Abstract
The advent of new technologies allowing the study of single biological molecules continues to have a major impact on studies of interacting systems as well as enzyme reactions. These approaches (fluorescence, optical and magnetic “tweezers”), in combination with ensemble methods, have been particularly useful for mechanistic studies of protein-nucleic acid interactions and enzymes that function on nucleic acids. We review progress in the use of single molecule methods to observe and perturb the activities of proteins and enzymes that function on flexible single stranded DNA. These include single stranded (ss)DNA binding (SSB) proteins, recombinases (RecA/Rad51) and helicases/translocases that operate as motor proteins and play central roles in genome maintenance. We emphasize methods that have been used to detect and study the movement of these proteins (both ATP-dependent directional and random movement) along the ssDNA and the mechanistic and functional information that can result from detailed analysis of such movement.
Keywords: FRET, helicases, RecA, Rad51, SSB, optical tweezers
INTRODUCTION
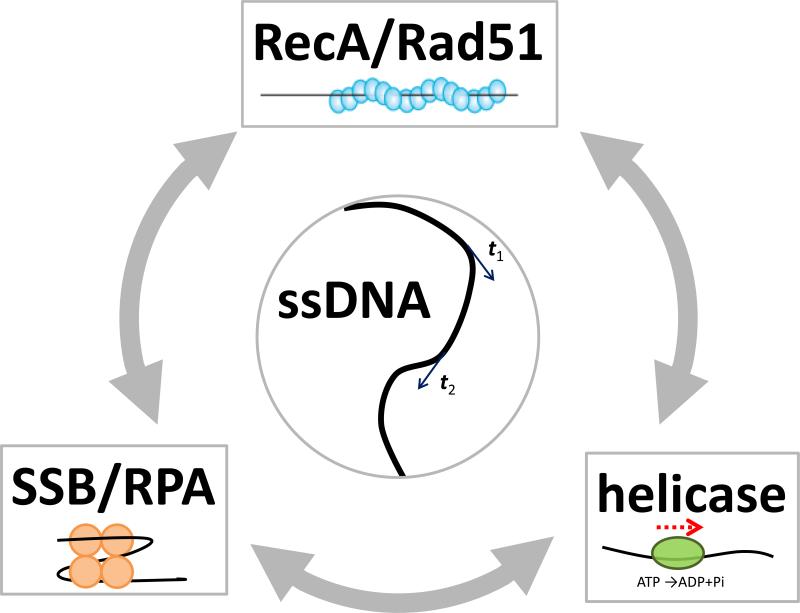
Single-stranded (ss)DNA is an essential transient intermediate in genome maintenance processes such as DNA replication, repair and recombination. In fact, the accumulation of ssDNA as marked by ssDNA-specific proteins is often a sign of trouble as observed in cancer cells. Multitudes of DNA binding proteins associate with ssDNA to carry out their functions and thus compete for access to ssDNA (Figure 1), and precise spatio-temporal coordination of their activities on ssDNA is likely to be essential in maintaining genome integrity.
Figure 1. Proteins on ssDNA.
ssDNA is depicted as a black line. (Center) tangential vectors (t1 and t2) along the contour of ssDNA are correlated if the two points are closer than the persistence length (1-3 nm for ssDNA). (Top) RecA or Rad51 forms a right-handed helical filament on ssDNA. ATP binds to the interface between two adjacent proteins. RecA/Rad51 filaments are negatively regulated by some helicases and can efficiently remove SSB proteins bound to ssDNA during their extension (gray arrows). (Bottom right) A helicase can use ATP hydrolysis to translocate directionally on ssDNA. (Bottom left) SSB/RPA binds and protects ssDNA. For E. coli SSB, ssDNA wraps around the tetrameric protein. SSB can stimulate RecA filament formation by removing DNA secondary structures. The consequence of an encounter between a translocating helicase and an SSB protein is an open question.
Single stranded DNA binding (SSB) proteins are the first responders when ssDNA is generated. They bind selectively and with high affinity to ssDNA, protecting ssDNA from degradation and facilitating recruitment of other proteins to their sites of function (121; 139). During homologous recombination, RecA/Rad51 proteins are recruited to SSB-coated ssDNA to form a filament, which then undergoes strand exchange with a homologous double stranded (ds)DNA(8; 65). DNA helicases are ATP-dependent motor proteins that unwind dsDNA into single strands and translocate on ssDNA to remove DNA-bound proteins such as RecA/Rad51(126). There are many elementary questions about the intermediate steps in the mechanisms of action of these proteins that remain unanswered. The traditional tools used to study these processes have not always been entirely adequate for the given task. For example, high-resolution structural tools provide only incomplete snapshots whereas ensemble biochemical and biophysical tools often do not possess sufficient resolution or clarity due to population averaging.
The ability to manipulate and monitor single biological molecules is revolutionizing modern biological inquiry, allowing us to look at elementary biochemical reactions with unprecedented precision and clarity (40; 60; 138). In this review, we discuss novel insights that single molecule measurements have provided on the properties of ssDNA and some proteins and enzymes that function on ssDNA including SSB/RPA, RecA/Rad51 and helicases, and the interplay among these proteins (Figure 1). There is a growing list of ssDNA-specific proteins beyond what is covered in this review, for example VirE2(21; 39) and MutL(99) which have also been studied at the single molecule level. The diverse single molecule methods that make these studies possible include single molecule fluorescence resonance energy transfer (smFRET)(44), optical tweezers(7), magnetic tweezers(127), tethered particle tracking(33), single fluorophore tracking(118), flow stretching(10) and nanopores (62). In most single molecule studies, DNA or proteins are immobilized to a surface to allow sufficiently long observation times and are often modified with fluorescent labels or other types of tethers. Therefore, it is essential to compare single molecule results with those obtained in bulk solution whenever possible. The information generated from ensemble studies is invaluable in designing and interpreting single molecule experiments.
SINGLE STRANDED DNA
Unlike dsDNA, ssDNA is highly flexible and does not generally form well-defined secondary structures by itself. In the absence of internal base pairing, ssDNA is a random polymer, most commonly modeled as a freely jointed chain or worm-like chain. In the worm-like chain model, angular correlation between tangential vectors, t1 and t2, at two different points along a polymer (Figure 1) is lost once the distance between the two points exceeds the persistence length, Lp. Lp for ssDNA is in the range of 1-3 nm, depending on salt concentration, and is much shorter than that of dsDNA which is about 50 nm. Three single molecule methods have been used to characterize the polymeric properties of ssDNA at the single molecule level. In the first method, a donor fluorophore and an acceptor fluorophore are attached to the ends of a ssDNA and FRET between the two fluorophores reports on the time-averaged distance between the two ends(90). This method can readily provide information on the properties of ssDNA as a function of sequence, ionic conditions, length of ssDNA and temperature (90). In general, ssDNA becomes more compact (higher FRET) as the salt concentration increases, due in part to the partial screening of electrostatic repulsion between the negatively charged phosphates in the backbone chain, in addition to direct binding of cations to the ssDNA due to its polyelectrolyte nature (106). Recently, a microfluidics based smFRET analysis was applied to look at ssDNA conformations under hundreds of different conditions (63). Fluorescence correlation spectroscopy, where the changes in diffusion coefficient are used as a readout, has also been used to examine ssDNA conformations (29).
In a second method, ssDNA is stretched using single molecule mechanical manipulation methods such as optical or magnetic tweezers (25; 141) and atomic force microscopy (17) so that the ssDNA extension is precisely controlled as a function of applied force. Secondary structures formed by internal base pairing within long heteropolymeric ssDNA makes it distinct from a classical self-avoiding polymer especially at low forces (25). Although earlier data were explained using a freely jointed chain model(85; 141), more recent studies are consistent with a worm like chain model (90; 117), which is also true for ssRNA(119).The difference in the polymeric properties of ssDNA vs. dsDNA can also be used to monitor the activities of enzymes that convert dsDNA into ssDNA (e.g., helicases(26), exonucleases(70; 136), displacement synthesis by a reverse transcriptase(80)) or ssDNA into dsDNA (DNA polymerases(72; 85; 141) and DNA annealing proteins (112)). The internal force in ssDNA can be measured by a combination of FRET and magnetic tweezers and it was proposed that ssDNA can be used as a force sensor (125). A third method uses atomic force microscopy to directly image ssDNA deposited on a mica surface to obtain information about flexibility, assuming that mica adsorption does not perturb the structure and captures the projection of its solution conformations(107). Direct imaging approaches have been used for many years in electron microscope studies of dsDNA conformation (34).
ssDNA with variable base composition can form secondary structures due to internal base pairing. Canonical Watson-Crick base pairing stabilizes hairpin structures, typically of limited duplex stem length, and the microsecond/millisecond dynamics of opening and closing have been studied using fluorescence correlation spectroscopy(12). For long stems, the hairpin is almost always in the closed state in solution and a force in the unzipping direction needs to be applied to observe the opening/closing dynamics at equilibrium (78; 140), which can reveal the properties of the transition state. Polyadenylate (both RNA and DNA) can also form secondary structures due to helical stacking of its adenine bases and its interconversion between its disordered and helical states can be slowed down sufficiently inside a biological nanopore so that the structural transitions of ss poly(rA) can be measured in real time via changes in current through the nanopore(76).
SINGLE STRANDED DNA BINDING PROTEINS
SSB proteins are essential in nearly all organisms, playing central roles in genome maintenance (16; 83; 88; 121). SSB proteins bind with high affinity and specificity to ssDNA intermediates to protect them from degradation and destabilize inhibitory secondary structures within the ssDNA. SSB proteins also regulate the activities of other proteins by direct binding to bring them to their sites of action on DNA (121).
