Abstract
Several microbial systems have been shown to yield advantageous mutations in slowly growing or nongrowing cultures. In one assay system, the stationary-phase mutation mechanism differs from growth-dependent mutation, demonstrating that the two are different processes. This system assays reversion of a lac frameshift allele on an F′ plasmid in Escherichia coli. The stationary-phase mutation mechanism at lac requires recombination proteins of the RecBCD double-strand-break repair system and the inducible error-prone DNA polymerase IV, and the mutations are mostly −1 deletions in small mononucleotide repeats. This mutation mechanism is proposed to occur by DNA polymerase errors made during replication primed by recombinational double-strand-break repair. It has been suggested that this mechanism is confined to the F plasmid. However, the cells that acquire the adaptive mutations show hypermutation of unrelated chromosomal genes, suggesting that chromosomal sites also might experience recombination protein-dependent stationary-phase mutation. Here we test directly whether the stationary-phase mutations in the bacterial chromosome also occur via a recombination protein- and pol IV-dependent mechanism. We describe an assay for chromosomal mutation in cells carrying the F′ lac. We show that the chromosomal mutation is recombination protein- and pol IV-dependent and also is associated with general hypermutation. The data indicate that, at least in these male cells, recombination protein-dependent stationary-phase mutation is a mechanism of general inducible genetic change capable of affecting genes in the bacterial chromosome.
Keywords: Escherichia coli, adaptive mutation, SOS response, DNA repair
Adaptive (or stationary-phase) mutation is a collection of phenomena in which mutations occur in populations of stressed, nongrowing, or slowly growing cells, and at least some of these mutations allow growth (reviewed by refs. 1–4). Stationary-phase mutation mechanisms may be important in development of antibiotic resistance mutations (5), phase variation in bacterial pathogens (3), and colonization of new bacterial hosts (6). Stationary-phase mutation mechanisms also may provide models for mutational escape of growth control, such as in oncogenesis, tumor progression, and resistance to chemotherapeutic drugs (7), and imply that genetic changes that fuel evolution may be accelerated during stress. Adaptive mutational processes contrast with the spontaneous mutation paradigm of Luria and Delbrück (8) in which mutations arise in growing cells, before cells are exposed to a selective environment, and more or less randomly in genomes. [These are the only mutations observed when the selection for mutants is lethal (reviewed in refs. 1 and 3).] It has been important to understand whether adaptive mutational processes represent departures in mechanism from spontaneous growth-dependent mutational processes, or whether they are merely growth-dependent mutations occurring in cell populations in which growth is difficult to measure. Stationary-phase mutations have been demonstrated to form via mechanisms unlike mutation in growing cells (and so, demonstrably, to be different processes) in only three experimental systems: (i) an assay that measures transposon-mediated deletions in Escherichia coli (9–11), (ii) an assay for substitution mutations in old E. coli colonies (12, 13), and (iii) the lac frameshift reversion assay in E. coli (14). The lac system measures reversion of a lac +1 frameshift mutation carried on an F′ conjugative plasmid in E. coli cells starved on lactose medium (14). The stationary-phase mutation mechanism at work in the lac system is the best characterized of any stationary-phase mutation mechanism and is the focus of this report.
The stationary-phase mutations at lac can be distinguished from growth-dependent Lac+ reversions as follows. The stationary-phase mutations occur in Lac− starving cells after exposure to lactose medium (15) at high frequency, accumulating over time (14). Unlike growth-dependent mutations, these require homologous recombination and double-strand break repair (DSBR) proteins RecA, RecBC, and RuvA, RuvB, and RuvC (16–18), implicating double-strand DNA breaks or double-strand ends as intermediates in mutation. F′ transfer functions, but not actual transfer, are required (19–21), suggesting that some aspect of the transfer process promotes mutation. An intact SOS response to DNA damage also is required for efficient stationary-phase mutation (14, 22), most of which requires the SOS error-prone DNA polymerase IV, encoded by dinB (23). The major replicative polymerase, DNA pol III, also has been implicated (23–25). The adaptive mutations are proposed to result from DNA polymerase errors accrued during replication primed by DSBR recombination (16) (although other models are possible; ref. 3 and discussed below). Finally, the cells that become Lac+, but not their similarly starved Lac− neighbors, carry high frequencies of additional, unselected mutations (26–29), but are not heritably mutator (26, 27, 30, 31). This finding suggests that some or all of the adaptive mutants arise in a transiently hypermutable subpopulation of cells (as proposed originally for recombination-independent stationary-phase mutations; refs. 32 and 33, and see refs. 29, 34, and 35, and below for further discussion regarding the Lac system). The additional mutations occur in all replicons of the cell, including the bacterial chromosome (26–29).
One minimalist model for Lac+ stationary-phase mutation (reviewed by refs. 2 and 3) follows: in a subpopulation of cells, double-strand breaks (DSBs) are formed. These are processed by the RecBCD enzyme, which begins recombinational DSBR. Recombination intermediates are used to prime DNA synthesis by using DNA pol IV and pol III, and errors in that synthesis can persist as mutations. This is a model with direct association of the action of the recombination proteins and the formation of mutations (recombination and mutation occur next to each other in the same piece of DNA). Indirect models in which action of the recombination proteins at one site promotes mutations elsewhere in the genome are also tenable (ref. 3 and below). For both direct and indirect models, a mechanism different from spontaneous mutation in growing cells generates the stationary-phase mutations: one specifically requiring recombination proteins and pol IV.
An important unanswered question regarding this novel, recombination protein-dependent mutation mechanism concerns the role(s) of the F′ conjugative plasmid. This question can be divided into two: are F′s (or other conjugative plasmids) required for the occurrence of the recombination protein-dependent mutation mechanism; and, in cells carrying the F′, do chromosomal sites experience recombination protein-dependent mutation? In this report we address the second question. Recombination-dependent mutation has been suggested to be confined to F′ plasmid DNA because of the following: first, the chromosomal lac operon in E. coli does not appear to undergo recombination-dependent mutation in cells lacking an F′ (20, 37); second, F′ transfer functions are required (although actual transfer is not) (19–21). Arguing against F′ specificity is the evidence that chromosomal sites show high mutability in Lac+ revertants (26–29). Moreover, this chromosomal mutability is not uniform: Loss-of-function mutations in a single gene, upp, were about 10 times more frequent than in the entire maltose regulon (>seven genes) (26, 36). Therefore, it seems possible that some chromosomal sites (cold ones, for example the chromosomal lac operon) might not be accessible to recombination protein-dependent mutation, whereas other (hotter) ones might be. Only one site (lac) in the E. coli chromosome has been examined for recombination protein-dependent stationary-phase mutability. We have examined a second locus in this study.
If a recombination protein-dependent stationary-phase mutation were F-specific, it might still contribute to bacterial evolution. Most wild E. coli carry conjugative plasmids and about 15% carry F-like conjugative plasmids (38). As suggested previously (39), an F′-specific mutation mechanism could be important to bacterial evolution because bacterial DNA is exchanged via recombination between episomes and chromosomes. The recombination protein-dependent mutation mechanism acting on the F′ would appear to be more obviously relevant to bacterial evolution if chromosomal genes also were affected.
