Abstract
Mec1, a member of the phosphoinositide three-kinase-related kinase (PIKK) family of proteins, is involved in the response to replicative stress and DNA damage and in telomere maintenance. An essential 30 to 35 residue, the FATC domain is found at the C-terminus of all PIKK family members. To investigate the roles of the C-terminal residues of Mec1, we characterized alleles of Saccharomyces cerevisiae mec1 that alter the FATC domain. A change of the terminal tryptophan to alanine resulted in temperature-sensitive growth, sensitivity to hydroxyurea, and diminished kinase activity in vitro. Addition of a terminal glycine or deletion of one, two, or three residues resulted in loss of cell viability and kinase function. Each of these Mec1 derivatives was less stable than wild-type Mec1, eluted abnormally from a size exclusion column, and showed reduced nuclear localization. We identified rpn3-L140P, which encodes a component of the 19S proteasomal regulatory particle of the 26S proteasome, as a suppressor of the temperature-sensitive growth caused by mec1-W2368A. The rpn3-L140P allele acted in a partially dominant fashion. It was not able to suppress the inviability of the C-terminal truncations or additions or the hydroxyurea sensitivity of mec1-W2368A. The rpn3-L140P allele restored Mec1-W2368A to nearly wild-type protein levels at 37°, an effect partially mimicked by the proteasome inhibitor MG-132. Our study supports a role for the C-terminus in Mec1 folding and stability, and suggests a role for the proteasome in regulating Mec1 levels.
Keywords: Mec1, FATC domain, PIKK proteins, Rpn3
Members of the phosphoinositide three-kinase-related kinase (PIKK) family of proteins are important in the cellular response to various forms of stress (Abraham 2004). The PIKK proteins are large (for example, Tra1 and Mec1 are 3744 and 2368 residues, respectively) and are characterized by a C-terminally positioned domain that resembles the phosphatidylinositol-3 kinases (PI3K; Abraham 2004; Lempiäinen and Halazonetis 2009; Lovejoy and Cortez 2009).
Three members of the PIKK family have principal roles in DNA damage-response pathways. Ataxia telangiectasia–mutated (ATM; Tel1 in Saccharomyces cerevisiae), ataxia telangiectasia and Rad3-related (ATR; Mec1 in S. cerevisiae), and the DNA-dependent protein kinase catalytic subunit (DNA-PKc) transmit and amplify the damage signal through the phosphorylation of target proteins. Recruitment to sites of DNA damage is critical for their activation and function in checkpoint signaling and DNA repair (Falck et al. 2005). ATM/Tel1 acts principally in response to double-strand breaks, whereas ATR/Mec1 responds to a number of DNA insults, particularly replicative stress caused by stalled replication forks (Labib and De 2011). Both have roles in the maintenance of stable telomeres (Tomita and Cooper 2008; Moser et al. 2011; Yamazaki et al. 2012). DNA-PKc acts with Ku70 and Ku80 in nonhomologous end-joining of double-strand breaks; a direct homolog has not been found in S. cerevisiae.
Mec1/ATR is an essential gene (Cimprich and Cortez 2008). It is recruited to stalled replication forks and sites of DNA damage through the direct interaction of its associated protein Lcd1/Ddc2 with RPA-coated single-strand DNA (Rouse and Jackson 2002; Zou and Elledge 2003; Katou et al. 2003; Osborn and Elledge 2003; Ball et al. 2005). This recruitment is part of a series of events that results in the phosphorylation of regulatory and effector molecules that activate a cell-cycle checkpoint or apoptotic signals. Mec1 target molecules include histone H2A (Downs et al. 2000; Cobb et al. 2005), RPA (Brush et al. 1996), components of the Mcm2-7 DNA helicase complex (Cortez et al. 2004; Yoo et al. 2004), the Ino80 subunit Ies4 (Morrison et al. 2007), and the effector kinases Rad53 and Chk1 (Pellicioli et al. 1999; Sweeney et al. 2005; Ma et al. 2006). Fork stabilization is the essential function of Mec1 (Desany et al. 1998; Friedel et al. 2009) and occurs through multiple mechanisms, including retaining DNA polymerase at the replication fork (Cobb et al. 2003). In addition, Mec1 is required to activate the expression of ribonucleotide reductase, thus enhancing deoxyribonucleotide synthesis after DNA damage and perhaps during normal S phase (Zegerman and Diffley 2009). This function can be bypassed by increasing deoxyribonucleotide levels either by overexpressing Rnr3, one of the catalytic subunits of ribonucleotide reductase, or by deletion of the ribonucleotide reductase inhibitor Sml1 (Desany et al. 1998; Zhao et al. 1998). Interestingly, Mec1 and ATR are required to prevent chromosome breaks even in the absence of genotoxic stress. Augmenting deoxyribonucleotide levels does not rescue the checkpoint defects or DNA damage sensitivity of cells lacking functional Mec1 (Zhao et al. 1998).
Other PIKK family members include SMG-1 (suppressor with morphological effect on genitalia family member), target of rapamycin (TOR), and transformation/transcription domain-associated protein (TRRAP). SMG-1 is found in metazoans. It has many similar roles related to genotoxic stress (Brumbaugh et al. 2004; Gehen et al. 2008) and telomere stability (Azzalin et al. 2007) as ATM and ATR, and it is also the key signaling molecule required for the nonsense-mediated decay pathway (Chang et al. 2007). TOR is found in two complexes, TORC1 and TORC2. TORC1 integrates nutrient and growth factor signals, inducing anabolic pathways including protein synthesis and inhibiting catabolic pathways (Loewith and Hall 2011). TORC2 is involved in spatial control of cell growth by regulating the actin cytoskeleton (Cybulski and Hall 2009). The only member of the PIKK family that is not a Ser/Thr kinase is the transcriptional cofactor TRRAP (McMahon et al. 1998; Saleh et al. 1998). The S. cerevisiae homolog Tra1 is an essential component of the multisubunit, multifunctional SAGA and NuA4 histone acetyltransferase complexes (Grant et al. 1998; Saleh et al. 1998). A key role for Tra1 is to recruit HAT complexes to promoters via its association with transcriptional activators (Brown et al. 2001; Bhaumik et al. 2004; Fishburn et al. 2005; Reeves and Hahn 2005).
As well as having the PI3K domain, the PIKK proteins share a number of other features. N-terminal to the PI3K domain is a FAT (FRAP-ATM-TRRAP) domain that consists largely of helical HEAT (Huntington, elongation factor 3, PR65/A, TOR) and tetratricopeptide (TPR) repeats; in fact, these repeats extend through to the N-termini, making most of the protein helical (Bosotti et al. 2000; Perry and Kleckner 2003; Sibanda et al. 2010; Knutson and Hahn 2011). The C-terminal to the PI3K domain is a less highly conserved PIKK regulatory domain (PRD; Mordes et al. 2008). At the C-terminus of the PIKK molecules is the 30 to 35 residue FATC domain (FAT C-terminal; Bosotti et al. 2000). The structure of the isolated FATC domain of S. cerevisiae Tor1 (Dames et al. 2005) is helical, with a C-terminal loop held in place by a disulphide linkage. Although the cysteines are not conserved and the cellular FATC domain is not likely to always exist in an oxidized state, the helical structure is likely conserved (Lempiäinen and Halazonetis 2009; Sturgill and Hall 2009). The crystal structure of mammalian target of rapamycin (mTOR) including the FAT through to the C-terminus reveals that the FATC domain is an integral part of the kinase, positioned adjacent to the activation loop (Yang et al. 2013). The substrate-binding groove also includes portions of the FATC domain. The importance of the FATC domain has been demonstrated with molecular studies. Deletion of the C-terminus of mTOR or mutation of a conserved tryptophan five residues from the C-terminus eliminates kinase activity (Takahashi et al. 2000). Similar mutations within the FATC domains of DNA-PKcs and SMG-1 cause a loss of kinase activity (Priestley et al. 1998; Beamish et al. 2000; Morita et al. 2007). As suggested by Lempiäinen and Halazonetis (2009), the FATC domain likely regulates the kinase domain through interactions with the activation loop similar to the helical domains of the PI3 kinases. The FATC domain is also critical for the function of Tra1. Addition of as little as a single glycine to the C-terminus of Tra1 results in loss of cellular viability (Hoke et al. 2010). A strain with a mutation of the terminal phenylalanine residue of Tra1 to alanine (tra1-F3744A) shows growth defects, including temperature sensitivity and slow growth on media containing ethanol, Calcofluor white, or rapamycin. The F3744A mutation also results in mislocalization of the protein to the cytoplasm, particularly under conditions of stress (Genereaux et al. 2012). We identified a partially dominant mutation within tti2 as a suppressor of tra1-F3744A (Genereaux et al. 2012). TTI2 encodes a component of the Tel2-Tti1-Tti2 complex that associates with chaperone proteins in the folding of PIKK proteins (Takai et al., 2007, 2010; Horejsí et al., 2010; Hurov et al., 2010; Kaizuka et al., 2010). This suggests that the C-terminal residue of the molecule is required for folding and was consistent with the tti2 suppressor mutation decreasing the proteolytic degradation of Tra1-F3744A and increasing its nuclear localization.
To determine if the C-terminus of the FATC domain is a general requirement for the folding and function of the PIKK proteins, we have examined alleles of mec1 that disrupt this domain. Addition or deletion of residues results in loss of viability and kinase activity. Proteins containing these insertions or deletions, as well as a C-terminal change of tryptophan to alanine, have reduced nuclear localization and elute from a gel filtration column with an abnormally high molecular mass. They are also less stable after isolation than the wild-type protein. A second site mutation of rpn3-L140P, which encodes a component of the 19S proteasomal regulatory particle, restores the levels of the Mec1-W2368A protein and growth of the mec1-W2368A strain at 37°, further supporting a role for the C-terminus in protein folding and stability.
Materials and Methods
Yeast strains and growth
Yeast strains are listed in Table 1 and are derivatives of the diploid strain BY4743 (Winzeler and Davis 1997). Strains containing one Flag5-tagged (CY6172) or eGFP-tagged (CY6295) MEC1 allele, marked with URA3, were made by one-step integration of the SphI-EcoRI fragments of pCB2363 and pCB2395, respectively. Expression of these alleles is driven by the TRA1 promoter. Integration of the mec1-W2368A allele has been described previously (Genereaux et al. 2012). This strain, strains with C-terminal truncations of one, two, or three residues (mec1-Δ1, Δ2, and Δ3), and the strain with the addition of a glycine residue (mec1-2369G) were similarly integrated as SphI-SacI fragments of the HIS3-tagged allele. Haploid strains containing mec1-W2368A (CY6175) or wild-type MEC1 (CY6194) were generated by sporulation of their respective diploid strains. Haploid strains containing mec1 deletion and addition alleles were made by integrating the mec1 allele into CY6319 or CY6321 MATa sml1::KanMX deletion strains derived from the consortium collection and containing eGFP-MEC1 or Flag5-MEC1, respectively. Diploid strains expressing eGFP-tagged Mec1, Mec1-W2388A, Mec1-Δ1, Mec1-Δ2, or Mec1-2369G, and RFP-tagged Nic96 were made by mating of CY6307, CY6302, CY6330, CY6344, and CY6342 with the RFP-tagged NIC96 strain in the EY0987 background (MATα his3Δ1 lys2Δ0 ura3Δ0; Huh et al. 2003; kindly provided by Peter Arvidson). CY6265 containing mec1-W2368A and rpn3-L140P was isolated in the selection scheme described. CY6398 (rpn3-L140P) was obtained after backcrossing with BY4741. The diploid strain CY6400 (MEC1/Flag5-mec1-W2368A rpn3-L140P/rpn3-L140P) was obtained by mating of CY6391 and CY6399. CY6418 (tra1-W3744A rpn3-L140P) was obtained after crossing with CY4350 (Genereaux et al. 2012) and selecting for spore colonies that grew in the absence of histidine and carried the rpn3-L140P allele as determined by sequencing. CY6465 was derived from CY6076 by integrating rpn3-L140P contained in the URA3-containing yeast integrating plasmid YIPlac211 (CB2457) after digestion with BsaB1.
