Abstract
Background
The Ice krill, Euphausia crystallorophias is one of the species at the base of the Southern Ocean food chain. Given their significant contribution to the biomass of the Southern Ocean, it is vitally important to gain a better understanding of their physiology and, in particular, anticipate their responses to climate change effects in the warming seas around Antarctica.
Methodology/Principal Findings
Illumina sequencing was used to produce a transcriptome of the ice krill. Analysis of the assembled contigs via two different methods, produced 36 new pre-pro-peptides, coding for 61 neuropeptides or peptide hormones belonging to the following families: Allatostatins (A, B et C), Bursicon (α and β), Crustacean Hyperglycemic Hormones (CHH and MIH/VIHs), Crustacean Cardioactive Peptide (CCAP), Corazonin, Diuretic Hormones (DH), the Eclosion Hormone (EH), Neuroparsin, Neuropeptide F (NPF), small Neuropeptide F (sNPF), Pigment Dispersing Hormone (PDH), Red Pigment Concentrating Hormone (RPCH) and finally Tachykinin. LC/MS/MS proteomics was also carried out on eyestalk extracts, which are the major site of neuropeptide synthesis in decapod crustaceans. Results confirmed the presence of six neuropeptides and six precursor-related peptides previously identified in the transcriptome analyses.
Conclusions
This study represents the first comprehensive analysis of neuropeptide hormones in a Eucarida non-decapod Malacostraca, several of which are described for the first time in a non-decapod crustacean. Additionally, there is a potential expansion of PDH and Neuropeptide F family members, which may reflect certain life history traits such as circadian rhythms associated with diurnal migrations and also the confirmation via mass spectrometry of several novel pre-pro-peptides, of unknown function. Knowledge of these essential hormones provides a vital framework for understanding the physiological response of this key Southern Ocean species to climate change and provides a valuable resource for studies into the molecular phylogeny of these organisms and the evolution of neuropeptide hormones.
Introduction
Krill (Eucarida, Malacostraca) are a keystone species of the Southern Ocean food chain. The best-known member of this family of crustaceans is the Antarctic krill stricto sensu Euphausia superba. It is also the most abundant, forming 50% of the Antarctic zooplankton biomass of the Southern Ocean. However this family of Euphausiidae comprises several members [1], all having circumpolar distributions, with their range largely governed by temperature tolerances. Within the Southern Ocean, there is a decrease in water surface temperature with higher latitudes and the different species are present in a latitudinal succession. Hence they present ideal models for phylogenetically controlled studies to understand the underlying mechanisms into species resilience/sensitivity to global warming. E. superba occurs mainly south of 60°S, but north of 74°S, which is co-incident with the shelf-break waters. There is virtually no geographical overlap with the distribution of the Ice krill, Euphausia crystallorophias. This species is epi-pelagic and restricted to the inshore waters of the continental plateau at depths of less than 300 metres [2], where it is the dominant euphausiid. Whilst E. superba has been found in water temperatures up to 3.9°C, E. crystallorophias is more stenothermal and only found in waters below 0°C.
These two euphausiids clearly represent sentinel species in the study of the impact of global warming in this region. Molecular studies were initiated in order to gain a better understanding of their physiology and anticipate their responses in the face of diverse environmental stresses. A comprehensive knowledge of these genomes at the DNA level is unlikely in the near future due to their large sizes (approximately 33pg for E. crystallorophias and 48pg for E. superba) [3]. However, transcriptome sequencing, particularly using HiSeq technologies constitutes a very attractive option because sequence quantity outputs are continually increasing and costs decreasing. E. superba was the first of these two species to be sequenced [4]. In that study, 454 sequencing allowed the preliminary characterisation of putative stress genes, to identify potential molecular markers for environmental change. A similar global approach was also taken in this study, with E. crystallorophias, using the Next Generation Sequencing Illumina Hi-Seq platform. With this data we have been able to undertake a detailed study into the principal neuropeptides and neurohormones of both of these species.
This paper describes the characterisation of these sequences in both E. crystallorophias and E. superba. The previously published 454 transcriptome of E. superba was developed from RNA extracted from whole animals, whilst the Illumina Hi-Seq dataset of E. crystallorophias was generated from whole animal RNA supplemented with RNA from eyestalks. This latter strategy increased the success of identifying relevant transcripts, as these peptides are produced in the major neuroendocrine organ of the crustaceans, the X organ-sinus gland complex, which is located in the eyestalks. The peptides were chosen because of their physiological importance in crustaceans and insects, many were thought (but not proven) to be present in the neuroendocrine system of the crustaceans. The pre-pro-peptides were annotated using Blast2GO and were confirmed by alignment with orthologous sequences already identified in other species of crustaceans and hexapods. The sequence data was further supplemented with proteomic mass spectrometry analysis on E. crystallorophias eyestalk extracts. The peptide sequencing results were used to validate the assemblies and also confirm the predicted gene structures of the neuropeptides and their associated cryptic peptides. The majority (90%) of these peptides have never been characterised in the Malacostraca outside of the Decapoda or within the Crustacea with the main exceptions of the daphnias, Daphnia magna [5] and D. pulex [6], [7] with genome data available for the latter (www.fleabase.org) (Table 1).
Table 1. List of mature peptides and precursor related peptides (PRP) of E. crystallorophias and E. superba.
| Peptide name | Peptide sequence | Previous identification in Arthropods | Pfam/Interpro accession N° | |
| Referenced peptides | Generic peptides | |||
| Corticotropin RF family | Daphnia | PF00473/IPR000187 | ||
| Euc-CRFLDH56 | GWRGLGARYARSRPQGLSLSIDASMKVLREALYLEIARKKQRQHQLRAAHNHQLLQ– | |||
| Neuropeptide F/Y family | Daphnia, decapod, insects | PF00159/IPR001955 | ||
| Euc-NPF1 | KPDPTQLAAMADAIKYLQELDKYYSQVARPRFa | Daphnia,Decapod (L. vannamei) | ||
| Euc-NPF1-L | KPDPTQLAAMADAIKYLQELDKYYSQVARPSTRSAPSTGAGKIDVLENTLKFLQLQELGKLYNVRARPRFa | Decapod (Litopenaeus vannamei) | ||
| Euc-NPF2 | - - - - - - - - - - - - - - - - -YLELLNRYYAIAGRPRFa | None | ||
| Euc-NPF1-PRP | SEYAMAPRDALMEASEKLMETFEHQR | Decapod (Litopenaeus vannamei) | ||
| Tachykinin Related Pept. | Daphnia, decapods and insects, | -/IPR013206 | ||
| Euc-TKRP | APSGFLGMRa | - | ||
| Euc-TKRP-PRP | pQVDPLSDALDQNQLAQTLYDYRD | None | - | |
| Arthropod specific peptides | ||||
| Allatostatins A family | Cirriped, copepod, daphnia, decapods and insects | PF05953/IPR010276 | ||
| Euc-AST A1 | IPGYSFGLa | |||
| Euc-AST A2 | QRQNKAYSFGLa | |||
| Euc-AST A3 | ARNYAFGIa | |||
| Euc-AST A4 | AKSYAFGLa | |||
| Euc-AST A5 | ANNMYSFGLa | |||
| Euc-AST A6 | GDGYNFGLa | |||
| Euc-AST A7 | DNSYGFGLa | |||
| Euc-AST A8 | GGNNMYGFGLa | |||
| Euc-AST A9 | GGKSYGFGLa | |||
| Euc-AST A10 | GKGAYSFGLa | |||
| Euc-AST A11 | GGDKMYGFGLa | |||
| Euc-AST A12 | ASDYGFGLa | |||
| Euc-AST A13 | VRDPYSFGLa | |||
| Euc-AST A14 | EPYAFGLa | |||
| Euc-AST A15 | SYDFGLa | |||
| Euc-AST A16 | SGPYSFGIa | |||
| Euc-AST A-PRP1 | QDTSSQIDTQQLLQALRELRELYSSRGYTFGN | Cirriped, copepod, decapods, insects | ||
| Euc-CCAP | PFCNAFTGCa | Ixod, daphnia, decapods and insects | PF11105/IPR024276 | |
| CHH family | Daphnia, isopod, decapods and insects | PF01147/IPR001166 | ||
| Euc-CHH | SIFDPSCKGFYNKEVFKKLNHICDDCYNLYRDASVAVKCKENCFGNPVFEQCIYELLIDDQVDELSKIVRTLa | |||
| Euc-MIH/VIH1 | NVAHLSSCGSLAGQRHIHRQVEQICLDCDNLYRQSRAGYNCRQSCYANPHFELCVHDLLLSHRVMEFRLLISMLQASL | Isopod, decapods | ||
| Euc-MIH/VIH2 | CGSIFGQRHIALKVEQVCRDCENLSRNYQTAFNCRKDCYTSETYTKCL – | Decapods (peneids) | ||
| Corazonin family | Ixod, daphnia, decapods and insects | -/IPR020190 | ||
| Euc-Arg7-CRZ1 | pQTFQYSRGWTNa | |||
| Eus-Arg7-CRZ1 | pQTFQYSRGWTNa | |||
| Euc- Ser4- Arg7 -CRZ2 | pQTFSYSRGWTNa | None | ||
| Eus- Ser4- Arg7 -CRZ2 | pQTFEYSRGWTNa | idem | ||
| Euc-Arg7-CRZ1-PRP3 | MMIQQSFEERIRNLEQELGDVY | Insects (A. gambiae, T. castaneum) | ||
| Euc-Eclosion Hormone | SYTGMCIRNCGQCKDMYGAYFNSQSCAESCIMSQGNSVPDCNNPSTFKNFL | Cirriped, daphnia, decapods and insects | -/IPR006825 | |
| Neuroparsin family | Copepod, daphnia and insects | PF07327/IPR010850 | ||
| Euc-NP1 | APNCDVGNDVNPETCKYGTVRNWCRHSVCAKGPSEVCGGRWMQHGTCGTGTRCNCNRCLGCSSFTLECYTGGQVC | |||
| Eus-NP1 | APNCDTDEGTDVNPETCKYGTVRNWCRHMVCAKGPGDVCGGRWMQHGSCGTGTRCNCNRCLGCSSTTLECYTGGQFC | idem | ||
| Euc-NP2 | GEPVNEAACKFGVAMDWCRQVCAKGPGETCGGRWMQHGQCGDGLRCSCSRCSGCSPVTLDCFYGQFC | Copepod, daphnia | ||
| Eus-NP2 | APNCDVAVGEPVNEKTCKFGVAMDWCRRQVCAKGPGESCGGHWMQHGQCSEGLRCSCNRCSGCSPVTLDCFYGQFC | idem | ||
| Pigment Dispersing Hormone family | Cirriped,daphnia, amphipod, isopod, decapods and insects | PF06324/IPR009396 | ||
| Euc-PDH-L α | NSGTINSMLGLPRTYNLRRMMMHAa | None | ||
| Eus-PDH-L α | NSGTINSMLGLPRTYNLRRMMMNAa | None | ||
| Euc-PDH-Lβ1 | NSELINSMLGLPQTLRAQKLMANMa | None | ||
| Euc-PDH-L β2 | NAETINTMLGLPQTLRAQKLMAKL | None | ||
| Euc-PDH α | NSELINSLLGLPKVMNDAa | - | ||
| Eus-PDH β | NSELINSLLGLPKVMNDAa | - | ||
| Euc-PDH-L β1-PRP | pQEDQERQAVGNLALDILRVVGRAPSAMQ | None | ||
| Euc-PDH-L β2-PRP | pQEDQERQVVGELALGILRIVGQESSGPQ | None | ||
| Euc-RPCH/AKH | pQLNFSPGWa | Daphnia, decapods and insects | PF06377/IPR010475 | |
| Unreferenced peptides | ||||
| Allatostatin B family | Shrimp | |||
| Euc-AST B1 | DLRSVSPRSTNWSSLRGAWa | - | ||
| Euc-AST B2 | GGPNNWSNLRGAWa | - | ||
| Euc-AST B3 | GGPGDWGSFRGSWa | - | ||
| Euc-AST B4 | GGPGDWSNFRGSWa | - | ||
| Euc-AST B5 | GGADTDWNSFRGSWa | - | ||
| Allatostatin C family | Cirriped, dapnia, decapods and insects | |||
| Euc-AST C | SYWKQCAFNAVSCFa | - | ||
| Eus-AST C | SYWKQCAFNA- - - - | - | ||
| Bursicon family | ||||
| Euc-Bursicon α | VDECSLTPVIHILSYPGCKSKPIPSFACQGRCTSYVQVSGSKIWQTERSCMCCQESGEREAAVTLNCPKARSGEPKLKKVLTRAPIDCMCRPCTEVEAGAVMAQEIANFIGSNNMGDVPFLK | Cirriped, daphnia, decapods and insects | - | |
| Eus-Bursicon α | VDECSLTPVIHILSYPGCKSKPIPSFACQGRCTSYVQVSGSKIWQTERSCMCCQESGEREAAVTLNCPKARSGEPKLKKVPARA-- | idem | - | |
| Euc-Bursicon β | KHYGTECETLPSTIHIVKEEFDDAGQVTVNCEEDIAVNKCEGACLSKVQPSVNTPSGFLKDCRCCRETHLRTREVTLNHCYDGDGNRLSGEKGKVQVKLREPADCQCYKCGDSNR | Cirriped, daphnia, decapods and insects | - | |
| Calcitonin-Like Diuretic H. | ||||
| Euc-CLDH31 | GLDLGLGRGFSGSQAAKHLMGMAAANFAGGPa | Ixod, Cirriped, copepod, daphnia, lobster and insects | - | |
| Euc-CLDH33 | VQMLDLGLGRGFSGAQAGKHLIGLLAASAAGGPa | None | - | |
| Euc-CHH PRP | RNIEPLNNDAMASLLSVANFKHVPAVS | Decapods | - | |
| Euc-SIFamide | GYRKPPFNGSIFa | Ixod, daphnia, decapods and insects | - | |
| Small Neuropeptide F | ||||
| Euc-sNPF1-1 | SPSMRLRFa | Ixod, daphnia, decapods and insects | - | |
| Euc-sNPF1-2 | DYWQVSQRSMPAVRLRFa | idem | - | |
| Euc-sNPF1-3 | NFQQIPMESSLINDKDTRSPQLRLRFa | idem | - | |
| Euc-sNPF1-4 | APDHGEILPFHELGTSSLGSEIYQKSIRSPQLRLRFa | Idem | - | |
| Euc-sNPF1-5 | EDDADQEWTREMSNAALLDELLAPKELRSPQLRLRFa | Idem | - | |
| Euc-sNPF2-5 | - - - - - - - - - - - - - - - - - - - - -DELMAPKALRSPQLRLRFa | Idem | - | |
| Euc-sNPF1-6 | EPDNQYEQLLDQIEQKDTRSPKLRLRFa | Idem | - | |
| Euc-sNPF2-6 | EPDNQYEQLPNQIEQKDTRSPKLRLRFa | Idem | - | |
| Euc-sNPF1-7 | DQQVEDFDNDSGLSDAVNQKSIRSPQLRLRFa | Idem | - | |
| Euc-sNPF2-7 | DQQIEDFDNDTGLADAVDQKSIWSPQLRLRFa | idem | - | |
The peptides of E. superba are in italic and underlined. The peptides and the PRPs identified in the peptidome of E. crystallorophias eyestalks are highlighted in bold. a = amide; amphipod = Talitrus saltator; cirriped = Amphibalanus amphitrite; daphnia = Daphnia pulex; decapods = identified in more than two species of decapods; insects = identified in more than two species of hexapods; isopod = Armadillidium vulgare; Ixod = Ixodus scapularis; lobster = Homarus americanus;
Preliminary identifications were carried out in E. crystallorophias, the results of which were then used as a database to interrogate the E. superba transcriptome.
