Abstract
MicroRNAs (miRs) have been shown to modulate critical gene transcripts involved in tumorigenesis, but their role in tumor-mediated immune suppression is largely unknown. On the basis of miRNA gene expression in gliomas using tissue microarrays, in situ hybridization, and molecular modeling, miR-124 was identified as a lead candidate for modulating signal transducer and activator of transcription 3 (STAT3) signaling, a key pathway mediating immune suppression in the tumor microenvironment. miR-124 is absent in all grades and pathological types of gliomas. Upon up regulating miR-124 in glioma cancer stem cells (gCSCs), the STAT3 pathway was inhibited, and miR-124 reversed gCSC-mediated immune suppression of T-cell proliferation and induction of Foxp3+ regulatory T-cells (Tregs). Treatment of T-cells from immunosuppressed glioblastoma patients with miR-124 induced marked effector response including up regulation of IL-2, IFN-γ, and tumor necrosis factor (TNF)-α. Both systemic administration of miR-124 or adoptive miR-124-transfected T-cell transfers exerted potent anti-glioma therapeutic effects in clonotypic and genetically engineered murine models of glioblastoma and enhanced effector responses in the local tumor microenvironment. These therapeutic effects were ablated in both CD4+ and CD8+ depleted mice and nude mouse systems, indicating that the therapeutic effect of miR-124 depends on the presence of a T-cell-mediated antitumor immune response. Our findings highlight the potential application of miR-124 as a novel immunotherapeutic agent for neoplasms and serve as a model for identifying miRNAs that can be exploited as immune therapeutics.
Keywords: microRNAs, miR-124, cancer stem cells, glioblastoma multiforme, immune suppression, signal transducer and activator of transcription 3, regulatory T-cells
Introduction
Glioblastoma multiforme (GBM) is one of the most aggressive primary brain tumors because of its rapid cell growth and immunosuppressive capabilities. Because of the ineffectiveness of many chemotherapy agents and drug treatments, immune therapeutic strategies are an appealing approach; however, they are limited by the profound immune suppression that these tumors mediate. Increased antitumor immune responses have been linked to enhanced survival in many cancers, including GBM (1-7). Signal transducer and activator of transcription 3 (STAT3) is a transcription factor that is a potent regulator of tumorigenesis and immune suppression (8, 9). STAT3 is upregulated in many cancers, including gliomas (10) and promotes tumorigenesis by preventing apoptosis and enhancing proliferation, angiogenesis, invasion, and metastasis (11, 12). The STAT3 pathway can also become active in tumor-infiltrating immune cells, markedly impairing their antitumor effector responses (9) while enhancing the functional activity of immune-suppressive cells (13, 14). Glioma cancer stem cells (gCSCs) demonstrate activation of the STAT3 pathway (12), which has been shown to modulate their profound immune-suppressive properties (13, 14).
Recent studies have demonstrated that the levels of distinct microRNAs (miRs) in the glioma environment differ from those in peritumor tissue. These miRs are non-coding molecules involved in posttranscriptional gene regulation, which have been shown to modulate tumor cell proliferation and apoptosis and to act as oncogenes or tumor-suppressor genes (15-18). Although miRs have been linked to tumor progression, the connection between tumor-mediated immune suppression and miRs has yet to be explored. It is plausible that they control the STAT3 pathway or are themselves regulated by STAT3, such as miR-21 (19). In addition, oncogenic miR inhibition in murine glioma models has resulted in in vivo growth inhibition (16). MiR-124, which is highly expressed in the central nervous system (CNS), including the cerebellum (20), plays a role in neurogenesis (21), and stimulates neuronal differentiation by antagonizing the transcriptional repressor element 1 silencing transcription factor (REST), which maintains embryonic stem cells’ self-renewal abilities and pluripotency (21, 22). In high-grade malignant gliomas and astrocytes, miR-124 is scarcely expressed or is absent (23, 24). Furthermore, the loss of miR-124 enhances stem-like traits and increases the invasion of glioma cells (25) whereas miR-124 is strongly induced during neural differentiation of embryonic stem cells (24, 26). miR-124 has been shown to inhibit GBM and medulloblastoma cell proliferation and differentiation (27, 28), and the expression of miR-124 in GBM cell lines results in decreased migration and invasion (23). Here we hypothesized that miR-124, by interacting with the STAT3 pathway, regulates the immune-suppressive properties of glioma cells and that miR-124 up regulation or administration in vivo will exert potent antitumor immune effects. To determine whether miRNAs can be exploited as immune therapeutics, we performed a tissue microarray analysis (TMA) to detect miRNAs preferentially absent in gliomas relative to miRNA expression in normal brain, selected lead candidates that could bind to key immune suppressive pathways, and then evaluated the therapeutic efficacy of these candidates in multiple murine glioma models.
Materials and Methods
miR comparison of GBM to normal brain tissue
This study was approved by the institutional review board at M.D. Anderson and conducted according to protocol #LAB03-0687. Tumors were pathologically confirmed as GBM (World Health Organization grade IV) by a board-certified neuropathologist. Tumors were washed in RPMI1640 medium and dissected to remove blood products and surrounding non-tumor brain tissue. The total tissue was broken down into smaller pieces and digested in digesting buffer from the cancer cell isolation kit (Panomics, Santa Clara, CA) for 2 hours. The cells were suspended in RNAlater solution (Ambion, Austin, TX) in Rnase-free tubes and stored at 4°C overnight; after 24 hours, they were transferred to −80°C until needed for total RNA extraction. Extraction was performed using the mirVana kit (Ambion). Once extracted, RNA levels were analyzed for concentrations and purity using UV/Vis spectroscopy at 230, 260, and 280 nm.
Total RNA extracted from patients was sent to Phalanx Biotech Group (Belmont, CA) for microRNA and mRNA-gene expression analyses. Total RNA from normal brain tissues was obtained from Biochain (Hayward, CA). The results of the GBM human miRNA OneArray Microarray v2 analysis were used to determine which miRs had significant differences in expression compared with normal donor miRs. Expressional differences in terms of multiples (- fold differences) were calculated with Microsoft Excel, and miRs with the most significant differences in expression levels were chosen for the miR target analysis using TargetScan (Release 5.1)(30). miRs of interest were selected on the basis of putative targets and the degree of deviation from normal brain.
Real-time PCR to confirm relative miRNA expression levels
Total RNA extracted from GBM cells or gCSCs was used as the template for reverse transcription using the TaqMan reverse transcription kit (Applied Biosystems, Carlsbad, CA) in a thermocycler, per the manufacturer’s instructions. Primers for reverse transcription and PCR were purchased for human miR-124, miR-21, U6 and U18 snRNAs (Applied Biosystems). U6 and U18 was used as an endogenous control. cDNA was used as the template for real-time PCR. U18 and miR-124 amplifications were run in triplicate using the TaqMan real-time PCR kit (Applied Biosystems) in the 7500 real-time PCR system (Applied Biosystems). Further reactions, substituting water for the cDNA template, were used as additional controls. Excel was used to calculate the mean levels of each miR and the U18 internal control. The relative expression levels of miR-124 were compared with those of the internal controls, and a bar graph was generated.
Glioma tissue microarray and in situ hybridization
See Supporting Information for details.
miR-124 transfection in gCSCs, astrocytes and T-cells
The precursor form of miR-124 (30 nM) and the scramble negative control were used to transfect gCSCs and T-cells using the siPORT NeoFX transfection agent (Applied Biosystems) or Nucleofector transfection kit (Lonza, Allendale, NJ). Cells were incubated for 72 hours at 37°C to determine cell surface marker expression and collect secreted cytokines. miR-124 expression was verified via RT-PCR after transfection. The morphologic characteristics of the gCSCs were documented at 48 hours after the transfection. A rescue experiment of miR-124 inhibition was accomplished by cotransfection with a plasmid expressing wild-type, constitutively active STAT3 without a miR-124 binding 3′ UTR site (kindly provided by Dr. Jinbo Yang).
