Abstract
Rhabdomyosarcoma (RMS) is a soft tissue sarcoma categorized into two major subtypes: alveolar RMS (ARMS) and embryonal RMS (ERMS). Most ARMS express the PAX3-FOXO1 (P3F) fusion oncoprotein generated by the 2;13 chromosomal translocation. In the present study, the downstream target genes of P3F were identified by analyzing two independent sets of gene expression profiles: primary RMS tumors and RD ERMS cells transduced with inducible P3F constructs. We found 34 potential target genes (27 upregulated and 7 downregulated) that were significantly and differentially expressed between P3F-positive and P3F-negative categories, both in primary RMS tumors and in the inducible P3F cell culture system. Gene ontology analysis of microarray data of the inducible P3F cell culture system employed indicated apoptosis, cell death, development, and signal transduction as overrepresented significant functional categories found in both upregulated and downregulated genes. Therefore, among the 34 potential target genes, the expression of cell death-related [Gremlin1, cysteine knot superfamily 1, BMP antagonist 1 (GREM1) and death-associated protein kinase 1 (DAPK1)] and development-related [myogenic differentiation 1 (MYOD1) and hairy/enhancer-of-split related with YRPW motif 1 (HEY1)] genes were further investigated. The differential expression of GREM1, DAPK1, MYOD1 and HEY1 was confirmed in independent tumors and inducible cell culture systems. The expression of GREM1, DAPK1 and MYOD1 were significantly upregulated; HEY1 was significantly downregulated in independent P3F-positive ARMS tumors and transcriptionally active P3F cells, compared to those in ERMS tumors and transcriptionally inactive P3F cells. This study identified target genes of P3F and suggested that four downstream targets (GREM1, DAPK1, MYOD1 and HEY1) can contribute to the biological activities of P3F involved in growth suppression or cell death and myogenic differentiation.
Keywords: rabdomyosarcoma, PAX3-FOXO1, gene expression, microarray
Introduction
Rhabdomyosarcoma (RMS), the most common type of pediatric soft tissue sarcoma, is associated with the skeletal muscle lineage. Pediatric RMS is categorized into two major subtypes: embryonal RMS (ERMS) and alveolar RMS (ARMS) based on their histologic appearance (1). ERMS shows a more favorable prognosis, while ARMS is more aggressive with a high frequency of metastases at initial diagnosis and a worse prognosis (2,3).
ERMS has not been found to be associated with any diagnostic genetic alterations, but loss of heterozygosity at 11p15 is a common finding (4,5). In contrast, ARMS is associated with recurrent chromosomal translocations. The translocation t(2;13)(q35;q14) leading to the PAX3-FOXO1 (P3F) gene fusion was found to be present in 55% of ARMS cases, while the translocation t(1;13)(q36;q14) leading to the PAX7-FOXO1 gene fusion was present in 22% of cases, and 23% of ARMS were fusion-negative (2). ARMS with the 2;13 translocation is characterized as having overexpression of P3F relative to wild-type PAX3, at both the RNA and protein levels (6). In both PAX3 and P3F, an alternative splice occurs at the intron 2/exon 3 junction resulting in inclusion or exclusion of a glutamine (Q) residue [PAX3(+Q) or (−Q) forms, respectively] (7). As a result, PAX3(+Q)-FOXO1 and PAX3(−Q)-FOXO1 are coexpressed in P3F-positive ARMS tumors.
Among RMS tumors, PAX3(+/−Q)-FOXO1-positive ARMS is the most clinically intractable fusion subtype of pediatric RMS (1,2,8,9). However, the histologic classification of RMS into ARMS and ERMS may be difficult in some cases (9) and there are no specific drugs for treating specific histologic or fusion subtypes. Therefore, there is substantial impetus to elucidate target genes of P3F, which can be used to identify therapeutic targets and markers for use in RMS diagnosis and management.
Although several recent studies utilized gene expression profiles to classify RMS and/or identify target genes of P3F and PAX7-FOXO1 or P3F only in ARMS (10–16), more research is needed to validate the function of genes that are biologically relevant in ARMS development.
In the present study, we identified potential target genes of P3F by analyzing gene expression profiles from two independent systems: primary tumors (P3F-positive ARMS and fusion-negative ERMS) and a cell culture system expressing the inducible PAX3(+/−Q)-FOXO1-estrogen receptor (ER) ligand binding domain construct in the pBabe (pB) retroviral vector. Among the potential target genes of P3F, we focused on cell death or apoptosis-related [Gremlin1, cysteine knot superfamily 1, BMP antagonist 1 (GREM1), death-associated protein kinase 1 (DAPK1)] and development-related [myogenic differentiation 1 (MYOD1) and hairy/enhancer-of-split related with YRPW motif 1 (HEY1)] genes since apoptosis and development were significantly overrepresented functional categories in our gene expression profiles.
Materials and methods
Tumor samples
The tumor specimens (15 ERMS and 16 P3F-positive ARMS) used for microarray were previously described (15). Independent tumor specimens (20 ERMS and 17 P3F-positive ARMS) examined for quantitative reverse-transcription PCR (qRT-PCR) in the validation studies were previously described (15). The presence of P3F in ARMS tumor specimens was determined and confirmed by RT-PCR and/or qRT-PCR (17).
Cell culture
RD ERMS cell line and RD-derived cells transduced with inducible PAX3(+/−Q)-FOXO1 in pB or pK1 (pK) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Grand Island, NY, USA) with 10% fetal bovine serum (FBS) (Thermo Scientific HyClone, Logan, UT, USA). The medium was supplemented with 1% penicillin/streptomycin (P/S) and 1% antibiotic-antimycotic (AM) (both from Invitrogen).
DNA constructs and construction of PAX3(−Q)-FOXO1-ER in pB retroviral vector
Retroviral constructs with several relevant DNA inserts [pK1-PAX3(+Q)-FOXO1-ER (pK-P3F-ER), pBabe-PAX2(+Q)-FOXO1-ER (pB-P3F-ER) and pCDNA3-PAX3(+Q)-FOXO1-ER (pCDNA-P3F-ER)] were previously generated by Dr Frederic G. Barr’s laboratory (18–21). A modified ER ligand-binding domain was provided by Dr G.I. Evan (22). Unless it is noted as PAX3(−Q)-FOXO1, the P3F constructs used in our studies refer to the PAX3(+Q)-FOXO1 isoform. In order to clone an inducible construct of PAX3(−Q)-FOXO1-ER into the pB retroviral vector, two consecutive mutations were introduced, using QuikChange site-directed mutagenesis (Stratagene; Agilent Technologies, Santa Clara, CA, USA) followed by multiple subcloning steps. Protocols are available upon request.
