Abstract
All organisms require S-adenosylmethionine (SAM) as a methyl group donor and cofactor for various biologically important processes. However, certain obligate intracellular parasitic bacteria and also the amoeba symbiont Amoebophilus asiaticus have lost the capacity to synthesize this cofactor and hence rely on its uptake from host cells. Genome analyses revealed that A. asiaticus encodes a putative SAM transporter. The corresponding protein was functionally characterized in Escherichia coli: import studies demonstrated that it is specific for SAM and S-adenosylhomocysteine (SAH), the end product of methylation. SAM transport activity was shown to be highly dependent on the presence of a membrane potential, and by targeted analyses, we obtained direct evidence for a proton-driven SAM/SAH antiport mechanism. Sequence analyses suggest that SAM carriers from Rickettsiales might operate in a similar way, in contrast to chlamydial SAM transporters. SAM/SAH antiport is of high physiological importance, as it allows for compensation for the missing methylation cycle. The identification of a SAM transporter in A. asiaticus belonging to the Bacteroidetes phylum demonstrates that SAM transport is more widely spread than previously assumed and occurs in bacteria belonging to three different phyla (Proteobacteria, Chlamydiae, and Bacteroidetes).
INTRODUCTION
Methylation occurs in all organismic groups, from bacteria to eukaryotes, and is involved in general processes, such as RNA metabolism and the regulation of gene expression and protein function, as well as in more specific mechanisms, like modification of neurotransmitters and detoxification of heavy metals, etc. (1–3). In various synthetic and regulatory methylation reactions, S-adenosylmethionine (SAM) acts as a methyl group donor, and specific methyltransferases mediate the transfer of the reactive methyl group to the respective acceptor molecule (4–6). Methyl group transfer from SAM results in the formation of S-adenosylhomocysteine (SAH). SAH is an efficient competitive inhibitor of methyltransferases, and accordingly, its removal by catabolizing enzymes (such as specific hydrolases or nucleosidases) is required to guarantee maintenance of methylation processes (7, 8). In addition to its role in methylation, SAM is also an important reagent for posttranscriptional modification of tRNAs and is used as a source of ribosyl groups in the biosynthesis of queuosine, a hypermodified tRNA nucleoside occurring in tRNAs coding for asparagine, aspartic acid, histidine, and tyrosine (3). Moreover, SAM acts as a precursor (amino carboxylpropyl group donor) in polyamine generation, in bacterial N-acetylhomoserine lactone synthesis, as well as in ethylene and nicotinamine production in plants and also plays an important role as a radical source in various biological transformations during, e.g., DNA precursor, vitamin, or cofactor synthesis (3).
Most organisms are able to generate SAM from ATP and methionine via the enzyme SAM synthetase (MetK [EC 2.5.1.6]) (9–13). The Escherichia coli genome encodes only one single SAM synthetase isoform (metK gene), and the incapability to obtain metK deletion mutants demonstrated that SAM formation and, consequently, methylation are essential for cellular viability and growth (14). Interestingly, several obligate intracellular bacteria belonging to the Rickettsiales and Chlamydiales apparently have lost the capacity to synthesize this important cofactor because they lack a functional metK gene (15, 16). In Rickettsia prowazekii and in related strains that cause spotted fever, the metK gene is defective due to internal stop codons or frameshifts, and SAH recycling also seems to be absent (16, 17). R. prowazekii harbors a drug metabolite transporter superfamily protein involved in SAM provision (RP076) (16). A possible H+/SAM symport was suggested to allow net uptake of SAM and compensation for the missing synthetic activity. Competition studies performed with the rickettsial carrier revealed that an excess of SAH caused significantly reduced SAM uptake, and therefore, SAH was discussed as a potential additional substrate of this transport protein (16). However, whether this carrier catalyzes SAM transport in exchange with SAH was not investigated in corresponding transport studies. A possible SAM/SAH antiport would supply SAM to the bacterium and synchronously facilitate the export of the demethylated backbone. Among the Chlamydiales, solely Parachlamydia acanthamoebae and Waddlia chondrophila harbor enzymes for SAM generation and SAH degradation and thus exhibit a complete methylation cycle (15, 18). Remarkably, SAM-dependent methylation (of 16S rRNA or class I release factors) is performed in these bacteria; however, methylation of DNA most likely seems to be of no relevance in chlamydiae due to the absence of DNA methyltransferase coding sequences in the corresponding genomes (15, 19, 20).
Recently, a carrier mediating SAM uptake (CTL0843) was also identified in Chlamydia trachomatis (15). Biochemical data led the authors of that study to the assumption that it exhibits diverse properties: the carrier is capable of proton-driven SAM net uptake as well as substrate counterexchange. It is noteworthy that significant counterexchange occurred in the presence or absence of a proton gradient. Moreover, slight SAM efflux was observed when both proton gradient and counterexchange substrates were missing (15). Although accepting identical/similar substrates and although belonging to the same transporter superfamily, the chlamydial and the rickettsial SAM carriers exhibit only low sequence similarities (approximately 20% amino acid sequence identity).
A reduced genome size accompanied by the loss of biosynthetic pathways and the recruitment of carriers for compensation for missing or truncated metabolic pathways is a characteristic common feature of Chlamydiales and Rickettsiales (21, 22): specific nucleotide transporters were shown to allow energy parasitism and complementation of missing purine and pyrimidine nucleotide or cofactor biosynthesis pathways (23–29). Deciphering of more and more genomes demonstrated that many intracellular bacteria are biosynthetically highly impaired. Moreover, several obligate endosymbionts and intracellular pathogens also contain a limited repertoire of transporters (30, 31). Analyses of the genome of the obligate intracellular Acanthamoeba symbiont Amoebophilus asiaticus strain 5a2, a member of the phylum Bacteroidetes, revealed an extraordinarily high degree of reduction in its biosynthetic capacities (32). The genome has a size of 1.89 Mbp, encodes 1,557 proteins, and is thus only moderately reduced in size compared to the sizes of many other obligate intracellular bacteria (33, 34). However, the biosynthetic capabilities of A. asiaticus are extremely limited; its genome does not encode pathways for de novo biosynthesis of cofactors, nucleotides, and almost all amino acids (32). Interestingly, its genome harbors one gene (Aasi_1859) with significant similarities to the rickettsial SAM carrier (45% amino acid similarity) (32). Characterization of the Aasi_1859 gene product in the heterologous host E. coli revealed that apart from rickettsial and chlamydial species, A. asiaticus also possesses a SAM transport protein. Its catalytic activity allows import of SAM from the host cell by the simultaneous removal of the end product of methyltransferase reactions.
MATERIALS AND METHODS
Sequence and phylogenetic analyses.
The genome sequence of A. asiaticus 5a2 has recently been determined and analyzed (32) and is available at GenBank under accession no. CP001102. SAM transporter amino acid sequences were retrieved using BLASTP against GenBank by using Aasi_1859 as a query, and only those sequences having more than 30% amino acid identity to Aasi_1859 were used for phylogenetic analyses. Amino acid sequences (101 in total) were aligned with MAFFT (35), and phylogenetic trees were reconstructed with MEGA (36) by using the neighbor-joining method and the Poisson correction, the parsimony bootstrap method, and the maximum likelihood method (using the Jones-Taylor-Thornton [JTT] amino acid substitution model); all trees were calculated with 1,000× bootstrapping. All positions containing gaps and missing data were eliminated from the data sets.
Transcriptional analysis.
Acanthamoeba sp. strain 5a2 (ATCC PRA-228) amoebae harboring A. asiaticus 5a2 cells were harvested by centrifugation (7,000 × g for 3 min at 27°C). The resulting cell pellet was resuspended in 750 μl TRIzol (Invitrogen Life Technologies), transferred into a Lysing Matrix A tube (MP Biomedicals), and homogenized by using a BIO101/Savant FastPrep FP120 instrument (speed, 4.5 m/s; 30 s). RNA was extracted by phase separation, precipitation, washing, and redissolving according to the recommendations of the manufacturer (TRIzol; Invitrogen Life Technologies). The remaining DNA was removed by using the Turbo DNA-free kit (Ambion). After DNase treatment, RNA was resuspended in double-distilled water (ddH2O) with diethyl pyrocarbonate (DEPC) and stored at −80°C until use. The absence of DNA contamination in the DNase-treated RNA was verified by performing a control PCR with 42 cycles by using primers targeting a 361-bp fragment of the Aasi_1859 gene (forward primer 5′-ATG GAG CCA GGG GAT TAA AG-3′ and reverse primer 5′-GTT GGT GGG AGT ACG CCA TA-3′) and an annealing temperature of 66.4°C. DNA-free total RNA (containing host and symbiont RNA) was used to synthesize cDNA by using the RevertAid first-strand cDNA synthesis kit (Fermentas) according to the recommendations of the manufacturer. cDNA was subsequently used as the template in standard PCRs (35 cycles and an annealing temperature of 66.4°C). Negative controls (no cDNA added) and positive controls (genomic DNA) were included in all PCRs. All experiments were performed in biologically independent triplicates.
