Abstract
A combinatorial library method was developed to systematically profile the substrate specificity of protein phosphatases toward phosphoseryl (pS) and phosphothreonyl (pT) peptides. Application of this method and a previously reported phosphotyrosyl (pY) library screening technique to dual-specificity phosphatase (DUSP) VH1 of vaccinia virus revealed that VH1 is highly active toward both pS/pT and pY peptides. VH1 exhibits different and more stringent sequence specificity toward pS/pT than pY substrates. Unlike previously characterized protein tyrosine phosphatases (PTPs), the activity and specificity of VH1 is primarily determined by the amino acid residues C-terminal to the pS, pT, or pY residue. In contrast, the mammalian VH1-related (VHR) DUSP has intrinsically low catalytic activity toward pS and pT substrates, suggesting that its primary physiological function is to dephosphorylate pY residues in substrate proteins. This method is applicable to other DUSPs and protein-serine/threonine phosphatases and the substrate specificity data will be useful for identifying the physiological substrates of these enzymes.
Keywords: Combinatorial peptide library, dual-specificity phosphatase, sequence specificity, phosphoserine, phosphothreonine
INTRODUCTION
Reversible phosphorylation of proteins on serine, threonine, and tyrosine residues mediates/regulates numerous physiological processes and is controlled by the opposing action of protein kinases and protein phosphatases. Unlike the protein kinases, which all belong to the same structurally conserved superfamily, protein phosphatases are classified into two distinct superfamilies, protein-serine/threonine phosphatases versus protein-tyrosine phosphatases (PTPs). The 107 human PTPs can be further grouped into four separate subfamilies on the basis of the amino acid sequences of their catalytic domains.1–3 The largest subfamily contains 38 classical PTPs, which are specific for phosphotyrosyl (pY) proteins, and 61 dual-specificity phosphatases (DUSPs), which can dephosphorylate both pY and phosphoserine (pS)/phosphothreonine (pT) residues within a protein substrate. Both classical PTPs and DUSPs share a highly conserved sequence motif (HCXXXXXR) in the active site and a common catalytic mechanism that involves the formation of a covalent phosphocysteinyl intermediate.4 First identified more than two decades ago,5 DUSPs have been implicated as major modulators of critical signaling pathways that are dysregulated in various diseases.1–3 However, identification of the physiological substrates of DUSPs and understanding the molecular basis of their function remain highly challenging. Because DUSPs have a relatively shallow active-site pocket6 and often bind weakly to their substrates, identification of DUSP substrates by “substrate trapping”7 has met with only limited success.8–10 For most of the DUSPs, the three MAPKs, Erk, JNK, and p38, are the only reported substrates, some of which remain controversial.2,3
PTPs are thought to have exquisite substrate specificity in vivo, which is combinatorially controlled by the sequence specificity of the PTP active site and protein-protein interaction via substrate recruiting domains/docking sites outside the PTP active site.11 Thus, determination of the sequence specificity of a PTP catalytic domain may provide important clues to its in vivo protein substrates. We recently developed a peptide library method for systematically profiling the sequence specificity of PTPs against pY peptides.12 Specificity profiling of a dozen or so PTPs revealed that each PTP has a unique specificity profile, although different PTPs exhibit varying levels of sequence specificity and there is significant overlap in specificity among some of the PTPs.13,14 In particular, we found that vaccinia virus VH1-related (VHR) phosphatase, a member of the atypical DUSP subgroup,2,3 has more stringent sequence specificity than the classical PTPs against pY peptides.15 The consensus sequences derived from the library screening closely resemble the reported VHR dephosphorylation sites in its protein substrates, providing further proof that the intrinsic sequence specificity of the PTP active site is a key determinant of its in vivo substrate specificity. In addition, by using the specificity profiles, we were able to predict the protein substrates of PTPs, some of which were subsequently confirmed experimentally.14,15
In principle, the above “reverse interactomics”16 approach should also be applicable to the identification of pS and pT protein substrates of protein serine/threonine phosphatases and DUSPs. Unfortunately, there has been no methodology available for comprehensive analysis of the substrate specificity of a phosphatase toward pS or pT substrates. The limited specificity data on several protein serine/threonine phosphatases were obtained by in-solution kinetic analysis of individually synthesized pS/pT peptides.17–19 To our knowledge, the substrate specificity of only one of the 61 human DUSPs (VHZ) has recently been evaluated by assaying against a small panel of pS and pT peptides.20 In this work, we report a novel library screening method for systematically profiling the specificity of DUSPs and protein serine/threonine phosphatases against pS/pT peptides and its application to VHR and vaccinia VH1 phosphatases. The specificity profiles of VH1 and VHR revealed several previously unrecognized properties of these two enzymes. Thus, the new method provides a powerful tool for studying the catalytic properties, in vivo substrates, and biological function of DUSPs and other protein phosphatases.
EXPERIMENTAL SECTION
Materials
Reagents for peptide synthesis were from Advanced ChemTech (Louisville, KY), Peptides International (Louisville, KY), or NovaBiochem (La Jolla, CA). N-(9-Fluorenylmethoxycarbonyloxy)succinimide (Fmoc-OSu) was from Advanced ChemTech. α-Cyano-4-hydroxycinnamic acid, phenyl isothiocyanate (PITC), 3-methyl-2-benzothiazolinonehydrazone (MBTH), and NBDH were obtained from Sigma-Aldrich (St. Louis, MO). Nα-Fmoc-O-t-butyl-3,5-difluorotyrosine [Fmoc-F2Y(tBu)-OH] was synthesized as previously described.21 Streptomyces antibioticus tyrosinase was purified as previously described.13 GST-VH1 and VHR were expressed in Escherichia coli and purified as previously described.5,22
Synthesis of Peptide Libraries
pY libraries (libraries III and IV) were synthesized as previously described.13 Library II [Alloc/Fmoc-XXXXX(pS/pT)AANNBBRM-resin, where B is β-alanine] was synthesized on 2 g of amino polyethylene glycol polyacrylamide (PEGA) resin (0.4 mmol/g, 150–300 μm in water). All manipulations were performed at room temperature unless otherwise noted. The linker sequence (AANNBBRM) was synthesized with 4 equiv of Fmoc-amino acids using HBTU/HOBt/N-methylmorpholine (NMM) as the coupling reagents, and the coupling reaction was terminated after negative ninhydrin tests. The resin was split into two equal portions and individually coupled with 4 equiv of Fmoc-Ser(HPO3Bzl)-OH and Fmoc-Thr(HPO3Bzl)-OH and HATU/HOAt/DIPEA for 3 h. For the synthesis of random residues, the resin was split into 18 equal portions, and each portion was coupled twice, each with 5 equiv of a different Fmoc-amino acid/HATU/DIPEA for 2 h (twice). To facilitate sequence determination by mass spectrometry,23 5% (mol/mol) CD3CO2D was added to the coupling reactions of leucine and lysine, whereas 5% CH3CD2CO2D was added to the coupling reaction of Nle. After the last random residue was added, the resin was combined, treated twice with 20% piperidine in DMF (5 + 15 min), and incubated with 0.5 equiv of N-(allyloxycarbonyloxy)succinimide (Alloc-OSu), 0.5 equiv of Fmoc-OSU and 1.5 equiv of DIPEA in NMP for 24 h. The library was deprotected with a modified reagent K [79:7.5:5:5:2.5:1 (v/v) trifluoroacetic acid(TFA)/phenol/H2O/thioanisole/ethanedithiol/anisole] for 2 h, washed extensively with TFA and dichloromethane, and stored at −20 °C until use. Libraries Ia and Ib were similarly synthesized.