SSB proteins from different organisms can differ in structure and assembly state, although all possess oligosaccharide/oligonucleotide binding folds (OB-folds) that contain the ssDNA binding site (91). The bacteriophage T4 gene 32 protein (gp32) (61; 120) and bacteriophage T7 gene 2.5 protein (gp2.5) (51) exist as monomers and dimers, respectively, whereas E. coli SSB is a stable homotetramer (103; 104). The main eukaryotic SSB, called RPA is a heterotrimer (11; 55; 139). Most bacterial SSB proteins are homotetramers containing one OB-fold per monomer (see for example (15; 115; 116)). The mitochondrial SSB proteins from yeast and human (133; 142) are structurally similar to E. coli SSB.
E. coli SSB protein
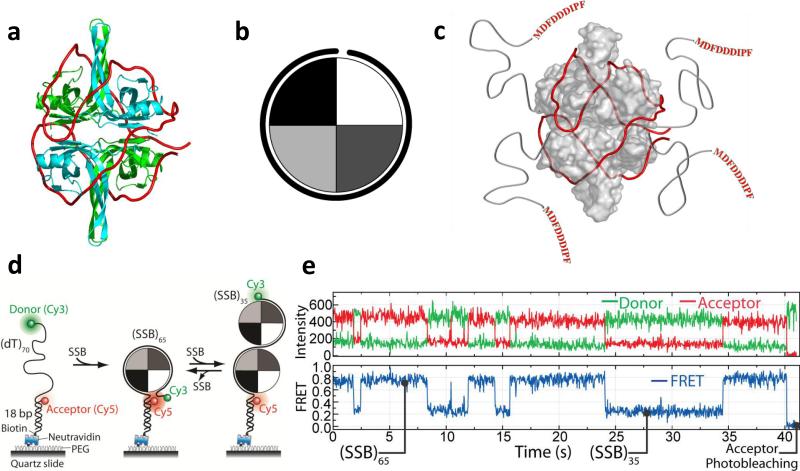
Due to its homotetrameric nature and the resulting four potential ssDNA binding sites E. coli SSB can bind to long ssDNA in multiple modes. Three major ssDNA binding modes, (SSB)35, (SSB)56 and (SSB)65, have been identified where the subscripts denote the average number of ssDNA nucleotides occluded per SSB tetramer (13). The relative stabilities of these binding modes depend on salt concentration, cation and anion type and valence (13; 81) as well as protein to DNA ratio (14; 20; 41; 113). In the (SSB)65 mode (Fig. 2a), ~65 nucleotides of ssDNA wrap around all four subunits of the tetramer. In the (SSB)35 mode, favored at low salt and high SSB to DNA ratios, ~35 nucleotides of ssDNA interact with an average of two subunits of the tetramer (13; 41; 81). The electron densities for the four C-terminal tails (residues 113-177) are not observable in any of the crystal structures (103; 104), suggesting that these are disordered. The last 8-10 amino acids in these C-termini serve as the primary sites to which more than a dozen other proteins bind to SSB (121) (see Fig. 2c).
Figure 2. Structure and dynamics of E. coli SSB/ssDNA complexes.
(a) Model of an SSB tetramer bound to 65 nt of ssDNA (SSB)65 mode) (red) based on a crystal structure(104). (b) Cartoon depiction of an SSB tetramer wrapped by ~ 65 nt of ssDNA. (c) SSB has four C-terminal extensions that bind other proteins. (d) smFRET scheme for monitoring SSB binding to a 70 nt ssDNA. When a single SSB tetramer binds in its (SSB)65 mode, the donor and the acceptor are in close proximity, yielding high FRET. When an additional SSB tetramer binds to the same ssDNA with both tetramers in the (SSB)35 mode, a lower FRET value results. The DNA is tethered to a polymer-passivated surface, and extensive comparison to bulk solution data showed that fluorescence labeling and surface tethering did not perturb the reaction. (e) smFRET time traces showing switching between (SSB)65 and (SSB)35 modes induced by dissociation and binding of a second SSB tetramer (Figures adapted from Ref. (113).
Single molecule force studies of SSB-ssDNA interactions
Single molecule force spectroscopy has been used to study a number of proteins that bind either dsDNA, ssDNA or both (24; 46; 47; 79; 97; 98; 123; 124), focusing on the ability of these proteins to destabilize duplex DNA.
Stretching of dsDNA molecules with optical tweezers has been used to study the kinetics and thermodynamics of gp32 and gp2.5 binding to ss and dsDNA (97; 98; 123; 124 ). In these experiments the force needed to melt dsDNA was monitored as a function of protein concentration and pulling rate and models were used to estimate site sizes, association rates and equilibrium constants for protein binding to ss and dsDNA. Single molecule magnetic tweezers experiments were used to study the ability of gp32 and E. coli SSB proteins to prevent rezipping of two complementary DNA strands (46; 47). It appears that both proteins act by binding to ssDNA and inhibiting reformation of duplex DNA (rezipping) (46). A recent study combining smFRET and optical tweezers showed that ssDNA unraveling from an E. coli SSB tetramer begins at forces ~1 pN, and dissociation of SSB from the ssDNA occurs in the range from 7 to 12 pN (148).
The eukaryotic replication protein A (RPA) is a heterotrimer that can potentially use its multiple ssDNA sites (up to 5 OB folds) to regulate its ssDNA binding activity (11; 55; 68; 139). The ability of human RPA (hRPA) to bind to transient ssDNA bubbles formed within negatively supercoiled dsDNA has been studied using magnetic tweezers (24). At constant force (0.5 pN) the association kinetics are strongly dependent on salt with binding rate decreasing five-fold as [NaCl] increases from 30 to 90 mM. No binding is observed above 120 mM NaCl, consistent with the idea that DNA breathing (ssDNA bubble formation), required for hRPA nucleation, is suppressed at this salt concentration. The association and dissociation kinetics were also studied as a function of supercoiling leading to the conclusion that hRPA can relieve the torsional stress on negatively supercoiled dsDNA.
smFRET studies E. coli SSB-ssDNA binding modes
At moderate to high NaCl concentrations SSB binds (dT)70 in its (SSB)65 mode forming a 1:1 molar complex in which ssDNA wraps around the tetramer interacting with all 4 subunits such that two ends of the ssDNA are in close proximity (shown in Fig. 2a and c and in cartoon form in Fig. 2b). At lower NaCl concentrations (< 0.2M) a single (dT)70 can bind two SSB tetramers each in the (SSB)35 binding mode. The end to end distance of the (dT)70 differs for each of these complexes as well as free (dT)70. Hence a (dT)70 labeled at each end with a pair of fluorophores (e.g., Cy3 and Cy5) for FRET measurements can be used to study the kinetics of SSB-(dT)70 binding and the transitions between its different binding modes (66; 67; 69; 113) (see Fig. 2d). The fully wrapped (SSB)65 mode is characterized by high FRET values (~0.8, Fig. 2e,) whereas the (SSB)35 mode displays intermediate FRET (~0.2, Fig. 2e). Analysis of smFRET traces of the type shown in Fig. 2e, showed that the interconversion rates between the two modes vary as much as 200-fold for only a four-fold change in NaCl concentration (113). Deletion of the acidic C termini on the SSB tetramer, the sites of binding of several proteins involved in DNA metabolism, shifts the equilibrium towards the (SSB)35 mode, suggesting that interactions of proteins with the SSB C terminus may regulate the binding mode transition and vice versa.
Direct observation of SSB diffusion along ssDNA
SSB proteins are believed to function mainly on the lagging strand during DNA replication, coating the ssDNA template before it is replicated. The fact that SSB proteins and E. coli SSB in particular bind with very high affinity to ssDNA with a very low rate of dissociation would appear to make it difficult for them to be recycled, moved or rearranged on the DNA. However, early ensemble studies (67; 69; 109) suggested that E. coli SSB may be capable of diffusive movement along ssDNA and direct transfer between DNA sites (67).
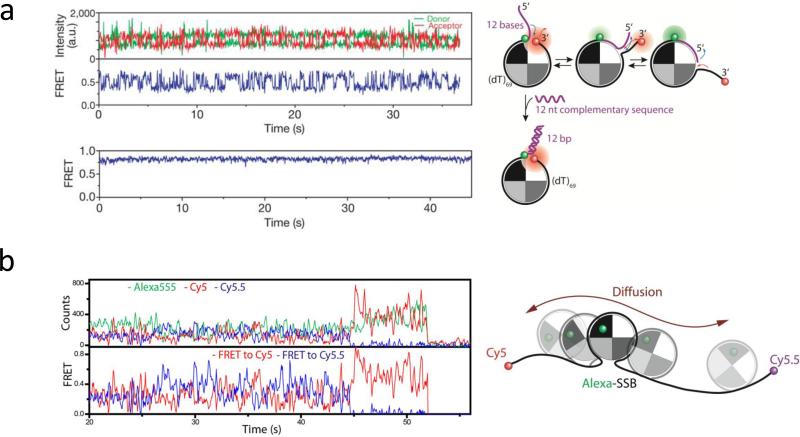
Diffusion along ssDNA of E. coli SSB in its (SSB)65 binding mode has been directly demonstrated using smFRET (114). SSB was pre-bound to ssDNA labeled with donor (Cy3) and acceptor (Cy5) fluorophores separated by 68 nucleotides (all dT), but containing a 12 nucleotide extension (see Fig. 3a). FRET fluctuations in the millisecond time range were observed even in the absence of free SSB in solution indicative of SSB movement over the 80 nucleotide ssDNA region. However, these FRET fluctuations were totally suppressed when the ssDNA region was limited to 68 nucleotides (Fig. 3a), the occluded site size of the SSB tetramer in its (SSB)65 binding mode. As a further demonstration of this diffusion, three color smFRET was employed where an SSB tetramer was labeled on one subunit with a fluorescence donor (one Alexa555 per SSB tetramer), whereas two different acceptor fluorophores, Cy5 and Cy5.5, were positioned at the ends of a (dT)130 ssDNA (Fig. 3b). Rapid and anti-correlated fluctuations in the Alexa555/Cy5 and Alexa555/Cy5.5 FRET efficiencies clearly demonstrated that an SSB tetramer could diffuse from one end of the (dT)130 to the other. Hidden Markov analysis of the data provided an estimate of an apparent step size of 3 nucleotides for SSB tetramer diffusion with a diffusion coefficient of 270 (nucleotides)2 s−1 (37°C).
