Abstract
Evolution hinges on the ability of organisms to adapt to their environment. A key regulator of adaptability is mutation rate, which must be balanced to maintain genome fidelity while permitting sufficient plasticity to cope with environmental changes. Multiple mechanisms govern an organism's mutation rate. Constitutive mechanisms include mutator alleles that drive global, permanent increases in mutation rates, but these changes are confined to the subpopulation that carries the mutator allele. Other mechanisms focus mutagenesis in time and space to improve the chances that adaptive mutations can spread through the population. For example, environmental stress can induce mechanisms that transiently relax the fidelity of DNA repair to bring about a temporary increase in mutation rates during times when an organism experiences a reduced fitness for its surroundings, as has been demonstrated for double-strand break repair in Escherichia coli. Still, other mechanisms control the spatial distribution of mutations by directing changes to especially mutable sequences in the genome. In eukaryotic cells, for example, the stress-sensitive chaperone Hsp90 can regulate the length of trinucleotide repeats to fine-tune gene function and can regulate the mobility of transposable elements to enable larger functional changes. Here, we review the regulation of mutation rate, with special emphasis on the roles of tandem repeats and environmental stress in genome evolution.
Key Words: Microsatellite repeats, Genome instability, Stress-induced mutagenesis, Evolution
Introduction
Inherited variation is the raw material upon which natural selection acts, allowing organisms in a population to survive changes in their environment. Mutation rate is an important determinant of this variation and is likely the product of factors beyond the simple mechanical limitations of DNA repair proteins. The discovery of mutator and anti-mutator alleles, polymerases with different fidelities, and inducible error-prone repair suggests mechanisms for regulating mutation rate [reviewed in Caporale, 2003]. In addition, mutational hotspots can bias mutagenesis to specific sites such as double-strand breaks (DSBs). This review focuses on one type of mutational hotspot: tandem repeats, which are an important source of genetic variation in most organisms and have been demonstrated to facilitate adaptability through the modulation of gene function [reviewed in Gemayel et al., 2010].
The modern evolutionary synthesis suggests mutations are random with respect to their effect on fitness, and that there is independence between mutation and selection. Consequently, selection would only operate on genome variation that exists within a population prior to the selection. This variation is proposed to accumulate at a constant rate, regardless of selective pressures and environmental stressors [Kimura, 1968]. These ideas have their genesis in August Weismann's proposal that somatic cells are derived from germline cells, and that while mutations in somatic cells could be expressed within an individual, these variations were not passed on to offspring. Conversely, he proposed that mutations in the germline, while not expressed in the individual, could be passed on to and expressed by an organism's offspring. This idea contradicted the Lamarckian theory that organisms adapted in direct response to their environment and subsequently passed those adaptations on to their progeny; therefore implying that mutation, environmental effects, and selection are separate phenomena [Kutschera and Niklas, 2004; Pigliucci, 2008].
This view was further supported by the classic fluctuation test experiments of Luria and Delbrück [1943]. This began with the observation that bacteria exposed to T1 bacteriophages occasionally displayed resistance to the phages. However, it was unclear whether this resistance was already present in the bacterial population, or arose in response to the selective pressure of the phages. When a number of independent bacterial colonies were plated, the number of phage-resistant mutants varied widely from colony to colony, suggesting that resistance-conferring mutations were already present in each population [Luria and Delbrück, 1943]. Thus, the experiment suggested that mutation and selection are separate processes.
However, the stress placed on the bacterial population in Delbrück and Luria's experiment was both sudden and extreme. Therefore, such an experiment might not comprehensively model all stresses facing evolving populations. Indeed, a growing body of evidence suggests that selection and mutation may not be entirely independent. For example, the offspring of male mice fed a low-protein diet exhibit elevated expression of genes for lipid and cholesterol biosynthesis in their livers, relative to offspring of males that receive a control diet. The altered gene expression is a result of epigenetic changes in the paternal mice, and therefore illustrates that a selection pressure in the environment could result in heritable changes to the genome [Carone et al., 2010]. Similarly, environmental stress increases the mutation rate in diverse species, including bacteria, yeast, and mammals [Cairns and Foster, 1991; Coyle and Kroll, 2008; Forche et al., 2011; Galhardo et al., 2007; McKenzie et al., 2000; Mittelman et al., 2010]. Such a connection between selection and mutation could even be evolutionarily advantageous, even if it was not evolved for that purpose, and provides a mechanism for organisms to rapidly adapt to environmental change [Galhardo et al., 2007; Mittelman et al., 2010; Rando and Verstrepen, 2007; Shee et al., 2011].
