Abstract
Profiling studies of mRNA and miRNA, particularly microarray-based studies, have been extensively used to create compendia of genes that are preferentially expressed in the immune system. In some instances, functional studies have been subsequently pursued. Recent efforts such as ENCODE have demonstrated the benefit of coupling RNA-Seq analysis with information from expressed sequence tags (ESTs) for transcriptomic analysis. However, the full characterization and identification of transcripts that function as modulators of human immune responses remains incomplete. In this study, we demonstrate that an integrated analysis of human ESTs provides a robust platform to identify the immune transcriptome. Beyond recovering a reference set of immune-enriched genes and providing large-scale cross-validation of previous microarray studies, we discovered hundreds of novel genes preferentially expressed in the immune system, including non-coding RNAs. As a result, we have established the Immunogene database, representing an integrated EST “road map” of gene expression in human immune cells, which can be used to further investigate the function of coding and non-coding genes in the immune system. Using this approach, we have uncovered a unique metabolic gene signature of human macrophages and identified PRDM15 as a novel overexpressed gene in human lymphomas. Thus we demonstrate the utility of EST profiling as a basis for further deconstruction of physiologic and pathologic immune processes.
Introduction
The immune system is our central defense against bacterial and viral pathogens. As such, misregulation of certain genes that are critical for normal immune function can result in immunodeficiency and autoimmune disorders. Furthermore, disorders such as diabetes, obesity, and cancer are increasingly viewed as having roots in immunological dysfunction (1). Despite this recognition of the immune system as a central player in health and disease, the full spectrum of genes and pathways capable of modulating immune responses is not known.
Analyzing the function of genes that are highly enriched or specific for immune cell/tissue expression may aid in capturing important regulators of immune function. It is important to note that this premise does not require that ubiquitously expressed genes cannot be critically important in immune function and development. However, considering examples such as B cell receptor/T cell receptor signaling cascade components, it is striking that genes that are expressed specifically in immune cells/tissues are often central players in immune physiology. In addition, genes responsible for Mendelian immunodeficiences or autommunity often show enriched expression patterns in the immune system. Examples include the causes of Omenn syndrome (RAG1/2, OMIM ID: 603554), hyper IgM syndrome (AICDA, OMIM ID: 308230), and X-linked lymphoproliferative disorder (SH2D1A, OMIM ID: 308240).
Historically, gene discovery in the immune system has relied on unbiased searches for genes exhibiting differential expression in cells/tissues of the immune system as compared to other cells/tissues. These approaches have used techniques such as differential display, microarray profiling, serial analysis of gene expression (SAGE), and most recently RNA-Seq (2–11). In the case of microarray profiling, experiments often reveal a large number of differentially expressed genes, making cross-validation by alternative approaches a challenge. In addition, microarrays suffer from difficulty in detecting low-abundance transcripts and may not contain valid probes for mRNAs, non-coding RNAs (ncRNAs), or miRNAs, since their design is based on human annotation. Recent studies have utilized RNA-Seq data to examine expression profiles of immune cells; however, this has not diminished the utility of expressed sequence tag (EST) data, as recently demonstrated by the ENCODE consortium’s extensive use of EST data (12–14). For example, EST data are often used to aid in deciphering expression of alternative 5′ UTR isoforms, since RNA-Seq alone is insufficient to define transcription start sites (TSSs) of genes.
With the goal of identifying genes enriched in the immune system, we developed a rigorous quantitative systems-level expression database called Immunogene, which harnesses the approximately 7 million human ESTs in the UniGene database. To date, the UniGene database has not been exploited in a systematic fashion for genome-wide immune system gene discovery, although it has successfully been used for retinal gene discovery (15, 16). Early studies employing UniGene for analysis of the bone marrow transcriptome in zebrafish further highlighted the value of this approach (17). Notably, the UniGene database has the benefit of not being biased toward human gene annotation, representing instead a resource of sequence data coupled with tissue/cell type annotation. Using this approach, we demonstrate unique transmembrane, enzymatic, and nutrient/ion transport network signatures in the immune system. We also demonstrate the utility of the Immunogene database as an aid in deciphering blood-related cancers, identifying PRDM15 as a novel overexpressed gene in human B cell lymphomas. Thus, we show that EST profiling can significantly enhance our understanding ofthe ensemble of genes and processes important for immune function.
Materials and Methods
Data acquisition and statistics
Human UniGene database build 202 and mouse build 163 were obtained from NCBI (ftp.ncbi.nih.gov/repository/UniGene). These databases contain mapping of each EST to a UniGene cluster and mapping of UniGene clusters to genes and tissue of origin for each EST. Upon analysis of library descriptors, we found that the human database contained 469 unique tissue/cell types, of which 91 were associated with the immune system, allowing each EST to be classified as either immune or non-immune according to its associated library descriptor. A normalized or weighted immune percentage (WIP) was calculated for each UniGene cluster using the following formula:
A = Number of ESTs from immune-related libraries in UniGene Cluster X
B = Total number of ESTs from immune-related libraries in entire UniGene database
C = Number of ESTs from non-immune-related libraries in UniGene Cluster X
D = Total number of ESTs from non-immune-related libraries in entire UniGene database
A P value for immune enrichment was computed for each UniGene cluster using a χ2 test in R (version 2.5.1). For this test, a 2 × 2 contingency table was created with ESTs derived from immune tissues versus non-immune tissues (shown below). The P value was calculated with respect to the number of ESTs in all clusters that had at least 10 ESTs.
| Individual cluster | Entire UniGene collection | |
|---|---|---|
|
|
||
| ESTs derived from immune tissues | X | 575,059 |
|
|
||
| ESTs derived from non-immune tissues | Y | 5,474,516 |
|
|
||
P values for all clusters with at least 10 ESTs were corrected for multiple hypotheses testing using the false discovery rate (FDR) of Benjamini and Hochberg (18).For Figure 5A, we identified the U133 Plus2/U133A quantile-normalized relative expression level of PRDM15 in various subtypes of lymphoma cell lines compared to median expression levels of PRDM15 in all lymphoma cell types in the body atlas database of NextBio. These normalized relative expression levels were used to generate a box plot to show the expression level of PRDM15 in different lymphoma subtypes. Non-lymphoma cell lines from lung and colon cancer were also used for comparison.
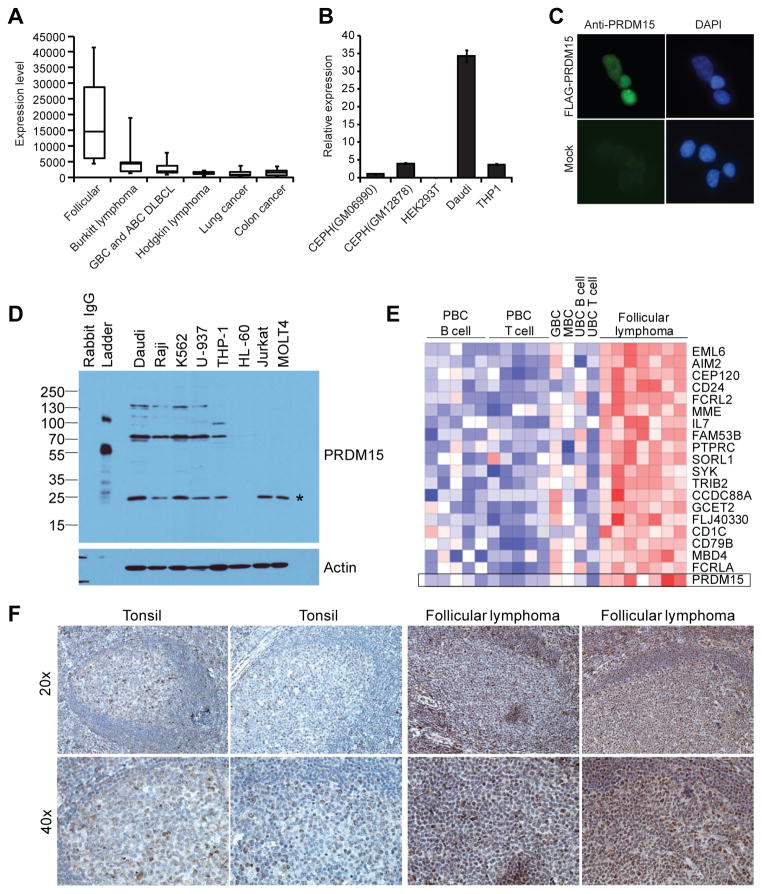
Figure 5. The Immunogene PRDM15 is overexpressed in human lymphomas.
