Abstract
Suppression of the cellular apoptotic program by the oncogenic herpesvirus Epstein-Barr virus (EBV) is central to both the establishment of latent infection and the development of EBV-associated malignancies. We have previously shown that expression of the EBV latent membrane protein 1 (LMP1) in Burkitt's lymphoma cell lines leads to increased mRNA levels from the cellular antiapoptotic bfl-1 gene (also known as A1). Furthermore, ectopic expression of Bfl-1 in an EBV-positive cell line exhibiting a latency type 1 infection protects against apoptosis induced by growth factor deprivation (B. N. D'Souza, M. Rowe, and D. Walls, J. Virol. 74:6652-6658, 2000). We now report that LMP1 drives bfl-1 promoter activity through interactions with components of the tumor necrosis factor receptor (TNFR)/CD40 signaling pathway. We present evidence that this process is NF-κB dependent, involves the recruitment of TNFR-associated factor 2, and is mediated to a greater extent by the carboxyl-terminal activating region 2 (CTAR2) relative to the CTAR1 domain of LMP1. Activation of CD40 receptor also led to increased bfl-1 mRNA levels and an NF-κB-dependent increase in bfl-1 promoter activity in Burkitt's lymphoma-derived cell lines. We have delineated a 95-bp region of the promoter that functions as an LMP1-dependent transcriptional enhancer in this cellular context. This sequence contains a novel NF-κB-like binding motif that is essential for transactivation of bfl-1 by LMP1, CD40, and the NF-κB subunit protein p65. These findings highlight the role of LMP1 as a mediator of EBV-host cell interactions and may indicate an important route by which it exerts its cellular growth transforming properties.
The Epstein-Barr virus (EBV) is a ubiquitous human herpesvirus that is associated with infectious mononucleosis and a spectrum of malignant diseases including African endemic Burkitt's lymphoma (BL), anaplastic nasopharyngeal carcinoma, Hodgkin's disease, and lymphoproliferative disorders in immunodeficient individuals (for reviews, see references 59 and 102). In vitro, EBV is exceptionally efficient at transforming and immortalizing resting human B lymphocytes leading to the outgrowth of transformed and immortalized lymphoblastoid cell lines (LCLs) displaying elevated levels of several cellular activation antigens and adhesion molecules (for a review, see reference 74). In an LCL, viral gene expression is generally restricted to a limited number of latent genes that encode six Epstein-Barr nuclear antigens (EBNA1, 2, 3A, 3B, 3C, and LP), three integral membrane proteins (LMP1, LMP2A, and 2B), and two small nuclear RNAs (for a review, see reference 37). Five of these (EBNA1, EBNA2, EBNA3A, EBNA3C, and LMP1) have been shown to be essential for the process of B-cell immortalization (for a review, see reference 62).
The frequent detection of LMP1 expression in many EBV-associated malignancies has led to the suggestion that this protein contributes to tumorigenesis. Several studies have shown that LMP1 possesses oncogenic properties. The expression of LMP1 leads to the transformation of rodent fibroblast cell lines and renders them tumorigenic in nude mice (118). In BL-derived cell lines, LMP1 induces many of the phenotypic changes observed in EBV infection including the upregulation of B-cell activation markers and cell adhesion molecules and an increased resistance to stimuli that induce apoptosis (74). In epithelial cells, LMP1 expression blocks the normal process of differentiation, reminiscent of the undifferentiated phenotype frequently observed in nasopharyngeal carcinoma (23). In keeping with these in vitro findings, targeted expression of LMP1 in the skin or B-cell compartment of transgenic mice leads to the induction of epithelial hyperproliferation and lymphomagenesis, respectively (77, 120). It has also recently been shown that LMP1 is critical for rendering LCLs tumorigenic in SCID mice (27).
The suppression of apoptotic death is a function of LMP1 that contributes to its oncogenicity. One well-documented mechanism by which LMP1 can protect against apoptosis is by upregulating the expression of several antiapoptotic proteins including Bcl-2, A20, and Mcl-1 (41, 53, 78, 91, 106, 119), thus raising the apoptotic threshold of the infected cell and also providing protection against a range of apoptosis-inducing stimuli. It was previously shown that elevated mRNA levels from an additional bcl-2 family member, bfl-1, are a feature of EBV-infected B lymphocytes exhibiting type 3 latency and that the expression of LMP1 in an EBV-negative BL cell line coincided with a dramatic increase in bfl-1 mRNA levels (30). In that study, Bfl-1 protected against serum depletion-induced apoptosis when expressed in the same cell context. Bfl-1 is an antiapoptotic protein whose preferential expression in hematopoietic and endothelial cells is controlled by inflammatory stimuli such as tumor necrosis factor (TNF) and interleukin-1 (IL-1) (17, 67, 68). bfl-1 is a mouse A1 homologue and encodes a 175-amino-acid protein that shares the highly conserved Bcl homology 1 (BH1), BH2, and BH3 domains with other Bcl-2 family members. Bfl-1 has been shown to suppress p53-mediated apoptosis and to possess cell proliferation and transforming activities in vitro (28, 29, 84). Elevated bfl-1 expression was reported in normal leukocytes and several cancer cell lines (73, 97).
LMP1, a member of the TNF receptor (TNFR)/CD40 superfamily (5) functions as a constitutively active receptor (46) and signals principally from intracellular compartments (80). Both oligomerization and localization within glycosphingolipid-rich membrane rafts are essential for the initiation of signaling (21, 36, 56). The cytoplasmic carboxyl-terminal tail of LMP1 contains two major effector domains, C-terminal activating region 1 (CTAR1), also known as transformation effector site 1 (TES1), and CTAR/TES2. CTAR1 is located proximal to the membrane, binds TNFR-associated factors (TRAFs) (26, 61), and is essential for EBV-mediated B-cell immortalization (65, 69, 70). CTAR2/TES2, which is located near the C terminus, supports the long-term growth of immortalized B cells (64) and recruits the TNFR-associated death domain (TRADD) protein and receptor-interacting protein (34, 61, 63). Consequently, LMP1 triggers several signaling pathways that lead to the activation of transcription factors, including NF-κB, STATs, AP-1, and ATF2 (8, 33, 35, 45, 61, 75, 101). NF-κB plays a key role in most LMP1-stimulated gene expression (25, 52, 88, 95, 121). Both LMP1 and activated CD40 receptor initiate overlapping signaling pathways that regulate major cell fate decisions including proliferation, differentiation, and apoptosis (1, 5, 46, 51, 72, 76, 114, 122). Despite many similarities, however, they also differ substantively (for a review, see reference 79). In this regard, it has been demonstrated that although LMP1 and CD40 can independently bind several of the same TRAF molecules (namely TRAF2, 3, and 5), TRAF6 binds to CD40 but not to LMP1, and TRAF1 and TRADD have been reported to bind to LMP1 but not to bind CD40 directly (10).
We now report for the first time that LMP1 stimulates bfl-1 promoter activity in EBV-negative BL-derived cell lines and that this process involves interactions with components of the TNFR/CD40 signaling pathway. We present evidence that this process is NF-κB dependent, involves the recruitment of TRAF2, and is mediated to a greater extent by CTAR2 than by CTAR1. Activation of CD40 receptor also led to increased bfl-1 mRNA levels and an NF-κB-dependent increase in bfl-1 promoter activity in the same cell context. We delineate a 95-bp region of the promoter that functions as an LMP1-dependent transcriptional enhancer, and we present evidence that this sequence contains a novel NF-κB-like binding site that is essential for transactivation of bfl-1 by LMP1, CD40, and the NF-κB subunit protein p65.
MATERIALS AND METHODS
Cell cultures.
DG75 and BL41 are EBV-negative BL cell lines (7, 104). BL41-B95-8 is a derivative of BL41 that was infected with the B95-8 strain of EBV (13). MUTU-I and Rael are group I/latency 1 cell lines that were established from EBV-positive BL biopsy specimens (48, 87). The DG75tTA-LMP1 transfectant of DG75 contains a tetracycline-regulated LMP1 expression plasmid and has been described previously (38). All of the cell lines were maintained as suspension cultures in RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (Gibco), 2 mM glutamine, 100 μg of streptomycin/ml, and 100 IU of penicillin/ml at 37°C in a humidified atmosphere containing 5% carbon dioxide. DG75-tTA-LMP1 cells were maintained under selection with 800 μg of hygromycin B (Roche Diagnostics)/ml and 2 mg of G418 (Roche Diagnostics)/ml as well as 1 μg of tetracycline (Sigma)/ml to repress LMP1 expression. For the induction of LMP1, cells were washed five times in phosphate-buffered saline and recultured in tetracycline-free medium. The establishment and culture of mouse fibroblastic L cell lines stably transfected with human CD32/Fc-gRII (CD32-L cells) or human CD40 ligand (CD40lig-L cells) have been described elsewhere (43, 98).
Plasmids.
The luciferase reporter constructs −1374/+81-Luc, −1374/+81(mκB-833), −1240/+81-Luc, −367/+81-Luc, and −129/+81-Luc were generated by subcloning bfl-1 promoter sequences from a corresponding series of chloramphenicol acetyltransferase reporter constructs (123). Thus, bfl-1 promoter sequences were amplified by PCR with a forward oligonucleotide primer (5′-TTGCATGCCTGCAGGTCGA-3′) from the multiple cloning site of the chloramphenicol acetyltransferase reporter vectors and a reverse primer (5′-gggaagcttCTAGAGCTGCCTGGTG-3′) from the bfl-1 promoter, which included a HindIII restriction site and digestion clamp at its 5′ end (shown in lowercase type). All PCR products therefore contained a common 3′ terminus at +81 bp relative to the transcription start site. The PCR products were then digested with SalI and HindIII and cloned between the XhoI and HindIII sites of the promoterless luciferase reporter construct pGL2Basic (Promega). The SalI site was present in the amplified portion of the polylinker at the 5′ end of the bfl-1 promoter sequence. To generate −57/+81-Luc, the forward primer 5′-ccgctcgagGAAGGATATTATATAAAG-3′ (which included an XhoI site and digestion clamp, shown in lowercase type) was used with the same reverse primer as above to amplify the bfl-1 promoter sequence between −57 bp and + 81 bp from −129/+81-Luc. The PCR product was digested with XhoI and HindIII and subcloned into pGL2Basic as described above.
To generate pGa(−129/+1), pGa(−129/−26), pGa(−129/−34), pGa(−129/−34mκB), pGa(−129/−54), and pGa(−34/−129mκB), portions of the bfl-1 promoter were amplified by PCR with forward and reverse primers that included BamHI sites at their 5′ ends (shown in lowercase type). The PCR primers used were as follows: −129forward, 5′-cgcggatccAAACTTTCTCTTTCATAC-3′; +1reverse, 5′-cgcggatccGAGCTTGACTGAGTTATG-3′; −26reverse, 5′-cgcggatccTGATACATGGAGGCTGGT-3′; −34reverse, 5′-cgcggatccGAGGCTGGTGGAATTTCTGTTTG-3′; −34reverse (NF-κB mutated), 5′-cgcggatccGAGGCTGGTGAGACTTCTGTTTG-3′; −54reverse, 5′-cgcggatccTTTGCATCACTTTATAAT-3′. In the case of −34reverse (NFκB mutated), the base changes made to the NF-κB-like site on the sequence subcloned into pGa(−129/−34mκB) are underlined in the primer sequence. All PCR products were digested with BamHI and subcloned into the BamHI site in the polylinker of pGa50-7 (81). Site-directed mutagenesis was performed with the QuikChange XL site-directed mutagenesis kit (Stratagene). The following pairs of complementary oligonucleotides were used to generate mutations (the introduced sequence changes are underlined): NF-κB-like site at −52, 5′-GTGATGCAAACAGAAGTCTCACCAGCCTCCATGTATCATC-3′ and 5′-GATGATACATGGAGGCTGGTGAGACTTCTGTTTGCATCAC-3′; AP1-like site at −104/−94, 5′-CTTTCTCTTTCATACATATGGTATAACACAGCCTACGCACG-3′ and 5′-CGTGCGTAGGCTGTGTTATACCATATGTATGAAAGAGAAAG-3′; AP1-like site at −74/−64, 5′-GCCTACGCACGAAAGAATCTAGGAGGAAGGATATTATAAAGTG-3′ and 5′-CACTTTATAATATCCTTCCTCCTAGATTCTTTCGTGCGTAGGC-3′. The 3Enh-Luc reporter construct contains three κB elements upstream of a minimal conalbumin promoter linked to the firefly luciferase gene (2). The following plasmids that are used have been published elsewhere: pEFCX, pEFCX-LMP1, and pEFCX-IκBαDN (83); pSFFVA20 and pcDNA3-TRAF2Δ(6-86) (32, 34); pSG5-LMP1 (61); pSG5-LMPAAA (35); pSG5-LMPG (40); pSG5-LMPAAAG (8); pcDNAp65, pcDNAp50, and pcDNAc-Rel (123).
Transfections and reporter assays.
