Abstract
The African malaria mosquito Anopheles gambiae is polymorphic for chromosomal inversion 2La, whose frequency strongly correlates with degree of aridity across environmental gradients. Recent physiological studies have associated 2La with resistance to desiccation in adults and thermal stress in larvae, consistent with its proposed role in aridity tolerance. However, the genetic basis of these traits remains unknown. To identify genes that could be involved in the differential response to thermal stress, we compared global gene expression profiles of heat hardened 2La or 2L+a larvae at three time points, for up to eight hours following exposure to the heat stress. Treatment and control time series, replicated four times, revealed a common and massive induction of a core set of heat shock genes regardless of 2La orientation. However, clear differences between the 2La and 2L+a arrangements emerged at the earliest (0.25 h) time point, in the intensity and nature of the stress response. Overall, 2La was associated with the more aggressive response: larger numbers of genes were heat responsive and up-regulated. Transcriptionally induced genes were enriched for functions related to ubiquitin-proteasomal degradation, chaperoning, and energy metabolism. The more muted transcriptional response of 2L+a was largely repressive, including genes involved in proteolysis and energy metabolism. These results may help explain the maintenance of the 2La inversion polymorphism in An. gambiae, as the survival benefits offered by high thermal sensitivity in harsh climates could be offset by the metabolic costs of such a drastic response in more equable climates.
Keywords: Anopheles gambiae, chromosomal inversion, heat hardening, malaria vector, microarray, thermal stress, transcriptional profiling
Introduction
Africa bears a high burden of morbidity and mortality due to the malignant human malaria parasite, Plasmodium falciparum (Rowe et al. 2006). The principal African mosquito vector responsible for its transmission is Anopheles gambiae, a species whose distribution spans much of the continent south of the Sahara, and includes a wide diversity of ecogeographic regions. This mosquito’s ability to adapt to seasonal and spatial environmental heterogeneities, notably those arising as a result of anthropogenic environmental modification, expands the scope and stability of malaria transmission in Africa (Coluzzi 1994; Coluzzi et al. 1979; della Torre et al. 2002).
The ecological breadth of An. gambiae sets it apart from all but one other member (Anopheles arabiensis) of the eponymous An. gambiae complex, comprising at least seven sibling species. This extraordinary ecological flexibility is associated with a very high degree of inversion polymorphism absent from less widespread members of the species complex. These observations suggest a causal link between inversion polymorphism and adaptive potential in this species group (Coluzzi et al. 2002; Coluzzi et al. 1979; Costantini et al. 2009; Pombi et al. 2008; Powell et al. 1999), as has been postulated in other species (Ayala et al. 2011; Hoffmann& Rieseberg 2008; Hoffmann et al. 2004; Olivera et al. 1979; Schaeffer 2008).
Evidence for selection on inversion polymorphisms comes from their nonrandom spatial and temporal distribution relative to environmental abiotic factors (Hoffmann& Rieseberg 2008; Krimbas& Powell 1992). In An. gambiae, climatic variables (e.g., mean annual precipitation, evapotranspiration, temperature) are significantly correlated with the distribution of chromosomal inversions (Bayoh et al. 2001; Coluzzi et al. 1979; Costantini et al. 2009; Lee et al. 2009; Simard et al. 2009). In particular, inversion 2La on the left arm of chromosome 2 is strongly linked to degree of aridity. Multiple studies have shown that the frequency of 2La (i) increases with aridity along climatic clines replicated across Africa, (ii) increases with aridity at microspatial scales related to indoor/outdoor resting behavior, and (iii) cycles between dry and rainy seasons (Bryan et al. 1982; Coluzzi 1992; Coluzzi et al. 1979; Petrarca et al. 1990; Rishikesh et al. 1985; Wondji et al. 2005). These observations suggest that the 2La arrangement confers a selective advantage in xeric habitats, while the alternative 2L+a arrangement is more beneficial in mesic habitats, resulting in the maintenance of the 2La/+a inversion polymorphism in the species as a whole. In An. gambiae laboratory colonies, the 2La/+a inversion polymorphism appears to persist indefinitely, possibly owing to heterosis (della Torre et al. 1997).
A variety of traits have been associated with inversions in organisms such as Drosophila, midges, blackflies, and the apple maggot Rhagoletis pomonella (Hoffmann& Rieseberg 2008). These include body size, fecundity, diapause, and resistance to heat and cold. In An. gambiae, recent physiological studies have associated inversion 2La with two traits consistent with a role in aridity tolerance. Under controlled laboratory conditions, adult females carrying the inverted arrangement were more resistant to desiccation, due to lower rates of water loss (at emergence) and higher initial body water content (at four days post-emergence) (Gray et al. 2009). Prior acclimation increased desiccation resistance for both inverted and standard arrangements, but the energy storage strategy apparently differed according to inversion orientation (Gray et al. 2009). For 2L+a, acclimation was associated with increased lipid and decreased glycogen content; the opposite was observed for 2La, with possible implications for fecundity, immunity, longevity and other fitness traits in carriers of alternative arrangements of 2La in natural populations. In addition to adult desiccation resistance, 2La also has been associated with superior resistance of larvae to an acute thermal stress, if the larvae were previously heat hardened at a sub-lethal temperature (Rocca et al. 2009). Overall, these results may be reflective of the trade-off between high energetic costs versus survival benefits of mounting a stress response, in habitats where the intensity and frequency of climatic stress varies from very high (in arid savanna and sahel environments associated with 2La) to low (in humid rainforest environments associated with 2L+a).
The genetic basis of the desiccation and heat response differences between alternative arrangements of 2La remain unknown. Using genomic DNA hybridizations to gene-based microarrays, White et al. (2007) identified two ~1.5 Mb regions within the inversion of significantly elevated sequence divergence between 2La and 2L+a arrangements, near but not adjacent to the inversion breakpoints. Together, these regions encompass 210 genes including a large cluster of cuticle protein genes and three tandem hsp83 heat shock genes. Persistent genetic association between sequence variants in these diverged regions and the 2La arrangement suggested that they could contain at least some of the stress responsive genes contributing to An. gambiae ecological adaptation in challenging arid habitats. However, additional experimental approaches are required to uncover the specific genes and molecular mechanisms underlying inversion-associated traits.
As part of the larger goal of identifying genes that could be involved in differential response to various stresses by alternative arrangements of 2La, here we conducted microarray analyses of the larval thermal stress response. Applying the same one-hour heat hardening treatment that elicited differential survival to subsequent heat shock in previous physiological studies (Rocca et al. 2009), we compared global gene expression profiles in 2La and 2L+a larvae at three time points, for up to eight hours following application of heat stress. Treatment and control time series, replicated four times, revealed a core set of heat shock protein (HSP) genes involved in a common and immediate response to thermal stress in An. gambiae regardless of 2La orientation, but they also suggest that the presence of the 2La inverted arrangement preconditions a much more aggressive response to stress, geared toward sharply increased proteolytic degradation and energy metabolism.
Materials and Methods
Mosquito colonies and maintenance
Experiments were conducted using two homokaryotypic sub-colonies of Anopheles gambiae M form (SUCAM 2La and SUCAM 2L+a) that originated from a parental colony polymorphic for 2La but fixed and standard for all other An. gambiae inversions (Rocca et al. 2009). Colonies were maintained in an insectary under controlled conditions of 27°C, 85% RH, and a 12 h:12 h light-dark cycle with 1 h crepuscular transitions. Each generation, eggs were placed in plastic trays (27 cm x 16 cm x 6.5 cm) containing 1 L of water purified by reverse osmosis. Larvae were reared at low density (~100 per pan) and fed daily with a mixture of 2:1 finely ground tropical fish pellet:bakers yeast. Pupae were transferred to 0.2 m3 emergence cages. Upon emergence, adult mosquitoes were supplied absorbent cotton saturated with 20% sucrose solution.