Cloning of Aasi_1859 and heterologous protein synthesis in E. coli.
Acanthamoeba sp. 5a2 (ATCC PRA-228) amoebae harboring A. asiaticus cells were used for DNA isolation with the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's recommendations. The Aasi_1859 gene, coding for the putative SAM transport protein, was amplified by using High Fidelity PCR enzyme mix (MBI-Fermentas) according to the instructions of the manufacturer. A forward primer (5′-CCT GCG CAT ATG TTG AAA TAT TTT AAA GCA-3′), introducing an NdeI restriction site before the start codon, and a reverse primer (5′-CCT CGC CTC GAG TCA AGC TTT AGG TTG ATT-3′), containing an XhoI restriction site after the stop codon, were used. PCR conditions were as follows: a denaturation step at 94°C for 3 min, followed by 35 cycles of (i) denaturation at 94°C for 30 s, (ii) annealing at 56°C for 40 s, and (iii) elongation at 68°C for 90 s and a final elongation step at 68°C for 10 min. The resulting amplification products were gel purified and cloned into the cloning vector pCR-XL-TOPO by using the TOPO XL cloning kit (Invitrogen Life Technologies). The resulting plasmid was digested with the restriction endonucleases NdeI and XhoI, gel purified, and inserted in frame into the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression vector pET16b containing a promoter site for the T7 RNA polymerase (Novagen). The newly constructed expression plasmid was transformed into and maintained in E. coli XL1-Blue cells (Stratagene). Integrity of the cloned gene was confirmed by sequencing on an ABI 3130 XL genetic analyzer using BigDye Terminator kit v3.1 (ABI). After the correctness of the insertion was proven, the construct was used for transformation of BLR(DE3) expression cells (Merck Biosciences). E. coli cells were cultured in standard yeast extract-tryptone (YT) medium at 37°C with vigorous shaking. Heterologous protein synthesis was induced by addition of 1 mM IPTG during exponential cell growth (at an optical density at 600 nm [OD600] of 0.5). One hour after induction, cells were concentrated to an OD600 of 5.0 by centrifugation (3,000 × g for 5 min at 8°C). Cells were either suspended in 50 mM potassium phosphate buffer (pH 7.0) (KPi) to an OD600 of 5.0 and directly used for import studies or applied for protein fractionation and immune detection.
Protein fractionation and immune detection of the recombinant SAM carrier.
Heterologous expression and insertion of the recombinant protein in the membrane fraction were analyzed by immune detection. First, cell wall integrity was reduced by freezing of the pellet in liquid nitrogen and subsequent thawing, and incubation for 5 to 10 min at 37°C resulted in release and activity of endogenous lysozyme of the BLR cells. Autolysis was conducted in the presence of the protease inhibitor phenylmethylsulfonyl fluoride (PMSF) (1 mM), and cell disruption was complemented by sonication (addition of RNase and DNase). In a first centrifugation step (20,000 × g for 15 min at 4°C), cell debris and incorrectly folded membrane protein aggregates, so-called inclusion bodies, were enriched from the homogenate. Membrane proteins of the supernatant were afterwards separated from soluble proteins by ultracentrifugation (100,000 × g for 30 min at 4°C). Proteins of the membrane fraction were analyzed by SDS-PAGE (3% stacking and 15% separating gel) (37). Following electrophoresis, proteins were Coomassie stained or transferred onto a nitrocellulose membrane in a wet-blotting apparatus. Expression of the recombinant protein was verified by Western blotting and immune detection with anti-poly-His IgG combined with a secondary alkaline phosphatase-conjugated anti-mouse IgG (Sigma). Alkaline phosphatase activity was demonstrated by nitroblue tetrazolium chloride–5-bromo-4-chloro-3′-indolyl phosphate toluidine staining. A prestained broad-range marker (7 to 175 kDa; New England BioLabs) was applied for estimation of the molecular protein masses.
Import studies with radioactively labeled SAM.
Transport studies with intact cells are well suited to investigate SAM import because E. coli does not possess endogenous SAM uptake systems (15, 16). Import of radioactively labeled SAM was determined with induced and noninduced (control) E. coli cells harboring the corresponding plasmids. For this, E. coli cells were incubated at 30°C in 50 mM potassium phosphate buffer complemented with the indicated concentrations of labeled SAM (NEN). Optionally, the transport medium was supplemented with the indicated concentrations of nonlabeled substrates or molecules. Termination of transport was achieved by removal of the external substrate due to application of the cells onto prewetted filters (mixed cellulose ester, 0.45-μm pore size; Whatman), vacuum filtration, and washing (three times with 4 ml of KPi buffer). Radioactivity of the cell samples at the filters was quantified with a scintillation counter (Beckman LS6500; Beckman Coulter).
RESULTS
Metabolic requirement of SAM import and SAH export in A. asiaticus.
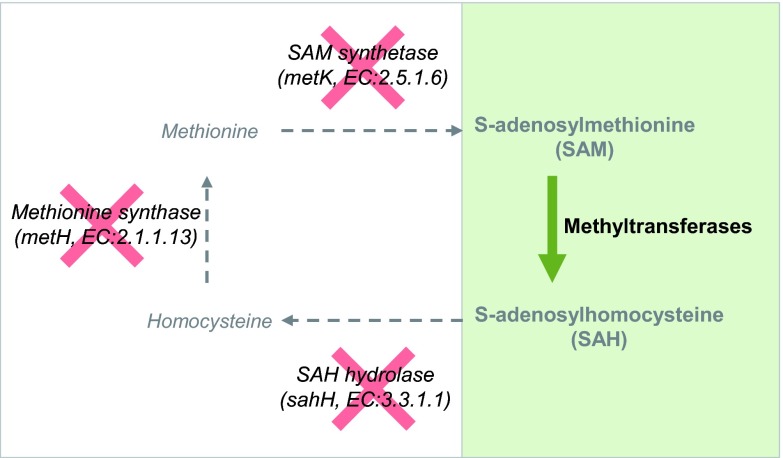
The methylation pathway, including the SAM synthetase MetK, the SAH hydrolase SahH, and the methionine synthetase MetH, is completely absent in A. asiaticus (Fig. 1). However, the A. asiaticus genome encodes 17 putative methyltransferases as well as a homologue of the SAM-tRNA ribosyltransferase-isomerase (QueA [Aasi_0780]), which is responsible for the transfer of the ribose moiety of SAM into the modified tRNA (see Table S1 in the supplemental material). Therefore, there is clearly a need for SAM as a cofactor of methylation reactions as well as a donor of ribosyl groups in tRNA synthesis in A. asiaticus. Moreover, because A. asiaticus apparently lacks SAH-degrading enzymes, specific removal of SAH is mandatory to prevent inhibition of methyltransferases by accumulating SAH (7, 8). Consequently, a SAM import and SAH export system is predicted for A. asiaticus.
Fig 1.
A. asiaticus harbors an incomplete methylation cycle. In total, 17 methyltransferases were identified in the A. asiaticus genome; the remaining enzymes of the methylation cycle required for SAM (re)generation are absent (crossed out in red). Methyl group transfer from SAM leads to the generation of SAH (shaded in green and in boldface type), whereas the capacities for SAH degradation and SAM regeneration are missing (italic type). Details on the methyltransferases present in A. asiaticus can be found in Table S1 in the supplemental material.
Comparative sequence analyses and phylogeny of SAM transport proteins.