Table 1. Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 | Winzeler and Davis (1997) |
| BY4741 | MATa ura3Δ0 met15Δ0 his3Δ0 leu2Δ0 | Winzeler and Davis (1997) |
| BY4742 | MATα ura3Δ0 lys2Δ0 his3Δ0 leu2Δ0 | Winzeler and Davis (1997) |
| CY4350 | MATa ura3Δ0 his3Δ0 leu2Δ0 tra1-F3744A-HIS3 | Hoke et al. (2010) |
| CY4353 | MATa ura3Δ0 his3Δ0 leu2Δ0 TRA1-HIS3 | Hoke et al. (2010) |
| CY6076 | MATa ura3Δ0 his3Δ0 leu2Δ0 mec1-W2368A-HIS3 | Genereaux et al. (2012) |
| CY6077 | MATα ura3Δ0 his3Δ0 leu2Δ0 mec1-W2368A-HIS3 | This work |
| CY6106 | MATa ura3Δ0 his3Δ0 leu2Δ0 mec1-W2368A-URA3 | This work |
| CY6172 | Isogenic to BY4743 except MEC1/URA3-Flag5-MEC1 | This work |
| CY6175 | MATa ura3Δ0 his3Δ0 leu2Δ0 URA3-Flag5-mec1-W2368A-HIS3 | This work |
| CY6192 | Isogenic to BY4743 except MEC1/URA3-Flag5-mec1-W2368A-HIS3 | This work |
| CY6184 | Isogenic to BY4743 except MEC1/URA3-Flag5-mec1-Δ1-HIS3 | This work |
| CY6194 | MATa ura3Δ0 his3Δ0 leu2Δ0 URA3-Flag5-MEC1 | This work |
| CY6203 | Isogenic to BY4743 except MEC1/URA3-Flag5-mec1-2369G-HIS3 | This work |
| CY6233 | Isogenic to BY4743 except MEC1/URA3-Flag5-mec1-Δ3-HIS3 | This work |
| CY6250 | Isogenic to BY4743 except MEC1/URA3-Flag5-mec1-Δ2-HIS3 | This work |
| CY6265 | MATα ura3Δ0 his3Δ0 leu2Δ0 mec1-W2368A-URA3 rpn3-L140P | This work |
| CY6291 | Isogenic to BY4743 except LEU2-MEC1/URA3-eGFP-MEC1-Δ1-HIS3 | This work |
| CY6292 | Isogenic to BY4743 except LEU2-MEC1/URA3-eGFP-MEC1-Δ3-HIS3 | This work |
| CY6293 | Isogenic to BY4743 except LEU2-MEC1/URA3-eGFP-mec1-2369G-HIS3 | This work |
| CY6294 | Isogenic to BY4743 except LEU2-MEC1/URA3-eGFP-MEC1-Δ2-HIS3 | This work |
| CY6295 | Isogenic to BY4743 except MEC1/URA3-eGFP-MEC1 | This work |
| CY6296 | Isogenic to BY4743 except MEC1/URA3-eGFP-mec1-W2368A-HIS3 | This work |
| CY6302 | MATa ura3Δ0 his3Δ0 leu2Δ0 URA3-eGFP-mec1-W2368A-HIS3 | This work |
| CY6303 | MATα ura3Δ0 his3Δ0 leu2Δ0 URA3-eGFP-mec1-W2368A-HIS3 | This work |
| CY6306 | MATα ura3Δ0 his3Δ0 leu2Δ0 URA3-eGFP-MEC1 | This work |
| CY6307 | MATa ura3Δ0 his3Δ0 leu2Δ0 URA3-eGFP-MEC1 | This work |
| CY6319 | MATa ura3Δ0 his3Δ0 leu2Δ0 sml1Δ0::KanMX URA3-eGFP-MEC1 | This work |
| CY6321 | MATa ura3Δ0 his3Δ0 leu2Δ0 sml1Δ0::KanMX URA3- Flag5-MEC1 | This work |
| CY6330 | MATa ura3Δ0 his3Δ0 leu2Δ0 sml1Δ0::KanMX URA3-eGFP-mec1-Δ1-HIS3 | This work |
| CY6342 | MATa ura3Δ0 his3Δ0 leu2Δ0 sml1Δ0::KanMX URA3-eGFP-mec1-2369G-HIS3 | This work |
| CY6344 | MATa ura3Δ0 his3Δ0 leu2Δ0 sml1Δ0::KanMX URA3-eGFP-mec1-Δ2-HIS3 | This work |
| CY6349 | MATa ura3Δ0 his3Δ0 leu2Δ0 sml1Δ0::KanMX URA3-Flag5-mec1-2369G-HIS3 | This work |
| CY6391 | MATα ura3Δ0 his3Δ0 leu2Δ0 mec1-W2368A-HIS3 rpn3-L140P | This work |
| CY6398 | MATa ura3Δ0 his3Δ0 leu2Δ0 rpn3-L140P | This work |
| CY6399 | MATα ura3Δ0 his3Δ0 leu2Δ0 rpn3-L140P | This work |
| CY6400 | MATa/α ura3Δ0/ura3Δ0 his3Δ0/his3Δ0 leu2Δ0/leu2Δ0 URA3- Flag5-mec1-W2368A/MEC1 rpn3-L140P/rpn3-L140P | This work |
| CY6418 | MATa ura3Δ0 his3Δ0 leu2Δ0 tra1-F3744A-HIS3 rpn3-L140P | This work |
| CY6449 | MATa ura3Δ0 his3Δ0 leu2Δ0 sml1Δ0::KanMX URA3-Flag5-mec1-2369G-HIS3 rpn3-L140P | This work |
Growth comparisons were performed on YP media containing 2% glucose (YPD) or selective plates after 3–5 days at 30° unless stated otherwise. Standard concentrations used for the selections were 0.03% methyl methanesulfonate (Sigma-Aldrich), 6% ethanol, and 200 mM hydroxyurea (Sigma-Aldrich). Growth on plates containing MG-132 (Calbiochem) was adapted from Liu et al. (2007) using conditions that permeabilize cells. Strains were grown overnight in synthetic complete media lacking uracil and containing 0.1% proline as the nitrogen source, then 4 hr in the same media also containing 0.003% sodium dodecyl sulfate (SDS). Serial dilutions were spotted onto synthetic complete media containing 0.003% sodium dodecyl sulfate and 10, 25, or 50 μM MG-132 and grown at 30°.
DNA molecules
An integrative vector to generate a Flag5 (pCB6192) fusion of Mec1 was constructed from pCB2143 (Genereaux et al. 2012) by replacing TRA1 flanking sequences with MEC1 as SphI-HindIII and NotI-EcoRI fragments using oligonucleotides 6288-1/6288-1 and 6288-3/6288-4 (Table 2). A 1.1-kbp HindIII genomic fragment encoding URA3 was inserted into this molecule to allow selection. The plasmid pCB6295 allowing integration of eGFP was created from pCB2143 by the replacement of the Flag cassette with a BamHI-Not1 cassette encoding eGFP (Hoke et al. 2010). The mec1 mutant alleles were constructed by polymerase chain reaction (PCR) using the oligonucleotides indicated in Table 2 and inserted into pCB2317 (mec1-W2368A). Myc9-tagged LCD1 and RPN3 were expressed from the DED1 promoter in YCplac111, a LEU2 centromeric plasmid, by inserting a NotI-SstI fragment amplified from genomic DNA using oligonucleotides 6486-1/6486-1 and 6518-1/6522-1, respectively, downstream of the DED1 promoter-myc9 cassette (Hoke et al. 2010). For integration in cells, rpn3-L140P including its native promoter was synthesized by PCR using oligonucleotides 6577-1 and 6510-2 and cloned as a HindIII-EcoR1 fragment into YIPlac211.
Table 2. Oligonucleotides used in this study.
| Number | Sequence | Description |
|---|---|---|
| 6288-1 | ATAAGGCGGCCGCCATGGAATCACACGTCAA-ATATCTTG | 5′ coding region of MEC1 to insert tags |
| 6288-2 | Atatgtcgaccgcctcataaaccatattctgtg | 5′ coding region of MEC1 to insert tags |
| 6288-3 | Ttcgcatgccttttcaaggctccataactat | MEC1 promoter region to insert tags |
| 6288-4 | Ggaaagcttggagcgtgcgttccatcta | MEC1 promoter region to insert tags |
| 6313-1 | AAGCTTGCATGCGTTGATGAATGTG | 3′ coding region of MEC1 for cloning of deletions |
| 6313-2 | Gcgtgatcaaaatggaagccaaccaatatac | mec1-Δ1 |
| 6338-1 | Gcgtgatcatggaagccaaccaatatacatc | mec1-Δ2 |
| 6338-2 | gcgtgatcaaccccaaaatggaagccaacc-aatatacatc | mec1-2369G |
| 6349-3 | Gcgtgatcaaagccaaccaatatacatcttgc | mec1-Δ3 |
| 6408-1 | Cccagtccgccctgagcaaag | eGFP to confirm integration of tag |
| 6408-2 | Ccgtaaaattcgacacatgctttg | 5′ coding of MEC1 to confirm integration of tags |
| 6486-1 | ATAAGAATGCGGCCGCGATGAGACGAGAAACGGTGGG | 5′ coding of LCD1 |
| 6486-2 | CGGAATTCCAAACCGGTTCTGCTAAG | 3′ coding of LCD1 |
| 6518-1 | ATAAGAATGCGGCCGCAATGGCTAGCACTGCAGTAAT | 5′ coding of RPN3 |
| 6522-1 | Cggaattcgcgcccttataagaatcccaaatcg | 3′ coding of RPN3 |
| 6577-1 | CCCAAGCTTCGGAGTACGACCAGACGCTGA | Promoter of RPN3 |
Fluorescence microscopy
Yeast cells expressing eGFP or RFP fusions were grown in synthetic complete media to stationary phase, then diluted 1:20 into synthetic complete media and grown for 6 hr with shaking. Fluorescent images were obtained using a Zeiss Axioskop 2 microscope driven by ImageJ 1.41 software (National Institutes of Health, Bethesda, MD) and a Scion CFW Monochrome CCD Firewire Camera (Scion Corporation, Frederick, MD) using RFP and GFP filter sets.
