Abstract
The FHIT gene is a tumor suppressor that is frequently inactivated by genomic alterations at chromosomal region 3p14.2. In the last few years, a considerable amount of data describing inactivation of FHIT in a variety of human malignancies and demonstrating the tumor suppressor potential of Fhit have been reported. Despite the demonstration that FHIT functions as a tumor suppressor, the pathway through which Fhit induces apoptosis and inhibits growth of cancer cells is not known. Our data demonstrate that Fhit is a target of tyrosine phosphorylation by the Src protein kinase. We show that Src phosphorylates Y114 of Fhit in vitro and in vivo, providing insight into a biochemical pathway involved in Fhit signaling.
The human FHIT gene at chromosome 3p14.2, spanning the constitutive chromosomal fragile site FRA3B, is often rearranged in the most common forms of cancer and is a tumor suppressor gene (1). The gene is ≈1.6 megabase in size and encodes a mRNA of 1.1 kb and a protein of 147 aa (1, 2). Expression studies have demonstrated that FHIT is a major target of chromosomal aberrations involving the short arm of human chromosome 3 and is inactivated in the majority of the most common human malignancies, including lung, head and neck, stomach, esophageal, kidney, and breast cancers (3-6). Over the last several years the evidence supporting a tumor suppressor function of FHIT has accumulated. For example, Ishii et al. treated esophageal cancer cell lines with an adenoviral vector expressing Fhit (7); adenovirus-mediated FHIT transduction caused suppression of cell growth in three cell lines. Two of these three cell lines exhibited caspase-dependent apoptosis, whereas the third cell line showed growth arrest and the accumulation of cells in G2-M and S phases (7). Another study evaluated effects of re-expression of Fhit in Fhit-negative pancreatic cancer cell lines by using both adenoviral and adeno-associated viral vectors (8). As in the previous study, Fhit re-expression resulted in growth inhibition and apoptosis. The final proof of the tumor suppressor function of FHIT is the formation of spontaneous tumors in a homozygous (-/-) knockout mouse. Our recent study evaluated the phenotype of Fhit -/- animals (9). We observed a much higher incidence of tumors in Fhit -/- and +/- mice than in wild type (WT) mice over a two-year period. These spontaneous tumors included lymphomas, sebaceous tumors, liver tumors, gastrointestinal tumors, and others. To determine the role of Fhit in carcinogen-induced tumors we treated heterozygous (+/-) Fhit deficient mice with the established gastric carcinogen N-nitrosomethylbenzylamine (NMBA) (10). Ten weeks after NMBA treatment, 100% of Fhit +/- mice had developed gastric tumors, including squamous papillomas, adenomas, and invasive carcinomas. In comparison, only 25% of WT mice developed tumors (10).
Despite the demonstration of the tumor suppressor potential of Fhit, the pathway through which Fhit induces apoptosis in cancer cells is still not known, although Siprashvili et al. (11) showed that the Ap3A hydrolase activity of Fhit was not required for its tumor suppressor activity. In another report, Chaudhuri et al. studied the interaction between Fhit and tubulin in vitro and found that Fhit is able to bind specifically to tubulin without causing nucleation or formation of microtubules and to promote assembly of microtubules more efficiently than did microtubule-associated proteins alone (12). Nevertheless, the physiological significance of these observations is unknown. Recently, the physical interaction between Fhit and human ubiquitin-conjugating enzyme 9 (hUBC9) has been reported (13). Because yeast UBC9 is involved in the regulation of M- and S-phase cyclins, these findings suggest that Fhit may play a role in cell cycle control through this interaction. The exact role Fhit in this pathway needs to be further clarified. Here we report that Fhit is a target of tyrosine phosphorylation by Src protein kinase. We show that Src phosphorylates Y114 of Fhit in vitro and in vivo and therefore provide important clues to biochemical mechanisms involved in Fhit signaling.