Induction of thermal stress
Experimental design entailed a 1 h heat treatment at 38°C (or 1 h untreated control at 27°C) followed by transfer back to 27°C until sample collection at three time points: 0.25 h, 2 h, and 8 h post heat-stress. Each sample consisted of 24 4th instar larvae, and each time series (treated and control) was replicated four times using larvae from different cohorts (Table 1). At the onset of each time series, 144 seven-day-old 4th instar larvae from each karyotype (SUCAM 2La and SUCAM 2L+a) were randomly selected (from three pans, to minimize the contribution of any one pan to variation between samples) and placed individually into 13×100 mm glass culture tubes containing 2 ml of water. For heat treatment, tubes containing larvae were placed in a 38°C water bath for 1 h, followed by transfer to a 27°C water bath, where they were maintained until sample collection at 0.25 h, 2 h, and 8 h post heat treatment. Untreated controls were handled identically, except that they were maintained at 27°C throughout. At each time point, pools of 24 heat-treated or control larvae for each karyotype were collected into a 1.5 ml microcentrifuge tube, frozen in liquid nitrogen, and stored at −80°C until RNA isolation.
Table 1.
Experimental design and number of microarrays hybridized (and analyzed)1.
| Karyotype | Treatment | Sampling Times (h)
|
||
|---|---|---|---|---|
| 0.25 | 2 | 8 | ||
| 2L+a | Heat stress | 4 (3) | 4 | 4 (3) |
| Control | 4 | 4 | 4 | |
| 2La | Heat stress | 4 | 4 | 4 |
| Control | 4 | 4 (3) | 4 | |
In three instances, poor data quality necessitated the omission of one of the four replicate hybridizations.
RNA Isolation
Total RNA was extracted from pools of 24 individuals using the RNeasy Mini Kit (QIAGEN). RNA quality was examined using the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies) at wavelengths of 230 nm, 260 nm, and 280 nm. The integrity of RNA was further interrogated by electrophoresis of a 1μl sample on 1.5% agarose gels. Total RNA was treated with DNase I (Invitrogen) to remove any residual DNA. RNA was quantified using RiboGreen (Molecular Probes/Invitrogen) and the SpectraMAX M2 microplate reader (Molecular Devices).
Custom array design
Custom arrays were designed based on the Roche NimbleGen 12-plex format (12 arrays per slide with 135,000 60-mer probes per array). The design includes at least five probes per gene, synthesized in duplicate, providing a total of 131,212 probes that interrogate the 13,254 genes annotated in the AgamP3.5 genebuild (www.vectorbase.org) (Lawson et al. 2009). This array is available through NimbleGen (catalog number OID22384; design name 090706_A_gambiae_NotreDame_BH_expr_HX12).
Microarray processing and analysis
Total RNA was amplified and converted into double-stranded cDNA using the TransPlex Whole Transcriptome Amplification Kit (Sigma-Aldrich). Quality and quantity of cDNA was assessed using the 2100 Bioanalyzer (Agilent) and the ND-1000 Spectrophotometer at wavelengths of 230 nm, 260 nm, and 280 nm. Using 1μg of cDNA, labeling and amplification employed validated Cy3 dye randomers (TriLink) and followed the standard sample labeling protocol recommended by the array manufacturer (Roche NimbleGen). After labeling, 6μg of product was hybridized per each array, using the NimbleGen Hybridization Kit as recommended by Roche NimbleGen. Arrays were washed after hybridization using the NimbleGen Wash Buffer Kit and scanned with a NimbleGen MS 200 Microarray Scanner at 2μm resolution. Hybridization and scanning were performed in the University of Notre Dame Genomics Core Facility. Array image data quality was assessed and raw fluorescence intensity values for each probe were obtained using NimbleScan v2.5 software (Roche NimbleGen). A total of four 12-plex chips, totaling 48 arrays (2 chromosomal arrangements × 2 treatments × 3 time points × 4 replicates) were run.
XYS files containing the raw intensity values were imported into Bioconductor (www.bioconductor.org), an open-source software project based on the R programming language (www.r-project.org). Using custom R scripts (available on request from CC), a filter was applied to remove probes affected by physical blemishes on the slide (small scratches, dust, and/or wash artifacts), and to account for incomplete annotation of the An. gambiae genome, under the assumption that some probes in a probe set (i.e., the set of probes designed to target the same gene) may interrogate misannotated target genes or unannotated genes not intentionally targeted. To mitigate these potential problems, the highest and lowest intensity probes were omitted from each probe set. The resulting set of filtered probe sets was reduced to those that corresponded to expressed genes. These were defined by calculating an average intensity value for each filtered probe set, and identifying those probe sets whose average intensity value exceeded a threshold of 2000 in any two replicates of at least one condition (i.e., karyotype x treatment x time). Only filtered, expressed probe sets were used for subsequent analysis. The final set of filtered, expressed probes was subjected to background subtraction, normalization, and summarization using the RMA function. Fully MIAME-compliant microarray data were submitted to ArrayExpress (accession number E-MEXP-3078). Transcriptional profiles of subsets of genes (see below) were displayed as a heat map using heatmaps.2 in the gplots package in R.
We constructed a three-way analysis of variance (ANOVA) model using the nlme package in R, to assess the impact on gene expression due to three main fixed factors (and their interactions): treatment (heat stressed or control), karyotype (2L+a or 2La), and time (0.25 h, 2 h, or 8h). Replicate was modeled as a random factor. Significance of each factor was defined at a false discovery rate (FDR) of 0.10, for this and all other analyses unless otherwise specified. As time was determined to be a significant factor in the overall model, post hoc tests for individual time points were conducted on the set of genes whose expression was significantly affected by treatment, karyotype, and/or their interaction in the model. Post hoc tests included two-way ANOVA models with fixed factors karyotype and treatment (and their interaction); replicates were a random factor. At each time point, candidate genes that were differentially expressed between karyotypes in response to heat stress were those with significant karyotype x treatment interactions. Candidate genes were categorized as differentially induced or repressed by heat treatment if their heat-responsive expression in one karyotype exceeded a (log2)fold-change threshold of 1.3 (Huggins et al. 2008) in all three pairwise comparisons: with the other treated karyotype and both sets of controls. Other post hoc Bayes-moderated t-tests were conducted for each karyotype, to identify the set of significantly heat-responsive genes in paired treated versus control samples at each time point. These tests were implemented using the limma package in R.
Functional annotation of An. gambiae genes is largely incomplete. Where possible, functional categorization of candidate genes was achieved based on Gene Ontology (GO) terms mapped to An. gambiae genes (AgamP3.5) supplemented (if available) by functional information from orthologs in model organisms such as Drosophila. Functional enrichment of GO and other annotation terms in candidate gene lists was explored using the DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/) (Huang et al. 2009). The enrichment score assigned each gene group (annotation cluster) represents the geometric mean of the EASE Scores (modified Fisher Exact) associated with each enriched annotation term in the gene group (Hosack et al. 2003; Huang et al. 2007), and is intended to order the relative importance of the groups as part of an exploratory rather than strictly statistical analysis. For this reason, enrichment scores are presented in the form of minus log transformed geometric means instead of an absolute P-value (Huang et al. 2007). DAVID also was used to identify enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (http://www.kegg.com/kegg/kegg1a.html).
Results
Beginning with a laboratory colony of An. gambiae polymorphic for inversion 2La, sub-colonies were established that carry alternative homokaryotypic arrangements (2L+a or 2La) on an otherwise shared genetic background (Rocca et al. 2009). Using 4th instar larvae from these subcolonies, Rocca et al (2009) showed that survivorship following exposure to an acute heat shock (40°C for 120 min) did not differ, but that prior heat hardening at a sub-lethal temperature (38°C for 60 min) improved 24 h survival of the 2La subpopulation significantly more than the 2L+a subpopulation, following the heat shock. To uncover molecular mechanisms underlying the different responses to heat hardening by alternative karyotypes, we used microarrays (Roche NimbleGen) custom-designed from the AgamP3.5 genebuild to compare the genome-wide transcriptomes of these two subpopulations at three time points (0.25 h, 2 h, and 8 h) following heat hardening (or an untreated control incubation). In total, 48 arrays were hybridized [2 karyotypes × 2 treatments (heat hardened or control) × 3 time points × 4 replicates], although data from three arrays were omitted prior to analysis due to poor data quality (Table 1).
