Arabidopsis plants exposed to triple stress are characterized by transcript responses not predictable from single stress treatments that significantly alter the expression of genes involved in signaling and defense processes.
Abstract
Considering global climate change, the incidence of combined drought and heat stress is likely to increase in the future and will considerably influence plant-pathogen interactions. Until now, little has been known about plants exposed to simultaneously occurring abiotic and biotic stresses. To shed some light on molecular plant responses to multiple stress factors, a versatile multifactorial test system, allowing simultaneous application of heat, drought, and virus stress, was developed in Arabidopsis (Arabidopsis thaliana). Comparative analysis of single, double, and triple stress responses by transcriptome and metabolome analysis revealed that gene expression under multifactorial stress is not predictable from single stress treatments. Hierarchical cluster and principal component analyses identified heat as the major stress factor, clearly separating heat-stressed from non-heat-stressed plants. We identified 11 genes differentially regulated in all stress combinations as well as 23 genes specifically regulated under triple stress. Furthermore, we showed that virus-treated plants displayed enhanced expression of defense genes, which was abolished in plants additionally subjected to heat and drought stress. Triple stress also reduced the expression of genes involved in the R-mediated disease response and increased the cytoplasmic protein response, which was not seen under single stress conditions. These observations suggested that abiotic stress factors significantly altered turnip mosaic virus-specific signaling networks, which led to a deactivation of defense responses and a higher susceptibility of plants. Collectively, our transcriptome and metabolome data provide a powerful resource to study plant responses during multifactorial stress and allow identifying metabolic processes and functional networks involved in tripartite interactions of plants with their environment.
Current climate prediction models indicate a gradual increase in ambient temperature and an enhancement of the frequency and amplitude of heat episodes (Ahuja et al., 2010; Mittler and Blumwald, 2010; Mittler et al., 2012; Li et al., 2013). Furthermore, global warming will be accompanied by drought, flood, and heat waves that drastically affect the conditions under which crop plants are grown (IPCC, 2008; Atkinson and Urwin, 2012). Due to their sessile lifestyle, plants are continuously exposed to a wide range of environmental stimuli and stresses. Several of the most frequently occurring abiotic stresses in nature, such as heat, cold, drought, salinity, solar radiation, and nutrient deficiency, affect many physiological processes in plants and have a large impact on world agriculture (Naika et al., 2013). Moreover, plants have to face attacks by various pests and pathogens, including bacteria, fungi, viruses, nematodes, and herbivores. Several environmental factors influence interactions between plants and pathogens and act on pathogenicity and host defense responses (Colhoun, 1973; Browder, 1985). Thus, abiotic stress may weaken or enhance plant defense responses, the outcome depending on the timing, nature, and severity of each stress (Atkinson and Urwin, 2012). An early study suggested that high temperatures suppress plant immunity (Dropkin, 1969), and low temperatures were found to impair gene silencing, a potent plant defense against viral pathogens (Szittya et al., 2003). More recent studies provided confirmation that increasing temperatures promote pathogen spread (Luck et al., 2011; Madgwick et al., 2011). Plant viruses benefit from elevated temperatures by an increased availability of insect vectors and possibly by weakened host resistance. Temperature-dependent suppression of host resistance has been reported for Tobacco mosaic virus (TMV) and Tomato spotted wilt virus (TSWV). TMV is able to overcome the N gene-mediated resistance at temperatures above 28°C in tobacco (Nicotiana tabacum; Kiraly et al., 2008), while TSWV is able to suppress the TSWV-mediated resistance in pepper (Capsicum annuum) plants at high temperatures (Moury et al., 1998). Although the first reports on the impact of heat stress on plant virus infection date back to 1931, little is known concerning the molecular basis of the observed suppression of host resistance. Only recently could it be demonstrated that the temperature sensitivity of the N gene-mediated TMV resistance of tobacco is caused by temperature-induced conformational changes of the R protein (Zhu et al., 2010). The temperature sensitivity of the R gene could be overcome by specific mutations in the R protein conferring heat-stable virus resistance. Since temperature-sensitive resistance is not only observed in plant-virus interactions but also in plant-bacteria, plant-fungi, and plant-nematode interactions, multiple cellular processes are likely to cause the observed phenotypes. In addition, opposite effects have been reported. One example is the high-temperature adult plant resistance to stripe rust (Puccinia striiformis f. sp. tritici) in spring wheat (Triticum aestivum; Carter et al., 2009). Therefore, studying the complex interrelationship between biotic and abiotic stress responses is required to understand the molecular basis of plant responses exposed to climate changes and to breed for durable plant resistance to viruses.
To date, most studies on plant responses to environmental changes include single or double stress scenarios, and experiments are generally short term (Garrett et al., 2006). Although comparing molecular responses to single stress conditions allowed the identification of genes that are up-regulated under more or less all stress situations (Swindell, 2006; Kilian et al., 2007; Weston et al., 2008), studies investigating stress factors in isolation fail to explain complex plant responses to more than one stress factor (Atkinson and Urwin, 2012). So far, only a few researchers have studied different stress factors occurring simultaneously and analyzed combined stress situations. Initial studies combining heat and drought stress revealed altered gene expression patterns when subjected to a combination of both (Rizhsky et al., 2004). The authors concluded that specific programs are activated and plant responses to multiple stresses differ from single stress applications. These differences are to be expected, since drought and heat result in partly opposing physiological changes that are reflected at the transcriptional level (Rizhsky et al., 2004). Under drought stress, plants tend to close their stomata to avoid unnecessary water loss. Under heat conditions, stomata are open to reduce leaf temperature. Therefore, simultaneously applied heat and drought must influence signals controlling gas exchange and, hence, will also play an important role in regulating the interaction of plants with bacteria or fungi entering leaves via stomata. In a recent article, plant responses to single stress factors (cold, heat, high light, salt, and flagellin) as well as their double combinations were studied (Rasmussen et al., 2013). Transcriptome changes in response to double stress revealed that 61% of the transcripts were not predictable from the responses to single stress treatments, and only for 6% of the transcripts did plants prioritize between antagonistic responses to stress combinations (Rasmussen et al., 2013). These data further support the need for additional studies concentrating on combined stress responses.
Plants are able to sense changes in ambient temperature via signaling pathways in order to adjust their metabolism and cellular functions to prevent heat-related damage (Mittler et al., 2012). Several studies indicated that many plant temperature responses share common signaling components, which are integrated in temperature signaling networks (for review, see Penfield, 2008). During heat stress, unfolded proteins accumulating in the cytosol, which is usually called cytoplasmic protein response (CPR) or the endoplasmic reticulum (ER; unfolded protein response [UPR]) induce specific heat shock responses (Howell, 2013). These include the induction of so-called heat shock proteins (HSPs), which help to bind and stabilize misfolded proteins. Thereby, HSPs function as molecular chaperones preventing protein aggregation (Jakob et al., 1993; Hartl and Hayer-Hartl, 2002). The expression of HSPs is known to be controlled by heat shock factors (HSFs). Compared with animals, plants possess a large number of HSF genes. This led to the hypothesis that HSFs have gained additional functions in plants (Von Koskull-Doring et al., 2007). Support for this hypothesis comes from the overexpression of HEAT SHOCK FACTOR A2 (HSFA2) in Arabidopsis (Arabidopsis thaliana), which resulted in a higher tolerance to combined light and heat stress of the transgenic plants as compared with the untransformed control (Nishizawa et al., 2006). In addition, different biotic stresses are known to induce HSF expression (Von Koskull-Doring et al., 2007), indicating that they may also play a role in pathogen defense. Since biotic- and abiotic-induced signaling pathways may act antagonistically (Anderson et al., 2004; Asselbergh et al., 2008), temporal and spatial control of these pathways will be essential for an effective plant response (Lee et al., 2012). Mutant analysis and integrated genome-wide network analysis have started to uncover significant parts of these complex networks (Ferrier et al., 2011; Gechev and Hille, 2012).
In this study, we established a multifactorial test system allowing simultaneous application of heat, drought, and virus stress. Transcriptome and metabolome analysis was applied for comparison of the molecular and biochemical responses of Arabidopsis plants exposed to these multifactorial stress scenarios. Here, we show that the number of differentially regulated features increased with the complexity and severity of stress. Comparative analysis of differentially expressed genes revealed stress-regulated candidate genes specifically expressed in one or more situations. Eleven genes were found to be differentially expressed under all experimental stress conditions, while 23 genes were specifically regulated under triple stress conditions. Moreover, virus infection combined with heat and drought stress revealed that basal and R-mediated disease resistance is compromised, which is paralleled by an increased CPR. Subsequently, different signaling coexpression networks have been found to be activated in single and combined stress situations. Our results show both interesting candidates involved in different stress conditions and changed plant defense mechanisms when virus infection is combined with other abiotic factors, indicating different regulatory components involved in single and multiple stress responses.
RESULTS AND DISCUSSION
Physiological Characterization of Arabidopsis Plants Subjected to Different Combinations of Abiotic and Biotic Stress Conditions
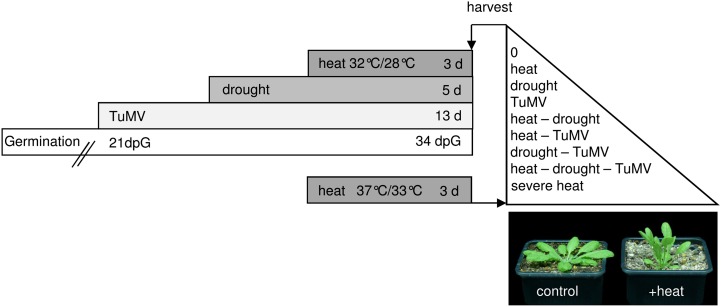
To study the impact of different stress combinations on metabolic and transcriptional plant responses, Arabidopsis plants were exposed to Turnip mosaic virus (TuMV), heat, and drought in single, double, and triple combinations. The optimal individual conditions were defined first and combined as shown in Figure 1. Mild conditions were chosen to simulate naturally occurring long-term stress and to make sure that combinations of different stresses were not lethal to plants. Drought stress was applied by withholding water and adjustment of the water status to a soil moisture content of 30% field capacity (Harb et al., 2010). To guarantee accurate experimental conditions, the water potential of plants was measured as well (Supplemental Table S1). For the application of heat stress in the initial experiment, plants were exposed to elevated temperatures (32°C day/28°C night) for 3 d, which represents a long-term stress under mild conditions compared with the heat shock used by other researchers in comparable studies (Rizhsky et al., 2004; Pecinka et al., 2010; Zhang et al., 2010). As the virus infection spreads rather slowly within Arabidopsis plants, inoculation was performed at 21 d after germination. The application of combined stress conditions is demonstrated in Figure 1. First, we inoculated plants with TuMV and then applied drought stress for 5 d followed by 3 d of heat stress. As a control, well-watered plants under ambient temperatures (22°C day/18°C night) were used. Furthermore, Arabidopsis plants were exposed to a single severe heat stress (37°C day/33°C night) to mimic the severity of the triple stress experiment. After the heat treatment, systemically infected leaves from plants treated either with a single stress or a combination of different stress conditions were sampled in both experiments. In the end, leaf samples from nine differentially treated plant groups were harvested (Fig. 1).
Figure 1.
Design of multifactorial stress experiments. The scheme illustrates the experimental design for exposing Arabidopsis plants to heat, drought, and viral stress (TuMV) separately or in the different combinations as described in “Materials and Methods.” To mimic the severity of the triple stress experiment, plants were additionally treated with severe heat in an independent experiment. Leaves of all conditions were harvested: heat, drought, TuMV, drought and heat, TuMV and heat, drought and TuMV, TuMV, drought, and heat, and severe heat. The photograph shows the phenotypes of control and heat-treated plants.
As a starting point for the analysis, fresh and dry weight as well as leaf number of stressed plants were determined. It was expected that stress treatments would result in growth retardation (Smith and Stitt, 2007; Bechtold et al., 2010; Hummel et al., 2010). At the time of harvest, a significant reduction in biomass was found for all single stress conditions, which was emphasized even more when different stresses were combined. This was mainly due to a reduced number of leaves, especially in combination with heat; the combination of virus and heat as well as the triple stress showed the lowest biomass and the lowest number of leaves (Table I). Leaves of plants exposed to heat stress stayed vertical, avoiding direct exposure to sunlight, overheating, and extended water loss (Fig. 1; Pastenes et al., 2005). The smallest leaves of infected plants already started to develop clear TuMV symptoms after 9 d, such as reduced growth; nevertheless, the application of TuMV had only minor influence on overall biomass production, may be due to the fact that the locally applied virus needed time to spread through the whole plant. Initial studies suggested that stomata are closed during combined heat and drought stress (Rizhsky et al., 2002), which is in line with our microscopy observations (Table I). Additionally, we found that stomata were closed during a combination of virus and drought as well as during triple stress. Strikingly, during a combination of virus and heat, stomata were also closed, while single heat or virus treatment resulted in stomata opening (Table I). This result strongly suggests that molecular and physiological responses of plants exposed to multifactorial stresses cannot be foreseen and need to be tested.