Materials and Methods
Materials. Antibodies used were anti-phospho-tyrosine RC20:HRPO antibody (Becton Dickinson), rabbit polyclonal anti-Fhit (Zymed), and mouse monoclonal anti-Src clone GD11 (Upstate Biotechnology, Lake Placid, NY). The MonoQ FPLC column was purchased from Amersham Pharmacia. Sypro Ruby gel stain was from Molecular Probes. Endoproteinase Glu-C (V8 protease) and trypsin were from Roche Molecular Biochemicals and Promega, respectively.
Purification of Different Forms of Fhit. E. coli SG100 transformed with pSGA02-FHIT was used to express human Fhit, and Fhit was purified as described (14). After purified Fhit was incubated in vitro in the absence or presence of Src kinase, we exchanged the reaction solutions into 1,3-bis[tris(hydroxymethyl)methylamino]propane (BTP), pH 6.8, buffer and subjected the solutions to anion-exchange chromatography on a MonoQ column using an Akta FPLC (Amersham Pharmacia) system at a flow rate of 1 ml/min at ≈25°C. Unphosphorylated, monophosphorylated, and diphosphorylated forms of Fhit were separated by using a linear gradient of 0-0.2 M NaCl in BTP, pH 6.8. Fractions corresponding to the putative different forms of Fhit were pooled and dialyzed into 50 mM Hepes, pH 6.8.
In Vitro Kinase Assay. 10-500 units of purified Src kinase or Lyn kinase (Biomol, Plymouth Meeting, PA) were incubated with 1 μg-1 mg of purified Fhit and 0.25 mM ATP in kinase buffer (50 mM Tris·HCl, pH 7.5/2 mM DTT/1 mM EGTA/0.01% Brij 35/10 mM MgCl2) for 0 or 30 min at 30°C. Fhit was also incubated in the absence of Src kinase as control. Phosphorylated proteins were separated by SDS/PAGE and analyzed by Western blot. Kinase reaction solutions were also subjected to chromatography as described above.
Cell Culture. Human embryonic kidney 293 and NIH-3T3 cells were grown in DMEM supplemented with 10% FBS and gentamicin (GIBCO/BRL, Invitrogen). Cells were maintained at 37°C in a water-saturated atmosphere of 5% CO2 in air.
Plasmid Constructs, Transient Transfections, Immunoprecipitations, and Immunoblot Analysis. WT Fhit, activated Src, and dominant negative (DN)-Src mutant plasmids were purchased from Upstate Biotechnology. FhitY114F, FhitY145F and FhitY114F, Y145F mutants were created in the same vector as WT Fhit. Transient transfections were carried out by using FuGENE 6 transfection reagent according to the manufacturer's instructions (Roche Molecular Biochemicals). Cells were lysed by using Nonidet P-40 lysis buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 10% glycerol, 0.5% Nonidet P-40, and protease inhibitors. Immunoprecipitations were carried out overnight in the same buffer using 0.5 mg of protein, 5 μg of antibody, and 40 μl of protein A/G PLUS agarose (Santa Cruz Biotechnology) and were washed four times with the same buffer containing 0.1% Nonidet P-40 and 0.1% SDS. Western blotting was performed under standard conditions by using 12% Tris-glycine PAGE gels.
Mass Spectrometry. Matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectra were acquired on an Applied Biosystems Voyager-Elite using a matrix of sinapinic acid (saturated solution in 50% acetonitrile containing 0.1% trifluoroacetic acid). Dialyzed MonoQ column fractions were subjected to reversed-phase chromatography on a C18 matrix (ZipTips from Millipore) and eluted with 50% acetonitrile-0.1% trifluoroacetic acid (TFA). One microliter of eluate was spotted on the MALDI target, overlayed with matrix, and allowed to dry at room temperature. Each MALDI-TOF mass spectrum represents the average of 64 laser shots acquired in linear mode over a mass range of m/z 4,000-28,000, by using delayed extraction and positive ion detection. Spectra were processed by the smoothing and noise reduction protocols of the data system software. Close external calibration based on myoglobin was used.
