Human parathyroid hormone related protein (hPTHrP), originally isolated from lung cancer cell lines in 1987, [1] is a 141-amino acid polypeptide widely found in both normal and tumor tissue cells. The N-terminal region of hPTHrP possesses a high degree of structural homology with human parathyroid hormone (hPTH), and both hormones effect the elevation of calcium levels in the blood.[2] Although PTH and PTHrP act through binding to the same receptor, the PTH-receptor type-1 (PTHR1), in vitro studies suggest that the two ligands may differ in the precise molecular modes of their receptor interactions.[3,4] Under normal conditions, hPTHrP, which is widely expressed in the tissues of embryos and adults, plays an essential role in a range of functions related to development and growth, including: fostering of the cartilaginous growth plate, [5] bone anabolism, [6] development of mammary gland, [7] transport of calcium ions across the placenta, [8] relaxation of smooth muscle, or vasodilatation, [9] and eruption of tooth.[10] In analogy to the related anti-osteoporosis therapeutic agent, PTH, researchers have found that PTHrP, administered daily, may induce anabolic effects on the skeleton. Interestingly, the risk of hypercalcemia associated with PTH–based therapeutics may be lowered with the use of PTHrP. These findings raise the possibility that PTHrP and/or congeners, thereof could offer an advantage over currently used PTH peptides in therapeutics related to osteoporosis.
A growing understanding of the role that hPTHrP may play in mediating the progression of cancer further enhances interest in this polypeptide. An intriguing property of hPTHrP is the finding that it exhibits anti-apoptotic and proliferation–promoting effects on tumor cells.[11–14] Recent studies have shown that antagonists of PTHR1 are able to remarkably inhibit the growth of tumors.[15–17]
The development of an efficient synthetic route to homogeneous hPTHrP, and analogs thereof, would facilitate the systematic study of the interaction between hPTHrP and its receptor, PTHR1. Such research would offer important insights into the structure-activity relationship (SAR) of the polypeptide, and could well facilitate the development of practical PTHR1 antagonists, to suppress the growth of tumors, or agonists, for the treatment of osteoporosis.[18] Certainly, one could imagine that a wisely crafted hPTHrP lookalike could have exploitable antiproliferative properties.
In our judgment, the synthesis of protein targets offers significant learning opportunities at the interface of chemistry, biology and medicine.[19] The advantage of pursuing chemistry based approaches to protein targets arises from the fact that this forum uniquely allows for the versatile design of unnatural probe structures possessing defined alterations of amino sequence and structure, including the incorporation of non-proteogenic amino acids.[20–22] Notwithstanding impressive accomplishments in protein engineering, which were enabled by spectacular advances in molecular biology, we have felt that chemical based synthesis, in principle, also has much to offer in terms of reaching a specific protein target, in reasonable research–level quantities (usually several milligrams), above all with very high levels of homogeneity. Thus, the purposes of this research were several. First, we hoped to reach hPTHrP by purely chemical means, and to show that it manifests full biological function. With this accomplished, the basis for an SAR program, involving alterations of primary structure (proteogenic and non-proteogenic amino acid substitutions) and molecular constraints, would be solidly in place. More broadly, we would be exploring, albeit in only a preliminary fashion, prospects for using chemistry as a major resource in protein discovery science.[23]
The field of protein chemical synthesis was greatly advanced with the discovery of cysteine-based native chemical ligation (NCL), by Kent and co-workers.[24–26] More recently, the scope of NCL has been expanded to encompass a wide range of non-cysteine amino acids, through methods developed in our laboratory and others.[27–33] As outlined in Figure 1, the general non-cysteine based NCL strategy adopted by our group involves the installation of a temporary thiol functionality on the N-terminal amino acid residue at the site of ligation. Following amide bond formation, the polypeptide or glycopeptide is exposed to mild, metal-free dethiylation conditions, resulting in the selective removal of the extraneous thiol functionality.
Figure 1.