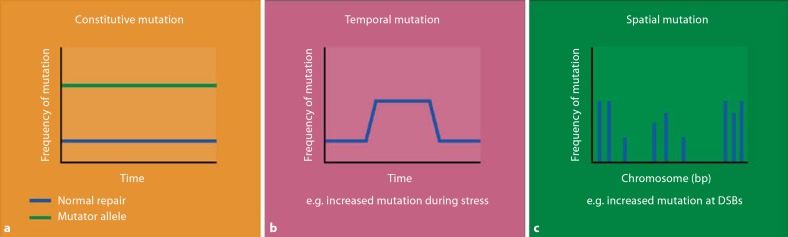
The ability of an organism to undergo mutation in response to environmental stresses suggests an ability to alter the rate of mutation, which could take effect globally or in specific genomic regions, and temporarily or permanently. Mutation rates could be altered globally and permanently by the presence of mutator alleles, temporarily due to transient events such as environmental stress, or locally at ‘hotspot’ locations in the genome (fig. 1). Certainly from an evolutionary perspective, it would be advantageous for mutation to be restricted in both time and space, since most functional mutations are likely to be deleterious. The focus of this review is to highlight findings on the effect of each of these factors on repeat mutation and genomic stability. Many tandem repeats are functionally important and can modulate morphological, behavioral and life history traits through quantitative effects on gene function. In addition, repeats are mutational hotspots and are further mutated by constitutive and stress-induced pathways of mutagenesis.
Fig. 1.
DNA mutagenesis can be regulated constitutively, temporally, and spatially. Constitutive mechanisms (a) include mutator alleles that drive global, permanent increases in mutation rates. Other mechanisms focus mutagenesis in time (b) and space (c) to improve the chances producing adaptive mutations, while minimizing deleterious mutations.
Repeats as Agents of Adaptability
Tandem repeats are dispersed throughout the genome, in and around gene regions. They are highly variable in most organisms and encode their own mutability through the unit size, length, and purity of the repeat tract [King and Kashi, 2007]. The high mutation rate of most repeat sequences led to initial assumptions that these sequences were ‘junk DNA’ or not functionally important. However, about 20 years ago, triplet repeats were identified as agents of disease. Since then, several microsatellite repeats (not all of which are triplets) have been identified as the underlying basis for a wide range of neurological and morphological disorders in humans and other mammals [Albrecht and Mundlos, 2005; Lopez Castel et al., 2010; Orr, 2009].
In addition to causing disease, microsatellites can exert subtle effects on gene function and quantitative traits. Coding microsatellites are enriched in transcription factors and other regulatory proteins, where changes in repeat length incrementally impact gene function [Albrecht et al., 2004; Gerber et al., 1994; Verstrepen et al., 2005]. Variations in the lengths of noncoding repeats in the promoters of genes have been shown to quantitatively affect transcription and can facilitate transcriptional plasticity [Vinces et al., 2009]. Emerging evidence implicates coding and noncoding microsatellites as important sources of functional genetic variation in most species [reviewed in Gemayel et al., 2010]. Particularly in mammals, repeats have been shown to affect morphological and behavioral traits [Fondon and Garner, 2004; Hammock and Young, 2005].
Mutator Alleles
Several genes involved in DNA metabolism are essential, reflecting a critical requirement for maintaining DNA fidelity for fitness. Mutations in genes involved in DNA repair, such as those required for repair of mismatched bases, can increase the global mutation rate (fig. 1a). This is particularly relevant for asexual organisms where mutations that affect the global mutation rate (termed ‘mutator alleles’) can give rise to hypermutable organisms within a population [Whittam et al., 1998]. Many mutator alleles that have been identified are in the mismatch DNA repair (MMR) pathway, impairing the cell's ability to recognize and correct mispaired bases. The presence of mutator alleles can lead to decreased stringency when an organism undergoes homologous recombination (HR), because HR is highly dependent on MMR to prevent recombination between divergent DNA sequences [Rayssiguier et al., 1989; Surtees et al., 2004]. Therefore, impairment of MMR can lead to mutations that arise from misincorporated bases during DNA replication that remain unrepaired, or through genomic rearrangements caused by aberrant recombination.
The absence of recombination in asexual organisms enables mutator alleles to ‘hitchhike’ to high frequencies via linkage to any beneficial mutations they produce. In contrast, mutator alleles would not be expected to persist in sexual populations because recombination separates them from any beneficial mutations, and the ability to hitchhike is lost. However, in humans, mutator alleles have been described in somatic cells and are associated with certain cancers [reviewed in Loeb, 2001]. In particular, tandem repeats are destabilized by mutator alleles, and this is a major source of genetic instability in MLH1-deficient colon cancers [Bacon et al., 2000; Simpkins et al., 1999].
Stress-Induced Mutation
Although an increased global mutation rate could give rise to advantageous mutations, its utility is confined to asexual populations and the rare advantage mutations come at an incredible cost or mutational load. An alternative mechanism for increased mutation rate is stress-induced, or adaptive mutation which transiently upregulates mutation in response to environmental stress (fig. 1b). Although it occurs in multiple organisms, stress-induced mutation has been most clearly described in bacteria using the lac+ frameshift reversion assay [Cairns and Foster, 1991], which has lead to the identification of two distinct mechanisms of adaptive mutation. First, after encountering a stressful environment (i.e. nutrient starvation), stationary phase cells (considered to be nondividing cells) can create an ‘adaptive’ frameshift reversion in an inactive lactose gene (a lac+ frameshift reversion) allowing the cells to utilize lactose in the environment as a carbon source. A second mechanism occurs via amplification of a low-expressing gene to high copy number in order to produce a sufficient amount of the protein for growth. The amplified copies of the gene allowing adaptation are unstable and are easily lost from a culture in the absence of selective stress. Therefore, adaptive amplification represents a method by which bacteria can transiently adapt to a stressful environment and then rapidly return to their original state if the environment changes again.
