Abstract
The background linkage disequilibrium (LD) in genetic isolates is of great interest in human genetics. Although many empirical studies have evaluated the background LD in European isolates, such as the Finnish and Sardinians, few data from other regions, such as Asia, have been reported. To evaluate the extent of background LD in East Asian genetic isolates, we analyzed the X chromosome in the Japanese population and in four Mongolian populations (Khalkh, Khoton, Uriankhai, and Zakhchin), the demographic histories of which are quite different from one another. Fisher’s exact test revealed that the Japanese and Khalkh, which are the expanded populations, had the same or a relatively higher level of LD than did the Finnish, European American, and Sardinian populations. In contrast, the Khoton, Uriankhai, and Zakhchin populations, which have kept their population size constant, had a higher background LD. These results were consistent with previous genetic anthropological studies in European isolates and indicate that the Japanese and Khalkh populations could be utilized in the fine mapping of both complex and monogenic diseases, whereas the Khoton, Uriankhai, and Zakhchin populations could play an important role in the initial mapping of complex disease genes.
There are many debates among geneticists, concerning the utility of genetically isolated populations for the mapping of disease genes (Wright et al. 1999; Shifman and Darvasi 2001). One type of genetic isolate—for example, the populations of Finland, Iceland, and Sardinia, which have undergone distinct population isolation and subsequent expansion—has modestly higher levels of background linkage disequilibrium (LD) on many chromosomal regions than do outbred populations (Dunning et al. 2000; Eaves et al. 2000; Taillon-Miller et al. 2000; Zavattari et al. 2000; Angius et al. 2001). In contrast, another type of isolate—for example, the Scandinavian Saami isolate, which is from a small population that remained constant in size after isolation—shows very strong and long-range background LD (Laan and Pääbo 1997; Lonjou et al. 1999). These types of genetic isolates have led to the identification of disease genes (Hästbacka et al. 1992; de la Chapelle and Wright 1998), and, therefore, the value of genetic isolates has increasingly been appreciated during recent attempts at whole-genome LD mapping and association mapping (Wright et al. 1999). However, most studies have mainly focused on particular populations from European regions, such as those from Finland and Sardinia. On the other hand, few data from Asian regions have been reported. It is necessary to investigate the properties of genetic isolates in East Asia and to compare these properties to those of genetic isolates in Europe.
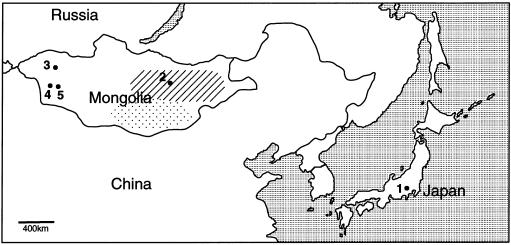
We focused on five populations from two regions of East Asia: Japan and Mongolia (fig. 1). All of the census data are available at the authors' Web site. The demographic history of the Japanese population has been described elsewhere (Koyama 1978; Hayami 2001; Statistics Bureau/Statistical Research and Training Institute 2001). The population size in Japan ∼2,000 years ago was estimated to be ∼600,000 (Koyama 1978). After several gradual expansions, the modern Japanese population consists of ∼120 million people (Statistics Bureau/Statistical Research and Training Institute 2001). Throughout the history of Japan, there seems to be no obvious recent admixture. The Mongolian population consists of ∼2.4 million people, which are divided into ∼20 ethnic groups (Tumen 1992). Among them, the Khalkh is the largest population, with ∼1.8 million people. The Khalkh group is considered to be the direct descendant of the core Mongol tribes, which have inhabited the present-day climatically mild geographical territory of eastern Mongolia (Badamkhatan 1987; Nyambuu 1992). In contrast, the Khoton, Uriankhai, and Zakhchin are regarded as the young, isolated subpopulations of western Mongolia, where the Altai Mountains are located. The Khoton are an extremely small population, with ∼6,000 people living in northwestern Mongolia (fig. 1). They are a Turkish descendant population that has not experienced significant admixture and that migrated into Mongolia in the 12th century (Nyambuu 1992). The Uriankhai and Zakhchin populations consist of ∼23,000 and ∼25,000 people, respectively. They are independent descendants of mixed populations formed by western tribes of Mongolian and Turkish origin (Nyambuu 1992; Badamkhatan 1996). These subpopulations, especially the Khoton, have been separated by severe geographic factors and have become isolates.
Figure 1.
Geographic map around East Asia. Sampling was performed on the following populations: Japanese in Kanagawa and Tokyo (1); Khalkh in Ulaanbaatar (2); Khoton in Tarialan, Uvs aimag (3); Uriankhai in Munkhkhairkhan and Duut, Khovd aimag (4); and Zakhchin in Mankhan and Zereg, Khovd aimag (5). Tarialan, Munkhkhairkhan, and Mankhan are bordered by the steep mountain district formed by part of the Altai Mountains. The hatched and dotted areas indicate the regions of the vast steppe and the Gobi desert, respectively. The Japanese population has the historical and demographic properties of an island population, which would make them a genetic isolate, like the Finnish (Graham and Thompson 1998). The Mongolian populations experienced a historical event in the 12th century. Their country, the “Mongol Empire,” expanded from the eastern steppe to the Near East, and then reduced to its modern size. In this century, although there were immigrations from Turkish and central Asian regions, these immigrants have settled in the mountainous district in western Mongolia (Nyambuu 1992; Badamkhatan 1996).
We collected samples from 182 healthy unrelated Japanese individuals from the Kanto region of the central part of the mainland, which includes Kanagawa and Tokyo (fig. 1). From Mongolia, we collected 146 Khalkh from eastern Mongolia, as well as 74 Khoton, 58 Uriankhai, and 59 Zakhchin from western Mongolia (fig. 1); all individuals were unrelated or at least second cousins. After the mtDNA analysis, we chose 100 Japanese, 83 Khalkh, 40 Khoton, 55 Uriankhai, and 59 Zakhchin male samples for the X-chromosome analysis, including the 49 European Americans (Human variation panel, Caucasian, HD100CAU) as a control population.
To verify the demographic history of our five tested populations, we first tested Tajima’s D statistic, using the nucleotide sequence variations of hypervariable region 1 (HVR1) in mtDNA; this statistic can identify a past expansion in population size (Tajima 1989a, 1989b). As is illustrated in table 1, this test showed that the values were significantly negative in the Japanese and Khalkh (P<.05), as well as in the Finnish and European Americans, indicating that these two East Asian populations had expanded in size in the past. In contrast, the D statistics did not significantly deviate from zero in the Khoton (P>.10), Uriankhai, and Zakhchin (P>.05) or in the Saami, indicating that these three young subpopulations have kept their sizes small and constant over time.
Table 1.