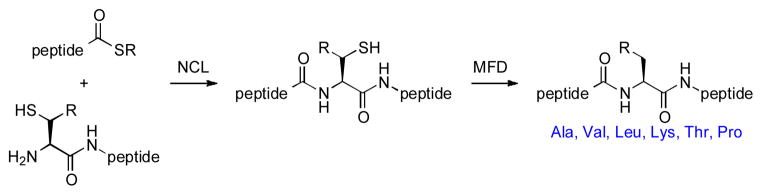
In a demonstration of the applicability of this ligation strategy to the assembly of challenging polypeptides lacking Cys residues, we recently disclosed total syntheses of hPTH, [34] and analogs thereof. Using these methods, we have now achieved the de novo total synthesis of hPTHrP (1–141).[35] We describe herein the synthesis and demonstration of biological activity of our synthetic hPTHrP (1– 141) polypeptide and a truncated analog, hPTHrP (1–37).
Synthetic Design
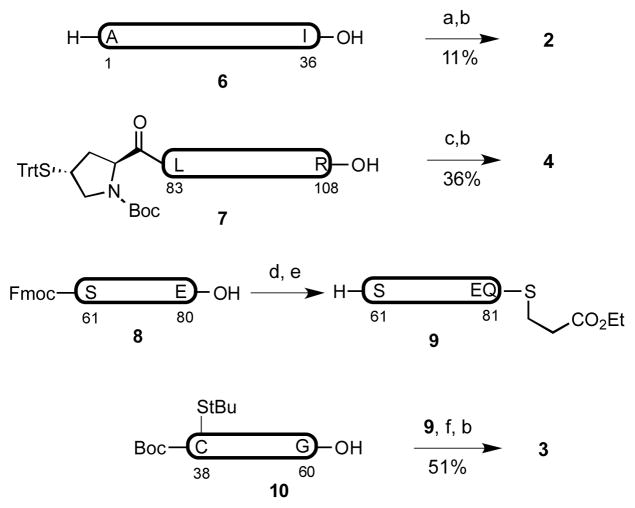
In our original synthetic route toward PTHrP(1–141), 1, we envisioned gaining access to four peptide segments of approximately equal size (2–5), through recourse to solid phase peptide synthesis (SPPS) methods (Figure 2). The component fragments, bearing temporary Cys residues at positions 38 and 110, and a thio-Pro surrogate at position 82, would then be iteratively merged through standard ligation protocols. Finally, the fully ligated peptide sequence would be subjected to MFD conditions to remove the three extraneous thiol groups, revealing Ala residues at 38 and 110, and the natural Pro residue at position 82.
Figure 2.
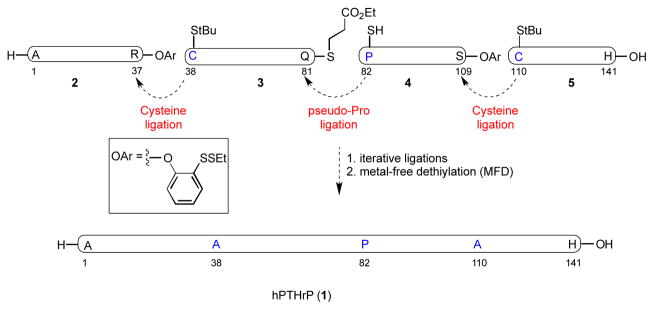
Initial synthetic plan toward hPTHrP(1–141).
Original Route to hPTHrP
The synthesis of hPTHrP commenced with the assembly of fragments 2–5, via Fmoc-based SPPS, on a 0.05–0.10 mmol scale (Figure 3).[36,37] The thio-proline surrogate of fragment 4 was manually appended at the N-terminus of the fully protected peptide via HATU-mediated coupling. Peptide segments bearing C-terminal thioesters (2–4) were prepared from the fully protected peptide precursors, through EDCI-mediated amide formation in the presence of HOOBt, under the epimerization-free conditions developed by Sakakibara.[38] The synthesis of peptide 3 via direct SPPS proved quite difficult. Ultimately, the fragment was further divided into smaller segments, 8 and 10, which were readily accessed through SPPS. Appendage of the thioester moiety to peptide 8, with subsequent removal of the N-terminal Fmoc group, yielded peptide 9. The fully protected peptides 9 and 10 were successfully connected through EDC coupling in CHCl3 [39] to deliver the target peptide, 3. With the four component peptide fragments in hand, we now sought to accomplish their merger.
