Abstract
Background
Salivary adenoid cystic carcinoma (ACC) is a rare relentlessly progressive malignant tumor. The molecular events associated with ACC tumorigenesis are poorly understood. Variable microRNAs (miRNA) have been correlated with tumorigenesis of several solid tumors but not in ACC. To investigate the association of miRNAs with the development and/or progression of ACC, we performed a comparative analysis of primary ACC specimens and matched normal samples and a pooled salivary gland standard and correlated the results with clinicopathologic factors and validated selected miRNAs in a separate set of 30 tumors.
Methods
MiRNA array platform was used for the identification of target miRNAs and the data was subjected to informatics and statistical interrelations. The results were also collected with the MYB-NFIB fusion status and the clinicopathologic features.
Results
Differentially dysregulated miRNAs in ACC were characterized in comparison to normal expression. No significant differences in miRNA expression were found between the MYB-NFIB fusion positive and -negative ACCs. Of the highly dysregulated miRNA in ACC, overexpression of the miR-17 and miR-20a were significantly associated with poor outcome in the screening and validation sets.
Conclusion
Our study indicates that the upregulation of miR-17-92 may play a role in the biology of ACC and could be potentially targeted in future therapeutic studies.
Introduction
Adenoid cystic carcinoma, an uncommon salivary gland malignancy, is characterized by histopathologic and cellular heterogeneity and a relentless progressive clinical course [1], [2]. The primary treatment of ACC is complete surgical excision with and without post-operative radiotherapy [3]. Patients with locally advanced primary, recurrent, and metastatic ACC have been treated experimentally with chemotherapy and targeted agents with minimal success [4], [5]. Several genomic investigations exclusive of miRNA analysis have been carried out in ACC to identify biological markers of therapeutic potential [6]–[9]. These efforts, however, have been largely unrewarding and additional investigations of new targets are needed.
MiRNAs, a new class of highly conserved, short (19-22-nucleotides) non-coding RNA molecules, are products of a highly coordinated processing of a long RNA sequence template by specific RNAase III endonucleases [10]. Most miRNAs' regulatory functions are achieved through binding to the 3′ untranslated sequence of the RNA target (3′-UTR) transcript. Complete complementarity of miRNA to their messenger RNA targets results in complete transcriptional repression, while imperfect matching, the most common occurrence lead to partial transcriptional dysregulation. Imperfect or partial base-pairing with target mRNAs, however, allows the miRNA to bind to a large number of coding genes. Moreover, multiple miRNAs can be produced from a single pre-miRNA transcript and these may act independently or in concert on a wide range of genes in both normal and tumorigenic status [11]–[15].
Except for a study of miRNA expression in pleomorphic adenoma, a common benign salivary gland tumor, little is known about the role of these molecules in malignant salivary tumors including ACC [16]. Recently, a t(6; 9) leading to a fusion between the MYB and NFIB genes and the MYB gene overexpression was reported in a large subset of ACCs [17], [18]. Interestingly, the upregulation of MYB in ACC has been suggested due to the disruption of the 3′ UTR (miRNA binding sites) by the translocation with the NFIB gene [19]–[21]. Furthermore, evidence for a regulatory effect of the MYB gene on other miRNAs has been shown [22]. Collectively, these findings suggest a role for certain miRNAs in ACC tumorigenesis.
We hypothesize that certain miRNAs play a role in the regulation of cellular pathways in the ACC tumorigenesis and this may be influenced by the fusion gene status. To test our hypothesis we performed miRNA analysis on normal salivary tissues and MYB-NFIB fusion positive and negative ACCs to determine differentially altered candidates of potential biological significance.
Materials and Methods
Ethics Statement
This study was approved by the MD Anderson Cancer Center Institutional Review Board (IRB protocol # Lab07-0382). Written informed consent was provided by all patients in this study to perform the subsequent analyses.
Tissue samples and RNA extractions
For the screening of miRNA expression profiling, fresh frozen tissue specimens from 30 primary ACCs and 4 matched normal salivary samples were collected initially. For the validation of identified miRNAs, 30 further ACC tumor samples were used. All tissue samples were accessioned at the head and neck section, MD Anderson Cancer Center, from 1989 to 2010, and formed the materials for this study. The clinicopathological features were described in Table 1. All tissues were harvested immediately in fresh state and placed in liquid nitrogen and stored at −80°C until used. Total RNA was extracted with the Trizol reagent (Invitrogen, Carlsbad, CA, USA), and then cleaned by RNeasy mini cleanup kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The quality of the total RNA was verified by an Agilent 2100 Bioanalyzer profile. All the samples had an RNA integrity number greater than 7.0.
Table 1. Demographic and clinicopathologic characteristics of the initial screening (n = 30) and the validation sets (n = 30) of salivary adenoid cystic carcinoma.
| Characteristic | Screening set (N, %) | Validation set (N, %) |
| Gender | ||
| Male | 20 (67) | 18 (60) |
| Female | 10 (33) | 12 (40) |
| Age (years) | ||
| <60 | 20 (67) | 22 (73) |
| ≥60 | 10 (33) | 8 (27) |
| Tumor site | ||
| Major | 6 (20) | 6 (20) |
| Minor | 24 (80) | 24 (80) |
| Tumor size | 22 (73) | |
| <4cm | 15 (50) | 8 (27) |
| ≥4cm | 15 (50) | |
| Histologic type | ||
| T/C | 16 (53) | 15 (50) |
| Any solid | 14 (47) | 15 (50) |
| PNIa | ||
| No | 1 (3) | 3 (10) |
| Yes | 28 (93) | 19 (63) |
| Not stated | 1 (3) | 8 (27) |
| Stage | ||
| I or II | 2 (7) | 10 (33) |
| III or IV | 23 (77) | 12 (40) |
| NA | 5 (17) | 8 (27) |
| Recurrence | ||
| No | 9 (30) | 12 (40) |
| Yes | 21 (70) | 18 (60) |
| Distant metastasis | ||
| No | 16 (53) | 15 (50) |
| Yes | 14 (47) | 15 (50) |
| MYB-NFIB fusionb | ||
| No | 10 (33) | 11 (37) |
| Yes | 20 (67) | 19 (63) |
N; Number. aPNI; Perineural invasion, T/C; Tubular/Cribriform. b MYB-NFIB fusion was identified by FISH (refs. 17 and 21).
