Abstract
Hermansky-Pudlak Syndrome (HPS) is a set of genetically heterogeneous diseases caused by mutations in one of nine known HPS genes. HPS patients display oculocutaneous hypopigmentation and bleeding diathesis and, depending on the disease subtype, pulmonary fibrosis, congenital nystagmus, reduced visual acuity, and platelet aggregation deficiency. Mouse models for all known HPS subtypes have contributed greatly to our understanding of the disease, but many of the molecular and cellular mechanisms underlying HPS remain unknown. Here, we characterize ocular defects in the zebrafish (Danio rerio) mutant snow white (snw), which possesses a recessive, missense mutation in hps5 (hps5I76N). Melanosome biogenesis is disrupted in snw/hps5 mutants, resulting in hypopigmentation, a significant decrease in the number, size, and maturity of melanosomes, and the presence of ectopic multi-melanosome clusters throughout the mutant retina and choroid. snw/hps5I76N is the first Hps5 mutation identified within the N-terminal WD40 repeat protein–protein binding domain. Through in vitro coexpression assays, we demonstrate that Hps5I76N retains the ability to bind its protein complex partners, Hps3 and Hps6. Furthermore, while Hps5 and Hps6 stabilize each other’s expression, this stabilization is disrupted by Hps5I76N. The snw/hps5I76N mutant provides a valuable resource for structure–function analyses of Hps5 and enables further elucidation of the molecular and cellular mechanisms underlying HPS.
Keywords: Hermansky-Pudlak syndrome, melanosome, Hps5, lysosome-related organelle, zebrafish
EUKARYOTIC cells contain lysosomes, membrane-bound organelles critical for the degradation of unneeded by-products of a cell. Some specialized cells also contain lysosome-related organelles (LROs), which are similar to lysosomes but are distinct in both morphology and function. LROs include melanosomes, found in melanocytes and the retinal pigmented epithelium (RPE), α- and δ-granules in platelet cells, and lytic granules in lymphocytes, among others. LROs are defined by having several characteristics common to the lysosome: they also form via the endocytic/secretory pathway, they have an acidic lumenal pH, and they contain lysosomal hydrolases and membrane proteins. However, LROs diverge from a typical lysosome in their patterns of morphogenesis, their unique contents, and their cell-specific functions. Studying their development can give insight into unique aspects of the endocytic/secretory pathway, as well as the specific pathologies underlying a variety of LRO-related diseases.
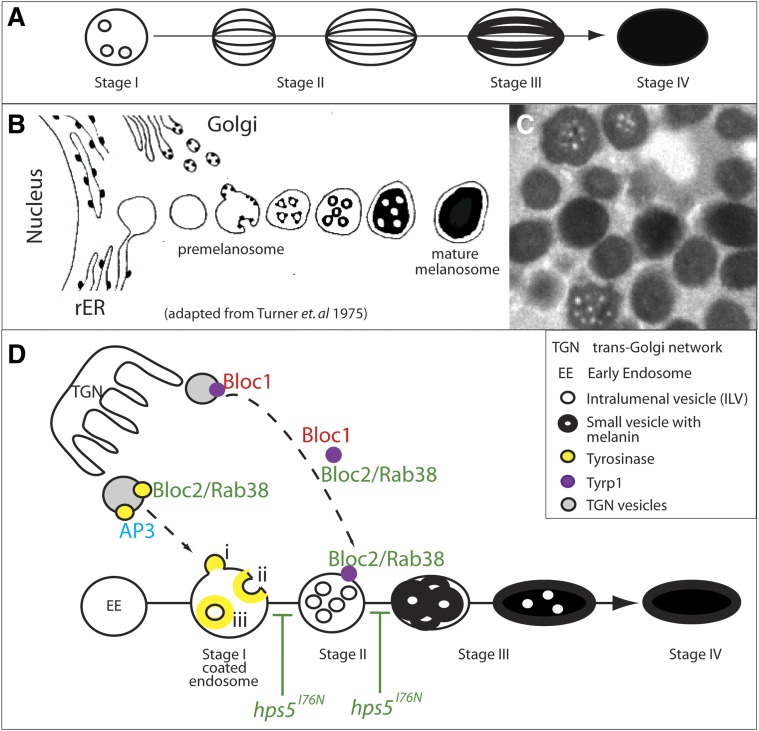
Melanosomes are the organelles responsible for the synthesis and storage of eumelanin and pheomelanin pigments within dermal melanocytes (melanophores in fish) and RPE cells. They have been one of the more extensively studied LROs, likely because of the ease of identifying hypopigmentation mutants (Swank et al. 1998; Huizing et al. 2008). Melanosomes are thought to arise from coated early endosomes, subsequently undergoing four stereotypic stages of maturation (Figure 7A) (Seiji et al. 1963a,b; see also Huizing et al. 2008; Sitaram and Marks 2012). A stage I premelanosome is thought to form when the limiting membrane of an early endosome invaginates, resulting in intralumenal vesicles (ILVs) (Berson et al. 2001; Raposo et al. 2001). Pmel17, a transmembrane glycoprotein, is then trafficked inside premelanosomes where it is cleaved and initiates the formation of fibrillar striations (Theos et al. 2005a). In stage II melanosomes, these Pmel17 striations arrange into sheets, forming an internal matrix. Enzymes to catalyze melanin synthesis are then trafficked to stage II melanosomes. Melanins are subsequently synthesized in stage III melanosomes and deposited upon the fibril striations, darkening the maturing organelle. By stage IV, the mature melanosome is fully pigmented. While mature melanosomes are transported out of melanocytes into keratinocytes, they remain permanent residents of RPE cells.
Figure 7.
Models of melanosome biogenesis. (A) Stereotypical stages of mammalian melanosome biogenesis. (B) Historical model of melanosome formation in goldfish based on EM data (adapted from Turner et al. 1975). (C) TEM image of melanosomes in a wild-type zebrafish RPE demonstrates that pigment deposition does not occur on fibrous striations as it does in mammals, but instead appears to collect outside internal vesicles. (D) Predicted function(s) of HPS complexes in trafficking Tyr and Tyrp1 to developing melanosomes (see Sitaram and Marks 2012 for review, but based on Seiji et al. 1963a,b; Raposo et al. 2001; Theos et al. 2005b; Di Pietro et al. 2006; Helip-Wooley et al. 2007; Setty et al. 2007; Huizing et al. 2008; Bultema et al. 2012). Bloc2-dependent transport of Tyr and/or Tyrp1 is possibly inhibited by hps5I76N at the indicated stages of melanosome development.
For melanin synthesis to occur, melanogenic enzymes must be trafficked from the Golgi to the developing melanosome. The most important of these enzymes are tyrosinase (Tyr), tyrosinase-related protein 1 (Tyrp1), and tyrosinase-related protein 2 (Tyrp2). Trafficking of organelle-specific proteins, such as these enzymes, into developing melanosomes requires precise and cell-type-specific regulation (Raposo et al. 2001). These trafficking pathways and their regulation have become better understood through the investigation of diseases affecting LRO biogenesis, including Chediak-Higashi syndrome, oculocutaneous albinism, and Hermansky-Pudlak syndrome, among others (see Sitaram and Marks 2012).
Hermansky-Pudlak syndrome (HPS) is a collection of phenotypically related but genetically heterogeneous human diseases all caused by defects in the synthesis, trafficking, or processing of LROs (Hermansky and Pudlak 1959). HPS is generally characterized by oculocutaneous albinism and bleeding diathesis and can also include pulmonary fibrosis, congenital nystagmus, reduced visual acuity, and platelet aggregation deficiency, depending on the disease subtype (Huizing et al. 2008). Nine human HPS-causing genes have been identified, each of which corresponds to a different HPS subtype (Oh et al. 1996; Dell’Angelica et al. 1999; Anikster et al. 2001; Suzuki et al. 2002; Li et al. 2003; Zhang et al. 2003; Morgan et al. 2006; Cullinane et al. 2011). Fifteen mouse mutant strains cause HPS-like phenotypes, 9 of which map to the mouse orthologues of human HPS genes (Wei 2006). Interestingly, HPS genes are ubiquitously expressed across all tissues, while the HPS phenotype is limited to the specialized cells containing LROs, suggesting that disease penetrance may be modified by the differential expression of cofactors or effector molecules (Wei 2006).
All HPS gene products identified thus far function as subunits in one of four multi-protein complexes, the three biogenesis of lysosome-related organelle complexes (Bloc1, Bloc2, and Bloc3) and the adaptor protein 3 (AP3) complex. These complexes function in trafficking enzymes and precursor molecules required for melanin synthesis to the forming melanosome (Huizing et al. 2008). While these factors are known to be critical for LRO biogenesis, their interactions and molecular functions within LROs are just beginning to be clarified.
With an interest in the critical role that the RPE plays in the development and function of the eye, we have studied several zebrafish pigmentation mutants (Ng et al. 2009; Nuckels et al. 2009). Here, we report the cloning and characterization of another pigmentation mutant, snow white (Stemple et al. 1996), in which we have identified a recessive, loss-of-function mutation in the Hermansky-Pudlak syndrome 5 (hps5) gene, hps5I76N. Hps5 functions in the Bloc2 complex, which also contains Hps3 and Hps6 (Zhang et al. 2003; Gautam et al. 2004), and the HPS5 gene is mutated in Hermansky-Pudlak syndrome type 5. A mutation in any one of the three Bloc2 subunits results in a relatively mild form of HPS, including partial albinism, mild nystagmus, a decrease in visual acuity, and symptoms of platelet aggregation defects, but no defects in pulmonary function (Wei 2006).