Transfections were performed by using the DEAE-dextran method. Briefly, 107 cells were washed once with phosphate-buffered saline and then resuspended in 1.2 ml of transfection solution (0.5 mg of DEAE-dextran [Sigma]/ml) and up to 16.0 μg of DNA in TBS (25 mM Tris-HCl [pH 7.4], 137 mM NaCl, 5 mM KCl, 0.7 mM CaCl2, 0.5 mM MgCl2, 0.6 mM Na2HPO4). In any transfection experiment, the total amount of DNA was kept constant with any deficiencies made up with the corresponding empty vector. The transfection reaction mixtures were incubated for 30 min at room temperature. Thereafter, the cells were washed twice with 10 ml of RPMI 1640-10% fetal bovine serum, resuspended in 10 ml of fresh growth medium, and cultured for 24 or 48 h. In some cases, cells were stimulated for the final 12 h with anti-CD40 antibodies (G28.5) before preparation of cellular extracts. Luciferase activity was determined from cell extracts by means of a luciferase assay system (Promega Corp.) and a luminometer (Berthold Analytical Instruments) essentially according to the manufacturer's specifications. Luciferase levels were normalized after determining β-galactosidase activity expressed from pCMVlacZ (1 μg), which was included in all transfections. β-Galactosidase assays were performed as previously described (107) by using the same extracts that were used for measuring luciferase activity.
Northern blot analysis.
Total cellular RNA was prepared by using RNA isolator solution (Genosys) essentially according to the manufacturer's specifications. Thirty-microgram samples of total RNA were size fractionated in 1.3% formaldehyde-agarose gels and then transferred to nitrocellulose filters (BDH). Antisense 32P-labeled riboprobes were synthesized by in vitro transcription (Riboquant Multiprobe RNase protection assay system, multiprobe hAPO-2 template set; Pharmingen) as described by the manufacturer. Labeled transcripts were size fractionated in denaturing polyacrylamide gels, and probes were located by autoradiography, excised, and then eluted in buffer containing 0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA (pH 8.0), and 0.1% sodium dodecyl sulfate (SDS) by the crush and soak method (107) followed by ethanol precipitation. Probes (106 cpm/ml) were hybridized to filters in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50% formamide, 1% SDS, 0.1% Tween 20, and 100 μg of Escherichia coli tRNA/ml for 16 to 24 h at 55°C. Filters were washed twice in 1× SSC-0.1% SDS at room temperature for 30 min and then twice in 0.1× SSC-0.1% SDS at 65°C for 30 min prior to exposure to X-ray film at −70°C.
Western Blot analysis.
To detect LMP1, protein lysates were prepared by boiling for 10 min in 2% SDS, 100 mM NaCl, 0.01 M Tris-HCl, 5% β-mercaptoethanol, 1 mM EDTA, 100 μg of phenylmethylsulfonyl fluoride/ml, and 2 μg of leupeptin/ml and briefly sonicated on ice. The lysates were then clarified by centrifugation at 13,000 rpm for 10 min at room temperature. Protein from 5 × 105 cells was separated by discontinuous SDS-5 to 10% polyacrylamide gel electrophoresis and blotted onto a nitrocellulose filter. Filters were probed with the anti-LMP1 CS 1-4 antibody cocktail (105) diluted to 1:100 in Blotto (5% skim milk and 0.1% Tween 20 in Tris-buffered saline) overnight at 4°C. Immunocomplexes were detected with alkaline phosphatase-conjugated sheep anti-mouse immunoglobulin G (IgG) (Promega) and visualized with 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium liquid substrate (Sigma).
RESULTS
LMP1 transactivates the bfl-1 promoter in EBV-negative BL-derived cell lines by a mechanism that is dependent upon the transcription factor NF-κB.
To determine whether LMP1 could transactivate the bfl-1 promoter, a suitable promoter-reporter construct was first generated. To this end, a 1.4-kb DNA sequence from the 5′ regulatory region of the bfl-1 gene (from −1374 to + 81 relative to the transcription initiation site) (123) was positioned upstream of the luciferase gene in the promoterless vector pGL2Basic (Promega) to generate the reporter construct −1374/+81-Luc. Initially, expression vectors derived from the plasmid pEFCX were used for the expression of LMP1 and other effector proteins in transient transfections. Recombinant genes expressed from pEFCX are driven by the promoter for the polypeptide chain elongation factor 1α in an NF-κB-independent manner (83). LMP1 levels expressed from pEFCX-LMP1 would not therefore be expected to fluctuate in the presence of molecules that regulate activation of this transcription factor. Transient transfections of the EBV-negative BL-derived cell lines DG75 and BL41 showed that the use of increasing amounts of cotransfected pEFCX-LMP1 led to a dose-dependent increase in luciferase activity at 24 h posttransfection when using −1374/+81-Luc as the reporter vector. Similar results were obtained with both cell lines used in this and subsequent experiments, and only the results for DG75 are presented (Fig. 1A). A maximal increase of 8.5-fold in luciferase values was detected with 2.5 μg of pEFCX-LMP1 (7.8-fold in BL41). The level of expression of LMP1 in transfected cell extracts was assessed by Western blotting, and as expected, this was seen to rise as the quantity of pEFCX-LMP1 was increased with no LMP1 being detected when pEFCX was used as the cotransfected plasmid (Fig. 1A). The effect of LMP1 was mediated by the bfl-1 promoter sequence, since LMP1 expression did not affect the corresponding promoterless vector pGL2Basic (not shown). These results indicate that LMP1 upregulates bfl-1 promoter activity in these EBV-negative BL-derived cell lines.
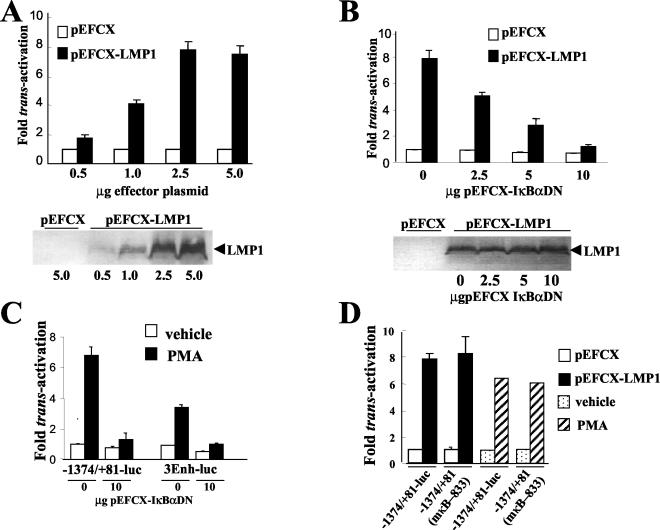
FIG.1.
LMP1 and PMA transactivate the bfl-1 promoter in DG75 cells by mechanism(s) that are dependent upon the transcription factor NFκB; transactivation does not require a previously identified NF-κB-binding site at position −833/−823 relative to the transcription start site. (A) Dose-dependent transactivation of the bfl-1 promoter by LMP1. DG75 cells were cotransfected with increasing amounts of either pEFCX or pEFCX-LMP1 (effector plasmids) and 2.5 μg of −1374/+81-Luc. Cells were harvested at 24 h posttransfection and analyzed for luciferase activities, which were then normalized for transfection efficiency (based on β-galactosidase activity measured from cotransfected pCMVlacZ reporter, which was included in all transfections). Luciferase values obtained by cotransfection with pEFCX were arbitrarily assigned a value of 1.0 and activation represents the relative normalized luciferase activities obtained upon cotransfection with the effector plasmids with the quantities indicated. Selected extracts from these transfections were analyzed for LMP1 expression by Western blotting (shown underneath). (B) Inhibition of LMP1-mediated activation of the bfl-1 promoter by overexpression of a superrepressor mutant form of IκBα (IκBαDN). DG75 cells were cotransfected with 2.5 μg of pEFCX-LMP1 or empty vector pEFCX and 2.5 μg of −1374/+81-Luc together with various amounts (0, 2.5, 5.0, or 10.0 μg) of pEFCX-IκBαDN. Cells were harvested at 24 h posttransfection, and luciferase values were measured, normalized as above, and presented as activation (n-fold) over control (empty vector in the absence of expression of IκBαDN equals 1). Selected extracts from these transfections were analyzed for LMP1 expression by Western blotting (shown underneath). (C) PMA activates the bfl-1 promoter in an NF-κB-dependent manner. DG75 cells were transfected with either 2.5 μg of −1374/+81-Luc or 3Enh-Luc in the presence or absence of 10.0 μg of pEFCX-IκBαDN and then treated with either 10−7 M PMA or vehicle control (0.062% ethanol) at 18 h posttransfection. Cells were harvested at 24 h posttreatment, and the normalized luciferase values obtained are presented as increases (n-fold) over control values. The values obtained with transfected cells that had been treated with vehicle control were arbitrarily assigned a value of 1. (D) Neither LMP1- nor PMA-mediated activation of the bfl-1 promoter requires the known NF-κB-binding site at position −833/−823 in the bfl-1 promoter. DG75 cells were cotransfected with 2.5 μg of pEFCX-LMP1 or empty vector pEFCX and 2.5 μg of −1374/+81-Luc or its NF-κB-mutated derivative −1374/+81(mκB-833)-Luc. In the PMA study, DG75 cells were transfected with 2.5 μg of −1374/+81-Luc or 1374/+81(mκB-833) and then treated with either 10−7 M PMA or vehicle control (0.062% ethanol) at 18 h posttransfection. Cells were harvested at 24 h either posttransfection or posttreatment (PMA or vehicle, respectively) and assayed for luciferase activity. Normalized luciferase values were expressed as activation (n-fold) over control empty vector or vehicle as appropriate.
Previous experiments in which the human bfl-1 promoter was shown to be responsive to TNF-α via an NF-κB-dependent pathway (123) suggested that transactivation by LMP1 might also be mediated by this transcription factor. To directly test this hypothesis, DG75 cells were cotransfected with −1374/+81-Luc and pEFCX-LMP1 together with pEFCX-IκBαDN, which expresses a superrepressor mutant form of IκBα in which the serine residues at positions 32 and 36 on that protein have been replaced with alanines. IκBαDN can no longer be phosphorylated and is consequently not proteolyzed upon treatment of cells with an NF-κB-inducing agent (9). This mutant efficiently retains NF-κB in the cytoplasm, thus blocking its function as a regulator of transcription by preventing it from translocating to the nucleus (83). It can be seen from this experiment that expression of IκBαDN efficiently inhibited LMP1-induced bfl-1 promoter transactivation in DG75 cells in a dose-dependent manner (Fig. 1B). The inhibitory action of IκBαDN was not the result of inhibition of LMP1 expression from pEFCX-LMP1, as can be seen from Western blot analysis of LMP1 levels in protein extracts from the transfected cells (Fig. 1B). Similar observations were made in BL41 cells; however, a lower amount (5 μg) of pEFCXIκBαDN was required to achieve complete inhibition of transactivation by LMP1 (data not shown). These results strongly suggest that activation of the bfl-1 promoter by LMP1 is NF-κB dependent. In DG75, overexpression of IκBαDN also inhibited the basal level of bfl-1 promoter activity (30% decrease in the presence of 10.0 μg of pEFCX-IκBαDN), suggesting that the latter may be NF-κB dependent, at least in part, in this cell line. In control transfection experiments, NF-κB activation was monitored with a known NF-κB-dependent reporter construct (3Enh-Luc). In these cases, using DG75 and BL41 cells, luciferase values increased by factors of 4 and 8, respectively, when this vector was cotransfected with pEFCX-LMP1, and an efficient dose-dependent inhibition of activation was observed in both cell lines in the presence of pEFCX-IκBαDN (data not shown).
Treatment of a variety of cell lines with the chemical agent phorbol-12-myristate 13-acetate (PMA), a well-known activator of NF-κB, has been shown to upregulate bfl-1 mRNA levels in BL cell lines (93). We therefore investigated whether PMA, when used at the same concentration (10−7 M) as that reported previously, could also activate the bfl-1 promoter. DG75 cells that had been transfected with either 3Enh-Luc or −1374/+81-Luc were treated with PMA, and luciferase activities were then determined from transfected cell extracts as before. In this experiment, PMA treatment was seen to activate NF-κB by approximately 3.5-fold (Fig. 1C). In the case of the bfl-1 promoter, PMA treatment resulted in a 6.8-fold increase in bfl-1 promoter activity (Fig. 1C). In both cases, promoter activation was demonstrated to be NF-κB dependent, since cotransfection with pEFCX-IκBαDN led to a significant decrease in luciferase values (Fig. 1C). An NF-κB binding site in the bfl-1 promoter (5′-GGGGATTTACC-3′ at positions −833 to −823 relative to the transcription start site) has previously been shown by site-directed mutagenesis to be essential for activation by c-Rel in the HeLa cell line (123). To investigate whether this NF-κB-binding motif played a role in LMP1-associated transactivation, a second bfl-1 reporter construct was generated which differed from −1374/+81-Luc in that it carried the same inactivating mutation in this particular motif [1374/+81(mκB-833)]. In transient-transfection assays, however, it can be seen that LMP1 activated the promoter fragments on both of these constructs to a similar extent in DG75 cells and that elimination of the NF-κB-binding motif at −833/−823 did not significantly affect the basal level of promoter activity (Fig. 1D), and the same result was achieved with BL41 (data not shown). In addition, PMA treatment also activated the mutated promoter to the same extent as its unmutated counterpart in both cell lines (Fig. 1D). The ensemble of these results clearly indicated that the bfl-1 promoter is transactivated by LMP1 in the EBV-negative BL cell lines used, that transactivation is NF-κB dependent, and that the NF-κB binding motif at position −833/−823 is not essential for activation of the promoter by LMP1 or PMA in this cell context.
LMP1-associated bfl-1 promoter activation is blocked by overexpression of A20 and occurs by a mechanism involving TRAF2.
