Abstract
Background
The role of mitochondria in cancer is poorly understood. Ulcerative colitis (UC) is an inflammatory bowel disease that predisposes to colorectal cancer and is an excellent model to study tumor progression. Our goal was to characterize mitochondrial alterations in UC tumorigenesis.
Methods
Nondysplastic colon biopsies from UC patients with high-grade dysplasia or cancer (progressors; n = 9) and UC patients dysplasia free (nonprogressors; n = 9) were immunostained for cytochrome C oxidase (COX), a component of the electron transport chain, and were quantified by multispectral imaging. For six additional progressors, nondysplastic and dysplastic biopsies were stained for COX and additional mitochondrial proteins including PGC1α, the master regulator of mitochondrial biogenesis. Mitochondrial DNA (mtDNA) copy number was determined by quantitative polymerase chain reaction. Generalized estimating equations with two-sided tests were used to account for correlation of measurements within individuals.
Results
Nondysplastic biopsies of UC progressors showed statistically significant COX loss compared with UC nonprogressors by generalized estimating equation (−18.5 units, 95% confidence interval = −12.1 to −24.9; P < .001). COX intensity progressively decreased with proximity to dysplasia and was the lowest in adjacent to dysplasia and dysplastic epithelium. Surprisingly, COX intensity was statistically significantly increased in cancers. This bimodal pattern was observed for other mitochondrial proteins, including PGC1α, and was confirmed by mtDNA copy number.
Conclusions
Mitochondrial loss precedes the development of dysplasia, and it could be used to detect and potentially predict cancer. Cancer cells restore mitochondria, suggesting that mitochondria are needed for further proliferation. This bimodal pattern might be driven by transcriptional regulation of mitochondrial biogenesis by PGC1α.
Mitochondrial dysfunction has long been suspected to be involved in tumorigenesis (1). Cancers harbor somatic mitochondrial DNA (mtDNA) mutations, which are believed to be the consequence of reactive oxygen species (2). However, little is known about how mitochondrial alterations contribute to tumorigenesis. Warburg first suggested that mitochondrial dysfunction triggered aerobic glycolysis in cancer cells (3). However, recent research has demonstrated that most tumor mitochondria are functional and their metabolism is reprogrammed to support rapid cell growth (4). Mitochondrial dysfunction in cancer has typically been analyzed at the DNA level (1,2), but studies in the aging colon have demonstrated that immunostaining of cytochrome C oxidase (COX) is a good surrogate of mitochondrial function (5). COX is composed of 13 subunits (3 mtDNA encoded and 10 nuclear DNA encoded) and forms complex IV of the electron transport chain. In the aging colon, COX loss involves whole crypts, presumably as a consequence of mtDNA mutations that occur in the stem cells and then propagate along the crypt (5,6). COX loss has been reported in the normal colonic epithelium from patients with sporadic colorectal cancer (7,8), which suggests that it might precede neoplasia.
Ulcerative colitis (UC) is an inflammatory bowel disease that predisposes to colorectal cancer and is an excellent model to study mitochondrial dysfunction for two reasons. First, dysplasia arises in precancerous fields of histologically normal tissue that harbors molecular alterations (9–12) and then it progresses from low-grade dysplasia (LGD) to high-grade dysplasia (HGD) and cancer. Reactive oxygen species secondary to inflammation contribute to this progression (13) and are likely to cause mitochondrial damage. Second, metabolic (14–16) and proteomic studies (17–19) have reported mitochondrial dysfunction associated with UC and its progression to cancer. Thus, we hypothesized that COX loss is associated with tumor progression and could be used to discriminate UC patients with HGD and cancer (progressors) from UC patients who are dysplasia-free (nonprogressors). This hypothesis is tested in the first part of the study. The second part aims to characterize the mitochondrial changes that occur in UC tumorigenesis.
Methods
Patients and Samples
This study included two sets of patients (Table 1). The first set was designed as a case–control study (part 1) and included nine UC nonprogressors and nine UC progressors. Two to four biopsies that were negative for dysplasia were analyzed for each patient. Five non-UC colon biopsies were included as normal controls. The second set of patients (part 2) included six UC progressors from whom multiple biopsies with different stages of dysplasia were available. All UC patients were recruited at the University of Washington Medical Center in Seattle, Washington, with approval from the Human Subjects Review Board and written informed consent. Non-UC control biopsies were collected at the Cleveland Clinic Foundation in Cleveland, Ohio, under an institutional review boards waiver of consent. See Supplementary Methods (available online) for more information on patients and samples.
Table 1.
