Abstract
Much more than ever, nucleic acids are recognized as key building blocks in many of life's processes, and the science of studying these molecular wonders at the single-molecule level is thriving. A new method of doing so has been introduced in the mid 1990's. This method is exceedingly simple: a nanoscale pore that spans across an impermeable thin membrane is placed between two chambers that contain an electrolyte, and voltage is applied across the membrane using two electrodes. These conditions lead to a steady stream of ion flow across the pore. Nucleic acid molecules in solution can be driven through the pore, and structural features of the biomolecules are observed as measurable changes in the trans-membrane ion current. In essence, a nanopore is a high-throughput ion microscope and a single-molecule force apparatus. Nanopores are taking center stage as a tool that promises to read a DNA sequence, and this promise has resulted in overwhelming academic, industrial, and national interest. Regardless of the fate of future nanopore applications, in the process of this 16-year-long exploration, many studies have validated the indispensability of nanopores in the toolkit of single-molecule biophysics. This review surveys past and current studies related to nucleic acid biophysics, and will hopefully provoke a discussion of immediate and future prospects for the field.
Keywords: Nanopores, Nanotechnology, DNA sequencing, Drug discovery, Single-molecule biophysics
1. Introduction
Biophysics is a branch of science that utilizes physical methods and principles for studying biological systems at various levels of organization, from molecules to cells to organs to organisms. Since the 1970's, advances in optics and miniaturization of mechanical systems have enabled new tools that have helped biophysicists to study biomolecular systems at the single-molecule level. These techniques, which primarily include atomic force microscopy, fluorescence-based methods, and optical/magnetic tweezers, have allowed for the first time the probing of structure and dynamics of systems in their functional environment. Whereas X-ray crystallography, nuclear magnetic resonance, and gel electrophoresis have provided decades of information on biomolecular structure and function with detail and resolution, single-molecule tools emerged to offer a peek into the inner workings of life's smallest machines, proteins and nucleic acids, while these are in action. Combined with the power of traditional structural methods and assays, the details unraveled by single-molecule techniques have already shed light on enigmatic topics such as DNA replication, the synthesis of adenosine triphosphate (ATP), and transport of various cargoes across the cell.
The nanopore sensor is perhaps the youngest single-molecule technique to be developed to date. In less than two decades, nanopore research has allowed a multitude of studies on small biomolecules, nucleic acids and proteins. Further, nanopores are on the verge of delivering new technologies that will undoubtedly improve health, the most important of them being DNA sequencing.
1.1. Brief history: From cells to molecules
Before we embark on a journey that describes the impact of nanopores on biophysics over the past two decades, it is important to succinctly note a key moment in history that preceded this exciting field. The first incarnation of nanopores took form at a much larger scale: in the late 1940's Wallace H. Coulter invented orifice-based resistive counters for counting and sizing blood cells. What started with a hole poked in a cellophane cigarette wrapper with a hot needle [1] has quickly turned out to be a transformative tool in hematology. In this method, patented in 1953 under the title “Means for Counting Particles Suspended in a Fluid” [2], a pair of electrodes is placed across an orifice filled with electrolyte solution, and the solution conductivity is measured as a function of time using an electrometer connected to the electrodes and a chart plotter. While the conductivity does not change for a pure electrolyte solution, pushing a suspension of cells or particles through the orifice results in discrete resistive spikes, or pulses, that stem from transient occlusions of the orifice. These pulses are called resistive because the conductivity of the orifice decreases upon entry of a cell into it. The frequency of these resistive pulses with time is related to the total amount of cells present in the sample, and the distribution of electrical pulse amplitudes is related to the cell size distribution.
The simplicity of Coulter's “hole-based” sensor concept was perceived as trivial, and Coulter's pleas for submitting a patent on these ideas were met with reluctance [1]. It wasn't until four years later that his patent was awarded. Yet, Coulter's patent is among only a few patents in science that revolutionized clinical practice to this very day. Traditional histology-based methods for counting blood cells involved preparing and staining microscope slides for each patient, followed by a tedious, manual process of counting and sizing that is prone to various types of errors. In contrast, the automated Coulter counters used by hospitals today can analyze cells with speed and accuracy that cannot be matched with traditional methods. This was the first revolution, and it happened circa 1950's.
In the 1970's, DeBlois and Bean refined the Coulter technique using sub-micrometer track-etched pores to achieve detection of nanoscale particles and viruses [3]. However, the 1990's marked a true reincarnation of the Coulter counter, and with it came the second revolution in hole-based sensing. Coupled to the four-decade time gap was the vast gap in target analyte size: the hole was not of millimeter but of nanometer dimensions, and the target analytes were not cells but ions, small molecules, and individual biomolecules. This exciting leap into the nanoscale has introduced another element that is inseparable from any nanoscale interface problem – the importance of chemistry. At the nanoscale, the chemical identity of a surface governs its interaction with any analyte, as highlighted in some reviews [4–7].
A critical push towards the birth of the nanopore field was made by ion-channel electrophysiologists, who have since the early 1970's been able to monitor protein ion channels in synthetic planar lipid bilayer geometries [8,9]. Ion-channel electrophysiology is a rich discipline in biophysics, actively used for screening peptide-binding drugs and for understanding microscopic cellular processes, particularly for studying neuronal and cardiac tissues. In addition, electrophysiology can be used for detecting extremely toxic proteins, such as the lethal factor of the anthrax protective antigen [10]. In this review, emphasis is placed on the use of nanopores for identification and investigation of biomolecules. Nanopore biophysics is a much younger branch of biophysics than electrophysiology, yet it has much grown since its inception in the early 1990's, mainly due to hefty investments in potential technology for sequencing individual nucleic acid molecules. The challenges in nanopore-based DNA sequencing are as grand as the potential rewards of its success, and this has propelled diverse research and immense creativity that has sparked interest in the field.
Before zooming in we must appreciate the fact that the principle of resistive sensing applies for such a vast range of sizes, ranging from cells and micron-scale particles to small molecules. When considering the physical properties of the species that are being measured, this phenomenon becomes trivial. Table 1 lists estimated Stokes–Einstein radii and corresponding diffusion coefficients in water for a red blood cell, 20-bp DNA fragment [11,12], and K+ and Cl− ions at 25 °C. Table 1 also lists typical experimental concentrations of each species.
Table 1.
Estimated hydrodynamic radii and Stokes diffusion coefficients of aqueous species used in Coulter counters and nanopore sensors. Typical experimental concentrations are also listed.
| Species | RH (nm) | D (μm2s–1)a | Typical conc. (M) |
|---|---|---|---|
| Red blood cell | 3500 | 0.06 | ~ 10–14 |
| 20-bp DNA fragment | 3 | 140 | 10–9–10–6 |
| K+,Cl– | 0.14 | 1600 | 0.1–1 |
Calculated from D = kBT/6πηRH using T = 298 K and η = 0.001 Pa s.
As evident from the table, the ions employed for pore-based sensing are smaller, faster, and more concentrated than the analytes that they are employed to detect. This yields three important consequences that form a solid basis for resistive sensing using pores: (1) For every analyte molecule that passes through a pore, many ions pass through the pore; (2) the pore must permit transport of ions for current to be passed and it must be wide enough to accommodate analyte molecules. Therefore, the pore size should be larger than the hydrodynamic radius of the ions that are transported, as well as larger than the critical dimension of any analyte that is passed through it (i.e., > 2 nm for a linear double-stranded DNA polymer); (3) since the smallest hydrodynamic particle is an ion (~ 0.1 nm), to a first approximation, flow of ions across the pore should be able to report on differences between two analytes that differ in cross section by the ion's hydrodynamic size. Nanopores can indeed discern very subtle differences, such as the presence and absence of methyl groups in a DNA base, and the binding–unbinding kinetics of protons or deuterons to a protein [13,14]! However, it is key to recall throughout the review that such sensitive detection requires the passage of many ions for each analyte molecule that is sensed. Borrowing from the Born–Oppenheimer approximation, during the analyte's passage through the pore the ions move much faster than the analyte, and therefore, the analyte is stationary with respect to the ions. To illustrate, during a 1-µs passage of a single DNA base through an α-hemolysin nanopore, a residual current of 20 pA corresponds to the passage of (1 × 10−6 s)(2 × 10−11 A)(6.02 × 1023 ions/mol)/(96,485 sA/mol) = 124 ions/μs. This is not unique in nanopore-based sensing: in single-molecule microscopy, a single photon is a poor indicator of a process, and often many photons are required to ascertain the identity and position of a fluorescent molecule in a sample.
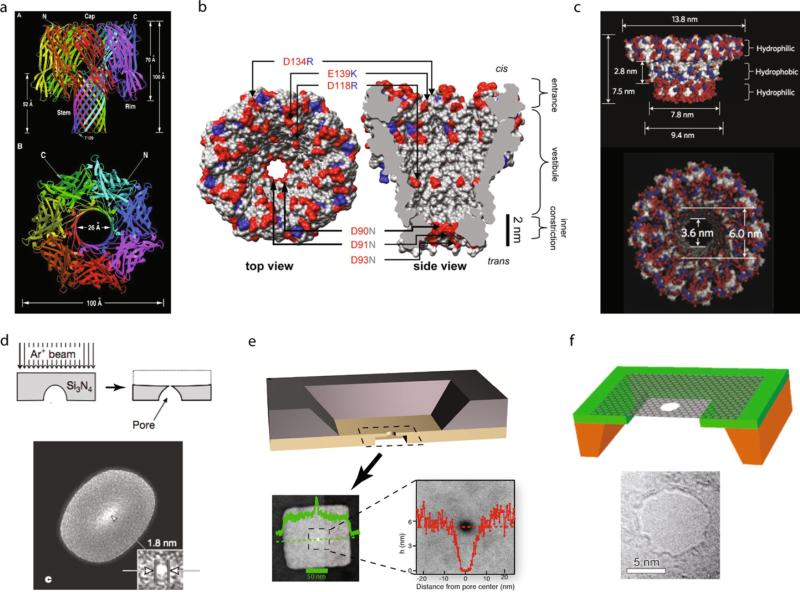
Fig. 1 shows some examples of nanopores that are discussed in this review. The toxin α-hemolysin secreted by Staphylococcus aureus is the trans-membrane channel that sparked the field of nanopore-based biomolecular analysis [15]. While it remains the most common biological pore in experiments, the channel is too long for recording the current from individual nucleotides as a DNA polymer threads through the pore. Recently, MspA from Mycobacterium smegmatis, shown in Fig. 1b, was engineered and cleverly used for discrimination among different DNA bases [16]. In contrast to α-hemolysin, the structural profile of MspA reveals a “sharp” inner constriction whose effective length is ~ 1 nm. A number of key amino-acid substitutions had to be made to the wild-type structure in order for the pore to not exhibit gating, a term used to describe a channel's inherent structural changes that are accompanied by stochastic changes in the ion current. A comparison between α-hemolysin and MspA follows later in the review. Fig. 1c shows a phi29 viral packaging motor, which was engineered to be reconstituted into lipid bilayers and has been used for transporting double-stranded nucleic acids [17].
Fig. 1. Some examples of biological and synthetic nanopores.
(a) The toxin α-hemolysin secreted by Staphylococcus aureus [15]. (b) MspA from Mycobacterium smegmatis [16]. (c) Engineered phi29 viral packaging motor [17]. (d) Ion-sculpted pores in silicon nitride membrane [18]. (e) Sub-10 nm thick solid-state pores generated by dry etching a selected area of a silicon nitride membrane and electron-beam pore drilling [20]. (f) Pores in a suspended single-layer graphene membrane [21]. Images were obtained with permission from the publishers.
Note that it is not straightforward to find non-gating biological pores that have larger apertures than α-hemolysin. To accommodate various biomolecules, synthetic nanopores of tunable dimensions are needed. Fig. 1d shows ion-sculpted pores in silicon nitride membranes, which have paved the way to an active decade of research in the solid-state nanopore field [18]. In this scheme, a bowl-shaped cavity is pre-fabricated on the back of a thin silicon nitride membrane, and bombardment using a beam of Ar+ ions is used to further thin the membrane from the top side, until a pore is formed (indicated by the ion flux). Two years later, a method was introduced that allowed pore fabrication inside a transmission electron microscope (TEM) [19]. This technique has many advantages over the ion-beam approach as it allows the pore dimensions to be tuned in real time during fabrication. Recently, this method was utilized to fabricate solid-state pores in sub-10-nm membranes, generated by dry etching a selected area of a silicon nitride membrane and TEM pore fabrication (see Fig. 1e) [20]. Fig. 1f shows a nanopore fabricated using the TEM in a single-layer graphene membrane [21]. Graphene, owing to its two-dimensional lattice of carbon atoms, is one of the most robust materials known to mankind, and thus also holds promise as a substrate for nanopore fabrication.
1.2. On signal and noise: The double-edged sword of ion-based detection
As mentioned earlier, ion transport through nanopores provides the baseline current signal that is detected by connecting the electrodes to an electrometer. When voltage is applied across a membrane that contains a pore, two reversible electrochemical reactions take place at the electrodes, which are typically Ag/AgCl (a non-polarizable electrode with fast kinetics). An oxidative electrochemical reaction Ag(s) + Cl− → AgCl(s) + e− occurs at the anode (+). The results of Cl− capture from solution at the electrode are twofold: an electron migrates through the wire to the electrometer, producing current and generating a charge imbalance at the electrode, which results in cation migration towards the membrane (typically a K+ or Na+ ion). The reverse reaction occurs at the cathode (−), AgCl(s) + e− → Ag(s) + Cl−, where the released chloride ion migrates towards the membrane and an electron is used from the circuit. In the bias window for which this reaction is the sole reaction that occurs (±1 V vs. Ag/AgCl for aq. KCl) [22], the resulting current–voltage response for a nanopore is Ohmic, or linear. At larger bias values, water and other species are more likely to be electrochemically active, resulting in nonlinear electrochemical processes and significant pH instability in weakly buffered solutions (less prevalent in smaller pores due to their high resistance). For this and other reasons (e.g., membrane instability), nanopore experiments are frequently performed under biases lower than 1 V.
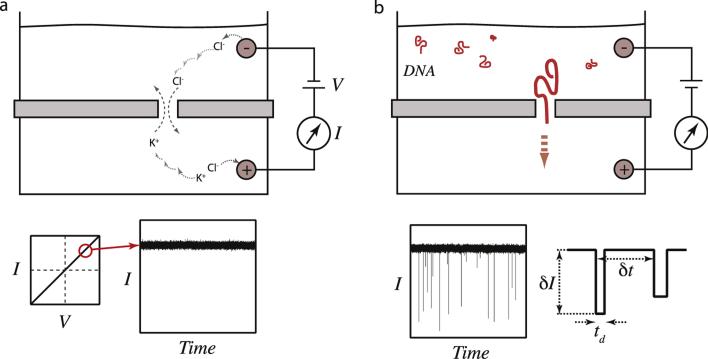
Fig. 2 shows a pore in a generic membrane of arbitrary dimensions, the electrochemical half-cells on either side of the membrane, and the current measuring device. In this scheme, application of bias result in a steady DC current that is constant for an open (unobstructed) pore, providing a baseline current signal. Entry and exit of individual molecules from the nanopore produces a set of resistive pulses, as shown in the figure. Analysis of a large set of spikes involves assignment of at least three important parameters of interest: the total dwell time of the molecule in the pore (td), the mean current amplitude of the current pulse (δI), and the waiting time between two successive events (δt). Respectively, these parameters report on a biopolymer's length and its interactions with the pore, its cross-sectional diameter, and its solution concentration.