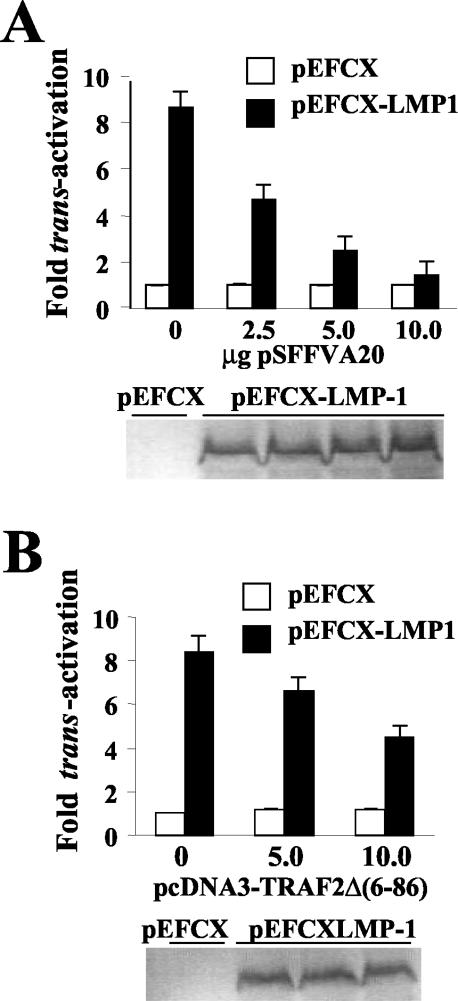
A20 is an antiapoptotic RING finger protein that interacts with TRAFs and efficiently inhibits NF-κB and JNK activation by LMP1 by displacing TRAF and TRADD molecules from LMP1-TRAF and LMP1-TRADD complexes (32, 34, 42). To investigate whether overexpression of A20 could inhibit transactivation of the bfl-1 promoter by LMP1, cotransfections were performed as before with the inclusion of various amounts of an A20 expression vector (pSFFVA20). It can be seen that A20 expression potently inhibited bfl-1 promoter activation by LMP1 in DG75 cells, with transfection of 5.0 and 10.0 μg of pSFFVA20 resulting in approximately 80 and 94% inhibition, respectively (Fig. 2A). A similar degree of inhibition of NF-κB activation was observed when using the 3Enh-Luc reporter (data not shown). A20 expression did not alter the basal levels of bfl-1 promoter activity or endogenously activated NF-κB, suggesting that A20 only affected LMP1-induced signaling. In addition, A20 expression did not interfere with LMP1 levels produced from cotransfected vector as assessed by Western blotting of lysates prepared from the transfected cell populations (Fig. 2A).
FIG. 2.
LMP1-associated bfl-1 promoter activation is blocked by overexpression of A20 and occurs by a mechanism involving TRAF2. DG75 cells were cotransfected with 2.5 μg of pEFCX-LMP1 or pEFCX and 2.5 μg of −1374/+81-Luc together with various amounts of plasmids expressing either A20 (0, 2.5, 5.0, or 10.0 μg of pSFFVA20) (A) or a dominant-negative mutant of TRAF2 [0, 5.0, or 10.0 μg of pcDNA3-TRAF2Δ(6-86)] (B). In each case, cells were harvested at 24 h posttransfection and analyzed for luciferase activity and LMP1 expression (shown below each graph), as described before. Normalized luciferase values were expressed as activation (n-fold) over the corresponding control empty vector.
TRAF2 has been shown to bind to the cytoplasmic tail of LMP1 and is an important mediator of NF-κB activation by LMP1. TRAF2 binds directly to the CTAR1 domain of LMP1 and binds to CTAR2 via the adaptor protein TRADD (58, 71, 103). The carboxyl-terminal domain of TRAF2 mediates TRAF2-binding to the LMP1 CTAR1 domain and TRADD, whereas the amino-terminal RING finger-containing domain is involved in homo- and heterodimerization of TRAF family members and binding to cellular inhibitors of apoptosis (c-IAPs) and is essential for TRAF2-mediated activation of NF-κB and JNK (58, 71, 103). A dominant-negative mutant of TRAF2 [TRAF2Δ(6-86)], which has a deletion of amino acids 6 to 86 in the N-terminal region, has been shown to inhibit activation of NF-κB and JNK by LMP1 (32). To determine whether TRAF2 had a direct role in mediating LMP1-associated bfl-1 promoter activation, we used a vector that expresses TRAF2Δ(6-86) in transient cotransfection assays. Coexpression of TRAF2Δ(6-86) was found to significantly reduce bfl-1 promoter activation by LMP1 in a dose-dependent manner in that 5.0 and 10 μg of pcDNA3-TRAF2Δ(6-86) decreased LMP1-associated transactivation by 25 and 48%, respectively (Fig. 2B). Western blotting again showed that the inhibitory effect of TRAF2Δ(6-86) was not due to inhibition of LMP1 expression (Fig. 2B). The effect of TRAF2Δ(6-86) expression on LMP1-mediated activation of NF-κB was also investigated with the 3Enh-Luc construct. In this experiment, cotransfection of 5 and 10 μg of pcDNA3-TRAF2Δ(6-86) resulted in decreases of 40 and 65%, respectively, in LMP1-mediated activation of NF-κB (data not shown). Similar results were obtained with regard to both LMP1 activation of the bfl-1 promoter and activation of NF-κB with A20 and TRAF2Δ(6-86) in transfections of the BL41 cell line (data not shown). Taken together, these results implicate the involvement of TRAF2 in LMP1-mediated transactivation of the bfl-1 promoter in these cells.
Identification of LMP1 signaling domains required for induction of bfl-1 promoter activity.
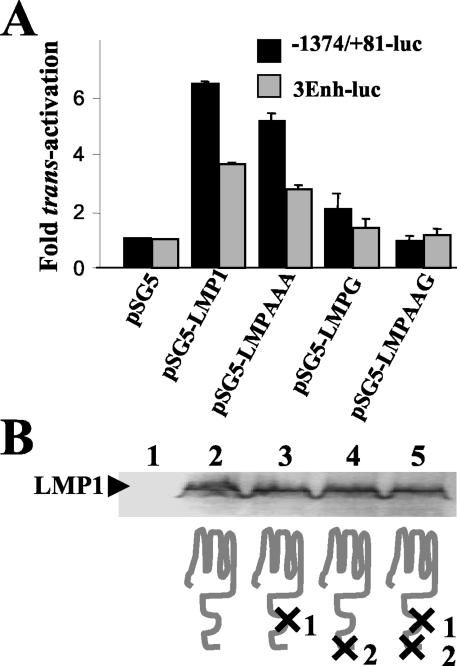
To identify the signaling domain(s) of LMP1 that are important for mediating bfl-1 promoter transactivation, plasmids expressing LMP1 molecules with amino acid substitutions at critical residues in CTAR1 and CTAR2 were used in comparative analyses with wild-type LMP1 in promoter-luciferase assays. Thus, LMPAAA contains a triple amino acid substitution in the CTAR1-TRAF interaction domain (P204xQ206xT208 → AxAxA) which has been shown to block NF-κB activation by this domain (26, 32, 34); LMPG contains a single point mutation in CTAR2 such that the tyrosine residue at position 384 has been replaced with a glycine, a modification that potently inhibits activation of NF-κB and AP-1 by inhibiting TRADD binding (34, 40); LMPAAAG contains the modifications from both LMPAAA and LMPG and is not able to activate NF-κB or AP-1 (8, 39). Both transactivation of the bfl-1 promoter and NF-κB activation by wild-type and signaling-defective LMP1 proteins were investigated by cotransfection of the corresponding pSG5-based effector plasmids with the −1374/+81-Luc and 3Enh-Luc constructs, respectively. It can be seen that expression of wild-type LMP1 activated the bfl-1 and control NF-κB-responsive promoters by approximately 6.4- and 3.6-fold, respectively, in DG75 cells (Fig. 3A). In comparison, activation of both promoters by LMPAAA and LMPG achieved levels of approximately 75 and 20%, respectively, when compared to values obtained with wild-type LMP1. LMPAAAG was effectively unable to activate either promoter (Fig. 3A). The observed effects were not due to differences in the levels of wild-type and mutant LMP1 proteins expressed during transfection (Fig. 3B). These results show that the CTAR1 and CTAR2 domains of LMP1 can independently contribute to bfl-1 promoter activation, with CTAR2 being the predominant contributor in this regard. Furthermore, both domains are required for maximal activation of the bfl-1 promoter and activation of NF-κB. The relative contribution of the individual domains of LMP1 to inducing bfl-1 promoter activity can be seen to correlate well with NF-κB activation, suggesting again that the latter is a key event in mediating the effect of LMP1 on bfl-1.
FIG. 3.
Comparative analysis of the effects of wild-type and functionally mutated LMP1 molecules on bfl-1 promoter activity. (A) DG75 cells were cotransfected with 2.5 μg of either pSG5, pSG5-LMP1 (expressing wild-type LMP1), pSG5-LMPAAA (expressing LMP1 containing a nonfunctional CTAR1 domain), pSG5-LMPG (expressing LMP1containing a nonfunctional CTAR2 domain), or pSG5-LMPAAAG (expressing LMP1 mutant that is nonfunctional in both CTAR1 and CTAR2) and 2.5 μg of either −1374/+81-Luc or 3Enh-Luc. Cells were harvested at 24 h posttransfection and analyzed for luciferase activity. Normalized luciferase values were expressed as activation (n-fold) over control (empty vector), as described before. (B) Western blot analysis of the level of expression of wild-type and mutated LMP1 molecules in transient-transfection assays in DG75 cells; protein lysates were prepared from the transfection shown in panel A and loaded (lanes 1 to 5) in the same order as the corresponding expression vectors shown in that figure (only cell extracts from the cotransfections with −1374/+81-Luc are shown). A schematic representation of the LMP1 protein and signaling-deficient derivatives thereof is shown underneath. Mutations are indicated as X in the cytoplasmic signaling domains CTAR1/TES-1 and CTAR2/TES-2, which are labeled as 1 and 2, respectively.
Activation of CD40 receptor leads to increased bfl-1 mRNA levels and an NF-κB-dependent increase in bfl-1 promoter activity in BL-derived cell lines.
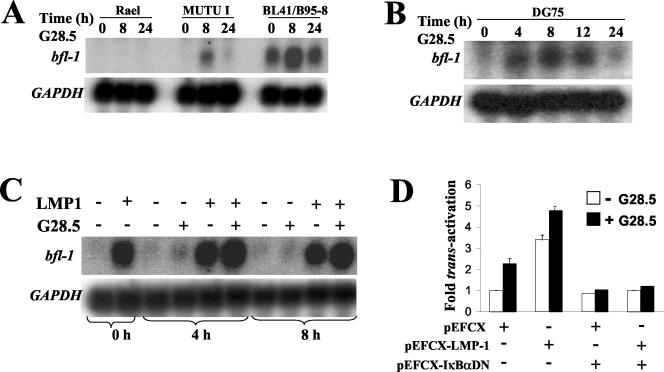
CD40 engagement on BL-derived cell lines has previously been shown to lead to increased bfl-1 mRNA levels (82), but the mechanism involved has not yet been investigated. Here, Northern blot analysis revealed that treatment of the Mutu I (EBV-positive latency type 1) and BL41-B95.8 (EBV-positive latency type 3) cell lines with the agonistic anti-CD40 antibody G28.5 (20) resulted in increased bfl-1 mRNA levels (Fig. 4A). Both cell lines express CD40 (data not shown), and in both cases, the effect was transient with significant upregulation detectable by 8 h followed by a return to the level seen in untreated cells by 24 h. The transient nature of this effect is consistent with observations made elsewhere (82). In contrast, treatment of the BL cell line Rael with G28.5 did not lead to detectable levels of bfl-1 mRNA (Fig. 4A). Although Rael expresses CD40, it has been shown by others to be unresponsive to several CD40-mediated effects on gene expression (54). A more detailed analysis of the kinetics of upregulation of bfl-1 mRNA by CD40 ligation was performed with DG75 cells (Fig. 4B). In this experiment, it can be seen that an increase in the level of bfl-1 mRNA was detected as early as 4 h posttreatment with anti-CD40 antibody. This level was maintained for at least 12 h and then decreased to near-basal levels by 24 h.
FIG. 4.
Activation of CD40 receptor leads to increased bfl-1 mRNA levels and an NF-κB-dependent increase in bfl-1 promoter activity in BL-derived cell lines. Rael, Mutu I, BL41-B95.8 (A), and DG75 (B) cells were left untreated or were treated with the CD40 agonistic antibody G28.5 either as a 1:4,000 dilution of ascites (Rael, Mutu I, and BL41-B95.8) or at a concentration of 1 μg of purified antibody (DG75)/ml. Total RNA samples were prepared from cells at 0, 8, and 24 h (Rael, Mutu I, and BL41-B95.8) or at 0, 4, 8, 12, and 24 h (DG75) posttreatment and subjected to Northern blot analysis with an antisense bfl-1 riboprobe (upper panels of panels A and B). The blots were then sequentially stripped and rehybridized with antisense riboprobe to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (lower panels of both panels A and B). (C) LMP1 and CD40 cooperate in upregulating bfl-1 mRNA in DG75tTA-LMP1 cells. Uninduced DG75tTA-LMP1 cells or DG75tTA-LMP1 cells induced to express LMP1 (36 h after tetracycline removal) were either left untreated or stimulated with 1 μg of the anti-CD40 antibody G28.5/ml. Total RNA extracted from cells at 0, 4, and 8 h after stimulation with G28.5 was used in Northern blot analysis with an antisense bfl-1 riboprobe (upper panel). The blots were sequentially stripped and reprobed with a GAPDH riboprobe (lower panel). (D) Expression of a superrepressor IκBα mutant inhibits bfl-1 promoter upregulation after activation of CD40 in the presence (+) or absence (−) of LMP1. DG75 cells were cotransfected with 2.5 μg of pEFCX-LMP1 or pEFCX and 2.5 μg of −1374/+81-Luc in the absence or presence of 10.0 μg of pEFCX-IκBαDN. At 36 h posttransfection, the cells were either left untreated or were treated with 1 μg of G28.5/ml for 12 h prior to harvesting for measurement of luciferase activity. Normalized luciferase values are expressed as activation (n-fold) over that of the control (empty vector in the absence of expression of IκBαDN or absence of treatment with G28.5 equals 1).
