Abstract
Background
Cysteine and methionine are the two sulfur containing amino acids in proteins. While the roles of protein-bound cysteinyl residues as endogenous antioxidants are well appreciated, those of methionine remain largely unexplored.
Scope
We summarize the key roles of methionine residues in proteins.
Major Conclusion
Recent studies establish that cysteine and methionine have remarkably similar functions.
General Significance
Both cysteine and methionine serve as important cellular antioxidants, stabilize the structure of proteins, and can act as regulatory switches through reversible oxidation and reduction.
1. Introduction
Methionine and cysteine are the two sulfur-containing amino acids that are present in peptides and proteins. It would not be difficult for most of us to list the functions of cysteine residues in proteins. Well-known roles include antioxidant defense, catalysis, protein structure, and redox sensing and regulation [1]. In contrast, we might have difficulty listing the functions of methionine (Met) residues, other than its well-known role in protein initiation. Biochemistry texts typically treat Met as a generic hydrophobic amino acid, readily interchangeable with other residues such as leucine or valine. This concept is woefully outdated. Over the last 15 years, research from a number of laboratories supports the concept that Met in proteins shares much of the same job description as cysteine, playing important roles in oxidant defense redox sensing and regulation as well as protein structure.
The most important common characteristic of cysteine and Met residues in proteins is that both are subject to reversible oxidation and reduction, mediated either enzymatically or non-enzymatically. While cysteine forms cystine through a disulfide linkage, Met forms methionine sulfoxide (MetO) by addition of oxygen to its sulfur atom. Disulfides may be reduced back to the thiol form by various reductases, often utilizing thioredoxin [2]. MetO is reduced back to Met by the methionine sulfoxide reductases, thioredoxin-dependent enzymes that are virtually universal among aerobic organisms [3, 4]. Oxidation of Met to MetO introduces a chiral center at the sulfur atom so there are two epimers of MetO; R-MetO and S-MetO. While an epimerase could theoretically exist that interconverts the forms, none has been found so far. Instead, organisms have two types of methionine sulfoxide reductases (Msr). MsrA specifically reduces S-MetO, but not R-MetO. Conversely, MsrB reduces R-MetO, but not S-MetO. The existence of MsrA has been appreciated for decades, while the existence of MsrB was only reported recently [5]. To date, there is substantial experimental evidence to support the importance of MsrA, both in vivo and in vitro. Knocking out MsrA caused increased susceptibility to oxidative stress in mice [6], yeast [7], and bacteria [8–10]. Conversely, overexpressing MsrA conferred increased resistance to oxidative stress in Drosophila [11], Saccharomyces [12], Arabidopsis [13], PC-12 cells [14], and human T cells [12]. Interestingly, overexpression in Drosophila doubled the lifespan of the flies [11]. Critical functions for MsrB remain to be defined given its more recent discovery.
While cysteine is well-recognized for the ease of its oxidation, it is often not appreciated that Met can be readily oxidized to MetO [15, 16]. Indeed, the standard redox potential for the two electron reduction of dimethyl sulfoxide is +160 mV [17] while that for cystine is +220 mV [18]. Cysteine is easily oxidized when ionized to its thiolate, but is difficult to oxidize when in the thiol form [19]. Cysteine residues at the active sites of enzymes such as phosphatases, dehydrogenases, reductases, and peroxidases generally have a low pKa which makes them readily oxidizable [19]. However, the majority of cysteine residues, including those in glutathione, have a pKa around 8.3–8.7 and are not easily oxidized at physiological pH, unless the oxidation is catalyzed by an enzyme. In contrast, oxidation of Met residues is essentially independent of pH [20]. In vitro, hypochlorous acid (HOCl), a major halogenated oxidant generated by leukocytes, reacts rapidly with Met at physiological pH [20, 21], but hydrogen peroxide does not, although the rate can be accelerated by the bicarbonate/carbon dioxide present in vivo [22]. The relative importance in vivo of cysteine and Met as antioxidants has not been established and most likely varies depending on the oxidizing agent.