Here we describe an assay allowing direct selection of chromosomal mutations in cells under lactose starvation. We test the idea that chromosomal genes are accessible to stationary-phase mutation via the recombination protein- and pol IV-dependent mechanism by examining the chromosomal upp gene (40), which is hypermutated in Lac+ stationary-phase mutants 10 times more frequently than two multigene regulons (26). We have inserted into upp a selectable marker, a tet+1 frameshift allele, which can confer tetracycline resistance (TetR) if reverted by a (net) −1 frameshift mutation. We find that TetR mutations at this chromosomal site accumulate under lactose selection conditions, independently of a cell becoming Lac+. TetR mutation displays genetic requirements indistinguishable from that of Lac+ mutation on the F′, requiring recombination proteins and pol IV. TetR mutants also show associated hypermutation. These observations demonstrate the occurrence of a recombination protein- and pol IV-dependent stationary-phase mutation mechanism in the E. coli chromosome.
Materials and Methods
Bacterial Strains and Growth Conditions.
The E. coli K-12 strains used in this study are listed in Table 1, and strain constructions are described below. P1 transduction was performed by using standard techniques (41). Antibiotics were used at the following concentrations: tetracycline, 10 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml.
Table 1.
E. coli K-12 strains
| Strain | Description | Reference or construction |
|---|---|---|
| BW229 | lac rpsL rfa-209∷Tn10 pyrE70 gltS10 metB thi | E. coli Genetic Stock Center |
| ES1481 | mutS215∷Tn10 | E. coli Genetic Stock Center |
| FC29 | Δ(lac-proAB)XIIIthi ara[F′ proAB+ Δ(lacI-lacZ)] | |
| FC40 | Δ(lac-proAB)XIIIthi ara RifR [F′ proAB+lacI33ΩlacZ] | (14) |
| FC526 | FC40 ΔrecG263∷kan | (17) |
| FC722 | FC40 [F′ zzf-1831∷Tn10dtet+1] | (39) |
| GM2159 | dam13∷Tn9 | M. Marinus (Univ. of Massachusetts, Worcester, MA) |
| GS1481 | ΔruvC64∷kan | (44) |
| SMR749 | FC40 mutS215∷Tn10 | FC40 × P1(ES1481) |
| SMR2597 | FC40 Δ(xseA-gua) zff-3139∷Tn10kan | Lab collection |
| SMR4251 | FC40 dam13∷Tn9 | FC40 × P1(SMR56, a derivative of GM2159) |
| SMR4576 | FC40 upp∷Tn10dtet+1 | This study* |
| SMR4608 | SMR4576 recA∷Tn10dcam | SMR4576 × P1(SMR4610) |
| SMR4610 | FC40 recA∷Tn10dcam | Lab collection |
| SMR4719 | FC40 recD1018 Δ(xseA-gua) zff-3139∷Tn10kan | Lab collection |
| SMR4721 | SMR4576 recD1018 | SMR4719 × P1(SMR4576) |
| SMR4727 | SMR4721 recA∷Tn10dcam | SMR4721 × P1(SMR4610) |
| SMR4733 | SMR4576 ΔrecG263∷kan | SMR4576 × P1(FC526) |
| SMR4758 | SMR4733 recA∷Tn10dcam | SMR4733 × P1(SMR4610) |
| SMR4822 | SMR4576 ΔruvC64∷kan | SMR4576 × P1(GS1481) |
| SMR4823 | SMR4721 ΔruvC64∷kan | SMR4721 × P1(GS1481) |
| SMR5499 | SMR4576 ΔrecG263∷kan rfa-209∷Tn10 pyrE70 | SMR4733 × P1(BW229) |
| SMR5522 | FC29 ΔruvC64∷kan | FC29 × P1(GS1481) |
| SMR5790 | SMR4576 radC102 | SMR5499 × P1(SR1187) |
| SMR5830 | FC40 dinB10 | (23) |
| SMR5969 | FC40 dinB10 Δ(xseA-gua) zff-3139∷Tn10kan | SMR5830 × P1(STL1605) |
| SMR6047 | SMR4576 ruvC53 eda∷Tn10dcam | This study* |
| SMR6048 | SMR4576 radC102 ruvC53 eda∷Tn10dcam | SMR5790 × P1(SMR6047) |
| SMR6049 | SMR4576 dinB10 | SMR5969 × P1(SMR4576) |
| SR1187 | argE3 hisG4 leuB6 Δ(gpt-proA)62 thr-1 thi-1 ara-14 galK2 lacY1 mtl-1 xyl-5 tsx-33 rfbD1 mgl-1 rpsL31 supE44 rac radC102(=recG) | (77, 78) |
| STL1605 | argE3 hisG4 leuB6 Δ(gpt-proA)62 thr-1 thi-1 ara-14 galK2 lacY1 mtl-1 xyl-5 tsx-33 rfbD1 mgl-1 rpsL31 supE44 Δ(xseA-gua)zff- 3139∷Tn10kan | S. T. Lovett (Brandeis Univ., Waltham, MA) |
See Materials and Methods for details of construction.
SMR4608, SMR4727, and SMR4758 were made by transducing recA∷Tn10dcam from SMR4610 into strains SMR4576, SMR4721, and SMR4733, respectively, selecting for chloramphenicol resistance (CamR) and confirming UV sensitivity. SMR4721 was constructed by transducing SMR4719 selecting for growth on minimal medium without guanosine (selecting Gua+) by using SMR4576 as the P1 donor, then screening for 5-fluoro-cytosine resistance, and kanamycin sensitivity. SMR4733 was constructed by transducing ΔrecG263∷kan from FC526 into SMR4576 selecting for KanR and screening for mild UV sensitivity. SMR4822 and SMR4823 were constructed by introducing ΔruvC64∷kan from GS1481, selecting KanR transductants, and confirming UV sensitivity. SMR5790 was constructed by transduction of SMR5499 with radC102 from SR1187, selecting Pyr+ and screening for Kan and Tet sensitivity. SMR6048 was constructed by introducing ruvC53 with selection for the linked marker eda∷Tn10dcam into SMR5790. The eda∷Tn10dcam allele was derived from eda-51∷Tn10 by using a short homology recombination method (42). A PCR product generated from Tn10dcam (43) with a primer to the very end of IS10R (5′-CTGATGAATCCCCTAATGATTTTGGTA-3′) was recombined into an eda-51∷Tn10 strain (replacing Tn10 with Tn10dcam) selecting for CamR. A CamR TetS isolate with eda∷Tn10dcam linked to ruvC53 was saved as SMR6047. All strains carrying a ruvC mutation were constructed and grown at 32°C (until assayed for mutation at 37°C) because this minimizes the accumulation of suppressors and revertants (18, 44). Minimal medium cultures of ruvC strains were given 72 h to reach saturation rather than the usual 48 h.
Construction of the upp∷Tn10dtet+1 Allele.