Screening of pS/pT Library
All steps were carried out at room temperature unless otherwise noted. For library I, a typical screening reaction involved 10 mg of library beads (~30,000 beads) placed in a 1-ml Bio-Rad Microspin column. The beads were washed with 50 mM NaCl plus 0.02% tween 20 (5x) and incubated with DUSP reaction buffer (100 mM Tris, 100 mM NaOAc, pH 7.4, 0.1% bovine serum albumin, 0.01% tween 20) for 30 min. After being washed three times with the reaction buffer, the beads were suspended in 400 μl of DUSP reaction buffer containing 500 nM GST-VH1 and 5 mM tris(carboxyethyl)phosphine and incubated for 2 h. After that, the beads were washed with 50 mM NaCl containing 0.02% tween 20 and anhydrous THF (5 times each). The resin was then treated with 0.9 ml of THF containing 5 mg of Pd(PPh3)4, 30 mg of PPh3, 30 μl of N–methylaniline for 3 h. The solution was drained and the beads were washed with DMF (5 × 0.9 ml) and incubated in 0.9 ml of DMF containing 0.5% dimethyldithiocarbamate for 15 min. The beads were again washed with DMF (5 × 0.9 ml) and 50 mM NaCl containing 0.02% tween 20 (5 × 0.9 ml) and treated with 900 μl of sodium phosphate (pH 7.0–7.4) containing 40 mM NaIO4 (freshly prepared in H2O). After incubation for 30 min, the solution was drained and the beads were washed with 50 mM NaCl containing 0.02% tween 20 (5 × 0.9 ml) and incubated in 1% ethylene glycol in H2O for 10 min. The beads were washed with 50 mM NaCl containing 0.02% tween 20 (5 × 0.9 ml) and dye labeling buffer [1:2 (v/v) mixture of 30 mM NaOAc pH 4.7 and MeCN] and incubated in 99 μl of the labeling buffer and 1 μl of 100 mM 4-hydrazino-7-nitro-2,1,3-benzoxadiazole (NBDH) freshly prepared in DMSO for 1 h. The beads were washed exhaustively with DMF (10 × 0.9 ml) and 50 mM NaCl containing 0.02% tween 20 (5 × 0.9 ml) and transferred to a Petri dish, where fluorescent beads were isolated with a micropipette under a fluorescence microscope. The positive beads were sequenced by the partial Edman degradation-mass spectrometry (PED-MS) method as previously described.23
The N-terminal library (library II, 20 mg) was similarly washed and treated with 50 nM GST-VH1 and 5 mM tris(carboxyethyl)phosphine in 800 μl of DUSP reaction buffer for 1 h. The beads were washed with 50 mM NaCl containing 0.02% tween 20 (5 × 0.9 ml) and DMF (6 × 0.9 ml) and treated twice with 20% piperidine in DMF (0.9 ml each for 5 and 15 min). The beads were washed with DMF (10 × 0.9 ml) and pyridine (5 × 0.9 ml) and suspended in 160 μl of pyridine. A 2:1 (v/v) mixture of pyridine/H2O containing 0.1% triethylamine (160 μl) was quickly mixed with 50 μl of PITC in a microcentrifuge tube and the resulting solution was immediately added to the beads. After incubation for 30 min, the solution was drained and the beads were washed with pyridine (3 × 900 μl), acetonitrile (5 × 900 μl) and TFA (3 × 900 μl). The beads were then treated twice with TFA (900 μl for 15 min each time). The beads were washed with dichloromethane (6 × 0.9 ml) and pyridine (3 × 0.9 ml) and the above Edman degradation procedure was repeated for four times. The library beads were washed with and incubated in 50% TFA in H2O for 1 h and the subjected to oxidation with sodium periodate and dye labeling as described above.
Screening of pY Peptide Libraries
This was carried out as previously described,13 except that the screening experiment involved incubation of the peptide library (~30,000 beads) with GST-VH1 (final 400 nM) in 50 mM Tris, 50 mM Bis-Tris pH 7.4, 100 mM NaOAc, 0.1% bovine serum albumin, 0.01% tween 20, 5 mM tris(carboxylethyl)phosphine at room temperature for 15 min.
Synthesis of Selected Peptides
Individual peptides were synthesized on 100 mg of CLEAR-amide resin using standard Fmoc/HBTU/HOBt chemistry. For the coupling reaction of pY, 2.0 equiv of Fmoc-amino acid was employed, whereas 4.0 equiv was used for all other amino acids. The peptides were cleaved from the resin and side-chain deprotected using the modified reagent K at room temperature for 2 h. The solvents were removed by evaporation under a N2 stream and the residue was triturated in cold diethyl ether. The precipitate was collected and dried under vacuum. The crude peptides were purified by reversed-phase HPLC on a semi-preparative C18 column. The identity of each peptide was confirmed by MALDI-TOF.
PTP Assays
VH1 activity assays were performed with synthetic pS or pT peptides as substrates in a 96-well plate. The buffer used was a three-component system consisting of 0.1 M sodium acetate, 0.05 M Tris, 0.05 M Bis-Tris (pH 7.4), and 1 mM tris(carboxylethyl)phosphine. The reaction (total volume 50 μl) contained 0–4 mM peptide substrate and was initiated by the addition of GST-VH1 (final concentration 50–100 nM). After 15–40 min, the reaction was quenched by the addition of 50 μl of a malachite green solution prepared as previously described.24 After incubation for 30 min at room temperature, the absorbance at 620 nm was measured on a FlexStation3-Multi-detection Reader (Molecular Devices). The initial rate was calculated from the absorbance value by comparison with a standard curve generated known concentrations of sodium phosphate. VH1 activity assay against pY peptides was performed in a quartz microcuvette. The reaction (total volume 120 μl) was initiated by the addition of GST-VH1 (final 800 nM) and monitored continuously at 282 nm (Δε = 1102 M−1cm−1) on a UV-vis spectrophotometer. The initial rate was calculated from the early region of the reaction progress curve (<1 min). The resulting initial rates were fitted to the Michaelis-Menten equation to obtain the kcat, KM, and/or kcat/KM value. A few substrates (e.g., Ac-AApYIDWTG) were also assayed in 50 mM citric acid (pH 7.4), 50 mM NaCl, and 1 mM tris(carboxylethyl)phosphine as buffer. Similar kcat/KM values were obtained (≤2-fold difference), demonstrating that the buffer components do not significantly affect VH1 catalysis under the conditions employed in this work.