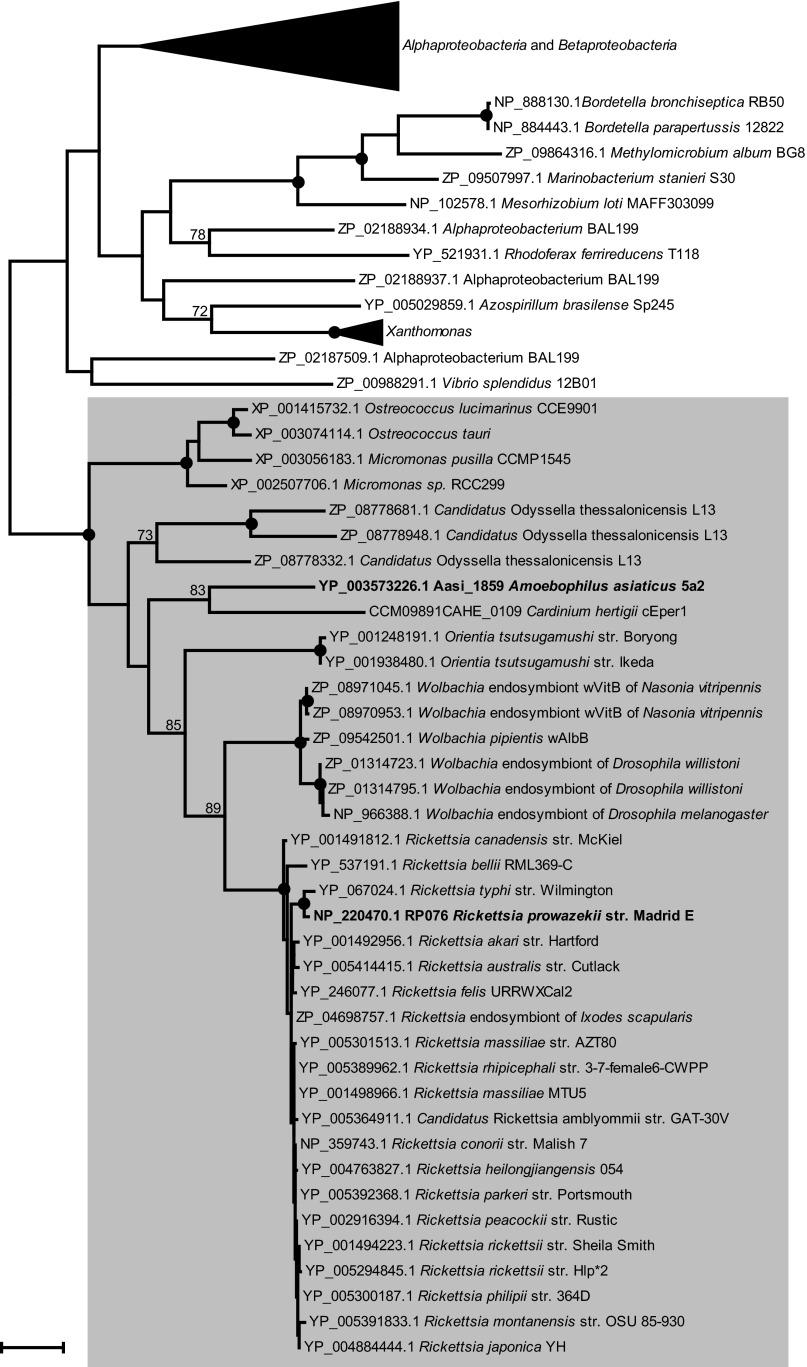
During analysis of the A. asiaticus genome, we identified a putative SAM transporter: Aasi_1859 is a 285-amino-acid protein with 10 predicted transmembrane helices and shows 45% amino acid sequence identity to the functionally characterized rickettsial SAM transporter encoded by the RP076 gene. Aasi_1859 and homologues contain a duplicated (functionally uncharacterized) EamA domain (Pfam accession no. PF00892) and belong to the drug-metabolite transporter (DMT) superfamily and the 10-transmembrane-segment (10-TMS) drug-metabolite exporter (DME) family (2.A.7.3) (38). Recently, a SAM transporter has also been identified in Chlamydia trachomatis (CTL0843) (15). Aasi_1859 and RP076 show only low amino acid sequence identity to CTL0843 (approximately 20%) (see Fig. S1 in the supplemental material). However, all carriers belong to the 10-TMS DME family. In addition, RP076 and Aasi_1859 homologues with more than 40% amino acid identity were also identified in other obligate intracellular bacteria belonging to the Rickettsiales and Bacteroidetes. The highest amino acid identity (47% amino acid identity) of Aasi_1859 is shared with a homologue found in “Candidatus Odyssella thessalonicensis,” an amoeba symbiont belonging to the Rickettsiales, as well as with “Candidatus Cardinium hertigii” cEper1, an obligate intracellular symbiont of parasitic wasps, representing the sister lineage of A. asiaticus (39). Interestingly, “Ca. Odyssella thessalonicensis” encodes at least three highly similar copies of Aasi_1859 homologues. Surprisingly, we also identified homologues in some members of the green algae (prasinophytes, order Mamiellales): Ostreococcus and Micromonas (40 to 42% amino acid sequence identity). We retrieved 101 homologues of Aasi_1859 and performed phylogenetic analyses of the (putative) SAM transport proteins. Among the Aasi_1859 and RP076 homologues used for phylogenetic analyses, no functionally characterized proteins were found; all homologues belong to the 10-TMS DME family. The application of maximum likelihood, neighbor-joining, and maximum parsimony treeing methods yielded stable phylogenetic relationships: all candidate SAM transporters and homologues of Aasi_1859 and RP076 clustered in a stable monophyletic lineage (Fig. 2). Due to the low sequence similarity, no calculation of phylogenetic relationships of Aasi_1859 and RP076 homologues with CTL0843 and homologues was possible.
Fig 2.
Phylogenetic relationships of Aasi_1859 and related characterized and putative SAM transport proteins. An amino-acid-based phylogenetic tree calculated with MEGA5 using the maximum likelihood algorithm with the JTT model is shown. Black dots indicate nodes which are supported by maximum likelihood, maximum parsimony, and neighbor-joining bootstrap values (1,000× resampling) greater than 90%. GenBank accession numbers are indicated. The bar represents 20% estimated evolutionary distance. Functionally characterized SAM transporters are shown in boldface type. The group comprising (putative and characterized) SAM transporters from Rickettsiales, prasinophytes, and Bacteroidetes is highlighted in gray.
Heterologous expression of Aasi_1859 stimulates [methyl-14C]SAM uptake into E. coli.
The high similarity to the rickettsial SAM carrier suggests that the homologue from A. asiaticus might act as a SAM transporter mediating the uptake of the essential cofactor into the endosymbiont. Reverse transcriptase PCR analysis with total RNA purified from amoebae harboring bacterial endosymbionts demonstrated transcription of Aasi_1859 during intracellular multiplication of A. asiaticus (see Fig. S2 in the supplemental material). Because functional analyses of carriers in A. asiaticus are hampered, if not impossible, due to its obligate intracellular lifestyle, we applied the heterologous E. coli expression system to investigate the biochemical properties of the Aasi_1859 gene product. Import measurements in intact E. coli cells synthesizing the recombinant carrier were previously successfully used to functionally characterize the rickettsial as well as the chlamydial SAM carriers (15, 16). To allow comparison of our results with published data, we also applied the E. coli expression system and used S-adenosyl-l-[methyl-14C]methionine ([methyl-14C]SAM) for the majority of import studies.
