Abstract
Since its discovery two decades ago, the activation of the JAK/STAT pathway by numerous cytokines and growth factors has resulted in it becoming one of the most well studied intracellular signalling networks. The field has progressed from the identification of the individual components, to high-resolution crystal structures of both JAK and STAT, and an understanding of the complexities of the molecular activation and deactivation cycle which results in a diverse, yet highly specific and regulated pattern of transcriptional responses. While there is still more to learn, we now appreciate how disruption and de-regulation of this pathway can result in clinical disease and look forward to adoption of the next generation of JAK inhibitors in routine clinical treatment.
Keywords: JAK, STAT, signalling, cytokine, receptor, SOCS
The canonical JAK/STAT pathway
No longer “Just another kinase”, the Janus Kinase (JAK) protein tyrosine kinases are now recognised as an integral component of the cytokine receptor subunits, and enzyme activation, as the initiating step in a signalling cascade required for embryonic development, tissue growth, haemopoietic development and differentiation, innate and adaptive immunity and the inflammatory response.
Upon ligand binding to the cognate transmembrane receptors, two or more receptor-associated JAKs are brought into close proximity through receptor oligomerization to allow auto-phosphorylation and/or trans-phosphorylation by the opposing JAK kinase. Once activated, JAKs then phosphorylate signature tyrosine residues in the cytoplasmic region of the receptors to create docking sites for members of the signal transducers and activators of transcription (STAT) family, so named for their dual ability to function as cytoplasmic signalling modules and transcription factors (1). Upon binding to the receptor through their Src homology 2 (SH2) domain, the STATs themselves become a JAK substrate. Phosphorylation of the conserved tyrosine residue located between the SH2 domain and the C-terminal transactivation domain results in the formation of parallel STAT dimers, which are stabilized by reciprocal phosphotyrosine and SH2 domain interactions (2,3). Dimer formation is followed by translocation to the nucleus (4–9) where the STAT dimers bind to specific palindromic sequences within the promoter regions of target genes to initiate the appropriate transcriptional response (10,11).
Just another kinase: an historical perspective
The JAK/STAT pathway has become one of the most well studied signalling cascades of the past two decades. There are four members of the mammalian JAK family, JAK1-3 and tyrosine kinase 2 (TYK2). JAK1 and JAK2 were discovered using a degenerate PCR screen based on conserved residues in the catalytic units of known protein tyrosine kinases (12–14). Tyk2 was identified by homology screening to the c-fms kinase domain (15) and JAK3 was discovered shortly thereafter (16–18). As their significance became apparent, the acronym transitioned from being “just another kinase” to “Janus” kinase, after the two-faced Roman god Janus (19), and in reflection of the characteristic tandem kinase domains.
The intimate connection between JAK and the cytokine receptors was first revealed by an elegant series of experiments by Ian Kerr, George Stark, Sandra Pellegrini and colleagues. The somatic cell genetic screen resulted in six different complementation groups (U1–U6), which were unresponsive to IFNα (1,20–22). U2–U6 were also unresponsive to IFNβ and U2–U4 were unresponsive to IFNγ, strongly suggesting that the complementation groups were deficient in components both unique to and shared between the different ligand responses. Indeed, genetic studies revealed that U1A cells were deficient in Tyk2, whereas the U4–U6 cells were eventually found to be deficient in other components of the IFNβ and IFNγ JAK-STAT signaling cascades (1,23–25). An analogous screen selecting for cells that were only unresponsive to IFN-γ identified a mutant cell line (γ1A) that could be complemented by JAK2 (26). These experiments also defined the pathway hierarchy, as expression of JAK1 in U4A cells restored phosphorylation of STAT1 (24), and conceptually suggested that the JAKs may be utilised by other cytokine receptors. Subsequently, JAK3 was shown to interact with the IL-2 receptor (IL-2R) common gamma chain (γc) in a region commonly mutated in patients suffering from severe combined immunodeficiency disease (SCID) (27).
The STATs were originally characterised as components of the IFNα/β and IFNγ transcriptional response (28,29) and this early data preceded the genetic complementation studies. STAT1 and STAT2 (together with IRF9) were found to form a three-protein transcription complex (ISGF3; interferon-stimulated gene factor-3), which bound to common promoter elements within IFNα-responsive genes (ISRE; interferon-stimulated response element) (5,30–33). Similarly, STAT1 complexes were found to bind promoter elements within IFNγ-responsive genes (GAS; IFNγ activation site) (34,35).
Numerous papers followed, identifying a bewildering array of JAKs and STATs that were activated in response to different cytokines/growth factors. The definitive papers utilised gene targeting to demonstrate a layer of specificity, which until that point, had not been fully appreciated.
Studies in non-mammalian species have also made important contributions to our understanding of JAK-STAT biology and its significance, for instance see (36–38). Due to space constraints, this review is limited to mammalian studies.
Non-redundant biological roles of JAK
JAK1 knockout mice die shortly after birth and analysis of JAK1-deficient cells revealed a requirement for JAK1 in signalling via the class II receptors (IFNα/β, IFNγ, IL-10), those cytokines which signal through the γc receptor (IL-2, IL-4, IL-7, IL-9, IL-15, IL-21), and those that utilise gp130 (39). JAK2-deficient mice were embryonic lethal due to a lack of definitive erythropoiesis and analysis of JAK2-deficient cells revealed non-redundant roles for JAK2 in regulating TPO, IL-3, GM-CSF and IFNγ (40,41). While the majority of JAKs are ubiquitously expressed (42–44), JAK3 expression is restricted to the haemopoietic lineages and vascular muscle cells (43,45,46). Gene targeting in the mouse confirmed a critical role for JAK3 in lymphocyte development and function, mediated through cytokines that share the IL-2R γc chain (47,48), and this was paralleled by human studies, which identified a JAK3 mutation in a patient with SCID (49). In contrast, deletion of the Tyk2 gene revealed a slightly different picture to that suggested by the genetic complementation experiments, with Tyk2-deficient mice displaying only a modest reduction in signalling capacity in response to IFNα. Responses to IL-12 however, were severely impaired (50).
Non-redundant biological roles of STAT
There are seven STATs in mammalian cells, STAT1-4, 5a, 5b, and 6 (31,33,51–55). STAT5a and STAT5b are closely related and result from a gene duplication event (56), while splice variants of STAT1, 3, 4 and 5 have been reported, which result in a truncated C-terminus (1,57–59). STAT1 is widely expressed, with high levels in heart, thymus and spleen, STAT4 is found mainly in testis, thymus and spleen (60,61), while STAT5a and 5b exhibit differential expression in muscle, brain, mammary gland and secretary organs (seminal vesicles and salivary gland) (56). STAT2, STAT3 and STAT6 are expressed in the majority of tissues (55,60,62).
Stat1-deficient mice display deficiencies in immune responses mediated by IFNα and IFNγ and are highly susceptible to infection with L. moncytogenes and vesicular stomatitis virus (VSV) (63,64). The complete lack of responsiveness to interferon confirmed the earlier studies, which had predicted a critical role for Stat1 in interferon signalling. Similarly, Stat2-knockout mice are also sensitive to viral infection (65), with analysis of Stat2-deficient mice and cells confirming a critical role in regulating signalling by the type I interferons (65).
In contrast, Stat3-deficient mice were embryonic lethal (E6.5–7.5), most likely due to a failure to form visceral endoderm (66). Conditional deletion of Stat3 in adult mouse tissues has demonstrated the importance of Stat3 for a wide range of physiological processes, with defects found in lung (67), bone (68), colon (69), heart (70), the nervous system (71) and skin (72), and are consistent with Stat3 inducing biological responses for a large variety of cytokines, including the IL-6/gp130 family (73–77), IL-10 (78,79), G-CSF (80), leptin (81) and IL-21 (82,83). Stat4-deficient mice fail to respond to IL-12 and to IL-23 (which shares the IL-12Rβ1) resulting in reduced Th1 differentiation and NK cell function (84,85).
Stat5a and Stat5b are required to elicit biological responses to IL-3, GM-CSF (86–88), γc cytokines (89–91), growth hormone and prolactin (86,87,92,93). Stat5a/Stat5b double deficient mice have impaired mammary gland development and growth retardation, consistent with the roles of STAT5a and STAT5b in mediating prolactin and growth hormone responses, respectively (86,93–95). Mice lacking both Stat5a and b are severely anaemic and the majority die perinatally (95), with an earlier study suggesting reduced erythropoiesis and increased cell death due to reduced EPO-STAT5 driven Bcl-XL expression (96). Most recently, STAT5 has been found to play additional roles in haemopoietic cell development, differentiation and survival (97–99). Stat6-deficient mice are refractory to IL-4 and IL-13 and as a consequence have defective Th2 polarisation, IgG1 and IgE class switching, and greater susceptibility to parasite infection (100–103).
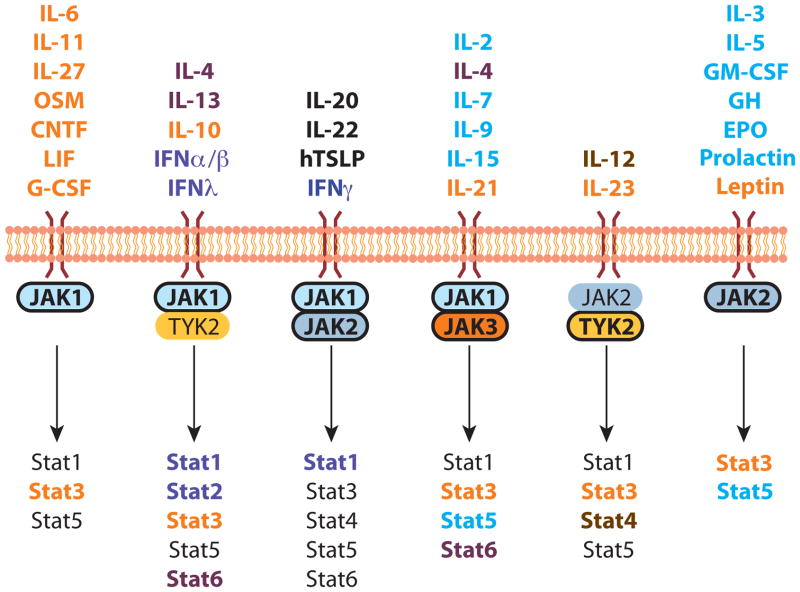
The requirement for individual JAK and Stat molecules is summarised in figure 1.
Figure 1. Non-redundant JAK/STAT signalling in mice.
Schematic showing the preferential cytokine/growth factor usage of different JAKs and STATs, as based on gene-targeting studies in mice. Emphasis in bold indicates the dominant JAK of the pair and colour coding links the individual cytokine/growth factors with their requisite STAT/s. See text for references.
Domain architecture and mechanism of action
JAK protein tyrosine kinases
Sequence alignment identified seven JAK homology (JH) regions (14) starting with a tyrosine kinase domain (JH1) at the carboxyl (C)-terminus, a pseudokinase domain (104–106) (JH2), an SH2-like domain (JH3-4) (107,108), and finally a divergent four-point-one, ezrin, radixin, moesin (FERM) homology domain (JH4-7) at the amino (N)-terminus (109), with JH4 crossing into both the SH2-like and FERM domains (Figure 2). The high-resolution crystal structures of all four active JAK JH1 domains have now been solved and reveal a highly conserved and typical bi-lobed kinase domain (110–112).
Figure 2. JAK and STAT domain organisation.
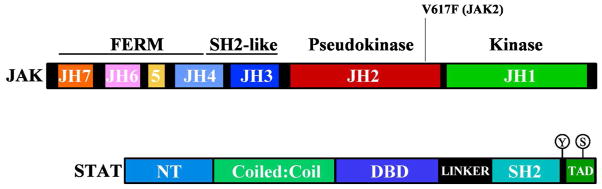
Schematic showing the domain organisation of JAK and STAT proteins. The valine 617 commonly mutated in JAK2 in myeloproliferative neoplasms is shown. NT: N-terminal region, DBD: DNA binding domain, TAD: transcriptional activation domain.
Until recently, the pseudokinase domain was believed to be catalytically inactive as it lacked the key amino acids required for enzyme function, and instead had an autoinhibitory role, with deletion or mutation of the domain resulting in enhanced JAK2 and JAK3 kinase activity (104,105,113). While it was known for some time that phosphorylation within the pseudokinase domain was required for inhibition (114–116), it now appears that the domain is a dual specificity kinase and that autophosphorylation of Ser523 and Tyr570 is required to maintain the inactive kinase in its basal state (117). This result implies that a key step in activation of the JAKs might be de-phosphorylation of Ser523, and the identification of the phosphatase/s responsible will be an intriguing piece of the puzzle. Understanding exactly how the pseudokinase domain mediates its inhibitory function will require the three dimensional structure of the JH2:JH1 complex. The biological significance of this domain is underscored by the acquired mutations found in human myeloproliferative neoplasms (discussed in more detail below), which are predicted to mitigate its autoinhibitory function and which include point mutation of valine 617.