Bloc2 is involved in the trafficking of cargo, namely Tyr and Tyrp1, likely from the trans-Golgi network to stage II melanosomes (Helip-Wooley et al. 2005, 2007; Richmond et al. 2005; Di Pietro et al. 2006; Setty et al. 2007). Within Bloc2, Hps5 binds directly to Hps6, and neither have been shown to bind directly to Hps3; it remains uncertain if an unidentified bridging cofactor links Hps3 to Bloc2 (Figure 6B′) (Zhang et al. 2003; Di Pietro et al. 2004; Gautam et al. 2004). The I76N mutation in snow white (snw) is located in the only functional domain described for Hps5: a conserved N-terminal WD40 repeat, classically involved in protein–protein binding (Stirnimann et al. 2010; Xu and Min 2011). While this type of domain lends itself to many plausible hypotheses as to the function of a protein involved in the trafficking of other proteins, the specific subcellular function of the Hps5 WD40 repeat is unknown. Here, we report the characterization of ocular defects in snw/hps5 mutants, and we utilize the mutant to begin to determine how Hps5 functions within the Bloc2 complex.
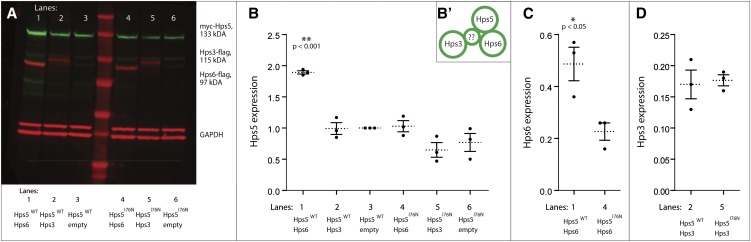
Figure 6.
Stabilization of expression resulting from Hps5/Hps6 cotransfection is disrupted by Hps5I76N. (A) COS7 cells were transiently cotransfected with Hps5WT (lanes 1, 2, and 3) or Hps5I76N (lanes 4, 5, and 6) as well as one of the following: Hps6 (lanes 1 and 4), Hps3 (lanes 2 and 5), or empty flag vector (lanes 3 and 6). On a fluorescent Western blot, Cy3-conjugated secondary antibody detects Hps5 in lanes 1–6 (green), and Cy5-conjugated secondary antibody detects Hps6 in lanes 1 and 4 and Hps3 in lanes 2 and 5, as well as GAPDH doublet loading control bands (red). (B) Quantification of Hps5 expression in A; dots represent n = 3 replicates normalized to Hps5WT/empty vector control (lane 3), mean ± SE. All expression values were initially background corrected and normalized to GAPDH-loading controls. Expression of Hps5WT is significantly increased when coexpressed with Hps6 (lane 1, **P < 0.001, one-way ANOVA and post-hoc Bonferroni multiple comparison tests), as compared to coexpression with either Hps3 or empty flag vector (lanes 2 or 3). In contrast, levels of Hps5I76N are not significantly different upon coexpression with either Hps6 or Hps3, as compared to coexpression with empty flag vector (lanes 4, 5, or 6). (B′) Within Bloc2, Hps5 has been shown to bind directly to Hps6 and possibly indirectly to Hps3 (Zhang et al. 2003; Gautam et al. 2004). (C) Expression of Hps6 is significantly increased when coexpressed with Hps5WT (lane 1, *P = 0.02, t-test), as compared to coexpression with Hps5I76N (lane 4). (D) Expression of Hps3 is not significantly different if coexpressed with either Hps5WT (lane 2) or Hps5I76N (lane 5, t-test).
Materials and Methods
Animals
Zebrafish (Danio rerio) were maintained at 28.5° on a 14-h light/10-h dark cycle. Embryos were obtained from the natural spawning of heterozygous carriers set up in pair-wise crosses. The snwm454 allele was used in this study (Stemple et al. 1996) and was obtained from the Zebrafish International Resource Center (ZIRC). Animals were treated in accordance with University of Texas at Austin Institutional Animal Care and Use Committee provisions.
Histology
Embryos were fixed in 1% paraformaldehyde (PFA)/2.5% gluteraldehyde/3% sucrose at 4°C overnight, washed 3× in PBS, fixed for 60 min in 1% osmium tetroxide in the dark at 4°C, washed 3× in PBS, and dehydrated (50, 70, 80, and 90% and 2 × 100% EtOH, 2× 100% propylene oxide). Embryos were gradually infiltrated with resin [20% Epon 812, 20% Araldyte 502, 60% dodecenylsuccinic anhydride (DDSA), 1% DMP-30] for 1 hr in 50% propylene oxide/50% resin and then overnight in 100% resin. Embryos were placed in fresh 100% resin and allowed to harden in a 60° oven for 3 days. Samples were sectioned on a Leica Ultracut UTC ultramicrotome at 1 μm and stained with 1% toluidine blue/1% Borax.
Melanin assay
The average amount of melanin in total fish was determined as described (Maldonado et al. 2006). Briefly, embryos were dechorionated, pooled (n = 30 embryos/stage, three biological replicates) in 350 μl homogenization buffer (20 mM Tris⋅HCl, 2 mM EGTA, 1 mM PMSF), and homogenized with a 25-gauge syringe. Following the addition of 500 μl, 2 M NaOH, and 100 μl DMSO, homogenates were incubated at 27° × 2 hr and centrifuged, and absorbance of the supernatant at 350 nm was measured on a Nanodrop spectrophotometer (Thermo Scientific). Samples were quantified using a melanin standard curve (0–100 μg melanin, Sigma Aldrich).
Immunohistochemistry
Embryos [5 days post fertilization (dpf)] were fixed in 4% PFA overnight, sucrose-protected, embedded in OCT tissue-freezing medium (TBS, Inc.), and sectioned at 12 μm on a Leica CM1850 cryostat. Following storage at −20°C, slides were thawed to room temperature (RT), and sections were circled with a PAP pen (Beckman Coulter). Sections were rehydrated in 1× PBS/0.1% tween 20/1% DMSO (PBTD) × 3 min and blocked in 5% normal goat serum/PBTD × 1 hr at RT in a humid chamber. Sections were incubated in primary antibody overnight at 4°, washed 3× for 10 min in PBTD at RT, and incubated in secondary antibody for 3 hr at RT in the dark. Slides were washed in PBTD for 5 min at RT and then stained with 1:4000 Sytox in blocking solution for 5 min at RT. Slides were washed 3× in PBTD for 10 min at RT, mounted with Vectashield (Vector Laboratories), and visualized on a LSM5 Pascal confocal microscope (Zeiss). Primary antibodies included 1:100 protein kinase (PKC) (bipolar cells, Santa Cruz Biotechnology), 1:200 Zpr1 (cone cells, ZIRC), 1:100 Zn8 (ganglion cells, ZIRC), and 1:200 Zpr3 (rod cells, ZIRC). Secondary antibodies included 1:200 Cy3-anti-rabbit or 1:200 Cy5-anti-mouse (Jackson Laboratories).
In situ hybridization
Hybridizations were performed essentially as described by Thisse and Thisse (2008) using digoxigenin-labeled antisense RNA probes. Genes were cloned from complementary DNA (cDNA), ligated into pGEM-T-Easy, and used for probe synthesis (cloning and probe details available upon request). Phenylthiourea-treated embryos [26 hours post fertilization (hpf)] from wild-type AB crosses were fixed overnight in 4% PFA at 4°, incubated in methanol (MeOH) for 15 min at RT, and then incubated overnight in fresh MeOH at −20°. Embryos were rehydrated for 5 min each in 75% MeOH, 25% 1× PBS/0.1% tween 20 (PBST), 50% MeOH/PBST, 25% MeOH/PBST, and washed 4× in PBST. Tissues were digested with 10 μg/ml proteinase K for 12 min, washed 2× in PBST, refixed in 4% PFA, washed 4× in PBST, and equilibrated in hybridization solution (50% formamide/5× SSC/0.1% Tween 20/5 mg/ml yeast transfer RNA/50 μg/ml heparin) for 3–5 hr at 60°. Embryos were incubated in antisense or sense probe (hps3, hps5, or hps6) overnight at 60°. Embryos were sequentially washed in 50% formamide/50% 2× sodium chloride/sodium citrate/tween 20 (SSCT) (20 min at 55°), 2× SSCT (3× 10 min at 37°), 0.2× SSCT (2 × 15 min at 55°), PBST (5 min at 37°), and PBST (5 min at RT); blocked in blocking solution (1% BSA/1% DMSO/0.3% Triton X100/0.25% Tween 20/2% normal goat serum/PBS) for 7–8 hr at RT; and incubated in 1:1000 anti-DIG antibody overnight at 4°. Embryos were washed in PBST, briefly incubated in staining buffer, and incubated in nitro blue tetrazolium/5-Bromo-4-Chloro-3-Indolyl-Phosphatase (NBT/BCIP) in the dark. When staining was complete, embryos were washed in PBST and imaged.
Transmission electron microscopy
Embryos were fixed in fresh 2% PFA/5% gluteraldehyde/0.1 M cacodylate at 4° overnight, stained with 2% OsO4 and 2% iron cyanide, and microwave-embedded in a SpurrEpon hard formulation using a BDMA accelerator (Electron Microscopy Services) via a 20, 40, 60, 80, 100, and 100% resin/acetone infiltration series. Samples were cut on a Leica Ultracut UTC ultramicrotome at a thickness of 70 nm and visualized on a FEI Tecnai transmission electron microscope at ×1250 magnification to locate the optic nerve for reference. Images (n = 3) of the RPE just dorsal to the optic nerve were collected from both eyes in n = 3 fish at ×11,500 magnification. Images were rotated to orient the RPE on a horizontal line. Melanosomes contained within RPE cells in an 800- × 600-pixel area (23 μm2) were counted and measured using Image-Pro Plus (Media Cybernetics); ectopic clusters of melanosomes were not included in counts.