HPLC-electrospray ionization (ESI) tandem mass spectra were acquired on a Thermo Finnigan LCQ Classic ion trap mass spectrometer connected via a home-built microspray interface to a Michrom BioResources MAGIC 2002 micro HPLC. For on-line HPLC separations, a New Objective PicoFrit was packed to 10 cm with Vydac 218MSB5 (5 μm, 300 Å) C18 reversed-phase matrix. HPLC conditions were as follows: mobile phase A, 0.5% acetic acid/0.005% TFA; mobile phase B, 90% acetonitrile/0.5% acetic acid/0.005% TFA; gradient, 2% B to 72% B in 30 min; initial flow rate, ≈0.4 μl/min. ESI/tandem MS (MS/MS) conditions were: electrospray voltage, 3 kV; heated capillary, 175°C; isolation window for collision-induced dissociation (CID), 2.5; relative collision energy, 35%; spectral acquisition, centroid mode. For initial on-line HPLC-ESI/MS/MS experiments, each scan segment consisted of a survey scan followed by acquisition of CID mass spectra of the four most abundant ions in the survey scan above a selected threshold. Subsequent HPLC-ESI/MS/MS analyses used a targeted scan strategy in which the specific m/z values for Fhit proteolytic peptides of interest were monitored.
Diphosphorylated Fhit was subjected to proteolysis by trypsin and by endoproteinase Glu-C for 16-18 h at ≈25°C in 40 mM ammonium bicarbonate, pH 7.6, and in 25 mM ammonium acetate, pH 4.0, respectively. The resulting peptides were analyzed by ESI mass spectrometry, as described above, to identify the specific residue(s) phosphorylated.
Results
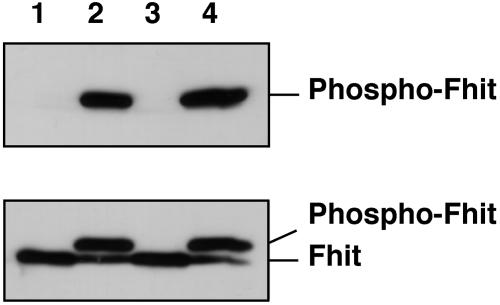
Src Phosphorylates Fhit in Vitro. To investigate whether Fhit could be a target of phosphorylation or other posttranslational modifications we analyzed its primary sequence. Src kinase has an optimal substrate sequence of (D/E)xIY(G/E)EF (15). Fhit contains two tyrosine residues, Y114 and Y145 (1). The sequence DSIY114EEL containing Y114 is a possible substrate sequence for Src kinase. The context of Y145 is not favorable for Src kinase. Therefore, we hypothesized that tyrosine 114 of Fhit could be a potential target of phosphorylation by Src kinase or its family members. To test this possibility we carried out in vitro phosphorylation reactions. Results are shown in Fig. 1. Recombinant Fhit was incubated with recombinant Src (lanes 1 and 2) or Lyn, a member of the Src family of tyrosine kinases (lanes 3 and 4), and immunoblotted with anti-phosphotyrosine (Fig. 1 Upper) or anti-Fhit (Fig. 1 Lower) antibodies. Significant phosphorylation of Fhit by both c-Src and Lyn (Fig. 1 Upper) was observed, and phosphorylation resulted in a significant shift of the Fhit protein position on the gel (Fig. 1 Lower).
Fig. 1.
Src phosphorylates Fhit in vitro. One microgram of recombinant Fhit was incubated with 10 units of recombinant Src (lanes 1 and 2) or Lyn (lanes 3 and 4) at 30°C and immunoblotted with anti-phosphotyrosine (Upper) or anti-Fhit (Lower) antibodies. Incubations were carried out for 0 (lanes 1 and 3) or 30 (lanes 2 and 4) min.
