Abstract
Because spontaneous mutation is the source of all genetic diversity, measuring mutation rates can reveal how natural selection drives patterns of variation within and between species. We sequenced eight genomes produced by a mutation-accumulation experiment in Drosophila melanogaster. Our analysis reveals that point mutation and small indel rates vary significantly between the two different genetic backgrounds examined. We also find evidence that ∼2% of mutational events affect multiple closely spaced nucleotides. Unlike previous similar experiments, we were able to estimate genome-wide rates of large deletions and tandem duplications. These results suggest that, at least in inbred lines like those examined here, mutational pressures may result in net growth rather than contraction of the Drosophila genome. By comparing our mutation rate estimates to polymorphism data, we are able to estimate the fraction of new mutations that are eliminated by purifying selection. These results suggest that ∼99% of duplications and deletions are deleterious—making them 10 times more likely to be removed by selection than nonsynonymous mutations. Our results illuminate not only the rates of new small- and large-scale mutations, but also the selective forces that they encounter once they arise.
Keywords: copy-number variation, natural selection, mutation rates
BECAUSE all genetic variation on which natural selection acts originates via spontaneous mutation, the rate at which various types of mutations appear in natural populations has important consequences for the manner in which organisms evolve. Once we determine the rate at which adaptive and deleterious alleles arise, we can understand how natural selection shapes the spectrum of variation within and between species (Lynch et al. 2008). Mutation-accumulation (MA) experiments are the most widely used method for directly measuring spontaneous mutation rates (Halligan and Keightley 2009). In these experiments genetically identical individuals are subdivided into initially homogeneous sublines, and these sublines are maintained over many generations. At each generation a minimum number of individuals are randomly selected to reproduce and propagate the MA subline, thereby limiting the ability of natural selection to purge new deleterious mutations or to favor new adaptive alleles. Because the MA sublines are initially identical (barring any contamination or residual heterozygosity), any genetic differences among lines must arise via mutations occurring during the course of the experiment.
Mutation-accumulation experiments were initially used to assess the phenotypic impacts of mutation (e.g., Mukai et al. 1972). More recently, mutation-accumulation has been used to assess the rate of spontaneous mutation on the genome at specific loci (Mukai and Cockerham 1977), selected genomic regions (Haag-Liautard et al. 2007), or, with the advent of next-generation sequencing technology, genome-wide. Mutation accumulation is now used to estimate genome-wide estimates of per-site mutation rates in a variety of eukaryotes, including Saccharomyces cerevisiae (Lynch et al. 2008), Drosophila melanogaster (Keightley et al. 2009), Caenorhabditis elegans (Denver et al. 2009, 2012), Chlamydomonas reinhardtii (Ness et al. 2012; Sung et al. 2012), and Arabidopsis thaliana (Ossowski et al. 2010). These studies do not suffer from the biases affecting indirect estimates of the mutation rate (e.g., based on the rate of synonymous divergence between species) or estimates based on only a few loci. However, the relatively small number of generations and/or mutations captured by some of these experiments limits their statistical power.
In addition, most of these studies used sequencing methods that do not allow for comprehensive detection of large genomic duplications and deletions. Recent studies have found that such large-scale mutations can have dramatic evolutionary and phenotypic consequences. Comparative genomic studies have revealed that changes in gene copy number caused by duplication and deletion events are often adaptive (Demuth et al. 2006) and suggest that gene duplication events can result in the evolution of new gene functions [e.g., the excess of dN/dS ratios >1 in young gene duplicates observed in Kondrashov et al. (2002) and Zhang et al. (2003)]. In addition, large duplications and deletions segregating within populations, often referred to as copy-number variants (CNVs), have been shown to be both beneficial (Perry et al. 2007; Turner et al. 2008; Kolaczkowski et al. 2011) and deleterious (McCarroll and Altshuler 2007; Stankiewicz and Lupski 2010; Girirajan et al. 2011); these polymorphisms are widespread in Drosophila (Emerson et al. 2008; Langley et al. 2012). It is therefore essential for our understanding of the rate of adaptive evolution and the origin of genomic disorders that we accurately measure the spontaneous rates of these mutations. Although several studies examining either MA lines (Lynch et al. 2008; Ossowski et al. 2010; Lipinski et al. 2011) or human parent-offspring trios (Itsara et al. 2010) have been able to estimate the rate of large duplication and deletion events, these studies were able to reliably detect events only several kilobases in length due to their reliance on low-resolution detection methods (e.g., microarray hybridization, sequence read depth, or pulsed-field gel electrophoresis).
Here, we present results from an MA experiment conducted in D. melanogaster. We performed mutation accumulation in two nearly homozygous lines obtained by brother–sister mating from a single outbred lab population (Houle and Nuzhdin 2004). The use of inbred lines can have effects on mutation rate either through the fixation of rare recessive alleles during the initial construction of the lines (perhaps more likely to increase mutation rates) or through the effects of homozygosity on the mutation process itself (perhaps more likely to decrease mutation rates; see the Appendix). The replicate sublines derived from each line were then maintained by close inbreeding with an effective population size slightly >2; population genetic theory suggests that our design will miss the small number of mutations with drastic fitness effects in the homozygous state, but will capture the vast majority of mutations that have small-to-negligible fitness effects. We discuss the nature of the biases introduced by our design in further detail below.
Our results capture dozens of large-scale copy-number changes and hundreds of point mutations and small indels. Roughly 1160 generations of mutation accumulation are assayed in this experiment—more than previous sequence-based MA studies in Drosophila. The resultant wealth of data reveals a substantial difference in point mutation rates between two inbred lines, each derived from the same outbred population, suggesting that mutation rates may vary substantially among individuals within a species. These data also confirm the previous finding that mutation events affecting several nonadjacent base pairs in close proximity to one another occur quite frequently (Schrider et al. 2011). In addition, the high quality of our sequence data allows for accurate and direct genome-wide inference of the spontaneous rates of large duplications and deletions. Although the mutation rates of small indels support previous claims that the D. melanogaster genome is experiencing mutational pressure toward contraction (Petrov et al. 1996, 1998; Petrov 2002; Leushkin et al. 2013), we observe more duplicated than deleted base pairs per generation when considering mutations of all sizes, suggesting the possibility of a mutational tendency toward net genomic expansion. By comparing our mutation data to polymorphism data from D. melanogaster, we estimate that ∼90% of nonsynonymous mutations and ∼99% of new duplications and deletions are deleterious enough to be quickly removed from natural populations by purifying selection.
Materials and Methods
DNA preparation, sequencing, and mapping
DNA was extracted from eight D. melanogaster MA sublines derived from two inbred lines originating from a laboratory population founded from flies captured in Massachusetts in 1975 [sublines 33-45, 33-27, 33-55, 33-5 derived from line 33 and sublines 39-58, 39-67, 39-51, and 39-18 derived from line 39 (Houle and Nuzhdin 2004)]. These sublines were among those examined by Haag-Liautard et al. (2007) at a limited number of loci. For each subline, the Qiagen DNEasy protocol entitled “Purification of Total DNA from Animal Tissues (Spin-Column Protocol)” was used, including the optional RNase A step. The DNA extracted from these sublines was then multiplexed and sequenced in one flow cell of the Illumina Genome Analyzer II at the Indiana University Center for Genomics and Bioinformatics. The average paired-end insert size was 175 bp, with 72 bp sequenced on each end after removing barcode sequences. These sequence data are available for download on the National Center for Biotechnology Information Short Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under experiment ID SRX285615. The reads were mapped to release 5 of the D. melanogaster reference genome assembly using version 0.6.2 of BWA (Li and Durbin 2009) with the mismatch parameter (-n) set to 0.1. This parameter sets the maximum fraction of mismatches between a mapped read and the reference genome for an alignment to be retained. These alignments were used to search for mutations and indels as described below. To facilitate detection of large duplications and deletions, in which case a larger gap between sequenced ends of the insert is preferable, we mapped the reads again with the same parameters after trimming the inner 36 bp of each read. For detecting single-nucleotide mutations and small indels, we mapped the entire read sequence.
Identifying single-nucleotide mutations
After mapping reads to the reference genome, SAM (sequence alignment map) format mapping output was run through version 0.1.9 of the SAMtools (Li et al. 2009) pipeline for calling variants using the pileup command. When the -c option is used, this command displays all potential differences between the sequenced individual and the reference genome. We did not impose a mapping quality cutoff for read alignments before running the pileup command to search for variants. Instead, we attempted to distinguish between true differences from the reference genome and false positives after using the pileup command by retaining only putative mutations with at least five reads mapped to the genome, having at least one read on each strand, and having <10% of reads exhibiting a nucleotide other than the putative mutant base. This approach is similar to that of Keightley et al. (2009), which was shown to have near-perfect accuracy.
We searched for mutations unique to one of the MA sublines, as these differences could represent mutations arising during mutation accumulation rather than genetic polymorphism present in the sequenced sublines prior to MA. This was done by examining all reads mapping to the mutant position in all other MA sublines derived from the same inbred line and by ensuring that each of these sublines had at least five reads mapped to the position, with no more than 10% of these reads exhibiting the mutant nucleotide. Additionally, if 10% or more of all reads from these sublines exhibited a base other than the ancestral nucleotide, the putative mutation was thrown out. To calculate the mutation rate for each of the two inbred lines, the number of mutations found in the four MA sublines derived from the same parental line was divided by the number of positions having at least five aligned reads in each of these four sublines. Since one of the MA sublines derived from line 39 (39-67) was highly similar to line 33 along the distal end of chromosome 2R and most of chromosome X due to contamination, we disregarded mutations in these regions in subline 39-67. To prevent contamination in other genomic regions from affecting our mutation rate estimates, we also disregarded all mutations for which the mutant base appeared in any other MA line (whether derived from the same ancestral line or not). These steps may have caused us to miss a very small fraction (<1%) of true mutation events: those occurring at sites already exhibiting the mutant base as a polymorphism in the other ancestral line. Confidence intervals on the mutation rate were estimated by assuming a binomial distribution.
