Background: Low density lipoprotein receptor (LDLR) mediates clearance of blood coagulation factor VIII (FVIII).
Results: The region of complement-type repeats 2–5 in LDLR was identified as the binding site for FVIII and for α-2-macroglobulin receptor-associated protein (RAP).
Conclusion: Binding sites of LDLR for FVIII, and also for RAP, were characterized.
Significance: This provides new data on LDLR structure and function and on FVIII catabolism.
Keywords: Blood Coagulation Factors, Factor VIII, Lipoprotein Receptor, Receptor Structure-Function, Recombinant Protein Expression, Hemophilia A
Abstract
Low density lipoprotein receptor (LDLR) was shown to mediate clearance of blood coagulation factor VIII (FVIII) from the circulation. To elucidate the mechanism of interaction of LDLR and FVIII, our objective was to identify the region of the receptor necessary for binding FVIII. Using surface plasmon resonance, we found that LDLR exodomain and its cluster of complement-type repeats (CRs) bind FVIII in the same mode. This indicated that the LDLR site for FVIII is located within the LDLR cluster. Similar results were obtained for another ligand of LDLR, α-2-macroglobulin receptor-associated protein (RAP), a common ligand of receptors from the LDLR family. We further generated a set of recombinant fragments of the LDLR cluster and assessed their structural integrity by binding to RAP and by circular dichroism. A number of fragments overlapping CR.2-5 of the cluster were positive for binding RAP and FVIII. The specificity of these interactions was tested by site-directed mutagenesis of conserved tryptophans within the LDLR fragments. For FVIII, the specificity was also tested using a single-chain variable antibody fragment directed against the FVIII light chain as a competitor. Both cases resulted in decreased binding, thus confirming its specificity. The mutagenic study also showed an importance of the conserved tryptophans in LDLR for both ligands, and the competitive binding results showed an involvement of the light chain of FVIII in its interaction with LDLR. In conclusion, the region of CR.2-5 of LDLR was defined as the binding site for FVIII and RAP.
Introduction
Factor VIII (FVIII)2 is a plasma protein (∼300 kDa) composed of a heavy chain and a light chain with the domain structures of A1-A2-B and A3-C1-C2, respectively (1). FVIII serves as a cofactor for activated factor IX in the intrinsic pathway of blood coagulation, and deficiency in FVIII results in a bleeding disorder known as hemophilia A. This disease is treated by FVIII products, which require frequent infusions due to the short half-life of FVIII in plasma (∼14 h). Despite the clinical significance of understanding mechanisms of FVIII clearance, the details of it are unknown.
One of receptors involved in clearance of FVIII is the low density lipoprotein receptor (LDLR). In this process, LDLR acts in concert with the low density lipoprotein receptor-related protein (LRP). Indeed, deficiency in either LDLR or LRP in mice prolonged the half-life of FVIII about 1.5-fold, whereas the combined deficiency resulted in ∼4.8-fold prolongation (2). Furthermore, inhibition of both receptors by their highly specific ligand α-2-macroglobulin receptor-associated protein (RAP) (3, 4) increased its half-life in mice (5, 6). In humans, polymorphism in either LDLR or LRP is associated with elevated levels of FVIII (7–10).
LDLR and LRP belong to a large group of endocytic receptors known as the LDLR family. Members of this family are structurally similar, yet have a diverse pattern of tissue expression and a broad spectrum of ligands. LDLR and LRP, expressed in the liver, serve to catabolize various plasma ligands (11, 12), in addition to FVIII. Two other receptors from the family, very low density lipoprotein receptor (VLDLR) and megalin, are also found in the circulation. Although both receptors bind to FVIII in a purified system (13, 14), they are likely not involved in its clearance in vivo (15, 16).
The ligand binding moiety of the LDLR family receptors is represented by highly homologous complement-type repeats (CRs). Each CR is composed of ∼40 amino acid residues and forms an autonomous domain. All CRs of LDLR were characterized for their tertiary structures (17–22), as well as some CRs of LRP (23). These data showed that each CR domain contains three internal disulfide bonds, formed by six conserved cysteines, and coordinates Ca2+ via several conserved acidic residues. During the interaction with a ligand, each CR domain “docks” a specific lysine via conserved tryptophan and acidic residues (18, 21, 24–26).
The CR domains are connected to each other by short flexible linkers (23) and are composed in clusters. LDLR contains seven CRs grouped in one cluster (11), whereas LRP contains 31 CRs grouped in four clusters (12). Typically, the binding sites of the ligands are formed by several adjacent CRs, among which a minimal binding unit is presented by a pair of CRs (CR doublet). Such organization of the sites was found in LDLR for binding RAP (22), apoE (27), and apoB (28) and found in LRP for a number of its ligands including FVIII (29–31).
For FVIII, LRP has two binding sites; each site is formed by 3–4 adjacent CRs and located in a separate CR cluster of the receptor (31, 32). At the same time, the FVIII-binding site in LDLR is unknown. In vitro, the affinity of FVIII to LDLR (KD of ∼200 nm) (14) was found to be less than to LRP (KD of ∼80 nm) (2, 5, 33). Such affinities are unlikely to provide effective direct interactions of FVIII with both receptors in vivo, considering the concentration of FVIII in plasma (∼0.3 nm). For LRP, its in vivo interaction with FVIII is facilitated by cell surface heparan sulfate proteoglycans (34). Whether this type of receptors serves a similar role for LDLR is unknown.
In the present work, we aimed to determine the specific CRs of LDLR responsible for FVIII binding. We generated a set of LDLR fragments and tested their ability to bind FVIII in a purified system. The specificity of these interactions was verified using an anti-FVIII antibody fragment and site-directed mutagenesis of the LDLR fragments. As a result, we identified specific CRs of the receptor that form a binding region for FVIII.
EXPERIMENTAL PROCEDURES
Reagents
FVIII products, Advate (Baxter, CA) and Xyntha (Wyeth, PA), corresponding to recombinant full size FVIII and BDD-FVIII, respectively, were purchased from the National Institutes of Health Pharmacy (Bethesda, MD). Plasma FVIII was isolated as described (35). Recombinant LDLR exodomain (expressed in mouse cells) and RAP (expressed in bacteria) were purchased from R&D Systems (Minneapolis, MN). Anti-FVIII ScFv iKM33 was produced as described (36). LDLR cDNA was obtained from Dr. G. Rudenko. Anti-myc tag mAb 9E10 was purchased from Sigma-Aldrich.
Generation of Constructs Coding LDLR Fragments
A modified pFastBac1 plasmid containing a melittin secretion signal, His6 tag, a multiple cloning site, c-myc tag, and a stop codon was used as a vector as described (37). The coding regions of the LDLR fragments were generated by PCR using LDLR cDNA as a template and corresponding primers. Point mutations of selected LDLR fragments were performed by overlapping PCR. All resulting PCR fragments were cloned into the modified pFastBac1 vector.