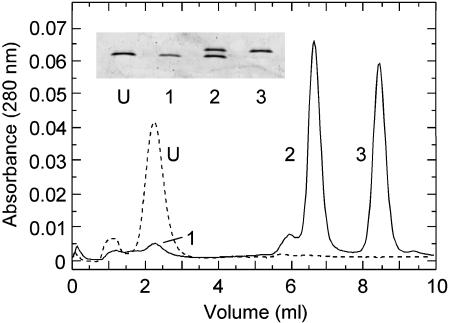
Because Fhit behaves as a homodimer in solution, with two tyrosine residues per monomer (16), 9 different forms, ranging from unphosphorylated to quadruply phosphorylated, could potentially occur on in vitro phosphorylation. Thus, the in vitro phosphorylated Fhit was run on a Mono Q column, as described in Materials and Methods, and three, well-separated protein fractions were obtained (Fig. 2). Isolation of only three forms indicated that only one residue per monomer was phosphorylated. Fhit incubated without Src kinase eluted at ≈2.3 min (Fig. 2, dashed line). Fhit incubated with Src kinase eluted as a minor form at 2.3 min. and two major forms at 6.7 min and 8.5 min (Fig. 2, solid line). The inset in Fig. 2 shows the SDS gel pattern of the MonoQ column fractions. We interpret fractions 1, 2, and 3 to be Fhit that is unphosphorylated, phosphorylated on one subunit, and phosphorylated on both subunits, respectively. The amount of residual, unphosphorylated Fhit in three independent, in vitro phosphorylation reactions was less than 10% of the two major phosphorylated forms, indicating that in vitro phosphorylation by Src kinase is highly efficient.
Fig. 2.
Chromatographic separation of unphosphorylated and phosphorylated forms of Fhit. Dashed line, the MonoQ column elution profile of Fhit incubated in buffer with ATP and without Src kinase; solid line, the MonoQ column elution profile of Fhit incubated in the presence of ATP and Src kinase. (Inset) SDS gel electrophoresis pattern of Fhit samples corresponding to the indicated column peaks: U, unphosphorylated Fhit incubated without Src kinase; 1, residual, unphosphorylated Fhit after incubation with Src kinase; 2, Fhit phosphorylated on one subunit; and 3, Fhit phosphorylated on both subunits. Approximately 0.2 μg of each sample was subjected to electrophoresis, and the gel was stained with Sypro Ruby.
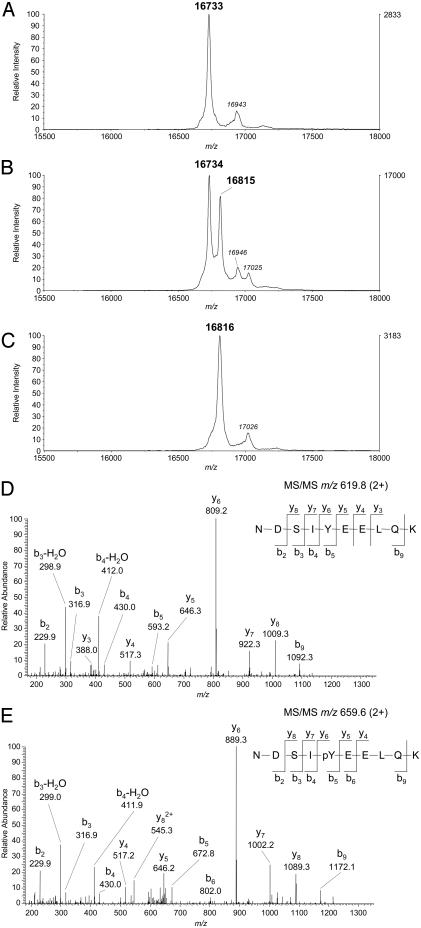
To confirm the identity of the putative forms of Fhit and to identify the phosphorylated residue(s), MonoQ column fractions were analyzed by mass spectrometry. MALDI-TOF analysis showed that fraction 1 had a mass of 16,733 Da, the mass expected for unphosphorylated Fhit monomer with the N-terminal methionine removed (Fig. 3A). Fraction 2 exhibited two peaks, one with a mass of 16,734 Da and one with a mass of 16,815 Da. The mass increment of +81 Da indicates that one residue is phosphorylated (Fig. 3B). Thus, fraction 2 was Fhit phosphorylated on only one subunit in the homodimer. Fraction 3 exhibited a single peak with a mass of 16,816 Da (Fig. 3C). Thus, fraction 3 was Fhit phosphorylated on both subunits of the homodimer with only one residue phosphorylated in each subunit.