Figure 3. smFRET fluctuations arising from diffusional migration of E. coli SSB on ssDNA.
(a) –(Top) Rapid fluctuations between several FRET states are observed due to diffusion of an SSB tetramer on an 81 nucleotide ssDNA; (bottom) when the available ssDNA is limited to 69 nucleotides by forming a duplex with the 12 nucleotide extension, the FRET fluctuations are severely reduced reflecting less protein diffusion; (b) – anti-correlated FRET time traces from a three color experiment showing SSB tetramer diffusion (Donor (Alexa555)-labeled SSB bound to a (dT)130 DNA labeled at each end with acceptors (Cy5 and Cy5.5) Figures adapted from Ref. (114).
The recognition that E. coli SSB can diffuse along ssDNA provides an explanation for its ability to transiently destabilize DNA secondary structures such as hairpins. A DNA hairpin within a ssDNA will transiently undergo spontaneous melting of the base pairs at the base of the hairpin. An SSB tetramer can then diffuse onto the transiently melted ssDNA regions and thus prevent reformation of the hairpin region. In fact, it has been known (64) that SSB can facilitate the formation of a stable filament of RecA protein on natural ssDNA (i.e., ssDNA capable of forming hairpin structures), yet the mechanism for this was unclear. smFRET experiments (114) showed that an SSB tetramer bound to a ssDNA ahead of a growing RecA filament could be pushed directionally and facilitate the melting of the hairpin and thus remove the inhibitory effects of the hairpin on RecA filament formation.
Mechanism of SSB diffusion along ssDNA
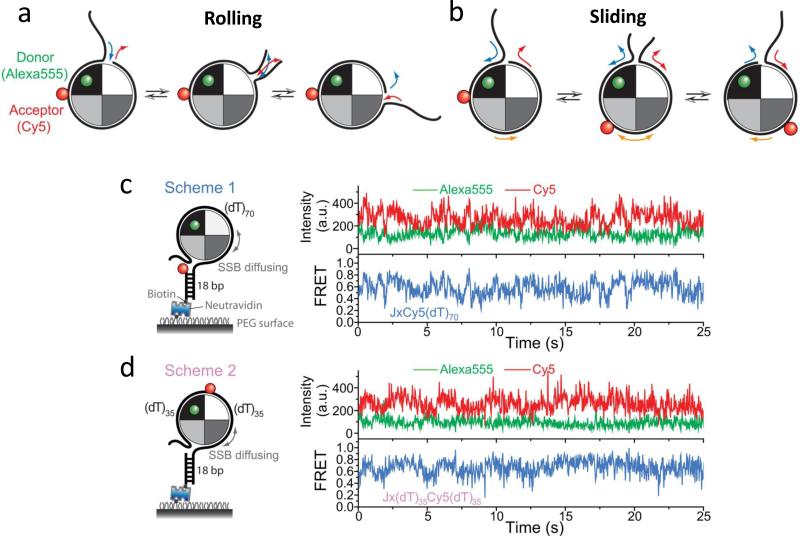
Although the ability of an SSB tetramer to diffuse along ssDNA was clearly demonstrated using single molecule approaches (114), the mechanism by which an SSB tetramer can undergo a rapid migration along ssDNA remained unclear when considering its extensive ssDNA interactions (~65 nt) and high DNA affinity. A rolling mechanism had been suggested (67; 69; 109) that utilizes the closed wrapping of ssDNA around the tetramer (see Fig. 4a), but an alternative possibility was that the ssDNA can “slide” relative to the SSB protein surface (see Fig. 4b).
Figure 4. smFRET studies support a sliding model for SSB diffusion on ssDNA.
(a) -Rolling mechanism for SSB diffusion. One end of the wrapped ssDNA could partially dissociate from the SSB resulting in an available site on the SSB to which the free ssDNA at the other end can bind. There would only be relative movement between the donor (green) on the SSB surface and the acceptor (red) on the ssDNA if the acceptor is near the DNA end, but not the midsection. (b) - Sliding mechanism for SSB diffusion. The entire ssDNA (65 nt) moves relative to the SSB protein surface during sliding, hence relative movement between the donor and acceptor should occur for all acceptor positions on the DNA. (c) and (d) - Representative single-molecule time traces of donor(Alexa555) and acceptor(Cy5) intensities and the corresponding FRET efficiencies show the same level of FRET fluctuations regardless of the acceptor (Cy5) position on the DNA. Figures adapted from Ref. (148).
Zhou et al. (148) used an approach that combined single molecule fluorescence with force measurements to differentiate between a rolling and a sliding mechanism. In these experiments an SSB tetramer labeled with Alexa555 (donor) was bound to a (dT)70 containing a Cy5 (acceptor) attached to either the end (Fig. 4c, scheme 1) or the middle (Fig. 4d, scheme 2). A DNA of this length (5 nucleotides longer than the SSB tetramer occluded site size) provided room for some SSB diffusion (114). If a rolling mechanism were operative, only the end segments of the DNA in scheme 1 would be expected to move relative to the SSB and show FRET fluctuations, whereas the middle regions of the DNA in scheme 2 should show little movement. However, FRET time traces for both experiments showed anti-correlated donor-acceptor fluctuations of similar amplitudes (Fig. 4c and Fig. 4d) strongly suggesting a sliding rather than a rolling model. At the same time no fluctuations were observed for similarly labeled ssDNA substrates with a total length of 40 or 51 nucleotides indicating that SSB diffusion is eliminated if the length of the ssDNA is shorter than the SSB tetramer binding site size (65 nucleotides).
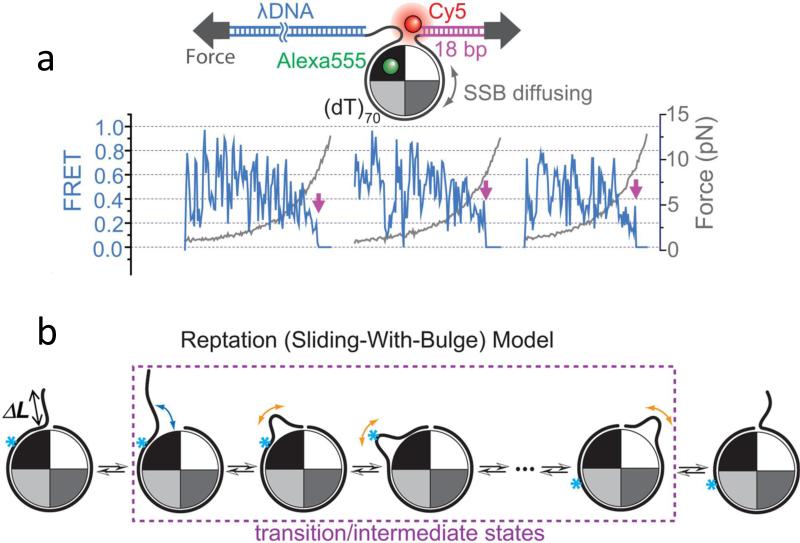
A further test for a sliding mechanism was performed by applying tension to the ends of the ssDNA in order to gradually disrupt the ssDNA wrapping around the SSB tetramer that is required if a rolling mechanism is operative. However, the diffusion-induced FRET fluctuations persisted even up to ~5 pN forces (Fig. 5a) where the ssDNA unwrapping is nearly complete. Further analysis suggested that a “reptation” mechanism is likely for SSB sliding. In this mechanism (see Fig. 5b) a thermally activated ssDNA bulge forms at one end of the ssDNA that enters the SSB tetramer and this bulge propagates along the SSB surface via a random walk until it either returns to where it started rendering no net movement, or emerges on the other side, leading to diffusion of the SSB tetramer by one step (3-4 nucleotides) along the ssDNA. Interestingly, the binding of RecO protein to the C-terminal tails of SSB did not eliminate the ability of SSB to diffuse along the ssDNA although it does reduce the diffusion rate.
Figure 5. A combined smFRET-force experiment supports a reptation mechanism for SSB sliding on ssDNA.
(a) - SSB diffusion persists even with ssDNA under tension. SSB diffusion-induced FRET fluctuations continue for forces up to 5 pN. Magenta arrows indicate SSBf dissociation events. (b) - a reptation (sliding-with-bulge) mechanism for SSB diffusion is proposed to occur by nucleating a thermally activated bulge that propagates via a random walk around the SSB surface. The bulge diffuses back and forth over the entire SSB surface, and if it emerges on the other side, the SSB moves along the ssDNA by one step (~3 nucleotides). The arrows represent the DNA movements and the cyan asterisk represents a single nucleotide position on the DNA. The asterisk-marked position on the ssDNA will move relative to the protein surface. Figures adapted from Ref. (148).