Fig. 2. The basics of nanopore measurements.
(a) Application of voltage across a pore triggers electrochemical half-reactions leading to ion migration towards the membrane. Transport of the ions across the pore leads to electric current that is measured using a high-bandwidth electrometer. Typically the current–voltage response of a symmetric pore is linear, and holding the voltage at a constant voltage results in a steady-state DC current signal. (b) When charged biopolymers are added to the chamber, such as DNA molecules, biopolymer molecules diffuse towards the pore and stochastically enter it, producing a measurable “resistive pulse”. First-order parameters that can help in characterizing a sample are the dwell time (td), the average event amplitude (δI), and the time between successive events (δt).
At the membrane, the ions contribute to both the signal (good) and to noise (bad). Nanopore sensing relies on ions going through the nanopore and completing an electrochemical circuit, which produces a measured DC current signal. However, ions also line up at the membrane opposite their counterions on the other side of the membrane, forming what can be modeled as a parallel-plate electrolytic capacitor. This membrane capacitance, while not producing an appreciable DC component, is haunting for ion-channel electrophysiologists and nanopore researchers because it produces noise fluctuations that increase with bandwidths, thereby the time resolution cannot be arbitrarily high in experiments. High-frequency noise contributes to the root-mean squared (RMS) noise about the DC current value, as seen in the ion current traces in Figs. 1a and 1b. The result is a perpetual experimental battle between signal quality and so needed time resolution.
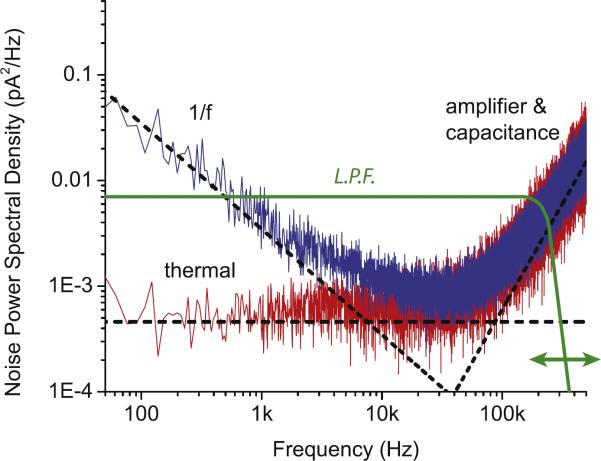
A common way to compensate for this deleterious noise has been to reduce the bandwidth of the measurements by attenuating the high-frequency components of the current signal. This is achieved using analog or digital low-pass filters. Fig. 3 shows current noise power spectra of a 4 nm nanopore when 0 V (red) and 300 mV voltage (blue) are applied. The plot identifies at least two extreme regimes of interest, the 1/f regime below 10 kHz, and the capacitance/amplifier noise regime at > 40 kHz. First we discuss the 1/f regime: under zero-bias the 1/f noise is flat, in which case the noise is thermal (flat response), under applied bias, there is a negative power dependence with noise on frequency. The magnitude of the 1/f slope is related to the ion flux and variability in ion flux through the nanopore. Driving current through contaminated pores typically results in increased 1/f noise, making its slope with frequency more negative. The second frequency regime where noise becomes pronounced is the high-frequency regime (> 40 kHz). In this regime, membrane capacitance governs the power spectrum, and it increases with the bandwidth of the measurement. Typically, the noise at high bandwidth can be reduced by the use of digital or analog low-pass filters, as seen by the green curve in Fig. 3. While this is an effective means of noise reduction, it is unavoidably accompanied by a degradation of the measurement time resolution, which distorts the signal and masks structural information that can reveal DNA sequence and other important molecular features that can be measured in an experiment. The behavior of noise in nanopores has been studied extensively [23–31], highlighting its importance for progress in the field.
Fig. 3. Noise in nanopores.
Current noise power spectra for a 4 nm diameter silicon nitride nanopore under no bias (red) and 300 mV bias (blue), measured in 1 M KCl with a custom high-bandwidth voltage-clamp amplifier. The signal was digitized at 4 MS/s, and the noise spectrum was estimated using Welch's method from three seconds of continuous data. (Courtesy of Jacob Rosenstein, Columbia University.) (For interpretation of the references to color in this figure, the reader is referred to the web version of this article.)
A hefty fraction of nanopore research in recent years has been devoted to improving the signal quality of biomolecule measurements through nanopores. From the noise perspective, this includes a reduction of the membrane capacitance (e.g., by improving the dielectric properties of the membrane support [32]) and optimization of the electrical shielding from external noise. In addition, efforts to optimize the signal from biomolecules include improvement of its shape, optimization of the used electrolyte, and increases in the applied bias, as discussed later in the review.
While a reduction of the membrane's capacitance can reduce the noise, it does not contribute to enhancement of the signal amplitude of a biomolecule, δI (see traces in Fig. 2). When voltage V is applied across a cylindrical pore with a diameter d and length h, the ion current Io through the pore is approximated in high ionic strength solutions (> 100 mM) by:
| (1) |
where n is the number density of the species in the electrolyte, μ is their respective electrophoretic mobility, and e is the elementary charge. The last term in the equation is necessary to explain ion transport through highly charged pore surfaces, where μ is the solution mobility of the counterion adsorbed on the charged pore, whose surface charge density is σ (opposite in sign to the charge of the counterion) [33]. This equation combines equations from prior works [20,33,34] in order to include the contribution of access resistance [35,36], which dominates the conductance in the limit of zero-thickness membranes, i.e., as h approaches 0. Fig. 4 summarizes various results obtained for various synthetic pores (see caption).
Fig. 4. Signal in nanopores.
(a) Current–voltage curves for a 10.2 nm diameter, 34 nm length silicon oxide pore in various KCl concentrations [33]. (b) Conductance (G) values for various 10 nm diameter pores in indicated KCl concentrations [33]. (c) Conductance (G) of various diameter nanopores in 50 nm thick silicon nitride membranes at 1 M KCl and 0.2 M KCl [34]. Solid lines are best fits to the data. (d) Experimental conductivities of different diameter graphene nanopores measured in a 1 M KCl solution [21]. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article.)
Fig. 4 summarizes the dependence of ionic strength and pore diameter on the conductance of synthetic nanopores of different sizes. In panel a, current–voltage curves are shown for a 10.2 nm diameter, 34 nm long silicon oxide pore in various KCl concentrations [33]. As the salt concentration increases, the pore conductance increases. In panel b, conductance (G) values are plotted for various 10 nm diameter pores and different KCl concentrations [33]. The results here clearly show that bulk solution conductance only scales with pore conductance for high ionic strengths (green curve). For salt concentrations smaller than 100 mM KCl, the conductance scales differently than for high ionic strengths, suggesting that surface charge plays an increasing role at low ionic strengths. Moreover, surface charge of a fixed magnitude cannot explain the results (blue curve), and only a model that takes into account the changing surface charge with ionic strength can fully account for the observed conductance behavior (see red curve). Panel c displays the conductance (G) of various diameter nanopores in 50 nm thick silicon nitride membranes at 1 M KCl and 0.2 M KCl [34]. Solid lines are best fits to the data, using an equation that takes into account the pore geometry approximated as an hourglass shape, but that neglects access resistance and surface charge (as apparently valid for high ionic strengths) [20,37]. Panel d shows experimental conductivities of different diameter nanopores in graphene membranes, measured in a 1 M KCl solution [21]. The data best fit the dark solid curve, modeled using a membrane thickness of L = 0.6 nm, highlighting that access resistance dominates ion transport behavior in such thin membranes. Note the difference between panels c and d: in thicker pores, the conductance scales with d2, whereas in very thin pores, the conductance scales linearly with d. Revisiting Eq. (1), note that in the limit of h → 0, the access resistance term dominates the conductance, and conductance is linear with d.
The effects of temperature and solution viscosity on pore conductance have also been studied. Fig. 5 shows the effect of electrolyte temperature and solution viscosity on the conductance through an α-hemolysin nanopore. In Fig. 4a, current is presented for a 1 M KCl solution at a trans-membrane voltage of 120 mV [38]. The line through the points reflects the bulk solution viscosity behavior as a function of temperature of water. The excellent fit suggests that the α-hemolysin channel does not have any structural transitions that critically affect the pore conductance in the range 2–50 °C. In Fig. 5b, the dependence of channel conductance on the bulk solution viscosity at 24 °C for α-hemolysin is presented, where glycerol was used to vary the solution's viscosity [39]. The reasonable fit to the data, particularly for viscosities in the range 1–5 mPa/s, suggests that viscosity impacts the mobility of the ions directly according to the Stokes–Einstein equation. This is an important finding that should not be understated: At high ionic strengths commonly used in nanopore experiments ([Salt] > 0.1 M), ion transport through channels as narrow as α-hemolysin (d ~ 1.8 nm) is governed by the ion's diffusion and migration dynamics, not by pore/ion interactions. This does not mean that surface effects are negligible. The surface is important, but as we have seen in Fig. 4b, its impact becomes pronounced only at ionic strengths where the Debye length is comparable with the pore dimensions.
Fig. 5. Viscosity effects on current through α-hemolysin channels.
(a) Experimental current values for an α-hemolysin channel as a function of temperature in the range 2–50 °C (1 M KCl, 120 mV applied voltage) [38]. (b) Log–log plot of the conductance of an α-hemolysin channel as a function of the solution viscosity, controlled by the addition of glycerol to the electrolyte solution (measured for 1 M KCl solutions at 120 mV voltage, 24 °C) [39]. Images obtained with permissions from the publishers.
1.3. On nanopore resolution: Time, amplitude, and geometry
Nanopore experiments yield at least three different types of merits of resolution that need to be clearly distinguished, despite their relationship to one another. The first is a temporal resolution. The fastest time resolution of a measurement is usually limited by the sampling rate, and limited to the maximum bandwidth of the measurement. For example, if a 10 kHz low-pass filter is used to collect current data for an experiment, the maximum time resolution of the experiment is ~ 50 μs. The second type of resolution is current amplitude resolution. This is determined by the current per digitized bit, and is typically dependent on the gain setting of the amplifier. For a 12-bit digitizer collecting current data at unity gain (1 V/nA) using a patch-clamp amplifier, the current resolution is ~1 pA. Since time resolution depends on the bandwidth, and bandwidth defines the fidelity at which current is recorded, these two resolutions are related (although this is beyond the scope of this review).
The third type of resolution is a geometric resolution. Geometric resolution is easier to describe, because it is defined simply by the sharpness profile of the smallest constriction of the pore. In 2000 Meller conducted an experiment that probed the geometric resolution of an α-hemolysin nanopore [40]. Oligonucleotides of varying lengths in the range 5–50 were driven through an α-hemolysin pore, and the findings, presented in Fig. 6a, show that the most probable residual current fraction, Ip, for different length DNA fragments is fairly constant when the number of nucleotides exceeds 12. For fragments smaller than 12 nucleotides the residual current is larger, suggesting that the pore constriction is not fully occupied during the fragment's transport. Based on these measurements it is reasonable to estimate that the β-barrel region of the channel spans the length of 12 nucleotides, matching well the crystal structure for α-hemolysin. As shown in Fig. 1, MspA channels exhibit a sharper pore structure, which can greatly assist in a direct readout of DNA sequence (discussed later). Decreasing the thickness of membranes in synthetic pores can also aid in improving the signal and resolution of DNA analysis. Fig. 6b illustrates a 4 nm diameter nanopore in silicon nitride membrane where the parameter varied is h, the membrane thickness [20]. The ion current traces for sets of 3 kbp DNA fragment translocations reveal that as the membrane thickness is decreased, the ion current of the pore increases and the amplitude of the resistive pulse increases, as consistent with Eq. (1).
Fig. 6. Geometric resolution of a pore.
(a) Resistive pulses that show ssDNA transport through an α-hemolysin channel [40]. The mean residual current for different ssDNA fragments (Ip) is constant for ssDNA with a contour length corresponding to N > 12 bases, consistent with the length of the channel's stem (see white ladder representing the 12 nucleotides). (b) Transport of dsDNA through nanopores fabricated in precision-thinned silicon nitride membranes [20]. Figure shows concatenated events during the transport of 3 kbp dsDNA for similar diameter pores fabricated in progressively thinner membranes, which resulted in increased baseline pore currents and increased pulse amplitudes (V = 300 mV). Images obtained with permissions from the publishers.
1.4. On capture of biomolecules into nanopores
The process of capturing a biomolecule into a nanopore has received a lot of attention because it is both intriguing and practical to understand how very long polymers find their way into a very narrow pore. Fig. 7a shows the mean capture rate as a function of voltage for dC40 DNA into an α-hemolysin pore [41]. In the low concentration limit, DNA molecules in the solution do not appreciably interact with one another, and thus, the process of capture is a classic arrival time Poisson process with no memory effect, for which the normalized probability of arrival time-delay has the form:
| (2) |
where δt is the measured time-delay between successive events and RC is the mean capture rate. The inset in Fig. 7a shows this general behavior for the capture of different ssDNA molecules into an α-hemolysin pore [41]. The distributions of δt fit Eq. (2) well for over two orders of magnitude. This process generally holds, except in situations where the mean wait time 1/RC is comparable to the transport time, the analyte molecules are interacting with one another, or the analyte solution is not homogeneously mixed.
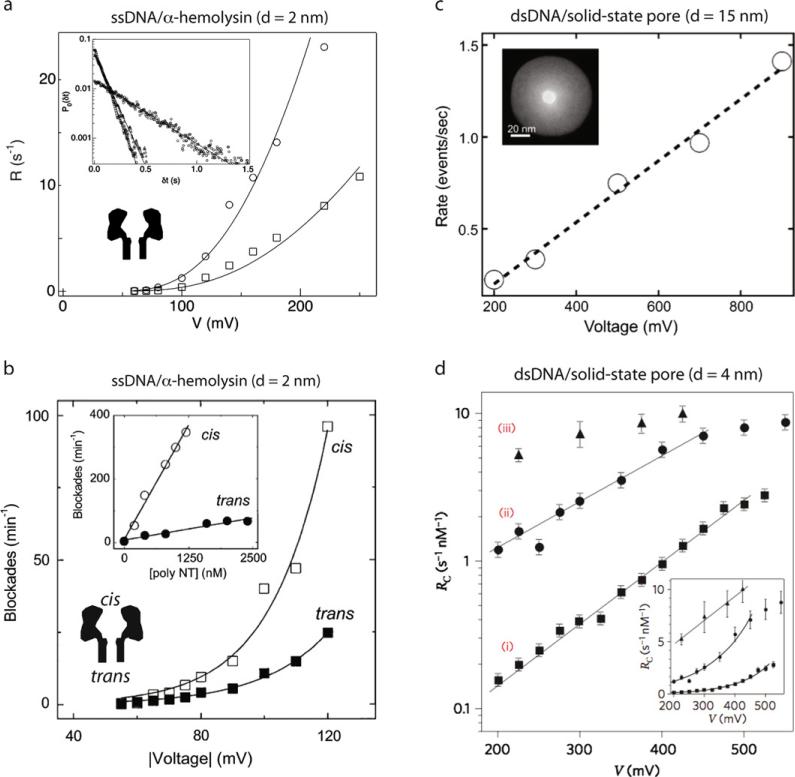
Fig. 7. DNA capture into a pore.