Of the 13,254 genes interrogated on the array, 888 were eliminated from subsequent analysis due to missing expression values (e.g., resulting from blemishes on the chip). An additional 1,980 genes were omitted because they were not detected as expressed in either karyotype at any of the three time points, in heat stressed or control samples. The remaining 10,386 expressed genes formed the basis of subsequent analyses.
To identify genes whose expression differs between samples as a function of karyotype, treatment, time, or the interactions of these factors, we applied a linear mixed model ANOVA, controlling for multiple testing by imposing an FDR of 0.10. Table 2 indicates that all three factors had a significant bearing on the pattern of gene expression; this was particularly true of the interaction of karyotype and either treatment or time, although the effect of a three-way interaction did not rise to the level of significance. In total, 8,931 genes (86% of all expressed genes) differed significantly among samples with respect to at least one of the three factors. The number of genes responsive to each factor or interaction is illustrated in Fig. 1, which emphasizes considerable overlap in treatment-, karyotype-, and time-dependent transcriptional response.
Table 2.
Analysis of variance (ANOVA) describing the effects of treatment, karyotype, time and their interaction on gene expression in An. gambiae fourth instar larvae.
| Factor(s) | d.f. | SS | MS | F | P |
|---|---|---|---|---|---|
| Treatment | 1 | 36 | 35.5 | 16.5 | 4.75E-05 |
| Karyotype | 1 | 19 | 18.5 | 8.6 | 0.003 |
| Time | 2 | 26 | 13.2 | 6.1 | 0.002 |
| Karyotype x Treatment | 1 | 100 | 100.4 | 46.8 | 7.98E-12 |
| Karyotype x Time | 2 | 134 | 66.9 | 31.2 | 2.94E-14 |
| Treatment x Time | 2 | 3 | 1.6 | 0.8 | 0.466 |
| Karyotype x Treatment x Time | 2 | 10 | 5.2 | 2.4 | 0.088 |
Figure 1.
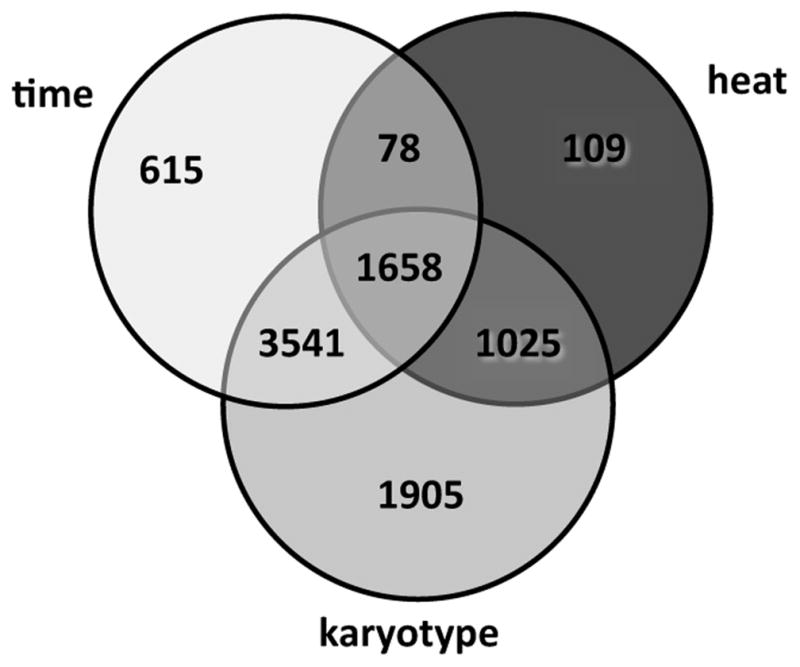
Venn diagram indicating the number of An. gambiae genes differentially expressed as a function of heat stress, karyotype, time, or their interactions, based on ANOVA (FDR < 0.1).
Heat shock protein genes most dramatically up-regulated by thermal stress are largely the same in both karyotypes
Thermal stress is well known to rapidly and massively increase the expression of many heat shock chaperone genes (hsps), within minutes of exposure (Feder& Hofmann 1999; Lindquist 1986). Ubiquitous and highly conserved, heat-inducible molecular chaperones prevent aggregation of non-native proteins and assist in their transport, refolding, or degradation, and as such, their involvement in the heat stress response is expected regardless of 2La karyotype. On the other hand, there is no a priori expectation that the same set of hsp genes will be induced, and to the same extent, in 2La versus 2L+a karyotype classes. To address this question, we focused on the initial time point (0.25 h post stress), which should capture the induced hsp genes. For each karyotype class, we ranked the set of heat stress responsive genes by the degree of (log2)fold-change increase between stressed and control groups at 0.25h. Of the 1105 total candidate genes that were significantly induced in a 2La background (FDR<0.1), 34 were hsps or encode products predicted to interact with HSPs, of which almost half (15) were among the 25 top-ranked candidates (i.e., those showing the greatest induction following heat stress) (Table 3). Although sharply fewer (only 49) candidate genes were significantly induced by heat in the alternative 2L+a karyotype, 18 of these were hsp or hsp-interacting and the vast majority (15) were ranked in the top 25. Notably, the top eight candidates in both karyotype classes are the same hsp genes and share nearly the same ranking, though the degree of up-regulation was generally lower for 2L+a. However, the 9th-ranked gene induced by heat stress in 2La, predicted to encode the HSP90 co-chaperone Aha1 (Activator of HSP90 ATPase), was not significantly induced by heat in 2L+a. Also missing among the heat responsive genes in 2L+a were half of those ranked from 12 to 25 in the alternative 2La karyotype, including two other co-chaperones. Transcript abundance of hsps and other top heat-responsive genes diminished rapidly with time—often back to levels indistinguishable from controls—by 8h post heat stress.
Table 3.
