Summary
Motile bacteria sense their physical and chemical environment through highly cooperative, ordered arrays of chemoreceptors. These signaling complexes phosphorylate a response regulator which in turn governs flagellar motor reversals, driving cells towards favorable environments. The structural changes that translate chemoeffector binding into the appropriate kinase output are not known. Here, we apply high-resolution electron cryotomography to visualize mutant chemoreceptor signaling arrays in well-defined kinase activity states. The arrays were well ordered in all signaling states, with no discernible differences in receptor conformation at 2-3 nm resolution. Differences were observed, however, in a keel-like density that we identify here as CheA kinase domains P1 and P2, which are the phosphorylation site domain and the binding domain for response regulator target proteins, respectively. Mutant receptor arrays with high kinase activities all exhibited small keels and high proteolysis susceptibility, indicative of mobile P1 and P2 domains. In contrast, arrays in kinase-off signaling states exhibited a range of keel sizes. These findings confirm that chemoreceptor arrays do not undergo large structural changes during signaling, and suggest instead that kinase activity is modulated at least in part by changes in the mobility of key domains.
Keywords: bacterial chemotaxis, signal transduction, electron cryotomography
Introduction
Motile bacteria track gradients of attractant and repellent chemicals with high sensitivity and wide dynamic range using arrays of transmembrane chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs) (Hazelbauer et al., 2008). The external sensing domains of MCP molecules monitor chemoeffector concentrations; their internal signaling domains, comprising extended four-helix coiled-coils, regulate the autophosphorylation activity of an associated kinase, CheA (Fig. 1A). In the extensively studied chemotaxis system of Escherichia coli, favorable stimuli, such as an increasing attractant level, down-regulate CheA, reducing the flow of phosphoryl groups to the cytoplasmic CheY response regulator, whose phosphorylation status regulates the direction of flagellar motor rotation. High CheA activity elicits clockwise (CW) motor rotation and frequent directional changes; low CheA activity promotes counter-clockwise (CCW) motor rotation and forward swimming. MCPs form trimers of dimers (Kim et al., 1999, Studdert & Parkinson, 2004) that assemble into extended, hexagonally-packed arrays (Briegel et al., 2012, Liu et al., 2012). Trimers are linked into the array at their cytoplasmic tips by CheA and the coupling protein CheW (Fig. 1C).
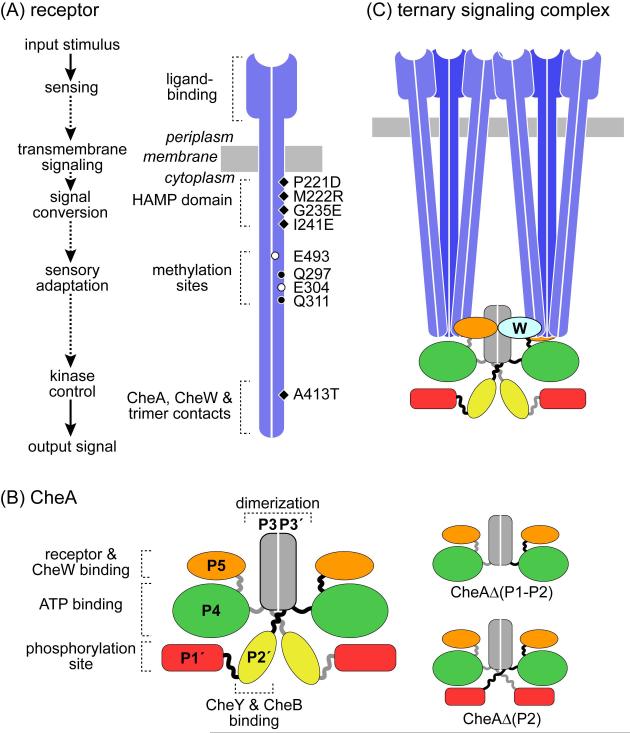
Fig. 1. Structures of MCP receptors, CheA kinase, and ternary signaling complexes.
(A) Input-output signaling in chemoreceptors of the MCP family. These molecules function as homodimers; the subunits are approximately 500 residues in length and mainly alpha-helical in secondary structure. The approximate positions of amino acid changes used to modify receptor output state in this work are shown.
(B) Domain organization of CheA. This autokinase functions as a homodimer. Autophosphorylation is a trans reaction, involving interaction of the phosphorylation site domain in one subunit with the ATP-binding domain of the other (Wolfe & Stewart, 1993). Two CheA variants used in the present work are shown at the right.
(C) The core signaling units of bacterial chemoreceptors. MCP molecules assemble into trimers of dimers through interactions between their highly conserved cytoplasmic tips (Kim et al., 1999, Studdert & Parkinson, 2004). Two trimers share and control one CheA dimer through binding interactions to its two P5 domains (one is hidden behind the trimer on the right) and to two P5-like CheW coupling proteins (one is hidden behind the trimer on the left). The CheW proteins each interact with a P5 domain, providing additional conformational control connections to the receptors. These core complexes assemble into higher order arrays through additional P5-CheW interactions (Briegel et al., 2012).
