Abstract
Objectives
We characterized human H1N1 influenza isolate A/Hokkaido/15/02, which has haemagglutinin and neuraminidase mutations that reduce drug susceptibility to oseltamivir, zanamivir and peramivir.
Methods
One wild-type and three mutant viruses were isolated by plaque purification. Viruses were tested in MUNANA-based enzyme assays, cell culture and receptor binding assays.
Results
Two viruses had a neuraminidase Y155H mutation that reduced susceptibility in the enzyme inhibition assay to all inhibitors by 30-fold to >100-fold. The Y155H mutation reduced plaque size and affected the stability, Km and pH activity profile of the enzyme. In contrast to previous mutants, this neuraminidase demonstrated a slower rate of inhibitor binding in the IC50 kinetics assay. One virus had both the Y155H mutation and a haemagglutinin D225G mutation that rescued the small-plaque phenotype of the Y155H virus and affected receptor binding and drug susceptibility in cell culture and binding assays. We also isolated a third mutant virus, with both neuraminidase V114I and haemagglutinin D225N mutations, which affected susceptibility in the enzyme inhibition assay and receptor binding, respectively, but to lesser extents than the Y155H and D225G mutations.
Conclusions
Neither Y155 nor V114 is conserved across neuraminidase subtypes. Furthermore, Y155 is not conserved even among avian and swine N1 viruses. Structurally, both residues reside far from the neuraminidase active site. D225 forms part of the receptor binding site of the haemagglutinin. We believe this is the first demonstration of a specific haemagglutinin mutation correlating with reduced drug susceptibility in plaque assays in both Madin Darby Canine Kidney and SIAT cells.
Keywords: oseltamivir, zanamivir, peramivir
Introduction
The influenza neuraminidase inhibitors (NAIs) zanamivir, oseltamivir and peramivir are effective inhibitors of all strains of influenza. The neuraminidase (NA) active site contains 9 conserved catalytic residues and a further 10 residues that form a second shell providing structural stability to the catalytic residues.1,2 Mutations conferring altered susceptibility to the NAIs mostly reside within these amino acids.3 The most common mutation in influenza N1 strains is H274Y, which confers resistance to both oseltamivir and peramivir. Many early studies showed that H274Y mutant viruses were compromised in their fitness. However, an A/Brisbane/59/2007-like H1N1 virus with an H274Y mutation that recently emerged has no reduced fitness, and rapidly spread globally.4,5 Studies on the evolution of this virus have revealed that non-active site residues (R222Q, V234M and D344N) have subtle but important effects on NA enzyme function, stability and levels of expression.6,7
The A/Hokkaido/15/02 virus was initially identified as an outlier during NAI susceptibility screening of global isolates through the Neuraminidase Inhibitor Susceptibility Network.8 The mutation conferred reduced susceptibility to both oseltamivir and zanamivir, with IC50 values in the chemiluminescence assay >100-fold higher than the wild-type. Sequencing identified a unique Y155H mutation in the NA. Searching through the sequence databases identified that Y155 is conserved in all human N1 viruses, but H155 is found in some wild-type swine and avian N1 viruses and in some earlier human N2 viruses. The NAs of the highly pathogenic H5N1 avian influenza viruses have H155 yet are susceptible to the inhibitors.9 Although we know that mutations conferring resistance vary between different influenza NA subtypes,3,10,11 these findings additionally demonstrate that resistance can vary even within a subtype.
Plaque assays of the amplified A/Hokkaido/15/02 sample revealed a heterogeneous mixture of large and small plaques. Plaque purification identified viruses with a mixture of wild-type and resistant phenotypes in enzyme assays and cell culture. In addition to viruses with the Y155H NA mutation, we identified another plaque isolate with a V114I NA mutation, also at a non-conserved residue, that reduced drug binding. Two isolates also contained D225 (H3 numbering) haemagglutinin (HA) mutations in addition to an NA mutation, which affected receptor binding and drug susceptibility. We have characterized the impact of the mutations on the properties of HA and NA, and on virus growth.
Materials and methods
Cells and viruses
The A/Hokkaido/15/02 and wild-type A/Hokkaido/6/02 viruses were originally isolated at the Public Health Institute of Hokkaido, and were obtained from the National Institute of Infectious Diseases (NIID) after one passage in Caco2 cells (P1) followed by three passages in Madin Darby Canine Kidney (MDCK) cells. Plaques were picked and replaqued prior to amplification in MDCK cells. SIAT MDCK cells, which express increased levels of the gene for 2,6-sialyltransferase, were kindly provided by Dr H. Klenk (Marburg, Germany). Reassortant viruses were selected from coinfection with the A/Hokkaido/15/02 wild-type virus and one of each of the two viruses that had both HA and NA mutations, to generate viruses with wild-type NA and each HA mutation.
Cells were grown in Dulbecco's modified Eagle's medium (DMEM)/F12 medium supplemented with 7.5% fetal calf serum, glutamine and antibiotics. SIAT cells were supplemented with Geneticin (Sigma, Australia) at 1 mg/mL. Viruses were amplified in maintenance medium (MM), minimal essential medium (MEM) and Liebovitz L15 at a ratio of 1 : 1, without serum, supplemented with 1 μg/mL trypsin (Worthington, USA). Plaque assays in both cells used an overlay of DMEM/F12 without serum, and 0.5% immunodiffusion grade agarose (MP Biomedicals, Australia) containing 1 μg/mL trypsin. For plaque reduction assays, zanamivir was incorporated into the agarose overlay to final concentrations in the range of 0.01 nM to 10 μM.
Kinetics of replication
MDCK and SIAT cells in 24-well dishes were infected at a multiplicity of infection of 1.0, and duplicate wells were harvested every 6 or 8 h, respectively. Supernatants were removed for titration of cell-free virus and cells were washed with PBS and scraped into the same volume of fresh MM, then frozen and thawed prior to titration of cell-associated virus.
Chemicals and inhibitors
Zanamivir and peramivir were synthesized by GlaxoSmithKline (Stevenage, UK). Oseltamivir carboxylate was prepared by Dr Keith Watson (Walter and Eliza Hall Institute, Australia) by dissolving oseltamivir phosphate powder in water, neutralizing with sodium bicarbonate, then extracting with ethyl acetate to produce free oseltamivir. This was then dissolved in tetrahydrofuran and treated with potassium hydroxide as per Kim et al.,12 to produce oseltamivir carboxylate. Dilutions of inhibitors were prepared in water, ranging from 0.001 nM to 100 000 nM. 4-Methylumbelliferyl N-acetyl-α-d-neuraminic acid (MUNANA) was obtained from Carbosynth, UK.
Enzyme assays
We used the MUNANA-based fluorescence assay13 for measuring NA activity and drug inhibition, as previously described.14 Final concentrations in the assay were 50 mM sodium acetate pH 5.5, 5 mM CaCl2 and 100 μM MUNANA. Fluorescence was measured using a BMG FLUOstar Optima reader with 355 nM excitation and 460 nM emission filters. Two different assays were carried out to determine the IC50 kinetics.15–17 The first assay used a 30 min pre-incubation of virus or NA with inhibitor prior to the addition of substrate. In the second assay, the NA, inhibitors and MUNANA were added simultaneously. Fluorescence for both assays was monitored immediately after substrate addition and thereafter at 1 min intervals for 60 min. Graphs of inhibitor concentration versus percentage enzyme inhibition compared with the control were plotted after 10, 20, 30, 40, 50 and 60 min. The IC50 was calculated as the concentration of inhibitor resulting in a 50% reduction in fluorescence units (FU) compared with the control. IC50 values were then plotted as bar graphs for each of the 10 min timepoints for both assays.