Expression and Purification of the LDLR Fragments
Recombinant baculoviruses for the wild-type and mutant fragments of LDLR were generated using the plasmid constructs and the Bac-to-Bac expression system (Invitrogen). Optimization of the expression of the proteins in Sf9 cells was performed as described (38). The proteins were expressed in 120 ml of the suspension culture. The culture supernatants were harvested 72 h after infection and exchanged into PBS, pH 7.4, buffer using a tangential flow filter Pellicon XL 5k (Millipore, Billerica, MA). The fragments were then purified on nickel-Sepharose 6 Fast Flow resin (GE Healthcare). Upon adjusting the solutions to contain 300 mm NaCl, 20 mm imidazole, they were passed through the column followed by washing the resin with PBS, pH 7.4, 300 mm NaCl, 30 mm imidazole and eluting the proteins with PBS, pH 7.4, 150 mm NaCl, 230 mm imidazole. The eluted product was concentrated and further purified using size-exclusion chromatography on either Superdex-75 or Superdex-200 (GE Healthcare) to isolate the monomeric forms in 20 mm HEPES, pH 7.4, 0.15 m NaCl, 0.005% Tween 20, and 5 mm CaCl2 (HBS/Ca). LDLR CR cluster (CR.1-7) and its mutant were refolded as described (29). All proteins were verified by PAGE with GelCode Blue Safe (Thermo Scientific) staining and Western blotting with anti-myc mAb9E10. The protein concentrations were measured by absorbance spectroscopy at 280 nm using calculated respective extinction coefficients.
Circular Dichroism Measurements
Far-UV CD measurements were performed on a Jasco J-815 spectropolarimeter (JASCO Co., Japan) at 25 ± 0.2 °C, as maintained by a PTC 423S/15 Peltier temperature controller (JASCO). The protein concentration in the samples was adjusted to ∼ 30 μm in HBS/Ca. Titration of the samples by EDTA was conducted stepwise by adding a calculated amount of the 0.5 m stock solution. The spectra were recorded between 180 and 260 nm in a 0.5-mm path length quartz cuvette using a scan speed of 20 nm/min, bandwidth of 1.0 nm, and resolution of 0.2 nm and accumulated in triplicate. An ellipticity of CD spectra was expressed in millidegrees.
Surface Plasmon Resonance Measurements
Prior to the measurements, FVIII products were reconstituted and dialyzed in a HBS/Ca buffer prepared by adjusting HBS-P buffer (GE Healthcare) to contain 5 mm of CaCl2. The concentration of the dialyzed FVIII was measured using a BCA kit (Thermo Scientific). Binding assays were performed in HBS-P/Ca using Biacore 3000 (GE Healthcare). LDLR fragments were immobilized on a CM5 chip using an amine coupling kit at ∼1000 resonance units for CR doublets and at ∼2000 resonance units for the CR cluster. Association of the fragments with RAP or with FVIII in the presence or absence of iKM33 was recorded at a flow rate of 10 μl/min for 3 min. The dissociations were recorded for 5 min in the running buffer. Regeneration of the sensors was performed by 0.1 m H3PO4. The estimated KD values were derived by fitting the association and dissociation curves with a 1:1 (Langmuir) model using the BIAevaluation 4.1.1 program.
RESULTS
Expression of LDLR Fragments
The generated LDLR fragments were its CR cluster and six overlapping CR doublets (Figs. 1 and 2). This strategy was assumed suitable, based on its previous use for mapping the RAP-binding site on LRP (29). The LDLR fragments were expressed in insect cells, which were previously shown capable of producing functionally active fragments of LRP and megalin (37, 39, 40). The LDLR fragments obtained were essentially pure (Fig. 1); the observed double banding for some of the CR doublets was attributed to a possible difference in glycosylation, similar to that found previously for expressed fragments of LRP (31).
FIGURE 1.
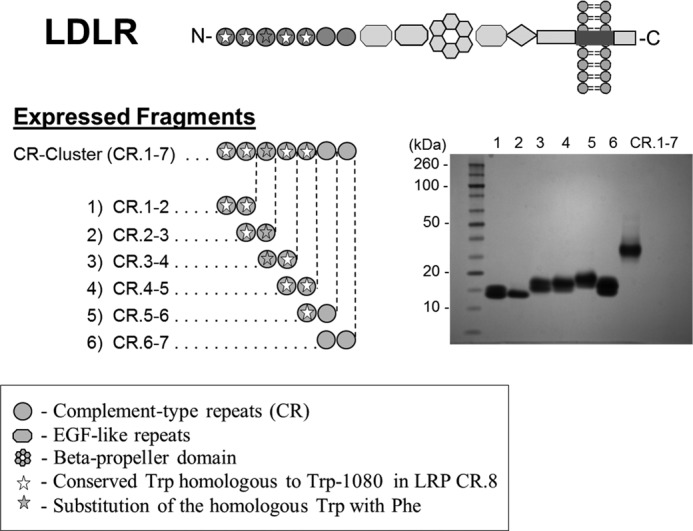
The domain structure of LDLR, strategy of expressing its fragments, and their analysis by PAGE. The inset shows an image of the GelCode Blue Safe-stained gel with the purified LDLR fragments.
FIGURE 2.
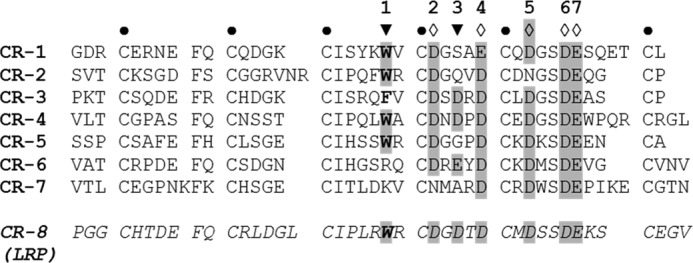
LDLR complement-type repeats. The sequences are aligned by the conservative cysteines (●). Other conservative residues (in gray) are aspartic (D) or glutamic (E) acid residues 2, 4–7 (♢) coordinating Ca2+ via side-chain carboxyl oxygens, and those, 1 and 3 (▾), coordinating Ca2+ via backbone carbonyl oxygens. Conservative residues 4 and 5 coordinate Ca2+ alternatively. Conservative tryptophan (W) at position 1 (in bold) also interacts with the aliphatic moiety of the lysine of the ligand. Shown at the bottom is LRP CR.8, which matches the consensus sequence (in italics).
Interaction of LDLR Fragments with RAP
Because RAP is known to inhibit interactions of the LDLR family with their ligands in vitro, the integrity of the LDLR fragments was assessed by testing them for binding to RAP. As a control, we used the LDLR exodomain (full size), expressed in mammalian cells. In preliminary experiments, we found that the preparation of the LDLR cluster (of CRs) was less active for binding than some CR doublets, likely due to a higher content of the misfolded form. Chemical refolding of this preparation resulted in an increase of its binding activity; therefore, this approach was employed in further studies.
In SPR, both the LDLR exodomain and its cluster bound RAP in a similar mode (Fig. 3, A and B). The assessed kinetic constants including both KD values (∼1 nm, supplemental Table S1) were similar. This indicated that the LDLR cluster contains all necessary elements of the receptor for binding RAP, similar to that previously found for LRP (29). Among the CR doublets overlapping the cluster, CR.2-3, CR.3-4, and CR.4-5 were able to bind RAP, whereas other doublets were inactive (Fig. 3, C–H). The assessment of the kinetic constants indicated higher affinity for CR.3-4 and CR.4-5 (KD values of 3–6 nm) than CR.2-3 (KD ∼29 nm). Although the binding signals did not fit well, several independent experiments produced essentially the same results. These data suggested that the LDLR region CR.2-5 forms a binding site for RAP.
FIGURE 3.