CheA, a homodimer, contains five domains in each subunit (Fig. 1B). During autophosphorylation, the ATP-binding domain (P4) from one subunit transfers a phosphoryl group to a histidine residue in the phosphorylation site domain (P1) of the other subunit (Wolfe & Stewart, 1993, Swanson et al., 1993a). Residues in both P4 and P1 probably play catalytic roles in the phosphorylation reaction (Stewart, 2010, Quezada et al., 2005). The P5 domain at the C-terminus of each subunit binds MCPs (Briegel et al., 2012, Wang et al., 2012) and CheW (Park et al., 2006, Zhao & Parkinson, 2006a, Zhao & Parkinson, 2006b) to form the ternary signaling complex that couples CheA activity to receptor control (Borkovich et al., 1989). The P2 domain provides a docking site for CheY and CheB (Swanson et al., 1993b), enhancing phosphotransfer efficiency to these response regulator targets of CheA (Stewart et al., 2000) (Fig. 1B). Flanking flexible linkers connect P2 to the P1 domain and to the P3 dimerization domain (Morrison & Parkinson, 1994) (Fig. 1B).
Despite a wealth of genetic, biochemical, and structural information about the E. coli chemotaxis machinery, a central question remains unanswered: how does chemoeffector binding at one end of the MCP molecule toggle the activity of a CheA molecule bound to its other end? One model proposes that kinase activity is controlled by large-scale changes in receptor packing (Borrock et al., 2008, Lamanna et al., 2005, Wu et al., 2011, Khursigara et al.), but no such rearrangements have been seen upon array stimulation in a number of studies (Lybarger & Maddock, 1999, Erbse & Falke, 2009, Homma et al., 2004, Liberman et al., 2004, Schulmeister et al., 2008). An alternative model suggests that CheA inactivation involves sequestration of its P1 phosphotransfer domain (Hamel et al., 2006).
To investigate these models, we used electron cryotomography (ECT) (Gan & Jensen, 2012) to visualize arrays of chemoreceptors mutationally locked in well-defined kinase activity states. We found that the order and packing of the arrays were independent of their activity, implying that signaling does not trigger large-scale reorganization. We did however observe differences in the CheA P1 and P2 densities suggesting that these domains are partially mobile in kinase-active arrays. These findings suggest that receptors control kinase activity at least in part by regulating CheA domain mobility.
Results
Mutant receptors and their kinase activities
To investigate the structural basis for CheA control in receptor signaling complexes, we imaged chemoreceptor arrays in E. coli strains containing different forms of the serine receptor, Tsr, as their only MCP (Table S1). We chose Tsr representatives known to form ternary signaling complexes with different kinase activity states (Fig. 1A; Table 1). The mutations targeted three functionally important regions of Tsr: the HAMP (histidine kinases, adenyl cyclases, MCPs and some phosphatases) domain involved in signal propagation (P221D, M222R, G235E, I241E); the methylation region involved in sensory adaptation (EEEE, QEQE, QQQQ); and the cytoplasmic tip, which binds CheA and CheW (A413T) (Fig. 1A). To prevent confounding effects of sensory adaptation, these Tsr variants were expressed in cells lacking the sensory adaptation enzymes CheR (methyltransferase) and CheB (methylesterase, deamidase). The cheA and cheW genes were wild-type.
Table 1.
Signaling properties of variant Tsr receptors
| Tsr variant |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| behavioral test | A413T | M222R | EEEE | P221D | QEQEg | QQQQ | G235E | I241E | comments / interpretation |
| % of flagellar rotation time spent in CW mode in Δ(cheRB) / (cheRB)+ strainsa |
0 / 0 | 2 / 0 | 27 / 28 | 0 / 28 | 75 / 27 | 65 / 24 | 91 / 84 | 88 / 80 | CW output directly reflects kinase activity level; sensory adaptation system cannot alter output signals of locked receptors |
| expression levelb | 1.5 | 1.1 | 0.6 | 1.4 | 1.0 | 0.9 | 1.2 | 0.9 | variant proteins have normal intracellular stabilities |
| receptor clusters and ternary complexesc |
YES | YES | YES | YES | YES | YES | YES | YES | normal array and ternary complex formation |
| jamming abilityd | YES | NO | na | YES | na | na | YES | YES | locked output receptors jam signaling by normal receptors |
| K1/2 [SER]e | NR | NR | NR | NR | 17 μM | 200 μM | NR | NR | no response (NR) is due to very low or locked-on kinase activity |
| kinase activityf SER / KCN |
– / 0 | – / 0 | – / 0 | – / 0 | 100 / 100 | 84 / – | – / 80 | – / 73 | |
| output state | locked OFF kinase activity |
low kinase activity; subject to sensory adaptation control |
high kinase activity; subject to stimulus and sensory adaptation control |
locked ON kinase activity |
|||||
Tsr expression plasmids were tested in host strains that either carried (cheRB)+ or lacked [Δ(cheRB)] sensory adaptation functions. The CheR enzyme adds methyl groups to the receptor’s adaptation sites; the CheB enzyme demethylates those residues. CheB also deamidates Q residues at the adaptation sites, converting them to E residues that are capable of accepting a methyl group. Receptor methylation augments the activity of CheA kinase molecules coupled to the receptor; demethylation and deamidation reduce the activity of receptor-coupled CheA molecules.
relative to wild-type Tsr (the QEQE form).
Approximately 75-85% of cells carrying wild-type Tsr exhibit one or more fluorescent polar spots in these tests (Mowery et al., 2008, Ames et al., 2002). All Tsr variants exhibited similar behaviors.
Tsr receptors that have locked signal outputs interfere with Tar signaling, thereby jamming aspartate chemotaxis (Ames et al., 2002). na = not applicable; jamming tests with methylation-state mimics are not meaningful in adaptation-proficient strains.
