Abstract
Methionine sulfoxide reductases (Msr’s) reduce methionine sulfoxide to methionine and protect bacteria against reactive oxygen intermediates (ROI) and reactive nitrogen intermediates (RNI). Many organisms express both MsrA, active against methionine-(S)-sulfoxide, and MsrB, active against methionine-(R)-sulfoxide. Mycobacterium tuberculosis (Mtb) expresses MsrA, which protects ΔmsrA-E. coli from ROI and RNI (St. John et al., 2001). However, the function of MsrA in Mtb has not been defined, and it is unknown whether Mtb expresses MsrB. We identified MsrB as the protein encoded by Rv2674 in Mtb and confirmed the distinct stereospecificities of recombinant Mtb MsrA and MsrB. We generated strains of Mtb deficient in MsrA, MsrB or both and complemented the mutants. Lysates of singly deficient strains displayed half as much Msr activity as wild type against N-acetyl methionine sulfoxide. However, in contrast to other bacteria, single mutants were no more vulnerable than wild type to killing by ROI/RNI. Only Mtb lacking both MsrA and MsrB was more readily killed by nitrite or hypochlorite. Thus, MsrA and MsrB contribute to the enzymatic defenses of Mtb against ROI and RNI.
Introduction
ROI are critical to the control of Mtb infection in humans, judging from the high incidence of tuberculosis in people with chronic granulomatous disease, a specific deficiency in phagocyte oxidase (NOX2), the principal source of ROI in macrophages (Bustamante et al., 2007). RNI are critical to the control of Mtb infection in mice, based on the failure of mice lacking nitric oxide synthase 2 (NOS2; iNOS) to restrain replication of Mtb or to survive Mtb infection for more than a few weeks (MacMicking et al., 1997). In addition, the administration of NOS2 inhibitors to clinically stable, Mtb-infected wild type mice leads to rapid progression of infection and early death (MacMicking et al., 1997; Scanga et al., 2001). NOS2 deficiency states have not been identified in humans, but human macrophages from Mtb-infected organs have been repeatedly found to express NOS2 (Nicholson et al., 1996; Wang et al., 1998; Facchetti et al., 1999; Wang and Kuo, 2001; Choi et al., 2002; Schon et al., 2004; Samarina, 2005), and killing of mycobacteria in vitro by NOS2-positive human macrophages was blocked by a NOS2 inhibitor (Nozaki et al., 1997). Thus, it is likely that both ROI and RNI contribute to host defense against Mtb.
The persistence of Mtb in many and perhaps most infected people with normal immunity suggests that the microbe has mechanisms to resist ROI and RNI. Many such mechanisms have been identified. Some microbes encode enzymes that break down both ROI and RNI, such as a peroxynitrite reductase and peroxidase comprised of four proteins: alkylhydroperoxide reductase subunit C (AhpC), a thioredoxin-related oxidoreductase called AhpC-neighboring protein D (AhpD), dihydrolipoamide acyltransferase (DlaT) (formerly SucB), and lipoamide dehydrogenase (Lpd) (Bryk et al., 2000; Bryk et al., 2002; Tian et al., 2005). Others help degrade macromolecules (or portions thereof) damaged by RNI, such as UV repair B (Darwin et al., 2003; Darwin et al., 2005) and, presumptively, the proteasomal protease (Darwin et al., 2003; Gandotra et al., 2007) aided by its associated ATPase, Mpa (Darwin et al., 2003; Darwin et al., 2005). Still another class of enzymatic defense of Mtb proteins against ROI and RNI can be postulated: the reversal of specific, potentially harmful oxidations by reduction. To our knowledge, the only precedent for this in Mtb lies in the cyclic oxidation and reduction of antioxidant enzymes themselves as part of their normal reaction mechanism.
Within proteins, the atom most susceptible to oxidation is sulfur, both in cysteine and methionine residues (Weissbach et al., 2002). Exposure to ROI leads to disulfide bonding, the formation of higher oxides of cysteinyl sulfur (sulfenes, sulfines and sulfones), and the generation of methionine sulfoxide. Less well appreciated is that aerobic exposure of bacteria to nitrite or S-nitrosothiols can also lead to disulfide formation (Rhee et al., 2005) and methionine oxidation, most likely via generation of peroxynitrite (St John et al., 2001).
The oxidation of methionine to methionine sulfoxide, whether in proteins or as a free amino acid, results in the formation of equimolar amounts of its two epimers, methionine-(S)-sulfoxide and methionine-(R)-sulfoxide (Weissbach et al., 2002). Many prokaryotes and eukaryotes express a family of methionine sulfoxide reductases (Msr), whose function is to reduce methionine sulfoxide to methionine using thioredoxin, thioredoxin reductase and NADPH as the reducing system (Boschi-Muller et al., 2005). It has been proposed (Levine et al., 1996) that the reversible oxidation of methionine acts as a sink for oxidants that might otherwise carry out less readily reversible reactions. Most organisms express at least one MsrA active on both free and peptidyl methionine-(S)-sulfoxide and one or more MsrBs active on peptidyl (but not free) methionine-(R)-sulfoxide. Additional Msr isoforms or activities have been described, some of which vary in subcellular localization and/or utilization of free and peptidyl methionine sulfoxide (Singh and Moskovitz, 2003; Lin et al., 2007).
Deficiency of one Msr was sufficient to render E. coli (St John et al., 2001), S. aureus (Singh and Moskovitz, 2003), Helicobacter pylori (Alamuri and Maier, 2004), and Mycobacterium smegmatis (Douglas et al., 2004) hypersusceptible to killing by ROI, and in some cases, to RNI. Strains of H. pylori (Alamuri and Maier, 2004) and M. smegmatis (Nino, 2007) deficient in both MsrA and MsrB had little additional phenotype. There have been no reports in which it was necessary to delete both msrA and msrB in one bacterium in order to observe such a phenotype.
Msrs from Mtb have only recently come under study. Recombinant Mtb MsrA was shown to reduce N-acetyl methionine sulfoxide (St John et al., 2001), and its crystal structure has been solved (Taylor et al., 2003). However, the generation of a strain of Mtb deficient in msrA has not been reported, and Mtb MsrB has not yet been identified or characterized. In this work, we have identifed Mtb MsrB as the product of the gene Rv2674, confirmed the stereospecificity of Mtb’s MsrA and MsrB as recombinant proteins and generated single and double deletion mutants of Msr A and Msr B.