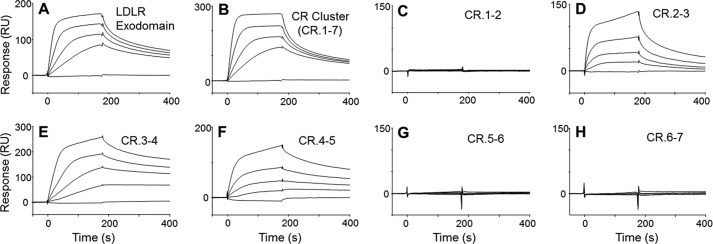
Binding of RAP to LDLR fragments. In SPR, the LDLR fragments were immobilized on a sensor at ∼2000 resonance units (RU) for the LDLR exodomain (A) and LDLR cluster (B) and at ∼1000 resonance units for CR doublets (C–H) and tested for binding with RAP (0, 2.5, 5, 10, and 20 nm). The RAP association was recorded for 3 min followed by measuring its dissociation in the running buffer. RU, resonance units.
Interaction of LDLR Fragments with FVIII
Under the same experimental conditions, the LDLR fragments were tested for binding to FVIII (plasma-derived). We found that FVIII bound to the LDLR exodomain and LDLR cluster in a similar fashion (Fig. 4, A and B). Although fitting of the signals was not optimal, similar to that found for FVIII and LRP (41), the respective kinetic constants including KD values (30–60 nm) were assessed as similar (supplemental Table S2). These data indicated that the LDLR cluster contains all required elements to bind FVIII, similar to that found for LRP clusters (31). Among the CR doublets, CR.2-3, CR.3-4, and CR.4-5 were found active for FVIII (Fig. 4, C–F) with similar KD values (40–90 nm). These data suggested that the LDLR region CR.2-5 forms a binding site for FVIII.
FIGURE 4.
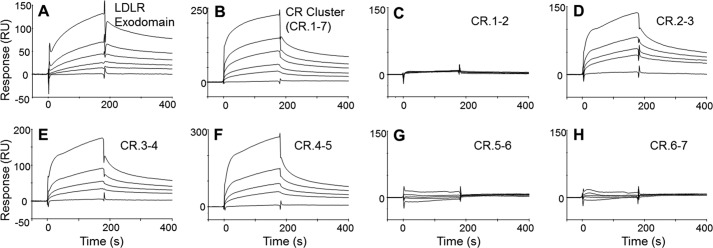
Binding of FVIII to LDLR fragments. In SPR, the LDLR fragments, LDLR exodomain (A), LDLR cluster (B), and CR doublets (C–H), were immobilized and tested for binding with various concentrations of FVIII (0, 56.3, 112.5, 225, and 450 nm). RU, resonance units.
To confirm the results, all the CR doublets were retested with two recombinant FVIII variants, full size FVIII and BDD-FVIII. We asked whether the same CR doublets bind to all FVIII variants. At a selected concentration, each FVIII preparation was tested with each CR doublet for the signal intensity in the association phase. We found that active binding LDLR fragments were indeed the same for all three FVIII variants (Fig. 5). At the same time, the intensity of the signals differed among the FVIII variants, possibly reflecting their variability in characteristics. In particular, BDD-FVIII has a lower molecular mass producing lower signals; therefore, a higher concentration of BDD-FVIII was used. More extensive pre-processing of plasma-derived FVIII possibly caused its lower signals as compared with those of the full size recombinant FVIII. Other characteristics include heterogeneity of full size FVIII due to the natural variability in the B-domain length and other properties, previously compared between various FVIII products (42–45). A residual binding of both recombinant FVIII variants to the CR doublets non-active for plasma-derived FVIII was attributed to a nonspecific component. Also, our data suggest that the B-domain of FVIII does not contribute to its binding to LDLR, similarly to that indicated for LRP (41). In conclusion, our data demonstrate that the LDLR-binding site is essentially the same for each FVIII variant, which supports the results of the mapping study.
FIGURE 5.
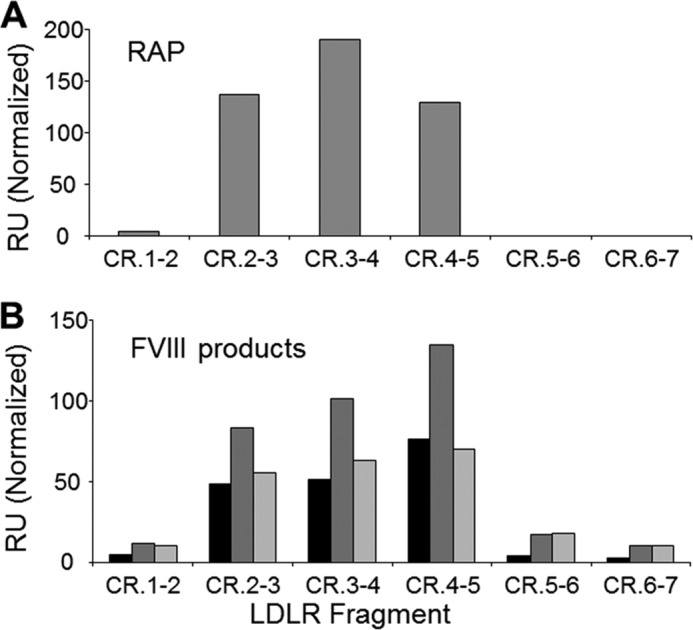
Binding of various FVIII products to CR doublets of LDLR. In SPR, the LDLR fragments were immobilized and tested for binding with RAP (32 nm) (A) and with FVIII variants plasma-derived FVIII (225 nm, dark gray), full size recombinant FVIII (275 nm, medium gray) and BDD-FVIII (400 nm, light gray) (B). The bars correspond to signals recorded after 3 min of the association. RU, resonance units.
Interaction of LDLR Fragments with FVIII in the Presence of an Anti-FVIII Antibody Fragment
The specificity of FVIII binding to the LDLR fragments was verified using a recombinant anti-FVIII ScFv iKM33 (36), which recognizes the C1-domain of FVIII and inhibits its binding to LDLR (46–48). We asked whether iKM33 had a similar effect on the binding of FVIII to the LDLR fragments. FVIII (full size recombinant) was incubated with increasing amounts of iKM33, and the samples were tested with the LDLR cluster and CR.2-3, CR.3-4, and CR.4-5. In all cases, iKM33 strongly inhibited the binding in a dose-dependent manner (Fig. 6), thus supporting the specificity of the interactions. Also, these data indicate involvement of the FVIII light chain, in particular its C1-domain, in the binding to LDLR.
FIGURE 6.
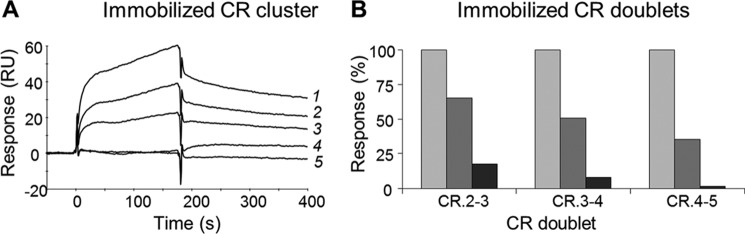
Binding of FVIII to LDLR fragments in the presence of ScFv iKM33. A, in SPR, immobilized LDLR cluster was tested for binding with FVIII (full size recombinant FVIII, 275 nm) in the absence of iKM33 (1) or in its presence at 275 nm (2), 550 nm (3), or 1375 nm (4). Curve 5 corresponds to injection of buffer. RU, resonance units. B, in a similar way, the immobilized CR doublets 2-3, 3-4, and 4-5 were tested for binding with FVIII (275 nm) in the absence of iKM33 (light gray bars) or in its presence at 275 nm (dark gray bars) or 1375 nm (black bars). The resulting signals were recorded after 3 min of the association and are expressed in percentages of those obtained in the absence of iKM33.