2. Methionine residues in proteins as antioxidants
2.1. α2macroglobulin
α2macroglobulin (A2M) is a high molecular weight (~725 kDa), physiologically important plasma proteinase inhibitor that targets a wide variety of proteinases [23, 24]. Acting in a “venus-flytrap”-like mode that serves to entrap proteinases in a molecular cage [25], A2M normally circulates as a homotetrameric molecule that is disulfide linked into a pair of dimers that are held in association by strong non-covalent forces. In its open conformation, target proteinases cleave an exposed “bait” region within the A2M tetramer that triggers the structural changes that result in the irreversible entrapment of the protease. Often acting at sites of inflammation where reactive oxygen and nitrogen species are at relatively high concentration, it was initially thought that A2M was resistant to oxidative modification [26]. However, studies by Weiss and colleagues demonstrated that the antiproteinase was sensitive to oxidative modification by activated neutrophils, HOCl or derivative chloramines (a natural byproduct of neutrophil-generated HOCl following its reaction with amines) [24, 26]. In the course of these reactions, Met residues in A2M readily react with chlorinated oxidants, consuming the reactive species while oxidizing Met to its corresponding sulfoxide [26]. These oxidations had previously not been observed because only the activity of A2M was monitored, and, at least initially, the oxidation of Met residues proceeds without loss of anti-proteinase activity [26]. Our detailed study of the oxidation reaction established that each subunit of A2M consumes 8 mol of chloramine without any loss of anti-proteinase function [26]. During a second phase of oxidative modification, the A2M is inactivated with loss of activity proceeding in a manner directly proportional to the consumption of chlorinated oxidants. At this point, each subunit consumed 16 mol of chloramine, but only 14 Met residues were oxidized. Further studies demonstrated that a single tryptophan residue in each subunit was being oxidized by the remaining chloramine and that the decrease in total tryptophan residues (from 11 to 10) was directly proportional to loss of anti-proteinase activity in tandem with the dissociation of the tetrameric A2M into dimers [26].
That the tryptophan became susceptible to oxidation only after conversion of more than 8 Met residues to their sulfoxides could be explained by perturbation of the normal A2M structure by the presence of the additional sulfoxides, leading to an inactive conformation with incidental exposure of the normally buried tryptophan. However, the ability of A2M to tolerate oxidation of 8 Met per subunit without loss of activity led to the proposal of an alternative hypothesis in which these residues functioned as antioxidants that protected the critical tryptophan residue from oxidation. Using high pressure liquid chromatography and mass spectrometry, we have determined that the oxidized Trp residue is Trp413. We then generated both recombinant wild-type and Trp413Ala and Trp413Phe site-specific mutants in order to test whether loss of Trp413 was sufficient to cause dissociation of the tetramer to the dimer. Native gel electrophoresis demonstrated that wild-type A2M is, as expected, a tetramer, but both Trp413 mutants are almost completely dissociated to the dimer (Fig. 1). Thus, Trp413 must be intact for normal subunit-subunit interaction. We therefore proposed that the purpose of the multiple, readily oxidized Met residues in A2M was to act as a last line of defense against reactive species that had evaded low molecular weight antioxidants and enzymatic antioxidant systems [26–28]. In other words, Met residues serve as innate antioxidants or “molecular bodyguards”, positioned to intercept reactive species to prevent damage to other residues critical to the function of the protein.
Fig. 1.
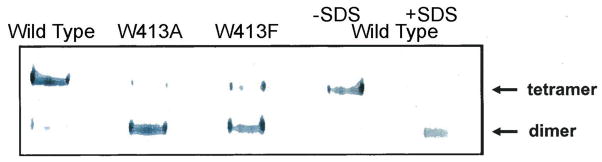
Native gel electrophoresis of wild-type and Trp413 mutant A2M. Samples were analyzed on a single native gel. The wild-type sample in the far right lane was made 2% in SDS to dissociate tetramers to disulfide-linked dimers.
The proteins were produced as follows: Chinese hamster ovary CHO-K1 cells were purchased from ATCC (catalog CCL-61, Manassas, Virginia, USA) and grown in DMEM (Gibco-BRL 10566-016, Life Technologies, Grand Island, New York, USA) containing 10% fetal calf serum. Cells were maintained in 5% CO2 and 5% oxygen. The human A2M gene sequence encoding wild type (WT), Trp413Ala, Trp413Phe was cloned into pcDNA 3.1 (+) (Invitrogen V790-20, Life Technologies, Grand Island, New York, USA). CHO-K1 cells were stably transfected with the expression vectors. The stably transfected cells were selected in DMEM with 10% FBS containing 3 mg/ml Geneticin (Invitrogen 10131-027). Selected cell lines were then grown in CHO serum-free culture medium (BioWhittaker 12-029Q, Walkersville, Maryland, USA) without Geneticin. After 73 h in culture, 30 ml of medium were collected and concentrated to 1.5 ml through a centrifugal filter YM10 (EMD Millipore, Billerica, Massachusetts, USA). The concentrated samples were subjected to native gel electrophoresis on a 6% Tris-glycine gel (EC6068, Invitrogen) run at 125 volts for 130 min at room temperature. Proteins were electroblotted to a polyvinylidene difluoride membrane (LC2002, Invitrogen), incubated with rabbit anti-human A2M as the primary antibody (DAKO A0033, DAKO, Carpinteria, California, USA) and an anti-rabbit IgG conjugated to alkaline phosphatase (475-1516, KPL, Gaithersburg, Maryland, USA) as the secondary antibody. A2M was visualized by incubation with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (50-81-08, KPL).