Table I. Influence of single and multifactorial stress on biomass, leaf number, and stomata.
The table shows if stomata are more opened or closed under single and multiple stress influence compared with control plants. Stomata on the abaxial side of Arabidopsis plants were analyzed using a Leica DMR light microscope. The fresh weight of whole rosettes of differentially treated plants was determined. In parallel, 10 plants of each condition were dried at 80°C for 3 d for determination of plant dry weight. As the control plants of the triple stress experiment and the severe heat experiment had comparable controls, data were combined. Data points (plant, leaf) represent averages from 10 or more plants ± sd. n.d., Not determined. On average, 12 or more stomata were analyzed per treatment. Asterisks indicate statistically significant differences from the wild type (P ≤ 0.05). Conditions are as follows: heat (h), drought (d), TuMV (v), drought and heat (dh), TuMV and heat (vh), drought and TuMV (dv), TuMV, drought, and heat (vdh), and severe heat (severe h).
| Condition | Plant Fresh Weight | Plant Dry Weight | Leaf No. | Stomata, Length-Width Ratio |
|---|---|---|---|---|
| mg | ||||
| c | 148.83 ± 26.45 | 11.86 ± 1.18 | 7.70 ± 0.73 | 1.36 ± 0.09 |
| h | 106.44 ± 15.81* | 8.16 ± 1.31* | 8.13 ± 0.87 | 1.18 ± 0.11* |
| d | 101.63 ± 16.23* | 9.09 ± 0.93* | 7.70 ± 0.67 | 1.53 ± 0.09* |
| v | 127.53 ± 11.72* | 9.92 ± 1.19* | 7.45 ± 0.83 | 1.27 ± 0.14* |
| vd | 86.93 ± 8.70* | 8.21 ± 1.00* | 7.66 ± 0.55 | 1.58 ± 0.09* |
| dh | 75.23 ± 4.79* | 8.53 ± 1.12* | 6.96 ± 0.60 | 1.51 ± 0.08* |
| vh | 113.03 ± 6.44* | 8.23 ± 1.00* | 6.79* ± 0.86 | 1.46 ± 0.10* |
| vdh | 76.99 ± 10.95* | 7.46 ± 0.84* | 6.19* ± 0.83 | 1.58 ± 0.15* |
| severe h | 95.80 ± 18.53* | 7.80 ± 0.72* | 7.76 ± 0.88 | n.d. |
Transcriptome and Metabolome Profiles of Plants Exposed to Multifactorial Stress
To further improve our understanding of the molecular responses of plants subjected to single or multifactorial stress, whole-genome Agilent microarray mRNA hybridizations were performed using RNA samples isolated from leaves harvested from plants grown under nine different environmental conditions (Fig. 1). After conducting an ANOVA test on all samples of the initial triple stress experiment, a condition tree based on the hierarchical clustering of expression values of the remaining 27,870 entities showed that replicates of differentially treated plants clustered together (Fig. 2A). In one case, principal component analysis (PCA) revealed an outlier (single virus stress), which was removed from the analysis (Supplemental Fig. S1). Moreover, replicate grouping was strongly influenced by different treatments, especially heat-stressed samples, which formed an isolated branch, indicating that heat treatment obviously had the greatest influence on gene expression under our experimental conditions. An earlier study focusing on a double combination of stresses also found responses of heat more dominant than salt stress (Rasmussen et al., 2013). Within this isolated branch, a separation of drought-stressed and virus-stressed plants can be observed. However, if no heat stress was applied, the separation of virus from virus in combination with drought stress was not that strong, possibly reflecting the mild nature of the applied stresses. To validate the data, a PCA was performed. The first principal component was heat (63.2%), followed by drought (14.8%), and virus (9.9%), supporting a clear influence of all three parameters on the microarray data set (Supplemental Fig. S2, A and B).
Figure 2.
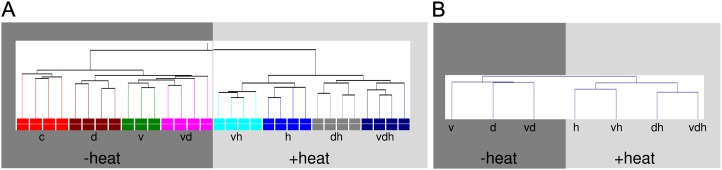
Transcriptome and metabolome profiles of triple-stressed plants. A, Condition tree based on hierarchical clustering of expression data separating replicates of each condition. For the analysis, four biological replicates were hybridized. Colored boxes at the bottom indicate different treatments. B, Condition tree based on hierarchical clustering of metabolome data separating treatments of each condition. Metabolome data are given as mean values with n = 3. Conditions are as follows: heat (h), drought (d), TuMV (v), drought and heat (dh), TuMV and heat (vh), drought and TuMV (dv), and TuMV, drought, and heat (vdh).
Additionally, metabolic profiling of different abiotic- and biotic-stressed plants was performed in parallel to the transcriptional analysis. Again, metabolome data of heat-treated samples could be clearly distinguished from samples without heat stress in a hierarchical clustering analysis (Fig. 2B; Supplemental Fig. S3). This was further supported by PCA, separating the different stress conditions (Supplemental Fig. S2, C and D). Thereby, one component, responsible for 47.9% of the variance, could be associated with heat stress. The most important metabolites for the loading included l-His as well as l-Tyr, which has already been described to increase under heat stress (Kaplan et al., 2004; Guy et al., 2008). The second component of the PCA, accounting for 21.5% of the variance, was drought treatment, and the biggest influences on that separation were l-Pro, l-Met, l-Ser, and l-Gly. Accumulation of Pro during drought stress is well known and has already been reviewed by Hanson and Hitz (1982). Pro acts as a compatible solute and protects plants against osmotic stress (Hare and Cress, 1996). To our knowledge, l-Met, l-Ser, and l-Gly, have not been shown to accumulate after drought stress, but they are known to be increased under cold shock (Kaplan et al., 2004), indicating that they may be stress responsive. Nevertheless, the PCA results of metabolites also demonstrated that our stress applications were adequate for studying plant responses under multiparallel stress treatments.
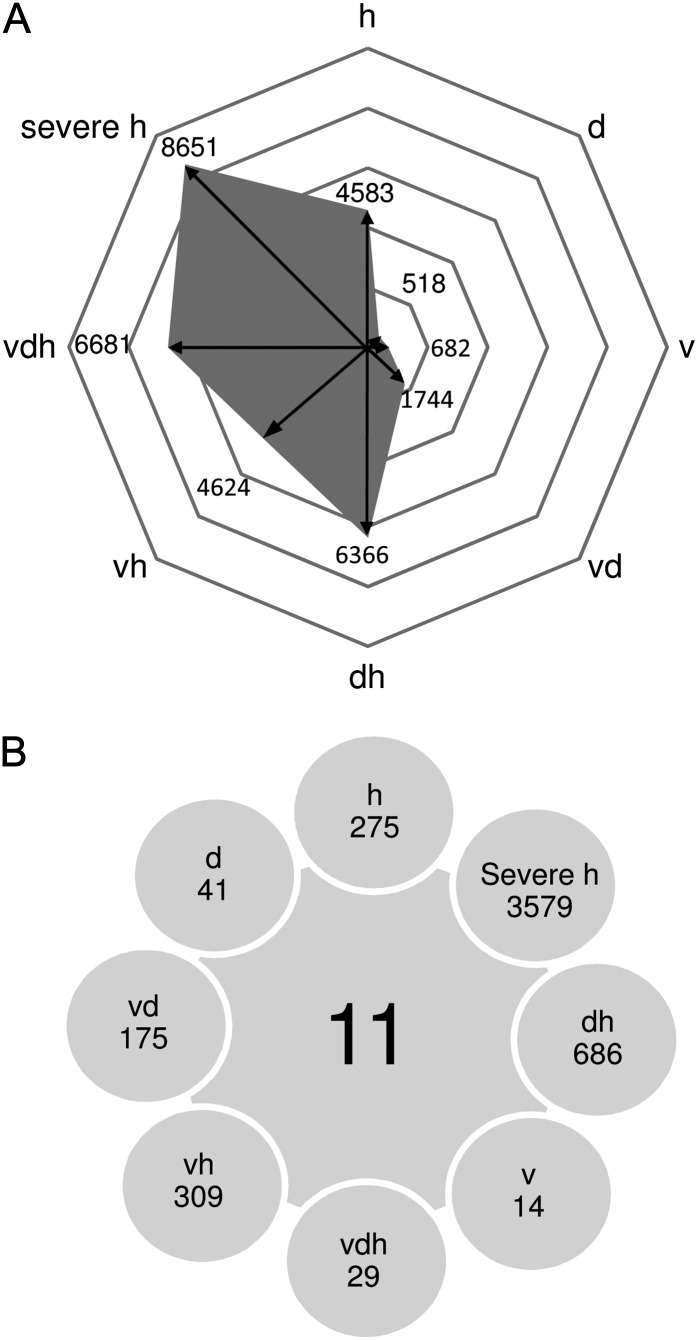
Next, we examined how many genes were specifically regulated in each stress condition compared with control plants using volcano-plot analysis. Severe heat stress samples were compared with their respective controls. Figure 3A illustrates the numbers of statistically significant identified entities of each stress condition, showing large differences between applied stresses (lists of all features are given in Supplemental Table S2). Remarkably, the number of regulated features increased with the complexity or severity of the stress applied. For instance, the number of regulated features in single drought and virus stress was 518 and 682, respectively, but rose to 1,744 when both stresses were applied in combination. This effect was even more pronounced if plants were additionally exposed to heat stress: the number of regulated genes was more than three times higher (6,681; Fig. 3A). In addition, a high number of significantly regulated genes (8,651) were also regulated by severe heat stress (37°C day/33°C night). For experimental reasons, heat stress and control treatments were carried out in two separate growth chambers. Therefore, the difference between the two chambers might have some influence on the number of differentially expressed genes. Based on this observation, we conclude that the number of regulated genes increases with (1) the severity and (2) the complexity of stress. Since we were particularly interested in multifactorial stress, the triple stress experiment was repeated twice in independent experiments. Combining data from all three experiments reduced the number of statistically significant genes to 2,715 features.
Figure 3.
Transcriptome profiles of multifactorially stressed plants. A, Number of statistically significant features for each stress condition identified by volcano-plot analysis. The significance threshold was set at change greater than 2-fold and false discovery rate less than 0.05. B, Intersection identifying at least 2-fold statistically significant entities present in all stress conditions as well as features unique to individual treatments. Conditions are as follows: heat (h), drought (d), TuMV (v), drought and heat (dh), TuMV and heat (vh), drought and TuMV (dv), TuMV, drought, and heat (vdh), and severe heat (severe h).
Intersections provide an overview showing the distribution of stress-specific and commonly regulated genes and allow the identification of genes important for the adaptation of plants to different stress conditions (lists of all features are given in Supplemental Table S3). Using our microarray data set, this analysis revealed 11 genes regulated under all the different stress conditions (Fig. 3B; Supplemental Fig. S4A). Interestingly, among these 11 genes were two transcription factors, Rap2.9 and G-BOX BINDING FACTOR3 (GBF3). Both have been suggested to be involved in stress responses.
Rap2.9, also called DEAR5, belongs to the DREB subfamily, characterized by a conserved WLG motif and an AP2 domain (Magnani et al., 2004), which has been described to be involved in stress responses (Sakuma et al., 2002; Shen et al., 2003). Rap2.9 is known to be a homolog of DEAR1, with significant homology in the DREB domain and EAR motif (Tsutsui et al., 2009). Using an overexpression construct, Tsutsui et al. (2009) have suggested that DEAR1 is a transcriptional repressor upstream of the freezing tolerance. Under normal conditions, DEAR1 is up-regulated to keep stress responses under tight control, whereas under stress, the repressor is down-regulated to avoid further inhibition of stress-related gene expression and corresponding tolerance mechanisms. This model would be in agreement with our data, as Rap2.9 was down-regulated under abiotic stress conditions. This indicated that Rap2.9 functions as a repressor under heat and drought stress. As the overexpression of DEAR1 showed pathogen resistance against Pseudomonas syringae DC3000, Tsutsui et al. (2009) concluded that DEAR1 is a positive regulator of pathogen defense. For DEAR5, the authors have shown pathogen inducibility against bacteria via reverse transcription (RT)-PCR analysis, assuming a similar function. However, TuMV infection results in a down-regulation of Rap2.9, suggesting different roles of Rap2.9 in stress adaption to bacteria and viruses. In a combination of biotic and abiotic stresses, Rap2.9 was also down-regulated, supporting a repressor function of defense responses of Rap2.9 under normal growth conditions.
