SUMMARY
Stress granules and P-bodies are conserved cytoplasmic aggregates of non-translating mRNPs implicated in the regulation of mRNA translation and decay, and are related to RNP granules in embryos, neurons and pathological inclusions in some degenerative diseases. In a genetic screen using bakers yeast, we identified 125 genes that affect the dynamics of P-bodies and/or stress granules. Analyses of additional mutants, including Cdc48 alleles, provide evidence that stress granules can be targeted to the vacuole by autophagy, in a process termed granulophagy. Moreover, stress granule clearance in mammalian cells is reduced by inhibition of autophagy or by depletion or pathogenic mutations in VCP, the human ortholog of Cdc48. Since mutations in VCP predispose humans to ALS, FTLD, IBM and MSP this work suggests that autophagic clearance of stress-granule related and pathogenic RNP granules that arise in degenerative diseases may be important in reducing their pathology.
INTRODUCTION
The control of mRNA translation, localization and degradation plays an important role in the proper modulation of gene expression. Such post-transcriptional control is emphasized in many biological conditions including stress responses, embryogenesis and in synaptic plasticity (Spriggs et al, 2010; Medioni et al, 2012; Doyle and Kiebler, 2011; Gkogkas et al, 2010). Key issues in understanding post-transcriptional control mechanisms include understanding the messenger ribonucleoprotein complexes (mRNPs) that form and how the cell controls the translation, localization and degradation of such mRNPs.
In the last decade it has become clear that non-translating mRNPs in eukaryotic cells often assemble into conserved and dynamic cytoplasmic mRNP granules known as P-bodies and stress granules (Erickson and Lykke-Andersen, 2011; Anderson and Kedersha, 2009; Buchan and Parker, 2009). Stress granules are typically observed when translation initiation is limiting and consist of mRNAs associated with some translation initiation factors and RNA binding proteins, and thus are thought to represent a pool of mRNPs stalled in the process of translation initiation (Anderson and Kedersha, 2009; Buchan and Parker, 2009). P-bodies consist of mRNAs associated with translation repressors and the mRNA decay machinery, and while typically present in cells at modest levels, they increase when the pool of non-translating mRNPs is larger (Parker and Sheth, 2007).
P-bodies and stress granules are of interest since they have been connected to a number of important cellular processes including normal mRNA degradation (Sheth and Parker, 2003), nonsense mediated decay (Sheth and Parker, 2006; Franks et al., 2010), miRNA function (Bhattacharyya et al, 2006; Leung et al, 2006), viral replication (Beckham and Parker 2008), and cell-signaling (Arimoto et al, 2008; Takahara and Maeda, 2012). In addition, P-bodies and stress granules are related to mRNP granules found in neurons, which are involved in mRNA transport and translational control at synapses, and to mRNP granules in embryogenesis where maternal mRNAs are stored (Anderson and Kedersha, 2009; Buchan and Parker, 2009).
More recently, stress granules have emerged as being involved in some degenerative diseases. For example, conditions such as amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD), fragile X syndrome, spinocerebellar ataxia-2, inclusion body myopathy (IBM) and multisystem proteinopathy (MSP) can result from mutations in known stress granule proteins which often increase their tendency to aggregate (Ito and Suzuki, 2011; Didiot et al, 2009; Nonhoff et al, 2007; Kim et al., 2013). Additionally, a hallmark of ALS, FTLD and some other degenerative diseases is the accumulation of cytoplasmic aggregates that contain several stress granule factors and RNA (Dewey et al, 2012; Ito and Suzuki, 2011; Ginsberg et al, 1998). This leads to the hypothesis that inappropriate formation or persistence of stress granules, or some related mRNP aggregate, might be related to the pathogenesis in these diseases. Interestingly, mutations in valosin-containing protein (VCP) cause ALS, FTLD and MSP which are all characterized by pathological accumulation of TDP-43 and in some cases other stress granule proteins in cytoplasmic aggregates (Johnson et al, 2010; Salajegheh et al. 2009; Kim et al., 2013), raising the possibility that VCP is involved in stress granule dynamics.
The formation of stress granules and P-bodies is based on two principles. First, they require non-translating RNA for their assembly. Second, individual mRNPs are brought together by dimerization or aggregation domains present on mRNP binding proteins. For example, the assembly of P-bodies in yeast is driven in part by a dimerization domain on the Edc3 protein and a “prion domain” present on the Lsm4 protein (Decker et al., 2007; Reijns et al., 2008). Similarly, stress granule formation in mammalian cells is promoted by a prion domain on the TIA1 protein (Gilks et al., 2004), and mRNA binding proteins frequently contain such aggregation prone prion-like or low-complexity domains (Decker et al., 2007; Kato et al., 2012; Kim et al., 2013). The prevalence of such aggregation domains in RNA binding proteins as part of their normal role in forming stress granules and P-bodies suggests they provide a significant target for mutations that create pathologically aggregated proteins.
Given the biological roles of stress granules and P-bodies, as well as their connection to degenerative diseases, an important goal is to understand the mechanisms the control stress granule and P-body assembly, disassembly and clearance from the cell. To identify proteins involved in modulating these mRNP granules, we performed a genetic screen in S.cerevisiae to identify mutations that altered stress granules or P-bodies. We examined yeast mutants under normal growth conditions rather than stress, as this allowed us to better distinguish mutations that give constitutive stress granules or elevated P-bodies, perhaps due to defects in disassembly or clearance mechanisms. We identified over 100 highly interconnected genes that affect stress granules and/or P-bodies. Additional analyses provided evidence that clearance of these mRNP granules is affected by both autophagy and Cdc48/VCP function, thus identifying a new mechanism for clearing these assemblies from the cell, which may also play a role in clearing toxic RNPs from mammalian cells to limit pathologies.
RESULTS
Identification of proteins affecting P-body and Stress Granule assembly
A microscopy-based screen of 4249 non-essential gene deletions (Table S2) in S. cerevisiae was conducted to determine genes whose absence alters P-body or stress granule assembly. For this screen the stress granule component, Pab1-GFP, and the P-body component, Edc3-mCh, were introduced into each strain on a centromere plasmid. Each mutant strain was examined microscopically under good growth conditions where P-bodies are small, allowing us to detect increases and decreases in P-bodies, and stress granules are almost completely absent, allowing us to identify mutants with constitutive stress granules.
