [1,5,8-10] and are known to play major roles in root-to-shoot transfer of phosphate, while Class Ι genes are important in biotic and/or abiotic stress responses [15-17]. Tandem gene arrangements and copy number variation of PHO1 Class ΙA genes may arise from a response to different environmental selections during evolution. This assumption was further supported by our expression analyses in soybeans.
The majority of conserved introns remain unchanged over time [34,36]. Thus, intron/exon changes could provide clues on evolutionary relationships [37,38]. The intronless PHO1 genes originated from a eukaryotic ancestor, evidence for which is provided by the structure of unicellular yeast and green algae (Figure 5). Once multicellular organisms (mammals and land plants) appeared, PHO1 genes became more complicated. Introns can probably be gradually acquired, or obtained once during the evolution of the multicellular organisms from the unicellular ancestors. Our gene structure reconstruction and exon-intron analysis with all available sequences revealed that the gene structure with 13 introns might be an original organization in land plants, and that PHO1 genes might have evolved along with multicellular organisms (Figure 5). We therefore proposed that the intron loss is a major event in the evolution of PHO1 genes of land plants. Most Class I members had fewer introns than their ancestors, due to intron loss accompanying the exon-fusion event. This event occurred less frequently in Class IIA and Class IIB members, therefore, most of them contained 13 or 14 introns and the 14-intron structure arose from an additional novel intron gain (Figure 5; see Additional file 6: Figure S2), which only occurred in Class IIA. Also, intron losses occurred independently, resulting in 12–14 introns. Consistent with the previous observation that intron phase change is rare [38], intron phases of adjacent exons were found to be almost conserved in PHO1 genes in different classes.
Furthermore, we found that 119 of 130 genes lost their first intron in Class I. Plant studies offer evidence of intron effects on gene expression [39-42]. The first intron of Arabidopsis histone H3 gene, Petunia actin-depolymerizing factor 1 (PhADF1) and maize shrunken-1 (Sh1) can alter gene expression levels [43-45]. Introns are also involved in the regulation of spatial and temporal gene expression patterns, such as photosystem I subunit II (PsaD), AGAMOUS (AG) and flowering locus C (FLC) [46-48]. In the dicot-specific Class IA, almost all genes lost their first introns. Indeed, the Arabidopsis Class I PHO1 genes have a boarder expression domain than Class II genes [7]. Our data support these finding: expression domains of the soybean genes in Class I are more diverse than those in Class II.
Functional diversification of the PHO1 gene family in soybean
We surveyed the genome draft of soybean Williams 82 and predicted 14 PHO1-like genes. They were found to be either tandemly or segmentally located on the chromosomes. We characterized the cDNA sequences from the Nannong1138-2 cultivars to validate 12 of the predicted sequences. These genes on the phylogenetic tree were classified into the Class I and Class II, suggesting their functional divergence in developmental processes and in response to abiotic stresses.
Transcripts of the predicated two genes (GmaPHO1; H11 and GmaPHO1; H13) in Class IA were not detected in Nannong 1138–2, indicating that either these genes were silenced or had extremely low levels. The expression profiles of soybean PHO1-like genes provided clues to their functional relevance and divergence. We found that the genes in the two classes were expressed with different tissue-specific patterns, while the genes in the same class showed a similar expression pattern. The genes in Class I had a broader expression domain than those in Class II and they were developmentally regulated during fruit development in soybean. These results are corroborated by previous studies on AthPHO1; H4 in Class I that plays a role in the response of hypocotyls to blue light [11], as well as seed size and flowering [12-14]. The genes in Class IIB were expressed in all tissues examined, while the genes in Class IIA seemed to be restricted to roots, stems and flowers. These findings, therefore, support a lineage-specific functional differentiation related to soybean plant and fruit development.
