Abstract
The eukaryotic family of ADP-ribosylation factor (Arf) GTPases plays a key role in the regulation of protein trafficking, and guanine-nucleotide exchange is crucial for Arf function. Exchange is stimulated by members of another family of proteins characterized by a 200-amino acid Sec7 domain, which alone is sufficient to catalyze exchange on Arf. Here, we analyzed the phylogeny of Sec7-domain–containing proteins in seven model organisms, representing fungi, plants, and animals. The phylogenetic tree has seven main groups, of which two include members from all seven model systems. Three groups are specific for animals, whereas two are specific for fungi. Based on this grouping, we propose a phylogenetically consistent set of names for members of the Sec7-domain family. Each group, except for one, contains proteins with known Arf exchange activity, implying that all members of this family have this activity. Contrary to the current convention, the sensitivity of Arf exchange activity to the inhibitor brefeldin A probably cannot be predicted by group membership. Multiple alignment reveals group-specific domains outside the Sec7 domain and a set of highly conserved amino acids within it. Determination of the importance of these conserved elements in Arf exchange activity and other cellular functions is now possible.
INTRODUCTION
Arf GTPases are conserved molecular switches that regulate vesicular transport in all eukaryotic cells (Donaldson and Jackson, 2000; Randazzo et al., 2000). In this process, membrane-bound vesicles bud from one intracellular compartment and fuse with another. These vesicles mediate the different protein transport steps of the exocytic and endocytic pathways. The Arf family has been studied extensively in two model systems: yeast and mammals (Table 1). Saccharomyces cerevisiae has five Arf or Arf-related (Arl) proteins. The Arf1/Arf2 pair is required for cell viability and is implicated in all the steps of the exocytic and endocytic pathways (Stearns et al., 1990; Gaynor et al., 1998; Yahara et al., 2001). The other three proteins, Arf3/Arl2, Arl1, and Arl3, are not required for cell viability, and their function is less clear. The mammalian Arfs include three groups: classes I–III. Although class I Arfs are involved in transport through the exocytic pathway, class III is implicated in endocytosis and in actin dynamics at the plasma membrane (D'Souza-Schorey et al., 1995; Peters et al., 1995; Radhakrishna and Donaldson, 1997). Little is known about the roles of the class II Arfs. In addition, a number of mammalian Arls share ∼35–55% identity with the Arfs (Proteome BioKnowledge Library. http://www.incyte.com/sequence/proteome/index.shtml on 5/1/2003). The most extensively studied Arfs, the yeast Arf1/2 and the mammalian class I Arfs, share ∼80% identity and are thought to be important for the process of vesicle budding (Randazzo et al., 2000).
Table 1.
Substrate specificity of Arf GEFs
| % Identity1 | Yeast | Mammals | GEFs |
|---|---|---|---|
| 100 | Arf1/2pa | GEA12, SEC73, SYT14 | |
| Class I: Arf1/3b | BIG1,25, CYH1,3,4,ARNO/CYH26 | ||
| 82 | Class II: Arf 4/5c | GBF7 | |
| 68 | Class III: Arf6 | BRAG2/ArfGEP1008, EFA69, ARNO/CYH210 | |
| 50-57 | Arf3p, Arl1p | Unknown | |
| 40-57 | Arl1-8 | Unknown | |
| 34-37 | Arl3p | ARP1 | Unknown |
| 28-35 | Sarlp | Sar1 | Sec12d, 11 |
Groups of Arfs from yeast and mammals are shown with respect to their sequence similarity and their GEF specificity. Percent identity shows each group's sequence similarity to either Arf1/2 (for yeast proteins) or to class I (for mammalian proteins). Yeast Arf1/2 and mammalian class I Arfs are 80% identical. The GEFs column shows known preferred nucleotide exchangers to each Arf group.
Arf1 and 2 are 96% identical
Arf1 and 3 are 96% identical
Arf4 and 5 are 90% identical
Sec12 is not a Sec7-domain–containing protein
Peyroche et al., (1996)
Chardin et al., 1996; Meacci et al., 1997; Franco et al., 1998; Klarlund et al., 1998; Ogasawara et al., 2000
Claude et al. (1999)
Switching between the GDP- and the GTP-bound forms is crucial for Arf function (Dascher and Balch, 1994; Kahn et al., 1995). The switch from the GDP to the GTP-bound form is stimulated by guanine-nucleotide exchange factors (GEFs). GTPase activating proteins stimulate the switch from the GTP- to the GDP-bound form of Arf. All Arf GEFs identified to date have a Sec7 domain (Jackson and Casanova, 2000), a 200-amino acid region that is sufficient for GEF activity (Chardin et al., 1996; Jones et al., 1999). The phylogeny of the Sec7-domain protein family is the subject of this study. The multiplicity of Arfs and Sec7-domain proteins suggests that different Sec7-domain proteins have substrate preference toward specific Arfs. In yeast, the substrate specificity question has not been addressed, because only Arf1/2 were tested as substrates for the Sec7-domain proteins. Mammalian Sec7-domain proteins have known substrate preference in vitro (Table 1). For example, BIG1 preferentially acts as a GEF on class I Arfs, Golgi-specific brefeldin A (BFA) resistance factor 1 (GBF1) on class II, whereas BRAG2/Arf-GEP100 and EFA6 act on class III Arf6. The cytohesins (CYH) can act on both class I and III Arfs, but the Sec7 domain of CYH1 and ARNO/CYH2 do not catalyze exchange on Arl1–3 and Arl3, respectively (Chardin et al., 1996; Pacheco-Rodriguez et al., 1998). It is not clear that this in vitro substrate preference is physiological. Different Sec7-domain proteins are known to exhibit different patterns of intracellular localization and biological function (Jackson and Casanova, 2000; Bonifacino and Jackson, 2003).
The three-dimensional structures of the Sec7 domains of several GEF proteins have been solved, both alone and in complex with Arf. The Arf and Arl proteins are unique among GTPases in having two regions of the protein that change conformation upon release of GDP. As in all GTPases, Switch I and Switch II undergo a major conformational change, resulting in effector binding to these regions. Unique to the Arf/Arl subfamily of GTPases is a second conformational change which extends to the myrisoylated N-terminal amphipathic helix and results in the attachment of the GTPase to membranes (Randazzo et al., 1995; Antonny et al., 1997; Pasqualato et al., 2002). Therefore, Arf nucleotide switching also determines the cycling of Arf between the cytoplasm and membranes. A possible exception to this rule is Arf6, which under certain conditions seems to be constitutively bound to membranes (Cavenagh et al., 1996; Yang et al., 1998; Gaschet and Hsu, 1999). If true, GTP-binding induces a conformational change on Arf6 without affecting its membrane association.
The 10 α-helices and connecting loops of the Sec7 domains of S. cerevisiae Gea2p and Homo sapiens ARNO/CYH2 are organized into two subdomains with a hydrophobic groove between them (Cherfils et al., 1998; Goldberg, 1998). In the complex of the Sec7 domain with nucleotide-free Arf, the two switch domains of Arf are positioned in the hydrophobic groove (Goldberg, 1998). Superimposing the unbound and Arf-bound Sec7 domain of Gea2p suggests that the two proteins are flexible and that they change each other's structure upon binding (Renault et al., 2002). Arf-GDP binding to the Sec7 domain results in the closure of the Sec7-domain hydrophobic groove. This closure triggers the protrusion of the catalytic glutamate from the Sec7 domain, which in turn causes Arf to lose its nucleotide. The nucleotide-free form of Arf is already in a conformation very close to that of the GTP-bound form. When in the GDP-bound form, Arf is largely soluble, with its amphipathic N-terminal helix tucked into a hydrophobic pocket of the protein. In the Arf–Sec7 domain complex, this pocket disappears due to movement of the β2 and β3 strands and the loop between them, forcing the N-terminal myrisoylated α-helix to swing out and become inserted into the membrane (Beraud-Dufour et al., 1999; Pasqualato et al., 2002).
BFA, a fungal metabolite, has been widely used as a drug for studying protein transport. The main cellular target of BFA has been identified as a subset of the Arf GEFs (Donaldson et al., 1992; Helms and Rothman, 1992; Randazzo et al., 1993; Peyroche et al., 1999). BFA blocks the GEF activity of the Sec7 domain itself (Sata et al., 1998; Jones et al., 1999) through a somewhat unusual mechanism in which BFA binds to both the Sec7 domain and Arf (Mansour et al., 1999; Peyroche et al., 1999). The target of BFA is an early intermediate in the exchange reaction, an Arf-GDP–Sec7 protein complex (Robineau et al., 2000). BFA biding interferes with the closure of the Sec7-domain groove, freezing the Sec7-domain Arf–GDP complex (Peyroche et al., 1999; Renault et al., 2002). The convention in the field is that small Sec7-domain proteins, e.g., H. sapiens CYH1, are BFA resistant, whereas large Sec7-domain proteins, e.g., BIG and GEA, are BFA sensitive (Jackson and Casanova, 2000). Analysis presented here challenges the generalization of this convention to other systems.
Phylogenetic analyses are now possible due to the increasing number of fully sequenced genomes. The protein transport machinery and mechanisms are highly conserved from yeast to mammals (Mellman and Warren, 2000). Therefore, a phylogenetic analysis of its components should provide a basis for extrapolation of what is known about the mechanisms in one model system to others. The phylogenetic analysis of the Sec7 proteins presented here suggests their distribution into seven groups. Based on the distribution of proteins in these groups and the known intracellular function of some Sec7-domain proteins, predictions can be made regarding the biological function of other members of the same group in other organisms. Multiple alignments of the Sec7 domain suggest that both Arf binding and the conserved structure are crucial for the function of this domain. Multiple alignments of whole proteins reveal group-specific as well as a number of cross-group homologies outside the Sec7 domain. The group-specific domains suggest that members of individual groups share group-specific functions that might include intracellular localization and intermolecular interactions in addition to their primary Arf-GEF activity.
MATERIALS AND METHODS
Databases
Sequences of completely finished and curated proteomes of eight eukaryotes, 16 archaea, and 77 bacteria were acquired from European Bioinformatics Institute in April 2003 (Pruess et al., 2003). The annotated protein sequences from assembly version 3 of the Neuorspora crassa genome were obtained from the Whitehead Institute Center for Genome Research (Galagan et al., 2003). Annotated protein sequences for the Legionella pneumophila genome were from the Columbia Genome Center (Russo, 2003). The whole genome nucleotide sequences for Drosophila melanogaster (Celniker et al., 2002), and Caenorhabditis elegans (C. elegans Sequencing Consortium, 1998), H. sapiens (Lander et al., 2001), and 136 microbial genomes were from National Center for Biotechnology Information. Additional names and annotations were derived from the nonredundant protein database at National Center for Biotechnology Information (Benson et al., 2003).
Identification of Sec7-Domain Proteins
Sec7-domain–containing proteins were identified in the annotated proteome databases by iteratively comparing this database to known Sec7-domain sequences by three blast (Altschul et al., 1997) rounds with an expectation value of 10-2. In the first round, protein blast detected 69 sequences homologous to the Sec7 domain of the S. cerevisiae protein Sec7 (183 amino acids, from 838-FNNK to HQAM-1020), which was shown to posses an Arf-GEF activity (Jones et al., 1999). Custom perl scripts stripped each of these blast hits to just the sequences homologous to the query sequence. The original sequence plus the homologous hits were the query sequences for a second round of protein blast, which revealed nine more hits, for a total of 79 Sec7-domain–containing proteins. Subsequent rounds of iterative blasting revealed no additional hits in the protein database. To be sure that no Sec7-domain–containing sequences were omitted during the proteome curation at EBI and elsewhere, the entire published genomes of D. melanogaster, C. elegans, and H. sapiens were translated in all six reading frames using the universal translation table. All open reading frames (ORFs) longer than 30 amino acids were blasted against the Sec7-domain–containing proteins defined above. None of the resultant hits from the translated ORFs revealed any novel sequences.
Full-length versions of each sequence were isolated from the protein databases for all further work. Inspection of these sequences revealed many to be truncated duplicates of other sequences. Only the longest sequences were retained. Sequences from the currently incomplete Mus musculus genome were removed, because they were sufficiently similar in subsequent phylogenetic analysis to H. sapiens sequences that they offered little additional information, except for the mouse FBX8 (see below). The remaining 52 protein sequences were compared against the nonredundant database at National Center for Biotechnology Information (Benson et al., 2003) by using protein blast to identify the most recent National Center for Biotechnology Information names, annotations, and alternative names, all of which are recorded in Table 2. All sequences in our database corresponded to an identical sequence (same length and residues) at National Center for Biotechnology Information.
Table 2.