Positional cloning
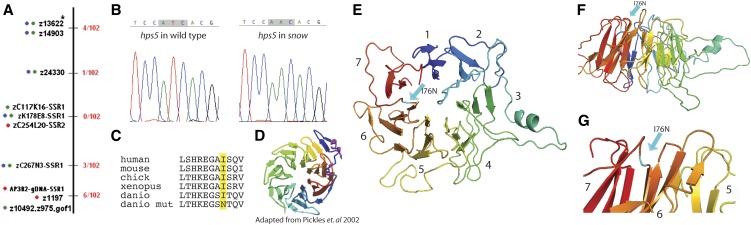
Bulked segregant analysis was performed using 96 snw mutants and 96 wild-type sibs (Willer et al. 2005). Linkage results positioned the snw mutation to chromosome 25 flanked by simple sequence-length polymorphism markers Z11092 and Z15260 (Knapik et al. 1998) at positions 5.1 and 15.2 cM, respectively. A critical interval likely to contain the snw mutation was determined to be between z24330 (1/102 recombinants) and newly designed simple sequence repeat (SSR) marker zC267N3-SSR1 (3/102 recombinants). Within this interval, newly designed SSR markers zC117K16-SSR1, zK178E8-SSR1, and zC254L20-SSR2 all showed 0/102 recombinants.
Protein modeling
Hps5 secondary and tertiary structures were predicted using Swiss-Model’s protein structure homology-modeling server (http://swissmodel.expasy.org/; Peitsch 1995; Arnold et al. 2006; Kiefer et al. 2009) in the automatic modeling mode. Both the full protein (1133 residues) and the N terminus (residues #1–370) of Hps5WT and Hps5I76N were modeled. Structures were visualized and rotated, and residue colors were adjusted using PyMol (http://www.pymol.org/). Quality assessment parameters were computed (see Supporting Information, Table S1, Table S2, Table S3, and Table S4).
Morpholino knockdown
A splice-blocking morpholino oligo (MO) targeting the exon 4/intron 4 boundary of hps5 was designed (5′-TGAAGGATTAAGAGGAATACCTGGT-3′, Gene Tools) and dissolved to a 1-mM stock solution in ddH2O. A standard control morpholino was used as a control (control MO). Morpholino (4 ng) plus phenol red and fluorescein dextran (tracking dyes) was injected into the yolk of one- to four-cell-stage embryos. Embryos were allowed to recover for 24 hr. Injection efficiency was detected by the presence of fluorescein dextran on a fluorescent microscope, and embryos with low or no fluorescence were removed. Embryos were collected at 24 and 48 hr, photographed, pooled in groups (uninjected, mild phenotype, severe phenotype), dissociated by syringe in 0.8 ml Trizol (Invitrogen), and stored at −80°. Subsequently, total RNA was extracted, and cDNA was synthesized using SuperScript reverse transcriptase II (Invitrogen). PCR analysis was performed to visualize hps5 transcript length (Forward: 5′-AGAGTTTGACTGTCTGGACCCCC-3′, Reverse: 5′-CCGCTCCTTATTACCCACACG-3′).
Messenger RNA injections
hps5WT and hps5snw sequences were amplified via PCR from cDNA derived from phenotypically wild-type and mutant snw embryos using proofreading 50× Advantage 2 polymerase mix (Clontech), cloned into pGEM-T Easy vector (Promega) and subsequently subcloned into pCS10R. hps5WT and hps5snw constructs were linearized, and messenger RNA (mRNA) was transcribed using mMessage mMachine (Invitrogen) and purified by phenol:chloroform extraction. hps5WT or hps5snw (4 ng) mRNA plus phenol red and fluorescein dextran was injected into the yolk of one-cell-stage embryos resulting from snw+/− × snw+/− crosses. Embryos were allowed to recover and grow for 48 hr; injection efficiency was detected by the presence of fluorescein dextran on a fluorescent microscope, and embryos with low or no fluorescence were removed. Remaining embryos were sorted into three categories: light, medium, or dark oculocutaneous pigmentation. Brightfield images were taken using a dissecting microscope; embryonic heads were subsequently processed for TEM analysis, while tails were individually genotyped (Forward: 5′-CGTGTGGAGTCCTTTCATCGG-3′, Reverse: 5′-CCAACAACCCCAAGTTTGTAATTG-3′).
Cell culture
COS7 cells were grown in culture in 10% FBS (Atlas Biologicals)/1% penicillin/streptomycin/DMEM (Gibco) in a dedicated incubator (5% CO2, 37°) according to standard procedures (see http://www.atcc.org, #CRL-1651).
DNA constructs
hps5WT, hps3, and hps6 were amplified via PCR from a wild-type D. rerio cDNA library using proofreading 50× Advantage 2 polymerase mix (Clontech), cloned into pGEM-T Easy, and subcloned into cytomegalovirus promoter-containing expression vectors (i) N-terminal 6× myc tag (a kind gift of John Wallingford) or (ii) C-terminal 3x flag tag (Sigma E4901, a kind gift of Steve Vokes). hps5I76N was created by site-directed mutagenesis of hps5WT (T227A) using the QuikChange Lightening kit (Agilent) according to the manufacturer’s instructions with the following primers: (Forward: 5′-ACAAAGAAGGCTCCAACACGCAGGTCTCCTG-3′, Reverse: 5′-CAGGAGACCTGCGTGTTGGAGCCTTCTTTGT-3′).
In vitro coexpression assays
COS7 cells (1.5 × 104 per well of six-well plates) grown overnight were transiently cotransfected with tagged hps DNA constructs (1 μg each) using 6 μl Xtreme Gene Transfection Reagent 9 (Roche) in 100 μl OptiMem serum-free media (Invitrogen). Transfection reactions were dripped onto cells in 1 ml fresh full serum media and grown overnight. Media was refreshed at 24 hr and cells were harvested at 48 hr. Cells were lysed in Zhang Lysis Buffer (see Zhang et al. 2003) supplemented with Complete Mini protease inhibitor cocktail (Roche) by pipetting and vortexing. Protein expression in whole-cell lysate was visualized by Western blot using a 4–12% Bis-Tris gel and transferred onto PVDF membrane (Invitrogen NuPage system), according to manufacturer’s instructions. Blots were incubated with three primary antibodies simultaneously: 1:2000 anti-flag (Sigma F1804), 1:2000 anti-myc (abcam ab9106), and 1:2000 anti-GAPDH (Santa Cruz sc-166545). Fluorescent-tagged secondary antibodies were utilized (1:2000 anti-mouse-Cy5 and 1:2000 anti-rabbit-Cy3), allowing detection and quantification of bands on a FluorChem Q machine (Protein Simple, Santa Clara, CA).
In vitro co-immunoprecipitation assays
COS7 cells (7 ×105 in 10-cm dish) grown overnight were transiently cotransfected with 5 ug of tagged hps DNA constructs in pairs using 30 μl Xtreme Gene Transfection Reagent 9 (Roche) in 1 ml OptiMem serum-free media (Invitrogen) for 5 min. Following transfection, full serum media (9 ml) was added, and cells were grown overnight. Media were refreshed at 24 hr and cells were harvested at 48 hr. Cells were lysed in Zhang Lysis Buffer (see Zhang et al. 2003) supplemented with Complete Mini protease inhibitor cocktail (Roche) by pipetting and vortexing. Protein G magnetic Sepharose 4 Fast Flow beads (GE Healthcare) were prepared by washing with lysis buffer. Cell lysate was precleared with an aliquot of beads, and 5 μg of either anti-Flag (Sigma F1804) or anti-Myc (Sigma M4439) antibody was used to pull down tagged proteins. Pull-downs were visualized by Western blot using a 4–12% Bis-Tris gel and transferred onto PVDF membrane (Invitrogen NuPage system) according to the manufacturer’s instructions and as described above. Empty vector transfections and no antibody pull-down controls were performed (data not shown).
Results
snw mutants display oculocutaneous albinism
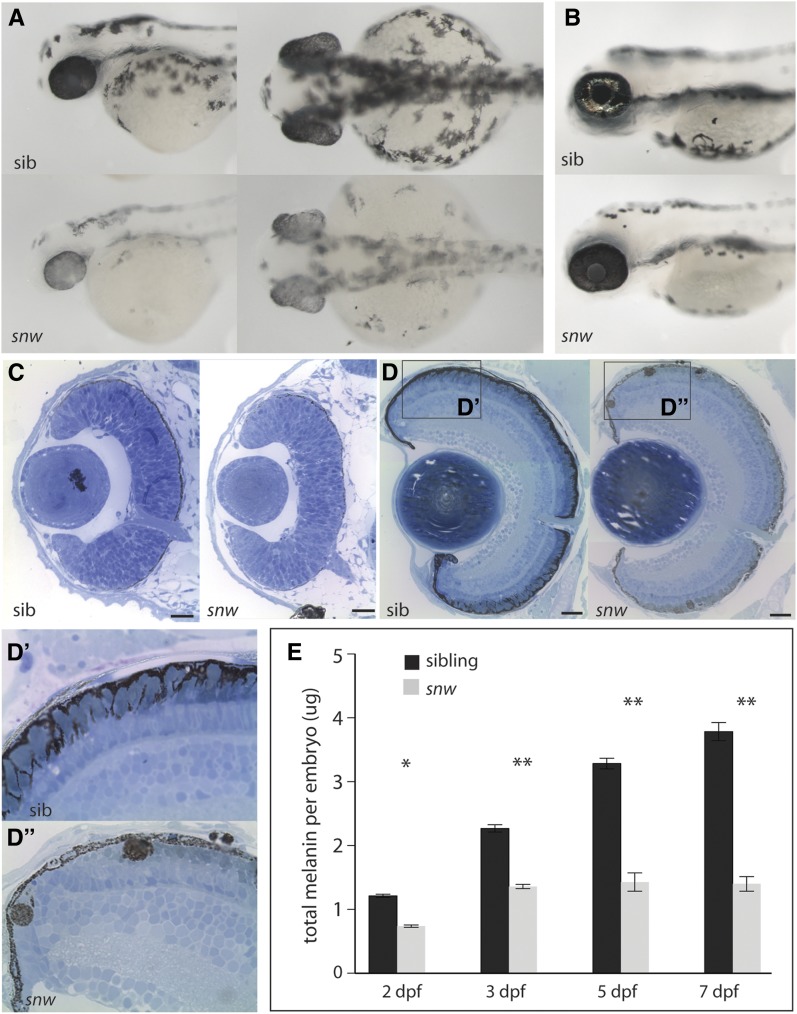
snw mutants display oculocutaneous hypopigmentation from 2 dpf through 7 dpf (Figure 1, A and B). snw embryos do not inflate their swim bladder and die between 7 and 12 dpf. As compared to sibling embryos, snw fish are variably microphthalmic at 2 dpf, with eye size in the mutants reaching that of wild-type siblings by 7 dpf, and they lack iridophores (Figure 1, A–C). Retinal morphology, lamination, and neuronal differentiation appears to be unaffected (Figure S1); however, subtle defects could be present that were not detected in our assays. The RPE in snw embryos appears hypopigmented in histological sections (Figure 1, C and D). snw mutants possess reduced pigmentation within the RPE cell body, and mature melanosomes were rarely observed in apical processes of RPE cells, as they were in phenotypically wild-type siblings (Figure 1, D′ and D′′). In snw mutants, melanosomes were also observed in ectopic, multi-melanosome clusters; these ectopic clusters were detected in the dorsal and central RPE (Figure 1D′′) throughout the retina (Figure 2) and in the choroid (Figure 1D′′). Quantification of total melanin in wild-type embryos revealed a steady increase from 2 to 7 dpf (Figure 1E). In contrast, melanin levels in snw embryos were significantly lower at all stages; there was a slight increase from 2 to 3 dpf, at which point levels plateaued.
