Phosphorylation of the scaffolding protein JIP1 serves as a molecular switch to coordinate anterograde and retrograde microtubule motor complexes involved in amyloid precursor protein transport.
Abstract
Regulation of the opposing kinesin and dynein motors that drive axonal transport is essential to maintain neuronal homeostasis. Here, we examine coordination of motor activity by the scaffolding protein JNK-interacting protein 1 (JIP1), which we find is required for long-range anterograde and retrograde amyloid precursor protein (APP) motility in axons. We identify novel interactions between JIP1 and kinesin heavy chain (KHC) that relieve KHC autoinhibition, activating motor function in single molecule assays. The direct binding of the dynactin subunit p150Glued to JIP1 competitively inhibits KHC activation in vitro and disrupts the transport of APP in neurons. Together, these experiments support a model whereby JIP1 coordinates APP transport by switching between anterograde and retrograde motile complexes. We find that mutations in the JNK-dependent phosphorylation site S421 in JIP1 alter both KHC activation in vitro and the directionality of APP transport in neurons. Thus phosphorylation of S421 of JIP1 serves as a molecular switch to regulate the direction of APP transport in neurons.
Introduction
Targeted long-distance transport of proteins and organelles is critical in neurons, which extend polarized axons of up to one meter long in humans. In axons, the family of anterograde kinesin motors and the retrograde dynein motor transport cargos on microtubule tracks of uniform polarity. These cargos include synaptic vesicles, signaling endosomes, lysosomes, RNA granules, and mitochondria (Hirokawa et al., 2010).
Constitutive transport of axonal cargos can either be bidirectional, characterized by saltatory or frequent back and forth movement, or highly processive, characterized by long run lengths and high speeds. For example, mitochondria (Morris and Hollenbeck, 1993) and late endosomes/lysosomes (Hendricks et al., 2010) often move bidirectionally along axons, with short runs in either direction punctuated by frequent directional switches. In contrast, autophagosomes display highly processive and unidirectional retrograde motility along axons (Maday et al., 2012).
Three models have been proposed to explain how net direction of microtubule-based transport is determined at a molecular level (Gross, 2004; Welte, 2004). In the first model, only anterograde or retrograde motors can bind to a cargo at any given time. However, both in vitro and cellular studies suggest that opposing motors can bind simultaneously to cargos (Soppina et al., 2009; Hendricks et al., 2010; Encalada et al., 2011; Maday et al., 2012). In a tug-of-war model, opposing kinesin and dynein motors can bind simultaneously to cargo and drive motility toward either the microtubule plus or minus end in a stochastic and unregulated manner (Müller et al., 2008; Hendricks et al., 2010). In this model, net direction of transport is determined by which set of motors exerts the most force at any given time; frequent directional switches are predicted, consistent with the motility of bidirectional cargos. In contrast, in the third, coordination model, a cargo-bound adaptor regulates the activity of one or both motors, leading to processive motility along the microtubule, with few directional changes.
To understand how the activity of opposing kinesin and dynein motors may be coordinated during axonal transport, we turned to the vesicular transmembrane protein amyloid precursor protein (APP). Axonal transport of APP is highly processive, with fast velocities and long run lengths in both anterograde and retrograde directions (Kaether et al., 2000; Falzone et al., 2009). Impaired axonal transport of APP correlates with increased production of amyloid-β, an APP cleavage product that aggregates to form senile plaques in Alzheimer’s disease (Stokin et al., 2005). Despite this relationship between dysfunctional APP trafficking and disease pathology, the molecular mechanisms that regulate APP transport in neurons are not yet understood.
Anterograde APP transport is mediated via direct binding (Matsuda et al., 2001; Scheinfeld et al., 2002) to the scaffolding protein JNK-interacting protein 1 (JIP1; Muresan and Muresan, 2005b). JIP1 was originally identified for its ability to recruit multiple kinases in the JNK pathway (Dickens et al., 1997). Genetic studies suggest that JIP1 regulates constitutive axonal transport (Horiuchi et al., 2005), whereas the structurally unrelated scaffolding protein JIP3 (Whitmarsh, 2006; Koushika, 2008) plays a role in injury signaling (Cavalli et al., 2005; Abe et al., 2009). Conventional Kinesin-1 is a heterotetramer consisting of the adaptor protein kinesin light chain (KLC) and the motor protein kinesin heavy chain (KHC or KIF5). JIP1 directly binds to KLC via a conserved 11-aa motif at the C terminus (Verhey et al., 2001). However, this binding domain is insufficient to activate KHC-mediated anterograde transport (Kawano et al., 2012), suggesting that additional interactions may be responsible for KHC activation in APP transport. Furthermore, though axonal transport of APP occurs in both anterograde and retrograde directions, neither the mechanism underlying its retrograde transport nor the switch regulating its directionality are currently known.
Here, we show that knockdown of JIP1 leads to severe deficits in both anterograde and retrograde axonal transport of APP in primary neurons. We identify novel, KLC-independent interactions between JIP1 and KHC and show via single molecule motility assays that JIP1 binding activates KHC motility in vitro. Furthermore, we identify another novel JIP1 interactor, p150Glued, a subunit of the retrograde dynein–dynactin complex. p150Glued competitively inhibits the JIP1-mediated enhancement of KHC processivity in vitro and disrupts anterograde APP axonal transport. Furthermore, mutations at a JNK-dependent phosphorylation site in JIP1 (S421) alter KHC activation in vitro and the directionality of APP transport in neurons. Together, these experiments establish JIP1 as a coordinator of anterograde and retrograde motor activity whose regulation by phosphorylation determines the directionality of the axonal transport of APP.
Results
JIP1 knockdown disrupts both anterograde and retrograde axonal transport of APP
To probe the role of JIP1 in the regulation of axonal transport, we depleted JIP1 expression using targeted siRNA. Because primary neurons have low levels of transfection, we first tested the efficiency of our siRNA in the CAD mouse neuronal cell line, whose ability to extend long neurites upon differentiation by serum deprivation has been exploited to study polarized neuronal transport (Blasius et al., 2007). At 48 h after transfection, our siRNA depleted endogenous JIP1 by more than 90% when assessed by immunostaining and Western blotting, with no compensatory changes in motor protein expression (Fig. S1, A and B). In addition, a sequence-specific scrambled siRNA had no off-target effects on APP transport (Fig. S1 C).
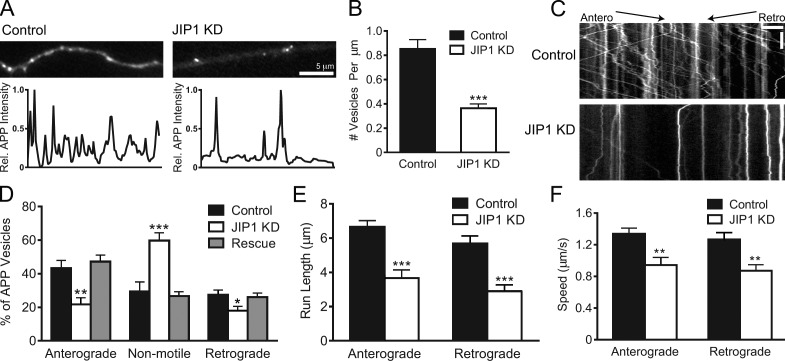
Next, we knocked down JIP1 in primary mouse dorsal root ganglion (DRG) sensory neurons that extend elongated axons with uniform microtubule polarity and have been previously used to study APP axonal function (Nikolaev et al., 2009). Using fluorescent siRNA to identify JIP1-depleted cells, we imaged APP-YFP–positive axons and observed a striking ∼60% decrease in the number of APP-positive vesicles (Fig. 1, A and B). This dearth of APP-positive vesicles likely represents the cumulative effect of a shift in the steady state of vesicles entering or exiting the axon, perhaps resulting from changes in anterograde and retrograde transport.
Figure 1.
JIP1 knockdown disrupts both anterograde and retrograde transport of APP-positive vesicles. (A) Representative images and line scans of APP-YFP intensity show that JIP1-knockdown DRGs contain fewer APP-positive vesicles in the axon than control DRGs. (B) JIP1 knockdown in DRGs significantly decreased the number of APP-positive vesicles in the axon. Control, 0.85 ± 0.08/µm; JIP1 siRNA, 0.36 ± 0.04/µm. (B–F) Data represent three independent experiments (n = 15–23 neurons). (C) Kymographs of APP-YFP motility in DRG transfected with siRNA against JIP1. Kymographs represent cumulative organelle movement (displacement on the x-axis) over time (y-axis). Arrested vesicles appear as vertical lines, whereas motile vesicles appear as diagonal lines toward either the right (anterograde) or left (retrograde). Bars: (horizontal) 5 µm; (vertical) 15 s. (D) JIP1 depletion significantly alters the directional distribution of APP-positive vesicles, causing decreases in the percentages of anterograde and retrograde vesicles and an increase in the percentage of arrested vesicles. Transport changes induced by JIP1 depletion are fully rescued by expression of a human JIP1 cDNA resistant to the siRNA. (E and F) JIP1 depletion significantly decreases mean run lengths and speeds of APP-positive vesicles in both anterograde and retrograde directions. Means represent only vesicles categorized as motile (i.e., anterograde or retrograde in D). Error bars show the mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, using kymograph analysis, we classified APP-positive vesicles as anterograde, retrograde, or non-motile. Whereas control neurons exhibited robust APP transport, JIP1-depleted neurons displayed decreases of >50% and 30% in anterograde and retrograde APP motility, respectively, with a doubling of the percentage of non-motile vesicles (Fig. 1, C and D; and Video 1). CAD cells depleted of JIP1 also showed similar changes in APP motility (Fig. S1 D). Importantly, these alterations in APP transport are a targeted and specific effect of endogenous mouse JIP1 knockdown as both anterograde and retrograde motility are fully rescued by a bicistronic construct coexpressing siRNA-resistant human JIP1 and the transfection marker BFP (Fig. 1 D). In addition, APP-positive vesicles that remain motile in JIP1-knockdown neurons exhibited decreases in both anterograde and retrograde run length and speed (Fig. 1, E and F). The commensurate shift to arrested motility as well as decreases in speed and run length in JIP1-depleted neurons indicate that APP-positive vesicles are impaired in their ability to sustain processive runs in the absence of JIP1.