Sec7-domain–containing proteins in seven eukaryotic model systems and bacteria
| Group | Species | Name | Synonyms | NCBI accession | Protein no. aa | Sec7D no. aa | GEF activity |
|---|---|---|---|---|---|---|---|
| CYH | Ce | CYH1 | YMX4, GRP-1 | NP_498764 | 393 | 181 | |
| CYH | Dm | CYH1 | Q9V9Q6, CG11628-PA, ARNO | NP_610120 | 410 | 181 | |
| CYH | Hs | CYH1 | B2-1, Q9V9Q6 | NP_004753 | 398 | 182 | 1 |
| CYH | Hs | CYH2 | Q99418, ARNO | NP_059431 | 400 | 182 | 2 |
| CYH | Hs | CYH3 | ARNO3, GRP1 | O43739 | 400 | 182 | 3 |
| CYH | Hs | CYH4 | Q9UIA0 | NP_037517 | 394 | 182 | 4 |
| BIG/SEC7 | Sc | SEC7 | Sec7p | NP_010454 | 2009 | 183 | 5 |
| BIG/SEC7 | Sp | SEC71 | Q9P7V5 | NP_594555 | 1822 | 183 | |
| BIG/SEC7 | Sp | SEC72 | Q9UT02 | NP_594954 | 1811 | 183 | |
| BIG/SEC7 | Nc | SEC7 | HP | EAA33549 | 1940 | 182 | |
| BIG/SEC7 | Ce | BIG1 | Q9XWG5 | NP_493386 | 1628 | 184 | |
| BIG/SEC7 | Dm | BIG1 | AAN10848, CG7578-PB | NP_723839 | 1614 | 185 | |
| BIG/SEC7 | Hs | BIG1 | p200, p200 ARF-GEP1 | NP_006412 | 1849 | 184 | 6 |
| BIG/SEC7 | Hs | BIG2 | Q9Y6D5, ARFGEF2 | NP_006411 | 1785 | 183 | 7 |
| BIG/SEC7 | At | BIG1 | HP-F20D10,320 | T05647 | 1643 | 181 | |
| BIG/SEC7 | At | BIG2 | At3g60860.1 | NP_191645 | 1793 | 181 | |
| BIG/SEC7 | At | BIG3 | HP-Atlg01960.1 | NP_171698 | 1750 | 181 | |
| BIG/SEC7 | At | BIG4 | At4g35380 | NP_195264 | 1711 | 181 | |
| BIG/SEC7 | At | BIG5 | At3g43300 | NP_189916 | 1669 | 179 | |
| GBF/GEA | Sc | GEA1 | P47102 | NP_012565 | 1408 | 206 | 8 |
| GBF/GEA | Sc | GEA2 | P39993 | NP_010892 | 1459 | 198 | 8 |
| GBF/GEA | Sp | GEA1 | Q9P7R8 | NP_596613 | 1462 | 185 | |
| GBF/GEA | Nc | GEA1 | HP-NCU080231, EAA33457 | EAA33457 | 1626 | 186 | |
| GBF/GEA | Ce | GBF1 | Q9XTF0, C24H11, 3M842 | NP_499522 | 1820 | 184 | |
| GBF/GEA | Dm | GBF1 | Q8ML25, CG8487-PA, garz | NP_725133 | 1740 | 188 | |
| GBF/GEA | Hs | GBF1 | Q92538 | NP_004184 | 1859 | 186 | 9 |
| GBF/GEA | At | GBF1 | GNL1, Q9FLY5, At5g39500.1 | NP_198766 | 1443 | 184 | |
| GBF/GEA | At | GBF2 | GNL2, At5g19610 | NP_197462 | 1375 | 185 | |
| GBF/GEA | At | GBF3 | EMB30, GNOM, Q42510, Atlg13980 | NP_172851 | 1451 | 184 | 10 |
| BRAG | Ce | BRAG1 | Q94287 | NP_500420 | 614 | 195 | |
| BRAG | Dm | BRAG1 | Q9VP80, CG32434-PB | NP_730594 | 1325 | 187 | |
| BRAG | Hs | BRAG1 | HP-O60275, KIAA0522, ArfGEP100 | BAA25448 | 1560 | 187 | 11 |
| BRAG | Hs | BRAG2 | HP-O94863, KIAA0763, GEP-100 | NP_055684 | 841 | 187 | |
| BRAG | Hs | BRAG3 | HP-Q9UPP2, KIAA1110 | BAA83062 | 740 | 187 | |
| SYT1 | Sc | SYT1 | Q06836 | NP_015420 | 1226 | 168 | 12 |
| SYT1 | Sp | SYT11 | YDG1, Q10491, C26F1.01 | Q10491 | 928 | 151 | |
| SYT1 | Sp | SYT12 | O13817, SEC73 | O13817 | 1082 | 155 | |
| SYT1 | Nc | SYT1 | HP-NCU014651 | EAA27506 | 1448 | 155 | |
| SYT2 | Sc | SYT2 | HP-Yb1060, YBG0, P34225, ArfGEF5 | NP_009493 | 687 | 155 | |
| SYT2 | Sp | SYT21 | YDH4, Q92349 | NP_594113 | 1318 | 107 | |
| SYT2 | Sp | SYT22 | O13690 | NP_594936 | 942 | 143 | |
| SYT2 | Nc | SYT2 | HP-NCU023471 | EAA30233 | 1607 | 142 | |
| EFA | Ce | EFA6 | Q95Q14, Y55D9A, 1b, 4N370 | NP_502416 | 818 | 167 | |
| EFA | Dm | EFA6 | Q9VCX8, CG31158-PA | NP_732769 | 1076 | 170 | |
| EFA | Hs | EFA6A | Q15673, EFA6R, TYL, PSD | G01205 | 645 | 169 | 13 |
| EFA | Hs | EFA6B | O95621, Tic | CAD30842 | 1056 | 168 | |
| EFA | Hs | EFA6C | HP-Q9BQ17, DKFZp761B0514 | NP_115665 | 771 | 169 | |
| EFA | Hs | EFA6D | Q9NY10, EFA6R, KIAA0942, hepatocellular carcinoma-associated antigen 67 | NP_056125 | 534 | 170 | |
| FBX | Hs | FBX8 | Q9NRD0, FBS | Q9NRD0 | 319 | 177 | |
| FBX | Mm | FBX8 | Fbxo8 | NP_056606 | 319 | 177 | |
| BACT | Lp | RalF | AAL23711 | 374 | 185 | 14 | |
| BACT | Rp | SEC7 | Q9ZDF5 | NP_220757 | 462 | 188 |
List of the 52 Sec7-domain proteins identified here organized by group: 49 in the seven model systems, the mouse FBX8, and two bacterial sequences. Group names correspond to those shown in Figure 1. HP, hypothetical protein. Synonyms list names found in databases and publications. The next two columns show the number of amino acids of the whole protein (protein aa) and the Sec7 domain (Sec7D aa). The last column indicates the references for experimental evidence for Arf GEF activity.
1Meacci et al., 1997; 2Chardin et al., 1996; Frank et al., 1998; 3Franco et al., 1998; Klarlund et al., 1998; 4Ogasawara et al., 2000; 5Sata et al., 1998; Jones et al., 1999; 6Morinaga et al., 1997; Mansour et al., 1999; 7Togawa et al., 1999; 8Peyroche et al., 1996; 9Claude et al., 1999; 10Steinmann et al., 1999; 11Someya et al., 2001; 12Jones et al., 1999; 13EFA6A Franco et al., 1999; Macia et al., 2001; 14L. pneumophila RalF Nagai et al., 2002.
Construction of the Phylogenetic Trees
The 51 core Sec7-domain protein sequences form the seven model systems and bacteria, were aligned by Clustal (Thompson et al., 1994), by using the default settings for slow/accurate alignments (gap penalty of 10, gap extension cost of 0.2, 30% delay for divergent sequences, four space gap separation distance, without end gap separation, with residue-specific penalties, and by using the Gonnet series protein weight matrix). The aligned core Sec7 sequences, defined by the iterative blasting described above, were manually trimmed on the N- and C-terminal ends to remove weak or ambiguous alignments. Phylogenetic analysis was performed using PAUP 4.0b10 (Swofford, 2003) as informed by a globular protein substitution matrix (Whelan and Goldman, 2001). Distance trees were created using the heuristic search for optimal trees. The initial tree was created by neighbor joining, and branch swapping used the TBR algorithm. To estimate the reliability of this tree, bootstrap analysis was performed with 1000 replicates of full heuristic searches, by using the same weighting parameters used in the initial analysis. The distance and bootstrap trees were drawn by TreeView version 1.6.6 (Page, 1996) and manually modified in a general-purpose graphics editor. For the bootstrap results, the branches were color coded from red to green to reflect their persistence in 50–100% of the bootstrapped trees.
Multiple Alignments within and across Groups
Alignment illustrations were based on the above-mentioned Clustal alignments and generated by a custom perl script to reflect comparisons of the qualities of alignments between and within groups of Sec7 family proteins. All Sec7 family groups were assigned a primary hue on the HSL color wheel from red to purple, the same as those seen in Figure 1. For a given group of proteins, the alignment was broken into segments according to cross-family alignments (e.g., a run of just CYH alignments, a run of CYH and BIG alignment, etc.). For regions with no alignment in at least half of the members of the group, gray boxes were drawn in the middle of the region reflecting the average size of the members that had unrelated sequences in that area.
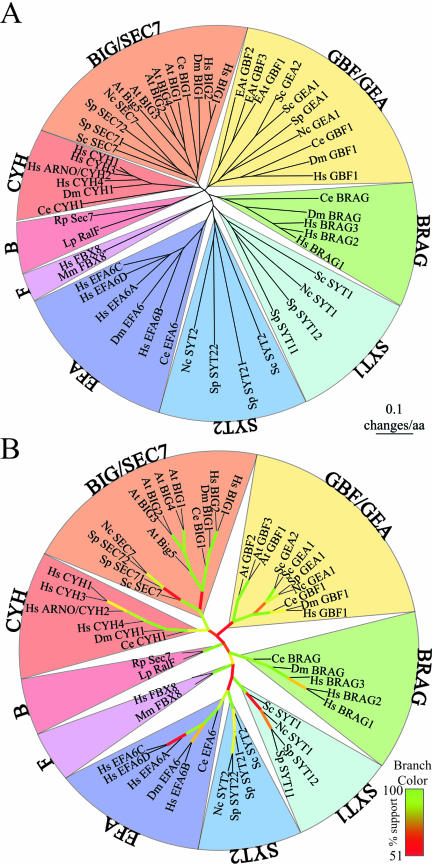
Figure 1.
Unrooted phylogenetic tree relating the Sec7-domain ARF-GEF family members. (A) Tree was constructed using neighbor-joining with core Sec7-domain gene sequences. Pie sections demarcate seven phylogenetically related groups of the Sec7 family plus one in mammals and one in bacteria. The bar on the lower left-hand shows distance. (B) Confidence measurements of phylogentic tree branches. Branches as in A, except that lengths no longer show phylogenetic distance and branch color indicates data from bootstrap analysis (1000 iterations). Colors range from red, indicating 50% bootstrap support, to green, indicating 100% support.
For each other region, both intra- and intergroup similarity scores were computed. Intragroup scores were the average PAM250 (Altschul et al., 1997) value, averaged over all possible pairings between members of the group. Intergroup similarities were computed as the average PAM250 score of each possible pair consisting of one member of each of the two groups. Each comparison generated a bar colored according to the primary hue of the target group but with an intensity reflecting the quality of the alignment. PAM250 scores of less than zero are drawn in black. Positive scores are drawn in the target group's hue but with a saturation scaled from 0 to 100% for PAM250 scores of 0–3, respectively. The pleckstrin homology (PH), coiled coil (CC), and F-box domains were identified by the hidden Markov models used by SMART (Schultz et al., 1998; Letunic et al., 2002).
RESULTS
Identification of Sec7-Domain–containing Proteins
We searched the fully sequenced and annotated genomes of seven eukaryotic model systems that span fungi, animals, and plants, for Sec7-domain–containing proteins. The fungi are S. cerevisiae (Sc), Schizosaccharomyces pombe (Sp), and N. crassa (Nc); Arabidopsis thaliana (At) represents the plants; and the animals are C. elegans (Ce), D. melanogaster (Dm), and H. sapiens (Hs). We chose only completely sequenced and well-curated genomes to be reasonably sure that the generated trees contain all Sec7-domain proteins from each organism. This allows us to make conclusions regarding the groups that are present or not present in those organisms. An iterated blasting approach detected all sequences containing a Sec7 domain present in the proteome databases, no matter how far diverged from one another. We began with the Sec7 domain of S. cerevisiae Sec7p as seed sequence because it was shown to posses an Arf-GEF activity on its own (Jones et al., 1999). Briefly, sequences homologous at a blast score of 10-2 to this seed were trimmed to just the core Sec7 domain and blasted against the databases, generating new hits to blast against the databases until no new sequences were found (see MATERIALS AND METHODS).
This method revealed a total of 49 Sec7-domain–containing protein sequences in the seven eukaryotic model systems (Table 2; http://www.uic.edu/~nava/papers/SEC7). Most discovered sequences were not well described in the literature. More than one-half of these are only automatically named. Of those well studied, most are from S. cerevisiae or mammals. Table 2 suggests a consistent set of names for all identified Sec7-domain proteins based on the phylogenetic analysis shown in Figure 1A, by using published names of the related well-studied members of each group as templates. All seven searched eukaryotic model systems have Sec7-domain proteins. Interestingly, one fully sequenced eukaryote genome, the endosymbiotic algal nucleus of Guillardia theta, has no recognizable Sec7-domain protein. However, evolutionary compaction eliminated the majority of the genome of this nucleomorph (Douglas et al., 2001).
The A. thaliana Big5 sequence found at European Bioinformatics Institute has a 26-amino acid deletion relative to the other A. thaliana BIGs. This deletion corresponds to most of helix α7 and includes the catalytic glutamate and three other highly conserved amino acids (sites h to j; Figure 4B). Such a deletion should cause a significant change in the structure of the Sec7 domain of this protein and loss of Arf GEF activity. In addition, the deletion results in an unexplainable positioning of A. thaliana Big5 in the phylogenetic tree. We found the nucleotide code for the missing 26 amino acids in the DNA sequence immediately after the intronexon border as defined by TAIR (The Arabidopsis Information Resource. www.arabidopsis.org on October 1, 2003). The missing sequence is almost perfectly conserved with that of the other A. thaliana BIGs. Even though its inclusion requires the use of an unconventional splice site, this region is immediately followed by a stop codon, as is often the case for used splice sites (Li et al., 2002). It is possible that the A. thaliana BIG5 evolved from a typical GEF, had a splice site changed to abolish the catalytic activity, and acts as a dominant negative protein. However, such a change must have happened very recently given the high sequence conservation of the 26 amino acids. Therefore, the corrected A. thaliana Big5 protein sequence was used for all further analyses. When these amino acids were included, the A. thaliana BIG5 groups together with the other BIGs on the phylogenetic tree.