Protein extracts and immunoprecipitation
Yeast strains were grown in YP media containing 2% glucose to an A600∼3.0. Extracts were prepared cryogenically as previously described (Saleh et al. 1997) . For immunoprecipitations, all steps were performed at 4°; 4.0 mg extract from the Mec1 derivative strains or 2 mg wild-type extract was suspended in immunoprecipitation (IP) buffer (50 mM HEPES, pH 7.5; 100 mM KCl; 0.1 mM EDTA; 0.2% Tween20; 1.0 mM dithiothreitol) containing protease inhibitors (1.0 mM phenylmethylsulfonyl fluoride, 1.0 mM benzamadine, 50 μg/ml trypsin inhibitor, 5 μg/ml pepstatin, and 5 μg/ml leupeptin; Sigma-Aldrich). Immunoprecipitations were performed with 100 μl 50% slurry of anti-Flag M2 magnetic beads (Sigma-Aldrich) and rotated for 2.5 hr. Beads were washed five times with 1.0 ml IP buffer. Protein was eluted in 1× SDS loading buffer (without reducing agent) at 65° for 6 min, then transferred to a fresh tube. Dithiothreitol was added to 20 mM and the sample was heated at 65° for 2 min.
Protein kinase assays
Proteins captured by immunoprecipitation as described above were processed as in Mallory and Petes (2000) with modifications. Washed anti-Flag M2 beads (30 μl) containing the immunoprecipitated proteins were resuspended at a ratio of 1:1 in kinase buffer (10 mM HEPES-NaOH, pH 7.4; 50 mM NaCl; 10 mM MnCl2; 1 mM DTT). Kinase reactions contained the washed beads in which buffer was removed and 11.5 μl kinase buffer, 1.5 μl 200-μM ATP, 10 μCi γ-32P–labeled ATP (4500 Ci/mmol; 1 Ci = 37 GBq), and 1 μl rat 4E-BP1 (1 μg/μl; Santa Cruz Biotechnology). The reactions were performed at 30° for 30 min. Protein was eluted in 1× SDS loading buffer, boiled for 2 min, and then separated on a 15% Tris-Tricine PAGE gel. The gel was stained with Coomassie brilliant blue, destained, and incubated 30 min in 20% polyethylene glycol 400 and 50% methanol. The gel was dried, exposed to a storage phosphor screen, and analyzed on a Storm 860 Phosphorimager (GE Healthcare Life Sciences). Densitometry analysis of the gel image was performed using ImageQuant 5.2 (Molecular Dynamics).
Gel filtration chromatography
Yeast extract (4.5 mg) prepared in 50 mM sodium phosphate (pH 7.0; 150 mM NaCl) was loaded at a flow rate of 0.3 ml/min onto a 24-ml FPLC Superose 6HR10/30 column (Amersham Pharmacia Biotech). Protein from 20-μl aliquots of 250-μl fractions was resolved by SDS-PAGE and proteins were detected by Western blotting. Densitometric scanning of films was performed using AlphaImager 3400 software (Alpha Innotech, San Leandro, CA).
Western blotting
Western blotting was performed using polyvinylidene fluoride membranes and anti-Flag (M2; Sigma-Aldrich) or anti-Myc antibodies as described by Mutiu et al. (2007) and Hoke et al. (2010).
Selection of suppressor strains
CY6106 (mec1-W2368A-URA3) was grown to stationary phase in YPD. In two separate experiments, 10 μl culture, approximately 2 million cells, was plated onto each of five YPD plates and ultraviolet-irradiated at a wavelength of 302 nm for 10 s. Survival was approximately 10%. Colonies growing at 37° were colony-purified under nonselective conditions and reanalyzed for growth at 37°. The suppressor strains were crossed with the HIS3-tagged mec1-W2368A strain, CY6077, to determine linkage of the suppressor with mec1-W2368A. In the two selections, nine strains had an unlinked suppressor mutation that segregated in a 2:2 fashion. Spore colonies of each were backcrossed with BY4741 or BY4742 six times and a nonmutated mec1-W2368A strain three times at each stage, selecting for temperature resistant spore colonies. The final isolates were sent for genomic sequencing as described. The rpn3 mutation was verified after isolation of genomic DNA, PCR with oligonucleotides 6510-1 and 6510-2 (Table 2), and sequencing of the PCR product using oligonucleotide 6510-1 as primer.
Genomic sequence analysis
Genomic DNAs were prepared from CY6076, CY6077, and four sister spore colonies from the final backcross of each of suppressor strains as described previously (Genereaux et al. 2012). The DNA from CY6076 and from CY6077 were pooled, as were those from each of the identical suppressor strains, and 5 µg DNA from each pooled sample was sent to the Biodiversity Research Centre (University of British Columbia, Vancouver, Canada) for DNA library construction and next-generation sequencing using 100-bp paired-end reads with the Illumina HiSequation 2000 platform. The S. cerevisiae genome sequence was downloaded from the Saccharomyces Genome Database (http://www.yeastgenome.org) on March 24, 2011. Custom bash and Perl scripts were written for the sequencing analysis. The program Bowtie (Langmead et al. 2009), allowing up to three mismatches per read, was used to map the reads to each chromosome of the yeast genome and output mapped reads in SAM format (Sequence Alignment/Map; Li et al. 2009). The variant call format from SAMtools (Li et al. 2009) was used to obtain a raw list of polymorphisms from the mapped reads. Those reads with a Phred quality score less than 20 were eliminated to obtain a filtered list of polymorphisms. A custom Perl script was written to identify those polymorphisms that were unique to the suppressor strain.
Results
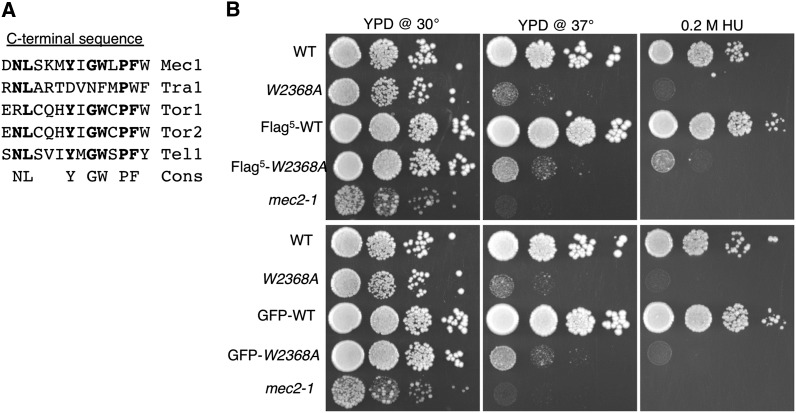
The penultimate and terminal amino acid residues of the PIKK molecules are hydrophobic in family members (Figure 1A). Changing the terminal phenylalanine of Tra1 to alanine (tra1-F3744A) results in stress-related phenotypes and partial mislocalization of the protein to the cytoplasm (Genereaux et al. 2012). This mutation was suppressible by a F328S change in the chaperone component Tti2. A similar mutation in the terminal tryptophan of Mec1 (Mec1-W2368A) results in temperature-sensitive growth but is not suppressible by tti2-F328S (Genereaux et al. 2012). Here, we characterize Mec1 derivatives altered at their C-terminus to determine if the FATC domain has a general role in stability of PIKK family members. For these studies we constructed strains that express a genomically encoded allele with either five tandem copies of the Flag epitope (Flag5) or eGFP tag positioned at the N-terminus. As shown in Figure 1B, neither the Flag5 nor the eGFP tag alters growth of a strain with the otherwise wild-type allele in YPD media at 30°, 37°, or in the presence of 0.2 M hydroxyurea, a competitive inhibitor of ribonucleotide reductase (compare wild-type with Flag5–wild-type and GFP–wild-type). Growth of a strain with the mec1-W2368A allele also was examined. The mec1-W2368A strain was sensitive to hydroxyurea and, as shown previously (Genereaux et al. 2012), was temperature-sensitive. Neither Flag5 nor eGFP tags decreased the growth of the mec1-W2368A strain under these conditions.
Figure 1.
Phenotype of Flag5 and GFP-tagged MEC1 strains. (A) Conservation of hydrophobic residues at the C-terminus of phosphoinositide three-kinase-related kinase (PIKK) family members. The C-terminal residues of the five PIKK family members found in S. cerevisiae are shown with the following consensus. (B) Yeast strains BY4741 [MEC1; wild-type (WT)], CY6076 (mec1-W2368A), CY6194 (Flag5-MEC1), CY6175 (Flag5-mec1-W2368A), a mec2-1 strain (Weinert et al. 1994; included to verify the hydroxyurea plates), CY6307 (GFP-MEC1), and CY6302 (GFP-mec1-W2368A) were grown to stationary phase diluted 1/102 and 10-fold serial dilutions were spotted onto selection plates as follows: YPD at 30°, YPD at 37°, and YPD at 30° containing 0.2 M hydroxyurea (HU).
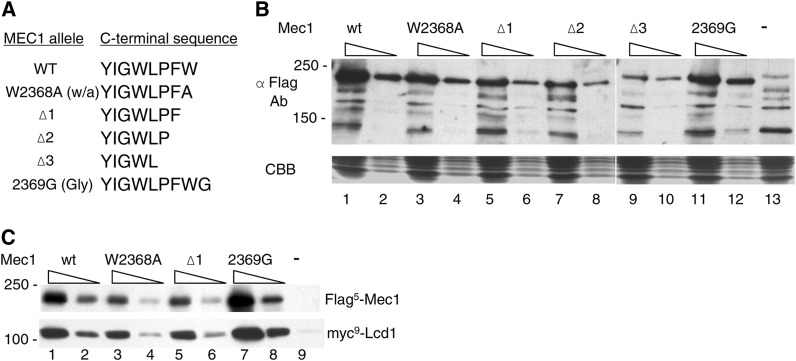
Four additional mec1 alleles were constructed and integrated into a diploid strain. mec1-Δ1, mec1-Δ2, and mec1-Δ3 contain deletions of one, two, or three residues at the C-terminus of Mec1 (Figure 2A). Three deletions were constructed to decrease the likelihood that the C-terminal carboxyl might be artificially repositioned in a hydrophobic pocket in all of the derivatives. Mec1-2369G contains an additional glycine residue at the C-terminus. A comparable addition to Tra1 results in inviability (Hoke et al. 2010). After sporulation, there was a 2:2 segregation of viable and inviable spore colonies for each, indicating that insertion or deletions to the C-terminus of Mec1 compromise viability. To determine if the C-terminal mutations affect expression of Mec1, diploid strains containing a single copy of Flag5-tagged wild-type or mutant Mec1, and an untagged wild-type allele were analyzed by Western blotting. As shown in Figure 2B, all of the mec1 alleles were expressed when cells were grown at 30°. Flag5-Mec1-W2368A and Flag5-Mec1-2369G were found at nearly wild-type levels. The deletion proteins were somewhat reduced, with the reduction paralleling the extent of the deletion. We also examined if the different mec1 alleles interacted with Lcd1/Ddc2 (ATRIP in mammalian cells), an association required for the checkpoint functions of Mec1 (Rouse and Jackson 2000; Paciotti et al. 2000; Wakayama et al. 2001). Immunoprecipitations were performed with anti-Flag antibody and extracts prepared from strains containing myc9-tagged Lcd1/Ddc2 and a Flag5-tagged Mec1 derivative. As shown in Figure 2C, the ratio of myc9-tagged Lcd1/Ddc2 to Flag5-Mec1 was similar for each of the derivatives, indicating that Lcd1/Ddc2 interacted with Mec1-W2368A, Mec1-Δ1, and Mec1-G2369 as efficiently as the wild-type protein.