Identifying small indels
To find small indel mutations occurring during the MA experiment, we ran all gapped read alignments through the program Dindel (Albers et al. 2011). As with single-nucleotide mutations, for both inbred lines we looked only at sites having at least five reads in each MA subline. Indels called by Dindel were counted as mutations occurring during MA if >80% of reads in the mutant subline supported the indel, if not all of these reads were on the same strand, and if no more than one of each of the seven other sublines exhibited any read alignments supporting an indel within 20 bp of the putative mutation. In addition, read alignments (in pileup format) in the vicinity of each putative mutation were examined in each of the eight MA sublines to ensure that the indel was unique to one subline. In some cases, a precipitous drop in read depth either at the precise location of the indel or within a couple of base pairs was observed in each of the other three sublines derived from the same inbred line. In these cases, read depth dropped to < ∼5× in each of the other sublines, and often to 0, 1, or 2×. This observation is consistent with reads containing the indel failing to be aligned rather than being aligned with a gap, and the candidate indel was rejected. To estimate the number of indels that we may have missed, we compared the ratio of single nucleotide mutations to indels in our study with the ratio found in these same lines using denaturing high-performance liquid chromatography (DHPLC), which detects indels with high sensitivity (Haag-Liautard et al. 2007).
Validating point mutations and indels with Sanger sequencing
Single-nucleotide mutations, indels, complex mutations, and multinucleotide mutations (MNMs) were amplified via PCR after designing primers using Primer3 (Rozen and Skaletsky 2000). PCR products were cleaned using an Exo-SAP protocol, and sequencing reactions were run using ABI BigDye Terminator Cycle Sequencing Kit, and sequences were produced at the Indiana Molecular Biology Institute using the ABI3730. Sequences were then mapped to the reference genome using BLAST, and alignments were examined to ensure that the mutant line possessed the putative mutant genotype. In some cases, small stretches of repetitive sequence near indel mutations resulted in alignments in which the indel mutation could have been placed in one of several locations. Thus, while our validation results suggest that the vast majority of our putative indels are true mutations, the reported coordinates of these mutations may be inexact.
Calculating the probability of observing apparent multinucleotide mutations and complex mutations caused by independent mutation events
To calculate the probability of observing apparent multinucleotide mutation events (two or more mutations found within 50 bp of one another within the same MA subline) under the assumption of independent mutation events, we randomly permuted the coordinates of the mutations observed in our study, counted the number of such clusters observed, and repeated this simulation 10,000 times. To perform a similar calculation for complex mutation events (defined as the observation of an indel located within 50 bp of another mutation occurring within the same subline), we performed 10,000 simulations permuting the locations of mutations and indels and counted the number of apparent complex mutations observed.
Identifying large deletion events
When paired-end sequencing is performed, the range of expected insert sizes is typically relatively narrow. However, when paired ends are derived from an individual harboring a deletion, read pairs spanning the deleted locus will appear much farther apart than expected from the insert-size distribution when mapped to the reference genome, provided the region deleted from the sequenced individual is present in the reference genome (see figure 1 in Tuzun et al. 2005). We used this approach to detect large deletions in our MA lines, first calculating each subline’s insert-size distribution by examining all read pairs mapped in proper orientation and within 500 bp of one another. Inserts in the 99th percentile of this distribution were kept as potentially indicative of deletions.
Figure 1.
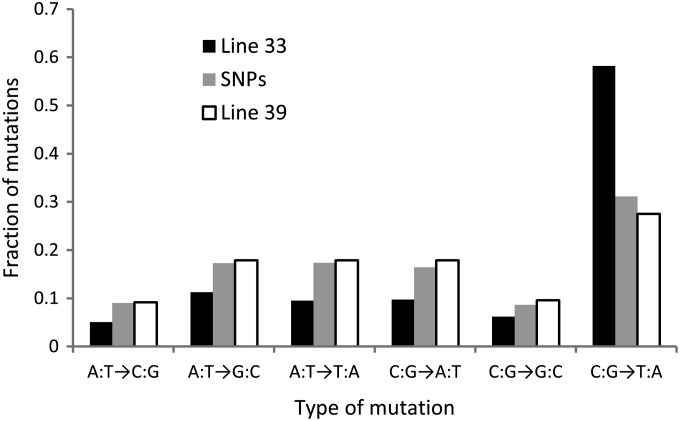
Spectra of point mutations in the two MA lines and low-frequency (<0.1) single-nucleotide polymorphisms from Langley et al. (2012).
To find deletions supported by multiple inserts (or read pairs), these “distant” inserts were clustered as follows. First, each insert was assigned to its own cluster. Then we searched for pairs of clusters, (Ci,Cj), that appeared to correspond to the same deletion. This was considered to be the case if at least 75% of all pairs of inserts from the two clusters (comparing the mapping coordinates of one insert from Ci to one from Cj) were close to one another (the sum of the distance between the leftmost ends of the two inserts and the distance between the rightmost ends of the two inserts must be <200 bp) and suggestive of a deletion of roughly the same size (<50 bp difference in the inferred sizes of the two inserts). These cutoffs were chosen because an examination of insert-size distributions revealed that few pairs of inserts spanning the same deletion would be expected to violate them. If these criteria were met for enough pairs of inserts from the two clusters, then the two clusters were combined into one new cluster. This process was repeated until no more clusters could be merged. Once clustering was completed, each cluster was examined to remove any read pairs that do not support the putative deletion. This was done by comparing the insert in the cluster with the median insert size to all other read pairs, and any read pairs that were mapped too far from this insert (again, if the sum of the distance between the leftmost ends of the inserts and the distances between the rightmost ends of the inserts ≥200 bp) or did not support a deletion of the same size (≥50-bp difference in insert sizes) were removed from the cluster.
Given our relatively high sequencing coverage, we would expect a deletion to be spanned by multiple inserts. We therefore removed all clusters with only one read pair from further consideration. Since we would expect all deletions occurring during mutation accumulation to be homozygous and therefore to have very low read depth, we calculated the average read depth of each possible deletion (examining all positions not masked by RepeatMasker between the end position of the rightmost forward-mapped read in the cluster and the starting position of the leftmost reverse-mapped read in the cluster) in each MA subline, after correcting read depth for a given subline according to the mean read depth across the genome relative to the genome-wide means across all eight MA sublines. We then removed all deletions for which the average read depth of the MA subline containing the putative deletion was greater than one-half of the average depth of the other seven MA sublines. To remove polymorphisms present prior to mutation accumulation, we removed all clusters from further analysis that closely matched two or more “distant” read pairs from the other seven MA sublines (using the same criteria for comparing read pairs during clustering). To ensure that our approach was not missing any potential new large deletions, we looked for regions containing stretches of zero read depth in only one of the MA sublines. This approach added two additional potential deletions >100 bp in length. The breakpoints of the final set of deletions were determined by manually examining read depth and noting where regions of zero read depth began and ended. The true breakpoints of these deletions are probably no more than a few base pairs away from these estimates.
Since our insert-size distributions had relatively low variance, we were able to detect a few deletions using this method that were as small as the largest indels detected by gapped alignment (two deletions <40 bp in length). One cannot conclude from this that we have adequate power to detect deletions of any size, however, as our insert-size-based method has lower power to detect events near or below 50 bp in length. In addition, several indels removed from our final set were deletions of several dozen base pairs present in multiple sublines (i.e., polymorphisms segregating prior to the experiment) but not called in these individuals and discovered only after manual inspection; this observation suggests that, given our read lengths, gapped alignment has reduced power to detect indels more than a dozen or so base pairs in length. Thus, we may not have adequate power to detect all deletions of between ∼10 and 50 bp.
Identifying large duplication events
Given that the vast majority of duplication polymorphisms in Drosophila are tandem (Emerson et al. 2008; Cridland and Thornton 2010), we used an approach to find tandem duplications using read pairs mapped near one another but in an incorrect orientation (−/+ instead of +/−; see figure 3 in Cooper et al. 2008). These read pairs were then clustered using an approach similar to that for deletions described above, with two clusters merged if there existed a pair of inserts, one from each cluster, such that the sum of the distances between the left reads and the distances between the right reads was <200 bp. We required only one pair of inserts to meet this criteria (instead of 75% of all pairs examined for deletions) because the paucity of −/+ (“everted”) inserts relative to distant read pairs in proper orientation implies that the false-discovery rate of such inserts is far lower than for distant read pairs and that the need for stringency is therefore reduced. Once clustering was completed, any such cluster of everted read pairs found to closely match a cluster from another MA subline was removed from further consideration to remove polymorphism present prior to MA. Two everted read-pair clusters from different MA sublines were considered to support the same duplication if the sum of the distance between the leftmost read mappings of the two clusters and the distance between the rightmost mappings of these clusters was <200 bp, and the difference in inferred lengths of the putative duplications (calculated by subtracting the leftmost mapping position of the read-pair cluster from the rightmost position in the cluster) was <100 bp.
Figure 3.
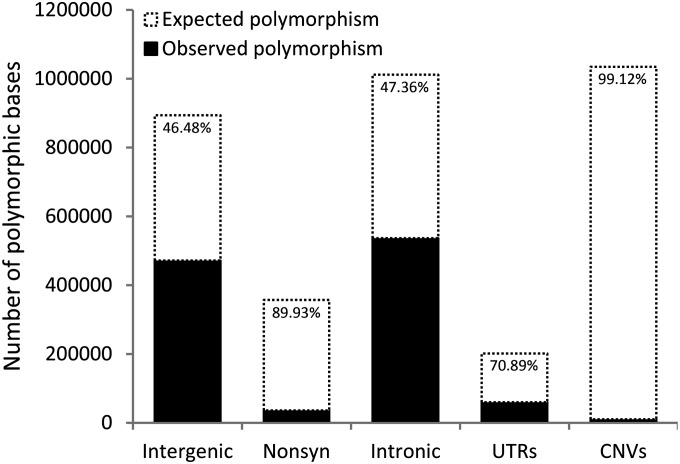
Expected and observed numbers of single-nucleotide polymorphisms and base pairs lying within CNVs. The solid bars show the amount of polymorphism observed within each category [polymorphisms with observed minor allele frequency ≥0.15 from Langley et al. (2012)], and the dashed-border bars show the expectation inferred from observed mutations and synonymous SNPs. The difference between the two bars reveals the fraction of mutations that are eliminated by purifying selection. The numbers of expected and observed nonsynonymous SNPs and SNPs within UTRs, introns, and intergenic regions are shown, as are the expected and observed numbers of base pairs varying in copy number (which have been rescaled by dividing by 100 for comparison with SNPs). The estimated fraction of new mutations removed by purifying selection in each category is shown within the dashed portions of the bars.
As a final filtering step for remaining clusters, we examined average read depths along the stretch of sequence spanned by the cluster in each of the MA sublines (as with deletions, ignoring regions masked by RepeatMasker and correcting read depths for a given subline based on its genome-wide average relative to the genome-wide average across all sublines). The ratio of the average read depth within the MA subline containing the putative duplication and the average read depth of all other sublines was taken. If the duplication were truly unique and homozygous, this ratio would be expected to be ∼2.0. Therefore, all unique everted clusters for which this ratio was <1.5 were removed from the set of potential duplications. The clusters that remained were subjected to PCR validation as described below.