First, an insertion of Tn10dtet into the chromosomal upp gene was isolated from a library of random Tn10dtet insertions into FC40 from λNK1323 (as per ref. 43). The library was screened for isolates with insertions in upp based on their resistance to 5-fluoro-cytosine and insertion into upp confirmed by linkage to xseA and gua genes. Second, the tet+ gene of the upp∷Tn10dtet allele was replaced with the tet+1 frameshift allele from FC722 (39) [Tn10dtet + 1 G at bp 331 (4G to 5G), hereafter referred to as Tn10dtet+1] by homologous recombination of the upp∷Tn10dtet transduced into FC722, then selected for TetS recombinant cells (45). The putative upp∷Tn10dtet+1 allele was transduced into SMR2597, selecting Gua+, to produce SMR4576, and the presence of tet+1 was confirmed by PCR and sequencing. Primers used for PCR were 5′-TACCACTCCCTATCAGTGATAG-3′ and 5′-ATAACATCATTTGGTGAC-3′. Sequencing primers were 5′-TACCACTCCCTATCAGTGATAG-3′, 5′-TGCTGTATTTAGGCCGTTTG-3′, and 5′-AAAGCGATCCCACCACCAG-3′. Sequencing was done by the Baylor College of Medicine Core Facility.
Stationary-Phase Mutation Assays.
TetR stationary-phase mutation assays were performed essentially as described (39). For each strain, six independent cultures were tested in parallel. Aliquots of cell suspensions were mixed with 0.1 ml of 20- to 25-fold concentrated scavenger cells [FC29 or SMR5522 when ruvC frameshift-bearing cells were tested (see ref. 18)] and 2.5 ml of M9 top agar with 0.1% lactose on M9 plates containing 0.1% lactose. All plates were overlaid with an additional 2.5 ml of M9 lactose top agar to prevent colony smearing during additional overlays. Four complete sets of plates were prepared for each culture. Every day for 4 days (including day 0, the day of plating) one complete set of plates was overlaid with 5 ml of M9 top agar containing 0.12 ml of 50% glycerol, 0.04 ml of tetracycline (10 mg/ml), and 0.01 ml of 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) (20 mg/ml). Plates then were incubated an additional 3 days, and the white (Lac-) TetR colonies were counted. Additional incubation of up to 2 more days yielded no more than an additional 10% increase in colony numbers regardless of strain genotype. In early experiments, Tet resistance was confirmed by patching from the colonies on the overlaid plates onto Luria–Bertani–Herskowitz (LBH) (e.g., ref. 26) Tet plates. Lac+ TetR colonies were excluded because the TetR mutation might have formed during the growth of the Lac+ colony and as such could be a growth-dependent mutation, and not of interest here. In every TetR mutation experiment, a Lac+ stationary-phase mutation assay (18) run in parallel confirmed that strains mutated as expected from previous results (16–18). Net cell growth or death on the lactose selection plates was monitored as described (18) and varied less than 2-fold for all experiments reported.
Secondary mutations in the TetR mutants were detected by patching day 3 TetR Lac− mutant (white) colonies from overlaid plates onto LBH Tet. The resulting TetR patches were replica-plated onto MacConkey Tet medium (41) containing either xylose or Mal to score for the presence of unselected mutations in genes required for Xyl and Mal fermentation, respectively. Putative TetR Mal− and TetR Xyl− mutants were purified, and their fermentation-defective phenotypes were confirmed before their use in tests for heritable mutator/nonmutator status (see Table 4).
Table 4.
Most chromosomal TetR mutants with secondary mutations are not heritably mutator
| Strain* | No. independent isolates tested | Mutant colonies observed, mean (range)†
|
|||
|---|---|---|---|---|---|
| Nalidixic acidR | StreptomycinR | SpectinomycinR | Phenotype | ||
| TetS | 10 | 0.2 (0–2) | 0.05 (0–1) | 1.1 (0–6) | Nonmutator |
| mutS | 10 | >25 (14–>50) | 0.95 (0–4) | 4.7 (2–9) | Mutator |
| dam | 10 | 4.3 (1–11) | 0.22 (0–1) | 0.28 (0–2) | Mutator |
| Day 0 TetR | 26 | 0.42 (0–3) | <0.04§ | <0.04§ | Nonmutator |
| Day 3 TetR‡ | 44 | 0.15 (0–4) | <0.01§ | <0.01§ | Nonmutator |
| Day 3 TetR Mal− or TetR Xyl−¶ | 11 | 0.04 (0–1) | 0.04 (0–1) | <0.04§ | Nonmutator |
Strains are TetS, SMR4576 (negative control); mutS, SMR749; dam, SMR4251, (positive controls), and then chromosomal TetR growth-dependent mutants (day 0), stationary-phase mutants (day 3), and 11 independent isolates of double mutants: stationary-phase TetR mutants carrying an unselected secondary mutation preventing fermentation of maltose or xylose. All strains not noted as Mal− or Xyl− were confirmed Mal+ or Xyl+, as well as Lac− (see Materials and Methods).
One saturated LBH culture was used per isolate and two 5-μl aliquots were spotted onto selective medium, and the numbers of mutant colonies were counted after incubation. The mean number (and range) of colonies per 5-μl spot are given. Antibiotic concentrations were: nalidixic acid, 40 μg/ml; streptomycin or spectinomycin, 100 μg/ml.
Note that TetR isolates from day 3 include some preexisting mutants (see Fig. 1), such that 25–50% are likely to be preexisting mutants.
In these cases, there were no mutant colonies detected and “less than” values were calculated as if a single colony had appeared in one of the spots being averaged.
Of these, seven are Mal−, three are Xyl−, and one is both Mal− and Xyl−. Two of the Xyl− isolates were derived from the same TetS culture. These two are potentially siblings and may be preexisting mutants, as discussed above.
Growth-Dependent Mutation Assay.
Growth-dependent mutation rates were determined by using 20-tube fluctuation tests selecting on LBH Tet plates and measuring total viable cells on LBH plates. To control for the possibility of postplating mutation events, the time to colony formation was determined for previously isolated TetR mutants of each genotype. A total of 50–100 cells from each of six TetR mutants were plated on LBH Tet with about 2 × 109 nonreverting FC29 cells. The colonies formed from the seeded TetR cells were counted at various times to determine the time required to form colonies (typically ≈90% emergence, relative to colony counts on LBH, by ca. 12 h for all genotypes). Counts of TetR colonies from the test cultures at additional times up to 72 h revealed that additional TetR colonies continued to appear each day (data not shown), indicating that TetR mutations continue to form on the Tet plates. Thus, the number of TetR colonies visible at 12 h was used to determine mutant frequencies (as mutants/cell plated), to exclude mutations occurring on the plate after selection. Mutant frequency values for each genotype were divided by the fraction of TetR colonies emerged for the TetR control isolates in each experiment (typically ≈0.9). Mutation rates were calculated by the method of the median (46, 47).
Results
Experimental System.
We wanted to assay chromosomal mutations at the upp locus during lactose selection in the lac frameshift-bearing cells in which adaptive mutation has been studied. To do this we inserted a selectable marker into the upp gene: a tetA +1 frameshift allele (Materials and Methods). Cells carrying this allele are sensitive to tetracycline (TetS), but a compensating frameshift mutation within a run of five Gs in this allele (39) restores tetracycline resistance (TetR). Mutation to TetR during lactose starvation is detected by assaying multiple sets of lactose selection plates and rescuing TetR cells from one set on each day of starvation on lactose. The plates are overlaid with agar containing glycerol (as a carbon source) and tetracycline, and incubated 3 additional days so that any TetR cells present can form colonies. Lac+ colonies are excluded because mutation to TetR can occur during the growth of the colony.
Chromosomal Mutation to TetR Occurs During Lactose Selection and Requires Recombination Proteins.
