Abstract
Sirtuins are a conserved family of deacetylases whose activities are dependent on nicotinamide adenine dinucleotide (NAD+). Sirtuins act in different cellular compartments, such as the nucleus where they deacetylate histones and transcriptional factors, in the cytoplasm where they modulate cytoskeletal and signaling molecules, and in the mitochondria where they engage components of the metabolic machinery. Collectively, they tune metabolic processes to energy availability, and modulate stress responses, protein aggregation, inflammatory processes, and genome stability. As such, they have garnered much interest and have been widely studied in aging and age-related neurodegeneration. In this chapter, we review the identification of sirtuins and their biological targets. We focus on their biological mechanisms of action and how they might be regulated, including via NAD metabolism, transcriptional and posttranscriptional control, and as targets of pharmacological agents. Lastly, we highlight the numerous studies suggesting that sirtuins are efficacious therapeutic targets in neurodegenerative disease and injury.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0214-5) contains supplementary material, which is available to authorized users.
Keywords: Sirtuin, SIRT, Neurodegenerative disease, Neuroprotection, Therapeutic targets
Introduction
Proteins may undergo posttranslational modifications to alter their activity, interaction with other proteins, or structural conformation. One such modification is the acetylation of lysine residues, mediated by the enzymatic activities of acetyltransferases [1, 2]. In contrast to acetylation, the removal of these acetyl groups, deacetylation, is carried out by a group of enzymes called deacetylases. This group of deacetylases is also referred to as histone deacetylases (HDACs) due to their capacity for deacetylating histone proteins, predominantly the ε-amino groups of specific lysines in N-terminal domains of histone H3 and histone H4 [3, 4]. The deacetylation of histone proteins within chromatin generally correlates with a more compacted and transcriptionally silent state [5, 6].
Deacetylases are classified into four families in mammals, many of which are conserved from yeast to humans. Class I HDACs are ubiquitously expressed and have high sequence similarity to yeast Rpd3, and include HDAC-1, HDAC-2, HDAC-3 and HDAC-8. Class II HDACs are expressed primarily in striated muscle and brain, are similar to yeast Hda1, and include HDAC-4, HDAC-5, HDAC-6, HDAC-7 and HDAC9 [3, 4]. Class III contains the sirtuin family of HDACs. They are similar to yeast Sir2, and include SIRT1 to SIRT7 [7]. The last group is class IV, which contains the solitary member, HDAC 11 [8]. The active sites of class I, II, and IV HDACs have a high degree of sequence similarity, and have a common zinc-dependent mechanism for deacetylation. In contrast to this, the class III HDACs, or sirtuins, differ in that they require nicotinamide adenine dinucleotide (NAD+) for catalysis [9]. As such, they are also referred to as the NAD+-dependent histone deacetylases.
Sirtuins were first identified in Saccharomyces cerevisiae as genetic silencing factors in which they were found to participate in heterochromatic silencing at mating-type loci [10, 11]. Later, in longevity studies, it was discovered that the silent information regulation (Sir) genes, particularly Sir2, were determinants of calorie induced replicative lifespan extension in S. cerevisiae, and that increased Sir2 or loss of Sir2 extend or reduced lifespan in wild-type cells, respectively [12, 13]. The role of Sir2 in this action was linked to its ability to suppress recombination in the rDNA region and consequently reduce the formation of extrachromosomal rDNA circles [12, 13]. In parallel, Sir2 was found to be NAD+-dependent deacetylase for histone proteins, consistent with its capacity for gene silencing, recombination suppression, and life span extension [14]. Given that calorie restriction induced lifespan elongation and appears to be a phylogenetically conserved phenomenon, observed in yeast, worm, fruit fly and rodents, these early findings have generated intense interest and research effort [12, 13, 15–17].
As discussed, seven sirtuins have been identified to date in mammals: SIRT1–SIRT7. Together they share significant homology in structure, particularly in their highly conserved catalytic and NAD+-binding domains [7]. Despite this, they have very distinct enzymatic activities, expression patterns, cellular localizations, and biological function (Table 1). SIRT1 is robust deacetylase that contains both nuclear localization and export sequences. It is predominantly localized in the nucleus, though studies have reported it as a nucleocytoplasmic shuttling protein [18–20]. Although the physiological relevance of this shuttling is unclear, it is possible that SIRT1 has either important cytosolic targets or that shuttling is another level of control on nuclear target proteins. SIRT2 is a robust deacetylase with a cytosolic localization [21–23]. However, it has been identified in the nucleus during the G2 to M phase transition of the cell cycle [22]. SIRT3, SIRT4 and SIRT5 are all mitochondrial sirtuins containing mitochondrial-targeting sequences [24–28]. Of these, SIRT3 is a robust deacetylase [28, 29], whereas SIRT4 and SIRT5 display weak deacetylase activity on substrates tested so far. Rather, SIRT4 appears to be better ADP-ribosyltransferases [30], and SIRT5 a very effective demalonylase and desuccinylase [24, 31]. SIRT6 and SIRT7 are both predominantly localized in the nucleus [32–34]. Both display weak deacetylase activity, and SIRT6, like SIRT4, displays ADP-ribosyltransferase activity [32, 33].
Table 1.
Mammalian sirtuin localization and function
| Sirtuin | Localization | Activity | Examples of targets |
|---|---|---|---|
| SIRT1 | Nucleus | Deacetylase | Histones [35], FoxO [36–38], PGC1α [39], p53 [41, 42], p300 [42], etc. |
| SIRT2 | Cytoplasm | Deacetylase | α-Tubulin [23], FoxO [46, 47], PEPCK1 [49] |
| SIRT3 | Mitochondria | Deacetylase | ACS2 [50], LCA [52], HMGCS2 [53], GDH [31], IDH2 [54], Ornithine transcarbamylase [50] |
| SIRT4 | Mitochondria | ADP-ribosyltransferase | GDH [25] |
| SIRT5 | Mitochondria | Demalonylase desuccinylase | CPS1 [27] |
| SIRT6 | Nucleus | Weak deacetylase ADP-ribosyltransferase | Histones [57], PARP1 [59], Hif1α [60] |
| SIRT7 | Nucleus | Weak deacetylase | Histones [62], p53 [61], Hif1α and Hif2α [63] |
Biological Targets of Sirtuin Activity
SIRT1
SIRT1 is the closest homolog of yeast Sir2, and the most-studied member of the mammalian sirtuin family. While it was originally described to deacetylate histone proteins [35], many non-histone targets have been identified for SIRT1, including forkhead box O (FoxO) transcription factors [36–38], peroxisome proliferator-activated receptor-γ co-activator 1α (PGC1α) [39], p53 [40, 41], and p300 [42]. Collectively, the enzymatic deacetylating activity of SIRT1 on these substrates appears to be important for metabolic adjustment, survival promotion, genome stability and autophagy, all of which are consistent with longevity. For example, both calorie restriction and cellular stress have been shown to facilitate binding, and subsequent deacetylation of FoxO3a by SIRT1 [36–38]. FoxO transcription factors are downstream targets of the insulin-phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathway and are important regulators of cell fate, due to their ability to control proliferation, differentiation, apoptosis, DNA repair, and defense against oxidative stress [43]. However, findings suggest that SIRT1-mediated deacetylation does not just activate or inhibit the FoxO3a but selectively directs it to certain targets, thus functions of FoxO3a, such as cell-cycle arrest and oxidative-stress resistance, can be enhanced while other functions, such as cell death are inhibited [36–38]. Another target of SIRT1 is the transcription factor PGC1α. SIRT1 deacetylation of PGC1α leads to its activation and to the induction of mitochondrial gene expression and downstream pathways that enhance mitochondrial biogenesis and activity [39, 44].
Several studies using mouse models provide evidence that SIRT1 may improve genetic stability and suppress tumor formation. SIRT1 null mice die perinatally due to developmental defects including chromosome abnormalities. The cause behind this genomic instability has been attributed to a defective signaling of DNA double-strand breaks (DSBs) since SIRT1 deacetylation of the repair factor NBS1 is a modification required for its subsequent phosphorylation by the Ataxia Telangiectasia Mutated (ATM) kinase in the first steps of the DNA damage response [45–47]. Furthermore, SIRT1 appears to be involved in multiple DNA-repair pathways, including homologous recombination (HR) repair or non-homologous end joining (NHEJ) of double strand breaks, and nucleotide excision repair (NER) of DNA single strand breaks [48–50]. The tumor suppressor, p53, which is induced in response to cellular stress, including oxidative and DNA damage, is also a target of SIRT1. In coordination with other cellular targets, SIRT1 can deacetylate p53, repressing its function, reducing its proapoptotic effects and promoting survival [40, 41]. Thus, it appears that SIRT1 has evolved to coordinate both proper genomic integrity and adequate metabolic adaptation, in this way allowing cells to adapt against stress.
