Abstract
Barrier to autointegration factor (BAF/BANF1) is a cellular DNA-binding protein found in the nucleus and cytoplasm. Cytoplasmic BAF binds to foreign DNA and can act as a defense against vaccinia DNA replication. To evade BAF, vaccinia expresses the B1 kinase, which phosphorylates BAF and blocks its ability to bind DNA. Interestingly, B1 is also needed for viral intermediate gene expression via an unknown mechanism. Therefore, we evaluated the impact of B1-BAF signaling on vaccinia transcription. Strikingly, the decrease in vaccinia transcription caused by loss of B1 can be rescued by depletion of BAF. The repressive action of BAF is greatest on a viral promoter, and is more modest when non-vaccinia promoters are employed, which suggests BAF acts in a gene specific manner. These studies expand our understanding of the role of the B1 kinase during infection and provide the first evidence that BAF is a defense against viral gene expression.
Keywords: vaccinia, transcription, virus host interaction, BAF, BANF1, barrier to autointegration
INTRODUCTION
Vaccinia virus is the prototypical member of the large DNA virus family Poxviridae. Like all poxviruses, vaccinia exhibits significant autonomy from the host cell and carries out its entire lifecycle in the cytoplasm. The fact that vaccinia virus is capable of both DNA replication and transcription in this location is unique among DNA viruses, and is achieved by a large repertoire of virus-encoded proteins including multisubunit DNA and RNA polymerases and multiple accessory factors (Ahn et al., 1990, Amegadzie et al., 1991, Challberg, MD., and Englund, PT, 1979, Condit et al., 1991, Earl et al., 1986, Klemperer et al., 2001, McDonald et al., 1997, Moss et al., 1991). Functional characterization of these proteins over the last three decades has provided broad mechanistic insights into genome replication and transcription, which are of interest to virology and cell biology alike. While poxviruses avoid obstacles such as chromatin which nuclear viruses face, cytoplasmic replication means that they are also challenged by host defenses specific to that environment. In turn, viruses employ countermeasures to inactivate the host defenses which are the most significant threat. Recent studies have revealed that the product of the vaccinia B1R gene is one such countermeasure.
Analysis of temperature sensitive mutant viruses, including the ts2 virus, which contain a point mutation within the B1 ORF (Condit et al., 1983, Rempel et al., 1990, Rempel and Traktman, 1992, Traktman et al., 1989) provided the first evidence of the essential role played by B1 during the vaccinia lifecycle (Banham and Smith, 1992, Lin et al., 1992, Rempel and Traktman, 1992). Subsequent genetic and biochemical analysis of B1 revealed it to be a Ser/Thr kinase essential for viral DNA replication as well as for optimal intermediate transcription (Banham and Smith, 1992, Kovacs et al., 2001, Lin et al., 1992), and also determined that the lesion found in the ts2 virus abrogates B1's kinase activity. The vaccinia B1 protein exhibits significant homology with a family of eukaryotic kinases now referred to as vaccinia-related kinases (VRKs) (Nezu et al., 1997, Nichols and Traktman, 2004). Importantly, Boyle and Traktman discovered that the cellular kinase VRK1, when expressed from the ts2 genome, rescued the DNA replication defect exhibited by this virus (Boyle and Traktman, 2004). This study suggested that B1 and VRK1 both target the same substrate needed to permit genome replication, and laid the foundation for the identification of the barrier to autointegration factor (BAF/BANF1) as a cellular substrate of B1 and VRK1 (Wiebe and Traktman, 2007).
BAF is a highly conserved 10-kDa DNA binding protein found in both the cytoplasm and the nucleus of many cell types, and has been found to be important for cell survival in multiple model systems (Cai et al., 1998, Cox et al., 2011, Furukawa et al., 2003, Lee and Craigie, 1998, Margalit et al., 2005, Margalit et al., 2007, Puente et al., 2011, Zheng et al., 2000). Specifically, BAF is needed for survival and differentiation of both human and mouse embryonic stem cells (Cox et al., 2011). Additionally, attempts to deplete or knock out BAF in Caenorhabditis elegans and Drosophila melanogaster resulted in lethal phenotypes early in embryogenesis (Furukawa et al., 2003, Margalit et al., 2007). Recent data has also provided new insight of the importance of BAF in human disease. Specifically, a point mutation within the BAF coding region causes Nestor-Guillermo syndrome, a hereditary progeroid syndrome (Puente et al., 2011).
Extensive in vitro biochemical and structural studies of BAF have advanced our understanding of BAF's function and regulation (Bradley et al., 2005, Haraguchi et al., 2001, Harris and Engelman, 2000, Ibrahim et al., 2011, Margalit et al., 2005, Segura-Totten et al., 2002, Segura-Totten and Wilson, 2004, Umland et al., 2000). BAF binds double-stranded DNA independent of sequence (Lee and Craigie, 1998, Umland et al., 2000, Zheng et al., 2000), but does not bind ssDNA, ssRNA, or dsRNA (Ibrahim et al., 2011, Lee and Craigie, 1998). Through its ability to homodimerize, BAF can cross-bridge DNA to form higher order nucleoprotein complexes (Lee and Craigie, 1998, Zheng et al., 2000). Phosphorylation of BAF by either the viral B1 or cellular VRK1 proteins strongly inhibits BAF's ability to bind DNA, and regulates BAF in both the cytoplasm and nucleus (Nichols et al., 2006). In the nucleus, BAF interacts with LEM-domain proteins, histones, and transcriptional regulators (Dechat et al., 2004, Furukawa, 1999, Haraguchi et al., 2001, Lee et al., 2001, Margalit et al., 2005, Montes de Oca et al., 2005, Montes de Oca et al., 2009, Segura-Totten et al., 2002, Segura-Totten and Wilson, 2004, Shumaker et al., 2001). These partners allow BAF to play an important role during nuclear reassembly, and likely allow BAF to modulate gene expression as well. In the cytoplasm BAF is capable of inhibiting viral DNA replication (Wiebe and Traktman, 2007). This host defense activity of BAF depends on its DNA binding and bridging properties and is blocked through phosphorylation in the presence of active B1 kinase (Ibrahim et al., 2011).
In this manuscript, we further explore the importance of the B1-BAF signaling axis during poxviral infection. Specifically, we test the hypothesis that the action of BAF explains why B1 kinase is needed for intermediate transcription as well as DNA replication. We have determined that depletion of BAF from mouse L929 cells rescues the transcriptional defect of the ts2 B1-deficient virus. Interestingly, the impact of BAF is greatest on the viral G8 promoter as compared with other non-vaccinia promoters examined. Thus, our data suggests that while BAF can bind any dsDNA sequences, its ability to repress transcription is specific to the promoter BAF is acting on. Together, these data reveal a novel property of BAF as a transcriptional inhibitor and demonstrate that vaccinia is an excellent model system for future characterization of BAF's function as a regulator of gene expression.
MATERIALS AND METHODS
Cell culture, transfection and infection
All chemicals were purchased from Fisher Scientific or Sigma-Aldrich unless otherwise stated. African green monkey BSC40 kidney epithelial cells, CV1, U2OS, and mouse fibroblast L929 cells were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals) and penicillin-streptomycin and incubated at 37°C in a 5% CO2 atmosphere.
Plasmids
For transfection experiments performed in the absence of vaccinia infection, two plasmids expressing firefly luciferase were used: CMV-Luc expresses luciferase under the CMV immediate early enhancer promoter (pGL4.51[luc2/CMV/Neo], Catalog# E1320), and minP-Luc expresses luciferase under a minimal promoter containing only a TATA box (pGL4.26[luc2/min/Hygro], Catalog# E8441). For transfection-infection experiments, pG8-luciferase expresses luciferase under the viral intermediate G8 promoter (a generous gift from B. Moss, NIAID, Bethesda, MD), and T7-luciferase (Promega) were used.
Plasmids containing A2, I1 and Consensus Intermediate promoter sequences upstream of the firefly luciferase gene were constructed as follows. PCR was performed using pG8-Luciferase plasmid as a template and primers designed to include sequences previously published of I1, A2 and Consensus promoters (Baldick et al., 1992, Knutson et al., 2006, Yang et al., 2012). All three promoters (in bold) were placed upstream of 18bp homologous to the 5’ end of firefly luciferase gene (in italics). Non-viral sequences including three nucleotides followed by EcoRI restriction sites were placed upstream of the promoter sequences (in lower case). The upstream primers are A2L-Luc (5’ccggaattcGCAACGTCTAGAAATAAAATGTTTTTATATAAAAatggaagatgccaaaaac-3’), I1L-Luc (5’-ccggaattcTTTGTATTTAAAAGTTGTTTGGTGAACTTAAATGGCagacgccaaaaacataaag-3’) and Consensus-Luc (5’-ccggaattcTAATATATTTAAAATAAAAATTAATATTATAAAatggaagatgccaaaaac-3’). The downstream primer, Universal-Luc-3’ (5’-caggaattcttacacggcgatcttTc-3’), is homologous to the 3’-end of the firefly luciferase ORF, and contains an EcoRI site (lowercase) as well. PCR products amplified using these primers were cloned into the pCRII-TOPO plasmid (Invitrogen) using the EcoRI restriction and promoter identity verified following DNA sequencing.