Serine responses of Tsr mutants were assessed with in vivo FRET-based kinase assays (Sourjik et al., 2007). K1/2 is the serine concentration that produced a 50% reduction in receptor-coupled CheA kinase activity. NR = no serine response detected (up to 10-100 mM serine).
Kinase activities (% of wild-type Tsr value) measured by in vivo FRET. SER: activity calculated from the FRET change elicited by a saturating serine stimulus; KCN: activity calculated from the FRET change elicited by 3 mM KCN.
wild-type Tsr.
Fluorescence light microscopy confirmed that all of the Tsr variants formed polar arrays (Table 1). Next the kinase activity of each array was quantified in two ways. First, because CheA autophosphorylation is the rate-limiting step in phosphorylation of CheY, and phospho-CheY promotes clockwise (CW) flagellar rotation, we simply counted the percentage of tethered cells rotating clockwise in each strain. Second, because phospho-CheY binds to its phosphatase CheZ, the fluorescence resonance energy transfer (FRET) between CheZ-CFP (FRET donor) and CheY-YFP (FRET acceptor) fusion proteins was monitored (Sourjik et al., 2007). The decrease in the FRET signal induced by exposure to a saturating attractant stimulus (defined as inhibiting all receptor-coupled CheA activity) was used as a background-corrected measure of CheA autophosphorylation rate. To measure receptor-coupled kinase activity in cells that could not respond to an attractant stimulus, we blocked ATP production in the cells with KCN, thereby stopping CheA autophosphorylation. The FRET change upon KCN treatment was then used as a measure of cellular CheA activity. Where both FRET-based measurements were possible, the resultant kinase activities were similar. As expected, the results of the flagellar rotation and FRET-based kinase assays agreed well and enabled us to place the mutants into two groups: “kinase-on” (QEQE, QQQQ, G235E, I241E) and “kinase-off” (P221D, M222R, EEEE, A413T) (Table 1, Fig. S1).
Receptor arrays in all signaling states are well-ordered
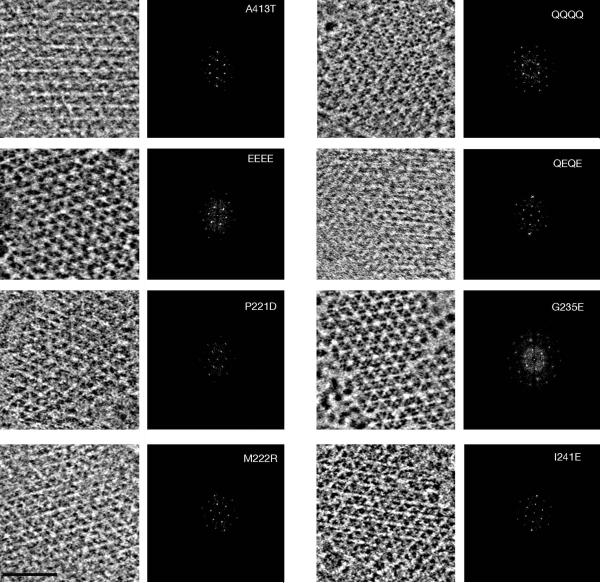
ECT of cells of each strain revealed that all eight of the mutant receptors formed well-ordered, extended arrays (Fig. 2). Some fragmentation of arrays into two or a few large patches was occasionally observed (Fig. S2), but because such fragmentation occurred in all mutant and wild-type strains, in both this study and in previous studies (Briegel et al., 2009), it was likely a result of cell lysis and flattening rather than signaling state. The array order and spacing (12 nm) of all mutant arrays were indistinguishable from one another and from previously reported chemoreceptor arrays (Fig. 2).
Fig. 2. No large-scale differences are evident in mutant receptor arrays.
Representative tomographic slices and corresponding power spectra of kinase-inactive (left column) and kinase-active receptor mutant arrays (right column). All arrays have comparable hexagonal organization with 12 nm spacing. Scale bar (bottom left panel): 30 nm.
The size of a keel-like density varies in different receptor signaling states
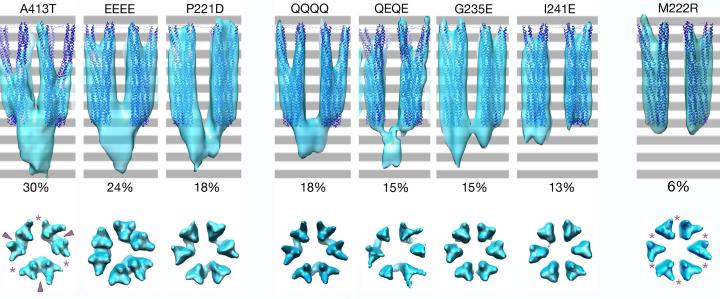
As before (Briegel et al., 2012), subtomogram averages corrected for the contrast transfer function (CTF) revealed a “two-on-two” organization of the receptor trimers-of-dimers in which pairs of dimers faced each other at each interface around the ring (Fig. 3). Although signal-related structural changes were not detected in the receptors themselves, keel-like densities of varying size were observed just below the receptor tips (i.e., closer to the cell center) (Fig. 3). Top views revealed that the keels connected opposing trimers, but on only three sides of each hexagon (Fig. 3 arrowheads), alternating with gaps (Fig. 3 asterisks).
Fig. 3. Keel electron density varies in ternary signaling complexes in different output states.