Figure 2.
Expression of Mec1 derivatives with C-terminal mutations. (A) Amino acid sequences of the Mec1 derivatives. (B) Yeast strains CY6172 (Flag5-MEC1; lanes 1 and 2), CY6192 (Flag5-mec1-W2368A; lanes 3 and 4), CY6184 (Flag5-mec1-Δ1; lanes 5 and 6), CY6250 (Flag5-mec1-Δ2; lanes 7 and 8), CY6233 (Flag5-mec1-Δ3; lanes 9 and 10), CY6203 (Flag5-mec1-2369G; lanes 11 and 12), and BY4743 (lane 13) were grown in YPD media to mid-log phase and extracts were prepared by grinding in liquid nitrogen. Extracts were solubilized in the presence of protease inhibitors (1.0 mM phenylmethylsulfonyl fluoride, 5 μg/ml pepstatin, 1.0 mM benzamadine, 50 μg/ml trypsin inhibitor, and 5 μg/ml leupeptin); 50 μg and 10 μg protein were separated by sodium dodecyl sulfate (SDS)-PAGE; 50 μg was used for BY4743. The top portion of the gel was Western-blotted with anti-Flag (M2) antibody; the bottom portion was stained with Coomassie brilliant blue. (C) Interaction with Lcd1/Ddc2; 3 mg protein extract from yeast strains CY6172 (Flag5-MEC1; lanes 1 and 2), CY6192 (Flag5-mec1-W2368A; lanes 3 and 4), CY6184 (Flag5-mec1-Δ1; lanes 5 and 6), CY6203 (Flag5-mec1-2369G; lanes 7 and 8), and BY4743 (lane 9) containing YCplac111-myc9-Lcd1 were immunoprecipitated with anti-Flag antibody; 20 μl (odd-numbered lanes) and 5 μl of the immunoprecipitates were separated by SDS-PAGE and Western-blotted with anti-Flag antibody (upper panel) or anti-myc antibody (lower panel).
Terminal residues of Mec1 are required for full kinase activity
Because the phenotypes of the Mec1 derivatives were not attributable to their lack of expression or reduced interaction with Lcd1, we performed in vitro kinase assays to determine if this activity was impaired. We assayed the activity of Flag5-tagged Mec1, Mec1-Δ1, Mec1-Δ2, Mec1-Δ3, Mec1-2369G, and Mec1-W2368A after isolating the proteins by immunoprecipitation on Flag-antibody resin from diploid yeast strains. The immunoprecipitates of the Flag5-tagged proteins used in the kinase assays are shown in Figure 3A. Kinase assays were performed with rat 4E-BP1 (PHAS-1) as the substrate (Lin et al. 1994; von Manteuffel et al. 1996). The results for an assay performed at 30° are shown in Figure 3B. Immunoprecipitates from a strain wild-type for MEC1, but without tagged protein (BY4743; lane 1), and from CY6172 (Flag5-tagged Mec1; lane 10) in the absence 4E-BP1 were performed to control for nonspecific phosphorylation and to identify 4E-BP1, respectively. Phosphorylation of 4E-BP1 was 7.5-fold greater than background with the wild-type protein (compare lanes 1 and 2). Interestingly, phosphorylation by Flag5-Mec1-W2368A was less than 10% of that found for the wild-type protein (compare lanes 2 and 9). Although 4E-BP1 is not necessarily indicative of all native Mec1 substrates, this result suggests that only low levels of kinase activity are essential for viability in rich media. Phosphorylation of 4E-BP1 was not above background levels for Mec1-2369G (lane 8), likely explaining the inviability of the strain containing this derivative. Similarly, the kinase activity of the Mec1 deletion derivatives (lanes 5–7) was not above background, although exact comparison with the wild-type protein was difficult because of their somewhat reduced abundance in the immunoprecipitates.
Figure 3.
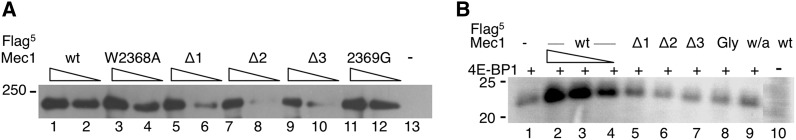
The terminal residues of Mec1 are required for its kinase activity. (A) Protein extracts were prepared from yeast strains CY6172 (Flag5-MEC1; lanes 1 and 2), CY6192 (Flag5-mec1-W2368A; lanes 3 and 4), CY6184 (Flag5-mec1-Δ1; lanes 5 and 6), CY6250 (Flag5-mec1-Δ2; lanes 7 and 8), CY6233 (Flag5-mec1-Δ3; lanes 9 and 10), CY6203 (Flag5-mec1-2369G; lanes 11 and 12), and BY4743 (lane 13) in the presence of protease inhibitors; 2 mg protein from CY6192 and 4 mg protein from the other strains were immunoprecipitated. One-third of the beads was suspended in 100 μl of 1× sodium dodecyl sulfate (SDS) loading buffer, boiled, and 10 μl or 5 μl were separated by SDS-PAGE (5% gel) and Western blotted with anti-Flag antibody. The BY4743 sample contained 10 μl bead suspension. (B) Phosphorylation assays. Two-thirds of the immunoprecipitate in (A) was suspended in 60 μl kinase buffer. Aliquots [8, 4, and 2 μl for wild-type (WT), 8 μl of the other samples] were used in kinase assays at 30° with 4E-BP1 as substrate (with the exception of lane 10 which contained buffered glycerol). Lane 1, BY4743; lanes 2–4 and 10, CY6172 (Flag5-MEC1); lane 5, CY6184 (Flag5-mec1-Δ1); lane 6, CY6250 (Flag5-mec1-Δ2); lane 7, CY6233 (Flag5-mec1-Δ3); lane 8, CY6203 (Flag5-mec1-2369G); and lane 9, CY6192 (Flag5-mec1-W2368A). Reactions were stopped with 3× SDS loading buffer and protein was separated on a 15% Tris-Tricine gel. The gel was fixed and processed as described in Materials and Methods. Lane 10 was analyzed on the same gel but was moved proximal to the other samples.
The terminal tryptophan residue is required for stability and localization of the protein
Based on the properties of Tra1 derivatives with C-terminal mutations (Genereaux et al. 2012), we predicted that the lack of kinase activity found for the Mec1 derivatives might result from improper folding of the molecules. The Mec1-W2368A, 2369G, and Δ1 derivatives were found within cells at levels approaching that of the wild-type when extracts were prepared with protease inhibitors and analyzed directly after isolation (Figure 2B). Predicting that the C-terminus was required for stability of the protein, we addressed whether the Mec1 derivatives would be susceptible to proteolysis in the absence of protease inhibitors. Protein was isolated from diploid strains containing Flag5 derivatives of Mec1, Mec1-2369G, Mec1-W2368A, and Mec1-Δ1. The extract was then incubated at 30° for 10 min or 2 hr, and aliquots were analyzed by Western blotting (Figure 4). The wild-type protein was stable in extracts with no obvious degradation in 2 hr. In contrast, each of the Mec1 derivatives showed some sign of degradation at the 10-min time point.
Figure 4.
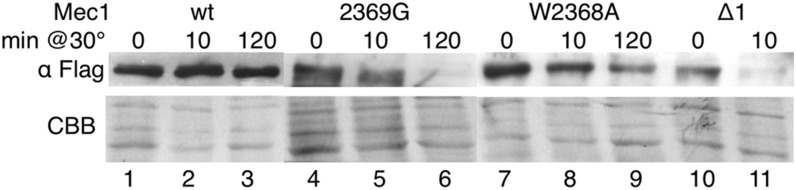
The terminal residues of Mec1 are required for its stability in extracts. Yeast strains CY6172 (Flag5-MEC1; lanes 1–3), CY6203 (Flag5-mec1-2369G; lanes 4–6), CY6192 (Flag5-mec1-W2368A; lanes 7–9), and CY6184 (Flag5-mec1-Δ1; lanes 10 and 11) were grown in YPD media to mid-log phase and extracts were prepared by grinding with glass beads in the absence of protease inhibitors. Extracts were then incubated at 30° for the indicated time period and 50 μg protein from each extract was separated by sodium dodecyl sulfate (SDS)-PAGE and Western-blotted with anti-Flag antibody. The lower panel is the lower portion of the gel after staining with Coomassie brilliant blue (CBB).
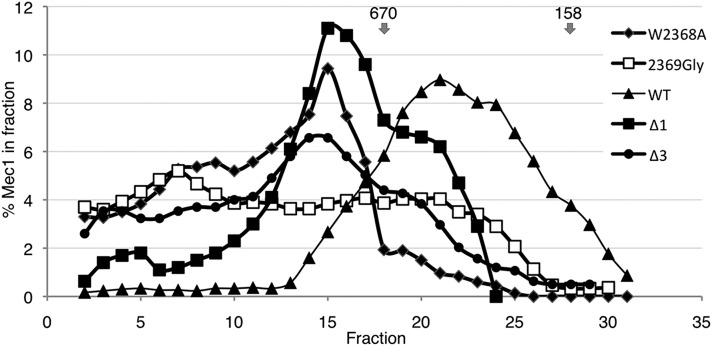
As another measure of whether the Mec1 derivatives were present in their functional forms, we performed size exclusion chromatography on extracts prepared from strains containing Flag5-tagged versions. As shown in Figure 5, wild-type Mec1 elutes from a Superose 6 column with an estimated mass less than ∼670 kD. A portion of each of the mutant versions eluted from the column in a fraction with a significantly greater estimated mass, with none of the profiles paralleling the wild-type. Mec1-2369G had a distinct profile in that it was dispersed almost equally across the high-molecular-mass fractions. The presence of the higher-mass forms suggests the possibility of improper folding of the Mec1 derivatives, or perhaps prolonged association with chaperone complexes that are required for the formation of functional PIKK proteins (Boulon et al. 2012; Makhnevych and Houry 2012).
Figure 5.
Resolution of Mec1 derivatives by size-exclusion chromatography. Protein extracts were prepared from yeast strains CY6172 (Flag5-MEC1), CY6192 (Flag5-mec1-W2368A), CY6184 (Flag5-mec1-Δ1), CY6233 (Flag5-mec1-Δ3), and CY6203 (Flag5-mec1-2369G); 4.5 mg protein from each strain was separated independently on a 24-ml FPLC Superose 6HR10/30 column (Amersham Pharmacia Biotech). Protein from 20 μl aliquots of 250 μl fractions was resolved by sodium dodecyl sulfate (SDS)-PAGE and proteins were detected by Western blotting. The amount of protein in each fraction was determined by densitometry and calculated as a ratio of the total protein. The plot shows the average of a three-fraction window. The arrows indicate the migration of 670 kD and 158 kD molecular mass standards.