Viruses
The following viruses were used: wild-type vaccinia virus (WR strain), the B1-deficient ts2 virus (Condit et al., 1983, Rempel et al., 1990, Rempel and Traktman, 1992), and vTF7.3 (Fuerst et al., 1986). Recombinant virus ts2/B1 (Boyle and Traktman, 2004) was provided by Dr. Traktman (Medical College of Wisconsin). Viruses ts42 (E9 mutant) and ts24 (D5 mutant) were gifts from Dr. Rich Condit (University of Florida). Stocks of all viruses were purified from cytoplasmic lysates of infected BSC40 cells by ultracentrifugation through 36% sucrose; and quantified by plaque assay titration on BSC40 cells.
Production of cells stably overexpressing BAF
The stable overexpression of BAF in L929 cells was performed by using a lentivirus expressing 3XFlag-BAF (plasmids were a kind gift from Dr. Paula Traktman (Molitor and Traktman, 2013)). Specifically, 293T cells were transfected with pHM-3XFlag-BAF or pHM-MCS plasmid (Mostoslavsky et al., 2006, Ory et al., 1996) with a combination of viral packaging plasmids pVSV-G, pTat, pREV and pGag/Pol. The next day, media was replaced with fresh media containing 5mM Sodium Butyrate. Eight hours later fresh media containing 10mM HEPES pH7.4 was added for additional 12h. Next, media containing lentivirus was filtered through a 0.45μm sterile filter, and polybrene (8μg/ml) was added and stored at −80°C. For transduction, L929 cells were seeded in 35 mm dishes at 1×106 per well. The next day, medium was replaced with 1 mL of lentivirus supernatant. After 24hr, medium was replaced with fresh media for additional 24h. Cells were then grown in media containing 200 μg/ml of hygromycin to select for stable lentiviral integration.
Production of cells stably depleted of BAF
The stable depletion of BAF was performed using a lentivirus expressing a BAF-specific short hairpin RNA (shRNA) or control (scrambled) shRNA as previously described (Ibrahim et al., 2011), with the exception that transduced cells were selected with 15 ug/ml of puromycin. To express shRNA specific to murine BAF mRNA, the primers mBAFsense (5’-TGGCTTATGTGGTCCTTGGCTTCAAGAGAGCCAAGGACCACATAAGCCTTTTTGGAAAC-3’) and mBAFantisense (5’-TCGAGTTTCCAAAAAGGCTTATGTGGTCCTTGGCTCTCTTGAAGCCAAGGACCACATAAGCCA -3’) were annealed and cloned into the pLL3.7 as previously described (Wiebe and Traktman, 2007).
Luciferase assays
Transfection-Infection experiments
L929 cells seeded at 2 × 105 per well of a 12-well plate were transfected using Lipofectamine 2000 (Invitrogen) with 10 ng of pG8-Luc DNA per well. The plasmid-Lipofectamine transfection complexes were prepared as follows:130ng of pG8-luciferase and 1 μl of Lipofectamine 2000 were combined in 2.3 mL of DMEM; and 200 μl of the transfection mixture was used per well. Cells were incubated at 37°C in a 5% CO2 atmosphere for 7h to allow for plasmid introduction into the cells, which were then infected with vaccinia virus at a multiplicity of infection (MOI) of 3 in media containing 50 μM AraC and placed back in the incubator for additional 18h. Cells were then washed twice with phosphate-buffered saline(PBS) (10 mM Na2HPO4-7H2O, 1 mM KH2PO4, 2mM KCl, 140 mM NaCl [pH 7.4]), then lysed in 300 μl of 1X Reporter Lysis buffer (Promega) by two freeze-thaw cycles. The luciferase activity was measured using 50 μl of lysate for 100 μl of luciferase assay substrate buffer in a Berthold multiwell Luminometer.
Transfection alone
L929 cells were seeded at 2 × 105 per well of a 12-well plate, and the next day they were transfected using Lipofectamine 2000 (2 μl per μg DNA; Invitrogen) with CMV-Luc or minP-luciferase for additional 12h. Cells were washed twice with PBS, and then lysed with 300 μl of 1X Reporter Lysis buffer (Promega), and freeze-thaw twice. The luciferase activity was measured using 50 μl of lysate and 100 μl of luciferase assay substrate buffer in a Berthold multiwell Luminometer. Firefly luciferase expression was normalized to relative total protein level in each lysate. Protein level in each lysate was quantified by BCA protein assay (Thermo Scientific).
Immunoblot Analysis
L929 and L929 stably expressing specific shRNA were freshly collected in 300 μl of SDS sample buffer (100 mM Tris pH6.8, 2% β-mercaptoethanol, 2% SDS, 32.5% glycerol, bromophenol blue) supplemented with 10 units of Benzonase. Lysate volume equivalent to 105 cells were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on a 18% gel, transferred to PVDF, and incubated with primary against BAF (Wiebe and Traktman, 2007), and rabbit secondary antibodies. Blots were developed with chemiluminescent reagents, and quantified by a Bio-Rad Chemidoc XRS instrument to verify that BAF expression had been depleted >85%.
B1 siRNA Transfection/Infection
B1-specific and control siRNAs were designed and ordered from Dharmacon. The B1-1siRNA sense sequence is 5’-CAAUAUGCACCUAGAGAAUUU-3’ and the B1-2 siRNA sense sequence is 5’-GCCCAAAGCUAACGGAUCAUU-3’. The siControl sense strand sequence is 5’-CAGUCGCGUUUGCGACUGGUU-3’. L-929 cells were transfected with 100 nM of siRNA Control, siB1R-1, or siB1R-2 using RNAimax (Life Technologies) as per manufacturer's protocol. 16-24 hours post transfection cells were infected with WT virus at an MOI of 3 and harvested for B1R mRNA at 4 hpi. RNA was extracted using TRIzol reagent, purified with Aurum Total RNA mini kit (Biorad), reverse transcribed to generate cDNA (Applied Biosystems, High Capacity cDNA Reverse Transcription Kit), and treated with 0.5 μg/ml RNase A prior to qPCR analysis.
qPCR
qPCR performed on B1R cDNA was done using TaqMan master mix (Applied Biosystems) and 900 nM of primers B1R F (5 -AATCAATGGGTCGTTGGACCAT-3 ) and B1R R (5 -AATACATCATTTTTATCTCGGGTTTCGATTGC-3 ), and 250 nM B1R probe (5 -56-FAM/AG GTG CAG ATC TAG ATG CGG TGA TCA /3IABkFQ-3 ), specific to the vaccinia virus B1R gene. To measure viral DNA replication, DNA was extracted from the cells with the QIAamp DNA Blood Mini Kit (Qiagen) and treated with 5 μg/ml RNase A. Viral DNA replication was quantified using SYBR green PCR mix (Applied Biosystems) with primers specific to the vaccinia virus HA gene at a concentration of 900 nM each (HA F: 5 -CATCATCTGGAATTGTCACTA CTAAA-3 and HA R: 5 -ACGGCCGACAATATAATTAATGC-3 ).
ChIP Assays
L929 cells (107) stably expressing specific shRNA were transfected with 150 ng pG8-Luc plasmid for 24 h. Cells were cross-linked with 1% formaldehyde for 10 min at room temperature followed by quenching with 0.75% glycine. Cell lysis was done using FA lysis buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8, 1% TritonX-100, 0.1% sodium deoxycholate, 0.1% SDS, and protease inhibitor (Complete, Roche)). Lysates were sonicated with Misonix XL-2000 Sonicator for 10 × 10 seconds at power setting 3 (output wattage 6) followed by centrifugation at 8000g for 1 min. A fraction (6%) of the supernatant was used as ‘input’. Clarified lysates were then incubated at 4°C overnight with 2 μg of anti-FLAG antibody (F7425, Sigma) and an equal volume of RIPA buffer (50 mM Tris.HCl pH 8, 150 mM NaCl, 2 mM EDTA pH 8, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor). To each sample, 10 μL of Dynabeads Protein G (10003D, Novex) were added and incubated on a rotator at 4°C for 2 h. Beads were washed 3 times with wash buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA pH 8, 1% Triton X-100, and 0.1% SDS), and once with final wash buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 2 mM EDTA pH 8, 1% Triton X-100, and 0.1% SDS). Bound complexes were eluted from the beads with elution buffer (100 mM NaHCO3 and 1% SDS) by incubating at 30°C for 20 min. Crosslinking was reversed by overnight incubation at 60–65°C in the presence of 10 μg/ml RNase A and 1 mg/ml Proteinase K. DNA was purified using Zymoclean Gel DNA Recovery Kit (D4002, Zymo Research). Immunoprecipitated and input material was analyzed by quantitative PCR (StepOne Plus Real Time PCR, Applied Biosystems) using SYBR green PCR mix (Applied Biosystems) with three different primers sets as followed: G8Pro ChIP Fwd ( 5’- CTTCGTGGATCCTGTAGAACG-3’) and G8Pro ChIP Rev ( 5’- CCATCTTCCAGCGGATAGAATG-3’) which flank the G8 promoter of pG8-Luc DNA; Neo Fwd (5’-CTTGCTCCTGCCGAGAAAGT-3’) and Neo Rev (5’-TTCGCTTGGTGGTCGAATG-3’); and ActinB Fwd (5’-GGTCATCACTATTGGCAACG-3’) and ActinB Rev (5’-CGTCACACTTCATGATGGAATTG-3’). Serial dilutions were also included in each qPCR run and used to develop a standard curve and determine the PCR efficiency of the primer sets in that experiment set. The efficiency (E) is calculated as such: E = 10(−1/slope). Fold enrichment was calculated relative to negative control sets as such: fold enrichment = E−(ΔCttarget – ΔCtcontrol).