The second commonly regulated transcription factor, GBF3, was strongly up-regulated under all stress conditions. Interestingly, GBF3 is abscisic acid (ABA) inducible, and it has been proposed that GBF3 is responsive to multiple stress conditions (Fujita et al., 2005). This view is supported by public microarray data (www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007) showing GBF3 to be up-regulated by osmotic stress, salt stress, cold stress, and after infiltration with P. syringae DC3000 after 24 h. A role in multifactorial stress adaptations has not been shown. Among the 11 commonly regulated genes were two genes annotated as hypothetical protein as well as two genes with hydrolase activity acting on O-glycosyl compounds, β-glucosidase, and a xyloglucan endotransglycosylase.
Besides these commonly regulated genes, closer inspection of the Venn diagram allows the identification of stress-specific genes, which were only significantly regulated under one stress situation (Fig. 3B). This analysis revealed 29 features, corresponding to 23 individual genes, specifically expressed under triple stress conditions (Supplemental Fig. S4B). Most of them could be associated with stress: among them were three transcription factors, including DREB2A, and two zinc finger proteins as well as other candidates that had already been described as stress associated, like COLD-REGULATED47, ABI5 BINDING PROTEIN (AFP1), a PENTATRICOPEPTIDE REPEATING-CONTAINING PROTEIN, and a gene classified as UNIVERSAL STRESS PROTEIN FAMILY PROTEIN. Induction of AFP1 is counterintuitive, since it is supposed to be a negative regulator of ABA by targeting ABI5 for proteolytic degradation (Lopez-Molina et al., 2003). This finding is even more perplexing, since another member of the ABA response network, ATL43, is up-regulated under triple stress conditions. This protein belongs to a gene family encoding RING-H2 finger proteins and is essential for ABA responses (Serrano et al., 2006). These contrasting results may reflect the complicated situation of plants treated with multiple and partly contrasting stress conditions and highlight the importance of analyzing complex interactions.
Altogether, the described comparative analysis reveals a small group of genes expressed under all stress conditions and genes that are only expressed under individual stress treatments of this study. General stress-responsive genes may be candidates to improve the stress tolerance of crop plants, while analysis of the stress-specific genes may shed some light into the complex adjustment of stress-specific signal transduction pathways under multifactorial stress conditions.
Down-Regulation of Primary Carbon Metabolism Is a General Stress Response
Beyond the analysis of single candidate genes, we were interested to determine processes mainly influenced under different stress situations. To get a comprehensive overview of gradual and complex changes in plant responses to altering environmental stimuli, differentially expressed features were categorized into functional groups using categories defined by MAPMAN (mapman.gabipd.org/web/guest/mapmanstore). For each category, a ratio comparing the number of significantly regulated features within a given treatment compared with the whole array was determined. The functional assignments and corresponding enrichments are given in Table II.
Table II. Functional assignment of up- and down-regulated features of plants exposed to individual and combined stress.
Transcripts found to be differentially regulated in each treatment in comparison with control plants were divided into up- and down-regulated groups and grouped according to a classification of features based on bins provided by MAPMAN. Numbers illustrate the percentage of significantly regulated features from a specific functional group relative to the percentage of features from that specific group to the entire chip. Up-regulated features have been marked as positive values and down-regulated ones as negative values. Conditions are as follows: heat (h), drought (d), TuMV (v), drought and heat (dh), TuMV and heat (vh), drought and TuMV (dv), TuMV, drought, and heat (vdh), and severe heat (severe h).
| Category | Relative Enrichment of Up-Regulated Features |
Relative Enrichment of Down-Regulated Features |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| h | d | v | vd | dh | vh | vdh | severe h | h | d | v | vd | dh | vh | vdh | severe h | |
| Photosynthesis | 0.04 | 0.00 | 0.00 | 0.09 | 0.21 | 0.14 | 0.21 | 0.31 | −7.04 | 0.00 | −8.07 | −5.87 | −5.54 | −7.73 | −5.15 | −3.56 |
| Major carbohydrates | 0.55 | 0.00 | 0.55 | 0.50 | 0.65 | 0.63 | 0.67 | 0.99 | −4.54 | −2.54 | 0.00 | −0.63 | −3.45 | −4.70 | −2.94 | −2.60 |
| Minor carbohydrates | 1.57 | 1.81 | 0.84 | 1.12 | 1.13 | 1.62 | 1.01 | 1.44 | −1.29 | 0.00 | 0.00 | −1.91 | −1.39 | −1.37 | −1.36 | −1.38 |
| Glycolysis | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.12 | 0.10 | −2.43 | 0.00 | 0.00 | −1.62 | −2.35 | −1.17 | −0.85 | −2.08 |
| Fermentation | 0.72 | 0.00 | 0.00 | 0.00 | 0.61 | 0.83 | 0.00 | 0.50 | −6.24 | 0.00 | 0.00 | 0.00 | −2.13 | −4.00 | −0.63 | −0.46 |
| Gluconeogenesis | 2.26 | 4.13 | 0.00 | 1.28 | 1.93 | 1.96 | 1.98 | 3.16 | −0.98 | 0.00 | 0.00 | 0.00 | −0.56 | 0.00 | −0.49 | 0.00 |
| Oxidative pentose phosphate pathway | 1.90 | 0.00 | 0.00 | 0.00 | 1.08 | 1.10 | 1.11 | 0.88 | −2.20 | 0.00 | 0.00 | −1.83 | −0.94 | −0.88 | −0.28 | −0.61 |
| TCA | 0.67 | 0.98 | 0.00 | 0.00 | 0.34 | 0.62 | 0.47 | 0.56 | −2.33 | 0.00 | 0.00 | 0.00 | −2.12 | −2.42 | −2.10 | −2.42 |
| ATP synthesis | 1.85 | 0.00 | 0.37 | 0.50 | 1.64 | 2.06 | 1.55 | 2.01 | −0.51 | 0.00 | 0.00 | −0.86 | −0.51 | −0.82 | −0.39 | −0.38 |
| Cell wall | 0.92 | 2.68 | 2.21 | 1.09 | 0.68 | 1.01 | 0.58 | 0.80 | −1.03 | −1.06 | −2.02 | −0.80 | −1.86 | −1.08 | −2.17 | −2.07 |
| Lipid metabolism | 0.86 | 0.81 | 0.42 | 0.82 | 1.16 | 1.06 | 1.09 | 0.89 | −1.11 | 0.00 | −0.61 | −0.96 | −1.40 | −1.16 | −1.40 | −1.72 |
| Nitrogen metabolism | 2.21 | 0.00 | 1.87 | 0.84 | 1.88 | 2.56 | 1.93 | 1.03 | 0.00 | 0.00 | 0.00 | −2.13 | −2.55 | −1.53 | −2.56 | −2.14 |
| Amino acid metabolism | 0.83 | 0.55 | 0.38 | 0.69 | 0.93 | 0.92 | 0.89 | 0.90 | −1.38 | −2.63 | 0.00 | −0.22 | −1.34 | −0.89 | −1.08 | −1.22 |
| Sulfur assimilation | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.58 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Metal handling | 1.53 | 0.86 | 2.39 | 1.34 | 0.81 | 1.78 | 0.72 | 0.58 | −3.48 | 0.00 | 0.00 | 0.00 | −2.34 | −2.63 | −2.26 | −1.83 |
| Secondary metabolism | 1.26 | 1.29 | 1.04 | 2.00 | 1.20 | 1.50 | 1.49 | 1.35 | −1.58 | −2.05 | 0.00 | −0.68 | −1.42 | −1.31 | −1.28 | −1.08 |
| Hormone metabolism | 2.09 | 3.05 | 2.34 | 2.10 | 1.67 | 1.73 | 1.33 | 1.27 | −1.60 | −3.58 | −6.80 | −3.56 | −2.28 | −1.53 | −1.93 | −1.56 |
| Cofactor | 0.00 | 0.00 | 0.00 | 0.00 | 0.19 | 0.00 | 0.20 | 0.16 | −0.78 | 0.00 | 0.00 | 0.00 | −0.67 | −0.63 | −0.79 | −0.58 |
| Tetrapyrrole synthesis | 0.22 | 0.00 | 1.13 | 0.00 | 0.19 | 0.00 | 0.00 | 0.00 | −7.35 | −15.55 | 0.00 | −11.61 | −5.95 | −6.82 | −4.84 | −4.46 |
| Stress | 1.45 | 1.38 | 1.86 | 2.03 | 1.58 | 1.50 | 1.54 | 1.20 | −1.62 | −2.07 | −0.88 | −1.32 | −1.25 | −1.60 | −1.33 | −1.32 |
| Redox regulation | 1.00 | 1.10 | 1.02 | 1.25 | 0.60 | 1.16 | 0.70 | 0.73 | −2.87 | −6.99 | 0.00 | −2.90 | −2.77 | −3.41 | −2.48 | −1.68 |
| Polyamine metabolism | 1.27 | 0.00 | 0.00 | 2.88 | 2.70 | 0.00 | 2.77 | 2.21 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | −1.10 | −0.82 |
| Nucleotide metabolism | 0.76 | 0.00 | 0.00 | 0.35 | 1.04 | 1.23 | 1.40 | 0.80 | −1.58 | −1.77 | 0.00 | −1.32 | −2.18 | −1.27 | −1.46 | −1.62 |
| Xenobiotics | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.40 | 2.21 | −0.78 | 0.00 | 0.00 | 0.00 | −0.45 | −0.63 | −0.39 | −0.87 |
| C1 metabolism | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.39 | −2.45 | 0.00 | 0.00 | 0.00 | −1.40 | −0.39 | −1.47 | −1.28 |
| Miscellaneous | 1.35 | 1.12 | 1.55 | 1.71 | 1.14 | 1.39 | 1.18 | 1.19 | −1.52 | −1.43 | −1.35 | −1.11 | −1.42 | −1.38 | −1.41 | −1.25 |
| RNA | 1.16 | 0.88 | 0.85 | 0.90 | 1.26 | 1.13 | 1.34 | 1.25 | −0.65 | −1.63 | −1.55 | −1.24 | −0.70 | −0.70 | −0.71 | −0.68 |
| DNA | 0.45 | 0.48 | 0.04 | 0.21 | 0.51 | 0.56 | 0.58 | 0.58 | −0.29 | −0.19 | −0.36 | −0.29 | −0.44 | −0.40 | −0.47 | −0.74 |
| Protein | 0.71 | 0.43 | 0.55 | 0.58 | 0.77 | 0.72 | 0.82 | 0.90 | −0.65 | −0.27 | −0.65 | −0.77 | −0.65 | −0.61 | −0.60 | −0.58 |
| Signaling | 1.12 | 2.21 | 3.40 | 2.69 | 1.05 | 0.89 | 0.66 | 0.77 | −0.75 | −0.64 | −0.41 | −0.69 | −0.93 | −0.82 | −1.21 | −1.67 |
| Cell | 0.72 | 0.66 | 0.91 | 0.85 | 0.82 | 0.68 | 0.70 | 0.71 | −0.60 | −0.35 | −0.66 | −0.52 | −0.99 | −0.81 | −0.85 | −1.34 |
| MicroRNA | 0.62 | 0.82 | 0.28 | 0.38 | 1.10 | 1.24 | 1.28 | 1.29 | −0.88 | −1.30 | −1.24 | −1.95 | −0.67 | −0.94 | −0.73 | −0.58 |
| Development | 1.45 | 0.62 | 0.94 | 1.00 | 1.34 | 1.33 | 1.56 | 1.31 | −0.79 | −0.39 | −1.12 | −0.68 | −1.05 | −1.06 | −1.14 | −0.86 |
| Transport | 1.03 | 0.64 | 1.66 | 1.51 | 1.05 | 1.09 | 1.14 | 1.29 | −1.47 | −2.79 | −0.72 | −1.45 | −1.52 | −1.36 | −1.48 | −1.29 |
| Not assigned | 1.00 | 1.10 | 0.85 | 0.85 | 0.99 | 0.97 | 0.98 | 0.98 | −0.83 | −0.70 | −0.84 | −0.82 | −0.78 | −0.83 | −0.83 | −0.85 |
Based on functional categorization, photosynthetic genes involved in PSI, PSII, and ATP synthesis were strongly down-regulated under almost all stress conditions (Supplemental Fig. S5). Under single drought stress, photosynthetic genes were less affected. The relative insensitivity of photosynthetic genes to drought stress had been described before (Chaves et al., 2009). The general down-regulation of photosynthetic genes is in agreement with other studies focusing on stress experiments (Law and Crafts-Brandner, 1999; Jung et al., 2003; Wong et al., 2006). In leaves, several thousand genes, including photosynthetic genes, are regulated by the circadian clock (Gibon et al., 2006). The clock itself is modulated by environmental factors, including water deficit, light, and temperature (Bieniawska et al., 2008; Baerenfaller et al., 2012). While the effects of low temperature on the expression of clock-regulated genes have been extensively studied, adaption of clock genes to elevated temperatures has received little attention so far. Gould et al. (2006) demonstrated in Arabidopsis that the amplitude of clock genes is changed under elevated temperatures. The authors showed increased TIMING OF CAB1 (TOC1) and decreased CIRCADIAN CLOCK ASSOCIATED1 (CCA1) amplitudes under elevated (27°C) compared with ambient (17°C) temperatures. Baerenfaller et al. (2012) nicely demonstrated that water deficit led to significant changes in TOC1 and CCA1 expression, with TOC1 being up-regulated at the end of the day and CCA1 being up-regulated at the end of the night. Interestingly, CCA1, the molecular oscillator component of the morning loop of the circadian clock (Alabadi et al., 2001; Yakir et al., 2007), showed highly increased expression under combined stress conditions in our microarray analysis (Supplemental Fig. S6). Inspired by this observation, the expression of clock-associated genes was studied in more detail in a time-course experiment. Columbia plants were exposed to severe heat stress (37°C), and leaf material was sampled at different time points during the day. Quantitative PCR (qPCR) analysis revealed a strong decline during the day for CCA1 and increased amounts of TOC1 mRNA levels for control plants (Supplemental Fig. S7). However, qPCR analysis of stressed plants compared with control plants showed a slower reduction of CCA1 and a dampened increase of TOC1 transcripts, which indicate an altered ratio of the main clock regulators under combined stress.