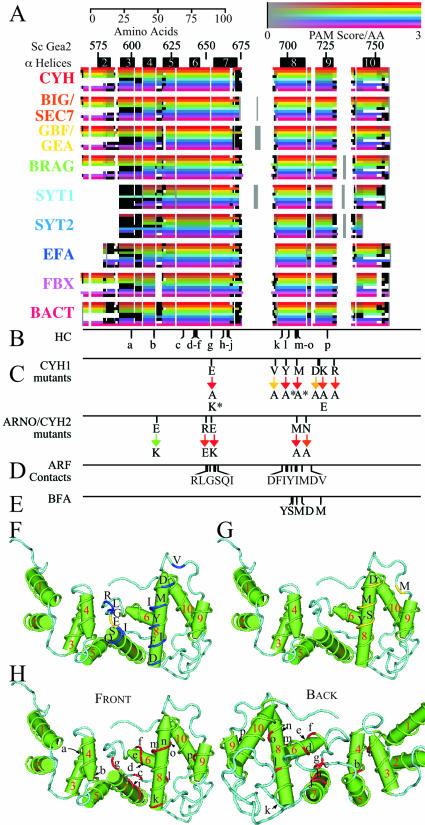
Figure 4.
Multiple alignments of the Sec7 domain arranged by group. (A) Visual representation of the multiple alignment strength in the Sec7 catalytic domain. As in Figure 3, except all cross-group homologies, including those with limited quality, are shown as colored areas. In addition, the gray areas indicating regions with no intragroup sequence similarity are embedded in white spaces whose width indicates the maximum insert size. (B) Positions of the HC amino acids identified here (further described in Table 4). (C) Positions of previously characterized mutations that affect Arf GEF activity. The top row indicates the wild-type sequence and the bottom row shows the target mutations. Color from red to green indicates the proportion (from 0 to 100%) of wild-type activity expressed by the mutant. Mutations that abolish Arf binding are indicated by *. CYH1 mutant coordinates: E157K, V179A, Y187A, M195A, D207A, K208A, K208E, R219A, and E157A (Betz et al., 1998). ARNO/CYH2 mutant coordinates: E117K, R152E, E156K, M194A, and N201A (Cherfils et al., 1998). (D) Arf contacts. The positions of the 15 amino acids that make contact with Arf in the cocrystal (Goldberg, 1998). See coordinates in F legend. (E) BFA sensitivity consensus. The positions of the amino acids, which are proposed here to define a consensus for BFA sensitivity (Table 5). See structural positions in G. (F) Arf contact sites. The Arf-binding surface in the hydrophobic groove is shown on the three-dimensional structure of the Sec7 domain of S. cerevisiae GEA2 (by x-ray crystallography). Numbered green solid arrows indicate the 10 predicted alpha helices. Light blue indicates predicted loops. Bright blue sequences correspond to the Arf-contact sites positioned in the hydrophobic grove as defined in the Arf–Sec7 domain complex (Renault et al., 2002). Yellow indicates the catalytic glutamic acid. The N-side of the groove is composed of helices α1–5, the glutamate finger loop, and N-the terminal half of helix α7. The C-terminal side includes helices α6, the C-terminal half of helix α7 and helices α8–10. Positions for amino acids are, in S. cerevisiae Gea2 coordinates: R650, L651, G653, S655, Q656, I658, D696, F699, I700, Y703, I706, M707, D711, and V717. (G) BFA sensitivity consensus. As in F, except that yellow sequences correspond to amino acids that are predicted to define BFA sensitivity shown in E and Table 5. Positions for consensus sites are in S. cerevisiae Gea2 coordinates: Y703, S704, M707, D711, and M721. (H) HC amino acids. HC amino acids are positioned on both sides of the Sec7 domain. Two 180° opposite views of the Sec7 domain are shown: front and back. Red indicates HC sites shown in B and Table 4. Limited overlap, 4/16, exists between HC amino acids and Arf contact sites of the Sec7 domain.
We also searched fully annotated proteomes of 77 bacteria and 16 archaea. The latter are considered to share a common ancestor with eukaryotes (Iwabe et al., 1989; Brown and Doolittle, 1995; Baldauf et al., 1996), which could suggest that they have related proteins. However, no archael Sec7-doain proteins were found, implying that the Sec7-domain family evolved in eukaryotes after their divergence from the archaea. Instead, two bacterial proteomes, both parasites of eukaryotes, each contain a single Sec7-domain protein: L. pneumophila (Lp) (Nagai et al., 2002) and Rickettsia prowazekii (Rp). Furthermore, a protein blast against 136 translated microbial genomes at National Center for Biotechnology Information as of May 2003 revealed no hits against the Sec7 domain of S. cerevisiae Sec7 with an e <10-2, except for the R. prowazekii protein already in our database (the L. pneumophila genome was not in National Center for Biotechnology Information database at the time). Both L. pneumophila and R. prowazekii belong to the Proteobacteria phylum. However, 23 other Proteobacteria, including, notably, a close relative of R. prowazekii, Rickettsia conorii, do not have Sec7-domain proteins. Based on these findings and the fact that the two are parasites of eukaryotes, it seems clear that the BACT group resulted from horizontal transfer, as was suggested previously for the L. pneumophila RalF (Nagai et al., 2002).
Phylogenetic Tree for Sec7-Domain Proteins
Figure 1A shows a distance tree of the 52 identified Sec7-domain proteins (49 from model eukaryotic organisms, a single mouse protein noted below, and two bacterial proteins) that was constructed using the core Sec7-domain protein sequences and the programs described in MATERIALS AND METHODS. We used only the core sequence because tree construction is based on sequence alignment, and there is very little cross-group sequence similarity outside the Sec7 domain (Figure 3). The tree is unrooted because we could not find any sequences that are related, even at an extremely lenient blast cutoff (e <10-2) and are unambiguously diverged from all seven model eukaryotes.
Figure 3.
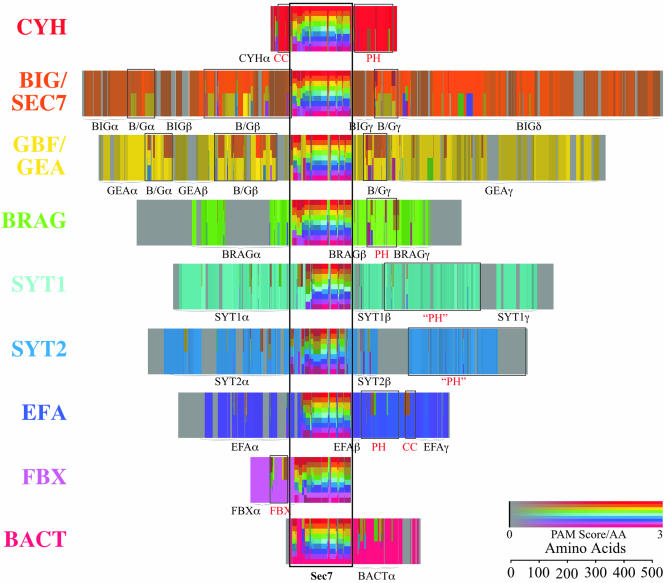
Multiple alignments of whole proteins arranged by group. Visual representation of the multiple alignment strength in the Sec7-domain–containing proteins. Each group is shown in a different color and row. For a given row, colored bands indicate the strength of the alignment between members of that group and each other group for which there is significant alignment. For example, the orange areas in the BIG/SEC7 row indicate intragroup homologies, whereas the yellow areas represent BIG/SEC7-GBF/GEA intergroup homologies. Color intensities indicate average PAM scores between all pairs of sequences in the comparison, with gray indicating no sequence similarity (0 or less) to bright colors indicating strong sequence similarity (3). Tall gray areas indicate regions with no sequence similarity even within a group. The width of these gray areas is proportional to the average insert size. Boxes mark areas of large cross-group sequence similarity, including the central Sec7 domain. Names are attached to regions of sequence similarity either within or across groups. Actual sequences corresponding to these regions can be viewed at http://www.uic.edu/~nava/papers/SEC7.
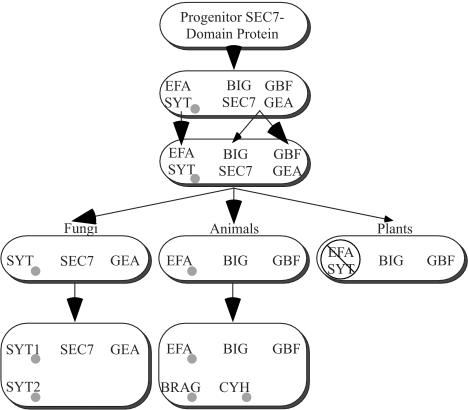
The colored pie sections delineate groups of the Sec7 family. The rationale for this grouping comes from the topology of the tree and is based on known phylogenetic relationships (e.g., all animal or fungi members of a particular group are clustered together). Further justification of this grouping comes from the findings that the branches at the base of each group, except GBF/GEA, are strong (Figure 1B) and that the members of each group, including GBF/GEA, share similar overall structure (Figure 3). Two groups, BIG/SEC7 and GBF/GEA, include members from fungi, plants, and animals. Three groups, CYT, BRAG, and EFA, are specific for animals. Two groups, SYT1 and SYT2, are specific for fungi. One outlier is the human FBX8, which is found at the base of the tree, between the BRAG and the SYT/EFA branches. This protein is not included in another group because it does not meet the above-mentioned three considerations: phylogenetic relationship, strong branch position, and similar structure (discussed further below with respect to Figure 2). An almost identical protein is present in M. musculus, shown in Figure 1 for comparison.
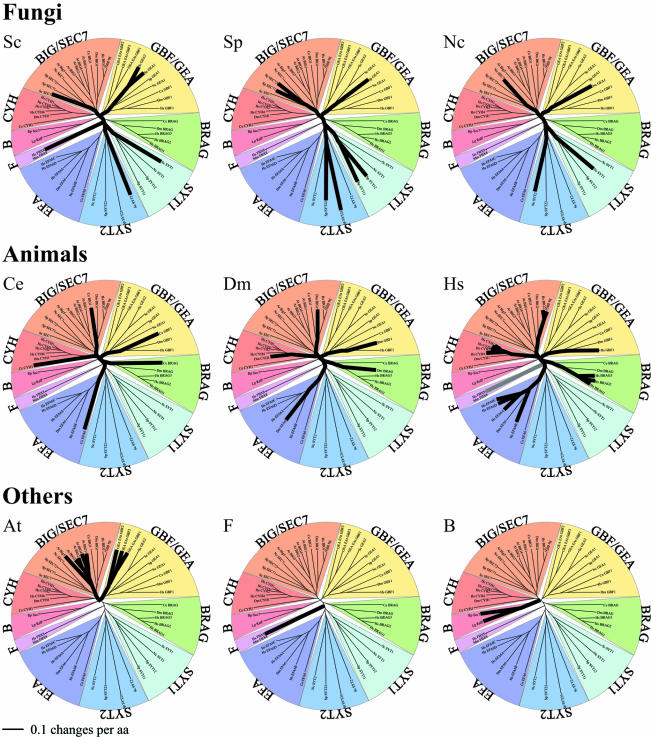
Figure 2.
Distribution of Sec7-domain Arf-GEFs in different organisms. Trees are as in Figure 1 except that the thick black lines show the members of the family in each organism. All fungi have Sec7 proteins in the same four groups. Animals each have members in five groups. Plants have members in just two groups. F, human and mouse genomes also contain a novel gene, FXB8, which is only weakly related to any other group. B, in addition to the eukaryotic sequences, two related bacterial sequences were also found, which are not closely related to any other group.
The two bacterial Sec7-domain proteins branch from the center of the tree, not especially close to any other group. In addition, these two proteins do not show substantial structural similarity to the eukaryotic Sec7-domain proteins outside the Sec7 domain, even though they do share extensive similarity between themselves (Figure 3). We have not used these bacterial sequences to root the eukaryotic tree because of the following two reasons. First, the lack of Sec7-domain proteins in most bacteria suggests horizontal transfer from eukaryotes, and not evolution of the eukaryotic Sec7-domain family from a bacterial progenitor (see above). Second, the fact that the bacterial Sec7-domain proteins are not close to any of the eukaryotic groups does not necessarily mean that the horizontal transfer occurred before the divergence of the eukaryotic groups, but rather that the evolution of bacterial and eukaryotic proteins could occur at very different rates. Therefore, an unrooted tree is the most reasonable representation of relationships among these Sec7-domain proteins.
Bootstrap Analysis of the Sec7-Domain Tree
To assess the reliability of the position of the branches, a distance-based bootstrap analysis in PAUP by using 1000 iterations was performed. This method computes many trees after randomly resampling individual amino acids. Branches that are supported by few characters, or by characters that change multiple times, will be present in only a small fraction of these trees, whereas branches with robust support will be present in a high percentage of all trees. Figure 1B graphically illustrates the strength of these branches in terms of the fraction of trees containing the same branch.
In the bootstrap analysis of the Sec7-domain protein tree, many branches, especially within groups, show substantial bootstrap support (>90%). Furthermore, the branches at the bases of the groups, except for GBF/GEA, all show 84–100% bootstrap support, implying that the definition of these groups is solid. Membership of the A. thaliana GBFs to the GBF/GEA group is only weakly supported (53%). However, sequence similarity outside the Sec7 domain (Figure 3) implies that they do belong to this group. We conclude that the definition of these seven groups shown in Figure 1 is solid.
The center of the tree has weak bootstrap support (28–78%). The center branches are relatively short, and their poor support probably reflects the fact that short internal branches can be difficult to resolve with a limited amount of data (e.g., Saitou and Nei, 1986). Consequently, the exact relationship between the groups remains uncertain. The branch that separates the mammalian EFA and the fungal SYT groups from the other groups has 78% bootstrap support, suggesting that these groups are related. However, we do not define animal EFA and fungal SYTs as one group, EFA/SYT, because unlike the other two groups that include fungal and animal members, BIG/SEC7 and GBF/GEA, the EFAs and the SYTs do not share sequence similarity either outside the Sec7 domain, or at the N terminus of the Sec7 domain (helices 2 and 3), where they diverge from the other groups (Figures 3 and 4A).
