Abstract
The use of nanopores is a powerful new frontier in single-molecule sciences. Nanopores have been used effectively in exploring various biophysical features of small polypeptides and proteins, such as their folding state and structure, ligand interactions, and enzymatic activity. In particular, the α-hemolysin protein pore (αHL) has been used extensively for the detection, characterization and analysis of polypeptides, because this protein nanopore is highly robust, versatile and tractable under various experimental conditions. Inspired by the mechanisms of protein translocation across the outer membrane translocases of mitochondria, we have shown the ability to use nanopore-probe techniques in controlling a single protein using engineered αHL pores. Here, we provide a detailed protocol for the preparation of αHL protein nanopores. Moreover, we demonstrate that placing attractive electrostatic traps is instrumental in tackling single-molecule stochastic sensing of folded proteins.
Keywords: α-hemolysin, nanopore, single-channel electrical recordings, protein engineering, biotechnology, biosensors
1. Introduction
The observation of the translocation of single-stranded DNA through the α-hemolysin (αHL) protein nanopore paved the way for the birth of single-molecule stochastic of small molecules (1). Since then, other research groups have explored and set up the principles for stochastic sensing of molecules (2–8).
α-Hemolysin is a monomeric, 293-residue long toxin protein that is secreted by the pathogenic Staphylococcal aureus (9). The synthesized monomers self-assemble on synthetic membranes to form a large aqueous protein pore. The protein pore complex is heptameric and features a β-barrel mushroom-shape (Fig. 1A). This pore has many advantages over other transmembrane protein nanopores, such as its the known crystal structure (9), amenability to molecular engineering and localized functionalization (10–14), and large single-channel conductance that renders high resolution, single-channel electrical recordings (1;15). Moreover, the pore remains open for long periods of time under various experimental conditions, such as pH (16), ionic strength (17), temperature (18;19), high transmembrane potential (20;21) and mild concentrations of denaturing chemical agents (22;23). Therefore, this protein has been wildly favored for use in single-molecule stochastic sensing using the resistive-pulse technique (24).
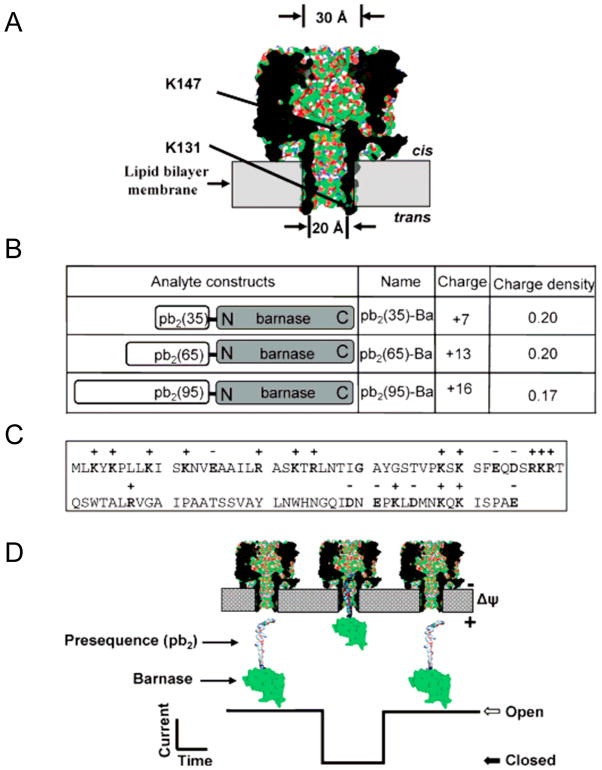
Fig. 1.
Engineered αHL protein pores and pb2-Ba proteins for protein sensing: (A) Strategic functionalization of the αHL protein pores. Two mutations that introduced two electrostatic traps (acidic rings of aspartates) are engineered at either the trans opening (K131) or the cis opening (K147) of the β barrel, as shown by arrows. PyMOL53 was used to generate the view using the coordinates from the crystal structure of the αHL pore (7ahl.pdb); (B) Molecular design of the analyte proteins (pb2-Ba). Three different lengths of Yeast pre-cytochrome b2 (pb2) with different charge densities were fused to the N-terminus of barnase (Ba); (C) The panel shows the amino acid sequence of the first 95 residues of the pb2 presequence; (D) Illustration of how pb2-Ba protein partitions into the αHL protein pore from the trans side of the bilayer. This single-molecule partitioning is monitored by a transient blockade in the single-channel electrical trace. Figure was reproduced, with permission, from Ref (27).
We were inspired by the mechanisms of the protein translocation system in outer membranes of mitochondria (25) to design engineered α-hemolysin protein pores for single-molecule stochastic sensing of folded proteins. The underlying mechanism of this biological system is that a single protein is recruited by its positively-charged presequence to the binding sites within the pore involved in the protein translocation machinery. Then, the protein is electrophoretically inserted into the pore by the transmembrane potential. According to this biological system, it may be possible to control a single protein using α-hemolysin pore imbedded in a synthetic membrane, in which the transmembrane potential will pull on the presequence in the protein analyte, assisting the partitioning of the protein into α-hemolysin pore lumen (Fig. 1A–D). To develop α-hemolysin protein pores for protein sensing, we engineered electrostatic traps within the pore lumen (Fig. 1A) as recruiting and binding sites for folded proteins, RNase fused to the signal polypeptide of the precytochrome b2 (pb2-Ba) (Fig. 1B–D). In this heterologous system, the fusion protein pb2-Ba is electrophoretically inserted into the engineered protein nanopore by the transmembrane potential (Fig. 1D). Our work demonstrated that engineering negatively-charged rings, placed at strategic locations in the pore lumen of the αHL protein, played a key role in recruiting positively-charged protein analytes. We were able to trap and control proteins at a single-molecule level (26;27). Further, we explored factors that altered the trapping of proteins, for example, the length of the pb2 presequence (Fig. 1B–C), location of the engineered trap, the transmembrane potential and the ionic strength. These studies revealed that trapping a single protein with a nanopore follows simple principles of physics; while the biology of protein translocation machineries is complex (26).
