Abstract
Myxobacteria are soil bacteria whose unusually social behavior distinguishes them from other groups of procaryotes. Perhaps the most remarkable aspect of their social behavior occurs during development, when tens of thousands of cells aggregate and form a colorful fruiting body. Inside the fruiting body the vegetative cells convert into dormant, resistant myxospores. However, myxobacterial social behavior is not restricted to the developmental cycle, and three other social behaviors have been described. Vegetative cells have a multigene social motility system in which cell-cell contact is essential for gliding in multicellular swarms. Cell growth on protein is cooperative in that the growth rate increases with the cell density. Rippling is a periodic behavior in which the cells align themselves in ridges and move in waves. These social behaviors indicate that myxobacterial colonies are not merely collections of individual cells but are societies in which cell behavior is synchronized by cell-cell interactions. The molecular basis of these social behaviors is becoming clear through the use of a combination of behavioral, biochemical, and genetic experimental approaches.
Full text
PDF
























Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. P., Roth J. R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- Arnold J. W., Shimkets L. J. Cell surface properties correlated with cohesion in Myxococcus xanthus. J Bacteriol. 1988 Dec;170(12):5771–5777. doi: 10.1128/jb.170.12.5771-5777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J. W., Shimkets L. J. Inhibition of cell-cell interactions in Myxococcus xanthus by congo red. J Bacteriol. 1988 Dec;170(12):5765–5770. doi: 10.1128/jb.170.12.5765-5770.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., Kaiser D. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol Gen Genet. 1983;191(1):99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- Bacon K., Clutter D., Kottel R. H., Orlowski M., White D. Carbohydrate accumulation during myxospore formation in Myxococcus xanthus. J Bacteriol. 1975 Dec;124(3):1635–1636. doi: 10.1128/jb.124.3.1635-1636.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E. P., Randall L. L. The requirement for energy during export of beta-lactamase in Escherichia coli is fulfilled by the total protonmotive force. EMBO J. 1984 Apr;3(4):895–900. doi: 10.1002/j.1460-2075.1984.tb01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre J. M., Ruiz-Vazquez R. M., Murillo F. J. Light induction of gene expression in Myxococcus xanthus. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2359–2362. doi: 10.1073/pnas.84.8.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. "Frizzy" genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. Analysis of the products of the Myxococcus xanthus frz genes. J Bacteriol. 1986 May;166(2):673–678. doi: 10.1128/jb.166.2.673-678.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. Cloning and complementation analysis of the "Frizzy" genes of Myxococcus xanthus. Mol Gen Genet. 1985;198(2):243–254. doi: 10.1007/BF00383002. [DOI] [PubMed] [Google Scholar]
- Breton A. M., Jaoua S., Guespin-Michel J. Transfer of plasmid RP4 to Myxococcus xanthus and evidence for its integration into the chromosome. J Bacteriol. 1985 Feb;161(2):523–528. doi: 10.1128/jb.161.2.523-528.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard R. P., Burchard A. C., Parish J. H. Pigmentation phenotype instability in Myxococcus xanthus. Can J Microbiol. 1977 Dec;23(12):1657–1662. doi: 10.1139/m77-238. [DOI] [PubMed] [Google Scholar]
- Burchard R. P., Dworkin M. Light-induced lysis and carotenogenesis in Myxococcus xanthus. J Bacteriol. 1966 Feb;91(2):535–545. doi: 10.1128/jb.91.2.535-545.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard R. P. Gliding motility mutants of Myxococcus xanthus. J Bacteriol. 1970 Nov;104(2):940–947. doi: 10.1128/jb.104.2.940-947.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J. M., Zusman D. R. Regulation of development in Myxococcus xanthus: effect of 3':5'-cyclic AMP, ADP, and nutrition. Proc Natl Acad Sci U S A. 1975 Feb;72(2):518–522. doi: 10.1073/pnas.72.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Keseler I. M., Shimkets L. J. Genome size of Myxococcus xanthus determined by pulsed-field gel electrophoresis. J Bacteriol. 1990 Aug;172(8):4206–4213. doi: 10.1128/jb.172.8.4206-4213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Kaiser D. dsg, a gene required for Myxococcus development, is necessary for cell viability. J Bacteriol. 1989 Jul;171(7):3727–3731. doi: 10.1128/jb.171.7.3727-3731.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Kaiser D. dsg, a gene required for cell-cell interaction early in Myxococcus development. J Bacteriol. 1989 Jul;171(7):3719–3726. doi: 10.1128/jb.171.7.3719-3726.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward G., Bremer H. Determination of deoxyribonucleic acid replication time in exponentially growing Escherichia coli B/r. J Bacteriol. 1977 Jun;130(3):1206–1213. doi: 10.1128/jb.130.3.1206-1213.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Cheng K. J., Geesey G. G., Ladd T. I., Nickel J. C., Dasgupta M., Marrie T. J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Cumsky M. G., Zusman D. R. Binding properties of myxobacterial hemagglutinin. J Biol Chem. 1981 Dec 10;256(23):12596–12599. [PubMed] [Google Scholar]
- Cumsky M. G., Zusman D. R. Purification and characterization of myxobacterial hemagglutinin, a development-specific lectin of Myxococcus xanthus. J Biol Chem. 1981 Dec 10;256(23):12581–12588. [PubMed] [Google Scholar]