Figure 1.
snow white mutants are hypopigmented and possess melanosomal defects. (A) snow white (snw) embryos present with oculocutaneous hypopigmentation beginning at 2 dpf and are microphthalmic: lateral and dorsal views. (B) At 4 dpf, hypopigmentation persists and no iridophores are apparent. (C) Histological sections through the 2 dpf snw eye (Bar, 20 nm). Dorsal is up in all histology sections. (D) At 7 dpf, the snw RPE remains less pigmented (Bar, 100 nm). (D′ and D′′) Melanosomes do not localize in the apical processes of snw RPE cells, as occurs in siblings. Instead, ectopic multi-melanosome clusters are evident in snw embryos throughout the retina and choroid. (E) Total melanin levels in snw embryos are significantly lower than in siblings from 2 through 7 dpf (two-way ANOVA, *P < 0.05, **P < 0.001; mean ± SE).
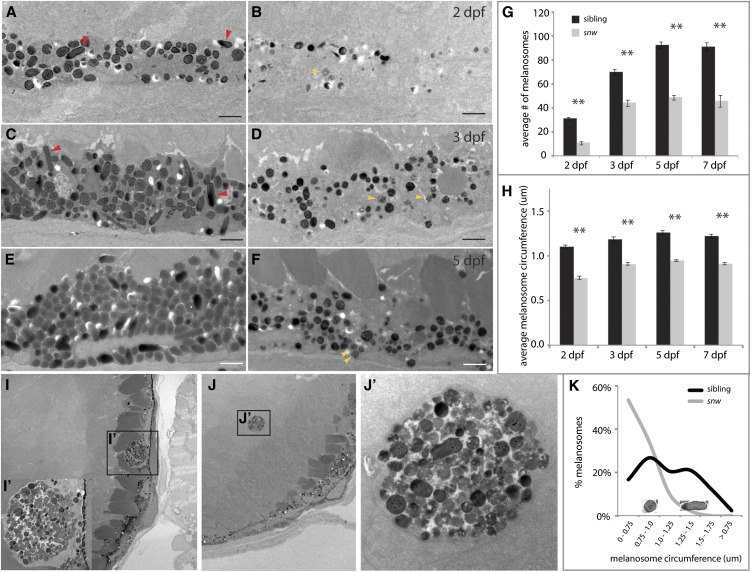
Figure 2.
The snw RPE contains fewer, smaller, and more immature melanosomes. (A–F) TEM reveals fewer melanosomes within the snw RPE (B, D, and F) as compared to phenotypically wild-type siblings (A, C, and E) throughout development (Bar, 1 μm). Red arrowheads indicate mature ellipsoid melanosomes; yellow arrowheads indicate immature melanosomes. (G) The average number of melanosomes in a 23-μm2 area of the RPE is significantly lower in snw embryos from 2 through 7 dpf (two-way ANOVA, **P < 0.001; mean ± SE). (H) The average circumference of snw melanosomes is also significantly smaller (two-way ANOVA, **P < 0.001; mean ± SE). (I and J) At 7 dpf, melanosomes mislocalize in ectopic clusters within the RPE (I and I′) and retina of snw mutants (J and J′). (K) The distribution of melanosome circumference is shifted toward smaller sizes in snw RPE as compared to siblings, and mature ellipsoid eumelanosomes are not observed. Images of round and ellipsoid melanosomes corresponding to size peaks in the graph are shown as an example; they are not to scale.
Melanosome density, size, shape, and localization are abnormal in snw mutants
To begin to elucidate the nature of the melanosome defects in snw mutants, the ultrastructure of the RPE was examined by TEM. TEM image analysis revealed that the density, size, shape, and localization of melanosomes were all altered in snw mutants (Figure 2). snw RPE cells appeared to contain substantially fewer melanosomes as compared to siblings (Figure 2, A–F). When quantified, the number of melanosomes in the wild-type RPE increased from 2 to 5 dpf and then remained level (Figure 2G), paralleling the increase in melanin (Figure 1E). In contrast, the number of melanosomes found within the snw RPE (but not including those in ectopic clusters) was significantly lower at all stages; an increase from 2 to 3 dpf was observed, after which the number of melanosomes did not change (Figure 2G). Furthermore, the average circumference of snw melanosomes was significantly smaller than sibling melanosomes at all time points examined (Figure 2H).
Eumelanosomes (containing eumelanin) complete the process of maturation by elongating to adopt an ellipsoid morphology; in contrast, pheomelanosomes (containing pheomelanin) remain round (Hirose and Matsumoto 1994; Kim and Choi 1998; Nguyen et al. 2002, Navarro et al. 2008). Both round and ellipsoid (red arrowheads) melanosomes were observed in the RPE of wild-type embryos, with a varying range of diameters (Figure 2, A, C, and E). Conversely, many melanosomes in the snw RPE remained immature, appearing as unpigmented stage I or II melanosomes or partially pigmented stage III melanosomes (yellow arrowheads, Figure 2, B, D, and F). Significantly fewer stage IV round pheomelanosomes were present in the snw RPE, and mature ellipsoid eumelanosomes were not detected. When melanosome circumference is viewed as a frequency distribution, distinct peaks corresponding to round and ellipsoid melanosomes are distinguishable in phenotypically wild-type sibling embryos (Figure 2K). However, in snw mutants, the peak corresponding to ellipsoid melanosomes is absent. Finally, as identified in histological sections, ectopic clusters of mislocalized multi-melanosomes in snw mutants are observed in TEM images (Figure 2, I, I′, J, and J′). Ultrastructural analysis reveals that, while some melanosomes in these clusters appear fully formed, others appear to be immature or possibly degrading (Figure 2J′).
Positional cloning identifies a mutation in hps5 as a candidate locus underlying the snw phenotype
To identify the affected locus in snw, an SSLP-based positional cloning strategy was utilized (see Materials and Methods for details). The snw mutation was mapped to a region of chromosome 25 between the markers z24330 and zC267N3-SSR1, which bracket a 1.26-Mb interval (25:1409767 to 25:2670814) (Figure 3A). This interval was predicted to contain 32 coding genes, and one of these, hps5, stood out as a significant potential candidate for underlying the snw phenotype. HPS5 is one of nine genes underlying Hermansky-Pudlak syndrome in humans, the phenotype of which includes hypopigmentation resulting from defects in melanosome biogenesis (Huizing et al. 2004). Thus, hps5 was cloned and sequenced from wild-type siblings and snw mutants; among 11 SNPs, a missense mutation was identified at base pair T227A. Base pairs 226–228 encode amino acid #76, an isoleucine (Ile) in wild-type embryos and an asparagine (Asn) in snw mutants (hps5I76N, Figure 3B). I76 is conserved in Hps5 orthologs across vertebrates (Figure 3C). The remaining 10 SNPs were found in zebrafish known to be genetically wild type at the snw locus, suggesting that they represent nonfunctional changes (Figure S2).
Figure 3.
hps5 is a candidate gene for the snow white locus. (A) Linkage mapping identified a 1.3-Mb region of chromosome 25 that contains the snw locus. Red numbers are number of recombination events/total embryos. (B) A candidate gene in this region, hps5, was sequenced and revealed a missense mutation in snw mutants, T227A (I76N). (C) Ile 76 resides in the predicted WD40 domain of Hps5 and is conserved across vertebrates. (D) The crystal structure of a classic WD40 repeat domain is a seven-bladed propeller, where each blade is made up of three to four β-sheets (adapted from Pickles et al. 2002). (E) The modeled protein structure of amino acids #31–358 of D. rerio Hps5. A seven-bladed propeller made up of β-sheets is predicted by sequence homology. The N terminus is in red, C terminus is in blue, and propeller blades are labeled #1–7 according to convention. Residue #76 is highlighted in teal and resides between blades #6 and #7 (arrow). (F) Side view of the D. rerio predicted Hps5 structure shown in E. Residue #76 is located on the top of the propeller core, outside of the β-sheets (teal arrow). (G) Zoomed-in view of F; residue I76N is highlighted by teal arrow; blades #5–7 are labeled for reference.