2. Materials
The performer of this protocol must have training in chemical hygiene and handling radioactive materials. The common chemicals that are used for making buffers are purchased from Sigma (St. Louis, MO) and stored at room temperature, unless otherwise mentioned. The buffers and solutions used throughout this protocol are made from distilled and deionized water.
2.1 Expression and purification of αHL heptameric protein pores
2.1.1 Preparation of lipid membranes from rabbit red blood cells
Rabbit red blood cells (HemoStat, Dixon, CA).
Washing buffer (isotonic): 150 mM NaCl, 10 mM Na-MOPS, 0.1% bovine serum albumin (BSA), pH 7.4. The buffer is filtered through a 0.2 μm-cut off cellulose membrane filter (Thermo Fisher Scientific, Rochester, NY) and stored at 4 °C.
Rupturing buffer (hypotonic): 5 mM Na2HPO4, 1mM EDTA, pH 8.0, store at 4 °C.
2.1.2 In vitro transcription-coupled translation (IVTT) reaction for expressing αHL pores
E. coli T7 S30 extract system kit for expressing proteins in vitro from circular DNA (Promega, Madison, WI). It includes the following reagents: amino acid mixture without methionine (relevant to this protocol), T7 S30 extract containing T7 RNA polymerase and other translation machinery components such as ribosomes, S30 premix without amino acids (see Note 1).
αHL genes cloned into plasmids containing a T7 promoter and a ribosome binding site (125–300 ng/ul).
[35S]methionine, specific activity > 1000 Ci/mmole, >37 TBq/mmole at 10 μCi/μl (MP Biomedical, Costa Mesa, CA) (see Note 2).
Rabbit red blood cell membranes (rRBCM) (subheading 3.1.1) (see Note 3).
RNase inhibitor (RNase out, 40 units/μl) (Invitogen, Carlsbad, CA) (see Note 4).
Autoradiography film (BioMax MR Film) (Kodak, Geneva, NY).
Gel dryer system (BioRad, Hercules, CA). It includes gel dryer platform model 583, vacuum pump and cellophane sheets.
Microcentrifuge membrane filters (Rainin, Woburn, MA).
Parafilm laboratory film (Richiney, Menasha, WI).
2.1.3 SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE) for purifying αHL pores
Resolving gel buffer: 1.5 M Tris-HCl, pH 8.8. Can be stored at room temperature.
Stacking gel buffer: 0.5 M Tris-HCl, pH 6.8. Can be stored at room temperature.
30% acrylamide/bis solution (37.5:1) (Sigma, St. Louis, MO) (see Note 5).
10% ammonium persulphate solution in water, made freshly for immediate use.
10% (w/v) SDS solution in water (see Note 6).
Tetramethylethylenediamine (TEMED) (Sigma, St. Louis, MO) (see Note 7).
Isopropanol (Sigma, St. Louis, MO).
Running buffer (10X): 250 mM Tris, 1920 mM glycine, 0.5% (w/v) SDS. Can be stored at room temperature.
2X SDS sample buffer (4.75 ml): mix the following: 1.8 ml water, 0.625 ml 0.5 M Tris-HCl (pH 6.8), 1.25 ml glycerol, 1.0 ml 10% (w/v) SDS, 0.1 ml 0.5% (w/v) Bromophenol Blue.
Molecular weight markers: SeeBlue 2, range from 250 to 4 kDa (Invitogen, Carlsbad, CA).
2.2 Electrophysiology
Silver wire, 1.5 mm diameter (Sigma, St. Louis, MO).
Polytetrafluoroethylene (PTFE) film, 0.025 mm in thickness (Goodfellow Corporation; Malvern, PA).
1.5 % agarose solution made from ultra pure DNA Grade agarose (Bio-Rad Laboratories; Hercules, CA) in 10 mM phosphate buffer, 3 M KCl, pH 8.0.
Copper wire, D-sub crimp style male and heat shrink tubing.
Manufactured in house two-compartment chamber (Fig. 2).
Electrophysiology instrument set up: an Axopatch 200B patch-clamp amplifier, a CV-203BU headstage, a DigiData 1322A A/D converter (Axon Instruments, Foster City, CA) in Faraday cage (A Faraday cage, or Faraday shield is an enclosure formed by conducting material or by a mesh of such material to block external electric interference), an 8-pole filter (Model 900, Frequency Devices; Haverhill, MA) and 20 MHz sweep/function generator (BK Precision, Yorba Linda, CA).
10% (v/v) hexadecane in high-purity n-pentane (Sigma, St. Louis, MO) (see Note 8).
10 mg/ml of 1,2-diphytanoyl-sn-glycerophosphocholine lipids (Avanti Polar Lipids; Alabaster, AL), in n-pentane (see Note 9).
Chamber buffer: 10 mM potassium phosphate, pH 7.4, which contains 1 M KCl or as desired.
Spark generator (Daedalon Corporation, Salem, MA).