Functional categorization further revealed that most enzymes of the Calvin-Benson cycle followed the same transcriptional trend in response to stress. By contrast, the expression of genes involved in photorespiration, for instance glycolate oxidase and Glu:glyoxylate aminotransferase, were up-regulated under multifactorial and severe stress (Supplemental Fig. S8). As all plants exposed to stress conditions showed reduced dry weight (Table I), this could indicate an up-regulation of photorespiration under stress, which has also been described by Rivero et al. (2009).
Further analysis of metabolites and genes encoding enzymes of primary metabolism, including glycolysis and the tricarboxylic acid (TCA) cycle, revealed a preferential down-regulation of both associated transcripts and metabolites (Supplemental Fig. S9). However, some TCA cycle intermediates and their derived amino acids accumulated under some stress conditions, which was paralleled by an increase in associated transcripts. For instance, Pro originated from Glu, which itself is derived from α-ketoglutarate. Under single drought stress, l-Pro is up-regulated. This is in agreement with many similar studies focusing on drought (for review, see Verbruggen and Hermans, 2008). TCA cycle genes encoding enzymes producing α-ketoglutarate were up-regulated during drought stress, namely aconitase, and isocitrate dehydrogenase. However, a combination of drought and heat showed no accumulation of Pro, which is in line with findings from Rizhsky et al. (2004). They found elevated Suc levels instead of Pro under combined drought and heat stress. Interestingly, a combination of drought and virus showed increased Pro amounts. Obviously, drought stress is responsible for increased contents of Pro, whereas heat, also in combination with virus infection, seems to change the energy and redox metabolism. With the exception of drought, virus, and combined virus and drought treatment, the expression of most genes encoding proteins of the mitochondrial electron transport chain was also down-regulated. This includes all complexes I to V. By contrast, alternative oxidase (AOX), a sole component of the alternative respiratory pathway, was slightly up-regulated (Supplemental Fig. S10). Although the exact role of AOX under stress conditions is under debate (for review, see Van Aken et al., 2009), the expression of AOX genes has been found to be increased under different stress conditions. For instance, elevated expression levels of AOX were determined under combined drought and heat stress in tobacco plants (Rizhsky et al., 2002). Interestingly, it has been assumed that AOX acts as negative regulator of induced resistance to viruses (Gilliland, 2003). The authors used transgenic tobacco plants with an increased capacity of the alternative pathway and showed that TMV resistance was impaired. The interaction of TuMV and plants exposed to multiparallel stress will be discussed in the following sections in more detail. The observation that drought stress leads to pronounced growth reductions in the absence of significant transcriptional changes in genes of the respiratory pathway is in agreement with previous observations that mild and severe drought stress do not alter dark respiration but plant growth (Hummel et al., 2010). This might indicate that carbon flux does not limit leaf growth under water deficit but does under combined stress conditions.
Altogether, pathway analysis shows that the expression of most genes encoding enzymes of the central carbon metabolism and respiration is reduced, and only a few differences in the response to single and multifactorial stress could be identified. This suggests that the expression of genes encoding enzymes of the central metabolism responds to general stress signals and/or altered demand rather than specific signals.
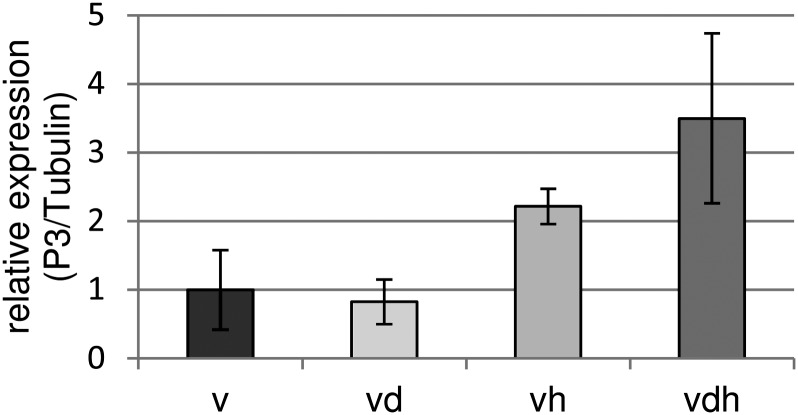
Combined Heat and Drought Stress Increases the Susceptibility of Arabidopsis Plants to Virus Infections
Previous studies have shown that R gene-mediated virus resistance can be disturbed under elevated temperatures. To the best of our knowledge, no data exist describing the effect of abiotic stress on the susceptible plant-virus interaction. To study the effect of heat and drought on the susceptible interaction, TuMV replication in differentially stressed plants was determined using quantitative RT-PCR. As a marker for viral replication, the P3 gene was chosen, and primers were used as described earlier (Kim et al., 2010). As illustrated in Figure 4, there was no difference detectable comparing single virus stress and the combination of virus and drought stress. However, combining virus infections with heat or heat and drought increased the level of detectable P3 2-fold or 3-fold, respectively. As discussed above, earlier studies have already indicated that abiotic stress applied for longer periods is able to compromise plant defense (Xiong, 2003; Goel et al., 2008). For avirulent P. syringae strains, enhanced replication on drought-stressed Arabidopsis plants has been reported (Mohr and Cahill, 2003). In another study, virus infection of tobacco plants led to an increased drought tolerance of infected plants as compared with the uninfected controls (Xu et al., 2008). In our study, plants simultaneously exposed to virus and drought stress showed significantly less biomass as compared with plants treated with either virus or drought alone. Therefore, we do not have evidence that virus infection would enhance the drought tolerance of infected plants. However, due to overlapping and possibly counterbalancing effects, a positive impact of virus infection on drought tolerance may have been overlooked.
Figure 4.
qPCR analysis of the viral P3 gene in TuMV-infected Arabidopsis plants. Accumulation of P3 mRNA was quantified by quantitative RT-PCR using primers published by Kim et al. (2010). Relative expression levels were normalized to tubulin expression. Error bars represent se (n = 3). v, Virus infection; vd, combined virus infection and drought stress; vh, combined virus infection and heat stress; vdh, combined virus infection, drought, and heat stress.
Nevertheless, our study clearly shows that heat or heat and drought increases the susceptibility of Arabidopsis plants to virus infections. The cause for this increased susceptibility may reside in an altered expression of components of the signal transduction pathway or in a modified metabolite composition leading to altered metabolite signaling.
Plant Defense Responses Are Altered under Multifactorial Stress
Elevated temperatures have been implicated in interfering with plant-pathogen interactions. In general, it is believed that high temperatures compromise R gene-mediated disease responses including the hypersensitive response, in different plant systems against biotrophic, hemibiotrophic, and necrotrophic microbes (Koeda et al., 2012). This view is supported by the observation that moderate increases in temperature inhibit the R gene-mediated hypersensitive response of Arabidopsis infected with the bacterial pathogen P. syringae (Wang et al., 2009). Plants exposed to 28°C compared with 22°C were more susceptible to the virulent bacteria strain, indicating that elevated temperatures influence defense responses. Mild increases in temperature also compromised the R gene-mediated hypersensitive response following potato virus X coat protein or TMV helicase expression in Nicotiana benthamiana (Wang et al., 2009). As TuMV propagated much better in plants exposed to heat and drought in our approach, the question arose whether plant defense mechanisms were influenced. In general, plant immunity occurs at different levels and can be divided into basal and R gene-mediated resistance (Chisholm et al., 2006; Jones and Dangl, 2006).
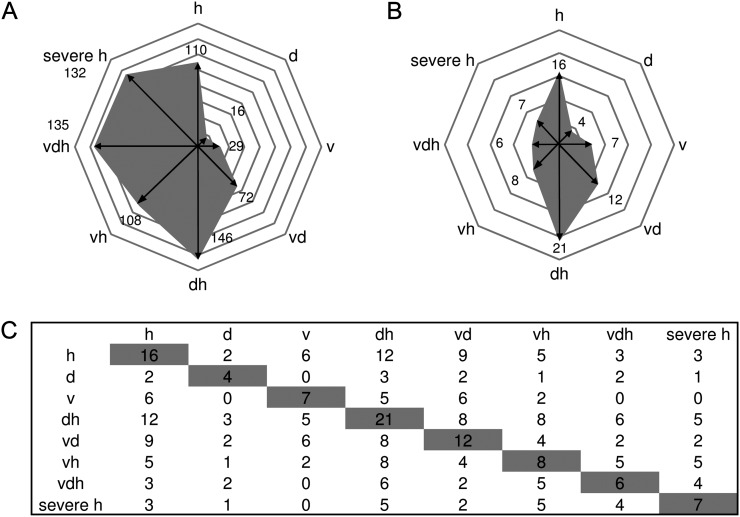
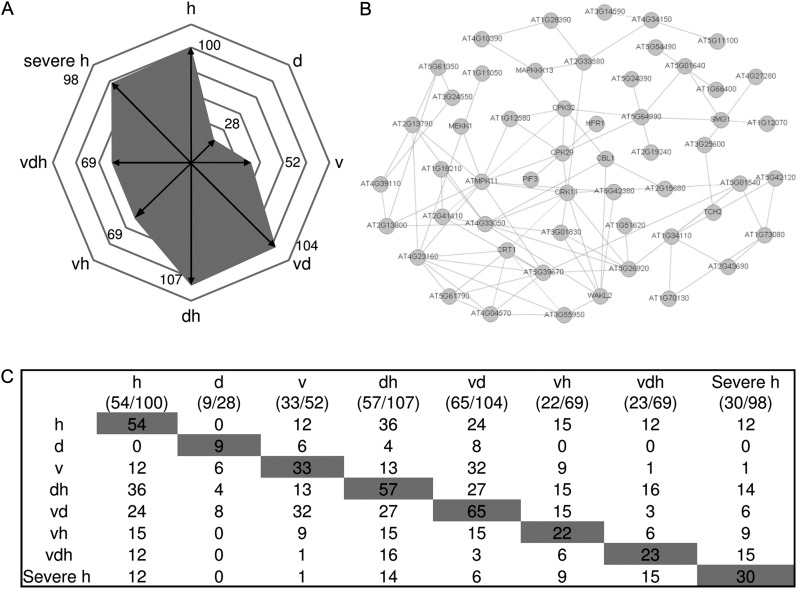
To investigate whether R gene-mediated resistance might be altered by the application of multifactorial stress, the functional group of stress-associated features (Table II) was analyzed in detail. Figure 5A illustrates the number of statistically significantly up-regulated genes according to functional group stress. This analysis showed that stress-associated features were strongly up-regulated under heat (110 up-regulated genes), virus and drought (72 up-regulated genes), drought and heat (146 up-regulated genes), heat and virus (108 up-regulated genes), triple stress (135 up-regulated genes), and severe heat stress (132 up-regulated genes). Single drought (16 up-regulated genes) or virus treatment (29 up-regulated genes) only moderately induced general stress features. The largest class of R proteins contained a nucleotide-binding site (NBS) and leucine-rich repeats (LRRs) and is termed NBS-HSFs (Dangl and Jones, 2001). There are two major subfamilies of plant NBS-LRR proteins, which can be distinguished by the presence of Toll/Interleukin1 receptor (TIR) or coiled-coil motifs in the N-terminal domain (Gohre and Robatzek, 2008).
