Abstract
Mitochondrial (mt) genome sequences have enabled comparison of population genetics and evolution for numerous free-living and parasitic nematodes. Here we define the complete mt genome of Wuchereria bancrofti through analysis of isolates from Papua New Guinea, India and West Africa. Sequences were assembled for each isolate and annotated with reference to the mt genome sequence for Brugia malayi. The length of the W. bancrofti mt genome is approximately 13,637 nucleotides, contains 2 ribosomal RNAs (rrns), 22 transfer RNAs (trns), 12 protein-coding genes, and is characterized by a 74.6% AT content. The W. bancrofti mt gene order is identical to that reported for Onchocerca volvulus, Dirofilaria immitis, Setaria digitata and B. malayi. In addition to using translational start codons identified previously in the mt protein-coding genes of other filarial nematodes, W. bancrofti appears to be unique in using TGT as a translational start codon. Similarly, use of incomplete stop codons in mt protein-coding genes appears to be more common in W. bancrofti than in other human filarial parasites. The complete mt genome sequence reported here provides new genetic markers for investigating phylogenetic and geographic relationships between isolates, and assessing population diversity within endemic regions. The sequence polymorphism enables new strategies to monitor the progress of public health interventions to control and eliminate this important human parasite. We illustrate the utility of this sequence and single nucleotide polymorphisms by inferring the divergence times between the three W. bancrofti isolates, suggesting predictions into their origin and migration.
1. Introduction
Wuchereria bancrofti, a filarial nematode of the order Spirurida is the primary causative agent of lymphatic filariasis, a deforming and debilitating disease estimated to affect 120 million people in 72 countries [57]. Mosquito species belonging to Anopheles, Aedes, Culex and Mansonia have been reported to be involved in the transmission of lymphatic filariasis [26]. While the Culex spp. are the principal vectors of bancroftian filariasis in India, the Anopheles spp. are important in the transmission of W. bancrofti in West Africa, Southern Asia and the island of New Guinea. Despite constituting a major public health problem in many tropical and subtropical regions, W. bancrofti remains poorly understood with respect to its genetics and genomics.
Mitochondrial (mt) DNA has provided a rich source of markers for taxonomic and population genetic studies of various species of parasitic nematodes. The mtDNA consists of a closed circular molecule ranging in size from approximately 13.6 to 14.3 kb. Most nematode mt genomes are smaller than those of other metazoans and encode 2 ribosomal RNA (rrn) genes, 22 transfer RNA (trn) genes, and 12 protein-coding genes [22]. Together, these genes play an important role in oxidative phosphorylation in nematode mitochondria. Prior to this study, the only W. bancrofti mtDNA sequence data available was a partial cox1 sequence reported in a molecular phylogenetic study of filarial parasites [6]. Because mutation and recombination events in nematode mt genomes are usually taxon-specific, mtDNA sequences are valuable for the study of genetic variation within and between nematode species [30]. Analysis of substitution rates at mt cox1 and nad4 loci in pairs of congeneric nematode species suggests that mtDNA markers are well suited for the detection of cryptic nematode species [4]. In another study, Hu and colleagues used complete mt genome sequences to estimate the magnitude of genetic variability between two geographically distinct isolates of hookworm Necator americanus [18]. Revealing substantial nucleotide differences between these isolates, this work demonstrated the potential usefulness of mtDNA markers in future studies aimed at understanding the genetic basis for different epidemiological patterns before and during attempts at disease control, or population stratification that may be associated with anthelmintic drug resistance [44], observed in W. bancrofti infection.
With the advent of new PCR and sequencing technologies [23,24], research aimed at determining the sequence and structure of nematode mt genomes has increased in recent years. To date, complete mt genome sequences have been reported for more than 12 species of parasitic nematodes, including four filarial species [13,18,25,56]. Genomic sequence data may provide the basis for both fundamental and applied studies of the biochemistry and molecular biology of W. bancrofti. These studies may lead to novel approaches to the diagnosis and treatment of filarial infection [41].
Here, we report the complete mt genome sequence of W. bancrofti from three different geographic locations namely India, West Africa and Papua New Guinea (PNG). In reporting these sequences, we describe their gene order, codon usage bias, nucleotide composition and genetic variability among the three isolates. Using mtDNA sequence polymorphisms we further attempt to reconstruct divergence dates among the three isolates and to infer the geographic origin of W. bancrofti. Through our efforts we aim to facilitate future epidemiological studies of bancroftian filariasis by identifying molecular polymorphisms for this important human parasite.
2. Materials and methods
2.1. Blood sample collection
We obtained whole blood samples from individuals living in a W. bancrofti endemic region of PNG, Dreikikir District, East Sepik Province. The W. bancrofti sample from South India was isolated from purified microfilariae obtained from microfilaremic subjects in South India (Chennai). The W. bancrofti sample from Mali was obtained from a single adult worm removed during fine needle aspiration during a diagnostic procedure. All blood samples were collected under clinical protocols approved by institutional review boards (IRBs) at University Hospitals Case Medical Center and the Papua New Guinea Institute of Medical Research. The microfilariae from South India were collected as part of a clinical protocol (NCT00342576) approved by the IRBs of the NIAID and the Tuberculosis Research Centre. The sample from Mali was obtained as part of a clinical trial (NCT00471666) that had been approved by the IRBs of the NIAID and the University of Bamako, Mali.
2.2. Genomic DNA extraction
We extracted genomic DNA (gDNA) from whole blood samples (200 µL) of Papua New Guinea study subjects using a QIAamp 96 DNA Blood Kit (QIAGEN, Valencia, CA). Genomic DNA for the Indian sample was extracted by phenol-chloroform extraction protocol and purified by CsCl density gradient centrifugation. Genomic DNA from the adult worm from Mali was isolated using the Qiagen genomic tip 100/g kit (QIAGEN).
After isolation all gDNA samples were stored at 4°C. W. bancrofti positive sample was confirmed by post-PCR ligase detection reaction-fluorescent microsphere assay (LDR- FMA) [33] and blood smear microscopy. Positive samples were kept at 4°C for PCR amplification while negative samples were stored at −80°C for future reference.
2.3. W. bancrofti mt genome amplification strategy
The current state of the W. bancrofti whole genome sequencing project (PRJNA37759) at the Broad Institute includes over 25,000 super-contigs and is working toward developing completed chromosome assemblies and annotation. We therefore sought to independently assemble the W. bancrofti mt genome. We designed our PCR primers by a BLAST search of the Broad Institute’s filarial worm sequencing database (www.broadinstitute.org/annotation/genome/filarial_worms), which contains the whole genome sequence for a W. bancrofti isolate from Mali, West Africa, using the assembled mt genome sequence of B. malayi (GenBank ID AF538716) as a reference. Our BLAST search returned 9725 base pairs of sequence that were then aligned to the B. malayi mt genome using MacVector 11.1 (MacVector, Cary, NC). The resulting sequence alignment produced preliminary mt DNA sequences with several large gaps. Next we designed 15 primer sets spanning the entire mt genome, using the draft assembly produced above (Table S1).
We performed PCR amplification on 3-µL aliquots DNA template found to be positive for W. bancrofti. PCR reactions varied in their use of buffer, where reactions involving primer sets 701, 1751, 2721, 3691, 5611, 6425, 7441, and 11420 used a 10-fold dilutions of a 10× buffer containing 16.6 mM (NH4)2SO4, 10 mM β-mercaptoethanol, 3.4 or 6.7 mM MgSO4, and 67 mM Tris-HCl (pH 8.0, 8.3, 8.5, or 8.8) and reactions involving primer sets 41, 4651, 8461, 9441, 10421, 12381, and 13119 used a 10-fold dilutions of a 10× buffer containing 500 mM KCl, 0.1% (w/v) gelatin, 7.5 or 15 mM MgCl2, and 100 mM Tris- HCl at pH 8.3. PCR master mixtures 3 µL pooled gDNA (22 ng), 0.6 µL each primer (6 pmol), 2 µL dNTPs (5 nmol each dNTP), 2.5 µL 10× buffer, 0.6 µL Taq DNA polymerase (2.5 units), and 18.7 µL nuclease-free water. We used an MJ Research PTC-225 thermocycler (MJ Research, Waltham, MA) under the thermocycling conditions: 2 min (initial denaturation) at 92°C, followed by 40 cycles of 30 s denaturation) at 92°C, 30 s (annealing) at 48°C, and 60 s (extension) at 72°C, for all reactions.