(A) Box plot of normalized microarray expression levels of PRDM15 across 744 cell lines from various cancer types with PRDM15 levels shown for B cell lymphoma subtypes, lung cancer, and colon cancer. (B) PRDM15 mRNA is overexpressed in Burkitt lymphoma. RT-qPCR of PRDM15 transcript levels in EBV-transformed B cell lymphoblastoid GM06990 and GM12878 cell lines, embryonic kidney HEK293T cells, Burkitt lymphoma Daudi cells, and monocytic THP-1 cells. Levels were normalized to S18 transcript and expressed relative to PRDM15 levels in GM06990 cells. (C) PRDM15 localizes to the nucleus. HEK293T cells were mock transfected or transfected with N-terminal FLAG-tagged PRDM15 and stained with anti-PRDM15 antibody and DAPI. (D) Western blot analysis of PRDM15 in multiple human cell lines including Daudi and Raji cells (Burkitt lymphomas), K562 cells (myelogenous leukemia, BCR-ABL positive), U-937 cells (histiocytic lymphoma, negative IgH, negative EBV, monocytic), THP-1 cells (acute monocytic leukemia), HL-60 cells (acute promyelocytic leukemia), Jurkat cells (acute T cell leukemia), and MOLT4 cells (acute T-lymphoblastic leukemia). * indicates a likely nonspecific band as it does not correspond to any PRDM15 predicted isoform weight. (E) PRDM15 mRNA is overexpressed in primary follicular B cell lymphomas. Normalized log2 median-centered expression of PRDM15 in human follicular lymphomas (n = 7) versus normal immune cell populations including peripheral blood B cells (PBC B cells, n = 5), peripheral blood CD4+ T cells (PBC T cells, n = 5), germinal center B cells (GBC, n = 1), memory B lymphocytes (MBC, n = 1), umbilical cord B lymphocytes (UBC B cell, n = 1), and umbilical cord T lymphocytes (UBC T cell, n = 1). (F) PRDM15 protein is expressed in normal germinal centers and overexpressed in primary human follicular lymphomas. Shown are the results comparing PRDM15 levels in normal human tonsils and two of seven representative independent primary human follicular lymphomas samples that were stained with anti-PRDM15 antibody by immunohistochemistry.
To identify potential divergent ncRNAs emanating near the TSS of immune-enriched protein-coding genes, we downloaded EST data from the University of California Santa Cruz genome browser. Using custom PERL scripts, we analyzed the data to identify sequences that met the following requirements: (1) at least one EST is spliced and transcribed on the opposite strand of a coding gene and has a start site within 5 kb of the TSS of the RefSeq Immunogene; (2) the spliced EST does not show evidence of splicing to a neighboring RefSeq gene; and (3) no open reading frame of >100 aa could be identified in the longest EST associated with the coding RefSeq, if there was more than one EST that met our criteria. We utilized the data feature “intronOrientation” downloaded from the University of California Santa Cruz Genome Browser to confirm EST strand orientation.
GO and KEGG pathway functional enrichment analysis
Enrichment analyses for gene ontology (GO) biological process and KEGG pathways were conducted based on various gene list categories including the EST-defined Immunogenes with Entrez IDs (n = 2,232) and the reference set of genes (n = 570) documented in the literature as being preferentially expressed in the immune system. These gene lists were submitted to the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) for enrichment analyses of GO biological processes and KEGG pathways, with the number of genes in each category and Bonferroni-corrected P values displayed (19).
Microarray analysis
GC robust multi-array average (GCRMA)-normalized gene expression profiles across 79 tissues were obtained from the Genomics Institute of the Novartis Research Foundation (GNF) consortium (20). Expression profiles from the Affymetrix U133A platform and GNF custom probes were used. In accordance with Affymetrix guidelines, probes with highest expression value below log2(100) across all tissues were removed. Expression profiles were clustered using Cluster 3 (21) and visualized using JavaTreeView (22). Immune enrichment was calculated with the R program (version 2.5.1) using the Wilcoxon rank sum test for each probe and P values were corrected using the FDR method (18). In cases where multiple probes were mapped to the same gene, the most significant P value was accepted. The following tissues in the GNF dataset were classified as immune and tested versus all other tissues: bone marrow, CD19 B cells, tonsils, lymph nodes, thymus, CD4 T cells, CD8 T cells, CD56 T cells, whole blood, CD33 myeloid cells, CD14 monocytes, dendritic cells, fetal liver, CD105 endothelial cells, leukemia cell lines, lymphoma cell lines, and erythroid cells. For Figure 5E,, we used the normalized log2 median-centered expression of PRDM15 and several co-regulated genes obtained from publishedmicroarray expression profiling (23).
Human-mouse homolog genes
For each human protein, the homolog mouse protein was identified using NCBI HomoloGene (24). In some cases, multiple mouse proteins matched a single human protein and vice versa. For the purpose of immune enrichment, at least one mouse homolog was required to be immune-enriched.
Cells, plasmids, and antibodies
Human PRDM15 [GenBank ID: BC067102] was obtained from Open Biosystems and subcloned into N-terminal and C-terminal FLAG-tagged pCMV-3xFLAG vectors using 5′ EcoRI and 3′ HindIII; vectors were generated by modifying ClonTech pCMV-Myc vector by introduction of appropriate epitope tags and modification of the multiple cloning site. Primary antibodies used were anti-PRDM15 (Santa Cruz; sc-83314), anti-FLAG M2 (Sigma; F1804), and anti-actin (Sigma; F2066). Secondary antibodies for ECL development were HRP-conjugated anti-mouse (Amersham; NXA931) and anti-rabbit (Amersham; NA934).
Western blotting, immunofluorescence, and immunohistochemistry
Human Cell Line Blot II (ProSci Inc; 1502) was obtained containing 15 μg of protein from various human cell lines. Anti-PRDM15 Western blotting was performed under standard procedures with 1:1000 primary antibody and 1:3000 secondary antibody. For immunofluorescence, HEK293T cells were transfected with Lipofectamine (Invitrogen) with PRDM15 FLAG-tagged constructs and stained with either anti-FLAG (1:400) or anti-PRDM15 (1:200) antibodies. Immunohistochemistry on human tonsil or follicular lymphomas obtained from the Pathology Core at Massachusetts General Hospital was performed as described using anti-PRDM15 antibody (1:100) (25).
RNA-Seq analysis
RPKM (reads per kilobase of exon model per million mapped reads) calls from an RNA-Seq atlas were downloaded from Medicalgenomics at http://medicalgenomics.org/rna_seq_atlas, which encompasses ribodepleted RNA-Seq data from 11 different tissues (colon, heart, hypothalamus, kidney, liver, lung, skeletal muscle, spleen, testes, ovary, and adipose tissue). Immune enrichment was calculated for each gene using the Wilcoxon signed rank test. In the RNA-Seq atlas, spleen was classified as an immune tissue and tested versus all other tissues. In this dataset, 21,725 RefSeq isoforms for mRNAs or ncRNAs were designated by NM or NR accessions, encompassing 14,713 unique gene symbols (10). Of the 26,552 UniGene clusters analyzed for their WIP score, there were RNA-Seq data for 11,449 corresponding genes. Furthermore, of the 3,352 Immunogene clusters identified in the Unigene database (see above analysis), there were RNA-Seq data for 1,336 corresponding genes. The sigma, W- and Z-score statistics were calculated for all 11,449 genes versus the 1,336 Immunogenes to test for the presence of a trend towards immune enrichment based on RNA-Seq data.