Figure 3.
a) H-Arg(Pbf)-O(2-EtSS)Ph•HCl, EDC, CHCl3/TFE 3:1 v/v. b) TFA/PhOH/iPr3SiH/H2O 88:2:6:4 v/v. c) H-Ser(tBu)-O(2- EtSS)Ph•HCl, EDC, CHCl3/TFE 3:1 v/v. d) H-Gln(Trt)-SCH2CH2CO2Et•HCl, EDC, CHCl3/TFE 3:1 v/v. e) piperidine, CH2Cl2. f) EDC, HOOBt, CHCl3. Pbf = 2,2,4,6,7-pentamethyl-2,3- dihydrobenzofuran-5-sulfonyl, EDC = N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide, TFA = trifluoroacetic acid, TFE = trifluoroethanol, Trt = triphenylmethyl, HOOBt = 3-hydroxy-1,2,3-benzotriazin-4(3H)- one. Peptide fragments 6–10 (in bold) contain protected amino acid residues.
As outlined in Figure 4, the hoped-for ligation between fragments 4 and 5 was accomplished in 6.0 M guanidinium pH 7.2 buffer system, to provide the target peptide 12 in 48% yield within 3h. Similarly, peptides 2 and 3 were ligated under NCL conditions to afford the desired peptide, 11, in 40% yield.
Figure 4.
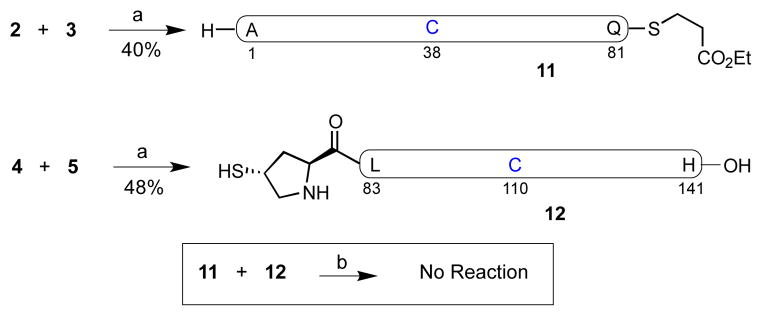
a) 6 M Gn•HCl, 100 mM Na2HPO4, 50 mM TCEP, pH 7.2. b) 6 M Gn•HCl, 300 mM Na2HPO4, 200 mM MPAA, 20 mM TCEP, pH 7.2. Gn = guanidine, TCEP = tris(2-carboxyethyl)phosphine hydrochloride, MPAA = 4-mercaptophenylacetic acid.
We next sought to merge fragments 11 and 12, en route to hPTHrP, through proline-based ligation.40 However, in the presence of 4-mercaptophenylacetic acid (MPAA) catalyst under our standard reaction conditions, no product was observed after 7h at room temperature. This result was somewhat surprising, given that the proposed ligation pattern; i.e. C-terminal Gln and N-terminal Pro, had been previously demonstrated in our laboratory, albeit in somewhat smaller peptide substrates.[41] Clearly, our initial synthetic plan toward hPTHrP would require reconfiguration.