Top 25 genes induced by heat stress in 2La and 2L+a genetic backgrounds
| Rank in 2La | (Rank in 2L+a) | Fold change
|
Gene ID | Putative Function | Rank in 2L+a | (Rank in 2La) | Fold change
|
Gene ID | Putative Function | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.25h | 2h | 8h | 0.25h | 2h | 8h | ||||||||
| 1 | 2 | 181.74 | 76.16 | 3.74 | AGAP007159 | Hsp20 | 1 | 2 | 144.26 | 63.46 | 2.15 | AGAP007158 | Hsp20 |
| 2 | 1 | 159.86 | 66.56 | 3.37 | AGAP007158 | Hsp20 | 2 | 1 | 131.03 | 53.82 | 2.81 | AGAP007159 | Hsp20 |
| 3 | 4 | 147.41 | 96.73 | --- | AGAP005547 | Hsp20 | 3 | 4 | 119.03 | 19.87 | 2.96 | AGAP005548 | Hsp20 |
| 4 | 3 | 91.74 | 2.59 | --- | AGAP005548 | Hsp20 | 4 | 3 | 101.98 | 46.01 | 3.84 | AGAP005547 | Hsp20 |
| 5 | 5 | 52.51 | 5.26 | --- | AGAP004581 | Hsp70 | 5 | 5 | 30.63 | 2.13 | --- | AGAP004581 | Hsp70 |
| 6 | 6 | 33.96 | --- | --- | AGAP004583 | Hsp70 | 6 | 6 | 14.84 | --- | --- | AGAP004583 | Hsp70 |
| 7 | 8 | 16.41 | 3.53 | --- | AGAP004582 | Hsp70 | 7 | 8 | 14.21 | 2.13 | --- | AGAP012891 | Hsp70 |
| 8 | 7 | 15.74 | 3.06 | 3.07 | AGAP012891 | Hsp70 | 8 | 7 | 13.54 | 1.93 | --- | AGAP004582 | Hsp70 |
| 9 | --- | 13.48 | 2.26 | --- | AGAP010514 | Aha1 | 9 | 10 | 7.60 | 3.60 | --- | AGAP003727 | Tom34 (hsp70- interacting) |
| 10 | 9 | 13.39 | 1.49 | --- | AGAP003727 | Tom34 (hsp70- interacting) | 10 | 24 | 5.50 | --- | --- | AGAP007107 | Hsp40/DnaJ |
| 11 | 11 | 10.29 | --- | --- | AGAP002107 | Calcyclin- binding/Siah- interacting (ubiquitin- mediated proteolysis) |
11 | 11 | 5.08 | --- | --- | AGAP002107 | calcyclin- binding/Siah- interacting (ubiquitin- mediated proteolysis) |
| 12 | --- | 9.05 | 2.77 | 3.12 | AGAP006187 | Unknown | 12 | 13 | 4.90 | 2.10 | --- | AGAP011762 | starvin (BAG3) |
| 13 | 12 | 8.51 | 3.70 | --- | AGAP011762 | starving (BAG3) | 13 | 14 | 4.61 | 2.91 | --- | AGAP009883 | unknown |
| 14 | 13 | 8.05 | 2.30 | --- | AGAP009883 | Unknown | 14 | 21 | 4.61 | --- | --- | AGAP006959 | Hsp90 |
| 15 | --- | 7.12 | --- | --- | AGAP002339 | Arsenite inducible RNA associated protein (AIP1) |
15 | 38 | 4.58 | --- | --- | AGAP006961 | Hsp90 |
| 16 | --- | 6.98 | --- | --- | AGAP011614 | Chitin- binding | 16 | 40 | 3.81 | --- | --- | AGAP001896 | unknown |
| 17 | --- | 6.74 | 2.66 | --- | AGAP005981 | Hsp40/DnaJ | 17 | 3.42 | --- | --- | AGAP006117 | Hsp20-like | |
| 18 | 6.63 | --- | --- | AGAP004426 | EMI/FAS1- containing | 18 | 25 | 3.28 | --- | --- | AGAP010848 | unknown | |
| 19 | 23 | 6.53 | --- | --- | AGAP001324 | Unknown | 19 | 78 | 3.22 | --- | --- | AGAP004428 | EMI/FAS1- containing |
| 20 | --- | 6.36 | --- | --- | AGAP011278 | Galectin | 20 | 207 | 2.64 | --- | --- | AGAP000601 | unknown |
| 21 | 14 | 6.05 | 2.62 | --- | AGAP006959 | Hsp90 | 21 | 202 | 2.13 | --- | --- | AGAP009616 | neurotransmitter receptor |
| 22 | 5.98 | 2.31 | --- | AGAP008615 | endoplasmic reticulum protein with retention signal |
22 | 811 | 2.08 | --- | --- | AGAP001730 | cyclin-like | |
| 23 | --- | 5.88 | --- | --- | AGAP010188 | Hsp70/Hsp90 organizing protein (Hop) |
23 | 19 | 2.04 | 1.65 | --- | AGAP001324 | unknown |
| 24 | 10 | 5.87 | --- | --- | AGAP007107 | Hsp40/DnaJ | 24 | 122 | 2.04 | --- | --- | AGAP002752 | Hsp40/DnaJ |
| 25 | 18 | 5.66 | --- | --- | AGAP010848 | Unknown | 25 | 703 | 2.01 | --- | --- | AGAP009144 | unknown |
Substantial karyotype-dependent differences in gene expression profiles immediately following heat stress
Although the pattern of transcriptional induction of the core hsp chaperones (families HSP20, HSP70, HSP90) was similar, other genes are involved in the stress response (Feder& Hofmann 1999; Sorensen et al. 2005; Young et al. 2004) and could be differentially expressed between the alternative karyotypes. To examine differential expression between heat-stressed karyotypes at individual time points, post-hoc analyses were performed on the subset of genes detected as significantly responsive to treatment, karyotype, or their interaction in the initial ANOVA (7,232 genes). At 0.25 h, 2 h, and 8 h post-heat stress, we identified the set of genes with a significant karyotype x treatment interaction (by two-way ANOVA). The results overwhelmingly support an immediate differential response to heat stress by the alternative karyotypes: we identified 1,175 genes with a significant condition x karyotype interaction at 0.25 h, but no (zero) genes at 2 h, and only three genes at 8 h. Because of the trivial number of differentially expressed genes detected beyond 0.25 h following exposure to heat stress, all subsequent analysis was confined to the 1,175 candidate genes identified at this “immediate” (i.e., 0.25 h) time point.
Divergent transcriptional response to heat stress largely owing to induction in the 2La karyotype
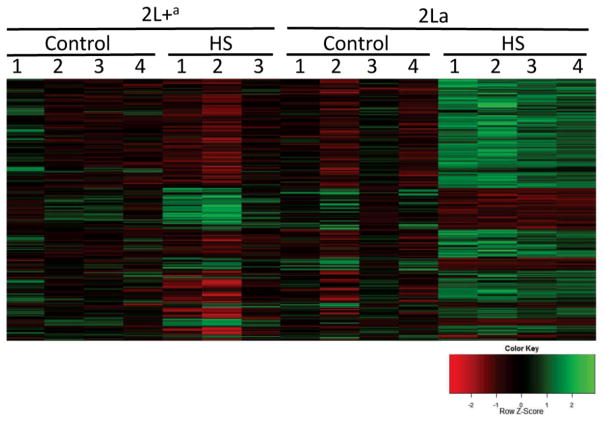
Transcriptional profiles of the 1,175 karyotype- and heat stress-responsive candidate genes are presented in Fig. 2 as a heat map. Three trends are apparent from informal appraisal of the heat map. First, the results are relatively consistent among replicates. Second, the main response to heat stress in the 2La background seems to be up-regulation of the majority of candidate genes. A much smaller fraction of genes are up-regulated in the 2L+a background in response to heat stress. Third, these two sets of up-regulated genes appear to be largely mutually exclusive; genes induced in one background in response to heat stress are unchanged or potentially repressed in the alternative karyotypic background.
Figure 2.
Transcriptional profile of the 1,175 genes that were differentially heat-responsive between alternative karyotypes at 0.25 h post heat stress (FDR < 0.1). Each row represents an individual gene; each column labeled 1–4 represents a replicate sample that was untreated (Control) or exposed to heat stress (HS) for each karyotype (2La or 2L+a). For each gene (row), the relative expression level in each sample (column) is represented by a color that reflects its row z-score (shown in the color key), calculated by subtracting the mean expression value for the row from individual sample values and dividing by the standard deviation of the row.
To more rigorously quantify these results, candidate genes were defined as differentially up- or down-regulated by heat stress in one karyotype if their expression exceeded a (log2)fold-change threshold of 1.3 (Huggins et al. 2008) relative to the other karyotype and both controls (assessed in separate pairwise comparisons). All except 207 of the 1175 candidate genes (i.e., 969 genes) could be classified in this fashion; the exceptions did not meet the minimum threshold of fold-change difference in one or more of the comparisons. The 969 candidate genes that are differentially expressed between karyotypes in response to heat are listed in Supplementary Table S1. Partitioning these genes by karyotype, most (833) were heat responsive (either up- or down-regulated) in 2La samples. Moreover, the 2La candidates are largely up-regulated (629 of 833 genes; ~76%). Not only are the heat responsive 2L+a candidates much fewer overall (452), but the majority are down-regulated (277; 61%).
Of note are the instances in which the same candidate gene responded to heat stress in an opposite fashion in alternative karyotypes. This was the case for one-third of the 969 differentially expressed heat-responsive genes, (indicated by ‘+’ and ‘#’ in Table S1). In particular, 30% of the genes up-regulated by thermal stress in 2La and 73% of those up-regulated in 2L+a also were repressed (relative to controls) in the alternative karyotype.
