Abstract
The San Nicolas Island fox (Urocyon littoralis dickeyi) is genetically the most monomorphic sexually reproducing animal population yet reported and has no variation in hypervariable genetic markers. Such low levels of variation imply lower resistance to pathogens, reduced fitness, and problems in distinguishing kin from non-kin. In vertebrates, the MHC contains genes that influence disease resistance and kin recognition and may be under intense balancing selection in some populations. Hence, genetic variation at the MHC might persist despite the extreme monomorphism shown by neutral markers. We examine variation of five loci within the MHC of San Nicolas Island foxes and find remarkably high levels of variation. Further, we show by simulation that genetic monomorphism at neutral loci and high MHC variation could arise only through an extreme population bottleneck of <10 individuals, ≈10–20 generations ago, accompanied by unprecedented selection coefficients of >0.5 on MHC loci. These results support the importance of balancing selection as a mechanism to maintain variation in natural populations and expose the difficulty of using neutral markers as surrogates for variation in fitness-related loci.
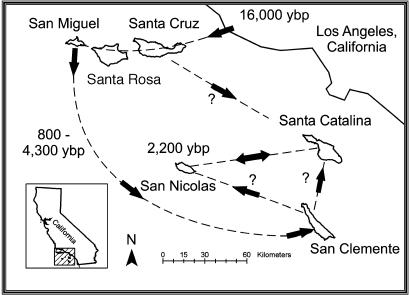
The island fox (Urocyon littoralis) is an endemic North American canid that inhabits six of the eight Channel Islands off the coast of southern California (Fig. 1). As suggested by the archeological record and molecular genetic data, foxes colonized the three northern Channel Islands (San Miguel, Santa Rosa, and Santa Cruz) ≈16,000 years ago and, subsequently, were transported by Native Americans to the three southern Channel Islands (San Nicolas, Santa Catalina, and San Clemente) 800 to 4,300 years ago (1–4) (Fig. 1). Effective population size varies with island area and ranges from <200 to ≈1,000 individuals (Table 1). Levels of genetic variation reflect population size and colonization history, with the San Nicolas Island population having the second smallest effective population size and a recent colonization history (4) (Fig. 1 and Table 1). No variation has been discovered for any of four independent genetic marker classes in the San Nicolas Island population, including supposedly neutral hypervariable microsatellite loci (5) and multilocus fingerprints (2), for which the probability of genetic identity is commonly <1 in several million (6). Recently, because of dramatic declines, populations on the three Northern Islands and Santa Catalina Island have been proposed for listing under the U.S. Endangered Species Act (7).
Fig. 1.
Location of the six southern California Channel Islands where island foxes are found. Dotted line indicates hypothesized colonization routes (3). Approximate colonization times in years before present (ybp) based on the archeological record are provided (4).
Table 1.
Effective population size (Ne) and observed mean heterozygosity and number of alleles (in parentheses where applicable) for genetic markers surveyed in island foxes (2,3,5)
| Ne | Allozymes | Minisatellites | Microsatellites | n | DRB | DQB | FH2202 | CFA12–4 | CFA12–13 | |
|---|---|---|---|---|---|---|---|---|---|---|
| San Miguel | 163 | 0.008 (1.1) | 0.13 | 0.11 (1.78) | 25.8 | 0.00 (2) | 0.00 (1) | 0.43 (6) | 0.33 (2) | 0.50 (4) |
| Santa Rosa | 955 | 0.055 (1.2) | 0.34 | 0.21 (2.56) | 29.0 | 0.16 (2) | 0.00 (1) | 0.59 (8) | 0.50 (7) | 0.44 (4) |
| Santa Cruz | 984 | 0.041 (1.1) | 0.19 | 0.22 (2.39) | 22.8 | 0.14 (2) | 0.21 (2) | 0.65 (6) | 0.64 (5) | 0.46 (3) |
| San Nicolas | 247 | 0.000 (1.0) | 0.00 | 0.00 (1.00) | 26.6 | 0.36 (2) | 0.00 (1) | 0.62 (3) | 0.57 (3) | 0.33 (2) |
| Santa Catalina | 979 | 0.000 (1.0) | 0.45 | 0.36 (2.61) | 29.0 | 0.36 (3) | 0.55 (4) | 0.63 (8) | 0.24 (5) | 0.37 (3) |
| San Clemente | 551 | 0.013 (1.1) | 0.25 | 0.26 (2.11) | 19.0 | 0.00 (1) | 0.00 (1) | 0.68 (5) | 0.50 (4) | 0.60 (3) |
Ne was estimated from population census sizes (3). Mean sample size (n) is indicated for the MHC assays.