Figure 5.
Shifts of gene expression patterns analyzing defense systems of plants exposed to single stress compared with multifactorial or severe heat stress. A, Number of differentially regulated genes according to the functional group “stress” is given for each treatment. B, Distribution of stress-regulated features annotated as TIR-NBS-LRR genes between different stress treatments. C, Table showing the number of treatment-specific TIR-NBS-LRR genes found to be coexpressed within one treatment (shaded) and the number of commonly shared genes compared with coexpressed TIR-NBS-LRR genes of other stress conditions (not shaded). Conditions are as follows: heat (h), drought (d), TuMV (v), drought and heat (dh), TuMV and heat (vh), drought and TuMV (dv), TuMV, drought, and heat (vdh), and severe heat (severe h).
Our analysis showed that TIR-NBS-LRR genes are differentially expressed under the different stress conditions (Fig. 5B; Supplemental Table S4). Intriguingly, the highest number of TIR-NBS-LRR genes was induced under heat stress (16) and in the combination of heat and drought (21), which can be explained by studies suggesting that the function of NBS-LRR proteins is regulated by HSPs such as HSP90 (for review, see Belkhadir et al., 2004). For instance, for HSP90, in vivo interactions with RPM1, a disease resistance protein, has been shown (Hubert et al., 2003). However, different TIR-NBS-LRR genes are regulated under the different treatments. Figure 5C shows that out of the 16 TIR-NBS-LRR genes regulated under moderate heat stress (32°C), only six TIR-NBS-LRR genes are differentially regulated under single virus stress. Interestingly, the overlap between mild heat stress and the combination of virus and heat stress still gives five differentially regulated TIR-NBS-LRR genes, but from these five, only two genes are shared with single virus stress (Fig. 5C). These data already showed that different TIR-NBS-LRR genes are activated under single and multifactorial stress situations. However, in the triple stress situation, not only a reduction of regulated TIR-NBS-LRR genes can be found (from 16 to six), but out of the six TIR-NBS-LRR genes regulated under triple stress, only three genes are common to mild heat stress. Moreover, these six TIR-NBS-LRR genes regulated under triple stress shared no commonly regulated genes between virus and triple stress, indicating changes in the defense programs, which could not be predicted from single-stressed plants (Fig. 5C).
Unfortunately, little information is available for most of the individual TIR-NBS-LRR genes. Interestingly, RPS6 can be found among the TIR-NBS-LRR genes only regulated under triple stress but not under mild heat and single virus stress. RPS6 has been shown to mediate resistance via EDS1 to the P. syringae pv syringae effector HopA1 (Kim et al., 2009). Furthermore, the deduced amino acid sequence of RPS6 shows highest similarity to the Toll/Interleukin-1 receptor (TIR)-NBS-LRR class resistance protein RAC1 that determines resistance to the oomycete pathogen Albugo candida (Borhan et al., 2004; Kim et al., 2009). Probably, RPS6 is an important component against many pathogens. For viruses, many resistance genes have been described to operate specifically, especially in crop plants (Palukaitis and Carr, 2008). Those resistance genes are assumed to operate as quantitative trait loci and thus function additively in conferring resistance (Maule et al., 2007). Turnip mosaic potyviruses systemically infect susceptible plants (Whitham et al., 2003). Therefore, specific resistance genes might play only a minor role in our virus-plant interaction under multiparallel stress. Nevertheless, as TIR-NBS-LRR genes showed altered transcript gene expression under multiparallel stress situations, they can be assumed to be important for general defense mechanisms against various pathogens. The still uncharacterized genes found to be regulated under different stress situations provide interesting candidates on the way to more resistant plants.
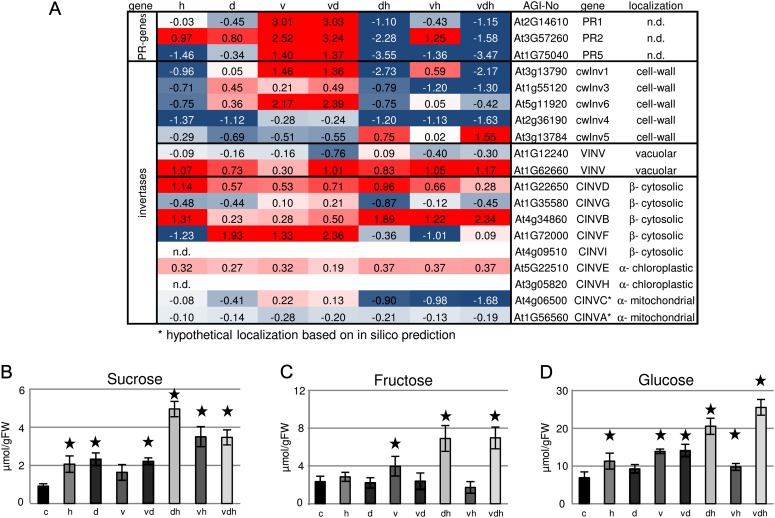
As the R gene-mediated resistance seems to be altered, it was interesting to see how basal defense mechanisms, which normally restrict the growth of virulent pathogens, are influenced under multiparallel stress. An interesting expression pattern for pathogen-related (PR) genes has been found: virus infection led to an up-regulation of PR gene transcripts, including PR1 (9-fold), PR2 (8-fold), and PR5 (3-fold), which was not altered by additional drought stress (Fig. 6A). By contrast, PR genes were down-regulated under combined heat stress situations, which suggest that basal defense is compromised under multiparallel stress conditions. As sugars (hexoses) are known to be signaling molecules leading to an up-regulation of defense gene expression (Herbers et al., 1996), the question arose if the accumulation of Suc and hexoses was involved in defense processes of multiparallel stressed Arabidopsis plants. Suc levels increased under all stress conditions with the exception of single virus treatment (Fig. 6B). In agreement with previous publications, hexoses accumulate in virus-infected leaves. This accumulation is not affected by additional drought stress. Combinations of virus with heat or heat and drought further increased the levels of hexoses (Fig. 6, C and D). Hexose accumulation in virus-infected leaves has been shown to correlate with the induction of cell wall-bound invertase expression and activity (Herbers et al., 2000). Parallel to the up-regulation of PR genes, transcripts encoding cell wall-bound invertase were found to be induced, which is in line with an earlier study speculating that apoplastically released hexoses are sensed and lead to enhanced expression of PR genes (Kocal et al., 2008). By contrast, cell wall-bound invertase transcripts were not up-regulated when heat or heat and drought were applied together with TuMV. This observation is in disagreement with the large amount of hexoses found in leaves harvested from plants subjected to double (heat and virus) and triple stress. However, Suc hydrolysis not only takes place in the cell wall but also occurs in the vacuole, cytosol, and plastids (Roitsch and Gonzalez, 2004). To test whether the failure of accumulated sugars to induce PR gene expression may be caused by their subcellular compartmentation, the expression of vacuolar and neutral invertases was investigated using a previously published classification of Arabidopsis invertase genes (Xiang et al., 2011). As shown in Figure 6A, heat application leads to a strong down-regulation of all major cell wall-bound invertase isoforms. In contrast, the expression of vacuolar and cytosolic invertases is up-regulated (Fig. 6A). This observation suggests that heat treatment alters Suc degradation in leaves and results in intracellular sugar hydrolysis. This would circumvent sugar signals generated in the cell wall compartment and, hence, might explain why heat treatment causes an increased susceptibility of infected plants.
Figure 6.
Altered sugar accumulation and expression of invertase isoforms in multifactorial stressed plants. A, Transcriptional changes of PR genes and invertases expressed in different compartments under different stress conditions. Categorization was done according to data from Xiang et al. (2011). B to D, Levels of Suc, Fru, and Glc in differentially treated plants compared with control plants. Values represent means ± sd of three to four replicates for log2 values of the fold change compared with control plants. FW, Fresh weight. The colors saturate at 1.3-fold change. Red represents an increase and blue represents a decrease in transcript levels. n.d., Not determined. Statistically significant differences from control plants were determined using a two-tailed t test assuming a normal distribution and are indicated by asterisks (P < 0.05). Conditions are as follows: heat (h), drought (d), TuMV (v), drought and heat (dh), TuMV and heat (vh), drought and TuMV (dv), and TuMV, drought, and heat (vdh).
In conclusion, our transcriptome data suggest that infected plants show altered defense mechanisms in combination with abiotic stress components. As basal defense and R-mediated disease resistance was compromised, this might have implications for other pathogens being recognized by R genes.
Different Coexpression Networks Are Active under Different Stress Conditions
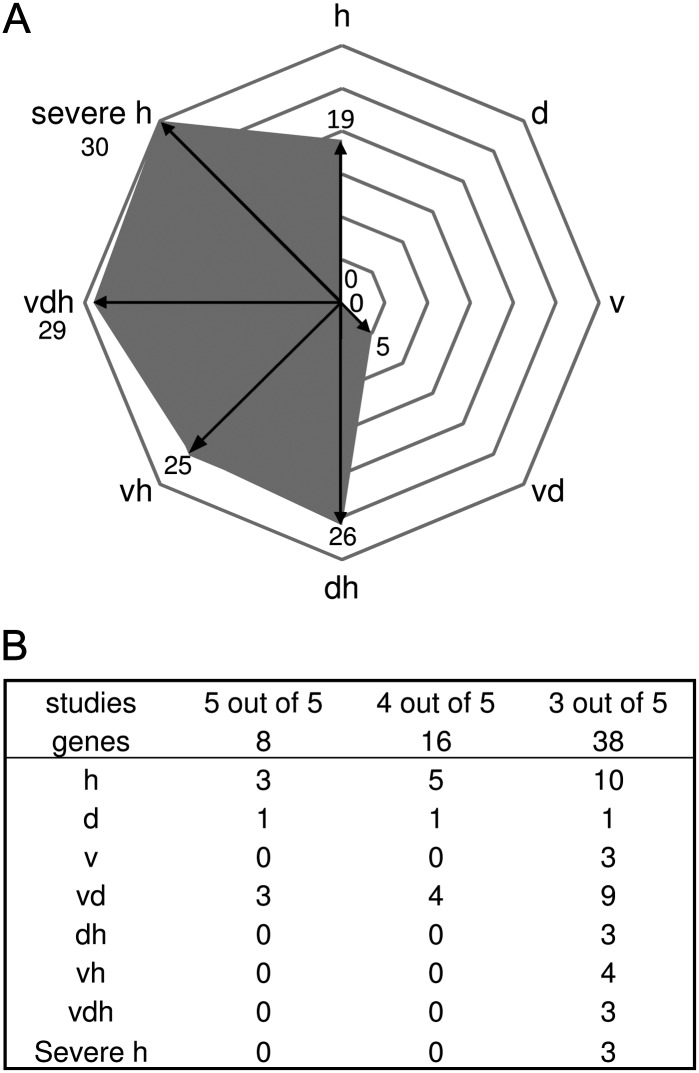
Plants continuously monitor environmental signals to allow adaptations to changing environments. However, little is known about the integration of different stress signals to allow optimal responses. To improve our understanding of signaling networks in plants exposed to multifactorial stress, transcriptional data of genes associated with signaling were compared between differentially treated plants. An enrichment of entities within the category signaling could be detected in the case of drought stress, virus stress, and the combination of both stresses (Table II). The total numbers of regulated genes involved in signaling for each treatment are shown in Figure 7A. Most genes were up-regulated under heat (100), the combination of drought and heat (107), and drought in combination with virus (104). In the case of drought as well as virus stress, still a large number of genes were regulated, 28 and 52, respectively, explaining the enrichment in the category signaling (Table II). Stress responses are often regulated by complex signaling networks (Balbi and Devoto, 2008). In order to illustrate the differences between single and combined stress situations in more detail, coexpression analysis of signaling genes using the ARANET tool (www.functionalnet.org/aranet/) was performed. As a starting point, out of the 100 signaling genes found to be differentially regulated under mild heat stress compared with control plants, quite a large number (54 genes) were found to be coexpressed in one network (Fig. 7B). Among the group of signaling genes in the other stress situations, a similar picture could be observed: quite a large number fell into a network within a given treatment; in the situation of severe heat stress, 31% (30 out of 98), up to 63% in the case of virus stress (33 out of 52) as well as in a combination of virus and drought stress (65 out of 104; Fig. 7C; Supplemental Table S5). Moreover, the overlap between the identified networks between treatments was limited, indicating that different signaling networks are activated. In this context, it is remarkable that between heat and virus networks, 12 genes overlap; however, triple stress remodels the inner network in a way that only one of the virus and only 12 of the heat network members are represented. Interestingly, two transcription factors were found in the overlap between signaling genes under heat and triple stress: LONG HYPOCOTYL IN FARRED (HFR1) and PHYTOCHROME-INTERACTING FACTOR (PIF3). It was recently demonstrated that PIF3 is at least partly responsible for the light-mediated induction of CCA1, through binding directly to the CCA1 promoter (Martinez-Garcia et al., 2000), giving a link for signaling and influence of the circadian clock rhythms. HFR1, a helix-loop-helix transcription factor, is known to be a light signaling component. But interestingly, HFR1 shows a light-dependent accumulation and activity, which is highly temperature dependent (Foreman et al., 2011). The authors showed that HFR1 acts to minimize the potentially devastating effects of elevated temperature on plant metabolism. Obviously, as HFR1 is found under single and multifactorial stress in our analysis, HFR1 probably serves in a general stress response. This is supported by Rasmussen et al. (2013) studying transcriptional changes among different double stresses occurring at the same time. In that study, HFR1 was found in a weighted gene co-expression analysis module described as cold and high light associated.