SIRT2
In mammals, SIRT2 is primarily present in the cytoplasm, where it co-localizes with microtubules and deacetylates the major component of microtubules, α-tubulin at lysine 40 [23]. However, SIRT2 has been found to transiently migrate to the nucleus during G2/M transition and deacetylate histone H4 at lysine 16, thereby modulating chromatin condensation during metaphase [51]. SIRT2, like SIRT1, can also deacetylate the FoxO transcription factors, FoxO1 and FoxO3a [52, 53], potentially linking its multiple cellular processes, including DNA repair, cell cycle, apoptosis, metabolism and ageing [54]. Another metabolic target of SIRT2 is phosphoenolpyruvate carboxykinase 1 (PEPCK1), the deacetylation of which prevents its ubiquitylation-dependent degradation [55]. Given that PEPCK1 is the rate-limiting enzyme in gluconeogenesis, deacetylation by SIRT2 serves to modulate cellular responses to glucose. SIRT2 has both roles in tumor suppression and promotion. SIRT2 is reported to be a tumor suppressor in several cancers [56, 57]. Direct support of SIRT2’s tumor suppressor role is demonstrated in studies showing that SIRT2 knock-out mice develop tumors in various organs due to abnormal chromosomal segregation and aneuploidy caused by increased expression of mitotic regulators including aurora kinases [58]. However, SIRT2 may promote oncogenic phenotypes. SIRT2 is increased in acute myeloid leukemia cells compared with normal bone marrow cells, and SIRT2 inhibition causes apoptosis of acute myeloid leukemia cells in vitro [59].
SIRT3, 4 and 5
SIRT3, SIRT4 and SIRT5 localize to, and function, primarily in the mitochondria. SIRT3 was first discovered to deacetylate and activate acetyl-CoA synthetase 2 (ACS2) [60], an enzyme that converts free acetate to acetyl-CoA, which can be oxidized in the citric acid cycle (TCA cycle) to produce energy. Subsequently, SIRT3 has been found to engage and regulate multiple metabolic components of the TCA cycle, fatty acid oxidation, and oxidative phosphorylation [61]. In particular, it has been shown to deacetylate and activate long-chain acyl–CoA dehydrogenase (LCAD) [62], 3-hydroxy-3-methylglutaryl–CoA synthase 2 (HMGCS2) [63], and glutamate dehydrogenase (GDH) [31]. As a result of SIRT3 deacetylation, LCAD, HMGCS2, and GDH promote fatty-acid oxidation, the formation of ketone body by-products from acetyl-CoA, and α-ketoglutarate from the amino acid glutamate, respectively [31, 62, 63]. SIRT3 also deacetylates superoxide dismutase-2 (SOD2), which is important for oxygen detoxification [64], and mitochondrial matrix protein isocitrate dehydrogenase 2 (IDH2), which through its oxidative decarboxylation of isocitrate to α-ketoglutarate is a major source of NADPH [65, 66]. Through these functions, SIRT3 is suggested to suppress reactive oxygen species (ROS) production, which promotes oxygen-dependent prolyl-hydroxylase domain-containing enzyme (PHD) activity and destabilizes the hypoxia-inducible factor, HIF1α [67]. SIRT3 is also important for the deacetylation and activation of ornithine transcarbamylase, a key component of the urea cycle, which is important for preventing the toxic build-up of ammonia from amino acid catabolism [60]. Thus taken together, it is hypothesized that SIRT3 is important for promoting metabolic processes and detoxification that are characteristic of a fasting state when activated by low nutrient levels.
SIRT4 is another mitochondrial sirtuin that primarily appears to have functions in metabolism. The first role identified for SIRT4 was the ADP-ribosylation of GDH, which represses its enzymatic activity and limits the metabolism of glutamate and glutamine to generate ATP [25]. Consistent with glutamate and glutamine promoting pancreatic β cells secretion of insulin via metabolism by GDH, the deletion of SIRT4 in mice results in a loss of regulated insulin secretion [25]. Thus, it is hypothesized that ADP-ribosylation of GDH may coordinate amino acid metabolism in different tissues with changes in diet. SIRT4 has also been reported to play a role in the regulation mitochondrial oxidative metabolism in hepatocytes and myocytes. In a study by Nasrin et al. [68], the genetic knockdown of SIRT4 was associated with the enhanced expression of genes that control fatty acid oxidation and mitochondrial oxidative capacity. However, this may be SIRT1 and SIRT3 mediated as both are increased with SIRT4 knockdown, and fatty acid oxidation enhancement is lost when SIRT1 is inhibited. Recent findings suggest that SIRT4 also plays a tumor suppressor role. SIRT4 is induced by genotoxic stress and is required for the repression of mitochondrial glutamine metabolism which contributes to the control of cell cycle progression and the maintenance of genomic integrity in response to DNA damage [69]. Indeed, loss of SIRT4 increased glutamine-dependent tumor cell proliferation and tumorigenesis. In mice, SIRT4 loss resulted in spontaneous tumor development [69].
So far, the only target described for SIRT5 deacetylase activity is carbamoyl phosphate synthetase 1 (CPS1), which is deacetylated and thereby activated to regulate entry into the urea cycle and promote ammonia detoxification during amino acid catabolism [27]. Given SIRT5’s weak deacetylase activity it is possible that it does not primarily act as a deacetylase for CPS1, but rather a desuccinylase [24]. Indeed, pyruvate dehydrogenase complex (PDC) and succinate dehydrogenase (SD) have been determined to be lysine desuccinylation targets of SIRT5, a modification that suppresses their biochemical activities and, consequently, mitochondrial respiration driven by these complexes [70].
SIRT6
Studies have demonstrated a link between SIRT6 expression and longevity, in which the overexpression of SIRT6 extends the lifespan of male mice [71]. In this study, SIRT6 overexpression lowered serum levels of insulin-like growth factor 1 (IGF1) and increased the expression of insulin-like growth factor-binding protein 1 in male mice, bringing the values closer to those observed in control female mice [71]. Our understanding of SIRT6 function also comes from its deletion in mice, which results in genomic instability, metabolic defects and degenerative pathologies associated with aging [34, 72]. SIRT6 mainly resides in the nucleus where it has been shown to specifically bind to the telomeric chromatin and deacetylate histone H3 at lysines 9 to modulate telomeric chromatin structure [73]. Loss of SIRT6 can lead to the dysfunction of telomeres similar to that of Werner syndrome, and can result in chromosome end fusion and cellular senescence [73]. With regard to DNA damage repair, SIRT6 has been implicated in the regulation of base excision repair (BER) either by modulating BER factors or regulating the density of chromatin thereby changing the accessibility of DNA damage sites to BER factors [34]. Studies suggest SIRT6 also impacts DNA double-strand break (DSB) repair by stabilizing DNA dependent protein kinase on the chromatin and facilitate its joining [74]. Lastly, SIRT6 has been shown to mono-ADP-ribosylate PARP1, thereby activating PARP1’s poly-ADP-ribosylase activity and enhancing DSB repair under conditions of oxidative stress [75]. In contrast to genome stability and DNA damage repair, studies on the metabolic defects in SIRT6 knockout mice have led to the proposal that SIRT6 functions as a co-repressor of Hif1α transcriptional activity, where it can deacetylate histone H3 at lysines 9 and 56 at Hif1α target gene promoters [76]. Thus, SIRT6, like SIRT3, is suggested to be a negative regulator of the hypoxia response pathway.
SIRT7
SIRT7 is arguably the least studied of the mammalian sirtuins. It has been reported to activate RNA polymerase I transcription, although the protein substrate for this action is still unknown [32]. It has been shown to interact with and deacetylate p53 in vitro, which has been suggested to correspond to the hyperacetylation of p53, cardiac hypertrophy, and inflammatory cardiomyopathy evident in mice that lack SIRT7 [77]. More recently it has been identified as a deacetylase of histone H3 lysine residue 18, important for stabilizing the transformed state of cancer cells [78]. Lastly, SIRT7 has been identified as a negative regulator of Hif1α and Hif2α, suggesting that, like SIRT6, SIRT7 may be a regulator of the hypoxia response pathway. Interestingly, the mechanism of action for this regulation appears to be independent of SIRT7’s catalytic activity [79].
Sirtuin Biochemical Mechanism
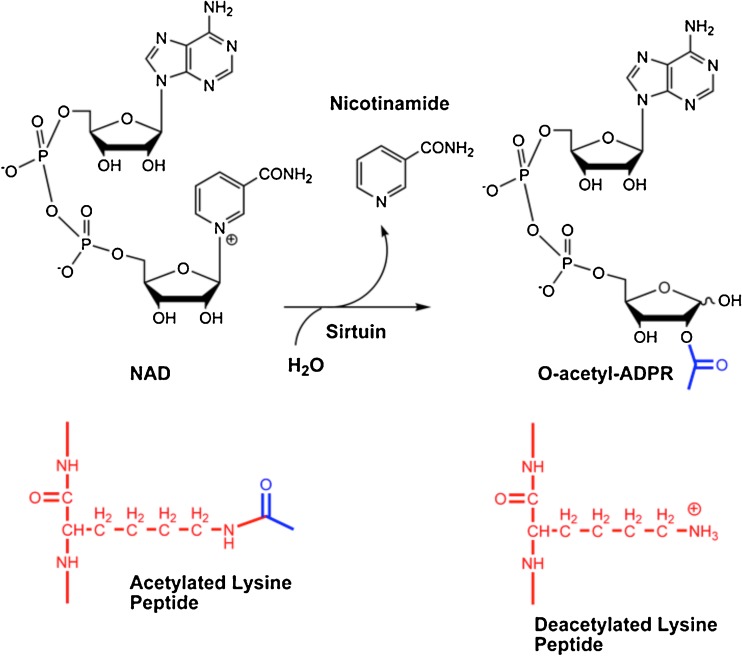
Sirtuins are NAD+-dependent deacetylases. Unlike class I, II and IV deacetylases, which are zinc dependent and deacetylate proteins by activation of a water molecule that hydrolyzes the acetyl-amide bond to form acetate in a reaction that is thermodynamically highly favorable [9], sirtuins catalyze NAD+-dependent deacetylation of acetyllysine, resulting in the production of deacetylated lysine, nicotinamide, and a third product 2′-O-acetyl-ADP-ribose (OAADPr) (Scheme 1), an unusual species, and still largely mysterious [80]. Moazed et al. showed this compound is a stabilizer of heterochromatin assemblies in yeast [81], with the caveat that yeast heterochromatin is not evolutionarily conserved, and so functions in humans are less clear. The possible roles of AADPR have been reviewed recently by Denu [82]. This complex chemistry, featuring three reactants and three products, is highly biologically conserved, and organisms widely distributed across all phyla of life encode sirtuins in their genomes. The production of O-acetyl-ADPR has been noted to be catalyzed by sirtuins from Archaea [83], eubacteria [83], protozoa, yeast [80, 84] and mammals [83].