Thermal stress differentially induces proteasomal, chaperone, and metabolic activity in the 2La karyotype
To guide biological interpretation, we partitioned the 969 differentially heat responsive genes into four lists (induced/repressed by thermal stress in 2La/2L+a), which were explored using the DAVID data-mining tool (Huang et al. 2009). Using the DAVID functional annotation clustering module set at the default (medium) stringency level, genes were classified into functional groups. This clustering method condenses the input gene list into functionally related genes (annotation clusters), taking into account the similarity of their annotation profiles based on multiple annotation sources (e.g., GO terms and Interpro keywords). The annotation clusters with significant enrichment scores (i.e., greater than 1.3 in minus log scale) are given in Table 4. No clusters with scores meeting this threshold were identified in the two smallest gene lists, containing genes down-regulated by thermal stress in 2La, or up-regulated in 2L+a. By contrast, 11 annotation clusters were identified among the large set of genes up-regulated by thermal stress in 2La, that we have interpreted in the context of three broad functions, discussed below: proteolytic, chaperone, and metabolic activity.
Table 4.
Functional annotation clusters of genes differentially expressed in response to thermal stress by alternative (2La or 2L+a) karyotypes
| Stress responsive genes | Annotation Cluster (representative annotation terms) | Gene count | Enrichment score |
|---|---|---|---|
| 2La/Up | 1 Proteasome complex | 45 | 4.47 |
| 2 Translation/translation initiation factor | 33 | 3.79 | |
| 3 Tetratricopeptide (TPR) repeat | 14 | 3.56 | |
| 4 Protein catabolism | 53 | 3.31 | |
| 5 Tricarboxylic acid (TCA) cycle/cellular respiration | 25 | 3.11 | |
| 6 PCI (proteasome, CSN, eIF3) domain | 9 | 2.94 | |
| 7 Proteosome complex/proteolysis | 50 | 2.92 | |
| 8 Chaperonin (Cpn60/TCP-1) | 10 | 2.77 | |
| 9 ATP binding | 75 | 1.97 | |
| 10 U-box/ubiquitin ligase | 19 | 1.92 | |
| 11 Beta-ketoacyl synthase (fatty acid biosynthesis) | 4 | 1.68 | |
| 2La/Down | --- | --- | --- |
| 2L+a/Up | --- | --- | --- |
| 2L+a/Down | 12 TCP-1/cpn60 chaperonin | 6 | 2.32 |
| 13 Proteasome/proteolysis | 36 | 2.28 | |
| 14 Protein transport/localization | 9 | 1.31 |
Six of the annotation clusters (#1, 2, 4, 6, 7, and 10 in Table 4) relate to protein life span and proteolysis. Many genes in these clusters encode regulatory and core components of the proteosome and the enzymes involved in ubiquitin modification (activation, conjugation and ligation), thus pointing to the ubiquitin proteasome pathway—the main pathway for elimination of misfolded proteins. Reinforcing this result, KEGG-based pathway analysis in DAVID identified “proteasome” as over-represented among the up-regulated genes in 2La samples (n = 20, P = 5.2E-10, Benjamini-Hochberg adjusted P = 3.5E-8); this pathway and affected genes are illustrated in Fig. S1. Other components of ubiquitin-mediated proteolysis are also present in this gene list, including the An. gambiae ortholog of a gene whose product has been proposed to mark ubiquitinated protein aggregates for autophagic degradation (the mammalian polyubiquitin binding protein p62, called Ref(2)P in Drosophila; Nezis et al. 2008). In addition, several genes potentially encoding ubiquitin-like proteins are represented (e.g., ubiquitin-fold modifier 1 [Ufm1], ubiquitin-fold conjugating enzyme [Ufc1], and NEDD8), as are genes whose products are involved in the COP9 signalosome and other protein degradation pathways: five putative autophagy-related (ATG) genes, and a gene encoding the highly conserved stress protein, Lon protease.
Although the core chaperone machinery is up-regulated by heat stress in both karyotypes, other molecular chaperones and co-chaperones contribute to differential enrichment of chaperone-related annotation terms in heat stressed 2La samples. The T-complex protein (TCP-1) family of chaperonins, highlighted in annotation cluster 8, is also known as ‘TCP1 ring complex’ (TRiC) or ‘chaperonin containing TCP1’ (CCT). These cytosolic chaperonins are responsible for folding nascent proteins including tubulin and actin, as well as WD40-repeat proteins and protein complexes (reviewed in Young et al. 2004), in cooperation with HSP70 and prefoldin (also up-regulated transcriptionally in 2La). Consistent with ongoing translation as hinted by annotation cluster 2, another chaperone pathway represented in the gene list is the nascent-polypeptide-associated complex (NAC), which protects polypeptides emerging from ribosomes (Wiedmann et al. 1994). The enrichment of the tetratricopeptide repeat (TPR) domain among annotation terms associated with heat stressed 2La samples (cluster 3) appears to be largely a consequence of the transcriptional up-regulation of HSP70/HSP90 co-chaperone genes containing this motif (including putative orthologs of mammalian CHIP, HOP, HIP, TPR2, and the FK506-binding proteins (FKBPs); Taipale et al. 2010; Young et al. 2004). These co-chaperones interact directly with HSP70 and/or HSP90 through the TPR domain (D’Andrea& Regan 2003) and modulate their activity, by influencing substrate binding and transfer of cargo between chaperones, and by recruiting chaperones to cellular processes distinct from protein folding, such as protein sorting to organelles or to the proteosome for degradation (Young et al. 2004). Indeed, the sorting of HSP70-HSP90 clients to the mitochondria is suggested by the presence of up-regulated TPR-containing genes that are orthologs of the mitochondrial import receptor (translocase of the outer membrane [TOM] complex). In mammals, the sorting of HSP90 clients to the proteasome for degradation is accomplished through the co-chaperones CHIP (carboxyl terminus of HSP70-interacting protein) and a BCL2-associated athanogene (BAG) (McClellan et al. 2005), both of which are induced transcriptionally in 2La samples.
Increased energy metabolism in 2La samples is suggested by enrichment for key enzymes in fatty acid biosynthesis and the tricarboxylic acid (TCA) cycle (annotation clusters 5, 11), including isocitrate dehydrogenase, citrate synthase, and phosphoenolpyruvate carboxykinase (PEPCK). The TCA cycle also was identified as an over-represented KEGG pathway (n = 11, P = 3.0E-4, B–H adjusted P = 1.0E-2), whose perturbed components are shown in Fig. S2. It has been suggested that induction of isocitrate dehydrogenase and citrate synthase, which contribute strongly to control of the TCA cycle, may be necessary for generating reducing equivalents (NADH, NADPH) needed for protection against oxidative damage during the stress response (Kultz 2005). The role of these metabolic pathways in energy generation is also likely to be important to compensate for the ATP-dependent requirements of protein chaperoning and degradation (Kultz 2005). In this regard, the up-regulation of PEPCK in 2La samples may also be important. This enzyme catalyzes the first committed and rate-limiting step of gluconeogenesis, another over-represented KEGG pathway (n = 7, P = 2.7E-2, B–H adjusted P = NS; Fig. S3). However, PEPCK also plays a key role in glyceroneogenesis, serine synthesis, and recycling of the carbon skeletons of amino acids back into the TCA cycle for subsequent oxidation or conversion to fatty acids (Yang et al. 2009). Notably, both PEPCK and isocitrate dehydrogenase are down-regulated in the alternative 2L+a samples.
Although not detected as significantly enriched in our analysis based on a threshold enrichment score ≥ 1.3, programmed cell death was recognized as an annotation cluster with a score of 0.8, and may still be potentially interesting. This cluster of six genes included the aforementioned BAG gene (a BAG3 homolog known as starvin in D. melanogaster), which is an inhibitor of apoptosis expressed during recovery from cold stress in D. melanogaster (Colinet& Hoffmann 2010). This gene, like other inhibitors of apoptosis in the annotation cluster, is up-regulated in 2La. Consistent with the hypothesis that negative regulation of apoptosis may be part of the heat stress response program in 2La, a gene with proapoptotic ability (Michelob_x; Zhou et al. 2005) is down-regulated in 2La (and up-regulated in 2L+a) samples.