Establishment of ERMS cell culture systems inducibly expressing P3F
Retroviral transduction was performed as described previously with modifications (20,21). Cells that were transduced with retroviral DNA constructs were selected with puromycin (BD Biosciences, San Jose, CA, USA) at 1 μg/ml for RD-derived cells with PAX3(+/−Q)-FOXO1-ER in pB or in pK1. Cells carrying inducible PAX3(+/−Q)-FOXO1-ER were treated with the ligand 4-hydroxytamoxifen (Tmf) (Sigma-Aldrich, St. Louis, MO, USA), which activates and induces transcriptional activity of PAX3(+/−Q)-FOXO1-ER by translocating PAX3(+/−Q)-FOXO1-ER to the nucleus.
Luciferase reporter assays
RD cells expressing PAX3(+/−Q)-FOXO1-ER in pB or pB alone were plated at 5×104 per well in 24-well plates. The cells were transfected with 0.3 μg firefly luciferase reporter DNA containing PAX3 DNA binding sites (6 × PRS9) and 0.015 μg pRL-TK Renilla luciferase DNA using FuGENE® 6 (both from Promega, Madison, WI, USA) and incubated for 2 days. The transcriptional activity of PAX3(+/−Q)-FOXO1 was measured by the dual-luciferase assay (Promega) and was normalized by transfection efficiency control (pRL-TK Renilla luciferase).
RNA extraction and microarray data analysis
For microarray analysis, total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). For other studies including qRT-PCR and RT-PCR, total RNA was extracted using RNA STAT-60 (Tel-Test, Inc., Friendswood, TX, USA). Microarray analysis of RNA isolated from RMS tumors and RD cell culture systems expressing inducible PAX3(+/−Q)-FOXO1-ER in pB was performed on Affymetrix arrays. Microarray analysis of 31 RNA tumor samples (15 ERMS and 16 P3F-positive ARMS) was described in our previous publication (15).
For microarray analysis of inducible cell culture systems, three RD cell populations were independently transduced with each construct [PAX3(+Q)-FOXO1-ER in pB; PAX3(−Q)-FOXO1-ER in pB; pB vector alone] and treated without or with 30 nM Tmf for 24 h to generate a total of 18 samples. RNA isolated from these cells was hybridized onto Affymetrix GeneChip Human Genome U133 Plus v.2.0 (HG U133+ v.2.0). After the expression levels were determined using Affymetrix Microarray Suite 5.0, the expression levels were normalized in GeneSpring (Agilent Technology). The probe-sets, whose expression levels were detected in at least 2 of the 18 samples (in inducible cell culture system) or 1 of the 31 samples (in tumors), were selected for further analysis. To compare the gene expression profiles from tumors with those from the inducible cell culture systems, the probes on the HG U133A array were used for GeneSpring analysis.
The Significance Analysis of Microarrays (SAM) (23) with two-class paired condition was used as the common method to identify differentially expressed genes between 16 P3F positive-ARMS and 15 fusion-negative ERMS tumors as well as between the RD cells expressing PAX3(+/−Q)-FOXO1-ER in pB (treated with Tmf; 6 samples) and the RD cells expressing PAX3(+/−Q)-FOXO1-ER in pB (not treated with Tmf; 6 samples). Data was filtered by a false discovery rate (FDR) at <5% (FDR 4.94% for cells; FDR 4.91% for tumors) and ≥1.5 fold-change. The PAX3(+Q)-FOXO1-ER and PAX3(−Q)-FOXO1-ER gene expression profiles were pooled since SAM did not reveal significant differentially expressed genes between these two groups. The genes that were differentially expressed between transcriptionally active PAX3(+/−Q)-FOXO1-ER (Tmf-treated) and inactive PAX3(+/−Q)-FOXO1-ER (untreated control) were then examined.
Finally, common genes that were present in both tumors and the inducible PAX3(+/−Q)-FOXO1-ER cell culture system were identified by Venn Diagram analysis. After this analysis, the probe-sets that encoded the same gene with similar expression profiles and showed raw expression values at <150 were removed. Classifications and functional annotations of genes were analyzed via web-accessible programs: Expression Analysis Systematic Explorer (EASE) 2010 and Database for Annotation, Visualization and Integrated Discovery (DAVID) 2010 (24).
Quantitative reverse transcription-PCR (qRT-PCR) analysis
The qRT-PCR assay was performed as described previously (15,16). Test gene assays were normalized to the expression of 18S rRNA. Taqman gene expression assays used (Applied Biosystems) were: DAPK1 (assay ID# Hs00234489_m1), GREM1 (assay ID# Hs00171951_m1) and MYOD1 (assay ID# Hs00159528_m1). The sequences of forward and reverse primers and probes of PAX3-FOXO1 and HEY1 are available upon request.
Extraction of cellular proteins and isolation of secreted proteins from the medium of cultured cells and immunoblot analysis
The cells were seeded at 106/100-mm dish in phenol red-free DMEM medium containing 10% FBS, 1% P/S and 1% AM for 24 h prior to Tmf treatment. The cells expressing inducible PAX3(+/−Q)-FOXO1-ER were then treated with 30 nM Tmf for various times. Cellular proteins were lysed as described previously (20). To assay the secreted GREM1 protein, the medium containing 10% FBS was replaced with FBS-free DMEM after 24 h of Tmf treatment, and cells were cultured for an additional 24 or 48 h with Tmf treatment. The medium of the cultured cells was collected and concentrated to <1,000 μl using Amicon Ultra-4 with Ultracel-10K (UFC80-1024; Millipore, Billerica, MA, USA). Protein was quantified using Coomassie Plus Protein Assay (Pierce Biotechnology, Inc., Rockford, IL, USA).
Immunoblot analysis was performed as described previously (25) with modifications. The primary antibodies used were: anti-PAX3 rabbit polyclonal, DAPK1 (ab10443; both from Abcam, Cambridge, MA, USA) and GREM1 (cat# AP6133a; Abgent, San Diego, CA, USA).
Statistical analysis
The qRT-PCR data of tumors (20 fusion-negative ERMS and 17 P3(+Q)F-positive ARMS) were analyzed by the Mann-Whitney Test (Wilcoxon Rank Sum Test) and differences between the two tumor groups were considered significant at a P-value <0.05. Statistical analysis of gene expression profiles are described in ‘RNA extraction and microarray data analysis’.
