Abstract
Mortierella alpina is a filamentous fungus commonly found in soil that is able to produce lipids in the form of triacylglycerols that account for up to 50% of its dry weight. Analysis of the M. alpina genome suggests that there is a phenylalanine-hydroxylating system for the catabolism of phenylalanine, which has never been found in fungi before. We characterized the phenylalanine-hydroxylating system in M. alpina to explore its role in phenylalanine metabolism and its relationship to lipid biosynthesis. Significant changes were found in the profile of fatty acids in M. alpina grown on medium containing an inhibitor of the phenylalanine-hydroxylating system compared to M. alpina grown on medium without inhibitor. Genes encoding enzymes involved in the phenylalanine-hydroxylating system (phenylalanine hydroxylase [PAH], pterin-4α-carbinolamine dehydratase, and dihydropteridine reductase) were expressed heterologously in Escherichia coli, and the resulting proteins were purified to homogeneity. Their enzymatic activity was investigated by high-performance liquid chromatography (HPLC) or visible (Vis)-UV spectroscopy. Two functional PAH enzymes were observed, encoded by distinct gene copies. A novel role for tetrahydrobiopterin in fungi as a cofactor for PAH, which is similar to its function in higher life forms, is suggested. This study establishes a novel scheme for the fungal degradation of an aromatic substance (phenylalanine) and suggests that the phenylalanine-hydroxylating system is functionally significant in lipid metabolism.
INTRODUCTION
The phenylalanine-hydroxylating system catalyzes the irreversible hydroxylation of phenylalanine to tyrosine, which is the rate-limiting step in animal phenylalanine catabolism and protein and neurotransmitter biosynthesis and results in the formation of one molecule of fumarate and one of acetyl-coenzyme A (CoA) from each molecule of phenylalanine (1, 2). The animal phenylalanine-hydroxylating system consists of several essential components: phenylalanine hydroxylase (PAH) (EC 1.14.16.1), pterin-4α-carbinolamine dehydratase (PCD) (EC 4.2.1.96), dihydropteridine reductase (DHPR) (EC 1.5.1.34), and the obligatory cofactors tetrahydrobiopterin (BH4) and molecular oxygen (3, 4) (Fig. 1). PAH is one of the BH4-dependent aromatic amino acid hydroxylases, which also include tryptophan hydroxylase (TrpOHase) (EC 1.14.16.4) and tyrosine hydroxylase (TyrOHase) (EC 1.14.16.2) (5). BH4 is regenerated following PAH-mediated phenylalanine hydroxylation by two additional enzymes, PCD and DHPR (Fig. 1) (6). The PAH, PCD, and DHPR genes responsible for the phenylalanine-hydroxylating system in animals have already been characterized (7, 8).
Fig 1.
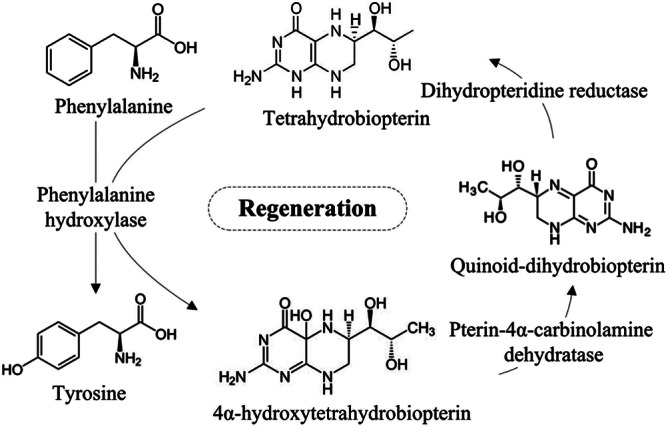
The animal phenylalanine-hydroxylating system.
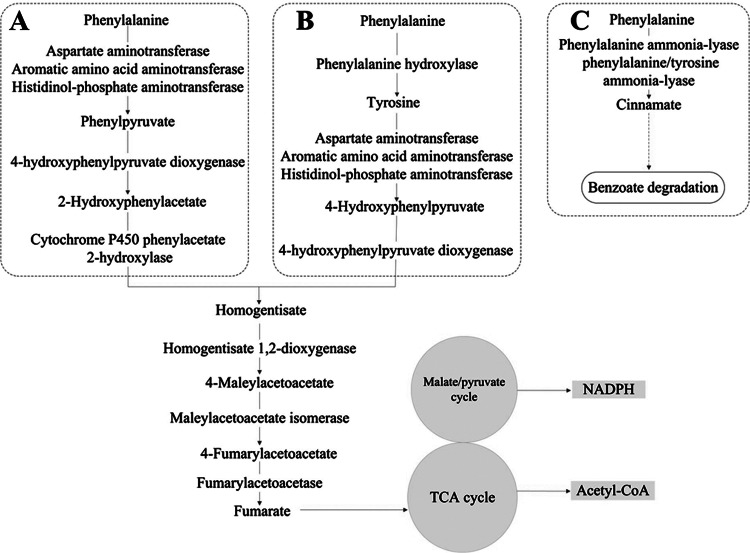
Phenylalanine is an important aromatic compound, one of the structurally diverse and second most abundant class of organic substrates (9). The greatest challenge for organisms using aromatic compounds as growth substrates is the stabilizing resonance energy of the aromatic ring system (10). This aromatic structure makes the substrate unreactive to oxidation or reduction and thus requires elaborate degradation strategies (9). The degradation of aromatic substances is dominated by aerobic and anaerobic bacteria and aerobic fungi, and the strategies used by these organisms are quite different from those of animals (10). Unlike higher organisms, most bacteria, fungi, and plants do not convert phenylalanine into tyrosine. The presence of PAH has been reported in only a few bacteria based on the identification of related genes (11–14). In the fungi Aspergillus nidulans and Aspergillus fumigatus, tyrosine was thought to be synthesized from phenylalanine by PAH (15, 16); however, no gene encoding PAH was found in their genomes (see Table S1 in the supplemental material). To date, no PAH, PCD, or DHPR involved in the phenylalanine-hydroxylating system has been identified in any fungi. Phenylalanine is converted into phenylacetate in Penicillium chrysogenum, A. nidulans, and Aspergillus niger (Fig. 2A) or into cinnamic acid in Bjerkandera adusta and Schizophyllum commune (Fig. 2C) (17–20). The cinnamate thus formed is converted to protocatechuate through benzoate, whereas phenylpyruvate is converted to homogentisate, which is catabolized by cleavage of the aromatic ring to yield fumarate and acetyl-CoA. Recently, we sequenced the whole genome of Mortierella alpina (ATCC 32222) (21), and our analysis suggested that there are two putative copies of the BH4-dependent PAH gene for the catabolism of phenylalanine in M. alpina. Genes encoding PCD and DHPR, which are required for the regeneration of BH4, an essential component of the phenylalanine-hydroxylating system (6), were also found in the M. alpina genome. The BH4 de novo synthesis pathway in M. alpina was first purified and characterized in our laboratory (22), but the function of BH4 in fungi is still not well understood. The presence of the phenylalanine-hydroxylating system in M. alpina suggests that fungi can make use of the system in their phenylalanine degradation strategy.
Fig 2.
Fungal degradation of an aromatic substance (phenylalanine) in P. chrysogenum, A. nidulans, and A. niger (A), M. alpina (B), and B. adusta and S. commune (C). TCA, tricarboxylic acid.
