Abstract
Background
Microarray studies have shown that the E2F transcription factor influences the expression of many genes but it is unclear how many of these targets are important for E2F-mediated control of cell proliferation.
Results
We assembled a collection of mutant alleles in 44 dE2F1-dependent genes and tested whether these could modify visible phenotypes caused by the tissue-specific depletion of dE2F1. More than half of the mutant alleles dominantly enhanced de2f1-dsRNA phenotypes suggesting that the in vivo functions of dE2F1 can be limited by the reduction in the level of expression of many different targets. Unexpectedly, several mutant alleles suppressed de2f1-dsRNA phenotypes. One of the strongest of these suppressors was Orc5. Depletion of ORC5 increased proliferation in cells with reduced dE2F1 and specifically elevated the expression of dE2F1-regulated genes. Importantly, these effects were independent of dE2F1 protein levels, suggesting that reducing the level of ORC5 did not interfere with the general targeting of dE2F1.
Conclusions
We propose that the interaction between ORC5 and dE2F1 may reflect a feedback mechanism between replication initiation proteins and dE2F1 that ensures that proliferating cells maintain a robust level of replication proteins for the next cell cycle.
Keywords: E2F, ORC, PCNA, Drosophila
INTRODUCTION
The temporal regulation of gene expression is an important component of cell cycle control. The periodic synthesis of Cyclins and the activation of Cyclin-dependent-kinases (Cdk's) promote the expression of genes that are needed for cell cycle progression and for cell proliferation. Studies of genome-wide patterns of gene expression in Saccharomyces cerevisiae have revealed that cell cycle progression involves at least two interconnected oscillators, the Cdk cycle and a transcriptional network of periodically expressed genes, which cycle in tandem with one another (Orlando et al., 2008).
In higher eukaryotes, the E2F transcription factor is one of the best-known regulators of cell cycle dependent gene expression (Blais and Dynlacht, 2004; Bracken et al., 2004). E2F-containing complexes repress gene expression in quiescent cells and in early G1-phase of the cell cycle. During G1- to S-phase progression the effects of repressor complexes are reversed by activator E2F's. This regulation allows a large number of E2F target genes to be coordinately expressed at specific stages of the cell cycle. E2F targets encode proteins that are needed for the replication of the genome, for mitosis, and include genes that control progression through most of the key cell cycle transitions (Muller et al., 2001; Ren et al., 2002). Accordingly, experiments in several different systems show that E2F activity is rate-limiting for cell proliferation: the genetic inactivation of activator E2F genes impairs cell proliferation, whereas the mis-expression of activator E2F's is sufficient to drive quiescent cells into S-phase (Johnson et al., 1993; Dobrowolski et al., 1994; Sellers et al., 1995; Asano et al., 1996; Du et al., 1996; Ishizaki et al., 1996; Wu et al., 1996; Lukas et al., 1997).
The Drosophila dE2F/RBF proteins provide many of the same biological functions as their mammalian counterparts but they form a network that is far less complex. This system can be used to ask questions about the role and regulation of E2F-dependent transcription that are difficult to address in mammalian cells. For example, genetic studies in Drosophila have shown that dE2F-regulation is not absolutely essential for cell division but cells lacking dE2F or dDP proteins are at a severe disadvantage when forced to compete with wild-type cells in mosaic animals (Frolov et al., 2005).
E2F targets include genes encoding the origin recognition complex (ORC) and mini chromosome maintenance (MCM) proteins (Ohtani et al., 1996). ORC is a six subunit complex, which functions as a platform for building replication initiation complexes (Dutta and Bell, 1997; Machida et al., 2005). Its function is important for the initiation of DNA synthesis, and studies in Drosophila have shown that Orc2 mutants disrupt DNA amplification (Landis et al., 1997). Interestingly, dE2F1, dDP and RBF1 have been shown to form a complex with Drosophila ORCs (Bosco et al., 2001; Ahlander et al., 2008). dE2F1 directly binds to chorion gene origins of replication in ovaries in vivo, raising the possibility that dE2F1 and RBF can function at replication origins to limit DNA replication (Bosco et al., 2001).
Precisely how many human or fly genes are controlled by E2F is unclear. E2F-binding sites are found in the promoters of hundreds of genes (Weinmann et al., 2002; Bracken et al., 2004). Estimates from microarray analyses of the transcriptional changes induced by E2F proteins suggest that mammalian cells contain several hundred to more than a thousand E2F-inducible genes. The extent of E2F regulation varies from gene to gene and the functional relevance of these changes is impossible to assess; the importance of gene function for cell proliferation, the threshold of gene expression that is needed for gene function, and the relative importance of transcriptional control versus other mechanisms of regulation are all poorly understood parameters that vary from gene to gene. Consequently, it is impossible to say, when looking at a list of E2F-dependent genes, which of the targets are key to E2F's ability to control cell proliferation and which targets are unimportant.
A central issue that has yet to be resolved is whether E2F control of cell proliferation results from its regulation of a small number of key targets, or whether this control is derived from integrated effects of E2F across hundreds of target genes. Surprisingly, there are few studies in the literature that have addressed this issue. One of the best-known targets of E2F is Cyclin E. Studies in both Drosophila and mammalian cells show that the ectopic expression of Cyclin E can transiently rescue DNA synthesis in cells that are mutant for de2f1, or in which E2F activity is inhibited (Duronio and O'Farrell, 1995; Lukas et al., 1997). A partial rescue in this type of experiments may indicate that Cyclin E is initially the rate-limiting target of E2F, and that other targets become limiting with time. Alternatively, the result may indicate that elevated levels of Cyclin E promote DNA replication in parallel to E2F and can bypass a requirement for E2F-inducible genes early in S-phase. The notion that Cyclin E may not be the only functionally significant target of dE2F is supported by studies of the Drosophila PCNA gene, in which mutation of dE2F binding sites in the PCNA promoter reduced the ability of PCNA transgenes to rescue null mutation of PCNA (Thacker et al., 2003). Analogous studies have yet to be reported for other target genes and currently, it is unknown how many of the E2F-inducible genes are rate-limiting for E2F-mediated control of cell proliferation.
de2f1 mutants are lethal at the early larval stages (Duronio et al., 1995; Brook et al., 1996; Royzman et al., 1997). To circumvent this problem, we have used dsRNA to reduce the expression of dE2F1 in a spatially restricted manner during fly development. de2f1-dsRNA transgenes knock-down the level of dE2F1 protein, reducing dE2F1-dependent transcription, and decreasing cell proliferation (Morris, 2008). Expression of de2f1-dsRNA transgenes with either GMR-GAL4 or patched-GAL4 (ptc-GAL4) drivers gives visible phenotypes in the wing and the eye that are dose-sensitive and amenable to modification. In this study we collected mutant alleles of known dE2F1 target genes and took advantage of de2f1-dsRNA transgenes to ask whether halving the gene dose of a few, or most, or all of these dE2F1-dependent genes would affect cell proliferation in settings where dE2F1 activity is limiting.
RESULTS
Mutant alleles of dE2F1-dependent genes modify de2f1-dsRNA phenotypes
Previous studies have provided lists of genes that are dependent on each of the Drosophila E2F or RBF proteins (Cayirlioglu et al., 2003; Dimova et al., 2003; Stevaux et al., 2005). Dimova at al., depleted S2 cells of each component of the E2F/RBF network and identified genes whose expression changed when either dE2F1 or dE2F2 were targeted. These E2F-regulated genes were subdivided into 5 groups (A-E) depending on the relative importance of dE2F1-mediated activation or dE2F2-mediated repression and included 119 genes (Groups A-C) that were dependent on dE2F1 (Dimova et al., 2003). Chromatin immunoprecipitation (ChIP) analysis of a representative set of promoters suggests that at least 70% of these targets are likely to be directly regulated by dE2F1 (Dimova et al., 2003). The list of dE2F1-dependent genes includes genes with known functions in DNA replication and mitosis. The caveat to this analysis is that the functional significance of these changes is unclear. Potentially, each one of these changes in gene expression might be sufficient to block cell proliferation. Alternatively, since many replication proteins are abundant and stable, one might predict that very few of these transcriptional changes would have a measurable effect on cell proliferation, even if sustained for a long period of time.