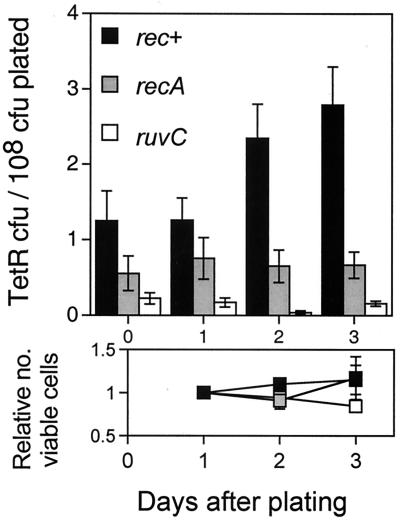
In Fig. 1, we see that TetR mutations accumulate over time, after exposure to lactose medium, increasing 2- to 3-fold, over 3 days on lactose. Like adaptive mutation at lac on the F′, we find that RecA and RuvC are required for accumulation of chromosomal TetR mutations over time (Fig. 1). This demonstrates recombination protein-dependent stationary-phase mutation in the bacterial chromosome.
Figure 1.
Chromosomal TetR mutation during lactose starvation requires RecA and RuvC. Values are the averaged mutation frequencies from multiple independent experiments (n = 9, 5, and 5 for rec+, recA, and ruvC, respectively). Daily measurements of lac− viable cells on the plates (below), shown normalized to the first day's count, show no net growth or death during the experiments (black, rec+; gray, recA; white, ruvC). Error bars represent one SEM. The strains are SMR4576, SMR4608, and SMR4822.
Elevation of TetR Mutation by recD and recG Alleles.
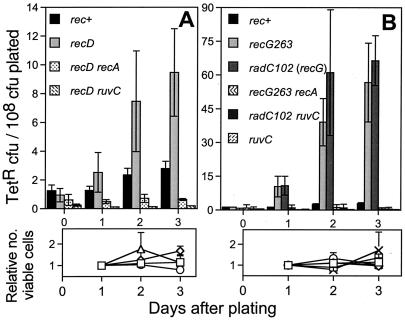
Cells lacking recD and recG have increased RecA-dependent Lac+ stationary-phase mutation (16–18, 29). This could be caused by hyperrecombination in the case of recD (16, 29) and stabilization of recombination intermediates in the case of recG (17, 18). Both recD and recG defects are thought to increase steady-state levels of strand exchange intermediates that prime synthesis leading to polymerase error and mutation. Alternatively, both recD and recG mutations promote the SOS response (48, 49), which is required for efficient Lac+ stationary-phase mutation (14, 22) and so might specifically increase stationary-phase mutation that way. We see that recD and recG mutations also increase chromosomal TetR mutation, recD by about 4-fold (Fig. 2A) and recG by 25-fold (Fig. 2B). These phenotypes are quantitatively nearly identical to the effects of recD and recG on Lac+ stationary-phase mutation (data not shown for Lac+ assays run in parallel and refs. 16–18). Importantly, RecA and RuvC are required for the increased chromosomal TetR mutation in recD and recG mutants (Fig. 2). Therefore, the increased mutation is recombination protein-dependent, and not via activation of some other, recombination protein-independent, stationary-phase mutation pathway.
Figure 2.
Chromosomal stationary-phase TetR mutation increases in recD and recG cells, RecA and RuvC dependently. (A) The strains are SMR4576, SMR4721, SMR4727, and SMR4823. (B) The strains are SMR4576, SMR4733, SMR5790, SMR4758, SMR6048, and SMR6047. radC102 is a point mutation in the recG gene (77, 78). Use of this allele was necessary to make the recG ruvC combination. Values are the averaged mutation frequencies from multiple independent experiments: (A) n = 9, 6, 3, and 3 for rec+, recD, recD recA, and recD ruvC, respectively; (B) n = 9, 4, 3, 2, 2, and 3 for rec+, recG263, radC102, recG263 recA, radC102 ruvC, and ruvC, respectively). The rec+ data are those shown in Figs. 1 and 4. Error bars represent one SEM except for those in which n = 2 in which the error bars show the range. Mutant frequencies were determined as described in Materials and Methods except for two differences for the radC102 ruvC strain. First, only five cultures were tested in each experiment rather than six, and second, no TetR colonies were observed in any of the five cultures tested on day 0 in one experiment. The same is true for days 1 and 3 in the other experiment. For those days the value was calculated as if a single colony had been observed in one of the five cultures and that value was used in calculating the average shown, which is thus a “less than” value. Daily measurements of lac− viable cells on the plates (below), shown normalized to the first day's count, show no net growth or death during the experiments: (A) □, rec+; ◊, recD; ○, recD ruvC; ▵, recD recA. (B) □, rec+; ◊, recG263; ○, recG ruvA; ▵, ruvC53; crossed squares, radC102; X, radC102 ruvC53.
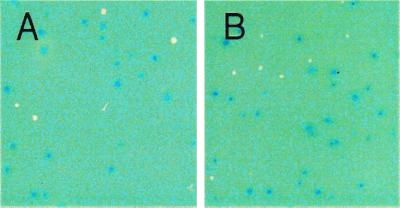
We have seen that numbers of chromosomal TetR mutants increase in recD or recG cells and decrease in recA and ruvC cells, just as Lac+ stationary-phase mutations on the F′ do (Figs. 1 and 2). A possible artifactual explanation for these apparently identical genetic requirements could be that growth of the Lac+ mutant colonies releases carbon sources into the medium nearby, allowing growth of neighboring cells that then generate growth-dependent TetR mutations (cross-feeding). We can rule out cross-feeding because it predicts that the TetR mutants should form only close to Lac+ mutant colonies, as satellite colonies, which we do not observe. A representative example is shown in Fig. 3 in which blue Lac+ colonies and white TetR colonies show no particular association. Also, we do not observe growth of the lac− cells on the plates in these experiments (Figs. 1, 2, and 4). Thus, the genetic requirements described here can be attributed to stationary-phase chromosomal TetR mutation.
Figure 3.
TetR and Lac+ stationary-phase mutant colonies are randomly distributed on plates. We find that TetR (white) colonies are not satellites of the Lac+ (blue) mutant colonies, as would have been predicted if the TetR mutations were formed during growth of cells next to Lac+ colonies (caused by cross-feeding, discussed in text). A and B are day 3 plates of rec+ (SMR4576) and recG (SMR4733), respectively.
Figure 4.
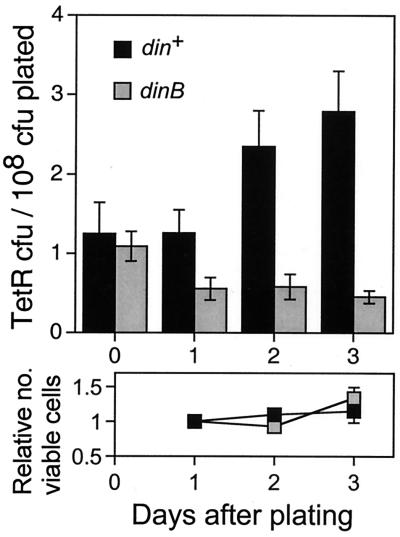
SOS-inducible DNA polymerase IV is required for TetR stationary-phase mutation in the bacterial chromosome. The strains are SMR4576 and SMR6049. Values are the averaged mutation frequencies from multiple experiments (n = 9 and 4 for din+ and dinB10, respectively). The din+ data are those shown in Figs. 1 and 2 (rec+). Daily measurements of lac− viable cells on the plates (below), shown normalized to the first day's count, show no net growth or death during the experiments (black, din+; gray, dinB10). Error bars represent one SEM.