Sequence Variation of HVR1 in mtDNA
| Population | Sample Size | No. of Haplotypes | Nucleotide Diversity | Tajima's D | P Valuea |
| Japanese | 182 | 123 | .018 ± .001 | −1.925 | <.05 |
| Khalkh | 146 | 106 | .018 ± .001 | −1.979 | <.05 |
| Khoton | 74 | 23 | .012 ± .001 | −1.491 | >.10 |
| Uriankhai | 58 | 38 | .017 ± .001 | −1.619 | >.05 |
| Zakhchin | 59 | 43 | .020 ± .001 | −1.608 | >.05 |
| European American | 50 | 44 | .014 ± .001 | −2.241 | <.05 |
| Finnishb | 50 | 35 | .011 ± .001 | −1.820 | <.05 |
| Saamib | 25 | 11 | .009 ± .001 | −.986 | >.10 |
| Western Pygmyc | 17 | 8 | .023 ± .003 | 1.219 | >.10 |
To compare these results with those from the European isolates studied elsewhere (Laan and Pääbo 1997; Zavattari et al. 2000), we analyzed the extent of background LD among the East Asian males through use of seven microsatellite markers on Xq13 (DXS983, DXS8092, DXS8037, DXS8082, DXS1225, DXS986, and DXS995). Detailed data for number of alleles in the populations are listed in table 2, and the distribution of haplotypes among populations is detailed in an online-only tableonline-only table. Fisher’s exact test was used to detect significant LD between every pair of the microsatellite markers. All of the uncorrected P values are listed in table 3. The results showed that the Japanese population exhibited a relatively higher level of background LD than did the Finnish, European American, and Sardinian populations. Before correction, the Japanese had 5 of 21 pairs with significant LD (P<.05), and the other 3 pairs had suggestive LD (.05⩽P<.10). The rapid expansion of the Japanese population to a size of 120 million occurred at a rate of 108%, which is slightly higher than that of the Finnish population (106%). Such a rapid expansion might reduce the LD level in the Japanese, compared with the Finnish. This inconsistency could be explained by population structure differences (Kittles et al. 1998; Jorde et al. 2001). Our Japanese samples were collected in the Kanto region, which is inhabited by the Japanese homogeneous majority. Our Japanese samples may show stronger LD, reflecting that genetic homogeneity. It should be noted that the Japanese sample size (100) was somewhat larger than that of the Finnish (80) and Sardinians (73). To avoid an overestimation due to the effect of sample size on LD, the equilibration of sample size by the randomization test described by Varilo et al. (2000) was applied to the Fisher’s exact test. The sample size equilibration in the Japanese showed that similar LD results were obtained between the original and adjusted Japanese population sizes in table 3, indicating that the larger sample size is not responsible for the strong LD in the Japanese.
Table 2.
Gene Diversity and Haplotype Mismatch Based on Seven Microsatellite Markers on Xq13
|
No. of Alleles (Diversity), Based on Marker |
||||||||||||
| Population | SampleSize | No. ofHaplotypes | DXS983 | DXS986 | DXS8092 | DXS8082 | DXS1225 | DXS8037 | DXS995 | Mean± SDGeneDiversity | MeanHaplotypeMIsmatch | Variance ofHaplotypeMismatch |
| Japanese | 100 | 94 | 5 (.45) | 14 (.90) | 16 (.91) | 8 (.7) | 7 (.69) | 3 (.06) | 3 (.51) | .60 ± .33 | 4.21 | 1.46 |
| Khalkh | 83 | 80 | 5 (.43) | 12 (.87) | 13 (.90) | 9 (.80) | 9 (.74) | 6 (.41) | 4 (.53) | .67 ± .37 | 4.67 | 1.78 |
| Khoton | 40 | 32 | 6 (.66) | 11 (.89) | 11 (.88) | 7 (.84) | 9 (.86) | 3 (.53) | 4 (.55) | .74 ± .41 | 5.21 | 2.21 |
| Uriankhai | 55 | 50 | 3 (.35) | 14 (.91) | 13 (.90) | 10 (.84) | 9 (.82) | 6 (.61) | 6 (.64) | .72 ± .40 | 5.07 | 1.51 |
| Zakhchin | 59 | 54 | 6 (.49) | 14 (.91) | 14 (.91) | 8 (.81) | 10 (.81) | 5 (.46) | 5 (.56) | .71 ± .39 | 4.95 | 1.43 |
| European American | 49 | 49 | 8 (.76) | 7 (.69) | 10 (.81) | 8 (.78) | 10 (.77) | 6 (.68) | 6 (.59) | .73 ± .40 | 5.09 | 1.54 |
| Finnisha | 80 | 75 | 6 (.70) | 11 (.80) | 12 (.85) | 9 (.75) | 9 (.73) | 8 (.71) | 6 (.63) | .74 | 5.15 | 1.38 |
| Saamia | 54 | 32 | 4 (.58) | 10 (.80) | 8 (.83) | 6 (.79) | 6 (.76) | 5 (.50) | 3 (.44) | .67 | 4.59 | 3.08 |
Data are from Laan and Pääbo (1997). SDs were unavailable.
Online-Only Table.
Haplotype Data for Six Populations
|
No. of Individualsb |
||||||||
| HaplotypeNumber | Haplotypea | Japanese | Khalkh | Khoton | Uriankhi | Zakhchin | EuropeanAmerican | Total |
| Hap001 | 168-159-277-228-198-247-193 | 1 | 1 | |||||
| Hap002 | 170-159-273-218-210-247-197 | 1 | 1 | |||||
| Hap003 | 170-161-273-218-214-247-199 | 1 | 1 | |||||
| Hap004 | 170-161-281-218-210-247-197 | 1 | 1 | |||||
| Hap005 | 170-163-277-218-210-249-197 | 1 | 1 | |||||
| Hap006 | 170-167-271-216-202-249-197 | 1 | 1 | |||||
| Hap007 | 172-147-271-218-210-247-197 | 1 | 1 | |||||
| Hap008 | 174-143-281-224-200-237-193 | 1 | 1 | |||||