Results
Identification of Mtb MsrB and construction of Msr mutants
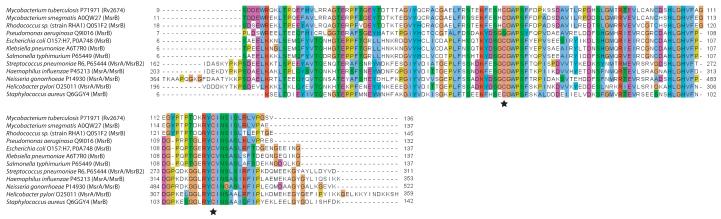
Rv2674 is a 411 bp gene annotated as encoding a conserved hypothetical protein. We performed a multiple sequence alignment against MsrB proteins from E. coli (Grimaud et al., 2001) and N. gonorrhoeae (Skaar et al., 2002) (both of which have been expressed and proven to have Msr activity) as well as putative MsrB sequences from numerous other bacteria (Fig. 1). Rv2674 is 87% identical in amino acid sequence to MsrB from M. smegmatis and 64% identical to MsrB from E. coli and includes two conserved cysteine residues (Cys 67 and Cys 123) as well as adjacent clusters of residues that form the presumptive active site (Lowther et al., 2002).
Fig. 1.
Partial amino acid sequence alignment of Rv2674 with MsrB proteins from selected bacteria.
Alignment was performed with ClustalW and colored following the ClustalX scheme (only partial alignment shown). Stars indicate conserved cysteine residues involved in the reaction mechanism; note the conservation of surrounding residues as well.
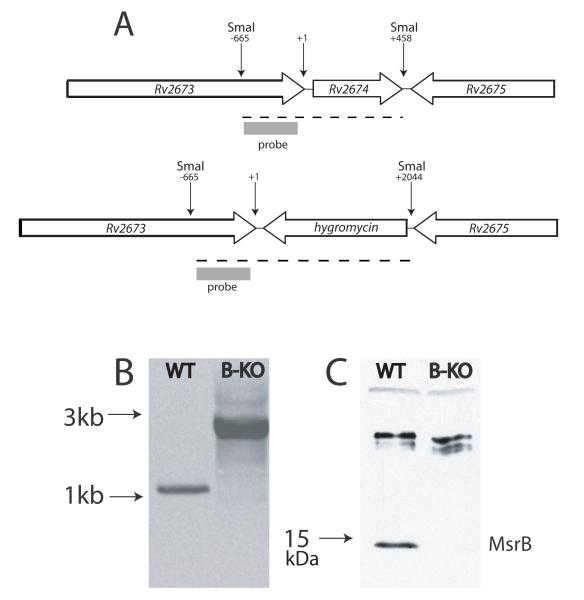
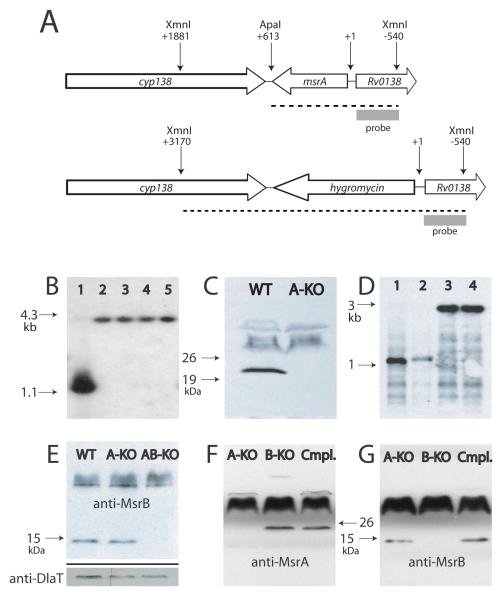
The Rv2674 (msrB) single knockout mutant (H37RvΔmsrB) was constructed in Mtb strain H37Rv by single-step homologous recombination using a specialized transducing phage system. Replacement of the gene by a hygromycin resistance cassette was verified by Southern blotting (Fig. 2A-B). The loss of protein expression was confirmed by Western blotting (Fig. 2C). Although Mtb msrA has been cloned (St John et al., 2001), the knockout has not been reported. Accordingly, the msrA single knockout mutant (H37RvΔmsrA) was constructed in Mtb strain H37Rv and confirmed by the same approaches (Fig. 3A-B). The loss of protein expression was confirmed by Western blotting (Fig. 3C).
Fig. 2.
Generation of MsrB-deficient Mtb.
A. Genomic context of wild type msrB and organization of the disrupted allele. Dashed lines indicate SmaI restriction products.
B. Southern blot of SmaI-restricted DNA from wild type (WT) and msrB-deficient (B-KO) strains.
C. Western blot of lysates of WT and MsrB-deficient strains using rabbit antiserum raised against recombinant MsrB (75 μg lysate/lane).
Figure 3.
Generation of MsrA- and MsrA, B-deficient Mtb.
A. Genomic context of wild type msrA and organization of the disrupted allele. Dashed lines indicate products of restriction with XmnI and ApaI.
B. Southern blot of DNA restricted with XmnI and ApaI from wild type (lane 1) and single msrA-knockout clones (lanes 2-5).
C. Western blot of lysates of wild type (WT) and an msrA-knockout clone (A-KO) (30 μg lysate/lane).
D. Southern blot of DNA restricted with SmaI from wild type Mtb (lane 1), an msrA single knockout (lane 2), an msrB single knockout (lane 3), and an msrA/msrB double knockout strain (lane 4).
E. Western blot of lysates of wild type (WT), MsrA-deficient (A-KO) and MsrA, B-deficient (AB-KO) strains using anti-MsrB antiserum. Lower panel shows a loading control after the membrane shown in the main panel was stripped and reblotted with antiserum against dihydrolipoamide acyltransferase (50 μg lysate/lane).
F., G. Western blots of lysates from MsrA-deficient (A-KO), MsrB-deficient (B-KO) and the doubly-deficient strain complemented with both wild type alleles (Cmpl.), using anti-MsrA (F) or anti-MsrB (G) antisera (65 μg lysate/lane).
A double knockout mutant lacking both msrA and msrB (H37RvΔmsrAB) was subsequently constructed in H37RvΔmsrA by single-step homologous recombination to replace msrB with a streptomycin resistance cassette. Deletion of msrB in H37RvΔmsrA was confirmed by both Southern and Western blotting (Fig. 3D-E). Transformation of H37RvΔmsrAB with an integrative plasmid containing both the msrA and msrB genes and their native promoters (pMV306-msrA/msrB) was successful in restoring expression of both proteins to approximately wild type levels (Fig. 3F-G).