The MHC contains the most variable set of coding genes in vertebrates, with as many as 349 alleles described for a single locus (8) and heterozygosity values that generally exceed those predicted by neutrality (9). Class I and II MHC molecules are responsible for the presentation to T cells of intracellular (endogenous) and extracellular (exogenous) peptides, respectively (10). High levels of heterozygosity at the MHC may be maintained by balancing selection through pathogen-mediated selection, negative assortative mating, and maternal–fetal interactions (9). Consequently, loss of MHC variation due to drift in small island fox populations may increase the risk of infection by pathogens or may reduce fecundity. Island foxes have tested positive for canine pathogens (11), and a recent canine distemper epidemic has decimated the Santa Catalina Island population (12). Further, because the island fox avoids mating with close kin and discriminates between kin and non-kin in territorial encounters (13), monomorphism at the MHC may influence the selectivity of mate choice and the degree of territoriality. To determine whether MHC variation has been maintained by natural selection despite the intense genetic drift implied by the genetic monomorphism of neutral genetic markers, we assess genetic variability at two class II MHC genes (DRB and DQB) and three class II MHC-linked microsatellite loci. We compare variation in San Nicolas Island foxes with those on the other Channel Islands to estimate levels of MHC variation in populations ancestral to the San Nicolas population and thus account for the influence of population history on levels of MHC variation.
Materials and Methods
Genotyping. Island foxes have been previously genotyped at 18 dinucleotide microsatellite loci (5) (Table 1). We genotyped island foxes from six island populations for sequence variation in exon 2 of class II DRB and DQB genes using single strand conformation polymorphism (SSCP) (14). Only predistemper outbreak individuals from Santa Catalina Island were used in this study. We used published PCR primers and protocols (15, 16) to amplify 267-bp (DRB) and 142-bp (DQB) fragments that were visualized by end-labeling one primer with 32P (17) followed by separation on 5%-acrylamide gels (with 5% glycerol vol/vol) for 5–9 h at room temperature. Gels were transferred to Whatman paper, dried, and exposed to autoradiographic film for 12–48 h. Unique alleles were reamplified and sequenced for each gel according to Sunnucks et al. (14). Sequencing was performed on a Beckman CEQ2000 as per manufacturer's protocols. Nonsynonymous (dN) and synonymous (dS) substitution rates were estimated for antigen-binding site (ABS) and non-ABS codons (18) for each locus by using the method of Nei and Gojobori (19) in the program mega 2 (20). Sequences have been deposited in GenBank (AY366482-AY366488).