Results
Development of inducible PAX3(+/−Q)-FOXO1 cell culture systems
RD ERMS cell line was transduced with PAX3(+/−Q)-FOXO1-ER in pB [pB-P3(+/−Q)F-ER] or pB alone and selected with puromycin for 2 weeks. The transduction of RD cells with these pB-P3(+/−Q)F-ER constructs resulted in the expression of PAX3(+/−Q)-FOXO1-ER at both the mRNA (Fig. 1A) and protein levels (Fig. 1B).
Figure 1.

Expression of inducible forms of PAX3(+/−Q)-FOXO1 in RD cells transduced with pBabe (pB), pB-PAX3(+Q)FKHR-ER [pB-P3(+Q)F-ER], pB-PAX3(−Q)FKHR-ER [pB-P3(−Q)F-ER], pK1 (pK), or pK-PAX3(+Q)-FOXO1-ER [pK-P3(+Q)F-ER]. (A) Total RNA was isolated and the expression of PAX3(+/−Q)-FOXO1 was determined by qRT-PCR and was normalized to 18S rRNA expression (means ± SD). (B) Total protein was extracted after 2 weeks of puromycin selection from RD ERMS cells transduced with pB-P3(+Q)F-ER (lanes 1–3), pB-P3(−Q)F-ER (lanes 4–6), pB vector alone (lanes 7–9).
In order to determine and confirm the inducible PAX3(+/−Q)-FOXO1 function, the RD-derived cells transduced with pB-P3(+/−Q)F-ER or pB were transiently transfected with a luciferase gene reporter construct containing PAX3 DNA binding sites and were treated with tamoxifen at various concentrations (0, 1, 3, 10, 30 and 100 nM) for 24 h (Fig. 2). The RD cells transduced with an empty vector pB alone (Fig. 2A) showed a very low (~0.02) transcriptional activity indicating only an endogenous PAX3 transcriptional activity. In contrast, RD cells transduced with pB-P3(+Q)F-ER (Fig. 2B) or pB-P3(−Q)F-ER (Fig. 2C) demonstrated much higher transcriptional activities as Tmf concentrations increased. The maximal transcriptional activity in RD-derived cells with pB-P3(+/−Q)F-ER was observed at 30 nM Tmf and showed a ~46- to 47-fold difference in luciferase gene transactivation between RD-P3(+/−Q)F-ER cells and RD-pB cells (Fig. 2B and C).
Figure 2.

Transcriptional activity of inducible forms of PAX3-FOXO1 (P3F) in RD cells transduced with (A) pBabe (pB), (B) pB-PAX3(+Q)FKHR-ER [pB-P3(+Q)F-ER], or (C) pB-PAX3(−Q)FKHR-ER [pB-P3(−Q)F-ER]. The cells were seeded at 5×104 cells/well, and transcriptional activity was determined by dual luciferase assay with a reporter containing PAX3 DNA binding sites (6 × PRS9) and then the cells were treated with tamoxifen at 1, 3, 10, 30 and 100 nM for 24 h. The luciferase activity was normalized by transfection efficiency control (pRL-TK-Renilla). Data shown are means ± SEM (n=3). Where an error bar is not shown, it lies within the dimensions of the symbol.
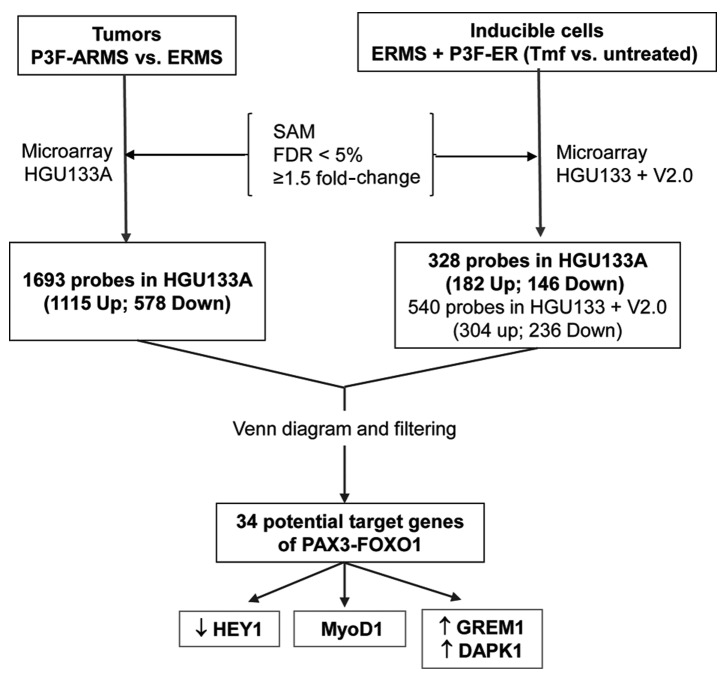
Microarray data analysis of tumors and inducible cell culture system identifies 34 potential target genes of P3F
To identify target genes of P3F in the present study, we utilized RD cells expressing inducible PAX3(+/−Q)-FOXO1-ER constructs in the pB vector (Figs. 1 and 2) based on the following rationale and significance. First, because the expression of PAX3(+/−Q)-FOXO1 in ARMS tumors varies, the utilization of cell culture models having various levels of PAX3(+/−Q)-FOXO1 can provide an unbiased approach for identifying potential target genes of PAX3(+/−Q)-FOXO1. Our previous gene expression profiling study investigated RD cells transduced with an inducible PAX3(+Q)-FOXO1-ER construct in the pK vector that enforces high expression of PAX3(+Q)-FOXO1-ER (16). In contrast, our present expression profiling study used the pB vector to express lower levels of inducible PAX3(+/−Q)-FOXO1-ER in RD cells. Second, a previous expression profiling analysis of the inducible cell system (16) was complicated by the confounding factor of a treatment-day effect. Thus, microarray data from inducible cells with PAX3(+Q)-FOXO1-ER in pK were analyzed by Mixed ANOVA, whereas primary RMS tumors were analyzed by SAM. In comparison, no such confounding factors were present in the present study and thus microarray data from both primary RMS tumors and inducible cells were both analyzed by SAM.