To analyze the effect of dual signaling from LMP1 and CD40 on bfl-1 mRNA levels, we used a stable transfectant of DG75 in which LMP1 expression can be regulated by tetracycline (DG75tTA-LMP1) (38). Northern blotting was used to monitor bfl-1 mRNA levels in DG75tTA-LMP1 cells before induction and at 36 h postinduction of LMP1 expression in the presence or absence of G28.5 (Fig. 4C). As expected, induction of LMP1 resulted in a marked increase in the level of bfl-1 mRNA. A small but consistent further increase in bfl-1 mRNA levels was detectable at 4 and 8 h post-CD40 stimulation of LMP1-expressing cells. The minor effect of CD40 ligation on bfl-1 mRNA levels in the absence of LMP1 may be a reflection of the low levels of CD40 expression in these cells (even lower than in untransfected DG75 cells) which are significantly elevated upon induction of LMP1 (38; also data not shown). To determine whether ligation of CD40 was leading to increased bfl-1 promoter activity, DG75 cells that had been transfected with −1374/+81-Luc were treated at 36 h posttransfection for a further 12 h with G28.5, and luciferase values were compared to those obtained with untreated transfected cells. In this experiment it can be seen that (i) stimulation of CD40 led to a 2.2-fold increase in bfl-1 promoter activity, (ii) transactivation by LMP1 was 3.4-fold, (iii) activation of CD40 in the presence of LMP1 led to an overall 4.8-fold increase, and (iv) expression of IκBαDN was seen to inhibit CD40-mediated promoter activation, indicating that the pathway is NF-κB dependent (Fig. 4D). In this case, the extent of activation as a result of dual signaling from CD40 and LMP1 is greater than that provided by each of the individual signals but is less than that expected of an additive effect from both signals. These findings suggest that the signaling pathways initiated from CD40 and LMP1 may converge when it comes to driving the bfl-1 promoter. It is also possible, however, that part of the effect of CD40 activation is due to the increased surface expression of CD40 in LMP1-transfected cells. In addition, it cannot be excluded that the dual effect of LMP1 and CD40, which is also seen to be NF-κB dependent, may be the result of an indirect effect on the NF-κB-mediated process of upregulating the level of CD40 (25). The lower extent of promoter activation by LMP1 in this experiment is likely to be due to the differences in the time of harvesting of the transfected cells for reporter assays (106, 119).
A 210-bp bfl-1 promoter fragment (−129/+81) mediates NF-κB-dependent transactivation by LMP1.
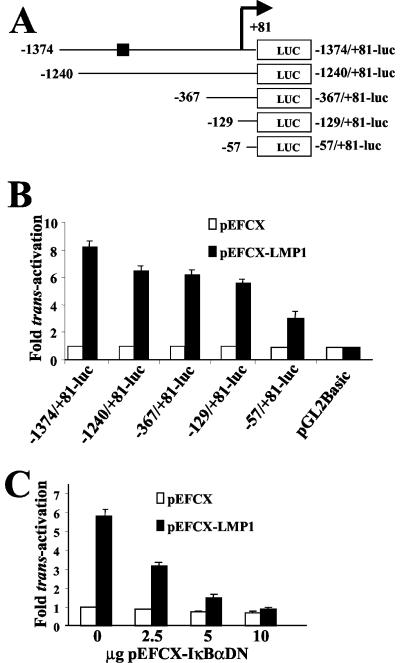
To identify DNA sequence elements that mediate transactivation by LMP1, a series of reporter constructs was generated in which progressive deletions were introduced from the 5′ end of the bfl-1 promoter sequence in −1374/+81-Luc (Fig. 5A). The ability of LMP1 to transactivate these truncated promoter fragments was investigated by cotransfections with pEFCX-LMP1, as before, with the DG75 and BL41 cell lines. It can be seen that in DG75 cells, LMP1 transactivated −1374/+81-Luc by a factor of 8.2 and that considerable transactivation was retained by the promoter fragments −1240/+81, −367/+81-Luc, and −129/+81-Luc (6.5-, 6.2-, and 5.6-fold, respectively), with a further decrease to 3.3-fold observed when only the −57/+81 promoter sequence was retained (Fig. 5B). The fact that similar transactivation values were observed with −1240/+81-Luc and −367/+81-Luc again indicated that the previously identified NF-κB binding motif at position −833/−823 does not play a significant role in LMP1-associated activation of the promoter in this cell context. The observation that −57/+81-Luc still retained a considerable degree of responsiveness to LMP1 (but significantly less than −129/+81-Luc) indicated that the −129 to −57 region of the promoter might contain DNA sequence elements that contributed to but were not essential for activation by LMP1. It can also be seen that coexpression of IκBαDN efficiently inhibited LMP1-induced transactivation of −129/+81-Luc in a dose-dependent manner (Fig. 5C) (with the levels of LMP1 expressed from pEFCX-LMP1 remaining constant in each transfection, as before) (data not shown). A significant NF-κB-dependent response was also seen with PMA with the −129/+81-Luc construct (68% of the activity observed with −1374/+81-Luc), and in all cases, similar results were observed when BL41 cells were used (data not shown). It can thus be concluded that the −129/+81 sequence from the bfl-1 transcriptional regulatory region contains DNA sequence elements that mediate a high proportion of the NF-κB-dependent activation of this gene by LMP1 in these BL-derived cell lines.
FIG. 5.
A 210-bp fragment (−129/+81) of the bfl-1 promoter is transactivated by LMP1 in an NF-κB-dependent manner. (A) Schematic representation of bfl-1 promoter-reporter constructs in which the promoter sequence has been progressively deleted from the 5′ end (coordinates of the 5′ ends are given to the left of each construct). They all share a common 3′ terminus 81 bp downstream from the transcription initiation site (designated by a bent arrow), at which point they are joined to the luciferase gene (LUC). The position of a previously identified NF-κB site at position −833 is indicated on the plasmid −1374/+81-Luc as a black box. (B) DG75 cells were transfected with 2.5 μg of either pEFCX-LMP1 or pEFCX together with 2.5 μg of the individual bfl promoter-luciferase reporter constructs shown in panel A. Cells were harvested 24 h posttransfection and assayed for luciferase activity as described before. Normalized luciferase values were expressed as activation (n-fold) relative to the corresponding value obtained for each reporter construct when cotransfected with control pEFCX. (C) LMP1-mediated activation of the −129/+81 region of the bfl-1 promoter is inhibited by expression of a superrepressor IκBα mutant (IκBαDN) in DG75 cells. DG75 cells were cotransfected with 2.5 μg of pEFCX-LMP1 or pEFCX and 2.5 μg of −129/+81-Luc together with the indicated amounts of pEFCX-IκBαDN. Cells were harvested at 24 h posttransfection and analyzed for luciferase activity. Normalized luciferase values are given as activation (n-fold) over those of the controls (empty vector in the absence of expression of IκBαDN equals 1).
Identification of NF-κB-like and AP-1-like binding sites on the bfl-1 promoter that mediate transactivation by LMP1.
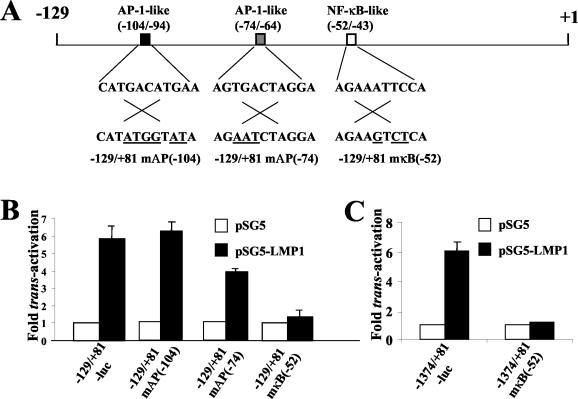
Using Transcription Element Search Software, we analyzed the −129/+81 region of the bfl-1 promoter for candidate sequence elements that may bind transcription factors known to play a role in LMP1-mediated regulation of gene expression. One NF-κB-like binding site (5′-AGAAATTCCA-3′ at −52 to −43) and two AP-1-like binding sites (5′-CATGACATGAA-3′ at −104 to −94 and 5′-AGTGACTAGGA-3′ at −74 to −64) were identified in the region 5′ to the transcription initiation site (Fig. 6A). Base substitutions were then introduced into the core of each of these motifs in the reporter plasmid −129/+81-Luc by site-directed mutagenesis to eliminate potential binding by the corresponding transcription factor (Fig. 6A). Transient transfections of these constructs with pSG5 or pSG5-LMP1 were then carried out with DG75 cells. It can be seen that mutation of the NF-κB-like binding site led to the nearly complete loss of transactivation by LMP1 (Fig. 6B), an effect that was reproduced when the same mutation was introduced into the promoter sequence in −1374/+81-Luc (Fig. 6C). Mutation of the AP-1-like binding site at −104/−94 did not impair transactivation (actually seen to marginally increase), whereas elimination of the site at −74/−64 resulted in a significant decrease (reduced by 34%) in promoter activity in the presence of LMP1 (Fig. 6B). The latter result correlates with the observation of a similar reduction in LMP1-associated transactivation upon deletion of the −129 to −57 region (Fig. 5B, compare −57/+81-Luc and −129/+81-Luc).
FIG. 6.
Identification of NF-κB-like and AP1-like binding sites on the bfl-1 promoter that mediate transactivation by LMP1. (A) Schematic representation of −129 to + 1 region of the bfl-1 promoter showing the relative locations and sequences of candidate binding sites for relevant transcription factors. Three further reporter constructs were made by introducing base changes (underlined) to each of the sequences shown by site-directed mutagenesis, thus eliminating each candidate site in turn. The name of the corresponding vector which carries the individual mutation is given underneath. In panels B and C, DG75 cells were transfected with 1 μg of either pSG-LMP1 or pSG5 together with 1 μg of the individual bfl promoter-luciferase reporter constructs shown underneath each graph. An identical sequence change to that indicated in panel A was made to the NF-κB-like motif (−52/−43) in the reporter construct with the longest bfl-1 promoter sequence available, thus generating −1374/+81mκB(−52). Cells were harvested 24 h posttransfection and assayed for luciferase activity as described before. Normalized luciferase values were expressed as activation (n-fold) relative to the corresponding value obtained for each reporter construct when cotransfected with control pEFCX.
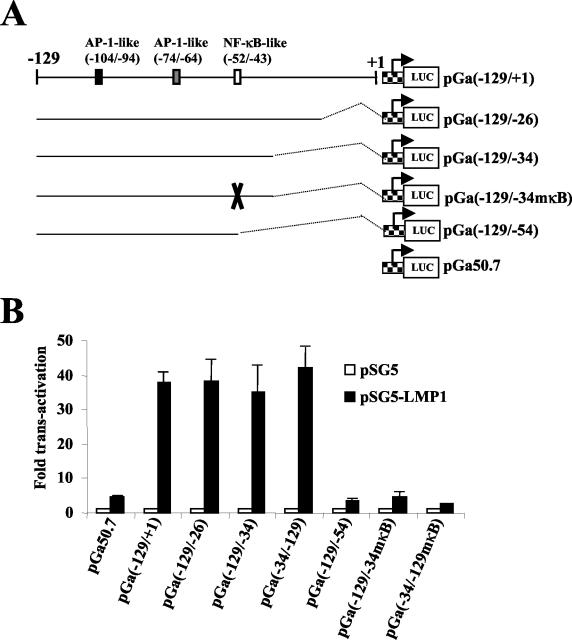
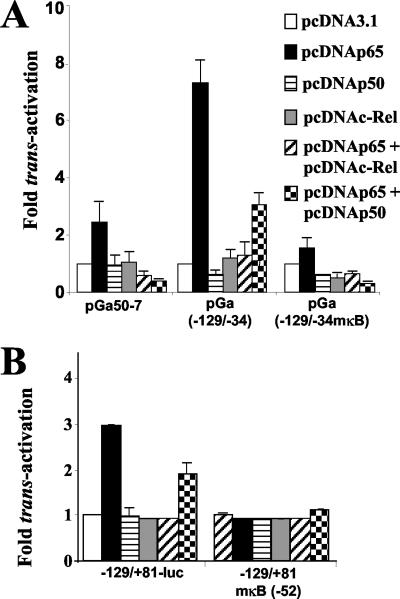
To determine whether sequences from this region of the bfl-1 promoter could confer LMP1 responsiveness on a heterologous promoter, portions of the −129/+1 sequence were subcloned upstream of the minimal β-globin promoter in the luciferase reporter construct pGa50.7 (Fig. 7A) (81). Transient cotransfections of these reporter plasmids with pSG5 or pSG5-LMP1 were then carried out with DG75 cells, and the results of this experiment are shown in Fig. 7B. It can be seen that a fivefold increase in luciferase values was consistently obtained when the basal vector (pGa50.7) was cotransfected with pSG5-LMP1 but that this increased by a further factor of 8 when the −129/+1 sequence was linked to the minimal promoter [Fig. 7B, compare pGa50-7 and pGa(−129/+1)]. Deletion of up to 35 bp from the 3′ end of the bfl-1 sequence, which still retained the NF-κB-like and AP-1-like binding sites, did not lead to any decrease in LMP1-associated transactivation [Fig. 7B, pGa(−129/−26) and pGa(−129/−34)]. It can also be seen that a construct in which the −129/−34 sequence had been inserted in the opposite orientation [pGa(−34/−129)] was transactivated by LMP1 at least as efficiently as pGa(−129/−34). However, deletion or mutation of the NF-κB-like binding site at −52/−43 led to the complete loss of LMP1-associated transactivation [pGa(−129/−54), pGa(−129/−34mκB), and pGa(−34/−129mκB)]. We conclude that the −129/−34 region of the bfl-1 promoter functions as an LMP1-dependent transcriptional enhancer and that the NF-κB-like binding site at position −52 to −43 is essential for this effect.