Class II microsatellite loci CFA12-4 (forward, GCA ATG GCA AGA CCT AAA GC; reverse, AGG GAG GAA AGT CTC CGT GT), CFA12-13 (forward, TGG GAG AGT TAG AGG AGG CA; reverse, GCC CAC CAC TCT CAC ACA TG), and FH2202 (21) were typed by using dye-labeled primers on a Beckman CEQ 2000. CFA12-4 is located in the 56.2-kb segment spanned by genes DLA-DQA1 and DLA-DRB1 and is ≈18.0 kb from the former in the domestic dog (S.D., unpublished data). CFA12-13 is located in intron 5 of the psm B8 genes in the domestic dog (S.D., unpublished data). FH2202 is located near the DLA-DRB2 pseudogene in the domestic dog (21) and is located in the 216.8-kb region spanned by CFA12-4 and CFA12-13 (S.D., unpublished data). The forward primers were augmented on the 5′ end with the addition of an M13 sequence, and a dye-labeled M13 primer was included in the PCR reaction (22). The hybrid forward-M13 primer and the labeled M13 primer were used in limiting concentration in the PCR (2.5 μm each). We performed a two-step PCR, which included an initial 3-min denaturation at 94°C followed by 22 cycles with the following conditions: 94°C for 45 sec, 56°C for 45 sec, and 72°C for 45 sec. An additional eight cycles were performed with an annealing temperature of 53°C, with the same time profiles, and was followed by a 72°C soak for 5 min. PCR products were run with a size standard (Beckman) on the CEQ 2000 following the manufacturer's protocols.
Allelic diversity (number of alleles per locus) and observed heterozygosity were measured for all island populations. Expected heterozygosity, linkage disequilibrium, and FST were calculated by using the program genetix (23). Bonferroni corrections were used to account for multiple comparisons. RST was computed by using the approach described by Goodman (24) with the program rstcalc.
Simulations. We conducted forward simulations to establish the intensity of selection (s) needed to maintain the observed heterozygosity at the MHC-linked microsatellite loci and the DRB locus in San Nicolas Island foxes. To estimate the selection coefficient, we first needed to establish the effective size of the population (Ne) and effective bottleneck size (Ne-b) needed to account for the lack of variation observed at 18 microsatellite loci for 29 individuals (5). Following Hoelzel et al. (25), we assumed that heterozygosity would have been detected in our sample if at least 1 individual of the 29 sampled was heterozygous on average at one locus. Consequently, the upper threshold of heterozygosity is 0.002 [1/(29·18)]. This threshold gives a probability of 0.35 (0.99818·29), for sampling 29 individuals that are all monomorphic at 18 loci. Each microsatellite locus was assumed to evolve in a step-wise manner (26) with a mutation rate of 10-2, 10-3, or 10-4. Mating was random with separate sexes and nonoverlapping generations. To maximize the diversity of starting simulation conditions, initial allele frequencies were set at 1/k, where k is the mean number of alleles observed for each microsatellite locus across all island fox populations. To allow allele frequencies to equilibrate, populations were allowed to reproduce for 2,000 generations, followed by the introduction of bottleneck events of varying intensity and duration. After the bottleneck, the population increased at an intrinsic rate of increase (r) of 0.28 per generation as observed in island fox populations (G.R., unpublished data) to a final effective population size of 125 and 250. These effective sizes were reached in 10 to 20 generations, depending on the bottleneck size and duration and were 0.25 and 0.50, respectively, of the census population size on San Nicolas Island (3, 27). We assumed 2 years per island fox generation (3). Five hundred simulations were performed. Initial simulations clearly suggested a severe bottleneck (to an effective size of 10 individuals or fewer for one or two generations, followed by ≈12 generations of population growth) was necessary to explain near monomorphism at the 18 loci. A severe bottleneck followed by a small number of generations was required to eliminate heterozygosity in the microsatellite loci yet prevent its restoration by mutation. All other less severe demographic scenarios had occurrence frequencies much less than 5% and hence were regarded as implausible. In the one-generation bottleneck scenario, with a mutation rate of 10-4, 16.2% (81 of 500 simulations) of the simulations possessed a mean heterozygosity for the 18 loci lower than 0.002 (Table 2, which is published as supporting information on the PNAS web site). A higher mutation rate, or larger effective bottleneck size (Ne-b) or effective size of the population (Ne) yielded <5% of simulations with this level of heterozygosity.