After analyzing microarray data from primary RMS tumors as well as inducible cells [PAX3(+/−Q)-FOXO1-ER in pB] by SAM, data were filtered for expression values, FDR, and fold-change (FDR at <5% and ≥1.5 fold-change). In the tumor microarray data, 1,693 probe-sets (1,115 upregulated and 578 downregulated) on the HG U133A platform were either significantly upregulated or downregulated in the PAX3(+/−Q)-FOXO1-positive ARMS tumors (n=16) compared to the fusion-negative ERMS tumors (n=15). Analysis of genes differentially expressed between induced (transcriptionally active) PAX3(+/−Q)-FOXO1-ER in pB (Tmf treated, n=6) and uninduced PAX3(+/−Q)-FOXO1-ER in pB (untreated control, n=6) revealed 540 probe sets (304 upregulated and 236 downregulated) on the HG U133+ v.2.0 array. Of these 540 probe sets, 328 were present on the HGU133A array (182 upregulated and 146 downregulated). Finally, 34 potential target genes (27 upregulated and 7 downregulated) were differentially expressed both in tumors and in the inducible cell culture system, using probes from the HG U133A array (Table I).
Table I.
Potential target genes of PAX3-FOXO1 as identified by microarray analysis.
| Fold-change | ||||
|---|---|---|---|---|
|
|
||||
| Affymetrix ID | Symbol | Gene name | Tumors (n=31) | Inducible cells (n=12) |
| Downregulated genes in P3F-positive categories of tumors and cells | ||||
| 218974_at | FLJ10159 | Hypothetical protein FLJ10159 | 2.85 | 1.82 |
| 218839_at | HEY1 | Hairy/enhancer-of-split related with YRPW motif 1 | 6.21 | 1.72 |
| 209030_s_at | IGSF4 | Cell adhesion molecule 1 (Immunoglobulin superfamily, member 4) | 2.86 | 1.53 |
| 203708_at | PDE4B | Phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) | 4.80 | 3.77 |
| 202732_at | PKIG | Protein kinase (cAMP-dependent, catalytic) inhibitor γ | 1.85 | 1.69 |
| 209875_s_at | SPP1 | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | 6.43 | 2.14 |
| 201368_at | ZFP36L2 | Human Tis11d gene, complete cds. | 2.60 | 1.56 |
| Upregulated genes in P3F-positive categories of tumors and cells | ||||
| 209459_s_at | ABAT | 4-Aminobutyrate aminotransferase | 10.52 | 5.84 |
| 206704_at | CLCN5 | Chloride channel 5 (nephrolithiasis 2, X-linked, Dent disease) | 2.68 | 2.01 |
| 203139_at | DAPK1 | Death-associated protein kinase 1 | 2.51 | 2.40 |
| 213712_at | ELOVL2 | Catenin (cadherin-associated protein), α-like 1 | 4.16 | 1.92 |
| 218469_at | GREM1 | Gremlin1, cysteine knot superfamily 1, BMP antagonist 1 | 5.34 | 2.13 |
| 203233_at | IL4R | Interleukin 4 receptor | 2.03 | 1.74 |
| 203126_at | IMPA2 | Inositol(myo)-1(or 4)-monophosphatase 2 | 2.90 | 1.92 |
| 202794_at | INPP1 | Inositol polyphosphate-1-phosphatase | 1.89 | 1.51 |
| 205902_at | KCNN3 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 | 6.40 | 1.97 |
| 204094_s_at | KIAA0669 | TSC22D2, TSC22 domain family, member 2 | 4.63 | 2.18 |
| 218829_s_at | KIAA1416 | CHD7, chromodomain helicase DNA binding protein 7 | 2.26 | 2.31 |
| 211042_x_at | MCAM | Melanoma cell adhesion molecule | 2.14 | 1.73 |
| 213256_at | MGC48332 | Hypothetical protein MGC48332 | 4.35 | 1.52 |
| 206657_s_at | MYOD1 | Myogenic differentiation 1 | 2.59 | 2.28 |
| 209106_at | NCOA1 | Nuclear receptor coactivator 1 | 3.15 | 1.66 |
| 209289_at | NFIB | Nuclear factor I/B | 1.97 | 1.51 |
| 205858_at | NGFR | Nerve growth factor receptor (TNFR superfamily, member 16) | 4.03 | 1.77 |
| 204105_s_at | NRCAM | Neuronal cell adhesion molecule | 6.62 | 2.36 |
| 209123_at | QDPR | Quinoid dihydropteridine reductase | 5.04 | 1.84 |
| 203217_s_at | SIAT9 | Sialyltransferase 9 (CMP-NeuAc:lactosylceramide α-2,3-sialyltransferase; GM3 synthase) | 2.43 | 1.84 |
| 203625_x_at | SKP2 | S-phase kinase-associated protein 2 (p45) | 2.64 | 1.91 |
| 213624_at | SMPDL3A | Sphingomyelin phosphodiesterase, acid-like 3A | 2.05 | 1.76 |
| 212761_at | TCF7L2 | Transcription factor 7-like 2 (T-cell specific, HMG-box) | 2.68 | 1.59 |
| 209656_s_at | TM4SF10 | Transmembrane 4 superfamily member 10 | 2.36 | 3.11 |
| 215389_s_at | TNNT2 | Troponin T2, cardiac | 5.39 | 1.86 |
| 219038_at | ZCWCC2 | Zinc finger, CW-type with coiled-coil domain 2 | 3.52 | 2.02 |
| 49111_at | DKFZp762M127 | MRNA; cDNA DKFZp762M127 (from clone DKFZp762M127) | 6.60 | 8.23 |
The 34 genes were significantly differentially expressed between P3F positive-ARMS and fusion-negative ERMS tumors and between induced (transcriptionally active) PAX3(+/−Q)-FOXO1-ER in pB and uninduced (inactive) PAX3(+/−Q)-FOXO1-ER in pB in RD-derived cells. The fold-change difference was calculated by dividing the expression levels of P3F-positive categories of tumors (ARMS) and inducible cells with those of P3F-negative categories of tumors (ERMS) and inducible cells. P3F, PAX3-FOXO1; ARMS, alveolar rhabdomyosarcoma; ERMS, embryonal rhabdomyosarcoma; pB, pBabe.
Many of the differentially expressed genes in the PAX3(+/−Q)-FOXO1-ER cell systems are related to apoptosis and development
To investigate biological consequences of the PAX3(+/−Q)-FOXO1 expression signature, the 540 significantly upregulated or downregulated probes identified in RD cells expressing the inducible PAX3(+/−Q)-FOXO1-ER in pB on the HG-U133+ v.2.0 platform were analyzed using EASE and DAVID (24). Notably, 10 out of the 15 most significant functional categories analyzed by EASE for the upregulated genes were apoptosis, cell death, or negative regulation of cell proliferation (Fig. 3).
Figure 3.