Image averages are shown for signaling complexes of eight Tsr variants with the amino acid changes listed across the top: wild-type receptor subunits (QEQE) have Q residues at two of the four methylation sites; the EEEE and QQQQ variants mimic the fully unmethylated or fully methylated forms of the receptor. The A413T, P221D, M222R, G235E and I241E mutant receptors are variants of the QEQE wild-type. Receptor crystal structures (purple) were fitted into sub-tomogram averages by MDFF for reference and alignment. An extra keel-like density, most prominent in the A413T complexes, is seen below the receptors in the side views (top row). The numbers below the keels give the volume of the keel density as a percentage of the total volume occupied by two trimers plus the keel at an appropriate threshold (shown in blue). Bottom row: top views showing keel densities connecting adjacent trimers (arrowheads), alternating with gaps around the hexagons (asterisks).
To compare the volume of the keel densities in different mutants, we first aligned the subtomogram averages by fitting a crystal structure of a Tsr receptor tip into each average with molecular dynamics flexible fitting (MDFF). We then selected a reasonable density threshold for each average that just enclosed at least one of the three receptor dimers. Because the averages were not identical in quality or resolution, to reduce errors introduced by the choice of different thresholds, we calculated the keel volume as a percentage of the total volume of two trimers of receptor dimers and their associated keel. The keel volumes of the kinase-active mutants fell between 13% and 18%. In contrast, kinase inactive mutants exhibited widely varying keel volumes. Two (A413T and EEEE) had large keel densities (30% and 24% respectively), one was similar to the kinase active mutants (P221D, 18%), and one had the smallest keel density (M222R, 6%).
The keel density comprises the P1 and P2 domains of CheA
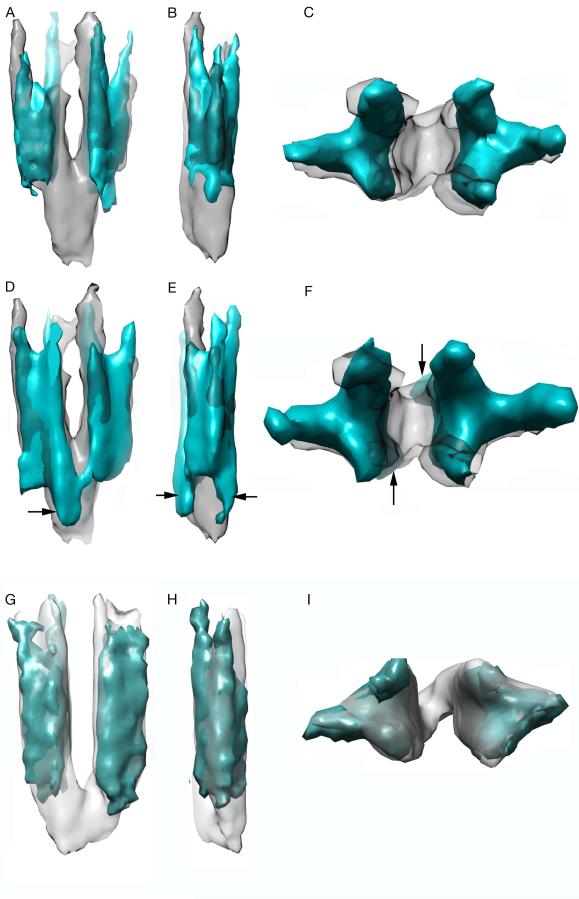
The position of the keel density matched that of CheA dimers in the array (Briegel et al., 2012, Liu et al., 2012), suggesting that it might correspond to one or more CheA domains. Liu et al. observed a similar structure in arrays of wild-type receptors and suggested that it represented P1 (Liu et al., 2012). We found that the keel comprises at least two CheA domains, P1 and P2 (Fig. 4). In a strain containing locked-off A413T receptors and CheA molecules that lacked both the P1 and P2 domains (see Fig. 1B), the keel density was completely missing (Fig. 4A-C; blue density). Images of CheAΔ(P1-P2) in arrays of a different kinase-off receptor mutant (P221D) produced an identical result (Fig. 4 G-I; cyan density). Previous characterization of this CheA mutant showed that it was still capable of forming ternary signaling complexes and, when supplied with P1 domains in trans, its phosphorylation activity still responded to stimulus control (Garzon & Parkinson, 1996). Because free P1 domains are not known to bind directly to receptors, this implies that receptor signaling must involve conformational changes in the P3-P4-P5 domains of CheA. In a strain with the A413T mutant receptor and CheA molecules lacking only the P2 domain, the keel was partially missing (Fig. 4D-F; blue density). The residual keel formed a clamp-like density (arrows in Fig. 4D-F) surrounding an empty center region. The P2-deleted CheA molecule is also known to form ternary signaling complexes and to respond to stimuli (Jahreis et al., 2004). Thus the partial keel formed by the CheAΔP2 molecule most likely corresponds to the P1 domain, possibly docked to its interaction partner, the ATP-binding P4 domain.
Fig. 4. Keel electron density comprises CheA subunits P1 and P2.
From left to right: side views, side views rotated 90°, top views.
A-F: Signaling complexes of the A413T receptor that compare the keels seen with wild-type CheA (gray) to those seen with CheA mutants (blue).
(A, B, C) wild-type CheA and CheAΔ(P1-P2) signaling complexes.
(D, E, F) wild-type CheA and CheAΔP2 signaling complexes. Black arrows point to clamp-like structures on each side of the keel that probably contain the P1 domain and possibly part of the P4 domain with which it interacts.