The I76N mutation occurs in a predicted WD40 repeat at the N terminus of Hps5. WD40 repeat domains typically form a seven- or eight-bladed propeller core, where each blade is made of three or four β-sheets (Figure 3D). D. rerio Hps5 sequence was modeled by homology using Swiss-Model to predict secondary and tertiary structure (Peitsch 1995; Arnold et al. 2006; Kiefer et al. 2009). Modeling of zebrafish Hps5 resulted in a classic WD40 propeller core structure at the N terminus (Figure 3E). The I76N residue was positioned at the top of the propeller, outside of the predicted β-sheets, between blades 6 and 7 (Figure 3, F and G). The opening at the top surface of WD40 propeller cores is thought to be where binding to other proteins occurs (Stirnimann et al. 2010; Xu and Min 2011). Model quality estimation parameters are given for reference (Table S1, Table S2, Table S3, and Table S4).
hps5, hps3, and hps6 are expressed in the zebrafish eye
Despite the fact that only a few specific cell types are affected in HPS patients and animal models, HPS gene products have been found to be expressed in all tissue types examined thus far (Wei 2006). To confirm the hypothesis that hps5, hps3, and hps6 are present in the developing zebrafish embryo as they are in mice and humans, we performed in situ hybridization on the three Bloc2 complex members. As expected, all three are expressed throughout the zebrafish, including in the developing eye and RPE (Figure S3).
Knockdown of hps5 by a morpholino antisense oligo recapitulates the snw phenotype
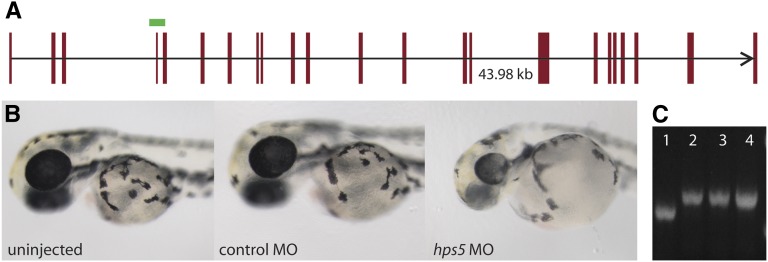
To determine if Hps5 is required for normal pigmentation, a splice-blocking morpholino designed against the hps5 exon 4/intron 4 boundary was injected into one-cell-stage wild-type embryos (Figure 4A). After 48 hr, embryos injected with hps5 MO resembled snw mutants (Figure 4B). Embryos injected with an equal amount of a standard control MO developed normally. Amplification of hps5 transcript from embryos injected with hps5 MO revealed the inclusion of the 84-bp intron 4 (Figure 4C), resulting in a translational frameshift and premature stop codon (data not shown).
Figure 4.
Morpholino knockdown of hps5 phenocopies snw. (A) The predicted genomic organization of hps5 (red bars) covers 43.98 kb on the reverse strand of chromosome 25 (oriented in the forward direction). A splice-blocking antisense MO targeting hps5 at the exon 4/intron 4 boundary (green bar) results in the inclusion of intron 4 in the hps5 transcript, a frameshift, and a premature stop codon. (B) Wild-type zebrafish injected with hps5 MO phenocopy snw embryos, displaying oculocutaneous hypopigmentation and microphthalmia at 48 hpf. (C) The hps5 transcript has incorporated the 84-bp intron 4 as a result of hps5 MO injection, demonstrated by RT-PCR. Lane 1, uninjected; lanes 2–4, three unique replicates of hps5 MO-injected pools of embryos.
snw phenotype is rescued by injection of hps5WT, but not hps5snw, mRNA
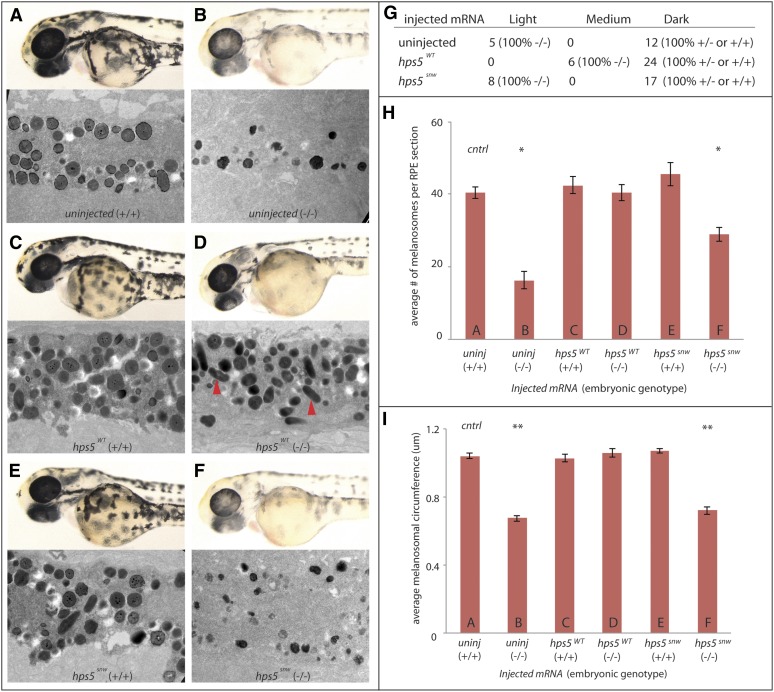
While morpholino injection data support a role for Hps5 in regulating pigmentation, these results do not unambiguously support the hypothesis that a loss of hps5 function is the mechanism underlying pigmentation defects in snw mutants. Thus, to further test this hypothesis, mRNA rescue experiments were performed to determine if exogenous hps5WT or hps5snw mRNA could rescue pigmentation defects in snw mutants. Embryos derived from a snw+/- incross were injected with either hps5WT or hps5snw mRNA and allowed to develop for 48 hr alongside uninjected controls. Embryos were then sorted into three pigment categories (light, medium, or dark), individually genotyped for the putative T227A SNP of hps5, and analyzed by TEM to determine the effects on melanosome phenotype in the RPE.
Uninjected embryos categorized as “dark” and genotyped as homozygous wild type or heterozygous for the hps5I76N SNP displayed expected pigmentation levels, densities of melanosomes within the RPE, and melanosome shapes and sizes (Figure 5, A, G, H, and I). Similarly, all uninjected control embryos categorized as “light” displayed expected hypopigmentation and melanosome defects and were confirmed as homozygous hps5I76N mutants (Figure 5, B, G, H, and I). Injection of hps5WT into homozygous wild-type or heterozygous embryos did not alter normal melanosome development, indicating that there was no overexpression phenotype (Figure 5, C, H, and I). Injection of hps5WT into homozygous mutant embryos, however, resulted in a substantial rescue in overall pigmentation of whole embryos and restored melanosome density, size, and shape to normal levels (Figure 5, D, H, and I). Mature ellipsoid eumelanosomes were observed in hsp5WT-injected mutant embryos (Figure 5D, red arrowheads). Injection of hps5snw into homozygous wild-type or heterozygous embryos did not result in any changes in pigmentation or melanosome phenotype (Figure 5, E, H, and I). Injection of hps5snw into mutant fish did not rescue pigmentation and had little effect on melanosome formation (Figure 5, F, H, and I). These data support a model in which hps5I76N underlies the snw phenotype and suggest that the WD40 domain of hps5 is required for normal melanosome biogenesis.
Figure 5.
hps5WT, but not hps5snw, rescues the snw phenotype. (A and B) Pigmentation and melanosome phenotype in uninjected control wild-type sibling and mutant embryos derived from snw+/- incrosses. Labels follow the convention type of mRNA injected (embryonic genotype). (C) Wild-type embryos (+/+) injected with hps5WT mRNA demonstrate that exogenous hps5 does not alter melanosome phenotype. (D) snw embryos (−/−) injected with hps5WT mRNA show marked rescue of hypopigmentation levels; examination of RPE ultrastructure by TEM highlights a rescue of melanosome formation, including the presence of mature ellipsoid eumelanosomes (red arrowheads). (E) hps5snw mRNA does not lead to a phenotype when injected in wild-type embryos (+/+). (F) snw embryos (−/−) injected with hps5snw mRNA show little effect on overall pigmentation or melanosome formation as compared to uninjected mutant controls. (G) At 48 hpf, zebrafish injected with hps5WT or hps5snw mRNA, as well as uninjected control embryos, were visually sorted into three pigmentation categories—light, medium, and dark—and individually genotyped. (H) The average number of melanosomes was significantly lower in uninjected snw-/- embryos (B) and snw-/- embryos injected with hps5mut mRNA (F) when compared to uninjected wild-type controls (A) (*P < 0.01, one-way ANOVA and post-hoc Dunnett’s multiple comparison test). Injection of hps5WT mRNA into snw-/- embryos rescued melanosome number (D). Labels A–F in the bar graphs in H and I refer to the samples shown in panels A–F (n = 3 embryos per condition, six eyes). (I) Average melanosome circumference in snw-/- embryos injected with hps5WT mRNA was also rescued to wild-type levels (D); this rescue did not occur with injected hps5mut mRNA (F) (**P < 0.001).
Hps5I76N binds to both Hps6 and Hps3 in vitro
Hps5 is known to function within the Bloc2 complex (Zhang et al. 2003). A parsimonious hypothesis to explain the deleterious effects of hps5I76N would be that the I76N mutation in the predicted WD40 domain prevents Hps5 from binding to Bloc2 complex partners, Hps3, and/or Hps6. We tested this hypothesis by transiently cotransfecting zebrafish hps3, hps6, hps5WT, hps5I76N, and/or empty vector DNA constructs in pairwise combinations into COS7 cells to determine the effect of the mutation on the ability of Hps5 to interact with its complex partners in vitro. Co-immunoprecipitation pull-down assays demonstrated that Hps5WT and Hps5I76N were each able to pull down Hps6 and Hps3 in an in vitro culture system (Figure S4). These data demonstrate that the I76N mutation does not prevent Hps5 from binding to these two complex partners.