RecA/Rad51
In bacteria, RecA forms a right-handed helical filament on ssDNA generated as an intermediate during repair of DNA breaks. The filament searches for a homologous dsDNA and catalyzes a strand exchange reaction where the original ssDNA within the RecA filament replaces the homologous strand in the duplex(8; 65). RecA-mediated recombination is a complex process with several transient reaction intermediates and single molecule assays are well-poised to probe such complex reactions. Most single molecule studies of RecA and its eukaryotic homolog, Rad51, have been carried out using dsDNA(23; 37; 48; 50; 75; 108) despite the fact that the physiologically relevant RecA/Rad51 filament assembles on ssDNA. This is partly because of the technical difficulty in preparing long ssDNA required for such studies and also because RecA filament formation on dsDNA recapitulates most properties of the ssDNA filament because RecA interacts primarily with one of the two strands in the duplex.
Filament dynamics
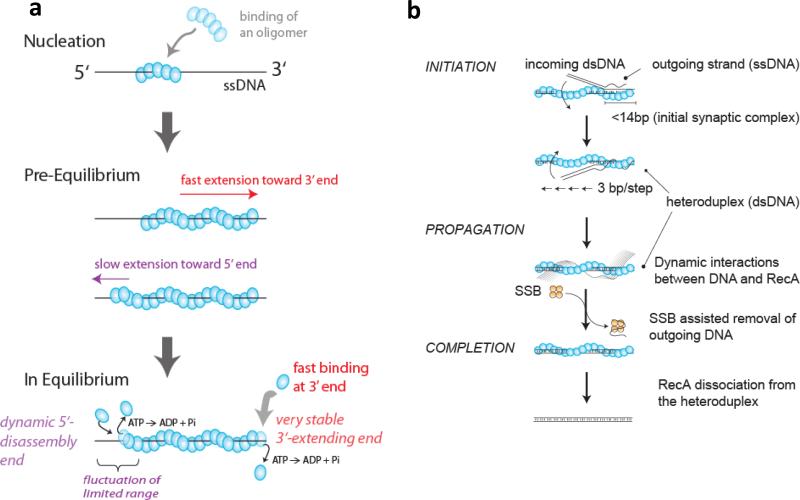
The rate-limiting step in RecA/Rad51 filament assembly is the nucleation process (Fig. 6a). Basic details such as the number of protein monomers that are needed to nucleate the filament, and also the unit of filament extension and disassembly, remained controversial. In recent smFRET experiments using various lengths of poly(dT), it was found that ssDNA of at least 17 nt is required for RecA filament formation(59). Combined with the binding stoichiometry of 3 nt per RecA monomer, it was proposed that 4-5 monomers form the nucleation cluster. Another study visualized the initial assembly of fluorescently labeled RecA protein on long dsDNA (~50 kbp)(37) and found that the initiation rate increases with the 4th power of the RecA concentration, also suggesting cooperative binding of 4-5 monomers is needed to nucleate a filament. Subsequent single molecule studies of both RecA and Rad51 also showed the nonlinear dependence on concentration, consistent with 2-5 monomers in a nucleation cluster(50; 89; 134). In the crystal structure of a ssDNA-bound RecA filament that was finally determined in 2008, researchers used the trick of covalently fusing 4 or 5 RecA monomers which gave rise to a stable crystal in complex with ssDNA, also supporting the notion that 4-5 RecA monomers are needed, and probably sufficient to form a filament-like structure(18). RecA filament assembly on short ssDNA has also been used to demonstrate novel single molecule confinement technologies such as a porous nanocontainer(22) and an electrokinetic trap (30). Confining RecA and ssDNA inside a small 100 nm diameter vesicle showed that RecA filament rebinding to ssDNA after its dissociation is two orders of magnitude faster than de novo nucleation, suggesting that when a RecA nucleation cluster dissociates from the DNA, it does not immediately break apart into monomers(22).
Figure 6. Dynamics of RecA filament formation and strand exchange reactions.
(a) RecA filament formations on ssDNA is nucleated by the binding of a RecA oligomer, likely between 4-5 monomers(37; 59), and is extended rapidly in monomer un units mainly in the 3’ direction with a 10 times slower extension in the 5’ direction(59). In the steady state, RecA monomers can dissociate from both ends as monomers upon ATP hydrolysis(59). (b) RecA-ssDNA filament binds to a homologous dsDNA forming an initial synaptic complex (< 14 bp in length), then propagates rapidly in 3 bp increments while undergoing base pair exchange. The outgoing ssDNA displays dynamic interactions with the resulting RecA-dsDNA filament and its removal is facilitated by SSB. Finally, RecA dissociates from the heteroduplex product, completing the reaction (Adapted from Ref. (105)).
Once a filament nucleates, the RecA filament grows primarily in the 5’ to 3’ direction while ATP hydrolysis causes RecA filaments to shrink from the 5’ end (Fig. 6a) (6). However, the unit of extension was not known (monomer, dimer or higher order assemblies). smFRET data and hidden Markov analysis (87) provided direct evidence that the unit for extension and shrinkage is one monomer. Moreover, it was shown that the filament directionality is caused by the 10 fold difference in the monomer binding rates at the two ends whereas the dissociation rate is the same(59). A single color fluorescence detection experiment using protein-induced fluorescence enhancement also supports RecA binding to and dissociation from a filament as a monomer (54). Although Rad51 is expected to behave in similar ways to RecA, there is presently no consensus in the directionality of Rad51 filament formation(49) but recent studies are more consistent with the 3’ to 5’ direction, which would be opposite to that of RecA(4; 110). Indirect evidence based on Rad51 filament dissociation from long dsDNA supports the notion that the unit for disassembly is a monomer(135), and magnetic tweezers studies of Rad51 filament formation directly detected monomeric extension through a stepwise twisting of the DNA(5). Although the emerging consensus supports the RecA/Rad51 monomer as the unit of filament growth and shrinkage, some single molecule studies suggested indirectly that the extension occurs in oligomers (89; 134).
RecA/Rad51 filaments undergo conformational changes during the ATP binding and hydrolysis cycle without protein dissociation and rebinding, and these changes have been visualized at the single DNA level by following the lengthening and shortening of dsDNA(50; 108) or ssDNA(59). In general, ADP filaments are shorter than ATP filaments.
Homology search
Once a RecA/Rad51 filament forms on ssDNA, it needs to find its target, the homologous sequence within a dsDNA (Fig. 6b). This process has drawn the interest of many researchers because the task is akin to finding a needle in a haystack and is further complicated by the mismatch in length: ATP-bound filaments are about 50% longer than dsDNA of the same number of base pairs. One attractive possibility is that the filament may scan the dsDNA by a sliding mechanism until the homologous sequence is found. In fact, many proteins are known to slide on dsDNA or ssDNA which may be important for their function. However, no such sliding has yet been observed during homology search by RecA/Rad51 and an experimental test of mechanisms for homology search does not support sliding(1). In a recent smFRET study measuring strand exchange of short DNA (tens of base pairs in length), it was found that the homology search was essentially complete within the time resolution of the experiment, 30 ms(105) but homology searches on long DNA have not been observed directly.
Strand exchange
Once a region of homology is located, the strand exchange reaction ensues whereby the ssDNA forms base pairs with its homologous strand within the duplex. Magnetic tweezers studies followed the real time progress of the reaction via the change in overall extension of the DNA (35; 132) yielding a key insight that the synapse size remains constant (~80 bp) during the reaction. This observation suggested that as additional segments of the incoming ssDNA are incorporated into the heteroduplex product, the RecA filament concurrently dissociates via ATP hydrolysis (132). A more recent smFRET study(105) examined the reaction with a higher spatio-temporal resolution (a few bp and 30 ms resolution) and provided kinetic evidence that the initial synapse between an incoming filament and dsDNA is about 14 bp in length and the base pairing exchange to form a longer joint molecule occurs in 3 bp increments (Fig. 6b), which matches the number of ssDNA nucleotides bound by each RecA monomer as seen in the crystal structure. The bp exchange propagated at a surprisingly high rate of ~ 180 bp/s, which would explain why the presumed 3 bp steps could not be seen. Although the reaction probed was at a local level of less than 20 bp, this was much faster than the 2 bp/s determined from the magnetic tweezers experiments (132). The difference may lie in the length scale: on a much longer length scale, the overall reaction slowed down substantially(105), potentially due to initial synapse formation occurring at multiple positions as suggested by a four-color smFRET study(71)
HELICASES
Helicases participate in virtually all cellular processes that involve DNA and RNA metabolism (56; 126). The most fundamental activity for all DNA helicases is ‘translocation’, the ability to move directionally along nucleic acids as a motor protein. Translocation is essential for many critical functions such as DNA unwinding, protein displacement form DNA, Holliday junction branch migration, and chromatin remodeling(84; 126).
Helicases/translocases are motor proteins since translocation is powered by ATP hydrolysis. For example, the non-hexameric helicases Rep, UvrD and PcrA translocate with 3’ to 5’ directionality along ssDNA(84). As a result, unwinding of double stranded (ds)DNA is facilitated if it has a 3’ ssDNA tail, although translocation alone is not always sufficient for DNA helicase activity(84). Most helicases that function to unwind DNA at a replication fork are hexameric (101). These ring-shaped helicases have six binding sites for ATP and their intersubunit coordination adds another layer of complexity that further challenges our abilities to understand their mechanisms. Rapid developments in single molecule tools have contributed to our understanding of how these enzymes catalyze DNA unwinding (10; 26; 28; 45; 58; 102; 128; 145). Here, we focus on several interesting findings concerning ssDNA translocation of DNA helicases and how translocation activity is coupled to DNA unwinding.
