Abstract
Here, we studied the effects of cytochrome P450 (CYP)3A deficiency on the mRNA expression of genes encoding regulators of hepatic cholesterol levels using Cyp3a-knockout (Cyp3a−/−) mice. The mRNA expression levels of genes encoding enzymes involved in cholesterol biosynthesis in the livers of Cyp3a−/− mice were higher than those of wild-type (WT) mice. Nuclear levels of sterol regulatory element-binding protein-2 (SREBP-2), which enhances cholesterol biosynthesis, were also higher in the livers of Cyp3a−/− mice. Binding of SREBP-2 to the Hmgcs1 gene promoter was more abundant in the livers of Cyp3a−/− mice. These results suggest that deficiency of CYP3A enzymes enhances transcription of genes encoding enzymes involved in cholesterol biosynthesis via activation of SREBP-2. On the other hand, hepatic cholesterol levels in Cyp3a−/− mice were 20% lower than those in WT mice. The mRNA expression levels of genes encoding enzymes involved in bile acid synthesis, plasma levels of 7α-hydroxy-4-cholesten-3-one and hepatic levels of total bile acid were significantly higher in Cyp3a−/− mice than in WT mice. These findings suggest that reduction of hepatic total cholesterol in Cyp3a−/− mice would be the consequence of enhanced bile acid synthesis. Therefore, CYP3A enzymes appear to play roles in the synthesis of cholesterol and bile acid in vivo.
Keywords: 25-hydroxy-cholesterol, HMG-CoA synthase 1, squalene epoxidase, sterol regulatory element-binding protein-2, cytochrome P450 3A
Cholesterol has multifunctional roles as a component of cellular membranes and a precursor of biological molecules such as steroid hormones, oxysterols, vitamins, and bile acids in humans and animals (1–4). The endogenous synthesis of cholesterol is catalyzed by many enzymes including HMG-CoA synthase 1 (HMGCS1), HMG-CoA reductase (HMGCR), and squalene epoxidase (SQLE). When cellular sterol levels are low, sterol regulatory element-binding protein-2 (SREBP-2) is processed by proteases and translocates into the nucleus. Nuclear SREBP-2 activates the transcription of genes encoding enzymes involved in the biosynthesis of cholesterol (5). Thus, SREBP-2 is considered as a master regulator of cholesterol biosynthesis (6).
Hepatic cholesterol levels are regulated by elaborate networks such as influx, efflux, and conversion into bile acids in addition to biosynthesis. Dietary cholesterol is absorbed from the jejunum and transported to the liver (7). A part of the cholesterol in the liver is enclosed in lipoprotein and excreted into blood circulation. Hepatic cholesterol is also secreted into the duodenum via the biliary canaliculus. Most of the biliary cholesterol is reabsorbed in the jejunum with dietary cholesterol and transported back to the liver (7). Such behavior of cholesterol is regulated by many transporters. For example, Niemann-Pick C1-like 1 (NPC1L1) imports cholesterol from the lumen into enterocytes of the intestine (8). At the basolateral membrane of the intestine, ATP-binding cassette (ABC)A1 transports cholesterol into the bloodstream. In the liver, ABCA1 also functions at the basolateral surface to transport cholesterol into the bloodstream (9). ABCG5 and ABCG8, form a functional heterodimer complex that mediates the secretion of cholesterol from hepatocytes into the biliary canaliculus and also the apical efflux of cholesterol from enterocytes into the lumen of the intestine (10, 11). Hepatic low density lipoprotein receptor (LDLR) is responsible for uptake of LDL cholesterol from blood circulation into hepatocytes (12). On the other hand, a part of the cholesterol in the liver is metabolized to bile acids through many steps catalyzed by enzymes such as cytochrome P450 (CYP)7A1 and CYP8B1 (13). Bile acids are excreted into the biliary canaliculus with cholesterol and then assist the absorption of biliary and dietary cholesterol in the intestine.
Members of the CYP3A subfamily are the most abundant CYP enzymes in the liver and small intestine. CYP3A enzymes metabolize over 50% of drugs in clinical use and are therefore closely involved in pharmacokinetics, drug interactions, drug efficacy, and drug toxicity (14). CYP3A enzymes metabolize not only xenobiotics but also many endogenous compounds such as cholesterol, bile acids, steroid hormones, and vitamins. The extremely wide substrate specificity of CYP3A enzymes has resulted in versatile physiological functions of CYP3A (15–18). As for cholesterol, it has been reported that human CYP3A4 mediates the conversion of cholesterol into 4β-hydroxy-cholesterol, an endogenous oxysterol (19, 20). 4β-Hydroxy-cholesterol is expected to be useful as an endogenous marker of CYP3A because it is found in human circulation (18). In addition, a recent study demonstrated that CYP3A was involved in 25-hydroxylation of cholesterol (21). 25-Hydroxy-cholesterol acts as not only a precursor of bile acids but also a physiologically active oxysterol that suppresses SREBP-2 and downregulates cholesterol biosynthesis (22–24). However, the roles of cholesterol metabolites formed by CYP3A in biosynthesis of cholesterol have not been fully elucidated, particularly in in vivo experiments.
The Cyp3a-knockout (KO) (Cyp3a−/−) mouse is a valuable research tool in studies on drug metabolism and drug-induced toxicity in vivo (25, 26). However, the physiological role of CYP3A has not been clearly elucidated in in vivo experiments. To reveal the role of CYP3A in cholesterol homeostasis, we investigated the effects of CYP3A deficiency on expression levels of genes responsible for influx, efflux, metabolism, and biosynthesis of cholesterol using Cyp3a−/− mice.
MATERIALS AND METHODS
Chemicals
7α-Hydroxy-4-cholesten-3-one (C4) was purchased from Santa Cruz (Santa Cruz, CA). 25-Hydroxy-cholesterol was purchased from Steraloids (Newport, RI). Lathosterol was purchased from Sigma (St. Louis, MO). Acetonitrile, chloroform, hexane, and methanol were HPLC grade (Wako, Tokyo, Japan).
Animals
C57BL/6JJcl mice (CLEA, Tokyo, Japan) were used as wild-type (WT) mice. Recently, we generated Cyp3a−/− mice by targeted deletion of endogenous Cyp3a genes (27). Briefly, Cyp3a13−/− lines and Cyp3a57-59−/− lines were generated by conventional gene targeting and Cre-loxP-mediated chromosomal deletion, respectively, and the two lines were mated to generate Cyp3a−/− mice lacking the mouse Cyp3a gene cluster. The distance between the Cyp3a57-59 gene cluster and a Cyp3a13−/− gene is ∼5 Mb and several genes are located between the loci, so the two KO lines were generated independently. It has been confirmed that the six genes on the mouse Cyp3a cluster (Cyp3a13/57/44/11/25/59) were expressed in the livers and intestines of WT mice but not in those of Cyp3a−/− mice (27). Fetal specific Cyp3a16 and female specific Cyp3a41 were not detected at mRNA levels because only adult male mice were used in this study (data not shown). In addition, α- and 4-hydroxylation activities of triazolam, which are catalyzed by mouse CYP3A (28) in the liver microsomes of Cyp3a−/− mice, were less than 7% of those in WT mice (supplementary Fig. I). These findings suggest the complete deletion of CYP3A functions in Cyp3a−/− mice used in the present study. The control genetic background is almost the same as Cyp3a−/− mice, because Cyp3a−/− mice were backcrossed to C57BL/6JJcl mice at least four times. All experiments were done using male mice between 10 and 11 weeks of age. Animals were kept in a temperature-controlled environment with a 12 h light/dark cycle. The light-cycle hours were between 7:00 AM and 7:00 PM. The mice received a standard diet containing 0.1% cholesterol (CE-2, CLEA, Tokyo, Japan, supplementary Table II). Animals were sacrificed between 10:00 AM and 11:00 AM without fast. The present study was conducted in accordance with the guidelines for the Care and Use of Laboratory Animals, as adopted by the Committee on Animal Research of Tottori University and Chiba University.