In vivo experiments
The miR-124 duplex that mimics pre-miR-124a (sense: 5′-UAAGGCACGCGGUGAAUGCCA-3′, antisense: 3′-UAAUUCCGUGCGCCACUUACG-5′) and the scramble control miRNA duplex (sense: 5′-AGUACUGCUUACGAUACGGTT-3′, antisense: 3′-TTUCAUGACGAAUGCUAUGCC-5′) were synthesized (SynGen, San Carlos, CA). The sequence of murine miR-124 is identical to human miR-124 on the basis of NCBI blast data (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The treatment cohorts consisted of 20 μg of the miR-124 duplex or scramble control in 10 μL of PBS mixed with the vehicle (80 μL PBS containing 10 μL lipofectamine 2000; Invitrogen) or the vehicle control (90 μl PBS + 10 μl lipofectamine 2000). The dosing was identical for intratumoral delivery or intravenous infusion. Mice were maintained in the M.D. Anderson Isolation Facility in accordance with Laboratory Animal Resources Commission standards and handled according to the approved protocol 08-06-11831.
See supporting information for syngeneic subcutaneous, intracranial, and genetically engineered murine glioma models.
Statistical analysis
The distribution of each continuous variable was summarized by its mean, standard deviation, and range. The distribution of each categorical variable was summarized in terms of its frequencies and percentages. Continuous variables were compared between treatment groups by a two-sample t test. In the case of comparing two paired groups, a paired t test is conducted. Kaplan-Meier curves were used to estimate unadjusted time to event variables. Log-rank tests were used to compare each time-to-event variable between groups. P values of less than 0.05 (two-sided) were considered statistically significant. All statistical analyses were performed using the Statistical Package for the Social Sciences v.12.0.0 (SPSS, Chicago, IL) and SAS v. 9.1 (SAS Institute, Cary, NC). Error bars represent SD.
Results
miR-124 expression in gliomas
To determine the pattern of miR expression in GBM relative to normal brain tissue, we used the Human miRNA OneArray Microarray v2. miR-124 emerged as a leading candidate, with a mean 24.6-fold decrease in expression from that seen in normal brain tissue (Supplementary Table 1). A subsequent analysis using reverse transcription-polymerase chain reaction (RT-PCR) confirmed that miR-124 was absent or minimally expressed in GBM specimens (n = 4), glioma cell lines (n = 2), and gCSCs (n = 4) compared with normal brain tissues (n = 3) (Fig. 1A). GBM-associated microglia/macrophages also have low or undetectable expression of miR-124 (Supplementary Fig. 1). When the gCSCs were placed under neural differentiation conditions, miR-124 expression levels were increased (Supplementary Fig. 1). Because miR-124 was a leading candidate down regulated in GBM, as observed by ourselves and others (27, 31), we determined whether it was decreased in other types of gliomas. Using a glioma tissue microarray and in situ hybridization, we found that all glioma grades and types lacked miR-124 expression (Fig. 1B and Table 1). All cortex-containing neurons demonstrated positive expression of miR-124 (n=19). No differences in survival time among GBM patients were found on the basis of the relative but negligible expression of miR-124 in The Cancer Genome Atlas data set (http://cancergenome.nih.gov).
Figure 1.
miR-124 expression is significantly reduced in GBMs and inhibits STAT3. (A) Relative expression levels of miR-124 were detected by TaqMan quantitative PCR. There was a significant difference in the relative expression levels of miR-124 between normal brain tissues, GBM specimens, and gCSCs. (B) Representative specimens of normal brain (I) demonstrating miR-124 expression (arrow) by in situ hybridization in neurons and in a GBM (II) lacking miR-124 expression (at 40x magnification). (C) Sequence of the predicted miR-124 binding site on the STAT3-3’UTR and the STAT3-3’UTR mutant that disrupted miR-124 binding. On the basis of predictive algorithms, there exists an 11-bp nucleotide binding site in the STAT3 3′-UTR that is imperfectly paired with miR-124, which is predicted to lead to STAT3 mRNA degradation. (D) Relative luciferase activity of HeLa cells after transfection with miR-124- or scramble control-expressing plasmids in conjunction with a wild type STAT3 3′-UTR or miR-124 binding site mutant reporter construct, demonstrating inhibition of STAT3. **P < 0.01. (E) Western blot analysis of the STAT3 signaling pathway gCSCs transfected with scramble control (S) compared with miR-124 (M).
Table 1.
miRNA expression incidence in gliomas
| Tumor pathologic type | WHO grade | % with miR-124 expression |
N |
|---|---|---|---|
| Glioblastoma | IV | 0 | 150 |
| Gliosarcoma | IV | 0 | 6 |
| Anaplastic astrocytoma | III | 0 | 24 |
| Anaplastic mixed oligodendroglioma |
III | 0 | 9 |
| Anaplastic oligodendroglioma | III | 0 | 16 |
| Mixed oligoastrocytoma | II | 0 | 5 |
| Oligodendroglioma | II | 0 | 24 |
| Low-grade astrocytoma | II | 0 | 1 |
| Subependymoma | I | 0 | 2 |
miR-124 interacts with the STAT3 pathway
To determine which miRs interact with STAT3, we used TargetScan to identify a group of miRs with conserved target sites in the STAT3 3′-UTR. Theoretically, these miRs can inhibit STAT3 expression and thus down regulate STAT3-mediated immune suppression in GBM. The top-rated candidates were miR-124, miR-17, miR-125, and miR-129, with aggregate PCT scores of 0.85, 0.85, 0.84, and 0.58 (respectively) (Supplementary Table 2). An additional analysis suggests that miR-124 is predicted to bind to STAT3 (http://cbcsrv.watson.ibm.com:8080/teriresias/RNA22). Therefore, on the basis of these cumulative bioinformatics data, we performed a mechanistic and therapeutic evaluation of miR-124. Because the predicted binding sites of miR-124 to STAT3 are in a conserved, homologous region (Fig. 1C), we determined whether miR-124 directly inhibits STAT3 protein expression by binding to the 3′-UTR. miR-124-negative HeLa cells were transfected with pre-miR-124 plasmid or pre-miR-control plasmid. The 3′-UTR reporter activities of STAT3 were assessed by luciferase assays. miR-124 inhibited STAT3 luciferase activity in cotransfected HeLa cells (Fig. 1D), whereas directed mutational alteration of the miR-124 3′-UTR STAT3 binding site (Fig. 1C) resulted in complete abolishment of miR-124 inhibition of luciferase activity in cotransfected HeLa cells (Fig. 1D). Subsequently, both STAT3 and pSTAT3 expression at the protein level were inhibited by miR-124 within gCSCs as detected by Western blot (Fig. 1E).
