Abstract
Subtelomeric regions in eukaryotic organisms are known for harboring species-specific tandemly repeated satellite sequences. However, studies on the molecular organization and evolution of subtelomeric repeats are scarce, especially in plants. Khipu is a satellite DNA of 528-bp repeat unit, specific of the Phaseolus genus, with a subtelomeric distribution in common bean, P. vulgaris. To investigate the genomic organization and the evolution of khipu, we performed genome-wide analysis on the complete genome sequence of the common bean genotype G19833. We identified 2,460 khipu units located at most distal ends of the sequenced regions. Khipu units are arranged in discrete blocks of 2–55 copies and are heterogeneously distributed among the different chromosome ends of G19833 (from 0 to 555 khipus units per chromosome arm). Phylogenetically related khipu units are spread between numerous chromosome ends, suggesting frequent exchanges between non-homologous subtelomeres. However, most subclades contain numerous khipu units from only one or few chromosome ends indicating that local duplication is also driving khipu expansion. Unexpectedly, we also identified 81 khipu units located at centromeres. All the centromeric khipu units belong to a single divergent clade also comprised of a few units from several subtelomeres, suggesting that a few sequence exchanges between centromeres and subtelomeres took place in the common bean genome. The divergence and low copy number of these centromeric units from the subtelomeric units could explain why they were not detected by FISH (Fluorescence in situ Hybridization) although it can not be excluded that these centromeric units may have resulted from errors in the pseudomolecule assembly. Altogether our data highlight extensive sequence exchanges in subtelomeres between non-homologous chromosomes in common bean and confirm that subtelomeres represent one of the most dynamic and rapidly evolving regions in eukaryotic genomes.
Keywords: Common bean, satellite DNA, tandem repeat, evolution, FISH, genome sequencing, centromere, subtelomere
Introduction
Common bean (Phaseolus vulgaris) is a major source of protein for human consumption in many parts of the world (FAO 1980), especially in developing countries such as tropical areas of Latin America and Eastern Africa where common bean is one of the major staple crops (Pastor-Corrales and Tu, 1989; Broughton et al., 2003). Together with sorghum, millet, groundnut, cowpea, chickpea, pigeonpea, cassava, yam, and sweet potato, common bean is often referred to as an “orphan crop.” Indeed, even if common bean is an important crop in developing countries, it is not extensively traded and receives less attention from researchers compared to crops such as maize, rice, and wheat (Varshney et al., 2012). Common bean has a small diploid genome (2n = 22) of 588 Mb (Bennett and Leitch, 1995) including a large amount of repeated sequences (Schlueter et al., 2008; Pedrosa-Harand et al., 2009) compared with other legume species with larger genome sizes, such as Trifolium repens (956 Mb; Bennett and Smith, 1991) and soybean (1,103 Mb; Bennett and Leitch, 1997). Recently, the revolution in sequencing technologies has allowed the establishment of full genome sequencing programs for orphan crops like common bean and the full genome is now available (since July 20121; Jackson et al., in preparation). The selected common bean genotype is “G19833,” an Andean landrace for which a BAC library was used to construct a draft physical map (Schlueter et al., 2008).
Satellite DNA can be defined as highly reiterated non-coding DNA sequences, organized as long arrays of head-to-tail linked repeats located in the constitutive heterochromatin (Plohl et al., 2008). Despite their ubiquity in eukaryotic genomes, the function of such repeats is poorly understood. Early hypotheses considered them to be non-functional “selfish” DNA that proliferate for their own sake or as useless genomic elements accumulated as “junk” with no selective advantage to the organism (Ohno, 1972; Orgel and Crick, 1980). More recently, identification of satellite DNA at structurally important parts of chromosomes, such as centromeres, has suggested functional roles of satellite DNA (Ma and Jackson, 2006).
Satellite DNA is an important component of the knobs, which are cytologically visible regions of highly condensed chromatin (heterochromatin) that are distinct from pericentromeric regions in pachytene chromosomes (Fransz et al., 2000). In common bean, a 528-bp subtelomeric satellite repeat, referred to as khipu, has been identified (David et al., 2009). Khipu is present on most chromosomal terminal knobs and is specific to the Phaseolus genus (David et al., 2009; Geffroy et al., 2009). Subtelomeric satellite repeats have been reported in different plant and animal species. Indeed, cytologically confirmed subtelomeric satellite repeats have been identified in various plant species, including potato (Torres et al., 2011), rice (Cheng et al., 2001), tomato (Lapitan et al., 1989), maize (Li et al., 2009), barley (Brandes et al., 1995), tobacco (Kenton et al., 1993; Chen et al., 1997), rye (Vershinin et al., 1995), Silene latifolia (Buzek et al., 1997), and Beta species (Dechyeva and Schmidt, 2006). The subtelomeric locations of these repeats were confirmed mostly by FISH (fluorescence in situ hybridization) experiments and were not based on sequence analysis. Except in rice, where sequencing and characterization of the structure of the subtelomeric TrsA sequences were conducted (Ohmido and Fukui, 1997; Mizuno et al., 2006, 2008), sequence-based analysis of the molecular organization and evolution of subtelomeric repeats is rare.
In the present paper, we conducted genome-wide analysis to investigate the physical organization and the evolution of khipu sequence based on the complete genome sequence of common bean genotype G19833.
Materials and Methods
Data sources
We used the “Phaseolus vulgaris v1.0” genome sequence of the Andean common bean genotype G198332 and sequenced BAC clones from G19833 reported in Innes et al. (2008) that correspond to ∼1 Mb of the Co-2 cluster, located at the end of the long arm of chromosome 11, and from Chen et al. (2010), corresponding to 239 kb located at one end of chromosome 5, referred to as PvA05A.
Khipu annotation
The khipu satellite DNA was recovered using hmmsearch3 (Eddy, 1998) with a khipu profile previously defined on 92 khipu (David et al., 2009). In order to work with a “clean” set of khipu, we excluded the first and last khipu element from blocks of tandemly organized khipu. In addition, we excluded khipu < 500 pb and having “n”s in their sequence. The resulting data was imported into the annotation platform Artemis for manual analysis (Rutherford et al., 2000).
Centromeric positions in individual pseudomolecules were identified by BLASTN using centromere satellite repeats; CentPv1 for chromosomes 01, 02, 03, 04, 07, 08, 09, and 10 and CentPv2 for chromosomes 05, 06, and 11 (Jackson et al., in preparation). Each khipu sequence extracted from the genome of G19833 was named Pvxxyk#####, with Pv referring to Phaseolus vulgaris, “xx” referring to pseudomolecule (01–11), “y” corresponding to the location on the chromosome (S for short arm, C for centromere, and L for long arm), “k” referring to “khipu,” and a 5-digit number referring to the khipu order on the pseudomolecules from the start (5′) to the end (3′). khipu elements on each pseudomolecule were sequentially numbered in increments of 10. Each khipu extracted from BAC sequences was named PvAxxzk##### (Axxz is the name of the contig), with Pv referring to Phaseolus vulgaris, A referring to Andean (these BAC come from the Andean genotype G19833), xx referring to chromosome (05 or 11), z is a letter given to the different contigs from the same chromosome, k referring to khipu, and 5-digit number referring to the khipu order on the contig in increments of 10. For example, khipu named Pv01Sk00010 is the first khipu unit on pseudomolecule 01, located on short arm and PvA05Ak00010 is the first khipu unit of the BAC contig A from chromosome 05.
To determine the coordinates of BACs on the pseudomolecules, we performed a BLASTN of the entire BAC sequence against the genome sequence. BLAST results were inspected manually to set the start and end positions of the BAC on the pseudomolecules. BAC sequences and pseudomolecules were aligned and visualized using Mauve, a genome alignment tool, using the minimal match seed weight value4 (Darling et al., 2004).