Materials and Methods
Animal collection
The ice krill, Euphausia crystallorophias were collected close to the Dumont d'Urville station in Terre Adélie (66°S411-102°E430), in the austral summer of 2010, by the deployment of an IKMT zooplankton net from the L'Astrolabe vessel. Krill swarms were located using an echo sounder and the sampling achieved by towing a net through the swarm for 10 to 15 min at a speed of 2 knots. The animals were flash frozen in liquid nitrogen and stored at −80°C. Six whole animals were used for the RNA extractions. These samples were supplemented withmaterial from eyestalks, one of the main organs for the production of neuropeptides. The eyestalks were partially dissected to remove the pigmented regions and then snap frozen in liquid nitrogen. Approximately 20 eyestalks were used to enrich the transcriptome, with a similar number used in the proteomic extractions. This project (IPEV-1039) was approved by IPEV (Institut Paul Emile Victor, the French polar institute) review committee and was declared to and approved by the “Terres Australes et Antarctiques Françaises” in 2009 according the Annex I of the Madrid Protocol and the French Decret No 2005-403. No endangered or protected species were used.
Illumina sequencing
The sequencing process included mRNA isolation from whole animals and 20 eyestalks using the SV Total RNA Isolation System (Promega, Madison, WI, USA). Sequencing was conducted by the McGill University and Génome Québec Innovation Centre (Montréal, Québec, Canada) following the manufacturer's instructions (Illumina, San Diego, CA). These data have been submitted to the SRA-EBI with Accession number (ERR264582.
RNA-Seq data sets
The RNA libraries yielded 15.3 million paired end reads with a maximum read length of 96 bp. Raw reads were filtered and trimmed using the FASTX-toolkit (Version 0.0.13 from Assaf Gordon Hannon lab) and rRNA contamination removed using ribopicker [8] and cutadapt (Version 1.1) [9], with a final quality check performed using fastQC (Version 0.10.0 http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/).
The contigs were assembled using two strategies leading to two transcriptome assemblies. The goal was to obtain the most comprehensive view of the whole transcriptome of E. crystallorophias. By using two assembly strategies and combining the results, our aim was to reduce the errors inherent in each of the technologies.
The first assembly was performed using Newbler (version 2.6) [10] and a paired end assembly strategy. Redundancy was determined by self-Blasting and the use of CD-HIT (95% similarity) [11].
The second assembly was performed using Trinity (release 2011-10-29) [12], the genome-independent transcriptome assembler. The assembler was run with following parameters (kmer size of 25 and minimum contig length of 300). Redundancy of the second assembly was reduced by using CD-HIT (95% similarity). The transcripts from both assemblies were processed through the Blast2GO pipeline [13] to produce putative annotations and functional classifications based on Blastx results against the GenBank NR database release 190 and InterProScan analysis against the InterPo database release 37.0.
Relative abundances of all the transcripts resulting from both assemblies were estimated by remapping the reads on each assembly with Bowtie [14] and performing RSEM abundance estimation [15].
Peptide selection
A local database of annotated peptides, with their corresponding sequences was developed. The peptides were chosen based on the most highly characterised neuropeptide and neurohormone sequences in the Arthropods [16]–[19], with particular reference to the D. pulex genome [7]. A maximum expected size for the mature peptide was set at 130aa. In the first instance, relevant Blast2Go annotations from the Trinity assembly were identified. Each identified contig was then Blast searched independently to confirm the annotation and further verified in the Newbler assembly. The contigs were then translated and the putative coding sequences delineated. These sequences were then subjected to a Blastp search and subsequently aligned with orthologous sequences from the Arthropods (mainly the insects and the crustaceans). A sequence identified in E. crystallorophias and not identified in the annotation of the E. superba transcriptome was then systematically searched by local Blast alignment against the E. superba transcriptome. All of the Blast search data and alignments were performed in CLC Main Workbench. The signal peptides were identified using SignalP.
Mass spectrometry
Two types of peptide extraction were applied. The first used cold 0.1% TFA, the second, a mixture of acetic acid, water and methanol (1:9:90). The extracts were pooled and then concentrated on a microTIP C18 (OMIX, VARIAN). The peptide eluate was either directly injected into a nano-LC and analysed by MS or was subjected to a trypsin digestion, with a separation stage in the nano-LC according to the following protocol: Endogenic peptides were reduced in 10 mM DTT at 60°C for 45 min and alkylated in 100 mM iodoacetamide for 30 min in the dark. Digestion was performed by adding 25 µl of 6 ng/µl trysin (Promega, USA) in 25 mM NH4HCO3. Samples were incubated at 37°C for 15 h. After digestion, peptides were collected and dried in a SpeedVac evaporator. Samples were resuspended in TFA 0.1% before nano-LC fractionation. The chromatography step was performed on a nano-LC system (Prominence, Shimadzu). Peptides were concentrated on a Zorbax 5×0.3mm C18 precolumn (Agilent) and separated onto a ACE 50×0.5mm C18 column (AIT, France). Mobile phases consisted of 0.1% acetic acid, 99.9% water (v/v) (A) and 0.1% acetic acid, 20% water in 79.9% ACN (v/v/v) (B). The nanoflow rate was set at 800 nl/min, and the gradient profile was as follows: constant 5% B for 5 min, from 5 to 100% B in 75 min, constant 100% B for 20 min, and return to 10% B. The 800 nl/min volume of the peptide solution was mixed with 1.6 µL/min volumes of solutions of 5mg/ml CHCA matrix prepared in a diluted solution of 50% CAN with 0.1% TFA. Fifteen second fractions were spotted by an AccuSpot spotter (Shimadzu) on a stainless steel Opti-TOF™ 384 target.
MS experiments were carried out on an AB Sciex 5800 proteomics analyzer equipped with TOF/TOF ion optics and an OptiBeam™ on-axis laser irradiation with 1000 Hz repetition rate. The system was calibrated immediately before analysis with a mixture of des-Arg-Bradykinin, Angiotensin I, Glu1-Fibrinopeptide B, ACTH (18–39), ACTH (7–38) and mass precision was greater than 50 ppm. All acquisitions were taken in automatic mode. A laser intensity of 3000 was typically employed for ionizing. MS spectra were acquired in the positive reflector mode by combining 1000 single spectra (5×200) in the mass range from 600 to 4000 Da. MS/MS spectra were acquired in the positive MS/MS reflector mode by combining a maximum of 2500 single spectra (10×250) with a laser intensity of 3900. For the tandem MS experiments, the acceleration voltage applied was 1 kV and air was used as the collision gas with gas pressure set to medium. The fragmentation pattern was used to determine the sequence of the peptide. Database searching was performed using the Mascot 2.2.04 program (Matrix Science) on the transcriptome database of E. crystallorophias (including the 42,632 contigs from the Trinity assemblies). The variable modifications allowed were as follows: C-terminal amidation, N-terminal pyroglutamate, N-terminal acetylation, methionine oxidation and dioxidation. “No enzyme” was selected. Mass accuracy was set to 100 ppm and 0.6 Da for MS and MS/MS mode respectively.
Results and Discussion
RNAseq assemblies
Quality processing of the raw reads led to the generation of 14.4 million good quality paired end reads. Two transcriptome assemblies were performed using the Newbler and Trinity assemblers. For the first assembly (Newbler), reads were assembled into 88,067 contigs with lengths varying from a minimum of 100 nt to 7,588 nt with a mean length of 289 nt, then reduced by CD-HIT clustering to a set of 21,425 contigs with lengths varying from a minimum of 300 nt to 7,588 nt with a mean length of 669 nt.
For the second assembly (Trinity), reads were assembled into 42,632 contigs of lengths varying from a minimum of 300 nt to 8,341 nt with a mean length of 760 nt, then rendundancy was reduced by clustering with CD-HIT to a set of 36,345 contigs with lengths varying from a minimum of 300 nt to 8,341 nt with a mean length of 698 nt.
Clustering of the 57,770 transcripts generated by the two approaches (36,345 Trinity transcripts and the 21,425 Newbler transcripts) at 95% of similarity indicated that 16,426 transcripts of the Newbler assembly (76.7%) clustered with 40.2% of the transcripts generated by Trinity (14,619/36,345) for a total of 14,178 mixed clusters. In 85% of the “mixed” clusters (2,128/14,178) the longest sequence was generated by the Trinity assembly. It was noteworthy that whilst there was greater diversity in the Trinity assembly, 4,999 contigs of the Newbler assembly were not detected (according to the CDHIT clustering) within the Trinity assembly. So to be as exhaustive as possible in cataloging the full transcriptome of Euphausia crystallorophias, annotations and further studies were performed on all the transcripts generated by both assemblers.
Expression levels of the transcripts in the assemblies were estimated by remapping the quality reads on both transcriptomes separately with Bowtie [14] and RSEM [15]. The metrics of TPM (Transcripts per million) and FPKM (Fragments Per Kilobase of exon per Million fragments mapped) were used. For each assembly, these data were: TPM mean: 23.46 and FPKM mean: 19.23; TPM median: 3.14 – FPKM median: 2.57 (Trinity) and TPM mean: 11.35 and FPKM mean: 47.75 – TPM median: 0.67; FPKM median: 2.82 (Newbler).
As expression measurements were not performed in replicate samples and because the samples were further enriched with eyestalk extracts, the measured expression values were relative and did not reflect the expression of the transcriptome at a specific time. The expression values were generally correlated between the two assemblies for similar transcripts (Table 2). The observed differences were related to the counting methods and could have been due to the large difference in the total number of transcripts (TPM) between the assemblies and/or the size of the transcripts (FPKM), with lower values obtained by the Trinity assembly since the transcripts were longer than in the Newbler assembly.
Table 2. Alphabetical list of peptide precursors, contig expression values and associated Blast matches.
| Peptide designation | Size (aa) | Comp ID Contig ID | Size (pb) | TPM | FPKM | BLAST matches |
| Allatostatin | 361 | 15895_c0_seq1 | 1444 | 5.01 | 4.11 | Type A pre-pro-allatostatin (Machrobrachium rosenbergii) |
| A precursor | 06348 | 760 | 0.93 | 3.9 | 4.4 e-51 | |
| Allatostatin | 98 | 40876_c0_seq1 | 424 | 1.8 | 1.48 | Type B pre-pro-allatostatin (Pandalopsis japonica) |
| B precursor | - | - | - | - | 2.0 e-10 | |
| Allatostatin | 106 | 7109_c0_seq1 | 589 | 11.08 | 9.08 | Type C pre-pro-allatostatin (Daphnia pulex) |
| C precursor | 10325 | 501 | 2.12 | 8.92 | 1.96 e-22 | |
| α Bursicon | 148 | 25818_c0_seq1 | 636 | 3.51 | 2.88 | Bursicon hormone alpha subunit (Callinectes sapidus) |
| precursor | 24449 | 273 | 0.58 | 2.42 | 9.35 e-72 | |
| β Bursicon | 137 | 17391_c0_seq1 | 701 | 4.1 | 3.36 | Bursicon hormone beta subunit (Homarus gammarus) |
| precursor | 10676 | 489 | 0.8 | 3.38 | 1.37 e-64 | |
| CCAP | 140 | 20513_c0_seq1 | 607 | 4.71 | 3.86 | Crustacean cardioactive peptide (Procambarus clarkii) |
| precursor | 20601 | 308 | 0.96 | 4.05 | 7.16 e-32 | |
| CHH | 131 | 4803_c0_seq2 | 493 | 21.75 | 17.83 | CPRP/cHH precursor (Charybdis japonica) |
| precursor | 17526 | 347 | 3.57 | 15 | 7.43 e-28 | |
| CLDH31 | 154 | 5480_c0_seq1 | 619 | 9.34 | 7.65 | Pre-pro-calcitonin-like diuretic hormone (Homarus |
| precursor | 24873 | 269 | 1.33 | 5.6 | americanus) 6.16 e-23 | |
| CLDH33 | 149 | 5480_c0_seq2 | 598 | 11.06 | 9.06 | Pre-pro-calcitonin-like diuretic hormone (Homarus |
| precursor | 17839 | 342 | 3.46 | 14.53 | americanus) 2.59 e-44 | |
| CLDH56 | 109 | 44788_c0_seq1 | 330 | 1.31 | 1.07 | corticotropin releasing factor-like protein (R. prolixus) |
| precursor | - | - | - | - | 7.61 e-7 | |
| Corazonin | 92 | 814_c1_seq4 | 429 | 176.02 | 144.28 | Pro-corazonin (Acromyrmex echinatior) |
| precursor 1 | 24335 | 274 | 35.51 | 149.34 | 4.66 e-7 | |
| Corazonin | 82 | 814_c1_seq2 | 667 | 7.65 | 6.27 | Corazonin preprohormone (Daphnia pulex) |
| precursor 2 | 05623 | 841 | 1.94 | 8.18 | 1.05 e-6 | |
| Eclosion | 92 | 673_c2_seq1 | 1179 | 121.27 | 99.4 | Eclosion hormone (Amphibalanus amphitrite) |
| hormone | 19560 | 320 | 6.82 | 28.68 | 1.80 e-13 | |
| MIH/VIH1 | 82 | 46742_c0_seq1 | 423 | 1.39 | 1.14 | Probable molt inhibiting hormone (Jasus lalandii) |
| - | - | - | - | 2.57 e-14 | ||
| MIH/VIH2 | 48 | - | - | - | - | Probable molt inhibiting hormone (Jasus lalandii) |
| 17459 | 347 | 2.23 | 9.36 | 1.93 e-8 | ||
| Neuroparsin | 103 | 18741_c0_seq1 | 382 | 5.96 | 4.88 | Neuroparsin-A precursor (Caligus rogercresseyi) |
| 1 precursor | 17227 | 351 | 1.16 | 4.86 | 2.63 e-17 | |
| Neuroparsin | 67 | - | - | - | - | Putative neuroparsin (Daphnia pulex) |
| 2 precursor | 16193 | 363 | 0.54 | 2.27 | 7.97 e-7 | |
| NPF1 | 90 | 2304_c0_seq1 | 732 | 31.94 | 26.18 | Preproneuropeptide F1 (Litopenaeus vannamei) |
| precursor | 31384 | 227 | 9.64 | 40.52 | 1.00 e-28 | |
| NPF1L | 128 | 2304_c0_seq2 | 734 | 10.68 | 8.75 | Preproneuropeptide F2 (Litopenaeus vannamei) |
| precursor | - | - | - | - | 2.27 e-47 | |
| NPF2 | 62 | 8750_c0_seq1 | 733 | 12.84 | 10.53 | NPF-like precursor (Nassonia vitripennis) |
| precursor | 07726 | 643 | 2.24 | 9.41 | 1.25 e-7 | |
| PDHLα | 87 | 4981_c0_seq1 | 415 | 23.89 | 19.58 | Pigment dispersing hormone (Litopenaeus vannamei) |
| precursor | 05401 | 870 | 2.65 | 11.15 | 5.09 e-5 | |
| PDHLβ1 | 78 | 1192_c0_seq2 | 562 | 37.32 | 30.59 | Pigment dispersing hormone (Litopenaeus vannamei) |
| precursor | 17681 | 344 | 30.05 | 58.16 | 1.65 e-10 | |
| PDHLβ2 | 56 | 1192_c0_seq3 | 405 | 53 | 43.44 | Pigment dispersing hormone (Litopenaeus vannamei) |
| precursor | 08747 | 576 | 15.73 | 66.14 | 1.65 e-10 | |
| PDHβ | 74 | - | - | - | - | Pigment dispersing hormone type 2 (Litopenaeus |
| precursor | 33371 | 218 | 111.78 | 470.11 | vannamei) 1.54 e-15 | |
| RPCH | 104 | 9506_c0_seq2 | 832 | 7.48 | 6.13 | Red pigment concentrating hormone (M. rosenbergii) |
| precursor | 06590 | 737 | 1.33 | 5.61 | 3.82 e-18 | |
| SIFamide | 69 | 2588_c0_seq1 | 1015 | 18.97 | 15.55 | SIFamide (Procambarus clarkii) |
| precursor | 03910 | 1128 | 3.92 | 16.5 | 5.75 e-25 | |
| sNPF1 | 257 | 7638_c0_seq1 | 502 | 13.49 | 11.05 | Short neuropeptide F precursor (Aedes aegypti) |
| precursor | 18326 | 335 | 2.35 | 9.89 | 1.77 e-13 | |
| sNPF2 | 137 | - | - | - | - | Short neuropeptide F precursor (Aedes aegypti) |
| precursor | 03109 | 791 | 2.19 | 9.21 | 9.38 e-11 | |
| Tachykinin | 205 | 4591_c0_seq3 | 1933 | 14.61 | 11.97 | Preprotachykinin (Panulirus interruptus) |
| precursor | 03739 | 1170 | 4.47 | 18.8 | 6.56 e-38 |
comp ID: sequences assembled with Trinity.contig ID (italic) sequences assembled with Newbler. Size (aa): deduced coding sequences. Size (pb): comp or contig sizes in pair bases. TPM = Transcripts Per Million. FPKM = Fragments Per Kilobase of exon per Million fragments mapped. Bold values are over the TPM or FPKM averages. Underlined peptides have been characterised by mass.