Sequencing of isolates
RNA for sequencing from the original P1 isolate was amplified using Access Quick RT–PCR (Promega, USA). RNA from plaque-purified viruses was extracted from viruses in cell culture media using the QIAamp Viral RNA Mini Kit and protocol (Qiagen, Australia). Then cDNA was prepared using the Phusion RT–PCR Kit and Phusion Hot Start High-Fidelity DNA Polymerase (Finnzymes, Finland) using specific primers binding to each end of the NA or HA genes to amplify the entire gene. Sequencing was carried out using Big Dye Terminator Cycle Sequencing by Micromon (Monash University, Victoria, Australia) using primers specific to either the NA or HA gene sequence.
Expression of recombinant NA
Since A/Hokkaido/15/02 contained mixed populations of wild-type and mutant viruses, we cloned the full-length NA from the A/Hokkaido/6/02 wild-type virus. The NA was cloned into pFastBac1 (Invitrogen) using the manufacturer's protocols. The Y155H mutant NA was generated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). Sf21 cells were infected with the recombinant viruses and after 3–4 days the cells containing the membrane-bound NA were pelleted and resuspended in Sf900II medium.
Stability of NA activity
Wild-type or Y155H viruses were diluted in parallel in PBS, tris-buffered saline (TBS) or 2-(N-morpholino)ethanesulfonic acid (MES) with or without 10 mM CaCl2, and MM or MM plus 1% CHAPS. Samples were incubated in the fluorimeter at 37°C for 0, 30, 60 and 90 min prior to the addition of MUNANA. Readings were taken at 1 min intervals from 0 min for 3 h. The mean activity in FU was calculated for triplicate samples after incubation with MUNANA for 60 min (wild-type) or 90 min (Y155H). The remaining activity was calculated as the percentage of the mean activity of the 0 min sample.
Effect of pH on enzyme activity
NA mutations can result in a change in the optimum pH for enzyme activity.18 We prepared a series of MUNANA reaction mixtures to cover the range from pH 4.5 to 8.5. We used citrate–phosphate and MES for pH 4–8 and acetate for pH 4–7.5. Samples of virus were incubated in each of the solutions in parallel for 60 min and the FU was read after the addition of Stop solution after 60 min. Relative FU were expressed as a percentage of the values for pH 6, as this was the middle of the pH range.
Km and Ki
The Michaelis constant, Km, for MUNANA was calculated by measuring the FU at 1 min intervals with MUNANA concentrations ranging from 6.25 to 3200 μM in acetate buffer. Km for the Y155H NA was also determined in a MES pH 7.0 buffer to optimize stability and activity. Values are the means from at least three assays. Initial velocities of the reactions were calculated by measuring the maximum slope plotted as a function of substrate concentration. The Km was calculated in GraphPad Prism using a non-linear regression function. The Ki was calculated using non-linear regression and one-site competitive binding in GraphPad Prism.15,19
Red blood cell (RBC) binding and elution
RBCs were obtained from chickens and turkeys. Serial 2-fold dilutions of virus were prepared in 50 μL of PBS, and 50 μL of a 1% RBC suspension was added. Plates were incubated at 4°C for 1 h and then scanned to record the HA titre. Plates were then incubated at 37°C and rescanned after 1–2 h, 4 h and overnight to monitor the rate at which elution occurred (haemagglutination elution). HA mutations reducing receptor affinity will lead to more rapid elution. Samples were then gently mixed and resuspended and reincubated at 4°C to determine whether elution appeared to be due to cleavage of receptors by the NA, in which case reagglutination would not occur, or due to weak HA affinity, in which case reagglutination should occur. A combination of both NA cleavage and weak HA affinity would result in only partial reagglutination.
We also investigated the susceptibility of virus elution to the NAIs, to determine the roles in elution of both the NA and HA.17 A dilution of virus that eluted in the initial haemagglutination elution assay was incubated with serial 2-fold dilutions of zanamivir and oseltamivir, ranging from 10 μM to 9 nM. Plates were incubated at 4°C to allow the initial agglutination to occur and were then incubated at 37°C to determine at what concentration of NAI elution could still occur.
Results
The A/Hokkaido/15/02 H1N1 virus provided by the NIID to Viromed was amplified by Viromed before testing.8 Genotypic analysis by the NIID confirmed that although the original clinical sample was not available, the Y155H mutation was present in the P1 isolate. However, it cannot be ruled out that the mutation arose in this first passage.
Plaque purification
Plaquing of the P1 stock and MDCK passaged virus revealed a mixture of large plaques (∼3 mm) and small plaques (∼1 mm; ∼10%–15% of the total plaques were small). Plaques of each type were picked and serially replaqued until the plaque-purified populations were homogeneous (Table 1).
Table 1.
Phenotypic and genotypic properties of Hokkaido plaque-purified and reassortant viruses
| Plaque phenotype | NA | HA | IC50 (nM)a |
|
|---|---|---|---|---|
| SIAT cells | MDCK cells | |||
| Large | wt | wt | 1–10 | 10 |
| Large | V114I | D225N | 10 | 10 |
| Large | Y155H | D225G | 10–100 | 100–1000 |
| Small | Y155H | wt | 10 | 10–100 |
| Large | wt | D225N | 10 | 10–100 |
| Large | wt | D225G | 10–100 | 10–100 |
wt, wild-type.
aSusceptibility to zanamivir in the plaque reduction assay. The IC50 is based on the zanamivir dilution resulting in a 50% reduction in plaque size. A range is provided when the 50% reduction in size fell between two drug concentrations. Results are based on duplicate assays.
Initial screening of the first large-plaquing virus against zanamivir and oseltamivir in the MUNANA-based enzyme inhibition fluorescence assay showed it was susceptible to both inhibitors. Sequencing of the NA identified Y155 (HAwtNAwt).
The small plaque had the same sequence as reported by Monto et al.,8 with wild-type HA and the unique NA Y155H mutation (HAwt NAY155H). Titration in an enzyme assay demonstrated it had lower NA activity per pfu (or per particle in an HA assay) compared with the wild-type virus, which may explain the smaller plaque size. Initial screening against zanamivir and oseltamivir in the MUNANA assay revealed it had reduced susceptibility to both NAIs, as was originally reported in the chemiluminescence assay.8 Further detailed analysis of drug susceptibility is described in the Enzyme inhibition assays section.
The second large-plaquing virus had not only the NA Y155H mutation but also an HA D225G (H3 numbering) mutation (HAD225G NAY155H), which rescued the small-plaque phenotype seen with the HAwtNAY155H virus. These viruses also had lower NA activity per pfu or particle. D225 is known to affect HA receptor binding,20–23 which would be consistent with previous observations of HA mutations compensating for decreased NA activity.24–26
The third large-plaquing virus had a novel V114I mutation in the NA and a D225N HA mutation (HAD225NNAV114I). Initial screening in the MUNANA assay showed this mutation reduced susceptibility to zanamivir and oseltamivir by 3- to 5-fold.
To investigate the role of the HA mutations without NA mutations we generated reassortants between the HAwtNAwt virus and either the HAD225NNAV114I or the HAD225GNAY155H viruses. We identified viruses that had the wild-type NA by their susceptibility in the enzyme inhibition assay, and the HA mutations were confirmed by reduced susceptibility in the plaque reduction assay with zanamivir. Genotypes were then confirmed by sequencing both the HA and NA.
Effects of mutations on replication and drug susceptibility in cell culture
Since HA mutations are known to affect the specificity or affinity of receptor binding, we compared infectivity and drug susceptibility in both MDCK and SIAT cells. The plaques in SIAT cells were smaller than in MDCK cells for all viruses, but titres in SIAT cells were about 50% higher for all viruses compared with titres in MDCK cells (Figure S1, available as Supplementary data at JAC Online), consistent with the preference of human-derived isolates for α2,6 receptors.