| Hap009 | 174-153-263-218-212-237-193 | 1 | 1 | |||||
| Hap010 | 174-153-273-218-202-237-193 | 1 | 1 | |||||
| Hap011 | 174-155-279-218-208-249-191 | 1 | 1 | |||||
| Hap012 | 174-157-271-218-206-247-197 | 1 | 1 | |||||
| Hap013 | 174-165-289-232-192-237-193 | 1 | 1 | |||||
| Hap014 | 174-167-267-218-210-249-197 | 1 | 1 | |||||
| Hap015 | 174-173-281-224-198-237-197 | 1 | 1 | |||||
| Hap016 | 174-173-283-218-202-237-193 | 1 | 1 | |||||
| Hap017 | 176-147-267-218-210-247-193 | 1 | 1 | |||||
| Hap018 | 176-155-267-216-216-245-193 | 1 | 1 | |||||
| Hap019 | 176-155-273-218-212-237-197 | 1 | 1 | |||||
| Hap020 | 176-155-275-224-198-237-193 | 1 | 1 | |||||
| Hap021 | 176-155-283-224-198-237-197 | 1 | 1 | |||||
| Hap022 | 176-159-271-218-210-249-193 | 1 | 1 | |||||
| Hap023 | 176-159-271-228-192-249-197 | 1 | 1 | |||||
| Hap024 | 176-159-273-218-216-249-193 | 1 | 1 | |||||
| Hap025 | 176-159-273-220-198-247-193 | 1 | 1 | |||||
| Hap026 | 176-159-273-222-210-247-193 | 1 | 1 | |||||
| Hap027 | 176-159-273-228-192-247-197 | 1 | 1 | |||||
| Hap028 | 176-159-277-218-210-247-197 | 1 | 1 | |||||
| Hap029 | 176-159-285-218-214-237-197 | 1 | 1 | |||||
| Hap030 | 176-161-271-218-206-245-197 | 1 | 1 | |||||
| Hap031 | 176-161-273-220-198-247-197 | 1 | 1 | |||||
| Hap032 | 176-161-275-224-192-245-197 | 1 | 1 | |||||
| Hap033 | 176-161-277-216-202-237-193 | 2 | 2 | |||||
| Hap034 | 176-161-277-218-210-249-193 | 1 | 1 | |||||
| Hap035 | 176-161-277-224-198-237-197 | 1 | 1 | |||||
| Hap036 | 176-161-279-216-202-245-197 | 1 | 1 | |||||
| Hap037 | 176-161-279-220-198-237-197 | 1 | 1 | |||||
| Hap038 | 176-161-281-210-202-237-193 | 1 | 1 | |||||
| Hap039 | 176-161-289-216-202-237-197 | 1 | 1 | |||||
| Hap040 | 176-163-279-224-198-237-197 | 1 | 1 | |||||
| Hap041 | 176-163-279-226-202-237-197 | 1 | 1 | |||||
| Hap042 | 176-163-285-224-200-237-197 | 1 | 1 | |||||
| Hap043 | 176-165-277-218-202-237-197 | 1 | 1 | |||||
| Hap044 | 176-165-293-216-202-237-197 | 1 | 1 | |||||
| Hap045 | 176-167-275-220-200-247-197 | 1 | 1 | |||||
| Hap046 | 176-167-275-224-200-237-197 | 1 | 1 | |||||
| Hap047 | 176-167-279-210-202-237-197 | 1 | 1 | |||||
| Hap048 | 176-167-279-224-200-237-197 | 1 | 1 | |||||
| Hap049 | 176-169-273-224-198-249-193 | 1 | 1 | |||||
| Hap050 | 176-169-279-224-198-237-197 | 1 | 1 | |||||
| Hap051 | 176-169-281-226-200-237-197 | 1 | 1 | |||||
| Hap052 | 176-169-285-224-198-237-201 | 1 | 1 | |||||
| Hap053 | 176-171-275-210-206-247-193 | 1 | 1 | |||||
| Hap054 | 176-171-277-224-196-237-193 | 1 | 1 | |||||
| Hap055 | 176-171-283-228-198-237-195 | 1 | 1 | |||||
| Hap056 | 176-171-287-216-204-237-193 | 1 | 1 | |||||
| Hap057 | 176-175-281-224-198-237-197 | 1 | 1 | |||||
| Hap058 | 176-175-283-216-202-237-197 | 1 | 1 | |||||
| Hap059 | 178-143-289-224-200-237-193 | 1 | 1 | |||||
| Hap060 | 178-147-269-216-210-247-197 | 1 | 1 | |||||
| Hap061 | 178-147-269-220-210-247-197 | 1 | 1 | |||||
| Hap062 | 178-153-267-210-204-247-197 | 1 | 1 | |||||
| Hap063 | 178-153-271-218-202-247-193 | 2 | 2 | |||||
| Hap064 | 178-153-271-218-206-237-193 | 1 | 1 | |||||
| Hap065 | 178-153-275-210-206-237-197 | 1 | 1 | |||||
| Hap066 | 178-153-277-224-212-237-197 | 1 | 1 | |||||
| Hap067 | 178-153-281-216-202-237-197 | 1 | 1 | |||||
| Hap068 | 178-153-281-224-200-237-197 | 1 | 1 | |||||
| Hap069 | 178-153-281-224-210-237-197 | 1 | 1 | |||||
| Hap070 | 178-153-283-220-202-237-197 | 1 | 1 | |||||
| Hap071 | 178-153-283-224-212-237-193 | 1 | 1 | |||||
| Hap072 | 178-153-285-224-212-237-193 | 1 | 1 | |||||
| Hap073 | 178-153-287-224-210-237-193 | 1 | 1 | |||||
| Hap074 | 178-155-269-210-204-237-197 | 1 | 1 | |||||
| Hap075 | 178-155-273-224-198-237-193 | 1 | 1 | |||||
| Hap076 | 178-155-275-218-210-247-197 | 1 | 1 | |||||
| Hap077 | 178-155-275-228-192-249-197 | 1 | 1 | |||||
| Hap078 | 178-155-277-228-198-237-193 | 1 | 1 | |||||
| Hap079 | 178-155-279-218-208-249-193 | 1 | 1 | |||||
| Hap080 | 178-155-279-226-198-237-197 | 1 | 1 | |||||
| Hap081 | 178-155-283-204-200-237-193 | 1 | 1 | |||||
| Hap082 | 178-155-283-218-202-237-193 | 1 | 1 | |||||
| Hap083 | 178-155-283-224-200-237-193 | 1 | 1 | |||||
| Hap084 | 178-155-291-224-200-237-193 | 1 | 1 | |||||
| Hap085 | 178-157-271-216-202-237-197 | 1 | 1 | |||||
| Hap086 | 178-157-271-218-206-247-197 | 1 | 1 | |||||
| Hap087 | 178-157-271-220-198-247-197 | 1 | 1 | |||||
| Hap088 | 178-157-271-220-210-247-197 | 1 | 1 | |||||
| Hap089 | 178-157-279-228-198-245-193 | 1 | 1 | |||||
| Hap090 | 178-157-283-218-212-237-197 | 1 | 1 | 2 | ||||
| Hap091 | 178-159-265-218-210-247-197 | 1 | 1 | |||||
| Hap092 | 178-159-267-218-210-245-193 | 1 | 1 | |||||
| Hap093 | 178-159-267-218-210-247-193 | 1 | 1 | |||||