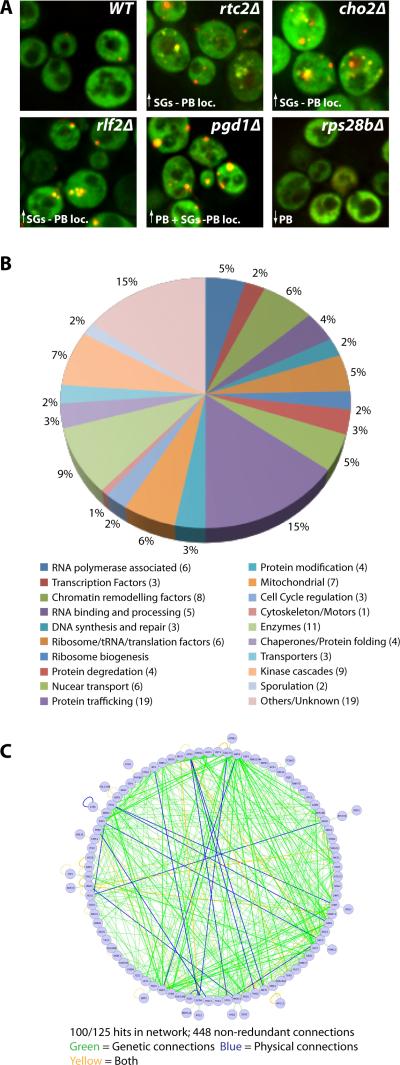
We identified 125 mutants that gave reproducible stress granules and/or P-body phenotypes (see below). For each mutant, the phenotype was verified by repeat experiments and the gene deletion verified by PCR and sequencing analysis. Expression levels of Pab1-GFP in a subset of the screen deletion hits showed only modest variation, thus the observed phenotypes are generally unlikely to be due to altered granule protein expression levels (Figure S1). Phenotypes varied as to the effect on stress granules and P-bodies, and the severity of the phenotype (summarized in Table S1 and Table S3; examples in Figure 1A) and included increased stress granules associated with P-bodies (81 genes), increased stress granules distinct from P-bodies (20 genes), increased P-bodies (28 genes), decreased P-bodies (23 genes), and intra-vacuolar accumulation of Pab1 and Edc3 (1 gene). These subcategories total to more than 125 since some mutants have more than one phenotype. In most cases, the observed Pab1-GFP foci contain another stress granule component, Pub1-mCh, indicating they represent stress granules (Figure S2).
Figure 1. Overview of screen for genes affecting stress granule and P-body assembly.
A.) Typical screen phenotypes (log. growth). Pab1-GFP and Edc3-mCh serve as stress granule and P-body markers respectively. Examples of P-body localized stress granules (rtc2Δ, cho2Δ, rlf2Δ), increased P-bodies (pgd1Δ) and decreased P-bodies (rp28bΔ) are shown. B.) 125 screen hits categorized by GO-terms. C.) Network analysis of known physical and genetic connections of screen hits. See also Figure S3 and Table S1.
The 125 genes identified in our screen exhibited diverse functions according to GO-term analysis (Figure 1B), but network analysis reveals a remarkably interconnected genetic and physical network formed by 100/125 of the screen hits, involving 448 non-redundant connections (Figure 1C; P-value of 5.34 ×10−6, see experimental procedures). These interconnections suggest these factors form a previously unknown network regulating mRNP granule formation.
Several mutants mapped to specific complexes or pathways in yeast cells, typically with similar phenotypes within each grouping (Figure S3A, Table S1). For example, multiple defects in the THO and TREX-2 complexes (hpr1Δ, mft1Δ, sac3Δ, thp1Δ, thp2Δ and tho2Δ), which function in coupling transcription to mRNA export, showed constitutive stress granules, and in a smaller subset of cells nuclear accumulation of Pab1. Similarly, mutations in three subunits of the prefoldin complex (gim4Δ, pfd1Δ, gim5Δ) gave decreased P-bodies (Figure S3A). In a final example, we observed that deletions of five subunits of the vacuolar ATPase complex (vma11Δ, vma16Δ, vma2Δ, vma4Δ and vma3Δ), and a chaperone require for its assembly (vma21Δ) gave increased stress granules (Figure S3A). The vacuolar ATPase might affect stress granule dynamics by direct contacts to mRNAs since the Vma1 subunit has recently been identified as an mRNP binding protein (Mitchell et al., 2013), and we observe that Vma2 co-localizes with P-bodies and stress granules under stress conditions (Figure S3B). Alternatively, the vacuolar ATPase may affect stress granules by altering membrane dynamics, which can affect autophagy (Yamamoto et al 1998; Mijaljica et al, 2011; see below).
Yeast stress granules and P-bodies can be targeted for autophagy
An important result was that several mutations that disrupt autophagy (atg8Δ, atg11Δ, atg18Δ, mon1Δ, and meh1Δ) gave an increase in constitutive stress granules, which were typically associated with P-bodies (Figure 2A, Table S1). In addition, we observed the accumulation of Pab1-GFP and Edc3-mCh in an intra-vacuolar compartment (referred to as the IVC) in atg15Δ mutants (Figure 2A). Moreover, this IVC seen in the atg15Δ also contains other components of stress granules including Pbp1, Ded1, and Pub1 (Figure S4). This is consistent with the fact that Atg15 is a vacuolar lipase that helps break open vesicles targeted to the vacuole from autophagic trafficking pathways (Teter et al, 2001). These observations argue that stress granules and P-bodies can be targeted to the vacuole by autophagy and therefore in strains defective in autophagy, stress granules and P-bodies accumulate in the cytosol, or within the vacuole when late steps in autophagic vesicle breakdown are defective as in the atg15Δ strain.
Figure 2. Several autophagy and Cdc48-complex factors exhibit stress granule phenotypes.
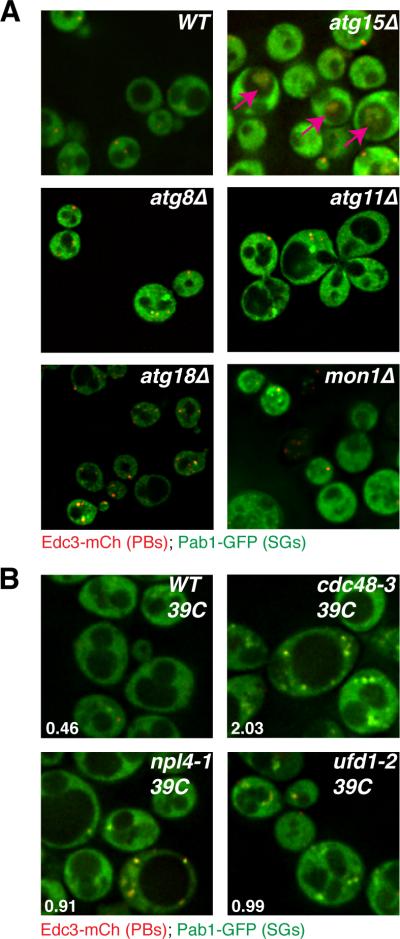
A.) Logarithmically growing strains lacking autophagy factors exhibit accumulation and/or mis-localization of stress granule and P-body proteins. B.) W303 strain background ts alleles of Cdc48, Ufd1 and Npl4 exhibit increased stress granules relative to WT at the non-permissive temperature. Numbers indicate the average number of stress granule foci/cell in each strain, based on 3 independent experiments. See also Figure S4 and Table S5.