FIG. 7.
The NF-κB-like binding site at −52/−43 is essential for conferring LMP1-responsiveness on a heterologous minimal promoter. (A) Schematic representation of luc reporter constructs generated after subcloning portions of the −129/+1 region of the bfl-1 promoter upstream of the β-globin minimal promoter (checkered box with bent arrow) in pGa50.7. The name of each reporter construct includes the relevant 5′ and 3′ ends of the bfl-1 sequence. The mutation introduced into the NF-κB-like site is indicated with an X. (B) DG75 cells were transfected with 1 μg of either pSG-LMP1 or pSG5 together with 1 μg of the luciferase reporter constructs as indicated underneath each graph. Cells were harvested at 24 h posttransfection and assayed for luciferase activity as described before. Normalized luciferase values were expressed as activation (n-fold) relative to the corresponding value obtained for each reporter construct when cotransfected with control pSG5.
Mutation of the NF-κB-like binding site at −52/−43 reduces bfl-1 promoter transcriptional activation upon engagement of CD40 receptor.
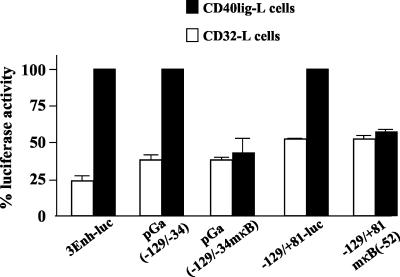
We then investigated whether the NF-κB-like binding site at −52/−43 in the bfl-1 promoter played a role in mediating transactivation by CD40. In these experiments, transiently transfected DG75 cells were cocultivated with either a mouse fibroblast L cell line that stably expressed recombinant human CD40 ligand (CD40lig-L cells), to activate CD40 signaling (43), or a human CD32/Fc-gRII (CD32-L cells)-expressing cell line, as a control (98). It can be seen that engagement of the CD40 receptor on transfected DG75 cells led to increases in luciferase values of approximately 3.4-, 2.7-, and 2.0-fold, respectively, when the 3Enh-Luc, pGa(−129/−34), and −129/+81-Luc vectors were used as reporters (Fig. 8). Mutation of the NF-κB-like binding site in both pGa(−129/−34) and −129/+81-Luc led to decreases of 66% ± 18% and 46% ± 5%, respectively, in the levels of promoter activity that were recorded upon engagement of CD40 receptor with CD40 ligand (Fig. 8). This experiment showed that the NF-κB-like binding site at −52/−43 plays a key role in CD40-mediated activation of the bfl-1 promoter. It also served to corroborate the finding presented in Fig. 4D, in which it is shown that bfl-1 promoter upregulation was triggered by an agonistic anti-CD40 antibody.
FIG. 8.
Mutation of the NF-κB-like binding site at −52/−43 reduces bfl-1 promoter transcriptional activation upon engagement of CD40 receptor. DG75 cells were transfected with 2 μg of reporter construct (indicated underneath the graph) and then cultured for 24 h on either CD40lig-L cells or CD32-L cells, which express human CD40 ligand and human CD32/Fc-gRII, respectively. DG75 cells were then harvested and analyzed for luciferase activities which were normalized for transfection efficiency (based on β-galactosidase activity measured from cotransfected pCMVlacZ reporter, which was included in all transfections). The absolute luciferase values obtained for 3Enh-Luc, pGa(−129/−34), and −129/+81-Luc after transfection and cocultivation with CD40lig-L cells were set at 100%. Percent values for pGa(−129/−34mκB) and −129/+81mκB are given relative to pGa(−129/−34) and −129/+81, respectively.
The NF-κB-like binding site at −52/−43 mediates transactivation by the NF-κB subunit protein p65 but not c-Rel.
The subunit composition of NF-κB can greatly influence not only its ability to bind to a particular DNA sequence motif but also the extent of promoter transactivation (99). To directly investigate the ability of Rel family members to drive transcription from the bfl-1 promoter, transient cotransfections were therefore performed with DG75 cells with vectors that express individual NF-κB subunit proteins. It can be seen that the pGa(−129/−34) reporter was transactivated approximately 3.0-fold (over pGa50-7) when cotransfected with p65 expression vector (Fig. 9A) and that transactivation was completely abolished upon mutation of the NF-κB-like binding site at −52/−43 [Fig. 9A, pGa(−129/−34mκB)]. Cotransfection of the same reporter with either a c-Rel or a p50 expression vector failed to significantly transactivate pGa(−129/−34) (Fig. 9A). It can also be seen that there was a lack of any synergistic effect when either p65/p50 or p65/c-Rel expression vector combinations were cotransfected with the same reporter construct (Fig. 9A). Similar results were obtained with the −129/+81-Luc reporter, in that cotransfection of p65 effector plasmid alone led to a significant increase (threefold) in bfl-1 promoter activity, and mutation of the NF-κB-like binding site again led to the loss of transactivation by p65 (Fig. 9B). These experiments showed that p65 directly transactivated the bfl-1 promoter and that the NF-κB-like binding site at −52/−43 was essential both for this effect and also for conferring p65 responsiveness on a heterologous minimal promoter in DG75 cells.
FIG. 9.
The NF-κB-like binding site at −52/−43 mediates transactivation by the NF-κB subunit protein p65 but not c-Rel. (A and B) DG75 cells were transfected with 2 μg of vector expressing various NF-κB subunit proteins together with 1 μg of the luciferase reporter constructs indicated underneath each graph. Cells were harvested at 48 h posttransfection and assayed for luciferase activity as described before. Normalized luciferase values were expressed as activation (n-fold) relative to the corresponding value obtained for each reporter construct when cotransfected with control vector pcDNA3.1.
DISCUSSION
It was already shown that expression of LMP1 in EBV-negative BL cell lines coincides with a dramatic increase in the level of bfl-1 mRNA (30). Here, we show for the first time that LMP1 transactivates the bfl-1 promoter in this cell context, and we present evidence that points to a key role for NF-κB in this process. The implication of a role for NF-κB is in keeping with other studies in which increases in bfl-1 mRNA levels have been demonstrated in response to agents such as lipopolysaccharide, TNF-α, or etoposide, all of which are known to lead to the activation of this transcription factor (60, 117, 123). NF-κB inhibition has been associated with decreased bfl-1 mRNA levels in an LCL (11). Furthermore, ectopic expression of the NF-κB subunit proteins c-Rel and p65 (but not p50) was also shown to independently upregulate bfl-1 mRNA levels in HeLa cells, and in the case of TNF-α, this was demonstrated to be due to an increase in bfl-1 promoter activity (123). In the same study, it was shown that c-Rel transactivated the bfl-1 promoter through its interaction with a consensus NF-κB binding site at position at −833/−823 relative to the known transcription start site of this gene. Elsewhere, the expression of mouse A1 (the murine bfl-1 homologue) was shown to be induced in response to mitogen stimulation of primary B cells in a c-Rel-dependent fashion (49).
In this study, the evidence of an important role for NF-κB in the regulation of bfl-1 promoter activity by LMP1 is based on several observations. First, coexpression of a superrepressor IκBα mutant inhibited transactivation. Second, inhibition of TRAF2-mediated activation of NF-κB by either coexpression of a dominant-negative TRAF2 molecule or overexpression of the TRAF2-binding zinc finger protein A20 impaired transactivation by LMP1. Third, treatment with PMA, a well-known activator of NF-κB in many cell types, also stimulated bfl-1 promoter activity. Fourth, expression of the NF-κB subunit protein p65 transactivates the bfl-1 promoter. In all cases, the anticipated inhibitory or stimulatory effects of these molecules were mirrored when monitored in parallel with an NF-κB-responsive reporter construct.
The loss of transactivation that is seen in response to TRAF2Δ(6-86) and A20 strongly implies that TRAF2 is a component of the signaling pathway involved in the activation of bfl-1 by LMP1. However, of the two, A20 was more efficient at inhibiting LMP1 activation of both the bfl-1 promoter and NF-κB. Similar findings with these TRAF2 inhibitors have been reported with other promoters in studies involving LMP1 and TNF-α (32, 34, 58, 71, 100). In the case of LMP1, TRAF2Δ(6-86) has been shown to only partially block the activation of NF-κB by CTAR2 (40%) while almost completely blocking that mediated by CTAR1 (>75%) (71). One interpretation of these observations is that although TRAF2 is a component of the signaling pathway involved in the activation of bfl-1 by LMP1, additional contributions from other CTAR1/2-associated proteins, such as other TRAFs, may also be involved (26). In this regard, the receptor-interacting protein has also been shown to interact directly with CTAR2 but does not mediate activation of NF-κB and is therefore not a likely candidate for inclusion in this case (63). Alternatively, it is possible that TRAF2 is bound in a stable complex with other proteins and that either insufficient quantities of TRAF2Δ(6-86) were expressed or the posttransfection incubation period was not long enough to permit the mutant to efficiently displace endogenous wild-type TRAF2. Indeed a number of TRAF2-interacting proteins have been identified, such as TRAF1, TANK/I-TRAF, and c-IAPs, which may affect TRAF2 heterocomplexes in terms of their stability and signaling potential (for a review, see reference 36). Consistent with this possibility is the observation that while CTAR1 can directly bind TRAF2, CTAR2 binds TRAF2 indirectly via TRADD (63). Hence, the differences in the stoichiometry of binding of TRAF2 to these two domains of LMP1 may account for the inefficiency of TRAF2Δ(6-86) inhibition of TRAF2-mediated signaling from the predominant signaling domain CTAR2, relative to CTAR1. A20, a zinc finger protein whose expression is induced by LMP1 in an NF-κB-dependent manner, can inhibit both LMP1-induced NF-κB and JNK activation, thereby acting in a negative feedback loop to control LMP1 signaling. Such control may be important in that overexpression of LMP1 can have toxic effects on the cell (42, 50, 86). Although this activity of A20 was initially presumed to be dependent upon its interaction with only TRAF2 (34), more-detailed studies suggest that A20 can function as a promiscuous inhibitor of the activities of other TRAFs and TRADD (42). In this context, the data presented in this study do not rule out an additional role for other LMP1-interacting signaling molecules in mediating activation of the bfl-1 promoter.
Several studies have indicated that CTAR1 and CTAR2 can independently mediate activation of NF-κB by LMP1; however, the extent of the contribution of each of these domains to the total activation of NF-κB varies between cell lines (40, 61, 92). In general, CTAR2 has been found to be a stronger activator of NF-κB than CTAR1. The NF-κB-dependent upregulation of A20 expression by LMP1 maps to both the CTAR1 and CTAR2 domains, and the contribution of each domain to this effect correlates with the relative extents of activation of NF-κB by the two domains (89, 90). A third possible activation domain, CTAR3, mapping to a region between CTAR1 and CTAR2 (residues 275 to 307) has been shown to mediate activation of JAK3 (45). In this study, we used LMP1 mutants that are defective in signaling from either CTAR1 (LMPAAA) or CTAR2 (LMPG) or both of these domains together (LMPAAAG) to show that both of these domains are required for maximal activation of the bfl-1 promoter in EBV-negative BL-derived cell lines. LMPAAAG has already been shown to function as a dominant-negative mutant and can efficiently inhibit the activation of NF-κB, JAK3, and Jun transcriptional activity by wild-type LMP1 (8). This mutant functions by interacting with wild-type LMP1 and interfering with its ability to bind TRAF2. The requirement for cooperation between the CTAR1 and CTAR2 domains on different LMP1 molecules within the same hetero-oligomeric complex is likely to form the basis for the high efficiency with which LMPAAAG can inhibit LMP1-mediated signaling. In this regard, only one LMPAAAG molecule within the oligomeric complex, which is at least trimeric, may be required to alter signaling from the complex (39, 40). Consistent with these findings, we observed that LMPAAAG can efficiently inhibit wild-type LMP1-mediated activation of the bfl-1 promoter and NF-κB in DG75 cells (data not shown).
The observation that the double mutation in CTAR1 and CTAR2 completely eliminates LMP1-mediated activation of the bfl-1 promoter does not necessarily exclude a role for CTAR3 (or JAK3), since the function of CTAR3 may be dependent on CTAR1 and/or CTAR2 (8). The existence of a cooperative function between CTAR1 and CTAR2 cautions against overinterpretation of data obtained with mutants nonfunctional for CTAR1 or CTAR2 but particularly with mutants containing large deletions that may adversely affect physical cooperation between CTAR1 and CTAR2 (26, 108).