2.4. Agarose gel electrophoresis and gel extraction
We confirmed PCR product amplification by electrophoresis through 2% (w/v) agarose gels in 1× TBE. Gels were then stained with SYBR Gold (Molecular Probes, Eugene, OR) and the embedded amplicons visualized using a Storm 860 molecular imaging system with ImageQuant 5.2 software Molecular Dynamics, Sunnyvale, CA). We confirmed product amplification by comparing observed length on the agarose gel with expected fragment length, as predicted by MacVector. We then extracted and purified observed products that met the predicted nucleotide length using the QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA). Purified PCR products were TA cloned into pCR2.1-TOPO vector and transformed into chemically competent E. coli TOP10 cells according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). Sequencing reactions of plasmid inserts using M13 universal primers and capillary electrophoresis were performed by Beckman Coulter Genomics (Danvers, MA).
2.5. Sequence assembly and gene annotation
We edited and assembled sequence read data using Sequencher 4.8 (Gene Codes, Ann Arbor, MI) and Geneious 5.3 (Biomatters, Auckland, NZ). These sequences were then assembled into contigs and aligned to the complete mt genome reported previously for B. malayi [13]; annotations from B. malayi were used as a template for annotation of the complete mt genome of Wb. The boundaries of the rrn and protein-coding genes, as well as those of the non-coding AT- rich region, were determined from the resulting sequence alignment. All trn genes were identified using either ARWEN or trnAscan- SE 1.21 [29,32,49].
Following de-novo amplification, assembly, and annotation of the PNG and Indian W. bancrofti mtDNA sequences, we verified our sequence by a BLAST search against the Broad Institute’s filarial worm sequencing database. The BLAST search returned the same result as noted above, which prompted us to reassemble the mtDNA sequence of the West African isolate from the whole genome sequence deposited in the Sequence Read Archive (SRA; accession number SRX004833). ABySS [51] was run using the following parameters: k-mer size=35, coverage=100. The two largest contigs (9kb and 5kb) were then annotated and aligned to both the B. malayi and the W. bancrofti consensus sequence from PNG and India using Geneious 5.3. The full-length mtDNA sequences of W. bancrofti-PNG, W. bancrofti-India, and W. bancrofti-West Africa have been deposited in GenBank under accession numbers JF775522, JQ316200, and JN367461, respectively.
We used Geneious 5.3 to determine nucleotide identities between the rrn sequences of W. bancrofti and those B. malayi. In addition, we also estimated patterns of codon usage bias for the 12-mt protein-coding genes using CodonO [1] (Table S2). Non-coding regions were analyzed for tandem and inverted repeats using Tandem Repeats Finder and EMBOSS, respectively [3,45]. Based on the results of these analyses, a secondary structure was predicted for a 46-nt non-coding sequence located between trnW and nad6 (Figure S1). Further secondary structures were also predicted for all trn genes using both ARWEN and tRNAscan-SE 1.21 [29,32] (Figure S2). The structures predicted by these programs were reformatted using RnaViz 2.0 [47,48].
2.6. Sequence Polymorphisms
We generated a nucleotide sequence alignment containing all three W. bancrofti isolates in Geneious 5.3 using the MUSCLE algorithm [10]. The nucleotide alignment was checked for translational reading frame shifts by translation and then visual inspection. The statistic π [36], pairwise nucleotide diversity, was calculated between the three isolates using a 100 base pair sliding window in 20 base pair steps implemented in the program DnaSP 5.0 [31] (Figure 1). Single nucleotide polymorphisms were calculated for each site with PNG as a reference by comparing alignments using the coding library Biopython (Figure 1). Percent identity and sequence length were calculated using Geneious 5.3, Ts:Tv, the transition to transversion ratio of nucleotide substitutions was calculated using Arlequin 3.0[11], and Dn/Ds ratios for each gene, which represents the ratio of non-synonymous substitution (those that change the amino acid sequence) to synonymous substitutions (those that do not change the amino acid sequence), were calculated in DnaSP 5.0 [31].
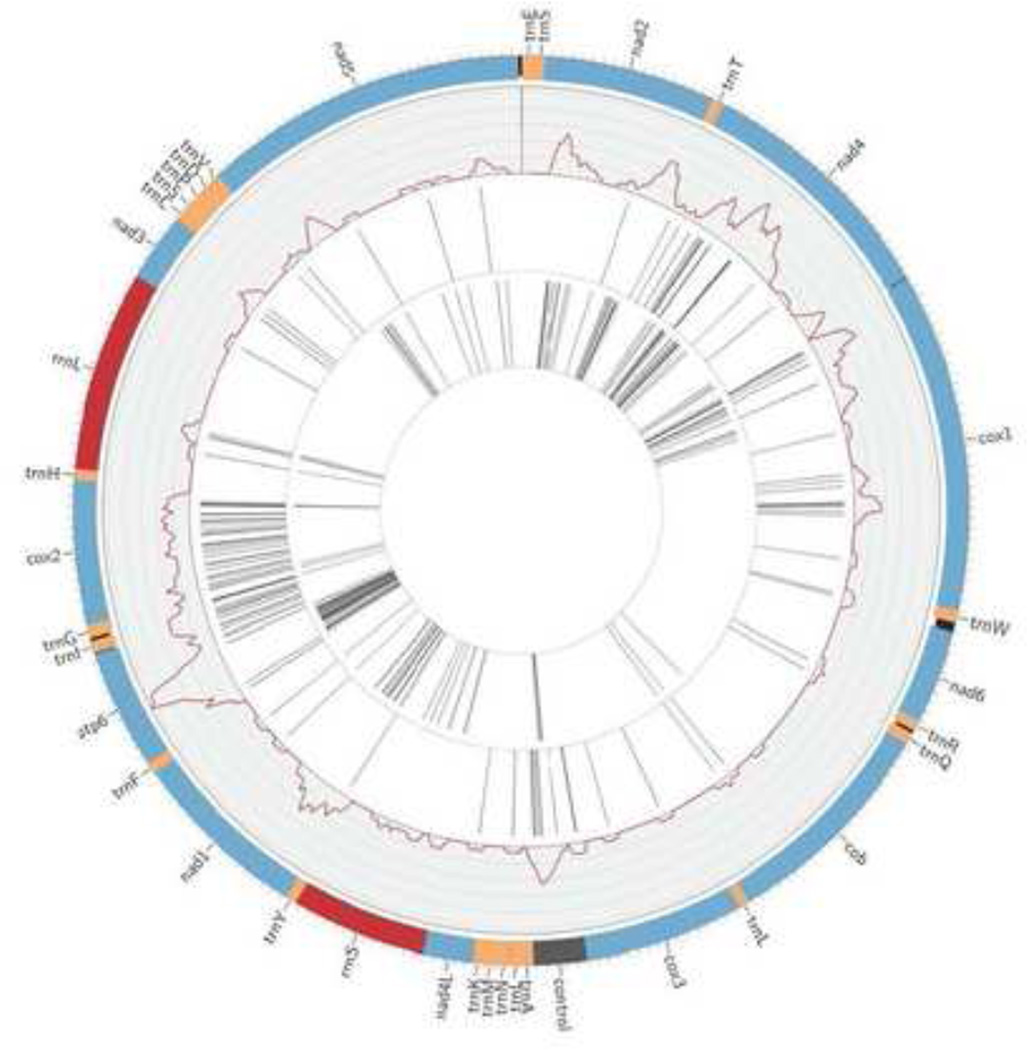
Figure 1.
Mt genome representation using the program circos [28]. Protein coding sequences (CDS) are represented in blue, tRNAs (trns) represented in orange, rRNAs (rrns) represented in red, AT control region represented in grey, and intergenic regions are black. The outermost inset represents the pairwise nucleotide diversity,π, between the three isolates; axis is scaled from 0 to a maximum value of 0.2. The second and third insets are a representation of nucleotide positions containing SNPs between PNG-West Africa and PNG-India, respectively.
