Abstract
Cross-fertilization is predicted to facilitate the short-term response and the long-term persistence of host populations engaged in antagonistic coevolutionary interactions. Consistent with this idea, our previous work has shown that coevolving bacterial pathogens (Serratia marcescens) can drive obligately selfing hosts (Caenorhabditis elegans) to extinction, while the obligately outcrossing and partially outcrossing populations persisted. We focused the present study on the partially outcrossing (mixed mating) and obligately outcrossing hosts, and analyzed the changes in the host resistance/avoidance (and pathogen infectivity) over time. We found that host mortality rates increased in the mixed mating populations over the first ten generations of coevolution when outcrossing rates were initially low. However, mortality rates decreased after elevated outcrossing rates evolved during the experiment. In contrast, host mortality rates decreased in the obligately outcrossing populations during the first ten generations of coevolution, and remained low throughout the experiment. Therefore, predominant selfing reduced the ability of the hosts to respond to coevolving pathogens compared to outcrossing hosts. Thus, we found that host-pathogen coevolution can generate rapid evolutionary change, and that host mating system can influence the outcome of coevolution at a fine temporal scale.
Keywords: Adaptation, Mating Systems, Parasitism, Experimental Selection
The nematode Caenorhabditis elegans has long been recognized as a model organism in genetics and developmental biology. Sydney Brenner first demonstrated the nematode’s utility in genetics to conduct mutant screens and map unprecedented numbers of phenotypes, by capitalizing on the general ease of maintaining large populations of the worm in the laboratory, their rapid generation time (~4 days at 20°C), and their ability to self-fertilize (Brenner 1974). Though perhaps a bit under-utilized in evolutionary biology for the first couple decades of its use as a model in genetics, C. elegans has quickly become a multi-purpose model system for use in experimental evolution studies over the last several years for many of the same reasons Sydney Brenner originally chose the species for his work.
Although it might seem that nematodes lack interesting phenotypes, evolutionary biologists have recently capitalized on the wealth of genetic tools available for manipulating phenotypes (from mating systems to mutation repair), and the ease of maintaining large populations of C. elegans, to test fundamental questions in evolutionary biology via experimental evolution. The nematode has been utilized in the laboratory to: (1) determine mutation rates (Vassilieva and Lynch 1999; Vassilieva et al. 2000; Estes and Lynch 2003; Denver et al. 2004; Estes et al. 2004; Ajie et al. 2005; Baer et al. 2005; Cutter 2005; Baer et al. 2006; Manoel et al. 2007; Ostrow et al. 2007; Morran et al. 2009; Morran et al. 2010; Estes et al. 2011), (2) to study life-history evolution (Anderson et al. 2011; Murray and Cutter 2011), (3) to examine adaptation to novel environments (Lopes et al. 2008; Morran et al. 2009; Wegewitz et al. 2009; Anderson et al. 2010; Teotonio et al. 2012), (4) to examine mating-system evolution (Chasnov and Chow 2002; Stewart and Phillips 2002; Cutter 2005; Manoel et al. 2007; Katju et al. 2008; Wegewitz et al. 2008; Morran et al. 2009; Wegewitz et al. 2009; Chandler et al. 2011; Morran et al. 2011; Teotonio et al. 2012), and finally (5) to experimentally evaluate host-pathogen coevolution (Schulte et al. 2010; Morran et al. 2011). Experimental evolution is a particularly valuable tool for investigating host-pathogen coevolutionary dynamics, because it permits the assessment of interactions between hosts and pathogens as they coevolve. This dissection of host-pathogen coevolutionary dynamics, rather than just the ultimate outcome of coevolution, is crucial for understanding many of the evolutionary implications of host-pathogen coevolution (Hall et al. 2011).
The C. elegans system may be especially valuable as an experimental model to understand the role of host-parasite coevolution on the evolution and maintenance of high levels of outcrossing. Where pathogens are common, highly virulent, and impose frequency-dependent selection, hosts are predicted to evolve greater levels of recombination and/or outcrossing (Hamilton 1980; Peters and Lively 1999; Gandon and Otto 2007). Recent empirical studies are consistent with these basic ideas on host-pathogen coevolutionary dynamics (e.g., Fischer and Schmid-Hempel 2005; Jokela et al. 2009; King et al. 2009; Schulte et al. 2010; Morran et al. 2011; Kerstes et al. 2012), which form the conceptual foundation of the Red Queen hypothesis as it relates to the maintenance of outcrossing (Jaenike 1978; Hamilton 1980; Bell 1982).
Van Valen’s macroevolutionary formulation of the Red Queen hypothesis did not specifically address mating-system evolution; however his view of the Red Queen hypothesis is still relevant to host-pathogen coevolutionary dynamics and perhaps the maintenance of outcrossing. The premise of Van Valen’s Red Queen hypothesis was that species engaging in coevolution must rapidly evolve or face the threat of extinction (Van Valen 1973). Under this scenario, host populations must continue to produce resistant types to evade the pathogens, and the pathogens must evolve to infect the newly resistant types.
Outcrossing could be the most effective mating system for enabling the persistence of hosts under both the microevolutionary and macroevolutionary variants of the Red Queen hypothesis, whether by producing offspring with novel genotypes that confer greater resistance, or by generating offspring with rare genotypes that avoid infection. Outcrossing has been shown to facilitate more rapid adaptation than self-fertilization to a novel environment. Presumably, mating systems that constrain alleles to specific lineages, like self-fertilization, ultimately slow host adaptation (Stebbins 1957; Maynard Smith 1978; Lande and Schemske 1985), and thus increase the probability of infection and potentially extinction. Therefore, the mating system of the host population should strongly influence the outcome of host-pathogen coevolution when the interaction between the two species generates strong selection.