Peptide families identified
Specific families of neuropeptide were analysed in this study, chosen on the basis of their putative physiological importance in insects and crustaceans. In this discussion, these peptide and peptidic hormone families have been grouped according to the existing entries in Pfam (http://pfam.sanger.ac.uk) or InterPro (http://www.ebi.ac.uk/interpro) and their arthropod specificity (Table 1).
Generic peptide families
These peptides are present in a wide range of species and are not restricted to arthropods or crustaceans.
Corticotropin-Releasing Factor-Like Diuretic Hormone (CRFLDH) – PF00473/IPR000187
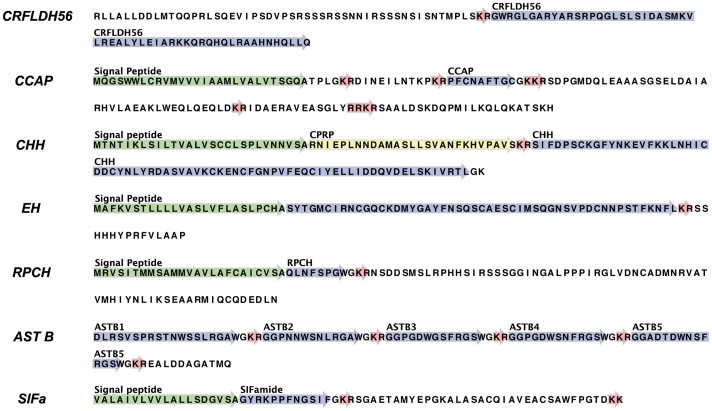
Contig 44788 exhibited sequence similarity to the family of corticotropin releasing factors (CRFLDH) (Table 1; Figure 1). The precursor, although only a partial sequence, contained almost the complete CRFLDH. This peptide has been isolated from a number of insects [20]–[23], but has yet to be described in the Crustacea, except in the daphnia [7]. Sequence alignments with other gene family members from different species suggested that the final 2aa were absent in the E. crystallorophias orthologue (Figure S1 in File S1). However, this still resulted in a putative mature protein of 56aa (amino acids) (DH56) and therefore represented one of the longer sequences of this family along with that of Ixodes (DH61) [24]. The function of this gene in the Crustacea is unknown, but CRFLDH is associated with post-prandial diuresis in insects [25], [26].
Figure 1. Complete and partial sequences from E. crystallorophias of the pre-pro-peptides containing the Corticotrophin Releasing Factor-like Diuretic hormone (CRFLDH), the Crustacean Cardio Active Peptide (CCAP), the Crustacean Hyperglycaemic Hormone (CHH), the Eclosion Hormone (EH), the Red Pigment Dispersing Hormone (RPCH), the Allatostatin Bs and the SIFamide.
The mature peptides are highlighted in blue. The potential bibasic cleavage site is highlighted in red. The CPRP (CHH PRP) is highlighted in yellow.
Neuropeptide F (NPF/Y) -PF00159/IPR001955
These peptides are between 28–45aa with a C-terminal motif of either -RPRFa or RVRFa. They are highly represented within the invertebrates, and in particular, the insects, but also a diverse array of arthropods including the crustaceans [6], [7], [17], [27]–[30]. In the E. crystallorophias assemblies, three precursors were identified (Table 1; Figure 2). The first of these encoded a putative 90aa protein, including a 29aa signal peptide. Euc-NPF1 was encoded immediately after the signal peptide and terminated at position 62 with a glycine, which permits amidation of the C-terminal with the production of a mature peptide of 32aa. The sequence corresponded to the consensus Xn-Y-L-X2-L-X2-Y-Y-X4-R-P-R-Fa proposed by Nässel and Wegener (2011) [31]. This sequence was identified in the eyestalk proteomic analysis. In addition, the sequence of the PRP, situated 3′ to Eu-NPF1 was also identified in the same analysis, thereby establishing that it is expressed. A sequence orthologous to this PRP also exists in the pro-peptide of the shrimp Penaeus vannamei (AEC12204, AEC12204) (Figure S2 in File S1) [30]. The second precursor represented the long form of the first transcript (NPF1-L) and apart from the extra insertion in NPF-1L, both transcripts were identical. NPF-1L appeared to be complete at a size of 70aa. It was interesting to note that the additional sequence is the same length in all species in which it has been identified to date (Figure S2 in File S1). Therefore it is difficult to determine whether it is indeed an alternative splice form or an incompletely processed intron [30]. However, this situation has previously been reported in the silk moth Bombyx mori [32]. The third transcript (NPF2), although only partial, was clearly different, especially in the N-terminus and contained a dibasic cleavage site upstream from the single tyrosine (Figure 2). This produced a short peptide of 17aa of YL/X5/YY/X2/AGRPRF. The sequence of this second form was validated by the discovery of one very similar in daphnia [7]. As with all other organisms, a single copy of NPF was present in the precursors. In contrast, the NPF characterised by a C-terminus of GX2RY and reported in the same cladoceran was not found in the transcriptome databases.
Figure 2. Complete protein sequences of the pre-pro-peptides of the Neuropeptides F1 and 2 from E. crystallorophias.
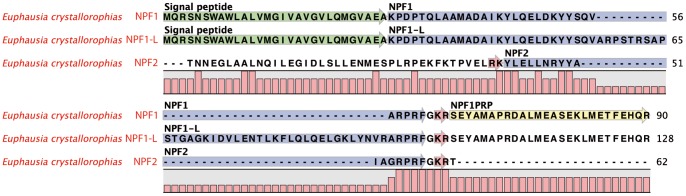
The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectroscopy.
Tachykinin-Related Peptide (TKRP) – IPR013206
The tachykinins (or neurokinins) represent another group of highly conserved peptides. The transcript in E. crystallorophias contained a precursor of 618bp, putatively encoding a 206aa protein (Tables 1 and 2; Figure 3). 6 identical sequences of TKRP were present in the pro-peptide with the sequence APSGFLGMR-NH2. This sequence corresponds to that originally identified in the nervous system of the crab Cancer borealis [33] and has subsequently been identified in other arthropods [34]–[37] (Figure S3a in File S1). The precursor of TKRP in lobster is very similar with a virtually identical structure, but this species also has an extra isoform slightly modified at TPSGFLGMR-NH2 [37]. The pro-peptide possessed an extra 5 sequences potentially coding for PRPs. The mass spectrometry analysis, not only enabled the confirmation of Euc-TKRP (Figure S3b in File S1), but also one of the precursor-related peptides (Figure S3c in File S1).
Figure 3. Complete protein sequence of the pre-pro-peptide containing the Tachykinins of E. crystallorophias.

The 6 identical examples of Tachykinin are highlighted in blue. The signal peptide is highlighted in green and the potential bibasic cleavage sites, in red. The latter delimit the 4 potential PRPs. The PRP highlighted in yellow was characterised by mass spectrometry.
Peptide families specific to arthropods
These peptides are present in the Pfam and InterPro databases, but to date, have only been characterised in arthropods.
Allatostatin A (AST-A or -FGL amide) – PF05953/IPR010276
The members of this first family are characterised by a C-terminus with the structure: F/Y-X-F-G-L/I-amide. A single contig in the E. crystallorophias database potentially encoded the complete precursor molecule of AST-A, with a putative peptide sequence of 361aa and a signal peptide of 31 residues (Table 1; Figure 4). 16 sequences containing the signature of AST-A were present in the precursor, each with a size between 6–11 residues. This represents a greater number of copies than has been found previously in the insects (at 13–14 forms) [38], but fewer than in the majority of the crustaceans, where between 30 to more than 40 sequences have been characterised [39]–[41]. Each of the sequences appeared to be of a unique origin, which is in contrast to analyses in Macrobrachium rosenbergi [40] and Procambarus clarkii [39] where two AST-A sequences are present numerous times, indicating multiple duplication events. All of those identified in E. crystallorophias took the form: Y-X-F-G-L/I, with the final amino acid in the majority of cases (14 out of 16) being a leucine. The AST-A3 was the only one family member characterised in eyestalk extracts (Figure S4 in File S1) with a cryptic peptide (PRP1 = Precursor-related peptide) localised immediately after the signal peptide and upstream of the pre-pro-peptide (Figure 4). These data, whilst hinting at the incomplete nature of the transcriptomes and mass spectrometry data, showed that allatostatin A is primarily produced in the eyestalks of E. crystallorophias.
Figure 4. Complete protein sequence of the pre-pro-peptide containing the Allatostatin type As of d'E. crystallorophias.

The 16 Allostatin As are highlighted in blue and numbered according to their position in the sequence. The signal peptide is highlighted in green and the potential bibasic cleavage sites, in red. The latter delimit the 8 potential PRPs. The PRP highlighted in yellow was characterised by mass spectrometry.
Crustacean Cardioactive Peptide (CCAP)- PF11105/IPR024276
The predicted sequence of the pre-pro-peptide in E. crystallorophias was composed of 140aa with a signal peptide of 28aa (Figure 1). The CCAP sequence started at position 47 and finished at position 56 with a glycine, which is implicated in the amidation of the peptide. This sequence was identical to that found in all the decapods (Figure S5 in File S1). Four CCAP-PRPs were present in the propeptide, one upstream of CCAP and three downstream. Their function is unknown.
Crustacean Hyperglycemic Hormone (CHH) family- PF01147/IPR001166: Type I peptide: Crustacean Hyperglycemic Hormone stricto sensu
The nucleotide sequence produced from E. crystallorophias assembly covered 396bp putatively coding for the 131 aa of the pre-pro-peptide (Figure 1). This pre-pro-peptide could be partitioned into a signal peptide of 28aa, followed by a CPRP (CHH precursor-related peptide) of 27aa and finally a mature peptide of 72aa. The latter was obtained after the cleavage of the two terminal amino acids Gly(130)-Lys(131) during the C-terminal amidation of the peptide (Figure S6a in File S1).
The gene itself has alternative splice forms, producing either a short or a long form (CHH-L), of which increased length is the main characteristic [42]. Given the number of amino acids in the putative translation of the mature Euc-CHH sequence, it was clearly not the long form of the peptide. The potential for amidation at the C-terminal is an additional diagnostic since the long form never has this modification. This sequence therefore represented a CHH peptide as either the product of a gene of three exons, similar to that which exists in the peneid shrimps, or the spliced isoform of a CHH comprising 4 exons, the latter of which is composed of exons 1, 2 and 4.
The Euc-CPRP with a size of 27aa, was below the average size of these peptide inserts identified so far in Decapods, the longest being 50aa in the hermit crab Pagurus bernhardus [43] and was one of the smallest for a gene of 4 exons (Figure S6b in File S1). In effect, there appears to be a correlation between the structure of CPRP and the structure of the CHH gene of either 3 or 4 exons in the Malacostraca. A short CPRP (less than 20aa) is characteristic of a pro-peptide translated from a gene of 3 exons. Although significantly different from the sequences of the decapods, Euc-CPRP did contain the unique signatures for the propeptides found in genes of 4 exons (R1 – I4E5-L7 – M11-S13L14L15S16), in particular between amino acids 11 and 16. It should be noted that the RNA used in the sequencing was enriched using material from eyestalks to maximise the chances of identifying neurohormones from the X-organ-sinus gland complex. The RNAs coding for the long forms generally originate from different tissues (ganglions of the pericardial and thoracic ventral nervous system [42]), which were probably not sufficiently represented in the original extractions due to the low numbers of animals used and therefore were not identified in this study.
Crustacean Hyperglycemic Hormone (CHH) family- PF01147/IPR001166: Type II peptide: Vitellogenesis/Moult/Mandibular Organ Inhibiting Hormone
In the E. crystallorophias transcriptome assembly, two partial sequences were identified with sequence similarity to type II peptides (Figure 5). A few publications have reported the presence of different isoforms of MIH/VIH, most notably in the peneid [44]–[48] and caridean shrimps [49], but not in any of the other decapods (Figure S7 in File S1). The sequence of the first form putatively encoded the complete mature peptide, comprising 78aa and the sequence signatures (Gly and Val) mentioned above. Although the signal peptide was incomplete, the sequence was similar to other type II family members, i.e. without an intermediate peptide, the mature peptide being produced directly from the signal peptide within the translated propeptide. The sequence of the second form was partial and comprised the amino acids between the first and last cysteine. There was a significant difference between the two forms, with only a sequence identity of 49% between the 47aa that both partial sequences had in common. This is in comparison with, for example, the 71% aa identity between isoforms in the peneid shrimp Litopenaeus vannamei. This large divergence at the sequence level implies a potential difference in function between the two forms, but this still has to be verified through functional studies.