Recombination Functions Do Not Affect Growth-Dependent Mutation Rates.
Importantly, growth-dependent mutation rates to TetR are unaffected by the rec and ruv gene mutations (Table 2). Therefore the recombination protein dependence of TetR chromosomal mutation is specific to stationary-phase mutation, as is recombination protein-dependent mutation at lac on the F′ (16–18).
Table 2.
Recombination-independence of growth-dependent TetR mutation
| rec genotype* | Exp. | TetR mutation rate, mutations/cell per generation | Mean (± SEM) |
|---|---|---|---|
| rec+ | 1 | 3.1 × 10−10 | |
| 2 | 3.9 × 10−10 | ||
| 3 | 2.9 × 10−10 | ||
| 4 | 2.2 × 10−10 | ||
| 5 | 5.1 × 10−10 | ||
| 6 | 3.5 × 10−10 | 3.5 (±0.41) × 10−10 | |
| recA | 1 | 5.9 × 10−10 | |
| 2 | 1.1 × 10−9 | ||
| 3 | 3.1 × 10−10 | 6.7 (±2.3) × 10−10 | |
| recD | 1 | 2.4 × 10−10 | |
| 2 | 4.0 × 10−10 | ||
| 3 | 1.1 × 10−9 | 5.8 (±2.6) × 10−10 | |
| recG | 1 | 2.7 × 10−10 | |
| 2 | 4.3 × 10−10 | ||
| 3 | 4.5 × 10−10 | 3.8 (±0.57) × 10−10 | |
| ruvC | 4 | 4.1 × 10−10 | |
| 5 | 4.4 × 10−10 | ||
| 6 | 4.6 × 10−10 | 4.4 (±0.15) × 10−10 |
Strains used are SMR4576, SMR4608, SMR4721, SMR4733, and SMR4822.
Requirement for DNA Pol IV.
DNA pol IV is an error-prone DNA pol encoded by the SOS-inducible dinB gene (50, 51). It is a member of the newly discovered DinB/UmuDC superfamily of DNA polymerases present in eubacteria, archaea, and eukaryotes, which includes tumor suppressor protein XP-V (xeroderma pigmentosum variant), many mammalian pols of unknown function, and also the E. coli UmuDC SOS mutator DNA pol V (reviewed in ref. 52, and see refs. 53 and 54). dinB is the first gene of an apparent four-gene operon, all of the genes of which are likely to be inactivated by the commonly used null alleles of dinB (reviewed in ref. 23). Using a nonpolar null allele as the sole dinB allele in E. coli, we found that pol IV is required for most stationary-phase, but not growth-dependent, Lac+ mutation on the F′ (23). Using the same nonpolar dinB10 allele, encoding a polymerase that is nonfunctional in vivo and in vitro (51), we find that pol IV also is required for accumulation of TetR stationary-phase mutants (Fig. 4). dinB10 has no effect on the growth-dependent mutation rate to TetR in this same strain (23), indicating a specific effect on mutation in stationary phase. Thus, stationary-phase mutation to Lac+ on the F′ and mutation to TetR at the chromosomal upp site share a specific requirement for the SOS-inducible DNA pol IV.
The TetR Mutants Experienced Hypermutation.
Lac+ stationary-phase mutants carry a high frequency of unselected secondary mutations at chromosomal sites, as if some or all of them have undergone a general genomewide hypermutation (26–29). We tested whether chromosomal TetR stationary-phase mutants also exhibit hypermutation by screening day 3 TetR mutants for unselected secondary mutations preventing the fermentation of maltose or xylose (Materials and Methods). We note that because these TetR mutants will include some TetR mutants present at the time of plating (about 25–50%, see Fig. 1). We observed secondary mutations at a high frequency (Table 3). The numbers of fermentation-defective mutants per TetR mutants screened were 18/869, 27/1,304, and 58/950 from the rec+, recD, and recG strains, respectively. This finding represents hypermutation relative to that observed in (nonmutated) Lac−-starved cells from the lactose selection plates [i.e., 3/70,000 (26) and 2/28,000 (29)]. As observed previously for Lac+ stationary-phase mutants carrying secondary mutations (26, 27), most of the TetR mutants with secondary mutations are not heritable mutator mutants (such as mismatch repair defective mutants) (Table 4), and so appear to have originated during a transient period of hypermutability.
Table 3.
Hypermutation in chromosomal TetR stationary-phase mutants
| rec genotype* | No. of Mal− and Xyl− mutants among TetR mutants, mutant colonies/colonies scored† |
|---|---|
| rec+ | 18/869 |
| recD | 27/1,304 |
| recG | 58/950 |
Strains are SMR4576, SMR4721, and SMR4733.
Mal− and Xyl− mutations are loss-of-function mutations in the chromosomal maltose and xylose regulons, conferring inability to ferment these sugars.
Discussion
We have described an assay for direct selection of mutations in the bacterial chromosome under adaptive mutation conditions in the well characterized, F′-based lac system. Using this assay, we see stationary-phase accumulation of chromosomal TetR mutations at a site in the upp gene. These accumulate to levels 10- to 30-fold lower than Lac+ or TetR mutations on the F′ (ref. 39, discussed below). However, like adaptive Lac+ mutation on the F′ (16–18, 23), formation of these chromosomal mutations requires recombination proteins RecA and RuvC, and the error-prone DNA polymerase, pol IV, and does so specifically for stationary-phase and not growth-dependent mutation. The requirement for SOS-inducible pol IV for chromosomal mutation also implicates the SOS response, which is required for efficient stationary-phase mutation at lac on the F′ (14, 22). Also, both mutation at lac on the F′ (16–18) and at upp∷tet+1 (Fig. 2) are stimulated by recD and recG mutations, and this stimulation is RecA- and RuvC-dependent. These data support the idea that the recombination-dependent stationary-phase mutation process is available to (at least some of) the bacterial chromosome and therefore is not confined to DNA in conjugative plasmids in E. coli. This conclusion is important to our understanding of mechanisms of genetic change that shape the bacterial genome and drive bacterial evolution.
How Many Mechanisms of Mutation in This System?
Do all of the stationary-phase mutations assayed in this system arise via this recombination protein-dependent mechanism? Four classes of stationary-phase mutations have been described, three of which have been shown directly to be recombination protein-dependent: (I) Lac+ adaptive mutations on the F′; (II) chromosomal stationary-phase mutations (described here), selected independently of Lac+ mutations; and (III) F′-located stationary-phase mutations, which also are selected independently of Lac+ but at a site on the F′ (39). We note that the chromosomal mutations (class II) form less frequently than the two F′-based classes (I and III) by about 10- to 30-fold (e.g., refs. 14, 16, and 39, and discussed below). However, their similar genetic requirements (Figs. 1, 2, and 4) and associated hypermutation (refs. 26–29 and Table 3) imply a common mechanism, albeit one that may occur less frequently on the chromosome (discussed below). (IV) The fourth class is “secondary” mutations detected in Lac+ adaptive mutants at non-lac (unselected) sites on the chromosome and other replicons (26–29), and which occur with nonuniform (“hot” and “cold” region) distribution (36). Like the other three classes, secondary mutations are increased in hyperRec, hyperadaptive mutation-enhancing recD and recG strains (29). The results presented here, that chromosomal mutations are recombination protein-dependent and pol IV-dependent, argue for a common mechanism underlying all of these classes of mutation.