Expression analysis systematic explorer (EASE) of gene ontology annotations in gene expression profiles of the inducible PAX3-FOXO1 (P3F) cell systems. Functional categories enriched in the significantly upregulated (□) and downregulated (■) genes in the gene expression profiles of inducible P3F cell systems are shown.
The most significant 20 overrepresented functional groups in the RD cells carrying inducible PAX3(+/−Q)-FOXO1-ER in pB identified by DAVID are listed in Table II. Among these 20 groups, five groups were commonly found in both upregulated and downregulated genes. Four out of these 5 functional groups were related to development (developmental process, multicellular organismal development, anatomical structure development, system development) and one group to signal transduction. MYOD1 (an upregulated gene) and HEY1 (a downregulated gene) were found in all of the 5 common functional groups.
Table II.
DAVID analysis of the microarray data of the inducible PAX3-FOXO1 cell culture system.
| Gene category | Counts (no. of genes) | No. of probes | P-value |
|---|---|---|---|
| Upregulated genes in transcriptionally active PAX3(+/−Q)-FOXO1 cells | |||
| Protein binding | 112 | 163 | 1.08E-08 |
| Developmental process | 62 | 91 | 3.04E-06 |
| Multicellular organismal development | 44 | 63 | 2.60E-04 |
| Ras GTPase binding | 6 | 7 | 4.32E-04 |
| Enzyme binding | 11 | 19 | 5.41E-04 |
| Anatomical structure development | 40 | 65 | 6.75E-04 |
| Small GTPase binding | 6 | 7 | 7.56E-04 |
| GTPase binding | 6 | 7 | 1.24E-03 |
| Regulation of cellular process | 67 | 96 | 1.48E-03 |
| Binding | 151 | 209 | 1.49E-03 |
| Cell communication | 63 | 94 | 1.88E-03 |
| Central nervous system development | 10 | 14 | 2.09E-03 |
| System development | 33 | 50 | 2.13E-03 |
| Splice variant | 62 | 96 | 3.27E-03 |
| Glycolipid metabolic process | 4 | 5 | 3.68E-03 |
| Regulation of biological process | 69 | 99 | 3.94E-03 |
| Transcription factor binding | 12 | 19 | 4.11E-03 |
| Signal transduction | 57 | 84 | 4.36E-03 |
| Apoptosis | 10 | 12 | 4.43E-03 |
| Transcription cofactor activity | 10 | 14 | 4.88E-03 |
| Downregulated genes in transcriptionally active PAX3(+/−Q)-FOXO1 cells | |||
| Immune system process | 26 | 36 | 5.41E-05 |
| Glycoprotein | 57 | 77 | 5.48E-05 |
| Anatomical structure morphogenesis | 25 | 35 | 9.55E-05 |
| Developmental process | 50 | 67 | 1.39E-04 |
| Anatomical structure development | 37 | 50 | 1.97E-04 |
| Multicellular organismal development | 39 | 53 | 2.46E-04 |
| Response to virus | 7 | 11 | 2.99E-04 |
| von Willebrand factor, type C | 5 | 8 | 3.21E-04 |
| Immune response | 21 | 31 | 3.59E-04 |
| 2-5-Oligoadenylate synthetase | 3 | 5 | 4.45E-04 |
| Negative regulation of biological process | 24 | 35 | 4.58E-04 |
| Signal | 44 | 60 | 5.68E-04 |
| Signal transduction | 53 | 68 | 6.54E-04 |
| 2-5-Oligoadenylate synthetase, ubiquitin like region | 3 | 5 | 7.37E-04 |
| VWC (von Willebrand factor (vWF) type C domain) | 5 | 8 | 8.24E-04 |
| Response to external stimulus | 16 | 23 | 8.50E-04 |
| PIRSF005680:Interferon-induced 56K protein | 3 | 5 | 9.61E-04 |
| Morphogenesis of an epithelium | 6 | 6 | 1.05E-03 |
| System development | 30 | 39 | 1.21E-03 |
| Multicellular organismal process | 52 | 70 | 1.22E-03 |
The 540 probe sets of significantly upregulated or downregulated probes identified in RD cells expressing pB-PAX3(+/−Q)-FOXO1-ER in the HG-U133+ v.2.0 platform were analyzed using DAVID. The most significant 20 functional groups for either upregulated or downregulate genes are listed. DAVID, Database for Annotation, Visualization and Integrated Discovery; pB, pBabe.
GREM1, DAPK1, MYOD1 and HEY1 are significantly and differentially expressed in both independent tumors and inducible PAX3-FOXO1-ER cell systems
Since gene ontology analysis EASE or DAVID indicated apoptosis (Fig. 3) and development (Table II) as significant functional categories, we validated the expression of apoptosis-related genes (GREM1, DAPK1) and development-related genes (MYOD1, HEY1) that were identified among the 34 potential target genes (Table I) differentially expressed in both primary tumors and the inducible cell culture system. These genes were analyzed in an independent panel of primary tumors (20 fusion-negative ERMS and 17 PAX3-FOXO1-positive ARMS) (Fig. 4; Table III) and RD cells carrying inducible PAX3(+/−Q)-FOXO1-ER in pB or pK (Tmf-treated versus untreated control) (Fig. 5; Table III). The validation was carried out at the RNA level using qRT-PCR for the expression of GREM1 (Figs. 4A and 5A), DAPK1 (Figs. 4B and 5B), MYOD1 (Figs. 4C and 5C) and HEY1 (Figs. 4D and 5D). Furthermore, the upregulation of GREM1 and DAPK1 at the protein level was validated using immunoblot analysis (Fig. 5E). These validation studies of GREM1, DAPK1, MYOD1 and HEY1 (Figs. 4 and 5) consistently demonstrated significant differential expression, which was comparable to that noted in the microarray expression signature (Table III).
Figure 4.
Validation of expression of genes in an independent set of tumors using qRT-PCR. The expression of GREM1, DAPK1, MYOD1 and HEY1 was determined by qRT-PCR in fusion-negative ERMS (n=20, □) and PAX3-FOXO1 (P3F)-positive ARMS (n=17, ■) primary tumors. Total RNA was isolated and the expression levels of these genes were normalized to 18S rRNA expression (mean ± SD). The mean expression for the 4 genes in P3F-positive ARMS were significantly different (P<0.005; see Table I) from those of the ERMS, respectively.
Table III.