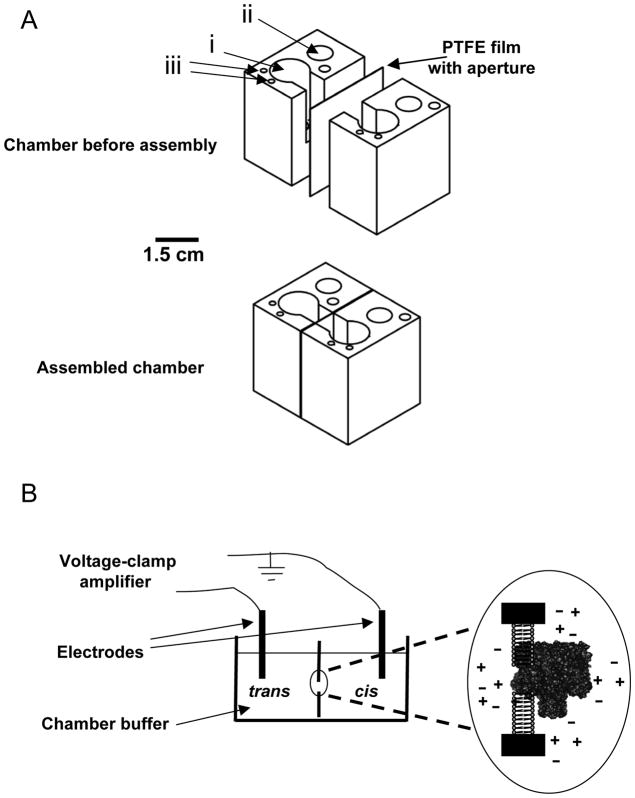
Fig. 2.
Single-channel recordings with a folded planar lipid bilayer. (A) Chamber made in house. Prior to complete assembly, the chamber consists of two compartments, cis and trans and a PTFE film containing an aperture for membrane formation. After assembly, the PTFE film is sandwiched between the two compartments, creating the chamber for the electrophysiology experiments. Holes i, ii and iii that are filled with chamber buffer and used for adding protein, submerging electrodes and buffer perfusion, respectively. Chamber is made from Delrin material and diagram is drawn to scale. The aluminum casting is not depicted in these drawings; (B) Illustration of the bilayer formation and the channel insertion. Chamber buffer is added to cis and trans compartments. Electrodes are submerged in both compartments. Application of the transmembrane voltage is controlled by a voltage-clamp amplifier. The αHL protein pore inserts into the formed bilayer after the protein sample is added to the cis compartment. This diagram is not drawn to scale.
3. Methods
The heptameric αHL pore readily inserts into the synthetic lipid bilayer, forming aqueous channels. We provide a simple method for obtaining satisfactory yield of pre-assembled αHL protein pores directly from SDS-PAGE (28;29). Further, using this protocol, we can obtain high-purity samples that lead to reliable and highly reproducible results. Single-channel recordings with the αHL protein pores feature high temporal resolution (~10 μs) and allow real-time detection and chemical sampling of various small molecules and biopolymers, including polypeptides (21;30). Nevertheless, protein engineering is required to enhance the sensitivity of single-molecule stochastic sensing (26;27;31).
3.1 Expression and purification of the αHL heptameric protein pores
3.1.1 Preparation of cell membranes from rabbit red blood cells (rRBCM)
Collecting the red blood cells: spin down 10–15 ml rabbit blood at ~ 600 xg for 20 minutes.
Washing the red blood cells: decant supernatant and re-suspend the pellet in 50 ml washing buffer.
Re-centrifuge at ~ 600 xg for 20 minutes.
Decant the supernatant.
Repeat steps 2 to 4 at least 6 times.
Rupturing the red blood cells: re-suspend the pellet in 60–80 ml rupturing buffer.
Centrifuge at 30,000 xg for 45 minutes. After centrifugation, the pellet will be loosely compacted and easily decanted with the supernatant. Exercise caution.
Decant the supernatant and re-suspend the pellet in the rupturing buffer.
Repeat steps 7–9 at least 8 times, or until the pellet color (pink) becomes very faint. The color might not totally disappear; however, the pellet will be washed again 4 times before use in IVTT reaction (see later).
Quantitate the membranes by DC assay (BioRad, Hercules, CA). Make the final concentration 0.2 – 1.0 mg/ml. Make 50 μl aliquots and freeze at −80 °C (see Note 10).
3.1.2 Expression of the αHL protein pores by in vitro transcription-coupled translation (IVTT) reaction
This protocol shows the critical steps for the expression and purification of the heptameric αHL protein pores. If the plan is to obtain only monomers, then do not add the red blood cell membranes (First step).
Pipette 5–10 μl (1–10 μg) of ruptured red blood cells into a 1.5 ml microcentrifuge tube. This quantity is sufficient for a 25–50 μl IVTT reaction.
Spin down at 16 000 xg for 5 minutes. Carefully pipette out the supernatant.
Add 1 ml of washing buffer. Mix well by vortexing and spin down at 16 000 xg for 5 minutes. Decant the supernatant and pipette out the remaining buffer. Repeat step 5 at least four times. The pink color of the ruptured membranes should disappear after these washes and becomes white.
Air dry the membranes for 10 minutes in a fume hood.
Add the following to the dried membranes: 10 μl S30 premix, 2.5 μl amino acid mixture without methionine, 1 μl [35S]methionine, 3 μl DNA, 1 μl RNase out and 7.5 μl rifampcin-treated T7 S30 extract. Mix by pipetting the reaction mixture up and down.
Incubate at 37°C for 1 hour (see Note 11).
Place on ice for 10 minutes to stop the reaction.
Spin down at 16 000 xg for 5 minutes. Pipette out the supernatant.
Add 1 ml washing buffer, tap the pellet to mix, and spin down again at 16 000 xg for 5 minutes. Repeat step 9 at least 3 times.
Add 25–50 μl 1X SDS sample buffer to solubilize the sample. DO NOT BOIL. Sample is ready to be loaded onto SDS-PAGE gel (see the following section).