2.2. Glutamine Synthetase
To date, there are no crystal structures for native A2M. While there is a low resolution structure of methylamine-reacted A2M (a form of A2M wherein the anti-proteinase undergoes conformational changes similar to those elicited by targeted proteinases) [29], this conformation is known to be very different than that of the native A2M [30, 31], thereby complicating efforts to characterize oxidative events at the structural level. We therefore turned our focus to glutamine synthetase from E. coli, for which several excellent crystal structures have been determined [32–34]. Exposure of the enzyme to varying concentrations of hydrogen peroxide generated a series of preparations with an increasing content of MetO; no other covalent modifications were detected [27]. Eight of the 16 Met residues could be oxidized without loss of catalytic activity. Mapping of the oxidizable Met residues revealed that all were surface exposed; conversely, the residues that remained unoxidized were buried. More detailed examination of the topographic distribution of the oxidizable Met residues was intriguing as these residues were found to line the bay leading from the surface of the enzyme to its active site (Fig. 4 in [27]). In other words, these Met residues are mustered in a phalanx guarding the active site where they too function as macromolecular bodyguards.
2.3. Enlistment of methionines as antioxidants is widespread
In addition to glutamine synthetase and A2M, many other proteins have likely evolved with similar placement of “guardian” Met residues. For example, mammalian 15-lipoxygenases undergo an apparently irreversible auto-inactivation during the catalytic cycle. The enzyme contains ~16 Met residues, and oxidation of a single Met near the active site correlates with inactivation [35]. However, Gan and colleagues reported that site-specific replacement of the oxidizable Met590 by a leucine residue yielded an enzyme that remained sensitive to auto-inactivation [36]. Thus, while Met590 is critical to in the regulation or catalytic activity of the lipoxygenase, it presumably functions as a guardian antioxidant for the active site.
Consistent with the proposition that Met residues can serve as endogenous antioxidants, it is interesting to note that the effective concentration of exposed Met residues is extremely high near the protein surface; greater than 1 M given certain assumptions [27]. Recognizing the ease of Met oxidation, surface-exposed Met residues may well constitute a formidable antioxidant defense mechanism, capable of protecting critical residues within the protein itself or even surrounding molecules. An elegant example of the latter comes from Stocker and colleagues who established that high density lipoproteins reduce cholesteryl ester hydroperoxides to alcohols, with the concomitant oxidation of two Met residues to the sulfoxides [37]. The apolipoprotein may also function in a catalytic manner as the oxidized apolipoprotein can be reduced by Msr [38] with the rate of repair rendered even more efficient with the myristoylated form of MsrA that is present in vivo [39].
Studies of changes that occur during evolution provide additional support for the importance of Met residues as endogenous antioxidants. Bender and colleagues focused on the evolution of the mitochondrial genetic code, taking note of the fact that it differs from the nuclear code in many animals [40]. In the nucleus, AUA codes for isoleucine, but it specifies Met in animals using the modified code. By analyzing a large number of species that do not use the modified mitochondrial code, they established that the average Met content in mitochondrially-encoded proteins is ~2%, which is the same as that for nuclear encoded in those organisms. However, in organisms whose mitochondrial code evolved to specify Met by AUA, the average mitochondrial Met content jumped 3-fold to ~6%. Moreover, the additional Met residues were again topographically arranged on the surface of the proteins, ideally positioned to intercept reactive oxygen species generated by mitochondrial respiration.
A direct test of the hypothesis that Met residues are in vivo antioxidants was performed by globally altering the Met content of proteins [41]. This can be done in microorganisms, such as E. coli, by culturing a Met auxotroph in medium rich in norleucine, but poor in Met [41]. Norleucine, being the carbon analogue of Met, cannot form a sulfoxide and thus, lacks the antioxidant potential of Met. When grown in the norleucine medium, 40% of the Met residues in E. coli were replaced by norleucine [42]. If the hypothesis that Met serves as an antioxidant hypothesis were correct, the Met-poor organisms should prove more sensitive to oxidative stress. Indeed, when left unstressed, both control and norleucine-substituted cells survived equally well in stationary phase for at least 32 hours. In contrast, when challenged by exposure to hypochlorous acid, hydrogen peroxide or ionizing radiation, the norleucine-substituted cells died more rapidly than the control cells [42]. To date, this is one of the few direct experimental tests of our operative hypothesis, so more studies are clearly needed.