Fig. 3.
Mass spectral analysis of Fhit. (A-C) MALDI-TOF spectra of unphosphorylated, monophosphorylated, and diphosphorylated forms of Fhit, respectively, were acquired on an Applied Biosystems Voyager Elite as described in Materials and Methods. The m/z values shown in bold represent the average of three spectra obtained from the same sample spot on the MALDI target. The lower intensity peaks in each panel represent matrix adducts. (D and E) ESI tandem mass spectra were acquired on a Thermo Finnigan LCQ Classic spectrometer as described in Materials and Methods. (D) Tandem mass spectrum of the 2+ ion (m/z 619.8) of tryptic peptide 109-118 from unphosphorylated Fhit. (E) Tandem mass spectrum from the 2+ ion (m/z 659.6) of tryptic peptide 109-118 from phosphorylated Fhit. Spectra were obtained by targeted scans of the indicated ions. Peptide fragments are denoted on the spectra using the nomenclature of Roepstorff and Fohlman (23). Identification of the site of phosphorylation was obtained by comparison of the y-series ions in the two spectra.
To identify which tyrosine is phosphorylated, a trypsin digest of fraction 3 was analyzed by HPLC-ion-trap mass spectrometry. Only peptide 110-119 was phosphorylated, and the fragmentation pattern of the peptide was compatible only with Y114 being phosphorylated and not S112 (Fig. 3E). There was a small amount of nonphosphorylated peptide 110-119, indicating that some dephosphorylation probably occurred during proteolysis, purification, and analysis (Fig. 3D). The tryptic peptide containing Y145 is composed of only four residues, which is too small for adequate recovery. Analysis of Y145 was achieved by endoproteinase Glu-C digestion. No phosphorylated form of peptide 138-146 was detected. The only phosphorylated peptide detected was 87-114 containing Y114 (data not shown). Thus, the mass spectrometry data demonstrated that fraction 3 is Fhit phosphorylated on Y114 on both subunits.
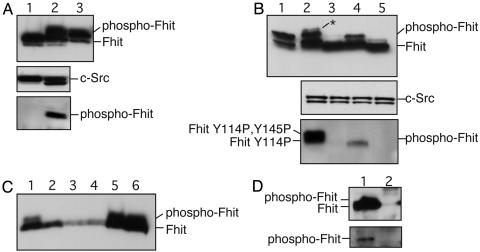
Src Phosphorylates Fhit in Vivo. Because Fhit was phosphorylated by Src in vitro, the next step was to determine whether phosphorylation also occurs in vivo. For this determination, 293 cells were cotransfected with FHIT and activated SRC expression constructs or with FHIT and a dominant negative mutant of SRC (SRCDN). Subsequent Western blot analysis demonstrated that activated Src but not SrcDN phosphorylated Fhit (Fig. 4A Top, lanes 2 versus 1). The same lysates were immunoprecipitated with anti-Fhit antibody and immunoblotted with anti-phosphotyrosine (Fig. 4A Bottom). Tyrosine phosphorylated Fhit was detected in lane 2, confirming phosphorylation by activated Src.
Fig. 4.