RNA isolation and cDNA synthesis
Total RNA was isolated from tissue pieces of livers and small intestines. Because most of cholesterol is reabsorbed in the jejunum (7), the upper parts of small intestines (1 cm segments collected at 10 cm from the pyloric region) were used. Total RNA was extracted using ISOGEN (Nippon Gene, Tokyo, Japan) and purified using RNeasy columns (Qiagen, Hilden, Germany) in accordance with the manufacturer's instructions, and then treated with RNase-free DNase I (Wako Pure Chemicals, Osaka, Japan). The cDNA was generated with a random hexamer by using a high-capacity cDNA reverse transcription kit (Applied Biosytems, Foster, CA).
Analysis of mRNA levels
Expression levels were measured by quantitative real-time PCR carried out on an Eco real-time PCR system (Illumina, Hayward, CA). Expression levels of HMGCS1, HMGCR, SQLE, sterol regulatory element-binding factor 2 (SREBF2), ABCG5, ABCG8, ABCA1, LDLR, NPC1L1, CYP7A1, CYP8B1, CYP27A1, CYP7B1, and small heterodimer partner (SHP) were measured by SYBR Green-based real-time PCR using KAPA SYBR Fast ABI Prism 1000 (KAPA Biosystems, Woburn, MA). Specific primers for real-time PCR are presented in supplementary Table II. The expression levels of mRNA were normalized with the expression level of mouse GAPDH as an endogenous control gene.
Preparation of nuclear extracts
Nuclear and cytosol fractions were prepared from liver tissues. Aliquots of frozen liver (about 100 mg) were homogenized in 1 ml of lysis buffer (10 mM HEPES adjusted to pH 7.9, 1.5 mM MgCl2, and 10 mM KCl) supplemented with protease inhibitor cocktail set III (Calbiochem, La Jolla, CA) and 1 mM DTT. The liver homogenate was centrifuged at 10,000 g for 5 min at 4°C. The supernatant (cytosol fraction) was removed and stored at −80°C. The nuclear pellet was resuspended in 0.15 ml extraction buffer (20 mM HEPES adjusted to pH 7.9, 2.5% glycerol, 0.42 M NaCl, and 1.5 mM MgCl2) supplemented with 1 mM DTT and protease inhibitor cocktail set III. The suspension was rotated at 4°C for 30 min and centrifuged at 20,000 g for 15 min at 4°C. The supernatant from this spin was designated as the nuclear extract and stored at −80°C until use.
Western blot analysis
Nuclear extracts (30 μg/lane) were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. After 1 h incubation with blocking buffer (0.05% Tween 20 in PBS containing 3% skim milk), the membrane was probed for 1 h at room temperature using a polyclonal anti-SREBP-2 rabbit antibody (1:500 dilution, Ab2848; Abcam, Cambridge, UK). A monoclonal anti-TATA-binding protein (TBP) mouse antibody (1:2,000 dilution, Ab818; Abcam) was used as an internal control. After washing, the membrane was incubated with a peroxidase-conjugated secondary antibody for 1 h at room temperature. The secondary antibodies used were anti-rabbit IgG (1:100,000 dilution; Sigma-Aldrich, St. Louis, MO) and anti-mouse IgG (1:5,000 dilution; Sigma-Aldrich) for anti-SREBP-2 and anti-TBP antibodies, respectively. The primary and secondary antibodies were diluted with 0.05% Tween 20 in PBS containing 3% BSA. Protein bands were visualized by using ECL Western blotting detection reagents (GE Healthcare, Buckingham, UK), ImmunoStar LD (Wako), and LAS-1000 plus (Fujifilm, Tokyo, Japan).
Chromatin immunoprecipitation assay
Liver tissue (100 mg) of WT and Cyp3a−/− mice was minced and formaldehyde was added to a final concentration of 1%. After rocking at room temperature for 15 min, glycine was added to a final concentration of 0.125 M to stop the cross-linking. The tissues were collected by centrifugation and were homogenized by a dounce homogenizer in cold cell lysis buffer (5 mM PIPES, pH 8.0, 85 mM KCl, 0.5% Nonidet P-40) supplemented with proteinase inhibitor cocktail set III (Calbiochem). Nuclei pellets were collected by centrifugation at 6,200 g for 10 min at 4°C and resuspended in 400 μl of nuclei lysis buffer (50 mM Tris-HCl, pH 8.1, 10 mM EDTA, 1% SDS) supplemented with proteinase inhibitor cocktail set III (Calbiochem). Nuclei were sonicated before immunoprecipitation with polyclonal anti-SREBP-2 rabbit antibody (ab28482; Abcam) or normal IgG (sc2027; Santa Cruz, CA) overnight at 4°C. DNA in the precipitated samples was reverse cross-linked at 65°C for 6 h, and the DNA was recovered by phenol/chloroform extraction and ethanol precipitation. DNA samples were measured by semi-quantitative PCR with GoTaq® Green Master Mix (Promega, Madison, WI) and quantitative PCR using SYBR Green based real-time PCR with KAPA SYBR Fast ABI Prism 1000 (KAPA Biosystems, Woburn, MA). PCR was carried out with Hmgcs1-specific primers encompassing the sterol regulatory element (SRE) site in the 5′-flanking region (−348 to −208 bp) and a region in intron 5 (+11,862 to +11,948 bp) as a negative control. The PCR conditions were one cycle of predenaturation at 95°C for 10 min, 45 cycles of denaturation at 95°C for 15 s, annealing at 58°C for 30 s, and elongation at 72°C for 15 s The primers used are as follows: Hmgcs1/SRE, 5′-ATTGGTCGGAGAACCTCTC-3′ and 5′-AGGGGTGGGAACAAAGTCC-3′ Hmgcs1/negative control, 5′-ATTAGACAGGATTTGGGGTCACT-3′ and 5′- GTCTACACTGCAAAGAGAGCAGG-3′.
Cholesterol analysis
Hepatic lipids were extracted according to the procedure of Bligh and Dyer (29) with slight modification. Briefly, frozen liver tissues (about 5 mg) were homogenized and extracted with a mixture of chloroform/isopropanol/Triton X-100 (7:11:0.1, v/v/v). The organic layer was subjected to cholesterol determination after evaporation and resuspension in methanol. Total cholesterol in liver and plasma was determined by use of a cholesterol E-test kit (Wako) according to the manufacturer's instructions.
Analysis of plasma lathosterol
Concentration of lathosterol in plasma was determined by an HPLC method. To extract lipids, plasma (0.1 ml) was diluted with 0.1 ml of distilled water and vigorously mixed with 0.8 ml of hexane for 1 min, and then centrifuged at 2,000 g for 5 min. The supernatant (hexane layer) was transferred to a new tube and further extracted once with 0.2 ml of water. Separately the water layer was further extracted twice with 0.8 ml of hexane. The hexane extracts were combined and dried under vacuum. The residue was reconstituted with 0.1 ml of methanol, and 50 μl of reconstituted sample was injected onto a Hitachi L-7000 HPLC system (Tokyo, Japan) and Poroshell120 EC-C18 (3.0 × 100 mm, 2.7 μm) column (Agilent Technology, Santa Clara, CA). The mobile phase consisted of acetonitrile/methanol (8:2, v/v) at a flow rate of 0.6 ml/min. The eluent was monitored at a wavelength of 235 nm.
Analysis of 25-hydroxy-cholesterol in the liver
Concentration of 25-hydroxy-cholesterol in the liver was determined by methods described elsewhere (21) with slight modifications. In brief, liver tissues (50 mg) were homogenized in 0.1 M potassium phosphate buffer (pH 7.4) containing 4 mM MgCl2, 1 mM EDTA, and 30 mM nicotinamide. The homogenates were incubated with 1 N ethanolic potassium hydroxide, extracted with hexane, and derivatized to picolinyl esters. The samples were analyzed by HPLC-MS/MS using Triple TOF 5600 System (AB SCIEX, Framingham, MA).
Analyses of total bile acids and their intermediary product C4
Bile acid levels in the liver were determined following bile acid extraction of liver tissues. Briefly, liver tissues (200 mg) were homogenized in 1 ml of ethanol and heated at 85°C for 1 min and then centrifuged at 1,000 g for 5 min. After the supernatant was isolated, the precipitate was extracted twice with 1 ml of ethanol and the combined extracts were dried under vacuum. The residue was resuspended in 0.2 ml of 75% methanol and centrifuged at 14,000 g for 10 min, filtered using a filter unit (0.45 μm; Millipore, Billerica, MA), and dried under vacuum. The residue was reconstituted with 10 μl of 75% methanol. After dilution with distilled water, the sample was analyzed by use of a total bile-test kit (Wako, Tokyo, Japan) according to the manufacturer's instructions.