3.1.3 Purification of αHL pores from SDS-PAGE
These instructions assume the use of Mini-PROTEAN Tetra Cell (BioRad, USA). However, they can be considered as general instructions for other gel apparatus with the care of scaling up or down the reagents to accommodate specific gel volumes.
Preparing the glass plates: clean the plates with laboratory detergent like Sparkclean (Fisher, Hampton, NH), rinse thoroughly with water, then finish cleaning with 95% ethanol and let dry.
Making 8% resolving gel (10 ml): mix 2.5 ml resolving gel buffer with 2.7 ml 30% acrylamide/bis solution, 4.7 ml water, 50 μl ammonium persulfate solution and 100 μl 10% SDS solution and 5 μl TEMED. Pour the gel while avoiding bubbles, leaving space for a stacking gel, and overlay immediately with isopropanol. Keep the remaining mixture (see next step). The gel should polymerize in about 30–45 minutes.
Check if the resolving gel is polymerized by examining the remaining of the mixture for polymerization.
Decant the isopropanol and rinse with water (2 to 4 times).
Making 4% stacking gel (5 ml): mix 1.25 ml of stacking gel buffer with 0.65 ml 30% acrylamide/bis solution, 3.05 ml water, 25 μl ammonium persulfate solution, 50 μl 10% SDS solution and 10 μl TEMED. Quickly pour the mixture on the top of the resolving gel and insert the comb. Keep the remaining mixture. The stacking gel should polymerize within 45 min.
Check if the staking gel is ready by examining the remaining of the mixture for polymerization. If polymerized, remove the comb slowly under running water to avoid the collapsing of wells.
Making the running buffer: dilute the 10X running buffer 1:10 in water, stir to mix.
Assemble the gel apparatus.
Add the running buffer to cathode and anode compartments of the gel unit. Clean the wells by pipetting the buffer up and down in the wells with micropipette loading tips.
Load the solubilized IVTT samples. Add one well for pre-stained molecular weight markers. Add 1X SDS sample buffer to the empty wells to avoid “smiling effect” during the run.
Finish the assembly of the apparatus and connect to a power supply. The completion of the run will take about 1.5 hours at 150 V (see Note 12).
Drying the gel: disassemble the gel apparatus and retrieve the gel. Equilibrate the gel in water for 10–15 minutes to prevent gel cracking during the drying process. Wet the cellophane with water. On a flat surface, lay the gel on a cellophane sheet and add water on the top of the gel and wells to help removing any bubbles that may have formed during the addition of the second sheet. Carefully, add the second sheet on the top starting at the edge of the sheets and slowly lower the sheet to cover the gel. Transfer the gel between the two sheets and lay it on a filter paper. Add more water to both sides of the gel, including the filter paper. All these careful steps are necessary to avoid gel cracking. Now, dry the gel for 2–3 hours at 50 °C.
Exposing the dried gel to the film: place the autoradiography film on the top of the dried gel, positioning the notch on the top right corner of the film (if you are using one-sided film). It is critical to know the precise locations of the bands on the gel from the autoradiography film. So, staple the dried gel to the film at two different corners of the film. Doing so will leave marks on the film that match the ones on the gel for the next step (see Note 13).
Developing the autoradiography film and cutting the band: remove the staples and develop the film. Fig. 3A represents a typical autoradiography film obtained routinely from the above procedure. To elute the desired band, align the film with dried gel by matching the staple marks. The band that corresponds to the heptameric αHL pores runs below 148 kDa molecular weight markers under our gel running conditions (Fig. 3A). Mark the band to be cut. Then separate the film from the gel and cut the band by scissors. Transfer the piece of the gel to 500 μl water to elute the protein by diffusion. After re-hydration (1–2 hours), remove the two cellophane pieces and then take the gel piece and smash it between parafilm sheets. Collect the smashed gel and put back into the same 500 μl water. The sample will become slurry. Incubate at 37°C for 1 hour, and then incubate overnight at 4°C while rotating.
Separation of diffused protein from gel pieces: pipette all sample and load it onto the membrane filter reservoir. Spin down using the microcentrifuge at 16 000 xg. The filtrate contains the αHL heptameric pores. Aliquot and store at −80 °C. The αHL protein pores can be highly active for many months. Samples are now ready to be used in electrophysiology.
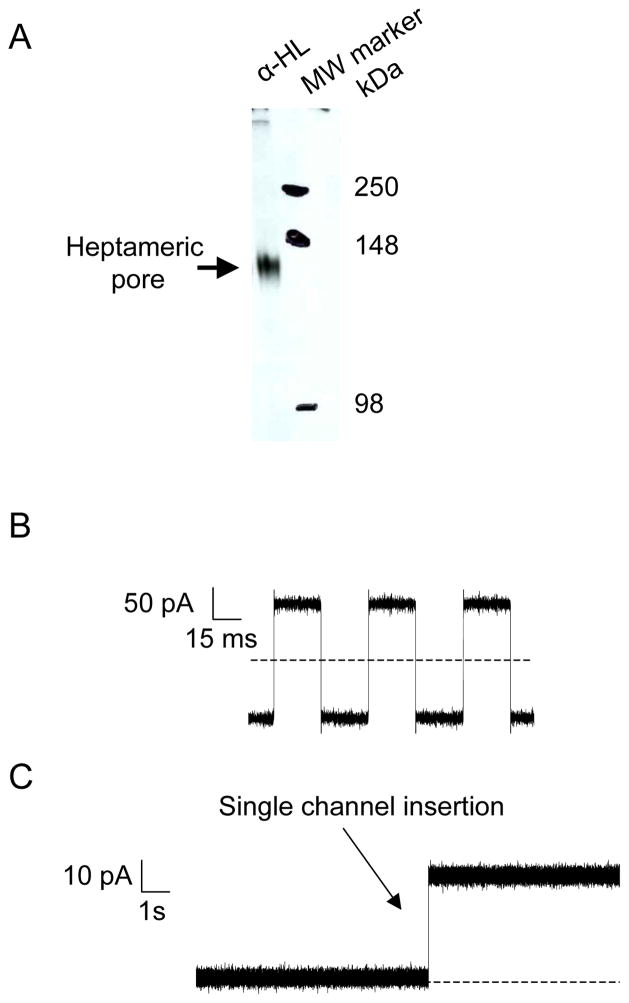
Fig. 3.
