Abstract
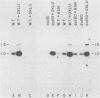
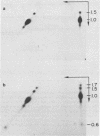
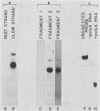
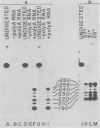
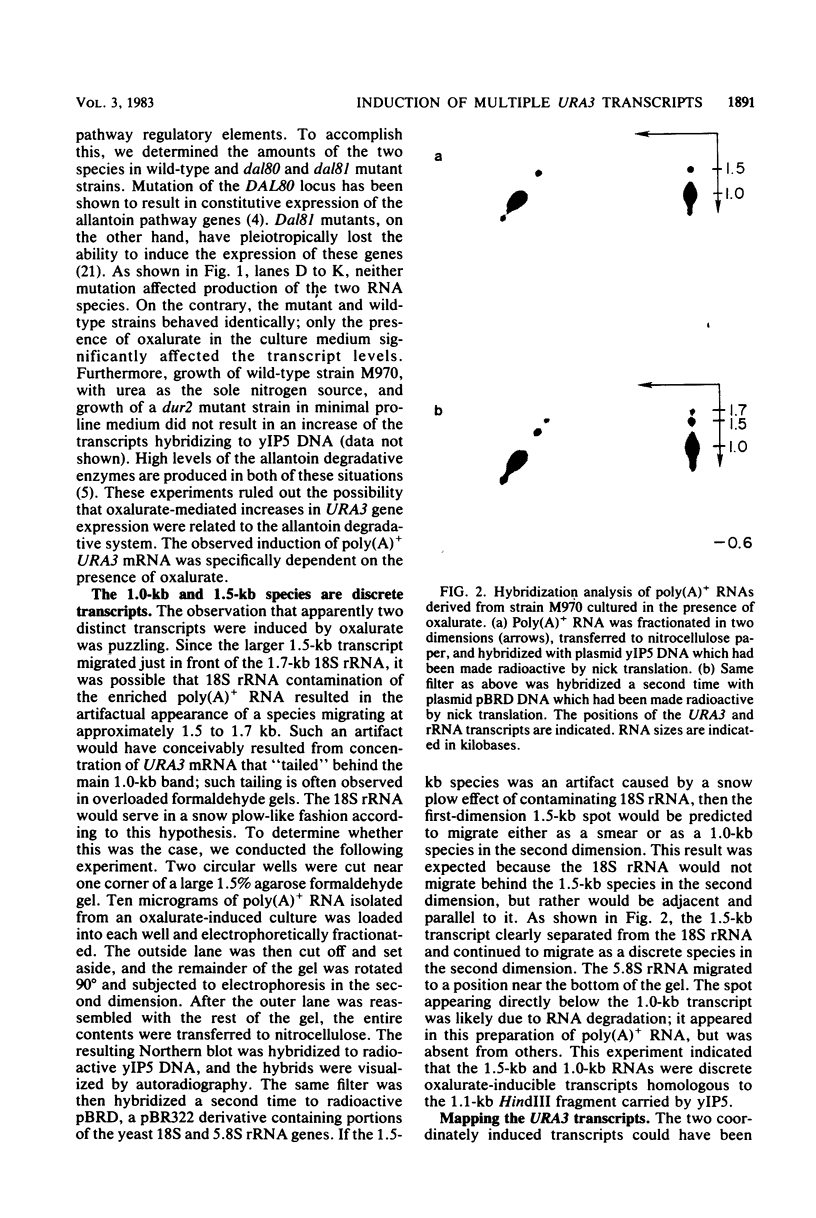
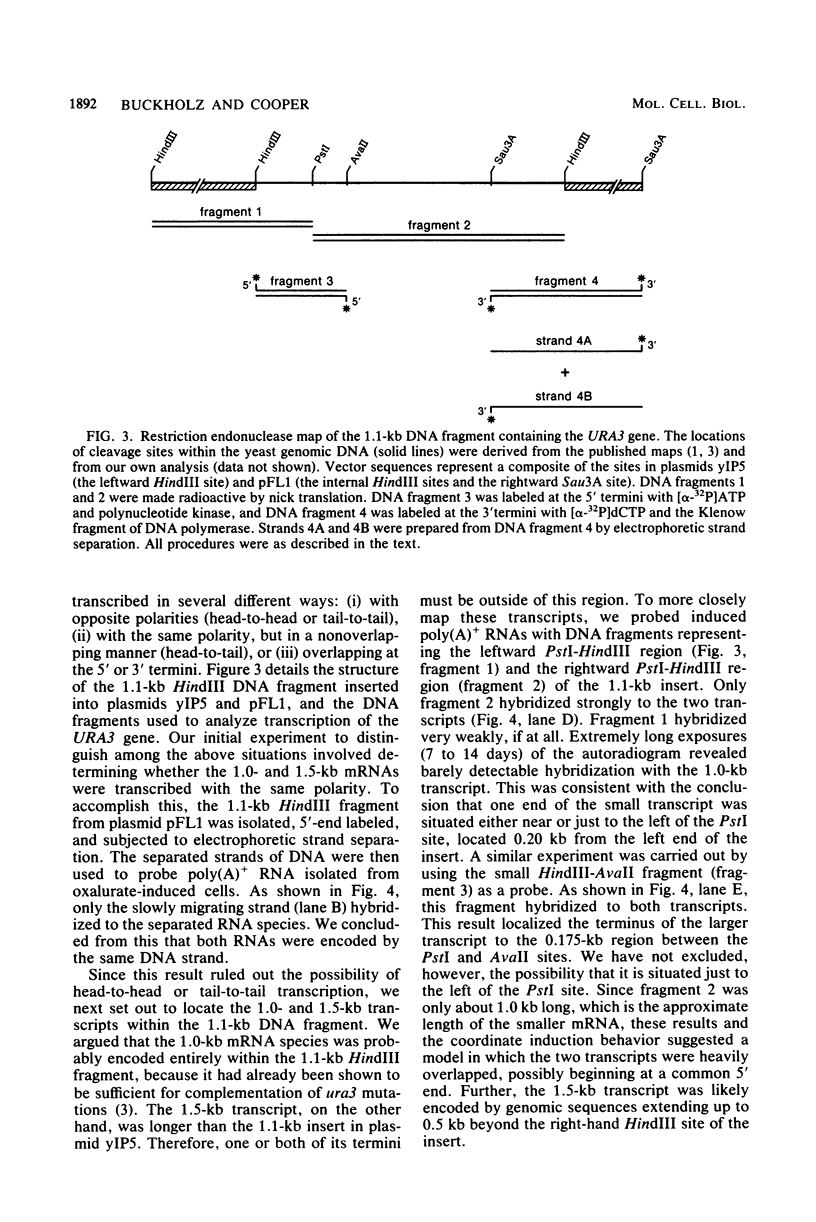
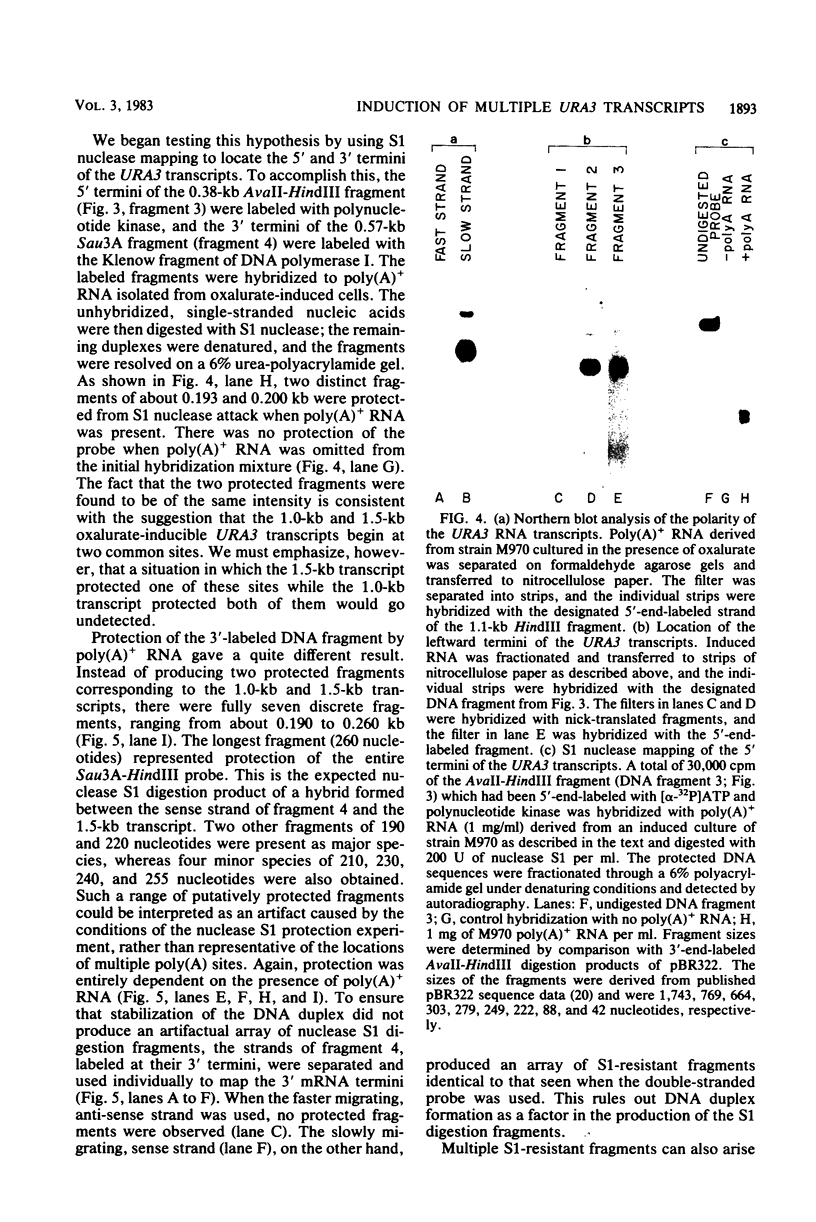
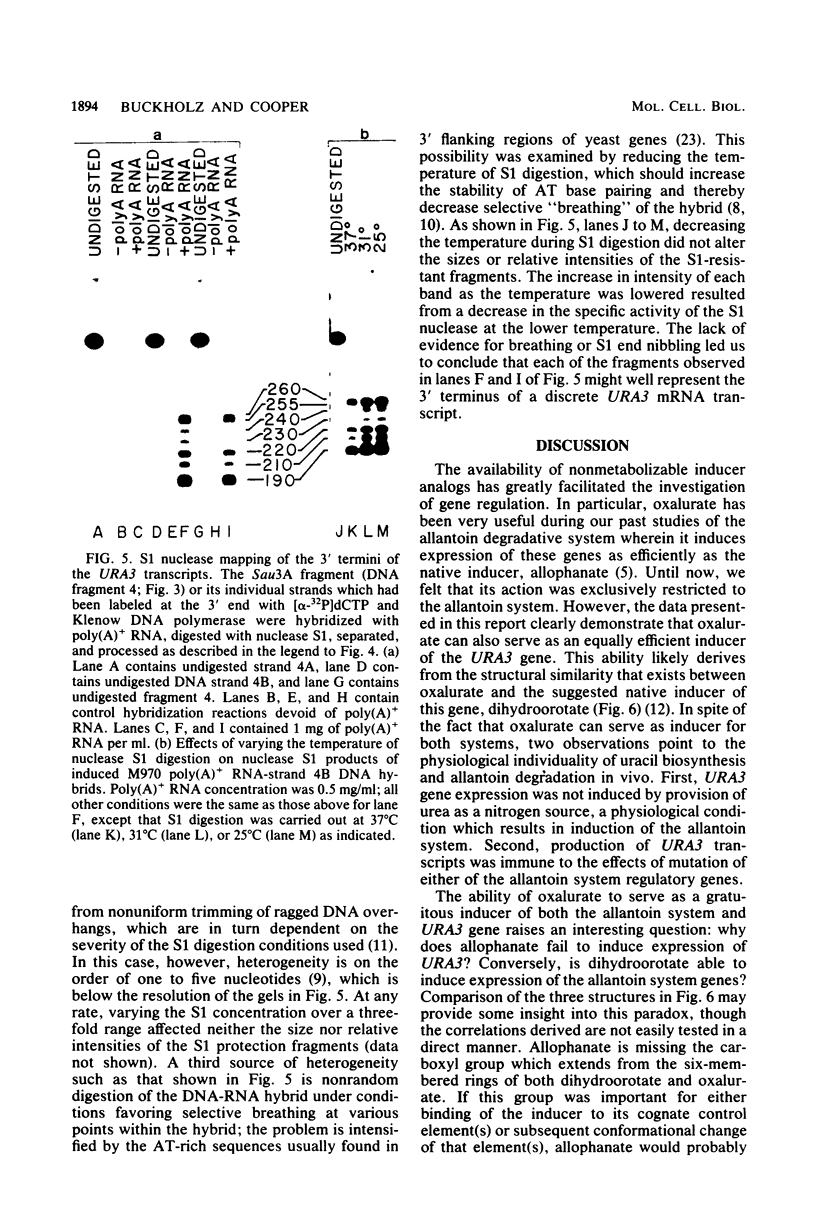
The URA3 gene from Saccharomyces cerevisiae is localized on a 1.1-kilobase (kb) DNA fragment. By using this fragment as a hybridization probe, we found that oxalurate, a gratuitous inducer of the allantoin degradative system, also serves to induce URA3 specific RNA. This response is restricted to oxalurate; other conditions which bring about high-level synthesis of the allantoin degradative enzymes did not produce the effect. Two classes of RNA (1.0 and 1.5 kb) were found to be oxalurate induced. Both classes are encoded by the URA3 gene, overlap, and probably do not significantly differ at their 5' termini. Northern blot mapping of the transcripts indicated that the 1.5-kb transcript was likely encoded by sequences extending up to 0.5 kb downstream from the 3' terminus of the 1.0-kb transcript. Analysis of the endpoints of the