In Vitro Dephosphorylation of Protein Substrates by VH1
Activated GST-Erk5 (from SignalChem, Richmond, BC, Canada; final concentration 19 μM) was incubated with GST-VH1 (final concentration 1.5 μM) in 100 mM Tris-HCl, pH 7.4, 50 mM NaCl, 2 mM EDTA, and 5 mM at 25 °C (total reaction volume 56 μL). At various time points (0–540 min), 7-μL aliquots were withdrawn and mixed with an equal volume of 2x SDS-PAGE loading buffer. The samples were boiled for 10 min, separated on 8% SDS-PAGE, transferred to PVDF membrane and immunoblotted with anti-Erk5 (pT219) antibody (Pierce Biotechnology, Rockford, IL). The membrane was washed in 200 mM glycine, pH 2.2, 3.5 mM SDS, 1% Tween 20, and reprobed with anti-pY (clone 4G10) antibody (Millipore, Temecula, CA) following manufacturer’s recommendations. The catalytic efficiency was estimated by quantifying the phosphorylated GST-Erk5 using a Molecular Dynamics Typhoon 8600 Phosphoimager and Imagequant software.
RESULTS
Design, Synthesis, and Screening of pS/pT Peptide Libraries
To determine the specificity of DUSPs toward pS/pT substrates, two one-bead-one-compound peptide libraries, Alloc-(pS/pT)XXXXXNNBBRM-resin [library I, where B is β-alanine and X is norleucine (Nle or M, used as a methionine analog) or any of the 18 proteinogenic amino acids excluding Met and Cys] and Fmoc/Alloc-XXXXX(pS/pT)AANNBBRM-resin [library II, where B is β-alanine and X is Nle, L-2-aminobutyrate (Abu or U, used as a Cys analog), or any of the 16 proteinogenic amino acids excluding Met, Cys, Ser, and Thr] were synthesized on amino polyethylene glycol polyacrylamide (PEGA) resin (0.4 mmol/g, 150–300 μm in water) (Figure 1). The linker sequence NNBBRM was added to enhance aqueous solubility, permit peptide release from the resin (after Met with CNBr), and facilitate mass spectrometric analysis (positively charged Arg).23 An Alloc and/or Fmoc group was added to the N-terminus of all library peptides to prevent any bias exerted by a positively charged N-terminus and facilitate library screening and sequencing (vide infra). Substitution of Nle for methionine in the random region prevents internal cleavage during peptide release with CNBr. For the N-terminal library (library II), two alanine residues C-terminal to pS/pT were added to minimize any bias caused by the C-terminal Arg residue; in addition, Ser and Thr were excluded from the random region, as any incomplete Edman degradation of Ser or Thr residue would generate false positive beads. Each library has a theoretical diversity of ~4 × 106.
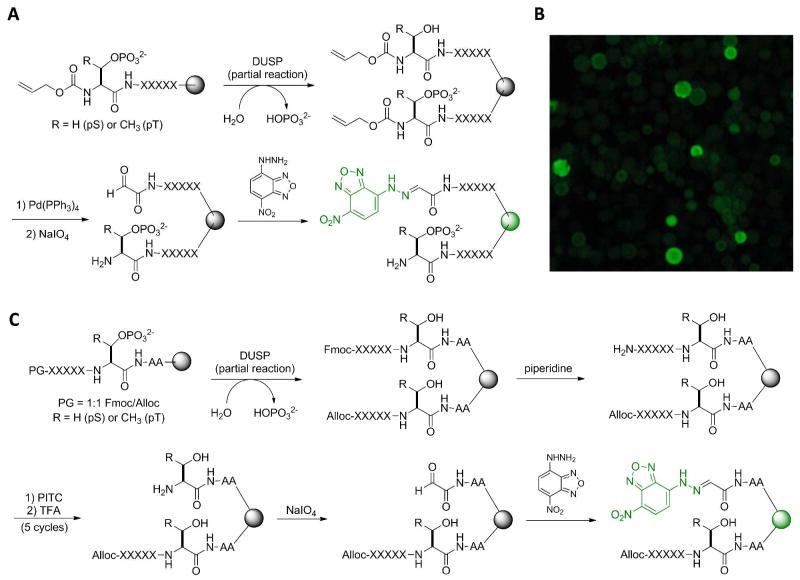
Figure 1.
Screening of DUSPs against pS/pT peptide libraries. (A and C) Reactions involved in the screening of C- (A) and N-terminal libraries (C). (B) A portion of the peptide library after DUSP and NBDH treatment (viewed under a fluorescent microscope). The positive beads show a green fluorescent ring on the surface because the surface peptides were more accessible to the 92-kD GST-VH1 dimer. Z, pS or pT.
Library screening involved treatment of the above libraries with a small amount of the DUSP of interest for a limited time, so that only the beads carrying the most efficient substrates underwent partial dephosphorylation of the pS/pT peptides (typically a few percent or less), while the rest of the beads had little or no reaction. To identify and isolate the reacted (positive) beads for sequence analysis, we developed two novel methods to selectively derivatize the DUSP reaction products with a fluorescent dye. For library I, which contains variable sequences C-terminal to pS/pT, the beads were treated with Pd(PPh3)4 and N-methylaniline to remove the N-terminal Alloc group (Figure 1A). The exposed N-terminal Ser (or Thr) on the positive beads was then converted into a glyoxyl group by oxidation with sodium periodate, whereas the N-terminal pS (or pT) residue on unreacted peptides was unaffected. Finally, the glyoxyl moiety was selectively reacted with NBDH, which is nonfluorescent due to photoelectron transfer,25 to produce an intensely green fluorescent dye covalently bound to the positive beads (Figure 1B). The fluorescent beads were manually isolated from the library under a fluorescent microscope and individually sequenced by the PED-MS method.23