Figure 5. Alignment of the partial protein sequences of the pro-peptides of MIH/VIH from d'E. crystallorophias.
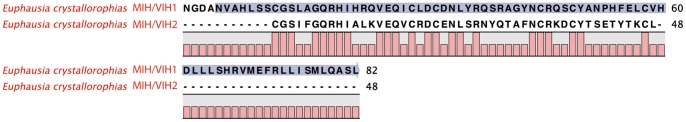
The complete mature peptide of MIH/VIH1 is highlighted in blue. Only a partial sequence for MIH/VIH2 was identified.
Corazonin (CRZ)-IPR020190
Corazonin is an 11aa peptide, blocked at both ends. The most common form of this peptide is: pQTFQYSRGWTNamide (Arg7-CRZ) [50],[51], In E. crystallorophias two different contigs coding for CRZ propeptides were identified (Table 1 and 2; Figure 6). The first pre-pro-Euc-CRZ1, comprised 92aa with a signal sequence estimated at 23 residues, cleavage of which could occur just upstream of the mature corazonin peptide (CRZ). This was identical to the form designated Arg7-CRZ, which has been identified in all other crustaceans (Figure S8a in File S1). This sequence was validated by the mass spectrometry data and confirmed the presence of a pyroGlu at the N-terminus and an amidation at the C-terminus (Figure S8b in File S1). Downstream of the CRZ1 pro-peptide were three further peptides (all essentially classified as Precursor Related Peptides (PRP's)) flanked with dibasic cleavage sites, the functions of which are unknown. The proteomic study enabled the characterisation of a third Euc-CRZ1-PRP in the eyestalks. However, whilst the CRZ peptides remain highly conserved between different taxa, the same is not true of the PRPs, which are extremely variable. To date, two PRPs have been described as being present in each pre-pro-protein of the insects [52]. An orthologue to the E. crystallorophias pre-pro-Euc-CRZ1 was identified in E. superba. However, the characterisation of the second form, pre-pro-Euc-CRZ2 produced a surprising result. It comprised 82aa with a signal peptide of a similar size to isoform 1, namely 23 residues. The downstream peptide was clearly a member of the corazonin family, but with a modification in position 4 with a serine replacing a glutamine (Ser4- Arg7-CRZ). Similarly to the pre-pro-EucCRZ1, the pre-pro-Euc-CRZ2 contained at least three other PRPs. The orthologue to the second isoform was also characterised in E. superba, but the Eus-CRZ2 was different to that in E. crystallorophias, with a glutamate in position 4 (Glu4- Arg7-CRZ). The number of CRZ-PRPs was identical. The existence of these two euphausiid orthologues of CRZ1 and 2 from different databases produced by different sequencing methods validated the existence of two isoforms in the same species. It is also worth noting that a second variant form exists in the daphnia (Gln3-Arg4-CRZ) (ACJ05606 and EFX86608).
Figure 6. Alignment of the protein sequences of the pre-pro-peptides for Corazonin 1 and 2 from E. crystallorophias (completes) and E. superba (partial).

The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectroscopy.
It is thought that the primary functions of CRZ are involved in myotropism and pigmentotrophism. However a specific isoform: His7-CRZ has been identified and associated with the gregarious phase of locust [53]–[55]. This function is potentially very interesting with regard to krill, as these animals are found in large swarms at incredible densities.
Eclosion Hormone (EH) – IPR006825
This neuropeptide has been primarily characterised in insects where it is implicated with Ecdysis Triggering Hormone (ETH), CCAP and Bursicon in the hormonal cascade following cuticle hardening post-ecdysis [56], [57]. In contrast to the insects, this gene has been rarely described in the Eucrustacea. An EH-like transcript was present in the assembly of E. crystallorophias. The pro-peptide putatively comprised 92 residues with a signal peptide of 25aa (Table 1: Figure 1). The peptide of 67aa contained a dibasic cleavage site, which implied that the mature peptide was actually 51aa. This recognition site for cleavage enzymes seems to be present, not only in the euphausiids, but also in the peneids and the chelicerates (Figure S9 in File S1). In contrast, it is absent in most insects except Tribolium castaneum. The percentage of amino acid identity for the mature peptide varies from 25-39% and exceeds 49–76% when considering only the sequence 3′ to the first cysteine, at the dibasic cleavage site. There are 6 cysteines, which are position-invariant in all sequences.
Neuroparsin – PF07327/IPR010850
In the E. crystallorophias assemblies, and also that of E. superba, two contigs with sequence similarity to neuroparsins were evaluated (Table 2; Figure 7). The precursors of the first isoform (NP1) comprised 103 aa and 101aa for each species respectively. The signal peptide was estimated to be 25aa for each of the two euphausiids. The propeptide of the second isoform (NP2) was incomplete in the N-terminal region for the E. crystallorophias transcript, but was full length in E. superba with 103aa and a signal peptide of 27 residues. Euc-NP1 possessed 15 cysteines as opposed to 14 for Eus-NP1, with 3 and 2 cysteines in the signal peptide respectively. Eus-NP2 comprised the same structure. It is worth noting that interrogating the assemblies using Blast sequence similarity searching revealed several different putative isoforms. Only the most highly represented isoforms, which were present in both euphausiid species were retained (Figure 7). Hence it is probable that there are more isoforms present than have been described here. The number of precursors is variable between taxons, although in the majority of cases, a single precursor has been characterised from sequence databases. Having said that the locust possesses four genes and the kissing bug Rhodnius prolixus, three (Figure S10 in File S1). In crustaceans, only a few ESTs have been characterised in decapods, a copepod and a cladoceran [7], [18], [19] (Figure S10 in File S1). A Blast search against the ESTs of Penaeus monodon revealed at least 3 isoforms of NP (GO071574, GW996622, GO080130).
Figure 7. Complete protein sequences of the pre-pro-peptides of the Neuroparsins 1 and 2 of E. crystallorophias and of E. superba.

The mature peptides are highlighted in blue. The signal peptides are highlighted in green.
Pigment Dispersing Hormones (PDH/PDF) – PF06324/IPR009396
A multiple sequence alignment of this family of sequences in crustaceans shows that they possess two types (Figure S11a in File S1). These are designated α and β and share a conserved structure with a mature peptide of around 18aa [58]. A first partial sequence for E. crystallorophias and a full length sequence for E. superba [4] were retrieved from the databases (Table 1; Figure 8). This Euc/Eus-PDH was completely identical in amino acid sequence to other sequences in the decapod crustaceans and was the β form (Figure S11a in File S1). A second form was identified in E. crystallorophias, which was classified as a duplicated β form (PDH-Lβ1), because of the presence of a glutamate in position 3. However, whilst the first 12 amino acids of this peptide were conserved and confirmed membership of the PDHs, this sequence possessed a longer 3′ end, with a further 11aa in the mature peptide, not the usual 6 (Figure 8). The Euc-PDH-Lβ1 was also identified in the mass spectrometry studies validating it as an original form of PDH with a C-terminal amidation (Figure S11b in File S1). The same study showed that the PRP upstream of PDH-Lβ1 in the pro-peptide was also expressed, as suggested by its position between the signal peptide and a dibasic cleavage site. Its function is, of course, unknown. In addition, an extra form of PDH-L, which resembles a β form due to the presence of a glutamine at position 3, was identified in the assembly and was confirmed by proteomic analysis. This peptide (Euc-PDH-L β2) was distinguished by the presence of an alanine in place of the serine, classically located in position 2 of the mature peptide. The size of the putative peptide was 24aa, similar to the other long forms of PDH also identified in this study. The transcript was incomplete, but appeared to start at the PRP, with the traditional signal peptide absent. The PRP of this latter isoform was also identified in the proteomic analyses.
Figure 8. Complete protein sequences of the pre-pro-peptides of the Pigment Dispersing Hormones (PDH) α and β of E. crystallorophias and E. superba.
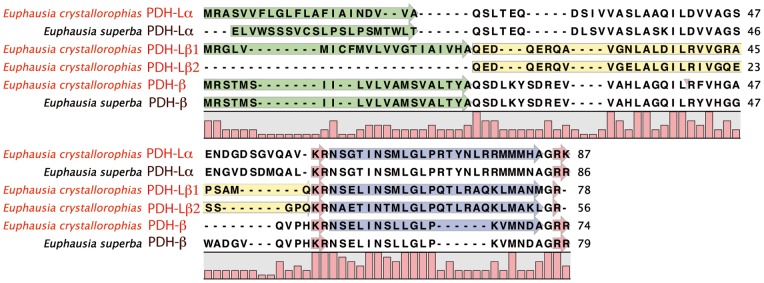
The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectroscopy.
The assemblies revealed the presence of a further sequence with high sequence identity to PDH-L, but containing the sequence signature of type α; a glycine in position 3 (Figure 8). No “short” sequences of PDHα have been identified in euphausiid species. Extensive Blast sequence similarity searching revealed no further PDH-L gene family members outside euphausians and therefore the sequences described above would appear to be krill-specific. Clearly, with only sequence data available, the exact function of these transcripts cannot be determined, however a strong role in circadian rhythms might be predicted. Similar to RPCH (Red Pigment Concentrating Hormone) and AKH (Adiopokinetic Hormone), where the crustacean and insect orthologues have different names, PDH in crustacean is the orthologue of PDF (Pigment Dispensing Factor) in insects [59]–[61]. PDF has recently been implicated in the control of circadian rhythms of some crustaceans as well as in the insects [62], [63]. Krill undergo diurnal vertical migrations, living at depth during the day and returning to the surface waters at night to feed, which are influenced by seasonality. Hence there is an important requirement for krill, in particular E. superba, to have control over their daily rhythm, with an internal synchronicity and compartmentalisation in their physiology and localisation within the water column [64], [65]. This may be supplied by the putative PDH transcripts identified here, but clearly more work is required on these candidate genes. Multiple isoforms of this gene in krill suggests that there has been an expansion of functions in this species.
Red Pigment Concentrating Hormone (RPCH/AKH) – PF06377/IPR010475
In crustaceans, the variation in colour is controlled primarily by PDH, but also by RPCH. Similar to the situation with PDH, there are numerous orthologues to RPCH in the insects, which are called adipokinetic hormone (AKH) [66]. In the decapods, this sequence is highly conserved and consists of an octopeptide amidated at the C-terminal, p-QLNFSPGW-NH2. The putative E. crystallorophias RPCH pre-pro-peptide comprised 315bp with a signal peptide of 25aa, a mature peptide consisting of 8 characteristic amino acids and a C-terminal glycine which enabled amidation, upstream of which, there was a dibasic -KR- cleavage site (Figure 1). The sequence which was identified in the genomic database of Daphnia pulex appeared to be more related to AKH, rather than the RPCH of the decapods, due to a number of important modifications (3 out of the 8aa in positions 2, 6, and 7). This first sequence of RPCH from the (non-decapod) Malacostraca was identical to those described in the decapods (Figure S12a in File S1), but only one isoform was present in the krill assembly, similar to the situation in Daphnia where a single gene was identified [7]. This differed from the situation in insects where up to 3 different isoforms have been characterised in the locust, Locusta migratoria [67] (Figure S12b in File S1).
Unreferenced peptide families
These were peptide families with no accession numbers or functional annotations in either Pfam or InterPro.
Allatostatin B (AST-B or-W(X6)Wamide)
Peptides belonging to the AST-B family have a tryptophan at the C-terminal and also at position 6. These peptides were primarily isolated in insects due to their myo-inhibitory effect and subsequently for their action on the prothoracic gland [68], [69]. The E. crystallorophias assembly contained several transcripts with sequence similarity to AST-B. The pre-pro-peptide sequence was not full-length, but 5 forms could be differentiated (Figure 1). Certain amino acids were conserved in all isoforms, in addition to the tryptophans previously mentioned. This permitted the development of a consensus sequence for the euphausiids: (G-G)-Xn-W-X3-RG-A/S-W-NH2. The two glycines in the C-terminus were present in 4 of the forms. There appeared to be fewer AST-B genes in E. crystallorophias than in other crustaceans, however this may simply be due to the partial nature of the transcriptome data. There are 13 of these sequences in Carcinus maenas [19], [70] and 9 in Cancer borealis [71]) where these peptides have been mainly characterised.
Allatostatin C (AST-C)
The type C allatostatins were originally described in Manduca sexta [72] and comprise a full-length peptide of 15 aa, with a non-amidated C-terminal with a common motif of -PISCF and a disulphide bridge between the cysteines at positions 7 and 14 [70]. These peptides have long been known in the Endopterygota (Holometabola) and have recently been described in the Crustacea [7], [18], [73]–[75]. A pre-pro-peptide of 106aa, with a signal sequence of 27 residues was present in the transcriptome assembly of E. crystallorophias (Figure 9). In common with previously characterised AST-C genes and in contrast to the A and B types, only a single copy of the mature peptide was present in the assembly. It was distinguished by the signature motif –AVSCF and possessed an identical sequence (SYWKQCAFNAVSCF) not only to those putatively identified in other crustaceans [73] but also to those in other species of insect (Locusta migratoria, Apis melifera, Laupala kohalensis): (Figure S13 in File S1). A partial sequence, which was highly similar to that identified in E. crystallorophias, was also found in the E. superba transcriptome. Despite reciprocal Blast searches, it was not possible to identify both isoforms in the same krill species. This is in line with the hypothesis of Dickinson et al, that each species only contains one form of this gene, which they proposed when they discovered a single peptide with the variant sequence –AVSCF in H. americanus [73]. However, more recently, an isoform with the –PISCF motif has since been discovered in the same species [76]. An identical situation (with 2 isoforms) has also been found in the crab Cancer borealis [77]. Therefore there is ample evidence to suggest that both forms are present, at least, within the decapods. To further underline the complexity found in this gene family, a study of the proteome of Daphnia pulex, not only demonstrated the presence of the two forms, but an additional one with the motif -AVSCF [7]. This last isoform was not present in either of the krill transcriptome assemblies.
Figure 9. Protein sequence alignment of the pre-pro-peptides of the Allatostatin Cs of E. crystallorophias (complete) and E. superba (partial).
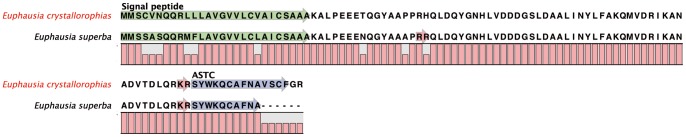
The Allatostatin C is highlighted in blue. The signal peptide is highlighted in green and the potential bibasic cleavage sites, in red.