Consistent with this interpretation, in wild type cells we observed a small degree of co-localization between both Pub1-mCh or Edc3-mCh foci with Atg11-GFP or Atg19-GFP (Figure S5-S6, Table S5). Atg11 and Atg19 both facilitate loading of cargo proteins into autophagic vesicles at the phagophore assembly site (Yorimitsu and Klionsky, 2005), whilst Atg19 also becomes incorporated in autophagosomes destined for the vacuole. Given that impairment of autophagy could lead to greater accumulation of stress granule/autophagy intermediates, we further examined co-localization of Pub1-mCh and Atg19-GFP in a mon1Δ strain (which should impair autophagy prior to autophagosome/vacuole fusion) and in the atg15Δ strain (which results in IVCs). Importantly, we saw an increased co-localization of Pub1 with Atg19 in the mon1Δ background, and could also observe clear co-localization of Atg19 with Pub1 in IVCs in the atg15Δ background (Figure S6, Table S5), arguing that co-localization is a consequence of a transient interaction of stress granule with autophagosomes for targeting to the vacuole.
We also observed that stress granules accumulated in an ubx2Δ strain, whilst the vms1Δ strain exhibited decreased P-bodies (Table S1). Ubx2 and Vms1 both interact with Cdc48, the yeast ortholog of VCP, which is an AAA-ATPase that can promote autophagy (Krick et al, 2010; Meyer et al, 2012), including targeting of large ribosomal subunits for ribophagy (Ossareh-Nazari et al, 2010). These observations suggested that Cdc48 might also be important in targeting stress granules and/or P-bodies for autophagy. To test this possibility, we examined the effects of a conditional allele of Cdc48 with a previously described autophagy defect (Krick et al, 2010) upon the accumulation of stress granules in yeast cells. We observed that at the restrictive temperature, Cdc48 inactivation led to the strong accumulation of stress granules, suggesting that Cdc48 plays a role in stress granule clearance (Figure 2B). Although the phenotype was not as strong as in the Cdc48 allele, we also observed increased stress granules in conditional alleles of the Cdc48 interacting proteins Ufd1 and Npl4 (Figure 2B; Table S5) suggesting these proteins can also affect the dynamics of stress granules.
Epistasis Tests define a pathway of targeting stress granules to vacuoles
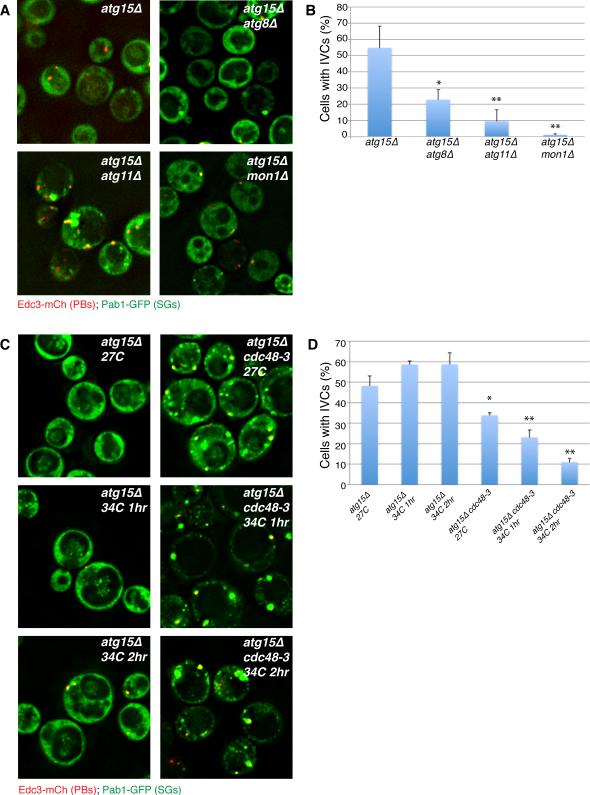
The Cdc48 protein could affect stress granule dynamics in multiple manners. Cdc48 uses the energy of ATP hydrolysis to rearrange complexes containing ubiquitinated proteins (Stoltz et al, 2011), and can function in targeting proteins to the proteasome (e.g. Meyer et al, 2012; Stoltz et al, 2011), as well as promoting autophagy (Krick et al, 2010; Meyer et al, 2012; Ossareh-Nazari et al, 2010). To determine if Cdc48 affected autophagy of mRNP granules, we investigated how inactivation of Cdc48, or other autophagy genes, affected the formation of IVCs containing stress granule or P-body markers in the atg15Δ strain. If Cdc48 is required for autophagy of stress granules and/or P-bodies we would expect a decrease in IVCs containing these components in an Cdc48-inactivated atg15Δ strain as compared to atg15Δ alone. Conversely, if Cdc48 affects stress granules by an unrelated mechanism, then the IVC compartments should be unchanged upon inactivation of Cdc48 activity.
This analysis of atg15Δ double mutants revealed two important observations. First, we observed that deletion of ATG8, ATG11 or MON1, which promote autophagy (Wang et al, 2002; Yorimitsu and Klionsky, 2005), significantly reduced the accumulation of stress granule markers within IVCs in the atg15Δ background (Figure 3A-B). Second, we observed that the temperature sensitive Cdc48-3 allele led to a significant and progressive decrease in IVC levels following a one or two-hour shift to the restrictive temperature, whereas IVC levels in the atg15Δ control cells actually elevated slightly under identical conditions (Figure 3C-D). IVC levels were already slightly reduced in the cdc48-3 atg15Δ strain prior to temperature shift, possibly suggesting partial loss of function, and stress granules were more abundant relative atg15Δ control cells, consistent with a role for Cdc48 in their clearance. We interpret these observations to indicate that stress granules can be targeted to the vacuole in a process that is dependent on the autophagy machinery and Cdc48. Moreover, since inactivation of Cdc48 activity reduces the accumulation of IVCs in the atg15Δ strain, this argues that Cdc48 enhances the targeting of stress granules to autophagy.
Figure 3. Atg15-induced IVCs are decreased by upstream mutations in autophagy pathways and in Cdc48.
A.) BY4741-background strains in early stationary phase. IVCs present in an atg15Δ strain are reduced by secondary mutations in autophagy factors acting at/prior to autophagosome-vacuolar membrane fusion. B.) Quantitation of data in A, mean values based on 3 independent experiments +/− SD. P-values (* = <0.05, ** = < 0.005) relative to atg15Δ indicated. C.) W303 strain background, examined in early stationary phase following 0, 1 or 2hrs of 34°C heatshock. Inactivation of Cdc48 results in decreased IVCs caused by atg15 deletion. D.) Quantitation of data in C, mean values based on a minimum of 3 replicate experiments +/− SD. P-values as above. See also Table S5.