A New Route to hPTHrP
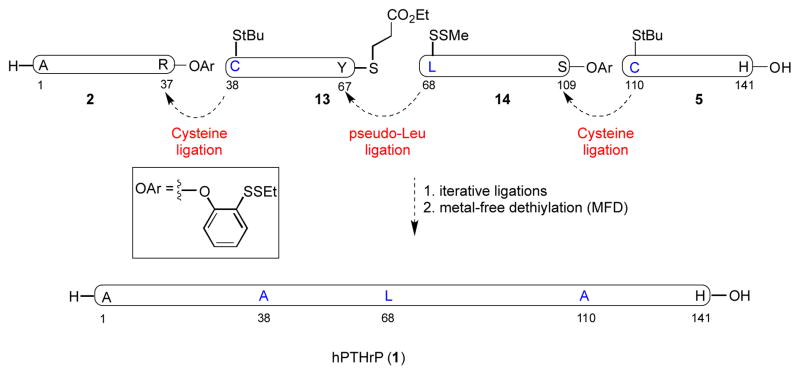
In re-examining the primary structure corresponding to hPTHrP, we elected to shift the site of the final ligation from Gln81–Pro82 to Tyr67–Leu68. We anticipated that application of our recently described formal leucine ligation protocol would enable this proposal.[42] This shift in the disconnection site would require the assembly of fragments 13 and 14, in place of peptides 3 and 4 (Figure 5). In fact, this alternative disconnection strategy proved quite advantageous from the standpoint of peptide synthesis. Thus, while the synthesis of the previous precursor fragment, 3, had required the coupling of two shorter peptides, the new substrates, 13 and 14, were both readily accessed through direct SPPS with high purity. Notably, although peptide 14 is quite long, the lysine rich regions of this segment serve to alleviate potential issues of peptide aggregation.
Figure 5.
Alternative retrosynthetic plan of hPTHrP (1–141)
As shown in Figure 6, the fully protected peptides, 15 and 16, were synthesized via Fmoc-based solid phase peptide synthesis (0.1 mmol scale, Figure 5). The pre-leucine surrogate was incorporated onto the N-terminus of the fully protected peptide through application of the previously described method. Finally, amino acid residues presenting C-terminal thioester moieties for NCL were attached to the peptides through recourse to standard protocols.
Figure 6.
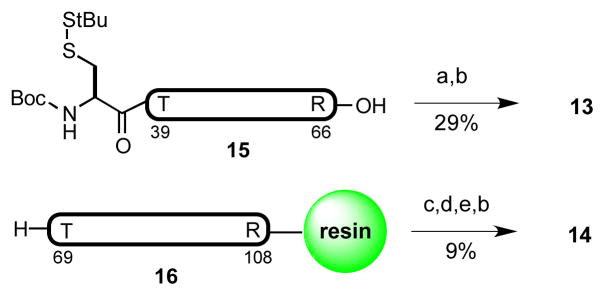
a) H-Tyr(tBu)-SCH2CH2CO2Et, EDC, HOOBt, CHCl3/TFE 3:1 v/v. b) TFA/PhOH/iPr3SiH/H2O 88:2:6:4 v/v. c) (3S)-N-Boc-3- CH3SS-Leu-OH, HATU, iPr2EtN, DMF. d) HOAc/TFE/DCM 1:1:8 v/v. e) H-Ser(tBu)-O-(2-EtSS)Ph•HCl, EDC, HOOBt, CHCl3/TFE 3:1 v/v. Boc = t-butoxycarbonyl, HATU = 1-[bis(dimethylamino)methylene]-1H- 1,2,3-triazolo[4,5-b]pyridinium 3-oxide hexafluorophosphate, DMF = N, N-dimethylformamide. Peptide fragments 15 and 16 (in bold) contain protected amino acid residues.
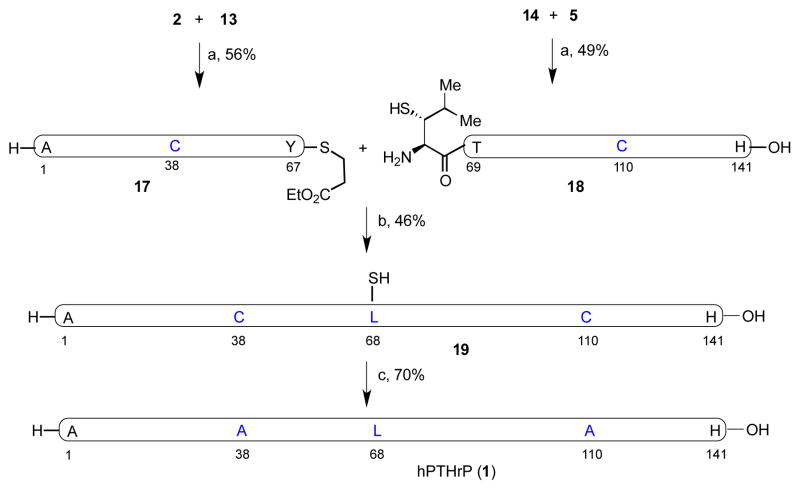
In the event, NCL of peptide segments 2 and 13 was conducted in a pH 7.2 buffer at room temperature, to give the desired peptide 17 in 56% yield after 3h (Figure 7). Segments 14 and 5 were similarly ligated to generate 18 in 49% isolated yield. In the key coupling event, peptide segments 17 and 18 smoothly underwent the hoped-for thio-Leu ligation, in the presence of 4-mercaptophenylacetic acid (MPAA) in pH 7.2 buffer, to yield peptide 19 in 46% isolated yield after 20 h. Finally, under the metal-free radical desulfurization conditions developed in our laboratory (VA-044, TCEP pH 7.2 buffer at 37 °C), peptide 19, bearing three extraneous thiol groups, was readily converted to the target compound, PTHrP(1–141) in 70% isolated yield (Figure 7 and Supporting Information). Under native conditions, the synthetic hPTHrP(1–141) folded spontaneously, [43,44] as demonstrated in the circular dichroism (CD) measurements (190–250 nm, Supporting Information).
