Abstract
Over the last decade, RNA interference technology has shown therapeutic promise in rodent models of dominantly inherited brain diseases, including those caused by polyglutamine repeat expansions in the coding region of the affected gene. For some of these diseases, proof-of concept studies in model organisms have transitioned to safety testing in larger animal models, such as the nonhuman primate. Here, we review recent progress on RNA interference-based therapies in various model systems. We also highlight outstanding questions or concerns that have emerged as a result of an improved (and ever advancing) understanding of the technologies employed.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0183-8) contains supplementary material, which is available to authorized users.
Keywords: RNAi, CNS, miRNA, shRNA, siRNA, polyQ
Introduction
The phenomenon of modulating gene expression by small RNAs is termed RNA interference (RNAi). Since the discovery of RNAi in plants and worms in the 1990s, scientists have made steady progress to understand the mechanisms of RNAi and use that knowledge to generate tools to study gene function. In addition, RNAi methodologies have been used to modulate the expression of disease genes with an intention of developing novel therapies. RNAi-based therapies are now in clinical trials for a variety of diseases, such as age-related macular degeneration, diabetic macular edema, solid tumors, and chronic myeloid leukemia (clinicaltrials.gov). For central nervous system (CNS) diseases in particular, early studies in model organisms and safety studies in nonhuman primates have shown that RNAi is an attractive therapy that warrants testing in patients. Initial pioneering studies using transgenic mice with inducible disease genes showed that it is possible to reverse disease phenotypes after onset [1–4], and set the stage for the therapeutic development of RNAi. In this review, we highlight recent discoveries in cell and animal models that have advanced the field of RNAi-based CNS therapeutics.
RNAi Mechanism and Function
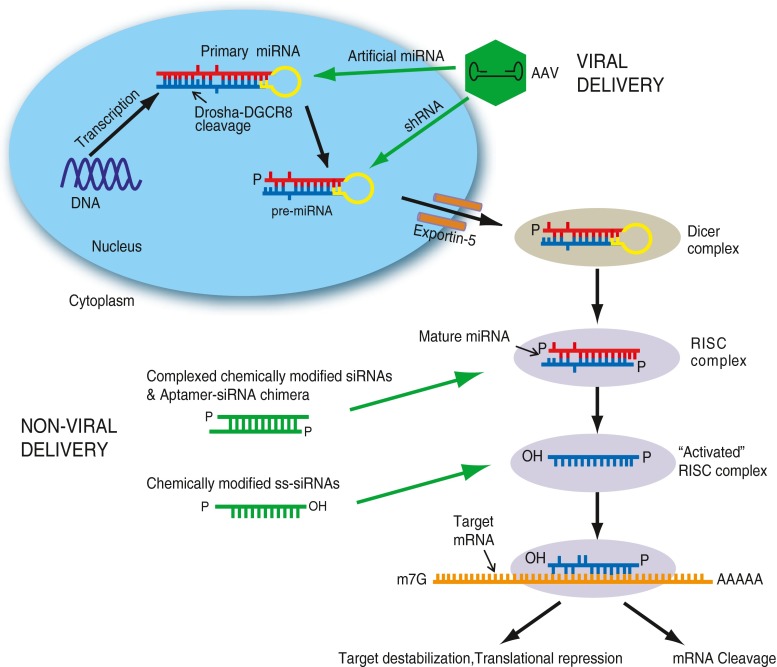
RNAi is an innate gene regulatory mechanism that is essential to many cellular processes, such as proliferation, differentiation, cell death, and remodeling [5]. RNAi also plays an important role in host defense by protecting against viral infection and transposable elements [6]. In RNAi, the cell makes use of double-stranded (ds)RNA molecules to silence the expression of an messenger (m)RNA molecule by complementary base pairing. One form of naturally-occurring dsRNA molecules are microRNAs (miRNAs), which are transcribed in the nucleus as stem loop structured “primary miRNAs” (pri-miRNAs) from pol II or pol III promoters [7]. Pri-miRNAs are cleaved by the Drosha-DGCR8 microprocessor complex in the nucleus to form ~60–70 nucleotide hairpin-like structures called precursor-miRNAs (pre-miRNAs) [8, 9]. The pre-miRNAs are then exported to the cytoplasm by Exportin-5 and further processed by the Dicer (an RNase III endonuclease)-containing complex, which cleaves the loop structure of the pre-miRNA to release short, ~21-nucleotide, mature miRNA sequences [10, 11]. Dicer also processes exogenous long dsRNAs (such as viral RNAs) into smaller, 21-nucleotide small interfering RNAs (siRNAs) [12]. One strand of the miRNA or siRNA duplex, known as the antisense or guide strand, is selectively loaded into the Ago-containing RNA-induced silencing complex (RISC). This process is known as strand biasing; the non-loaded strand is referred to as the passenger strand [13]. This “activated” RISC carries out gene silencing, either by Ago2-mediated cleavage of the complementary target mRNA (in the case of siRNAs) or by target destabilization or translational repression (in the case of miRNAs) after imperfect base pairing to the 3’ untranslated region (UTR) (Fig. 1) [14–16].
Fig. 1.
Co-opting the microRNA (miRNA) pathway for delivery of RNA interference triggers to the central nervous system (CNS). Primary miRNAs are transcribed in the nucleus and are processed by the Drosha-DGCR complex to give rise to precursor (Pre)-miRNAs. Pre-miRNAs are exported out of the nucleus by Exportin-5 and undergo further processing by Dicer in the cytoplasm to give rise to mature miRNAs. The mature miRNA is then loaded into RNA-induced silencing complex (RISC) to carry out silencing by binding to complementary messenger RNA (mRNA) sequences. Artificial miRNAs or short hairpin RNAs (shRNAs) can be delivered via an adeno-associated virus (AAV) and enter the miRNA pathway at different stages of the miRNA pathway. Small interfering RNAs (siRNAs) that are complexed or delivered directly into cells, enter the pathway at the Dicer-to-RISC stage or can be incorporated directly into RISC to carry out gene silencing
RNAi as a Tool for Directed Gene Silencing
RNAi has evolved rapidly as a tool for directed gene silencing. The RNAi machinery can be co-opted in many ways to achieve gene expression knockdown of a select target (Fig. 1). Synthetic siRNAs (~21 nucleotides) can be introduced into cells, which are loaded directly into RISC or, in the case of longer dsRNAs (25–27 nucleotides), first processed by Dicer and then loaded into the RISC to achieve gene silencing [17]. Gene-targeting siRNA duplexes can also be embedded in hairpin-based structures made to mimic the pri-miRNA (called artificial miRNAs) or the pre-miRNA (called short hairpin or shRNAs); when placed into expression vectors, they are transcribed in the nucleus and processed by the endogenous RNAi pathway to achieve gene silencing. shRNAs are typically expressed from strong Pol III promoters (such as U6 or H1), while artificial miRNAs can be expressed from pol II or pol III promoters. While shRNAs may have more potent silencing capability, they are often expressed at very high levels and can saturate the RNAi machinery, which disrupts endogenous miRNA processing and can induce toxicity [18, 19]. Artificial miRNAs, however, are generally safer and less toxic, and they do not appear to disrupt endogenous miRNA processing [18–23]. Although artificial miRNAs are less toxic, their safety profile is also dictated by the design of the RNAi sequence.
Designing RNAi Sequences
Designing a siRNA sequence to reduce target gene expression with efficacy and specificity is one of the key factors to achieve successful RNAi. Important steps in designing an efficient siRNA have been described by several groups [24–26]. Two important criteria include designing sequences for proper strand biasing and minimizing off-targeting.
To favor antisense strand incorporation into RISC, the antisense strand of the siRNA should have strong G–C base pairing at the 3’ end and weak base pairing (A–U or G–U) at the 5’ end, as RISC loads the strand with the lowest 5’ thermodynamic stability [13]. There are many online tools and guidelines that help to design siRNA sequences to a gene target of interest [27–30]. Newer online tools also incorporate the very important aspect of siRNA off-targeting [31–33].