Targeting of RNP granules by autophagy increases when decapping is inhibited
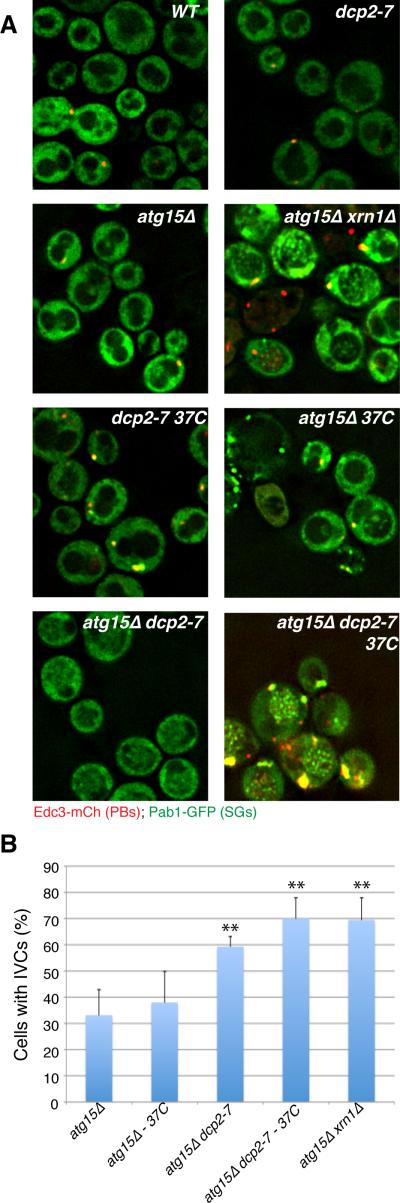
The targeting of stress granules and possibly P-bodies to autophagy provides another mechanism regulating the clearance of these structures. This is in addition to translation initiation, which can reduce P-bodies and stress granules by promoting the return of mRNAs to translation (Brengues et al., 2005; Bhattacharyya et al, 2006), as well as decapping and 5' to 3' degradation, which can reduce the mRNAs present within P-bodies (Sheth and Parker, 2003). Since these RNP granules can have multiple fates, it suggests that decreases in one of these pathways would increase others. For example, we hypothesized that decreases in decapping or 5' to 3' exonucleolytic degradation, which lead to the accumulation of P-bodies, stress granules and a pool of non-degraded mRNAs (Sheth and Parker, 2003; Buchan et al., 2008) might lead to an increased flux of mRNPs into autophagy. To test this prediction, we analyzed the amount of IVC accumulating in an atg15Δ strain carrying the xrn1Δ deletion or a temperature-sensitive allele of the decapping enzyme (dcp2-7). Strikingly, compared to the atg15Δ strain, we observed an increase in both the number of cells with IVC compartments and the relative size and intensity of the IVC in xrn1Δ atg15Δ strains, and in the dcp2-7atg15Δ strain after a shift to the restrictive temperature (Figure 4). This observation provides additional evidence that stress granules can be targeted for autophagy and suggests that this pathway increases when mRNA decapping is blocked and the cell has accumulated a larger pool of RNP granules.
Figure 4. Impairment of cytoplasmic mRNA decay increases accumulation of granule proteins in IVCs.
A.) RP840-background strains in early stationary phase. Temperature shifted strains were subject to 1hr at 37°C (lower row). B.) Quantitation of data in A, mean values based on a minimum of 3 replicate experiments +/− SD. P-values (* = <0.05, ** = < 0.005) relative to atg15Δ indicated. See also Table S5.
Autophagy affects stress granule clearance in mammalian cells
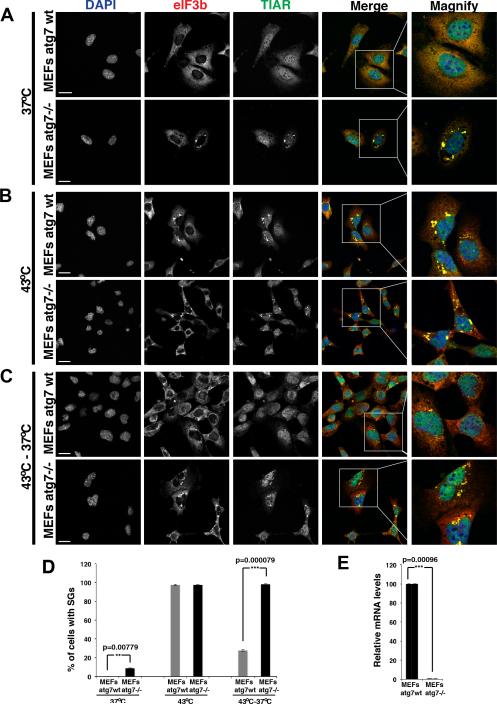
The conservation of P-bodies, stress granules, and the autophagic process implies that mRNP granules would also be targeted for autophagy in other eukaryotic cells. This model predicts that inhibiting autophagy in mammalian cells would lead to the accumulation of constitutive stress granules, and would reduce the rate at which they are cleared following removal of a stress. To test this possibility we examined stress granules in MEFs bearing homozygous deletion of ATG7 both in the absence of stress and following relief of a stress.
Examination of endogenous stress granule components revealed two observations arguing that autophagy affects the rate of stress granule clearance. First, in the absence of any stress we observed that atg7−/− MEFs showed a small but consistent percentage of cells with clearly visible stress granules, while corresponding wild-type MEFs cells showed no cells with stress granules (Figure 5A, D-E); a similar result was also observed in atg3−/− MEF lines (Figure S7C). Second, we observed that while stress granules formed robustly in both wild-type and atg7−/− MEFs in response to a heat shock (Figure 5B, D), the majority of the stress granules were cleared from wild-type MEFs upon return to 37°C, while persisting in atg7−/− MEFs for up to 2 hours (Figure 5C-D). These observations argue that at least a subset of stress granules are cleared by autophagy in mammalian cells. Consistent with that interpretation, we observe that autophagy-promoting drugs (either rapamycin or 3-MA; Wu et al, 2010) enhance the clearance of arsenite-induced stress granules (Figure S7A, B). By contrast, the autophagy inhibitor wortmannin attenuates stress granule clearance (Figure S7A, B).
Figure 5. Mammalian stress granule clearance is impaired by defective autophagy.
A.) atg7−/− MEFs showed a low level of cells with clearly visible stress granules even in the absence of exogenous stressors. B.) Heat shock for 1hr at 43°C resulted in robust stress granule formation in both WT and atg7−/− MEFs. C.) After shift from 43°C back to 37°C stress granules clear within minutes in wild type MEFs, but persist for the duration of the assay (2hrs) in atg7−/− MEFs. Scale bar equals 10μm. D) Quantitation of the data in A-C, mean values based on a minimum of 3 replicate experiments +/− SEM. E) Real time quantitative PCR verifies the expression levels of atg7 in wild type and atg7−/− MEFs. See also Figure S7.