Human Disease SNP analysis
The genome-wide association study (GWAS) single nucleotide polymorphism (SNP) catalogwas downloaded from http://www.genome.gov/gwastudies/. SNPs were considered “immune disease/trait-associated” if they associated with the following diseases/traits: atopic dermatitis, systematic lupus erythematosus, Behcet’s disease, primary biliary cirrhosis, rheumatoid arthritis, psoriasis, psoriatic arthritis, vitiligo, sarcoidosis, inflammatory bowel disease, Crohn’s disease, ulcerative colitis, primary sclerosing cholangitis, type I diabetes, type I diabetes auto-antibodies, multiple sclerosis, ankylosing spondylitis, juvenile idiopathic arthritis, IgE levels, IgA levels, allergic rhinitis, leprosy, IgE grass sensitization, cytomegalovirus antibody response, Graves disease, or celiac disease. These diseases/traits were associated with 1,102 unique entries in the catalog, encompassing 808 unique immune disease/trait-associated SNPs given overlapping identification of SNPs among diseases and studies. The remaining diseases/traits were classified as “non-immune” and were associated with 8,795 entries encompassing 7,355 unique SNPs. We next calculated the distance from each immune disease/trait-associated SNP, as well as each non-immune disease/trait-associated SNP, to the nearest TSS of a coding RefSeq. We then calculated the frequency of immune disease/trait-associated SNPs within binned distances from EST-defined Immunogenes versus the frequency of non-immune disease/trait-associated SNPs within the same binned distances from EST-defined Immunogenes (Fig. 4C). We computed a P value by permutation testing, selecting 808 SNPs at random from the non-immune disease/trait-associated SNP pool 10,000 times and calculating the percentage of SNPs with a nearest EST-defined Immunogene each time.
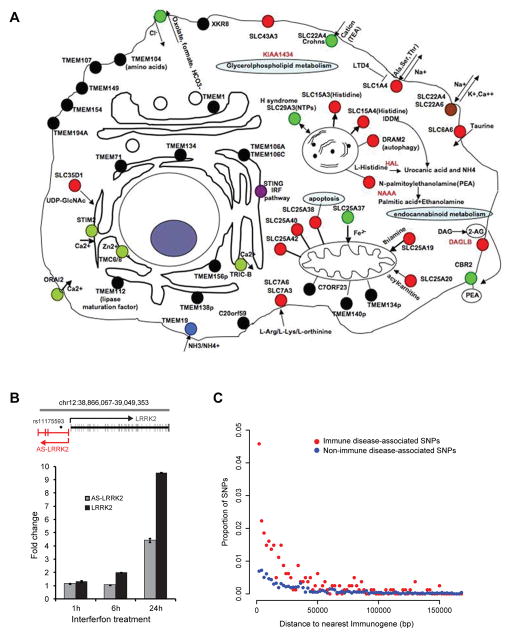
Figure 4. Immune cell transporters and enzymes identified using Immunogene, lncRNA co-regulated with LRRK2, and proximity of immune disease/trait GWAS SNPs to Immunogenes.
(A) Graphical representation of known or predicted locations and functions of transmembrane proteins and/or genes related to metabolism or nutrient/ion transport in the Immunogene dataset. Green circles indicate transmembrane proteins or transporters associated with complex or Mendelian diseases; red circles indicate transporters with known function; black circles indicate transmembrane proteins with predicted localization and no known function. (B) A divergently transcribed lncRNA is co-regulated with LRRK2. Top: Schematic of exon structure of LRRK2 and the divergent antisense (AS)-LRRK2 transcript(s), with location of SNP rs11175593 associated with Crohn’s disease (37). Bottom: The THP-1 monocytic cell line was stimulated with γ-interferon for the indicated time. RT-qPCR of LRRK2 and AS-LRRK2 transcript levels are shown normalized to S18 transcript and relative to expression levels of LRRK2 in resting THP-1 cells. Data shown are representative of one of two biological replicates with errors bars representing standard deviation of triplicate technical replicates. (C) The distance (2-kb bins) between SNPs associated by GWAS with immune diseases/traits (red) and the nearest Immunogene is shown in comparison to the distance between SNPs not associated with immune diseases/traits (blue) and the nearest Immunogene.
Gold standard immune-enriched database annotation
We compiled a list of genes documented in the literature to be preferentially expressed in the immune system. We defined evidence for immune enrichment based on RT-PCR or Northern blots. To remove selection biases as much as possible, we catalogued the expression pattern of sets of genes that would be generally regarded as preferentially expressed in the immune system (e.g. cluster of differentiation antigens, cytokine genes and receptors, and Toll-like receptor signaling components) using various web-based resources that catalog these genes. We consider this very stringent approach necessary, since not all Toll-like receptor genes (e.g. TLR5) or CD antigens (e.g. CD56) are enriched in the immune system.
RT-PCR analysis
First-strand cDNA from human spleen was obtained from OriGene. Reactions were performed in 25 μl volumes with 40 ng of sample first-strand cDNA and 640 nM concentrations of both forward and reverse primers. PCR was performed under the following conditions: initial denaturation of 95°C for 5 minutes followed by 34 cycles of 95°C for 30 seconds and 60°C for 30 seconds. To control for specificity of PCR reactions and primer-dimers, the same PCR reaction was also conducted with no input cDNA. All primers were designed to be exon-spanning. PCR products were cloned and sequenced to verify amplification of expected target lncRNA. A complete list of UniGene clusters and associated PCR primers used to amplify spliced ncRNAs is shown below. For RT-qPCR experiments, total RNA was extracted from the indicated cell lines; the RT step was performed with Superscript and the qPCR step was performed with SYBR green.
| Unigene cluster (primer name) | Sequence | Product | EST Accession | NR Accession |
|---|---|---|---|---|
| Hs.662020 (ncRNA4) | F: GGAGGATATGGCTCAGGACA R: TGGGCCACCAAGTTGTTTAT |
346 bp | AK097922 | NR_045486.1 |
| Hs.690944 (ncRNA6) | F: CACCTCATTCAGTGGCTCCT R: GGCAATGTCAGGGAAAAGAA |
353 bp | DA953527 | |
| Hs.439791 (ncRNA12) | F: TGCCTCA0CCACTTGTCTCAG R: CAGGATGACTCAGTGGAGCA |
823 bp | CR598049 | NR_024464.1 |
| Hs.667175 (ncRNA16) | F: CCAACCCAGGAGACAGTGTT R: CGTTCTCCCCATGTCTCTGT |
275 bp | AL832284 | |
| Hs.657722 (ncRNA17) | F: CCTCCTGCCTCCTTGGTAAT R: TGGTATGTTCACGGCTGAAA |
399 bp | CD367998 | NR_026544.1 |
| Hs.130180 (ncRNA18) | F: GCCATCTTGACTCACTGCAA R: AGGATGTGTGAGCCATGACA |
332 bp | DA937565 | |
| Hs.672896 (ncRNA20) | F: AGAGATGGAACCAGCTCGAA R: GATTCCCTCCAGCTCTCAAA |
366 bp | DA468624 | |
| (AS-LRRK2) | F: CTTCAACCCAGCAAATCTCC R: ATCAGCCCTTCCAATGTCAC |
158 bp | DA673065 | |
| (LRRK2) | F: TGACAGCACAGCTAGGAAGC R: TGGAAGATTGATGTCCCAAA |
128 bp | NM_198578.3 | |
| (PRDM15) | F: CGCAGTACAGAGCATTCAGC R: AGCTGTAACTGGCGTTCAGG |
186 bp | NM_022115.3 |
Results
EST profiling is a robust method to measure mRNA immune enrichment
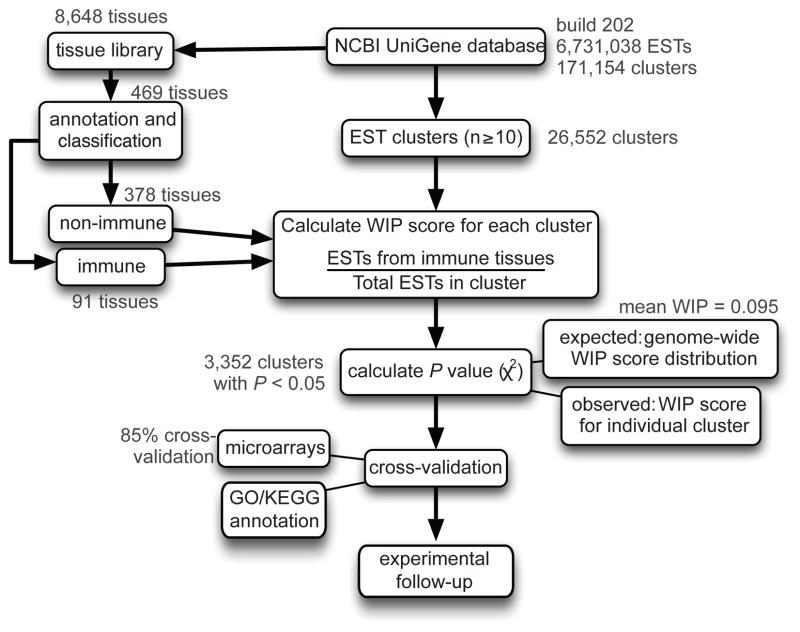
To identify immune-enriched genes using EST datasets, we developed a quantitative schema outlined in Figure 1. We use the term “immune-enriched” to indicate higher mRNA transcript levels in immune tissues compared to non-immune tissues.