PCR validation of large duplications and deletions
PCR validation was performed using DNA extracted from the eight MA sublines in the same manner as described above to confirm or reject 8 putative deletions and 19 putative duplications. To confirm deletions, primers were designed outside of the span of the deletion, facing inward, so that individuals containing the deletion would yield a small band and individuals lacking the deletion would yield a notably larger band. To confirm duplications, we designed outward-facing primers within the duplicated region (following Emerson et al. 2008), which would yield a small band for a tandem duplication and no band if the duplication were absent. Primer pairs were designed by extracting sequence from the appropriate regions of the reference genome and placed in the desired order, and then Primer3 (Rozen and Skaletsky 2000) with the Drosophila mispriming library was used to select optimal primers from this sequence.
We attempted to amplify each putative mutation in both the MA subline believed to possess the mutation and a control subline derived from the same inbred line. Deletions that yielded the same length band for the putative mutant sample and the control sample were rejected, while those that yielded a larger band for the control were considered to be confirmed. Duplications were considered to be confirmed if the mutant MA subline yielded an ∼300-bp band and the control subline yielded no band. For both duplications and deletions, if neither the mutant subline nor the control yielded a band, the experiment was repeated. Duplications failing to yield a band for either sample after two attempts could represent nontandem duplications. However, since such duplications are relatively uncommon in Drosophila (Emerson et al. 2008), and we predicted that these were all tandem based on everted paired-end reads, we rejected these.
Assessing evidence for mutational bias between X and autosomes
To determine the largest difference in mutation rates between the X chromosome and the autosomes that we could detect given our data, we performed χ2 tests assessing the goodness of fit of the total number of single-nucleotide mutations that we observed to null models with varying degrees of bias. The degrees of bias tested ranged from a 0.1% increase or decrease of the mutation rate on the X chromosome to a 100% increase or decrease in mutation rate on the X. The largest mutational bias (either an excess or a deficit) to yield a P-value >0.05 was considered to be the largest mutational bias consistent with our data. This approach was also used to determine the largest deficit of mutations occurring within exonic sequence that remained consistent with the observed distribution of single-nucleotide mutations.
Correction for missed small indels
We observed that the ratio of small indels to single-nucleotide mutations was 0.24 for Haag-Liautard et al. (2007), but only 0.082 in our study. This suggests either that we may have missed some small indels or that previous estimates were erroneously high. In either case, to be conservative when comparing the net number of base pairs added by large duplications and deletions to the net number of base pairs removed by small indels each generation, we assumed that we missed two-thirds of all small indels and corrected for this by multiplying the net number of base pairs removed by such mutations each generation by three.
While this correction is quite crude, we feel it nonetheless yields a useful first approximation to the number of base pairs affected by small indels over the course of our MA experiment, especially given that the length distribution of small indel events reported here appears quite similar to that of Haag-Liautard et al. (2007). This correction is primarily performed to compare the number of base pairs affected by small indels to the number of base pairs affected by large duplications and deletions. Given that even after this correction we find that the net effect of large duplications and deletions adds >15 times as many base pairs as are removed by small indels, it seems quite unlikely that the correction is inaccurate enough to qualitatively change the result of that comparison (see below).
Testing the significance of the observed excess of duplicated bases
To assess the significance of the observed excess of duplicated base pairs over deleted base pairs during the course of the MA experiment, we simulated sets of duplications and deletions with the numbers of each type of event drawn from a Poisson distribution with a mean equal to the observed number of events of that type. Once the number of each type of event was selected, lengths were randomly selected with replacement from the observed length distributions of the two types of mutations, and the total number of base pairs within each type of mutation was calculated.
Results and Discussion
Single-nucleotide mutation and small indel rates
To estimate the mutation rate in D. melanogaster, we sequenced genomic DNA from eight MA sublines descended from two inbred lines, from which 20 Mbp were previously examined by Haag-Liautard et al. (2007). The inbred lines (33 and 39) were derived from a long-term laboratory population, IV, originally collected from Amherst, Massachusetts, in 1975 (Houle and Rowe 2003). Four MA sublines were founded from each of these inbred lines (yielding eight sublines in total), and then each was subjected to 145–149 generations of mutation accumulation (Houle and Nuzhdin 2004). Sequence was obtained using the Illumina Genome Analyzer II. We mapped reads from these sublines to the D. melanogaster reference genome (Adams et al. 2000) and searched for mutations present in only one of the four sublines derived from each inbred line (Materials and Methods). Mutations occurring during mutation accumulation would be expected to meet this criterion, while polymorphism segregating prior to the MA experiment (and differing from the reference genome) would probably be fixed during the inbreeding that preceded MA and would therefore be present in all sublines.
In total, 21.4 Gbp of paired-end sequence summed across all sublines was mapped to the five major chromosome arms, for an average of 22.5× sequencing coverage per subline. Our approach (verified here with PCR and Sanger sequencing) allowed us to accurately detect point mutations, as well as almost all duplications and deletions that occurred during the course of the MA experiment (Materials and Methods). From examining the 96.5% of the major chromosome arms covered by at least five reads in each of the four MA sublines derived from each progenitor (114,966,662 positions in line 33 and 114,955,079 positions in line 39), we estimate that the rate of single-nucleotide mutation averaged over both progenitor lines is 5.49 × 10−9 per site per generation (95% confidence interval (C.I.): 5.10 × 10−9 to 5.90 × 10−9). This estimate is based on 732 observed single-nucleotide mutations (Supporting Information, Table S1). We performed Sanger sequencing to validate 14 of these mutations picked at random and confirmed all of them, implying that very few of our mutations are false positives due to sequencing error or misalignment and that our rate estimates are accurate. Previous estimates of the per-site mutation rate in selected genomic regions in these same MA lines using denaturing high-performance liquid chromatography rather than high-throughput sequencing (Haag-Liautard et al. 2007) did not differ significantly from those reported here.
In addition to point mutations, we were able to detect small (≤40 bp) indels occurring during mutation accumulation (Materials and Methods). The 60 indels that we detect are listed in Table S2. Again, we Sanger-sequenced 26 of these indels, confirming all of them. Consistent with previous results in D. melanogaster (Haag-Liautard et al. 2007), we find that small deletions arise at a significantly higher rate than small insertions (P = 1.6 × 10−7). The approximately fivefold excess of small deletions relative to small insertions is also in agreement with previous studies finding evidence for a deletion bias in dead-on-arrival transposable elements in the D. melanogaster genome (e.g., Petrov et al. 1996).
We do not find any variation in mutation rates among chromosome arms (P = 0.269 for point mutations; P = 0.106 for indels; χ2 tests). There is also no significant difference in mutation rates between the X chromosome and the autosomes (5.09 × 10−9 on the X vs. 5.58 × 10−9 on the autosomes for single-nucleotide mutations; P = 0.367; 1.82 × 10−10 on the X vs. 2.35 × 10−10 on the autosomes for indels; P = 0.618; Fisher’s exact tests), although we can confidently rule out only an elevation in mutation rate on the X >10.3% or a deficit >24.7% (the limit of our statistical power; Materials and Methods). In addition to our relatively low power, even a relatively large sex bias in mutation rates will result in a more subtle bias when comparing mutation bias between the X and the autosomes. For example, if the mutation rate in males were double the rate in females, then the overall mutation rate on the autosomes would be 1.5-fold greater than the rate measured from females alone, while the overall rate on the X would be 1.33-fold greater than that measured from females alone; this corresponds to a rate difference of only 11.3% between the X and autosomes. In addition, it is likely that the average effect of a deleterious mutation on the X chromosome is slightly magnified in hemizygous males, potentially biasing our recovery of mutations on the X. We therefore cannot rule out sex-specific mutation rates in D. melanogaster (Bachtrog 2008), especially given that our experimental design forced mating at a young age, and the disparity in mutation rates between the sexes could increase with age.
Genetic variation in the mutation rate
Because our eight MA lines were derived from two different ancestral lines, we are able to compare mutation rates between these two genetic backgrounds. We estimate that the rate of single-nucleotide mutations in line 33 is 7.71 × 10−9 per site per generation (95% C.I.: 7.06 × 10−9 to 8.40 × 10−9) and the rate in line 39 is 3.27 × 10−9 (95% C.I.: 2.85 × 10−9 to 3.73 × 10−9). A similar trend was observed in a previous study using denaturing high-performance liquid chromatography rather than high-throughput sequencing (Haag-Liautard et al. 2007). However, this previous study detected only 37 mutations and therefore did not have enough power to confirm the significance of the difference in mutation rates between these lines. We now have adequate power to conclude with high confidence that line 33 has a mutation rate at least twice as high as line 39 (P = 2.71 × 10−4; goodness-of-fit test based on binomial distribution). Indels also occur at a significantly higher rate in line 33 than in line 39 (4.95 × 10−10 for deletions and 1.20 × 10−10 for insertions in line 33; 2.55 × 10−10 for deletions and 0.30 × 10−10 for insertions in line 39; P = 0.0062).
The difference in mutation rate between the two ancestral lines is largely due to G:C→A:T transitions, which occurred over five times as often in line 33 as in line 39; this disparity is highly significant (P < 2.2 × 10−16; Table 1). The difference in the rate of G:C→A:T mutations was also seen in the previous study of these lines that used a different mutation-detection technology (Haag-Liautard et al. 2007). Given that our estimate of the mutation rate for line 39 is also very similar to estimates previously derived from unrelated MA lines (Haag-Liautard et al. 2007; Keightley et al. 2009), it seems likely that one or more alleles conferring an elevated rate of G:C→A:T transitions—and segregating in the base population prior to the MA experiment—are present in line 33 but absent from line 39.
Table 1. Six types of point mutations occurring during mutation accumulation.
| Mutations | Line 33 | Line 39 |
|---|---|---|
| A:T→C:G | 26 | 20 |
| C:G→G:C | 32 | 21 |
| A:T→T:A | 49 | 39 |
| C:G→A:T | 50 | 39 |
| A:T→G:C | 58 | 39 |
| C:G→T:A | 299 | 60 |
To search for mutations conferring such a change in G:C→A:T transition rates, we examined all differences between line 33 and line 39 likely to affect the function of genes involved in DNA replication and repair. Given that any genetic determinant of the difference in mutation rates between line 33 and line 39 was almost certainly present prior to mutation accumulation, we examined sites exhibiting the same allele in each MA subline from line 33 and an alternative allele in each subline from line 39. In particular, we looked for polymorphisms likely to affect the function of genes that could affect mutation rates. To do this, we examined mutations in genes identified as being important for DNA damage repair (Ravi et al. 2009), particularly those mutations resulting in premature termination codons, frameshifts, and deletions of large portions of coding sequence. While we found many genes having different nonsynonymous alleles in the two inbred lines, we did not find any inactivating mutations in genes known to be involved in processes related to G:C→A:T transitions.