Distribution of the Sec7-Domain Proteins in the Seven Model Systems
Figure 2 and Table 3 show the occurrence of the Sec7-domain proteins with respect to the groups defined in Figure 1, in the seven model systems analyzed here. Two groups are present in all the model systems: BIG/SEC7 and GBF/GEA. The three fungi have four to seven Sec7-domain proteins that are representatives of four groups, two of which are fungal specific, SYT1, and SYT2. Animals have five to 15 Sec7-domain proteins from five groups of which three are animal specific, CYH, BRAG, and EFA. N. crassa, C. elegans, and D. melanogaster each has single representatives for the groups common to their respective kingdoms, but S. cerevisiae, S. pombe, and H. sapiens show some apparently recent duplication. The A. thaliana plant genome seems to have specialized only in BIG/SEC7 and GBF/GEA GEFs, making multiple duplicates of each. The two bacterial sequences form a separate group as judged by the blast, bootstrapping, and structural analysis.
Table 3.
Summary of group members present in each organism
| Sc | Sp | Nc | Hs | Dm | Ce | At | Sum | |
|---|---|---|---|---|---|---|---|---|
| CYH | 4 | 1 | 1 | 6 | ||||
| BIG/SEC7 | 1 | 2 | 1 | 2 | 1 | 1 | 5 | 13 |
| GBF/GEA | 2 | 1 | 1 | 1 | 1 | 1 | 3 | 10 |
| BRAG | 3 | 1 | 1 | 5 | ||||
| SYT1 | 1 | 2 | 1 | 4 | ||||
| SYT2 | 1 | 2 | 1 | 4 | ||||
| EFA | 4 | 1 | 1 | 6 | ||||
| FBX | 1 | 1 | ||||||
| Sum | 5 | 7 | 4 | 15 | 5 | 5 | 8 | 49 |
Groups and key as in Table 2.
In addition to the normal five groups found in all animals, H. sapiens contains an additional novel member, FBX8. Almost identical sequences for FBX8 are found in the human and mouse genomes, implying that FBX8 is not a sequencing mistake of the H. sapiens genome. However, it not found in other model systems, including the animals C. elegans and D. melanogaster, in spite of a thorough search for potentially missed genes in unannotated ORFs (see MATERIALS AND METHODS). The two FBX8 proteins lie between the BRAG and EFA/SYT groups by PAUP analysis (Figure 1). However, because they are not present in lower animals, and because they are different from all Sec7-domain proteins, including the BRAG, EFA, SYT1, and SYT2 groups, by structure (Figure 3), we define them as a separate group. Sequencing genomes of organisms between D. melanogaster and H. sapiens should help determine when the FBX group occurred in evolution.
Identification of Common and Group-specific Domains of Sec7-Domain Proteins
Multiple alignments of whole Sec7-domain proteins arranged by group are shown in Figure 3. Each group has extensive regions of sequence similarity shared by all members of the group, across all model systems indicated by color (as opposed to gray). A few domains besides the Sec7 domain are common to several groups. Regions of sequence similarity that are common to all members of at least two groups are boxed, whereas regions of sequence similarity that are common to all members of one group are marked as group-specific domains. Sequences corresponding to these regions are viewable at http://www.uic.edu/~nava/papers/SEC7.
The seven eukaryotic groups of Sec7-domain proteins have a centrally located Sec7 domain flanked by varying cross-groups and group-specific domains. The PH domain occurs in five of the seven eukaryotic groups, as determined by hidden Markov models at SMART (Schultz et al., 1998). PH domains are important for the interaction of proteins with inositol phosphates and are thought to have a role in the association of proteins with specific membranes, and/or for protein–protein interactions (Blomberg et al., 1999; Lemmon and Ferguson, 2001). However, although there is sequence similarity within each group in the PH domain, there is no significant blast sequence similarity between PH domains of the five groups (at e < 10-2). The PH domains of representatives from the CYH and EFA groups were shown to be functional (Chardin et al., 1996; Derrien et al., 2002). The putative PH domains in BRAG, SYT1, and SYT2 do not share significant sequence similarity between themselves or with those of the CYH or the EFA groups. Therefore, further functional characterization of these domains is required. SMART also recognizes CC motifs in CYH and EFA, although these regions show no cross-group sequence similarity. The BIG/SEC7 and GBF/GEA groups share common B/G α,β,γ domains. In addition, there are many group-specific domains. Besides the PH and CC domains, these other cross-group and group-specific domains have no similarity to known motifs. Potential functions and binding partners for the cross-group and group-specific domains are discussed below.
The FBX and BACT groups have a C- and N-terminal Sec7 domain, respectively. The FBX proteins contain an F-BOX domain (determined by SMART), which is known to interact with ubiquitin ligases (Kipreos and Pagano, 2000). The BACTαdomain is on the carboxy side of the Sec7 domain. It is not significantly homologous (e < 10-2) to any sequence in the database (National Center for Biotechnology Information databases as of May 2003). However, BLAST does find weak alignments with several bacterial proteins with a variety of identified functions, suggesting that this domain is of a bacterial origin.
The group-specific and cross-group domains allow us to speculate about the evolution of the Sec7-domain family and the importance of these domains for their function (see DISCUSSION).
Sequence Similarity within the Sec7 Domain
A detailed multiple alignment of the Sec7 domains of the 52 identified Sec7-domain proteins arranged by group is shown in Figure 4A. For comparison, the crystal structure of the Sec7 domain of S. cerevisiae GEA2 is shown in Figure 4F (Renault et al., 2002). The domain contains 10 α-helices, although there is little or no sequence similarity in the first α-helix between different Sec7 groups. Interestingly, the break points in the alignment shown in Figure 4A almost exactly correspond to borders between α-helical domains (see DISCUSSION). The multiple alignments show excellent conservation in helices α4-α10 between all groups. Helices α2 and α3 are conserved across all groups, including FBX and BACT, except as follows. Helix α2 is not conserved for EFA and the SYTs. Helix α3 is not conserved across groups for SYT1 and is weakly conserved for SYT2. However, the latter is conserved within each group, namely between all members of the SYT1 and SYT2 groups (see DISCUSSION).
Sixteen amino acids that are highly conserved between at least 50 of the 52 Sec7-domain proteins identified here are shown in Figure 4B and Table 4. These highly conserved (HC) sites are labeled a to p. Also shown are amino acids that were previously identified by mutagenesis to be important for the interaction of the Sec7 domain of H. sapiens ARNO/CYH2 with Arf in helices α6-α9 and for catalysis of exchange (Figure 4C) (Beraud-Dufour et al., 1998; Cherfils et al., 1998). Arf-contact sites, as identified by the Arf-Sec7 domain cocrystal structure derived from S. cerevisiae GEA2 (Goldberg, 1998) are shown in Figure 4, D and F (coordinates are derived from Renault et al., 2002). Most of the HC amino acids defined here (HC sites g to p) are within the region of Sec7-domain-Arf contact as delimited by the mutations and the cocrystal. One of these HC residues, site g (E654), is the catalytic glutamate that has been shown to play a critical role in the exchange reaction, by destabilizing Mg2+ and GDP from Arf1 (Beraud-Dufour et al., 1998). Only one of the GEFs, S. pombe SYT21, contains a change in this amino acid to aspartic acid (see DISCUSSION).
Table 4.
HC amino acids in the Sec7 domain in all Sec7-domain proteins
| HC Site | Gea2 Pos | Highly no. | Conserved aa | Region | Sc SYT1 | Sc SYT2 | Sp SYT12 | Sp SYT21 | Lp RalF | Rp Sec7 |
|---|---|---|---|---|---|---|---|---|---|---|
| a | 600 | 50 | (34 L, 8 F, 6 I, 2 M) | α3 | K | Y | ||||
| b | 615 | 51 | (43 L, 5 I, 2 V, 1 M) | α4 | ||||||
| c | 633 | 51 | (48 F, 2 L, 1 I) | 5-6 | P | |||||
| d | 641 | 50 | (44 A, 5 S, 1 T) | α6 | C | G | ||||
| e | 642 | 52 | (43 L, 8 I, 1 F) | α6 | ||||||
| f | 643 | 51 | (50 R, 1 Q) | α6 | F | |||||
| g* | 654 | 52 | (50 E, 1 D) | 6-7 | ||||||
| h | 662 | 52 | (19 M, 18 L, 10 I, 4 V) | α7 | ||||||
| i | 665 | 52 | (50 F, 1 L) | α7 | ||||||
| j | 666 | 50 | (28 S, 21 A, 1 T) | α7 | G | |||||
| k* | 696 | 50 | (49 D, 1 Q) | α8 | K | |||||
| l | 701 | 52 | (45 L, 5 I, 2 V) | α8 | ||||||
| m* | 706 | 52 | (38 I, 8 M, 5 L, 1 V) | α8 | ||||||
| n* | 707 | 52 | (32 M, 17 L, 3 I) | α8 | ||||||
| o | 708 | 52 | (50 L, 1 I, 1 V) | α8 | ||||||
| p | 726 | 50 | (41 F, 9 Y) | α9 | L | L |
Shown are amino acids that are conserved in at least 50 of the 52 sequences identified here and their position in the protein is shown with S. cerevisiae GEA2 coordinates. Region shows their position relative to the structural elements identified for S. cerevisiae GEA2 and H. sapiens CYH1 and 2 (Betz et al., 1998; Cherfils et al., 1998; Goldberg, 1998). Also shown are the few exceptions to the conservation found exclusively in the SYTs or the BACTs. The four HC residues that overlap with Arf-contact are indicated by *.
Four of the 16 HC residues (sites g, k, m, and n) are involved in binding directly to Arf, as determined by the cocrystal structure. Mutations in two of these sites, in addition to the catalytic Glu residue, have been shown to significantly decrease Arf exchange activity. However, there is an overlap of only four amino acids between the 16 HC amino acids identified here, and the 15 Arf-contact sites of the Sec7 domain identified by the cocrystal structure. The remaining 11 contact sites are conserved in at least 38 of the 52 sequences. This indicates that the majority of the amino acids in the Sec7 domain-Arf interface are not as highly conserved and that most of the HC amino acids (12/16) fulfill other roles. In agreement with the latter point, the HC sites are not all on one surface of the Sec7 domain (Figure 4H). These HC amino acids are very likely to be crucial for the structure of the Sec7 domain as a whole (see DISCUSSION).
Eight of the 16 highly conserved amino acids are conserved in all 52 Sec7-domain proteins identified here: HC site e in helix α6, site g in the 6–7 loop (the catalytic site), sites h to i in helix α7, and sites l to o in helix α8 (Table 4). The other eight highly conserved amino acids have one or two exceptions to the conservation as shown in Table 4. All the exceptions are in the fungi SYTs, or BACT. However, the SYTs and BACTs are probably functional Arf-GEFs, because such an activity was shown for one member of the SYT1 and BACT groups (Jones et al., 1999; Nagai et al., 2002). The definition of HC sites by conservation in 50/52 Sec7-domain proteins is not arbitrary. For amino acids that were conserved in <50 proteins, the exceptions are more randomly distributed in groups other than the SYTs and the BACTs (our unpublished data).
BFA Sensitivity of the Different Sec7-Domain Protein Groups
The secretion-inhibiting drug BFA acts as an uncompetitive inhibitor of the exchange reaction, binding to a normally short-lived Arf–GDP–Sec7 domain protein complex (Mansour et al., 1999; Peyroche et al., 1999). However, only some Sec7 domains are direct targets of BFA (Chardin et al., 1996; Jones et al., 1999; Moss and Vaughan, 2002). Previous studies have identified amino acids within the Sec7 domain that are responsible for conferring BFA sensitivity or resistance. Five residues were identified that converted a BFA sensitive Arf GEF into a BFA-resistant one, or vise versa, both in vitro and in vivo. In a BFA-sensitive GEF, these residues are YS-M-DM (Y703 S704-M707-D711 M721, S. cerevisiae Gea2p coordinates) in helix α8 and loop α8 to α9 (Figure 4, E and G). We suggest a consensus for BFA sensitivity of the Sec7 domain based on mutations in the Sec7 domain (Table 5A). The sensitivity consensus is: the middle M and either the N-terminal YS or the C-terminal DM (Table 5A).
Table 5.