Figure 7.
a) 6 M Gn•HCl, 100 mM Na2HPO4, 50 mM TCEP, pH 7.2. b) 6 M Gn•HCl, 300 mM Na2HPO4, 200 mM MPAA, 20 mM TCEP, pH 7.2. c) TCEP, tBuSH, VA-044, 37 °C. VA-044 = 2,2′-azobis[2-(2- imidazolin-2-yl)propane]dihydrochloride.
Biological Evaluation
The functional properties of the PTHrP(1–141) polypeptide were evaluated in vitro using cells expressing the hPTHR1. When assessed in an HEK-293-derived cell line that stably expresses the hPTHR1 along with the Glosensor cAMP reporter gene construct, [45] the PTHrP(1–141) peptide induced the formation of cAMP with the same potency (EC50) and efficacy (Emax) as did PTHrP(1–37) (Figure 8A; Table 1).[46] The affinity with which these ligands bound to the hPTHR1 was assessed in membrane-based competition assays designed to assess binding to two pharmacologically distinct, high affinity PTHR conformations: a G protein-independent, conformation, R0, and a G protein-dependent conformation, RG.[47] Assays for R0 were performed using 125I-PTH(1–34) as a tracer radioligand, and an excess of GTPγS, which uncouples receptor-G protein complexes, was added to the reactions. Assays for RG binding were performed using 125I-M-PTH(1–15) tracer radioligand and membranes from cells expressing a high affinity, Gαs mutant. Under either of these conditions, the PTHrP(1–141) peptide bound with an affinity that was sufficient to fully compete with the tracer radioligand for binding to the receptor, but nevertheless was moderately weaker than that of PTHrP(1–37) (Figure 8B and C; Table 1). The reasons for this weaker apparent binding of the full-length peptide to the receptor, as compared to the shorter-length N-terminal fragment peptide, despite a similar potency for cAMP formation, is not clear at present. Indeed, crystal structure analysis of the complex formed between a PTHrP peptide and the N-terminal binding domain of the PTHR1 indicates that most, if not all, of the key binding interactions that occur between the ligand and this region of the receptor involve residues limited to the (20–34) region of the ligand.[48]
Figure 8.
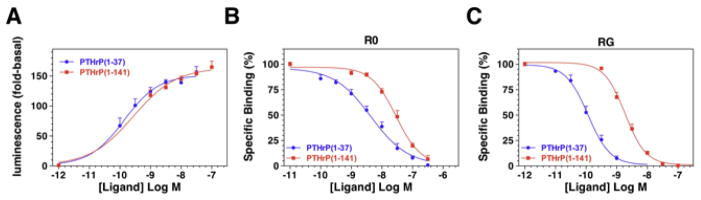
The Biological properties of hPTHrP(1–37) and hPTHrP(1–141) assessed in vitro.
Table 1.