Figure 1. Schematic of the human EST-based profiling analysis pipeline.
Starting with the NCBI UniGene database, 26,552 clusters with at least 10 ESTs were profiled for their expression in immune tissues. Based on the genome-wide distribution of WIP scores, a P value was calculated for each gene. A significant P value (< 0.05) indicated that there were enough ESTs to conclude that a particular WIP score was higher than average. Clusters identified as immune-enriched were cross-validated using a microarray data platform and GO/KEGG annotation with subsequent experimental follow-up. Text in red indicates the results of analysis for the human EST library build 202.
The NCBI human UniGene database (build 202) contained 6,731,038 ESTs derived from 8,648 human cDNA libraries. These ESTs were clustered by sequence alignment with no genomic scaffold into 171,154 clusters. Of the resulting clusters, we focused on the 26,552 that had at least 10 ESTs, encompassing a total of 6,049,575 ESTs. The UniGene tissue library descriptors were annotated and standardized, resulting in 469 tissues/cell types. Of these tissues/cell types, 91 (~20%) were classified as immune tissues, with the remaining defined as non-immune tissues. Since each EST in a particular UniGene cluster has an associated library ID, we were able to create a binary classification scheme, categorizing each EST as to whether it derived from immune or non-immune tissues (Supplemental Fig. 1A).
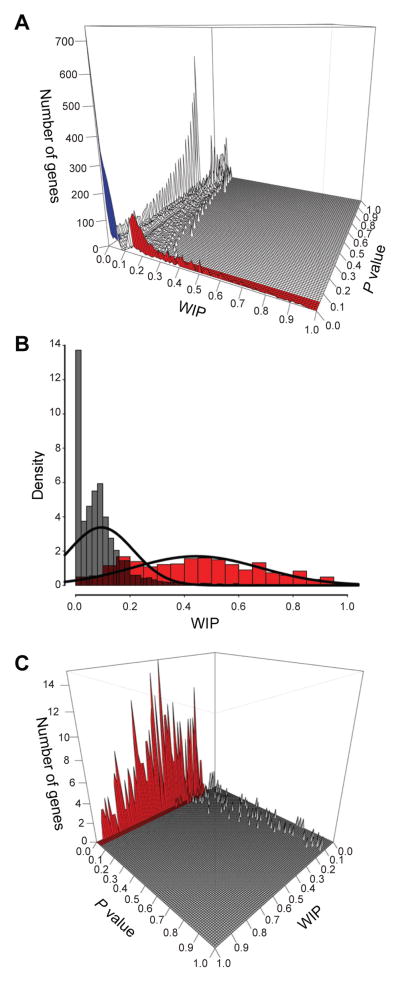
Next, we introduced a scoring system to quantify the level of immune expression for each of the 26,552 clusters. Specifically, we applied a weighted immune percentage (WIP) score, which was calculated as the percentage of ESTs derived from immune tissues. To assess whether the WIP score for each UniGene cluster was significantly enriched in immune expression, we applied a χ2 statistic and corrected for multiple hypothesis testing using the FDR method (Materials and Methods). The distribution of the WIP score (mean = 0.095) and P values for all 26,552 UniGene clusters are shown in Figure 2A and Supplemental Table I.
Figure 2. Quantitative EST-based immune expression enrichment analysis utilizing 26,552UniGene clusters.
(A) The frequency distribution of the WIP score (mean = 0.095) and P values(χ2 metric with FDR correction) for 26,552 UniGene clusters with ≥10 ESTs is shown. Red indicates clusters/genes significantly enriched in immune expression (WIP > 0.095 and P < 0.05) while blue indicates clusters/genes significantly underexpressed in the immune system (WIP < 0.05 and P < 0.05). (B) The frequency distribution (density) of WIP scores for 26,552 UniGene clusters (gray, mean = 0.095) versus the WIP scores of the 570 known immune-enriched reference gene set (red, mean 0.44). (C) The distribution of WIP scores and P values for the 570 known immune-enriched reference gene set showing that 85% (red) of this gold standard gene set have significant WIP and P values.
To evaluate whether our EST immune-based expression profiling was sufficiently sensitive and specific to discover immune-related genes, we compiled a reference set of genes (n = 570) documented in the literature as being preferentially expressed in the immune system as defined by RT-PCR or Northern blot analysis (Supplemental Table I). To cross-validate our literature annotation for this immune-enriched gold standard set of genes, we performed Gene Ontology (GO) biological processes, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and microarray enrichment analyses on this gene set. GO enrichment analysis revealed enrichment in categories such as immune system process, leukocyte activation, T cell activation, and inflammatory response. (Supplemental Fig. 2A). All KEGG pathways identified as being significantly over-represented were immune pathways (Supplemental Fig. 2B). Microarray expression analysis of our compiled immune-enriched reference set showed that these genes are preferentially expressed in human immune tissues, thus providing a high-confidence set of immune-enriched genes against which we could benchmark our EST profiling results (Supplemental Fig. 2C, 2D).
We identified 3,352 UniGene clusters with a significant P value (P < 0.05) and a WIP score greater than the average (0.095). Importantly, the average WIP scores for the gold standard genes were significantly higher, with a mean score of 0.44 and a standard deviation of 0.235, suggesting that our EST profiling methodology was robust (Fig. 2B). In addition, the vast majority of our gold standard genes had significant P values (P < 0.05), indicating the specificity of our methods (Fig. 2C). Using a conservative WIP score threshold of 0.15 and P < 0.05, we identified a total of 2,834 (10.6%) clusters matching 2,232 unique NCBI Gene LocusLink IDs as being immune-enriched. We termed these 2,232 genes “Immunogenes.” Although we focus on the results of human immune profiling in this report, application of our methodology to the mouse UniGene database ultimately identified 3,108 unique mouse genes as immune-enriched (Supplemental Fig. 1B, 1C; Supplemental Table I).
Microarray, GO, KEGG and RNA-Seq signatures of immune-enriched genes
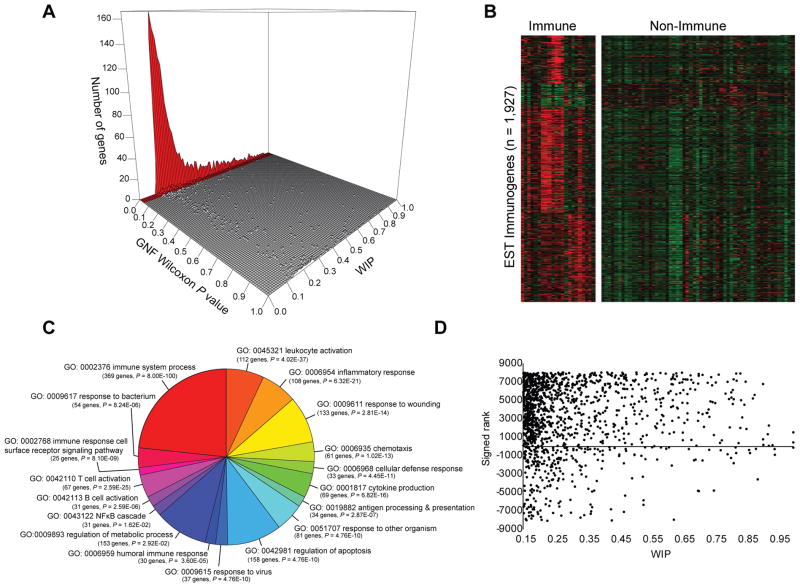
We next assessed the strength of EST profiling against microarray profiling in order to provide (1) an independent method of cross-validation of each platform and (2) increased cellular resolution within the immune system of our EST-derived dataset. We quantified immune enrichment by calculating a Wilcoxon rank sum test and adjusted for multiple hypothesis testing (FDR corrected P < 0.05) using the publicly available BioGPS atlas (9). This atlas is a broad compendium of human tissues, with 22 of 79 profiled tissues categorized as immune tissues. Of the 2,232 Immunogenes, 1,927 had representative probes on the microarray. Strikingly, 1,522/1,927 (79%) of the genes found to be immune-enriched based on EST profiling WIP score (WIP > 0.15) also exhibited significantly higher immune expression by microarray (P < 0.05; FDR microarray Wilcoxon test) (Fig. 3A). Hierarchical clustering and heat map representation of the 1,927 EST Immunogene profiles separated by immune versus non-immune tissue is shown in Figure 3B, revealing enrichment of EST-derived Immunogenes in the BioGPS compendia and providing further cross-validation of our methodology.