In a previous study, we exposed obligate selfing, mixed mating (i.e., a mixture of selfing and outcrossing individuals), and obligately outcrossing Caenorhabditis elegans hosts to coevolving populations of the bacterial pathogen, Serratia marcescens, for thirty generations. We found that the obligately outcrossing populations evolved reduced host mortality rates, and that the mixed mating populations maintained equivalent levels of host mortality, relative to their respective ancestral populations. Conversely, the obligately self-fertilizing populations exhibited substantial increases in host mortality rates. Both the mixed mating and obligately outcrossing hosts persisted throughout the experiment, but the obligately selfing hosts went extinct, which is consistent with the macroevolutionary version of the Red Queen. Further, we found that exposure to the coevolving pathogens selected for greater rates of outcrossing in the mixed mating hosts, which is consistent with the microevolutionary version of the Red Queen (Morran et al. 2011).
In the present study, we capitalized on the ability to freeze and revive C. elegans to assess the temporal dynamics of host-pathogen coevolution in host and pathogen populations periodically preserved during our previous study (Morran et al. 2011). We compared the infectivity of the coevolving bacterial pathogens over time on host populations with different mating systems: mixed mating vs. obligately outcrossing. In particular, we documented the change in pathogen infectivity as outcrossing rates increased in the mixed mating populations over time. The results demonstrate that host-pathogen coevolution generates rapid evolutionary change, and provide direct empirical support for the idea that outcrossing permits hosts to keep pace with their coevolving pathogens, while predominant self-fertilization leads to greater infection.
Methods
HOST AND PATHOGEN POPULATIONS
C. elegans is an androdioecious nematode species composed of hermaphrodites that can either reproduce by self-fertilization or by outcrossing with a male (Brenner 1974). Therefore, C. elegans populations are naturally mixed mating, but are often heavily reliant on selfing as a means of reproduction (Barriere and Felix 2005; Haber et al. 2005; Sivasundar and Hey 2005; Barriere and Felix 2007; Rockman and Kruglyak 2009). C. elegans lines used in this study were derived from the CB4856 strain (originally from Hawaii, USA), obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). In order to generate an obligately outcrossing line, we used the fog-2 mutant allele, which inhibits sperm production in C. elegans, rendering them incapable of self-fertilization (Schedl and Kimble 1988). This allele was backcrossed into an inbred CB4856 genetic background (PX382) to generate the PX386 strain (Morran et al. 2009).. For the parasite, we used S. marcescens, which is a virulent gram-negative soil bacterium that rapidly induces high rates of host mortality when consumed by C. elegans (Schulenburg and Ewbank 2004). We used S. marcescens strain Sm2170 as it exhibits the highest levels of virulence against C. elegans, relative to other S. marcescens strains.
Experimental evolution
Because all host populations were derived from a single inbred strain (PX382), and therefore had little genetic variation, C. elegans were mutagenized with EMS prior to experimental evolution (Anderson 1995; Morran et al. 2011). Five replicate populations of mixed mating C. elegans (PX382), and five replicate populations of obligately outcrossing C. elegans (PX386), were independently mutagenized, then designated as populations # 1 through # 5. These populations were frozen as the “ancestor” populations. To start the experiment, a portion of these ancestral populations were thawed and allowed two generations for recovery; and then each population was divided into three separate treatments for experimental evolution: control (hosts were exposed to heat-killed S. marcescens), fixed pathogen (hosts were exposed to a genetically fixed population of S. marcescens), and recycled pathogen (hosts were exposed to potentially coevolving populations of S. marcescens). Note that the fixed pathogen treatment was previously referred to as the “Evolution” treatment and the recycled pathogen treatment referred to as the “Coevolution” treatment in Morran et. al (2011). All obligately outcrossing hosts designated as “Population # 1” shared the same ancestral population no matter which treatment they experienced. Additionally, five populations of S. marcescens, derived from the same ancestral colony as the populations in the recycled pathogen treatment, were evolved and transferred in same manner as the populations in the recycled pathogen treatment but without exposure to C. elegans, and thus serve as a control with regards to the pathogen.
Hosts were exposed to the pathogen on “Serratia Selection Plates” (SSP) (Morran et al. 2009). Only the offspring from surviving hosts were transferred to a fresh SSP each generation. Under the recycled-pathogen treatment, only S. marcescens that successfully infected and killed C. elegans within 24 hours were transferred to SSP plates, upon which the next generation of hosts would be placed. Selected colonies of S. marcescens were isolated by harvesting and crushing C. elegans carcasses 24 hours after exposure. The isolated pathogens were then grown on agar plates (Nematode Growth Medium-Lite, US Biological, Swampscott, MA, USA); ten colonies were then randomly picked, grown overnight in Luria Broth (LB), and then used to seed the subsequent SSP (Morran et al. 2011).
The experiment was run for 30 host generations, during which both the host and pathogen populations were frozen every ten host generations. The outcrossing rate of the mixed mating populations was estimated using male frequency counts every 4 generations (Stewart and Phillips 2002). The outcrossing rate for the obligately outcrossing populations was assumed to be one (Morran et al. 2011).