Functional Properties of intact and N-terminal PTHrP peptide ligands
| ligand | Binding | n | cAMP (Glosensor assay) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R0 | P | RG | |||||||||||
| pIC50 | pIC50 | P | Emax | p | pEC50 | p | n | ||||||
| nM | fold | nM | fold | fold-basala | nM | fold | |||||||
| PTHrP(1–37)-OH | 8.40 ± 0.12 | 1.00 | 9.98 ±0.10 | 1.00 | 4 | 161 ± 21 | 1.000 | 9.68 ± 0.37 | 1.000 | 3 | |||
| 3.98 | 1.0 | 0.105 | 1.0 | 0.21 | 1.0 | ||||||||
| PTHrP(1–141)-OH | 7.53 ± 0.11 | 0.002 | 8.66 ± 0.08 | 0.0001 | 4 | 175 ± 20 | 0.670 | 9.39 ± 0.39 | 0.610 | 3 | |||
| 29.51 | 7.4 | 2.19 | 21 | 0.41 | 2.0 | ||||||||
Basal = 1.223±145 cps, n=3
Conclusion
In summary, the first total chemical synthesis of hPHTrP(1–141) has been achieved. This highly convergent route features iterative, non-cysteine based native chemical ligations, and metal-free desulfurization. The synthesis was accomplished in a convergent fashion. The total yield was 16% from peptide 14. This efficient synthetic strategy will now be used as a means by which to produce significant quantities of homogeneous hPTHrP(1–141), and congeners thereof, to facilitate hPTHrP- and PTHR1- directed research in the fields of oncology and osteoporosis therapeutics.
More broadly, it seems to be the case that, from a pharma-type protein discovery perspective, chemical synthesis may well provide a faster first–time synthesis of a significant sized all-proteogenic protein in higher levels of purity than that available through molecular biology means of expression. This is even more the case with significant size glycoproteins, let alone protein carrying unnatural amino acids or specifically designed implements for controlling secondary structure.
Supplementary Material
Footnotes
This research was supported by NIH grant HL25848 (S.J.D.). S.D.T. is grateful to Weill Cornell Medical School for an NIH postdoctoral fellowship (CA062948).
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Dr. Jianfeng Li, Laboratory for Bioorganic Chemistry, Sloan-Kettering Institute for Cancer Research, 1275 York Avenue, New York, NY 10065 (USA)
Dr. Suwei Dong, Laboratory for Bioorganic Chemistry, Sloan-Kettering Institute for Cancer Research, 1275 York Avenue, New York, NY 10065 (USA)
Dr. Steven D. Townsend, Laboratory for Bioorganic Chemistry, Sloan-Kettering Institute for Cancer Research, 1275 York Avenue, New York, NY 10065 (USA)
Dr. Thomas Dean, Endocrine Unit, Massachusetts General Hospital and Harvard Medical School, 51 Blossom Street, Boston, MA (USA)
Prof. Thomas J. Gardella, Endocrine Unit, Massachusetts General Hospital and Harvard Medical School, 51 Blossom Street, Boston, MA (USA)
Prof. Samuel J. Danishefsky, Email: s-danishefsky@ski.mskcc.org, Laboratory for Bioorganic Chemistry, Sloan-Kettering Institute for Cancer Research, 1275 York Avenue, New York, NY 10065 (USA). Department of Chemistry, Columbia University, 3000 Broadway, New York, NY 10027
References
- 1.Moseley JM, Kubota M, Diefenbach-Jagger H, Wettenhall REH, Kemp BE, Suva LJ, Rodda CP, Ebeling PR, Hudson PJ, Zajac JD, Martin TJ. Proc Natl Acad Sci USA. 1987;84:5048–5052. doi: 10.1073/pnas.84.14.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suva LJ, Winslow GA, Wettenhall REH, Hammonds RG, Moseley JM, Diefenbach-Jagger H, Rodda CP, Kemp BE, Rodriguez H, Chen EY, Hudson PJ, Martin TJ, Wood WI. Science. 1987;237:893–896. doi: 10.1126/science.3616618. [DOI] [PubMed] [Google Scholar]