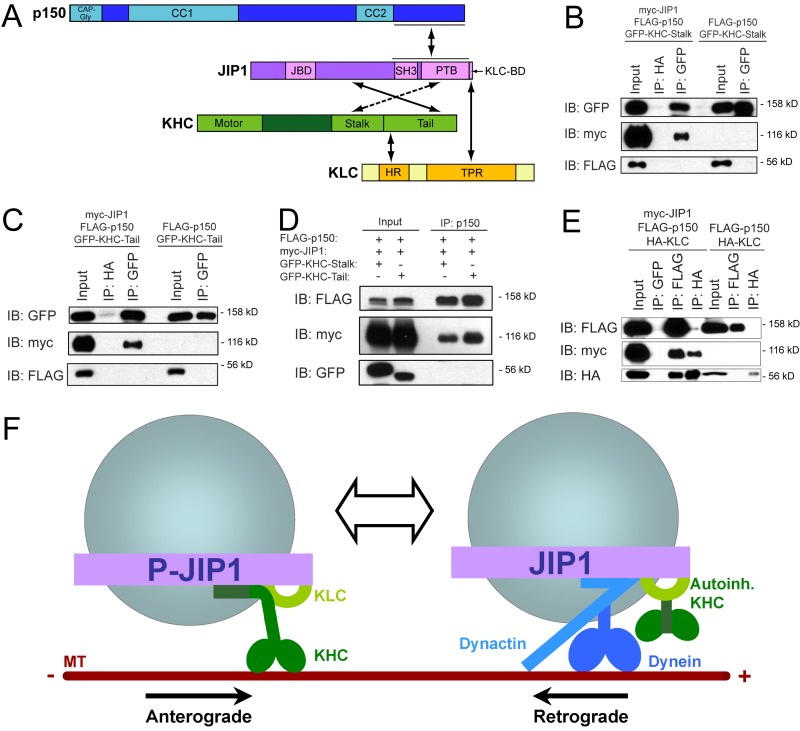
Direct binding of JIP1 to KHC stalk and tail is independent of KLC
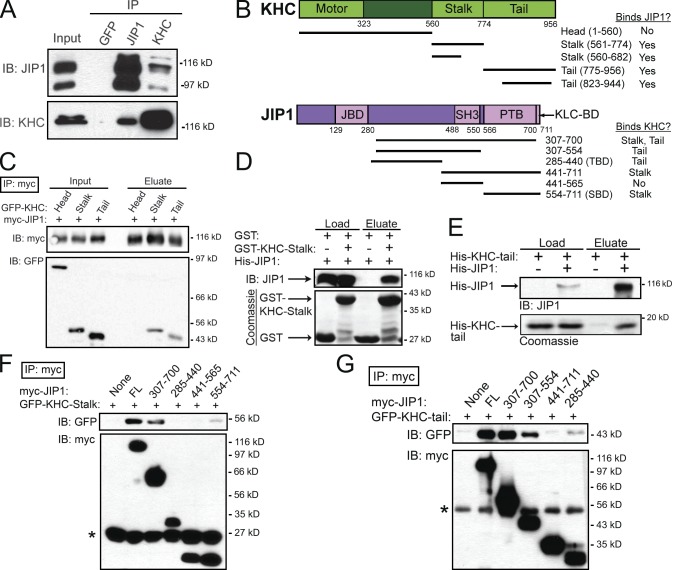
The transport changes that we observed upon JIP1 depletion were consistent with a role for JIP1 in the formation and maintenance of a functional transport complex. Interestingly, recent experiments suggest that binding of KLC to JIP1 is insufficient for transport initiation (Kawano et al., 2012). Thus, we tested for additional interactions between JIP1 and Kinesin-1 by performing coimmunoprecipitations using mouse brain homogenate. An anti-JIP1 antibody coimmunoprecipitated KHC and a monoclonal anti-KHC antibody that recognizes the KHC head region robustly coimmunoprecipitated JIP1 (Fig. 2 A). Interestingly, the anti-KHC immunoprecipitation concentrated an ∼120-kD JIP1 band, which is found at very low levels in the brain homogenate and may represent a posttranslationally modified form of JIP1.
Figure 2.
JIP1 binds directly to both stalk and tail domains of KHC independently of KLC. (A) JIP1 coimmunoprecipitates with KHC in mouse brain homogenate. A monoclonal JIP1 antibody immunoprecipitates the expected 110-kD band as well as a 90-kD band that likely represents a splice isoform. Both bands are also recognized by multiple JIP1 monoclonal antibodies (R&D Systems and BD). (B) Schematics of KHC (KIF5C) and JIP1 constructs and summary of mapping results. JIP1 contains JBD (JNK-binding domain), SH3 (src homology), and PTB domains. (C) JIP1 binds to both stalk and tail domains of KHC. Lysates from COS7 cells cotransfected with myc-JIP1 and GFP-KHC fragments were immunoprecipitated with an anti-myc antibody. (D) JIP1 binds directly to KHC stalk. Purified His-JIP1 incubated with glutathione beads bound to either GST or GST-KHC-stalk (aa 560–682) selectively bound to GST-KHC-stalk but not GST. (E) JIP1 binds directly to KHC tail. Purified His-KHC-tail (KIF5B aa 823–944) with or without His-JIP1 was coincubated with anti-JIP1 antibody, which specifically coimmunoprecipitated His-KHC-tail. (F) KHC stalk binds to the C terminus of JIP1 independently of KLC. Lysates from COS7 cells cotransfected with GFP-KHC-stalk and myc-JIP1 fragments were immunoprecipitated with an anti-myc antibody. KHC stalk coimmunoprecipitated with myc-JIP1[554–711] (myc-JIP1-SBD). Asterisk shows antibody light chain bands. (G) KHC tail binds to JIP1 independently of KLC. Lysates from COS7 cells transfected with GFP-KHC-tail and myc-JIP1 fragments were immunoprecipitated with an anti-myc antibody. KHC tail coimmunoprecipitated with myc-JIP1[285–440] (myc-JIP1-TBD). Asterisk shows antibody heavy chain bands.
To map the KHC domains that interact with JIP1, we cotransfected full-length myc-JIP1 along with GFP-KHC fragments (Konishi and Setou, 2009) into COS7 cells. Of the three mammalian KHCs, KIF5B is ubiquitously expressed, whereas KIF5A and KIF5C are enriched in neurons (Kanai et al., 2000); thus we used KIF5C constructs in this study. Immunoprecipitations revealed that both KHC stalk and tail regions can bind independently to JIP1, whereas the KHC head region containing the motor domain does not (Fig. 2, B and C). Using purified recombinant proteins, we demonstrated these interactions are direct, and further refined the KHC binding domains. Full-length His-JIP1 (Fig. S2 A) binds independently to both GST-KHC-stalk (aa 560–682) and His-KHC-tail (aa 823–944; Fig. 2, B, D, and E). Moreover, neither of the recombinant KHC stalk or tail constructs include the KLC-binding domain of KHC (aa 682–810; Verhey et al., 1998), further indicating that the binding of JIP1 to KHC is independent of KLC.
To map the KHC binding sites within JIP1, we transfected either GFP-KHC-stalk or -tail along with myc-JIP1 fragments into COS7 cells. Immunoprecipitations against myc-JIP1[307–700], a truncated JIP1 construct missing the 11-aa C-terminal KLC-binding domain (KLC-BD), demonstrate robust binding to both KHC stalk and tail (Fig. 2, F and G), confirming that KHC binds JIP1 independently of KLC. In addition, GFP-KHC-stalk bound to myc-JIP1[554–711], a C-terminal JIP1 fragment containing the phosphotyrosine binding (PTB) domain (Fig. 2 F), whereas GFP-KHC-tail bound to myc-JIP1[285–440] (Fig. 2 G). Further experiments with an N-terminal fragment, JIP1[1–390], demonstrated no binding (Fig. S2 B), which effectively restricts the minimal KHC-tail–binding domain to aa 391–440. These distinct binding sites confirm that KHC stalk and tail interact with separate regions of JIP1 and suggest that dual interactions may function to enhance the association of JIP1 to KHC.
JIP1 relieves KHC autoinhibition and activates KHC motility in vitro
In the cell, KHC tail binds to the KHC motor head domain to autoinhibit its ATPase activity, which likely prevents wasteful ATP hydrolysis and microtubule track congestion (Verhey and Hammond, 2009). This well-characterized interaction occurs via hydrogen bonding between the basic IAK motif in the tail and acidic residues on the motor head (Kaan et al., 2011). Interestingly, the minimal KHC-tail–binding domain in JIP1 (aa 391–440) contains 22% acidic residues (Fig. S2 C) and may compete against KHC head for binding to KHC tail. Hence, we hypothesized that binding of KHC tail to JIP1 relieves KHC autoinhibition and activates KHC motility.
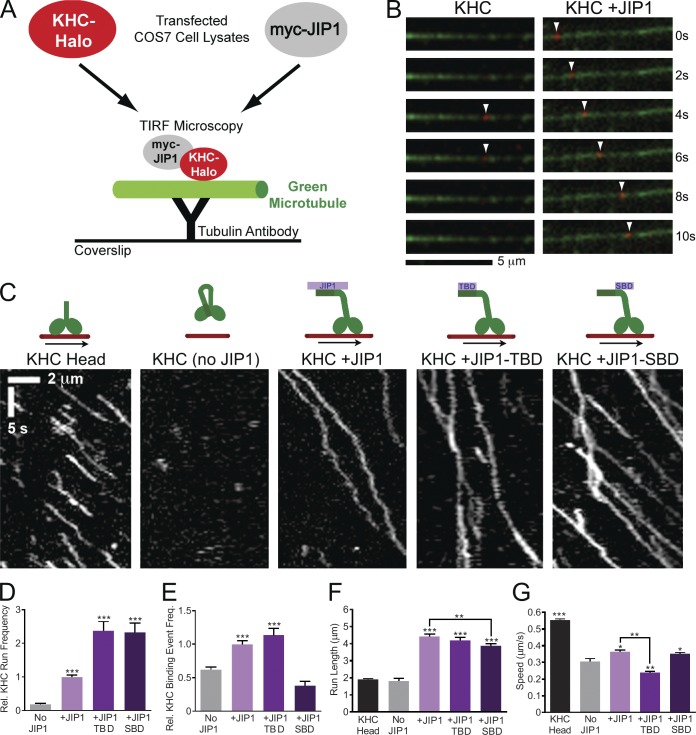
To test this idea functionally, we used an in vitro single molecule motility assay (Blasius et al., 2007). COS7 cells transfected with full-length KHC containing a C-terminal HaloTag (KHC-Halo) were incubated with membrane-permeable red TMR-conjugated HaloTag ligand and lysed. When applied to flow chambers containing immobilized green microtubules, fluorescent KHC-Halo from COS7 lysates can be visualized by total internal reflection fluorescence (TIRF) microscopy (Fig. 3, A–C). Consistent with the established mechanism of full-length KHC autoinhibition, KHC-Halo alone showed only rare non-motile microtubule binding events and runs (Fig. 3, D and E; and Video 2), which may be attributed to stochastic KHC unfolding or activation by endogenous KHC-binding adaptors that are present at low levels in the cell lysate. As a positive control, we also imaged the motility of KHC-Head-Halo (aa 1–560), which exhibited a mean run length of ∼2 µm and mean speed of ∼0.55 µm/s (Fig. 3, F and G), values comparable to those of recombinant KHC head (Dixit et al., 2008).