Src phosphorylates Fhit in vivo. (A) 293 cells were cotransfected with WT FHIT and SRCDN (lane 1) or activated SRC (lane 2) and immunoblotted with anti-Fhit (Top), anti-Src (Middle), or anti-phosphotyrosine (Bottom) antibodies. Lysates were immunoprecipitated with anti-Fhit before Western blotting with anti-phosphotyrosine. Lane 3, an aliquot of in vitro phosphorylated Fhit was loaded for comparison. (B) 293 cells were cotransfected with SRCDN and WT FHIT (lane 2), FHITY114F (lane 3), FHITY145F (lane 4), and FHITY114F, Y145F (lane 5). Lysates were immunoblotted as in A. Lane 1, an aliquot of in vitro phosphorylated Fhit was loaded for comparison. (C) Western blot analysis of normal human tissues with anti-Fhit antibody; 100 μg of each lysate were loaded in each lane: lane 1, fetal liver; lane 2, fetal brain; lane 3, fetal lung; lane 4, lung; lane 5, fetal kidney; and lane 6, kidney. (D) One milligram of normal human kidney lysate was immunoprecipitated with anti-Fhit antibody (lane 1) or with rabbit IgG (lane 2) and immunoblotted with anti-Fhit (Upper) or with anti-phosphotyrosine (Lower).
To determine which tyrosine is phosphorylated in vivo by Src, several Fhit mutants were created, FhitY114F, FhitY145F and FhitY114F, Y145F, individually cotransfected with the activated SRC construct, and checked for tyrosine phosphorylation by Western blotting. Phosphorylated Fhit was detected on expression of WT Fhit or FhitY145F (Fig. 4B Top, lanes 2 and 4) but not FhitY114F or FhitY114F, Y145F (Fig. 4B Top, lanes 3 and 5). The tyrosine phosphorylation was confirmed by using anti-phosphotyrosine antibody (Fig. 4B Bottom). These results confirmed that Y114 of Fhit is a target of Src phosphorylation in vivo.
Interestingly, Western blot analysis of exogenous phosphorylated Fhit in 293 cells revealed the presence of a second phospho-Fhit band (Fig. 4B Top, lane 2, marked as *). This band was detected on expression of WT Fhit but not FhitY145F and appeared less intense than the major phospho-Fhit band. We concluded that this band represents double phosphorylated Fhit at residues Y114 and Y145. This conclusion is supported by the fact that this weaker band, as detected by anti-Fhit antibody (Fig. 4B Top, lane 2), had the same intensity as the FhitY114 band when detected by anti-phosphotyrosine antibody (Fig. 4B Bottom, lane 2). Because Y145 is not phosphorylated by Src in vitro and FhitY114F is not phosphorylated by Src in 293 cells (Fig. 4B, lane 3), we conclude that Y145 is phosphorylated by a tyrosine kinase other than Src upon Y114 phosphorylation. To determine whether phosphorylation of Fhit by Src would affect the intracellular localization of either protein, we cotransfected NIH-3T3 cells with WT FHIT and activated SRC and carried out immunofluorescence experiments. Coexpression of both proteins in NIH-3T3 cells did not change their generally cytoplasmic localization (data not shown).
To evaluate whether Y114 phosphorylation is physiologically relevant we analyzed cell lysates of normal and tumor tissues for the presence of phosphorylated Fhit. Normal human kidney, fetal kidney, and fetal liver exhibited significant phospho-Fhit (Fig. 4C), although several other human tissues did not show expression of phospho-Fhit (Fig. 4C). To confirm that the upper band on Fig. 4C is indeed tyrosine-phosphorylated Fhit, we immunoprecipitated Fhit from normal human kidney lysate and checked for tyrosine phosphorylation by Western blotting (Fig. 4D). Tyrosine-phosphorylated Fhit was detected on lane 1 (Fig. 4D Lower). We were unable to detect endogenous phospho-Fhit in any of the human tumor cell lines we examined (data not shown).
Discussion
The data clearly show that Fhit is phosphorylated on tyrosine 114 by Src kinase both in vitro and in vivo. One possibility is that phosphorylation results in a gain of function of Fhit with respect to stimulation of apoptosis. If this hypothesis is correct, one would predict that phospho-Fhit would be higher in normal tissues during normal waves of apoptosis than in tumor tissue, and that phospho-Fhit levels would decrease as a function of tumor progression. The data shown in Fig. 4 demonstrating the presence of phospho-Fhit in some normal tissues generally support the first part of this prediction.