Patients and samples*
| Patient group | Diagnosis | Age, years | Sex | Disease duration, years | Age of onset, years | No. of biopsies analyzed, all negative for dysplasia |
|||
|---|---|---|---|---|---|---|---|---|---|
| Set 1 | |||||||||
| non-UC | Constipation | 20 | F | — | — | 1 | |||
| non-UC | Constipation | 26 | F | — | — | 1 | |||
| non-UC | Diverticulitis | 36 | M | — | — | 1 | |||
| non-UC | Diverticulitis | 43 | M | — | — | 1 | |||
| non-UC | Diverticulitis | 50 | M | — | — | 1 | |||
| UC NP | Negative | 20 | M | 12 | 8 | 4 | |||
| UC NP | Negative | 33 | F | 8 | 25 | 2 | |||
| UC NP | Negative | 43 | M | 13 | 30 | 2 | |||
| UC NP | Negative | 46 | F | 17 | 29 | 3 | |||
| UC NP | Negative | 51 | F | 20 | 31 | 3 | |||
| UC NP | Negative | 52 | F | 18 | 16 | 3 | |||
| UC NP | Negative | 57 | F | 8 | 49 | 4 | |||
| UC NP | Negative | 60 | M | 44 | 16 | 2 | |||
| UC NP | Negative | 77 | F | 24 | 53 | 2 | |||
| UC P | Cancer | 35 | M | 14 | 21 | 4 | |||
| UC P | Cancer | 40 | M | 20 | 20 | 4 | |||
| UC P | HGD | 47 | M | 20 | 27 | 4 | |||
| UC P | Cancer | 47 | F | 14 | 33 | 2 | |||
| UC P | HGD | 51 | M | 15 | 36 | 2 | |||
| UC P | HGD | 54 | M | 20 | 34 | 4 | |||
| UC P | Cancer | 61 | M | 8 | 53 | 3 | |||
| UC P | Cancer | 63 | M | >8 | NA | 3 | |||
| UC P | Cancer | 70 | F | 52 | 18 | 3 | |||
| Total | 59 | ||||||||
| No. of biopsies analyzed, highest dysplastic grade | |||||||||
| Negative | LGD | HGD | Cancer | ||||||
| Set 2 | |||||||||
| UC P | HGD | 48 | M | 10 | 38 | 3 | 1 | 1 | |
| UC P | HGD | 58 | M | 29 | 29 | 5 | 1 | 1 | |
| UC P | Cancer | 31 | M | 4 | 27 | 11 | 1 | NA | 2 |
| UC P | Cancer | 33 | F | 22 | 11 | 1 | 1 | 1 | 3 |
| UC P | Cancer | 36 | M | 8 | 28 | 3 | 1 | 1 | 1 |
| UC P | Cancer | 51 | F | 13 | 38 | 4 | 1 | 2 | 2 |
| Total | 27 | 6 | 6 | 8 | |||||
* F = female; HGD = high-grade dysplasia; LGD = low-grade dysplasia; M = male; NA = not available; NP = nonprogressor; P = progressor; UC = ulcerative colitis.
Immunohistochemistry and Quantification
Consecutive 5-micron sections were obtained from formalin-fixed, paraffin-embedded (FFPE) biopsies and stained with the appropriate primary antibody (Table 2) following standard procedures. Images were processed and quantified with a Nuance Multispectral Imaging System (CRI, Woburn, MA) and in-house software. See Supplementary Methods and Supplementary Figure 1 (available online).
Table 2.
Antibodies*
| Antibody name | Gene name | Gene symbol (HUGO ID) | Function | DNA encoded | Host species/ type |
Company | Catalog number | Dilution |
|---|---|---|---|---|---|---|---|---|
| Complex II subunit 30 kDa | Succinate dehydrogenase complex, subunit B, iron sulfur (Ip) | SDHB (10681) | ETC: succinate ubiquinone oxidoreductase | Nuclear | Mouse monoclonal | Invitrogen | 459230 | 1:100 |
| Complex III subunit Core 2 | Ubiquinol-cytochrome c reductase core protein II | UQCRC2 (12586) | ETC: ubiquinol cytochrome c oxidoreductase | Nuclear | Mouse monoclonal | Invitrogen | 459220 | 1:2000 |
| Complex IV subunit I (COX) | Mitochondrially encoded cytochrome c oxidase I | MT-CO1 (7419) | ETC: cytochrome c oxidase | Mitochondrial | Mouse monoclonal | Invitrogen | 459600 | 1:200 |
| Complex IV subunit II | Mitochondrially encoded cytochrome c oxidase II | MT-CO2 (7421) | ETC: cytochrome c oxidase | Mitochondrial | Mouse monoclonal | Invitrogen | A-6404 | 1:400 |
| Complex IV subunit Va | Cytochrome c oxidase subunit Va | COX5A (2267) | ETC: cytochrome c oxidase | Nuclear | Mouse monoclonal | Invitrogen | 459120 | 1:200 |
| Prohibitin | Prohibitin | PHB (8912) | Mitochondrial chaperone and scaffold | Nuclear | Rabbit polyclonal | Novus Biologicals | NBP1-45816 | 1:400 |
| Opa1 | Optic atrophy 1 | OPA1 (8140) | Mitochondrial fission and biogenesis | Nuclear | Rabbit polyclonal | Novus Biologicals | NB110-55290 | 1:200 |
| PGC1α | Peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | PPARGC1A (9237) | Master regulator of mitochondrial biogenesis | Nuclear | Rabbit polyclonal | Gift from Dr Moschetta | NA | 1:1000 |
| MCT4 | Solute carrier family 16, member 3 (monocarboxylic acid transporter 4) | SLC16A3 (10924) | Lactate transporter in cytoplasmic membrane | Nuclear | Rabbit polyclonal | Santa Cruz Biotech | sc-50329 | 1:150 |
* ETC = electron transport chain; NA = not applicable.