Scheme 1.
Depiction of stoichiometry of sirtuin catalyzed deacetylation
Recent work has identified additional substrates for sirtuins, including acylated substrates bearing side chains such as propionyl [85], butyryl [85], succinyl [86], malonyl [86] and myristoyl [87]. These modifications hint at broad spectra of acylations with potential regulatory impact in biologic significance, beyond those already well known, such as acetylation.
Regulation of Sirtuins by NAD Metabolism
The biochemical requirement of NAD+ by sirtuin enzymes provides a basic source of sirtuin activity regulation, through modulating NAD level. The premise that sirtuin activity regulation is dependent on NAD levels was first established in yeast, wherein genetic modifications of the NAD biosynthetic pathway that altered NAD levels, affected measurable biological activities associated with Sir2 [88, 89]. For example, reduced NAD levels caused phenotypic changes in yeast, which included a loss of gene silencing at heterochromatin [88], increased recombination frequency, and decreased replicative lifespan [88, 90]. As discussed in this section, the notion that sirtuins are regulated by NAD metabolism in general and appears conserved across a wide breadth of phylogenetic space is broadly supported by a wealth of experimental data.
The key dynamics of NAD metabolism that affect sirtuin activity are changes in NAD and nicotinamide levels [91–93]. NAD is the reactant of the sirtuin reaction and provides stimulation via increased probability of maintenance of the Michaelis complex. Nicotinamide is a product of sirtuin chemistry [83], and causes decreased activity of sirtuins via chemical reaction with the first intermediate formed on the enzyme from NAD, called an ADPR-peptidyl imidate [80]. This intermediate is formed by a chemical reaction of NAD with an acetyllysine substrate bound on the enzyme active site [94].
Evidence that nicotinamide affects sirtuin function was first obtained in the yeast, where the overexpression of an enzyme called nicotinamidase (pnc1), known to convert nicotinamide to nicotinic acid, caused increased replicative lifespan and increased gene silencing [92]. Conversely, deletion of pnc1 caused loss of replicative lifespan and reduced gene silencing in heterochromatin, consistent with a negative effect on Sir2 catalytic function in yeast [91, 95, 96]. Further, Sauve et al. have shown by isotope dilution and mass-spectrometry that nicotinamide levels increase 10-fold in pnc1Δ yeast [97]. Consistently, nicotinamide added exogenously to yeast causes a loss of gene silencing and replicative lifespan [91].
In mammalian systems there appears to be an analogous sensitivity of sirtuins to NAD levels and NAD metabolism. First, sirtuins such as SIRT1 have K m values in the 100–500 μM range [98], consistent with regulation by physiological NAD levels, which are reportedly 300 μM in several rodent tissues [93, 99, 100]. This biochemical data predicts that NAD modulation would be able to biochemically alter sirtuin signaling strength. Indeed, numerous studies now support this point of view. For example, PARP1 knockouts cause systemic NAD increases in animals, suggesting that PARP1 competes for a limited NAD pool [99]. The increase in NAD concentration in tissues is associated with increased SIRT1 activity and correlates with decreased FoxO1 and PGC1α acetylation [99]. More recently, it has been shown that the small molecule NAD precursor nicotinamide riboside can enhance NAD levels in several peripheral tissues including muscle [100]. This increased NAD is associated with decreased FoxO1 and PGC1α, acetylation, and consistent with an observed increased mitochondrial activity [100]. In this study, a decreased acetylation of SOD2 is also noted, suggesting an increased SIRT3 activity [100]. These observations are consistent with increased NAD levels in mitochondria isolated from tissue.
Less is known about how nicotinamide concentrations affect the activities of sirtuins in mammalian cells. Nicotinamide is a fairly potent inhibitor of SIRT1 and SIRT2, as determined from reported biochemical data, and consequently, has been widely applied to provide pharmacological sirtuin inhibition. However, data on nicotinamide inhibition of sirtuin isoforms is rather limited beyond SIRT1 and SIRT2, and it is currently unclear how effective nicotinamide is as an inhibitor of the sirtuin isoforms SIRT3–7. On the other hand, evidence reported from multiple studies suggests that endogenous nicotinamide concentrations are likely to provide physiological regulation of sirtuin isoforms, particularly SIRT1 and SIRT2. Work done by the Sauve laboratory, for example, has not only provided Ki values for SIRT1 [101], but also nicotinamide concentrations in brain ranging from 100 to 300 μM [93]. This concentration range is high enough to cause inhibition of sirtuin isoforms. Nevertheless, the effects of nicotinamide as an endogenous repressor of sirtuin activity have been difficult to quantify.
Regulation of Sirtuins by Transcriptional and Posttrancriptional Control
There is evidence that sirtuins are not only regulated at the level of metabolism but also at the transcriptional and possibly translational levels. Early speculations conceived that human sirtuins might positively respond to low calorie stress, such as those elicited by fasting. This hypothesis has been confirmed for several sirtuin isoforms, including SIRT1, SIRT3, SIRT5 and SIRT6. The transcriptional upregulation of sirtuin isoforms has encouraged further studies to determine if sirtuin isoforms are key to cellular and tissue specific adaptations associated with low calorie diets.
The transcriptional upregulation of SIRT1 has been noted to be a direct consequence of low calorie conditions or fasting in rodents. This transcriptional upregulation has been observed in several tissues [102], including fat, liver, brain and kidney [93]. It is probable that some of the effects of SIRT1 in a biological setting is driven by absolute SIRT1 abundance, a concept that is supported by numerous biological studies. For instance, overexpression of SIRT1 via expression vectors in cell-based systems shows that SIRT1 activity is typically limiting, and increasing SIRT1 abundance is key for its biological effects. These dosage effects associated with SIRT1 amount, as well as evidence that its biological activity is typically subsaturated under many physiologic conditions, has stimulated efforts to target SIRT1 with small molecules that could stimulate SIRT1 activity ([103, 104]; and discussed below).
At the level of posttranslational modification, Puigserver et al. [105] identified a key serine phosphorylation site on SIRT1 (S434), which is sensitive to β-adrenergic signaling. This modification leads to an apparent increase in SIRT1 activity via decreased Km (55 % decrease) and increased Vmax (250 % increase) [105]. Conversely, another study has suggested that a threonine residue (T344) within SIRT1 can be phosphorylated through the activation of AMPKinase signaling, which leads to its inactivation [106]. The mechanisms by which these phosphorylations occur are still unclear, although it appears that independent kinases could be involved. For the SIRT1 S434 site, at least, Gerhart-Hines et al. has suggested that cAMP is crucial for establishing phosphorylation, and it can be stimulated by clenbuterol, forskolin or Br-cAMP [105]. This activating modification suggests that SIRT1 activity may normally be reduced from a maximal level, and higher levels can be unleashed by key structural changes to the SIRT1 protein. Indeed, other authors have reported that sumoylation of SIRT1 on Lys 734 can modify SIRT1 enzymatic activity [107].
Several studies have noted that SIRT3 levels are increased at the transcriptional level by CR [108, 109] implying that SIRT3 transcription is responsive to nutrition and consistent with its role of metabolic adaption. Among these is an apparent requirement of SIRT3 in CR-induced ROS detoxification, as measured by glutathione GSSH/GSSG ratio and by accumulation of 4-hydroxy-2-nonenal. As discussed, downstream of CR-induced SIRT3 are the deacetylation targets SOD2, which are important for oxygen detoxification [64], and the mitochondrial matrix protein IDH2, which through its oxidative decarboxylation of isocitrate to α-ketoglutarate is a major source of NADPH [65].
Sirtuins 5 and 6 are less well characterized than SIRT1 or SIRT3, and less is known about their biological functions, including how they are regulated. Nevertheless, both SIRT5 [110] and SIRT6 [111] are sensitive to CR conditions and are transcriptionally induced by low calorie stress. SIRT5 is mitochondrial and appears to regulate urea cycle function, via upregulation of the activity of carbamoyl phosphate synthetase 1 (CPS1) [27]. Thus, upregulated SIRT5 activity in liver, during CR stress, accommodates increased physiologic needs for nitrogen disposal, since amino acids become more readily metabolized under these conditions [27]. Mice lacking SIRT5 experience hyperammonemia during fasting, apparently due to failure of the urea cycle to be upregulated appropriately [27].
SIRT6 is a potent repressor of several transcription factor activities central to metabolism, including Hif1α [76, 112]. The fact that Hif1α controls flux through glycolysis and pyruvate dehydrogenase implicates SIRT6 as a central regulator of glucose homeostasis. Thus, a presumptive role of SIRT6 activation during CR may be to reduce glucose demand in cells and tissues in which SIRT6 activity is upregulated.
The role of sirtuins in mediating CR effects is difficult to fully define, and the fact that several mammalian sirtuins are upregulated by CR does not necessarily mean that sirtuins are required for all physiologic effects observed in calorie-restricted animals. This caveat is mentioned here to provide balance to the discussions that follow where effects of CR in neuropathology are described.