Figure 7.
Coexpression analysis of signaling genes under single and combined stress conditions. A, Number of differentially regulated genes according to the functional group “signaling” is given for each stress condition. B, Significant regulated signaling genes within the heat network from A visualized in a network of coexpressed genes provided by the ARANET Web tool (www.functionalnet.org/aranet/). C, Table shows the number of treatment-specific signaling genes found to be coexpressed within one treatment (shaded) and the number of commonly shared genes compared with coexpressed signaling genes of other stress conditions (not shaded). Conditions are as follows: heat (h), drought (d), TuMV (v), drought and heat (dh), TuMV and heat (vh), drought and TuMV (dv), TuMV, drought, and heat (vdh), and severe heat (severe h).
As virus-induced signaling genes were not enriched under triple stress and an enhanced amount of viral RNA has been detected (see above), TuMV-specific networks were obviously deactivated, which might have caused the down-regulation of signaling components relevant for innate immunity. Interestingly, no signaling gene was shared between all treatments (Fig. 7C). By contrast, we also detected genes in the overlap of virus and different abiotic stress combinations, indicating a common role in both biotic and abiotic stress signaling.
Further distribution analysis between signaling genes under severe heat stress revealed that 50% of the genes are shared with that of triple stress, but the others are unique to severe heat application, indicating partially shared genes but also specific signaling processes between triple and severe stress. Our results suggest that each stress treatment activates a distinct signaling network, which changes according to the specific conditions. These changes may explain why specific combinations of abiotic or biotic stress act synergistically or antagonistically, respectively.
The Expression of Genes Indicative of a Cytosolic Protein Response Is Elevated under Triple and Severe Heat Stress
Heat stress results in altered protein homeostasis, which leads to the induction/activation of complex regulatory networks of transcription factors (HSFs). This causes the induction of molecular chaperones that, among other effects, prevent protein aggregation and target misfolded proteins for degradation (Alzhanova et al., 2001; Sugio et al., 2009). Molecular triggers for these heat-shock responses are unfolded proteins that accumulate in the cytosol or ER, leading to CPR or UPR, respectively. Transcriptional changes induced by CPR are mainly regulated by HSFA2. Overexpression of HSFA2 in transgenic Arabidopsis plants revealed 46 up-regulated target genes compared with untransformed control plants under ambient growth conditions (Nishizawa et al., 2006). These target genes can be taken as molecular markers for CPR. To investigate whether CPR marker genes are up-regulated under the different stress conditions applied in our study, the expression of HSFA2 target genes was analyzed (Supplemental Table S6). This analysis revealed only a few of the CPR marker genes being up-regulated under mild heat stress. The expression of CPR marker genes increased under combined or severe heat stress conditions (Fig. 8A). Triple stress as well as severe heat stress led to the up-regulation of 29 and 30 out of 46 CPR marker genes, respectively. An earlier study revealed that combinations of drought and virus infections resulted in additive effects (Clover et al., 1999). Nevertheless, heat is obviously the major trigger for elevated CPR, but the level of CPR seems to be enhanced by the severity of stress. TuMV replication occurs in the cytosol of infected plant cells and is dependent on different host factors, including host chaperones (Ahlquist et al., 2003). In previous studies, we could show that TuMV and other potyviruses require functional HSP40/HSP70 complexes for replication (Hofius et al., 2007; Hafren et al., 2010; Jungkunz et al., 2011). Interestingly, virus infection triggers the induction of HSPs in the absence of heat stress (Aranda et al., 1996; Whitham et al., 2003). It has been speculated that this induction is due to the massive protein production observed during late stages of viral infection. These newly synthesized proteins compete for endogenous chaperones, probably resulting in an increased accumulation of unfolded proteins triggering CPR (Mayer and Bukau, 2005). In a combination of heat and virus infections, enhanced chaperone expression might be expected, providing a potential advantage for viral replication. This is further supported by data provided from Aparicio et al. (2005). The authors showed that a generic response to ecotypic protein expression and accumulation, involving chaperones, is induced by high-level production of proteins in the cytosol. This response can be due to enhanced viral replication and the production of virus-specific proteins or other processes promoting increased protein expression. Systemic viral spread requires cell-to-cell transport via plasmodesmata. Since chaperones are also involved in protein transport processes (Sullivan et al., 2000), the induction of these proteins might accelerate viral movement and, hence, systemic infections.
Figure 8.
Expression pattern of CPR and UPR genes of plants exposed to single stress compared with multifactorial or severe heat stress. A, Diagram shows the distribution of HSFA2-regulated genes identified by Nishizawa et al. (2006) under single and multifactorial stress. B, Distribution of tunicamycin-regulated genes under different stress situations. Genes of five studies describing tunicamycin-regulated genes (Martinez and Chrispeels, 2003; Noh et al., 2003; Kamauchi et al., 2005; Iwata et al., 2008, 2010) were compared with genes differentially regulated under single and combined stress. Conditions are as follows: heat (h), drought (d), TuMV (v), drought and heat (dh), TuMV and heat (vh), drought and TuMV (dv), TuMV, drought, and heat (vdh), and severe heat (severe h).
Based on the above described results, CPR is enhanced under combined or severe stress conditions. Heat shock responses are not only triggered by CPR but may also occur due to ER stress. ER stress results from the accumulation of misfolded proteins in the ER (Howell, 2013), which induce the UPR. By treating plants with chemicals influencing protein folding in the ER, the UPR can be induced (Patil and Walter, 2001). One such agent is tunicamycin, which is known to be an inhibitor of N-glycosylation (Koizumi et al., 1999). Previously, tunicamycin-induced transcriptional changes have been studied in Arabidopsis using an Affymetrix GeneChip (Martinez and Chrispeels, 2003; Noh et al., 2003), a fluid microarray (Kamauchi et al., 2005), and Agilent Arabidopsis 2-Oligo microarrays (Iwata et al., 2008, 2010). We compared these studies and identified eight commonly tunicamycin-regulated genes (Supplemental Table S7). As the overlap between these studies was quite low, it is uncertain whether these genes can be classified as UPR markers. Therefore, we looked at genes only found in some of the considered studies and compared the expression of up-regulated genes under the different stress conditions applied in our study (Fig. 8B; Supplemental Table S7). Of the eight conserved tunicamycin-inducible genes, three (38%) were found to be up-regulated under moderate heat stress and the combination of virus and drought stress. By contrast, none of the conserved tunicamycin-inducible genes could be found under multifactorial or severe heat stress. An analysis of genes found to be regulated in three out of five tunicamycin studies revealed 10 out of 38 genes to be up-regulated under mild heat stress. This number decreased when moderate heat stress was combined with drought (three up-regulated genes), virus (four up-regulated genes), or in the triple stress experiment (three up-regulated genes). Interestingly, also severe heat stress reduced the number of tunicamycin-induced genes (three up-regulated genes). The same tendency can be found analyzing only two out of five studies, indicating a general trend of reduction from mild heat stress-induced UPR genes (approximately 30%) to only approximately 10% regulated genes in the case of multifactorial or severe heat stress. Heat stress-induced UPR has been reported by Liu and Howell (2010). So far, no correlation between the expression of UPR genes and the severity of the applied heat stress has been published. Although it seems difficult to define UPR marker genes, these data give, to our knowledge, a first hint that moderate heat stress results primarily in the UPR, while severe heat stress shifts the stress response to the CPR. The observed up-regulation of UPR genes by TuMV is in agreement with earlier studies showing that potato virus X infection of N. benthamiana results in mild ER stress, resulting in an enhanced UPR (Ye et al., 2011).
Depending on the severity of stress, the UPR can either lead to autophagy or cell death (for review, see Howell, 2013). In our study, few autophagy genes were found to be up-regulated, indicating that this process may not be triggered under our experimental conditions. In this context, it is worth mentioning that senescence-regulated genes were also not enriched under any stress situation (Supplemental Fig. S11).
In summary, the expression of UPR-related genes seems to be negatively affected by combined stress conditions, while the expression of marker genes of CPR is strongly induced under multifactorial and severe stress conditions, which might support viral replication and increased susceptibility of host plants.
CONCLUSION
In this study, we developed a controlled experimental system to apply single, double, and triple stress to Arabidopsis plants. To the best of our knowledge, this is this first study investigating molecular responses of plants to triple stress. This study revealed an inhibition of heat-induced stomata opening by viruses, suggesting that viruses might negatively affect heat responses in Arabidopsis. Transcript and metabolite profiling allowed us to distinguish all stress treatments. Thus, molecular and biochemical responses to single, double, and triple stress are specific. From this analysis, specific and common molecular processes can be identified. Down-regulation of primary carbon metabolism was observed under all stress conditions and seems to be a general stress response. By contrast, genes involved in plant defense showed significantly different expression patterns when compared between the different treatments. Especially, the expression of TIR-NBS-LRR genes differed in number and isoform specificity. Interestingly, multiple stresses led to a shift in expression of Suc hydrolytic enzymes, which potentially interfered with sugar signaling. Analysis of genes encoding signaling components revealed a stress-specific regulation of gene networks. Combined heat and drought stress as well as severe heat stress led to enhanced expression of CPR genes, whereas the expression of UPR-related genes declined with increasing stress. In multifactorial stressed plants, fewer TIR-NBS-LRR genes were found to be differentially expressed, indicating different regulatory mechanisms induced under single or combined stresses. Additionally, signaling components were found to be reduced under multiparallel stress, probably deactivating TuMV-specific signaling networks.
MATERIALS AND METHODS
Plants and Growth Conditions
Three days after vernalization in darkness at 4°C, Arabidopsis (Arabidopsis thaliana) plants (ecotype Columbia) were grown on soil under short-day conditions (8 h of light, 16 h of dark), 60% humidity, and 22°C (day) or 18°C/19°C (night). Twelve days after germination, young seedlings were transferred to single plant pots containing 160 g of soil. Plants were watered with defined volumes of water (see below). Further plant cultivation was implemented in a plant climate chamber (Plant-Master PGR 3045; CLF Plant Climatics) that guarantees a diurnal rhythm of 12 h of light at approximately 80 µmol m−2 s−1 and exactly 60% humidity at day and night. Growth chambers used in this experiment were in a similar state, same time and age. The temperature regime followed the light/dark cycle with 22°C and 18°C. Three weeks after germination, plants were treated with the virus TuMV (see below). Control plants were treated with water to equalize damaging effects on locally infected leaves. Eight days later, approximately 85% of the young Arabidopsis leaves, which had been infected systemically, showed typical virus symptoms. The obviously infected plants were exposed to drought stress by reducing water amounts (see below). After 2 d of mild drought influence, heat stress was applied by increasing temperature for 3 d (see below). Plants exposed to the different stress treatments were randomly mixed and distributed throughout the growth chamber. Harvest of plant material started 4 h before the end of the light period and was completed before the start of the dark period. To avoid systematic errors, plants of different stress treatments were sampled in a randomized fashion. The triple stress experiment has been repeated two times in independent plant cultivations. For the time-course experiment, Arabidopsis plants were exposed to severe heat stress.