MiRNA array profiling
One µg total RNA from tumor and normal tissue samples and a pooled normal salivary gland standard (Clontech Laboratories) was labelled with Hy3™ and Hy5™ fluorescent label, respectively, using the miRCURY™ LNA Array power labelling kit (Exiqon, Vedbaek, Denmark) following the procedure described by the manufacturer. The Hy3™-labeled samples and a Hy5™-labeled reference RNA sample were mixed pair-wise and hybridized to the miRCURY™ LNA Array version 5th Generation (Exiqon), which contains capture probes targeting all miRNAs for human, mouse or rat registered in the miRBASE version 15.0 at the Sanger Institute. The hybridization was performed according to the miRCURY™ LNA array manual using a Tecan HS4800 hybridization station (Tecan, Männedorf, Switzerland). After hybridization, the microarray slides were scanned and stored in an ozone free environment (ozone level below 2.0 ppb) in order to prevent potential bleaching of the fluorescent dyes. The miRCURY™ LNA array microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Santa Clara, CA, USA) and the image analysis was carried out using the ImaGene 8.0 software (BioDiscovery, El Segundo, CA, USA). The quantified signals were background corrected (Normexp with offset value −10) [23] and normalized using the global Lowess (Locally Weighted Scatterplot Smoothing) regression algorithm.
MiRNA array data and statistical analysis
The ratios of median values for expression of each miRNA tumor/normal tissues were determined and compared using Mann-Whitney U tests. A cut off of values <0.05 (under-expression) or >2 (over-expression), coupled with a p-values of <0.05 by Mann-Whitney U tests, were considered significant. To investigate the effect of the MYB-NFIB fusion, the miRNA expression in fusion positive and negative tumors were compared. Fusion status was decided by using fluorescence in-situ hybridization (FISH) on touch preparation of fresh tissues as previously described [17], [21]. For association of miRNAs with clinicopathological parameter and histological patterns of ACC, the median ratio values of each miRNA were compared by Mann-Whitney U test. The significant analysis of microarrays (SAM) algorithm were performed [24] and all miRNAs listed in the false discovery rate (FDR<0.05%) were calculated. For visualization, the linkage clustering with centrered Peason correlation was performed by Multiexperiment Viewer (MeV, http://www.tm4.org/mev.html) tool.
For assessing the correlation of individual miRNAs with survival outcomes, the median ratio values of each miRNA from tumors of patients who had died and who were still living at last contact were compared by Mann-Whitney U test. The direction of the difference in median ratios of those miRNAs that showed statistical significance was determined from the ratios of the median values from tumors of dead/living patients. If the median ratio value of the miRNAs of patients who had died was higher than that from living patients, then the highest quartile of values of all 30 tumor patients were tested against the lower quartile values by the log rank test for survival plots. If the median ratio value of miRNA from patients who had died was lower than that from patients who are still living, then the lowest quartile of values from all 30 tumor patients were similarly tested for correlation with lower survival curves. Those miRNAs that showed statistically significant correlations (p<0.05) in both Mann-Whitney U tests and log rank tests were considered to be significantly correlated with unfavorable survival outcomes. To help determine the magnitude of the effect, Risk Ratios were calculated by Cox regression analysis.
Quantitative RT-PCR of miRNA
For validation, we selected highly dysregulated miRNAs to be tested by quantitative RT-PCR in a separate set of 30 tumors. The miRUCURY LNATM Universal RT miRNA PCR assays (Exiqon) for hsa-miR-455-3p, miR-455-5p, miR-375, miR-142-3p, miR-17, and miR-20a were used for their quantification. Five ng of total RNA was used for reverse transcription by Universal cDNA synthesis kit (Exiqon) according to the manufacturer's instructions. Quantitative RT-PCR reactions were run using the 7900HT Fast Real-Time PCR System (Applied Biosystems) with SYBR® Green Master Mix Universal RT (Exiqon) in the Quantitative Genomics Core Laboratory (The University of Texas Health Science Center at Houston). Hsa-miR-99b* was selected as a normalisation primer by NormFinder software [25]. The mean Ct was determined from duplicate reactions. The expression of each miRNAs were determined by the ΔCT method (Average CT-target miRNA – Average CT-miR-99*).