Bursicon
Bursicon is a hormone well known in arthropods for its role in the hardening of the cuticle after moulting [78]–[80]. It is comprised of two sub-units, α and β which form a heterodimer [81]–[84]. Two distinct sequences were present in the assemblies of E. crystallorophias (Figure 10). The alignments with sequences from crustaceans and insects showed that the two sequences represented each one of the dimers, with 11 highly conserved and characteristic cysteines. The sequence of the bursicon α chain putatively comprised 146aa with a signal peptide of 26 residues. That of the β form was predicted at 137aa and included a signal peptide of 22 residues. Sequence conservation is important with each isoform, in particular for the sequences between the cysteines. The sequence of α in E. crystallorophias was practically identical to the orthologue in E. superba [18] and similar to the two daphnia species and also the other crustaceans, notably the decapods (Figure S14a in File S1). This conservation was also found in the β form (Figure S14b in File S1).
Figure 10. Protein sequence alignment of the pre-pro-peptides of Bursicon α and β of E. crystallorophias (complete) and Bursicon α of E. superba (partial).

The mature peptides are highlighted in blue. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. Only the sequences of E. crystallorophias are annotated.
Calcitonin-Like Diuretic Hormones (CLDH)
Originally isolated from the nervous tissue of Diploptera punctata [85], CLDH or DH31 has only been characterised in three crustaceans Homarus americanus [86], Daphnia pulex [7], [74] and Caligus clemensi [18]. Similar to the situation in these three species, a contig with sequence similarity to DH31 was identified in E. crystallorophias (Table 1; Figure 11). It is possible to distinguish it from the DH31s of Homarus and Tribolium [22] by the exchange of L22 for M22, and Acyrthosiphon by the change of S7 to G7 [87]. The E. crystallorophias precursor was predicted to be 149aa long, encoding a signal peptide of 26aa, a PRP of 47aa separated from the mature peptide by a dibasic cleavage site and finally a second PRP of 38aa. Whilst DH31 is highly conserved within the different species of insect, crustaceans and chelicerates, the other parts of the precursor molecule are less well conserved (Figure S15 in File S1). The majority of CLDHs are 31aa, however, another form has been identified in the chelicerate Ixodes scapularis, which is extended by the addition of 3aa at the start of the sequence [24]. A second form with sequence similarity to CLDH with an extra 2aa was identified in E. crystallorophias (Figure S16 in File S1). If confirmed, this represents the first example of two CLDH isoforms within the same species. This second longer sequence was more variable than the first with 6aa substitutions in 31aa. The very high level of sequence conservation of this peptide within the Pancrustacea and to a similar extent in the insects suggests the action of a strong evolutionary selection pressure to maintain a major function, potentially that of a cardioactive peptide [86]. The number of important amino acid modifications found in the second contig could be explained by weaker selection pressure on this second copy, as a result of neofunctionalisation, enabling both gene copies to be retained in the genome.
Figure 11. Alignment of the complete protein sequences of the pre-pro-peptides of the Calcitonin-like Diuretic hormones (CLDH 31 and 33) from E. crystallorophias.

The mature peptides are highlighted in blue. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red.
SIFamide
The SIFamides represent a family of neuropeptides, which are highly conserved within the arthropods [89]. Since their first characterisation in the flesh fly Neobelleria bullata (as a myotropic agent) [90], an increasing number of these peptides have also been identified in the Hexapoda [91]–[94], in the Crustacea [7], [19], [34], [74], [88], [95]–[97] and in the Chelicerates [24]. They are highly abundant peptides and therefore relatively easy to isolate using techniques such as HPLC, MALDI and QTOF, but are also present in the molecular databases of genome sequences, ESTs and transcriptomes. The dodecapeptide contains the consensus sequence of the type XYRKPPFNGSIFamide where X1 can be R, G, V or T. Two exceptions exist for this 11aa structure (Figure S16 in File S1): Daphnia pulex where the Y2 is missing and the P4 is replaced with a leucine [7] and the aphid Acyrthosiphon pisum where the G1 seems to remain within the signal peptide and the mature peptide starting not with a tyrosine, but a phenylalanine [94]. It is interesting to note that the dibasic couplet within this sequence does not constitute a cleavage site [98] and to date the putative peptide PPFNGSIFamide has not been detected [91], [97]. Although there have been a limited number of precursor molecules identified, the structure is well defined. The SIFamide is positioned directly downstream of the signal peptide and upstream of a PRP. The function of the latter is unknown, but is characterised by the presence of 2 cysteines separated by 6 amino acids. The SIFamide has been implicated as a neuromodulator of reproduction [89], [91] and olfaction [39], [99].
The sequence of the putative precursor in the E. crystallorophias transcriptome (Figure 1) was truncated at the N-terminus and therefore the final pre-pro-peptide length of 77aa was missing 8aa from the signal peptide, the final sequence of which could therefore, not be confirmed. The Euc-SIFamide was present in full, as was the PRP. The mature peptide GYRKPPFNGSIFamide was identical to those in three other crustaceans Cancer borealis, Carcinus maenas and Procambarus clarkii, but also an insect, Culex quinquefasciatus (Figure S16 in File S1). This G1-SIFamide was the only such peptide identified in this analysis. Numerous isoforms have been identified in other crustaceans, such as C. maenas [19] and also H. americanus [97] in which a V1-SIFamide is present and was originally thought to be specific to lobsters. These two peptides have been identified in different tissues of the crab, with the isoform G1-SIFamide present within the brain, the sinus glands and the thoracic ganglions, whilst the second form V1-SIFamide is restricted to the thoracic ganglions. This regionalisation of expression could explain the varying success with which the different hormones have been isolated from krill. That the original RNA was enriched with extra eyestalks, which are a major source of the G1-SIFamide isoform, may have contributed to this finding.
Small Neuropeptide F
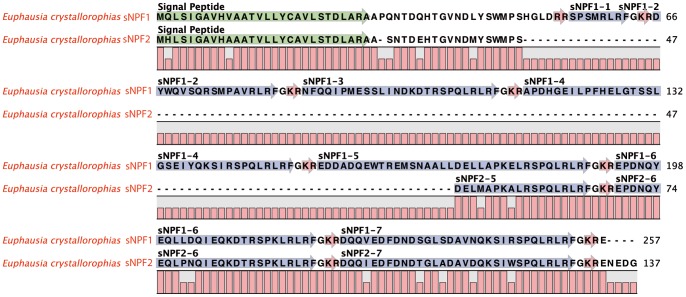
Contrary to what their name might imply the small neuropeptide F (sNPF) is not a short alternative spliced form of NPF as there are important differences in the precursors of the two types of peptides. There is considerable information on these peptides in crustaceans and insects. Although they are easy to characterise biochemically due to their small size, there is little information available on the structure of the pre-pro-peptides [31]. It seems that these peptides may be restricted to the arthropods, as they have yet to be identified in other taxa. Several isoforms are known in Homarus americanus (5) [88] and Penaeus monodon (4) [27]. In the E. crystallorophias transcriptome, two partial pre-pro-peptides were identified (Figure 12). The first, Euc-sPNF1putatively comprised 257aa, but there was no terminal stop codon. The signal peptide was 28aa. The characteristic C-terminus of Xn-P-X2-R-L-R-Fa was found in 7 peptides distinguished by dibasic cleavage sites. The size of each of these sNPFs was completely different, between 8–36aa. The second pre-pro-peptide Euc-sNPF2 was incomplete compared to the previous sequence, but aligned exactly with the N-terminal region (amino acids 1-48) and then less so up to position 172 of Euc-sNPF1. This precursor contained paralogues corresponding to sNPF1-5, 6 and 7, with peptide 5 truncated. In light of such results it was possible to hypothesize that two genes exist, coding for two different precursors, probably the result of a gene duplication, with, given the incomplete nature of the contigs, the potential to produce at least 14 different Euc-sNPFs. This number is larger than that reported for other species, but requires more verification from expression analyses. The sizes of some of these peptides were larger than those generally attributed to members of this peptide family. They reached the size range of those designated as NPFs and render the term “small” somewhat ineffective, especially since short forms of NFP were reported. To date, the existence of two genes has only been established in the mosquito Aedes, with a proposal for delineation into two families of peptides (head peptides versus sNPFs). However, it was not possible to make this distinction in the ice krill. The terminal structure of (L)RLRFa was remarkably conserved. Functional studies in Drosophila show that these peptides have functions in regulatory cascades controlling biological clock, feeding regulation, learning and memory [31].
Figure 12. Complete protein sequences of the pre-pro-peptides of the small Neuropeptides F1 and 2 from E. crystallorophias.
The mature peptides are highlighted in blue and numbered according to their position in the sequence. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red.
Levels of expression and characterisation by mass spectrometry
It was not possible to obtain an accurate overview of expression levels of each of the peptides identified in this study, due to the lack of replicates, but some clues could be provided by the relative expression levels in the assemblies and between the different techniques. The addition of RNA extracted from the eyestalks included in the RNA samples obtained from whole animals, prior to sequencing, meant that identification of peptides located to the X organ-sinus gland complex was reliant upon an over-representation of transcripts with regard to FPKM values. The proteomic analysis of the peptides obtained from eyestalk material confirmed both the presence of certain peptides predicted from the transcript assemblies and also their sequence. Neuropeptide transcripts with the highest FPKM values were also, for the most part, those which were characterised by mass spectrometry, indicating their importance in the initial mix of material. However, the reverse was not always true (Table 2).
The most highly represented peptides were those belonging to the PDH family, in particular PDHβ. This peptide possesses a particularly conserved structure within the decapod crustaceans and the importance of its expression and translation indicates one or more major biological functions. Although members of this family have been characterised in numerous species of arthropods, crustaceans and insects, the function, or functions, of these remain largely unknown and are potentially different between species: either neurohormone regulation of pigment movement and/or neurotransmission/modulation [100]. Given that there is clearly a structural relationship between PDH family members and those of PDF in insects, it is possible to infer that there might be homologous functions, in particular, in the modulation of circadian rhythms which are very important in krill. The most highly expressed members of PDH in other species are also PDHβ. Two isoforms exist, but these were not identified in krill [63], [101]–[105]. However, in E. crystallorophias, two long isoforms (PDHLβ I and II) co-exist with an α long form. These seem to be specific to the Euphausiacea, and are described for the first time in this study. Their expression levels were significantly lower than those for PDHβ [100], [105], [106].
If the peptides above, identified by the proteomic analyses, showed high expression levels, this was not necessarily the case in all peptides identified by the two techniques. Tachykinin and allatostatin A (represented by a single mature sequence (AST-A3) out of the 16 potential coding sequences) and their PRPs were characterised in both the eyestalk extracts and in the transcript assemblies. However, their FPKM values were much lower than those listed above (Table 2). This observation suggests that care has to be taken with the importance accorded to the relative expression levels of the PDHs. It is also interesting to note that the PRPs of the two precursor molecules were characterised by mass spectrometry, as was the case with those of PDHLβ, but not with PDHLα, however most abundant. Similarly, it was surprising that a single form of AST-A3 was characterised by mass spectrometry. It was clear that there was the potential for other forms to be expressed and translated due to the presence of dibasic cleavage sites in the precursor molecule, as evidenced by sequence data in the transcriptome. However, the lack of identification of these additional forms in the mass spectrometry data may have been for several reasons: the different extraction methods, the quality of the assemblies or the reality of very different translation levels for each of the AST-A transcripts. A potentially similar phenomenon is observed in D. pulex where out of 6 potential isoforms of AST-A, to date, only 4 have been confirmed by mass spectrometry [7].
CHH was the first peptide purified and sequenced from the eyestalks of the crab Carcinus maenas, largely as a result of its very high abundance [107]. It has not been characterised by mass spectrometry in this study due to the size filter that was applied (600–4,000Da), as its theorical mass is 8,389.5Da. The FPKM values of this peptide in this study, were lower than those of PDH and corazonin, underlining its lesser expression levels. It should be noted that CHH is functionally associated with stress, important for increasing the level of glucose in the circulation, by the glycolytic activation of glycogen stores in the midgut gland and the abdominal muscle [108]. The neuropeptide is primarily stored in the sinus gland and is released into the circulating haemolymph when stimulated. The quantity of RNA is therefore, not necessarily representative of the level of the available peptide and the expression is regulated via a feed-back mechanism [109].
In contrast, MIH/VIH1 was particularly weakly expressed, as evidenced by the low FPKM values. Most crustaceans stop or reduce the frequence of moulting and also their growth during their reproductive period, due to their respective energetic costs. In the krill, gravid females can moult and reproduce in parallel, which constitutes an adaptation to the very short productive period of the Antarctic summer [110]. The animals in this study were sampled in January and therefore were in their main period of maturation due to the very short window of opportunity for growth and reproduction in this region for phytoplankton feeders [111]. The weak expression levels of MIH/VIH1, which showed an inhibiting action in crustaceans tested to date, in either moulting or vitellogenesis, agree with the results here relating to the seasonality of the sampling period. The synergy between growth and moulting in the krill, does not allow for differentiation between the functions of VIH or MIH of the characterised peptide. It is also possible to consider the action of CHH on the secretion of moulting hormone and therefore its possible MIH function, as it has been demonstrated on the peneid shrimp Penaeus vannamei [112], which belongs to the decapods and is phylogenetically closest to the Euphasiacea. The possible functions of MIH/VIH2 are unknown.
Two other peptides were particularly represented in the assemblies: corazonin (CZN) and eclosion hormone (EH). The sequence of corazonin 1 was confirmed by mass spectrometry, and also had a high FPKM value in each assembly (Table 2). It was not possible to confirm the second isoform using this approach and the low FPKM value indicated a relatively low expression level in the eyestalks and potentially a different cellular origin. The two peptides are found in the insects and have been implicated in the regulation and development of the cuticle associated with moulting. They are upstream signalling molecules, activating the hormonal cascade responsible for initiating the moult. Corazonin is the most upstream molecule in this cascade, inducing the release of ecdysis triggering hormone (ETH), which was not identified in the assemblies. EH is expressed in response to ETH and is implicated in the pre-ecdysial phase. CCAP and bursicon are also implicated in the pre-ecdysis and post ecdysis phases respectively [57], [113]. The FPKM values for these last two peptides were lower than those for corazonin and EH. The mass spectrometry results for corazonin prove that corazonin is present in the eyestalks, as previously demonstrated in the lobster [88] and this localisation potentially explains its strong expression. The localisation of EH, is to date unknown in the crustaceans. This peptide was not identified in the mass spectrometry analyses, but the sequence of 118aa (12.9k Da) would exclude it from identification via this type of analysis on the basis of size (similarly to CHH and VIH/MIH) and this absence, therefore is not proof of absence in eyestalks. In contrast, its high expression level, without being definitive proof, supports the hypothesis of the site of production being in the eyestalks.
CCAP and bursicon, respectively, represent the subsequent molecules in the ecdysis cascade. They are not localised in the sinus gland of the eyestalks, but are known to be present in the same neuronal ganglions of the thorax and abdomen [114]–[116]. This simple difference explains their weak FPKM values. These two peptides are implicated in the final stages of moulting, as confirmed recently in the crab Caninus maenas [116], where, although they are the products of the same neurons in the pericardiac organ, they do not have the same expression kinetics, allied with important variations in the levels of CCAP during the moult cycle. This variant in expression was not observed for bursicon. The links in the chain controlling the moult cycle in krill were therefore clearly present, similar to most of the arthropods, in significant quantities, but their different localisation does not allow a full interpretation with regard to relating their variation in expression levels with the moult cycle.