Figure 3.
JIP1 binding relieves KHC autoinhibition in in vitro TIRF motility assays. (A) Schematic of in vitro TIRF motility assay. Lysate from COS7 cells transfected with KHC-Halo and incubated with red fluorescent TMR ligand was combined with lysate from cells expressing myc-JIP1 constructs and applied to flow chambers containing green fluorescent microtubules, which were immobilized on glass coverslips with anti-tubulin antibody. KHC-Halo motility was imaged using a TIRF microscope. (B) Time-lapsed images acquired from a flow chamber containing KHC-Halo (red) lysate alone (left) show a brief non-motile binding event (arrowheads) to a microtubule (green). Images from a flow chamber containing KHC-Halo and myc-JIP1 lysates (right) show processive movement (arrowheads) along the microtubule. (C) Representative kymographs show activation of KHC-Halo by full-length JIP1, JIP1-TBD, or JIP1-SBD. 100 total frames (∼33 s) are shown. (D) Addition of full-length JIP1, JIP1-TBD, and JIP1-SBD increases the run frequency of full-length KHC-Halo. The absolute number of runs per 10 µm of microtubule was normalized to the KHC-Halo +JIP1 condition for each experiment. (D–G) Data represent three or more independent experiments per condition (n = 52–181 microtubules and n = 109–758 runs) and statistical comparisons were made relative to the KHC-Halo alone (no JIP1) condition unless otherwise indicated. (E) Addition of full-length JIP1 or JIP1-TBD increases the relative frequency of non-motile microtubule-binding events by full-length KHC-Halo. The number of non-motile binding events per 10 µm microtubule length was normalized relative to the KHC-Halo +JIP1 condition for each independent experiment. (F) Addition of full-length JIP1, JIP1-TBD or JIP1-SBD increases KHC-Halo run lengths. (G) Addition of full-length JIP1 or JIP1-SBD but not JIP1-TBD increases speed of KHC-Halo runs. Error bars show the mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
When combined with myc-JIP1 lysate, the frequency of KHC-Halo processive runs increased by more than fivefold (Fig. 3 D and Video 2). Addition of myc-JIP1 also significantly increases the number of stationary binding events (Fig. 3 E), indicating that JIP1 binding increased the probability of KHC unfolding. Moreover, KHC-Halo runs in the presence of myc-JIP1 were significantly faster and had longer run lengths, which doubled to ∼5 µm (Fig. 3, F and G) with ∼15% of motile events reaching run lengths >8 µm (Fig. S3).
To explore the functional consequence of distinct JIP1 interactions with KHC stalk and tail, we tested the effects of JIP1 tail-binding domain (JIP1-TBD; aa 285–440) or JIP1 stalk-binding domain (SBD; aa 554–711) on KHC motility. Addition of JIP1-TBD or JIP1-SBD increased the number of motile KHC runs (Fig. 3 D), indicating that either fragment is sufficient to activate KHC. Interestingly, when compared with the KHC-Halo alone, addition of JIP1-TBD increased the number of non-motile microtubule binding events, but JIP1-SBD did not (Fig. 3 E), suggesting that a greater percentage of KHC unfolding events are converted into runs in the presence of JIP1-SBD.
However, neither JIP1-TBD nor JIP1-SBD was able to fully recapitulate the processive properties of KHC-Halo in the presence of full-length JIP1. Though addition of either JIP-TBD or JIP1-SBD increased KHC-Halo run length relative to the condition lacking JIP1, run lengths in the presence of JIP1-SBD were significantly shorter (by ∼12%) than those in the presence of full-length JIP1 (Fig. 3 F). Histograms of run length distribution indicate that this lower mean is the result of a higher proportion of short runs (<2 µm) and not because of an inability of JIP1-SBD to sustain long runs (Fig. S3). Moreover, addition of JIP1-SBD, but not JIP1-TBD, increased KHC-Halo speed relative to KHC alone (Fig. 3 G), suggesting that binding of KHC stalk to JIP1 may function to enhance KHC speed. Thus, although either JIP1-SBD or JIP1-TBD is sufficient to activate KHC motility in vitro, binding of JIP1 to both KHC stalk and tail likely amplifies the fidelity of the JIP1–KHC interaction, allowing KHC to remain unfolded and process along the microtubule more quickly and for longer run lengths.
JIP1 associates with retrograde motors via direct binding to the p150Glued subunit of dynactin
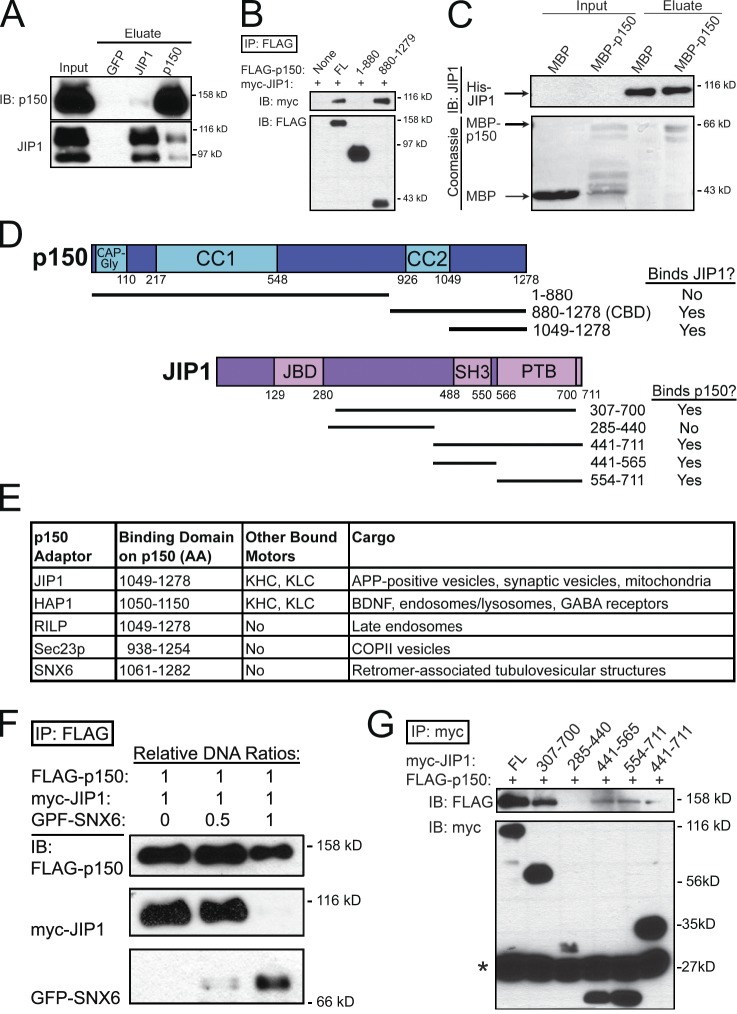
The ability of JIP1 to activate KHC motility is consistent with the observed disruption of anterograde APP transport upon JIP1 depletion in DRGs (Fig. 1, D–F), but these knockdown results also induced deficits in retrograde APP transport. To investigate whether JIP1 associates with the retrograde motor complex, we performed coimmunoprecipitations using mouse brain homogenate and detected an interaction between JIP1 and the p150Glued subunit of the dynein activator dynactin. An anti-p150Glued antibody coimmunoprecipitates both 110- and 90-kD bands of JIP1; an anti-JIP1 antibody also coimmunoprecipitates p150Glued, although to a lesser extent (Fig. 4 A), perhaps because of the additional nonmotor scaffolding functions of JIP1 in the JNK signaling pathway (Dickens et al., 1997). In addition to this biochemical interaction, both JIP1 and p150Glued are enriched at the distal axon tip (Dajas-Bailador et al., 2008; Moughamian and Holzbaur, 2012).
Figure 4.
JIP1 binds directly to the p150Glued subunit of dynactin. (A) Endogenous JIP1 and p150Glued coimmunoprecipitate from mouse brain homogenate. (B) JIP1 binds to the C-terminal p150Glued-CBD. Lysates from COS7 cells cotransfected with myc-JIP1 and FLAG-p150Glued fragments were immunoprecipitated with an anti-FLAG antibody. JIP1 coimmunoprecipitated with C-terminal p150Glued[880–1278], but not with N-terminal p150Glued[1–880]. (C) JIP1 binds directly to C-terminal p150Glued. When applied to His-Bind resin bound to His-JIP1, MBP-p150Glued[1049–1278] selectively bound whereas MBP did not. (D) Summary of p150Glued and JIP1 mapping results. (E) A diverse set of cargo adaptors binds to p150Glued-CBD. (F) SNX6 and JIP1 bind competitively to p150Glued. Lysates from COS7 cells transfected with fixed amounts of FLAG-p150 and myc-JIP1 DNA and progressively increasing amounts of GFP-SNX6 DNA were immunoprecipitated with anti-FLAG antibody. FLAG-p150 coimmunoprecipitates predominantly with myc-JIP1 in the low GFP-SNX6 condition, but mostly with GFP-SNX6 in the high GFP-SNX6 condition. (G) C-terminal JIP1 binds to p150Glued. Lysates from COS7 cells cotransfected with FLAG-p150Glued and myc-JIP1 fragments were immunoprecipitated with an anti-myc antibody. p150Glued coimmunoprecipitated with both myc-JIP1[441–565] and myc-JIP1[554–711]; the presence of both these domains in JIP1 (myc-JIP1[441–711]) did not strengthen the interaction. Asterisk shows antibody light chain bands.
To further define the interaction between p150Glued and JIP1, we performed a series of coimmunoprecipitations using lysates from COS7 cells cotransfected with full-length JIP1 and truncated p150Glued. These experiments revealed that JIP1 does not bind to an N-terminal p150Glued fragment (aa 1–880) containing both the microtubule-binding CAP-Gly domain and the dynein-binding CC1 domain. Rather, JIP1 bound robustly to a C-terminal p150Glued construct containing aa 880–1278 (Fig. 4 B), which will henceforth be referred to as the p150Glued cargo-binding domain (p150Glued-CBD). We further refined this binding domain using recombinant purified full-length His-JIP1 and a C-terminal p150Glued fragment that excludes the CC2 region (aa 1049–1278; Fig. 4 D). When applied to a column with bound His-JIP1, MBP did not bind to His-JIP1, whereas MBP-p150Glued[1049–1278] was specifically retained (Fig. 4 C). Interestingly, smaller copurifying fragments of MBP-p150Glued[1049–1278] (Fig. 4 C, second lane) did not bind to His-JIP1 (fourth lane), suggesting that the last ∼100 aa of the C terminus of p150Glued are essential for JIP1 binding.
