Abstract
Sister chromatid separation depends on the activity of separase, which in turn requires the proteolysis of its inhibitor, securin. It has been speculated that securin also supports the activation of separase. In this study, we found that PTTG1 was the major securin isoform expressed in most normal and cancer cell lines. Remarkably, a highly homologous isoform called PTTG2 was unable to interact with separase. Using chimeras between PTTG1 and PTTG2 and other approaches, we pinpointed a single amino acid that accounted for the loss of securin function in PTTG2. Mutation of the homologous position in PTTG1 (H134) switched PTTG1 from an inhibitor into an activator of separase. In agreement with this, PTTG1 lacking H134 was able to trigger premature sister chromatid separation. Conversely, introduction of H134 into PTTG2 is sufficient to allow it to bind separase. These data demonstrate that while the motif containing H134 has a strong affinity for separase and is involved in inhibiting it, another domain(s) is involved in activating separase and has a weaker affinity for it. Although PTTG2 lacks securin function, its differences from PTTG1 provide evidence of independent inhibitory and activating functions of PTTG1 on separase.
INTRODUCTION
Sister chromatid cohesion is established during DNA replication and maintained throughout the G2 phase and early mitosis by cohesin complexes (1). Separase is responsible for cleaving the cohesin in the centromeric region, allowing sister chromatid separation at the onset of anaphase (2). Timely activation of separase is controlled by binding to an inhibitor called securin. At the end of mitosis, securin is degraded by the anaphase-promoting complex or cyclosome (APC/C) proteasome pathway, thereby allowing separase to fulfill its function (3). Mammalian securin was initially isolated from rat pituitary tumor cells as pituitary tumor-transforming gene 1 (PTTG1) (4) and was subsequently demonstrated to function as a securin (5). Human PTTG1 is located on chromosome 5q35.1 (6). A homologous intronless gene, PTTG2, was found to be located at 4p12 (7). A third intronless isoform, PTTG3, is located at 8q13.1 (8) and should be a pseudogene (9).
Given that securin is a relatively unconserved protein, it is not surprising that its mode of binding to separase differs considerably in different organisms. In budding yeast, securin prevents the interaction between the N-terminal domain and the C-terminal domain of separase, inhibiting both protease activity and access to substrates (10). In Drosophila, separase (SSE) is a much smaller protein and securin (PIM) binds to the N terminus of SSE, as well as to THR, which may functionally correspond to the N terminus of separase in other organisms (11). Biochemical studies of human securin suggest that it inhibits separase by preventing the access of substrates to the active site of separase (12).
Evidence from different organisms indicates that securin not only may inhibit separase, but is also required for proper separase activation. In budding yeast, securin mutants display retarded anaphase entry and synthetic lethality with separase mutations (13). In fission yeast, the loss of securin produces the same block to chromosome segregation as the loss of separase (14). In these systems, securin is implicated in the nuclear targeting of separase (10, 15, 16). Although securin is less conserved in higher eukaryotes, a role of securin in activating separase has also been suggested. Similar to separase mutants, securin mutants in Drosophila fail to initiate anaphase (17). Human HCT116 cells lacking PTTG1 lose chromosomes at a high frequency due to defective anaphase (18). Deletion of just one copy of the ESP1 gene is lethal to mouse embryos lacking PTTG1, indicating that deletion of PTTG1 reduces separase activity (19). As the nuclear envelope is dismantled during mitosis in higher eukaryotes, how securin promotes separase activity in organisms other than yeast remains unknown.
In this study, we showed that PTTG2 does not interact with separase. Using this as a starting point, we then discovered a single residue in PTTG1 that is indispensable for inhibition of separase without affecting the activation of separase. These results for the first time demonstrated separable inhibitory and activating functions of PTTG1.
MATERIALS AND METHODS
Cell culture.
HeLa (cervical carcinoma), Hep3B (hepatocellular carcinoma), HepG2 (hepatoblastoma), and IMR90 (normal fibroblasts) were obtained from the American Type Culture Collection (Manassas, VA, USA). The HeLa cell line used in this study was a clone that expressed the tTA tetracycline repressor chimera (20). HeLa cells expressing an APC/C biosensor were as described previously (21). NP361, NP460, NP550 (immortalized normal nasopharyngeal epithelial), and C666-1 (nasopharyngeal carcinoma) cells were gifts from George Tsao (University of Hong Kong). CNE2, HONE1, and HNE1 (nasopharyngeal carcinoma) cells were obtained from AoE NPC Research Tissue Bank (Hong Kong). LO2 and MIHA (immortalized normal liver) cells were gifts from Irene Ng (University of Hong Kong).
Cells were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) calf serum (Life Technologies, Carlsbad, CA, USA) (for HeLa cells) or fetal bovine serum (Life Technologies) (for Hep3B, HepG2, HONE1, LO2, and MIHA cells) and 50 U/ml penicillin-streptomycin (Life Technologies) in a humidified incubator at 37°C in 5% CO2. IMR90 cells were cultured in Eagle minimum essential medium (Life Technologies) supplemented with 2× essential and nonessential amino acids and 15% fetal bovine serum. NP361, NP460, and NP550 cells were cultured in Epilife plus defined keratinocyte serum-free medium (DKSFM) (1:1) with 0.75% penicillin-streptomycin. Supplements were also added according to the manufacturer's instructions. C666-1 cells were grown in RPMI 1640 medium (Life Technologies) supplemented with 10% fetal bovine serum. CNE2 and HNE1 cells were propagated in DMEM supplemented with 5% (vol/vol) calf serum and 5% (vol/vol) fetal bovine serum.
Stable inducible cell lines expressing PTTG1 (or PTTG2) were produced by transfection of enhanced green fluorescent protein (EGFP)-tagged PTTG1 (or PTTG2) in pUHD-P1/PUR into HeLa cells. Two days after transfection, the cells were selected in medium supplemented with puromycin and doxycycline. A mixed population was isolated after about 2 weeks of selection. The expression of EGFP-tagged proteins was analyzed after the cells were cultured in medium in the presence or absence of doxycycline for 24 h. The established cell lines were propagated in the presence of doxycycline but without puromycin. HeLa cells that stably expressed histone H2B-GFP (22) were used for live-cell imaging.
Unless otherwise stated, cells were treated with the following reagents at the indicated final concentrations: blasticidin (5 μg/ml), cycloheximide (10 μg/ml), doxycycline (1 μg/ml), hydroxyurea (1.5 mM), MG132 (10 μM), nocodazole (NOC) (0.1 μg/ml), puromycin (1 μg/ml), RO3306 (Alexis) (10 μM), and thymidine (2 mM).
Cells were transfected with plasmids and small interfering RNAs (siRNAs) (10 nM unless otherwise stated), using a calcium phosphate precipitation method (23) and Lipofectamine RNAiMAX (Life Technologies), respectively. Cell extracts were prepared as described previously (24). Cell cycle synchronization by double-thymidine block or NOC block was described previously (25). Mitotic chromosome spread was performed as described previously (26).
Plasmid constructs.
PTTG1 in pEF-plink2 and PTTG1DΔ (with mutations in both the destruction box [D box] and the KEN box) in pEF-plink2 were generous gifts from Michael Brandeis (Hebrew University of Jerusalem, Jerusalem, Israel). The coding region of PTTG1 (or the DΔ mutant) was amplified by PCR with 5′-AGAATTCATGGCTACTCTGATCTATGT-3′ and 5′-AGAATTCTTAAATATCTATGTCACAGC-3′; the PCR product was cut with EcoRI and ligated into pUHD-P1 (27) to generate FLAG-PTTG1 (or DΔ) in pUHD-P1.
PTTG2 cDNA (IMAGE clone ID 7262134) was amplified by PCR with 5′-AGAATTCATGGCTACTCTGATCTATGT-3′ and 5′-CCGAATTCTCAACATCCAGGGTCGACAGA-3′; the PCR product was cut with EcoRI and ligated into pUHD-P1 to obtain FLAG-PTTG2 in pUHD-P1.
Site-directed mutagenesis was carried out with a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) using the following primers. PTTG1H134R, 5′-AAGAGCGTCAGATTGCGCACCTCCC-3′ and 5′-AATCTGACGCTCTTCAGGCAGGTCA-3′; PTTG1H134A, 5′-AAGAGGCCCAGATTGCGCACCTCCC-3′ and 5′-AATCTGGGCCTCTTCAGGCAGGTCA-3′; PTTG1R153G, 5′-CGAGGAGGGAGAGCTTGAAAAGCTG-3′ and 5′-AGCTCTCCCTCCTCGTCAAGGATCA-3′; DΔ version of PTTG2 (changing the D-box motif RKAL to AKAA), 5′-AGCTACCGCAAAGGCTGCGGGCACTGT-3′ and 5′-CAGTGCCCGCAGCCTTTGCGGTAGCTT-3′; PTTG2R134H, 5′-TGAAGAGCACCAGATTGCACACCTC-3′ and 5′-AATCTGGTGCTCTTCAGGCAGGTCA-3′; PTTG2G153R, 5′-AGGAGAGAGAGCTTGAAAAGCTGTT-3′ and 5′-CTCTCTCTCCTCATCAAGGATCATG-3′.
PTTG1 was amplified by PCR with a forward primer in the vector and 5′-CGAATTCAAACAGCTTTTCAAGCTC-3′; the PCR product was cut with EcoRI-BamHI and ligated into pUHD-P1 to generate FLAG-PTTG1CΔ159 in pUHD-P1. FLAG-PTTG1CΔ123 in pUHD-P1 was obtained by removing the XbaI fragment from FLAG-PTTG1 in pUHD-P1 (the product contained an extra 20-amino-acid [aa] artifact at the C terminus). FLAG-PTTG1CΔ174 in pUHD-P1 was constructed by ligating the NcoI fragment from FLAG-PTTG1 in pUHD-P1 into pUHD-P1 (the product contained an extra 32-aa artifact at the C terminus). DΔ versions of the C-terminally truncated PTTG1 mutants were generated exactly the same way by using PTTG1DΔ as the parental clone.
The FLAG-PTTG1 (or -PTTG2) was cut with EcoR I and ligated into pGEX-6P-1 (GE Healthcare, United Kingdom) to create GST-PTTG1 (or -PTTG2) in pGEX-6P-1. The EcoRI fragment of FLAG-PTTG1H134R was ligated into pGEX-6P-1 to create GST-PTTG1H134R.
PTTG1 (or PTTG2) was released with EcoRI from FLAG-PTTG1 (or -PTTG2) in pUHD-P1 and put into FLAG-3C-EGFP/PUR (a gift from Ken Ma, Hong Kong University of Science and Technology) to obtain FLAG-3C-PTTG1 (or -PTTG2)-EGFP in FLAG-3C-EGFP/PUR. The FLAG-PTTG1 (or -PTTG2) was cleaved with EcoRI and subcloned into EcoRI-cut pcDNA3.1(−) (Life Technologies) to produce PTTG1 (or PTTG2) in pcDNA3.1(−).