Inflammation Quantification
FFPE slides were stained with hematoxylin and eosin and examined under a light microscope for chronic inflammation (lymphocytes in lamina propria) and acute inflammation (neutrophils in epithelium and cryptitis).
mtDNA Copy Number Quantification
mtDNA was extracted from frozen UC colon biopsies from the six UC progressor patients in Table 1, set 2. The maximum histological grade of each biopsy was determined by pathological examination of a paired FFPE biopsy collected at the same location. Most paired FFPE biopsies were previously analyzed for COX. The number of biopsies in each histological grade were as follows: 10 negative located 10cm or more from dysplasia, 14 negative located less than 10cm from dysplasia, 9 LGD, 6 HGD, and 5 cancers. The number of mtDNA copies in total DNA extracted from epithelial cells was measured by quantitative polymerase chain reaction (PCR) as the difference in amplification cycles between a mitochondrial PCR and a control nuclear PCR (see Supplementary Methods, available online).
Telomere Fluorescence In Situ Hybridization
Paraffin-embedded, lightly paraformaldehyde-fixed slides were hybridized with a telomere probe conjugated to fluorescein isothiocyanate, as previously described [(20) and Supplementary Methods, available online]. Consecutive pictures of adjacent colon crypts were taken at ×63 with a Zeiss 510 META confocal microscope (Carl Zeiss, Germany). In Photoshop, these pictures were compiled into a single image that reconstructed the crypt morphology of the tissue to facilitate the comparison of telomeres with COX staining at the crypt level. For each nuclei, telomere mean intensity was quantified using previously described algorithms (20,21). For each crypt, telomere length was calculated as the mean intensity from all nuclei divided by the mean intensity of the adjacent stroma.
Statistical Analysis
The method of generalized estimating equations (22) was used to account for correlation of measurements within individuals (lack of independence) when assessing the magnitude and statistical significance of differences in COX values by patient diagnostic group, distance to dysplasia, inflammation, and stage of progression (see Supplementary Methods, available online). Generalized estimating equations analyses were done using R with “geepack” functions (http://www.r-project.org). All the tests were two-sided at an alpha level (type 1 error rate) of 0.05.
Results
Comparison of COX Staining in Nondysplastic Biopsies of UC Progressors and Nonprogressors
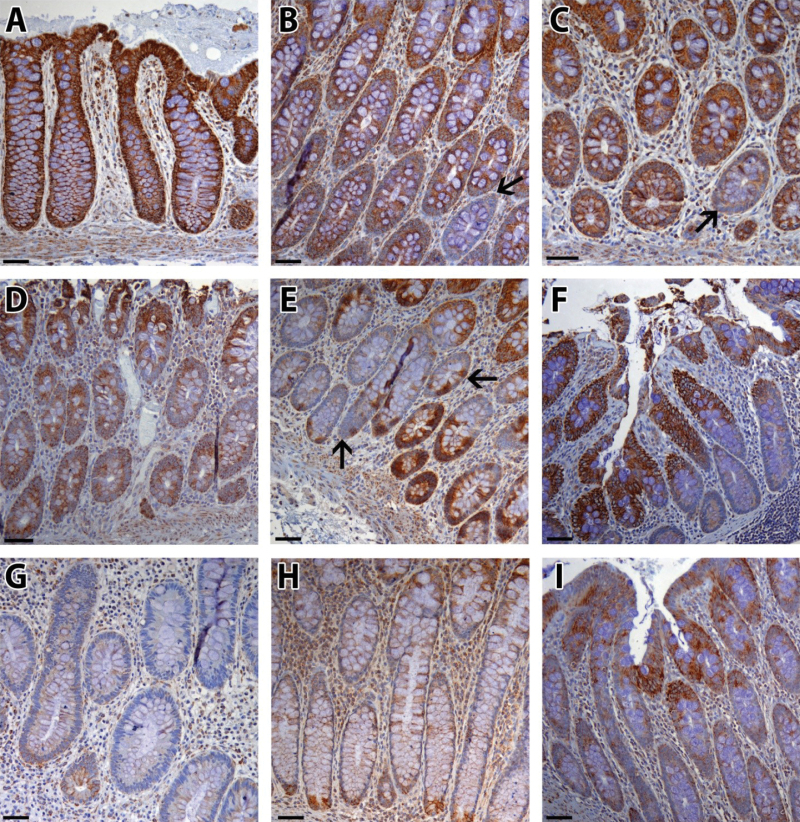
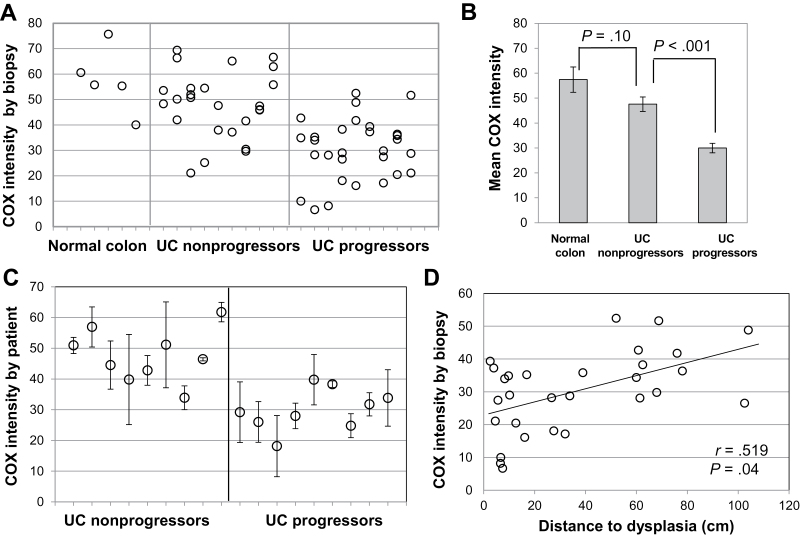
Colon biopsies from UC progressors, nonprogressors, and non-UC patients were stained for COX subunit I (Table 1). COX expression was high and uniform in normal colon epithelium, indicating intact mitochondrial function (Figure 1, A–C). Sporadically, we observed rare whole crypts completely devoid of COX (Figure 1, B and C, arrows), in agreement with findings from aging colon (5). By comparison, in UC epithelium, most COX loss followed patterns not previously described. In UC nonprogressors, COX expression