The phylogenetic analysis showed that GmaPHO1; H1, GmaPHO1; H4, GmaPHO1; H5 and GmaPHO1; H8 are grouped with AthPHO1 and OsaPHO1; H2 that play important roles in Pi transference [7,9], hinting that these genes might have related functions. These genes indeed showed a similar tissue-specific expression pattern with little variation. Most GmaPHO1 genes were expressed in roots and responded to Pi deficient or sufficient treatments in complex manners. Surprisingly, each gene in the different Pi stresses had a similar responding mode in most treated-time points. This is not well understood yet. Nonetheless, our observations suggest that they might function in Pi sensing or transporting in soybean as their homologs AthPHO1 in Arabidopsis and OsaPHO1; H2 in rice [1,7,9].
AthPHO1; H10 in Arabidopsis is activated by a diversity of stimuli including salinity, osmotic, pathogens and even by wounding [15], thus suggesting that some PHO1-like genes might be involved in these physiological processes. GmaPHO1 genes exhibited varied expression patterns under abiotic stresses. The genes, GmaPHO1; H3 in Class IA, GmaPHO1; H1/H4, GmaPHO1; H5, GmaPHO1; H8 and GmaPHO1; H12/H14 in Class IIA were rapidly and constantly inhibited by osmotic stress, while all genes in Class IA and GmaPHO1; H12/H14 in Class IIB were found to be strongly induced by salt stress, suggesting that they are likely associated with the processes that respond to these stresses. Inspection of chromosomal localization of PHO1 gens in soybean and other species genomes suggests that almost 50% of the genes in Class IA are arranged tandemly on the chromosomes. Our study combined with the evidences from Arabidopsis supports the deduction that tandem duplication and variation in copy number of PHO1 gene in Class ΙA might result from a response to different environmental selection during evolution. During evolution, modulating gene expression and increasing gene numbers possibly allows the different PHO1genes to act synergistically to promote plant development and to reinforce tolerance to environment stresses. These genes can greatly contribute to plant adaptation and survival in adverse conditions through offsetting the effects of mutations.
Conclusions
Using comparative genomic and phylogenetic analyses we identified two major Classes (I and II) of the PHO1 gene family in land plants. Class I genes were expanded in dicotyledonous plants and Selaginella moellendorffii, while Class II genes were found in all land plants. Class IA subfamily gene expansion in dicots and the loss of the subfamily genes in monocots were also documented. Intron loss was found to be an active force in the evolution of gene structure since the origin of a complicated structure (14-EXONS) in multicellular plants. The functional divergences among PHO1 genes in soybean suggested that significant subclass-specific functional evolution took place after their phylogenetic diversification. Our results suggest that the expansion and sequence variation accompanying dramatic functional diversifications of the plant PHO1 gene family are associated with the origin of diversification of land plants. Our findings provided valuable information and new insights into the molecular and functional evolutionary pattern of the plant gene family during evolution of plants. Evaluating the function of this gene family could provide valuable insights into their involvement in metabolic and biochemical processes.
Methods
Identification and physical locations of PHO1 genes in soybean
Arabidopsis PHO1 orthologs were used to search the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) and Phytozome (http://www.phytozome.net) databases. The primers used for cloning PHO1s were designed from the 5′ends and 3′ ends of putative coding regions according to their linear alignment with Arabidopsis genes. For those cDNA fragments lacking 5′ and/or 3′ ends of the coding regions, 5′-RACE and/or 3′-RACE was performed to obtain the missing regions (Roche, Mannheim, Germany). PHO1-like cDNAs were amplified by PCR from a combination of cDNAs from different soybean tissues using gene specific primers as described in Additional file 11: Table S7. Genes were mapped on chromosomes by identifying their chromosomal position provided in the Phytozome database. Protein subcellular localization was predicted using PSORT software (http://wolfpsort.org).