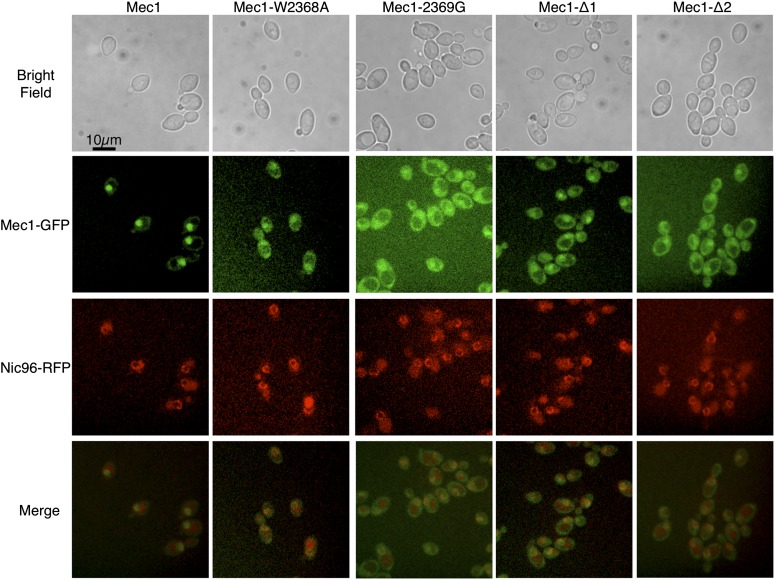
We also examined the localization of N-terminally eGFP-tagged versions of the proteins. Diploid strains expressing wild-type Mec1 and a single copy of eGFP-tagged Mec1 or a derivative were grown at 30°. RFP-tagged Nic96 was used to mark the nuclear periphery. As shown in Figure 6, eGFP-tagged wild-type Mec1 is found principally in the nucleus. The distributions of each of Mec1-W2368A, Mec1-2369G, Mec1-Δ1, and Mec1-Δ2 are more disperse, with both nuclear and cytoplasmic localization apparent. The experiment also was performed for cells grown at 37°, but the signal was significantly reduced for the Mec1 derivatives.
Figure 6.
Localization of eGFP-Mec1. Diploid yeast strains containing an eGFP-tagged Mec1 derivative (CY6307, Mec1; CY6302, Mec1-W2368A; CY6330, Mec1-Δ1; CY6344, Mec1-Δ2; and CY6342, Mec1-2369G) were mated to a strain containing RFP-tagged Nic96 (Huh et al. 2003). Diploid strains were grown at 30° in synthetic complete (SC) media to stationary phase, diluted 1:20 in SC, grown another 6 hr, and visualized by fluorescence microscopy. A representative field of cells is shown.
rpn3-L140P suppresses the temperature-sensitive growth of mec1-W2368A
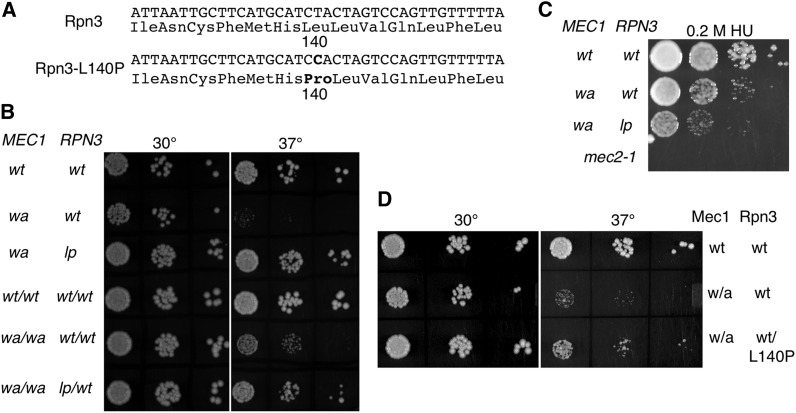
Together these results suggested that the C-terminal residues of Mec1 were required for the folding/stability of the molecule. If this is the case, then we predict that it should be possible to identify second site suppressor mutations that restore the levels of the protein and allowed growth of the mec1-W2368A strain at 37°. We plated approximately 20 million cells at a density of 1 million cells per plate, subjected them to a low dose of ultraviolet radiation, and selected colonies that grew at 37°. Nine colonies were identified that carried a single extragenic suppressor mutation. These were backcrossed an additional nine times to eliminate variations unlinked to the suppressor mutation, and then their genomic sequence was determined. Unique mutations were identified in the following four strains: rpn3 (L140P; a T-to-C transition at bp 419; Figure 7A); rfx1 (S213N); rfx1 (S462L); and sml1 (L62ochre). Loss of function of Sml1 and loss of function of Rfx1 are known to suppress nonfunctional alleles of mec1 (Weinert et al. 1994; Desany et al. 1998; Huang et al. 1998). Consistent with these being causative mutations, we found that our sml1 and rfx1 strains also would suppress mec1-Δ1. In contrast, the rpn3-L140P allele would not suppress mec1-Δ1, mec1-Δ2, or mec1-2369G.
Figure 7.
Suppression of the temperature-sensitive growth resulting from mec1-W2368A by rpn3-L140P. (A) Sequence of RPN3 and the rpn3-L140P allele. (B) Yeast strains BY4742 (MEC1 RPN3), CY6077 (mec1-W2368A RPN3), CY6265 (mec1-W2368A rpn3-L140P), BY4743 (MEC1/MEC1 RPN3/RPN3), CY6368 (mec1-W2368A/mec1-W2368A), and CY6385 (mec1-W2368A/mec1-W2368A RPN3/rpn3-L140P) were grown to stationary phase, diluted 1/102, and 10-fold serial dilutions were spotted onto YPD plates at 30° and 37°. (C) Serial dilutions of BY4742, CY6077, and CY6265 were spotted onto a YPD plate containing 0.2 M hydroxyurea. A mec2-1 strain was included to verify the quality of the hydroxyurea. (D) Serial dilutions of BY4741, CY6076 (mec1-W2368A RPN3), and CY6465 (mec1-W2368A RPN3/rpn3-L140P-URA3) were spotted onto YPD plates at 30° and 37°.
Because Rpn3 encodes a component of the 19S proteasomal regulatory particle (Kominami et al. 1997), thus potentially linking it to the stability of Mec1, we chose to examine rpn3-L140P in greater detail. The strain containing this mutation grew at 37°, at a level comparable with a wild-type strain, and the mutation responsible did so in a partially dominant fashion (Figure 7B). The suppressor mutation did not facilitate growth on plates containing 0.2 M hydroxyurea, but rather resulted in a further reduction of growth (Figure 7C). Two approaches were used to demonstrate that suppression of mec1-W2368A was the result of rpn3-L140P. First, we compared the partially dominant effect of rpn3-L140P/RPN3 as shown in Figure 7B, with the ability of rpn3-L140P inserted into CY6076 (mec1-W2368A) on an integrating plasmid (Figure 7D). As was found for the heterozygous diploid, addition of the rpn3-L140P–containing plasmid partially, but not completely, reversed the slow growth at 37° because of mec1-W2368A. Second, we analyzed independent spore colonies from a cross of CY6265 (rpn3-L140P mec1-W2368A) and CY6076 (RPN3mec1-W2368A). The RPN3 allele from each spore colony was isolated by PCR and sequenced. For 14 alleles (seven fast-growing strains and seven slow-growing strains), the fast growth predicted the presence of the rpn3-L140P allele.
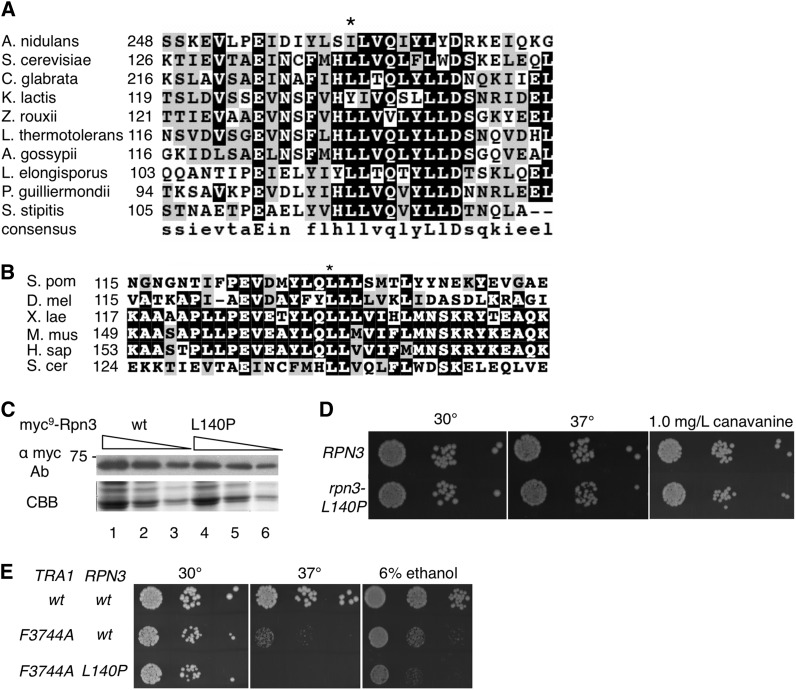
Rpn3 is an essential 524-amino-acid residue protein that contains PCI/PINT-associated module (PAM) and winged helix domains located C-terminally distal to residue 140. As shown in Figure 8, A and B, L140 is in a hydrophobic region conserved in fungal species and more broadly in eukaryotes. The L140P mutation does not alter the level of Rpn3 found in the cell (Figure 8C). To determine the effect of Rpn3-L140P in isolation, yeast strain CY6398 (MEC1rpn3-L140P) was engineered by backcrossing CY6265 with BY4741. As shown in Figure 8D, the rpn3-L140P allele had no detectable effect on growth of cells in rich media at 30°. A slight reduction in growth was observed for cells grown at 37° and in media containing the arginine analog canavanine.
Figure 8.
Leucine 140 is found in a conserved hydrophobic region. (A) Multiple sequence alignment of Rpn3 from a variety of fungal species. Leucine 140 (S. cerevisiae) is indicated by an asterisk. The alignment was performed with MUSCLE (Edgar 2004). In order, the proteins are as follows: Aspergillus nidulans, XP_660371.1; Saccharomyces cerevisiae, NP_010938.1; Candida glabrata, XP_447606.1; Kluyveromyces lactis, XP_453544.1; Zygosaccharomyces rouxii, XP_002496514.1; Lachancea thermotolerans, XP_002554976.1; Ashbya gossypii, NP_983112.1; Lodderomyces elongisporus, XP_001526739.1; Pichia guilliermondii, XP_001487379.1; and Scheffersomyces stipitis, XP_001383755.2. (B) Multiple sequence alignment of Rpn3 from a variety of eukaryotes. The proteins are as follows: Schizosaccharomyces pombe, NP_595282.2; Drosophila melanogaster, NP_477300; Xenopus laevis, NP_001085955.1; Mus musculus, NP_033465.1; and Homo sapiens, NP_002800.2. (C) Expression of Rpn3-L140P. Extracts were prepared from BY4741 expressing myc9-Rpn3 (lanes 1-3) or myc9-Rpn3-L140P from a centromeric plasmid; 50, 25, and 10 μg of protein was separated by sodium dodecyl sulfate (SDS)-PAGE and Western-blotted with anti-Myc antibody (upper panel). The lower section of the gel was stained with Coomassie brilliant blue (lower panel). (D) Phenotype of rpn3-L140P in isolation. Yeast strains BY4741 (RPN3) and CY6398 (rpn3-L140P) were grown to stationary phase and serial dilutions were spotted onto YPD plates at 30° and 37° and onto a minimal plate containing 1.0 mg/l canavanine. (E) rpn3-L140P does not suppress tra1-F3744A. Yeast strains CY4353 (TRA1 RPN3), CY4350 (tra1-F3744A RPN3), and CY6418 (tra1-F3744A rpn3-L140P) were grown to stationary phase and serial dilutions were spotted onto YPD plates at 30° and 37° or a YPD plate containing 6% ethanol at 30°.
