Abstract
Replication of plasmid deoxyribonucleic acid (DNA) is dependent on three stages: initiation, elongation, and termination. The first stage, initiation, depends on plasmid-encoded properties such as the replication origin and, in most cases, the replication initiation protein (Rep protein). In recent years the understanding of initiation and regulation of plasmid replication in Escherichia coli has increased considerably, but it is only for the ColE1-type plasmids that significant biochemical data about the initial priming reaction of DNA synthesis exist. Detailed models have been developed for the initiation and regulation of ColE1 replication. For other plasmids, such as pSC101, some hypotheses for priming mechanisms and replication initiation are presented. These hypotheses are based on experimental evidence and speculative comparisons with other systems, e.g., the chromosomal origin of E. coli. In most cases, knowledge concerning plasmid replication is limited to regulation mechanisms. These mechanisms coordinate plasmid replication to the host cell cycle, and they also seem to determine the host range of a plasmid. Most plasmids studied exhibit a narrow host range, limited to E. coli and related bacteria. In contrast, some others, such as the IncP plasmid RK2 and the IncQ plasmid RSF1010, are able to replicate in nearly all gram-negative bacteria. This broad host range may depend on the correct expression of the essential rep genes, which may be mediated by a complex regulatory mechanism (RK2) or by the use of different promoters (RSF1010). Alternatively or additionally, owing to the structure of their origin and/or to different forms of their replication initiation proteins, broad-host-range plasmids may adapt better to the host enzymes that participate in initiation. Furthermore, a broad host range can result when replication initiation is independent of host proteins, as is found in the priming reaction of RSF1010.
Full text
PDF




















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Austin S. J. P1 plasmid replication requires methylated DNA. EMBO J. 1987 Oct;6(10):3185–3189. doi: 10.1002/j.1460-2075.1987.tb02630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles A. L., Snyder K. M., Chattoraj D. K. P1 plasmid replication: replicon structure. J Mol Biol. 1984 Mar 5;173(3):307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- Armstrong K. A., Acosta R., Ledner E., Machida Y., Pancotto M., McCormick M., Ohtsubo H., Ohtsubo E. A 37 X 10(3) molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J Mol Biol. 1984 May 25;175(3):331–348. doi: 10.1016/0022-2836(84)90352-8. [DOI] [PubMed] [Google Scholar]
- Ataai M. M., Shuler M. L. Mathematical model for the control of ColE1 type plasmid replication. Plasmid. 1986 Nov;16(3):204–212. doi: 10.1016/0147-619x(86)90058-2. [DOI] [PubMed] [Google Scholar]
- Austin S. J. Plasmid partition. Plasmid. 1988 Jul;20(1):1–9. doi: 10.1016/0147-619x(88)90001-7. [DOI] [PubMed] [Google Scholar]
- Backman K., Betlach M., Boyer H. W., Yanofsky S. Genetic and physical studies on the replication of ColE1-type plasmids. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):69–76. doi: 10.1101/sqb.1979.043.01.012. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M., Timmis K. N. Host: vector systems for gene cloning in Pseudomonas. Curr Top Microbiol Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- Baker T. A., Funnell B. E., Kornberg A. Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J Biol Chem. 1987 May 15;262(14):6877–6885. [PubMed] [Google Scholar]
- Baker T. A., Sekimizu K., Funnell B. E., Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986 Apr 11;45(1):53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- Balbás P., Soberón X., Merino E., Zurita M., Lomeli H., Valle F., Flores N., Bolivar F. Plasmid vector pBR322 and its special-purpose derivatives--a review. Gene. 1986;50(1-3):3–40. doi: 10.1016/0378-1119(86)90307-0. [DOI] [PubMed] [Google Scholar]
- Banner D. W., Kokkinidis M., Tsernoglou D. Structure of the ColE1 rop protein at 1.7 A resolution. J Mol Biol. 1987 Aug 5;196(3):657–675. doi: 10.1016/0022-2836(87)90039-8. [DOI] [PubMed] [Google Scholar]
- Barbour A. G., Garon C. F. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987 Jul 24;237(4813):409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- Barth P. T., Ellis K., Bechhofer D. H., Figurski D. H. Involvement of kil and kor genes in the phenotype of a host-range mutant of RP4. Mol Gen Genet. 1984;197(2):236–243. doi: 10.1007/BF00330969. [DOI] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Figurski D. H. Map location and nucleotide sequence of korA, a key regulatory gene of promiscuous plasmid RK2. Nucleic Acids Res. 1983 Nov 11;11(21):7453–7469. doi: 10.1093/nar/11.21.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Kornacki J. A., Firshein W., Figurski D. H. Gene control in broad host range plasmid RK2: expression, polypeptide product, and multiple regulatory functions of korB. Proc Natl Acad Sci U S A. 1986 Jan;83(2):394–398. doi: 10.1073/pnas.83.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
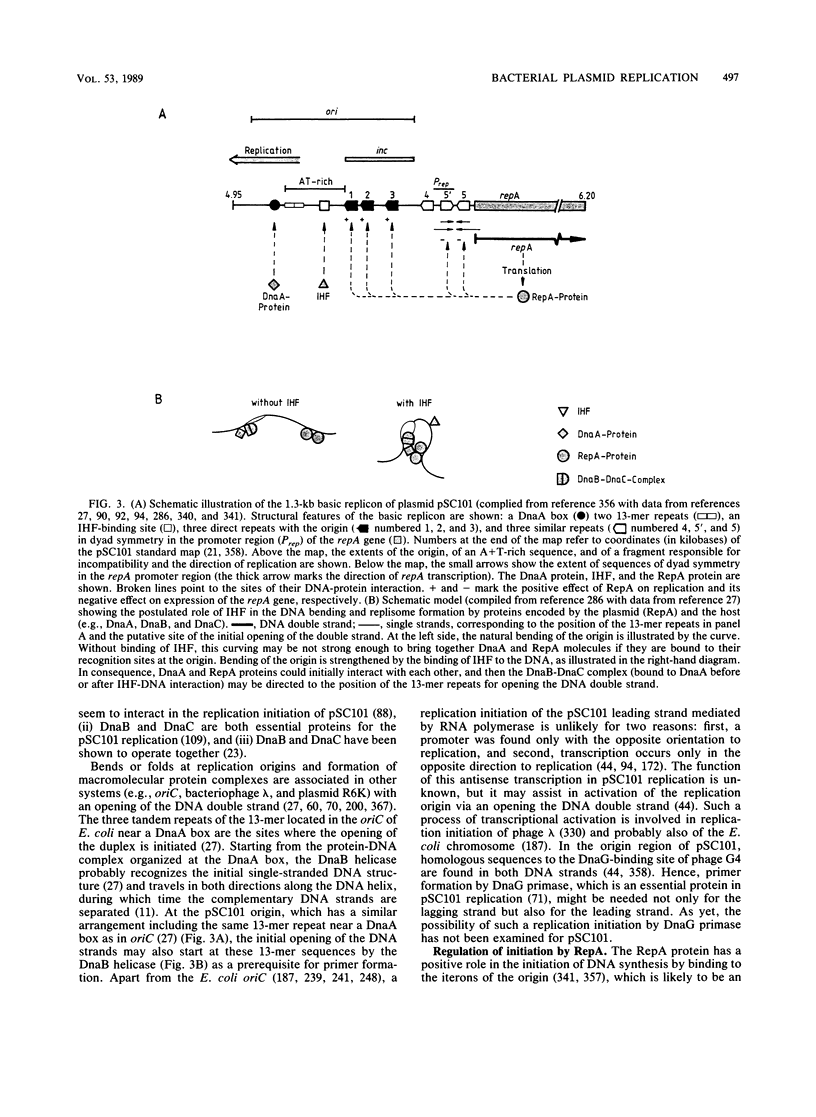
- Bernardi A., Bernardi F. Complete sequence of pSC101. Nucleic Acids Res. 1984 Dec 21;12(24):9415–9426. doi: 10.1093/nar/12.24.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bex F., Piérard P., Desmyter A., Drèze P., Colet M., Couturier M. Mini-F E protein: the carboxy-terminal end is essential for E gene repression and mini-F copy number control. J Mol Biol. 1986 May 20;189(2):293–303. doi: 10.1016/0022-2836(86)90511-5. [DOI] [PubMed] [Google Scholar]
- Biswas S. B., Biswas E. E. Regulation of dnaB function in DNA replication in Escherichia coli by dnaC and lambda P gene products. J Biol Chem. 1987 Jun 5;262(16):7831–7838. [PubMed] [Google Scholar]
- Bonekamp F., Jensen K. F. The AGG codon is translated slowly in E. coli even at very low expression levels. Nucleic Acids Res. 1988 Apr 11;16(7):3013–3024. doi: 10.1093/nar/16.7.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Brasch M. A., Meyer R. J. Genetic organization of plasmid R1162 DNA involved in conjugative mobilization. J Bacteriol. 1986 Aug;167(2):703–710. doi: 10.1128/jb.167.2.703-710.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawerman G. Determinants of messenger RNA stability. Cell. 1987 Jan 16;48(1):5–6. doi: 10.1016/0092-8674(87)90346-1. [DOI] [PubMed] [Google Scholar]
- Broyles S. S., Pettijohn D. E. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J Mol Biol. 1986 Jan 5;187(1):47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- Burkardt H. J., Riess G., Pühler A. Relationship of group P1 plasmids revealed by heteroduplex experiments: RP1, RP4, R68 and RK2 are identical. J Gen Microbiol. 1979 Oct;114(2):341–348. doi: 10.1099/00221287-114-2-341. [DOI] [PubMed] [Google Scholar]
- Böldicke T. W., Hillenbrand G., Lanka E., Staudenbauer W. L. Rifampicin-resistant initiation of DNA synthesis on the isolated strands of ColE plasmid DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5215–5231. doi: 10.1093/nar/9.20.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello F., Timmis K., Cohen S. N. Replication control in a composite plasmid constructed by in vitro linkage of two distinct replicons. Nature. 1976 Jan 29;259(5541):285–290. doi: 10.1038/259285a0. [DOI] [PubMed] [Google Scholar]
- Campbell A. Evolutionary significance of accessory DNA elements in bacteria. Annu Rev Microbiol. 1981;35:55–83. doi: 10.1146/annurev.mi.35.100181.000415. [DOI] [PubMed] [Google Scholar]
- Castagnoli L. Characterization of a promoter up mutation in the -35 region of the promoter of the primer for ColE1 replication. Mol Gen Genet. 1987 Jan;206(1):178–180. doi: 10.1007/BF00326555. [DOI] [PubMed] [Google Scholar]
- Castagnoli L., Lacatena R. M., Cesareni G. Analysis of dominant copy number mutants of the plasmid pMB1. Nucleic Acids Res. 1985 Jul 25;13(14):5353–5367. doi: 10.1093/nar/13.14.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesareni G., Cornelissen M., Lacatena R. M., Castagnoli L. Control of pMB1 replication: inhibition of primer formation by Rop requires RNA1. EMBO J. 1984 Jun;3(6):1365–1369. doi: 10.1002/j.1460-2075.1984.tb01978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. T., Ohmori H., Tomizawa J., Lebowitz J. Nucleotide sequence and gene organization of ColE1 DNA. J Biol Chem. 1985 Jul 25;260(15):8925–8935. [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoraj D. K., Abeles A. L., Yarmolinsky M. B. P1 plasmid maintenance: a paradigm of precise control. Basic Life Sci. 1985;30:355–381. doi: 10.1007/978-1-4613-2447-8_27. [DOI] [PubMed] [Google Scholar]
- Chattoraj D. K., Snyder K. M., Abeles A. L. P1 plasmid replication: multiple functions of RepA protein at the origin. Proc Natl Acad Sci U S A. 1985 May;82(9):2588–2592. doi: 10.1073/pnas.82.9.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah U. E., Weigand W. A., Stark B. C. Effects of recombinant plasmid size on cellular processes in Escherichia coli. Plasmid. 1987 Sep;18(2):127–134. doi: 10.1016/0147-619x(87)90040-0. [DOI] [PubMed] [Google Scholar]