An important aspect of CD40 signaling in B cells is its ability to control apoptosis. Isolated germinal center cells, which spontaneously undergo apoptosis in culture, are capable of surviving for extended periods when treated with anti-CD40 antibodies or CD40 ligand (4, 72). In addition, CD40 is known to rescue immature B cells and mature B cells from surface IgM and Fas-mediated apoptosis (47). Several studies have indicated that the antiapoptotic function of CD40 is mediated by upregulation of bcl-xL and A20 (82, 109, 113). The results presented in this study support the addition of bfl-1 to this list. Our finding that Rael cells do not show increased bfl-1 mRNA levels in response to CD40 activation is in accordance with the findings of others in that this latency type 1 BL-derived cell line is nonresponsive to CD40 activation with respect to other parameters as well (54). It has already been reported that activation of CD40 coincides with a transient increase in bfl-1 mRNA levels in B lymphoma-derived cell lines (22, 82). Here, we corroborate these findings and additionally show that signaling through CD40 receptor, when triggered either by engagement with its ligand or through cross-linking with an anti-CD40 antibody, leads to transactivation of the bfl-1 promoter by an NF-κB-dependent pathway.
If LMP1 and CD40 utilize different signaling pathways to transactivate the bfl-1 promoter, then at least an additive effect would have been expected. The finding of a less than additive effect but greater than an individual effect of signaling from the two membrane molecules can be interpreted to mean that both LMP1 and CD40 share components of the same signaling pathway. Both membrane proteins have been reported to cooperate in an additive or synergistic manner in upregulating cell surface molecules such as B7.1 and IgM as well as in the induction of IL-6 secretion in mouse B cell lines (10). However, although CD40 and LMP1 could individually induce ICAM-1 and LFA-1 expression, a clear cooperative effect was not evident in that study. In BL-derived cell lines, dual signaling from CD40 and LMP1 has been shown not to result in a cooperative effect on the induction of an NF-κB reporter construct in transient-transfection assays (39). Thus, cooperation between CD40 and LMP1 in regulating gene expression may not be a global phenomenon. At the mRNA level, it is further complicated by the fact that LMP1 can upregulate CD40 expression in BL-derived cell lines. Furthermore, it has already been shown that bfl-1 mRNA is more stable in BL cell lines with a latency group 3 phenotype and that this effect is mediated at least in part by LMP1 (30). However, our subsequent half-life studies showed no evidence of an increase in the stability of this transcript following the treatment of the EBV-negative BL cell line BJAB with G28.5 antibody (data not shown). This cell line has detectable levels of bfl-1 transcript in the absence of CD40 activation. This result was somewhat surprising, since mRNA stabilization is an effect that is often found associated with genes that show a rapid response to signals that modulate gene expression (14). It remains possible, however, that the lack of an effect of CD40 activation on bfl-1 mRNA stability may be specific to this cell line.
We have demonstrated for the first time that that the −129/−34 region of the bfl-1 promoter can function as an LMP1-dependent transcriptional enhancer in EBV-negative BL-derived cell lines. Furthermore, we have shown that transactivation of this enhancer by LMP1 is NF-κB dependent. Subsequent deletion and mutational analyses lead to the conclusion that a novel NF-κB-like binding site at position −52 to −43 is essential for transactivation by LMP1 and also implicate a significant role for a nearby AP-1-like binding site (−74/−64). Mutation of the NF-κB-like binding site in the context of a 1.4-kb bfl-1 promoter fragment led to the nearly complete loss of transactivation by LMP1. Classic NF-κB (p65/p50) binds the sequence 5′-GGGRNNYYCC-3′, whereas the RelA/cRel dimer binds to the sequence 5′-HGGARNYYCC-3′ (H indicates A, C, or T; R is purine [A or G]; and Y is pyrimidine [T or C]) (6). The κB-like site at position −52/−43 on the bfl-1 promoter differs from the p65/p50 and p65/c-Rel consensus binding sites at nucleotide positions 3 and 2, respectively. NF-κB-like sites that diverge from the consensus have been identified in the promoters of several apoptosis-related genes such as bcl-x, c-IAP1, c-IAP2, and mouse A1 as well as cytokine genes such as IL-8, and the functionality of these sites in promoter activation has been demonstrated in several of these cases (15, 49, 57, 94). Significantly, the −52/−43 site on the bfl-1 promoter is 100% complementary to a CD28-responsive κB site in the IL-2 promoter (18, 19, 115) and both c-Rel and p65 have been shown to play roles in mediating this response (44).
We have not been able to detect the interaction of NF-κB subunit proteins with the NF-κB-like binding site at −52/−43 in electrophoretic mobility shift assays (EMSA). In control experiments, however, p65-containing complexes were detectable when a probe which contained the κB enhancer element from the Ig(κ) light chain gene was incubated with nuclear protein extracts from DG75 cells (no EMSA data are shown). The presence of p65 was confirmed, as preincubation of nuclear extracts with anti-p65 antibody led to the supershifting of specific DNA-protein complexes. This finding is consistent with the high constitutive levels of activated NF-κB known to be present in this cell context (61, 106). When nuclear protein extracts from DG75-tTA-LMP1 were used, the induction of LMP1 led to increases in the levels of these two p65-containing complexes without the formation of any new complex. When various DNA fragments (ranging in size from 14 bp up to the complete −129/+81 fragment) that encompassed the bfl-1 −52/−43 sequence motif were used as probes, however, neither the profile nor the intensity of DNA-protein complex formation was altered in response to LMP1 induction. If NF-κB subunits do bind to this motif in vivo, then our inability to detect binding in vitro may be the result of a low-affinity interaction due to the fact that it is not a consensus NF-κB-binding site. LMP1-associated upregulation may then be occurring by the phosphorylation of a weakly bound p65-containing complex. In this regard, IL-1 (a member of the TNFR superfamily) has been shown to stimulate the transactivation potential of p65 by inducing the phosphorylation of its transactivation domain via a phosphoinositide-3-OH kinase-Akt-dependent pathway (85, 110), and the latter has recently been shown to be activated by LMP1 (24). Alternatively, NF-κB-associated transactivation of the bfl-1 promoter in DG75 and BL41 may be indirect and mediated by a transcription factor that is induced or activated by NF-κB and which failed to bind under the EMSA conditions used.
A sole NF-κB DNA binding motif (−833/−823) has previously been implicated in regulating the c-Rel-dependent transactivation of the bfl-1 promoter in the epithelial cell line HeLa (123). More recently, chromatin immunoprecipitation assays showed that the in vivo interaction of c-Rel and p50 (but not p65) with this site promotes the cooperative binding of C/EBPβ and AP-1 in an enhanceosome-like complex in PMA-ionomycin-treated Jurkat T cells (31). We observed that in the context of DG75 or BL41 cells, removal of this site by deletion or mutation did not affect LMP1-associated activation of the bfl-1 promoter. Although it is clear that LMP1 activates different subsets of NF-κB subunit proteins in a cell-type dependent manner (16, 55, 96), c-Rel is known to be activated by LMP1 in a lymphocyte cellular context, including DG75 cells (16, 83). In addition to being nonresponsive to LMP1, we observed that this NF-κB-binding site does not play a role in mediating transactivation of bfl-1 by either PMA or p65 in EBV-negative BL-derived cell lines. This, coupled with the fact that c-Rel failed to activate luciferase expression via the −52/−43 site, may indicate that p65, and not c-Rel, is the predominant subunit involved in regulating the bfl-1 promoter in EBV-negative BL cell lines in response to LMP1. The failure of LMP1 to transactivate via the −833/−823 site might therefore be due to its low affinity for p65. It is also possible, however, that access to this site may be obstructed in EBV-negative BL-derived cell lines by lymphocyte-specific nuclear proteins binding in its vicinity.
LMP1-associated signaling also activates the JNK pathway, which results in the induction of the transcription factor AP1. We have shown that an AP1-like binding site at −74 to −64 also plays a role in LMP1-mediated transactivation of the bfl-1 promoter. However, in this case, the contribution of this sequence motif is dependent on the NF-κB-like binding site at −52/−43, since any promoter fragments that lack the latter are not responsive to LMP1. It has also been shown that the mitogen-activated protein kinase p38 and NF-κB contribute to the transactivation of certain LMP-1-inducible genes, such as IL-6, IL-8, and IL-10, through coregulation of their promoter sequences (33, 116). In a preliminary experiment, however, a well-established and specific inhibitor of p38 activation (SB203580) (112) did not affect transactivation of the bfl-1 promoter by LMP1 in DG75 cells (result not shown).
In lymphoid follicles, the bfl-1 transcript has been detected in the germinal centers which are the sites for B-cell proliferation and differentiation (66). CD40 activation is necessary for rescuing germinal center B cells from spontaneous apoptosis, and long-lived B cells are distinguished by elevated expression of bfl-1 (111). In this regard, Bfl-1 might be viewed as a key determinant of cell fate for immature peripheral B cells. The strong induction of bfl-1 transcripts by proinflammatory cytokines in endothelial, leukemic, and hemopoietic cells is consistent with a role for protecting them from apoptotic extinction (17, 67, 68, 84, 93). Consistent with a role for NF-κB in this process is the reduction in constitutive bfl-1 mRNA levels in B cell lines derived from c-Rel−/− mice (49). The ability of LMP1 to provide cell survival signals by upregulating the expression of antiapoptotic members of the bcl-2 family of proteins, including bfl-1, is very significant from the standpoint of the mechanism of persistence of EBV in the memory B cell in peripheral blood. In vivo, EBV-positive tonsillar memory B cells express a restricted pattern of latent gene transcripts which resembles the pattern of latent gene expression detected in EBV-associated tumors (3). The actions of LMP1 and LMP2A provide the surrogate T-cell help and B-cell receptor signals that memory B cells require for long-term survival (12). Since BL lines exhibit phenotypic features reminiscent of germinal center B cells, and as Bfl-1 was shown previously to cooperate with the adenovirus E1A protein in inducing cell transformation (28, 29), the NF-κB-dependent activation of bfl-1 by LMP1 may be a key event in the rescue of EBV-infected B cells from apoptosis and an important route by which LMP1 functions in oncogenesis.
Acknowledgments
We gratefully acknowledge the following gifts of plasmids: pEFCX, pEFCX-LMP1, and pEFCX-IkBαDN from P. Brodin; pcDNA3TRAF2Δ(6-86) and pSFFVA20 from L. Young and A. Eliopoulos. The anti-CD40 monoclonal antibody G28.5 was a generous gift from E. Clark. We are grateful to P. Garrone for the CD40lig-L and CD32-L cell lines. We thank P. Cahill, K. Fitzgerald, and P. Brennan for helpful discussions and advice.
This work was supported by a research grant from the Health Research Board (HRB; Dublin, Ireland) (D.W.). We also acknowledge support from National Institutes of Health grant CA83937 (C.G.). B.N.D. was supported by a postdoctoral fellowship from Enterprise Ireland.