Implications for the Hypermutable Subpopulation.
Our data support the idea that adaptive mutations in this system arise from a condition of transient, genomewide hypermutability as follows. Lac+ adaptive mutants have high frequencies of unselected secondary mutations at chromosomal and other sites (e.g., 10−3 at one chromosomal site whereas Lac−-starved cells from the same plate show <10−5) (26–28). This was taken as evidence that the Lac+ mutants descend from a hypermutable subpopulation of cells in which lac and unrelated genes are mutated. An alternative to this view is that those Lac+ mutants that have a detectable secondary mutation arose from a hypermutable subpopulation, but that most Lac+ did not (27). Those authors argue that only about 10% of Lac+ adaptive mutants arose from hypermutable cells (those presumably having 10 times more secondary mutations per Lac+ cell, e.g., 10−2), but that the rest arose from a different population of cells in which lac mutates but other genes do not. The implication of this idea is that, whereas sites on the F′ experience the major recombination-dependent mutation route (applying to 90% of Lac+ mutants), all of the secondary chromosomal mutations and the Lac+ mutations that they are associated with (10%) arise by some mechanism not relevant to most adaptive mutation (presumably not recombination protein-dependent). Our data fail to support this model because we find that chromosomal unselected mutations are also recombination protein-dependent (Figs. 1 and 2). Instead, the data support models in which chromosomal and F′ mutations arise via a common mechanism. This is easiest to imagine occurring in a common subpopulation. In supporting a one-mechanism (one-subpopulation) model, the data also discourage the idea that the adaptive mutation mechanism is directed to DNA adjacent to the selected (lac) gene or replicon (e.g., ref. 55), and favor random, nongene-directed mutation models.
Hypermutation in Chromosomal Mutants.
The TetR mutants that arise during extended lactose selection have a very high frequency of unselected secondary mutations (Table 3). Whereas 53/41,238 (0.1%) of Lac+ adaptive mutants have lost the ability to ferment maltose or xylose (26), 18/869 (2.1%) of TetR stationary-phase mutants have (Table 3). These values are different with a contingency χ2 value of 198 (P < 0.00001). A possible explanation is that cells that become Lac+ are likely to be released immediately from the starvation conditions. In contrast, cells subjected to lactose starvation that become TetR (and remain Lac−) may tend to be trapped for longer periods in the hypermutable state, and thus continue to accumulate additional mutations until rescued via addition of glycerol and tetracycline. Extended lactose selection is reported to increase the frequency of unselected mutation, supporting this idea (28).
This process might occur via a “sliding scale” model as follows: After exposure to selection, entry into the hypermutable state may be random (with respect to time), but the exit time may be fixed as the time of mutation to Lac+, which would end the stress condition. This set of rules would generate a population of Lac+ mutants that had spent variable lengths of time in the state of hypermutability, and so had varying probabilities of having accumulated secondary mutations. If the rate of mutation in the hypermutable state is constant with time, then the frequency of mutation in any given Lac+ isolate will be proportional to the time spent in the hypermutable state. In this model, Lac+ colonies that are found to carry a secondary mutation are likely to have spent a longer time than average in the hypermutable state. For this reason, they are also more likely than average to carry additional unselected mutations. Thus, a sliding scale model can explain the observation that the frequency of Lac+ mutant cells with two unselected mutations (triple mutants) is higher than expected from the frequency of cells that are Lac+ with a single unselected mutation (double mutants) (if a single mutation rate is assumed). This observation was used to suggest that two different populations yield Lac+ mutants (27). We are cautious about the reality of this observation because it is based on very few data (five triple mutants in ref. 26, only four triple mutants in ref. 27). However, should the collection of more data indicate that the frequency of triple mutants is indeed disproportionately high, then the sliding scale model could accommodate that finding without having to invoke a second mutable population. The triple mutants would represent cells that spent longer time in a hypermutable condition. This model also seems more economical in view of the data presented here and previously (29) supporting similar genetic requirements for chromosomal and most F′ Lac+ mutations—the two classes that would be assigned to two different populations in the two-population model (27).
Role(s) of the F′.
The F′ appears not to be a passive component in recombination protein-dependent stationary-phase mutation. First, F-plasmid-encoded transfer (Tra) functions promote adaptive mutation at F′ lac, although actual conjugative transfer is not required (19–21). Second, recombination protein-dependent Lac+ (16–18) and TetR mutations on the F′ (39) are 10- to 30-fold more frequent than recombination protein-dependent mutation of the same tet+1 allele in the chromosomal upp location (Figs. 1, 2, and 4). In one model for the contribution of the F′ to mutation, the F Tra functions provide a “programmed” DSB. Nicking of the origin of transfer (oriT) by Tra proteins could promote DSBs or double-strand ends. These are known to be required for mutation in this system because of the RecBC dependence, which demands linear DNA (16). This model is harmonious with our discovery of recombination protein-dependent mutations in the chromosome (here), which could occur by the same mechanism, but less frequently, because the DSBs that initiate them may not be F-programmed.
The role of the F′ in mutation in the chromosome remains to be determined. The F′ could potentially stimulate chromosomal mutation in trans (28) or in cis because the F′ can integrate into the chromosome (called formation of an Hfr). Two studies imply that the chromosomal stationary-phase mutations form in trans to the F′, and not in cis, in cells that are Hfrs. First, although Hfr formation occurs measurably in this system, Lac+ adaptive mutants carrying a chromosomal mutation are not enriched for Hfrs (56). Thus, there is no correlation between integrating the F′ and acquiring a chromosomal mutation. Second, chromosomal mutations coincident with Lac+ adaptive mutation behave as independent events from Lac adaptive reversion (29). This is seen in the following way: the recD and recG mutations, which increase recombination-dependent adaptive mutation, presumably by increasing levels of recombination intermediates that promote mutation, increase the frequency of Lac+ mutants with a coincident chromosomal mutation, but not with an F′-located coincident mutation (29). These results imply that the coincident F′ mutations were not independent of formation of Lac+ (and happen at a constant frequency per Lac+ event), but that the coincident chromosomal mutations are independent and were stimulated in independent events from those leading to Lac+ on the F′ (29).
Regardless of how the F′ may affect mutation in the chromosome, the end result is that the bacterial genome is susceptible to recombination protein-dependent stationary-phase mutation, an inducible genetic change mechanism that uses recombination proteins and DNA synthesis. Because most wild E. coli and related bacteria carry conjugative plasmids with about 15% carrying F-like plasmids (38, 57), effects of F-like plasmids are likely to be relevant to genetic change in these species.
Further Discussion
What is the significance of the recombination protein-dependent stationary-phase mutation mechanism, shown here to act in the bacterial chromosome, for bacteria, possibly for other organisms, and for our understanding of the connection between DNA replication and recombination?