- Chikami G. K., Guiney D. G., Schmidhauser T. J., Helinski D. R. Comparison of 10 IncP plasmids: homology in the regions involved in plasmid replication. J Bacteriol. 1985 May;162(2):656–660. doi: 10.1128/jb.162.2.656-660.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward G., Linder P., Caro L. The nucleotide sequence of replication and maintenance functions encoded by plasmid pSC101. Nucleic Acids Res. 1983 Aug 25;11(16):5645–5659. doi: 10.1093/nar/11.16.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Revised interpretation of the origin of the pSC101 plasmid. J Bacteriol. 1977 Nov;132(2):734–737. doi: 10.1128/jb.132.2.734-737.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier M., Bex F., Bergquist P. L., Maas W. K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988 Sep;52(3):375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan P., Krishnapillai V. Tn7 insertion mutations affecting the host range of the promiscuous IncP-1 plasmid R18. Plasmid. 1982 Sep;8(2):164–174. doi: 10.1016/0147-619x(82)90054-3. [DOI] [PubMed] [Google Scholar]
- Crosa J. H. Three origins of replication are active in vivo in the R plasmid RSF1040. J Biol Chem. 1980 Dec 10;255(23):11075–11077. [PubMed] [Google Scholar]
- Cross M. A., Warne S. R., Thomas C. M. Analysis of the vegetative replication origin of broad-host-range plasmid RK2 by transposon mutagenesis. Plasmid. 1986 Mar;15(2):132–146. doi: 10.1016/0147-619x(86)90049-1. [DOI] [PubMed] [Google Scholar]
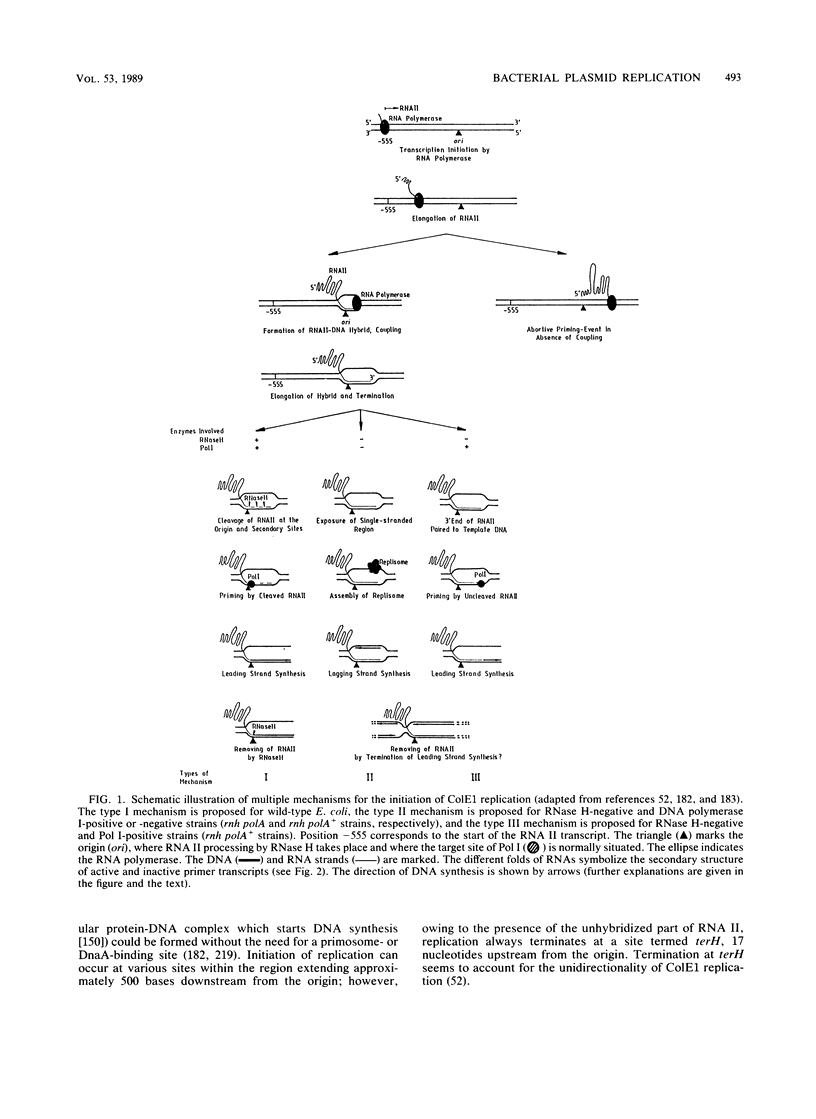
- Dasgupta S., Masukata H., Tomizawa J. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell. 1987 Dec 24;51(6):1113–1122. doi: 10.1016/0092-8674(87)90597-6. [DOI] [PubMed] [Google Scholar]
- Davison J. Mechanism of control of DNA replication and incompatibility in ColE1-type plasmids--a review. Gene. 1984 Apr;28(1):1–15. doi: 10.1016/0378-1119(84)90082-9. [DOI] [PubMed] [Google Scholar]
- Diaz R., Staudenbauer W. L. Replication of the broad host range plasmid RSF1010 in cell-free extracts of Escherichia coli and Pseudomonas aeruginosa. Nucleic Acids Res. 1982 Aug 11;10(15):4687–4702. doi: 10.1093/nar/10.15.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S. DNA methylation can enhance or induce DNA curvature. EMBO J. 1987 Dec 20;6(13):4213–4217. doi: 10.1002/j.1460-2075.1987.tb02769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon N. E., Kornberg A. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jan;81(2):424–428. doi: 10.1073/pnas.81.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M., Echols H., Wickner S., Alfano C., Mensa-Wilmot K., Gomes B., LeBowitz J., Roberts J. D., McMacken R. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: localized unwinding of duplex DNA by a six-protein reaction. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7638–7642. doi: 10.1073/pnas.83.20.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M., Roberts J., McMacken R., Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: complexes with lambda O protein and with lambda O, lambda P, and Escherichia coli DnaB proteins. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4678–4682. doi: 10.1073/pnas.82.14.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. N., Womble D. D., Rownd R. H. Transcriptional pausing in a region important for plasmid NR1 replication control. J Bacteriol. 1987 Dec;169(12):5353–5363. doi: 10.1128/jb.169.12.5353-5363.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Womble D. D., Luckow V. A., Rownd R. H. Regulation of transcription of the repA1 gene in the replication control region of IncFII plasmid NR1 by gene dosage of the repA2 transcription repressor protein. J Bacteriol. 1985 Feb;161(2):544–551. doi: 10.1128/jb.161.2.544-551.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue D. J., Sharp P. A. Replication of colicin E1 plasmid DNA in vivo requires no plasmid-encoded proteins. J Bacteriol. 1978 Mar;133(3):1287–1294. doi: 10.1128/jb.133.3.1287-1294.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley T. P., Polisky B. Suppression of ColE1 RNA-RNA mismatch mutations in vivo by the ColE1 Rop protein. Plasmid. 1987 Jul;18(1):24–34. doi: 10.1016/0147-619x(87)90075-8. [DOI] [PubMed] [Google Scholar]
- Dooley T. P., Tamm J., Polisky B. Isolation and characterization of mutants affecting functional domains of ColE1 RNAI. J Mol Biol. 1985 Nov 5;186(1):87–96. doi: 10.1016/0022-2836(85)90259-1. [DOI] [PubMed] [Google Scholar]
- Dreiseikelmann B., Riedel H. D., Schuster H. A dnaB-like protein of Pseudomonas aeruginosa. Nucleic Acids Res. 1987 Jan 26;15(2):385–395. doi: 10.1093/nar/15.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
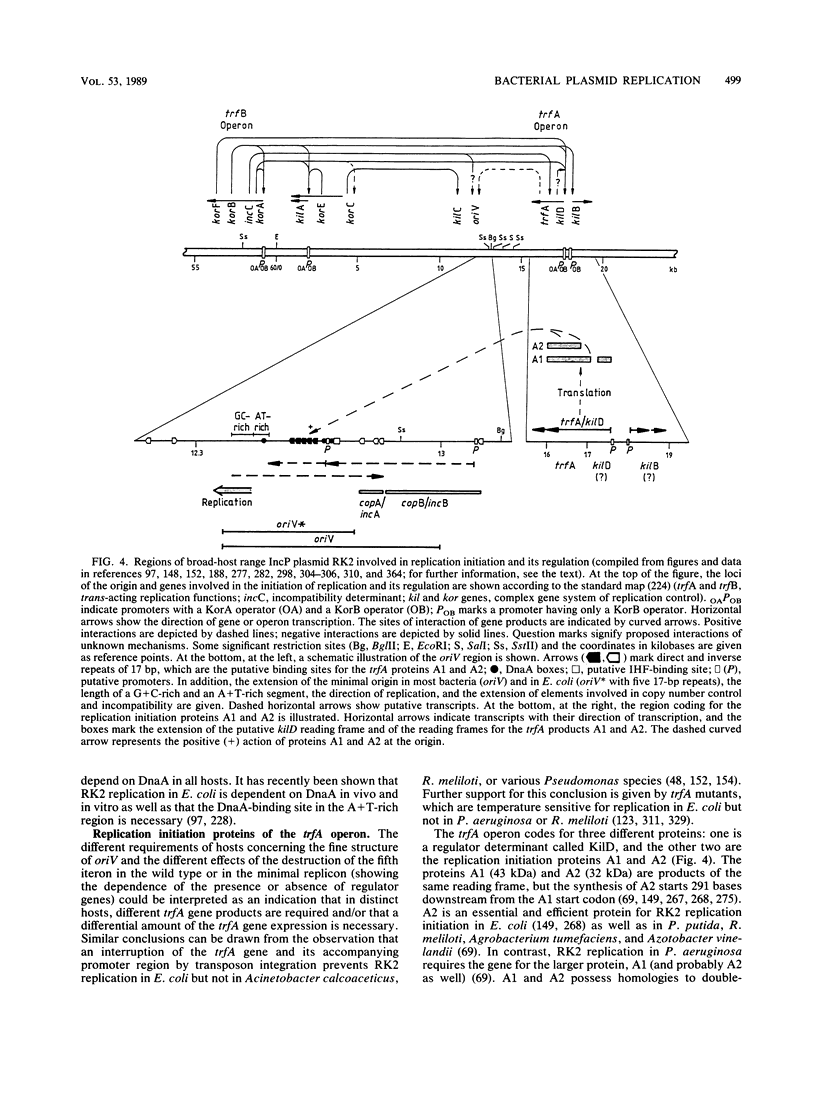
- Durland R. H., Helinski D. R. The sequence encoding the 43-kilodalton trfA protein is required for efficient replication or maintenance of minimal RK2 replicons in Pseudomonas aeruginosa. Plasmid. 1987 Sep;18(2):164–169. doi: 10.1016/0147-619x(87)90044-8. [DOI] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986 Sep 5;233(4768):1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Ely S., Wright A. Maintenance of plasmid pSC101 in Escherichia coli requires the host primase. J Bacteriol. 1985 Oct;164(1):484–486. doi: 10.1128/jb.164.1.484-486.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J. D., von Hippel P. H. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J Biol Chem. 1978 Feb 10;253(3):927–934. [PubMed] [Google Scholar]
- Engleberg N. C., Cianciotto N., Smith J., Eisenstein B. I. Transfer and maintenance of small, mobilizable plasmids with ColE1 replication origins in Legionella pneumophila. Plasmid. 1988 Jul;20(1):83–91. doi: 10.1016/0147-619x(88)90010-8. [DOI] [PubMed] [Google Scholar]
- Felton J., Wright A. Plasmid pSC101 replication in integratively suppressed cells requires dnaA function. Mol Gen Genet. 1979 Sep;175(2):231–233. doi: 10.1007/BF00425541. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Meyer R. J., Helinski D. R. Suppression of Co1E1 replication properties by the Inc P-1 plasmid RK2 in hybrid plasmids constructed in vitro. J Mol Biol. 1979 Sep 25;133(3):295–318. doi: 10.1016/0022-2836(79)90395-4. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Pohlman R. F., Bechhofer D. H., Prince A. S., Kelton C. A. Broad host range plasmid RK2 encodes multiple kil genes potentially lethal to Escherichia coli host cells. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1935–1939. doi: 10.1073/pnas.79.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M., Appelt K. The integration host factor of Escherichia coli binds to multiple sites at plasmid R6K gamma origin and is essential for replication. Nucleic Acids Res. 1988 May 11;16(9):3829–3843. doi: 10.1093/nar/16.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M., McEachern M. J., Helinski D. R. Positive and negative roles of an initiator protein at an origin of replication. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9645–9649. doi: 10.1073/pnas.83.24.9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M., McEachern M. J., Mukhopadhyay P., Greener A., Yang S. L., Helinski D. R. DNA and protein interactions in the regulation of plasmid replication. J Cell Sci Suppl. 1987;7:15–31. doi: 10.1242/jcs.1987.supplement_7.2. [DOI] [PubMed] [Google Scholar]
- Filutowicz M., McEachern M., Greener A., Mukhopadhyay P., Uhlenhopp E., Durland R., Helinski D. Role of the pi initiation protein and direct nucleotide sequence repeats in the regulation of plasmid R6K replication. Basic Life Sci. 1985;30:125–140. doi: 10.1007/978-1-4613-2447-8_13. [DOI] [PubMed] [Google Scholar]
- Firshein W., Caro L. Detection of displacement ("D") loops with the properties of a replicating intermediate synthesized by a DNA/membrane complex derived from the low-copy-number plasmid RK2. Plasmid. 1984 Nov;12(3):227–232. doi: 10.1016/0147-619x(84)90051-9. [DOI] [PubMed] [Google Scholar]
- Firshein W., Strumph P., Benjamin P., Burnstein K., Kornacki J. Replication of a low-copy-number plasmid by a plasmid DNA-membrane complex extracted from minicells of Escherichia coli. J Bacteriol. 1982 Jun;150(3):1234–1243. doi: 10.1128/jb.150.3.1234-1243.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzwater T., Tamm J., Polisky B. RNA1 is sufficient to mediate plasmid ColE1 incompatibility in vivo. J Mol Biol. 1984 May 25;175(3):409–417. doi: 10.1016/0022-2836(84)90357-7. [DOI] [PubMed] [Google Scholar]
- Fitzwater T., Zhang X. Y., Elble R., Polisky B. Conditional high copy number ColE1 mutants: resistance to RNA1 inhibition in vivo and in vitro. EMBO J. 1988 Oct;7(10):3289–3297. doi: 10.1002/j.1460-2075.1988.tb03196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouser L., Bird R. E. Accumulation of ColE1 early replicative intermediates catalyzed by extracts of Escherichia coli dnaG mutant strains. J Bacteriol. 1983 Jun;154(3):1174–1183. doi: 10.1128/jb.154.3.1174-1183.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey J., Chandler M., Caro L. Overinitiation of chromosome and plasmid replication in a dna Acos mutant of Escherichia coli K12. Evidence for dnaA-dnaB interactions. J Mol Biol. 1984 Oct 25;179(2):171–183. doi: 10.1016/0022-2836(84)90464-9. [DOI] [PubMed] [Google Scholar]
- Frey J., Chandler M., Caro L. The effects of an Escherichia coli dnaAts mutation on the replication of the plasmids colE1 pSC101, R100.1 and RTF-TC. Mol Gen Genet. 1979 Jul 13;174(2):117–126. doi: 10.1007/BF00268349. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. R., Georgopoulos C., Tilly K., Herskowitz I., Banuett F. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol Rev. 1984 Dec;48(4):299–325. doi: 10.1128/mr.48.4.299-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Kornberg A. Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5817–5821. doi: 10.1073/pnas.80.19.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell B. E., Baker T. A., Kornberg A. Complete enzymatic replication of plasmids containing the origin of the Escherichia coli chromosome. J Biol Chem. 1986 Apr 25;261(12):5616–5624. [PubMed] [Google Scholar]