Interaction of Mutated CR Doublets with FVIII and RAP
To further verify specificity of the interactions of the LDLR fragments with FVIII, we asked whether specific mutations of the fragments have an effect on the binding. As shown previously, a conserved tryptophan in position 1 (Fig. 2) of a CR from the LDLR family interacts with the aliphatic chain of a lysine residue of the ligand (18, 21, 24–26). A replacement of such tryptophan by serine in either CR of CR.5-6 of LRP significantly reduced its binding to RAP, thus confirming specificity of this interaction (30).
Within the CR.2-5 region, the conserved tryptophan is presented in CR.2 (Trp-66), CR.4 (Trp-144), and CR.5 (Trp-193), whereas CR.3 contains a phenylalanine (Phe-105) at this position (Fig. 2). Using the above mutagenic approach (30), we targeted these tryptophans in all active binding CR doublets in a way to affect one CR per doublet. The respective replacements (Trp → Ser) were made in CR.2 of CR.2-3, in CR.4 of CR.3-4, and in CR.5 of CR.4-5, resulting in generation of CR.2-3 (W66S), CR.3-4 (W144S), and CR.4-5 (W193S), respectively. Hereafter, the CR domains affected by the mutagenesis are marked with an asterisk (*).
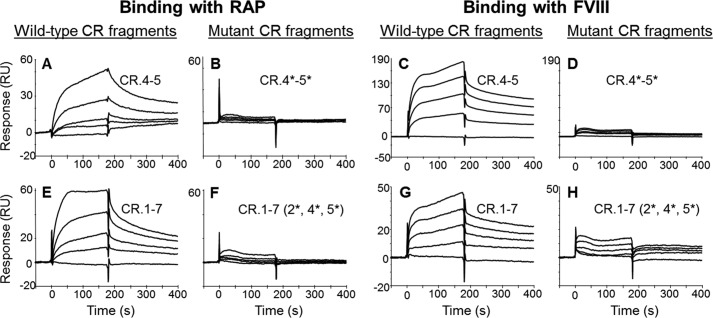
In SPR, we found that binding of RAP and FVIII (full size recombinant) to CR.2*-3 and CR.3-4* was significantly reduced (supplemental Fig. S1). At the same time, the binding of both ligands to CR.4-5* was not affected (data not shown). The additional mutation (Trp → Ser) introduced into CR.4 of this doublet completely abolished binding of the resulting double mutant (CR.4*-5*) to both RAP (Fig. 7, A and B) and FVIII (Fig. 7, C and D). Altogether, these data indicated that the interactions of the doublets CR.2-3, CR.3-4, and CR.4-5 with RAP and FVIII are specific.
FIGURE 7.
Binding of RAP and FVIII to the mutants of CR.4-5 and LDLR cluster. In SPR, CR.4-5 (A and C), CR.4-5 W144S/W193S (B and D), LDLR cluster (E and G), and its triple-mutant W66S/W144S/W193S (F and H) were immobilized and tested for binding with RAP (0.6, 1.3, 2.5, and 5 nm) (A, B, E, and F) and FVIII (full size recombinant FVIII, 50, 100, 150, and 200 nm) (C, D, G, and H). Injections of the buffer only were used as controls. For each pair of a particular LDLR fragment and its mutant tested versus a given ligand, the signals are shown in the same scale. CR domains affected by mutagenesis are marked by *. RU, resonance units.
Assessment of the Conformation of CR Doublets and Their Mutants
Using far-UV CD, the conformation of all CR doublets and their mutants was assessed for the presence of bound Ca2+, which is essential for proper folding. All the initial CD spectra (190–260 nm) were highly similar, suggesting similarity in the secondary structure content. Upon the titration of a CR doublet with a Ca2+-chelating agent (EDTA), we observed both a dose-dependent shift of the negative maximum (∼200 nm) to a longer wavelength (∼206 nm) and a decrease of the signal intensities by 34–43%. These changes were highly similar among all preparations of the CR doublets. The spectra for a representative doublet CR.4-5 and its double mutant (CR.4*-5*) are shown in Fig. 8. For other active binding doublets, CR.2-3, CR.3-4, and their mutants, the spectra are shown in supplemental Fig. S2, and not shown for the non-binders.
FIGURE 8.
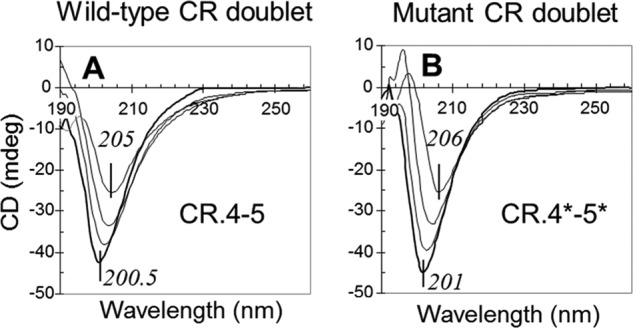
Far-UV CD spectra of CR.4-5 and its mutant upon the titration by EDTA. CR.4-5 (A) and CR.4-5 W144S/W193S (B) were taken at concentrations of 30 μm. The spectra correspond to the absence of EDTA (bottom curves in bold) and its increase as 1×, 2×, and 3× over molar equivalent of Ca2+ in the solutions, shown as respective increase in the signals (upper curves). mdeg, millidegrees.
Based on previous studies (19, 20, 49–51), the observed spectra changes reflect a gradual loss of the secondary structures of the proteins due to progressive removal of the bound Ca2+. Therefore, the similarity of the spectra is indicative of the correctness and similarity of the folding of all the CR doublets including their mutants. Due to the structural independence of CRs (23), it is likely that each individual CR domain is correctly folded within both the respective CR doublet and the LDLR cluster. Thus, the CD data further supported the results of our binding studies.
Interaction of the Mutated LDLR Cluster with FVIII and RAP
Finally, we reproduced all the above mutations within the LDLR cluster. The resulting mutant CR.1-7 W66S/W144S/W193S (CR.1-2*-3-4*-5*-6-7) had all active CR pairs affected. Based on our CD data, we assumed similarity of the folding of both the wild type and the mutant variants. In SPR, the binding of the mutated cluster to RAP (Fig. 7, E and F) and to FVIII (full size recombinant) was diminished (Fig. 7, G and H). The residual interaction of the mutated cluster with FVIII, also observed for the doublets CR.2*-3 and CR.3-4* (supplemental Fig. S1), was likely supported by the non-mutated CR.3, which was presented in each of these fragments.
Thus, specific point mutations in all active binding LDLR fragments did affect their interactions with RAP and FVIII. This confirms the specificity of the interactions and validates the mapping results. These data also demonstrate an important role of the LDLR tryptophans 66 (CR.2), 144 (CR.4), and 193 (CR.5) for the interactions of LDLR with RAP and FVIII.
DISCUSSION
In the present study, we demonstrate that the second through fifth complement-type repeats of LDLR provide a binding site for both FVIII and RAP. These conclusions are based on results from testing the binding of these ligands to various fragments of LDLR in the presence or absence of an anti-FVIII antibody fragment and to the fragments containing mutations of selected conserved residues.