M. alpina is a well-known polyunsaturated fatty acid (PUFA)-producing oleaginous fungus commonly found in soil (23). Some of the genes necessary for lipid synthesis in M. alpina have been cloned and partially characterized (24–28), and several biochemical reactions have been studied in detail (29, 30). However, the molecular mechanism of efficient lipid biosynthesis is still not well understood in oleaginous fungi in general and in M. alpina in particular. PAH and its cofactor BH4 have been suggested to be essential for lipid metabolism in higher organisms (31–36), and some amino acid metabolism pathways have been postulated to be involved in fatty acid biosynthesis in oleaginous fungi (37, 38). However, the functional significance of the phenylalanine-hydroxylating system in the biosynthesis of lipids and closely related compounds has yet to be fully elucidated. In higher organisms, the phenylalanine-hydroxylating system is inhibited by p-chlorophenylalanine or esculin. Both are complete and irreversible inhibitors of PAHs in vivo (39, 40). M. alpina is noteworthy for its production of PUFA de novo and, due to its high lipid content (23), provides an interesting model for studying the relationship between the phenylalanine-hydroxylating system and lipid metabolism.
In this study, we investigated the role of the phenylalanine-hydroxylating system in lipid metabolism and probed its possible function in the degradation of aromatic substances. We established a novel role for BH4 in fungi similar to that known to exist in higher life forms. We characterized the genes encoding PAH, PCD, and DHPR and their functions in the phenylalanine-hydroxylating system in vitro. We identified two functional PAH genes (the PAH-1 and PAH-2 genes) and measured kinetic parameters and the effects of temperature, pH, and aromatic amino acids on PAH activity. Multiple-sequence alignment and phylogenetic analysis of the PAH proteins with other, homologous proteins were also performed.
MATERIALS AND METHODS
Gene search.
Predicted proteins in the M. alpina (ATCC 32222) genome (GenBank accession number ADAG00000000) were annotated by BLAST (41) searches against the following protein databases with the E value 1E−5: NR (http://www.ncbi.nlm.nih.gov), KOGs and COGs (42), KEGG (43), Swiss-Prot and UniRef100 (44), and BRENDA (45) and by InterProScan (46) using the default parameter settings. Pathway mapping was conducted by associating the EC assignment and KO assignment with the KEGG metabolic pathways based on the BLAST search results. The predicted PAH-1, PAH-2, PCD, and DHPR proteins in the M. alpina genome were searched against predicted proteins of sequenced fungal genomes by BLAST with the E value 1E−5.
Strains and growth conditions.
M. alpina (ATCC 32222) was cultured on potato dextrose agar at 25°C for 5 to 7 days. The fungal cultures were initially cultivated in 200 ml of Kendrick medium (47) and incubated at 25°C for 6 days. The mycelia were then collected by filtration through sterile cheesecloth and frozen immediately in liquid nitrogen for RNA extraction.
Aromatic compound degradation test.
The indicated sole carbon sources (glucose at 3%, phenylalanine at 25 mM, phenylacetate at 10 mM, and tyrosine at 25 mM) (15) were added to minimal medium to test the aromatic compound degradation in M. alpina. The minimal medium contained 0.3% diammonium tartrate, 0.7% KH2PO4, 0.2% Na2HPO4, 0.15% MgSO4 · 7H2O, 0.67% yeast nitrogen base, and 2% agarose, pH 6.0. The plates were photographed after 3 days of incubation at 25°C. The mycelia of M. alpina were stained with 0.01% triphenyltetrazolium chloride (TTC) to facilitate growth measurements (48). This experiment was replicated three times.
Effect of a PAH inhibitor on lipid synthesis.
For the whole-cell inhibition studies, cultures were grown in 50 ml of Kendrick medium (50 g/liter glucose, 2 g/liter diammonium tartrate, 7.0 g/liter KH2PO4, 2.0 g/liter Na2HPO4, 1.5 g/liter MgSO4 · 7H2O, 1.5 g/liter Bacto yeast extract, 0.1 g/liter CaCl22H2O, 8 mg/liter FeCl3 · 6H2O, 1 mg/liter ZnSO4 · 7H2O, 0.1 mg/liter CuSO4 · 5H2O, 0.1 mg/liter Co(NO3)2 · 6H2O, and 0.1 mg/liter MnSO4 · 5H2O, pH 6.0) with the addition of 5 mM p-chlorophenylalanine. The medium and incubation protocols were as previously described (21). The lipid composition of M. alpina was also investigated when p-chlorophenylalanine (5 mM) and tyrosine (5 mM) were both added to the medium. M. alpina cultures were incubated at 25°C for 6 days. Aspergillus oryzae is another oleaginous fungus (49, 50), and genome analysis suggests there is no phenylalanine-hydroxylating system in A. oryzae (see Table S2 in the supplemental material). The addition of PAH inhibitor in A. oryzae was included to verify whether there were any changes in lipids in the absence of a phenylalanine-hydroxylating system. A. oryzae was incubated at 25°C for 6 days. The mycelia were then collected by filtration, and approximately 20 mg was used for each lipid extraction. Accurately weighed portions of pulverized mycelium were extracted using the method of Bligh and Dyer (51) under acidified conditions with pentadecanoic acid and heneicosanoic acid added as internal standards (21). This experiment was replicated 3 times.
Cloning and plasmid construction.
Total RNA extraction was performed using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA was subjected to RNase-free DNase digestion and then purified using the RNeasy Minikit (Qiagen). The quantity and quality of the total RNA were evaluated using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc.). The total RNA was reverse transcribed with the PrimeScript RT reagent kit (TaKaRa Bio, Inc.) following the manufacturer's instructions before PCR amplification of PAH-1, PAH-2, PCD, and DHPR genes using the primer pairs shown in Table 1. The PCR conditions used were as follows: denaturation at 95°C for 30 s, annealing at 50°C for 45 s, and extension at 72°C for 1 min (for 25 cycles in total). The final volume in each well was 50 μl. The amplified product was cloned into pET28a+ to construct pwl47864 (containing PAH-1), pwl47866 (containing PAH-2), pwl39696 (containing PCD), or pwl39698 (containing DHPR). The presence of inserts in the plasmids was confirmed by sequencing using an ABI 3730 sequencer.
Table 1.
Primers used in the study
| Primer | Primer sequence used for cloninga |
|---|---|
| PAH-1 (forward) | CGGAATTCATGTCTGCTGCTGCCAACCACCT |
| PAH-1 (reverse) | CCCAAGCTTTTACGCCAGCTTCTGCAGGGC |
| PAH-2 (forward) | CGGAATTCATGTCCTCCACCCCAGGCC |
| PAH-2 (reverse) | CCCAAGCTTTTACATCTTCTGCAGGGCATCGAC |
| PCD (forward) | CGGAATTCATGACTCTCAAGAAGCTCGATG |
| PCD (reverse) | CCGCTCGAGTTATTGCTTGAAGATCTCATCG |
| DHPR (forward) | CGGAATTCATGGTTTCTAGCATCGTCGTCTAT |
| DHPR (reverse) | CCGCTCGAGTTACTTCTCGGTGTACGTGGTGT |
Restriction enzyme cleavage sites are underlined.
Protein expression and purification.