- Funnell B. E., Baker T. A., Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987 Jul 25;262(21):10327–10334. [PubMed] [Google Scholar]
- Gamas P., Burger A. C., Churchward G., Caro L., Galas D., Chandler M. Replication of pSC101: effects of mutations in the E. coli DNA binding protein IHF. Mol Gen Genet. 1986 Jul;204(1):85–89. doi: 10.1007/BF00330192. [DOI] [PubMed] [Google Scholar]
- Gamas P., Chandler M. G., Prentki P., Galas D. J. Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hot-spot in pBR322. J Mol Biol. 1987 May 20;195(2):261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- Gayle R. B., 3rd, Vermersch P. S., Bennett G. N. Construction and characterization of pBR322-derived plasmids with deletions of the RNA I region. Gene. 1986;41(2-3):281–288. doi: 10.1016/0378-1119(86)90108-3. [DOI] [PubMed] [Google Scholar]
- Gaylo P. J., Turjman N., Bastia D. DnaA protein is required for replication of the minimal replicon of the broad-host-range plasmid RK2 in Escherichia coli. J Bacteriol. 1987 Oct;169(10):4703–4709. doi: 10.1128/jb.169.10.4703-4709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Rasmussen P. B., Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A. 1986 May;83(10):3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germino J., Bastia D. The replication initiator protein of plasmid R6K tagged with beta-galactosidase shows sequence-specific DNA-binding. Cell. 1983 Jan;32(1):131–140. doi: 10.1016/0092-8674(83)90503-2. [DOI] [PubMed] [Google Scholar]
- Givskov M., Stougaard P., Light J., Molin S. Identification and characterization of mutations responsible for a runaway replication phenotype of plasmid R1. Gene. 1987;57(2-3):203–211. doi: 10.1016/0378-1119(87)90123-5. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L., Dice J. F. Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem. 1974;43(0):835–869. doi: 10.1146/annurev.bi.43.070174.004155. [DOI] [PubMed] [Google Scholar]
- Grinter N. J., Barth P. T. Characterization of SmSu plasmids by restriction endonuclease cleavage and compatibility testing. J Bacteriol. 1976 Oct;128(1):394–400. doi: 10.1128/jb.128.1.394-400.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter N. J. Replication control of IncP plasmids. Plasmid. 1984 Jan;11(1):74–81. doi: 10.1016/0147-619x(84)90009-x. [DOI] [PubMed] [Google Scholar]
- Guerry P., van Embden J., Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974 Feb;117(2):619–630. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Hasegawa P., Davis C. E. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen E. B., Yarmolinsky M. B. Host participation in plasmid maintenance: dependence upon dnaA of replicons derived from P1 and F. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4423–4427. doi: 10.1073/pnas.83.12.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring V., Scholz P., Scherzinger E., Frey J., Derbyshire K., Hatfull G., Willetts N. S., Bagdasarian M. Protein RepC is involved in copy number control of the broad host range plasmid RSF1010. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6090–6094. doi: 10.1073/pnas.82.18.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma K., Sekiguchi M. Effect of dna mutations on the replication of plasmid pSC101 in Escherichia coli K-12. J Bacteriol. 1979 Mar;137(3):1095–1099. doi: 10.1128/jb.137.3.1095-1099.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma K., Sekiguchi M. Replication of plasmid pSC101 in Escherichia coli K12: requirement for dnaA function. Mol Gen Genet. 1977 Sep 9;154(3):225–230. doi: 10.1007/BF00571277. [DOI] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M., Schroeter A., Mach F. Physiological studies on pBR 322 DNA amplification in an Escherichia coli relA strain. J Basic Microbiol. 1986;26(6):329–333. doi: 10.1002/jobm.3620260605. [DOI] [PubMed] [Google Scholar]
- Hecker M., Schroeter A., Mach F. Replication of pBR322 DNA in stringent and relaxed strains of Escherichia coli. Mol Gen Genet. 1983;190(2):355–357. doi: 10.1007/BF00330665. [DOI] [PubMed] [Google Scholar]
- Helmer-Citterich M., Anceschi M. M., Banner D. W., Cesareni G. Control of ColE1 replication: low affinity specific binding of Rop (Rom) to RNAI and RNAII. EMBO J. 1988 Feb;7(2):557–566. doi: 10.1002/j.1460-2075.1988.tb02845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenbrand G., Staudenbauer W. L. Discriminatory function of ribonuclease H in the selective initiation of plasmid DNA replication. Nucleic Acids Res. 1982 Feb 11;10(3):833–853. doi: 10.1093/nar/10.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S., Jaffé A., Ogura T., Mori H., Takahashi H. F plasmid ccd mechanism in Escherichia coli. J Bacteriol. 1986 Apr;166(1):100–104. doi: 10.1128/jb.166.1.100-104.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., Nakamura K., Sakaguchi K. A linear DNA plasmid from Streptomyces rochei with an inverted terminal repetition of 614 base pairs. EMBO J. 1984 Apr;3(4):761–766. doi: 10.1002/j.1460-2075.1984.tb01881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogrefe C., Friedrich B. Isolation and characterization of megaplasmid DNA from lithoautotrophic bacteria. Plasmid. 1984 Nov;12(3):161–169. doi: 10.1016/0147-619x(84)90040-4. [DOI] [PubMed] [Google Scholar]
- Holm L. Codon usage and gene expression. Nucleic Acids Res. 1986 Apr 11;14(7):3075–3087. doi: 10.1093/nar/14.7.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda Y., Sakai H., Komano T. Two single-strand DNA initiation signals located in the oriV region of plasmid RSF1010. Gene. 1988 Sep 7;68(2):221–228. doi: 10.1016/0378-1119(88)90024-8. [DOI] [PubMed] [Google Scholar]
- Hooykaas P. J., den Dulk-Ras H., Schilperoort R. A. Phenotypic expression of mutations in a wide-host-range R plasmid in Escherichia coli and Rhizobium meliloti. J Bacteriol. 1982 Apr;150(1):395–397. doi: 10.1128/jb.150.1.395-397.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Asai Y., Nakazawa A., Nakazawa T. Nucleotide sequence of a DNA segment promoting transcription in Pseudomonas putida. J Bacteriol. 1986 Jun;166(3):739–745. doi: 10.1128/jb.166.3.739-745.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inselburg J. Replication of colicin E1 plasmid DNA in minicells from a unique replication initiation site. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2256–2259. doi: 10.1073/pnas.71.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980 May;77(5):2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Initiation of replication of plasmid ColE1 DNA by RNA polymerase, ribonuclease H, and DNA polymerase I. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):409–417. doi: 10.1101/sqb.1979.043.01.047. [DOI] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Purification of ribonuclease H as a factor required for initiation of in vitro Co1E1 DNA replication. Nucleic Acids Res. 1982 Oct 11;10(19):5949–5965. doi: 10.1093/nar/10.19.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y., Watson J. M., Haas D., Leisinger T. Genetic and molecular characterization of the Pseudomonas plasmid pVS1. Plasmid. 1984 May;11(3):206–220. doi: 10.1016/0147-619x(84)90027-1. [DOI] [PubMed] [Google Scholar]
- Ivanov I., Yavashev L., Gigova L., Alexciev K., Christov C. A conditional high-copy-number plasmid derivative of pBR322. Microbiologica. 1988 Apr;11(2):95–99. [PubMed] [Google Scholar]
- Jaoua S., Guespin-Michel J. F., Breton A. M. Mode of insertion of the broad-host-range plasmid RP4 and its derivatives into the chromosome of Myxococcus xanthus. Plasmid. 1987 Sep;18(2):111–119. doi: 10.1016/0147-619x(87)90038-2. [DOI] [PubMed] [Google Scholar]
- Kamio Y., Tabuchi A., Itoh Y., Katagiri H., Terawaki Y. Complete nucleotide sequence of mini-Rts1 and its copy mutant. J Bacteriol. 1984 Apr;158(1):307–312. doi: 10.1128/jb.158.1.307-312.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley W., Bastia D. Replication initiator protein of plasmid R6K autoregulates its own synthesis at the transcriptional step. Proc Natl Acad Sci U S A. 1985 May;82(9):2574–2578. doi: 10.1073/pnas.82.9.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Meyer R. J. Copy number of the broad host-range plasmid R1162 is determined by the amounts of essential plasmid-encoded proteins. J Mol Biol. 1985 Oct 20;185(4):755–767. doi: 10.1016/0022-2836(85)90060-9. [DOI] [PubMed] [Google Scholar]
- Kim K., Meyer R. J. Copy-number of broad host-range plasmid R1162 is regulated by a small RNA. Nucleic Acids Res. 1986 Oct 24;14(20):8027–8046. doi: 10.1093/nar/14.20.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Lin L. S., Meyer R. J. Two domains at the origin are required for replication and maintenance of broad-host-range plasmid R1162. J Bacteriol. 1987 Dec;169(12):5870–5872. doi: 10.1128/jb.169.12.5870-5872.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. DNA polymerase as a requirement for the maintenance of the bacterial plasmid colicinogenic factor E1. Biochem Biophys Res Commun. 1970 Dec 24;41(6):1538–1544. doi: 10.1016/0006-291x(70)90562-0. [DOI] [PubMed] [Google Scholar]
- Kline B. C. A review of mini-F plasmid maintenance. Plasmid. 1985 Jul;14(1):1–16. doi: 10.1016/0147-619x(85)90027-7. [DOI] [PubMed] [Google Scholar]
- Kline B. C. Aspects of plasmid F maintenance in Escherichia coli. Can J Microbiol. 1988 Apr;34(4):526–535. doi: 10.1139/m88-090. [DOI] [PubMed] [Google Scholar]