- 3.Dean T, Vilardaga JP, Potts JT, Jr, Gardella TJ. Mol Endocrinol. 2008;22:156–166. doi: 10.1210/me.2007-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pioszak AA, Parker NR, Gardella TJ, Xu HE. J Biol Chem. 2009;284:28382–28391. doi: 10.1074/jbc.M109.022905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amizuka N, Warshawsky H, Henderson JE, Goltzman D, Karaplis AC. J Cell Biol. 1994;126:1611–1623. doi: 10.1083/jcb.126.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao D, He B, Jiang Y, Kobayashi T, Soroceanu MA, Zhao J, Su H, Tong X, Amizuka N, Gupta A, Genant HK, Kronenberg HM, Goltzman D, Karaplis AC. J Clin Invest. 2005;115:2402–2411. doi: 10.1172/JCI24918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foley J, Dann P, Hong J, Cosgrove J, Dreyer B, Rimm D, Dunbar M, Philbrick W, Wysolmerski J. Development. 2001;128:513–525. doi: 10.1242/dev.128.4.513. [DOI] [PubMed] [Google Scholar]
- 8.Kovacs CS, Lanske B, Hunzelman JL, Guo J, Karaplis AC, Kronenberg HM. Proc Natl Acad Sci USA. 1996;93:15233–15238. doi: 10.1073/pnas.93.26.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schordan E, Welsch S, Rothhut S, Lambert A, Barthelmebs M, Helwig JJ, Massfelder T. J Am Soc Nephrol. 2004;15:3016–3025. doi: 10.1097/01.ASN.0000145529.19135.EF. [DOI] [PubMed] [Google Scholar]
- 10.Philbrick WM, Dreyer BE, Nakchbandi IA, Karaplis AC. Proc Natl Acad Sci USA. 1998;95:11846–11851. doi: 10.1073/pnas.95.20.11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen X, Falzon M. Exp Cell Res. 2006;312:3822–3834. doi: 10.1016/j.yexcr.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Downs TM, Burton DW, Araiza FL, Hastings RH, Deftos LJ. Cancer Lett. 2011;306:52. doi: 10.1016/j.canlet.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 13.Deftos LJ, Barken I, Burton DW, Hoffman RM, Geller J. Biochem Biophys Res Commun. 2005;327:468–472. doi: 10.1016/j.bbrc.2004.11.162. [DOI] [PubMed] [Google Scholar]
- 14.Dittmer A, Schunke D, Dittmer J. Cancer Lett. 2008;260:56–61. doi: 10.1016/j.canlet.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Karaplis AC, Huang DC, Siegel PM, Camirand A, Yan XF, Muller WJ, Kremer R. J Clin Invest. 2011;121:4655–4669. doi: 10.1172/JCI46134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hastings RH, Burton DW, Nefzi A, Montgrain PR, Quintana R, Deftos LJ. Cancer Biol & Ther. 2010;10:1067–1075. doi: 10.4161/cbt.10.10.13374. [DOI] [PubMed] [Google Scholar]
- 17.Safina A, Sotomayor P, Limoge M, Morrison C, Bakin AV. Mol Cancer Res. 2011;9:1042–1053. doi: 10.1158/1541-7786.MCR-10-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horwitz MJ, Tedesco MB, Garcia-Ocana A, Sereika SM, Prebehala L, Bisello A, Hollis BW, Gundberg CM, Stewart AF. Clin Endocrinol Metab. 2010;95:1279–1287. doi: 10.1210/jc.2009-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid RE. Peptide and protein drug analysis. M. Dekker; New York: 2000. pp. 1–885. [Google Scholar]
- 20.Borgia JA, Fields GB. Trends Biotechnol. 2000;18:243–251. doi: 10.1016/s0167-7799(00)01445-1. [DOI] [PubMed] [Google Scholar]
- 21.Casi G, Hilvert D. Curr Opin Struct Biol. 2003;13:589–594. doi: 10.1016/j.sbi.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Kent S. J Pept Sci. 2003;9:574–593. doi: 10.1002/psc.475. [DOI] [PubMed] [Google Scholar]
- 23.Liu CC, Schultz PG. Ann Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 24.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 25.Kent SBH. Chem Soc Rev. 2009;38:338–351. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- 26.For a review on the chemical synthesis of proteins, see: Nilsson BL, Soellner MB, Raines RT. Annu Rev Biophys Struct. 2005:34:91–118. doi: 10.1146/annurev.biophys.34.040204.144700.