major 1.0-kb URA3 transcript by S1 nuclease mapping revealed the existence of two 5' termini, separated by 5 to 10 nucleotides, and seven 3' termini, separated by 5 to 20 nucleotides each, over a range of about 70 bases.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach M. L., Lacroute F., Botstein D. Evidence for transcriptional regulation of orotidine-5'-phosphate decarboxylase in yeast by hybridization of mRNA to the yeast structural gene cloned in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):386–390. doi: 10.1073/pnas.76.1.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chevallier M. R., Bloch J. C., Lacroute F. Transcriptional and translational expression of a chimeric bacterial-yeast plasmid in yeasts. Gene. 1980 Oct;11(1-2):11–19. doi: 10.1016/0378-1119(80)90082-7. [DOI] [PubMed] [Google Scholar]
- Chisholm G., Cooper T. G. Isolation and characterization of mutants that produce the allantoin-degrading enzymes constitutively in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Sep;2(9):1088–1095. doi: 10.1128/mcb.2.9.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Lawther R. P. Induction of the allantoin degradative enzymes in Saccharomyces cerevisiae by the last intermediate of the pathway. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2340–2344. doi: 10.1073/pnas.70.8.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Gregory S. P., Dillon N. O., Butterworth P. H. The localisation of the 5'-termini of in vivo transcripts of a cloned rainbow trout protamine gene. Nucleic Acids Res. 1982 Dec 11;10(23):7581–7592. doi: 10.1093/nar/10.23.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld G. C., de Boer E., Shewmaker C. K., Flavell R. A. DNA sequences necessary for transcription of the rabbit beta-globin gene in vivo. Nature. 1982 Jan 14;295(5845):120–126. doi: 10.1038/295120a0. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Lacroute F. Regulation of pyrimidine biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1968 Mar;95(3):824–832. doi: 10.1128/jb.95.3.824-832.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Parnes J. R., Robinson R. R., Seidman J. G. Multiple mRNA species with distinct 3' termini are transcribed from the beta 2-microglobulin gene. 1983 Mar 31-Apr 6Nature. 302(5907):449–452. doi: 10.1038/302449a0. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Williamson V. M., Young E. T. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem. 1983 Feb 25;258(4):2674–2682. [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982 Dec;2(12):1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R., Cooper T. G. Oxaluric acid: a non-metabolizable inducer of the allantoin degradative enzymes in Saccharomyces cerevisiae. J Bacteriol. 1974 Mar;117(3):1240–1247. doi: 10.1128/jb.117.3.1240-1247.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Turoscy V., Cooper T. G. Pleiotropic control of five eucaryotic genes by multiple regulatory elements. J Bacteriol. 1982 Sep;151(3):1237–1246. doi: 10.1128/jb.151.3.1237-1246.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickerham L. J. A Critical Evaluation of the Nitrogen Assimilation Tests Commonly Used in the Classification of Yeasts. J Bacteriol. 1946 Sep;52(3):293–301. [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zehner Z. E., Paterson B. M. Characterization of the chicken vimentin gene: single copy gene producing multiple mRNAs. Proc Natl Acad Sci U S A. 1983 Feb;80(4):911–915. doi: 10.1073/pnas.80.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]