Phylogenetic analysis
Multiple sequence alignments of khipu sequences were generated using Muscle (Edgar, 2004a,b) with gapopen = −1000 and MaxIter = 3. Optimized alignement is provided in Figure S1 in Supplementary Material. Graphical representation of the khipu alignment was visualized using the WebLogo server5 (Crooks et al., 2004) (Figure S2 in Supplementary Material). Recombination among loci was assessed using several methods implemented in RDP v.3.15 (Martin et al., 2005b): RDP (Martin and Rybicki, 2000), Geneconv (Padidam et al., 1999), Chimera (Posada and Crandall, 2001), and Bootscan (Martin et al., 2005a). Default parameter settings were used for each method except as follows: RDP (internal reference sequence), Bootscan (window = 150, step = 20, NJ trees, 200 replicates, 95% cutoff, J&N model with Ti:Tv = 2, coefficient of variation = 2). The maximum p-value for accepting recombination was set at 0.001 (after Bonferroni correction).
A Maximum-Likelihood tree was made with FastTree 2.1.3 program (Price et al., 2010) with the Jukes–Cantor model of nucleotide evolution. Bootstrap values were computed with the consensus of 100 random trees using the Phylip’s consense program (Felsenstein, 1989) and the random trees were calculated with the “−n” option of the FastTree program over a list of bootstrapped sequences generated from the original sequence alignment using Seqboot in the PHYLIP package. The resulting phylogenetic tree (Figure S3 in Supplementary Material) was displayed using MEGA version 5 (Tamura et al., 2011). One representative khipu sequence from each major clade of the phylogenetic tree, is provided in Figure S2 in Supplementary Material.
Cytogenetic analysis
The khipu probe was generated using a pool of five subclones from BAC clones (Table A1 in Appendix). Three subclones come from different khipu blocks spread over sequenced BAC clones from the long arm of chromosome 11 (Innes et al., 2008), one additional subclone come from a subtelomeric BAC from the short arm of chromosome 5 (Chen et al., 2010), and the 1H04 subclone come from the B4 locus (short arm of chromosome 4 from the Mesoamerican BAT93 genotype) described in David et al. (2009). Pachytene chromosomes were prepared from young flower buds of G19833 and JaloEEP558 fixed in ethanol:acetic acid (3:1, v/v). Buds were macerated in 2% cellulase/2% pectolyase/2% cytohelicase in 0.01 M citric acid-sodium citrate buffer, pH 4.8, for 3 h at 37°C, incubated in 60% acetic acid up to 2 h, and squashed after removal of petals and sepals and flaming. Slide selection and pretreatment, chromosome and probe denaturation and hybridization, posthybridization washes, detection, and image analyses were performed according to Fonseca et al. (2010).
Results and Discussion
Genome-wide identification of the khipu satellite repeat in the pseudomolecules of common bean G19833
To identify khipu sequences in the common bean genome, we used the P. vulgaris genome v1.0 from Phytozome6. A total of 2766 khipu units were identified. After size selection of khipu units > 500 bp and discarding khipu units with “n”s, we had 2460 khipu units for further analysis. The distribution of these 2460 khipu units in the common bean genome is presented in Table 1. The short arm of chromosome 01 (chr01S), Chr04S, Chr04L (L = long arm), Chr05S, Chr10L, and Chr11L, had the largest number of khipu units, with 193, 349, 261, 169, 435, and 555, respectively. Chr08S and Chr09L, however, had fewer than 10 khipu units and Chr06S and Chr09S were devoid of khipu. These data are in general agreement with the cytogenetic distribution of BAC 63H6, which contains khipu (K.G.B. dos Santos, personal communication) in G19833 (Altrock et al., 2011), except that Chr10S and Chr11S seem to have large amounts of khipu (as estimated based on the intensity of FISH signals) and no khipu signal could be detected in Chr09L. In agreement with previous FISH analysis on BAT93 showing the subtelomeric distribution of khipu (David et al., 2009), khipu units were mainly located in the first or last five megabase-pairs of the pseudomolecules. These khipu were organized in tandem arrays with varying numbers units, referred to as khipu blocks. For example, chr04S had 28 khipu blocks containing fewer than 13 units and nine blocks containing more than 13 units, within the first 4.7 Mb of the pseudomolecule. In rice, similar organization in discrete clusters of 3–103 copies in a chromosome specific manner was also observed for the TrsA subtelomeric repeats (Mizuno et al., 2008). In common bean, the largest khipu blocks were found on Chr04S and Chr11L, where khipu blocks had 45 and 55 khipu units, respectively. Notably, Chr04S and Chr11L contain the B4 (David et al., 2009) and Co-2 disease resistance gene clusters (Innes et al., 2008; David et al., 2009; Chen et al., 2010) suggesting a possible link between the evolution of resistance clusters and the khipu sequences. As expected for a subtelomeric repeat, few or no khipu (<13) were identified in centromeric regions, with the exception of chromosome 8 centromere which had 55 khipu sequences.