Although the induction of an adaptive mutation is one possible outcome of stress, stressed cells also accumulate nonadaptive mutations at a higher rate than nonstressed cells, indicating that cells experiencing stress have an increased mutation rate genome-wide [Gonzalez et al., 2008; Torkelson et al., 1997]. This increase in mutation rate is not a general property of the cells that accumulate mutation. Instead, it is limited to times of stress, as shown by a requirement for the activation of at least three different stress responses: the cell envelope stress response [Gibson et al., 2010], the general stress response [Layton and Foster, 2003; Lombardo et al., 2004] and the SOS DNA damage stress response [McKenzie et al., 2000]. The mutation rate is increased 1,000-fold by the presence of a DSB in DNA [Ponder et al., 2005] and the mechanism underlying the increase in mutation rate appears to be a switch from high fidelity to error-prone repair of the DSB. The error prone polymerases DinB and Pol II are responsible for introducing point mutations during double-strand break repair (DSBR) in stressed cells [Frisch et al., 2010; Galhardo et al., 2009] because they are able to outcompete the non-error-prone polymerases at their stress-induced levels [Hastings et al., 2010]. Interestingly, recent work has indicated that the switch from high fidelity to error-prone DSBR is not necessary for the resolution of the DSB [Shee et al., 2011]. These results are very provocative in that they indicate stress-induced mutagenesis at DSBs is not simply a product of defective DNA repair. The opposition to the hypothesis of an optimized mutation rate has long been based on the argument that mutation is a ‘mechanical inevitability’, or the byproduct of physical limitations or defects in the fidelity of DNA repair processes, and not the product of natural selection.
In contrast to point mutation mechanisms in which polymerase errors are responsible for an increased mutation rate, adaptive amplification is hypothesized to occur via a transcription-coupled, microhomology-mediated, break-induced replication mechanism [Hastings et al., 2009a]. In humans, it is also hypothesized to underlie copy-number variation in humans [Hastings et al., 2009b]. Adaptive amplification is restricted temporally to times of stress, and through coupling mutation to transcription, amplification is also restricted spatially to those regions of the genome experiencing high transcriptional activity; the very regions that have the best potential for yielding an adaptive advantage. The ability to restrict mutations not only temporally but also spatially within the genome is critical for minimizing the accumulation of deleterious mutation, and is not unique to bacteria as multiple organisms have specific genomic regions that are more mutable and therefore classified as mutational hotspots (fig. 1c).
Repeats as Mutational Hotspots
Mutational hotspots are governed by a number of factors including the sequence and structure of the DNA itself [Wang et al., 2008]. Pathogenic bacteria have numerous hypervariable loci, termed contingency loci, which contain repetitive DNA elements and encode virulence factors critical for host-pathogen interaction. The hypervariability of these loci comes from slippage of the repetitive DNA, and creates variability in the population [Field et al., 1999; Moxon et al., 2006]. Tandem repeats are highly variable and prone to expansion and contraction mutations that result in the insertion or deletion of a repeated unit sequence. Several features of repeats including the purity and size of the repeated unit, as well as the length of the repeat tract, affect the mutation rate of repeats [Fondon et al., 1998; Legendre et al., 2007]. Microsatellite instability has also been shown to be induced by nearly any aspect of DNA metabolism including transcription [Lin and Wilson, 2007], methylation [Gorbunova et al., 2004], mismatch repair [Jaworski et al., 1995; Schweitzer and Livingston, 1997], nucleotide excision repair [Panigrahi et al., 2002] and base-excision repair [Kovtun et al., 2007]. Many of these processes have been proposed to affect repeat stability through ‘correction’ of slipped-strand structures that can arise during DNA metabolism [Parniewski et al., 2000]. Repeats are therefore mutational hotspots because the repair or even transcription of a repeat sequence is very likely to induce mutations in the sequence.
DSBR in Repeat Regions
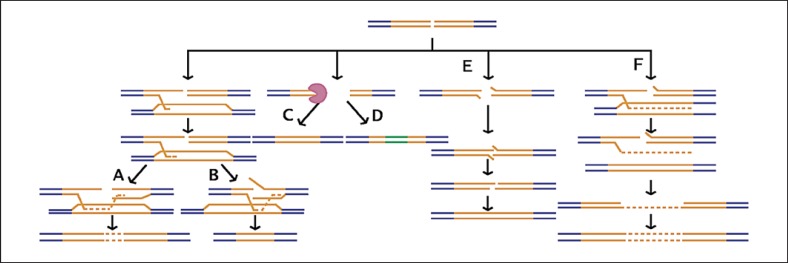
Repetitive DNA sequences are prone to DSBs [Cleary et al., 2002; Jankowski et al., 2000; Marcadier and Pearson, 2003; Nag and Kurst, 1997], and error-prone repair of these breaks is one mechanism by which repeats can mutate. DSBs in repeat regions can be repaired by diverse mechanisms including HR, nonhomologous end joining (NHEJ), and RAD51-independent strand annealing pathways (single-strand annealing, SSA; synthesis-dependent strand annealing) of the broken DNA ends (fig. 2). The majority of breaks are improperly repaired and result in the addition or loss of complete repeat units. But some breaks in repetitive DNA are repaired by insertion of nonrepeat bases by the mutation-prone DNA polymerase kappa in a way that disrupts the repeat [Hile and Eckert, 2008]. The exact contribution of DSBR pathways to repeat mutation is not yet clear, and is an area of active study. In one study, microsatellite repeats were destabilized by the impairment of RAD51 [Mittelman et al., 2010]. Since SSA increases in the absence of RAD51 [Mansour et al., 2008], it is likely that SSA contributes to the increased microsatellite instability in the absence of RAD51.