The size of the bile acid pool was expressed as the sum of total bile acid content in the liver, small intestine, and gallbladder per 100 g body weight. Bile acid content in the small intestine was determined after extraction of small intestine tissues (200 mg) as described for bile acid content in the liver. For the gallbladder, bile drawn from the gallbladder was diluted with distilled water (2 ml/mg tissue of empty gallbladder) and analyzed as described above.
Concentration of C4, an intermediary product in the synthesis of bile acids, in plasma was determined by a HPLC method of Lenicek et al. (30) with slight modifications. Plasma (0.2 ml) was vigorously mixed with 2.5 ml of chloroform/methanol (2:1, v/v), and centrifuged at 2,000 g for 3 min. After the upper phase was discarded, the lower phase was mixed with 1 ml of 125 mM NaCl in 50% methanol, and centrifuged as above. The lower phase was dried under vacuum. The residue was reconstituted with 0.2 ml of acetonitrile. Eighty microliters of reconstituted sample was injected onto a Hitachi L-7000 HPLC system (Tokyo, Japan) and a CAPCELL PAK C18 UG120 column (4.6 mm × 250 mm, 5 μm; Shiseido, Tokyo, Japan). The mobile phase consisted of acetonitrile/water (92:8, v/v) at a flow rate of 1.0 ml/min. The eluent was monitored at a wavelength of 241 nm.
Statistical analysis
The number of mice in each group is described in the figure and table legends. Data are shown as means ± SEM or SD. Statistical analyses were performed by using Statcel (OMS, Saitama, Japan). Comparison was made with Student's t-test or Welche's t-test. P < 0.05 was considered statistically significant.
RESULTS
Effects of CYP3A deficiency on expression levels of enzymes involved in cholesterol biosynthesis
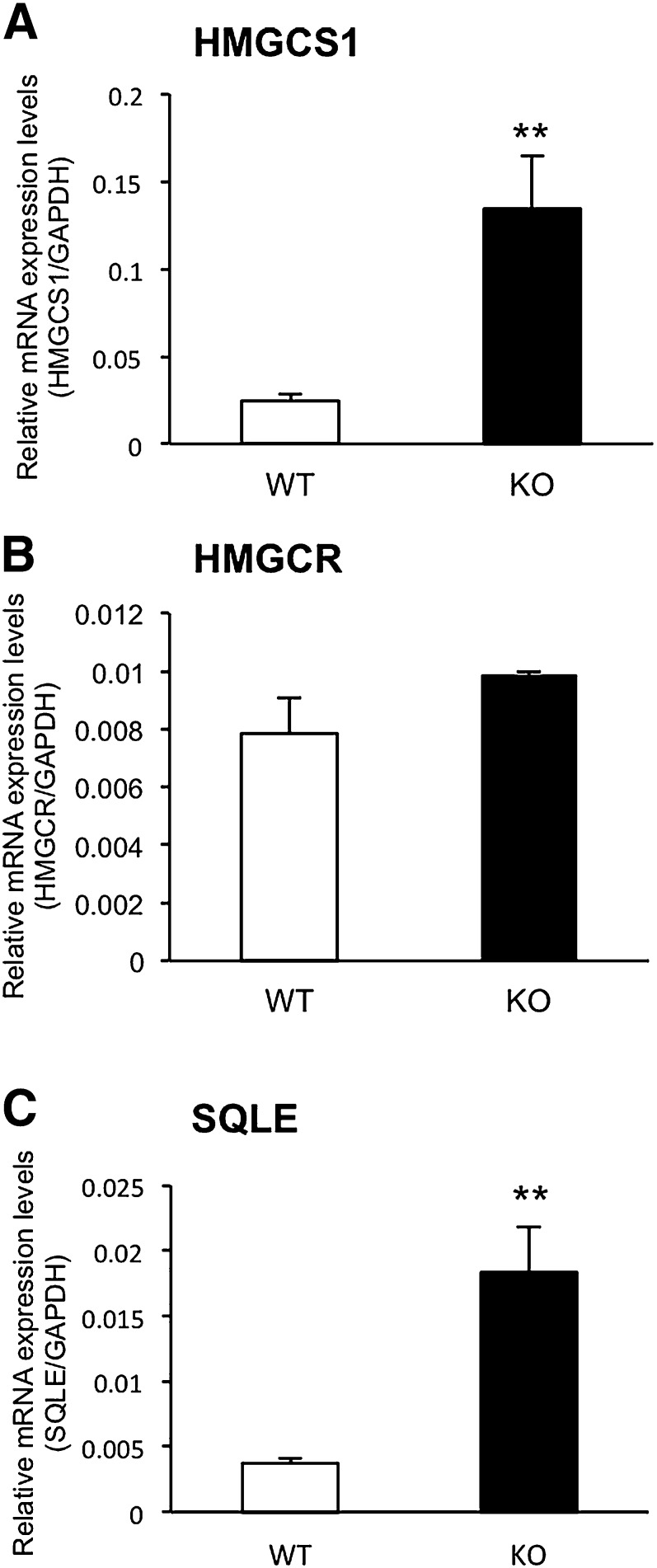
We measured the mRNA levels of HMGCS1, HMGCR, and SQLE in the livers of WT and Cyp3a−/− mice in order to investigate the effects of CYP3A deficiency on mRNA expression of enzymes involved in cholesterol biosynthesis. As shown in Fig. 1, expression levels of HMGCS1 and SQLE mRNAs in the livers of Cyp3a−/− mice were significantly higher than those of WT mice (5.4- and 4.9-fold, respectively). Expression levels of HMGCR mRNAs in the livers of Cyp3a−/− mice showed a trend toward higher levels than those of WT mice, although the differences were not significant. Expression levels of HMGCS1 and SQLE mRNAs in the small intestine were similar between WT and Cyp3a−/− mice. Expression levels of HMGCR mRNAs in the small intestines of Cyp3a−/− mice were 1.4-fold higher than those of WT mice (supplementary Fig. II).
Fig. 1.
Expression levels of HMGCS1 (A), HMGCR (B), and SQLE (C) mRNAs in the livers of WT and Cyp3a−/− (KO) mice. Expression levels were normalized by expression levels of GAPDH and are shown as means ± SEM of six mice in each group. cDNAs prepared from each mouse were used for triplicate determination. **P < 0.01 versus WT mice.
Effects of CYP3A deficiency on expression of SREBP-2
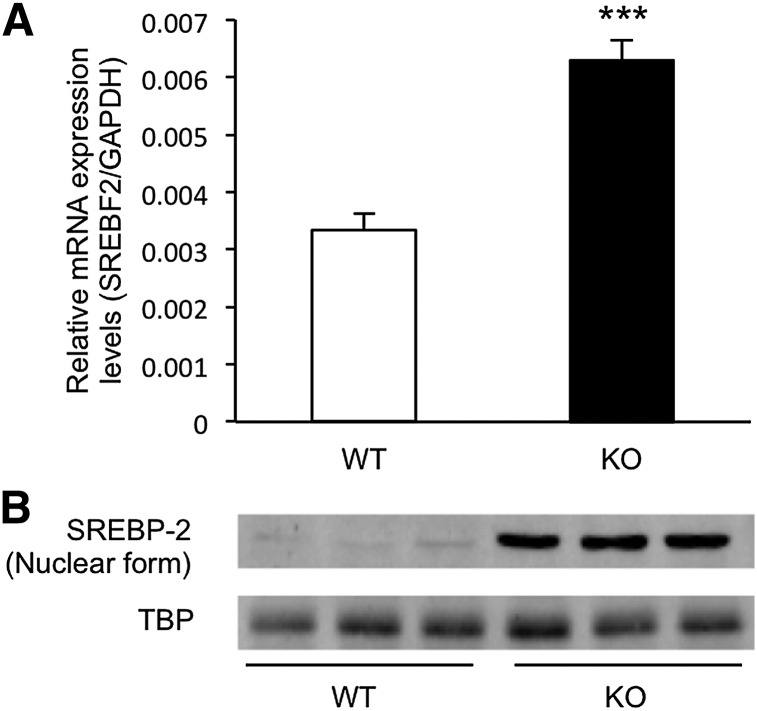
The Srebf2 gene encoding SREBP-2 is also known as a target gene of SREBP-2 (6). As shown in Fig. 2A, expression levels of SREBF2 mRNA in the livers of Cyp3a−/− mice were 1.9-fold higher than those of WT mice (P < 0.001). In the small intestine, expression levels of SREBF2 mRNAs were similar between WT and Cyp3a−/− mice (supplementary Fig. II). Because mRNA expression levels of HMGCS1, SQLE, and SREBF2 in the livers of Cyp3a−/− mice were significantly higher than those of WT mice, we determined nuclear levels of SREBP-2 protein in the livers of WT and Cyp3a−/− mice. As shown in Fig. 2B, nuclear levels of SREBP-2 protein in the livers of Cyp3a−/− mice were markedly higher than those in WT mice.