Table 1.
Number of complete khipu units in each pseudomolecule of Phaseolus vulgaris.
| Pseudomolecules | Short arm | Centromere | Long arm | Total |
|---|---|---|---|---|
| Chr01 | 193 | 0 | 19 | 212 |
| Chr02 | 46 | 0 | 39 | 85 |
| Chr03 | 60 | 3 | 17 | 80 |
| Chr04 | 349 | 13 | 261 | 623 |
| Chr05 | 169 | 0 | 33 | 202 |
| Chr06 | 0 | 0 | 17 | 17 |
| Chr07 | 17 | 7 | 14 | 38 |
| Chr08 | 7 | 55 | 51 | 113 |
| Chr09 | 0 | 2 | 10 | 12 |
| Chr10 | 71 | 1 | 435 | 507 |
| Chr11 | 16 | 0 | 555 | 571 |
| Total | 81 | 2460 |
Spread of khipu to non-homologous chromosome ends
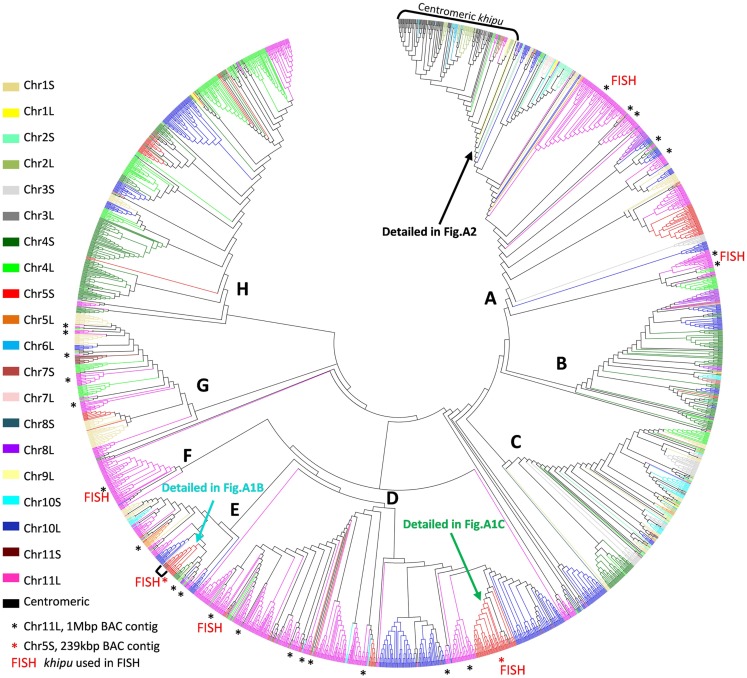
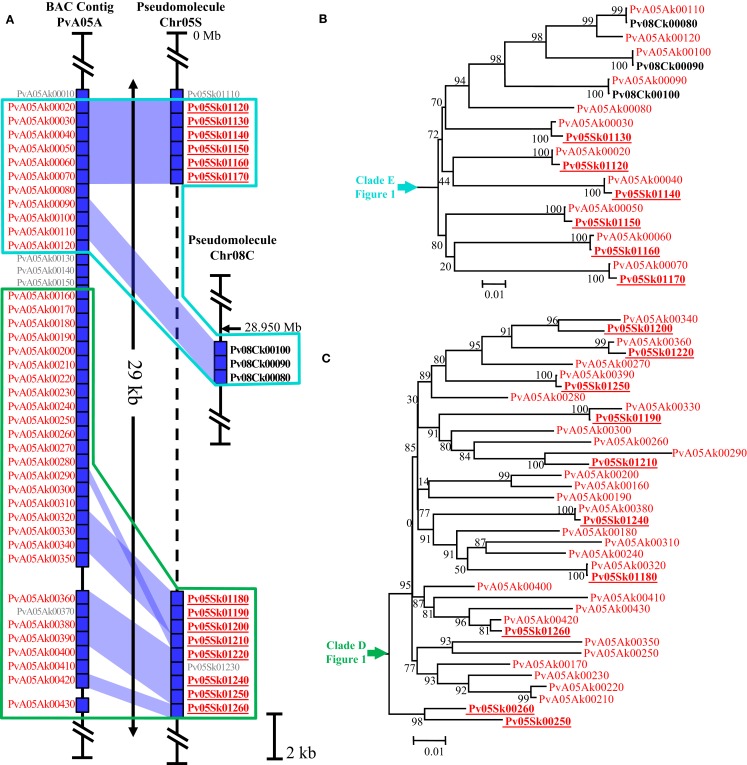
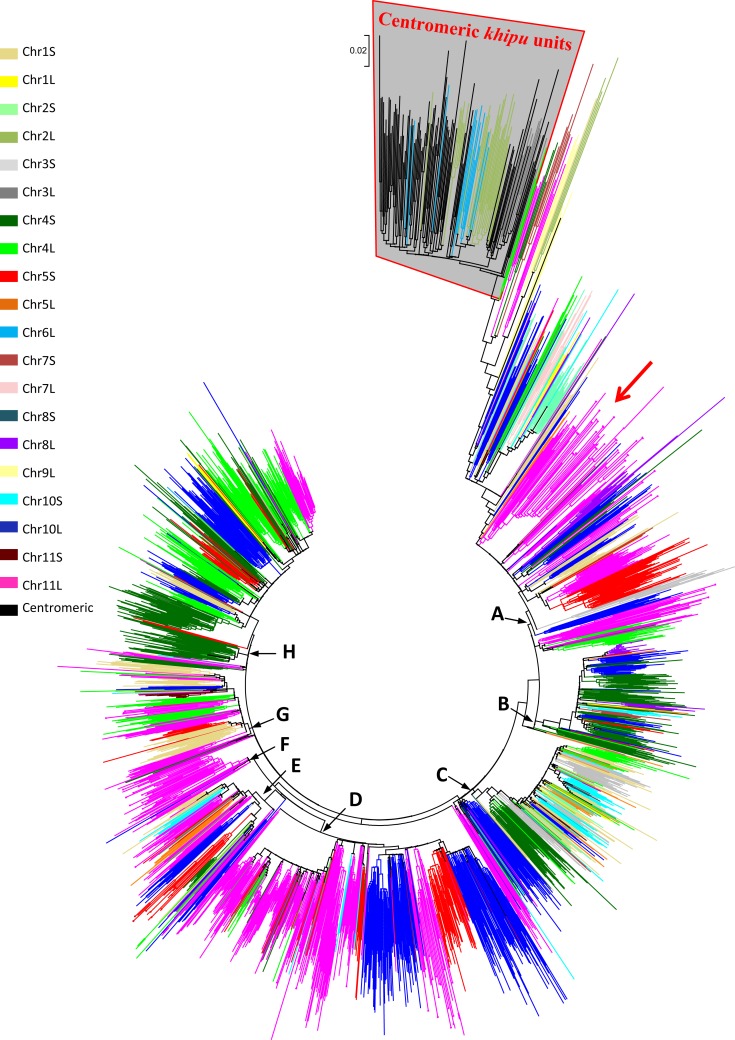
To study khipu satellite evolution in the common bean genome, a phylogenetic tree was constructed (Figure 1). A multiple alignment including the 2460 khipu units from the G19833 genome plus 201 units from G19833 BACs (Innes et al., 2008; Chen et al., 2010) were first screened for recombination events using RDP4 and recombinant sequences removed (76 and 4 khipu units from G19833 genome and BACs, respectively). Khipu units fell into eight major clades (A–H) with Bootstrap support >75% (Figure 1). Except for the small clade F that contains only khipu units from Chr11L, Chr04S and Chr10L, each major clade contains khipu units from most chromosome ends. This indicates that phylogenetically related khipu units were spread among the chromosome ends, suggesting frequent exchanges between non-homologous subtelomeres. Within a chromosomal cluster, khipu units were found across the tree, indicating that phylogenetically distant khipu units are physically close to each other. This is particularly striking for Chr10L (blue) where khipu elements are spread across the eight clades. The distribution of khipu units coming from BAC clones reinforces these results. For example, khipu units coming from the BAC from Chr11L (∼1 Mbp; Innes et al., 2008) are spread across 24 subclades belonging to five of the eight major clades (black asterisks in Figure 1). Thus, almost the entire khipu diversity is represented in a single genomic region as small as ∼1 Mbp. Additionally, BAC PvA05A (239kbp; Chen et al., 2010) from Chr05S contains a khipu block bearing phylogenetically distant khipu units (Figure 1; Figure A1 in Appendix). Indeed, this khipu block is composed of 11 complete units from major clade E (light blue, PvA05Ak00020–PvA05Ak00120) followed by an array of 27 khipu units from major clade D (green, PvA05Ak00160–PvA05Ak00430).
Figure 1.
Comparison between phylogeny and physical distribution of khipu repeats within the G19833 genome. Phylogenetic tree of the 2460 khipu repeats from the G19833 genome sequence. The eight major clades are indicated with bold letters (A–H). Each color corresponds to a chromosome arm subtelomeric region named by a number, corresponding to the chromosome number, followed by the letter L (long arm) or S (short arm). Note that centromeric khipu repeats (black) are found only in two small subclades highlighted by brackets. Clades comprising khipu repeats from previously sequenced BAC contigs from chromosome 11 long arm or chromosome 5 short arm are highlighted by black or red asterisks, respectively. Clades comprising khipu repeats used for FISH experiments are highlighted by FISH written in red.
Together these results indicate that each subtelomere contains a patchwork of phylogenetically distant khipu units that is likely the result of shuffling between non-homologous chromosome ends. However, most subclades contain numerous khipu units from only one or few chromosome ends (Figure 1). Thus, in addition to khipu spreading between non-homologous loci, local duplication is driving khipu expansion. In the human genome, extensive cytogenetic and sequence analyses revealed that subtelomeres are hot spots of interchromosomal recombination and segmental duplications (Linardopoulou et al., 2005). This exceptional dynamic activity of subtelomeres has been reported in such diverse organisms as yeast and the malaria parasite Plasmodium (Louis, 1995; Freitas-Junior et al., 2000, 2005). As expected for a plastic region of the genome subject to reshuffling through recombination events, subtelomeres exhibit unusually high levels of within-species structural and nucleotide polymorphism (Mefford and Trask, 2002). In plants, this plasticity of subtelomeres was not found in Arabidopsis thaliana (Heacock et al., 2004; Kuo et al., 2006) and, to our knowledge, has not been reported for any other sequenced plant species.