Biochemical characterization of Msr proteins and mutants
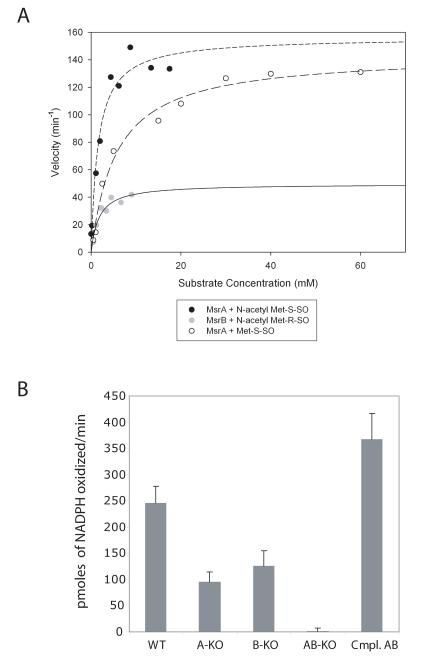
We expressed and purified recombinant MsrB from Mtb and compared it to the MsrA previously purified (St John et al., 2001). MsrA was active against the S epimer of free methionine sulfoxide (but not the R epimer) with Michaelis-Menten kinetics, with a Km of 4.3 ± 0.6 mM and a Vmax of 2.4 ± 0.1 secm=-1 (Vmax/Km 558 ± 81 M−1 s−1) (Fig. 4A). This is comparable to a Km of 1.9 mM reported for MsrA from E. coli (Boschi-Muller et al., 2005). MsrB displayed no activity against either epimer of free methionine sulfoxide either in the presence or absence of 10 μM zinc (data not shown). MsrA and MsrB each displayed activity against N-acetyl methionine sulfoxide, used as a surrogate for peptidyl methionine sulfoxide, in a stereospecific manner. Thus, MsrA reduced N-acetyl methionine-(S)-sulfoxide with a Km value of 1.6 ± 0.4 mM and a Vmax of 2.6 ± 0.2 s−1 (Vmax/Km 1625 ± 425 M−1 s−1), while MsrB reduced N-acetyl methionine-(R)-sulfoxide with a Km of 1.7 ± 0.6 mM and a Vmax of 0.8 ± 0.1 s−1 (Vmax/Km 470.6 ± 176 M−1s−1) (Fig. 4A). The stereospecificity of MsrA and MsrB for peptidyl methionine sulfoxide was confirmed using the oxidized Met(R,S)O form of the opioid pentapeptide Met-enkephalin as a substrate (Supplemental Figure 1).
Fig. 4.
Enzyme activity of recombinant Msrs and cell lysates.
A. Kinetic analysis of recombinant MsrA and MsrB against peptidyl and free methionine sulfoxide epimers. Results are representative of 3 independent experiments.
B. Activity (pmoles of NADPH oxidized/min) of 200 μg of lysate from indicated strains against N-acetyl methionine-(R,S)-sulfoxide. Cmpl. AB refers to the strain deficient in both msrA and msrB after complementation with both genes. Results are the mean ± standard error from 3 independent experiments.
To determine if native MsrA and MsrB also functioned to reduce oxidized methionine and if they accounted for all detectable Msr activity in Mtb lysates, we tested lysates from wild type and mutant strains for activity against N-acetyl methionine sulfoxide. The single knockout strains had intermediate levels of Msr activity, while activity was undetectable in the strain deficient in both MsrA and MsrB. Transformation of the double knockout strain with msrA and msrB restored Msr activity to approximately wild type levels (Fig. 4B).
In vitro growth and susceptibility of Msr mutants to oxidative and nitrosative stresses
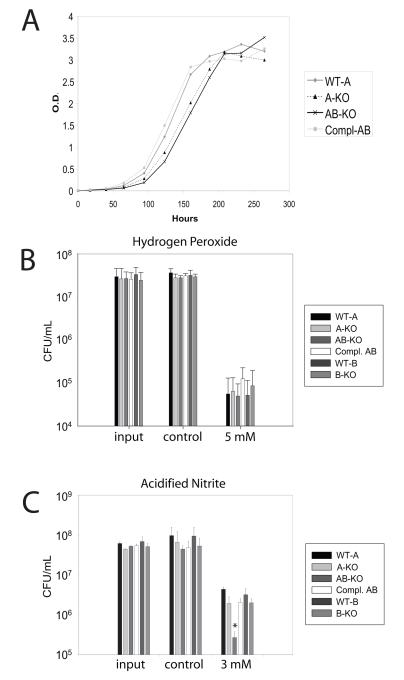
H37RvΔmsrB displayed a rate of growth indistinguishable from wild type (not shown). The growth of H37RvΔmsrA displayed a mild lag, while H37RvΔmsrAB exhibited a greater lag in growth than H37RvΔmsrA. This defect was fully corrected in the complemented strain (H37RvΔmsrAB-pMV306:msrAB) (Fig. 5A). All strains exhibited similar post-lag growth rates, grew to the same final optical density, and upon plating, yielded approximately the same number of colonies (not shown). The minimal growth phenotypes suggested that the mutants were not severely compromised under standard growth conditions.
Fig. 5.
Phenotypes of Msr-deficient strains of Mtb.
A. Growth curve of indicated strains. Compl. AB refers to the strain deficient in both msrA and msrB after complementation with both genes. Curve is representative of 3 independent experiments.
B. Indicated strains were exposed to 5 mM H2O2 or medium alone for 24 hours then plated for colony forming units (cfu). Data are means ± standard error from 4 independent experiments.