| Hap094 | 178-159-269-226-198-245-197 | 1 | 1 | |||||
| Hap095 | 178-159-273-210-202-247-197 | 1 | 1 | |||||
| Hap096 | 178-159-273-210-202-253-197 | 1 | 1 | |||||
| Hap097 | 178-159-273-216-210-247-197 | 1 | 1 | |||||
| Hap098 | 178-159-273-218-210-247-197 | 1 | 1 | |||||
| Hap099 | 178-159-273-220-210-245-197 | 1 | 1 | |||||
| Hap100 | 178-159-273-224-198-245-195 | 1 | 1 | |||||
| Hap101 | 178-159-273-228-192-247-193 | 1 | 1 | |||||
| Hap102 | 178-159-275-218-210-251-197 | 1 | 1 | |||||
| Hap103 | 178-159-275-220-210-247-199 | 1 | 1 | |||||
| Hap104 | 178-159-277-210-202-249-193 | 1 | 1 | |||||
| Hap105 | 178-159-277-218-210-247-193 | 1 | 1 | |||||
| Hap106 | 178-159-277-226-198-237-197 | 3 | 3 | |||||
| Hap107 | 178-159-277-228-192-249-197 | 1 | 1 | |||||
| Hap108 | 178-159-279-210-202-245-197 | 1 | 1 | |||||
| Hap109 | 178-159-279-210-202-249-193 | 1 | 1 | |||||
| Hap110 | 178-159-279-218-210-247-193 | 1 | 1 | |||||
| Hap111 | 178-159-281-216-198-237-193 | 1 | 1 | |||||
| Hap112 | 178-159-283-224-200-237-193 | 1 | 1 | |||||
| Hap113 | 178-159-285-220-210-243-197 | 1 | 1 | |||||
| Hap114 | 178-161-267-230-198-249-197 | 1 | 1 | |||||
| Hap115 | 178-161-269-218-210-249-197 | 1 | 1 | |||||
| Hap116 | 178-161-269-226-198-247-193 | 1 | 1 | |||||
| Hap117 | 178-161-271-218-210-249-193 | 1 | 1 | |||||
| Hap118 | 178-161-273-216-202-247-193 | 1 | 1 | |||||
| Hap119 | 178-161-273-216-202-249-193 | 1 | 1 | |||||
| Hap120 | 178-161-273-226-192-247-191 | 1 | 1 | |||||
| Hap121 | 178-161-275-210-206-247-197 | 1 | 1 | |||||
| Hap122 | 178-161-275-216-202-237-193 | 1 | 1 | |||||
| Hap123 | 178-161-275-216-202-249-193 | 1 | 1 | |||||
| Hap124 | 178-161-275-224-192-245-197 | 1 | 1 | |||||
| Hap125 | 178-161-275-224-200-237-193 | 1 | 1 | |||||
| Hap126 | 178-161-275-228-192-249-193 | 1 | 1 | |||||
| Hap127 | 178-161-275-228-198-249-193 | 1 | 1 | |||||
| Hap128 | 178-161-277-210-202-237-199 | 1 | 1 | |||||
| Hap129 | 178-161-277-216-190-249-193 | 1 | 1 | |||||
| Hap130 | 178-161-277-216-202-237-193 | 2 | 2 | |||||
| Hap131 | 178-161-277-216-202-237-197 | 1 | 1 | |||||
| Hap132 | 178-161-277-216-204-237-195 | 1 | 1 | |||||
| Hap133 | 178-161-277-224-192-245-197 | 1 | 1 | |||||
| Hap134 | 178-161-277-224-198-237-193 | 1 | 1 | 2 | ||||
| Hap135 | 178-161-277-224-200-237-197 | 1 | 1 | |||||
| Hap136 | 178-161-277-226-198-237-197 | 1 | 1 | |||||
| Hap137 | 178-161-279-216-202-237-193 | 1 | 2 | 3 | ||||
| Hap138 | 178-161-279-216-202-247-197 | 1 | 1 | |||||
| Hap139 | 178-161-281-210-202-237-193 | 1 | 1 | |||||
| Hap140 | 178-161-281-216-202-237-193 | 1 | 1 | 2 | ||||
| Hap141 | 178-161-283-216-202-237-193 | 1 | 1 | |||||
| Hap142 | 178-161-283-216-202-237-197 | 1 | 1 | |||||
| Hap143 | 178-161-283-220-200-237-199 | 1 | 1 | |||||
| Hap144 | 178-161-285-216-202-237-195 | 1 | 1 | |||||
| Hap145 | 178-161-285-218-206-237-193 | 1 | 1 | |||||
| Hap146 | 178-161-285-220-202-237-193 | 1 | 1 | |||||
| Hap147 | 178-161-287-216-202-237-193 | 1 | 1 | |||||
| Hap148 | 178-161-287-216-202-237-197 | 1 | 1 | |||||
| Hap149 | 178-161-287-224-214-237-193 | 1 | 1 | |||||
| Hap150 | 178-161-289-216-202-237-193 | 1 | 1 | |||||
| Hap151 | 178-161-289-216-202-237-197 | 1 | 1 | |||||
| Hap152 | 178-161-291-216-202-237-193 | 1 | 1 | |||||
| Hap153 | 178-163-267-216-202-237-197 | 1 | 1 | |||||
| Hap154 | 178-163-267-218-210-249-193 | 1 | 1 | |||||
| Hap155 | 178-163-271-216-202-237-197 | 1 | 1 | |||||
| Hap156 | 178-163-277-210-202-243-193 | 1 | 1 | |||||
| Hap157 | 178-163-277-216-202-237-197 | 1 | 1 | |||||
| Hap158 | 178-163-277-224-200-239-193 | 1 | 1 | |||||
| Hap159 | 178-163-277-228-198-237-197 | 1 | 1 | |||||
| Hap160 | 178-163-279-214-202-237-197 | 1 | 1 | |||||
| Hap161 | 178-163-279-226-198-247-193 | 1 | 1 | |||||
| Hap162 | 178-163-279-226-202-237-193 | 1 | 1 | |||||
| Hap163 | 178-163-281-210-202-237-197 | 1 | 1 | |||||
| Hap164 | 178-163-283-216-202-237-197 | 1 | 1 | |||||
| Hap165 | 178-163-283-216-204-237-197 | 1 | 1 | |||||
| Hap166 | 178-163-283-226-202-237-197 | 1 | 1 | |||||
| Hap167 | 178-163-285-216-202-237-193 | 1 | 1 | |||||
| Hap168 | 178-163-285-224-198-237-197 | 2 | 2 | |||||
| Hap169 | 178-163-285-226-198-237-197 | 1 | 1 | |||||
| Hap170 | 178-163-287-216-202-237-193 | 1 | 1 | |||||
| Hap171 | 178-163-287-228-202-237-193 | 1 | 1 | |||||
| Hap172 | 178-163-289-216-200-237-193 | 1 | 1 | |||||
| Hap173 | 178-163-289-216-202-237-197 | 1 | 1 | |||||
| Hap174 | 178-163-289-216-204-239-197 | 1 | 1 | |||||
| Hap175 | 178-165-259-216-202-237-195 | 1 | 1 | |||||
| Hap176 | 178-165-271-216-202-237-197 | 1 | 1 | |||||
| Hap177 | 178-165-271-218-210-237-197 | 1 | 1 | |||||
| Hap178 | 178-165-273-216-202-237-197 | 1 | 1 | |||||
| Hap179 | 178-165-273-224-198-237-197 | 1 | 1 | |||||