Quality of khipu sequences within the G19833 genome
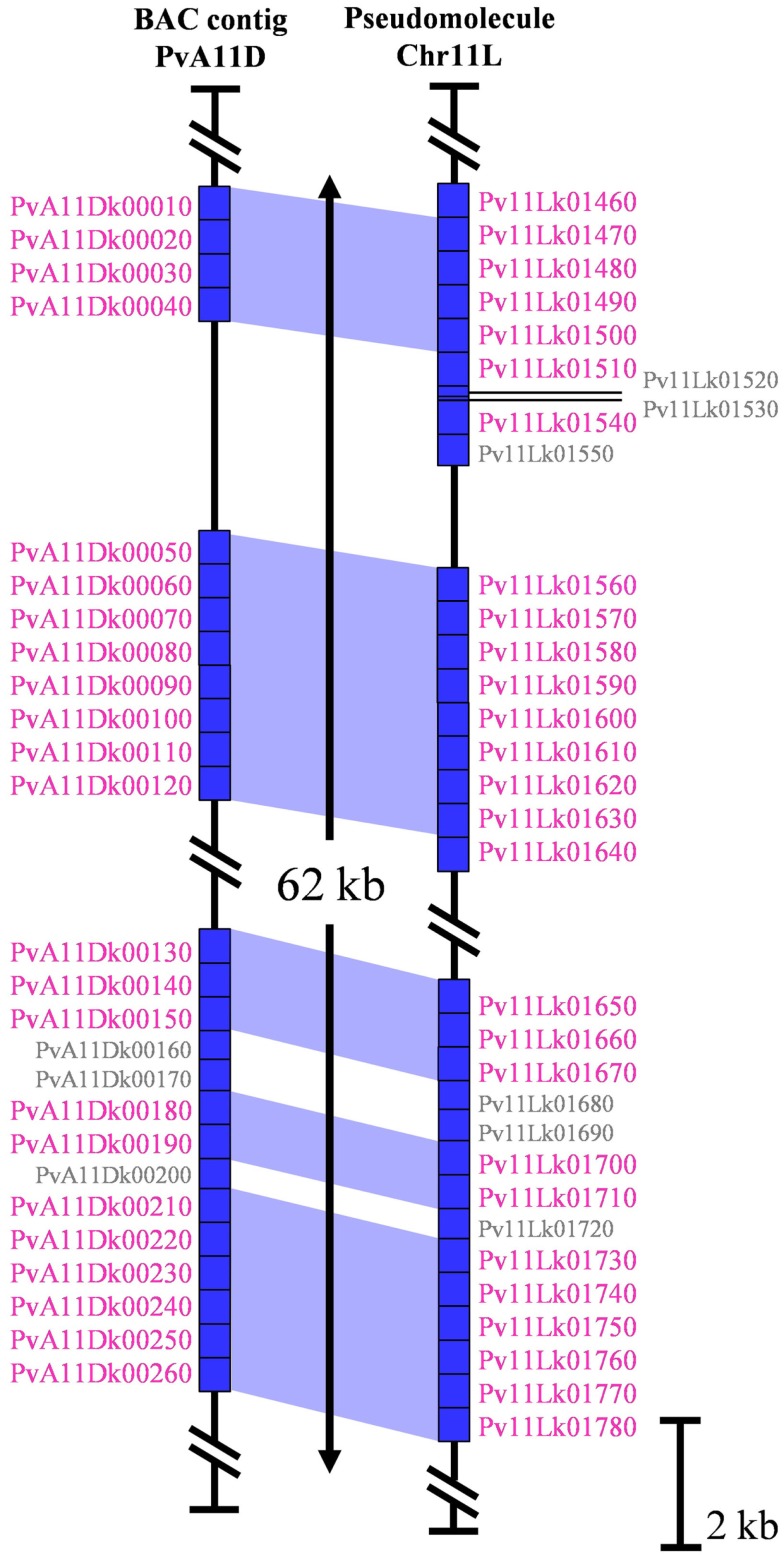
Genomic regions containing highly repetitive sequences, especially tandem arrays of satellite repeats, constitute a challenge for short-read, whole-genome shotgun sequencing and are thus considered to be error-prone regions of whole-genome sequencing projects (Jackson et al., 2011). In order to check the quality and the consistency of the G19833 pseudomolecules for khipu, we compared pseudomolecules sequence data with six G19833 BACs (sequenced by the classical Sanger method) containing khipu sequences and coming from two distinct genomic regions (Innes et al., 2008; Chen et al., 2010, unpublished data). Here we present in details the results from two BAC contigs: PvA11D is a 200 kbp clone corresponding to the coordinates 47.3–47.5 Mb of the long arm of pseudomolecule 11 (Chr11L) (Figure 2) while PvA05A is a 239 kbp clone corresponding to the coordinates 1.0–1.2 Mb of the pseudomolecule 5 (Chr05S) (Figure A1A in Appendix). In these regions, apart from khipu blocks, sequences share over 98% nucleic identity between BAC contigs and pseudomolecule sequences (data not shown). A combination of phylogenetic and genomic analyses shows that most khipu units are identical in position and sequence in both the pseudomolecules and BACs (blue areas in Figure 2; Figure A1A in Appendix). For example, khipus from PvA11D and its pseudomolecule counterpart are nearly identical (Figure 2). Similar results were obtained with four other BAC contigs containing khipu units (data not shown), confirming that genome data is of high quality even for khipu containing regions. However, PvA11D contains khipu blocks of only 14 khipu units (∼7400 bp) and a different situation is observed for PvA05A which bears a larger khipu block of more than 35 khipu units (Figure A1 in Appendix). Of these, six (PvA05Ak00020–PvA05Ak00070) are completely identical with corresponding region of Chr05 pseudomolecule, but 21 (PvA05Ak00080, PvA05Ak00120, PvA05Ak00300, PvA05Ak00310, PvA05Ak00350, PvA05Ak00400, PvA05Ak00410, PvA05Ak00430, and PvA05Ak00160–PvA05Ak00280) are completely absent in the whole-genome sequence, resulting in a ∼10 kbp gap (Figure A1A in Appendix). Moreover, three khipu units (PvA05Ak00090, PvA05Ak00100, and PvA05Ak00110) from this Chr05 BAC contig share 100% nucleotide identity with three khipu units from pseudomolecule 8 centromeric region (Pv08Ck00100, Pv08Ck00090, and Pv08Ck00080) (Figures A1A,B in Appendix). Because we were unable to find corresponding khipu units from Chr05S, it is possible that these three Chr08 centromeric khipu are the result of errors in the assembly. Even though BACs are considered to be stable cloning vectors, it has been reported that tandem repeats can be unstable in BAC clones (Song et al., 2001). In conclusion, comparisons between BACs and the genome sequence data suggest that small arrays of satellite units can be well-resolved, while larger satellite blocks may be slightly more error prone. However, based on six independent BACs/genome comparisons, we conclude that genome data is of high quality to conduct genome-wide analysis o khipu sequences.
Figure 2.
Schematic representation of 62 kb of PvA11D (200 kb), containing khipu, with the corresponding region of pseudomolecule 11. Sequences of these regions are represented by vertical black lines. Blue rectangle represent annotated khipu of the region with their name written on the side of each one. Khipu with name written in small gray correspond to khipu shorter than 500 bp discarded from the analyses. The blue areas represent correspondences between khipu from BAC sequence and khipu from genome sequence according to the phylogenetic tree.
Centromeric khipu units: Divergent khipu sequence and/or erroneous assembly?