The SH2-like domain has many of the structural and sequence-related hallmarks of a classic SH2 module, yet lacks some of the key residues normally conserved within the SH2 domain family, with experimental mutation of the critical arginine further suggesting that an ability to bind phosphotyrosine is not required for its function (118). Instead, the SH2-like domain is likely to have a structural role and is for instance, required for JAK1 binding to the OSM-R (119) and for TYK2 to maintain surface expression of IFNAR1 (120,121).
FERM domains classically form a three-lobed structure; encompassing a ubiquitin-like fold (F1), an acyl-coenzyme A binding-like fold (F2) and a pleckstrin homology domain fold (F3) (122). The JAK FERM domain mediates binding to the receptor cytoplasmic domains (123–125) and with the suggestion that the FERM domain also contributes to kinase integrity, may be involved in a more complex structural interplay with other JH domains (126,127). The FERM F1 and F2 subdomains (JH6-7) are the minimum requirement for interaction with the membrane-proximal, proline-rich Box 1 region of the receptor cytoplasmic domains (120,124,128–130), although in some instances, other receptor residues such as those within the hydrophobic Box 2 region of the G-CSF-R and EPO-R, are required for both interaction and full JAK activation (131,132).
Careful biochemical studies by Claude Haan and colleagues (133,134) would suggest that the JAKs are an integral component of the receptor subunit with very little release or exchange into the cytoplasm and as such are located primarily at the plasma membrane. This relationship may be functionally required even before the receptors reach the cell surface, with data showing that an interaction with the JAK2 FERM domain is required for proper processing of the immature EPO-R in the endoplasmic reticulum and its subsequent surface expression (135).
STATs
Each STAT has seven conserved features: an N-terminal domain (NT), a coiled-coil domain (CC), a central DNA-binding domain (DBD), a linker region, an SH2 domain followed by a single conserved tyrosine residue, and a C-terminal transactivation domain (TAD). Prior to cytokine stimulation, non-phosphorylated STATs exist as anti-parallel dimers, formed through reciprocal interactions between the N-terminal domains (136–140), which continuously shuttle between the cytoplasm and nucleus. Upon cytokine stimulation, the STATs are localised to the receptor complex by interaction of the SH2 domain with the receptor phosphotyrosine residues (Table 1). JAK phosphorylation of the STAT proteins then results in a spatial reorganisation of the dimer complex, to form an active, parallel dimer stabilised by reciprocal SH2 and phosphotyrosine interactions, which disengages from the receptor and translocates to the nucleus. STAT1, 3, 4, 5 and 6 form homodimeric complexes, while STAT1 and STAT3, at least in vitro, can also form a heterodimeric complex. STAT2 primarily functions as a heterodimer with STAT1 (together with IRF9), but can also act independently of STAT1 (141,142).
Table 1.
Phosphotyrosyl sites shown to bind STAT-SH2 domains
| SH2 Binding1 | Receptor | References | Dimerization motif2 | |
|---|---|---|---|---|
| STAT1 | PYXPQ, PYDXXH | IFNγRα | (300,301) | TGPYIKT |
| STAT2 | PYVXXXS | IFNαR1 | (302,303) | RKPYLKH |
| STAT3 | PYXXQ | GP130, LIFR, G-CSFR | (268,304) | APPYLKT |
| STAT4 | TXXGPYLXX | IL-12Rβ2 | (305,306) | KGPYVPS |
| STAT5 | DXPYXXL/F | EPO, IL-2Rβ, IL-7R | (307–309) | DGPYVKP |
| STAT6 | PYKXF | IL-4R | (55) | RGPYVPA |
Known consensus sites for STAT-SH2 domain binding together with the relevant receptor subunit, where pY indicates phosphorylated tyrosine and X is any amino acid.
Sequences surrounding the C-terminal STAT phosphotyrosine that mediates STAT dimerization. Sequences are identical in mouse and human STATs, with the exception of STAT6 (RGPYVST in mouse)
To enter into the nucleus, STAT dimers need to traverse the nuclear pore complex (NPC); a bi-directional transport channel embedded in the nuclear envelope and composed of nucleoporins (143,144). Importin α5 binds to the nuclear localising signal (NLS) of phosphorylated and dimerized STAT1 and 2, and acts as a chaperone to actively traffic the STATs into the nucleus (6,7,145). In the non-phosphorylated state, the NLS is masked and nuclear translocation of STAT1 and 2 is thought to be facilitated by direct binding to nucleoporins; alternatively the STAT protein may be transported by binding to heterologous NLS-containing proteins (146,147). Interestingly, another mechanism has been described for STAT1 during monocyte differentiation, whereby STAT1 binds to nucleolin and it is the NLS of nucleolin that facilitates STAT1 nuclear translocation (148).
In contrast to STAT1 and 2, STAT3 contains a constitutive form of the NLS that allows nuclear accumulation of both phosphorylated and non-phosphorylated STAT3 through association with importin α3 and in a tissue-dependent manner, with importin α6 (149,150). Less is known about nuclear trafficking of other STATs although the importin system has been implicated for both STAT5A and STAT6 (9,151). Rac1 and Rac GTPase-activating protein (MgcRacGAP) have been reported to enhance the nuclear accumulation of STAT3 and STAT5A and subsequent transcriptional activity (8,9).
Once in the nucleus, STAT dimers or higher order complexes, are stabilised by NT:NT interactions and bind cooperatively to tandem sequence elements within promoter regions, (often referred to as gamma-activated sequence (GAS) elements) (152,153), to activate the transcription of specific gene subsets. All STATs bind to similar palindromic elements represented by a core TTCN2-4GAA consensus sequence (154). STAT1, 3 and 4 prefer a sequence separated by 3 nucleotides, whilst STAT6 prefers a sequence separated by 4 nucleotides (155). The crystal structure of the phosphodimer core (coiled:coil, DBD and SH2 domain) bound to DNA revealed a clamp-like configuration stabilised by the reciprocal SH2-phosphotyrosine interactions and the interface of the DNA-binding domains with DNA, with the four alpha-helices of the coiled-coil domain projecting outward (156,157).
Serine phosphorylation of a conserved MAPK consensus sequence within the transactivation domain (PMSP within STAT1, 3 and 4 and PLSP within STAT5), (158–161) by various serine kinases (e.g. MAPK, p38, JNK, PKCδ, mTOR, PI3K) (162–166) greatly increases STAT-mediated gene transcription (159,160,162,167,168) and the truncated STAT isoforms, which lack the TAD are largely thought to act as dominant negatives (4,169,170). To add another layer of complexity, it has been suggested that unphosphorylated STAT3b dimers are also transcriptionally active, and initiate a distinct set of genes from that of the phosphorylated STAT (171,172).
Once released from DNA, the STAT dimer is thought to undergo a conformational change (parallel back to antiparallel, stabilised by NT:NT and CC:DBD interfaces), which exposes the C-terminal phosphotyrosines to phosphatase activity. Dephosphorylation and detachment of DNA are required for STATs to exit the nucleus (173,174) via a process that is dependent on the nuclear export signal (NES) of the STAT molecules and the nuclear export factor, chromosome region maintenance 1 (CRM1) (146,147,175–181); thus completing the activation and deactivation cycle.
In addition to tyrosine and serine phosphorylation, a number of post-translational modifications have been reported to regulate STAT activity. Acetylation of STAT3 on Lys685 by p300/CBP contributes to stable dimer formation and to transcriptional activation (182–186); controversially, acetylation has also been suggested to facilitate dephosphorylation and latency of STAT1 (187). While STAT1 may be acetylated in some circumstances, others cannot reproduce the link between acetylation and dephosphorylation (188). The role of PIAS1 in regulating STAT1 activity by small ubiquitin-like modifier (SUMO) conjugation of Lys703 has also been controversial (189–192). PIAS1 and the impact of SUMO-conjugation are discussed in more detail in the following section.
Negative regulation of JAK-STAT signalling
Given that JAK-STAT signalling is the universal and essential intracellular pathway for cytokine action, a number of regulatory mechanisms have evolved to control the magnitude and duration of signalling. This allows fine-tuning of cytokine-mediated cellular effects and prevents the inappropriate activity often associated with disease development. There are three major mechanisms for negative regulation: receptor internalization, de-phosphorylation by phosphotyrosine phosphatases (PTPs), and direct inhibition by protein inhibitors of STATs (PIAS) and suppressor of cytokine signalling (SOCS) proteins. The importance of receptor internalization by endocytic vesicles and subsequent receptor degradation by proteasomal and/or lysosomal pathways (193–200) is classically illustrated by the mutations acquired in the cytoplasmic domain of the G-CSF receptor and found in patients with severe congenital neutropenia. These mutations result in truncation of the cytoplasmic tail of the G-CSFR, simultaneously blocking maturation signalling by G-CSF and leading to defective ligand-induced internalization, as a consequence, the strong proliferative signal predisposes these patients to acute myeloid leukemia (AML) (201–203).
Since tyrosine phosphorylation is integral to JAK-STAT signal transduction, various tyrosine phosphatases such as SH2 domain-containing phosphatase (SHP) 1, SHP2, protein tyrosine phosphatase (PTP) 1B, T cell PTP (TC-PTP) and CD45 are involved in attenuation of signalling, acting either at the membrane to target the receptor-kinase complex, or in the nucleus to target STAT. SHP1 can directly interact with a number of receptors (including the EPOR and IFNα-receptor complex) and can inhibit JAK1 and JAK2 phosphorylation (204) (205). SHP2 has been shown to prevent JAK1 (206), STAT5α (207), and STAT1 phosphorylation (the latter at both tyrosine and serine residues) (208). PTP1B dephosphorylates JAK1 and TYK2 (209), while TC-PTP targets JAK1 and JAK3 (210). TC45, a nuclear isoform of TC-PTP, dephosphorylates STAT1 and STAT3 (211). Unlike other PTPs, expression of CD45 is restricted to haemopoietic cells and has been demonstrated to indiscriminately dephosphorylate the various JAKs (212).
The PIAS family of E3 SUMO ligases consists of four members, PIAS1, PIASx, PIAS3 and PIASy, and various alternatively spliced isoforms. They were originally named as protein inhibitors of activated STAT because of the observation that PIAS1 and PIAS3 could block STAT1 and STAT3 DNA binding activity when over-expressed (213,214). PIASx and PIASy were also shown to inhibit STAT4 and STAT1-mediated transcription, respectively, but without affecting DNA binding, presumably by recruiting co-repressors such as histone deacetylases (215,216). The role of PIAS1-mediated SUMO-conjugation of STAT1 has been controversial; SUMOylation of Lys703 has been suggested to selectively inhibit a subset of STAT1-responsive genes (189), while contradictory results suggesting that it is unlikely to have an effect on STAT1 transcription (191). Apart from PIAS1, which is indeed a partial physiological regulator of STAT1 (217), knockouts of other PIAS members revealed relatively little role in STAT-dependent pathways (218–220). It is now clear that the primary E3 activity of PIAS regulates proteins other than the STATs, and may in fact, regulate a general cellular process that impacts on many proteins (221–223).
A new twist on the SUMOylation story has recently emerged from the Vinkemeier group with evidence that SUMO-conjugation obstructs tyrosine phosphorylation of STAT1, resulting in “semi-phosphorylated” dimers and preventing the polymerization and assembly of STAT1 into paracrystalline arrays in the nucleus, enhancing STAT1 de-phosphorylation (224). The physiological consequences of SUMO-conjugation were explored with a “knock-in” mutation (Glu705-Gln), which abolished SUMO-conjugation of Lys730 and resulted in enhanced IFN-γ signalling, suggesting that SUMOylation of STAT1 may be a unique mechanism that has evolved to negatively regulate this pathway. The identity of the E3 SUMO ligase responsible remains unclear (225).
The suppressors of cytokine signalling (SOCS)
Perhaps the most studied inhibitors of JAK-STAT signalling are the SOCS proteins (226). This family of small, cytokine-inducible proteins inhibits signal transduction by blocking JAK and STAT activation and phosphorylation, creating a negative feedback loop. Their induction by cytokines or other stimuli can also cross-regulate signals downstream of other cytokines (227,228). There are eight family members, SOCS1-7 and CIS (cytokine-inducible SH2-containing protein) (226,229); each containing an N-terminal region of variable length with little sequence conservation, a central SH2 domain and a conserved C-terminal SOCS box motif, which interacts with elongins B and C, recruiting Cullin5, and RING-box2 (Rbx2) to form an E3 ubiquitin ligase complex (230). The SOCS proteins therefore function as adaptors to bring the E3 ligase into close proximity with its substrate, promoting the ubiquitination and subsequent proteasomal degradation of SOCS binding partners (231–233).
In addition to their role as E3 ligases, SOCS1 and SOCS3 are able to directly inhibit JAK enzymatic activity. SOCS1 and SOCS3 have a 12 amino acid region adjacent to the SH2 domain, known as the kinase inhibitory region (KIR), which was originally thought to act as a pseudo-substrate blocking the enzymatic activity of JAK by binding to its catalytic cleft (234–236). We now know that the SOCS3 KIR binds directly to a conserved “GQM” motif located within the atypical insertion loop of the JAK1, JAK2 and TYK2, but not JAK3 kinase domains, with the KIR binding in a manner that doesn’t compete with either substrate or ATP binding (237).