Repetitive helicases
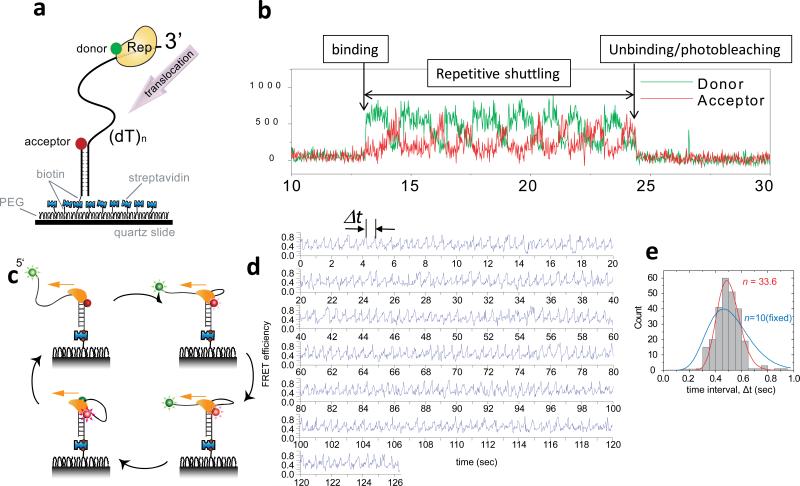
In 2005, smFRET analysis of the E. coli Rep helicase, labeled with Cy3, moving on a ssDNA labeled with Cy5 (Fig. 7a), showed that the monomeric translocase can repetitively translocate on the same stretch of ssDNA without full dissociation and rebinding (92) (Fig. 7b). This repetitive shuttling may keep the ssDNA free of unwanted proteins such as RecA and prevent potentially unregulated recombination events. In further studies this repetitive behavior was observed during translocation or unwinding on both RNA and DNA for helicases from various organisms including bacterial, human and viral. Examples include PcrA translocation on ssDNA(100), human BLM unwinding of dsDNA(144) and HCV NS3 unwinding dsDNA(93). In addition, the human viral RNA sensor RIG-I, which possesses an RNA helicase domain, can translocate repetitively on dsRNA, the first example of a helicase-like motor protein moving on dsRNA in the absence of unwinding(94). These findings show an amazing capability of these enzymes to undergo movement along and transfer between different positions along a nucleic acid substrate without full dissociation. There are exceptions however. No evidence of repetition has been found for hexameric helicases so far, and archaeal XPD helicase did not show repetition during ssDNA translocation(52). Table 1 summarizes the helicases for which repetitive translocation has been observed and their potential biological functions.
Figure 7. Repetitive ssDNA translocation of Rep and PcrA.
(a) A donor-labeled Rep monomer translocates on an acceptor-labeled DNA in the 3’ to 5’ direction. (b) Initial binding of Rep is detected as a sudden increase in fluorescence. This is followed by a gradual increase in the acceptor intensity (red) and a concomitant decrease in the donor intensity (green) due to Rep translocation on ssDNA. The protein then snaps back to the initial low FRET state and repeats the cycle until protein unbinding or photobleaching of the donor terminates the fluorescence signal. (c) A cartoon showing how a PcrA monomer can anchor itself to a ds/ssDNA junction and use its ssDNA translocation activity to reel in a 5’ ssDNA tail. Once it runs off the tail end it can re-initiate the repetitive looping cycle from the junction. (d) Representative smFRET time traces of repetitive ssDNA looping (256 cycles) induced by a single molecule of PcrA. Δt denotes the time interval of each cycle. (e) Δt histogram from a single PcrA molecule showing 256 cycles of looping. A fit to the Gamma distribution gives the number of hidden steps, n=33.6, for 40 nt ssDNA (red), supporting a single nt kinetic step size. A forced fit with a fixed n value of 10 to mimic a 4 nt kinetic step size gives a much poorer fit (blue). Figures were adapted from Refs (92; 100).
Table 1.
Helicases showing Repetitive translocation/unwinding
| origin | Repetition observed | Repetition mechanism | Functional implications | |
|---|---|---|---|---|
| Rep | Bacterial | ssDNA translocation | Blockade-induced 3’ capture | Prevention of RecA filament formation |
| UvrD | Bacterial | ssDNA translocation | Duplex-anchored looping and release | Removal of RecA filament |
| PcrA | Bacterial | ssDNA translocation | Duplex-anchored looping and release | Removal of RecA filament |
| NS3 | Viral | dsDNA unwinding | Holding on to unwound strand | Unknown |
| RIG-I | Human | dsRNA translocation | unknown | Viral RNA recognition and signaling |
| BLM | Human | dsDNA unwinding | Strand switching | Unknown |
A helicase reels in DNA
PcrA is a 3’ to 5’ helicase so most previous studies were performed on duplex DNA with a 3’ ssDNA tail. However, when PcrA and ATP were added to a surface-tethered duplex DNA with a 5’ ssDNA tail, PcrA translocated in the 3’ to 5’ direction while maintaining contact with the duplex DNA, thereby reeling in the 5’ ssDNA tail and extruding a ssDNA loop (100) (Fig. 7c). This activity could be seen as a gradual increase in FRET between the donor and acceptor fluorophores linked to the ends of ssDNA followed by a sudden FRET decrease, and many repetitions of such cycles were observed (Fig. 7d). This structure-specific binding and translocation-coupled looping resulted in efficient dismantling of RecA filaments on ssDNA even at nanomolar PcrA concentrations. A similar binding to a 5’ tailed dsDNA and translocation-coupled ssDNA looping, although less robust, was also observed for UvrD (131). Despite the similarity in sequence and structure, the repetitive translocation mechanism for PcrA appears different from that of Rep, which may be due to the differences in affinities for the duplex DNA.
ssDNA Translocation mechanism: step size
An understanding of the mechanism of ssDNA translocation requires knowledge of the step-size of the translocating enzyme. There have been several definitions of step sizes (84). The chemical step size is the number of nucleotides translocated per ATP hydrolyzed, whereas the kinetic step size (2) is defined as the average number of nucleotides moved between repeated rate-limiting steps in the translocation cycle. A chemical translocation step size of 1 nucleotide per ATP has been directly measured for PcrA (27), UvrD (130) and BLM (43) and is consistent with inferences from structural data (95). Basically, the enzyme can be viewed as an inchworm with two moving parts, i.e., two RecA-like domains. When an ATP binds to the crevice between the two domains, the inchworm contracts. When the product of ATP hydrolysis is released, the inchworm expands. If the relative DNA binding affinities of the two RecA-like domains can be precisely coordinated during contraction and expansion, one can achieve directional translocation. Such coordination was demonstrated computationally based on the PcrA-DNA co-crystal structures with and without ATP(146) but thus far no experiments have resolved these single nucleotide steps directly during translocation. Ensemble transient kinetic studies have led to the proposal that UvrD and PcrA monomers translocate with a kinetic step size of ~ 4 nt (96; 130). Combined with the chemical step size of 1 ATP/nucleotide translocated it was proposed that PcrA and UvrD translocate via a non-uniform stepping mechanism where it moves about 4 nt very rapidly using 1 ATP per nt, then pauses momentarily before taking the next four steps(130).
The kinetic step size could be examined using single molecule methods for PcrA due to its unusual propensity to undergo multiple rounds of repetitive looping during translocation. Since the looping repeated hundreds of times without full dissociation of PcrA (Fig. 7d), a histogram of time periods Δt of each cycle, could be compiled from one molecule only (Fig. 7e). By fitting the Δt histogram to the Gamma distribution, (Δt)n–1e–kΔt, Park et al could determine n, the number of steps needed to complete one cycle of translocation over N nt of ssDNA, and k, the rate of each step(100). n values exceeding 30 were obtained when N was 40, supporting a kinetic step size smaller than 1.5. Interestingly, a significant variation in the values of k was observed between single PcrA molecules that persisted during the observation time of several minutes (termed ‘static disorder’). As a result, the Δt histogram built from multiple single molecules is significantly broader, causing a decrease in the apparent n value, leading to an inflation of the kinetic step size to ~ 3 nt. Therefore, it is possible that the apparent larger kinetic step size determined from analysis of ensemble kinetic studies, which explicitly assumed that all enzymes have the same reaction rate, is an overestimation due to the presence of ‘static disorder’. Future ensemble studies need to consider the consequences of molecular heterogeneity which appears ubiquitous in single molecule observations. It should be noted, however, that the effect of rate heterogeneity on the estimation of a kinetic step size should be negligible if the true number of steps, n, is small, for example less than 7 or so. This is because the Δt histogram is already broad even without heterogeneity and the effect of molecular heterogeneity on the relative width becomes insignificant. Hence, it remains that reports of kinetic step sizes larger than unity performed on short DNA substrates need further tests using high-resolution single-molecule tools.
Coupling between translocation and unwinding
How is the translocation activity coupled to DNA unwinding? ssDNA translocation activity alone is not necessarily sufficient for DNA unwinding as demonstrated for Rep, UvrD and PcrA helicases (84). One classification of unwinding mechanisms is as either passive or active (82) (9). By definition, a purely passive helicase does not interact with the duplex DNA but rather uses its directional translocation activity to stabilize the transient ssDNA formed through thermal fraying of the duplex end and thus prevent reannealing of the open base-pairs. In contrast, an active helicase participates in the destabilization of the DNA duplex. As a result, a fully active helicase should be able to unwind a dsDNA substrate as fast as it translocates on ssDNA.
Recent single-molecule mechanical studies examined this issue and found that HCV NS3 (19) and E. coli RecQ helicases (86) are mostly active, T4 hexameric helicase mostly passive (77) and T7 hexameric helicase somewhere in between(58). In these studies, unwinding rate as a function of applied force was a main observable and its comparison to the translocation rate was a critical element of the mathematical models used. Although ssDNA translocation itself was not directly observed, the available data indicate that translocation rate is independent of the force applied on ssDNA. A potential factor that may influence the coupling between translocation and unwinding is the interaction between the helicase and the excluded strand, i.e. the strand that is complementary to the tracking strand on which the helicase translocates. smFRET studies revealed dynamic interactions between the excluded strand and the archael MCM helicase(111), which likely involves ssDNA wrapping on the surface of this hexameric enzyme(38), akin to ssDNA wrapping around heptameric Rad52 (42; 53). Interactions with the excluded strand were also reported for E. coli hexameric helicase DnaB(36).