Phylogenetic tree constructions
To analyze the sequence features of the 223 typical identified PHO1 genes in plants, a multiple sequence alignment (MSA) was carried by the GUIDANCE web-server using the MSA algorithm MAFFT (http://guidance.tau.ac.il/index.html) [49], and eliminated the poorly aligned positions were further using the G-blocks server (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) [50,51]. ProtTest version 2.4 [52] was used to conclude that JTT model was the best-fit model for amino acid evolution according to both Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC). To obtain optimized alignment, the deduced amino acid sequences were adjusted manually using BioEdit (v7.0.5) [53]. The ML phylogenetic tree was constructed using PhyML (v3.0) [54] under the JTT amino acid substitution model, with 100 replicates of bootstrap analysis, estimated gamma distribution parameter, and optimized starting BIONJ tree. The tree was deposited in the TreeBASE (S14085 Matrix ID). ML trees generated using both WAG and LG models had a topology similar to the JTT model, and these are presented in Additional file 2: Figure S1. The phylogenetic tree was visualized using the TreeView1.6.6 tool with a 50% threshold of branch value [55]. Notung software (version 2.6) was used for tree reconciliation [56,57]. The species tree used to reconcile the gene tree was based on taxonomic information from the NCBI database. Correlations between gene copy numbers and genome size as well as whole-genome duplication events timing was estimated using the Pearson correlation with a two tailed significance test. Statistical analysis was performed in SPSS 15.0 for Windows.
Gene structure analyses
For intron/exon structure analysis, the DNA and cDNA sequences corresponding to each predicted gene from the Phytozome database annotation were downloaded, and their intron distribution patterns and splicing phases were analyzed manually using BioEdit (v7.0.5). Intron phases were determined as follows: splicing occurring after the third nucleotide of the codon as phase 0, splicing occurring after the first nucleotide of the codon as phase 1 and splicing occurring after the second nucleotide as phase 2. To trace the plant PHO1 gene structures evolution, we conducted the character state reconstruction using Mesquite version 2.72 (http://mesquiteproject.org) [58]. The structure of the PHO1 genes possesses 19 character states. They are a)14-EXON type , b) 1st-Intron loss type c) 2nd-Intron loss type, d) 3rd-Inron absent type, e) 4th-Intron absent type, f) 5th-Intron absent type, g) 6th-Intron absent type, h) 7th-Intron absent type, i) 8th-Intron absent type, j) 9th-Intron absent type, k) 10th-Inrton absent type, l) 11th-Intron absent type, m) 12th-Intron absent type, n) 13th-Intron absent type, o) Intronless type, p) 1st-Intron insertion type, q) Animal-14-Intron type, r) Animal-15-Intron type, and s) unknown type. A total of 227 matrices reflecting the gene structures were built, and the topology reflecting the phylogenetic tree of PHO1 genes was used as the input tree. Reconstruction of the ancestral state was performed using MP criterion. The unordered model in which all state changes are treated equally was applied for the parsimony analyses.
Plant growth and stress treatments
Soybean (Glycine max L.) Nannong1138-2 cultivars were grown in vermiculite in a greenhouse at 22±2°C with a 16/10 h (light/dark) photoperiod. Roots, leaves, and stems were harvested from 3-week-old seedlings. Mature flowers and the fruits of 1-, 3-, 5-, and 7-days after fertilization were sampled in the greenhouse. For stress treatments, 3-week-old seedlings were grown in a 0.5×Hoagland solution (pH5.8). Drought stress was mimicked by submerging seedlings in Hoagland solution containing 15% (w/v) PEG 6000 for 4 and 8 h. The seedlings were subjected to salt stress by growing them in Hoagland solution containing 150 mM NaCl for 4 and 8 h. We designed 2 experiments for Pi stress: (1) first subjecting the seedlings to low Pi stress (LP, 0 mM phosphorus) for 24 h, and then high Pi stress (HP, 5 mM phosphorus) for 24 h. The second treatment approach was to reverse the stresses, presenting HP first for 24 h and then LP for 24 h. Untreated seedlings in Hoagland solution were used as controls for all samples. Roots were harvested at the indicated times. Collected plant materials were flash-frozen in liquid nitrogen and then stored at −80°C for RNA isolation.
