Abstract
SUMMARY
Although Escherichia coli can be an innocuous resident of the gastrointestinal tract, it also has the pathogenic capacity to cause significant diarrheal and extraintestinal diseases. Pathogenic variants of E. coli (pathovars or pathotypes) cause much morbidity and mortality worldwide. Consequently, pathogenic E. coli is widely studied in humans, animals, food, and the environment. While there are many common features that these pathotypes employ to colonize the intestinal mucosa and cause disease, the course, onset, and complications vary significantly. Outbreaks are common in developed and developing countries, and they sometimes have fatal consequences. Many of these pathotypes are a major public health concern as they have low infectious doses and are transmitted through ubiquitous mediums, including food and water. The seriousness of pathogenic E. coli is exemplified by dedicated national and international surveillance programs that monitor and track outbreaks; unfortunately, this surveillance is often lacking in developing countries. While not all pathotypes carry the same public health profile, they all carry an enormous potential to cause disease and continue to present challenges to human health. This comprehensive review highlights recent advances in our understanding of the intestinal pathotypes of E. coli.
INTRODUCTION
Theodor Escherich first reported the isolation and characterization of slender short rods from infant stool, which he named Bacterium coli commune, in his 1885 publication (reprinted in English [1]). Although the organism was later described under multiple synonyms and iterations by other researchers, the name Escherichia coli was not fully recognized until 1954 (2). Over 125 years later, E. coli is known as a harmless commensal of the gastrointestinal tract in warm-blooded animals and is used as the colloquial laboratory workhorse. However, there is an alternate side to E. coli afforded through gene gain and loss that enable it to become a highly diverse and adapted pathogen. Pathogenic E. coli can cause a broad range of human diseases that span from the gastrointestinal tract to extraintestinal sites such as the urinary tract, bloodstream, and central nervous system (3, 4).
Diarrheal illness causes much mortality worldwide, particularly in children under the age of 5 (5) (Fig. 1) and particularly in countries in sub-Saharan Africa and South Asia, whose children suffer many diarrhea-related deaths. While there are many etiological agents responsible for diarrhea, pathogenic E. coli is a major contributor. Recent data from the Global Enteric Multi-Center Study (GEMS), one of the largest case-control studies aiming to understand the burden of pediatric diarrheal disease in sub-Saharan Africa and South Asia (6), illustrate that enterotoxigenic E. coli and Shigella are among two of the four main causative agents of moderate to severe diarrhea among children in these areas (7). In addition, increased fatality rates are associated with enteropathogenic E. coli and certain enterotoxigenic E. coli strains, thus underlining the significant role of pathogenic E. coli in the global health burden of diarrheal disease. The E. coli scientific and clinical communities have made great strides in understanding E. coli microbiology, pathogenesis, ecology, and interactions with its host. These advances are essential for novel approaches to vaccines and treatments that prevent some of the serious sequelae and complications associated with E. coli-induced diarrheal illness.
Fig 1.
Global mortality from diarrhea in children under the age of 5 in 2010. Estimates of diarrhea-specific mortality among children under 5 for each country reflect high mortality in developing countries, with the highest tolls present in countries in sub-Saharan Africa and South Asia. Many etiological agents, including pathogenic E. coli, are responsible for diarrhea-related mortality in these children. Recent work published by GEMS found significant child mortality associated with EPEC and ETEC infections in developing countries (7). Source data for the map: World Health Organization (5).
While various pathotypes contribute to diarrhea, the clinical symptoms and outcomes, site and mechanism of colonization, and disease can differ (Table 1), exemplifying the diversity of E. coli. In this review, we focus on advancements in our understanding of enteric pathogenic E. coli since the comprehensive 1998 review by James Nataro and James Kaper (8). We discuss the six major diarrheagenic E. coli pathotypes: enteropathogenic E. coli (EPEC), Shiga toxin-producing E. coli (STEC) (e.g., enterohemorrhagic E. coli [EHEC]), Shigella/enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), diffusely adherent E. coli (DAEC), and enterotoxigenic E. coli (ETEC), as well as a new pathotype, adherent invasive E. coli (AIEC), in the context of detection, diagnosis, epidemiology, public health, pathogenesis, and human disease.
Table 1.
General overview of enteric E. coli pathotypes
| Pathotype | Host(s) | Site of colonization | Disease(s) | Known reservoir(s)/source(s) of contamination | Treatment | Adhesiona | Genetic identifiers |
|---|---|---|---|---|---|---|---|
| tEPEC | Children <5 yr, adults at high inocula | Small intestine | Profuse watery diarrhea | Humans | Oral rehydration, antibiotics for persistent cases | Attaching and effacing | eae+, bfp+, stx− |
| aEPEC | Humans, animals | eae+, stx− | |||||
| STEC | Adults, children | Distal ileum, colon | Watery diarrhea, hemorrhagic colitis, HUS | Humans, animals, food, water | Hydration, supportive for HUS | Attaching and effacingb | eae+/−, stx+ |
| EIEC/Shigella | Children <5 yr, adults, travelers, immunocompromised persons | Colon | Shigellosis/bacillary dysentery, potential HUS | Humans, animals, food, water | Oral rehydration, antibiotics | NA (invasive) | ipaH+, ial+, stx+ (S. dysenteriae) |
| EAEC | Adults | Small intestine and/or colon | Traveler's diarrhea, HUS (stx+) | Food, occasionally adult carriers | Antibiotics, oral rehydration | Stacked brick and/or invasive | aatA+, aaiC+ (535), other candidates (585) |
| Children | Persistent diarrhea | Antibiotics, oral rehydration, potentially probiotics | |||||
| Immunocompromised persons | Persistent diarrhea | Fluoroquinolones (685) | |||||
| ETEC | Children <5 yr, travelers | Small intestine | Watery diarrhea | Food, water, humans, animals | Rehydration, antibiotics | CF mediated | CFs, LT, ST |
| DAEC | Children (increasing in severity from 18 mo to 5 yr), adults | Intestine (uncharacterized location) | Persistent watery diarrhea in children, speculated to contribute to Crohn's disease in adults (809) | Unknown | Rehydration | Diffuse adherent and/or invasive | No uniform markers (802) |
| AIEC | Adults, children | Small intestine | Crohn's disease | Unknown | Antibiotics, surgical resection | NA (invasive) | Uncharacterized |
NA, not applicable.
Only for LEE-positive STEC, not for LEE-negative STEC.
MICROBIOLOGY, ISOLATION, AND TYPING OF E. COLI
E. coli is a Gram-negative, oxidase-negative, rod-shaped bacterium from the family Enterobacteriaceae. It is able to grow both aerobically and anaerobically, preferably at 37°C, and can either be nonmotile or motile, with peritrichous flagella. E. coli is readily isolated from fecal samples by plating on selective media. The change in pH due to lactose fermentation can be used to differentiate between lactose-fermenting and non-lactose-fermenting strains, as lactose-positive E. coli colonies will appear red or pink on media such as MacConkey agar. Not all E. coli strains, particularly most EIEC and Shigella strains, ferment lactose, so caution must be used when using this diagnostic. While this selective plating can aid in isolating E. coli from Gram-positive bacteria and some other Enterobacteriaceae members, further morphological, phenotypic, and genotypic characteristics need to be tested for further identification and verification of pathotypes. Traditional culture techniques for pathogenic E. coli can be time-consuming and laborious. The adoption of molecular techniques (Fig. 2) has allowed for more rapid detection and identification of the different pathotypes. Current methods of identification of each pathotype are discussed in the appropriate sections below.
Fig 2.
Diagnostic tools for intestinal pathogenic E. coli. E. coli causes a variety of diarrheal diseases in humans owing to specific colonization and virulence factors associated with each pathotype. As no single method can be used to detect and diagnose all pathogenic E. coli strains, a number of biochemical tests, typing methods, and molecular approaches have been developed to isolate E. coli from other enteric bacteria as well as to differentiate between particular pathotypes. Prospective methods such as whole-genome sequencing or high-throughput sequencing are becoming fast and affordable, providing much information about the pathogen that may be useful to clinicians, epidemiologists, and public health workers.
Classic serotyping is based on the Kauffman classification scheme, where the O (somatic) polysaccharides and H (flagellar) surface antigens are determined (8). Molecular methods such as PCR of genes involved in O-antigen biogenesis (e.g., wzx and wzy genes) and of fliC for the H antigen, can also be used to identify the serotype (9, 10). A designation of NM or H− indicates an absence of the H antigen, and that the isolate is nonmotile. Currently, there are 174 E. coli O (10) and 53 E. coli H (9) antigens recognized; however only a small subset of O:H combinations are associated with disease.
While serotyping is informative for certain pathotypes (e.g., STEC O157:H7), it is not always useful for others due to isolates being untypeable or cross-reactivity between antigens. Other methods have been developed to type isolates for phylogenetic analysis, outbreaks, and surveillance investigations. Pulsed-field gel electrophoresis (PFGE) is considered the gold standard for typing and is applied in epidemiological investigations to discriminate between outbreak isolates (11). However, PFGE is laborious and time-consuming, requires technical expertise, and is not portable to laboratories that are not well equipped. An alternative method, called multilocus variable-number tandem repeat analysis (MVLA), has been shown to discriminate between sporadic isolates and outbreaks (12) and may be more portable than PFGE.
Multilocus sequence typing (MLST) has become a common method for typing pathogenic E. coli strains (13–18) and establishing their relatedness. A small number of housekeeping genes are sequenced and assigned a unique allele, and the allelic profile of the housekeeping genes can be used to give an isolate a sequence type (e.g., E. coli O104:H4 is ST678 and many STEC O157:H7 isolates are ST11, based on the MLST databases [see below]). Sequence types can be further grouped into clonal complexes based on their similarity. However, genetic diversity can be found within strains of a similar sequence type, so higher-resolution typing may be required to understand evolutionary relationships (19).
Currently three MLST schemes exist in publically curated databases: the EcMLST (http://www.shigatox.net), Institut Pasteur Escherichia coli MLST database (http://www.pasteur.fr/mlst), and MLST databases (20; http://www.mlst.net) enabling comparative results between laboratories. Recently, a public server that uses whole-genome sequencing to identify sequence types was set up (21; http://cge.cbs.dtu.dk/services/MLST). For more information on the use of MLST for molecular epidemiology, see a recent, thorough review dealing with this subject (22).
EVOLUTION OF PATHOGENIC E. COLI
As a population, E. coli strains can be assigned phylogenetically to 5 main groups, i.e., A, B1, B2, D, and E, with Shigella forming different groups (23) (Fig. 3). Commensal isolates mostly group in phylogroup A; however, not all E. coli pathotypes group together. For example ETEC was found to group with phylogenetic groups A and B1 (24), while EAEC grouped in phylogenetic groups A, B1, B2, and D (16), demonstrating the disparate nature of pathogenic E. coli genotypes. The population genetics and evolution of commensal and pathogenic E. coli strains have been recently reviewed (25).
Fig 3.
Phylogenetic tree of intestinal pathogenic E. coli. E. coli strains can be grouped into 5 main phylogenetic groups: A (blue), B1 (green), B2 (brown), D (pink), and E (red). Shigella/EIEC also form additional phylogroups (black). Pathotypes do not always group together in the same phylogroup. The hybrid EAEC and STEC strains are denoted with both an open square and open circle. Unmarked strains are either commensal, extraintestinal pathogenic E. coli (ExPEC), or avian-pathogenic E. coli (APEC). ETEC strains are isolated from both humans and animals, while DAEC is not represented in the phylogenetic tree. (Adapted from reference 19, which was published under a Creative Commons license.)