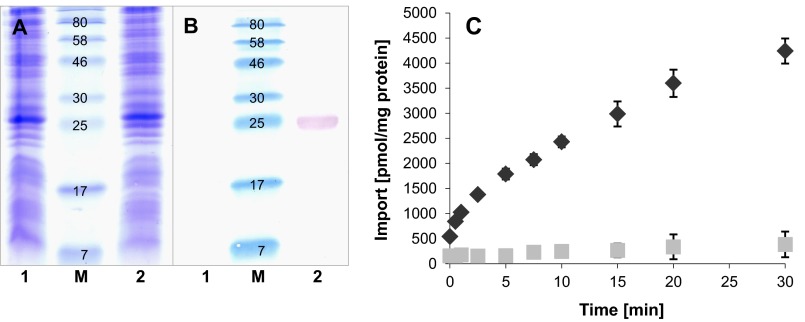
Heterologous expression in E. coli and membrane insertion of the recombinant protein were analyzed by SDS-PAGE, Western blotting, and immunostaining (Fig. 3A and B). Induction of expression resulted in a significant accumulation of the recombinant carrier in the membrane fraction, whereas noninduced cells showed no comparable heterologous protein synthesis (Fig. 3B). Moreover, import studies with radioactively labeled [methyl-14C]SAM demonstrated that the carrier from A. asiaticus mediates a time-dependent uptake of radioactivity into induced cells (Fig. 3C). Import was linear for the first 2.5 min and slowly approached saturation of about 4,500 pmol mg protein−1 at 30 min. Noninduced (control) cells showed no or comparatively low import of radioactivity, with maximal values of about 400 pmol mg protein−1 (Fig. 3C). These data demonstrate that (i) Aasi_1859 is heterologously expressed, (ii) the recombinant carrier is functional in the context of the E. coli membrane, and (iii) it accepts SAM as a substrate and thus might act as a SAM transporter in A. asiaticus. Here, we refer to this protein as AaSAMT.
Fig 3.
Heterologous expression and verification of functional membrane insertion of AaSAMT. (A and B) Proteins of the E. coli membrane fraction (25 μg per lane) were separated by SDS-PAGE (A), and the presence of recombinant AaSAMT was verified by immunodetection (B). Lanes: M, molecular mass marker (in kDa); 1, total membrane proteins of noninduced E. coli cells harboring the AaSAMT-pET16b expression vector; 2, total membrane proteins from E. coli expressing AaSAMT. (C) Time dependency of AaSAMT-catalyzed uptake of radioactively labeled [methyl-14C]SAM. IPTG-induced (◆) and noninduced (▩) E. coli cells were incubated with 10 μM 14C-labeled SAM, and import was stopped at the indicated times by removal of external substrates via vacuum filtration and washing. Data are the means of data from three independent experiments, each with two technical replicates. Standard errors are displayed.
SAM transport depends on the presence of a proton gradient.
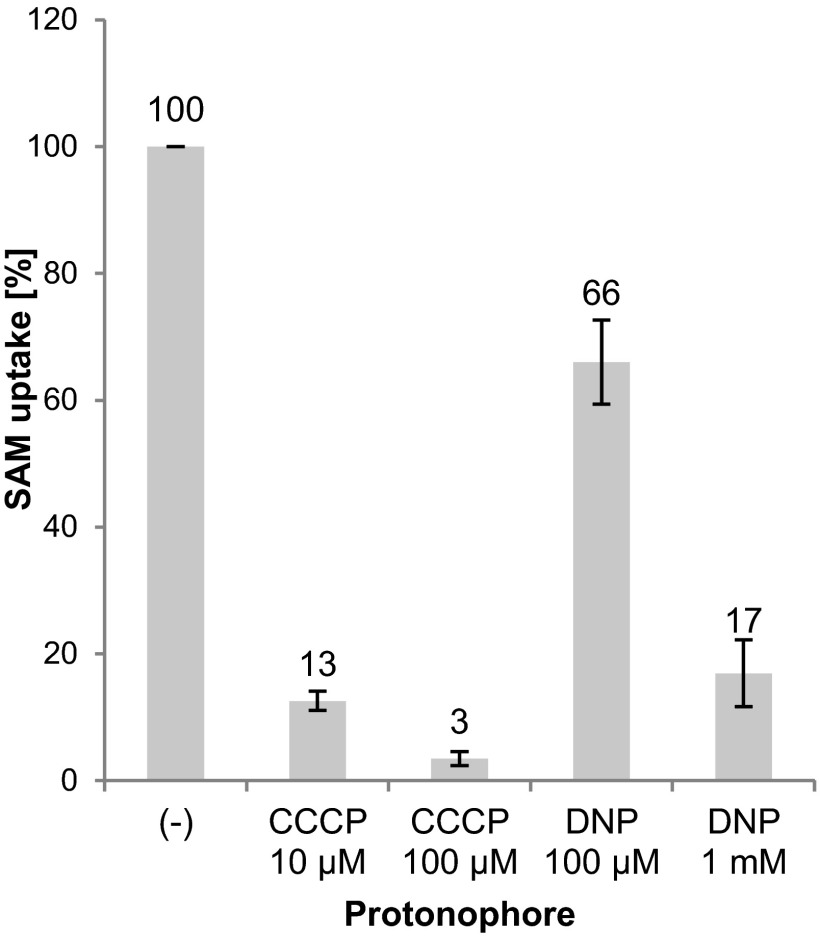
SAM uptake via the rickettsial carrier was shown to be highly reduced by addition of the protonophore DNP (2,4-dinitrophenol) (1 mM) (16). Accordingly, rickettsial SAM translocation was suggested to be a proton gradient-dependent process. SAM import via the chlamydial carrier was also affected by protonophore addition (50 μM CCCP [cyanide m-chlorophenylhydrazone]) but to a lesser extent (50% residual activity) (15). To elucidate the transport mode of AaSAMT and to identify whether AaSAMT function is also influenced by the proton gradient, transport studies were performed in the presence of protonophores. Application of 10 μM CCCP already inhibited [methyl-14C]SAM uptake to a residual rate of about 13%, and the presence of 100 μM CCCP nearly completely abolished SAM accumulation (3.5% residual activity) compared to unaffected transport (set to 100%) (Fig. 4). In contrast to CCCP, higher concentrations of DNP (1 mM) were required to significantly reduce SAM import into E. coli cells expressing AaSAMT (17% residual activity). This is because CCCP is known to be a more efficient protonophore than DNP, and comparably low concentrations of CCCP are sufficient to deplete the proton gradient across the E. coli membrane (40). The pronounced inhibitory effect of protonophores on SAM transport demonstrates that AaSAMT operates not identically to the chlamydial SAM carrier but rather acts like the rickettsial SAM carrier.
Fig 4.
Analysis of the proton dependency of AaSAMT. Shown are the effects of the protonophores CCCP and DNP on [methyl-14C]SAM uptake (5 min, 10 μM SAM). Transport rates are given as a percentage of nonaffected transport (−), which was set to 100%. Net values (minus control, noninduced cells) were used for calculation. Error bars are indicated. The values of the transport rates are displayed above the corresponding bars.
Determination of the substrate specificity of AaSAMT.
The rickettsial as well as the chlamydial SAM transporters were shown to be specific for SAM uptake, and SAH was also suggested to be a potential substrate of these carriers (15, 16). To obtain a preliminary indication of the substrate spectrum of AaSAMT, we performed competition experiments with molecules highly as well as distantly related structurally to SAM (see Fig. S3 in the supplemental material). Import of [methyl-14C]SAM was measured in the presence of 14 different nonlabeled tested molecules applied in a 10-fold excess. The corresponding import was calculated in relation to nonaffected SAM uptake (set to 100%). A large reduction of the import rate might be indicative of transport inhibition or competition of the added compound with SAM during translocation. SAM uptake by recombinant AaSAMT was highly reduced by addition of SAM or SAH (<10% residual activity). A significant decrease of SAM import was also obtained by addition of the aminopropyl transferase inhibitor S-(5′-adenosyl)-3-thiopropylamine (dc-SAH) (∼34% residual activity), whereas the methyltransferase inhibitor sinefungin caused only a slight reduction (∼69% residual activity). All remaining tested molecules had no or comparably small effects (>80% residual activity) (Table 1). The observed effects suggest that the presence of the sulfur atom but not of the methyl and carboxyl group is required for substrate (or inhibitor) recognition. Apparently, AaSAMT exhibits quite high specificity for SAM uptake, but SAH might also represent another important substrate.
Table 1.