Macromolecular Regulators of Sirtuins
Consistent with the view that sirtuin activity is involved in regulation of various parts of transcription and chromatin, it is not surprising that endogenous macromolecular mediators of sirtuin activity have been identified. Among these are macromolecular regulators of SIRT1. DBC1 [113, 114] and AROS [115] have both been tied to SIRT1 activity regulation, with repression or derepression of SIRT1 function featuring in these activities. The presumption that SIRT1 activity is typically suboptimal underscores the importance of these regulators as well as the effects of metabolic or other activating effects on SIRT1 activity.
Pharmacological Control of Sirtuin Activity
The starting point for sirtuin pharmacology can be thought of as nicotinamide and NAD, wherein nicotinamide is a natural inhibitor of sirtuin activity and NAD is a natural activator (see Fig. 1). NADH has also been suggested to be a sirtuin modulator [116]. The role of NADH has been controversial and biochemical studies by the Denu laboratory have suggested that NADH might not directly influence sirtuin activities as physiological concentrations [117]. NAD and nicotinamide are metabolites that are putative pharmacologic endpoints for modulation, which can provide a basis for sirtuin activity manipulation, but of course only in a crude sense. That is because increased NAD or increased nicotinamide provides broad non-specific changes in sirtuin activities, not specific to a single subcellular compartment or sirtuin isoform. A case in point is the small molecule nicotinamide riboside, which reportedly activates both SIRT1 and SIRT3 activities, presumably by increased NAD levels in cytoplasm and mitochondria [100]. Interestingly enough, there is still limited knowledge about how NAD and nicotinamide changes occurring physiologically affect isoform activities in tissues, and this is particularly true in brain, where the effects of NAD metabolism changes might be a fundamental part of the aging process and could participate in the development of various pathologies.
Fig. 1.
Reaction of sirtuin mediated deacetylation and role of NAD metabolites nicotinamide and NAD in modulating chemistry occurring on sirtuin enzymes (discussed in text)
Resveratrol and STACS
Sirtuins are pleiotropic but central regulators of numerous physiologic processes, which are amplified by low calorie stresses and associated with beneficial health effects in mammals, including humans. These findings have underscored the potential behind modulating the sirtuin activities as a means to not only study them, but to evaluate and exploit these enzymes as potentially interesting therapeutic targets.
Considerable attention and controversy has developed around the compounds resveratrol and other so-called Sirtuin Activating Compounds “STACs” [118] as potential activators of sirtuins, particularly SIRT1. This attention has been due largely to very interesting biological effects of these compounds. For example, resveratrol has been shown to have potent effects in mitigating the toxic effects of high fat diet in mice [119]. Two independent laboratories established that resveratrol increases mitochondrial biogenesis in resveratrol treated animals exposed to high fat diet [44, 119]. Compounds of independent structure from resveratrol, discovered by similar assays to that which obtained resveratrol as a STAC, have also provided impressive insulin sensitizing effects and positive health outcome in high fat diet challenged rodents [120]. Obviously, these impressive effects also extend to neuroprotection, in which resveratrol has been demonstrated to have mitigating effects in a number of neurodegeneration and neural injury models, as discussed in the next section.
The effects of resveratrol and “STACs” were initially interpreted to be from direct binding to SIRT1, as shown by assays wherein they were shown to activate SIRT1. This interpretation of their mode of action has been subject to intense scrutiny and other interpretations have emerged. Pacholec et al. concluded that resveratrol and STACs do not act upon SIRT1 with whole protein physiologic substrates such as p53 or acetylCoA synthetase, and concluded that the effects of STACs and resveratrol were instead attributable to off-target activities [121]. In fact, resveratrol has been widely acknowledged to have a number of additional targets, in addition to SIRT1, such as AMPK [122]. In addition, resveratrol targets include cAMP phosphodiesterases, which modulates cAMP signaling strengths in cells [123]. The lack of clarity regarding mechanism of action of the “STACs” has hindered further development of these compounds, and has created some confusion about their effects and mode of action.
Sirtuins as Therapeutic Targets in Neurodegenerative Disease and Injury
To date, the most studied sirtuins in the context of neurodegeneration and injury are SIRT1 and SIRT2. Accumulating evidence suggests they may play different roles in neurodegeneration, with different protein targets, and have different potentials for the development of therapeutic applications. Additionally, as awareness grows for the roles that the mitochondrial sirtuins, SIRT3, SIRT4, and SIRT5 play in metabolic regulation and adaption, so too does their potential as therapeutic targets in the nervous system. In this section, we discuss the effects of sirtuins on the outcomes of a number of important neurological disorders, including Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis, multiple sclerosis, and ischemic stroke.
Alzheimer’s Disease
Alzheimer’s disease (AD) is a progressive, degenerative disorder causing neurological damage and brain atrophy resulting in memory loss, cognitive and functional decline, and death. It is the most common neurodegenerative disorder [124]. The disease presents itself in two variants: the first is the familial form, which accounts for a small percentage of all AD patients, and is induced by dominant mutations in the amyloid precursor protein (APP), presenilin-1 (PS1) or PS2 genes [125, 126]; the second is the aging-associated sporadic or late-onset form that is characterized by the early presence of inflammatory mediators both in plasma and in the brain [127, 128]. Importantly, a major risk factor for both forms of the disease is the inheritance of the ApoE ε4 allele [129]. The pathologic hallmarks of Alzheimer's disease are neurofibrillary tangles and neuritic plaques [124]. Neurofibrillary tangles consist largely of hyperphosphorylated tau and are located within cell bodies of affected neurons. The principal component of plaques is amyloid-β, or Aβ, a peptide derived from the amyloid precursor protein by β- and γ-secretase in the amyloidogenic pathway (reviewed in [130, 131]).
The indication that sirtuins might play a protective role against AD initially came from caloric restriction (CR) studies, in which CR reduced Aβ generation and Aβ plaques in AD-transgenic mice [132, 133]. In support of this, CR studies in AD-type brain amyloidosis in Squirrel monkeys were similarly found to result in reduced Aβ. Importantly, CR and reduced Aβ are correlated with increased SIRT1 levels in both in monkeys and mice [93, 134]. CR mediated SIRT1 expression appears to reduce Aβ peptide generation through mechanisms that favor a α-secretase non-amyloidogenic processing of APP. For example, SIRT1 has been demonstrated to inhibit the expression of the serine/threonine Rho kinase, ROCK1, a protein known to inhibit α-secretase processing of the amyloid precursor protein [93]. In another study, SIRT1 has been shown to deacetylate and activate the retinoic acid receptor β (RARβ), stimulating the expression of ADAM10, a membrane-tethered protease belonging to the A disintegrin and metalloprotease family and implicated in α-secretase cleavage of APP [135]. In addition to reducing Aβ plaques, SIRT1 may also impact neurofibrillary tangle pathology. An early p300-mediated event that precedes the accumulation of neurofibrillary tangles is the acetylation of tau, which slows its degradation by inhibiting ubiquitination. Studies have shown that SIRT1 activity can reduce the acetylation level of tau, thereby promoting its degradation and clearance [136].
Parkinson’s Disease
Parkinson’s disease (PD) is a progressive neurodegenerative disease characterized by movement disorders, resulting from damage or destruction of dopaminergic neurons in the substantia nigra [137, 138]. It is the second most common neurodegenerative disorder. Although numerous genes responsible for familial PD have been identified, the etiology of sporadic PD, which accounts for the majority of PD cases, is still unknown [139, 140]. Detailed studies in PD pathology suggest that the degeneration of neurons involve several cellular and molecular events, including mitochondrial dysfunction, oxidative stress, microglia-mediated inflammation, and the misfolding and aggregation of proteins [141, 142]. Lewy bodies that contain α-synuclein aggregates are present in both familial and sporadic forms of PD.
Like AD, CR has shown protection in models of PD. In rhesus monkeys CR was shown to reduce nigro-striatal dopaminergic neuron vulnerability to the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) [143]. Similarly, mice subjected to CR or 2-deoxy-D-glucose have been shown to exhibit reduced dopaminergic neuron damage in the substantia nigra and improved behavioral outcome following MPTP treatment [144]. More direct evidence has come from studies showing that SIRT1 is necessary and sufficient for increasing lifespan in a mutant α-synuclein mouse model of PD. In this study, in response to α-synuclein aggregation, SIRT1 was found to deacetylate heat shock factor 1 (HSF1) and increase the expression of the molecular chaperone, HSP70 [145]. In contrast to this however, Kakefuda et al. [146] have shown that neuron-specific overexpression of SIRT1 in mice is not sufficient to protect against MPTP toxicity, and a further study by Park et al. [147] has shown that SIRT1 knockdown actually attenuates 1-methyl-4-phenylpyridinium ion (MPP(+))-induced cytotoxicity in the SH-SY5Y dopaminergic cell line.
SIRT2 has also been identified as a therapeutic target in PD. For example, the inhibition or knockdown of SIRT2 has been shown to rescue alpha-synuclein toxicity and modify inclusion morphology in cellular models of PD, and protect against dopaminergic cell death in a Drosophila model of PD [148]. Confirming these findings, a more recent study has shown that the genetic deletion of SIRT2 in mice can reduce MPTP-induced nigro-striatal damage [148]. The proposed mechanism for this protection is that the loss of SIRT2 activity prevents MPTP stress-induced FoxO3a deacetylation and subsequent increased levels of the pro-apoptotic mediator Bim [149].