2.7. Phylogenetic reconstruction and estimation of the divergence times between the W. bancrofti species in India, PNG and West Africa
For phylogenetic analysis the protein coding sequence for the three W. bancrofti isolates and 4 other species (B. malayi (GenBank ID NC004298), O. volvulus (GenBank ID NC001861), D immitis (GenBank ID NC005305), and S. digitata (GenBank ID NC014282) were extracted and aligned using the MUSCLE algorithm in the program Geneious 5.3. The sequence alignment was then used to infer the best-fit model of nucleotide substitution using the program jModelTest [43]. jModelTest constructs a likelihood ratio test to compare different models of nucleotide substitutions by using an information criteria framework. We selected the nucleotide substitution model that had the best Akaike information criterion (AIC) for our dataset.
We used BEAST [9] to construct a Bayesian phylogeny using the aligned data set outlined above. Model parameters in BEAST were set to the following values: W. bancrofti was treated as a monophyletic group with S. digitata acting as an out-group sequence; we set the uncorrelated log-normal mutation rate prior with a log-normal distribution around a mean rate as given by the mutation accumulation experiment in Pristionchus pacificus [35]; we set GTR +I +G, the general time reversible model with I proportion of invariant sites and rates between sites described by a gamma distribution G [54], as the nucleotide substitution model which was supported by the best AIC-corrected likelihood using jModelTest; we also assumed a random starting tree with a Yule prior [9] for species tree construction. The compiled model was run 3 independent times for 20 million states e ach with a burn-in of 500k states. We ensured convergence of each independent run by examining the autocorrelation between parameters, trace file of the Markov chains, and only accepted parameter estimates with an effective sample size of greater than 200.
In addition to BEAST we inferred divergence times using IMa2, an Isolation-Migration model framework [15] that allows for more accurate estimates of divergence times when there is concurrent gene flow. We used the same alignment as the Bayesian analysis but chose to use the substitution model, Hasegawa, Kishino, and Yano model allows for different rates of transitional and transversional substitutions as well as taking into account nucleotide frequencies [14], as the GTR model was not an available option. The choice of HKY, instead of the previously supported GTR, has been shown to have little effect on mean parameter estimates but may increase the variance around these estimates [53]. Populations were compared in pairwise models with the following priors: upper bound of divergence time was set at=200, which equals approximately 200 kya; migration was represented by a uniform distribution from 0–1; upper bound for population size, which was 5 times the geometric mean of the population mutation rate for the locus, was set at 800. We ran each model 3 separate times for 20 million states each, with sampling occurring every 1000 states. Each model included 10 independent Markov chains that followed a geometric heating model to increase mixing among chains[12].
3. Results and discussion
3.1. General features of the mt genome
The complete mt genome sequence of W. bancrofti is 13,637 nt in length (Figure 1), shorter than the mt genomes of B. malayi (13,657 nt), O. volvulus (13,747 nt), D. immitis (13,814 nt), and S. digitata (13,839 nt). Although it is shorter than the mt genomes of other filarial nematodes, the mt genome of W. bancrofti exhibits the same gene order, including the positions of the trn, rrn and non-coding regions. Though the genome organization is distinctly different from other secernentean nematodes; the gene and gene-block translocations resulting due to mt rearrangements do not disrupt the mt function [5].
The mt genome of W. bancrofti contains 2 rrns, 22 trns, 12 protein-coding genes (cox1-cox3, nad1-nad6, nad4L, atp6, and cob), and an AT-rich region. Similar to the mt genomes of other filarial nematodes, the W. bancrofti mt genome lacks the atp8 gene. The overall nucleotide composition is 20.1% A, 54.5% T, 18.1% G, and 7.2% C (Table S3).
Short intergenic regions ranging from 1 to 46 nt in length are interspersed throughout the W. bancrofti mt genome. Six protein-coding genes are found to overlap with adjacent trn genes by 1 to 3 nt. One of these, cob, overlaps both its 5’- and 3’-adjacent trns, sharing 1 nt with trnQ at its 5’-end and 2 nt with trnL at its 3’-end. The nad1 and trnF genes of W. bancrofti mtDNA are not predicted to overlap. This situation contrasts starkly with that observed in D. immitis, O. volvulus, and S. digitata, where the mt nad1 and trnF genes are predicted to overlap by 21, 23, and 26 nt, respectively. Overlap of adjacent genes is common in nematode mtDNA and it has been suggested that these overlaps are resolved by transcript editing [19].
3.2. Protein-coding genes
The mt12 protein-coding genes are all transcribed in the same direction. Four of these genes use ATT as the translation initiation codon (cob and cox1-cox3) and two of them use TTG (nad1 and nad4). The other translation initiation codons used are TTA (nad2), TGT (nad6), GTA (nad4L), ATA (atp6), CTT (nad3), and TTT (nad5) (Table 1). Whereas TTT has been reported as the initiation codon for nad5 in B. malayi and S. digitata, CTT has been reported as the initiation codon for nad3 in B. malayi, O. volvulus, and D. immitis. TTA, TAT, and GTA are rarely used as initiation codons in nematodes, yet they have been reported as initiation codons for the nad4L gene in B. malayi and D. immitis. Use of TGT as an initiation codon appears to be unique to the mt genome of W. bancrofti.
Table 1.
Nucleotide positions and lengths of each gene encoded by PNG W. bancrofti isolate mitochondrial DNA, as well as start and stop codons and lengths of translation products inferred for each protein-coding gene. The numbers in parenthesis represent differences between the PNG W. bancrofti isolate and the W. African and Indian isolate, respectively.
| Gene/region | nt start position |
nt stop position |
Length | Codon | ||
|---|---|---|---|---|---|---|
| No. of nt | No. of aa | Initiation | Termination | |||
| trnE | 1 | 59 | 59 | |||
| trnS(AGN) | 59 | 112 | 54 | |||
| nad2 | 112 | 955 | 844 | 281 | TTA | T |
| trnT | 956 | 1013 (+2/+2) | 58 (+2/+2) | |||
| nad4 | 1013 (+2/+2) | 2242 (+2/+2) | 1230 | 409 | TTG | TAA |
| cox1 | 2250 (+2/+2) | 3896 (+2/+2) | 1647 | 548 | ATT | TAA |
| trnW | 3899 (+2/+2) | 3956(+1/+2) | 58(−1/−) | |||
| nad6 | 4000 (+1/+2) | 4455 (+1/+2) | 456 | 150 | TGT | TAA |
| trnR | 4454 (+1/+2) | 4508 (+1/+3) | 55(−/+2) | |||
| trnQ | 4526 (+1/+3) | 4581 (+1/+3) | 56 | |||
| cob | 4581 (+1/+3) | 5668 (+1/+3) | 1088 | 362 | ATT | TA |
| trnL(CUN) | 5667 (+1/+3) | 5722 (+1/+3) | 56 | |||
| cox3 | 5722 (+1/+3) | 6501 (+1/+3) | 780 | 259 | ATT | TAA |
| AT-rich region | 6502 (+1/+3) | 6768 (−2/+3) | 267(−3/−) | |||
| trnA | 6769 (−2/+3) | 6825 (−2/+3) | 57 | |||
| trnL(UUR) | 6828 (−2/+3) | 6880 (−2/+3) | 53 | |||
| trnN | 6884 (−2/+3) | 6940 (−2/+3) | 57 | |||
| trnM | 6943 (−2/+3) | 7001 (−2/+3) | 59 | |||
| trnK | 7001 (−2/+3) | 7058 (−2/+3) | 58 | |||
| nad4L | 7059 (−2/+3) | 7301 (−2/+3) | 243 | 80 | GTA | TAA |
| rrnS | 7302 (−2/+3) | 7972 (−2/+3) | 671 (+1/−) | |||
| trnY | 7973 (−1/+3) | 8026 (−1/+3) | 54 | |||
| nad1 | 8024 (−1/+3) | 8900(−1/+3) | 877 | 292 | TTG | T |
| trnF | 8901 (−1/+3) | 8956 (−1/+3) | 56 | |||
| atp6 | 8957 (−1/+3) | 9538 (−1/+3) | 582 | 193 | ATA | TAG |
| trnI | 9546 (−1/+3) | 9603 (−1/+3) | 58 | |||
| trnG | 9621(−1/+3) | 9678 (−1/+3) | 58 | |||
| cox2 | 9681 (−1/+3) | 10380 (−1/+3) | 700 | 233 | ATT | T |
| trnH | 10381 (−1/+3) | 10434 (−1/+4) | 54 (−/+5) | |||
| rrnL | 10436 (−1/+4) | 11407 (−1/+5) | 972(−/+1) | |||
| nad3 | 11409 (−1/+5) | 11745 (−1/+5) | 337 | 112 | CTT | T |
| trnC | 11746 (−1/+5) | 11801 (−1/+5) | 56 | |||
| trnS(UCN) | 11802 (−1/+5) | 11857 (−1/+5) | 56 | |||
| trnP | 11858 (−1/+5) | 11915 (−1/+5) | 58 | |||
| trnD | 11918 (−1/+5) | 11976 (−1/+5) | 59 | |||
| trnV | 11976 (−1/+5) | 12031 (−1/+5) | 56 | |||
| nad5 | 12032 (−1/+5) | 13627 (−1/+5) | 1596 | 531 | TTT | TAG |
While 7 of the 12 protein-coding genes use TAG/TAA as translation termination codons, the other 5 genes (cob, cox2, and nad1-nad3) use abbreviated termination codons of the form T or TA (Table 1). Interestingly, trn genes follow all genes that employ an incomplete stop codon. It has been suggested that these incomplete stop codons are converted to TAA post-transcriptionally [21].