Results
MiRNA expression profiles in ACC tumor
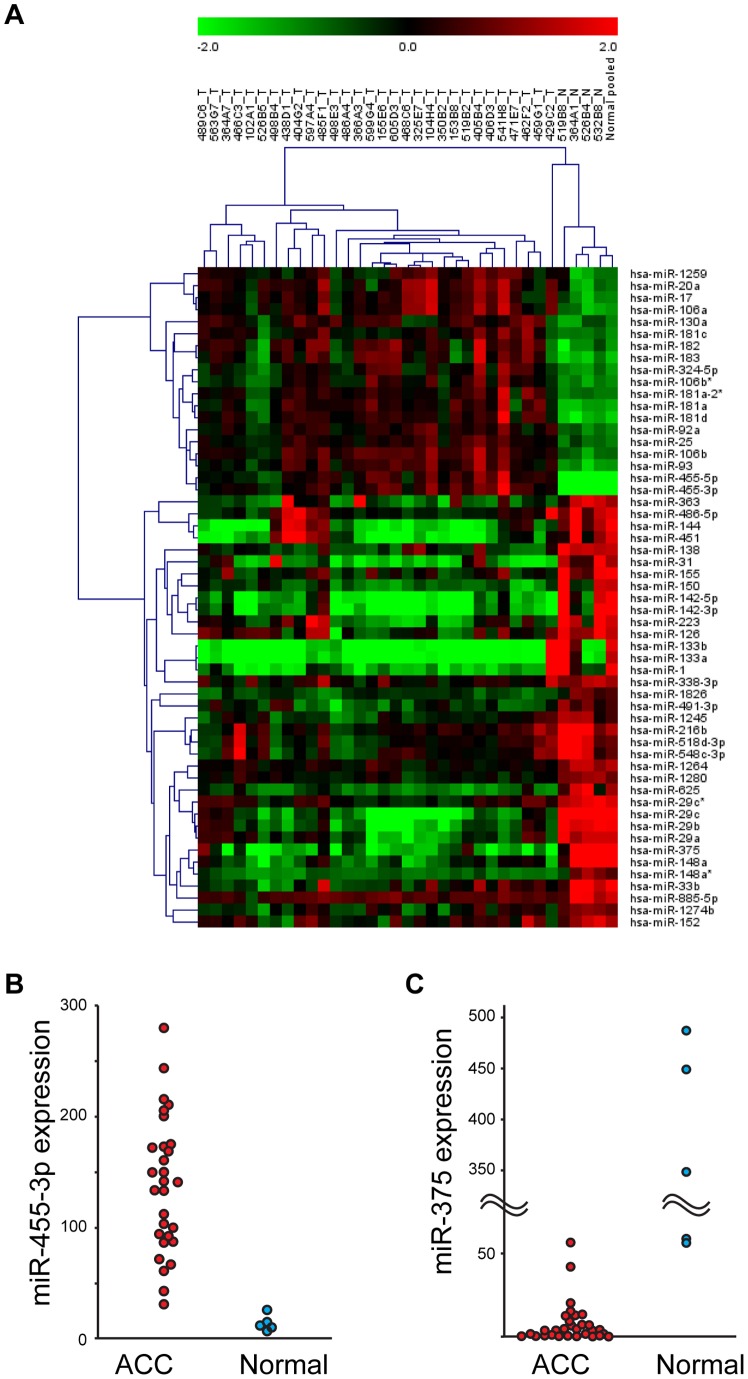
Table 1 presents the clinicopathologic characteristics of the 30 ACCs from the screening set and figure 1A, the differentially expressed miRNAs and the heat map diagram of the two-way hierarchical clustering of tumor and normal specimens. The results showed 55 miRNAs to be significantly different between ACCs and normal specimens by both Mann-Whitney U test and SAM algorithm. Nineteen miRNAs were up-regulated (Table 2) and 36 were down-regulated (Table 3) in tumor in comparison to normal and standard. Eight of the 19 up-regulated miRNAs in ACCs represented the miR17-92 cluster and its paralogs, miR106 b-25 and miR106a-363. All highly dysregulated miRNAs were selected for validation by quantitative RT-PCR. Figure 1B and 1C presents the expression of miR-455-3p (up-regulated) and miR-375 (down-regulated) in both normal and tumor specimens; miR455-3p shows high levels of expression in tumor specimens in contrast to normal tissues (p<0.001), paradoxically, miR-375 (p<0.001) was markedly down regulated in tumors in contrast to normal tissues (Mann-Whitney U test).
Figure 1. MiRNA expression differences in salivary ACCs and normals.
(A) A heat map of the differential miRNA expression in normal salivary tissues and control versus tumor samples. Note the clean distinction between normal tumors. Quantitative RT-PCR validation of miRNA expression, miR-455-3p (B) and miR-375 (C).
Table 2. Upregulated miRNAs in salivary adenoid cystic carcinoma in comparison to normal salivary gland.
| miRNAs | p a | T/N ratiob | Chrc | Host Gened |
| hsa-miR-455-3p | 0.001 | 10.75 | 9q32 | COL27A1 |
| hsa-miR-455-5p | 0.001 | 7.11 | 9q32 | COL27A1 |
| hsa-miR-181d | 0.001 | 3.63 | 13p.13 | intergenic |
| hsa-miR-183 | 0.001 | 3.53 | 7q32.2 | intergenic |
| hsa-miR-181a | 0.001 | 3.04 | 9q33.3 | NR6A1 |
| ehsa-miR-93 | 0.001 | 2.96 | 7q22.1 | MCM7 |
| hsa-miR-182 | 0.001 | 2.86 | 7q32.2 | intergenic |
| ehsa-miR-106a | 0.001 | 2.80 | Xq26.2 | intergenic |
| ehsa-miR-17 | 0.001 | 2.66 | 13q31.3 | MIR17HG |
| hsa-miR-130a | 0.001 | 2.56 | 11q12.1 | AP000662.4 |
| ehsa-miR-20a | 0.001 | 2.43 | 13q31.3 | MIR17HG |
| hsa-miR-324-5p | 0.001 | 2.35 | 17p13.1 | ACADVL |
| ehsa-miR-106b | 0.001 | 2.29 | 7q22.1 | MCM7 |
| hsa-miR-181a-2* | 0.001 | 2.21 | 9q33.3 | MIR181A2HG, NR6A1 |
| hsa-miR-181c | 0.001 | 2.16 | 19p13.13 | intergenic |
| hsa-miR-1259 | 0.004 | 2.11 | NA | NA |
| ehsa-miR-106b* | 0.001 | 2.09 | 7q22.1 | MCM7 |
| ehsa-miR-25 | 0.001 | 2.07 | 7q22.1 | MCM7 |
| ehsa-miR-92a | 0.001 | 2.01 | 13q31.3 | MIR17HG |
NA; no information. miRNA(*); miRNA star strand.