| Hap180 | 178-165-273-224-200-237-193 | 1 | 1 | |||||
| Hap181 | 178-165-275-224-200-237-197 | 1 | 1 | |||||
| Hap182 | 178-165-277-216-202-237-193 | 2 | 2 | |||||
| Hap183 | 178-165-277-216-202-237-197 | 1 | 1 | |||||
| Hap184 | 178-165-279-216-202-237-193 | 1 | 1 | |||||
| Hap185 | 178-165-279-216-202-237-197 | 1 | 1 | |||||
| Hap186 | 178-165-279-224-202-237-193 | 1 | 1 | |||||
| Hap187 | 178-165-279-226-202-237-197 | 1 | 1 | |||||
| Hap188 | 178-165-281-210-202-237-197 | 1 | 1 | |||||
| Hap189 | 178-165-281-216-198-237-193 | 1 | 1 | |||||
| Hap190 | 178-165-281-216-202-237-197 | 1 | 3 | 1 | 5 | |||
| Hap191 | 178-165-281-224-200-237-193 | 1 | 1 | |||||
| Hap192 | 178-165-283-216-202-237-193 | 2 | 2 | |||||
| Hap193 | 178-165-285-216-202-237-193 | 1 | 1 | 1 | 3 | |||
| Hap194 | 178-167-269-216-202-237-197 | 3 | 3 | |||||
| Hap195 | 178-167-273-216-200-237-197 | 2 | 2 | |||||
| Hap196 | 178-167-273-226-210-237-193 | 1 | 1 | |||||
| Hap197 | 178-167-275-210-206-247-193 | 1 | 1 | |||||
| Hap198 | 178-167-275-216-198-237-193 | 2 | 2 | |||||
| Hap199 | 178-167-275-224-192-237-193 | 2 | 2 | |||||
| Hap200 | 178-167-277-216-202-237-197 | 1 | 1 | |||||
| Hap201 | 178-167-277-224-198-237-197 | 3 | 3 | |||||
| Hap202 | 178-167-279-224-198-237-193 | 1 | 1 | 2 | ||||
| Hap203 | 178-167-281-210-200-237-191 | 1 | 1 | |||||
| Hap204 | 178-167-281-216-202-237-193 | 1 | 1 | |||||
| Hap205 | 178-167-281-216-202-237-197 | 1 | 1 | |||||
| Hap206 | 178-167-281-224-200-237-197 | 1 | 1 | |||||
| Hap207 | 178-167-281-226-202-237-197 | 1 | 1 | |||||
| Hap208 | 178-167-283-216-202-237-193 | 1 | 1 | |||||
| Hap209 | 178-167-283-224-198-237-193 | 1 | 1 | |||||
| Hap210 | 178-167-283-224-198-237-197 | 1 | 1 | |||||
| Hap211 | 178-167-287-216-202-237-197 | 1 | 1 | |||||
| Hap212 | 178-167-287-222-198-237-197 | 1 | 1 | |||||
| Hap213 | 178-167-289-224-198-237-197 | 1 | 1 | |||||
| Hap214 | 178-167-289-224-200-237-193 | 1 | 1 | |||||
| Hap215 | 178-167-291-216-202-237-201 | 1 | 1 | |||||
| Hap216 | 178-169-265-224-212-237-193 | 1 | 1 | |||||
| Hap217 | 178-169-269-218-210-237-193 | 1 | 1 | |||||
| Hap218 | 178-169-269-224-198-237-197 | 1 | 1 | |||||
| Hap219 | 178-169-273-210-204-247-193 | 1 | 1 | |||||
| Hap220 | 178-169-273-218-210-249-197 | 1 | 1 | |||||
| Hap221 | 178-169-273-220-198-247-197 | 1 | 1 | |||||
| Hap222 | 178-169-277-218-210-249-197 | 1 | 1 | |||||
| Hap223 | 178-169-279-216-202-237-197 | 1 | 1 | |||||
| Hap224 | 178-169-281-224-200-237-193 | 1 | 1 | |||||
| Hap225 | 178-169-281-226-200-237-193 | 1 | 1 | |||||
| Hap226 | 178-169-283-216-202-237-193 | 1 | 1 | |||||
| Hap227 | 178-169-283-222-198-237-197 | 1 | 1 | |||||
| Hap228 | 178-169-285-216-202-237-197 | 1 | 1 | |||||
| Hap229 | 178-169-285-216-204-237-201 | 1 | 1 | |||||
| Hap230 | 178-169-285-226-200-239-197 | 1 | 1 | |||||
| Hap231 | 178-169-287-218-202-237-193 | 1 | 1 | |||||
| Hap232 | 178-169-289-228-198-237-197 | 1 | 1 | |||||
| Hap233 | 178-171-271-216-202-237-197 | 1 | 1 | |||||
| Hap234 | 178-171-273-210-204-237-193 | 1 | 1 | |||||
| Hap235 | 178-171-273-224-198-237-197 | 3 | 3 | |||||
| Hap236 | 178-171-275-210-202-247-199 | 2 | 2 | |||||
| Hap237 | 178-171-275-210-206-247-193 | 1 | 1 | |||||
| Hap238 | 178-171-275-214-210-247-197 | 1 | 1 | |||||
| Hap239 | 178-171-275-224-198-237-193 | 1 | 1 | |||||
| Hap240 | 178-171-275-224-200-237-197 | 1 | 1 | |||||
| Hap241 | 178-171-275-224-200-247-197 | 1 | 1 | |||||
| Hap242 | 178-171-277-210-198-237-193 | 1 | 1 | |||||
| Hap243 | 178-171-277-216-202-237-197 | 1 | 1 | |||||
| Hap244 | 178-171-277-218-202-237-197 | 1 | 2 | 3 | ||||
| Hap245 | 178-171-277-218-210-237-193 | 1 | 1 | |||||
| Hap246 | 178-171-277-224-196-237-197 | 1 | 1 | |||||
| Hap247 | 178-171-277-224-200-237-197 | 1 | 1 | 2 | ||||
| Hap248 | 178-171-279-216-202-237-197 | 1 | 1 | |||||
| Hap249 | 178-171-281-224-198-237-197 | 1 | 1 | 2 | ||||
| Hap250 | 178-171-281-228-202-237-197 | 1 | 1 | |||||
| Hap251 | 178-171-283-216-202-237-193 | 1 | 1 | 2 | ||||
| Hap252 | 178-171-283-218-202-237-197 | 1 | 1 | |||||
| Hap253 | 178-171-283-228-198-237-193 | 1 | 1 | |||||
| Hap254 | 178-171-285-224-196-237-197 | 1 | 1 | |||||
| Hap255 | 178-171-285-224-210-237-193 | 1 | 1 | |||||
| Hap256 | 178-171-289-218-212-237-197 | 1 | 1 | |||||
| Hap257 | 178-171-289-224-198-237-197 | 1 | 1 | |||||
| Hap258 | 178-171-293-216-202-237-193 | 1 | 1 | |||||
| Hap259 | 178-173-265-224-198-237-195 | 1 | 1 | |||||
| Hap260 | 178-173-265-224-198-237-197 | 1 | 1 | |||||
| Hap261 | 178-173-273-224-200-247-193 | 1 | 1 | |||||
| Hap262 | 178-173-275-224-198-237-193 | 1 | 1 | |||||
| Hap263 | 178-173-277-224-200-237-193 | 1 | 1 | |||||
| Hap264 | 178-173-277-226-198-237-197 | 1 | 1 | |||||