Huntington’s Disease
Huntington's disease (HD) is an autosomal dominant neurodegenerative disorder characterized by motor, cognitive and behavioral dysfunction. It is caused by an unstable expansion of CAG repeats in the coding region of the Huntingtin gene IT15 [150], which generates a stretch of glutamine residues spanning the N-terminus of the Huntingtin protein (HTT). In general, individuals with ≥40 repeats are at risk of developing HD as they age [151, 152]. Studies suggest that the aggregation of mutant HTT fragments is the major cause of toxicity, specifically damaging cortical and striatal medium spiny neurons in HD patients [152–156].
Early studies in mutant HTT transgenic mice (N171-82Q) showed that CR can delay the onset of motor dysfunction and prolong lifespan [157]. However, the first report demonstrating a direct connection between SIRT1 and HD came from Parker et al. [158], who found that overexpression of Sir2 or treatment with resveratrol can rescue neuronal dysfunction phenotypes induced by mutant polyglutamine in Caenorhabditis elegans. Contrary to these findings though, Pallos et al. [159] have used a Drosophila melanogaster model of HD to show that a 50 % reduction in Sir2 expression extends survival of photoreceptor neurons expressing mutant Htt [159]. Overexpression of Sir2 neither had a deleterious nor beneficial effect on mutant HTT photoreceptor neurons. In mouse models of HD, the role of SIRT1 in mutant HTT neurotoxicity has been more apparent. In one study that crossed a N171-82Q HD mouse line with a brain SIRT1 overexpression mouse line, an attenuation in brain atrophy, delayed onset, and a slowing of motor deficit progression was observed [160]. Similarly, in a different HD mouse model, the R6/2 line, in which a N-terminal huntingtin fragment containing an expanded polyglutamine tract is overexpressed, high levels of SIRT1 expressed from an endogenous β-actin promoter was able to attenuate brain pathology, reduce protein aggregation and improve (in males) survival. In contrast to this, brain-specific deletion of SIRT1 exacerbated HD brain pathology [161]. Several mechanisms for SIRT1 protection have been proposed from these studies. One mechanism is that SIRT1 deacetylates and activates CREB-regulated transcription coactivator 1 (TORC1), a brain-specific modulator of CREB activity, which rescues mutant-HTT-mediated interference of TORC1 activity, facilitates its interaction with CREB, and promotes the transcriptional activation of brain-derived neurotrophic factor (BDNF) [161]. Another mechanism is that through its deacetylase activity, SIRT1 can correct a hyperacetylation of its substrates, which occurs in mutant HTT expressing cells. In particular, Jiang et al. [161] demonstrate that SIRT1 can reduce mutant HTT-induced FoxO3a acetylation and ameliorate mutant HTT-induced deficits of dopamine- and cAMP-regulated phosphoprotein, 32 kDa (DARPP32) and BDNF expression.
SIRT2 has also been studied with regard to its potential as a therapeutic target in HD. In one study, the genetic reduction of SIRT2 in the Drosophila melanogaster HD model was found to lead to greater survival of photoreceptor neurons, although it did not suppress overall fly lethality [159]. SIRT2 inhibition has also shown protection in primary neuronal HD models. This protection was attributed to a reduction in mutant huntingtin aggregates and the downregulation of genes responsible for cholesterol biosynthesis, a pathway which is dysregulated in HD patients and HD mouse models [162]. In contrast to this, however, studies looking at SIRT2 reduction or knockout in the mouse R6/2 HD model were not found to be neuroprotective, nor did they affect polyglutamine aggregation and cholesterol biosynthesis [163].
Amyotrophic Lateral Sclerosis
Amyotrophic lateral sclerosis (ALS; also known as Lou Gehrig's disease) is a progressive and fatal neurodegenerative disease that primarily affects motor neurons [164]. A hallmark of ALS is the appearance of cytoplasmic protein inclusions in affected motor neurons and glial cells [164, 165]. To date, well over 100 independent mutations in the copper/zinc superoxide dismutase 1 (SOD1) have been reported to be causative of ALS, accounting for ∼ 20 % of familial ALS [164, 166–168].
SIRT1 has been studied in the context of mutant SOD1 (SOD1-G37R) overexpression in cells and transgenic mice. Of interest, these studies have found SIRT1 to be increased in the spinal cords of mutant mice, particularly when severe neurodegeneration is evident [169]. Furthermore, lentiviral-mediated infection and overexpression of SIRT1, but not a non-functional SIRT1, can protect cultured primary neurons that express a toxic mutant SOD [169]. In support of this, resveratrol has been shown to protect neurons in cell-based models of ALS [170, 171] and in the mutant SOD1-G93A mouse model [172, 173]. Proposed mechanisms for this protection include the SIRT1 mediated deacetylation of HSF1, which induces transcription of the molecular chaperones HSP70 and HSP25, promoting motor neuron survival [173].
SIRT3 has also been identified as a potential therapeutic target in ALS. In an ALS model that uses SOD1-G93A-expressing rat spinal cord motor neurons, SIRT3 overexpression can rescue mutant SOD1 induced defects in mitochondrial dynamics [174]. While the mechanism underlying this protection is not well understood, it may involve the deacetylation of SOD2 and isocitrate dehydrogenase 2 (IDH2), which increases their enzymatic activities. Additionally, cyclophilin D can also be deacetylated by SIRT3 and may be important for the prevention of mitochondrial permeability transition [174].
Multiple Sclerosis
Multiple sclerosis (MS) is a severe human neurological disorder that involves inflammation at multiple foci throughout the central nervous system. The pathogenic mechanism of MS is regarded to be autoimmune-mediated demyelination, which leaves axons vulnerable to degeneration [175]. Relapsing-remitting MS, the most common form of the disease, is marked by intermittent episodes of focal neurologic dysfunction that partially recovers as acute inflammation resolves. Other MS patients follow a primary or secondary progressive disease course marked by a slow neurologic decline due to axonal damage and loss of neurons without discrete episodes of inflammation and demyelination [175].
SIRT1 has been identified as a therapeutic target in MS through studies in mice where MS is modeled by experimental autoimmune encephalomyelitis (EAE). In one study, two structurally distinct SIRT1 activators, SRT647 and SRT501, were shown to be neuroprotective in retinal ganglion cells during EAE-induced acute optic neuritis. While the specific substrates of SIRT1 that mediate survival were not identified, it was determined that SIRT1 activation did not prevent inflammation, suggesting SIRT1 is neuroprotective even in the presence of active inflammation [176]. Contrary to this study, a recent publication has demonstrated that SIRT1 inactivation increases the production of new oligodendrocyte progenitor cells in the adult mouse brain, which can ameliorate remyelination and delay paralysis in a mouse EAE model [177].
Neurological Injury
Stroke is one of the leading causes of death and adult disability worldwide. The majority of strokes are ischemic and result from an occlusion of a major cerebral artery by a thrombus or embolism. The other major type of stroke is hemorrhagic, which results from a blood vessel rupture either in the brain or on its surface [178]. The consequence of both of these events is a significant reduction in blood flow and nutrients critical for neural function and survival. The pathophysiology of the affected area includes energy failure, excitotoxicity, free radical generation and elevation of intracellular calcium, which culminate in cell death or dysfunction [178, 179]. Depending on the location and magnitude of damage, stroke may impact movement, sensation, vision, speech, and cognition.
The first indications that sirtuins might represent therapeutic targets for stroke came from neuroprotection studies in cellular and animal models showing that resveratrol could protect from brain injury [180–182] and cerebral ischemia [183–185]. Importantly, additional studies went on to show that the blockade of SIRT1 activation by sirtinol abolishes the resveratrol neuroprotection [186]. In addition to resveratrol, overexpression of nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme of the NAD+ salvage pathway, has also been shown to offer protection against stroke, while NAMPT inhibition by FK866 exacerbated ischemic injury [187]. In this study, SIRT1 was shown to be essential for the neuroprotective effect of NAMPT overexpression since protection was blocked by SIRT1 deletion [187]. Despite these positive findings, studies that have used SIRT1 gain-of-function or loss-of-function have yielded mixed results. In one study, using SIRT1 transgenic mice in which human SIRT1 was overexpressed under the control of the rat neuron-specific enolase (NSE) promoter, no neuroprotection was observed against stroke [146]. Consistently, another study reported that SIRT1 inhibition by nicotinamide had a positive effect on outcome after cerebral ischemia [188]. One explanation for these findings is that because SIRT1 is a NAD+ consuming enzyme, the detrimental effects of its high-energy consumption outweigh its beneficial effects during periods of nutrient deficiency such as stroke. In spite of this explanation, a more recent report has demonstrated that SIRT1 knockout mice have larger infarct volumes and worse neurological outcome than their wild-type littermates after permanent focal ischemia [189].
Conclusions
In recent years it has become clear that the modulation of sirtuin activity may be a valuable therapeutic strategy for ameliorating or delaying the functional and pathological deficits associated with neurodegenerative disease or injury. However, research into sirtuin function and neurodegeneration has largely focused on SIRT1 and SIRT2. Insights into the metabolic roles of SIRT3–SIRT5 in the mitochondria, and SIRT6’s role in genome stability, DNA repair, and metabolism, highlight their potential as similar therapeutic targets. Similarly, the biological targets and possible roles of SIRT7 in neurodegeneration are largely unexplored.