Plaque reduction assay
Since HA mutations confer resistance to all NAIs, and resistance was higher to zanamivir in the enzyme inhibition assay, all viruses were screened for susceptibility to zanamivir in plaque reduction assays in SIAT and MDCK cells (Table 1). The plaque reduction assay IC50 is based on the drug dilution resulting in a 50% reduction in plaque size. A range is provided where there is a 50% reduction in size between two drug concentrations. Except for the HAD225NNAV114I virus, all viruses were more susceptible to zanamivir in SIAT cells than MDCK cells. Others have also reported an increase in oseltamivir susceptibility in SIAT cells.27 The NA Y155H mutation conferred a ≤10-fold reduction in susceptibility, with the greatest reduction in susceptibility of 10- to 100-fold seen for the HAD225GNAY155H virus. The HA D225G and NA Y155H mutations appeared to be acting synergistically when compared with viruses with only one of the mutations (HAwtNAY155H and HAD225GNAwt). The HAD225NNAV114I virus showed a ≤10-fold reduction in susceptibility only in SIAT cells, compared with the HAwtNAwt control. Susceptibility of the HAD225NNAwt virus with the single HA D225N mutation was the same or less than that of the double mutant, suggesting a minimal role for the V114I NA mutation in drug susceptibility in cell culture.
Kinetics of replication
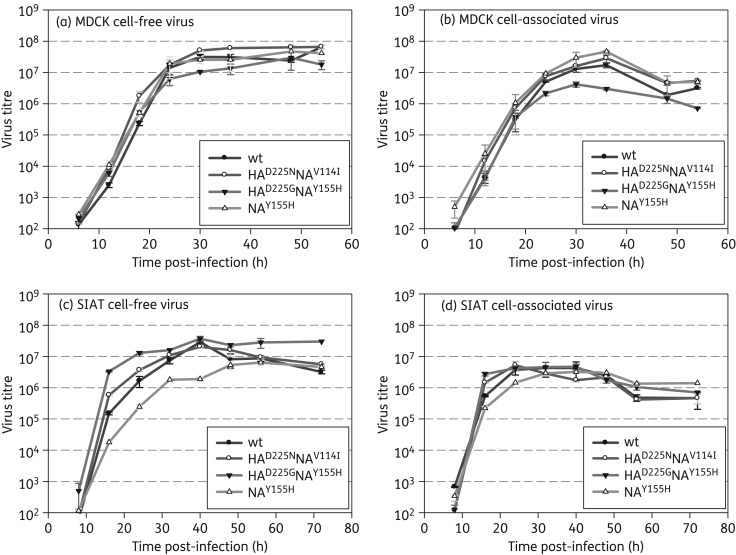
We next investigated whether the mutations affected the kinetics of replication in MDCK and SIAT cells (Figure 1). Initial replication of all viruses was more rapid in SIAT cells, although, despite higher plaquing efficiency, the maximum yields were lower than in the MDCK cells. Yields for the HAwtNAY155H cell-free virus in SIAT cells were 10-fold lower compared with the other viruses by 40 h post-infection. Although the HAwtNAY155H virus had small plaques in both cell lines, this mutation had no impact on HAwtNAY155H replication in liquid culture in MDCK cells.
Figure 1.
Kinetics of replication in MDCK and SIAT cells. Cells were infected at a multiplicity of infection of 1.0 and samples were harvested from duplicate wells every 6–8 h. Yields for both cell-free virus in supernatants and cell-associated virus were titrated in MDCK cells. (a) MDCK cell free. (b) MDCK cell associated. (c) SIAT cell free. (d) SIAT cell associated. The Y155H NA mutation decreased replication in SIAT cells, but not in MDCK cells, whereas the combined HA D225G and NA Y155H mutations decreased replication in MDCK cells; D225G rescued the poorer growth of the Y155H NA mutant in SIAT cells.
For the HAD225GNAY155H virus, the D225G mutation rescued yields especially of cell-free virus compared with the HAwtNAY155H virus in SIAT cells. This could correlate with reduced affinity of the D225G HA facilitating virus release. Yields for the HAD225GNAY155H double mutant were 3-fold lower in MDCKs compared with the other viruses by 30 h post-infection. While lower affinity could rescue plaque size, it may also lead to less efficient infection of cells and hence lower yields.
Enzyme inhibition assays
We have recently developed a real-time IC50 kinetics assay15–17 to identify slow and fast binding of NAIs to wild-type and mutant viruses. If the inhibitor is slow binding then pre-incubation enhances occupancy of the enzyme active site, leading to a lower IC50 than without pre-incubation. Conversely, without pre-incubation the IC50 decreases with time, as the inhibitor gradually occupies the active site. In these previous papers we saw slow binding to the wild-type viruses. We saw loss of slow binding in the mutants, as demonstrated by similar IC50 values with or without pre-incubation and hence a ratio of ∼1 for the two IC50 values.
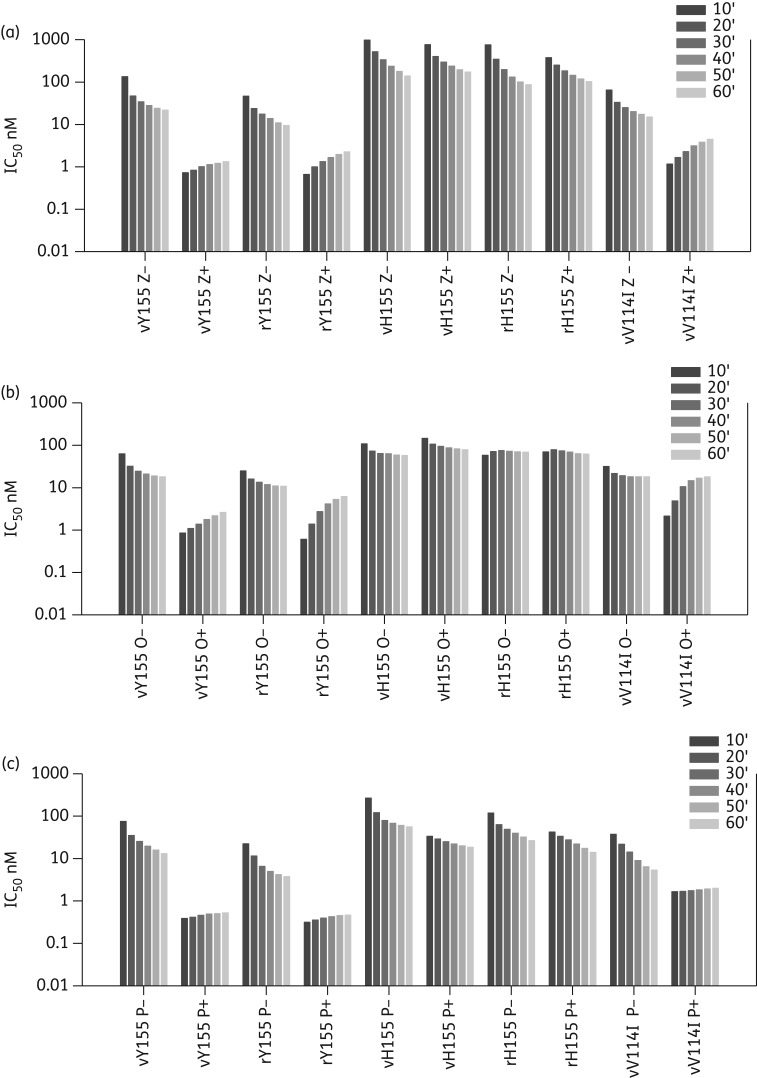
To confirm the role of the Y155H mutation in resistance we also expressed recombinant full-length wild-type and mutant NAs in insect cells.28 We confirmed that in the fluorescence assay the Y155H mutant virus and the recombinant Y155H NA had reduced susceptibility to both zanamivir and oseltamivir, with IC50 values of ≥100 and 60 nM, respectively. IC50 values for the mutant virus were comparable to those previously observed in the chemiluminescence assay (150 and 69 nM, respectively).8 Further testing also demonstrated that both virus and recombinant Y155H NAs had about 30-fold reduced susceptibility to peramivir compared with the wild-type NAs, (Figure 2 and Table 2).