Effects of various metabolites/inhibitors on [methyl-14C]SAM import by AaSAMTa
| Effector | SAM import (%) | SE (%) |
|---|---|---|
| None | 100 | ±7 |
| SAM | 8 | ±2 |
| SAH | 6 | ±5 |
| dc-SAH | 34 | ±4 |
| Sinefungin | 69 | ±5 |
| S-Adenosylcysteine | 83 | ±3 |
| ATP | 97 | ±3 |
| ADP | 100 | ±5 |
| AMP | 102 | ±4 |
| Methylthioadenosine | 95 | ±3 |
| Adenosine | 130 | ±6 |
| Adenine | 95 | ±5 |
| Cystathionine | 101 | ±4 |
| Methionine | 98 | ±5 |
| Homocysteine | 99 | ±6 |
| Cysteine | 98 | ±4 |
Uptake of [methyl-14C]SAM by recombinant AaSAMT was measured at a substrate concentration of 10 μM, and nonlabeled effectors were present in a 10-fold excess. Structures of the tested molecules are shown in Fig. S3 in the supplemental material. Import was stopped after 2 min. Rates of SAM uptake are net values (minus control, noninduced E. coli cells) given as percentage of nonaffected transport (set to 100%). Data are the means of data from four independent experiments. Standard errors are given.
AaSAMT catalyzes counterexchange of SAM and SAH.
We investigated the possible counterexchange capacity of AaSAMT to clarify its transport mode. Simultaneously, the effect of CCCP on the maintenance of the intracellular label was also analyzed. So-called chase or efflux experiments allow determination of whether external substrates/effectors can induce the release of labeled substrates previously loaded into E. coli cells. Because SAH might represent an additional substrate of AaSAMT, we also focused on its role during counterexchange transport.
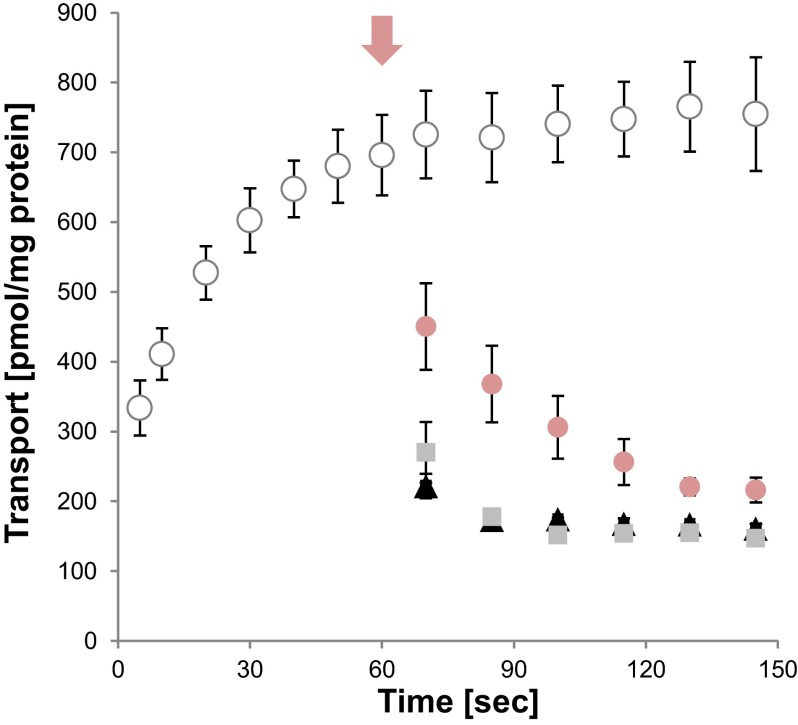
It is very likely that viable recombinant E. coli cells exhibit efficient methylation activity and transfer the labeled methyl group of imported [methyl-14C]SAM to the diverse substrates of methyltransferases. Therefore, label becomes at least partially “fixed” in the cell during transport measurements conducted with [methyl-14C]SAM. As a consequence, externally added substrates/effectors might be unable to induce complete efflux of radioactivity from the cells, and also, export of demethylated SAH cannot be monitored when cells are loaded with [methyl-14C]SAM. Therefore, we performed a chase experiment with S-adenosyl-l-[carboxy-14C]methionine ([carboxy-14C]SAM) carrying a labeled carboxyl group. Because the 14C label is not removed during endogenous methylation processes, the use of [carboxy-14C]SAM enables detection of SAM plus SAH export. We determined time-dependent uptake of [carboxy-14C]SAM (loading of radioactivity into the cell) and analyzed whether the addition of a 20-fold excess of nonlabeled SAM or SAH induces the efflux of label. Nonlabeled SAM as well as nonlabeled SAH caused fast and approximately complete efflux of radioactivity (Fig. 5). Therefore, it becomes evident that AaSAMT acts in an antiport manner and that both SAM and SAH represent efficient counterexchange substrates.
Fig 5.
AaSAMT-driven import of [carboxy-14C]SAM and efflux analysis. Shown is the time-dependent import of 10 μM [carboxy-14C]SAM into IPTG-induced E. coli cells. Possible efflux of internal radioactivity was induced by addition of 200 μM nonlabeled substrates or 100 μM CCCP at the end of the linear phase of import (time point of addition is marked with a pink arrow). Shown are the time course of SAM import in the absence of effectors/nonlabeled substrates (white circles) and the time course of reduction of interior radioactivity after addition of nonlabeled SAM (black triangles), SAH (gray squares), or CCCP (pink circles). Data are the means of data from three independent experiments and represent net values of transport (calculated by subtraction of uptake rates of noninduced cells from import into cells expressing AaSAMT). Standard errors are displayed.
Addition of CCCP also led to a considerable depletion of cellular radioactivity. However, uncoupling of the proton gradient caused a slower decrease, and the plateau phase was reached at a slightly higher residual activity than the reduction of interior label by SAM or SAH addition (Fig. 5). This observation suggests that transport/maintenance of interior label is dependent on the proton gradient.
A closer examination of the time kinetics reveals that application of [carboxy-14C]SAM results in faster saturation and lower maximal transport rates than the time-dependent accumulation of [methyl-14C]SAM (compare Fig. 5, white circles, and 3C, black diamonds). Knowledge about the counterexchange activity of AaSAMT helps to interpret these differences. Reduced accumulation of [carboxy-14C]SAM might result from the simultaneous export of label, more precisely from the export of [carboxy-14C]SAM and [carboxy-14C]SAH. Whereas uptake of [methyl-14C]SAM is not accompanied by a comparably high loss of interior label, apparently small amounts of [methyl-14C]SAM but large amounts of nonlabeled SAH are exported. Therefore, we propose that E. coli efficiently converts SAM into SAH, the methyl group becomes trapped in the cell, and significant amounts of SAH are exported during counterexchange.
AaSAMT exhibits high affinity for SAM and SAH import.
Finally, we determined the biochemical parameters of AaSAMT for SAM and SAH import. The apparent affinities and maximal velocities for [methyl-14C]SAM and for [carboxy-14C]SAM import were analyzed by application of increasing exterior substrate concentrations. Because the Km values for import of [methyl-14C]SAM and [carboxy-14C]SAM are quite similar (13.9 ± 0.8 μM and 10.8 μM ± 1.2 μM, respectively), the affinity of the carrier for SAM apparently is not or is only marginally influenced by the position of the label (Table 2). Moreover, due to the low SAM Km value, AaSAMT is considered, like its chlamydial and rickettsial homologues, to be a high-affinity SAM transporter. AaSAMT imported [methyl-14C]SAM with about a 2-fold-higher Vmax (104.8 nmol mg protein−1 h−1) than that of [carboxy-14C]SAM (46.6 ± 3.2 nmol mg protein−1 h−1) (Table 2), demonstrating that the observed difference in the time courses of [methyl-14C]SAM (Fig. 3C) and [carboxy-14C]SAM (Fig. 5) uptake resulted from different maximal import velocities of the corresponding transport processes. Generally, Vmax values are influenced by the amount of functional recombinant transport protein in the E. coli membrane. However, import studies for Vmax determination were performed in parallel with exactly the same E. coli cells, which guarantees that identical amounts of recombinant carriers were analyzed. A plausible explanation for the different Vmax values is that AaSAMT mediates measurable SAM counterexchange with demethylated products. As mentioned above, intact, metabolically active E. coli cells are capable of using SAM as a methyl group donor. Accordingly, endogenous methylation processes at least partially trap the methyl group of [methyl-14C]SAM in the cell, and export of SAH is not accompanied by a loss of label. However, after demethylation of [carboxy-14C]SAM, the 14C label still remains at SAH, and counterexchange of SAM and SAH causes a greater loss of internal radioactivity. Accordingly, import and export of labeled substrates ([carboxy-14C]SAM versus [carboxy-14C]SAH) result in a faster equilibrium, a faster saturation, and, thus, a lower apparent Vmax value of the corresponding transport.