Escherichia coli BL21 carrying pwl47864, pwl47866, pwl39696, or pwl39698 was grown overnight at 37°C with shaking in LB medium containing 50 μg/ml of kanamycin. The overnight culture (10 ml) was inoculated into 500 ml of fresh medium and grown until the optical density at 600 nm (OD600) reached 0.6. PAH-1 expression was induced by the addition of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 30°C for 20 h, PAH-2 expression was induced by 0.01 mM IPTG at 30°C for 20 h, and PCD or DHPR expression was induced by 0.5 mM IPTG at 37°C for 16 h. After the IPTG induction, cells were harvested by centrifugation, washed with binding buffer (50 mM Tris-HCl, 300 mM NaCl, and 10 mM imidazole; pH 8.0), resuspended in the same buffer (supplemented with 1 mM phenylmethanesulfonyl fluoride and 1 mg/ml lysozyme), and then sonicated. Cell debris was removed by centrifugation, and the supernatants (containing the soluble proteins) were collected. The His-tagged fusion proteins were purified by nickel ion affinity chromatography using a Chelating Sepharose Fast Flow column (GE Healthcare) according to the manufacturer's instructions. Unbound proteins were washed through with 100 ml of wash buffer (50 mM Tris-HCl, 300 mM NaCl, and 25 mM imidazole, pH 8.0). Fusion proteins were eluted with 3 ml of elution buffer (50 mM Tris-HCl, 300 mM NaCl, and 250 mM imidazole, pH 8.0) and dialyzed overnight at 4°C against 50 mM Tris-HCl buffer containing 20% glycerol (pH 7.4). The protein concentrations were determined by the Bradford method. The purified proteins were stored at −80°C.
Enzyme activity assays.
The PAH activity was assayed using the method of Bailey and Ayling (52) with minor modifications. The reaction mixture for PAH contained 100 mM Tris-HCl (pH 7.0), 1 mM phenylalanine, 1 mg/ml catalase, and 0.14 μg/ml purified PAH-1 protein or 0.22 μg/ml purified PAH-2 protein and was maintained at 25°C for 5 min. Then, 100 μM ferrous ammonium sulfate was added and allowed to incubate for 1 min. The reaction was started by the addition of 100 μM BH4 (Sigma) or 6-methyltetrahydropterin (Sigma) and 5 mM dithiothreitol (DTT) in a total volume of 50 μl. After a period of 15 min, the reaction was stopped by adding 50 μl 2 M trichloroacetic acid. Samples were prepared for high-performance liquid chromatography (HPLC) after centrifugation. The HPLC was performed on a Shimadzu model LC-20AT equipped with a Rheodyne (IDEX Health and Science) loop of 20 μl, an Inertsil ODS-3 column (5 μm; 150 by 4.6 mm; GL Science), and a Shimadzu model RF-20A fluorescence detector. The mobile phase consisted of 0.1 M NH4OH adjusted to pH 4.6 with acetic acid at a flow rate of 1.5 ml/min. To observe the separation of phenylalanine and tyrosine, the excitation and emission wavelengths of the fluorescence detector were set at 260 and 282 nm, respectively (53). To determine the amount of tyrosine, the excitation and emission wavelengths of the fluorescence detector were set at 274 and 304 nm, respectively (52). Quantitation was based on external calibration using a standard curve prepared with different amounts of tyrosine.
The reaction mixture for DHPR contained 100 mM Tris-HCl (pH 7.0), 10 μg/ml peroxidase, 10 mM hydrogen peroxide, 10 μM BH4, and 0.1 mM NADH. After an incubation period of 4.5 min at 25°C, the reaction was started by adding 3.99 μg/ml purified DHPR protein in a final volume of 1 ml. The initial rates were obtained from the initial rate of decrease of A340 (E340 for NADH, 6,200 M−1 cm−1) (54, 55). The background reactions were eliminated when the reaction was catalyzed by peroxidase or DHPR alone.
The 50-μl PCD assay reaction mixture contained 100 mM Tris-HCl (pH 7.0), 1 mg/ml catalase, 1 mM l-phenylalanine, and 0.14 μg/ml purified PAH-1 protein. The mixture was incubated for 2 min at 25°C, and 20 μM BH4 and 5 mM DTT were added. After a 5-hour PAH reaction, 0.1 mM NADH, 6.24 μg/ml purified PCD protein, and 3.99 μg/ml purified DHPR protein were added. The amount of tyrosine formed after 24 h was determined fluorometrically and corrected for the tyrosine formed in controls without PCD (56, 57). The background reactions were eliminated when the reaction was catalyzed by PCD or DHPR alone. These experiments were replicated 3 times.
Determination of the temperature and pH optima and effects of aromatic amino acids on PAH activity.
In order to determine the optimum temperature, PAH reactions were carried out at 5°C, 15°C, 25°C, 37°C, 50°C, and 60°C for 15 min at pH 7.0. For determinations of optimum pH, three different buffers were used over a range of pH 3 to 11, including 100 mM sodium acetate-acetic acid (pH 3 to 6), 100 mM Tris-HCl (pH 7 to 8), and 100 mM glycine-NaOH (pH 10 to 12). These reactions were carried out for 15 min at 25°C. A range of amino acid concentrations (0.5 μM to 5 mM) were used to test their effects on the PAH activity, including tryptophan, glycine, alanine, p-chlorophenylalanine, and esculin. These reactions were carried out for 15 min at 25°C, pH 7.0. The experiments were replicated 3 times.
Measurement of kinetic parameters.
To measure the Km and Vmax values for both PAHs using phenylalanine, reactions were carried out using various concentrations of phenylalanine (0.5 to 2 mM) in conjunction with a fixed concentration of BH4 (1 mM). To measure the Km and Vmax values for both PAHs acting on BH4, reactions were carried out using various concentrations of BH4 (70 to 400 μM) but with a fixed concentration of phenylalanine (1 mM). The PAH reactions were performed in Tris-HCl, pH 7.5, at 25°C for 15 min in a final volume of 50 μl. The conversion of phenylalanine to tyrosine was monitored by HPLC. The kinetic parameters were obtained by fitting the initial rates of reaction measured at various concentrations of one substrate at a fixed concentration of the other to the Michaelis-Menten equation using nonlinear regression. This experiment was replicated 3 times.
RESULTS
Gene search.
Putative PAH-1, PAH-2, PCD, and DHPR genes were found in the M. alpina genomes (GenBank accession numbers JN982953, JN982954, JQ281474, and JQ281475), suggesting the presence of a phenylalanine-hydroxylating system in the fungus. The search results for all of the sequenced fungal genomes are shown in Table S1 in the supplemental material.
Aromatic compound degradation test.
The growth phenotypes of M. alpina are shown in Fig. S1 in the supplemental material. M. alpina showed fine hyphal growth on medium containing phenylalanine (see Fig. S1B in the supplemental material) or tyrosine (see Fig. S1E) as the sole carbon source. The growth of M. alpina was repressed by the addition of p-chlorophenylalanine to medium containing phenylalanine as the sole source of carbon (see Fig. S1D). M. alpina was unable to grow on medium containing phenylacetate as the sole carbon source (see Fig. S1C), even if it was incubated for over 2 weeks or at a higher phenylacetate concentration (1 M), which is in contrast to other fungi, such as P. chrysogenum, A. nidulans, and A. niger (17–20). Thus, these results show that M. alpina can degrade phenylalanine and tyrosine, but not phenylacetate.
Effects of p-chlorophenylalanine on PUFA synthesis.
To explore the relationship between the phenylalanine-hydroxylating system and lipid synthesis, we investigated the effects of 5 mM p-chlorophenylalanine, an inhibitor of the phenylalanine-hydroxylating system in higher organisms, on the fatty acid status (see Table S2 in the supplemental material). The M. alpina cultures grown in medium plus inhibitor showed reduced total saturated fatty acids (SAFA), ω6 PUFA, ω3 PUFA, and total fatty acid (FA) accumulation levels by about 30%, 50%, 20%, and 30%, respectively, while the level of monounsaturated fatty acid (MUFA) was increased by about 30% (Fig. 3). The addition of tyrosine did not change the inhibitory effects of p-chlorophenylalanine on lipids when M. alpina was grown in medium plus inhibitor and tyrosine (Fig. 3). The lipid status of A. oryzae grown on PAH inhibitor medium did not change compared to medium without inhibitor (see Table S2 in the supplemental material), suggesting that the PAH inhibitor used in our experiment is selective.