Statistics
Error bars shown represent one standard deviation from the mean. The p-values indicated were calculated using the Students t-test.
RESULTS
The regulation of viral intermediate gene expression requires the B1 kinase independently of its role in DNA replication
Temperature sensitive vaccinia viruses with lesions in the B1 kinase (ts2 and ts25) display a primary block at the stage of DNA replication at nonpermissive temperature (Condit et al., 1983). The requirement for B1 at this stage is linked to its ability to inactivate the cellular factor BAF via phosphorylation. If left unphosphorylated, BAF relocalizes to viral factories and impairs viral genome replication via its ability to bind and crossbridge DNA (Ibrahim et al., 2011, Nichols et al., 2006, Wiebe and Traktman, 2007). Interestingly, prior to the discovery of the B1-BAF signaling axis, Kovacs et al have shown that B1 is also required for at least one post replicative stage in the viral life cycle. Specifically, using a plasmid transfection / infection approach to examine intermediate gene expression independent of DNA replication, it was discovered that intermediate promoter activity is impaired during infection with ts25 (Kovacs et al., 2001). However, the mechanism through which B1 contributes to gene expression has yet to be determined.
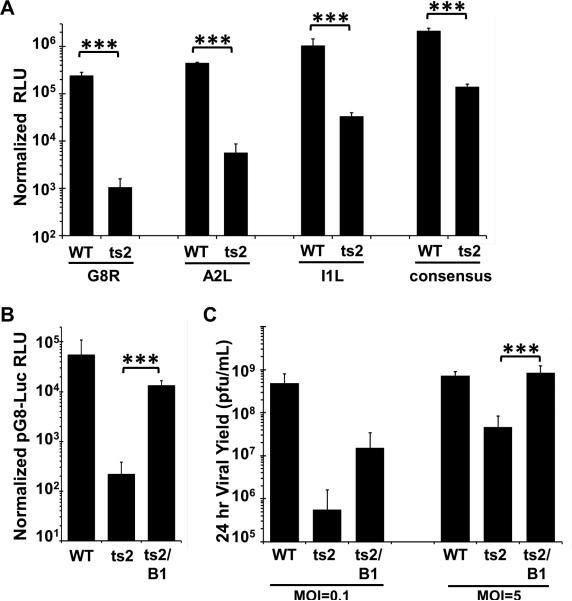
The goal of this present study was to further examine the role of B1 during vaccinia intermediate gene expression and test the hypothesis that without an active B1 kinase the cellular factor BAF is not only able to impede DNA replication, but viral transcription as well. To test this hypothesis, we first worked to confirm the role of B1 in intermediate gene expression as previously reported (Kovacs et al., 2001). L929 fibroblasts were transfected with plasmids expressing firefly luciferase under the intermediate promoters of the G8R, A2L, I1L genes or a synthetic intermediate promoter based on the consensus sequence described by Yang et al. (Yang et al., 2012). These cells were then infected with WT or ts2 viruses at an MOI of 3 and incubated at 37°C overnight before harvesting and measurement of luciferase activity. A temperature of 37°C was chosen for our studies because we found that L929 cells are highly sensitive to heat induced stress at 39.7°C. As we found that multiple temperature-sensitive viruses retain their ts phenotype at 37°C in L929 cells (see Fig. 2 below), we chose to employ this temperature in our studies to minimize the impact of unknown stress responses on the interpretation of our data. To examine viral intermediate gene expression independent of DNA replication, the nucleoside analog cytosine arabinoside (AraC) was added during the infection, as previous studies have shown that the expression of vaccinia genes from a plasmid introduced into infected cells does not require viral DNA replication (Keck et al., 1990, Vos, JC. and Stunnenberg, HG., 1988). As shown in Figure 1A, expression from pG8-Luciferase (pG8-Luc) is inhibited in ts2-infected cells over 200-fold compared to WT-infected cells. This is in line with the previous observation by Kovacs et al that the presence of a functional B1 kinase is necessary for the viral intermediate gene expression. Likewise, activation of each of the A2L, I1L, and consensus intermediate promoters was significantly decreased in the ts2 infected cells as compared to WT infected cells, demonstrating that B1 is important for expression of intermediate genes in general. As the pG8-Luc construct exhibited the greatest fold difference between WT and ts2 infections, we focused on it for the remainder of our study to allow for the greatest sensitivity in our assays.
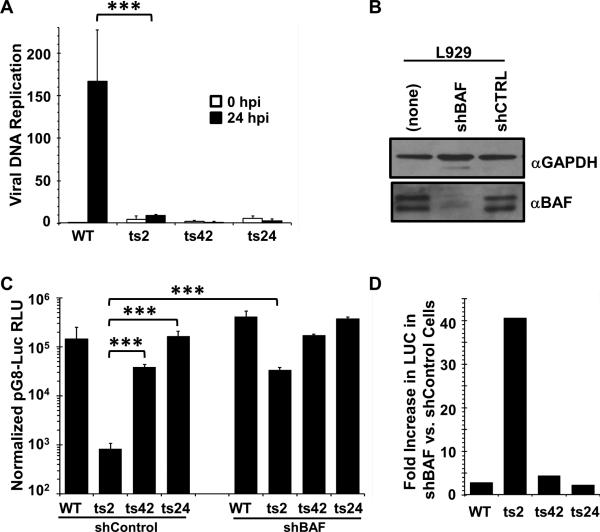
Figure 2. The defect in intermediate transcription is specific to ts2 and is rescued by depletion of BAF.
(A). DNA replication assays. Total DNA was isolated at 30 minutes and 24 hours after infection (MOI=3) with WT, ts2, ts42, or ts24 at 37°C, then quantified by qPCR. Data was obtained from two independent experiments, PCR amplified in duplicate. Data is shown as a fold difference compared to the WT sample at 0.5 hpi. Error bars represent standard deviation. (B). Immunoblot analysis of BAF expression in L929 cells stably depleted of BAF using specific shRNA. Lysates from equivalent numbers of cells were collected and analyzed using antibody against BAF. The total amounts of BAF in each lane were quantified using a Bio-Rad Chemidoc XRS instrument. GAPDH level was used as loading control. (C) L929 cells stably expressing shBAF or shControl were transfected with pG8-Luciferase for 7hr, then infected at 37°C with WT, ts2, ts42 or ts24 at MOI=3 in the presence of 50 μM AraC. Lysates were prepared at 16h after infection and assayed for luciferase activity. RLU shown is normalized to total protein measured by BCA assay. Data were obtained from three independent experiments performed in triplicate wells. Data from a representative experiment is shown. Error bars represent standard deviation. D) Data from the luciferase expression shown in (C) was replotted as a fold difference between the shControl and shBAF cell lines for each virus. (*** indicates a p-value <0.05)
Figure 1. The ts2 defect in intermediate transcription is observed on multiple promoters and is rescued by B1 expression in L929 cells.