Conclusions
Although the peptides of arthropods have been the subject of numerous studies, including transcriptomic approaches [7], [17], [117], [118], these have been largely focussed on hexapods and decapod crustaceans. Besides the interest in krill, as polar sentinels for climate change, these eucarids are also at the base of the decapod lineage [119] and therefore represent an important taxa for evolutionary studies. This study presents, for the first time an overview of neuropeptides in euphasiids. These have been principally described in E. crystallorophias, but also to a lesser extent in E. superba, some of which, to date, have only been characterised in a handful of arthropods. This study characterised new mature peptide sequences in krill (61 and 8, from E. crystallorophias and E. superba respectively) including, in most cases, the encoding pre-pro-peptide sequence.
In this paper, we describe neuropeptides belonging to several gene families encoded by 36 different precursor molecules. Within the E. crystallorophias peptides, 6 were confirmed by mass spectrometry on eyestalk extracts along with 6 precursor-related-peptides (Table 1). Numerous other peptides obtained by mass spectrometry were also identified in the transcriptome assemblies. However, when these sequences were searched against the databases, the matches retrieved classified them as “unknown”, even if the orthologues were found in arthropods with a complete genome sequence. Therefore, the description of this group of neuropeptides is almost certainly not exhaustive and underestimates the information available within both the sequence and protein data.
The transcriptomes in this study were produced from two sister species of krill, the ice krill and the Antarctic krill using two types of Next Generation Sequencing (454 for E. Superba and Illumina for E. crystallorophias) and analysed via two different assembly packages (Newbler and Trinity). The use of these two assemblers aided sequence validation, as did the comparative approach of using two species, which aided delineation of transcript structure, as there were notable sequence differences to those previously identified in other crustaceans or hexapods. The prime example of this was the characterisation of PDH-L, which to date has not been identified in any other species. Additionally, the presence of the long forms of PDHα in the proteome provided final proof of their translation and underlined the diversity of PDH isoforms and their probable physiological diversity.
The proteomic analysis carried out on the extracts of eyestalks of E. crystallorophias provided a final confirmation of sequence. This analysis also proved, as in the example of PDH-Lα, the existence of biologically active peptides (PRPs: Pro-peptide-Related Peptides), which were previously only predicted, based on the sequence of the pro-peptides. These data attest to the high level of expression and translation of mRNAs coding for these pre-pro-peptides in eyestalks as seen in the pro-peptides of allatostatin A, Arg7-corazonin 1, neuropeptide F1, PDH-Lβ1and 2, and tachykinin.
It was not surprising that most of the neuropeptides identified in E. crystallorophias were identified by sequence database searching, as all the peptides in this group are remarkably conserved. This included the PRPs present in certain pro-peptides indicating a strong selection pressure, at least on one of the paralogues, even if multiple copies of the gene were encoded in the genome. The sequences described here, were often highly similar to those of peneid shrimps, confirming phylogenetic studies which show them to share a common ancestor with the decapods [119].
The functionality of the peptides characterised in this study, must remain speculative, as these data are reliant upon predictions from studies carried out in neighbouring taxa, such as the hexapods or decapods. However, the sequences reported here have been identified as key molecules in the life history traits of a wide range of species, including the Crustacea. Hence a detailed understanding of these hormones will provide valuable biomarkers for studies into the effects of climate change and the potential trade-offs that occur between life history traits in a changing world. They will also enable more in-depth studies into the molecular evolution and phylogeny of krill.
Supporting Information
File includes Figures S1–S12. Figure S1. Alignment of the peptide sequences of the pre-pro-peptides of Corticotropin Releasing Factor Like Diuretic Hormone (CRFLDH) identified in the Pancrustacea. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S2. Alignment of the peptide sequences of the pre-pro-peptides of The Neuropeptide Fs (NPF) identified in the Pancrustacea. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectrometry during the course of this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S3. Alignment and Maldi TOF MS/MS spectra of the Tachykinin Related Peptides. Figure S3a: Alignment of the peptide sequences of the pre-pro-peptides of the Tachykinin Related Peptides (TKRP) identified in the Pancrustacea. The mature peptides are highlighted in blue. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectrometry during the course of this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S3b: Maldi TOF MS/MS spectra of MH+ APSGFLGMRa (Euc-TKRP); Figure S3c: Maldi TOF MS/MS spectra of MH+ pQVDPLSDALDQNQLAQTLYDYRD (Euc-TKRP-PRP). Figure S4. Maldi TOF MS/MS spectra of MH+ ARNYAFGIa (Euc-AST A3). Figure S5. Alignment of the peptide sequences from the pre-pro-peptides of the crustacean cardioactive peptide (CCAP) identified in the Eucrustacea. The mature peptide highlighted in blue belongs to E. crystallorophias. The signal peptides are highlighted in green and the potential dibasic cleavage sites in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S6. Alignments of the CHH and CHH precursor-related peptides (CPRP) identified in Eucrustacea. Figure S6a: Alignment of the CHH peptide sequences identified in the Eucrustacea. The mature peptide highlighted in blue belongs to E. crystallorophias. The six characteristic cysteines of the peptide are highlighted in yellow in this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S6b: Alignment of the peptide sequences of the CHH precursor-related peptides (CPRP) identified in the Eucrustacea. The CPRP highlighted in yellow belongs to E. crystallorophias. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S7. Alignment of the peptide sequences of VIH/MIH identified in the Eucrustacea. The mature peptide highlighted in blue belongs to E. crystallorophias. The six characteristic cysteines are highlighted in yellow. The glycine highlighted in red is equally characteristic of members of the CHH type II peptides. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S8. Alignment and Maldi TOF MS/MS spectra of Euc-Corazonins. Figure S8a: Alignment of the peptide sequences of the pre-pro-peptides of Corazonin (CRZ) identified in the Arthropods. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectroscopy in this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S8b: Maldi TOF MS/MS spectra of MH+ pQTFQYSRGWTNa (Euc-Arg7-CRZ1). Figure S9. Alignment of the peptide sequences of the pre-pro-peptides of the Eclosion Hormone (EH) identified in the Arthropods. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S10. Alignment of the peptide sequences of the pre-pro-peptides of the Neuroparsins (NP) identified in the Pancustacea. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S11. Alignment and Maldi TOF MS/MS spectra of PDH. Figure S11a: Alignment of the peptide sequences of the pre-pro-peptides of the Pigment Dispersing Hormones (PDH) and Pigment Dispersing Factors identified in the Pancrustacea. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectrometry during the course of this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S11b: Maldi TOF MS/MS spectra of MH+NSELINSMLGLPQTLRAQKLMANMa (Euc-PDH-L β1). Figure S12. Alignments of the peptide sequences of Red Pigment Concentrating Hormones (RPCH) identified in the Eucrustacea and Pancrustacea. Figure S12a: Alignment of the peptide sequences of the pre-pro-peptides of Red Pigment Concentrating Hormones (RPCH) identified in the Eucrustacea. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S12b: Alignment of the peptide sequences of the Red Pigment Concentrating Hormones (RPCH) and AKH identified in the Pancrustacea. The mature peptides highlighted in blue belong to the Eucrustacea. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S13. Alignment of the mature peptide sequences of the Allatostatin Cs identified in arthropods. The mature peptide highlighted in blue belongs to E. crystallorophias. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S14. Alignment of the peptide sequences of the pre-pro-peptides of Bursicon α and β identified in the Eucrustacea. Figure S14a: Alignment of the peptide sequences of the pre-pro-peptides of Bursicon α identified in the Eucrustacea. The mature peptide highlighted in blue belongs to E. crystallorophias. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S14b: Alignment of the peptide sequences of the pre-pro-peptides of Bursicon β identified in the Eucrustacea. The mature peptide highlighted in blue belongs to E. crystallorophias. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S15. Alignment of the peptide sequences of the pre-pro-peptides of Calcitonin-like Diuretic Hormone (CLDH) identified in the Arthropods. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectroscopy in this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S16. Alignment of the peptide sequences of the pre-pro-peptides of SIFamides identifed in the Arthropods. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences.
(PDF)
Acknowledgments
The authors would like to thank all the staff at IPEV and the team on the ship Astrolabe for their help during different Antarctic campaigns in Terre Adélie.
Funding Statement
KC received a PhD grant by the Émergence-UPMC 2011 research program and the «Région Bretagne». JYT benefits from funding provided by Institut Paul Emile Victor (KREVET program) and also by la Région Bretagne (SAD-1 – DRAKAR program). MSC and MAST were funded by Natural Environment Research Council core funding to the British Antarctic Survey. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jarman S, Elliott N, Nicol S, McMinn A (2000) Molecular phylogenetics of circumglobal Euphausia species (Euphausia: Crustacea). Canadian Journal of Fisheries and Aquatic Sciences 57 (suppl 3)51–58. [Google Scholar]
- 2.Kirkwood (1982) A guide to the Euphasiacea of the Southern Ocean. ANARE Research Notes 1: Published by AAD.
- 3.Jeffery NW (2011) The first genome size estimates for six species of krill (Malacostraca, Euphausiidae): large genomes at the north and south poles. Polar Biology.
- 4.Clark MS, Thorne MAS, Toullec JY, Meng Y, Guan LL, et al.. (2011) Antarctic Krill 454 Pyrosequencing Reveals Chaperone and Stress Transcriptome. Plos One 6. [DOI] [PMC free article] [PubMed]
- 5.Montagné N, Desdevises Y, Soyez D, Toullec JY (2010) Molecular evolution of the crustacean hyperglycemic hormone family in ecdysozoans. BMC Evolutionary Biology 10: –. [DOI] [PMC free article] [PubMed]
- 6. Christie AE, McCoole MD, Harmon SM, Baer KN, Lenz PH (2011) Genomic analyses of the Daphnia pulex peptidome. General and Comparative Endocrinology 171: 131–150. [DOI] [PubMed] [Google Scholar]
- 7. Dircksen H, Neupert S, Predel R, Verleyen P, Huybrechts J, et al. (2011) Genomics, Transcriptomics, and Peptidomics of Daphnia pulex Neuropeptides and Protein Hormones. Journal of Proteome Research 10: 4478–4504. [DOI] [PubMed] [Google Scholar]
- 8. Schmieder R, Lim YW, Edwards R (2012) Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics 28: 433–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17: 10–12. [Google Scholar]
- 10. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- 12. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 14.Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10. [DOI] [PMC free article] [PubMed]
- 15.Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12. [DOI] [PMC free article] [PubMed]
- 16. Christie AE, Nolan DH, Ohno P, Hartline N, Lenz PH (2011) Identification of chelicerate neuropeptides using bioinformatics of publicly accessible expressed sequence tags. General and Comparative Endocrinology 170: 144–155. [DOI] [PubMed] [Google Scholar]
- 17. Ma MM, Gard AL, Xiang F, Wang JH, Davoodian N, et al. (2010) Combining in silico transcriptome mining and biological mass spectrometry for neuropeptide discovery in the Pacific white shrimp Litopenaeus vannamei . Peptides 31: 27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Christie AE, Durkin CS, Hartline N, Ohno P, Lenz PH (2010) Bioinformatic analyses of the publicly accessible crustacean expressed sequence tags (ESTs) reveal numerous novel neuropeptide-encoding precursor proteins, including ones from members of several little studied taxa. General and Comparative Endocrinology 167: 164–178. [DOI] [PubMed] [Google Scholar]
- 19. Ma MM, Bors EK, Dickinson ES, Kwiatkowski MA, Sousa GL, et al. (2009) Characterization of the Carcinus maenas neuropeptidome by mass spectrometry and functional genomics. General and Comparative Endocrinology 161: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kay I, Patel M, Coast GM, Totty NF, Mallet AI, et al. (1992) Isolation, characterization and biological activity of a CRF-related diuretic peptide from Periplaneta americana L. Regul Pept. 42: 111–122. [DOI] [PubMed] [Google Scholar]
- 21. Coast GM (1996) Neuropeptides implicated in the control of diuresis in insects. Peptides 17: 327–336. [DOI] [PubMed] [Google Scholar]
- 22. Li B, Predel R, Neupert S, Hauser F, Tanaka Y, et al. (2008) Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum . Genome Research 18: 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Te Brugge V, Paluzzi JP (2011) Schooley DA, Orchard I (2011) Identification of the elusive peptidergic diuretic hormone in the blood-feeding bug Rhodnius prolixus: a CRF-related peptide. J Exp Biol 214: 371–381. [DOI] [PubMed] [Google Scholar]