This extreme scenario is consistent with the recent population crash of island foxes on the east end of Santa Catalina Island where the population was reduced from >1,000 to 10 individuals in a single generation due to a canine distemper epidemic in 1999. A similar event may have occurred on San Nicolas Island because fox numbers were low in the 1970s (28) and increased rapidly during the 1980s (29) to 500 individuals by 1988 (2). Considering the epidemic on Santa Catalina Island, and the results of our simulation, we used a single generation bottleneck with effective population size of 5 (half the census population size of 10 on Santa Catalina Island) in our final simulation to assess the intensity of selection.
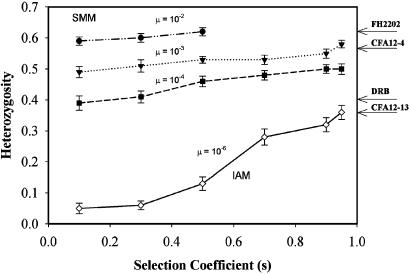
Selection was introduced to the simulation by adding an additional locus experiencing symmetrical overdominant selection. Heterozygotes had a fitness of 1 and homozygotes a fitness of 1 - s, where s is the selection coefficient against homozygotes. After each mating, an individual was evaluated for heterozygosity at this locus; if the individual was homozygous, it had a (1 - s) probability of survival to the next generation. The locus was modeled as evolving under the stepwise mutation model (SMM) for microsatellite loci with mutation rates of 10-2,10-3, and 10-4 (30) or the infinite alleles model (IAM) for the DRB gene with mutation rates of 10-6 (31). Initial allele frequencies for the IAM locus were assumed to be 1/k, where k equals the maximum number of DRB alleles observed in a population (three on Santa Catalina; Table 1). The approach assumes close linkage of these loci to the locus under selection. A selection coefficient of 0.08 was used before the bottleneck and after the selection event (9). We varied the selection coefficient for the one-generation bottleneck event and for two generations that followed in each simulation experiment of 500 runs, until average heterozygosity was near that observed in the San Nicolas Island population (see Fig. 2). For the SMM simulation, the observed values of heterozygosity were obtained with a periodic s of 0.5 and 0.95 for mutation rates of 10-2 and 10-4, respectively (Fig. 2). For the IAM locus, a periodic s of 0.95 was needed to achieve a heterozygosity of 0.36. Conceivably, selection might be maintained for more than two generations after the bottleneck, and a maximum of 11 generations is consistent with our results given the presence of Hardy–Weinberg equilibrium at MHC loci in the extant population (the twelfth generation). We cannot envision a scenario in which this intensity of selection might be maintained; however, in this extreme scenario, if we impose a single-generation bottleneck followed by 11 generations of selection and one generation of random breeding, selection coefficients of 0.5 (SMM - μ = 10-3) are required to maintain a heterozygosity of 0.62, and a selection coefficient of 0.5 (IAM - μ = 10-6) is needed to maintain the heterozygosity of 0.36.
Fig. 2.
The relationship between heterozygosity and the selection coefficient (s) as determined by demographic simulations for the IAM (DRB, μ = 10-6) and the SMM (FH2202, CFA12-4, and CFA12-13, μ = 10-2, 10-3, and 10-4). Mean heterozygosity and SE are reported for 500 simulations (see Supporting Text).