(G, H, I) Signaling complexes of the P221D receptor that compare the keels seen with wild-type CheA (light gray) to those seen with CheAΔ(P1-P2) (cyan).
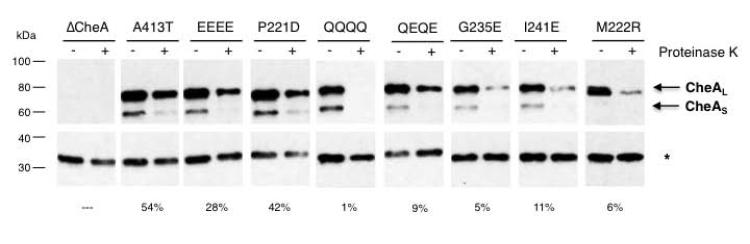
Sequestered CheA molecules resist proteolysis
Because densities in sub-tomogram averages depend on the consistent localization of array components, we hypothesized that the size of the keels might reflect the mobility of CheA domains P1 and P2. To test this idea, we looked for differences in protease sensitivity of CheA molecules in the mutant receptor arrays. In CheA molecules that are not associated with receptors in signaling complexes, the flexible linkers flanking the P2 domain are preferred targets of proteolysis (Morrison & Parkinson, 1994). Accordingly, we exposed arrays in gently lysed cells to proteinase K briefly and then analyzed the CheA degradation products by western blot. The antibody used recognizes epitopes primarily in P1 and therefore detects cleavage events that separate this domain from intact CheA. We found that CheA was more rapidly cleaved in strains with small keel volumes than in strains with large keel volumes (Fig. 5), consistent with increased mobility of P1 and P2.
Fig. 5. CheA protease sensitivity corresponds to keel size.
Western blots of mutant strains without (−) or with (+) treatment with proteinase K. Top: α-CheA. Arrows indicate long and short isoforms of CheA. Bottom: loading control (proteinase K-resistant background band (*) detected by α-β-lactamase). Values at bottom indicate percent of CheAL signal remaining after treatment for each strain.
Discussion
Whether or not a chemotactic stimulus alters the gross structure of the chemoreceptor array has been a contentious topic in the chemotaxis literature. While some studies have found signaling-dependent changes in receptor packing (Borrock et al., 2008, Lamanna et al., 2005, Wu et al., 2011, Khursigara et al.), others have not (Lybarger & Maddock, 1999, Erbse & Falke, 2009, Homma et al., 2004, Liberman et al., 2004, Schulmeister et al., 2008). In the present study, ECT images showed that eight E. coli strains with widely variant kinase activity all contained well-ordered, hexagonally packed chemoreceptor arrays with identical 12-nm lattice spacing. This confirms the findings of a previous ECT study of Caulobacter crescentus before and after exposure to attractant (Briegel et al., 2011). We conclude that receptor arrays do not undergo large rearrangements between signaling states. We did, however, find that CheA domains P1 and P2 were more mobile in kinase-active arrays than in arrays containing the A413T and EEEE receptors, which are locked in a kinase-off state. Although we cannot be certain that Tsr-A413T is locked in a physiologically-relevant signaling state, the unmethylated EEEE form of Tsr is a component of native signaling complexes in adaptation-proficient strains. Because removal of methyl groups drives receptors toward the attractant-induced kinase-off state, the EEEE form most likely represents this physiologically relevant signaling state (Hazelbauer et al., 2008).
Sequestration of the P1 domain was previously proposed as a mechanism that prevents productive interactions with P4 (Hamel et al., 2006), but the data presented here are the first direct evidence for such a model. We suggest that stimulus inputs promote structural changes along the length of the receptors, which in turn impart conformational changes to CheA through receptor-P5 and receptor-CheW-P5 connections (Fig. 1C). Changes in the conformation or orientation of the ATP-binding CheA-P4 domain then modulate its interaction with the CheA-P1 phosphorylation site domain. We further propose that in the kinase-on signaling state, increased mobility of the P1 and P2 domains allows for cycles of P1 engagement, phosphorylation, and release. Trapping the P1 and P2 domains in a static, nonproductive complex with P4 (manifest here as a large keel density with low susceptibility to proteolysis) appears to be one way to deactivate the kinase.
Although we were unable to resolve conformational differences along the receptors themselves, the idea that the kinase is controlled by changes in the mobility of its domains supports an important principle of the dynamic-bundle (Zhou et al., 2009) and yin-yang (Swain et al., 2009) models of receptor signaling. The dynamic-bundle model asserts that the packing stabilities of the HAMP bundle and the neighboring methylation bundle are in opposition: structural changes (induced by stimuli, adaptational modifications or amino acid replacements) that destabilize one element will stabilize the other. The yin-yang model postulates a similar opposing stability interaction between the methylation bundle and the receptor tip. Here we have shown that at least the downstream target of these signals, CheA, does in fact have states with different dynamic properties.