SOCS1 and SOCS3 can be recruited to the receptor complex through the SH2 domain binding directly to JAK or to the receptor, although the former probably only occurs with SOCS over-expression (238–244). It is most likely that SH2 binding to phosphorylated receptor tyrosine residues brings the KIR into close proximity with the JAK kinase domain, and indeed SOCS3 can bind simultaneously to a gp30 phosphopeptide via its SH2 domain and to JAK via the KIR (237). SOCS1 and SOCS3 can also attenuate signal transduction by targeting the receptor and/or JAK for ubiquitination and proteasomal degradation, although gene-targeting studies in which only the SOCS box was deleted, suggest that regulation by the SOCS box is the lesser component of SOCS1 and SOCS3 activity (233,245). Other SOCS proteins such as SOCS2 and CIS also appear to have a dual inhibitory function, blocking signalling by competitive binding to STAT docking sites on the receptor (246–249) and via SOCS box-dependent mechanisms (246,250–252).
SOCS1, SOCS2, SOCS3 and CIS are the best characterised of the SOCS family proteins. As revealed by gene-targeting experiments, loss of SOCS1, SOCS2 or SOCS3, results in excessive STAT activity in response to IFNγ, IL-4 (SOCS1), growth hormone (SOCS2), G-CSF and those cytokines which signal through gp130 (SOCS3) (253–255). SOCS1-deficient mice die shortly after birth, due to widespread inflammation and SOCS3-deficient mice are embryonic lethal, due to inappropriate LIF signaling (256–258), while SOCS2-deficiency leads to abnormal postnatal somatic growth (259). In contrast, CIS-deficient mice reportedly have no overt phenotype (256), despite overexpression of CIS implicating it in regulation of growth hormone, prolactin and IL-2 signalling (260,261). The pathways regulated by SOCS4-7 are not as well characterized, with these SOCS proteins functioning largely outside of the JAK-STAT paradigm.
Methylation of the SOCS genes provides another layer of regulation within the JAK-STAT pathway, with hypermethylation of CpG islands within the SOCS1 and SOCS3 promoters correlating with transcriptional silencing in various tumors (262–265).
While much of the JAK/STAT signalling cascade is well understood, surprisingly, some important aspects remain unknown, resulting no doubt from the difficulties in producing recombinant full-length JAK protein, the low levels of endogenous proteins and a lack of quality, high-affinity antibodies. Some questions remain, such as: how the signal is initiated, which JAK initiates the signal, and what is the sequence of phosphorylation events required for activation (and deactivation)? Complete structural information will no doubt clarify how JAK interacts with the receptor and the interplay between the different JAK domains. Similarly, the structure of JAK bound to its negative regulators, the SOCS proteins, is likely to reveal how this family of small molecules controls the extent and magnitude of signalling.
Specificity and diversity of action
Specificity and diversity is introduced into the JAK-STAT signalling pathway by a number of mechanisms. It is initiated by ligand binding to specific receptors, with the differential expression of various receptor subunits determining the cell types that will respond and the magnitude of the response. The receptor-JAK complex is determined by the sequence of the receptor cytoplasmic domains and although this results in different combinations of JAKs (Figure 1), it is not yet clear whether the JAKs display selectivity for phosphorylation of individual STAT or receptor tyrosine motifs and indeed, a comparison of the JAK2 and JAK3 JH1 domains suggests a significant overlap in substrate specificity (266). Rather, it is the STAT-SH2 domain, which drives specificity of signalling, with the sequences surrounding the phosphorylated receptor tyrosine residues determining STAT recruitment to the receptor (Table 1). Similarly, the combination of STATs and the pairing of SH2 domain with phosphotyrosine ligand, determines formation of the homo or heterodimeric complexes (267,268). The relative stabilities of STAT homodimers and heterodimers and their association with other transcription factors (e.g. IRF, Sp1, Jun, Fos, NF-kB, glucocorticoid receptor) and/or coactivators (e.g. p300/CBP, PCAF, GCN5, BRG1, HDAC) further broadens the range of STAT/DNA-binding complexes and transcriptional activities, contributing to the biological diversity (183–186).
In addition to tyrosine and serine phosphorylation, various post-translational modifications (as discussed earlier) provide another level of regulation to modulate STAT transcriptional responses. The negative regulators of JAK-STAT signalling are also important determinants of specificity. A typical example of this is SOCS3 regulation of IL-6 signalling, where expression of SOCS3 prevents IFNγ-like STAT1 transcriptional responses (254). In fact, the differential expression of downstream components such as the JAKS, STATS or SOCS can also contribute to the specificity of the cytokine response. Limited proteolytic processing of the C-terminus has been described for a number of the STAT proteins and is generally thought to generate a dominant-negative protein, reviewed in (269). However, a recent study suggests that estrogen-induced proteolytic cleavage of STAT1 may enhance, rather than inhibit, inflammatory responses (270).
Role of the JAK/STAT pathway in clinical disease
There have been many papers examining the role of STATs in malignancy and transformation, reviewed in (271–274). In this context, studies expressing mutant JAK (275) and STAT proteins (either dominant-negative or constitutively active) have been particularly informative (276–280). Here we have chosen to focus on the role of the JAK kinases in clinical disease.
Loss of function mutations
Inactivating JAK3 mutations have been documented in humans with severe combined immunodeficiency disease (SCID), characterized by loss of T and NK cells, abnormal B cell function and hypoplasia of lymphoid tissues (49,281). The clinical phenotypes induced by JAK3 mutations are indistinguishable from those resulting from loss-of-function mutations in the γc (282), indicating that JAK3 is indispensable for signal transduction from these receptors and hence indispensible for lymphoid development. Similarly, a homozygous mutation in TYK2 that resulted in the generation of a premature stop codon and subsequent loss of protein expression has been reported in a patient with hyper-IgE syndrome. This patient was highly susceptible to infections by multiple microorganisms and the patient’s cells showed defective responses to IL-12, type I IFNs, IL-6, IL-23 and IL-10, demonstrating the importance of TYK2 for both innate and acquired immunity in humans (283). This study also highlights some of the species differences that exist between mouse and humans as for instance the Tyk2−/− mice do not appear to have defective IL-6 signalling (283,284).
Gain-of-function mutations
While loss-of-function mutations in JAK3 and TYK2 are associated with immunodeficiency, the majority of naturally occurring mutations in JAK2, including chromosomal translocation, point mutations, insertions and deletions, are gain-of-function mutations and are associated with acute leukemia or myeloproliferative disorders (MPDs). The chromosomal translocations of JAK2 loci (e.g. TEL-JAK2, PCM1-JAK2, BCR-JAK2 and PAX-JAK2) lead to the development of both myeloid and lymphoid hematological malignancies (285). These translocations result in fusion of the JAK2 catalytic kinase (JH1) domain with multimerization subunits of partner proteins leading to constitutive tyrosine kinase activity and transformation.
Interestingly, point mutations, deletions and insertions in JAK2 are localized to the pseudokinase (JH2) domain and are associated with patients with MPDs. Since its discovery in 2005, most of the research has focused on the JAK2 V617F mutation (286–289). It is a somatic, gain-of-function mutation that has been frequently found in classic Philadelphia chromosome-negative myeloproliferative neoplasms (MPNs), in more than 90% of patients with polycythemia vera (PV), and in over 50% of patients with essential thrombocythemia (ET) and primary myelofibrosis (PMF) (290). Valine 617 lies within the pseudokinase (JH2) domain, and substitution of the valine with phenylalanine reduces the ability of JH2 to repress kinase activity, leading to constitutive tyrosine phosphorylation of JAK2. As a result, it confers cytokine independence and/or hypersensitivity to the mutated cells giving them a survival advantage.
It is believed that STAT5 is required to mediate altered gene expression and subsequent transformation by mutant JAK2 (291). However, recent findings suggest that JAK2 can also bypass STAT transcriptional activity by acting as an epigenetic modulator. Dawson et al., observed nuclear localization of JAK2 in haemopoietic cells, where it phosphorylated histone 3 at tyrosine 41 (H3Y41) to inhibit binding of the transcriptional repressor heterochromatin protein-1α (HP1α), enhancing the expression of genes that are not necessarily under direct STAT-mediated control (292). Similarly, in ES cells JAK2V617F was able to bypass Stat3 activation to maintain pluripotency, again correlating with an increase in H3Y41 phosphorylation (293). Liu et al., found that JAK2 interaction with and phosphorylation of a type II arginine methyltransferase, PRMT5, inhibited its methyltransferase activity and consequently, histone methylation (294). While some controversy still surrounds JAK localization in the nucleus (295), these studies demonstrate the ability of JAK2 to disrupt chromatin stability and potentiate the oncogenic properties of the V617F mutation.
Given the involvement of the V617F mutation in MPNs, most of the therapeutic development has focused on identifying low molecular mass ATP-competitive JAK2 inhibitors, reviewed in (296,297). TG101348 and Ruxolitinib (or INCB018424) (298,299) are two of many selective and potent JAK inhibitors currently in use in phase II and III MPN clinical trials, respectively. Ruxolitinib has a strong inhibitory effect on both JAK1 and JAK2, while TG101348 has greater selectivity for JAK2. Both of these inhibitors effectively attenuate downstream signalling, including phosphorylation of STAT and ERK1/2, and induce apoptosis in vitro. MPN patients treated with these inhibitors demonstrated a dramatic improvement in constitutional symptoms and a reduction in spleen size. Although unpleasant side-effects and a less satisfactory improvement in cytopaenia, marrow fibrosis and JAKV617F burden were also observed, inhibition of JAK2 has already proven to be an excellent target for therapeutic intervention in MPNs.
What the future holds.
The past 20 years have seen the JAK/STAT field progress from the discovery of the individual components and delineation of the pathway, to an understanding of the role of the JAKs and STATs in human disease, which coupled with resolution of the molecular structures should result in JAK inhibitors becoming a routine part of clinical treatment.
Acknowledgments
There have been many papers published in the past 20 years that have contributed to our understanding of the JAK/STAT pathway. Due to space constraints and in the interests of clarity we have been unable to cite all relevant references. We apologise to the authors of those papers that have not been included and do not mean to infer that their contribution has been of any less value. We thank Nicos Nicola for reviewing this manuscript and Peter Maltezos for figure production.
Footnotes
Declaration of interest
The authors were supported by the National Health and Medical Research Council (NHMRC), Australia (Program grant 461219, fellowship to SEN) and the National Institutes of Health, USA (Grant CA022556-33). This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS.