As a first step towards understanding the significance of these targets for the in vivo functions of de2f1, we assembled a collection of mutant alleles from public stock centers that represented 44 of the 119 dE2F1-regulated genes identified by Dimova et al. We selected previously characterized loss-of-function alleles, where possible, or chose P-element insertions in the 5’ region of the genes. We used de2f1-dsRNA transgenes to reduce the level of dE2F1 in either the eye (using GMR-GAL4 to drive expression of the dsRNA) or the wing (using ptc-GAL4, (Morris, 2008)) and then asked, in this context where dE2F1 activity is limiting for cell proliferation, whether halving the gene dose of any of the 44 dE2F1-dependent genes would modify the phenotype. Reducing the copy number of a rate-limiting target of dE2F is expected to further reduce cell proliferation, and would therefore enhance de2f1-dsRNA-induced phenotypes. In contrast, if the expression of a dE2F1-regulated gene was in excess of that needed for cell proliferation then halving the gene dose would not be expected to affect the phenotype. As a control, none of the heterozygous mutant alleles gave a rough eye phenotype or disrupted wing development in the absence of de2f1-dsRNA. In addition, the de2f1-dsRNA-induced phenotypes used here were fully suppressed by co-expression of dE2F1 from UAS-de2f1 transgenes (data not shown), indicating that the phenotypes are solely due to loss of E2F1 expression.
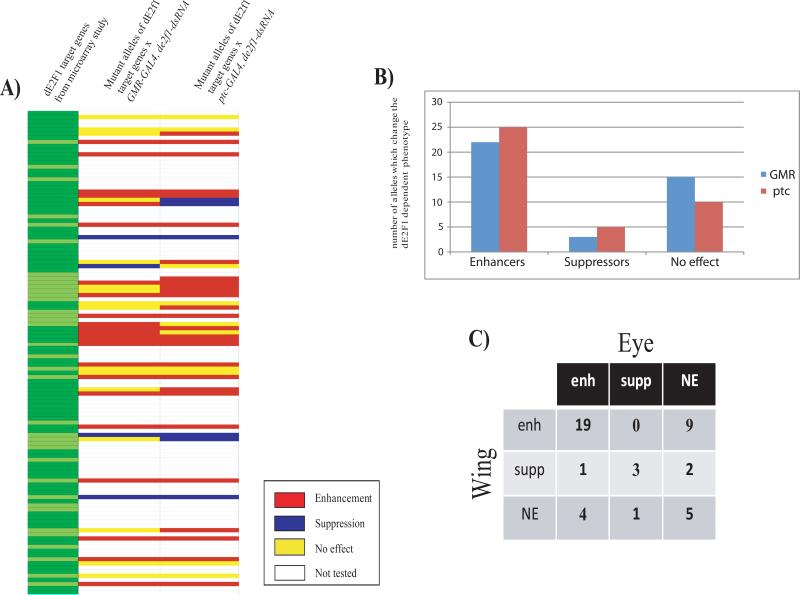
Nineteen of the alleles tested enhanced de2f1-dsRNA-induced phenotypes in both the eye and the wing (Figure 1 and Table 1). For example and as expected, multiple alleles of Cyclin E enhanced both GMR-GAL4, de2f1-dsRNA and ptc-GAL4, de2f1-dsRNA phenotypes, consistent with the idea that the level of Cyclin E expression is rate-limiting when dE2F1 is needed to drive cell proliferation in vivo. ptc-GAL4 is expressed in cells that will form the L3/L4 intervein region of the adult wing and each intervein cell is marked with a hair making it possible to distinguish between effects on cell number and cell size. ptc-GAL4 driven expression of de2f1-dsRNA reduced cell proliferation and mutant alleles enhanced this phenotype, which can be seen by the reduced number of hair in a marked area when compared to the control (Figure 2C). This suggests that enhancement was primarily due to a change in cell number.
FIGURE 1. Mutant alleles of many dE2F1-dependent genes modify dE2F1-dependent phenotypes.
1A) A heat map summarizing the genetic interactions observed when mutant alleles of dE2F1-regulated genes were tested for their ability to dominantly modify visible phenotypes caused by dsRNA-mediated targeting of dE2F1 in the eye (GMR-GAL4;de2f1-dsRNA) or the wing (ptc-GAL4;de2f1-dsRNA). Alleles were tested for genes of dE2F1 targets that were identified in previous microarray studies (Dimova et al., 2003). Enhancers are shown in red, suppressors in blue, mutant alleles that had no effect are shown in yellow and alleles which where not tested in white. For details of the alleles used and the strength of the interaction see Table 1.
1B) Bar graph illustrating the number of mutant alleles that enhanced, suppressed, or had no effect on the eye phenotype (GMR, blue) or the wing phenotype (ptc, red).
1C) Table showing the actual number of mutants which either affected both eye and wing phenotype, which affected one but not the other, or which had no effect on either of the two phenotypes. Abbreviations: enhanced (enh), suppressed (supp) or no effect (NE).
TABLE 1.
Table showing the tested alleles from Figure 1 and Dimova et. al. compared with the effects of halving the dosage of each of the dE2F1 target genes on the dE2F1 dependent phenotype in either the eye (GMR-GAL4) or wing (ptc-GAL4) with the following abbreviations: enhanced (enh), suppressed (supp) or no effect (NE).
| dE2F1 target gene | Bloomington number and allele description | effect with GMR-GAL4 | effect with ptc-GAL4 |
|---|---|---|---|
| NetA | #12856, w1118 P{GT1}NetA, BG02298 | NE | NE |
| mus209 | #11192, cn[1] P{ry[+t7.2]=PZ}mus209[02448]/CyO; ry[506] | 20% enh | NE |
| Df31 | #10579, y[1] w[67c23]; P{w[+mC]=lacW}Df31[k05815]/CyO | NE | 20% enh |
| stg | #2500, ru1 h1 th1 st1 cu1 sr1 es stg4 ca1/TM3, Sb1 Ser1 | 40% enh | 20% enh |