In addition to STAT3, TargetScan and other online software also suggest that miR-124 maytarget other components of the STAT3 signaling pathways including IL6Rα, Tyk2, Src homology 2 domain-containing transforming protein 1 (Shc1) and MAPK1 (Supplementary Table 3). In order to test whether miR-124 suppresses these predicted targets, we investigated the effect of forced expression of miR-124 in five gCSCs isolated from different glioblastoma patients (Fig. 1E). Shc1 is also a preferred target of miR-124 in gCSCs and is down regulated in all gCSCs treated with miR-124. pMAPK1/3 is down modulated in one gCSC cell line but this line is notable for the lack of IL-6Rα expression, suggesting that p-STAT3 activation may be due to alternative EGFR/MAPK1/3 dominant signaling. Although IL-6Rα, Tyk2, and MAPK1/3 are not preferred targets of miR-124 in gCSCs by Western blot, miR-124 can target at least two key components in the STAT3 signaling pathway (Supplementary Fig. 2). To determine if miR-124 can down modulate targets downstream of p-STAT3 such as miR-21, we measured miR-21 level in miR-124 overexpressing gCSCs by RT-PCR and found that miR-21 expression was inhibited by miR-124 (Supplementary Fig. 3).
miR-124 reverses gCSC-mediated immune suppression
To determine the phenotypic consequences of up regulating miR-124, we transiently transfected human immune-suppressive gCSCs (13) with precursor miRs and confirmed the up regulation of miR-124 by RT-PCR. The miR-124 expression was increased in the range of 5- 20,000 fold among different gCSCs. After 24 hours, gCSCs demonstrated increased adherence to the bottom of the plate, which was more pronounced after 48 hours. Specifically, the typical neurosphere morphology of the gCSCs was altered to become petri dish-attached with an elongated configuration and with contact inhibition (Fig. 2A). In contrast, transfection of astrocytes with miR-124 did not alter morphology, proliferation, apoptosis or cell cycle status (data not shown). To characterize their immunological phenotype, gCSCs were assessed for their expression of major histocompatibility complex (MHC) I, MHC II, CD40, CD80, CD86, and B7-H1, by RT-PCR and flow cytometry after transfection with miR-124. No changes were found in MHC I, MHC II, CD40, CD80, B7-H1 or CD86 mRNA and protein expression levels (data not shown). To determine what immune-suppressive soluble factors are affected by miR-124, we analyzed the conditioned medium of miR-124- or scramble (control)-transfected gCSCs using enzyme-linked immunosorbent assays (ELISAs) and cytokine and chemokine arrays. Here, we found lower levels of IL-8 (scramble: 5844 ± 1108 pg/ml versus miR-124: 2115 ± 672 pg/ml; n=4, P < 0.05), galectin-3 (scramble: 933 ± 214 pg/ml versus miR-124: 555 ± 72 pg/ml; n=4, P < 0.01), and MIC-1 (scramble: 13 ± 4 pg/ml versus miR-124: 4 ± 1 pg/ml; n=4, P < 0.05) (Supplementary Fig. 1) but not of VEGF. Cytokine and chemokine array data revealed a modest decrease in levels of TGF-β2, macrophage migration inhibitory factor, Serpin E1, CX3CL1, CXCL10, CXCL16, and chemokine C-C motif-2, when miR-124 was overexpressed in gCSCs, but these findings were not statistically significant.
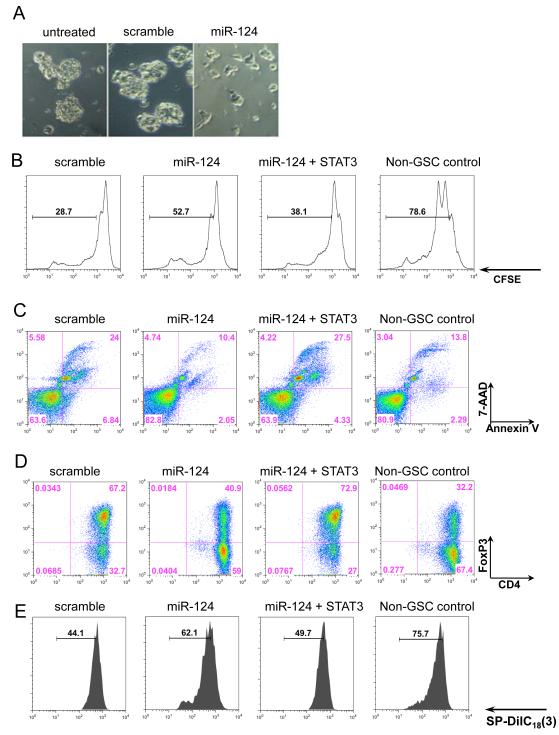
Figure 2.
miR-124 reverses glioma-mediated immunosuppression. (A) The gCSC phenotype was markedly altered by miR-124, as photographed 48 hours after transfection. (B) T-cell proliferation was detected by flow cytometry analysis with CFSE staining after 3 days of treatment with medium alone, gCSC-scramble-transfected conditioned medium, gCSC-miR-124-transfected conditioned medium, or gCSC-miR-124 + STAT3-transfected conditioned medium. miR-124 up regulation caused a reversal of inhibition in T-cell proliferation compared with the gCSC-scramble-transfected conditioned medium. STAT3 overexpression restored gCSC inhibition on T-cell proliferation. (C) T-cell apoptosis was measured by the percentage of annexin V+ 7-AAD+ cells. Similarly, miR-124 up regulation decreased gCSC-induced T-cell apoptosis, whereas STAT3 overexpression reversed the miR-124 effect. (D) T-cells were analyzed on the basis of CD4 and FoxP3 expression levels, by flow cytometry analysis. Up regulation of miR-124 caused a decline in Treg induction compared with the scrambled gCSC control, and STAT3 addition restored the ability of the gCSCs to induce Tregs. (E) The functional suppressive activity of FoxP3+ Tregs induced in (D) was verified by autologous coculture with CD4+ T-cells. The experiments were repeated three times with similar results, and one representative set of data is shown.
To determine whether miR-124 transfection reverses the functional gCSC-mediated immune inhibition of T-cells, we activated with anti-CD3/CD28 naïve CD4+ T-cells from healthy donors’ PBMCs in the presence of gCSC medium, 3-day gCSC-conditioned medium from gCSCs transfected with scramble control, miR-124, and miR-124 plus STAT3. The medium from scrambled miRNA-transfected gCSCs inhibited T-cell proliferation by 63.5 ± 13.8% versus 33.0 ± 10.1% in miR-124-transfected gCSCs (n=4, P = 0.023) (Fig. 2B). Moreover, fewer apoptotic T-cells were induced by medium from miR124-tranfected gCSCs than by medium from scramble-transfected gCSCs (Fig. 2C). Next, we determined whether miR-124 could diminish forkhead box P3 (FoxP3)+ Treg generation induced from naïve CD4+ T-cells, mediated by gCSC-conditioned medium. Indeed, the medium from miR-124-transfected gCSCs led to decreased FoxP3+ T-cell generation compared with scrambled miRNA-transfected gCSCs (Fig. 2D). Moreover, these were functional Tregs, as assessed by autologous CD4+ T-cell proliferation in coculture assays (Fig. 2E). Furthermore, all the effects mediated by miR-124 were reversed by cotransfection of wild-type, constitutively active STAT3 lacking a miR-124 sensitive 3′-UTR fragment (Fig. 2B, C, D, E). In contrast, miR-21 enhanced gCSC-mediated immune suppression as assessed by suppression of T-cell proliferation (Supplementary Fig. 3).
Because miR-124 can modify the immune-suppressive function of gCSCs, we determined whether it could exert a direct effect on the immune effector function in immunosuppressed GBM patients. PBMCs were obtained from patients newly diagnosed with GBM during tumor resection. The baseline miR-124 expression in GBM patient’s T-cells (n=4) and normal donors (n=4) is undetectable when determined by RT-PCR (data not shown). The T-cells were stimulated and simultaneously transfected with the scrambled control oligonucleotides or with miR-124. Levels of IL-2, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ were significantly increased in miR-124-transfected CD4+ T-cells and CD8+ T-cells (Fig. 3). In parallel, we also observed that miR-124 overexpression in healthy donor peripheral blood T-cells enhances production of effector cytokines, such as IFN-γ, TNF-α, and IL-12, from CD4+ and CD8+ T-cells (data not shown).