References
- 1.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 3.Meyer T, Hendry L, Begitt A, John S, Vinkemeier U. A single residue modulates tyrosine dephosphorylation, oligomerization, and nuclear accumulation of stat transcription factors. J Biol Chem. 2004;279:18998–19007. doi: 10.1074/jbc.M400766200. [DOI] [PubMed] [Google Scholar]
- 4.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 5.Kessler DS, Veals SA, Fu XY, Levy DE. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 1990;4:1753–1765. doi: 10.1101/gad.4.10.1753. [DOI] [PubMed] [Google Scholar]
- 6.Fagerlund R, Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin alpha 5. J Biol Chem. 2002;277:30072–30078. doi: 10.1074/jbc.M202943200. [DOI] [PubMed] [Google Scholar]
- 7.Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. Embo J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawashima T, Bao YC, Nomura Y, Moon Y, Tonozuka Y, Minoshima Y, Hatori T, Tsuchiya A, Kiyono M, Nosaka T, Nakajima H, Williams DA, Kitamura T. Rac1 and a GTPase-activating protein, MgcRacGAP, are required for nuclear translocation of STAT transcription factors. J Cell Biol. 2006;175:937–946. doi: 10.1083/jcb.200604073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawashima T, Bao YC, Minoshima Y, Nomura Y, Hatori T, Hori T, Fukagawa T, Fukada T, Takahashi N, Nosaka T, Inoue M, Sato T, Kukimoto-Niino M, Shirouzu M, Yokoyama S, Kitamura T. A Rac GTPase-activating protein, MgcRacGAP, is a nuclear localizing signal-containing nuclear chaperone in the activation of STAT transcription factors. Mol Cell Biol. 2009;29:1796–1813. doi: 10.1128/MCB.01423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schindler C, Darnell JE., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 11.Ihle JN. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 12.Wilks AF. Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc Natl Acad Sci U S A. 1989;86:1603–1607. doi: 10.1073/pnas.86.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilks AF, Harpur AG, Kurban RR, Ralph SJ, Zurcher G, Ziemiecki A. Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol Cell Biol. 1991;11:2057–2065. doi: 10.1128/mcb.11.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harpur AG, Andres AC, Ziemiecki A, Aston RR, Wilks AF. JAK2, a third member of the JAK family of protein tyrosine kinases. Oncogene. 1992;7:1347–1353. [PubMed] [Google Scholar]
- 15.Firmbach-Kraft I, Byers M, Shows T, Dalla-Favera R, Krolewski JJ. tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene. 1990;5:1329–1336. [PubMed] [Google Scholar]
- 16.Witthuhn BA, Silvennoinen O, Miura O, Lai KS, Cwik C, Liu ET, Ihle JN. Involvement of the Jak-3 Janus kinase in signalling by interleukins 2 and 4 in lymphoid and myeloid cells. Nature. 1994;370:153–157. doi: 10.1038/370153a0. [DOI] [PubMed] [Google Scholar]
- 17.Cance WG, Liu ET. Protein kinases in human breast cancer. Breast Cancer Res Treat. 1995;35:105–114. doi: 10.1007/BF00694751. [DOI] [PubMed] [Google Scholar]
- 18.Rane SG, Reddy EP. JAK3: a novel JAK kinase associated with terminal differentiation of hematopoietic cells. Oncogene. 1994;9:2415–2423. [PubMed] [Google Scholar]
- 19.Encyclopedia Brittanica. 1989. [Google Scholar]
- 20.Pellegrini S, John J, Shearer M, Kerr IM, Stark GR. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol Cell Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKendry R, Pellegrini S, Kerr IM, Stark GR. Constitutive production of alpha and beta interferons in mutant human cell lines. J Virol. 1994;68:4057–4062. doi: 10.1128/jvi.68.6.4057-4062.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.John J, McKendry R, Pellegrini S, Flavell D, Kerr IM, Stark GR. Isolation and characterization of a new mutant human cell line unresponsive to alpha and beta interferons. Mol Cell Biol. 1991;11:4189–4195. doi: 10.1128/mcb.11.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 24.Muller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, Harpur AG, Barbieri G, Witthuhn BA, Schindler C, et al. The protein tyrosine kinase JAK1 complements defects in interferon-alpha/beta and -gamma signal transduction. Nature. 1993;366:129–135. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 25.Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell JE, Jr, Stark GR, Kerr IM. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. Embo J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn BA, Quelle FW, Rogers NC, Schindler C, Stark GR, Ihle JN, et al. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-gamma signal transduction pathway. Nature. 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 27.Russell SM, Johnston JA, Noguchi M, Kawamura M, Bacon CM, Friedmann M, Berg M, McVicar DW, Witthuhn BA, Silvennoinen O, et al. Interaction of IL-2R beta and gamma c chains with Jak1 and Jak3: implications for XSCID and XCID. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 28.Levy DE, Kessler DS, Pine R, Reich N, Darnell JE., Jr Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 29.Levy DE, Kessler DS, Pine R, Darnell JE., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 30.Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci U S A. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu XY, Schindler C, Improta T, Aebersold R, Darnell JE., Jr The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci U S A. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veals SA, Schindler C, Leonard D, Fu XY, Aebersold R, Darnell JE, Jr, Levy DE. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci U S A. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 35.Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 36.Hombria JC, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr Biol. 2002;12:R569–575. doi: 10.1016/s0960-9822(02)01057-6. [DOI] [PubMed] [Google Scholar]
- 37.Brown S, Zeidler MP. Unphosphorylated STATs go nuclear. Curr Opin Genet Dev. 2008;18:455–460. doi: 10.1016/j.gde.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Hou SX, Zheng Z, Chen X, Perrimon N. The Jak/STAT pathway in model organisms: emerging roles in cell movement. Dev Cell. 2002;3:765–778. doi: 10.1016/s1534-5807(02)00376-3. [DOI] [PubMed] [Google Scholar]
- 39.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM, Jr, Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 40.Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 41.Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 42.Kawamura M, McVicar DW, Johnston JA, Blake TB, Chen YQ, Lal BK, Lloyd AR, Kelvin DJ, Staples JE, Ortaldo JR, et al. Molecular cloning of L-JAK, a Janus family protein-tyrosine kinase expressed in natural killer cells and activated leukocytes. Proc Natl Acad Sci U S A. 1994;91:6374–6378. doi: 10.1073/pnas.91.14.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musso T, Johnston JA, Linnekin D, Varesio L, Rowe TK, O’Shea JJ, McVicar DW. Regulation of JAK3 expression in human monocytes: phosphorylation in response to interleukins 2, 4, and 7. J Exp Med. 1995;181:1425–1431. doi: 10.1084/jem.181.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tortolani PJ, Lal BK, Riva A, Johnston JA, Chen YQ, Reaman GH, Beckwith M, Longo D, Ortaldo JR, Bhatia K, McGrath I, Kehrl J, Tuscano J, McVicar DW, O’Shea JJ. Regulation of JAK3 expression and activation in human B cells and B cell malignancies. J Immunol. 1995;155:5220–5226. [PubMed] [Google Scholar]
- 45.Gurniak CB, Berg LJ. Murine JAK3 is preferentially expressed in hematopoietic tissues and lymphocyte precursor cells. Blood. 1996;87:3151–3160. [PubMed] [Google Scholar]
- 46.Verbsky JW, Bach EA, Fang YF, Yang L, Randolph DA, Fields LE. Expression of Janus kinase 3 in human endothelial and other non-lymphoid and non-myeloid cells. J Biol Chem. 1996;271:13976–13980. doi: 10.1074/jbc.271.24.13976. [DOI] [PubMed] [Google Scholar]
- 47.Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 48.Nosaka T, van Deursen JM, Tripp RA, Thierfelder WE, Witthuhn BA, McMickle AP, Doherty PC, Grosveld GC, Ihle JN. Defective lymphoid development in mice lacking Jak3. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- 49.Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, Aman MJ, Migone TS, Noguchi M, Markert ML, Buckley RH, O’Shea JJ, Leonard WJ. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 50.Shimoda K, Kato K, Aoki K, Matsuda T, Miyamoto A, Shibamori M, Yamashita M, Numata A, Takase K, Kobayashi S, Shibata S, Asano Y, Gondo H, Sekiguchi K, Nakayama K, Nakayama T, Okamura T, Okamura S, Niho Y. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity. 2000;13:561–571. doi: 10.1016/s1074-7613(00)00055-8. [DOI] [PubMed] [Google Scholar]
- 51.Wakao H, Harada N, Kitamura T, Mui AL, Miyajima A. Interleukin 2 and erythropoietin activate STAT5/MGF via distinct pathways. Embo J. 1995;14:2527–2535. doi: 10.1002/j.1460-2075.1995.tb07250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mui AL, Wakao H, Harada N, O’Farrell AM, Miyajima A. Interleukin-3, granulocyte-macrophage colony-stimulating factor, and interleukin-5 transduce signals through two forms of STAT5. J Leukoc Biol. 1995;57:799–803. doi: 10.1002/jlb.57.5.799. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto K, Quelle FW, Thierfelder WE, Kreider BL, Gilbert DJ, Jenkins NA, Copeland NG, Silvennoinen O, Ihle JN. Stat4, a novel gamma interferon activation site-binding protein expressed in early myeloid differentiation. Mol Cell Biol. 1994;14:4342–4349. doi: 10.1128/mcb.14.7.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 55.Hou J, Schindler U, Henzel WJ, Ho TC, Brasseur M, McKnight SL. An interleukin-4-induced transcription factor: IL-4 Stat. Science. 1994;265:1701–1706. doi: 10.1126/science.8085155. [DOI] [PubMed] [Google Scholar]
- 56.Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci U S A. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle JN. Naturally occurring dominant negative variants of Stat5. Mol Cell Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 59.Hoey T, Zhang S, Schmidt N, Yu Q, Ramchandani S, Xu X, Naeger LK, Sun YL, Kaplan MH. Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. Embo J. 2003;22:4237–4248. doi: 10.1093/emboj/cdg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong Z, Wen Z, Darnell JE., Jr Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc Natl Acad Sci U S A. 1994;91:4806–4810. doi: 10.1073/pnas.91.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herrada G, Wolgemuth DJ. The mouse transcription factor Stat4 is expressed in haploid male germ cells and is present in the perinuclear theca of spermatozoa. J Cell Sci. 1997;110 (Pt 14):1543–1553. doi: 10.1242/jcs.110.14.1543. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Q, Ekhterae D, Kim KH. Molecular cloning and characterization of P113, a mouse SNF2/SWI2-related transcription factor. Gene. 1997;202:31–37. doi: 10.1016/s0378-1119(97)00446-0. [DOI] [PubMed] [Google Scholar]
- 63.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 64.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 65.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 66.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hokuto I, Ikegami M, Yoshida M, Takeda K, Akira S, Perl AK, Hull WM, Wert SE, Whitsett JA. Stat-3 is required for pulmonary homeostasis during hyperoxia. J Clin Invest. 2004;113:28–37. doi: 10.1172/JCI200419491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Z, Welte T, Troiano N, Maher SE, Fu XY, Bothwell AL. Osteoporosis with increased osteoclastogenesis in hematopoietic cell-specific STAT3-deficient mice. Biochem Biophys Res Commun. 2005;328:800–807. doi: 10.1016/j.bbrc.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 69.Welte T, Zhang SS, Wang T, Zhang Z, Hesslein DG, Yin Z, Kano A, Iwamoto Y, Li E, Craft JE, Bothwell AL, Fikrig E, Koni PA, Flavell RA, Fu XY. STAT3 deletion during hematopoiesis causes Crohn’s disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci U S A. 2003;100:1879–1884. doi: 10.1073/pnas.0237137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hilfiker-Kleiner D, Hilfiker A, Drexler H. Many good reasons to have STAT3 in the heart. Pharmacol Ther. 2005;107:131–137. doi: 10.1016/j.pharmthera.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 71.Okada S, Nakamura M, Katoh H, Miyao T, Shimazaki T, Ishii K, Yamane J, Yoshimura A, Iwamoto Y, Toyama Y, Okano H. Conditional ablation of Stat3 or Socs3 discloses a dual role for reactive astrocytes after spinal cord injury. Nat Med. 2006;12:829–834. doi: 10.1038/nm1425. [DOI] [PubMed] [Google Scholar]
- 72.Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, Yoshikawa K, Akira S, Takeda J. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. Embo J. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 74.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. Embo J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 75.Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161:4652–4660. [PubMed] [Google Scholar]