(a) Capture rate for 2.6 μM (circles) and 0.9 μM (squares) dC40 DNA into α-hemolysin as a function of voltage, measured at 2 °C. Inset shows distributions of the wait time (δt) between successive events for (dA)20 at 2.3 μM (squares); (dCdCdTdCdC)6 at 1.8 μM (triangles); (dC)40 at 0.5 μM (circles) [41]. (b) Capture rate of an end-biotinylated poly(dC)30 as a function of voltage when the nucleotide is introduced to the cis or trans chamber of an α-hemolysin channel [44]. (c) Capture rate as a function of voltage for λ-DNA into a 15 nm diameter solid-state nanopore as a function of voltage [45]. Inset shows a TEM image of the nanopore used in this study. (d) Semi-log plot of the capture rate of (i) 400 bp dsDNA, (ii) 3500 bp DNA, (iii) 48.5 kbp λ-DNA into a 4 nm diameter silicon nitride pore (inset shows linear plot) [42]. Images obtained with permissions from the publishers.
Another revealing parameter to probe is the dependence of capture rate on the applied voltage. In Fig. 7a, the exponential dependence of ssDNA capture on voltage reveals that capture is not a diffusion-limited process. That is, not every molecule that attempts to enter the pore is captured. To explain, consider the voltage profile outside the pore mouth, which decays with radial distance from the pore r as follows [42,43]:
| (3) |
where d is the pore diameter, h is the pore height, and ΔV is the applied voltage. When a negatively charged biopolymer such as DNA diffuses towards the pore, at some distance r, its diffusion becomes biased and it migrates towards the pore. Since this biased diffusion is a function of the electric force and diffusion, the distance r is dependent on the DNA length and is dictated by its overall charge and diffusion coefficient. Therefore, the radius of capture should depend linearly with applied voltage, if capture is a diffusion-limited process. However, as Fig. 7a shows, capture of ssDNA into α-hemolysin depends exponentially on voltage, which suggests that capture is not perfect at the pore, i.e., the pore rejects a fraction of the molecules that approach its mouth. One interpretation is related to an entropic barrier for capture: an ssDNA polymer can assume many configurations in solution, and a polymer threaded into a pore is geometrically constrained, reducing the number of degrees of freedom. This entropic barrier for stretching the DNA into the pore is reduced by increasing the applied voltage, and a reduction of the voltage barrier exponentially increases the capture rate. Beyond some critical voltage where the entropic barrier is negligible the process is expected to be diffusion-limited, and we can expect a linear dependence on capture rate with voltage.
Pore geometry also affects the capture rate of biopolymers. In Fig. 7b, the frequency of current blockade spikes upon introduction of poly(dC)30 DNA into an α-hemolysin pore is plotted as a function of applied voltage, when the DNA was introduced into the cis or trans chamber [44]. The data reveals two important features: First, for both cis and trans experiments, DNA capture into the pore is exponentially dependent on voltage, which suggests a voltage-reduced barrier for capture (as shown in Fig. 7a). Second, the rate of capture is vastly different for cis and trans experiments, with cis capture being pronouncedly more favorable than trans capture at any given voltage. These results suggest that the wider opening of the α-hemolysin vestibule improves capture, which the authors ascribe to a reduced entropic barrier for DNA entry. Since ssDNA can enter the vestibular cis side in many orientations while only capable of entering from the trans side in an extended conformation, DNA entry into the cis chamber is energetically favorable.
Chen and co-workers studied the capture of λ-DNA (48.5 kbp) as a function of voltage applied to a 15 nm diameter solid-state pore (see Fig. 7c) [45]. A linear dependence was observed in the voltage range 200–900 mV, suggesting that capture is diffusion-limited. The fact that no barrier was observed for capture can be rationalized by the large diameter of the pore and the stiffness of the DNA molecules, which can flexibly allow entry of various configurations of the DNA [46–48]. In contrast, a barrier was observed for capture of dsDNA into 4 nm diameter solid-state pores, as shown in Fig. 7d [42]. The semi-log plot of capture rate vs. voltage for 400 bp DNA, 3000 bp DNA, and 48,500 bp λ-DNA contains two noteworthy features: for short DNA fragments (400 bp and 3000 bp) capture is exponentially dependent on voltage, implying that capture is not a diffusion-limited process. Second, for very long DNA capture becomes a diffusion-limited process. A more detailed study shows a transition from a barrier-limited to a diffusion-limited process occurring at ~ 10 kbp DNA (for 300 mV voltage, 4 nm silicon nitride pore, 1 M KCl) [42]. This is counterintuitive because one would expect the increased coil entropy and flexibility to present barriers for threading into the pore. Despite this, the increasing coil size presents advantages for capture, the details of which are presented elsewhere [42,43].
While nanopores are capable of detecting single molecules, it is the rate of capture of an analyte that determines the minimum practical quantity of nucleic acids that can be analyzed. Therefore, there is wide interest in controlling and enhancing the rate at which nucleic acids are captured. Several approaches have been used for increasing the capture rate: (1) increasing the applied voltage draws molecules further away from the pore [41,45], (2) modifying the surface charge of the pore induces electro-osmotic flow that increases the capture rate [49], and (3) introducing a trans-membrane salt gradient where the salt concentration is low in the nucleic acid chamber and high in the collection chamber allows picomolar levels of DNA to be detected at a practical rate [42].
1.5. On transport dynamics of biomolecules through nanopores
One of the most important processes to understand in biopolymer analysis using nanopores is the microscopic detail of the transport process. Since nanopores offer the hope of revealing sub-molecular features in a biopolymer, it is hoped that the residual current signal recorded during a polymer's transport reports on some key structural elements of a polymer. The required information boils down to knowledge of where a polymer is along the translocation trajectory, and what is its velocity. Towards this, several groups have studied in detail the process of nucleic acid translocation.
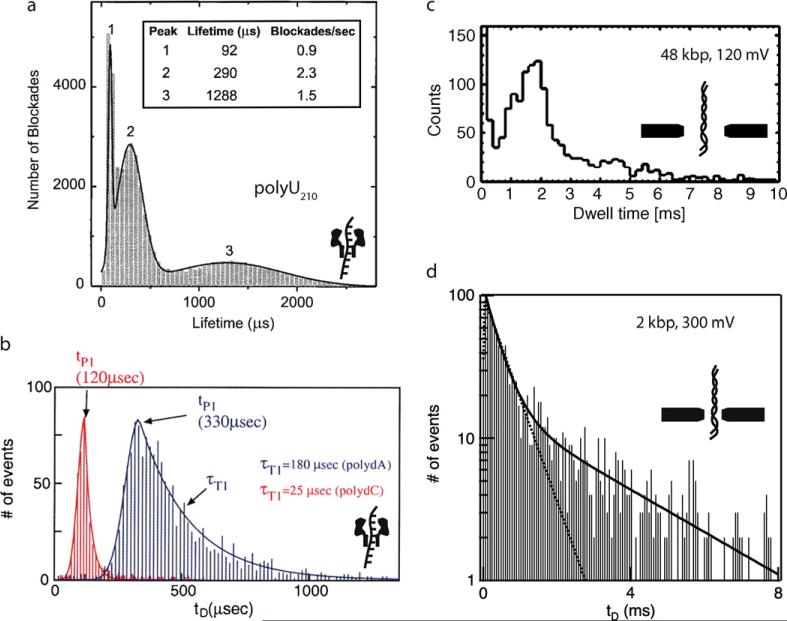
Revealing information about the translocation process is found by compiling a distribution of pulse durations for a given sample. Fig. 8 shows a selection of pulse duration distributions for selected pore/nucleic acid combinations. From the onset of research in this area, the work of Kasianowicz and co-workers [50] elucidated several populations for a uniform sample of nucleic acid allowed to interact with a pore. It is logical to assume that each population is a distinct process with a characteristic timescale, and that the shape of each distribution reveals the mechanism of the process. Kasianowicz and co-workers studied polyU210 RNA interactions with an α-hemolysin pore (see Fig. 8a) [50], attributing the faster event population (1) to collisions, and the slower two populations (2 + 3) to translocations of the RNA through the pore with different entry orientations (as later showed by Mathe and+co-workers, see next section). Meller and co-workers later showed that for different DNA homopolymers the shape of the distributions was not perfectly Gaussian, exhibiting a fast rise for short times and a tail of varying slopes that presumably depend on interactions of the molecules within the pore (see Fig. 8b) [51]. For a distribution of this type, the mean time is not the peak time, but a longer time than the peak time. Thus, the homopurine poly(dA) interacts more with the pore than the polypyrimidine poly(dC), and this results in different peak pulse durations (tP) and tail shapes. The behavior is somewhat similar for the transport of long dsDNA through 10 nm diameter SiO2 pores, as revealed by Storm and co-workers (see Fig. 8c) [47]. More complex pulse duration distributions were obtained for dsDNA interactions with 4 nm diameter silicon nitride pores (twice the cross-sectional diameter of dsDNA) [52]. In the limit of small pores a broad-tailed distribution is observed that best fits a two-exponential decay (see Fig. 8d). Moreover, the relative fraction of events in each population is length-dependent, and the longer the DNA molecule, the more events in the long timescale are found.
Fig. 8. Pulse duration distributions for various nucleic acid/pore combinations.
(a) Distribution for polyU210 ssRNA through an α-hemolysin pore shows 3 distinct populations, which were attributed to nucleic acid/pore collisions (1) and translocations with either of two entry orientations (2 + 3) [50]. (b) Distributions for poly(dA) and poly(dC) ssDNA homopolymers through α-hemolysin, which exhibit different mean timescales (tP) and tail shapes due to different interactions with the pore [51]. (c) Distributions for linear λ-DNA dsDNA through a 10 nm SiO2 pore reveal a very fast population in the first bin at < 0.1 ms and a tailing distribution that peaks at 2 ms [47]. (d) Distributions (semi-log) for 2 kbp linear dsDNA through a 4 nm silicon nitride pore, showing distinct fast and slow populations that were attributed to presence of interactions with the pore and the membrane [52]. Images obtained with permissions from the publishers.
A quick comparison of the data in Figs. 8c and 8d shows that shrinking the pore size results in an effective means to increase DNA interactions with the pore, with mean transit times of ~ 1 ms for 2000 bp DNA and ~ 2 ms for 48,000 bp DNA in 4 nm and 10 nm pores, respectively. Another explanation for the reduced average speed is an increased electro-osmotic ion current along the DNA backbone (in the direction opposite to DNA motion), which generates hydrodynamic drag [53].
The dependence of mean pulse durations on the DNA length is key for understanding the transport process. In Fig. 9a, average velocities of different lengths of the ssDNA polymer poly(dA)N are plotted, calculated from the ratio of the ssDNA contour length to the mean dwell time [40]. A constant average velocity was found for DNA translocation for polymers above 12 nucleotides, as indicated by the observed independence of the mean velocities on ssDNA length. Both the linear dependence of transport times on DNA length and asymmetric pulse duration distributions were predicted by Lubensky and Nelson, who noted that the first-passage time distribution for polymer translocation is given by a non-Gaussian form for a strongly interacting pore [54]. The asymmetric distribution shape was modeled as the ssDNA polymer encountering a series of potential barriers during its passage. The result was that the height of the barriers altered the distribution shape, but not the dependence of the mean velocity on DNA length (i.e., td ~ L).
Fig. 9. Various scaling exponents with length for DNA translocation through nanopores.
(a) Mean velocity of poly(dA)N translocation through α-hemolysin is constant for N > 12 (recorded at 2 °C and 120 mV) [51]. Inset shows a typical, asymmetric dwell-time distribution. (b) Mean dwell time vs. DNA length for dsDNA through a 10 nm SiO2 pore (recorded at 21 °C and 120 mV) [47,55]. Inset shows log-normal distributions for different DNA lengths. (c) Mean dwell time vs. DNA length for dsDNA through a 4 nm diameter pore (recorded at 21 °C and 300 mV) [52]. Cartoons illustrate the possible interactions that lead to the transition from t1- to t2-dominated dynamics. Images obtained with permissions from the publishers.
In contrast to ssDNA through α-hemolysin, the transport of dsDNA through large solid-state nanopores obeys different scaling laws. When studying the length dependence, Storm et al. found that a nonlinear dependence on DNA length, where td ~ L1.27 (see Fig. 9b) [47,55]. Rather than attributing the behavior to interactions within the pore, the nonlinear scaling was attributed to hydrodynamics of the shrinking Gaussian DNA coil outside the pore during translocation, which yielded , where Rg is the radius of gyration that scales with N0.6 (0.6 is the Flory exponent). It is possible that this explanation overlooks the notion that in the fast translocation regime, the DNA coil does not equilibrate during its translocation, and therefore the “coil” approximation is not valid (i.e., the translocation time is faster than the Zimm relaxation time). Despite this obscurity, the reasoning that hydrodynamics play a role is still plausible because of the large pore size, stiffer DNA, and faster transport times.
In contrast to large pores, in small pores dsDNA obeys a more complex length scaling (see Fig. 9c) [52]. For a DNA molecule shorter than 3500 bp, the prominent timescale (t1) follows a power-law dependence on N, where the power is equal to 1.40. For longer polymer lengths where t2 dominates, a much stronger power-law dependence emerges: t2 ~ N2.3. The striking shift from t1-dominated to t2-dominated transport dynamics can be attributed to additional mechanisms of external DNA interactions with the silicon nitride membrane, interactions of the DNA coil with itself, or a coil-dependent electric field profile that is affected by co-ion exclusion by the DNA coil. In support of these results, Luan and co-workers observed using molecular dynamics that redistribution of ions around the negatively charged DNA surface results in an unbalanced total electrolyte charge that generates electro-osmotic flow of water near the DNA [56].
A recent review by Venkatesan and Bashir [57] summarizes the mean translocation velocities accumulated over the past 15 years for synthetic and protein pores. This compilation provides compelling evidence that the process of DNA translocation is poorly understood to date. The common presumption that DNA traverses a nanopore with a constant or otherwise known speed is incorrect, and should be avoided. An important experiment that can help to elucidate DNA transport dynamics is to label DNA with multiple labels at known intervals. Until this experiment is performed, the spatial trajectory of the DNA as a function of time during a translocation event remains obscure.