| CycE | #11396, P{ry[+t7.2]=PZ}CycE[05206] cn[1]/CyO; ry[506] | 100% enh | 40-60% enh |
| dup | #7275, dup[a1]/CyO | 100% enh | 80% enh |
| ncd | #1720, Df(3R)ca[nd1], Sb[1] ss[1] Ubx[bx-34e] e[s] ncd[1] ca[nd1]/TM3 | 80% enh | NE-20enh |
| Mcm2 | #12122, ry[506] P{ry[+t7.2]=PZ}Mcm2[rL074]/TM3, Sb[1] | NE | 20% supp |
| CG7530 | #6766, y[1] w[*]; P{y[+t7.7]=Mae-UAS.6.11}CG7530[UY713] | 20% enh | 20% supp |
| Cap-G | #15944 (EP), y[1] w[67c23]; P{w[+mC] y[+mDint2]=EPgy2}CG12400[EY04850]/CyO | 20% enh | 40-60% enh |
| Orc2 | #9014 (ems), mwh[1] Orc2[1] e[1]/TM1; #15940 (EY), y1 w67c23; P{EPgy2}Orc2EY04752 | 9014: 80% supp, 15940 100% supp | 9014: 20% supp, 15940 NE |
| RnrL | #10644, y[1] w[67c23]; P{w[+mC]=lacW}RnrL[k06709]/CyO | NE | 40% enh |
| CG13345, tumbleweed | #15072, y[1] w[67c23]; P{w[+mC] y[+mDint2]=EPgy2}tum[EY01491b] P{EPgy2}shot[EY01491a] | 100% supp | NE |
| nod | #7252, nod[DR3] f[1]/C(1)DX, y[1] f[1]/Dp(1;Y)B[S] | variable phenotype | female lethal |
| CG31453 pch2 | #15536 (EP), y[1] w[67c23]; P{w[+mC] y[+mDint2]=?EPgy2}pch2[EY01788a] P{EPgy2}EY01788b | 60% enh | 20% enh |
| Hrb87F | #14414, y[1] w[67c23]; ry[506] P{y[+mDint2] w[BR.E.BR]=SUPor-P}Hrb87F[KG02089] | NE | 60% enh |
| CG15500, mod | #10312, y[1] w[1118]; P{w[+mC]=lacW}mod(mdg4)[L3101]/TM3, Ser[1] | NE | 40% enh |
| CG10712, chro | #12932, y[1]; P{y[+mDint2] w[BR.E.BR]=SUPor-P}Chro[KG03258] ry[506]/TM3, Sb[1] Ser[1] | 40-60% enh | 20% enh |
| cdc2c | #6632, w[*]; cdc2c[2]/TM3, Sb[1] P{w[+mC]=35UZ}2 and #6636, w[*]; cdc2c[3]/TM3, Sb[1] P{w[+mC]=35UZ}2 | NE | NE |
| pav | #4384, y[1] w[1]; P{w[+mC]=lacW}64A pav[B200] th[1] st[1] cu[1] sr[1] e[s] ca[1]/TM6B, P{w[+mC]=iab-2(1.7)lacZ}6B, Tb[1] | NE | 40-60% enh |
| CG1888 | #15570 (EP), y[1] w[67c23]; P{w[+mC] y[+mDint2]=EPgy2}CG1888[EY02539] | 40% enh | 40% enh |
| Map60 | #14880, y[1]; P{y[+mDint2] w[BR.E.BR]=SUPor-P}Map60[KG00506]/SM6a; ry[506] | 100% enh | NE |
| polo | #13941, y[1]; P{y[+mDint2] w[BR.E.BR]=SUPor-P}polo[KG03033] ry[506]/TM3, Sb[1] Ser[1] | 13941: NE, 11543:80% enh | 13941: 20% enh, 11543: NE |
| and #11543, P{ry[+t7.2]=PZ}polo[01673] ry[506]/TM3, ry[RK] Sb[1] Ser[1] | |||
| CG6874, neo | #10264, mwh[1] P{hsneo}l(3)neo26[1] red[1] e[1]/TM3, ry[RK] Sb[1] Ser[1] | 40-60% enh | NE |
| CG9273, RPA2 | #12882, y[1] P{SUPor-P}rdgA[KG00771a]; P{SUPor-P}RPA2[KG00771b] | 50-70% enh | 40-60% enh |
| Chrac-14 | #13190, y[1] w[67c23]; P{y[+mDint2] w[BR.E.BR]=SUPor-P}mus201[KG01051] Chrac-14[KG01051] | 100% enh | 20-40% enh |
| Hel25E | #11043, y[1] w[67c23]; P{w[+mC]=lacW}Hel25E[k11511]/CyO | 50-80% enh | 20% enh |
| CG10522, sticky | #13471, y[1]; P{y[+mDint2] w[BR.E.BR]=SUPor-P}sti[KG01697] ry[506]/TM3, Sb[1] Ser[1] | 20-50% enh | 60-80% enh |
| dap | #11377, cn[1] P{ry[+t7.2]=PZ}dap[04454]/CyO; ry[506] | NE | NE-40% enh |
| Sd | #101498, Hex-C[nNC1] / SM1 | NE | NE |
| CG1558, Kmn1 | #11877, w[67c23] P{w[+mC]=lacW}Kmn1[G0237]/FM7c | 20% enh | 60% enh |
| DNAprim | #12108, y[1] w[*]; P{w[+mC]=lacW}DNApol-alpha60[j10B2]/TM3, Sb[1] | NE | 20% enh |
| His2Av | #11650, ry[506] P{ry[+t7.2]=PZ}His2Av[05146]/TM3, ry[RK] Sb[1] Ser[1] | 80% enh | 20% enh |
| CG3183, geminin | #11118, y[1] w[67c23]; P{w[+mC]=lacW}geminin[k14019]/CyO | 40% enh | 60 - 100% enh |
| RfC40 | #5157, bw[1]; RfC4[A18] st[1]/TM6B, Tb[1]and #5158, bw[1]; RfC4[B6] st[1]/TM6B, Tb[1] | 5157:100% supp, 5158:NE | 5157: 20% supp, 5158: NE |
| Orc5 | #3593, b[1] Orc5[2] elA[1] rd[s] pr[1] cn[1]/CyO | variable: 100% supp/NE | 60% supp |
| CG10336 | #15234, y[1]; P{y[+mDint2] w[BR.E.BR]=SUPor-P}CG10336[KG10159]/CyO; ry[506] | 30% enh | 40% enh |
| CG5949, DNApol-delta | #17109 (EP), w[1118]; P{w[+mC]=EP}DNApol-delta[EP3292] Hip14[EP3292] | 100% supp | 20% supp |
| Rrp1 | #13700, y[1] w[67c23]; P{y[+mDint2] w[BR.E.BR]=SUPor-P}Rrp1[KG01159] | NE | 60% enh |
| CG10489, pole 2 | #13486, y[1] w[67c23]; P{y[+mDint2] w[BR.E.BR]=SUPor-P}Pole2[KG02588] ry[506] | 60-80% enh | 40% enh |
| Bub1 | #10526, y[1] w[67c23]; P{w[+mC]=lacW}BubR1[k03113]/CyO | 60-80% enh | 20% enh |
| CG17064, mars | #19094, w1118; PBac{WH}marsf07689 | NE | NE |
| neb | #10391, y[1] w[67c23]; P{w[+mC]=lacW}neb[k05702]/CyO | variable, NE & enh | NE |
| CG7341 | #15952 (EP), y[1] w[67c23]; P{w[+mC] y[+mDint2]=EPgy2}CG32195[EY05483] CG7341[EY05483] | 20% enh | 40% enh |
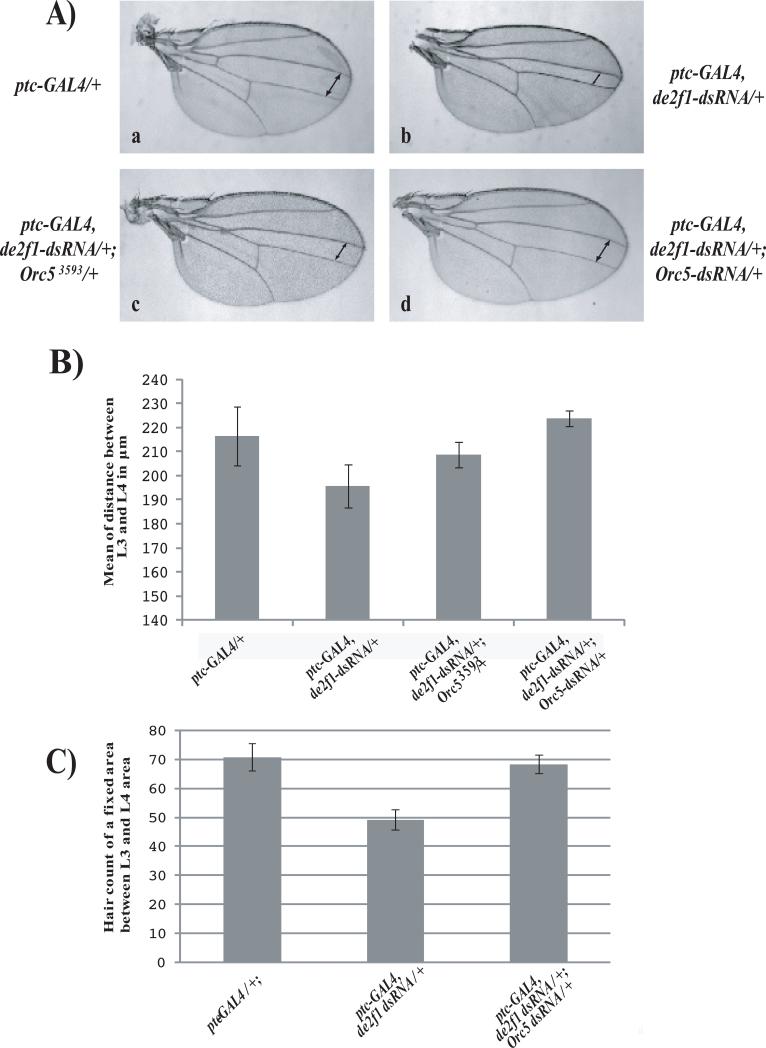
FIGURE 2. ORC5 reduction suppresses the de2f1-dsRNA phenotype in the wing.