Figure 3. Cross-validation of EST profiling methodology with microarray profiling, GO pathway analysis, and epigenetic signatures.
(A) Of the 1,927 genes found to be significantly enriched by EST profiling, 1,522 genes (79%) exhibited significantly higher expression by Wilcoxon rank sum test and FDR (P < 0.05) in the immune system based on analysis of the GNF BioGPS microarray data tissue compendium. (B) Microarray profile clustering in GNF tissue/cell compendium of 1,927 immune-enriched genes defined by EST profiling segregating immune versus non-immune tissue types. (C) Examples of GO biological process terms statistically enriched (P < 0.05, Bonferroni corrected) for all EST Immunogenes. (D) The Wilcoxon signed rank (Y axis) for 1,336 Immunogenes compared to their WIP score (X axis) showing that most Immunogenes show a positively signed rank.
We also reasoned that if expression is an indicator of potential function, then enrichment would also be found in GO biological process categories and KEGG pathways relevant to the immune system, which was indeed the case (Fig. 3C, Supplemental Fig. 1D). In addition, analysis of publicly available RNA-Seq data comparing spleen to ten other non-immune tissues revealed that the list of Immunogenes defined by EST profiling showed a significantly higher tendency to be expressed in spleen versus non-immune tissues (Z = 7.03 for all genes, Z = 151.42 for Immunogenes, Wilcoxon signed ranked test) (Fig. 3D). KIAA0226L (WIP = 0.37; P = 1.65 × 10−38) is an example of a novel immune-enriched gene identified by EST profiling. Interestingly, KIAA0226L has been implicated via proteomics to be involved in autophagy (26) and RNA-Seq has demonstrated expression of KIAA0226L in human spleen, mouse C19+ B cells, but not in mouse CD4+ T cells (3, 10). As the ability of EST analysis to spotlight immune-enriched genes was confirmed by multiple analyses, we designed a browseable web-based portal of Immunogenes accessible at http://xavierlab2.mgh.harvard.edu/ESTProfiler/index.html.
EST profiling reveals immune-enriched transmembrane, metabolic, and enzymatic signatures
To fully characterize the Immunogene dataset, we took advantage of structure-function relationships. For this approach, we analyzed each Immunogene systematically by (1) determining the presence of potential protein domains using SMART (Simple Modular Architecture Research Tool) and PFAM database criteria, (2) checking for homology to any known or predicted proteins in multiple species using BLASTP alignments, and (3) identifying predicted or known transmembrane regions using the TMHMM prediction algorithm. Structural annotation/prediction revealed that 20% of Immunogenes (440 of 2,232) have known or predicted enzymatic functions utilizing an enzymology classification system or homology modeling. These observations led us to question whether such a large fraction of enzymes in the Immunogene geneset was a reflection of the unique metabolic pathways and needs of immune cells. We used a similar number of UniGenes (460 of 2,611) that were significantly underrepresented in immune tissues (WIP < 0.05; P < 0.05) as a control set of enzymes. Using two independent expression maps, BioGPS (P = 1.4 × 10−15) and NCBI SAGE tags (P = 1.2 × 10−20), we observed that our EST immune enzyme inventory was most enriched in monocytes/dendritic cells compared to other cell types. In contrast, the control set of enzymes was enriched in the nervous system (P = 1 × 10−13). These results strongly support our conclusion that EST profiling has revealed a genome-wide compendium of enzymes that are likely to be particularly relevant to immune system function and development. For example, one novel potential enzyme identified as being immune-enriched was ARL11 (WIP = 0.3; P = 0.028), which is predicted to encode an ADP-ribosylation factor.
In other instances, we identified enzymes that have clear enzymatic function, but which have only been evaluated to a limited degree in immune system function. One such enzyme is fructose 1,6-diphosphatase (FBP1), which is known to be critical in gluconeogenesis in the liver, but was revealed by EST profiling to be enriched in macrophages. FBP1 was previously found to be a vitamin D-responsive gene in monocytes (27). Thus our data highlight how EST profiling can also help focus attention on potential intriguing links between metabolism and immune function (27, 28).
In addition to the enzymatic footprints of immune cells, our structure-function analysis demonstrated highly significant enrichment of genes encoding transmembrane proteins (n = 215; P = 7 × 10−10 “integral to membrane” GO annotation) with an even greater number of genes (n = 285) identified by transmembrane prediction algorithms. This result correlated with our finding that the largest PFAM and InterPro classes of protein domains in the Immunogene set were immunoglobulin domains (n = 87; P = 3.9 × 10−12). These transmembrane proteins included cytokine receptors, ion channels such the calcium channel ORAI2, as well as novel transmembrane proteins (n = 69) and predicted secreted proteins (n = 22). One example of a transmembrane protein that we identified as being immune-enriched is the nucleotide transporter SLC29A3, which causes H syndrome, Faisalabad histiocytosis, and Rosai-Dorfman disease (OMIM ID: 602782) when mutated. Patients with H syndrome have overlapping features of hemophagocytic syndromes, suggesting that functional characterization of SCL29A3 is likely to yield insights into immune function (29). The strong representation of known or predicted nutrient and ion transporters is also consistent with the unique metabolic needs of the immune system. Our results provide a guide to further dissect the links between metabolism and immune function. An integrated portrait of immune cell transporters and enzymes generated using Immunogene is shown in Figure 4A.
Discovery and verification of lncRNAs using EST profiling
Given that our EST profiling is agnostic to the type of transcripts it identifies, we also identified several immune-enriched microRNAs, such as miR-155 (BIC) (P = 2.33 × 10−14). Further inspection of the human EST dataset revealed that 30% (n = 388) of the UniGene clusters with WIP > 0.20 and P < 0.05 were not annotated as hypothetical or validated reference protein-coding genes at NCBI. Manual curation of these clusters revealed that a large fraction was likely composed of genomic contaminants, alternative exons, or untranslated regions (UTRs) of known protein-coding genes (Materials and Methods). However, 46 UniGene clusters demonstrated evidence of splicing and did not show any significant similarity to known protein-coding genes using BLASTX, suggesting that these clusters likely represent long (> 200 bp) non-coding RNAs (lncRNAs). Using RT-PCR, we confirmed expression of 10 of 17 (59%) randomly selected clusters in human spleen (see Supplemental Fig. 3A, 3B for examples). One such human-specific lncRNA identified as an Immunogene was AK056817 (WIP = 0.34; P = 7.0 × 10−9)). Although the functions of the ncRNAs are largely unknown, analysis of ENCODE ChIP-Seq data in leukemic K562 cells revealed interferon-inducible binding of STAT1 to the promoter region of AK056817, suggesting that this lncRNA may function in interferon response pathways (30). Thus, we suggest that integrating additional genome-wide datasets can be used to refine potential functional pathways of Immunogenes.
AK056817 represents a likely lncRNA that is transcribed in an intergenic region away from other known genes. Yet many coding genes have recently been found to exist as pairs with lncRNAs that are divergently transcribed on the opposite strand with a TSS within a few kb of the known mRNA TSS (31). We therefore asked how many protein-coding Immunogenes harbored a divergently transcribed lncRNA based on the presence of a divergent spliced EST. For this analysis, we relaxed our stringency requirements such that a single EST from any tissue was sufficient to be counted, since most lncRNAs are transcribed at significantly lower levels than mRNAs. Interestingly, we identified 188 Immunogenes with divergently transcribed lncRNAs, including multiple susceptibility genes for immune disorders such as LRRK2, CYLD, and MFHAS1 (Supplemental Fig. 3C). We had previously shown that LRRK2 is a γ-interferon responsive gene. Strikingly, we show that the antisense LRRK2 lncRNA, which we term AS-LRRK2, is coordinately up-regulated with LRRK2 upon γ-interferon stimulation (Fig. 4B). Thus,EST profiling can also be used to reveal novel lncRNAs responsive to specific immune signalingpathways (30, 32, 33).