SDS-PAGE-purified αHL proteins form an aqueous channel in an artificial lipid bilayer. (A) Expression and assembly of the αHL protein pore. WT-αHL proteins were translated in IVTT reactions in the presence of red blood cell membranes. Proteins were separated on an 8% SDS-polyacrylamide gel. Molecular-weight markers are indicated on the right-hand side; (B) Application of external pulses with which the membrane capacitance of the bilayer can be measured; (C) Capture of the channel insertion during single-channel electrical recording. SDS-PAGE purified proteins were added to the cis chamber, stirred for few seconds and a single αHL protein pore was allowed to insert without further stirring. Single-channel recordings were carried out at room temperature in 1 M KCl, 10 mM potassium phosphate, pH 7.4 with an applied transmembrane potential of +40 mV. Dashed lines represent zero current in (B) and (C). The single-channel electrical traces were low-pass Bessel filtered at 2 kHz.
3.2 Electrophysiology
These instructions are specific to our in house-manufactured chamber (Fig. 2) (32;33). However, the first steps can be considered as a general method for the use in other chamber designs (34;35).
3.2.1 Forming the synthetic bilayer and obtaining single-channel insertion
Making the electrodes: expose 0.25 inch at both ends of the 5 inch long copper wire by removing the insulation. Weld the 0.5 inch silver wire to one exposed end of the copper wire making sure that the majority of silver wire is free, then clamp and weld the other end with D-sub crimp. Cover the welded parts of the wires with the heat-shrink tubing. Submerge the silver wire end in bleach for 1 hour to overnight to make Ag/AgCl2 wire. Prepare the agarose solution and heat it to boil within the microwave oven. Before it cools down, and by using a transfer pipette, fill a pipette tip with the agarose solution (20–200 μl tips), then insert the Ag/AgCl2 wire into the tip and submerge the tip in water immediately so that the agarose solidifies faster and to reduce leakage from the other end of the tip. Cut the end of the tip to expose the agarose. Store in 3 M KCl solution. The electrodes are ready to use.
The preparation of the aperture: cut a 5 × 3.0 cm piece of PTFE film, place it between the generator plates and apply 4–6 sparks at 1 Hz at the maximum voltage setting. Inspect under light microscope to assure that you have an aperture without rough edges. The aperture should be 50–80 μm in diameter.
Assembly of the chamber: make a very thin layer of silicone sealing on one half of the chamber (Fig. 2A). Lay the PTFE film on the silicone layer, making sure that the aperture is not covered by the chamber wall. Prepare another very thin layer of silicone on the other half of the chamber, and adhere both of chamber halves together. Put them in the aluminum cast and tighten the screw. During tightening, pay extra attention so as not to have the silicone spread over the film and block the aperture. Let sit over night, then the chamber will be ready for use in electrophysiology experiments.
Set the 8-pole filter devise at 10 kHz and the sweep/function generator device to triangle wave function with a frequency of 15 Hz and an output level of 200 peak to peak (the sweep/function generator should be connected to the Axon 200B’s external command input).
Add 0.75 ml chamber buffer to each side of the chamber (Fig. 2A, hole i). Let ~ 10 μl of hexadecane solution slide on each side of the PTFE film, this will create a hydrophobic environment on the edges of the aperture to initiate bilayer formation. Add 10 μl of the lipid solution to both sides of the chamber. DO NOT MIX. Let set for 3 minutes giving time for the pentane to evaporate. Meanwhile, insert the electrodes (Fig. 2A, hole ii) and connect to the headstage in the Faraday cage. The current should read zero. Otherwise there is a leak in the chamber and the chamber has to be reassembled.
Forming the bilayer: add 0.75 ml chamber buffer to both sides. At this time, the current read by the amplifier might be large, indicating that the bilayer is not formed yet. To form the bilayer, pipette one side of the chamber up and down (Fig. 2A, hole iii) until you see ~ 0 current. (see Note 14).
Apply the external command function from the patch-clamp amplifier to monitor the capacitance of the bilayer (see Note 15). Expected results for a good bilayer are illustrated in Fig. 3B.
Add 1–2.5 μl αHL protein to the cis side of the chamber (cis side is the grounded side). Apply positive or negative voltages to monitor the channel insertion (see Note 16). Fig. 3C represents a typical single-channel insertion of the wild-type αHL pore.