There were only three annotation clusters significantly enriched based on the set of genes differentially down-regulated in 2L+a samples. Of these, two coincide with annotation clusters identified among genes up-regulated in 2La and they evoke the functional themes of chaperonin and proteolytic activity. The third, and one other below the cut-off (with enrichment score 1.18; not shown), relate to protein translation and protein localization or transport. Further reinforcing the pattern of opposed responses to heat stress in the two karyotype classes, over-represented KEGG pathways included “proteasome” (n = 11, P = 2.0E-6, BH P = 5.1E-5; Fig. S1) and “oxidative phosphorylation” (n = 10, P = 9.6E-3, BH P = NS; Fig S4).
Heat-responsive genes are not overrepresented within the 2La inversion
We searched for nonrandom patterns in the genomic distribution of the 2,870 genes identified from the initial linear mixed model ANOVA (P < 0.01) as heat-responsive. As a first step, we examined whether heat-responsive genes were disproportionately represented on any chromosome arm relative to the number expected given the total number of expressed genes per arm. By χ2 test, no significant deviation from a uniform distribution was detected. In particular, there was no apparent excess of heat-responsive genes on 2L.
The 2La inversion spans ~21.6 Mb (Sharakhov et al. 2006) and contains 937 genes represented and detected on the microarray. Of the 1,175 candidate genes that were significantly heat-responsive at 0.25h, 105 candidates lie within the inversion breakpoints. Overall, there was no significant excess of candidate genes within the rearranged region relative to those in the rest of the genome, given the respective numbers of genes interrogated and detected by microarray. In fact, there were significantly fewer candidate genes than expected inside the inversion that were either up-regulated in the 2La samples at 0.25h (37 versus 59 expected; P = 0.002) or down-regulated in the corresponding 2L+a samples (13 versus 26 expected; P = 0.007).
Although there was no departure from expectation in terms of the number of candidate genes inside versus outside the 2La rearrangement, there may exist non-random clusters of genes. We tested for this by sliding window analysis, using 100 gene windows and 10 gene steps, arm-by-arm and within the rearranged region. Comparing observed versus expected numbers in each window by χ2 test with Bonferroni correction for multiple tests, no significant clusters were identified.
Discussion
The strong correlation between 2La inversion frequency and degree of aridity across environmental gradients presumably reflects spatially and seasonally varying selection that is responsible for the maintenance of the 2La/+a inversion polymorphism in An. gambiae. However, the genetic basis of environmental adaptations conferred by these (or any) chromosomal rearrangements is poorly understood (Hoffmann& Rieseberg 2008). Under the assumption that changes in the transcriptome are a major component of phenotypic evolution (Laayouni et al. 2007; Rifkin et al. 2003; Wray et al. 2003), we compared global gene expression profiles between alternative 2La arrangements in response to an abiotic stress already known to differentially affect 2La and 2L+a physiology (Rocca et al. 2009). Although the two karyotypes do not differ in survivorship following acute heat shock, 24 h survival of 2La is significantly better than 2L+a given the same heat hardening prior to heat shock. The present study provides the first account of the transcriptional response to heat hardening in An. gambiae, and the results uncover clear differences between the alternative 2La/+a karyotypes that may provide some mechanistic insight.
The two karyotypes shared a massive induction of the same core hsp machinery (hsp20, hsp70, and hsp90 families) following heat stress, although the induction was more extreme and generally lasted longer in the 2La background. However, nearly 1,000 genes were differentially responsive to heat between the two karyotypes at the earliest time point (0.25 h). The transcriptional response by 2La involved many more genes, and was largely characterized by induction of additional chaperone machinery, proteolysis functions, and energy metabolism (under the important assumption that increased transcript abundance presages protein abundance). This pattern deviates from gene expression studies of the stress response in Drosophila melanogaster, which typically report a higher proportion of down- versus up-regulated genes (e.g., Landis et al. 2004; Sorensen et al. 2005), as observed for the alternative 2L+a karyotype. The latter karyotype did not appear to respond as robustly to an identical heat stress, and most of the response comprised down-regulation of many of the same functions up-regulated in the 2La samples, including proteasomal degradation and energy metabolism. Many of the hsp70/hsp90 co-chaperone genes strongly induced in 2La samples were unchanged in heat stressed 2L+a samples relative to the corresponding untreated controls. One of the most dramatic differences between the karyotypes concerned a gene with apparent homology to the HSP90 co-chaperone Aha1, exclusively up-regulated in 2La samples with a (log2)fold change exceeding 13 (Table 2, AGAP010514). In yeast and humans, the protein product of this stress-regulated gene binds directly to HSP90 and accelerates its inherently low ATPase activity, suggesting that Aha1 contributes to HSP90 efficiency under stressful conditions (Panaretou et al. 2002). Although speculative, a possible connection between up-regulation of Aha1 and enrichment of the ubiquitin-proteasome degradation pathway in 2La samples may exist. Molecular chaperones like HSP90 not only participate in protein folding and re-folding, but also in the degradation of mis-folded proteins when folding attempts are aborted (McClellan& Frydman 2001; McClellan et al. 2005). The molecular switch that directs the balance between chaperone-mediated folding versus degradation remains unclear, but one model suggests that Aha1 may play a role by regulating the “dwell time” of HSP90 with client proteins, thereby impacting the stability of folding intermediates (Koulov et al. 2010).
Whatever the specific mechanism, our results lead to a working model in which the 2La background has much higher thermal sensitivity than 2L+a, in that it responds much more drastically to the same heat stress, as if preparing “for the worst”. This strategy may be essential to survival under the more hostile climatic conditions in which 2La prevails, characterized by greater extremes and longer bouts of heat and aridity. Although adult mosquitoes may be able to limit their exposure to some extent behaviorally, this is not the case for An. gambiae larvae confined to the typical aquatic habitat: shallow, stagnant pools fully exposed to sunlight and devoid of emergent vegetation, whose temperature can reach or exceed 40°C (Paaijmans et al. 2008). In dry-season Sudan, An. gambiae s.l. larvae and pupae have been found living in pools whose recorded temperature varied between 40.5°C and 41.8°C, close to the thermal death point of 42°C (Holstein 1954). Under these extreme conditions, the accumulation of misfolded and aggregated proteins could overwhelm the chaperone system and lead to death of cells or the whole organism unless they were eliminated. However, under more benign conditions, any benefit of this aggressive degradation strategy is probably outweighed by the heavy metabolic cost, depleting energy reserves that could leave 2La carriers at a competitive disadvantage in terms of development, fertility, fecundity and/or immunity (Feder& Hofmann 1999; Sorensen et al. 2003). Reciprocal transplantation experiments in the field represent one approach to test the predictions of this model.
The experimental design was founded on two subcolonies derived from the same parental colony. The subcolonies differ only in the arrangement of 2La—homozygous inverted in one and uninverted (standard) in the other—and are assumed to share an otherwise common genetic background. As such, the differential responses to an acute heat stress should stem from genetic differences inside the rearranged region. Interestingly, there was no excess of differentially expressed heat-responsive genes mapping within this ~22 Mb region. Several non-exclusive explanations for this outcome can be proposed. First, the differential transcriptional response could be a consequence of only one or very few “master regulator” transcription factors situated inside the inversion but acting on any number of genes elsewhere in the genome. Based on the genes annotated inside 2La in the current AgamP3.6 gene set, 27 may function in the regulation of transcription according to their associated GO terms and/or the KEGG transcription factor database (Table S2). Of these, none was detected as significantly differentially expressed between alternative arrangements. However, two (AGAP006923 and AGAP013107) map within the distal ~1.5 Mb region of significantly elevated sequence divergence implicated in the maintenance of 2La in An. gambiae populations (White et al. 2007). It may be the case that differences in the coding regions of one or both of these putative transcription factors are responsible for differential expression of their target genes. More generally, any functional differences between arrangements owing to coding sequence rather than transcript abundance would be missed by focusing on the transcriptome, as would differences at the translational or post-translational level. In this regard, it is worth noting that two of three tandem HSP90-family genes (hsp83) located within the same distal region of elevated sequence divergence were both induced by heat stress in the two karyotype classes, raising the possibility that nonsynonymous mutational differences in the coding sequence could alter HSP90 function, thereby contributing to the observed phenotypic variation (Jarosz& Lindquist 2010).