Genome sizes of E. coli can differ by a million base pairs between commensals and pathogenic variants, and this extra genetic content can contain virulence and fitness genes. Comparative genomics have shown that E. coli genomes are split between a shared, conserved set of genes, called the core genome, and a flexible gene pool. A recent comparison of 186 E. coli genomes found approximately 1,700 homolog gene clusters shared in all genomes and a pangenome of about 16,400 gene clusters (26). The pathogenic ability of E. coli is therefore largely afforded by the flexible gene pool through the gain and loss of genetic material at a number of hot spots throughout the genome (23) (Fig. 4). For example, nearly one-quarter of the EAEC strain 042 genomic content is made up of genomic islands (27), similar to the percentage of unique genomic islands found in STEC O157:H7 strain EDL933 (28). DNA can be moved between prokaryotic hosts through mechanisms such as conjugation, transformation, and transduction (reviewed in reference 29), encoded by mobile genetic elements, resulting in horizontal gene transfer (HGT). Mobile genetic elements, such as transposons, insertion sequences, bacteriophages, and plasmids, can exist either integrated into the chromosome or through self-replication within the new host to provide new traits and fitness advantages. Most definable virulence factors found in pathogenic E. coli are derived from genetic mobile elements. For example, most of the genes for toxins and colonization factors (CFs) required for the pathogenesis of ETEC are found almost exclusively on plasmids (30), while EPEC and some STEC strains share a 35-kb cluster of virulence genes on a chromosomal pathogenicity island (PAI) called the locus of enterocyte effacement (LEE), which is required for the attaching and effacing (A/E) phenotype that is necessary for virulence (31). It should be noted that genes encoding products referred to as virulence factors in humans (e.g., LEE or Shiga toxins) may in fact contribute to survival in the environment or commensalism in other hosts and may provide a driving adaptive factor for retention of certain traits (32).
Fig 4.
General overview of pathogenic gene acquisition and loss for different pathotypes. Gene gain and loss afford pathogenic traits to E. coli and ultimately lead to the pathotypes discussed in this review. Acquisition of genes is generally from mobile elements such as transposons, prophages, and plasmids. Typical EPEC carries the LEE and bundle-forming pilus gene (bfp), while most LEE-positive STEC strains (such as EHEC) also carry the LEE as well as Shiga toxin genes (stx1, stx2, or a combination). ETEC isolates carry enterotoxins LT and ST solely or together on plasmids, as well as colonization factors (CFs). Some DAEC isolates have acquired fimbriae that enhance adherence, called the Afa/Dr, while many virulence determinants for EAEC for some isolates are found on the pAA plasmid. Additionally, the O104:H4 serotype of EAEC, which was involved in the recent outbreak in Germany, acquired the stx2 gene. EIEC/Shigella gained the ability to invade cells mainly through the pINV plasmid and acquired additional virulence traits in the form of chromosomal pathogenicity islands (PAIs). Subsequent pathoadaptation, including loss of antivirulence factors and motility, potentiate its virulence. Genes involved in the pathogenesis of AIEC are unclear. For more details about these genetic determinants, see the text.
Bacteriophages play a large part in the genome plasticity of E. coli. Although many of these phages are seemingly defective, some still form infectious particles. A study of 18 phages found in an STEC O157:H7 isolate determined that some of these phages, including two phages that carry the Shiga toxin (Stx), can infect other E. coli strains, thus contributing further to HGT (33). While a large number of virulence factors can be phage associated, accessory genes can also be acquired on phages. In STEC isolates, tRNA genes for rare codons were introduced that are frequently used by foreign genes (34, 35).
The acquisition of new genes through HGT provides bacteria with a variety of new traits; however, gene loss can also favor the fitness or adaptation of a pathogen in a particular niche. There is a higher rate of gene loss in Shigella than in other pathogenic E. coli strains, which may be due to its restricted host range and lifestyle (36). Interestingly, Shigella has anywhere from 447 to 978 pseudogenes and gene deletions throughout its genome, which is more than any other pathogenic E. coli strains that were included in the study (37). This loss of gene function has been described as pathoadaptivity, and it may be an underappreciated mechanism of pathogenesis that allows the survival of the pathogen in the host. Just a few examples of genes whose loss has helped Shigella adapt to an intracellular lifestyle are genes for lysine decarboxylase (LDC) (cadA), surface protease (ompT), and amino acid transport (argT) (reviewed in reference 38). Pathoadaptive events of EIEC and Shigella are discussed in more detail below. Pathoadaptation is not limited to Shigella, as loss of lysine decarboxylase activity has been shown in EPEC, ETEC, STEC, and EAEC (39, 40), and these pathoadaptive lesions may contribute to enhanced virulence in STEC O157:H7 and outbreak strains of EAEC (39, 41).
The description of pathotypes has usually relied on a signature set of genotypic and phenotypic traits. Because of the plasticity of the E. coli genome and increased sequencing and genomic studies, the designation of certain isolates into a pathotype becomes complicated. Prior to 2011, there were only been a few reports of Shiga toxin-producing EAEC causing bloody diarrhea and hemolytic uremic syndrome (HUS) (42, 43), but these are now more appreciated due to the E. coli O104:H4 outbreak in Germany (44). Characterization of isolates from this outbreak identified key virulence features belonging to different pathotypes, such as an aggregative adhesive phenotype in vitro, lack of the LEE PAI, and expression of a Shiga toxin (45). Therefore, these isolates can be considered a hybrid of both EAEC and EHEC (a subset of STEC), and it has been suggested that the STEC O104:H4 strain associated with the 2011 German outbreak be called enteroaggregative hemorrhagic E. coli (EAHEC) (46). Additionally, genome analysis of LEE-negative STEC has uncovered homologs and subunits of ETEC toxins in some isolates (47), further demonstrating the potential for the emergence of novel pathogenic E. coli hybrids.
As more genome sequences of pathogenic E. coli are being completed, it is clear that the genetic diversity of this organism is vast. As such, defining a pathotype of E. coli based on a small set of features is challenging, as many of these defining genes may not be restricted to a particular pathotype. However, it is still informative to consider the main pathotypes as a framework to an overview of enteric E. coli.
ENTEROPATHOGENIC E. COLI
Enteropathogenic E. coli (EPEC) belongs to a group of bacteria collectively known as attaching and effacing (A/E) pathogens based on their ability to form distinctive lesions on the surfaces of intestinal epithelial cells (IECs). Of the diarrheagenic E. coli, EPEC was the first pathotype to be identified. The term “EPEC” was first used in 1955 (48) to describe a number of E. coli strains epidemiologically related to a series of outbreaks of infantile diarrhea in the 1940s and 1950s (49, 50). Originally defined by serotype, EPEC strains are now classified based on pathogenic characteristics (8).
Classification
Atypical versus typical isolates.
Most EPEC isolates correspond to conventional O serogroups (20, 51), but advances in molecular and cellular detection of EPEC have expanded the known repertoire of EPEC lineages to include strains that would not have been considered EPEC based on serotype alone (52). EPEC, unlike LEE-positive STEC, does not produce Shiga toxin and is distinguished from other diarrheagenic E. coli by the ability to form A/E lesions in the small intestine, a phenotype afforded to it by genes of the LEE (3). EPEC is further classified into “typical” and “atypical” subtypes based on the presence or absence of the E. coli adherence factor plasmid (pEAF) (53). Based on MLST, strains of EPEC fall into 4 clonal lineages, designated EPEC1 to EPEC4, that seem to have evolved through independent acquisition of the LEE and pEAF (15). Despite their formal designation as EPEC, typical and atypical isolates constitute two distinct groups of organisms, with some atypical EPEC (aEPEC) strains being more closely related to LEE-positive STEC in serotypes, genetic characteristics, virulence properties, and reservoirs (53). In contrast to typical EPEC (tEPEC), aEPEC is a highly heterogenous group, and despite its relatively high prevalence in asymptomatic children, aEPEC is now thought to be important in endemic diarrhea in children as well as in outbreaks (54). The highly pathogenic STEC serotype O157:H7 is closely related to aEPEC O55:H7 (55, 56), and recent comparative genomic and proteomic analyses indicate that STEC O157:H7 evolved from an O55:H7 ancestor, with an approximate divergence time of 400 years (57). Among aEPEC, strains of the O51 serogroup are the most frequently isolated, followed by O145, O26, O55, O111, and O119; however, many aEPEC strains are O/H-antigen nontypeable (58). Additionally, some aEPEC lineages have evolved from typical strains that have lost plasmid-carried virulence genes (59, 60), rendering lineage designations based on serotype alone inaccurate. Table 2 shows frequently isolated aEPEC and tEPEC serotypes, including common motile and nonmotile strains.
Table 2.
Classification of frequently isolated typical and atypical EPEC serotypes and common motility phenotypes
| Group | Serotypes |
|
|---|---|---|
| Motile | Nonmotile | |
| Typical | O86:H34, O114:H2, O127:H6, O127:H40, O142:H6, O142:H34 | O55:H6, O55:NM, O111:NM, O111:H2, O119:H6 |
| Atypical | O55:H34, O86:H8, O111:H25, O119:H2, O125ac:H6, O128ab:H2 | O26:H11, O55:H7, O111:H8, O111:H9 |
Epidemiology
Incidence.
The epidemiology of EPEC infection has shifted since these strains were first identified in the 1940s and 1950s. Initially, EPEC was an important cause of infantile diarrhea in the developed world, but over the years it became much more prevalent in developing countries (8). The prevalence of EPEC infection varies between epidemiological studies based on differences in study populations, age distributions, and methods used for detection and diagnosis (61). In addition, geographic region and socioeconomic class may also contribute to the epidemiology of EPEC-induced diarrheal disease (62). In order to understand these issues, large case studies like GEMS are needed to more accurately assess the etiology and population-based burden of EPEC-induced diarrheal disease. Based on recent GEMS data, tEPEC was significantly associated with moderate to severe diarrhea in children under 2 years of age in Kenya, whereas aEPEC was not associated with this type of diarrhea at any of the GEMS sites (7). Overall, tEPEC was not strongly associated with cases of moderate to severe diarrhea, but when present, it was associated with an increased risk of death in patients aged 0 to 11 months (7). Recent estimates from the Centers for Disease Control and Prevention (CDC) on food-related illness in the United States listed only 4 hospitalizations as a result of EPEC infection (63); however, this pathogen continues to persist in other parts of the world and continues to be regarded as a serious threat to children under the age of 2.
The occurrence of diarrhea due to tEPEC decreases with age, and infections in adults are rarely reported. This apparent resistance in adults and older children has been attributed to the loss of specific EPEC receptors with age or development of immunity (8). For many years, infections with aEPEC were thought to predominate in industrialized nations while being relatively rare in the developing world (53, 64); however, recent data indicate that infections with aEPEC exceed those with tEPEC in both developing and developed countries (58, 61). For example, a 5-year study of children under 12 years of age in Thailand found that 71.8% of EPEC isolates were atypical in nature (65). Likewise, 92% of EPEC isolates collected from children in Brazil between 2001 and 2002 were atypical (66), compared to 38% in a 1998-1999 study (52). More recently, 39.3% of EPEC strains isolated from children with diarrhea in Iran tested positive for the bfp gene, while the remaining 61.7% appeared as aEPEC, which lacks bfp (67). Studies from Norway (68) and Australia (69) also suggest that aEPEC isolates are more commonly found among persistent cases of diarrhea than typical isolates. However, this notion cannot be generalized, as other studies still report tEPEC being more prevalent than aEPEC as a cause of diarrhea (70). Although many countries no longer consider tEPEC strains to be an important cause of acute diarrhea, the occurrence of severe disease outcomes associated with these infections has reemerged.