REFERENCES
- 1.Aicher, A., G. L. Shu, D. Magaletti, T. Mulvania, A. Pezzutto, A. Craxton, and E. A. Clark. 1999. Differential role for p38 mitogen-activated protein kinase in regulating CD40-induced gene expression in dendritic cells and B cells. J. Immunol. 163:5786-5795. [PubMed] [Google Scholar]
- 2.Arenzana-Seisdedos, F., B. Fernandez, I. Dominguez, J. M. Jacque, D. Thomas, M. T. Diaz-Meco, J. Moscat, and J. L. Virelizier. 1993. Phosphatidylcholine hydrolysis activates NF-κB and increases human immunodeficiency virus replication in human monocytes and T lymphocytes. J. Virol. 67:6596-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babcock, G. J., and D. A. Thorley-Lawson. 2000. Tonsillar memory B cells, latently infected with Epstein-Barr virus, express the restricted pattern of latent genes previously found only in Epstein-Barr virus-associated tumors. Proc. Natl. Acad. Sci. USA 97:12250-12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, M., A. Eliopoulos, L. Young, R. Armitage, C. Gregory, and J. Gordon. 1998. Prolonged phenotypic, functional and molecular change in group I Burkitt lymphoma cells on short-term exposure to CD40 ligand. Blood 92:2830-2843. [PubMed] [Google Scholar]
- 5.Baker, S. J., and E. P. Reddy. 1996. Transducers of life and death: TNF receptor superfamily and associated proteins. Oncogene 12:1-9. [PubMed] [Google Scholar]
- 6.Baldwin, A. 1996. The NF-κB and IκB proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-683. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Bassat, H. N., S. Goldblum, T. Mitrani, J. M. Goldblum, M. M. Yoffey, Z. Cohen, B. Bentwich, E. Ramot, E. Klein, and G. Klein. 1977. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D. G.-75). Int. J. Cancer 19:27-33. [DOI] [PubMed] [Google Scholar]
- 8.Brennan, P., J. E. Floettmann, A. Mehl, M. Jones, and M. Rowe. 2001. Mechanism of action of a novel latent membrane protein-1 dominant negative. J. Biol. Chem. 276:1195-1203. [DOI] [PubMed] [Google Scholar]
- 9.Brown, K., S. Gerstberger, L. Carlson, G. Franzoso, and U. Siebenlist. 1995. Control of IκBα-proteolysis by site-specific, signal-induced phosphorylation. Science 267:1485-1488. [DOI] [PubMed] [Google Scholar]
- 10.Busch, L., and G. Bishop. 1999. The EBV transforming protein latent membrane protein 1 mimics and cooperates with CD40 signaling in B lymphocytes. J. Immunol. 162:2555-2561. [PubMed] [Google Scholar]
- 11.Cahir-Mcfarland, E., D. M. Davidson, S. L. Schauer, J. Duong, and E. Kieff. 2000. NF-κB inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 97:6055-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caldwell, R. G., J. B. Wilson, S. J. Anderson, and R. Longnecker. 1998. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 9:405-411. [DOI] [PubMed] [Google Scholar]
- 13.Calendar, A., M. Billaud, J. Aubry, J. Banchereau, M. Vuillaume, and G. Lenoir. 1987. Epstein-Barr virus induces expression of B cell activation markers on in vitro infection of Epstein-Barr virus negative B lymphoma cells. Proc. Natl. Acad. Sci. USA 84:8060-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, C.-Y. A., and A.-B. Shyu. 1995. AU-rich elements: characterisation and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 15.Chen, F., L. M. Demers, V. Vallyathan, Y. Lu, V. Castranova, and X. Shi. 1999. Involvement of 5′-flanking κB-like sites within bcl-x gene in silica-induced Bcl-x expression. J. Biol. Chem. 274:35591-35595. [DOI] [PubMed] [Google Scholar]
- 16.Chien, M.-L., and M.-L. Hammarskjold. 2000. Epstein Barr virus latent membrane protein (LMP1) induces specific NF-κB complexes in human T-lymphoid cells. Virus Res. 67:17-30. [DOI] [PubMed] [Google Scholar]
- 17.Choi, S. S., I. C. Park, J. W. Yun, Y. C. Sung, S. I. Hong, and H. S. Shin. 1995. A novel Bcl-2 related gene, Bfl-1, is overexpressed in stomach cancer and preferentially expressed in bone marrow. Oncogene 11:1693-1698. [PubMed] [Google Scholar]
- 18.Civil, A., A. Bakker, I. Rensink, S. Doerre, L. Aarden, and C. Verweij. 1996. Nuclear appearance of a factor that binds the CD28 response element within the interleukin 2 enhancer correlates with interleukin 2 production. J. Biol. Chem. 271:8321-8327. [DOI] [PubMed] [Google Scholar]
- 19.Civil, A., I. Rensink, L. Aarden, and C. Verweij. 1999. Functional disparity of distinct CD28 response elements toward mitogenic responses. J. Biol. Chem. 274:34369-34374. [DOI] [PubMed] [Google Scholar]
- 20.Clark, E. A., and J. A. Ledbetter. 1986. Activation of human B cells through two distinct cell surface differentiation antigens, Bp35 and Bp50. Proc. Natl. Acad. Sci. USA 83:4494-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clausse, B., K. Fizazi, V. Walczak, C. Tetaud, J. Wiels, T. Tursz, and P. Busson. 1997. High concentration of the EBV latent membrane protein 1 in glycosphingolipid-rich complexes from both epithelial and lymphoid cells. Virology 228:285-293. [DOI] [PubMed] [Google Scholar]
- 22.Craxton, A., P. I. Chuang, G. Shu, J. M. Harlan, and E. A. Clark. 2000. The CD40-inducible Bcl-2 family member A1 protects B cells from antigen receptor-mediated apoptosis. Cell. Immunol. 200:56-62. [DOI] [PubMed] [Google Scholar]
- 23.Dawson, C. W., A. B. Rickinson, and L. S. Young. 1990. Epstein-Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature 344:777-780. [DOI] [PubMed] [Google Scholar]
- 24.Dawson, C. W., G. Tramountanis, A. G. Eliopoulos, and L. S. Young. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J. Biol. Chem. 278:3694-3704. [DOI] [PubMed] [Google Scholar]
- 25.Devergne, O., E. D. Cahir McFarland, G. Mosialos, K. M. Izumi, C. F. Ware, and E. Kieff. 1998. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 72:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devergne, O., E. Hatzivassiliou, K. M. Izumi, K. M. Kaye, M. F. Kleijnen, E. Kieff, and G. Mosialos. 1996. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol. Cell. Biol. 16:7098-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dirmeier, U., B. Neuhierl, E. Kilger, G. Reisbach, M. L. Sandberg, and W. Hammerschmidt. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res. 63:2982-2989. [PubMed] [Google Scholar]
- 28.D'Sa-Eipper, C., and G. Chinnadurai. 1998. Functional dissection of bfl-1, a bcl-2 homolog: anti-apoptosis, oncogene cooperation and cell proliferation activities. Oncogene 16:3105-3114. [DOI] [PubMed] [Google Scholar]
- 29.D'Sa-Eipper, C., T. Subramanian, and G. Chinnadurai. 1996. bfl-1, a bcl-2 homologue, suppresses p53-induced apoptosis and exhibits potent cooperative transforming activity. Cancer Res. 56:3879-3882. [PubMed] [Google Scholar]
- 30.D'Souza, B. N., M. Rowe, and D. Walls. 2000. The bfl-1 gene is transcriptionally upregulated by the Epstein-Barr virus LMP1, and its expression promotes the survival of a Burkitt's lymphoma cell line. J. Virol. 74:6652-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edelstein, L. C., L. Lagos, M. Simmons, H. Tirumalai, and C. Gelinas. 2003. NF-κB-dependent assembly of an enhanceosome-like complex on the promoter region of apoptosis inhibitor Bfl-1. Mol. Cell. Biol. 23:2749-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eliopoulos, A. G., M. Stack, C. W. Dawson, K. M. Kaye, L. Hodgkin, S. Sihota, M. Rowe, and L. S. Young. 1997. Epstein-Barr virus-encoded LMP1 and CD40 mediate IL-6 production in epithelial cells via an NF-κB pathway involving TNF-receptor-associated factors. Oncogene 14:2899-2916. [DOI] [PubMed] [Google Scholar]
- 33.Eliopoulos, A. G., N. J. Gallagher, S. M. Blake, C. W. Dawson, and L. S. Young. 1999. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J. Biol. Chem. 274:16085-16096. [DOI] [PubMed] [Google Scholar]
- 34.Eliopoulos, A. G., S. M. Blake, J. E. Floettmann, M. Rowe, and L. S. Young. 1999. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J. Virol. 73:1023-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eliopoulos, A. G., and L. S. Young. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1731-1742. [DOI] [PubMed] [Google Scholar]
- 36.Eliopoulos, A. G., and L. S. Young. 2001. LMP1 structure and signal transduction. Semin. Cancer Biol. 11:435-444. [DOI] [PubMed] [Google Scholar]
- 37.Farrell, P. 1995. Epstein-Barr virus immortalizing genes. Trends Microbiol. 3:105-109. [DOI] [PubMed] [Google Scholar]
- 38.Floettmann, J. E., K. Ward, A. B. Rickinson, and M. Rowe. 1996. Cytostatic effect of Epstein-Barr virus latent membrane protein 1 analysed using tetracycline-regulated expression in B cell lines. Virology 223:29-40. [DOI] [PubMed] [Google Scholar]
- 39.Floettmann, J. E., A. G. Eliopoulos, M. Jones, L. S. Young, and M. Rowe. 1998. Epstein-Barr virus latent membrane protein-1 (LMP1) signaling is distinct from CD40 and involves physical cooperation of its two C-terminus functional regions. Oncogene 17:2383-2392. [DOI] [PubMed] [Google Scholar]
- 40.Floettmann, J. E., and M. Rowe. 1997. Epstein-Barr virus latent membrane protein 1 (LMP1) C-terminus activation region 2 (CTAR-2) maps to the far C-terminus and requires oligomerisation for NF-κB activation. Oncogene 15:1851-1858. [DOI] [PubMed] [Google Scholar]
- 41.Fries, K. L., W. E. Miller, and N. Raab-Traub. 1996. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J. Virol. 70:8653-8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fries, K. L., W. E. Miller, and N. Raab-Traub. 1999. The A20 protein interacts with the Epstein-Barr virus latent membrane protein 1 (LMP1) and alters the LMP1/TRAF1/TRADD complex. Virology 264:159-166. [DOI] [PubMed] [Google Scholar]
- 43.Garrone, P., E. M. Neidhardt, E. Garcia, C. Galibert, C. van Kooten, and J. Banchereau. 1995. Fas ligand induces apoptosis of CD40-activated human B lymphocytes. J. Exp. Med. 182:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh, P., T. Tan, N. Rice, A. Sica, and H. Young. 1993. The interleukin 2 CD28-responsive complex contains at least three members of the NF-κB family: c-Rel, p50 and p65. Proc. Natl. Acad. Sci. USA 90:1696-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gires, O., U. Zimber-Strobl, R. Gonnella, M. Ueffing, G. Marschall, R. Zeidler, D. Pich, and W. Hammerschmidt. 1997. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 16:6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory, C. D. 1995. Control of apoptosis in human B cells: implications for neoplasia, p. 257-311. In C. D. Gregory (ed.), Apoptosis and the immune response. Wiley-Liss Inc., New York, N.Y.
- 48.Gregory, C. D., M. Rowe, and A. B. Rickinson. 1990. Different Epstein-Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt's lymphoma cell line. J. Gen. Virol. 71:1481-1495. [DOI] [PubMed] [Google Scholar]
- 49.Grumont, R., I. Rourke, and S. Gerondakis. 1999. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation induced apoptosis. Genes Dev. 13:400-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammerschmidt, W., B. Sugden, and V. Baichwal. 1989. The transforming domain alone of the latent membrane protein of Epstein Barr virus is toxic to cells when expressed at high levels. J. Virol. 63:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatzivassilliou, E., W. Miller, N. Raab-Traub, E. Kieff, and G. Mosialos. 1998. A fusion of the EBV latent membrane protein 1 (LMP1) transmembrane domains to the CD40 cytoplasmic domain is similar to LMP1 in constitutive activation of epidermal growth factor receptor expression, nuclear factor kappa-B, and stress activated protein kinase. J. Immunol. 160:1116-1121. [PubMed] [Google Scholar]
- 52.He, Z., B. Xin, X. Yang, C. Chan, and L. Cao. 2000. Nuclear factor-kappaB activation is involved in LMP1-mediated transformation and tumorigenesis of rat-1 fibroblasts. Cancer Res. 60:1845-1848. [PubMed] [Google Scholar]
- 53.Henderson, S., M. Rowe, C. D. Gregory, D. Croom-Carter, F. Wang, R. Longnecker, E. Kieff, and A. B. Rickinson. 1991. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell 65:1107-1115. [DOI] [PubMed] [Google Scholar]
- 54.Henriquez, N. V., E. Floettmann, M. Salmon, M. Rowe, and A. B. Rickinson. 1999. Differential responses to CD40 ligation among Burkitt lymphoma lines that are uniformly responsive to Epstein-Barr virus latent membrane protein 1. J. Immunol. 162:3298-3307. [PubMed] [Google Scholar]
- 55.Herrero, J. A., P. Mathew, and C. V. Paya. 1995. LMP1 activates NF-κB by targeting the inhibitory molecule IκBα. J. Virol. 69:2168-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Higuchi, M., K. M. Izumi, and E. Kieff. 2001. Epstein-Barr virus latent-infection membrane proteins are palmitoylated and raft-associated: protein 1 binds to the cytoskeleton through TNF receptor cytoplasmic factors. Proc. Natl. Acad. Sci. USA 98:4675-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong, S.-Y., W.-H. Yoon, J.-H. Park, S.-G. Kang, J.-H. Ahn, and T. H. Lee. 2000. Involvement of two NF-κB binding elements in tumour necrosis factor, CD40-, and Epstein-Barr virus latent membrane protein-1-mediated induction of the cellular inhibitor of apoptosis protein 2 gene. J. Biol. Chem. 275:18022-18028. [DOI] [PubMed] [Google Scholar]
- 58.Hsu, H., H. B. Shu, M. G. Pan, and D. V. Goeddel. 1996. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84:299-308. [DOI] [PubMed] [Google Scholar]
- 59.Hsu, J. L., and S. L. Glaser. 2000. Epstein-Barr virus-associated malignancies: epidemiologic patterns and etiologic implications. Crit. Rev. Oncol. Hematol. 34:27-53. [DOI] [PubMed] [Google Scholar]
- 60.Hu, X., E. Yee, J. M. Harlan, F. Wong, and A. Karsan. 1998. Lipopolysaccharide induces the antiapoptotic molecules, A1 and A20, in microvascular endothelial cells. Blood 92:2759-2765. [PubMed] [Google Scholar]
- 61.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10:549-560. [PubMed] [Google Scholar]
- 62.Izumi, K. M. 2001. Identification of EBV transforming genes by recombinant EBV technology. Semin. Cancer Biol. 11:407-414. [DOI] [PubMed] [Google Scholar]
- 63.Izumi, K. M., E. Cahir-Mcfarland, A. Ting, E. Riley, B. Seed, and E. Kieff. 1999. The Epstein Barr virus oncoprotein latent membrane protein 1 engages the tumour necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-κB activation. Mol. Cell. Biol. 19:5759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Izumi, K. M., K. M. Kaye, and E. D. Kieff. 1997. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 94:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jung-Ha, H., D. Kim, S.-B. Lee, S.-I. Hong, S.-Y. Park, J. Huh, C.-W. Kim, S.-S. Kim, Y. Lee, S. S. Choi, and H.-S. Shin. 1998. Expression of Bfl-1 in normal and tumour tissues: Bfl-1 overexpression in cancer is attributable to its preferential expression in infiltrating inflammatory cells. Hum. Pathol. 29:723-728. [DOI] [PubMed] [Google Scholar]
- 67.Karsan, A., E. Yee, K. Kaushansky, and J. M. Harlan. 1996. Cloning of a human Bcl-2 homologue: inflammatory cytokines induce human A1, in cultured endothelial cells. Blood 87:3089-3096. [PubMed] [Google Scholar]
- 68.Karsan, A., E. Yee, and J. M. Harlan. 1996. Endothelial cell death induced by tumour necrosis factor-α is inhibited by the Bcl-2 family member, A1. J. Biol. Chem. 271:27201-27204. [DOI] [PubMed] [Google Scholar]
- 69.Kaye, K. M., K. M. Izumi, G. Mosialos, and E. Kieff. 1995. The Epstein-Barr virus LMP1 cytoplasmic carboxy terminus is essential for B-lymphocyte transformation; fibroblast cocultivation complements a critical function within the terminal 155 residues. J. Virol. 69:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaye, K. M., K. M. Izumi, H. Li, E. Johannsen, D. Davidson, R. Longnecker, and E. Kieff. 1999. An Epstein Barr virus that expresses only the first 231 LMP1 amino acids efficiently initiates primary B-lymphocyte growth transformation. J. Virol. 73:10525-10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaye, K. M., O. Devergne, J. Harada, K. M. Izumi, R. Yalamanchili, E. Kieff, and G. Mosialos. 1996. Tumour necrosis factor receptor associated factor 2 is a mediator of NF-κB activation by latent infection membrane protein 1, the Epstein Barr virus transforming protein. Proc. Natl. Acad. Sci. USA 93:11085-11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kehry, M. 1996. CD40-mediated signaling in B cells-balancing cell survival, growth and death. J. Immunol. 156:2345-2348. [PubMed] [Google Scholar]
- 73.Kenny, J. J., T. J. Knobloch, M. Augustus, K. C. Carter, C. A. Rosen, and J. C. Lang. 1997. GRS, a novel member of the Bcl-2 gene family, is highly expressed in multiple cancer cell lines and in normal leukocytes. Oncogene 14:997-1001. [DOI] [PubMed] [Google Scholar]
- 74.Kieff, E., and A. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Virology, 4th ed. Lippincott, Williams, and Wilkins, Philadelphia, Pa.