Figure 3.
miR-124 enhances T-cell effector cytokine production in GBM patients. PBMCs were isolated from the freshly drawn blood of GBM patients undergoing resection, transfected with miR-124 or scramble control oligonucleotides, and sequentially stimulated with anti-CD3/anti-CD28 antibodies for T-cell proliferation. miR-124 administration significantly enhanced effector cytokine responses, such as IFN-γ (n=10) (A), IL-2 (n=10) (B), and TNF-α (n=7) (C), in both CD4+ T and CD8+ T-cells of PBMCs from GBM patients. miR-124 up regulation suppressed pSTAT3 activity in CD8+ T-cells (n=9) and to a lesser degree in the CD4+ T-cells (n=9) (D). Each symbol represents PBMCs from different patients without transfection or with transfection with either the scramble control or miR-124.
miR-124 inhibits in vivo glioma growth
Given miR-124’s role in modulating the STAT3 pathway and immune responses, we next determined whether miR-124 could exert a therapeutic effect in vivo. To assess the in vivo antitumor efficacy of miR-124, we implanted GL261 murine glioma cells into immune competent C57BL/6 mice and treated them with miR-124 or scramble control (n=10 per group). After the subcutaneous GL261 tumors had grown to a palpable size, miR-124 duplex or scramble control was administered. Subcutaneous tumor growth progressed in all the C57BL/6J mice treated with the scramble control. In contrast, in the miR-124-treated group, the tumor volume was markedly suppressed (P = 0.01) (Fig. 4A). Gliomas started to shrink as soon as miR-124 was administered; moreover, the tumors continued to regress even after miR-124 treatment was discontinued. In contrast, tumors kept growing aggressively in scramble microRNA-treated and untreated tumor-bearing mice groups. An immunohistochemical analysis revealed that p-STAT3 glioma expression levels were markedly inhibited in the miR-124-treated cohort (P = 0.0039) (Fig. 4B).
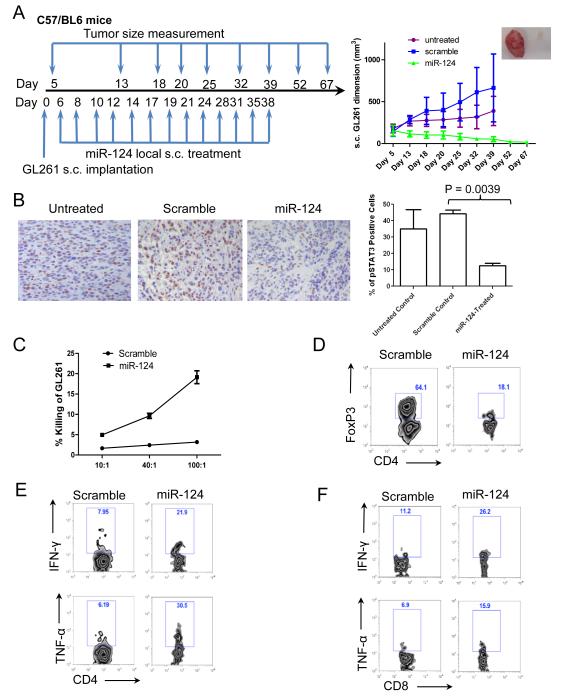
Figure 4.
miR-124 exerts potent efficacy to suppress subcutaneous GL261 tumors in a syngeneic mouse model. (A) The treatment schema and the volumes of subcutaneous GL261 tumors in C57BL/6J mice treated intratumorally with either miR-124 or scramble control, or left untreated starting on day 6 (n=10/group/experiment). The figure is the result of a single experiment. In the miR-124 group, * denotes a P value of <0.01 compared with both the untreated and scramble control tumors. Standard deviations are shown. Arrows indicate days of treatment and tumor size measurements. The inset photo on the right is of a representative ex vivo GL261 tumor comparison between miR-124- and scramble control-treated tumor-bearing mice. (B) Ex vivo immunohistochemical analysis of gliomas, untreated (n=2), or treated with a scramble control (n=5), or miR-124 (n=3), which demonstrates a marked decrease in p-STAT3 expression in the local tumor microenvironment (P = 0.0039). (C) Splenocytes from miR-124 intratumorally treated GL261 mice (n=3) have markedly increased cytotoxicity at 48 hours after coculture with GL261 cells compared with that in splenocytes from scramble control oligonucleotide-treated tumor-bearing mice (P < 0.001). The ratios of splenocytes to GL261 cells are 10:1, 40:1, and 100:1. Error bars in the curve represent the standard deviation in the data from 3 mice. (D) Decreased tumor-infiltrating FoxP3+ Tregs in miR-124-treated GL261 mice. A single example is shown, but identical results were obtained in two other tumor bearing animals. Furthermore, miR-124 administration enhances IFN-γ and TNF-α production in CD4 T-cells (E) and CD8 T-cells (F) in the tumor local microenvironment. One set of representative FACS plots is shown. Similar results were obtained from another two tumor-bearing mice treated with miR-124 or scramble control, respectively.
To determine whether enhanced immunological tumor cytotoxicity was correlated with miR-124’s efficacy in vivo, we evaluated the immune cytotoxic responses directed toward GL261 glioma cells. Splenocytes from tumor-bearing mice treated with miR-124 duplex or scramble miRNA were isolated and cocultured with CFSE-labeled GL261 target cells for 48 hours. The immune cells from the tumor-bearing mice treated with miRNA-124 increased the cytotoxic clearance of the GL261 target cells relative to that in scramble-treated mice (P < 0.05) (Fig. 4C and Supplementary Fig. 4). We next analyzed ex vivo GL261 tumor tissues from miR-124- or scramble microRNA-treated tumor-bearing mice and found that the percentage of FoxP3+ Tregs in the tumor microenvironment was reduced to 19.0 ± 8.8% in the miR-124- treated group (n=3) compared with 64.7± 5.4 % in the scramble-treated group (n=3) (P = 0.0015) (one representative FACS plot shown as Fig. 4D). We observed no significant decrease in the number of FoxP3+ Tregs in the spleen or lymph nodes of miR-124-treated tumor-bearing mice relative to control-treated mice (data not shown), indicating that miR-124’s Treg modulatory effects were confined to the tumor. To determine whether miR-124 mediates an enhanced immune activation of effector T-cells in the tumor microenvironment, we determined the production of effector cytokines such as IFN-γ and TNF-α in tumor-infiltrating T-cells. Consistent with the enhanced antitumor activity in the miR-124-treated group, a marked increase in effector cells (i.e., producing IFN-γ or TNF-α) was found in the glioma microenvironment, including CD4+ T-cells (Fig. 4E; IFN-γ: from 7.7 ± 2.0% to 21.6 ± 3.3%, P = 0.0032; TNF-α: from 6.4 ± 1.7% to 29.1± 7.4%, P = 0.0066 ) and CD8+ T-cells (Fig. 4F; IFN-γ: from 10.9 ± 3.3% to 26.0 ± 4.0%, P = 0.007; TNF-α: from 6.4 ± 1.7% to 16.4 ± 1.7%, P = 0.0019).