It has recently been suggested that the accumulation of deleterious mutations, as occurs during MA, could increase mutation rates (Sharp and Agrawal 2012). To rule out the possibility that the observed more than twofold difference in mutation rates between the two lines is the result of changes in mutation rates occurring during MA, we compared each subline from line 33 to each subline from line 39. We found that each subline derived from line 33 mutates significantly faster than each MA line derived from line 39 (P < 0.05 for each comparison; Fisher’s exact test; Figure S1). Thus, while rate changes during MA may upwardly bias rate estimates from each of our sequenced sublines, they do not explain the difference in mutation rates between line 33 and line 39, as it seems unlikely that deleterious mutator alleles having roughly the same effect would have independently arisen in all four sublines from line 33 at a higher rate than in line 39 by chance. Consistent with our interpretation that the difference in mutation rates is not a result of changes during MA, Keightley et al. (2009) did not observe any difference among lines despite sequencing three MA lines undergoing, on average, nearly 120 more generations of MA than ours. Therefore, we infer that a mutator allele (or alleles) resulting in an elevated mutation rate in line 33 must have been present prior to mutation accumulation.
The evolutionary importance of the mutator allele(s) responsible for the difference in mutation rates between lines depends on how typical such variation is. Because our mutation accumulation took place in inbred lines, it is possible that the differences in mutation rate that we observed are atypical of those among genotypes in natural populations; the differences may be caused by a rare recessive allele. We therefore examined low-frequency SNPs from a natural population of D. melanogaster, finding that the spectrum of these SNPs lies in between the spectra of mutations from lines 33 and 39 with respect to G:C→A:T transitions, although the difference between the SNP data and line 39 is not significant. This comparison is shown in Figure 1, which was generated using polarized SNPs from Langley et al. (2012) with derived allele frequencies <0.1. Because natural selection rarely allows deleterious mutations to reach high derived allele frequencies, low-frequency alleles should represent the class of polymorphisms least biased by natural selection; they therefore should give a more accurate picture of the relative rates of different types of single-nucleotide mutations (Messer 2009). To remove false positives from the polymorphism data, only nonsingleton polymorphisms from Langley et al. (2012) were included in this comparison. Including singletons does not qualitatively affect the results, nor does examining singletons exclusively: the fraction of G:C→A:T transitions in the SNP data remains intermediate between the mutation data from line 33 and line 39. Similarly, we compared our estimates to mutational parameters inferred by Zeng (2010) from synonymous polymorphism data while taking codon bias into account, finding that rate of G:C→A:T mutations inferred from polymorphism data lie in between those estimated from lines 33 and 39 [36.5% of mutations according to Zeng (2010) vs. 27.5% for line 39 and 58.2% for line 33].
The possibility of genetic polymorphism causing substantial variation in mutation rates among individuals within a population is supported by the observation of different rates of lethal and visible mutations among D. melanogaster lines (Ives 1945), as well as the recent observation by Conrad et al. (2011) of different numbers of mutations occurring in two human parent-offspring trios. Moreover, the striking difference in the rate of G:C→A:T transitions between our lines raises the possibility that the expected genomic GC content (the fraction of base pairs that are Gs or Cs) of a species may be shaped by a distribution of mutation rates and spectra segregating within species. Further studies are necessary to reveal whether such variation in mutation rates is widespread in Drosophila and other eukaryotes.
MNM events are common in D. melanogaster
We observed 10 groups of two or three mutations occurring within 50 bp of one another (Figure S2), far more than are expected under the assumption that mutations occur independently of one another (P < 1.0 × 10−4; Materials and Methods). We confirmed each of these groups of mutations using Sanger sequencing, showing that these are true events and not either sequencing errors or mismapped reads from paralogous loci (see below). As shown previously (Averof et al. 2000; Drake 2007; Schrider et al. 2011), this pattern is most likely caused by multinucleotide mutation (MNM) events, in which two or more closely spaced single-nucleotide mutations occur simultaneously. We estimate that these events occur roughly once per haploid genome every 116 generations, averaged across all MA lines. Indeed, we find that 2.79% of single-nucleotide mutations in D. melanogaster are caused by MNMs, consistent with previous estimates from human polymorphism data and MA experiments from a variety of eukaryotes (Denver et al. 2004, 2009; Lynch et al. 2008; Keightley et al. 2009; Ossowski et al. 2010; Schrider et al. 2011). These MNMs are listed in Table S3.
In addition, seven indels occurring during mutation accumulation were found near other indels or nucleotide mutation events—another observation that cannot be explained by independent mutations (P < 1 × 10−4). These seven events (Table S4), each confirmed by Sanger sequencing, are probably the result of so-called “complex mutations,” in which a stretch of nucleotides is replaced by a different stretch of seemingly random nucleotides that differs in length, creating the appearance of multiple indels or a combination of indels and nucleotide changes. Complex mutations are also frequently observed as polymorphisms (Levy et al. 2007) and as new mutations arising during MA experiments (Haag-Liautard et al. 2007) or human parent-offspring trios (Lynch 2010). As with MNMs, the multiple apparent mutations that compose a complex mutation are probably the result of a single event (Schrider et al. 2011). We estimate that these events occur once per haploid genome every 166 generations on average.
Notably, all MNMs and complex mutations were confirmed by Sanger sequencing. Because the Sanger reads confirming these mutations map uniquely to the reference genome, these mutations are not false positives that were called from the Illumina data due to mismapping of paralogous regions in the reference. Such mismapping could also occur if paralogous sequences absent from the reference were present in these lines (i.e., CNVs). If this were the case, then we would expect roughly twice the expected read depth at these sites. However, read depth at these sites appeared to be relatively normal compared to genome-wide averages, with only one mutation cluster exhibiting more than 1.5× the mutant line’s genome-wide average read depth, and none exhibiting at least twice the genome-wide average. Moreover, if these events were artifacts of mismapping from paralogous sequences rather than mutations occurring during MA, then we would expect to observe them in more than one MA line. However, as with all mutations reported in this study, these MNMs and complex mutations were detected in only one MA line. It therefore appears that none of these MNMs or complex mutations are artifacts of mismapping from paralogs.
Both MNMs and complex mutations in principle have the potential to facilitate complex adaptation (Lynch and Abegg 2010) by allowing multiple mutations that are individually deleterious but beneficial to the organism in concert to occur simultaneously. Our results confirm that MNMs and complex mutations occur at a relatively high rate—a necessary (but not sufficient) condition for these mutations to play an important role in adaptive evolution.
Mutation rate estimates for large duplications and deletions
Because we sequenced our MA sublines to high coverage and used paired-end technology, we have the ability to detect deletions and tandem duplications of almost any size with high accuracy; we did not attempt to detect nontandem duplications, as these are relatively rare in Drosophila (Emerson et al. 2008). We found 19 candidate duplications supported by paired-end data with a ratio of read depth of the mutant line to the average read depth of the sublines lacking the putative mutation >1.5 (Materials and Methods); the expectation for this ratio within a duplication is 2.0. We used PCR amplification to determine which of these candidates were true de novo mutations and which were false positives: we confirmed 7 and rejected the remaining 12 after two attempts to amplify the duplication failed (Materials and Methods). Notably, all of the confirmed duplications had read-depth ratios >1.7 and were supported by at least four read pairs. On the other hand, all of the rejected duplications were supported by only a single insert, and only four relatively small candidates (<300 bp) had read-depth ratios >1.7.
Note that for duplications our validation experiments cannot distinguish negative controls lacking the duplication from PCR failures. Thus, our PCR results alone are not sufficient to rule out with certainty the possibility that one or more of the seven duplications in our final set may be false positives. However, the fact that none of these putative duplications were found in the control lines via either PCR or everted read pairs suggests that they are all true new mutations. It is also worth noting that all of the duplications that we confirmed were >900 bp in length. We did detect several putative duplications only a few hundred base pairs in length, but these were all rejected by PCR. This is not surprising, given that smaller stretches are more likely to exhibit a read-depth ratio >1.5 by chance. By the same token, a small true duplication could have a read-depth ratio <1.5 by chance. This could imply that we have a “blind spot” for duplications only several hundred base pairs in length. However, there were only a handful of everted read-pair clusters spanning <1 Kbp and supported by multiple read pairs, so we may not be greatly underestimating the duplication rate.
Similarly, we found 26 candidate deletions supported by paired-end data and having low read depth. We performed PCR validation on two groups of putative deletions: four potential deletions with approximately zero read depth, and another four that had a read depth that was lower than expected (<0.5 times the nonmutant lines) but substantially greater than zero. All four of the deletions with zero read depth were confirmed by PCR, while three of the other potential deletions were rejected, although one could not be amplified after two attempts. Given our rejection of the non-zero-depth deletions, and the low likelihood of new deletions exhibiting such read depth (due to the homozygosity of the sublines), we believe it is reasonable to conclude that apparent deletions with non-zero read depth are all false positives. To ensure that we were not missing any deletions, we searched for stretches of the genome >100 bp in length with zero read depth, finding only two of these not already present in our set of candidate deletions. We added these two events to our final set, although removing them does not qualitatively change any of the results presented below. All together, the results of our experimental validation suggest that we can confidently discriminate between true and false positives for both large deletions and duplications and that our final set contains the majority of these events occurring during MA.
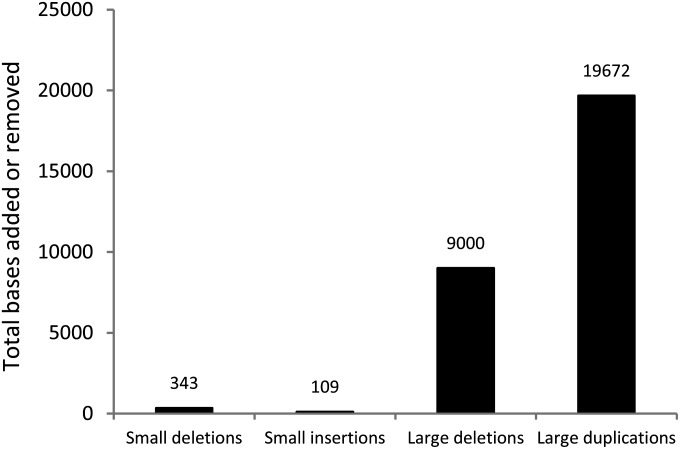
Our final data set consists of 22 large deletions (0.019 events per genome per generation) and 7 duplications (0.00603 events per genome per generation). In contrast to the apparently higher rate at which deletions arise during MA, we found that the average duplication length was much longer than that for deletions (2810 bp vs. 409 bp; P < 2.2 × 10−16). In fact, more than twice as many base pairs were added to the genome via duplication events during the MA experiment (seven events listed in Table 2 duplicating 19,672 bp in total; 16.96 bp per subline per generation) than were removed by deletion (22 events listed in Table 3 deleting 9000 bp in total; 7.76 bp per subline per generation).