was usually high (Figure 1D), but occasionally patches of COX loss were found that affected only portions of adjacent crypts (Figure 1E, arrows) or the bottom half of crypts (Figure 1F). In UC progressors, there were large areas of COX loss (Figure 1G), but sometimes the base of the crypts (Figure 1H) or the surface epithelium (Figure 1I) remained COX positive. COX loss in UC progressors compared with nonprogressors was obvious by visual examination of the slides and was confirmed by quantification of COX intensity (Figure 2). Three representative pictures from each biopsy were quantified, and the average COX intensity for each biopsy was plotted for each patient (Figure 2A). The average COX intensity was statistically significantly lower among UC progressors than among nonprogressors (−18.5 units; 95% confidence interval [CI] = −12.1 to −24.9; P < .001) (Figure 2B). Although UC nonprogressors had some COX loss compared with normal (non-UC) colons, that difference was not statistically significant (P = .10). In spite of the large variation of COX staining between biopsies from the same UC patient, the mean COX intensity for each patient could distinguish UC progressors from nonprogressors with 100% sensitivity and 77.7% specificity (9 of 9 progressors and 2 of 9 nonprogressors had mean COX intensity < 40) (Figure 2C). Interestingly, COX intensity and distance to dysplasia were statistically significantly correlated (slope = 0.18 units/cm, 95% CI = 0.01 to 0.34; P = .04) (Figure 1D), which indicates a progressive defect of loss of mitochondria that might precede dysplasia. There was no association between COX intensity and distance to rectum for either nonprogressors or progressors. COX intensity was not statistically significantly associated with either chronic or acute inflammation.
Figure 1.
Representative cytochrome C oxidase (COX) staining patterns. A–C) Normal colon. Black arrows in (B) and (C) indicate crypts with complete COX loss, consistent with previous findings in aging colon. D–F) Colon biopsies from ulcerative colitis (UC) nonprogressors. Black arrows in (C) show COX-positive cells alternating with COX-negative cells. G–I) Nondysplastic biopsies from UC progressors. Scale bar = 100 µm.
Figure 2.
Quantification of cytochrome C oxidase (COX) staining in nondysplastic biopsies from ulcerative colitis (UC) progressors and nonprogressors. A) COX intensity by biopsy. Each dot represents a single biopsy. Biopsies in the same column are from the same patient. Two to four biopsies from nine UC nonprogressors and nine UC progressors are shown, together with five normal colon control biopsies. B) Group means for COX intensity. Bars indicate generalized estimating equations standard deviation (SD). P values correspond to two-sided mean comparisons from generalized estimating equations. C) COX intensity by patient. Each dot represents the mean COX intensity for all the biopsies analyzed for each patient. Bars indicate SD. D) Correlation between COX intensity and distance to dysplasia for nondysplastic biopsies from UC progressors. Pearson correlation coefficient and two-sided P value are shown.