Nucleotide compositions of the protein-coding genes a re shown in Table S4. These nucleotide compositions reflect the strong overall T bias of the W. bancrofti mt genome. The mt protein-coding genes of W. bancrofti are characterized by a high AT content ranging from 67.8 to 81%. Relatively long polyT tracts (8–15 Ts) are found in abundance in these genes; the longest tract within the protein coding genes (15 Ts) is found within the nad5 gene. In addition, the third codon position of these genes has a higher T bias (68.5%) than either the first (47.3%) or the second (51%) codon positions (Table S3). Though the overall genome is biased against C, there is selection for a higher C content (7.9 –12%) in the first and second codon positions, indicative of the mutational pattern in the mt genome [50]. Similar results have been observed in other nematodes [19,21,25,56].
Of the 64 possible codons, all but all but 6 codons (CTC, CCC, ACA, ACG, GCG and CGC) are used in the W. bancrofti mt genome (Table S2). The most frequently used codons are T-rich: TTT (17.9%), GTT (7.9%), TTG (7.1%), TAT (6.2%), ATT (5.4%), TTA (4.9%), and TCT (4.6%). Although serine, leucine, and phenylalanine are among the most frequently used amino acids in W. bancrofti mt proteins (Table S5), codons TTC (Phe), CTN (Leu), TCC (Ser), TCA (Ser), and TCG (Ser) are seldom used due to the strong bias against C in this genome. Within each codon family, T is preferred in the third position over G, A, and C, highlighting the strong overall T bias of the genome Table S3). Glutamine (CAR) and histidine (CAY) are the amino acids least used in W. bancrofti mt proteins (Table S5).
The predicted lengths of all the genes in W. bancrofti are very similar to those of B. malayi (≤ 1 amino acid difference), D. immitis and O. volvulus (≤ 5 amino acid differences). With the exception of the cox1 gene, the predicted lengths are similar to those of Necator americanus, Ancylostoma duodenale, Caenorhabditis elegans (≤8 amino acid differences), Ascaris suum and Strongyloides stercoralis (≤6 amino acid differences). The length of the cox1 gene is highly variable (≤35 amino acid differences); however W. bancrofti, B. malayi, D. immitis and O. volvulus have an upstream TA that may terminate the protein, resulting in a protein of length 535 amino acids).
Pair-wise comparisons of all the protein coding genes were made with other secernentean nematodes and it was observed that cox1 and cob are the most conserved proteins while nad6 and atp6 are least conserved among the three isolates (±1% w.r.t W. bancrofti PNG isolate). However, variation of atp6 (±5% w.r.t W. bancrofti PNG isolate) is observed among the three isolates (Table 2).
Table 2.
Identities of each of the 12 mitochondrial proteins of the PNG, Indian and W. African W. bancrofti isolates with Brugia malayi (Bm),Onchocerca volvulus (Ov) and Caenorhabditis elegans (Ce), respectively.
| Protein | Amino acid identity (%) | ||
|---|---|---|---|
| Wb/Bm | Wb/Ov | Wb/Ce | |
| NAD2 | 86/86/85 | 75/76/75 | 36/36/35 |
| NAD4 | 91/91/90 | 85/85/84 | 46/46/45 |
| COX1 | 96/96/96 | 90/90/90 | 53/53/52 |
| NAD6 | 86/86/86 | 61/61/61 | 28/28/28 |
| COB | 90/90/90 | 84/84/84 | 51/51/51 |
| COX3 | 92/92/92 | 79/79/79 | 33/33/33 |
| NAD4L | 90/90/90 | 81/81/81 | 37/37/37 |
| NAD1 | 87/86/86 | 82/82/82 | 50/51/51 |
| ATP6 | 87/86/82 | 80/79/73 | 19/20/19 |
| COX2 | 88/91/91 | 84/85/86 | 39/40/40 |
| NAD3 | 88/88/88 | 71/71/71 | 38/38/38 |
| NAD5 | 91/89/90 | 83/81/82 | 40/39/39 |
3.3. Transfer RNA genes
A total of 22 trn genes (53–59 nt in length) were identified in the W. bancrofti mt genome sequence (Figure S2). The secondary structures predicted for the transcripts of these genes are similar to those predicted for the mt trn transcripts of other nematode studied to date [18,20,21,25,56]. Twenty of the trn structures lack both a TΨC arm and variable loop, possessing instead a TV-replacement loop that varies in length from 5 to 8 nt. The remaining two trns structures lack a DHU arm, but possess a DHU-replacement loop of either 6 or 9 nt. Ohtsuki and colleagues have proposed that loss of the TΨC and DHU arms from these trns has resulted in the emergence of two forms of nematode mt elongation factor Tu [37,38]. Indeed, these researchers have identified two distinct mt elongation factors (EF-Tu1 and EF-Tu2) in C. elegans and have shown that these proteins exhibit different modes of trn binding. Whereas EF-Tu1 has been found to bind specifically to trns lacking a TΨC arm, EF-Tu2 has been shown to bind specifically to trns lacking a DHU arm. Distinct from translation elongation factors observed in other species, nematode mt EF-Tu proteins represent attractive targets for future anthelmintic drug discovery.
All trns are predicted to have 5 to 8 base pairings in the aminoacyl acceptor stem and 4 to 6 such pairings in the anticodon stem. All, these trns are also predicted to have 2 to 4 base pairings in the DHU stem or theTΨC arm (trn S). Mismatches in these stems are supposedly corrected by RNA editing [21].
3.4. Ribosomal RNA genes
The mt rrnS and rrnL genes of W. bancrofti were identified by sequence comparison with those of B. malayi. The precise boundaries of these genes have not been verified by primer extension analysis, a method used previously to define the 5’ termini of the mt rrn genes of C. elegans [40]. Consequently, the W. bancrofti rrnS and rrnL genes are assigned tentative lengths of 672 and 972 nt, respectively (Table 1). These lengths are similar to those described previously for the mt rrn genes of other filarial nematodes [13,21,25,56].
Whereas the rrnS gene is located between nad4L and trnY, the rrnL gene is located between trnH and nad3. The combined AT content of the W. bancrofti PNG isolate rrn genes is 77.6% (Table S3). In agreement with this high AT content, the rrnL gene encodes the longest polyT tract (20 Ts) found in any coding region of the W. bancrofti mt genome. Pairwise nucleotide identity between the rrnS sequence of W. bancrofti and those of other filarial nematodes ranges from 89% for O. volvulus, D. immitis, and S. digitata to 94.4% for B. malayi. Similarly, pairwise nucleotide identity between the rrnL sequence of W. bancrofti and those of other filarial nematodes ranges from 83.4% for O. volvulus to 92.6% for B. malayi.
Secondary structure models for the rrn genes (Figures S3a, S3b, S3c) of W. bancrofti were constructed based on models of filarial nematodes that have been previously reported [19,20,21,25,39]. The mt rrnS gene of W. bancrofti is similar to other filarial nematodes in that it has an aberrant secondary structure lacking several elements present in the E. coli 16S rRNA[7]. W. bancrofti contains secondary structure elements corresponding to domains 1–4, 19–21, 23, 25, 29–34, 36–38, 43, 45–48 in the E. coli model and is similar in topology to the models described for N. americanus, A. duodenale, D. immitis and S. stercoralis [19,20,21,25].