Helicase conformational changes
Various structural changes, large and small, have been observed in crystal structures of helicases but how binding of DNA controls these movements and ATP (and its hydrolysis) during helicase function are only beginning to emerge(57; 74). Thus far, two types of helicase conformational changes have been visualized at the single molecule level. The first is movement of a regulatory subdomain called 2B in Rep, UvrD and PcrA. The 2B subdomain has been observed in two major orientations called open and closed in various crystal structures and subsequently in ensemble studies (summarized in Ref. (57)) and functional roles of these conformations remain controversial (57; 74). In smFRET studies, the 2B and another domain called 1B were stochastically labeled with a donor and an acceptor such that the closed form shows high FRET and the open form low FRET. During repetitive shuttling by Rep, the 2B subdomain was observed to close gradually when a Rep monomer translocated along ssDNA toward a physical blockade such as streptavidin or a dsDNA that it cannot unwind as a monomer(92). In contrast, the monomeric PcrA translocase maintains an open conformation while it is reeling ssDNA on a 5’-tailed DNA(100), suggesting that the 2B subdomain can access different conformations that affect its function. Second, relative movements between two RecA-like domains were visualized also using smFRET for an RNA helicase(129). Interestingly, the closure of the crevice between the two domains required simultaneous binding of an RNA strand and an ATP, showing cooperativity.
INTERPLAY AMONG PROTEINS
RecA vs helicase
DNA helicases can function as anti-recombinases, by removing the recombinases (Rad51 and RecA) from ssDNA (4; 137). Repetitive shuttling of a helicase on the same stretch of ssDNA is an attractive mechanism to enhance the anti-recombinase activity because a single binding event can result in an efficient localization of the helicase to a DNA structure and can keep the region clear of recombinases. smFRET studies indeed showed that a Rep translocase shuttling on ssDNA can prevent RecA filament formation(92) and that a PcrA translocase reeling in a 5’ ssDNA tail can remove a pre-formed RecA filament(100). In these studies, it was critical to include free RecA in solution to examine helicase-driven RecA removal because a RecA filament can spontaneously disassemble upon ATP hydrolysis. A curious observation that an ATPase-deficient mutant of PcrA can remove a RecA filament from ssDNA may be attributed to the fact that in their studies free RecA was not included(3).
SSB vs. RecA
smFRET data showed that a growing RecA filament can push SSB along ssDNA very efficiently, and in fact at the speed of filament growth, showing that a bound SSB does not present a significant barrier(59). The demonstration of SSB diffusion on ssDNA provides an elegant mechanism wherein a small stretch of ssDNA freed up by SSB diffusion might allow RecA monomer binding at the end of an extending RecA filament(114). In this way, RecA filament growth can rectify a random walk by SSB, making SSB repositioning and removal much more rapid.
Helicase vs. SSB
In principle, a similar rectification of a diffusing SSB may apply to a translocating helicase. However, this has not been experimentally tested yet for E. coli SSB. For an archael RPA and an XPD helicase, single molecule fluorescence analysis indicated that a translocating helicase may bypass an RPA bound to ssDNA without full dissociation of RPA(52). Further studies are necessary to test if this type of interplay operates for other helicase/SSB pairs and to understand the mechanism of bypass or removal. On the methodological side, accumulation of fluorescently labeled SSB has been used to monitor helicase unwinding activities from single DNA molecules (31).
FINAL THOUGHTS
Observing movements on long ssDNA
Technical difficulties have limited direct fluorescence imaging of proteins on DNA to dsDNA or short ssDNA. In particular, single protein movements on ssDNA have been imaged almost exclusively using smFRET. However, FRET is insensitive to distances beyond about 10 nm and, for this reason, is not adequate for studies of protein movements on longer ssDNA. Because thousands of nucleotides of ssDNA can be generated during DNA replication and repair in vivo, there is a need to develop methods that can image multiple proteins on a long ssDNA as has been demonstrated for proteins on dsDNA(32). Recent technical developments in multi-color fluorescence imaging in combination with optical tweezers should facilitate research in this direction and should enable visualization of the consequences of an encounter between two diffusing SSB tetramers or between a translocating helicase and an SSB protein.
Proteins on G-rich DNA
G-rich sequences can form various G-quadruplex structures in vitro and in vivo. There are many single molecule studies that revealed the conformational diversity and mechanical stabilities of G-rich DNA (21; 73; 122; 143; 147). Many proteins that can recognize G-rich DNA in its single stranded form or in its folded G-quadruplex form have been identified. Because G-rich DNA displays dynamic transitions between the folded and disordered forms which may be bound by proteins such as SSB/RPA, RecA/Rad51 and helicases, it will be exciting to probe protein movements on G-rich DNA.
SUMMARY POINTS.
-Wealth of novel insights is emerging from single molecule studies of proteins that function on single stranded DNA.
-SSB protein's diffusion on single stranded DNA was directly observed and its sliding-via-reptation mechanism was proposed.
-SSB protein's diffusion can melt secondary structures transiently and provide a mechanism for rapid reposition and removal by other proteins moving on DNA.
-Single molecule mechanical studies have revealed the detailed kinetic mechanism of SSB and RecA/Rad51 proteins binding and dissociation from double stranded and single stranded DNA.
-Single molecule studies revealed the size of the nucleation cluster for RecA/Rad51 filaments.
-Emerging consensus is that RecA/Rad51 filaments extend and shrink in monomeric units.
-Single molecule studies have revealed repetitive movements of helicases on the same stretch of DNA.
-Molecular heterogeneity can mask the true kinetic step size of DNA translocation or unwinding.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (GM030498, GM045948, GM065367), the National Science Foundation (0822613 and 0646550). We thank all members of the Ha and the Lohman lab for their input.
Footnotes
Acronyms
FRET: Fluorescence (Förster) resonance energy transfer
SSB: Single stranded DNA binding protein
RPA: Replication Protein A
smFRET: FRET at the single molecule level
dsDNA: double stranded DNA
ssDNA: single stranded DNA
Key Terms
Helicase translocation: ATP powered directional motion of a helicase protein on DNA or RNA lattice
Kinetic step size: the average number of nucleotides moved between repeated rate-limiting steps in the translocation cycle
LITERATURE CITED
- 1.Adzuma K. No sliding during homology search by RecA protein. J Biol Chem. 1998;273:31565–73. doi: 10.1074/jbc.273.47.31565. [DOI] [PubMed] [Google Scholar]
- 2.Ali JA, Lohman TM. Kinetic Measurement of the Step Size of DNA Unwinding By Escherichia Coli Uvrd Helicase. Science. 1997;275:377–80. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 3.Anand SP, Zheng H, Bianco PR, Leuba SH, Khan SA. DNA helicase activity of PcrA is not required for the displacement of RecA protein from DNA or inhibition of RecA-mediated strand exchange. J Bacteriol. 2007;189:4502–9. doi: 10.1128/JB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–15. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arata H, Dupont A, Mine-Hattab J, Disseau L, Renodon-Corniere A, et al. Direct observation of twisting steps during Rad51 polymerization on DNA. Proc Natl Acad Sci U S A. 2009;106:19239–44. doi: 10.1073/pnas.0902234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arenson TA, Tsodikov OV, Cox MM. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J Mol Biol. 1999;288:391–401. doi: 10.1006/jmbi.1999.2705. [DOI] [PubMed] [Google Scholar]
- 7.Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. Observation of a single-beam gradient force optical trap for dielectric particles. Optics Lett. 1986;11:288–90. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 8.Bell CE. Structure and mechanism of Escherichia coli RecA ATPase. Mol Microbiol. 2005;58:358–66. doi: 10.1111/j.1365-2958.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 9.Betterton MD, Julicher F. A motor that makes its own track: helicase unwinding of DNA. Phys Rev Lett. 2003;91:258103. doi: 10.1103/PhysRevLett.91.258103. [Theoretical framework for understanidng the active and passive mechanisms of DNA unwinding by helicases.] [DOI] [PubMed] [Google Scholar]
- 10.Bianco PR, Brewer LR, Corzett M, Balhhorn R, Yeh Y, et al. Processive translocation and DN A unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–8. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 11.Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Bonnet G, Krichevsky O, Libchaber A. Kinetics of Conformational Fluctuations in DNA Hairpin-Loops. Proc Natl Acad Sci U S A. 1998;95:8602–6. doi: 10.1073/pnas.95.15.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 14.Bujalowski W, Overman LB, Lohman TM. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. J Biol Chem. 1988;263:4629–40. [PubMed] [Google Scholar]
- 15.Chan KW, Lee YJ, Wang CH, Huang H, Sun YJ. Single-stranded DNA-binding protein complex from Helicobacter pylori suggests an ssDNA-binding surface. J Mol Biol. 2009;388:508–19. doi: 10.1016/j.jmb.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 16.Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–36. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 17.Chen WS, Chen WH, Chen Z, Gooding AA, Lin KJ, Kiang CH. Direct observation of multiple pathways of single-stranded DNA stretching. Phys Rev Lett. 2010;105:218104. doi: 10.1103/PhysRevLett.105.218104. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–4. doi: 10.1038/nature06971. [First RecA crystal structures in complex with DNA (both single and double standed DNA).] [DOI] [PubMed] [Google Scholar]