Results and Discussion
Levels of MHC Variation. We found surprisingly high heterozygosity in four of five surveyed MHC loci (Table 1). The three MHC-linked microsatellites (FH2202, CFA12-4, and CFA12-13) had observed heterozygosity values of 0.62, 0.57, and 0.33, respectively, whereas the DRB locus had a heterozygosity value of 0.36. The DQB locus was monomorphic in the San Nicolas population. These values of heterozygosity are similar to those in larger populations of island foxes and suggest the action of intense balancing selection over a sizable genomic interval within the fox MHC class II. However, this selection is unlikely to be ongoing because a predicted excess of heterozygotes (32) is found for only one locus, FH2202 (Ho = 0.62; He = 0.54), and even this difference was not statistically significant (P = 0.094). Consequently, this result implies that negative assortative mating, maternal–fetal interactions, or intense pathogen-based selection in the current population is not the cause of high levels of heterozygosity at the MHC. Finally, we found linkage disequilibrium between all three MHC-linked microsatellite loci in the San Nicolas, Santa Rosa, and Santa Catalina Island populations (P < 0.016 with Bonferroni correction) and an absence of disequilibrium between these microsatellite loci and the DRB locus. Consequently, this result and preliminary mapping information suggest two regions of <1 cM may be influenced by selection (S.D., unpublished data).
Conceivably, gene duplication might be a contributing cause to high heterozygosity because simultaneous amplification of alleles from duplicated MHC loci by PCR would inflate heterozygosity. However, this result is not likely for the following reasons: (i) the gene phylogeny of DRB does not support a duplication event (See Figs. 3 and 4 and Supporting Text, which are published as supporting information on the PNAS web site); (ii) all five loci with one possible exception fit Hardy–Weinberg expectations; (iii) no more than two alleles at a locus were ever observed in a single individual, a result which is unlikely for duplicated microsatellite loci (see Supporting Text); and (iv) pedigree data show Mendelian inheritance of the four loci, a result that is unlikely if a duplication had occurred (A.A., unpublished data).
The other island populations also show high levels of variation at the MHC relative to background levels of variation at neutral loci (Table 1 and Supporting Text); and Tables 3 and 4, which are published as supporting information on the PNAS web site. However, the influence of colonization history is evident in patterns of heterozygosity and monomorphism. For example, the class II DQB gene is monomorphic in San Nicolas, San Clemente, and San Miguel Island populations. Based on previous molecular genetic data, the latter island was the source of migrants that colonized San Clemente and San Nicolas Islands (refs. 2, 3, and 5 and Fig. 1). Similarly, based on mitochondrial DNA polymorphisms, the Santa Catalina Island population was thought to have received foxes from both San Clemente and the northern Channel Islands (3). High levels of variation at DRB and DQB in the Santa Catalina Island population (Table 1 and Supporting Text), which are monomorphic in the San Clemente Island population, are consistent with multiple migration events. However, in this regard, high levels of heterozygosity and allelic diversity of DRB in the San Nicolas population are surprising given that the locus is monomorphic in the putative ancestral populations of San Miguel and San Clemente Islands. Either San Nicolas Island was colonized multiple times, although this is not evidenced by the unique mtDNA haplotype found there (2), or subsequent drift in the ancestral populations caused the loss of alternative alleles.
Balancing Selection at the MHC. Under balancing selection, the level of heterozygosity is related to the product of effective population size, mutation rate, and the selection coefficient (33). Consequently, balancing selection at the MHC must have been intense in San Nicolas Island foxes to maintain high levels of variation, given the small effective population size suggested by monomorphism at hypervariable loci. To estimate an effective population size consistent with the absence of variation in the 18 microsatellite loci that we surveyed (5), we performed simulations in which we varied population size and the intensity of population bottlenecks. We found that only scenarios incorporating a one- or several-generation population bottleneck as recently as 10–20 generations ago, and that consisted of 10 or fewer individuals, resulted in at least 15% of simulations with levels of heterozygosity consistent with that observed. We explored numerous other less extreme demographic scenarios, but none approached observed levels of variation in >5% of simulations, and hence we regard them as implausible. Further, the possibility of an extreme bottleneck is consistent with that observed on Santa Catalina Island where, in a recent distemper epidemic afflicting 80% of the island area, the fox population declined to only 10 individuals observed alive (12). Given our demographic scenario for San Nicolas Island foxes, we found that periodic selection coefficients for the microsatellite loci as high as 0.5–0.95 are required to maintain heterozygosity values near 0.62 at microsatellite locus FH2202 and 0.36 at the DRB locus (Fig. 2 and Supporting Text). These selection coefficients are much larger than those reported in natural (34–36) (range: 0.05–0.15) and human populations (37, 38) (range: 0.19–0.39) for a locus under balancing selection.