Fig 3.
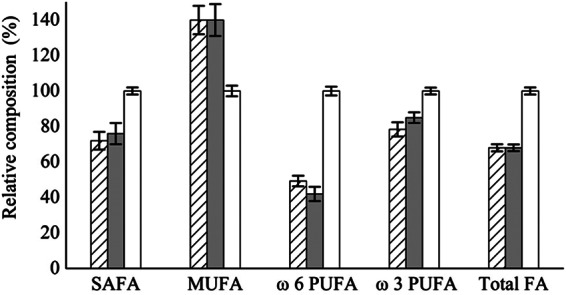
Effects of an inhibitor (p-chlorophenylalanine) of the phenylalanine-hydroxylating system on fatty acid synthesis in M. alpina. The hatched bars, shaded bars, and open bars represent M. alpina grown in inhibitor medium, inhibitor and tyrosine medium, and inhibitor-free medium, respectively. The data were normalized to the concentration of fatty acids (mg/g [cell dry weight]) from M. alpina grown on inhibitor-free medium, defined as 100%. The data shown are the averages (± standard deviations) of three independent experiments.
Expression and purification of PAHs, PCD, and DHPR.
PAH-1, PAH-2, PCD, and DHPR were expressed as His-tagged fusion proteins in E. coli BL21 by IPTG induction and purified to near homogeneity by nickel ion affinity chromatography (see Fig. S2 in the supplemental material). The molecular masses estimated from the SDS-PAGE analysis were 54 kDa for PAH-1 and PAH-2, 14 kDa for PCD, and 27 kDa for DHPR, which correspond well to the calculated masses (excluding the His tag, these were 50 kDa, 50 kDa, 11 kDa, and 23 kDa, respectively). The yields of PAH-1, PAH-2, PCD, and DHPR were 1.40 μg/ml, 2.24 μg/ml, 62.44 μg/ml, and 39.88 μg/ml, respectively.
Activities of PAHs, PCD, and DHPR.
Phenylalanine was incubated with the purified PAH proteins, and the reaction products were identified by HPLC, as shown in Fig. S3 in the supplemental material. Phenylalanine was observed at 6.5 min, corresponding to the position of the phenylalanine standard (see Fig. S3B in the supplemental material) in the sample incubated in the absence of the PAHs (see Fig. S3C). Phenylalanine was converted to a peak that eluted at 2.6 min, corresponding to the position of the tyrosine standard (see Fig. S3A) in the reaction catalyzed by PAH-1 (see Fig. S3D) or PAH-2 (see Fig. S3E). These results confirm that the recombinant proteins show PAH activity, which is dependent on the presence of all of the substrates (including BH4 and 6-methyltetrahydropterin). To examine whether the purified DHPR protein showed DHPR activity, we determined the initial velocity of its NADH consumption. When the reaction was catalyzed by peroxidase or DHPR alone, it had no effect on the rate of NADH oxidation. The initial rate of NADH oxidation was dramatically increased by the addition of the recombinant protein, indicating that the purified protein possesses DHPR activity (see Fig. S4 in the supplemental material). The specific activity of the recombinant DHPR protein was 29.92 μmol/min/mg. One unit of DHPR activity was defined as being the amount of enzyme that caused the oxidation of 1 μmol NADH per minute under the above conditions. PCD activity was assayed by the ability to stimulate PAH activity in the presence of BH4. When the reaction was catalyzed by PCD or DHPR alone, it had no effect on the rate of tyrosine production. The amount of tyrosine formed with the addition of PCD was 0.17 mmol, and the tyrosine formed in controls without PCD was 0.13 mmol. Thus, the specific activity of the recombinant PCD protein was 0.35 μmol/min/mg. One unit of PCD activity is defined as 1 μmol of tyrosine formed in 30 min under the above conditions. The data shown are the averages of three independent experiments.
Effects of temperature, pH, and aromatic amino acids on PAH activity.
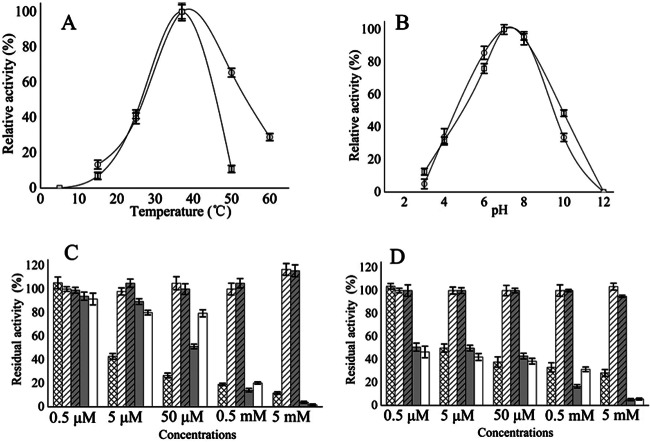
PAH-1 and PAH-2 were active at each of the temperatures tested (in the range of 15°C to 65°C), with the greatest activity detected at 37°C and 40°C (Fig. 4A). One hundred percent activity in the PAH-1 and the PAH-2 reactions occurred at 37°C under the aforementioned conditions. The optimal temperatures for PAH-1 and PAH-2 are in accordance with data previously obtained for humans and Dictyostelium discoideum (58, 59) but are different from data obtained for Chromobacterium violaceum, Colwellia psychrerythraea, and Chloroflexus aurantiacus (13, 60, 61). PAH-1 activity decreased quickly at higher temperatures and was limited at 50°C (Fig. 4A). For PAH-2, high activity (65%) was observed at temperatures up to 50°C, and activity was still found at 60°C (Fig. 4A). Therefore, PAH-2 was more heat stable than PAH-1.
Fig 4.
Effects of temperature (A), pH (B), and aromatic amino acids (C and D) on PAH-1 (A and B, open circles, and C) and PAH-2 (A and B, open squares, and D) activity. The crosshatched bars, open hatched bars, shaded hatched bars, shaded bars, and open bars show the effects of tryptophan, glycine, alanine, p-chlorophenylalanine, and esculin, respectively, on PAH activity. Each point represents the average (± standard deviation) of three independent measurements.
PAH-1 and PAH-2 both showed optimal activity at pH 7.5 in 100 mM Tris-HCl buffer (Fig. 4B), which is similar to the optimum pH for recombinant human and C. violaceum PAHs (59, 60). Maximal activity for both PAH-1 and PAH-2 occurred at pH 7.0 under the aforementioned conditions. The PAHs were active over a broad range of pH values (4 to 10), which contrasts with the significantly decreased activity of human PAH below pH 5.5. PAH-1 and PAH-2 exhibited a very similar pH sensitivity profile.
Some aromatic amino acids react with the substrate binding site of PAH and can be hydroxylated under appropriate conditions in vitro or can competitively inhibit the hydroxylation of phenylalanine (59). PAH-1 activity was significantly inhibited by 5 mM concentrations of a variety of amino acids, including tryptophan (90% inhibition) and p-chlorophenylalanine (96% inhibition) (Fig. 4C). PAH-1 activity was also significantly inhibited by 5 mM esculin (98% inhibition) (Fig. 4C). The inhibition profile of PAH-2 activity was similar: tryptophan (70% inhibition), p-chlorophenylalanine (95% inhibition), and esculin (95% inhibition) (Fig. 4D). However, low concentrations of p-chlorophenylalanine and esculin were more effective in reducing PAH-2 than PAH-1 activity. Glycine and alanine had little effect on PAH-1 and PAH-2 activity. One hundred percent activity was defined as a PAH reaction without competing amino acids under the previous PAH activity assay conditions. The inhibitory effects of the aromatic amino acids and esculin were also observed for PAH in humans, and this inhibition is one of the hallmarks of PAH activity (59).