The therapeutic effect of miR-124 is immune mediated
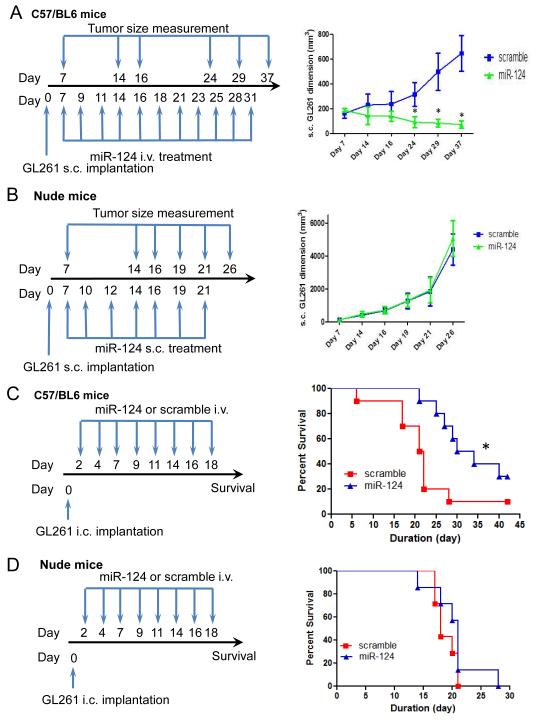
Although we found that miR-124 had a therapeutic effect when injected directly into the tumor, this is unlikely to be a viable therapeutic approach for patients. Therefore, we tested intravenous miR-124 administration in established murine glioma models. Confirming the results of the direct delivery approach, intravenous administration of miR-124 led to marked inhibition of glioma growth in vivo (Fig. 5A). To determine whether this therapeutic effect was secondarily mediated by the immune system, we implanted GL261 murine glioma cells in immune-incompetent (nude) mice and treated them with miR-124 or scramble control. Intratumoral treatment was initiated when the tumors grew to a palpable size. In the immune-incompetent animal background, miR-124 failed to exert a therapeutic effect, indicating that miR-124 mediates in vivo activity via the immune system (Fig. 5B).To determine whether treatment with miR-124 was effective against established intracerebral tumors, we administered miR-124 to C57BL/6J mice with intracerebral tumors from GL261 cells, starting after tumor cell implantation. The median survival duration for the scramble control group was 21.5 days. For mice treated with miR-124, the median survival duration was 32 days (P = 0.02) (Fig. 5C). When the experiment was repeated in an immune-incompetent model system, therapeutic efficacy was once again lost (Fig. 5D).
Figure 5.
The therapeutic effect of miR-124 is lost in immune-incompetent models. (A) The treatment schema and the volumes of subcutaneous GL261 tumors in C57BL/6J mice treated intravenously with miR-124 or scramble control starting on day 7 (n=10/group/experiment). In the miR-124 group, * denotes a P value of <0.01 compared with scramble control tumors. Standard deviations are shown. The arrows indicate days of treatment and tumor size measurements. (B) The treatment schema and volumes of subcutaneous GL261 tumors in nude mice treated intratumorally with miR-124 or scramble control starting on day 7 (n=10/group/experiment). Standard deviations are shown. Arrows indicate days of treatment and tumor size measurements. (C) Treatment schema and graph of the Kaplan-Meier estimate, demonstrating better survival in C57BL/6J mice with miR-124-treated intracerebral GL261 gliomas (n=10/group/experiment) than in scrambled controls. * denotes a P value = 0.02 compared with scramble control gliomas. (D) Graph of the Kaplan-Meier estimate demonstrates miR-124’s lack of therapeutic effect in nude mice with intracerebral GL261 gliomas (n=10/group/experiment, P = 0.24).
The immune therapeutic efficacy of miR-124 depends on T-cells
To further investigate which T-cell compartment mediates miR-124’s in vivo antitumor activity, we depleted CD4+ or CD8+ T-cells in GL261 tumor-bearing mice with neutralizing antibodies while treating those mice with miR-124 or scramble RNA oligonucleotides. We found that depletion of both CD4+ T-cells and CD8+ T-cells completely abrogated the anti-glioma efficacy of miR-124 (Fig. 6A), indicating that CD4+ and CD8+ T-cells are critical immune cell components mediating miR-124 therapeutic efficacy in vivo. In order to determine whether CD3+ T-cells are directly targeted during intravenous administration of miR-124, we isolated CD3+ T-cells from the peripheral blood, spleens and GL261 tumors and measured the expression of miR-124 by quantitative RT-PCR. There is minimal baseline expression of miR-124 in the T-cells from non-tumor bearing and GL261-bearing mice (Supplementary Fig. 5). After in vivo miR-124 treatment, there is an increase in the miR-124 expression levels in both the peripheral blood T-cells and within the glioma-infiltrating T-cells. This coincided with decreased intracellular p-STAT3 expression (Supplementary Fig. 5).
Figure 6.
T-cells mediate the antiglioma immune therapeutic efficacy of miR-124. (A) Histogram shows miR-124 transfection inhibited p-STAT3 activity in adoptively transferred T cells. (B) The treatment schema and volumes of subcutaneous GL261 tumors in C57BL/6J mice treated intratumorally with miR-124 or scramble control subcutaneously on day 7 (n=8/group/experiment) in the setting of in vivo CD4+ T-cells or CD8+ T-cell depletion. Arrows indicate days of treatment and tumor size measurements. *denotes a P value of <0.01 comparing the anti-CD4 or anti-CD8 depletion group with the isotype and miR-124-treated group. Standard deviations are shown. (C) The treatment schema and volumes of subcutaneous GL261 tumors in C57BL/6J mice treated intravenously with miR-124 or scramble control transfected CD3+ T-cells on day 14 (n=10/group/experiment). *denotes a P value of <0.01 comparing the miR-124 transfected T-cell treated group with the scramble control and untreated group.
Next, we isolated CD3+ T-cells, transfected them with miR-124 or scramble control, and expanded their numbers in vitro for 48 hours before adoptively transferring these cells into GL261 tumor-bearing mice. This miR-124 transfection inhibited p-STAT3 activity in the adoptively transferred T cells (Fig. 6B). Consistent with miR-124-enhanced T-cell effector function (as shown in Fig. 3) and the miR-124 therapeutic effects relying on T-cells (as shown in Fig. 6A), we found that GL261 gliomas regressed upon adoptive transfer of miR-124-transfected T-cells but not with control scramble-transfected T-cells (Fig. 6C), further demonstrating the pivotal role of the immune system in miR-124-mediated antitumor effects. To investigate the in vivo cellular mechanisms of adoptively miR-124-transfected T-cell treatment, we determined the percentage of infiltrating CD4+ T-cells, CD8+ T-cells and FoxP3+ Tregs in the GL261 tumors 6 days after treatment with the miRNA-transfected CD3+ T-cells. Within the glioma microenvironment, there was an increase in the CD4+ T-cell infiltration from 2.6 ± 0.9% in the scramble-control transfected CD3+ T-cell treated group to 7.4 ± 1.9% in the miR-124- transfected CD3+ T-cell treated group (P = 0.04, n=3 per group), a decrease in FoxP3+ Tregs from 26.9 ± 5.9% to 7.0 ± 0.3% in the respective groups (P = 0.014), but no change in the absolute numbers of CD8+ T-cell infiltration. Similar to the findings in Fig. 4E and F, there was a marked increase in immune effector cells within the glioma microenvironment after treatment with the miR-124-transfected T-cells; specifically, in the CD4+ T-cell compartment (IFN-γ: from 3.7 ± 2.2% in the scramble-control transfected CD3+ T-cells to 22.5 ± 6.2% in the miR-124- transfected CD3+ T-cells, P = 0.023; TNF-α: from 4.1 ± 1.9% to 17.2 ± 2.6%, P = 0.0076). Although there was no increase in the absolute number of CD8+ T-cells, the effector status of the CD8+ T-cells within the glioma microenvironment was enhanced (IFN-γ: from 1.4 ± 0.7% to 7.3 ± 1.8%, P = 0.0018; TNF-α: from 5.2 ± 0.8% to 15 ± 4.4%, P = 0.043).