The HindIII fragment from FLAG-PTTG1 (or -PTTG2) in pUHD-P1 was put into HindIII-cut FLAG-PTTG2 (or -PTTG1) in pUHD-P1 to generate FLAG-PTTG2CΔ86-PTTG1NΔ87 (or FLAG-PTTG1CΔ86-PTTG2NΔ87) in pUHD-P1. Similarly, the PvuII fragment from FLAG-PTTG1 (or -PTTG2) was put into PvuII-cut FLAG-PTTG2 (or -PTTG1) in pUHD-P1 to generate FLAG-PTTG2CΔ160-PTTG1NΔ161 (or FLAG-PTTG1CΔ160-PTTG2NΔ161) in pUHD-P1.
ESP1-V5 in pEF6-V5-H6-TOPO was a gift from David Pellman (Harvard University, Cambridge, MA, USA). The cDNA was amplified by PCR with 5′-AGCCATGGATTCCAGCAAGAAGAAGCTG-3′ and 5′-TCGAATTCTTACCGCAGAGAGACAGGCA-3′; the PCR product was then cut with NcoI-EcoRI and ligated into pUHD-P1 to create FLAG-ESP1NΔ1537 in pUHD-P1. The resulting construct was cut with NheI-EcoRI and ligated into pcDNA3.1(−) (Life Technologies) to create ESP1NΔ1537 in pcDNA3.1(−). A C2029A mutation was introduced into this construct using 5′-TGCTGTTTGGCGCTAGCAGTGCGGC-3′ and 5′-GCCGCACTGCTAGCGCCAAACAGCA-3′. The XbaI fragment from ESP1 was put into XbaI-cut pUHD-P1 to create ESP1NΔ1150 in pUHD-P1. The PvuI-SphI fragment from FLAG-ESP1NΔ1537+C2029A was ligated into PvuI-SphI-cut ESP1NΔ1150 in pUHD-P1. The XbaI fragment released from the resulting construct was religated to XbaI-cut ESP1 in pEF6-V5-H6-Topo to generate catalytically inactive ESP1C2029A.
RNA interference (RNAi).
PTTG1 short hairpin RNA (shRNA) was created by annealing the primers 5′-TTTGCCAGGCACCCGTGTGGTTGTTCAAGAGACAACCACACGGGTGCCTGGTTTTT-3′ and 5′-CTAGAAAAACCAGGCACCCGTGTGGTTGTCTCTTGAACAACCACACGGGTGCCTGG-3′ into BbsI- and XbaI-cut mU6pro (28) (a gift from David Turner, University of Michigan, Ann Arbor, MI, USA). PTTG1 resistant to the shRNA was created by introducing silent mutations using the oligonucleotide 5′-GAGAACCCGGGACTAGGGTGGTTGC-3′ and its antisense oligonucleotide. Stealth siRNA targeting PTTG1 (HSS167058) and control siRNA were obtained from Life Technologies; siRNA against ESP1 (GCAGGUUCUGUUCUUGCUU) was obtained from RiboBio (Guangzhou, China).
Live-cell imaging.
The setup and conditions of time-lapse microscopy of living cells were as previously described (29).
Flow cytometry analysis.
Flow cytometry analysis after propidium iodide staining was performed as described previously (30).
Antibodies and immunological methods.
Antibodies against cyclin B1 (22), β-actin (31), FLAG tag (32), and phospho-histone H3S10 (21) were obtained from sources described previously. Monoclonal antibodies to securin (sc-56207; Santa Cruz Biotechnology, Santa Cruz, CA, USA), ESP1 (ab16170; Abcam, Cambridge, United Kingdom), and V5 tag (R960-25; Life Technologies) were obtained from the indicated manufacturers. Immunoblotting was performed as described previously (24). Where indicated, signals on immunoblots were quantified using the Odyssey infrared imaging system (Li-Cor Biosciences) according to the manufacturer's instructions.
Immunoprecipitation was performed using 500 μg of cell lysates. The lysates were diluted to 250 μl with wash buffer (25 mM Tris-HCl, pH 7.4, 2.5 mM NaF, 130 mM NaCl, 2.5 mM EDTA, 2.5 mM EGTA, 0.05% Nonidet P-40, 2 μg/ml aprotinin, 15 μg/ml benzamidine, 1 μg/ml leupeptin, 10 μg/ml soy bean trypsin inhibitor, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and added to 15 μl of protein A-Sepharose or protein G-Sepharose prebound with 3 μl of antiserum or 1 μg of FLAG monoclonal antibody, respectively.
For other immunoprecipitation experiments, 500 μg of cell lysates was diluted to 250 μl with wash buffer and added to 10 μl of M2 beads (Sepharose beads coated with anti-FLAG monoclonal antibodies; Sigma-Aldrich, St. Louis, MO, USA). After incubation at 4°C for 120 min with end-to-end rotation, the supernatant was removed, and the beads were washed three times with wash buffer (250 μl each time). The FLAG-tagged proteins were then eluted by incubation with 100 μg/ml of FLAG peptides (Sigma-Aldrich) in 50 μl of wash buffer at 4°C for 90 min. The supernatant was dissolved in 10 μl SDS sample buffer, and 15 μl was applied to SDS-PAGE. Total cell lysates (10 μg) were applied to indicate the input.
For immunofluorescence microscopy, cells grown on poly-l-lysine-treated coverslips were fixed with freshly made 3% formaldehyde and 2% sucrose in phosphate-buffered saline (PBS) at 25°C for 5 min. The cells were then washed three times with PBS for 5 min each time, blocked and permeabilized with 3% bovine serum albumin (BSA) and 0.4% Triton X-100 in PBS at 25°C for 30 min, and washed three times with wash buffer (2% BSA and 0.2% Triton X-100 in PBS) for 5 min each time. The cells were incubated with antibodies against FLAG at 4°C for 16 h. After being washed three times with wash buffer, the cells were incubated with Alexa Fluor–594 goat anti-rabbit IgG secondary antibodies (Life Technologies) for 2 h at 25°C. After being washed three times with wash buffer, the coverslips were mounted with 2% (wt/vol) N-propylgallate in glycerol.
In vitro ESP1 autocleavage assay.
The autocleavage assay was performed as described previously (33) with minor modifications. Briefly, the ESP1-V5-transfected and NOC-arrested HeLa cells were lysed in lysis buffer (20 mM Tris-HCl, pH 7.7, 100 mM NaCl, 1 mM NaF, 20 mM β-glycerophosphate, 5 mM MgCl2, 0.1% Triton X-100, and 1 μM microcystin-LR). The supernatant was incubated with 15 μl of protein A-Sepharose and protein G-Sepharose mixture prebound with 1 μg V5 antibody at 4°C for 90 min. The beads were washed three times with wash buffer. For cleavage assays, 5 μl of beads from the immunoprecipitation was incubated with 1 μg of glutathione S-transferase (GST)–PTTG1 or –PTTG1H134R in 10 μl of cleavage buffer (30 mM HEPES-KOH, pH 7.7, 30% glycerol, 25 mM NaF, 25 mM KCl, 5 mM MgCl2, 1.5 mM ATP, and 1 mM EGTA) at 37°C for 1 h.
RESULTS
PTTG2 is a minor isoform of securin.
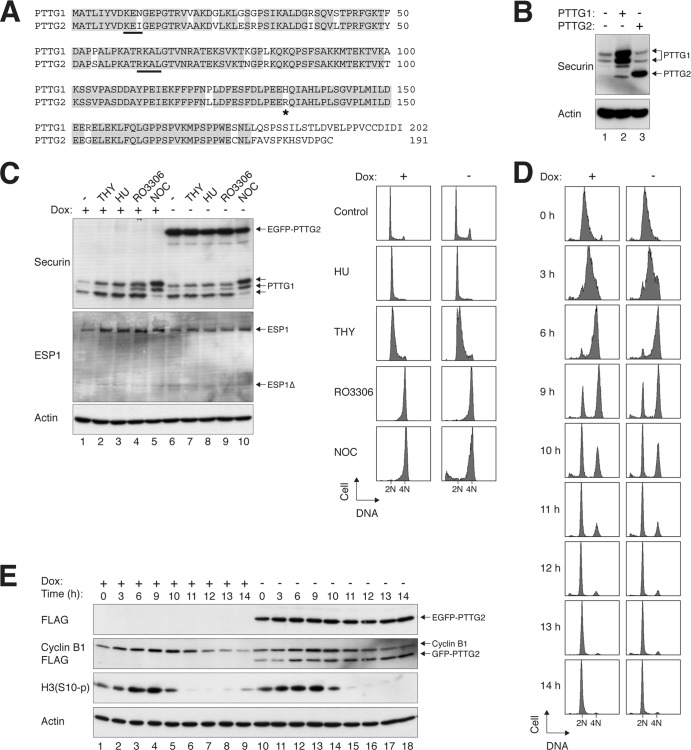
Human PTTG1 and PTTG2 are highly homologous, sharing >80% amino acid sequence identity (Fig. 1A). The major difference is at the C terminus, which is due to a frameshift caused by a base deletion in PTTG2.
Fig 1.
PTTG2 is a minor isoform of securin. (A) Sequence alignment of human PTTG1 and PTTG2. PTTG1 and PTTG2 share >80% identity at the amino acid level (>90% if the C terminus is excluded). Identical residues are highlighted. The destruction signals KEN box and D box (RXXL) of PTTG1 are underlined. The asterisk indicates the position of H134 in PTTG1 (or R134 in PTTG2). (B) PTTG2 is usually not present in HeLa cells. Lysates of asynchronously growing HeLa cells were prepared and analyzed with a PTTG1/2 monoclonal antibody (lane 1). Recombinant (untagged) PTTG1 and PTTG2 were expressed in HeLa cells and loaded side by side as standards. Note that PTTG1 appeared as a doublet and PTTG2 as a single band under these conditions. The overexpressed PTTG1 also contained some degradative products. (C) Ectopic expression of PTTG2 does not affect the cell cycle. Stable EGFP-PTTG2-expressing HeLa cells were cultured in the absence or presence of doxycycline (Dox) to turn on or off the expression of EGFP-PTTG2, respectively. The cells were exposed to thymidine (THY), hydroxyurea (HU), RO3306, or NOC for 16 h. Lysates were prepared and subjected to immunoblotting to detect the expression of endogenous PTTG1 and to confirm the expression of EGFP-PTTG2. Uniform loading of lysates was confirmed by immunoblotting for actin. The cells were also fixed, and the cell cycle profiles were analyzed by flow cytometry. (D) Cell cycle progression is not affected by PTTG2. HeLa cells that expressed FLAG-EGFP-tagged PTTG2 were generated. The cells were cultured continuously in the presence or absence of doxycycline to turn the PTTG2 off or on, respectively. After being synchronously released by a double-thymidine procedure, the cells were harvested at the indicated time points and analyzed by flow cytometry. The positions of 2N and 4N DNA contents are indicated. (E) Expression of mitotic markers is not affected by PTTG2. Cells were synchronized and harvested exactly as for panel D. Lysates were prepared and analyzed by immunoblotting. Equal loading of lysates was confirmed by immunoblotting for actin.