Mann-Whitney U test. bTumor/Normal median ratio >2 classified as significant. cChromosomal location. dThese information obtained from miRBase database (http://www.mirbase.org). eThe miR-17-92 cluster and its paralogs.
Table 3. Downregulated miRNAs in salivary adenoid cystic carcinoma in comparison to normal salivary gland.
| miRNAs | p a | T/N ratiob | Chrc | Host Gened |
| hsa-miR-375 | 0.001 | 0.042 | 2q35 | MIR375 |
| hsa-miR-142-3p | 0.002 | 0.084 | 17q22 | intergenic |
| hsa-miR-142-5p | 0.002 | 0.096 | 17q22 | intergenic |
| hsa-miR-148a | 0.001 | 0.098 | 7p15.2 | intergenic |
| hsa-miR-29c | 0.001 | 0.102 | 1q32.2 | intergenic |
| hsa-miR-133b | 0.003 | 0.112 | 6p12.2 | RP11-771D21 |
| hsa-miR-144 | 0.002 | 0.138 | 17q11.2 | intergenic |
| hsa-miR-133a | 0.004 | 0.138 | 18q11.2 | MIB1 |
| hsa-miR-29b | 0.001 | 0.158 | 7q32.3 | AC016831.7 |
| hsa-miR-31 | 0.001 | 0.161 | 9p21.3 | RP11-354P17.9 |
| hsa-miR-451 | 0.007 | 0.174 | 17q11.2 | intergenic |
| hsa-miR-216b | 0.001 | 0.186 | 2p16.1 | AC011306.2 |
| hsa-miR-33b | 0.001 | 0.193 | 17p11.2 | SREBF1 |
| hsa-miR-150 | 0.008 | 0.212 | 19q13.33 | intergenic |
| ehsa-miR-363 | 0.003 | 0.214 | Xq26.2 | intergenic |
| hsa-miR-223 | 0.008 | 0.221 | Xq12 | AL034397.1 |
| hsa-miR-155 | 0.001 | 0.234 | 21q21.3 | MIR155HG |
| hsa-miR-625 | 0.001 | 0.235 | 14q23.3 | FUT8 |
| hsa-miR-29a | 0.001 | 0.254 | 7q32.3 | MIR29A, AC016831.7 |
| hsa-miR-138 | 0.001 | 0.259 | 16q13 | intergenic |
| hsa-miR-518d-3p | 0.002 | 0.303 | 19q13.42 | intergenic |
| hsa-miR-1 | 0.002 | 0.305 | 18q11.2 | MIB1 |
| hsa-miR-548c-3p | 0.005 | 0.313 | 12q14.2 | RASSF3 |
| hsa-miR-486-5p | 0.005 | 0.318 | 8p11.21 | ANK1 |
| hsa-miR-148a* | 0.001 | 0.319 | 7p15.2 | intergenic |
| hsa-miR-29c* | 0.001 | 0.322 | 1q32.2 | intergenic |
| hsa-miR-1245 | 0.001 | 0.335 | 2q32.2 | COL3A1 |
| hsa-miR-126 | 0.001 | 0.372 | 9q34.3 | EGFL7 |
| hsa-miR-1264 | 0.001 | 0.395 | Xq23 | HTR2C |
| hsa-miR-1274b | 0.001 | 0.449 | NA | NA |
| hsa-miR-338-3p | 0.002 | 0.451 | 17q25.3 | AATK |
| hsa-miR-491-3p | 0.001 | 0.466 | 9p21.3 | KIAA1797 |
| hsa-miR-152 | 0.002 | 0.470 | 17q21.32 | COPZ2 |
| hsa-miR-1826 | 0.001 | 0.480 | NA | NA |
| hsa-miR-1280 | 0.001 | 0.486 | 3q21.3 | EEFSEC |
| hsa-miR-885-5p | 0.005 | 0.498 | 3p25.3 | ATP2B2 |
NA; no information. miRNA(*); miRNA star strand.
Mann-Whitney U test, bTumor/Normal median ratio <0.5 classified as significant. cchromosomal location. dThese information obtained from miRBase database (http://www.mirbase.org). eThe miR-17-92 cluster and its paralogs.
Correlation of miRNA levels and clinicopathologic parameters
Correlation with clinicopathologic factors revealed 108 dysregulated miRNAs to be correlated significantly with tumor size (T), 18 with tumor stage (T), 13 with lymph node metastasis (N), and 39 with tumor recurrence (p<0.05, Mann-Whitney U test). Only two miRNAs, let-7a and miR-150, were significantly correlated with T or N and Stage (Table 4); over-expression of miR-let-7a was also correlated with tumor recurrence (Table 4).
Table 4. Correlation of miRNAs and Tumor size, Lymph node status, Stage, and Recurrence in patients with adenoid cystic carcinoma.
| miRNAs | T | N | Stage | Recurrence |
| hsa-let-7a | 0.012 | ns | 0.027 | 0.04 |
| hsa-miR-150 | ns | 0.019 | 0.013 | ns |
P-value by Mann-Whitney U tests. T; Tumor size, N; Lymph node status, ns; Not significant.
We also noted that the expression of 133 miRNAs to be significantly associated with tumor that contained solid component. Fifty-five of these were significant in 2-tailed Fisher exact tests and thirty by SAM algorithm (Table 5), including miR-20a and miR-17 (T/N ratio; miR-20a, 1.34, miR-17a, 1.41).