3.2.2 Sensing a single protein by engineered αHL pores
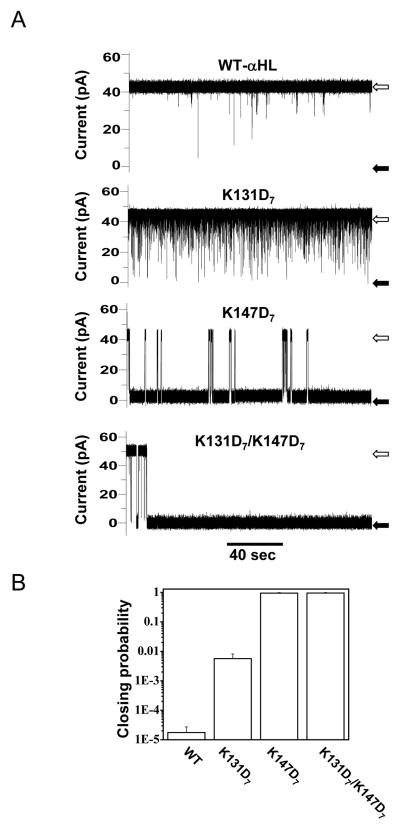
To demonstrate the power of the nanopore technique to sense proteins and to investigate factors that affect their interactions with nanopores, we have used wild-type and engineered αHL pores: K131D7, K147D7 and K131D7/ K147D7, where the electrostatic traps are at the trans opening, cis opening and both, respectively (Fig. 1A). In addition, we used pb2-Ba proteins (36;37), which contain positively-charged leading presequence pb2 of varying length, as analytes for protein sensing (Fig. 1B and C). Fig. 4 shows an example of the effect of pb2(95)-Ba protein analyte when added to the trans side of the chamber (Fig. 4A). Here, the pb2(95) leading sequence contains 95 amino acids. The top trace shows the interaction of the pb2(95)-Ba with the wild-type αHL protein pores. Short-lived and long-lived current blockades were observed with K131D7 and K147D7 protein pores, respectively (Fig. 4A). Remarkably, a single pb2(95)-Ba protein is arrested within the pore lumen of the K131D7/ K147D7 protein. We find that engineering electrostatic traps within the nanopore lumen increased the capability of αHL pores to tackle protein sensing (Fig. 4B).
Fig. 4.
Different single-channel electrical signatures given by the interactions of the pb2(95)-Ba protein with various engineered αHL protein pores. (A) single-channel electrical recordings shows that pb2(95)-Baproteins interact more strongely with the pore (channel almost closed, black arrows) when the two electrostatic traps are present in the pore. The wild-type αHL protein pore shows the weakest interaction (channel mostly open, white arrows), as also indicated by the residence probability; (B) The residence probability is obtained by adding the channel closure times and dividing by the total recording time. 200 nM pb2(95)-Ba was added to the trans chamber. Single-channel recordings were performed as in Fig. 3C. Figure was reproduced from Ref. (27).
Footnotes
This work was supported in part by grants from the US National Science Foundation (DMR-0706517) and National Institutes of Health (R01 GM088403) to LM.
The amount of T7 S30 extract can be reduced and still produce approximately the same expression level. But, we have to warn that following this recommendation has to be approached empirically. In theory, this IVTT expression can work well for other transmembrane proteins as long as rRBCM are supplemented in the reaction and the desired protein is SDS stable (38).
It is critical that the plasmid DNA is clean for the IVTT reaction to work properly (Generally, spectrometer absorbance of A260/280 ratio should be between 1.8 and 2.0). If you are using DNA prepared in large volumes, we recommend re-purifying DNA by phenol/chloroform extraction method and then precipitating DNA by ethanol. We found that cleaning DNA is needed in many DNA preparations.
We recommend that you do not store the blood for a long time. Prepare the membranes as soon as you receive the blood. Once the blood starts to coagulate, the membrane preparations will become difficult to achieve and produce membranes that are not suitable for the IVTT reaction.
The addition of RNase out might not be needed in all labs. However, molecular biology laboratories often use RNases; for example, the RNases found in kits for DNA preparations. In this case, the addition of the RNase out might be necessary to reduce the degradation of RNAs in the IVTT reaction. In our case, the addition of RNase out helped in increasing the expression of protein pores. Further, we do not recommend the use of autoclaved tips when setting up the IVTT reaction, since RNase can survive the autoclave process. Therefore, use aerosol resistant tips, or use the tips directly from the manufacturer, if they come as RNase- and DNase-free.
Acrylamide solution is a neurotoxin, so exercise caution. Use proper protective gloves and goggles when pouring the gels, since some of the solution may splash upon inserting the comb.
Avoid breathing directly above SDS when you are weighing.
TEMED has an unpleasant smell. If you can, open the bottle of TEMED in the fume hood.
This chemical is not expensive, we recommend making the solution every week or so. If you do not make it fresh, store the solution in the −20°C freezer.
Lipids are expensive and lipid solution can be stored in the −20°C freezer. However, we do not recommend making high quantity of this solution. We routinely ask the provider (Avanti) to ship aliquots of 25 mg in air-tide glass container. Therefore, we only make 25 mg lipids at time.
We have found that when we replaced rRBCM (see above) with microsomes (Promega, Madison, WI), the heptamer αHL protein pores were formed in the IVTT reaction. If you do not wish to work with rRBCM, use 0.5–1 μl microsomes in 25 μl IVTT reaction. Also, we have tested the membrane vesicles (MVs) that are exported to the growth media by Pseudomonas aeruginosa (39) and have found that αHL monomers form heptameric proteins on these vesicles as well. If you use microsomes or MVs, the rest of the protocol will remain the same as for rRBCM.
For expression of αHL proteins in this protocol, the reaction does not need to be incubated more than one hour at 37°C. However, if you are expressing different engineered αHL pores, or even expressing different membrane proteins and the standard reaction at 37°C for 1 hour does not produce the expected results, you may need to try the reactions at lower temperatures. According to the manufacturer, lower temperature produces a slower rate of translation, yet often extends the time of the linear rate. Normally, the fastest linear rate occurs at 37°C for approximately 2 hours. Temperature and time of the reactions should be determined empirically.
We normally apply lower voltage at the beginning of the gel run (70 V) until the samples pass the stacking gel. Once the samples enter the resolving gel, we increase the voltage to 150 V. Doing so allows compact protein bands during the run. During the run, you will notice a reduction in the amperage; you should not attempt to increase the voltage to compensate the loss of amps. Higher voltage will result in heating the gel and consequently breaking the glass plates. If you decide to run the gel at constant amperage, you need to have a cooling system connected to your gel apparatus to avoid plate breakage.