HOST MORTALITY RATE AND OUTCROSSING RATE MEASUREMENTS
Mortality assays of coevolving pathogens on contemporary host populations
Samples of each host and pathogen population were thawed in the recycled pathogen treatment at host-generation time points 0 (ancestor), 10, 20, and 30. Host populations were then cycled for two generations under standard laboratory conditions to recover from thawing (i.e., 10cm Petri dishes filled with NGM Lite seeded with 10μL of OP50 stored at 20°C). The thawed pathogen populations were grown overnight in LB at 28°C; they were then used to seed 10cm Petri dishes filled with NGM-Lite (100 μL per dish) and grown at 28°C overnight.
After these steps, each host population was exposed to its corresponding pathogen population from the same specific time point (the contemporary pathogen). These host populations were also exposed the ancestral pathogen population, as well as to a corresponding control pathogen population. For each host population, 200 L4 C. elegans individuals were suspended in M9 buffer and transferred to a 10 cm Petri dish (NGM-Lite agar), which had been seeded with a lawn of S. marcescens (approximately 6 cm in diameter). After twenty-four hours of exposure, we counted the total number of dead nematodes. We replicated each host x pathogen combination at least three times at each host generation time point (some host x pathogen combinations have more than three replicate measurements). We attempted to measure five replicates of some mixed mating populations, because the mixed mating populations exhibited the greatest level of variance. In several cases, a replicate was found to contain a contaminating bacteria, and it was not measured. Twenty-four hour mortality rates were calculated by dividing the number of dead nematodes by the total transferred. Data points for hosts at generation zero and generation thirty were previously reported in Morran et al. (2011).
A separate ANOVA was performed in JMP-IN 8.0 for each mating type (obligate outcrossing and mixed mating) testing the effects of (1) generation of coevolution, (2) replicate population, (3) and the state of the pathogen (contemporary, ancestral, control), as well the interaction between main effects, excluding the interaction between replicate population and pathogen. The dependent variable was the observed mean mortality rate. Replicate population was treated as a random effect, with generation as a fixed effect. Least squared mean contrast tests were performed to test for differences between specific means within each mating type. The mortality rates for obligately outcrossing hosts deviated significantly from normality and exhibited unequal variances. Although ANOVA is robust to deviations from normality and violations of the assumption of homogeneity of variance at our sample size (Lindman 1974), we nonetheless utilized nonparametric Wilcoxon tests for the mortality rate data in JMP-IN 8.0 to confirm the results of each least squared mean contrast test we reported for the obligately outcrossing populations in the text. The statistics we report are from the least squared mean contrast tests, though we obtained qualitatively equivalent results using the Wilcoxon tests.
Mortality assays of coevolving pathogens on ancestral host populations
Samples of each pathogen population in the recycled pathogen treatment at host generation time points 0 (ancestor), 10, 20, and 30 were thawed, along with all of the ancestral host populations. The host and pathogen populations were maintained in the same manner as described in the previous morality assay description. Each ancestral host population was exposed to the corresponding coevolving pathogen population at each time point throughout the experiment. This allowed us to measure the change in host mortality induced by the pathogen population over time against a fixed host population. For each host population, approximately 200 L4 C. elegans individuals were suspended in M9 buffer and transferred to a 10 cm Petri dish (NGM-Lite agar) seeded with a lawn of S. marcescens (approximately 6 cm in diameter). Due to clumping in the nematode populations, we specifically counted the number of nematodes transferred in an aliquot to more accurately measure mortality rates. After twenty-four hours of exposure, we counted the total number of dead nematodes. We replicated each pathogen population at least four times at each time point. Twenty-four hour mortality rates were calculated by dividing the number of dead nematodes by the total transferred (Morran et al. 2011).
Separate ANOVAs were performed in JMP-IN 8.0 for each mating system group testing the effects of generation of coevolution and replicate population on mean mortality rates. Replicate population was treated as a random effect with generation and as a fixed effect. Least squared mean contrast tests were performed to test differences between specific means.
Outcrossing rates
Male frequency was measured every four generations until generation twenty-eight, and then again at generation thirty. All mature worms were sexed and counted after transfer. Outcrossing rates were extrapolated from male frequency data (Morran et al. 2011). Outcrossing events in C. elegans produce equal proportions of males and hermaphrodites. However, males are also spontaneously produced through hermaphroditic self-fertilization if X chromosome nondisjunction occurs during meiosis (Brenner 1974). By correcting for the number of males produced through nondisjunction and multiplying the remaining male frequency by two (to account for both males and hermaphrodites produced via outcrossing), we calculated the outcrossing rate for each replicate population (Stewart and Phillips 2002). We report the arithmetic mean outcrossing rate of all replicate populations in the mixed mating and obligately outcrossing groups approximately ever four generations over the span of experimental coevolution. Arithmetic mean outcrossing rates in the mixed mating populations were previously reported in Morran et al. (2011). Further, we determined the harmonic mean outcrossing rate of each replicate population over three ten-generation intervals (generations 0–10, 10–20, and 20–30). The harmonic mean is more sensitive to low values and therefore is a more accurate representation of the genetic dynamics underlying outcrossing rates. A nonparametric Wilcoxon test was performed in JMP 8.0 to test the effect of host generation block (generations 0–10 vs. generations 10–20) on the harmonic mean outcrossing rates of the mixed mating host populations. We also used JMP 8.0 to perform linear regression analysis on the relationship between the change in the observed change in host mortality rate and harmonic mean outcrossing for the five replicate populations in the mixed mating group over the first twenty generations of coevolution.