As mentioned, rpn3-L140P does not suppress mec1-Δ1, mec1-Δ2, or mec1-2369G. To address whether rpn3-L140P would suppress a mutation similar to mec1-W2368A in a related PIKK protein, we introduced rpn3-L140P into a strain containing a Phe-to-Ala change of the terminal phenylalanine of Tra1 (tra1-F3744A). As shown in Figure 8E, rpn3-L140P did not suppress the slow growth at 37° or in ethanol-containing media caused by tra1-F3744A, and, in fact, resulted in synthetic slow growth in this context.
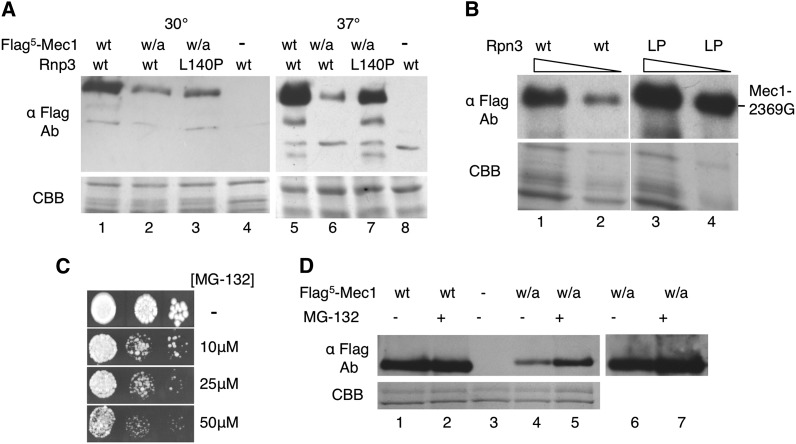
The terminal tryptophan-to-alanine mutation in Mec1 reduced the stability of the protein (Figure 4). To investigate the effect of rpn3-L140P on Mec1-W2368A, the diploid strain CY6400 with the genotype Flag5-mec1-W2368A/MEC1rpn3-L140P/rpn3-L140P was engineered. The level of Flag5-Mec1 in this strain was compared to that found in a wild-type RPN3/RPN3 background (Figure 9A). For extracts prepared from cells grown at 30° and in the absence of protease inhibitors, the level of Mec1-W2368A was somewhat reduced relative to the wild-type (Figure 9A, compare lanes 1 and 2). The rpn3-L140P resulted in an increase in Mec1-W2368A (Figure 9A, compare lanes 2 and 3). For extracts prepared from cells grown at 37°, the level of Mec1-W2368A was further reduced relative to the wild-type (Figure 9A, compare lanes 5 and 6), with rpn3-L140P resulting in a dramatic increase (Figure 9A, compare lanes 6 and 7). We conclude that rpn3-L140P increases the level of Mec1-W2368A, thus facilitating growth at elevated temperatures. As shown in Figure 9B, rpn3-L140P acts more broadly, increasing the level of Mec1-2369G when cells are grown at 37°.
Figure 9.
rpn3-L140P increases the level of C-terminally altered Mec1 derivatives. (A) Mec1-W2368A. Protein extracts were prepared by bead lysis in buffer lacking protease inhibitors from diploid yeast strains CY6172 (MEC1/Flag5-MEC1 RPN3/RPN3), CY6192 (MEC1/Flag5-mec1-W2368A RPN3/RPN3), CY6400 (MEC1/Flag5-mec1-W2368A rpn3-L140P/rpn3-L140P), and BY4743 (MEC1/MEC1 RPN3/RPN3) grown at 30° or 37°; 50 μg of protein was separated by sodium dodecyl sulfate (SDS)-PAGE and the upper portion of the gel was Western-blotted with anti-Flag antibody. The lower part of the gel was stained with Coomassie brilliant blue (CBB). (B) Mec1-2369G. Yeast strains CY6349 (mec1-2369G RPN3 sml1Δ0::KanMX; lanes 1 and 2) and CY6449 (mec1-2369G rpn3-L140P sml1Δ0::KanMX; lanes 3 and 4) were grown in YPD media for 8 hr at 37°; 40 μg (odd lanes) and 20 μg (even lanes) of protein extract was separated by SDS-PAGE and Western-blotted with anti-Flag antibody or stained with CBB. (C) BY4742 was grown in media containing proline as the nitrogen source and 0.003% SDS for 3 hr, then serial dilutions were plated onto identical synthetic complete media with the indicated amount of MG-132. (D) CY6172 (MEC1/Flag5-MEC1; lanes 1 and 2), BY4743 (lane 3), and CY6192 (MEC1/Flag5-mec1-W2368A; lanes 4–7) were grown in media containing proline as the nitrogen source and 0.003% SDS for 3 hr at 37°. MG-132 was added to a final concentration of 75 μM (+) or the equivalent volume of dimethylsulfoxide (−) and the cells were grown for an additional 2 hr. Extracts were prepared by glass bead lysis and 30 μg was separated by SDS-PAGE. The upper portion of the gel was Western-blotted with anti-Flag antibody. The lower portion was stained with CBB. Lanes 6 and 7 are an overexposure of lanes 4 and 5.
The suppression of mec1-W2368A by rpn3-L140P introduces the possibility that Mec1 turnover is mediated by its degradation by the proteasome. To begin to address this issue, we analyzed Flag5-tagged Mec1 and Mec1-W2368A in cells treated with the proteasome inhibitor MG-132. Cells were treated under conditions described by Liu et al. (2007) to increase their permeability to MG-132. As shown in Figure 9C, when grown in media containing proline as the nitrogen source and 0.003% SDS, yeast is sensitive to MG-132 in micromolar concentrations. For the analysis of Mec1 and Mec1-W2368A, extracts were prepared from cells grown at 37° in the presence or absence of 75 μM MG-132 for 2 hr. Wild-type Mec1 was not altered by the MG-132 treatment (Figure 9D, compare lanes 1 and 2). In contrast, Mec1-W2368A levels increased approximately two-fold on treatment (Figure 9D, compare lanes 4 and 5), suggesting that the proteasome contributes, at least in part, to the turnover of the protein. For the wild-type and Mec1-W2368A proteins, there was no evidence of higher-molecular-mass forms that would be indicative of ubiquitylation of the protein.
Discussion
The FATC domain is found at the C-terminus of all the PIKK proteins. In each, the two most C-terminal residues are large and hydrophobic. We have shown that the terminal tryptophan residue of Mec1 is required for the function of the protein. Deletion of this residue results in a loss of viability; conversion to alanine results in temperature-sensitive growth and reduced growth in hydroxyurea, a condition in which deoxyribonucleotides are depleted. Moreover, the position of the terminal tryptophan is essential. Addition of a single glycine residue also causes loss of viability. Because these results parallel those found for Tra1 (Genereaux et al. 2012), we conclude that the terminal residues are likely a key feature of all the PIKK proteins, a conclusion consistent with the positioning of the FATC domain in the structure of mTOR (Yang et al. 2013).
We propose that at least one role for the terminal residue is assisting in the folding of the protein. The C-terminally deleted Mec1 proteins were less abundant in crude protein lysates, and all of the derivatives analyzed were unstable in cell extracts prepared in the absence of protease inhibitors. This suggests that the temperature-sensitive phenotype of the mec1-W2368A strain may result from decreased stability of the protein at elevated temperatures, a conclusion supported by the increased levels found in the rpn3-L140P strain or in the presence of MG-132. Also suggesting the possibility of misfolding, we found that whereas wild-type Mec1 was found almost exclusively in the nucleus, the partially active and inactive forms of the protein showed partial cytoplasmic localization. In addition, each of the mutant forms of Mec1 chromatographed aberrantly on a size-exclusion column. Structural models (Lempiäinen and Halazonetis 2009; Sturgill and Hall 2009; Yang et al. 2013) position the FATC domain in a hydrophobic pocket that in porcine PI3K-γ interacts with helical domains (Walker et al. 1999). Loss of hydrophobic interactions may decrease the stability of the PI3K domain, whereas addition of a glycine residue may not permit packing into the pocket. In both cases, the FATC domain and the core PI3K domain would be susceptible to proteolysis.
Our results with Mec1 closely parallel what we have observed with Tra1 (Hoke et al. 2010). In the case of Tra1, converting the terminal phenylalanine to alanine results in mislocalization, decreased protein levels, and a number of stress-related phenotypes. Consistent with a role for the FATC domain in folding, alleles of tti2, whose product with Tel2 and Tti1 is proposed to act as a chaperone (Horejsi et al. 2010; Hurov et al. 2010; Kaizuka et al. 2010; Takai et al. 2010), suppress tra1-F3744A in a partially dominant fashion (Genereaux et al. 2012). It was interesting that both Tra1-F3744A and Mec1-W2368A are mislocalized. Whether the other properties of the Mec1 and Tra1 derivatives result from or cause the mislocalization will require additional experimentation; however, our working model is that the molecules need to be specifically localized in the cytoplasm in complex with chaperones until they achieve a conformation that will support nuclear import and, if diverted from this pathway, are at risk of being degraded.
Mutation of the terminal residue of Mec1 to alanine diminished kinase activity to a level approximately 10-fold less than that of the wild-type protein. The decrease in kinase activity of Mec1-W2368A and the other Mec1 derivatives can be explained by a role for the terminal residues in folding and their possible participation in substrate recognition as found for mTOR (Yang et al. 2013). Because mec1-W2368A will support viability, minimal kinase activity must be sufficient for growth under optimal conditions. In media containing hydroxyurea, where replication fork collapse will increase, this same level of activity was insufficient. It is also this requirement for maximal levels of Mec1 activity during replicative stress that likely explains why rpn3-L140P would not suppress slow growth of the mec1-W2368A strain in hydroxyurea. We note that interpretation of this experiment is somewhat complicated because of the role of Rpn3 in aspects of cell-cycle control (Bailly and Reed 1999).
Lcd1/Ddc2 interacts with the N-terminal HEAT domain sequences of Mec1 (Wakayama et al. 2001), thus providing a rationale for why it interacted with each of the altered Mec1 proteins with approximately the same efficiency. Because some of these derivatives are partially excluded from the nucleus, this result suggests that the Mec1-Lcd1/Ddc2 interaction takes place in the cytoplasm.