We expose our films at room temperature and we usually get sufficient signal to cut the proteins bands for elution. However, if you experience otherwise, expose your film at ~−80 °C. The cold temperature will substantially enhance the signal (40;41).
If you could not form the bilayer within a few trials of pipetting up and down, add a drop or two from the lipid solution. Let sit for 3 minutes for the pentane to evaporate and try again. If you could not form the bilayer, wash the chamber with water, then 95% ethanol and try again by starting from step 4. Forming the bilayer requires diligence; it might take a couple of trials to familiarize yourself with this technique.
Since the sweep/function generator is set for triangle wave, you expect that the capacitance of the bilayer to give electrical signature as in Fig. 3B with amplitude between 100–200 pA. Low capacitance bilayers, or “leaky” (the electric signature is tilted), should not be used in the experiments. For reproducible results, you need to aim at an ideal bilayer with capacitance between 100–120 pF with no leak. Also, once you have formed the bilayer, apply higher voltage (not more than 200 mV) to check for the stability of the bilayer and any potential current leak at higher voltages.
It is possible that you will have many channels insert in the synthetic bilayer with the amount of the proteins added to the chamber. If so, take some of the chamber buffer from the cis side of the chamber and then add the same amount of the new buffer back to cis side. This will dilute the proteins in the cis chamber to give a better chance for getting a single channel. This will damage the bilayer. Reform the bilayer as in steps 6 and 7, and wait for the insertion. Stirring is not necessary, but the channel insertions may take longer.
References
- 1.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayley H. Membrane-Protein Structure Piercing Insights. Nature. 2009;459:651–652. doi: 10.1038/459651a. [DOI] [PubMed] [Google Scholar]
- 3.Bayley H, Cremer PS. Stochastic sensors inspired by biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 4.Movileanu L. Interrogating single proteins through nanopores: challenges and opportunities. Trends in Biotechnology. 2009;27:333–341. doi: 10.1016/j.tibtech.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Movileanu L. Squeezing a single polypeptide through a nanopore. Soft Matter. 2008;4:925–931. doi: 10.1039/b719850g. [DOI] [PubMed] [Google Scholar]
- 6.Howorka S, Siwy Z. Nanopore analytics: sensing of single molecules. Chemical Society Reviews. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 7.Siwy ZS, Howorka S. Engineered voltage-responsive nanopores. Chemical Society Reviews. 2010;39:1115–1132. doi: 10.1039/b909105j. [DOI] [PubMed] [Google Scholar]
- 8.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang XH, Jovanovich SB, Krstic PS, Lindsay S, Ling XSS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. The potential and challenges of nanopore sequencing. Nature Biotechnology. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song LZ, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 10.Movileanu L, Cheley S, Howorka S, Braha O, Bayley H. Location of a constriction in the lumen of a transmembrane pore by targeted covalent attachment of polymer molecules. Journal of General Physiology. 2001;117:239–251. doi: 10.1085/jgp.117.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe AJ, Mohammad MM, Cheley S, Bayley H, Movileanu L. Catalyzing the translocation of polypeptides through attractive interactions. Journal of the American Chemical Society. 2007;129:14034–14041. doi: 10.1021/ja0749340. [DOI] [PubMed] [Google Scholar]
- 12.Movileanu L, Howorka S, Braha O, Bayley H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nature Biotechnology. 2000;18:1091–1095. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- 13.Movileanu L, Bayley H. Partitioning of a polymer into a nanoscopic protein pore obeys a simple scaling law. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10137–10141. doi: 10.1073/pnas.181089798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howorka S, Movileanu L, Lu XF, Magnon M, Cheley S, Braha O, Bayley H. A protein pore with a single polymer chain tethered within the lumen. Journal of the American Chemical Society. 2000;122:2411–2416. [Google Scholar]
- 15.Bezrukov SM, Vodyanoy I, Brutyan RA, Kasianowicz JJ. Dynamics and free energy of polymers partitioning into a nanoscale pore. Macromolecules. 1996;29:8517–8522. [Google Scholar]
- 16.Bezrukov SM, Kasianowicz JJ. Current Noise Reveals Protonation Kinetics and Number of Ionizable Sites in An Open Protein Ion Channel. Physical Review Letters. 1993;70:2352–2355. doi: 10.1103/PhysRevLett.70.2352. [DOI] [PubMed] [Google Scholar]
- 17.Krasilnikov OV, Rodrigues CG, Bezrukov SM. Single polymer molecules in a protein nanopore in the limit of a strong polymer-pore attraction. Physical Review Letters. 2006;97 doi: 10.1103/PhysRevLett.97.018301. [DOI] [PubMed] [Google Scholar]
- 18.Jung Y, Bayley H, Movileanu L. Temperature-responsive protein pores. Journal of the American Chemical Society. 2006;128:15332–15340. doi: 10.1021/ja065827t. [DOI] [PubMed] [Google Scholar]
- 19.Kang XF, Gu LQ, Cheley S, Bayley H. Single protein pores containing molecular adapters at high temperatures. Angewandte Chemie-International Edition. 2005;44:1495–1499. doi: 10.1002/anie.200461885. [DOI] [PubMed] [Google Scholar]