Fig. 2.
Multiple DSBR pathways may contribute to the mutagenesis of tandem repeats. A: repair by HR with the insertion of repeat units; B: repair by HR with the deletion of repeat units; C: repair by NHEJ with a deletion of any number of bases that can disrupt the repeated unit pattern; D: repair by NHEJ with the insertion of nonrepeat bases; E: repair by SSA with the loss of a single or few repeat units; F: repair by synthesis-dependent strand annealing with the replication of additional repeat units. The orange lines indicate the repeat region, the blue lines indicate flanking nonrepetitive sequence, and the green lines indicate inserted nonrepeat sequence. The dashed orange lines indicate new synthesis of repeat region and the purple pac-man represents degradation of DNA during NHEJ.
HSP90 and Stress
Genetic variation is easily described in terms of mutation and mutation rates. In contrast, phenotypic variation is easily visible, but is much more difficult to define because its sources can include multiple genetic and epigenetic factors. In eukaryotes, HSP90 is a molecular chaperone that functions to repair or activate proteins, but, in addition, can buffer genetic variation by forcing proteins to fold properly in spite of small changes to their sequence. During times of stress, such as heat shock, the number of proteins that require assistance in folding is increased, diverting HSP90 from its normal functions and revealing the cryptic genetic variation in the form of novel phenotypes. Titration of HSP90 during environmental stress is one proposed mechanism by which plants and animals can rapidly adapt to their environment.
Impairment of HSP90 also results in an increase in mutation rate of CAG repeats [Mittelman et al., 2010], suggesting an additional mechanism by which HSP90 can facilitate adaptation to stress: by mediating a switch from stable to unstable microsatellites. In bacteria, stress-mediated expansion of a triplet repeat resulted in a switch in the reductive function of a protein, possibly facilitating adaptation to oxidative stress [Ritz et al., 2001]. Although a beneficial role for stress-induced repeat instability has not been clearly demonstrated in eukaryotic cells, it is possible that this is one mechanism by which cells could facilitate adaptation to environmental stress. At minimum, Hsp90-induced repeat mutation might underlie some of the novel animal and plant phenotypes reported following Hsp90 impairment [Mittelman and Wilson, 2010]. The discovery that Hsp90 plays a role in the maintenance of genome suggests the environment can modulate genome variation, further connecting the forces of selection and mutation.
DSBR and HSP90
The HSP90 chaperone has now been demonstrated to affect many aspects of DNA repair and many of these interactions are summarized in table 1. HSP90 is a key component of HR, possibly through its regulation of BRCA2 folding. BRCA2 is an essential HR protein and mediates RAD51 filament formation [Jensen et al., 2010; Liu et al., 2010]. Inhibition of HSP90 has been shown to lead to altered RAD51 activity and also the degradation of BRCA2 [Noguchi et al., 2006]. Significantly, depletion of HSP90 and RAD51 using siRNA led to similar levels of repeat instability [Mittelman et al., 2010]. The increased instability in the absence of Rad51 likely indicates that breaks in repeats are normally repaired by HR in a conservative and faithful manner, but in the absence of RAD51, DSBR activity is altered allowing an alternative, error-prone pathway of repair. For example, in the presence of RAD51, strand invasion during HR may use a longer region for the homology search, having a greater chance for unique sequence flanking the repetitive DNA to be included in the alignment prior to recombination, while a shorter region of homology may lead to misalignment of repetitive DNA and expansion or contraction.
Table 1.