Figure 2.
IC50 kinetics of NAI binding for wild-type and mutant NAs. Comparison of IC50 after each 10 min without pre-incubation with inhibitor (−) and with a 30 min pre-incubation (+) with inhibitor. Results are the means of duplicate assays. (a) Zanamivir. (b) Oseltamivir. (c) Peramivir. If the inhibitor is slow binding there is a decrease in IC50 over the 60 min in the reaction without pre-incubation (−) as more inhibitor binds. Additionally, pre-incubation with drug enhances binding, so that a lower initial IC50 is seen in the (+) reaction compared with the (−) reaction. If NAs have lost slow binding, the pre-incubation does not enhance inhibitor binding and hence similar IC50 values are seen in both assays. vY155, virus wild-type NA; rY155, recombinant wild-type NA; vH155, virus mutant NA; rH155, recombinant mutant NA; vV114I, virus mutant NA.
Table 2.
IC50 values (nM) calculated 60 min after addition of MUNANA for virus-associated and recombinant NAs
| Inhibitor | NA sequence | Virus-associated NA |
Recombinant NA |
||||
|---|---|---|---|---|---|---|---|
| no prea | prea | ratio no pre:pre | no prea | prea | ratio no pre:pre | ||
| Zanamivir | Y155 | 21.9 | 1.3 | 16.8 | 9.5 | 2.3 | 4.1 |
| Y155H | 139.5 | 172.2 | 0.8b | 86.9 | 102.8 | 0.8b | |
| V114I | 15.1 | 4.5 | 3.4 | NA | NA | — | |
| Oseltamivir | Y155 | 18.1 | 2.6 | 7.0 | 10.9 | 6.16 | 1.8 |
| Y155H | 57.7 | 78.6 | 0.7b | 68.6 | 62.5 | 1.1b | |
| V114I | 18.1 | 17.9 | 1.0b | NA | NA | — | |
| Peramivir | Y155 | 13.1 | 0.5 | 26.2 | 3.8 | 0.5 | 7.6 |
| Y155H | 56.1 | 18.5 | 3.0 | 26.6 | 14 | 1.9 | |
| V114I | 5.3 | 2 | 2.7 | NA | NA | — | |
NA, not applicable; no pre, virus, NAI and MUNANA all co-incubated; pre, virus and NAI pre-incubated for 30 min prior to the addition of MUNANA.
aValues are means of duplicate samples.
bWhere the pre and no pre 60 min IC50 values are similar (ratio ∼1) this indicates loss of slow binding.
In the IC50 kinetics assay, all three NAIs demonstrated slow binding to the wild-type virus and recombinant NA (Figure 2), with pre-incubation enhancing binding to the virus NA by 7- to 26-fold (Table 2) and by 1.8- to 7.6-fold for the recombinant NA. Additionally, there was an increase in IC50 after the addition of substrate in the pre-incubation reaction, as seen previously with other wild-type NAs (Figure 2).16
We observed a novel pattern of behaviour for the NAIs binding to the Y155H mutant NA. There was sometimes enhancement of enzyme activity in 1–10 nM concentrations of the NAIs, so activity was higher than the control (Figure S2, available as Supplementary data at JAC Online). Pre-incubation of recombinant or virus NA with peramivir enhanced binding by 2-fold and 3-fold, respectively, but there was no enhancement for zanamivir and oseltamivir (60 min IC50 ratio ≤1), suggesting loss of slow binding. However, analysing the kinetics graphs showed an unexpected decrease in IC50 in the pre-incubation assay from 10–60 min (Figure 2). This novel behaviour, seen with both virus and recombinant NAs, was more apparent for zanamivir and peramivir, although there was also a small decrease for oseltamivir. This suggests that decreased susceptibility of the H155 is related to extremely slow binding of the NAIs.
We also investigated the IC50 kinetics of the HAD225NNAV114I virus, which had a small reduction in susceptibility to all three NAIs of 3.5- to 7-fold. Pre-incubation with both zanamivir and peramivir still decreased the IC50 values, indicating slow binding (Table 2). However, there was a greater increase in the zanamivir IC50 from 10 to 60 min in the pre-incubation reaction compared with the wild-type, indicating a faster dissociation rate. The IC50 for peramivir changed little over time, but started and ended higher than that of the wild-type. Although the oseltamivir IC50 values for the early timepoints showed enhancement of binding with pre-incubation, there was little difference in the final oseltamivir IC50 values with or without pre-incubation. This indicates faster dissociation compared with the wild-type (8.5-fold change from 10 to 60 min compared with 3.3-fold for the wild-type). Thus, even though the NA V114I mutation only had a small effect on susceptibility, it led to detectable differences in the kinetics of inhibitor binding.
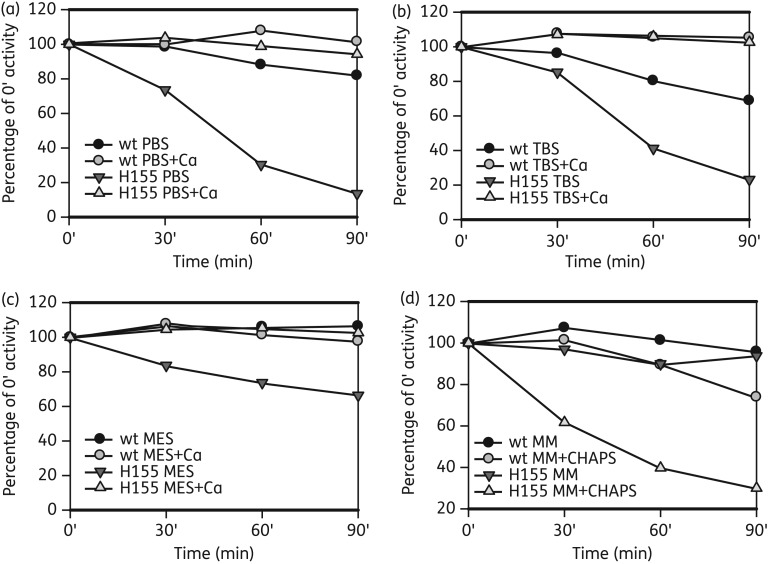
Stability of the wild-type and Y155H NA
We often observed a loss of enzyme activity of either virus or recombinant Y155H samples. We therefore compared the stability of the activity of the wild-type and Y155H mutant virus NAs after incubation in PBS, TBS or MES with or without CaCl2, and MM with or without CHAPS detergent, which we often use to increase NA activity (Figure 3). Compared with the wild-type, the Y155H NA was very unstable in PBS and TBS, losing up to 80% of activity in 90 min. There was only a 30% loss of activity in MES, and none in MM. CaCl2 stabilized both the wild-type and mutant activities, which may account for the stability in MM. CHAPS also dramatically reduced the Y155H NA activity compared with the wild-type NA. The recombinant mutant NA was also unstable in CHAPS (data not shown). Thus, the Y155H mutation led to instability of the enzyme, but this was buffer dependent. Activity was also buffer dependent (Figure S3, available as Supplementary data at JAC Online), with the maximum activity for the wild-type in MM; however, the mutant had the lowest activity in MM, despite this being the best buffer for stability, and the highest activity in MES.
Figure 3.