Fig. 2.
Expression levels of SREBP-2 in the livers of WT and KO mice. A: Expression levels of SREBF2 mRNAs were normalized by expression levels of GAPDH and are shown as means ± SEM of six mice in each group. cDNAs prepared from each mouse were used for triplicate determination. ***P < 0.001 versus WT mice. B: SREBP-2 protein was assessed using Western blotting of isolated hepatic nuclear protein. TBP was used as a loading control.
Effects of CYP3A deficiency on SREBP-2 binding to the SRE motif
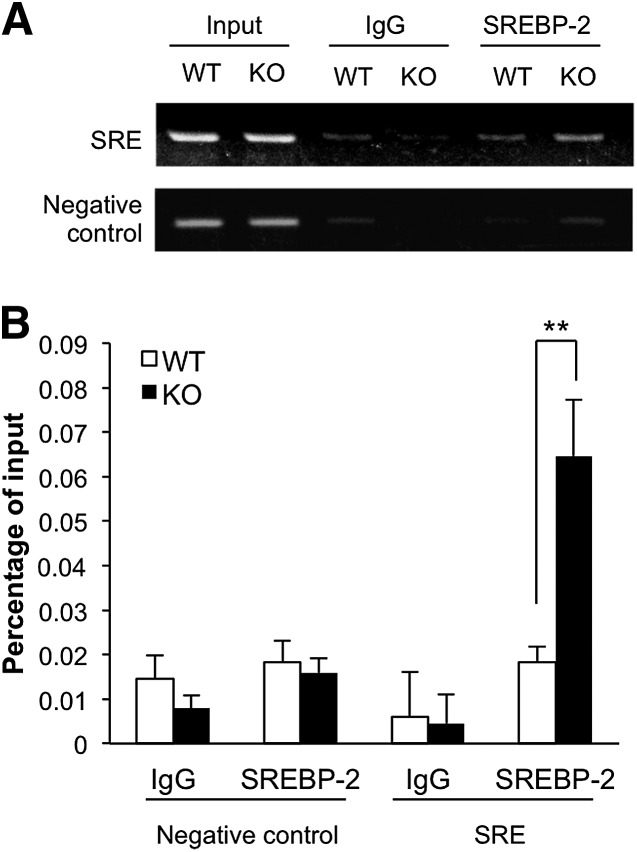
To verify whether CYP3A deficiency affected binding of SREBP-2 to an SRE motif in a genomic context, chromatin immunoprecipitation (ChIP) assays were performed with livers of WT and Cyp3a−/− mice. The Hmgcs1 promoter contains a known SRE motif (31). The specific binding of SREBP-2 to the SRE-containing region (−348 to −208 bp) was significantly more abundant in the livers of Cyp3a−/− mice than in the livers of WT mice (P < 0.01, Fig. 3). The degree of binding of SREBP-2 to the negative control region was low and showed no difference between WT and Cyp3a−/− mice.
Fig. 3.
SREBP-2 binding to Hmgcs1 promoter (−348 to −208 bp) in the livers of WT and KO mice. ChIP assay was performed using anti-SREBP-2 antibody. Liver tissues from two mice in each group were pooled and subjected to the assay. The immunoprecipitated DNA, along with the DNA isolated before immunoprecipitation (Input), were analyzed by semi-quantitative PCR (A) and quantitative PCR (B). Signals were normalized to input DNA. Data are means ± SD of three independent determinations, each performed in duplicate. **P < 0.01 versus WT mice.
Effects of CYP3A deficiency on cholesterol biosynthesis
Because transcription of genes encoding enzymes involved in cholesterol biosynthesis is enhanced in Cyp3a−/− mice, we examined total cholesterol levels in the plasma and livers, lathosterol levels in the plasma, and 25-hydroxy-cholesterol levels in the livers of WT and Cyp3a−/− mice. As shown in Table 1, body weight and liver weight of Cyp3a−/− mice were higher than those of WT mice. Total cholesterol levels in the livers of Cyp3a−/− mice were about 20% lower than those in the livers of WT mice (P < 0.05), although no significant difference was found in plasma total cholesterol levels between WT and Cyp3a−/− mice. Plasma lathosterol levels, as an indicator of cholesterol biosynthesis (32), showed a trend toward higher than those of WT mice, although the difference was not significant. 25-Hydroxy-cholesterol levels in the livers of Cyp3a−/− mice were significantly lower than those in the livers of WT mice (P < 0.05).
TABLE 1.
Parameters of cholesterol metabolism in WT and Cyp3a−/− mice
| Genotype |
||
| Parameter | WT | Cyp3a−/− |
| Body weight (g) | 23.7 ± 0.3 | 27.7 ± 1.1a |
| Liver weight (g) | 1.0 ± 0.03 | 1.5 ± 0.07b |
| Plasma lathosterol (ng/ml) | 32.1 ± 2.0 | 41.3 ± 7.9 |
| Plasma total cholesterol (mg/dl) | 63.4 ± 9.8 | 82.6 ± 7.5 |
| Liver total cholesterol (mg/g) | 2.8 ± 0.14 | 2.2 ± 0.03c |
| Liver 25-hydroxy-cholesterol (ng/g) | 20.0 ± 1.4 | 14.9 ± 1.3c |
Different groups of animals were used for measurements of plasma lathosterol, plasma cholesterol, liver cholesterol, and liver 25-hydroxy-cholesterol. Data for body and liver weight represent the mean ± SEM of nine mice in each group. Data for cholesterol and 25-hydroxy-cholesterol levels are shown as means ± SEM of three and six mice in each group, respectively. Data for lathosterol levels are expressed as a mean ± SD of three independent determinations, using pooled plasma from six mice.
P < 0.01 versus WT mice.
P < 0.001 versus WT mice.
P < 0.05 versus WT mice.
Effects of CYP3A deficiency on expression levels of cholesterol transporters
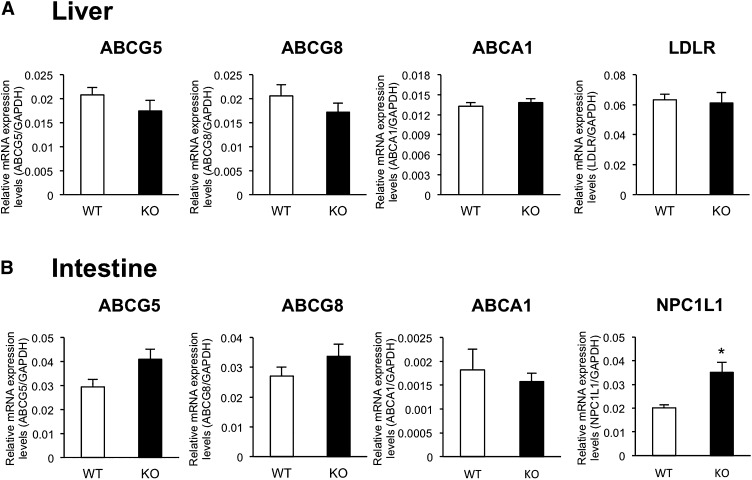
To investigate the mechanism of decreased hepatic total cholesterol in Cyp3a−/− mice, we determined the mRNA expression levels of genes involved in the influx and efflux of cholesterol. As shown in Fig. 4A, no significant differences in expression levels of ABCG5, ABCG8, ABCA1, and LDLR mRNAs in the liver were found between WT and Cyp3a−/− mice. As shown in Fig. 4B, expression levels of ABCG5, ABCG8, and ABCA1 mRNAs in the small intestine were also similar between WT and Cyp3a−/− mice. Expression levels of NPC1L1 mRNA in the small intestines of Cyp3a−/− mice were 1.7-fold higher than those of WT mice.