3. Methionine residues and protein structure
Valley and colleagues recently established that Met residues in proteins are frequently positioned so that they establish a hydrophobic bond between their sulfur atoms and the rings of aromatic residues, including tryptophan, phenylalanine and tyrosine [43]. These hydrophobic sulfur-ring bonds are very common and contribute to the structural stability of proteins with a bond energy of 1.0–1.5 kcal/mol each -- approximately equal to that of an ionic salt bridge [43]. The aromatic amino acids are among the most susceptible to oxidation by reactive species [44], so that interaction with the Met establishes the optimal positioning needed to provide antioxidant protection. Of course, conversion of the Met to MetO will eliminate the hydrophobic bond, and it is likely to perturb the normal 3-dimensional folding of the protein. These perturbations may expose otherwise normally buried residues, explaining the association of Met oxidation with increased surface hydrophobicity of proteins [27, 45]. This effect may be substantial during aging in which progressive increases in the surface hydrophobicity of proteins correlate with an age-related increase in MetO content [45]. Many other reports of increased MetO content in aging tissues have appeared, but the changes are rather modest and no studies have yet validated total MetO content as a marker of biological aging. Progress in developing MetO as a useful marker is severely hampered by the lack of analytical tools for detecting and quantitating MetO content of individual proteins in complex mixtures such as those found in tissue homogenates. The absence of immunochemical or chemical methods for detecting and quantitating MetO is particularly vexing [46], although efforts to circumvent this problem may be forthcoming [47].
Met oxidation has usually been considered in terms of pathological, oxidative events. For example, Johnson and Travis established that the oxidative inactivation of alpha-1 proteinase inhibitor (α1-antitrypsin) was due to oxidation of a critical Met, Met358 [48], and many reports implicate this oxidative inactivation as contributing to pulmonary diseases [49–51]. In an unusual example of physiologic Met oxidation, Hudson and colleagues [52] have demonstrated a key role for this event in stabilizing type IV collagen structure (a key component of basement membranes, a specialized form of extracellular matrix that subtends all endothelial and epithelial cells). In a mechanism that is conserved from flies to humans, the carboxyl-terminal Met of one type IV collagen subunit is covalently linked to a lysine of another subunit through a novel sulfilimine (S=N) bond [52]. Formation of the sulfilimine is catalyzed by a specific peroxidase, termed peroxidasin, that appears to generate the sulfilimine by formation of hypohalous acids as a reactive intermediate [53]. Drosophila with mutant peroxidasin fail to generate sulfilimine cross-links and display disorganized collagen IV networks with associated defects in basement membrane structure [53].
4. Regulation by oxidation and reduction of methionine residues
Like phosphorylation, Met oxidation is a reversible covalent modification. Thus, cyclic oxidation and reduction of Met residues could function as regulatory processes, including cell signaling [54, 55]. Ciorba and colleagues reported that the inactivation of a potassium channel by nitric oxide was likely due to oxidation of an essential Met residue in the channel [56]. Similarly, Sroussi et al presented evidence that the ability of the calcium-binding proteins to direct leukocyte migration was abolished by oxidation of specific Met residues [57]. Interestingly, consistent with the notion that Met oxidation does not invariably link to enzyme inactivation, Erickson and collaborators convincingly identified a calcium-independent pathway for activation of Ca2+/calmodulin-dependent protein kinase that was mediated by oxidation of specific Met residue in vitro and in vivo [58]. In yet another example of a MetO “activation” process, oxidation of Met80, an iron ligand in cytochrome c, increases cytoplasmic translocation of the cytochrome as part of a potential defense system against nitrative stress in non-apoptotic cells [59]. Likewise, in plants, hydrogen peroxide-triggered protein phosphorylation can be regulated by oxidation of a specific Met residue in the substrate recognition site of kinases [60]. Alternatively, Met oxidation may lead to enhanced function via indirect routes. For example, the blood clotting protein, von Willebrand factor, undergoes HOCl-dependent Met oxidation that renders the protein resistant to proteolysis by the metalloprotease, ADAMTS123, thereby endowing it with increased activity [61]. Although most examples of Met oxidation are dependent on the generation of reactive oxidants, the Terman laboratory has recently identified an NADPH oxidoreductase, Mical, that specifically oxidizes a Met residue in actin that induces filament severing and decreases actin polymerization [62].