miR-124 modulates T helper cell differentiation
To further investigate whether Th1 and Th17 differentiation are responsive to modulation with miR-124, we activated CD4+CD45RA+CD45RO-naïve T-cells with plate-bound anti-CD3 and soluble anti-CD28 under Th1, Th17, and inducible Treg polarization conditions before miR-124 transfection. IL-17A+ Th17 cells and FoxP3+ Treg induction was inhibited when miR-124 was overexpressed, whereas miR-124 promoted differentiation of IFN-γ+ Th1 cells (Supplementary Fig. 6).
miR-124 exerts a therapeutic effect in STAT3-expressing genetically engineered murine models
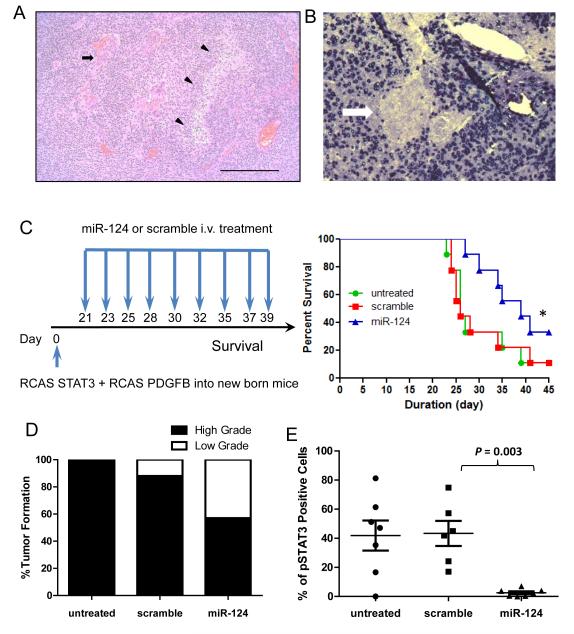
The limitation of evaluating therapeutic strategies in clonotypic models has been previously noted (32); we created a genetically engineered murine model that expresses STAT3 (11). We injected newborn Ntv-a mice with RCAS-STAT3 and RCAS-PDGFB vectors to reproducibly and consistently obtain high-grade gliomas, with the defining histologic features of microvascular proliferation, necrosis, and invasion (Fig. 7A) and lacking miR-124 expression (Fig. 7B). Similar to the findings in glioma patients, miR-124 expression in these induced gliomas was also markedly diminished. To determine whether treatment with miR-124 was also efficacious in this model system, we treated Ntv-a mice with miR-124, starting on day 21 after tumor induction. No behavioral or neurological abnormalities of the mice were noted during treatment. The median survival duration in the control group was 26 days. In mice treated with miR-124, the median survival duration was 39 days (P = 0.04) (Fig. 7C). Necropsies of glioma-bearing Ntv-a mice revealed that the miR-124-treated cohort had a lower incidence of high-grade gliomas, as determined by the study neuropathologist, on the basis of the characteristic features of necrosis and neovascular proliferation (Fig. 7D). Furthermore, there was no evidence of demyelination, macrophage infiltration, or lymphocytic infiltration in the non-tumor bearing areas of the CNS that would indicate the induction of autoimmunity (data not shown). Systemic administration of miR-124 resulted in lower p-STAT3 expression in the gliomas than in scrambled miRNA and untreated controls (Fig. 7E).
Figure 7.
miR-124 exerts a therapeutic effect in Ntv-a mice. (A) Representative hematoxylin and eosin staining of a high-grade glioma induced in Ntv-a mice transfected with RCAS-PDGFB and RCAS-STAT3 transgenes demonstrates neovascular proliferation (arrow) and pseudopallisading necrosis (arrowhead) at 100x magnification. (B) Representative specimen from the brain of an Ntv-a mouse transfected with the RCAS-PDGFB and RCAS-STAT3 transgenes demonstrates miR-124 expression by in situ hybridization in neurons surrounding a glioma devoid of miR-124 expression (arrow) at 400x magnification. (C) Treatment schema and graph of the Kaplan-Meier estimate demonstrates improved survival in miR-124-treated Ntv-a mice transfected with the RCAS-PDGFB and RCAS-STAT3 transgenes (n=9/group) compared with scramble control and untreated mice (lipofectamine 2000 vehicle only). *denotes a P value of 0.04. (D) Summary graph demonstrates the incidence of high- and low-grade gliomas on the basis of hematoxylin and eosin staining features of necrosis and neovascular proliferation in miR-124-treated Ntv-a mice transfected with RCAS-PDGFB and RCAS-STAT3 transgenes (n=7) compared with scrambled controls (n=8) and untreated mice (n=7) (P < 0.0001). (E) An ex vivo immunohistochemical analysis of gliomas, untreated (n=7) or treated with a scramble control (n=6) or miR-124 (n=7), demonstrates a marked decrease in p-STAT3 expression in the local tumor microenvironment of the miR-124-treated group (scramble vs. miR-124: P = 0.003; untreated vs miR-124: P = 0.007; untreated vs scramble: P = 0.87). Quantification of p-STAT3 expression was obtained by averaging the number of nuclear positive p-STAT3 cells by immunohistochemistry from 10 non-overlapping high-power microscopic fields (magnification × 400) of the gliomas obtained from either untreated RCAS-PDGF-B + RCAS-STAT3 mice or mice treated with the scramble control or miR-124. Each dot represents the analysis of one mouse glioma.
DISCUSSION
To our knowledge, this is the first study to demonstrate that miRNA approaches can be exploited for immune therapeutic purposes against malignancies. A significant confounding factor in the translational implementation of miR-based approaches has been adequate delivery to the target tumor cells. To circumvent this, we selected a miRNA that could reverse tumor-mediated immune suppression—specifically, a key molecular hub, STAT3—resulting in immunological recognition and clearance of the malignancy. We have also provided a strategy for identifying potential miRNA immune therapeutics that may be applicable to other types of malignancies by sequentially: 1) screening for down-modulated miRNAs using tumor microarrays; 2) determining the scope of potential use in humans by in situ hybridization of tissue microarrays; 3) screening and selecting the miRNA candidates that target immunosuppressive pathways and/or mechanisms; and 4) evaluating mechanism and therapeutic effect within immune competent model systems. Although we used the STAT3 target as proof of principal, other immunosuppressive targets such as CTLA-4, PD-1, and transforming growth factor-β could be used. Several other candidate miRNAs identified in the human glioblastoma miRNA microarray expression library likely target several of these as well and are also being evaluated for their potential as therapeutic agents in a complementary or alternative fashion with miR-124.