Table 2. Duplications occurring during mutation accumulation.
| Mutant subline | Chromosome | Duplication start | Duplication end | Duplication length | Genes completely duplicated | Genes partially duplicated |
|---|---|---|---|---|---|---|
| 33-45 | 2L | 19,885,354 | 19,887,108 | 1755 | sick | |
| 33-27 | 3R | 5,871,296 | 5,874,483 | 3188 | CG12811, mtTFB2 | |
| 33-27 | 3R | 17,830,172 | 17,834,456 | 4285 | Eip93F | |
| 33-27 | X | 16,714,088 | 16,715,991 | 1904 | CG4829 | |
| 33-55 | X | 20,825,924 | 20,826,862 | 939 | ||
| 39-51 | 3R | 23,543,430 | 23,546,747 | 3318 | ||
| 39-51 | X | 2,911,681 | 2,915,963 | 4283 | kirre |
Table 3. Deletions occurring during mutation accumulation.
| Mutant subline | Chromosome | Deletion start | Deletion end | Deletion length | Whole genes deleted | Genes partially deleted |
|---|---|---|---|---|---|---|
| 33-45 | 2R | 3,796,256 | 3,796,317 | 62 | CG30377 | |
| 33-45 | 3L | 3,417,350 | 3,417,631 | 282 | sty | |
| 33-27 | 3L | 14,411,558 | 14,411,626 | 69 | bbg | |
| 33-27 | 3R | 6,264,619 | 6,265,964 | 1346 | ||
| 33-27 | 3R | 6,498,265 | 6,498,580 | 316 | ||
| 33-55 | 2R | 7,443,243 | 7,443,616 | 374 | ||
| 33-55 | 3R | 4,382,412 | 4,382,479 | 68 | ||
| 33-55 | 3R | 24,200,482 | 24,201,119 | 638 | ||
| 33-5 | 2L | 11,219,248 | 11,219,776 | 529 | ab | |
| 33-5 | 2R | 6,803,669 | 6,803,750 | 82 | ||
| 33-5 | 3R | 7,933,175 | 7,935,816 | 2642 | dpr17 | |
| 33-5 | X | 8,157,581 | 8,157,611 | 31 | CG1632 | |
| 33-5 | X | 10,746,256 | 10,746,281 | 26 | CG2202 | |
| 39-58 | 3L | 14,551,937 | 14,551,993 | 57 | ||
| 39-58 | X | 1,337,087 | 1,337,482 | 396 | futsch | |
| 39-58 | X | 3,586,099 | 3,586,393 | 295 | Mnt | |
| 39-51 | 2L | 1,767,992 | 1,768,181 | 190 | ||
| 39-51 | 3L | 3,591,610 | 3,592,045 | 436 | Eip63E | |
| 39-51 | 3L | 4,898,131 | 4,898,234 | 104 | CG13705 | |
| 39-51 | 3L | 9,500,895 | 9,501,385 | 491 | CG3408 | |
| 39-51 | 3L | 14,831,422 | 14,831,703 | 282 | CG17839 | |
| 39-18 | 2L | 2,355,809 | 2,356,092 | 284 | eys, CG9967 |
The net loss of nucleotides due to the excess of small deletions over small insertions discussed above is far too small to overcome the net gain of DNA from large duplications and deletions (Figure 2); even after correcting for any small indels that we may have missed (Materials and Methods), we estimate that the net number of base pairs added by large duplications and deletions each generation is over an order of magnitude greater than the net number of base pairs removed by small indels. This finding suggests that the net loss of base pairs exhibited by smaller indels—first described by Petrov et al. (1996) and reproduced here—could be reversed when larger duplications and deletions are included. Such an excess of duplicated sequence would have been missed by earlier approaches: by examining the mutations accumulating within dead-on-arrival transposable elements, Petrov et al. (1996) were limited to seeing only mutations that were smaller than the transposable element itself. However, the excess of duplicated base pairs that we observed is based on a relatively small number of events and is not statistically significant (P = 0.10; one-tailed test described in Materials and Methods); more data are required to confirm this observation. We may also be underestimating the rate of DNA gain per generation as we do not detect new transposable element insertions, and we cannot detect mutations that occur and then subsequently revert to their original state. This could downwardly bias our estimates of the rate of duplications relative to deletions, as the latter have little opportunity for reversion.
Figure 2.
The total number of bases added or removed over the entire course of the MA experiment by indels, large duplications, and large deletions.
As noted above, the fixation probability of new variants is strongly affected by their effects on fitness. While the vast majority of single base-pair mutations can be assumed to have small-to-negligible fitness effects, the same cannot be said for larger duplications and deletions. Consequently, if the fitness consequences of duplications differ from deletions, our estimates could be biased. Thus, an alternative explanation for the apparent difference in rate would be that deletions are more likely to have large deleterious fitness effects than duplications. For example, because mutations that result in sterility in either sex or lethality are not detected in our experiment, our comparison of deletions and duplications will be biased if deletions are more likely to have these effects. While it is estimated that at least 500 genes can cause male sterility in D. melanogaster (Wakimoto et al. 2004), it is not known if deletions are more likely to cause sterility than duplications. On the other hand, it is well known that deletions have more severe viability effects on D. melanogaster than duplications, that larger duplications have stronger effects on viability than smaller duplications, and that homozygous deletions of genes tend to be nonviable (Ashburner et al. 2012). These possible differences must be kept in mind when interpreting our results because new mutations are maintained in the homozygous state, so even recessive mutations with large harmful effects will be missed.
Recent mutation rate estimates in humans (Itsara et al. 2010), S. cerevisiae (Lynch et al. 2008), and C. elegans (Lipinski et al. 2011) all show an excess of duplicated relative to deleted base pairs. CNVs with allele frequencies <0.1 from a natural population of D. melanogaster from Raleigh, North Carolina, also reveal a similar 2:1 excess of duplicated sequence (Langley et al. 2012). Similar to our examination of low-frequency SNPs above, these low-frequency CNV data should more closely mirror patterns of spontaneous mutation. These data include 1305 duplications and 2067 deletions, such that the relatively smaller number but greater length of individual duplicated sequences match the result of our MA experiment. A similar pattern is observed when all CNVs from Langley et al. (2012) are included, again with twice as many base pairs encompassed by duplications as deletions. These CNV data therefore suggest that the D. melanogaster genome is expanding, regardless of biases in the MA experiment. It is of course also possible that small deletions have a disproportionate effect on fixed differences between species, such that there is no net growth. This cannot be resolved by comparing the size of the assembled D. melanogaster genome to that of Drosophila simulans (Hu et al. 2013), as there are several reasons why the D. melanogaster estimate could be inflated relative to D. simulans—especially the inclusion of heterochromatic regions in the former. Furthermore, even similar euchromatic genome sizes would not be informative if both species were experiencing similar mutational and selective pressures affecting genome-size evolution; there is currently no evidence for or against this possibility. Considering all lines of evidence, it is not clear whether our observed 2:1 excess of duplicated bases is due to the small number of events detected, a result of stronger purifying selection against deletions, or an indication of true mutational patterns in Drosophila.
Large duplications and deletions containing coding sequence or entire genes occur quite frequently in the MA data (9.37 × 10−7 partially deleted genes/gene/generation; 3.75 × 10−7 partially duplicated genes/gene/generation; 1.25 × 10−7 new whole-gene duplicates/gene/generation). We compared these results to those from two previous studies that were able to estimate the rates of gene duplications and deletions with low-resolution array-based techniques, one in C. elegans (Lipinski et al. 2011) and one in S. cerevisiae (Lynch et al. 2008). Our estimates of these rates in Drosophila are somewhat higher than those observed in C. elegans (2.2 × 10−7 partially deleted genes/gene/generation; 3.4 × 10−7 partially duplicated genes/gene/generation, according to Lipinski et al. 2011), but lower than the rates estimated in S. cerevisiae (3.4 × 10−6 duplicated genes/gene/generation; 2.1 × 10−6 deleted genes/gene/generation, according to Lynch et al. 2008). In all three data sets, however, the rate at which new genic copy-number mutations arise is quite high, with approximately one whole-gene duplicate arising per haploid genome every 50–500 generations.
Estimating the fraction of new mutations that are deleterious
We compared the locations of point mutations occurring during mutation accumulation to the locations of SNPs segregating in the North American D. melanogaster population (Langley et al. 2012). We used synonymous polymorphisms as the baseline against which other categories are compared. The evidence for weak selection on synonyomous sites in D. melanogaster suggests that our estimates of selection on single-nucleotide mutations in other categories, selection against large duplications and deletions and of U, the genome-wide rate of deleterious mutation, shown below are biased downward (Zeng and Charlesworth 2010). We also assume that the mutations that we recovered during MA are not biased substantially by purifying selection (which generally appears to be the case; see Appendix). We calculated the expected number of SNPs within introns, intergenic regions, UTRs, and at nonsynonymous sites using the total number of mutations observed in each of these four categories, and the ratio of the number of mutations to the number of polymorphisms found at synonymous sites; in the absence of natural selection, the ratio of the number of mutations to the number of polymorphisms for synonymous sites should equal that for any other category. This approach is similar to comparisons of nucleotide diversity (π) at silent sites to π in other categories, but does not require the assumption of identical mutation rates across categories.
Previous estimates of the fraction of mutations that are deleterious from MA sequence data were based on comparisons with divergence data (e.g., Denver et al. 2004; Haag-Liautard et al. 2007) rather than polymorphism data. We utilized polymorphism data as described above because a substantial fraction of nucleotide differences among Drosophila species is fixed by positive selection (Langley et al. 2012), and as a result levels of divergence are inflated relative to polymorphism. This can be corrected if the fraction of substitutions driven by positive selection is known, but to our knowledge this fraction has not been estimated for all noncoding regions of the genome. Therefore, comparing mutation data to divergence to infer the rate of deleterious mutation could in principle be more error prone than comparing mutation data to polymorphism. Furthermore, by comparing mutation data to polymorphism in various sequence categories as described above, we are able to use copy-number variation data to infer the fraction of large deletions and duplications that are deleterious. Without a comprehensive catalog of fixed duplications and deletions among Drosophila species, this estimate cannot be obtained from divergence data. An approach using polymorphism could be extended to infer the strength of selection for other cases where polymorphism data exist but divergence data are difficult to obtain, either due to inherent difficulties in calculating the divergence level for a particular class of mutation (e.g., CNVs, transposable elements) or because an MA experiment is performed in a species with no close relatives having a high-quality genome assembly.