- Cumsky M., Zusman D. R. Myxobacterial hemagglutinin: a development-specific lectin of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5505–5509. doi: 10.1073/pnas.76.11.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DWORKIN M., GIBSON S. M. A SYSTEM FOR STUDYING MICROBIAL MORPHOGENESIS: RAPID FORMATION OF MICROCYSTS IN MYXOCOCCUS XANTHUS. Science. 1964 Oct 9;146(3641):243–244. doi: 10.1126/science.146.3641.243. [DOI] [PubMed] [Google Scholar]
- DWORKIN M. NUTRITIONAL REGU.ATION OF MORPHOGENESIS IN MYXOCOCCUS XANTHUS. J Bacteriol. 1963 Jul;86:67–72. doi: 10.1128/jb.86.1.67-72.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date T., Goodman J. M., Wickner W. T. Procoat, the precursor of M13 coat protein, requires an electrochemical potential for membrane insertion. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4669–4673. doi: 10.1073/pnas.77.8.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhundale A. R., Furuichi T., Inouye S., Inouye M. Distribution of multicopy single-stranded DNA among myxobacteria and related species. J Bacteriol. 1985 Nov;164(2):914–917. doi: 10.1128/jb.164.2.914-917.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhundale A., Furuichi T., Inouye M., Inouye S. Mutations that affect production of branched RNA-linked msDNA in Myxococcus xanthus. J Bacteriol. 1988 Dec;170(12):5620–5624. doi: 10.1128/jb.170.12.5620-5624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhundale A., Inouye M., Inouye S. A new species of multicopy single-stranded DNA from Myxococcus xanthus with conserved structural features. J Biol Chem. 1988 Jun 25;263(18):9055–9058. [PubMed] [Google Scholar]
- Dhundale A., Lampson B., Furuichi T., Inouye M., Inouye S. Structure of msDNA from Myxococcus xanthus: evidence for a long, self-annealing RNA precursor for the covalently linked, branched RNA. Cell. 1987 Dec 24;51(6):1105–1112. doi: 10.1016/0092-8674(87)90596-4. [DOI] [PubMed] [Google Scholar]
- Downard J. S. Identification of the RNA products of the ops gene of Myxococcus xanthus and mapping of ops and tps RNAs. J Bacteriol. 1987 Apr;169(4):1522–1528. doi: 10.1128/jb.169.4.1522-1528.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downard J. S., Kim S. H., Kil K. S. Localization of the cis-acting regulatory DNA sequences of the Myxococcus xanthus tps and ops genes. J Bacteriol. 1988 Oct;170(10):4931–4938. doi: 10.1128/jb.170.10.4931-4938.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downard J. S., Kupfer D., Zusman D. R. Gene expression during development of Myxococcus xanthus. Analysis of the genes for protein S. J Mol Biol. 1984 Jun 5;175(4):469–492. doi: 10.1016/0022-2836(84)90180-3. [DOI] [PubMed] [Google Scholar]
- Downard J. S., Zusman D. R. Differential expression of protein S genes during Myxococcus xanthus development. J Bacteriol. 1985 Mar;161(3):1146–1155. doi: 10.1128/jb.161.3.1146-1155.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin M., Eide D. Myxococcus xanthus does not respond chemotactically to moderate concentration gradients. J Bacteriol. 1983 Apr;154(1):437–442. doi: 10.1128/jb.154.1.437-442.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin M., Kaiser D. Cell interactions in myxobacterial growth and development. Science. 1985 Oct 4;230(4721):18–24. doi: 10.1126/science.3929384. [DOI] [PubMed] [Google Scholar]
- Dworkin M. Tactic behavior of Myxococcus xanthus. J Bacteriol. 1983 Apr;154(1):452–459. doi: 10.1128/jb.154.1.452-459.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easson D. D., Jr, Sinskey A. J., Peoples O. P. Isolation of Zoogloea ramigera I-16-M exopolysaccharide biosynthetic genes and evidence for instability within this region. J Bacteriol. 1987 Oct;169(10):4518–4524. doi: 10.1128/jb.169.10.4518-4524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enequist H. G., Hirst T. R., Harayama S., Hardy S. J., Randall L. L. Energy is required for maturation of exported proteins in Escherichia coli. Eur J Biochem. 1981 May 15;116(2):227–233. doi: 10.1111/j.1432-1033.1981.tb05323.x. [DOI] [PubMed] [Google Scholar]
- Filer D., Kindler S. H., Rosenberg E. Myxospore coat synthesis in Myxococcus xanthus: enzymes associated with uridine 5'-diphosphate-N-acetylgalactosamine formation during myxospore development. J Bacteriol. 1977 Sep;131(3):745–750. doi: 10.1128/jb.131.3.745-750.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filer D., White D., Kindler S. H., Rosenberg E. Myxospore coat synthesis in Myxococcus xanthus: in vivo incorporation of acetate and glycine. J Bacteriol. 1977 Sep;131(3):751–758. doi: 10.1128/jb.131.3.751-758.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J. M., Kalos M., Zissler J. F. Isolation of cell surface antigen mutants of Myxococcus xanthus by use of monoclonal antibodies. J Bacteriol. 1989 Apr;171(4):2033–2041. doi: 10.1128/jb.171.4.2033-2041.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J. M., Zissler J. F. Characterization of lipopolysaccharide from Myxococcus xanthus by use of monoclonal antibodies. J Bacteriol. 1989 Apr;171(4):2028–2032. doi: 10.1128/jb.171.4.2028-2032.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J. M., Zissler J. F. Defects in motility and development of Myxococcus xanthus lipopolysaccharide mutants. J Bacteriol. 1989 Apr;171(4):2042–2048. doi: 10.1128/jb.171.4.2042-2048.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi T., Dhundale A., Inouye M., Inouye S. Branched RNA covalently linked to the 5' end of a single-stranded DNA in Stigmatella aurantiaca: structure of msDNA. Cell. 1987 Jan 16;48(1):47–53. doi: 10.1016/0092-8674(87)90354-0. [DOI] [PubMed] [Google Scholar]
- Furuichi T., Inouye S., Inouye M. Biosynthesis and structure of stable branched RNA covalently linked to the 5' end of multicopy single-stranded DNA of Stigmatella aurantiaca. Cell. 1987 Jan 16;48(1):55–62. doi: 10.1016/0092-8674(87)90355-2. [DOI] [PubMed] [Google Scholar]