VCP affects stress granule clearance in mammalian cells
The effect of Cdc48 on stress granules in yeast suggested that the mammalian ortholog VCP might play a role in stress granule clearance. To test this hypothesis, we depleted VCP function in mammalian cells either by siRNA, or by chemical inhibition of VCP activity and examined the formation and clearance of stress granules during heat shock and recovery. We observed that either siRNA knockdown (Figure 6A-D) or chemical inhibition of VCP in HeLa cells (Figure 6E-F) led to significantly reduced stress granule clearance, indicating that VCP is required for efficient stress granule clearance in mammalian cells. Moreover, because endogenous VCP accumulates in stress granules during different stress conditions (heat stress, osmotic stress, oxidative stress; Figure 7A), the simplest model is that VCP directly acts on stress granules to promote their clearance.
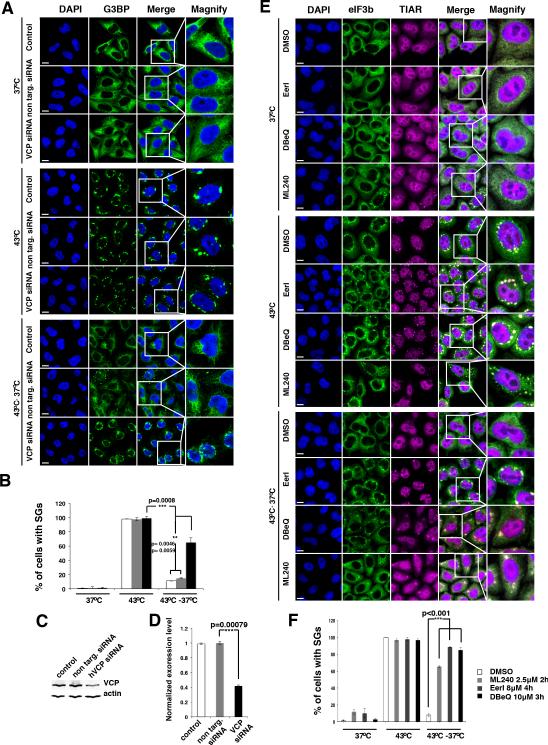
Figure 6. VCP is essential for stress granule clearance.
A.) HeLa cells examined by immunofluorescence for the stress granule marker G3BP. Cells treated with non-targeting siRNA show stress granule assembly upon heat shock at 43°C (2hrs) and rapid clearance of stress granules upon return to 37°C. Cells treated with VCP-targeting siRNA show normal stress granule assembly, but these stress granules fail to clear following return to 37°C (2hrs). Scale bar equals 10μm. B.) Quantification of data in A, mean values based on a minimum of 3 replicate experiments +/− SEM. C). and D.) Western blot and quantification of VCP protein levels in cells from A, mean values based on a minimum of 3 replicate experiments +/− SEM. E.) HeLa cells examined by immunofluorescence for the stress granule markers TIAR and eIF3b. Pre-treatment with chemical inhibitors of VCP impairs clearance of stress granules. Cells were treated with vehicle (DMSO) or specific inhibitors Eer1 (4hrs pre-treatment with 8μM Eeyarestatin I, per Wang et al, 2010); DBeQ (3hrs pre-treatment with 10μM DBeQ, per Chou et al, 2011); ML240 (2hrs pre-treatment with 2.5μM ML240, per Chou et al, 2013). Scale bar equal 10μm. F.) Quantification of the data in E, mean values based on a minimum of 3 replicate experiments +/− SEM.
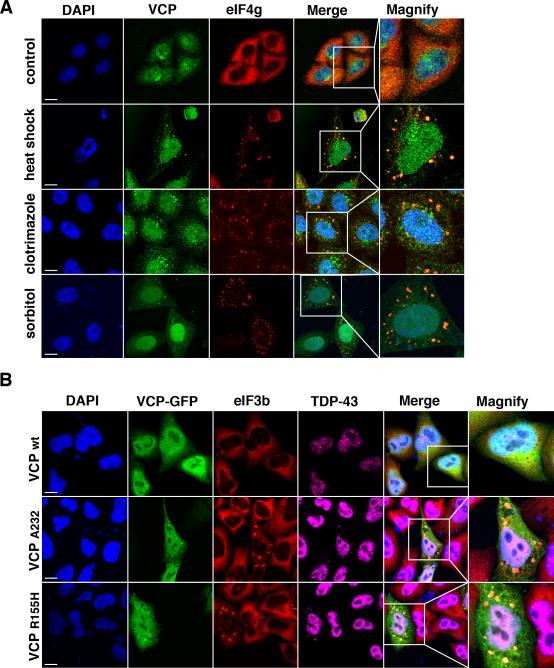
Figure 7. VCP is recruited to stress granules and disease-causing mutations in VCP induce constitutive stress granules that contain the disease protein TDP-43.
A.) HeLa cells stained for endogenous VCP and the stress granule marker eIF4G. VCP is recruited to stress granules induced by three distinct stimuli: heat shock at 43°C for 2hrs, incubation with 20μM clotrimazole for 1hr, or incubation with 0.6M sorbitol for 1hr. Scale bar equal 10μm. B.) HeLa cells transfected with plasmid expressing wild type or mutant (A232 and R155H) VCP-GFP and stained for eIF3b and TDP-43. Over-expression of mutant but not wild type VCP results in the formation of constitutive stress granules that contain the disease protein TDP-43. Scale bar equals 10μm.
The requirement of VCP for stress granule clearance in mammalian cells raised the possibility that disease-causing mutations in VCP might impair stress granule dynamics. Indeed, VCP-related diseases, including ALS, FTLD and MSP, are all characterized by accumulation of cytoplasmic inclusions of stress granule constituents TDP-43, hnRNPA1, and/or hnRNPA2 (Neumann et al., 2006; Neumann et al., 2007; Salajegheh et al., 2009; Kim et al., 2013). Strikingly, we observed that HeLa cells overexpressing the VCP disease-related mutations A232E and R155H showed the constitutive appearance of stress granules containing eIF3 subunits, TDP-43 and VCP itself (Figure 7D). The accumulation of constitutive stress granules in these VCP pathological alleles is consistent with the hypothesis that impaired stress granule dynamics may be a shared defect underlying the pathogenesis of disease initiated by mutations in VCP as well as disease initiated by stress granule-promoting mutations in RNA-binding proteins TDP-43, FUS, hnRNPA1 and hnRNPA2B1.