We have shown that a mutation resulting in a change of leucine 140 to proline in Rpn3 suppresses the temperature-sensitive growth of a mec1-W2368A strain. Rpn3 is a component of the 19S regulatory particle of the 26S proteasome (Beck et al. 2012; Lander et al. 2012; Lasker et al. 2012; Kish-Trier and Hill 2013). The regulatory particle can be subdivided into a base and lid. The base, composed of 10 components, interacts directly with the 20S proteolytic core particle and contains ATPases (Rpt1-6) required for the unfolding of substrate proteins (Smith et al. 2007; Rabl et al. 2008; Tomko et al. 2010). In addition to Rpn3, the lid contains Rpn5–Rpn9, Rpn11, Rpn12, and Sem1. The 26S proteasome is the key enzymatic machinery required to degrade ubiquitylated proteins, either misfolded or short-lived, in the cell (Finley 2009). Although the exact role of Rpn3 is unknown, the lid is involved in recognizing ubiquitylated target proteins (van Nocker et al. 1996; Husnjak et al. 2008; Schreiner et al. 2008) and removing the ubiquitin chains before the degradation of the protein by the 20S proteolytic core particle (Verma et al. 2002). Also of note, independent functions for the 19S regulatory particle have been observed in nucleotide excision repair (Russell et al. 1999; Gillette et al. 2001) and transcription (Ferdous et al. 2001; Gonzalez et al. 2002; Lee et al. 2005; Uprety et al. 2012).
We have considered possible models for how rpn3-L140P may suppress mec1-W2368A. It is unlikely that Rpn3-L140P acts indirectly by altering the DNA damage response, as is seen with loss of function of Sml1 or Rfx1, because of the observed allele specificity and the increased levels of Mec1-W2368A in the rpn3-L140P background. Models for how rpn3-L140P suppresses the temperature sensitivity of mec1-W238A need to reflect the role of the terminal sequences in the folding, localization, or stability of the protein. We present two models that are not mutually exclusive. Key in these is that rpn3-L140P increases the level of Mec1-W2368A, and does so more for Mec1-W2368A than for wild-type Mec1, and also more at 37° than 30°. First, the proteasomal regulatory particle has chaperone activities that normally participate in the delivery of the protein target to the 20S core particle (Braun et al. 1999). Independently, this activity could aid in the folding of Mec1-W2368A at elevated temperature and could be enhanced by Rpn3-L140P. This model would account for the partial dominance of the rpn3-L140P allele and would be analogous to the suppression of tra1-F3744A by alleles encoding the chaperone component Tti2. Interestingly, the same tti2 alleles do not suppress mec1-W2368A (Genereaux et al. 2012), and loss of interactions with the Tel2-Tti1-Tti2 component Tel2 only modestly affects Mec1 function (Anderson and Blackburn 2008), leaving the possibility that another chaperone may replace or augment their activity. In the second model, the proteasome is involved in the turnover of improperly folded or mislocalized Mec1-W2368A. Subtle changes in targeting or activity of the proteasome caused by the L140P substitution in Rpn3 could decrease the rate of turnover, affording additional time for Mec1-W2368A to fold into a stable conformation or to be correctly localized. The rpn3 allele would have partial dominance by altering the activity of half of the proteasome particles. We favor this model because inhibiting the proteolytic activity of the proteasome with MG-132 also increased the level of Mec1-W2368A. In addition, the L140P mutation is in a highly conserved region of the protein, suggesting a functional role. Although the phenotypes attributable to rpn3-L140P were not severe under most conditions, it does result in synthetic slow growth in combination with tra1-F3744A and reduced growth in combination with mec1-W2368A in media containing hydroxyurea. We did not observe an obvious loss of mobility of Mec1-W2368A in any of our Western blots that would be indicative of Mec1 polyubiquitylation, the general signal for targeting to the proteasome. The model is actually simplified in the absence of Mec1-W2368A ubiquitylation because it is less likely that polyubiquitylated Mec1-W2368A could be restored to an active form. The question would remain, however, of how Mec1-W2368A might be targeted to the proteasome. Examples of proteasomal targeting not requiring direct ubiquitylation do exist, this targeting is mediated by a second molecule, which in some cases is ubiquitylated (Bercovich et al. 1989; Sheaff et al. 2000; Perrotti et al. 2000; Sdek et al. 2005; Isono et al. 2008). Because of the nature of the role of Rpn3 in the proteasome, we cannot exclude a third model whereby rpn3-L140P acts indirectly by altering the level or activity of an intermediary protein.
The rpn3-L140P mutation suppressed some (growth at 37°) but not all (growth in hydroxyurea) phenotypes resulting from mec1-W2368A. We believe that this can be attributed to the threshold of Mec1 activity required for growth in hydroxyurea being greater than that for growth at 37°. In turn, this suggests that Rpn3-L140P is not sufficient to increase the activity of Mec1-W2368A above this threshold. We have considered whether Rpn3-L140P is able to increase the kinase activity of Mec1-W2368A by analyzing the activity of Flag5-tagged Mec1-W2368A isolated by immunoprecipitation from wild-type RPN3 and rpn3-L140P strains. The experiment was performed with cells grown at 30°, at which temperature the control mec1-W2368A RPN3 strain is able to grow. After normalizing to the amount of Mec1-W2368A present, an increase in activity was not observed for the protein from the rpn3-L140P strain (Supporting Information, Figure S1). This result is consistent with rpn3-L140P suppressing mec1-2368A by increasing the level and localization of Mec1-W2368A protein rather than by increasing the per-molecule activity.
As indicated, the 19S regulatory particle has a role in transcriptional regulation. Rpn3-L140P thus could increase the level of active Mec1-W2368A by activating its transcription; however, several points argue against this. Of particular note, the suppression by rpn3-L140P was observed when Mec1-W2368A was expressed from both its native promoter, the case for the original selections, and from the TRA1 promoter, the case for the Flag-tagged derivatives. Furthermore, a role in transcription would be expected to be manifest by increased protein levels of approximately the same magnitude at both 30° and 37°. Increased expression was greater for cells grown at 37°, at which temperature Mec1-W2368A would have greater tendency to misfold. The finding that MG-132 mimics rpn-L140P also supports a role for the mutation in altering protein turnover rather than transcription. Finally, unlike the ATPase encoding genes RPT6 (SUG1) and RPT4 (SUG2), alleles of RPN3 were not identified in screens for altered transcriptional regulation (Swaffield et al. 1992; Xu et al. 1995; Russell et al. 1996).
In summary, we have shown that the C-terminus of Mec1 is important for the stability of the protein, a feature likely common to the PIKK family. The reduced kinase activity of Mec1-W2368A suggests the C-terminal residues participate in the activity of the protein, with the viability of the mec1-W2368A strain indicating that minimal kinase activity is required for growth in rich media. Furthermore, suppression of the temperature sensitivity of mec1-W2368A by an allele encoding the proteasome component Rpn3 implicates the proteasome in regulating Mec1 levels, and implicates Rpn3 as having a key role in this function.
Supplementary Material
Acknowledgments
We thank Dr. David Edgell and Dr. Darah Christie for critically reading the manuscript, Peter Arvidson for providing RFP-tagged strains, David Litchfield and Laszlo Gyenis for reagents, and Bernard Duncker for yeast strains. This work was supported by Canadian Institutes of Health Research grant MOP10845 (to C.J.B.). L.F.D. was supported by a Canadian Institutes of Health Research grant to Megan Davey.
Footnotes
Communicating editor: C. S. Hoffman
Literature Cited
- Abraham R. T., 2004. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst.) 3: 883–887 [DOI] [PubMed] [Google Scholar]
- Anderson C. M., Blackburn E. H., 2008. Mec1 function in the DNA damage response does not require its interaction with Tel2. Cell Cycle 7: 3695–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J., 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bailly E., Reed S. I., 1999. Functional characterization of rpn3 uncovers a distinct 19S proteasomal subunit requirement for ubiquitin-dependent proteolysis of cell cycle regulatory proteins in budding yeast. Mol. Cell. Biol. 19: 6872–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball H. L., Myers J. S., Cortez D., 2005. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell 16: 2372–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamish H. J., Jessberger R., Riballo E., Priestley A., Blunt T., et al. , 2000. The C-terminal conserved domain of DNA-PKcs, missing in the SCID mouse, is required for kinase activity. Nucleic Acids Res. 28: 1506–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck F., Unverdorben P., Bohn S., Schweitzer A., Pfeifer G., et al. , 2012. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. USA 109: 14870–14875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovich Z., Rosenberg-Hasson Y., Ciechanover A., Kahana C., 1989. Degradation of ornithine decarboxylase in reticulocyte lysate is ATP-dependent but ubiquitin-independent. J. Biol. Chem. 264: 15949–15952 [PubMed] [Google Scholar]
- Bhaumik S. R., Raha T., Aiello D. P., Green M. R., 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18: 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosotti R., Isacchi A., Sonnhammer E. L., 2000. FAT: a novel domain in PIK-related kinases. Trends Biochem. Sci. 25: 225–227 [DOI] [PubMed] [Google Scholar]
- Boulon S., Bertrand E., Pradet-Balade B., 2012. HSP90 and the R2TP co-chaperone complex: building multi-protein machineries essential for cell growth and gene expression. RNA Biol. 9: 148–154 [DOI] [PubMed] [Google Scholar]
- Braun B. C., Glickman M., Kraft R., Dahlmann B., Kloetzel P. M., et al. , 1999. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1: 221–226 [DOI] [PubMed] [Google Scholar]
- Brown C. E., Howe L., Sousa K., Alley S. C., Carrozza M. J., et al. , 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333–2337 [DOI] [PubMed] [Google Scholar]
- Brumbaugh K. M., Otterness D. M., Geisen C., Oliveira V., Brognard J., et al. , 2004. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol. Cell 14: 585–598 [DOI] [PubMed] [Google Scholar]
- Brush G. S., Morrow D. M., Hieter P., Kelly T. J., 1996. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc. Natl. Acad. Sci. USA 93: 15075–15080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. F., Imam J. S., Wilkinson M. F., 2007. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Cimprich K. A., Cortez D., 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J. A., Bjergbaek L., Shimada K., Frei C., Gasser S. M., 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J. A., Schleker T., Rojas V., Bjergbaek L., Tercero J. A., et al. , 2005. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Glick G., Elledge S. J., 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA 101: 10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski N., Hall M. N., 2009. TOR complex 2: a signaling pathway of its own. Trends Biochem. Sci. 34: 620–627 [DOI] [PubMed] [Google Scholar]
- Dames S. A., Mulet J. M., Rathgeb-Szabo K., Hall M. N., Grzesiek S., 2005. The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J. Biol. Chem. 280: 20558–20564 [DOI] [PubMed] [Google Scholar]
- Desany B. A., Alcasabas A. A., Bachant J. B., Elledge S. J., 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12: 2956–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs J. A., Lowndes N. F., Jackson S. P., 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S. P., 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611 [DOI] [PubMed] [Google Scholar]
- Ferdous A., Gonzalez F., Sun L., Kodadek T., Johnston S. A., 2001. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell 7: 981–991 [DOI] [PubMed] [Google Scholar]
- Finley D., 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J., Mohibullah N., Hahn S., 2005. Function of a eukaryotic transcription activator during the transcription cycle. Mol. Cell 18: 369–378 [DOI] [PubMed] [Google Scholar]
- Friedel A. M., Pike B. L., Gasser S. M., 2009. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 21: 237–244 [DOI] [PubMed] [Google Scholar]