- GILLESPIE D. C., COOK F. D. EXTRACELLULAR ENZYMES FROM STRAINS OF SORANGIUM. Can J Microbiol. 1965 Feb;11:109–118. doi: 10.1139/m65-014. [DOI] [PubMed] [Google Scholar]
- Gelvan I., Varon M., Rosenberg E. Cell-density-dependent killing of Myxococcus xanthus by autocide AMV. J Bacteriol. 1987 Feb;169(2):844–848. doi: 10.1128/jb.169.2.844-848.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J. S., Dworkin M. Cell surface antigens during submerged development of Myxococcus xanthus examined with monoclonal antibodies. J Bacteriol. 1986 Nov;168(2):505–511. doi: 10.1128/jb.168.2.505-511.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J. S., Dworkin M. Isolation of additional monoclonal antibodies directed against cell surface antigens of Myxococcus xanthus cells undergoing submerged development. J Bacteriol. 1988 Dec;170(12):5953–5955. doi: 10.1128/jb.170.12.5953-5955.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J. S., Jarvis B. W., Dworkin M. Inhibition of development in Myxococcus xanthus by monoclonal antibody 1604. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4505–4508. doi: 10.1073/pnas.84.13.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R. E., Bornemann M. C. Identification and characterization of the Myxococcus xanthus bsgA gene product. J Bacteriol. 1988 Nov;170(11):5289–5297. doi: 10.1128/jb.170.11.5289-5297.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R. E., Cull M. G. Control of developmental gene expression by cell-to-cell interactions in Myxococcus xanthus. J Bacteriol. 1986 Oct;168(1):341–347. doi: 10.1128/jb.168.1.341-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R. E., Cull M. G., Fly S. Genetic identification and cloning of a gene required for developmental cell interactions in Myxococcus xanthus. J Bacteriol. 1988 Nov;170(11):5279–5288. doi: 10.1128/jb.170.11.5279-5288.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore D. F., White D. Energy-dependent cell cohesion in myxobacteria. J Bacteriol. 1985 Jan;161(1):113–117. doi: 10.1128/jb.161.1.113-117.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnosspelius G. Myxobacterial slime and proteolytic activity. Arch Microbiol. 1978 Jan 23;116(1):51–59. doi: 10.1007/BF00408733. [DOI] [PubMed] [Google Scholar]
- Gnosspelius G. Purification and properties of an extracellular protease from Myxococcus virescens. J Bacteriol. 1978 Jan;133(1):17–25. doi: 10.1128/jb.133.1.17-25.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
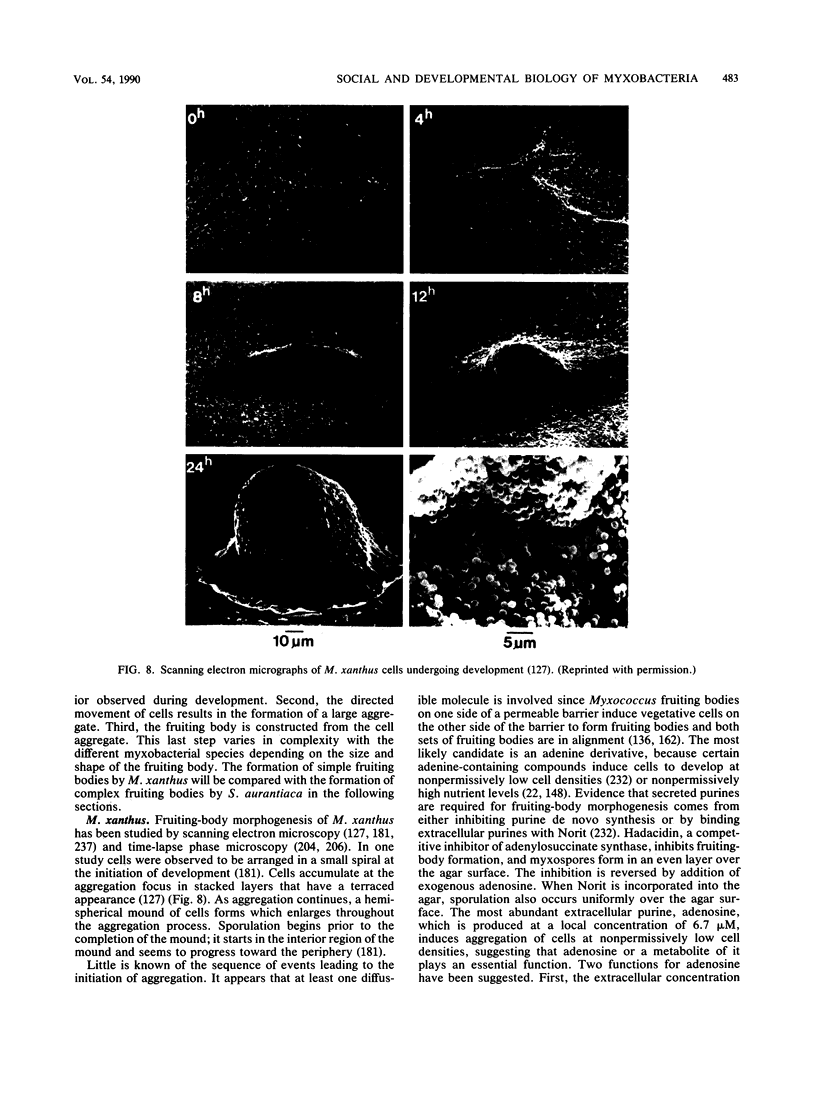
- Grilione P. L., Pangborn J. Scanning electron microscopy of fruiting body formation by myxobacteria. J Bacteriol. 1975 Dec;124(3):1558–1565. doi: 10.1128/jb.124.3.1558-1565.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Hagen T. J., Shimkets L. J. Nucleotide sequence and transcriptional products of the csg locus of Myxococcus xanthus. J Bacteriol. 1990 Jan;172(1):15–23. doi: 10.1128/jb.172.1.15-23.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler C. H., Brown R. M., Jr, Benziman M. Calcofluor white ST Alters the in vivo assembly of cellulose microfibrils. Science. 1980 Nov 21;210(4472):903–906. doi: 10.1126/science.7434003. [DOI] [PubMed] [Google Scholar]
- Hall J. C., Rosbash M. Mutations and molecules influencing biological rhythms. Annu Rev Neurosci. 1988;11:373–393. doi: 10.1146/annurev.ne.11.030188.002105. [DOI] [PubMed] [Google Scholar]
- Hamada H., Kakunaga T. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982 Jul 22;298(5872):396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- Herth W. Calcofluor white and Congo red inhibit chitin microfibril assembly of Poterioochromonas: evidence for a gap between polymerization and microfibril formation. J Cell Biol. 1980 Nov;87(2 Pt 1):442–450. doi: 10.1083/jcb.87.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M. Y., Inouye S., Inouye M. Structural requirements of the RNA precursor for the biosynthesis of the branched RNA-linked multicopy single-stranded DNA of Myxococcus xanthus. J Biol Chem. 1989 Apr 15;264(11):6214–6219. [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Gene expression during development of Myxococcus xanthus: pattern of protein synthesis. Dev Biol. 1979 Feb;68(2):579–591. doi: 10.1016/0012-1606(79)90228-8. [DOI] [PubMed] [Google Scholar]
- Inouye S., Franceschini T., Inouye M. Structural similarities between the development-specific protein S from a gram-negative bacterium, Myxococcus xanthus, and calmodulin. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6829–6833. doi: 10.1073/pnas.80.22.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Harada W., Zusman D., Inouye M. Development-specific protein S of Myxococcus xanthus: purification and characterization. J Bacteriol. 1981 Nov;148(2):678–683. doi: 10.1128/jb.148.2.678-683.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Herzer P. J., Inouye M. Two independent retrons with highly diverse reverse transcriptases in Myxococcus xanthus. Proc Natl Acad Sci U S A. 1990 Feb;87(3):942–945. doi: 10.1073/pnas.87.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Hsu M. Y., Eagle S., Inouye M. Reverse transcriptase associated with the biosynthesis of the branched RNA-linked msDNA in Myxococcus xanthus. Cell. 1989 Feb 24;56(4):709–717. doi: 10.1016/0092-8674(89)90593-x. [DOI] [PubMed] [Google Scholar]
- Inouye S., Ike Y., Inouye M. Tandem repeat of the genes for protein S, a development-specific protein of Myxococcus xanthus. J Biol Chem. 1983 Jan 10;258(1):38–40. [PubMed] [Google Scholar]