Results
Mortality assays of coevolving pathogens on ancestral host populations
The pathogens that coevolved with the mixed mating and obligately outcrossing hosts were sampled at host-generations 0, 10, 20, and 30, and then exposed the to ancestral host populations, to gauge their general infectivity over time. We found that, regardless of host mating system, the pathogen populations induced significantly greater host mortality rates after passage with their respective hosts (Fig. 1; generation 0 versus all subsequent generations: mixed mating, F1,12 = 13.69, p = 0.003; obligately outcrossing, F1,12 = 79.79, p < 0.0001). Host mortality rates serve as a measure of pathogen infectivity within this system, because host death was required for successful infection and transmission during experimental evolution (Morran et al. 2011). Therefore, the pathogen populations that were passaged with either the mixed-mating or obligately outcrossing hosts evolved greater infectivity during experimental evolution (Fig 1).
Figure 1.
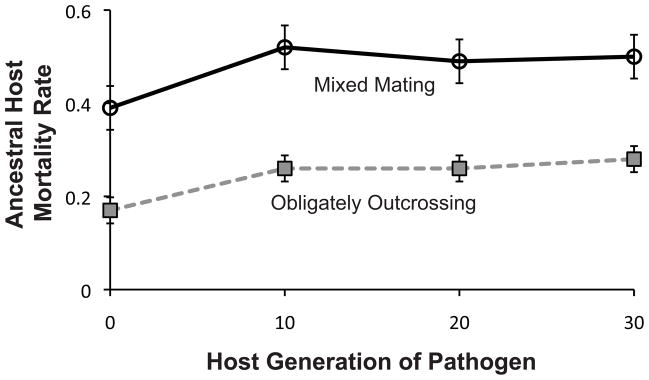
Pathogen infectivity over time. S. marcescens populations coevolved with either mixed mating or obligately outcrossing C. elegans were sampled every ten generations of coevolution. These pathogens were used to infect ancestral populations of the hosts with which they were coevolving, thus measuring their infectivity over time. The pathogen populations show an initial increase and then maintenance of infectivity throughout the experiment, as evidenced by the increase (generation 0 to generation 10 ; mixed mating, F1,12= 13.69, p = 0.003; obligately outcrossing, F1,12 = 79.79, p < 0.0001) and maintenance (generation 10 to generation 30; mixed mating, F1,12 = 0.4547, p = 0.5129; obligately outcrossing, F1,12 = 0.6826, p = 0.4248) of host mortality rates against the same ancestral hosts. Error bars indicate ± 1 S.E.M.
Mortality assays of coevolving pathogens on contemporary host populations
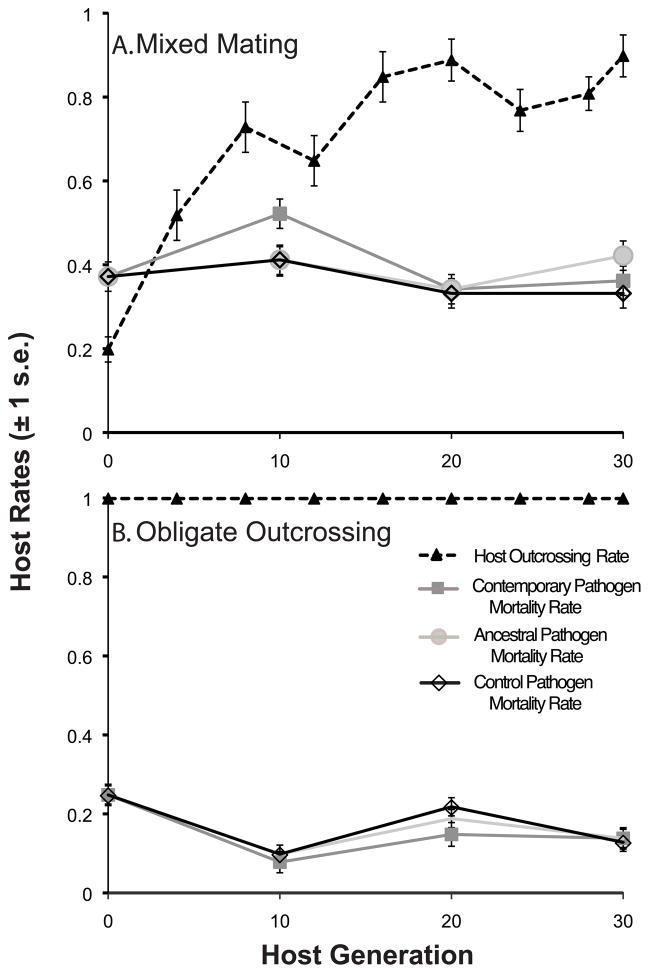
Although patterns for the evolution of infectivity were similar for pathogens coevolving with the mixed mating and obligately outcrossing hosts (Fig. 1), the host-pathogen coevolutionary dynamics ultimately showed marked differences between host mating systems. After only ten generations of coevolution, contemporary pathogens induced greater levels of mortality against the mixed mating hosts compared to the ancestral generation (Fig. 2A; ancestral generation 0 vs. contemporary generation 10; F1,8 = 26.16, p = 0.0009). This increase in infectivity was predominantly the result of evolution on the part of the contemporary pathogens (Fig. 1), and not decreased resistance on the part of the hosts. Specifically, the contemporary pathogens from generation ten were significantly more infective than their respective control and ancestral pathogens (Fig. 2A; contemporary generation 10 vs. control generation 10; F1,8 = 14.03, p = 0.005; contemporary generation 10 vs. ancestral generation 10; F1,8 = 14.48, p = 0.006), which did not significantly differ from generation zero measurements (Fig. 2A; control generation 10 vs. ancestral generation 0; F1,8 = 1.876, p = 0.208; ancestral generation 10 vs. ancestral generation 0; F1,8 = 1.717, p = 0.2265). These results suggest that, during the first ten generations of the experiment, the mixed mating hosts did not evolve resistance rapidly enough to prevent significant increases in mortality induced by the more rapidly evolving pathogens.