Table 2.
Determination of kinetic parameters of AaSAMTa
| Substrate | Km (μM) (SE) | Vmax (nmol mg protein−1 h−1) (SE) |
|---|---|---|
| [methyl-14C]SAM | 13.89 (0.81) | 104.80 (6.49) |
| [carboxy-14C]SAM | 10.78 (1.21) | 46.59 (3.20) |
For determination of the respective Km and Vmax values, import was measured with increasing concentrations of [methyl-14C]SAM or [carboxy-14C]SAM. Determination of [methyl-14C]SAM uptake (5 μM) was performed in the presence of increasing concentrations of unlabeled SAH for determination of the Ki value (8.10 ± 1.20 μM) and IC90 (concentration resulting in 90% inhibition of SAM uptake) (66.67 ± 7.45 μM). Transport of [methyl-14C]SAM was allowed for 2 min and transport of [carboxy-14C]SAM was allowed for 1 min and was stopped by vacuum filtration and washing. Data represent net values (minus control, noninduced E. coli cells) and are means of data from at least three independent experiments; standard errors are also indicated.
Two important observations indicate that SAH represents an additional substrate of AaSAMT: first, it competes with SAM for import (Fig. 5), and second, SAH—just like SAM—induces the efflux of label from E. coli cells loaded with [carboxy-14C]SAM (Table 1). Because radioactively labeled SAH is not commercially available, we applied increasing concentrations of unlabeled SAH to [methyl-14C]SAM import to get an idea about the affinity of AaSAMT for SAH. By this approach, we identified an apparent Ki value of about 8.1 ± 1.0 μM, and 90% SAM transport inhibition was obtained by addition of 66.7 ± 7.5 μM SAH. Therefore, AaSAMT exhibits a high apparent affinity for SAH import, quite similar to that of SAM uptake. The determined characteristics suggest that AaSAMT can efficiently mediate SAM/SAH exchange.
DISCUSSION
Obligate intracellular bacteria are generally characterized by a highly reduced genome size and an impaired metabolic capacity (21, 22, 33, 34, 41, 42). In these organisms, essential metabolic pathways are often truncated or missing completely, and hence, import of intermediates or products is of high physiological importance. In the past years, several carrier proteins that mediate the provision of diverse metabolically relevant molecules and thus compensate for the reduced biosynthetic activity in intracellular bacteria have been identified (15, 16, 23–29, 43). Analysis of the genome of A. asiaticus revealed that its size is comparable to those of other intracellular bacteria; however, it encodes an unusually small number of proteins involved in metabolic processes (32). A. asiaticus lacks the oxidative pentose phosphate pathway and is impaired in ATP regeneration via the electron transport chain, the tricarboxylic acid cycle, and glycolysis. Moreover, in A. asiaticus, pathways for the de novo synthesis of purine and pyrimidine nucleotides, cofactors, and almost all amino acids are absent (32). This metabolic reduction necessitates the uptake of diverse metabolites from the amoeba host.
A. asiaticus encodes 17 putative methyltransferases (see Table S1 in the supplemental material), and thus, methylation apparently still takes place in this endosymbiont. This observation, combined with the fact that A. asiaticus does not possess SAM-generating and SAH-degrading enzymes, implies that corresponding reactions have to be performed by the host cell and that SAM and SAH have to be shuttled across the bacterial membrane.
Our analyses suggest that the protein AaSAMT, encoded by the Aasi_1859 gene, possesses the biochemical prerequisites required to fulfill SAM and SAH exchange in A. asiaticus. The recombinant carrier mediates significant import of radioactively labeled SAM when heterologously expressed in E. coli (Fig. 3C). A proton gradient across the E. coli membrane was shown to be required for accumulation (Fig. 4) and maintenance (Fig. 5) of interior label. At first glance, the proton dependency suggested that SAMT from A. asiaticus catalyzes a secondary active H+/SAM symport and thus might be capable of net SAM supply. However, effector studies, application of differentially labeled SAM, and efflux studies demonstrated that AaSAMT mediates counterexchange of SAM and SAH (Table 1 and Fig. 3C and 5). Both substrates are transported with comparably high affinities (Km of ∼12 μM for SAM and Ki of ∼8 μM for SAH upon SAM import) (Table 2).
The absence of the methylation cycle (Fig. 1) necessitates SAM import and SAH export in A. asiaticus. Because SAM and SAH represent import and export substrates of AaSAMT, it is important to check which physiological conditions allow SAM exploitation of the host and removal of bacterial SAH. Eukaryotic organisms generally exhibit higher SAM than SAH concentrations, and even under conditions of methyl deficiency, cellular SAM/SAH ratios higher than 1 were still identified in almost all investigated tissues (except from kidney [ratio of 0.61]) of mice (44). Accordingly, a balanced nutrient supply guarantees that more SAM than SAH is available in the host amoeba and that mainly SAM enters the bacterium. Moreover, methylation in A. asiaticus results in SAM consumption and fuels the carrier with SAH at the bacterial inner face. Interestingly, a decrease in SAM content was observed in Physarum flavicomum amoebae during the developmental transition from a growing state to dormant cysts (45, 46). Transferring this situation to the host of A. asiaticus, efficient SAM exploitation by the endosymbiont is rather restricted to the growing state of the amoeba. A decrease of the SAM/SAH ratio in the host (methyl deficiency and transition to dormancy) will cause SAH uptake into the endosymbiont. As a consequence, endosymbiotic methylation processes will slow down due to inhibition of methyltransferases by excess SAH (7, 8) and/or due to substrate deprivation.
Moreover, decreased metabolic activity of the host generally affects metabolite provision to the symbiotic bacterium, resulting in alteration of physiological processes and most likely in an insufficient membrane potential (47). The proposed reduction of the bacterial proton gradient inactivates H+ symport and influences H+-regulated carriers, including AaSAMT.
Interestingly, Aasi_1859 is located within a cluster of three genes involved in tRNA modification: mnmA (Aasi_1200), mnmE (Aasi_1201), and tilS (Aasi_1198) (see Fig. S4 in the supplemental material). This might suggest a role of Aasi_1859—more precisely, of its substrate SAM—in tRNA modification (3). The corresponding ribosyl group transfer results in formation of methionine. The incapability of methionine to compete with SAM for import (Table 1) suggests that methionine is no substrate of AaSAMT, and therefore, AaSAMT apparently does not catalyze SAM uptake in exchange with methionine.
To establish a basic pool of SAM and to fuel nonmethylation processes with this cofactor, net uptake of SAM or at least SAM exchange with substrates different from SAH is required in A. asiaticus. Remarkably, addition of CCCP resulted in a significant loss of radioactively labeled SAM and SAH from E. coli cells expressing AaSAMT (Fig. 5). The corresponding substrate flux is most likely driven by the concentration gradient across the bacterial membrane. Accordingly, net provision of SAM or SAH to the bacterium by AaSAMT is imaginable, at least under conditions of a reduced membrane potential. SAM uptake in exchange with other, not-yet-identified substrates might provide SAM nonmethylation processes. However, it is also possible that further SAM import systems exist in A. asiaticus.
In 2003, the first bacterial SAM transporter was identified in R. prowazekii (16), and recently, SAM transport was also clarified for C. trachomatis (15). These carriers showed high affinities for SAM import (Km values of ∼2.5 μM for R. prowazekii RP076 and ∼6 μM for C. trachomatis CTL0843) and also a quite low SAH Ki of SAM transport (Ki values of ∼14.3 μM for R. prowazekii RP076 and ∼4.2 μM for C. trachomatis CTL0843), and thus, these parameters are comparable to those of AaSAMT. The rickettsial SAM transporter was proposed to act as an H+/SAM symporter because its activity was highly influenced by the proton gradient (16). The capacity of the rickettsial SAM carrier to perform counterexchange was not investigated. Not only does R. prowazekii lack functional SAM synthetase, SAH degradation and SAM recycling are also missing in all sequenced Rickettsia species based on analyses using the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (http://www.genome.jp/kegg/) (48). Therefore, SAM import and SAH export are also required in R. prowazekii. In fact, SAH was shown to efficiently compete with SAM for uptake and therefore might represent an additional substrate of the rickettsial SAM carrier (16). Moreover, the close phylogenetic relationship of AaSAMT and the rickettsial SAM carrier (Fig. 2) is indicative of their common evolutionary origin and horizontal gene transfer. The high amino acid sequence identity of these carriers (see Fig. S1 in the supplemental material) suggests that both proteins still possess similar biochemical properties. Therefore, it is conceivable that the rickettsial SAM carrier does not represent an H+/SAM symporter but facilitates SAM/SAH counterexchange that is regulated by the proton gradient.