- Inouye S., Inouye M., McKeever B., Sarma R. Preliminary crystallographic data for protein S, a development-specific protein of Myxococcus xanthus. J Biol Chem. 1980 Apr 25;255(8):3713–3714. [PubMed] [Google Scholar]
- Inouye S., White D., Inouye M. Development of Stigmatella aurantiaca: effects of light and gene expression. J Bacteriol. 1980 Mar;141(3):1360–1365. doi: 10.1128/jb.141.3.1360-1365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irschik H., Jansen R., Gerth K., Höfle G., Reichenbach H. The sorangicins, novel and powerful inhibitors of eubacterial RNA polymerase isolated from myxobacteria. J Antibiot (Tokyo) 1987 Jan;40(1):7–13. doi: 10.7164/antibiotics.40.7. [DOI] [PubMed] [Google Scholar]
- Janssen G. R., Dworkin M. Cell-cell interactions in developmental lysis of Myxococcus xanthus. Dev Biol. 1985 Nov;112(1):194–202. doi: 10.1016/0012-1606(85)90133-2. [DOI] [PubMed] [Google Scholar]
- Jarvis B. W., Dworkin M. Purification and properties of Myxococcus xanthus cell surface antigen 1604. J Bacteriol. 1989 Sep;171(9):4655–4666. doi: 10.1128/jb.171.9.4655-4666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis B. W., Dworkin M. Role of Myxococcus xanthus cell surface antigen 1604 in development. J Bacteriol. 1989 Sep;171(9):4667–4673. doi: 10.1128/jb.171.9.4667-4673.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Ordal E. J. Deoxyribonucleic acid homology in bacterial taxonomy: effect of incubation temperature on reaction specificity. J Bacteriol. 1968 Mar;95(3):893–900. doi: 10.1128/jb.95.3.893-900.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. Y., White D. Myxospore formation in Myxococcus xanthus: chemical changes in the cell wall during cellular morphogenesis. J Bacteriol. 1972 Nov;112(2):849–855. doi: 10.1128/jb.112.2.849-855.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. Control of multicellular development: Dictyostelium and Myxococcus. Annu Rev Genet. 1986;20:539–566. doi: 10.1146/annurev.ge.20.120186.002543. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil K. S., Brown G. L., Downard J. S. A segment of Myxococcus xanthus ops DNA functions as an upstream activation site for tps gene transcription. J Bacteriol. 1990 Jun;172(6):3081–3088. doi: 10.1128/jb.172.6.3081-3088.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen K. P., Nelson D. R. Acceleration of starvation- and glycerol-induced myxospore formation by prior heat shock in Myxococcus xanthus. J Bacteriol. 1988 Nov;170(11):5200–5207. doi: 10.1128/jb.170.11.5200-5207.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
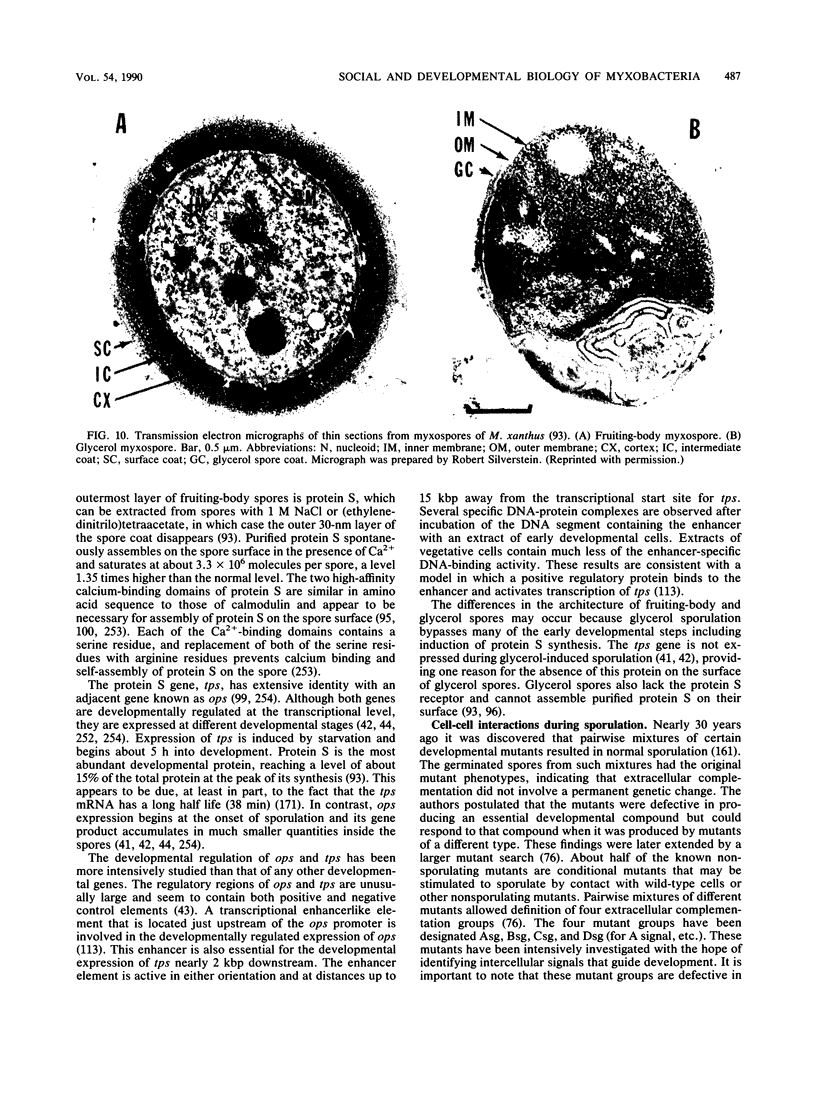
- Kim S. K., Kaiser D. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990 Apr 6;61(1):19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- Kim S. K., Kaiser D. Cell alignment required in differentiation of Myxococcus xanthus. Science. 1990 Aug 24;249(4971):926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- Kim S. K., Kaiser D. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 1990 Jun;4(6):896–904. doi: 10.1101/gad.4.6.896. [DOI] [PubMed] [Google Scholar]
- Kim S. K., Kaiser D. Purification and properties of Myxococcus xanthus C-factor, an intercellular signaling protein. Proc Natl Acad Sci U S A. 1990 May;87(10):3635–3639. doi: 10.1073/pnas.87.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A., Schnorr M. Genome complexity of methanogenic bacteria. J Bacteriol. 1984 May;158(2):628–631. doi: 10.1128/jb.158.2.628-631.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottel R. H., Bacon K., Clutter D., White D. Coats from Myxococcus xanthus: characterization and synthesis during myxospore differentiation. J Bacteriol. 1975 Oct;124(1):550–557. doi: 10.1128/jb.124.1.550-557.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottel R., White D. Autolytic activity associated with myxospore formation in Myxococcus xanthus. Arch Mikrobiol. 1974 Mar 1;95(1):91–95. [PubMed] [Google Scholar]
- Kroos L., Hartzell P., Stephens K., Kaiser D. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev. 1988 Dec;2(12A):1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987 Oct;1(8):840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kuspa A., Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- Kubitschek H. E., Newman C. N. Chromosome replication during the division cycle in slowly growing, steady-state cultures of three Escherichia coli B/r strains. J Bacteriol. 1978 Oct;136(1):179–190. doi: 10.1128/jb.136.1.179-190.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner J. M., Kaiser D. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J Bacteriol. 1982 Jul;151(1):458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze B., Bedorf N., Kohl W., Höfle G., Reichenbach H. Myxochelin A, a new iron-chelating compound from Angiococcus disciformis (Myxobacterales). Production, isolation, physico-chemical and biological properties. J Antibiot (Tokyo) 1989 Jan;42(1):14–17. doi: 10.7164/antibiotics.42.14. [DOI] [PubMed] [Google Scholar]
- Kuspa A., Kaiser D. Genes required for developmental signalling in Myxococcus xanthus: three asg loci. J Bacteriol. 1989 May;171(5):2762–2772. doi: 10.1128/jb.171.5.2762-2772.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A., Kroos L., Kaiser D. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- LaRossa R., Kuner J., Hagen D., Manoil C., Kaiser D. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J Bacteriol. 1983 Mar;153(3):1394–1404. doi: 10.1128/jb.153.3.1394-1404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampky J. R. Ultrastructure of Polyangium cellulosum. J Bacteriol. 1976 Jun;126(3):1278–1284. doi: 10.1128/jb.126.3.1278-1284.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson B. C., Inouye M., Inouye S. Reverse transcriptase with concomitant ribonuclease H activity in the cell-free synthesis of branched RNA-linked msDNA of Myxococcus xanthus. Cell. 1989 Feb 24;56(4):701–707. doi: 10.1016/0092-8674(89)90592-8. [DOI] [PubMed] [Google Scholar]