DISCUSSION
In this work we identified over one hundred genes that are involved in the modulation of stress granules and/or P-bodies in yeast. These proteins show a dense network of physical and genetic interactions indicating they reveal a previously unknown network of proteins controlling mRNP granule dynamics. Given the role of RNP granules in translation and mRNA degradation, it was not unexpected that this set of proteins would contain multiple factors involved in mRNA biogenesis and function (Edc2, Lsm6, Mlp1, Pap2, Rnh70, Rpl35b, Rpl39, Rpl42a, Rps28b, Sto1, Tif3, Trm12, Sro9 and Xrn1). Surprisingly, we also observed that defects in the THO or TREX-2 complexes led to the constitutive formation of stress granules, perhaps because some newly exported mRNAs are deficient at entering translation. Another interesting concentration of mutants included defects in the prefoldin complex, which decreased P-bodies, and could reflect a failure to properly assemble cytoskeletal structures to which mRNP granule assembly and trafficking has been linked (Aizer et al, 2008; Loschi et al, 2009). Mutants affecting various signaling pathways (e.g. Kcs1, Cmk1, Mds3) and intriguing cellular processes (e.g. Chromatin modification/assembly – Gcn5, Hst1, Iki3, Isw1, Rlf2, Set2, Swc5, Swd3 and Swr1) were also observed. An important area of future research will be to understand how these proteins affect RNA granule dynamics in molecular detail, and the physiological consequences of such aberrant assembly.
Analysis of these mutants provided several lines of evidence that yeast stress granules and P-bodies can be targeted to the vacuole by autophagy (Figure S8), a process we refer to as granulophagy. First, mutations that inhibit autophagy at stages prior to autophagosome-vacuolar membrane fusion accumulated stress granules (Figure 2), which is consistent with stress granules continually being cleared by an autophagic process. Second, strains deficient in the Atg15 lipase, which breaks down autophagic vesicles within the vacuole, accumulate stress granule and P-body markers in IVCs, consistent with the accumulation of autophagic vesicles in the atg15Δ strain (Figure 2). Third, combining the atg15Δ deletion with earlier blocks to autophagy decreases the accumulation of stress granule and P-body markers in IVCs and often increases the cytoplasmic accumulation of stress granules (Figure 3). Fourth, when decapping or 5' to 3' mRNA degradation is inhibited, which leads to increased levels of mRNP granules, we observed an even greater accumulation of stress granule markers within IVCs in atg15Δ strains (Figure 4). This argues that the targeting of RNP granules to autophagy is increased in the absence of normal levels of mRNA degradation. Taken together, we conclude that stress granules and P-bodies, and presumably the mRNAs within them can be targeted to vacuoles by autophagy.
Several observations suggest that yeast stress granules are more commonly targeted for autophagy than P-bodies. First, mutants defective in autophagy show a greater increase in stress granules than P-bodies (Table S1). Second, in dcp2-7 and xrn1Δ mutants, which show an accumulation of both P-bodies and stress granules we see a greater accumulation of the stress granule marker Pab1 in vacuoles than the P-body component Edc3 (Figure 4). Third, our analyses to date have determined four known stress granule factors within atg15Δ IVCs (Pab1, Pub1, Pbp1 and Ded1 – Figure S5), whereas only Edc3 has been observed at a modest level; Dcp2 and Lsm1 are not detected in IVCs (data not shown). One possible model to explain the bias in stress granules in targeting for autophagy is that mRNPs within P-bodies can also either be degraded, or can undergo mRNP remodeling to enter stress granules (Buchan et al., 2008). Thus, if autophagy is generally slow compared to these events, then most mRNPs will transition to a stress granule type of mRNP before being targeted for autophagy.
Several observations suggest that the targeting of stress granules to degradative organelles by autophagy is conserved in eukaryotes including mammals. First, MEFs defective in autophagy show a low level of constitutive stress granules, consistent with a defect in stress granule resolution (Figure 5A, Figure S7C). Second, atg7−/− MEFs are defective in clearing stress granules after relief of heat stress (Figure 5B-D). Third, we observed that 3-MA and to a lesser extent rapamycin, which can stimulate autophagy (Wu et al, 2010), increased the rate at which stress granules were cleared following the relief of oxidative stress (Figure S7A, B). In contrast, inhibition of autophagy by wortmannin slowed the rate of stress granule clearance (Figure S7A, B). Fourth, inhibition of the vacuolar ATPase function, either by mutations in yeast or pharmacologically in mammalian cells, lead to increases in SG (Table S1, Table S5, Figure S7D). This is relevant since vacuolar ATPases have been implicated in a wide array of vesicular trafficking events, often independent of proton-pumping activity, including maturation and fusion of autophagosomes with the lysosome in mammals (Klionsky et al, 2008), vacuole-vacuole fusion (Peters et al, 2001), and phagosome-lysosome fusion (Peri and Nusslein-Volhard, 2008). Finally, in C. elegans it has been suggested that P-granules, which are RNP granules related to stress granules and P-bodies can be cleared from blastomeres by autophagy (Zhang et al, 2009).
The autophagic degradation of stress granules and P-bodies provides an additional fate for the mRNPs that accumulate within these structures. Such a fate might be particularly important for mRNPs within stress granules, which can also return to translation, and might serve an important role in modulating the stress response, particularly during prolonged stress, where stress granule associated mRNPs unable to return to translation might instead be targeted for vacuole/lysosome based degradation. Moreover, this pathway provides a novel mechanism by which eukaryotic mRNAs could degraded and one anticipates that for a subset of mRNAs this is a prevalent pathway by which they are degraded. Indeed, such a pathway could explain why miRNA based mRNA degradation is affected by alterations in membrane flow given that miRNA based repression machinery associates with endosomal compartments (Gibbings et al, 2009). Related to this, Ago2 and Dicer appear to be targeted by selective autophagy, with resulting consequences on miRNA-based repression (Gibbings et al, 2012).
Several observations also argue that Cdc48 and its mammalian ortholog VCP function in stress granule clearance. First, temperature sensitive alleles of Cdc48, as well as mutations in the associated Ubx2, Npl4, and Ufd1 proteins, showed an accumulation of stress granules (Figure 2, Table S1). Second, depletion of VCP activity in tissue culture cells with either siRNAs or VCP inhibitors leads to a defect in the clearance of stress granules (Figure 6). Third, pathogenic mutations in VCP lead to constitutive accumulation of stress granules in cells in culture (Figure 7), which contain TDP-43, the major defining constituent of pathological cytoplasmic inclusions in patients with ALS, FTLD, IBM and MSP (Neumann et al., 2006; Neumann et al., 2007; Salajegheh et al., 2009; Kim et al., 2013). Taken together, these observations demonstrate CDC48/VCP is required for efficient stress granule clearance and this function may be relevant to its role in pathologies.