We analyzed the expression of PTTG1 and PTTG2 in HeLa cells using a monoclonal antibody raised against full-length PTTG1. PTTG1 and PTTG2 are recognized equally by the antibody (indicated by experiments when equal amounts of FLAG-tagged standards were used [our unpublished data]). As indicated by the nontagged standards, PTTG1 and PTTG2 could be distinguished by their gel mobility (Fig. 1B). Although endogenous PTTG1 was readily detectable, no PTTG2 could be detected in HeLa cells. Similar results were obtained with other cancer cell lines, immortalized epithelial cells, or normal fibroblasts (see Fig. 4F) or when cells were trapped at different phases of the cell cycle (Fig. 1C). These data indicate that PTTG1 is the major isoform of securin expressed in most cell lines.
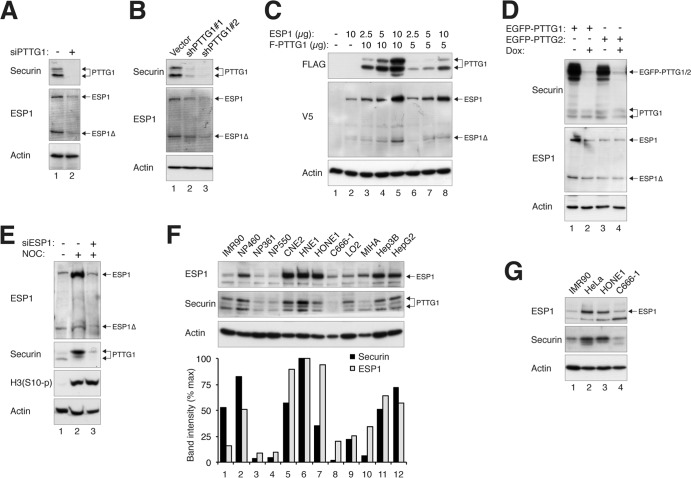
Fig 4.
Reciprocal regulation of the expression of PTTG1 and ESP1. (A) Depletion of PTTG1 reduces endogenous ESP1. HeLa cells were transfected with control or siRNA against PTTG1 (siPTTG1). The cells were harvested 24 h after transfection. Lysates were prepared, and the indicated proteins were detected by immunoblotting. Uniform loading was confirmed by immunoblotting for actin. (B) Depletion of PTTG1 decreases the expression of ESP1. HeLa cells were transfected with control vector or two PTTG1 shRNA-expressing plasmids. A plasmid expressing histone H2B-GFP and a blasticidin resistance gene was cotransfected. After enriching the transfected cells by selection with blasticidin, cell extracts were prepared and subjected to immunoblotting for PTTG1 and ESP1. (C) Ectopic expression of ESP1 increases the expression of PTTG1. HeLa cells were transfected with different amounts of plasmids expressing FLAG-PTTG1 (10 and 5 μg), together with different amounts of ESP1-expressing plasmids (10, 5, and 2.5 μg). Lysates were prepared, and the indicated proteins were detected by immunoblotting. Lysates from vector control (lane 1) and ESP1 alone (lane 2) were loaded as controls. (D) ESP1 is upregulated in PTTG1-overexpressing cells. Cell lines expressing EGFP-PTTG1 or EGFP-PTTG2 were cultured in medium without or supplemented with doxycycline to turn the recombinant protein on or off, respectively. Lysates were prepared, and the expression of PTTG1, PTTG2, and ESP1 was detected by immunoblotting for securin and ESP1. Uniform loading of lysates was confirmed by immunoblotting for actin. (E) Downregulation of ESP1 reduces PTTG1. HeLa cells were transfected with control or siRNA against ESP1. NOC was added for 16 h to trap the cells in mitosis, when the expression of PTTG1 is usually highest. Mitotic cells were collected by mechanical shake off. Lysates were prepared and analyzed by immunoblotting. The increase in histone H3S10 phosphorylation [H3(S10-p)] confirmed the mitotic block. Equal loading of lysates was confirmed by immunoblotting for actin. Control extracts from growing cells were loaded in lane 1. (F) Reciprocal expression of ESP1 and PTTG1 in normal and cancer cell lines. Lysates of various cell lines were prepared and analyzed by immunoblotting for ESP1 and PTTG1. Normal fibroblasts (IMR90) and immortalized nasopharyngeal epithelial (NP460, NP361, and NP550), nasopharyngeal carcinoma (CNE2, HNE1, HONE1, and C666-1), immortalized liver epithelial (LO2 and MIHA), and liver cancer (Hep3B and HepG2) cells were included in the analysis. The ESP1 and PTTG1 signals on the immunoblots were quantified using ImageJ software (National Institutes of Health). (G) Reciprocal expression of ESP1 and PTTG1 in different cell lines during mitosis. The indicated cell lines were treated with nocodazole for 16 h before mitotic cells were isolated by mechanical shake off. Lysates were prepared and analyzed by immunoblotting for ESP1 and PTTG1. Normal fibroblasts (IMR90) and cervical carcinoma (HeLa) and nasopharyngeal carcinoma (HONE1 and C666-1) cells were included in the analysis.
PTTG2 does not have securin function in mitosis.
Given that PTTG2 is normally not expressed, we next investigated if ectopic expression of PTTG2 affects the cell cycle. First, we generated cell lines that expressed FLAG- and EGFP-tagged PTTG2 under the control of doxycycline. Even when expressed to a level higher than the endogenous PTTG1, PTTG2 did not significantly affect the cell cycle profiles in growing cells or cells treated with various cell cycle blockers (Fig. 1C).
To detect more detailed cell cycle changes, PTTG2-expressing cells were synchronized using a double-thymidine block method. Flow cytometry analysis indicated that PTTG2-expressing cells progressed from early S through G2 to mitosis at a rate similar to that of control cells (Fig. 1D). In agreement with this, reduction of mitotic markers, including cyclin B1 and phosphohistone H3S10, occurred at similar times in control and PTTG2-expressing cells (Fig. 1E).
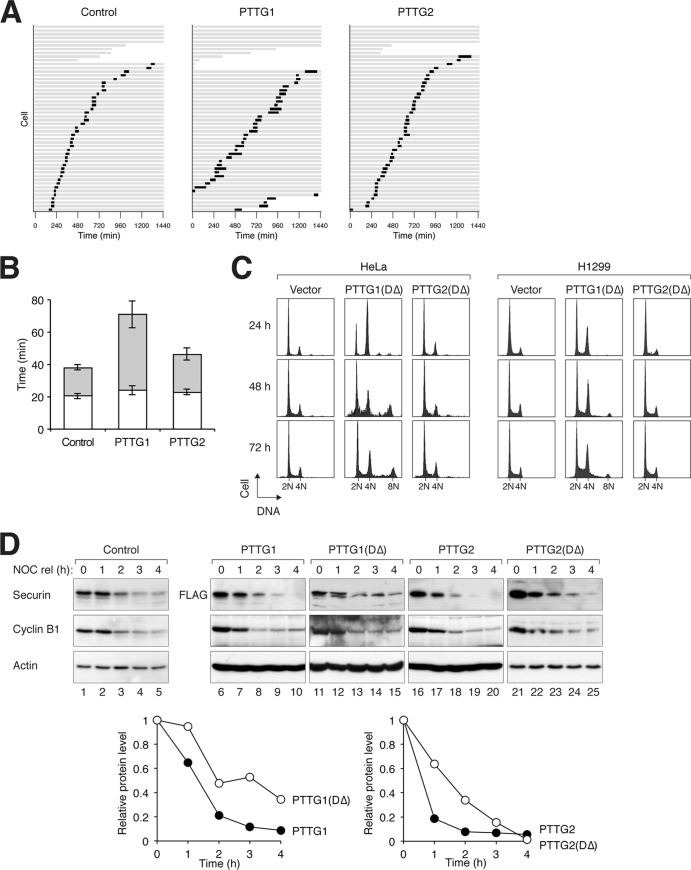
Since PTTG1 is a key player in mitosis, we next analyzed the effects of PTTG2 on mitosis using time-lapse microscopy (Fig. 2A). While the duration from DNA condensation to anaphase was ∼40 min in control cells, it was extended to ∼70 min in PTTG1-expressing cells. Most of the extension was due to a delay from metaphase to anaphase (Fig. 2B). Mitosis in typical control and PTTG1-expressing cells is shown in Videos S1 and S2 in the supplemental material, respectively. In marked contrast, mitotic progression was not affected in PTTG2-expressing cells (Fig. 2B).
Fig 2.
Ectopic expression of PTTG2 does not affect the cell cycle. (A) Overexpression of PTTG1 but not PTTG2 prolongs mitosis. HeLa cells expressing histone H2B-GFP were transfected with control vector or plasmids expressing PTTG1 or PTTG2. Histone H2B-monomeric red fluorescent protein (mRFP) was used as a cotransfection marker. Individual cells were then tracked for 24 h with time-lapse microscopy. Each horizontal bar represents one cell (n = 50). Gray, interphase; black, mitosis (from DNA condensation to anaphase or cell death); truncated bars, cell death. (B) Anaphase onset is delayed in PTTG1-overexpressing cells. PTTG1- and PTTG2-transfected cells were analyzed as described for panel A. The duration of mitosis was quantified (mean ± 90% confidence interval). Mitosis was subdivided into two periods: from DNA condensation to metaphase (white) and from metaphase to anaphase (gray). (C) Nondegradable PTTG2 does not interfere with sister chromatid separation. PTTG1DΔ or PTTG2DΔ was expressed in HeLa or H1299 cells (a Tet-off clone). Histone H2B-GFP was used as a cotransfection marker. The cells were harvested at the indicated time points and analyzed by flow cytometry (only transfected cells were analyzed). (D) Mutation of the destruction sequences delays the degradation of PTTG1 and PTTG2. PTTG1DΔ was generated by mutation of the D box and the KEN box sequences of PTTG1. PTTG2DΔ was similarly generated by mutation of the D box (there is no KEN box in PTTG2) (Fig. 1A). FLAG-tagged PTTG1, PTTG1DΔ, PTTG2, or PTTG2DΔ was expressed in HeLa cells. The cells were synchronized at mitosis with a NOC block and released into the cell cycle by shaking off and washing. Doxycycline and cycloheximide were added to turn off the expression of the recombinant proteins. The cells were then harvested at the indicated time points. Lysates were prepared and analyzed by immunoblotting. Actin analysis was included to assess protein loading and transfer. The relative amounts of the proteins were also quantified using the Odyssey infrared imaging system (bottom). Actin signals were used to normalize the FLAG-tagged recombinant protein signals.
PTTG1 is targeted to degradation by ubiquitination at the end of mitosis (3). To ensure that the lack of function of PTTG2 was not due to untimely degradation, we also generated a PTTG2 mutant that lacked the D box (DΔ). We confirmed that both PTTG1DΔ and PTTG2DΔ were more stable than the respective wild-type proteins during mitotic exit (Fig. 2D). Unlike the overexpression of PTTG1, which only delayed anaphase (Fig. 2B), PTTG1DΔ interfered with sister chromatid separation and prevented anaphase onset (see below). This resulted in an accumulation of tetraploids with 4N or larger DNA contents (Fig. 2C). In contrast, PTTG2DΔ-expressing cells displayed a DNA profile identical to that of control cells. Similar results were obtained using H1299 cells, indicating that the effects of PTTG1DΔ were not merely specific to HeLa cells (Fig. 2C).
Taken together, these data indicate that the isoform PTTG2 does not affect mitosis even when expressed at levels considerably higher than those of the endogenous PTTG1.
PTTG2 does not interact with separase.
