Abstract
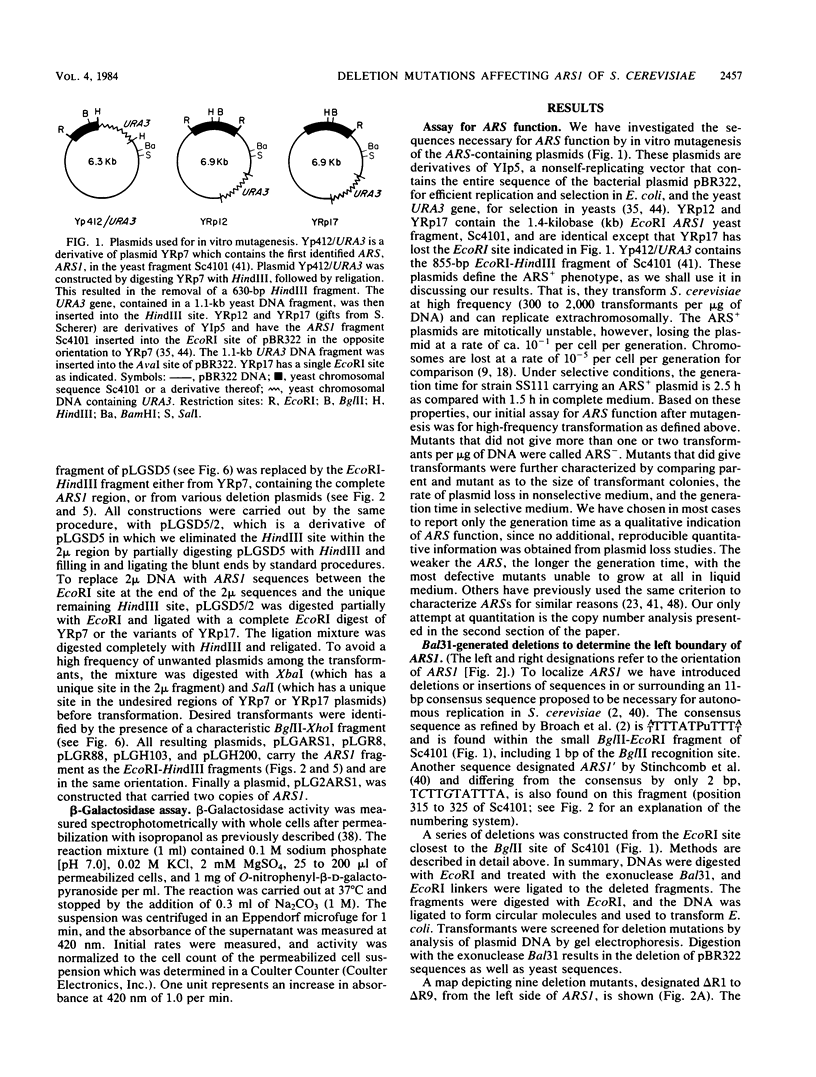
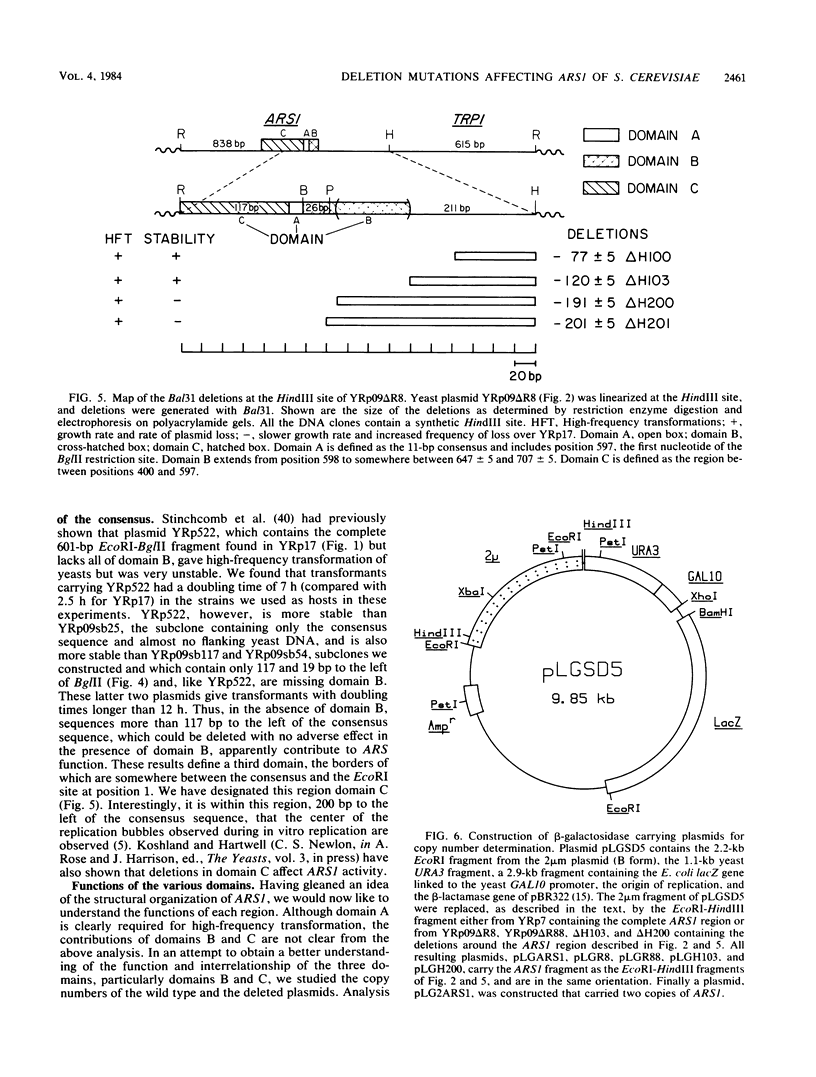
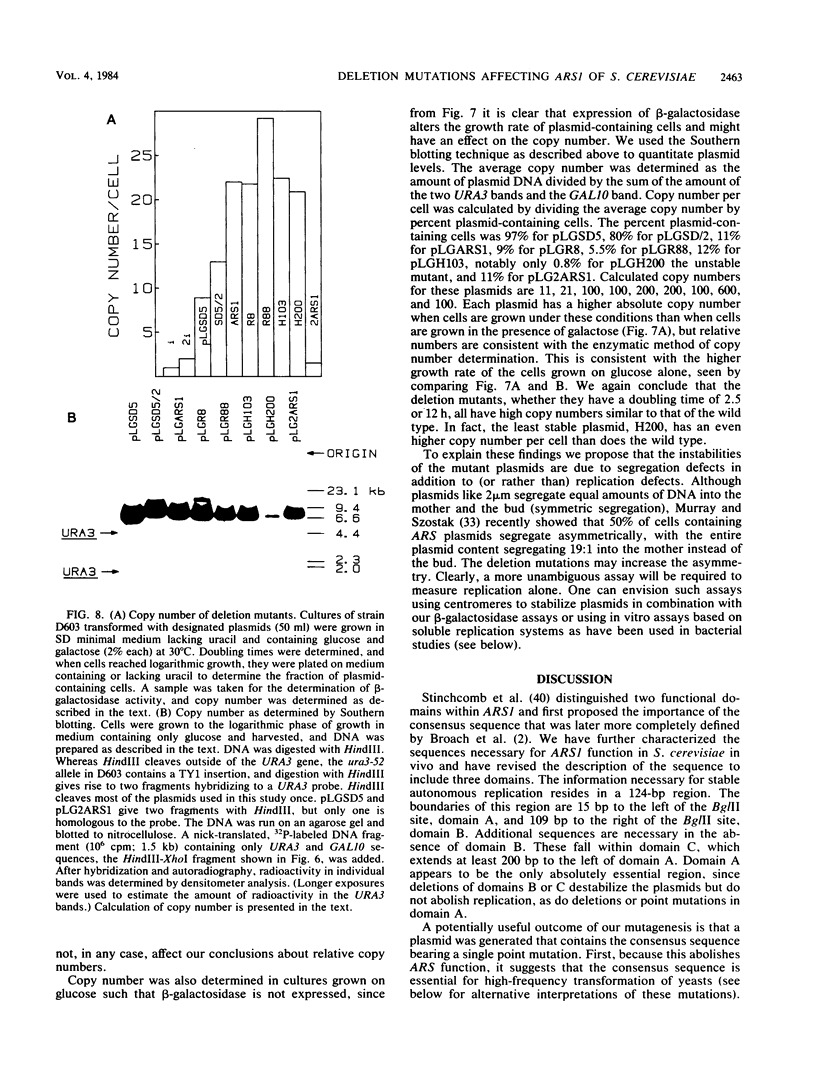
DNAs that contain specific yeast chromosomal sequences called ARSs transform Saccharomyces cerevisiae at high frequency and can replicate extrachromosomally as plasmids when introduced into S. cerevisiae by transformation. To determine the boundaries of the minimal sequences required for autonomous replication in S. cerevisiae, we have carried out in vitro mutagenesis of the first chromosomal ARS described, ARS1. Rather than identifying a distinct and continuous segment that mediates the ARS+ phenotype, we find three different functional domains within ARS1. We define domain A as the 11-base-pair (bp) sequence that is also found at most other ARS regions. It is necessary but not sufficient for high-frequency transformation. Domain B, which cannot mediate high-frequency transformation, or replicate by itself, is required for efficient, stable replication of plasmids containing domain A. Domain B, as we define it, is continuous with domain A in ARS1, but insertions of 4 bp between the two do not affect replication. The extent of domain B has an upper limit of 109 bp and a lower limit of 46 bp in size. There is no obvious sequence homology between domain B of ARS1 and any other ARS sequence. Finally, domain C is defined on the basis of our deletions as at least 200 bp flanking domain A on the opposite side from domain B and is also required for the stability of domain A in S. cerevisiae. The effect of deletions of domain C can be observed only in the absence of domain B, at least by the assays used in the current study, and the significance of this finding is discussed.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beach D., Piper M., Shall S. Isolation of chromosomal origins of replication in yeast. Nature. 1980 Mar 13;284(5752):185–187. doi: 10.1038/284185a0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Li Y. Y., Feldman J., Jayaram M., Abraham J., Nasmyth K. A., Hicks J. B. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chan C. S., Tye B. K. Autonomously replicating sequences in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6329–6333. doi: 10.1073/pnas.77.11.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Tye B. K. Organization of DNA sequences and replication origins at yeast telomeres. Cell. 1983 Jun;33(2):563–573. doi: 10.1016/0092-8674(83)90437-3. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature. 1983 Sep 1;305(5929):23–28. doi: 10.1038/305023a0. [DOI] [PubMed] [Google Scholar]
- Conrad S. E., Campbell J. L. Characterization of an improved in vitro DNA replication system for Escherichia coli plasmids. Nucleic Acids Res. 1979 Jul 25;6(10):3289–3304. doi: 10.1093/nar/6.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W. L., Hice R. H., Chlebowicz-Sledziewska E. ARS replication during the yeast S phase. Cell. 1983 Mar;32(3):831–838. doi: 10.1016/0092-8674(83)90069-7. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Kornberg A. Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5817–5821. doi: 10.1073/pnas.80.19.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. N. Electrophoresis of ribonucleic acid on a polyacrylamide gel which contains disulfide cross-linkages. Anal Biochem. 1976 Nov;76(50):37–44. doi: 10.1016/0003-2697(76)90261-x. [DOI] [PubMed] [Google Scholar]
- Harland R. M., Laskey R. A. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980 Oct;21(3):761–771. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- Hsiao C. L., Carbon J. High-frequency transformation of yeast by plasmids containing the cloned yeast ARG4 gene. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski S. M., Niedzwiecka A., Edelman G. M. In vitro association of a replication complex with a yeast chromosomal replicator. J Biol Chem. 1983 Mar 10;258(5):2754–2757. [PubMed] [Google Scholar]