Off-targeting is a phenomenon by which a siRNA binds to and represses unintended targets owing to complementarity with the siRNA ‘seed’ sequence. In 2006, Fedorov et al. [34] reported toxic effects due to off-target effects of siRNAs. As miRNAs primarily target the 3’UTRs of genes, it was found that seed complementarity to hexamers in the 3’UTR’s of genes is proportional to the number of off-target effects [35, 36]. Limiting off-targeting is particularly critical when designing siRNAs for therapeutic purposes. Our laboratory has designed a web-based program that designs highly-specific siRNA sequences to a target of interest by taking into consideration the off-targets of an siRNA seed sequence, and gives each siRNA a score depending on the number of potential off-targets [31]. Researchers can thus pick siRNA sequences with low potential off-targets and screen them in vitro to identify candidate sequences for delivery in vivo. Alternatively, the siRNA sequence can be incorporated into a miRNA or shRNA backbone for delivery in vitro and in vivo. The process of designing and screening of hairpin-based RNAi sequences (shRNAs and artificial miRNAs) has been described in detail previously [37].
Delivering RNAi Sequences to the CNS
RNAi delivery to the CNS faces unique challenges. For effective RNAi delivery via the blood, the presence of the blood–brain barrier (BBB) is an obstacle that must be overcome, while, for direct injection into the brain, steps must be taken to avoid toxic or inflammatory reactions. An ideal delivery system for RNAi to the CNS should be minimally immunogenic, nontoxic, target specific cells of the CNS, knockdown the specific target mRNA efficiently, and be easy to manufacture [38]. Two major types of delivery systems have emerged over the years, differing in production, safety, and efficacy. These are broadly classified as viral and non-viral delivery systems.
Non-viral Delivery
siRNAs can be delivered directly in vitro or in vivo to achieve knockdown of a gene of interest. However, exogenously delivered siRNAs are natural substrates for nucleases, which makes them inherently unstable. This can be overcome by chemically modifying siRNAs to resist nucleases. In addition, chemical modifications can increase cell uptake, strand biasing, and efficiency of gene knockdown, while reducing immunogenicity and off-target effects [39]. Different types of chemical modifications have been introduced into the backbone of the siRNA, the most popular of which are 2’-Fluoro, 2’-O-methyl and locked nucleic acids [39]. A recent study demonstrated allele-specific silencing of mutant Htt in Huntington disease (HD) mice using chemically-modified single-stranded siRNA (ss-siRNA) molecules [40, 41]. ss-siRNAs are advantageous as they avoid off-target effects associated with passenger strand loading, although some other issues remain [42]. In addition to chemical modifications, siRNAs can be complexed into liposomes or nanoparticles, or can be incorporated into aptamer structures for delivery in vivo. Owing to their ability to cross the BBB and their small size, nanoparticles delivered intravenously or via carotid artery can gain entry to the CNS [43, 44]. Recently, aptamers functionalized with nanoparticles targeted glioblastomas for potential therapy [45]. Nanoparticles can target specific cell types in tissues, but have low transfection efficiency [46].
While non-viral methods have been used in vivo, they are generally less efficacious and are, by nature, transient, requiring repeated delivery. This transient nature can be advantageous for therapies that do not require long-term treatment, such as in antitumor and antiviral therapies. In addition, this provides an important safety measure; in the case of adverse side effects, treatment can simply be terminated.
While methods to improve the efficacy of non-viral molecules are currently under development, viral vectors have been used successfully from mouse models to human studies in gene replacement strategies.
Viral Delivery
There are a number of viral vectors that can be used for gene delivery to the CNS, as discussed in a prior review [38]. The two main viral vector systems that are used to transduce the CNS are lentiviruses (LV) and adeno-associated viruses (AAV). Both viruses are minimally immunogenic and can transduce a number of CNS cell types. Recombinant lentiviruses are pseudotyped with various glycoproteins that can impart different tropisms after directed delivery into brain [47, 48], and they have been used successfully in gain-of-function [49] and loss of function studies [50–53]. One difference between lentivirus and AAV or adenovirus-based systems is the level of expression. This is due, in part, because LV-mediated transduction often results in low copy numbers of transgene/cell. Also, the placement of the expression cassette in the LV genome can affect expression levels [54]. Most LV vectors integrate unless the integrase activity has been inactivated. As integrase-deficient vectors often have low titers compared with their integrase competent counterparts, their production for use for therapeutic applications is impractical. Integration competency for CNS applications may be less of an issue than in the setting of stem cell transduction (most cells in the CNS are not dividing), where integration and activation of an oncogenic gene provides a growth advantage for the transformed cell [55, 56].
AAV belongs to the genus Dependovirus and in its wild type state requires a helper virus, such as adenovirus, to replicate. A number of factors make AAV suitable for gene delivery in vivo. AAV can be manufactured easily and it is scalable for human use [57, 58], particularly for the relatively low volumes needed for brain-expressed targets. Additionally, AAV rarely integrates into the genome. In general, AAVs are non-pathogenic and have low immunogenic properties, which make them ideal for gene delivery in vivo [38]. AAVs confer robust expression, efficiently transduce neurons and other cell types, and, in the absence of an immune response to what is being expressed, can afford long-term expression [59–61].
Tissue tropism of AAV is dictated by the capsid serotype. AAV capsids with different cell/tissue tropisms have been identified and, depending on the capsid serotype, AAV can transduce neurons, astrocytes, glia, and ependymal cells with a high transduction efficiency [62–66]. The AAV capsid can be modified to alter its tropism in several ways, including directed evolution, capsid shuffling, and incorporation of targeting peptides. Directed evolution involves mutagenesis of the capsid, which may alter tropism [67, 68]. Capsid shuffling involves the assembly of variant capsid sequences to give rise to recombinant capsids with tropisms to different cell types [69–71]. AAV tropism can also be altered by the incorporation of targeting ligand into the capsid, to mediate ligand specific receptor binding [72–74].
Gene transfer after direct delivery of AAV vectors by intraparenchymal, intraventricular, or intrathecal injections to target cells of the brain is used for RNAi delivery. Direct delivery by intraparenchymal injections has proven effective for targeting neurons in various neurodegenerative diseases and it limits transduction to those tissues most relevant to disease. Widespread exposure of a transgene product occurs after intraventricular or intrathecal delivery of AAVs, when the transgene product is a secreted molecule [75]. Recently, intrathecal injection of AAV9 or AAV2.5 showed robust transduction of the brain and spinal cord in nonhuman primates [76].
Peripheral delivery of AAVs for brain targeting has also been used [72–74, 77–79]. Concerns about this approach for clinical application are the high doses needed, the transduction of peripheral organs (which may not be desirable), and the induction of a robust anti-AAV and likely anti-transgene response. Nonetheless, using vectors that can cross the BBB may be beneficial for some applications. Intravenous delivery of AAV9, a recently identified serotype, can cross the BBB after, and transduce neurons in neonatal mice, and astrocytes and scattered neurons in adult mice and rhesus macaques [77, 78]. Also, variants of AAV9 transduce motor neurons and astrocytes after systemic delivery by intravenous injection to adult mice [78, 79].
Although AAVs have a small packaging capacity (~4.7 kb), they are ideally suited to deliver the small RNAi expression cassette. RNAi sequences delivered to the brain after AAV injection have shown therapeutic promise in mouse models of dominantly inherited polyglutamine (polyQ) diseases and other neurological disorders, as discussed in the following.
Emerging Therapies
RNAi therapy is well suited for diseases where the disease-causing gene acquires a toxic ‘gain of function’ effect. The identification of such disease-causing genes (modifiers or mutant genes) has allowed researchers to design RNAi molecules to target the disease-causing allele and demonstrate therapeutic potential (Table 1).
Table 1.