C. Indicated strains were exposed to 3 mM sodium nitrite or medium alone (control) at pH 5.5 for 4 days, then plated for cfu. Data are representative of 3 independent experiments. WT-A represents the wild-type strain from which the ΔmsrA strain was derived; WT-B represents the wild-type strain from which the ΔmsrB strain was derived. *p<0.01, AB-KO vs. WT-A
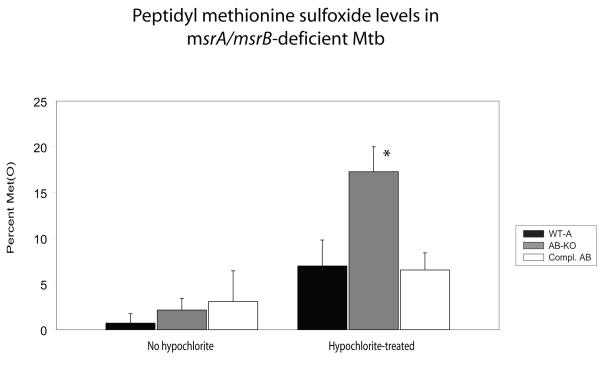
Next, we tested the resistance of the mutant strains to oxidant stresses. Hypochlorite is thought to preferentially oxidize methionine residues in proteins (Vogt, 1995). For H37RvΔmsrA, the minimal inhibitory concentration (MIC) of sodium hypochlorite (NaOCl) was identical to that of the wild type (0.094%). Deficiency of msrB alone caused a two-fold decrease in MIC to NaOCl, Lack of both msrA and msrB caused further sensitivity, with a four-fold decrease in MIC compared to wild type (Table 1). Nonetheless, none of the knockout strains displayed increased sensitivity to H2O2 or cumene hydroperoxide (Fig. 5B and data not shown). To test if hypersusceptibility of H37RvΔmsrAB to hypochlorite was associated with accumulation of peptidyl methionine sulfoxide, we exposed Mtb strains to NaOCl for 15 minutes, washed them and allowed 2 hours for recovery before preparing lysates and quantifying the amount of peptidyl methionine sulfoxide. In keeping with the MIC data, we observed a signficant increase in methionine sulfoxide levels only in proteins from the double knockout strain relative to the wild type and complemented strains (Figure 6).
Table 1.
| M.I.C. to NaOCl | |
|---|---|
| WT-A | 0.094% |
| WT-B | 0.094% |
| A-KO | 0.094% |
| B-KO | 0.047% |
| AB-KO | 0.023% |
| Compl. AB | 0.094% |
Figure 6.
Quantitation of methionine sulfoxide in proteins from Mtb
Wild-type (WT-A), MsrA/MsrB-deficient (AB-KO), and the MsrA/MsrB-deficient strain of Mtb complemented with both genes (Compl. AB) were exposed to sodium hypochlorite (or vehicle control) for 10 minutes then allowed to recover for 2 hours. Lysates from the different strains were analyzed for Met(O) levels. Values are means and standard deviations from two independent experiments, each in duplicate. *p <0.05 for comparison of hypochlorite-treated AB-KO and WT strains
To address the hypothesis that Msr enzymes protect Mtb against nitroxidative stress, we exposed wild type and mutant strains to acidified nitrite. H37RvΔmsrAB was killed to a 40-fold greater extent by 3 mM nitrite (pH 5.5) than wild type H37Rv, while sensitivity of the single knockout strains was similar to wild type. Complementation of msrA and msrB restored the sensitivity of the double knockout strain to wild type levels (Fig. 5C).
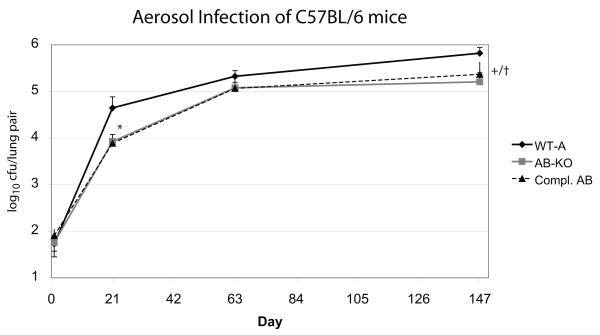
In order to study the in vivo effect of the double mutation, groups of five C57BL/6 mice per strain were infected by inhalation. The number of colony forming units recovered from the lungs 21 weeks later was 0.75 log10 lower for the Mtb H37RvΔmsrAB than for wild type H37Rv, but this phenotype was not complemented (Fig. 7). Failure of complementation was not due to loss of the plasmid, because Mtb grown from the lungs of mice infected for 21 weeks with the complemented strain were fully Msr-positive by western blot (data not shown). Thus we can draw no conclusions about the role of MsrA and B in the pathogenesis of Mtb in the mouse.
Figure 7.
Aerosol infection of C57BL/6 mice with MsrA/MsrB-deficient Mtb.
C57BL/6 mice were infected with 100 CFU of wild-type (WT-A, diamonds) Mtb, MsrA/MsrB-deficient (AB-KO, grey squares) Mtb, or the MsrA/MsrB-deficient strain of Mtb complemented with both genes (Compl. AB, triangles). At the indicated times, five mice were sacrified per group and lung homogenates were plated for CFU. Error bars indicate standard deviations. *p <0.05 for AB-KO vs. WT-A and for complemented strain (Compl. AB) vs. WT-A; +p<0.01 for AB-KO vs. WT-A, †p<0.05 for complemented strain vs. WT-A
Discussion
This work establishes that Mtb, like many other organisms, expresses at least two Msrs, each stereospecific for one of the epimers of methionine sulfoxide. Unlike in other organisms, the collective contribution of MsrA and MsrB to the resistance of Mtb to ROI is limited, in that their dual deficiency increased sensitivity to ROI only four-fold and only to a very strong oxidant, hypochlorite, but not to hydrogen peroxide or an organic peroxide. Deficiency of MsrB alone only mildly increased sensitivity to hypochlorite and the MsrA-deficient strain was indistinguishable in this respect from wild type. The ability of MsrA and MsrB to protect Mtb from hypochlorite may help the organism resist being killed by myeloperoxidase-expressing cells, such as neutrophils and monocytes (Borelli et al., 1999). The lack of sensitivity of Msr-deficient Mtb to cumene hydroperoxide is in contrast to the marked vulnerability to this oxidant exhibited by MsrA-deficient Mycobacterium smegmatis (Douglas et al., 2004). Differences between the antioxidant defenses of Mtb and non-pathogenic mycobacteria are not without precedent (Lama et al., 2006; Stewart, 2000) and may reflect adaptations to the different levels of oxidative stress faced by the organisms or difficulties in using gene disruption to evaluate roles of pathways that are to some extent redundant. The same explanation may account for the greater sensitivity of MsrA-deficient E. coli to oxidants, which was rescued by complementation with msrA from Mtb (St John et al., 2001). In contrast to other organisms (including E. coli and M. smegmatis), Mtb persists inside phagosomes which are highly oxidative and nitrosative environments (Schnappinger et al., 2003). Thus, the lack of impact of Msr deficiency on susceptibility to peroxides may reflect the diversity and effectiveness of other antioxidant defenses of Mtb. These include catalase (Ng et al., 2004), mycothiol and mycothiol reductase (Buchmeier et al., 2003), several thioredoxins (Jaeger et al., 2004) and the peroxiredoxin system (Bryk et al., 2002). In previous work, neither msrA nor msrB were induced in Mtb upon exposure to various stresses, including hydrogen peroxide, hypoxia, thermal stress, and nitric oxide (Schnappinger et al., 2003).