Comparative analysis of M. alpina PAH proteins and other homologous proteins.
The kinetic parameters of PAH-1 and PAH-2 (for phenylalanine and BH4) were measured. The kinetics of the reactions catalyzed by these enzymes present a good fit with the Michaelis-Menten model, and nonlinear regression of the experimental data to the Michaelis-Menten equation yielded the kinetic parameters shown in Table 2. PAH-1 and PAH-2 share 12.7 to 55.3% identity with other characterized PAH proteins and are most similar to those from D. discoideum. Multiple-sequence alignment of the M. alpina PAHs and other characterized PAH proteins revealed the presence of certain conserved motifs and residues (see Fig. S5 in the supplemental material), including two motifs, Gly-Ala-Leu (residues 46 to 48 in human PAH) and Ile-Glu-Ser-Arg-Pro (residues 65 to 69 in human PAH), that form a phenylalanine-binding pocket (62); a Gly-Leu-Leu-Ser-Ser motif (residues 247 to 251 in human PAH) involved in BH4 cofactor binding (13, 63, 64); two motifs, Tyr-Thr-Pro-Glu-Pro (residues 277 to 281 in human PAH) and Gly-Ala-Gly-Leu-Leu-Ser (residues 344 to 350 in human PAH), involved in substrate binding (13, 64); 4 iron-binding residues (human His146, Arg158, His285, and His290) (65–67); 4 residues (human Arg270, Trp326, Phe331, and Glu353) involved in substrate binding (13, 64); 6 residues (human Phe254, Leu255, His264, Glu286, Ala322, and Tyr325) involved in BH4 cofactor binding (13, 63, 64); and 6 residues (human Pro122, Leu128, Phe131, Ala132, Arg158, and His146) involved in the N-terminal regulatory domain. The residues of the pterin binding loop, the N-terminal loop, and the loop region between residues 272 and 282 differ from bacterial and human versions of PAH, creating a 10-fold-higher specific activity for BH4 in C. violaceum than in humans (65, 68). These residues in human PAH are almost all conserved in M. alpina PAH, except for human Ile125, Ile135, and Gly272 and the N-terminal autoregulatory sequence (NARS) (human residues 19 to 33) (65), which is consistent with the similar activity observed in human PAH and M. alpina PAHs. Residues Asp116, Lys274, and Ser439, which are responsible for dimer and tetramer formation, substrate binding, and deamidation rates, respectively (65, 67), are not conserved in PAH-1 but do occur in PAH-2. The increased Km of M. alpina PAH-1 compared with PAH-2 may be due to the loss of one or several of these residues, which may consequently impair substrate binding and decrease catalytic activity. The other enzymatic properties of PAH-1 and PAH-2 are generally similar, which is consistent with the data observed for eukaryotes (58, 59).
Table 2.
Kinetic parametersa
| Enzyme | Substrate | Km (mM) | Vmax (μM/min) |
|---|---|---|---|
| PAH-1 | Phenylalanine | 0.1750 ± 0.003962 | 3.827 ± 0.1342 |
| PAH-1 | BH4 | 0.1253 ± 0.002518 | 2.176 ± 0.03927 |
| PAH-2 | Phenylalanine | 0.06885 ± 0.002018 | 11.21 ± 0.2164 |
| PAH-2 | BH4 | 0.04778 ± 0.001889 | 5.879 ± 0.06391 |
The data shown are the averages of three independent experiments.
Phylogenetic analysis of PAH.
A phylogenetic tree was generated with amino acid hydroxylase (AAH) sequences from fungi, animals, protists, plants, and bacteria (see Fig. S6 in the supplemental material). The tree is rooted with PAHs from plant species as an outgroup and further recognizes PAH as an ancestral AAH function and gives insight into the evolution of AAHs (58, 69). Based on the search results in the fungal genomes (see Table S1 in the supplemental material), M. alpina, Allomyces macrogynus, Batrachochytrium dendrobatidis, Rhizopus oryzae, Spizellomyces punctatus, Mucor circinelloides, and Phycomyces blakesleeanus are fungi that contain a putative PAH gene. The BH4 de novo pathway is conserved in all of these fungi except A. macrogynus, due to the absence of the 6-pyruvoyltetrahydropterin synthase gene (EC 4.2.3.12) (see Table S1 in the supplemental material). PAH cannot work in A. macrogynus due to the lack of cofactor BH4, which may be why A. macrogynus PAH and other fungal PAHs cluster as a distinct group in the phylogenetic tree. Our phylogenetic analysis of fungal PAH sequences and animal, protist, plant, and bacterial AAHs of known specificity places the fungal proteins closest to those of animals. The tree illustrates the clear distinction between fungi that contain the phenylalanine-hydroxylating system and those that do not and also shows the close relationship between M. alpina and animal PAH (see Fig. S6 in the supplemental material). Based on sequence comparisons and enzymatic properties, the PAHs from M. alpina and animals are thought to have evolved from the same ancestor.
DISCUSSION
The fungal BH4 synthesis pathway has been characterized at the molecular level only in M. alpina (22), and BH4 acts as a cofactor in nitric oxide synthesis only in the fungi P. blakesleeanus and Neurospora crassa (70, 71). Because most fungi do not synthesize BH4 and lack BH4-dependent monooxygenases, the function of BH4 in fungi is unclear. The BH4-dependent PAH is present in only about 7% of the fungal genomes sequenced so far (see Table S1 in the supplemental material), and other BH4-dependent enzymes have not been discovered in M. alpina. This work provides direct biochemical evidence that BH4 can function as a biological cofactor for PAH in fungi, based on the characterization of the BH4 synthesis pathway and PAH homologues in M. alpina. This function has never been demonstrated in fungi before, and thus, we hypothesize that BH4 plays a novel role in fungi similar to that known to exist in higher life forms.
In most fungi, phenylalanine is converted into phenylacetate. The conventional aerobic route for phenylacetate, which leads to homogentisate, occurs in fungi (Fig. 2) (17–20). However, the M. alpina genome sequence lacks an ortholog of phenylacetate 2-hydroxylase (EC 1.14.13.-) to catalyze the hydroxylation of phenylacetate to homogentisate (Fig. 2B), which is consistent with the inability of M. alpina to grow on medium containing phenylacetate as the sole carbon source (see Fig. S1C in the supplemental material). This is in contrast to the fact that most fungi can grow on phenylacetate medium (15). Because this step is needed for the complete oxidation of phenylalanine to carbon dioxide and water (14), conventional fungal phenylalanine catabolism is blocked in M. alpina. However, M. alpina can grow on phenylalanine as the sole carbon source (see Fig. S1B in the supplemental material), which means that it has an unconventional strategy for phenylalanine metabolism. In this report, we have shown that in this organism, phenylalanine is hydroxylated to tyrosine by the phenylalanine-hydroxylating system; tyrosine can then be converted into 4-hydroxyphenylpyruvate, followed by hydroxylation to homogentisate, the same intermediate as in other fungi (Fig. 2). The putative genes needed for tyrosine oxidation are all present in the M. alpina genome, which was proved by the fact that M. alpina could grow on tyrosine as the sole carbon source (see Fig. S1E in the supplemental material). Our characterization of the phenylalanine-hydroxylating system provides the molecular basis for a novel scheme for the degradation of an aromatic substance (phenylalanine) in fungi. This paradigm for phenylalanine catabolism differs significantly from the established knowledge for other fungi. Conserved genes of the phenylalanine-hydroxylating system have been found in 7% of the fungal species sequenced thus far (see Table S1 in the supplemental material). These species belong to the groups of mucoromycotina (M. alpina, R. oryzae, M. circinelloides, and P. blakesleeanus) and chytridiomycota (B. dendrobatidis and S. punctatus). Thus, this scheme for aromatic degradation is of major importance but is not widespread in fungi.