Table 5. Correlations of miRNAs with histologic pattern of adenoid cystic carcinoma.
| miRNAs | Mann-Whitney U p a | Fisher Exact p a | S/N ratiob |
| hsa-miR-205 | 0.009 | 0.002 | 0.15 |
| hsa-miR-381 | 0.005 | 0.039 | 0.28 |
| hsa-miR-143 | 0.013 | 0.002 | 0.33 |
| hsa-miR-145 | 0.009 | 0.002 | 0.34 |
| hsa-miR-376c | 0.001 | 0.039 | 0.35 |
| hsa-miR-145* | 0.009 | 0.002 | 0.35 |
| hsa-miR-34c-5p | 0.002 | 0.039 | 0.37 |
| hsa-miR-376a* | 0.001 | 0.039 | 0.38 |
| hsa-miR-136 | 0.001 | 0.039 | 0.39 |
| hsa-miR-143* | 0.007 | 0.002 | 0.39 |
| hsa-miR-127-3p | 0.001 | 0.039 | 0.41 |
| hsa-miR-409-3p | 0.001 | 0.039 | 0.41 |
| hsa-miR-654-3p | 0.001 | 0.039 | 0.41 |
| hsa-miR-154 | 0.001 | 0.039 | 0.42 |
| hsa-miR-410 | 0.001 | 0.039 | 0.42 |
| hsa-miR-411 | 0.003 | 0.039 | 0.43 |
| hsa-miR-379 | 0.001 | 0.039 | 0.45 |
| hsa-miR-382 | 0.001 | 0.039 | 0.45 |
| hsa-miR-136* | 0.001 | 0.039 | 0.46 |
| hsa-miR-377 | 0.002 | 0.039 | 0.46 |
| hsa-miR-329 | 0.001 | 0.039 | 0.50 |
| hsa-miR-432 | 0.001 | 0.039 | 0.52 |
| hsa-miR-22 | 0.005 | 0.039 | 0.53 |
| hsa-miR-495 | 0.001 | 0.039 | 0.56 |
| hsa-miR-127-5p | 0.001 | 0.039 | 0.59 |
| hsa-miR-376b | 0.001 | 0.039 | 0.59 |
| hsa-miR-20a | 0.013 | 0.002 | 1.34 |
| hsa-miR-17 | 0.010 | 0.002 | 1.41 |
| hsa-miR-9* | 0.003 | 0.002 | 4.49 |
| hsa-miR-9 | 0.002 | 0.002 | 4.59 |
miRNA(*); miRNA star strand.
P-values were calculated using the Mann-Whitney U test and 2-tailed Fisher exact test for solid and non-solid types. bS/N ratio; Solid/non-solid median ratio.
Correlation of MiRNA levels and survival
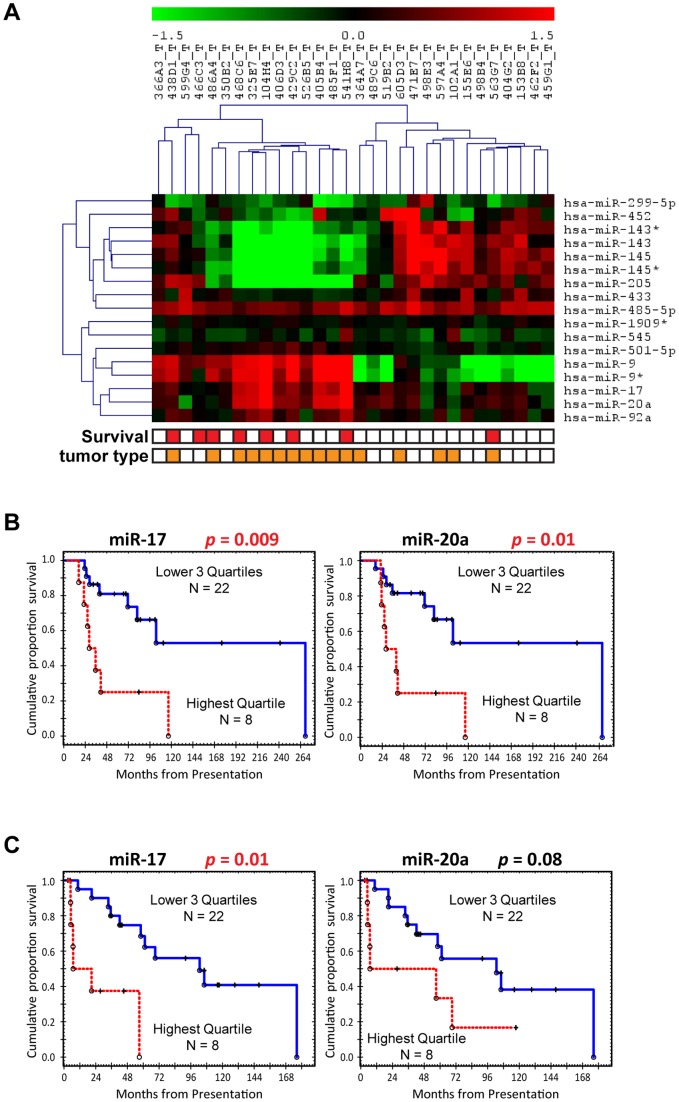
Figure 2A presents the unsupervised hierarchical clustering of miRNAs expression and patients' survival and Table 6 presents the Log Rank test and Cox proportional hazards regression analysis. Seventeen of the highly dysregulated miRNAs were significantly correlated with poor survival outcomes (Hazard ratios of 2.9 and 4.4). The most significantly correlated miRNAs with survival were the miR-17-92 cluster in a screening set by quantitative RT-PCR (miR-17, and -20a; Figure 2B, Log-rank test, p = 0.009 and p = 0.01 respectively).