Balancing selection at the MHC is predicted to result in lower levels of differentiation among island populations relative to levels based on neutral loci (39). To measure population differentiation, we used RST and FST for microsatellite and sequence information, respectively (24). Mean RST for the unlinked microsatellite loci was 0.72 (0.71–0.74; 95% CI) and significantly higher than 0.53 (0.48–0.58; 95% CI) for MHC-linked microsatellites. Similarly, FST was elevated for the DRB (0.58) and DQB (0.55); however, these values are comparable to that for allozyme loci (0.56) (2). Balancing selection over evolutionary time periods is expected to maintain corrected ratios of nonsynonymous (replacement) to synonymous (silent) substitutions of greater than one (40). We assessed nonsynonymous (dN) to synonymous (dS) substitution ratios in the antigen binding sites (ABS) and found them significantly greater than one for the two class II MHC genes (1.94 and 1.34 for DRB and DQB, respectively) and elevated relative to the non-ABS portions of the gene (0.646 and 0.012 for DRB and DQB, respectively). Consequently, these results provide independent support for the action of balancing selection at the MHC loci that we surveyed.
Conclusions and Conservation Implications. Our empirical and simulation results suggest that intense periodic balancing selection at the MHC may have allowed the persistence of variation in San Nicolas foxes despite strong genetic drift. This result adds significantly to the small number of studies showing evidence for balancing selection as a mechanism to maintain genetic variation (41–45). In effect, periodic selection has rescued genetic variation at the MHC and, potentially, other fitness-related genes. Consequently, San Nicolas Island foxes may not have suffered some of the predicted fitness and kin recognition effects of extreme genetic monomorphism. Such periodic selection may explain the inconsistent relationship between MHC heterozygosity and neutral loci in other species (46). Therefore, although neutral loci may be useful in predicting inbreeding depression and loss of genetic variation due to drift (47), new molecular tools are needed to directly assess the fitness decline of small populations. Either a panel of candidate genes influencing fitness should be considered (48) or loci under selection should be identified directly through genomic scans (49).
Finally, our results rekindle the debate concerning the use of MHC and other fitness-related genes in the management of captive and endangered wild populations. Currently, neutral markers are used exclusively in the absence of pedigree information to minimize inbreeding in small populations. With the discovery of the function and importance of MHC genes in natural populations, it was suggested that maintaining allelic diversity at MHC should be an additional goal of genetic management (50). This notion was debated and largely discredited (51, 52). Our results, and those showing an absence of association between variation in neutral and fitness-related traits in natural populations (53), suggest reconsideration of this idea. Preservation of a diverse array of fitness-related genes (54, 55), along with neutral variation, might be the key to the long-term survival of endangered populations.
Supplementary Material
Acknowledgments
We thank S. Edward, P. Hedrick, G. Luikart, P. Morin, C. Taylor, and G. Shin for helpful comments on the manuscript and A. Selaya for technical assistance. This project was funded in part by the National Science Foundation and the Academic Senate of the University of California.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: s, selection coefficient; SMM, stepwise mutation model; IAM, infinite alleles model.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY366482–AY366488).
References
- 1.Collins, P. W. (1982) Master's thesis (Univ. of California, Santa Barbara).