In higher organisms, PAH and its cofactor BH4 are suggested to be essential for lipid metabolism. Mutations in the human PAH gene result in the metabolic disorder known as phenylketonuria (PKU) (67), and PKU patients present with lower concentrations of long-chain polyunsaturated fatty acids (LCPUFA), such as arachidonic acid (AA) and eicosapentaenoic acid, and higher concentrations of oleic acid 18:1 (31, 32). It has also been suggested that the PAH cofactor BH4 is essential for the desaturation or omega oxidation of long-chain fatty acids and the incorporation of unsaturated fatty acids into phospholipids (33–36). Significant changes were observed in the concentration of total ω6 PUFA and AA in M. alpina grown on PAH inhibitor medium compared to M. alpina grown on medium without inhibitor (Fig. 3). Of all the fatty acids, AA—the main commercial product of M. alpina (23)—showed the most pronounced change. Interestingly, the trends in the lipid status of M. alpina grown on PAH inhibitor medium parallel those observed in PKU patients. These results suggest that the phenylalanine-hydroxylating system may control lipid accumulation, either on its own or via interactions with other as yet unrecognized regulatory factors in M. alpina. Acetyl-CoA and NADPH are two key factors in lipid metabolism. Acetyl-CoA is the essential building block and necessary precursor of fatty acids (30), and NADPH is the critical reducing agent and limiting factor in fatty acid synthesis (72). Based on the genome information, the phenylalanine catabolism pathway in M. alpina was constructed to map the utilization of phenylalanine (Fig. 2). The M. alpina phenylalanine-hydroxylating system is supposed to be blocked by PAH inhibitor, which may interfere with fatty acid synthesis by reducing the production of NADPH and acetyl-CoA through the tricarboxylic acid cycle. We hypothesize that the function of the phenylalanine-hydroxylating system in M. alpina, including PAH and BH4, is to contribute NADPH and acetyl-CoA for lipid accumulation by the utilization of phenylalanine and tyrosine, and this may also explain in part how M. alpina achieves its high level of lipid synthesis. Moreover, as there are two copies of PAH in M. alpina and PAH is the rate-limiting factor in animal phenylalanine catabolism (1, 2), ectopic overexpression of PAH should enhance the ability of M. alpina to use phenylalanine for lipid synthesis. RNA interference (RNAi) of PAH genes should be utilized in future work to explore the relationship between the phenylalanine-hydroxylating system and lipid synthesis.
It was suggested that NADPH produced by malic enzymes was the unique source of reducing power for fatty acid synthesis in fungi like M. circinelloides and M. alpina and that malic enzyme is consequently the rate-limiting step for fatty acid biosynthesis (73). Recently, leucine metabolism was found to be another bottleneck in lipid accumulation in the oleaginous fungus M. circinelloides (37). In that work, the overexpression of the LEU2 gene (a gene encoding β-isopropylmalate dehydrogenase, involved in leucine biosynthesis) significantly increased the lipid content, in accordance with previous observations in Saccharomyces cerevisiae (74). Comparative genome analysis of the nonoleaginous and oleaginous strains points to a relationship between lipid and amino acid metabolism, including leucine and lysine degradation and lysine biosynthesis, in the biosynthesis of the important intermediate metabolite acetyl-CoA, which might be related to the capability of the oleaginous microorganisms to accumulate high levels of lipids (38). Leucine and lysine metabolic pathways were all conserved in the M. alpina genome. Our study suggests that phenylalanine metabolism in M. alpina may also be important for lipid accumulation. It may support previous data postulating amino acid metabolism as one of the pathways involved in the generation of the acetyl-CoA required for fatty acid biosynthesis (38, 74). The relationship between lipid and amino acid metabolism is thus of major importance and is probably widespread in oleaginous fungi. Our identification of a phenylalanine-hydroxylating system involved in amino acid metabolism and lipid accumulation extends the range of target candidates for genetic manipulation in M. alpina for obtaining strains with increased amounts of lipids.
In conclusion, the phenylalanine-hydroxylating system has been characterized at the molecular level in a fungus, M. alpina. Genes encoding PAH, PCD, and DHPR were functionally characterized in vitro. Two functional PAH genes (the PAH-1 and PAH-2 genes) were observed. A novel role for tetrahydrobiopterin in fungi as a cofactor for PAH similar to that known to exist in higher life forms is suggested. This work establishes a novel scheme for the fungal degradation of an aromatic substance (phenylalanine) and suggests that the phenylalanine-hydroxylating system is functionally significant in lipid metabolism.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Fund for Distinguished Young Scholars (31125021), the National Natural Science Foundation of China (21276108, 31271812, 31171636, and 20836003), the National High Technology Research and Development Program of China (2011AA100905), the National Basic Research Program of China 973 Program (2012CB720802), the 111 project B07029, Fundamental Research Funds for the Central Universities (JUSRP51320B), and the Doctor Candidate Foundation of Jiangnan University (JUDCF11026).
Footnotes
Published ahead of print 15 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00238-13.
REFERENCES
- 1. Kappock TJ, Caradonna JP. 1996. Pterin-dependent amino acid hydroxylases. Chem. Rev. 96: 2659–2756 [DOI] [PubMed] [Google Scholar]
- 2. Neckameyer WS, Coleman CM, Eadie S, Goodwin SF. 2007. Compartmentalization of neuronal and peripheral serotonin synthesis in Drosophila melanogaster. Genes Brain Behav. 6: 756–769 [DOI] [PubMed] [Google Scholar]
- 3. Ding Z, Harding CO, Rebuffat A, Elzaouk L, Wolff JA, Thony B. 2008. Correction of murine PKU following AAV-mediated intramuscular expression of a complete phenylalanine hydroxylating system. Mol. Ther. 16: 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaufman S. 1971. The phenylalanine hydroxylating system from mammalian liver. Adv. Enzymol. Relat. Areas Mol. Biol. 35: 245–319 [DOI] [PubMed] [Google Scholar]
- 5. Werner-Felmayer G, Golderer G, Werner ER. 2002. Tetrahydrobiopterin biosynthesis, utilization and pharmacological effects. Curr. Drug Metab. 3: 159–173 [DOI] [PubMed] [Google Scholar]
- 6. Thony B, Auerbach G, Blau N. 2000. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 347: 1–16 [PMC free article] [PubMed] [Google Scholar]
- 7. Shiman R, Gray DW, Hill MA. 1994. Regulation of rat liver phenylalanine hydroxylase. I. Kinetic properties of the enzyme's iron and enzyme reduction site. J. Biol. Chem. 269: 24637–24646 [PubMed] [Google Scholar]