- Kogoma T. Absence of RNase H allows replication of pBR322 in Escherichia coli mutants lacking DNA polymerase I. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7845–7849. doi: 10.1073/pnas.81.24.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T., Kline B. C. Integrative suppression of dnaA(Ts) mutations mediated by plasmid F in Escherichia coli is a DnaA-dependent process. Mol Gen Genet. 1987 Dec;210(2):262–269. doi: 10.1007/BF00325692. [DOI] [PubMed] [Google Scholar]
- Kogoma T. RNase H-defective mutants of Escherichia coli. J Bacteriol. 1986 May;166(2):361–363. doi: 10.1128/jb.166.2.361-363.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Kohwi Y. Poly(dG)-poly(dC) sequences, under torsional stress, induce an altered DNA conformation upon neighboring DNA sequences. Cell. 1985 Nov;43(1):199–206. doi: 10.1016/0092-8674(85)90024-8. [DOI] [PubMed] [Google Scholar]
- Kolter R., Helinski D. R. Regulation of initiation of DNA replication. Annu Rev Genet. 1979;13:355–391. doi: 10.1146/annurev.ge.13.120179.002035. [DOI] [PubMed] [Google Scholar]
- Kontomichalou P., Mitani M., Clowes R. C. Circular R-factor molecules controlling penicillinase synthesis, replicating in Escherichia coli under either relaxed or stringent control. J Bacteriol. 1970 Oct;104(1):34–44. doi: 10.1128/jb.104.1.34-44.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kornacki J. A., Balderes P. J., Figurski D. H. Nucleotide sequence of korB, a replication control gene of broad host-range plasmid RK2. J Mol Biol. 1987 Nov 20;198(2):211–222. doi: 10.1016/0022-2836(87)90307-x. [DOI] [PubMed] [Google Scholar]
- Kornacki J. A., Firshein W. Replication of plasmid RK2 in vitro by a DNA-membrane complex: evidence for initiation of replication and its coupling to transcription and translation. J Bacteriol. 1986 Jul;167(1):319–326. doi: 10.1128/jb.167.1.319-326.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornacki J. A., West A. H., Firshein W. Proteins encoded by the trans-acting replication and maintenance regions of broad host range plasmid RK2. Plasmid. 1984 Jan;11(1):48–57. doi: 10.1016/0147-619x(84)90006-4. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnapillai V., Nash J., Lanka E. Insertion mutations in the promiscuous IncP-1 plasmid R18 which affect its host range between Pseudomonas species. Plasmid. 1984 Nov;12(3):170–180. doi: 10.1016/0147-619x(84)90041-6. [DOI] [PubMed] [Google Scholar]
- Krishnapillai V., Wexler M., Nash J., Figurski D. H. Genetic basis of a Tn7 insertion mutation in the trfA region of the promiscuous IncP-1 plasmid R18 which affects its host range. Plasmid. 1987 Mar;17(2):164–166. doi: 10.1016/0147-619x(87)90022-9. [DOI] [PubMed] [Google Scholar]
- Lacatena R. M., Banner D. W., Castagnoli L., Cesareni G. Control of initiation of pMB1 replication: purified Rop protein and RNA I affect primer formation in vitro. Cell. 1984 Jul;37(3):1009–1014. doi: 10.1016/0092-8674(84)90435-5. [DOI] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981 Dec 17;294(5842):623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Interaction between RNA1 and the primer precursor in the regulation of Co1E1 replication. J Mol Biol. 1983 Nov 5;170(3):635–650. doi: 10.1016/s0022-2836(83)80125-9. [DOI] [PubMed] [Google Scholar]
- Lane H. E. Replication and incompatibility of F and plasmids in the IncFI Group. Plasmid. 1981 Jan;5(1):100–126. doi: 10.1016/0147-619x(81)90079-2. [DOI] [PubMed] [Google Scholar]
- Lanka E., Barth P. T. Plasmid RP4 specifies a deoxyribonucleic acid primase involved in its conjugal transfer and maintenance. J Bacteriol. 1981 Dec;148(3):769–781. doi: 10.1128/jb.148.3.769-781.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanka E., Lurz R., Fürste J. P. Molecular cloning and mapping of SphI restriction fragments of plasmid RP4. Plasmid. 1983 Nov;10(3):303–307. doi: 10.1016/0147-619x(83)90047-1. [DOI] [PubMed] [Google Scholar]
- Lanka E., Lurz R., Kröger M., Fürste J. P. Plasmid RP4 encodes two forms of a DNA primase. Mol Gen Genet. 1984;194(1-2):65–72. doi: 10.1007/BF00383499. [DOI] [PubMed] [Google Scholar]
- LeBowitz J. H., McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986 Apr 5;261(10):4738–4748. [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S., Lesser C. F., Youderian P., Susskind M. M., Landy A. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem. 1985 Apr 10;260(7):4468–4477. [PubMed] [Google Scholar]
- Lin-Chao S., Bremer H. Activities of the RNAI and RNAII promoters of plasmid pBR322. J Bacteriol. 1987 Mar;169(3):1217–1222. doi: 10.1128/jb.169.3.1217-1222.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. S., Kim Y. J., Meyer R. J. The 20 bp, directly repeated DNA sequence of broad host range plasmid R1162 exerts incompatibility in vivo and inhibits R1162 DNA replication in vitro. Mol Gen Genet. 1987 Jul;208(3):390–397. doi: 10.1007/BF00328129. [DOI] [PubMed] [Google Scholar]
- Lin L. S., Meyer R. J. DNA synthesis is initiated at two positions within the origin of replication of plasmid R1162. Nucleic Acids Res. 1987 Oct 26;15(20):8319–8331. doi: 10.1093/nar/15.20.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. S., Meyer R. J. Directly repeated, 20-bp sequence of plasmid R1162 DNA is required for replication, expression of incompatibility, and copy-number control. Plasmid. 1986 Jan;15(1):35–47. doi: 10.1016/0147-619x(86)90012-0. [DOI] [PubMed] [Google Scholar]
- Lin L. S., Meyer R. J. Nucleotide sequence and functional properties of DNA encoding incompatibility in the broad host-range plasmid R1162. Mol Gen Genet. 1984;194(3):423–431. doi: 10.1007/BF00425554. [DOI] [PubMed] [Google Scholar]
- Linder P., Churchward G., Caro L. Plasmid pSC101 replication mutants generated by insertion of the transposon Tn1000. J Mol Biol. 1983 Oct 25;170(2):287–303. doi: 10.1016/s0022-2836(83)80149-1. [DOI] [PubMed] [Google Scholar]
- Linder P., Churchward G., Xia G. X., Yu Y. Y., Caro L. An essential replication gene, repA, of plasmid pSC101 is autoregulated. J Mol Biol. 1985 Feb 5;181(3):383–393. doi: 10.1016/0022-2836(85)90227-x. [DOI] [PubMed] [Google Scholar]
- Looman A. C., van Knippenberg P. H. Effects of GUG and AUG initiation codons on the expression of lacZ in Escherichia coli. FEBS Lett. 1986 Mar 3;197(1-2):315–320. doi: 10.1016/0014-5793(86)80349-0. [DOI] [PubMed] [Google Scholar]
- Lundquist P. D., Levin B. R. Transitory derepression and the maintenance of conjugative plasmids. Genetics. 1986 Jul;113(3):483–497. doi: 10.1093/genetics/113.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann N. H. Plasmid partitioning in Escherichia coli. Microbiol Sci. 1985 Oct;2(10):299–302. [PubMed] [Google Scholar]
- Masai H., Arai K. Initiation of lagging-strand synthesis for pBR322 plasmid DNA replication in vitro is dependent on primosomal protein i encoded by dnaT. J Biol Chem. 1988 Oct 15;263(29):15016–15023. [PubMed] [Google Scholar]
- Masai H., Arai K. RepA and DnaA proteins are required for initiation of R1 plasmid replication in vitro and interact with the oriR sequence. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4781–4785. doi: 10.1073/pnas.84.14.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Bond M. W., Arai K. Cloning of the Escherichia coli gene for primosomal protein i: the relationship to dnaT, essential for chromosomal DNA replication. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1256–1260. doi: 10.1073/pnas.83.5.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson L., Ray D. S. Mechanism of autonomous control of the Escherichia coli F plasmid: different complexes of the initiator/repressor protein are bound to its operator and to an F plasmid replication origin. Nucleic Acids Res. 1986 Jul 25;14(14):5693–5711. doi: 10.1093/nar/14.14.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masukata H., Dasgupta S., Tomizawa J. Transcriptional activation of ColE1 DNA synthesis by displacement of the nontranscribed strand. Cell. 1987 Dec 24;51(6):1123–1130. doi: 10.1016/0092-8674(87)90598-8. [DOI] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell. 1986 Jan 17;44(1):125–136. doi: 10.1016/0092-8674(86)90491-5. [DOI] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Effects of point mutations on formation and structure of the RNA primer for ColE1 DNA replication. Cell. 1984 Feb;36(2):513–522. doi: 10.1016/0092-8674(84)90244-7. [DOI] [PubMed] [Google Scholar]
- Mermod N., Lehrbach P. R., Reineke W., Timmis K. N. Transcription of the TOL plasmid toluate catabolic pathway operon of Pseudomonas putida is determined by a pair of co-ordinately and positively regulated overlapping promoters. EMBO J. 1984 Nov;3(11):2461–2466. doi: 10.1002/j.1460-2075.1984.tb02156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merryweather A., Barth P. T., Wilkins B. M. Role and specificity of plasmid RP4-encoded DNA primase in bacterial conjugation. J Bacteriol. 1986 Jul;167(1):12–17. doi: 10.1128/jb.167.1.12-17.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W. Initiation of DNA replication in Escherichia coli. J Bacteriol. 1987 Aug;169(8):3395–3399. doi: 10.1128/jb.169.8.3395-3399.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. J., Helinski D. R. Unidirectional replication of the P-group plasmid RK2. Biochim Biophys Acta. 1977 Sep 6;478(1):109–113. doi: 10.1016/0005-2787(77)90249-0. [DOI] [PubMed] [Google Scholar]
- Meyer R. J., Lin L. S., Kim K., Brasch M. A. Broad host-range plasmid R1162: replication, incompatibility, and copy-number control. Basic Life Sci. 1985;30:173–188. doi: 10.1007/978-1-4613-2447-8_16. [DOI] [PubMed] [Google Scholar]