Immunogenes are enriched in primary immunodeficiency genes and in genomic regions associated with immune diseases/traits in GWAS studies
We found that the Immunogene compendium is significantly enriched in the KEGG category “primary immunodeficiency” (hsa05340; n = 28 of 34; P = 2.03 × 10−14). This KEGG pathway does not include other primary immunodeficiency genes that we identified as Immunogenes such as CARD9, CLEC7A, IL17RA (familial candidiasis 2, 4, 5; OMIM IDs: 212050, 613108, 613953), CXCR4 (WHIM syndrome; OMIM ID:193670), DOCK8 (hyper-IgE syndrome; OMIM ID: 243700), WAS (Wiskott-Aldrich syndrome; OMIM ID: 301000), and SH2D1A (X-linked lymphoproliferative disease; OMIM ID: 308240) (34) (Supplemental Table I). These results demonstrate the ability of our approach to highlight genes responsible for monogenic immune-related human diseases.
In terms of complex traits, GWAS have identified thousands of genetic loci related to disease phenotypes or traits, although identification of the exact causative polymorphism and mechanism(s) of action including relevant target gene(s) remains a challenge. We postulated that EST-defined Immunogenes may be significantly more enriched at loci implicated in immune-related diseases/traits (e.g. inflammatory bowel disease) given their expression in disease/trait-relevant tissue/cells (see Materials and Methods for details of immune-related diseases/traits analyzed). To test this possibility, we plotted the frequency of SNPs associated with human immune diseases/traits relative to the distance to the nearest Immunogene. The distance distribution of SNPs associated with non-immune-related diseases/traits to the nearest Immunogene was used as a comparison. As shown in Figure 4C, there is a higher frequency of SNPs associated with immune-related diseases/traits near Immunogenes, as compared to SNPs not associated with immune-related SNPs. Strikingly, we found that 254 of 808 (31.44%) immune-related disease/trait-associated SNPs had their closest RefSeq coding gene as an EST-defined Immunogene versus 783 of 7355 (10.65%) non-immune disease/trait-associated SNPs (P<10−4 based on permutation testing).
A recent GWAS study identified the locus tagging SNP rs140522 as associated with multiple sclerosis (MS). In this study, the authors cited the nearby gene encoding cytochrome oxidase deficient homolog 2 (SCO2) as the MS candidate gene within the locus (35). However, the nearest gene to rs140522 is not SCO2, but rather outer dense fiber of sperm tails 3B (ODF3B), which previously has not been implicated in immune function. Our approach identified ODF3B as novel Immunogene (Hs.531314; WIP = 0.229, P = 0.005). Thus, ODF3B is an alternative candidate gene for further testing in the pathophysiology of MS, illustrating the utility of integrating our EST expression profiling results with GWAS studies.
Identification of PRDM15 as a novel B cell gene overexpressed in human B cell lymphomas
We identified PRDM15, a zinc finger and SET domain-encoding gene, as one of the most highly immune-enriched (WIP = 0.71; P = 5.1 × 10−19) genes with little known function(s). We chose to focus on PRDM15 given our interest in B cell lymphoma pathogenesis coupled with our observation that a high fraction of the PRDM15-derived ESTs were from either normal germinal center B cells (BGC) or from B cell lymphoma cell line cDNA libraries (36). BGCs are thought to be the normal counterpart of some human B cell malignancies, including follicular lymphoma, Burkitt lymphoma, and diffuse large B cell lymphoma. To refine and further corroborate our EST data, we therefore analyzed the relative expression pattern of PRDM15 among various types of B cell lymphomas and non-B cell cancer-derived cell lines using published microarray data (23). As shown in Figure 5A, PRDM15 showed the highest expression in cell lines derived from follicular lymphomas (n = 9) and the second highest expression level in Burkitt lymphomas (n=17), compared to germinal B cell- and activated B cell-type non-Hodgkin diffuse large B cell lymphoma (n = 11), Hodgkin lymphomas (n = 9), lung cancers (n = 99), and colon cancers (n = 52). These results supported the observation that PRDM15 is specifically overexpressed in human B cell lymphomas. These data are consistent with RT-qPCR data showing that the Burkitt lymphoma cell line, Daudi, has 8–35 times more PRDM15 mRNA expression than several other cell lines/types (Fig. 5B). To examine PRDM15 expression at the protein level, we confirmed the nuclear localization of PRDM15 in HEK293T cells (Fig. 5C) and used Western blot analysis to demonstrate that PRDM15 protein is expressed in multiple Burkitt lymphomas and some monocytic leukemias, but not in T cell leukemias, suggesting that PRDM15 is expressed at higher levels in Burkitt B cell lymphomas not only at the mRNA level, but also at the protein level (Fig. 5D). In terms of follicular lymphoma expression, additional microarray analysis confirmed that PRDM15 was not only overexpressed in follicular lymphoma cell lines, but also in primary follicular lymphomas compared to primary normal B cell populations (Fig. 5E). Strikingly, immunohistochemistry analyses also revealed that PRDM15 was overexpressed in 5 of 7 independent human follicular lymphoma samples examined, as compared to normal tonsillar germinal centers (Fig. 5F). Thus, our results demonstrate how initial insights gained by EST profiling can be used to identify a novel transcription factor overexpressed in human B celllymphomas.
Discussion
In this study, we present the application of a quantitative genome-wide analysis of the human/mouse dbEST database composed of more than 6 million data points to elucidate a collection of genes enriched in immune expression, which we term Immunogenes. Using this database, we were able to identify an immune transmembrane and enzymatic signature. Furthermore, we implicated a novel transcription factor in B cell oncogenesis. Our results provide an important complement to other studies aimed at understanding the immune system including the RefDIC, ImmGen, and IRIS databases, all of which largely used microarray profiling (2–5).
Having a genome-wide set of genes with preferential immune expression, we undertook a systems-level analysis of the Immunogene compendium, finding a unique enzymatic signature of immune cells. The study of the large set of known or predicted enzymes, transporters, and corresponding metabolites is likely to yield critical insights into the metabolism of immunity. Furthermore, the Immunogene database provides an enhanced view of the potential spectrum of potential transmembrane cell surface markers expressed on various immune cells. These novel transmembrane Immunogenes represent potential candidates for targeted therapeutics or cell purification over the long-term via the development of antibodies directed against these antigens. We demonstrated how the Immunogene approach can be used to identify genes relevant to the pathogenesis of lymphomas. Along these lines, we dissected the expression pattern of PRDM15, finding it to be overexpressed at the level of mRNA and protein in human follicular lymphomas. Thus, delineation of PRMD15 function may prove to be important in understanding BGCphysiology and B cell malignancies.
The identification of genes involved in both Mendelian and complex traits encompassed within the dataset support the utility of the Immunogene database as an aid in genetic studies. In this regard, we demonstrate how the Immunogene dataset can be utilized to spotlight candidate genes where GWAS studies, genetic linkage studies, quantitative trait locus analysis, or mouse mutagenesis screens have identified genomic intervals containing numerous genes. In addition, the Immunogene database can be harnessed to probe the variation of immune system responses through focused analysis of SNPs and copy number variations of the identified Immunogenes. The analysis of genetic variation in the Immunogene dataset, through methods such as coding variant SNP analysis or resequencing, is poised to yield a pool of human genes at the cornerstone of variation in human immune responses, including susceptibility to autoimmune diseases such as lupus, inflammatory bowel disease, and diabetes. In addition, we demonstrated how EST data can be used to identify lncRNAs in autoimmune disease loci, with the identification of interferon-inducible AS-LRRK2 lncRNA in the Crohn’s disease LRRK2 locus. Our data suggest that Crohn’s disease-associated SNPs such as rs11175593, which lies in an intron of the AS-LRRK2 lncRNA, may modulate the function and/or expression of not only LRRK2, but also its co-regulated partner AS-LRRK2. Furthermore, extension of the Immunogene paradigm to other tissues may aid in deciphering human genes likely to be uniquely associated with other systems, such as the nervous system. We anticipate that by starting with the Immunogene dataset as a robust input, accompanied by multiple new layers of annotation, this resource will provide a dynamic platform to generate hypotheses to study known and novel genes in a variety of immunepathways.
Supplementary Material
Acknowledgments
This work was supported by grants DK083756, DK086502, HL088297, and DK043351 from the National Institutes of Health (to R.J.X.).
We thank Natalia Nedelsky for substantive editorial assistance.