| Hap265 | 178-173-279-224-198-237-193 | 1 | 1 | |||||
| Hap266 | 178-173-279-224-198-237-197 | 1 | 1 | |||||
| Hap267 | 178-173-279-224-200-237-197 | 1 | 1 | |||||
| Hap268 | 178-173-279-224-202-237-197 | 1 | 1 | |||||
| Hap269 | 178-173-279-226-198-237-197 | 1 | 1 | |||||
| Hap270 | 178-173-281-216-202-237-193 | 1 | 1 | |||||
| Hap271 | 178-173-281-224-200-237-193 | 1 | 1 | |||||
| Hap272 | 178-173-283-224-198-237-193 | 1 | 1 | |||||
| Hap273 | 178-173-285-216-202-237-193 | 1 | 1 | |||||
| Hap274 | 178-173-285-224-198-237-193 | 1 | 1 | |||||
| Hap275 | 178-173-285-224-200-237-197 | 1 | 1 | |||||
| Hap276 | 178-173-285-228-198-237-193 | 1 | 1 | |||||
| Hap277 | 178-173-287-226-202-237-197 | 1 | 1 | |||||
| Hap278 | 178-173-289-226-198-237-197 | 1 | 1 | |||||
| Hap279 | 178-175-279-216-202-237-193 | 1 | 1 | |||||
| Hap280 | 178-175-283-210-202-237-193 | 1 | 1 | |||||
| Hap281 | 178-175-283-226-198-237-197 | 1 | 1 | |||||
| Hap282 | 178-175-285-216-202-237-193 | 1 | 1 | |||||
| Hap283 | 178-177-271-222-200-237-197 | 1 | 1 | |||||
| Hap284 | 178-179-283-210-202-237-193 | 1 | 1 | |||||
| Hap285 | 178-197-291-224-200-237-195 | 1 | 1 | |||||
| Hap286 | 180-153-271-218-202-247-193 | 1 | 1 | |||||
| Hap287 | 180-153-273-218-202-237-193 | 1 | 1 | |||||
| Hap288 | 180-153-279-224-200-237-193 | 1 | 1 | |||||
| Hap289 | 180-153-279-224-210-237-197 | 1 | 1 | |||||
| Hap290 | 180-155-267-220-198-249-193 | 1 | 1 | |||||
| Hap291 | 180-155-273-218-210-249-197 | 1 | 1 | |||||
| Hap292 | 180-155-287-216-202-237-193 | 1 | 1 | |||||
| Hap293 | 180-157-265-220-208-247-197 | 1 | 1 | |||||
| Hap294 | 180-157-277-228-192-245-197 | 1 | 1 | |||||
| Hap295 | 180-159-267-228-196-249-197 | 1 | 1 | |||||
| Hap296 | 180-159-269-218-208-247-193 | 1 | 1 | |||||
| Hap297 | 180-159-269-218-210-251-193 | 1 | 1 | |||||
| Hap298 | 180-159-269-220-198-247-193 | 1 | 1 | |||||
| Hap299 | 180-159-273-218-210-247-193 | 1 | 1 | |||||
| Hap300 | 180-159-275-218-210-247-197 | 1 | 1 | |||||
| Hap301 | 180-159-275-218-210-255-193 | 1 | 1 | |||||
| Hap302 | 180-159-275-228-192-247-197 | 1 | 1 | |||||
| Hap303 | 180-159-277-226-198-237-197 | 1 | 1 | |||||
| Hap304 | 180-159-285-228-192-247-193 | 1 | 1 | |||||
| Hap305 | 180-161-267-228-198-247-193 | 1 | 1 | |||||
| Hap306 | 180-161-273-218-210-249-199 | 1 | 1 | |||||
| Hap307 | 180-161-273-228-198-247-197 | 1 | 1 | |||||
| Hap308 | 180-161-277-210-198-247-193 | 1 | 1 | |||||
| Hap309 | 180-161-279-216-202-237-193 | 1 | 1 | |||||
| Hap310 | 180-161-283-220-202-237-193 | 1 | 1 | |||||
| Hap311 | 180-161-283-224-200-237-193 | 1 | 1 | |||||
| Hap312 | 180-161-287-216-202-237-197 | 1 | 1 | |||||
| Hap313 | 180-163-273-216-202-237-193 | 1 | 1 | |||||
| Hap314 | 180-163-277-210-202-237-197 | 1 | 1 | |||||
| Hap315 | 180-165-271-216-202-237-197 | 1 | 1 | |||||
| Hap316 | 180-167-273-218-208-245-197 | 1 | 1 | |||||
| Hap317 | 180-167-275-216-204-249-197 | 1 | 1 | |||||
| Hap318 | 180-167-279-216-202-237-197 | 1 | 1 | |||||
| Hap319 | 180-167-279-224-198-237-193 | 1 | 1 | |||||
| Hap320 | 180-167-283-220-204-237-197 | 1 | 1 | |||||
| Hap321 | 180-169-277-216-202-237-197 | 1 | 1 | |||||
| Hap322 | 180-169-277-224-198-237-197 | 1 | 1 | |||||
| Hap323 | 180-169-277-226-198-237-197 | 1 | 1 | |||||
| Hap324 | 180-169-279-220-198-247-193 | 1 | 1 | |||||
| Hap325 | 180-171-271-228-192-245-193 | 2 | 1 | 3 | ||||
| Hap326 | 180-171-275-218-210-247-193 | 1 | 1 | |||||
| Hap327 | 180-171-275-224-198-247-197 | 1 | 1 | |||||
| Hap328 | 180-171-277-224-200-237-197 | 1 | 1 | |||||
| Hap329 | 180-175-273-216-202-237-197 | 1 | 1 | |||||
| Hap330 | 180-175-275-216-202-237-193 | 1 | 1 | |||||
| Hap331 | 180-177-273-224-198-247-193 | 1 | 1 | |||||
| Hap332 | 182-159-271-218-210-251-201 | 1 | 1 | |||||
| Hap333 | 182-159-275-218-210-247-193 | 1 | 1 | |||||
| Hap334 | 182-161-275-218-210-247-197 | 1 | 1 | |||||
| Hap335 | 182-161-277-210-202-237-197 | 1 | 1 | |||||
| Hap336 | 182-161-283-228-200-247-193 | 5 | 5 | |||||
| Hap337 | 182-163-283-228-200-247-193 | 1 | 1 | |||||
| Hap338 | 182-163-287-226-202-237-197 | 1 | 1 | |||||
| Hap339 | 182-165-273-224-198-237-193 | 1 | 1 | |||||
| Hap340 | 182-167-287-224-198-237-193 | 1 | 1 | |||||
| Hap341 | 182-167-293-216-202-237-197 | 1 | 1 | |||||
| Hap342 | 182-175-279-226-198-237-193 | 1 | 1 | |||||
| Hap343 | 184-155-277-210-202-245-191 | 1 | 1 | |||||
| Hap344 | 184-161-285-224-198-237-197 | 1 | 1 | |||||
| Hap345 | 184-167-287-224-200-237-193 | 1 |
1 |
|||||
| Total Haplotypes | 94 | 80 | 32 | 50 | 54 | 49 | 359 | |
| Total Individuals | 100 | 83 | 40 | 55 | 59 | 49 | 386 | |
The composition of each haplotype is DXS983-DXS986-DXS8092-DXS8082-DXS1225-DXS8037-DXS995.
The shared haplotypes are double-counted.
Table 3.