- 75.Kieser, A., E. Kilger, O. Gires, M. Ueffing, W. Kolch, and W. Hammerschmidt. 1997. Epstein-Barr virus latent membrane protein 1 triggers AP-1 activity via the c-jun N-terminal kinase cascade. EMBO J. 16:6478-6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kilger, E., A. Kieser, M. Baumann, and W. Hammerschmidt. 1998. Epstein-Barr virus-mediated B cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 17:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kulwichit, W., R. Edwards, E. Davenport, J. Baskar, V. Godfrey, and N. Raab-Traub. 1998. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc. Natl. Acad. Sci. USA 95:11963-11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laherty, C. D., H. M. Hu, A. W. Opipari, F. Wang, and V. M. Dixit. 1992. The Epstein-Barr virus LMP-1 gene product induces A20 zinc finger protein expression by activating nuclear factor-κB. J. Biol. Chem. 267:24157-24160. [PubMed] [Google Scholar]
- 79.Lam, N., and B. Sugden. 2003. CD40 and its viral mimic, LMP1: similar means to different ends. Cell. Signal. 15:9-16. [DOI] [PubMed] [Google Scholar]
- 80.Lam, N., and B. Sugden. 2003. LMP1, a viral relative of the TNF receptor family, signals principally from intracellular compartments. EMBO J. 22:3027-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laux, G., F. Dugrillon, C. Eckert, B. Adam, U. Zimber-Strobl, and G. W. Bornkamm. 1994. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis vlement in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J. Virol. 68:6947-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee, H. H., H. Dadgostar, Q. Cheng, J. Shu, and G. Cheng. 1999. NF-κB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl. Acad. Sci. USA 96:9136-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liljeholm, S., K. Hughes, T. Grundstorm, and P. Brodin. 1998. NF-kappaB only partially mediates Epstein-Barr virus latent membrane protein 1 activation of B cells. J. Gen. Virol. 79:2117-2125. [DOI] [PubMed] [Google Scholar]
- 84.Lin, E. Y., A. Orlofsky, M. S. Berger, M. B. Prystowsky. 1996. Characterization of A1, a novel hemopoietic-specific early-response gene with sequence similarity to bcl-2. J. Immunol. 151:1979-1988. [PubMed] [Google Scholar]
- 85.Madrid, L. V., C. Wang, D. Guttridge, A. Schottelius, A. Baldwin, and M. Mayo. 2000. Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-κB. Mol. Cell. Biol. 20:1626-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin, J. M., D. Veis, S. J. Korsmeyer, and B. Sugden. 1993. Latent membrane protein of Epstein-Barr virus induces cellular phenotypes independently of expression of BclII. J. Virol. 67:5269-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Massuci, M. G., B. Contreras-Salazar, E. Ragnar, K. Falk, J. Minarovits, I. Ernberg, and G. Klein. 1989. 5-Azacytidine upregulates the expression of Epstein-Barr virus nuclear antigen 2 (EBNA-2) through EBNA-6 and latent membrane protein in the Burkitt's lymphoma cell line Rael. J. Virol. 63:3135-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mehl, A. M., J. E. Floettmann, M. Jones, P. Brennan, and M. Rowe. 2001. Characterization of intercellular adhesion molecule-1 regulation by Epstein-Barr virus-encoded latent membrane protein-1 identifies pathways that cooperate with nuclear factor kappa B to activate transcription. J. Biol. Chem. 276:984-992. [DOI] [PubMed] [Google Scholar]
- 89.Miller, W. E., G. Mosialos, E. Kieff, and N. Raab-Traub. 1997. Epstein-Barr virus LMP1 induction of epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J. Virol. 71:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miller, W. E., H. Earp, and N. Raab-Traub. 1995. The Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor. J. Virol. 69:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Milner, A. E., G. D. Johnson, and C. D. Gregory. 1992. Prevention of programmed cell death in Burkitt lymphoma cell lines by bcl-2-dependent and -independent mechanisms. Int. J. Cancer 52:636-644. [DOI] [PubMed] [Google Scholar]
- 92.Mitchell, T., and B. Sugden. 1995. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J. Virol. 69:2968-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moreb, J. S., and M. Schweder. 1997. Human A1, a bcl-2-related gene, is induced in leukemic cells by cytokines as well as differentiating factors. Leukemia 11:998-1004. [DOI] [PubMed] [Google Scholar]
- 94.Okamoto, S., N. Mukaida, K. Yasumoto, N. Rice, Y. Ishikawa, H. Horiguchi, S. Murakami, and K. Matsushima. 1994. The interleukin 8 AP-1 and κB-like sites are genetic end targets of FK506-sensitive pathway accompanied by calcium mobilisation. J. Biol. Chem. 269:8582-8589. [PubMed] [Google Scholar]
- 95.Pai, S., B. J. O'Sullivan, L. Cooper, R. Thomas, and R. Khanna. 2002. RelB nuclear translocation mediated by C-terminal activator regions of Epstein-Barr virus-encoded latent membrane protein 1 and its effect on antigen-presenting function in B cells. J. Virol. 76:1914-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paine, E., R. Scheinman, A. Baldwin, and N. Raab-Traub. 1995. Expression of LMP1 in epithelial cells leads to activation of a select subset of NF-κB/Rel family proteins. J. Virol. 69:4572-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park, I.-C., S. H. Lee, D. Y. Whang, W. S. Hong, S. S. Choi, H. S. Shin, T.-B. Choe, and S. I. Hong. 1997. Expression of a novel bcl-2 related gene, bfl-1, in various human cancers and cancer cell lines. Anticancer Res. 17:4619-4622. [PubMed] [Google Scholar]
- 98.Peltz, G. A., M. L. Trounstine, and K. W. Moore. 1988. Cloned and expressed human Fc receptor for IgG mediates anti-CD3 dependent lymphoproliferation. J. Immunol. 141:1891-1896. [PubMed] [Google Scholar]
- 99.Perkins, N. D., R. M. Schmid, C. S. Duckett, K. Leung, N. Rice, and G. Nabel. 1992. Distinct combinations of NF-κB subunits determine the specificity of transcriptional activation. Proc. Natl. Acad. Sci. USA 89:1529-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reinhard, C., B. Shamoon, V. Shyamala, and L. T. Williams. 1997. Tumour necrosis factor α induced activation of c-jun N-terminal kinase is mediated by TRAF2. EMBO J. 16:1080-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Richardson, C., C. Fielding, M. Rowe, and P. Brennan. 2003. Epstein-Barr virus regulates STAT1 through latent membrane protein 1. J. Virol. 77:4439-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rickinson, A., and E. Kieff. 2001. Epstein-Barr virus, p. 2573-2627. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Virology, 4th ed. Lippincott, Williams, and Wilkins, Philadelphia, Pa.
- 103.Rothe, M., V. Sarma, V. Dixit, and D. Goeddel. 1995. TRAF2 mediated activation of NF-κB by TNF receptor 2 and CD40. Science 269:1424-1427. [DOI] [PubMed] [Google Scholar]
- 104.Rowe, M., C. M. Rooney, C. F. Edwards, G. M. Lenoir, and A. B. Rickinson. 1986. Epstein-Barr virus status and tumour cell phenotype in sporadic Burkitt's lymphoma. Int. J. Cancer 37:367-373. [DOI] [PubMed] [Google Scholar]
- 105.Rowe, M., D. T. Rowe, C. D. Gregory, L. S. Young, P. J. Farrell, H. Rupani, and A. B. Rickson. 1987. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J. 6:2743-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rowe, M., M. Peng-Pilon, D. S. Huen, R. Hardy, D. Croom-Carter, E. Lundgren, and A. B. Rickinson. 1994. Upregulation of bcl-2 by Epstein-Barr virus latent membrane protein LMP1: a B-cell specific response that is delayed relative to NF-κB activation and to induction of cell surface markers. J. Virol. 68:5602-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 108.Sandberg, M., W. Hammerschmidt, and B. Sugden. 1997. Characterization of LMP1's association with TRAF1, TRAF2, and TRAF3. J. Virol. 71:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sarma, V., Z. Lin, L. Clark, B. Rust, M. Tewari, R. Noelle, and V. Dixit. 1995. Activation of B-cell surface receptor CD40 induces A20, a novel zinc finger protein that inhibits apoptosis. J. Biol. Chem. 270:12343-12346. [DOI] [PubMed] [Google Scholar]
- 110.Sizemore, N., S. Leung, and G. Stark. 1999. Activation of phosphatidyl 3-kinase in response to interleukin-1 leads to phosphorylation and activation of the NF-κB p65/RelA subunit. Mol. Cell. Biol. 19:4798-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tomayko, M. M., and M. P. Cancro. 1998. Long-lived B cells are distinguished by elevated expression of A1. J. Immunol. 160:107-111. [PubMed] [Google Scholar]
- 112.Tong, L., S. Pav, D. M. White, S. Rogers, K. M. Crane, C. L. Cywin, M. L. Brown, and C. A. Pargellis. 1997. A highly specific inhibitor of human p38 MAP kinase binds in the ATP pocket. Nat. Struct. Biol. 4:311-316. [DOI] [PubMed] [Google Scholar]
- 113.Tuscano, J. M., K. M. Druey, A. Riva, J. Pena, C. B. Thompson, and J. H. Kehrl. 1996. Bcl-x rather than Bcl-2 mediates CD40-dependent centrocyte survival in the germinal center. Blood 4:1359-1364. [PubMed] [Google Scholar]
- 114.van Kooten, C., and J. Banchereau. 1997. Functions of CD40 on B cells, dendritic cells and other cells. Curr. Opin. Immunol. 9:330-337. [DOI] [PubMed] [Google Scholar]
- 115.Verweij, C., M. Geerts, and L. Aarden. 1991. Activation of interleukin 2 gene transcription via the T cell surface molecule CD28 is mediated through an NF-κB-like response element. J. Biol. Chem. 266:14179-14182. [PubMed] [Google Scholar]
- 116.Vockerodt, M., B. Haier, P. Buttgereit, H. Tesch, and D. Kube. 2001. The EBV LMP1 induces interleukin-10 in Burkitt's lymphoma cells but not in Hodgkin's cells involving the p38/SAPK2 pathway. Virology 280:183-198. [DOI] [PubMed] [Google Scholar]
- 117.Wang, C.-Y., D. C. Guttridge, M. W. Mayo, and A. S. Baldwin. 1999. NF-κB induces expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol. Cell. Biol. 19:5923-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang, D., D. Leibowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalised lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 119.Wang, S., M. Rowe, and E. Lundgren. 1996. Expression of the Epstein-Barr virus transforming protein LMP1 causes a rapid and transient stimulation of the bcl-2 homologue mcl-1 levels in B cell lines. Cancer Res. 56:4610-4613. [PubMed] [Google Scholar]
- 120.Wilson, J., W. Weinberg, R. Johnson, S. Yuspa, and A. J. Levine. 1990. Expression of the BNLF-1 oncogene of Epstein-Barr virus in the skin of transgenic mice induces hyperplasia and aberrant expression of keratin 6. Cell 61:1315-1327. [DOI] [PubMed] [Google Scholar]
- 121.Zhang, L., L. Wu, K. Hong, and J. S. Pagano. 2001. Intracellular signaling molecules activated by Epstein-Barr virus for induction of interferon regulatory factor 7. J. Virol. 75:12393-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zimber-Strobl, U., B. Kempkes, G. Marschall, R. Zeidler, C. van Kooten, J. Banchereau, G. W. Bornkamm, and W. Hammerschmidt. 1996. Epstein-Barr virus latent membrane protein (LMP1) is not sufficient to maintain proliferation of B cells but both it and activated CD40 can prolong their survival. EMBO J. 15:7070-7078. [PMC free article] [PubMed] [Google Scholar]
- 123.Zong, W.-X., L. C. Edelstein, C. Chen, J. Bash, and C. Gelinas. 1999. The pro-survival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-κB that blocks TNF-α-induced apoptosis. Genes Dev. 13:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]