Progress in RNA inteference (RNAi) therapeutics demonstrating therapeutic potential in cell and animal models
| Disease | Target gene | Approach used | Delivery vehicle | Study demonstrating therapeutic potential | References |
|---|---|---|---|---|---|
| Huntington’s Disease (HD) | HTT | Allele-specific (AS) and nonallele- specific (NAS) | AAV1, AAV2, chemically modified ss-siRNAs | Silencing of endogenous HTT by shRNAs and artificial miRNAs in rhesus is tolerated up to 6 months without toxicity. Potent allele-specific silencing of mutant HTT is demonstrated in HD mice using chemically modified ss-siRNAs targeting expanded CAG repeats. | [40, 41, 80–82, 97] |
| SCA1 | ATXN1 | NAS | AAV1 | Silencing of ATXN1 using shRNAs and, more recently, artificial miRNAs in SCA1 transgenic and knock-in mouse models show improvement of motor coordination without toxicity. | [94] |
| SCA2 | ATXN2 | AAV1 | Partial suppression of InsP3R in the cerebellum improved motor coordination, reduced Purkinje cell degeneration in SCA2 transgenic mice. | [97] | |
| SCA3 | ATXN3 | NAS, AS | Lentivirus | Nonallele-specific silencing and allele-specific silencing of mutant ATXN3 was well tolerated and reduced neuropathology in a rat model of SCA3. | [104, 105] |
| SCA6 | CACNA1A | AS | Splice-isoform specific RNAi using artificial miRNAs demonstrated allele-specific silencing of mutant CACNA1A in vitro | [113] | |
| SCA7 | ATXN7 | AS | Allele-specific silencing of mutant ATXN7 is demonstrated in vitro using shRNAs. | [117] | |
| Parkinson’s disease | SNCA | Lentivirus, AAV2 | Allele-specific silencing of α-syn using shRNAs was observed in the rat brain and ameliorated behavioral deficits, but was also toxic in dopamine neurons. | [22, 132] | |
| LRRK2 | AS | Allele-specific silencing of mutant α-syn and LRRK2 was achieved in vitro using artificial mirtron mimics. | [134] | ||
| Alzheimer’s disease | BACE1 | Lentivirus | shRNAs silence BACE1 to reduce amyloid production and behavioral deficits in a transgenic mouse model. | [122] | |
| Tau | AS | Allele-specific silencing of mutant Tau demonstrated using shRNAs in vitro. | [127] | ||
| APP | AS | AAV5 | Allele-specfic silencing of APP in a transgenic AD mouse model mitigated phenotypic progression. | [124] | |
| PS1 | AS | Allele-specific siRNAs silence mutant PS1 in vitro and reduced amyloid β42 production. | [124, 125] | ||
| CDK5 | Lentivirus | shRNAs targeting CDK5 reduced neurofibrillary tangles in a transgenic mouse model. | [126] | ||
| PLK1 | Lentivirus | RNAi silencing of Plk1 in vitro reduced amyloid β-induced cell death. | [129] | ||
| MSUT2 | Silencing of MSUT2 using siRNAs decreased tau aggregation in vitro. | [128] | |||
| Dystonia | TOR1A | AS | Lentivirus | Allele specific silencing of TorsinA(ΔE) by shRNAs worked well in vitro, but when moved into a mouse model, the shRNAs proved to be toxic. | [21, 53] |
| SBMA | CELF2 | AAV9 | Overexpression of naturally occurring miR-196a indirectly enhances decay of androgen receptor through silencing of CELF2 in vivo. | [141] | |
| ALS | SOD1 | AS | Lentivirus | Silencing SOD1 slows progression and extends survival in rodent models of ALS | [51, 52, 147, 148] |
ss-siRNA = single-stranded small interfering RNA; shRNA = short hairpin RNA; miRNA = microRNA; SCA1 (2, 3, 6, 7) = spinocerebellar ataxia type 1 (2, 3, 6, 7); InsP3 = inositol 1,4,5 phosphate receptor; APP = amyloid precursor protein; siRNA = small interfering RNA; PS1 = presenilin-1; CDK5 = cyclin-dependent kinase 5; SBMA = spinobulbar muscular atrophy; ALS = amyotrophic lateral sclerosis
Huntington’s Disease
Huntington’s Disease (HD) is caused by a polyQ expansion in the coding region of the gene Huntingtin (HTT). HD is a gain-of-function autosomal dominant disease with neuronal dysfunction occurring prior to cell death in medium spiny neurons within the striatum, as well as other brain regions. Patients exhibit involuntary hyperkinetic movements, coordination difficulties, and cognitive disturbances [50, 80]. Both nonallele-specific (targeting the mutant and wild type alleles) and allele-specific (targeting only the mutant allele) approaches for HD therapy are under development. It has been shown recently that nonallele-specific silencing, using AAV-mediated delivery of RNAi, provides benefits in a mouse model of HD and 2 studies assessing the effect of knockdown of endogenous HTT in rhesus found no adverse effects up to 6 months post-injection [81, 82]. However, HTT is necessary for embryonic development and is involved in cellular pathways in differentiated neurons [83–88]. Thus, long-term therapies may require targeting only the mutant allele. This has led to the development of strategies for targeting only mutant HTT [83–88]. An example is the work done by Hu et al. [89] targeting the CAG repeat sequence expansion. Their target strategy capitalized on the concept that siRNAs bind to their targets with full complementarity, while miRNAs exhibit one or more mismatches to their targets. Mimicking the miRNA mechanism, one or two mismatched bases were introduced into therapeutic RNAi sequences. These miRNA-like sequences showed greater inhibition of the mutant HTT in patient-derived cells whereas the siRNAs showed little selectivity between wild type and mutant alleles. This study also looked at the CAG containing mRNAs (TBP and FOXP2) and found no off-target silencing [89]. Other allele-specific techniques have targeted single nucleotide polymorphisms (SNPs) occurring in some, but not all, mutant HTT alleles [90–92].
Spinocerebellar Ataxia Type 1
Spinocerebellar ataxia type 1 (SCA1) is a late onset, autosomal dominant neurodegenerative disease caused by a polyQ expansion in Ataxin1 (ATXN1), which encodes the ATXN1 protein. The average age of onset is within the fourth decade of life, although juvenile cases have been documented [93]. Symptoms include loss of coordination, dysarthria, and cognitive impairment. Purkinje cell (PC) death and brain stem neuronal death is characteristic of SCA1 [93]. In 2004, RNAi was established as a potential therapy for SCA1 after successful rescue of the disease phenotype in SCA1 transgenic mice expressing the human ATXN1 protein containing 82 pathogenic polyQ repeats. AAVs expressing shRNAs against human 82Q ATXN1 were injected to SCA1 mice cerebellar cortices, improving molecular and behavioral phenotypes [94]. Further testing of RNAi triggers using artificial miRNAs is being pursued.
Spinocerebellar Ataxia Type 2
Spinocerebellar ataxia type 2 (SCA2) is a polyQ disease caused by an expansion of >31 CAG repeats in the coding region of Ataxin2 (ATXN2), while normal individuals have ~20 CAG repeats (CAG8-CAA-CAG4-CAA-CAG8) [95]. SCA2 is characterized by initial hyper-reflexia followed by hyporeflexia with disease progression, ophthalmoplegia, dysphagia, ataxia, and, occasionally, symptoms associated with Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and multiple system atrophy [95]. RNAi against the mutant allele may be beneficial as in other polyQ diseases (HD, SCA1, SCA3, SCA6). Nonallele-specific silencing will likely be tolerable as SCA2 knockout mice are viable and fertile [96]. In addition to targeting the mutant allele, modifier genes can also be targeted for treatment. Studies in the SCA2-58Q transgenic mouse showed an increase in inositol 1,4,5 phosphate (InsP3)-mediated calcium release in the PCs resulting in dysregulated PC function [97]. When presymptomatic SCA2 mice were treated with a calcium stabilizer drug (Dantrolene), or by partial suppression of the InsP3 receptor, PC degeneration was significantly reduced and significant motor improvement was seen [97, 98]. These studies are promising and suggest that suppressing InsP3-mediated calcium release by targeting the InsP3 receptor in PCs by RNAi may be therapeutic, particularly if regulated RNAi systems are developed. Dantrolene was also found to be neuroprotective in a HD mouse model (YAC128) and in an SCA3 mouse model [99, 100]. Dysregulated calcium signaling has emerged as a theme in many ataxias and other neurodegenerative diseases, and could provide a novel therapeutic target for RNAi therapies [101, 102].
Spinocerebellar Ataxia Type 3
Spinocerebellar ataxia type 2 (SCA3), also known as Machado–Joseph disease, is a dominant polyQ disease caused by a CAG expansion in the coding region of Ataxin3 (ATXN3). SCA3 is characterized by impairment of gait, vision, and speech [103]. The therapeutic utility of RNAi for SCA3 was first tested in the rat by targeting a SNP in the mutant allele. Reducing levels of the mutant allele rescued diseased phenotypes [104]. Later, work from the same group showed that shRNAs designed to be nonallele-specific, silenced both wild type and mutant ataxin-3, and rescued SCA3 phenotypes. Thus, nonallele-specific silencing for SCA3 may be a safe and effective treatment [105]. Alternate methods of gene silencing used peptide nucleic acid conjugates and achieved mutant ATXN3 silencing in vitro [106].