However, our data demonstrate that MsrA and MsrB collectively contributed to the defense mechanisms of Mtb against mildly acidified nitrite. This was so notwithstanding that Mtb encodes many other potential defenses against RNI, including those cited in the introduction and a truncated hemoglobin (Pathania et al., 2002). Unexpectedly, only the absence of both MsrA and MsrB resulted in sensitivity to RNI. If RNI such as peroxynitrite oxidize methionine residues with a potentially lethal effect on Mtb, it would be expected that the resulting methionine sulfoxide would be comprised of 50% of each epimer. In Mtb lacking only MsrA, the methionine-(R)-sulfoxides should be reduced but the methionine-(S)-sulfoxides should accumulate, impacting protein function (Sun et al., 1999). Conversely, in Mtb lacking only MsrB, the methionine-(S)-sulfoxides should be reduced but the methionine-(R)-sulfoxides should accumulate, again with potentially adverse functional consequences. In both cases, sensitivity to RNI should be greater than in wild type Mtb. That it required deletion of both msrA and msrB to see this phenotype suggests that the two enzymes are functionally redundant for defence against nitrite, although we have demonstrated that they act on stereochemically distinct substrates. Perhaps a phenotype resulting from deficiency of only one Msr escaped detection under the conditions of our tests. As a related possibility, there may be a threshold of functionally consequential damage resulting from the oxidation of methionine in proteins that was only exceeded in Mtb when both Msr enzymes were deficient.
Experimental procedures
Mycobacteria and culture conditions
Wild-type M. tuberculosis H37Rv and its mutants were grown at 37°C in Middlebrook 7H9 broth containing 0.2% glycerol, 0.5% bovine serum albumin, 0.05% Tween 80, 0.2% dextrose, and 0.085% sodium chloride (7H9-ADNaCl) or on Middlebrook 7H11 agar containing 10% oleic acid-albumin-dextrose-catalase (7H11-OADC). The antibiotics used were 50 μg/ml hygromycin for the H37RvΔmsrA and ΔmsrB strains and 50 μg/ml hygromycin plus 30 μg/ml streptomycin for the H37RvΔmsrA/ΔmsrB double knockout strain. Kanamycin (30 μg/ml) was added for the H37RvΔmsrA/ΔmsrB double knockout strain that was complemented with pMV306-msrA/msrB.
Sequence alignment of Rv2674 (msrB)
Multiple sequence alignment of Rv2674 with the sequences of MsrB proteins from representative bacteria was performed using ClustalW and analyzed using Jalview (Clamp et al., 2004). Coloring of the alignment was performed following the ClustalX scheme for percent consensus for given residues or sets of residues.
Construction, complementation and confirmation of mutants
H37RvΔmsrA and H37RvΔmsrB strains were created by single step homologous recombination using the temperature-sensitive mycobacteriophage phAE87 (Bardarov et al., 2002). The single knockouts of ΔmsrA and ΔmsrB were generated in H37Rv wild type strains obtained from two different sources (designated WT-A [from ATCC] and WT-B [from the Trudeau Institute], respectively) while the double knockout, ΔmsrA/ΔmsrB, was generated in the ΔmsrA background. Flanking regions of msrA and msrB were amplified from Mtb genomic DNA by PCR. For msrA, primer pairs GSJ16 5′TCGGTACCAAGCCGTTCGCGCCAATGAC and GSJ17 5′GTACTAGACTGTGAGCGCTTCGGAGTTCTC and GSJ18 5′CTGAAGCTTCGTCGGCGCATCACCAC and GCJ 19 5′GTA GATCTGCCGGGCACGAAACCAC were used to amplify the upstream and downstream flanking regions, respectively. For msrB, primer pairs msrB15+ 5′ GTCCACTAGTTTCTCAAACCCCTGCTCGG and msrB16-5′GTCCAAGCTTCGGACAGTTCTAGCTTTGGGC and msrB17+ 5′GTCCTCTAGATATTGCATCAACTCCATTTCGC and msrB18-GTCCGGTACCGAGCAGTTGGTGCACTCGC were used to amplify the upstream and downstream flanking regions, respectively.
Fragments were cloned into pJSC284 such that they flanked the hygromycin cassette, generating the allelic exchange substrate (AES). For the msrB AES, SpeI and HindIII were used for the upstream amplicon and KpnI and XbaI for the downstream segment; for the msrA AES, KpnI/XbaI and HindIII/BglII were used. For generation of the ΔmsrA/ΔmsrB strain, a variant of pJSC284 containing a streptomycin cassette in place of hygromycin was used to create the msrB AES. The resulting cloning vector was digested with PacI and inserted into the PacI site of phAE287. After in vitro packaging and selective pressure, cosmids were used to transform M. smegmatis. Candidate phages were amplified and screened for the allelic exchange substrate by PCR and by restriction digest. Phage containing the AES was used to infect Mtb (wild type or ΔmsrA) at a multiplicity of infection of 10:1, which was then plated on antibiotic-impregnated agar for selection. Colonies of Mtb were screened by PCR and verification of the knockout of msrA or msrB (or both) was performed by Southern blotting, using an Amersham ECL kit in accordance with the manufacturer’s instructions.
For complementation, the H37RvΔmsrA/ΔmsrB strain was transformed with the plasmid pMV306 containing the Mtb msrA gene and a 351-bp upstream DNA sequence which may contain the promoter of the gene as well as the msrB gene and a 250-bp upstream sequence that may contain its promoter (pMV306-msrA/msrB). The H37RvΔmsrA/ΔmsrB strain was grown to mid-log phase, collected by centrifugation, washed three times with 10% glycerol, and transformed by electroporation with pMV306-msrA/msrB. Transformants were selected on 7H11-OADC containing 50 μg/ml hygromycin plus 30 μg/ml kanamycin and streptomycin.