Our findings support the immune modulatory effects of miR-124. First, in vivo therapeutic efficacy was ablated in immune-incompetent murine model systems. Second, miR-124 transfection reduced the immune-suppressive properties in gCSCs, including inhibiting secretion of immune-suppressive cytokines such as galectin-3 (which is downstream from the STAT3 pathway (33) and induces T-cell apoptosis, promotes tumor growth, and induces Tregs), MIC-1 (which inhibits macrophage production of antitumor TNF-α), and IL-8 (which induces immune chemotaxis and is a potent promoter of angiogenesis). Furthermore, inhibition of T-cell proliferative responses and effector functions by the gCSCs was reversed upon transfection with miR-124. The restoration of T-cell TNF-α effector functions with miR-124 is consistent with a previous report that STAT3 negatively regulates TNF-α (34). Third, miR-124 treatment of T-cells from immune-suppressed GBM patients induced potent effector responses, including IL-2 and IFN-γ production. Fourth, the immune responses in the glioma microenvironment in miR-124-treated murine models demonstrated an enhancement of proinflammatory effector CD4 and CD8 T-cells, with diminished Treg intratumoral trafficking. Finally, ex vivo glioma cytotoxicity assays from miR-124-treated mice demonstrated enhanced glioma killing. Cumulatively, these data are consistent with those of previous studies that demonstrated that modulating the STAT3 pathway in the immune cell population is sufficient to mediate efficacious antitumor immune responses (35).
STAT3 signaling has been shown to be a key regulator of microglia/macrophage – mediated immune-suppression (14). MiR-124 is low or undetectable in these cells ; thus miR-124 administration may abolish or reverse their immune suppression by down-regulating STAT3 activity. Although this study focused on adaptive anti-tumor immune responses, we can’t exclude that part of the therapeutic effect was mediated via innate immunity. Other investigators have shown that the peripheral administration of miR-124 in an experimental murine autoimmune encephalomyelitis model caused deactivation of macrophages, reduced activation of myelin-specific T cells and markedly suppressed the disease (36). This discrepancy can be explained by the contextual target – i.e. miR-124 targets overactive C/EBP-α-PU.1 signaling in the context of induced autoimmunity versus STAT3 signaling in the glioma microenvironment with the resulting contrasting immune functional differences. Alternatively, one could hypothesize that the GBM is negatively regulating the expression of miR-124 in the surrounding microglia/macrophage population.
When data from The Cancer Genome Atlas were used to compare miR-124 expression and survival in patients with GBM, no differences in patient outcome were identified; however, the miR-124 expression levels were negligible in these patients, and the marginal differences are probably attributable to the submitted specimens containing intervening miR-124-expressing infiltrating neurons. Given miR-124’s role in neuronal development, we were not surprised to find that it was expressed in the normal CNS as assessed by in situ hybridization. miR-124 expression was lost across all grades and types of gliomas, suggesting not only that this loss is an early event in glioma initiation and development but also that miR-124 therapeutic approaches will be useful in a variety of gliomas.
On the basis of multiple predictive binding algorithms, luciferase expression assays, and mutational analyses, miR-124 appears to down regulate the expression of STAT3, including the activated form, p-STAT3. This finding was further supported by the results of in vitro studies that demonstrated p-STAT3 inhibition in human gCSCs and immune cells and in vivo in the local glioma microenvironment. These data are also consistent with a recent publication demonstrating that miR-124 binds to the STAT3 3′-UTR in the rat cardiomyocyte (37). However, miR-124 also targets other components of the STAT3 signaling pathway such as Shc1. Although IL-6Rα has been proved to be a target of miR-124 in hepatocarcinoma cell lines (38), this was not the case in any of the gCSCs, indicating that miR-124 has differential targets in various cells or tissues. Shc1 is not present in normal brain but is expressed in all grades of gliomas (39). In glioma patients, the loss of miR-124 may result in the expression of Shc1, which assembles the EGFR/MAPK1/3 signaling complex, thereby enhancing the activation of this signaling pathway. Because Shc1 is upstream of MAPK1/3 in the EGFR/MAPK1/3 signaling pathway, the reduced p-MAPK1/3 level might be due to the down regulation of Shc1 by miR-124. However, down modulation of Shc1 in most of the gCSCs did not correlate with down modulation of p-MAPK1/3. In the one gCSC that demonstrated reduced p-MAPK1/3 expression, the IL-6Rα expression was absent, indicating a potential greater reliance on the EGFR/MAPK1/3 signaling pathway and illustrates that although miR-124 inhibits p-STAT3, inhibition of other components of the signaling axis are contextual and hierarchical.
Other than by immune regulation, miR-124 may also reduce gliomagenesis via multiple mechanisms, including inducing gCSC differentiation, targeting multiple oncogenic signaling pathways (such as NFATc and PIK3CA), and repressing tumor cell proliferation (40), if sufficient levels of miR-124 are able to enter the CNS. Our data in the miR-124-treated Ntv-a glioma model demonstrated a decreased incidence of high-grade glioma, probably secondary to the diminished p-STAT3 expression in the local tumor microenvironment. This finding confirms those of previous studies that have linked miR-124 to gliomagenesis.
An advantage of intravenous administration of miR-124 is the ease of translational implementation as opposed to siRNA approaches that have required ex vivo transduction of the cancer cells (41), direct tumor delivery (42), knock-out in the hematopoietic cell population (35), or conjugation to CpG to target the immune population (43). Moreover, it is possible that the physiological expression of miR-124 in normal brain tissues confers tolerance to exogenous administration of this miRNA, thus minimizing toxicity. Indeed, we did not observe any evidence of CNS toxicity or induced autoimmunity in treated mice. Alternatively, because miR targets “networks” as opposed to a singular target, as is the case with siRNA, other unidentified therapeutic targets may be contributing to the beneficial in vivo effects observed with the miR-124. Specifically, miR-124 has been previously shown to target a variety of mRNAs (44) and we found it can also target miR-21, which is regulated by STAT3 (19). miR-21 has been shown to be significantly elevated in GBMs and can regulate multiple genes associated with preventing glioma cell apoptosis (45) and enhancing migration and invasion (15). miR-21 inhibition can inhibit the growth of GBM cells in vitro (18) and in vivo (16, 46). Thus, a component of the observed in vivo therapeutic effect could be secondary to the modulation of miR-21 by miR-124.
In summary, these findings provide proof-of-concept support for the systemic delivery of immune-modulatory miRNAs as a powerful and specific anticancer therapeutic modality. In the future, immune modulatory miRNAs could be used in combination and delivered in the context of nanoparticles, liposomes or exosomes or used to modify cellular vaccine strategies. Because the STAT3 pathway has been shown to mediate resistance to chemotherapeutics by modulating miR-17 (47), miR-124 may also have a therapeutic role in the setting of treatment failure. Screening miRNA expression in tumors could ultimately lead to a personalized medicine approach. Ultimately, this novel immunotherapeutic approach has the potential to not only overcome immune quiescence and resistance but also to overcome the vexing issue of miRNA delivery by exploiting the immune system as an antitumor “Trojan horse.”
Supplementary Material
Acknowledgments
The contents of this manuscript were a platform presentation at the annual meeting of the Society for Immunotherapy of Cancer in Bethesda, Maryland, October, 2012. We thank Dr. Patrick Hwu and Dr. Elizabeth Grimm for insightful commentary, Dr. Jun Yao for helping to generate heatmaps, and Audria Patrick, Ann Sutton, and Dr. David M. Wildrick for editorial assistance.
Grant support These studies were supported by the Anthony Bullock III Foundation (ABH), Cynthia and George Mitchell Foundation (ABH), the Dr. Marnie Rose Foundation (ABH), the Dr. Silverman Foundation (ABH), the Vaughn Foundation (ABH), and the National Institutes of Health CA120813-01, P50 CA127001 (ABH), MDACC Brain SPORE Career Developmental Grant (JW) and K08 NS070928 (GR).