Role of Recombination-Dependent Mutation in Bacterial Evolution.
The mutations selected in the assay system used here are frameshift mutations (58, 59). Frameshifts usually are considered as destructive of gene function, not as mutations likely to be adaptive. So is this process relevant to bacterial evolution? It may be. First, only frameshift mutations are selected in this assay, but substitution mutations also may be promoted. This possibility seems likely because DNA pol IV, which is implicated in this process (ref. 23 and Fig. 4), promotes substitutions in addition to frameshift mutations (51, 60, 61). Second, frameshift mutations themselves appear to be highly relevant to bacterial evolution. Many pathogenic bacteria regulate the expression of “contingency genes” (used during their battles against host defenses) by frequent frameshift mutations in simple repeated sequences, turning genes on and off by switching reading frames, and also activating promoters (62, 63). These bacteria might use adaptive mutation strategies similar to the mechanism discussed here.
Relevance to Other Organisms and Replication Primed by Recombination.
Recombination-promoted mutation in yeast (64) resembles recombination-dependent stationary-phase mutation in E. coli in at least two respects. First, both are associated with DSBR. In yeast a direct association is known. The mutations occur next to the site of an induced DSB that has been repaired and in the chromosome that was broken (64–66). In E. coli such direct association between mutation and recombination has not yet been demonstrated, but rather the involvement of DSBs and DSBR is inferred from the RecBC dependence of the mutagenesis (16, and discussed further below). In E. coli, indirect action of DSBR proteins in promoting mutation (for example by activating the SOS response, see ref. 3) is also possible.
Second, and very interestingly, both the bacterial and yeast mutations require DinB/UmuDC superfamily DNA polymerases. Most (about 85%) of recombination-dependent stationary-phase point mutation in E. coli requires pol IV (ref. 23 and Fig. 4), and the substitution component of yeast DSBR-promoted mutation requires the REV3-encoded pol zeta, another DinB/UmuDC polymerase superfamily member (67). There is also a frameshift mutation component of yeast DSBR-promoted mutation that does not require pol zeta, suggesting that another polymerase, not yet identified is also involved (64, 67). Thus, DNA synthesis promoted by DSBR may be one of the functions of these new DNA polymerases. It will be interesting to see whether they also function in somatic hypermutation of Ig genes (52), which has been suggested to proceed via a similar mechanism of pol errors made during recombinational break repair (68, 69). The abundance of the DinB/pol IV-ortholog, DinB1 or pol kappa, in germ-line tissues (52) also raises the issue of promotion of heritable (germ-line) mutations by these polymerases, even in multicellular eukaryotes (3, 23).
For stationary-phase mutation in E. coli, the proteins used in DSBR are required, but a direct demonstration of mutations adjacent to DSBR has not yet been made. We note, however, that the direct connection between DSBR and replication in E. coli in vivo has been made. Newly replicated DNA has been demonstrated in molecules that have experienced RecBCD-mediated DSBR (70). Measurements of new (unlabeled) nucleotides incorporated into recombining density-labeled DNA in vivo revealed two DSBR mechanisms that run concurrently: a break-join mechanism in which little or no new DNA synthesis accompanies recombinational DSBR leading to crossing over (observed previously, refs. 71 and 72), and which requires the Holliday junction processing proteins (RuvC and/or RecG) (70); and a replicative DSBR pathway, which might be break-copy (although “join-cut-copy”, discussed by Mosig in ref. 73, and see ref. 74, is also possible). The replicative DSBR mechanism requires the major replicative polymerase, pol III, and results in incorporation of unlabeled DNA isotopes in an amount proportional to the distance between the site of crossing over and the end of the chromosome, in both strands of DNA. This finding is compatible with break-copy models (75) (also called break-induced replication, see ref. 76) in which the act of strand invasion during DSBR primes a replication fork, and in which the two new DNA strands segregate conservatively.
Acknowledgments
We thank P. J. Hastings and J. F. Petrosino for comments on the manuscript; P. Foster, R. G. Lloyd, S. T. Lovett, R. Maurer, and the E. coli Genetic Stock Center (Yale University) for providing bacterial strains; G. J. McKenzie for providing the dinB10 allele; and M. Price for excellent medium preparation. This work was supported by National Institutes of Health Grants F32-GM19909 (to M.-J.L.), R01-GM53158, and R01-CA85777.
Abbreviations
- DSB
double-strand break
- DSBR
DSB repair
- LBH
Luria–Bertani–Herskowitz medium
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Foster P L. Annu Rev Genet. 1999;33:57–88. doi: 10.1146/annurev.genet.33.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lombardo M-J, Rosenberg S M. J Genet. 1999;78:13–21. [Google Scholar]
- 3.Rosenberg, S. M. (2001) Nat. Rev. Genet., in press. [DOI] [PubMed]
- 4.Lombardo M-J, Harris R S, Rosenberg S M. In: Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization. Lerner H R, editor. New York: Dekker; 1999. pp. 71–90. [Google Scholar]
- 5.Martinez J L, Baquero F. Antimicrob Agents Chemother. 2000;44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- 7.Strauss B S. Cancer Res. 1992;52:249–253. [PubMed] [Google Scholar]
- 8.Luria S E, Delbrück M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maenhaut-Michel G, Shapiro J A. EMBO J. 1994;13:5229–5239. doi: 10.1002/j.1460-2075.1994.tb06854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maenhaut-Michel G, Blake C E, Leach D R, Shapiro J A. Mol Microbiol. 1997;23:133–145. doi: 10.1046/j.1365-2958.1997.3031666.x. [DOI] [PubMed] [Google Scholar]
- 11.Lamrani S, Ranquet C, Gama M J, Nakai H, Shapiro J A, Toussaint A, Maenhaut-Michel G. Mol Microbiol. 1999;32:327–343. doi: 10.1046/j.1365-2958.1999.01352.x. [DOI] [PubMed] [Google Scholar]
- 12.Taddei F, Matic I, Radman M. Proc Natl Acad Sci USA. 1995;92:11736–11740. doi: 10.1073/pnas.92.25.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taddei F, Halliday J A, Matic I, Radman M. Mol Gen Genet. 1997;256:277–281. doi: 10.1007/s004380050570. [DOI] [PubMed] [Google Scholar]
- 14.Cairns J, Foster P L. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenzie G J, Lombardo M-J, Rosenberg S M. Genetics. 1998;149:1163–1165. doi: 10.1093/genetics/149.2.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris R S, Longerich S, Rosenberg S M. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 17.Foster P L, Trimarchi J M, Maurer R A. Genetics. 1996;142:25–37. doi: 10.1093/genetics/142.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris R S, Ross K J, Rosenberg S M. Genetics. 1996;142:681–691. doi: 10.1093/genetics/142.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galitski T, Roth J R. Science. 1995;268:421–423. doi: 10.1126/science.7716546. [DOI] [PubMed] [Google Scholar]