- Meyer R., Hinds M., Brasch M. Properties of R1162, a broad-host-range, high-copy-number plasmid. J Bacteriol. 1982 May;150(2):552–562. doi: 10.1128/jb.150.2.552-562.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R., Hinds M. Multiple mechanisms for expression of incompatibility by broad-host-range plasmid RK2. J Bacteriol. 1982 Dec;152(3):1078–1090. doi: 10.1128/jb.152.3.1078-1090.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R., Laux R., Boch G., Hinds M., Bayly R., Shapiro J. A. Broad-host-range IncP-4 plasmid R1162: effects of deletions and insertions on plasmid maintenance and host range. J Bacteriol. 1982 Oct;152(1):140–150. doi: 10.1128/jb.152.1.140-150.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minden J. S., Marians K. J. Escherichia coli topoisomerase I can segregate replicating pBR322 daughter DNA molecules in vitro. J Biol Chem. 1986 Sep 5;261(25):11906–11917. [PubMed] [Google Scholar]
- Minden J. S., Marians K. J. Replication of pBR322 DNA in vitro with purified proteins. Requirement for topoisomerase I in the maintenance of template specificity. J Biol Chem. 1985 Aug 5;260(16):9316–9325. [PubMed] [Google Scholar]
- Morita M., Oka A. The structure of a transcriptional unit on colicin E1 plasmid. Eur J Biochem. 1979 Jul;97(2):435–443. doi: 10.1111/j.1432-1033.1979.tb13131.x. [DOI] [PubMed] [Google Scholar]
- Morlon J., Chartier M., Bidaud M., Lazdunski C. The complete nucleotide sequence of the colicinogenic plasmid ColA. High extent of homology with ColE1. Mol Gen Genet. 1988 Feb;211(2):231–243. doi: 10.1007/BF00330599. [DOI] [PubMed] [Google Scholar]
- Moser D. R., Ma D., Moser C. D., Campbell J. L. cis-acting mutations that affect rop protein control of plasmid copy number. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4465–4469. doi: 10.1073/pnas.81.14.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser D. R., Moser C. D., Sinn E., Campbell J. L. Suppressors of a temperature-sensitive copy-number mutation in plasmid NTP1. Mol Gen Genet. 1983;192(1-2):95–100. doi: 10.1007/BF00327652. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Erickson H., Bastia D. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell. 1988 Feb 12;52(3):375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Patel I., Bastia D. Conformational changes in a replication origin induced by an initiator protein. Cell. 1985 Nov;43(1):189–197. doi: 10.1016/0092-8674(85)90023-6. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Ohmori H., Yura T., Nagata T. Requirement of the Escherichia coli dnaA gene function for ori-2-dependent mini-F plasmid replication. J Bacteriol. 1987 Apr;169(4):1724–1730. doi: 10.1128/jb.169.4.1724-1730.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito S., Kitani T., Ogawa T., Okazaki T., Uchida H. Escherichia coli mutants suppressing replication-defective mutations of the ColE1 plasmid. Proc Natl Acad Sci U S A. 1984 Jan;81(2):550–554. doi: 10.1073/pnas.81.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito S., Uchida H. Initiation of DNA replication in a ColE1-type plasmid: isolation of mutations in the ori region. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6744–6748. doi: 10.1073/pnas.77.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito S., Uchida H. RNase H and replication of ColE1 DNA in Escherichia coli. J Bacteriol. 1986 Apr;166(1):143–147. doi: 10.1128/jb.166.1.143-147.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash J., Krishnapillai V. DNA sequence analysis of host range mutants of the promiscuous IncP-1 plasmids R18 and R68 with Tn7 insertions in oriV. Plasmid. 1987 Jul;18(1):35–45. doi: 10.1016/0147-619x(87)90076-x. [DOI] [PubMed] [Google Scholar]
- Nijkamp H. J., de Lang R., Stuitje A. R., van den Elzen P. J., Veltkamp E., van Putten A. J. The complete nucleotide sequence of the bacteriocinogenic plasmid CloDF13. Plasmid. 1986 Sep;16(2):135–160. doi: 10.1016/0147-619x(86)90072-7. [DOI] [PubMed] [Google Scholar]
- Nomura N., Low R. L., Ray D. S. Identification of ColE1 DNA sequences that direct single strand-to-double strand conversion by a phi X174 type mechanism. Proc Natl Acad Sci U S A. 1982 May;79(10):3153–3157. doi: 10.1073/pnas.79.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Aagaard-Hansen H. Maintenance of bacterial plasmids: comparison of theoretical calculations and experiments with plasmid R1 in Escherichia coli. Mol Gen Genet. 1984;197(1):1–7. doi: 10.1007/BF00327915. [DOI] [PubMed] [Google Scholar]
- Nordström K. Control of plasmid replication: theoretical considerations and practical solutions. Basic Life Sci. 1985;30:189–214. doi: 10.1007/978-1-4613-2447-8_17. [DOI] [PubMed] [Google Scholar]
- Nordström K., Molin S., Light J. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984 Sep;12(2):71–90. doi: 10.1016/0147-619x(84)90054-4. [DOI] [PubMed] [Google Scholar]
- Nordström M., Nordström K. Control of replication of FII plasmids: comparison of the basic replicons and of the copB systems of plasmids R100 and R1. Plasmid. 1985 Mar;13(2):81–87. doi: 10.1016/0147-619x(85)90060-5. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Plasmid incompatibility. Microbiol Rev. 1987 Dec;51(4):381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent M. E., Smith T. J., Tacon W. C. Characterization and incompatibility properties of ROM- derivatives of pBR322-based plasmids. J Gen Microbiol. 1986 Apr;132(4):1021–1026. doi: 10.1099/00221287-132-4-1021. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., von Meyenburg K., Hansen F. G., Yoshikawa H. Conservation of genes and their organization in the chromosomal replication origin region of Bacillus subtilis and Escherichia coli. EMBO J. 1985 Dec 1;4(12):3345–3350. doi: 10.1002/j.1460-2075.1985.tb04087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Baker T. A., van der Ende A., Kornberg A. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: contributions of RNA polymerase and primase. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3562–3566. doi: 10.1073/pnas.82.11.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Okazaki T. Function of RNase H in DNA replication revealed by RNase H defective mutants of Escherichia coli. Mol Gen Genet. 1984;193(2):231–237. doi: 10.1007/BF00330673. [DOI] [PubMed] [Google Scholar]
- Ohmori H., Murakami Y., Nagata T. Nucleotide sequences required for a ColE1-type plasmid to replicate in Escherichia coli cells with or without RNase H. J Mol Biol. 1987 Nov 20;198(2):223–234. doi: 10.1016/0022-2836(87)90308-1. [DOI] [PubMed] [Google Scholar]
- Oka A., Nomura N., Morita M., Sugisaki H., Sugimoto K., Takanami M. Nucleotide sequence of small ColE1 derivatives: structure of the regions essential for autonomous replication and colicin E1 immunity. Mol Gen Genet. 1979 May 4;172(2):151–159. doi: 10.1007/BF00268276. [DOI] [PubMed] [Google Scholar]
- Orr E., Staudenbauer W. L. An Escherichia coli mutant thermosensitive in the B subunit of DNA gyrase: effect on the structure and replication of the colicin E1 plasmid in vitro. Mol Gen Genet. 1981;181(1):52–56. doi: 10.1007/BF00339004. [DOI] [PubMed] [Google Scholar]
- Ortega S., Lanka E., Diaz R. The involvement of host replication proteins and of specific origin sequences in the in vitro replication of miniplasmid R1 DNA. Nucleic Acids Res. 1986 Jun 25;14(12):4865–4879. doi: 10.1093/nar/14.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotatos N. DNA replication regulated by the priming promoter. Nucleic Acids Res. 1984 Mar 26;12(6):2641–2648. doi: 10.1093/nar/12.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Lanka E. Conservation of a common 'backbone' in the genetic organization of the IncP plasmids RP4 and R751. Nucleic Acids Res. 1987 Mar 11;15(5):2385–2385. doi: 10.1093/nar/15.5.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel I., Bastia D. A replication initiator protein enhances the rate of hybrid formation between a silencer RNA and an activator RNA. Cell. 1987 Nov 6;51(3):455–462. doi: 10.1016/0092-8674(87)90641-6. [DOI] [PubMed] [Google Scholar]
- Persson C., Nordström K. Control of replication of the broad host range plasmid RSF1010: the incompatibility determinant consists of directly repeated DNA sequences. Mol Gen Genet. 1986 Apr;203(1):189–192. doi: 10.1007/BF00330402. [DOI] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: kinetics of in vitro interaction between the antisense RNA, CopA, and its target, CopT. EMBO J. 1988 Oct;7(10):3279–3288. doi: 10.1002/j.1460-2075.1988.tb03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkney M., Diaz R., Lanka E., Thomas C. M. Replication of mini RK2 plasmid in extracts of Escherichia coli requires plasmid-encoded protein TrfA and host-encoded proteins DnaA, B, G DNA gyrase and DNA polymerase III. J Mol Biol. 1988 Oct 20;203(4):927–938. doi: 10.1016/0022-2836(88)90118-0. [DOI] [PubMed] [Google Scholar]
- Pinkney M., Theophilus B. D., Warne S. R., Tacon W. C., Thomas C. M. Analysis of transcription from the trfA promoter of broad host range plasmid RK2 in Escherichia coli, Pseudomonas putida, and Pseudomonas aeruginosa. Plasmid. 1987 May;17(3):222–232. doi: 10.1016/0147-619x(87)90030-8. [DOI] [PubMed] [Google Scholar]
- Pinkney M., Thomas C. M. Replication and maintenance of promiscuous plasmids of gram-negative bacteria. Microbiol Sci. 1987 Jun;4(6):186–191. [PubMed] [Google Scholar]
- Pohlman R. F., Figurski D. H. Conditional lethal mutants of the kilB determinant of broad host range plasmid RK2. Plasmid. 1983 Jul;10(1):82–95. doi: 10.1016/0147-619x(83)90060-4. [DOI] [PubMed] [Google Scholar]