2A) The distance between the L3 and L4 interveins of wild-type wings (a, indicated by arrows) is reduced by the expression of the de2f1-dsRNA transgene (b) from a ptc-GAL4 driver. Note that this effect is significantly suppressed by either halving the dosage of OCR5 (c, orc53593) or by expression of an Orc5-dsRNA transgene (d).
2B) Quantification of L3-L4 intervein distance (μm) for each genotype depicted in a-d (mean ± SD, n = 10 individual females per genotype).
2C) Barr graph illustrating wing hair counts from adult wings of a fixed area between L3 and L4 showing that reducing the levels of dE2F1 resulted in a decrease in cell number when compared to wild-type counts and this reduction was suppressed by the simultaneous expression of an Orc5-dsRNA transgene. (mean ± SD, n = 10 individual females per genotype).
These interactions indicate that the reduced expression of many different dE2F1-dependent genes is sufficient to reduce cell proliferation in vivo in contexts where the levels of dE2F1 are limiting; i.e. many different dE2 F-regulated genes can be rate-limiting for dE2F1-dependent cell proliferation. Interestingly, some mutant alleles enhanced the de2f1-dsRNA-induced phenotype in either the eye or the wing, but not both tissues. For example, recombination repair protein 1 (Rrp1), a gene involved in DNA repair, strongly enhanced the wing phenotype but had no effect in the eye. In total, sixteen mutants had different effects on the two phenotypes (36%, Figure 1C). This data suggests that in different tissues different dE2F1-dependent genes are important for dE2F1-dependent cell proliferation. Remarkably, just five alleles had no effect on either phenotype, indicating that for these genes, the level of gene expression was not close to an important threshold.
Reduced expression of ORC5 suppresses de2f1-dsRNA-induced phenotypes
A very curious feature of the results shown in Table 1 is that several mutant alleles were found to suppress, rather than to enhance, the de2f1-dsRNA-induced phenotypes. This type of interaction is unexpected given the well-established model that E2F co-ordinates the expression of genes that are needed for cell proliferation. Suppression might be expected if a dE2F1 target gene were to encode a negative regulator of cell proliferation, and were part of a negative feedback loop. However, somewhat paradoxically, the suppressor alleles included mutations in genes that are needed for cell proliferation. For example, one of the strongest suppressors of the ptc-GAL4, de2f1-dsRNA phenotype was an allele of Orc5, a gene that is needed for DNA replication in imaginal discs (Pflumm and Botchan, 2001). Importantly, this suppression was evident using two independent alleles: a loss-of-function mutant allele of Orc5 and a transgene expressing Orc5-dsRNA. Suppression was quantified by measuring the distance between veins L3 and L4 in the adult wing. Reducing the levels of ORC5 with the two independent Orc5 alleles returned the L3/L4 intervein distance to nearly wild-type levels (Figure 2A and 2B). Cell counts of a specific area between L3 and L4 confirmed that the number of cells generated when dE2F1 and ORC5 were both depleted was greater than when dE2F1 was targeted alone (Figure 2C).
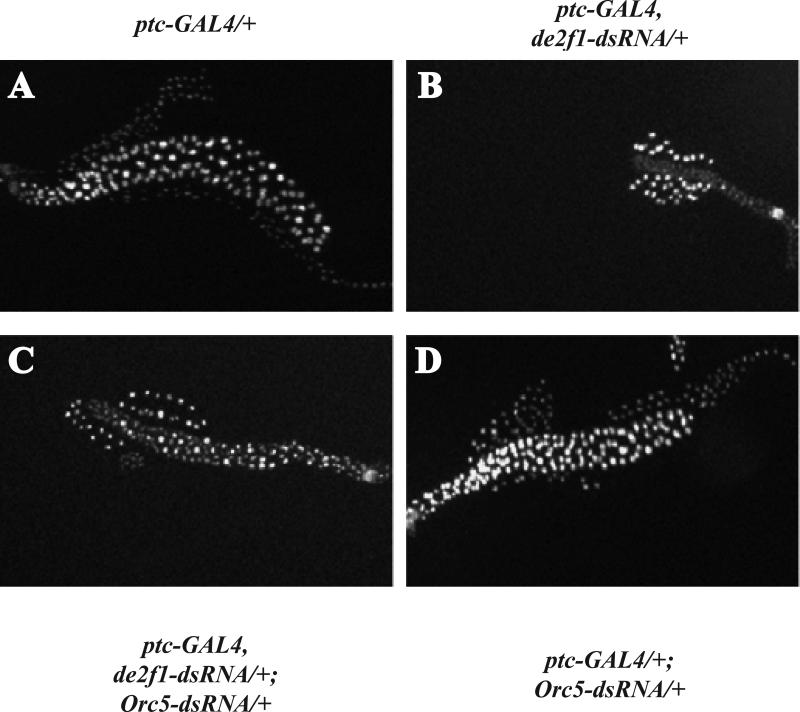
Similar genetic interactions between de2f1 and Orc5-dsRNA were seen in the eye and also in salivary glands. Drosophila salivary glands contain giant chromosomes that are formed by approximately 10 successive rounds of replication without any further cell division during embryonic and larval development; hence, salivary glands grow by increasing the volume of individual cells (Andrew et al., 2000). ptc-GAL4 drives expression in both the developing wing and the salivary gland, making it possible to examine the effects of de2f1-dsRNA expression in two different tissues within the same animal. Consistent with previous studies (Royzman et al., 1997), reducing the levels of dE2F1 during salivary gland development strongly decreased the overall size of the gland, producing cells and nuclei that were much smaller than wild-type glands or surrounding fat body cells (Figure 3b). When squashed, these small and fragile glands gave very thin chromosomal spreads that stained weakly for DAPI and appeared to contain under-replicated regions (data not shown). Importantly, these abnormalities were largely suppressed when ORC5 was co-depleted with dE2F1 (Figure 3c). Taken together, these results indicate that reducing ORC5 levels suppresses the effects of reducing dE2F1 in multiple tissues and different developmental contexts.
FIGURE 3. Reducing ORC5 levels suppress the size defects caused by ectopic expression of de2f1-dsRNA in salivary glands.
Hoechst staining of whole salivary glands from wandering third instar larvae demonstrates that knock-down of dE2F1 results in a strong size reduction (b) when compared to wild-type (a). This phenotype is strongly suppressed when ORC5 levels are reduced at the same time (c). Expression of Orc5-dsRNA alone had no detectable effect on salivary gland development (d).
Reducing the levels of ORC5 improves S-phase progression in cells with low levels of dE2F1
To better understand the genetic interaction between Orc5 and de2f1, we examined wing discs of third instar larvae, which are the precursors of the adult wing.