- Kearsey S. Structural requirements for the function of a yeast chromosomal replicator. Cell. 1984 May;37(1):299–307. doi: 10.1016/0092-8674(84)90326-x. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y. Yeast plasmid requires a cis-acting locus and two plasmid proteins for its stable maintenance. Cell. 1983 Dec;35(2 Pt 1):487–493. doi: 10.1016/0092-8674(83)90182-4. [DOI] [PubMed] [Google Scholar]
- Kuo C. L., Campbell J. L. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8-1 mutation. Mol Cell Biol. 1983 Oct;3(10):1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerski R. J., Hodnett J. L., Gray H. B., Jr Extracellular nucleases of pseudomonas BAL 31. III. Use of the double-strand deoxyriboexonuclease activity as the basis of a convenient method for the mapping of fragments of DNA produced by cleavage with restriction enzymes. Nucleic Acids Res. 1978 May;5(5):1445–1464. doi: 10.1093/nar/5.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L. Functional relationships between transcriptional control signals of the thymidine kinase gene of herpes simplex virus. Cell. 1982 Dec;31(2 Pt 1):355–365. doi: 10.1016/0092-8674(82)90129-5. [DOI] [PubMed] [Google Scholar]
- Montiel J. F., Norbury C. J., Tuite M. F., Dobson M. J., Mills J. S., Kingsman A. J., Kingsman S. M. Characterization of human chromosomal DNA sequences which replicate autonomously in Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Jan 25;12(2):1049–1068. doi: 10.1093/nar/12.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins J. I., Casey J. W., Nicolson M. O., Burck K. B., Davidson N. Sequence arrangement and biological activity of cloned feline leukemia virus proviruses from a virus-productive human cell line. J Virol. 1981 May;38(2):688–703. doi: 10.1128/jvi.38.2.688-703.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Pedigree analysis of plasmid segregation in yeast. Cell. 1983 Oct;34(3):961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Reed S. I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson D. C., Gaarder M. E., Livingston D. M. Characterization of the transmission during cytoductant formation of the 2 micrometers DNA plasmid from Saccharomyces. Mol Gen Genet. 