Hsp90 interacts with proteins involved in diverse pathways of DNA repair
| Protein/complex | Role | Method of interaction | Reference |
|---|---|---|---|
| ATR | CHK1 activation and DNA damage response | direct | Ha et al. [2011] |
| BRCA2 | HR of DSBs | direct | Noguchi et al. [2006] Dungey et al. [2009] |
| CHK1 | cell cycle checkpoint and DNA damage response | indirect through ATR | Ha et al. [2011] Arlander et al. [2003] |
| DNA-PKcs/ERBB1 | damage response to radiation | direct through ERBB1; contributes to HSP90 stability | Dote et al. [2006] Kang et al. [2008] |
| FANC complex | DNA crosslink repair | direct through FANCA | Oda et al. [2007] |
| MLH1 | mismatch DNA repair | possible functional interaction | Fedier et al. [2005] |
| MRE11/RAD50/NBS1 | MRN complex, repair of DSBs | direct through NBS1 | Dote et al. [2006] |
| PIDD | p53-induced, NF-κB activation | Hsp90 binds cytoplasmic PIDD | Tinel et al. [2011] |
| Polymerase eta | translesion synthesis polymerase | direct; disrupts interaction with PCNA | Sekimoto et al. [2010] |
| RAD51 | HR | indirect through BRCA2; possibly other interactions | Noguchi et al. [2006] Mittelman et al. [2010] |
| REV-1 polymerase | translesion synthesis polymerase | direct; disrupts interaction with PCNA | Mayca Pozo et al. [2011] |
| USP50/WEE1 | cell cycle inhibition during DNA damage | direct through USP50 | Aressy et al. [2010] |
Although the role of RAD51 in HR at DNA breaks is well established, recent work has indicated that BRCA2 and RAD51 also play an important role in protecting stalled replication forks from degradation [Schlacher et al., 2011]. Replication forks may stall at secondary structures that form in repetitive DNA. The absence or alteration of RAD51 activity at these replication forks may allow for slippage structures to form, resulting in repeat instability.
Furthermore, the instability induced by HSP90 inhibition may not be completely due to HR and RAD51. HSP90 has been implicated in several different DNA repair pathways as it has been shown to interact with the MRN complex [Dote et al., 2006]. The MRN complex recruits both HR and NHEJ proteins to DNA breaks as well as regulates cell cycle proteins [Kim et al., 2005]. HSP90 has also been shown to interact with polymerase η [Sekimoto et al., 2010] and REV [Mayca Pozo et al., 2011], translesion synthesis polymerases which may contribute to repeat instability. The induction of repeat instability by HSP90 impairment or stress likely involves a switch to a more error-prone repair in at least the DSBR pathway. However, it is also possible that other stress-sensitive proteins regulate genome stability as well and that these stress-induced pathways might operate through multiple DSBR pathways as well as other repair pathways. This is supported by recent studies that show stress-induced mutagenesis in animals targets more than just tandem repeats.
Stress-Induced Transposon Activation
HSP90 has recently been shown to have a broader impact on genomic stability through regulation of the Piwi-interacting RNA pathway [Gangaraju et al., 2011; Specchia et al., 2010]. piRNAs are a special class of siRNAs whose main role is transposon silencing in the germline. In E. coli, stress-induced transposon activation can inactivate genes and activate otherwise cryptic operons, potentially providing an adaptive advantage to cells [Hall, 1999; Zhang and Saier, 2009]. Although most TE families are not mobile in human genomes, some long interspersed nuclear element 1 (LINE1 or L1) retrotransposons are still active. piRNA has been shown to silence L1 retrotransposition [Siomi et al., 2011]. Interestingly, transposons have been shown to integrate into repetitive DNA [Mancuso et al., 2010; Pan et al., 2010]. Retrotransposition of L1 into the genome may create insertions or deletions, affect gene expression, or alter the splicing of genes. Transposon integration into or excision from repeats might stimulate recombination, increasing microsatellite instability as well [discussed in Yant et al., 2005]. L1 mRNA is most commonly found in meiotic cells, but L1 insertions have been identified in neuronal progenitor cells, and have been shown to influence cell fate [Muotri et al., 2005]. It will be interesting in the future to examine whether activation of L1 or other transposons in neurons can be linked to microsatellite instability and/or neurological disorders. Microsatellite instability can impact social behavior [Hammock and Young, 2005], but whether such a case can be demonstrated for L1 retrotransposition remains to be seen.
Conclusion
Although there was an initial separation established between the forces of selection and mutation, emerging studies now suggest that the genome is more sensitive to the environment than previously suspected. The genome is dynamic and likely responsive to environmental stressors that might include heat, infection, inflammation, toxins, and changes in diet. There are many classes of mutation, all with different properties and rates of change; and some sites in the genome are more error prone than others (such as regions that accumulate DSBs).
Tandem repeats are an ideal model for studying the stability of the genome since they are mutational hotspots. Furthermore, repeat mutation is influenced by mutator alleles that cause variation on a genome-wide scale and is responsive to environmental stress. Importantly, repeats are also sources of functional variation upon which selection can act. The connection between HSP90 and microsatellites is particularly intriguing since it provides a possible mechanism to differentially mutate genes containing microsatellites. One area of future exploration will be to examine microsatellite instability on a genome-wide scale to determine the extent to which HSP90 inhibition induces changes to tandem repeats. The exact pathways that facilitate mutagenesis at repeats and other sites in the genome are of significant interest and will be an active area of research for the future as well.
Finally, repeats are likely just the first of many targets of stress-induced mutagenesis. So far, the additional targets include transposable elements and epigenetic patterns. Fortunately, fast and inexpensive sequencing methods such as next-generation sequencing technologies are poised to enable the comprehensive monitoring of genomes under stress. Next-generation sequencing has been used to document the genetic changes that underlie the transition from a normal cell to a malignant tumor cell [Ley et al., 2010; TCGA, 2008]. Future studies using whole-genome sequencing will dramatically enhance our understanding of stress-induced mutagenesis and reveal just how malleable the human genome really is.