For library II, which features variable sequences N-terminal to pS/pT and a 1:1 mixture of Alloc and Fmoc groups at the N-terminus, the beads after DUSP reaction were treated with piperidine to remove the Fmoc group from 50% of the peptides and subjected to 5 cycles of Edman degradation reaction (Figure 1C). This procedure removed the five random residues from 50% of the peptides on each bead, exposing an N-terminal Ser/Thr (on positive beads) or pS/pT residue (on both positive and negative beads). The other 50% of the peptides contained an N-terminal Alloc group and were protected from Edman degradation. The exposed N-terminal Ser/Thr residue on positive beads was similarly oxidized with sodium periodate and derivatized with NBDH. The resulting fluorescent beads were isolated from the library, treated with Pd(PPh3)4 and N-methylaniline to remove the N-terminal Alloc group from the remaining 50% peptides, and sequenced by the PED-MS method.23
VH1 Specificity toward pS/pT Peptides
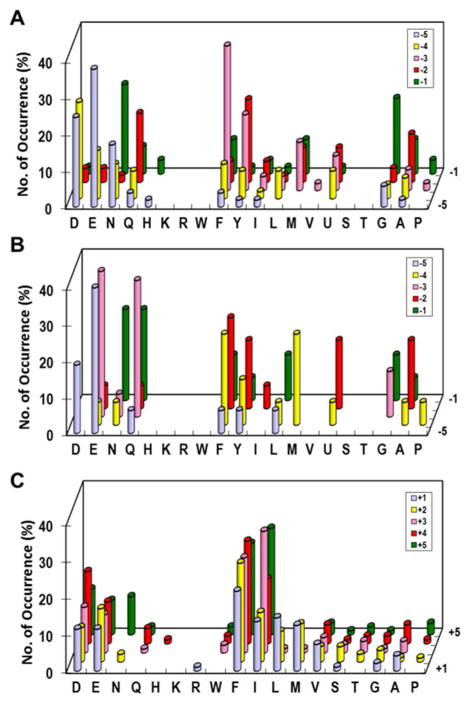
Screening of ~25 mg of the N-terminal library (dry weight, ~75,000 beads) against 50 nM VH1 resulted in 20 intensely fluorescent beads and 85 lightly colored beads. Sequencing of the most intensely colored beads produced 18 unambiguous sequences (Table 1), which should represent the most efficient substrates of VH1. Sequencing of the lightly colored beads gave 51 complete sequences. These sequences are separated into two different classes on the basis of sequence similarities, both of which contain an acidic residue (Asp or Glu) at the P−5 position and less frequently at the P−4 position (relative to pS/pT, which is defined as the P0 position) (Figure 2A and 2B). VH1 prefers a small, neutral, and hydrophilic residue (especially Gly and Asn) and tolerates larger hydrophobic residues, but disfavors charged residues (Asp, Glu, Arg, and Lys) at the P−1 position; out of the 69 selected sequences, only one (1.4%) contains an Asp at this position (EFYQDpT). Both types of substrates contain at least one large, hydrophobic residue (especially Phe and Tyr) but at different positions. The class I substrates typically have a Phe or Tyr at the P−3 position and frequently a second hydrophobic residue at the P−2 (or occasionally P−4) position, whereas the class II peptides contain a hydrophilic residue (e.g., Gln and Glu) at the P−3 position and usually two hydrophobic residues at P−2 and P−4 positions. All of the selected sequences contain a pT at the P0 position, although pS and pT were equally populated in the original library. Screening the N-terminal library under less stringent conditions (at 0.1 and 1 μM VH1) produced progressively more diverse sequences (Table S1 and S2), but the same overall specificity profile was observed.
Table 1.
Preferred VH1 Substrates Selected from Library II [XXXXX(pS/pT)AA] a
| Intensely Colored | Lightly Colored | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Class I | Class II | Class I | Class II | |||||||
| EEFMNt | DLMQGt | EFQAGt | EDFYPt | DYFDGt | ADFIAt | EDMQNt | DLMQUt | * NLLQNt | EMEYNt | EYGUFt |
| EDYMHt | DYVYGt | EFQYNt | EEFYPt | ENFYNt | QDYYQt | DFAAFt | EAMMGt | NQLQYt | DMEUNt | EEGIQt |
| DQFIQt | EQIINt | DMQYAt | DDYYNt | EFYQDt | NEFAFt | EFAYGt | YEMQAt | NNUUNt | DMEAGt | ENNFLt |
| DGFQMt | EAMYGt | * EYQFNt | DDYYNt | ELYQMt | GDFUHt | EFMAQt | GEUUFt | NNUNGt | EFEUQt | ELQFLt |
| DUFYGt | * NDFAIt | EDYFAt | ENFLGt | HDFUNt | DYAANt | FDUGFt | FPEEYt | LUQQQt | ||
| EFYEAt | NDYFIt | EQFQFt | EGYGLt | QEFUGt | EYPYMt | IIUFIt | QFEFQt | |||
| DUFMNt | GEFMQt | DUFYAt | EUYENt | NNFDNt | FDFAGt | NAIAMt | YAEAFt | |||
M, Nle; U, Abu; t, pT;
sequences selected for kinetic assays.
Figure 2.
Sequence specificity of VH1 toward pS/pT peptides. (A) Preferred class I substrates selected from the N-terminal library (X5pS/pT). (B) Preferred class II substrates selected from the N-terminal library. (C) Preferred substrates selected from the C-terminal library (pS/pTX5). The histograms represent the amino acids identified at each position from P−5 to P+5 (z-axis). Frequency of occurrence on the y-axis represents the percentage of selected sequences that contained a particular amino acid at a certain position. M, Nle; U, Abu.
In light of the overwhelming selection of pT sequences from the N-terminal library, we synthesized and screened the C-terminal library containing pS (library Ia) or pT (library Ib) at the P0 position in separate experiments. Screening of libraries Ia and Ib (20 and 10 mg, respectively) resulted in 82 and 41 fluorescent beads, which were sequenced to give 63 and 34 complete sequences, respectively (Table S3). Similar sequences were selected from both libraries, which are rich in large, hydrophobic residues at all five positions (Figure 2C). Most of the sequences also contain an acidic residue (Asp or Glu), which is randomly distributed among the five positions. Overall, the class I and II VH1 substrates have the consensus sequences of (E/D/N)(D/E/N)(F/Y/M)X(G/N)(pS/pT)φφφφ and (E/D)(F/M/Y)(Q/E)(F/Y/φ)(N/Q)(pS/pT)φφφφ (where X is any except for basic amino acid and φ is a hydrophobic amino acid), respectively.