C-terminal p150Glued also associates with other cargo adaptors, including huntingtin-associated protein 1 (HAP1; Engelender et al., 1997), Rab7-interacting lysosomal protein (RILP; Johansson et al., 2007), Sec23p (Watson et al., 2005), and the retromer subunit SNX6 (Hong et al., 2009; Wassmer et al., 2009). A comparison of binding studies shows that they all bind to the p150Glued region spanning aa 1049–1278 (Fig. 4 E). If multiple cargo adaptors share this common binding domain, then they are expected to compete for binding to p150Glued. To test this idea, we transfected COS7 cells with fixed amounts of JIP1 and p150Glued DNA and increasing amounts of SNX6 DNA. Communoprecipitations showed that at lower levels of SNX6 expression, p150Glued predominantly binds to JIP1, but that at higher levels of SNX6 expression, p150Glued predominantly binds to SNX6 (Fig. 4 F). This competitive binding between SNX6 and JIP1 suggests that they share a binding site on C-terminal p150Glued.
Next, we performed a series of coimmunoprecipitations using COS7 cells cotransfected with full-length p150Glued and truncated JIP1. These experiments show that the interaction between p150Glued and JIP1 is KLC independent, because FLAG-p150Glued robustly binds myc-JIP1[307–700], which lacks the C-terminal KLC-BD. Interestingly, FLAG-p150Glued binds to both myc-JIP1[441–565] and myc-JIP1[554–711] (Fig. 4 G). The myc-JIP1[441–565] fragment contains the SH3 dimerization domain of JIP1 (Kristensen et al., 2006) and thus may bind to endogenous full-length JIP1.
Anterograde and retrograde JIP1 motile complexes are mutually exclusive
In addition to the known interaction of JIP1 with KLC, we have now identified three novel interactions of JIP1 with KHC stalk, KHC tail, and p150Glued-CBD (Fig. 5 A). These interactions establish JIP1 as a scaffolding protein that binds to both anterograde and retrograde motor complexes; other proteins with this ability include the huntingtin–HAP1 complex (Engelender et al., 1997; McGuire et al., 2006; Caviston et al., 2007; Twelvetrees et al., 2010), JIP3 (Cavalli et al., 2005; Arimoto et al., 2011; Sun et al., 2011), and Milton/TRAK (Glater et al., 2006; van Spronsen et al., 2013).
Figure 5.
Anterograde and retrograde JIP1 motor complexes are mutually exclusive. (A) Summary schematic of direct binding interactions between JIP1, Kinesin-1, and dynactin. (B) JIP1 cannot bind simultaneously to both p150Glued and KHC stalk. Lysates from COS7 cells triple transfected with myc-JIP1, FLAG-p150Glued, and GFP-KHC-stalk were immunoprecipitated with an anti-GFP antibody. FLAG-p150Glued and GFP-KHC-stalk do not interact with each other either in the absence or presence of myc-JIP1. (C) JIP1 cannot bind simultaneously to both p150Glued and KHC tail. Lysates from COS7 cells triple transfected with myc-JIP1, FLAG-p150Glued, and GFP-KHC-tail were immunoprecipitated with an anti-GFP antibody. FLAG-p150Glued and GFP-KHC-tail do not interact with each other either in the absence or presence of myc-JIP1. (D) JIP1 cannot bind simultaneously to p150Glued and KHC. Lysates from COS7 cells triple transfected with myc-JIP1, FLAG-p150Glued, and either GFP-KHC-tail or GFP-KHC-stalk were immunoprecipitated with an anti-p150Glued antibody. Though robust levels of FLAG-p150Glued and associated myc-JIP1 are coimmunoprecipitated, no interacting GFP-KHC-stalk or GFP-KHC-tail can be detected. (E) JIP1 can bind simultaneously to both p150Glued and KLC. Lysates from COS7 cells triple transfected with myc-JIP1, FLAG-p150Glued, and HA-KLC were immunoprecipitated with either an anti-FLAG or anti-HA antibody. In the absence of myc-JIP1, FLAG-p150Glued and HA-KLC do not interact. The addition of myc-JIP1 facilitates the indirect interaction between p150Glued and KLC as both FLAG and HA antibodies immunoprecipitate triple complexes of FLAG-p150Glued, myc-JIP1, and HA-KLC. (F) Model of two mutually exclusive JIP1 motile complexes. The anterograde JIP1 complex activates KHC motility via direct binding to both stalk and tail domains (left) but cannot bind simultaneously to p150Glued; KLC may remain bound via the C-terminal tail of JIP1 (Verhey et al., 2001). The retrograde JIP1 complex binds directly to p150Glued to facilitate dynein-mediated transport and may retain autoinhibited KHC via simultaneous binding to KLC (right).
However, it is unclear whether anterograde and retrograde motor complexes interact with scaffolding proteins simultaneously or alternatingly. Because KHC stalk and p150Glued share a binding domain at the C-terminal PTB region of JIP1 (Figs. 2 F and 4 G), we hypothesized that KHC and p150Glued compete for binding to JIP1. Thus, we asked whether p150Glued and KHC stalk can simultaneously bind to JIP1 by coexpressing myc-JIP1, FLAG-p150Glued, and GFP-KHC-stalk in COS7 cells. Immunoprecipitated GFP-KHC-stalk pulls down myc-JIP1, but no associated FLAG-p150Glued (Fig. 5 B); in the complementary experiment, immunoprecipitated FLAG-p150Glued robustly pulled down myc-JIP1, but no associated GFP-KHC-stalk (Fig. 5 D). This suggests that KHC stalk and p150Glued cannot form a tripartite complex with JIP1, consistent with KHC stalk and p150Glued sharing a binding domain at the PTB region in JIP1.
A similar triple transfection was performed using GFP-KHC-tail; coimmunoprecipitation against KHC tail also revealed no coeluted FLAG-p150Glued (Fig. 5 C). Likewise, the complementary immunoprecipitation against FLAG-p150Glued pulled down myc-JIP1, but no associated KHC tail (Fig. 5 D). Though p150Glued and KHC tail do not share a common binding domain, KHC tail binding to JIP1 may sterically hinder p150Glued binding in the three-dimensional structure of full-length JIP1, which remains unsolved. The results of these experiments are consistent with the exclusion of p150Glued from the JIP1–KHC complex and the exclusion of KHC from the JIP1–p150Glued complex.
Because the KLC-BD and the p150Glued-binding domain on JIP1 do not overlap, we hypothesized that KLC and p150Glued would be able to form a tripartite complex with JIP1. To address this, we performed triple transfections in COS7 cells with HA-KLC, FLAG-p150Glued, and myc-JIP1 and immunoprecipitated against the tags for KLC and p150Glued. Although HA-KLC and FLAG-p150Glued do not coimmunoprecipitate in control lysates lacking exogenous JIP1, cotransfection of myc-JIP1 leads to formation of a tripartite complex containing KLC, p150Glued, and JIP1 (Fig. 5 E), which is consistent with the colocalization of KLC and dynein on APP vesicles in neurons (Szpankowski et al., 2012).
Collectively, these binding experiments suggest that the JIP1 motile complex exists in two mutually exclusive states. In one conformation, JIP1 binds directly to both KHC stalk and tail and excludes p150Glued from binding to JIP1. This JIP1 complex likely mediates anterograde transport, consistent with the ability of JIP1 to activate full-length KHC motility. In another conformation, JIP1 binds directly to p150Glued to mediate retrograde transport and can simultaneously bind to KLC (Fig. 5 E). However, because KHC cannot directly bind to this p150Glued-associated JIP1 complex, simultaneous binding of KLC may function to retain autoinhibited KHC on the vesicle. This model is consistent with previous studies showing that KLC is inhibitory to microtubule binding (Verhey et al., 1998) and KHC motility in vitro (Friedman and Vale, 1999), and that the JIP1 KLC-BD is sufficient for KHC recruitment to vesicles, but not for activation of motility (Kawano et al., 2012).
Furthermore, we affirmed that these JIP1 complexes indeed associate with APP. Immunoprecipitation of JIP1 and APP from mouse brain homogenate pulls down the expected set of associated motors, including KHC, p150Glued, and dynein intermediate chain (Fig. S4 A). Immunostaining of nontransfected cultured DRGs shows that endogenous APP and JIP1 colocalize on puncta along the axon (Fig. S4 B). Moreover, these complexes are functional, as fluorescently tagged JIP1 and APP comigrate on both anterograde and retrograde moving vesicles along DRG axons (Fig. S4, C and D).
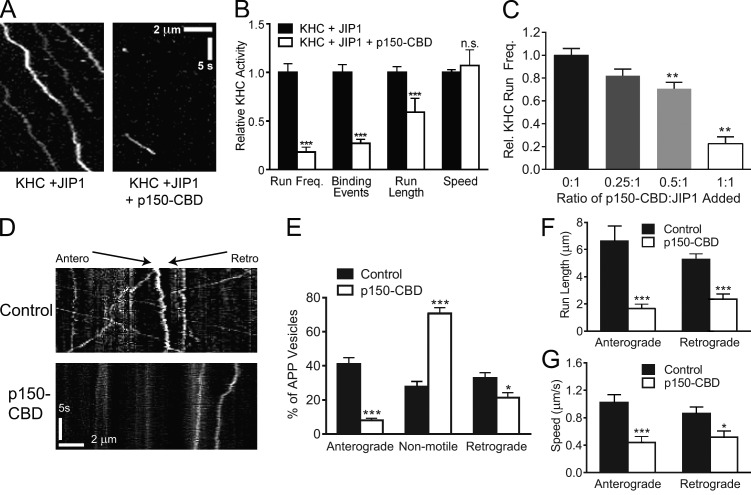
p150Glued binding competitively inhibits activation of KHC by JIP1 and disrupts anterograde APP transport
Binding experiments show that KHC and p150Glued cannot bind simultaneously to JIP1 and likely compete for binding to JIP1. Thus, we hypothesized that p150Glued binding to JIP1 will functionally disrupt KHC activation in vitro. To this end, we performed motility experiments in which p150Glued-CBD lysates were mixed first with JIP1 lysates and then KHC-Halo lysates. The addition of p150Glued-CBD severely disrupted the ability of JIP1 to activate KHC motility and only rare short runs could be observed (Fig. 6 A and Video 3). When normalized to KHC-Halo motility in the presence of JIP1, addition of p150Glued-CBD significantly decreases both the relative frequency of stationary microtubule-binding events and runs as well as run length with no significant changes in speed (Fig. 6 B). Moreover, at constant levels of JIP1, addition of increasing amounts of p150Glued-CBD resulted in incremental decreases in KHC run frequency (Fig. 6 C), suggesting that inhibition of JIP1-mediated KHC motility by p150Glued occurs in a competitive manner.