An unresolved role is the specific function of Cdc48/VCP in stress granule clearance. Because inhibition of Cdc48 function reduces the presence of stress granules in the IVCs in the atg15Δ background (Figure 3), we suggest that at least part of Cdc48's function in yeast is to promote autophagy of these mRNP granules. Ubiquitination is likely to be involved in modulating stress granules and P-body dynamics since Cdc48/VCP utilizes ATP hydrolysis to segregate ubiquitinated proteins from a variety of cellular complexes (Stolz et al, 2011), stress granules in mammalian cells are heavily ubiquitinated (Kwon et al, 2007), and we observed that deletion of an E3 ubiquitin ligase of unknown function, Hel2, leads to increased stress granules (Table S1). In addition, autophagic clearance of ubiquitinated protein aggregates by VCP is facilitated by its known binding partner HDAC6, which itself binds ubiquitinated proteins, and also regulates stress granule assembly in mammalian cells (Ju et al, 2008; Kwon et al, 2007). Since VCP accumulates within stress granules (Figure 7), one possible model is that the Cdc48/VCP complex works on some ubiquitinated component of stress granules to alter the mRNP complexes in a manner that promotes disassembly and/or targeting of the RNP granule to autophagy. However, ubiquitin may also affect stress granule dynamics through the proteasome since we identified mutations in the PRE9 and POC4 genes, which affect proteasome function, as increasing constitutive stress granules (Table S1), and inhibiting proteasome activity in mammals induces stress granules (Mazroui et al., 2007).
Our observations have important implications for the understanding of the role of RNP aggregates in some degenerative diseases. First, the targeting of stress granules to vacuoles/lysosome for degradation implies that aberrant forms of these granules that accumulate in some degenerative diseases, such as ALS, FTLD, IBM and MSP, may also be cleared by this process. Notably, TDP-43, the most consistently observed component of pathological cytoplasmic inclusions in ALS, FTLD and MSP, is preferentially cleared by autophagy (Wang et al, 2010; 2012). Consistent with this notion, Cdc48 inactivation was also recently shown to enhance TDP-43 toxicity in a yeast neurodegenerative model system (Armakola et al, 2012). Thus, mechanisms to enhance autophagy of stress granules and related RNP aggregates may have potential as therapies to treat various degenerative diseases.
These results also strengthen the hypothesis that ALS, FTLD, and some related pathologies arise due to hyper-formation or stabilization of stress granules. This was first suggested by the observations that mutations in the stress granule components TDP-43, FUS, hnRNPA1, hnRNPA2, and Atx2 that enhanced their self-assembly or aggregation of could be causative in these diseases as well as causing the accumulation of stress granules, or related RNP aggregates in model systems as well as patient tissue (Kim et al, 2013; Dewey et al, 2012; Ito and Suzuki 2011). Strikingly, VCP mutations lead to the same spectrum of pathologies as the aggregation prone mutations in these RNA binding proteins. Given a role for VCP/Cdc48 in stress granule clearance, and pathological mutations in VCP leading to the constitutive appearance of stress granules, we suggest that the pathologies of VCP mutations and hyper-aggregation mutations in RNA binding proteins are similar because they both lead to the accumulation and/or persistence of stress granules. This also raises the exciting possibility that mutations we have identified in yeast that lead to the formation of constitutive stress granules may identify candidate genes that, when mutated in humans, contribute to the formation of both constitutive stress granules in mammalian cells and degenerative disease.
EXPERIMENTAL PROCEDURES
Yeast Microscopy
For the screen, deletion strain and WT control transformants were inoculated overnight in 300μl of selective minimal media in 96-well plates, and agitated at 30°C for optimal growth. The following morning, staggered re-inoculation at OD600s of < 0.1, followed by growth to an approximate OD600 of 0.5 was conducted. Samples were spotted onto 8-well Teflon-coated slides (Tekdon inc), coverslips were sealed with nail-varnish, and cells were imaged using a Deltavision RT microscope with 100x objective (Applied Biosystems). 2 images were collected for each strain at 2×2 binning, resulting in the capture of approximately 50-100 cells. Manual imaging, blind to gene identity, ensured both high quality datasets and helped detect phenotypes that are hard to quantify with automated algorithms. Since non-stressed yeast cells exhibit virtually no stress granules and have reasonable numbers of semi-bright P-bodies, deletion strains exhibiting stress granule induction or P-body induction/reduction phenotypes were easily distinguished. Strains identified with a phenotype were re-screened blindly as above, and only those with repeating phenotypes made the final list (more detail on phenotype classification in supplemental methods).
Additional microscopy analysis was conducted using the above system, a Deltavision Elite system with 100x objective (Applied Biosystems), and a Nikon A1R Confocal operating in wide field mode, also with a 100x objective (Nikon). Yeast strains were examined either in logarithmic grow (OD600 0.4-0.5) or in early stationary phase (inoculation at OD600 0.2 followed by growth for 18-24 hours). Entry into early stationary phase was ensured for each strain with OD600 measurements and prior knowledge of a given strain's maximal OD600 in minimal media culture. Examination under such conditions maximized detection of IVCs in an atg15Δ background. Details of yeast transformation, strain identification and image analysis are in supplemental methods.
Bioinformatic analysis
Cytoscape v 2.8.2 (Smoot et al, 2011), and the S.cerevisiae Biogrid interaction dataset 3.1.86 (thebiogrid.org), were used to plot network and sub-network analyses of the known physical and genetic interactions of the 125 screen hits. Significance of the resulting 100 node, 448 non-redundant interaction network was determined by simulation analysis using 125 randomly selected genes from the 4249 strains transformed in the screen, and plotting networks using identical parameters to the above. The average values for 100 random networks exhibited a Gaussian distribution, with means of 47.8 +/− 11.8 nodes and 77.6 +/− 25.7 non-redundant interactions. Z-score analysis confirmed our screen network to be significant (P-value 5.34 ×10−6). GO-term classification was obtained at the S.cerevisiae database (www.yeastgenome.org).
HeLa and MEF cell growth conditions and fixation
HeLa cells, incubated in a 37°C incubator with a 5% CO2 concentration, were grown in DMEM media (Gibco or Hyclone) supplemented with 10% FBS (Atlas Biological or Hyclone), 2mM L-GlutaMAX (Gibco), and Pen-Strep (100 U/ml penicillin, 100μg/ml streptomycin; Gibco). Prior to analysis, cells were passaged from flasks onto 8-well chamber slides (Nalge Nunc Intl.) or 4-well and 8-well slides (Millipore), and grown for 1.5-2 days to reach 60–70% confluency. For details on HeLa cell drug additions, see supplemental methods. During stress granule clearance experiments, cells were subject to a 1hr arsenite stress. This was followed by washing in fresh media, followed by re-suspension in fresh media alone, or in the additional presence of a drug, and fixation at 20 min, 1hr, 2hr and 3hr time points. Paraformaldehyde fixation of cells was as described in Kedersha and Anderson (2007).
Atg3 −/− MEF cells were grown similarly to HeLa, but in media containing sodium pyruvate (Gibco) and non-essential amino acids (Sigma), whilst lacking GlutaMAX. Fixation protocols were as above. Atg7−/− MEF cells were grown in DMEM (Hyclone) 10% FBS (Hyclone) and GlutaMax-1x (Gibco). The cells were fixed in 4% Paraformaldehyde (Electron Microscopy Science). Transfection details are in supplemental methods.