MsrA and MsrB are stereospecific reductases, but a stereospecific peroxidase is also required to complete the enzymatic requirements for a reversible covalent modification. We and others had searched for such a stereospecific methionine peroxidase, until recently without success. What we had not realized was that we already had a peroxidase in hand: It is methionine sulfoxide reductase A, which we now recognize as a bifunctional enzyme capable of both stereospecific oxidation of Met residues to S-MetO as well as reduction back to Met [63]. The enzyme has an active site cysteine, Cys72, located in the NH2-terminal domain of the protein. When functioning as a reductase, Cys72 reduces MetO and is itself oxidized to the sulfenic acid. The sulfenic acid is then reduced back to the thiol by “resolving” cysteine residues located close to the carboxyl-terminus of the protein. In turn, the resolving cysteines form a disulfide bond that is reduced back to free thiols by thioredoxin, for which a binding site exists in the carboxyl terminal domain of MsrA. Conversely, when functioning as a peroxidase, the Cys72 residue in MsrA is also oxidized to the sulfenic acid, but instead of transferring the oxidizing equivalents to the resolving cysteines, the sulfenic acid oxidizes a Met in a protein substrate. Either hydrogen peroxide or MetO can serve as the donor of the oxidizing equivalents [64]. Dual-function enzymes that catalyze both the forward and reverse reactions of reversible covalent modifications are well-described in the literature [65, 66]. Futile cycles are prevented by mechanisms that inactivate either the forward or reverse reactions, generally by a covalent modification of the bifunctional enzyme such as phosphorylation or by binding of a regulatory protein [66]. We have suggested that the latter mechanism is the more likely means of directing MsrA to function as a peroxidase or a reductase. To direct MsrA to function as a peroxidase, the putative regulatory protein needs only bind to the carboxyl domain of MsrA to block access to the resolving cysteines. In this fashion, dissociation of the proteins would switch MsrA to the reductase mode [63].
The finding that MsrA is a stereospecific methionine peroxidase establishes the potential of cyclic oxidation and reduction of Met residues to function as regulatory switches. However, until proteins that undergo such redox cycles in vivo are identified, the proposition remains hypothetical. Intriguing possibilities include reversal of the Mical oxidation of actin mentioned above [62] and in the insulin signaling pathway wherein Styskal and colleagues have reported that MsrA knockout mice display reduced insulin responses and are prone to develop insulin resistance [67]. Efforts to characterize MsrA function in these mice may lead to insights into Met redox cycling.
5. Conclusion
The two sulfur containing amino acids, cysteine and Met, share common functions. Increasing evidence indicates that both amino acids are important antioxidants that contribute to the structure and stability of proteins. Their relative ease of reversible oxidation and reduction imbues both amino acids with the ability to serve as regulatory switches and signals in increasingly broad venues.
Highlights.
The roles of methionine in proteins are not well understood.
Methionine can act as antioxidant, protein stabilizer, or regulatory switch.
We conclude that cysteine and methionine have remarkably similar functions.
Acknowledgments
Funding: This study was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute.
Abbreviations
- A2M
α2macroglobulin
- MetO
methionine sulfoxide
- Msr
methionine sulfoxide reductase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, Stamler JS, Rhee SG, van der Vliet A. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic Biol Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. European journal of biochemistry / FEBS. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 3.Brot N, Weissbach H. Biochemistry and physiological role of methionine sulfoxide reductase in proteins. Arch Biochem Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 4.Zhang XH, Weissbach H. Origin and evolution of the protein-repairing enzymes methionine sulphoxide reductases. Biol Rev Camb Philos Soc. 2008;83:249–257. doi: 10.1111/j.1469-185X.2008.00042.x. [DOI] [PubMed] [Google Scholar]