- 76.Gao Q, Wolfgang MJ, Neschen S, Morino K, Horvath TL, Shulman GI, Fu XY. Disruption of neural signal transducer and activator of transcription 3 causes obesity, diabetes, infertility, and thermal dysregulation. Proc Natl Acad Sci U S A. 2004;101:4661–4666. doi: 10.1073/pnas.0303992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Owaki T, Asakawa M, Morishima N, Mizoguchi I, Fukai F, Takeda K, Mizuguchi J, Yoshimoto T. STAT3 is indispensable to IL-27-mediated cell proliferation but not to IL-27-induced Th1 differentiation and suppression of proinflammatory cytokine production. J Immunol. 2008;180:2903–2911. doi: 10.4049/jimmunol.180.5.2903. [DOI] [PubMed] [Google Scholar]
- 78.Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, Akira S. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 79.Blumberg H, Conklin D, Xu WF, Grossmann A, Brender T, Carollo S, Eagan M, Foster D, Haldeman BA, Hammond A, Haugen H, Jelinek L, Kelly JD, Madden K, Maurer MF, Parrish-Novak J, Prunkard D, Sexson S, Sprecher C, Waggie K, West J, Whitmore TE, Yao L, Kuechle MK, Dale BA, Chandrasekher YA. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell. 2001;104:9–19. doi: 10.1016/s0092-8674(01)00187-8. [DOI] [PubMed] [Google Scholar]
- 80.Shimozaki K, Nakajima K, Hirano T, Nagata S. Involvement of STAT3 in the granulocyte colony-stimulating factor-induced differentiation of myeloid cells. J Biol Chem. 1997;272:25184–25189. doi: 10.1074/jbc.272.40.25184. [DOI] [PubMed] [Google Scholar]
- 81.Bates SH, Stearns WH, Dundon TA, Schubert M, Tso AW, Wang Y, Banks AS, Lavery HJ, Haq AK, Maratos-Flier E, Neel BG, Schwartz MW, Myers MG., Jr STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature. 2003;421:856–859. doi: 10.1038/nature01388. [DOI] [PubMed] [Google Scholar]
- 82.Zhou L, Ivanov, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 83.Avery DT, Deenick EK, Ma CS, Suryani S, Simpson N, Chew GY, Chan TD, Palendira U, Bustamante J, Boisson-Dupuis S, Choo S, Bleasel KE, Peake J, King C, French MA, Engelhard D, Al-Hajjar S, Al-Muhsen S, Magdorf K, Roesler J, Arkwright PD, Hissaria P, Riminton DS, Wong M, Brink R, Fulcher DA, Casanova JL, Cook MC, Tangye SG. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J Exp Med. 2010;207:155–171. doi: 10.1084/jem.20091706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DA, Doherty PC, Grosveld GC, Ihle JN. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 85.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 86.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 87.Ilaria RL, Jr, Hawley RG, Van Etten RA. Dominant negative mutants implicate STAT5 in myeloid cell proliferation and neutrophil differentiation. Blood. 1999;93:4154–4166. [PubMed] [Google Scholar]
- 88.Feldman GM, Rosenthal LA, Liu X, Hayes MP, Wynshaw-Boris A, Leonard WJ, Hennighausen L, Finbloom DS. STAT5A-deficient mice demonstrate a defect in granulocyte-macrophage colony-stimulating factor-induced proliferation and gene expression. Blood. 1997;90:1768–1776. [PubMed] [Google Scholar]
- 89.Imada K, Bloom ET, Nakajima H, Horvath-Arcidiacono JA, Udy GB, Davey HW, Leonard WJ. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J Exp Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moriggl R, Topham DJ, Teglund S, Sexl V, McKay C, Wang D, Hoffmeyer A, van Deursen J, Sangster MY, Bunting KD, Grosveld GC, Ihle JN. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–259. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 91.Moriggl R, Sexl V, Piekorz R, Topham D, Ihle JN. Stat5 activation is uniquely associated with cytokine signaling in peripheral T cells. Immunity. 1999;11:225–230. doi: 10.1016/s1074-7613(00)80097-7. [DOI] [PubMed] [Google Scholar]
- 92.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 93.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boghaert ER, Sells SF, Walid AJ, Malone P, Williams NM, Weinstein MH, Strange R, Rangnekar VM. Immunohistochemical analysis of the proapoptotic protein Par-4 in normal rat tissues. Cell Growth Differ. 1997;8:881–890. [PubMed] [Google Scholar]
- 95.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction. Cell. 1999;98:181–191. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- 97.Kieslinger M, Woldman I, Moriggl R, Hofmann J, Marine JC, Ihle JN, Beug H, Decker T. Antiapoptotic activity of Stat5 required during terminal stages of myeloid differentiation. Genes Dev. 2000;14:232–244. [PMC free article] [PubMed] [Google Scholar]
- 98.Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, Hennighausen L, O’Shea JJ. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Z, Li G, Tse W, Bunting KD. Conditional deletion of STAT5 in adult mouse hematopoietic stem cells causes loss of quiescence and permits efficient nonablative stem cell replacement. Blood. 2009;113:4856–4865. doi: 10.1182/blood-2008-09-181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takeda K, Kamanaka M, Tanaka T, Kishimoto T, Akira S. Impaired IL-13-mediated functions of macrophages in STAT6-deficient mice. J Immunol. 1996;157:3220–3222. [PubMed] [Google Scholar]
- 101.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 102.Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, Chu C, Quelle FW, Nosaka T, Vignali DA, Doherty PC, Grosveld G, Paul WE, Ihle JN. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 103.Urban JF, Jr, Noben-Trauth N, Donaldson DD, Madden KB, Morris SC, Collins M, Finkelman FD. IL-13, IL-4Ralpha, and Stat6 are required for the expulsion of the gastrointestinal nematode parasite Nippostrongylus brasiliensis. Immunity. 1998;8:255–264. doi: 10.1016/s1074-7613(00)80477-x. [DOI] [PubMed] [Google Scholar]
- 104.Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20:3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Saharinen P, Vihinen M, Silvennoinen O. Autoinhibition of Jak2 tyrosine kinase is dependent on specific regions in its pseudokinase domain. Mol Biol Cell. 2003;14:1448–1459. doi: 10.1091/mbc.E02-06-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yeh TC, Dondi E, Uze G, Pellegrini S. A dual role for the kinase-like domain of the tyrosine kinase Tyk2 in interferon-alpha signaling. Proc Natl Acad Sci U S A. 2000;97:8991–8996. doi: 10.1073/pnas.160130297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bernards A. Predicted tyk2 protein contains two tandem protein kinase domains. Oncogene. 1991;6:1185–1187. [PubMed] [Google Scholar]
- 108.Kampa D, Burnside J. Computational and functional analysis of the putative SH2 domain in Janus Kinases. Biochem Biophys Res Commun. 2000;278:175–182. doi: 10.1006/bbrc.2000.3757. [DOI] [PubMed] [Google Scholar]
- 109.Yamaoka K, Saharinen P, Pesu M, Holt VE, 3rd, Silvennoinen O, O’Shea JJ. The Janus kinases (Jaks) Genome Biol. 2004;5:253. doi: 10.1186/gb-2004-5-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Williams NK, Bamert RS, Patel O, Wang C, Walden PM, Wilks AF, Fantino E, Rossjohn J, Lucet IS. Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains. J Mol Biol. 2009;387:219–232. doi: 10.1016/j.jmb.2009.01.041. [DOI] [PubMed] [Google Scholar]
- 111.Lucet IS, Fantino E, Styles M, Bamert R, Patel O, Broughton SE, Walter M, Burns CJ, Treutlein H, Wilks AF, Rossjohn J. The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood. 2006;107:176–183. doi: 10.1182/blood-2005-06-2413. [DOI] [PubMed] [Google Scholar]
- 112.Chrencik JE, Patny A, Leung IK, Korniski B, Emmons TL, Hall T, Weinberg RA, Gormley JA, Williams JM, Day JE, Hirsch JL, Kiefer JR, Leone JW, Fischer HD, Sommers CD, Huang HC, Jacobsen EJ, Tenbrink RE, Tomasselli AG, Benson TE. Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP-690550 and CMP-6. J Mol Biol. 2010;400:413–433. doi: 10.1016/j.jmb.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 113.Saharinen P, Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J Biol Chem. 2002;277:47954–47963. doi: 10.1074/jbc.M205156200. [DOI] [PubMed] [Google Scholar]
- 114.Feener EP, Rosario F, Dunn SL, Stancheva Z, Myers MG., Jr Tyrosine phosphorylation of Jak2 in the JH2 domain inhibits cytokine signaling. Mol Cell Biol. 2004;24:4968–4978. doi: 10.1128/MCB.24.11.4968-4978.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Argetsinger LS, Kouadio JL, Steen H, Stensballe A, Jensen ON, Carter-Su C. Autophosphorylation of JAK2 on tyrosines 221 and 570 regulates its activity. Mol Cell Biol. 2004;24:4955–4967. doi: 10.1128/MCB.24.11.4955-4967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ishida-Takahashi R, Rosario F, Gong Y, Kopp K, Stancheva Z, Chen X, Feener EP, Myers MG., Jr Phosphorylation of Jak2 on Ser(523) inhibits Jak2-dependent leptin receptor signaling. Mol Cell Biol. 2006;26:4063–4073. doi: 10.1128/MCB.01589-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, Xu CF, Neubert TA, Skoda RC, Hubbard SR, Silvennoinen O. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kohlhuber F, Rogers NC, Watling D, Feng J, Guschin D, Briscoe J, Witthuhn BA, Kotenko SV, Pestka S, Stark GR, Ihle JN, Kerr IM. A JAK1/JAK2 chimera can sustain alpha and gamma interferon responses. Mol Cell Biol. 1997;17:695–706. doi: 10.1128/mcb.17.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Radtke S, Haan S, Jorissen A, Hermanns HM, Diefenbach S, Smyczek T, Schmitz-Vandeleur H, Heinrich PC, Behrmann I, Haan C. The Jak1 SH2 domain does not fulfill a classical SH2 function in Jak/STAT signaling but plays a structural role for receptor interaction and up-regulation of receptor surface expression. J Biol Chem. 2005;280:25760–25768. doi: 10.1074/jbc.M500822200. [DOI] [PubMed] [Google Scholar]
- 120.Richter MF, Dumenil G, Uze G, Fellous M, Pellegrini S. Specific contribution of Tyk2 JH regions to the binding and the expression of the interferon alpha/beta receptor component IFNAR1. J Biol Chem. 1998;273:24723–24729. doi: 10.1074/jbc.273.38.24723. [DOI] [PubMed] [Google Scholar]
- 121.Ragimbeau J, Dondi E, Alcover A, Eid P, Uze G, Pellegrini S. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. Embo J. 2003;22:537–547. doi: 10.1093/emboj/cdg038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 123.Girault JA, Labesse G, Mornon JP, Callebaut I. Janus kinases and focal adhesion kinases play in the 4.1 band: a superfamily of band 4.1 domains important for cell structure and signal transduction. Mol Med. 1998;4:751–769. [PMC free article] [PubMed] [Google Scholar]
- 124.Cacalano NA, Migone TS, Bazan F, Hanson EP, Chen M, Candotti F, O’Shea JJ, Johnston JA. Autosomal SCID caused by a point mutation in the N-terminus of Jak3: mapping of the Jak3-receptor interaction domain. Embo J. 1999;18:1549–1558. doi: 10.1093/emboj/18.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Haan C, Is’harc H, Hermanns HM, Schmitz-Van De Leur H, Kerr IM, Heinrich PC, Grotzinger J, Behrmann I. Mapping of a region within the N terminus of Jak1 involved in cytokine receptor interaction. J Biol Chem. 2001;276:37451–37458. doi: 10.1074/jbc.M106135200. [DOI] [PubMed] [Google Scholar]
- 126.Zhou YJ, Chen M, Cusack NA, Kimmel LH, Magnuson KS, Boyd JG, Lin W, Roberts JL, Lengi A, Buckley RH, Geahlen RL, Candotti F, Gadina M, Changelian PS, O’Shea JJ. Unexpected effects of FERM domain mutations on catalytic activity of Jak3: structural implication for Janus kinases. Mol Cell. 2001;8:959–969. doi: 10.1016/s1097-2765(01)00398-7. [DOI] [PubMed] [Google Scholar]
- 127.Haan S, Margue C, Engrand A, Rolvering C, Schmitz-Van de Leur H, Heinrich PC, Behrmann I, Haan C. Dual role of the Jak1 FERM and kinase domains in cytokine receptor binding and in stimulation-dependent Jak activation. J Immunol. 2008;180:998–1007. doi: 10.4049/jimmunol.180.2.998. [DOI] [PubMed] [Google Scholar]
- 128.Lebrun JJ, Ali S, Ullrich A, Kelly PA. Proline-rich sequence-mediated Jak2 association to the prolactin receptor is required but not sufficient for signal transduction. J Biol Chem. 1995;270:10664–10670. doi: 10.1074/jbc.270.18.10664. [DOI] [PubMed] [Google Scholar]
- 129.Tanner JW, Chen W, Young RL, Longmore GD, Shaw AS. The conserved box 1 motif of cytokine receptors is required for association with JAK kinases. J Biol Chem. 1995;270:6523–6530. doi: 10.1074/jbc.270.12.6523. [DOI] [PubMed] [Google Scholar]
- 130.Zhao Y, Wagner F, Frank SJ, Kraft AS. The amino-terminal portion of the JAK2 protein kinase is necessary for binding and phosphorylation of the granulocyte-macrophage colony-stimulating factor receptor beta c chain. J Biol Chem. 1995;270:13814–13818. doi: 10.1074/jbc.270.23.13814. [DOI] [PubMed] [Google Scholar]
- 131.Barge RM, de Koning JP, Pouwels K, Dong F, Lowenberg B, Touw IP. Tryptophan 650 of human granulocyte colony-stimulating factor (G-CSF) receptor, implicated in the activation of JAK2, is also required for G-CSF-mediated activation of signaling complexes of the p21ras route. Blood. 1996;87:2148–2153. [PubMed] [Google Scholar]
- 132.Witthuhn BA, Quelle FW, Silvennoinen O, Yi T, Tang B, Miura O, Ihle JN. JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-l. [DOI] [PubMed] [Google Scholar]