- 8. Martinez A, Knappskog PM, Olafsdottir S, Doskeland AP, Eiken HG, Svebak RM, Bozzini M, Apold J, Flatmark T. 1995. Expression of recombinant human phenylalanine hydroxylase as fusion protein in Escherichia coli circumvents proteolytic degradation by host cell proteases. Isolation and characterization of the wild-type enzyme. Biochem. J. 306: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. U. S. A. 107: 14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds—from one strategy to four. Nat. Rev. Microbiol. 9: 803–816 [DOI] [PubMed] [Google Scholar]
- 11. Zhao G, Xia T, Song J, Jensen RA. 1994. Pseudomonas aeruginosa possesses homologues of mammalian phenylalanine hydroxylase and 4 alpha-carbinolamine dehydratase/DCoH as part of a three-component gene cluster. Proc. Natl. Acad. Sci. U. S. A. 91: 1366–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakata H, Yamauchi T, Fujisawa H. 1979. Phenylalanine hydroxylase from Chromobacterium violaceum. Purification and characterization. J. Biol. Chem. 254: 1829–1833 [PubMed] [Google Scholar]
- 13. Leiros HK, Pey AL, Innselset M, Moe E, Leiros I, Steen IH, Martinez A. 2007. Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. J. Biol. Chem. 282: 21973–21986 [DOI] [PubMed] [Google Scholar]
- 14. Arias-Barrau E, Olivera ER, Luengo JM, Fernandez C, Galan B, Garcia JL, Diaz E, Minambres B. 2004. The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 186: 5062–5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fernandez-Canon JM, Penalva MA. 1995. Fungal metabolic model for human type I hereditary tyrosinaemia. Proc. Natl. Acad. Sci. U. S. A. 92: 9132–9136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmaler-Ripcke J, Sugareva V, Gebhardt P, Winkler R, Kniemeyer O, Heinekamp T, Brakhage AA. 2009. Production of pyomelanin, a second type of melanin, via the tyrosine degradation pathway in Aspergillus fumigatus. Appl. Environ. Microbiol. 75: 493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodriguez-Saiz M, Barredo JL, Moreno MA, Fernandez-Canon JM, Penalva MA, Diez B. 2001. Reduced function of a phenylacetate-oxidizing cytochrome p450 caused strong genetic improvement in early phylogeny of penicillin-producing strains. J. Bacteriol. 183: 5465–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kishore G, Sugumaran M, Vaidyanathan CS. 1976. Metabolism of DL-(+/−)-phenylalanine by Aspergillus niger. J. Bacteriol. 128: 182–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lapadatescu C, Ginies C, Le Quere JL, Bonnarme P. 2000. Novel scheme for biosynthesis of aryl metabolites from L-phenylalanine in the fungus Bjerkandera adusta. Appl. Environ. Microbiol. 66: 1517–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rao PV, Moore K, Towers GH. 1967. Degradation of aromatic amino acids by fungi. II. Purification and properties of phenylalanine ammonia-lyase from Ustilago hordei. Can. J. Biochem. 45: 1863–1872 [DOI] [PubMed] [Google Scholar]
- 21. Wang L, Chen W, Feng Y, Ren Y, Gu Z, Chen H, Wang H, Thomas MJ, Zhang B, Berquin IM, Li Y, Wu J, Zhang H, Song Y, Liu X, Norris JS, Wang S, Du P, Shen J, Wang N, Yang Y, Wang W, Feng L, Ratledge C, Chen YQ. 2011. Genome characterization of the oleaginous fungus Mortierella alpina. PLoS One 6: e28319 doi:10.1371/journal.pone.0028319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang H, Yang B, Hao G, Feng Y, Chen H, Feng L, Zhao J, Zhang H, Chen YQ, Wang L, Chen W. 2011. Biochemical characterization of the tetrahydrobiopterin synthesis pathway in the oleaginous fungus Mortierella alpina. Microbiology 157: 3059–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sakuradani E, Ando A, Ogawa J, Shimizu S. 2009. Improved production of various polyunsaturated fatty acids through filamentous fungus Mortierella alpina breeding. Appl. Microbiol. Biotechnol. 84: 1–10 [DOI] [PubMed] [Google Scholar]
- 24. Michaelson LV, Lazarus CM, Griffiths G, Napier JA, Stobart AK. 1998. Isolation of a Delta5-fatty acid desaturase gene from Mortierella alpina. J. Biol. Chem. 273: 19055–19059 [DOI] [PubMed] [Google Scholar]
- 25. Sakuradani E, Kobayashi M, Shimizu S. 1999. Delta6-fatty acid desaturase from an arachidonic acid-producing Mortierella fungus. Gene cloning and its heterologous expression in a fungus, Aspergillus. Gene 238: 445–453 [DOI] [PubMed] [Google Scholar]
- 26. Sakuradani E, Kobayashi M, Shimizu S. 1999. Delta 9-fatty acid desaturase from arachidonic acid-producing fungus. Unique gene sequence and its heterologous expression in a fungus, Aspergillus. Eur. J. Biochem. 260: 208–216 [DOI] [PubMed] [Google Scholar]
- 27. Sakuradani E, Kobayashi M, Ashikari T, Shimizu S. 1999. Identification of Delta12-fatty acid desaturase from arachidonic acid-producing mortierella fungus by heterologous expression in the yeast Saccharomyces cerevisiae and the fungus Aspergillus oryzae. Eur. J. Biochem. 261: 812–820 [DOI] [PubMed] [Google Scholar]
- 28. Sakuradani E, Abe T, Iguchi K, Shimizu S. 2005. A novel fungal omega3-desaturase with wide substrate specificity from arachidonic acid-producing Mortierella alpina 1S-4. Appl. Microbiol. Biotechnol. 66: 648–654 [DOI] [PubMed] [Google Scholar]
- 29. Ratledge C. 2002. Regulation of lipid accumulation in oleaginous micro-organisms. Biochem. Soc. Trans. 30: 1047–1050 [DOI] [PubMed] [Google Scholar]
- 30. Ratledge C. 2004. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie 86: 807–815 [DOI] [PubMed] [Google Scholar]
- 31. Moseley K, Koch R, Moser AB. 2002. Lipid status and long-chain polyunsaturated fatty acid concentrations in adults and adolescents with phenylketonuria on phenylalanine-restricted diet. J. Inherited Metab. Dis. 25: 56–64 [DOI] [PubMed] [Google Scholar]
- 32. Giovannini M, Biasucci G, Agostoni C, Luotti D, Riva E. 1995. Lipid status and fatty acid metabolism in phenylketonuria. J. Inherited Metab. Dis. 18: 265–272 [DOI] [PubMed] [Google Scholar]
- 33. Kaufman S. 1993. New tetrahydrobiopterin-dependent systems. Annu. Rev. Nutr. 13: 261–286 [DOI] [PubMed] [Google Scholar]
- 34. Forrest HS, Van Baalen C. 1970. Microbiology of unconjugated pteridines. Annu. Rev. Microbiol. 24: 91–108 [DOI] [PubMed] [Google Scholar]
- 35. Kaufman S. 1967. Pteridine cofactors. Annu. Rev. Biochem. 36: 171–184 [DOI] [PubMed] [Google Scholar]
- 36. Rudzite V, Jurika E, Baier-Bitterlich G, Widner B, Reibnegger G, Fuchs D. 1998. Pteridines and lipid metabolism. Pteridines 9: 103–112 [Google Scholar]
- 37. Rodriguez-Frometa RA, Gutierrez A, Torres-Martinez S, Garre V. 2012. Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biotechnol. [Epub ahead of print.] doi:10.1007/s00253-012-4432-2 [DOI] [PubMed] [Google Scholar]
- 38. Vorapreeda T, Thammarongtham C, Cheevadhanarak S, Laoteng K. 2012. Alternative routes of acetyl-CoA synthesis identified by comparative genomic analysis: involvement in the lipid production of oleaginous yeast and fungi. Microbiology 158: 217–228 [DOI] [PubMed] [Google Scholar]
- 39. Valdivieso F, Gimenez C, Mayor F. 1975. In vivo inhibition of rat liver phenylalanine hydroxylase by p-chlorophenylalanine and esculin. Experimental model of phenylketonuria. Biochem. Med. 12: 72–78 [DOI] [PubMed] [Google Scholar]