One of the challenges moving forward will be to understand how each of the sirtuins might best be targeted. As described in this Chapter, sirtuin activity may be controlled at multiple levels: by their expression, localization, substrate availability, NAD or nicotinamide level, posttranslational modifications, and by small molecule activators, e.g. STACS. With regard to neurodegeneration and injury, a number of these have been exploited to modulate sirtuin function. For example, a number of STACs, including resveratrol have shown impressive efficacy in multiple models of neurodegeneration. Similarly, genetic manipulation of sirtuin levels, modulation of NAD by nicotinamide riboside, exogenous NAD administration, or inhibiting PARP, have similarly shown efficacy. However, a greater understanding of sirtuin regulation and NAD metabolism is needed. For example, little is known about how NAD and nicotinamide pools differ in different cellular compartments and how these regulate certain sirtuin family members or are impacted during disease or pharmacological intervention. Moreover, our ability to measure the activities of individual sirtuin family members in situ, especially in neurons, limits identification of precise and disease-relevant molecular targets, particularly given the large number of biological substrates affected by sirtuins.
Finally, neurodegenerative disease and injury represent disorders with complex etiologies and multiple perturbed factors and pathways. Given that sirtuins have numerous cellular target substrates can significantly impact cellular and mitochondrial metabolism and promote longevity, it is reasonable to anticipate that they would represent valuable therapeutic targets. Further understanding of their precise roles, substrates, activity and regulation will undoubtedly lead to improved therapeutic strategies for combating neurodegenerative disease and injury.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 1225 kb)
Acknowledgments
This work was supported by grants from the New York State Spinal Cord Injury Research Program (CO19772), the National Institutes of Health (NS071056), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and the Burke Medical Research Foundation.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Contributor Information
Brett Langley, Phone: +1-914-3683152, FAX: +1-914-3683142, Email: blangley@burke.org.
Anthony Sauve, Phone: +1-212-7466224, Email: aas2004@med.cornell.edu.
References
- 1.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Ann Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 2.Mellert HS, McMahon SB. Biochemical pathways that regulate acetyltransferase and deacetylase activity in mammalian cells. Trends Biochem Sci. 2009;34(11):571–8. doi: 10.1016/j.tibs.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp Cell Res. 2001;262(2):75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 4.Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann NY Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 5.Hake SB, Xiao A, Allis CD. Linking the epigenetic 'language' of covalent histone modifications to cancer. Br J Cancer. 2004;90(4):761–9. doi: 10.1038/sj.bjc.6601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404(1):1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Bio Chem. 2002;277(28):25748–55. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 9.Hernick M, Fierke CA. Zinc hydrolases: the mechanisms of zinc-dependent deacetylases. Arch Biochem Biophys. 2005;433(1):71–84. doi: 10.1016/j.abb.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Klar AJ, Fogel S, Macleod K. MAR1-a Regulator of the HMa and HMalpha Loci in SACCHAROMYCES CEREVISIAE. Genetics. 1979;93(1):37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rine J, Strathern JN, Hicks JB, Herskowitz I. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics. 1979;93(4):877–901. doi: 10.1093/genetics/93.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Devel. 1999;13(19):2570–80. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S, Benguria A, Lai CY, Jazwinski SM. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10(10):3125–36. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 15.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(45):15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410(6825):227–30. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 17.Weindruch R, Walford RL. Dietary restriction in mice beginning at 1 year of age: effect on life-span and spontaneous cancer incidence. Science. 1982;215(4538):1415–8. doi: 10.1126/science.7063854. [DOI] [PubMed] [Google Scholar]
- 18.Byles V, Chmilewski LK, Wang J, Zhu L, Forman LW, Faller DV, et al. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. Int J Biol Sci. 2010;6(6):599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Q, Yan T, Ge X, Sun C, Shi X, Zhai Q. Cytoplasm-localized SIRT1 enhances apoptosis. J Cell Physiol. 2007;213(1):88–97. doi: 10.1002/jcp.21091. [DOI] [PubMed] [Google Scholar]
- 20.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD + -dependent histone deacetylase SIRT1. J Bio Chem. 2007;282(9):6823–32. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 21.Afshar G, Murnane JP. Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene. 1999;234(1):161–8. doi: 10.1016/s0378-1119(99)00162-6. [DOI] [PubMed] [Google Scholar]
- 22.Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, et al. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2007;26(7):945–57. doi: 10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- 23.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD + -dependent tubulin deacetylase. Molec Cell. 2003;11(2):437–44. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 24.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334(6057):806–9. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa T, Guarente L. Urea cycle regulation by mitochondrial sirtuin, SIRT5. Aging. 2009;1(6):578–81. doi: 10.18632/aging.100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137(3):560–70. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc National Academy of Sciences of the United States of America. 2002;99(21):13653–8. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27(24):8807–14. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Bio Chem. 2007;282(46):33583–92. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 31.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Molec Biol. 2008;382(3):790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 32.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Gene Devel. 2006;20(9):1075–80. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Bio Chem. 2005;280(22):21313–20. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 34.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–29. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 35.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, et al. A phylogenetically conserved NAD + -dependent protein deacetylase activity in the Sir2 protein family. Proc National Academy of Sciences of the United States of America. 2000;97(12):6658–63. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Bio Chem. 2004;279(28):28873–9. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 37.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116(4):551–63. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 38.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 39.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 40.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 41.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 42.Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, et al. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Bio Chem. 2005;280(11):10264–76. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 43.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annual review of biochemistry. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 44.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127(6):1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 45.Yuan Z, Seto E. A functional link between SIRT1 deacetylase and NBS1 in DNA damage response. Cell Cycle. 2007;6(23):2869–71. doi: 10.4161/cc.6.23.5026. [DOI] [PubMed] [Google Scholar]
- 46.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Molec Cell. 2007;27(1):149–62. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14(4):312–23. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li K, Casta A, Wang R, Lozada E, Fan W, Kane S, et al. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Bio Chem. 2008;283(12):7590–8. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- 49.Fan W, Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Molecular cell. 2010;39(2):247–58. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 50.Ming M, Shea CR, Guo X, Li X, Soltani K, Han W, et al. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc National Academy of Sciences of the United States of America. 2010;107(52):22623–8. doi: 10.1073/pnas.1010377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Devel. 2006;20(10):1256–61. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging cell. 2007;6(4):505–14. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 53.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metabol. 2007;6(2):105–14. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27(16):2276–88. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 55.Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Molecular cell. 2011;43(1):33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, et al. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Comm. 2003;309(3):558–66. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 57.Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc National Academy of Sciences of the United States of America. 2005;102(44):16013–8. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20(4):487–99. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dan L, Klimenkova O, Klimiankou M, Klusman JH, van den Heuvel-Eibrink MM, Reinhardt D, et al. The role of sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica. 2012;97(4):551–9. doi: 10.3324/haematol.2011.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc National Academy of Sciences of the United States of America. 2006;103(27):10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35(12):669–75. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–5. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12(6):654–61. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12(6):662–7. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 65.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143(5):802–12. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Bio Chem. 2012;287(17):14078–86. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30(26):2986–96. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, et al. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Bio Chem. 2010;285(42):31995–2002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23(4):450–63. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Molec Cell. 2013;50(6):919–30. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483(7388):218–21. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 72.Jia G, Su L, Singhal S, Liu X. Emerging roles of SIRT6 on telomere maintenance, DNA repair, metabolism and mammalian aging. Molec Cell Biochem. 2012;364(1-2):345–50. doi: 10.1007/s11010-012-1236-8. [DOI] [PubMed] [Google Scholar]