BFA sensitivity as determined by alignment
| Sensitivity consensus
|
||||||
|---|---|---|---|---|---|---|
| A Group | Species | Name | YS | M | D-M | BFA phenotype (Reference) |
| CYH | Hs | CYH1 | FA | M | S-P | R (1) |
| CYH | Hs | mCYH1 | FA | M | D-M | S (2) |
| CYH | Hs | ARNO/CYH2 | FA | M | S-P | R (3) |
| CYH | Hs | mARNO/mCYH2 | YS | M | S-P | S (4) |
| GEA | Sc | GEA1 | YS | M | D-M | S (5) |
| GEA | Sc | mGEA1 | YS | L | D-M | R (4) |
| Sensitivity consensus
|
BFA phenotype
|
||||||
|---|---|---|---|---|---|---|---|
| B Group | Species | Name | YS | M | D-M | Obs (Ref) | Pre |
| CYH | Ce | CYH1 | YS | M | L-P | S | |
| CYH | Dm | CYH1 | FA | M | S-P | R | |
| CYH | Hs | CYH1 | FA | M | S-P | R (1) | R |
| CYH | Hs | ARNO/CYH2 | FA | M | S-P | R (3) | R |
| CYH | Hs | CYH3 | FA | M | S-P | R (6,7) | R |
| CYH | Hs | CYH4 | FS | M | S-P | R | |
| BIG/SEC7 | Sc | SEC7 | YS | M | D-M | S (8) | S |
| BIG/SEC7 | Sp | SEC71 | YS | L | D-M | R | |
| BIG/SEC7 | Sp | SEC72 | YS | M | D-M | S | |
| BIG/SEC7 | Nc | SEC7 | YS | M | D-M | S | |
| BIG/SEC7 | Ce | BIG1 | FS | M | D-M | S | |
| BIG/SEC7 | Dm | BIG1 | FS | M | D-M | S | |
| BIG/SEC7 | Hs | BIG1 | YS | M | D-M | S (9,10) | S |
| BIG/SEC7 | Hs | BIG2 | YS | M | D-M | S (11) | S |
| BIG/SEC7 | At | BIG1 | YS | M | D-M | S | |
| BIG/SEC7 | At | BIG2 | YS | M | D-M | S | |
| BIG/SEC7 | At | BIG3 | YS | L | D-M | R | |
| BIG/SEC7 | At | BIG4 | YS | M | D-M | S | |
| BIG/SEC7 | At | BIG5 | YA | M | D-M | S | |
| GBF/GEA | Sc | GEA1 | YS | M | D-M | S (5) | S |
| GBF/GEA | Sc | GEA2 | YS | M | D-M | S (5) | S |
| GBF/GEA | Sp | GEA1 | YS | M | D-M | S | |
| GBF/GEA | Nc | GEA1 | YA | M | D-M | S | |
| GBF/GEA | Ce | GBF1 | YA | M | D-M | S | |
| GBF/GEA | Dm | GBF1 | YA | M | D-M | S | |
| GBF/GEA | Hs | GBF1 | YA | M | D-M | R (12) | S |
| GBF/GEA | At | GBF1 | YS | L | D-M | R | |
| GBF/GEA | At | GBF2 | YS | M | D-M | S | |
| GBF/GEA | At | GBF3 | YS | M | D-M | S (13) | S |
| BRAG | Ce | BRAG | FA | M | D-M | S | |
| BRAG | Dm | BRAG | FA | M | D-M | S | |
| BRAG | Hs | BRAG1 | FA | L | D-M | R | |
| BRAG | Hs | BRAG2 | FA | L | D-M | R (14) | R |
| BRAG | Hs | BRAG3 | FA | L | D-M | R | |
| SYT1 | Sc | SYT1 | FS | M | D-M | S | |
| SYT1 | Sp | SYT11 | FS | I | D-M | R | |
| SYT1 | Sp | SYT12 | FS | M | D-M | S | |
| SYT1 | Nc | SYT1 | FS | I | D-M | R | |
| SYT2 | Sc | SYT2 | FS | I | D-F | R | |
| SYT2 | Sp | SYT21 | VK | L | D- | R | |
| SYT2 | Sp | SYT22 | FS | L | D-M | R | |
| SYT2 | Nc | SYT2 | YS | L | D-M | R | |
| EFA | Ce | EFA6 | CA | L | D-M | R | |
| EFA | Dm | EFA6 | CA | L | D-M | R | |
| EFA | Hs | EFA6A | CA | L | D-M | R (15) | R |
| EFA | Hs | EFA6B | CA | L | D-M | R | |
| EFA | Hs | EFA6C | CA | L | D-M | R | |
| EFA | Hs | EFA6D | CA | L | D-M | R | |
| FBX8 | Hs | FBX8 | YS | L | D-M | R | |
| FBX8 | Mm | FBX8 | YS | L | D-M | R | |
| BACT | BLp | RalF | FQ | M | D-M | S | |
| BACT | BRp | SEC7 | YQ | M | D-M | S | |
(A) Mutations in the Sec7 domain that change the BFA phenotype. Based on these mutations, we propose the BFA sensitivity consensus: YS-M-DM. (B) Observed (Obs) and predicted (Pre) BFA phenotypes for the 52 Sec7 domain proteins identified here. Groups and names are as in Table 2. Sensitivity Consensus shows comparison of aligned residues with the predicted sequences required for BFA sensitivity. These sequences are present in helix α8 and the following loop of the Sec7 domain as shown in Figure 4G. Bold letters indicate conformance to this sequence, whereas italic letters indicated divergence. The observed BFA phenotype column shows known sensitivity (S) or resistance (R) as determined in an in vitro exchange assay. The last column indicates the predicted BFA phenotype, based on this rule.
This consensus is based on the following findings. First, the importance of the middle M residue for BFA sensitivity of Gea2 and the Sec7-domain protein of the malaria parasite Plasmodium falciparum was shown in screens for BFA-resistant mutants (M was changed to L or I). Moreover, the M-to-L mutant of the Gea1p Sec7 domain was resistant to BFA in vitro (Peyroche et al., 1999; Baumgartner et al., 2001). These findings indicate that the middle M residue is important both in vivo and in vitro for BFA sensitivity. Second, the roles of the N-terminal YS and the C-terminal DM were shown in a number of studies with the BFA-resistant CYHs that normally have the sequence FA-M-SP (F190 A191-M194-S198 P208, ARNO/CYH2 coordinates). Mutating either the N-terminal FA to YS of H. sapiens ARNO/CYH2, or the C-terminal SP to DM of H. sapiens CYH1 converts these Sec7-domain proteins from BFA resistant to BFA sensitive (Peyroche et al., 1999; Sata et al., 1999). The strength of BFA binding to the ARNO/CYH2 Sec7 domain complex was determined using wild-type and mutant Sec7 domains. Binding of BFA to the wild-type ARNO/CYH2 Sec7 domain was undetectable, whereas each of the double mutants FA-to-YS and SP-to-DM exhibited similar and higher levels of BFA binding. The quadruple mutant bound twofold more BFA than either double mutant alone, and the dissociation rate of BFA from the complex was 10-fold slower for the quadruple mutant compared with each single mutant (Robineau et al., 2000).
Interestingly, the five amino acids of this consensus are positioned on one surface of the Sec7 domain, and three of them, Y703, M707, and D711, are also Arf contact sites (Figure 4, F and G). BFA is large enough to cover all three contact points on helix α8 (Suh et al., 2002). Although it is certainly possible that other residues of the Sec7 domain contribute to the BFA sensitivity or resistance of a given Arf GEF, the importance of these five residues is well documented. Hence, we used the above-mentioned consensus to predict the BFA sensitivity or resistance of the 52 Sec7-domain proteins identified here (Table 4B). It should be kept in mind that this analysis is a simplification of the situation, because there is a range of BFA sensitivities in any given assay, and “resistant” or “sensitive” represent points chosen near each end of the distribution.
Eleven of the 12 observed BFA phenotypes are as predicted by our consensus. However, in one case our prediction does not match the reported phenotype: H. sapiens GBF1 has YA-M-DM and is predicted to be BFA sensitive because it has the Y-M-DM BFA-sensitive residues, but it was reported to be resistant for GEF activity on Arf5 (Claude et al., 1999). One possible explanation for this discrepancy is that other amino acids are important for the BFA phenotype besides the consensus defined here. Alternatively, because BFA interacts not only with the Sec7 domain but also with Arf (Robineau et al., 2000), it is possible that BFA has specificity toward Arf1/3, but not toward Arf5. Testing the BFA sensitivity of the GBF1 GEF activity on Arf1/3 might help resolve this issue.
Our analysis predicts that the convention that members of the GEA/GBF and SEC7/BIG groups are BFA sensitive, whereas members of the CYH, EFA, and BRAG groups are BFA resistant, seems to be valid for human Sec7-domain proteins. This convention, however, breaks down in other organisms, where group membership does not seem to be a predictor for BFA sensitivity or resistance.
DISCUSSION
The phylogenetic analysis of the Sec7-domain protein family presented here identified 49 proteins in seven eukaryotic model systems spanning the fungi, plant, and animal kingdoms. Three additional sequences used in this study are the mouse homolog of H. sapiens FBX8, and two bacterial Sec7-domain proteins. These 52 sequences were used for generating multiple alignments and a phylogenetic tree. The tree and the alignments were in turn used for four purposes. First, we defined seven family groups and suggested a consistent set of names for family members, which should also help naming newly identified members in other organisms. Second, alignments of the Sec7 domain itself allowed us to identify elements that might be important for the Arf-GEF activity of the domain and its sensitivity to BFA. Third, alignments of entire proteins identified common as well as group-specific domains, which are likely to be important for aspects of the Sec7-protein family function other than Arf GEF activity. Fourth, the tree and the alignments form the basis for speculation about the evolution of the Sec7 domain family.
Sec7-Domain Family Groups
The phylogenetic tree and the multiple alignments of the 52 proteins formed the basis for defining the seven eukaryotic Sec7-domain protein groups. Three criteria were used for group definition: known phylogenetic relationships (e.g., all animal or fungi in a group are clustered together), strength of branches at the bases of groups, and similar structure within and outside the Sec7 domain. Two groups include members from fungi, animals and plants: BIG/SEC7 and GBF/GEA. The other groups are specific to fungi or to animals. FBX and BACT are defined as separate groups according to the above three criteria. These groups form the basis for consistent naming of the Sec7-domain protein family (Table 2).
We propose that all Sec7-domain proteins identified here possess an Arf GEF activity based on the following two lines of evidence. First, most of the Sec7 domain is highly conserved in all identified family members (Figure 4A), and this domain acts as an Arf GEF by itself. Second, at least one representative from each group, except for the SYT2 group, was shown to have such an activity. Group members that were shown to act as Arf GEFs are indicated in Table 2.
Members of the GBF/GEA and BIG/SEC7 groups, which act on class I Arfs, localize and function at the early and late Golgi, respectively (Franzusoff and Schekman, 1989; Peyroche et al., 2001; Spang et al., 2001; Kawamoto et al., 2002; Shinotsuka et al., 2002a,b; Zhao et al., 2002). EFA group members and their substrate class III Arf6 reside on the plasma membrane (PM), and function in PM ruffling (Franco et al., 1999; Derrien et al., 2002). BRAGs also act on class III Arf6, and at least one group member, BRAG2/GEP100, was suggested to localize to an endosomal compartment (Someya et al., 2001). The CYHs, which can act on class I and class III Arfs, are largely cytoplasmic in resting cells, but are recruited to the PM upon PI-3 kinase stimulation and have also been localized to the Golgi (Jackson and Casanova, 2000; Klarlund et al., 2000; Lee et al., 2000; Claing et al., 2001). The S. cerevisiae SYT1 and SYT2 are not required for cell viability (Jones et al., 1999; Dolinski et al., 2003), and their localization and function are still unknown. Although the cellular localization and function are conserved from yeast to human for the GBF/GEA and BIG/SEC7 groups, it does not necessarily mean they are conserved between all members of each group. For example, in the plant A. thaliana, which has representatives only from the GBF/GEA and BIG/SEC7 groups, one member of the GBF/GEA group, GBF3/GNOM, localizes to endosomes (Geldner et al., 2003), not the Golgi. Therefore, probably all members of each group have some, but not all, functions in common, those mediated by the group-specific domains.
Why are BIG/SEC7 and GBF/GEA conserved from yeast to plants and humans, whereas the other groups are not? One explanation is that at least some members of these two subfamilies serve an essential function in transport through the secretory pathway, which is essential to all eukaryotic cells. Members of the other groups function in endocytic pathways or in transport between the Golgi and endosomal/lysosomal compartments, which are more specialized in each organism. For example, certain animal cells can be stimulated to become highly mobile, a change that results in dramatic PM alterations that require Arf6 and its GEFs (Donaldson, 2003). Yeast cells, on the other hand, are nonmotile, but have an elaborate machinery to establish and maintain polarity during bud formation, a process that requires Arf3, and presumably its GEFs as well (Huang et al., 2003).
The Sec7 Domain and Arf GEF Activity
Alignments of the Sec7 domains of 52 proteins identified here provide insights regarding elements that are important for the Arf GEF activity of the domain. Breakpoints in the conserved alignment almost precisely correspond to known structural borders of the Sec7 domain. The correspondence between structural and alignment data suggests that the three-dimensional structure of the Sec7 domain of S. cerevisiae GEA2 is conserved not only with that of H. sapiens CYH1/2 (Betz et al., 1998; Cherfils et al., 1998; Goldberg, 1998) but also between all family members. It also implies that the α-helices have a positive selective value presumably due to conserved biological roles but that the loops (except for the catalytic glutamate) are merely spacers. Inspection of the core Sec7 domain identified a set of HC amino acids that are likely to have a crucial role in molecular shape integrity, interaction with Arf, Arf GEF activity, and/or interaction with other proteins. Most of the HC amino acids are not involved in Arf binding, as determined by previous structural and mutagenesis studies. Together, the breakpoints positioning and the HC amino acids suggest that conservation of the structure of the whole Sec7 domain is as important for its Arf GEF function as the Arf contact points. Such conservation probably reflects the importance of the flexibility of the Sec7-domain groove and the spatial arrangement necessary for the complex interactions with Arf during nucleotide exchange.
There are two possible cases where family members might have lost an Arf GEF activity. First, S. pombe SYT21 probably has a significantly reduced Arf GEF activity because it contains a change in the catalytic glutamate. Although a conserved change, E to D, the analogous mutation in H. sapiens ARNO/CHY2 resulted in 400 times lower exchange activity (Beraud-Dufour et al., 1998). This suggests that S. pombe SYT21 is either a weak Arf GEF or that this is a sequencing mistake (possible with a single nucleotide substitution). If the former is correct, its cellular role should be explored. Second, the Sec7 domain of A. thaliana BIG5 either uses an unconventional splice site, which is our favored explanation, or contains a 26-amino acid deletion that includes the catalytic glutamate. Such a deletion is expected to yield a protein that cannot act as an Arf GEF. It is not clear what might be the biological function of a Sec7-domain protein that has a significantly reduced Arf GEF activity or has lost it completely. One possibility is that it acts as a dominant inhibiting protein that sequesters Arf or other binding proteins and thereby hinders transport steps that are dependent on their function.