(A-B) L929 cells were transfected with the reporter construct shown for 7hr, then infected at 37°C with WT, ts2, or ts2/B1 as indicated (MOI=3) in the presence of 50 μM AraC. Lysates were prepared at 16h after infection and assayed for luciferase activity. RLU shown is normalized to total protein measured by BCA assay. Data were obtained from three independent experiments performed in triplicate wells. Data from a representative experiment is shown. Error bars represent the standard deviation. (C) L929 cells were infected with WT, ts2, or ts2/B1 at MOI= 0.1 or MOI=5 for 24h at 37°C. After lysates were collected virus yield was determined by a plaque titration on monolayers of BSC40 cells at 32°C. Data were obtained from triplicate experiments, and standard deviation is presented by the error bars. (*** indicates a p-value <0.05)
Next, we tested whether the defect in intermediate gene expression of ts2 can be rescued by expression of B1 from the viral TK locus of the ts2 virus. Using recombinant ts2 viruses, Boyle and Traktman demonstrated that expression of B1 from the TK locus of the ts2 genome rescues the DNA replication defect of this virus (Boyle and Traktman, 2004). Using the same virus, we verified that in L929 cells the expression of B1 from the ts2 virus rescued viral yield compared to ts2 alone at either low or high MOI (Fig. 1C), as was previously observed in BSC40 cells (Boyle and Traktman, 2004). We also found that luciferase activity measured in ts2/B1 was 50-fold higher than that found in ts2-infected cells (Fig. 1B). These results support the role of the B1 kinase during viral intermediate gene expression, and demonstrate that expression of B1 from the ts2 genome can both rescue viral yield and enhance viral intermediate gene expression in L929 cells.
BAF affects viral intermediate gene expression in a B1 dependent manner
The above data indicate that the B1 kinase is required not only for DNA replication, but intermediate gene expression as well. To determine whether B1 is unique in functioning at both of these stages of the viral lifecycle, we examined whether intermediate gene expression was decreased during infection with other ts viruses displaying blocks in DNA replication. Specifically, two ts mutants with DNA replication defects were employed: ts42 virus carries a mutation in the viral catalytic DNA polymerase (E9) (McDonald and Traktman, 1994, Sridhar P and Condit, R.C., 1983) and ts24 carries a mutation in the D5 primase/helicase protein (De Silva et al., 2007, De Silva et al., 2009, Evans, 1992, Evans et al., 1995). First, we verified that viral DNA replication was impaired at 37°C during infection with these viruses. L929 cells were infected (MOI=3) with WT, ts2, ts42, or ts24 and lysates were collected at 0.5 and 24hpi for DNA quantification using quantitative PCR. As shown in Figure 2A, while during WT infection viral DNA increased more than 150-fold in 24 hours, ts2 DNA was increased only ~3-fold compared to input and no increase in DNA was detected during ts42 and ts24 infections. These data demonstrate that in L929 cells, 37°C is a nonpermissive temperature for each of these mutant viruses.
Next, these viruses were used to begin our examination of the role of BAF in intermediate gene expression. First, we transduced L929 cells with replication-incompetent lentiviral vectors expressing either a BAF-specific or control (CTRL) shRNA. Immunoblot analysis of lysates from these cells revealed that expression of BAF in L929-shBAF cells is decreased to 15-20% compared to that from the untransduced L929 cells, while no impact on BAF expression was detectable in the shCTRL cells (Fig. 2B). Next, L929-Control and -shBAF cells were transfected with pG8-Luc, then infected with WT, ts2, ts42 or ts24 viruses (MOI=3) in the presence of AraC for 16-20 hours prior to harvest and measurement of luciferase activity. In L929-shControl cells (Fig. 2C, left), we found that pG8-Luc reporter activity in cells infected with ts24 was identical to WT infection, and ts42 were only slightly (2.5-fold) reduced. This was in clear contrast to pG8-Luc activity during ts2 infection, which was more than two orders of magnitude lower. This result indicates that E9 and D5 do not affect intermediate gene expression and that the importance of B1 at this stage is not common among viral DNA replication proteins.
Next, the same transfection/infection protocol was performed in L929-shBAF cells where BAF expression is stably depleted. Interestingly, in these cells reporter gene expression was enhanced to at least some degree regardless of the virus used (Fig. 2C). Luciferase activity in WT, ts42, and ts24 infected L929-shBAF cells was 2-3 folds greater than that observed for each of those viruses in L929-shControl cells (Fig. 2D). Strikingly however, luciferase activity in ts2 infected L929-shBAF cells was close to 40-fold greater than that in L929-shControl cells (Fig. 2C and D). This specific rescue of ts2-mediated intermediate gene expression in BAF depleted cells suggests that the B1-BAF signaling axis is important at this stage of the life cycle as well as during DNA replication.
Depletion of B1 during WT vaccinia infection also impedes transcription in a BAF-dependent manner
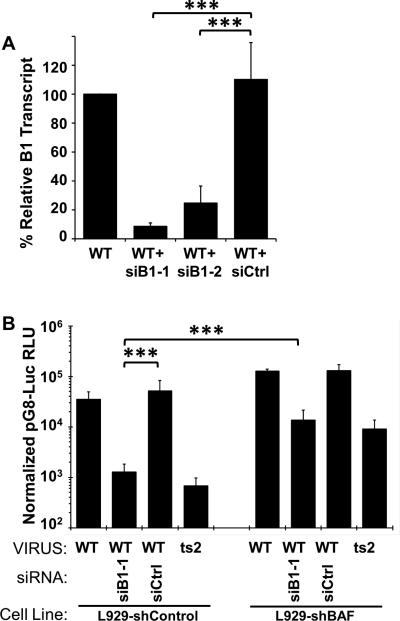
To further characterize the role of B1 during intermediate gene regulation, we examined whether siRNA-mediated depletion of B1 would yield results similar to those obtained above with the ts2 virus. To establish a method for depletion B1 mRNA, L929 cells were transfected with 100nM of B1-specific or control siRNA for 24h, followed by an infection with WT virus at MOI=3 at 37C for 4 hours. Two B1-specific siRNA were tested. Lysates were collected and RNA extracted for subsequent qPCR using primers specific to the B1 ORF. Based on RT-qPCR results (Fig. 3A), the accumulation of B1-specific transcripts was substantially reduced in siB1-pretreated cells compared non-treated cells. The decrease in B1-specific transcripts was 90% with siB1-1 and 70% with siB1-2. Due to the greater depletion by siB1-1, we selected that siRNA for use in subsequent studies.
Figure 3. Depletion of B1 impairs intermediate transcription following WT virus infection, and can be rescued by BAF depletion.
(A) L-929 cells were transfected with 100 nM of siRNA siControl, siB1R-1, or siB1R-2. At 24 hours post transfection, cells were infected with WT virus at a MOI of 3 and total RNA harvested at 4hpi. Following reverse transcription, cDNA was quantified by qPCR. (B) L929 stably expressing shBAF or shControl were transfected with 100nM siRNA specific to B1 kinase (siB1-1) or siControl for 12h, and then transfected with pG8-Luciferase for 7hr, then infected at 37°C with WT virus at MOI=3 in the presence of 50 uM AraC. An infection of L929 with ts2 at MOI=3 in the presence of AraC at 37°C was also performed. Lysates were prepared at 12h after infection and assayed for luciferase activity, and RLU normalized to protein level. Data were obtained from three independent experiments performed in triplicate wells. Data from a representative experiment is shown. Error bars represent standard deviation. (*** indicates a p-value <0.05)
To determine whether depletion of B1 results in diminished intermediate gene expression, L929 were transfected with siB1 or siControl for 12h, then transfected with pG8-luc for 7h before infecting with WT vaccinia in the presence of AraC. Cells transfected only with pG8-Luc and infected with ts2 + AraC were included for comparison purposes. To simultaneously investigate the involvement of BAF in these studies, both L929-shControl and L929-shBAF cells were employed. In L929-shControl, we found that depletion of B1 leads to a substantial decrease in the activity of the reporter gene when compared to siControl-transfected cells or cells not treated with any siRNA (Fig. 3B). Specifically, we observed an average decrease of ~40-fold in luciferase activity, bringing the reporter activity to a level very similar to that obtained during the ts2 infection. In L929-shBAF cells, the depletion of BAF was able to rescue the loss of intermediate gene expression caused by siB1 treatment as well as during ts2 infection. As in Fig. 2C-D, the fact that the loss of BAF specifically enhances gene expression when B1 has been depleted further supports our model that B1 is needed to repress the inhibitory activity of BAF.