For confirmation of the targeted deletion of msrB, genomic DNAs were prepared from wild-type H37Rv and candidate strains, digested with SmaI, separated by agarose gel electrophoresis, and transferred to nylon membranes. The membrane was probed with a 425-bp fragment containing the upstream msrB 5′ flanking sequence, revealing a 1.1 kbp fragment for wild type Mtb and a 2.7 kbp fragment for the H37RvΔmsrB mutant. For H37RvΔmsrA, genomic DNA was digested with XmnI and ApaI and the membrane was hybridized with an upstream flank probe, revealing a 1.2 kbp fragment for the wild type strain and 3.7 kbp hybridization product for the ΔmsrA mutant.
In addition to Southern blotting, western blotting was used to confirm deletion and complementation, using the anti-MsrA antiserum reported (St John et al., 2001) and anti-MsrB antiserum prepared analogously. Strains were grown to log phase and lysed by bead beating in cold buffer containing 50 mM Tris (pH 8.0) and 1 tablet of complete protease inhibitor cocktail (Roche) per 10 mL of buffer. Proteins were separated by 15% SDS-PAGE and transferred to nitrocellulose membranes. After blocking with 5% low-fat milk, membranes were probed with 1:500 anti-MsrA or 1:1000 anti-MsrB. HRP-conjugated secondary antibodies were used at 1:10000 and blots were developed using the ECL kit from Amersham.
Expression and purification of recombinant MsrA and MsrB
MsrB (Mtb Rv2674) was amplified from Mtb chromosomal DNA using primers PR5 5′GTCCCATATGACGCGCCCAAAGCTAGAAC and PR7 5′ GTCCCTCGAGCACGCTACCGGGGACCAGG and cloned into the NdeI-XhoI sites of pET30b which adds 2 amino acids to the C-terminus and then a tag containing 6 histidine residues. Mtb MsrA was amplified from Mtb chromosomal DNA using primers PR10 5′GTCCCATATGACGAGCAATCAGAAAGCG and PR11 5′GTCCCTCGAGCCCGAGTTCGGGTGAAAGGC and also cloned into the NdeI-XhoI of pET30b. The final construct for MsrA expression contains a deletion of the final two amino acids, which did not affect the catalytic activity of the purified protein and was used for these studies. Both recombinant MsrA and MsrB were overexpressed in E. coli BL21(DE3) cells after a 4 hour induction with 1 mM IPTG. Approximately 15 mg/L each of MsrA and MsrB were obtained and purified to >95% purity by nickel affinity agarose chromatography.
Substrate preparation
The 2 epimers of methionine sulfoxide were separated from free l-methionine-(R,S)-sulfoxide (Sigma) by a reported method (Lavine, 1947). Their purity was determined by HPLC; methionine-(R)-sulfoxide contained 6% contamination with the S epimer, while the S epimer contained no detectable R epimer.
N-acetylation of methionine-(R,S)-sulfoxide was based on the published procedure (Vieira Dos Santos et al., 2005). One mmol of methionine sulfoxide was shaken with 4 mL of acetic glacial acid and 4 mL of acetic anhydride for 2 hours at room temperature. The reaction was quenched with 8 mL of water. The dried product was dissolved in water and applied to a Dowex 50H + cation exchange column to remove unacetylated products. The column was washed with 1 mL of water and the eluate combined with the flow through. The solution was lyophilized and the lyophilate dissolved in 500 μL of water. We used NMR analysis to confirm acetylation of methionine sulfoxide and an enzymatic assay to determine the concentration of N-acetyl methionine sulfoxide. We assumed a 1:1 stoichiometric relationship between the amount of N-acetyl methionine sulfoxide added and the amount of N-acetyl-Met formed and that the N-acetyl methionine sulfoxide was a mixture containing 50% S and 50% R epimers. A fixed volume of substrate was added to a 150 μL reaction mixture containing 50 mM Tris, pH 8.0, 40 μM E. coli thioredoxin (Trx) (Km= 8.9 μM ± 1.4) (Promega), 340 nM E. coli thioredoxin reductase (TrxR) (Sigma), 300 μM NADPH (Sigma), and 40 nM of MsrA. Reactions were run to completion and the change in absorbance was used to determine the quantity of N-acetyl methionine sulfoxide converted to N-acetyl methionine and back-calculations were done to determine the starting concentration of N-acetyl methionine sulfoxide. As a control, known concentrations of methionine sulfoxide were used to confirm the validity of the assay.
Kinetic parameters and enzyme activity in Mtb lysates
Msr activity was measured by following the decrease in absorbance at 340 nm due to oxidation of NADPH at 37 °C. For every mole of methionine produced from N-acetyl methionine-(R,S)-sulfoxide, one mole of NADPH is oxidized. A typical reaction mixture (150 μl) contained 50 mM Tris, pH 8.0, 40 μM E. coli thioredoxin (Trx) (Km= 8.9 μM ± 1.4) (Promega), 340 nM E. coli thioredoxin reductase (TrxR) (Sigma), 300 μM NADPH (Sigma), 40 nM purified recombinant Mtb MsrA or MsrB, and varying concentrations of substrate. Under these conditions, initial rate measurements were proportional to the amount of enzyme added. Initial rate measurements were performed on an Uvikon XL spectrophotometer by monitoring the decrease in absorbance at 340 nm due to oxidation of NADPH (ε=6220 M−1 cm−1). Initial rates were fitted to: ν = (Vmax [S])/( Km + [S]). Background NADPH oxidation was subtracted from the values obtained with methionine-(R,S)-sulfoxide. Vmax and Km values were determined using nonlinear least-squares curve fitting program of SigmaPlot 2002 for Windows, version 8.0. To determine the amount of MsrA and B activity present in Mtb lysates, 200 μg of lysate (Bradford assay) and a saturating concentration of N-acetyl methionine-(R,S)-sulfoxide (20 mM) was added to the reaction mixture.