The SAM transporter from C. trachomatis, although belonging to the same transporter class, exhibits comparably low sequence similarities to the SAM carriers from R. prowazekii and A. asiaticus (see Fig. S1 in the supplemental material). Destruction of the proton gradient across the membrane of E. coli expressing the chlamydial SAM carrier resulted in partial reduction of SAM uptake and induced slight SAM efflux in the absence of suitable counterexchange substrates but apparently did not affect counterexchange transport (15). Therefore, the chlamydial carrier was assumed to catalyze a proton gradient-independent SAM/SAH exchange in addition to a proton-driven net SAM import. In this context, it is difficult to understand why significant counterexchange also occurs in the presence of a proton gradient. The postulated transport mode would imply that the recombinant chlamydial SAM carrier proteins act partially as symporters and partially as SAM/SAH counterexchangers when a proton gradient exists and hence presupposes a heterogeneous and inconsistent regulation.
The chlamydial SAM transporter differs from the rickettsial and A. asiaticus SAM carriers, at least in the regulatory impact of the proton gradient on SAM counterexchange. Establishment from different ancestral carriers of the DMT group might explain the functional differences of the chlamydial SAM carrier and the SAM transporter from R. prowazekii and A. asiaticus. In this context, it is interesting to note that chlamydiae are located within vacuoles (the so-called inclusion) inside their host cells (49), whereas rickettsiae and A. asiaticus as well as its relative “Ca. Cardinium hertigii” are located directly inside the host cytoplasm (32, 50, 51). This fundamental difference in subcellular location might explain the presence of different SAM transporters and thus different transport modes in the rickettsial/Bacteroidetes group of SAM transporters and chlamydiae.
Obviously, SAM transporter genes were spread due to horizontal gene transfer. However, until now, it has been impossible to determine the direction of transfer unambiguously (Fig. 2). Most likely, Aasi_1859 and RP076-like SAM transporters were invented in an intracellular ancestor of the Rickettsiales and then transferred to A. asiaticus, “Ca. Cardinium hertigii” (Bacteroidetes), and some members of the prasinophytes. Horizontal gene transfer between intracellular bacteria, including rickettsiae and A. asiaticus, has been suggested previously (32, 52–54). Due to the high level of divergence of SAM transport proteins from their nearest neighbors, these horizontal gene transfer events are most likely evolutionarily ancient.
In parasitic or endosymbiotic bacteria, the establishment of SAM transporters apparently is tightly associated with the intracellular lifestyle and particularly with the loss of the SAM biosynthesis capacity. However, until now, it has been completely unclear why Prasinophyceae, comparably primitive, mainly marine green algae, harbor carrier proteins highly related to the bacterial SAM transporters. The absence of homologues in other algae and higher plants points to a special function of the Aasi_1859 and RP076 homologues in Prasinophyceae. Gene transfer from intracellular bacteria (particularly from chlamydiae) to plants has been suggested by several studies (21, 55–57). In higher plants, members of the mitochondrial carrier family (MCF) (which are unrelated to Aasi_1859 and homologues) were shown to catalyze SAM provision to mitochondria and plastids (58, 59). Whether the DME-type carriers in addition to or instead of MCF-type carriers mediate mitochondrial or plastidial SAM transport and whether they might act in another compartment or accept substrates other than SAM and SAH in Prasinophyceae are open questions to be investigated in further studies. However, a role of the DME-type SAM transporters in uptake of extracellular SAM into corresponding algae can most likely be ruled out because SAM is not freely available in their habitat.
Conclusion.
Our analyses indicate that the SAM transporter of A. asiaticus operates as a proton gradient-dependent SAM/SAH antiporter and thus perfectly complements the restricted metabolic capabilities of A. asiaticus. Our results expand previous studies characterizing SAM transporters in R. prowazekii and C. trachomatis. The presence of functionally different SAM transporters in Bacteroidetes and Rickettsiales on the one hand and in chlamydiae on the other hand might be the result of different functional constraints due to their different intracellular localizations. Interestingly, SAM transporter-like genes were horizontally transferred between rickettsiae and Bacteroidetes and some members of the prasinophytes.
The analysis of additional bacterial SAM transporters, including homologues of prasinophytes, might help us to gain insights into structure-function relationships of this carrier subgroup.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a grant from the Austrian Science Fund (FWF) (project no. P22703-B17) to S.S.-E. and M.H. and the Deutsche Forschungsgemeinschaft (DFG) (project no. HA 5423/1-1 and SPP 1580) to the laboratory of I.H. M.H. acknowledges support from the European Research Council (ERC StG EvoChlamy).
Footnotes
Published ahead of print 10 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00195-13.
REFERENCES
- 1. Cantoni GL. 1975. Biological methylation: selected aspects. Annu. Rev. Biochem. 44:435–451 [DOI] [PubMed] [Google Scholar]
- 2. Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. 1996. S-Adenosylmethionine and methylation. FASEB J. 10:471–480 [PubMed] [Google Scholar]
- 3. Fontecave M, Atta M, Mulliez E. 2004. S-Adenosylmethionine: nothing goes to waste. Trends Biochem. Sci. 29:243–249 [DOI] [PubMed] [Google Scholar]
- 4. Cheng X, Roberts RJ. 2001. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 29:3784–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martin JL, McMillan FM. 2002. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12:783–793 [DOI] [PubMed] [Google Scholar]
- 6. Schubert HL, Blumenthal RM, Cheng X. 2003. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28:329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borchardt RT, Huber JA, Wu YS. 1976. Potential inhibitors of S-adenosylmethionine-dependent methyltransferases. 4. Further modifications of the amino and base portions of S-adenosyl-L-homocysteine. J. Med. Chem. 19:1094–1099 [DOI] [PubMed] [Google Scholar]
- 8. Borchardt RT, Shiong Y, Huber JA, Wycpalek AF. 1976. Potential inhibitors of S-adenosylmethionine-dependent methyltransferases. 6. Structural modifications of S-adenosylmethionine. J. Med. Chem. 19:1104–1110 [DOI] [PubMed] [Google Scholar]
- 9. Markham GD, DeParasis J, Gatmaitan J. 1984. The sequence of metK, the structural gene for S-adenosylmethionine synthetase in Escherichia coli. J. Biol. Chem. 259:14505–14507 [PubMed] [Google Scholar]
- 10. Thomas D, Surdin-Kerjan Y. 1987. SAM1, the structural gene for one of the S-adenosylmethionine synthetases in Saccharomyces cerevisiae. Sequence and expression. J. Biol. Chem. 262:16704–16709 [PubMed] [Google Scholar]
- 11. Cherest H, Surdin-Kerjan Y. 1978. S-Adenosyl methionine requiring mutants in Saccharomyces cerevisiae: evidences for the existence of two methionine adenosyl transferases. Mol. Gen. Genet. 163:153–167 [DOI] [PubMed] [Google Scholar]
- 12. Peleman J, Boerjan W, Engler G, Seurinck J, Botterman J, Alliotte T, Van MM, Inze D. 1989. Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell 1:81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mato JM, Corrales FJ, Lu SC, Avila MA. 2002. S-Adenosylmethionine: a control switch that regulates liver function. FASEB J. 16:15–26 [DOI] [PubMed] [Google Scholar]
- 14. Hafner EW, Tabor CW, Tabor H. 1977. Isolation of a metK mutant with a temperature-sensitive S-adenosylmethionine synthetase. J. Bacteriol. 132:832–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Binet R, Fernandez RE, Fisher DJ, Maurelli AT. 2011. Identification and characterization of the Chlamydia trachomatis L2 S-adenosylmethionine transporter. mBio 2(3):e00051–11. 10.1128/mBio.00051-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tucker AM, Winkler HH, Driskell LO, Wood DO. 2003. S-Adenosylmethionine transport in Rickettsia prowazekii. J. Bacteriol. 185:3031–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andersson JO, Andersson SG. 1999. Genome degradation is an ongoing process in Rickettsia. Mol. Biol. Evol. 16:1178–1191 [DOI] [PubMed] [Google Scholar]