Previous work has suggested, however, that there are more than just two simple “kinase-on” and “kinase-off” states. Instead, both HAMP-stabilizing and HAMP-destabilizing lesions were found to deactivate the kinase, leading to a biphasic output model in which intermediate HAMP stabilities result in kinase activation, whereas stability perturbations to either extreme result in deactivation (Zhou et al., 2011). Our observations echo these ideas, because the mutant arrays with intermediate keel sizes had active kinases, whereas arrays with both the largest (A413T and EEEE) and smallest (M222R) keels were kinase-off. The M222R lesion in particular was previously suggested to drive receptor output to a kinase-off state [CCW(B)] that differs from the physiologically relevant, attractant-induced kinase-off state [CCW(A)] (Zhou et al., 2011). We assume that in the M222R receptor complex additional conformational changes in P4 or other components block CheA activity despite its mobile P1 and P2 domains. The P221D mutant is more difficult to interpret, because it exhibited an intermediate P1-P2 dynamic state but was still kinase-off. Conceivably, as with M222R, structural changes other than P1 and P2 immobilization can block kinase activity. Further imaging of additional mutant receptors and signaling complexes, and to higher resolution, should clarify these issues.
Experimental Procedures
Bacterial strains
Strains used in this study (Table S1) were isogenic derivatives of RP437, a wild-type chemotaxis derivative of E. coli K-12 (Parkinson & Houts, 1982). Mutant alleles of tsr and cheA were crossed from PCR-amplified DNA fragments into the corresponding E. coli chromosomal loci by λ Red-mediated homologous recombination as described (Ames et al., 2008). Genotypes were verified by sequencing PCR-amplified chromosomal DNA.
Plasmids
Regulatable Tsr expression plasmids [pRR53 (Studdert & Parkinson, 2005); pPA114 (Ames et al., 2002)] were used to characterize signaling properties of mutant receptor alleles in the tests described below. For in vivo kinase assays, FRET reporters were expressed from regulated plasmids that were compatible with the mutant Tsr plasmids: pVS88 (Sourjik et al., 2007) (compatible with pPA114) and pRZ30 (this work, compatible with pRR53).
Flagellar rotation assays
Strains containing Tsr expression plasmids were grown to mid-exponential phase in tryptone broth (TB) at 30°C and tethered to microscope cover slips with anti-flagellum antibodies to measure clockwise (CW) flagellar rotation time (Zhou et al., 2011, Ames et al., 2002).
Receptor expression levels
The intracellular levels of the variant Tsr proteins expressed from regulatable plasmids were measured as described (Ames & Parkinson, 2006). The inducer concentrations used produced an expression level of wild-type Tsr comparable to that of chromosomally-encoded Tsr in plasmid-free cells. Cell lysates were subjected to denaturing gel electrophoresis and Tsr molecules detected by western blotting with a rabbit polyclonal antiserum directed against the highly conserved protein interaction region of the Tsr molecule.
Cluster and ternary complex formation assays
The ability of variant receptors to form macroscopic clusters, characteristic of receptor arrays, was determined by fluorescence microscopy, as described (Mowery et al., 2008, Ames et al., 2002). Direct tests of clustering ability used a YFP-CheR reporter, which binds directly to the C-terminus of Tsr molecules (Wu et al., 1996). Ternary complex formation was assessed in a similar manner with a YFP-CheZ reporter, which binds to CheAS molecules that lack the P1 domain (Cantwell et al., 2003).
Jamming assays
Tsr plasmids were expressed in a strain that was wild-type for aspartate chemotaxis and tested for ability to block or jam Tar receptor function, as described (Ames et al., 2002).
In vivo FRET kinase assays
Sensitivity of mutant Tsr receptors to a serine stimulus was assessed with in vivo FRET-based kinase assays (Sourjik et al., 2007). Plasmids expressing Tsr variants were tested in strain UU2567 that also carried a compatible plasmid expressing the CheY-YFP and CheZ-CFP FRET reporters.
Electron cryotomography
E. coli strains were grown to mid-exponential phase at 30°C in TB. Cells were concentrated and incubated with 0.3 μg ml−1 penicillin for 1 hour at 30°C with shaking. Lysed cells were kept on ice until freezing. Immediately before plunge freezing, the cell lysates were mixed with colloidal gold pretreated with BSA to avoid particle aggregation (Iancu et al., 2007).
3 μl gold-lysate mixture were applied to R2/2 copper/rhodium Quantifoil ™ grids (Quantifoil Micro Tools), or R2/2 C-flat™ grids (Protochips). Excess liquid was blotted and the grids were plunge frozen in liquid ethane or ethane propane mixture (Tivol et al., 2008, Iancu et al., 2007).
Images were collected using an FEI Polara ™ (FEI) 300 kV field emission gun transmission electron microscope equipped with a Gatan energy filter and a lens-coupled 4,000 × 4,000 Ultracam (Gatan) or a direct detector in counting mode using the ‘K2 Summit’ detector (Gatan). Tilt series from −65° to 65° in 1 degree increments and an underfocus of −8 to −10 μm, and effective pixel sizes after 2 × 2 binning between 6.4 and 6.6 Å were recorded using Leginon or UCSFtomo (Zheng et al., 2007, Suloway et al., 2009). A cumulative dose of ~160 electrons/ Å2 was used for each tilt series. Tilt series were aligned using the IMOD software package (Kremer et al., 1996), CTF corrected (Kremer et al., 1996, Fernandez et al., 2006), and SIRT reconstructed using TOMO3D (Agulleiro & Fernandez, 2011). Subvolume averaging and 3-fold symmetrizing was done using PEET (Nicastro et al., 2006). Visualization and volume measurements were done using Chimera software (Pettersen et al., 2004).