Fig. 4.
Expression levels of ABCG5, ABCG8, ABCA1, LDLR, and NPC1L1 mRNAs in the livers (A) and intestines (B) of WT and KO mice. Expression levels were normalized by expression levels of GAPDH and are shown as means ± SEM of six mice in each group. cDNAs prepared from each mouse were used for triplicate determination. *P < 0.05 versus WT mice.
Effects of CYP3A deficiency on bile acid biosynthesis
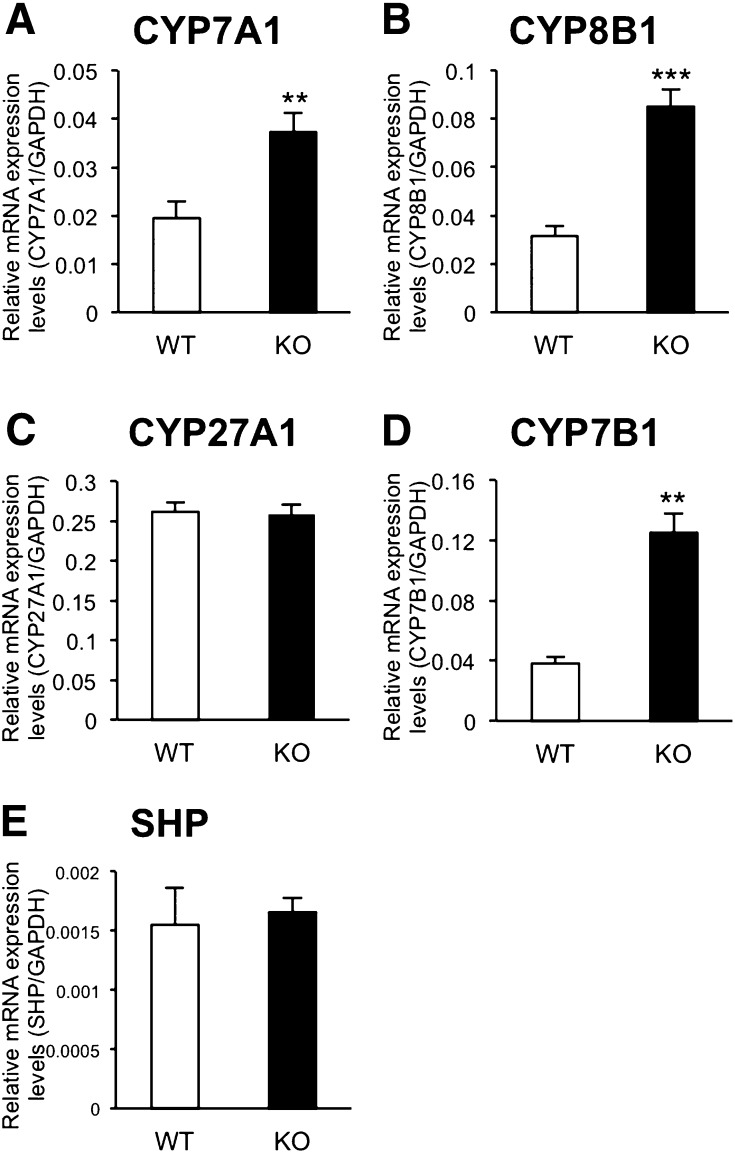
Synthesis and secretion of bile acids constitute a major route for elimination of cholesterol from the liver (13). To try to determine the mechanism of reduction of hepatic cholesterol in Cyp3a−/− mice, we investigated expression levels of four enzymes involved in bile acid biosynthesis. The classical pathway of bile acid synthesis is initiated by 7α-hydroxylation of cholesterol catalyzed by CYP7A1, which is a rate-limiting enzyme of this pathway (13). The next product, C4, is metabolized to 7α,12α-dihydroxy-4-cholesten-3-one by CYP8B1. Then the resulting product is converted ultimately into cholic acid (13). An alternative pathway of bile acid synthesis is initiated by 27-hydroxylation of cholesterol catalyzed by CYP27A1 (13). Subsequently, 27-hydroxy-cholesterol is metabolized to 7α,27-dihydroxy-cholesterol by CYP7B1 (13) and 3β-hydroxy-5-cholestenoic acid by CYP27A1 (13). As shown in Fig. 5, the expression levels of CYP7A1, CYP8B1, and CYP7B1 mRNAs were significantly higher in the livers of Cyp3a−/− mice than those of WT mice (1.9-, 2.7-, and 3.3-fold, respectively). Expression levels of CYP27A1 and SHP mRNAs in the liver were similar between WT and Cyp3a−/− mice.
Fig. 5.
Expression levels of CYP7A1 (A), CYP8B1 (B), CYP27A1 (C), CYP7B1 (D), and SHP (E) mRNAs in the livers of WT and KO mice. Expression levels were normalized by expression levels of GAPDH and are shown as means ± SEM of six mice in each group. cDNAs prepared from each mouse were used for triplicate determination. **P < 0.01 and ***P < 0.001 versus WT mice.
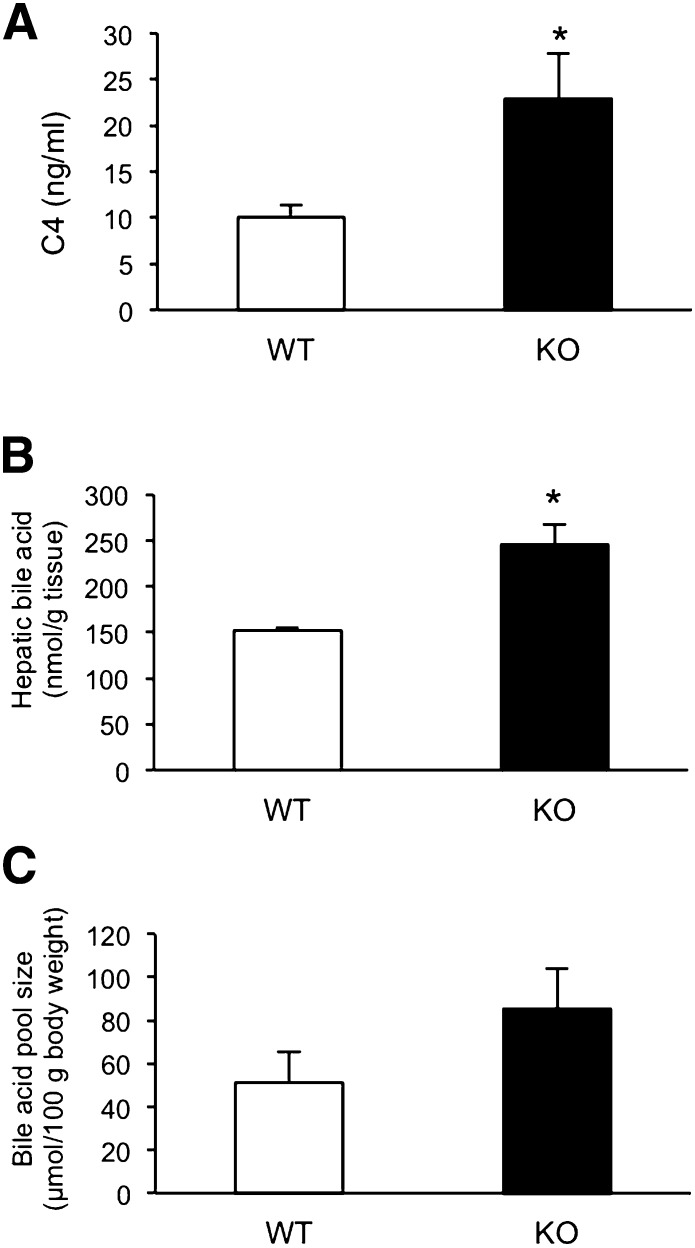
Because the expression levels of CYP7A1, CYP8B1, and CYP7B1 were higher in the livers of Cyp3a−/− mice compared with WT mice, we studied whether bile acid synthesis was enhanced in Cyp3a−/− mice. As shown in Fig. 6A, plasma concentration of C4 was 1.6-fold higher in Cyp3a−/− mice than in WT mice (P < 0.05). As shown in Fig. 6B, total bile acid levels in the livers of Cyp3a−/− mice were also significantly higher than those of WT mice (P < 0.05). Bile acid pool size of Cyp3a−/− mice showed a trend toward higher levels than those of WT mice, although differences were not significant (Fig. 6C).