Acknowledgements
We would like to thank Dr. John Wilson, Zalman Vaksman, John McCormick, and members of the Mittelman lab for helpful comments and manuscript suggestions. This work was supported by a grant from the NIH/NINDS (NS064762) to D.M., and an award through the NVIDIA Foundation's ‘Compute the Cure’ program to D.M.
References
- Albrecht A, Mundlos S. The other trinucleotide repeat: polyalanine expansion disorders. Curr Opin Genet Dev. 2005;15:285–293. doi: 10.1016/j.gde.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Albrecht AN, Kornak U, Boddrich A, Suring K, Robinson PN, Stiege AC, Lurz R, Stricker S, Wanker EE, Mundlos S. A molecular pathogenesis for transcription factor associated poly-alanine tract expansions. Hum Mol Genet. 2004;13:2351–2359. doi: 10.1093/hmg/ddh277. [DOI] [PubMed] [Google Scholar]
- Aressy B, Jullien D, Cazales M, Marcellin M, Bugler B, Burlet-Schiltz O, Ducommun B. A screen for deubiquitinating enzymes involved in the G/M checkpoint identifies USP50 as a regulator of HSP90-dependent Wee1 stability. Cell Cycle. 2010;9:3815–3822. doi: 10.4161/cc.9.18.13133. [DOI] [PubMed] [Google Scholar]
- Arlander SJ, Eapen AK, Vroman BT, McDonald RJ, Toft DO, Karnitz LM. Hsp90 inhibition depletes Chk1 and sensitizes tumor cells to replication stress. J Biol Chem. 2003;278:52572–52577. doi: 10.1074/jbc.M309054200. [DOI] [PubMed] [Google Scholar]
- Bacon AL, Farrington SM, Dunlop MG. Sequence interruptions confer differential stability at microsatellite alleles in mismatch repair-deficient cells. Hum Mol Genet. 2000;9:2707–2713. doi: 10.1093/hmg/9.18.2707. [DOI] [PubMed] [Google Scholar]
- Cairns J, Foster PL. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporale LH. Natural selection and the emergence of a mutation phenotype: an update of the evolutionary synthesis considering mechanisms that affect genome variation. Annu Rev Microbiol. 2003;57:467–485. doi: 10.1146/annurev.micro.57.030502.090855. [DOI] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, Weng Z, Hofmann HA, Friedman N, Rando OJ. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- Coyle S, Kroll E. Starvation induces genomic rearrangements and starvation-resilient phenotypes in yeast. Mol Biol Evol. 2008;25:310–318. doi: 10.1093/molbev/msm256. [DOI] [PubMed] [Google Scholar]
- Dote H, Burgan WE, Camphausen K, Tofilon PJ. Inhibition of Hsp90 compromises the DNA damage response to radiation. Cancer Res. 2006;66:9211–9220. doi: 10.1158/0008-5472.CAN-06-2181. [DOI] [PubMed] [Google Scholar]
- Dungey FA, Caldecott KW, Chalmers AJ. Enhanced radiosensitization of human glioma cells by combining inhibition of poly(ADP-ribose) polymerase with inhibition of heat shock protein 90. Mol Cancer Ther. 2009;8:2243–2254. doi: 10.1158/1535-7163.MCT-09-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedier A, Stuedli A, Fink D. Presence of MLH1 protein aggravates the potential of the HSP90 inhibitor radicicol to sensitize tumor cells to cisplatin. Int J Oncol. 2005;27:1697–1705. [PubMed] [Google Scholar]
- Field D, Magnasco MO, Moxon ER, Metzgar D, Tanaka MM, Wills C, Thaler DS. Contingency loci, mutator alleles, and their interactions: synergistic strategies for microbial evolution and adaptation in pathogenesis. Ann NY Acad Sci. 1999;870:378–382. doi: 10.1111/j.1749-6632.1999.tb08907.x. [DOI] [PubMed] [Google Scholar]
- Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondon JW, 3rd, Mele GM, Brezinschek RI, Cummings D, Pande A, Wren J, O’Brien KM, Kupfer KC, Wei MH, Lerman M, Minna JD, Garner HR. Computerized polymorphic marker identification: experimental validation and a predicted human polymorphism catalog. Proc Natl Acad Sci USA. 1998;95:7514–7519. doi: 10.1073/pnas.95.13.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Abbey D, Pisithkul T, Weinzierl MA, Ringstrom T, Bruck D, Petersen K, Berman J. Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio. 2011;2:e00129-11. doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch RL, Su Y, Thornton PC, Gibson JL, Rosenberg SM, Hastings PJ. Separate DNA Pol II- and Pol IV-dependent pathways of stress-induced mutation during double-strand-break repair in Escherichia coli are controlled by RpoS. J Bacteriol. 2010;192:4694–4700. doi: 10.1128/JB.00570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Do R, Yamada M, Friedberg EC, Hastings PJ, Nohmi T, Rosenberg SM. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics. 2009;182:55–68. doi: 10.1534/genetics.109.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, Lin H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat Genet. 2011;43:153–158. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, Schaffner W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- Gibson JL, Lombardo MJ, Thornton PC, Hu KH, Galhardo RS, Beadle B, Habib A, Magner DB, Frost LS, Herman C, Hastings PJ, Rosenberg SM. The sigma(E) stress response is required for stress-induced mutation and amplification in Escherichia coli. Mol Microbiol. 2010;77:415–430. doi: 10.1111/j.1365-2958.2010.07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C, Hadany L, Ponder RG, Price M, Hastings PJ, Rosenberg SM. Mutability and importance of a hypermutable cell subpopulation that produces stress-induced mutants in Escherichia coli. PLoS Genet. 2008;4:e1000208. doi: 10.1371/journal.pgen.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Mittelman D, Wilson JH. Genome-wide demethylation destabilizes CTG.CAG trinucleotide repeats in mammalian cells. Hum Mol Genet. 2004;13:2979–2989. doi: 10.1093/hmg/ddh317. [DOI] [PubMed] [Google Scholar]