VH1 Specificity toward pY peptides
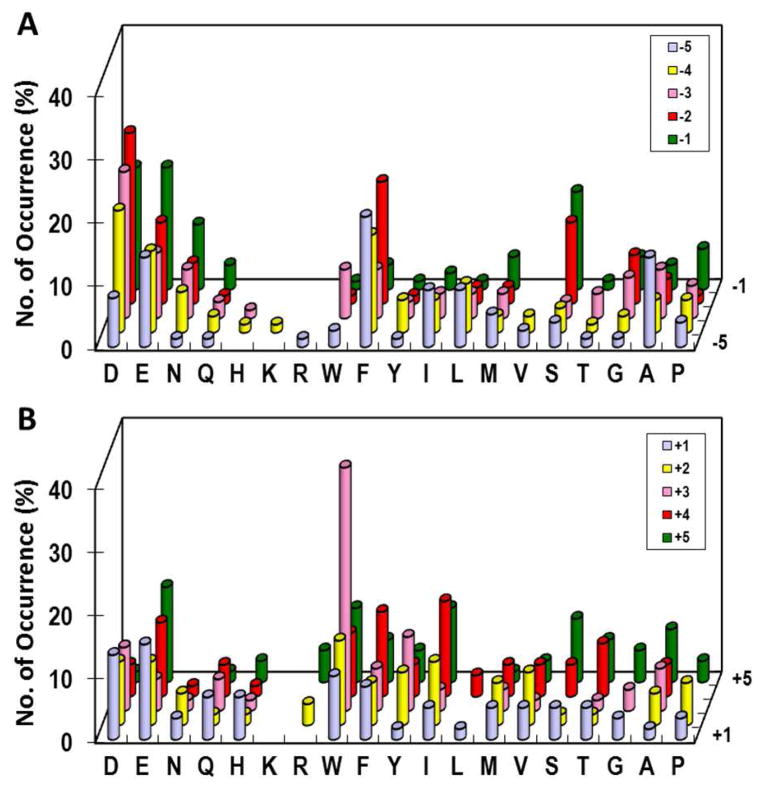
The ability of DUSPs to dephosphorylate both pT and pY residues of the pTXpY motif in MAPKs26,27 suggests that DUSPs recognize different sequences surrounding pS/pT vs pY residues. The availability of methods for screening both pY and pS/pT libraries permitted us to rigorously test this notion. To determine the specificity of VH1 toward pY substrates, we screened two pY peptide libraries, N-terminal library Alloc-ASXXXXXpYAABBRM-resin (library III) and C-terminal library Alloc-AApYXXXXXNNBBRM-resin (library IV), where B is β-alanine and X is 3,5-difluorotyrosine (F2Y), Nle, or any of the 17 proteinogenic amino acids excluding Met, Cys and Tyr. Library screening was based on selective oxidation of the Tyr side chain exposed by VH1 into an orthoquinone by tyrosinase reaction and reaction of the orthoquinone with 3-methyl-2-benzothiazolinonehydrazone to form a red pigment on positive beads.12–15 F2Y was used as a Tyr analog21 to avoid false positives during library screening. Screening of libraries III and IV (~90,000 beads each) gave 78 and 60 complete sequences, respectively (Table S4 and S5). VH1 has broad specificity N-terminal to pY, with a general preference for acidic (but not basic) residues at positions P−1 to P−3 and hydrophobic residues at P−4 and P−5 positions (Figure 3A). Most of the selected substrates have a hydrophilic or small residue at the P−1 position and contain at least one hydrophobic residue somewhere between P−2 and P−5 positions. On the C-terminal side of pY, VH1 strongly prefers an aromatic or other hydrophobic residues at the P+3 position; out of the 60 selected peptides, 34 contain Trp, Tyr, or Phe and another 5 peptides have Nle, Ile, or Val at this position (Figure 3B). The rest of the peptides have aromatic and aliphatic hydrophobic residues at P+2 and/or P+4 positions. In addition, VH1 has a slight preference for acidic residues at the P+1 position.
Figure 3.
Sequence specificity of VH1 toward pY peptides on the N- (A) and C-terminal side of pY (B). The histograms represent the amino acids identified at each position from P−5 to P+5 (z-axis). Frequency of occurrence on the y-axis represents the percentage of selected sequences that contained a particular amino acid at a certain position. M, Nle; Y, F2Y.
Kinetic Properties of VH1 toward Selected Peptide Substrates
To confirm the library screening results, three class I pT peptides (Table 2, peptides 1, 5, and 9) and one class II peptide (peptide 10) were resynthesized, purified, and assayed against VH1 in solution. All four peptides are relatively efficient substrates, with kcat/KM values of 180–870 M−1 s−1. Replacement of the acidic residue at P−4 or P−5 position with an alanine reduced the activity by ~2-fold (Table 2, compare peptides 1 and 2 or 5 and 6). Removal of Phe side chain at position P−3 of peptide 1 decreased the catalytic activity by 20-fold (Table 2, compare peptides 1 and 3). Replacement of the P−1 Gly of peptide 5 with an Asn had minimal effect, whereas conversion of the P−2 Ala into Tyr increased its activity by 1.6-fold (Table 2, compare peptides 5, 7, and 8). This is in agreement with the observation that Gly and Asn were selected at position P−1 with equal frequencies during library screening, while Tyr was more frequently selected than Ala at position P−2 (especially among the most colored beads) (Table 1). The ~2-fold difference in activity between peptides 1 and 4 suggests that VH1 is generally more active toward pT than pS substrates, offering a partial explanation for the overwhelming selection of pT sequences from the N-terminal library (Table 1), although the lower stability of pS (relative to pT) is likely another contributing factor (see Discussion).
Table 2.
Catalytic Activity of VH1 toward pS/pT Peptides
| Peptide No | Peptide sequencea | kcat (s−1) | KM (mM) | kcat/KM (M−1 s−1) |
|---|---|---|---|---|
| 1 | YNDFAIpTAA | 0.4 ± 0.2 | 2.1 ± 1.0 | 180 ± 9 |
| 2 | YNAFAIpTAA | NDb | >2 | 93 ± 3 |
| 3 | YNDAAIpTAA | ND | >2 | 9 ± 1 |
| 4 | YNDFAIpSAA | 0.2 ± 0.1 | 2.3 ± 0.3 | 95 ± 10 |
| 5 | YEFFAGpTAA | 0.6 ± 0.1 | 0.8 ± 0.1 | 700 ± 16 |
| 6 | YAFFAGpTAA | ND | >0.5 | 350 ± 13 |
| 7 | YEFFANpTAA | 1.4 ± 0.1 | 2.9 ± 0.1 | 500 ± 7 |
| 8 | YEFFYGpTAA | ND | >0.5 | 1100 ± 37 |
| 9 | YNLLQNpTAA | 1.8 ± 0.1 | 4.6 ± 0.4 | 390 ± 18 |
| 10 | YEYQFNpTAA | 1.1 ± 0.1 | 1.2 ± 0.2 | 870 ± 18 |
| 11 | YNpTDFIQF | ND | >0.5 | 500 ± 23 |
| 12 | YNpTFDIQF | ND | >0.5 | 300 ± 24 |
| 13 | YNpTFSDFI | ND | >1 | 350 ± 9 |
| 14 | YNpTFIADF | 0.8 ± 0.1 | 0.8 ± 0.1 | 1020 ± 42 |
| 15 | YNpTFLIVE | 2.3 ± 0.5 | 0.6 ± 0.2 | 3800 ± 450 |
| 16 | YEYQFNpTDFIQF | 0.5 ± 0.1 | 0.03 ± 0.01 | 16300 ± 6600 |
All peptides contained an acetyl group at the N-terminus and a C-terminal amide. ND, kcat and KM values could not be accurately determined due to limited peptide solubility.
Five representative sequences selected from the C-terminal library (library Ib), which each contain an acidic residue at a different position, were resynthesized and tested against VH1 (Table 2, peptides 11–15). The results show that as the acidic residue moves away from pT, there is a general increase in VH1 activity. This, together with the random distribution of the acidic residues, suggests that VH1 prefers hydrophobic residues C-terminal to pS/pT and the acidic residues were likely selected due to their ability to increase VH1 accessibility to the resin-bound peptides (see Discussion). Note that the most active C-terminal peptide (peptide 15, with a kcat/KM value of 3800 M−1 s−1) has significantly higher activity than the most reactive N-terminal peptide (peptide 8, with a kcat/KM value of 1100 M−1 s−1). This indicates that unlike most of the other PTPs, the primary specificity determinant of VH1 resides on the C-terminal side of the phosphoamino acid. Finally, peptides containing preferred sequences on both sides of pT are very efficient substrates (kcat/KM value of 1.6 × 104 M−1 s−1 for peptide 16).