The secondary structure of the mt rrnL gene is also similar in structure to the nematodes that have been reported earlier [19,20,21,25,40]. All the secondary structure elements and sites involved in the binding of the tRNA to the aminoacyl site, peptidyl transferase site, or both, in the E. coli 23S rRNA are conserved between W. bancrofti and other nematode species. However, several nucleotides associated with the proposed exit sites in the E. coli model [34] are absent in W. bancrofti and other filarial nematodes that have been reported, suggesting that the nematode mt may lack these exit sites [19,20,21,25].
3.5. Non-coding regions
Only 2.8% of the W. bancrofti mt genome represents non-coding sequence. Located between the cox3 and trnA genes, the longest non-coding region (267 nt) is characterized by a high AT content (83.9%). This AT-rich region is shorter than those described previously for B. malayi (283 nt), D.immitis (362 nt), and S. digitata (506 nt) [13,21,56]. Although this region in W. bancrofti lacks the 43-nt tandem repeat motifs (CR1-CR6) observed in C. elegans [40], as well as the runs of AT dinucleotide observed in both A. suum and C. elegans, it does contain two copies of a 10-nt tandem repeat that display 100% sequence identity (Figure S4). The AT-rich region of W. bancrofti mtDNA also contains 7 pairs of inverted repeats ranging from 5 to 28 nt in length. The presence of these repeats suggests that this region assumes a certain secondary structure that is important for its presumed function in initiating replication and transcription.
Although the long non-coding region located between the nad4 and cox1 genes (276 bp of A. suum and C. elegans mtDNA is absent from the mt genome of W. bancrofti, the latter contains a 46-nt intergernic region between its trnW and nad6 genes that may be folded into a stem-loop structure (Figure S1). This structure may provide a RNA processing signal in much the same way that trn secondary structure may direct cleavage of polycistronic transcripts in mammalian mitochondria [40].
3.6. Defining Sequence Polymorphism
We examined single nucleotide polymorphisms (SNPs) in the three isolates of W. bancrofti from PNG, West Africa, and India and the resulting pairwise nucleotide diversity (π). We found highly variable regions throughout the genome with most substitutions at the 3rd codon position, 4-fold degeneracy, in coding sequences (Table 3, Figure 1) and higher sequence similarity between rrn and trn genes.
Table 3.
Nucleotide alignment statistics between the three isolates where each comparison assumes PNG as the reference sequence. Ts:Tv is the transition:transversion ratio, Ds/Dv is the ratio non-synonymous to synonymous nucleotide substitutions during amino acid translation; substitutions at first, second, third protein-coding sequence.
| Gene/region | Nucleotide Sequence Length |
Nucleotide Difference (%) |
Ts:Tv | 1st | 2nd | 3rd | Dn/Ds |
|---|---|---|---|---|---|---|---|
| atp6 | 582 | ||||||
| W. Africa | 2.2 | 3.3:1 | 3 | 2 | 8 | 0.25 | |
| India | 6.1 | 1.4:1 | 7 | 9 | 21 | 0.94 | |
| cob | 1088 | ||||||
| W. Africa | 0.18 | 1:0 | 0 | 0 | 2 | 0 | |
| India | 0.25 | 1:0 | 1 | 0 | 2 | 0 | |
| cox1 | 1647 | ||||||
| W. Africa | 1 | 7.5:1 | 6 | 0 | 11 | 0.23 | |
| India | 1.6 | 1:1 | 6 | 2 | 19 | 0.24 | |
| cox2 | 700 | ||||||
| W. Africa | 3.7 | 7.6:1 | 3 | 2 | 21 | 0.32 | |
| India | 0.57 | 1:0 | 0 | 0 | 4 | 0 | |
| cox3 | 780 | ||||||
| W. Africa | 0.51 | 3:1 | 0 | 1 | 3 | 0.33 | |
| India | 0.51 | 0:1 | 0 | 1 | 3 | 0.33 | |
| nad1 | 877 | ||||||
| W. Africa | 0.34 | 1:0 | 1 | 0 | 2 | 0.5 | |
| India | 1.9 | 4.6:1 | 4 | 0 | 13 | 0.23 | |
| nad2 | 844 | ||||||
| W. Africa | 0.12 | 1:0 | 0 | 0 | 1 | 0 | |
| India | 2.7 | 2.3:1 | 4 | 3 | 16 | 0.42 | |
| nad3 | 337 | ||||||
| W. Africa | 1.2 | 1:1 | 0 | 1 | 3 | 0.5 | |
| India | 1.2 | 0:1 | 0 | 1 | 3 | 0.5 | |
| nad4 | 1230 | ||||||
| W. Africa | 1.5 | 3.5:1 | 9 | 6 | 18 | 1.5 | |
| India | 2.5 | 2.1:1 | 5 | 5 | 21 | 0.55 | |
| nad4L | 243 | ||||||
| W. Africa | 0.41 | 1:0 | 0 | 0 | 1 | 0 | |
| India | 0.41 | 0:1 | 0 | 0 | 1 | 0 | |
| nad5 | 1596 | ||||||
| W. Africa | 0.25 | 3:1 | 0 | 2 | 2 | 1 | |
| India | 0.88 | 0.4:1 | 4 | 5 | 5 | 1.6 | |
| nad6 | 453 | ||||||
| W. Africa | 0.22 | 0:1 | 0 | 0 | 1 | 0 | |
| India | 0.22 | 0:1 | 0 | 0 | 1 | 0 | |
| All 22 trns | 1,245 | ||||||
| W. Africa | (+1) | -- | -- | -- | -- | -- | -- |
| India | (+4) | -- | -- | -- | -- | -- | -- |
| rrnS | 672 | ||||||
| W. Africa | (+1) | -- | -- | -- | -- | -- | -- |
| India | -- | -- | -- | -- | -- | -- | |
| rrnL | 972 | ||||||
| W. Africa | -- | -- | -- | -- | -- | -- | |
| India | (+1) | -- | -- | -- | -- | -- | -- |
| ATrich region | 267 | ||||||
| W. Africa | (−3) | -- | -- | -- | -- | -- | -- |
| India | -- | -- | -- | -- | -- | -- | |
These polymorphisms identified among worms from three geographical locations, provide candidate loci for population level studies and reference data for regional comparisons of W. bancrofti. The presence of unique differences among regions, due to higher rates of substitutions and drift at each locus, will allows us to track the spread of specific W. bancrofti strains. For example if a strain emerges carrying a drug resistant allele in the nuclear genome we can then track the resistant strain using the associated mt haplotype. Unique polymorphisms allow us to differentiate among multiple regions as an origin for new outbreaks of W. bancrofti. Though the focus of our study was not to provide population level data we believe that loci such as atp6 and nad4 would prove good candidate loci for future population level studies on W. bancrofti since these two protein-coding loci had the most variation.
The ratio of non-synonymous substitutions to synonymous substitution is used to determine if natural selection has had any influence on variation on a given locus [36]. The mt genome of W. bancrofti reflects an overall trend of purifying selection for the protein coding sequences in the genome (Table 3). Though we might expect the mitochondria to behave neutrally in most circumstances, a signal of purifying selection is common because any change in the protein coding sequence may completely debilitate the cellular mitochondria [52].