Susceptibility of Mtb strains to in vitro stresses
Strains were grown to log phase, centrifuged at 800 rpm for 10 min at room temperature to remove clumps, diluted in medium and exposed to 1-10 mM H2O2 for 1-24 hours, followed by plating of serial dilutions on 7H11 agar. Alternatively, strains were grown to log phase, then washed with 7H9 medium that had been acidified to pH 5.5 with HCl, depleted of clumps as above, and diluted in 7H9-ADNaCl (pH 5.5) with or without 3.0 mM NaNO2. Strains were then incubated at 37 °C for 4 days, serially diluted and plated. CFU were counted after 3 weeks. Where indicated, strains were grown to log phase, diluted in 7H9-ADNaCl to an optical density of 0.01 in a 96-well plate and exposed to serial (1:1) dilutions of sodium hypochlorite (0.00075%-0.375%) in medium. After 2-3 weeks, the lowest concentration of NaOCl that completely inhibited growth (measured by optical density at 600 nm) was recorded.
Quantitation of peptidyl methionine sulfoxide from Mtb exposed to sodium hypochlorite
The various strains of Mtb were grown in 7H9 medium to to late log phase. The cultures were centrifuged and washed twice with PBS plus 0.02% Tyloxapol and resuspended in 10 mL of PBS plus 0.02% Tyloxapol to a final OD of 1 in the absence or presence of 250 μM NaOCl. Treated and untreated cultures were incubated for 15 min at 37°C with shaking. Cold PBS (5mL) was added to each culture to dilute the NaOCl and the cultures were then centrifuged, washed twice with 5 mL of 7H9 medium, resuspended in 10 mL of 7H9 medium and allowed to recover for 2 hours at 37 °C with shaking. Cell lysates were prepared as previously described, but in the absence of protease inhibitor. An aliquot containing about 100 μg of protein was dried in a vacuum centrifuge and the residue incubated for 1 hr at 80°C in a solution containing 0.1M CNBr in 70% formic acid. A duplicate sample was removed which was not subjected to CNBr treatment. All samples were hydrolyzed in vacuo in 6N HCl containing 1mM mercaptoethanol, for 24 hrs at 110°C, dried by vacuum centrifugation and the amino acids analyzed on a Beckman model 7300 amino acid analyzer. Under these conditions any methionine sulfoxide in the sample will be quantitatively converted to methionine during the hydrolysis. Because CNBr destroys only methionine, any methionine sulfoxide in the sample will be determined as methionine. The duplicate tube that did not contain CNBr will yield the total methionine content of the sample. Thus % Met(O) is calculated as the methionine recovered after CNBr/total methionine. All samples were normalized by the recovery of Glu, Ala, Leu, Tyr and Phe.
Mouse aerosol infections with wild-type and mutant M. tuberculosis
C57BL/6 mice were infected with logarithmic-phase cultures of M. tuberculosis by the aerosol route, using a Glas-Col inhalation exposure system (Glas-Col Inc., Terre Haute, IN), as described (Shi and Ehrt, 2006). Briefly, twenty mice were infected per strain of Mtb, and five mice were sampled at each of four time points. Animals were exposed for 40 min to an aerosol produced by nebulizing 5 ml of a bacterial suspension in phosphate-buffered saline at a concentration of 2 × 107 bacilli/ml. This resulted in an inoculum of 100 CFU per lung, as determined by plating homogenized lungs onto enriched 7H11 plates at 24 h after the infection.
Mice were sacrificed by inhalation of CO2 on day 1 and at 3, 9, and 21 weeks after the initial infection. Lungs were aseptically removed and homogenized in 4 ml phosphate-buffered saline containing 0.05% Tween 80. Serial dilutions of the lung homogenates were plated on 7H11-OADC plates. CFU were enumerated after 2 to 3 weeks incubation at 37 °C.
Statistical analysis
Student’s unpaired, two-tailed t test was used to compare results for different strains of Mtb. P <0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Jeffrey A. Cox for collaborating in making H37RvΔmsrA and Jean Schneider for technical assistance. W.L.L. was supported by the Canadian Institutes for Health Research and a Detweiler Travelling Fellowship of the Royal College of Physicians and Surgeons of Canada. Part of this work was supported by NIH grant RO1 HL61241 to CN. The Department of Microbiology & Immunology is supported by the William Randolph Hearst Foundation.
References
- Alamuri P, Maier RJ. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004;53:1397–1406. doi: 10.1111/j.1365-2958.2004.04190.x. [DOI] [PubMed] [Google Scholar]
- Bardarov S, Bardarov S, Jr., Pavelka MS, Jr., Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR., Jr Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. Jr. Jr. Jr. [DOI] [PubMed] [Google Scholar]
- Borelli V, Banfi E, Perrotta MG, Zabucchi G. Myeloperoxidase exerts microbicidal activity against Mycobacterium tuberculosis. Infect Immun. 1999;67:4149–4152. doi: 10.1128/iai.67.8.4149-4152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschi-Muller S, Azza S, Branlant G. E. coli methionine sulfoxide reductase with a truncated N terminus or C terminus, or both, retains the ability to reduce methionine sulfoxide. Protein Sci. 2001;10:2272–9. doi: 10.1110/ps.10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschi-Muller S, Olry A, Antoine M, Branlant G. The enzymology and biochemistry of methionine sulfoxide reductases. Biochim Biophys Acta. 2005;1703:231–238. doi: 10.1016/j.bbapap.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Bryk R, Griffin P, Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 2000;407:211–215. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- Buchmeier NA, Newton GL, Koledin T, Fahey RC. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol Microbiol. 2003;47:1723–1732. doi: 10.1046/j.1365-2958.2003.03416.x. [DOI] [PubMed] [Google Scholar]
- Bustamante J, Aksu G, Vogt G, de Beaucoudrey L, Genel F, Chapgier A, Filipe-Santos O, Feinberg J, Emile JF, Kutukculer N, Casanova JL. BCG-osis and tuberculosis in a child with chronic granulomatous disease. J Allergy Clin Immunol. 2007;120:32–38. doi: 10.1016/j.jaci.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Choi HS, Rai PR, Chu HW, Cool C, Chan ED. Analysis of nitric oxide synthase and nitrotyrosine expression in human pulmonary tuberculosis. Am J Respir Crit Care Med. 2002;166:178–186. doi: 10.1164/rccm.2201023. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- Douglas T, Daniel DS, Parida BK, Jagannath C, Dhandayuthapani S. Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J Bacteriol. 2004;186:3590–3598. doi: 10.1128/JB.186.11.3590-3598.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchetti F, Vermi W, Fiorentini S, Chilosi M, Caruso A, Duse M, Notarangelo LD, Badolato R. Expression of inducible nitric oxide synthase in human granulomas and histiocytic reactions. Am J Pathol. 1999;154:145–152. doi: 10.1016/S0002-9440(10)65261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, Derrick PJ, Barras F. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- Jaeger T, Budde H, Flohe L, Menge U, Singh M, Trujillo M, Radi R. Multiple thioredoxin-mediated routes to detoxify hydroperoxides in Mycobacterium tuberculosis. Arch Biochem Biophys. 2004;423:182–191. doi: 10.1016/j.abb.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Lama A, Pawaria S, Dikshit KL. Oxygen binding and NO scavenging properties of truncated hemoglobin, HbN, of Mycobacterium smegmatis. FEBS Lett. 2006;580:4031–41. doi: 10.1016/j.febslet.2006.06.037. [DOI] [PubMed] [Google Scholar]