- 24. Christie AE (2008) Neuropeptide discovery in Ixodoidea: An in silico investigation using publicly accessible expressed sequence tags. General and Comparative Endocrinology 157: 174–185. [DOI] [PubMed] [Google Scholar]
- 25. Coast GM, TeBrugge VA, Nachman RJ, Lopez J, Aldrich JR, et al. (2010) Neurohormones implicated in the control of Malpighian tubule secretion in plant sucking heteropterans: The stink bugs Acrosternum hilare and Nezara viridula . Peptides 31: 468–473. [DOI] [PubMed] [Google Scholar]
- 26. Paluzzi JP, Naikkhwah W, O'Donnell MJ (2012) Natriuresis and diuretic hormone synergism in R. prolixus upper Malpighian tubules is inhibited by the anti-diuretic hormone, RhoprCAPA-alpha2. Journal of Insect Physiology 58: 534–542. [DOI] [PubMed] [Google Scholar]
- 27. Sithigorngul P, Pupuem H, Krungkolsem C, Longyant S, Panchan N, et al. (2002) Four novel PYFs: members of NPY/PP peptide superfamily from the eyestalk of the giant tiger prawn Penaeus monodon . Peptides 23: 1895–1906. [DOI] [PubMed] [Google Scholar]
- 28. Stemmler EA, Bruns EA, Gardner NP, Dickinson PS, Christie AE (2007) Mass spectrometric identification of pEGFYSQRYamide: A crustacean peptide hormone possessing a vertebrate neuropeptide Y (NPY)-like carboxy-terminus. General and Comparative Endocrinology 152: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Christie AE, Cashman CR, Brennan HR, Ma MM, Sousa GL, et al. (2008) Identification of putative crustacean neuropeptides using in silico analyses of publicly accessible expressed sequence tags. General and Comparative Endocrinology 156: 246–264. [DOI] [PubMed] [Google Scholar]
- 30. Christie AE, Chapline MC, Jackson JM, Dowda JK, Hartline N, et al. (2011) Identification, tissue distribution and orexigenic activity of neuropeptide F (NPF) in penaeid shrimp. Journal of Experimental Biology 214: 1386–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nassel DR, Wegener C (2011) A comparative review of short and long neuropeptide F signaling in invertebrates: Any similarities to vertebrate neuropeptide Y signaling? Peptides 32: 1335–1355. [DOI] [PubMed] [Google Scholar]
- 32. Roller L, Yamanaka N, Watanabe K, Daubnerova I, Zitnan D, et al. (2008) The unique evolution of neuropeptide genes in the silkworm Bombyx mori . Insect Biochemistry and Molecular Biology 38: 1147–1157. [DOI] [PubMed] [Google Scholar]
- 33. Christie AE, Lundquist CT, Nassel DR, Nusbaum MP (1997) Two novel tachykinin-related peptides from the nervous system of the crab Cancer borealis . Journal of Experimental Biology 200: 2279–2294. [DOI] [PubMed] [Google Scholar]
- 34. Christie AE, Kutz-Naber KK, Stemmler EA, Klein A, Messinger DI, et al. (2007) Midgut epithelial endocrine cells are a rich source of the neuropeptides APSGFLGMRamide (Cancer borealis tachykinin-related peptide Ia) and GYRKPPFNGSIFamide (Gly(1)-SIFamide) in the crabs Cancer borealis, Cancer magister and Cancer productus . Journal of Experimental Biology 210: 699–714. [DOI] [PubMed] [Google Scholar]
- 35. Stemmler EA, Peguero B, Bruns EA, Dickinson PS, Christie AE (2007) Identification, physiological actions, and distribution of TPSGFLGMRamide: a novel tachykinin-related peptide from the midgut and stomatogastric nervous system of Cancer crabs. Journal of Neurochemistry 101: 1351–1366. [DOI] [PubMed] [Google Scholar]
- 36. Sousa GL, Lenz PH, Hartline DK, Christie AE (2008) Distribution of pigment dispersing hormone- and tachykinin-related peptides in the central nervous system of the copepod crustacean Calanus finmarchicus . General and Comparative Endocrinology 156: 454–459. [DOI] [PubMed] [Google Scholar]
- 37. Christie AE, Cashman CR, Stevens JS, Smith CM, Beale KM, et al. (2008) Identification and cardiotropic actions of brain/gut-derived tachykinin-related peptides (TRPs) from the American lobster Homarus americanus . Peptides 29: 1909–1918. [DOI] [PubMed] [Google Scholar]
- 38. Belles X, GRAHAM LA, BENDENA WG, DING Q, EDWARDS JP, et al. (1999) The molecular evolution of the allatostatin precursor in cockroaches. Peptides 20: 11–22. [DOI] [PubMed] [Google Scholar]
- 39. Yasuda-Kamatani Y, Yasuda A (2006) Characteristic expression patterns of allatostatin-like peptide, FMRFamide-related peptide, orcokinin, tachykinin-related peptide, and SIFamide in the olfactory system of crayfish Procambarus clarkii . Journal of Comparative Neurology 496: 135–147. [DOI] [PubMed] [Google Scholar]
- 40. Yin GL, Yang JS, Cao JX, Yang WJ (2006) Molecular cloning and characterization of FGLamide allatostatin gene from the prawn, Macrobrachium rosenbergii . Peptides 27: 1241–1250. [DOI] [PubMed] [Google Scholar]
- 41. Christie AE, Sousa GL, Rus S, Smith CM, Towle DW, et al. (2008) Identification of A-type allatostatins possessing -YXFGI/Vamide carboxy-termini from the nervous system of the copepod crustacean Calanus finmarchicus . Gen Comp Endocrinol 155: 526–533. [DOI] [PubMed] [Google Scholar]
- 42. Dircksen H, Bocking D, Heyn U, Mandel C, Chung JS, et al. (2001) Crustacean hyperglycaemic hormone (CHH)-like peptides and CHH-precursor-related peptides from pericardial organ neurosecretory cells in the shore crab, Carcinus maenas, are putatively spliced and modified products of multiple genes. Biochem J 356: 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Montagne N, Soyez D, Gallois D, Ollivaux C, Toullec JY (2008) New insights into evolution of crustacean hyperglycaemic hormone in decapods-first characterization in Anomura. FEBS J 275: 1039–1052. [DOI] [PubMed] [Google Scholar]
- 44. Gu PL, Tobe SS, Chow BK, Chu KH, He JG, et al. (2002) Characterization of an additional molt inhibiting hormone-like neuropeptide from the shrimp Metapenaeus ensis . Peptides 23: 1875–1883. [DOI] [PubMed] [Google Scholar]
- 45. Krungkasem C, Ohira T, Yang WJ, Abdullah R, Nagasawa H, et al. (2002) Identification of two distinct molt-inhibiting hormone-related peptides from the giant tiger prawn Penaeus monodon . Mar Biotechnol (NY) 4: 132–140. [DOI] [PubMed] [Google Scholar]
- 46. Wang ZZ, Xiang JH (2003) [Cloning and analysis of three genes encoding type II CHH family neuropeptides from Fenneropenaeus chinensis]. Yi Chuan Xue Bao 30: 961–966. [PubMed] [Google Scholar]
- 47. Ohira T, Katayama H, Tominaga S, Takasuka T, Nakatsuji T, et al. (2005) Cloning and characterization of a molt-inhibiting hormone-like peptide from the prawn Marsupenaeus japonicus . Peptides 26: 259–268. [DOI] [PubMed] [Google Scholar]
- 48. Chen HY, Watson RD, Chen JC, Liu HF, Lee CY (2007) Molecular characterization and gene expression pattern of two putative molt-inhibiting hormones from Litopenaeus vannamei . Gen Comp Endocrinol 151: 72–81. [DOI] [PubMed] [Google Scholar]
- 49. Yang WJ, Rao KR (2001) Cloning of precursors for two MIH/VIH-related peptides in the prawn, Macrobrachium rosenbergii . Biochem Biophys Res Commun 289: 407–413. [DOI] [PubMed] [Google Scholar]
- 50. Veenstra JA (1989) Isolation and Structure of Corazonin, a Cardioactive Peptide from the American Cockroach. Febs Letters 250: 231–234. [DOI] [PubMed] [Google Scholar]
- 51. Predel R, Neupert S, Russell WK, Scheibner O, Nachman RJ (2007) Corazonin in insects. Peptides 28: 3–10. [DOI] [PubMed] [Google Scholar]
- 52. Boerjan B, Verleyen P, Huybrechts J, Schoofs L, De Loof A (2010) In search for a common denominator for the diverse functions of arthropod corazonin: A role in the physiology of stress? General and Comparative Endocrinology 166: 222–233. [DOI] [PubMed] [Google Scholar]
- 53. Tawfik AI, Tanaka S, De Loof A, Schoofs L, Baggerman G, et al. (1999) Identification of the gregarization-associated dark-pigmentotropin in locusts through an albino mutant. Proceedings of the National Academy of Sciences of the United States of America 96: 7083–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoste B, Simpson SJ, Tanaka S, Zhu DH, De Loof A, et al. (2002) Effects of [His(7)]-corazonin on the phase state of isolated-reared (solitarious) desert locusts, Schistocerca gregaria. . Journal of Insect Physiology 48: 981–990. [DOI] [PubMed] [Google Scholar]
- 55. Hoste B, Simpson SJ, Tanaka S, De Loof A, Breuer M (2002) A comparison of phase-related shifts in behavior and morphometrics of an albino strain, deficient in [His(7)]-corazonin, and a normally colored Locusta migratoria strain. Journal of Insect Physiology 48: 791–801. [DOI] [PubMed] [Google Scholar]
- 56. Davis MM, O'Keefe SL, Primrose DA, Hodgetts RB (2007) A neuropeptide hormone cascade controls the precise onset of post-eclosion cuticular tanning in Drosophila melanogaster . Development 134: 4395–4404. [DOI] [PubMed] [Google Scholar]
- 57. Arakane Y, Li B, Muthukrishnan S, Beeman RW, Kramer KJ, et al. (2008) Functional analysis of four neuropeptides, EH, ETH, CCAP and bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum . Mechanisms of Development 125: 984–995. [DOI] [PubMed] [Google Scholar]
- 58. Rao KR (2001) Crustacean pigmentary-effector hormones: Chemistry and functions of RPCH, PDH, and related peptides. American Zoologist 41: 364–379. [Google Scholar]
- 59. Helfrichforster C, Homberg U (1993) Pigment-Dispersing Hormone-Immunoreactive Neurons in the Nervous-System of Wild-Type Drosophila melanogaster and of Several Mutants with Altered Circadian Rhythmicity. Journal of Comparative Neurology 337: 177–190. [DOI] [PubMed] [Google Scholar]
- 60. Nassel DR, Shiga S, Mohrherr CJ, Rao KR (1993) Pigment-Dispersing Hormone-Like Peptide in the Nervous-System of the Flies Phormia and Drosophila – Immunocytochemistry and Partial Characterization. Journal of Comparative Neurology 331: 183–198. [DOI] [PubMed] [Google Scholar]
- 61. Shiga S, Rao KR, Nassel DR (1993) Pigment-Dispersing Hormone Immunoreactive Neurons in the Blowfly Nervous-System. Acta Biologica Hungarica 44: 55–59. [PubMed] [Google Scholar]
- 62.Strauss J, Dircksen H (2010) Circadian clocks in crustaceans: identified neuronal and cellular systems. Frontiers in Bioscience-Landmark 15: 1040–+. [DOI] [PubMed]
- 63. Strauss J, Zhang Q, Verleyen P, Huybrechts J, Neupert S, et al. (2011) Pigment-dispersing hormone in Daphnia interneurons, one type homologous to insect clock neurons displaying circadian rhythmicity. Cellular and Molecular Life Sciences 68: 3403–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Teschke M, Kawaguchi S, Meyer B (2007) Simulated light regimes affect feeding and metabolism of Antarctic krill, Euphausia superba . Limnology and Oceanography 52: 1046–1054. [Google Scholar]
- 65.Teschke M, Wendt S, Kawaguchi S, Kramer A, Meyer B (2011) A Circadian Clock in Antarctic Krill: An Endogenous Timing System Governs Metabolic Output Rhythms in the Euphausid Species Euphausia superba. Plos One 6. [DOI] [PMC free article] [PubMed]
- 66. Gade G (2009) Peptides of the Adipokinetic Hormone/Red Pigment-Concentrating Hormone Family A New Take on Biodiversity. Trends in Comparative Endocrinology and Neurobiology 1163: 125–136. [DOI] [PubMed] [Google Scholar]
- 67. Bogerd J, Kooiman FP, Pijnenburg MAP, Hekking LHP, Oudejans RCHM, et al. (1995) Molecular-Cloning of 3 Distinct Cdnas, Each Encoding a Different Adipokinetic Hormone Precursor, of the Migratory Locust, Locusta migratoria – Differential Expression of the Distinct Adipokinetic Hormone Precursor Genes during Flight Activity. Journal of Biological Chemistry 270: 23038–23043. [DOI] [PubMed] [Google Scholar]
- 68. Lorenz MW, Kellner R, Hoffmann KH (1995) IDENTIFICATION OF 2 Allatostatins from the cricket, Gryllus bimaculatus Degeer (Ensifera, Gryllidae) – Additional members of a family of neuropeptides inhibiting juvenile-hormone biosynthesis. Regulatory Peptides 57: 227–236. [DOI] [PubMed] [Google Scholar]
- 69. Lorenz MW, Kellner R, Hoffmann KH (1999) Allatostatins in Gryllus bimaculatus (Ensifera: Gryllidae): New structures and physiological properties. European Journal of Entomology 96: 267–274. [Google Scholar]
- 70. Stay B, Tobe SS (2007) The role of allatostatins in juvenile hormone synthesis in insects and crustaceans. Annual Review of Entomology 52: 277–299. [DOI] [PubMed] [Google Scholar]
- 71. Szabo TM, Chen R, Goeritz ML, Maloney RT, Tang LS, et al. (2011) Distribution and physiological effects of B-type allatostatins (myoinhibitory peptides, MIPs) in the stomatogastric nervous system of the crab Cancer borealis . The Journal of Comparative Neurology 519: 2658–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kramer SJ, Toschi A, Miller CA, Kataoka H, Quistad GB, et al. (1991) Identification of an Allatostatin from the Tobacco Hornworm Manduca sexta . Proceedings of the National Academy of Sciences of the United States of America 88: 9458–9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dickinson PS, Wiwatpanit T, Gabranski ER, Ackerman RJ, Stevens JS, et al. (2009) Identification of SYWKQCAFNAVSCFamide: a broadly conserved crustacean C-type allatostatin-like peptide with both neuromodulatory and cardioactive properties. J Exp Biol 212: 1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gard AL, Lenz PH, Shaw JR, Christie AE (2009) Identification of putative peptide paracrines/hormones in the water flea Daphnia pulex (Crustacea; Branchiopoda; Cladocera) using transcriptomics and immunohistochemistry. General and Comparative Endocrinology 160: 271–287. [DOI] [PubMed] [Google Scholar]
- 75. Dircksen H, Neupert S, Predel R, Verleyen P, Huybrechts J, et al. (2011) Genomics, Transcriptomics, and Peptidomics of Daphnia pulex Neuropeptides and Protein Hormones. Journal of Proteome Research 10: 4478–4504. [DOI] [PubMed] [Google Scholar]
- 76. Stemmler EA, Bruns EA, Cashman CR, Dickinson PS, Christie AE (2010) Molecular and mass spectral identification of the broadly conserved decapod crustacean neuropeptide pQIRYHQCYFNPISCF: The first PISCF-allatostatin (Manduca sexta- or C-type allatostatin) from a non-insect. General and Comparative Endocrinology 165: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ma M, Szabo TM, Jia C, Marder E, Li L (2009) Mass spectrometric characterization and physiological actions of novel crustacean C-type allatostatins. Peptides 30: 1660–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vincent JF (1969) Bursicon in Locusta, Schistocerca and Periplaneta. Release and Effects of Hormone in Locusta. General and Comparative Endocrinology 13: 538–&.