- 5.Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, Derrick PJ, Barras F. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 6.Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci USA. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moskovitz J, Berlett BS, Poston JM, Stadtman ER. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci USA. 1997;94:9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moskovitz J, Rahman MA, Strassman J, Yancey SO, Kushner SR, Brot N, Weissbach H. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas T, Daniel DS, Parida BK, Jagannath C, Dhandayuthapani S. Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J Bacteriol. 2004;186:3590–3598. doi: 10.1128/JB.186.11.3590-3598.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, Nathan C. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci USA. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruan H, Tang XD, Chen ML, Joiner ML, Sun G, Brot N, Weissbach H, Heinemann SH, Iverson L, Wu CF, Hoshi T, Chen ML, Joiner MA, Heinemann SH. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moskovitz J, Flescher E, Berlett BS, Azare J, Poston JM, Stadtman ER. Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc Natl Acad Sci USA. 1998;95:14071–14075. doi: 10.1073/pnas.95.24.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romero HM, Berlett BS, Jensen PJ, Pell EJ, Tien M. Investigations into the role of the plastidial peptide methionine sulfoxide reductase in response to oxidative stress in Arabidopsis. Plant Physiol. 2004;136:3784–3794. doi: 10.1104/pp.104.046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yermolaieva O, Xu R, Schinstock C, Brot N, Weissbach H, Heinemann SH, Hoshi T. Methionine sulfoxide reductase A protects neuronal cells against brief hypoxia/reoxygenation. Proc Natl Acad Sci USA. 2004;101:1159–1164. doi: 10.1073/pnas.0308215100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavine TF. The formation, resolution, and optical properties of the diasteriomeric sulfoxides derived from L-methionine. J Biol Chem. 1947;169:477–491. [PubMed] [Google Scholar]
- 16.Vogt W. Oxidation of methionine residues in proteins: Tools, targets, and reversal. Free Rad Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 17.Wood PM. The redox potential for dimethyl sulphoxide reduction to dimethyl sulphide: evaluation and biochemical implications. FEBS Lett. 1981;124:11–14. doi: 10.1016/0014-5793(81)80042-7. [DOI] [PubMed] [Google Scholar]
- 18.Jocelyn PC. The Standard Redox Potential of Cysteine-Cystine from the Thiol-Disulphide Exchange Reaction with Glutathione and Lipoic Acid. Eur J Biochem. 1967;2:327–331. doi: 10.1111/j.1432-1033.1967.tb00142.x. [DOI] [PubMed] [Google Scholar]
- 19.Barton JP, Packer JE, Sims RJ. Kinetics of reaction of hydrogen-peroxide with cysteine and cysteamine. J Chem Soc Perkin Trans2. 1973:1547–1549. [Google Scholar]
- 20.Peskin AV, Winterbourn CC. Kinetics of the reactions of hypochlorous acid and amino acid chloramines with thiols, methionine, and ascorbate. Free Radic Biol Med. 2001;30:572–579. doi: 10.1016/s0891-5849(00)00506-2. [DOI] [PubMed] [Google Scholar]
- 21.Winterbourn CC. Comparative reactivities of various biological compounds with myeloperoxidase-hydrogen peroxide-chloride, and similarity of the oxidant to hypochlorite. Biochim Biophys Acta. 1985 Jun 18;840:204–210. doi: 10.1016/0304-4165(85)90120-5. [DOI] [PubMed] [Google Scholar]
- 22.Richardson DE, Regino CA, Yao H, Johnson JV. Methionine oxidation by peroxymonocarbonate, a reactive oxygen species formed from CO2/bicarbonate and hydrogen peroxide. Free Radic Biol Med. 2003;35:1538–1550. doi: 10.1016/j.freeradbiomed.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Sottrup-Jensen L. Alpha-macroglobulins: structure, shape, and mechanism of proteinase complex formation. J Biol Chem. 1989;264:11539–11542. [PubMed] [Google Scholar]
- 24.Travis J, Salvesen GS. Human plasma proteinase inhibitors. Annu Rev Biochem. 1983;52:655–709. doi: 10.1146/annurev.bi.52.070183.003255. [DOI] [PubMed] [Google Scholar]
- 25.Barrett AJ, Starkey PM. The interaction of alpha 2-macroglobulin with proteinases. Characteristics and specificity of the reaction, and a hypothesis concerning its molecular mechanism. Biochem J. 1973;133:709–724. doi: 10.1042/bj1330709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy VY, Desrochers PE, Pizzo SV, Gonias SL, Sahakian JA, Levine RL, Weiss SJ. Oxidative dissociation of human α2macroglobulin tetramers into dysfunctional dimers. J Biol Chem. 1994;269:4683–4691. [PubMed] [Google Scholar]
- 27.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine RL, Berlett BS, Moskovitz J, Mosoni L, Stadtman ER. Methionine residues may protect proteins from critical oxidative damage. Mech Ageing Dev. 1999;107:323–332. doi: 10.1016/s0047-6374(98)00152-3. [DOI] [PubMed] [Google Scholar]
- 29.Marrero A, Duquerroy S, Trapani S, Goulas T, Guevara T, Andersen GR, Navaza J, Sottrup-Jensen L, Gomis-Ruth FX. The crystal structure of human alpha2-macroglobulin reveals a unique molecular cage. Angew Chem Int Ed Engl. 2012;51:3340–3344. doi: 10.1002/anie.201108015. [DOI] [PubMed] [Google Scholar]