The overlap of FVIII- and RAP-binding sites on LDLR is in accordance with other studies. In LRP-deficient mice, adenovirus-mediated overexpression of RAP resulted in an increase of FVIII level (6). Most likely, this was due to blocking of LDLR by RAP, and not other RAP-sensitive determinants. Among them, VLDLR (expressed in endothelium) does not contribute to FVIII clearance in mice (15); also, such a role of megalin cannot be justified due to its location in the renal proximal tubule, accessible only to small plasma ligands (16). In turn, upon injection of [125I]FVIII in mice, most of the radioactivity was accumulated in the liver (5), which further supports the role of LDLR in the above study (6).
Our data support the general model of ligand recognition by the LDLR family, previously defined in a number of studies (17, 21, 22, 24, 29, 52–54). According to this model, a CR consensus sequence contains a conserved tryptophan at position 1 and acidic residues at positions 2, 6, 7, and either 4 or 5, and predominantly an acidic residue at position 3 (Fig. 2). These residues coordinate Ca2+ via backbone carbonyls (positions 1 and 3) and side-chain carboxyls (positions 2, 6, 7, and either 4 or 5). The side chain of the conserved tryptophan also interacts with the aliphatic moiety of the lysine of the ligand. In our mutagenic study, the conserved tryptophans of CR.2, CR.4, and CR.5 were found to be important for binding RAP and FVIII. Moreover, our study indicated that the less conserved homologous phenylalanine (Phe-105) of CR.3 for both ligands serves the same role as the tryptophan. This observation supports the data of Fisher et al. (22), which proposed the same role of this phenylalanine (in CR.3) for the interaction with RAP.
In accordance with previous studies (17–21, 50, 55, 56), our CD study showed that all seven CR domains of LDLR coordinate Ca2+. Importantly, this study showed that the folding of all expressed CR doublets and their mutants was similar, thus supporting the validity of our mapping results. At the same time, the folding of the purified LDLR cluster was less efficient based on its low binding activity. This was most likely due to more complexity of the cluster as compared with a CR doublet. Upon refolding of the preparation of the cluster, its binding activity was restored to the level comparable with the LDLR exodomain (expressed in mammalian cells).
In regard to the affinity of RAP for LDLR, their KD was previously assessed as 50–250 nm in solid phase- and cell culture-based assays (57). In our study, based on real-time binding, the assessed KD values for both the LDLR exodomain and the LDLR cluster (∼1 nm) were comparable with RAP KD values for other receptors from the LDLR family: VLDLR (KD = 0.7 nm), megalin (KD = 8 nm), and LRP (KD = 18 nm) (58–60). The discrepancy between our result and that of Medh et al. (57) was likely due to differences in the experimental conditions. More importantly, the similarity of the affinities of both LDLR fragments in our study showed that the RAP-binding site of LDLR is located within its cluster.
Our mapping of the RAP-binding site of LDLR is in agreement with previous studies, which showed that the isolated CR.3-4 and CR.4-5 bind RAP (22, 56). Adding CR.2-3 to this set defines the whole RAP-binding region of LDLR as CR.2-5 with a possible preference for CR.3-5. As RAP is a common ligand of the LDLR family, it is worthwhile to compare its binding sites between the receptors. In LRP cluster II, the RAP-binding region is presented by seven adjacent CRs and comprises the sites for other ligands (30, 31, 37, 61). In LDLR, the RAP-binding region comprises the sites for FVIII (present study) and apoE (CR.4-5) (27), whereas a longer region involving CR.3-7 and an EGF-like repeat is required for apoB-100 (28, 62). In contrast, proprotein convertase subtilisin/kexin type 9 (PCSK9) has an atypical site on the receptor, which is formed by the EGF-like domain (63). Thus, at least for LDLR and LRP, the binding sites for RAP typically comprise or overlap sites for other ligands.
The comparison of the LDLR-binding sites for its ligands shows that CR.4-5 is involved in the majority of these interactions. Notably, this doublet has an unusually long linker between the domains that provides more flexibility to adapt to the ligands, and the module CR.5 has significantly higher affinity for Ca2+ than any other CR of the receptor (56). In contrast to other CR doublets forming the known binding sites of LDLR, CR.4-5 has the conserved tryptophans in both CRs, and our study required mutation of both to decrease the binding activity of the doublet.
The following summarizes the major data for the interactions of FVIII with LDLR and LRP. In a series of studies by Mertens and co-workers (2, 64, 65), it was shown that the CR clusters of LDLR and LRP compete for binding to FVIII. Within each of the LRP clusters II and IV, approximately four adjacent CRs form a binding site for FVIII (31, 33), whereas the matching epitope of FVIII involves multiple lysines on all three domains of its light chain (65–68). Consistent with this, the anti-FVIII ScFv KM33, which recognizes the C1-domain of the light chain, interferes with FVIII binding to both LDLR and LRP (46, 48, 69). Our present study shows that similarly to LRP, four adjacent CRs of LDLR form a site for FVIII, and on the FVIII side, the contact interface involves the C1-domain. Altogether, these data indicate that the FVIII-binding sites for both receptors are similar and involve the A3, C1, and C2 domains. Based on these considerations, we propose a model of interaction of LDLR and FVIII in which the CR.2-5 region of the receptor matches the respective site of FVIII, as shown for the particular orientation of these molecules on Fig. 9.
FIGURE 9.
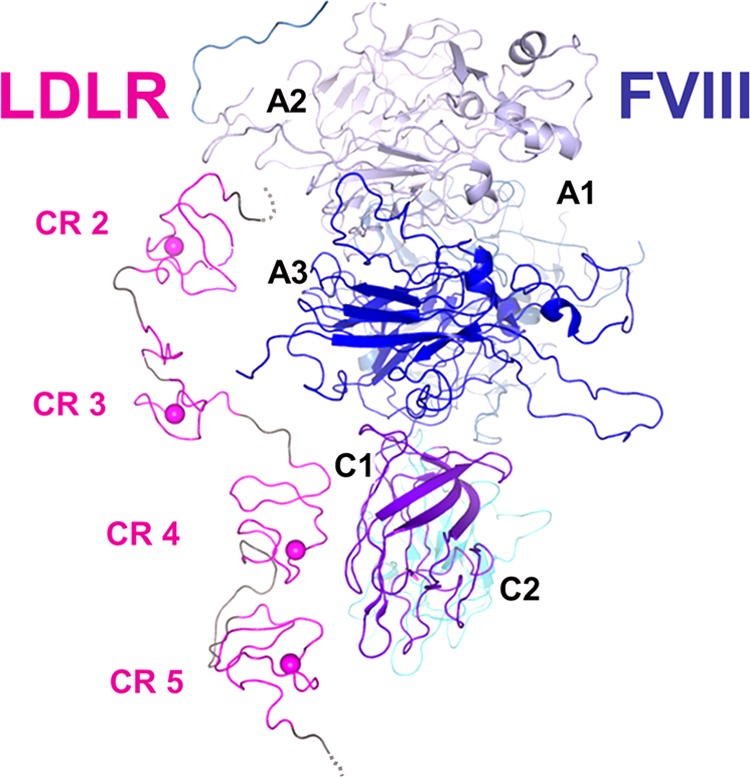
Proposed interaction of LDLR with FVIII. The model is based on atomic coordinates of crystal structures of the LDLR exodomain (Protein Data Bank (PDB) ID 1N7D) and FVIII (PDB ID 2R7E). For the LDLR part, only the CR.2-5 region is shown (magenta and gray). In each CR domain, the coordinated Ca2+ is depicted as a sphere. The model was constructed using PyMOL.