Fig. 6.
Concentration of C4 in plasma (A), hepatic bile acid (B), and bile acid pool size (C) of WT and KO mice. Plasma C4 concentrations are expressed as a mean ± SD of triplicate determinations, using pooled plasma from six mice. Hepatic bile acid levels were expressed as means ± SEM of three mice in each group. Bile acid pool size is determined as total bile acid from liver, gallbladder, and intestine. The values of bile acid pool size were shown as means ± SEM of three mice in each group. *P < 0.05 versus WT mice.
Effects of CYP3A deficiency on expression of target genes of pregnane X receptor
Because activated pregnane X receptor (PXR) could suppress transcription of the mouse Cyp7a1 gene (33, 34), we measured the mRNA expression levels of target genes of PXR in the livers of WT and Cyp3a−/− mice. The mRNA expression levels of Mdr1a in the livers of Cyp3a−/− mice were lower than those of WT mice, whereas expression levels of OATP2, SULT2A1, CYP2C55, and CYP2B10 mRNAs in the livers of Cyp3a−/− mice were higher than those of WT mice (supplementary Fig. III).
DISCUSSION
In this study, we showed for the first time that mRNA expression levels of enzymes (i.e., HMGCS1 and SQLE) involved in cholesterol biosynthesis were higher in the livers of Cyp3a−/− mice than in the livers of WT mice (Fig. 1). We also found that expression levels of SREBP-2, which activates the transcription of these genes (5, 6), were increased in the livers of Cyp3a−/− mice compared with those in the livers of WT mice at the levels of mRNA and nuclear protein (Fig. 2). In addition, results of the ChIP assay showed that binding of SREBP-2 to the promoter region of the Hmgcs1 gene was greater in the livers of Cyp3a−/− mice than in the livers of WT mice (Fig. 3). These results clearly indicated that SREBP-2 was activated in the livers of Cyp3a−/− mice, resulting in enhancing transcription of genes encoding enzymes involved in cholesterol biosynthesis. Furthermore, plasma levels of lathosterol in Cyp3a−/− mice showed a trend toward higher than those of WT mice, although the difference was not significant (Table 1). Therefore, deficiency of CYP3A enzymes may increase cholesterol synthesis.
Despite the fact that cholesterol biosynthesis was activated in Cyp3a−/− mice, total cholesterol levels in the liver were decreased in Cyp3a−/− mice compared with those in WT mice (Table 1). Because hepatic cholesterol levels are regulated by influx, efflux, and conversion into bile acids of cholesterol, we investigated mRNA expression levels of cholesterol transporters and a rate-limiting enzyme in bile acid biosynthesis in WT and Cyp3a−/− mice. Results of the present study showed that deficiency of CYP3A enzymes did not affect mRNA expression levels of ABCG5, ABCG8, ABCA1, and LDLR in the liver (Fig. 4A). In the intestine, mRNA expression levels of ABCG5, ABCG8, and ABCA1 were similar between WT and Cyp3a−/− mice, and deficiency of CYP3A enzymes did not decrease mRNA levels of cholesterol influx transporter NPC1L1 (Fig. 4B). On the other hand, mRNA levels of CYP7A1, CYP8B1, and CYP7B1 were significantly higher in Cyp3a−/− mice than in WT mice (Fig. 5). Plasma concentration of C4 and total bile acid levels in the liver were also significantly higher in Cyp3a−/− mice than in WT mice (Fig. 6A, B). Therefore, it is thought that reduction of hepatic total cholesterol in Cyp3a−/− mice was due to enhanced bile acid synthesis rather than alteration in cholesterol transport in the intestine and liver. This idea is supported by results of a previous study showing that hepatic total cholesterol in rat CYP7A1 transgenic hamsters maintained on a Western-type diet was decreased because of enhancement of bile acid synthesis (35). Conversely, CYP7A1 mutation, which leads to loss of active sites and function, resulted in suppression of the classical pathway of bile acid synthesis and substantial cholesterol accumulation in the human liver (36). These findings suggest that reduction of hepatic total cholesterol in Cyp3a−/− mice would be the consequence of enhanced bile acid synthesis by CYP7A1, CYP8B1, and CYP7B1.
In this study, we found that deficiency of CYP3A enzymes enhanced expression of CYP7A1 and CYP8B1 (Fig. 5A, B). It is well known that the Cyp7a1 gene is transcriptionally activated by liver X receptor α (LXRα) (37). However, no difference between WT and Cyp3a−/− mice was observed in the expression levels of ABCG5, ABCG8, and ABCA1, which were typical target genes of LXRα (Fig. 4) (9, 38). Therefore, it was not suggested that deficiency of CYP3A enzymes was involved in the activation of LXRα. Expression of CYP7A1 and CYP8B1 is regulated by farnesoid X receptor (FXR), which suppresses the transcription of genes through activation of SHP (39, 40). However, deficiency of CYP3A enzymes did not affect the expression of SHP mRNA (Fig. 5E). Expression of CYP7A1 is also suppressed through activation of PXR (33, 34). If deficiency of CYP3A enzymes could lead to deactivation of PXR through decreased formation of PXR ligands, expression of CYP7A1 might be increased. However, deficiency of CYP3A enzymes increased the expression of four target genes of PXR except for Mdr1 (supplementary Fig. III). Therefore, it is hard to consider that PXRs were deactivated in the livers of Cyp3a−/− mice compared with those of WT mice. Taken together with these findings, enhanced expression of CYP7A1 and CYP8B1 in Cyp3a−/− mice may be regulated by factors except for LXRα, FXR, and PXR. Further studies are needed to clarify the mechanism.
Several studies have shown that CYP3A enzymes are involved in cholesterol metabolism. It has been reported that CYP3A enzymes catalyze 4β- and 25-hydroxylation of cholesterol (18, 21). 4β-Hydroxy-cholesterol was recently shown as an LXR activator (41), whereas its role in the cholesterol synthesis is unclear. On the other hand, 25-hydroxy-cholesterol is well known to be a negative regulator in cholesterol synthesis (23). 25-Hydroxy-cholesterol and other oxysterols suppress cholesterol biosynthesis by suppressing proteolytic activation of SREBP-2 (23). Although 25-hydroxy-cholesterol is also produced by microsomal cholesterol 25-hydroxylase (42), mitochondrial CYP27A1 (43, 44), and nonenzymatic autoxidation of cholesterol (45), results of the present study showed that levels of 25-hydroxy-cholesterol in the livers of Cyp3a−/− mice were significantly lower than those in WT mice (Table 1). Therefore, it is suggested that deficiency of CYP3A leads to reduction of 25-hydroxy-cholesterol synthesis, resulting in activation of SREBP-2.
25-Hydroxy-cholesterol is metabolized to 7α,25-dihydroxy-cholesterol by CYP7A1 and CYP7B1 (46). Results of the present study showed that the expression of CYP7A1 and CYP7B1 in the livers of Cyp3a−/− mice were higher than those of WT mice (Fig. 5). Therefore, upregulation of these genes may also relate to the reduction of 25-hydroxy-cholesterol and the activation of SREBP-2 in the livers of Cyp3a−/− mice. In addition, it is possible that deficiency of CYP3A enzymes enhanced bile acid synthesis with consumption of cholesterol in the liver, resulting in compensatory upregulation of cholesterol synthesis.
Interestingly, it has been reported that mice fed a 25-hydroxy-cholesterol-containing diet (0.25%) show decreased body weight relative to control mice fed the basal diet (47). In this study, deficiency of CYP3A enzymes caused an increase of body and liver weight (Table 1). Taken together, decreased oxysterols appear to increase body and liver weight of Cyp3a−/− mice, although the mechanism is unclear.
Activated SREBP-2 upregulates the expression of numbers of genes involved in cholesterol synthesis and uptake. However, there are large differences in the effects of SREBP-2 on the expression of mRNA (48). Therefore, no change of the expression of LDLR mRNA in the livers of Cyp3a−/− mice may be due to the small response to SREBP-2 activation.