BAF immunoprecipitates pG8-Luc DNA and depletion of BAF rescues viral intermediate gene expression at the transcriptional level
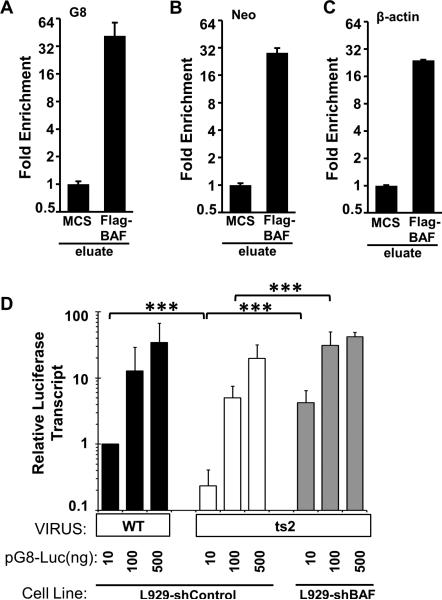
Although we have previously shown that BAF relocalizes to transfected dsDNA by immunofluorescence (Ibrahim et al., 2011) and other studies have also shown BAF's DNA-binding capability in vitro via biochemical and structural studies (Bradley et al., 2005, Cai et al., 1998, Umland et al., 2000, Zheng et al., 2000), BAF's direct interaction with DNA in an intact cell has yet to be demonstrated by co-immunoprecipitation. Therefore, to validate that BAF impairs gene expression through its direct DNA-binding activity, we performed chromatin immunoprecipitation analysis in L929 cells stably expressing MCS empty vector (control) or FLAG-BAF. Cells were transfected with 150 ng pG8-Luc plasmid for 24 h followed by immunoprecipitation of BAF-DNA complexes. Bound pG8-Luc DNA was analyzed by qPCR analysis using a pG8-Luc-specific primer set which flanks the G8 promoter region. In FLAG-BAF-expressing cells, we observed an increase in pG8-Luc DNA binding compared to control cells . Specifically, while the plasmid DNA harvested for the input samples was similar between the MCS and Flag-BAF expressing cells, following immunoprecipitation the average difference in Ct was 5.3 Ct lower (Supp Fig 2) in the Flag-BAF cells and fold enrichment ~40 fold higher than the control cells (Fig. 4A). As all previous studies of BAF have shown that BAF binds dsDNA in a sequence independent manner we were curious as to whether BAF is preferentially binding the G8 promoter. Therefore, in addition to G8 promoter-specific primers, we also utilized primers specific to the Neomycin resistance gene on the opposite site of the plasmid and the cellular beta-actin gene. We found that the Neo region of the plasmid and beta-actin gene were immunoprecipitated with an efficiency almost equal to the G8 promoter (Fig. 4B-C). These data support a model in which BAF directly interacts with many, if not all, regions of the plasmid construct during its impairment of gene expression. Such a model would also be consistent with previous in vitro DNA binding data and structural studies demonstrating that BAF binds dsDNA in a sequence independent manner.
Figure 4. BAF immunoprecipitates pG8-Luc DNA in L929 cells and depletion of BAF rescues viral intermediate gene expression at the transcriptional level.
(A) ChIP assays were performed to determine the binding of BAF to DNA. L929 cells stably expressing MCS plasmid (control) or 3xFlag-BAF were transfected with 150 ng pG8-luciferase plasmid for 24h. Following fixation and sonication, BAF-DNA complex was immunoprecipitated using anti-FLAG antibody and analyzed by quantitative PCR using primers flanking the G8 promoter (A) or Neomycin resistance gene (B) of pG8-Luc DNA or the beta-actin cellular gene (C). Data shown is a representative from three independent experiments, each performed in duplicate. Error bars represent standard deviation. Data from the Raw Ct values (shown in Supp Fig. 2) was plotted as fold enrichment relative to MCS. (D) L929 cells stably expressing shBAF or shControl were transfected with 10, 100, and 500ng of pG8-luciferase for 7h, then infected with 37°C with WT or ts2 at MOI=3 in the presence of AraC. Lysates were collected for RNA extraction and luciferase transcripts quantified by RT-qPCR. (*** indicates a p-value <0.05)
Kovacs et al have previously shown that B1 impacts intermediate gene expression at the level of transcription (Kovacs et al., 2001). To confirm and extend those results, we sought to determine whether B1's impact on transcript levels is BAF-dependent. For these studies, L929-shControl and shBAF cells were transfected with increasing amounts of pG8-Luc plasmid, then infected with WT or ts2 virus (MOI=3) and incubated at 37°C in the presence of AraC. At 4 hpi, lysates were collected and RNA extracted for RT-qPCR analysis using a luciferase-specific primer/probe set. In L929-shControl cells, the accumulation of transcripts in ts2 infected cells was reduced compared to WT infected cells (Fig. 4D). This difference between ts2 and WT transcription was observed at all plasmid concentrations, but was greatest at lower plasmid amounts. As higher amount of plasmids were used, the difference in transcripts between WT and ts2 infection shrinks. Next, upon examining the impact of BAF depletion on transcripts synthesized under the viral G8 promoter, we found that ts2-mediated transcription was greater in L929-shBAF cells than in L929-shControl cells. Most significantly, when using 10 ng of DNA there was a >10-fold increase in transcript accumulation in L929-shBAF cell as compared to ts2-infected L929-shControl cells. Together, these data are consistent with our model that BAF can be a repressor of viral gene expression at the transcriptional level if B1 is not present to phosphorylate it.
The impact of BAF on gene expression is promoter dependent
In light of the ability of BAF to bind to dsDNA in a sequence independent manner, we hypothesized that it may be capable of impacting transcription of genes other than the pG8-Luc reporter construct. To address this hypothesis, we began by asking 1) whether the transcriptional role of BAF during gene expression is promoter dependent and 2) whether the effect of B1 is also promoter dependent. For these studies, plasmids were employed that express firefly luciferase under a T7, CMV or a ‘minimal’ (minP) promoter, which contains primarily a TATA box. T7-Luc was selected to be used in conjunction with the recombinant vaccinia virus vTF7.3, which expresses a wild-type B1 protein as well as the T7 polymerase. Thus, transcription from the T7-Luc/vTF7.3 system will allow us to measure reporter gene expression from transcription in the cytoplasm, but in a manner independent of the vaccinia RNA polymerase and transcription factors. The CMV-Luc and minP-Luc constructs contain a strong and weak promoter, respectively, and are both activated by cellular factors and therefore result in nuclear transcription. As BAF is present in both the cytoplasm and nucleus, its impact on transcription in both locations is of interest.
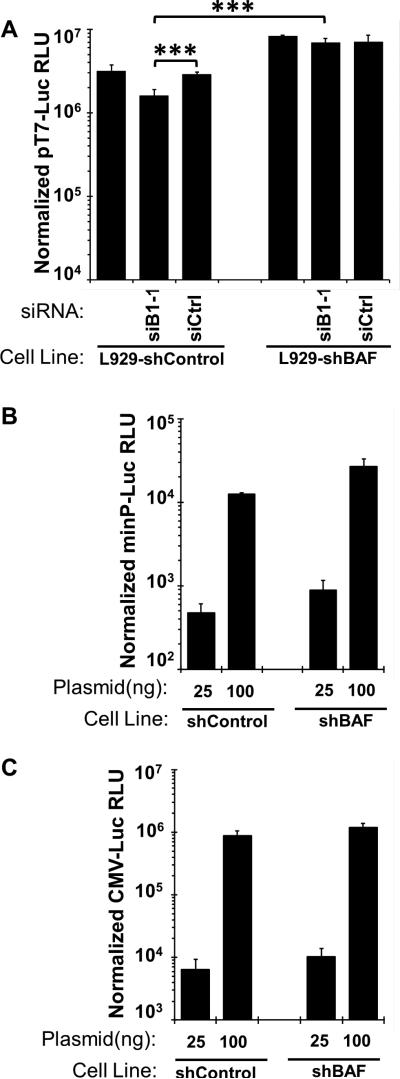
We began by examining the impact of the B1-BAF axis on T7-mediated gene expression. L929-shBAF and shControl cells were transfected as indicated with siB1 or siControl, then transfected with T7-luciferase, and later infected with vTF7.3 virus at MOI=3 in the presence of AraC for an additional 16 hours before harvest. As shown in Figure 5A (left), siB1 treatment of L929-shControl cells infected with vTF7.3 resulted in only a minor (2.5-fold) reduction in T7-Luc reporter gene expression, contrary to what was observed for pG8-Luc expression during WT infection (Fig. 3). When the same transfection / infection study using T7-Luc was performed in L929-shBAF cells, transcription activity rose modestly and was similar in all cases regardless of B1 siRNA treatment regimen. While these data suggest the B1-BAF axis contribute somewhat to the level of T7-driven reporter activity observed, the magnitude of their contribution is far less than what was observed when studying the pG8-Luc reporter.
Figure 5. The role of B1 kinase and BAF in viral intermediate gene expression is promoter dependent.