Figure 2.
Host-pathogen infection dynamics over time. Mean host mortality and mean outcrossing rates were measured over thirty generations of coevolution between C. elegans hosts and S. marcescens pathogens. Host mortality rates were measured on hosts sampled every ten generations. Each host population at each time point was infected with the ancestral pathogen, the contemporary pathogen, and a non-coevolving control pathogen, each corresponding to the particular host population being infected. Host outcrossing rates were estimated from male frequency data collected every four host generations (and at generation 30) throughout the experiment, and value of the mixed mating populations were previously reported in Morran et al. 2011. (A.) Mixed mating hosts. (B.) Obligately outcrossing hosts.
In contrast, the obligately outcrossing populations exhibited significantly lower mortality rates over the first ten generations of coevolution (Fig. 2B; generation 0 vs. generation 10; F1,8 = 189.3, p < 0.0001), even though their coevolving pathogens evolved greater infectivity (Fig. 1). Therefore, the obligately outcrossing hosts evolved resistance (or avoidance) rapidly enough to actually reduce mortality as their pathogens became more infective (as measured by infectivity on ancestral hosts).
Outcrossing Rates
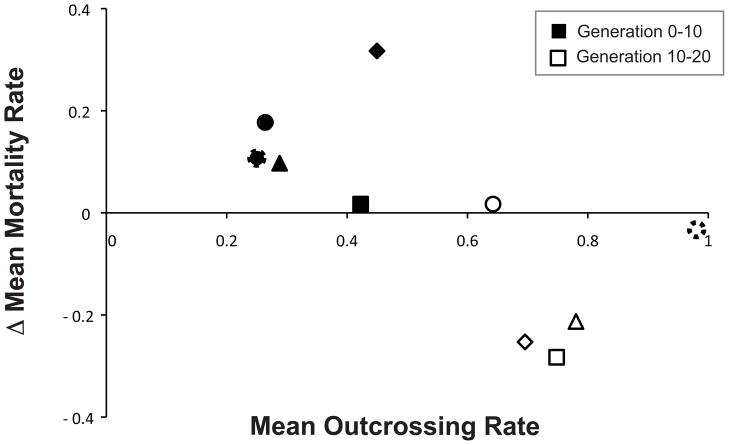
Interestingly, selfing was the most prevalent form of reproduction over the first ten generations of coevolution in the mixed mating populations (Fig. 2A). Then, after twenty generations of coevolution, after the evolution of significantly greater rates of outcrossing (generation 0–10 vs. generation 10–20 harmonic mean outcrossing rates; X2= 6.82, df = 1, p = 0.009), the mixed-mating populations exhibited reduced levels of host mortality with regards to their coevolving pathogens (contemporary generation 10 vs. contemporary generation 20; F1,8 = 43.97, p = 0.0002). This reduction in mortality rates for mixed-mating hosts, is correlated with elevated outcrossing over the first twenty generations of coevolution (Fig. 3; R2 = 0.435, F1,8 = 6.166, p = 0.0379). Therefore, the mixed mating hosts evolved greater resistance and/or avoidance to their coevolving pathogens after evolving elevated outcrossing rates (Fig. 2).
Figure 3.
Negative correlation between host outcrossing rate and change in host mortality in mixed mating populations. Elevated levels of outcrossing are generally correlated with decreases in host mortality rates (R2 = 0.435, F1,8 = 6.166, p = 0.0379). The harmonic mean outcrossing rate of each replicate population was plotted against the mean change in host mortality over two time periods, generations 0 to 10 (filled symbols) and generations 10 to 20 (open symbols), matching symbols indicate the same population measured at the different time points.
Although the mixed mating hosts evolved greater resistance following the increase in outcrossing, ultimately there was no net change in the host mortality rates exhibited by the mixed mating hosts after thirty generations of coevolution relative to their ancestral hosts (Fig. 2A; generation 0 vs. generation 30; F1,8 = 0.0033, p = 0.9954). However, the obligately outcrossing hosts exhibited a net decrease in host mortality rates of approximately 44% over the course of thirty generations of coevolution (Fig. 2B; generation 0 vs. generation 30; F1,8 = 99.17, p < 0.0001). Therefore, the outcome and dynamics of the host-pathogen coevolutionary interactions were greatly influenced by the host mating system, and more specifically the outcrossing rates exhibited by the host population.
Discussion
Our previous work demonstrated that coevolving pathogens are capable of rapidly driving obligately selfing populations to extinction, while mixed-mating hosts and obligately outcrossing hosts persisted under those same conditions (Morran et al. 2011); hence the results suggested that outcrossing allowed for persistence in the macroevolutionary game envisioned by Van Valen’s view of the Red Queen. In addition, the mixed mating populations responded to selection from coevolving pathogens by evolving and maintaining greater outcrossing rates, which is consistent with the microevolutionary view of the Red Queen, envisioned by Jaenike (1978), Hamilton (1980), Lloyd (1980), and Bell (1982). In the present study, we found that, within those mixed mating populations, relatively low levels of outcrossing were not sufficient to escape the evolution of greater rates of infection by the pathogen over the first ten generations of coevolution (Fig. 2A). However, host mortality rates declined in the mixed mating host populations after high levels of outcrossing evolved (Fig. 2A). Therefore, parasite infectivity decreased as outcrossing became more prevalent in the mixed mating populations. It thus appears that predominant selfing in the host allowed the pathogen to adapt to the host population faster than the host population could respond; but the evolution of higher levels of outcrossing allowed the mixed mating hosts to keep pace with their coevolving pathogens and to persist throughout the experiment.