Figure 6.
p150Glued-CBD disrupts JIP1-mediated KHC motility in vitro and anterograde APP-positive vesicle transport in DRGs. (A) Representative kymographs show that addition of p150Glued-CBD disrupts enhancement of KHC-Halo motility by JIP1. Lysate from COS7 cells transfected with myc-JIP1 were combined with FLAG-p150Glued-CBD lysate and immediately combined with KHC-Halo lysate. This lysate mixture was applied to flow chambers containing immobilized fluorescent microtubules and imaged. 100 total frames (∼33 s) are shown. (B) Addition of p150Glued-CBD decreases the number of motile KHC events mediated by JIP1. Motility measurements in the presence of FLAG-p150Glued-CBD were normalized to the condition containing only myc-JIP1 and KHC-Halo and represent three independent experiments (n = 60–100 microtubules and n = 23–214 runs). (C) p150Glued-CBD competitively inhibits JIP1-mediated KHC motility in vitro. At constant levels of myc-JIP1 lysate, addition of incrementally higher levels of p150Glued-CBD lysate leads to complementary decreases in relative KHC-Halo run frequency. Data represents three independent experiments (n = 6–52 microtubules). (D) Kymographs of APP-DsRed motility in DRGs transfected with a bicistronic construct coexpressing FLAG-p150Glued-CBD and GFP. Approximately 80 total frames (∼20 s) are shown. (E) Expression of p150Glued-CBD significantly decreases the percentage of anterograde APP-positive vesicles and correspondingly increases the percentage of arrested vesicles. (E–G) Data represent four independent experiments (n = 12–14 neurons). (F and G) Expression of p150Glued-CBD significantly decreases run length and speed of both anterograde and retrograde APP-positive vesicles. Means represent only vesicles categorized as motile (i.e., anterograde or retrograde in E). Error bars show the mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To detect whether p150Glued binding to JIP1 also disrupts motility in neurons, we imaged APP-DsRed transport in DRGs cotransfected with a bicistronic vector coexpressing p150Glued-CBD and a GFP transfection marker. The overall number of APP-positive vesicles in the axon did not change significantly upon p150Glued-CBD expression (control, 0.37 ± 0.05/µm; p150Glued-CBD, 0.42 ± 0.08/µm). However, neurons expressing p150Glued-CBD showed dramatic inhibition of APP transport in both anterograde and retrograde directions, with a majority of arrested APP-positive vesicles (Fig. 6, D and E; and Video 4). Moreover, p150Glued-CBD expression in DRGs also decreased the run lengths and speeds of both anterograde and retrograde APP-positive vesicles (Fig. 6, F and G). In the retrograde direction, p150Glued-CBD likely acts as dominant negative by competing against endogenous full-length p150Glued for JIP1 binding, whereas in the anterograde direction p150Glued-CBD likely prevents formation of the anterograde JIP1 motile complex by disrupting the binding of KHC to JIP1.
JIP1 phosphorylation enhances KHC activation in vitro and promotes anterograde APP transport
To further validate the mechanism regulating switching between the anterograde and retrograde JIP1 motile complexes, we endeavored to identify a regulatory mechanism controlling JIP1 binding activity. A previous study in Drosophila melanogaster suggests a role for JNK in the regulation of JIP1-mediated transport of synaptic vesicles (Horiuchi et al., 2007). Immunoprecipitation of KHC from mouse brain homogenate preferentially pulls down an ∼120-kD JIP1 band (Fig. 2 A). This ∼120-kD JIP1 band as well as a lower molecular mass JIP1 band are phosphoproteins, as they are dephosphorylated upon λ phosphatase treatment (Fig. S5 A). These observations suggest that phospho-JIP1 preferentially binds to KHC. In addition, expression of truncated JIP1[307–554], which overlaps with JIP1-TBD, results in an additional phosphorylated band, which selectively binds to KHC tail; further, this binding is significantly disrupted in the absence of phosphatase inhibitors (Fig. S5 B).
Thus, to identify JIP1 phosphorylation sites that enhance binding to KHC tail, we superimposed a map of known JIP1 phosphorylation sites (Nihalani et al., 2003; D’Ambrosio et al., 2006) onto our map of motor binding domains (Fig. S5 C). Though no known phosphorylation sites are in C-terminal JIP1 where KLC and KHC stalk, the minimal KHC-tail–binding domain of JIP1 (aa 391–440) contains a proline-directed site, S421, previously demonstrated to be directly phosphorylated by JNK in vitro (Nihalani et al., 2003). This region of JIP1 is heavily conserved in humans and rodents (Fig. S5 D) and may represent a conserved KHC-tail–binding motif, as a similar region is found within the KHC-tail–binding domain of JIP3 (Fig. S5 E).
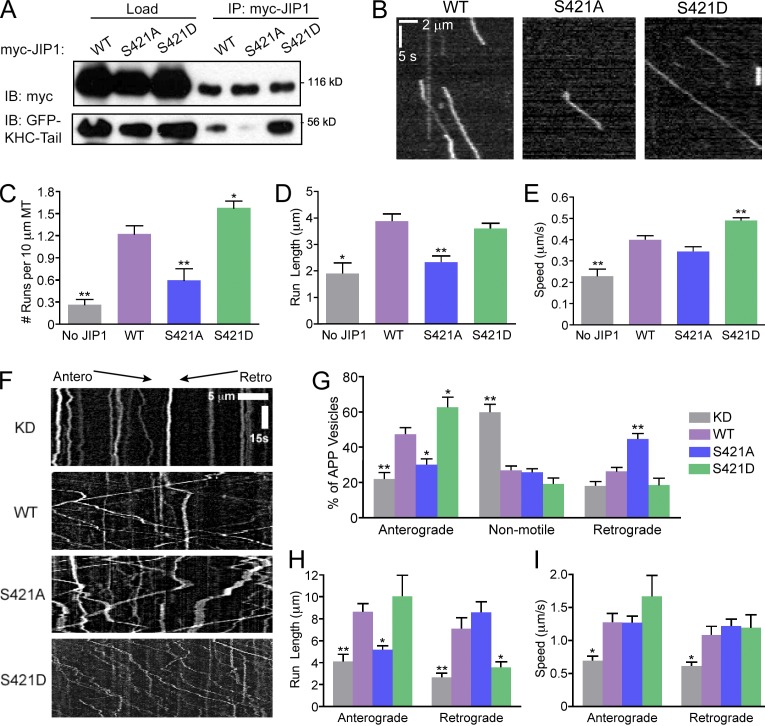
Because the minimal KHC-tail–binding domain of JIP1 contains a high percentage of negatively charged residues (Fig. S2 C), we hypothesized that phosphorylation in this region would further strengthen the interaction of JIP1 with the positively charged IAK region of KHC tail responsible for autoinhibition. Initially, we tested the ability of JIP1 phosphomutants to bind to KHC tail in COS7 lysates; whereas phosphodeficient JIP1-S421A binds weakly to KHC tail, phosphomimetic JIP1-S421D binds more robustly to KHC tail than wild-type JIP1 (Fig. 7 A). Next, we confirmed that JIP1-S421 phosphomutants also have altered ability to activate full-length KHC motility in vitro (Fig. 7 B and Video 5). Relative to wild-type JIP1, JIP1-S421A activated fewer KHC runs with shorter run lengths and no change in speed, whereas JIP1-S421D activated more runs with faster speed and no change in run length (Fig. 7, C–E).
Figure 7.
Mutations of the JNK phosphorylation site S421 in JIP1 alter KHC activation in vitro and APP directionality in neurons. (A) Mutations at JIP1-S421 alter KHC-tail–binding ability. COS7 cells were cotransfected with GFP-KHC-tail and wild-type or mutant myc-JIP1 and immunoprecipitated with anti-myc antibody. (B) Representative kymographs of KHC-Halo motility show weak activation by myc-JIP1-S421A and enhanced activation by myc-JIP1-S421D in in vitro motility assays. 100 total frames (∼33 s) are shown. (C) KHC-Halo run frequencies in vitro decrease in the presence of JIP1-S421A and increase in the presence of JIP1-S421D. (C–E) Data from three independent experiments (n = 48–101 microtubules and n = 18–254 runs) are shown and statistical comparisons were made versus the wild-type JIP1 condition. (D) KHC-Halo run length is decreased with addition of JIP1-S421A. (E) KHC-Halo speed is increased with addition of JIP1-S421D. (F) Representative kymographs of APP-YFP motility in DRGs transfected with siRNA targeted to mouse JIP1 and rescued with a bicistronic construct coexpressing human wild-type or mutant JIP1 as well as the fluorescent transfection marker BFP. (G) DRGs expressing JIP1-S421D have increased percentages of anterograde APP vesicles, whereas DRGs expressing JIP1-S421A have increased percentages of retrograde APP vesicles. (G–I) Data from three independent experiments (n = 7–9 neurons and n = 78–224 runs) are shown with statistical comparisons made against the wild-type rescue condition. (H) APP-positive vesicles in DRGs expressing JIP1-S421A have decreased anterograde run length, whereas those expressing JIP1-S421D have decreased retrograde run length. (I) No significant differences are observed in APP speeds in DRGs expressing JIP1-S421 phosphomutants. Error bars show the mean ± SEM; *, P < 0.05; **, P < 0.01.
Finally, we tested the effect of JIP1 phosphorylation on APP transport in DRGs by knockdown of endogenous JIP1 and rescue with a bicistronic vector coexpressing human wild-type or mutant JIP1 and a BFP transfection marker (Fig. 7 F and Video 6). When compared with neurons rescued with wild-type JIP1, neurons expressing JIP1-S421D have increased anterograde APP transport. Conversely, neurons rescued with JIP1-S421A have decreased anterograde and increased retrograde APP transport (Fig. 7 G). These shifts in direction of APP transport are consistent with the association of phospho-JIP1 in the anterograde JIP1 motile complex and of nonphosphorylated JIP1 in the retrograde JIP1 motile complex (Fig. 5 F).
When compared with neurons rescued with wild-type JIP1, neurons expressing JIP1-S421A exhibited decreased anterograde run length, whereas JIP1-S421D exhibited decreased retrograde run length (Fig. 7 H). This indicates that nonphosphorylated JIP1 cannot sustain long anterograde runs whereas phospho-JIP1 cannot sustain long retrograde runs. Though no significant changes in APP speed were observed when rescuing with either JIP1 phosphomutant (Fig. 7 I), rescue with JIP1-S421D doubled the percentage of anterograde APP runs with speeds higher than 2.5 µm/s (Fig. S5 F). Interestingly, trends in anterograde APP motility in neurons closely parallel KHC activation measurements in vitro (Fig. 7, C–E), consistent with our model that facilitation of an anterograde motile complex by JIP1 phosphorylation may underlie these observed changes in APP axonal transport.