(A) L929 stably expressing shBAF or shControl were transfected with siRNA specific to B1 kinase (siB1-1) or siControl for 12h, and then transfected with 1 ng of T7-Luciferase for 7hr, then infected with 37°C with Wt-VTF7.3 at MOI=3 in the presence of AraC. Lysates were prepared at 12h after infection and assayed for luciferase activity, with RLU shown normalized to protein level. (B-C) L929 stably expressing shBAF or shControl were transfected with the indicated amounts of minP-Luc (B) or CMV-Luc (C). Lysates were prepared at 24h after transfection and assayed for luciferase activity, with RLU shown normalized to protein level. For all graphs, data were obtained from three independent experiments performed in triplicate wells. Data from a representative experiment is shown. Error bars represent standard deviation. (*** indicates a p-value <0.05)
Next, we wanted to test whether the depletion of BAF can enhance the expression of a reporter gene under a promoter transcribed in the nucleus. L929-shControl and shBAF were transfected with two different amount of CMV-Luc (immediate early promoter) or minP-Luc plasmid for 24h at 37°C; then lysates were collected and analyzed for luciferase activity. As shown in Figure 5B, at both DNA concentrations minP promoter-mediated luciferase expression was enhanced only 2-fold in L929-shBAF cells, and the CMV promoter-mediated luciferase expression (Fig. 5C) was similar regardless of BAF level of expression. Based on the contrast between these data with the T7, CMV and minP promoters and the earlier data using the G8 promoter, it is evident that the B1-BAF axis affects transcription in a gene specific manner. In our view, this specificity may be due to differences in transcription factors and polymerases in play and / or subcellular localization of the DNA in question and is discussed in more detail below.
DISCUSSION
Vaccinia virus possesses multiple sophisticated mechanisms to manipulate cellular signaling pathways and subvert host defenses (Bowie and Haga, 2005, Moss and Shisler, 2001, Perdiguero and Esteban, 2009). Studies on vaccinia-host interactions continue to provide novel insights into the mechanisms employed by both virus and host alike to achieve their respective goals during the course of an infection. For example, the vaccinia B1 Ser/Thr kinase phosphorylates the cellular factor BAF, which leads to the potent inhibition of BAF's ability to bind and crossbridge viral DNA (Ibrahim et al., 2011, Wiebe and Traktman, 2007). VRK1, a cellular homolog of B1, targets BAF on the same residues as B1 (Nichols et al., 2006). It is now known that the VRK1-BAF axis plays an important role in mitotic regulation (reviewed in Valbuena et al., 2011), while the B1-BAF axis is needed for viral DNA replication (Ibrahim et al., 2011, Wiebe and Traktman, 2007). Importantly, these studies revealed a novel function for BAF as an antiviral effector capable of targeting vaccinia DNA in the cytoplasm. Currently, much remains to be learned about the contributions of the B1 kinase to the viral lifecycle and specifically whether its phosphorylation of BAF is needed to promote more than just viral DNA replication.
The requirement of B1 for viral DNA replication, which itself is needed for intermediate and late gene expression, is an obstacle to the study of B1's contribution to processes beyond DNA replication. However, as reported by Kovacs et al. (2001), by using plasmid reporters containing viral promoters to avoid the need for genome replication, B1 was indeed found to be needed beyond DNA replication (Kovacs et al., 2001). However, the mechanism underlying B1's contribution to gene expression was not fully defined. Therefore, in light of the importance of the B1-BAF axis for DNA replication, herein we examined whether B1 was acting through this same pathway to facilitate intermediate gene expression. As expected, the B1-deficient ts2 virus was only capable of activating the luciferase reporter gene under the G8R promoter to <1% to that of the WT virus in L929 cells (Fig. 1). The ts2 virus also exhibited a marked reduction of expression from three other promoters in this study, demonstrating that B1's role in transcription likely extends to intermediate gene expression in general. A reduction in ts2-driven reporter activation as compared to WT virus was also observed in other cell types including CV1, U2OS, and BSC40 cells (Supp Fig 1A). The fact that L929 cells display the greatest sensitivity in this assay was also found by Kovacs et al, and is the reason why they were employed for the remainder of the study. However, it is interesting to note that all of these cell lines express similar amounts of BAF protein (Supp Fig 1B) indicating that either the ts2-B1 protein or BAF exhibits cell type specific activity. Efforts to tease out the mechanisms underlying these cell type specific differences is currently underway.
To directly test BAF's involvement in the transcriptional downregulation seen during ts2 infection we constructed L929 cells stably depleted of BAF and performed luciferase reporter assays in those cells (Fig. 2). Strikingly, cells with decreased levels of BAF exhibited a 40-fold increase in ts2-mediated gene expression as compared to control L929 cells (Fig. 2D). While it is interesting to note that the other viruses tested in this study (WT, ts42, and ts24) also exhibited modestly increased reporter activation, the fact that ts2 was more dramatically affected than other viruses demonstrates that the B1-BAF axis impacts expression of the reporter gene. This conclusion is also supported by our complementary data that siRNA-mediated depletion of B1 from WT virus leads to a decrease in reporter activation which can be rescued by the depletion of BAF (Fig. 3B). Together, these data strongly suggest that in the absence of the B1 kinase, BAF is capable of impairing viral gene expression.
Finally, we have made progress towards determining the mechanism underlying BAF's impact on viral gene expression. First, we demonstrated that multiple regions of the plasmid co-immunoprecipitates with BAF, supporting a model in which BAF binds foreign DNA in a sequence independent manner, including interaction with the promoter region of the reporter gene. Second, we directly tested whether BAF was affecting transcription of the reporter gene by measuring luciferase transcript accumulation during WT and ts2 infection. Using RT-qPCR, we observed that decreased transcript levels observed during ts2 infection could be rescued by BAF depletion, indicating that BAF functions at the level of gene transcription. The specificity of BAF's action against transcription was also examined using promoter-reporter constructs able to be expressed independently of the vaccinia RNA polymerase. Interestingly, both BAF and B1 depletions had little impact on a reporter under a T7 promoter during infection with vTF7.3, which expresses the T7 polymerase. Similarly, BAF depletion had little if any impact on either minP or CMV driven reporter gene expression, both of which are promoters responsive to RNA polymerase II and cellular transcription factors. Together, these results provide the first evidence that BAF can function as a gene-dependent inhibitor of transcription, which can itself be inactivated by the B1 kinase.
In regard to the mechanism of BAF's inhibition of viral gene expression, previous studies have shown that BAF's ability to block viral DNA replication requires its DNA binding and dimerization activity (Ibrahim et al., 2011). These properties allow BAF to bind and crossbridge/aggregate DNA (Bradley et al., 2005, Skoko et al., 2009, Zheng et al., 2000), and also likely allow BAF to impair transcription in the absence of B1. Precedent for this model can be found in prokaryotes, where DNA crossbridging by dimeric proteins such as H-NS is known to be a potent transcriptional silencer of foreign DNA (Lucchini, S. Rowley, G. et al., 2006). Interestingly, the potency of silencing due to H-NS binding is a function of the DNA binding affinity or other attributes of the transcription factors attempting to gain access to the promoter bound by H-NS (Navarre et al., 2006, Navarre et al., 2007), consistent with a model in which the competition of repressor and activator proteins determine the level of gene activation.
We posit that if BAF functions via a similar mechanism to H-NS and is attempting to compete for DNA binding with transcriptional activators it may, in part, explain the promoter-specific repression we are observing. Following this logic, in the case of the vaccinia promoters, the presence of BAF may be sufficient to hinder recruitment of some of the activators found to contribute to vaccinia intermediate transcription. For example, the action of viral proteins such as A8 and A23 (Sanz and Moss, 1999) as well as cellular factors including G3BP/p137 (Katsafanas and Moss, 2004), TBP (Knutson et al., 2006), and Yin-Yang 1 (YY1) (Broyles et al., 1999, Knutson et al., 2009) have been linked to intermediate transcription and their access to the promoter may be affected by BAF. In contrast, the fact that gene expression from the T7, minP, and CMV promoters is similar in the presence or absence of BAF suggests that binding of the transcription factors and / or the polymerases to those promoters may not be greatly impeded by BAF. Additionally, as the minP and CMV promoters are transcribed in the nucleus, BAF's activity may be regulated differently in that environment (such as by the nuclear kinase VRK1), thus reducing BAF's ability to repress foreign DNA. This possibility might also explain why we have found that the beta-actin gene could be co-immunoprecipitated with BAF (Fig 4C), while beta-actin transcript levels remain unchanged in control or BAF-depleted cells (data not shown). Future work to determine the relative contribution of each of these possible regulatory mechanisms will be of great interest.
Finally, we would be remiss if we did not point out that in each of our experiments using L929-shBAF cells, the depletion of BAF never led to a complete recovery of the transcriptional activity lost due to impairment of the B1 kinase. We would suggest two possible explanations for these observations. First, while BAF depletion is significant, it is still incomplete. The remaining BAF may be capable of inhibiting intermediate gene expression. Secondly, we posit that B1 may have substrates other than BAF that are also needed for optimal transcription, which are mediating this difference in gene expression. In this regard one possible candidate is the viral H5 protein, which is known to be phosphorylated by B1 and has been linked to transcriptional regulation in other studies (Black et al., 1998, Brown et al., 2000, D'Costa et al., 2008, D'Costa et al., 2010, Kovacs and Moss, 1996). Additional studies will be needed to distinguish between these two possibilities.