Future studies would involve more detailed characterization of the molecular interface between LDLR and FVIII and investigation of a possible role of heparan sulfate proteoglycans (HSPGs) or other intermediates in this interaction. These directions could facilitate generation of new FVIII products with better sustainability in the circulation.
Supplementary Material
Acknowledgments
We are thankful to Dr. Gabby Rudenko for providing us with a construct for LDLR expression and to our colleagues from FDA/CBER, Dr. Michael Kennedy and Malgorzata Norton, for support in the SPR, and to Dr. Pei Zhang, Laura Wood, and Samuel Woodle for help in the manuscript preparation.
This work was supported by funds from the Center for Biologics Evaluation and Research (CBER) of the United States Food and Drug Administration (FDA) and also by the Research Participation Program at the CBER administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the United States Department of Energy and the United States Food and Drug Administration. This work was initially presented at the 54th American Society of Hematology Annual Meeting, December 8–11, 2012, in Atlanta, GA.

This article contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- FVIII
- blood coagulation factor VIII
- BDD-FVIII
- B-domain-deleted recombinant FVIII
- LDLR
- low density lipoprotein receptor
- LRP
- low density lipoprotein receptor-related protein
- LDLR cluster
- cluster of complement-type repeats of LDLR
- VLDLR
- very low density lipoprotein receptor
- CR
- complement-type repeat
- CR doublet
- a pair of adjacent CRs
- RAP
- α-2-macroglobulin receptor-associated protein
- ScFv
- single-chain variable antibody fragment
- HBS
- HEPES-buffered saline.
REFERENCES
- 1. Fay P. J. (2006) Factor VIII structure and function. Int. J. Hematol. 83, 103–108 [DOI] [PubMed] [Google Scholar]
- 2. Bovenschen N., Mertens K., Hu L., Havekes L. M., van Vlijmen B. J. (2005) LDL receptor cooperates with LDL receptor-related protein in regulating plasma levels of coagulation factor VIII in vivo. Blood 106, 906–912 [DOI] [PubMed] [Google Scholar]
- 3. Strickland D. K., Ashcom J. D., Williams S., Battey F., Behre E., McTigue K., Battey J. F., Argraves W. S. (1991) Primary structure of α2-macroglobulin receptor-associated protein. Human homologue of a Heymann nephritis antigen. J. Biol. Chem. 266, 13364–13369 [PubMed] [Google Scholar]
- 4. Lazic A., Dolmer K., Strickland D. K., Gettins P. G. (2003) Structural organization of the receptor associated protein. Biochemistry 42, 14913–14920 [DOI] [PubMed] [Google Scholar]
- 5. Saenko E. L., Yakhyaev A. V., Mikhailenko I., Strickland D. K., Sarafanov A. G. (1999) Role of the low density lipoprotein-related protein receptor in mediation of factor VIII catabolism. J. Biol. Chem. 274, 37685–37692 [DOI] [PubMed] [Google Scholar]
- 6. Bovenschen N., Herz J., Grimbergen J. M., Lenting P. J., Havekes L. M., Mertens K., van Vlijmen B. J. (2003) Elevated plasma factor VIII in a mouse model of low-density lipoprotein receptor-related protein deficiency. Blood 101, 3933–3939 [DOI] [PubMed] [Google Scholar]
- 7. Martinelli N., Girelli D., Lunghi B., Pinotti M., Marchetti G., Malerba G., Pignatti P. F., Corrocher R., Olivieri O., Bernardi F. (2010) Polymorphisms at LDLR locus may be associated with coronary artery disease through modulation of coagulation factor VIII activity and independently from lipid profile. Blood 116, 5688–5697 [DOI] [PubMed] [Google Scholar]
- 8. Mello T. B., Siqueira L. H., Montavão S. A., Ozello M. C., Annichino-Bizzacchi J. M. (2008) Low density lipoprotein receptor-related protein polymorphisms are not risk factors for venous thromboembolism. Thromb. Res. 121, 625–629 [DOI] [PubMed] [Google Scholar]
- 9. Pocathikorn A., Granath B., Thiry E., Van Leuven F., Taylor R., Mamotte C. (2003) Influence of exonic polymorphisms in the gene for LDL receptor-related protein (LRP) on risk of coronary artery disease. Atherosclerosis 168, 115–121 [DOI] [PubMed] [Google Scholar]
- 10. Vormittag R., Bencur P., Ay C., Tengler T., Vukovich T., Quehenberger P., Mannhalter C., Pabinger I. (2007) Low-density lipoprotein receptor-related protein 1 polymorphism 663 C → T affects clotting factor VIII activity and increases the risk of venous thromboembolism. J. Thromb. Haemost. 5, 497–502 [DOI] [PubMed] [Google Scholar]
- 11. Jeon H., Blacklow S. C. (2005) Structure and physiologic function of the low-density lipoprotein receptor. Annu. Rev. Biochem. 74, 535–562 [DOI] [PubMed] [Google Scholar]
- 12. Strickland D. K., Ranganathan S. (2003) Diverse role of LDL receptor-related protein in the clearance of proteases and in signaling. J. Thromb. Haemost. 1, 1663–1670 [DOI] [PubMed] [Google Scholar]
- 13. Ananyeva N. M., Makogonenko Y. M., Kouiavskaia D. V., Ruiz J., Limburg V., Meijer A. B., Khrenov A. V., Shima M., Strickland D. K., Saenko E. L. (2008) The binding sites for the very low density lipoprotein receptor and low-density lipoprotein receptor-related protein are shared within coagulation factor VIII. Blood Coagul. Fibrinolysis 19, 166–177 [DOI] [PubMed] [Google Scholar]
- 14. Ananyeva N. M., Makogonenko Y. M., Sarafanov A. G., Pechik I. V., Gorlatova N., Radtke K. P., Shima M., Saenko E. L. (2008) Interaction of coagulation factor VIII with members of the low-density lipoprotein receptor family follows common mechanism and involves consensus residues within the A2 binding site 484–509. Blood Coagul. Fibrinolysis 19, 543–555 [DOI] [PubMed] [Google Scholar]
- 15. Bovenschen N., van Dijk K. W., Havekes L. M., Mertens K., van Vlijmen B. J. (2004) Clearance of coagulation factor VIII in very low-density lipoprotein receptor knockout mice. Br. J. Haematol. 126, 722–725 [DOI] [PubMed] [Google Scholar]
- 16. Christensen E. I., Birn H., Storm T., Weyer K., Nielsen R. (2012) Endocytic receptors in the renal proximal tubule. Physiology (Bethesda) 27, 223–236 [DOI] [PubMed] [Google Scholar]
- 17. Fass D., Blacklow S., Kim P. S., Berger J. M. (1997) Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388, 691–693 [DOI] [PubMed] [Google Scholar]
- 18. Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., Deisenhofer J. (2002) Structure of the LDL receptor extracellular domain at endosomal pH. Science 298, 2353–2358 [DOI] [PubMed] [Google Scholar]
- 19. Daly N. L., Scanlon M. J., Djordjevic J. T., Kroon P. A., Smith R. (1995) Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 92, 6334–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daly N. L., Djordjevic J. T., Kroon P. A., Smith R. (1995) Three-dimensional structure of the second cysteine-rich repeat from the human low-density lipoprotein receptor. Biochemistry 34, 14474–14481 [DOI] [PubMed] [Google Scholar]