To estimate U, we first calculated the percentage of new mutations that are purged by purifying selection. We did this by comparing mutation data to polymorphism data for single-nucleotide changes that are nonsynonymous, synonymous, or within UTRs, introns, or intergenic regions according to version 32 of the FlyBase gene annotation (McQuilton et al. 2012). The polymorphism data used for this comparison was the set of all Q20 SNPs in each of these sequence categories found in 37 Raleigh genomes sequenced for the Drosophila Population Genomics Project (Langley et al. 2012) and having a minor allele frequency ≥0.15. This allele-frequency cutoff removes the majority of deleterious SNPs (Charlesworth and Eyre-Walker 2008), the presence of which would downwardly bias our estimate of the fraction of new mutations removed by purifying selection. We then compared the ratio of synonymous polymorphisms in the Drosophila Population Genomics Project data to synonymous mutations in the MA data to the ratios for the other categories of mutation. This ratio is lower in all other categories, implying that new nonsynonymous mutations and mutations in UTRs, introns, and intergenic regions are more deleterious than synonymous mutations on average. This also implies that the ratio of synonymous polymorphisms to mutations can be multiplied by the number of mutations in other categories to get the expected number of polymorphisms and that this expectation can be compared to the number of observed polymorphisms to determine the fraction of new mutations that are removed by purifying selection. To estimate U, this fraction is multiplied by the mutation rate and the number of euchromatic base pairs in the genome (119,029,689). Our estimate of U is the expected number of mutations occurring in a diploid genome each generation that purifying selection will not allow to reach a minor allele frequency ≥0.15. This estimate includes all mutations that natural selection will purge from the population before they are able to fix.
We also used the ratio of synonymous SNPs to synonymous mutations to predict the expected number of duplication and deletion polymorphisms and again compared this expectation to observed numbers of polymorphisms (CNVs with minor allele frequency ≥0.15) from Langley et al. (2012). This method was used in the same manner described above to estimate the fraction of copy-number mutations removed by purifying selection. Similarly, we compared the expected number of base pairs differing in copy number in the absence of selection to the observed number of base pairs within CNVs and used this number to predict reduction in the rate of addition and subtraction of base pairs in the D. melanogaster genome by duplication and deletion caused by purifying selection, respectively. This approach was used to determine the predicted net growth rate of the genome caused by mutations allowed to reach appreciable frequencies in natural populations. One limitation of this approach is that it does not take positive selection into account. This method also does not explicitly account for the recent bottleneck experienced by North American D. melanogaster. However, this nonequilibrium demographic history will only downwardly bias our estimate of U to the extent that it results in deleterious polymorphisms with minor allele frequency ≥0.15.
As shown in Figure 3, we find a large deficit of nonsynonymous polymorphisms and polymorphisms within UTRs, introns, and intergenic regions relative to expectations if all mutations were neutral (1,230,302 observed polymorphisms vs. 2,588,433 expected; P < 2.2 × 10−16). This deficit represents the 52.5% of mutations that are removed by purifying selection before they can reach moderate frequencies within natural populations and translates to a deleterious mutation rate of U = 0.69 mutations per diploid genome per generation. Like previous estimates of U from sequence data, ours is probably an underestimate, due to the omission of all mutations in heterochromatic regions of the genome and the inability to account for slightly deleterious but effectively neutral mutations (s < 1/2N). Our method is also downwardly biased by the omission of indels (for which polymorphism data were not available) and selection against synonymous polymorphisms as stated above, although selection against such polymorphisms is weak in D. melanogaster (Akashi 1995). However, it is important to note that, unlike previous estimates of U from sequence data, our estimate is directly inferred from the proportion of mutations prevented from becoming polymorphic, rather than an indirect estimate based on divergence (e.g., Denver et al. 2004; Haag-Liautard et al. 2007), which is likely more affected by positive selection than polymorphism. Our estimate of U is lower than that of Haag-Liautard et al. (2007), who estimate that U = 1.2 (95% C.I.: 0.51–2.28). This difference is not significant, as the Haag-Liautard estimate was derived from a much smaller number of mutations and therefore has a very wide confidence interval; in addition, this previous estimate includes indels, while our estimate does not. After removing indels from the Haag-Liautard et al. (2007) data, their estimate of U becomes 0.80 (95% C.I.: 0.29–3.19), which is much more similar to ours (U = 0.69; 95% C.I.: 0.64–0.74). Finally, our estimate of the fraction of single-nucleotide mutations that are deleterious is 52.5%, while Haag-Liautard et al. (2007) gave 58% as their estimate. Our estimate may be downwardly biased because of selection against synonymous mutations as discussed above and also because the rate of synonymous mutation is elevated in line 33 due to the excess of G:C→A:T mutations in this line; transitions are more often synonymous than are transversions. However, given that our estimate of the fraction of deleterious mutations does not differ greatly from that of Haag-Liautard et al. (2007), we feel that our polymorphism-based approach is a valid one.
We estimate that 98.8% of duplications and 99.2% of deletions in D. melanogaster are removed by purifying selection (Figure 3). The fractions of duplications and deletions that are deleterious are remarkably high in comparison to other types of mutations. For example, while the vast majority (90%) of nonsynonymous mutations are deleterious, we infer that they are 10 times less likely to be eliminated by purifying selection than duplications or deletions. Duplications and deletions could be deleterious because they affect gene-expression levels or remove or disrupt functional sequence (e.g., tandem duplications occurring within genes; Emerson et al. 2008). Our results suggest that mutations resulting in large duplications and deletions are relatively frequent and that many of these events affect genes. While this mutational input of gene copy-number polymorphisms has likely been extremely important for adaptation in Drosophila (Hahn et al. 2007), our results imply that most of these mutations are quickly eliminated by purifying selection. In addition, the lower allele frequencies and deficit of genic relative to intergenic copy-number polymorphisms (Langley et al. 2012) suggests that this selection is especially strong against genic duplications and deletions. In other words, while many adaptive gene gains and even gene losses may arise over evolutionary time scales, the fate of the vast majority of new gene CNVs is elimination.
Conclusions
The results of our examination of both small- and large-scale mutations in the euchromatic regions of the D. melanogaster genome have several important implications for evolution. First, we have shown that mutation rates can vary substantially among inbred genotypes in D. melanogaster. Second, we find evidence supporting previous claims that the appearance of multiple single-nucleotide mutations and/or indels in close proximity to one another can often be explained by a single mutation event (Averof et al. 2000; Drake 2007; Schrider et al. 2011). Third, we present estimates of the rates at which large duplications and deletions occur in highly inbred (homozygous) D. melanogaster lines, showing that the number of base pairs added and removed by duplication and deletion events is over an order of magnitude greater than the number of nucleotides affected by point mutations and small indels. Contrary to previous results examining only small indels, we find evidence suggesting that mutational forces alone may result in net expansion rather than contraction of the Drosophila genome, although this result does not achieve statistical significance. Future studies with more statistical power are required to confirm these results and determine whether they hold for natural outbred populations as well. We also find that 99% of large duplications and deletions are deleterious, making these mutations ∼10 times more likely to be purged by natural selection than nonsynonymous mutations. This rampant purifying selection would radically reduce any impact of de novo duplication and deletion rates on genome-size changes relative to that expected from mutational pressure alone. Given the expanding number of CNVs implicated in genomic disorders in humans, it seems plausible that our finding of widespread purifying selection against these mutations could hold for a wide range of eukaryotes. Indeed, even many duplications and deletions that are allowed to reach appreciable frequencies are subject to purifying selection (Emerson et al. 2008; Conrad et al. 2010; Langley et al. 2012), with stronger selection against larger variants (Langley et al. 2012). As noted throughout the text, there are multiple possible biases that could affect our mutation rate estimates, including selection during MA, the effect of homozygosity on mutation rate, read-mapping biases, repetitive elements, and residual heterozygosity. We address each of these in the Appendix.
Our study illustrates how mutation-accumulation studies combined with polymorphism data can reveal the fraction of new mutations that are deleterious as well as the rates at which these mutations arise. Future studies accurately measuring the rates of additional types of events (e.g., transposable element insertion, gene duplication via retrotransposition, gene conversion, and inversions) and comparing these patterns to polymorphism data will therefore not only facilitate estimates of the rate of change in genome content, size, and structure, but also will reveal the selective forces acting on these mutations that are invisible to studies of polymorphism and divergence alone.
Supplementary Material
Acknowledgments
We thank Kristi Montooth and members of her lab for assistance and Kevin Cook for pointing us to relevant literature. D.R.S. is supported by the Indiana University Genetics, Molecular and Cellular Sciences Training Grant T32-GM007757. D.H. is supported by the National Sciences and Engineering Research Council and by National Science Foundation (NSF) grant DEB-0950002. M.L. is supported by National Institutes of Health grant R01GM036827. M.W.H. is supported by NSF grant DBI-0845494 and by a fellowship from the Alfred P. Sloan Foundation.
Appendix: Assessing the Impact of Experimental Biases on the Mutation Rate
Natural Selection
As noted above, our experiment cannot recover mutations that are lethal or sterile in homozygotes and is less likely to recover mutations with large deleterious effects. Mutation accumulation took place in sublines with an effective size slightly >2. To quantify the extent to which natural selection could prevent us from measuring deleterious mutations, we modeled the fixation or elimination of new mutations occurring during MA in a population of size 2 as a Markov process with six states: the mutation is eliminated (homozygous absence in both flies), the new mutation is present in the heterozygous state in one fly, the mutation is heterozygous in both flies, the mutation is homozygous in one fly, the mutation is homozygous in one fly and heterozygous in the other, or the mutation is fixed (homozygous in both flies). The probability of a heterozygous parent passing on the mutant allele is given by , and the transition probabilities between states are given by the following matrix:
The probability of fixing a new mutation with some selection coefficient s can be obtained by multiplying the initial state vector [0 1 0 0 0 0 ] with the above matrix repeatedly until the resulting probability vector converges. Under this model, we find that a fitness disadvantage of 1% (i.e., s = 0.01) is only ∼6% less likely to fix as one with s = 0, while mutations with s = 0.05, s = 0.01, and s = 0.5 are ∼28%, ∼50%, and ∼99.5% less likely to fix, respectively. Population surveys and previous MA results in D. melanogaster using balancer chromosomes show that fitness effects of deleterious mutations have a probability distribution with two modes—a small mode of recessive lethals occurring with a probability of a few percent per generation and a much larger class of mutations with fitness effects <10% (e.g., Mukai et al. 1972). Taken together, these empirical and theoretical considerations suggest that we are likely to have recovered the vast majority of mutations and the majority of deleterious mutations in particular.