Various ways to slow DNA transport velocities have been demonstrated, from reducing the voltage [50], the solution temperature [51], increasing the solution viscosity [58], and changing the bath salt concentrations [42]. For example, reducing the bath temperature from 21 °C to 0 °C decreases mean dsDNA velocities through 4 nm pores by a greater factor than expected from the increased solution viscosity [52], which can be explained by considering interactions with the pore walls as thermally-activated energy barriers to translocation.
2. Biophysical studies
Now that we've explored the basics of ion transport through various pores and its dependence on some key parameters, we are in position to highlight some experiments that studied the behavior of biomolecules in nanopores.
2.1. Entry orientation of single-stranded nucleic acids into α-hemolysin
In 1996, Kasianowicz et al. demonstrated the transport of RNA through α-hemolysin channels [50]. A single channel was reconstituted in a lipid bilayer, providing a baseline current of 120 pA at 120 mV applied voltage. Then, single-stranded poly(U) RNA homopolymers were added to the negative chamber, which resulted in resistive spikes (see Fig. 10a). Since this is the first experiment, one can naively expect to observe a single current amplitude level that corresponds to the fraction of ions blocked by the RNA molecule in the pore. However, three distinct current levels were clearly observed, as shown in Fig. 10a. Based on intuition, Kasianowicz and co-workers attributed the observed signal to collisions (population “1” in distribution of Fig. 10a) and translocations of the RNA with different entry orientation (populations “2” and “3”), since a nucleic acid strand can enter the pore either with its 3′ or 5′ terminus.
Fig. 10. Nucleic acid entry orientation matters.
(a) In pioneering experiments, Kasianowicz et al. passed poly(U) RNA homopolymers through an α-hemolysin channel. Based on the three distinct signal amplitude distributions, they hypothesized that events 1–3 corresponded to RNA collisions, passage of RNA in one orientation, and passage of RNA in another orientation, respectively [50]. (b) Mathe et al. later proved that the distributions of a DNA homopolymer, poly(dA), produce different signals by measuring the residual current in the presence of immobilized hairpins [59]. (c) Molecular dynamics simulations show that the mechanism for the orientation selectivity involves preferential base orientation up or down at the smallest pore constriction, depending on the orientation entry see dashed circles [59]. Images obtained with permissions from the publishers.
In 2005, Mathe and co-workers tested the hypothesis by Kasianowicz by cleverly designing DNA hairpins that contain single-stranded poly(dA) overhangs with both 3′ and 5′ orientations [59]. Hairpins were designed in order to immobilize the DNA within the pore with a specific orientation, as well as to keep the DNA immobilized in the pore long enough that the current level can be recorded. Comparison of the current amplitude distributions for a single-stranded DNA and the two immobilized hairpins is shown in Fig. 10b. It is clearly seen that the hairpins produce distinct levels that are based on orientation, while the single-stranded DNA produces a mixture of the two levels, based on the orientations of entry. In Fig. 10c, molecular dynamics simulations of DNA entry with both orientations show that the bases prefer to align themselves in different directions, depending on the orientation of entry. Another finding that the MD simulations reproduced well (not shown here) is the magnitude of the current levels for both DNA entry orientations. These results represent an important finding for the nanopore field, illustrating that the current signal and the probability of capture of a specific nucleic acid orientation is highly dependent on microscopic interactions of the molecules with the pore.
2.2. Discrimination among nucleic acid polymers
Along with pioneering results, the 1996 work by Kasianowicz [50] contained a meaningful observation: if the each base along a DNA/RNA polymer can produce a distinct current signal as it traversed a pore, the time-domain current signal recorded during DNA/RNA transport could in principle contain DNA sequence information. Again, a naive expectation is that the purines (adenine and guanine) would provide a deeper signal than the pyrimidines (cytosine and thymine for DNA or uracil for RNA). A priori, a logical experiment to test this hypothesis would be to pass different homopolymers through a pore and record the signal.
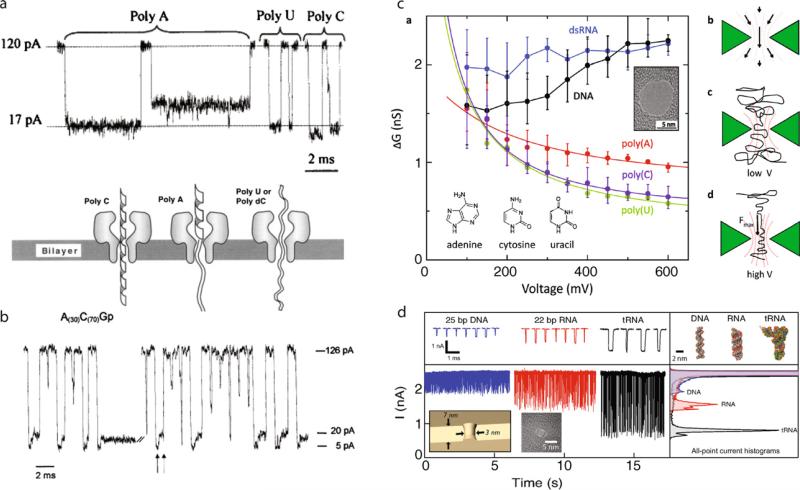
In 1999, Akeson and co-workers studied in detail the signal from different RNA and DNA polymers in α-hemolysin nanopores. Example current traces for different RNA polymers are shown in Fig. 11a. In a counterintuitive result, purine poly(A) molecules blocked less pore current than the poly-pyrimidines poly(C) and poly(U). Akeson and co-workers concluded from this study that the secondary structure of a homopolymer is responsible for the observed signals, not its primary structure (i.e., its base content). Since poly(A) forms a bulkier secondary structure, the structure has to be unstable within the pore, and therefore it must disassemble. In contrast, poly(C) and poly(U) have narrower secondary structure, and therefore, they can block as much, or more ions than poly(A). Later experiments by Meller and co-workers showed differences among DNA homopolymers and heteropolymers of dA and dC, in which both the timescale and pulse height were clearly distinguishable [51]. This was later studied using immobilized hairpins, in which Purnell and co-workers showed an improved current resolution of homopolymer orientation of entry for different bases [60].
Fig. 11. Discrimination among RNA homopolymers.
(a) Typical resistive pulses during the passage of different RNA homopolymers through an α-hemolysin channel [61]. The scheme below attributes differences in the depth of the pulses to secondary structure, as opposed to bulkiness of the bases. (b) Resistive pulses during translocation of the block co-polymer A30C70 show two distinct levels, corresponding to the sequential occupancy of each type of base in the pore [61]. (c) Various types of single-stranded and double-stranded nucleic acids discriminated among by the change in pore conductance (ΔG) they induce upon entry into a ~ 10 nm diameter silicon nitride nanopore [62]. (d) Resistive pulses of short DNA and RNA fragments using 3 nm diameter pores in ultrathin silicon nitride membranes, showing the sensitivity of small pores to bent structures such as transfer-RNA (V = 500 mV, T = 0 °C) [20]. Images obtained with permissions from the publishers.
Perhaps the first compelling evidence that motivates the use of nanopores for DNA sequencing is shown in Fig. 11b [61]. Akeson and co-workers took a block co-polymer of RNA that contains 70 C-bases and 30 A-bases, and found that the resistive pulses produced contained two levels that are in sequence. While the authors attribute the different signal levels to the variation in the polymer's secondary structure, the presence of discrete levels that can be temporally resolved suggests that given the right pore geometry and translocation dynamics, a DNA sequence could be directly read out from the current through a pore, as will be discussed later.
Later experiments using solid-state nanopores demonstrated the discrimination among different nucleic acid polymers. In Fig. 11c, mean conductance changes (ΔG) are plotted as a function of voltage for entry of different types of nucleic acids into 10 nm diameter silicon nitride pores [62]. It is apparent that the resolving ability of the nanopore is superior at intermediate voltages (~ 300 mV), in which the difference in ΔG is the largest for all polymers. Differential behavior of ΔG for the different polymers was attributed to different hydrodynamic profiles that result upon changing the voltage. In Fig. 11d, continuous current traces are shown for short fragments of double-stranded DNA, RNA, and for transfer-RNA, through a 3 nm pore fabricated in a 7 nm thick silicon nitride membrane (500 mV and 0 °C) [20]. Based on their different secondary structures, namely, width of the molecule and bent structure vs. linear structure, the resistive spikes were significantly different for all three polymers.
2.3. Force-induced duplex unzipping
In addition to the rich set of pioneering results by Kasianowicz in 1996 [50], the perceptive set of implications and hypotheses reported in that work have produced even more interest that fueled the nanopore field since. First, translocation of polymers was hypothesized to enable direct readout of a DNA sequence, which in essence makes a nanopore a nucleic acid scanner. Second, the duration of an event and the rate of events are related to the polymer's overall length and its concentration, making it an analytical tool. Third, varying the voltage applied to the nanopores enables the force applied to a nucleic acid to be tuned, as seen by the reduced polymer transit times across the pore. This last property is extremely intriguing from biophysics and biotechnology perspectives, and has received considerable attention since. Since a nucleic acid is uniformly charged along its backbone, interactions of the electric field inside the pore with these charges results in migration of the polymer with a force that is related to the voltage, and therefore, voltage is a force knob.
This has been used to unzip DNA duplexes, which reveals information that is related to the duplex free energy. Fig. 12 highlights various types of DNA unzipping experiments. Sauer-Budge et al. compared the unzipping process of perfectly complementary vs. mismatched DNA by hybridizing two different 50-mers to a 100-mer strand that contained a Watson–Crick complementary region, as shown in Fig. 12a (the other 50-nucleotide sequence was used to pull on the DNA in the pore) [63]. In these experiments, voltage of ≥ 120 mV was applied and the duration of the events was recorded, followed by PCR experiments on the collection chamber to demonstrate which DNA oligo was transported across the pore. The results show that the DNA hybridized to the perfect complement takes significantly longer to traverse the pore (mean = 435 ms) than the same DNA hybridized to a mismatched sequence (mean = 185 ms). Gel electrophoresis, as shown in the figure, demonstrates that the 100-mer DNA oligo indeed was transported through the nanopore. These two observations, namely, faster transport of the DNA hybridized to a mismatched strand and presence of the 100-mer DNA oligo on the other side of the membrane following the application of trans-membrane voltage, strongly support the mechanism shown in the figure in which the single-stranded overhang enters the pore first for unzipping to occur. The first rate of the process can be defined as an equilibrium unzipping/rezipping which yields locally-melted DNA, a process which has corresponding rates k1 and k−1, while the second process, namely, complete unzipping, is irreversible because the two strands become separated and physically inaccessible to each other.
Fig. 12. Nucleic acid unzipping.
(a) Unzipping time distributions for two different 50-bp duplex DNA molecules through an α-hemolysin pore, one of which contains a few mismatches (highlighted in sequence below the distributions). Gel images of PCR amplification products of the different strands following an unzipping experiment. Bottom shows the proposed two-step model of reversible unzipping/rezipping, followed by an irreversible unzipping process [63]. (b) Unzipping 10-bp DNA at different temperatures and voltage ramp rates reveals a transition between the reversible and the irreversible process (dashed line), both of which exhibit different kinetics [64]. (c) Unzipping duplex DNA molecules using a 2 nm diameter silicon nitride nanopore [68]. Scheme of the setup and TEM image of the pore are shown on top, while bottom shows pulse duration distributions for an ssDNA (dA120), a blunt 24-bp dsDNA hairpin (24B), and a 50-bp dsDNA with a 50-mer ssDNA overhang (HP50). The substantially longer time scales for HP50 suggests that unzipping requires an ssDNA as an electrophoretic “rope” for unzipping the DNA. (d) Multi-nanopore force spectroscopy [69]. Step a: Probe DNA molecules are threaded and anchored at the pore using a streptavidin molecule. Step b: Complementary probes are hybridized to the probe DNA at the trans chamber. Step c: Unhybridized probes are allowed to escape. Step d: Unzipping voltage is applied and current is recorded, providing the complete unzipping probability plot in one experiment. Step e: Reset step. Images obtained with permissions from the publishers. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article.)
Mathe and co-workers investigated the reversibility of this unzipping process in detail, as shown in Fig. 12b [64]. Dynamic voltage control programs were developed that can apply a variable force ramp to the nanopore, allowing true force spectroscopy to be conducted on individual untethered DNA molecules at the pore [65,66]. Using a linear voltage ramp, Mathe and co-workers extracted mean critical voltages at which unzipping occurred as a function of the ramp rate and the temperature. The results showed a clear transition that occurs from a reversible zipping/unzipping transition at low ramp rates to an irreversible unzipping process at high voltages (red dashed line marks the transition). Theoretical details regarding this and other unzipping experiments are discussed elsewhere [67].
Unzipping was also found to occur in solid-state nanopores, an important discovery that is relevant for next-generation DNA sequencing (discussed later) [68]. As shown in Fig. 12c, the resistive pulse distributions for an ssDNA (dA120), a blunt 24-bp dsDNA hairpin (24B), and a 50-bp dsDNA with a 50-mer ssDNA overhang (HP50), clearly show that blunt duplexes cannot penetrate the 2 nm pore, and a single-stranded overhang is required. Moreover, the longer timescale exhibited by HP50 as compared with dA120 (a similar overall length) suggests that the rate limiting step is unzipping of the duplex region in HP50.
In order to carry out unzipping experiments in single-channel measurements, one has to wait for each molecule to arrive and unzip and then trap another. However, this process can be performed in parallel by recording simultaneous data from many nanopores, given that the process of unzipping can be synchronized. Tropini and co-workers utilized the strength of the biotin–streptavidin conjugate in order to investigate a synchronous, multi-nanopore unzipping process (see Fig. 12d) [69]. In this system, biotin-terminated single-stranded DNA is bound to streptavidin in the cis chamber of an array of α-hemolysin pores, and voltage is applied to thread DNA molecules through, where they are prevented from complete translocation by the streptavidin endings. Hybridization to complementary DNA probes on the trans side, followed by a reversal of the voltage, affords a current signal that increases in magnitude with time until reaching a steady-state maximum, in which all of the hybridized DNA molecules have been unzipped. The obtained current trajectory (see inset) is the time probability that the DNA stays zipped in the pore, given by (after normalization):
| (4) |
where PU(t) is the normalized unzipping time distribution, for example, like the one shown in Fig. 12a. Therefore, while PU(t) is traditionally obtained by measuring many individual events in series using a single pore, PS can be elegantly obtained in a single, parallel experiment that employs many pores.