Validation of the expression of 4 target genes (GREM1, DAPK1, MYOD1 and HEY1).
| Fold-change | |||||
|---|---|---|---|---|---|
|
|
|||||
| Microarray | qRT-PCR | ||||
|
|
|
||||
| Tumors | Inducible cells | Tumors | P-values (tumors) | Inducible cells | |
| Downregulated genes | |||||
| HEY1 | 6.21 | 1.72 | 4.83 | 0.0000 | 2.14 |
| Upregulated genes | |||||
| GREM1 | 5.34 | 2.13 | 6.82 | 0.0000 | 1.76 |
| DAPK1 | 2.51 | 2.40 | 4.26 | 0.0000 | 2.17 |
| MYOD1 | 2.59 | 2.28 | 1.86 | 0.0033 | 2.11 |
The expression of genes was determined by qRT-PCR in independent tumors (20 ERMS; 17 P3F-positive ARMS) and RD ERMS cells transduced with inducible PAX3(+/−Q)-FOXO1-ER in pB. For microarray, 31 tumors (15 ERMS; 16 P3F-positive ARMS) and 12 inducible cell culture samples (6 Tmf-treated, 6 untreated) were analyzed. The fold-change difference was calculated by dividing the expression levels of P3F-positive categories of tumors (ARMS) and inducible cells with those of P3F-negative categories of tumors (ERMS) and inducible cells. GREM1, gremlin1, cysteine knot superfamily 1, BMP antagonist 1; DAPK1, death-associated protein kinase 1; MYOD1, myogenic differentiation 1; HEY1, hairy/enhancer-of-split related with YRPW motif 1; qRT-PCR, quantitative reverse-transcription PCR; ERMS, embryonal rhabdomyosarcoma; P3F, PAX3-FOXO1; ARMS, alveolar rhabdomyosarcoma; pB, pBabe; Tmf, 4-hydroxytamoxifen.
Figure 5.
Validation of the expression of genes in the RD cells transduced with PAX3(+/−Q)-FOXO1-ER using qRT-PCR and immunoblot assays. (A-D) RD cells transduced with +Q or −Q isoforms of PAX3(+/Q)-FOXO1-ER [P3(+/−Q)F-ER] (■) in pBabe (pB) or P3(+Q)F-ER in pK1 (pK) (■) or retroviral vectors (□) alone (pB or pK), were treated without or with 4-hydroxytamoxifen (Tmf) at 30 nM for 24 h. Total RNA was isolated and the expression of these genes was determined by qRT-PCR. The relative expression levels of the 4 genes were normalized to 18S rRNA (mean ± SD; n=3). (E) Cells were cultured for 48 h with 30 nM Tmf. Total proteins were extracted and protein expression was determined by immunoblot analysis. The protein expression levels of GREM1 and DAPK1 were normalized by β-actin protein expression for densitometry graphs. ND, not determined.
Discussion
In the present study, we developed the RD ERMS cell culture system expressing the inducible PAX3(+/−Q)-FOXO1-ER in pB, which is useful for determining the early short-term effects of PAX3(+/−Q)-FOXO1 and for regulating the strength and duration of transcriptional activity of PAX3-FOXO1. We analyzed two independent sets of gene expression profiles: primary RMS tumors and RD ERMS cells transduced with inducible PAX3(+/−Q)-FOXO1 constructs. We found 34 potential target genes (27 upregulated and 7 downregulated) that were significantly and differentially expressed between PAX3(+/−Q)-FOXO1-positive and fusion-negative categories, in both primary tumors and the inducible PAX3(+/−Q)-FOXO1 cell culture system. Among the 34 genes, we investigated cell death or apoptosis-related (GREM1, DAPK1) and development-related genes (MYOD1, HEY1).
Target genes of PAX3-FOXO1 and PAX7-FOXO1, or PAX3-FOXO1 only, were previously reported, based on gene expression profiling in both primary tumors and cells transduced with constitutive PAX3/PAX7-FOXO1 (13) or inducible PAX3-FOXO1-ER (16) constructs. Constitutive PAX3-FOXO1 cell system represents relatively longer (later) effects of PAX3-FOXO1 compared to the inducible PAX3-FOXO1-ER cell systems. These two studies (13,16) reported 81 and 39 target genes, respectively. Compared with these two previous studies, 11 and 12 target genes, respectively, were found in common with the present study (Table IV). Six genes (KCNN3, MCAM, MYOD1, TCF7L2, TM4SF10, ZFP36L2) were identified as targets in all three studies (Table IV) (13,16, present study).
Table IV.
Comparison of the potential target genes of PAX3-FOXO1 in three studies (13,16, present study).
| 6 common genes in all three studies [Davicioni et al(13), Mercado et al(16) and present study] | 12 common genes in present study and Mercado et al(16) | 11 common genes in present study and Davicioni et al(13) | 16 common genes in Davicioni et al(13) and Mercado et al(16) |
|---|---|---|---|
| KCNN3 | DAPK1 | ABAT | DCX |
| MCAM | DKFZp762M127 | IL4R | DUSP4 (↓) |
| MYOD1 | GREM1 | INPP1 | GADD45A |
| TCF7L2 | HEY1 (↓) | KCNN3 | IGFBP3 (↓) |
| TM4SF10 | KCNN3 | MCAM | KCNN3 |
| ZFP36L2 (↓) | MCAM | MYOD1 | MARCH3 |
| MYOD1 | NRCAM | MCAM | |
| NCOA1 | SKP2 | MEG3 | |
| QDPR | TCF7L2 | MET | |
| TCF7L2 | TM4SF10 | MYCN | |
| TM4SF10 | ZFP36L2 (↓) | MYOD1 | |
| ZFP36L2 (↓) | NEBL | ||
| PRKAR2B | |||
| TCF7L2 | |||
| TM4SF10 | |||
| ZFP36L2 (↓) |
The potential target genes of P3F were identified by microarray analysis in both primary RMS tumors and cells transduced with constitutive PAX3/PAX7-FOXO1 (13) or inducible P3F-ER (16, present study) constructs. Unless noted as (↓) for downregulated genes, all other genes were upregulated genes.
We demonstrated that higher expression of MYOD1 and lower expression of HEY1 were consistently observed in all PAX3-FOXO1-expressing ARMS primary tumors and cells, in comparison to fusion-negative ERMS primary tumors and cells. These findings of MYOD1 upregulation are in accord with results reported in previous studies of MYOD1 expression in human ARMS (26,27), PAX3/PAX7-FOXO1-positive cells (13), mesenchymal stem cells transfected with PAX3-FOXO1 (28), and NIH3T3 fibroblasts transduced with PAX3-FOXO1 (10,29). In a previous study (30), HEY1 overexpression was found to inhibit MYOD1, an early myogenic differentiation marker, which indicates that HEY1 downregulation found in our study is consistent with this PAX3-FOXO1-induced myogenic developmental program including MYOD1 upregulation. It is hypothesized that PAX3-FOXO1 simultaneously reinforces myogenic determination by upregulating MYOD1, while suppressing terminal myogenic differentiation (31,32).