Although numerous studies have determined the in vitro substrate preference of various Sec7-domain proteins, little is known about substrate specificity of the Sec7-domain proteins in cells. An important question is whether substrate specificity observed in vitro is determined by the Sec7 domain itself or by other domains of these proteins. One study indicates that domains flanking the Sec7 domain are important for substrate specificity of CHY1 (Pacheco-Rodriguez et al., 1998). A multiple alignment of the Sec7 domains shows significant consensus in the α4-α10 helices and some diversity in the α2-α3 helices. These latter helices are conserved among most groups, but differ in SYT1, SYT2, and EFA (Figure 4A). This diversity cannot explain the in vitro specificity of EFA to class III Arfs, because BRAG and CYH can also act on class III Arfs, and they share sequence similarity in helices α2-α3 with the GBF and GEA groups, which act on class I Arfs (Table 1; see DISCUSSION under The Role of Common and Group-specific Domains). However, the Sec7 domain of the SYTs might be important for their substrate preference because in addition to the lack of conservation in helices α2-α3 with those of the other groups, four of the eight SYTs have nonconserved substitutions in the HC sites (Table 4). Helices α2 and α3 are not next to the Arf-contact points in the cocrystal structure. However, it is possible that they are important indirectly for Arf binding specificity. For example, these helices could be involved in the localization of the GEFs or in their interaction with other proteins, which in turn might be important for substrate specificity. Therefore, the Sec7 domain itself can bind Arf, act as its GEF, and might also play a role in substrate preference.
Sec7-domain proteins were shown to act as GEFs for Arfs that share >65% identity among themselves (yeast Arfs1/2 and mammalian classes I-III) (Table 1). Most studies do not address the question of substrate specificity outside the Arf group, namely, Arls, even in vitro, although the CYHs do not seem to act on Arl1–3 (Chardin et al., 1996; Pacheco-Rodriguez et al., 1998). We suggest that Sec7-domain proteins probably cannot act on Arls that share only 35% similarity with Arfs. This suggestion stems from the fact that a distant relative of the Arfs, Sar1, which shares 35% identity with the Arfs, has its own GEF, Sec12 (Barlowe and Schekman, 1993). Sec12 has no sequence similarity to the Sec7 domain even at e <10-2. Therefore, the limit of the substrate recognition of the Sec7-domain family is probably ≥35% identity to the Arfs. Further in vitro and in vivo analyses are required to address the issue of substrate preference of Sec7 domain proteins toward Arls that share 35–65% identity with Arfs. Subsequent structure–function analyses would determine the role played by specific Sec7 subdomains in this characteristic.
BFA Sensitivity
BFA binds both to the Sec7 domain and to Arf and can inhibit the GEF activity of some Sec7-domain proteins on class I Arfs. Multiple alignments of the Sec7 domain was used to predict the BFA sensitivity of members of the family. We used information regarding three Sec7 domain mutations that change the BFA phenotype to define a BFA-sensitivity consensus and to predict the BFA phenotypes of all family members identified here. With the caveat of Arf specificity, it seems that group membership cannot be a predictor for BFA sensitivity. This will be important to keep in mind as new Arf GEFs are identified.
Although the BFA sensitivity phenotype has been used for research purposes, it is not clear whether it also has a biological significance. BFA is a natural fungal drug, probably produced to inhibit certain Arf GEFs regulating key transport steps. If true, coevolution of BFA and the Sec7 domain family could determine the current BFA sensitivity phenotype of family members. Because the C-terminal region containing the BFA sensitivity/resistance residues is ideally placed to play a regulatory role in GEF activity, it will be interesting to determine whether regulatory proteins or lipids bind to this region of Sec7 domain proteins.
The Role of Common and Group-specific Domains
The common and group-specific domains of the Sec7 domain proteins are probably important for interaction with partners other than Arfs. These interactions might determine specific membrane localization, substrate specificity, and upstream regulation of family members. Group-specific domains suggest similar interactions, and therefore function, for all members of the group. Determining the cellular function and identification of interactors of at least some members of each Sec7 domain group will allow further structure–function analysis to elucidate the roles of the various domains identified here. Although the precise functions of most of the cross-group domains flanking the Sec7 domain of the Arf GEFs are unknown, clues to function for certain domains are beginning to emerge.
One domain common to five of the seven groups is a PH domain, although there is no significant sequence similarity between the PH domains of the five groups. PH domains are involved in binding phosphoinositides (PIs). Therefore, one possible role of the domain in all Sec7 domain proteins that contain it is specific membrane attachment of the Sec7 domain proteins, as has been demonstrated for certain members of the CYH and EFA6 groups. The lack of sequence similarity between the PH domains of the Sec7 domain groups probably reflects binding specificity to various PIs and thereby to different cellular membranes. Interestingly, the presence of a PH domain distinguishes CYH, EFA, and BRAG, which act on class III Arfs, from BIG and GBF, which do not (Table 1). Therefore, it is possible that the PH domain determines the specificity of the Sec7 domain proteins toward class III Arfs indirectly. Because PH domains interact with PIs (Lemmon and Ferguson, 2000), substrate specificity can be mediated through the effect of PIs on membrane localization of the GEFs, to areas where Arf6 resides.
The PI binding specificity of the PH domains and the cellular localization were determined for members of the EFA and CYH groups. Specifically, the PH domains of EFA6A and B probably interact with phosphatidylinositol bisphosphate and determine their colocalization with Arf6 on actin-rich membrane ruffles and microvilli-like projections on the cell surface (Derrien et al., 2002). Deletion of the PH domain prevented membrane localization of EFA6A (Franco et al., 1999), and isolated PH domain of EFA6A localizes to the same membranes as does the whole protein (Derrien et al., 2002). The PH domains of CYHs interact with phosphatidylinositol bisphosphate and phosphatidylinositol trisphosphate, and mediate their recruitment to the PM and possibly the Golgi (Jackson and Casanova, 2000; Klarlund et al., 2000; Lee et al., 2000; Claing et al., 2001).
We cannot make predictions regarding the PI binding specificity and the localization of the BRAG, SYT1, and SYT2 groups based on the sequence of their putative PH domains for the following reason. The best hit of any of the PH domain of any SYT1, SYT2, or BRAG members to the Conserved Domain Database is Sp SYT21 (at 3e-8) (Marchler-Bauer et al., 2003). This sequence is weakly homologous to the PH domain of Hs EFA6A and Hs betaIII spectrin (at e = 0.002 for both). These two proteins are known to localize to different compartments: plasma membrane (Derrien et al., 2002) and Golgi (Stankewich et al., 1998), respectively. Therefore, one cannot draw any credible conclusion about the membrane localization of Sp SYT21. The PH domains of all other members of the SYT1, SYT2, and BRAG groups have an even poorer homology to the PH domain consensus as determined by Conserved Domain Database and SMART. Importantly, even a few modified amino acids can change the PI binding specificity of PH domains and thereby their intracellular localization (Klarlund et al., 2000). Therefore, the PI binding specificity and localization of the PH domains of BRAG, SYT1, and SYT2 need to be defined empirically, as was done for the known PH domains.
The coiled-coil domain at the N-terminal region of the CYH proteins seems to function as a protein–protein interaction domain in a number of the CYHs. Five partners have been identified for various members of the CYH family, all of which interact with the CC domain of these GEFs. These cytohesin partners are the presynaptic Munc13-1 (Neeb et al., 1999), GRP1-associated scaffold protein (Nevrivy et al., 2000), GRP1-binding partner (GRSP1) (Klarlund et al., 2001), the scaffolding protein CASP/Cybr (cytohesin binder and regulator) (Mansour et al., 2002; Tang et al., 2002), and interaction protein for cytohesin exchange factors 1 (IPCEF1) (Venkateswarlu, 2003). Two partners, GRSP1 and CASP/Cybr, have a coiled coil domain themselves, which is responsible for interaction with the CYH family members (Klarlund et al., 2001; Mansour et al., 2002; Tang et al., 2002). The functions of these partners are not known, although two, CASP/Cybr and IPCEF1, have been shown to stimulate Arf GEF activity (Tang et al., 2002; Venkateswarlu, 2003).
Members of the GBF/GEA and BIG/SEC7 groups share three common domains, B/Gα-γ, suggesting that these two families have at least partial overlapping interactions and/or cellular localization. Interacting proteins have been identified for all three B/Gα-γ domains. The N-terminal 331 amino acid region of BIG1 has been shown to interact with FKBP13 (Padilla et al., 2003). This portion of BIG1 contains the B/Gαregion that shares homology with members of both BIG/SEC7 and GBF/GEA subfamilies. B/Gα is also present in the first 246-aa region of Arabidopsis GNOM/GBF3, which has been dubbed the “DBC” domain, for dimerization and cyclophilin binding (Grebe et al., 2000). This region interacts with cyclophilin 5 and was also demonstrated to be essential for interaction between GNOM/GBF3 monomers. Although the functional significance of these two interactions is not understood, it is interesting that both FKBP13 and cyclophilin are targets of immunosuppressive drugs FK506 and cyclosporin A, respectively. A likely hypothesis is that these interactions are involved in regulating GEF function.
Another possible regulatory interaction has been uncovered for BIG2, which binds the regulatory subunit of protein kinase A (PKA). Three potential PKA binding motifs were identified in the N-terminal region of BIG2 (Li et al., 2003). Two of these motifs are in poorly conserved regions with homologies only among animal BIG proteins, whereas the third is present in region B/Gβ, which is highly conserved between all GBF/GEA and BIG/SEC7 family members. It will be interesting to determine which of these potential interaction sites act as PKA binding sites in cells and to investigate PKA regulation of Arf GEF activity. The third region of homology common to both GBF/GEA and BIG/SEC7 subfamilies, B/Gγ, is involved in the binding of Gea2p to the transmembrane protein Gmh1p (Chantalat et al., 2003). Gmh1p has homologues in all eukaryotic cells examined, and it will be interesting to determine whether other GBF/GEA and BIG/SEC7 proteins interact with Gmh1p family members, and whether they serve to recruit these GEF proteins to membranes. A proline-rich region at the C-terminus of GBF1 was found to interact with the vesicle tethering factor p115. This interaction is likely unique to higher eukaryotes, as the proline-rich C-terminal region is present in only animal GBF1 proteins (Garcia-Mata et al., 2003).
Structural analysis of the FBX group outside the Sec7 domain suggests a dominant negative role in protein transport. The FBX group has an F-Box domain, which was shown to interact with ubiquitin ligases that mark proteins for degradation (Kipreos and Pagano, 2000). A protein that can interact through one domain with ubiquitin ligase, and through another domain with Arfs, might have a role in specific degradation of Arfs, thereby blocking Arf-dependent protein transport steps.
At least one of the bacterial proteins has a functional Arf GEF activity (Nagai et al., 2002), but they both lack the other domains required for proper localization and function of the eukaryotic Arf-GEFs. This suggests that the bacterial Sec7 domain proteins allow the parasite to alter the host protein transport pathway. Indeed, the BACT RalF protein is required for the localization of Arf to phagosomes containing L. pneumophila. Presumably, the bacterium then uses Arf to redirect host protein traffic to create a vacuole that supports it own replication (Nagai et al., 2002). This is an exploitation of the normal usage of Sec7 domain proteins. The role of the BACTαspecific domain in this process is not known, but it might be important for the translocation of RalF through the phagosomal membrane, which was shown to be dependent on the bacterial Dot/Icm transporter (Nagai et al., 2002).
Evolution of the Eukaryotic Sec7 Domain Family
The topology of the phylogenetic tree within the groups is consistent with accepted notions of evolution (Figure 1). For example, in all five groups that include animals, the animals are always arranged in order of worms (Ce), flies (Dm), and mammals (Hs). The evolution of the groups themselves is ambiguous partly because the tree is unrooted, which among other things, obscures the minimal number of Sec7-domain proteins in the progenitor species. However, rooting the tree requires the identification of another group of proteins that have homology to the Sec7-domain family but clearly diverged from the family before the divergence of the eukaryotic groups. We suggest that the bacterial Sec7-domain proteins do not meet this requirement (see below). Therefore, information about genomes between archaea, which do not have their own Sec7-domain proteins, and fungi, which do, might provide the missing information.
Based on the phylogenetic tree (Figure 1) and the multiple alignments (Figures 3 and 4), we propose a scheme for the evolution of the seven Sec7-domain groups (Figure 5). First, the two groups that contain members in all the seven model systems, BIG/SEC7 and GBF/GEA, probably diverged from a common progenitor, because they share sequence similarity boxes outside the Sec7 domain. This event probably happened before the divergence of fungi, animals, and plants, because they all contain both groups. Second, an EFA/SYT progenitor was probably also present before the divergence of fungi and animals. This idea is based on the fact that the fungal SYTs and the animal EFAs are closer to each other than to the other groups (Figure 1A). Although we did not combine them into one group, due to lack of similarity outside the Sec7 domain, the phylogenetic tree suggests that they are related. They all contain a PH domain, and they are the only groups in which helix α2 of the Sec7 domain is not conserved with those of the other groups. The divergence of the SYT1, SYT2, and EFA groups, and the apparent elimination of this group from plants, probably occurred when fungi, animals, and plants diverged.
Figure 5.
Model for the evolution of the Sec7-domain protein family. Based on the phylogenetic analysis of the eukaryotic Sec7-domain proteins, we propose a model for the evolution of the seven groups in fungi, animals, and plants, from a single Sec7 domain progenitor (see text for DISCUSSION). The PH domain is shown as a gray circle.
The two animal-specific groups, CYH and BRAG, probably evolved from one of the other Sec7-domain groups. This could happen by deletion of all sequences but the Sec7 domain and recombination with other genes. CYH probably diverged from BIG/SEC7 based on its branching close to the BIG/SEC7 branch, with 60% bootstrap support. The mammalian FBX group probably also evolved by a rearrangement that yielded a protein with a Sec7 and F-box domains. Filling in the vast phylogenetic distances between the seven model organisms with other sequenced genomes should help to resolve the relationships between the different groups of the tree and their evolution. For example, to understand the origin of the FBX group, sequences between D. melanogaster and H. sapiens are needed.