Transmission and reservoirs.
As with other diarrheagenic E. coli, EPEC is transmitted from host to host via the fecal-oral route through contaminated surfaces, weaning fluids, and human carriers (71) (Fig. 5). Although rare, outbreaks among adults seem to occur through ingestion of contaminated food and water; however, no specific environmental reservoir has been identified as the most likely source of infection (8). The infectious dose in adult volunteers is high, at 108 to 1010 organisms (72, 73), while the actual dose required to cause disease during natural infection is unknown. EPEC outbreaks have been reported to show a seasonal distribution with peaks during the warm months (64, 68, 74, 75). In Norwegian children, 47.7% of EPEC isolates were identified between July and September (68). Humans are the only known reservoir for tEPEC, with symptomatic and asymptomatic children and asymptomatic adults being the most likely source (71). In contrast, atypical strains have been isolated from human and animal sources, including dogs, rabbits, monkeys, and sheep (76, 77). Many human and animal EPEC species are clonally related (77), sharing many virulence properties. A recent survey of laboratory rabbits obtained from commercial vendors and presenting with acute diarrhea found a disease-associated strain of aEPEC in animal stools and a significant association between diarrheic animals and the presence of aEPEC (78). These data suggest interspecies transmission as a means for human infection with aEPEC and rabbits as a possible animal reservoir. Strategies to prevent transmission and spread of EPEC include proper hand washing and improvements in sanitary conditions and freshwater supplies, as well as instruction on food, domestic, and personal hygiene (62).
Fig 5.
General overview of potential reservoirs and modes of transmission for pathogenic E. coli. Pathogenic E. coli strains can be found in various animal reservoirs and can spread between these and other animals. Fecal matter can contaminate food, irrigation water, or recreational/drinking water. Humans can become exposed following the ingestion of contaminated food or water or through direct contact with colonized animals. Secondary transmission can occur between humans, commonly in day care centers or nursing homes. Food can become contaminated through poor cooking practice, where, for example, uncooked meat could come in contact with other food. Additionally, symptomatic or asymptomatic food handlers can contaminate food, particularly when hand hygiene is inadequate. Contamination of recreational or drinking water can occur through exposure of human sewage.
Pathogenesis
In general, EPEC is a noninvasive organism and does not produce heat-labile (LT) or heat-stable (ST) enterotoxins. EPEC belongs to a group of pathogenic bacteria capable of causing A/E lesions on the surface of the host's intestinal epithelium (Fig. 6). EPEC is the prototype organism for strains causing A/E histopathology; however, other human pathogens, including LEE-positive STEC and Escherichia albertii, as well as several animal pathogens, such as rabbit-enteropathogenic E. coli (REPEC)/RDEC-1, porcine-enteropathogenic E. coli (PEPEC), dog-enteropathogenic E. coli (DEPEC), and Citrobacter rodentium (mouse), are also members of the A/E pathogen family (79). The A/E lesion is a hallmark of EPEC pathogenesis, characterized by effacement of brush border microvilli at the site of bacterial attachment (80). The dissolution of the intestinal brush border is accompanied by formation of actin pedestals that extend from the surface of the epithelium into the lumen (81). These pedestal-like structures are produced through secretion of a conserved bacterial receptor protein, Tir, via a type III secretion system (T3SS) that is absolutely required for EPEC pathogenesis (82). A three-stage model of EPEC pathogenesis was first described in the early 1990s by Donnenberg and Kaper, including localized adherence to host cells, signal transduction, and intimate attachment (83). Concomitant with intimate attachment, a series of bacterial effector proteins are injected into host cells, where they subvert actin dynamics and other host cellular processes (84).
Fig 6.
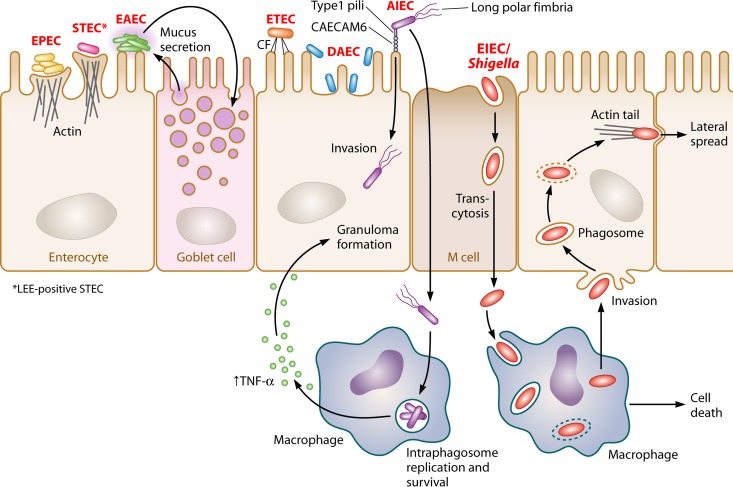
Adherence patterns of enteric E. coli. Pathogenic E. coli requires adherence to the host epithelium. Enteropathogenic E. coli (EPEC) (represented in yellow) and LEE-positive Shiga toxin-producing E.coli (STEC) (represented in pink) are extracellular pathogens that attach to the intestinal epithelium and efface microvilli, forming characteristic A/E lesions. Due to the presence of bundle-forming pili, EPEC is capable of forming microcolonies, resulting in a localized adherence (LA) pattern. Enterotoxigenic E. coli (ETEC) (represented in orange) uses colonization factors (CFs) for attachment to host intestinal cells. Enteroaggregative E. coli (EAEC) (represented in green) forms biofilms on the intestinal mucosa, and bacteria adhere to each other as well as to the cell surface to form an aggregative adherence pattern (AA) known as “stacked brick.” Diffusely adherent E. coli (DAEC) (represented in blue) is dispersed over the surfaces of intestinal cells, resulting in a diffuse adherence (DA) pattern. Adherent invasive E. coli (AIEC) (represented in purple) colonizes the intestinal mucosae of patients with Crohn's disease and is capable of invading epithelial cells as well as replicating within macrophages. AIEC uses type I pili to adhere to intestinal cells and long polar fimbriae that contribute to invasion. Enteroinvasive E. coli (EIEC)/Shigella (represented in red) are intracellular pathogens that penetrate the intestinal epithelium through M cells to gain access to the submucosa. EIEC/Shigella escape submucosal macrophages by induction of macrophage cell death followed by basolateral invasion of colonocytes and lateral spread.
Adherence.
Initial attachment of tEPEC to the surface of the host intestinal epithelium is mediated by the bundle-forming pili (BFP) (85). BFP are type IV pili that tether individual bacteria to one another, producing a localized adherence (LA) pattern in the form of compact three-dimensional microcolonies that can be seen on HEp-2/HeLa cells within 3 h of infection (86). The LA phenotype correlates with EPEC1 and EPEC2 clonal-lineage strains carrying large EAF plasmids (87). The self-transmissible EAF plasmid pMAR2 is found among strains of the EPEC1 lineage and contains an intact transfer region, unlike pB171, which is more common among EPEC2 strains (88). Both plasmids share a pEAF backbone and, in addition to the bfp operon, carry a second virulence-related operon known as perABC (89). Between pMAR2 and pB171, the bfp and per loci share 99% sequence similarity (88), and both BFP and PerA have been shown to contribute to virulence in human volunteers (72). PerABC are plasmid-encoded regulators required for expression of the bfp and perABC operons as well as the LEE-encoded global regulator Ler (reviewed in reference 90), thereby linking BFP expression to expression of the LEE in tEPEC strains. In contrast, aEPEC strains do not harbor pEAF and thus do not produce BFP, resulting in the formation of loose clusters of bacteria on tissue culture cells. This pattern, known as “localized adherence-like” (LAL) (91), is slower to establish and can take up to 6 h to form (58). LAL is the most common pattern seen among aEPEC strains; however, some strains display alternate adherence phenotypes such as diffuse adherence (DA) and aggregative adherence (AA) (53) (Fig. 7). In addition to BFP, tEPEC strains encode a large surface protein, lymphocyte inhibitory factor (LifA), that contributes to epithelial cell adherence in vitro (92) and is required for intestinal colonization of mice by the related A/E pathogen C. rodentium (93). LifA has recently been identified as the largest secreted effector for any pathogen possessing a T3SS (94), and the gene encoding LifA has been found in the genomes of several A/E pathogens, including many isolates of aEPEC (92). The lifA gene is more commonly found among tEPEC rather than aEPEC strains (95); however, aEPEC strains harboring lifA have a significant association with diarrhea in children under 5 years of age (96). The E. coli common pilus (ECP) is an additional adherence factor, which is involved in epithelial cell colonization of commensal and pathogenic E. coli strains (97). In EPEC, ECP has been shown to act as an accessory adherence factor, playing a role during cell adherence and/or in bacterium-bacterium interactions (98). Although mutation of pilus genes in STEC O157:H7 leads to a substantially reduced ability to adhere to epithelial cells, the significance of ECP to EPEC pathogenesis has not been determined (97). The T3SS filament EspA has also been shown to play a role in initial brush border attachment. While BFP has been shown to be the predominant factor required for initial attachment of tEPEC (99), EspA filaments do promote attachment, albeit in a less efficient manner, and could mediate adherence of strains lacking BFP.
Fig 7.
Adherence patterns of enteropathogenic E. coli (EPEC) on tissue culture cells. (a) Localized adherence (LA); (b) localized adherence like (LAL); (c) diffuse adherence (DA); (d) aggregative adherence (AA). (Reprinted from reference 58 with permission [© 2009 Federation of European Microbiological Societies].)
Signal transduction and intimate attachment.
Formation of the A/E lesion occurs by subversion of actin dynamics within host cells and is mediated by the interaction between intimin and the bacterial translocated intimin receptor, Tir (100–102). Intimin is a 94-kDa protein encoded by the eae gene, which is found in all strains capable of inducing A/E histopathology (8). The N terminus of intimin is highly conserved among A/E pathogens, whereas the C terminus shows much less homology (103). Differences in the C-terminal region of intimin have been used as a basis for classification into several distinct subtypes, with the α and β subtypes more commonly found among tEPEC strains and the α, β, γ, ζ, δ, and ε subtypes being found among aEPEC strains worldwide (104). Intimate attachment of EPEC to intestinal cells induces diverse signal transduction pathways within the host, leading to subversion of many cellular processes for the benefit of the pathogen. The genes required for signal transduction and production of the distinctive A/E lesion are carried on the conserved 35-kb LEE PAI that is found in all A/E pathogens (31, 105). The core LEE encodes a T3SS, the main function of which is to secrete protein components of the translocon (EspA, EspB, and EspD) and to drive effectors directly into host cells. There is a high degree of conservation in the genes encoding the T3SS apparatus itself, whereas the genes encoding effector proteins show considerable variation (106). In addition to the T3SS apparatus, the LEE carries regulatory genes and genes for secreted effectors and their related chaperones (107–109). In some tEPEC, aEPEC, and STEC strains, the presence of additional genes in the LEE flanking regions can extend the LEE up to 110 kb (106), although the role of these additional genes in pathogenesis is variable or unknown.
Secreted proteins.