- Lampson B. C., Sun J., Hsu M. Y., Vallejo-Ramirez J., Inouye S., Inouye M. Reverse transcriptase in a clinical strain of Escherichia coli: production of branched RNA-linked msDNA. Science. 1989 Feb 24;243(4894 Pt 1):1033–1038. doi: 10.1126/science.2466332. [DOI] [PubMed] [Google Scholar]
- Levine H. B., Ringel S. M., Cobb J. M. Therapeutic properties of oral ambruticin (W7783) in experimental pulmonary coccidioidomycosis of mice. Chest. 1978 Feb;73(2):202–206. doi: 10.1378/chest.73.2.202. [DOI] [PubMed] [Google Scholar]
- Lim D., Maas W. K. Reverse transcriptase-dependent synthesis of a covalently linked, branched DNA-RNA compound in E. coli B. Cell. 1989 Mar 10;56(5):891–904. doi: 10.1016/0092-8674(89)90693-4. [DOI] [PubMed] [Google Scholar]
- MCVITTIE A., MESSIK F., ZAHLER S. A. Developmental biology of Myxococcus. J Bacteriol. 1962 Sep;84:546–551. doi: 10.1128/jb.84.3.546-551.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae T. H., Dobson W. J., McCurdy H. D. Fimbriation in gliding bacteria. Can J Microbiol. 1977 Aug;23(8):1096–1108. doi: 10.1139/m77-165. [DOI] [PubMed] [Google Scholar]
- Maeba P. Y. Iodination of Myxococcus xanthus during development. J Bacteriol. 1983 Sep;155(3):1033–1041. doi: 10.1128/jb.155.3.1033-1041.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeba P. Y. Isolation of a surface glycoprotein from Myxococcus xanthus. J Bacteriol. 1986 May;166(2):644–650. doi: 10.1128/jb.166.2.644-650.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Leadbetter E. R. Deoxyribonucleic acid base composition of myxobacteria. J Bacteriol. 1965 Dec;90(6):1795–1796. doi: 10.1128/jb.90.6.1795-1796.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Kaiser D. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J Bacteriol. 1980 Jan;141(1):297–304. doi: 10.1128/jb.141.1.297-304.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Kaiser D. Guanosine pentaphosphate and guanosine tetraphosphate accumulation and induction of Myxococcus xanthus fruiting body development. J Bacteriol. 1980 Jan;141(1):305–315. doi: 10.1128/jb.141.1.305-315.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Kaiser D. Purine-containing compounds, including cyclic adenosine 3',5'-monophosphate, induce fruiting of Myxococcus xanthus by nutritional imbalance. J Bacteriol. 1980 Jan;141(1):374–377. doi: 10.1128/jb.141.1.374-377.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Laborda A., Murillo F. J. Genic and allelic interactions in the carotenogenic response of myxococcus xanthus to blue light. Genetics. 1989 Jul;122(3):481–490. doi: 10.1093/genetics/122.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson P. J., Guespin-Michel J. F. An extracellular blood-anticoagulant glycopeptide produced exclusively during vegetative growth by Myxococcus xanthus and other myxobacteria is not co-regulated with other extracellular macromolecules. J Gen Microbiol. 1988 Mar;134(3):801–806. doi: 10.1099/00221287-134-3-801. [DOI] [PubMed] [Google Scholar]
- Mayo K. A., Kaiser D. asgB, a gene required early for developmental signalling, aggregation, and sporulation of Myxococcus xanthus. Mol Gen Genet. 1989 Sep;218(3):409–418. doi: 10.1007/BF00332403. [DOI] [PubMed] [Google Scholar]
- McBride M. J., Weinberg R. A., Zusman D. R. "Frizzy" aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci U S A. 1989 Jan;86(2):424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride M. J., Zusman D. R. Trehalose accumulation in vegetative cells and spores of Myxococcus xanthus. J Bacteriol. 1989 Nov;171(11):6383–6386. doi: 10.1128/jb.171.11.6383-6386.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., McBride M. J., Zusman D. R. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990 Sep;172(9):4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., Zusman D. R. FrzE of Myxococcus xanthus is homologous to both CheA and CheY of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5898–5902. doi: 10.1073/pnas.87.15.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurdy H. D. Studies on the taxonomy of the Myxobacterales. I. Record of Canadian isolates and survey of methods. Can J Microbiol. 1969 Dec;15(12):1453–1461. doi: 10.1139/m69-259. [DOI] [PubMed] [Google Scholar]
- McCurdy H. D., Wolf S. Deoxyribonucleic acid base compositions of fruiting Myxobacterales. Can J Microbiol. 1967 Dec;13(12):1707–1708. doi: 10.1139/m67-222. [DOI] [PubMed] [Google Scholar]
- Mitchell R. M., Loeblich L. A., Klotz L. C., Loeblich A. R., 3rd DNA organization of Methanobacterium thermoautotrophicum. Science. 1979 Jun 8;204(4397):1082–1084. doi: 10.1126/science.377486. [DOI] [PubMed] [Google Scholar]
- Moore R. L., McCarthy B. J. Base sequence homology and renaturation studies of the deoxyribonucleic acid of extremely halophilic bacteria. J Bacteriol. 1969 Jul;99(1):255–262. doi: 10.1128/jb.99.1.255-262.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J., Kushner S. R., Ivarie R. The simple repeat poly(dT-dG).poly(dC-dA) common to eukaryotes is absent from eubacteria and archaebacteria and rare in protozoans. Mol Biol Evol. 1986 Jul;3(4):343–355. doi: 10.1093/oxfordjournals.molbev.a040399. [DOI] [PubMed] [Google Scholar]
- Morrison C. E., Zusman D. R. Myxococcus xanthus mutants with temperature-sensitive, stage-specific defects: evidence for independent pathways in development. J Bacteriol. 1979 Dec;140(3):1036–1042. doi: 10.1128/jb.140.3.1036-1042.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R., Cumsky M. G., Zusman D. R. Localization of myxobacterial hemagglutinin in the periplasmic space and on the cell surface of Myxococcus xanthus during developmental aggregation. J Biol Chem. 1981 Dec 10;256(23):12589–12595. [PubMed] [Google Scholar]