Spinocerebellar Ataxia Type 6
Spinocerebellar ataxia type 6 (SCA6) is caused by a polyQ expansion of the CACNA1A gene, which encodes the α1A (Cav2.1) subunit [107]. SCA6 requires a relatively small expansion of glutamine (19–33 CAGs) to manifest disease. SCA6 is characterized by progressive ataxia, dysarthria, and nystagmus. Onset generally occurs in the fifth decade of life and lifespan is not shortened [108, 109]. Neurodegeneration occurs selectively in the PCs of the cerebellum with no neuropathy in other neurons, making it a pure cerebellar ataxia [110]. Unlike other polyQ diseases, only certain isoforms of Cav2.1 contain the expanded glutamine tract [111, 112]. A recent experiment took advantage of this isoform-specific mutation. Tsou et al. [113] used a novel splice isoform-specific RNAi strategy to target the polyQ calcium channel splice-variant. The splice isoform-specific miRNA mimics achieved allele selective silencing in vitro [113]. These results suggest a potential therapy for SCA6 that could be tested in vivo.
Spinocerebellar Ataxia Type 7
Spinocerebellar ataxia type 7 (SCA7) is unique among the SCAs, as patients experience vision loss in addition to ataxia. A polyQ expansion of >32 CAG repeats in the Ataxin7 (ATXN7) gene causes the slowly progressing ataxia, while > 52 CAG repeats in ATXN7 results in retinal degeneration in addition to cerebellar ataxia. ATXN7 is present in a transcriptional coactivator complex, STAGA or TFTC, which modulates Gcn5 histone acetyl transferase activity and deubiquitinase activity [114]. Mutant polyQ ATXN7 alters STAGA recruitment to target genes, altering its activity [115, 116]. The exact function of ATXN7 in the STAGA complex is unknown. As there is no knockout model for SCA7, the consequences of complete or partial ATXN7 knockdown by RNAi remain to be tested. Recently, shRNA sequences were designed to target a SNP found linked to mutant ATXN7 in an affected South African population, resulting in allele-specific silencing in vitro [117]. This SNP (G to A) is found in the 3’ region of the mutant gene in 50% of South African SCA7 patients. However, neither allele-specific nor nonallele-specific approaches for SCA7 have been investigated in vivo.
Alzheimer’s Disease
Alzheimer’s disease (AD) is the most common neurodegenerative dementia. AD is characterized by the accumulation of extracellular amyloid plaques and intracellular neurofibrillary tangles that cause brain atrophy [118]. Amyloid plaques are created by the misfolding of amyloid-β (Aβ) peptides, while neurofibrillary tangles are comprised of hyperphosporylated-tau. AD pathogenesis is not completely understood; Aβ aggregation or hyperphosphorylation of tau may be the primary cause of AD pathogenesis [119, 120]. The majority of work using RNAi as a therapy for AD has focused on proteins involved in amyloid plaque formation. Aβ is formed by proteolytic processing of amyloid precursor protein (APP) by β-secretase (BACE1) and γ-secretase, a large enzyme complex containing presenilin-1 (PS1) [121]. APP, BACE1, and PS1 have all been targeted in studies to prevent Aβ formation. In 2005, siRNAs designed to suppress BACE1 reduced APP cleavage and lowered Aβ formation, improving neuropathy in APP transgenic mice [122]. Allele-specific shRNAs designed to silence APP directly also improved phenotypes and decreased levels of soluble Aβ in transgenic mice [123]. Other studies targeting the amyloid cascade have focused on PS1. Both nonallele-specific and allele-specific silencing of PS1 decreased toxic Aβ formation in cell culture providing a possible in vivo strategy [124, 125]. Piedrahita et al. [126] targeted cyclin-dependent kinase 5, an enzyme required for the phosphorylation of tau using RNAi. Also, studies have targeted tau specifically [127]. cyclin-dependent kinase 5 suppression reduced phosphorylated tau levels and blocked neurofibrillary tangle formation in transgenic AD mice. Additional novel studies have targeted PLK1 and MSUT2, two genes whose suppression by RNAi reduces toxic aggregation in vitro [128, 129]. Thus, both the Aβ and the tau hyperphosphorylation pathways provide many potential targets for RNAi therapy for AD.
Parkinson’s Disease
Parkinson’s disease (PD) is a neurodegenerative movement disorder characterized by resting tremor, bradykinesia, and rigidity. Predominant cell death is seen in dopaminergic neurons of the substantia nigra pars compacta. Two of the known genes associated with PD are the aggregate-forming alpha-synuclein (α-syn) seen in Lewy Bodies, and leucine-rich repeat kinase-2 (LRRK2) [130, 131]. Most RNAi studies in PD have focused on targeting SNPs in α-syn or LRRK2. In 2006, Sapru et al. [22, 132] designed shRNAs targeting a SNP in mutant α-syn. Allele-specific silencing was achieved in vitro and in the rat brain. Sibley et al. [133] have also achieved efficient allele-specific silencing of LRRK2 using shRNAs or using an RNAi trigger mimicking miR-1224 in vitro [134].
Dystonia
Primary early-onset dystonia (DYT1) is the most common form of inherited dystonia. The cause of DYT1 in most patients is the result of a glutamic acid deletion in the torsinA protein [135]. TorsinA is an ATPase associated with diverse cellular activities [136], whose mutant form (TorsinA(ΔE)) is thought to have a dominant negative effect [137, 138]. Allele-specific TorsinA(ΔE) suppression was first achieved by shRNA delivery in vitro [53]. However, when AAV expressing the shRNAs were delivered into the striatum of transgenic DYT1 mice, the shRNAs induced lethal neurotoxicity [21]. This study is intriguing in that it raises the question as to whether the DYT1 brain is intolerant to the exogenous expression of RNAi triggers or if the particular sequences tested were toxic. Mutant TorsinA forms perinuclear aggregates that may contribute to this lack of tolerability. Importantly, this work was done prior to general utility of artificial miRNAs for directed gene silencing in brain. However, RNAi sequences to target the common TorsinA(ΔE) mutant allele are confined to those surrounding the mutation, and may, by nature, induce deleterious off-target effects, regardless of the platform used to elaborate the final siRNA product.
Spinobulbar Muscular atrophy
Spinobulbar muscular atrophy (SBMA) is caused by a polyQ expansion of the X-linked androgen receptor (AR) gene [139]. SBMA is characterized by proximal muscular weakness and atrophy, and facial muscle fasciculations, difficulty in speech, and swallowing [140]. Recently, Miyazaki et al. [141] demonstrated a potential SBMA therapy using a novel approach exploiting the miRNA pathway. They identified miR-196a expression as significantly upregulated in the spinal cord of transgenic mice at advanced disease stages. miR-196a does not target the AR gene directly, but rather silences CUGBP, Elav-like family member 2 (CELF2). CELF2 is responsible for the stability of AR transcripts by binding the CUG repeat sequence upstream of the polyQ expansion in AR mRNA. AAV delivery to hindlimb skeletal muscle for miR-196a overexpression decreased CELF2 expression in SBMA transgenic mice. This, in turn, enhanced decay of AR mRNA and ameliorated disease phenotypes. With a clinical setting in mind, experiments were done in human patient fibroblasts. miR-196a overexpression in an SBMA mouse model significantly downregulated both CELF2 and AR mRNA levels, improving SBMA phenotypes, suggesting miR-196a-mediated treatment as a potential clinical therapy for SBMA patients [141].
Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS) is a dominant neurodegenerative disease characterized by the progressive loss of motor neurons. Although many genes contribute to ALS pathogenesis, as reviewed by Strong [142], RNAi for ALS therapy has focused on gain-of-function mutations in the Cu/Zn superoxide dismutase (SOD1) gene [143, 144]. Because wild type SOD1 is important for normal cell function allele-specific silencing is important [145, 146]. Allele-specific shRNAs targeting mutant SOD1 improved disease phenotypes in transgenic mice [51, 52, 147]. More recently, asymmetric siRNAs with mismatched base pairs achieved allele-specific silencing in vitro, but these have not been tested in animal models [148]. While SOD1 therapies show promise for ALS, other target genes have been documented. SNPs within OPTN [149], TARDBP [150–156], FUS [157–160], and ANG/VEGF [161–165] provide additional opportunities for potential allele-specific ALS therapies.
Taking RNAi to the Clinic
As RNAi therapies for CNS diseases approach the clinic, there are obvious considerations to moving each potential drug forward.