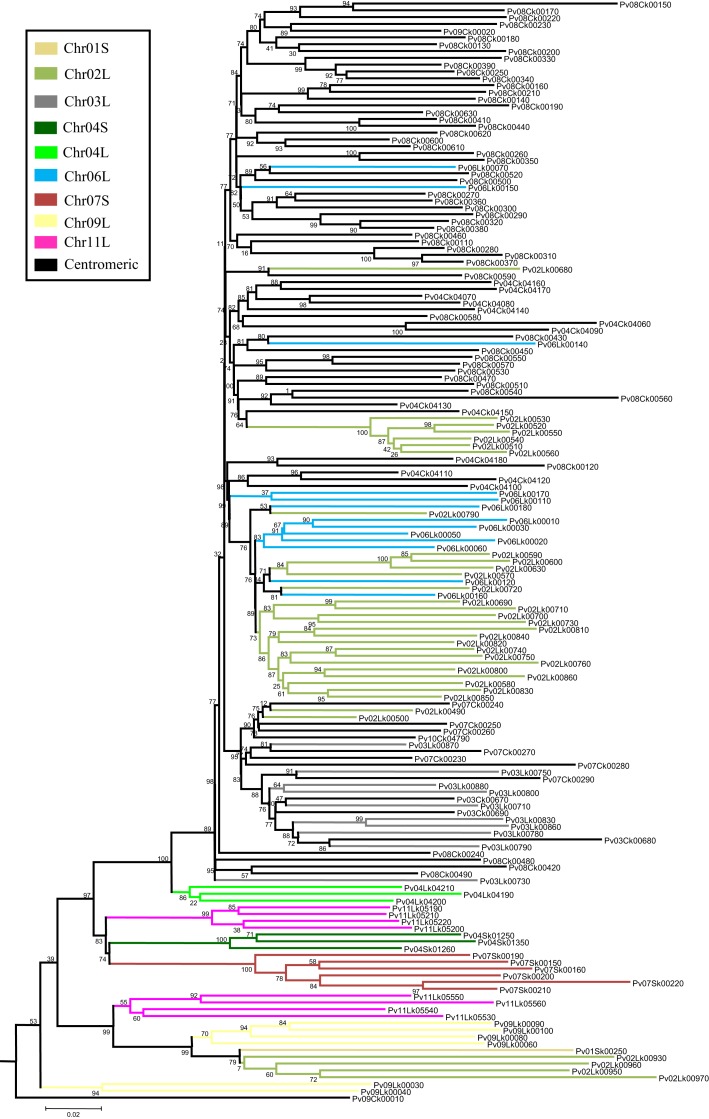
Except for the three potentially false centromeric khipu units (identical to subtelomeric khipu units from Chr05S, Figures A1A,B in Appendix), all 78 centromeric khipu units belong to a single well-defined subclade that also includes subtelomeric units from Chr06L, Chr03L, and Chr02L (Figure 1; Figure A2 in Appendix) which indicates that they are less diverse than the subtelomeric units. It is surprising that no centromeric signals were found in previous FISH experiments on mitotic chromosomes, using either a khipu-specific probe in the BAT93 common bean genome (David et al., 2009; Geffroy et al., 2009) or a khipu-bearing subtelomeric BAC clone in G19833 (Altrock et al., 2011). These conflicting results raise the question of whether the centromeric khipu units are real centromeric sequences or misassembled sequences.
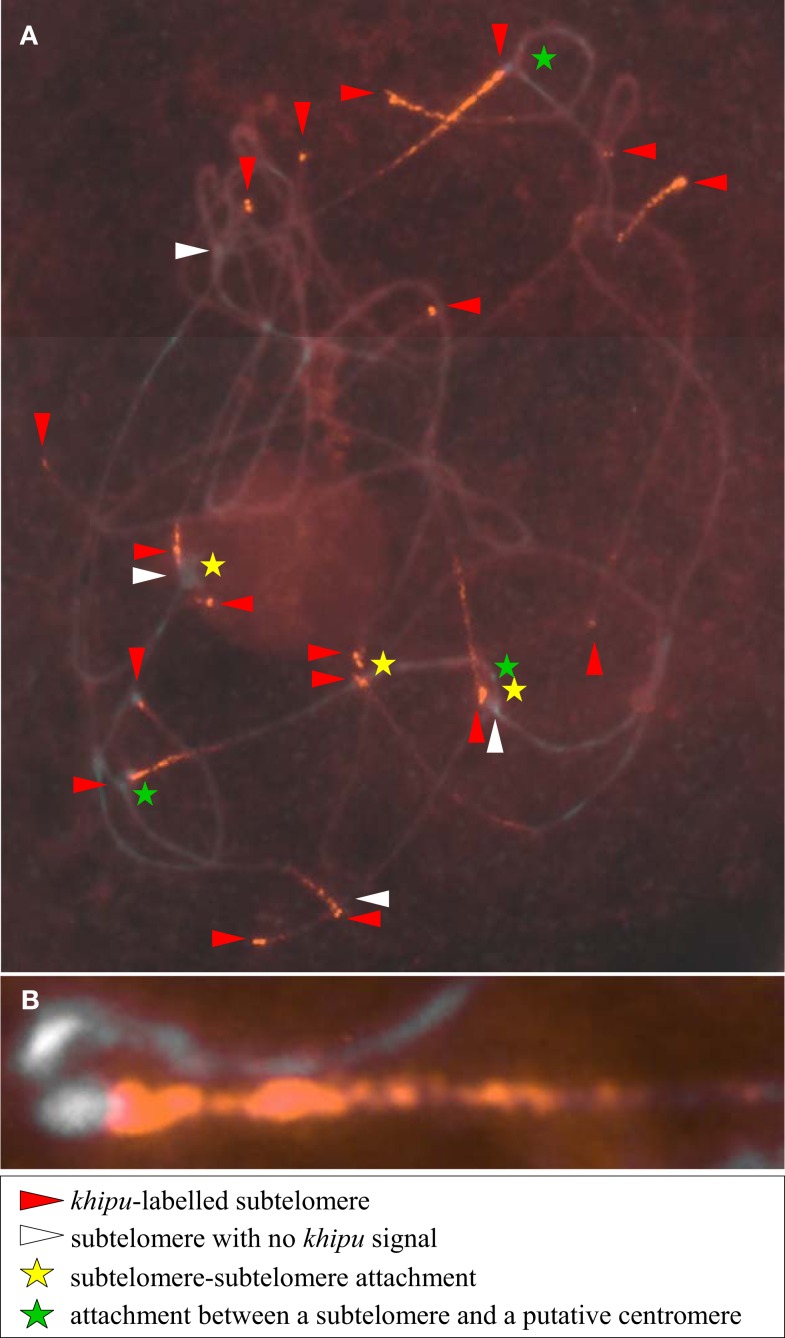
To try to solve this puzzle, we performed FISH on pachytene chromosomes from G19833, using a khipu FISH probe containing a wide diversity of khipu units (highlighted by “FISH” written in red in Figure 1; Table A1 in Appendix). We detected khipu signals on 17 chromosome ends, but found no evidence of centromeric signals in G19833 chromosomes (Figure A3 in Appendix). This result is similar to khipu distribution at 17 chromosome ends in the BAT93 genotype (David et al., 2009) suggesting that khipu distribution has been stable since the split between Andean and Mesoamerican gene pools. Interestingly, during these FISH experiments heterochromatin connections between non-homologous chromosomes were observed (Figure A4 in Appendix), providing indirect evidence that subtelomeres are hot spot of interchromosomal recombination. Similar attachments were observed in rye (Gonzalez-Garcia et al., 2006). In common bean, according to whole-genome data Chr08C comprises 55 khipu (Table 1). This begs the question: why was not it possible to detect these 55 centromeric khipu units by FISH?