Evolution of the Bacterial Sec7-Domain Proteins
We propose that the evolution of the bacterial Sec7-domain proteins of L. pneumophila and R. prowazekii occurred by two horizontal transfer events; the first from eukaryotes to bacteria and the second between the two bacteria. The suggestion that the two bacteria acquired the Sec7-domain proteins through horizontal transfer from eukaryotes is based on the fact that no bacterial genome currently present in databases, except for two eukaryotic parasites, carries a Sec7 domain protein. The second horizontal transfer from one bacterium to the other is also supported by two facts. The two bacterial Sec7 proteins share similarity both in the N-terminal Sec7 domain (Figure 4A), and in the C-terminal bacterial-specific domain (Figure 3). The uniqueness of this structural similarity suggests that the horizontal transfer from a eukaryotic host was not independent. In addition, 23 other bacteria in the Proteobacteria, the phylum that includes Legionella and Rickettsia, do not contain a Sec7-domain protein in their genome, arguing against the alternative possibility that the horizontal transfer from eukaryotes occurred before the divergence of the two bacteria. However, it is currently unclear whether the horizontal transfer from eukaryotes to bacteria occurred before or after the divergence of the eukaryotic groups, although the absence in other close members of the bacteria argues against the transfer being ancient. Therefore, the bacterial sequences were not used for rooting the phylogenetic tree.
Perspectives
The phylogenetic analysis presented here allowed us to propose testable hypotheses regarding the structure and function relationship in Sec7-domain proteins. It also formed the basis for speculations regarding the evolution of the Sec7-domain family discussed above. One idea proposed here is that structural elements, besides the Arf-contact sites, are important for the GEF activity of the Sec7 domain. This idea is based on the finding that helices α4-α10 of the domain show excellent consensus, and on the identification of HC amino acids that do not seem to make a contact with Arf, as previously defined by structural and mutagenesis studies. This idea can be tested by studying the effect of mutations in the HC sites on Arf binding and GEF activity.
Two other questions regarding the Arf GEF activity of the Sec7-domain family that need to be addressed are the specificity of the various Sec7-domain proteins toward their Arf substrates and the structural basis of this specificity. Determination of substrate preference, both in vitro and in vivo, is a prerequisite for subsequent structure–function analysis aimed to identify domains important for this preference. Substrate specificity in vivo probably also involves domains outside the Sec7 domain, which are important for intracellular localization of the proteins.
The common and group-specific domains defined here provide a basis for analysis of functions of Sec7 domain proteins beyond their ARF GEF activity. One goal is to understand the roles of these domains in the specific cellular localization or protein interaction and to determine whether they are conserved in the different model systems. The cellular role of most of the Sec7-domain proteins is still not clear, and loss-of-function mutations or inhibitory RNA experiments should provide such information. Subsequent structure–function analysis of specific domains in more than one model system will help determining the roles of specific domains and whether the roles are conserved. Different in vivo roles have been proposed for the BIG/SEC7 and GBF/GEA groups, and these roles are conserved between S. cerevisiae and mammals. Because of structure similarity between these two groups, domain swapping can help to define the roles of the B/G common and group-specific domains. In addition, information regarding binding partners for specific domains will also help to resolve the roles played by the different common and group-specific domains. Finally, the idea that some members of the Sec7-domain family may act in a dominant negative manner (e.g., FBX8, S. pombe SYT21, and A. thaliana BIG5) should also be explored.
To gain better insight regarding the evolution of the Sec7-domain family, both within and among the six major groups, phylogenetic analysis of additional proteomes is needed. For example, more plant proteomes should help confirm that plants do not have SYTs, EFA, BRAG, or CYH groups and resolve the relationship of its GBFs to the other members of the GBF group, more proteomes between fungi and C. elegans, and between D. melanogaster and H. sapiens, should help resolve evolution within the different groups. Finally, one of the most difficult phylogenetic problems remaining is the lack of an appropriate outgroup; hypotheses about evolution among the major Sec7 domain groups will be greatly influenced by the placement of the root of the tree. Although no likely candidate has yet been found, the analysis of more proteomes might identify early diverging eukaryotes, whose origin predates the divergence of the major Sec7 groups. Sec7 sequences from such basal eukaryotes might provide several benefits, including clarification of the origin of the Sec7 family, rooting the Sec7 tree, and elucidation of the relationships between the Sec7-family groups.
Acknowledgments
Special thanks to E. Kaufman for kind support and advice. This research was supported by grant GM-45444 from National Institutes of Health (to N.S.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–06–0443. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–06–0443.
Abbreviations used: BFA, brefeldin A; GEF, guanine nucleotide exchange factor; CYH, cytohesin; HC, highly conserved; PH, plekstrin homology; PI, phosphoinositide.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny, B., Beraud-Dufour, S., Chardin, P., and Chabre, M. (1997). N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 36, 4675-4684. [DOI] [PubMed] [Google Scholar]
- Baldauf, S.L., Palmer, J.D., and Doolittle, W.F. (1996). The root of the universal tree and the origin of eukaryotes based on elongation factor phylogeny. Proc. Natl. Acad. Sci. USA 93, 7749-7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C., and Schekman, R. (1993). SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365, 347-349. [DOI] [PubMed] [Google Scholar]
- Baumgartner, F., Wiek, S., Paprotka, K., Zauner, S., and Lingelbach, K. (2001). A point mutation in an unusual Sec7 domain is linked to brefeldin A resistance in a Plasmodium falciparum line generated by drug selection. Mol. Microbiol. 41, 1151-1158. [DOI] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., and Wheeler, D.L. (2003). GenBank. Nucleic Acids Res. 31, 23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraud-Dufour, S., Paris, S., Chabre, M., and Antonny, B. (1999). Dual interaction of ADP ribosylation factor 1 with Sec7 domain and with lipid membranes during catalysis of guanine nucleotide exchange. J. Biol. Chem. 274, 37629-37636. [DOI] [PubMed] [Google Scholar]
- Beraud-Dufour, S., Robineau, S., Chardin, P., Paris, S., Chabre, M., Cherfils, J., and Antonny, B. (1998). A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J. 17, 3651-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz, S.F., Schnuchel, A., Wang, H., Olejniczak, E.T., Meadows, R.P., Lipsky, B.P., Harris, E.A., Staunton, D.E., and Fesik, S.W. (1998). Solution structure of the cytohesin-1 (B2–1) Sec7 domain and its interaction with the GTPase ADP ribosylation factor 1. Proc. Natl. Acad. Sci. USA 95, 7909-7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg, N., Baraldi, E., Nilges, M., and Saraste, M. (1999). The PH super-fold: a structural scaffold for multiple functions. Trends Biochem. Sci. 24, 441-445. [DOI] [PubMed] [Google Scholar]
- Bonifacino, J.S., and Jackson, C.L. (2003). Endosome-specific localization and function of the ARF activator GNOM. Cell 112, 141-142. [DOI] [PubMed] [Google Scholar]
- Brown, J.R., and Doolittle, W.F. (1995). Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc. Natl. Acad. Sci. USA 92, 2441-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavenagh, M.M., Whitney, J.A., Carroll, K., Zhang, C., Boman, A.L., Rosenwald, A.G., Mellman, I., and Kahn, R.A. (1996). Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J. Biol. Chem. 271, 21767-21774. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012-2018. [DOI] [PubMed] [Google Scholar]
- Celniker, S.E., et al. (2002). Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3, RESEARCH0079.1-14. [DOI] [PMC free article] [PubMed]
- Chantalat, S., Courbeyrette, R., Senic-Matuglia, F., Jackson, C.L., Goud, B., and Peyroche, A. (2003). A novel Golgi membrane protein is a partner of the ARF exchange factors Gea1p and Gea2p. Mol. Biol. Cell 14, 2357-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin, P., Paris, S., Antonny, B., Robineau, S., Beraud-Dufour, S., Jackson, C.L., and Chabre, M. (1996). A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature 384, 481-484. [DOI] [PubMed] [Google Scholar]
- Cherfils, J., Menetrey, J., Mathieu, M., Le Bras, G., Robineau, S., Beraud-Dufour, S., Antonny, B., and Chardin, P. (1998). Structure of the Sec7 domain of the Arf exchange factor ARNO. Nature 392, 101-105. [DOI] [PubMed] [Google Scholar]
- Claing, A., Chen, W., Miller, W.E., Vitale, N., Moss, J., Premont, R.T., and Lefkowitz, R.J. (2001). beta-Arrestin-mediated ADP-ribosylation factor 6 activation and beta 2-adrenergic receptor endocytosis. J. Biol. Chem. 276, 42509-42513. [DOI] [PubMed] [Google Scholar]
- Claude, A., Zhao, B.P., Kuziemsky, C.E., Dahan, S., Berger, S.J., Yan, J.P., Armold, A.D., Sullivan, E.M., and Melancon, P. (1999). GBF 1, A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J. Cell Biol. 146, 71-84. [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., Li, G., Colombo, M.I., and Stahl, P.D. (1995). A regulatory role for ARF6 in receptor-mediated endocytosis. Science 267, 1175-1178. [DOI] [PubMed] [Google Scholar]
- Dascher, C., and Balch, W.E. (1994). Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J. Biol. Chem. 269, 1437-1448. [PubMed] [Google Scholar]
- Derrien, V., Couillault, C., Franco, M., Martineau, S., Montcourrier, P., Houlgatte, R., and Chavrier, P. (2002). A conserved C-terminal domain of EFA6-family ARF6-guanine nucleotide exchange factors induces lengthening of microvilli-like membrane protrusions. J. Cell Sci. 115, 2867-2879. [DOI] [PubMed] [Google Scholar]
- Dolinski, K., et al. (2003). Saccharomyces Genome Database. http://www.yeastgenome.org/ on 5/1.
- Donaldson, J.G. (2003). Multiple roles for Arf 6, sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278, 41573-41576. [DOI] [PubMed] [Google Scholar]
- Donaldson, J.G., Finazzi, D., and Klausner, R.D. (1992). Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature 360, 350-352. [DOI] [PubMed] [Google Scholar]
- Donaldson, J.G., and Jackson, C.L. (2000). Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 12, 475-482. [DOI] [PubMed] [Google Scholar]
- Douglas, S., et al. (2001). The highly reduced genome of an enslaved algal nucleus. Nature 410, 1091-1096. [DOI] [PubMed] [Google Scholar]
- Franco, M., Boretto, J., Robineau, S., Monier, S., Goud, B., Chardin, P., and Chavrier, P. (1998). ARNO3, a Sec7-domain guanine nucleotide exchange factor for ADP ribosylation factor 1, is involved in the control of Golgi structure and function. Proc. Natl. Acad. Sci. USA 95, 9926-9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco, M., Peters, P.J., Boretto, J., van Donselaar, E., Neri, A., D'Souza-Schorey, C., and Chavrier, P. (1999). EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 18, 1480-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, S., Upender, S., Hansen, S.H., and Casanova, J.E. (1998). ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J. Biol. Chem. 273, 23-27. [DOI] [PubMed] [Google Scholar]
- Franzusoff, A., and Schekman, R. (1989). Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 8, 2695-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan, J.E., et al. (2003). The genome sequence of the filamentous fungus Neurospora crassa. Nature 422, 859-868. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata, R., Szul, T., Alvarez, C., and Sztul, E. (2003). ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol. Biol. Cell 14, 2250-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschet, J., and Hsu, V.W. (1999). Distribution of ARF6 between membrane and cytosol is regulated by its GTPase cycle. J. Biol. Chem. 274, 20040-20045. [DOI] [PubMed] [Google Scholar]
- Gaynor, E.C., Chen, C.Y., Emr, S.D., and Graham, T.R. (1998). ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol. Biol. Cell 9, 653-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N., et al. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219-230. [DOI] [PubMed] [Google Scholar]
- Goldberg, J. (1998). Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell 95, 237-248. [DOI] [PubMed] [Google Scholar]
- Grebe, M., Gadea, J., Steinmann, T., Kientz, M., Rahfeld, J.U., Salchert, K., Koncz, C., and Jurgens, G. (2000). A conserved domain of the arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12, 343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms, J.B., and Rothman, J.E. (1992). Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature 360, 352-354. [DOI] [PubMed] [Google Scholar]
- Hodges, P.E., Carrico, P.M., Hogan, J.D., O'Neill, K.E., Owen, J.J., Mangan, M., Davis, B.P., Brooks, J.E., and Garrels, J.I. (2002). Annotating the human proteome: the Human Proteome Survey Database (HumanPSD) and an indepth target database for G protein-coupled receptors (GPCR-PD) from Incyte Genomics. Nucleic Acids Res. 30, 137-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges, P.E., McKee, A.H., Davis, B.P., Payne, W.E., and Garrels, J.I. (1999). The Yeast Proteome Database (YPD): a model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 27, 69-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.F., Liu, Y.W., Tung, L., Lin, C.H., and Lee, F.J. (2003). Role for Arf3p in development of polarity, but not endocytosis, in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 3834-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabe, N., Kuma, K., Hasegawa, M., Osawa, S., and Miyata, T. (1989). Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl. Acad. Sci. USA 86, 9355-9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C.L., and Casanova, J.E. (2000). Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10, 60-67. [DOI] [PubMed] [Google Scholar]