Allele-specific versus non-allele-specific silencing. While targeting the mutant allele is always desirable, it may not be necessary for some diseases, as RNAi reduces, but does not fully remove, the targeted gene product. Moreover, wild type levels of the gene being targeted may not be required for maintenance of cell viability. For example, SCA1, SCA2, and SCA3 knockout mice are viable and fertile, indicating that knockdown of the wild type allele function may be tolerable [96, 166, 167]. Nonallele-specific silencing of HTT in HD mice resulted in a significant rescue of the HD phenotype, and 2 studies have shown that reducing levels of wild type HTT in the adult rhesus macaque striatum is safe and well tolerated for at least 6 months [80–82]. However, as the HD null mice are embryonic lethal, and the levels of HTT required for cell viability of adult neurons is unknown, researchers are also investigating allele-specific silencing options [83]. For every disease being tested by nonallele-specific silencing, it is important to consider whether partial loss of function of the wild type allele is sufficient to retain function long-term.
Dose, delivery, and distribution of RNAi in the human brain. Ultimately, the goal of developing RNAi therapies and testing them in animal models is for treatment in humans. Thus, it is important to understand what kind of dose may be appropriate and how long the RNAi efficacy is retained in the human brain. A number of studies have focused on determining an appropriate dose of RNAi that is efficacious and safe in the primate brain using viral and nonviral methods [81, 82, 168, 169]. Nonviral delivery systems used cannulas implanted in the brain or convection enhanced delivery systems that use flow pressure to increase the volume of distribution. For long-term effects, repeated dosing would be required. For AAVs, directed delivery can be done, or, if the targeted structure is larger, convection enhanced delivery is effective [168]. Various methods are used to inform investigators about the distribution of the drug after delivery. To detect siRNAs after delivery, Stiles et al. [168] used radiolalebeled siRNAs, which allowed comparison of the volume of brain for which there was target suppression, as a function of dose and spread. For viral vectors, a common strategy to determine the distribution of transduced cells takes advantage of reporter genes, such as eGFP [80, 82]. In the future, targeting the brain after systemic delivery may be possible. Alternatively, researchers may be able to take advantage of the impaired BBB in some neurological diseases, allowing for diffusion of viruses, drugs, and other small molecules into the brain that are delivered systemically [67, 170].
Duration of silencing. For non-viral methods of delivery, targeting the brain with RNAi molecules will require repeated dosing as their effect will eventually wane. Thus, indwelling ports for brain access will be required. However, expression of transgenes after AAV delivery has been observed to last many years (>8 years) in the primate brain [60]. Whether these same platforms can provide for lasting expression of RNAi triggers sufficient to last the life of the patient, or at the least, many years, is not yet known. The longevity of expression from viral vectors may vary depending on the vector type, the promoter, the cell types transduced, and the pathology of the particular disease. While longevity of the transgene expression by viral vectors is important, it is also important to consider regulating its expression in the case of adverse effects from off-targeting. In this regard, regulating expression of the transgene by exogenous factors such as the erythromycin based on–off system may be an important parameter to consider. This system has the benefit of using the clinically approved drug erythromycin and uses erythromycin-responsive Escherichia coli operator or repressor elements to either turn on or off a transgene [171].
In summary, cumulative and ongoing studies with RNAi delivery for CNS therapies are encouraging. RNAi activity in neurons continues to show efficacy in animal models for treatment of “gain-of-function” neurodegenerative diseases, and with careful choice of siRNA to avoid off-target effects, holds much promise for translation to the clinic.
Electronic supplementary material
(PDF 499 kb)
Acknowledgments
We thank members of the Davidson Lab for critical comments, and the NIH, NAF, HDF, and Roy J Carver Trust for funding this work.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
P. S. Ramachandran and M. S. Keiser contributed equally to this work.
References
- 1.Zu T, Duvick LA, Kaytor MD, et al. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J Neurosci. 2004;24:8853–8861. doi: 10.1523/JNEUROSCI.2978-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 3.Diaz-Hernandez M, Torres-Peraza J, Salvatori-Abarca A, et al. Full motor recovery despite striatal neuron loss and formation of irreversible amyloid-like inclusions in a conditional mouse model of Huntington's disease. J Neurosci. 2005;25:9773–9781. doi: 10.1523/JNEUROSCI.3183-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engel T, Hernandez F, Avila J, Lucas JJ. Full reversal of Alzheimer's disease-like phenotype in a mouse model with conditional overexpression of glycogen synthase kinase-3. J Neurosci. 2006;26:5083–5090. doi: 10.1523/JNEUROSCI.0604-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 6.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 7.Monteys AM, Spengler RM, Wan J, et al. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 9.Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 10.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 13.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 14.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eulalio A, Huntzinger E, Nishihara T, et al. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Kim DH, Behlke MA, Rose SD, et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 18.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride JL, Boudreau RL, Harper SQ, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y, Khodr CE, Sapru MK, Pedapati J, Bohn MC. A microRNA embedded AAV alpha-synuclein gene silencing vector for dopaminergic neurons. Brain Res. 2011;1386:15–24. doi: 10.1016/j.brainres.2011.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin JN, Wolken N, Brown T, et al. Lethal toxicity caused by expression of shRNA in the mouse striatum: implications for therapeutic design. Gene Ther. 2011;18:666–673. doi: 10.1038/gt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khodr CE, Sapru MK, Pedapati J, et al. An alpha-synuclein AAV gene silencing vector ameliorates a behavioral deficit in a rat model of Parkinson's disease, but displays toxicity in dopamine neurons. Brain Res. 2011;1395:94–107. doi: 10.1016/j.brainres.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehlert EM, Eggers R, Niclou SP, Verhaagen J. Cellular toxicity following application of adeno-associated viral vector-mediated RNA interference in the nervous system. BMC Neurosci. 2010;11:20. doi: 10.1186/1471-2202-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birmingham A, Anderson E, Sullivan K, et al. A protocol for designing siRNAs with high functionality and specificity. Nat Protoc. 2007;2:2068–2078. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- 25.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 26.Matveeva O, Nechipurenko Y, Rossi L, et al. Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res. 2007;35:e63. doi: 10.1093/nar/gkm088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong W, Ren Y, Zhou H, et al. siDRM: an effective and generally applicable online siRNA design tool. Bioinformatics 2008;24:2405–2406. [DOI] [PubMed]
- 28.Lu ZJ, Mathews DH. OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 2008;36:W104–108. doi: 10.1093/nar/gkn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naito Y, Yamada T, Ui-Tei K, Morishita S, Saigo K. siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res. 2004;32:W124–129. doi: 10.1093/nar/gkh442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Mu FY. A Web-based design center for vector-based siRNA and siRNA cassette. Bioinformatics. 2004;20:1818–1820. doi: 10.1093/bioinformatics/bth164. [DOI] [PubMed] [Google Scholar]
- 31.Boudreau RL, Spengler RM, Hylock RH, et al. siSPOTR: a tool for designing highly specific and potent siRNAs for human and mouse. Nucleic Acids Res 2013;41:e9. [DOI] [PMC free article] [PubMed]
- 32.Naito Y, Ui-Tei K. Designing functional siRNA with reduced off-target effects. Methods Mol Biol. 2013;942:57–68. doi: 10.1007/978-1-62703-119-6_3. [DOI] [PubMed] [Google Scholar]