- 30.Kolodziej SJ, Wagenknecht T, Strickland DK, Stoops JK. The three-dimensional structure of the human alpha 2-macroglobulin dimer reveals its structural organization in the tetrameric native and chymotrypsin alpha 2-macroglobulin complexes. J Biol Chem. 2002;277:28031–28037. doi: 10.1074/jbc.M202714200. [DOI] [PubMed] [Google Scholar]
- 31.Van Leuven F, Cassiman JJ, Van den Berghe H. Functional modifications of alpha 2-macroglobulin by primary amines. I. Characterization of alpha 2 M after derivatization by methylamine and by factor XIII. J Biol Chem. 1981;256:9016–9022. [PubMed] [Google Scholar]
- 32.Almassy RJ, Janson CA, Hamlin R, Xuong NH, Eisenberg D. Novel subunit-subunit interactions in the structure of glutamine synthetase. Nature. 1986;323:304–309. doi: 10.1038/323304a0. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita MM, Almassy RJ, Janson CA, Cascio D, Eisenberg D. Refined atomic model of glutamine synthetase at 3.5 A resolution. J Biol Chem. 1989;264:17681–17690. doi: 10.2210/pdb2gls/pdb. [DOI] [PubMed] [Google Scholar]
- 34.Liaw SH, Eisenberg D. Structural model for the reaction mechanism of glutamine synthetase, based on five crystal structures of enzyme-substrate complexes. Biochemistry. 1994;33:675–681. doi: 10.1021/bi00169a007. [DOI] [PubMed] [Google Scholar]
- 35.Rapoport S, Hartel B, Hausdorf G. Methionine sulfoxide formation: the cause of self-inactivation of reticulocyte lipoxygenase. Eur J Biochem. 1984;139:573–576. doi: 10.1111/j.1432-1033.1984.tb08043.x. [DOI] [PubMed] [Google Scholar]
- 36.Gan QF, Witkop GL, Sloane DL, Straub KM, Sigal E. Identification of a specific methionine in mammalian 15- lipoxygenase which is oxygenated by the enzyme product 13-HPODE: dissociation of sulfoxide formation from self-inactivation. Biochemistry. 1995;34:7069–7079. doi: 10.1021/bi00021a019. [DOI] [PubMed] [Google Scholar]
- 37.Garner B, Waldeck AR, Witting PK, Rye KA, Stocker R. Oxidation of high density lipoproteins. II. Evidence for direct reduction of lipid hydroperoxides by methionine residues of apolipoproteins AI and AII. J Biol Chem. 1998;273:6088–6095. doi: 10.1074/jbc.273.11.6088. [DOI] [PubMed] [Google Scholar]
- 38.Sigalov AB, Stern LJ. Enzymatic repair of oxidative damage to human apolipoprotein AI. FEBS Lett. 1998;433:196–200. doi: 10.1016/s0014-5793(98)00908-9. [DOI] [PubMed] [Google Scholar]
- 39.Lim JC, Gruschus JM, Ghesquiere B, Kim G, Piszczek G, Tjandra N, Levine RL. Characterization and solution structure of mouse myristoylated methionine sulfoxide reductase A. J Biol Chem. 2012;287:25589–25595. doi: 10.1074/jbc.M112.368936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bender A, Hajieva P, Moosmann B. Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc Natl Acad Sci USA. 2008;105:16496–16501. doi: 10.1073/pnas.0802779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cowie DB, Cohen GN, Bolton ET, De Robichon-Szulmajster H. Amino acid analog incorporation into bacterial proteins. Biochim Biophys Acta. 1959;34:39–46. doi: 10.1016/0006-3002(59)90230-6. [DOI] [PubMed] [Google Scholar]
- 42.Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J. 2009;23:464–472. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valley CC, Cembran A, Perlmutter JD, Lewis AK, Labello NP, Gao J, Sachs JN. The Methionine-aromatic Motif Plays a Unique Role in Stabilizing Protein Structure. J Biol Chem. 2012;287:34979–34991. doi: 10.1074/jbc.M112.374504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liebster J, Kopoldova J. The radiation chemistry of amino acids. Adv Rad Biol. 1964;1:157–226. [Google Scholar]
- 45.Chao CC, Ma YS, Stadtman ER. Modification of protein surface hydrophobicity and methionine oxidation by oxidative systems. Proc Natl Acad Sci USA. 1997;94:2969–2974. doi: 10.1073/pnas.94.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wehr NB, Levine RL. Wanted and wanting: antibody against methionine sulfoxide. Free Radic Biol Med. 2012;53:1222–1225. doi: 10.1016/j.freeradbiomed.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghesquiere B, Jonckheere V, Colaert N, Van Durme J, Timmerman E, Goethals M, Schymkowitz J, Rousseau F, Vandekerckhove J, Gevaert K. Redox proteomics of protein-bound methionine oxidation. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M1110.006866-006861-006812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson D, Travis J. The oxidative inactivation of human alpha-1-proteinase inhibitor. Further evidence for methionine at the reactive center. JBiol Chem. 1979;254:4022–4026. [PubMed] [Google Scholar]