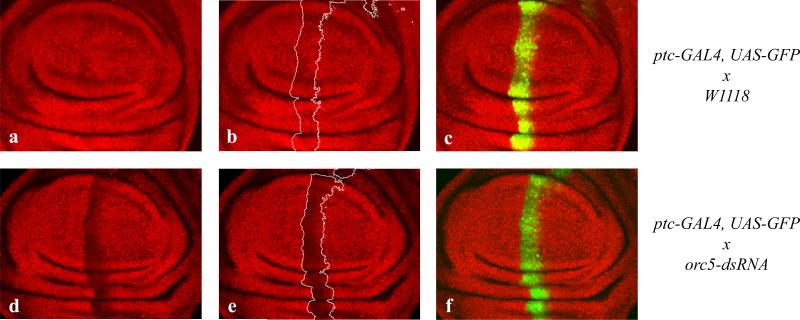
Expression of de2f1-dsRNA reduced the number of cells that incorporate BrdU, as expected (Figure 4Ab). Some BrdU positive cells remained, suggesting that dE2F1 levels were reduced by the dsRNA treatment, but that a level of dE2F1 function was still present. No change in BrdU incorporation was observed in the ptc-GAL4 stripe following the expression of Orc5-dsRNA (Figure 4Ad), even though the levels of ORC5 protein were clearly reduced by the treatment when we stained wing discs with an antibody against ORC5 (Figure 5). Studies have shown that the homozygous mutation of Orc5 strongly inhibits DNA replication in imaginal discs (Pflumm and Botchan, 2001). We infer that the expression of Orc5-dsRNA reduces, but does not eliminate ORC5, and that the residual level of ORC5 protein is sufficient for DNA replication. However, reducing the levels of ORC5 ameliorated the effects of depleting dE2F1 as the number of BrdU positive cells seen in ptc-GAL4, de2f1-dsRNA-expressing cells increased when ORC5 levels were reduced (Figure 4Ac) and it appears that the intensity of BrdU labeling is reduced in the de2f1- Orc5-dsRNA double knock-down. Although this is consistent with our model, we believe that additional experiments would be necessary to make a general statement from this observation. This effect was also very clear in the eye disc. In this tissue GMR-GAL4 driven expression of de2f1-dsRNA broadened the stripe of BrdU-positive cells in the second mitotic wave, an effect that was strongly suppressed by the expression of Orc5-dsRNA (Figure 4B).
FIGURE 4. Decreasing ORC5 levels suppresses de2f1-dsRNA effects on S-phase progression.
4A) Immunohistochemical staining of BrdU (red) of ptc-GAL4-driven de2f1-dsRNA expressing 3rd-instar wing discs showed a significant reduction in BrdU positive cells (b, b’) when compared to wild-type discs (a, a’). Knock-down of ORC5 at the same time restored the normal BrdU pattern (c, c’) and expression of Orc5-dsRNA alone had no detectable effect on S-phase progression (d, d’). The ptc-expressing stripe is indicated by expression of the UAS-GFP transgene (a”-d”).
4B) BrdU incorporation in red in third instar larvae eye discs. GMR-GAL4 driven expression of de2f1-dsRNA broadens the stripe of S-phase cells and this effect is suppressed by co-expression of Orc5-dsRNA
FIGURE 5. Knock-down of ORC5 causes a strong decrease in ORC5 protein levels.
Immunohistochemical staining of ORC5 (red) in ptc-GAL4-driven Orc5-dsRNA expressing 3rd-instar wing discs show a strong reduction in ORC5 positive cells (d, f) when compared to wild-type discs (a, b). c-f show a merge between ORC5 (red) and GFP (green) to mark the ptc-GAL4 stripe.
Reducing the levels of ORC5 selectively increases transcription of dE2F1-regulated genes
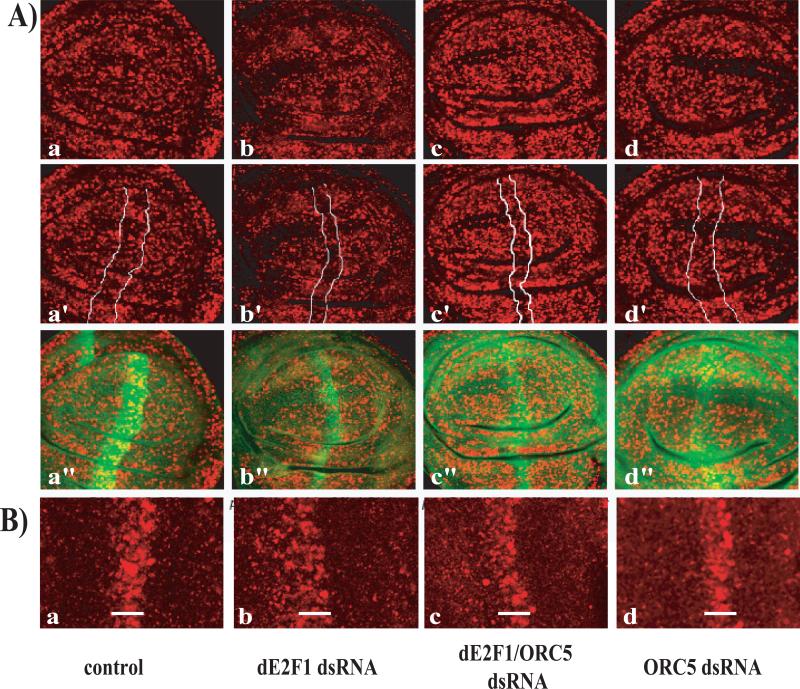
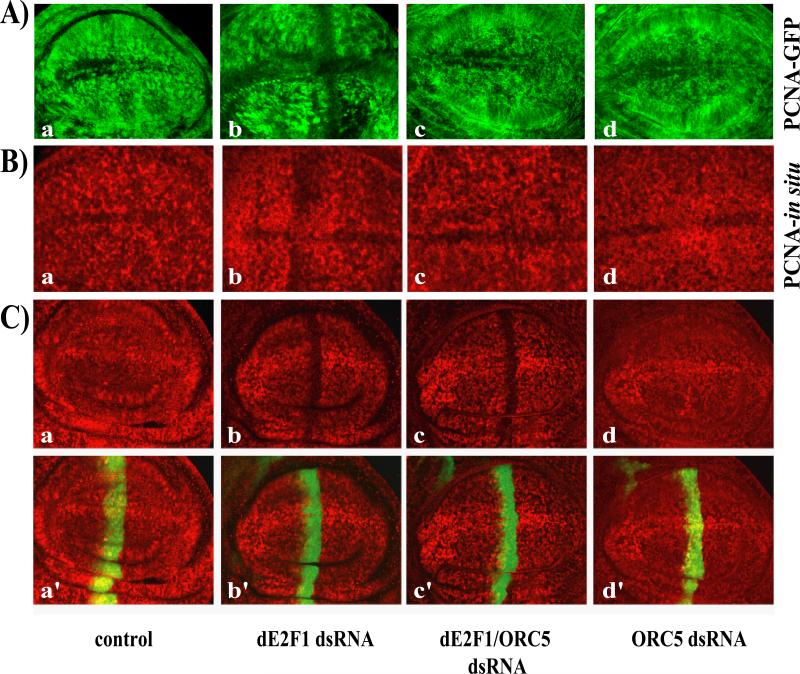
We next examined the effects of ORC5-depletion on the transcriptional activity of dE2F1. The PCNA promoter is one of the best-characterized dE2F1-regulated targets and PCNA-GFP has been used in several studies to provide an in vivo readout of dE2F-activity (Thacker et al., 2003). de2f1-dsRNA expression strongly decreased the expression of the PCNA-GFP reporter (Figure 6Ab and(Morris, 2008), an effect that could be suppressed by over-expressing dE2F1 (data not shown). Expression of Orc5-dsRNA had no effect on PCNA-GFP expression when tested alone (Figure 6Ad), but restored PCNA-GFP expression in discs expressing de2f1-dsRNA (Figure 6Ac), suggesting that reducing ORC5 levels increases the activity of dE2F1. A similar result was obtained using in situ hybridization to monitor the expression of the endogenous PCNA gene: PCNA expression was reduced by expression of de2f1-dsRNA with ptc-GAL4 (Figure 6Bb) and this decrease was suppressed by the co-expression of Orc5-dsRNA (Figure 6Bc). These results suggest that reduced ORC5 levels suppress the effects of dE2F1 depletion by enhancing, or sustaining, dE2F1 activity.
FIGURE 6. Reducing the levels of ORC5 restores dE2F1 activity but this is independent of dE2F1 protein levels.