Several other considerations also argue that the list of candidate genes inside of 2La is almost certainly incomplete, even if attention is limited to transcriptional differences. Biologically relevant but very small changes in transcript abundance may be overlooked or go undetected for technical or statistical reasons. Important differences may occur over time periods or developmental stages not sampled, or at the level of tissue and organ, which whole body measurements may not detect. Moreover, for logistical reasons we have focused on only one isolated stress—an acute sub-lethal thermal stress—rather than the more realistic pileup of repeated or chronic environmental stresses encountered in the field. In nature, heat and desiccation stress are rarely uncoupled, and it has already been demonstrated in the laboratory that 2La confers greater resistance to desiccation in adult An. gambiae (Gray et al. 2009). While some genes may protect against multiple stresses (e.g., core hsps; Benoit et al. 2010), others are likely to be stress-specific and will not have been revealed by the experimental design. Finally, our experimental design did not include other inversions segregating within natural populations of An. gambiae. In particular, significant genetic association between the 2La and 2Rb arrangements are often observed despite their locations on opposite arms of chromosome 2 (Costantini et al. 2009; Simard et al. 2009), suggesting that additive and/or epistatic interactions between genes located within these inversions may be at work. Ongoing transcriptional profiling experiments that account for multiple chromosomal rearrangements and additional stresses (e.g., adult desiccation) in will begin to shed light on these questions.
Aside from its evolutionary importance, inversion 2La polymorphism also has significance for malaria transmission and control in Africa. At broad geographic and temporal scales, 2La has facilitated the successful exploitation of xeric habitats and seasons, but even at the very fine scale of a village, the 2La inversion system has been linked to differences in adult biting and resting behavior (Coluzzi et al. 1979), Plasmodium infection rate (Petrarca& Beier 1992), and non-uniform exposure of An. gambiae to indoor residual insecticides (Molineaux& Grammicia 1980). Detailed understanding of the genetic and molecular basis of these inversion-associated differences and their relationship to behavioral traits will allow more accurate predictions of how An. gambiae populations are likely to respond to anti-vector malaria interventions, and will uncover novel and specific molecular targets for such interventions. Toward that end, future studies will need to extend transcriptional profiling to proteomic and phospho-proteomic approaches, and equally important, will need to evaluate these patterns and their impact on fitness, in field populations.
Supplementary Material
Figure S1. Proteosome components from the KEGG/DAVID database. Boxes with green/white fill indicate genes with/without identified homologs in An. gambiae. Genes differentially responsive to heat stress are outlined by green circles (up-regulated in 2La samples) or blue boxes (up-regulated in 2La and down-regulated in 2L+a). Corresponding An. gambiae gene identifiers are shown adjacent to the candidate genes; only the numerical part of the gene ID is indicated, after omitting the leading letters (AGAP) and zeros.
Figure S2. TCA cycle pathway from the KEGG/DAVID database. Boxes with green/white fill indicate genes with/without identified homologs in An. gambiae. Genes differentially responsive to heat stress are outlined by green circles (up-regulated in 2La samples) or blue boxes (up-regulated in 2La and down-regulated in 2L+a). Enzyme names and corresponding An. gambiae gene identifiers are shown adjacent to the candidate genes; only the numerical part of the gene ID is indicated, after omitting the leading letters (AGAP) and zeros.
Figure S3. Glycolysis/gluconeogenesis pathway from the KEGG/DAVID database. Boxes with green/white fill indicate genes with/without identified homologs in An. gambiae. Genes differentially responsive to heat stress are outlined by green circles (up-regulated in 2La samples) or blue boxes (up-regulated in 2La and down-regulated in 2L+a). Enzyme names and corresponding An. gambiae gene identifiers are shown adjacent to the candidate genes; only the numerical part of the gene ID is indicated, after omitting the leading letters (AGAP) and zeros.
Figure S4. Oxidative phosphorylation pathway from the KEGG/DAVID database. Boxes with green/white fill indicate genes with/without identified homologs in An. gambiae. Genes differentially responsive to heat stress are outlined by green circles (up-regulated in 2La samples), red circles (down-regulated in 2L+a) or blue boxes (up-regulated in 2La and down-regulated in 2L+a). Enzyme names and corresponding An. gambiae gene identifiers are shown adjacent to the candidate genes; only the numerical part of the gene ID is indicated, after omitting the leading letters (AGAP) and zeros.
Acknowledgments
We thank Marcia Kern for insectary assistance, Erliang Zeng for discussion, and John Duman for critical review. Microarray hybridizations were performed in the Notre Dame Genomics Core Facility. Funding for this work was provided by National Institutes of Health grant R01 AI076584 to NJB. MJM received support from the Glynn Family Honors Program and the College of Science, University of Notre Dame.
Footnotes
Data Accessibility
Microarray data: ArrayExpress accession E-MEXP-3078
References
- Ayala D, Fontaine MC, Cohuet A, et al. Chromosomal inversions, natural selection and adaptation in the malaria vector Anopheles funestus. Mol Biol Evol. 2011;28:745–758. doi: 10.1093/molbev/msq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayoh MN, Thomas CJ, Lindsay SW. Mapping distributions of chromosomal forms of Anopheles gambiae in West Africa using climate data. Med Vet Entomol. 2001;15:267–274. doi: 10.1046/j.0269-283x.2001.00298.x. [DOI] [PubMed] [Google Scholar]
- Benoit JB, Lopez-Martinez G, Phillips ZP, Patrick KR, Denlinger DL. Heat shock proteins contribute to mosquito dehydration tolerance. J Insect Physiol. 2010;56:151–156. doi: 10.1016/j.jinsphys.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan JH, Di Deco MA, Petrarca V, Coluzzi M. Inversion polymorphism and incipient speciation in Anopheles gambiae s. str. in The Gambia, West Africa. Genetica. 1982;59:167–176. [Google Scholar]
- Colinet H, Hoffmann A. Gene and protein expression of Drosophila Starvin during cold stress and recovery from chill coma. Insect Biochem Mol Biol. 2010;40:425–428. doi: 10.1016/j.ibmb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Coluzzi M. Malaria vector analysis and control. Parasitology Today. 1992;8:113–118. doi: 10.1016/0169-4758(92)90277-9. [DOI] [PubMed] [Google Scholar]
- Coluzzi M. Malaria and the Afrotropical ecosystems: impact of man’made environmental changes. Parassitologia. 1994;36:223–227. [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73:483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- Costantini C, Ayala D, Guelbeogo WM, et al. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 2009;9:16. doi: 10.1186/1472-6785-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- della Torre A, Costantini C, Besansky NJ, et al. Speciation within Anopheles gambiae--the glass is half full. Science. 2002;298:115–117. doi: 10.1126/science.1078170. [DOI] [PubMed] [Google Scholar]
- della Torre A, Merzagora L, Powell JR, Coluzzi M. Selective introgression of paracentric inversions between two sibling species of the Anopheles gambiae complex. Genetics. 1997;146:239–244. doi: 10.1093/genetics/146.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Gray EM, Rocca KA, Costantini C, Besansky NJ. Inversion 2La is associated with enhanced desiccation resistance in Anopheles gambiae. Malar J. 2009;8:215. doi: 10.1186/1475-2875-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Ann Rev Ecol Evol Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Sgro CM, Weeks AR. Chromosomal inversion polymorphisms and adaptation. Trends Ecol Evol. 2004;19:482–488. doi: 10.1016/j.tree.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Holstein MH. Biology of Anopheles gambiae. World Health Organization; Geneva: 1954. [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins CE, Domenighetti AA, Ritchie ME, et al. Functional and metabolic remodelling in GLUT4-deficient hearts confers hyper-responsiveness to substrate intervention. J Mol Cell Cardiol. 2008;44:270–280. doi: 10.1016/j.yjmcc.2007.11.020. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulov AV, Lapointe P, Lu B, et al. Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol Biol Cell. 2010;21:871–884. doi: 10.1091/mbc.E09-12-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas CB, Powell JR. Introduction. In: Krimbas CB, Powell JR, editors. Drosophila inversion polymorphism. CRC Press; Boca Raton: 1992. pp. 1–52. [Google Scholar]