- 19.Cheng W, Dumont S, Tinoco I, Jr., Bustamante C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc Natl Acad Sci U S A. 2007;104:13954–9. doi: 10.1073/pnas.0702315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc Natl Acad Sci U S A. 1982;79:5803–7. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu JF, Chang TC, Li HW. Single-molecule TPM studies on the conversion of human telomeric DNA. Biophys J. 2010;98:1608–16. doi: 10.1016/j.bpj.2009.12.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cisse I, Okumus B, Joo C, Ha T. Fueling protein DNA interactions inside porous nanocontainers. Proc Natl Acad Sci U S A. 2007;104:12646–50. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conover AJ, Danilowicz C, Gunaratne R, Coljee VW, Kleckner N, Prentiss M. Changes in the tension in dsDNA alter the conformation of RecA bound to dsDNA-RecA filaments. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Vlaminck I, Vidic I, van Loenhout MT, Kanaar R, Lebbink JH, Dekker C. Torsional regulation of hRPA-induced unwinding of double-stranded DNA. Nucleic Acids Res. 2010;38:4133–42. doi: 10.1093/nar/gkq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dessinges MN, Maier B, Zhang Y, Peliti M, Bensimon D, Croquette V. Stretching single stranded DNA, a model polyelectrolyte. Phys Rev Lett. 2002;89:248102. doi: 10.1103/PhysRevLett.89.248102. [DOI] [PubMed] [Google Scholar]
- 26.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci U S A. 2004;101:6439–44. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: Measurement of step size and translocation speed. Biochemistry. 2000;39:205–12. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 28.Dohoney KM, Gelles J. Chi-Sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature. 2001;409:370–4. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 29.Doose S, Barsch H, Sauer M. Polymer properties of polythymine as revealed by translational diffusion. Biophys J. 2007;93:1224–34. doi: 10.1529/biophysj.107.107342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fields AP, Cohen AE. Electrokinetic trapping at the one nanometer limit. Proc Natl Acad Sci U S A. 2011;108:8937–42. doi: 10.1073/pnas.1103554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fili N, Mashanov GI, Toseland CP, Batters C, Wallace MI, et al. Visualizing helicases unwinding DNA at the single molecule level. Nucleic Acids Res. 2010;38:4448–57. doi: 10.1093/nar/gkq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finkelstein IJ, Visnapuu ML, Greene EC. Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature. 2010;468:983–7. doi: 10.1038/nature09561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finzi L, Gelles J. Measurement of Lactose Repressor-Mediated Loop Formation and Breakdown in Single DNA Molecules. Science. 1995;267:378–80. doi: 10.1126/science.7824935. [DOI] [PubMed] [Google Scholar]
- 34.Frontali C, Dore E, Ferrauto A, Gratton E, Bettini A, et al. An absolute method for the determination of the persistence length of native DNA from electron micrographs. Biopolymers. 1979;18:1353–73. doi: 10.1002/bip.1979.360180604. [DOI] [PubMed] [Google Scholar]
- 35.Fulconis R, Mine J, Bancaud A, Dutreix M, Viovy JL. Mechanism of RecA-mediated homologous recombination revisited by single molecule nanomanipulation. EMBO J. 2006;25:4293–304. doi: 10.1038/sj.emboj.7601260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: the effect of the 3' arm and the stability of the dsDNA on the unwinding activity of the Escherichia coli DnaB helicase. J Mol Biol. 2004;343:101–14. doi: 10.1016/j.jmb.2004.07.056. [DOI] [PubMed] [Google Scholar]
- 37.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–8. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 38.Graham BW, Schauer GD, Leuba SH, Trakselis MA. Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res. 2011;39:6585–95. doi: 10.1093/nar/gkr345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grange W, Duckely M, Husale S, Jacob S, Engel A, Hegner M. VirE2: a unique ssDNA-compacting molecular machine. PLoS Biol. 2008;6:e44. doi: 10.1371/journal.pbio.0060044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–90. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffith JD, Harris LD, Register J., 3rd. Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb Symp Quant Biol. 1984;49:553–9. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 42.Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, et al. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 2010;38:2917–30. doi: 10.1093/nar/gkp1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gyimesi M, Sarlos K, Kovacs M. Processive translocation mechanism of the human Bloom's syndrome helicase along single-stranded DNA. Nucleic Acids Res. 2010;38:4404–14. doi: 10.1093/nar/gkq145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules - fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A. 1996;93:6264–8. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss G, et al. Initiation and reinitiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–41. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 46.Hatch K, Danilowicz C, Coljee V, Prentiss M. Direct measurements of the stabilization of single-stranded DNA under tension by single-stranded binding proteins. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:021916. doi: 10.1103/PhysRevE.76.021916. [DOI] [PubMed] [Google Scholar]
- 47.Hatch K, Danilowicz C, Coljee V, Prentiss M. Measurement of the salt-dependent stabilization of partially open DNA by Escherichia coli SSB protein. Nucleic Acids Res. 2008;36:294–9. doi: 10.1093/nar/gkm1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hegner M, Smith SB, Bustamante C. Polymerization and mechanical properties of single RecA-DNA filaments. Proc Natl Acad Sci U S A. 1999;96:10109–14. doi: 10.1073/pnas.96.18.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heyer WD. Biochemistry of eukaryotic homologous recombination. Top Curr Genet. 2007;17:95–133. doi: 10.1007/978-3-540-71021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci U S A. 2009;106:361–8. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hollis T, Stattel JM, Walther DS, Richardson CC, Ellenberger T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc Natl Acad Sci U S A. 2001;98:9557–62. doi: 10.1073/pnas.171317698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Honda M, Park J, Pugh RA, Ha T, Spies M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Mol Cell. 2009;35:694–703. doi: 10.1016/j.molcel.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Honda M, Okuno Y, Yoo J, Ha T, Spies M. Tyrosine phosphorylation enhances RAD52-mediated annealing by modulating its DNA binding. EMBO J. 2011;30:3368–82. doi: 10.1038/emboj.2011.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci U S A. 2011;108:7414–8. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;34:141–80. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 56.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia H, Korolev S, Niedziela-Majka A, Maluf NK, Gauss GH, et al. Rotations of the 2B Sub domain of E. coli UvrD Helicase/Translocase Coupled to Nucleotide and DNA Binding. J Mol Biol. 2011;411:633–48. doi: 10.1016/j.jmb.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–27. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 60.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 61.Karpel RL. In: In The Biology of Non-Specific DNA-Protein Interactions. Revzin A, editor. CRC Press; Boca Raton: 1990. pp. 103–30. Number of 103-30. [Google Scholar]
- 62.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996;93:13770–3. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim S, Streets AM, Lin RR, Quake SR, Weiss S, Majumdar DS. High-throughput single-molecule optofluidic analysis. Nat Methods. 2011;8:242–5. doi: 10.1038/nmeth.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kowalczykowski SC, Krupp RA. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein. Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J Mol Biol. 1987;193:97–113. doi: 10.1016/0022-2836(87)90630-9. [DOI] [PubMed] [Google Scholar]
- 65.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–65. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 66.Kozlov AG, Lohman TM. Kinetic mechanism of direct transfer of Escherichia coli SSB tetramers between single-stranded DNA molecules. Biochemistry. 2002;41:11611–27. doi: 10.1021/bi020361m. [DOI] [PubMed] [Google Scholar]
- 67.Kozlov AG, Lohman TM. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry. 2002;41:6032–44. doi: 10.1021/bi020122z. [DOI] [PubMed] [Google Scholar]
- 68.Kumaran S, Kozlov AG, Lohman TM. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–73. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuznetsov SV, Kozlov AG, Lohman TM, Ansari A. Microsecond dynamics of protein-DNA interactions: Direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J Mol Biol. 2006;359:55–65. doi: 10.1016/j.jmb.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 70.Lee G, Yoo J, Leslie BJ, Ha T. Single-molecule analysis reveals three phases of DNA degradation by an exonuclease. Nat Chem Biol. 2011;7:367–74. doi: 10.1038/nchembio.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J, Lee S, Ragunathan K, Joo C, Ha T, Hohng S. Single-Molecule Four-Color FRET. Angew Chem Int Ed Engl. 2010;49:9922–5. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–4. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 73.Lee JY, Okumus B, Kim DS, Ha T. Extreme conformational diversity in human telomeric DNA. Proc Natl Acad Sci U S A. 2005;102:18938–43. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–60. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leger JF, Robert J, Bourdieu L, Chatenay D, Marko JF. RecA Binding to a Single Double-Stranded DNA Molecule - a Possible Role of DNA Conformational Fluctuations. Proc Natl Acad Sci U S A. 1998;95:12295–9. doi: 10.1073/pnas.95.21.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin J, Kolomeisky A, Meller A. Helix-coil kinetics of individual polyadenylic acid molecules in a protein channel. Phys Rev Lett. 2010;104:158101. doi: 10.1103/PhysRevLett.104.158101. [DOI] [PubMed] [Google Scholar]
- 77.Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc Natl Acad Sci U S A. 2007;104:19790–5. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liphardt J, Onoa B, Smith SB, Tinoco I, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–7. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 79.Liu N, Bu T, Song Y, Zhang W, Li J, et al. The nature of the force-induced conformation transition of dsDNA studied by using single molecule force spectroscopy. Langmuir. 2010;26:9491–6. doi: 10.1021/la100037z. [DOI] [PubMed] [Google Scholar]
- 80.Liu S, Abbondanzieri EA, Rausch JW, Le Grice SF, Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–7. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–603. [PubMed] [Google Scholar]