Representative pY peptides selected from libraries III and IV, as well as a panel of artificially designed substrates were tested against VH1 in solution (Table 3, peptides 17–31). In keeping with the broad N-terminal specificity as revealed by library screening, all of the N-terminal peptides have similar kcat/KM values (102–103 M−1 s−1), despite their very different sequences (Table 3, peptides 17–23). Again, peptides selected from the C-terminal library are generally more reactive (kcat/KM values of 103 – 104 M−1 s−1 for peptides 24–25). Peptide 26 (YDEDFpYDYEF), which contains preferred sequences both N- and C-terminal to pY, is an excellent substrate for VH1, with a kcat value of 2.6 s−1, a KM value of 11 μM, and a kcat/KM value of 2.3 × 105 M−1 s−1. Consistent with the screening data, substitution of Ala for Tyr at position P+2 decreased VH1 activity by 10-fold, whereas replacement of other C-terminal residues with Ala had smaller effects (Table 3, compare peptides 27–30). Finally, the 500-fold difference in activity between peptides 26 and 31 confirmed that VH1 has an overall preference for acidic over basic residues.
Table 3.
Catalytic Activity of VH1 toward pY Peptides
| Peptide No | Peptide Sequencea | kcat/KM (M−1 s−1) |
|---|---|---|
| 17 | EDAFSpYAA | 3100 ± 400 |
| 18 | ADTDFpYAA | 2600 ± 100 |
| 19 | AQDFDpYAA | 1600 ± 200 |
| 20 | FVTDDpYAA | 900 ± 200 |
| 21 | AFGDSpYAA | 300 ± 100 |
| 22 | YSFDPpYAA | 3300 ± 100 |
| 23 | DTADVpYAA | 1200 ± 100 |
| 24 | AApYQWDSF | 16000 ± 1000 |
| 25 | AApYIDWTG | 4000 ± 100 |
| 26 | YDEDFpYDYEF | 230000 ± 4000 |
| 27 | YDEDFpYAYEF | 120000 ± 2000 |
| 28 | YDEDFpYDAEF | 24000 ± 1000 |
| 29 | YDEDFpYDYAF | 120000 ± 2000 |
| 30 | YDEDFpYDYEA | 260000 ± 7000 |
| 31 | YRKRFpYRYKF | 500 ± 100 |
All peptides contained an acetyl group at the N-terminus and a C-terminal amide.
VHR Has Intrinsically Low Activity toward pS/pT Peptides
We have previously shown that VHR recognizes distinct sequence motifs N-terminal to pY.15 It was also reported that VHR is very active toward pY peptides but has only weak activity toward a pT peptide derived from p38.22,28 To determine whether this is a general property of VHR, we screened VHR against peptide libraries I and II. Incubation of the libraries with high concentrations of VHR (up to 20 μM) for extended periods of time (2 h) resulted in no fluorescence on any of the library beads. This indicates that VHR has low intrinsic activity toward pS and pT peptides.
Database Search and In Vitro Dephosphorylation of Protein Substrates by VH1
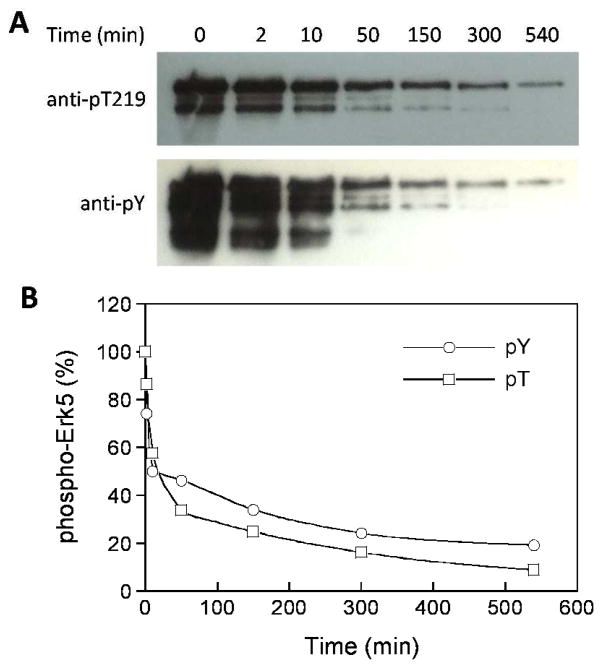
We searched the PhosphoSite database (http://www.phosphosite.org) against the sequence motif (F/Y)xx(pS/pT) (where x is any amino acid) and the resulting hits were manually filtered by removing any sequences that contained basic amino acids at P−1 to P−5 positions or ≥2 acidic residues at the P+1 to P+5 positions. This exercise resulted in 18 phosphoproteins as potential VH1 substrates (Table S6). Two of the proteins, extracellular signal-regulated kinase 5 (Erk5) and c-Jun N-terminal protein kinase 1 (Jnk1) were selected for testing because they contain sequences that closely match the VH1 class I substrate motifs (Erk5, HQYFMpT219EpYVAT; Jnk1, TSFMMpT183PpYVVT), are involved in multiple signaling pathways, and are commercially available in their phosphorylated forms. Thus, recombinant phospho-Erk5 was treated with GST-VH1 for varying periods of time and the reaction products were analyzed by SDS-PAGE and western blotting with antibodies specific for Erk5 pT219 and phosphotyrosine (Figure 4A). Based on the reaction progress curves (Figure 4B), we estimated kcat/KM values of 740 and 1410 M−1 s−1 for VH1-mediated dephosphorylation of pT219 and pY residues of Erk5, respectively. In comparison, synthetic peptides corresponding to the Erk5 motif, HQYFMpTEYVAT and HQYFMpTEpYVAT, were dephosphorylated by VH1 with kcat/KM values of 490 and 3200 M−1 s−1, respectively. Similar experiments with Jnk1 were unsuccessful because the commercially available Jnk1 could not be detected by either anti-Jnk1 pT183 or anti-pY (4G10) antibody.
Figure 4.
In vitro dephosphorylation of Erk5 (19 μM) by VH1 (1.5 μM). (A) Western blots showing the remaining pT219 and pY levels after different reaction times (0–540 min). (B) Reaction progress curves for VH1-mediated Erk5 dephosphorylation at pT and pY sites. The y axis values were derived from part A by phosphoimaging analysis and relative to that at reaction time 0 (100%).