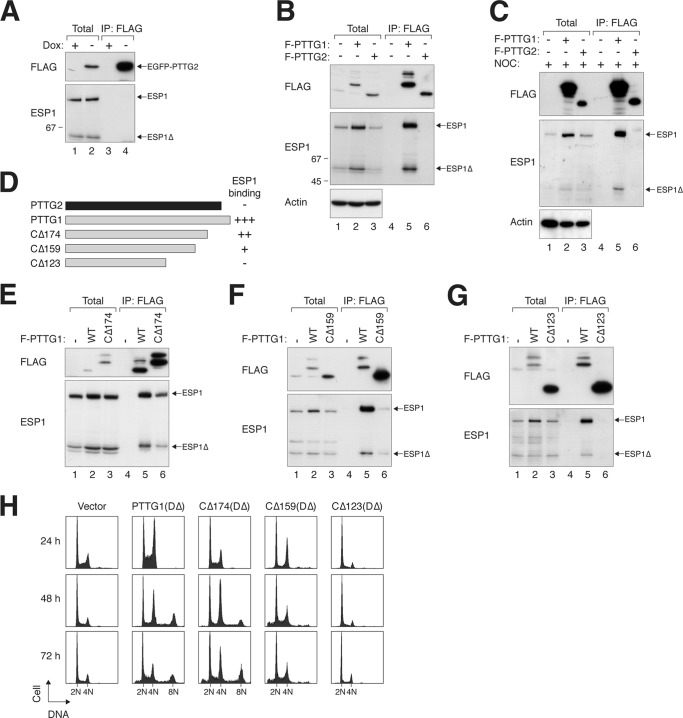
We next addressed whether PTTG2 could interact with separase (ESP1). When PTTG2 (tagged with both FLAG and EGFP) was isolated from lysates, no ESP1 was coimmunoprecipitated (Fig. 3A). In contrast, ESP1 was able to interact robustly with FLAG-tagged PTTG1 (Fig. 3B). A key characteristic of activated human ESP1 is its autocleavage activity (12, 34, 35). Notably, both full-length ESP1 and the cleaved product (ESP1Δ) were able to bind PTTG1 (Fig. 3B). PTTG2 also did not interact with ESP1 during mitosis (Fig. 3C).
Fig 3.
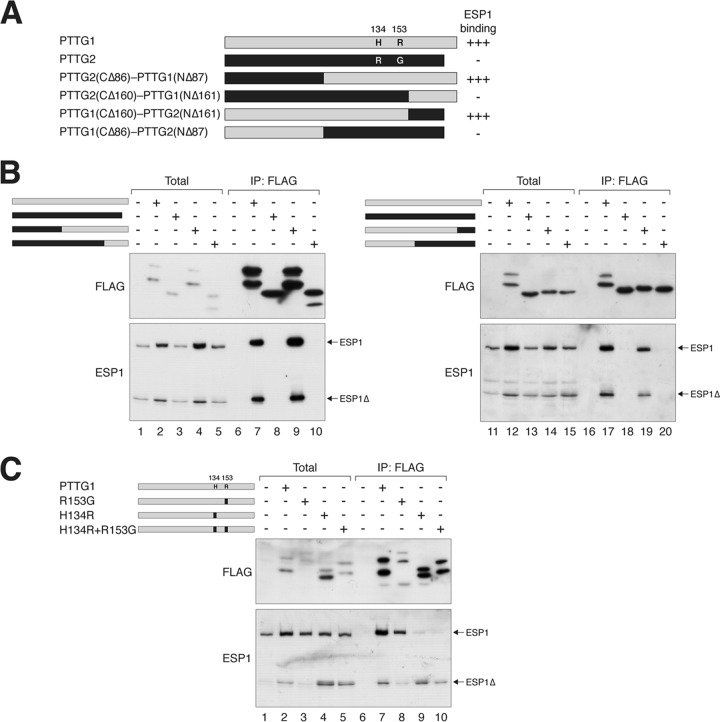
A C-terminal region of PTTG1 is required for interaction with separase. (A) PTTG2 does not interact with ESP1. A cell line expressing PTTG2 (FLAG-EGFP tagged) was transfected with ESP1-expressing plasmids. The cells were cultured in the absence or presence of doxycycline to turn PTTG2 on or off, respectively. Lysates were prepared and subjected to immunoprecipitation (IP) with FLAG antibodies. After elution with FLAG-tagged peptides, the supernatant (together with total lysates) was analyzed by immunoblotting. The positions of full-length and cleaved ESP1 (ESP1Δ) are indicated. (B) PTTG1, but not PTTG2, interacts with ESP1. Cells were transfected with ESP1- and FLAG-tagged PTTG1 (F-PTTG1)- or PTTG2-expressing plasmids as indicated. Immunoprecipitation and subsequent immunoblotting were performed as described for panel A. As a control, no ESP1 was immunoprecipitated when it was expressed in the absence of FLAG-containing proteins. (C) PTTG2 does not interact with ESP1 during mitosis. ESP1was coexpressed with FLAG-tagged PTTG1 or PTTG2 as indicated. The cells were treated with NOC for 16 h to trap them in mitosis. Lysates were prepared and subjected to immunoprecipitation with FLAG antibodies. After elution with FLAG-tagged peptides, the supernatant (together with total lysates) was analyzed by immunoblotting. The positions of full-length and cleaved ESP1 (ESP1Δ) are indicated. (D) The C-terminal region of PTTG1 is involved in interacting with ESP1. A schematic diagram of PTTG1 deletion constructs is shown (to scale). The numbers indicate the positions beyond which the amino acid residues of PTTG1 were deleted (full-length PTTG1 is 202 residues long). A summary of ESP1 binding is also shown. (E to G) The C-terminal region of PTTG1 is required for interaction with separase. FLAG-tagged full-length PTTG1 and PTTG1CΔ174 (E), PTTG1CΔ159 (F), or PTTG1CΔ123 (G) were coexpressed in HeLa cells. Lysates were prepared and subjected to immunoprecipitation with FLAG antibodies. After elution with FLAG-tagged peptides, the supernatant (together with total lysates) was analyzed by immunoblotting. PTTG1CΔ174 migrated more slowly than PTTG1 because of extra sequences from a cloning artifact at the C terminus. WT, wild type. (H) The C-terminal region of PTTG1 is required to promote mitotic defects. HeLa cells were transfected with plasmids expressing DΔ versions of PTTG1 or C-terminally truncated PTTG1. Histone H2B-GFP was used as a cotransfection marker. The cells were harvested at the indicated time points and analyzed by flow cytometry (only transfected cells were analyzed). Cells with impaired sister chromatid separation underwent cytokinesis failure and became tetraploids.
To demarcate the ESP1-interacting region in PTTG1, we constructed several C-terminal-truncation mutants (Fig. 3D). ESP1 binding was only slightly reduced when PTTG1 was deleted up to residue 174 (CΔ174) (Fig. 3E), with further impairment in binding in CΔ159 (Fig. 3F). A more extensive deletion (CΔ123), however, abolished binding to ESP1 (Fig. 3G). Note that due to the stabilization of ESP1 by PTTG1 (see below), the expression of ESP1 was higher in the presence of interacting PTTG1 constructs. Nevertheless, the large difference in binding affinity was unlikely to be explained by the ESP1 level alone.
Consistent with the binding results, expression of DΔ versions of CΔ174 and CΔ159 increased the number of cells with 4N and 8N DNA contents, suggesting interference with mitosis similar to that of PTTG1DΔ (Fig. 3H). In accordance with the lack of ESP1 binding, the cell cycle profile was not affected by CΔ123DΔ.
Collectively, these results indicate that, unlike PTTG1, PTTG2 does not physically associate with ESP1.
Reciprocal regulation of the stability of PTTG1 and ESP1.
As there was good correlation between the expression of ESP1 and PTTG1 that could bind ESP1, we next characterized their relationship in more detail. Downregulation of endogenous PTTG1 with siRNA resulted in a concomitant reduction in endogenous ESP1 (Fig. 4A). The result was reproducible using two shRNAs against different regions of PTTG1 (Fig. 4B), supporting the specificity of the effects. Conversely, overexpression of PTTG1 increased the abundance of cotransfected ESP1 (Fig. 4C). Expression of PTTG1 but not PTTG2 in stable cell lines also increased the level of endogenous ESP1 (Fig. 4D), further supporting the idea that PTTG1 influenced the abundance of ESP1. In the reciprocal experiment, overexpression of ESP1 enhanced the expression of cotransfected PTTG1 (Fig. 4C). Finally, depletion of ESP1 with siRNA significantly reduced the abundance of endogenous PTTG1 (Fig. 4E).
We also examined the endogenous ESP1 and PTTG1 in other normal and cancer cell lines. In particular, we analyzed a number of cell lines from nasopharyngeal carcinoma versus immortalized normal nasopharyngeal epithelial cells and liver cancer cells versus immortalized normal liver epithelial cells, as well as a normal fibroblast strain (Fig. 4F). Cancer cell lines typically contained higher concentrations of ESP1 than cells derived from normal tissues. Importantly, the expression levels of PTTG1 and ESP1 were closely related in all cell lines. Similar results were obtained when only mitotic cells were analyzed, indicating that the relative expression levels of PTTG1 and ESP1 in different cell lines were not due to differences in cell cycle distributions (Fig. 4G).
Collectively, these experiments demonstrate a tight relationship between ESP1 and PTTG1. Together with the fact that ESP1 is not increased by PTTG2 and is only increased by PTTG1 constructs that can bind ESP1, these results establish that the binding of PTTG1 to ESP1 stabilizes the complex.
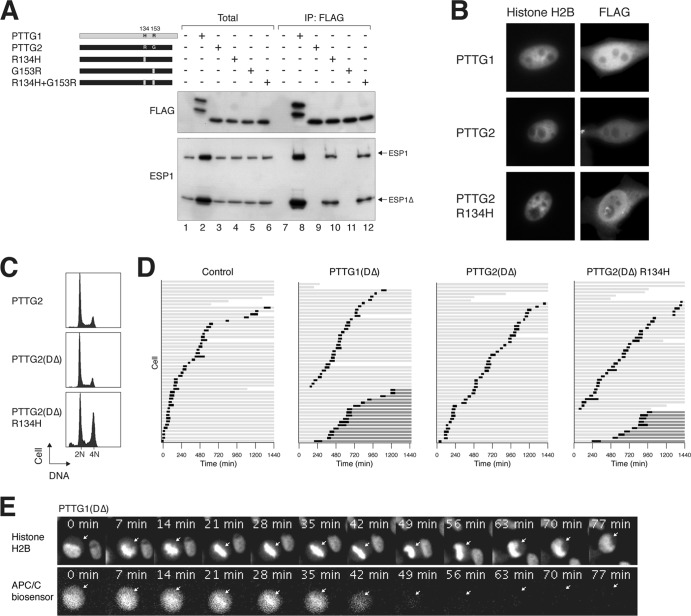
A single-amino-acid difference between PTTG1 and PTTG2 underlies the lack of separase-inhibitory function in PTTG2.