Figure 2. Identification of dysregulated miRNAs for overall survival in ACCs.
(A) Hierarchical clustering of the dysragulated miRNAs related to the patients' survival (Table 6) in ACCs. The samples marked by red color in the survival section indicate that the patient died within 3 years. The samples marked by orange color in the tumor type indicate the solid ACC. (B) Kaplan-Meier analysis of quantitative RT-PCR using the screening set of ACCs. High expression of miR-17 and miR-20a is associated with the poor survival significantly (p = 0.009 and p = 0.01, respectively). The cercle and cross marks indicate the date of death and last contact, respectively. (C) Kaplan-Meier analysis of quantitative RT-PCR using the validation set of ACCs. Further validation confirmed that high expression of miR-17 is correlated significantly with the poor survival (p = 0.01).
Table 6. Correlation between dysregulated miRNAs and clinical outcome in adenoid cystic carcinoma patients.
| miRNAs | Log rank p a | Expression | HR (95% Cl)b | Cox regression p a |
| hsa-miR-433 | 0.025 | Low | 4.43 (1.34–14.7) | 0.015 |
| hsa-miR-143 | 0.008 | Low | 4.37 (1.51–12.7) | 0.007 |
| hsa-miR-145 | 0.008 | Low | 4.37 (1.51–12.7) | 0.007 |
| hsa-miR-501-5p | 0.008 | High | 4.37 (1.51–12.7) | 0.007 |
| hsa-miR-483-5p | 0.035 | Low | 3.93 (1.19–12.9) | 0.024 |
| hsa-miR-545 | 0.025 | High | 3.92 (1.31–11.8) | 0.015 |
| hsa-miR-9 | 0.012 | High | 3.87 (1.34–11.2) | 0.012 |
| hsa-miR-9* | 0.012 | High | 3.87 (1.34–11.2) | 0.012 |
| hsa-miR-143* | 0.014 | Low | 3.70 (1.28–10.7) | 0.016 |
| hsa-miR-145* | 0.014 | Low | 3.70 (1.28–10.7) | 0.016 |
| hsa-miR-17 | 0.014 | High | 3.65 (1.27–10.5) | 0.016 |
| hsa-miR-20a | 0.014 | High | 3.65 (1.27–10.5) | 0.016 |
| hsa-miR-1909* | 0.035 | High | 3.56 (1.17–10.8) | 0.025 |
| hsa-miR-299-5p | 0.036 | Low | 3.47 (1.15–10.5) | 0.027 |
| hsa-miR-92a | 0.040 | High | 3.21 (1.11–9.34) | 0.032 |
| hsa-miR-205 | 0.046 | Low | 3.08 (1.06–8.99) | 0.039 |
| hsa-miR-452 | 0.039 | Low | 2.88 (1.00–8.32) | 0.049 |
miRNA(*); miRNA star strand.
P-values were calculated by Log rank test and Cox regression analysis. bHR; Hazard ratio, CI; confidence interval.
MiRNA and MYB-NFIB fusion in ACCs
To test whether the fusion status affects miRNA expression, comparative analysis of fusion positive and fusion negative ACC specimens was performed on the 30 screening set; 20 tumors were fusion positive and 10 were fusion negative. The results showed no significant difference in miRNA expression between fusion positive and negative ACCs by SAM. We also found no association between the expression of miR-15a, miR-16 and miR-150 reported to regulate the MYB gene through their binding to its 3′UTR [18].
Validation of miR-17-92 cluster as prognostic miRNAs
We selected members of the miR-17-92 cluster, miR-17 and -20a, for validation in a separate set of 30 tumors by quantitative RT-PCR analysis. In the validation set, no significant correlation was found between the expression levels of both miR-17- and -20a and tumors with solid component. This may be explained by the variable and subjective nature of assessing this component in tumors. Kaplan-Meier analysis shows that there was a significant correlation between miR-17 (Figure 2C, Log-rank test, p = 0.01) and strong association between miR-20a expression and poor outcome (Figure 2C, Log-rank test, p = 0.08). In combined analysis sets of the 60 ACCs (screening and validation sets), significant statistical association of high expression of miR-17 and miR-20a and poor survival was found (Log-rank test, p<0.01). As to the fusion status of the 30 validation set, 19 were fusion positive and 11 were fusion negative, there was no significant difference in the miRNA expression between fusion positive and negative tumors.
Discussion
As the first miRNA study of ACC, we identified differentially expressed miRNAs that distinctly separated normal salivary tissue and standard from ACC tumors. The most significantly over-expressed miRNAs in tumors were the miR-17-92 cluster and its paralogs including miR-455-3p, -455-5p, -181 and miR-183. In this study, upregulation of miR-17 and miR-20a, members of miR-17-92 cluster genes, was found to be significantly associated with the poor outcome by two different statistical methods. [26]–[37]. Interestingly, evidence for an association between these miRNAs and salivary gland development has been reported [26] suggesting a tissue/tumor context association. This is further supported by the finding that several of the highly dysregulated miRNAs in ACC were found to be involved in head and neck squamous carcinoma biology [38]. The miR-17-92 cluster are encoded by a polycistronic gene on chromosome 13q31 region and are conserved in all vertebrates and play a fundamental regulatory function in the development and progression of several tumor types [26]–[28], [34], [39]–[41]. Surprisingly, two of the highly upregulated miRNAs, (miR-455-3p and miR-455-5p) in our study are of an unknown function. Although the oncogenic and/or functional role of these miRNAs in ACC tumorgenesis and progression are currently unclear, future investigations are needed to determine their biological role in ACC.