Mitochondrial Changes in UC Tumorigenesis
In the second part of this study, we investigated COX expression in dysplasia and cancer. Figure 3, A–D, shows representative pictures from increasingly dysplastic tissue from one UC progressor. The nondysplastic epithelium adjacent to cancer and the epithelium with LGD had very low levels of COX expression; however, there was a clear increase of COX expression in cancer. To assess these changes quantitatively, a total of 20 dysplastic biopsies (n = 6 LGD; n = 6 HGD; n = 8 cancers, based on highest grade) from six progressor patients were stained (Table 1, set 2). Each biopsy contained areas with different histologic grades, and representative pictures were taken from each, including 34 areas of negative epithelium adjacent to dysplasia, 40 areas of LGD, 30 areas of HGD, and 28 areas of cancer. In addition, 79 areas of nondysplastic epithelium were also quantified (average of 3 random pictures from 27 nondysplastic biopsies) and were grouped according to their distance to dysplasia (≥10 cm: 39 areas; <10 cm: 40 areas). The cutoff point of 10cm was based on Figure 2D and previous data showing increased molecular alterations at 10cm from dysplasia (12). Thus, for these progressors, COX was analyzed across the full spectrum of tumor progression, represented by six categories: negative 10cm or more from dysplasia, negative less than 10cm from dysplasia, negative adjacent to dysplasia, LGD, HGD, and cancer. The result was a bimodal distribution of COX intensity (Figure 3E). In agreement with Figure 2D, in this independent set of progressors, we observed a decrease of COX with increasing proximity to dysplasia. There was a statistically significant decrease in COX across histologic grades from negative greater than 10cm to LDG (slope = −3.04 intensity units decrease per unit increase in progression; P = .002). After adjusting for the random effect of each patient (ie, the individual-level COX value), the average decrease of COX within patient was larger and more statistically significant than the unadjusted effect (−3.8 per unit increase in progression; P < .001), indicating a strong decrease within each patient but different intercepts for each. Although Figure 3C (HGD) shows more COX than Figure 3B (LGD), there was variability in the levels of COX staining in both stages. However, cancers had statistically significantly higher levels of COX than both LGD (P = .04) and HGD (P = .03). The individual plots for each patient are shown in Supplementary Figure 2 (available online). It is remarkable that the two patients with HGD as their highest degree of neoplasia at colectomy showed mostly a decrease in COX across progression (Supplementary Figure 2, A and B), whereas the four patients that developed cancer showed an initial decrease and a later increase (Supplementary Figure 2, C–F, available online).
Figure 3.
Mitochondrial changes in ulcerative colitis (UC) tumorigenesis. A–D) Representative example of increased cytochrome C oxidase (COX) staining with tumor progression. Brown: COX; blue: hematoxylin. Scale bar = 100 µm. E.)Quantification of COX intensity in nondysplastic and dysplastic biopsies from six progressor patients (set 2). Nondysplastic (negative) biopsies were divided by distance to dysplasia: equal to or greater than 10cm and less than 10cm. In dysplastic biopsies, all histological grades present in a given biopsy were quantified (negative adjacent [Neg adj], low-grade dysplasia [LGD], high-grade dysplasia [HGD], and cancer). The number of pictures analyzed for each histological grade is indicated in parenthesis. F) Quantification of mitochondrial DNA (mtDNA) copy number in nondysplastic and dysplastic biopsies from six progressor patients [same as in (E)]. The number of mtDNA copies was estimated by quantitative polymerase chain reaction using total DNA extracted from frozen biopsies whose maximum histological grade was previously determined by pathological examination of adjacent formalin-fixed, paraffin-embedded biopsies. Numbers in parentheses indicate number of biopsies of each given grade. For (E) and (F), box and whisker plots depict the median (black line), interquartile range (box), and maximum and minimum (ends of whiskers). The trend for the decrease of COX and mtDNA was quantified and tested for a difference from slope of zero using the two-sided Wald test from generalized estimating equations; the same test was applied to test for nonzero differences in mean between LGD, HGD, and cancer. Neg = Negative for dysplasia.
To investigate whether this bimodal pattern affected only COX or the whole mitochondria, we quantified mtDNA copy number in paired frozen biopsies adjacent to the FFPE biopsies used for COX staining. mtDNA was quantified by quantitative PCR from total DNA extracted from epithelial cells. In full agreement with COX data, we observed a statistically significant decrease of mtDNA copy number in negative greater than 10cm, negative less than 10cm, and LGD biopsies (P trend = .02). After adjusting for the random effect of each patient, the within-patient trend was still statistically significant (P = .03). Cancer biopsies showed a highly statistically significant increase of mtDNA molecules compared with LGD biopsies (mean difference = 299.1, 95% CI = 199.7 to 398.4; P < .001) and HGD biopsies (mean difference = 404.1, 95% CI = 321.7 to 486.3; P < .001), adding strong statistical evidence to the previous COX results.
Other Mitochondrial Markers and PGC1α in UC Tumorigenesis
To further confirm the preneoplastic loss and neoplastic gain of mitochondria, we stained serial sections of biopsies with the following antibodies: prohibitin, a mitochondrial chaperone (23); Opa1, a GTPase that regulates mitochondrial shape and fusion (24); and four proteins of the electron transport chain, both nuclear and mtDNA encoded (Table 2). Prohibitin and Opa1 showed a staining pattern very similar to that of COX (Supplementary Figure 3, available online), as well as the four electron transport chain proteins, regardless of whether they were nuclear or mitochondrial encoded (Supplementary Figure 4, available online).