- 73.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452(7186):492–6. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging. 2009;1(1):109–21. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332(6036):1443–6. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140(2):280–93. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102(6):703–10. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 78.Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487(7405):114–8. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hubbi ME, Hu H, Kshitiz NF, Gilkes DM, Semenza GL. Sirtuin-7 inhibits the activity of hypoxia-inducible factors. J Bio Chem. 2013. [DOI] [PMC free article] [PubMed]
- 80.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Ann Rev Biochem. 2006;75:435–65. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 81.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121(4):515–27. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 82.Tong L, Denu JM. Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. Biochim Biophys Acta. 2010;1804(8):1617–25. doi: 10.1016/j.bbapap.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD + -dependent deacetylation reactions. Biochemistry. 2001;40(51):15456–63. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 84.Jackson MD, Denu JM. Structural identification of 2'- and 3'-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD + -dependent histone/protein deacetylases. J Bio Chem. 2002;277(21):18535–44. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 85.Cheng Z, Tang Y, Chen Y, Kim S, Liu H, Li SS, et al. Molecular characterization of propionyllysines in non-histone proteins. Mol Cell Proteomics. 2009;8(1):45–52. doi: 10.1074/mcp.M800224-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10(12):M111 012658. [DOI] [PMC free article] [PubMed]
- 87.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496(7443):110–3. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289(5487):2126–8. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 89.Carri MT, Ferri A, Cozzolino M, Calabrese L, Rotilio G. Neurodegeneration in amyotrophic lateral sclerosis: the role of oxidative stress and altered homeostasis of metals. Brain Res Bull. 2003;61(4):365–74. doi: 10.1016/s0361-9230(03)00179-5. [DOI] [PubMed] [Google Scholar]
- 90.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, et al. Manipulation of a nuclear NAD + salvage pathway delays aging without altering steady-state NAD + levels. J Bio Chem. 2002;277(21):18881–90. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 91.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Bio Chem. 2002;277(47):45099–107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 92.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423(6936):181–5. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, et al. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J Bio Chem. 2006;281(31):21745–54. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- 94.Hawse WF, Hoff KG, Fatkins DG, Daines A, Zubkova OV, Schramm VL, et al. Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure. 2008;16(9):1368–77. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gallo CM, Smith DL, Jr, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24(3):1301–12. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McClure JM, Gallo CM, Smith DL, Jr, Matecic M, Hontz RD, Buck SW, et al. Pnc1p-mediated nicotinamide clearance modifies the epigenetic properties of rDNA silencing in Saccharomyces cerevisiae. Genetics. 2008;180(2):797–810. doi: 10.1534/genetics.108.091090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Molecular cell. 2005;17(4):595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 98.Sauve AA. Sirtuins. Biochim Biophys Acta. 2010;1804(8):1565–6. doi: 10.1016/j.bbapap.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 99.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metabol. 2011;13(4):461–8. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metabol. 2012;15(6):838–47. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochem. 2003;42(31):9249–56. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 102.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 103.Sauve AA. Pharmaceutical strategies for activating sirtuins. Curr Pharm Des. 2009;15(1):45–56. doi: 10.2174/138161209787185797. [DOI] [PubMed] [Google Scholar]
- 104.Baur JA. Biochemical effects of SIRT1 activators. Biochim Biophys Acta. 2010;1804(8):1626–34. doi: 10.1016/j.bbapap.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gerhart-Hines Z, Dominy JE, Jr, Blattler SM, Jedrychowski MP, Banks AS, Lim JH, et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Molecular Cell. 2011;44(6):851–63. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee CW, Wong LL, Tse EY, Liu HF, Leong VY, Lee JM, et al. AMPK promotes p53 acetylation via phosphorylation and inactivation of SIRT1 in liver cancer cells. Cancer Res. 2012;72(17):4394–404. doi: 10.1158/0008-5472.CAN-12-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9(11):1253–62. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL, 3rd, et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging. 2009;1(9):771–83. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Bio Chem. 2005;280(14):13560–7. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 110.Geng YQ, Li TT, Liu XY, Li ZH, Fu YC. SIRT1 and SIRT5 activity expression and behavioral responses to calorie restriction. J Cell Biochem. 2011;112(12):3755–61. doi: 10.1002/jcb.23315. [DOI] [PubMed] [Google Scholar]
- 111.Kanfi Y, Shalman R, Peshti V, Pilosof SN, Gozlan YM, Pearson KJ, et al. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008;582(5):543–8. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151(6):1185–99. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451(7178):583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 114.Escande C, Chini CC, Nin V, Dykhouse KM, Novak CM, Levine J, et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest. 2010;120(2):545–58. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Molec Cell. 2007;28(2):277–90. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 116.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Develop. 2004;18(1):12–6. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Bio Chem. 2004;279(38):40122–9. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- 118.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 119.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444(7117):337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450(7170):712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Bio Chem. 2010;285(11):8340–51. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc National Academy of Sciences of the United States of America. 2007;104(17):7217–22. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148(3):421–33. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Castellani RJ, Rolston RK, Smith MA. Alzheimer disease. Disease-a-month: DM. 2010;56(9):484–546. doi: 10.1016/j.disamonth.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cotman CW, Su JH. Mechanisms of neuronal death in Alzheimer's disease. Brain Pathol. 1996;6(4):493–506. doi: 10.1111/j.1750-3639.1996.tb00878.x. [DOI] [PubMed] [Google Scholar]
- 126.Harrington CR. The molecular pathology of Alzheimer's disease. Neuroimaging clinics of North America. 2012;22(1):11–22. doi: 10.1016/j.nic.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 127.Eikelenboom P, Veerhuis R, van Exel E, Hoozemans JJ, Rozemuller AJ, van Gool WA. The early involvement of the innate immunity in the pathogenesis of late-onset Alzheimer's disease: neuropathological, epidemiological and genetic evidence. Curr Alzheimer Res. 2011;8(2):142–50. doi: 10.2174/156720511795256080. [DOI] [PubMed] [Google Scholar]
- 128.Holmes C, Cunningham C, Zotova E, Woolford J, Dean C, Kerr S, et al. Systemic inflammation and disease progression in Alzheimer disease. Neurology. 2009;73(10):768–74. doi: 10.1212/WNL.0b013e3181b6bb95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, et al. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Molec Psych. 2011;16(9):903–7. doi: 10.1038/mp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001;81(2):741–66. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 131.Vassar R. BACE1: the beta-secretase enzyme in Alzheimer's disease. J Molec Neuroscie: MN. 2004;23(1-2):105–14. doi: 10.1385/JMN:23:1-2:105. [DOI] [PubMed] [Google Scholar]
- 132.Patel NV, Gordon MN, Connor KE, Good RA, Engelman RW, Mason J, et al. Caloric restriction attenuates Abeta-deposition in Alzheimer transgenic models. Neurobiol Aging. 2005;26(7):995–1000. doi: 10.1016/j.neurobiolaging.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 133.Wang J, Ho L, Qin W, Rocher AB, Seror I, Humala N, et al. Caloric restriction attenuates beta-amyloid neuropathology in a mouse model of Alzheimer's disease. FASEB. 2005;19(6):659–61. doi: 10.1096/fj.04-3182fje. [DOI] [PubMed] [Google Scholar]
- 134.Qin W, Chachich M, Lane M, Roth G, Bryant M, de Cabo R, et al. Calorie restriction attenuates Alzheimer's disease type brain amyloidosis in Squirrel monkeys (Saimiri sciureus) JAD. 2006;10(4):417–22. doi: 10.3233/jad-2006-10411. [DOI] [PubMed] [Google Scholar]
- 135.Kojro E, Fahrenholz F. The non-amyloidogenic pathway: structure and function of alpha-secretases. Sub-cell Biochem. 2005;38:105–27. doi: 10.1007/0-387-23226-5_5. [DOI] [PubMed] [Google Scholar]
- 136.Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67(6):953–66. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jankovic J, Aguilar LG. Current approaches to the treatment of Parkinson's disease. Neuropsy Dis Treat. 2008;4(4):743–57. doi: 10.2147/ndt.s2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Samii A, Nutt JG, Ransom BR. Parkinson's disease. Lancet. 2004;363(9423):1783–93. doi: 10.1016/S0140-6736(04)16305-8. [DOI] [PubMed] [Google Scholar]
- 139.Jain S, Wood NW, Healy DG. Molecular genetic pathways in Parkinson's disease: a review. Clin Sci (Lond). 2005;109(4):355–64. doi: 10.1042/CS20050106. [DOI] [PubMed] [Google Scholar]
- 140.Morris HR. Genetics of Parkinson's disease. Annals of medicine. 2005;37(2):86–96. doi: 10.1080/07853890510007269. [DOI] [PubMed] [Google Scholar]
- 141.Dauer W, Przedborski S. Parkinson's disease: mechanisms and models. Neuron. 2003;39(6):889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 142.von Bohlen Und Halbach O, Krieglstein K, Schober A, Schulz JB. The dopaminergic nigrostriatal system: development, physiology, disease. Cell Tissue Res. 2004;318(1):3. doi: 10.1007/s00441-004-0963-x. [DOI] [PubMed] [Google Scholar]