- 2.Gilbert, D., Lehman N., O'Brien, S. J. & Wayne, R. K. (1990) Nature, 344, 764-761. [DOI] [PubMed] [Google Scholar]
- 3.Wayne, R. K., George, S. B., Gilbert, D., Collins, P. W., Kovach, S. D., Girman, D. & Lehman, N. (1991) Evolution 45, 1849-1868. [DOI] [PubMed] [Google Scholar]
- 4.Collins, P. W. (1991) J. Ethnobiol. 11, 51-81. [Google Scholar]
- 5.Goldstein, D. B., Roemer, G. W., Smith, D. A., Reich, D. E., Bergman, A. & Wayne, R. K. (1999) Genetics 151, 797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Risch, N. J. & Devlin, B. (1992) Science 255, 717-720. 7. [DOI] [PubMed] [Google Scholar]
- 7.66 Federal Register 5 (2001), pp. 1295-1300.
- 8.Robinson, J., Malik, A., Parham, P., Bodmer, J. G. & Marsh, S. G. (2000) Tissue Antigens 55, 280-287. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, S. V. & Hedrick, P. W. (1998) Trends Ecol. Evol. 13, 305-311. [DOI] [PubMed] [Google Scholar]
- 10.Klein, J. (1986) Natural History of the Major Histocompatability Complex (Wiley, New York).
- 11.Garcelon, D. K., Wayne, R. K. & Gonzales, B. J. (1992) J. Wildl. Dis. 28, 223-229. [DOI] [PubMed] [Google Scholar]
- 12.Timm, S. F., Barker, W. D., Johnson, S. A., Sewell, J. H., Sharpe, P. B., Schmidt, G. A. & Garcelon, D. K. (2002) Island Fox Recovery Efforts on Santa Catalina Island, California (Inst. for Wildlife Studies, Arcata, CA).
- 13.Roemer, G. W., Smith, D. A., Garcelon, D. K. & Wayne, R. K. (2001) J. Zool. 255, 1-14. [Google Scholar]
- 14.Sunnucks, P., Wilson, A. C. C., Beheregary, L. B., Zenger, K., French J. & Taylor, A. C. (2000) Mol. Ecol. 9, 1699-1710. [DOI] [PubMed] [Google Scholar]
- 15.Wagner J. L., Burnett, R. C., DeRose, S. A., Francisco L. V., Storb, R. & Ostrander, E. A. (1996) Transplantation 62, 876-877. [DOI] [PubMed] [Google Scholar]
- 16.Hoelzel, A. R., Stephens, J. C. & O'Brien, S. J. (1999) Mol. Biol. Evol. 16, 611-618. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 18.Brown, J. H. (1993) Nature 364, 33-39. [DOI] [PubMed] [Google Scholar]
- 19.Nei, M. & Gojobori, T. (1986) Mol. Biol. Evol. 3, 418-426. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 21.Wagner, J. L., Burnett, R. C., Works, J. D. & Storb, R. (1996) Tissue Antigens 48, 554-561. [DOI] [PubMed] [Google Scholar]
- 22.Boutin-Ganache I., Raposo, M., Raymond, M. & Descepper, C. F. (2001) Biotechniques 31, 1-3. [PubMed] [Google Scholar]
- 23.Belkhir, K. (1999) genetix, A Windows Program for Population Genetic Analysis (Universite Montpellier II, Montpellier, France), Version 4.04.
- 24.Goodman, S. J. (1997) Mol. Ecol. 6, 881-885. [DOI] [PubMed] [Google Scholar]
- 25.Hoelzel, A. R. (1999) Biol. J. Linn. Soc. 68, 23-39. [Google Scholar]
- 26.Ohta, T. & Kimura, M. (1973) Genet. Res. 22, 201-204. [DOI] [PubMed] [Google Scholar]
- 27.Frankham, R. (1995) Genet. Res. 66, 95-107. [Google Scholar]
- 28.Laughrin, L. (1980) in The California Islands: Proceedings of a Multidisciplinary Symposium, ed. Powers, D. M. (Santa Barbara Museum of Natural History, Santa Barbara, CA.), pp. 745-750.
- 29.Kovach, S. D. & Dow, R. J. (1985) Annual Report: Island Fox Research on San Nicolas Island (Dept. of the Navy Pacific Missile Test Center, San Diego, CA.).