Discussion
Here, we examine the coordinated regulation of APP axonal transport by the scaffolding protein JIP1, which mediates the activity of both anterograde and retrograde motors via direct binding to KHC stalk and tail and the p150Glued subunit of dynactin. In vitro motility assays demonstrate that binding of JIP1 relieves autoinhibition of full-length KHC and enhances KHC processivity. Coimmunoprecipitations indicating that KHC and p150Glued cannot simultaneously bind to JIP1 led us to posit a model whereby JIP1 switches between two mutually exclusive conformations—an anterograde KHC-bound state and a retrograde p150Glued-bound state. Indeed, binding of p150Glued-CBD to JIP1 competitively disrupts enhancement of KHC motility by JIP1 in vitro and perturbs anterograde APP transport in neurons. Further, direct phosphorylation by JNK likely regulates the directional switching of JIP1, as phosphomimetic JIP1-S421D enhances KHC tail binding and promotes anterograde APP axonal transport.
Direct binding of JIP1 to KHC stalk represents a novel mechanism for KHC activation by an adaptor protein. The flexibility of the stalk or hinge region of KHC was first observed in electron micrographs showing folding or bending of KHC (Hirokawa et al., 1989). A hingeless KHC mutant displays weak autoinhibition, moving with higher speeds than full-length KHC (Friedman and Vale, 1999), suggesting that the stalk is necessary for KHC bending and tail-to-head binding. Indeed, the minimal JIP1-binding domain that we identified for KHC stalk (aa 560–682) overlaps with the truncated region in this hingeless KHC mutant (aa 505–610). In vitro, binding of JIP1-SBD to KHC stalk enhances run frequency and speed. Thus, binding of JIP1 to KHC stalk may conformationally restrain this flexible hinge region and hold KHC head and tail apart, effectively preventing KHC autoinhibition.
We find that both KHC stalk and tail interactions with JIP1 are necessary for maximal enhancement of KHC processivity, as addition of JIP1-TBD in vitro cannot increase KHC speed and JIP1-SBD does not enhance run lengths as efficiently as full-length JIP1. Consistent with the established stoichiometry that one KHC tail is sufficient to autoinhibit a motor head dimer (Hackney et al., 2009), the redundancy of multiple KHC binding sites on JIP1 may function to decrease the likelihood of full-length KHC returning to its autoinhibited conformation once bound to cargo. Moreover, the ability of JIP1 to dimerize via its SH3 domain (Kristensen et al., 2006) may allow recruitment of multiple Kinesin-1 motors onto a single APP-positive vesicle. This is consistent with the observation that long run lengths observed for APP in D. melanogaster are dependent on the activity of multiple KHCs (Reis et al., 2012) and with in vitro observations that increasing KHC motor number on a DNA scaffold correlates with increased run length (Derr et al., 2012; Furuta et al., 2013).
Though we have shown that direct binding of JIP1 to KHC is sufficient to activate motility in vitro, the majority of soluble Kinesin-1 in cells exists as a tetramer of KHC and KLC. Binding of KLC to KHC likely provides an additional layer of inhibition as addition of KLC decreases both microtubule binding (Blasius et al., 2007) and motility (Friedman and Vale, 1999) of KHC. In the presence of KLC, JIP1 is insufficient to activate KHC motility; activation of tetrameric Kinesin-1 requires an additional KHC-tail–binding partner, FEZ1 (Blasius et al., 2007). JIP1 and FEZ1 may cooperate in a stepwise manner where initial binding of JIP1 to KLC and binding of FEZ1 to KHC tail overcomes KHC autoinhibition, thus priming KHC for binding to JIP1, which then sustains anterograde transport via dual interactions with KHC stalk and tail.
Binding of JIP1 to KLC may also function to recruit Kinesin-1 to cargos. Several scaffolding proteins initially identified as KLC binding partners, including JIP3 (Bowman et al., 2000) and HAP1 (McGuire et al., 2006), also bind directly to KHC (Twelvetrees et al., 2010; Sun et al., 2011). Our biochemical observation that KLC and p150Glued can bind simultaneously to JIP1 is consistent with colocalization of KLC and dynein on APP vesicles in neurons (Szpankowski et al., 2012). Thus we propose that KLC may function to retain autoinhibited KHC on the organelle during retrograde transport (Fig. 5 F), fitting recent experiments that show JIP1 KLC-BD can recruit Kinesin-1 to vesicles but cannot activate transport (Kawano et al., 2012). The positioning of oppositely directed motors on the same scaffolding protein complex may function as a primed state that is poised for rapid transitions between retrograde and anterograde transport.
Together, our data support a model in which JIP1 phosphorylation regulates transport by switching between two distinct motile states. In contrast to an unregulated tug of war between opposing motors, mutually exclusive JIP1 motile complexes allow only one motor type to be active at any given time. Advantages to this regulatory scheme are twofold and particularly relevant in the extended axon. First, by avoiding frequent back-and-forth saltatory movement that is characteristic of the tug-of-war model, JIP1-coordinated transport can be sustained over long distances. Second, distinct anterograde and retrograde complexes confer directional bias for JIP1-associated cargos, promoting efficient transport in both directions. These attributes are fully consistent with the observed transport of APP, as this cargo moves quickly for long distances in both anterograde and retrograde directions.
Previous studies have also correlated posttranslational modifications of scaffolding proteins with directional transport changes, including phosphorylation of huntingtin in brain-derived neurotrophic factor–containing vesicles (Colin et al., 2008) and sumoylation of La in mRNA transport (van Niekerk et al., 2007). Here, we propose that direct phosphorylation of JIP1 by JNK acts as a molecular switch at the cargo level and affects changes in directionality of transport via direct alteration of motor binding affinities. Indeed, phosphomimetic JIP1-S421D shows enhanced binding to KHC tail and promotes anterograde APP transport, whereas phosphodeficient JIP1-S421A has reduced binding to KHC tail and likely associates with p150Glued to promote retrograde APP transport. Surprisingly, JIP1-S421A, which has an intact SBD, is unable to increase KHC run frequency and run length, in marked contrast to the robust effects of the JIP1-SBD fragment on KHC activation. This suggests phosphorylation of S421 may also indirectly regulate availability of C-terminal JIP1 for binding to KHC stalk, perhaps by inducing a conformation change that reveals the SBD. Though JIP1 S421 is directly phosphorylated by JNK in vitro (Nihalani et al., 2003) and synaptic vesicle transport in D. melanogaster relies on JNK and upstream kinases such as DLK (Horiuchi et al., 2007), further work will be required to directly correlate changes in JNK activity with changes in APP transport.
Finally, multiple levels of regulation may modulate APP transport in vivo. JIP1 can oligomerize and cotransport with JIP3 (Hammond et al., 2008), which does not directly bind to APP, but may facilitate APP transport by enhancing APP phosphorylation (Muresan and Muresan, 2005a), which in turn enhances the binding of APP to JIP1 (Muresan and Muresan, 2005b). Furthermore, reduction of endogenous GSK3 levels can enhance both anterograde and retrograde APP transport, likely via changes in microtubule acetylation and stability (Weaver et al., 2013).
The molecular mechanism described here for the regulation of APP transport by JIP1 raises many further questions. Does JIP1 regulate the transport of other cargos along the axon? How might the mechanism for JIP1-mediated transport of APP described here affect Aβ production and Alzheimer’s disease pathology? Although further work will be required to address these questions, our observations at both cellular and single molecule levels establish JIP1 as a coordinator of axonal transport that regulates transport directionality by alternating between anterograde and retrograde motile states. These new mechanistic insights further support a critical role for scaffolding proteins in the coordination of kinesin and dynein motor activity in the cell.
Materials and methods
Cell culture and transfection
Dissected adult mouse DRGs were treated with papain, collagenase, and dispase II, and then centrifuged through a 20% Percoll gradient (Perlson et al., 2009). Isolated DRGs were transfected using Amaxa Nucleofector SCN Program 6 (Lonza) and plated on glass-bottom microwell dishes (FluoroDish; World Precision Instruments) that were precoated with poly-l-lysine and laminin. DRGs were maintained in F12 media (Invitrogen) with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. For knockdown experiments, DRGs were transfected with fluorescent red DY-547–conjugated siRNA (Thermo Fisher Scientific) and either APP-YFP or EGFP-Rab7. For p150Glued-CBD overexpression experiments, neurons were transfected with APP-DsRed and pBI-CMV2(AcGFP)-FLAG-p150Glued-CBD. For JIP1 knockdown and rescue experiments, neurons were transfected with DY-547–conjugated siRNA, APP-YFP, and pBI-CMV2(BFP)-JIP1(WT, S421A, or S421D).
CAD cells were maintained in DMEM/F12 media supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. CAD cells were differentiated by serum deprivation and transfected with siRNA using RNAiMAX (Lipofectamine), and then transfected with APP-YFP using Fugene6 (Roche) 24 h later.
Live-cell imaging
Cultured DRGs were imaged at 2 DIV in Hibernate A low-fluorescence medium (Brain Bits) inside a 37°C imaging chamber. Double- or triple-fluorescent neurons were observed at 63× using an epifluorescence microscope (DMI6000B; Leica) with a CTR7000 HS control box run by AF6000 software (Leica) and an Orca-R2 camera (C10600; Hamamatsu Photonics). Images of APP transport were acquired at 250 ms per frame for 1 min. CAD cells were imaged using the same conditions 48 h after differentiation.
Vesicle tracking and analysis
APP-positive vesicles were analyzed by generating 50-µm kymographs (at least 200 µm from the soma) using Metamorph software. Motile particles (e.g., anterograde or retrograde) were defined as particles with net displacement >1 µm. Individual runs were defined as a run with constant velocity; in other words, one vesicle can have several runs within the duration of a movie if it pauses or changes speed or direction. F-tests confirmed that neurons in different dishes are not significantly different; thus the neuron was defined as the biologically relevant unit and motility parameters were averaged for each neuron and subject to subsequent statistical analysis.
Immunofluorescence
Differentiated CAD cells or cultured DRGs were grown on glass coverslips and fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, incubated 1 h in blocking solution (5% goat serum and 1% BSA), and then incubated with primary antibodies against JIP1 (B7; Santa Cruz Biotechnology, Inc.), p150Glued (BD), and/or APP (EMD Millipore) followed by incubation with species-specific fluorescent secondary antibodies.
Coimmunoprecipitations
COS7 cells transfected using Fugene6 (Roche) according to the manufacturer’s instructions were harvested 18–24 h after transfection and lysed using 0.5% Triton X-100 (in HEM buffer). Lysates were incubated with Protein G–Dynabeads (Invitrogen) and coimmunoprecipitations were performed following the manufacturer’s instructions using the following antibodies: anti-JIP1 (B7; Santa Cruz Biotechnology, Inc.), anti-p150Glued (BD), anti-KHC (1614; EMD Millipore), anti-FLAG (Sigma-Aldrich), anti-GFP (Takara Bio Inc.), anti-HA (Covance), and anti-myc (Invitrogen). All coimmunoprecipitations represent at least three independent experiments.