- 49.Carp H, Miller F, Hoidal JR, Janoff A. Potential mechanism of emphysema: alpha 1-proteinase inhibitor recovered from lungs of cigarette smokers contains oxidized methionine and has decreased elastase inhibitory capacity. Proc Natl Acad Sci USA. 1982;79:2041–2045. doi: 10.1073/pnas.79.6.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGuire WW, Spragg RG, Cohen AB, Cochrane CG. Studies on the pathogenesis of the adult respiratory distress syndrome. J Clin Invest. 1982;69:543–553. doi: 10.1172/JCI110480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogden BE, Murphy SA, Saunders GC, Pathak D, Johnson JD. Neonatal lung neutrophils and elastase/proteinase inhibitor imbalance. Am Rev Respir Dis. 1984;130:817–821. doi: 10.1164/arrd.1984.130.5.817. [DOI] [PubMed] [Google Scholar]
- 52.Vanacore R, Ham AJ, Voehler M, Sanders CR, Conrads TP, Veenstra TD, Sharpless KB, Dawson PE, Hudson BG. A sulfilimine bond identified in collagen IV. Science. 2009;325:1230–1234. doi: 10.1126/science.1176811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhave G, Cummings CF, Vanacore RM, Kumagai-Cresse C, Ero-Tolliver IA, Rafi M, Kang JS, Pedchenko V, Fessler LI, Fessler JH, Hudson BG. Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat Chem Biol. 2012;8:784–790. doi: 10.1038/nchembio.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoshi T, Heinemann S. Regulation of cell function by methionine oxidation and reduction. J Physiol. 2001;531:1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bigelow DJ, Squier TC. Redox modulation of cellular signaling and metabolism through reversible oxidation of methionine sensors in calcium regulatory proteins. Biochim Biophys Acta. 2005;1703:121–134. doi: 10.1016/j.bbapap.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 56.Ciorba MA, Heinemann SH, Weissbach H, Brot N, Hoshi T. Regulation of voltage-dependent K+ channels by methionine oxidation: effect of nitric oxide and vitamin C. FEBS Lett. 1999;442:48–52. doi: 10.1016/s0014-5793(98)01616-0. [DOI] [PubMed] [Google Scholar]
- 57.Sroussi HY, Berline J, Palefsky JM. Oxidation of methionine 63 and 83 regulates the effect of S100A9 on the migration of neutrophils in vitro. J Leukoc Biol. 2007;81:818–824. doi: 10.1189/jlb.0706433. [DOI] [PubMed] [Google Scholar]
- 58.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O’Donnell SE, Aykin-Burns N, Zimmerman MC, Zimmerman K, Ham AJ, Weiss RM, Spitz DR, Shea MA, Colbran RJ, Mohler PJ, Anderson ME. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Godoy LC, Munoz-Pinedo C, Castro L, Cardaci S, Schonhoff CM, King M, Tortora V, Marin M, Miao Q, Jiang JF, Kapralov A, Jemmerson R, Silkstone GG, Patel JN, Evans JE, Wilson MT, Green DR, Kagan VE, Radi R, Mannick JB. Disruption of the M80-Fe ligation stimulates the translocation of cytochrome c to the cytoplasm and nucleus in nonapoptotic cells. Proc Natl Acad Sci USA. 2009;106:2653–2658. doi: 10.1073/pnas.0809279106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hardin SC, Larue CT, Oh MH, Jain V, Huber SC. Coupling oxidative signals to protein phosphorylation via methionine oxidation in Arabidopsis. Biochem J. 2009;422:305–312. doi: 10.1042/BJ20090764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu X, Chen J, Gallagher R, Zheng Y, Chung DW, Lopez JA. Shear stress-induced unfolding of VWF accelerates oxidation of key methionine residues in the A1A2A3 region. Blood. 2011;118:5283–5291. doi: 10.1182/blood-2011-01-331074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lim JC, You Z, Kim G, Levine RL. Methionine sulfoxide reductase A is a stereospecific methionine oxidase. Proc Natl Acad Sci USA. 2011;108:10472–10477. doi: 10.1073/pnas.1101275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim JC, Gruschus JM, Kim G, Berlett BS, Tjandra N, Levine RL. A Low pKa cysteine at the active site of mouse methionine sulfoxide reductase A. J Biol Chem. 2012;275:25596–25601. doi: 10.1074/jbc.M112.369116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shapiro BM, Stadtman ER. The regulation of glutamine synthesis in microorganisms. Annu Rev Microbiol. 1970;24:501–524. doi: 10.1146/annurev.mi.24.100170.002441. [DOI] [PubMed] [Google Scholar]
- 66.Anderson WB, Hennig SB, Ginsburg A, Stadtman ER. Association of ATP: glutamine synthetase adenylyltransferase activity with the P1 component of the glutamine synthetase deadenylylation system. Proc Natl Acad Sci USA. 1970;67:1417–1424. doi: 10.1073/pnas.67.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Styskal J, Nwagwu FA, Watkins YN, Liang H, Richardson A, Musi N, Salmon AB. Methionine sulfoxide reductase A affects insulin resistance by protecting insulin receptor function. Free Radic Biol Med. 2013;56:123–132. doi: 10.1016/j.freeradbiomed.2012.10.544. [DOI] [PMC free article] [PubMed] [Google Scholar]