To directly ask whether there is an under-representation of point mutations in coding regions indicative of natural selection, we compared the numbers of mutations appearing within exonic, intronic, and intergenic regions to expectations based on the fractions of the Drosophila genome in each category and found no evidence of bias with respect to these categories (P = 0.496; χ2 test). Given our data, we can confidently reject any deficit >7.9% in exonic single-nucleotide mutations (Materials and Methods). This finding, combined with the large fraction of observed coding mutations that are nonsynonymous (73.7%), suggests that the patterns of point mutation that we observe are not significantly biased by purifying selection.
We also asked whether large duplications and deletions are under-represented in coding regions, as would be expected if they often result in lethality or sterility. We randomly assigned new coordinates to the deletions that we observed while preserving their lengths, finding that, on average, protein-coding sequences of 6.16 genes are affected (after 100,000 iterations) vs. 4 in our observed set. While this difference is not significant (P = 0.11; one-tailed permutation test), it is consistent with purifying selection removing approximately one-third of deletions that inactivate a single gene. This fraction is also consistent with the ∼33% of genes that result in lethality when knocked down in D. melanogaster (Chen et al. 2010). However, it should be noted that we do not know what proportion of genes result in lethality or sterility when present as homozygous deletions (Ashburner et al. 2012). If we assume that the observed distribution of deletion lengths is approximately correct, conservatively assume that all deletions affecting coding sequence are inactivating mutations, and assume that one-third of these are lethal, then the results of the permutation suggest that we may be underestimating the number of deletions of coding sequence by about two events, corresponding to ∼1816.5 additional bp deleted during the course of the MA experiment (based on the average lengths of deletions affecting coding sequence) or 1.57 bp per generation per subline. For duplications, the protein-coding sequences of 4.52 genes are affected on average, similar to the 4 observed in the real data (P = 0.47). These data are consistent with homozygous deletions having stronger fitness effects than homozygous duplications and suggest that purifying selection may bias our deletion-rate estimate more than our duplication-rate estimate.
Homozygosity and Mutation Rate
D. melanogaster is an outbreeding species with a large population size, so mutations will typically occur in genomes that are highly heterozygous. Our experiment studies the rate of mutation in homozygous genotypes. As noted above, it is possible that the inbreeding process has generated genotypes with atypical mutation spectra. Such changes might be expected to raise mutation rates on average if recessive mutator alleles are segregating at low frequency in nature. Although we cannot be sure that the absolute rates that we report are unbiased, the fact that the patterns of mutation observed in our lines closely match the observed patterns of natural variation suggests that our results are not atypical on average. However, it is possible that the variation in mutation rate found between lines is not typically found in natural outbred populations.
A second possible bias of our use of homozygous lines is that homozygosity itself alters the mutational process. For example, Montgomery et al. (1991) found that large duplications and deficiencies arose at a much higher rate in heterozygous genotypes than in homozygous ones. If heterozygosity engenders mutational events and/or makes their repair more difficult, then the mutation rate in our lines may be atypically low.
These possibilities can be resolved only with experiments in outbred flies. Two such designs might be possible, but they share a need for a great deal more sequencing and validation. A “middle-class-neighborhood” design has been used to study the phenotypic effects of mutation in an outbred background (Shabalina et al. 1997). Mutation has also been measured in parent-offspring trios in humans (e.g., Conrad et al. 2011; Kong et al. 2012) using naturally outbred individuals. However, the parent-offspring trio design would require sequencing ∼1800 genomes (one trio for every two generations of mutation accumulation) to obtain as many generations of mutation as is captured here. A middle-class neighborhood design would also be impractical for genome-wide detection of new mutations, as the entire starting population would need to be sequenced. Perhaps a better design would be to perform mutation accumulation while maintaining new mutations in heterozygous state using balancer chromosomes (e.g., Mukai and Cockerham 1977). Such a study could substantially reduce the impacts of homozygosity and purifying selection on observed patterns of mutation and would therefore address many of the questions raised by our work.
Read Mapping Biases and Depth of Coverage
Because genotyping accuracy increases with increased read coverage, we examined the effect of increasing the minimum read depth required to call a new mutation on our mutation rate estimates. When we increase the number of required reads in each subline from 5 to 10, the mutation rate in line 33 changes from 7.71 × 10−9 (514 mutations in 114,966,662 bases) to 7.83 × 10−9 (454 mutations at 100,023,281 covered bases), a difference that is not significant (P = 0.822; Fisher’s exact test). Similarly, the rate estimate in line 39 does not change significantly (from 218 mutations at 114,955,079 covered bases for a rate of 3.27 × 10−9 to 193 mutations at 100,621,822 covered bases for a rate of 3.31 × 10−9; P = 0.921). Again, the rate estimates obtained from requiring 20× coverage at each mutant site in each subline do not differ from those estimated requiring 5× coverage: a rate of 7.61 × 10−9 is estimated from line 33 (115 mutations at 26,042,397 covered sites; P = 0.959 when compared to the 5× coverage data set), and 3.26 × 10−9 is estimated from line 39 (60 mutations at 31,718,552 covered sites; P = 1). Table S5 shows mutation rate estimates for all minimum read-depth cutoffs between 5 and 20. At no point does the mutation rate in either line differ significantly from the ≥5× estimate.
We did observe a small but nearly significant reduction in read depth at mutant sites within the mutant lines vs. read depth in nonmutant lines (mean mutant read depth = 22.64; mean of median read depths from nonmutant sublines from the same inbred line = 23.12; P = 0.053; Wilcoxon signed-rank test), suggesting that reads with mismatches are slightly less likely to be mapped to the reference genome. Such a bias could result in mutations dropping below the threshold in read depth, in which case mutation rate estimates would be downwardly biased as mutant sites are more likely to be eliminated by the coverage cutoff than nonmutant sites (e.g., Keightley et al. 2009). However, because our average read depth (∼22.5) is far higher than our minimum read-depth cutoff (five reads), very few mutations had read depth near the minimum cutoff; only three mutations had a read depth of 6 in the mutant, and only one had a read depth of 5. Thus, this mapping bias probably caused us to miss very few, if any, mutations and resulted in a negligible reduction in our mutation rate estimates.
Because GC content is correlated with read depth (Bentley et al. 2008), it is possible that mutation-rate estimates could be affected. However, any such impact would be mediated through decreased read depth, and we did not find evidence for an effect of read depth on our power to detect mutations as discussed above. Thus, it does not appear that heterogeneity in GC content across the genome has an appreciable affect on our mutation rate estimates.
Another potential mapping-related source of bias is inadequate alignment sensitivity in the presence of small indels. While the aligner that we used, BWA, performs gapped alignment, more sensitive aligners such as Stampy (Lunter and Goodson 2011) or Novoalign (http://www.novocraft.com) recover small indels more reliably. Thus, we may have underestimated the rate at which small indels occurred during our MA experiment. We correct for this as described in Materials and Methods.
Repetitive Regions
To ensure that our mutation-rate estimates were not significantly altered by reduced accuracy in repetitive regions, we calculated mutation rates in the two lines after omitting regions masked by RepeatMasker. After omitting masked regions, the mutation rate in line 33 only changed to 8.00 × 10−9 (492 mutations at 106,053,201 covered bases), a difference that was not significant (P = 0.570; Fisher’s exact test). Similarly, the change in the rate estimate for line 39 after omitting masked regions was subtle (3.38 × 10−9; 208 mutations at 106,014,189 covered sites) and not statistically significant (P = 0.735).
Residual Heterozygosity
It is possible that residual heterozygosity present in one of the ancestral lines prior to mutation accumulation could appear in one subline and be erroneously classified as a mutation. Our approach to detect new mutations hinges on the assumption that alleles present in only one of the four sublines of a given MA line are not relics of polymorphism present in the ancestral line that the inbreeding process failed to remove prior to mutation accumulation. Given that ∼1 in every 100 bp differs between any two chromosomes in natural populations of D. melanogaster (Langley et al. 2012), a sizable window of residual heterozygosity would result in stretches of dense polymorphism among the four sublines derived from either inbred line, with either one subline exhibiting a different haplotype from the other three within this stretch or each line sharing a haplotype with one line but differing from the other two. Whenever one line exhibits a haplotype differing from the other three, each polymorphism within this stretch of residual heterozygosity would appear to be a new mutation by our criteria, resulting in a chromosomal stretch densely packed with apparent new mutations. An examination of all mutations in Table S1 reveals no such windows: with the exception of 10 MNMs, which are very small windows with mutations more densely packed than expected in the case of residual heterozygosity, only three pairs of mutations occur within 1 kb of each other. (Direct evidence that the 10 MNMs are not artifacts of residual heterozygosity is presented below.)
We also searched for regions of residual heterozygosity with two haplotypes each shared between two of the four sublines within a mutation-accumulation line. Although SNPs within these regions would not be called as mutations according to our criteria, identifying and comparing such regions to our list of new mutations could strengthen our confidence that residual heterozygosity does not affect the latter. Very few such alleles were found, except for a few large blocks on chromosome arm 3R and a smaller block on chromosome 3L. These blocks, which were present in both lines 39-67 and 39-18, exhibited the genotypes present in line 33, perhaps due to a contamination event prior to mutation accumulation. The blocks were typically dozens of kilobases in length, containing hundreds of polymorphisms, with a density of polymorphism always greater than 0.001 per base pair. We manually examined mutations appearing in these regions to ensure that they were not SNPs present in two sublines but miscalled as being present only in one and therefore inferred to be a mutation. In several cases where we observed a mutation at one of the two contaminated sublines, the other contaminated subline exhibited multiple reads supporting the mutation, but the other two sublines did not. We reasoned that these mutations were probably false positives due to contamination and removed them from our final set. Again, no clusters of single-nucleotide mutations with density approaching that seen in these regions of residual heterozygosity appear in our final set of mutations; if they did, we would have reported far more than the 732 mutations listed in Table S1. Indeed, we would also likely greatly overestimate mutation rates, while the rates that we report here are largely concordant with those published previously. However, because these blocks of residual heterozygosity may have been caused by a contamination event occurring shortly before mutation accumulation, they could be larger than blocks of residual heterozygosity that recombination has had more opportunities to reduce in size.