Hps5WT expression is stabilized by Hps6, while Hps5I76N is not
Because Hps5I76N was able to co-immunoprecipitate with both Hps6 and Hps3 under the given in vitro conditions, we tested the complementary hypothesis that hps5I76N expression instead affects the overall stability or expression level of either itself or other components within the Bloc2 complex. cDNA constructs encoding zebrafish hps3, hps6, hps5WT, hps5I76N, and/or empty vector were again transiently cotransfected in pairwise combinations into COS7 cells, this time to determine the effect of the hps5I76N mutation on in vitro levels of expression (Figure 6). Expression levels were quantified from fluorescent Western blots (Figure 6A) and graphed for comparison and statistical analysis (Figure 6, B, C, and D).
When cotransfected with empty flag vector, Hps5WT and Hps5I76N were detected at similar levels; the I76N mutation did not significantly alter Hps5 levels, in vitro (Figure 6, A and B). Similarly, the levels of Hps5WT and Hps5I76N were not significantly different from each other when coexpressed with Hps3, nor were they altered from coexpression levels with empty vector. However, Hps5WT levels were nearly double those of Hps5I76N when each was coexpressed with Hps6. Thus, it appears that Hps5WT expression is stabilized by the presence of Hps6 and in a way that Hps5I76N is not.
Hps6 expression is stabilized by Hps5WT, but not by Hps5I76N
Expression levels of Bloc2 complex members Hps6 and Hps3 were also examined to determine if Hps5I76N affected the levels of its binding partners. Hps6 levels were significantly higher when coexpressed with Hps5WT, as compared to Hps6 levels when coexpressed with Hps5I76N (Figure 6, A and C). In contrast, Hps3 levels remained unchanged when coexpressed with either version of Hps5 (Figure 6, A and D). These data indicate that, while the I76N mutation does not prevent the ability of Hps5 to interact with Hps6 or Hps3, it does decrease the stability of the Hps5–Hps6 interaction in vitro.
Discussion
Melanosome maturation is disrupted in snw/hps5 mutants
Here, we report identification of the first zebrafish model of Hermansky-Pudlak syndrome to date, snow white, which contains a missense mutation in the predicted WD40 domain at the N terminus of Hps5. snw/hps5 mutants display oculocutaneous hypopigmentation, resulting from decreased total levels of melanin, and melanosome defects characterized by significantly reduced numbers, smaller sizes, and immaturity (Figure 1). Mutants possess an increase in early stage melanosomes, with a substantial reduction in the number of mature stage IV melanosomes (Figure 2). Moreover, ectopic multi-melanosome clusters were detected throughout snw/hps5 mutant eyes, further indication of defects in melanosome biogenesis.
Defects in snw/hps5 mutants are nearly identical to those observed in mouse Hps5 mutants and human HPS5 patients (Nguyen et al. 2002; Zhang et al. 2003; Gautam et al. 2004; Huizing et al. 2004; Korswagen et al. 2008; Hirobe et al. 2011). In the mouse model of Hps5 (ruby-eye 2), ellipsoid eumelanosomes are also absent (Nguyen et al. 2002). Moreover, multi-melanosomes are detected in the choroid of mice possessing mutations in the genes encoding any of the three Bloc2 subunits (Zhang et al. 2003; Gautam et al. 2004). Interestingly, multi-melanosomes are restricted to Bloc2 mutants; they are not detected in animals with mutations in the genes encoding Bloc1, Bloc3, or AP3 subunits (Zhang et al. 2003; Dell’Angelica 2004; Wei 2006). While giant or macromelanosomes have been seen in other HPS mouse models (e.g., Hps1 and Hps4), these structures are quite different from the multi-melanosomes described here and in other Bloc2 mutants and are presumably produced by a different cellular mechanism (Gardner et al. 1997; Horikawa et al. 2000; Suzuki et al. 2002). Not found in mammals, iridophores are pigment cells containing crystallized purines, likely guanine, which stack into plates and reflect light (Fujii 2000). The biochemical and molecular underpinnings of iridophore formation and pigmentation are not well understood; however, it is possible that the iridophore defects observed in hps5I76N embryos reflect a role for Hps5 in this process.
hps5I76N underlies the snw phenotype
Positional cloning identified hps5 as a candidate underlying the snow white phenotype (Figure 3), and a missense mutation, I76N, was found within the predicted N-terminal WD40 repeat domain of Hps5 in snw embryos. hps5WT injection into snw embryos rescued overall RPE pigmentation, melanosome size, density, maturation, and shape, while injection of hps5snw did not rescue pigmentation or melanosome formation. Additionally, overexpression of hps5 in phenotypically wild-type embryos did not result in hyperpigmentation. Thus, while we cannot rule out the possibility that hps5WT overexpression acts epistatically to the mutation in snw, the more parsimonious explanation for these data is that they support a model in which hps5I76N is the mutation underlying pigmentation defects observed in snw mutants.
hps5I76N is the first nontruncation mutation characterized in the hps5 WD40 domain
Mutations in HPS5 have been identified in human HPS patients (Zhang et al. 2003; Huizing et al. 2004; Korswagen et al. 2008; Carmona-Rivera et al. 2011), and several mutant alleles exist in mouse (ruby-eye 2, Nguyen et al. 2002; Zhang et al. 2003; Hirobe et al. 2011) and in Drosophila (pink, Falcón -Perez et al. 2007; Syrzycka et al. 2007). Importantly, almost all of the characterized mutations in mouse or Drosophila result in a frameshift and premature truncation of Hps5. The only two exceptions, to our knowledge, are two ENU-induced recessive mutations in mouse, ru2Btlr and ru22Btlr (Hirobe et al. 2011). These each result in a transition and transversion mutation, respectively, located towards the C terminus of Hps5 and outside of the predicted WD40 repeat. While deletion mutants have been useful thus far in characterizing the functions of Hps5 in pigmentation and melanosome biogenesis in mouse and Drosophila, they do not allow a mechanistic biochemical analysis of Bloc2 complex formation or function. Here, we begin to investigate this question and reveal that at least part of the WD40 domain of Hps5 is required for stabilization of the interaction between Hps5 and Hps6. The I76N mutation identified here in snw represents a unique in vivo model through which structure–function analyses of Hps5 can now be performed.
Hps5/Hps6 interaction is destabilized by I76N
WD40 protein domains are characterized by a seven- or eight-bladed propeller core, where each blade is made up of β-sheets (Smith et al. 1999; Stirnimann et al. 2010; Xu and Min 2011). The core has a top surface, characterized by a narrow opening, and a second, wider opening at the bottom surface. It is thought that the top surface of the core is primarily the functional part of the WD40 domain, where protein–protein binding occurs (Smith et al. 1999; Stirnimann et al. 2010; Xu and Min 2011). Modeling the tertiary structure of D. rerio Hps5 revealed a predicted propeller core (Figure 3). Isoleucine 76 was predicted to localize to the top of the propeller, presumably where Hps5 interacts with other proteins. This is consistent with a model in which Hps5 interactions with its Bloc2 partners (Hps3, Hps6) might be prevented by the I76N mutation; however, data from in vitro co-immunoprecipitation assays demonstrated that at least some binding occurs (Figure S4).
A second hypothesis was thus tested: perhaps the expression of Bloc2 subunits would be affected by the I76N mutation. Through yeast two-hybrid studies, it is known that Hps5 and Hps6 bind directly to one another (Zhang et al. 2003; Gautam et al. 2004). Interestingly, the levels of both Hps5WT and Hps6 in COS7 cells were significantly increased by the presence of the other protein (Figure 6). In contrast, Hps5I76N and Hps6 were not able to stabilize each other, despite the fact that they co-immunoprecipitated in a complex (Figure S4). The change from the nonpolar, strongly hydrophobic isoleucine (I) to the polar amide asparagine (N) at position #76 could alter the conformation or the interaction between the complex, leading to a more transient Hps5/Hps6 association. Furthermore, although Hps5I76N can still bind to Hps6 and allow co-immunoprecipitation, perhaps the integrity of the complex is altered such that it is now susceptible to degradation. In summary, these data are consistent with the hypothesis that the interaction between these proteins (and/or additional proteins) is mediated by the WD40 repeat of Hps5 and is necessary for Bloc2 stability.
Protein transport to the developing melanosome may be disrupted in hps5I76N mutants
Maturation of the developing melanosome follows stereotypical stages, beginning with the formation of ILVs via invagination of the coated endosome limiting membrane; in mammals, these vesicles then mediate formation of fibrillar striations enriched with Pmel17 (Figure 7A) (Berson et al. 2001; Raposo et al. 2001; Marks and Seabra 2001). Pmel17 and the melanosomal matrix subsequently facilitate melanin synthesis. Following classic studies in mammalian melanocytes (Seiji et al. 1963a,b), early ultrastructural studies of the goldfish eye produced data describing melanosome maturation (Turner et al. 1975; Abramowitz et al. 1977; Kajishima and Takeuchi 1977). Interestingly, while many components of this pathway appear to be conserved across vertebrates, it has been noted that fibrillar striations do not develop in fish; instead, ILVs appear to remain intact inside the developing melanosome, acting as a substrate for melanin deposition themselves (Figure 7, B and C) (Navarro et al. 2008).
The synthesis of melanin pigments within the melanosome requires the highly regulated trafficking of specific precursor molecules and catalytic enyzmes to the developing organelle. For example, transport of the tyrosinase family of enzymes (Tyr, Tyrp1, and Tyrp2 most commonly) from the Golgi into vesicles and onward to maturing melanosomes requires the function of AP1, AP3, Bloc1, and Bloc2 (for review, see Huizing et al. 2008; Sitaram and Marks 2012). It is currently thought that Bloc1/Bloc2 and Bloc1/AP3 function in parallel Tyrp1 trafficking pathways, while Bloc1/AP1 and Bloc2/AP3 mediate parallel Tyr pathways (Figure 7D) (Sitaram and Marks 2012; Bultema and Di Pietro 2013). However, many of the specific molecular interactions between these Bloc trafficking complexes, their cargo, and downstream fusion events to the developing melanosome remain unclear.