- 33.Naito Y, Yoshimura J, Morishita S, Ui-Tei K. siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics. 2009;10:392. doi: 10.1186/1471-2105-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fedorov Y, Anderson EM, Birmingham A, et al. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birmingham A, Anderson EM, Reynolds A, et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 36.Jackson AL, Burchard J, Schelter J, et al. Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boudreau RL, Davidson BL. Generation of hairpin-based RNAi vectors for biological and therapeutic application. Methods Enzymol. 2012;507:275–296. doi: 10.1016/B978-0-12-386509-0.00014-4. [DOI] [PubMed] [Google Scholar]
- 38.Lentz TB, Gray SJ, Samulski RJ. Viral vectors for gene delivery to the central nervous system. Neurobiol Dis. 2012;48:179–188. doi: 10.1016/j.nbd.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rettig GR, Behlke MA. Progress toward in vivo use of siRNAs-II. Mol Ther. 2012;20:483–512. doi: 10.1038/mt.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu D, Pendergraff H, Liu J, et al. Single-Stranded RNAs Use RNAi to Potently and Allele-Selectively Inhibit Mutant Huntingtin Expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lima WF, Prakash TP, Murray HM, et al. Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 42.Davidson BL, Monteys AM. Singles engage the RNA interference pathway. Cell. 2012;150:873–875. doi: 10.1016/j.cell.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Vergoni AV, Tosi G, Tacchi R, et al. Nanoparticles as drug delivery agents specific for CNS: in vivo biodistribution. Nanomedicine. 2009;5:369–377. doi: 10.1016/j.nano.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Tahara K, Miyazaki Y, Kawashima Y, Kreuter J, Yamamoto H. Brain targeting with surface-modified poly(D,L-lactic-co-glycolic acid) nanoparticles delivered via carotid artery administration. Eur J Pharm Biopharm. 2011;77:84–88. doi: 10.1016/j.ejpb.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Gao H, Qian J, Yang Z, et al. Whole-cell SELEX aptamer-functionalised poly(ethyleneglycol)-poly(epsilon-caprolactone) nanoparticles for enhanced targeted glioblastoma therapy. Biomaterials. 2012;33:6264–6272. doi: 10.1016/j.biomaterials.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 46.Liu C, Zhang N. Nanoparticles in gene therapy principles, prospects, and challenges. Prog Mol Biol Transl Sci. 2011;104:509–562. doi: 10.1016/B978-0-12-416020-0.00013-9. [DOI] [PubMed] [Google Scholar]
- 47.Stein CS, Martins I, Davidson BL. The lymphocytic choriomeningitis virus envelope glycoprotein targets lentiviral gene transfer vector to neural progenitors in the murine brain. Mol Ther. 2005;11:382–389. doi: 10.1016/j.ymthe.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Lattanzi A, Neri M, Maderna C, et al. Widespread enzymatic correction of CNS tissues by a single intracerebral injection of therapeutic lentiviral vector in leukodystrophy mouse models. Hum Mol Genet. 2010;19:2208–2227. doi: 10.1093/hmg/ddq099. [DOI] [PubMed] [Google Scholar]
- 49.Di Domenico C, Villani GR, Di Napoli D, et al. Gene therapy for a mucopolysaccharidosis type I murine model with lentiviral-IDUA vector. Hum Gene Ther. 2005;16:81–90. doi: 10.1089/hum.2005.16.81. [DOI] [PubMed] [Google Scholar]
- 50.Drouet V, Perrin V, Hassig R, et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 51.Raoul C, Abbas-Terki T, Bensadoun JC, et al. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 52.Ralph GS, Radcliffe PA, Day DM, et al. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat Med. 2005;11:429–433. doi: 10.1038/nm1205. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Alegre P, Bode N, Davidson BL, Paulson HL. Silencing primary dystonia: lentiviral-mediated RNA interference therapy for DYT1 dystonia. J Neurosci. 2005;25:10502–10509. doi: 10.1523/JNEUROSCI.3016-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harper SQ, Staber PD, Beck CR, et al. Optimization of feline immunodeficiency virus vectors for RNA interference. J Virol. 2006;80:9371–9380. doi: 10.1128/JVI.00958-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 56.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 57.Vandenberghe LH, Xiao R, Lock M, et al. Efficient serotype-dependent release of functional vector into the culture medium during adeno-associated virus manufacturing. Hum Gene Ther. 2010;21:1251–1257. doi: 10.1089/hum.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson RD, Haskell RE, Xia H, Roessler BJ, Davidson BL. A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 2000;7:1034–1038. doi: 10.1038/sj.gt.3301197. [DOI] [PubMed] [Google Scholar]
- 59.Mittermeyer G, Christine CW, Rosenbluth KH, et al. Long-term evaluation of a phase 1 study of AADC gene therapy for Parkinson's disease. Hum Gene Ther. 2012;23:377–381. doi: 10.1089/hum.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hadaczek P, Eberling JL, Pivirotto P, et al. Eight years of clinical improvement in MPTP-lesioned primates after gene therapy with AAV2-hAADC. Mol Ther. 2010;18:1458–1461. doi: 10.1038/mt.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petit L, Lheriteau E, Weber M, et al. Restoration of vision in the pde6beta-deficient dog, a large animal model of rod-cone dystrophy. Mol Ther. 2012;20:2019–2030. doi: 10.1038/mt.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia H, Anderson B, Mao Q, Davidson BL. Recombinant human adenovirus: targeting to the human transferrin receptor improves gene transfer to brain microcapillary endothelium. J Virol. 2000;74:11359–11366. doi: 10.1128/jvi.74.23.11359-11366.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alisky JM, Hughes SM, Sauter SL, et al. Transduction of murine cerebellar neurons with recombinant FIV and AAV5 vectors. Neuroreport. 2000;11:2669–2673. doi: 10.1097/00001756-200008210-00013. [DOI] [PubMed] [Google Scholar]
- 64.Davidson BL, Stein CS, Heth JA, et al. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc Natl Acad Sci U S A. 2000;97:3428–3432. doi: 10.1073/pnas.050581197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang C, Wang CM, Clark KR, Sferra TJ. Recombinant AAV serotype 1 transduction efficiency and tropism in the murine brain. Gene Ther. 2003;10:1528–1534. doi: 10.1038/sj.gt.3302011. [DOI] [PubMed] [Google Scholar]
- 66.Liu G, Martins IH, Chiorini JA, Davidson BL. Adeno-associated virus type 4 (AAV4) targets ependyma and astrocytes in the subventricular zone and RMS. Gene Ther. 2005;12:1503–1508. doi: 10.1038/sj.gt.3302554. [DOI] [PubMed] [Google Scholar]
- 67.Gray SJ, Blake BL, Criswell HE, et al. Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood–brain barrier (BBB) Mol Ther. 2010;18:570–578. doi: 10.1038/mt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koerber JT, Klimczak R, Jang JH, et al. Molecular evolution of adeno-associated virus for enhanced glial gene delivery. Mol Ther. 2009;17:2088–2095. doi: 10.1038/mt.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koerber JT, Jang JH, Schaffer DV. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol Ther. 2008;16:1703–1709. doi: 10.1038/mt.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li W, Asokan A, Wu Z, et al. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol Ther. 2008;16:1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- 71.Grimm D, Lee JS, Wang L, et al. in vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Munch RC, Janicki H, Volker I, et al. Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Mol Ther. 2013;21:109–118. doi: 10.1038/mt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi W, Bartlett JS. RGD inclusion in VP3 provides adeno-associated virus type 2 (AAV2)-based vectors with a heparan sulfate-independent cell entry mechanism. Mol Ther. 2003;7:515–525. doi: 10.1016/s1525-0016(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 74.Stachler MD, Chen I, Ting AY, Bartlett JS. Site-specific modification of AAV vector particles with biophysical probes and targeting ligands using biotin ligase. Mol Ther. 2008;16:1467–1473. doi: 10.1038/mt.2008.129. [DOI] [PubMed] [Google Scholar]
- 75.Liu G, Martins I, Wemmie JA, Chiorini JA, Davidson BL. Functional correction of CNS phenotypes in a lysosomal storage disease model using adeno-associated virus type 4 vectors. J Neurosci. 2005;25:9321–9327. doi: 10.1523/JNEUROSCI.2936-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gray SJ, Nagabhushan Kalburgi S, McCown TJ, Jude Samulski R. Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther 2013;20:450–459. [DOI] [PMC free article] [PubMed]