Pairwise LD Based on Fisher's Exact Test Using Seven Microsatellite Markers on Xq13
|
Uncorrected P Valueb in |
|||||||||||
| Locus Pair | Distancea(cM) | Japanese(n=100) | Japanese(n=80)c | Khalkh(n=83) | Khoton(n=40) | Uriankhai(n=55) | Zakhchin(n=59) | EuropeanAmerican(n=49) | Finnish(n=80)a | Saami(n=54)a | Sardinian(n=73)d |
| DXS995-DXS983 | 4.0 | .480 | .291 | 1.000 | .003 | .097 | .927 | .030 |
.508 | .012 |
.394 |
| DXS995-DXS986 | 2.5 | .068 | .051 | .326 | .011 |
.482 | .402 | .328 | .729 | .000 | .482 |
| DXS995-DXS8092 | 2.4 | .120 | .055 | .076 | .047 |
.185 | .625 | .755 | .115 | .104 | .829 |
| DXS995-DXS8082 | 2.3 | .034 |
.033 |
.095 | .013 |
.579 | .021 |
.662 | .128 | .000 | .430 |
| DXS1225-DXS983 | 2.0 | .227 | .151 | .165 | .000 | .773 | .449 | .243 | .630 | .000 | .169 |
| DXS995-DXS8037 | 2.0 | .517 | .310 | .072 | .070 | .563 | .907 | .182 | .874 | .124 | .650 |
| DXS8037-DXS983 | 2.0 | .515 | .342 | .449 | .000 | .019 |
.521 | .604 | .683 | .300 | .036 |
| DXS995-DXS1225 | 2.0 | .910 | .388 | .062 | .002 | .654 | .469 | .282 | .154 | .001 | .355 |
| DXS8082-DXS983 | 1.7 | .064 | .096 | .556 | .002 | .922 | .970 | .448 | .565 | .000 | .142 |
| DXS8092-DXS983 | 1.6 | .383 | .065 | .318 | .051 | .154 | .796 | .458 | .314 | .000 | .876 |
| DXS986-DXS983 | 1.5 | .276 | .071 | .407 | .011 |
.835 | .210 | .530 | .829 | .000 | .825 |
| DXS8037-DXS986 | .5 | .139 | .179 | .150 | .035 |
.046 |
.190 | .929 | .620 | .000 | .302 |
| DXS1225-DXS986 | .5 | .003 |
.002 | .000 | .000 | .019 |
.098 | .233 | .393 | .000 | .166 |
| DXS1225-DXS8092 | .4 | .002 | .004 |
.105 | .000 | .047 |
.211 | .583 | .283 | .000 | .921 |
| DXS8037-DXS8092 | .4 | .426 | .294 | .000 | .086 | .025 |
.000 | .723 | .180 | .000 | .285 |
| DXS8037-DXS8082 | .3 | .373 | .353 | .008 |
.000 | .000 | .001 | .219 | .238 | .012 |
.630 |
| DXS1225-DXS8082 | .3 | .000 | .000 | .000 | .000 | .000 | .000 | .000 | .000 | .000 | .000 |
| DXS8082-DXS986 | .2 | .000 | .000 | .001 | .000 | .001 | .004 |
.524 | .092 | .000 | .322 |
| DXS8092-DXS986 | .1 | .612 | .003 |
.161 | .000 | .021 |
.004 |
.502 | .331 | .000 | .125 |
| DXS8082-DXS8092 | .1 | .051 | .014 |
.910 | .000 | .017 |
.300 | .115 | .044 |
.000 | .319 |
| DXS8037-DXS1225 | .0 | .372 | .301 | .000 | .000 | .000 | .000 | .198 | .836 | .091 | .710 |
Data are from Laan and Pääbo (1997).
The values that are significant only before correction are underlined, whereas the values that are significant even after the step-down Holm-Sidack correction (Ludbrook 1998) are given in boldface italics. The correction of P values in Finnish, Saami, and Sardinian was recalculated to amend the mistakes by Zavattari et al. (2000).
Reduced sample was obtained by the randomization test described by Varilo et al. (2000).
Data are from Zavattari et al. (2000).
The Khalkh showed a relatively higher level of LD than the Japanese. The largest genetic distance for the significant LD was 2.3 cM, and that for the suggestive LD was 2.5 cM. Three pairs still showed the significant LD even after correction, with a largest distance of 0.4 cM (DXS1225-DXS8092). Of the three Mongolian subpopulations, the Khoton had the largest number of pairs, almost all of which showed significant LD even after the correction, with an extent similar in magnitude to that in the Saami population. Before the correction, 18 of 21 pairs showed significant LD, with the remaining pairs showing suggestive LD. After the correction, 13 pairs remained significant for the largest distance reached, 4.0 cM (DXS995-DXS983). In contrast, the two remaining subpopulations, the Uriankhai and Zakhchin, had four pairs showing significant LD after correction, with largest distances of 0.3 cM (DXS8037-DXS8082) and 0.4 cM (DXS8037-DXS8092), respectively.
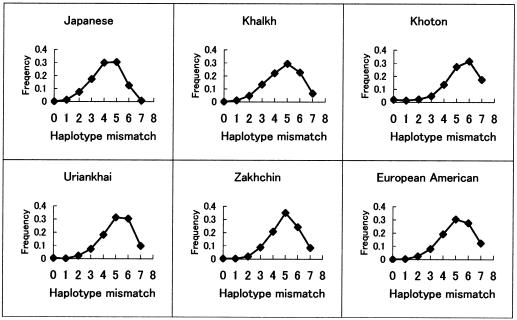
The strongest statistical significance and largest distance for LD in this study was observed in the Khoton. The background LD in the Khoton could be explained by strong isolation and constant population size. On the other hand, it should be noted that the microsatellite allelic diversity was generally greater in the Khoton than in the other Asian populations and that the average haplotype mismatch distance in the Khoton was also high, with a large variance (table 2). Moreover, the FST values obtained using SNPs on mtDNA and microsatellites on the X chromosome showed the close relationship between the Khoton and either the Europeans and Asians (see Appendix 4 at the authors' Web site). Therefore, the long-range LD in the Khoton may be the result of admixture between the Asian and European populations. However, many data from historical, ethnological, and anthropological studies previously performed in Mongolia suggest that the Khoton population was of Turkish origin and that there was no obvious admixture between the Khoton and other Mongolian populations (Batsuuri 1977; Nyambuu 1992). It is worth mentioning that, in contrast to the high allelic diversity of microsatellites, the mtDNA nucleotide diversity in the Khoton was the smallest of the values among the Asian populations (table 1). Taken together with the similar propensity for these diversities in the Saami, the inconsistency could be explained by the difference between the marker types. In addition, the average pairwise mismatch of haplotypes in the Khoton was not a bimodal distribution, which is the distribution that would be predicted for an admixed population (see online-only figureonline-only figure). The Khoton population was known to have the closest genetic relationship with the Turkish Kyrgyz (Batsuuri 1978). The bilaterally close relationship in the Khoton that is implied by the FST value may reflect the original character of an ancestral population, such as the Kyrgyz, rather than admixture in the Khoton. Moreover, although the Uriankhai and Zakhchin are obviously admixed populations with constant sizes, the extent of their LD was smaller than that of the Khoton. These facts suggest that the strong LD with long distance in the Khoton was contributed mainly by the isolation and constant size rather than by the admixture, even if there was some minor admixture.