- Pohlman R. F., Figurski D. H. Essential genes of plasmid RK2 in Escherichia coli: trfB region controls a kil gene near trfA. J Bacteriol. 1983 Nov;156(2):584–591. doi: 10.1128/jb.156.2.584-591.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Caro L. Replication of prophage P1 during the cell cycle of Escherichia coli. Mol Gen Genet. 1977 Mar 28;152(1):71–76. doi: 10.1007/BF00264942. [DOI] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Galas D. J. Escherichia coli integration host factor bends the DNA at the ends of IS1 and in an insertion hotspot with multiple IHF binding sites. EMBO J. 1987 Aug;6(8):2479–2487. doi: 10.1002/j.1460-2075.1987.tb02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen P. B., Gerdes K., Molin S. Genetic analysis of the parB+ locus of plasmid R1. Mol Gen Genet. 1987 Aug;209(1):122–128. doi: 10.1007/BF00329846. [DOI] [PubMed] [Google Scholar]
- Reddy P., Peterkofsky A., McKenney K. Translational efficiency of the Escherichia coli adenylate cyclase gene: mutating the UUG initiation codon to GUG or AUG results in increased gene expression. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5656–5660. doi: 10.1073/pnas.82.17.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkel L. J., van der Marel G. A., van Boom J. H., Altona C. Influence of N6-methylation of residue A(5) on the conformational behaviour of d(C-C-G-A-A-T-T-C-G-G) in solution studied by 1H-NMR spectroscopy. 2. The hairpin form. Eur J Biochem. 1987 Mar 2;163(2):287–296. doi: 10.1111/j.1432-1033.1987.tb10799.x. [DOI] [PubMed] [Google Scholar]
- Robinson M., Lilley R., Little S., Emtage J. S., Yarranton G., Stephens P., Millican A., Eaton M., Humphreys G. Codon usage can affect efficiency of translation of genes in Escherichia coli. Nucleic Acids Res. 1984 Sep 11;12(17):6663–6671. doi: 10.1093/nar/12.17.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokeach L. A., Kassavetis G. A., Zyskind J. W. RNA polymerase pauses in vitro within the Escherichia coli origin of replication at the same sites where termination occurs in vivo. J Biol Chem. 1987 May 25;262(15):7264–7272. [PubMed] [Google Scholar]
- Rokeach L. A., Søgaard-Andersen L., Molin S. Two functions of the E protein are key elements in the plasmid F replication control system. J Bacteriol. 1985 Dec;164(3):1262–1270. doi: 10.1128/jb.164.3.1262-1270.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokeach L. A., Zyskind J. W. RNA terminating within the E. coli origin of replication: stringent regulation and control by DnaA protein. Cell. 1986 Aug 29;46(5):763–771. doi: 10.1016/0092-8674(86)90352-1. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Ohtsubo H., Ohtsubo E. Role of RNA transcripts in replication incompatibility and copy number control in antibiotic resistance plasmid derivatives. Nature. 1981 Apr 30;290(5809):794–797. doi: 10.1038/290794a0. [DOI] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Yaniv M., Germond J. E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979 Jun;17(2):265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Zinder N. D. Hemimethylation prevents DNA replication in E. coli. Cell. 1987 Sep 25;50(7):1071–1079. doi: 10.1016/0092-8674(87)90173-5. [DOI] [PubMed] [Google Scholar]
- Saadi S., Maas W. K., Hill D. F., Bergquist P. L. Nucleotide sequence analysis of RepFIC, a basic replicon present in IncFI plasmids P307 and F, and its relation to the RepA replicon of IncFII plasmids. J Bacteriol. 1987 May;169(5):1836–1846. doi: 10.1128/jb.169.5.1836-1846.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saano A. K., Zinchenko V. V. A new IncQ plasmid R89S: properties and genetic organization. Plasmid. 1987 May;17(3):191–201. doi: 10.1016/0147-619x(87)90027-8. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y., Tomizawa J. Replication of colicin E1 plasmid DNA in cell extracts. II. Selective synthesis of early replicative intermediates. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1403–1407. doi: 10.1073/pnas.71.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauzu M. A., Kücherer C., Kölling R., Messer W., Lother H. Transcripts within the replication origin, oriC, of Escherichia coli. Nucleic Acids Res. 1987 Mar 25;15(6):2479–2497. doi: 10.1093/nar/15.6.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Bagdasarian M. M., Scholz P., Lurz R., Rückert B., Bagdasarian M. Replication of the broad host range plasmid RSF1010: requirement for three plasmid-encoded proteins. Proc Natl Acad Sci U S A. 1984 Feb;81(3):654–658. doi: 10.1073/pnas.81.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser T. J., Filutowicz M., Helinski D. R. Replication of derivatives of the broad host range plasmid RK2 in two distantly related bacteria. Plasmid. 1983 May;9(3):325–330. doi: 10.1016/0147-619x(83)90010-0. [DOI] [PubMed] [Google Scholar]
- Schmidhauser T. J., Helinski D. R. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J Bacteriol. 1985 Oct;164(1):446–455. doi: 10.1128/jb.164.1.446-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnos M., Zahn K., Inman R. B., Blattner F. R. Initiation protein induced helix destabilization at the lambda origin: a prepriming step in DNA replication. Cell. 1988 Feb 12;52(3):385–395. doi: 10.1016/s0092-8674(88)80031-x. [DOI] [PubMed] [Google Scholar]
- Scholz P., Haring V., Scherzinger E., Lurz R., Bagdasarian M. M., Schuster H., Bagdasarian M. Replication determinants of the broad host-range plasmid RSF1010. Basic Life Sci. 1985;30:243–259. doi: 10.1007/978-1-4613-2447-8_20. [DOI] [PubMed] [Google Scholar]
- Scholz P., Haring V., Wittmann-Liebold B., Ashman K., Bagdasarian M., Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989 Feb 20;75(2):271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- Schreiner H. C., Bechhofer D. H., Pohlman R. F., Young C., Borden P. A., Figurski D. H. Replication control in promiscuous plasmid RK2: kil and kor functions affect expression of the essential replication gene trfA. J Bacteriol. 1985 Jul;163(1):228–237. doi: 10.1128/jb.163.1.228-237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab H., Saurugger P. N., Lafferty R. M. Occurrence of deletion plasmids at high rates after conjugative transfer of the plasmids RP4 and RK2 from Escherichia coli to Alcaligenes eutrophus H16. Arch Microbiol. 1983 Nov;136(2):140–146. doi: 10.1007/BF00404789. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Regulation of plasmid replication. Microbiol Rev. 1984 Mar;48(1):1–23. doi: 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimizu K., Bramhill D., Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987 Jul 17;50(2):259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- Selzer G., Som T., Itoh T., Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983 Jan;32(1):119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- Selzer G., Tomizawa J. I. Specific cleavage of the p15A primer precursor by ribonuclease H at the origin of DNA replication. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7082–7086. doi: 10.1073/pnas.79.23.7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W., Dobrinski B., Lurz R., Messer W. Functionality of the dnaA protein binding site in DNA replication is orientation-dependent. J Biol Chem. 1988 Feb 25;263(6):2719–2723. [PubMed] [Google Scholar]
- Seufert W., Messer W. DnaA protein binding to the plasmid origin region can substitute for primosome assembly during replication of pBR322 in vitro. Cell. 1987 Jan 16;48(1):73–78. doi: 10.1016/0092-8674(87)90357-6. [DOI] [PubMed] [Google Scholar]
- Seufert W., Messer W. Start sites for bidirectional in vitro DNA replication inside the replication origin, oriC, of Escherichia coli. EMBO J. 1987 Aug;6(8):2469–2472. doi: 10.1002/j.1460-2075.1987.tb02527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24(1-2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. Codon usage in regulatory genes in Escherichia coli does not reflect selection for 'rare' codons. Nucleic Acids Res. 1986 Oct 10;14(19):7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Shinger V., Thomas C. M. Transcription in the trfA region of broad host range plasmid RK2 is regulated by trfB and korB. Mol Gen Genet. 1984;195(3):523–529. doi: 10.1007/BF00341457. [DOI] [PubMed] [Google Scholar]
- Shingler V., Thomas C. M. Analysis of the trfA region of broad host-range plasmid RK2 by transposon mutagenesis and identification of polypeptide products. J Mol Biol. 1984 May 25;175(3):229–249. doi: 10.1016/0022-2836(84)90346-2. [DOI] [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc Natl Acad Sci U S A. 1980 Feb;77(2):799–803. doi: 10.1073/pnas.77.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Getty C., Gardner J. F., Salyers A. A. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J Bacteriol. 1986 Mar;165(3):929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovgaard O., Hansen F. G. Comparison of dnaA nucleotide sequences of Escherichia coli, Salmonella typhimurium, and Serratia marcescens. J Bacteriol. 1987 Sep;169(9):3976–3981. doi: 10.1128/jb.169.9.3976-3981.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Shingler V., Thomas C. M. The trfA and trfB promoter regions of broad host range plasmid RK2 share common potential regulatory sequences. Nucleic Acids Res. 1984 Apr 25;12(8):3619–3630. doi: 10.1093/nar/12.8.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Thomas C. M. Comparison of the nucleotide sequences of the vegetative replication origins of broad host range IncP plasmids R751 and RK2 reveals conserved features of probable functional importance. Nucleic Acids Res. 1985 Jan 25;13(2):557–572. doi: 10.1093/nar/13.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Thomas C. M. Comparison of the organisation of the genomes of phenotypically diverse plasmids of incompatibility group P: members of the IncP beta sub-group are closely related. Mol Gen Genet. 1987 Mar;206(3):419–427. doi: 10.1007/BF00428881. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Thomas C. M. Deletion mapping of kil and kor functions in the trfA and trfB regions of broad host range plasmid RK2. Mol Gen Genet. 1983;190(2):245–254. doi: 10.1007/BF00330647. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Thomas C. M. Molecular gentic analysis of the trfB and korB region of broad host range plasmid RK2. J Gen Microbiol. 1984 Jul;130(7):1651–1663. doi: 10.1099/00221287-130-7-1651. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Thomas C. M. Narrow-host-range IncP plasmid pHH502-1 lacks a complete IncP replication system. J Gen Microbiol. 1987 Aug;133(8):2247–2252. doi: 10.1099/00221287-133-8-2247. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Thomas C. M. Nucleotide sequence of the trfA gene of broad host-range plasmid RK2. J Mol Biol. 1984 May 25;175(3):251–262. doi: 10.1016/0022-2836(84)90347-4. [DOI] [PubMed] [Google Scholar]