Molecular dynamics flexible fitting (MDFF)
As a starting point for modeling a hexamer of trimeric receptors, a previously constructed model of the Tsr receptor up to the level of the HAMP domain was used (Briegel et al., 2012). MDFF, a method that permits one to flexibly fit atomic structures in lower resolution density maps while preserving the structures’ stereochemical accuracy (Trabuco et al., 2008), was then used on each map to generate unique fitted structures in full atomistic detail. The fitting simulations were carried out with the molecular dynamics program NAMD 2.9 (Phillips et al., 2005). Restraints were applied to maintain the proteins’ secondary structure and chirality, as well as to prevent the formation of cis peptide bonds (Schreiner et al., 2011). Additional restraints to limit distortions of tertiary structure in the individual trimers and to enforce hexagonal symmetry between the distinct trimers were also utilized (Chan et al., 2011). Fitting was carried out in multiple stages over 1.5 ns for each map in order to optimize the position of the hexamer in the density and then the fit of the trimers themselves.
CheA proteolysis in receptor arrays
E. coli strains were grown to mid-exponential phase in TB at 30°C with shaking. Cells were lysed by incubation with 0.3 μg ml−1 penicillin for 1 hour at 30°C with shaking and collected by centrifugation. Lysates were divided into two sets; one set was untreated and the other was exposed to 10 μg ml−1 Proteinase K for 30 minutes at 37°C. Proteinase K was then inactivated by addition of 5 mM phenylmethylsulfonyl fluoride; after 2 minutes, Laemmli sample buffer (Laemmli, 1970) was added and samples were boiled for 10 minutes and analyzed by western blotting. CheA-derived bands were visualized with polyclonal rabbit anti-CheA serum (1:1,500) and donkey anti-rabbit-horseradish peroxidase (HRP) secondary antibody (1:60,000; Pierce). To correct for loading differences, normalization was carried out using an unknown protease-insensitive protein detected by β-lactamase antiserum (1:2,000).
Supplementary Material
Acknowledgements
This work was supported in part by NIGMS grants GM101425 (to GJJ and JCG) and GM19559 (to JSP), as well as a gift to Caltech from the Gordon and Betty Moore Foundation. The Protein-DNA Core Facility at the University of Utah receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute. The authors declare no conflicts of interest.
Footnotes
Accession Numbers The EMDB accession numbers for the subvolume averages reported in this paper are as follows (strains in parentheses):
EMD-5541 (A413T), EMD-5542 (P221D), EMD-5543 (EEEE), EMD-5545 (QQQQ), EMD-5546 (I241E), EMD-5547 (G235E), EMD-5548 (QEQE), EMD-5549 (A413TΔP1P2), EMD-5550 (A413TΔP2).
References
- Agulleiro JI, Fernandez JJ. Fast tomographic reconstruction on multicore computers. Bioinformatics. 2011;27:582–583. doi: 10.1093/bioinformatics/btq692. [DOI] [PubMed] [Google Scholar]
- Ames P, Parkinson JS. Conformational suppression of inter-receptor signaling defects. Proc Natl Acad Sci USA. 2006;102:9292–9297. doi: 10.1073/pnas.0602135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P, Zhou Q, Parkinson JS. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 2008;190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Kaplan N, Hess JF, Simon MI. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrock MJ, Kolonko EM, Kiessling LL. Chemical Probes of Bacterial Signal Transduction Reveal That Repellents Stabilize and Attractants Destabilize the Chemoreceptor Array. ASC Chem Biol. 2008;3:101–109. doi: 10.1021/cb700211s. [DOI] [PubMed] [Google Scholar]
- Briegel A, Beeby M, Thanbichler M, Jensen GJ. Activated chemoreceptor arrays remain intact and hexagonally packed. Mol Microbiol. 2011;82:748–757. doi: 10.1111/j.1365-2958.2011.07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci USA. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Müller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell BJ, Draheim RR, Weart RB, Nguyen C, Stewart RC, Manson MD. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. Journal of Bacteriology. 2003;185:2354–2361. doi: 10.1128/JB.185.7.2354-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K-Y, Gumbart J, McGreevy R, Watermeyer JM, Sewell BT, Schulten K. Symmetry-restrained flexible fitting for symmetric EM maps. Structure. 2011;19:1211–1218. doi: 10.1016/j.str.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbse AH, Falke JJ. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochem. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez JJ, Li S, Crowther RA. CTF determination and correction in electron cryotomography. Ultramicroscopy. 2006;106:587–596. doi: 10.1016/j.ultramic.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Gan L, Jensen GJ. Electron tomography of cells. Quart Rev Biophys. 2012;45:27–56. doi: 10.1017/S0033583511000102. [DOI] [PubMed] [Google Scholar]
- Garzon A, Parkinson JS. Chemotactic signaling by the P1 phosphorylation domain liberated from the CheA histidine kinase of Escherichia coli. J Bacteriol. 1996;178:6752–6758. doi: 10.1128/jb.178.23.6752-6758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel DJ, Zhou H, Starich MR, Byrd RA, Dahlquist FW. Chemical-shift-perturbation mapping of the phosphotransfer and catalytic domain interaction in the histidine autokinase CheA from Thermotoga maritima. Biochemistry. 2006;45:9509–9517. doi: 10.1021/bi060798k. [DOI] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, Shiomi D, Homma M, Kawagishi I. Attractant binding alters arrangement of chemoreceptor dimers within its cluster at the cell pole. PNAS. 2004;101:3462–3467. doi: 10.1073/pnas.0306660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, Wright ER, Li Z, Yu Z, Briegel A, Gan L, He Y, Jensen GJ. Electron cryotomography sample preparation using the Vitrobot. Nat Protoc. 2007;1:2813–2819. doi: 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- Jahreis K, Morrison TB, Garzon A, Parkinson JS. Chemotactic signaling by an Escherichia coli CheA mutant that lacks the binding domain for phosphoacceptor partners. J Bacteriol. 2004;186:2664–2672. doi: 10.1128/JB.186.9.2664-2672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara CM, Lan G, Neumann S, Wu X, Ravindran S, Borgia MJ, Sourjik V, Milne JLS, Tu Y, Subramaniam S. Lateral density of receptor arrays in the membrane plane influences sensitivity of the E. coli chemotaxis response. EMBO J. 2011;30:1719–1729. doi: 10.1038/emboj.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional data using Imod. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamanna AC, Ordal GW, Kiessling LL. Large increases in atteactant concentration disrupt the polar localization of bacterial chemoreceptors. Molecular Microbiology. 2005;57:774–785. doi: 10.1111/j.1365-2958.2005.04728.x. [DOI] [PubMed] [Google Scholar]