- 82.Lohman TM. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 83.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein - multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–70. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 84.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 85.Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc Natl Acad Sci U S A. 2000;97:12002–7. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Manosas M, Xi XG, Bensimon D, Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38:5518–26. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McKinney SA, Joo C, Ha T. Analysis of Single-Molecule FRET Trajectories Using Hidden Markov Modeling. Biophys J. 2006;91:1941–51. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–80. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mine J, Disseau L, Takahashi M, Cappello G, Dutreix M, Viovy JL. Real-time measurements of the nucleation, growth and dissociation of single Rad51-DNA nucleoprotein filaments. Nucleic Acids Res. 2007;35:7171–87. doi: 10.1093/nar/gkm752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86:2530–7. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–7. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–5. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 93.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–6. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, et al. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–4. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Myong S, Ha T. Stepwise translocation of nucleic acid motors. Curr Opin Struct Biol. 2010;20:121–7. doi: 10.1016/j.sbi.2009.12.008. [A review on the strucutral, biochemical and single molecule data on the single basepair or nucleotide unwinding/translocation of various DNA/RNA motors.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J Biol Chem. 2007;282:27076–85. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 97.Pant K, Karpel RL, Rouzina I, Williams MC. Mechanical measurement of single-molecule binding rates: kinetics of DNA helix-destabilization by T4 gene 32 protein. J Mol Biol. 2004;336:851–70. doi: 10.1016/j.jmb.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 98.Pant K, Karpel RL, Rouzina I, Williams MC. Salt dependent binding of T4 gene 32 protein to single and double-stranded DNA: single molecule force spectroscopy measurements. J Mol Biol. 2005;349:317–30. doi: 10.1016/j.jmb.2005.03.065. [DOI] [PubMed] [Google Scholar]
- 99.Park J, Jeon Y, In D, Fishel R, Ban C, Lee JB. Single-molecule analysis reveals the kinetics and physiological relevance of MutL-ssDNA binding. PLoS One. 2010;5:e15496. doi: 10.1371/journal.pone.0015496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–55. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–97. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 102.Perkins TT, Li HW, Dalal RV, Gelles J, Block SM. Forward and reverse motion of single RecBCD molecules on DNA. Biophys J. 2004;86:1640–8. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal Structure of the Homo-Tetrameric DNA Binding Domain of Escherichia Coli Single-Stranded DNA-Binding Protein Determined By Multiwavelength X-Ray Diffraction On the Selenomethionyl Protein At 2.9-Angstrom Resolution. Proc Natl Acad Sci U S A. 1997;94:6652–7. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–52. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 105.Ragunathan K, Joo C, Ha T. Real-time observation of strand exchange reaction with high spatiotemporal resolution. Structure. 2011;19:1064–73. doi: 10.1016/j.str.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Record MT, Jr., Anderson CF, Lohman TM. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978;11:103–78. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 107.Rivetti C, Walker C, Bustamante C. Polymer Chain Statistics and Conformational Analysis of DNA Molecules With Bends or Sections of Different Flexibility. J Mol Biol. 1998;280:41–59. doi: 10.1006/jmbi.1998.1830. [DOI] [PubMed] [Google Scholar]
- 108.Robertson RB, Moses DN, Kwon Y, Chan P, Chi P, et al. Structural transitions within human Rad51 nucleoprotein filaments. Proc Natl Acad Sci U S A. 2009;106:12688–93. doi: 10.1073/pnas.0811465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Romer R, Schomburg U, Krauss G, Maass G. Escherichia coli single-stranded DNA binding protein is mobile on DNA: 1H NMR study of its interaction with oligo- and polynucleotides. Biochemistry. 1984;23:6132–7. doi: 10.1021/bi00320a036. [DOI] [PubMed] [Google Scholar]
- 110.Rossi MJ, Mazina OM, Bugreev DV, Mazin AV. The RecA/RAD51 protein drives migration of Holliday junctions via polymerization on DNA. Proc Natl Acad Sci U S A. 2011;108:6432–7. doi: 10.1073/pnas.1016072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rothenberg E, Trakselis MA, Bell SD, Ha T. MCM forked substrate specificity involves dynamic interaction with the 5'-tail. J Biol Chem. 2007;282:34229–34. doi: 10.1074/jbc.M706300200. [DOI] [PubMed] [Google Scholar]
- 112.Rothenberg E, Grimme JM, Spies M, Ha T. Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc Natl Acad Sci U S A. 2008;105:20274–9. doi: 10.1073/pnas.0810317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J Mol Biol. 2007;369:1244–57. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–7. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saikrishnan K, Jeyakanthan J, Venkatesh J, Acharya N, Sekar K, et al. Structure of Mycobacterium tuberculosis single-stranded DNA-binding protein. Variability in quaternary structure and its implications. J Mol Biol. 2003;331:385–93. doi: 10.1016/s0022-2836(03)00729-0. [DOI] [PubMed] [Google Scholar]
- 116.Saikrishnan K, Manjunath GP, Singh P, Jeyakanthan J, Dauter Z, et al. Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association. Acta Crystallogr D Biol Crystallogr. 2005;61:1140–8. doi: 10.1107/S0907444905016896. [DOI] [PubMed] [Google Scholar]
- 117.Saleh OA, McIntosh DB, Pincus P, Ribeck N. Nonlinear low-force elasticity of single-stranded DNA molecules. Phys Rev Lett. 2009;102:068301. doi: 10.1103/PhysRevLett.102.068301. [DOI] [PubMed] [Google Scholar]
- 118.Schmidt T, Schutz GJ, Baumgartner W, Gruber HJ, Schindler H. Imaging of Single Molecule Diffusion. Proc Natl Acad Sci U S A. 1996;93:2926–9. doi: 10.1073/pnas.93.7.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seol Y, Skinner GM, Visscher K. Elastic properties of a single-stranded charged homopolymeric ribonucleotide. Phys Rev Lett. 2004;93:118102. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
- 120.Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–6. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 121.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shim JW, Tan Q, Gu LQ. Single-molecule detection of folding and unfolding of the G-quadruplex aptamer in a nanopore nanocavity. Nucleic Acids Res. 2009;37:972–82. doi: 10.1093/nar/gkn968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shokri L, Marintcheva B, Richardson CC, Rouzina I, Williams MC. Single molecule force spectroscopy of salt-dependent bacteriophage T7 gene 2.5 protein binding to single-stranded DNA. J Biol Chem. 2006;281:38689–96. doi: 10.1074/jbc.M608460200. [DOI] [PubMed] [Google Scholar]
- 124.Shokri L, Marintcheva B, Eldib M, Hanke A, Rouzina I, Williams MC. Kinetics and thermodynamics of salt-dependent T7 gene 2.5 protein binding to single- and double-stranded DNA. Nucleic Acids Res. 2008;36:5668–77. doi: 10.1093/nar/gkn551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shroff H, Reinhard BM, Siu M, Agarwal H, Spakowitz A, Liphardt J. Biocompatible force sensor with optical readout and dimensions of 6 nm3. Nano Lett. 2005;5:1509–14. doi: 10.1021/nl050875h. [DOI] [PubMed] [Google Scholar]
- 126.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 127.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The Elasticity of a Single Supercoiled DNA Molecule. Science. 1996;271:1835–7. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 128.Sun B, Wei KJ, Zhang B, Zhang XH, Dou SX, et al. Impediment of E. coli UvrD by DNA-destabilizing force reveals a strained-inchworm mechanism of DNA unwinding. EMBO J. 2008;27:3279–87. doi: 10.1038/emboj.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Theissen B, Karow AR, Kohler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc Natl Acad Sci U S A. 2008;105:548–53. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–47. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tomko EJ, Jia H, Park J, Maluf NK, Ha T, Lohman TM. 5'-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocase. EMBO J. 2010;29:3826–39. doi: 10.1038/emboj.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.van der Heijden T, Modesti M, Hage S, Kanaar R, Wyman C, Dekker C. Homologous recombination in real time: DNA strand exchange by RecA. Mol Cell. 2008;30:530–8. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 133.Van Dyck E, Foury F, Stillman B, Brill SJ. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 1992;11:3421–30. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van Loenhout MT, van der Heijden T, Kanaar R, Wyman C, Dekker C. Dynamics of RecA filaments on single-stranded DNA. Nucleic Acids Res. 2009;37:4089–99. doi: 10.1093/nar/gkp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJ, Wuite GJ. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–8. doi: 10.1038/nature07581. [A study of Rad51 filament dynamics using advanced combination of optical tweezes and fluorescence imaging.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–8. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 137.Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, et al. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–9. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Walter NG, Huang CY, Manzo AJ, Sobhy MA. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Methods. 2008;5:475–89. doi: 10.1038/nmeth.1215. [A comprehensive review of diverse single molecule techniques available.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 140.Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci U S A. 2006;103:6190–5. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wuite GJL, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–6. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 142.Yang C, Curth U, Urbanke C, Kang C. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat Struct Biol. 1997;4:153–7. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 143.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 2003;100:14629–34. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 2009;28:405–16. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yodh JG, Schlierf M, Ha T. Insight into helicase mechanism and function revealed through single-molecule approaches. Q Rev Biophys. 2010;43:185–217. doi: 10.1017/S0033583510000107. [A comprehensive review on single molecule studies of helicase activities.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yu J, Ha T, Schulten K. How directional translocation is regulated in a DNA helicase motor. Biophys J. 2007;93:3783–97. doi: 10.1529/biophysj.107.109546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yu Z, Schonhoft JD, Dhakal S, Bajracharya R, Hegde R, et al. ILPR G-quadruplexes formed in seconds demonstrate high mechanical stabilities. J Am Chem Soc. 2009;131:1876–82. doi: 10.1021/ja806782s. [DOI] [PubMed] [Google Scholar]
- 148.Zhou R, Kozlov AG, Roy R, Zhang J, Korolev S, et al. SSB Functions as a Sliding Platform that Migrates on DNA via Reptation. Cell. 2011;146:222–32. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]