2.4. Probing intermolecular reactions and dynamics
Two key advantages to resistive sensing using nanopores are that the analyte molecules are sampled rapidly in solution, and no chemical labeling/tagging is required for attachment to optical probes or surfaces. This can be exploited for dynamically probing biochemical reactions in a solution [70]. Fig. 13a illustrates the use of a conical nanopore fabricated in a track-etched polymer film for probing complexation of bovine serum albumin (BSA) with anti-BSA-Fab antibody [71]. Upon the addition of 270 nM anti-BSA-Fab to the solution containing 100 nM BSA, the distribution of dwell times and current amplitudes changed dramatically. An interesting observation is that the amplitude of the events is shallower for the complex than it is for free BSA, which can be attributed to charge effects near the pore or to incomplete entry of the complex into the 17 nm pore aperture. The interactions of viruses with anti-virus antibodies have also been investigated using sub-micrometer pores, as shown in Fig. 13b [72]. Paramecium bursaria Chlorella virus (PBCV-1) is a ~ 190 nm icosahedrical virus, and upon addition of anti-PBCV-1 to the solution events of deeper amplitudes were observed, signaling antibody binding to the capsid. Other reactions monitored using nanopores are amyloid peptide aggregation into fibrils, as shown in Fig. 13c [73]. Amyloid-beta (Aβ) peptides aggregate to form fibrils in solution, and the authors employed nanopores to detect the aggregates formed after prolonged incubation of the peptide in solution. A silicon nitride pore was coated with a lipid bilayer prior to detection, which allowed prolonged measurements of the fibril solution over time. In contrast, uncoated pores performed poorly, as indicated by their clogging over the first 10 minutes. Apart from their anti-fouling properties, the lipid-coated solid-state nanopores also allow measurements of protein diffusion across a lipid bilayer, serving as a new motif in surface-diffusion-based stochastic sensing [73]. Finally, the interaction of DNA with small molecule ligands was investigated (Fig. 13d) [74]. SYBR Green II is a cyanine dye used to stain single-stranded RNA and DNA. However, it does not bind to poly(dA) homopolymers because of their formation of a secondary structure. Addition of SGII to an ssDNA polymer that contains a poly(dA) segment and a random sequence segment results in binding only to the random sequence, as seen by the increased current amplitudes in the second portion of the translocation events. The shallower portion of the blockade corresponds to the level of the poly(dA) polymer, whereas the deeper portion corresponds to the dN50 random sequence that contains the intercalating SGII. Notably, in the vast majority of the events, the shallow event preceded the deeper event, which suggests that intercalation with the positively charged SGII helps to orient the DNA entry with the more negative side first.
Fig. 13. Probing intermolecular reactions in solution using nanopores.
(a) Scatter plots for 100 nM BSA only (black) and 100 nM BSA plus 270 nM anti-BSA-Fab (red) for a 17 nm conical pore at 1 V applied bias [71]. (b) Probing reaction between the PBCV-1 virus and an antibody specific to it using a sub-micrometer pore in glass [72]. (c) Use of lipid-coated solid-state nanopores for sensing in real time the evolution of amyloid-beta (Aβ) peptides. The lipids serve an important function in preventing clogging of the pores during transport of the peptides through the pore [73]. (d) Detection of small-molecule binding to DNA. SYBR Green II (SGII) is a dye that binds to random ssDNA sequences but not to poly(dA) [74]. Images obtained with permissions from the publishers. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article.)
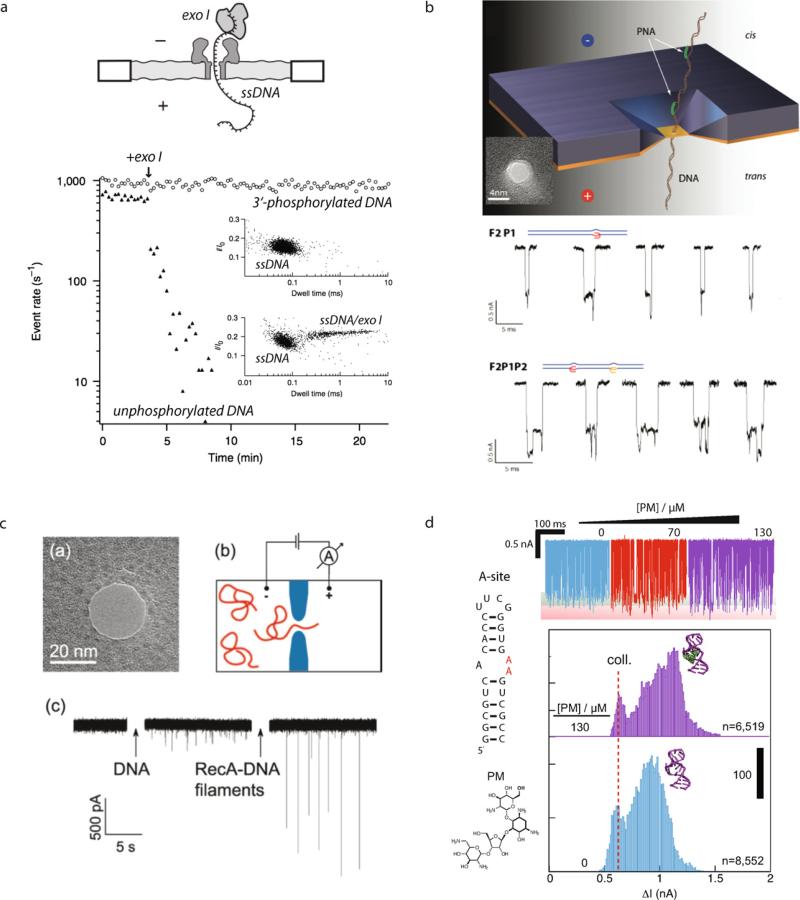
Other types of reactions monitored using nanopores include enzyme binding to DNA, DNA/PNA (peptide nucleic acid) complexes, DNA/protein complexes, and RNA/ligand interactions. The DNA-binding enzyme exo I binds to the 3′ terminus of ssDNA in the presence of Mg2+ ions, although it does not bind the 3′ phosphorylated state. As Fig. 14a shows, the addition of exo I to a chamber containing unphosphorylated or phosphorylated DNA results in vastly different behavior – the phosphorylated DNA remains intact, giving rise to the same frequency of events, whereas the unphosphorylated DNA binds exo I, resulting in a regular decrease in the event frequency [75]. This is a good method for establishing the activity of DNA-binding proteins in real time in solution without any modification of the DNA and/or protein. DNA interactions with sequence-specific peptide nucleic acids can also be examined using a 4.5 nm diameter solid-state pore [76]. Peptide nucleic acids bind to ssDNA or dsDNA with higher affinity than DNA oligonucleotides because the peptide backbone is neutral, leading to a reduced electrostatic repulsion. Addition of PNA to the duplex DNA sequences effectively “tags” the DNA in the region that contains the sequence of interest, and therefore the DNA sequence can be barcoded with one or several PNAs. The signatures of the DNA clearly show that the number of PNA molecules can be deduced, although the exact location cannot be determined due to insufficient resolution. Filamentation of DNA with RecA has also been studied using 20 nm diameter nanopores [77–79]. RecA can polymerize, forming long filaments on both dsDNA and ssDNA molecules. Therefore, the addition of RecA greatly enhances the signal from the DNA molecule, allowing its detection using larger pores. Finally, the 1:1 binding of a drug molecule to a clinical RNA target has recently been studied using 3 nm diameter ultrathin solid-state nanopores, as shown in Fig. 14d [80]. The A-site of the prokaryotic ribosome is a canonical drug target, because when a drug binds to it protein synthesis is prevented. By driving a truncated 27-mer RNA hairpin that contains the A-site in its middle, through the nanopore and titrating drug molecules, binding of aminoglycoside drugs was observed, and further, the binding fraction correlates with the amount of drug in the solution, affording binding isotherms.
Fig. 14. Probing nucleic acid complexes using nanopores.
(a) Probing DNA/enzyme complexes using an α-hemolysin pore [75]. The enzyme exo I binds to the 3′ terminus of ssDNA, although it does not bind a 3′-phosphorylated site, as seen by the event frequency as a function of time. (b) Probing DNA complexation with sequence-specific peptide nucleic acids (PNA) using a 4.5 nm solid-state pore [76]. Binding of the PNA to the DNA molecule results in an additional negative spike upon the complex's passage through the pore. (c) Probing DNA/RecA interactions using a 20 nm solid-state pore [79]. Addition of RecA dramatically enhances the signal from the DNA molecule, allowing its detection using larger pores. (d) Detecting RNA/drug complexation and affinity using a 3 nm ultrathin pore [80]. An excised RNA from the A-site of the prokaryotic ribosome produces a signal that changes upon binding aminoglycoside drugs such as paromomycin. Images obtained with permissions from the publishers.
The “mushroom-like” vestibule-barrel structure of α-hemolysin is amenable for various modes of probing inter-molecular reactions. Howorka and colleagues tethered a nucleic acid oligo into the vestibule region of an α-hemolysin channel in order to probe hybridization/de-hybridization dynamics with a Watson–Crick complementary oligo (see Fig. 15a) [81]. A complementary oligo that was added to the cis chamber solution was found to interact with the vestibule-tethered oligo with long “on” times (“b” in figure). The addition of a competing oligo scavenged the complementary oligo from binding to the vestibule, resulting in the disappearance of long-lived binding times. Fig. 15b shows the studies of Vercoutere and co-workers that probed the dynamics of short duplex DNA hairpins [82]. Rather than chemically modifying the vestibule of α-hemolysin, the group investigated short hairpin lifetimes in the vestibule and found a good correlation between the log of the residence times and the standard free energy of the duplex. Finally, the residence times of a protein in a nanopore were modulated by elegant engineering of an α-hemolysin pore, as done by Mohammad and co-workers (see Fig. 15c) [83,84]. The inner lumen of wild-type α-hemolysin is typically weakly charged, which does not allow long-lived interactions of a protein of interest with the channel. The group modified the charges within the nanopore at two strategic sites in order to provide an effective electrostatic trap, which was shown to enable to hold and interrogate a single pb2-Ba protein molecule at the pore for minutes.
Fig. 15. Probing intermolecular dynamics using nanopores.
(a) Ion current traces for an α-hemolysin pore that is tethered with oligo A in the vestibule region, before and after the addition of the complementary oligo B and following the subsequent addition of oligo A to compete with the hemolysin-bound oligo [81]. (b) Probing the dynamics of short DNA hairpins. The residence time of hairpins in the vestibule is plotted as a function of the duplex length, showing a logarithmic dependence of the event duration with the standard free energy of the duplex [82]. (c) Interrogating protein structure by electrostatically trapping the protein inside an α-hemolysin pore [83,84]. Engineering of the channel to contain aspartate residues on both ends of the β-barrel promoted the long-lived interactions of pb2-Ba with the channel, enabling its interrogation. Images obtained with permissions from the publishers.
2.5. Probing internal nucleic acid structure and modified bases
Nanopores can also be used to probe changes to the subtle changes to the internal structure of nucleic acids. Analysis of base modifications to native DNA is important because it provides information on epigenetic mechanisms in gene regulation pathways and environmental factors that may lead to deleterious DNA damage. Both of these types of modifications are implicated in cancer, aging, and other diseases. Fig. 16a shows work by Wallace and co-workers in which a DNA strand being pulled and held at a nanopore by a strongly bound protein [85], and the residual current can report on presence of various cytosine modifications that are present in humans [86]. These results are important because state-of-the-art biochemical assays lack the ability to resolve 5-methylcytosine (mC) from 5-hydroxymethylcytosine (hmC). Schibel and co-workers recently studied damaged DNA bases using the same principle of trapping a DNA molecule at the pore and measuring the residual current [87,88]. In their study, shown in Fig. 16b, modifications such as 8-oxo-7,8-dihydroguanine and guanine provide different residual currents, which facilitates their label-free detection. The impact of DNA base modifications on the global polymer structure of DNA was also studied, as shown in Fig. 16c [89]. DNA sequences that contain normal C showed different transport behavior through 4 nm solid-state nanopores than identical sequences that contain either mC or hmC instead of C. A temperature dependence study showed that differences in transport are attributed to the DNA flexibility: DNA containing the more polar cytosine hmC is most flexible, and DNA containing the least polar cytosine mC is the stiffest. This agrees with earlier results with methylated DNA sequences through small pores investigated using PCR, as well as with previous molecular dynamics simulations [90].
Fig. 16. Probing DNA internal structure and modified DNA bases.
(a) Detection of DNA base modifications using α-hemolysin. The residual current IRES for various base modifications reports on their presence in a sequence [86]. (b) Detection of damaged DNA bases on a tethered DNA molecule using α-hemolysin [87,88]. Scheme shows trapping of a molecule, measurement of the residual current, followed by its release. The technique can discriminate among normal and damaged guanines. (c) Analysis of base-modified DNA properties using a 4 nm diameter solid-state pore [89]. Concatenated ion current traces of identical 3 kbp sequences that contain different cytosines show different transport dynamics, related to the DNA flexibility. (d) Detecting the perturbation of secondary DNA structure by urea [91]. Scatter plots show that ssDNA is more likely to thread into an α-hemolysin pore when urea is added, pointing to the destabilization of secondary structure. (e) Detection of pH-induced destabilization of secondary structure using 4 nm diameter solid-state nanopores [93]. A 3000 nucleotide long ssDNA translocates through a 4 nm diameter pore differently at pH 7 and pH 13, attributed to the denaturing ability of alkaline solutions. Images obtained with permissions from the publishers.
The effects of denaturants such as urea, which perturb secondary DNA structure by a competitive binding with hydrogen bonding sites of the bases, were also studied using nanopores. Japrung and co-workers have recently studied the translocation of long ssDNA through α-hemolysin nanopores, as shown in Fig. 16d [91]. The addition of urea was found to facilitate ssDNA transport because it “unties” intramolecular secondary structures within the ssDNA molecules, thereby the pore can admit more molecules than it rejects. Using solid-state nanopores, Singer and coworkers also showed that a transition to a partly denatured dsDNA state could be observed by monitoring changes in the current through the pore [92]. Finally, in Fig. 16e, ssDNA denaturation and passage through 4 nm diameter solid-state nanopores was studied [93]. It was found that a 3000 nucleotide long ssDNA translocates through the 4 nm pore differently at pH 7 and pH 13, attributed to the denaturing ability of alkaline solutions, which melts secondary structures.
2.6. Engineered pores for biomolecular sensing and transport selectivity
Apart from passing nucleic acids, knowledge of the crystal structure of the protein α-hemolysin presented many opportunities for biochemical sensing, and since then Bayley and co-workers have developed elegant and impressive stochastic sensors based on chemically modified α-hemolysin channels. Fitting α-hemolysin with a chiral adapter β-cyclodextrin molecule [94,95] has proven useful for improving the channel's discrimination ability among enantiomer drugs, explosives, and nucleotides. The heptameric β-cyclodextrin structure forms a remarkably long-lived complex with the inner lumen of the heptameric channel α-hemolysin (~ 0.1 s), the lifetime of which can be further extended by modifying inner peptide residues in wild-type α-hemolysin. Because the adapter is chiral, interactions of the inner carbohydrate cavity with small biomolecules, organic molecules, and drugs, result in the transient formation of diastereomeric complexes. Since different diastereomeric complexes have different thermodynamic properties, measurements of the lifetime of each complex at the pore can report on the molecule's handedness. Moreover, other chemical tags applied to the channel's lumen can sense a variety of analytes, ranging from molecules to ions. Due to space limitations, the reader is referred to other recent reviews for a rich palette of examples of chemical and biochemical sensing using these adapted pores [5,96,97].