- Ha K, Fiskus W, Rao R, Balusu R, Venkannagari S, Nalabothula NR, Bhalla KN. Hsp90 inhibitor-mediated disruption of chaperone association of ATR with Hsp90 sensitizes cancer cells to DNA damage. Mol Cancer Ther. 2011;10:1194–1206. doi: 10.1158/1535-7163.MCT-11-0094. [DOI] [PubMed] [Google Scholar]
- Hall BG. Transposable elements as activators of cryptic genes in E. coli. Genetica. 1999;107:181–187. [PubMed] [Google Scholar]
- Hammock EA, Young LJ. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science. 2005;308:1630–1634. doi: 10.1126/science.1111427. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Hersh MN, Thornton PC, Fonville NC, Slack A, Frisch RL, Ray MP, Harris RS, Leal SM, Rosenberg SM. Competition of Escherichia coli DNA polymerases I, II and III with DNA Pol IV in stressed cells. PLoS One. 2010;5:e10862. doi: 10.1371/journal.pone.0010862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009a;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009b;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hile SE, Eckert KA. DNA polymerase kappa produces interrupted mutations and displays polar pausing within mononucleotide microsatellite sequences. Nucleic Acids Res. 2008;36:688–696. doi: 10.1093/nar/gkm1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski C, Nasar F, Nag DK. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc Natl Acad Sci USA. 2000;97:2134–2139. doi: 10.1073/pnas.040460297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski A, Rosche WA, Gellibolian R, Kang S, Shimizu M, Bowater RP, Sinden RR, Wells RD. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proc Natl Acad Sci USA. 1995;92:11019–11023. doi: 10.1073/pnas.92.24.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Jung SM, Kim MJ, Bae JH, Kim HB, Kim JY, Park SJ, Song HS, Kim DW, Kang CD, Kim SH. DNA-dependent protein kinase is involved in heat shock protein-mediated accumulation of hypoxia-inducible factor-1alpha in hypoxic preconditioned HepG2 cells. FEBS J. 2008;275:5969–5981. doi: 10.1111/j.1742-4658.2008.06725.x. [DOI] [PubMed] [Google Scholar]
- Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- King DG, Kashi Y. Indirect selection for mutability. Heredity. 2007;99:123–124. doi: 10.1038/sj.hdy.6800998. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U, Niklas KJ. The modern theory of biological evolution: an expanded synthesis. Naturwissenschaften. 2004;91:255–276. doi: 10.1007/s00114-004-0515-y. [DOI] [PubMed] [Google Scholar]
- Layton JC, Foster PL. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol Microbiol. 2003;50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Pochet N, Pak T, Verstrepen KJ. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 2007;17:1787–1796. doi: 10.1101/gr.6554007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61:3230–3239. [PubMed] [Google Scholar]
- Lombardo MJ, Aponyi I, Rosenberg SM. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics. 2004;166:669–680. doi: 10.1534/genetics.166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M, Sammarco MC, Grabczyk E. Transposon Tn7 preferentially inserts into GAA*TTC triplet repeats under conditions conducive to Y*R*Y triplex formation. PLoS One. 2010;5:e11121. doi: 10.1371/journal.pone.0011121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour WY, Schumacher S, Rosskopf R, Rhein T, Schmidt-Petersen F, Gatzemeier F, Haag F, Borgmann K, Willers H, Dahm-Daphi J. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008;36:4088–4098. doi: 10.1093/nar/gkn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcadier JL, Pearson CE. Fidelity of primate cell repair of a double-strand break within a (CTG).(CAG) tract. Effect of slipped DNA structures. J Biol Chem. 2003;278:33848–33856. doi: 10.1074/jbc.M304284200. [DOI] [PubMed] [Google Scholar]