- 40. Guroff G. 1969. Irreversible in vivo inhibition of rat liver phenylalanine hydroxylase by p-chlorophenylalanine. Arch. Biochem. Biophys. 134: 610–611 [DOI] [PubMed] [Google Scholar]
- 41. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O'Donovan C, Redaschi N, Suzek B. 2006. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 34: D187–D191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. 2009. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 37: D588–D592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33: W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kendrick A, Ratledge C. 1992. Desaturation of polyunsaturated fatty acids in Mucor circinelloides and the involvement of a novel membrane-bound malic enzyme. Eur. J. Biochem. 209: 667–673 [DOI] [PubMed] [Google Scholar]
- 48. Zhu M, Yu LJ, Liu Z, Xu HB. 2004. Isolating Mortierella alpina strains of high yield of arachidonic acid. Lett. Appl. Microbiol. 39: 332–335 [DOI] [PubMed] [Google Scholar]
- 49. Meng X, Yang JM, Xu X, Zhang L, Nie QJ, Xian M. 2009. Biodiesel production from oleaginous microorganisms. Renewable Energy 34: 1–5 [Google Scholar]
- 50. Shi S, Valle-Rodriguez JO, Siewers V, Nielsen J. 2011. Prospects for microbial biodiesel production. Biotechnol. J. 6: 277–285 [DOI] [PubMed] [Google Scholar]
- 51. Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- 52. Bailey SW, Ayling JE. 1980. An assay for picomole levels of tyrosine and related phenols and its application to the measurement of phenylalanine hydroxylase activity. Anal. Biochem. 107: 156–164 [DOI] [PubMed] [Google Scholar]
- 53. Kand'ar R, Zakova P. 2009. Determination of phenylalanine and tyrosine in plasma and dried blood samples using HPLC with fluorescence detection. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 3926–3929 [DOI] [PubMed] [Google Scholar]
- 54. Armarego WL, Cotton RG, Dahl HH, Dixon NE. 1989. High-level expression of human dihydropteridine reductase (EC 1.6.99.7), without N-terminal amino acid protection, in Escherichia coli. Biochem. J. 261: 265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Craine JE, Hall ES, Kaufman S. 1972. The isolation and characterization of dihydropteridine reductase from sheep liver. J. Biol. Chem. 247: 6082–6091 [PubMed] [Google Scholar]
- 56. Davis MD, Kaufman S. 1989. Evidence for the formation of the 4a-carbinolamine during the tyrosine-dependent oxidation of tetrahydrobiopterin by rat liver phenylalanine hydroxylase. J. Biol. Chem. 264: 8585–8596 [PubMed] [Google Scholar]
- 57. Hauer CR, Rebrin I, Thony B, Neuheiser F, Curtius HC, Hunziker P, Blau N, Ghisla S, Heizmann CW. 1993. Phenylalanine hydroxylase-stimulating protein/pterin-4 alpha-carbinolamine dehydratase from rat and human liver. Purification, characterization, and complete amino acid sequence. J. Biol. Chem. 268: 4828–4831 [PubMed] [Google Scholar]
- 58. Siltberg-Liberles J, Steen IH, Svebak RM, Martinez A. 2008. The phylogeny of the aromatic amino acid hydroxylases revisited by characterizing phenylalanine hydroxylase from Dictyostelium discoideum. Gene 427: 86–92 [DOI] [PubMed] [Google Scholar]
- 59. Ledley FD, Grenett HE, Woo SL. 1987. Biochemical characterization of recombinant human phenylalanine hydroxylase produced in Escherichia coli. J. Biol. Chem. 262: 2228–2233 [PubMed] [Google Scholar]
- 60. Zoidakis J, Loaiza B, Vu K, Abu-Omar MM. 2005. Effect of temperature, pH, and metals on the stability and activity of phenylalanine hydroxylase from Chromobacterium violaceum. J. Inorg. Biochem. 99: 771–775 [DOI] [PubMed] [Google Scholar]
- 61. Pey AL, Martinez A. 2009. Iron binding effects on the kinetic stability and unfolding energetics of a thermophilic phenylalanine hydroxylase from Chloroflexus aurantiacus. J. Biol. Inorg. Chem. 14: 521–531 [DOI] [PubMed] [Google Scholar]
- 62. Gjetting T, Petersen M, Guldberg P, Guttler F. 2001. Missense mutations in the N-terminal domain of human phenylalanine hydroxylase interfere with binding of regulatory phenylalanine. Am. J. Hum. Genet. 68: 1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Andersen OA, Flatmark T, Hough E. 2001. High resolution crystal structures of the catalytic domain of human phenylalanine hydroxylase in its catalytically active Fe(II) form and binary complex with tetrahydrobiopterin. J. Mol. Biol. 314: 279–291 [DOI] [PubMed] [Google Scholar]
- 64. Andersen OA, Flatmark T, Hough E. 2002. Crystal structure of the ternary complex of the catalytic domain of human phenylalanine hydroxylase with tetrahydrobiopterin and 3-(2-thienyl)-L-alanine, and its implications for the mechanism of catalysis and substrate activation. J. Mol. Biol. 320: 1095–1108 [DOI] [PubMed] [Google Scholar]
- 65. Erlandsen H, Kim JY, Patch MG, Han A, Volner A, Abu-Omar MM, Stevens RC. 2002. Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates. J. Mol. Biol. 320: 645–661 [DOI] [PubMed] [Google Scholar]
- 66. Kobe B, Jennings IG, House CM, Michell BJ, Goodwill KE, Santarsiero BD, Stevens RC, Cotton RG, Kemp BE. 1999. Structural basis of autoregulation of phenylalanine hydroxylase. Nat. Struct. Biol. 6: 442–448 [DOI] [PubMed] [Google Scholar]
- 67. Fusetti F, Erlandsen H, Flatmark T, Stevens RC. 1998. Structure of tetrameric human phenylalanine hydroxylase and its implications for phenylketonuria. J. Biol. Chem. 273: 16962–16967 [DOI] [PubMed] [Google Scholar]
- 68. Erlandsen H, Bjorgo E, Flatmark T, Stevens RC. 2000. Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase. Biochemistry 39: 2208–2217 [DOI] [PubMed] [Google Scholar]
- 69. Pribat A, Noiriel A, Morse AM, Davis JM, Fouquet R, Loizeau K, Ravanel S, Frank W, Haas R, Reski R, Bedair M, Sumner LW, Hanson AD. 2010. Nonflowering plants possess a unique folate-dependent phenylalanine hydroxylase that is localized in chloroplasts. Plant Cell 22: 3410–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Maier J, Hecker R, Rockel P, Ninnemann H. 2001. Role of nitric oxide synthase in the light-induced development of sporangiophores in Phycomyces blakesleeanus. Plant Physiol. 126: 1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ninnemann H, Maier J. 1996. Indications for the occurrence of nitric oxide synthases in fungi and plants and the involvement in photoconidiation of Neurospora crassa. Photochem. Photobiol. 64: 393–398 [DOI] [PubMed] [Google Scholar]
- 72. Zhang Y, Adams IP, Ratledge C. 2007. Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153: 2013–2025 [DOI] [PubMed] [Google Scholar]
- 73. Wynn JP, bin Abdul Hamid A, Ratledge C. 1999. The role of malic enzyme in the regulation of lipid accumulation in filamentous fungi. Microbiology 145: 1911–1917 [DOI] [PubMed] [Google Scholar]
- 74. Kamisaka Y, Tomita N, Kimura K, Kainou K, Uemura H. 2007. DGA1 (diacylglycerol acyltransferase gene) overexpression and leucine biosynthesis significantly increase lipid accumulation in the Deltasnf2 disruptant of Saccharomyces cerevisiae. Biochem. J. 408: 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.