Real-time RT-PCR analysis
Total RNAs was extracted with the Trizol Reagent (Invitrogen, Carlsbad, USA) and treated with the RQ1 RNase-free DNase (Promega, Madison, WI) to avoid DNA contamination. Then, 2.0 μg total RNA was used to synthesize cDNA with the oligo (dT)18 primer following the instructions of the M-MLV cDNA synthesis kit (Invitrogen, Carlsbad, CA). Real time RT-PCR analysis was performed on an Mx3000P QPCR system (Stratagene, Germany) using SYBR Premix Ex Taq (TaKaRa, Tokyo, Japan). Gene-specific primers are listed in Additional file 11: Table S7. Primers specificity was verified using the BLAST tool from the NCBI and a dissociation curve analysis was performed after the PCR reaction. The efficiency E value was calculated for all primer pairs individually by plotting the relationship between Ct value (threshold cycle) and log [cDNA]. PCRs were done using the following thermal cycle conditions: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 40 s. Controls without template were included for each primer pair and all reactions were repeated three times for 3 independent biological samples. Relative gene expression was analyzed using the cycle threshold (Ct) 2-ΔΔCt method [59] and normalized using the housekeeping gene actin. Two-tailed Student’s t-test was used to determine the statistical significance of gene expression variation between different tissues and in response to stresses (*P < 0.05, **P < 0.01 and ***P < 0.001). Statistical analysis was performed in SPSS 15.0 for Windows.
Abbreviations
ML: Maximum-likelihood; PHO1: PHOSPHATE1; RT-PCR: Reverse transcription-polymerase chain reaction amplification; RACE: Rapid amplification of cDNA ends.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CYH designed the study and conceived the experiments. LLH characterized the sequences and carried out most of the experiments. YW participated expression analyses. LLH and MZ performed evolutionary analyses. LLH and CYH analyzed data. JYG and CYH coordinated the work. LLH and CYH drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Information of the PHO1 gene family used in the present work.
ML trees generated using both WAG and LG models. (a) ML tree generated under LG model. (b) MLtree generatedunder WAG model. Bootstrap values (>50%) for this tree are shown on each branch. The different classes are marked with different colored backgrounds. The genes from the basal land plants are indicated in pink; dicot in red, monocots in blue and gymnosperm in green, respectively.
Copy numbers of PHO1 genes in different classes in 30 land plant species.
The presence of tandem duplications in Class IA.
Percent of members with different numbers of introns in each class.
Evolution of intron gain and loss events of the plant PHO1 gene family. ML tree of the plant PHO1 genes is identical to Figure1. Ancestral state reconstruction of PHO1 gene structure was conducted by Mesquite. Color scale depicts 19 different intron states of gene structures. Pie charts linked with the internal branches represent probability of ancestral states. Solid (one color) node indicates that the particular state is significantly better than all other possible states. Gene structures are based on the predicted sequences from the Phytozome database. The intron numbers corresponding to the assumed ancestral PHO1 genes are shown at the top. Numbers in boxes represent intron phases 0, 1 or 2, respectively. Black boxes indicate conserved introns, white boxes indicate lost introns, and gray boxes indicate inserted introns.
Chromosomal location of GmaPHO1 genes. The horizontal line represents the chromosome and the vertical lines represent the GmaPHO1 genes. Arrows indicate the transcriptional directions of the corresponding genes. H1-H14 are abbreviated for GmaPHO1; H1-H14 genes.
Variation in exon number and length of GmaPHO1 genes.
Multiple sequence alignment of PHO1 proteins in soybean, Arabidopsis and rice. The characteristic domains of the PHO1 proteins are highlighted in red (SPX, Pfam PF03105) and pink (EXS, Pfam PF03124) lines, respectively.
Pairwise comparison of the GmaPHO1 protein family.
Primer sequences of GmaPHO1 genes used in the present work.
The reciprocal significance of gene expression variation among tissues. (a-e) Significance of organ-specific expressions the Class I genes. (f-i) Significance of organ-specific expressions of the Class II genes.
Expression of GmaPHO1 genes under Pi deficient or sufficient treatments. (a-d) Root expression of the Class I genes in response to Pi stresses. (e-h) Root expression of the Class II genes in response to Pi stresses. The gene is indicated in each graph. Light grey lines represent the untreated controls; dark gray lines represent the treatment of LP-HP; black lines represent the treatment of HP-LP (see Methods). Actin was used as an internal control. The experiments were performed based on three independent biological samples. Error bars=standard deviations. Asterisks indicate significance of gene expression variation under each treatment in comparison to the untreated control (*P < 0.05, **P < 0.01 and ***P < 0.001).