- Nelson D. R., Killeen K. P. Heat shock proteins of vegetative and fruiting Myxococcus xanthus cells. J Bacteriol. 1986 Dec;168(3):1100–1106. doi: 10.1128/jb.168.3.1100-1106.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R., Zusman D. R. Evidence for long-lived mRNA during fruiting body formation in myxococcus xanthus. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1467–1471. doi: 10.1073/pnas.80.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R., Zusman D. R. Transport and localization of protein S, a spore coat protein, during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1983 May;154(2):547–553. doi: 10.1128/jb.154.2.547-553.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Coliphage P1-mediated transduction of cloned DNA from Escherichia coli to Myxococcus xanthus: use for complementation and recombinational analyses. J Bacteriol. 1983 Jul;155(1):317–329. doi: 10.1128/jb.155.1.317-329.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Genetic analysis of tag mutants of Myxococcus xanthus provides evidence for two developmental aggregation systems. J Bacteriol. 1990 Jul;172(7):3868–3878. doi: 10.1128/jb.172.7.3868-3878.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Patterns of cellular interactions during fruiting-body formation in Myxococcus xanthus. J Bacteriol. 1989 Nov;171(11):6013–6024. doi: 10.1128/jb.171.11.6013-6024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Reexamination of the role of autolysis in the development of Myxococcus xanthus. J Bacteriol. 1988 Sep;170(9):4103–4112. doi: 10.1128/jb.170.9.4103-4112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Wilson A. C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26(1-2):74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- Orlowski M., Martin P., White D., Wong M. C. Changes in activity of glyoxylate cycle enzymes during myxospore development in Myxococcus xanthus. J Bacteriol. 1972 Sep;111(3):784–790. doi: 10.1128/jb.111.3.784-790.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orndorff P. E., Dworkin M. Separation and properties of the cytoplasmic and outer membranes of vegetative cells of Myxococcus xanthus. J Bacteriol. 1980 Feb;141(2):914–927. doi: 10.1128/jb.141.2.914-927.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orndorff P. E., Dworkin M. Synthesis of several membrane proteins during developmental aggregation in Myxococcus xanthus. J Bacteriol. 1982 Jan;149(1):29–39. doi: 10.1128/jb.149.1.29-39.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford A. E. Observations Concerning the Growth and Metabolic Activities of Myxococci in a Simple Protein-free Liquid Medium. J Bacteriol. 1947 Feb;53(2):129–138. doi: 10.1128/jb.53.2.129-138.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko S. M., Jann B., Jann K. Novel change in the carbohydrate portion of Myxococcus xanthus lipopolysaccharide during development. J Bacteriol. 1989 Apr;171(4):1835–1840. doi: 10.1128/jb.171.4.1835-1840.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko S. M. Methylation of macromolecules during development in Myxococcus xanthus. J Bacteriol. 1985 Nov;164(2):495–500. doi: 10.1128/jb.164.2.495-500.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh C. S. ON TEMPERATURE INDEPENDENCE IN THE CLOCK SYSTEM CONTROLLING EMERGENCE TIME IN DROSOPHILA. Proc Natl Acad Sci U S A. 1954 Oct;40(10):1018–1029. doi: 10.1073/pnas.40.10.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postgate J. R., Kent H. M., Robson R. L., Chesshyre J. A. The genomes of Desulfovibrio gigas and D. vulgaris. J Gen Microbiol. 1984 Jul;130(7):1597–1601. doi: 10.1099/00221287-130-7-1597. [DOI] [PubMed] [Google Scholar]
- Qualls G. T., Stephens K., White D. Light-stimulated morphogenesis in the fruiting myxobacterium Stigmatella aurantiaca. Science. 1978 Aug 4;201(4354):444–445. doi: 10.1126/science.96528. [DOI] [PubMed] [Google Scholar]
- Qualls G. T., Stephens K., White D. Morphogenetic movements and multicellular development in the fruiting Myxobacterium, Stigmatella aurantiaca. Dev Biol. 1978 Sep;66(1):270–274. doi: 10.1016/0012-1606(78)90291-9. [DOI] [PubMed] [Google Scholar]
- Razin S., Barile M. F., Harasawa R., Amikam D., Glaser G. Characterization of the mycoplasma genome. Yale J Biol Med. 1983 Sep-Dec;56(5-6):357–366. [PMC free article] [PubMed] [Google Scholar]
- Reichenbach H., Dworkin M. Induction of myxospore formation in Stigmatella aurantiaca (Myxobacterales) by monovalent cations. J Bacteriol. 1970 Jan;101(1):325–326. doi: 10.1128/jb.101.1.325-326.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach H. The myxobacteria: common organisms with uncommon behaviour. Microbiol Sci. 1986 Sep;3(9):268–274. [PubMed] [Google Scholar]
- Rhie H. G., Shimkets L. J. Developmental bypass suppression of Myxococcus xanthus csgA mutations. J Bacteriol. 1989 Jun;171(6):3268–3276. doi: 10.1128/jb.171.6.3268-3276.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M., Solomon L., Zipkas D. Relationship between gene function and gene location in Escherichia coli. J Mol Evol. 1978 May 12;11(1):47–56. doi: 10.1007/BF01768024. [DOI] [PubMed] [Google Scholar]
- Ringel S. M., Greenough R. C., Roemer S., Connor D., Gutt A. L., Blair B., Kanter G., von Strandtmann Ambruticin (W7783), a new antifungal antibiotic. J Antibiot (Tokyo) 1977 May;30(5):371–375. doi: 10.7164/antibiotics.30.371. [DOI] [PubMed] [Google Scholar]