The EPEC genome contains seven LEE-encoded effector genes in addition to several non-LEE (Nle)-encoded effector genes, all of which exploit the LEE T3SS for delivery into host cells (107, 109, 110). Of the seven LEE-encoded effectors, one of the best studied is the translocated receptor, Tir. Tir is found at the tip of the pedestal, where it acts as a cellular receptor for the bacterial transmembrane protein intimin (102). In addition to its receptor function, Tir is also involved in effacement of microvilli and recruitment of cytoskeletal proteins for pedestal formation (79, 84). Recently, Tir has also been shown to inhibit NF-κB activity through tumor necrosis factor alpha (TNF-α) receptor-associated factors (111). The remaining six LEE-encoded effectors, Map, EspF, EspG, EspZ, EspH, and EspB, all have physiological roles relevant to A/E pathogen infection. Map stimulates formation of membrane filopodia and epithelial barrier disruption as well as mitochondrial dysfunction (112). Multifunctional properties have also been reported for EspF and EspG, both of which affect aquaporin localization, leading to diarrhea (113). Like Map, EspF localizes to mitochondria (114) and has been shown to disrupt tight junctions (115), while EspG alters host cytoskeletal components through its interaction with tubulin (116). EspZ promotes host cell survival (117), whereas EspH affects filopodium formation, participates in actin signaling during pedestal formation (118), and acts as a RhoGEF inhibitor (119). Both EspH and EspB are capable of inhibiting phagocytosis of EPEC by macrophages (120, 121). There is considerable variation in the number and type of Nle-encoded effectors among A/E pathogens (84). EPEC E2348/69 encodes at least 23 Nle effectors (94, 109), many of which are involved in dampening the host immune response. NleB, NleC, NleD, NleE, and NleH have all been shown to inhibit NF-κB activation through a variety of different mechanisms (122–129). In addition to immunomodulatory functions, Nle effectors such as EspJ have antiphagocytic properties (130), while NleA alters host protein secretion (131) and tight junction integrity (132) and inhibits vesicle trafficking (133). NleH is capable of modulating apoptotic response (134). For a thorough review of EPEC effector functions and host cellular targets, see references 84 and 135).
In addition to T3S effectors, some EPEC strains also encode the type V secreted virulence-associated protein EspC. EspC is a Per-activated serine protease autotransporter that acts as an enterotoxin, causing cytopathic effects on tissue culture cells (136) and rat jejunal segments (137). Cytotoxicity of EspC depends on a conserved serine protease motif as well as on epithelial cell internalization (138, 139). EspC has been shown to enter intestinal epithelial cells through a cooperative mechanism involving both the type V secretion system (T5SS) and the T3SS (138). Upon delivery to the exterior of the cell via the T5SS, EspC interacts with EspA and is internalized during pedestal formation via the T3SS translocon (138, 139). EspC confers enhanced lysozyme resistance to EPEC (140), and purified EspC has been shown to interact with and degrade hemoglobin (141) and to hydrolyze other proteins such as pepsin, factor V, and spectrin (142). Additionally, oligomerization of EspC gives rise to rope-like structures that serve as a substratum for adherence and biofilm formation as well as to protect bacteria from antimicrobial compounds (143). While the role of EspC in EPEC pathogenesis in vivo remains unclear, it has been suggested that EspC plays a significant role in EPEC survival (141, 143).
Mechanism of diarrhea production.
Unlike that induced by ETEC, EPEC-induced diarrhea is not mediated by toxin production. However, the enteroaggregative heat-stable toxin 1 (EAST1) gene, astA, has been found in aEPEC strains significantly associated with diarrhea in Brazil (144). The astA gene has also been found in aEPEC O39:NM, O111, and ONT:H45, which have been associated with outbreaks in the United States (145), Finland (146), and Japan (147), respectively. The EAST1-encoding gene has been detected in many diarrheagenic E. coli strains as well as in nonpathogenic commensal strains (148); however, whether or not astA is expressed in aEPEC and triggers diarrhea has yet to be determined (58). EPEC persists in the small bowel by resisting phagocytosis (149) and dampening the host immune response (150). The exact mechanism of diarrhea production is not fully understood and likely involves a combination of different mechanisms (79). The speed of diarrhea onset implies a secretory mechanism rather than malabsorption as a more likely cause of diarrhea. Nonetheless, the effacement of microvilli at the sites of bacterial attachment could lead to a decrease in absorptive surfaces, thereby contributing to diarrhea through interference with proper absorptive channels. A number of T3S effectors are known to impact various water and ion channels of the intestinal epithelia and are thought to contribute collectively to EPEC-induced diarrhea but the exact contribution of each mechanism is unknown. Tir, Map, EspF, and EspH inhibit the sodium–d-glucose transporter (SGLT1), which is responsible for fluid uptake from the intestine (84, 151), while EspF and EspG alter localization of aquaporins, which also play a role in water transport (113). EspG and EspG2 are also responsible for disruption of chloride transport across the apical membrane resulting in decreased Cl−/OH− exchange activity (152). Additionally, disruption of tight junctions by EspF (153), EspG (154), and Map (155) leads to increased intestinal permeability, which could also contribute to EPEC-induced diarrhea.
Clinical Considerations
Symptoms.
EPEC is a significant cause of infectious diarrhea that is often accompanied by fever, vomiting, and dehydration in children under 2 years of age (3, 8, 79). In human volunteers, the onset of diarrhea due to EPEC is fairly rapid, occurring as early as 2.9 h after ingestion of wild-type bacteria (156). Acute diarrhea is the most likely result of EPEC infection, but persistent cases, lasting more than 2 weeks, have also been reported (8, 71). In comparison to infection with other diarrheal pathogens such as adenovirus, rotavirus, Campylobacter, and Salmonella, infection with EPEC is more likely to lead to development of persistent diarrhea and hospitalization (69). Other clinical features associated with EPEC-induced diarrhea include intolerance to cow's milk and an increase in failure to respond to oral rehydration therapy (61).
Detection.
The traditional diagnosis of EPEC, involving O:H serotyping, has changed over the years as knowledge of this pathogen has increased. Most EPEC strains fall into well-established serogroups, but serotype designation is no longer a necessary trait for a strain to be considered “EPEC” (8). Diagnosis of EPEC is now made based on pathogenic characteristics that distinguish it from other E. coli species and subdivide EPEC into “typical” and “atypical” categories. EPEC is defined based on phenotypic properties, such as LA and A/E histopathology, that can be readily assessed using microscopy and cell culture techniques where tissue culture facilities are available, as well as by the presence or absence of genetic elements such as eae, bfp, and stx (8). The fluorescent actin staining (FAS) test, originally described by Knutton et al. (157), uses fluorescein isothiocyanate (FITC)- or rhodamine-conjugated phalloidin to label concentrated patches of filamentous actin beneath A/E bacteria on the surfaces of cultured epithelial cells. These concentrated spots of intense fluorescence are indicative of pedestal formation and signify a positive FAS test for A/E lesion formation (158, 159). In addition to the FAS test, a HEp-2 or HeLa cell adherence assay is another phenotypic test that can be used to distinguish tEPEC from aEPEC and other diarrheagenic E. coli such as DAEC and EAEC (86, 160, 161). Many clinical laboratories, however, do not have tissue culture facilities, and thus neither the HEp-2 adherence assay nor the FAS test can be routinely used for diagnosis in these settings. For this reason, genotypic tests for detection of EPEC are the preferred method of identification and generally involve DNA probe hybridization or PCR-based screens targeting eae, bfp, and EAF sequences. Genotypic detection methods have been adopted by clinical and research laboratories for the identification of EPEC strains; however, general concerns with respect to allelic variability in the eae and bfpA genes has generated interest in refining tests to include new genetic targets. For example, differentiating typical from atypical strains can be difficult based on bfpA alone. Under the current definition, isolates containing deletions in essential genes of the bfp operon outside bfpA would be classified as tEPEC while phenotypically behaving as atypical strains. Furthermore, certain isolates that produce BFP do not react with the EAF probe (53), while others that react with the EAF probe do not form BFP (59). Additional genome sequence data will be required to identify new genetic targets that can be used to correctly distinguish between tEPEC and aEPEC as well as between strains that cause acute diarrhea and those that do not.
Treatment.
In most cases, EPEC-induced diarrhea is self-limiting and can be effectively treated with oral rehydration therapy. Persistent infections may require the use of antimicrobials; however, the antibiotic resistance profile of EPEC is an important factor in determining the success of treatment in response to infection, as several clinical isolates exhibiting a high degree of resistance to standard antibiotics have been reported (162). In addition to resistance patterns, other factors limiting the effectiveness of antibiotics in the treatment of EPEC infections are cost and supply in developing countries, where EPEC infections are common (163).
Antibiotic resistance.
The prevalence of antibiotic-resistant EPEC in developing and developed countries is increasing (162). Antibiotic-resistant EPEC has been found across many continents, with reported cases in the United States (164), the United Kingdom (165–167), Brazil (52), Iran (168), and Singapore (169). EPEC displays resistance to a range of antibiotics, including penicillins, cephalosporins, and aminoglycosides (162). A recent study of 149 EPEC strains isolated from children in Brazil found that resistance was more common among tEPEC strains than among aEPEC strains (52). In addition, markers for a conjugative multidrug resistance plasmid were detected in 30% of atypical isolates, compared to only 4% of typical strains (52).
Vaccines.
The spread of infections due to EPEC has been well documented with numerous case studies in hospitals and nurseries (8, 71); however, no vaccines are currently available to control its spread. Antibodies against EPEC O antigens and outer membrane proteins have been found in breast milk (170, 171), and protection from mother to infant can be transmitted through colostrum IgA (172, 173). Antibodies from maternal colostrum and serum samples have been shown to recognize EPEC surface antigens such as Bfp and intimin, as well as the secreted proteins EspA and EspB, albeit with various degrees of reactivity that are likely dependent on antigen type and sample source (174). Therefore, several candidate vaccines based on conserved virulence proteins such as EspB (175, 176), BfpA (176, 177), and intimin (178) have been explored. Purified recombinant versions of EspB and BfpA were capable of eliciting an antibody response in rabbits and showed antigenic potential in humans when reacted with secretory IgA (sIgA) present in the stools of diarrheic pediatric patients (175), indicating that an immune response to these potential vaccine subunits can be produced at an early age (176). REPEC Δeae mutants show reduced adherence to intestinal brush borders and protection from homologous rechallenge using a rabbit model of infection (178). Conversely, human volunteer studies have shown that 36% of individuals who ingested an EPEC Δeae strain still developed diarrhea (156), indicating that live attenuated mutants of this nature may not be viable candidates for human vaccine development. In adult volunteers, diarrhea developed in 10% of individuals who consumed the ΔespB mutant, compared to 100% who consumed wild-type EPEC (179). Similarly, a Δbfp mutant was significantly less virulent to human volunteers than wild-type EPEC but was still able to colonize and cause diarrhea in some cases (72). These data suggest that more than one antigenic factor may be required for successful vaccine development. Recently, bacterial ghosts devoid of cytoplasmic contents but expressing all EPEC surface components were constructed and used in vaccination challenge experiments with mice (180). Vaccinated mice showed 84 to 90% protection when challenged with wild-type EPEC, compared to no protection in control mice (180). Homologous rechallenge with wild-type EPEC resulted in a reduced severity of disease but had no effect on incidence of diarrhea (181).
SHIGA TOXIN-PRODUCING E. COLI
The presence of the Shiga toxin 1 or 2 gene (stx1 or stx2), typically acquired by a lambdoid bacteriophage, in an isolate of E. coli qualifies it as Shiga toxin-producing E. coli (STEC) or verocytotoxin-producing E. coli (VTEC). Despite there being over 400 STEC serotypes identified, only a subset of these have been correlated to illness in humans (182). STEC encompasses a diverse pathotype that can cause mild to bloody diarrhea and HUS.