In this study, marked down-regulation of miR-375, -142-3p, 142-5p, -148, -155, -33b, and miR-29 family members in ACC was noted. Interestingly, low expression of these miRNAs was also found to be associated with aggressive behavior in several solid neoplastic entities [29], [31], [42]–[50]. Although no correlation between the down regulation of these miRNAs and adverse features of ACC was found, we contend that larger cohorts with long term follow-up is needed to determine their biological role.
Our results show several miRNAs to be correlated with the solid component and poor outcome in patients with ACC. However, our validation analysis in a separate cohort confirmed only the association of the selected miRNAs upregulation with the outcome. The data collectively suggest that the presence of the solid component and poor outcome are not mutually exclusive and these miRNAs play a role in the biological progression of ACC. The data also highlights the difficulties in assessing the extent and the contribution of the solid feature in this entity. Of particular interest, however, is the finding of significant correlation between low miR-205 expression and poor survival in patients with ACC. This miRNA has been reported to be exclusively expressed in the cytoplasm of myoepithelial cells in normal and hyperplastic breast tissues and its loss or down-regulation was significantly associated with progression to ductal carcinoma [51], [52]. We, therefore, contend that the loss of this miRNA in ACC could be attributed to the loss of myoepithelial cells in the solid form.
In this study, no significant correlation between miRNA expression and the fusion status of ACCs was found. We also found no dysregulation of miR-16, 16 and 150 in this cohort. These findings are at variance with previous studies implicating these miRNAs in the regulation of the MYB gene [18], [53], [54]. Since one of the major targets of this miRNA is the MYB gene, our results suggest that mechanisms other than 3′ UTR deletion are involved in its regulation in ACC [54]. Recently, copy number abnormalities and genomic rearrangements have been shown to influence miRNA in different tumor types; similar integrative analysis may also be needed in ACC [10], [55]. In conclusion, our data shows that upregulation of the miR-17-92 cluster was associated with the aggressive behavior of ACC tumors.
Acknowledgments
We thank Ms. Ann Sutton, Mr. Jason Martinez and Ms. Lora Lothringer for their editorial and administrative efforts. We also would like to thank Dr. Gregory L. Shipley (The University of Texas Health Science Center at Houston) for supporting the miRNA validation data collection and experiment.
Funding Statement
The study is supported in part by the NIH National Institute of Dental and Craniofacial Research and the NIH Office of Rare Diseases Research grant number U01DE019765, the Head and Neck SPORE program grant number P50 CA097007, The Kenneth D. Muller professorship and an NCI CA-16672 grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chomette G, Auriol M, Tranbaloc P, Vaillant JM (1982) Adenoid cystic carcinoma of minor salivary glands. Analysis of 86 cases. Clinico-pathological, histoenzymological and ultrastructural studies. Virchows Arch A Pathol Anat Histol 395: 289–301. [DOI] [PubMed] [Google Scholar]
- 2. Batsakis JG, Luna MA, El-Naggar A (1990) Histopathologic grading of salivary gland neoplasms: III. Adenoid cystic carcinomas. Ann Otol Rhinol Laryngol 99: 1007–1009. [DOI] [PubMed] [Google Scholar]
- 3. Spiro RH (1986) Salivary neoplasms: overview of a 35-year experience with 2,807 patients. Head Neck Surg 8: 177–184. [DOI] [PubMed] [Google Scholar]
- 4. Fordice J, Kershaw C, El-Naggar A, Goepfert H (1999) Adenoid cystic carcinoma of the head and neck: predictors of morbidity and mortality. Arch Otolaryngol Head Neck Surg 125: 149–152. [DOI] [PubMed] [Google Scholar]
- 5.El-Naggar AK, Huvos AG (2005) Adenoid cystic carcinoma. In: Barnes L, Eveson JW, Reichart P, Sidransky D, editors. World Health Organization Classification of Tumors Pathology and Genetics of Head and Neck Tumors: IARC Press, Lyon, France. 221–222.