Contributor Information
Lingli He, Email: linglihe@ibcas.ac.cn.
Man Zhao, Email: mzhao@ibcas.ac.cn.
Yan Wang, Email: ywang@ibcas.ac.cn.
Junyi Gai, Email: sri@njau.edu.cn.
Chaoying He, Email: chaoying@ibcas.ac.cn.
Acknowledgements
The authors want to thank Dr. Patrick Matheny and multiple anonymous reviewers for their valuable suggestion and comments to improve the manuscript. This work was supported by the Hundred Talent Program of the Chinese Academy of Sciences to CYH and by the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-J-1).
References
- Hamburger D, Rezzonico E, MacDonald-Comber Petetot J, Somerville C, Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 2002;14(4):889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell. 2009;21(11):3554–3566. doi: 10.1105/tpc.108.064980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Stefanovic A, Secco D, Arpat AB, Gout E, Bligny R, Poirier Y. Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J. 2011;65(4):557–570. doi: 10.1111/j.1365-313X.2010.04442.x. [DOI] [PubMed] [Google Scholar]
- Stefanovic A, Arpat AB, Bligny R, Gout E, Vidoudez C, Bensimon M, Poirier Y. Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J. 2011;66(4):689–699. doi: 10.1111/j.1365-313X.2011.04532.x. [DOI] [PubMed] [Google Scholar]
- Arpat AB, Magliano P, Wege S, Rouached H, Stefanovic A, Poirier Y. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant J. 2012;71(3):479–491. doi: 10.1111/j.1365-313X.2012.05004.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 2004;135(1):400–411. doi: 10.1104/pp.103.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J, Belbahri L, Delessert S, Poirier Y. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 2007;50(6):982–994. doi: 10.1111/j.1365-313X.2007.03108.x. [DOI] [PubMed] [Google Scholar]
- Secco D, Baumann A, Poirier Y. Characterization of the rice PHO1 gene family reveals a key role for OsPHO1; 2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol. 2010;152(3):1693–1704. doi: 10.1104/pp.109.149872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Secco D, Poirier Y. Characterization of the PHO1 gene family and the responses to phosphate deficiency of Physcomitrella patens. Plant Physiol. 2008;146(2):646–656. doi: 10.1104/pp.107.108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Ni M. Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell. 2006;18(4):921–934. doi: 10.1105/tpc.105.037879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ni M. SHB1 plays dual roles in photoperiodic and autonomous flowering. Dev Biol. 2009;331(1):50–57. doi: 10.1016/j.ydbio.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhao XY, Kang X, Zhang XS, Ni M. SHORTHYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell. 2009;21(1):1–12. doi: 10.1105/tpc.109.210180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ni M. SHORT HYPOCOTYL UNDER BLUE1 truncations and mutations alter its association with a signaling protein complex in Arabidopsis. Plant Cell. 2010;22(3):703–715. doi: 10.1105/tpc.109.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot C, Zimmerli C, Farmer E, Reymond P, Poirier Y. Induction of the Arabidopsis PHO1; H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1-dependent pathway. Plant Physiol. 2008;147(2):696–706. doi: 10.1104/pp.108.119321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot C, Wang Y, Poirier Y. Expression analyses of three members of the AtPHO1 family reveal differential interactions between signaling pathways involved in phosphate deficiency and the responses to auxin, cytokinin, and abscisic acid. Planta. 2008;227(5):1025–1036. doi: 10.1007/s00425-007-0677-x. [DOI] [PubMed] [Google Scholar]
- Zimmerli C, Ribot C, Vavasseur A, Bauer H, Hedrich R, Poirier Y. PHO1 expression in guard cells mediates the stomatal response to abscisic acid in Arabidopsis. Plant J. 2012;72(2):199–211. doi: 10.1111/j.1365-313X.2012.05058.x. [DOI] [PubMed] [Google Scholar]
- Battini JL, Rasko JEJ, Miller AD. A human cell surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc Natl Acad Sci USA. 1999;96:1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain BH, Koo D, Ramakrishnan M, Dzudzor B, Colicelli J. Truncated forms of a novel yeast protein suppress the lethality of a G protein subunit deficiency by interacting with the subunit. J Biol Chem. 1995;270:25435–25444. doi: 10.1074/jbc.270.43.25435. [DOI] [PubMed] [Google Scholar]