The above-mentioned deletion studies suggested that the C-terminal region of PTTG1 is involved in ESP1 binding (Fig. 3D). A shortcoming of deletion analysis is the likelihood of major alterations of secondary structures. Given the high homology between PTTG1 and PTTG2, we generated domain-swapping chimeras between PTTG1 and PTTG2 (Fig. 5A), which are less likely to give rise to major structural defects. The interaction of the chimeras with ESP1 was analyzed with coimmunoprecipitation (Fig. 5B). As before, blank vector served as a negative control and wild-type PTTG1 served as a positive control. These experiments revealed that binding was disrupted whenever a chimera contained residues 86 to 161 from PTTG2, indicating that the corresponding region in PTTG1 was critical for ESP1 binding.
Fig 5.
H134 of PTTG1 is critical for securin function. (A) Chimeras between PTTG1 and PTTG2 reveal the importance of H134 or R153 for securin function. A schematic diagram of domain-swapping constructs between PTTG1 (gray) and PTTG2 (black) is shown to scale. A summary of the ESP1 binding is shown. Together with the data from the C-terminally deleted mutants, the region between 123 and 161 is likely to be critical for ESP1 binding. The positions and amino acids of the only 2 residues that are different between PTTG1 and PTTG2 within this region are indicated. (B) PTTG1-PTTG2 chimeras define a region critical for ESP1 binding. ESP1- and FLAG-tagged PTTG1-PTTG2 chimeras were expressed in HeLa cells as indicated (cf. panel A). Binding to ESP1 was detected by immunoprecipitation with FLAG antibodies, followed by elution and immunoblotting. (C) H134 of PTTG1 is crucial for ESP1 binding. H134 and R153 of PTTG1 (gray) were mutated to the corresponding sequence in PTTG2 (black) either individually or together. ESP1- and the indicated FLAG-tagged PTTG1 constructs were expressed in HeLa cells. Binding to ESP1 was assayed by immunoprecipitation with FLAG antibodies, followed by elution and immunoblotting. The expression of PTTG1R153G was lower in this experiment, which may explain the lower level of coimmunoprecipitated ESP1. There is no evidence from other experiments that PTTG1R153G binds ESP1 more weakly than wild-type PTTG1.
Since ESP1 binding was lost in PTTG1CΔ123 (Fig. 3D), we examined the importance of the region between residues 123 and 161. Remarkably, only 2 residues are different between PTTG1 and PTTG2 within the region. While PTTG1 contains H134 and R153, the corresponding positions in PTTG2 are R134 and G153 (Fig. 5A). To test their importance, H134 and R153 were mutated to the corresponding sequence in PTTG2 either individually or together (Fig. 5C). Overall, PTTG1H134R associated with less ESP1 than wild-type PTTG1. More significantly, however, PTTG1H134R induced the cleavage of ESP1. In the absence of PTTG1, ESP1 was present mainly in the uncleaved, inactive form. In marked contrast, ESP1 was present mainly in the cleaved form in the presence of PTTG1H134R, suggesting that PTTG1H134R actively promotes ESP1 activity (Fig. 5C, compare lanes 1 and 4). The change in the ratio of cleaved versus full-length ESP1 was not simply caused by the stabilization of ESP1 (similar results were obtained when more ESP1 was transfected in the control than in the PTTG1H134R-cotransfected cells [data not shown]). Although ESP1 cleavage was apparent in total lysates, it was more prominent in the ESP1 binding to PTTG1H134R. ESP1 cleavage was also induced by the H134R-plus-R153G double mutant but not by R153G alone. These results indicate that mutation of H134 reduced ESP1 binding and stimulated ESP1 proteolytic activity.
H134R switches PTTG1 from an inhibitor into an activator of ESP1.
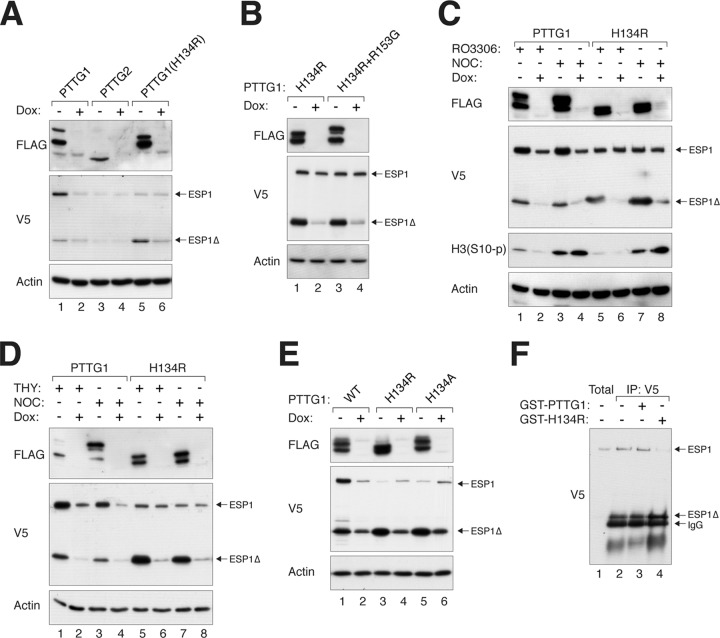
To ensure that the effects of PTTG1 constructs on coexpressed ESP1 were not simply due to different transfection efficiencies, they were turned on or off using an inducible system after transfection (Fig. 6A). These experiments confirmed that while PTTG1 (but not the negative control, PTTG2) mainly stabilized full-length ESP1, PTTG1H134R increased the cleaved ESP1Δ. The double mutant containing both H134R and R153G behaved identically to the single H134R mutant (Fig. 6B). By using the CDK1 inhibitor RO3306 (Fig. 6C), thymidine (Fig. 6D), and NOC (Fig. 6C and D), we also showed that PTTG1H134R induced ESP1 cleavage independently of the cell cycle.
Fig 6.
PTTG1 containing an H134 mutation activates ESP1. (A) PTTG1H134R promotes autocleavage of ESP1. HeLa cells were cotransfected with plasmids expressing ESP1 and FLAG-tagged PTTG1, PTTG2, or PTTG1H134R. The FLAG-tagged constructs were turned on or off with doxycycline as indicated. Lysates were prepared, and the expression of ESP1 was detected with an antibody against the V5 tag. Uniform loading was confirmed by immunoblotting for actin. (B) Both PTTG1H134R and PTTG1H134R+R153G promote cleavage of ESP1. HeLa cells were cotransfected with plasmids expressing ESP1 and the indicated FLAG-tagged PTTG1 proteins. The FLAG-tagged constructs were turned on or off with doxycycline as indicated. Lysates were prepared, and the expression of ESP1 was detected with an antibody against the V5 tag. Uniform loading was confirmed by immunoblotting for actin. (C) PTTG1H134R induces ESP1 cleavage in both G2 phase and mitosis. HeLa cells were cotransfected with plasmids expressing ESP1 and FLAG-tagged PTTG1 or PTTG1H134R. The FLAG-tagged constructs were turned on or off with doxycycline as indicated. The cells were incubated with RO3306 or NOC for 16 h to enrich the cells in G2 phase and mitosis, respectively. Lysates were prepared and analyzed by immunoblotting. Mitosis was confirmed by immunoblotting for phospho-histone H3S10. (D) PTTG1H134R stimulates ESP1 cleavage in both S phase and mitosis. HeLa cells were cotransfected with plasmids expressing ESP1 and FLAG-tagged PTTG1 or PTTG1H134R. The FLAG-tagged constructs were turned on or off with doxycycline as indicated. The cells were incubated with thymidine or NOC for 16 h to trap cells in S phase and mitosis, respectively. Lysates were prepared and analyzed by immunoblotting. (E) H134 is required to inhibit ESP1 cleavage. Cells were cotransfected with plasmids expressing ESP1 and FLAG-tagged PTTG1, PTTG1H134R, or PTTG1H134A. The PTTG1 constructs were controlled with doxycycline as indicated. Lysates were prepared, and the expression of ESP1 was detected with an antibody against the V5 tag. Uniform loading was confirmed by immunoblotting for actin. (F) PTTG1H134R facilitates ESP1 cleavage in vitro. HeLa cells depleted of PTTG1 with siRNA were transfected with plasmids expressing V5-tagged ESP1. The cells were treated with NOC for 16 h before mitotic cells were collected by mechanical shake off. Lysates were prepared and subjected to immunoprecipitation with V5 antibodies. The ESP1 immunoprecipitates were then incubated with bacterially expressed GST-PTTG1 or GST-PTTG1H134R at 37°C for 1 h. Autocleavage of ESP1 was detected by immunoblotting. The positions of ESP1, cleaved ESP1, and IgG from the immunoprecipitation are indicated.
PTTG1H134R was designed by changing H134 to the equivalent residue in PTTG2. Whether R134 is actually required for activating ESP1 was explored by mutating H134 to a different residue. Figure 6E shows that ESP1 cleavage was also induced by PTTG1H134A. These results indicate that PTTG1H134R was a loss-of-function mutation and that H134 is required to prevent ESP1 activation.
To obtain more direct evidence that PTTG1H134R could facilitate ESP1 activation, we incubated ESP1 immunoprecipitates with bacterially expressed PTTG1 in vitro (Fig. 6F). Although it was difficult to preserve uncleaved ESP1 following immunoprecipitation, we found that GST-PTTG1H134R was able to induce more efficient cleavage of ESP1 than GST-PTTG1.
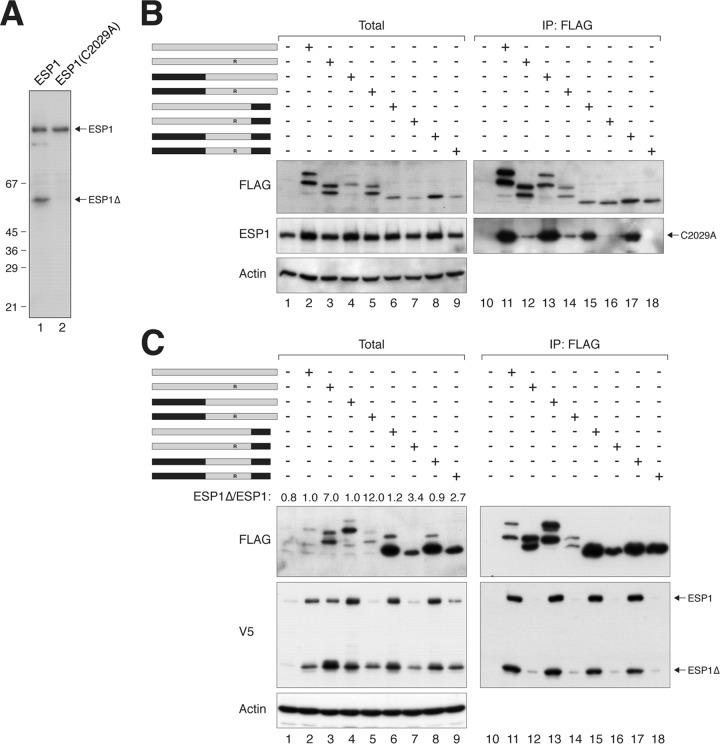
To further support the idea that PTTG1 normally inhibits ESP1 autocleavage, we generated a catalytically dead version of ESP1 (C2029A). As expected, ESP1C2029A abolished autocleavage activity (Fig. 7A). In contrast to ESP1, ESP1C2029A was only marginally stabilized when coexpressed with PTTG1 or PTTG1H134R (Fig. 7B, lanes 1 to 3), confirming that the normal increase in ESP1 caused by PTTG1 was due to inhibition of autocleavage. Collectively, these data suggest that mutation of H134 attenuates the ability of PTTG1 to inhibit ESP1.