Enterohemorrhagic E. coli (EHEC) is a subset of STEC and was originally described by its association with hemorrhagic colitis (HC), which was clinically distinct from shigellosis, and had genotypic and phenotypic features that differed from those of EPEC (8, 183). Generally, EHEC is LEE positive and forms A/E lesions as does EPEC. However, the term EHEC has also been used in the literature to describe LEE-negative STEC strains, such as serotypes O91:H21, O104:H4, and O113:H21, that have caused HC and HUS. STEC O104:H4 can be considered a hybrid of EHEC and EAEC and is discussed both in this section and in the section on enteroaggregative E. coli below, as the genetic backbone is more closely related to that of an stx-negative EAEC O104:H4 isolate from Central Africa (44, 45). It has been suggested in the literature that this strain be called EAHEC (46).
The most common EHEC serogroup is O157:H7 and has been the subject of many studies, especially for molecular mechanisms of pathogenesis. While STEC O157:H7 has been classified as an adulterant in beef since 1994, the U.S. Department of Agriculture (USDA) has recently declared 6 more EHEC serogroups, i.e., O26, O45, O103, O111, O121, and O145 (also known as the “Big 6”), to be adulterants (184), as they are the most commonly found non-O157 STEC strains associated with severe illness in humans. It should be noted, however, that the prevalence of non-O157 STEC differs geographically (185).
There are important implications of STEC in veterinary microbiology (animals as reservoirs), environmental microbiology (land and water), food microbiology (contamination, handling, and preparation), and clinical microbiology (public health, surveillance, and human illness). The wealth and diversity of literature on STEC exemplify its impact on many fields of study.
Classification
Common serotypes.
In most parts of the world, STEC O157:H7 is the most common serotype that causes human illness. However, it is becoming evident that non-O157 STEC strains also cause significant human illness (Table 3). A study of non-O157 STEC human illness in the United States between 1983 and 2002 found that the most common serogroups were the “Big 6” described above (186). In Australia, non-O157 STEC strains made up 42% of all typeable STEC isolates, with O111 and O26 being the most common serogroups (187). Likewise, these serotypes are common in many parts of the world, but the overall prevalence differs geographically (reviewed in reference 185). Sorbitol-fermenting O157:NM (SFO157:NM) was found to be isolated in approximately 17% of HUS cases from Germany and Austria between 1996 and 2006 (188).
Table 3.
Common STEC serotypes discussed in this reviewa
Lineages.
There are four different clonal lineages of STEC: EHEC 1, which includes O157:H7 and SFO157:NM; EHEC 2, which contains non-O157 STEC serotypes such as O111:H8 and O26:H11; STEC 1, which contains LEE-negative STEC serotypes such as O113:H21 and O91:H21; and STEC 2, which includes serotypes O45:H2 and O103:H2/H6 (55; http://www.shigatox.net). STEC O157:H7 strains have been further classified into 2 different lineages (lineages I and II), based on lineage-specific polymorphism assay 6 (LPSA-6) (189), and 9 different clades, based on single nucleotide polymorphism (SNP) analysis (190). The phylogeny of LEE-negative STEC strains is disparate, as they appear to be more related to other E. coli pathotypes than LEE-positive STEC strains and may have evolved on multiple occasions (47).
Epidemiology
The surveillance and control of STEC have become a major focus of public health authorities. While the primary focus was placed on the detection of E. coli O157:H7, it has become apparent that non-O157 STEC is also a major contributor to sporadic cases and outbreaks in North America, Australia, and Europe. A specific program called the Foodborne Diseases Active Surveillance Network (FoodNet) provides active surveillance of food-borne illness in the United States (191, 192). It should be noted that FoodNet provides surveillance for 15% of the population of the United States. Additionally, PulseNET was created to provide a database of standardized PFGE fingerprints, allowing for comparisons to aid in epidemiological investigations during outbreaks (11). Since its introduction, PulseNET networks have been set up internationally, except in sub-Saharan Africa.
Incidence.
In the United States, there were 463 STEC O157:H7 (0.97 per 100,000 population) and 521 non-O157 STEC (1.10/100,000) cases reported by FoodNet in 2011 (193). Hospitalization rates due to these cases were over 2-fold higher for STEC O157:H7 (43.4%) than for non-O157 STEC (18%). Similarly, the case-fatality rate for O157:H7 was about 2-fold higher than that for non-O157 STEC. What is promising is the fact that the incidence of STEC O157:H7 has dropped 42% in 2011 compared to the incidence in 1996 to 1998 (193). Increased incidences of non-O157 STEC were identified in a statewide study in Washington between 2005 and 2010; the authors suggest that this increase may be due to changes in testing (194). The incidence in Canada, reported by the National Enteric Surveillance Program (NESP), has also improved for STEC O157:H7 cases in 2010 (1.18/100,000) compared to the incidences in 2005 (2.28/100,000) and 2006 (3.0/100,000) (195). NESP reported that the non-O157 STEC incidence has not changed in the last 10 years. In Australia, the overall incidence of STEC illness from 2000 to 2010 was reported to be 0.4/100,000, with a slight increase in incidence over this time (187). In Europe, the European Center for Disease Prevention and Control (ECDC) and European Food Safety Authority (EFSA) report on STEC incidences from 24 European Union member states; they reported that the overall incidence of STEC in 2009 for the European Union was 0.75/100,000. Ireland and Denmark had incidence rates significantly higher than those in other countries (5.33 and 2.90/100,000, respectively) (196). It should be noted that the ECDC/EFSA recommends not comparing incidences between countries due to differences in detection.
STEC is also prevalent in developing countries such as Argentina, which has been described in the literature to have the highest worldwide incidence of HUS in children under the age of 5 (197). This may be due to excessive exposure to known risk factors associated with STEC infections, including meat consumption, playing in recreational water, and poor personal hygiene (198). In contrast, neighboring Brazil has low incidences of HUS (199), and cases of STEC O157:H7 are uncommon (200). While STEC infections are identified in other developing countries (201, 202), overall surveillance and clinical diagnostics are lacking.
Transmission, reservoirs, and sources.
Transmission of STEC is fecal-oral (Fig. 5), and the infectious dose is thought to be very low. For one STEC O157:H7 investigation, it was determined that the beef patty with the highest level of contaminated had only 675 organisms, and it was suggested that the infectious dose could be lower (203). Similarly, another study following an outbreak estimated a dose as small as one STEC O111:NM organism per 10 g of sausage (204). Low doses such as these were also estimated in sausage in the United States (205). These estimates of infectious dose may be further complicated by the ability of STEC to form viable-but-nonculturable (VBNC) cells when faced with stress in the environment. VBNC cells have been shown to form on food and are still able to produce the Shiga toxin (206); however, it is still unknown what impact VBNC cells have on human illness.
Ruminants such as cattle (both meat and dairy) are widely known to be major reservoirs for pathogenic STEC, and exposure to their fecal matter represents an important source of human illness (reviewed in reference 207). A recent survey of ground beef in the United States showed that approximately 24% of samples tested were positive for stx genes by PCR, with only a small proportion of isolates being potentially pathogenic to humans (208). STEC O157:H7 has been isolated from other animals and insects, which include but are not limited to swine, sheep, deer, wild boars, rabbits, birds, dogs, rodents, and insects (recently reviewed in reference 209). This demonstrates the capacity for human exposure to STEC or for further dissemination of STEC to other animals. Certain STEC serotypes, such as SFO157:NM, and O104:H4, have rarely been or have not been reported as being isolated from animals (210, 211).
Exposure by direct contact with animals or with their feces from petting zoos and farms is an important route of exposure of STEC. It is estimated that animal contact constitutes 8% of non-O157 and 6% of O157:H7 STEC illnesses in the United States (212). A study in Scotland showed that cattle feces can contain from 100 to over 106 CFU of STEC O157:H7 per gram of feces (213). Increased shedding was associated with STEC mucosal colonization at the rectoanal junction (213, 214). High-level shedding has implications for within-herd and between-farm transmission, as well as direct-contact and potential environmental exposure of humans, and has been discussed in more detail by Chase-Topping et al. (215).
In Scotland, it was estimated that 54% of STEC O157:H7 outbreaks were due to environmental exposure (216). STEC has been shown to survive in the soil for up to several months (217, 218), whereas other studies have shown survival for up to a year in nonaerated sheep manure (219) and for at least 3 weeks in various farm animal feces, including that of cattle and swine (220). For different water sources, survival of STEC O157:H7 for at least 2 months under laboratory conditions has been reported (221). The ability of E. coli to survive in the open environment is complex, as it must face many factors, such as temperature, nutrient availability, osmolarity, pH, moisture, and microbial communities (reviewed in references 222 and 223).
Contaminated food and water are responsible for many sporadic and outbreak-related illnesses due to STEC. In the United States, it is estimated that 68% of O157:H7 and 82% of non-O157 STEC illnesses are food related (224). Meat can become contaminated through contact with animal feces during slaughter and processing of colonized animals, whereas vegetables may become contaminated through the use of manure as fertilizer or through contaminated irrigation water (225). Contaminated runoff and irrigation water can also taint nearby water sources, affecting rivers, lakes, and private drinking water wells. Foods that have been involved in human illness include uncooked hamburger, sausage, raw milk and dairy products, apple cider, lettuce, spinach, and sprouts (226). Similar to its environmental survival, STEC has been shown to replicate and survive for long periods of time on various food sources (reviewed in reference 226). Washing of food may also become more difficult for certain foods, as both STEC O157:H7 and O26:H11/NM were shown to attach to spinach using the virulence factor EspA (described in the section on enteropathogenic E. coli above) (227), while Saldaña et al. reported internalization inside spinach leaf tissue (228). Lettuce that is damaged, from shredding or bruising, also provides an opportunity for STEC O157:H7 to replicate more rapidly than on intact leaf surfaces (229).
Nonsource exposure, such as person-to-person transmission, is thought to contribute to approximately 19% of cases during an STEC O157:H7 outbreak (230). Additionally, asymptomatic shedders may also be a source of person-to-person transmission, especially in the case of food handlers or when highly susceptible recipients are involved (231).
Outbreaks.
Outbreaks have continued to be reported for O157:H7 and non-O157 STEC worldwide. Over 1,000 cases and 2 deaths were reported in New York in 1999 due to water well contamination by cattle manure runoff that contained STEC O157:H7 (232). The following year, STEC O157:H7 and Campylobacter contaminated a municipal water source in Ontario, Canada, where there were over 2,000 suspected illnesses (233). STEC O157:H7 has also been detected in outbreaks of bloody diarrhea in countries in sub-Saharan Africa, such as Cameroon (234) and the Republic of Congo (235). An STEC O111:NM strain was the source of illness for 341 people in Oklahoma in 2008, which was reported as being the largest O111 outbreak in the United States (236, 237). Norway was hit with a food-borne outbreak of a rare serotype, STEC O103:H25, that resulted in an abnormally high incidence of HUS (238). The largest reported U.S. STEC O26:H11 outbreak occurred in 2010 at a child care center, where shedding was detected for up to a month (239). SFO157:NM caused an outbreak in Germany in 2002 that had an 11% case-fatality rate and an outbreak in Scotland, in which 8 of 18 patients developed HUS (240, 241).
While these are only a few examples of outbreaks and their sources, many more occur annually worldwide, demonstrating the hazardous effect that STEC has on human health. The large STEC O104:H4 outbreak in Germany will be addressed in section on enteroaggregative E. coli below.