In summary, these studies reveal that the B1-BAF signaling axis impacts both DNA replication and intermediate gene expression. Our observations demonstrate for the first time that BAF can act as a transcriptional repressor and provide the first evidence that this inhibitory activity can be regulated by phosphorylation of BAF. Although this study focuses on vaccinia transcription in the cytoplasm, the fact that a large fraction of BAF exists in the nucleus and is targeted there by VRK1, suggest that post-translational regulation of BAF probably has implications for cellular gene expression as well.
Supplementary Material
Research Highlights.
- The role of the B1-BAF axis in the vaccinia lifecycle is poorly understood
- Vaccinia B1 kinase is needed for viral intermediate transcription
- Depletion of BAF strongly rescues the transcriptional defect caused by the loss of B1
- Depletion of BAF has only a modest effect on non-vaccinia promoters
- BAF can act as a transcriptional repressor, but is inactivated by the B1 kinase
ACKNOWLEDGMENTS
This research was supported through NIH grants to M.W. (1K22 AI080941, 1R56 AI099062) and the Nebraska Center for Virology (1P20 RR15635). N.I. and A.J. were partially supported by a Ruth L. Kirschstein NRSA (1T32 AIO60547). We also thank Drs. Paula Traktman and Kathy Boyle at the Medical College of Wisconsin for editorial suggestions and helpful scientific discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahn BY, Gershon PD, Jones EV, Moss B. Identification of rpo30, a vaccinia virus RNA polymerase gene with structural similarity to a eucaryotic transcription elongation factor. Molecular and Cellular Biology. 1990;10:5433–5441. doi: 10.1128/mcb.10.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amegadzie BY, Ahn BY, Moss B. Identification, sequence, and expression of the gene encoding a Mr 35,000 subunit of the vaccinia virus DNA-dependent RNA polymerase. Journal of Biological Chemistry. 1991;266:13712–13718. [PubMed] [Google Scholar]
- Baldick CJ, Keck JG, Moss B. Mutational analysis of the core, spacer, and initiator regions of vaccinia virus intermediate-class promoters. Journal of Virology. 1992;66:4710–4719. doi: 10.1128/jvi.66.8.4710-4719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banham AH, Smith GL. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology. 1992;191:803–812. doi: 10.1016/0042-6822(92)90256-o. [DOI] [PubMed] [Google Scholar]
- Black EP, Moussatche N, Condit RC. Characterization of the interactions among vaccinia virus transcription factors G2R, A18R, and H5R. Virology. 1998;245:313–322. doi: 10.1006/viro.1998.9166. [DOI] [PubMed] [Google Scholar]
- Bowie AG, Haga IR. The role of Toll-like receptors in the host response to viruses. Mol.Immunol. 2005;42:859–867. doi: 10.1016/j.molimm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Boyle KA, Traktman P. Members of a novel family of mammalian protein kinases complement the DNA-negative phenotype of a vaccinia virus ts mutant defective in the B1 kinase. J.Virol. 2004;78:1992–2005. doi: 10.1128/JVI.78.4.1992-2005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley CM, Ronning DR, Ghirlando R, Craigie R, Dyda F. Structural basis for DNA bridging by barrier-to-autointegration factor. Nat.Struct.Mol.Biol. 2005;12:935–936. doi: 10.1038/nsmb989. [DOI] [PubMed] [Google Scholar]
- Brown NG, Nick Morrice D, Beaud G, Hardie G, Leader DP. Identification of sites phosphorylated by the vaccinia virus B1R kinase in viral protein H5R. BMC Biochem. 2000;1:2. doi: 10.1186/1471-2091-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles SS, Liu X, Zhu M, Kremer M. Transcription Factor YY1 Is a Vaccinia Virus Late Promoter Activator. Journal of Biological Chemistry. 1999;274:35662–35667. doi: 10.1074/jbc.274.50.35662. [DOI] [PubMed] [Google Scholar]
- Cai M, Huang Y, Zheng R, Wei SQ, Ghirlando R, Lee MS, Craigie R, Gronenborn AM, Clore GM. Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nat.Struct.Biol. 1998;5:903–909. doi: 10.1038/2345. [DOI] [PubMed] [Google Scholar]
- Challberg MD, Englund PT. Purification and properties of the deoxyribonucleic acid polymerase induced by vaccinia virus. 1979;254:7812–7819. [PubMed] [Google Scholar]
- Condit RC, Motyczka A, Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. 1983;128:429–243. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- Condit RC, Easterly R, Pacha RF, Fathi Z, Meis RJ. A Vaccinia virus isatin-β-thiosemicarbazone resistance mutation maps in the viral gene encoding the 132-kDa subunit of RNA polymerase. Virology. 1991;185:857–861. doi: 10.1016/0042-6822(91)90559-t. [DOI] [PubMed] [Google Scholar]
- Cox JL, Mallanna SK, Ormsbee BD, Desler M, Wiebe MS, Rizzino A. Banf1 is required to maintain the self-renewal of both mouse and human embryonic stem cells. Journal of Cell Science. 2011;124:2654–2665. doi: 10.1242/jcs.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Costa SM, Bainbridge TW, Condit RC. Purification and properties of the vaccinia virus mRNA processing factor. J.Biol.Chem. 2008;283:5267–5275. doi: 10.1074/jbc.M709258200. [DOI] [PubMed] [Google Scholar]
- D'Costa SM, Bainbridge TW, Kato SE, Prins C, Kelley K, Condit RC. Vaccinia H5 is a multifunctional protein involved in viral DNA replication, postreplicative gene transcription, and virion morphogenesis. Virology. 2010;401:49–60. doi: 10.1016/j.virol.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Lewis W, Berglund P, Koonin EV, Moss B. Poxvirus DNA primase. Proc.Natl.Acad.Sci.U.S.A. 2007;104:18724–18729. doi: 10.1073/pnas.0709276104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Paran N, Moss B. Products and substrate/template usage of vaccinia virus DNA primase. Virology. 2009;383:136–141. doi: 10.1016/j.virol.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Gajewski A, Korbei B, Gerlich D, Daigle N, Haraguchi T, Furukawa K, Ellenberg J, Foisner R. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J.Cell.Sci. 2004;117:6117–6128. doi: 10.1242/jcs.01529. [DOI] [PubMed] [Google Scholar]
- Earl PL, Jones EV, Moss B. Homology between DNA polymerases of poxviruses, herpesviruses, and adenoviruses: nucleotide sequence of the vaccinia virus DNA polymerase gene. Proceedings of the National Academy of Sciences. 1986;83:3659–3663. doi: 10.1073/pnas.83.11.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E.a.T.P. Characterization of vaccinia virus DNA replication mutants with lesions in the D5 gene. 1992;102:S72–S82. doi: 10.1007/BF02451789. [DOI] [PubMed] [Google Scholar]
- Evans E, Klemperer N, Ghosh R, Traktman P. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleoside triphosphatase. Journal of Virology. 1995;69:5353–5361. doi: 10.1128/jvi.69.9.5353-5361.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Traktman P. Molecular genetic analysis of a vaccinia virus gene with an essential role in DNA replication. Journal of Virology. 1987;61:3152–3162. doi: 10.1128/jvi.61.10.3152-3162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc.Natl.Acad.Sci.U.S.A. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J.Cell.Sci. 1999;112(Pt 15):2485–2492. doi: 10.1242/jcs.112.15.2485. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Sugiyama S, Osouda S, Goto H, Inagaki M, Horigome T, Omata S, McConnell M, Fisher PA, Nishida Y. Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. Journal of Cell Science. 2003;116:3811–3823. doi: 10.1242/jcs.00682. [DOI] [PubMed] [Google Scholar]
- Haraguchi T, Koujin T, Segura-Totten M, Lee KK, Matsuoka Y, Yoneda Y, Wilson KL, Hiraoka Y. BAF is required for emerin assembly into the reforming nuclear envelope. J.Cell.Sci. 2001;114:4575–4585. doi: 10.1242/jcs.114.24.4575. [DOI] [PubMed] [Google Scholar]
- Harris D, Engelman A. Both the structure and DNA binding function of the barrier-to-autointegration factor contribute to reconstitution of HIV type 1 integration in vitro. J.Biol.Chem. 2000;275:39671–39677. doi: 10.1074/jbc.M002626200. [DOI] [PubMed] [Google Scholar]
- Ibrahim N, Wicklund A, Wiebe MS. Molecular characterization of the host defense activity of the barrier to autointegration factor against vaccinia virus. J.Virol. 2011;85:11588–11600. doi: 10.1128/JVI.00641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsafanas GC, Moss B. Vaccinia Virus Intermediate Stage Transcription Is Complemented by Ras-GTPase-activating Protein SH3 Domain-binding Protein (G3BP) and Cytoplasmic Activation/Proliferation-associated Protein (p137) Individually or as a Heterodimer. Journal of Biological Chemistry. 2004;279:52210–52217. doi: 10.1074/jbc.M411033200. [DOI] [PubMed] [Google Scholar]
- Keck JG, Baldick CJ, Jr., Moss B. Role of DNA replication in vaccinia virus gene expression: A naked template is required for transcription of three late trans-activator genes. Cell. 1990;61:801–809. doi: 10.1016/0092-8674(90)90190-p. [DOI] [PubMed] [Google Scholar]