Indeed, the mechanisms by which Hps5 and Bloc2 function during melanosome formation and melanogenesis are unknown, as is true for almost all HPS proteins. Current data suggest that Bloc1 is involved in the sorting and budding of Tyr- and Tyrp1-containing vesicles from the trans-Golgi network, while Bloc2 may be involved in targeting or mediating fusion between small vesicles containing Tyr and/or Tyrp1 in the forming melanosome (Figure 7D) (Wei 2006; Di Pietro et al. 2006; Helip-Wooley et al. 2007; Setty et al. 2007; Huizing et al. 2008; Sitaram and Marks 2012). Interestingly, eumelanin production requires Tyr, Tyrp1, and Tyrp2, while pheomelanogenesis requires only Tyr (Prota et al. 1995). Although untested, our data are consistent with the hypothesis that, in the snw/hps5I76N mutant, reduced Bloc2-dependent transport of Tyrp1 inhibits eumelanosome biogenesis (Figure 7D).
Using the zebrafish to study HPS and LRO biogenesis
Zebrafish are an excellent model system to investigate the molecular underpinnings of Hermansky-Pudlak syndrome and elucidate the biology of LROs. Pigment is first observed in the embryo at ∼24 hpf, and hypopigmentation mutants are easily identified soon thereafter. To this end, several zebrafish mutants, including fading vision (fdv; pmel17), fade out (fad; unknown locus), platinum (plt; vps11), vps18, and leberknodel (lbk; vps39), have already been characterized for LRO dysfunction; Pmel17 is critical for melanosome formation, as described above, and vps11, vps18, and vps39 are part of the HOPS complex required for vesicle fusion and tethering (Schonthaler et al. 2005, 2008; Bahadori et al. 2006; Maldonado et al. 2006; Thomas et al. 2011; see Navarro et al. 2008 for review). Given the ability to perform forward genetic screens in zebrafish, the relative ease of cloning mutations from these screens (Bibliowicz et al. 2011; Gestri et al. 2012; Obholzer et al. 2012), and the ability to generate targeted mutations (Cade et al. 2012; Dahlem et al. 2012; Moore et al. 2012; Blackburn et al. 2013; Hwang et al. 2013), the identification and generation of additional zebrafish models of human albinism disorders should provide a promising complement through which the molecular and cellular underpinnings of these disorders can be elucidated.
Supplementary Material
Acknowledgments
We thank Haley Tucker and Melissa Popowski for assistance with cell culture, Dwight Romanovicz for assistance with TEM, Andrea Hartsock for helpful discussions, Diana Ross for technical assistance, Jeremy O’Connell for protein modeling expertise, Jiwoon Lee for in situ hybridization assistance, and John Wallingford and Steven Vokes for vectors. C.M.S.D. thanks John Dowling for additional support during this project. This work was supported by a CAREER Award from the National Science Foundation to J.M.G. (IOS-0745782), a fellowship grant from the Knights Templar Eye Foundation to C.M.S.D., and National Institutes of Health (NIH) grant RR020357 to R.G. Antibodies were obtained from Zebrafish International Resource Center, which is supported by NIH–National Center for Research Resources grant P40 RR012546.
Footnotes
Communicating editor: M. Halpern
Literature Cited
- Abramowitz J., Turner W. A., Chavin W., Tayler J. D., 1977. Tyrsinase positive oculocutaneous albinism in the goldfish, Carassius auratus l., and ultrastructural and biochemical study of the eye. Cell Tissue Res. 182(3): 409–419. [DOI] [PubMed] [Google Scholar]
- Anikster Y., Huizing M., White J., Shevchenko Y. O., Fitzpatrick D. L., et al. , 2001. Mutation of a new gene causes a unique form of Hermansky–Pudlak syndrome in a genetic isolate of central Puerto Rico. Nat. Genet. 28: 376–380. [DOI] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T., 2006. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- Bahadori R., Rinner O., Schonthaler H. B., Biehlmaier O., Makhankov Y. V., et al. , 2006. The zebrafish fade out mutant: a novel genetic model for Hermansky-Pudlak syndrome. Invest. Ophthalmol. Vis. Sci. 47: 4523–4531. [DOI] [PubMed] [Google Scholar]
- Berson J. F., Harper D. C., Tenza D., Raposo G., Marks M. S., 2001. Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol. Biol. Cell 12: 3451–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibliowicz J., Tittle R. K., Gross J. M., 2011. Toward a better understanding of human eye disease insights from the zebrafish, Danio rerio. Prog. Mol. Biol. Transl. Sci. 100: 287–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn P. R., Campbell J. M., Clark K. J., Ekker S. C., 2013. The CRISPR system: keeping zebrafish gene targeting fresh. Zebrafish 10(1): 116–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultema J., Di Pietro S., 2013. Cell type-specific Rab32 and Rab38 cooperate with the ubiquitous lysosome biogenesis machinery to synthesize specialized lysosome-related organelles. Small GTPases 4(1): 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultema J., Ambrosio A., Burek C., Di Pietro S., 2012. BLOC-2, AP-3, and AP-1 proteins function in concert with Rab38 and Rab32 proteins to mediate protein trafficking to lysosome-related organelles. J. Biol. Chem. 287(23): 19550–19563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade L., Reyon D., Hwang W. Y., Tsai S. Q., Patel S., et al. , 2012. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 40(16): 8001–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Rivera C., Golas G., Hess R. A., Cardillo N. D., Martin E. H., et al. , 2011. Clinical, molecular, and cellular features of non-Puerto Rican Hermansky-Pudlak syndrome patients of Hispanic descent. J. Invest. Dermatol. 131(12): 2394–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinane A. R., Curry J. A., Carmona-Rivera C., Summers C. G., Ciccone C., et al. , 2011. BLOC-1 mutation screen reveals PLDN (Pallidin) is mutated in Hermansky–Pudlak syndrome type 9 (HPS-9). Am. J. Hum. Genet. 88: 778–787. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., et al. , 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8(8): e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica E. C., 2004. The building BLOC(k)s of lysosomes and related organelles. Curr. Opin. Cell Biol. 16(4): 458–464. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica E. C., Shotelersuk V., Aguilar R. C., Gahl W. A., Bonifacino J. S., 1999. Altered trafficking of lysosomal proteins in Hermansky–Pudlak syndrome due to mutations in the b3A subunit of the AP-3 adaptor. Mol. Cell 3: 11–21. [DOI] [PubMed] [Google Scholar]
- Di Pietro S. M., Falcón-Pérez J. M., Dell’Angelica E. C., 2004. Characterization of BLOC-2, a complex containing the Hermansky-Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic 5(4): 276–283. [DOI] [PubMed] [Google Scholar]
- Di Pietro S. M., Falcón-Pérez J. M., Tenza D., Setty S. R., Marks M. S., et al. , 2006. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol. Biol. Cell 17(9): 4027–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón-Pérez J. M., Romero-Calderon R., Brooks E. S., Krantz D. E., Dell’Angelica E. C., 2007. The Drosophila pigmentation gene pink (p) encodes a homologue of human Hermansky-Pudlak syndrome 5 (HPS5). Traffic 8(2): 154–168. [DOI] [PubMed] [Google Scholar]
- Fujii R., 2000. The regulation of motile activity in fish chromatophores. Pigment Cell Res. 13: 300–319. [DOI] [PubMed] [Google Scholar]
- Gardner J. M., Wildenberg S. C., Keiper N. M., Novak E. K., Rusiniak M. E., et al. , 1997. The mouse pale ear (ep) mutation is the homologue of human Hermansky–Pudlak syndrome. Proc. Natl. Acad. Sci. USA 94: 9238–9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam R., Chintala S., Li W., Zhang Q., Tan J., et al. , 2004. The Hermansky-Pudlak syndrome 3 (cocoa) protein is a component of the biogenesis of lysosome-related organelles complex-2 (BLOC-2). J. Biol. Chem. 279(13): 12935–12942. [DOI] [PubMed] [Google Scholar]
- Gestri G., Link B. A., Neuhauss S. C., 2012. The visual system of zebrafish and its use to model human ocular diseases. Dev. Neurobiol. 72(3): 302–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helip-Wooley A., Westbroek W., Dorward H., Mommaas M., Boissy R. E., et al. , 2005. Association of the Hermansky-Pudlak syndrome type-3 protein with clathrin. BMC Cell Biol. 6: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helip-Wooley A., Westbroek W., Dorward H. M., Koshoffer A., Huizing M., et al. , 2007. Improper trafficking of melanocyte-specific proteins in Hermansky-Pudlak syndrome type-5. J. Invest. Dermatol. 127(6): 1471–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansky F., Pudlak P., 1959. Albinism associated with hemorrhagic diathesis and unusual pigmented reticular cells in the bone marrow: report of two cases with histochemical studies. Blood 14(2): 162–169. [PubMed] [Google Scholar]
- Hirobe T., Yoshihara C., Takeuchi S., Wakamatsu K., Ito S., et al. , 2011. A novel deletion mutation of mouse ruby-eye 2 named ru2d/Hps5ru2-d inhibits melanocyte differentiation and its impaired differentiation is rescued by L-tyrosine. Zoolog. Sci. 28: 790–801. [DOI] [PubMed] [Google Scholar]
- Hirose E., Matsumoto J., 1994. Differentiation of pigmented tissues in the eyes of medaka (Oryzias latipes) embryos: an electron microscopy study. Fish Biol. J. Medaka 6: 25–34. [Google Scholar]
- Horikawa T., Araki K., Fukai K., Ueda M., Ueda T., et al. , 2000. Heterozygous HPS1 mutations in a case of Hermansky-Pudlak syndrome with giant melanosomes. Br. J. Derm. 143: 635–640. [DOI] [PubMed] [Google Scholar]