To ensure that smaller regions of residual heterozygosity, even those consisting of a single SNP, had not qualitatively affected our results, we searched sequence data from a nearby population of D. melanogaster (Raleigh, NC) (Langley et al. 2012) to determine the fraction of our 732 single-nucleotide mutations that are segregating in natural populations. We examined all 2,924,713 SNP calls in this data set having a quality score ≥30 in at least one individual not matching the reference genome, finding that only 12 of the 732 (1.64%) mutations matched a nonreference allele present in one or more of the 37 sequenced lines from the Raleigh data set. Only 7 of the 732 (0.956%) mutations matched a nonreference allele present in 2 or more of the 37 lines. Given the possibility of sequencing error introducing low-frequency polymorphisms, and the possibility of new mutations occurring at sites already polymorphic in natural populations—over 2.5% of all sites in the Raleigh data set are polymorphic according to the criterion described above—it is fair to conclude that few, if any, of the mutations reported in Table S1 are actually SNPs segregating prior to mutation accumulation. In contrast, of the 5975 polymorphisms present in the large blocks of residual heterozygosity shared between two sublines described above, 5307 (88.8%) exhibit the nonreference allele in at least one of the sequenced Raleigh lines, and 5048 (84.5%) exhibit the nonreference allele in at least two lines. Because residual heterozygosity would result in false-positive “mutations” that appear far closer together than expected, we searched for evidence that any of the 10 MNMs were present in the Raleigh polymorphism data. We did not find a single SNP call from the 37 Raleigh genomes matching the mutant genotype at any of the sites within any of these MNMs. Moreover, none of the MNMs or complex mutations that we found occurred within the contaminated lines. We conclude from this that it is highly unlikely that any of these 10 MNMs was erroneously detected due to residual heterozygosity. The stark contrast between the fractions of SNPs within putative regions of residual heterozygosity and our list of mutations implies that vanishingly few (and possibly zero) of our putative mutations are a result of residual heterozygosity or any contamination event.
Footnotes
Communicating editor: C. H. Langley
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Akashi H., 1995. Inferring weak selection from patterns of polymorphism and divergence at silent sites in Drosophila DNA. Genetics 139: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers C. A., Lunter G., MacArthur D. G., McVean G., Ouwehand W. H., et al. , 2011. Dindel: accurate indel calls from short-read data. Genome Res. 21: 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Golic K. G., Hawley R. S., 2012. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Averof M., Rokas A., Wolfe K. H., Sharp P. M., 2000. Evidence for a high frequency of simultaneous double-nucleotide substitutions. Science 287: 1283–1286 [DOI] [PubMed] [Google Scholar]
- Bachtrog D., 2008. Evidence for male-driven evolution in Drosophila. Mol. Biol. Evol. 25: 617–619 [DOI] [PubMed] [Google Scholar]
- Bentley D. R., Balasubramanian S., Swerdlow H. P., Smith G. P., Milton J., et al. , 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth J., Eyre-Walker A., 2008. The McDonald-Kreitman test and slightly deleterious mutations. Mol. Biol. Evol. 25: 1007–1015 [DOI] [PubMed] [Google Scholar]
- Chen S., Zhang Y. E., Long M., 2010. New genes in Drosophila quickly become essential. Science 330: 1682–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D. F., Pinto D., Redon R., Feuk L., Gokcumen O., et al. , 2010. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D. F., Keebler J. E. M., DePristo M. A., Lindsay S. J., Zhang Y., et al. , 2011. Variation in genome-wide mutation rates within and between human families. Nat. Genet. 43: 712–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G. M., Zerr T., Kidd J. M., Eichler E. E., Nickerson D. A., 2008. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat. Genet. 40: 1199–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland J. M., Thornton K. R., 2010. Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol. Evol. 2: 83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth J. P., De Bie T., Stajich J. E., Cristianini N., Hahn M. W., 2006. The evolution of mammalian gene families. PLoS ONE 1: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D. R., Morris K., Lynch M., Thomas W. K., 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430: 679–682 [DOI] [PubMed] [Google Scholar]
- Denver D. R., Dolan P. C., Wilhelm L. J., Sung W., Lucas-Lledo J. I., et al. , 2009. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc. Natl. Acad. Sci. USA 106: 16310–16314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D. R., Wilhelm L. J., Howe D. K., Gafner K., Dolan P. C., et al. , 2012. Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biol. Evol. 4: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., 2007. Too many mutants with multiple mutations. Crit. Rev. Biochem. Mol. Biol. 42: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J. J., Cardoso-Moreira M., Borevitz J. O., Long M., 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320: 1629–1631 [DOI] [PubMed] [Google Scholar]
- Girirajan S., Campbell C. D., Eichler E. E., 2011. Human copy number variation and complex genetic disease. Annu. Rev. Genet. 45: 203–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard C., Dorris M., Maside X., Macaskill S., Halligan D. L., et al. , 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445: 82–85 [DOI] [PubMed] [Google Scholar]
- Hahn M. W., Han M. V., Han S. G., 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3: e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan D. L., Keightley P. D., 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40: 151–172 [Google Scholar]
- Houle D., Rowe L., 2003. Natural selection in a bottle. Am. Nat. 161: 50–67 [DOI] [PubMed] [Google Scholar]
- Houle D., Nuzhdin S. V., 2004. Mutation accumulation and the effect of copia insertions in Drosophila melanogaster. Genet. Res. 83: 7–18 [DOI] [PubMed] [Google Scholar]
- Hu T. T., Eisen M. B., Thornton K. R., Andolfatto P., 2013. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 23: 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A., Wu H., Smith J. D., Nickerson D. A., Romieu I., et al. , 2010. De novo rates and selection of large copy number variation. Genome Res. 20: 1469–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives P. T., 1945. The genetic structure of American populations of Drosophila melanogaster. Genetics 30: 167–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov F. A., I. B. Rogozin, Y. I. Wolf, and E. V. Koonin, 2002 Selection in the evolution of gene duplications. Genome Biol. 3. Available at: www.http://genomebiology.com/content/3/2/RESEARCH0008 [DOI] [PMC free article] [PubMed]
- Kong A., Frigge M. L., Masson G., Besenbacher S., Sulem P., et al. , 2012. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488: 471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., K. Stevens, C. Cardeno, Y. C. Lee, D. R. Schrider et al, 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leushkin E. V., Bazykin G. A., Kondrashov A. S., 2013. Strong mutational bias toward deletions in the Drosophila melanogaster genome is compensated by selection. Genome Biol. Evol. 5: 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S., Sutton G., Ng P. C., Feuk L., Halpern A. L., et al. , 2007. The diploid genome sequence of an individual human. PLoS Biol. 5: e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski K. J., Farslow J. C., Fitzpatrick K. A., Lynch M., Katju V., et al. , 2011. High spontaneous rate of gene duplication in Caenorhabditis elegans. Curr. Biol. 21: 306–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G., Goodson M., 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21: 936–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2010. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA 107: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Abegg A., 2010. The rate of establishment of complex adaptations. Mol. Biol. Evol. 27: 1404–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S. A., Altshuler D. M., 2007. Copy-number variation and association studies of human disease. Nat. Genet. 39: S37–S42 [DOI] [PubMed] [Google Scholar]
- McQuilton P., St Pierre S. E., Thurmond J.; FlyBase Consortium, 2012. FlyBase 101: the basics of navigating FlyBase. Nucleic Acids Res. 40: D706–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer P. W., 2009. Measuring the rates of spontaneous mutation from deep and large-scale polymorphism data. Genetics 182: 1219–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery E. A., Huang S. M., Langley C. H., Judd B. H., 1991. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: genome structure and evolution. Genetics 129: 1085–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Cockerham C. C., 1977. Spontaneous mutation rates at enzyme loci in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 74: 2514–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Chigusa S. I., Crow J. F., Mettler L. E., 1972. Mutation rate and dominance of genes affecting viability in Drosophila melanogaster. Genetics 72: 335–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness R. W., Morgan A. D., Colegrave N., Keightley P. D., 2012. Estimate of the spontaneous mutation rate in Chlamydomonas reinhardtii. Genetics 192: 1447–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledo J. I., Warthmann N., Clark R. M., et al. , 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G. H., Dominy N. J., Claw K. G., Lee A. S., Fiegler H., et al. , 2007. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39: 1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D. A., 2002. DNA loss and evolution of genome size in Drosophila. Genetica 115: 81–91 [DOI] [PubMed] [Google Scholar]
- Petrov D. A., Lozovskaya E. R., Hartl D. L., 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384: 346–349 [DOI] [PubMed] [Google Scholar]
- Petrov D. A., Chao Y. C., Stephenson E. C., Hartl D. L., 1998. Pseudogene evolution in Drosophila suggests a high rate of DNA loss. Mol. Biol. Evol. 15: 1562–1567 [DOI] [PubMed] [Google Scholar]
- Ravi D., Wiles A. M., Bhavani S., Ruan J., Leder P., et al. , 2009. A network of conserved damage survival pathways revealed by a genomic RNAi screen. PLoS Genet. 5: e1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Schrider D. R., Hourmozdi J. N., Hahn M. W., 2011. Pervasive multinucleotide mutational events in eukaryotes. Curr. Biol. 21: 1051–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina S. A., Yampolsky L. Y., Kondrashov A. S., 1997. Rapid decline of fitness in panmictic populations of Drosophila melanogaster maintained under relaxed natural selection. Proc. Natl. Acad. Sci. USA 94: 13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp N. P., Agrawal A. F., 2012. Evidence for elevated mutation rates in low-quality genotypes. Proc. Natl. Acad. Sci. USA 109: 6142–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J. R., 2010. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61: 437–455 [DOI] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Miller S. F., Doak T. G., Lynch M., 2012. Drift-barrier hypothesis and mutation-rate evolution. Proc. Natl. Acad. Sci. USA 109: 18488–18492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Levine M. T., Eckert M. L., Begun D. J., 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179: 455–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E., Sharp A. J., Bailey J. A., Kaul R., Morrison V. A., et al. , 2005. Fine-scale structural variation of the human genome. Nat. Genet. 37: 727–732 [DOI] [PubMed] [Google Scholar]
- Wakimoto B. T., Lindsley D. L., Herrera C., 2004. Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics 167: 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., 2010. A simple multiallele model and its application to identifying preferred-unpreferred codons using polymorphism data. Mol. Biol. Evol. 27: 1327–1337 [DOI] [PubMed] [Google Scholar]
- Zeng K., Charlesworth B., 2010. The effects of demography and linkage on the estimation of selection and mutation parameters. Genetics 186: 1411–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Gu Z. L., Li W. H., 2003. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 4: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.