- Kultz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- Laayouni H, Garcia-Franco F, Chavez-Sandoval BE, et al. Thermal evolution of gene expression profiles in Drosophila subobscura. BMC Evol Biol. 2007;7:42. doi: 10.1186/1471-2148-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis GN, Abdueva D, Skvortsov D, et al. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Cornel AJ, Meneses CR, et al. Ecological and genetic relationships of the Forest-M form among chromosomal and molecular forms of the malaria vector Anopheles gambiae sensu stricto. Malar J. 2009;8:75. doi: 10.1186/1475-2875-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Frydman J. Molecular chaperones and the art of recognizing a lost cause. Nat Cell Biol. 2001;3:E51–53. doi: 10.1038/35055162. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Tam S, Kaganovich D, Frydman J. Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol. 2005;7:736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- Molineaux L, Grammicia G. The Garki Project. Research on the Epidemiology and Control of Malaria in the Sudan Savanna of West Africa. World Health Organization; Geneva, Switzerland: 1980. [Google Scholar]
- Nezis IP, Simonsen A, Sagona AP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera O, Powell JR, De La Rosa ME, et al. Population genetics of Mexican Drosophila VI. cytogenetic aspects of the inversion polymorphism in Drosophila pseudoobscura. Evolution. 1979;33:381–395. doi: 10.1111/j.1558-5646.1979.tb04691.x. [DOI] [PubMed] [Google Scholar]
- Paaijmans KP, Takken W, Githeko AK, Jacobs AF. The effect of water turbidity on the near-surface water temperature of larval habitats of the malaria mosquito Anopheles gambiae. Int J Biometeorol. 2008;52:747–753. doi: 10.1007/s00484-008-0167-2. [DOI] [PubMed] [Google Scholar]
- Panaretou B, Siligardi G, Meyer P, et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- Petrarca V, Beier JC. Intraspecific chromosomal polymorphism in the Anopheles gambiae complex as a factor affecting malaria transmission in the Kisumu area of Kenya. Am J Trop Med Hyg. 1992;46:229–237. doi: 10.4269/ajtmh.1992.46.229. [DOI] [PubMed] [Google Scholar]
- Petrarca V, Sabatinelli G, Di Deco MA, Papakay M. The Anopheles gambiae complex in the Federal Islamic Republic of Comoros (Indian Ocean): some cytogenetic and biometric data. Parassitologia. 1990;32:371–380. [PubMed] [Google Scholar]
- Pombi M, Caputo B, Simard F, et al. Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions. BMC Evol Biol. 2008;8:309. doi: 10.1186/1471-2148-8-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Petrarca V, della Torre A, Caccone A, Coluzzi M. Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia. 1999;41:101–113. [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Rishikesh N, Di Deco MA, Petrarca V, Coluzzi M. Seasonal variations in indoor resting Anopheles gambiae and Anopheles arabiensis in Kaduna, Nigeria. Acta Trop. 1985;42:165–170. [PubMed] [Google Scholar]
- Rocca KA, Gray EM, Costantini C, Besansky NJ. 2La chromosomal inversion enhances thermal tolerance of Anopheles gambiae larvae. Malar J. 2009;8:147. doi: 10.1186/1475-2875-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe AK, Rowe SY, Snow RW, et al. The burden of malaria mortality among African children in the year 2000. Int J Epidemiol. 2006;35:691–704. doi: 10.1093/ije/dyl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW. Selection in heterogeneous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura. Evolution. 2008;62:3082–3099. doi: 10.1111/j.1558-5646.2008.00504.x. [DOI] [PubMed] [Google Scholar]
- Sharakhov IV, White BJ, Sharakhova MV, et al. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc Natl Acad Sci U S A. 2006;103:6258–6262. doi: 10.1073/pnas.0509683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard F, Ayala D, Kamdem GC, et al. Ecological niche partitioning between the M and S molecular forms of Anopheles gambiae in Cameroon: the ecological side of speciation. BMC Ecol. 2009;9:17. doi: 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen JG, Kristensen TN, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecology Letters. 2003;6:1025–1037. [Google Scholar]
- Sorensen JG, Nielsen MM, Kruhoffer M, Justesen J, Loeschcke V. Full genome gene expression analysis of the heat stress response in Drosophila melanogaster. Cell Stress Chaperones. 2005;10:312–328. doi: 10.1379/CSC-128R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- White BJ, Hahn MW, Pombi M, et al. Localization of candidate regions maintaining a common polymorphic inversion (2La) in Anopheles gambiae. PLoS Genet. 2007;3:e217. doi: 10.1371/journal.pgen.0030217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- Wondji C, Frederic S, Petrarca V, et al. Species and populations of the Anopheles gambiae complex in Cameroon with special emphasis on chromosomal and molecular forms of Anopheles gambiae s.s. J Med Entomol. 2005;42:998–1005. doi: 10.1093/jmedent/42.6.998. [DOI] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Yang J, Kalhan SC, Hanson RW. What is the metabolic role of phosphoenolpyruvate carboxykinase. J Biol Chem. 2009;284:27025–27029. doi: 10.1074/jbc.R109.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Zhou L, Jiang G, Chan G, et al. Michelob_x is the missing inhibitor of apoptosis protein antagonist in mosquito genomes. EMBO Rep. 2005;6:769–774. doi: 10.1038/sj.embor.7400473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Proteosome components from the KEGG/DAVID database. Boxes with green/white fill indicate genes with/without identified homologs in An. gambiae. Genes differentially responsive to heat stress are outlined by green circles (up-regulated in 2La samples) or blue boxes (up-regulated in 2La and down-regulated in 2L+a). Corresponding An. gambiae gene identifiers are shown adjacent to the candidate genes; only the numerical part of the gene ID is indicated, after omitting the leading letters (AGAP) and zeros.
Figure S2. TCA cycle pathway from the KEGG/DAVID database. Boxes with green/white fill indicate genes with/without identified homologs in An. gambiae. Genes differentially responsive to heat stress are outlined by green circles (up-regulated in 2La samples) or blue boxes (up-regulated in 2La and down-regulated in 2L+a). Enzyme names and corresponding An. gambiae gene identifiers are shown adjacent to the candidate genes; only the numerical part of the gene ID is indicated, after omitting the leading letters (AGAP) and zeros.
Figure S3. Glycolysis/gluconeogenesis pathway from the KEGG/DAVID database. Boxes with green/white fill indicate genes with/without identified homologs in An. gambiae. Genes differentially responsive to heat stress are outlined by green circles (up-regulated in 2La samples) or blue boxes (up-regulated in 2La and down-regulated in 2L+a). Enzyme names and corresponding An. gambiae gene identifiers are shown adjacent to the candidate genes; only the numerical part of the gene ID is indicated, after omitting the leading letters (AGAP) and zeros.
Figure S4. Oxidative phosphorylation pathway from the KEGG/DAVID database. Boxes with green/white fill indicate genes with/without identified homologs in An. gambiae. Genes differentially responsive to heat stress are outlined by green circles (up-regulated in 2La samples), red circles (down-regulated in 2L+a) or blue boxes (up-regulated in 2La and down-regulated in 2L+a). Enzyme names and corresponding An. gambiae gene identifiers are shown adjacent to the candidate genes; only the numerical part of the gene ID is indicated, after omitting the leading letters (AGAP) and zeros.