- Huizing M., Hess R., Dorward H., Claassen D. A., Helip-Wooley A., et al. , 2004. Cellular, molecular and clinical characterization of patients with Hermansky-Pudlak syndrome type 5. Traffic 5(9): 711–722. [DOI] [PubMed] [Google Scholar]
- Huizing M., Helip-Wooley A., Westbroek W., Gunay-Aygun M., Gahl W. A., 2008. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu. Rev. Genomics Hum. Genet. 9: 359–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31(3): 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajishima T., Takeuchi I. K., 1977. Ultrastructural analysis of gene interaction and melanosome differentiation in the retinal pigment cells of the albino goldfish. J. Exp. Zool. 200(3): 349–357. [DOI] [PubMed] [Google Scholar]
- Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T., 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37: D387–D392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I. T., Choi J. B., 1998. Melanosomes of retinal pigment epithelium: distribution, shape, and acid phosphatase activity. Korean J. Ophthalmol. 12: 85–91. [DOI] [PubMed] [Google Scholar]
- Knapik E. W., Goodman A., Ekker M., Chevrette M., Delgado J., et al. , 1998. A microsatellite genetic linkage map for zebrafish (Danio rerio). Nat. Genet. (4): 338–343. [DOI] [PubMed] [Google Scholar]
- Korswagen L. A., Huizing M., Simsek S., Janssen J. J., Zweegman S., 2008. A novel mutation in a Turkish patient with Hermansky-Pudlak syndrome type 5. Eur. J. Haematol. 80(4): 356–360. [DOI] [PubMed] [Google Scholar]
- Li W., Zhang Q., Oiso N., Novak E. K., Gautam R., et al. , 2003. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat. Genet. 35: 84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E., Hernandez F., Lozano C., Castro M. E., Navarro R. E., 2006. The zebrafish mutant vps18 as a model for vesicle-traffic related hypopigmentation diseases. Pigment Cell Res. 19: 315–326. [DOI] [PubMed] [Google Scholar]
- Marks M. S., Seabra M. C., 2001. The melanosome: membrane dynamics in black and white. Nat. Rev. Mol. Cell Biol. 2(10): 738–748. [DOI] [PubMed] [Google Scholar]
- Moore F. E., Reyon D., Sander J. D., Martinez S. A., Blackburn J. A., et al. , 2012. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs). PLoS ONE 7(5): e37877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan N. V., Pasha S., Johnson C. A., Ainsworth J. R., Eady R. A., et al. , 2006. A germline mutation in BLOC1S3/reduced pigmentation causes a novel variant of Hermansky-Pudlak syndrome (HPS8). Am. J. Hum. Genet. 78(1): 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro R. E., Ramos-Balderas J. L., Guerrero I., Pelcastre V., Maldonado E., 2008. Pigment dilution mutants from fish models with connection to lysosome-related organelles and vesicular traffic genes. Zebrafish 5: 309–318. [DOI] [PubMed] [Google Scholar]
- Ng A., Uribe R. A., Yieh L., Nuckels R., Gross J. M., 2009. Zebrafish mutations in gart and paics identify crucial roles for de novo purine synthesis in vertebrate pigmentation and ocular development. Development 136(15): 2601–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Novak E. K., Kermani M., Fluhr J., Peters L. L., et al. , 2002. Melanosome morphologies in murine models of Hermansky-Pudlak syndrome reflect blocks in organelle development. J. Invest. Dermatol. 119(5): 1156–1164. [DOI] [PubMed] [Google Scholar]
- Nuckels R. J., Ng A., Darland T., Gross J. M., 2009. The vacuolar-ATPase complex regulates retinoblast proliferation and survival, photoreceptor morphogenesis, and pigmentation in the zebrafish eye. Invest. Ophthalmol. Vis. Sci. 50(2): 893–905. [DOI] [PubMed] [Google Scholar]
- Obholzer N., Swinburne I. A., Schwab E., Nechiporuk A. V., Nicolson T., et al. , 2012. Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development 139(22): 4280–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh T., Bailin K., Fukai G. H., Feng L., Ho J. I., et al. , 1996. Positional cloning of a gene for Hermansky–Pudlak syndrome, a disorder of cytoplasmic organelles. Nat. Genet. 14: 300–306. [DOI] [PubMed] [Google Scholar]
- Peitsch M. C., 1995. Protein modeling by E-mail. Nat Biotechnol. 13: 658–660. [Google Scholar]
- Pickles L. M., Roe S. M., Hemingway E. J., Stifani S., Pearl L. H., 2002. Crystal structure of the C-terminal WD40 repeat domain of the human Groucho/TLE1 transcriptional corepressor. Structure 10(6): 751–761. [DOI] [PubMed] [Google Scholar]
- Prota G., Lamoreux M. L., Muller J., Kobayashi T., Napolitano A., et al. , 1995. Comparative analysis of melanins and melanosomes produced by various coat color mutants. Pigment Cell Res. 8(3): 153–163. [DOI] [PubMed] [Google Scholar]
- Raposo G., Tenza D., Murphy D. M., Berson J. F., Marks M. S., 2001. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J. Cell Biol. 152(4): 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond B., Huizing M., Knapp J., Koshoffer A., Zhao Y., et al. , 2005. Melanocytes derived from patients with Hermansky–Pudlak Syndrome types 1, 2, and 3 have distinct defects in cargo trafficking. J. Invest. Dermatol. 124: 420–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonthaler H. B., Lampert J. M., von Lintig J., Schwarz H., Geisler R., et al. , 2005. A mutation in the silver gene leads to defects in melanosome biogenesis and alterations in the visual system in the zebrafish mutant fading vision. Dev. Biol. 284: 421–436. [DOI] [PubMed] [Google Scholar]
- Schonthaler H. B., Fleisch V. C., Biehlmaier O., Makhankov Y., Rinner O., et al. , 2008. The zebrafish mutant lbk/vam6 resembles human multisystemic disorders caused by aberrant trafficking of endosomal vesicles. Development 135: 387–399. [DOI] [PubMed] [Google Scholar]
- Seiji M., Fitzpatrick T. B., Simpson R. T., Birbeck M. S., 1963a Chemical composition and terminology of specialized organelles (melanosomes and melanin granules) in mammalian melanocytes. Nature 197: 1082–1084. [DOI] [PubMed] [Google Scholar]
- Seiji M., Shimao K., Birbeck M. S., Fitzpatrick T. B., 1963b Subcellular localization of melanin biosynthesis. Ann. N. Y. Acad. Sci. 100: 497–533. [PubMed] [Google Scholar]
- Setty S. R., Tenza D., Truschel S. T., Chou E., Sviderskaya E. V., et al. , 2007. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol. Biol. Cell 18(3): 768–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram A., Marks M. S., 2012. Mechanisms of protein delivery to melanosomes in pigment cells. Physiology (Bethesda) 27: 85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Gaitatzes C., Saxena K., Neer E. J., 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24(5): 181–185. [DOI] [PubMed] [Google Scholar]
- Stemple D. L., Solnica-Krezel L., Zwartkruis F., Neuhauss S. C. F., Schier A. F., et al. , 1996. Mutations affecting development of the notochord in zebrafish. Development 123: 117–128. [DOI] [PubMed] [Google Scholar]
- Stirnimann C. U., Petsalaki E., Russell R. B., Muller C. W., 2010. WD40 proteins propel cellular networks. Trends Biochem. Sci. 35: 565–574. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Li W., Zhang Q., Karim A., Novak E. K., et al. , 2002. Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat. Genet. 30: 321–324. [DOI] [PubMed] [Google Scholar]
- Swank R. T., Novak E. K., McGarry M. P., Rusiniak M. E., Feng L., 1998. Mouse models of HPS: a review. Pigment Cell Res. 11(2): 60–80. [DOI] [PubMed] [Google Scholar]
- Syrzycka M., McEachern L. A., Kinneard J., Prabhu K., Fitzpatrick K., et al. , 2007. The pink gene encodes the Drosophila orthologue of the human Hermansky-Pudlak syndrome 5 (HPS5) gene. Genome 50(6): 548–556. [DOI] [PubMed] [Google Scholar]
- Theos A. C., Truschel S. T., Raposo G., Marks M. S., 2005a The Silver locus product Pmel17/gp100/Silv/ME20: controversial in name and function. Pigment Cell Res. 18(5): 322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos A. C., Tenza D., Martina J. A., Hurbain I., Peden A. A., et al. , 2005b Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol. Biol. Cell 16(11): 5356–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Thisse B., 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3(1): 59–69. [DOI] [PubMed] [Google Scholar]
- Thomas J. L., Vihtelic T. S., Dendekker A. D., Willer G., Luo X., et al. , 2011. The loss of vacuolar protein sorting 11 (vps11) causes retinal pathogenesis in a vertebrate model of syndromic albinism. Invest. Ophthalmol. Vis. Sci. 52(6): 3119–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner W. A., Taylor J. D., Tchen T. T., 1975. Melanosome formation in the goldfish: the role of multivesicular bodies. J. Ultrastruct. Res. 51(1): 16–31. [DOI] [PubMed] [Google Scholar]
- Wei M. L., 2006. Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment Cell Res. 19(1): 19–42. [DOI] [PubMed] [Google Scholar]
- Willer G. B., Lee V. M., Gregg R. G., Link B. A., 2005. Analysis of the zebrafish perplexed mutation reveals tissue-specific roles for de novo pyrimidine synthesis during development. Genetics 170(4): 1827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Min J., 2011. Structure and function of WD40 domain proteins. Protein Cell 2(3): 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhao B., Li W., Oiso N., Novak E. K., et al. , 2003. Ru2 and Ru encode mouse orthologs of the genes mutated in human Hermansky-Pudlak syndrome types 5 and 6. Nat. Genet. 33(2): 145–153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.