The results of the present study showed that SREBP-2 is activated in the livers of Cyp3a−/− mice possibly due to reduction of the formation of 25-hydroxy-cholesterol and/or other cholesterol metabolites, which may result in enhancement of the transcription of genes encoding enzymes involved in cholesterol biosynthesis. Interestingly, it has been reported that feeding a low-cholesterol diet to mice attenuates hepatic Cyp3a11 expression by activating hepatic SREBP-2 (49). Activated SREBP-2 has been suggested to interact with peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) in the mouse liver, resulting in reduced PGC1α recruitment to hepatocyte nuclear factor-4α on the Cyp3a11 promoter and subsequent downregulation of Cyp3a11 expression (49). Conversely, feeding high-cholesterol diets to rats has been reported to increase expression and activity of CYP3A (50). Similarly, a cholesterol and cholic acid-supplemented diet increased mRNA expression levels of mouse CYP3A11 (51). These findings are considered to be important, being coupled with the findings in the present study that transcription of genes encoding enzymes responsible for the synthesis of cholesterol was enhanced by activation of SREBP-2 in Cyp3a−/− mice. We are tempted to speculate that in the environment of a low-cholesterol diet, CYP3A enzymes are downregulated, and downregulated CYP3A enzymes attenuate the formation of oxysterols such as 25-hydroxy-cholesterol. This reduction of oxysterols would activate SREBP-2 and enhance expression of genes encoding enzymes involved in cholesterol biosynthesis, which may increase hepatic cholesterol biosynthesis to restore cholesterol levels. In the environment of a high-cholesterol diet, a reverse phenomenon may occur. Thus, CYP3A enzymes may play an important role in maintaining a constant level of hepatic cholesterol, irrespective of environmental changes in dietary cholesterol. However, this is only a hypothesis. Further studies are needed to clarify the role of CYP3A enzymes in the homeostasis of cholesterol in the liver.
In summary, the results of the present study showed that expression of genes encoding enzymes involved in cholesterol biosynthesis is enhanced via the activation of SREBP-2 in the livers of Cyp3a−/− mice, possibly due to the decreased formation of oxysterols. Because dietary cholesterol has been reported to affect gene expression of CYP3A enzymes (49–51), the present observations suggest that CYP3A enzymes play a role in maintaining cholesterol levels at a constant level in the liver. The results of the present study also showed that total cholesterol levels were also decreased by 20% in the livers of Cyp3a−/− mice, possibly due to enhanced bile acid synthesis by elevation of CYP7A1 in Cyp3a−/− mice. Further studies are required to clarify the role of CYP3A enzymes in the homeostasis of cholesterol in the liver.
Supplementary Material
Footnotes
Abbreviations:
- ChIP
- chromatin immunoprecipitation
- CYP
- cytochrome P450
- C4
- 7α-hydroxy-4-cholesten-3-one
- FXR
- farnesoid X receptor
- HMGCR
- HMG-CoA reductase
- HMGCS1
- HMG-CoA synthase 1
- KO
- knockout
- LDLR
- low density lipoprotein receptor
- LXRα
- liver X receptor α NPC1L1, Niemann-Pick C1-like 1
- PGC1α
- peroxisome proliferator-activated receptor γ coactivator 1α PXR, pregnane X receptor
- SHP
- small heterodimer partner
- SQLE
- squalene epoxidase
- SRE
- sterol regulatory element
- SREBF2
- sterol regulatory element-binding factor 2
- SREBP-2
- sterol regulatory element-binding protein-2
- TBP
- TATA-binding protein
- WT
- wild type
This work was supported in part by a Grant-in-Aid for Scientific Research (B, 21390040; C, 23590170), Global COE Program (Global Center for Education and Research in Immune System Regulation and Treatment), Special Funds for Education and Research (Development of SPECT Probes for Pharmaceutical Innovation) from the Ministry of Education, Culture, Sports, Science and Technology, and Funding Program for Next Generation World-Leading Researchers (NEXT Program) from Japan Society for the Promotion of Science, Japan.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures and two tables.
REFERENCES
- 1.Miller W. L. 1988. Molecular biology of steroid hormone synthesis. Endocr. Rev. 9: 295–318 [DOI] [PubMed] [Google Scholar]
- 2.Schroepfer G. J. 2000. Oxysterols: modulators of cholesterol metabolism and other processes. Physiol. Rev. 80: 361–554 [DOI] [PubMed] [Google Scholar]
- 3.Bloch K., Berg B. N., Rittenberg D. 1943. The biological conversion of cholesterol to cholic acid. J. Biol. Chem. 149: 511–517 [Google Scholar]
- 4.Wiseman H. 1993. Vitamin D is a membrane antioxidant. Ability to inhibit iron-dependent lipid peroxidation in liposomes compared to cholesterol, ergosterol and tamoxifen and relevance to anticancer action. FEBS Lett. 326: 285–288 [DOI] [PubMed] [Google Scholar]
- 5.Brown M. S., Goldstein J. L. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 89: 331–340 [DOI] [PubMed] [Google Scholar]
- 6.Horton J. D., Goldstein J. L., Brown M. S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D. Q. 2007. Regulation of intestinal cholesterol absorption. Annu. Rev. Physiol. 69: 221–248 [DOI] [PubMed] [Google Scholar]
- 8.Davis H. R., Zhu L. J., Hoos L. M., Tetzloff G., Maguire M., Liu J., Yao X., Iyer S. P., Lam M. H., Lund E. G., et al. 2004. Niemann-Pick C1 Like 1 (NPC1L1) is the intestinal phytosterol and cholesterol transporter and a key modulator of whole-body cholesterol homeostasis. J. Biol. Chem. 279: 33586–33592 [DOI] [PubMed] [Google Scholar]
- 9.Attie A. D. 2007. ABCA1: at the nexus of cholesterol, HDL and atherosclerosis. Trends Biochem. Sci. 32: 172–179 [DOI] [PubMed] [Google Scholar]
- 10.Berge K. E., Tian H., Graf G. A., Yu L., Grishin N. V., Schultz J., Kwiterovich P., Shan B., Barnes R., Hobbs H. H. 2000. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 290: 1771–1775 [DOI] [PubMed] [Google Scholar]
- 11.Wittenburg H., Carey M. C. 2002. Biliary cholesterol secretion by the twinned sterol half-transporters ABCG5 and ABCG8. J. Clin. Invest. 110: 605–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietschy J. M., Turley S. D., Spady D. K. 1993. Role of liver in the maintenance of cholesterol and low density lipoprotein homeostasis in different animal species, including humans. J. Lipid Res. 34: 1637–1659 [PubMed] [Google Scholar]
- 13.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174 [DOI] [PubMed] [Google Scholar]
- 14.Guengerich F. P. 1999. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 39: 1–17 [DOI] [PubMed] [Google Scholar]
- 15.Deb S., Pandey M., Adomat H., Guns E. S. 2012. Cytochrome P450 3A-mediated microsomal biotransformation of 1α,25-dihydroxyvitamin D3 in mouse and human liver: drug-related induction and inhibition of catabolism. Drug Metab. Dispos. 40: 907–918 [DOI] [PubMed] [Google Scholar]
- 16.Huang P., Zhu B., Wang L. S., Ouyang D. S., Huang S. L., Chen X. P., Zhou H. H. 2003. Relationship between CYP3A activity and breast cancer susceptibility in Chinese Han women. Eur. J. Clin. Pharmacol. 59: 471–476 [DOI] [PubMed] [Google Scholar]
- 17.Honda A., Salen G., Matsuzaki Y., Batta A. K., Xu G., Leitersdorf E., Tint G. S., Erickson S. K., Tanaka N., Shefer S. 2001. Side chain hydroxylations in bile acid biosynthesis catalyzed by CYP3A are markedly up-regulated in Cyp27-/- mice but not in cerebrotendinous xanthomatosis. J. Biol. Chem. 276: 34579–34585 [DOI] [PubMed] [Google Scholar]