6A) Third instar larvae wing discs expressing a PCNA-GFP reporter construct were immunostained with antibodies against GFP (6Aa-d) or probed for PCNA expression by in situ hybridization (6Ba-d). Larvae expressing de2f1-dsRNA driven by ptc-GAL4 (Ab and Bb) exhibit a reduction in PCNA in the area of the ptc-GAL4 stripe (arrow) when compared to wild-type (Aa, Ba). The reduction in PCNA staining was suppressed when Orc5-dsRNA was expressed concomitantly (Ac, Bc). Discs expressing Orc5-dsRNA alone showed no effect on PCNA expression (Ad, Bd).
6C) Immunofluorescent staining of third instar wing discs using antibodies against dE2F1 (red) and GFP (green, to mark the ptc-GAL4 stripe, a’-d’) demonstrate that larvae which express de2f1-dsRNA show a reduction of dE2F1 protein levels (b), when compared to wild-type (a). Reducing the levels of ORC5 simultaneously did not interfere with the targeting of dE2F1 (c) and expression of Orc5-dsRNA alone did not change the overall levels of dE2F1 (d).
Importantly, the restoration of dE2F1 activity seems to be independent of dE2f1 protein levels. Immunostaining experiments showed that ptc-GAL4 driven expression ofde2f1-dsRNA strongly reduced the level of dE2F1 protein in the stripe of ptc-GAL4-expressing cells marked by GFP (Figure 6Cb and (Morris, 2008) and this decrease was not overtly changed in animals that are heterozygous for Orc5 or in cells co-expressing Orc5-dsRNA and e2f1-dsRNA (Figure 6Cd, 6Cc). With the caveat that we have only qualitatively measured E2F1 levels, and do not distinguish between transcriptionally active and inactive forms of the dE2F1, this indicates that reducing the level of ORC5 did not interfere with the general targeting of dE2F1.
DISCUSSION
These experiments were inspired by microarray studies showing that E2F regulates the expression of hundreds of genes, thereby coordinating the elevated transcription of a broad set of targets with the G1/S transition (Ishida et al., 2001; Muller et al., 2001; Stanelle et al., 2002). Gene expression profiles raise the thorny issue of functional significance. It is unlikely that all of the E2F targets are equally important for the control of cell proliferation, but it is unclear which transcriptional changes are the most relevant. The impact of E2F regulation on an individual target gene depends on many parameters including the action of other transcription factors on the promoter, the basal level of gene expression, and the fold-increase in transcription that is needed for gene function. The significance of an E2F-induced transcriptional change also depends on the overall importance of the encoded protein and the extent of post-transcriptional regulation. Inevitably, some of these parameters will vary in different cellular environments, and even if one could answer this question, it is unclear which cellular context would provide a complete answer. The idea that E2F regulates cell proliferation is well established but it remains unclear which, or how many, of the E2F-dependent changes in gene expression that have been observed have any impact on cell proliferation.
To shed light on this question we took advantage of the availability of Drosophila mutants in dE2F1-regulated genes, and the recent development of transgenic lines that allow the levels of dE2F1 to be reduced by the expression of dsRNA. Insufficient dE2F1 causes visible, dosage-sensitive phenotypes. We have used these to compare the effects of halving the gene dosage of many different dE2F1-regulated genes in contexts where dE2F1 activity is limiting for cell proliferation (Morris, 2008). Halving the gene dose is thought, in most cases, to reduce gene expression and a genetic interaction in this type of assay identifies dE2F1-target genes that are close to a critical threshold. We were unable to obtain mutant alleles for all of the genes that are known to be dependent on dE2F1, but we have examined a large enough number for a clear picture to emerge: in this sensitized genetic background most of the mutant alleles tested modified a dE2F1-dependent phenotype in at least one of the two tissues examined. This implies that many dE2F1-dependent genes are within a two-fold range of being limiting for dE2F1-dependent cell proliferation and suggests that there is likely to be a strong selective pressure to retain dE2F1 regulation at most of its targets. This may explain why similar sets of genes are controlled by E2F in species as diverse as flies, mammals and plants.
This survey of dE2F1 target genes gave a second and surprising insight. It is widely accepted that E2F promotes the expression of genes that are needed for the cell division cycle. Therefore; loss of function mutations in key E2F target genes are expected to enhance the effects of reduced E2F activity (Blais and Dynlacht, 2004; Bracken et al., 2004; Dimova and Dyson, 2005; DeGregori and Johnson, 2006). Unexpectedly, however, we found mutant alleles in a subset of E2F-target genes that had the converse effect and suppressed the effects of reduced dE2F1. We focused on one of the strongest suppressors of the de2f1-dsRNA phenotypes, Orc5.
While Orc5 alleles and Orc5-dsRNA gave the strongest suppression of the de2f1-dsRNA phenotypes in our assays, mutant alleles affecting several different replication functions also partly suppressed one or both phenotypes (Table 1). These results suggest that the mechanism underlying this interaction can be triggered by reducing the level of replication proteins in general rather than by ORC5 in particular. Given that dE2F1 is transiently activated during G1/S progression, the most likely explanation is that reducing the levels of ORC proteins prolongs the period of the time that cells spend in the window of the cell cycle where dE2F1 is active, giving more time for transcription of dE2F1-target genes.
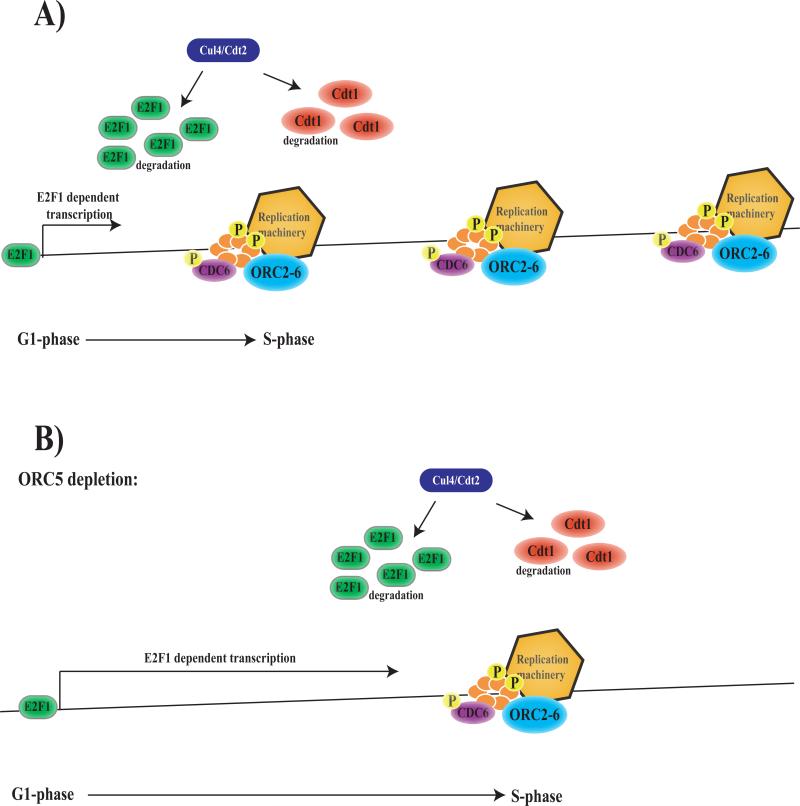
Recent work has shown that dE2F1 degradation during S-phase requires a PCNA-interacting-protein (PIP) motif in dE2F1 and the Cul4/Cdt2 E3 ubiquitin ligase (Figure 7A) (Shibutani et al., 2008) in a manner similar to that described for the pre-replication complex protein, Cdt1 (Hu et al., 2004; Arias and Walter, 2005; Arias and Walter, 2006; Hu and Xiong, 2006; Jin et al., 2006; Senga et al., 2006). The idea that E2F proteins activate transcription for a limited period of time during the G1 to S transition, and the finding that active dE2F1 protein is degraded by a mechanism that is coupled to the initiation of DNA replication, provide a simple model to explain these genetic interactions. The onset of dE2F1-dependent transcription is triggered by the activation of G1 Cdk's. The cessation of dE2F-dependent transcription may be determined, at least in part, by replication-coupled turnover of dE2F1. In cells where origins of replication are widely spaced, or where DNA synthesis is slow, it may take longer to remove the pool of DNA-bound and active dE2F1 (Figure 7B). Hence, the depletion of proteins like ORC5, that reduce the density or slow the firing of replication origins, has the consequence of enhancing the level of dE2F1-dependent transcription without altering the levels of dE2F1 protein. It is clear from this model why changes in only a subset of E2F-regulated genes will give this effect, since any mutations or dsRNA treatments that influence cell cycle progression earlier, or later, in the cell cycle would not have the same consequence. In addition, any changes that severely block replication or that affect the fidelity of DNA-synthesis would enhance de2f1-dsRNA phenotypes. Consistent with this model, we found that the homozygous mutation of Orc5 did not extend the development of de2f1 null mutant animals (data not shown), suggesting that some level of normal ORC5/dE2F1 function is necessary for the genetic interaction to occur.