- 133.Behrmann I, Smyczek T, Heinrich PC, Schmitz-Van de Leur H, Komyod W, Giese B, Muller-Newen G, Haan S, Haan C. Janus kinase (Jak) subcellular localization revisited: the exclusive membrane localization of endogenous Janus kinase 1 by cytokine receptor interaction uncovers the Jak. receptor complex to be equivalent to a receptor tyrosine kinase. J Biol Chem. 2004;279:35486–35493. doi: 10.1074/jbc.M404202200. [DOI] [PubMed] [Google Scholar]
- 134.Giese B, Au-Yeung CK, Herrmann A, Diefenbach S, Haan C, Kuster A, Wortmann SB, Roderburg C, Heinrich PC, Behrmann I, Muller-Newen G. Long term association of the cytokine receptor gp130 and the Janus kinase Jak1 revealed by FRAP analysis. J Biol Chem. 2003;278:39205–39213. doi: 10.1074/jbc.M303347200. [DOI] [PubMed] [Google Scholar]
- 135.Huang LJ, Constantinescu SN, Lodish HF. The N-terminal domain of Janus kinase 2 is required for Golgi processing and cell surface expression of erythropoietin receptor. Mol Cell. 2001;8:1327–1338. doi: 10.1016/s1097-2765(01)00401-4. [DOI] [PubMed] [Google Scholar]
- 136.Lackmann M, Harpur AG, Oates AC, Mann RJ, Gabriel A, Meutermans W, Alewood PF, Kerr IM, Stark GR, Wilks AF. Biomolecular interaction analysis of IFN gamma-induced signaling events in whole-cell lysates: prevalence of latent STAT1 in high-molecular weight complexes. Growth Factors. 1998;16:39–51. doi: 10.3109/08977199809017490. [DOI] [PubMed] [Google Scholar]
- 137.Ndubuisi MI, Guo GG, Fried VA, Etlinger JD, Sehgal PB. Cellular physiology of STAT3: Where’s the cytoplasmic monomer? J Biol Chem. 1999;274:25499–25509. doi: 10.1074/jbc.274.36.25499. [DOI] [PubMed] [Google Scholar]
- 138.Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem. 2003;278:34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- 139.Ota N, Brett TJ, Murphy TL, Fremont DH, Murphy KM. N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat Immunol. 2004;5:208–215. doi: 10.1038/ni1032. [DOI] [PubMed] [Google Scholar]
- 140.Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, McMurray JS, Demeler B, Darnell JE, Jr, Chen X. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–771. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 141.Hahm B, Trifilo MJ, Zuniga EI, Oldstone MB. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 2005;22:247–257. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 142.Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011;7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Onischenko E, Weis K. Nuclear pore complex-a coat specifically tailored for the nuclear envelope. Curr Opin Cell Biol. 2011;23:293–301. doi: 10.1016/j.ceb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Meyer T, Vinkemeier U. Nucleocytoplasmic shuttling of STAT transcription factors. Eur J Biochem. 2004;271:4606–4612. doi: 10.1111/j.1432-1033.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- 145.Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J Biol Chem. 2001;276:16447–16455. doi: 10.1074/jbc.M008821200. [DOI] [PubMed] [Google Scholar]
- 146.Marg A, Shan Y, Meyer T, Meissner T, Brandenburg M, Vinkemeier U. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J Cell Biol. 2004;165:823–833. doi: 10.1083/jcb.200403057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Banninger G, Reich NC. STAT2 nuclear trafficking. J Biol Chem. 2004;279:39199–39206. doi: 10.1074/jbc.M400815200. [DOI] [PubMed] [Google Scholar]
- 148.Jerke U, Tkachuk S, Kiyan J, Stepanova V, Kusch A, Hinz M, Dietz R, Haller H, Fuhrman B, Dumler I. Stat1 nuclear translocation by nucleolin upon monocyte differentiation. PLoS One. 2009;4:e8302. doi: 10.1371/journal.pone.0008302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc Natl Acad Sci U S A. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kohler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- 151.Chen HC, Reich NC. Live cell imaging reveals continuous STAT6 nuclear trafficking. J Immunol. 2010;185:64–70. doi: 10.4049/jimmunol.0903323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu X, Sun YL, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 153.Vinkemeier U, Cohen SL, Moarefi I, Chait BT, Kuriyan J, Darnell JE., Jr DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. Embo J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 154.Horvath CM, Wen Z, Darnell JE., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 155.Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 156.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 157.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 158.Frank DA, Mahajan S, Ritz J. B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Invest. 1997;100:3140–3148. doi: 10.1172/JCI119869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kovarik P, Stoiber D, Novy M, Decker T. Stat1 combines signals derived from IFN-gamma and LPS receptors during macrophage activation. Embo J. 1998;17:3660–3668. doi: 10.1093/emboj/17.13.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Visconti R, Gadina M, Chiariello M, Chen EH, Stancato LF, Gutkind JS, O’Shea JJ. Importance of the MKK6/p38 pathway for interleukin-12-induced STAT4 serine phosphorylation and transcriptional activity. Blood. 2000;96:1844–1852. [PubMed] [Google Scholar]
- 161.Yamashita H, Xu J, Erwin RA, Farrar WL, Kirken RA, Rui H. Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. J Biol Chem. 1998;273:30218–30224. doi: 10.1074/jbc.273.46.30218. [DOI] [PubMed] [Google Scholar]
- 162.Goh KC, Haque SJ, Williams BR. p38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. Embo J. 1999;18:5601–5608. doi: 10.1093/emboj/18.20.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 164.Chendil D, Das A, Dey S, Mohiuddin M, Ahmed MM. Par-4, a pro-apoptotic gene, inhibits radiation-induced NF kappa B activity and Bcl-2 expression leading to induction of radiosensitivity in human prostate cancer cells PC-3. Cancer Biol Ther. 2002;1:152–160. doi: 10.4161/cbt.61. [DOI] [PubMed] [Google Scholar]
- 165.Rhee SH, Jones BW, Toshchakov V, Vogel SN, Fenton MJ. Toll-like receptors 2 and 4 activate STAT1 serine phosphorylation by distinct mechanisms in macrophages. J Biol Chem. 2003;278:22506–22512. doi: 10.1074/jbc.M208633200. [DOI] [PubMed] [Google Scholar]
- 166.Deb DK, Sassano A, Lekmine F, Majchrzak B, Verma A, Kambhampati S, Uddin S, Rahman A, Fish EN, Platanias LC. Activation of protein kinase C delta by IFN-gamma. J Immunol. 2003;171:267–273. doi: 10.4049/jimmunol.171.1.267. [DOI] [PubMed] [Google Scholar]
- 167.Kovarik P, Stoiber D, Eyers PA, Menghini R, Neininger A, Gaestel M, Cohen P, Decker T. Stress-induced phosphorylation of STAT1 at Ser727 requires p38 mitogen-activated protein kinase whereas IFN-gamma uses a different signaling pathway. Proc Natl Acad Sci U S A. 1999;96:13956–13961. doi: 10.1073/pnas.96.24.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Boulton TG, Zhong Z, Wen Z, Darnell JE, Jr, Stahl N, Yancopoulos GD. STAT3 activation by cytokines utilizing gp130 and related transducers involves a secondary modification requiring an H7-sensitive kinase. Proc Natl Acad Sci U S A. 1995;92:6915–6919. doi: 10.1073/pnas.92.15.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yoo JY, Huso DL, Nathans D, Desiderio S. Specific ablation of Stat3beta distorts the pattern of Stat3-responsive gene expression and impairs recovery from endotoxic shock. Cell. 2002;108:331–344. doi: 10.1016/s0092-8674(02)00636-0. [DOI] [PubMed] [Google Scholar]
- 170.Caldenhoven E, van Dijk TB, Solari R, Armstrong J, Raaijmakers JA, Lammers JW, Koenderman L, de Groot RP. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J Biol Chem. 1996;271:13221–13227. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 171.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- 172.Cheon H, Yang J, Stark GR. The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J Interferon Cytokine Res. 2011;31:33–40. doi: 10.1089/jir.2010.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, ten Hoeve J, Ren Z, Mao X, Chen X, Shuai K, Darnell JE., Jr Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc Natl Acad Sci U S A. 2005;102:3966–3971. doi: 10.1073/pnas.0501063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mertens C, Zhong M, Krishnaraj R, Zou W, Chen X, Darnell JE., Jr Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev. 2006;20:3372–3381. doi: 10.1101/gad.1485406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Haspel RL, Salditt-Georgieff M, Darnell JE., Jr The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. Embo J. 1996;15:6262–6268. [PMC free article] [PubMed] [Google Scholar]
- 176.Meyer T, Marg A, Lemke P, Wiesner B, Vinkemeier U. DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev. 2003;17:1992–2005. doi: 10.1101/gad.268003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Begitt A, Meyer T, van Rossum M, Vinkemeier U. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc Natl Acad Sci U S A. 2000;97:10418–10423. doi: 10.1073/pnas.190318397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.McBride KM, McDonald C, Reich NC. Nuclear export signal located within theDNA-binding domain of the STAT1 transcription factor. Embo J. 2000;19:6196–6206. doi: 10.1093/emboj/19.22.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mowen K, David M. Regulation of STAT1 nuclear export by Jak1. Mol Cell Biol. 2000;20:7273–7281. doi: 10.1128/mcb.20.19.7273-7281.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bhattacharya S, Schindler C. Regulation of Stat3 nuclear export. J Clin Invest. 2003;111:553–559. doi: 10.1172/JCI15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zeng R, Aoki Y, Yoshida M, Arai K, Watanabe S. Stat5B shuttles between cytoplasm and nucleus in a cytokine-dependent and -independent manner. J Immunol. 2002;168:4567–4575. doi: 10.4049/jimmunol.168.9.4567. [DOI] [PubMed] [Google Scholar]
- 182.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 183.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston DM. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 184.Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell JE., Jr Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci U S A. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J Biol Chem. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 186.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 187.Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH, Stauber RH, Bohmer FD, Heinzel T. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23:223–235. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Antunes F, Marg A, Vinkemeier U. STAT1 signaling is not regulated by a phosphorylation-acetylation switch. Mol Cell Biol. 2011;31:3029–3037. doi: 10.1128/MCB.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ungureanu D, Vanhatupa S, Gronholm J, Palvimo JJ, Silvennoinen O. SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood. 2005;106:224–226. doi: 10.1182/blood-2004-11-4514. [DOI] [PubMed] [Google Scholar]
- 190.Rogers RS, Horvath CM, Matunis MJ. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J Biol Chem. 2003;278:30091–30097. doi: 10.1074/jbc.M301344200. [DOI] [PubMed] [Google Scholar]
- 191.Song L, Bhattacharya S, Yunus AA, Lima CD, Schindler C. Stat1 and SUMO modification. Blood. 2006;108:3237–3244. doi: 10.1182/blood-2006-04-020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ungureanu D, Vanhatupa S, Kotaja N, Yang J, Aittomaki S, Janne OA, Palvimo JJ, Silvennoinen O. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood. 2003;102:3311–3313. doi: 10.1182/blood-2002-12-3816. [DOI] [PubMed] [Google Scholar]
- 193.Dittrich E, Haft CR, Muys L, Heinrich PC, Graeve L. A di-leucine motif and an upstream serine in the interleukin-6 (IL-6) signal transducer gp130 mediate ligand-induced endocytosis and down-regulation of the IL-6 receptor. J Biol Chem. 1996;271:5487–5494. doi: 10.1074/jbc.271.10.5487. [DOI] [PubMed] [Google Scholar]
- 194.Thiel S, Dahmen H, Martens A, Muller-Newen G, Schaper F, Heinrich PC, Graeve L. Constitutive internalization and association with adaptor protein-2 of the interleukin-6 signal transducer gp130. FEBS Lett. 1998;441:231–234. doi: 10.1016/s0014-5793(98)01559-2. [DOI] [PubMed] [Google Scholar]
- 195.Uotani S, Bjorbaek C, Tornoe J, Flier JS. Functional properties of leptin receptor isoforms: internalization and degradation of leptin and ligand-induced receptor downregulation. Diabetes. 1999;48:279–286. doi: 10.2337/diabetes.48.2.279. [DOI] [PubMed] [Google Scholar]
- 196.Yen CH, Yang YC, Ruscetti SK, Kirken RA, Dai RM, Li CC. Involvement of the ubiquitin-proteasome pathway in the degradation of nontyrosine kinase-type cytokine receptors of IL-9, IL-2, and erythropoietin. J Immunol. 2000;165:6372–6380. doi: 10.4049/jimmunol.165.11.6372. [DOI] [PubMed] [Google Scholar]
- 197.van Kerkhof P, Strous GJ. The ubiquitin-proteasome pathway regulates lysosomal degradation of the growth hormone receptor and its ligand. Biochem Soc Trans. 2001;29:488–493. doi: 10.1042/bst0290488. [DOI] [PubMed] [Google Scholar]
- 198.Walrafen P, Verdier F, Kadri Z, Chretien S, Lacombe C, Mayeux P. Both proteasomes and lysosomes degrade the activated erythropoietin receptor. Blood. 2005;105:600–608. doi: 10.1182/blood-2004-03-1216. [DOI] [PubMed] [Google Scholar]