Stability of NA enzyme activity after incubation at 37°C in different buffers. Samples of wild-type and mutant Y155H virus were incubated at 37°C in (a) PBS, (b) TBS, (c) MES and (d) MM. Enzyme activity was determined in the MUNANA assay after 30, 60 and 90 min and the remaining activity was expressed as a percentage of that of the control sample assayed at 0 min. Values are based on the means of triplicate samples. The Y155H NA was much more unstable in all buffers and with CHAPS detergent compared with the wild-type. CaCl2 stabilized the activity in all buffers.
Effect of pH on enzyme activity
Mutations in the active site can affect the pH optima for enzyme activity.18 Because different buffers affected the stability and activity of the mutant NA, we compared activity in MES, acetate and citrate–phosphate buffers from pH 4 to pH 8.
In MES, both the wild-type and Y155H mutant NAs had high activity across a broad pH range, from pH 4 to pH 8 (Figure 4), with optimal activity at pH 6.5. Although the optimal activities for the wild-type and mutant viruses were both around pH 6.5, what was strikingly different between the wild-type and mutant pH profiles was that the wild-type still showed good activity in both acetate and citrate–phosphate buffers across a relatively broad pH range, from pH 5 to pH 7.5, whereas the mutant NA only had activity across a very narrow pH range. The effect of the citrate–phosphate buffer was the most dramatic, further demonstrating the critical nature of the buffer used to evaluate stability and enzyme activity.
Figure 4.
Effect of pH on NA enzyme activity. Wild-type and Y155H mutant viruses were incubated in MUNANA prepared in buffers ranging from pH 4 to pH 8. Total activity based on FU after 1 h was compared, and results expressed as a percentage of the pH 6.0 fluorescence. (a and b) MES. (c and d) Acetate. (e and f) Citrate–phosphate. The wild-type virus had activity across a broad pH range in each of the buffers tested. The mutant had activity across a broad pH range in MES, but only across a very narrow range in acetate and citrate–phosphate solutions.
Enzyme kinetics
To further understand the impacts of these mutations on enzyme function we calculated the affinity (Km values) of the NAs for the MUNANA substrate (Table 3). We initially carried out the Km calculations in our standard acetate buffer. The Km for the NA Y155H mutant was around 1000 μM (data not shown), compared with the wild-type value of 20 μM. As our other results suggested that a MES pH 7 buffer may be better for stability and activity of the Y155H mutant NA, we also repeated the mutant NA Km assays in MES buffer. While the Km was lower in MES buffer (222 μM), it was still almost 10 times that of the wild-type virus. We used 222 μM for determining the Ki values (Table 3). Also, and unexpectedly, despite the small reduction in susceptibility to the NAIs, the Km for the V114I NA was increased by more than three times that of the wild-type NA.
Table 3.
Km and Ki for wild-type and mutant NAs
| Kma (μM) MUNANA |
Kib (nM) |
|||
|---|---|---|---|---|
| zanamivir | oseltamivir | peramivir | ||
| Wild-type | 20.3 ± 1.6 | 0.31 | 1.2 | 0.11 |
| V114I | 64.8 ± 2.8 | 3.4 | 8.5 | 0.81 |
| Y155H | 222 ± 33.1 | 56.0 | 44.8 | 8.7 |
aKm values for wild-type and V114I in acetate buffer and Y155H in MES pH 7 buffer.
bKi is calculated from rates in the 50–60 min period of the pre-incubation reaction from duplicate assays.
We also calculated Ki values for each of the wild-type and mutant NAs as previously described.16 We have previously shown that since the Ki value depends on the rate of the reaction, it also changes with time.16 The wild-type and V114I NA Ki values increased with time if calculated from the pre-incubation reaction, whereas the Y155H Ki decreased with time (Figure S4, available as Supplementary data at JAC Online). The Ki values for the final 50–60 min time period of the pre-incubation reaction are shown in Table 3, as this approaches the most constant rate. The Ki values were elevated for both mutant viruses for all the NAIs, thus confirming decreased affinity for the NAIs.
RBC binding and elution assays
The plaque reduction assays suggested that the HA mutations affected receptor binding. We therefore investigated haemagglutination of viruses using chicken and turkey RBCs and subsequent elution at 37°C to look for differences in binding specificity or relative affinity (Table 4) that might contribute to drug resistance.17
Table 4.
Virus binding and elution from RBCs
| Virus | Sequential incubation conditions | HA titre |
|
|---|---|---|---|
| chicken RBCs | turkey RBCs | ||
| HAwtNAwt | 4°C/1–2 h | 256 | 128–256 |
| 37°C/1–2 h | 256 | 128–256 | |
| 37°C O/N | 256 | 128–256 | |
| remix 4°C | 256 | 128 | |
| HAwtNAY155H | 4°C/1–2 h | 128 | 32–64 |
| 37°C/1–2 h | 64–128 | 32–64 | |
| 37°C O/N | 64–128 | 32–64 | |
| remix 4°C | 64 | 32 | |
| HAD225NNAV114I | 4°C/1–2 h | 128 | 512–1024 |
| 37°C/1–2 h | 32 | 512 | |
| 37°C O/N | 0 | 512 | |
| remix 4°C | 128 | 256–512 | |
| HAD225GNAY155H | 4°C/1–2 h | 16 | 128–256 |
| 37°C/1–2 h | 0 | 128 | |
| 37°C O/N | 0 | 64–128 | |
| remix 4°C | 16 | 64–128 | |
| HAD225NNAwt | 4°C/1–2 h | 16–32 | 128 |
| 37°C/1–2 h | 0 | 128 | |
| 37°C O/N | 0 | 0 | |
| remix 4°C | 0 | 128 | |
| HAD225GNAwt | 4°C/1–2 h | 8–16 | 128–256 |
| 37°C/1–2 h | 0 | 0 | |
| 37°C O/N | 0 | 0 | |
| remix 4°C | 0 | 64–128 | |
O/N, overnight.
Assays were performed in duplicate.
While weak HAs facilitated elution, some NA activity was still needed. For chicken RBCs, binding of the wild-type HA was too strong for any elution to occur. However, both HAD225NNAV114I and HAD225GNAY155H viruses fully eluted, indicating a lower affinity compared with the wild-type HA, correlating with their reduced sensitivity in the plaque reduction assay. These viruses reagglutinated after remixing, indicating that elution was facilitated due to low affinity of their HAs, and that the low level of NA activity needed was not sufficient to deplete the receptors on the cells. By contrast, in the reassortant viruses with the wild-type NA and mutant HAs, neither of the viruses reagglutinated, suggesting that with the weaker-binding HAs the wild-type NA efficiently cleaved the majority of the receptors to which they bound.
Assays with turkey RBCs demonstrated further differences in the receptor binding properties of the virus panel. For the two viruses with the wild-type HA (HAwtNAwt and HAwtNAY155H) the HA titre with turkey RBCs was slightly less than with chicken RBCs, suggesting a lower affinity or different specificity compared with chicken RBC receptors. However, the wild-type HA still had a high affinity for the receptors, since neither virus eluted from the turkey RBCs. By contrast, the four viruses with the HA D225 mutations all had around a 10-fold higher HA titre with turkey RBCs compared with chicken RBCs, suggesting more/different target receptors compared with chicken RBCs. But only the reassortant viruses with the mutant HA and wild-type NAs (HAD225NNAwt and HAD225GNAwt) eluted from turkey RBCs, with faster elution for the HAD225GNAwt virus. The HAD225GNAY155H virus with the same HA D225G but the Y155H NA did not elute, suggesting that in addition to the weaker HA facilitating dissociation from the receptors (unlike the chicken RBCs, where either wild-type or mutant NAs were sufficient for elution), for turkey RBCs higher wild-type NA activity was needed. Neither reassortant virus reagglutinated at high virus concentrations (data not shown), indicating removal of all the receptors by excess virus. However, at lower virus concentrations there was reagglutination, suggesting that although significant NA activity was needed for elution, there was not sufficient activity to deplete the receptors.