- 20.Foster P L, Trimarchi J M. Proc Natl Acad Sci USA. 1995;92:5487–5490. doi: 10.1073/pnas.92.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster P L, Trimarchi J M. J Bacteriol. 1995;177:6670–6671. doi: 10.1128/jb.177.22.6670-6671.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenzie G J, Harris R S, Lee P L, Rosenberg S M. Proc Natl Acad Sci USA. 2000;97:6646–6651. doi: 10.1073/pnas.120161797. . (First Published May 30, 2000, 10.1073/pnas.120161797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenzie G J, Lee P L, Lombardo M-J, Hastings P J, Rosenberg S M. Mol Cell. 2001;7:571–579. doi: 10.1016/s1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- 24.Foster P L, Gudmundsson G, Trimarchi J M, Cai H, Goodman M F. Proc Natl Acad Sci USA. 1995;92:7951–7955. doi: 10.1073/pnas.92.17.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris R S, Bull H J, Rosenberg S M. Mutat Res. 1997;375:19–24. doi: 10.1016/s0027-5107(96)00244-8. [DOI] [PubMed] [Google Scholar]
- 26.Torkelson J, Harris R S, Lombardo M-J, Nagendran J, Thulin C, Rosenberg S M. EMBO. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosche W A, Foster P L. Proc Natl Acad Sci USA. 1999;96:6862–6867. doi: 10.1073/pnas.96.12.6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godoy V G, Gizatullin F S, Fox M S. Genetics. 2000;154:49–59. doi: 10.1093/genetics/154.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bull H J, McKenzie G J, Hastings P J, Rosenberg S M. Genetics. 2000;154:1427–1437. doi: 10.1093/genetics/154.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longerich S, Galloway A M, Harris R S, Wong C, Rosenberg S M. Proc Natl Acad Sci USA. 1995;92:12017–12020. doi: 10.1073/pnas.92.26.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg S M, Thulin C, Harris R S. Genetics. 1998;148:1559–1566. doi: 10.1093/genetics/148.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall B G. Genetics. 1990;126:5–16. doi: 10.1093/genetics/126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall B G. J Mol Evol. 1995;40:86–93. doi: 10.1007/BF00166599. [DOI] [PubMed] [Google Scholar]
- 34.Cairns J. Genetics. 2000;156:923. doi: 10.1093/genetics/156.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bull H J, McKenzie G J, Hastings P J, Rosenberg S M. Genetics. 2000;156:925–926. doi: 10.1093/genetics/154.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberg S M. Curr Opin Genet Dev. 1997;7:829–834. doi: 10.1016/s0959-437x(97)80047-0. [DOI] [PubMed] [Google Scholar]
- 37.Radicella J P, Park P U, Fox M S. Science. 1995;268:418–420. doi: 10.1126/science.7716545. [DOI] [PubMed] [Google Scholar]
- 38.Boyd E F, Hill C W, Rich S M, Hartl D L. Genetics. 1996;143:1091–1100. doi: 10.1093/genetics/143.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster P L. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neuhard J, Kelln R A. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F C, Curtis R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 580–599. [Google Scholar]
- 41.Miller J H. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 42.Datsenko K A, Wanner B L. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. . (First Published May 30, 2000, 10.1073/pnas.120163297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleckner N, Bender J, Gottesman S. Methods Enzymol. 1991;204:140–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 44.Mandal T N, Mahdi A A, Sharples G J, Lloyd R G. J Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maloy S R, Nunn W D. J Bacteriol. 1981;145:1110–1112. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lea D E, Coulson C A. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 47.von Borstel R C. Methods Cell Biol. 1978;20:1–24. doi: 10.1016/s0091-679x(08)62005-1. [DOI] [PubMed] [Google Scholar]
- 48.Chaudhury A M, Smith G R. Mol Gen Genet. 1985;201:525–528. doi: 10.1007/BF00331350. [DOI] [PubMed] [Google Scholar]
- 49.Lloyd R G, Buckman C. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kenyon C J, Walker G C. Proc Natl Acad Sci USA. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner J, Gruz P, Kim S R, Yamada M, Matsui K, Fuchs R P, Nohmi T. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 52.Friedberg E C, Feaver W J, Gerlach V L. Proc Natl Acad Sci USA. 2000;97:5681–5683. doi: 10.1073/pnas.120152397. . (First Published May 16, 2000, 10.1073/pnas.120152397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pham P, Rangarajan S, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 2001;98:8350–8354. doi: 10.1073/pnas.111007198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Washington M T, Johnson R E, Prakash L, Prakash S. Proc Natl Acad Sci USA. 2001;98:8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cairns J, Overbaugh J, Miller S. Nature (London) 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- 56.Lombardo M-J, Torkelson J, Bull H J, McKenzie G J, Rosenberg S M. Ann NY Acad Sci. 1999;870:275–289. doi: 10.1111/j.1749-6632.1999.tb08888.x. [DOI] [PubMed] [Google Scholar]
- 57.Boyd E F, Hartl D L. J Bacteriol. 1997;179:1622–1627. doi: 10.1128/jb.179.5.1622-1627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenberg S M, Longerich S, Gee P, Harris R S. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 59.Foster P L, Trimarchi J M. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim S R, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner J, Nohmi T. J Bacteriol. 2000;182:4587–4595. doi: 10.1128/jb.182.16.4587-4595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moxon E R, Rainey P B, Nowak M A, Lenski R E. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 63.Deitsch K W, Moxon E R, Wellems T E. Microbiol Mol Biol Rev. 1997;61:281–293. doi: 10.1128/mmbr.61.3.281-293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strathern J N, Shafer B K, McGill C B. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McGill C B, Holbeck S L, Strathern J N. Genetics. 1998;148:1525–1533. doi: 10.1093/genetics/148.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holbeck S L, Strathern J N. Ann NY Acad Sci. 1999;870:375–377. doi: 10.1111/j.1749-6632.1999.tb08906.x. [DOI] [PubMed] [Google Scholar]
- 67.Holbeck S L, Strathern J N. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maizels N. Cell. 1995;83:9–12. doi: 10.1016/0092-8674(95)90227-9. [DOI] [PubMed] [Google Scholar]
- 69.Harris R S, Kong Q, Maizels N. Mutat Res. 1999;436:157–178. doi: 10.1016/s1383-5742(99)00003-4. [DOI] [PubMed] [Google Scholar]
- 70.Motamedi M, Szigety S K, Rosenberg S M. Genes Dev. 1999;13:2889–2903. doi: 10.1101/gad.13.21.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McMilin K D, Russo V E A. J Mol Biol. 1972;68:49–55. doi: 10.1016/0022-2836(72)90261-6. [DOI] [PubMed] [Google Scholar]
- 72.McMilin K D, Stahl M M, Stahl F W. Genetics. 1974;77:409–423. doi: 10.1093/genetics/77.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mosig G, Gewin J, Luder A, Colowick N, Vo D. Proc Natl Acad Sci USA. 2001;98:8306–8311. doi: 10.1073/pnas.131007398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mosig G. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 75.Meselson M, Weigle J. Proc Natl Acad Sci USA. 1961;47:857–868. doi: 10.1073/pnas.47.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kraus E, Leung W-y, Haber J E. Proc Natl Acad Sci USA. 2001;98:8255–8262. doi: 10.1073/pnas.151008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Felzenszwalb I, Sargentini N J, Smith K C. Radiat Res. 1984;97:615–625. [PubMed] [Google Scholar]
- 78.Lombardo M-J, Rosenberg S M. J Bacteriol. 2000;182:6287–6291. doi: 10.1128/jb.182.22.6287-6291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]