We propose two possible modes of action for phospho-Fhit as the active form. The tumor suppressor activity of Fhit is independent of the hydrolysis of Ap3A (11), and tumor suppression with a FhitH96N mutant led to the proposal that a Fhit-Ap3A complex is the active form in tumor suppression (16). A recent study with FhitH96N, H96D and other site-directed mutants demonstrated an inverse correlation between the value of the Km for Ap3A and the induction of apoptosis, thereby supporting the proposal that substrate binding was a critical factor (17). However, such a complex presumably must have a sufficiently slow turnover for it to interact with a target protein, as noted (18). Potentially, phosphorylation of Y114 may function by trapping a substrate-bound complex and increasing its lifetime. If phospho-Fhit-Ap3A is the active complex, phosphorylated Fhit should exhibit a markedly decreased kcat for Ap3A hydrolysis in comparison to unphosphorylated Fhit. Alternatively, phosphorylated Fhit may interact directly with target proteins independently of bound Ap3A. We have been unsuccessful in identifying target protein(s) of Fhit despite repeated attempts using yeast two-hybrid, immunoprecipitation, and immunoaffinity techniques (unpublished results). The inability to identify target proteins may be due to Fhit not being in the requisite phosphorylated form for formation of interactive complexes.
The role of Src kinase in apoptosis is not well defined. For example, recent reports demonstrate that Src activation is associated with the induction of apoptosis in thymocytes and 293 cells (19, 20), whereas in fibroblasts and gall bladder epithelial cells Src exhibits anti-apoptotic effects (21, 22). Therefore, an alternative hypothesis is that Src inhibits Fhit by phosphorylation, with phosphorylation of Fhit leading to loss of function as a tumor suppressor and as apoptosis inducer. If this hypothesis is correct, a mutant that cannot be phosphorylated at Y114 (such as Y114F) should be a better inducer of apoptosis and a better tumor suppressor than WT Fhit.
Acknowledgments
We thank Dr. Charles Brenner for his critical reading of the manuscript, Angela K. Robinson and Jean Letofsky for expert technical assistance, and Dr. Susan Weintraub and Chris Carroll from the Center for Mass Spectrometry at the University of Texas Health Science Center for their assistance and expertise. The Center for Mass Spectrometry is supported by National Cancer Institute (NCI) Grant P30 CA54174 through the San Antonio Cancer Institute. This work was supported by grants from the National Science Foundation (MCB-9982645, to L.D.B.) and NCI (CA77738 and CA56036, to C.M.C. and K.H.) and by a Kimmel Scholar Award (to Y.P.).
Abbreviations: MALDI-TOF, matrix-assisted laser desorption/ionization-time-of-flight; ESI, electrospray ionization.
References
- 1.Ohta, M., Inoue, H., Cotticelli, M. G., Kastury, K., Baffa, R., Palazzo, J., Siprashvili, Z., Mori, M., McCue, P., Druck, T. et al. (1996) Cell 84, 587-597. [DOI] [PubMed] [Google Scholar]
- 2.Matsuyama, A., Shiraishi, T., Trapasso, F., Kuroki, T., Alder, H., Mori, M., Huebner, K. & Croce, C. (2003) Proc. Natl. Acad. Sci. USA (2003) 100, 14988-14993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sozzi, G., Tornielli, S., Tagliabue, E., Sard, L., Pezzella, F., Pastorino, U., Minoletti, F., Pilotti, S., Ratcliffe, C., Veronese, M. L., et al. (1997) Cancer Res. 57, 5207-5712. [PubMed] [Google Scholar]