Pathogenesis
Despite the separation of LEE-positive STEC strains into distinct lineages (EHEC 1 and EHEC 2), acquisition of major virulence factors appears to have occurred in parallel in both lineages (51). Analysis of single nucleotide polymorphisms (SNPs) in STEC O157:H7 showed that they could be further divided into 9 clades (190), where more virulent isolates grouped together in clade 8. Although the majority of STEC pathogenesis studies have been done with serotype O157:H7 (EHEC 1 lineage), the shared virulence factors found in both EHEC lineages suggests that they are likely similar mechanistically. A/E lesion formation is caused by the T3SS encoded by the LEE, which injects small effector proteins into the host cell (3, 4), and thus are grouped as LEE-positive STEC. LEE-negative STEC strains have also been isolated in cases of HC and HUS, but since they lack the T3SS, colonization and pathogenesis are likely caused by a unique set of virulence factors.
Many STEC virulence factors are found on large virulence plasmids or genomic islands. Genomic islands unique to STEC O157:H7 strain EDL933 were designated O islands (OIs) (28), and this nomenclature has been used to describe islands in other strains. STEC O157:H7 strains carry a plasmid called pO157 that contains a catalase-peroxidase gene (katP) and genes for other virulence factors such as an enterohemolysin (ehx) and toxB and espP (242, 243). SFO157:NM strains carry a significantly larger virulence plasmid, pSFO157, which lacks katP, toxB, and espP but carries a locus for an adhesin, sfp (244). Plasmids in non-O157 STEC strains, such as serotypes O26, O103, O111, and O145 have gene contents similar to that of pO157, with some variances (35, 245, 246). Interestingly, an exceptionally large plasmid, pO26-Vir, from an O26:H11 isolate, also carries a cluster that encodes a type IV pilus (245).
In contrast to virulence plasmids found in LEE-positive STEC, pO113 from the LEE-negative STEC O113:H21 contains many different virulence factors, including a subtilase (subAB), mucinase (epeA), and an adhesin (saa), but carries espP and ehx (247).
Seropathotypes.
A classification system was created to group STEC strains based on their association and frequency of detection with the incidence and severity of human illness (248). This system included 5 seropathotype classifications (A through E), with the strains most frequently causing severe illness falling in seropathotype A (exclusively STEC O157:H7 and SFO157:NM) and serotypes such as O26:H11, O103:H2, O111:NM, O121:H19, and O145:NM falling in seropathotype B. LEE-negative STEC strains, such as O91:H21 and O113:H21, were grouped in seropathotype C (Table 3). Virulence gene profiling in conjunction with seropathotype classifications has been used to assess the potential of STEC pathogenicity in humans (249, 250); however, a recent opinion published by the EFSA discussed limitations of seropathotype classifications, and proposed a new modified scheme called the HUS-associated serotype(s) (HAS) (251).
Virulence.
With the variety of serotypes and virulence factors found among STEC strains, it is of little surprise that the severity of disease can differ. While stx1-containing STEC can lead to HUS, the presence of stx2 is associated with more severe human disease than that of stx1 (252). In one study, stx2 and a variant, stx2c, were the only subtypes found from HUS cases (253). Recently, it has been noted that there has been an increase in stx2-carrying STEC O26:H11/NM relative to stx1 in Europe, which has been associated with higher rates of severe disease outcomes (254). Additionally, the presence of the LEE also correlated with disease severity (252). OI-122 has been correlated with severity of disease in non-O157 STEC, and in particular, certain nle genes carried by this island may be indicative of higher virulence (248, 255). Further analysis by Coombes et al. looked at the molecular risk associated with nle genes found on different OIs and demonstrated their prevalence in isolates that cause HUS (250).
Manning et al. contrasted a spinach outbreak and a lettuce outbreak of STEC O157:H7 from 2006 with the average of 350 STEC O157:H7 North American outbreaks and noted that the two outbreaks had much higher HUS rates (13% versus 4%) (190, 256). These more virulent isolates clustered in clade 8. The reasons for the increased virulence are unclear; however, these clade 8 isolates frequently carry both stx2 and stx2c, have a higher basal level of stx2 expression, are more adherent to epithelial cells in vitro, and have higher expression of virulence factors (190, 257–259). A recent comparison of a clade 8 genome to that of the STEC O157:H7 outbreak strain from Sakai, Japan, identified putative genes that may contribute to higher pathogenicity (260).
Mechanisms of disease.
Following ingestion, STEC must survive the low acidity of the stomach; like other E. coli strains, STEC strains have the ability to survive in low pH, which has been recently discussed by Hong et al. (261). In order to colonize the intestinal mucosa, STEC must attach to epithelial cells, which is achieved through a myriad of pili and fimbria (reviewed in reference 262). Below we discuss these mechanisms of pathogenesis depending on LEE status, including colonization factors and the contribution of other virulence factors to disease.
(i) Shiga toxin.
Shiga toxin is the main characteristic that defines STEC and is the key virulence factor in STEC causing HUS. Due to its clinical significance and ability to cause disease, it has been the subject of many investigations. Several in-depth reviews have been recently published regarding the mechanism of action in renal cells (positive for cell surface globotriaosylceramide [Gb3+]) (263), the intestinal epithelium (Gb3−) (264), and the vascular endothelium (Gb3+) (265) and its contribution to the pathophysiology of HUS (266).
Shiga toxins can be classed into two types, Stx1 and Stx2, with Stx1 having 3 subtypes (a, c, and d), and Stx2 having 7 (a to g) (267). STEC can carry a single variant, either stx1 or stx2, both stx1 and stx2, or a combination of stx2 subtypes (e.g., stx2a and stx2c). Both stx1- and stx2-containing STEC can lead to HUS; however, stx2 is more often associated with severe disease (252). It should be noted that not all subtypes have been isolated from human disease.
Both Stx1 and Stx2 are encoded on prophages that are integrated into the chromosome. Shiga toxin-carrying phages can become lytic during bacterial stress, and it is believed that Stx1/Stx2 is released from lysed bacterial cells during the lytic cycle of the phage (268). Clinically, the use of antibiotics to treat STEC infections has become contentious due to the stimulation of the lytic cycle and concomitant toxin release through the bacterial SOS response. Studies have shown that fluoroquinolones increased Stx2 production in STEC O157:H7 (269) and that subinhibitory concentrations of fluoroquinolones and trimethoprim induced the lytic cycle, while other antibiotics such as azithromycin did not (270).
Loss of the stx-containing phage has been reported in humans. Initially stx-positive STEC O26:H11/NM and SFO157:NM strains were isolated from patients; however, the isolates were stx negative when isolated several days later from the same patient (271, 272). Since stx-negative STEC fits the definition of aEPEC, Bielaszewska and colleagues looked at isolates that were recovered from bloody diarrhea and suggested that they are not aEPEC, and they coined the term EHEC that lost the Shiga toxin (EHEC-LST) (273). Thus, the stx status of STEC can fluctuate (274). While initial work was done mostly on STEC O26:H11 and O157:H7/NM, additional serotypes, such as O103:H2/NM and O145:H28/NM, could also be isolated as EHEC-LST (188).
The mechanism of Stx delivery and trafficking through endothelial cells, such as renal cells, has been thoroughly described by Lee et al. (275). Briefly, Stx binds to Gb3 on the surface of endothelial cells (276, 277) and is internalized and trafficked through the retrograde pathway from the Golgi apparatus and endoplasmic reticulum (ER) and eventually to the host cell cytoplasm. The A subunit is an RNA-glycosidase that removes an adenine from 28S rRNA, thereby inhibiting protein synthesis and causing cell death. The mechanisms of how Shiga toxins causes cell death are complex, and involve several stress pathways that have been recently reviewed (278). Mechanisms of Stx transport from the intestinal lumen across the epithelium are unknown. It is hypothesized that STEC-induced inflammation may provide the toxin an opportunity to breach the epithelial barrier, and there are other possible mechanisms (discussed in reference 264). A recent study proposed that STEC is able to cross the intestinal epithelium through microfold cells (M cells) and survive in macrophages, and this may be a way for the Shiga toxin to be released into the bloodstream, where it can target other organs (279).
(ii) Cytolethal distending toxin.
The cluster encoding the cytolethal distending toxin (cdtABC) can be found in many E. coli strains. It is found frequently in SFO157:NM isolates but not as often in STEC O157:H7 (280), where it has been associated with certain STEC O157:H7 phage types (281). Sequence differences between the four EPEC cdt variants relative to the one found in SFO157:NM have led it to be designated cdt-V (282). Once delivered into the cell, the enzymatically active CdtB is thought to trigger cell arrest by damaging host DNA (283). Bielaszewska et al. (282) confirmed that the Cdt-V variant from SFO157:NM was able to induce cell arrest in endothelial cells and may contribute to HUS (284). Cdt-V can be found in LEE-negative STEC serotypes O91:H21 and O113:H21.
(iii) EHEC hemolysin.
The EHEC hemolysin (EHEC-hlyA or ehx) is a pore-forming toxin that lyses sheep erythrocytes (285, 286). Although the role of Ehx in virulence has been unclear, several investigations have led to some insight on its contribution to disease. These toxins have been shown to be associated as cargo with outer membrane vesicles (OMVs), prolonging its activity (287). Additionally, Ehx was found to be cytotoxic to endothelial cells and may contribute to the development of HUS (288). More recently, Ehx was shown to be inactivated by another STEC virulence factor, EspP (289). In STEC O113:H21, ehx is found on the large pO113 plasmid (247).
(iv) Autotransporters.
The best studied autotransporter in STEC is the serine protease EspP. Of the four subtypes of EspP, only EspPα and EspPγ are secreted and active (290). EspPα is more common in STEC serotypes correlated with severe disease, such as STEC O157:H7, O26:H11/NM, O111:H8/NM, and O145:H25/H28/NM (290). EspP is a multifunctional protease that cleaves human coagulation factor V, pepsin A (291), complement (292), and EHEC hemolysin, inactivating its hemolytic activity (289).
(v) LEE-positive STEC.
Undoubtedly, LEE-positive STEC has been the best characterized subset of STEC, and these strains form A/E lesions similar to those formed by EPEC (Fig. 6). Studies on pathogenesis have been focused on STEC O157:H7, with extrapolation of some LEE and Nle function derived from the EPEC literature. One striking difference between LEE-positive STEC and EPEC is the effector repertoire found in LEE-positive STEC; genome searches of the STEC O157:H7 genome identified 62 possible effector genes, with 13 thought to be pseudogenes (293), compared to the ca. 30 effectors from EPEC (94). While further studies demonstrated that only 33 of the STEC O157:H7 effectors were likely functional, many serotypes, such as O26, O103, and O111, have more effectors than O157:H7 (35). Generally, most effectors are conserved among all LEE-positive STEC strains, with only minor differences in repertoire.
The regulation of LEE expression is complex and is actively being studied in STEC O157:H7 (reviewed in reference 90). Of particular interest are the stimuli that LEE-positive STEC encounters during the course of infection to strategically control LEE genes. It has been shown that LEE-positive STEC is able to gauge its environment in different hosts through the detection of hormones and signaling molecules from both the host and the microbial flora and to regulate the LEE accordingly (reviewed in references 294 and 295). LEE-positive STEC has also been demonstrated to sense short-chain fatty acids, ethanolamine, and fucose to regulate LEE expression (296–298), further adding to the complexity of LEE regulation during colonization and pathogenesis.
(a) Attachment.
The numerous adhesins encoded in the LEE-positive STEC genome likely contribute to colonization of the intestinal epithelium and other surfaces such as foods. Recent progress has been made in defining the contributions of these various structures and their potential in colonization. Fimbriae such as the E. coli YcbQ laminin-binding fimbria (ELF) and long polar fimbria (Lpf) have been shown to attach to the extracellular matrix (ECM) protein laminin (299, 300). Two lpf operons have been found in STEC O157:H7: lpf1, found on OI-141 (lpfO157/OI-141), and lpf2, found on OI-154 (lpfO157/OI-154) (301, 302). Interestingly, lpf mutants showed a change in tissue tropism and were able to colonize the small intestine in a human in vitro organ culture (IVOC) model (303). More broadly, lpf1 or lpf2 can be detected across all diarrheagenic E. coli pathotypes (304) as well as AIEC (305). Due to a number of lpf variants, a new classification scheme has been suggested by Torres et al. (306). Recently, a minor fimbrial protein, YadK, was shown to be involved in adherence to epithelial cells following exposure to acid (307).