- 77.Dehay B, Dalkara D, Dovero S, Li Q, Bezard E. Systemic scAAV9 variant mediates brain transduction in newborn rhesus macaques. Sci Rep. 2012;2:253. doi: 10.1038/srep00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Foust KD, Nurre E, Montgomery CL, et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duque S, Joussemet B, Riviere C, et al. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther. 2009;17:1187–1196. doi: 10.1038/mt.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boudreau RL, McBride JL, Martins I, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grondin R, Kaytor MD, Ai Y, et al. Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain. 2012;135:1197–1209. doi: 10.1093/brain/awr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McBride JL, Pitzer MR, Boudreau RL, et al. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington's disease. Mol Ther. 2011;19:2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nasir J, Floresco SB, O'Kusky JR, et al. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 84.Ho LW, Brown R, Maxwell M, Wyttenbach A, Rubinsztein DC. Wild type Huntingtin reduces the cellular toxicity of mutant Huntingtin in mammalian cell models of Huntington's disease. J Med Genet. 2001;38:450–452. doi: 10.1136/jmg.38.7.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 86.Trushina E, Dyer RB, Badger JD, 2nd, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zuccato C, Tartari M, Crotti A, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 88.Rigamonti D, Bauer JH, De-Fraja C, et al. Wild-type huntingtin protects from apoptosis upstream of caspase-3. J Neurosci. 2000;20:3705–3713. doi: 10.1523/JNEUROSCI.20-10-03705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu J, Liu J, Corey DR. Allele-selective inhibition of huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem Biol. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carroll JB, Warby SC, Southwell AL, et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gagnon KT, Pendergraff HM, Deleavey GF, et al. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pfister EL, Kennington L, Straubhaar J, et al. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zoghbi HY, Orr HT. Spinocerebellar ataxia type 1. Semin Cell Biol. 1995;6:29–35. doi: 10.1016/1043-4682(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 94.Xia H, Mao Q, Eliason SL, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 95.Auburger GW. Spinocerebellar ataxia type 2. Handb Clin Neurol. 2012;103:423–436. doi: 10.1016/B978-0-444-51892-7.00026-7. [DOI] [PubMed] [Google Scholar]
- 96.Kiehl TR, Nechiporuk A, Figueroa KP, et al. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem Biophys Res Commun. 2006;339:17–24. doi: 10.1016/j.bbrc.2005.10.186. [DOI] [PubMed] [Google Scholar]
- 97.Kasumu AW, Liang X, Egorova P, Vorontsova D, Bezprozvanny I. Chronic suppression of inositol 1,4,5-triphosphate receptor-mediated calcium signaling in cerebellar purkinje cells alleviates pathological phenotype in spinocerebellar ataxia 2 mice. J Neurosci. 2012;32:12786–12796. doi: 10.1523/JNEUROSCI.1643-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu J, Tang TS, Tu H, et al. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 2. J Neurosci. 2009;29:9148–62. doi: 10.1523/JNEUROSCI.0660-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen X, Wu J, Lvovskaya S, et al. Dantrolene is neuroprotective in Huntington's disease transgenic mouse model. Mol Neurodegener. 2011;6:81. doi: 10.1186/1750-1326-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen X, Tang TS, Tu H, et al. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 3. J Neurosci. 2008;28:12713–12724. doi: 10.1523/JNEUROSCI.3909-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kasumu A, Bezprozvanny I. Deranged calcium signaling in Purkinje cells and pathogenesis in spinocerebellar ataxia 2 (SCA2) and other ataxias. Cerebellum. 2012;11:630–639. doi: 10.1007/s12311-010-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Supnet C, Bezprozvanny I. The dysregulation of intracellular calcium in Alzheimer disease. Cell Calcium. 2010;47:183–189. doi: 10.1016/j.ceca.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paulson HL. Dominantly inherited ataxias: lessons learned from Machado-Joseph disease/spinocerebellar ataxia type 3. Semin Neurol. 2007;27:133–142. doi: 10.1055/s-2007-971172. [DOI] [PubMed] [Google Scholar]
- 104.Alves S, Nascimento-Ferreira I, Auregan G, et al. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS One. 2008;3:e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alves S, Nascimento-Ferreira I, Dufour N, et al. Silencing ataxin-3 mitigates degeneration in a rat model of Machado-Joseph disease: no role for wild-type ataxin-3? Hum Mol Genet. 2010;19:2380–2394. doi: 10.1093/hmg/ddq111. [DOI] [PubMed] [Google Scholar]
- 106.Hu J, Gagnon KT, Liu J, et al. Allele-selective inhibition of ataxin-3 (ATX3) expression by antisense oligomers and duplex RNAs. Biol Chem. 2011;392:315–325. doi: 10.1515/BC.2011.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhuchenko O, Bailey J, Bonnen P, et al. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat Genet. 1997;15:62–69. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]
- 108.Gomez CM, Thompson RM, Gammack JT, et al. Spinocerebellar ataxia type 6: gaze-evoked and vertical nystagmus, Purkinje cell degeneration, and variable age of onset. Ann Neurol. 1997;42:933–950. doi: 10.1002/ana.410420616. [DOI] [PubMed] [Google Scholar]
- 109.Jodice C, Mantuano E, Veneziano L, et al. Episodic ataxia type 2 (EA2) and spinocerebellar ataxia type 6 (SCA6) due to CAG repeat expansion in the CACNA1A gene on chromosome 19p. Hum Mol Genet. 1997;6:1973–1978. doi: 10.1093/hmg/6.11.1973. [DOI] [PubMed] [Google Scholar]
- 110.Kordasiewicz HB, Gomez CM. Molecular pathogenesis of spinocerebellar ataxia type 6. Neurotherapeutics. 2007;4:285–294. doi: 10.1016/j.nurt.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 111.Bourinet E, Soong TW, Sutton K, et al. Splicing of alpha 1A subunit gene generates phenotypic variants of P- and Q-type calcium channels. Nat Neurosci. 1999;2:407–415. doi: 10.1038/8070. [DOI] [PubMed] [Google Scholar]
- 112.Tsunemi T, Ishikawa K, Jin H, Mizusawa H. Cell-type-specific alternative splicing in spinocerebellar ataxia type 6. Neurosci Lett. 2008;447:78–81. doi: 10.1016/j.neulet.2008.09.065. [DOI] [PubMed] [Google Scholar]
- 113.Tsou WL, Soong BW, Paulson HL, Rodriguez-Lebron E. Splice isoform-specific suppression of the Cav2.1 variant underlying spinocerebellar ataxia type 6. Neurobiol Dis. 2011;43:533–542. doi: 10.1016/j.nbd.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Helmlinger D, Hardy S, Sasorith S, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- 115.Palhan VB, Chen S, Peng GH, et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Helmlinger D, Hardy S, Abou-Sleymane G, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scholefield J, Greenberg LJ, Weinberg MS, et al. Design of RNAi hairpins for mutation-specific silencing of ataxin-7 and correction of a SCA7 phenotype. PLoS One. 2009;4:e7232. doi: 10.1371/journal.pone.0007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.LaFerla FM, Oddo S. Alzheimer's disease: Abeta, tau and synaptic dysfunction. Trends Mol Med. 2005;11:170–176. doi: 10.1016/j.molmed.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 119.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 120.Arriagada PV, Growdon JH, Hedley-Whyte ET, Hyman BT. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer's disease. Neurology. 1992;42:631–639. doi: 10.1212/wnl.42.3.631. [DOI] [PubMed] [Google Scholar]
- 121.Wolfe MS. The gamma-secretase complex: membrane-embedded proteolytic ensemble. Biochemistry. 2006;45:7931–7939. doi: 10.1021/bi060799c. [DOI] [PubMed] [Google Scholar]
- 122.Singer O, Marr RA, Rockenstein E, et al. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- 123.Rodriguez-Lebron E, Gouvion CM, Moore SA, Davidson BL, Paulson HL. Allele-specific RNAi mitigates phenotypic progression in a transgenic model of Alzheimer's disease. Mol Ther. 2009;17:1563–1573. doi: 10.1038/mt.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kandimalla RJ, Wani WY, Binukumar BK, Gill KD. siRNA against presenilin 1 (PS1) down regulates amyloid beta42 production in IMR-32 cells. J Biomed Sci 2012;19:2. [DOI] [PMC free article] [PubMed]