Application of Mild Drought Stress
Mild drought stress was controlled by using soil moisture content measurements. To determine the volumetric water content, a Decagon Devices sensor was used to measure the dielectric constant of water. This constant is very different from the surrounding soil components, which allows a direct correlation between dielectric constant and water content and was subsequently used for field capacity measurements. The correlation between certain amounts of soil and the corresponding field capacity values resulted in a calibration curve. To define an upper limit, the field capacity of soil moisture content was set to 100%. Therefore, plant pots were watered to saturation and measured. By contrast, the lowest field capacity was given by drying 160 g of soil overnight, reweighing, and measuring field capacity values. Field capacity values in between were determined starting from 160 g of soil and watering with 10 mL. According to the calibration curve, well-watered plants were watered according to 55% of field capacity with a deviation of 10%, whereas mild drought stress was given by 30% of field capacity with a deviation of 10%. A progressive drought stress procedure was initiated by withholding water. Water loss was monitored by weighing each pot every day and watering after the lower limit of mild drought stress was achieved. Control plants were watered daily to maintain the soil humidity at the field capacity.
Application of Heat Stress
Mild heat stress was applied by increasing air temperature to 32°C during the day and 28°C during the night for 3 d. Increased air temperature of 37°C during the day and 33°C during the night for 3 d guaranteed severe heat stress in an independent experiment. Humidity of 60% was not changed, and water was not restricted for plants without drought stress.
Application of Mild TuMV Stress
TuMV infection was performed by inoculating the two largest leaves with TuMV-infected plant material ground in 5 mm sodium phosphate buffer (pH 7.2). In addition, silicon carbide powder was used to damage the leaf surface, ensuring a successful virus penetration. After a few minutes, the leaves were washed with water.
Sampling and RNA Extraction
Thirty-four days after germination, the fresh weights of whole rosettes of differentially treated plants were determined, and individual systemically infected Arabidopsis leaves were harvested and immediately frozen in liquid nitrogen. Leaves from five to eight plants were grouped for one biological replicate. For each treatment, four biological replicates were sampled. After sampling, material was stored at −80°C. In parallel, 10 plants of each condition were dried at 80°C for 3 d for determination of plant dry weight. As the control plants of the triple stress experiment and the severe heat experiment had comparable controls, data have been combined in Table I. For the time-course experiment, leaves of stressed and control plants were harvested at the beginning of the day (0 h), followed by sampling leaves every 4 h and at the end of the day (12 h). Isolation of total RNA from 80 mg of frozen leaf material was performed after the method published by Logemann et al. (1987). Four biological replicates were used for both microarray hybridization and metabolite analysis.
Microarray Hybridization
RNA quality and quantity were tested with an Agilent RNA 6000 Nano Chip on an Agilent 2100 BioAnalyzer (version B.02.03 BSI307) following by Agilent RNA 6000 Nano Assay Protocol2. The synthesis of complementary DNA (cDNA) and following antisense copy RNA were performed according to the one-color microarray-based gene expression analysis protocol provided by Agilent including the one-color RNA spike-in kit (Agilent Technologies). After fragmentation, Cy3-labeled samples were loaded on the array (Agilent Arabidopsis V4, design no. 21169) and hybridized overnight (17 h/65°C). After washing, slides were scanned on the Agilent Microarray Scanner with extended dynamic range at high resolution.
Data Extraction and Analysis
After importing text files via feature extraction software (version 11.7.1; Agilent Technologies) into GeneSpring GX 11.0 (Silicon Genetics), microarray data were log2 transformed followed by normalization to the 75th percentile and corrected to the median of all samples. Only one virus-treated sample did not pass the quality control parameter and was excluded from further analysis. A one-way ANOVA (P ≤ 0.05; variance assumed as equal) enabled us to select genes that show differential behavior and also pronounced similarities within the groups. Finally, hierarchical clustering of genes was performed using a Euclidian single algorithm. A total of 27,870 genes remained, and a volcano plot was applied to identify statistically significant (P ≤ 0.05; equal variances assumed), more than 2-fold differentially expressed genes between two conditions, including the multiple test correction of Benjamini and Hochberg (1995). The selected features were divided into up- and down-regulated groups. Up-regulated features have been marked as positive values, and down-regulated ones as negative values. Functional category assignment was performed by using MAPMAN bins (http://mapman.gabipd.org/web/guest/mapman). Therefore, the percentage of significantly regulated features from a specific functional group was determined relative to the percentage of features from that specific group to the entire chip. Venn diagrams containing up to three lists were created by the Agilent GeneSpring GX11.0 program, whereas higher numbers of lists were compared using a software provided by the University of Ghent (http://bioinformatics.psb.ugent.be/webtools/Venn/). Data were visualized by VANTED (Junker et al., 2006) and the Multiexperiment Viewer (Saeed et al., 2003). PCA of transcriptome data was performed by Analyst 1.5 software using the Pareto algorithm. For coexpression analysis, lists of significantly regulated genes were filtered for categories of specific interest (e.g. signaling), and networks were determined by input of these selected lists into the ARANET Web tool (http://www.functionalnet.org/aranet/; Lee et al., 2010).
qPCR
For quantitative detection of transcripts, real-time PCR analyses were performed using the Mx3000P qPCR system (Stratagene) using Brilliant II SYBR Green QPCR master mix (Stratagene). Template cDNA was diluted. Primers and temperature cycles for the detection of viral mRNA were used as described by Kim et al. (2010). Expression values were normalized to the expression of tubulin as an internal control. TOC1 and CCA1 transcript abundance was measured in each sample relative to an ACTIN control. Gene-specific primers used in this study were as follows: CCA1 forward primer, 5′-ACGGGTGTGAATGATGGAA-3′; CCA1 reverse primer, 5′-TGCTTGCGTTTGATGTCTCT-3′; TOC1 forward primer, 5′-ACCAACCCACAGAGAGGAAA-3′; TOC1 reverse primer, 5′-GGTGAGACCCAGCAAGATG-3′; ACTIN forward primer, 5′-GCCAACAGAGAGAAGATGACCCAGA-3′; and ACTIN reverse primer, 5′-ACACCATCACCAGAGTCCAACACAAT-3′. The efficiency value of amplification for each set of primers was determined beforehand by measuring the abundance of transcripts from a cDNA dilution series. Relative expression of transcripts was calculated by MxPro version 4.10 software (Stratagene).
Measurement of Water Potential
Arabidopsis leaf water potential was measured with a C-52 sample chamber plus the PSYPRO eight-channel water potential datalogger (WESCOR). Leaf discs from the differentially stressed plants were placed in the sample chamber, and after 10 min of temperature and water vapor equilibrium, measurements were done by the dew-point method using the following parameters: cooling time, 10 s; plateau delay, 5.6 s; read average time, 2.4 s; measurement period, 10 s.
Metabolite Extraction, Measurement, and Analysis
Organic acids and phosphorylated intermediates were extracted and measured after a protocol previously published by Horst et al. (2010). Measurements of major soluble sugars and starch were performed after the method described by Jones et al. (1977). For PCA, the log2 fold change, relative to the control, was determined and used for further analysis. Statistical analysis of metabolites was performed with the VANTED software (Junker et al., 2006) using the integrated Welch-Satterthwaite t test. PCA of metabolite data was performed by Analyst 1.5 software using the Pareto algorithm.
Array data from this article can be found in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE46760.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PCA of transcriptome profiles of triple stressed plants.
Supplemental Figure S2. PCA of transcript and metabolite data.
Supplemental Figure S3. Metabolome profiles of multifactorial stressed plants.
Supplemental Figure S4. Identification of stress-responsive genes.
Supplemental Figure S5. Transcriptional changes involved in the light reaction of photosynthesis under stress conditions.
Supplemental Figure S6. Transcriptional changes involved the circadian clock under different stress conditions.
Supplemental Figure S7. qPCR analysis of CCA1 and TOC1 in Arabidopsis plants exposed to severe heat during the day.
Supplemental Figure S8. Effects of single and multifactorial stress situations on primary carbon and nitrogen metabolism.
Supplemental Figure S9. Transcriptional and metabolite changes of the TCA cycle under different stress treatments.
Supplemental Figure S10. Transcriptional changes of genes associated with the electron transport chain in mitochondria under different stress conditions.
Supplemental Figure S11. Transcripts with involvement in senescence in plants exposed to multiparallel stress.
Supplemental Table S1. Influence of single and multifactorial stress in Arabidopsis plant water potential.
Supplemental Table S2. Numbers of statistically significant identified entities of each stress condition in the multiparallel stress experiment and the severe heat stress experiment compared with their respective controls.
Supplemental Table S3. Specific regulated features for each stress situation.
Supplemental Table S4. TIR-NBS-LRR genes differentially regulated in each stress situation in the multiparallel stress experiment.
Supplemental Table S5. Signaling genes differentially regulated in each stress situation.
Supplemental Table S6. Differentially regulated HSF2A regulated in each stress situation.
Supplemental Table S7. Tunicamycin-regulated differentially regulated genes for each stress situation.
Acknowledgments
We thank Dr. Sophia Sonnewald and Stephen Reid for realization of the microarray experiments as well as Dr. Joerg Hofmann and Alfred Schmiedl for giving excellent advice regarding metabolite and amino acid measurements. We also thank Kirsten Ott and Martina Lang for performing qPCR analysis. We are grateful to Dr. Urte Schlueter for excellent advice in the transcriptional analysis and to Anita Reiser for supporting plant cultivation.
Glossary
- TMV
Tobacco mosaic virus
- TSWV
Tomato spotted wilt virus
- HSP
heat shock protein
- HSF
heat shock factor
- TuMV
Turnip mosaic virus
- PCA
principal component analysis
- RT
reverse transcription
- ABA
abscisic acid
- qPCR
quantitative PCR
- TCA
tricarboxylic acid
- AOX
alternative oxidase
- TIR
Toll/Interleukin1 receptor
- NBS
nucleotide-binding site
- LRR
leucine-rich repeat
- PR
pathogen-related
- UPR
unfolded protein response
- cDNA
complementary DNA
References
- Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. (2003) Host factors in positive-strand RNA virus genome replication. J Virol 77: 8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja I, de Vos RC, Bones AM, Hall RD. (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15: 664–674 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Alzhanova DV, Napuli AJ, Creamer R, Dolja VV. (2001) Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO J 20: 6997–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio F, Thomas CL, Lederer C, Niu Y, Wang D, Maule AJ. (2005) Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol 138: 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda MA, Escaler M, Wang D, Maule AJ. (1996) Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc Natl Acad Sci USA 93: 15289–15293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio-Ibáñez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L. (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148: 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, De Vleesschauwer D, Höfte M. (2008) Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol Plant Microbe Interact 21: 709–719 [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Urwin PE. (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63: 3523–3543 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Massonnet C, Walsh S, Baginsky S, Bühlmann P, Hennig L, Hirsch-Hoffmann M, Howell KA, Kahlau S, Radziejwoski A, et al. (2012) Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol Syst Biol 8: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbi V, Devoto A. (2008) Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol 177: 301–318 [DOI] [PubMed] [Google Scholar]
- Bechtold U, Lawson T, Mejia-Carranza J, Meyer RC, Brown IR, Altmann T, Ton J, Mullineaux PM. (2010) Constitutive salicylic acid defences do not compromise seed yield, drought tolerance and water productivity in the Arabidopsis accession C24. Plant Cell Environ 33: 1959–1973 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Subramaniam R, Dangl JL. (2004) Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr Opin Plant Biol 7: 391–399 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA. (2008) Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol 147: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhan MH, Holub EB, Beynon JL, Rozwadowski K, Rimmer SR. (2004) The arabidopsis TIR-NB-LRR gene RAC1 confers resistance to Albugo candida (white rust) and is dependent on EDS1 but not PAD4. Mol Plant Microbe Interact 17: 711–719 [DOI] [PubMed] [Google Scholar]
- Browder LE. (1985) Parasite-host-environment specificity in the cereal rusts. Annu Rev Phytopathol 23: 201–222 [Google Scholar]
- Carter AH, Chen XM, Garland-Campbell K, Kidwell KK. (2009) Identifying QTL for high-temperature adult-plant resistance to stripe rust (Puccinia striiformis f. sp. tritici) in the spring wheat (Triticum aestivum L.) cultivar ‘Louise’. Theor Appl Genet 119: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Chaves MM, Flexas J, Pinheiro C. (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot (Lond) 103: 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Clover GRG, Smith HG, Azam-Ali SN, Jaggard KW (1999) The effects of drought on sugar beet growth in isolation and in combination with beet yellows virus infection. J Agric Sci 133: 251–261 [Google Scholar]
- Colhoun J. (1973) Effects of environmental factors on plant disease. Annu Rev Phytopathol 11: 343–364 [Google Scholar]