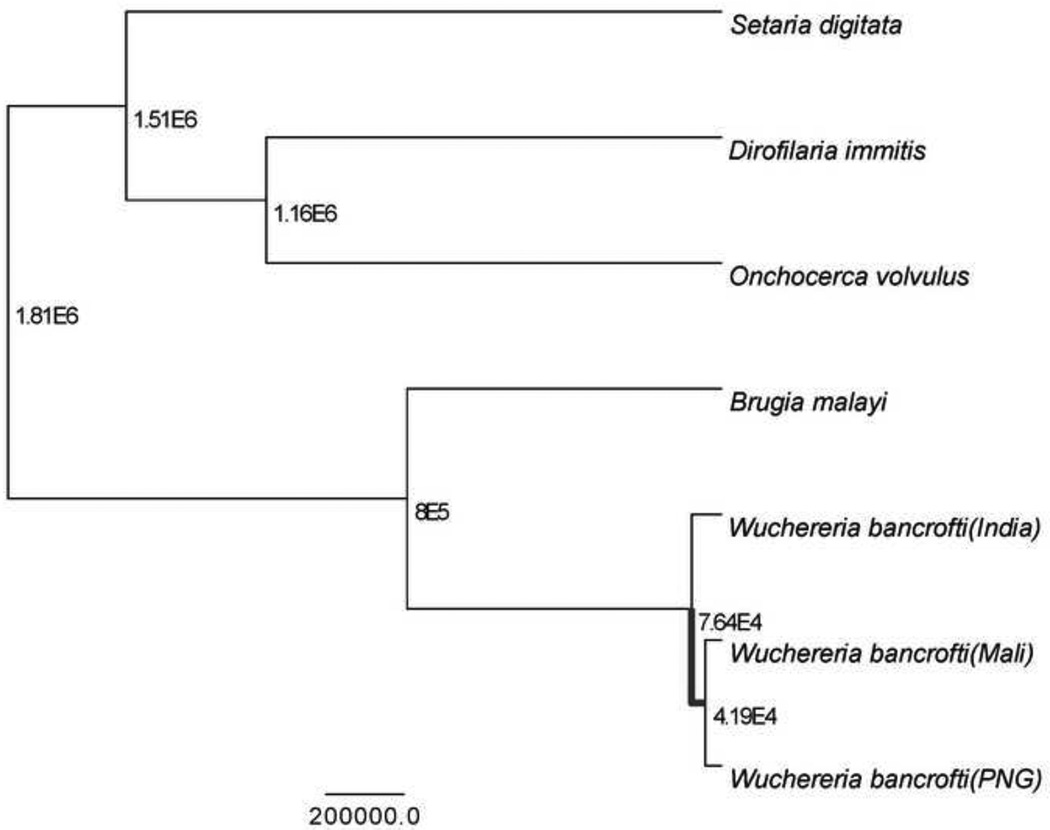
3.7. Phylogenetic reconstruction and divergence times
We found that our Bayesian phylogeny converged after 20 million states, with three independent runs converging to the same parameter estimates. Auto-correlation between parameters was low and the chain showed good mixing as observed in a graph of the trace file. The effective sample size for each parameter was greater than 500 providing a sufficient sample from the posterior distribution for parameter estimation. The topology of the phylogenetic tree (Figure 2) supported a monophyletic grouping of the W. bancrofti isolates with the PNG and West African isolates being more similar to each other than either one to the Indian isolate. B. malayi was monophyletic with the W. bancrofti clade which is concordant with other published phylogenetic analysis for nematodes [6]. All posterior probabilities were greater than 95% with only S. digitata having support less than 95% (Figure 2). Higher support for the placement of S. digitata using mt protein coding sequence is resolved when other nematode species such as A. simplex are included in the analysis with an out-group species of R. rotaria Small ST, unpublished results]. Substitution rates across the tree were best represented by the uncorrelated log-normal relaxed clock [8], which allows each branch to have a variable substitution rate independent of shared ancestry. Median divergence dates are shown for each node in the phylogenetic tree where the time is represented in generations. For example, if it we let W. bancrofti have a generation time of 1 year than all estimates for W. bancrofti from the phylogenetic analysis would be scaled in years, likewise fewer or more generations per year would scale times accordingly. Divergence dates between W. bancrofti isolates from PNG and West Africa are on the order of 47 (1.28–302) kga (median and 95% credible intervals in thousands of generations; kga = thousands of generation ago), while the clade of PNG-West Africa and India is on the order of 87 (2.14–519) kga. The median divergence date between the monophyletic W. bancrofti clade and B. malayi, another agent of LF, is on the order of 675 (19.0–3,879) kga providing evidence that these two agents of LF are relatively young species compared to the rest of the Onchocercidae, with a median divergence of 1785 (65.0–9,688) kga. We expect future studies to reduce variation in divergence time estimates by increasing the number of independently segregating loci.
Figure 2.
A Bayesian phylogeny of the filarioidea. Samples are presented at tips with >95% posterior support for all nodes except for S. digitata which was <70%. Branch lengths are scaled in generations with the scale axis representing 20,000 generations. Median estimates of node divergence time are given in scientific notation in generations.
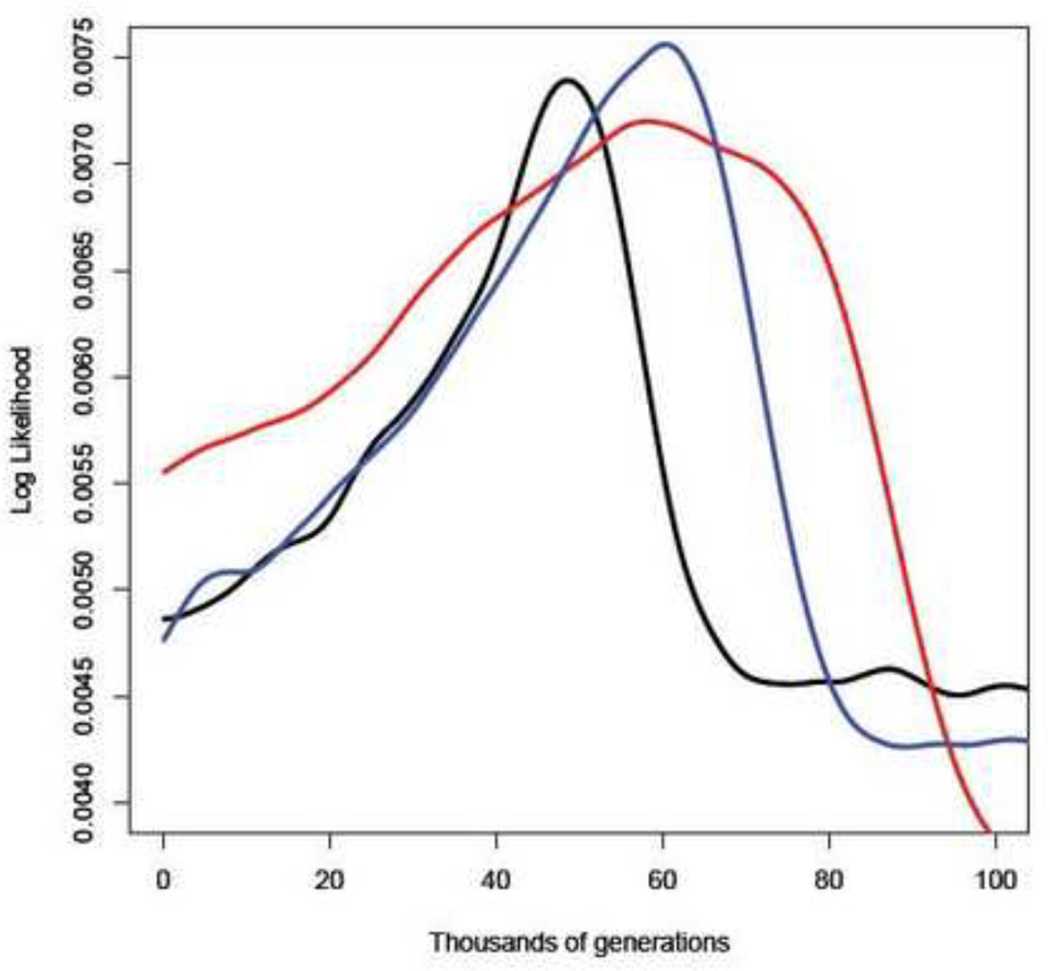
The Isolation-Migration analysis, which allows for migration between diverging populations, produced values similar to the phylogenetic analysis indicating that there was probably minimal migration between geographic isolates. Divergence dates were once again the youngest between isolates from PNG and West Africa with a median divergence time of 49 (5.1–187) kga compared to older estimates between PNG and India at 62 (4.9–186) kga and West Africa and India at 76 (4.90–186) kga (Figure 3). Estimation of migration rates between populations was symmetrical with a mean rate of 0.44±0.02 (0.023–0.975) genes per generation. Migration estimates are dependent on shared polymorphisms, which can be further verified by increasing sample size within region. IMa2 also allows the estimation of ancestral effective population sizes between populations. The ancestral population gives rise to the contemporary populations at the specific divergence time estimated for each population pair. The median ancestral effective population size was largest between the PNG and Indian isolates with approximately 9,500 individuals, which is not significantly different to the West African and Indian isolates with approximately 7,600 individuals. The smallest ancestral effective population size was between the PNG and West African isolates with approximately 2,600 individuals. In the future we would like to estimate the ancestral population size between the ancestor of the West African and PNG isolates versus the Indian isolate as well as migration rates. However, models with more than 2 populations require more data than is currently available for W. bancrofti isolates due to the exponential increase in the number of parameters to estimate [15]. Models comparing only 2 populations, pairwise comparisons, have been shown to produce robust parameter estimates even for data sets containing a single non-recombining locus (such as a mt genome) and small sample sizes [16].