Fig 7.
Evidence of an ESP1-activating domain in PTTG1. (A) ESP1C2029A abolishes autocleavage. HeLa cells were transfected with either wild-type ESP1 or catalytically inactive ESP1C2029A. Lysates were prepared and subjected to immunoblotting with ESP1 antibodies to detect the full-length and cleaved ESP1. The positions of molecular size standards (in kDa) are indicated. (B) PTTG1 does not strongly affect the expression of catalytically inactive ESP1. HeLa cells were cotransfected with plasmids expressing ESP1C2029A and FLAG-tagged PTTG1 or PTTG1-PTTG2 chimeras. A schematic diagram of the chimeras between PTTG1 (gray) and PTTG2 (black) is shown. Each pair of proteins consisted of a wild-type H134 and an H134R substitution. Lysates were prepared, subjected to immunoprecipitation with FLAG antibodies, and analyzed by immunoblotting. (C) The C-terminal region of PTTG1 is important for activation of ESP1. HeLa cells were cotransfected with plasmids expressing ESP1 and FLAG-tagged PTTG1 or PTTG1-PTTG2 chimeras. A schematic diagram of the chimeras between PTTG1 (gray) and PTTG2 (black) is shown. Each pair of proteins consisted of a wild-type H134 and an H134R substitution. Lysates were prepared, subjected to immunoprecipitation with FLAG antibodies, and analyzed by immunoblotting. The ratios between the cleaved ESP1 (ESP1Δ) and full-length ESP1 in the total lysates were quantified using serially diluted standards.
Evidence of an ESP1-activating domain in PTTG1.
Given that PTTG1H134R can promote the activation of ESP1, we predicted that PTTG1 should contain an ESP1-activating element that is absent in PTTG2. To test this hypothesis, we generated several chimeras between PTTG1 and PTTG2 that contain the H134R mutation (Fig. 7C). Substitution of the N-terminal region of PTTG2 into PTTG1H134R did not affect the promotion of ESP1 cleavage. In contrast, substitution of the C-terminal region of PTTG2 into PTTG1H134R impaired ESP1 cleavage (see quantification of the ESP1Δ/ESP1 ratios in Fig. 7C), suggesting that activation of ESP1 may in part involve the C terminus of PTTG1.
The ESP1 binding of various PTTG1-PTTG2 chimeras was also examined using catalytically inactive ESP1C2029A to circumvent the complication of autocleavage (Fig. 7B). Compared to PTTG1, the affinity of PTTG1H134R for ESP1C2029A was significantly reduced. Although substitution of the C terminus appears to further reduce binding, this was relatively minor in comparison to the contribution from H134.
Collectively, these results suggest that the H134-containing region has a strong affinity for ESP1 and is involved in inhibiting ESP1. On the other hand, the C terminus of PTTG1 is involved in activating ESP1 and has a relatively weak binding affinity for ESP1. However, the C terminus of PTTG1 is clearly not the only region involved in activating ESP1, because putting only the C terminus of PTTG1 into PTTG2 did not allow the chimera to activate ESP1 (Fig. 5B).
Mutation of H134 promotes premature activation of ESP1 and sister chromatid separation.
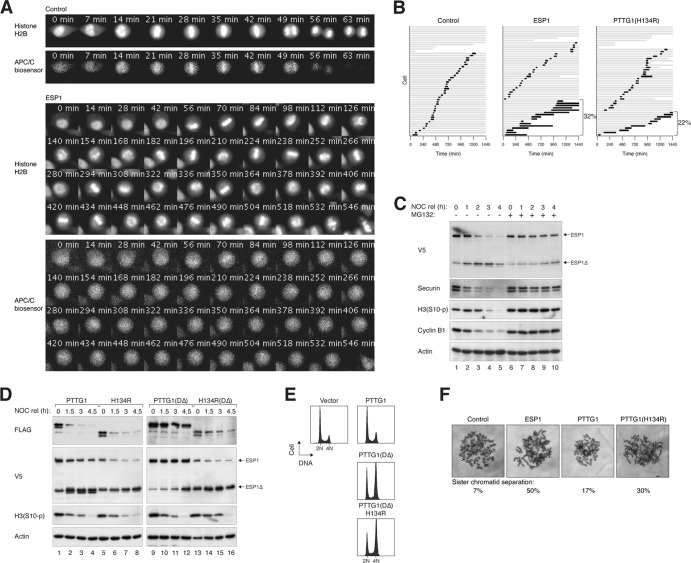
Given that PTTG1H134R can promote ESP1 activation, its effects on mitosis were next examined with live-cell imaging. We used a HeLa cell line that expressed both histone H2B-GFP and an APC/C biosensor (21). The time from DNA condensation to anaphase, when the APC/C biosensor was degraded, was ∼40 min in control cells (Fig. 8A; see Video S3 in the supplemental material). Overexpression of ESP1 dramatically extended the mitosis (Fig. 8B). Although metaphase plates could form, anaphase onset was inhibited, together with signs of premature loss of sister chromatid adhesion as chromosomes became more scattered at later time points. The continued presence of the APC/C biosensor indicated that the spindle assembly checkpoint remained active during the mitotic trap (Fig. 8A; see Video S4 in the supplemental material). These results indicate that when ectopically expressed, some ESP1 escaped normal control and caused premature sister chromatid separation. Notably, PTTG1H134R induced mitotic defects similar to those of ESP1 overexpression (Fig. 8B; see Video S5 in the supplemental material). As a control, wild-type PTTG1 exerted only a minor effect on mitosis (Fig. 2A; see Video S2 in the supplemental material).
Fig 8.
Mutation of His134 promotes premature activation of separase and sister chromatid separation. (A) Dysregulation of ESP1 induces defective mitotic exit. HeLa cells expressing histone H2B-GFP and mRFP-APC/C biosensor were transfected with control or ESP1- or PTTG1H134R-expressing plasmids. Transfected cells were distinguished from nontransfected cells by cotransfection of a yellow fluorescent protein (YFP) plasmid. At 24 h after transfection, cells were analyzed by time-lapse microscopy for 24 h. Representative still images of control or ESP1-expressing cells are shown. Channels of histone H2B-GFP and mRFP-APC/C biosensor are shown. The full videos can be found in the supplemental material: control, Video S3; ESP1, Video S4; and PTTG1H134R, Video S5. (B) Ectopic expression of ESP1 or PTTG1H134R induces prolonged mitosis and cell death. Cells were transfected and analyzed by time-lapse microscopy exactly as described for panel A. Individual cells were then tracked for 24 h. Each horizontal bar represents one cell (n = 50). Gray, interphase; black, mitosis (from DNA condensation to anaphase or cell death); truncated bars, cell death. The percentages of cells that died during the prolonged mitosis are shown. (C) Proteasome-dependent degradation of PTTG1 and activation of ESP1 during mitotic exit. HeLa cells were transfected with ESP1-expressing plasmids. The cells were synchronized with NOC and released (rel). The cells were incubated with doxycycline and cycloheximide (to stop protein synthesis) in either the presence or absence of the proteasome inhibitor MG132 and harvested at the indicated time points. Lysates were prepared and analyzed by immunoblotting. (D) PTTG1H134R induces ESP1 activation before mitotic exit. HeLa cells were transfected with plasmids expressing ESP1 and the indicated PTTG1 constructs. The cells were synchronized with NOC and released in the presence of doxycycline and cycloheximide. The cells were harvested at the indicated time points and analyzed by immunoblotting. (E) PTTG1H134R interferes with normal mitosis. HeLa cells were transfected with plasmids expressing the indicated constructs. Transfected cells were differentiated by virtue of a cotransfected plasmid expressing histone H2B-GFP. The cells were harvested at the indicated time points and analyzed by flow cytometry. (F) Ectopic expression of ESP1 or PTTG1H134R induces premature sister chromatid separation. HeLa cells were transfected with control or ESP1-, PTTG1-, or PTTG1H134R-expressing plasmids. After 24 h, the cells were treated with NOC for 6 h to enrich mitotic cells. Chromosome spreads were prepared and visualized after Giemsa staining under a light microscope. Representative images and the percentage of mitotic cells that contained separated sister chromatids (n = 50) are shown. Note that there was no selection for transfected cells.
Mitotic defects induced by PTTG1H134R were further investigated by its effect on cell cycle distribution. During normal mitosis, endogenous PTTG1 was destroyed by the APC/C-proteasome pathway, resulting in ESP1 activation and normal mitotic exit (Fig. 8C). However, the stable PTTG1DΔ prevented ESP1 activation (Fig. 8D) and promoted an accumulation of tetraploids (Fig. 8E). Although PTTG1H134R+DΔ also promoted tetraploidization (Fig. 8E), it was likely caused by premature ESP1 activation (Fig. 8D). To verify this hypothesis, mitotic chromosomes were examined with mitotic spread. While most control mitotic cells contained paired sister chromatids, a high percentage of ESP1- and PTTG1H134R-expressing cells contained separated sister chromatids (Fig. 8F).
Collectively, these data indicate that PTTG1H134R can induce precocious sister chromatid separation similarly to ectopic expression of ESP1 and further support the idea that PTTG1H134R could activate ESP1.
Introduction of H134 is sufficient to allow PTTG2 to bind separase.
Given that H134 of PTTG1 is critical for inhibition of ESP1, we next tested if H134 is sufficient to turn PTTG2 into a functional securin. R134 and G153 of PTTG2 were mutated to the corresponding sequences in PTTG1 either individually or together. While PTTG2 did not associate with ESP1, PTTG2R134H was able to bind ESP1, albeit with a slightly lower affinity than PTTG1 (Fig. 9A). In contrast, PTTG2G153R remained incapable of binding ESP1.
Fig 9.
Introduction of H134 is sufficient to allow PTTG2 to bind separase. (A) PTTG2R134H gains ESP1 binding capability. A schematic diagram of PTTG2 point mutants used in this study is shown. R134 and G153 of PTTG2 (black) were mutated to the corresponding sequences in PTTG1 (gray) either individually or together. HeLa cells were transiently transfected with ESP1- and FLAG-tagged wild-type or mutant PTTG2, as indicated. Binding to ESP1 was detected by immunoprecipitation with FLAG antibodies, followed by elution and immunoblotting. (B) PTTG1 and PTTG2 are differently located. HeLa cells stably expressing histone H2B-GFP were cotransfected with plasmids expressing the indicated FLAG-tagged constructs and histone H2B-GFP. Cells were processed for immunostaining with antibodies against FLAG. The histone H2B signals indicate the nucleus. (C) PTTG2R134H interferes with normal mitosis. HeLa cells were transfected with plasmids expressing the indicated constructs. Transfected cells were differentiated by virtue of a cotransfected histone H2B-GFP plasmid. The cells were harvested at the indicated time points and analyzed by flow cytometry. (D) Nondegradable PTTG2R134H prevents sister chromatid separation. HeLa cells expressing histone H2B-GFP and APC/C biosensor were transfected with plasmids expressing a DΔ version of PTTG1, PTTG2, or PTTG2R134H. A histone H2B-mRFP-expressing construct was used as a cotransfection marker. Individual cells were then tracked for 24-h time-lapse microscopy. Each horizontal bar represents one cell (n = 50). Light gray, interphase; black, mitosis (from DNA condensation to anaphase or cell death or DNA decondensation without anaphase); dark gray, interphase after DNA decondensation without anaphase; truncated bars, cell death. (E) Still images from a representative PTTG1DΔ-expressing mitotic cell. Channels of histone H2B-GFP and mRFP-APC/C biosensor are shown. The full video can be found as Video S6 in the supplemental material.