- Gehen S. C., Staversky R. J., Bambara R. A., Keng P. C., O’Reilly M. A., 2008. hSMG-1 and ATM sequentially and independently regulate the G1 checkpoint during oxidative stress. Oncogene 27: 4065–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genereaux J., Kvas S., Dobransky D., Karagiannis J., Gloor G. B., et al. , 2012. Genetic evidence links the ASTRA protein chaperone component Tti2 to the SAGA transcription factor Tra1. Genetics 191: 765–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette T. G., Huang W., Russell S. J., Reed S. H., Johnston S. A., et al. , 2001. The 19S complex of the proteasome regulates nucleotide excision repair in yeast. Genes Dev. 15: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F., Delahodde A., Kodadek T., Johnston S. A., 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296: 548–550 [DOI] [PubMed] [Google Scholar]
- Grant P. A., Schieltz D., Pray-Grant M. G., Yates J. R., III, Workman J. L., 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2: 863–867 [DOI] [PubMed] [Google Scholar]
- Hoke S. M., Irina M. A., Genereaux J., Kvas S., Buck M., et al. , 2010. Mutational analysis of the C-terminal FATC domain of Saccharomyces cerevisiae Tra1. Curr. Genet. 56: 447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horejsi Z., Takai H., Adelman C. A., Collis S. J., Flynn H., et al. , 2010. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol. Cell 39: 839–850 [DOI] [PubMed] [Google Scholar]
- Huang M., Zhou Z., Elledge S. J., 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605 [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., et al. , 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Hurov K. E., Cotta-Ramusino C., Elledge S. J., 2010. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 24: 1939–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., et al. , 2008. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453: 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono O., Ohshima T., Saeki Y., Matsumoto J., Hijikata M., et al. , 2008. Human T-cell leukemia virus type 1 HBZ protein bypasses the targeting function of ubiquitination. J. Biol. Chem. 283: 34273–34282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka T., Hara T., Oshiro N., Kikkawa U., Yonezawa K., et al. , 2010. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J. Biol. Chem. 285: 20109–20116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., et al. , 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Kish-Trier E., Hill C. P., 2013. Structural Biology of the Proteasome. Annu.Rev.Biophys 42: 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson B. A., Hahn S., 2011. Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol. Cell. Biol. 31: 818–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami K., Okura N., Kawamura M., DeMartino G. N., Slaughter C. A., et al. , 1997. Yeast counterparts of subunits S5a and p58 (S3) of the human 26S proteasome are encoded by two multicopy suppressors of nin1–1. Mol. Biol. Cell 8: 171–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K., De P. G., 2011. Surviving chromosome replication: the many roles of the S-phase checkpoint pathway. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 3554–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., et al. , 2012. Complete subunit architecture of the proteasome regulatory particle. Nature 482: 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K., Forster F., Bohn S., Walzthoeni T., Villa E., et al. , 2012. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. USA 109: 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Ezhkova E., Li B., Pattenden S. G., Tansey W. P., et al. , 2005. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123: 423–436 [DOI] [PubMed] [Google Scholar]
- Lempiäinen H., Halazonetis T. D., 2009. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 28: 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. A., Kong X., Haystead T. A., Pause A., Belsham G., et al. , 1994. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266: 653–656 [DOI] [PubMed] [Google Scholar]
- Liu C., Apodaca J., Davis L. E., Rao H., 2007. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques 42: 158–162 [DOI] [PubMed] [Google Scholar]
- Loewith R., Hall M. N., 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy C. A., Cortez D., 2009. Common mechanisms of PIKK regulation. DNA Repair (Amst.) 8: 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. L., Lee S. J., Duong J. K., Stern D. F., 2006. Activation of the checkpoint kinase Rad53 by the phosphatidyl inositol kinase-like kinase Mec1. J. Biol. Chem. 281: 3954–3963 [DOI] [PubMed] [Google Scholar]
- Makhnevych T., Houry W. A., 2012. The role of Hsp90 in protein complex assembly. Biochim. Biophys. Acta 1823: 674–682 [DOI] [PubMed] [Google Scholar]
- Mallory J. C., Petes T. D., 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. USA 97: 13749–13754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon S. B., Van Buskirk H. A., Dugan K. A., Copeland T. D., Cole M. D., 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94: 363–374 [DOI] [PubMed] [Google Scholar]
- Mordes D. A., Glick G. G., Zhao R., Cortez D., 2008. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 22: 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Yamashita A., Kashima I., Ogata K., Ishiura S., et al. , 2007. Distant N- and C-terminal domains are required for intrinsic kinase activity of SMG-1, a critical component of nonsense-mediated mRNA decay. J. Biol. Chem. 282: 7799–7808 [DOI] [PubMed] [Google Scholar]
- Morrison A. J., Kim J. A., Person M. D., Highland J., Xiao J., et al. , 2007. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 130: 499–511 [DOI] [PubMed] [Google Scholar]
- Moser B. A., Chang Y. T., Kosti J., Nakamura T. M., 2011. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat. Struct. Mol. Biol. 18: 1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutiu A. I., Hoke S. M., Genereaux J., Hannam C., MacKenzie K., et al. , 2007. Structure/function analysis of the phosphatidylinositol-3-kinase domain of yeast tra1. Genetics 177: 151–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn A. J., Elledge S. J., 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V., Clerici M., Lucchini G., Longhese M. P., 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14: 2046–2059 [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lucca C., Liberi G., Marini F., Lopes M., et al. , 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18: 6561–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti D., Iervolino A., Cesi V., Cirinna M., Lombardini S., et al. , 2000. BCR-ABL prevents c-jun-mediated and proteasome-dependent FUS (TLS) proteolysis through a protein kinase CbetaII-dependent pathway. Mol. Cell. Biol. 20: 6159–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J., Kleckner N., 2003. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112: 151–155 [DOI] [PubMed] [Google Scholar]
- Priestley A., Beamish H. J., Gell D., Amatucci A. G., Muhlmann-Diaz M. C., et al. , 1998. Molecular and biochemical characterisation of DNA-dependent protein kinase-defective rodent mutant irs-20. Nucleic Acids Res. 26: 1965–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J., Smith D. M., Yu Y., Chang S. C., Goldberg A. L., et al. , 2008. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30: 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves W. M., Hahn S., 2005. Targets of the Gal4 transcription activator in functional transcription complexes. Mol. Cell. Biol. 25: 9092–9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J., Jackson S. P., 2000. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 19: 5801–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J., Jackson S. P., 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9: 857–869 [DOI] [PubMed] [Google Scholar]
- Russell S. J., Reed S. H., Huang W., Friedberg E. C., Johnston S. A., 1999. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol. Cell 3: 687–695 [DOI] [PubMed] [Google Scholar]
- Russell S. J., Sathyanarayana U. G., Johnston S. A., 1996. Isolation and characterization of SUG2. A novel ATPase family component of the yeast 26 S proteasome. J. Biol. Chem. 271: 32810–32817 [DOI] [PubMed] [Google Scholar]
- Saleh A., Lang V., Cook R., Brandl C. J., 1997. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J. Biol. Chem. 272: 5571–5578 [DOI] [PubMed] [Google Scholar]
- Saleh A., Schieltz D., Ting N., McMahon S. B., Litchfield D. W., et al. , 1998. Tra1p is a component of the yeast Ada.Spt transcriptional regulatory complexes. J. Biol. Chem. 273: 26559–26565 [DOI] [PubMed] [Google Scholar]
- Schreiner P., Chen X., Husnjak K., Randles L., Zhang N., et al. , 2008. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453: 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdek P., Ying H., Chang D. L., Qiu W., Zheng H., et al. , 2005. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell 20: 699–708 [DOI] [PubMed] [Google Scholar]
- Sheaff R. J., Singer J. D., Swanger J., Smitherman M., Roberts J. M., et al. , 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5: 403–410 [DOI] [PubMed] [Google Scholar]
- Sibanda B. L., Chirgadze D. Y., Blundell T. L., 2010. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 463: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., et al. , 2007. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 27: 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill T. W., Hall M. N., 2009. Activating mutations in TOR are in similar structures as oncogenic mutations in PI3KCalpha. ACS Chem. Biol. 4: 999–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffield J. C., Bromberg J. F., Johnston S. A., 1992. Alterations in a yeast protein resembling HIV Tat-binding protein relieve requirement for an acidic activation domain in GAL4. Nature 357: 698–700 [DOI] [PubMed] [Google Scholar]
- Sweeney F. D., Yang F., Chi A., Shabanowitz J., Hunt D. F., et al. , 2005. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 15: 1364–1375 [DOI] [PubMed] [Google Scholar]
- Takahashi T., Hara K., Inoue H., Kawa Y., Tokunaga C., et al. , 2000. Carboxyl-terminal region conserved among phosphoinositide-kinase-related kinases is indispensable for mTOR function in vivo and in vitro. Genes Cells 5: 765–775 [DOI] [PubMed] [Google Scholar]
- Takai H., Wang R. C., Takai K. K., Yang H., de Lange T., 2007. Tel2 regulates the stability of PI3K-related protein kinases. Cell 131: 1248–1259 [DOI] [PubMed] [Google Scholar]
- Takai H., Xie Y., de Lange T., Pavletich N. P., 2010. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 24: 2019–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., Cooper J. P., 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 22: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko R. J., Jr, Funakoshi M., Schneider K., Wang J., Hochstrasser M., 2010. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol. Cell 38: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uprety B., Lahudkar S., Malik S., Bhaumik S. R., 2012. The 19S proteasome subcomplex promotes the targeting of NuA4 HAT to the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for transcriptional initiation in vivo. Nucleic Acids Res. 40: 1969–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S., Sadis S., Rubin D. M., Glickman M., Fu H., et al. , 1996. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 16: 6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., III, et al. , 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 611–615 [DOI] [PubMed] [Google Scholar]
- von Manteuffel S. R., Gingras A. C., Ming X. F., Sonenberg N., Thomas G., 1996. 4E–BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc. Natl. Acad. Sci. USA 93: 4076–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T., Kondo T., Ando S., Matsumoto K., Sugimoto K., 2001. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E. H., Perisic O., Ried C., Stephens L., Williams R. L., 1999. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature 402: 313–320 [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H., 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8: 652–665 [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Davis R. W., 1997. Functional analysis of the yeast genome. Curr. Opin. Genet. Dev. 7: 771–776 [DOI] [PubMed] [Google Scholar]
- Xu Q., Singer R. A., Johnston G. C., 1995. Sug1 modulates yeast transcription activation by Cdc68. Mol. Cell. Biol. 15: 6025–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Tarumoto Y., Ishikawa F., 2012. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 26: 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Rudge D. G., Koos J. D., Vaidialingam B., Yang H. J., et al. , 2013. mTOR kinase structure, mechanism and regulation. Nature 497: 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. Y., Shevchenko A., Shevchenko A., Dunphy W. G., 2004. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J. Biol. Chem. 279: 53353–53364 [DOI] [PubMed] [Google Scholar]
- Zegerman P., Diffley J. F., 2009. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst.) 8: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Zhao X., Muller E. G., Rothstein R., 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2: 329–340 [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S. J., 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.