Figure 3.
Maximum likelihood of the divergence times from the IMa2 output. Vertical axis is the log likelihood of the parameter estimate and the horizontal axis is the divergence time in generations.
3.8 Demographic history of W. bancrofti
To estimate the divergence times between the geographic isolates of W. bancrofti we were forced to make assumptions about generation time and mutation rate. In the above analyses we assumed that W. bancrofti had a generation time of 1 year and a mutation rate comparable to P. pacificus. The mutation rate data collected for P. pacificus did not significantly differ from rates estimated from C. elegans, giving confidence that these rate estimates are robust across nematode taxa for the mitochondria [35]. However, both P. pacificus and C. elegans are free-living nematodes and are not parasitic. A parasitic life history should skew substitution rates to be slightly faster due to evolutionary pressure to avoid host immune defenses [2,17,27]. In both analyses above we accounted for variation in mutation rate by allowing rates to vary across branches independent of shared ancestry (relaxed clock) and placing a prior distribution centered on the mean rate estimate in P. pacificus. We can capture the effect of mutation rate variation on our divergence times by including the uncertainty, 95% credible intervals, around the estimate (Figure 3). The second source of variation is the difficulty in estimating generation time for W. bancrofti, here we define generation time as the average age at which a female gives birth [46]. Molnar et al. 2011 used an upper bound of 100 generations per year when estimating most recent common ancestor of P. pacificus isolates, here we use the upper bound of 5 generations per year (2.4 months to reproductive maturity) due to the necessity of both vector and host life stages in W. bancrofti isolates [42,55].
Since W. bancrofti demography has not been previously explored, we hereby propose several hypotheses of W. bancrofti origins and migration using our genetic data. We begin by estimating the lineage of W. bancrofti to be at least 675,000 years old with a lower estimate of 135,000 years using an estimate of 5 generations per year. The common ancestor of B. malayi and W. bancrofti clade do not overlap with the appearance of modern humans on the continent of Africa, which suggests that the common ancestor may have targeted another species from the genus Homo or another mammalian host. However, the diversification of the lineage leading to W. bancrofti and possibly B. malayi is well within the proposed timeline for the emergence of modern humans (200k) and their migration out of the African continent. The origin of W. bancrofti cannot be known with certainty from the current data set (see below) but we can exclude some alternative hypotheses based on the expected pattern of divergence in the data.
Our apriori hypothesis is to assume that infected individuals transferred W. bancrofti leaving a geographic pattern of divergence along human migration routes. If Africa were the origin of W. bancrofti then we would expect the West African isolate to fall outside of the grouping of India and PNG on a phylogenetic tree. This would support a hypothesis that the contemporary distribution of W. bancrofti was due to the migration of humans out of Africa. This hypothesis fits well with an estimated divergence time of 60,000–70,000 years between West Africa and India but does not account for the more recent divergence of West Africa and PNG. It is possible that the true history of W. bancrofti is convoluted by a recent migration of infected individuals to PNG, which would make it appear that West Africa and PNG were more closely related. Information on ancestral effective population size supports more recent movement of a smaller population, approximately 2,600, responsible for founding the West African and PNG isolates, making it probable that a direct migration event rather than a historical migration connected West Africa and PNG. In the future we can test this hypothesis by including more population data from each region to account for shared polymorphisms that could only come from recent admixture. A PNG origin would create the opposite pattern than above.
It is unlikely that India is the origin of W. bancrofti even though it roots both West Africa and PNG in the phylogeny. In Figure 2 India could not be the origin and still maintain the West Africa-PNG clade unless the same mt haplotype spread from India to PNG and also from India to West Africa. This is an unlikely scenario due to the chance effects of only a single strain being carried [Ramesh A, unpublished results] within a group of migrating humans and that genetic drift favored the same strain in both localities. However, India could still be a considered a possible origin i f there was recent migration of infected individuals from West Africa to PNG.
Reconstructing the true origins of W. bancrofti and its associated disease, LF, requires a large data set encompassing the endemic range of W. bancrofti as well as a moderate population sample (10–20 individuals) from each geographic region for multiple independently segregating genetic loci. The limitations of our study include the lack of population level polymorphism data for both India and West Africa, which limits our inference about shared ancestral polymorphism or recent migration events. In the future, adding more independently segregating loci will decrease the variance in our estimates of divergence times. Depending on the genealogical history of each sampled locus, increasing sample size will reduce the variance in the estimate of divergence times but probably not shift the mean value [53].
4. Conclusions
We have characterized genetic diversity among three separate isolates of W. bancrofti from three disparate geographic regions. By summarizing genetic variation we have now made it possible for future studies to understand the population dynamics and life history of W. bancrofti. A better understanding of W. bancrofti life history will allow us to predict how populations will respond to transmission interruption efforts or drug treatment regiments. Studies of this nature will also allow us to assess the impact of new outbreaks. Consideration of W. bancrofti on a larger geographic scale allows us to test different hypotheses of the demographic history of W. bancrofti and LF. Understanding the origins of W. bancrofti will allow us to predict and test for the emergence of drug resistance strains by understanding how drug resistance develops and if it utilizes different molecular pathways in different regions. Understanding the history and contemporary population demographics can help design programs aimed at the elimination of LF and more efficiently constrain the distribution of this debilitating disease.
Supplementary Material
Highlights.
The complete mitochondrial genome sequence of several isolates of the filarial nematode Wuchereria bancrofti.
Comparison of geographic isolates from three important endemic regions West Africa, India and Papua New Guinea
Identification of mtDNA single nucleotide polymorphisms to enable population genetic and phylogenetic analyses
Acknowledgements
We thank study participants in PNG, as well as Dr. S. Bennuru for his assistance with primer design. We also thank Dr. R. Gutell for generating the rrnS and rrnL secondary structure diagrams (Figure S3a–3c) and the Broad Institute for access to the W. bancrofti mtDNA sequence data. We acknowledge financial support for this study through grants from the National Institutes of Health (AI065717 to J.W. Kazura and P.A. Zimmerman), the Fogarty International Center (Ecology of Infectious Diseases, TW007872 to P.A. Zimmerman, a T32 Postdoctoral Fellowship to S.T. Small (AI007024), and support from the Division of Intramural Research (DIR) of the National Institute of Allergy and Infectious Diseases (NIAID) to Dr. T.B. Nutman.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Angellotti MC, Bhuiyan SB, Chen G, Wan X-F. CodonO: codon usage bias analysis within and across genomes. Nucleic acids research. 2007;35:W132–W136. doi: 10.1093/nar/gkm392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babayan Sa, Read AF, Lawrence Ra, Bain O, Allen JE. Filarial parasites develop faster and reproduce earlier in response to host immune effectors that determine filarial life expectancy. PLoS biology. 2010;8:e1000525. doi: 10.1371/journal.pbio.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic acids research. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blouin MS. Molecular prospecting for cryptic species of nematodes: mitochondrial DNA versus internal transcribed spacer. International journal for parasitology. 2002;32:527–531. doi: 10.1016/s0020-7519(01)00357-5. [DOI] [PubMed] [Google Scholar]
- 5.Brown WM. No Title. Mechanisms of evolution in animal mitochondrial DNA. Ann NY Acad Sci. 1981:119–134. doi: 10.1111/j.1749-6632.1981.tb46515.x. [DOI] [PubMed] [Google Scholar]
- 6.Casiraghi M, Anderson TJ, Bandi C, Bazzocchi C, Genchi C. A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology. 2001;122(Pt 1):93–103. doi: 10.1017/s0031182000007149. [DOI] [PubMed] [Google Scholar]