Recombinant protein binding assays
For KHC-stalk–binding assays, mouse KIF5C stalk (aa 560–682) was subcloned into the pGEX6p-1 vector (GE Healthcare), expressed in BL21 (DE3) Escherichia coli (EMD Millipore), and induced at an OD600 of ∼0.6 for 2 h with 0.4 mM IPTG. E. coli were lysed with lysozyme and treated with DNAase I and RNase A and the resulting supernatant was purified by binding to glutathione Sepharose-4B (GE Healthcare). For KHC-stalk–binding experiments, pRSETA-His-JIP1 (Nihalani et al., 2003) was expressed in BL21 E. coli and purified using His-Bind Resin (EMD Millipore) under denaturing conditions using urea as previously described (Karki and Holzbaur, 1995). Glutathione beads bound with either GST or GST-KHC-stalk[560–682] were incubated for 30 min at room temperature with purified His-JIP1, washed, and eluted with denaturing buffer.
For KHC-tail–binding assays, His-JIP1 and His-KHC-tail[823–944] (Dietrich et al., 2008) were expressed in Rosetta E. coli and purified using His-Bind Resin following the manufacturer’s protocol. Purified His-KHC-tail[823–944] with or without purified His-JIP1 was incubated with Protein G–Dynabeads bound to anti-JIP1 antibody, washed, and eluted with denaturing buffer.
For p150Glued-binding assays, MBP-p150Glued[1049–1278] (Johansson et al., 2007) was expressed in Rosetta E. coli, purified using amylose resin (New England Biolabs, Inc.) following the manufacturer’s protocol. Purified His-JIP1 (from Rosetta E. coli) and MBP-p150Glued[1049–1278] were buffer exchanged into HEM buffer with 25 mM NaCl using PD10 columns (GE Healthcare). His-Bind resin bound to His-JIP1 was incubated for 30 min at room temperature with purified MBP or MBP-p150Glued[1049–1278], washed, and eluted with denaturing buffer. All binding assays were performed at least two times.
In vitro COS7 lysate motility assay
This assay was adapted from Blasius et al. (2007). Transfected COS7 cells expressing KHC-head-Halo (KIF5C[1–560]) or KHC-Halo were incubated with TMR ligand (Promega) following the manufacturer’s instructions. COS7 cells were lysed in P12 buffer (12 mM Pipes, 2 mM MgCl2, and 1 mM EGTA, pH 6.8) with 0.1% Triton X-100 and cleared by centrifugation at 1,000 g and then 100,000 g. Flow chambers were constructed using two strips of double-sided tape between a slide and a coverslip and lined with vacuum grease to yield a chamber volume of ∼10 µl. Four solutions were flowed sequentially into the chamber and incubated at room temperature for 5 min each: anti-tubulin antibody (Takara Bio Inc.), Pluronic F-127 (50 mg/ml), Taxol-stabilized fluorescent microtubules (HiLyte 488 tubulin; Cytoskeleton), and COS7 cell lysates in activation buffer (Cai et al., 2007). For each set of experiments, we conducted at least three independent trials, each with fresh lysate. To minimize variation between chambers, each chamber contained equal amounts of microtubules, KHC-Halo lysate, and total lysate (by adding nontransfected lysate).
For each condition, we acquired multiple videos for each trial. Videos were acquired at three frames per second for 1 min at room temperature using an Ultraview Vox spinning disk TIRF system (PerkinElmer) on an inverted Ti microscope (Nikon) with the 100× objective and an ImagEM C9100-13 camera (Hamamatsu Photonics) controlled by Volocity software. Kymographs of microtubules with lengths >10 µm were analyzed for stationary binding events and runs. Run frequency measurements were normalized with respect to microtubule length. Individual runs were measured at the level of each motile particle (i.e., each particle has one run length or net displacement and one speed measurement).
Statistical analysis
Statistical analysis was performed using Student’s t test or one-way analysis of variance with post-hoc Tukey’s test. For JIP1 mutant experiments, we used post-hoc Dunnett’s Test, comparing against the control wild-type JIP1 rescue condition. Bar graphs were plotted as mean ± SEM and the following denotations for statistical significance were used: *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
Online supplemental material
Fig. S1 shows disruption of APP transport by JIP1 knockdown in the CAD neuronal cell line. Fig. S2 includes information on His-JIP1 purification and characterization of the minimal KHC-tail–binding domain. Fig. S3 is a histogram of KHC run length distribution upon addition of full-length and truncated JIP1. Fig. S4 demonstrates that APP and JIP1 form functional transport complexes using coimmunoprecipitations, immunostaining, and live-cell comigration studies. Fig. S5 shows characterization of JIP1 phosphorylation in the brain and identification of the S421 phosphorylation site. In Video 1, JIP1 depletion disrupts APP-YFP motility in DRGs. In Video 2, JIP1 activates KHC-Halo motility in an in vitro single molecule TIRF assay. In Video 3, addition of FLAG-p150Glued-CBD inhibits JIP1-mediated enhancement of KHC-Halo motility in vitro. In Video 4, expression of p150Glued-CBD disrupts APP-DsRed transport in DRGs. In Video 5, phosphorylation mutants at JIP1-S421 alter KHC activation in vitro. In Video 6, phosphorylation mutants at JIP1-S421 alter directionality of APP-YFP transport in DRGs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201302078/DC1.
Supplementary Material
Acknowledgments
We gratefully acknowledge technical assistance from Mariko Tokito, Adam Hendricks, and Karen Wallace and helpful discussion and encouragement from members of the Holzbaur Laboratory, as well as Drs. Steve Scherer, Virginia Lee, and Dennis Kolson.
National Institutes of Health gave funding to M.M. Fu (T32GM7517 and F31NS073262) and E.L.F. Holzbaur (GM48661).
Footnotes
Abbreviations used in this paper:
- APP
- amyloid precursor protein
- DRG
- dorsal root ganglion
- HAP1
- huntingtin-associated protein 1
- JIP
- JNK-interacting protein
- JIP1-TBD
- JIP1 tail-binding domain
- KHC
- kinesin heavy chain
- KLC
- kinesin light chain
- p150Glued-CBD
- p150Glued cargo-binding domain
- PTB
- phosphotyrosine binding
- SBD
- stalk-binding domain
- TIRF
- total internal reflection fluorescence
References
- Abe N., Almenar-Queralt A., Lillo C., Shen Z., Lozach J., Briggs S.P., Williams D.S., Goldstein L.S., Cavalli V. 2009. Sunday driver interacts with two distinct classes of axonal organelles. J. Biol. Chem. 284:34628–34639 10.1074/jbc.M109.035022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto M., Koushika S.P., Choudhary B.C., Li C., Matsumoto K., Hisamoto N. 2011. The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. J. Neurosci. 31:2216–2224 10.1523/JNEUROSCI.2653-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius T.L., Cai D., Jih G.T., Toret C.P., Verhey K.J. 2007. Two binding partners cooperate to activate the molecular motor Kinesin-1. J. Cell Biol. 176:11–17 10.1083/jcb.200605099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A.B., Kamal A., Ritchings B.W., Philp A.V., McGrail M., Gindhart J.G., Goldstein L.S. 2000. Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell. 103:583–594 10.1016/S0092-8674(00)00162-8 [DOI] [PubMed] [Google Scholar]
- Cai D., Verhey K.J., Meyhöfer E. 2007. Tracking single Kinesin molecules in the cytoplasm of mammalian cells. Biophys. J. 92:4137–4144 10.1529/biophysj.106.100206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli V., Kujala P., Klumperman J., Goldstein L.S.B. 2005. Sunday Driver links axonal transport to damage signaling. J. Cell Biol. 168:775–787 10.1083/jcb.200410136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J.P., Ross J.L., Antony S.M., Tokito M., Holzbaur E.L. 2007. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc. Natl. Acad. Sci. USA. 104:10045–10050 10.1073/pnas.0610628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X.J., Saudou F., Humbert S. 2008. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 27:2124–2134 10.1038/emboj.2008.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio C., Arena S., Fulcoli G., Scheinfeld M.H., Zhou D., D’Adamio L., Scaloni A. 2006. Hyperphosphorylation of JNK-interacting protein 1, a protein associated with Alzheimer disease. Mol. Cell. Proteomics. 5:97–113 [DOI] [PubMed] [Google Scholar]
- Dajas-Bailador F., Jones E.V., Whitmarsh A.J. 2008. The JIP1 scaffold protein regulates axonal development in cortical neurons. Curr. Biol. 18:221–226 10.1016/j.cub.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr N.D., Goodman B.S., Jungmann R., Leschziner A.E., Shih W.M., Reck-Peterson S.L. 2012. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 338:662–665 10.1126/science.1226734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens M., Rogers J.S., Cavanagh J., Raitano A., Xia Z., Halpern J.R., Greenberg M.E., Sawyers C.L., Davis R.J. 1997. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 277:693–696 10.1126/science.277.5326.693 [DOI] [PubMed] [Google Scholar]
- Dietrich K.A., Sindelar C.V., Brewer P.D., Downing K.H., Cremo C.R., Rice S.E. 2008. The kinesin-1 motor protein is regulated by a direct interaction of its head and tail. Proc. Natl. Acad. Sci. USA. 105:8938–8943 10.1073/pnas.0803575105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Ross J.L., Goldman Y.E., Holzbaur E.L. 2008. Differential regulation of dynein and kinesin motor proteins by tau. Science. 319:1086–1089 10.1126/science.1152993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encalada S.E., Szpankowski L., Xia C.H., Goldstein L.S. 2011. Stable kinesin and dynein assemblies drive the axonal transport of mammalian prion protein vesicles. Cell. 144:551–565 10.1016/j.cell.2011.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelender S., Sharp A.H., Colomer V., Tokito M.K., Lanahan A., Worley P., Holzbaur E.L., Ross C.A. 1997. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum. Mol. Genet. 6:2205–2212 10.1093/hmg/6.13.2205 [DOI] [PubMed] [Google Scholar]
- Falzone T.L., Stokin G.B., Lillo C., Rodrigues E.M., Westerman E.L., Williams D.S., Goldstein L.S.B. 2009. Axonal stress kinase activation and tau misbehavior induced by kinesin-1 transport defects. J. Neurosci. 29:5758–5767 10.1523/JNEUROSCI.0780-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D.S., Vale R.D. 1999. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat. Cell Biol. 1:293–297 10.1038/13008 [DOI] [PubMed] [Google Scholar]
- Furuta K., Furuta A., Toyoshima Y.Y., Amino M., Oiwa K., Kojima H. 2013. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc. Natl. Acad. Sci. USA. 110:501–506 10.1073/pnas.1201390110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater E.E., Megeath L.J., Stowers R.S., Schwarz T.L. 2006. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 173:545–557 10.1083/jcb.200601067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.P. 2004. Hither and yon: a review of bi-directional microtubule-based transport. Phys. Biol. 1:R1–R11 10.1088/1478-3967/1/2/R01 [DOI] [PubMed] [Google Scholar]