- Lavine TF. The formation, resolution, and optical properties of the diastereoisomeric sulfoxides derived from L-methionine. J. Biol. Chem. 1947;169:477–492. [PubMed] [Google Scholar]
- Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci U S A. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Johnson LC, Weissbach H, Brot N, Lively MO, Lowther WT. Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function. Proc. Natl. Acad. Sci. 2007;104:9597–9602. doi: 10.1073/pnas.0703774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther WT, Weissbach H, Etienne F, Brot N, Matthews BW. The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB. Nat Struct Biol. 2002;9:348–352. doi: 10.1038/nsb783. [DOI] [PubMed] [Google Scholar]
- MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol. 2004;52:1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- Nicholson S, Bonecini-Almeida Mda G, Lapa e Silva JR, Nathan C, Xie QW, Mumford R, Weidner JR, Calaycay J, Geng J, Boechat N, Linhares C, Rom W, Ho JL. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J Exp Med. 1996;183:2293–2302. doi: 10.1084/jem.183.5.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nino CJ,C, Dhandayuthapani S. Role of Methionine-R-sulfoxide reductase (MsrB) in the intracellular survival of Mycobacterium smegmatis. Abstract Book, Keystone Symposia on Tuberculosis: From Lab Research to Field Trials. 2007:83. [Google Scholar]
- Nozaki Y, Hasegawa Y, Ichiyama S, Nakashima I, Shimokata K. Mechanism of nitric oxide-dependent killing of Mycobacterium bovis BCG in human alveolar macrophages. Infect. Immun. 1997;65:3644–3647. doi: 10.1128/iai.65.9.3644-3647.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania R, Navani NK, Gardner AM, Gardner PR, Dikshit KL. Nitric oxide scavenging and detoxification by the Mycobacterium tuberculosis haemoglobin, HbN in Escherichia coli. Mol Microbiol. 2002;45:1303–1314. doi: 10.1046/j.1365-2958.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: Enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci U S A. 2005;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarina A. Thesis. Karolinska Institutet; 2005. [Google Scholar]
- Scanga CA, Mohan VP, Tanaka K, Alland D, Flynn JL, Chan J. The inducible nitric oxide synthase locus confers protection against aerogenic challenge of both clinical and laboratory strains of Mycobacterium tuberculosis in mice. Infect Immun. 2001;69:7711–7717. doi: 10.1128/IAI.69.12.7711-7717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan I, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik G. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon T, Elmberger G, Negesse Y, Pando RH, Sundqvist T, Britton S. Local production of nitric oxide in patients with tuberculosis. Int J Tuberc Lung Dis. 2004;8:1134–1137. [PubMed] [Google Scholar]
- Shi S, Ehrt S. Dihydrolipoamide acyltransferase is critical for Mycobacterium tuberculosis pathogenesis. Infect Immun. 2006;74:56–63. doi: 10.1128/IAI.74.1.56-63.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Moskovitz J. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology. 2003;149:2739–2747. doi: 10.1099/mic.0.26442-0. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Tobiason DM, Quick J, Judd RC, Weissbach H, Etienne F, Brot N, Seifert HS. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc Natl Acad Sci U S A. 2002;99:10108–10113. doi: 10.1073/pnas.152334799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, Nathan C. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci U S A. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Gao J, Ferrington DA, Biesiada H, Williams TD, Squier TC. Repair of oxidized calmodulin by methionine sulfoxide reductase restores ability to activate the plasma membrane Ca-ATPase. Biochemistry. 1999;38:105–112. doi: 10.1021/bi981295k. [DOI] [PubMed] [Google Scholar]
- Taylor AB, Benglis DM, Jr., Dhandayuthapani S, Hart PJ. Structure of Mycobacterium tuberculosis methionine sulfoxide reductase A in complex with protein-bound methionine. J. Bacteriol. 2003;185:4119–4126. doi: 10.1128/JB.185.14.4119-4126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Bryk R, Shi S, Erdjument-Bromage H, Tempst P, Nathan C. Mycobacterium tuberculosis appears to lack alpha-ketoglutarate dehydrogenase and encodes pyruvate dehydrogenase in widely separated genes. Mol Microbiol. 2005;57:859–868. doi: 10.1111/j.1365-2958.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- Vieira Dos Santos C, Cuine S, Rouhier N, Rey P. The Arabidopsis plastidic methionine sulfoxide reductase B proteins. Sequence and activity characteristics, comparison of the expression with plastidic methionine sulfoxide reductase A, and induction by photooxidative stress. Plant Physiol. 2005;138:909–922. doi: 10.1104/pp.105.062430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt W. Oxidation of methionyl residues in proteins: tools, targets, and reversal. Free Radic Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- Wang CH, Liu CY, Lin HC, Yu CT, Chung KF, Kuo HP. Increased exhaled nitric oxide in active pulmonary tuberculosis due to inducible NO synthase upregulation in alveolar macrophages. Eur Respir J. 1998;11:809–815. doi: 10.1183/09031936.98.11040809. [DOI] [PubMed] [Google Scholar]
- Wang CH, Kuo HP. Nitric oxide modulates interleukin-1beta and tumour necrosis factor-alpha synthesis, and disease regression by alveolar macrophages in pulmonary tuberculosis. Respirology. 2001;6:79–84. doi: 10.1046/j.1440-1843.2001.00302.x. [DOI] [PubMed] [Google Scholar]
- Weissbach H, Etienne F, Hoshi T, Heinemann SH, Lowther WT, Matthews B, St John G, Nathan C, Brot N. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch Biochem Biophys. 2002;397:172–178. doi: 10.1006/abbi.2001.2664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.