- 20.Goodrich CP, Kirmizialtin S, Huyghues-Despointes BM, Zhu AP, Scholtz JM, Makarov DE, Movileanu L. Single-molecule electrophoresis of beta-hairpin peptides by electrical recordings and Langevin dynamics simulations. Journal of Physical Chemistry B. 2007;111:3332–3335. doi: 10.1021/jp071364h. [DOI] [PubMed] [Google Scholar]
- 21.Movileanu L, Schmittschmitt JP, Scholtz JM, Bayley H. Interactions of peptides with a protein pore. Biophysical Journal. 2005;89:1030–1045. doi: 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oukhaled G, Mathe J, Biance AL, Bacri L, Betton JM, Lairez D, Pelta J, Auvray L. Unfolding of proteins and long transient conformations detected by single nanopore recording. Physical Review Letters. 2007;98 doi: 10.1103/PhysRevLett.98.158101. [DOI] [PubMed] [Google Scholar]
- 23.Pastoriza-Gallego M, Oukhaled G, Mathe J, Thiebot B, Betton JM, Auvray L, Pelta J. Urea denaturation of alpha-hemolysin pore inserted in planar lipid bilayer detected by single nanopore recording: Loss of structural asymmetry. Febs Letters. 2007;581:3371–3376. doi: 10.1016/j.febslet.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 24.Bezrukov SM. Ion channels as molecular Coulter counters to probe metabolite transport. Journal of Membrane Biology. 2000;174:1–13. doi: 10.1007/s002320001026. [DOI] [PubMed] [Google Scholar]
- 25.Geissler A, Chacinska A, Truscott KN, Wiedemann N, Brandner K, Sickmann A, Meyer HE, Meisinger C, Pfanner N, Rehling P. The mitochondrial presequence translocase: An essential role of Tim50 in directing preproteins to the import channel. Cell. 2002;111:507–518. doi: 10.1016/s0092-8674(02)01073-5. [DOI] [PubMed] [Google Scholar]
- 26.Mohammad MM, Movileanu L. Excursion of a single polypeptide into a protein pore: simple physics, but complicated biology. European Biophysics Journal with Biophysics Letters. 2008;37:913–925. doi: 10.1007/s00249-008-0309-9. [DOI] [PubMed] [Google Scholar]
- 27.Mohammad MM, Prakash S, Matouschek A, Movileanu L. Controlling a single protein in a nanopore through electrostatic traps. Journal of the American Chemical Society. 2008;130:4081–4088. doi: 10.1021/ja710787a. [DOI] [PubMed] [Google Scholar]
- 28.Walker B, Krishnasastry M, Zorn L, Bayley H. Assembly of the Oligomeric Membrane Pore Formed by Staphylococcal Alpha-Hemolysin Examined by Truncation Mutagenesis. Journal of Biological Chemistry. 1992;267:21782–21786. [PubMed] [Google Scholar]
- 29.Gouaux JE, Braha O, Hobaugh MR, Song LZ, Cheley S, Shustak C, Bayley H. Subunit Stoichiometry of Staphylococcal Alpha-Hemolysin in Crystals and on Membranes - A Heptameric Transmembrane Pore. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:12828–12831. doi: 10.1073/pnas.91.26.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stefureac R, Long Yt, Kraatz HB, Howard P, Lee JS. Transport of +¦-Helical Peptides through +¦-Hemolysin and Aerolysin PoresΓÇá. Biochemistry. 2006;45:9172–9179. doi: 10.1021/bi0604835. [DOI] [PubMed] [Google Scholar]
- 31.Maglia G, Restrepo MR, Mikhailova E, Bayley H. Enhanced translocation of single DNA molecules through alpha-hemolysin nanopores by manipulation of internal charge. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miles G, Movileanu L, Bayley H. Subunit composition of a bicomponent toxin: Staphylococcal leukocidin forms an octameric transmembrane pore. Protein Science. 2002;11:894–902. doi: 10.1110/ps.4360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Movileanu L, Cheley S, Bayley H. Partitioning of Individual Flexible Polymers into a Nanoscopic Protein Pore. Biophysical Journal. 2003;85:897–910. doi: 10.1016/S0006-3495(03)74529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Q, Sigalov G, Dimitrov V, Dorvel B, Mirsaidov U, Sligar S, Aksimentiev A, Timp G. Detecting SNPs using a synthetic nanopore. Nano Letters. 2007;7:1680–1685. doi: 10.1021/nl070668c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guiard B. Structure, Expression and Regulation of A Nuclear Gene Encoding A Mitochondrial Protein - the Yeast L(+)-Lactate Cytochrome-C Oxidoreductase (Cytochrome-B2) Embo Journal. 1985;4:3265–3272. doi: 10.1002/j.1460-2075.1985.tb04076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matouschek A, Azem A, Ratliff K, Glick BS, Schmid K, Schatz G. Active unfolding of precursor proteins during mitochondrial protein import. Embo Journal. 1997;16:6727–6736. doi: 10.1093/emboj/16.22.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shim JW, Yang M, Gu LQ. In vitro synthesis, tetramerization and single channel characterization of virus-encoded potassium channel Kcv. Febs Letters. 2007;581:1027–1034. doi: 10.1016/j.febslet.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Kadurugamuwa JL, Beveridge TJ. Virulence Factors Are Released from Pseudomonas-Aeruginosa in Association with Membrane-Vesicles During Normal Growth and Exposure to Gentamicin - A Novel Mechanism of Enzyme-Secretion. Journal of Bacteriology. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohammad M, York RD, Hommel J, Kapler GM. Characterization of a novel origin recognition complex-like complex: Implications for DNA recognition, cell cycle control, and locus-specific gene amplification. Molecular and Cellular Biology. 2003;23:5005–5017. doi: 10.1128/MCB.23.14.5005-5017.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohammad M, Saha S, Kapler GM. Three different proteins recognize a multifunctional determinant that controls replication initiation, fork arrest and transcription in Tetrahymena. Nucleic Acids Research. 2000;28:843–851. doi: 10.1093/nar/28.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]