- Hackney D.D., Baek N., Snyder A.C. 2009. Half-site inhibition of dimeric kinesin head domains by monomeric tail domains. Biochemistry. 48:3448–3456 10.1021/bi8022575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.W., Griffin K., Jih G.T., Stuckey J., Verhey K.J. 2008. Co-operative versus independent transport of different cargoes by Kinesin-1. Traffic. 9:725–741 10.1111/j.1600-0854.2008.00722.x [DOI] [PubMed] [Google Scholar]
- Hendricks A.G., Perlson E., Ross J.L., Schroeder H.W., III, Tokito M., Holzbaur E.L. 2010. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 20:697–702 10.1016/j.cub.2010.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Pfister K.K., Yorifuji H., Wagner M.C., Brady S.T., Bloom G.S. 1989. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 56:867–878 10.1016/0092-8674(89)90691-0 [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Niwa S., Tanaka Y. 2010. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 68:610–638 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- Hong Z., Yang Y., Zhang C., Niu Y., Li K., Zhao X., Liu J.J. 2009. The retromer component SNX6 interacts with dynactin p150(Glued) and mediates endosome-to-TGN transport. Cell Res. 19:1334–1349 10.1038/cr.2009.130 [DOI] [PubMed] [Google Scholar]
- Horiuchi D., Barkus R.V., Pilling A.D., Gassman A., Saxton W.M. 2005. APLIP1, a kinesin binding JIP-1/JNK scaffold protein, influences the axonal transport of both vesicles and mitochondria in Drosophila. Curr. Biol. 15:2137–2141 10.1016/j.cub.2005.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi D., Collins C.A., Bhat P., Barkus R.V., Diantonio A., Saxton W.M. 2007. Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr. Biol. 17:1313–1317 10.1016/j.cub.2007.06.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V.M., Neefjes J. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7–RILP–p150Glued, ORP1L, and the receptor βlll spectrin. J. Cell Biol. 176:459–471 10.1083/jcb.200606077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaan H.Y., Hackney D.D., Kozielski F. 2011. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 333:883–885 10.1126/science.1204824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaether C., Skehel P., Dotti C.G. 2000. Axonal membrane proteins are transported in distinct carriers: a two-color video microscopy study in cultured hippocampal neurons. Mol. Biol. Cell. 11:1213–1224 10.1091/mbc.11.4.1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y., Okada Y., Tanaka Y., Harada A., Terada S., Hirokawa N. 2000. KIF5C, a novel neuronal kinesin enriched in motor neurons. J. Neurosci. 20:6374–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S., Holzbaur E.L. 1995. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J. Biol. Chem. 270:28806–28811 10.1074/jbc.270.48.28806 [DOI] [PubMed] [Google Scholar]
- Kawano T., Araseki M., Araki Y., Kinjo M., Yamamoto T., Suzuki T. 2012. A small peptide sequence is sufficient for initiating kinesin-1 activation through part of TPR region of KLC1. Traffic. 13:834–848 10.1111/j.1600-0854.2012.01350.x [DOI] [PubMed] [Google Scholar]
- Konishi Y., Setou M. 2009. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci. 12:559–567 10.1038/nn.2314 [DOI] [PubMed] [Google Scholar]
- Koushika S.P. 2008. “JIP”ing along the axon: the complex roles of JIPs in axonal transport. Bioessays. 30:10–14 10.1002/bies.20695 [DOI] [PubMed] [Google Scholar]
- Kristensen O., Guenat S., Dar I., Allaman-Pillet N., Abderrahmani A., Ferdaoussi M., Roduit R., Maurer F., Beckmann J.S., Kastrup J.S., et al. 2006. A unique set of SH3-SH3 interactions controls IB1 homodimerization. EMBO J. 25:785–797 10.1038/sj.emboj.7600982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S., Wallace K.E., Holzbaur E.L. 2012. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196:407–417 10.1083/jcb.201106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S., Yasukawa T., Homma Y., Ito Y., Niikura T., Hiraki T., Hirai S., Ohno S., Kita Y., Kawasumi M., et al. 2001. c-Jun N-terminal kinase (JNK)-interacting protein-1b/islet-brain-1 scaffolds Alzheimer’s amyloid precursor protein with JNK. J. Neurosci. 21:6597–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire J.R., Rong J., Li S.H., Li X.J. 2006. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J. Biol. Chem. 281:3552–3559 10.1074/jbc.M509806200 [DOI] [PubMed] [Google Scholar]
- Morris R.L., Hollenbeck P.J. 1993. The regulation of bidirectional mitochondrial transport is coordinated with axonal outgrowth. J. Cell Sci. 104:917–927 [DOI] [PubMed] [Google Scholar]
- Moughamian A.J., Holzbaur E.L. 2012. Dynactin is required for transport initiation from the distal axon. Neuron. 74:331–343 10.1016/j.neuron.2012.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.J., Klumpp S., Lipowsky R. 2008. Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc. Natl. Acad. Sci. USA. 105:4609–4614 10.1073/pnas.0706825105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan Z., Muresan V. 2005a. c-Jun NH2-terminal kinase-interacting protein-3 facilitates phosphorylation and controls localization of amyloid-beta precursor protein. J. Neurosci. 25:3741–3751 10.1523/JNEUROSCI.0152-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan Z., Muresan V. 2005b. Coordinated transport of phosphorylated amyloid-β precursor protein and c-Jun NH2-terminal kinase–interacting protein-1. J. Cell Biol. 171:615–625 10.1083/jcb.200502043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihalani D., Wong H.N., Holzman L.B. 2003. Recruitment of JNK to JIP1 and JNK-dependent JIP1 phosphorylation regulates JNK module dynamics and activation. J. Biol. Chem. 278:28694–28702 10.1074/jbc.M304212200 [DOI] [PubMed] [Google Scholar]
- Nikolaev A., McLaughlin T., O’Leary D.D., Tessier-Lavigne M. 2009. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 457:981–989 10.1038/nature07767 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Perlson E., Jeong G.B., Ross J.L., Dixit R., Wallace K.E., Kalb R.G., Holzbaur E.L. 2009. A switch in retrograde signaling from survival to stress in rapid-onset neurodegeneration. J. Neurosci. 29:9903–9917 10.1523/JNEUROSCI.0813-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis G.F., Yang G., Szpankowski L., Weaver C., Shah S.B., Robinson J.T., Hays T.S., Danuser G., Goldstein L.S. 2012. Molecular motor function in axonal transport in vivo probed by genetic and computational analysis in Drosophila. Mol. Biol. Cell. 23:1700–1714 10.1091/mbc.E11-11-0938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeld M.H., Roncarati R., Vito P., Lopez P.A., Abdallah M., D’Adamio L. 2002. Jun NH2-terminal kinase (JNK) interacting protein 1 (JIP1) binds the cytoplasmic domain of the Alzheimer’s beta-amyloid precursor protein (APP). J. Biol. Chem. 277:3767–3775 10.1074/jbc.M108357200 [DOI] [PubMed] [Google Scholar]
- Soppina V., Rai A.K., Ramaiya A.J., Barak P., Mallik R. 2009. Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc. Natl. Acad. Sci. USA. 106:19381–19386 10.1073/pnas.0906524106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokin G.B., Lillo C., Falzone T.L., Brusch R.G., Rockenstein E., Mount S.L., Raman R., Davies P., Masliah E., Williams D.S., Goldstein L.S. 2005. Axonopathy and transport deficits early in the pathogenesis of Alzheimer’s disease. Science. 307:1282–1288 10.1126/science.1105681 [DOI] [PubMed] [Google Scholar]
- Sun F., Zhu C., Dixit R., Cavalli V. 2011. Sunday Driver/JIP3 binds kinesin heavy chain directly and enhances its motility. EMBO J. 30:3416–3429 10.1038/emboj.2011.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpankowski L., Encalada S.E., Goldstein L.S. 2012. Subpixel colocalization reveals amyloid precursor protein-dependent kinesin-1 and dynein association with axonal vesicles. Proc. Natl. Acad. Sci. USA. 109:8582–8587 10.1073/pnas.1120510109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twelvetrees A.E., Yuen E.Y., Arancibia-Carcamo I.L., MacAskill A.F., Rostaing P., Lumb M.J., Humbert S., Triller A., Saudou F., Yan Z., Kittler J.T. 2010. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron. 65:53–65 10.1016/j.neuron.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niekerk E.A., Willis D.E., Chang J.H., Reumann K., Heise T., Twiss J.L. 2007. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc. Natl. Acad. Sci. USA. 104:12913–12918 10.1073/pnas.0611562104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen M., Mikhaylova M., Lipka J., Schlager M.A., van den Heuvel D.J., Kuijpers M., Wulf P.S., Keijzer N., Demmers J., Kapitein L.C., et al. 2013. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron. 77:485–502 10.1016/j.neuron.2012.11.027 [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Hammond J.W. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Lizotte D.L., Abramson T., Barenboim L., Schnapp B.J., Rapoport T.A. 1998. Light chain–dependent regulation of Kinesin’s interaction with microtubules. J. Cell Biol. 143:1053–1066 10.1083/jcb.143.4.1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K.J., Meyer D., Deehan R., Blenis J., Schnapp B.J., Rapoport T.A., Margolis B. 2001. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 152:959–970 10.1083/jcb.152.5.959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer T., Attar N., Harterink M., van Weering J.R., Traer C.J., Oakley J., Goud B., Stephens D.J., Verkade P., Korswagen H.C., Cullen P.J. 2009. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev. Cell. 17:110–122 10.1016/j.devcel.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P., Forster R., Palmer K.J., Pepperkok R., Stephens D.J. 2005. Coupling of ER exit to microtubules through direct interaction of COPII with dynactin. Nat. Cell Biol. 7:48–55 10.1038/ncb1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver C., Leidel C., Szpankowski L., Farley N.M., Shubeita G.T., Goldstein L.S. 2013. Endogenous GSK-3/shaggy regulates bidirectional axonal transport of the amyloid precursor protein. Traffic. 14:295–308 10.1111/tra.12037 [DOI] [PubMed] [Google Scholar]
- Welte M.A. 2004. Bidirectional transport along microtubules. Curr. Biol. 14:R525–R537 10.1016/j.cub.2004.06.045 [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J. 2006. The JIP family of MAPK scaffold proteins. Biochem. Soc. Trans. 34:828–832 10.1042/BST0340828 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.