The oncogenic activity of PAX3-FOXO1 is characterized in part by its stimulatory effects on cell proliferation (33–35) and cell survival/anti-apoptosis (36,37) as well as its inhibitory role on terminal myogenic differentiation (31,32). However, in addition to these effects, PAX3-FOXO1 can also cause growth suppression and cell death in other settings (20,21,38). These paradoxical features of PAX3-FOXO1 being both oncogenic and growth-suppressive were demonstrated in previous studies with immortalized murine fibroblasts (20,21) and human myoblasts (38). We identified and validated that tumor-suppressor genes, GREM1 (39) and DAPK1 (40), are upregulated at both RNA and protein levels in PAX3-FOXO1-positive ARMS tumors and PAX3-FOXO1-ER inducible cell culture systems. Our study suggests that GREM1 and DAPK1 tumor suppressor genes can be potential target genes contributing to this growth-suppressive activity of high PAX3-FOXO1 expression.
In conclusion, we identified 34 potential downstream target genes of PAX3(+/−Q)-FOXO1 by analyzing two independent sets of gene expression profiles: primary RMS tumors and RD ERMS cells transduced with inducible PAX3-FOXO1 constructs. Our study can serve as a basis to propose the 4 genes (GREM1, DAPK1, MYOD1 and HEY1) as targets that function in growth suppression or myogenic differentiation downstream of PAX3-FOXO1 in ARMS (Fig. 6).
Figure 6.
Identification of GREM1, DAPK1, and MYOD1 and HEY1 as potential target genes of PAX3-FOXO1 (P3F) in alveolar rhabdomyosarcoma (ARMS) based on two independent sets of gene expression profiles: primary RMS tumors and RD ERMS cells transduced with inducible P3F constructs.
Acknowledgements
This research was supported by NIH grants CA064202-13 (F.G. Barr), CA104896-03 (F.G. Barr), CA087812-06 (F.G. Barr, G.E. Mercado), and CA106450 (M. Ladanyi). We thank Dr Frederic G. Barr for his support and advice; Donna M. Gustafson for technical assistance; Dr Shujuan J. Xia for technical advice; Juseong Lee for editorial assistance; and Dr Lawrence A. Loeb (CA077852-14) for encouragement and comment.
Abbreviations
- RMS
rhabdomyosarcoma
- Q
glutamine
- ARMS
alveolar rhabdomyosarcoma
- ERMS
embryonal rhabdomyosarcoma
- ER
estrogen receptor
- pK
pK1
- P3F
PAX3-FOXO1
- P3F-ER
PAX3-FOXO1-ER
- Tmf
4-hydroxytamoxifen
References
- 1.Linardic CM. PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett. 2008;270:10–18. doi: 10.1016/j.canlet.2008.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorensen PH, Lynch JC, Qualman SJ, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the Children’s Oncology Group. J Clin Oncol. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 3.Stevens MC. Treatment for childhood rhabdomyosarcoma: the cost of cure. Lancet Oncol. 2005;6:77–84. doi: 10.1016/S1470-2045(05)01733-X. [DOI] [PubMed] [Google Scholar]
- 4.Scrable HJ, Witte DP, Lampkin BC, Cavenee WK. Chromosomal localization of the human rhabdomyosarcoma locus by mitotic recombination mapping. Nature. 1987;329:645–647. doi: 10.1038/329645a0. [DOI] [PubMed] [Google Scholar]
- 5.Visser M, Sijmons C, Bras J, et al. Allelotype of pediatric rhabdomyosarcoma. Oncogene. 1997;15:1309–1314. doi: 10.1038/sj.onc.1201302. [DOI] [PubMed] [Google Scholar]
- 6.Davis RJ, Barr FG. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proc Natl Acad Sci USA. 1997;94:8047–8051. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du S, Lawrence EJ, Strzelecki D, Rajput P, Xia SJ, Gottesman DM, Barr FG. Co-expression of alternatively spliced forms of PAX3, PAX7, PAX3-FKHR and PAX7-FKHR with distinct DNA binding and transactivation properties in rhabdomyosarcoma. Int J Cancer. 2005;115:85–92. doi: 10.1002/ijc.20844. [DOI] [PubMed] [Google Scholar]
- 8.Kelly KM, Womer RB, Sorensen PHB, Xiong QB, Barr FG. Common and variant gene fusions predict distinct clinical phenotypes in rhabdomyosarcoma. J Clin Oncol. 1997;15:1831–1836. doi: 10.1200/JCO.1997.15.5.1831. [DOI] [PubMed] [Google Scholar]
- 9.Anderson J, Gordon T, McManus A, et al. Detection of the PAX3-FKHR fusion gene in paediatric rhabdomyosarcoma: a reproducible predictor of outcome? Br J Cancer. 2001;85:831–835. doi: 10.1054/bjoc.2001.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan J, Bittner ML, Saal LH, et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci USA. 1999;96:13264–13269. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wachtel M, Dettling M, Koscielniak E, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;q23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64:5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 12.Wachtel M, Runge T, Leuschner I, et al. Subtype and prognostic classification of rhabdomyosarcoma by immunohistochemistry. J Clin Oncol. 2006;24:816–822. doi: 10.1200/JCO.2005.03.4934. [DOI] [PubMed] [Google Scholar]
- 13.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcoma. Cancer Res. 2006;66:6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 14.De Pittà C, Tombolan L, Albiero G, et al. Gene expression profiling identifies potential relevant genes in alveolar rhabdomyosarcoma pathogenesis and discriminates PAX3-FKHR positive and negative tumors. Int J Cancer. 2006;118:2772–2781. doi: 10.1002/ijc.21698. [DOI] [PubMed] [Google Scholar]
- 15.Laé M, Ahn EH, Mercado GE, et al. Global gene expression profiling of PAX-FKHR fusion-positive alveolar and PAX-FOXO1 fusion-negative embryonal rhabdomyosarcomas. J Pathol. 2007;212:143–151. doi: 10.1002/path.2170. [DOI] [PubMed] [Google Scholar]