DISCUSSION
VH1 is a prototypical DUSP in vaccinia virus and it plays an essential role in directing the transcription of early viral genes and controls viral infectivity.29 VHR was the first mammalian DUSP discovered and its role in modulating MAPKs has been well established.8,27 The specificity profile of VH1 revealed three interesting features. First, VH1 recognizes different sequences surrounding the pY vs pS/pT residue. For example, the preferred pY substrates are rich in acidic residues at the P−1 and P−2 positions, whereas its pS/pT substrates are devoid of Asp and Glu at these positions (Figure 2 and 3). On the other hand, the vast majority of preferred pS/pT substrates contain an acidic residue at position P−5 (or P−4), but the pY substrates have mostly hydrophobic residues at the positions. On the C-terminal side, the pS/pT substrates consist of very hydrophobic residues such as Phe, Ile, Leu, and Nle, whereas the pY substrates are rich in Trp and Tyr, which are capable of both hydrophobic interaction and hydrogen bonding. This is likely a common property of DUSPs, since several DUSPs have been shown to dephosphorylate both the pT and pY residues of the pTXpY motif in MAPKs.2,3 It remains to be determined how the same active site accommodates these very different sequences, but they may utilize different binding surfaces to recognize the pY vs pS/pT substrates.15
Second, VH1 has more stringent sequence specificity toward pS/pT than pY substrates, especially on the N-terminal side of the phosphoamino acid. This is evident from the kinetic data in Table 2 and 3; while pY peptides of different N-terminal sequences have similar kcat/KM values, alteration of a single residue in pT peptides resulted in large changes in kinetic activity (up to 20-fold). Presumably, pY, which has an extra hydrophobic phenyl group as compared to pS/pT, binds to the VH1 active-site pocket more tightly than the latter. As a result, binding of pY substrates to VH1 is energetically dominated by the pY residue and less dependent upon the surrounding amino acids. In contrast, productive interaction between VH1 and the amino acids adjacent to pS/pT is likely necessary to provide sufficient binding energy for pS/pT catalysis. In this regard, we note that several specific, high-affinity monoclonal antibodies have been developed against the pY residue, but no corresponding antibody against pS or pT is yet available. Instead, all of the specific anti-pS/pT antibodies recognize pS/pT in the context of a peptide sequence. Third, VH1 differs from other PTPs in that its specificity is primarily determined by residues C-terminal to the pS/pT/pY residue, although the N-terminal residues also make important contributions (especially for pS/pT substrates).
A main motivation for profiling the substrate specificity of DUSPs is to utilize the specificity information to predict their protein substrates. Based on the specificity profile of VH1, we predicted 18 pS/pT proteins as potential VH1 class I substrates (Table S6). We subsequently showed that one of these proteins, Erk5, is an in vitro substrate of VH1. Although the kcat/KM values of ~1000 M−1 s−1 are modest, this level of activity is theoretically sufficient to alter the phosphorylation status of a signaling protein in vivo. For example, if a vaccinia virus-infected cell produces 100 nM VH1 (which is the average cellular concentration of signaling proteins30), half of the intracellular Erk5 would be dephosphorylated in ~1.5 h. Additional studies will be necessary to test whether this indeed occurs in vivo. However, infection by other viruses can cause either up- or down-regulation of the Erk5 signaling pathway.31,32 VH1 has been reported to dephosphorylate vaccinia proteins A14 (at VIHTNHpS85DISMN)33 and A17 (at SLNTDDpY203- CO2H)33 and mammalian protein STAT1 (at GPKGTGpY701IKTEL)34 in vivo. According to our specificity profile, peptides corresponding to these pS/pY motifs are expected to be less efficient VH1 substrates. Analysis of the immunoblot data of Koksal and Cingolani35 using the method described in Figure 4 revealed a kcat/KM value of ~50 M−1 s−1 for VH1-mediated in vitro dephosphorylation of STAT1 at pY701. These data suggest a correlation between the catalytic efficiency of VH1 towards protein substrates and the corresponding peptide substrates. This in turn suggests that the amino acid sequence flanking pS/pT/pY is a key determinant of the kinetic activity of a protein substrate against VH1. Other factors, such as substrate recruiting via protein-protein interaction, may enhance the catalytic efficiency of VH1 in vivo. Proteins other than those listed in Table S6 (e.g., class II substrates) may also be VH1 substrates in vivo.
During the course of this work, we observed two factors that may potentially bias the library screening results. First, screening of the N-terminal pS/pT library gave overwhelmingly pT peptides. The higher activity of VH1 toward pT peptides (~2-fold) is clearly a factor. Another likely factor is the lower stability of pS (relative to pT), which may undergo β-elimination (to form dehydroalanine) and other side reactions during library synthesis and screening.36,37 As we have demonstrated with the C-terminal libraries, this problem is easily addressed by screening pS and pT libraries separately. Our data show that VH1 exhibits the same specificity profile for pS and pT peptides. Second, the kinetic data indicate that VH1 prefers hydrophobic amino acids at all positions C-terminal to pS/pT, and yet the selected sequences each contained at least one acidic residue. The random distribution of the acidic residue at positions P+1 to P+5 suggests that it unlikely reflects the specificity of VH1, but rather a bias of the on-bead screening procedure. This is because beads carrying very hydrophobic peptides undergo “hydrophobic collapse” in the aqueous reaction buffer (which is analogous to the protein folding process), rendering the peptides inaccessible to the PTP.13 Acidic residues increase aqueous solubility and prevent hydrophobic collapse due to repulsive interactions with each other and the negatively charged pS/pT residue. Such “solubility” effects can be easily identified by the random distribution of the acidic residues over several positions, whereas acidic residues specifically recognized by a PTP are likely found at a specific position(s). For example, the vast majority of the peptides selected from the N-terminal library contained an acidic residue(s) at the P−5 and/or P−4 positions, even though an acidic residue at these positions only enhanced the catalytic efficiency by 2-fold (Tables 1 and 2). Note that an optimal PTP substrate consisting of a highly hydrophobic sequence is unlikely to be physiologically relevant, as a protein containing such a sequence on its surface would not be stable. The “selection” of acidic residues by VH1 indicates that VH1 can tolerate acidic and other types of hydrophilic residues at some of the C-terminal positions.
In conclusion, we have developed a robust library screening method to determine the sequence specificity of DUSPs and other serine/threonine phosphatases. Together with our previous method for pY library screening,12 we can now systematically characterize the specificity of all protein phosphatases. Application of the methods to VH1 and VHR has already provided significant insight into their catalytic properties. The specificity data will be useful for identifying the protein substrates as well as developing robust assays for these enzymes.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (CA132855 and GM062820). R. L. was supported by a predoctoral fellowship from the Minister of Science and Technology, Thailand.