We found that while PTTG2 was evenly distributed between the nucleus and cytosol, PTTG1 was more concentrated in the nucleus (Fig. 9B). It is possible that while PTTG2 could shuttle in and out of the nucleus in equilibrium, PTTG1 was retained inside the nucleus after forming a complex with ESP1. In agreement with this, PTTG2R134H was also more concentrated in the nucleus (Fig. 9B).
To validate the gain of securin function of PTTG2R134H in cells, we generated a DΔ version of PTTG2R134H. As before, expression of PTTG1DΔ but not PTTG2DΔ promoted the accumulation of tetraploids (Fig. 9C). PTTG2R134H+DΔ, however, behaved similarly to PTTG1DΔ, suggesting that the R134H mutation was sufficient to allow PTTG2 to inhibit sister chromatid separation. In support of this, live-cell imaging revealed that both PTTG1DΔ and PTTG2R134H+DΔ prevented timely anaphase onset (Fig. 9D). Although cells entered mitosis normally, they were frequently unable to undergo anaphase, returning to interphase without proper chromosome segregation or cytokinesis (examples are shown in Fig. 9E and Videos S6 and S7 in the supplemental material). The APC/C biosensor was destroyed normally, indicating that switching off the spindle assembly checkpoint and APC/C activation were not affected by PTTG2R134H+DΔ.
Taken together, these results indicate that a single-amino-acid change in PTTG2 to create H134 is sufficient to convert an inactive isoform into one that can bind ESP1 and prevent anaphase.
DISCUSSION
PTTG1 was initially identified as an oncogene in a rat pituitary tumor (4). Human PTTG1 was subsequently found to be overexpressed in pituitary and many other cancers, often associated with a poor prognosis (36). PTTG1 is also one the 17 key signature genes associated with metastasis (37). Our results also indicate that PTTG1 is expressed at a higher level in hepatocellular carcinoma and nasopharyngeal carcinoma cell lines than in normal epithelial cells from the same tissues (Fig. 4F). In contrast, the expression of PTTG2 appeared to be extremely low, if present at all (Fig. 1B and 4F). Since PTTG2 is an intronless gene, it is possible that it is derived from a retroposition (38). Nevertheless, low levels of PTTG2 mRNA have been detected in some normal tissues and some cancer cell lines by reverse transcription (RT)-PCR (7, 8), but the expression of PTTG2 protein has not been reported.
PTTG1 can induce transformation in both mouse fibroblasts (39) and human embryonic kidney cells (40). Its oncogenic potential is probably based on the uncoupling of mitotic exit from ESP1 activation, resulting in aberrant sister chromatid separation and aneuploidy (41–43). Accordingly, anaphase was delayed by PTTG1 overexpression (Fig. 2A and B), which was accentuated when the more stable PTTG1DΔ was used (Fig. 9D). The inhibition of sister chromatid separation led to cytokinesis failure and tetraploidization (Fig. 2C and 9D). In contrast, PTTG2DΔ did not affect the timing of cell cycle progression (Fig. 1D), anaphase onset (Fig. 9D), or tetraploidization (Fig. 2C). All these data suggest that PTTG2 probably does not have the same oncogenic potential as PTTG1.
Nevertheless, it is conceivable that PTTG2 possesses functions other than acting as a securin. PTTG1 has been implicated in multiple processes, including DNA damage repair, signal transduction, and transcription. For example, PTTG1 can bind Ku, the regulatory subunit of the DNA-PK in the nonhomologous end-joining repair pathway (44). PTTG1 can also associate with p53 and block its binding to DNA (45). There is a school of thought that suggests the transcription factor function of PTTG1 contributes to its oncogenic activities (reviewed in reference 36). The transactivation domain of PTTG1 is believed to be in the acidic C-terminal region. Two PXXP motifs in this region, which are predicted SH3-interacting domains, are important to the transforming activity of PTTG1. Although both of these PXXP motifs are conserved in PTTG2, a recent study suggested that PTTG2 does not contain transactivation activity (46). Furthermore, PTTG1 enhances the invasive properties of breast cancer cells by inducing epithelial-to-mesenchymal transition and expansion of the cancer stem cell population (47). However, studies with shRNA against PTTG2 suggest that downregulation of PTTG2 stimulates epithelial-to-mesenchymal transition and apoptosis (46). Finally, there is also a possibility that PTTG2 possesses other functions not attributed to PTTG1. For example, a genome-wide RNAi screen in human cells identified PTTG2 as a regulator of the circadian clock (48).
PTTG1 affects ESP1 in several ways. (i) PTTG1 stabilizes ESP1. As demonstrated with approaches including siRNAs (Fig. 4A and E) and overexpression of PTTG1 or ESP1 (Fig. 4C), PTTG1 was also reciprocally stabilized by ESP1. This probably explains the good correlation between the expression levels of PTTG1 and ESP1 in different cell lines (Fig. 4F). A simple explanation may be the inhibition of autocleavage of ESP1. As PTTG1 itself is believed to be an unfolded protein (49), the structure induced by binding to ESP1 may prevent PTTG1 from degradation by machineries that target unfolded proteins. (ii) PTTG1 inhibits ESP1 activity. Hence, nondegradable PTTG1DΔ prevented ESP1 activation during mitotic exit (Fig. 8D), and the effects of PTTG1DΔ on mitosis (Fig. 9E) were similar to those when ESP1 was deleted (19). (iii) PTTG1 activates ESP1. This was apparent in the presence of PTTG1H134R, which lost the inhibitory activity but retains the activating activity.
Our studies indicated that H134 is critically important for PTTG1 to inhibit ESP1. In support of this, mutation of H134 (to either Arg or Ala) attenuated the ability to inhibit ESP1 (Fig. 6A and E). The affinity for ESP1 was also reduced considerably (Fig. 5C). This was shown more clearly when ESP1 autocleavage was abolished with a mutation that disrupted its catalytic activity (Fig. 7B). Accordingly, PTTG2, which contains R134, was unable to interact with ESP1 (Fig. 3A and B). Unlike PTTG2, however, PTTG1H134R did not simply fail to interact with ESP1, because it was able to promote ESP1 activation (Fig. 5C and 6A). Our conclusion is that the motif containing H134 has a strong affinity for ESP1 and is involved in inhibiting ESP1. A separate region(s), possibly involving the C terminus of PTTG1, is involved in activating ESP1 and has a weaker affinity for ESP1.
In support of this model, both deletion of the C terminus (Fig. 3D) and its replacement with PTTG2's C terminus (Fig. 7B) reduced binding to ESP1. Although the presence of PTTG2's C terminus significantly reduced ESP1 activation, it was not completely abolished (Fig. 7C), suggesting a complex picture of ESP1 activation in PTTG1. A rigorous test of the idea was the mutation of PTTG2 back to a PTTG1-like protein. Indeed, a single-amino-acid change in PTTG2 to create H134 is sufficient to promote ESP1 binding (Fig. 9A) and to prevent anaphase onset (Fig. 9D). However, although PTTG2R134H could inhibit ESP1, whether it was able to activate ESP1 is unclear. The possibility that the activation of ESP1 by PTTG1 involves only a transient interaction suggests that many ESP1 molecules could be activated by a few PTTG1 molecules. This may explain why overexpression of ESP1 alone could induce premature sister chromatid separation (Fig. 8B and F) and act as an oncogene (50).
Given that PTTG1 may be involved in activating ESP1, an interesting question is whether sister chromatid separation can occur in the absence of PTTG1. The question is, of course, relevant to whether PTTG1 is essential for mitosis. In particular, our data indicate that PTTG2 should not be able to compensate for PTTG1. In fact, unlike the essential function of securin in fission yeast and Drosophila, PTTG1-deficient mice are both viable and fertile (51). Similarly, although disruption of PTTG1 in HCT116 cells results in aberrant anaphase, most of the cells eventually are able to divide and remain viable (18). This was attributed to a backup mechanism in which separase is inhibited by CDK1-dependent phosphorylation and binding to cyclin B1-CDK1 (52, 53). While this explains how separase can be inhibited before anaphase onset, it does not explain how separase is activated in the absence of PTTG1.
In fission yeast securin (Cut2p), a region containing 127DIE129 is involved in binding to separase by acting as a pseudosubstrate (54). The fact that human securin inhibits separase by preventing access of substrates to separase's active site is also consistent with securin acting as a pseudosubstrate (12). Whether the PTTG1 domain containing H134, identified in this study as essential for binding to separase, acts as a pseudosubstrate is not immediately obvious but is an intriguing possibility to explore.
The structure of the securin-separase complex has so far evaded elucidation. Part of the problem is that PTTG1 is a natively unfolded protein (49). Nevertheless, protonless nuclear magnetic resonance (NMR) analysis indicates that PTTG1 contains some transient local structural elements (55). In particular, the fragment D150-F159 appears to be important for separase association. Our data were consistent with this, because while PTTG1CΔ159 could still associate with ESP1, further deletion abolished the interaction (Fig. 3E to G). The finding of the importance of H134 further refines the interacting region and should help future structural analysis. We further noted that H134 is also within the region that binds PBF (56), encoded by a potential proto-oncogene that is upregulated in several cancers (reviewed in reference 36).
In conclusion, these results illustrate an interesting example of the loss of function of an isoform that probably is not expressed in vivo. Despite the similarity between the two isoforms, PTTG2 is unable to interact with ESP1. The differences between the isoforms led us to pinpoint a residue that is critical for the inhibition of ESP1 by PTTG1 that is distinct from the region required for ESP1 activation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ken Ma for the APC/C reporter cell line, as well as help with preparing lysates from different cell lines, and Wing Yu Man for technical assistance.
This work was supported in part by Research Grants Council grants AOE-MG/M-08/06 and HKU7/CRG/09.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 24 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00057-13.