References
- 1.Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18:363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- 2.Hijikata A, Kitamura H, Kimura Y, Yokoyama R, Aiba Y, Bao Y, Fujita S, Hase K, Hori S, Ishii Y, Kanagawa O, Kawamoto H, Kawano K, Koseki H, Kubo M, Kurita-Miki A, Kurosaki T, Masuda K, Nakata M, Oboki K, Ohno H, Okamoto M, Okayama Y, Saito OWJH, Saito T, Sakuma M, Sato K, Seino K, Setoguchi R, Tamura Y, Tanaka M, Taniguchi M, Taniuchi I, Teng A, Watanabe T, Watarai H, Yamasaki S, Ohara O. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics. 2007;23:2934–2941. doi: 10.1093/bioinformatics/btm430. [DOI] [PubMed] [Google Scholar]
- 3.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 4.Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, Fong S, van Lookeren Campagne M, Godowski P, Williams PM, Chan AC, Clark HF. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 5.Hyatt G, Melamed R, Park R, Seguritan R, Laplace C, Poirot L, Zucchelli S, Obst R, Matos M, Venanzi E, Goldrath A, Nguyen L, Luckey J, Yamagata T, Herman A, Jacobs J, Mathis D, Benoist C. Gene expression microarrays: glimpses of the immunological genome. Nat Immunol. 2006;7:686–691. doi: 10.1038/ni0706-686. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman BG, Williams KL, Tien AH, Lu V, de Algara TR, Ting JP, Helgason CD. Identification of novel genes and transcription factors involved in spleen, thymus and immunological development and function. Genes Immun. 2006;7:101–112. doi: 10.1038/sj.gene.6364270. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto SI, Suzuki T, Nagai S, Yamashita T, Toyoda N, Matsushima K. Identification of genes specifically expressed in human activated and mature dendritic cells through serial analysis of gene expression. Blood. 2000;96:2206–2214. [PubMed] [Google Scholar]
- 8.Castle JC, Armour CD, Lower M, Haynor D, Biery M, Bouzek H, Chen R, Jackson S, Johnson JM, Rohl CA, Raymond CK. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krupp M, Marquardt JU, Sahin U, Galle PR, Castle J, Teufel A. RNA-Seq Atlas--a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics. 2012;28:1184–1185. doi: 10.1093/bioinformatics/bts084. [DOI] [PubMed] [Google Scholar]
- 11.Gu J, He T, Pei Y, Li F, Wang X, Zhang J, Zhang X, Li Y. Primary transcripts and expressions of mammal intergenic microRNAs detected by mapping ESTs to their flanking sequences. Mamm Genome. 2006;17:1033–1041. doi: 10.1007/s00335-006-0007-9. [DOI] [PubMed] [Google Scholar]
- 12.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Dunham I, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Khatun J, Kheradpour P, Kundaje A, Lassman T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LA, Adams LB, Kelly CJ, Zhang J, Wexler JR, Green ED, Good PJ, Feingold EA, Bernstein BE, Birney E, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Gerstein M, Giddings MC, Gingeras TR, Green ED, Guigo R, Hardison RC, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Snyder M, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Khatun J, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Giddings MC, Bernstein BE, Epstein CB, Shoresh N, Ernst J, Kheradpour P, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Ward LD, Altshuler RC, Eaton ML, Kellis M, Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena HP, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Risk BA, Robyr D, Ruan X, Sammeth M, Sandu KS, Schaeffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Hayashizaki Y, Harrow J, Gerstein M, Hubbard TJ, Reymond A, Antonarakis SE, Hannon GJ, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR, Rosenbloom KR, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kent WJ, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Furey TS, Song L, Grasfeder LL, Giresi PG, Lee BK, Battenhouse A, Sheffield NC, Simon JM, Showers KA, Safi A, London D, Bhinge AA, Shestak C, Schaner MR, Kim SK, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Birney E, Iyer VR, Lieb JD, Crawford GE, Li G, Sandhu KS, Zheng M, Wang P, Luo OJ, Shahab A, Fullwood MJ, Ruan X, Ruan Y, Myers RM, Pauli F, Williams BA, Gertz J, Marinov GK, Reddy TE, Vielmetter J, Partridge EC, Trout D, Varley KE, Gasper C, Bansal A, Pepke S, Jain P, Amrhein H, Bowling KM, Anaya M, Cross MK, King B, Muratet MA, Antoshechkin I, Newberry KM, McCue K, Nesmith AS, Fisher-Aylor KI, Pusey B, DeSalvo G, Parker SL, Balasubramanian S, Davis NS, Meadows SK, Eggleston T, Gunter C, Newberry JS, Levy SE, Absher DM, Mortazavi A, Wong WH, Wold B, Blow MJ, Visel A, Pennachio LA, Elnitski L, Margulies EH, Parker SC, Petrykowska HM, Abyzov A, Aken B, Barrell D, Barson G, Berry A, Bignell A, Boychenko V, Bussotti G, Chrast J, Davidson C, Derrien T, Despacio-Reyes G, Diekhans M, Ezkurdia I, Frankish A, Gilbert J, Gonzalez JM, Griffiths E, Harte R, Hendrix DA, Howald C, Hunt T, Jungreis I, Kay M, Khurana E, Kokocinski F, Leng J, Lin MF, Loveland J, Lu Z, Manthravadi D, Mariotti M, Mudge J, Mukherjee G, Notredame C, Pei B, Rodriguez JM, Saunders G, Sboner A, Searle S, Sisu C, Snow C, Steward C, Tanzer A, Tapanan E, Tress ML, van Baren MJ, Walters N, Washieti S, Wilming L, Zadissa A, Zhengdong Z, Brent M, Haussler D, Kellis M, Valencia A, Gerstein M, Raymond A, Guigo R, Harrow J, Hubbard TJ, Landt SG, Frietze S, Abyzov A, Addleman N, Alexander RP, Auerbach RK, Balasubramanian S, Bettinger K, Bhardwaj N, Boyle AP, Cao AR, Cayting P, Charos A, Cheng Y, Cheng C, Eastman C, Euskirchen G, Fleming JD, Grubert F, Habegger L, Hariharan M, Harmanci A, Iyenger S, Jin VX, Karczewski KJ, Kasowski M, Lacroute P, Lam H, Larnarre-Vincent N, Leng J, Lian J, Lindahl-Allen M, Min R, Miotto B, Monahan H, Moqtaderi Z, Mu XJ, O’Geen H, Ouyang Z, Patacsil D, Pei B, Raha D, Ramirez L, Reed B, Rozowsky J, Sboner A, Shi M, Sisu C, Slifer T, Witt H, Wu L, Xu X, Yan KK, Yang X, Yip KY, Zhang Z, Struhl K, Weissman SM, Gerstein M, Farnham PJ, Snyder M, Tenebaum SA, Penalva LO, Doyle F, Karmakar S, Landt SG, Bhanvadia RR, Choudhury A, Domanus M, Ma L, Moran J, Patacsil D, Slifer T, Victorsen A, Yang X, Snyder M, White KP, Auer T, Centarin L, Eichenlaub M, Gruhl F, Heerman S, Hoeckendorf B, Inoue D, Kellner T, Kirchmaier S, Mueller C, Reinhardt R, Schertel L, Schneider S, Sinn R, Wittbrodt B, Wittbrodt J, Weng Z, Whitfield TW, Wang J, Collins PJ, Aldred SF, Trinklein ND, Partridge EC, Myers RM, Dekker J, Jain G, Lajoie BR, Sanyal A, Balasundaram G, Bates DL, Byron R, Canfield TK, Diegel MJ, Dunn D, Ebersol AK, Ebersol AK, Frum T, Garg K, Gist E, Hansen RS, Boatman L, Haugen E, Humbert R, Jain G, Johnson AK, Johnson EM, Kutyavin TM, Lajoie BR, Lee K, Lotakis D, Maurano MT, Neph SJ, Neri FV, Nguyen ED, Qu H, Reynolds AP, Roach V, Rynes E, Sabo P, Sanchez ME, Sandstrom RS, Sanyal A, Shafer AO, Stergachis AB, Thomas S, Thurman RE, Vernot B, Vierstra J, Vong S, Wang H, Weaver MA, Yan Y, Zhang M, Akey JA, Bender M, Dorschner MO, Groudine M, MacCoss MJ, Navas P, Stamatoyannopoulos G, Kaul R, Dekker J, Stamatoyannopoulos JA, Dunham I, Beal K, Brazma A, Flicek P, Herrero