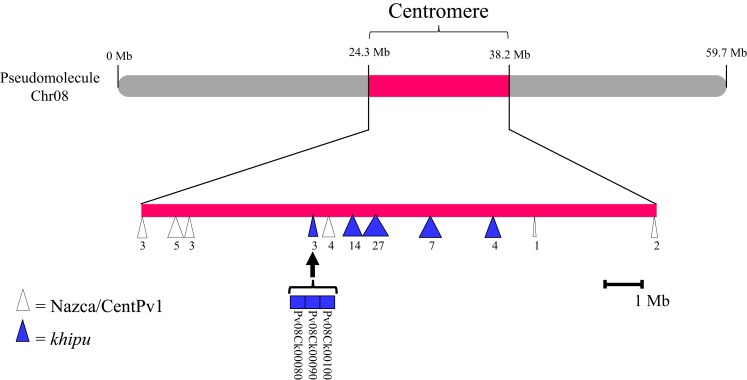
There are four potential explanations. The first hypothesis is that khipu units were grouped at Chr08C due to biases during assembly of the pseudomolecule. A comparison to BAC contig PvA05A allowed us to find three khipu units from Chr05S subtelomeric region that were assembled at Chr08C by mistake during pseudomolecules assembling (Figures A1A,B in Appendix). What about the other centromeric khipu? As previously discussed, the genome assembly is of high quality and the major differences to BAC sequences are numbers of khipu units within a khipu block rather than a wrong location of khipu blocks (Figure 2; Figure A1 in Appendix). In addition to Chr08C, we also found khipu units in centromeres of Chr03, Chr04, Chr07C, Chr09, and Chr10 (Figure A2 in Appendix). It is unlikely that these khipu units, clustered in a single subclade, would be misassembled at the centromere of various pseudomolecules. Moreover, it is not unusual to find non-centromeric defined repeats in centromeric regions in eukaryotes. Notably, detection of DNA sequences that are partially homologous to telomeric repeats in centromeres has been reported in many species including Drosophila melanogaster (Abad et al., 2004; Mendez-Lago et al., 2009), maize (Alfenito and Birchler, 1993; Jin et al., 2005), and in potato (Tek and Jiang, 2004; He et al., 2013). Moreover, in rice, a satellite DNA present both in subtelomeric and centromeric regions has also been identified (Lee et al., 2005; Bao et al., 2006). A second hypothesis to explain the absence of khipu centromeric FISH signal is that khipu units from Chr08C are spread across a wide region of the centromere (13.8 Mb), thus diluting the signal in FISH. However, according to the pseudomolecule, the khipu units are grouped at the very center of Chr08C, with one block of 27 khipu units which should have been detected by FISH (Figure A5 in Appendix). The third hypothesis is that centromeric khipu are too divergent from the khipu units used for FISH experiments (Figure A6 in Appendix). Even if our FISH probe presents a wide diversity of khipu units (as shown in Figure 1 with “FISH” written in red), the closest khipu unit used in FISH (red arrow in Figure A6 in Appendix) shares only 72% nucleotide identity with the khipu units from the subclade comprising centromeric repeats, while it shares more than 85% identity with the khipu units from other major clades (data not shown). Consequently, it is possible that our probe missed the centromeric khipu units considering the stringency used. The fourth hypothesis is that the chromatin architecture in the centromeric region may make these sequences inaccessible to FISH.
Conclusion
Satellite DNA is an enigmatic part of eukaryotic genomes. Subtelomeric satellite repeats have been reported in many eukaryotic chromosomes but their function remains largely unknown. A genome-wide analysis based on the complete genome sequence of the common bean genotype G19833 of the subtelomeric khipu satellite, revealed extensive sequence exchanges between non-homologous chromosomes in subtelomeric regions and also suggests sequence exchange between subtelomere and centromere.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online http://www.frontiersin.org/journal/10.3389/fpls.2013.00109/abstract
Khipu sequence alignement.
WebLogo representation of the consensus sequence derived from the multiple alignment of the khipu units and sequence of one representative khipu unit from each major clade.
Phylogenetic tree of whole-genome khipu sequence.
Acknowledgments
We thank Mireille Sévignac for stimulating discussions. The research was supported by INRA, CNRS, IFR87, and IDEEV, France. Andrea Pedrosa-Harand was supported by a grant from CNPq, Brazil.
Appendix
Figure A1.
Comparison of khipu containing region of the genome with the corresponding BAC contig and phylogenetic analysis of khipu sequence. (A) Schematic representation of 29 kb of PvA05A (239 kb), containing khipu, with the corresponding region of pseudomolecule 5 and with part of the centromeric region of chromosome 8 (coordinates: 28.950 Mb). Sequences of these regions are represented by vertical black lines with dotted line represent a gap in the sequence. Blue rectangle represent annotated khipu of the region with their name written on each side, in red for khipu from PvA05A sequence, bold red and underlined for khipu from the pseudomolecule 5 and bold black for centromeric khipu of pseudomolecule 8. Khipu in small gray type correspond to khipu shorter than 500 bp that were discarded from analysis. The blue area represents the correspondence between khipu from BAC sequence and khipu from genome sequence according to the phylogenetic tree. (B) and (C) Phylogenetic tree clades of khipu present on (A). (B) Part of the clade called “E” (cf. Figure 1) (C) Part of the clade called “D” (cf. Figure 1)
Figure A2.
Detailed view of a subtree from the phylogenetic tree presented in Figure 1 containing centromeric khipu units. Branch colors correspond to the colors defined in Figure 1. Note cases of subtelomeric khipu units from Chr02L, Chr03L, and Chr06L interspersed with centromeric khipu units (black), indicating movement of khipu units between subtelomeres and centromeres.
Figure A3.
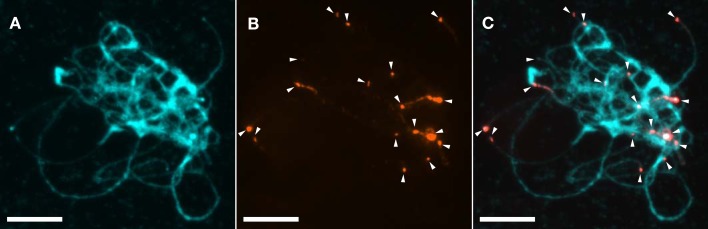
Physical distribution of khipu on G19833 pachytene chromosomes using FISH. FISH to G19833 pachytene chromosomes. Khipu was used as a probe in (B) and (C). Signals are in red, and chromosomes in blue in (A) and (C). The 17 chromosome termini with khipu signals are indicated by white arrowheads. Bars = 10 mm.
Figure A4.
Localization of khipu satellite DNA on JaloEEP558 pachytene chromosomes and evidences of chromosome attachments. Chromosomes are counterstained with DAPI (gray), and khipu signals are in red (original picture). (A) Chromosome termini with or without khipu labeling are indicated by red or white arrowheads, respectively. Stars indicate putative chromosome attachments. (B) Magnified view of an attachment between two non-homologous chromosomes.
Figure A5.
Organization of satellite DNA Nazca/CentPv1 and khipu in the centromeric region of chromosome 08. Chromosome 08 is represented in gray and the centromeric region in pink. The centromeric part is enlarged for positioning satellite DNA. Nazca/CentPv1 (centromeric repeat, Iwata et al., submitted) and khipu are represented by white and blue triangles, respectively. The number of repeated units inside each block is indicated below. The three khipu of the first block are detailed with their names indicated below. These are khipu phylogenetically clustered with three khipu of the BAC contig PvA05A presented in Figure A1.
Figure A6.
Phylogenetic tree presented in Figure 1 with branch length. Clade names (A–H) correspond to the clades defined in Figure 1 and branch colors correspond to the colors defined in Figure 1. The subclade containing the centromeric khipu units is boxed in gray. From this clade, the closest khipu unit used for FISH experiments is indicated by a red arrow.
Table A1.