We then investigated whether the various mutations enabled the viruses to elute in the presence of zanamivir and oseltamivir. Results for both drugs were similar and demonstrated that both NA and HA could contribute to drug resistance. Zanamivir results are shown in Table 5. For chicken RBCs the D225G HA mutation facilitated more rapid elution and in higher concentrations of inhibitors than the D225N HA. Either wild-type or drug-resistant NAs could cleave the receptors, but the combination of the weaker D225G HA and the mutant Y155H NA was the most drug resistant (HAD225GNAY155H). For turkey RBCs, both the reassortant viruses were more susceptible to the inhibitors than with chicken RBCs. As seen with the chicken RBCs, the D225G HA mutation enabled the reassortant virus to elute in higher concentrations of inhibitor compared with the D225N HA. Thus, the lower the affinity of the HA the less NA activity is needed and hence the greater drug resistance.
Table 5.
Concentration of zanamivir (nM) at which elution of viruses from RBCs was inhibited
| Virus | Sequential incubation conditions | Chicken RBCs | Turkey RBCs |
|---|---|---|---|
| HAwtNAwt | 37°C O/N | no elution | no elution |
| HAwtNAY155H | 37°C O/N | no elution | no elution |
| HAD225NNAV114I | 37°C/1–2 h | ≤10 | no elution |
| 37°C O/N | 20–40 | ||
| HAD225GNAY155H | 37°C/1–2 h | 10 000 | no elution |
| 37°C O/N | 10 000 | ||
| HAD225NNAwt | 37°C/1–2 h | 10–20 | no elution |
| 37°C O/N | 2500 | 10 | |
| HAD225GNAwt | 37°C/1–2 h | 20–40 (+/− to 1250)a | no elution |
| 37°C O/N | 5000 | 37.5 |
O/N, overnight.
Assays were performed in duplicate.
aFull elution at 40 nM, but partial elution through to 1250 nM.
Discussion
We describe here the isolation and identification of a number of mutant viruses with reduced susceptibility to the NAIs from a sample cultured in vitro from an untreated patient. The sample contained large and small plaques, which were identified as a wild-type and three mutants with HA and/or NA mutations after plaque purification. One large-plaquing virus had wild-type susceptibility in all assays. One virus that formed small plaques had a single Y155H NA mutation, but there was also a second virus with the Y155H NA mutation, which had a D225G HA mutation that rescued the small plaque phenotype. The Y155H mutation in either virus or recombinant NA led to a reduction in susceptibility to all NAIs tested (oseltamivir, peramivir and zanamivir) in the MUNANA-based enzyme inhibition assay. A third large-plaquing virus had V114I NA and D225N HA mutations. The V114I mutation led to a small but distinct reduction in susceptibility to all NAIs. Neither the Y155H nor the V114I mutation has been described previously as affecting drug susceptibility. Neither Y155 nor V114 is conserved across subtypes nor directly interacts with the substrate or inhibitor in the enzyme active site, yet both altered inhibitor and substrate binding properties of the mutant enzymes.
Mutations in the HA are also known to affect susceptibility to the NAIs by decreasing affinity for the cell receptors, reducing the need for NA activity for elution of progeny virions. While both the D225G and D225N mutations affected receptor binding and decreased NAI susceptibility in cell-based assays, the D225G HA mutation resulted in lower affinity and consequently had a greater impact on drug susceptibility. The HAD225GNAY155H virus was the most resistant in the plaque reduction assay and eluted from RBCs in the highest NAI concentration. However, the effects of the mutations on replication varied between MDCK and SIAT cells. The D225G mutation facilitated release of virus from SIAT cells, rescuing the poorer replication ability of the HAwtNAY155H virus to more than wild-type. By contrast, reduced affinity of the D225G NA resulted in poorer replication in MDCK cells.
The mutations in the HA seen here at amino acid 225 form part of the 220 loop in the HA receptor binding site (220–229).29,30 Mutations at 225 have been associated with changes in specificity of receptor binding of H1N1 viruses, with D225G enhancing binding to α2,3-linked avian-like receptors.31–33 Two recent reports suggest that D225G mutations may alter the drug susceptibility of pandemic viruses.34,35 There have been many reports of the effects of HA mutations on reduced drug susceptibility in cell culture,10,14,17,24,25,36,37 however, since the altered receptor specificity is dependent upon the cells in which the mutant virus is tested, their clinical relevance is still poorly understood.38 Our report here of the effect of D225 mutations on decreasing drug susceptibility in both MDCK and SIAT cells is, we believe, the first to demonstrate a reduction in both cell lines. The role of HA mutations in altered NAI susceptibility remains an important area for further investigation.
Surprisingly, although Y155 is at the outer edge of the NA monomer, at the tetramer interface, the Y155H mutation had dramatic effects on enzyme function and drug sensitivity and stability. The Y155H mutant virus had smaller plaques in both MDCK and SIAT cells, which would correlate with its apparent lower NA activity. In contrast to other drug-resistant NA mutations that lead to loss of slow binding15–17 the Y155H NA demonstrated slower binding than the wild-type NA. The mutation also had a dramatic effect on substrate affinity, with an increase in Km of between 10-fold and 50-fold, depending on the buffer, and a consequent effect on the Ki for all NAIs.
Despite the small effect on drug susceptibility, the V114I NA mutation had a very significant impact on Km, resulting in a >3-fold increase compared with the wild-type NA, and a consequent effect on the Ki values. This mutation led to partial loss of slow binding of oseltamivir. Why both mutations have a much greater impact on Km than inhibitor binding is unexpected. However, we have recently shown that while the H274Y mutation affects the Km by <2-fold (15 μM for wild-type and 27.6 μM for the H274Y mutant),16 an N146S mutation, which decreases drug susceptibility by 2-fold, doubles the Km.17
We have previously demonstrated that instability of mutant NAs can lead to loss of enzyme activity.39,40 We noticed loss of enzyme activity with the Y155H mutant. Since Y155 is located at the tetramer interface it is possible that its main effect is through destabilizing the tetramers, resulting in lower enzyme activity and an effect on drug susceptibility. Evaluating the effects of incubation at 37°C on the Y155H mutant enzyme function demonstrated that the NA was more unstable in PBS, TBS and CHAPS detergent compared with the wild-type NA. The pH activity profile of the mutant NA was also very different from that of the wild-type NA. The wild-type enzyme had activity across a broad pH range in MES, citrate–phosphate and acetate buffers. By contrast, the Y155H mutant NA had activity across a broad pH range in MES buffer, but only across a very narrow range in both acetate and citrate–phosphate buffers. An effect of specific buffer on pH optimum has not previously been reported, and would easily be missed if multiple buffers were not tested.
Structural analysis of the avian N1 NA (2HU4 pdb) shows that residues 155 and 114 are >12 Å and >17 Å, respectively, from the active site (Figure S5, available as Supplementary data at JAC Online). Residue 155 is located in the middle of a loop between D151 to R156, both of which extend into the active site. It is also close to the ‘150 loop’ formed by residues 149–152, which has been shown to adopt closed and open positions in crystallographic studies of type 1 influenza NAs.41–46 Interestingly, van der Vries et al.47 recently reported that the use of a phosphate or acetate buffer for crystallization of the pandemic N1 NA determined whether the 150 loop was in the open or closed conformation, respectively, which may contribute to the different effect of pH and buffers on the activity of the Y155H NA. Mutations at D151,48–50 R15237,51 and R15652 have also all been shown to result in reduced drug susceptibility, and R152K and R156K lead to reduced enzyme activity.