- Clemente TE, Cahoon EB. Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiol. 2009;151(3):1030–1040. doi: 10.1104/pp.109.146282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RC, Polzin K, Labate J, Specht J, Brummer EC, Olson T, Young N, Concibido V, Wilcox J, Tamulonis JP, Kochert G, Boerma HR. Genome duplication in soybean (Glycine subgenus soja) Genetics. 1996;144(1):329–338. doi: 10.1093/genetics/144.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8(2):135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16(7):1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Pattyn P, Gerats T, van de Peer Y. Journey through the past: 150 million years of plant genome evolution. Plant J. 2011;66(1):58–65. doi: 10.1111/j.1365-313X.2011.04521.x. [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol. 2011;62:185–206. doi: 10.1146/annurev-arplant-042110-103849. [DOI] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008;54:965–975. doi: 10.1111/j.1365-313X.2008.03460.x. [DOI] [PubMed] [Google Scholar]
- Secco D, Wang C, Arpat AB, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 2012;193(4):842–851. doi: 10.1111/j.1469-8137.2011.04002.x. [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Scheffler BE, Schlueter SD, Shoemaker RC. Sequence conservation of homeologous bacterial artificial chromosomes and transcription of homeologous genes in soybean (Glycine max L. Merr.) Genetics. 2006;174(2):1017–1028. doi: 10.1534/genetics.105.055020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;2(11):97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3(11):827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008;148:993–1003. doi: 10.1104/pp.108.122457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarrio R, Ayala FJ. Origins and evolution of spliceosomal introns. Annu Rev Genet. 2006;40:47–76. doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Logsdon JMJ. The recent origins of introns. Curr Opin Genet Dev. 1991;1(4):470–477. doi: 10.1016/S0959-437X(05)80194-7. [DOI] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. Rates of intron loss and gain: implications for early eukaryotic evolution. Proc Natl Acad Sci USA. 2005;102(16):5773–5778. doi: 10.1073/pnas.0500383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Lyons-Weiler J, Koonin EV. Intron sliding in conserved gene families. Trends Genet. 2000;16(10):430–432. doi: 10.1016/S0168-9525(00)02096-5. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Carmel L, Csuros M, Koonin EV. Origin and evolution of spliceosomal introns. Biol Direct. 2012;7:11. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman R, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Fromm M, Walbot D. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Rose AB, Korf I. Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res. 2011;39(13):5328–5337. doi: 10.1093/nar/gkr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XY, Vorst O, Fiers MW, Stiekema WJ, Nap JP. In plants, highly expressed genes are the least compact. Trends Genet. 2006;22:528–532. doi: 10.1016/j.tig.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Chaubet-Gigot N, Kapros T, Flenet M, Kahn K, Gigot C, Waterborg JH. Tissue-specific enhancement of transgene expression by introns of replacement histone H3 genes of Arabidopsis. Plant Mol Biol. 2001;45:17–30. doi: 10.1023/A:1006487023926. [DOI] [PubMed] [Google Scholar]
- Clancy M, Hannah LC. Splicing of the maize Sh1 first intron is essential for enhancement of gene expression, and a T-rich motif increases expression without affecting splicing. Plant Physiol. 2002;130:918–929. doi: 10.1104/pp.008235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JH, Lee SY, Yu HJ, Jeong YM, Shin MY, Kim H, Lee I, Kim SG. Petunia actin depolymerizing factor is mainly accumulated in vascular tissue and its gene expression is enhanced by the first intron. Gene. 2002;292:233–243. doi: 10.1016/S0378-1119(02)00646-7. [DOI] [PubMed] [Google Scholar]