- 16.Mercado GE, Xia SJ, Zhang C, et al. Identification of PAX3-FKHR-regulated genes differentially expressed between alveolar and embryonal rhabdomyosarcoma: focus on MYCN as a biologically relevant target. Genes Chromosomes Cancer. 2008;47:510–520. doi: 10.1002/gcc.20554. [DOI] [PubMed] [Google Scholar]
- 17.Barr FG, Smith LM, Lynch JC, Strzelecki D, Parham DM, Qualman SJ, Breitfeld PP. Examination of gene fusion status in archival samples of alveolar rhabdomyosarcoma entered on the Intergroup Rhabdomyosarcoma Study-III trial: a report from the Children’s Oncology Group. J Mol Diagn. 2006;8:202–208. doi: 10.2353/jmoldx.2006.050124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennicelli JL, Edwards RH, Barr FG. Mechanism for transcriptional gain of function resulting from chromosomal translocation in alveolar rhabdomyosarcoma. Proc Natl Acad Sci USA. 1996;93:5455–5459. doi: 10.1073/pnas.93.11.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomescu O, Xia SJ, Strezlecki D, Bennicelli JL, Ginsberg J, Pawel B, Barr FG. Inducible short-term and stable long-term cell culture systems reveal that the PAX3-FKHR fusion oncoprotein regulates CXCR4, PAX3, and PAX7 expression. Lab Invest. 2004;84:1060–1070. doi: 10.1038/labinvest.3700125. [DOI] [PubMed] [Google Scholar]
- 20.Xia SJ, Barr FG. Analysis of the transforming and growth suppressive activities of the PAX3-FKHR oncoprotein. Oncogene. 2004;23:6864–6871. doi: 10.1038/sj.onc.1207850. [DOI] [PubMed] [Google Scholar]
- 21.Xia SJ, Rajput P, Strzelecki DM, Barr FG. Analysis of genetic events that modulate the oncogenic and growth suppressive activities of the PAX3-FKHR fusion oncoprotein. Lab Invest. 2007;87:318–325. doi: 10.1038/labinvest.3700521. [DOI] [PubMed] [Google Scholar]
- 22.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Ahn EH, Schroeder JJ. Sphinganine causes early activation of JNK and p38 MAPK and inhibition of AKT in HT-29 human colon cancer cells. Anticancer Res. 2006;26:121–127. [PubMed] [Google Scholar]
- 26.Tonin PN, Scrable H, Shimada H, Cavenee WK. Muscle-specific gene expression in rhabdomyosarcomas and stages of human fetal skeletal muscle development. Cancer Res. 1991;51:5100–5106. [PubMed] [Google Scholar]
- 27.Tapscott SJ, Thayer MJ, Weintraub H. Deficiency in rhabdomyosarcomas of a factor required for MyoD activity and myogenesis. Science. 1993;259:1450–1453. doi: 10.1126/science.8383879. [DOI] [PubMed] [Google Scholar]
- 28.Ren YX, Finckenstein FG, Abdueva DA, et al. Mouse mesenchymal stem cells expressing PAX-FKHR form alveolar rhabdomyosarcomas by cooperating with secondary mutations. Cancer Res. 2008;68:6587–6597. doi: 10.1158/0008-5472.CAN-08-0859. [DOI] [PubMed] [Google Scholar]
- 29.Graf Finckenstein F, Shahbazian V, Davicioni E, Ren YX, Anderson MJ. PAX-FKHR function as pangenes by simultaneously inducing and inhibiting myogenesis. Oncogene. 2008;27:2004–2014. doi: 10.1038/sj.onc.1210835. [DOI] [PubMed] [Google Scholar]
- 30.Sun J, Kamei CN, Layne MD, Jain MK, Liao JK, Lee ME, Chin MT. Regulation of myogenic terminal differentiation by the hairy-related transcription factor CHF2. J Biol Chem. 2001;276:18591–18596. doi: 10.1074/jbc.M101163200. [DOI] [PubMed] [Google Scholar]
- 31.Epstein JA, Lam P, Jepeal L, Maas RL, Shapiro DN. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J Biol Chem. 1995;270:11719–11722. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- 32.Ebauer M, Wachtel M, Niggli FK, Schäfer BW. Comparative expression profiling identifies an in vivo target gene signature with TFAP2B as a mediator of the survival function of PAX3/FKHR. Oncogene. 2007;26:7267–7281. doi: 10.1038/sj.onc.1210525. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi K, Tsuchiya K, Otabe O, et al. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Biophys Res Commun. 2008;365:568–574. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 34.Collins MH, Zhao H, Womer RB, Barr FG. Proliferative and apoptotic differences between alveolar rhabdomyosarcoma subtypes: a comparative study of tumors containing PAX3-FKHR or PAX7-FKHR gene fusions. Med Pediatr Oncol. 2001;37:83–89. doi: 10.1002/mpo.1174. [DOI] [PubMed] [Google Scholar]
- 35.Anderson J, Ramsay A, Gould S, Pritchard-Jones K. PAX3-FKHR induces morphological change and enhances cellular proliferation and invasion in rhabdomyosarcoma. Am J Pathol. 2001;159:1089–1096. doi: 10.1016/S0002-9440(10)61784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernasconi M, Remppis A, Fredericks WJ, Rauscher FJ, III, Schäfer BW. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc Natl Acad Sci USA. 1996;93:13164–13169. doi: 10.1073/pnas.93.23.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayyanathan K, Fredericks WJ, Berking C, Herlyn M, Balakrishnan C, Gunther E, Rauscher FJ., III Hormone-dependent tumor regression in vivo by an inducible transcriptional repressor directed at the PAX3-FKHR oncogene. Cancer Res. 2000;60:5803–5814. [PubMed] [Google Scholar]
- 38.Xia SJ, Holder DD, Pawel BR, Zhang C, Barr FG. High expression of the PAX3-FKHR oncoprotein is required to promote tumorigenesis of human myoblasts. Am J Pathol. 2009;175:2600–2608. doi: 10.2353/ajpath.2009.090192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen B, Athanasiou M, Gu Q, Blair DG. Drm/Gremlin transcriptionally activates p21(Cip1) via a novel mechanism and inhibits neoplastic transformation. Biochem Biophys Res Commun. 2002;295:1135–1141. doi: 10.1016/s0006-291x(02)00828-8. [DOI] [PubMed] [Google Scholar]
- 40.Bialik S, Kimchi A. DAP-kinase as a target for drug design in cancer and diseases associated with accelerated cell death. Semin Cancer Biol. 2004;14:283–294. doi: 10.1016/j.semcancer.2004.04.008. [DOI] [PubMed] [Google Scholar]