Similarly, residue 114 forms part of a chain from 114–119 extending into the active site. Several residues on this chain are critical to substrate and inhibitor binding. R118 is one of the catalytic triad of arginines, and mutations at V116,53 I11753 and E1193,10,14,18,54,55 all alter drug susceptibility. There has also been a recent report of an E105K mutation in an influenza B NA that affects drug susceptibility.56 It is also located at the tetramer interface, and it was hypothesized that its mode of action is also through destabilization of the tetramers. Residues 155 and I108 (the equivalent location in N1 to E105) are on opposing sides of the tetramer interface (Figure S5). Since both 155 and 114 are remote from the active site, the effects of both mutations on substrate and inhibitor binding may be through perturbation of the chains extending into the active site, in addition to destabilization by the Y155.
Interestingly, since other N1 wild-type viruses have H155 there must be compensating mutations. Movements of both the 150 loop and an adjacent 430 loop41,46,57 demonstrate the dynamic nature of the NA and hence it is possible that compensating mutations could reside well away from the 155 residue.
Although our results demonstrate that both the NA and HA mutations can contribute to reduced NAI susceptibility, it is difficult to predict whether they would result in reduced clinical efficacy. However, our results emphasize the need for phenotypic testing to detect novel, non-conserved NA residues that may confer NAI resistance.
Funding
This work was supported by GlaxoSmithKline (UK and Australia) and the National Institutes of Health NIAID (5R01AI62721).
Transparency declarations
J. L. M.-B. has received honoraria and/or travel assistance from GlaxoSmithKline and Hoffman La-Roche for participation in advisory groups and scientific meetings. All other authors: none to declare.
Supplementary data
Figures S1 to S5 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
Parts of this work were presented at the ISIRV Antivirals Group conference entitled Influenza Antivirals: Efficacy and Resistance, Rio de Janeiro, Brazil, 8–11 November 2011, O33, p. 66 and the XIV International Conference on Negative Strand Viruses, Brugge, Belgium, 20–25 June 2010, Abstract 282, p. 188.
We thank Rebecca Attwood for technical assistance and Victor Streltsov for NA modelling.
References
- 1.Burmeister WP, Ruigrok RW, Cusack S. The 2.2 Å resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J. 1992;11:49–56. doi: 10.1002/j.1460-2075.1992.tb05026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varghese JN, McKimm-Breschkin JL, Caldwell JB, et al. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins. 1992;14:327–32. doi: 10.1002/prot.340140302. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen HT, Fry AM, Gubareva LV. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir Ther. 2012;17:159–73. doi: 10.3851/IMP2067. [DOI] [PubMed] [Google Scholar]
- 4.Hurt AC, Ernest J, Deng YM, et al. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res. 2009;83:90–3. doi: 10.1016/j.antiviral.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Meijer A, Lackenby A, Hungnes O, et al. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg Infect Dis. 2009;15:552–60. doi: 10.3201/eid1504.081280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abed Y, Pizzorno A, Bouhy X, et al. Role of permissive neuraminidase mutations in influenza A/Brisbane/59/2007-like (H1N1) viruses. PLoS Pathog. 2011;7:e1002431. doi: 10.1371/journal.ppat.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010;328:1272–5. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monto AS, McKimm-Breschkin JL, Macken C, et al. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother. 2006;50:2395–402. doi: 10.1128/AAC.01339-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macdonald SJ, Cameron R, Demaine DA, et al. Dimeric zanamivir conjugates with various linking groups are potent, long-lasting inhibitors of influenza neuraminidase including H5N1 avian influenza. J Med Chem. 2005;48:2964–71. doi: 10.1021/jm040891b. [DOI] [PubMed] [Google Scholar]
- 10.McKimm-Breschkin JL. Resistance of influenza viruses to neuraminidase inhibitors—a review. Antiviral Res. 2000;47:1–17. doi: 10.1016/s0166-3542(00)00103-0. [DOI] [PubMed] [Google Scholar]
- 11.McKimm-Breschkin JL. Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance. Influenza Other Respi Viruses. 2013;7(Suppl 1):25–36. doi: 10.1111/irv.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim CU, Lew W, Williams MA, et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J Am Chem Soc. 1997;119:681–90. doi: 10.1021/ja963036t. [DOI] [PubMed] [Google Scholar]
- 13.Potier M, Mameli L, Belisle M, et al. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-d-N-acetylneuraminate) substrate. Anal Biochem. 1979;94:287–96. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 14.Blick TJ, Tiong T, Sahasrabudhe A, et al. Generation and characterization of an influenza virus neuraminidase variant with decreased sensitivity to the neuraminidase-specific inhibitor 4-guanidino-Neu5Ac2en. Virology. 1995;214:475–84. doi: 10.1006/viro.1995.0058. [DOI] [PubMed] [Google Scholar]
- 15.Oakley AJ, Barrett S, Peat TS, et al. Structural and functional basis of resistance to neuraminidase inhibitors of influenza B viruses. J Med Chem. 2010;53:6421–31. doi: 10.1021/jm100621s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett S, Mohr PG, Schmidt PM, et al. Real time enzyme inhibition assays provide insights into differences in binding of neuraminidase inhibitors to wild type and mutant influenza viruses. PLoS One. 2011;6:e23627. doi: 10.1371/journal.pone.0023627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKimm-Breschkin JL, Rootes C, Mohr PG, et al. In vitro passaging of a pandemic H1N1/09 virus selects for viruses with neuraminidase mutations conferring high-level resistance to oseltamivir and peramivir, but not to zanamivir. J Antimicrob Chemother. 2012;67:1874–83. doi: 10.1093/jac/dks150. [DOI] [PubMed] [Google Scholar]
- 18.Gubareva LV, Robinson MJ, Bethell RC, et al. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J Virol. 1997;71:3385–90. doi: 10.1128/jvi.71.5.3385-3390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Y, Prusoff WH. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 20.Govorkova EA, Matrosovich MN, Tuzikov AB, et al. Selection of receptor-binding variants of human influenza A and B viruses in baby hamster kidney cells. Virology. 1999;262:31–8. doi: 10.1006/viro.1999.9892. [DOI] [PubMed] [Google Scholar]
- 21.Gubareva LV, Kaiser L, Matrosovich MN, et al. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J Infect Dis. 2001;183:523–31. doi: 10.1086/318537. [DOI] [PubMed] [Google Scholar]
- 22.Zheng B, Chan KH, Zhang AJ, et al. D225G mutation in hemagglutinin of pandemic influenza H1N1 (2009) virus enhances virulence in mice. Exp Biol Med (Maywood) 2010;235:981–8. doi: 10.1258/ebm.2010.010071. [DOI] [PubMed] [Google Scholar]
- 23.Tambunan US, Ramdhan Identification of sequence mutations affecting hemagglutinin specificity to sialic acid receptor in influenza A virus subtypes. Bioinformation. 2010;5:244–9. doi: 10.6026/97320630005244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKimm-Breschkin JL, Blick TJ, Sahasrabudhe A, et al. Generation and characterization of variants of NWS/G70C influenza virus after in vitro passage in 4-amino-Neu5Ac2en and 4-guanidino-Neu5Ac2en. Antimicrob Agents Chemother. 1996;40:40–6. doi: 10.1128/aac.40.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blick TJ, Sahasrabudhe A, McDonald M, et al. The interaction of neuraminidase and hemagglutinin mutations in influenza virus in resistance to 4-guanidino-Neu5Ac2en. Virology. 1998;246:95–103. doi: 10.1006/viro.1998.9194. [DOI] [PubMed] [Google Scholar]