- Bolle C, Herrmann RG, Oelmuller R. Intron sequences are involved in the plastid- and light-dependent expression of the spinach PsaD gene. Plant J. 1996;10:919–924. doi: 10.1046/j.1365-313X.1996.10050919.x. [DOI] [PubMed] [Google Scholar]
- Deyholos MK, Sieburth LE. Separable whorl-specific expression and negative regulation by enhancer element within the AGAMOUS second intron. Plant Cell. 2000;12:1799–1810. doi: 10.1105/tpc.12.10.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell. 2002;14:2527–2537. doi: 10.1105/tpc.004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 2010;38(Web Server issue):W23–W28. doi: 10.1093/nar/gkq443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56(4):564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Chen K, Durand D, Farach-Colton M. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol. 2000;7:429–447. doi: 10.1089/106652700750050871. [DOI] [PubMed] [Google Scholar]
- Durand D, Halldórsson BV, Vernot B. A hybrid micro-macroevolutionary approach to gene tree reconstruction. J Comput Biol. 2006;13(2):320–335. doi: 10.1089/cmb.2006.13.320. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.7. 2008. http://mesquiteproject.org.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information of the PHO1 gene family used in the present work.
ML trees generated using both WAG and LG models. (a) ML tree generated under LG model. (b) MLtree generatedunder WAG model. Bootstrap values (>50%) for this tree are shown on each branch. The different classes are marked with different colored backgrounds. The genes from the basal land plants are indicated in pink; dicot in red, monocots in blue and gymnosperm in green, respectively.
Copy numbers of PHO1 genes in different classes in 30 land plant species.
The presence of tandem duplications in Class IA.
Percent of members with different numbers of introns in each class.
Evolution of intron gain and loss events of the plant PHO1 gene family. ML tree of the plant PHO1 genes is identical to Figure1. Ancestral state reconstruction of PHO1 gene structure was conducted by Mesquite. Color scale depicts 19 different intron states of gene structures. Pie charts linked with the internal branches represent probability of ancestral states. Solid (one color) node indicates that the particular state is significantly better than all other possible states. Gene structures are based on the predicted sequences from the Phytozome database. The intron numbers corresponding to the assumed ancestral PHO1 genes are shown at the top. Numbers in boxes represent intron phases 0, 1 or 2, respectively. Black boxes indicate conserved introns, white boxes indicate lost introns, and gray boxes indicate inserted introns.
Chromosomal location of GmaPHO1 genes. The horizontal line represents the chromosome and the vertical lines represent the GmaPHO1 genes. Arrows indicate the transcriptional directions of the corresponding genes. H1-H14 are abbreviated for GmaPHO1; H1-H14 genes.
Variation in exon number and length of GmaPHO1 genes.
Multiple sequence alignment of PHO1 proteins in soybean, Arabidopsis and rice. The characteristic domains of the PHO1 proteins are highlighted in red (SPX, Pfam PF03105) and pink (EXS, Pfam PF03124) lines, respectively.
Pairwise comparison of the GmaPHO1 protein family.
Primer sequences of GmaPHO1 genes used in the present work.
The reciprocal significance of gene expression variation among tissues. (a-e) Significance of organ-specific expressions the Class I genes. (f-i) Significance of organ-specific expressions of the Class II genes.
Expression of GmaPHO1 genes under Pi deficient or sufficient treatments. (a-d) Root expression of the Class I genes in response to Pi stresses. (e-h) Root expression of the Class II genes in response to Pi stresses. The gene is indicated in each graph. Light grey lines represent the untreated controls; dark gray lines represent the treatment of LP-HP; black lines represent the treatment of HP-LP (see Methods). Actin was used as an internal control. The experiments were performed based on three independent biological samples. Error bars=standard deviations. Asterisks indicate significance of gene expression variation under each treatment in comparison to the untreated control (*P < 0.05, **P < 0.01 and ***P < 0.001).