Characteristics of the probes sed for FISH experiments.
| Probe | BAC clone | Contig | Genotype | Subclone | Length (bp) | Coordinates on BAC | Coordinates on contig | Notes and reference |
|---|---|---|---|---|---|---|---|---|
| khipu | Pva1-105k5 | PvAO5A | Gl9833 | TE0AAFA1Y119 | 6135 | 64475..70609 | 64475..70610 | Chen et al. (2010) |
| Pva1-68o6 | PvA11B | G19833 | TE0AAAA3YM09 | 5268 | 10430..15697 | 10430..15698 | Innes et al. (2008) | |
| Pval-34g17 | PvA11C | G19833 | TE0AANA1YI09 | 7692 | 14329..22020 | 165163..172854 | Innes et al. (2008) | |
| Pva1-119e5 | PvA11E | G19833 | TE0AAEA2YP22 | 5628 | 163980..169607 | 348510..354137 | Innes et al. (2008) | |
| – | Pv410-kb | BAT93 | 1H04 | 2410 | – | 388326..390736 | David et al. (2009) |
Footnotes
References
- Abad J. P., De Pablos B., Agudo M., Molina I., Giovinazzo G., Martin-Gallardo A., et al. (2004). Genomic and cytological analysis of the Y chromosome of Drosophila melanogaster: telomere-derived sequences at internal regions. Chromosoma 113, 295–304 10.1007/s00412-004-0318-0 [DOI] [PubMed] [Google Scholar]
- Alfenito M. R., Birchler J. A. (1993). Molecular characterization of a maize B-chromosome centric sequence. Genetics 135, 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altrock S., Fonseca A., Pedrosa-Harand A. (2011). Chromosome identification in the Andean common bean accession G19833 (Phaseolus vulgaris L., Fabaceae). Genet. Mol. Biol. 34, 459–463 10.1590/S1415-47572011005000029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W. D., Zhang W. L., Yang Q. Y., Zhang Y., Han B., Gu M. H., et al. (2006). Diversity of centromeric repeats in two closely related wild rice species, Oryza officinalis and Oryza rhizomatis. Mol. Genet. Genomics 275, 421–430 10.1007/s00438-006-0103-2 [DOI] [PubMed] [Google Scholar]
- Bennett M. D., Leitch I. J. (1995). Nuclear-DNA amounts in angiosperms. Ann. Bot. 76, 113–176 10.1006/anbo.1995.1085 [DOI] [Google Scholar]
- Bennett M. D., Leitch I. J. (1997). Nuclear DNA amounts in angiosperms – 583 new estimates. Ann. Bot. 80, 169–196 10.1006/anbo.1997.0415 [DOI] [Google Scholar]
- Bennett M. D., Smith J. B. (1991). Nuclear-DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 334, 309–345 10.1098/rstb.1991.0120 [DOI] [PubMed] [Google Scholar]
- Brandes A., Roder M. S., Ganal M. W. (1995). Barley telomeres are associated with 2 different types of satellite DNA sequences. Chromosome Res. 3, 315–320 10.1007/BF00713070 [DOI] [PubMed] [Google Scholar]
- Broughton W. J., Hernandez G., Blair M., Beebe S., Gepts P., Vanderleyden J. (2003). Beans (Phaseolus spp.) – model food legumes. Plant Soil 252, 55–128 10.1023/A:1024179717780 [DOI] [Google Scholar]
- Buzek J., Koutnikova H., Houben A., Riha K., Janousek B., Siroky J., et al. (1997). Isolation and characterization of X chromosome-derived DNA sequences from a dioecious plant Melandrium album. Chromosome Res. 5, 57–65 10.1023/A:1011693603279 [DOI] [PubMed] [Google Scholar]
- Chen C. M., Wang C. T., Wang C. J., Ho C. H., Kao Y. Y., Chen C. C. (1997). Two tandemly repeated telomere-associated sequences in Nicotiana plumbaginifolia. Chromosome Res. 5, 561–568 10.1023/A:1018449920968 [DOI] [PubMed] [Google Scholar]
- Chen N. W. G., Sevignac M., Thareau V., Magdelenat G., David P., Ashfield T., et al. (2010). Specific resistances against Pseudomonas syringae effectors AvrB and AvrRpm1 have evolved differently in common bean (Phaseolus vulgaris), soybean (Glycine max), and Arabidopsis thaliana. New Phytol. 187, 941–956 10.1111/j.1469-8137.2010.03337.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z. K., Stupar R. M., Gu M. H., Jiang J. M. (2001). A tandemly repeated DNA sequence is associated with both knob-like heterochromatin and a highly decondensed structure in the meiotic pachytene chromosomes of rice. Chromosoma 110, 24–31 10.1007/s004120000126 [DOI] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. C. E., Mau B., Blattner F. R., Perna N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David P., Chen N. W. G., Pedrosa-Harand A., Thareau V., Sevignac M., Cannon S. B., et al. (2009). A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol. 151, 1048–1065 10.1104/pp.109.142109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechyeva D., Schmidt T. (2006). Molecular organization of terminal repetitive DNA in Beta species. Chromosome Res. 14, 881–897 10.1007/s10577-006-1096-8 [DOI] [PubMed] [Google Scholar]
- Eddy S. R. (1998). Hidden Markov models and genome sequence analysis. FASEB J. 12, A1327–A1327 [Google Scholar]
- Edgar R. C. (2004a). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004b). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1989). PHYLIP – Phylogeny Inference Package (Version 3.2). Cladistics 5, 164–166 [Google Scholar]
- Fonseca A., Ferreira J., Dos Santos T. R., Mosiolek M., Bellucci E., Kami J., et al. (2010). Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosome Res. 18, 487–502 10.1007/s10577-010-9129-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P. F., Armstrong S., De Jong J. H., Parnell L. D., Van Drunen G., Dean C., et al. (2000). Integrated cytogenetic map of chromosome arm 4S of A-thaliana: a structural organization of heterochromatic knob and centromere region. Cell 100, 367–376 10.1016/S0092-8674(00)80672-8 [DOI] [PubMed] [Google Scholar]
- Freitas-Junior L. H., Bottius E., Pirrit L. A., Deitsch K. W., Scheidig C., Guinet F., et al. (2000). Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P-falciparum. Nature 407, 1018–1022 10.1038/35039531 [DOI] [PubMed] [Google Scholar]
- Freitas-Junior L. H., Hernandez-Rivas R., Ralph S. A., Montiel-Condado D., Ruvalcaba-Salazar O. K., Rojas-Meza A. P., et al. (2005). Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121, 25–36 10.1016/j.cell.2005.01.037 [DOI] [PubMed] [Google Scholar]
- Geffroy V., Macadre C., David P., Pedrosa-Harand A., Sevignac M., Dauga C., et al. (2009). Molecular analysis of a large subtelomeric Nucleotide-Binding-Site-Leucine-Rich-Repeat family in two representative genotypes of the major gene pools of Phaseolus vulgaris. Genetics 181, 405–419 10.1534/genetics.108.093583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia M., Gonzalez-Sanchez M., Puertas M. J. (2006). The high variability of subtelomeric heterochromatin and connections between nonhomologous chromosomes, suggest frequent ectopic recombination in rye meiocytes. Cytogenet. Genome Res. 115, 179–185 10.1159/000095240 [DOI] [PubMed] [Google Scholar]
- He L., Liu J., Torres G. A., Zhang H., Jiang J., Xie C. (2013). Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosome Res. 21, 5–13 10.1007/s10577-012-9332-x [DOI] [PubMed] [Google Scholar]
- Heacock M., Spangler E., Riha K., Puizina J., Shippen D. E. (2004). Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 23, 2304–2313 10.1038/sj.emboj.7600236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes R. W., Ameline-Torregrosa C., Ashfield T., Cannon E., Cannon S. B., Chacko B., et al. (2008). Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol. 148, 1740–1759 10.1104/pp.108.127902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. A., Iwata A., Lee S. H., Schmutz J., Shoemaker R. (2011). Sequencing crop genomes: approaches and applications. New Phytol. 191, 915–925 10.1111/j.1469-8137.2011.03804.x [DOI] [PubMed] [Google Scholar]