Abbreviations
- APC
allophycocyanin
- CFSE
carboxyfluorescein diacetate succinimidyl ester
- CNS
central nervous system
- ELISA
enzyme-linked immunosorbent assay
- FITC
fluorescein isothiocyanate
- FoxP3
forkhead box P3
- GBM
glioblastoma multiforme
- IFN
interferon
- IL
interleukin
- MHC
major histocompatibility complex
- PBMCs
peripheral blood mononuclear cells
- PE
phycoerythrin
- STAT3
signal transducer and activator of transcription 3
- TMA
tissue microarray
- Tregs
regulatory T-cells
Footnotes
Disclosure of Potential Conflicts of Interest The authors declare no competing financial interests.
Author Contributions Conception and design: JW, GAC, ABH
Data acquisition: JW, FW, L-YK, SX, TD, SDF, YY, KM, KJ, OG, WQ, NBL, FFL, GR, GNF, GAC, ABH
Analysis and interpretation: JW, FW, L-YK, SX, WQ, GNF, GAC, ABH
Writing of manuscript: JW, ABH
Study supervision: ABH
References
- 1.Brooks WH, Markesbery WR, Gupta GD, Roszman TL. Relationship of lymphocyte invasion and survival of brain tumor patients. Ann Neurol. 1978;4:219–24. doi: 10.1002/ana.410040305. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs JF, Idema AJ, Bol KF, Grotenhuis JA, de Vries IJ, Wesseling P, et al. Prognostic significance and mechanism of Treg infiltration in human brain tumors. J Neuroimmunol. 2010;225:195–9. doi: 10.1016/j.jneuroim.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 3.Kong LY, Wu AS, Doucette T, Wei J, Priebe W, Fuller GN, et al. Intratumoral mediated immunosuppression is prognostic in genetically engineered murine models of glioma and correlates to immunotherapeutic responses. Clin Cancer Res. 2010;16:5722–33. doi: 10.1158/1078-0432.CCR-10-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palma L, Di Lorenzo N, Guidetti B. Lymphocytic infiltrates in primary glioblastomas and recidivous gliomas. Incidence, fate, and relevance to prognosis in 228 operated cases. J Neurosurg. 1978;49:854–61. doi: 10.3171/jns.1978.49.6.0854. [DOI] [PubMed] [Google Scholar]
- 5.Safdari H, Hochberg FH, Richardson EP., Jr Prognostic value of round cell (lymphocyte) infiltration in malignant gliomas. Surg Neurol. 1985;23:221–6. doi: 10.1016/0090-3019(85)90086-2. [DOI] [PubMed] [Google Scholar]
- 6.von Hanwehr RI, Hofman FM, Taylor CR, Apuzzo ML. Mononuclear lymphoid populations infiltrating the microenvironment of primary CNS tumors. Characterization of cell subsets with monoclonal antibodies. J Neurosurg. 1984;60:1138–47. doi: 10.3171/jns.1984.60.6.1138. [DOI] [PubMed] [Google Scholar]
- 7.Yang I, Tihan T, Han SJ, Wrensch MR, Wiencke J, Sughrue ME, et al. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J Clin Neurosci. 2010;17:1381–5. doi: 10.1016/j.jocn.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 9.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 10.Abou-Ghazal M, Yang DS, Qiao W, Reina-Ortiz C, Wei J, Kong LY, et al. The incidence, correlation with tumor-infiltrating inflammation, and prognosis of phosphorylated STAT3 expression in human gliomas. Clin Cancer Res. 2008;14:8228–35. doi: 10.1158/1078-0432.CCR-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doucette TA, Kong LY, Yang Y, Ferguson SD, Yang J, Wei J, et al. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro Oncol. 2012;14:1136–45. doi: 10.1093/neuonc/nos139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–92. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei J, Barr J, Kong LY, Wang Y, Wu A, Sharma AK, et al. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol Cancer Ther. 2010;9:67–78. doi: 10.1158/1535-7163.MCT-09-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu A, Wei J, Kong LY, Wang Y, Priebe W, Qiao W, et al. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12:1113–25. doi: 10.1093/neuonc/noq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaur AB, Holbeck SL, Colburn NH, Israel MA. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro Oncol. 2011;13:580–90. doi: 10.1093/neuonc/nor033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–55. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 19.Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 20.Weng H, Shen C, Hirokawa G, Ji X, Takahashi R, Shimada K, et al. Plasma miR-124 as a biomarker for cerebral infarction. Biomed Res. 2011;32:135–41. doi: 10.2220/biomedres.32.135. [DOI] [PubMed] [Google Scholar]
- 21.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–9. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–7. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fowler A, Thomson D, Giles K, Maleki S, Mreich E, Wheeler H, et al. miR-124a is frequently down-regulated in glioblastoma and is involved in migration and invasion. Eur J Cancer. 2011;47:953–63. doi: 10.1016/j.ejca.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 24.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–77. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 25.Xia H, Cheung WK, Ng SS, Jiang X, Jiang S, Sze J, et al. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–71. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silber J, Hashizume R, Felix T, Hariono S, Yu M, Berger MS, et al. Expression of miR-124 inhibits growth of medulloblastoma cells. Neuro Oncol. 2013;15:83–90. doi: 10.1093/neuonc/nos281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia W, Li H, Yang X, Wong KB, Sun H. Metallo-GTPase HypB from Helicobacter pylori and its interaction with nickel chaperone protein HypA. J Biol Chem. 2012;287:6753–63. doi: 10.1074/jbc.M111.287581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–30. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 32.Huse JT, Holland EC. Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol. 2009;19:132–43. doi: 10.1111/j.1750-3639.2008.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei J, Barr J, Kong LY, Wang Y, Wu A, Sharma AK, et al. Glioma-associated cancer-initiating cells induce immunosuppression. Clin Cancer Res. 2010;16:461–73. doi: 10.1158/1078-0432.CCR-09-1983. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Murray PJ. STAT3-mediated anti-inflammatory signalling. Biochem Soc Trans. 2006;34:1028–31. doi: 10.1042/BST0341028. [DOI] [PubMed] [Google Scholar]
- 35.Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–21. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 36.Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai B, Li J, Wang J, Luo X, Ai J, Liu Y, et al. microRNA-124 regulates cardiomyocyte differentiation of bone marrow-derived mesenchymal stem cells via targeting STAT3 signaling. Stem Cells. 2012;30:1746–55. doi: 10.1002/stem.1154. [DOI] [PubMed] [Google Scholar]
- 38.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–47. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magrassi L, Conti L, Lanterna A, Zuccato C, Marchionni M, Cassini P, et al. Shc3 affects human high-grade astrocytomas survival. Oncogene. 2005;24:5198–206. doi: 10.1038/sj.onc.1208708. [DOI] [PubMed] [Google Scholar]
- 40.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–9. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 41.Scuto A, Kujawski M, Kowolik C, Krymskaya L, Wang L, Weiss LM, et al. STAT3 inhibition is a therapeutic strategy for ABC-like diffuse large B-cell lymphoma. Cancer Res. 2011;71:3182–8. doi: 10.1158/0008-5472.CAN-10-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bjorge JD, Pang AS, Funnell M, Chen KY, Diaz R, Magliocco AM, et al. Simultaneous siRNA targeting of Src and downstream signaling molecules inhibit tumor formation and metastasis of a human model breast cancer cell line. PLoS One. 2011;6:e19309. doi: 10.1371/journal.pone.0019309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kortylewski M, Swiderski P, Herrmann A, Wang L, Kowolik C, Kujawski M, et al. In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat Biotechnol. 2009;27:925–32. doi: 10.1038/nbt.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 45.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 46.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 47.Dai B, Meng J, Peyton M, Girard L, Bornmann WG, Ji L, et al. STAT3 mediates resistance to MEK inhibitor through microRNA miR-17. Cancer Res. 2011;71:3658–68. doi: 10.1158/0008-5472.CAN-10-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.