- Liberman L, Berg HC, Sourjik V. Effect of chemoreceptor modification on assembly and activity of the receptor-kinase complex in Escherichia coli. J Bacteriol. 2004;186:6643–6646. doi: 10.1128/JB.186.19.6643-6646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci USA. 2012;109:E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybarger SR, Maddock JR. Clustering of the chemoreceptor complex in Escherichia coli is independent of the methyltransferase CheR and the methylesterase CheB. Journal of Bacteriology. 1999;181:5527–5529. doi: 10.1128/jb.181.17.5527-5529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TB, Parkinson JS. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of E. coli. Proc Natl Acad Sci USA. 1994;91:5485–5489. doi: 10.1073/pnas.91.12.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowery P, Ostler JB, Parkinson JS. Different signaling roles of two conserved residues in the cytoplasmic hairpin tip of Tsr, the E. coli serine chemoreceptor. J Bacteriol. 2008;190:8065–8074. doi: 10.1128/JB.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz CL, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Park S-Y, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-A visualization system for exploratory research analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, D. SR, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada CM, Hamel DJ, Gradinaru C, Bilwes AM, Dahlquist FW, Crane BR, Simon MI. Structural and chemical requirements for histidine phosphorylation by the chemotaxis kinase CheA. The Journal of Biological Chemistry. 2005;280:30581–30585. doi: 10.1074/jbc.M505316200. [DOI] [PubMed] [Google Scholar]
- Schreiner E, Trabuco LG, Freddolino PL, Schulten K. Stereochemical errors and their implications for molecular dynamics simulations. BMC Bioinformatics. 2011;12:190. doi: 10.1186/1471-2105-12-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. Protein exchange dynamics at chemoreceptor clusters in E. coli. Proc Natl Acad Sci USA. 2008;105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Vaknin A, Shimizu TS, Berg HC. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol. 2007;423:365–391. doi: 10.1016/S0076-6879(07)23017-4. [DOI] [PubMed] [Google Scholar]
- Stewart RC. Protein histidine kinases: assembly of active sites and their regulation in signaling pathways. Curr Opin Microbiol. 2010;13:133–141. doi: 10.1016/j.mib.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RC, Jahreis K, Parkinson JS. Rapid phosphotransfer to CheY from a CheA protein lacking the CheY-binding domain. Biochem. 2000;39:13157–13165. doi: 10.1021/bi001100k. [DOI] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci U S A. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc Natl Acad Sci U S A. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochem. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson RV, Bourret RB, Simon MI. Intermolecular complementation of the kinase activity of CheA. Mol Microbiol. 1993a;8:435–441. doi: 10.1111/j.1365-2958.1993.tb01588.x. [DOI] [PubMed] [Google Scholar]
- Swanson RV, Schuster SC, Simon MI. Expression of CheA fragments which define domains encoding kinase, phosphotransfer, and CheY binding activities. Biochem. 1993b;32:7623–7629. doi: 10.1021/bi00081a004. [DOI] [PubMed] [Google Scholar]
- Tivol W, Briegel A, Jensen GJ. An Improved Cryogen for Plunge Freezing. Microsc Micoanal. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Vu A, Lee K, Dahlquist FW. CheA-receptor interaction sites in bacterial chemotaxis. J Mol Biol. 2012;422:282–290. doi: 10.1016/j.jmb.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Stewart RC. The short form of the CheA protein restores kinase activity and chemotactic ability to kinase-deficient mutants. Proc Natl Acad Sci USA. 1993;90:1518–1522. doi: 10.1073/pnas.90.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li J, Li G, Long DG, Weis RM. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochem. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- Wu K, Walukietwicz HE, Glekas GD, Ordal GW, Rao CV. Attractant binding induces distinct structural changes to the polar and lateral signaling clusters in Bacillus subtilis chemotaxis. J Biol Chem. 2011;286:2587–2595. doi: 10.1074/jbc.M110.188664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Parkinson JS. Cysteine-scanning analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signalling kinase CheA. J Bacteriol. 2006a;188:4321–4330. doi: 10.1128/JB.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Parkinson JS. Mutational analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. Journal of Bacteriology. 2006b;188:3299–3307. doi: 10.1128/JB.188.9.3299-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QS, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA. UCSF tomography: An integrated software suite for real-time electron microscopic tomographic data collection, alignment and reconstruction. J Struct Biol. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP signaling in the Escherichia coli serine chemoreceptor. Mol Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.