FIGURE 7. Model.
7A) Transition from G1- to S-phase is at least in part dependent on the degradation of E2F1 in a replication dependent manner, thereby coupling the transcription of replication proteins to the initiation of replication (Shibutani et al., 2008).
7B) Reducing the levels of ORC5 might increase the distance between replication or slow the firing of replication origins. This would delay the degradation of DNA-bound and active E2F1, thereby prolonging the period of E2F1 dependent transcription, causing an increase in the levels of replication proteins available for the next cell cycle.
Why does this arrangement exist? Pre-RC complexes are licensed early in G1, long before the G1/S transition (Dutta and Bell, 1997; DePamphilis, 2003; Prasanth et al., 2004; Machida et al., 2005). Hence dE2F1-mediated induction of Orc gene expression at G1/S is unlikely to have any functional impact until the G1 phase of the subsequent cell cycle. We suggest that cells entering S-phase with relatively low levels of replication proteins experience a prolonged period of dE2F1 activity, and that this ensures that the levels of replication proteins are elevated in preparation for the next cell cycle. In this way, the genetic interaction between Orc5-dsRNA and de2f1-dsRNA may illustrate a homeostatic mechanism that occurs during cell proliferation to ensure that cells maintain a robust level of replication proteins.
It is important to note that there might be additional connections between E2F and ORC proteins that, conceivably, could also contribute to these genetic interactions. Previous studies have shown that dE2F1 and RBF are important in follicle cells for the selective re-replication of regions of the Drosophila genome (Bosco et al., 2001) raising the possibility that E2F/RBF proteins may act directly at origins of replication. We have been unable to co-immunoprecipitate ORC5 with dE2F1, however we cannot completely exclude the possibility that these proteins might physically interact. A previous study mapped the position of origins of replication, and the location of ORC-binding sites, in a significant portion of the Drosophila genome (MacAlpine et al., 2004; Celniker et al., 2009). Analysis of the ORC binding sites relative to the position of dE2F1-regulated genes revealed that the distance from the promoters of dE2F1-regulated genes to the nearest ORC binding sites varied from 2bp to 29kbp. Although many of the dE2F1-regulated genes were proximal to an ORC binding site, there was no clear correlation between E2F regulation and ORC binding.
It is also conceivable that ORC5 depletion could activate dE2F1 as part of a (hypothetical) replication checkpoint triggered by incompletely formed pre-RC complexes or stalled replication forks. We have crossed dsRNA lines targeting the Drosophila orthologs of Chk1, ATM and ATR, proteins that are components of known S-phase checkpoint pathways to the ptc-Gal4; dE2F1-dsRNA lines (Abraham, 2001; Syljuasen et al., 2005). Rather than acting as suppressors, these alleles enhanced the adult wing phenotype and had no effect on PCNA-GFP expression. Nevertheless it remains possible that ORC5 depletion might activate a novel checkpoint signal that links to dE2F1.
Finally, it is possible that dE2F1 activity may be influenced in ORC-depleted cells by additional, indirect effects. Studies in viral systems have shown that transcription factors can activate replication origins by assisting the binding of an initiator (for example ORCs), either through direct recruitment of the initiator protein and/or by changing the chromatin structure to reduce the inhibitory effects of chromatin (for review see (Kohzaki and Murakami, 2005). There is evidence that the impact of ORC proteins can extend beyond replication, an example of this is in the establishment of transcriptionally repressed domains at the S.cerevisiae silent mating type loci (for review see (Shore, 2001). Indeed, genetic studies have identified domains in ORC proteins that can separate functions in DNA replication and transcriptional silencing (Bell et al., 1995; Fox et al., 1995; Dillin and Rine, 1997). Drosophila ORC proteins promote the establishment or maintenance of heterochromatin (Pak et al., 1997; Huang et al., 1998) and have been shown to directly interact with the heterochromatin protein 1 (HP1). Mutations in ORCs suppress heterochromatin-dependent transcriptional repression (Prasanth et al.; Pak et al., 1997). While we saw no changes in the position or type of histone modifications in the ORC5-depleted S2 cells (data not shown), we cannot completely exclude the possibility ORC5-depleted cells have an altered chromatin structure that is more permissive for dE2F1-dependent transcription.
EXPERIMENTAL PROCEDURES
Fly stocks
The following alleles were used in this study: ptc-GAL4, e2f1-dsRNA (II) and GMR-GAL4, e2f1-dsRNA (II) (Morris, 2008); PCNA-GFP was a gift from B. Duronio (Thacker et al., 2003); Orc5-dsRNA was obtained from the National Institute of Genetics in Japan (www.shigen.nig.ac.jp); various mutant alleles were obtained from the Bloomington Stock Center.
Immunohistochemistry and in situ hybridization
The following antibodies were used in this study: anti-e2f1 (guinea pig, 1:100, a gift from T. Orr-Weaver), anti-BrdU (mouse, 1:50, Becton Dickson Bioscience), anti-GFP (rabbit, 1:1000, Invitrogen), anti-ORC5 (mouse, 1:500, a gift from M. Gossen). The following secondary antibodies were used: appropriate anti-species Alexa 488 and anti-species Cy3 used at 1:500 (Jackson ImmunoResearch). For immunostaining, the discs were fixed in 4% Formaldehyde/PBS-0.1% Triton 100 (PBT) for 20 min at room temperature (RT), washed three times with PBT for 20 min at RT and blocked in 5% goat serum/PBT for 1 h at RT. Discs were incubated with primary antibody in 5% goat serum/PBT over night at 4°C, followed by three washes with PBT for 20 min at RT. Discs were then incubated with the secondary antibody in 5% goat serum/PBT for 3 h at RT, followed by three washes in PBT for 20 min each at RT. The discs were mounted in Vectashield (Vector laboratories) for confocal microscopy. BrdU labeling: Wing discs from third instar larvae were labeled in Schneider's medium with 0.2mg/ml BrdU (Sigma) at RT for 30 min. Wing discs were fixed in 4% Formaldehyde/PBT for 20 min at RT and washed three times in PBT for 20 min each at RT. Following 2M HCl acid treatment for 30 min at RT, additional washes in PBT were performed and discs were rehydration in MeOH/PBT (3:1, 1:1, 1:3), BrdU was detected with a mouse α-BrdU antibody (1:5, Becton Dickinson) and GFP with rabbit α-GFP (1:1000, Invitrogen). Cy3 donkey α-mouse (1:500, Jackson Immunolaboratories) was used as a secondary antibody. DNA staining was observed with Hoechst dye (1:1000, 10mg/ml). Images were collected on a Zeiss LSM510 confocal microscope or on a Nikon 90-I Fluorescent microscope. For in situ hybridization samples were prepared as described (Du, 2000) and the PCNA riboprobes were obtained by in vitro transcription using a DIG RNA labeling kit (Roche). The PCNA probe was detected using mouse anti-DIG-AP Fab fragment antibody (1:150, Roche) and anti-mouse Cy3 (1:500, Jackson ImmunoResearch) as a secondary antibody.