- 30.Ellegren, H. (2000) Trend. Genet. 16, 551-558. [DOI] [PubMed] [Google Scholar]
- 31.Kimura, M. & Crow, J. F. (1964) Genetics 49, 725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartl, D. L. & Clark A. G. (1997) Principles of Population Genetics (Sinauer, Sunderland, MA).
- 33.Maruyama, T. & Nei, M. (1981) Genetics 98, 441-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satta, Y., O'hUigin, C., Takahata, N. & Klein, J. (1994) Proc. Natl. Acad. Sci. USA. 91, 7184-7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thursz, M. R., Thomas, H. C., Greenwood, B. M. & Hill, A. V. S. (1997) Nat. Genet. 17, 11-12. [DOI] [PubMed] [Google Scholar]
- 36.Cavalli-Sforza, L. L. & Bodmer, W. F. (1999) The Genetics of Human Populations (Dover, New York,).
- 37.Markow, T., Hedrick, P. W., Zuerlin, K., Danilovs, J., Martin, J., Vyvial, T. & Armstrong C. (1993) Am. J. Hum. Genet. 53, 943-952. [PMC free article] [PubMed] [Google Scholar]
- 38.Black, F. L. & Hedrick, P. W. (1997) Proc. Natl. Acad. Sci USA 94, 12452-12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schierup, M. H., Vekemans, X. & Charlesworth, D. (2000) Genet. Res. 76, 51-62. [DOI] [PubMed] [Google Scholar]
- 40.Hughes, A. L. & Nei, M. (1988) Nature 352, 167-170. [DOI] [PubMed] [Google Scholar]
- 41.Van Delden, W. (1982) Evol. Biol. 15, 187-222. [Google Scholar]
- 42.Allison, A. C. (1964) Cold Spring Harbor Symp. Quant. Genet. 29, 139-149. [DOI] [PubMed] [Google Scholar]
- 43.Richman, A. D. & Kohn, J. R. (1996) Trends Ecol. Evol. 11, 479-502. [DOI] [PubMed] [Google Scholar]
- 44.Greaves, J. H., Redfern, R., Ayres, P. B. & Gill, J. E. (1977) Genet. Res. 30, 257-264. [DOI] [PubMed] [Google Scholar]
- 45.Garrigan, D. & Hedrick, P. W. (2003) Evolution 57, 1707-1722. [DOI] [PubMed] [Google Scholar]
- 46.Bernatchez, L. & Landry, C. (2003) J. Evol. Biol. 16, 363-377. [DOI] [PubMed] [Google Scholar]
- 47.Falconer, D. S. & MacKay, T. F. C. (1996) Introduction to Quantitative Genetics (Addison–Wesley, London). [DOI] [PMC free article] [PubMed]
- 48.Purugganan, M. & Gibson, G. (2003) Mol. Ecol. 12, 1109-1112. [DOI] [PubMed] [Google Scholar]
- 49.Schlötterer, C. (2002) Genetics 160, 753-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hughes, A. L. (1991) Conserv. Biol. 5, 249-251. [Google Scholar]
- 51.Vrijenhoek, R. C. & Leberg, P. L. (1991) Conserv. Biol. 5, 252-254. [Google Scholar]
- 52.Miller, P. S. & Hedrick, P. W. (1991) Conserv. Biol. 5, 556-558. [Google Scholar]
- 53.Reed, D. H. & Frankham, R. (2001) Evolution 55, 1095-1103. [DOI] [PubMed] [Google Scholar]
- 54.Crandall Bininda-Emonds, O. R. P., Mace, G. M. & Wayne, R. K. (2000) Trends Ecol. Evol. 15, 290-295. [DOI] [PubMed] [Google Scholar]
- 55.Luikart, G., England, P. R., Tallmon, D., Jordan, S. & Taberlet, P. (2003) Nat. Rev. Genet. 4, 981-994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.