- Romeo J. M., Esmon B., Zusman D. R. Nucleotide sequence of the myxobacterial hemagglutinin gene contains four homologous domains. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6332–6336. doi: 10.1073/pnas.83.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo J. M., Zusman D. R. Cloning of the gene for myxobacterial hemagglutinin and isolation and analysis of structural gene mutations. J Bacteriol. 1987 Aug;169(8):3801–3808. doi: 10.1128/jb.169.8.3801-3808.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Keller K. H., Dworkin M. Cell density-dependent growth of Myxococcus xanthus on casein. J Bacteriol. 1977 Feb;129(2):770–777. doi: 10.1128/jb.129.2.770-777.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh A., Nir R., Sahar E., Rosenberg E. Cell-density-dependent lysis and sporulation of Myxococcus xanthus in agarose microbeads. J Bacteriol. 1989 Sep;171(9):4923–4929. doi: 10.1128/jb.171.9.4923-4929.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh A., Rosenberg E. Autocide AMI rescues development in dsg mutants of Myxococcus xanthus. J Bacteriol. 1989 Mar;171(3):1513–1518. doi: 10.1128/jb.171.3.1513-1518.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh A., Rosenberg E. Sporulation of Myxococcus xanthus in liquid shake flask cultures. J Bacteriol. 1989 Aug;171(8):4521–4524. doi: 10.1128/jb.171.8.4521-4524.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier P., Hanquier J., Jaoua S., Reichenbach H., Guespin-Michel J. F. Utilization of IncP-1 plasmids as vectors for transposon mutagenesis in myxobacteria. J Gen Microbiol. 1988 Nov;134(11):2889–2895. doi: 10.1099/00221287-134-11-2889. [DOI] [PubMed] [Google Scholar]
- Scott M. P., O'Farrell P. H. Spatial programming of gene expression in early Drosophila embryogenesis. Annu Rev Cell Biol. 1986;2:49–80. doi: 10.1146/annurev.cb.02.110186.000405. [DOI] [PubMed] [Google Scholar]
- Shilo M. Lysis of blue-green algae by myxobacter. J Bacteriol. 1970 Oct;104(1):453–461. doi: 10.1128/jb.104.1.453-461.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J., Asher S. J. Use of recombination techniques to examine the structure of the csg locus of Myxococcus xanthus. Mol Gen Genet. 1988 Jan;211(1):63–71. doi: 10.1007/BF00338394. [DOI] [PubMed] [Google Scholar]
- Shimkets L. J. Control of morphogenesis in myxobacteria. Crit Rev Microbiol. 1987;14(3):195–227. doi: 10.3109/10408418709104439. [DOI] [PubMed] [Google Scholar]
- Shimkets L. J. Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus. J Bacteriol. 1986 Jun;166(3):837–841. doi: 10.1128/jb.166.3.837-841.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J., Dworkin M. Excreted adenosine is a cell density signal for the initiation of fruiting body formation in Myxococcus xanthus. Dev Biol. 1981 May;84(1):51–60. doi: 10.1016/0012-1606(81)90369-9. [DOI] [PubMed] [Google Scholar]
- Shimkets L. J., Gill R. E., Kaiser D. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J., Kaiser D. Induction of coordinated movement of Myxococcus xanthus cells. J Bacteriol. 1982 Oct;152(1):451–461. doi: 10.1128/jb.152.1.451-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J., Kaiser D. Murein components rescue developmental sporulation of Myxococcus xanthus. J Bacteriol. 1982 Oct;152(1):462–470. doi: 10.1128/jb.152.1.462-470.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J., Rafiee H. CsgA, an extracellular protein essential for Myxococcus xanthus development. J Bacteriol. 1990 Sep;172(9):5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J. Role of cell cohesion in Myxococcus xanthus fruiting body formation. J Bacteriol. 1986 Jun;166(3):842–848. doi: 10.1128/jb.166.3.842-848.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J. The role of the cell surface in social and adventurous behaviour of myxobacteria. Mol Microbiol. 1989 Sep;3(9):1295–1299. doi: 10.1111/j.1365-2958.1989.tb00280.x. [DOI] [PubMed] [Google Scholar]
- Shimkets L., Seale T. W. Fruiting-body formation and myxospore differentiation and germination in Mxyococcus xanthus viewed by scanning electron microscopy. J Bacteriol. 1975 Feb;121(2):711–720. doi: 10.1128/jb.121.2.711-720.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit G., Kijne J. W., Lugtenberg B. J. Involvement of both cellulose fibrils and a Ca2+-dependent adhesin in the attachment of Rhizobium leguminosarum to pea root hair tips. J Bacteriol. 1987 Sep;169(9):4294–4301. doi: 10.1128/jb.169.9.4294-4301.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y. A Note on Elasticotaxis in Myxobacteria. J Bacteriol. 1942 Oct;44(4):405–412. doi: 10.1128/jb.44.4.405-412.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens K., Hartzell P., Kaiser D. Gliding motility in Myxococcus xanthus: mgl locus, RNA, and predicted protein products. J Bacteriol. 1989 Feb;171(2):819–830. doi: 10.1128/jb.171.2.819-830.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens K., Hegeman G. D., White D. Pheromone produced by the myxobacterium Stigmatella aurantiaca. J Bacteriol. 1982 Feb;149(2):739–747. doi: 10.1128/jb.149.2.739-747.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens K., White D. Morphogenetic effects of light and guanine derivatives on the fruiting myxobacterium Stigmatella aurantiaca. J Bacteriol. 1980 Oct;144(1):322–326. doi: 10.1128/jb.144.1.322-326.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo S. Z., Dworkin M. Resistance of vegetative cells and microcysts of Myxococcus xanthus. J Bacteriol. 1969 Jun;98(3):883–887. doi: 10.1128/jb.98.3.883-887.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo S., Dworkin M. Bacteriolytic enzymes produced by Myxococcus xanthus. J Bacteriol. 1972 Apr;110(1):236–245. doi: 10.1128/jb.110.1.236-245.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977 Mar;56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sutherland I. W., Thomson S. Comparison of polysaccharides produced by Myxococcus strains. J Gen Microbiol. 1975 Jul;89(1):124–132. doi: 10.1099/00221287-89-1-124. [DOI] [PubMed] [Google Scholar]