The hemorrhagic coli pilus (HCP) is a type IV pilus found in STEC O157:H7 that forms long bundled fibers that are able to attach to extracellular matrix proteins on epithelial cells (308, 309). This pilus was also found to be multifunctional and contribute to phenotypes such as biofilm formation, twitching motility, and in vitro cell invasion (309), and it may trigger host immune responses (310). In SFO157:NM, the sfp operon is found on the pSFO157 virulence plasmid and was found to be involved in autoagglutination (311). Expression of the sfp under laboratory conditions required anaerobic conditions (312). Although sfp is generally thought to be restricted to SFO157:NM isolates, it has been found in STEC O165:H25/NM (313).
Other proteins that are thought to be involved in LEE-positive STEC adherence are Efa-1/LifA and its homolog, ToxB (314, 315). Unlike the EPEC efa-1/lifA products, STEC O157:H7 Efa-1 and ToxB do not have lymphostatin activity (316). Finally, the IrgA homolog adhesin (Iha), found in an OI with tellurite resistance, has adhesive properties and can also be found in LEE-negative STEC (317).
The autotransporter calcium-binding antigen 43 homolog (Cah) can promote cell-to-cell aggregation and biofilm formation (318). Similarly, the autotransporters EhaA and EhaB were also found to be involved in biofilm formation (319, 320). Contributions of Cah, EhaA, and EhaB in intestinal colonization remain unknown.
Intimate attachment is afforded by the interaction of intimin and Tir, similar to the case for EPEC as described above, except for some fundamental differences involving a separate effector that binds Tir and recruits host cell components that mediate actin assembly (reviewed in reference 321).
(b) StcE.
StcE is a large metalloprotease that is secreted by the type II secretion system (encoded by etp), also found on the pO157 virulence plasmid (322). The first target identified for StcE was the C1 esterase inhibitor; cleavage of C1 esterase inhibitor is thought to prevent the activation of the classic complement pathway (323). Other targets for StcE include glycoprotein 340, mucin 7, and CD43 and CD45 on neutrophils (324, 325). Interestingly, the mucinase activity of StcE is hypothesized to protect STEC O157:H7 in the oral cavity by reducing the viscosity of saliva and preventing mucin-bacterial aggregates (324). Additionally, these authors propose that StcE activity aids in intimate attachment of LEE-positive STEC to intestinal epithelial cells by reducing the mucus barrier on the host cells.
(c) HPI.
Originally identified in Yersinia spp., a high-pathogenicity island (HPI) was also discovered in the chromosome of STEC O26:H11/NM (326) and encodes the siderophore yersiniabactin. HPIs can be found infrequently in other pathotypes and commonly in EAEC (327).
(vi) LEE-negative STEC.
It is becoming clear that not all STEC strains that cause HC and HUS are LEE positive, and thus their molecular mechanisms of disease are likely fundamentally different from those of their LEE-positive counterparts. Various LEE-negative STEC strains have been isolated from HUS patients, such as serotypes O91:H21 (328), O104:H4 (44), and O113:H21 (329). Analysis of the German STEC O104:H4 outbreak strain has shown that it is more closely related to EAEC (330), and the pathogenic mechanisms are likely similar to those of EAEC. Indeed, there have been reports of stx-expressing EAEC, and this is further discussed below in the section on enteroaggregative E. coli.
(a) Attachment.
The mechanisms of epithelial cell attachment of LEE-negative STEC have not been studied as extensively as those of LEE-positive STEC. Nevertheless, several unique adhesins have been identified. Some LEE-negative STEC strains carry a single lpf2 homolog, which is commonly referred to as lpfO113 due to its original discovery in STEC O113:H21 (331). STEC autoagglutinating adhesin (encoded by saa) was identified in an STEC O113:H21 isolate (332) and was subsequently determined to be present in various other LEE-negative STEC strains that have been isolated from patients with diarrhea or HUS (332, 333). The autotransporter protein Sab (an STEC autotransporter contributing to biofilm formation) was shown to contribute to adherence to epithelial cells and biofilm formation when expressed in laboratory E. coli (334). Like Saa, Sab was also found in other LEE-negative STEC strains and was also suggested to contribute to attachment and colonization of LEE-negative STEC strains (334). Finally, E. coli immunoglobulin-binding (Eib) protein G was identified near phage genes in the chromosomes of certain STEC O91 serotypes and contributes to a chain-like adhesion pattern (CLAP), which can vary in length, depending on the EibG subtype (335, 336).
(b) Invasion.
Several other LEE-negative STEC serotypes, such as O113:H21, are able to be internalized and exist within a vacuole of epithelial cells in vitro (337). The flagellar antigen H21 contributes to invasion, and the invasive process may involve lipid rafts (338, 339).
(c) Activatable Stx2d.
Some LEE-negative isolates, such as STEC O91:H21, carry a variant of Stx2 called Stx2dactivatable, which becomes more cytotoxic in the presence of mucus (340). In a potency study, Stx2dactivatable was among the most cytotoxic Stx2 variants studied (341). A survey of different STEC serotypes found that Stx2dactivatable was carried only by LEE-negative STEC and was associated with HC and HUS (342).
(d) Subtilase cytotoxin.
The genes encoding the subtilase cytotoxin (subAB) were originally identified on a virulence plasmid in STEC O113:H21 (343); however, a variant can also be found chromosomally on a PAI in other STEC strains (344, 345). In Vero cell assays, recombinant SubAB was found to be more toxic than Stx1 or Stx2 (343). Pure SubAB given to mice is lethal and causes hemorrhaging in the small intestine and a disease similar to HUS, including renal damage, thrombotic microangiopathy, intestinal hemorrhages, hemolytic anemia, and thrombocytopenia (346, 347). Damage was also seen in different organs, such as the liver, spleen, and brain (347), and has been reported to cause leukocytosis (348).
The mechanism of SubAB toxicity and trafficking has been recently reviewed (349). Briefly, SubAB is internalized and trafficked to the endoplasmic reticulum. The catalytic A subunit is a serine protease that cleaves BiP/GRP78, a regulator of the stress response in the ER, and ultimately leads to cell death. SubAB may also be involved in altering host immune responses, as it was shown to prevent antibody secretion from B cells (350) and to suppress NO in macrophages, leading to the hypothesis that it may promote the survival of STEC in macrophages (351).
Clinical Considerations
Symptoms and onset of disease.
Infections with STEC can range from mild watery diarrhea to bloody diarrhea (hemorrhagic colitis) and present a risk for the development of HUS. Both O157 and non-O157 STEC can have similar clinical presentations, but disease severity can differ between different serotypes, and hence a seropathotype classification was proposed (248). Typically, the incubation period before the onset of symptoms for STEC O157:H7 is about 3 days (352), which is similar to what was reported for an outbreak of STEC O111:NM (236). The first symptom is usually diarrhea, which can be accompanied by fever, abdominal cramping, or vomiting (reviewed in reference 353). Patients may experience hemorrhagic colitis in the following days after the initial onset of diarrhea. It is generally thought that STEC O157:H7 results in higher rates of bloody diarrhea than non-O157 STEC. A study in Montana found that bloody diarrhea occurred in 81% of STEC O157:H7 cases, compared to 58% of non-O157 STEC cases (354). A review by Johnson et al. also discussed the diversity of clinical data regarding rates of bloody diarrhea related to non-O157 illness (185). It has been recommended that patients with bloody diarrhea be hospitalized to help manage the diarrhea and prevent further communicable spread (353). Diarrhea caused by STEC O157:H7 generally resolves in about a week following initial onset, while non-O157 STEC diarrhea may persist longer (355). During the STEC O104:H4 German outbreak, the median shedding time was about 2 weeks in adults and more than a month in children under the age of 15 (356).
Despite the resolution of diarrhea, STEC illnesses are of particular concern due to the association of STEC infections with HUS (357). While STEC O157:H7 is a major cause of HUS worldwide, SFO157:NM and non-O157 serotypes such as O26, O103, O104:H4, O111, O121, and O145 have also lead to cases of HUS (44, 185, 186, 240). HUS involves thrombocytopenia, hemolytic anemia, and acute renal failure and can develop between days 5 and 13 after the initial onset of diarrhea (reviewed in reference 353). A 20-year epidemiological survey of 350 outbreaks in the United States determined that STEC O157:H7-associated HUS occurred in about 4% of cases and had a mortality rate of 0.5% (256). These numbers are much lower than the estimated HUS rate of 14% in children under the age of 10 (358), with a mortality rate of about 1 to 4% (359). Perhaps not surprisingly, the diversity of non-O157 STEC leads to variable HUS rates, albeit lower than those for STEC O157:H7 (185). In the unprecedented O104:H4 outbreak, HUS developed in about 20% of cases (44), while an SFO157:NM outbreak had an 11% mortality rate in children (240). Further long-term sequelae (in 20 to 40% of patients) from HUS include cardiac complications, gastrointestinal complications, neurological disorders, hypertension, chronic renal disease, cognition and behavior changes, and diabetes mellitus and have been recently reviewed elsewhere (359). It has been recommended that follow-up assessments for late-onset sequelae be done for at least 5 years (360).
Detection.
Culture techniques have been previously described (8, 361), and the CDC has outlined methods of STEC isolation and identification along with recommendations in recent publications (362, 363), so these will only be briefly discussed here. Since most O157:H7 isolates are unable to ferment sorbitol within a 24-h period, they are easily distinguishable as clear colonies on sorbitol-MacConkey (SMAC) plates (364), although other chromogenic selective media have been shown to have fewer false-positives and may reduce operating laboratory costs (365). These identified colonies can then be checked for the O157 and H7 antigens using latex agglutination assays (366), which are available as commercial kits. Although uncommon in North America, SFO157:NM may not readily be identified from other sorbitol-fermenting E. coli strains and could be easily missed on SMAC plates. Additionally, the use of cefixime-tellurite (CT)–SMAC plates may also miss SFO157:NM isolates due to their sensitivity to tellurite (367) or may miss STEC O157:H7 in situations where OI-43 or OI-48 is lost due to deletions (368). For a strain to be classified as STEC, the Stx1 or Stx2 toxin must be detected, and this can be done through commercially available enzyme immunoassay (EIA) kits (362, 363). In serotypes O26, SFO157:NM, O103, and O145, it has been reported that stx can be lost from in vivo samples, and this may impact diagnostics (272, 274, 369). Because of the possibility of an stx-negative phenotype, a secondary target, such as intimin or, to differentiate SFO157:NM from O157:H7, sfp genes may be useful (188).
While STEC O157:H7 can usually be identified phenotypically on media, non-O157 STEC strains are not readily distinguishable from other E. coli strains because they are often able to ferment sorbitol, thus appearing as pink colonies on SMAC plates (363). Commercial enzymatic assays have been developed to increase the sensitivity of detecting both O157:H7 and non-O157 STEC compared to that with SMAC (370, 371). Although non-O157 STEC will be Stx1/Stx2 positive with EIA kits, further isolation is not routinely done in clinical laboratories (363).