Conceptually similar results were obtained from the outcrossing host populations. These populations were outcrossed throughout the entire experiment, and they rapidly responded to selection from the coevolving pathogens populations (Fig. 2B). Despite the fact that the pathogens evolved greater infectivity against ancestral hosts (Fig. 1), host mortality rates in the obligately outcrossing hosts were reduced by ~50% within just ten generations, and these reduced levels were maintained throughout the experiment (Fig. 2B). Conversely, the mixed mating populations, even after exhibiting high outcrossing rates for prolonged periods of time, did not exhibit significant reductions in host mortality relative to their ancestral starting point. Presumably the loss of genetic variation due to initially high selfing rates ultimately limited the number of novel genotypes in the mixed mating population. Therefore, although low levels of outcrossing are often thought to provide many of the benefits of obligate outcrossing, obligate outcrossing was more effective at facilitating rapid adaptation to a pathogen and decreasing host mortality rates.
The adaptive value of outcrossing, and genetic exchange between lineages in general, has also been demonstrated under various selective regimes. Directional selection resulting from exposure to novel environments is capable of selecting for temporarily increased sexual over asexual reproduction in rotifers (Becks and Agrawal 2012), in yeast (Goddard et al. 2005), and in green algae (Colegrave 2002). Further, directional selection imposed by a novel environment can select for increased outcrossing over self-fertilization in C. elegans populations (Lopes et al. 2008; Morran et al. 2009; Teotonio et al. 2012). Similarly, genetic exchange in bacterial populations can facilitate adaptation under directional selection (Cooper 2007; Baltrus et al. 2008). Further, sexual reproduction in the rotifer, Brachionus calyciflorus, is also favored by fluctuating selection generated by periodic environmental changes (Becks and Agrawal 2010). And, although the mode of selection is unclear, outcrossing seemed to facilitate C. elegans host adaptation during experimental coevolution with the bacterial pathogen Bacillus thuringiensis (H. Schulenburg, personal communication).
Within the context of our experiment, it is clear that high levels of self-fertilization were detrimental, and that predominant outcrossing was beneficial to host fitness. It is presently unclear if greater levels of outcrossing were beneficial due to fluctuating selection (i.e., Red Queen dynamics), or if outcrossing facilitated the response to directional selection leading to repeated selective sweeps (reviewed in Woolhouse et al. 2002). We suspect that both beneficial aspects of outcrossing may have played a role in our experiment. Given that these hosts and pathogen populations did not have a long-term evolutionary history prior to experimental coevolution, it is likely that outcrossing facilitated selective sweeps as hosts adapted to the novel pathogen.
Although the prospect of selective sweeps appears likely, it seems that the consistent shuffling of host genotypes may have also been favored by selection. Hall et al. (2011) recently found that arms race dynamics slowed over time, and a fluctuating selection dynamic became more prevalent during experimental coevolution of bacteria and phage. Therefore a combination of directional selection and frequency-dependent selection, particularly as coevolution proceeded, would not be surprising. Further, we found that high levels of outcrossing were maintained in the mixed mating populations, and low rates of outcrossing were not sufficient to maintain levels of host resistance or avoidance as the pathogens evolved (Fig. 2A). Even low levels of outcrossing can rapidly reduce linkage disequilibrium and presumably permit selective sweeps (Nordborg 1997; Barriere and Felix 2005; Anderson et al. 2010). So, the maintenance of high outcrossing rates suggests that the consistent production of recombinant genotypes was favored. Although it is possible that successive selective sweeps fueled the maintenance of elevated outcrossing, such sweeps could rapidly erode genome-wide diversity and consequently reduce the likelihood of further sweeps (Andolfatto 2001; Andersen et al. 2012) and the benefits of outcrossing.
In summary host-pathogen coevolution can impose strong selection and promote rapid evolution in both the host and pathogen populations. We observed substantial changes in host mortality rates within just ten host generations of coevolution, driven by both the host and pathogen populations. Further, the host mating system, or more specifically the outcrossing rate in the host population, can determine the nature of the coevolutionary dynamics. And, as expected under both formulations of the Red Queen hypothesis, predominant self-fertilization limited the ability of the hosts to respond to their coevolving pathogens, while frequent outcrossing permitted the hosts to keep pace with their pathogens.
Acknowledgments
We thank Sinead Collins for organizing and editing this issue. Additionally, we thank H. Hundley for the use of her laboratory space and equipment. We also thank F. Bashey, O. Schmidt, M. Allen, and R. Matteson for logistical support and beneficial discussion. And we thank the members of the Lively, Hall, and Wade labs at Indiana University, and two anonymous reviewers for helpful comments and discussion pertaining to this work. Funding was provided by the NSF (DEB-0640639 to CML) and the NIH (1F32GM096482-01 to LTM). The nematode strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). LTM and CML conceived and designed the experiments. LTM, RCP, and IAG performed the experiments. LTM and CML analyzed the data. LTM and CML wrote the paper.