- Teintze M., Inouye M., Inouye S. Characterization of calcium-binding sites in development-specific protein S of Myxococcus xanthus using site-specific mutagenesis. J Biol Chem. 1988 Jan 25;263(3):1199–1203. [PubMed] [Google Scholar]
- Teintze M., Thomas R., Furuichi T., Inouye M., Inouye S. Two homologous genes coding for spore-specific proteins are expressed at different times during development of Myxococcus xanthus. J Bacteriol. 1985 Jul;163(1):121–125. doi: 10.1128/jb.163.1.121-125.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomchik K. J., Devreotes P. N. Adenosine 3',5'-monophosphate waves in Dictyostelium discoideum: a demonstration by isotope dilution--fluorography. Science. 1981 Apr 24;212(4493):443–446. doi: 10.1126/science.6259734. [DOI] [PubMed] [Google Scholar]
- Torti S., Zusman D. R. Genetic characterization of aggregation-defective developmental mutants of Myxococcus xanthus. J Bacteriol. 1981 Sep;147(3):768–775. doi: 10.1128/jb.147.3.768-775.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon M., Tietz A., Rosenberg E. Myxococcus xanthus autocide AMI. J Bacteriol. 1986 Jul;167(1):356–361. doi: 10.1128/jb.167.1.356-361.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon M., Tietz A., Rosenberg E. Myxococcus xanthus autocide AMI. J Bacteriol. 1986 Jul;167(1):356–361. doi: 10.1128/jb.167.1.356-361.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez G. M., Qualls F., White D. Morphogenesis of Stigmatella aurantiaca fruiting bodies. J Bacteriol. 1985 Aug;163(2):515–521. doi: 10.1128/jb.163.2.515-521.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M., Inouye M., Inouye S. Myxococcus xanthus msDNA.Mx162 exists as a complex with proteins. J Biol Chem. 1989 Aug 15;264(23):13665–13671. [PubMed] [Google Scholar]
- Voelz H., Reichenbach H. Fine structure of fruiting bodies of Stigmatella aurantiaca (Myxobacterales). J Bacteriol. 1969 Sep;99(3):856–866. doi: 10.1128/jb.99.3.856-866.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOLFF S. Delay of chromosome rejoining in Vicia faba induced by irradiation. Nature. 1954 Mar 13;173(4402):501–502. doi: 10.1038/173501b0. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C., Morowitz H. J. Genome size and evolution. Chromosoma. 1973;40(2):121–126. doi: 10.1007/BF00321457. [DOI] [PubMed] [Google Scholar]
- Weinberg R. A., Zusman D. R. Alkaline, acid, and neutral phosphatase activities are induced during development in Myxococcus xanthus. J Bacteriol. 1990 May;172(5):2294–2302. doi: 10.1128/jb.172.5.2294-2302.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. A., Zusman D. R. Evidence that the Myxococcus xanthus frz genes are developmentally regulated. J Bacteriol. 1989 Nov;171(11):6174–6186. doi: 10.1128/jb.171.11.6174-6186.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D., Dworkin M., Tipper D. J. Peptidoglycan of Myxococcus xanthus: structure and relation to morphogenesis. J Bacteriol. 1968 Jun;95(6):2186–2197. doi: 10.1128/jb.95.6.2186-2197.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D., Shropshire W., Jr, Stephens K. Photocontrol of development by Stigmatella aurantiaca. J Bacteriol. 1980 Jun;142(3):1023–1024. doi: 10.1128/jb.142.3.1023-1024.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wireman J. W., Dworkin M. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1977 Feb;129(2):798–802. doi: 10.1128/jb.129.2.798-802.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wireman J. W., Dworkin M. Morphogenesis and developmental interactions in myxobacteria. Science. 1975 Aug 15;189(4202):516–523. doi: 10.1126/science.806967. [DOI] [PubMed] [Google Scholar]
- Wireman J. Developmental induction of Myxococcus xanthus myxospores. J Bacteriol. 1979 Oct;140(1):147–153. doi: 10.1128/jb.140.1.147-153.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womack B. J., Gilmore D. F., White D. Calcium requirement for gliding motility in myxobacteria. J Bacteriol. 1989 Nov;171(11):6093–6096. doi: 10.1128/jb.171.11.6093-6096.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee T., Furuichi T., Inouye S., Inouye M. Multicopy single-stranded DNA isolated from a gram-negative bacterium, Myxococcus xanthus. Cell. 1984 Aug;38(1):203–209. doi: 10.1016/0092-8674(84)90541-5. [DOI] [PubMed] [Google Scholar]
- Yee T., Inouye M. Reexamination of the genome size of myxobacteria, including the use of a new method for genome size analysis. J Bacteriol. 1981 Mar;145(3):1257–1265. doi: 10.1128/jb.145.3.1257-1265.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee T., Inouye M. Two-dimensional DNA electrophoresis applied to the study of DNA methylation and the analysis of genome size in Myxococcus xanthus. J Mol Biol. 1982 Jan 15;154(2):181–196. doi: 10.1016/0022-2836(82)90059-6. [DOI] [PubMed] [Google Scholar]
- Zusman D. R. "Frizzy" mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J Bacteriol. 1982 Jun;150(3):1430–1437. doi: 10.1128/jb.150.3.1430-1437.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman D. R., Krotoski D. M., Cumsky M. Chromosome replication in Myxococcus xanthus. J Bacteriol. 1978 Jan;133(1):122–129. doi: 10.1128/jb.133.1.122-129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman D., Rosenberg E. DNA cycle of Myxococcus xanthus. J Mol Biol. 1970 May 14;49(3):609–619. doi: 10.1016/0022-2836(70)90285-8. [DOI] [PubMed] [Google Scholar]
- Zusman D., Rosenberg E. Deoxyribonucleic acid synthesis during microcyst germination in Myxococcus xanthus. J Bacteriol. 1968 Oct;96(4):981–986. doi: 10.1128/jb.96.4.981-986.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]