- 6. Ambros V (2008) The evolution of our thinking about microRNAs. Nat Med 14: 1036–1040. [DOI] [PubMed] [Google Scholar]
- 7. Kim VN (2005) MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385. [DOI] [PubMed] [Google Scholar]
- 8. Rao PH, Roberts D, Zhao YJ, Bell D, Harris CP, et al. (2008) Deletion of 1p32-p36 is the most frequent genetic change and poor prognostic marker in adenoid cystic carcinoma of the salivary glands. Clin Cancer Res 14: 5181–5187. [DOI] [PubMed] [Google Scholar]
- 9. Rigoutsos I (2009) New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res 69: 3245–3248. [DOI] [PubMed] [Google Scholar]
- 10. van Kouwenhove M, Kedde M, Agami R (2011) MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11: 644–656. [DOI] [PubMed] [Google Scholar]
- 11. Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866. [DOI] [PubMed] [Google Scholar]
- 12. Ma L, Weinberg RA (2008) MicroRNAs in malignant progression. Cell Cycle 7: 570–572. [DOI] [PubMed] [Google Scholar]
- 13. Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z (2009) Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA 15: 1443–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, et al. (2006) Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20: 2202–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101: 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X, Cairns M, Rose B, O'Brien C, Shannon K, et al. (2009) Alterations in miRNA processing and expression in pleomorphic adenomas of the salivary gland. Int J Cancer 124: 2855–2863. [DOI] [PubMed] [Google Scholar]
- 17. Mitani Y, Li J, Rao PH, Zhao YJ, Bell D, et al. (2010) Comprehensive analysis of the MYB-NFIB gene fusion in salivary adenoid cystic carcinoma: Incidence, variability, and clinicopathologic significance. Clin Cancer Res 16: 4722–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Persson M, Andren Y, Mark J, Horlings HM, Persson F, et al. (2009) Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A 106: 18740–18744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harper ME, Franchini G, Love J, Simon MI, Gallo RC, et al. (1983) Chromosomal sublocalization of human c-myb and c-fes cellular onc genes. Nature 304: 169–171. [DOI] [PubMed] [Google Scholar]
- 20. Ramsay RG, Gonda TJ (2008) MYB function in normal and cancer cells. Nat Rev Cancer 8: 523–534. [DOI] [PubMed] [Google Scholar]
- 21. Mitani Y, Rao PH, Futreal PA, Roberts DB, Stephens PJ, et al. (2011) Novel chromosomal rearrangements and break points at the t(6;9) in salivary adenoid cystic carcinoma: association with MYB-NFIB chimeric fusion, MYB expression, and clinical outcome. Clin Cancer Res 17: 7003–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee J, Li Z, Brower-Sinning R, John B (2007) Regulatory circuit of human microRNA biogenesis. PLoS Comput Biol 3: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, et al. (2007) A comparison of background correction methods for two-colour microarrays. Bioinformatics 23: 2700–2707. [DOI] [PubMed] [Google Scholar]
- 24. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 26. Jevnaker AM, Khuu C, Kjole E, Bryne M, Osmundsen H (2011) Expression of members of the miRNA17-92 cluster during development and in carcinogenesis. J Cell Physiol 226: 2257–2266. [DOI] [PubMed] [Google Scholar]
- 27. Mendell JT (2008) miRiad roles for the miR-17-92 cluster in development and disease. Cell 133: 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olive V, Jiang I, He L (2010) mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 42: 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hiroki E, Akahira J, Suzuki F, Nagase S, Ito K, et al. (2010) Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci 101: 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ujifuku K, Mitsutake N, Takakura S, Matsuse M, Saenko V, et al. (2010) miR-195, miR-455-3p and miR-10a(*) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett 296: 241–248. [DOI] [PubMed] [Google Scholar]
- 31. Wald AI, Hoskins EE, Wells SI, Ferris RL, Khan SA (2011) Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck 33: 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, et al. (2006) Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res 66: 11590–11593. [DOI] [PubMed] [Google Scholar]
- 33. Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, et al. (2010) Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer 126: 1166–1176. [DOI] [PubMed] [Google Scholar]
- 34. Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, et al. (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tchernitsa O, Kasajima A, Schafer R, Kuban RJ, Ungethum U, et al. (2010) Systematic evaluation of the miRNA-ome and its downstream effects on mRNA expression identifies gastric cancer progression. J Pathol 222: 310–319. [DOI] [PubMed] [Google Scholar]
- 36. Cho WC, Chow AS, Au JS (2009) Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer 45: 2197–2206. [DOI] [PubMed] [Google Scholar]
- 37. Abraham D, Jackson N, Gundara JS, Zhao J, Gill AJ, et al. (2011) MicroRNA profiling of sporadic and hereditary medullary thyroid cancer identifies predictors of nodal metastasis, prognosis, and potential therapeutic targets. Clin Cancer Res 17: 4772–4781. [DOI] [PubMed] [Google Scholar]
- 38. Tran N, O'Brien CJ, Clark J, Rose B (2010) Potential role of micro-RNAs in head and neck tumorigenesis. Head Neck 32: 1099–1111. [DOI] [PubMed] [Google Scholar]
- 39. Huang G, Nishimoto K, Zhou Z, Hughes D, Kleinerman ES (2012) miR-20a encoded by the miR-17-92 cluster increases the metastatic potential of osteosarcoma cells by regulating Fas expression. Cancer Res 72: 908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, et al. (2011) The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res 17: 1875–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. De Preter K, Mestdagh P, Vermeulen J, Zeka F, Naranjo A, et al. (2011) miRNA expression profiling enables risk stratification in archived and fresh neuroblastoma tumor samples. Clin Cancer Res 17: 7684–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, et al. (2010) Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res 70: 9175–9184. [DOI] [PubMed] [Google Scholar]
- 43. Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, et al. (2010) Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res 16: 1129–1139. [DOI] [PubMed] [Google Scholar]
- 44. Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, et al. (2009) MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res 15: 6192–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang H, Garzon R, Sun H, Ladner KJ, Singh R, et al. (2008) NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Duursma AM, Kedde M, Schrier M, le Sage C, Agami R (2008) miR-148 targets human DNMT3b protein coding region. RNA 14: 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Haflidadottir BS, Bergsteinsdottir K, Praetorius C, Steingrimsson E (2010) miR-148 regulates Mitf in melanoma cells. PLoS One 5: e11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, et al. (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065–7070. [DOI] [PubMed] [Google Scholar]
- 49. Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, et al. (2010) Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res 70: 4163–4173. [DOI] [PubMed] [Google Scholar]
- 50. Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, et al. (2008) A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 105: 13556–13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, et al. (2007) Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 67: 11612–11620. [DOI] [PubMed] [Google Scholar]
- 52. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, et al. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601. [DOI] [PubMed] [Google Scholar]
- 53. Lin YC, Kuo MW, Yu J, Kuo HH, Lin RJ, et al. (2008) c-Myb is an evolutionary conserved miR-150 target and miR-150/c-Myb interaction is important for embryonic development. Mol Biol Evol 25: 2189–2198. [DOI] [PubMed] [Google Scholar]
- 54. Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, et al. (2007) MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131: 146–159. [DOI] [PubMed] [Google Scholar]
- 55. Mi S, Li Z, Chen P, He C, Cao D, et al. (2010) Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci U S A 107: 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]