LITERATURE CITED
- Ajie BC, Estes S, Lynch M, Phillips PC. Behavioral degradation under mutation accumulation in Caenorhabditis elegans. Genetics. 2005;170:655–660. doi: 10.1534/genetics.104.040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Felix MA, Kruglyak L. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012;44:285–290. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JL, Morran LT, Phillips PC. Outcrossing and the maintenance of males within C. elegans populations. J Hered. 2010;101(Suppl 1):S62–74. doi: 10.1093/jhered/esq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JL, Reynolds RM, Morran LT, Tolman-Thompson J, Phillips PC. Experimental evolution reveals antagonistic pleiotropy in reproductive timing but not life span in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2011;66:1300–1308. doi: 10.1093/gerona/glr143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. Mutagenesis. In: Epstein H, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Academic Press; London: 1995. pp. 31–54. [Google Scholar]
- Andolfatto P. Adaptive hitchhiking effects on genome variability. Curr Opin Genet Dev. 2001;11:635–641. doi: 10.1016/s0959-437x(00)00246-x. [DOI] [PubMed] [Google Scholar]
- Baer CF, Phillips N, Ostrow D, Avalos A, Blanton D, Boggs A, Keller T, Levy L, Mezerhane E. Cumulative effects of spontaneous mutations for fitness in Caenorhabditis: role of genotype, environment and stress. Genetics. 2006;174:1387–1395. doi: 10.1534/genetics.106.061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer CF, Shaw F, Steding C, Baumgartner M, Hawkins A, Houppert A, Mason N, Reed M, Simonelic K, Woodard W, Lynch M. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc Natl Acad Sci U S A. 2005;102:5785–5790. doi: 10.1073/pnas.0406056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus D, Guillemin K, Phillips P. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution. 2008;62:39–49. doi: 10.1111/j.1558-5646.2007.00271.x. [DOI] [PubMed] [Google Scholar]
- Barriere A, Felix MA. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr Biol. 2005;15:1176–1184. doi: 10.1016/j.cub.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Barriere A, Felix MA. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics. 2007;176:999–1011. doi: 10.1534/genetics.106.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becks L, Agrawal AF. Higher rates of sex evolve in spatially heterogeneous environments. Nature. 2010;468:89–93. doi: 10.1038/nature09449. [DOI] [PubMed] [Google Scholar]
- Becks L, Agrawal AF. The evolution of sex is favoured during adaptation to new environments. PLoS Biol. 2012;10:e1001317. doi: 10.1371/journal.pbio.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. The Evolution and Genetics of Sexuality. University of California Press; Berkley, CA: 1982. The Masterpiece of Nature. [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler CH, Chadderdon GE, Phillips PC, Dworkin I, Janzen FJ. Experimental evolution of the Caenorhabditis elegans sex determination pathway. Evolution. 2011;66:82–93. doi: 10.1111/j.1558-5646.2011.01420.x. [DOI] [PubMed] [Google Scholar]
- Chasnov JR, Chow KL. Why are there males in the hermaphroditic species Caenorhabditis elegans? Genetics. 2002;160:983–994. doi: 10.1093/genetics/160.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrave N. Sex releases the speed limit on evolution. Nature. 2002;420:664–666. doi: 10.1038/nature01191. [DOI] [PubMed] [Google Scholar]
- Cooper TF. Recombination speeds adaptation by reducing competition between beneficial mutations in populations of Escherichia coli. PLoS Biol. 2007;5:e225. doi: 10.1371/journal.pbio.0050225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. Mutation and the experimental evolution of outcrossing in Caenorhabditis elegans. J Evol Biol. 2005;18:27–34. doi: 10.1111/j.1420-9101.2004.00804.x. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Thomas WK. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430:679–682. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- Estes S, Lynch M. Rapid fitness recovery in mutationally degraded lines of Caenorhabditis elegans. Evolution. 2003;57:1022–1030. doi: 10.1111/j.0014-3820.2003.tb00313.x. [DOI] [PubMed] [Google Scholar]
- Estes S, Phillips PC, Denver DR. Fitness recovery and compensatory evolution in natural mutant lines of C. elegans. Evolution. 2011;65:2335–2344. doi: 10.1111/j.1558-5646.2011.01276.x. [DOI] [PubMed] [Google Scholar]
- Estes S, Phillips PC, Denver DR, Thomas WK, Lynch M. Mutation accumulation in populations of varying size: the distribution of mutational effects for fitness correlates in Caenorhabditis elegans. Genetics. 2004;166:1269–1279. doi: 10.1534/genetics.166.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer O, Schmid-Hempel P. Selection by parasites may increase host recombination frequency. Biol Lett. 2005;1:193–195. doi: 10.1098/rsbl.2005.0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandon S, Otto SP. The evolution of sex and recombination in response to abiotic or coevolutionary fluctuations in epistasis. Genetics. 2007;175:1835–1853. doi: 10.1534/genetics.106.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard MR, Charles H, Godfray J, Burt A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005;434:636–640. doi: 10.1038/nature03405. [DOI] [PubMed] [Google Scholar]
- Haber M, Schungel M, Putz A, Muller S, Hasert B, Schulenburg H. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Mol Biol Evol. 2005;22:160–173. doi: 10.1093/molbev/msh264. [DOI] [PubMed] [Google Scholar]
- Hall AR, Scanlan PD, Morgan AD, Buckling A. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett. 2011;14:635–642. doi: 10.1111/j.1461-0248.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. Sex Versus Non-Sex Versus Parasite. Oikos. 1980;35:282–290. [Google Scholar]