- 79.Mills RR (1967) Control of Cuticular Tanning in Cockroach – Bursicon Release by Nervous Stimulation. Journal of Insect Physiology 13: 815–&. [DOI] [PubMed]
- 80.Fraenkel G, Hsiao C, Seligman M (1966) Properties of Bursicon – an Insect Protein Hormone That Controls Cuticular Tanning. Science 151: 91–&. [DOI] [PubMed]
- 81. Sharp J, Wilcockson D, Webster S (2008) Identification and expression of mRNAs encoding bursicon in the central nervous system of decapod crustaceans. Comparative Biochemistry and Physiology a-Molecular & Integrative Physiology 150: S101–S101. [Google Scholar]
- 82. Luo CW, Dewey EM, Sudo S, Ewer J, Hsu SY, et al. (2005) Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proceedings of the National Academy of Sciences of the United States of America 102: 2820–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mendive FM, Van Loy T, Claeysen S, Poels J, Williamson M, et al. (2005) Drosophila molting neurohormone bursicon is a heterodimer and the natural agonist of the orphan receptor DLGR2. Febs Letters 579: 2171–2176. [DOI] [PubMed] [Google Scholar]
- 84. Honegger HW, Dewey EM, Ewer J (2008) Bursicon, the tanning hormone of insects: recent advances following the discovery of its molecular identity. Journal of Comparative Physiology A-Neuroethology Sensory Neural and Behavioral Physiology 194: 989–1005. [DOI] [PubMed] [Google Scholar]
- 85. Furuya K, Milchak RJ, Schegg KM, Zhang JR, Tobe SS, et al. (2000) Cockroach diuretic hormones: Characterization of a calcitonin-like peptide in insects. Proceedings of the National Academy of Sciences of the United States of America 97: 6469–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Christie AE, Stevens JS, Bowers MR, Chapline MC, Jensen DA, et al. (2010) Identification of a calcitonin-like diuretic hormone that functions as an intrinsic modulator of the American lobster, Homarus americanus, cardiac neuromuscular system. J Exp Biol 213: 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Huybrechts J, Bonhomme J, Minoli S, Prunier-Leterme N, Dombrovsky A, et al. (2010) Neuropeptide and neurohormone precursors in the pea aphid, Acyrthosiphon pisum . Insect Molecular Biology 19: 87–95. [DOI] [PubMed] [Google Scholar]
- 88. Ma MM, Chen RB, Sousa GL, Bors EK, Kwiatkowski MA, et al. (2008) Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the American lobster Homarus americanus . General and Comparative Endocrinology 156: 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Verleyen P, Huybrechts J, Schoofs L (2009) SIFamide illustrates the rapid evolution in Arthropod neuropeptide research. General and Comparative Endocrinology 162: 27–35. [DOI] [PubMed] [Google Scholar]
- 90. Janssen I, Schoofs L, Spittaels K, Neven H, VandenBroeck J, et al. (1996) Isolation of NEB-LFamide, a novel myotropic neuropeptide from the grey fleshfly. Molecular and Cellular Endocrinology 117: 157–165. [DOI] [PubMed] [Google Scholar]
- 91. Verleyen P, Huybrechts J, Baggerman G, Van Lommel A, De Loof A, et al. (2004) SIFamide is a highly conserved neuropeptide: a comparative study in different insect species. Biochemical and Biophysical Research Communications 320: 334–341. [DOI] [PubMed] [Google Scholar]
- 92. Audsley N, Weaver RJ (2006) Analysis of peptides in the brain and corpora cardiaca-corpora allata of the honey bee, Apis mellifera using MALDI-TOF mass spectrometry. Peptides 27: 512–520. [DOI] [PubMed] [Google Scholar]
- 93. Weaver RJ, Audsley N (2008) Neuropeptides of the beetle, Tenebrio molitor identified using MALDI-TOF mass spectrometry and deduced sequences from the Tribolium castaneum genome. Peptides 29: 168–178. [DOI] [PubMed] [Google Scholar]
- 94. Christie AE (2008) In silico analyses of peptide paracrines/hormones in Aphidoidea. General and Comparative Endocrinology 159: 67–79. [DOI] [PubMed] [Google Scholar]
- 95. Sithigorngul P, Pupuem J, Krungkasem C, Longyant S, Chaivisuthangkura P, et al. (2002) Seven novel FMRFamide-like neuropeptide sequences from the eyestalk of the giant tiger prawn Penaeus monodon . Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology 131: 325–337. [DOI] [PubMed] [Google Scholar]
- 96. Stemmler EA, Cashman CR, Messinger DI, Gardner NP, Dickinson PS, et al. (2007) High-mass-resolution direct-tissue MALDI-FTMS reveals broad conservation of three neuropeptides (APSGFLGMRamide, GYRKPPFNGSIFamide and pQDLDHVFLRFamide) across members of seven decapod crustaean infraorders. Peptides 28: 2104–2115. [DOI] [PubMed] [Google Scholar]
- 97. Dickinson PS, Stemmler EA, Cashman CR, Brennan HR, Dennison B, et al. (2008) SIFamide peptides in clawed lobsters and freshwater crayfish (Crustacea, Decapoda, Astacidea): A combined molecular, mass spectrometric and electrophysiological investigation. General and Comparative Endocrinology 156: 347–360. [DOI] [PubMed] [Google Scholar]
- 98. Veenstra JA (2000) Mono- and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Archives of Insect Biochemistry and Physiology 43: 49–63. [DOI] [PubMed] [Google Scholar]
- 99. Yasuda A, Yasuda-Kamatani Y, Nozaki M, Nakajima T (2004) Identification of GYRKPPFNGSIFamide (crustacean- SIFamide) in the crayfish Procambarus clarkii by topological mass spectrometry analysis. General and Comparative Endocrinology 135: 391–400. [DOI] [PubMed] [Google Scholar]
- 100. Hsu YWA, Stemmler EA, Messinger DI, Dickinson PS, Christie AE, et al. (2008) Cloning and differential expression of two beta-pigment-dispersing hormone (beta-PDH) isoforms in the crab Cancer productus: Evidence for authentic beta-PDH as a local neurotransmitter and beta-PDH II as a humoral factor. Journal of Comparative Neurology 508: 197–211. [DOI] [PubMed] [Google Scholar]
- 101. Verde MA, Barriga-Montoya C, Fuentes-Pardo B (2007) Pigment dispersing hormone generates a circadian response to light in the crayfish, Procambarus clarkii . Comparative Biochemistry and Physiology a-Molecular & Integrative Physiology 147: 983–992. [DOI] [PubMed] [Google Scholar]
- 102. Sullivan JM, Genco MC, Marlow ED, Benton JL, Beltz BS, et al. (2009) Brain Photoreceptor Pathways Contributing to Circadian Rhythmicity in Crayfish. Chronobiology International 26: 1136–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Helfrich-Forster C (2009) Neuropeptide PDF plays multiple roles in the circadian clock of Drosophila melanogaster . Sleep and Biological Rhythms 7: 130–143. [Google Scholar]
- 104. Harzsch S, Dircksen H, Beltz B (2009) Development of pigment-dispersing hormone-immunoreactive neurons in the American lobster: homology to the insect circadian pacemaker system? Cell and Tissue Research 335: 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wilcockson DC, Zhang L, Hastings MH, Kyriacou CP, Webster SG (2011) A Novel Form of Pigment-Dispersing Hormone in the Central Nervous System of the Intertidal Marine Isopod, Eurydice pulchra (Leach). Journal of Comparative Neurology 519: 562–575. [DOI] [PubMed] [Google Scholar]
- 106. Hsu YWA, Stemmler EA, Messinger DI, Dickinson PS, Christie AE, et al. (2010) Cloning and Differential Expression of Two beta-Pigment-Dispersing Hormone (beta-PDH) Isoforms in the Crab Cancer Productus: Evidence for Authentic beta-PDH as a Local Neurotransmitter and beta-PDH II as a Humoral Factor (vol 508, pg 197, 2008). Journal of Comparative Neurology 518: 4674–4674. [DOI] [PubMed] [Google Scholar]
- 107. Kegel G, Reichwein B, Weese S, Gaus G, Peter-Katalinic J, et al. (1989) Amino acid sequence of the crustacean hyperglycemic hormone (CHH) from the shore crab, Carcinus maenas . FEBS Lett 255: 10–14. [DOI] [PubMed] [Google Scholar]
- 108. Webster SG, Keller R, Dircksen H (2012) The CHH-superfamily of multifunctional peptide hormones controlling crustacean metabolism, osmoregulation, moulting, and reproduction. Gen Comp Endocrinol 175: 217–233. [DOI] [PubMed] [Google Scholar]
- 109. Glowik RM, Golowasch J, Keller R, Marder E (1997) D-glucose-sensitive neurosecretory cells of the crab Cancer borealis and negative feedback regulation of blood glucose level. J Exp Biol 200: 1421–1431. [DOI] [PubMed] [Google Scholar]
- 110. Buchholz F, Watkins JL, Priddle J, Morris DJ, Ricketts C (1996) Moult in relation to some aspects of reproduction and growth in swarms of Antarctic krill, Euphausia superba . Marine Biology 127: 201–208. [Google Scholar]
- 111. Cuzin-Roudy J (2000) Seasonal reproduction, multiple spawning, and fecundity in northern krill, Meganyctiphanes norvegica, and Antarctic krill, Euphausia superba . Canadian Journal of Fisheries and Aquatic Sciences 57: 6–15. [Google Scholar]
- 112. Sefiani M, LeCaer JP, Soyez D (1996) Characterization of hyperglycemic and molt-inhibiting activity from sinus glands of the penaeid shrimp Penaeus vannamei . General and Comparative Endocrinology 103: 41–53. [DOI] [PubMed] [Google Scholar]
- 113. Gammie SC, Truman JW (1999) Eclosion hormone provides a link between ecdysis-triggering hormone and crustacean cardioactive peptide in the neuroendocrine cascade that controls ecdysis behavior. Journal of Experimental Biology 202: 343–352. [DOI] [PubMed] [Google Scholar]
- 114. Sharp JH, Wilcockson DC, Webster SG (2010) Identification and expression of mRNAs encoding bursicon in the plesiomorphic central nervous system of Homarus gammarus . General and Comparative Endocrinology 169: 65–74. [DOI] [PubMed] [Google Scholar]
- 115. Wilcockson DC, Webster SG (2008) Identification and developmental expression of mRNAs encoding putative insect cuticle hardening hormone, bursicon in the green shore crab Carcinus maenas . General and Comparative Endocrinology 156: 113–125. [DOI] [PubMed] [Google Scholar]
- 116. Webster S, Wilcockson D, Mrinalini, sharp JH (2013) Bursicon and neuropeptide cascades during the ecdysis program of the shore crab, Carcinus maenas . Gen Comp Endocrinol 182: 54–64. [DOI] [PubMed] [Google Scholar]
- 117. Bissinger BW, Donohue KV, Khalil SMS, Grozinger CM, Sonenshine DE, et al. (2011) Synganglion transcriptome and developmental global gene expression in adult females of the American dog tick, Dermacentor variabilis (Acari: Ixodidae). Insect Molecular Biology 20: 465–491. [DOI] [PubMed] [Google Scholar]
- 118.Badisco L, Huybrechts J, Simonet G, Verlinden H, Marchal E, et al. (2011) Transcriptome Analysis of the Desert Locust Central Nervous System: Production and Annotation of a Schistocerca gregaria EST Database. Plos One 6. [DOI] [PMC free article] [PubMed]
- 119. Johansson ML, Sremba AL, Feinberg LR, Banks MA, Peterson WT (2012) The mitochondrial genomes of Euphausia pacifica and Thysanoessa raschii sequenced using 454 next-generation sequencing, with a phylogenetic analysis of their position in the Malacostracan family tree. Mol Biol Rep 39: 9009–9021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File includes Figures S1–S12. Figure S1. Alignment of the peptide sequences of the pre-pro-peptides of Corticotropin Releasing Factor Like Diuretic Hormone (CRFLDH) identified in the Pancrustacea. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S2. Alignment of the peptide sequences of the pre-pro-peptides of The Neuropeptide Fs (NPF) identified in the Pancrustacea. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectrometry during the course of this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S3. Alignment and Maldi TOF MS/MS spectra of the Tachykinin Related Peptides. Figure S3a: Alignment of the peptide sequences of the pre-pro-peptides of the Tachykinin Related Peptides (TKRP) identified in the Pancrustacea. The mature peptides are highlighted in blue. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectrometry during the course of this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S3b: Maldi TOF MS/MS spectra of MH+ APSGFLGMRa (Euc-TKRP); Figure S3c: Maldi TOF MS/MS spectra of MH+ pQVDPLSDALDQNQLAQTLYDYRD (Euc-TKRP-PRP). Figure S4. Maldi TOF MS/MS spectra of MH+ ARNYAFGIa (Euc-AST A3). Figure S5. Alignment of the peptide sequences from the pre-pro-peptides of the crustacean cardioactive peptide (CCAP) identified in the Eucrustacea. The mature peptide highlighted in blue belongs to E. crystallorophias. The signal peptides are highlighted in green and the potential dibasic cleavage sites in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S6. Alignments of the CHH and CHH precursor-related peptides (CPRP) identified in Eucrustacea. Figure S6a: Alignment of the CHH peptide sequences identified in the Eucrustacea. The mature peptide highlighted in blue belongs to E. crystallorophias. The six characteristic cysteines of the peptide are highlighted in yellow in this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S6b: Alignment of the peptide sequences of the CHH precursor-related peptides (CPRP) identified in the Eucrustacea. The CPRP highlighted in yellow belongs to E. crystallorophias. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S7. Alignment of the peptide sequences of VIH/MIH identified in the Eucrustacea. The mature peptide highlighted in blue belongs to E. crystallorophias. The six characteristic cysteines are highlighted in yellow. The glycine highlighted in red is equally characteristic of members of the CHH type II peptides. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S8. Alignment and Maldi TOF MS/MS spectra of Euc-Corazonins. Figure S8a: Alignment of the peptide sequences of the pre-pro-peptides of Corazonin (CRZ) identified in the Arthropods. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectroscopy in this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S8b: Maldi TOF MS/MS spectra of MH+ pQTFQYSRGWTNa (Euc-Arg7-CRZ1). Figure S9. Alignment of the peptide sequences of the pre-pro-peptides of the Eclosion Hormone (EH) identified in the Arthropods. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S10. Alignment of the peptide sequences of the pre-pro-peptides of the Neuroparsins (NP) identified in the Pancustacea. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S11. Alignment and Maldi TOF MS/MS spectra of PDH. Figure S11a: Alignment of the peptide sequences of the pre-pro-peptides of the Pigment Dispersing Hormones (PDH) and Pigment Dispersing Factors identified in the Pancrustacea. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectrometry during the course of this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S11b: Maldi TOF MS/MS spectra of MH+NSELINSMLGLPQTLRAQKLMANMa (Euc-PDH-L β1). Figure S12. Alignments of the peptide sequences of Red Pigment Concentrating Hormones (RPCH) identified in the Eucrustacea and Pancrustacea. Figure S12a: Alignment of the peptide sequences of the pre-pro-peptides of Red Pigment Concentrating Hormones (RPCH) identified in the Eucrustacea. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S12b: Alignment of the peptide sequences of the Red Pigment Concentrating Hormones (RPCH) and AKH identified in the Pancrustacea. The mature peptides highlighted in blue belong to the Eucrustacea. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S13. Alignment of the mature peptide sequences of the Allatostatin Cs identified in arthropods. The mature peptide highlighted in blue belongs to E. crystallorophias. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S14. Alignment of the peptide sequences of the pre-pro-peptides of Bursicon α and β identified in the Eucrustacea. Figure S14a: Alignment of the peptide sequences of the pre-pro-peptides of Bursicon α identified in the Eucrustacea. The mature peptide highlighted in blue belongs to E. crystallorophias. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S14b: Alignment of the peptide sequences of the pre-pro-peptides of Bursicon β identified in the Eucrustacea. The mature peptide highlighted in blue belongs to E. crystallorophias. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S15. Alignment of the peptide sequences of the pre-pro-peptides of Calcitonin-like Diuretic Hormone (CLDH) identified in the Arthropods. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The PRP highlighted in yellow was characterised by mass spectroscopy in this study. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences. Figure S16. Alignment of the peptide sequences of the pre-pro-peptides of SIFamides identifed in the Arthropods. The mature peptides are highlighted in blue only for E. crystallorophias. The signal peptides are highlighted in green and the potential bibasic cleavage sites, in red. The histogram at the base of the figure indicates the level of conservation of each amino acid in the sequences.
(PDF)