- Klemperer N, McDonald W, Boyle K, Unger B, Traktman P. The A20R protein is a stoichiometric component of the processive form of vaccinia virus DNA polymerase. J.Virol. 2001;75:12298–12307. doi: 10.1128/JVI.75.24.12298-12307.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson BA, Liu X, Oh J, Broyles SS. Vaccinia virus intermediate and late promoter elements are targeted by the TATA-binding protein. J.Virol. 2006;80:6784–6793. doi: 10.1128/JVI.02705-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson BA, Oh J, Broyles SS. Downregulation of vaccinia virus intermediate and late promoters by host transcription factor YY1. Journal of General Virology. 2009;90:1592–1599. doi: 10.1099/vir.0.006924-0. [DOI] [PubMed] [Google Scholar]
- Kovacs GR, Moss B. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning, and overexpression. J.Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs GR, Vasilakis N, Moss B. Regulation of viral intermediate gene expression by the vaccinia virus B1 protein kinase. J.Virol. 2001;75:4048–4055. doi: 10.1128/JVI.75.9.4048-4055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Haraguchi T, Lee RS, Koujin T, Hiraoka Y, Wilson KL. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J.Cell.Sci. 2001;114:4567–4573. doi: 10.1242/jcs.114.24.4567. [DOI] [PubMed] [Google Scholar]
- Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proceedings of the National Academy of Sciences. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Chen W, Broyles SS. The vaccinia virus B1R gene product is a serine/threonine protein kinase. J.Virol. 1992;66:2717–2723. doi: 10.1128/jvi.66.5.2717-2723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S, Rowley G, et al. H-NS Mediates the Silencing of Laterally Acquired Genes in Bacteria. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit A, Neufeld E, Feinstein N, Wilson KL, Podbilewicz B, Gruenbaum Y. Barrier to autointegration factor blocks premature cell fusion and maintains adult muscle integrity in C. elegans. J.Cell Biol. 2007;178:661–673. doi: 10.1083/jcb.200704049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit A, Segura-Totten M, Gruenbaum Y, Wilson KL. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc.Natl.Acad.Sci.U.S.A. 2005;102:3290–3295. doi: 10.1073/pnas.0408364102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WF, Traktman P. Vaccinia virus DNA polymerase. In vitro analysis of parameters affecting processivity. Journal of Biological Chemistry. 1994;269:31190–31197. [PubMed] [Google Scholar]
- McDonald WF, Klemperer N, Traktman P. Characterization of a Processive Form of the Vaccinia Virus DNA Polymerase. Virology. 1997;234:168–175. doi: 10.1006/viro.1997.8639. [DOI] [PubMed] [Google Scholar]
- Molitor TP, Traktman P. Molecular genetic analysis of VRK1 in mammary epithelial cells: depletion slows proliferation in vitro and tumor growth and metastasis in vivo. Oncogenesis. 2013;2:e48. doi: 10.1038/oncsis.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R, Lee KK, Wilson KL. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J.Biol.Chem. 2005;280:42252–42262. doi: 10.1074/jbc.M509917200. [DOI] [PubMed] [Google Scholar]
- Montes de Oca R, Shoemaker CJ, Gucek M, Cole RN, Wilson KL. Barrier-to-autointegration factor proteome reveals chromatin-regulatory partners. PLoS One. 2009;4:e7050. doi: 10.1371/journal.pone.0007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B, Ahn BY, Amegadzie B, Gershon PD, Keck JG. Cytoplasmic transcription system encoded by vaccinia virus. Journal of Biological Chemistry. 1991;266:1355–1358. [PubMed] [Google Scholar]
- Moss B, Shisler JL. Immunology 101 at poxvirus U: immune evasion genes. Semin.Immunol. 2001;13:59–66. doi: 10.1006/smim.2000.0296. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng H, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic Instability and Aging-like Phenotype in the Absence of Mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Nezu J, Oku A, Jones MH, Shimane M. Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics. 1997;45:327–331. doi: 10.1006/geno.1997.4938. [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Traktman P. Characterization of three paralogous members of the Mammalian vaccinia related kinase family. J.Biol.Chem. 2004;279:7934–7946. doi: 10.1074/jbc.M310813200. [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Wiebe MS, Traktman P. The vaccinia-related kinases phosphorylate the N’ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol.Biol.Cell. 2006;17:2451–2464. doi: 10.1091/mbc.E05-12-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proceedings of the National Academy of Sciences. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero B, Esteban M. The interferon system and vaccinia virus evasion mechanisms. J.Interferon Cytokine Res. 2009;29:581–598. doi: 10.1089/jir.2009.0073. [DOI] [PubMed] [Google Scholar]
- Puente XS, Quesada V, Osorio FG, Cabanillas R, Cadinanos J, Fraile JM, Ordonez GR, Puente DA, Gutierrez-Fernandez A, Fanjul-Fernandez M, Levy N, Freije JM, Lopez-Otin C. Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am.J.Hum.Genet. 2011;88:650–656. doi: 10.1016/j.ajhg.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel RE, Anderson MK, Evans E, Traktman P. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J.Virol. 1990;64:574–583. doi: 10.1128/jvi.64.2.574-583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel RE, Traktman P. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J.Virol. 1992;66:4413–4426. doi: 10.1128/jvi.66.7.4413-4426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P, Moss B. Identification of a transcription factor, encoded by two vaccinia virus early genes, that regulates the intermediate stage of viral gene expression. Proceedings of the National Academy of Sciences. 1999;96:2692–2697. doi: 10.1073/pnas.96.6.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Totten M, Kowalski AK, Craigie R, Wilson KL. Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J.Cell Biol. 2002;158:475–485. doi: 10.1083/jcb.200202019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Totten M, Wilson KL. BAF: roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol. 2004;14:261–266. doi: 10.1016/j.tcb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Shumaker DK, Lee KK, Tanhehco YC, Craigie R, Wilson KL. LAP2 binds to BAF.DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 2001;20:1754–1764. doi: 10.1093/emboj/20.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoko D, Li M, Huang Y, Mizuuchi M, Cai M, Bradley CM, Pease PJ, Xiao B, Marko JF, Craigie R, Mizuuchi K. Barrier-to-autointegration factor (BAF) condenses DNA by looping. Proc.Natl.Acad.Sci.U.S.A. 2009;106:16610–16615. doi: 10.1073/pnas.0909077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar P, Condit RC. Selection for temperature-sensitive mutations in specific vaccinia virus genes: isolation and characterization of a virus mutant which encodes a phosphonoacetic acid-resistant, temperature-sensitive DNA polymerase. 1983;128:444–457. doi: 10.1016/0042-6822(83)90269-6. [DOI] [PubMed] [Google Scholar]
- Taddie JA, Traktman P. Genetic characterization of the vaccinia virus DNA polymerase: cytosine arabinoside resistance requires a variable lesion conferring phosphonoacetate resistance in conjunction with an invariant mutation localized to the 3'-5' exonuclease domain. J.Virol. 1993;67:4323–4336. doi: 10.1128/jvi.67.7.4323-4336.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddie JA, Traktman P. Genetic characterization of the vaccinia virus DNA polymerase: identification of point mutations conferring altered drug sensitivities and reduced fidelity. J.Virol. 1991;65:869–879. doi: 10.1128/jvi.65.2.869-879.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktman P, Anderson MK, Rempel RE. Vaccinia virus encodes an essential gene with strong homology to protein kinases. J.Biol.Chem. 1989;264:21458–21461. [PubMed] [Google Scholar]
- Umland TC, Wei SQ, Craigie R, Davies DR. Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry. 2000;39:9130–9138. doi: 10.1021/bi000572w. [DOI] [PubMed] [Google Scholar]
- Valbuena A, Sanz-García M, López-Sánchez I, Vega FM, Lazo PA. Roles of VRK1 as a new player in the control of biological processes required for cell division. Cell.Signal. 2011;23:1267–1272. doi: 10.1016/j.cellsig.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Vos JC, Stunnenberg HG. Derepression of a novel class of vaccinia virus genes upon DNA replication. 1988;7:3487–3492. doi: 10.1002/j.1460-2075.1988.tb03224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebe MS, Traktman P. Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell.Host Microbe. 2007;1:187–197. doi: 10.1016/j.chom.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Martens CA, Bruno DP, Porcella SF, Moss B. Pervasive initiation and 3'-end formation of poxvirus postreplicative RNAs. J.Biol.Chem. 2012;287:31050–31060. doi: 10.1074/jbc.M112.390054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc.Natl.Acad.Sci.U.S.A. 2000;97:8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.