Analysis of wings
Wings were dissected, deposited on a slide and isopropanol was pipetted onto the slide. After evaporation of the isopropanol, Canada balm was pipetted around the wings and a cover slip was gently laid over the wing preparation. Microscopic images were captured with a Nikon CCD camera. To analyze wing hair numbers, hair were counted in a defined area of constant size. The values obtained for wings from flies from the genotype ptc-GAL4/ + were set as 100%.
Bullet points.
We have used a list of E2F-regulated genes that has been generated by microarray analysis for a genetic approach to ask which of these targets are important for E2F function in vivo.
Our results show that more than half of the genes tested are able to dominantly modify dE2F1-dependent phenotypes, indicating that the expression levels of most dE2F1-regulated genes are rate-limiting for dE2F1 function.
We show that the depletion of Orc5 elevates the expression of dE2F1-regulated genes.
Our results reveal the existence of an important feedback mechanism that can elevate dE2F1-dependent transcription in cells where ORC levels are low.
ACKNOWLEDGMENTS
We thank Terry Orr-Weaver and Bob Duronio for dE2F1 antibodies, Manfred Gossen for the OCR5 antibody, Bob Duronio for the PCNA-GFP transgenic line used in this study and for sharing unpublished data. We also thank the members of the Dyson laboratory for advice and technical assistance. M.S.L. was supported by a Fellowship Award from the Charles A. King Trust and J.J. was supported in part by a Tosteson Postdoctoral Fellowship from the MGH. This study was supported by NIH grant HG004279 to D.M.M. and NIH grants CA64402, GM53203 and GM81607 to N.D.
References
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Ahlander J, Chen XB, Bosco G. The N-terminal domain of the Drosophila retinoblastoma protein Rbf1 interacts with ORC and associates with chromatin in an E2F independent manner. PLoS One. 2008;3:e2831. doi: 10.1371/journal.pone.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew DJ, Henderson KD, Seshaiah P. Salivary gland development in Drosophila melanogaster. Mech Dev. 2000;92:5–17. doi: 10.1016/s0925-4773(99)00321-4. [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 2005;19:114–126. doi: 10.1101/gad.1255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- Asano M, Nevins JR, Wharton RP. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev. 1996;10:1422–1432. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr Opin Genet Dev. 2004;14:527–532. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Bosco G, Du W, Orr-Weaver TL. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat Cell Biol. 2001;3:289–295. doi: 10.1038/35060086. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Brook A, Xie JE, Du W, Dyson N. Requirements for dE2F function in proliferating cells and in post-mitotic differentiating cells. Embo J. 1996;15:3676–3683. [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P, Ward WO, Silver Key SC, Duronio RJ. Transcriptional repressor functions of Drosophila E2F1 and E2F2 cooperate to inhibit genomic DNA synthesis in ovarian follicle cells. Mol Cell Biol. 2003;23:2123–2134. doi: 10.1128/MCB.23.6.2123-2134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML. The ‘ORC cycle’: a novel pathway for regulating eukaryotic DNA replication. Gene. 2003;310:1–15. doi: 10.1016/s0378-1119(03)00546-8. [DOI] [PubMed] [Google Scholar]
- Dillin A, Rine J. Separable functions of ORC5 in replication initiation and silencing in Saccharomyces cerevisiae. Genetics. 1997;147:1053–1062. doi: 10.1093/genetics/147.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 2003;17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolski SF, Stacey DW, Harter ML, Stine JT, Hiebert SW. An E2F dominant negative mutant blocks E1A induced cell cycle progression. Oncogene. 1994;9:2605–2612. [PubMed] [Google Scholar]
- Du W. Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain. Development. 2000;127:367–379. doi: 10.1242/dev.127.2.367. [DOI] [PubMed] [Google Scholar]
- Du W, Xie JE, Dyson N. Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. Embo J. 1996;15:3684–3692. [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH. Developmental control of the G1 to S transition in Drosophila: cyclin Eis a limiting downstream target of E2F. Genes Dev. 1995;9:1456–1468. doi: 10.1101/gad.9.12.1456. [DOI] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH, Xie JE, Brook A, Dyson N. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 1995;9:1445–1455. doi: 10.1101/gad.9.12.1445. [DOI] [PubMed] [Google Scholar]
- Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- Fox CA, Loo S, Dillin A, Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- Frolov MV, Moon NS, Dyson NJ. dDP is needed for normal cell proliferation. Mol Cell Biol. 2005;25:3027–3039. doi: 10.1128/MCB.25.8.3027-3039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- Hu J, Xiong Y. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem. 2006;281:3753–3756. doi: 10.1074/jbc.C500464200. [DOI] [PubMed] [Google Scholar]
- Huang DW, Fanti L, Pak DT, Botchan MR, Pimpinelli S, Kellum R. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J Cell Biol. 1998;142:307–318. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki J, Nevins JR, Sullenger BA. Inhibition of cell proliferation by an RNA ligand that selectively blocks E2F function. Nat Med. 1996;2:1386–1389. doi: 10.1038/nm1296-1386. [DOI] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1- interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- Kohzaki H, Murakami Y. Transcription factors and DNA replication origin selection. Bioessays. 2005;27:1107–1116. doi: 10.1002/bies.20316. [DOI] [PubMed] [Google Scholar]
- Landis G, Kelley R, Spradling AC, Tower J. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc Natl Acad Sci U S A. 1997;94:3888–3892. doi: 10.1073/pnas.94.8.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Herzinger T, Hansen K, Moroni MC, Resnitzky D, Helin K, Reed SI, Bartek J. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Ji J-Y, Yang F, Di Stefano L, Herr A, Moon N-S, Kwon E-J, Haigis KM, Naar AM, Dyson NJ. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008 doi: 10.1038/nature07310. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, DeGregori J, Leone G, Herendeen DR, Kelly TJ, Nevins JR. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Wang JY, Socolar JE, Iversen ES, Hartemink AJ, Haase SB. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature. 2008;453:944–947. doi: 10.1038/nature06955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Pflumm MF, Botchan MR. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–1707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Mendez J, Prasanth KV, Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Philos Trans R Soc Lond B Biol Sci. 2004;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth SG, Shen Z, Prasanth KV, Stillman B. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci U S A. 107:15093–15098. doi: 10.1073/pnas.1009945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I, Whittaker AJ, Orr-Weaver TL. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev. 1997;11:1999–2011. doi: 10.1101/gad.11.15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers WR, Rodgers JW, Kaelin WG., Jr. A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci U S A. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem. 2006;281:6246–6252. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ, 3rd, Reis T, Edgar BA, Duronio RJ. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell. 2008;15:890–900. doi: 10.1016/j.devcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D. Transcriptional silencing: replication redux. Curr Biol. 2001;11:R816–819. doi: 10.1016/s0960-9822(01)00493-6. [DOI] [PubMed] [Google Scholar]
- Stanelle J, Stiewe T, Theseling CC, Peter M, Putzer BM. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 2002;30:1859–1867. doi: 10.1093/nar/30.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O, Dimova DK, Ji JY, Moon NS, Frolov MV, Dyson NJ. Retinoblastoma family 2 is required in vivo for the tissue-specific repression of dE2F2 target genes. Cell Cycle. 2005;4:1272–1280. doi: 10.4161/cc.4.9.1982. [DOI] [PubMed] [Google Scholar]
- Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker SA, Bonnette PC, Duronio RJ. The contribution of E2F-regulated transcription to Drosophila PCNA gene function. Curr Biol. 2003;13:53–58. doi: 10.1016/s0960-9822(02)01400-8. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CL, Classon M, Dyson N, Harlow E. Expression of dominant-negative mutant DP-1 blocks cell cycle progression in G1. Mol Cell Biol. 1996;16:3698–3706. doi: 10.1128/mcb.16.7.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]