- 4.Baffa, R., Veronese, M. L., Santoro, R., Mandes, B., Palazzo, J. P., Rugge, M., Santoro, E., Croce, C. M. & Huebner, K. (1998) Cancer Res. 58, 4708-4714. [PubMed] [Google Scholar]
- 5.Campiglio, M., Pekarsky, Y., Menard, S., Tagliabue, E., Pilotti, S. & Croce, C. M. (1999) Cancer Res. 59, 3866-3869. [PubMed] [Google Scholar]
- 6.Druck, T., Hadaczek, P., Fu, T. B., Ohta, M., Siprashvili, Z., Baffa, R., Negrini, M., Kastury, K., Veronese, M. L., Rosen, D., et al. (1997) Cancer Res. 57, 504-512. [PubMed] [Google Scholar]
- 7.Ishii, H., Dumon, K. R., Vecchione, A., Trapasso, F., Mimori, K., Alder, H., Mori, M., Sozzi, G., Baffa, R., Huebner, K. & Croce, C. M. (2001) Cancer Res. 61, 1578-1584. [PubMed] [Google Scholar]
- 8.Dumon, K. R., Ishii, H., Vecchione, A., Trapasso, F., Baldassarre, G., Chakrani, F., Druck, T., Rosato, E. F., Williams, N. N., Baffa, R., et al. (2001) Cancer Res. 61, 4827-4836. [PubMed] [Google Scholar]
- 9.Zanesi, N., Fidanza, V., Fong, L. Y., Mancini, R., Druck, T., Valtieri, M., Rudiger, T., McCue, P. A., Croce, C. M. & Huebner, K. (2001) Proc. Natl. Acad. Sci. USA 98, 10250-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong, L. Y., Fidanza, V., Zanesi, N., Lock, L. F., Siracusa, L. D., Mancini, R., Siprashvili, Z., Ottey, M., Martin, S. E., Druck, T., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 4742-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siprashvili, Z., Sozzi, G., Barnes, L. D., McCue, P., Robinson, A. K., Eryomin, V., Sard, L., Tagliabue, E., Greco, A., Fusetti, L., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 13771-13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhuri, A. R., Khan, I. A., Prasad, V., Robinson, A. K., Luduena, R. F. & Barnes, L. D. (1999) J. Biol. Chem. 274, 24378-24382. [DOI] [PubMed] [Google Scholar]
- 13.Shi, Y., Zou, M., Farid, N. R. & Paterson, M. C. (2000) Biochem. J. 352 Pt 2, 443-448. [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner, C., Pace, H. C., Garrison, P. N., Robinson, A. K., Rosler, A., Liu, X. H., Blackburn, G. M., Croce, C. M., Huebner, K. & Barnes, L. D. (1997) Protein Eng. 10, 1461-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Songyang, Z. & Cantley, L. C. (1995) Trends Biochem. Sci. 20, 470-475. [DOI] [PubMed] [Google Scholar]
- 16.Pace, H. C., Garrison, P. N., Robinson, A. K., Barnes, L. D., Draganescu, A., Rosler, A., Blackburn, G. M., Siprashvili, Z., Croce, C. M., Huebner, K. & Brenner, C. (1998) Proc. Natl. Acad. Sci. USA 95, 5484-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapasso, F., Krakowiak, A., Cesari, R., Arkles, J., Yendamuri, S., Ishii, H., Vecchione, A., Kuroki, T., Bieganowski, P., Pace, H. C., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 1592-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy, G. A., Halliday, D. & McLennan, A. G. (2000) Cancer Res. 60, 2342-2344. [PubMed] [Google Scholar]
- 19.Marchetti, M. C., Di Marco, B., Cifone, G., Migliorati, G. & Riccardi, C. (2003) Blood 101, 585-593. [DOI] [PubMed] [Google Scholar]
- 20.Lavoie, J. N., Champagne, C., Gingras, M. C. & Robert, A. (2000) J. Cell Biol. 150, 1037-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, D., Agochiya, M., Samejima, K., Earnshaw, W., Frame, M. & Wyke, J. (2000) Cell Death Differ. 7, 685-696. [DOI] [PubMed] [Google Scholar]
- 22.Boudny, V. & Nakano, S. (2002) Br. J. Cancer 86, 463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roepstorff, P. & Fohlman, J. (1984) Biomed. Mass Spectrom. 11, 601 (lett.). [DOI] [PubMed] [Google Scholar]