ABBREVIATIONS
- Alloc
allyloxycarbonyl
- DUSP
dual-specificity phosphatase
- Fmoc
(9-fluorenyl)methoxycarbonyl
- HBTU
N,N,N′,N′-tetramethyl-O-(1H-benzotriazol-1-yl)uronium hexafluorophosphate
- HOBt
1-Hydroxybenzotriazole
- HATU
N-[(Dimethylamino)-1H-1,2,3-triazolo-[4,5-b]pyridin-1- ylmethylene]-N-methylmethanaminium hexafluorophosphate N-oxide
- DIPEA
N,N-Diisopropylethylamine
- CLEAR
cross-linked ethoxylate acrylamide resin
- MAPK
mitogen-activated protein kinase
- NBDH
4-hydrazino-7-nitro-2,1,3-benzoxadiazole
- PED-MS
partial Edman degradation-mass spectrometry
- PEGA
polyethylene glycol polyacrylamide
- PTP
protein tyrosine phosphatase
- pS
phosphoserine
- pT
phosphothreonine
- pY
phosphotyrosine
- VHR
vaccinia VH1 related
Footnotes
Preferred VH1 Substrates Selected from library screening and predicted VH1 protein substrates. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Patterson KI, Brummer T, O’Brien PM, Daly RJ. Biochem J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 3.Bayon Y, Alonso A. Emerging Signaling Pathways in Tumor Biology. Transworld Research Network; Kerala, India: 2010. pp. 185–208. [Google Scholar]
- 4.Zhang ZY. Crit Rev Biochem Mol Biol. 1998;33:1–52. doi: 10.1080/10409239891204161. [DOI] [PubMed] [Google Scholar]
- 5.Guan KL, Broyles SS, Dixon JE. Nature. 1991;350:359–362. doi: 10.1038/350359a0. [DOI] [PubMed] [Google Scholar]
- 6.Yuvaniyama J, Denu JM, Dixon JE, Saper MA. Science. 1996;272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 7.Flint AJ, Tiganis T, Barford D, Tonks NK. Proc Natl Acad Sci USA. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todd JL, Tanner KG, Denu JM. J Biol Chem. 1999;274:13271–13280. doi: 10.1074/jbc.274.19.13271. [DOI] [PubMed] [Google Scholar]
- 9.Bloom J, Cristea IM, Procko AL, Lubkov V, Chait BT, Snyder M, Cross FR. J Biol Chem. 2011;285:5434–5445. doi: 10.1074/jbc.M110.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinney CM, Chandrasekharan UM, Yang L, Shen J, Kinter M, McDermott MS, DiCorleto PE. Am J Physiol Cell Physiol. 2009;296:C242–C249. doi: 10.1152/ajpcell.00492.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tonks NK, Neel BG. Curr Opin Cell Biol. 2001;13:182–195. doi: 10.1016/s0955-0674(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 12.Garaud M, Pei D. J Am Chem Soc. 2007;129:5366–5367. doi: 10.1021/ja071275i. [DOI] [PubMed] [Google Scholar]
- 13.Ren L, Chen X, Luechapanichkul R, Selner NG, Meyer TM, Wavreille A, Chan R, Iorio C, Zhou X, Neel BG, Pei D. Biochemistry. 2011;50:2339–2356. doi: 10.1021/bi1014453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Ren L, Kim S, Carpino N, Daniel JL, Kunapuli SP, Tsygankov AY, Pei D. J Biol Chem. 2010;285:31268–31276. doi: 10.1074/jbc.M110.114181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luechapanichkul R, Chen X, Taha HA, Vyas S, Guan X, Freitas MA, Hadad CM, Pei D. J Biol Chem. 2013;288:6498–6510. doi: 10.1074/jbc.M112.449611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei D, Wavreille AS. Mol Bio Syst. 2007;3:536–541. doi: 10.1039/b706041f. [DOI] [PubMed] [Google Scholar]
- 17.Donelladeana A, Krinks MH, Ruzzene M, Klee C, Pinna LA. Euro J Biochem. 1994;219:109–117. doi: 10.1111/j.1432-1033.1994.tb19920.x. [DOI] [PubMed] [Google Scholar]
- 18.Donelladeana A, Meyer HE, Pinna LA. Biochim Biophys Acta. 1991;1094:130–133. doi: 10.1016/0167-4889(91)90034-u. [DOI] [PubMed] [Google Scholar]
- 19.Donella Deana A, MacGowan CH, Cohen P, Marchiori F, Meyer HE, Pinna LA. Biochim Biophys Acta. 1990;1051:199–202. doi: 10.1016/0167-4889(90)90194-i. [DOI] [PubMed] [Google Scholar]
- 20.Kuznetsov VI, Hengge AC, Johnson SJ. Biochemistry. 2012;51:9869–9879. doi: 10.1021/bi300908y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gopishetty B, Ren L, Waller TM, Wavreille A, Lopez M, Thakkar A, Zhu J, Pei D. Org Lett. 2008;10:4605–4608. doi: 10.1021/ol801868a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denu JM, Zhou G, Wu L, Zhao R, Yuvaniyama J, Saper MA, Dixon JE. J Biol Chem. 1995;270:3796–3803. doi: 10.1074/jbc.270.8.3796. [DOI] [PubMed] [Google Scholar]
- 23.Thakkar A, Wavreille A, Pei D. Anal Chem. 2006;78:5935–5939. doi: 10.1021/ac0607414. [DOI] [PubMed] [Google Scholar]
- 24.Baykov AA, Evtushenko OA, Avaeva SM. Anal Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 25.Uzu S, Kanda S, Imai K, Nakashima K, Akiyama S. Analyst. 1990;115:1477–1482. [Google Scholar]
- 26.Todd JL, Rigas JD, Rafty LA, Denu JM. Oncogene. 2002;21:2573–2583. doi: 10.1038/sj.onc.1205344. [DOI] [PubMed] [Google Scholar]
- 27.Slack DN, Seternes OM, Gabrielsen M, Keyse SM. J Biol Chem. 2001;276:16491–16500. doi: 10.1074/jbc.M010966200. [DOI] [PubMed] [Google Scholar]
- 28.Schumacher MA, Todd JL, Rice AE, Tanner KG, Denu JM. Biochemistry. 2002;41:3009–3017. doi: 10.1021/bi015799l. [DOI] [PubMed] [Google Scholar]
- 29.Liu K, Lemon B, Traktman P. J Virol. 1995;69:7823–7834. doi: 10.1128/jvi.69.12.7823-7834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran U, Phillips R, Milo R. Cell. 2010;141:1262–1262. doi: 10.1016/j.cell.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 31.Katsarou K, Tsitoura P, Georgopoulou U. Biochim Biophys Acta. 2011;1813:1854–1862. doi: 10.1016/j.bbamcr.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 32.Ludwig S. Signal Transduction. 2007;7:81–88. [Google Scholar]
- 33.Derrien M, Punjabi A, Khanna M, Grubisha O, Traktman P. J Virol. 1999;73:7287–7296. doi: 10.1128/jvi.73.9.7287-7296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Najarro P, Traktman P, Lewis JA. J Virol. 2001;75:3185–3196. doi: 10.1128/JVI.75.7.3185-3196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koksal AC, Cingolani G. J Biol Chem. 2011;286:14373–14382. doi: 10.1074/jbc.M111.226357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacombe JM, Andriamanampisoa F, Pavia A. A Int J Pept Protein Res. 1990;36:275–280. doi: 10.1111/j.1399-3011.1990.tb00978.x. [DOI] [PubMed] [Google Scholar]
- 37.Paquet A, Johns M. Int J Pept Protein Res. 1990;36:97–103. doi: 10.1111/j.1399-3011.1990.tb00951.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.