- Jin W. W., Lamb J. C., Vega J. M., Dawe R. K., Birchler J. A., Jiang J. (2005). Molecular and functional dissection of the maize B chromosome centromere. Plant Cell 17, 1412–1423 10.1105/tpc.104.030643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenton A., Parokonny A. S., Gleba Y. Y., Bennett M. D. (1993). Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics. AIDS Res. Hum. Retroviruses 240, 159–169 [DOI] [PubMed] [Google Scholar]
- Kuo H. F., Olsen K. M., Richards E. J. (2006). Natural variation in a subtelomeric region of Arabidopsis: implications for the genomic dynamics of a chromosome end. Genetics 173, 401–417 10.1534/genetics.105.055202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapitan N. L. V., Ganal M. W., Tanksley S. D. (1989). Somatic chromosome karyotype of tomato based on in situ hybridization of the TGRI satellite repeat. Genome 32, 992–998 10.1139/g89-543 [DOI] [Google Scholar]
- Lee H. R., Zhang W. L., Langdon T., Jin W. W., Yan H. H., Cheng Z. K., et al. (2005). Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. U.S.A. 102, 11793–11798 10.1073/pnas.0507651102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang F., Zhu J., He S., Li L. (2009). Characterization of a tandemly repeated subtelomeric sequence with inverted telomere repeats in maize. Genome 52, 286–293 10.1139/G09-005 [DOI] [PubMed] [Google Scholar]
- Linardopoulou E. V., Williams E. M., Fan Y. X., Friedman C., Young J. M., Trask B. J. (2005). Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437, 94–100 10.1038/nature04029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E. J. (1995). The chromosome ends of Saccharomyces cerevisiae. Yeast 11, 1553–1573 10.1002/yea.320111604 [DOI] [PubMed] [Google Scholar]
- Ma J. X., Jackson S. A. (2006). Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Res. 16, 251–259 10.1101/gr.4583106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D., Rybicki E. (2000). RDP: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563 10.1093/bioinformatics/16.6.562 [DOI] [PubMed] [Google Scholar]
- Martin D. P., Posada D., Crandall K. A., Williamson C. (2005a). A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retroviruses 21, 98–102 10.1089/aid.2005.21.98 [DOI] [PubMed] [Google Scholar]
- Martin D. P., Williamson C., Posada D. (2005b). RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21, 260–262 10.1093/bioinformatics/bti671 [DOI] [PubMed] [Google Scholar]
- Mefford H. C., Trask B. J. (2002). The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 3, 91–102 10.1038/nrn725 [DOI] [PubMed] [Google Scholar]
- Mendez-Lago M., Wild J., Whitehead S. L., Tracey A., De Pablos B., Rogers J., et al. (2009). Novel sequencing strategy for repetitive DNA in a Drosophila BAC clone reveals that the centromeric region of the Y chromosome evolved from a telomere. Nucleic Acids Res. 37, 2264–2273 10.1093/nar/gkp085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H., Wu J. Z., Kanamori H., Fujisawa M., Namiki N., Saji S., et al. (2006). Sequencing and characterization of telomere and subtelomere regions in rice chromosomes 1S, 2S, 2L, 6L, 7S, 7L and 8L. Plant J. 46, 206–217 10.1111/j.1365-313X.2006.02684.x [DOI] [PubMed] [Google Scholar]
- Mizuno H., Wu J. Z., Katayose Y., Kanamori H., Sasaki T., Matsumoto T. (2008). Characterization of chromosome ends on the basis of the structure of TrsA subtelomeric repeats in rice (Oryza sativa L.). Mol. Genet. Genomics 280, 19–24 10.1007/s00438-008-0341-6 [DOI] [PubMed] [Google Scholar]
- Ohmido N., Fukui K. (1997). Visual verification of close disposition between a rice A genome-specific DNA sequence (TrsA) and the telomere sequence. Plant Mol. Biol. 35, 963–968 10.1023/A:1005822504690 [DOI] [PubMed] [Google Scholar]
- Ohno S. (1972). So much “junk” DNA in our genome. Brookhaven Symp. Biol. 23, 366–370 [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. C. (1980). Selfish DNA: the ultimate parasite. Nature 284, 604–607 10.1038/284604a0 [DOI] [PubMed] [Google Scholar]
- Padidam M., Sawyer S., Fauquet C. M. (1999). Possible emergence of new geminiviruses by frequent recombination. Virology 265, 218–225 10.1006/viro.1999.0056 [DOI] [PubMed] [Google Scholar]
- Pastor-Corrales M. A., Tu J. C. (1989). “Anthracnose,” in Bean Production Problems in the Tropics, 2nd Edn, eds Schwartz H. F., Pastor-Corrales M. A. (Cali: Centro Internacional de Agricultura Tropical (CIAT)), 77–104 [Google Scholar]
- Pedrosa-Harand A., Kami J., Gepts P., Geffroy V., Schweizer D. (2009). Cytogenetic mapping of common bean chromosomes reveals a less compartmentalized small-genome plant species. Chromosome Res. 17, 405–417 10.1007/s10577-009-9031-4 [DOI] [PubMed] [Google Scholar]
- Plohl M., Luchetti A., Mestrovic N., Mantovani B. (2008). Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409, 72–82 10.1016/j.gene.2007.11.013 [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K. A. (2001). Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. U.S.A. 98, 13757–13762 10.1073/pnas.241370698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2-approximately Maximum-Likelihood trees for large alignments. PLoS ONE 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- Schlueter J. A., Goicoechea J. L., Collura K., Gill N., Lin J.-Y., Yu Y., et al. (2008). BAC-end Sequence analysis and a draft physical map of the common bean (Phaseolus vulgaris L.) genome. Trop. Plant Biol. 1, 40–48 10.1007/s12042-007-9003-9 [DOI] [Google Scholar]
- Song J. Q., Dong F. G., Lilly J. W., Stupar R. M., Jiang J. M. (2001). Instability of bacterial artificial chromosome (BAC) clones containing tandemly repeated DNA sequences. Genome 44, 463–469 10.1139/g01-029 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular Evolutionary Genetics Analysis using Maximum Likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tek A. L., Jiang J. M. (2004). The centromeric regions of potato chromosomes contain megabase-sized tandem arrays of telomere-similiar sequence. Chromosoma 113, 77–83 10.1007/s00412-004-0297-1 [DOI] [PubMed] [Google Scholar]
- Torres G. A., Gong Z. Y., Iovene M., Hirsch C. D., Buell C. R., Bryan G. J., et al. (2011). Organization and evolution of subtelomeric satellite repeats in the potato genome. G3 (Bethesda) 1, 85–92 10.1534/g3.111.000125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R. K., Ribaut J. M., Buckler E. S., Tuberosa R., Rafalski J. A., Langridge P. (2012). Can genomics boost productivity of orphan crops? Nat. Biotechnol. 30, 1172–1176 10.1038/nbt.2440 [DOI] [PubMed] [Google Scholar]
- Vershinin A. V., Schwarzacher T., Heslopharrison J. S. (1995). The large-scale genomic organization of repetitive DNA families at the telomeres of rye chromosomes. Plant Cell 7, 1823–1833 10.1105/tpc.7.11.1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Khipu sequence alignement.
WebLogo representation of the consensus sequence derived from the multiple alignment of the khipu units and sequence of one representative khipu unit from each major clade.
Phylogenetic tree of whole-genome khipu sequence.