- 21. North C. L., Blacklow S. C. (2000) Solution structure of the sixth LDL-A module of the LDL receptor. Biochemistry 39, 2564–2571 [DOI] [PubMed] [Google Scholar]
- 22. Fisher C., Beglova N., Blacklow S. C. (2006) Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol. Cell 22, 277–283 [DOI] [PubMed] [Google Scholar]
- 23. Beglova N., North C. L., Blacklow S. C. (2001) Backbone dynamics of a module pair from the ligand-binding domain of the LDL receptor. Biochemistry 40, 2808–2815 [DOI] [PubMed] [Google Scholar]
- 24. Jensen G. A., Andersen O. M., Bonvin A. M., Bjerrum-Bohr I., Etzerodt M., Thøgersen H. C., O'Shea C., Poulsen F. M., Kragelund B. B. (2006) Binding site structure of one LRP-RAP complex: implications for a common ligand-receptor binding motif. J. Mol. Biol. 362, 700–716 [DOI] [PubMed] [Google Scholar]
- 25. Verdaguer N., Fita I., Reithmayer M., Moser R., Blaas D. (2004) X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat. Struct. Mol. Biol. 11, 429–434 [DOI] [PubMed] [Google Scholar]
- 26. Yasui N., Nogi T., Takagi J. (2010) Structural basis for specific recognition of reelin by its receptors. Structure 18, 320–331 [DOI] [PubMed] [Google Scholar]
- 27. Fisher C., Abdul-Aziz D., Blacklow S. C. (2004) A two-module region of the low-density lipoprotein receptor sufficient for formation of complexes with apolipoprotein E ligands. Biochemistry 43, 1037–1044 [DOI] [PubMed] [Google Scholar]
- 28. Russell D. W., Brown M. S., Goldstein J. L. (1989) Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J. Biol. Chem. 264, 21682–21688 [PubMed] [Google Scholar]
- 29. Andersen O. M., Christensen L. L., Christensen P. A., Sørensen E. S., Jacobsen C., Moestrup S. K., Etzerodt M., Thogersen H. C. (2000) Identification of the minimal functional unit in the low density lipoprotein receptor-related protein for binding the receptor-associated protein (RAP): a conserved acidic residue in the complement-type repeats is important for recognition of RAP. J. Biol. Chem. 275, 21017–21024 [DOI] [PubMed] [Google Scholar]
- 30. Andersen O. M., Petersen H. H., Jacobsen C., Moestrup S. K., Etzerodt M., Andreasen P. A., Thøgersen H. C. (2001) Analysis of a two-domain binding site for the urokinase-type plasminogen activator-plasminogen activator inhibitor-1 complex in low-density-lipoprotein-receptor-related protein. Biochem. J. 357, 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meijer A. B., Rohlena J., van der Zwaan C., van Zonneveld A. J., Boertjes R. C., Lenting P. J., Mertens K. (2007) Functional duplication of ligand-binding domains within low-density lipoprotein receptor-related protein for interaction with receptor associated protein, α2-macroglobulin, factor IXa, and factor VIII. Biochim. Biophys. Acta 1774, 714–722 [DOI] [PubMed] [Google Scholar]
- 32. Neels J. G., van Den Berg B. M., Lookene A., Olivecrona G., Pannekoek H., van Zonneveld A. J. (1999) The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J. Biol. Chem. 274, 31305–31311 [DOI] [PubMed] [Google Scholar]
- 33. Lenting P. J., Neels J. G., van den Berg B. M., Clijsters P. P., Meijerman D. W., Pannekoek H., van Mourik J. A., Mertens K., van Zonneveld A. J. (1999) The light chain of factor VIII comprises a binding site for low density lipoprotein receptor-related protein. J. Biol. Chem. 274, 23734–23739 [DOI] [PubMed] [Google Scholar]
- 34. Sarafanov A. G., Ananyeva N. M., Shima M., Saenko E. L. (2001) Cell surface heparan sulfate proteoglycans participate in factor VIII catabolism mediated by low density lipoprotein receptor-related protein. J. Biol. Chem. 276, 11970–11979 [DOI] [PubMed] [Google Scholar]
- 35. Saenko E. L., Shima M., Gilbert G. E., Scandella D. (1996) Slowed release of thrombin-cleaved factor VIII from von Willebrand factor by a monoclonal and a human antibody is a novel mechanism for factor VIII inhibition. J. Biol. Chem. 271, 27424–27431 [DOI] [PubMed] [Google Scholar]
- 36. Kurasawa J. H., Shestopal S. A., Jha N. K., Ovanesov M. V., Lee T. K., Sarafanov A. G. (2013) Insect cell-based expression and characterization of a single-chain variable antibody fragment directed against blood coagulation factor VIII. Protein. Expr. Purif. 88, 201–206 [DOI] [PubMed] [Google Scholar]
- 37. Sarafanov A. G., Makogonenko E. M., Andersen O. M., Mikhailenko I. A., Ananyeva N. M., Khrenov A. V., Shima M., Strickland D. K., Saenko E. L. (2007) Localization of the low-density lipoprotein receptor-related protein regions involved in binding to the A2 domain of coagulation factor VIII. Thromb. Haemost. 98, 1170–1181 [PubMed] [Google Scholar]
- 38. Sarafanov A., Saenko E. (2004) High-throughput optimization of protein expression in the baculovirus system based on determination of relative expression efficiency of viral stocks. Anal. Biochem. 328, 98–100 [DOI] [PubMed] [Google Scholar]
- 39. Orlando R. A., Exner M., Czekay R. P., Yamazaki H., Saito A., Ullrich R., Kerjaschki D., Farquhar M. G. (1997) Identification of the second cluster of ligand-binding repeats in megalin as a site for receptor-ligand interactions. Proc. Natl. Acad. Sci. U.S.A. 94, 2368–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sato A., Shimada Y., Herz J., Yamamoto T., Jingami H. (1999) 39-kDa receptor-associated protein (RAP) facilitates secretion and ligand binding of extracellular region of very-low-density-lipoprotein receptor: implications for a distinct pathway from low-density-lipoprotein receptor. Biochem. J. 341, 377–383 [PMC free article] [PubMed] [Google Scholar]
- 41. Christiansen M. L., Balling K. W., Persson E., Hilden I., Bagger-Sørensen A., Sørensen B. B., Viuff D., Segel S., Klausen N. K., Ezban M., Lethagen S., Steenstrup T. D., Kjalke M. (2010) Functional characteristics of N8, a new recombinant FVIII. Haemophilia 16, 878–887 [DOI] [PubMed] [Google Scholar]
- 42. Butenas S., Parhami-Seren B., Gissel M. T., Gomperts E. D., Fass D. N., Mann K. G. (2009) Potency and mass of factor VIII in FVIII products. Haemophilia 15, 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin Y., Yang X., Chevrier M. C., Craven S., Barrowcliffe T. W., Lemieux R., Ofosu F. A. (2004) Relationships between factor VIII:Ag and factor VIII in recombinant and plasma-derived factor VIII concentrates. Haemophilia 10, 459–469 [DOI] [PubMed] [Google Scholar]
- 44. Ofosu F. A., Tse H., Naqvi A., Bhakta H., Song Y. (2012) The fraction of recombinant factor VIII:Ag unable to bind von Willebrand factor has no FVIII coagulant activity: studies in vitro. Haemophilia 18, 917–925 [DOI] [PubMed] [Google Scholar]