- Dangl JL, Jones JD. (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Dropkin VH. (1969) Necrotic reaction of tomatoes and other hosts resistant to Meloidogyne: reversal by temperature. Phytol 59: 1632–1637 [Google Scholar]
- Ferrier T, Matus JT, Jin J, Riechmann JL. (2011) Arabidopsis paves the way: genomic and network analyses in crops. Curr Opin Biotechnol 22: 260–270 [DOI] [PubMed] [Google Scholar]
- Foreman J, Johansson H, Hornitschek P, Josse EM, Fankhauser C, Halliday KJ. (2011) Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J 65: 441–452 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett KA, Dendy SP, Frank EE, Rouse MN, Travers SE. (2006) Climate change effects on plant disease: genomes to ecosystems. Annu Rev Phytopathol 44: 489–509 [DOI] [PubMed] [Google Scholar]
- Gechev TS, Hille J. (2012) Molecular basis of plant stress. Cell Mol Life Sci 69: 3161–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland A, Singh DP, Hayward JM, Moore CA, Murphy AM, York CJ, Slator J, Carr JP. (2003) Genetic modification of alternative respiration has differential effects on antimycin A-induced versus salicylic acid-induced resistance to Tobacco mosaic virus. Plant Physiol 132: 1518–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel AK, Lundberg D, Torres MA, Matthews R, Akimoto-Tomiyama C, Farmer L, Dangl JL, Grant SR. (2008) The Pseudomonas syringae type III effector HopAM1 enhances virulence on water-stressed plants. Mol Plant Microbe Interact 21: 361–370 [DOI] [PubMed] [Google Scholar]
- Göhre V, Robatzek S. (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol 46: 189–215 [DOI] [PubMed] [Google Scholar]
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. (2008) Metabolomics of temperature stress. Physiol Plant 132: 220–235 [DOI] [PubMed] [Google Scholar]
- Hafrén A, Hofius D, Rönnholm G, Sonnewald U, Mäkinen K. (2010) HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Hitz WD. (1982) Metabolic responses of mesophytes to plant water deficits. Annu Rev Plant Physiol Plant Mol Biol 33: 163–203 [Google Scholar]
- Harb A, Krishnan A, Ambavaram MM, Pereira A. (2010) Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol 154: 1254–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare PD, Cress WA. (1996) Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul 21: 79–102 [Google Scholar]
- Hartl FU, Hayer-Hartl M. (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Herbers K, Meuwly P, Frommer WB, Metraux JP, Sonnewald U. (1996) Systemic acquired resistance mediated by the ectopic expression of invertase: possible hexose sensing in the secretory pathway. Plant Cell 8: 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K, Takahata Y, Melzer M, Mock HP, Hajirezaei M, Sonnewald U. (2000) Regulation of carbohydrate partitioning during the interaction of potato virus Y with tobacco. Mol Plant Pathol 1: 51–59 [DOI] [PubMed] [Google Scholar]
- Hofius D, Maier AT, Dietrich C, Jungkunz I, Börnke F, Maiss E, Sonnewald U. (2007) Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus Y infection in tobacco plants. J Virol 81: 11870–11880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst RJ, Doehlemann G, Wahl R, Hofmann J, Schmiedl A, Kahmann R, Kämper J, Sonnewald U, Voll LM. (2010) Ustilago maydis infection strongly alters organic nitrogen allocation in maize and stimulates productivity of systemic source leaves. Plant Physiol 152: 293–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SH. (2013) Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol 64: 2.1–2.23 [DOI] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J 22: 5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel I, Pantin F, Sulpice R, Piques M, Rolland G, Dauzat M, Christophe A, Pervent M, Bouteillé M, Stitt M, et al (2010) Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol 154: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC (2008) Climate change and water. In BC Bates, ZW Kundzewicz, J Palutikof, S Wu, eds, Technical Paper of the Intergovernmental Panel on Climate Change. IPCC Secretariat, Geneva, p 210 [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N. (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Nishino T, Takayama S, Koizumi N. (2010) Characterization of a plant-specific gene induced by endoplasmic reticulum stress in Arabidopsis thaliana. Biosci Biotechnol Biochem 74: 2087–2091 [DOI] [PubMed] [Google Scholar]
- Jakob U, Gaestel M, Engel K, Buchner J. (1993) Small heat shock proteins are molecular chaperones. J Biol Chem 268: 1517–1520 [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones MG, Outlaw WH, Lowry OH. (1977) Enzymic assay of 10−7 to 10−14 moles of sucrose in plant tissues. Plant Physiol 60: 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SH, Lee JY, Lee DH. (2003) Use of SAGE technology to reveal changes in gene expression in Arabidopsis leaves undergoing cold stress. Plant Mol Biol 52: 553–567 [DOI] [PubMed] [Google Scholar]
- Jungkunz I, Link K, Vogel F, Voll LM, Sonnewald S, Sonnewald U. (2011) AtHsp70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J 66: 983–995 [DOI] [PubMed] [Google Scholar]
- Junker BH, Klukas C, Schreiber F. (2006) VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamauchi S, Nakatani H, Nakano C, Urade R. (2005) Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J 272: 3461–3476 [DOI] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim BM, Suehiro N, Natsuaki T, Inukai T, Masuta C. (2010) The P3 protein of turnip mosaic virus can alone induce hypersensitive response-like cell death in Arabidopsis thaliana carrying TuNI. Mol Plant Microbe Interact 23: 144–152 [DOI] [PubMed] [Google Scholar]
- Kim SH, Kwon SI, Saha D, Anyanwu NC, Gassmann W. (2009) Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol 150: 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Király L, Hafez YM, Fodor J, Király Z. (2008) Suppression of tobacco mosaic virus-induced hypersensitive-type necrotization in tobacco at high temperature is associated with downregulation of NADPH oxidase and superoxide and stimulation of dehydroascorbate reductase. J Gen Virol 89: 799–808 [DOI] [PubMed] [Google Scholar]
- Kocal N, Sonnewald U, Sonnewald S. (2008) Cell wall-bound invertase limits sucrose export and is involved in symptom development and inhibition of photosynthesis during compatible interaction between tomato and Xanthomonas campestris pv vesicatoria. Plant Physiol 148: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeda S, Hosokawa M, Kang BC, Tanaka C, Choi D, Sano S, Shiina T, Doi M, Yazawa S. (2012) Defense response of a pepper cultivar cv. Sy-2 is induced at temperatures below 24°C. J Plant Res 125: 137–145 [DOI] [PubMed] [Google Scholar]
- Koizumi N, Ujino T, Sano H, Chrispeels MJ. (1999) Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol 121: 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RD, Crafts-Brandner SJ. (1999) Inhibition and acclimation of photosynthesis to heat stress is closely correlated with activation of ribulose-1,5-bisphosphate Carboxylase/Oxygenase. Plant Physiol 120: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Ambaru B, Thakkar P, Marcotte EM, Rhee SY. (2010) Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat Biotechnol 28: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Yun HS, Kwon C. (2012) Molecular communications between plant heat shock responses and disease resistance. Mol Cells 34: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lin X, Chen A, Peterson T, Ma K, Bertzky M, Ciais P, Kapos V, Peng C, Poulter B. (2013) Global priority conservation areas in the face of 21st century climate change. PLoS ONE 8: e54839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163: 16–20 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH. (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck J, Spackman M, Freeman A, Trebicki P, Griffiths W, Finlay K, Chakraborty S. (2011) Climate change and diseases of food crops. Plant Pathol 60: 113–121 [Google Scholar]
- Madgwick JW, West JS, White RP, Semenov MA, Townsend JA, Turner JA, Fitt BDL. (2011) Impacts of climate change on wheat anthesis and fusarium ear blight in the UK. Eur J Plant Pathol 130: 117–131 [Google Scholar]
- Magnani E, Sjölander K, Hake S. (2004) From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell 16: 2265–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez IM, Chrispeels MJ. (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH. (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Maule AJ, Caranta C, Boulton MI. (2007) Sources of natural resistance to plant viruses: status and prospects. Mol Plant Pathol 8: 223–231 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters HG, Devlin PF. (2011) Timing in plants—a rhythmic arrangement. FEBS Lett 585: 1474–1484 [DOI] [PubMed] [Google Scholar]
- Mittler R, Blumwald E. (2010) Genetic engineering for modern agriculture: challenges and perspectives. Annu Rev Plant Biol 61: 443–462 [DOI] [PubMed] [Google Scholar]
- Mittler R, Finka A, Goloubinoff P. (2012) How do plants feel the heat? Trends Biochem Sci 37: 118–125 [DOI] [PubMed] [Google Scholar]
- Mohr PG, Cahill DM. (2003) Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct Plant Biol 30: 461–469 [DOI] [PubMed] [Google Scholar]
- Moury B, Selassie KG, Marchoux G, Daubèze A, Palloix A. (1998) High temperature effects on hypersensitive resistance to Tomato spotted wilt tospovirus (TSWV) in pepper (Capsicum chinense Jacq.). Eur J Plant Pathol 104: 489–498 [Google Scholar]
- Naika M, Shameer K, Mathew OK, Gowda R, Sowdhamini R. (2013) STIFDB2: an updated version of plant stress-responsive transcription factor database with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in Arabidopsis and rice. Plant Cell Physiol 54: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Noh S-J, Kwon CS, Oh D-H, Moon JS, Chung W-I. (2003) Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311: 81–91 [DOI] [PubMed] [Google Scholar]
- Palukaitis P, Carr JP. (2008) Plant resistance to viruses. J Plant Pathol 90: 153–171 [Google Scholar]
- Pastenes C, Pimentel P, Lillo J. (2005) Leaf movements and photoinhibition in relation to water stress in field-grown beans. J Exp Bot 56: 425–433 [DOI] [PubMed] [Google Scholar]
- Patil C, Walter P. (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol 13: 349–355 [DOI] [PubMed] [Google Scholar]
- Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O. (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S. (2008) Temperature perception and signal transduction in plants. New Phytol 179: 615–628 [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J. (2013) Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol 161: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero RM, Shulaev V, Blumwald E. (2009) Cytokinin-dependent photorespiration and the protection of photosynthesis during water deficit. Plant Physiol 150: 1530–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R. (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 130: 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R. (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T, González MC. (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci 9: 606–613 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Serrano M, Parra S, Alcaraz LD, Guzmán P. (2006) The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J Mol Evol 62: 434–445 [DOI] [PubMed] [Google Scholar]
- Shen YG, Zhang WK, He SJ, Zhang JS, Liu Q, Chen SY. (2003) An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor Appl Genet 106: 923–930 [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M. (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Sugio A, Dreos R, Aparicio F, Maule AJ. (2009) The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21: 642–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Cantalupo P, Pipas JM. (2000) The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol Cell Biol 20: 6233–6243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR. (2006) The association among gene expression responses to nine abiotic stress treatments in Arabidopsis thaliana. Genetics 174: 1811–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Silhavy D, Molnár A, Havelda Z, Lovas A, Lakatos L, Bánfalvi Z, Burgyán J. (2003) Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J 22: 633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T, Kato W, Asada Y, Sako K, Sato T, Sonoda Y, Kidokoro S, Yamaguchi-Shinozaki K, Tamaoki M, Arakawa K, et al. (2009) DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J Plant Res 122: 633–643 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Giraud E, Clifton R, Whelan J. (2009) Alternative oxidase: a target and regulator of stress responses. Physiol Plant 137: 354–361 [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C. (2008) Proline accumulation in plants: a review. Amino Acids 35: 753–759 [DOI] [PubMed] [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L. (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Wang Y, Bao Z, Zhu Y, Hua J. (2009) Analysis of temperature modulation of plant defense against biotrophic microbes. Mol Plant Microbe Interact 22: 498–506 [DOI] [PubMed] [Google Scholar]
- Weston DJ, Gunter LE, Rogers A, Wullschleger SD. (2008) Connecting genes, coexpression modules, and molecular signatures to environmental stress phenotypes in plants. BMC Syst Biol 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham SA, Quan S, Chang HS, Cooper B, Estes B, Zhu T, Wang X, Hou YM. (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J 33: 271–283 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CE, Li Y, Labbe A, Guevara D, Nuin P, Whitty B, Diaz C, Golding GB, Gray GR, Weretilnyk EA, et al (2006) Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol 140: 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Le Roy K, Bolouri-Moghaddam MR, Vanhaecke M, Lammens W, Rolland F, Van den Ende W. (2011) Exploring the neutral invertase-oxidative stress defence connection in Arabidopsis thaliana. J Exp Bot 62: 3849–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Yang Y. (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Chen F, Mannas JP, Feldman T, Sumner LW, Roossinck MJ. (2008) Virus infection improves drought tolerance. New Phytol 180: 911–921 [DOI] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Hassidim M, Green RM. (2007) CIRCADIAN CLOCK ASSOCIATED1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol 145: 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Dickman MB, Whitham SA, Payton M, Verchot J. (2011) The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol 156: 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wise RR, Struck KR, Sharkey TD. (2010) Moderate heat stress of Arabidopsis thaliana leaves causes chloroplast swelling and plastoglobule formation. Photosynth Res 105: 123–134 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qian W, Hua J. (2010) Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog 6: e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]