- 143.Maswood N, Young J, Tilmont E, Zhang Z, Gash DM, Gerhardt GA, et al. Caloric restriction increases neurotrophic factor levels and attenuates neurochemical and behavioral deficits in a primate model of Parkinson's disease. Proc National Academy of Sciences of the United States of America. 2004;101(52):18171–6. doi: 10.1073/pnas.0405831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Holmer HK, Keyghobadi M, Moore C, Menashe RA, Meshul CK. Dietary restriction affects striatal glutamate in the MPTP-induced mouse model of nigrostriatal degeneration. Synapse. 2005;57(2):100–12. doi: 10.1002/syn.20163. [DOI] [PubMed] [Google Scholar]
- 145.Donmez G, Arun A, Chung CY, McLean PJ, Lindquist S, Guarente L. SIRT1 protects against alpha-synuclein aggregation by activating molecular chaperones. J Neurosci. 2012;32(1):124–32. doi: 10.1523/JNEUROSCI.3442-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 146.Kakefuda K, Fujita Y, Oyagi A, Hyakkoku K, Kojima T, Umemura K, et al. Sirtuin 1 overexpression mice show a reference memory deficit, but not neuroprotection. Biochem Biophys Res Comm. 2009;387(4):784–8. doi: 10.1016/j.bbrc.2009.07.119. [DOI] [PubMed] [Google Scholar]
- 147.Park G, Jeong JW, Kim JE. SIRT1 deficiency attenuates MPP + -induced apoptosis in dopaminergic cells. FEBS Lett. 2011;585(1):219–24. doi: 10.1016/j.febslet.2010.11.048. [DOI] [PubMed] [Google Scholar]
- 148.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317(5837):516–9. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 149.Liu L, Arun A, Ellis L, Peritore C, Donmez G. Sirtuin 2 (SIRT2) enhances 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced nigrostriatal damage via deacetylating forkhead box O3a (Foxo3a) and activating Bim protein. J Bio Chem. 2012;287(39):32307–11. doi: 10.1074/jbc.C112.403048. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 150.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72(6):971-83. [DOI] [PubMed]
- 151.Ruocco HH, Lopes-Cendes I, Li LM, Santos-Silva M, Cendes F. Striatal and extrastriatal atrophy in Huntington's disease and its relationship with length of the CAG repeat. Brazil J Med Bio Res. 2006;39(8):1129–36. doi: 10.1590/s0100-879x2006000800016. [DOI] [PubMed] [Google Scholar]
- 152.Gil JM, Rego AC. Mechanisms of neurodegeneration in Huntington's disease. The European J Neurosci. 2008;27(11):2803–20. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 153.Li SH, Li XJ. Huntingtin and its role in neuronal degeneration. Neuroscientist. 2004;10(5):467–75. doi: 10.1177/1073858404266777. [DOI] [PubMed] [Google Scholar]
- 154.Davies S, Ramsden DB. Huntington's disease. MP. 2001;54(6):409–13. doi: 10.1136/mp.54.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Imarisio S, Carmichael J, Korolchuk V, Chen CW, Saiki S, Rose C, et al. Huntington's disease: from pathology and genetics to potential therapies. Biochem J. 2008;412(2):191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 156.Johnson CD, Davidson BL. Huntington's disease: progress toward effective disease-modifying treatments and a cure. Human Molec Genet. 2010;19(R1):R98–R102. doi: 10.1093/hmg/ddq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Duan W, Guo Z, Jiang H, Ware M, Li XJ, Mattson MP. Dietary restriction normalizes glucose metabolism and BDNF levels, slows disease progression, and increases survival in huntingtin mutant mice. Proc National Academy of Sciences of the United States of America. 2003;100(5):2911–6. doi: 10.1073/pnas.0536856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nature Genet. 2005;37(4):349–50. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 159.Pallos J, Bodai L, Lukacsovich T, Purcell JM, Steffan JS, Thompson LM, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington's disease. Human Molec Genet. 2008;17(23):3767–75. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jiang M, Wang J, Fu J, Du L, Jeong H, West T, et al. Neuroprotective role of Sirt1 in mammalian models of Huntington's disease through activation of multiple Sirt1 targets. Nature Med. 2012;18(1):153–8. doi: 10.1038/nm.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jeong H, Cohen DE, Cui L, Supinski A, Savas JN, Mazzulli JR, et al. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nature Med. 2012;18(1):159–65. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Luthi-Carter R, Taylor DM, Pallos J, Lambert E, Amore A, Parker A, et al. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc National Academy of Sciences of the United States of America. 2010;107(17):7927–32. doi: 10.1073/pnas.1002924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bobrowska A, Donmez G, Weiss A, Guarente L, Bates G. SIRT2 ablation has no effect on tubulin acetylation in brain, cholesterol biosynthesis or the progression of Huntington's disease phenotypes in vivo. PloS One. 2012;7(4):e34805. doi: 10.1371/journal.pone.0034805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wijesekera LC, Leigh PN. Amyotrophic lateral sclerosis. Orphanet J Rare Dis. 2009;4:3. doi: 10.1186/1750-1172-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Watanabe M, Dykes-Hoberg M, Culotta VC, Price DL, Wong PC, Rothstein JD. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol Dis. 2001;8(6):933–41. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- 166.Chattopadhyay M, Valentine JS. Aggregation of copper-zinc superoxide dismutase in familial and sporadic ALS. Antiox Redox Signal. 2009;11(7):1603–14. doi: 10.1089/ars.2009.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364(6435):362. doi: 10.1038/364362c0. [DOI] [PubMed] [Google Scholar]
- 168.Valentine JS, Doucette PA, Zittin PS. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annual review of biochemistry. 2005;74:563–93. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 169.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. The EMBO journal. 2007;26(13):3169–79. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang J, Zhang Y, Tang L, Zhang N, Fan D. Protective effects of resveratrol through the up-regulation of SIRT1 expression in the mutant hSOD1-G93A-bearing motor neuron-like cell culture model of amyotrophic lateral sclerosis. Neurosci Lett. 2011;503(3):250–5. doi: 10.1016/j.neulet.2011.08.047. [DOI] [PubMed] [Google Scholar]
- 171.Yanez M, Galan L, Matias-Guiu J, Vela A, Guerrero A, Garcia AG. CSF from amyotrophic lateral sclerosis patients produces glutamate independent death of rat motor brain cortical neurons: protection by resveratrol but not riluzole. Brain Res. 2011;1423:77–86. doi: 10.1016/j.brainres.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 172.Markert CD, Kim E, Gifondorwa DJ, Childers MK, Milligan CE. A single-dose resveratrol treatment in a mouse model of amyotrophic lateral sclerosis. J Med Food. 2010;13(5):1081–5. doi: 10.1089/jmf.2009.0243. [DOI] [PubMed] [Google Scholar]
- 173.Han S, Choi JR, Soon Shin K, Kang SJ. Resveratrol upregulated heat shock proteins and extended the survival of G93A-SOD1 mice. Brain Res. 2012;1483:112–7. doi: 10.1016/j.brainres.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 174.Song W, Song Y, Kincaid B, Bossy B, Bossy-Wetzel E. Mutant SOD1G93A triggers mitochondrial fragmentation in spinal cord motor neurons: neuroprotection by SIRT3 and PGC-1alpha. Neurobiol Dis. 2013;51:72–81. doi: 10.1016/j.nbd.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Noseworthy JH, Lucchinetti C, Rodriguez M, Weinshenker BG. Multiple sclerosis. New England J Med. 2000;343(13):938–52. doi: 10.1056/NEJM200009283431307. [DOI] [PubMed] [Google Scholar]
- 176.Shindler KS, Ventura E, Rex TS, Elliott P, Rostami A. SIRT1 activation confers neuroprotection in experimental optic neuritis. Investig Ophthalmol Vis Sci. 2007;48(8):3602–9. doi: 10.1167/iovs.07-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rafalski VA, Ho PP, Brett JO, Ucar D, Dugas JC, Pollina EA, et al. Expansion of oligodendrocyte progenitor cells following SIRT1 inactivation in the adult brain. Nat Cell Biol. 2013;15(6):614–24. doi: 10.1038/ncb2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Della-Morte D, Guadagni F, Palmirotta R, Testa G, Caso V, Paciaroni M, et al. Genetics of ischemic stroke, stroke-related risk factors, stroke precursors and treatments. Pharmacogenomics. 2012;13(5):595–613. doi: 10.2217/pgs.12.14. [DOI] [PubMed] [Google Scholar]
- 179.Ratan RR, Siddiq A, Smirnova N, Karpisheva K, Haskew-Layton R, McConoughey S, et al. Harnessing hypoxic adaptation to prevent, treat, and repair stroke. J Mol Med (Berl). 2007;85(12):1331–8. doi: 10.1007/s00109-007-0283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Virgili M, Contestabile A. Partial neuroprotection of in vivo excitotoxic brain damage by chronic administration of the red wine antioxidant agent, trans-resveratrol in rats. Neurosci Lett. 2000;281(2-3):123–6. doi: 10.1016/s0304-3940(00)00820-x. [DOI] [PubMed] [Google Scholar]
- 181.Bastianetto S, Zheng WH, Quirion R. Neuroprotective abilities of resveratrol and other red wine constituents against nitric oxide-related toxicity in cultured hippocampal neurons. British J Pharmacol. 2000;131(4):711–20. doi: 10.1038/sj.bjp.0703626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang MJ, Huang HM, Hsieh SJ, Jeng KC, Kuo JS. Resveratrol inhibits interleukin-6 production in cortical mixed glial cells under hypoxia/hypoglycemia followed by reoxygenation. J Neuroimmun. 2001;112(1-2):28–34. doi: 10.1016/s0165-5728(00)00374-x. [DOI] [PubMed] [Google Scholar]
- 183.Huang SS, Tsai MC, Chih CL, Hung LM, Tsai SK. Resveratrol reduction of infarct size in Long-Evans rats subjected to focal cerebral ischemia. Life Sci. 2001;69(9):1057–65. doi: 10.1016/s0024-3205(01)01195-x. [DOI] [PubMed] [Google Scholar]
- 184.Wang Q, Xu J, Rottinghaus GE, Simonyi A, Lubahn D, Sun GY, et al. Resveratrol protects against global cerebral ischemic injury in gerbils. Brain Res. 2002;958(2):439–47. doi: 10.1016/s0006-8993(02)03543-6. [DOI] [PubMed] [Google Scholar]
- 185.Sinha K, Chaudhary G, Gupta YK. Protective effect of resveratrol against oxidative stress in middle cerebral artery occlusion model of stroke in rats. Life Sci. 2002;71(6):655–65. doi: 10.1016/s0024-3205(02)01691-0. [DOI] [PubMed] [Google Scholar]
- 186.Raval AP, Dave KR, Perez-Pinzon MA. Resveratrol mimics ischemic preconditioning in the brain. J Cereb Blood Flow Metabol. 2006;26(9):1141–7. doi: 10.1038/sj.jcbfm.9600262. [DOI] [PubMed] [Google Scholar]
- 187.Wang P, Xu TY, Guan YF, Tian WW, Viollet B, Rui YC, et al. Nicotinamide phosphoribosyltransferase protects against ischemic stroke through SIRT1-dependent adenosine monophosphate-activated kinase pathway. Ann Neurol. 2011;69(2):360–74. doi: 10.1002/ana.22236. [DOI] [PubMed] [Google Scholar]
- 188.Liu D, Gharavi R, Pitta M, Gleichmann M, Mattson MP. Nicotinamide prevents NAD + depletion and protects neurons against excitotoxicity and cerebral ischemia: NAD + consumption by SIRT1 may endanger energetically compromised neurons. Neuromolec Med. 2009;11(1):28–42. doi: 10.1007/s12017-009-8058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hernandez-Jimenez M, Hurtado O, Cuartero MI, Ballesteros I, Moraga A, Pradillo JM, et al. Silent information regulator 1 protects the brain against cerebral ischemic damage. Stroke. 2013. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1225 kb)