HeLa and MEFs immunofluorescence and microscopy
Antibodies and concentrations used in HeLa and MEF cell immunofluorescence are detailed in supplemental methods. Slides were mounted using Vectashield with DAPI (Vector Laboratories) or ProLong Gold Antifade Reagent with DAPI (Invitrogen; P3691). Images were captured using a Nikon A1R Confocal microscope (Nikon) using a 100x objective or a LSM510 (Zeiss) confocal microscope with a 63x objective. All images are collapsed Z-stacks (7uM depth, 0.25uM slices). Stress granules were quantified on a percentage cell basis, whilst cells deemed to have P-bodies required foci to be above a threshold intensity value. Atg3 −/− MEFs were stained as above, but imaged on a Deltavision Elite system (Applied Biosystems) using a 100x objective.
Supplementary Material
HIGHLIGHTS.
Network of 125 genes identified that affect stress granule and P-body dynamics
Stress granules are cleared by autophagy in eukaryotes
Cdc48/VCP facilitates stress granule clearance
Autophagy of related mRNP aggregates may help avoid degenerative pathology
ACKNOWLEDGEMENTS
We thank the Chen lab (Institute of Molecular Biology, Taiwan) for Cdc48, Ufd1 and Npl4 conditional allele strains, the Klionsky lab (University of Michigan) for a GFP-Atg19 expressing plasmid, the Komatsu lab (Tokyo Metropolitan Institute of Medical Sciences) for Atg 3−/− MEF cell lines and the Green lab (St. Jude Children's Research Hospital) for Atg7−/− cell lines. We also thank the Cell and Tissue Imaging Core Facility at St. Jude Children's Research Hospital, and all Parker lab members for feedback, particularly Saumya Jain for bioinformatics assistance. This work was supported by funds from the ALS Association to RMK, the Packard Center for ALS Research to JPT, and the Howard Hughes Medical Institute to JRB and RP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Bio. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, Diaz Z, Fang X, Shorter J, Krogan NJ, Finkbeiner S, et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat. Genet. 2012;44:1302–1309. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham CJ, Parker R. P bodies, stress granules, and viral life cycles. Cell host & microbe. 2008;3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harbor Symposia Quant. Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, et al. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc. Natl. Acad. Sci USA. 2011;108:4834–4839. doi: 10.1073/pnas.1015312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TF, Li K, Frankowski KJ, Schoenen FJ, Deshaies RJ. Structure activity relationship reveals ML240 and ML241 as potent and selective inhibitors of p97 ATPase. ChemMedChem. 2013 doi: 10.1002/cmdc.201200520. 10.1002/cmdc.201200520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Johnson BA, Herz J, Yu G. TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res. 2012;1462:16–25. doi: 10.1016/j.brainres.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot MC, Subramanian M, Flatter E, Mandel JL, Moine H. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol. Biol. Cell. 2009;20:428–437. doi: 10.1091/mbc.E08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 2011;30:3540–3552. doi: 10.1038/emboj.2011.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson SL, Lykke-Andersen J. Cytoplasmic mRNP granules at a glance. J. Cell Sci. 2011;124:293–297. doi: 10.1242/jcs.072140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010;143:938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat. Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Galvin JE, Chiu TS, Lee VM, Masliah E, Trojanowski JQ. RNA sequestration to pathological lesions of neurodegenerative diseases. Acta Neuropath. 1998;96:487–494. doi: 10.1007/s004010050923. [DOI] [PubMed] [Google Scholar]
- Gkogkas C, Sonenberg N, Costa-Mattioli M. Translational control mechanisms in long-lasting synaptic plasticity and memory. J. Biol. Chem. 2010;285:31913–31917. doi: 10.1074/jbc.R110.154476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Suzuki N. Conjoint pathologic cascades mediated by ALS/FTLD-U linked RNA-binding proteins TDP-43 and FUS. Neurology. 2011;77:1636–1643. doi: 10.1212/WNL.0b013e3182343365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Miller SE, Hanson PI, Weihl CC. Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease. J Biol. Chem. 2008;283:30289–30299. doi: 10.1074/jbc.M805517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Kim NC, Tresse E, Kolaitis RM, Molliex A, Thomas RE, Alami NH, Wang B, Joshi A, Smith RB, Ritson GP, et al. VCP Is Essential for Mitochondrial Quality Control by PINK1/Parkin and this Function Is Impaired by VCP Mutations. Neuron. 2013 doi: 10.1016/j.neuron.2013.02.029. 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–950. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen EL, Thumm M. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J. Cell Biol. 2010;190:965–973. doi: 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes & development. 2007;21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M, Leishman CC, Berardone N, Boccaccio GL. Dynein and kinesin regulate stress-granule and P-body dynamics. J. Cell Sci. 2009;122:3973–3982. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Di Marco S, Kaufman RJ, Gallouzi IE. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol. Biol. Cell. 2007;18:2603–2618. doi: 10.1091/mbc.E06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medioni C, Mowry K, Besse F. Principles and roles of mRNA localization in animal development. Development. 2012;139:3263–3276. doi: 10.1242/dev.078626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Mijaljica D, Prescott M, Devenish RJ. V-ATPase engagement in autophagic processes. Autophagy. 2011;7:666–668. doi: 10.4161/auto.7.6.15812. [DOI] [PubMed] [Google Scholar]
- Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2012 doi: 10.1038/nsmb.2468. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science (New York, NY. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Neumann M, Kwong LK, Sampathu DM, Trojanowski JQ, Lee VM. TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J Neuropathol. Exp. Neurol. 2007;66:152–157. doi: 10.1097/nen.0b013e31803020b9. [DOI] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van Dorsselaer A, Dargemont C. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO reps. 2010;11:548–554. doi: 10.1038/embor.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Peri F, Nusslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–927. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- Peters C, Bayer MJ, Buhler S, Andersen JS, Mann M, Mayer A. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 2001;409:581–588. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salajegheh M, Pinkus JL, Taylor JP, Amato AA, Nazareno R, Baloh RH, Greenberg SA. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle & nerve. 2009;40:19–31. doi: 10.1002/mus.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol. Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Stolz A, Hilt W, Buchberger A, Wolf DH. Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 2011;36:515–523. doi: 10.1016/j.tibs.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Takahara T, Maeda T. Transient sequestration of TORC1 into stress granules during heat stress. Mol. Cell. 2012;47:242–252. doi: 10.1016/j.molcel.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM, Klionsky DJ. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol Chem. 2001;276:2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Stromhaug PE, Shima J, Klionsky DJ. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J. Biol Chem. 2002;277:47917–47927. doi: 10.1074/jbc.M208191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IF, Guo BS, Liu YC, Wu CC, Yang CH, Tsai KJ, Shen CK. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc. Natl. Acad. Sci USA. 2012;109:15024–15029. doi: 10.1073/pnas.1206362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fan H, Ying Z, Li B, Wang H, Wang G. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci. Let. 2010;469:112–116. doi: 10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.