2.7. Sophisticated and emerging pore systems
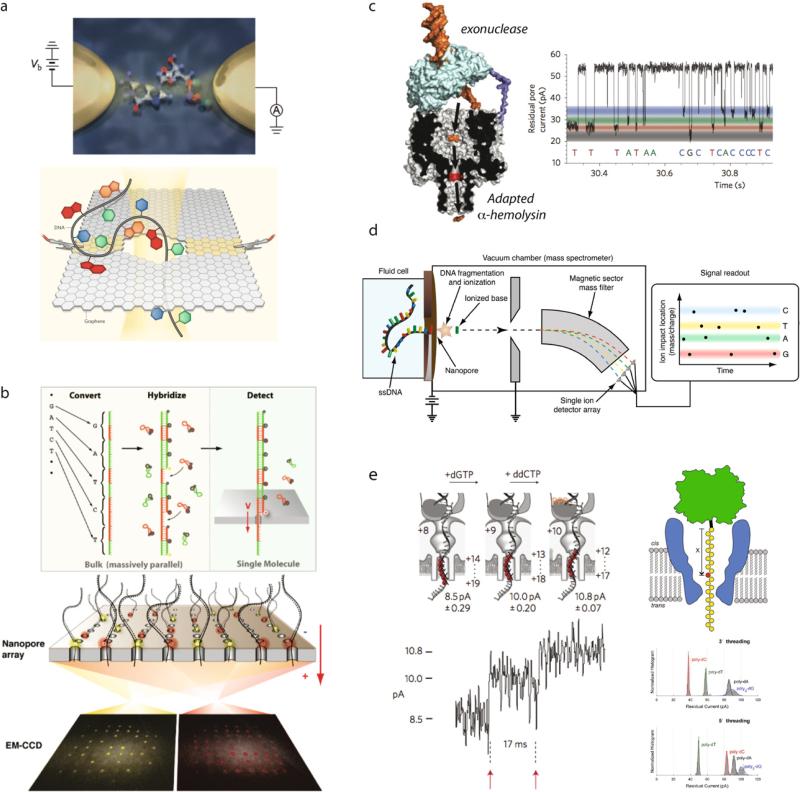
A pore–cavity–pore device was recently introduced by Pedone and co-workers, as depicted in Fig. 17a [98]. This device was fabricated by producing a small hole in silicon nitride deposited on a Si wafer, followed by anisotropically etching a thin silicon film through it. With this femtoliter-scale cavity that has a front entrance and a back exit, one can trap charged molecules, observe them using fluorescence microscopy, and investigate macromolecular escape times from the pore [99]. This device is particularly useful for fluorescence because the silicon on one of its sides is opaque, which allows selective illumination of the cavity without fluorescence background from and/or bleaching of the bulk sample. Recently, such a pore–cavity–pore device was used in order to perform a time-of-flight experiment on DNA molecules, where two serial resistive pulses reported on entry and exit of the molecule from the cavity [100].
Fig. 17. Exotic pore systems.
(a) Pore–cavity–pore system for trapping and manipulating single biomolecules [98,99]. A TEM image of the device is shown, and a fluorescence study of molecular capture/release is shown, as well as an ion current trace collected from a λ-DNA molecule time-of-flight experiment [100]. (b) Biomimetic nuclear pore complex [103]. Modification of silicon nitride nanopores with nucleoporins allows protein selectivity, which simulates a function of the nuclear envelope. (c) Hybrid solid-state/α-hemolysin pore [105]. Ion current traces are shown during transport of the DNA tail (pink), partial docking (yellow), and full docking (green). (d) Tunneling detector at a nanopore [108]. The device is fabricated using ion and electron beams, and allowed transverse electronic detection of DNA molecules as they are electrophoretically driven through a nanopore. (e) Receptor-modified solid-state pores [112]. A gold-coated SiN pore is used to introduce a single receptor via self-assembly of a functional thiol, followed by specific receptor binding (e.g., Ni-NTA/histidine tag chemistry). (f) Mechanical manipulation at a pore [117]. Combining various force apparatuses allows biomolecules to be interrogated with better control at a nanopore. Images obtained with permissions from the publishers. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article.)
Nanopores in natural living systems have properties that are not well understood at the atomic level. One such pore system is the nuclear pore complex, which contains species-selective elements that help to maintain specific protein gradients across the nucleus [101]. A biomimetic version of the nuclear pore complex was recently produced in bulk [102], and recently Kowalczyk and co-workers fabricated a single biomimetic pore complex [103] by chemically modifying a solid-state nanopore with nucleoporins tethered to polyethylene-glycol brushes using organosilane chemistry [104]. Functionalization of the pore demonstrated selectivity towards one particular protein over another (see Fig. 17b). By employing various chemical strategies to tailor chemical interactions of target macromolecules with the pore lumen, one can engineer the pore selectivity to optimize its detection and separation capabilities. Another interesting pore device is the hybrid solid-state/α-hemolysin device shown in Fig. 17c [105], which offers the robustness of a solid-state substrate combined with the chemical versatility and reproducibility of a protein channel. To produce such devices, a polyanionic DNA tail was tethered to the heptameric form of α-hemolysin, enabling its electrophoretic-based guidance and docking into a solid-state pore. A similar electrophoretic guidance approach was used to position an engineered DNA origami into a solid-state nanopore [106,107], offering chemical flexibility in a robust solid-state framework.
Another attractive device is the DNA tunneling detector embedded in a nanopore, as depicted in Fig. 17d [108]. The device consists of a nanopore fabricated through a nanoscale gap between two Pt electrodes. When voltage is applied across the electrode gap and biomolecules are driven through the pore using independent circuitry, biomolecules such as DNA provide a disruptive current that signals their occupancy [109,110]. Such devices are, at least theoretically, conceived as candidates for future nanopore-based DNA sequencing platforms [111]. However, these devices are also appealing for integration of nanopore array sensors, since each nanopore/gap zone can be addressed with independent circuitry and therefore parallelization can be achieved.
A challenging yet potentially versatile approach to protein sensing involves the immobilization of a single receptor inside of a robust solid-state pore. As shown in Fig. 17e, Rant and co-workers achieved this by coating a solid-state pore fabricated in silicon nitride with gold, so that alkanethiol chemistry can then be used to modify the gold surface with functional termini onto which a receptor can be specifically bound [112]. In the example shown in the figure, a complex of nickel with nitrilotriacetic acid (NTA) was immobilized on the pore surface, followed by specific binding of a histidine tagged protein receptor that enabled the specific detection of immunoglobulins (IgGs) at the single-molecule level based on their patterns of binding and unbinding kinetics at the receptor-modified pore.
Finally, it is of great interest to mechanically manipulate an individual biopolymer near a pore using either optical tweezers [113,114], magnetic tweezers [115], or an atomic force microscope tip [116] (see Fig. 17f, image taken from a recent review by Keyser [117]). The various possibilities afforded by combining these force apparatuses with nanopores include probing nucleic acid interactions (e.g., for unraveling RNA secondary structure), DNA/protein interactions, and for probing unfolding of proteins. This selection is by no means exhaustive, but rather it is intended to highlight recent creativity in the nanopore field.
3. Towards DNA sequencing using nanopores
As hopefully conveyed in the previous section, nanopores are an essential component in the growing toolbox of single-molecule biophysics. Much of the growth of the nanopore field can be attributed to their fascinating prospects as DNA sequencing platforms. Fig. 18 highlights some of the current directions of nanopore-based DNA sequencing. Fig. 18a shows two schemes for electronic readout of a DNA sequence by transverse electronic detection. The first picture, taken from a recent perspective by Bayley [118], shows the translocation of DNA through a graphene pore while electrons are passed through the graphene ribbon [119,120]. In this case, the current signal through the ribbon is affected by the identity of the base. In the second case, DNA is passed through an electrode gap and tunneling currents are recorded [121–123]. Fig. 18b shows optipore-based DNA sequencing, which is based on optical detection of fluorophores at a nanopore. In this scheme, a DNA molecule that is to be sequenced is first converted to a larger sequence and hybridized with reporter probes. Next, the constructs are unzipped using an array of small nanopores mounted on a cleverly designed fluorescence microscope [124], and each time a reporter is stripped off of the sequence, it emits photons of the color that corresponds to the identity of the reporter. The sequence of all four bases has been already measured using the device [125]. Although the base conversion steps are somewhat tedious, they can be performed in parallel for many sequences. In Fig. 18c, an exonuclease is tethered to an adapter-fitted α-hemolysin channel. This complex is powerful because the exonuclease can degrade a strand near the pore, while it can be detected in the cyclodextrin-adapted α-hemolysin channel. The current trace shows the great discrimination ability of the adapted α-hemolysin channel, which exhibits four characteristic levels corresponding to the four nucleotides [126,127]. In Fig. 18d, another degradation-based nucleotide detection method is discussed in which a DNA molecule is allowed to translocate across a nanopore into vacuum, where it is chemically fragmented using some irradiation-based method (e.g., photofragmentation) and then detected using a mass spectrometer [128]. Fig. 18e shows ion current based sequencing using a polymerase motor. This scheme is based on the ability to read base information using an immobilized strand at the pore [60,85,129]. A complex of a DNA polymerase and a template strand/primer duplex is first sucked into a pore by application of voltage. Each time a base is incorporated, the template strand is moved upwards [130], which provides a mechanism for driving a strand through a pore one base at a time. Also shown in the figure is an MspA channel, which can assist in the readout when combined with the polymerase motor approach by providing a better resolution [131,132].
Fig. 18. DNA sequencing using nanopores.
(a) Transverse electronic sequencing. Two approaches: cartoon [118] shows a single DNA strand being passed through a single-layer graphene nanopore while electrons are passed through the graphene ribbon [119,120], or, DNA is passed through an electrode gap and tunneling currents are recorded [121–123]. (b) Optipore-based DNA sequencing. In this scheme biochemical preparation of the target DNA molecules converts each base into a longer sequence equipped with optical reporters, and unzipping of the sequence provides an array-based readout. Both the biochemical conversion and readout steps have been demonstrated [125]. (c) Exonuclease/pore sequencing. In this scheme, an exonuclease tethered to an adapter-fitted α-hemolysin channel is used to degrade the strand one base at a time, while each base is attracted to the pore, where the residual ion current is measured as a function of time. The traces show a real current trace from mononucleotides in the solution (not the actual sequence of a DNA molecule), which exhibits four characteristic levels corresponding to the four nucleotides [126, 127]. (d) Nanopore/mass-spectrometer apparatus for single-molecule DNA sequencing. In this concept, a DNA strand passing through an orifice is subjected to irradiation-induced fragmentation under vacuum, resulting in a sequence of cleaved nucleotides that are detected using a mass-spectrometer. (e) Ion current based sequencing using a polymerase motor. A complex of a DNA polymerase and a template strand/primer duplex is sucked into a pore, and each time a base is incorporated, the template strand is moved upwards [130]. Improved current resolution of a single base by a protein-tethered MspA channel can assist in the readout when combined with the polymerase motor approach [131,132]. Images obtained with permissions from the publishers.
While the merits and drawbacks of the approaches shown in Fig. 18 are contemplated, a particular approach has gained a significant leap forward with the announcement of Oxford Nanopore Technologies (ONT) integrated DNA sequencing platform. While the exact details of the approach is yet unpublished, at the annual Advances in Genome Biology and Technology (AGBT) meeting in mid-February, 2012, ONT presented an abstract entitled “Single Molecule ‘Strand’ Sequencing Using Protein Nanopores and Scalable Electronic Devices”. In this presentation, ONT claimed that they are closer than ever to commercializing a sequencing device based on moving a DNA strand through a protein pore. Most likely, their approach to DNA sequencing originated from two main experimental breakthroughs: (1) Work that probed and mapped the sensitivity of the current through protein channels to the position and identity of immobilized bases along a DNA strand [85,129,131,133–135]. (2) The demonstration that DNA-processing enzymes (i.e., a DNA polymerase) can mediate base-by-base DNA translocation across a nanopore by processing a DNA template [129,133,134,136–139]. The first crucial feat of having mapped the identity and position of a nucleobase in a DNA strand immobilized at the pore allowed one to probe the structure of the protein channel, as well as its sensitivity to the relative positions of different sequences. The second crucial feat is the development of a practical method for moving bases across the pore one base at a time using a polymerase. In this translocation mode, the long residence time of each base sampled in the pore (> 1 ms) allows more precise readout, because capacitive current noise sources can be effectively filtered out using a low-pass filter. As demonstrated by Kasianowicz and co-workers, long residence times at the pore of neutral polymers allow increased current sensitivity that can report on the molecular mass and polydispersity of a polymer sample with sub-picoampere precision [140–142]. Similarly, the ONT team and others can in principle read out information from multiple bases in the pore simultaneously and extract their sequence, given no degeneracy (for reading 3 bases simultaneously, 43 non-degenerate current levels are necessary). This multiple-base readout can minimize the error rates and improve the analysis of base repeats, particularly since DNA-processing enzymes can occasionally pause or process DNA backwards.
4. Future directions of nanopores: Points for debate
4.1. Regulation of nucleic acid transport: Voltage- or enzyme-driven?
Let us assume an ideal, atomically thin nanopore is available for measuring the translocation of a single DNA strand. This pore does not interact very strongly with DNA, is just wide enough to accommodate a single DNA strand, and is robust and reproducible to the atomic level. Moreover, each nucleotide provides a different ion current level when in the pore. Going forward with this assumption we can pose the question: could a nanopore directly sequence a DNA strand?
Voltage-driven translocation of polynucleotides through a pore is extremely simple, yet the microscopic details of the transport are not well controlled because of intrinsic limitations of the translocation process. The Achilles heel of nanopores has been the inability to control or define the motion of DNA through a pore during voltage-driven translocation. As discussed earlier, both entropic (coil entropy) and enthalpic (interactions with pore) contributions dictate a random (diffusional) translocation process that is at best poorly defined. Additionally, voltage-driven translocations are often too fast for measuring an appreciable amount of ions during the translocation of a single base (detection of 1 μs of current is quite challenging for a small pore because noise is overwhelming). Reducing the voltage increases the average translocation times but results in more diffusive transport, which is slower but less regulated. This result leaves us with insufficient information for sequencing a DNA strand, for reasons that are beyond our control.
Voltage-induced unzipping duplex DNA at a nanopore has been used to slow down DNA transport. The kinetics of this process may be more regulated by the appropriate choice of voltage and duplex length/sequence. However, unzipping will not drive the DNA strand base-by-base into the pore, because duplex sets composed of several basepairs are needed to provide a thermally stable duplex. This necessitates expanding the nucleic acid of interest into a larger sequence, a task that can be challenging, particularly for harnessing the potential of nanopores for reading long read lengths.