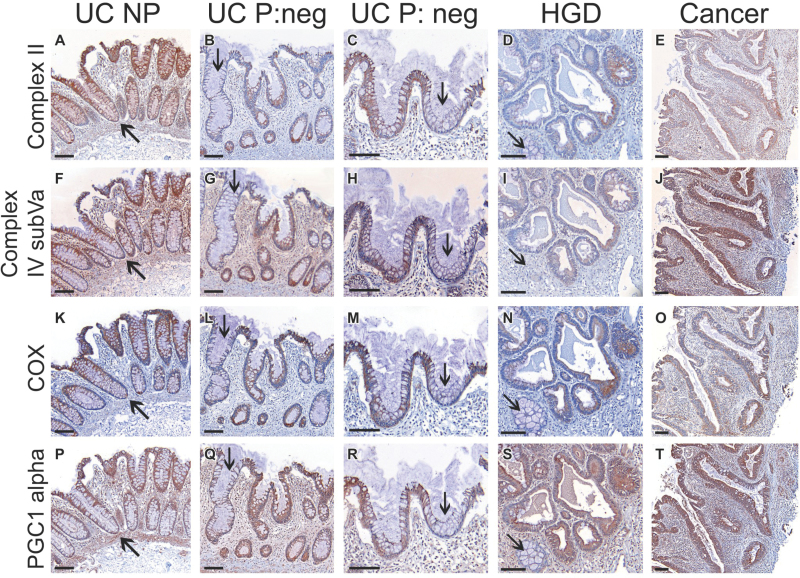
Mitochondrial biogenesis is regulated by peroxisome proliferator-activated receptor gamma coactivator 1alpha (PGC1α or PPARGC1A), which is a transcription factor that controls production of mitochondrial proteins (25). To determine whether the bimodal mitochondrial pattern was the result of PGC1α action, we stained serial sections of representative biopsies for COX, two electron transport chain proteins (complex II and complex IV subVa), and PGC1α (Figure 4). Again, we observed striking similarities in the staining patterns of the four markers, suggesting that PGC1α could be the driver of the mitochondrial changes observed here. Additional images showing evidence of concordant staining between COX and PGC1α are shown in Supplementary Figure 5 (available online). Note that we observed infrequent exceptions to this pattern, which corresponded to isolated crypts with complete COX loss in spite of high PCG1α expression (Supplementary Figure 5, A and B, black arrows). As mentioned before, these sporadic crypts are in agreement with the findings of clonally expanded mtDNA mutations in aging colon (5,6). In UC patients, these age-related, COX-negative crypts appear to be superimposed on a more global pattern of COX deficiency that appears to be driven by PGC1α.
Figure 4.
Serial staining of mitochondrial markers in representative biopsies of ulcerative colitis (UC) nonprogressors (NP) and progressors (P). A–E) Complex II subunit 30KDa (nuclear encoded). F–J) Complex IV subunit Va (nuclear encoded). K–O) Complex IV subunit I (cytochrome C oxidase [COX], mitochondrial encoded). P–T) PGC1α. Black arrows indicate concordant regions of COX loss. Scale bar = 100 µm. HGD = high-grade dysplasia; Neg = negative for dysplasia.
Other Cellular Mechanisms Associated With Mitochondrial Loss: Telomere Shortening and Glycolysis
Mitochondrial loss could be the consequence of shorter telomeres, which indirectly repress PGC1α (26).We previously demonstrated that, in UC tumorigenesis, telomeres first shorten and then lengthen (12) in a pattern strikingly similar to the mitochondrial results described here. Thus, we investigated a potential association between telomere shortening and mitochondrial loss by performing telomere fluorescence in situ hybridization in serial sections from biopsies previously stained and quantified for COX. We confirmed that mitochondria and telomeres follow the same U-shaped pattern in tumor progression (Supplementary Figure 6, A and B, available online). However, the association was not always true at the crypt level. Whereas some crypts contained very little COX (Supplementary Figure 6C, crypt 1, available online) and extremely short telomeres (Supplementary Figure 6, E and G, crypt 1, available online), others had low COX (Supplementary Figure 6D, crypt 2, available online) but long telomeres (Figure 5, F and H, crypt 2).
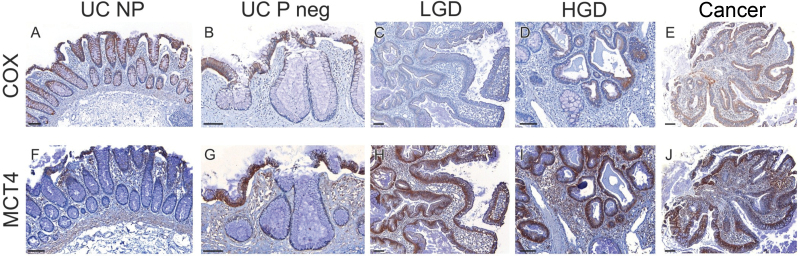
Figure 5.
Comparison of respiration and glycolysis in ulcerative colitis (UC) tumorigenesis. Serial staining in representative biopsies of UC nonprogressors (NP) and progressors (P) for cytochrome C oxidase (COX) (A–E) and MCT4, a marker of glycolysis (F–J). Scale bar = 100 µm. HGD = high-grade dysplasia; Neg = negative for dysplasia.
Historically, glycolysis in tumors has been attributed to mitochondrial dysfunction (Warburg effect) (3), which disagrees with our findings of mitochondria recovery in cancer. Thus, we explored glycolysis at the different stages of UC tumorigenesis. Serial sections of representative biopsies were stained with COX and with MCT4 (Table 2), a lactate exporter considered to be a reliable marker of glycolysis (Figure 5) (27). Lactate is an end-product of glycolysis that cells export by MCT4 to avoid acidification of the cytoplasm. Nondysplastic epithelium from UC nonprogressors and progressors was negative for MCT4, except at the surface epithelium. MCT4, however, was highly expressed in most epithelial cells from LGD, HGD, and cancer. This is in contrast to COX, which was negative in LGD, only partially positive in HGD, and fully positive in cancer.