- 199.Kindwall-Keller TL, Druhan LJ, Ai J, Hunter MG, Massullo P, Loveland M, Avalos BR. Role of the proteasome in modulating native G-CSFR expression. Cytokine. 2008;43:114–123. doi: 10.1016/j.cyto.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Martinez-Moczygemba M, Huston DP, Lei JT. JAK kinases control IL-5 receptor ubiquitination, degradation, and internalization. J Leukoc Biol. 2007;81:1137–1148. doi: 10.1189/jlb.0706465. [DOI] [PubMed] [Google Scholar]
- 201.Ward AC, van Aesch YM, Schelen AM, Touw IP. Defective internalization and sustained activation of truncated granulocyte colony-stimulating factor receptor found in severe congenital neutropenia/acute myeloid leukemia. Blood. 1999;93:447–458. [PubMed] [Google Scholar]
- 202.Hunter MG, Avalos BR. Deletion of a critical internalization domain in the G-CSFR in acute myelogenous leukemia preceded by severe congenital neutropenia. Blood. 1999;93:440–446. [PubMed] [Google Scholar]
- 203.Dong F, van Paassen M, van Buitenen C, Hoefsloot LH, Lowenberg B, Touw IP. A point mutation in the granulocyte colony-stimulating factor receptor (G-CSF-R) gene in a case of acute myeloid leukemia results in the overexpression of a novel G-CSF-R isoform. Blood. 1995;85:902–911. [PubMed] [Google Scholar]
- 204.Klingmuller U, Lorenz U, Cantley LC, Neel BG, Lodish HF. Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 205.David M, Chen HE, Goelz S, Larner AC, Neel BG. Differential regulation of the alpha/beta interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol Cell Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.You M, Yu DH, Feng GS. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol Cell Biol. 1999;19:2416–2424. doi: 10.1128/mcb.19.3.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen Y, Wen R, Yang S, Schuman J, Zhang EE, Yi T, Feng GS, Wang D. Identification of Shp-2 as a Stat5A phosphatase. J Biol Chem. 2003;278:16520–16527. doi: 10.1074/jbc.M210572200. [DOI] [PubMed] [Google Scholar]
- 208.Wu TR, Hong YK, Wang XD, Ling MY, Dragoi AM, Chung AS, Campbell AG, Han ZY, Feng GS, Chin YE. SHP-2 is a dual-specificity phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues in nuclei. J Biol Chem. 2002;277:47572–47580. doi: 10.1074/jbc.M207536200. [DOI] [PubMed] [Google Scholar]
- 209.Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien JP, Salmeen A, Barford D, Tonks NK. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276:47771–47774. doi: 10.1074/jbc.C100583200. [DOI] [PubMed] [Google Scholar]
- 210.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, McGlade CJ. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr Biol. 2002;12:446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 211.ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu Y, Zhu W, Tremblay M, David M, Shuai K. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Irie-Sasaki J, Sasaki T, Matsumoto W, Opavsky A, Cheng M, Welstead G, Griffiths E, Krawczyk C, Richardson CD, Aitken K, Iscove N, Koretzky G, Johnson P, Liu P, Rothstein DM, Penninger JM. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature. 2001;409:349–354. doi: 10.1038/35053086. [DOI] [PubMed] [Google Scholar]
- 213.Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 214.Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci U S A. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Liu B, Gross M, ten Hoeve J, Shuai K. A transcriptional corepressor of Stat1 with an essential LXXLL signature motif. Proc Natl Acad Sci U S A. 2001;98:3203–3207. doi: 10.1073/pnas.051489598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Arora T, Liu B, He H, Kim J, Murphy TL, Murphy KM, Modlin RL, Shuai K. PIASx is a transcriptional co-repressor of signal transducer and activator of transcription 4. J Biol Chem. 2003;278:21327–21330. doi: 10.1074/jbc.C300119200. [DOI] [PubMed] [Google Scholar]
- 217.Liu B, Mink S, Wong KA, Stein N, Getman C, Dempsey PW, Wu H, Shuai K. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat Immunol. 2004;5:891–898. doi: 10.1038/ni1104. [DOI] [PubMed] [Google Scholar]
- 218.Santti H, Mikkonen L, Anand A, Hirvonen-Santti S, Toppari J, Panhuysen M, Vauti F, Perera M, Corte G, Wurst W, Janne OA, Palvimo JJ. Disruption of the murine PIASx gene results in reduced testis weight. J Mol Endocrinol. 2005;34:645–654. doi: 10.1677/jme.1.01666. [DOI] [PubMed] [Google Scholar]
- 219.Wong KA, Kim R, Christofk H, Gao J, Lawson G, Wu H. Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Mol Cell Biol. 2004;24:5577–5586. doi: 10.1128/MCB.24.12.5577-5586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Roth W, Sustmann C, Kieslinger M, Gilmozzi A, Irmer D, Kremmer E, Turck C, Grosschedl R. PIASy-deficient mice display modest defects in IFN and Wnt signaling. J Immunol. 2004;173:6189–6199. doi: 10.4049/jimmunol.173.10.6189. [DOI] [PubMed] [Google Scholar]
- 221.Kotaja N, Vihinen M, Palvimo JJ, Janne OA. Androgen receptor-interacting protein 3 and other PIAS proteins cooperate with glucocorticoid receptor-interacting protein 1 in steroid receptor-dependent signaling. J Biol Chem. 2002;277:17781–17788. doi: 10.1074/jbc.M106354200. [DOI] [PubMed] [Google Scholar]
- 222.Schmidt D, Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 224.Droescher M, Begitt A, Marg A, Zacharias M, Vinkemeier U. Cytokine-induced Paracrystals Prolong the Activity of Signal Transducers and Activators of Transcription (STAT) and Provide a Model for the Regulation of Protein Solubility by Small Ubiquitin-like Modifier (SUMO) J Biol Chem. 2011;286:18731–18746. doi: 10.1074/jbc.M111.235978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Begitt A, Droescher M, Knobeloch KP, Vinkemeier U. SUMO conjugation of STAT1 protects cells from hyperresponsiveness to IFNgamma. Blood. 2011;118:1002–1007. doi: 10.1182/blood-2011-04-347930. [DOI] [PubMed] [Google Scholar]
- 226.Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc Natl Acad Sci U S A. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Bode JG, Nimmesgern A, Schmitz J, Schaper F, Schmitt M, Frisch W, Haussinger D, Heinrich PC, Graeve L. LPS and TNFalpha induce SOCS3 mRNA and inhibit IL-6-induced activation of STAT3 in macrophages. FEBS Lett. 1999;463:365–370. doi: 10.1016/s0014-5793(99)01662-2. [DOI] [PubMed] [Google Scholar]
- 228.Kiu H, Hilton DJ, Nicola NA, Ernst M, Marquez R, Alexander WS, Roberts AW, McManus EJ. Mechanism of crosstalk inhibition of IL-6 signaling in response to LPS and TNFalpha. Growth Factors. 2007;25:319–328. doi: 10.1080/08977190701830151. [DOI] [PubMed] [Google Scholar]
- 229.Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 230.Babon JJ, Sabo JK, Zhang JG, Nicola NA, Norton RS. The SOCS box encodes a hierarchy of affinities for Cullin5: implications for ubiquitin ligase formation and cytokine signalling suppression. J Mol Biol. 2009;387:162–174. doi: 10.1016/j.jmb.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kamura T, Sato S, Haque D, Liu L, Kaelin WG, Jr, Conaway RC, Conaway JW. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhang JG, Farley A, Nicholson SE, Willson TA, Zugaro LM, Simpson RJ, Moritz RL, Cary D, Richardson R, Hausmann G, Kile BJ, Kent SB, Alexander WS, Metcalf D, Hilton DJ, Nicola NA, Baca M. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci U S A. 1999;96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhang JG, Metcalf D, Rakar S, Asimakis M, Greenhalgh CJ, Willson TA, Starr R, Nicholson SE, Carter W, Alexander WS, Hilton DJ, Nicola NA. The SOCS box of suppressor of cytokine signaling-1 is important for inhibition of cytokine action in vivo. Proc Natl Acad Sci U S A. 2001;98:13261–13265. doi: 10.1073/pnas.231486498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle JN, Yoshimura A. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. Embo J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Nicholson SE, Willson TA, Farley A, Starr R, Zhang JG, Baca M, Alexander WS, Metcalf D, Hilton DJ, Nicola NA. Mutational analyses of the SOCS proteins suggest a dual domain requirement but distinct mechanisms for inhibition of LIF and IL-6 signal transduction. Embo J. 1999;18:375–385. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sasaki A, Yasukawa H, Suzuki A, Kamizono S, Syoda T, Kinjyo I, Sasaki M, Johnston JA, Yoshimura A. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells. 1999;4:339–351. doi: 10.1046/j.1365-2443.1999.00263.x. [DOI] [PubMed] [Google Scholar]
- 237.Babon JJ, Kershaw NJ, Murphy JM, Varghese LN, Laktyushin A, Young SN, Lucet SL, Norton RS, Nicola NA. SOCS3 binds to a site unique to JAKs and inhibits their kinase activity via a novel, non-competitive mechanism. Immunity. 2012 In Press. [Google Scholar]
- 238.Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, Miyazaki T, Leonor N, Taniguchi T, Fujita T, Kanakura Y, Komiya S, Yoshimura A. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 239.Bousquet C, Susini C, Melmed S. Inhibitory roles for SHP-1 and SOCS-3 following pituitary proopiomelanocortin induction by leukemia inhibitory factor. J Clin Invest. 1999;104:1277–1285. doi: 10.1172/JCI7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bjorbaek C, El-Haschimi K, Frantz JD, Flier JS. The role of SOCS-3 in leptin signaling and leptin resistance. J Biol Chem. 1999;274:30059–30065. doi: 10.1074/jbc.274.42.30059. [DOI] [PubMed] [Google Scholar]
- 241.Pezet A, Favre H, Kelly PA, Edery M. Inhibition and restoration of prolactin signal transduction by suppressors of cytokine signaling. J Biol Chem. 1999;274:24497–24502. doi: 10.1074/jbc.274.35.24497. [DOI] [PubMed] [Google Scholar]
- 242.Fenner JE, Starr R, Cornish AL, Zhang JG, Metcalf D, Schreiber RD, Sheehan K, Hilton DJ, Alexander WS, Hertzog PJ. Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat Immunol. 2006;7:33–39. doi: 10.1038/ni1287. [DOI] [PubMed] [Google Scholar]
- 243.Qing Y, Costa-Pereira AP, Watling D, Stark GR. Role of tyrosine 441 of interferon-gamma receptor subunit 1 in SOCS-1-mediated attenuation of STAT1 activation. J Biol Chem. 2005;280:1849–1853. doi: 10.1074/jbc.M409863200. [DOI] [PubMed] [Google Scholar]
- 244.Nicholson SE, De Souza D, Fabri LJ, Corbin J, Willson TA, Zhang JG, Silva A, Asimakis M, Farley A, Nash AD, Metcalf D, Hilton DJ, Nicola NA, Baca M. Suppressor of cytokine signaling-3 preferentially binds to the SHP-2-binding site on the shared cytokine receptor subunit gp130. Proc Natl Acad Sci U S A. 2000;97:6493–6498. doi: 10.1073/pnas.100135197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Boyle K, Egan P, Rakar S, Willson TA, Wicks IP, Metcalf D, Hilton DJ, Nicola NA, Alexander WS, Roberts AW, Robb L. The SOCS box of suppressor of cytokine signaling-3 contributes to the control of G-CSF responsiveness in vivo. Blood. 2007;110:1466–1474. doi: 10.1182/blood-2007-03-079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ram PA, Waxman DJ. Role of the cytokine-inducible SH2 protein CIS in desensitization of STAT5b signaling by continuous growth hormone. J Biol Chem. 2000;275:39487–39496. doi: 10.1074/jbc.M004755200. [DOI] [PubMed] [Google Scholar]
- 247.Ram PA, Waxman DJ. SOCS/CIS protein inhibition of growth hormone-stimulated STAT5 signaling by multiple mechanisms. J Biol Chem. 1999;274:35553–35561. doi: 10.1074/jbc.274.50.35553. [DOI] [PubMed] [Google Scholar]
- 248.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- 249.Greenhalgh CJ, Bertolino P, Asa SL, Metcalf D, Corbin JE, Adams TE, Davey HW, Nicola NA, Hilton DJ, Alexander WS. Growth enhancement in suppressor of cytokine signaling 2 (SOCS-2)-deficient mice is dependent on signal transducer and activator of transcription 5b (STAT5b) Mol Endocrinol. 2002;16:1394–1406. doi: 10.1210/mend.16.6.0845. [DOI] [PubMed] [Google Scholar]
- 250.Greenhalgh CJ, Rico-Bautista E, Lorentzon M, Thaus AL, Morgan PO, Willson TA, Zervoudakis P, Metcalf D, Street I, Nicola NA, Nash AD, Fabri LJ, Norstedt G, Ohlsson C, Flores-Morales A, Alexander WS, Hilton DJ. SOCS2 negatively regulates growth hormone action in vitro and in vivo. J Clin Invest. 2005;115:397–406. doi: 10.1172/JCI22710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Verdier F, Chretien S, Muller O, Varlet P, Yoshimura A, Gisselbrecht S, Lacombe C, Mayeux P. Proteasomes regulate erythropoietin receptor and signal transducer and activator of transcription 5 (STAT5) activation. Possible involvement of the ubiquitinated Cis protein. J Biol Chem. 1998;273:28185–28190. doi: 10.1074/jbc.273.43.28185. [DOI] [PubMed] [Google Scholar]
- 252.Landsman T, Waxman DJ. Role of the cytokine-induced SH2 domain-containing protein CIS in growth hormone receptor internalization. J Biol Chem. 2005;280:37471–37480. doi: 10.1074/jbc.M504125200. [DOI] [PubMed] [Google Scholar]