- Jaenike J. An hypothesis to account for the maintenance of sex within populations. Evol Theory. 1978;3:191–194. [Google Scholar]
- Jokela J, Dybdahl MF, Lively CM. The maintenance of sex, clonal dynamics, and host-parasite coevolution in a mixed population of sexual and asexual snails. Am Nat. 2009;174(Suppl 1):S43–53. doi: 10.1086/599080. [DOI] [PubMed] [Google Scholar]
- Katju V, LaBeau EM, Lipinski KJ, Bergthorsson U. Sex change by gene conversion in a Caenorhabditis elegans fog-2 mutant. Genetics. 2008;180:669–672. doi: 10.1534/genetics.108.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstes NAG, Berenos C, Schmid-Hempel P, Wegner KM. Antagonistic experimental coevolution with a parasite increases host recombination frequency. BMC Evol Biol. 2012;12:18. doi: 10.1186/1471-2148-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KC, Delph LF, Jokela J, Lively CM. The Geographic Mosaic of Sex and the Red Queen. Curr Biol. 2009;19:1438–1441. doi: 10.1016/j.cub.2009.06.062. [DOI] [PubMed] [Google Scholar]
- Lande R, Schemske DW. The evolution of self-fertilization and inbreeding depression in plants.1. genetic models. Evolution. 1985;39:24–40. doi: 10.1111/j.1558-5646.1985.tb04077.x. [DOI] [PubMed] [Google Scholar]
- Lindman HR. Analysis of Variance in Complex Experimental Designs. W.H. Freeman & Co; San Francisco: 1974. [Google Scholar]
- Lloyd DG. Benefits and handicaps of sexual reproduction. Evol Biol. 1980;13:69–106. [Google Scholar]
- Lopes PC, Sucena E, Santos ME, Magalhaes S. Rapid experimental evolution of pesticide resistance in C. elegans entails no costs and affects the mating system. PLoS ONE. 2008;3:e3741. doi: 10.1371/journal.pone.0003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoel D, Carvalho S, Phillips PC, Teotonio H. Selection against males in Caenorhabditis elegans under two mutational treatments. Proc R Soc B. 2007;274:417–424. doi: 10.1098/rspb.2006.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. The Evolution of Sex. Cambridge University Press; Cambridge, UK: 1978. [Google Scholar]
- Morran LT, Ohdera AH, Phillips PC. Purging deleterious mutations under self fertilization: paradoxical recovery in fitness with increasing mutation rate in Caenorhabditis elegans. PLoS One. 2010;5:e14473. doi: 10.1371/journal.pone.0014473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran LT, Parmenter MD, Phillips PC. Mutation load and rapid adaptation favor outcrossing over self-fertilization. Nature. 2009;462:350–352. doi: 10.1038/nature08496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran LT, Schmidt OG, Gelarden IA, Parrish RC, II, Lively CM. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science. 2011;333:216–218. doi: 10.1126/science.1206360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RL, Cutter AD. Experimental evolution of sperm count in protandrous self-fertilizing hermaphrodites. J Exp Biol. 2011;214:1740–1747. doi: 10.1242/jeb.053181. [DOI] [PubMed] [Google Scholar]
- Nordborg M. Structured coalescent processes on different time scales. Genetics. 1997;146:1501–1514. doi: 10.1093/genetics/146.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrow D, Phillips N, Avalos A, Blanton D, Boggs A, Keller T, Levy L, Rosenbloom J, Baer CF. Mutational bias for body size in Rhabditid nematodes. Genetics. 2007;176:1653–1661. doi: 10.1534/genetics.107.074666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AD, Lively CM. The red queen and fluctuating epistasis: a population genetic analysis of antagonistic coevolution. Am Nat. 1999;154:393–405. doi: 10.1086/303247. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T, Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics. 1988;119:43–61. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H, Ewbank JJ. Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol Biol. 2004;4:49. doi: 10.1186/1471-2148-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc Natl Acad Sci U S A. 2010;107:7359–7364. doi: 10.1073/pnas.1003113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasundar A, Hey J. Sampling from natural populations with RNAi reveals high outcrossing and population structure in Caenorhabditis elegans. Curr Biol. 2005;15:1598–1602. doi: 10.1016/j.cub.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Self-fertilization and population variation in higher plants. Am Nat. 1957;91:337–354. [Google Scholar]
- Stewart AD, Phillips PC. Selection and maintenance of androdioecy in Caenorhabditis elegans. Genetics. 2002;160:975–982. doi: 10.1093/genetics/160.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotonio H, Carvalho S, Manoel D, Roque M, Chelo IM. Evolution of outcrossing in experimental populations of Caenorhabditis elegans. PLoS One. 2012;7:e35811. doi: 10.1371/journal.pone.0035811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. A new evolutionary law. Evol Theory. 1973;1:1–30. [Google Scholar]
- Vassilieva LL, Hook AM, Lynch M. The fitness effects of spontaneous mutations in Caenorhabditis elegans. Evolution. 2000;54:1234–1246. doi: 10.1111/j.0014-3820.2000.tb00557.x. [DOI] [PubMed] [Google Scholar]
- Vassilieva LL, Lynch M. The rate of spontaneous mutation for life-history traits in Caenorhabditis elegans. Genetics. 1999;151:119–129. doi: 10.1093/genetics/151.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegewitz V, Schulenburg H, Streit A. Experimental insight into the proximate causes of male persistence variation among two strains of the androdioecious Caenorhabditis elegans (Nematoda) BMC Ecol. 2008;8:12. doi: 10.1186/1472-6785-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegewitz V, Schulenburg H, Streit A. Do males facilitate the spread of novel phenotypes within populations of the androdioecious nematode Caenorhabditis elegans? J Nematol. 2009;41:247–254. [PMC free article] [PubMed] [Google Scholar]
- Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]