- Som T., Tomizawa J. Regulatory regions of ColE1 that are involved in determination of plasmid copy number. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3232–3236. doi: 10.1073/pnas.80.11.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker D. M., Kolter R., Helinski D. R. Plasmid R6K DNA replication. I. Complete nucleotide sequence of an autonomously replicating segment. J Mol Biol. 1982 Oct 15;161(1):33–43. doi: 10.1016/0022-2836(82)90276-5. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., Thomas C. M., Helinski D. R. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):8–12. doi: 10.1007/BF00338997. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L. Replication of the ampicillin resistance plasmid RSF1030 in extracts of Escherichia coli: separation of the replication cycle into early and late stages. Mol Gen Genet. 1977 Nov 4;156(1):27–34. doi: 10.1007/BF00272248. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L., Scherzinger E., Lanka E. Replication of the colicin E1 plasmid in extracts of Escherichia coli: uncoupling of leading strand from lagging strand synthesis. Mol Gen Genet. 1979;177(1):113–120. doi: 10.1007/BF00267260. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Moore R. J., Krishnapillai V. Complementation analysis in Pseudomonas aeruginosa of the transfer genes of the wide host range R plasmid R18. Plasmid. 1981 Mar;5(2):202–212. doi: 10.1016/0147-619x(81)90021-4. [DOI] [PubMed] [Google Scholar]
- Stueber D., Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1(11):1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. K., Sherratt D. J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984 Apr;36(4):1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- Swack J. A., Pal S. K., Mason R. J., Abeles A. L., Chattoraj D. K. P1 plasmid replication: measurement of initiator protein concentration in vivo. J Bacteriol. 1987 Aug;169(8):3737–3742. doi: 10.1128/jb.169.8.3737-3742.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi A. Nucleotide sequence of the replication region of plasmid R401 and its incompatibility function. Microbiol Immunol. 1985;29(5):383–393. doi: 10.1111/j.1348-0421.1985.tb00839.x. [DOI] [PubMed] [Google Scholar]
- Tait R. C., Close T. J., Rodriguez R. L., Kado C. I. Isolation of the origin of replication of the IncW-group plasmid pSa. Gene. 1982 Nov;20(1):39–49. doi: 10.1016/0378-1119(82)90085-3. [DOI] [PubMed] [Google Scholar]
- Tait R. C., Kado C. I., Rodriguez R. L. A comparison of the origin of replication of pSa with R6K. Mol Gen Genet. 1983;192(1-2):32–38. doi: 10.1007/BF00327643. [DOI] [PubMed] [Google Scholar]
- Tamm J., Polisky B. Characterization of the ColE1 primer-RNA1 complex: analysis of a domain of ColE1 RNA1 necessary for its interaction with primer RNA. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2257–2261. doi: 10.1073/pnas.82.8.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm J., Polisky B. Structural analysis of RNA molecules involved in plasmid copy number control. Nucleic Acids Res. 1983 Sep 24;11(18):6381–6397. doi: 10.1093/nar/11.18.6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R. K., Harvey S. C. A comparison of six DNA bending models. J Biomol Struct Dyn. 1987 Dec;5(3):497–512. doi: 10.1080/07391102.1987.10506410. [DOI] [PubMed] [Google Scholar]
- Tardif G., Grant R. B. Transfer of plasmids from Escherichia coli to Pseudomonas aeruginosa: characterization of a Pseudomonas aeruginosa mutant with enhanced recipient ability for enterobacterial plasmids. Antimicrob Agents Chemother. 1983 Aug;24(2):201–208. doi: 10.1128/aac.24.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theophilus B. D., Cross M. A., Smith C. A., Thomas C. M. Regulation of the trfA and trfB promoters of broad host range plasmid RK2: identification of sequences essential for regulation by trfB/korA/korD. Nucleic Acids Res. 1985 Nov 25;13(22):8129–8142. doi: 10.1093/nar/13.22.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theophilus B. D., Thomas C. M. Nucleotide sequence of the transcriptional repressor gene korB which plays a key role in regulation of the copy number of broad host range plasmid RK2. Nucleic Acids Res. 1987 Sep 25;15(18):7443–7450. doi: 10.1093/nar/15.18.7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M. Complementation analysis of replication and maintenance functions of broad host range plasmids RK2 and RP1. Plasmid. 1981 May;5(3):277–291. doi: 10.1016/0147-619x(81)90005-6. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Cross M. A., Hussain A. A., Smith C. A. Analysis of copy number control elements in the region of the vegetative replication origin of the broad host range plasmid RK2. EMBO J. 1984 Jan;3(1):57–63. doi: 10.1002/j.1460-2075.1984.tb01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M. Evidence for the involvement of the incC locus of broad host range plasmid RK2 in plasmid maintenance. Plasmid. 1986 Jul;16(1):15–29. doi: 10.1016/0147-619x(86)90075-2. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Hussain A. A., Smith C. A. Maintenance of broad host range plasmid RK2 replicons in Pseudomonas aeruginosa. Nature. 1982 Aug 12;298(5875):674–676. doi: 10.1038/298674a0. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Hussain A. A. The korB gene of broad host range plasmid RK2 is a major copy number control element which may act together with trfB by limiting trfA expression. EMBO J. 1984 Jul;3(7):1513–1519. doi: 10.1002/j.1460-2075.1984.tb02004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M. Instability of a high-copy-number mutant of a miniplasmid derived from broad host range IncP plasmid RK2. Plasmid. 1983 Sep;10(2):184–195. doi: 10.1016/0147-619x(83)90071-9. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Meyer R., Helinski D. R. Regions of broad-host-range plasmid RK2 which are essential for replication and maintenance. J Bacteriol. 1980 Jan;141(1):213–222. doi: 10.1128/jb.141.1.213-222.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M. Molecular genetics of broad host range plasmid RK2. Plasmid. 1981 Jan;5(1):10–19. doi: 10.1016/0147-619x(81)90074-3. [DOI] [PubMed] [Google Scholar]
- Thomas C. M. Recent studies on the control of plasmid replication. Biochim Biophys Acta. 1988 Mar 31;949(3):253–263. doi: 10.1016/0167-4781(88)90150-9. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Smith C. A. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Smith C. A. The trfB region of broad host range plasmid RK2: the nucleotide sequence reveals incC and key regulatory gene trfB/korA/korD as overlapping genes. Nucleic Acids Res. 1986 Jun 11;14(11):4453–4469. doi: 10.1093/nar/14.11.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Stalker D. M., Helinski D. R. Replication and incompatibility properties of segments of the origin region of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):1–7. doi: 10.1007/BF00338996. [DOI] [PubMed] [Google Scholar]
- Tokino T., Murotsu T., Matsubara K. Purification and properties of the mini-F plasmid-encoded E protein needed for autonomous replication control of the plasmid. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4109–4113. doi: 10.1073/pnas.83.12.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomcsányi T., Apirion D. Processing enzyme ribonuclease E specifically cleaves RNA I. An inhibitor of primer formation in plasmid DNA synthesis. J Mol Biol. 1985 Oct 20;185(4):713–720. doi: 10.1016/0022-2836(85)90056-7. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Itoh T. The importance of RNA secondary structure in CoIE1 primer formation. Cell. 1982 Dec;31(3 Pt 2):575–583. doi: 10.1016/0092-8674(82)90313-0. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Ohmori H., Bird R. E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A. 1977 May;74(5):1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. I., Sakakibara Y., Kakefuda T. Replication of colicin E1 plasmid DNA added to cell extracts. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1050–1054. doi: 10.1073/pnas.72.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
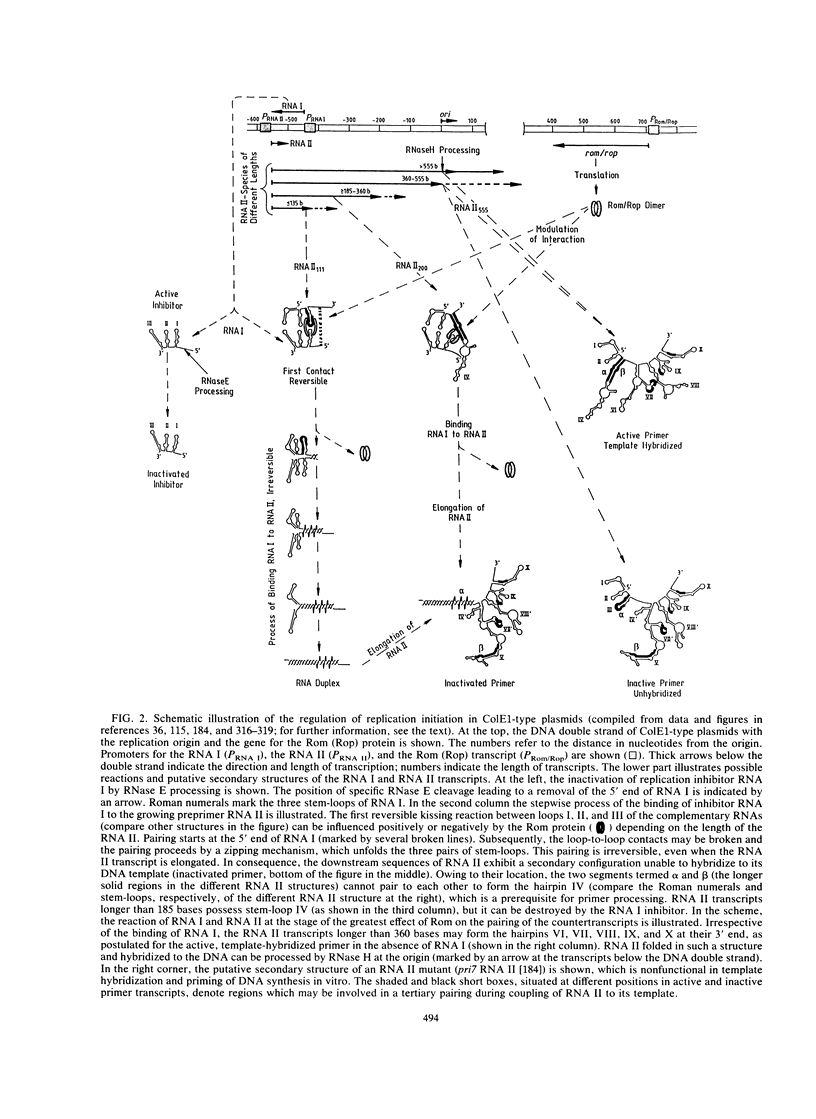