REFERENCES
- 1.Michaelis C, Ciosk R, Nasmyth K. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35–45 [DOI] [PubMed] [Google Scholar]
- 2.Uhlmann F, Lottspeich F, Nasmyth K. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37–42 [DOI] [PubMed] [Google Scholar]
- 3.Zur A, Brandeis M. 2001. Securin degradation is mediated by fzy and fzr, and is required for complete chromatid separation but not for cytokinesis. EMBO J. 20:792–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pei L, Melmed S. 1997. Isolation and characterization of a pituitary tumor-transforming gene (PTTG). Mol. Endocrinol. 11:433–441 [DOI] [PubMed] [Google Scholar]
- 5.Zou H, McGarry TJ, Bernal T, Kirschner MW. 1999. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285:418–422 [DOI] [PubMed] [Google Scholar]
- 6.Kakar SS. 1999. Molecular cloning, genomic organization, and identification of the promoter for the human pituitary tumor transforming gene (PTTG). Gene 240:317–324 [DOI] [PubMed] [Google Scholar]
- 7.Prezant TR, Kadioglu P, Melmed S. 1999. An intronless homolog of human proto-oncogene hPTTG is expressed in pituitary tumors: evidence for hPTTG family. J. Clin. Endocrinol. Metab. 84:1149–1152 [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Puri R, Lefkowitz EJ, Kakar SS. 2000. Identification of the human pituitary tumor transforming gene (hPTTG) family: molecular structure, expression, and chromosomal localization. Gene 248:41–50 [DOI] [PubMed] [Google Scholar]
- 9.Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. 2005. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 3:e357. 10.1371/journal.pbio.0030357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornig NC, Knowles PP, McDonald NQ, Uhlmann F. 2002. The dual mechanism of separase regulation by securin. Curr. Biol. 12:973–982 [DOI] [PubMed] [Google Scholar]
- 11.Jager H, Herzig A, Lehner CF, Heidmann S. 2001. Drosophila separase is required for sister chromatid separation and binds to PIM and THR. Genes Dev. 15:2572–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waizenegger I, Gimenez-Abian JF, Wernic D, Peters JM. 2002. Regulation of human separase by securin binding and autocleavage. Curr. Biol. 12:1368–1378 [DOI] [PubMed] [Google Scholar]
- 13.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93:1067–1076 [DOI] [PubMed] [Google Scholar]
- 14.Funabiki H, Kumada K, Yanagida M. 1996. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 15:6617–6628 [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen S, Segal M, Clarke DJ, Reed SI. 2001. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J. Cell Biol. 152:27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumada K, Nakamura T, Nagao K, Funabiki H, Nakagawa T, Yanagida M. 1998. Cut1 is loaded onto the spindle by binding to Cut2 and promotes anaphase spindle movement upon Cut2 proteolysis. Curr. Biol. 8:633–641 [DOI] [PubMed] [Google Scholar]
- 17.Stratmann R, Lehner CF. 1996. Separation of sister chromatids in mitosis requires the Drosophila pimples product, a protein degraded after the metaphase/anaphase transition. Cell 84:25–35 [DOI] [PubMed] [Google Scholar]
- 18.Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, Kinzler KW, Vogelstein B, Lengauer C. 2001. Securin is required for chromosomal stability in human cells. Cell 105:445–457 [DOI] [PubMed] [Google Scholar]
- 19.Wirth KG, Wutz G, Kudo NR, Desdouets C, Zetterberg A, Taghybeeglu S, Seznec J, Ducos GM, Ricci R, Firnberg N, Peters JM, Nasmyth K. 2006. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J. Cell Biol. 172:847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yam CH, Siu WY, Lau A, Poon RY. 2000. Degradation of cyclin A does not require its phosphorylation by CDC2 and cyclin-dependent kinase 2. J. Biol. Chem. 275:3158–3167 [DOI] [PubMed] [Google Scholar]
- 21.Ma HT, Tsang YH, Marxer M, Poon RY. 2009. Cyclin A2-cyclin-dependent kinase 2 cooperates with the PLK1-SCFbeta-TrCP1-EMI1-anaphase-promoting complex/cyclosome axis to promote genome reduplication in the absence of mitosis. Mol. Cell. Biol. 29:6500–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan YW, Ma HT, Wong W, Ho CC, On KF, Poon RY. 2008. CDK1 inhibitors antagonize the immediate apoptosis triggered by spindle disruption but promote apoptosis following the subsequent rereplication and abnormal mitosis. Cell Cycle 7:1449–1461 [DOI] [PubMed] [Google Scholar]
- 23.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1995. Current protocols in molecular biology. John Wiley & Sons, New York, NY [Google Scholar]
- 24.Poon RY, Toyoshima H, Hunter T. 1995. Redistribution of the CDK inhibitor p27 between different cyclin CDK complexes in the mouse fibroblast cell cycle and in cells arrested with lovastatin or ultraviolet irradiation. Mol. Biol. Cell 6:1197–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma HT, Poon RY. 2011. Synchronization of HeLa cells. Methods Mol. Biol. 761:151–161 [DOI] [PubMed] [Google Scholar]
- 26.Woo RA, Poon RY. 2004. Activated oncogenes promote and cooperate with chromosomal instability for neoplastic transformation. Genes Dev. 18:1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yam CH, Ng RW, Siu WY, Lau AW, Poon RY. 1999. Regulation of cyclin A-Cdk2 by SCF component Skp1 and F-box protein Skp2. Mol. Cell. Biol. 19:635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu JY, DeRuiter SL, Turner DL. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 99:6047–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.On KF, Chen Y, Ma HT, Chow JP, Poon RY. 2011. Determinants of mitotic catastrophe on abrogation of the G2 DNA damage checkpoint by UCN-01. Mol. Cancer Ther. 10:784–794 [DOI] [PubMed] [Google Scholar]
- 30.Siu WY, Yam CH, Poon RY. 1999. G1 versus G2 cell cycle arrest after adriamycin-induced damage in mouse Swiss3T3 cells. FEBS Lett. 461:299–305 [DOI] [PubMed] [Google Scholar]
- 31.Siu WY, Lau A, Arooz T, Chow JP, Ho HT, Poon RY. 2004. Topoisomerase poisons differentially activate DNA damage checkpoints through ataxia-telangiectasia mutated-dependent and -independent mechanisms. Mol. Cancer Ther. 3:621–632 [PubMed] [Google Scholar]
- 32.Fung TK, Siu WY, Yam CH, Lau A, Poon RY. 2002. Cyclin F is degraded during G2-M by mechanisms fundamentally different from other cyclins. J. Biol. Chem. 277:35140–35149 [DOI] [PubMed] [Google Scholar]
- 33.Fan HY, Sun QY, Zou H. 2006. Regulation of Separase in meiosis: Separase is activated at the metaphase I-II transition in Xenopus oocytes during meiosis. Cell Cycle 5:198–204 [DOI] [PubMed] [Google Scholar]
- 34.Chestukhin A, Pfeffer C, Milligan S, DeCaprio JA, Pellman D. 2003. Processing, localization, and requirement of human separase for normal anaphase progression. Proc. Natl. Acad. Sci. U. S. A. 100:4574–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou H, Stemman O, Anderson JS, Mann M, Kirschner MW. 2002. Anaphase specific auto-cleavage of separase. FEBS Lett. 528:246–250 [DOI] [PubMed] [Google Scholar]
- 36.Smith VE, Franklyn JA, McCabe CJ. 2010. Pituitary tumor-transforming gene and its binding factor in endocrine cancer. Expert Rev. Mol. Med. 12:e38. 10.1017/S1462399410001699 [DOI] [PubMed] [Google Scholar]
- 37.Ramaswamy S, Ross KN, Lander ES, Golub TR. 2003. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33:49–54 [DOI] [PubMed] [Google Scholar]
- 38.Long M, Betran E, Thornton K, Wang W. 2003. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 4:865–875 [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Horwitz GA, Prezant TR, Valentini A, Nakashima M, Bronstein MD, Melmed S. 1999. Structure, expression, and function of human pituitary tumor-transforming gene (PTTG). Mol. Endocrinol. 13:156–166 [DOI] [PubMed] [Google Scholar]
- 40.Hamid T, Malik MT, Kakar SS. 2005. Ectopic expression of PTTG1/securin promotes tumorigenesis in human embryonic kidney cells. Mol. Cancer 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagting A, Den Elzen N, Vodermaier HC, Waizenegger IC, Peters JM, Pines J. 2002. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J. Cell Biol. 157:1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu R, Heaney AP, Lu W, Chen J, Melmed S. 2000. Pituitary tumor transforming gene causes aneuploidy and p53-dependent and p53-independent apoptosis. J. Biol. Chem. 275:36502–36505 [DOI] [PubMed] [Google Scholar]
- 43.Yu R, Lu W, Chen J, McCabe CJ, Melmed S. 2003. Overexpressed pituitary tumor-transforming gene causes aneuploidy in live human cells. Endocrinology 144:4991–4998 [DOI] [PubMed] [Google Scholar]
- 44.Romero F, Multon MC, Ramos-Morales F, Dominguez A, Bernal JA, Pintor-Toro JA, Tortolero M. 2001. Human securin, hPTTG, is associated with Ku heterodimer, the regulatory subunit of the DNA-dependent protein kinase. Nucleic Acids Res. 29:1300–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernal JA, Luna R, Espina A, Lazaro I, Ramos-Morales F, Romero F, Arias C, Silva A, Tortolero M, Pintor-Toro JA. 2002. Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat. Genet. 32:306–311 [DOI] [PubMed] [Google Scholar]
- 46.Mendez-Vidal C, Gamez-Del Estal MM, Moreno-Mateos MA, Espina-Zambrano AG, Torres B, Pintor-Toro JA. 2013. PTTG2 silencing results in induction of epithelial-to-mesenchymal transition and apoptosis. Cell Death Dis. 4:e530. 10.1038/cddis.2013.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon CH, Kim MJ, Lee H, Kim RK, Lim EJ, Yoo KC, Lee GH, Cui YH, Oh YS, Gye MC, Lee YY, Park IC, An S, Hwang SG, Park MJ, Suh Y, Lee SJ. 2012. PTTG1 oncogene promotes tumor malignancy via epithelial to mesenchymal transition and expansion of cancer stem cell population. J. Biol. Chem. 287:19516–19527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss W, Jr, Janes J, Su AI, Hogenesch JB, Kay SA. 2009. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139:199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez-Puig N, Veprintsev DB, Fersht AR. 2005. Human full-length Securin is a natively unfolded protein. Protein Sci. 14:1410–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang N, Ge G, Meyer R, Sethi S, Basu D, Pradhan S, Zhao YJ, Li XN, Cai WW, El-Naggar AK, Baladandayuthapani V, Kittrell FS, Rao PH, Medina D, Pati D. 2008. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 105:13033–13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Yu R, Melmed S. 2001. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol. Endocrinol. 15:1870–1879 [DOI] [PubMed] [Google Scholar]
- 52.Gorr IH, Boos D, Stemmann O. 2005. Mutual inhibition of separase and Cdk1 by two-step complex formation. Mol. Cell 19:135–141 [DOI] [PubMed] [Google Scholar]
- 53.Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. 2001. Dual inhibition of sister chromatid separation at metaphase. Cell 107:715–726 [DOI] [PubMed] [Google Scholar]
- 54.Nagao K, Yanagida M. 2006. Securin can have a separase cleavage site by substitution mutations in the domain required for stabilization and inhibition of separase. Genes Cells 11:247–260 [DOI] [PubMed] [Google Scholar]
- 55.Csizmok V, Felli IC, Tompa P, Banci L, Bertini I. 2008. Structural and dynamic characterization of intrinsically disordered human securin by NMR spectroscopy. J. Am. Chem. Soc. 130:16873–16879 [DOI] [PubMed] [Google Scholar]
- 56.Chien W, Pei L. 2000. A novel binding factor facilitates nuclear translocation and transcriptional activation function of the pituitary tumor-transforming gene product. J. Biol. Chem. 275:19422–19427 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.