- Mayca Pozo F, Oda T, Sekimoto T, Murakumo Y, Masutani C, Hanaoka F, Yamashita T. Molecular chaperone Hsp90 regulates REV1-mediated mutagenesis. Mol Cell Biol. 2011;31:3396–3409. doi: 10.1128/MCB.05117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie GJ, Harris RS, Lee PL, Rosenberg SM. The SOS response regulates adaptive mutation. Proc Natl Acad Sci U S A. 2000;97:6646–6651. doi: 10.1073/pnas.120161797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelman D, Sykoudis K, Hersh M, Lin Y, Wilson JH. Hsp90 modulates CAG repeat instability in human cells. Cell Stress Chaperones. 2010;15:753–759. doi: 10.1007/s12192-010-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelman D, Wilson JH. Stress, genomes, and evolution. Cell Stress Chaperones. 2010;15:463–466. doi: 10.1007/s12192-010-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon R, Bayliss C, Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet. 2006;40:307–333. doi: 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Nag DK, Kurst A. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:835–847. doi: 10.1093/genetics/146.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi M, Yu D, Hirayama R, Ninomiya Y, Sekine E, Kubota N, Ando K, Okayasu R. Inhibition of homologous recombination repair in irradiated tumor cells pretreated with Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Biochem Biophys Res Commun. 2006;351:658–663. doi: 10.1016/j.bbrc.2006.10.094. [DOI] [PubMed] [Google Scholar]
- Oda T, Hayano T, Miyaso H, Takahashi N, Yamashita T. Hsp90 regulates the Fanconi anemia DNA damage response pathway. Blood. 2007;109:5016–5026. doi: 10.1182/blood-2006-08-038638. [DOI] [PubMed] [Google Scholar]
- Orr HT. Unstable nucleotide repeat minireview series: a molecular biography of unstable repeat disorders. J Biol Chem. 2009;284:7405. doi: 10.1074/jbc.R800067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Liao Y, Liu Y, Chang P, Liao L, Yang L, Li H. Transcription of AAT*ATT triplet repeats in Escherichia coli is silenced by H-NS and IS1E transposition. PLoS One. 2010;5:e14271. doi: 10.1371/journal.pone.0014271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi GB, Cleary JD, Pearson CE. In vitro (CTG)*(CAG) expansions and deletions by human cell extracts. J Biol Chem. 2002;277:13926–13934. doi: 10.1074/jbc.M109761200. [DOI] [PubMed] [Google Scholar]
- Parniewski P, Jaworski A, Wells RD, Bowater RP. Length of CTG.CAG repeats determines the influence of mismatch repair on genetic instability. J Mol Biol. 2000;299:865–874. doi: 10.1006/jmbi.2000.3796. [DOI] [PubMed] [Google Scholar]
- Pigliucci M. Is evolvability evolvable? Nat Rev Genet. 2008;9:75–82. doi: 10.1038/nrg2278. [DOI] [PubMed] [Google Scholar]
- Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128:655–668. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Rayssiguier C, Thaler DS, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- Ritz D, Lim J, Reynolds CM, Poole LB, Beckwith J. Conversion of a peroxiredoxin into a disulfide reductase by a triplet repeat expansion. Science. 2001;294:158–160. doi: 10.1126/science.1063143. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer JK, Livingston DM. Destabilization of CAG trinucleotide repeat tracts by mismatch repair mutations in yeast. Hum Mol Genet. 1997;6:349–355. doi: 10.1093/hmg/6.3.349. [DOI] [PubMed] [Google Scholar]
- Sekimoto T, Oda T, Pozo FM, Murakumo Y, Masutani C, Hanaoka F, Yamashita T. The molecular chaperone Hsp90 regulates accumulation of DNA polymerase eta at replication stalling sites in UV-irradiated cells. Mol Cell. 2010;37:79–89. doi: 10.1016/j.molcel.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Shee C, Gibson JL, Darrow MC, Gonzalez C, Rosenberg SM. Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc Natl Acad Sci USA. 2011;108:13659–13664. doi: 10.1073/pnas.1104681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpkins SB, Bocker T, Swisher EM, Mutch DG, Gersell DJ, Kovatich AJ, Palazzo JP, Fishel R, Goodfellow PJ. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum Mol Genet. 1999;8:661–666. doi: 10.1093/hmg/8.4.661. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Specchia V, Piacentini L, Tritto P, Fanti L, D’Alessandro R, Palumbo G, Pimpinelli S, Bozzetti MP. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- Surtees JA, Argueso JL, Alani E. Mismatch repair proteins: key regulators of genetic recombination. Cytogenet Genome Res. 2004;107:146–159. doi: 10.1159/000080593. [DOI] [PubMed] [Google Scholar]
- TCGA Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A, Eckert MJ, Logette E, Lippens S, Janssens S, Jaccard B, Quadroni M, Tschopp J. Regulation of PIDD auto-proteolysis and activity by the molecular chaperone Hsp90. Cell Death Differ. 2011;18:506–515. doi: 10.1038/cdd.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkelson J, Harris RS, Lombardo MJ, Nagendran J, Thulin C, Rosenberg SM. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Carbajal S, Vijg J, DiGiovanni J, Vasquez KM. DNA structure-induced genomic instability in vivo. J Natl Cancer Inst. 2008;100:1815–1817. doi: 10.1093/jnci/djn385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam TS, Reid SD, Selander RK. Mutators and long-term molecular evolution of pathogenic Escherichia coli O157:H7. Emerg Infect Dis. 1998;4:615–617. doi: 10.3201/eid0404.980411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Saier MH., Jr A novel mechanism of transposon-mediated gene activation. PLoS Genet. 2009;5:e1000689. doi: 10.1371/journal.pgen.1000689. [DOI] [PMC free article] [PubMed] [Google Scholar]