- 45. Pahl S., Pavlova A., Driesen J., Müller J., Pötzsch B., Oldenburg J. (2013) In vitro characterization of recombinant factor VIII concentrates reveals significant differences in protein content, activity, and thrombin activation profile. Haemophilia 19, 392–398 [DOI] [PubMed] [Google Scholar]
- 46. Limburg V., van der Zwaan C., Boertjes R. C., Bovenschen N., Mertens K., Meijer A. (2005) An antibody fragment against factor VIII that inhibits factor VIII assembly with von Willebrand factor and LDL receptor family members. J. Thromb. Haemost. 3, Suppl. 1, OR162 [Google Scholar]
- 47. Meijer. A., Limburg V., van der Zwaan C., Mertens K. (2007) The factor VIII C1 domain contributes to efficient LRP/LDL receptor binding. J. Thromb. Haemost. 5, Suppl. 2, P-M-040 [Google Scholar]
- 48. Meems H., Meijer A. B., Cullinan D. B., Mertens K., Gilbert G. E. (2009) Factor VIII C1 domain residues Lys 2092 and Phe 2093 contribute to membrane binding and cofactor activity. Blood 114, 3938–3946 [DOI] [PubMed] [Google Scholar]
- 49. Bieri S., Atkins A. R., Lee H. T., Winzor D. J., Smith R., Kroon P. A. (1998) Folding, calcium binding, and structural characterization of a concatemer of the first and second ligand-binding modules of the low-density lipoprotein receptor. Biochemistry 37, 10994–11002 [DOI] [PubMed] [Google Scholar]
- 50. Atkins A. R., Brereton I. M., Kroon P. A., Lee H. T., Smith R. (1998) Calcium is essential for the structural integrity of the cysteine-rich, ligand-binding repeat of the low-density lipoprotein receptor. Biochemistry 37, 1662–1670 [DOI] [PubMed] [Google Scholar]
- 51. Croy J. E., Brandon T., Komives E. A. (2004) Two apolipoprotein E mimetic peptides, ApoE(130–149) and ApoE(141–155)2, bind to LRP1. Biochemistry 43, 7328–7335 [DOI] [PubMed] [Google Scholar]
- 52. Huang W., Dolmer K., Gettins P. G. (1999) NMR solution structure of complement-like repeat CR8 from the low density lipoprotein receptor-related protein. J. Biol. Chem. 274, 14130–14136 [DOI] [PubMed] [Google Scholar]
- 53. Dolmer K., Huang W., Gettins P. G. (2000) NMR solution structure of complement-like repeat CR3 from the low density lipoprotein receptor-related protein. Evidence for specific binding to the receptor binding domain of human α2-macroglobulin. J. Biol. Chem. 275, 3264–3269 [DOI] [PubMed] [Google Scholar]
- 54. Guo Y., Yu X., Rihani K., Wang Q. Y., Rong L. (2004) The role of a conserved acidic residue in calcium-dependent protein folding for a low density lipoprotein (LDL)-A module: implications in structure and function for the LDL receptor superfamily. J. Biol. Chem. 279, 16629–16637 [DOI] [PubMed] [Google Scholar]
- 55. Simonovic M., Dolmer K., Huang W., Strickland D. K., Volz K., Gettins P. G. (2001) Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor. Biochemistry 40, 15127–15134 [DOI] [PubMed] [Google Scholar]
- 56. Guttman M., Komives E. A. (2011) The structure, dynamics, and binding of the LA45 module pair of the low-density lipoprotein receptor suggest an important role for LA4 in ligand release. Biochemistry 50, 11001–11008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Medh J. D., Fry G. L., Bowen S. L., Pladet M. W., Strickland D. K., Chappell D. A. (1995) The 39-kDa receptor-associated protein modulates lipoprotein catabolism by binding to LDL receptors. J. Biol. Chem. 270, 536–540 [DOI] [PubMed] [Google Scholar]
- 58. Strickland D. K., Ashcom J. D., Williams S., Burgess W. H., Migliorini M., Argraves W. S. (1990) Sequence identity between the α2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J. Biol. Chem. 265, 17401–17404 [PubMed] [Google Scholar]
- 59. Battey F. D., Gåfvels M. E., FitzGerald D. J., Argraves W. S., Chappell D. A., Strauss J. F., 3rd, Strickland D. K. (1994) The 39-kDa receptor-associated protein regulates ligand binding by the very low density lipoprotein receptor. J. Biol. Chem. 269, 23268–23273 [PubMed] [Google Scholar]
- 60. Kounnas M. Z., Argraves W. S., Strickland D. K. (1992) The 39-kDa receptor-associated protein interacts with two members of the low density lipoprotein receptor family, α2-macroglobulin receptor and glycoprotein 330. J. Biol. Chem. 267, 21162–21166 [PubMed] [Google Scholar]
- 61. Dolmer K., Gettins P. G. (2006) Three complement-like repeats compose the complete α2-macroglobulin binding site in the second ligand binding cluster of the low density lipoprotein receptor-related protein. J. Biol. Chem. 281, 34189–34196 [DOI] [PubMed] [Google Scholar]
- 62. Esser V., Limbird L. E., Brown M. S., Goldstein J. L., Russell D. W. (1988) Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J. Biol. Chem. 263, 13282–13290 [PubMed] [Google Scholar]
- 63. Zhang D. W., Garuti R., Tang W. J., Cohen J. C., Hobbs H. H. (2008) Structural requirements for PCSK9-mediated degradation of the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 105, 13045–13050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lenting P. J., Donath M. J., van Mourik J. A., Mertens K. (1994) Identification of a binding site for blood coagulation factor IXa on the light chain of human factor VIII. J. Biol. Chem. 269, 7150–7155 [PubMed] [Google Scholar]
- 65. Bovenschen N., Boertjes R. C., van Stempvoort G., Voorberg J., Lenting P. J., Meijer A. B., Mertens K. (2003) Low density lipoprotein receptor-related protein and factor IXa share structural requirements for binding to the A3 domain of coagulation factor VIII. J. Biol. Chem. 278, 9370–9377 [DOI] [PubMed] [Google Scholar]
- 66. Bovenschen N., van Stempvoort G., Voorberg J., Mertens K., Meijer A. B. (2006) Proteolytic cleavage of factor VIII heavy chain is required to expose the binding-site for low-density lipoprotein receptor-related protein within the A2 domain. J. Thromb. Haemost. 4, 1487–1493 [DOI] [PubMed] [Google Scholar]
- 67. Meems H., van den Biggelaar M., Rondaij M., van der Zwaan C., Mertens K., Meijer A. B. (2011) C1 domain residues Lys 2092 and Phe 2093 are of major importance for the endocytic uptake of coagulation factor VIII. Int. J. Biochem. Cell Biol. 43, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 68. van den Biggelaar M., van der Zwaan C., Mertens K., Meijer A. (2011) Low density lipoprotein receptor-related protein (LRP) cluster II interacts with the factor VIII light chain via an extended surface comprising multiple lysine residues. J. Thromb. Haemost. 9, Suppl. 2, 821 [Google Scholar]
- 69. van den Brink E. N., Turenhout E. A., Bovenschen N., Heijnen B. G., Mertens K., Peters M., Voorberg J. (2001) Multiple VH genes are used to assemble human antibodies directed toward the A3-C1 domains of factor VIII. Blood 97, 966–972 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.