- Tomizawa J. Control of ColE1 plasmid replication: binding of RNA I to RNA II and inhibition of primer formation. Cell. 1986 Oct 10;47(1):89–97. doi: 10.1016/0092-8674(86)90369-7. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: initial interaction of RNA I and the primer transcript is reversible. Cell. 1985 Mar;40(3):527–535. doi: 10.1016/0092-8674(85)90201-6. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984 Oct;38(3):861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Masukata H. Factor-independent termination of transcription in a stretch of deoxyadenosine residues in the template DNA. Cell. 1987 Nov 20;51(4):623–630. doi: 10.1016/0092-8674(87)90131-0. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Sakakibara Y., Kakefuda T. Replication of colicin E1 plasmid DNA in cell extracts. Origin and direction of replication. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2260–2264. doi: 10.1073/pnas.71.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Som T. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell. 1984 Oct;38(3):871–878. doi: 10.1016/0092-8674(84)90282-4. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Harayama S., Iino T. Tn501 insertion mutagenesis in Pseudomonas aeruginosa PAO. Mol Gen Genet. 1984;196(3):494–500. doi: 10.1007/BF00436198. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Matsubara K. Replication of bacteriophage lambda DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):681–691. doi: 10.1101/sqb.1983.047.01.079. [DOI] [PubMed] [Google Scholar]
- Tsutsui H., Fujiyama A., Murotsu T., Matsubara K. Role of nine repeating sequences of the mini-F genome for expression of F-specific incompatibility phenotype and copy number control. J Bacteriol. 1983 Jul;155(1):337–344. doi: 10.1128/jb.155.1.337-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. A runaway-replication mutant of plasmid R1drd-19: temperature-dependent loss of copy number control. Mol Gen Genet. 1978 Oct 4;165(2):167–179. doi: 10.1007/BF00269904. [DOI] [PubMed] [Google Scholar]
- Van der Hoeven N. Coexistence of incompatible plasmids in a bacterial population living under a feast and famine regime. J Math Biol. 1986;24(3):313–325. doi: 10.1007/BF00275640. [DOI] [PubMed] [Google Scholar]
- Varenne S., Buc J., Lloubes R., Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984 Dec 15;180(3):549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Villarroel R., Hedges R. W., Maenhaut R., Leemans J., Engler G., Van Montagu M., Schell J. Heteroduplex analysis of P-plasmid evolution: the role of insertion and deletion of transposable elements. Mol Gen Genet. 1983;189(3):390–399. doi: 10.1007/BF00325900. [DOI] [PubMed] [Google Scholar]
- Vocke C., Bastia D. DNA-protein interaction at the origin of DNA replication of the plasmid pSC101. Cell. 1983 Dec;35(2 Pt 1):495–502. doi: 10.1016/0092-8674(83)90183-6. [DOI] [PubMed] [Google Scholar]
- Vocke C., Bastia D. Primary structure of the essential replicon of the plasmid pSC101. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6557–6561. doi: 10.1073/pnas.80.21.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocke C., Bastia D. The replication initiator protein of plasmid pSC101 is a transcriptional repressor of its own cistron. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2252–2256. doi: 10.1073/pnas.82.8.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A., Suyama A. Local stability of DNA and RNA secondary structure and its relation to biological functions. Prog Biophys Mol Biol. 1986;47(2):113–157. doi: 10.1016/0079-6107(86)90012-x. [DOI] [PubMed] [Google Scholar]
- Wada C., Akiyama Y., Ito K., Yura T. Inhibition of F plasmid replication in htpR mutants of Escherichia coli deficient in sigma 32 protein. Mol Gen Genet. 1986 May;203(2):208–213. doi: 10.1007/BF00333956. [DOI] [PubMed] [Google Scholar]
- Wada C., Imai M., Yura T. Host control of plasmid replication: requirement for the sigma factor sigma 32 in transcription of mini-F replication initiator gene. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8849–8853. doi: 10.1073/pnas.84.24.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. G., von Heijne J., Nordström K. Control of replication of plasmid R1: translation of the 7k reading frame in the RepA mRNA leader region counteracts the interaction between CopA RNA and CopT RNA. EMBO J. 1987 Feb;6(2):515–522. doi: 10.1002/j.1460-2075.1987.tb04783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Palchaudhuri S. Incompatibility repressor in a RepA-like replicon of the IncFI plasmid ColV2-K94. J Bacteriol. 1986 Jun;166(3):1106–1112. doi: 10.1128/jb.166.3.1106-1112.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Oyaizu Y., Oyaizu H., Woese C. R. Natural relationship between bacteroides and flavobacteria. J Bacteriol. 1985 Oct;164(1):230–236. doi: 10.1128/jb.164.1.230-236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R. D. Unusual DNA structures. J Biol Chem. 1988 Jan 25;263(3):1095–1098. [PubMed] [Google Scholar]
- West S. E., Iglewski B. H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Oct 11;16(19):9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S. H., Chattoraj D. K. Replication of mini-P1 plasmid DNA in vitro requires two initiation proteins, encoded by the repA gene of phage P1 and the dnaA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3668–3672. doi: 10.1073/pnas.84.11.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S. H., Zahn K. Characterization of the DNA binding domain of bacteriophage lambda O protein. J Biol Chem. 1986 Jun 5;261(16):7537–7543. [PubMed] [Google Scholar]
- Winans S. C., Walker G. C. Identification of pKM101-encoded loci specifying potentially lethal gene products. J Bacteriol. 1985 Jan;161(1):417–424. doi: 10.1128/jb.161.1.417-424.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. M., Polisky B. Alternative conformations of the ColE1 replication primer modulate its interaction with RNA I. Cell. 1985 Oct;42(3):959–966. doi: 10.1016/0092-8674(85)90292-2. [DOI] [PubMed] [Google Scholar]
- Yakobson E., Guiney G. Homology in the transfer origins of broad host range IncP plasmids: definition of two subgroups of P plasmids. Mol Gen Genet. 1983;192(3):436–438. doi: 10.1007/BF00392187. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Masamune Y. Autogenous regulation of synthesis of the replication protein in plasmid pSC101. Mol Gen Genet. 1985;200(3):362–367. doi: 10.1007/BF00425718. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Yamaguchi M. The replication origin of pSC101: the nucleotide sequence and replication functions of the ori region. Gene. 1984 Jul-Aug;29(1-2):211–219. doi: 10.1016/0378-1119(84)90181-1. [DOI] [PubMed] [Google Scholar]
- Yamaki H., Ohtsubo E., Nagai K., Maeda Y. The oriC unwinding by dam methylation in Escherichia coli. Nucleic Acids Res. 1988 Jun 10;16(11):5067–5073. doi: 10.1093/nar/16.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavachev L., Ivanov I. What does the homology between E. coli tRNAs and RNAs controlling ColE1 plasmid replication mean? J Theor Biol. 1988 Mar 21;131(2):235–241. doi: 10.1016/s0022-5193(88)80240-6. [DOI] [PubMed] [Google Scholar]
- Young C., Bechhofer D. H., Figurski D. H. Gene regulation in plasmid RK2: positive control by korA in the expression of korC. J Bacteriol. 1984 Jan;157(1):247–252. doi: 10.1128/jb.157.1.247-252.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C., Burlage R. S., Figurski D. H. Control of the kilA gene of the broad-host-range plasmid RK2: involvement of korA, korB, and a new gene, korE. J Bacteriol. 1987 Mar;169(3):1315–1320. doi: 10.1128/jb.169.3.1315-1320.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C., Prince A. S., Figurski D. H. korA function of promiscuous plasmid RK2: an autorepressor that inhibits expression of host-lethal gene kilA and replication gene trfA. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7374–7378. doi: 10.1073/pnas.82.21.7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985 Oct 3;317(6036):451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]
- Zipursky S. L., Marians K. J. Identification of two Escherichia coli factor Y effector sites near the origins of replication of the plasmids (ColE1 and pBR322. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6521–6525. doi: 10.1073/pnas.77.11.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zverev V. V., Khmel I. A. The nucleotide sequences of the replication origins of plasmids ColA and ColD. Plasmid. 1985 Nov;14(3):192–199. doi: 10.1016/0147-619x(85)90002-2. [DOI] [PubMed] [Google Scholar]
- Zverev V. V., Kuzmin N. P., Zuyeva L. A., Burova E. I., Alexandrov A. A., Khmel I. A. Regions of homology in small colicinogenic plasmids. Plasmid. 1984 Nov;12(3):203–205. doi: 10.1016/0147-619x(84)90045-3. [DOI] [PubMed] [Google Scholar]
- Zyskind J. W., Smith D. W. The bacterial origin of replication, oriC. Cell. 1986 Aug 15;46(4):489–490. doi: 10.1016/0092-8674(86)90873-1. [DOI] [PubMed] [Google Scholar]
- Zünd P., Lebek G. Generation time-prolonging R plasmids: correlation between increases in the generation time of Escherichia coli caused by R plasmids and their molecular size. Plasmid. 1980 Jan;3(1):65–69. doi: 10.1016/s0147-619x(80)90034-7. [DOI] [PubMed] [Google Scholar]
- de Graaff J., Crosa J. H., Heffron F., Falkow S. Replication of the nonconjugative plasmid RSF1010 in Escherichia coli K-12. J Bacteriol. 1978 Jun;134(3):1117–1122. doi: 10.1128/jb.134.3.1117-1122.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Putten A. J., de Lang R., Veltkamp E., Nijkamp H. J., Van Solingen P., van den Berg J. A. Methylation-dependent transcription controls plasmid replication of the CloDF13 cop-1(Ts) mutant. J Bacteriol. 1986 Nov;168(2):728–733. doi: 10.1128/jb.168.2.728-733.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven N. Evolution of bacterial surface exclusion against incompatible plasmids. J Theor Biol. 1985 Dec 7;117(3):431–452. doi: 10.1016/s0022-5193(85)80153-3. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]