Online-Only Figure .
Mismatch distributions among pairs of haplotypes for seven X-chromosome loci
Our analysis is, to our knowledge, the first report on genetic isolates in East Asia. Although our results were restricted to the X chromosome and mtDNA, they were consistent with demographic, historical, ethnological, and anthropological studies of these populations. Studies of the Khoton, Uriankhai, and Zakhchin populations could play an important role in initial gene mapping of complex diseases, and studies of the Japanese and Khalkh populations could be applied to the fine mapping of either complex or monogenic diseases (Yu et al. 1996; Graham and Thompson 1998).
Acknowledgments
We would like to thank R. Sato, A. Yamamoto, S. Adachi, M. Tomizawa, and A. Denda, in our laboratory, for technical assistance. We also thank Dr. N. Saito, in the National Institute for Genetics, for his valuable suggestions. This work was partly supported by a Grant-in-Aid for Scientific Research on Priority Areas (C) “Medical Genome Science,” from the Ministry of Education, Science, Sports, and Culture of Japan.
Electronic-Database Information
The URL for data presented herein is as follows:
- Authors' Web site, http://locus1.med.u-tokai.ac.jp/~xchr/x-ld.html (for details of the material and methods of the present study, the census data, the FST values, and the complete haplotype data set)
References
- Angius A, Melis PM, Morelli L, Petretto E, Casu G, Maestrale GB, Fraumene C, Bebbere D, Forabosco P, Pirastu M (2001) Archival, demographic and genetic studies define a Sardinian sub-isolate as a suitable model for mapping complex traits. Hum Genet 109:198–209 [DOI] [PubMed] [Google Scholar]
- Badamkhatan S (ed) (1987) Ethnography of Mongolia. Vol 1. Mongolian Academy of Sciences, Ulaanbaatar, Mongolia [Google Scholar]
- ——— (1996) Ethnography of Mongolia. Vols 2 and 3. Mongolian Academy of Sciences, Ulaanbaatar, Mongolia [Google Scholar]
- Batsuuri J (1977) Anthropological characteristics of the ethnic group “Khoton.” Proceedings of the Institute of General and Experimental Biology, Mongolian Academy of Sciences, Ulaanbaatar, Mongolia 12:111–127 [Google Scholar]
- ——— (1978) Genetic structure of the ethnic group “Khoton.” Proceedings of the Institute of General and Experimental Biology, Mongolian Academy of Sciences, Ulaanbaatar, Mongolia 13:115–129 [Google Scholar]
- de la Chapelle A, Wright FA (1998) Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc Natl Acad Sci USA 95:12416–12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning AM, Durocher F, Healey CS, Teare MD, McBride SE, Carlomagno F, Xu CF, Dawson E, Rhodes S, Ueda S, Lai E, Luben RN, Van Rensburg EJ, Mannermaa A, Kataja V, Rennart G, Dunham I, Purvis I, Easton D, Ponder BA (2000) The extent of linkage disequilibrium in four populations with distinct demographic histories. Am J Hum Genet 67:1544–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves IA, Merriman TR, Barber RA, Nutland S, Tuomilehto-Wolf E, Tuomilehto J, Cucca F, Todd JA (2000) The genetically isolated populations of Finland and Sardinia may not be a panacea for linkage disequilibrium mapping of common disease genes. Nat Genet 25:320–323 [DOI] [PubMed] [Google Scholar]
- Graham J, Thompson EA (1998) Disequilibrium likelihoods for fine-scale mapping of a rare allele. Am J Hum Genet 63:1517–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hästbacka J, de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E (1992) Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat Genet 2:204–211 [DOI] [PubMed] [Google Scholar]
- Hayami A (2001) The historical demography of pre-modern Japan. University of Tokyo Press, Tokyo [Google Scholar]
- Jorde LB, Watkins WS, Bamshad MJ (2001) Population genomics: a bridge from evolutionary history to genetic medicine. Hum Mol Genet 10:2199–2207 [DOI] [PubMed] [Google Scholar]
- Kittles RA, Perola M, Peltonen L, Bergen AW, Aragon RA, Virkkunen M, Linnoila M, Goldman D, Long J (1998) Dual origins of Finns revealed by Y chromosome haplotype variation. Am J Hum Genet 62:1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama S (1978) Jomon subsistence and population. Senri Ethnol Stud 2:1–65 [Google Scholar]
- Laan M, Pääbo S (1997) Demographic history and linkage disequilibrium in human populations. Nat Genet 17:435–438 [DOI] [PubMed] [Google Scholar]
- Lonjou C, Collins A, Morton NE (1999) Allelic association between marker loci. Proc Natl Acad Sci USA 96:1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludbrook J (1998) Multiple comparison procedures updated. Clin Exp Pharmacol Physiol 25:1032–1037 [DOI] [PubMed] [Google Scholar]
- Nyambuu K (1992) Introduction to the ethnography of Mongolia. Mongolian State Press, Ulaanbaatar, Mongolia [Google Scholar]
- Sajantila A, Lahermo P, Anttinen T, Lukka M, Sistonen P, Savontaus ML, Aula P, Beckman L, Tranebjaerg L, Godde-Dahl T, Issel-Tarver L, DiRienzo A, Pääbo S. (1995) Genes and languages in Europe: an analysis of mitochondrial lineages. Genome Research 5:42–52 [DOI] [PubMed] [Google Scholar]
- Shifman S, Darvasi A (2001) The value of isolated populations. Nat Genet 28:309–310 [DOI] [PubMed] [Google Scholar]
- Statistics Bureau/Statistical Research and Training Institute (ed) (2001) Japan statistical yearbook 2002. Japan Statistical Association, Tokyo [Google Scholar]
- Taillon-Miller P, Bauer-Sardina I, Saccone NL, Putzel J, Laitinen T, Cao A, Kere J, Pilia G, Rice JP, Kwok PY (2000) Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat Genet 25:324–328 [DOI] [PubMed] [Google Scholar]
- Tajima F (1989a) The effect of change in population size on DNA polymorphism. Genetics 123:597–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1989b) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumen D (1992) Anthropology of the contemporary population of Mongolian People’s Republic. PhD thesis, MV Lomonosov Moscow State University, Moscow [Google Scholar]
- Varilo T, Laan M, Hovatta I, Wiebe V, Terwilliger JD, Peltonen L (2000) Linkage disequilibrium in isolated populations: Finland and a young sub-population of Kuusamo. Eur J Hum Genet 8:604–612 [DOI] [PubMed] [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507 [DOI] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M (1999) Population choice in mapping genes for complex diseases. Nat Genet 23:397–404 [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD (1996) Positional cloning of the Werner's syndrome gene. Science 272:258–262 [DOI] [PubMed] [Google Scholar]
- Zavattari P, Deidda E, Whalen M, Lampis R, Mulargia A, Loddo M, Eaves I, Mastio G, Toddo JA, Cucca F (2000) Major factors influencing linkage disequilibrium in distinct populations: demography, chromosome recombination frequency and selection. Hum Mol Genet 9:2947–2957 [DOI] [PubMed] [Google Scholar]