- 125.Sierant M, Paduszynska A, Kazmierczak-Baranska J, et al. Specific silencing of L392V PSEN1 mutant allele by RNA interference. Int J Alzheimers Dis. 2011;2011:809218. doi: 10.4061/2011/809218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Piedrahita D, Hernandez I, Lopez-Tobon A, et al. Silencing of CDK5 reduces neurofibrillary tangles in transgenic alzheimer's mice. J Neurosci. 2010;30:13966–13976. doi: 10.1523/JNEUROSCI.3637-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miller VM, Gouvion CM, Davidson BL, Paulson HL. Targeting Alzheimer's disease genes with RNA interference: an efficient strategy for silencing mutant alleles. Nucleic Acids Res. 2004;32:661–668. doi: 10.1093/nar/gkh208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Guthrie CR, Greenup L, Leverenz JB, Kraemer BC. MSUT2 is a determinant of susceptibility to tau neurotoxicity. Hum Mol Genet. 2011;20:1989–1999. doi: 10.1093/hmg/ddr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Song B, Davis K, Liu XS, et al. Inhibition of Polo-like kinase 1 reduces beta-amyloid-induced neuronal cell death in Alzheimer's disease. Aging (Albany NY) 2011;3:846–851. doi: 10.18632/aging.100382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Singleton AB, Farrer M, Johnson J, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science 2003;302:841. [DOI] [PubMed]
- 131.Shen J. Protein kinases linked to the pathogenesis of Parkinson's disease. Neuron. 2004;44:575–577. doi: 10.1016/j.neuron.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 132.Sapru MK, Yates JW, Hogan S, et al. Silencing of human alpha-synuclein in vitro and in rat brain using lentiviral-mediated RNAi. Exp Neurol. 2006;198:382–390. doi: 10.1016/j.expneurol.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 133.Sibley CR, Wood MJ. Identification of allele-specific RNAi effectors targeting genetic forms of Parkinson's disease. PLoS One. 2011;6:e26194. doi: 10.1371/journal.pone.0026194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sibley CR, Seow Y, Curtis H, Weinberg MS, Wood MJ. Silencing of Parkinson's disease-associated genes with artificial mirtron mimics of miR-1224. Nucleic Acids Res. 2012;40:9863–9875. doi: 10.1093/nar/gks712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ozelius LJ, Hewett J, Kramer P, et al. Fine localization of the torsion dystonia gene (DYT1) on human chromosome 9q34: YAC map and linkage disequilibrium. Genome Res. 1997;7:483–494. doi: 10.1101/gr.7.5.483. [DOI] [PubMed] [Google Scholar]
- 136.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 137.Breakefield XO, Kamm C, Hanson PI. TorsinA: movement at many levels. Neuron. 2001;31:9–12. doi: 10.1016/s0896-6273(01)00350-6. [DOI] [PubMed] [Google Scholar]
- 138.Torres GE, Sweeney AL, Beaulieu JM, Shashidharan P, Caron MG. Effect of torsinA on membrane proteins reveals a loss of function and a dominant-negative phenotype of the dystonia-associated DeltaE-torsinA mutant. Proc Natl Acad Sci U S A. 2004;101:15650–15655. doi: 10.1073/pnas.0308088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 140.Sobue G, Hashizume Y, Mukai E, et al. X-linked recessive bulbospinal neuronopathy. A clinicopathological study. Brain. 1989;112:209–232. doi: 10.1093/brain/112.1.209. [DOI] [PubMed] [Google Scholar]
- 141.Miyazaki Y, Adachi H, Katsuno M, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012;18:1136–1141. doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]
- 142.Strong MJ. The evidence for altered RNA metabolism in amyotrophic lateral sclerosis (ALS) J Neurol Sci. 2010;288:1–12. doi: 10.1016/j.jns.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 143.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 144.Bendotti C, Atzori C, Piva R, et al. Activated p38MAPK is a novel component of the intracellular inclusions found in human amyotrophic lateral sclerosis and mutant SOD1 transgenic mice. J Neuropathol Exp Neurol. 2004;63:113–119. doi: 10.1093/jnen/63.2.113. [DOI] [PubMed] [Google Scholar]
- 145.Shefner JM, Reaume AG, Flood DG, et al. Mice lacking cytosolic copper/zinc superoxide dismutase display a distinctive motor axonopathy. Neurology. 1999;53:1239–1246. doi: 10.1212/wnl.53.6.1239. [DOI] [PubMed] [Google Scholar]
- 146.Flood DG, Reaume AG, Gruner JA, et al. Hindlimb motor neurons require Cu/Zn superoxide dismutase for maintenance of neuromuscular junctions. Am J Pathol. 1999;155:663–672. doi: 10.1016/S0002-9440(10)65162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xia X, Zhou H, Huang Y, Xu Z. Allele-specific RNAi selectively silences mutant SOD1 and achieves significant therapeutic benefit in vivo. Neurobiol Dis. 2006;23:578–586. doi: 10.1016/j.nbd.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 148.Geng CM, Ding HL. Design of functional small interfering RNAs targeting amyotrophic lateral sclerosis-associated mutant alleles. Chin Med J (Engl) 2011;124:106–110. [PubMed] [Google Scholar]
- 149.Del Bo R, Tiloca C, Pensato V, et al. Novel optineurin mutations in patients with familial and sporadic amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2011;82:1239–1243. doi: 10.1136/jnnp.2011.242313. [DOI] [PubMed] [Google Scholar]
- 150.Lee EB, Lee VM, Trojanowski JQ, Neumann M. TDP-43 immunoreactivity in anoxic, ischemic and neoplastic lesions of the central nervous system. Acta Neuropathol. 2008;115:305–311. doi: 10.1007/s00401-007-0331-5. [DOI] [PubMed] [Google Scholar]
- 151.Yokoseki A, Shiga A, Tan CF, et al. TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann Neurol. 2008;63:538–542. doi: 10.1002/ana.21392. [DOI] [PubMed] [Google Scholar]
- 152.Gitcho MA, Baloh RH, Chakraverty S, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kabashi E, Valdmanis PN, Dion P, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 154.Winton MJ, Van Deerlin VM, Kwong LK, et al. A90V TDP-43 variant results in the aberrant localization of TDP-43 in vitro. FEBS Lett. 2008;582:2252–2256. doi: 10.1016/j.febslet.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Van Deerlin VM, Leverenz JB, Bekris LM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rutherford NJ, Zhang YJ, Baker M, et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 158.Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Abalkhail H, Mitchell J, Habgood J, Orrell R, de Belleroche J. A new familial amyotrophic lateral sclerosis locus on chromosome 16q12.1-16q12.2. Am J Hum Genet. 2003;73:383–389. doi: 10.1086/377156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ruddy DM, Parton MJ, Al-Chalabi A, et al. Two families with familial amyotrophic lateral sclerosis are linked to a novel locus on chromosome 16q. Am J Hum Genet. 2003;73:390–396. doi: 10.1086/377157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Greenway MJ, Andersen PM, Russ C, et al. ANG mutations segregate with familial and 'sporadic' amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 163.Gellera C, Colombrita C, Ticozzi N, et al. Identification of new ANG gene mutations in a large cohort of Italian patients with amyotrophic lateral sclerosis. Neurogenetics. 2008;9:33–40. doi: 10.1007/s10048-007-0111-3. [DOI] [PubMed] [Google Scholar]
- 164.Corrado L, Battistini S, Penco S, et al. Variations in the coding and regulatory sequences of the angiogenin (ANG) gene are not associated to ALS (amyotrophic lateral sclerosis) in the Italian population. J Neurol Sci. 2007;258:123–127. doi: 10.1016/j.jns.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 165.Lambrechts D, Storkebaum E, Morimoto M, et al. VEGF is a modifier of amyotrophic lateral sclerosis in mice and humans and protects motoneurons against ischemic death. Nat Genet. 2003;34:383–394. doi: 10.1038/ng1211. [DOI] [PubMed] [Google Scholar]
- 166.Matilla A, Roberson ED, Banfi S, et al. Mice lacking ataxin-1 display learning deficits and decreased hippocampal paired-pulse facilitation. J Neurosci. 1998;18:5508–5516. doi: 10.1523/JNEUROSCI.18-14-05508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Switonski PM, Fiszer A, Kazmierska K, et al. Mouse ataxin-3 functional knock-out model. Neuromolecular Med. 2011;13:54–65. doi: 10.1007/s12017-010-8137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stiles DK, Zhang Z, Ge P, et al. Widespread suppression of huntingtin with convection-enhanced delivery of siRNA. Exp Neurol. 2012;233:463–471. doi: 10.1016/j.expneurol.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 169.McCormack AL, Mak SK, Henderson JM, et al. Alpha-synuclein suppression by targeted small interfering RNA in the primate substantia nigra. PLoS One. 2010;5:e12122. doi: 10.1371/journal.pone.0012122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sarin H, Kanevsky AS, Wu H, et al. Effective transvascular delivery of nanoparticles across the blood–brain tumor barrier into malignant glioma cells. J Transl Med. 2008;6:80. doi: 10.1186/1479-5876-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Weber W, Fux C, Daoud-el Baba M, et al. Macrolide-based transgene control in mammalian cells and mice. Nat Biotechnol. 2002;20:901–907. doi: 10.1038/nbt731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 499 kb)