Discussion
The analysis of COX staining in UC biopsies has been exceptionally informative at two different levels. At the biomarker level, COX loss precedes tumor progression in UC, and thus it could potentially be used as a biomarker to distinguish UC patients with cancer. At the biological level, the loss of COX represents a reduction of the number of mitochondria in preneoplasia, which, surprisingly, is restored in cancers. This early decrease and later increase of mitochondria can also be detected by quantification of mtDNA copy number and by immunohistochemistry of other mitochondrial proteins, and it appears to be driven by PGC1α, the master regulator of mitochondrial biogenesis.
Regarding the potential of COX as a useful cancer biomarker, is worth noting that, in this preliminary study, we could discriminate progressors from nonprogressors with 100% sensitivity and 77.7% specificity by analyzing only two to four colonic biopsies per patient. The value of this approach is that these biopsies are random, histologically normal biopsies, which can detect the presence of HGD or cancer elsewhere in the colon because most of the colon is affected with mitochondrial dysfunction (field effect) in patients that have developed cancer. Actually, mitochondrial dysfunction is more prominent closer to dysplasia; therefore, by analyzing a larger number of biopsies, the ability to distinguish progressors based on random biopsies should increase even further.
The main limitation of this study is the small number of patients. This was an exploratory study, which will have to be validated in larger cohorts of patients. Another limitation is its cross-sectional design. Ideally, UC cancer biomarkers should be evaluated in retrospective or prospective studies so that longitudinal information about the progression status of the patient is available. In this study, two nonprogressors showed low levels of COX. Longitudinal information would allow to determine whether these two patients are truly nonprogressors with low COX (false positive) or patients that eventually developed cancer (future progressors). The advantage of COX is that it can be tested in FFPE biopsies, which are the archival material clinically available for retrospective studies. This makes it an ideal biomarker for cancer prediction in UC.
The pattern of COX loss observed in UC is different from the pattern described in aging colon (5,6) and sporadic colorectal cancer (7,8). These studies reported loss of COX in segments of crypts, whole crypts, and groups of crypts. This was attributed to mtDNA mutations that occur in colonic stem cells and propagate through the crypt and then the crypts expand by fission to generate groups of crypts that are completely COX negative. In our study, we observed loss of COX in irregular patches that do not fit with a crypt-based boundary. Moreover, we provided evidence that COX loss reflects mitochondrial loss, as demonstrated by mtDNA quantification and immunohistochemistry staining of multiple mitochondrial proteins (both nuclear and mtDNA encoded). In addition, mitochondrial loss appears to be the consequence of downregulation by PGC1α, the master regulator of mitochondrial biogenesis (25). Sporadically, we observed crypts with complete COX loss, which closely resembled the ones described in aging colon. Serial staining with PGC1α showed normal expression, indicating that the loss of COX in these rare crypts is not due to mitochondrial downregulation. MtDNA sequencing is currently under way to confirm COX mutations in those crypts. Rare colon crypts with mutated COX could also account for crypt to crypt discrepancies between telomere length and mitochondrial loss. The fact that both cellular events show identical bimodal distributions raises the possibility of a potential association, which is strengthened by the recently discovered link between short telomeres and mitochondrial dysfunction in telomerase knockout mice (28).Our study had limited amount of samples to explore this association in detail, but it will be the subject of future research.
A striking finding of this study is the recovery of mitochondria in tumors, which occurs in addition to the acquisition of glycolysis in LGD. Interestingly, cells in LGD typically show low mitochondrial content, which might trigger glycolysis as an alternative mechanism of ATP production. At the cancer stage, however, glycolysis is still present, but the number of mitochondria dramatically increases, which suggests a positive selection of mitochondria to allow further cell proliferation. This is in agreement with the concept that cancer metabolism is not the consequence of mitochondrial dysfunction but the result of mitochondrial reprogramming to meet the challenges of macromolecular synthesis (4). The current view is that cancer metabolism is a dynamic process (29) in which waves of gene regulation might initially suppress mitochondrial biogenesis and increase glycolysis but eventually restore oxidative phosphorylation in cancer cells (30). Our results offer unique in vivo data that fully supports this view.
Funding
This work was supported by the National Institutes of Health (K07 CA137136 to RAR and R01 CA068124 to TAB) and the Crohn’s and Colitis Foundation of America.
Supplementary Material
The authors are solely responsible for the study design, data collection, analysis and interpretation of the data, writing the manuscript, and decision to submit the manuscript for publication.
The authors have declared that no competing interests exist.
We thank Arielle Samuelson, Trang Lee, and Tara McIntyre for technical assistance; and Yasuko Tamura for assistance with tissue retrieval. We are grateful to Dr Mary Bronner for providing normal colon biopsies and to Dr Antonio Moschetta for providing the PGC1α antibody. We gratefully acknowledge the use of the Nuance microscope system (Cambridge Research Instrumentation), which is under the supervision of Dr Lawrence True. Finally, we thank Dr Jason Bielas for his helpful advice and Dr Lisa Lai, Dr Jesse Salk, and Dr Lawrence Loeb for their critical review of the manuscript.