J, Johnson N, Keefe D, Lukk M, Luscombe NM, Sobral D, Vaquerizas JM, Wilder SP, Batzoglou S, Sidow A, Hussami N, Kyriazopoulou-Panagiotopoulou S, Libbrecht MW, Schaub MA, Kundaje A, Hardison RC, Miller W, Giardine B, Harris RS, Wu W, Bickel PJ, Banfai B, Boley NP, Brown JB, Huang H, Li Q, Li JJ, Noble WS, Bilmes JA, Buske OJ, Hoffman MM, Sahu AO, Kharchenko PV, Park PJ, Baker D, Taylor J, Weng Z, Iyer S, Dong X, Greven M, Lin X, Wang J, Xi HS, Zhuang J, Gerstein M, Alexander RP, Balasubramanian S, Cheng C, Harmanci A, Lochovsky L, Min R, Mu XJ, Rozowsky J, Yan KK, Yip KY, Birney E Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O’Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsanis N, Worley KC, Gonzalez G, Ansley SJ, Lupski JR. A computational/functional genomics approach for the enrichment of the retinal transcriptome and the identification of positional candidate retinopathy genes. Proc Natl Acad Sci U S A. 2002;99:14326–14331. doi: 10.1073/pnas.222409099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang S, Zhao S, Mu X, Thomas T, Klein WH. Novel retinal genes discovered by mining the mouse embryonic RetinalExpress database. Mol Vis. 2004;10:773–786. [PubMed] [Google Scholar]
- 17.Song HD, Sun XJ, Deng M, Zhang GW, Zhou Y, Wu XY, Sheng Y, Chen Y, Ruan Z, Jiang CL, Fan HY, Zon LI, Kanki JP, Liu TX, Look AT, Chen Z. Hematopoietic gene expression profile in zebrafish kidney marrow. ProNatl Acad Sci U S A. 2004;101:16240–16245. doi: 10.1073/pnas.0407241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- 19.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and IntegratedDiscovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 20.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clusteringsoftware. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 22.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 23.Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, Yang L, Pickeral OK, Rassenti LZ, Powell J, Botstein D, Byrd JC, Grever MR, Cheson BD, Chiorazzi N, Wilson WH, Kipps TJ, Brown PO, Staudt LM. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Ostell J, Miller V, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu CL, Kirley SD, Xiao H, Chuang Y, Chung DC, Zukerberg LR. Cables enhances cdk2 tyrosine 15 phosphorylation by Wee1, inhibits cell growth, and islost in many human colon and squamous cancers. Cancer Res. 2001;61:7325–7332. [PubMed] [Google Scholar]
- 26.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization ofthe human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solomon DH, Raynal MC, Tejwani GA, Cayre YE. Activation of the fructose 1,6-bisphosphatase gene by 1,25-dihydroxyvitamin D3 during monocytic differentiation. Proc Natl Acad Sci U S A. 1988;85:6904–6908. doi: 10.1073/pnas.85.18.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muindi JR, Peng Y, Wilson JW, Johnson CS, Branch RA, Trump DL. Monocyte fructose 1,6-bisphosphatase and cytidine deaminase enzyme activities: potential pharmacodynamic measures of calcitriol effects in cancer patients. Cancer Chemother Pharmacol. 2007;59:97–104. doi: 10.1007/s00280-006-0247-0. [DOI] [PubMed] [Google Scholar]
- 29.Morgan NV, Morris MR, Cangul H, Gleeson D, Straatman-Iwanowska A, Davies N, Keenan S, Pasha S, Rahman F, Gentle D, Vreeswijk MP, Devilee P, Knowles MA, Ceylaner S, Trembath RC, Dalence C, Kismet E, Koseoglu V, Rossbach HC, Gissen P, Tannahill D, Maher ER. Mutations in SLC29A3, encoding an equilibrative nucleoside transporter ENT3, cause a familial histiocytosis syndrome (Faisalabad histiocytosis) and familial Rosai-Dorfman disease. PLoS Genet. 2010;6:e1000833. doi: 10.1371/journal.pgen.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring ofChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals globalproperties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. AmJ Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, Dilthey A, Su Z, Freeman C, Hunt SE, Edkins S, Gray E, Booth DR, Potter SC, Goris A, Band G, Oturai AB, Strange A, Saarela J, Bellenguez C, Fontaine B, Gillman M, Hemmer B, Gwilliam R, Zipp F, Jayakumar A, Martin R, Leslie S, Hawkins S, Giannoulatou E, D’Alfonso S, Blackburn H, Boneschi FMartinelli, Liddle J, Harbo HF, Perez ML, Spurkland A, Waller MJ, Mycko MP, Ricketts M, Comabella M, Hammond N, Kockum I, McCann OT, Ban M, Whittaker P, Kemppinen A, Weston P, Hawkins C, Widaa S, Zajicek J, Dronov S, Robertson N, Bumpstead SJ, Barcellos LF, Ravindrarajah R, Abraham R, Alfredsson L, Ardlie K, Aubin C, Baker A, Baker K, Baranzini SE, Bergamaschi L, Bergamaschi R, Bernstein A, Berthele A, Boggild M, Bradfield JP, Brassat D, Broadley SA, Buck D, Butzkueven H, Capra R, Carroll WM, Cavalla P, Celius EG, Cepok S, Chiavacci R, Clerget-Darpoux F, Clysters K, Comi G, Cossburn M, Cournu-Rebeix I, Cox MB, Cozen W, Cree BA, Cross AH, Cusi D, Daly MJ, Davis E, de Bakker PI, Debouverie M, D’Hooghe MB, Dixon K, Dobosi R, Dubois B, Ellinghaus D, Elovaara I, Esposito F, Fontenille C, Foote S, Franke A, Galimberti D, Ghezzi A, Glessner J, Gomez R, Gout O, Graham C, Grant SF, Guerini FR, Hakonarson H, Hall P, Hamsten A, Hartung HP, Heard RN, Heath S, Hobart J, Hoshi M, Infante-Duarte C, Ingram G, Ingram W, Islam T, Jagodic M, Kabesch M, Kermode AG, Kilpatrick TJ, Kim C, Klopp N, Koivisto K, Larsson M, Lathrop M, Lechner-Scott JS, Leone MA, Leppa V, Liljedahl U, Bomfim IL, Lincoln RR, Link J, Liu J, Lorentzen AR, Lupoli S, Macciardi F, Mack T, Marriott M, Martinelli V, Mason D, McCauley JL, Mentch F, Mero IL, Mihalova T, Montalban X, Mottershead J, Myhr KM, Naldi P, Ollier W, Page A, Palotie A, Pelletier J, Piccio L, Pickersgill T, Piehl F, Pobywajlo S, Quach HL, Ramsay PP, Reunanen M, Reynolds R, Rioux JD, Rodegher M, Roesner S, Rubio JP, Ruckert IM, Salvetti M, Salvi E, Santaniello A, Schaefer CA, Schreiber S, Schulze C, Scott RJ, Sellebjerg F, Selmaj KW, Sexton D, Shen L, Simms-Acuna B, Skidmore S, Sleiman PM, Smestad C, Sorensen PS, Sondergaard HB, Stankovich J, Strange RC, Sulonen AM, Sundqvist E, Syvanen AC, Taddeo F, Taylor B, Blackwell JM, Tienari P, Bramon E, Tourbah A, Brown MA, Tronczynska E, Casas JP, Tubridy N, Corvin A, Vickery J, Jankowski J, Villoslada P, Markus HS, Wang K, Mathew CG, Wason J, Palmer CN, Wichmann HE, Plomin R, Willoughby E, Rautanen A, Winkelmann J, Wittig M, Trembath RC, Yaouanq J, Viswanathan AC, Zhang H, Wood NW, Zuvich R, Deloukas P, Langford C, Duncanson A, Oksenberg JR, Pericak-Vance MA, Haines JL, Olsson T, Hillert J, Ivinson AJ, De Jager PL, Peltonen L, Stewart GJ, Hafler DA, Hauser SL, McVean G, Donnelly P, Compston A International Multiple Sclerosis Genetics, C., C. Wellcome Trust Case Control. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, Neuberg D, Monti S, Giallourakis CC, Gostissa M, Alt FW. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Belgian-French IBDC, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ NIG Consortium, C Wellcome Trust Case Control. Genome-wide association defines more than 30 distinctsusceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.