Enzyme-driven motion is yet another option for regulating transport. In this scheme, a DNA polymerase is used as fuel to drive an ssDNA template strand through the pore, one base at a time, while the DNA strand is pulled at the pore using voltage. Each time the enzyme incorporates a nucleotide triphosphate (NTP), a single base is puled across the pore. Regulating the enzyme can then be achieved by changing the concentrations of NTPs in the electrolyte, changing the voltage, or by choosing/engineering the appropriate polymerase. Possible drawbacks of this approach: (1) the enzyme will not be as active as it is in free solution, resulting in short processivity, (2) distinguishing base repeats may be difficult because the enzyme incorporation kinetics are stochastic. Despite these drawbacks, advantages of the enzymatic “stepper-motor” include compatibility with pore arrays, recyclability of the pore when enzyme is inactive, and compatibility with using pre-formed enzyme-DNA complexes.
4.2. Protein pores vs. solid-state pores: The tortoise vs. the hare
Just over a decade ago, solid-state nanopores have taken stage as a new type of pores that promises better capability and flexibility. Solid-state pores allow a tunable pore size and shape, stability, and integration with compact electronic/optical sensor modalities. In addition, many of the biophysical studies carried out using solid-state nanopores would not have been possible using protein pores. Finally, experiments with solid-state devices are in many ways easier than handling fragile lipid bilayers and waiting for single-channel reconstitution. However, when comparing the quality of results accrued over the past 15 years with α-hemolysin, it is clear that the atomic-level precision and control over functionality is a winning combination. The main drawback here is the relatively low precision with which pores are fabricated, as compared with the atomic precision of proteins. While techniques that offer better control over nanopore shape are developed, perhaps the questions should be: can we design protein channels with a more flexible size, and can we design more stable bilayers? When evaluating the state-of-the-art in nanopore fabrication, there's no comparison to protein channels. For the time being, protein channels are superior. As for the tortoise vs. hare analogy: solid-state pores have come on strong in the early 2000's as the pores that may be suitable for replacing protein channels in technological applications: “Robust, solid-state nanopores, fashioned to any desired diameter, could yield new data and understanding of transport in confined spaces, and will make it possible to create robust single-molecule-sensing devices to characterize molecules of DNA and other biopolymers at unprecedented speeds” [18]. However, since then, we have yet to witness a solid-state nanopore as perfect, precise, and reproducible as a protein channel. As proof that the tortoise won the marathon, ONT's recent announcement unfolds a nanopore-based device that can sequence DNA, and that device does not have a single hare in it.
5. Conclusions
In conclusion, the nanopore field is emerging with gusto. In just 15 years, the molecular scale Coulter counter has performed many tasks and is promising to perform many more. On the curvy road towards DNA sequencing, nanopores have sparked the imagination of scientists as tools that can help in solving problems in biophysics. Similar to atomic force microscopy, optical/magnetic tweezers, and fluorescence microscopy, nanopores are taking stage as one of a handful of main tools for investigating individual protein and DNA molecules. It is important to understand the drawbacks of any technique in order to advance it. For example, an obvious drawback is the inability of nanopores to provide any spectroscopic information about the identity of a molecule. Ion current from a nanopore cannot determine the identity of a molecule, and therefore, properties of the molecule are inferred. Another misconception comes from the tendency to assume that translocation occurs with constant speed. That may be the case, but there is no proof for it. A third assumption is that nanopores inherently have commercial value because of single-molecule detection capabilities. While nanopores do detect individual molecules, far more than a handful of molecules are required for detection in solution. Also, one should not overlook the possibility of pore clogging when discussing a platform that is based on information from individual pores. Therefore, a platform that does not provide a continuous source of renewable pores in a device is too consuming and in my opinion, will not reach commercialization. A commercial filter contains ca. 1010 pores on a ½ inch disc, and can tolerate 5% of ineffective pores. In contrast, a nanopore is only useful when perfectly open and clean. This is not so easy to achieve without foolproof ways to regenerate nanopores at the touch of a button. Since both lipid-embedded protein channels and solid-state nanopores have drawbacks, it will be interesting to see which device, or what combination of devices, will be available in years to come, if any. Quite a bit we've covered in this review, and quite a bit not. I hope that in this excursion the “potential” of the nanopore technique for probing nucleic acids and other biomolecules has emerged within the reader, as well as the inspiration to consider a nanopore experiment in his/her future research.
Acknowledgements
I sincerely thank my group and colleagues for valuable comments on this manuscript. Financial support has been provided by NIH grant HG6321-01 and Northeastern University's Start-up funds.
References
- 1.Graham MD. The Coulter principle: Foundation of an industry. J Lab Autom. 2003;8:72–81. [Google Scholar]
- 2.Coulter WH. 1953 US Patent #2,656,508.
- 3.Deblois RW, Bean CP, Wesley RKA. Electrokinetic measurements with submicron particles and pores by resistive pulse technique. J Colloid Interface Sci. 1977;61:323–35. [Google Scholar]
- 4.Bayley H, Martin CR. Resistive-pulse sensing – From microbes to molecules. Chem Rev. 2000;100:2575–94. doi: 10.1021/cr980099g. [DOI] [PubMed] [Google Scholar]
- 5.Bayley H, Jayasinghe L. Functional engineered channels and pores (Review). Molec Mem Bio. 2004;21:209–20. doi: 10.1080/09687680410001716853. [DOI] [PubMed] [Google Scholar]
- 6.Kasianowicz JJ, Robertson JWF, Chan ER, Reiner JE, Stanford VM. Annual review of analytical chemistry. Annual Reviews. 2008;1:737–66. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 7.Howorka S, Siwy Z. Nanopore analytics: sensing of single molecules. Chem Soc Rev. 2009;38:2360–84. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 8.Sakmann B, Neher E. Single-channel recording. Springer-Verlag; New York: 1995. [Google Scholar]
- 9.Hille B. Ion channels of excitable membranes. Sinauer. 2001 [Google Scholar]
- 10.Halverson KM, et al. Anthrax biosensor, protective antigen ion channel asymmetric blockade. J Biol Chem. 2005;280:34056–62. doi: 10.1074/jbc.M507928200. [DOI] [PubMed] [Google Scholar]
- 11.Eimer W, Pecora R. Rotational and translational diffusion of short rodlike molecules in solution – oligonucleotides. J Chem Phys. 1991;94:2324–9. [Google Scholar]
- 12.Stellwagen NC, Magnusdottir S, Gelfi C, Righetti PG. Measuring the translational diffusion coefficients of small DNA molecules by capillary electrophoresis. Biopolymers. 2001;58:390–7. doi: 10.1002/1097-0282(20010405)58:4<390::AID-BIP1015>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 13.Bezrukov SM, Kasianowicz JJ. Current noise reveals protonation kinetics and number of ionizable sites in an open protein ion channel. Phys Rev Lett. 1993;70:2352–5. doi: 10.1103/PhysRevLett.70.2352. [DOI] [PubMed] [Google Scholar]
- 14.Kasianowicz JJ, Bezrukov SM. Protonation dynamics of the alpha-toxin ion-channel from spectral-analysis of pH-dependent current fluctuations. Biophys J. 1995;69:94–105. doi: 10.1016/S0006-3495(95)79879-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song LZ, et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–66. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 16.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc Natl Acad Sci USA. 2008;105:20647–52. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wendell D, et al. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nano. 2009;4:765–72. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, et al. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–9. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 19.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Fabrication of solid-state nanopores with single-nanometre precision. Nat Mater. 2003;2:537–40. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 20.Wanunu M, et al. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nano. 2010;5:807–14. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 21.Garaj S, et al. Graphene as a subnanometre trans-electrode membrane. Nature. 2010;467:190–3. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris DC. Quantitative chemical analysis. W.H. Freeman; 1995. [Google Scholar]
- 23.Tabard-Cossa V, Trivedi D, Wiggin M, Jetha NN, Marziali A. Noise analysis and reduction in solid-state nanopores. Nanotechnology. 2007;18:305505. [Google Scholar]
- 24.Smeets RMM, Keyser UF, Dekker NH, Dekker C. Noise in solid-state nanopores. Proc Natl Acad Sci USA. 2008;105:417–21. doi: 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uram JD, Ke K, Mayer M. Noise and bandwidth of current recordings from submicrometer pores and nanopores. ACS Nano. 2008;2:857–72. doi: 10.1021/nn700322m. [DOI] [PubMed] [Google Scholar]
- 26.Powell MR, Vlassiouk I, Martens C, Siwy ZS. Nonequilibrium 1/f noise in rectifying nanopores. Phys Rev Lett. 2009;103:248104. doi: 10.1103/PhysRevLett.103.248104. [DOI] [PubMed] [Google Scholar]
- 27.Hoogerheide DP, Garaj S, Golovchenko JA. Probing surface charge fluctuations with solid-state nanopores. Phys Rev Lett. 2009;102:256804. doi: 10.1103/PhysRevLett.102.256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smeets RMM, Dekker NH, Dekker C. Low-frequency noise in solid-state nanopores. Nanotechnology. 2009;20:095501. doi: 10.1088/0957-4484/20/9/095501. [DOI] [PubMed] [Google Scholar]
- 29.Powell MR, Martens C, Siwy ZS. Asymmetric properties of ion current 1/f noise in conically shaped nanopores. Chem Phys. 2010;375:529–35. [Google Scholar]
- 30.Tasserit C, Koutsioubas A, Lairez D, Zalczer G, Clochard MC. Pink noise of ionic conductance through single artificial nanopores revisited. Phys Rev Lett. 2010;105:260602. doi: 10.1103/PhysRevLett.105.260602. [DOI] [PubMed] [Google Scholar]
- 31.Powell MR, et al. Noise properties of rectifying nanopores. J Phys Chem C. 2011;115:8775–83. [Google Scholar]
- 32.Ervin EN, White RJ, White HS. Sensitivity and signal complexity as a function of the number of ion channels in a stochastic sensor. Anal Chem. 2009;81:533–7. doi: 10.1021/ac801104v. [DOI] [PubMed] [Google Scholar]
- 33.Smeets RMM, et al. Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 2006;6:89–95. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
- 34.Kim MJ, Wanunu M, Bell DC, Meller A. Rapid fabrication of uniformly sized nanopores and nanopore arrays for parallel DNA analysis. Adv Mater. 2006;18:3149–55. [Google Scholar]
- 35.Hall JE. Access resistance of a small circular pore. J Gen Physiol. 1975;66:531–2. doi: 10.1085/jgp.66.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vodyanoy I, Bezrukov SM. Sizing of an ion pore by access resistance measurements. Biophys J. 1992;62:10–1. doi: 10.1016/S0006-3495(92)81762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowalczyk SW, Grosberg AY, Rabin Y, Dekker C. Modeling the conductance and DNA blockade of solid-state nanopores. Nanotechnology. 2011;22:315101. doi: 10.1088/0957-4484/22/31/315101. [DOI] [PubMed] [Google Scholar]
- 38.Meller A, Branton D. Single molecule measurements of DNA transport through a nanopore. Electrophoresis. 2002;23:2583–91. doi: 10.1002/1522-2683(200208)23:16<2583::AID-ELPS2583>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 39.Kawano R, Schibel AEP, Cauley C, White HS. Controlling the translocation of single-stranded DNA through alpha-hemolysin ion channels using viscosity. Langmuir. 2009;25:1233–7. doi: 10.1021/la803556p. [DOI] [PubMed] [Google Scholar]
- 40.Meller A, Nivon L, Branton D. Voltage-driven DNA translocations through a nanopore. Phys Rev Lett. 2001;86:3435–8. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 41.Meller A. Dynamics of polynucleotide transport through nanometre-scale pores. J Phys Cond Matt. 2003;15:R581–607. [Google Scholar]
- 42.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat Nano. 2010;5:160–5. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grosberg AY, Rabin Y. DNA capture into a nanopore: interplay of diffusion and electrohydrodynamics. J Chem Phys. 2010;133:165102. doi: 10.1063/1.3495481. [DOI] [PubMed] [Google Scholar]
- 44.Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Driven DNA transport into an asymmetric nanometer-scale pore. Phys Rev Lett. 2000;85:3057–60. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 45.Chen P, et al. Probing single DNA molecule transport using fabricated nanopores. Nano Lett. 2004;4:2293–8. doi: 10.1021/nl048654j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li JL, Gershow M, Stein D, Brandin E, Golovchenko JA. DNA molecules and configurations in a solid-state nanopore microscope. Nat Mater. 2003;2:611–5. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 47.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys Rev E. 2005;71:051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 48.Fologea D, Brandin E, Uplinger J, Branton D, Li J. DNA conformation and base number simultaneously determined in a nanopore. Electrophoresis. 2007;28:3186–92. doi: 10.1002/elps.200700047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu LQ, Cheley S, Bayley H. Electroosmotic enhancement of the binding of a neutral molecule to a transmembrane pore. Proc Natl Acad Sci USA. 2003;100:15498–503. doi: 10.1073/pnas.2531778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93:13770–3. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA. 2000;97:1079–84. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wanunu M, Sutin J, McNally B, Chow A, Meller A. DNA translocation governed by interactions with solid-state nanopores. Biophys J. 2008;95:4716–25. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Long D, Viovy J-L, Ajdari A. Simultaneous action of electric fields and nonelectric forces on a polyelectrolyte: motion and deformation. Phys Rev Lett. 1996;76:3858–61. doi: 10.1103/PhysRevLett.76.3858. [DOI] [PubMed] [Google Scholar]
- 54.Lubensky DK, Nelson DR. Driven polymer translocation through a narrow pore. Biophys J. 1999;77:1824–38. doi: 10.1016/S0006-3495(99)77027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Storm AJ, et al. Fast DNA translocation through a solid-state nanopore. Nano Lett. 2005;5:1193–7. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 56.Luan B, Aksimentiev A. Electro-osmotic screening of the DNA charge in a nanopore. Phys Rev E. 2008;78:021912. doi: 10.1103/PhysRevE.78.021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nat Nano. 2011;6:615–24. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 58.Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Slowing DNA translocation in solid-state nanopore. Nano Lett. 2005;5:1734–7. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathe J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Orientation discrimination of single-stranded DNA inside the alpha-hemolysin membrane channel. Proc Natl Acad Sci USA. 2005;102:12377–82. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purnell RF, Mehta KK, Schmidt JJ. Nucleotide identification and orientation discrimination of DNA homopolymers immobilized in a protein nanopore. Nano Lett. 2008;8:3029–34. doi: 10.1021/nl802312f. [DOI] [PubMed] [Google Scholar]