- 27.Wan Q, Danishefsky SJ. Angew Chem. 2007;119:9408. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2007;46:9248–9252. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Wan Q, Yuan Y, Zhu J, Danishefsky SJ. Angew Chem. 2008;120:8649. doi: 10.1002/anie.200803523. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:8521–8524. doi: 10.1002/anie.200803523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Wang P, Zhu J, Wan Q, Danishefsky SJ. Tetrahedron. 2010;66:2277–2283. doi: 10.1016/j.tet.2010.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan LZ, Dawson PE. J Am Chem Soc. 2001;123:526–533. doi: 10.1021/ja003265m. [DOI] [PubMed] [Google Scholar]
- 31.Botti P, Tchertchian S. Patent No. WO2006133962. 2006
- 32.Crich D, Banerjee A. J Am Chem Soc. 2007;129:10064–10065. doi: 10.1021/ja072804l. [DOI] [PubMed] [Google Scholar]
- 33.Haase C, Rohde H, Seitz O. Angew Chem. 2008;120:6912. doi: 10.1002/anie.200801590. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:6807–6810. doi: 10.1002/anie.200801590. [DOI] [PubMed] [Google Scholar]
- 34.Shang S, Tan Z, Danishefsky SJ. Proc Natl Acad Sci USA. 2011;108:5986–5989. doi: 10.1073/pnas.1103118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.For the recombinant method of hPTHrP(1–141) synthesis, see: Rian E, Jemtland R, Olstad OK, Gordeladze JO, Gautvik KM. Eur J Biochem. 1993;213:641–648. doi: 10.1111/j.1432-1033.1993.tb17803.x.
- 36.Merrifield RB. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- 37.Lloydwilliams P, Albericio F, Giralt E. Tetrahedron. 1993;49:11065–11133. [Google Scholar]
- 38.Sakakibara S. Biopolymers. 1995;37:17–28. doi: 10.1002/bip.360370105. [DOI] [PubMed] [Google Scholar]
- 39.It was found that the co-solvent trifluoroethanol can react with the Cterminus of peptide 3a, therefore it was not used in this situation.
- 40.Johnson EC, Kent SB. J Am Chem Soc. 2006;128:6640–6646. doi: 10.1021/ja058344i. [DOI] [PubMed] [Google Scholar]
- 41.Shang S, Tan Z, Dong S, Danishefsky SJ. J Am Chem Soc. 2011;133:10784–10786. doi: 10.1021/ja204277b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan Z, Shang S, Danishefsky SJ. Angew Chem. 2010;122:9690. doi: 10.1002/anie.201005513. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2010;49:9500–9503. doi: 10.1002/anie.201005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zull JE, Smith SK, Wiltshire R. J Biol Chem. 1990;265:5671–5676. [PubMed] [Google Scholar]
- 44.Alexander P, Orban J, Bryan P. Biochemistry. 1992;31:7243–7248. doi: 10.1021/bi00147a006. [DOI] [PubMed] [Google Scholar]
- 45.Binkowski BF, Butler BL, Stecha PF, Eggers CT, Otto P, Zimmerman K, Vidugiris G, Wood MG, Encell LP, Fan F. ACS Chem Biol. 2011;6:1193–1197. doi: 10.1021/cb200248h. [DOI] [PubMed] [Google Scholar]
- 46.Martin TJ, Moseley JM, Gillespie MT. Crit Rev Biochem Mol Biol. 1991;26:377–395. doi: 10.3109/10409239109114073. [DOI] [PubMed] [Google Scholar]
- 47.Dean T, Linglart A, Mahon MJ, Bastepe M, Juppner H, Potts JT, Jr, Gardella TJ. Mol Endocrinol. 2006;20:931–942. doi: 10.1210/me.2005-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pioszak AA, Parker NR, Gardella TJ, Xu HE. J Biol Chem. 2009;284:28382–28391. doi: 10.1074/jbc.M109.022905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.