- 253.Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, Williams E, Zhang JG, Hilton DJ, Nicola NA, Alexander WS, Roberts AW. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 254.Croker BA, Krebs DL, Zhang JG, Wormald S, Willson TA, Stanley EG, Robb L, Greenhalgh CJ, Forster I, Clausen BE, Nicola NA, Metcalf D, Hilton DJ, Roberts AW, Alexander WS. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4:540–545. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 255.Kimura A, Kinjyo I, Matsumura Y, Mori H, Mashima R, Harada M, Chien KR, Yasukawa H, Yoshimura A. SOCS3 is a physiological negative regulator for granulopoiesis and granulocyte colony-stimulating factor receptor signaling. J Biol Chem. 2004;279:6905–6910. doi: 10.1074/jbc.C300496200. [DOI] [PubMed] [Google Scholar]
- 256.Marine JC, McKay C, Wang D, Topham DJ, Parganas E, Nakajima H, Pendeville H, Yasukawa H, Sasaki A, Yoshimura A, Ihle JN. SOCS3 is essential in the regulation of fetal liver erythropoiesis. Cell. 1999;98:617–627. doi: 10.1016/s0092-8674(00)80049-5. [DOI] [PubMed] [Google Scholar]
- 257.Starr R, Metcalf D, Elefanty AG, Brysha M, Willson TA, Nicola NA, Hilton DJ, Alexander WS. Liver degeneration and lymphoid deficiencies in mice lacking suppressor of cytokine signaling-1. Proc Natl Acad Sci U S A. 1998;95:14395–14399. doi: 10.1073/pnas.95.24.14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Roberts AW, Robb L, Rakar S, Hartley L, Cluse L, Nicola NA, Metcalf D, Hilton DJ, Alexander WA. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signalling (SOCS)-3. PNAS. 2001;98:9324–9329. doi: 10.1073/pnas.161271798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Metcalf D, Greenhalgh CJ, Viney E, Willson TA, Starr R, Nicola NA, Hilton DJ, Alexander WS. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature. 2000;405:1069–1073. doi: 10.1038/35016611. [DOI] [PubMed] [Google Scholar]
- 260.Li S, Chen S, Xu X, Sundstedt A, Paulsson KM, Anderson P, Karlsson S, Sjogren HO, Wang P. Cytokine-induced Src homology 2 protein (CIS) promotes T cell receptor-mediated proliferation and prolongs survival of activated T cells. J Exp Med. 2000;191:985–994. doi: 10.1084/jem.191.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Matsumoto A, Seki Y, Kubo M, Ohtsuka S, Suzuki A, Hayashi I, Tsuji K, Nakahata T, Okabe M, Yamada S, Yoshimura A. Suppression of STAT5 functions in liver, mammary glands, and T cells in cytokine-inducible SH2-containing protein 1 transgenic mice. Mol Cell Biol. 1999;19:6396–6407. doi: 10.1128/mcb.19.9.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Yoshikawa H, Matsubara K, Qian GS, Jackson P, Groopman JD, Manning JE, Harris CC, Herman JG. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet. 2001;28:29–35. doi: 10.1038/ng0501-29. [DOI] [PubMed] [Google Scholar]
- 263.Galm O, Yoshikawa H, Esteller M, Osieka R, Herman JG. SOCS-1, a negative regulator of cytokine signaling, is frequently silenced by methylation in multiple myeloma. Blood. 2003;101:2784–2788. doi: 10.1182/blood-2002-06-1735. [DOI] [PubMed] [Google Scholar]
- 264.He B, You L, Uematsu K, Zang K, Xu Z, Lee AY, Costello JF, McCormick F, Jablons DM. SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc Natl Acad Sci U S A. 2003;100:14133–14138. doi: 10.1073/pnas.2232790100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Liu TC, Lin SF, Chang JG, Yang MY, Hung SY, Chang CS. Epigenetic alteration of the SOCS1 gene in chronic myeloid leukaemia. Br J Haematol. 2003;123:654–661. doi: 10.1046/j.1365-2141.2003.04660.x. [DOI] [PubMed] [Google Scholar]
- 266.Sanz A, Ungureanu D, Pekkala T, Ruijtenbeek R, Touw IP, Hilhorst R, Silvennoinen O. Analysis of Jak2 catalytic function by peptide microarrays: the role of the JH2 domain and V617F mutation. PLoS One. 2011;6:e18522. doi: 10.1371/journal.pone.0018522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Heim MH, Kerr IM, Stark GR, Darnell JE., Jr Contribution of STAT SH2 groups to specific interferon signaling by the Jak-STAT pathway. Science. 1995;267:1347–1349. doi: 10.1126/science.7871432. [DOI] [PubMed] [Google Scholar]
- 268.Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JE, Jr, Yancopoulos GD. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 269.Hendry L, John S. Regulation of STAT signalling by proteolytic processing. Eur J Biochem. 2004;271:4613–4620. doi: 10.1111/j.1432-1033.2004.04424.x. [DOI] [PubMed] [Google Scholar]
- 270.Dai R, Phillips RA, Karpuzoglu E, Khan D, Ahmed SA. Estrogen regulates transcription factors STAT-1 and NF-kappaB to promote inducible nitric oxide synthase and inflammatory responses. J Immunol. 2009;183:6998–7005. doi: 10.4049/jimmunol.0901737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Santos CI, Costa-Pereira AP. Signal transducers and activators of transcription-from cytokine signalling to cancer biology. Biochim Biophys Acta. 2011;1816:38–49. doi: 10.1016/j.bbcan.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 272.Ferbeyre G, Moriggl R. The role of Stat5 transcription factors as tumor suppressors or oncogenes. Biochim Biophys Acta. 2011;1815:104–114. doi: 10.1016/j.bbcan.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 273.Li N, Grivennikov SI, Karin M. The unholy trinity: inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell. 2011;19:429–431. doi: 10.1016/j.ccr.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Putoczki T, Ernst M. More than a sidekick: the IL-6 family cytokine IL-11 links inflammation to cancer. J Leukoc Biol. 2010;88:1109–1117. doi: 10.1189/jlb.0410226. [DOI] [PubMed] [Google Scholar]
- 275.Schwaller J, Frantsve J, Aster J, Williams IR, Tomasson MH, Ross TS, Peeters P, Van Rompaey L, Van Etten RA, Ilaria R, Jr, Marynen P, Gilliland DG. Transformation of hematopoietic cell lines to growth-factor independence and induction of a fatal myelo- and lymphoproliferative disease in mice by retrovirally transduced TEL/JAK2 fusion genes. Embo J. 1998;17:5321–5333. doi: 10.1093/emboj/17.18.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 277.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Onishi M, Nosaka T, Misawa K, Mui AL, Gorman D, McMahon M, Miyajima A, Kitamura T. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol Cell Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Moriggl R, Sexl V, Kenner L, Duntsch C, Stangl K, Gingras S, Hoffmeyer A, Bauer A, Piekorz R, Wang D, Bunting KD, Wagner EF, Sonneck K, Valent P, Ihle JN, Beug H. Stat5 tetramer formation is associated with leukemogenesis. Cancer Cell. 2005;7:87–99. doi: 10.1016/j.ccr.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 281.Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, Ugazio AG, Johnston JA, Candotti F, O’Shea JJ, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 282.Matthews DJ, Clark PA, Herbert J, Morgan G, Armitage RJ, Kinnon C, Minty A, Grabstein KH, Caput D, Ferrara P, et al. Function of the interleukin-2 (IL-2) receptor gamma-chain in biologic responses of X-linked severe combined immunodeficient B cells to IL-2, IL-4, IL-13, and IL-15. Blood. 1995;85:38–42. [PubMed] [Google Scholar]
- 283.Minegishi Y, Saito M, Morio T, Watanabe K, Agematsu K, Tsuchiya S, Takada H, Hara T, Kawamura N, Ariga T, Kaneko H, Kondo N, Tsuge I, Yachie A, Sakiyama Y, Iwata T, Bessho F, Ohishi T, Joh K, Imai K, Kogawa K, Shinohara M, Fujieda M, Wakiguchi H, Pasic S, Abinun M, Ochs HD, Renner ED, Jansson A, Belohradsky BH, Metin A, Shimizu N, Mizutani S, Miyawaki T, Nonoyama S, Karasuyama H. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 284.Karaghiosoff M, Neubauer H, Lassnig C, Kovarik P, Schindler H, Pircher H, McCoy B, Bogdan C, Decker T, Brem G, Pfeffer K, Muller M. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13:549–560. doi: 10.1016/s1074-7613(00)00054-6. [DOI] [PubMed] [Google Scholar]
- 285.Jatiani SS, Baker SJ, Silverman LR, Reddy EP. JAK/STAT Pathways in Cytokine Signaling and Myeloproliferative Disorders: Approaches for Targeted Therapies. Genes Cancer. 2010;1:979–993. doi: 10.1177/1947601910397187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 287.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN, Green AR. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 288.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D’Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 289.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 290.Campbell PJ, Green AR. The myeloproliferative disorders. N Engl J Med. 2006;355:2452–2466. doi: 10.1056/NEJMra063728. [DOI] [PubMed] [Google Scholar]
- 291.Funakoshi-Tago M, Tago K, Abe M, Sonoda Y, Kasahara T. STAT5 activation is critical for the transformation mediated by myeloproliferative disorder-associated JAK2 V617F mutant. J Biol Chem. 2010;285:5296–5307. doi: 10.1074/jbc.M109.040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Griffiths DS, Li J, Dawson MA, Trotter MW, Cheng YH, Smith AM, Mansfield W, Liu P, Kouzarides T, Nichols J, Bannister AJ, Green AR, Gottgens B. LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat Cell Biol. 2011;13:13–21. doi: 10.1038/ncb2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Liu F, Zhao X, Perna F, Wang L, Koppikar P, Abdel-Wahab O, Harr MW, Levine RL, Xu H, Tefferi A, Deblasio A, Hatlen M, Menendez S, Nimer SD. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell. 2011;19:283–294. doi: 10.1016/j.ccr.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Girodon F, Steinkamp MP, Cleyrat C, Hermouet S, Wilson BS. Confocal imaging studies cast doubt on nuclear localization of JAK2V617F. Blood. 2011;118:2633–2634. doi: 10.1182/blood-2011-02-336479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Quintas-Cardama A, Kantarjian H, Cortes J, Verstovsek S. Janus kinase inhibitors for the treatment of myeloproliferative neoplasias and beyond. Nat Rev Drug Discov. 2011;10:127–140. doi: 10.1038/nrd3264. [DOI] [PubMed] [Google Scholar]
- 297.Haan C, Behrmann I, Haan S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases. J Cell Mol Med. 2010;14:504–527. doi: 10.1111/j.1582-4934.2010.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, Estrov Z, Fridman JS, Bradley EC, Erickson-Viitanen S, Vaddi K, Levy R, Tefferi A. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363:1117–1127. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Pardanani A, Gotlib JR, Jamieson C, Cortes JE, Talpaz M, Stone RM, Silverman MH, Gilliland DG, Shorr J, Tefferi A. Safety and efficacy of TG101348, a selective JAK2 inhibitor, in myelofibrosis. J Clin Oncol. 2011;29:789–796. doi: 10.1200/JCO.2010.32.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Greenlund AC, Farrar MA, Viviano BL, Schreiber RD. Ligand-induced IFN gamma receptor tyrosine phosphorylation couples the receptor to its signal transduction system (p91) Embo J. 1994;13:1591–1600. doi: 10.1002/j.1460-2075.1994.tb06422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Gerhartz C, Heesel B, Sasse J, Hemmann U, Landgraf C, Schneider-Mergener J, Horn F, Heinrich PC, Graeve L. Differential activation of acute phase response factor/STAT3 and STAT1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. I. Definition of a novel phosphotyrosine motif mediating STAT1 activation. J Biol Chem. 1996;271:12991–12998. doi: 10.1074/jbc.271.22.12991. [DOI] [PubMed] [Google Scholar]
- 302.Yan H, Krishnan K, Greenlund AC, Gupta S, Lim JT, Schreiber RD, Schindler CW, Krolewski JJ. Phosphorylated interferon-alpha receptor 1 subunit (IFNaR1) acts as a docking site for the latent form of the 113 kDa STAT2 protein. Embo J. 1996;15:1064–1074. [PMC free article] [PubMed] [Google Scholar]
- 303.Krishnan K, Singh B, Krolewski JJ. Identification of amino acid residues critical for the Src-homology 2 domain-dependent docking of Stat2 to the interferon alpha receptor. J Biol Chem. 1998;273:19495–19501. doi: 10.1074/jbc.273.31.19495. [DOI] [PubMed] [Google Scholar]
- 304.de Koning JP, Dong F, Smith L, Schelen AM, Barge RM, van der Plas DC, Hoefsloot LH, Lowenberg B, Touw IP. The membrane-distal cytoplasmic region of human granulocyte colony-stimulating factor receptor is required for STAT3 but not STAT1 homodimer formation. Blood. 1996;87:1335–1342. [PubMed] [Google Scholar]
- 305.Naeger LK, McKinney J, Salvekar A, Hoey T. Identification of a STAT4 binding site in the interleukin-12 receptor required for signaling. J Biol Chem. 1999;274:1875–1878. doi: 10.1074/jbc.274.4.1875. [DOI] [PubMed] [Google Scholar]
- 306.Yao BB, Niu P, Surowy CS, Faltynek CR. Direct interaction of STAT4 with the IL-12 receptor. Arch Biochem Biophys. 1999;368:147–155. doi: 10.1006/abbi.1999.1302. [DOI] [PubMed] [Google Scholar]
- 307.Gobert S, Chretien S, Gouilleux F, Muller O, Pallard C, Dusanter-Fourt I, Groner B, Lacombe C, Gisselbrecht S, Mayeux P. Identification of tyrosine residues within the intracellular domain of the erythropoietin receptor crucial for STAT5 activation. Embo J. 1996;15:2434–2441. [PMC free article] [PubMed] [Google Scholar]
- 308.Lin JX, Migone TS, Tsang M, Friedmann M, Weatherbee JA, Zhou L, Yamauchi A, Bloom ET, Mietz J, John S, et al. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 309.Damen JE, Wakao H, Miyajima A, Krosl J, Humphries RK, Cutler RL, Krystal G. Tyrosine 343 in the erythropoietin receptor positively regulates erythropoietin-induced cell proliferation and Stat5 activation. Embo J. 1995;14:5557–5568. doi: 10.1002/j.1460-2075.1995.tb00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]