- 7.Dams Erna, Hendriks Lydia, Peer Yves Van de, Neefs Jean-Marc, Smits Geert, Vandenbempt Isidore WRD. Compilation of smal rinbosoal subunit RNA sequences. Nucleic Acids Research. 1988;16:87–173. doi: 10.1093/nar/16.suppl.r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC evolutionary biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular ecology resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 12.Geyer CJ, Chain M, Carlo M. Markov Chain Monte Carlo Maximum Likelihood. Methods [Google Scholar]
- 13.Ghedin E, Wang S, Spiro D, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science (New York, N.Y.) 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa M, Kishino H, Yano T-aki. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 15.Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hey J. Isolation with migration models for more than two populations. Molecular biology and evolution. 2010;27:905–920. doi: 10.1093/molbev/msp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holterman M, van der Wurff A, van den Elsen S, et al. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown Clades. Molecular biology and evolution. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- 18.Hu M, Chilton NB, Abs El-Osta YG, Gasser RB. Comparative analysis of mitochondrial genome data for Necator americanus from two endemic regions reveals substantial genetic variation. International Journal for Parasitology. 2003;33:955–963. doi: 10.1016/s0020-7519(03)00129-2. [DOI] [PubMed] [Google Scholar]
- 19.Hu M, Chilton NB, Gasser RB. The mitochondrial genomes of the human hookworms, Ancylostoma duodenale and Necator americanus (Nematoda: Secernentea) International journal for parasitology. 2002;32:145–158. doi: 10.1016/s0020-7519(01)00316-2. [DOI] [PubMed] [Google Scholar]
- 20.Hu M, Chilton NB, Gasser RB. The mitochondrial genome of Strongyloides stercoralis (Nematoda) – idiosyncratic gene order and evolutionary implications. International Journal for Parasitology. 2003;33:1393–1408. doi: 10.1016/s0020-7519(03)00130-9. [DOI] [PubMed] [Google Scholar]
- 21.Hu M, Gasser RB, Abs El-Osta YG, Chilton NB. Structure and organization of the mitochondrial genome of the canine heartworm, Dirofilaria immitis. Parasitology. 2003;127:37–51. doi: 10.1017/s0031182003003275. [DOI] [PubMed] [Google Scholar]
- 22.Hu M, Gasser RB. Mitochondrial genomes of parasitic nematodes--progress and perspectives. Trends in parasitology. 2006;22:78–84. doi: 10.1016/j.pt.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Hu M, Jex AR, Campbell BE, Gasser RB. Long PCR amplification of the entire mitochondrial genome from individual helminths for direct sequencing. Nature protocols. 2007;2:2339–2344. doi: 10.1038/nprot.2007.358. [DOI] [PubMed] [Google Scholar]
- 24.Jex AR, Littlewood DTJ, Gasser RB. Toward next-generation sequencing of mitochondrial genomes--focus on parasitic worms of animals and biotechnological implications. Biotechnology advances. 2010;28:151–159. doi: 10.1016/j.biotechadv.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Keddie EM, Higazi T, Unnasch TR. The mitochondrial genome of Onchocerca volvulus: sequence, structure and phylogenetic analysis. Molecular and biochemical parasitology. 1998;95:111–127. doi: 10.1016/s0166-6851(98)00102-9. [DOI] [PubMed] [Google Scholar]
- 26.King C, Freedman D. Filariasis. Hunter’s tropical medicine and emerging infectious diseases. (8th ed) 2000 [Google Scholar]
- 27.Kochin BF, Bull JJ, Antia R. Parasite evolution and life history theory. PLoS biology. 2010;8:e1000524. doi: 10.1371/journal.pbio.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laslett D, Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics (Oxford, England) 2008;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 30.Le TH, Blair D, McManus DP. Mitochondrial genomes of human helminths and their use as markers in population genetics and phylogeny. Acta tropica. 2000;77:243–256. doi: 10.1016/s0001-706x(00)00157-1. [DOI] [PubMed] [Google Scholar]
- 31.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics (Oxford, England) 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 32.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids research. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehlotra RK, Gray LR, Blood-Zikursh MJ, et al. Molecular-based assay for simultaneous detection of four Plasmodium spp. and Wuchereria bancrofti infections. The American journal of tropical medicine and hygiene. 2010;82:1030–1033. doi: 10.4269/ajtmh.2010.09-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moazed Danesh NH. Intermediate states in the movement of transfer RNA in the ribosome. nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 35.Molnar RI, Bartelmes G, Dinkelacker I, Witte H, Sommer RJ. Mutation Rates and Intraspecific Divergence of the Mitochondrial Genome of Pristionchus pacificus. Molecular biology and evolution. 2011;28:2317–2326. doi: 10.1093/molbev/msr057. [DOI] [PubMed] [Google Scholar]
- 36.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular biology and evolution. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 37.Ohtsuki T, Sato A, Y-ichi A, Watanabe K. A unique serine-specific elongation factor Tu found in nematode mitochondria. Nature structural biology. 2002;9:669–673. doi: 10.1038/nsb826. [DOI] [PubMed] [Google Scholar]
- 38.Ohtsuki T, Watanabe Yi, Takemoto C, et al. An “elongated” translation elongation factor Tu for truncated tRNAs in nematode mitochondria. The Journal of biological chemistry. 2001;276:21571–21577. doi: 10.1074/jbc.M011118200. [DOI] [PubMed] [Google Scholar]
- 39.Okimoto R, Macfarlane JL, Clary D, Wolstenholme DR. of Two. Library. 1992 doi: 10.1093/genetics/130.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okimoto R, Macfarlane JL, Wolstenholme DR. of Molecular Evolution The Mitochondrial Ribosomal RNA Genes of the Nematodes Caenorhabditis elegans and Ascaris suum : Consensus Secondary-Structure Models and Conserved Nucleotide Sets for Phylogenetic Analysis. 1994 doi: 10.1007/BF00160405. [DOI] [PubMed] [Google Scholar]
- 41.Omura S, Miyadera H, Ui H, et al. An anthelmintic compound, nafuredin, shows selective inhibition of complex I in helminth mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:60–62. doi: 10.1073/pnas.011524698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paily AKP, Hoti SL, Balaraman K. Development of Lymphatic Filarial Parasite Wuchereria bancrofti (Spirurida : Onchocercidae) in Mosquito Species (Diptera : Culicidae) Fed Artificially on Microfilaremic Blood Development of Lymphatic Filarial Parasite Wuchereria bancrofti (Spirurida. America. 2006;43:1222–1226. doi: 10.1603/0022-2585(2006)43[1222:dolfpw]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 43.Posada D. jModelTest: phylogenetic model averaging. Molecular biology and evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 44.Prichard RK. Markers for benzimidazole resistance in human parasitic nematodes? Parasitology. 2007;134:1087–1092. doi: 10.1017/S003118200700008X. [DOI] [PubMed] [Google Scholar]
- 45.Rice P. The European Molecular Biology Open Software Suite EMBOSS : The European Molecular Biology Open Software Suite. Science. 2000;16:2–3. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 46.Ricklefs RE, GM . Ecology. fourth edition. New York: W.H. Freeman; 1999. [Google Scholar]
- 47.De Rijk P, De Wachter R. RnaViz, a program for the visualisation of RNA secondary structure. Nucleic acids research. 1997;25:4679–4684. doi: 10.1093/nar/25.22.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Rijk P, Wuyts J, De Wachter R. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics (Oxford, England) 2003;19:299–300. doi: 10.1093/bioinformatics/19.2.299. [DOI] [PubMed] [Google Scholar]
- 49.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic acids research. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharp PM, Matassi G. Codon usage and genome evolution. Current opinion in genetics & development. 1994;4:851–860. doi: 10.1016/0959-437x(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 51.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. ABySS: a parallel assembler for short read sequence data. Genome research. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart JB, Freyer C, Elson JL, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS biology. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strasburg JL, Rieseberg LH. How robust are “isolation with migration” analyses to violations of the im model? A simulation study. Molecular biology and evolution. 2010;27:297–310. doi: 10.1093/molbev/msp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tavare S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lectures on Mathematics in the Life Sciences. 1986:57–86. [Google Scholar]
- 55.Vanamail P, Ramaiah KD, Pani SP, Das PK, Grenfell BT, Bundy Da. Estimation of the fecund life span of Wuchereria bancrofti in an endemic area. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1996;90:119–121. doi: 10.1016/s0035-9203(96)90106-6. [DOI] [PubMed] [Google Scholar]
- 56.Yatawara L, Wickramasinghe S, Rajapakse RPVJ, Agatsuma T. The complete mitochondrial genome of Setaria digitata (Nematoda: Filarioidea): Mitochondrial gene content, arrangement and composition compared with other nematodes. Molecular and biochemical parasitology. 2010;173:32–38. doi: 10.1016/j.molbiopara.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 57.Meeting of the International Task Force for Disease Eradication. Wkly Epidemiol Rec. 2011;86:53–59. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.