- 26.McKimm-Breschkin JL, Sahasrabudhe A, Blick TJ, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol. 1998;72:2456–62. doi: 10.1128/jvi.72.3.2456-2462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matrosovich M, Matrosovich T, Carr J, et al. Overexpression of the α-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol. 2003;77:8418–25. doi: 10.1128/JVI.77.15.8418-8425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mather KA, White JF, Hudson PJ, et al. Expression of influenza neuraminidase in baculovirus-infected cells. Virus Res. 1992;26:127–39. doi: 10.1016/0168-1702(92)90152-y. [DOI] [PubMed] [Google Scholar]
- 29.Gamblin SJ, Haire LF, Russell RJ, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–42. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 30.Nobusawa E, Aoyama T, Kato H, et al. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–85. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 31.Belser JA, Jayaraman A, Raman R, et al. Effect of D222G mutation in the hemagglutinin protein on receptor binding, pathogenesis and transmissibility of the 2009 pandemic H1N1 influenza virus. PLoS One. 2011;6:e25091. doi: 10.1371/journal.pone.0025091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chutinimitkul S, Herfst S, Steel J, et al. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol. 2010;84:11802–13. doi: 10.1128/JVI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Childs RA, Matrosovich T, et al. Altered receptor specificity and cell tropism of D222G hemagglutinin mutants isolated from fatal cases of pandemic A(H1N1) 2009 influenza virus. J Virol. 2010;84:12069–74. doi: 10.1128/JVI.01639-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Triana-Baltzer GB, Gubareva LV, Klimov AI, et al. Inhibition of neuraminidase inhibitor-resistant influenza virus by DAS181, a novel sialidase fusion protein. PLoS One. 2009;4:e7838. doi: 10.1371/journal.pone.0007838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan PK, Lee N, Joynt GM, et al. Clinical and virological course of infection with haemagglutinin D222G mutant strain of 2009 pandemic influenza A (H1N1) virus. J Clin Virol. 2011;50:320–4. doi: 10.1016/j.jcv.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 36.Ison MG, Gubareva LV, Atmar RL, et al. Recovery of drug-resistant influenza virus from immunocompromised patients: a case series. J Infect Dis. 2006;193:760–4. doi: 10.1086/500465. [DOI] [PubMed] [Google Scholar]
- 37.Gubareva LV, Matrosovich MN, Brenner MK, et al. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J Infect Dis. 1998;178:1257–62. doi: 10.1086/314440. [DOI] [PubMed] [Google Scholar]
- 38.Tisdale M. Monitoring of viral susceptibility: new challenges with the development of influenza NA inhibitors. Rev Med Virol. 2000;10:45–55. doi: 10.1002/(sici)1099-1654(200001/02)10:1<45::aid-rmv265>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 39.McKimm-Breschkin JL, McDonald M, Blick TJ, et al. Mutation in the influenza virus neuraminidase gene resulting in decreased sensitivity to the neuraminidase inhibitor 4-guanidino-Neu5Ac2en leads to instability of the enzyme. Virology. 1996;225:240–2. doi: 10.1006/viro.1996.0595. [DOI] [PubMed] [Google Scholar]
- 40.Sahasrabudhe A, Lawrence L, Epa VC, et al. Substrate, inhibitor, or antibody stabilizes the Glu 119 Gly mutant influenza virus neuraminidase. Virology. 1998;247:14–21. doi: 10.1006/viro.1998.9222. [DOI] [PubMed] [Google Scholar]
- 41.Amaro RE, Cheng X, Ivanov I, et al. Characterizing loop dynamics and ligand recognition in human- and avian-type influenza neuraminidases via generalized Born molecular dynamics and end-point free energy calculations. J Am Chem Soc. 2009;131:4702–9. doi: 10.1021/ja8085643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amaro RE, Minh DD, Cheng LS, et al. Remarkable loop flexibility in avian influenza N1 and its implications for antiviral drug design. J Am Chem Soc. 2007;129:7764–5. doi: 10.1021/ja0723535. [DOI] [PubMed] [Google Scholar]
- 43.Amaro RE, Swift RV, Votapka L, et al. Mechanism of 150-cavity formation in influenza neuraminidase. Nat Commun. 2011;2:388. doi: 10.1038/ncomms1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang M, Qi J, Liu Y, et al. Influenza A virus N5 neuraminidase has an extended 150-cavity. J Virol. 2011;85:8431–5. doi: 10.1128/JVI.00638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, Zhu X, Dwek RA, et al. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J Virol. 2008;82:10493–501. doi: 10.1128/JVI.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q, Qi J, Zhang W, et al. The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat Struct Mol Biol. 2010;17:1266–8. doi: 10.1038/nsmb.1909. [DOI] [PubMed] [Google Scholar]
- 47.van der Vries E, Collins PJ, Vachieri SG, et al. H1N1 2009 pandemic influenza virus: resistance of the I223R neuraminidase mutant explained by kinetic and structural analysis. PLoS Pathog. 2012;8:e1002914. doi: 10.1371/journal.ppat.1002914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheu TG, Deyde VM, Okomo-Adhiambo M, et al. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother. 2008;52:3284–92. doi: 10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKimm-Breschkin J, Trivedi T, Hampson A, et al. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob Agents Chemother. 2003;47:2264–72. doi: 10.1128/AAC.47.7.2264-2272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okomo-Adhiambo M, Nguyen HT, Sleeman K, et al. Host cell selection of influenza neuraminidase variants: implications for drug resistance monitoring in A(H1N1) viruses. Antiviral Res. 2010;85:381–8. doi: 10.1016/j.antiviral.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Zurcher T, Yates PJ, Daly J, et al. Mutations conferring zanamivir resistance in human influenza virus N2 neuraminidases compromise virus fitness and are not stably maintained in vitro. J Antimicrob Chemother. 2006;58:723–32. doi: 10.1093/jac/dkl321. [DOI] [PubMed] [Google Scholar]
- 52.Ilyushina NA, Bovin NV, Webster RG. Decreased neuraminidase activity is important for the adaptation of H5N1 influenza virus to human airway epithelium. J Virol. 2012;86:4724–33. doi: 10.1128/JVI.06774-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hurt AC, Selleck P, Komadina N, et al. Susceptibility of highly pathogenic A(H5N1) avian influenza viruses to the neuraminidase inhibitors and adamantanes. Antiviral Res. 2007;73:228–31. doi: 10.1016/j.antiviral.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Gubareva LV, Bethell RC, Penn CR, et al. In vitro characterization of 4-guanidino-Neu5Ac2en-resistant mutants of influenza A virus. In: Brown LE, Hampson AW, Webster RG, editors. Options for the Control of Influenza III. Amsterdam: Elsevier Science; 1996. pp. 753–60. [Google Scholar]
- 55.Okomo-Adhiambo M, Demmler-Harrison GJ, Deyde VM, et al. Detection of E119V and E119I mutations in influenza A (H3N2) viruses isolated from an immunocompromised patient: challenges in diagnosis of oseltamivir resistance. Antimicrob Agents Chemother. 2010;54:1834–41. doi: 10.1128/AAC.01608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujisaki S, Takashita E, Yokoyama M, et al. A single E105K mutation far from the active site of influenza B virus neuraminidase contributes to reduced susceptibility to multiple neuraminidase-inhibitor drugs. Biochem Biophys Res Commun. 2012;429:51–6. doi: 10.1016/j.bbrc.2012.10.095. [DOI] [PubMed] [Google Scholar]
- 57.Landon MR, Amaro RE, Baron R, et al. Novel druggable hot spots in avian influenza neuraminidase H5N1 revealed by computational solvent mapping of a reduced and representative receptor ensemble. Chem Biol Drug Des. 2008;71:106–16. doi: 10.1111/j.1747-0285.2007.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.