- 18. Sanchez-Perez GF, Bautista JM, Pajares MA. 2004. Methionine adenosyltransferase as a useful molecular systematics tool revealed by phylogenetic and structural analyses. J. Mol. Biol. 335:693–706 [DOI] [PubMed] [Google Scholar]
- 19. Binet R, Maurelli AT. 2009. The chlamydial functional homolog of KsgA confers kasugamycin sensitivity to Chlamydia trachomatis and impacts bacterial fitness. BMC Microbiol. 9:279. 10.1186/1471-2180-9-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pannekoek Y, Heurgué-Hamard V, Langerak AA, Speijer D, Buckingham RH, van der Ende AJ. 2005. The N5-glutamine S-adenosyl-L-methionine-dependent methyltransferase PrmC/HemK in Chlamydia trachomatis methylates class 1 release factors. J. Bacteriol. 187:507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Collingro A, Tischler P, Weinmaier T, Penz T, Heinz E, Brunham RC, Read TD, Bavoil PM, Sachse K, Kahane S, Friedman MG, Rattei T, Myers GS, Horn M. 2011. Unity in variety—the pan-genome of the Chlamydiae. Mol. Biol. Evol. 28:3253–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fuxelius HH, Darby A, Min CK, Cho NH, Andersson SG. 2007. The genomic and metabolic diversity of Rickettsia. Res. Microbiol. 158:745–753 [DOI] [PubMed] [Google Scholar]
- 23. Tjaden J, Winkler HH, Schwöppe C, van der Laan M, Möhlmann T, Neuhaus HE. 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for the transport of energy. J. Bacteriol. 181:1196–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haferkamp I, Schmitz-Esser S, Linka N, Urbany C, Collingro A, Wagner M, Horn M, Neuhaus HE. 2004. A candidate NAD+ transporter in an intracellular bacterial symbiont related to Chlamydiae. Nature 432:622–625 [DOI] [PubMed] [Google Scholar]
- 25. Audia JP, Winkler HH. 2006. Study of the five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J. Bacteriol. 188:6261–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schmitz-Esser S, Linka N, Collingro A, Beier CL, Neuhaus HE, Wagner M, Horn M. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to chlamydiae and rickettsiae. J. Bacteriol. 186:683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haferkamp I, Schmitz-Esser S, Wagner M, Neigel N, Horn M, Neuhaus HE. 2006. Tapping the nucleotide pool of the host: novel nucleotide carrier proteins of Protochlamydia amoebophila. Mol. Microbiol. 60:1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Knab S, Mushak TM, Schmitz-Esser S, Horn M, Haferkamp I. 2011. Nucleotide parasitism by Simkania negevensis (Chlamydiae). J. Bacteriol. 193:225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krause DC, Winkler HH, Wood DO. 1985. Cloning and expression of the Rickettsia prowazekii ADP/ATP translocator in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 82:3015–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ren Q, Paulsen IT. 2007. Large-scale comparative genomic analyses of cytoplasmic membrane transport systems in prokaryotes. J. Mol. Microbiol. Biotechnol. 12:165–179 [DOI] [PubMed] [Google Scholar]
- 31. Moya A, Peretó J, Gil R, Latorre A. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 9:218–229 [DOI] [PubMed] [Google Scholar]
- 32. Schmitz-Esser S, Tischler P, Arnold R, Montanaro J, Wagner M, Rattei T, Horn M. 2010. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J. Bacteriol. 192:1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. 2009. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol. Direct 4:13. 10.1186/1745-6150-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wernegreen JJ. 2005. For better or worse: genomic consequences of intracellular mutualism and parasitism. Curr. Opin. Genet. Dev. 15:572–583 [DOI] [PubMed] [Google Scholar]
- 35. Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9:286–298 [DOI] [PubMed] [Google Scholar]
- 36. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 38. Saier MH., Jr 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Penz T, Schmitz-Esser S, Kelly SE, Cass BN, Muller A, Woyke T, Malfatti SA, Hunter MS, Horn M. 2012. Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 8:e1003012. 10.1371/journal.pgen.1003012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jana B, Panja S, Saha S, Basu T. 2009. Mechanism of protonophores-mediated induction of heat-shock response in Escherichia coli. BMC Microbiol. 9:20. 10.1186/1471-2180-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Casadevall A. 2008. Evolution of intracellular pathogens. Annu. Rev. Microbiol. 62:19–33 [DOI] [PubMed] [Google Scholar]
- 42. Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, Rattei T, Mewes HW, Wagner M. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728–730 [DOI] [PubMed] [Google Scholar]
- 43. Schwöppe C, Winkler HH, Neuhaus HE. 2002. Properties of the glucose 6-phosphate transporter from Chlamydia pneumoniae (HPTcp) and the glucose 6-phosphate sensor from Escherichia coli (UhpC). J. Bacteriol. 184:2108–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, Santos-Guzman J, Swendseid ME, Cogger EA, James SJ. 2001. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J. Nutr. 131:2811–2818 [DOI] [PubMed] [Google Scholar]
- 45. Zhu CM, Cumaraswamy A, Henney HR., Jr 1989. Comparison of polyamine and S-adenosylmethionine contents of growing and encysted Acanthamoeba isolates. Mol. Cell. Biochem. 90:145–153 [DOI] [PubMed] [Google Scholar]
- 46. Cumaraswamy A, Henney H., Jr 1990. S-Adenosylmethionine and S-adenosylhomocysteine transitions in encysting Physarum flavicomum amoebae. Biochem. Cell Biol. 68:769–777 [DOI] [PubMed] [Google Scholar]
- 47. Lopez-Amoros R, Comas J, Vives-Rego J. 1995. Flow cytometric assessment of Escherichia coli and Salmonella typhimurium starvation-survival in seawater using rhodamine 123, propidium iodide, and oxonol. Appl. Environ. Microbiol. 61:2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saka HA, Valdivia RH. 2010. Acquisition of nutrients by Chlamydiae: unique challenges of living in an intracellular compartment. Curr. Opin. Microbiol. 13:4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hackstadt T. 1998. The diverse habitats of obligate intracellular parasites. Curr. Opin. Microbiol. 1:82–87 [DOI] [PubMed] [Google Scholar]
- 51. Zchori-Fein E, Perlman SJ, Kelly SE, Katzir N, Hunter MS. 2004. Characterization of a ‘Bacteroidetes ’ symbiont in Encarsia wasps (Hymenoptera: Aphelinidae): proposal of ‘Candidatus Cardinium hertigii’. Int. J. Syst. Evol. Microbiol. 54:961–968 [DOI] [PubMed] [Google Scholar]
- 52. Bertelli C, Greub G. 2012. Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms. Front. Cell. Infect. Microbiol. 2:110. 10.3389/fcimb.2012.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, Fournier PE, Claverie JM, Raoult D. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2:e76. 10.1371/journal.pgen.0020076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, Shukla M, Walenz B, Hill CA, Nene VM, Azad AF, Sobral BW, Caler E. 2012. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J. Bacteriol. 194:376–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tyra HM, Linka M, Weber AP, Bhattacharya D. 2007. Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 8:R212. 10.1186/gb-2007-8-10-r212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang J, Gogarten JP. 2007. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 8:R99. 10.1186/gb-2007-8-6-r99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moustafa A, Reyes-Prieto A, Bhattacharya D. 2008. Chlamydiae has contributed at least 55 genes to Plantae with predominantly plastid functions. PLoS One 3:e2205. 10.1371/journal.pone.0002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bouvier F, Linka N, Isner JC, Mutterer J, Weber AP, Camara B. 2006. Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 18:3088–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Palmieri L, Arrigoni R, Blanco E, Carrari F, Zanor MI, Studart-Guimaraes C, Fernie AR, Palmieri F. 2006. Molecular identification of an Arabidopsis S-adenosylmethionine transporter. Analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol. 142:855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.