References
- 1. Chatterjee A, Dasgupta S, Sidransky D. Mitochondrial subversion in cancer. Cancer Prev Res (Phila). 2011;4(5):638–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yu M. Somatic mitochondrial DNA mutations in human cancers. Adv Clin Chem. 2012;57:99–138 [DOI] [PubMed] [Google Scholar]
- 3. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314 [DOI] [PubMed] [Google Scholar]
- 4. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor RW, Barron MJ, Borthwick GM, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112(9):1351–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Greaves LC, Preston SL, Tadrous PJ, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci U S A. 2006;103(3):714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bernstein C, Facista A, Nguyen H, et al. Cancer and age related colonic crypt deficiencies in cytochrome c oxidase I. World J Gastrointest Oncol. 2010;2(12):429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Payne CM, Holubec H, Bernstein C, et al. Crypt-restricted loss and decreased protein expression of cytochrome C oxidase subunit I as potential hypothesis-driven biomarkers of colon cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14(9):2066–2075 [DOI] [PubMed] [Google Scholar]
- 9. Brentnall TA, Crispin DA, Rabinovitch PS, et al. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology. 1994;107(2):369–378 [DOI] [PubMed] [Google Scholar]
- 10. Burmer GC, Rabinovitch PS, Haggitt RC, et al. Neoplastic progression in ulcerative colitis: histology, DNA content, and loss of a p53 allele. Gastroenterology. 1992;103(5):1602–1610 [DOI] [PubMed] [Google Scholar]
- 11. Rabinovitch PS, Dziadon S, Brentnall TA, et al. Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res. 1999;59(20):5148–5153 [PubMed] [Google Scholar]
- 12. Risques RA, Lai LA, Himmetoglu C, et al. Ulcerative colitis-associated colorectal cancer arises in a field of short telomeres, senescence, and inflammation. Cancer Res. 2011;71(5):1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roessner A, Kuester D, Malfertheiner P, et al. Oxidative stress in ulcerative colitis-associated carcinogenesis. Pathol Res Pract. 2008;204(7):511–524 [DOI] [PubMed] [Google Scholar]
- 14. Santhanam S, Rajamanickam S, Motamarry A, et al. Mitochondrial electron transport chain complex dysfunction in the colonic mucosa in ulcerative colitis. Inflamm Bowel Dis. 2012;18(11):2158–2168 [DOI] [PubMed] [Google Scholar]
- 15. Santhanam S, Venkatraman A, Ramakrishna BS. Impairment of mitochondrial acetoacetyl CoA thiolase activity in the colonic mucosa of patients with ulcerative colitis. Gut. 2007;56(11):1543–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sifroni KG, Damiani CR, Stoffel C, et al. Mitochondrial respiratory chain in the colonic mucosal of patients with ulcerative colitis. Mol Cell Biochem. 2010;342(1–2):111–115 [DOI] [PubMed] [Google Scholar]
- 17. Brentnall TA, Pan S, Bronner MP, et al. Proteins that underlie neoplastic progression of ulcerative colitis. Proteomics Clin Appl. 2009;3(11):1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsieh SY, Shih TC, Yeh CY, et al. Comparative proteomic studies on the pathogenesis of human ulcerative colitis. Proteomics. 2006;6(19):5322–5331 [DOI] [PubMed] [Google Scholar]
- 19. May D, Pan S, Crispin DA, et al. Investigating neoplastic progression of ulcerative colitis with label-free comparative proteomics. J Proteome Res. 2011;10(1):200–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O’Sullivan JN, Bronner MP, Brentnall TA, et al. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat Genet. 2002;32(2):280–284 [DOI] [PubMed] [Google Scholar]
- 21. O’Sullivan JN, Finley JC, Risques RA, et al. Telomere length assessment in tissue sections by quantitative FISH: image analysis algorithms. Cytometry. 2004;58A(2):120–131 [DOI] [PubMed] [Google Scholar]
- 22. Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73(1):13–22 [Google Scholar]
- 23. Osman C, Merkwirth C, Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J Cell Sci. 2009;122(Pt 21):3823–3830 [DOI] [PubMed] [Google Scholar]
- 24. Zorzano A, Liesa M, Sebastian D, et al. Mitochondrial fusion proteins: dual regulators of morphology and metabolism. Semin Cell Dev Biol. 2010;21(6):566–574 [DOI] [PubMed] [Google Scholar]
- 25. Aquilano K, Vigilanza P, Baldelli S, et al. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285(28):21590–21599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464(7288):520–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dimmer KS, Friedrich B, Lang F, et al. The low-affinity monocarboxylate transporter MCT4 is adapted to the export of lactate in highly glycolytic cells. Biochem J. 2000;350(Pt 1):219–227 [PMC free article] [PubMed] [Google Scholar]
- 28. Sahin E, Colla S, Liesa M, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor’s dilemma? Biochim Biophys Acta. 2011;1807(6):552–561 [DOI] [PubMed] [Google Scholar]
- 30. Smolkova K, Plecita-Hlavata L, Bellance N, et al. Waves of gene regulation suppress and then restore oxidative phosphorylation in cancer cells. Int J Biochem Cell Biol. 2011;43(7):950–968 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.