- 18.Diczfalusy U., Nylén H., Elander P., Bertilsson L. 2011. 4β-Hydroxycholesterol, an endogenous marker of CYP3A4/5 activity in humans. Br. J. Clin. Pharmacol. 71: 183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodin K., Andersson U., Rystedt E., Ellis E., Norlin M., Pikuleva I., Eggertsen G., Björkhem I., Diczfalusy U. 2002. Metabolism of 4 beta-hydroxycholesterol in humans. J. Biol. Chem. 277: 31534–31540 [DOI] [PubMed] [Google Scholar]
- 20.Bodin K., Bretillon L., Aden Y., Bertilsson L., Broomé U., Einarsson C., Diczfalusy U. 2001. Antiepileptic drugs increase plasma levels of 4beta-hydroxycholesterol in humans: evidence for involvement of cytochrome p450 3A4. J. Biol. Chem. 276: 38685–38689 [DOI] [PubMed] [Google Scholar]
- 21.Honda A., Miyazaki T., Ikegami T., Iwamoto J., Maeda T., Hirayama T., Saito Y., Teramoto T., Matsuzaki Y. 2011. Cholesterol 25-hydroxylation activity of CYP3A. J. Lipid Res. 52: 1509–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swell L., Schwartz C. C., Gustafsson J., Danielsson H., Vlahcevic Z. R. 1981. A quantitative evaluation of the conversion of 25-hydroxycholesterol to bile acids in man. Biochim. Biophys. Acta. 663: 163–168 [DOI] [PubMed] [Google Scholar]
- 23.Adams C. M., Reitz J., De Brabander J. K., Feramisco J. D., Li L., Brown M. S., Goldstein J. L. 2004. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem. 279: 52772–52780 [DOI] [PubMed] [Google Scholar]
- 24.Radhakrishnan A., Ikeda Y., Kwon H. J., Brown M. S., Goldstein J. L. 2007. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. USA. 104: 6511–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Herwaarden A. E., Wagenaar E., van der Kruijssen C. M., van Waterschoot R. A., Smit J. W., Song J. Y., van der Valk M. A., van Tellingen O., van der Hoorn J. W., Rosing H., et al. 2007. Knockout of cytochrome P450 3A yields new mouse models for understanding xenobiotic metabolism. J. Clin. Invest. 117: 3583–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engels F. K., Sparreboom A., Mathot R. A., Verweij J. 2005. Potential for improvement of docetaxel-based chemotherapy: a pharmacological review. Br. J. Cancer. 93: 173–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazuki Y., Kobayashi K., Aueviriyavit S., Oshima T., Kuroiwa Y., Tsukazaki Y., Senda N., Kawakami H., Ohtsuki S., Abe S., et al. 2013. Trans-chromosomic mice containing a human CYP3A cluster for prediction of xenobiotic metabolism in humans. Hum. Mol. Genet. 22: 578–592 [DOI] [PubMed] [Google Scholar]
- 28.Perloff M. D., von Moltke L. L., Court M. H., Kotegawa T., Shader R. I., Greenblatt D. J. 2000. Midazolam and triazolam biotransformation in mouse and human liver microsomes: relative contribution of CYP3A and CYP2C isoforms. J. Pharmacol. Exp. Ther. 292: 618–628 [PubMed] [Google Scholar]
- 29.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- 30.Lenicek M., Juklova M., Zelenka J., Kovar J., Lukas M., Bortlik M., Vitek L. 2008. Improved HPLC analysis of serum 7alpha-hydroxycholest-4-en-3-one, a marker of bile acid malabsorption. Clin. Chem. 54: 1087–1088 [DOI] [PubMed] [Google Scholar]
- 31.Dooley K. A., Millinder S., Osborne T. F. 1998. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J. Biol. Chem. 273: 1349–1356 [DOI] [PubMed] [Google Scholar]
- 32.Miettinen T. A., Tilvis R. S., Kesäniemi Y. A. 1990. Serum plant sterols and cholesterol precursors reflect cholesterol absorption and synthesis in volunteers of a randomly selected male population. Am. J. Epidemiol. 131: 20–31 [DOI] [PubMed] [Google Scholar]
- 33.Li T., Chiang J. Y. 2005. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 288: G74–G84 [DOI] [PubMed] [Google Scholar]
- 34.Ma Y., Liu D. 2012. Activation of pregnane X receptor by pregnenolone 16 α-carbonitrile prevents high-fat diet-induced obesity in AKR/J mice. PLoS ONE. 7: e38734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spady D. K., Cuthbert J. A., Willard M. N., Meidell R. S. 1995. Adenovirus-mediated transfer of a gene encoding cholesterol 7 alpha-hydroxylase into hamsters increases hepatic enzyme activity and reduces plasma total and low density lipoprotein cholesterol. J. Clin. Invest. 96: 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pullinger C. R., Eng C., Salen G., Shefer S., Batta A. K., Erickson S. K., Verhagen A., Rivera C. R., Mulvihill S. J., Malloy M. J., et al. 2002. Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J. Clin. Invest. 110: 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang J. Y., Kimmel R., Stroup D. 2001. Regulation of cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRalpha). Gene. 262: 257–265 [DOI] [PubMed] [Google Scholar]
- 38.Repa J. J., Berge K. E., Pomajzl C., Richardson J. A., Hobbs H., Mangelsdorf D. J. 2002. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J. Biol. Chem. 277: 18793–18800 [DOI] [PubMed] [Google Scholar]
- 39.Forman B. M., Goode E., Chen J., Oro A. E., Bradley D. J., Perlmann T., Noonan D. J., Burka L. T., McMorris T., Lamph W. W., et al. 1995. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 81: 687–693 [DOI] [PubMed] [Google Scholar]
- 40.Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., Mangelsdorf D. J. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 6: 507–515 [DOI] [PubMed] [Google Scholar]
- 41.Nury T., Samadi M., Varin A., Lopez T., Zarrouk A., Boumhras M., Riedinger J. M., Masson D., Vejux A., Lizard G. 2013. Biological activities of the LXRα and β agonist, 4β-hydroxycholesterol, and of its isomer, 4α-hydroxycholesterol, on oligodendrocytes: effects on cell growth and viability, oxidative and inflammatory status. Biochimie. 95: 518–530 [DOI] [PubMed] [Google Scholar]
- 42.Lund E. G., Kerr T. A., Sakai J., Li W. P., Russell D. W. 1998. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J. Biol. Chem. 273: 34316–34327 [DOI] [PubMed] [Google Scholar]
- 43.Li X., Pandak W. M., Erickson S. K., Ma Y., Yin L., Hylemon P., Ren S. 2007. Biosynthesis of the regulatory oxysterol, 5-cholesten-3beta, 25-diol 3-sulfate, in hepatocytes. J. Lipid Res. 48: 2587–2596 [DOI] [PubMed] [Google Scholar]
- 44.Lund E., Björkhem I., Furster C., Wikvall K. 1993. 24-, 25- and 27-hydroxylation of cholesterol by a purified preparation of 27-hydroxylase from pig liver. Biochim. Biophys. Acta. 1166: 177–182 [DOI] [PubMed] [Google Scholar]
- 45.Smith L. L. 1981. Cholesterol Autoxidation. Plenum Press, New York. [Google Scholar]
- 46.Schwarz M., Lund E. G., Lathe R., Björkhem I., Russell D. W. 1997. Identification and characterization of a mouse oxysterol 7alpha-hydroxylase cDNA. J. Biol. Chem. 272: 23995–24001 [DOI] [PubMed] [Google Scholar]
- 47.Kandutsch A. A., Heiniger H. J., Chen H. W. 1977. Effects of 25-hydroxycholesterol and 7-ketocholesterol, inhibitors of sterol synthesis, administered orally to mice. Biochim. Biophys. Acta. 486: 260–272 [DOI] [PubMed] [Google Scholar]
- 48.Horton J. D., Shimomura I., Brown M. S., Hammer R. E., Goldstein J. L., Shimano H. 1998. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Invest. 101: 2331–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inoue S., Yoshinari K., Sugawara M., Yamazoe Y. 2011. Activated sterol regulatory element-binding protein-2 suppresses hepatocyte nuclear factor-4-mediated Cyp3a11 expression in mouse liver. Mol. Pharmacol. 79: 148–156 [DOI] [PubMed] [Google Scholar]
- 50.Hunt C. M., Westerkam W. R., Stave G. M., Wilson J. A. 1992. Hepatic cytochrome P-4503A (CYP3A) activity in the elderly. Mech. Ageing Dev. 64: 189–199 [DOI] [PubMed] [Google Scholar]
- 51.Sonoda J., Chong L. W., Downes M., Barish G. D., Coulter S., Liddle C., Lee C. H., Evans R. M. 2005. Pregnane X receptor prevents hepatorenal toxicity from cholesterol metabolites. Proc. Natl. Acad. Sci. USA. 102: 2198–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.