- Jones, S., Jedd, G., Kahn, R.A., Franzusoff, A., Bartolini, F., and Segev, N. (1999). Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics 152, 1543-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, R.A., Clark, J., Rulka, C., Stearns, T., Zhang, C.J., Randazzo, P.A., Terui, T., and Cavenagh, M. (1995). Mutational analysis of Saccharomyces cerevisiae ARF1. J. Biol. Chem. 270, 143-150. [DOI] [PubMed] [Google Scholar]
- Kawamoto, K., Yoshida, Y., Tamaki, H., Torii, S., Shinotsuka, C., Yamashina, S., and Nakayama, K. (2002). GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic 3, 483-495. [DOI] [PubMed] [Google Scholar]
- Kipreos, E.T., and Pagano, M. (2000). The F-box protein family. Genome Biol. REVIEWS3002.1-7. [DOI] [PMC free article] [PubMed]
- Klarlund, J.K., Guilherme, A., Holik, J.J., Virbasius, J.V., Chawla, A., and Czech, M.P. (1997). Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science 275, 1927-1930. [DOI] [PubMed] [Google Scholar]
- Klarlund, J.K., Holik, J., Chawla, A., Park, J.G., Buxton, J., and Czech, M.P. (2001). Signaling complexes of the FERM domain-containing protein GRSP1 bound to ARF exchange factor GRP1. J. Biol. Chem. 276, 40065-40070. [DOI] [PubMed] [Google Scholar]
- Klarlund, J.K., Rameh, L.E., Cantley, L.C., Buxton, J.M., Holik, J.J., Sakelis, C., Patki, V., Corvera, S., and Czech, M.P. (1998). Regulation of GRP1-catalyzed ADP ribosylation factor guanine nucleotide exchange by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273, 1859-1862. [DOI] [PubMed] [Google Scholar]
- Klarlund, J.K., Tsiaras, W., Holik, J.J., Chawla, A., and Czech, M.P. (2000). Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J. Biol. Chem. 275, 32816-32821. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- Lee, S.Y., Mansour, M., and Pohajdak, B. (2000). B2–1, a Sec7- and pleckstrin homology domain-containing protein, localizes to the Golgi complex. Exp. Cell Res. 256, 515-521. [DOI] [PubMed] [Google Scholar]
- Lemmon, M.A., and Ferguson, K.M. (2000). Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350 Pt 1, 1-18. [PMC free article] [PubMed] [Google Scholar]
- Lemmon, M.A., and Ferguson, K.M. (2001). Molecular determinants in pleckstrin homology domains that allow specific recognition of phosphoinositides. Biochem. Soc. Trans. 29, 377-384. [DOI] [PubMed] [Google Scholar]
- Letunic, I., et al. (2002). Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30, 242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Wachtel, C., Miriami, E., Yahalom, G., Friedlander, G., Sharon, G., Sperling, R., and Sperling, J. (2002). Stop codons affect 5′ splice site selection by surveillance of splicing. Proc. Natl. Acad. Sci. USA 99, 5277-5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Adamik, R., Pacheco-Rodriguez, G., Moss, J., and Vaughan, M. (2003). Protein kinase A-anchoring (AKAP) domains in brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2). Proc. Natl. Acad. Sci. USA 100, 1627-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia, E., Chabre, M., and Franco, M. (2001). Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6. J. Biol. Chem. 276, 24925-24930. [DOI] [PubMed] [Google Scholar]
- Mansour, M., Lee, S.Y., and Pohajdak, B. (2002). The N-terminal coiled coil domain of the cytohesin/ARNO family of guanine nucleotide exchange factors interacts with the scaffolding protein CASP. J. Biol. Chem. 277, 32302-32309. [DOI] [PubMed] [Google Scholar]
- Mansour, S.J., Skaug, J., Zhao, X.H., Giordano, J., Scherer, S.W., and Melancon, P. (1999). p200 ARF-GEP 1, a Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc. Natl. Acad. Sci. USA 96, 7968-7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer, A., et al. (2003). CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31, 383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacci, E., Tsai, S.C., Adamik, R., Moss, J., and Vaughan, M. (1997). Cytohesin-1, a cytosolic guanine nucleotide-exchange protein for ADP-ribosylation factor. Proc. Natl. Acad. Sci. USA 94, 1745-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman, I., and Warren, G. (2000). The road taken: past and future foundations of membrane traffic. Cell 100, 99-112. [DOI] [PubMed] [Google Scholar]
- Morinaga, N., Adamik, R., Moss, J., and Vaughan, M. (1999). Brefeldin A inhibited activity of the sec7 domain of p200, a mammalian guanine nucleotide-exchange protein for ADP-ribosylation factors. J. Biol. Chem. 274, 17417-17423. [DOI] [PubMed] [Google Scholar]
- Morinaga, N., Moss, J., and Vaughan, M. (1997). Cloning and expression of a cDNA encoding a bovine brain brefeldin A-sensitive guanine nucleotide-exchange protein for ADP-ribosylation factor. Proc. Natl. Acad. Sci. USA 94, 12926-12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, J., and Vaughan, M. (1998). Molecules in the ARF orbit. J. Biol. Chem. 273, 21431-21434. [DOI] [PubMed] [Google Scholar]
- Moss, J., and Vaughan, M. (2002). Cytohesin-1 in 2001. Arch. Biochem. Biophys. 397, 156-161. [DOI] [PubMed] [Google Scholar]
- Nagai, H., Kagan, J.C., Zhu, X., Kahn, R.A., and Roy, C.R. (2002). A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679-682. [DOI] [PubMed] [Google Scholar]
- Neeb, A., Koch, H., Schurmann, A., and Brose, N. (1999). Direct interaction between the ARF-specific guanine nucleotide exchange factor msec7–1 and presynaptic Munc13-1.Eur. J. Cell Biol. 78, 533-538. [DOI] [PubMed] [Google Scholar]
- Nevrivy, D.J., Peterson, V.J., Avram, D., Ishmael, J.E., Hansen, S.G., Dowell, P., Hruby, D.E., Dawson, M.I., and Leid, M. (2000). Interaction of GRASP, a protein encoded by a novel retinoic acid-induced gene, with members of the cytohesin family of guanine nucleotide exchange factors. J. Biol. Chem. 275, 16827-16836. [DOI] [PubMed] [Google Scholar]
- Ogasawara, M., Kim, S.C., Adamik, R., Togawa, A., Ferrans, V.J., Takeda, K., Kirby, M., Moss, J., and Vaughan, M. (2000). Similarities in function and gene structure of cytohesin-4 and cytohesin-1, guanine nucleotide-exchange proteins for ADP-ribosylation factors. J. Biol. Chem. 275, 3221-3230. [DOI] [PubMed] [Google Scholar]
- Pacheco-Rodriguez, G., Meacci, E., Vitale, N., Moss, J., and Vaughan, M. (1998). Guanine nucleotide exchange on ADP-ribosylation factors catalyzed by cytohesin-1 and its Sec7 domain. J. Biol. Chem. 273, 26543-26548. [DOI] [PubMed] [Google Scholar]
- Padilla, P.I., Chang, M.J., Pacheco-Rodriguez, G., Adamik, R., Moss, J., and Vaughan, M. (2003). Interaction of FK506-binding protein 13 with brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1): effects of FK506. Proc. Natl. Acad. Sci. USA 100, 2322-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R.D. (1996). TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357-358. [DOI] [PubMed] [Google Scholar]
- Pasqualato, S., Renault, L., and Cherfils, J. (2002). Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for `front-back'communication. EMBO Rep. 3, 1035-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, P.J., Hsu, V.W., Ooi, C.E., Finazzi, D., Teal, S.B., Oorschot, V., Donaldson, J.G., and Klausner, R.D. (1995). Overexpression of wild-type and mutant ARF1 and ARF 6, distinct perturbations of nonoverlapping membrane compartments. J. Cell Biol. 128, 1003-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche, A., Antonny, B., Robineau, S., Acker, J., Cherfils, J., and Jackson, C.L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3, 275-285. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Courbeyrette, R., Rambourg, A., and Jackson, C.L. (2001). The ARF exchange factors Gea1p and Gea2p regulate Golgi structure and function in yeast. J. Cell Sci. 114, 2241-2253. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Paris, S., and Jackson, C.L. (1996). Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 384, 479-481. [DOI] [PubMed] [Google Scholar]
- Pruess, M., et al. (2003). The Proteome Analysis database: a tool for the in silico analysis of whole proteomes. Nucleic Acids Res. 31, 414-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna, H., and Donaldson, J.G. (1997). ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 139, 49-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo, P.A., Nie, Z., Miura, K., and Hsu, V.W. (2000). Molecular aspects of the cellular activities of ADP-ribosylation factors. Sci. STKE 2000, RE1. [DOI] [PubMed] [Google Scholar]
- Randazzo, P.A., Terui, T., Sturch, S., Fales, H.M., Ferrige, A.G., and Kahn, R.A. (1995). The myristoylated amino terminus of ADP-ribosylation factor 1 is a phospholipid- and GTP-sensitive switch. J. Biol. Chem. 270, 14809-14815. [DOI] [PubMed] [Google Scholar]
- Randazzo, P.A., Yang, Y.C., Rulka, C., and Kahn, R.A. (1993). Activation of ADP-ribosylation factor by Golgi membranes. Evidence for a brefeldin A- and protease-sensitive activating factor on Golgi membranes. J. Biol. Chem. 268, 9555-9563. [PubMed] [Google Scholar]
- Renault, L., Christova, P., Guibert, B., Pasqualato, S., and Cherfils, J. (2002). Mechanism of domain closure of Sec7 domains and role in BFA sensitivity. Biochemistry 41, 3605-3612. [DOI] [PubMed] [Google Scholar]
- Robineau, S., Chabre, M., and Antonny, B. (2000). Binding site of brefeldin A at the interface between the small G protein ADP-ribosylation factor 1 (ARF1) and the nucleotide-exchange factor Sec7 domain. Proc. Natl. Acad. Sci. USA 97, 9913-9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, J. (2003). Legionella Genome. http://genome3.cpmc.columbia.edu/~legion.
- Saitou, N., and Nei, M. (1986). The number of nucleotides required to determine the branching order of three species, with special reference to the human-chimpanzee-gorilla divergence. J. Mol. Evol. 24, 189-204. [DOI] [PubMed] [Google Scholar]
- Sata, M., Donaldson, J.G., Moss, J., and Vaughan, M. (1998). Brefeldin A-inhibited guanine nucleotide-exchange activity of Sec7 domain from yeast Sec7 with yeast and mammalian ADP ribosylation factors. Proc. Natl. Acad. Sci. USA 95, 4204-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sata, M., Moss, J., and Vaughan, M. (1999). Structural basis for the inhibitory effect of brefeldin A on guanine nucleotide-exchange proteins for ADP-ribosylation factors. Proc. Natl. Acad. Sci. USA 96, 2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95, 5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinotsuka, C., Waguri, S., Wakasugi, M., Uchiyama, Y., and Nakayama, K. (2002a). Dominant-negative mutant of BIG2, an ARF-guanine nucleotide exchange factor, specifically affects membrane trafficking from the trans-Golgi network through inhibiting membrane association of AP-1 and GGA coat proteins. Biochem. Biophys. Res. Commun. 294, 254-260. [DOI] [PubMed] [Google Scholar]
- Shinotsuka, C., Yoshida, Y., Kawamoto, K., Takatsu, H., and Nakayama, K. (2002b). Overexpression of an ADP-ribosylation factor-guanine nucleotide exchange factor, BIG2, uncouples brefeldin A-induced adaptor protein-1 coat dissociation and membrane tubulation. J. Biol. Chem. 277, 9468-9473. [DOI] [PubMed] [Google Scholar]
- Someya, A., Sata, M., Takeda, K., Pacheco-Rodriguez, G., Ferrans, V.J., Moss, J., and Vaughan, M. (2001). ARF-GEP(100), a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc. Natl. Acad. Sci. USA 98, 2413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang, A., Herrmann, J.M., Hamamoto, S., and Schekman, R. (2001). The ADP ribosylation factor-nucleotide exchange factors Gea1p and Gea2p have overlapping, but not redundant functions in retrograde transport from the Golgi to the endoplasmic reticulum. Mol. Biol. Cell 12, 1035-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankewich, M.C., Tse, W.T., Peters, L.L., Ch'ng, Y., John, K.M., Stabach, P.R., Devarajan, P., Morrow, J.S., and Lux, S.E. (1998). A widely expressed betaIII spectrin associated with Golgi and cytoplasmic vesicles. Proc. Natl. Acad. Sci. USA 95, 14158-14163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns, T., Willingham, M.C., Botstein, D., and Kahn, R.A. (1990). ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc. Natl. Acad. Sci. USA 87, 1238-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Galweiler, L., Palme, K., and Jurgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316-318. [DOI] [PubMed] [Google Scholar]
- Suh, Y.G., Jung, J.K., Seo, S.Y., Min, K.H., Shin, D.Y., Lee, Y.S., Kim, S.H., and Park, H.J. (2002). Total synthesis of (+)-Brefeldin A. J. Org. Chem. 67, 4127-4137. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2003). Phylogenetic Analysis Using Parsimony (*and Other Methods). Sunderland, MA: Sinauer Associates.
- Tang, P., et al. (2002). Cybr, a cytokine-inducible protein that binds cytohesin-1 and regulates its activity. Proc. Natl. Acad. Sci. USA 99, 2625-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa, A., Morinaga, N., Ogasawara, M., Moss, J., and Vaughan, M. (1999). Purification and cloning of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J. Biol. Chem. 274, 12308-12315. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu, K. (2003). Interaction protein for cytohesin exchange factors 1 (IPCEF1) binds cytohesin 2 and modifies its activity. J. Biol. Chem. 278, 43460-43469. [DOI] [PubMed] [Google Scholar]
- Whelan, S., and Goldman, N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691-699. [DOI] [PubMed] [Google Scholar]
- Yahara, N., Ueda, T., Sato, K., and Nakano, A. (2001). Multiple roles of Arf1 GTPase in the yeast exocytic and endocytic pathways. Mol. Biol. Cell 12, 221-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.Z., Heimberg, H., D'Souza-Schorey, C., Mueckler, M.M., and Stahl, P.D. (1998). Subcellular distribution and differential expression of endogenous ADP-ribosylation factor 6 in mammalian cells. J. Biol. Chem. 273, 4006-4011. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Lasell, T.K., and Melancon, P. (2002). Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic. Mol. Biol. Cell 13, 119-133. [DOI] [PMC free article] [PubMed] [Google Scholar]