1981;183(1):59–65. doi: 10.1007/BF00270139. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Mann C., Davis R. W. Centromeric DNA from Saccharomyces cerevisiae. J Mol Biol. 1982 Jun 25;158(2):157–190. doi: 10.1016/0022-2836(82)90427-2. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Struhl K., Davis R. W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979 Nov 1;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Thomas M., Kelly J., Selker E., Davis R. W. Eukaryotic DNA segments capable of autonomous replication in yeast. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4559–4563. doi: 10.1073/pnas.77.8.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The new yeast genetics. 1983 Sep 29-Oct 5Nature. 305(5933):391–397. doi: 10.1038/305391a0. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Blackburn E. H. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982 May;29(1):245–255. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- Taketo M., Jazwinski S. M., Edelman G. M. Association of the 2-micron DNA plasmid with yeast folded chromosomes. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3144–3148. doi: 10.1073/pnas.77.6.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Delta sequences and double symmetry in a yeast chromosomal replicator region. J Mol Biol. 1982 Apr 5;156(2):293–307. doi: 10.1016/0022-2836(82)90330-8. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Zakian V. A. Origin of replication from Xenopus laevis mitochondrial DNA promotes high-frequency transformation of yeast. Proc Natl Acad Sci U S A. 1981 May;78(5):3128–3132. doi: 10.1073/pnas.78.5.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian V. A., Scott J. F. Construction, replication, and chromatin structure of TRP1 RI circle, a multiple-copy synthetic plasmid derived from Saccharomyces cerevisiae chromosomal DNA. Mol Cell Biol. 1982 Mar;2(3):221–232. doi: 10.1128/mcb.2.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]