- 61.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77:3227–33. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skinner GM, van den Hout M, Broekmans O, Dekker C, Dekker NH. Distinguishing single- and double-stranded nucleic acid molecules using solid-state nanopores. Nano Lett. 2009;9:2953–60. doi: 10.1021/nl901370w. [DOI] [PubMed] [Google Scholar]
- 63.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in a nanopore. Phys Rev Lett. 2003;90:238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 64.Mathe J, Arinstein A, Rabin Y, Meller A. Equilibrium and irreversible unzipping of DNA in a nanopore. Europhys Lett. 2006;73:128–34. [Google Scholar]
- 65.Bates M, Burns M, Meller A. Dynamics of DNA molecules in a membrane channel probed by active control techniques. Biophys J. 2003;84:2366–72. doi: 10.1016/S0006-3495(03)75042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakane JJ, Akeson M, Marziali A. Nanopore sensors for nucleic acid analysis. J Phys Cond Matt. 2003;15:R1365–93. [Google Scholar]
- 67.Dudko OK, Mathe J, Szabo A, Meller A, Hummer G. Extracting kinetics from single-molecule force spectroscopy: Nanopore unzipping of DNA hairpins. Biophys J. 2007;92:4188–95. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McNally B, Wanunu M, Meller A. Electromechanical unzipping of individual DNA molecules using synthetic sub-2 nm pores. Nano Lett. 2008;8:3418–22. doi: 10.1021/nl802218f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tropini C, Marziali A. Multi-nanopore force spectroscopy for DNA analysis. Biophys J. 2007;92:1632–7. doi: 10.1529/biophysj.106.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kasianowicz JJ, Henrickson SE, Weetall HH, Robertson B. Simultaneous multianalyte detection with a nanometer-scale pore. Anal Chem. 2001;73:2268–72. doi: 10.1021/ac000958c. [DOI] [PubMed] [Google Scholar]
- 71.Sexton LT, et al. Resistive-pulse studies of proteins and protein/antibody complexes using a conical nanotube sensor. J Am Chem Soc. 2007;129:13144–52. doi: 10.1021/ja0739943. [DOI] [PubMed] [Google Scholar]
- 72.Uram JD, Ke K, Hunt AJ, Mayer M. Submicrometer pore-based characterization and quantification of antibody–virus interactions. Small. 2006;2:967–72. doi: 10.1002/smll.200600006. [DOI] [PubMed] [Google Scholar]
- 73.Yusko EC, et al. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat Nano. 2011;6:253–60. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wanunu M, Sutin J, Meller A. DNA profiling using solid-state nanopores: detection of DNA-binding molecules. Nano Lett. 2009;9:3498–502. doi: 10.1021/nl901691v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hornblower B, et al. Single-molecule analysis of DNA–protein complexes using nanopores. Nat Meth. 2007;4:315–7. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 76.Singer A, et al. Nanopore based sequence specific detection of duplex DNA for genomic profiling. Nano Lett. 2010;10:738–42. doi: 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smeets RMM, Kowalczyk SW, Hall AR, Dekker NH, Dekker C. Translocation of RecA-coated double-stranded DNA through solid-state nanopores. Nano Lett. 2009;9:3089–95. doi: 10.1021/nl803189k. [DOI] [PubMed] [Google Scholar]
- 78.Sischka A, et al. Dynamic translocation of ligand-complexed DNA through solid-state nanopores with optical tweezers. J Phys Cond Matt. 2010;22:454121. doi: 10.1088/0953-8984/22/45/454121. [DOI] [PubMed] [Google Scholar]
- 79.Kowalczyk SW, Hall AR, Dekker C. Detection of local protein structures along DNA using solid-state nanopores. Nano Lett. 2010;10:324–8. doi: 10.1021/nl903631m. [DOI] [PubMed] [Google Scholar]
- 80.Wanunu M, et al. Nanopore analysis of individual RNA/antibiotic complexes. ACS Nano. 2011;5:9345–53. doi: 10.1021/nn203764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Howorka S, Movileanu L, Braha O, Bayley H. Kinetics of duplex formation for individual DNA strands within a single protein nanopore. Proc Natl Acad Sci USA. 2001;98:12996–3001. doi: 10.1073/pnas.231434698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vercoutere W, et al. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat Biotech. 2001;19:248–52. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 83.Mohammad MM, Prakash S, Matouschek A, Movileanu L. Controlling a single protein in a nanopore through electrostatic traps. J Am Chem Soc. 2008;130:4081–8. doi: 10.1021/ja710787a. [DOI] [PubMed] [Google Scholar]
- 84.Movileanu L. Interrogating single proteins through nanopores: challenges and opportunities. Trends Biotech. 2009;27:333–41. doi: 10.1016/j.tibtech.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 85.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. Recognizing a single base in an individual DNA strand: A step toward DNA sequencing in nanopores. Angew Chem. 2005;44:1401–4. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wallace EVB, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun. 2010;46:8195–7. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schibel AEP, et al. Nanopore detection of 8-oxo-7,8-dihydro-2′-deoxyguanosine in immobilized single-stranded DNA via adduct formation to the DNA damage site. J Am Chem Soc. 2010;132:17992–5. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schibel AEP, et al. Sequence-specific single-molecule analysis of 8-oxo-7,8-dihydroguanine lesions in DNA based on unzipping kinetics of complementary probes in ion channel recordings. J Am Chem Soc. 2011;133:14778–84. doi: 10.1021/ja205653v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wanunu M, et al. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J Am Chem Soc. 2011;133:486–92. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mirsaidov U, et al. Nanoelectromechanics of methylated DNA in a synthetic nanopore. Biophys J. 2009;96:L32–4. doi: 10.1016/j.bpj.2008.12.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Japrung D, Henricus M, Li QH, Maglia G, Bayley H. Urea facilitates the translocation of single-stranded DNA and RNA through the alpha-hemolysin nanopore. Biophys J. 2010;98:1856–63. doi: 10.1016/j.bpj.2009.12.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singer A, Kuhn H, Frank-Kamenetskii M, Meller A. Detection of urea-induced internal denaturation of dsDNA using solid-state nanopores. J Phys Cond Matt. 2010;22:454111. doi: 10.1088/0953-8984/22/45/454111. [DOI] [PubMed] [Google Scholar]
- 93.Fologea D, et al. Detecting single stranded DNA with a solid state nanopore. Nano Lett. 2005;5:1905–9. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature. 1999;398:686–90. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 95.Gu LQ, Cheley S, Bayley H. Capture of a single molecule in a nanocavity. Science. 2001;291:636–40. doi: 10.1126/science.291.5504.636. [DOI] [PubMed] [Google Scholar]
- 96.Bayley H. Designed membrane channels and pores. Curr Opin Biotech. 1999;10:94–103. doi: 10.1016/s0958-1669(99)80017-2. [DOI] [PubMed] [Google Scholar]
- 97.Maglia G, Heron AJ, Stoddart D, Japrung D, Bayley H. Walter NG, editor. Single molecule tools, pt B: super-resolution, particle tracking, multiparameter, and force based methods. Methods in enzymology. 2010;475:591–623. doi: 10.1016/S0076-6879(10)75022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pedone D, et al. Fabrication and electrical characterization of a pore–cavity–pore device. J Phys Cond Matt. 2010;22:454115. doi: 10.1088/0953-8984/22/45/454115. [DOI] [PubMed] [Google Scholar]
- 99.Pedone D, Langecker M, Abstreiter G, Rant U. A pore–cavity–pore device to trap and investigate single nanoparticles and DNA molecules in a femtoliter compartment: confined diffusion and narrow escape. Nano Lett. 2011;11:1561–7. doi: 10.1021/nl104359c. [DOI] [PubMed] [Google Scholar]
- 100.Langecker M, Pedone D, Simmel FC, Rant U. Electrophoretic time-of-flight measurements of single DNA molecules with two stacked nanopores. Nano Lett. 2011;11:5002–7. doi: 10.1021/nl2030079. [DOI] [PubMed] [Google Scholar]
- 101.Hoelz A, Debler EW, Blobel G. Kornberg RD, Raetz CRH, Rothman JE, Thorner JW, editors. Annual review of biochemistry. Annual Reviews. 2011;80:613–43. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 102.Jovanovic-Talisman T, et al. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2009;457:1023–7. doi: 10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kowalczyk SW, et al. Single-molecule transport across an individual biomimetic nuclear pore complex. Nat Nano. 2011;6:433–8. doi: 10.1038/nnano.2011.88. [DOI] [PubMed] [Google Scholar]
- 104.Wanunu M, Meller A. Chemically modified solid-state nanopores. Nano Lett. 2007;7:1580–5. doi: 10.1021/nl070462b. [DOI] [PubMed] [Google Scholar]
- 105.Hall AR, et al. Hybrid pore formation by directed insertion of alpha-haemolysin into solid-state nanopores. Nat Nano. 2010;5:874–7. doi: 10.1038/nnano.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wei R, Martin TG, Rant U, Dietz H. DNA origami gatekeepers for solid-state nanopores. Angew Chem. 2012 doi: 10.1002/anie.201200688. http://dx.doi.org/10.1002/anie.201200688. [DOI] [PubMed]
- 107.Bell NAW, et al. DNA origami nanopores. Nano Lett. 2012;12:512–7. doi: 10.1021/nl204098n. [DOI] [PubMed] [Google Scholar]
- 108.Ivanov AP, et al. DNA tunneling detector embedded in a nanopore. Nano Lett. 2011;11:279–85. doi: 10.1021/nl103873a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liang XG, Chou SY. Nanogap detector inside nanofluidic channel for fast real-time label-free DNA analysis. Nano Lett. 2008;8:1472–6. doi: 10.1021/nl080473k. [DOI] [PubMed] [Google Scholar]
- 110.Tsutsui M, Taniguchi M, Yokota K, Kawai T. Identifying single nucleotides by tunnelling current. Nat Nano. 2010;5:286–90. doi: 10.1038/nnano.2010.42. [DOI] [PubMed] [Google Scholar]
- 111.Lagerqvist J, Zwolak M, Di Ventra M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 2006;6:779–82. doi: 10.1021/nl0601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wei R, Gatterdam V, Wieneke R, Tampe R, Rant U. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat Nano. 2012 doi: 10.1038/nnano.2012.24. Advance online publication. [DOI] [PubMed] [Google Scholar]
- 113.Keyser UF, van der Does J, Dekker C, Dekker NH. Optical tweezers for force measurements on DNA in nanopores. Rev Sci Instrum. 2006;77:105105. [Google Scholar]
- 114.Keyser UF, et al. Direct force measurements on DNA in a solid-state nanopore. Nat Phys. 2006;2:473–7. [Google Scholar]
- 115.Peng HB, Ling XSS. Reverse DNA translocation through a solid-state nanopore by magnetic tweezers. Nanotechnology. 2009;20:185101. doi: 10.1088/0957-4484/20/18/185101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.King GM, Golovchenko JA. Probing nanotube–nanopore interactions. Phys Rev Lett. 2005;95:216103. doi: 10.1103/PhysRevLett.95.216103. [DOI] [PubMed] [Google Scholar]
- 117.Keyser UF. Controlling molecular transport through nanopores. J Roy Soc Interface. 2011;8:1369–78. doi: 10.1098/rsif.2011.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bayley H. Nanotechnology – holes with an edge. Nature. 2010;467:164–5. doi: 10.1038/467164a. [DOI] [PubMed] [Google Scholar]
- 119.Nelson T, Zhang B, Prezhdo OV. Detection of nucleic acids with graphene nanopores: ab initio characterization of a novel sequencing device. Nano Lett. 2010;10:3237–42. doi: 10.1021/nl9035934. [DOI] [PubMed] [Google Scholar]
- 120.Saha KK, Drndic M, Nikolic BK. DNA base-specific modulation of microampere transverse edge currents through a metallic graphene nanoribbon with a nanopore. Nano Lett. 2012;12:50–5. doi: 10.1021/nl202870y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zwolak M, Di Ventra M. Electronic signature of DNA nucleotides via transverse transport. Nano Lett. 2005;5:421–4. doi: 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]
- 122.He H, et al. Functionalized nanopore-embedded electrodes for rapid DNA sequencing. J Phys Chem C. 2008;112:3456–9. [Google Scholar]
- 123.Taniguchi M, Tsutsui M, Yokota K, Kawai T. Fabrication of the gating nanopore device. Appl Phys Lett. 2009;95:123701. [Google Scholar]
- 124.Soni GV, et al. Synchronous optical and electrical detection of biomolecules traversing through solid-state nanopores. Rev Sci Instrum. 2010;81:014301. doi: 10.1063/1.3277116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McNally B, et al. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 2010;10:2237–44. doi: 10.1021/nl1012147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: Direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J Am Chem Soc. 2006;128:1705–10. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 127.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nano. 2009;4:265–70. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 128.Stein D. Systems and methods for determining molecules using mass spectrometry and related techniques. U.S. Patent Application Number 13/056,755.
- 129.Cockroft SL, Chu J, Amorin M, Ghadiri MR. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J Am Chem Soc. 2008;130:818–20. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lieberman KR, et al. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J Am Chem Soc. 2010;132:17961–72. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Manrao EA, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH. Nucleotide discrimination with DNA immobilized in the MspA nanopore. PLoS One. 2011;6:e25723. doi: 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Derrington IM, et al. Nanopore DNA sequencing with MspA. Proc Natl Acad Sci USA. 2010;107:16060–5. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gyarfas B, et al. Mapping the position of DNA polymerase-bound DNA templates in a nanopore at 5 angstrom resolution. ACS Nano. 2009;3:1457–66. doi: 10.1021/nn900303g. [DOI] [PubMed] [Google Scholar]
- 134.Chu J, Gonzalez-Lopez M, Cockroft SL, Amorin M, Ghadiri MR. Real-time monitoring of DNA polymerase function and stepwise single-nucleotide DNA strand translocation through a protein nanopore. Angew Chem. 2010;49:10106–9. doi: 10.1002/anie.201005460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stoddart D, et al. Nucleobase recognition in ssDNA at the central constriction of the alpha-hemolysin pore. Nano Lett. 2010;10:3633–7. doi: 10.1021/nl101955a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wilson NA, et al. Electronic control of DNA polymerase binding and unbinding to single DNA molecules. ACS Nano. 2009;3:995–1003. doi: 10.1021/nn9000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Olasagasti F, et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nano. 2010;5:798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Garalde DR, et al. Distinct complexes of DNA polymerase I (Klenow fragment) for base and sugar discrimination during nucleotide substrate selection. J Biol Chem. 2011;286:14480–92. doi: 10.1074/jbc.M111.218750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision. Nat Biotech. 2012 doi: 10.1038/nbt.2147. http://dx.doi.org/10.1038/nbt.2147. Advance online publication. [DOI] [PMC free article] [PubMed]
- 140.Robertson JWF, et al. Single-molecule mass spectrometry in solution using a solitary nanopore. Proc Natl Acad Sci USA. 2007;104:8207–11. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reiner JE, Kasianowicz JJ, Nablo BJ, Robertson JWF. Theory for polymer analysis using nanopore-based single-molecule mass spectrometry. Proc Natl Acad Sci USA. 2010;107:12080–5. doi: 10.1073/pnas.1002194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Robertson JWF, Kasianowicz J, Reiner JE. Changes in ion channel geometry resolved to sub-angstrom precision via single molecule mass spectrometry. J Phys Cond Matt. 2010;22:454108. doi: 10.1088/0953-8984/22/45/454108. [DOI] [PubMed] [Google Scholar]