Many methods have been developed for rapid identification of STEC from food sources and stool samples. Depending on the technique, these methods are able to indicate the serogroup and possible virulence factors held by particular isolates. With the extensive sequencing of the O-antigen-coding locus of various E. coli strains, serogroup-specific oligonucleotide pairs can be used in PCR assays (reviewed in reference 10). Serotyping methods have been developed to detect STEC using microarrays, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), and microbeads (372–374). Quantitative PCR (qPCR) panels have also been developed to look at multiple genes, such as rfbE (O157 antigen), stx, eae, ehx, fliC, and O-antigen genes to profile for certain STEC isolates (375). These methods are generally not approved for diagnosis from human samples but may be useful for epidemiological and outbreak studies by public health laboratories.
Recently, the CDC recommended Shiga toxin testing for the detection of non-O157 STEC and cultures for STEC O157:H7 of all stools submitted from patients with community-acquired diarrhea (362, 363). These recommendations, however, have not been adopted by all (376).
Treatment.
The course of STEC infection usually is self-limiting and resolves after a week (355). Currently, there is no way to prevent the development of HUS following an STEC infection, and management of HUS has been discussed elsewhere (reviewed in reference 377). A recent publication by Wong et al. has identified risk factors that are associated with the development of HUS in children, including vomiting, a high leukocyte count, and use of antibiotics (358). Recent work has shown improved renal protection in children under the age of 18 against oligoanuria HUS when given intravenous fluids (378). Suggestions for clinical management of STEC infections were made by Holtz et al. (379), which include the use of intravenous fluids, renal function and platelet monitoring, and discouragement of the use of antimotility drugs, pain relief drugs, and antibiotics.
Alternative and experimental treatments.
With no clear treatment of HUS, scientists and clinicians have been developing alternate methods that may provide useful treatment in the future, which have been recently reviewed by Goldwater and Bettelheim (380).
Monoclonal antibodies (MAbs) have been shown to reduce the cytotoxicity of Stx1 and Stx2 in vitro (381), while Urtoxazumab, a humanized MAb against Stx2, was shown to be safe in a phase 1 study (382). Similarly, phase 1 clinical trials of chimeric monoclonal Stx1- and Stx2-neutralizing antibodies were also shown to be generally well tolerated in injected volunteers (383, 384). Alternatively, short peptides may block Stx2 transport through epithelial cells (385). These cell-permeative peptides were also protective in a baboon model following Stx2 injection, as renal injury was abated (386). Cytotoxicity of both Stx1 and Stx2 could be neutralized with globotriose conjugated to the polysaccharide chitosan, by binding the toxin (387). This globotriose conjugate was also able to protect mice against STEC O157:H7 challenge. Synsorb-PK, a receptor analog, was designed to eliminate Stx1/2 from the intestine and was tested in clinical trials. Rates of HUS did not differ from those for controls, and the authors suggest that treatment may have been given too late in the progression of disease (388).
Other strategies that are being evaluated in the laboratory involve probiotics, such as secreted molecules from Lactobacillus acidophilus, that were able to inhibit STEC O157:H7 from adhering to epithelial cells in vitro and reduce shedding in mice (389). Bifidobacterium spp. and Lactobacillus spp. also were shown to be capable of inhibiting STEC O157:H7 growth in the laboratory (390). Manganese ions were shown to prevent cell death by Stx1 by preventing trafficking through the Golgi apparatus and protected mice against fatal Stx1 injections (391). A more recent paper, however, has challenged the use of manganese as a potential therapeutic against Shiga toxin (392).
Antibiotic resistance.
Although antibiotics are not recommended for the treatment of STEC, many isolates carry resistance to various antimicrobials. Resistance to sulfonamide and tetracycline is common among O157:H7 and non-O157 STEC from both human and bovine sources (393). Furthermore, there is significant resistance to ampicillin, streptomycin, and trimethoprim, which can be more frequent in non-O157 STEC isolates. Plasmids have been found to be a source of antimicrobial resistance genes in STEC isolates (35, 394). Recently, an extended-spectrum β-lactamase (ESBL) plasmid, rarely found in STEC O157:H7, was isolated from a 2-year-old child in Denmark (395). An ESBL plasmid was also present in the STEC O104:H4 German outbreak strain (396). Tellurite resistance is also common in STEC O157:H7 and certain serotypes such as O26:H11/NM, O111:H8/NM, and O145:NM but is rare in SFO157:NM (317, 367, 397).
Vaccines.
Vaccines are being developed against STEC (reviewed in reference 380). Some of these strategies have focused on reducing carriage and shedding in reservoir hosts such as cattle (398). Many studies have focused on protein fusions of different virulence factors. Some recent examples include fusions of Stx1B and an inactivated Stx2A (SAmB), Stx2B-Tir-Stx1B-zot (zot; mucosal delivery protein), Stx2B-Stx1B-initimin (SSI), and EspA-initimin-Stx2 (EIS), all of which have shown either reduced STEC shedding or protected against STEC or Stx1/Stx2 challenge in mice (399–402). Different delivery mechanisms for potential vaccines have also been explored, such as the EspA-intimin-Tir fusion that is expressed in tobacco leaves and canola seeds, Salmonella expressing intimin or EIS, and Stx2B expressed in the Mycobacterium bovis BCG vaccine strain (401, 403–405). Additionally, nontoxic Stx2A DNA vaccines have also been shown to provide some protection in mice (406).
ENTEROINVASIVE E. COLI/SHIGELLA
Enteroinvasive E. coli (EIEC) and Shigella spp. are facultative intracellular pathogens and the etiological agents of bacillary dysentery or shigellosis. Shigella, originally known as Bacillus dysenteriae, was first described by Kiyoshi Shiga in 1897 during an epidemic in Japan, where it infected more than 91,000 people and caused a mortality rate of greater than 20% (407, 408). EIEC was discovered about 50 years later and shares biochemical, genetic, and pathogenetic properties with Shigella (8, 409). In general, Shigella is nonmotile, lysine decarboxylase negative, and unable to ferment lactose (except for S. sonnei, a slow lactose fermenter). These and other characteristics also apply for most EIEC, with some exceptions that are described in more detail in “Detection” below (410, 411). Due to their high similarity, EIEC and Shigella are discussed here in parallel.
Classification
Common serotypes.
EIEC comprises 21 major serotypes, which are typically defined by their O-antigen pattern, with a few exceptions that also present H antigens (410) (Table 4). Some of the EIEC O antigens are identical or closely related to Shigella O antigens and thus complicate the discrimination between EIEC and Shigella in conventional serotyping (412, 413) (Table 4). Traditionally, Shigella is classified based on biochemical, serological, and clinical phenotypes, not on phylogenetic correlation (411, 414, 415). It includes 49 sero- and subserotypes that are further clustered into the four species S. dysenteriae (serogroup A, 15 serotypes), S. flexneri (serogroup B, 14 sero- and subserotypes), S. boydii (serogroup C, 19 serotypes), and S. sonnei (serogroup D, 1 serotype) (Table 5). Until recently, S. boydii was subdivided into 20 serotypes; however, phylogenetic analysis revealed that S. boydii 13 belongs to the E. albertii lineage, which is distinct from typical Shigella (416).
Table 4.
Serotypes of EIEC
| O serogroupa | H antigen(s) |
|---|---|
| O28ac | NM |
| O29 | NM |
| O112ac (D2, B15) | NM |
| O115 | NM |
| O121 | NM |
| O124 (D3) | NM, H7, H30, H32 |
| O135 (F2a) | NM |
| O136 | NM |
| O143 (B8) | NM |
| O144 | NM, H25 |
| O152 (D12) | NM |
| O159 | H2 |
| O164 | NM |
| O167 (B3) | NM, H4, H5 |
| O173 | NM |
Shigella serotypes that share the same or a closely related O antigen are indicated in parentheses. B, S. boydii; D, S. dysenteriae; F, S. flexneri.
Table 5.
Serotypes and subserotypes of Shigella
| Shigella species (subgroup) | Serotypes and/or subserotypes |
|---|---|
| S. dysenteriae (A) | 1–15 |
| S. flexneri (B) | 1a, 1b, 2a, 2b, 3a, 3b, 4a, 4b, 4c, 5a, 5b, 6, X, Y |
| S. boydii (Ca) | 1–20 |
| S. sonnei (D) | NAb |
S. boydii type 13 was recently reclassified as E. albertii (416).
NA, not applicable.
Serotype classification provides the basis for the current Shigella and EIEC nomenclature. However, recent advances in phylogenetic analysis challenge this traditional classification scheme, which will be further discussed in the subsequent paragraph.
Shigella/EIEC: a distinct E. coli pathotype.
The genus name Shigella is still used, mainly for historical reasons and its association with the disease shigellosis. However, diverse phylogenetic studies demonstrated that Shigella clearly belongs to the species E. coli (417–420). As exemplified by an analysis of 36 housekeeping genes from E. coli K-12, STEC O157:H7, and S. flexneri 2a, Shigella and the commensal K-12 strain showed an average sequence divergence of only 1.12% (417). In the same study, Shigella proved to be even more similar to E. coli K-12 than STEC O157:H7. Multilocus and whole-genome analyses revealed that EIEC and Shigella form a single pathotype within E. coli and emerged in several independent events from multiple ancestral origins (20, 409, 419, 421, 422). In a phylogenetic comparison study of 32 EIEC strains, 46 Shigella strains, and 20 E. coli reference strains of the E. coli Collection of Reference (ECOR) group, Shigella and EIEC strains, with few exceptions, formed three and four distinct clusters, respectively (409) (Fig. 8). Three out of four EIEC clusters contained strains with different O antigens, and strains sharing the same O antigen have been observed in more than one cluster. For Shigella, all clusters comprised strains of various traditionally defined serogroups. Thus, genetic relationships between strains are not comprehensively reflected by the commonly used traditional surface antigen profile-based classification scheme. EIEC showed less divergence from commensal E. coli than Shigella, which implies that EIEC arose later than Shigella and represents either a precursor of fully evolved Shigella or a form distinct from Shigella. This is in line with the reduced divergence observed within phylogenetic EIEC clusters compared to Shigella clusters (409).
Fig 8.
Phylogenetic tree of Shigella, EIEC, and nonpathogenic E. coli strains, showing the evolutionary relationship between 32 EIEC strains, 46 Shigella strains, and 20 E. coli reference strains of the ECOR group (EcoR strains) based on comparison of selected housekeeping gene sequences. Shigella groups in 3 phylogenetic clusters (C1 to C3) and EIEC in 4 phylogenetic clusters (C4 to C7), with broad distribution of traditionally surface antigen profile-classified strains. EIEC and Shigella strains outside clusters C1 to C7 are depicted in bold. Both EIEC and Shigella arose several times from multiple ancestral origins. The EIEC strain in C2 is assumed to be misclassified. The number of strains in a cluster is shown in parentheses. The Salmonella LT2 strain serves as outgroup. (Adapted from reference 409 with permission.)
So far, no common nomenclature exists for Shigella and EIEC that also incorporates the phylogenetic relationship between different lineages. Therefore, the traditional serotype-based nomenclature will be used for the rest of this section.
Pathoadaptation.
Both the acquisition and loss of genes were necessary for Shigella and EIEC to evolve from being commensal E. coli with an extracellular lifestyle to being an intracellular pathogen that is fully adapted to the diverse environmental challenges within its host. These events have been well researched for Shigella and therefore are addressed here for Shigella in more detail. The transition from commensal E. coli to pathogen required the acquisition of novel virulence-promoting factors and the depletion or suppression of unnecessary and virulence-opposing genetic elements (Table 6). The extensive virulence toolbox of EIEC and Shigella derived mainly from the acquisition of the invasion plasmid pINV and multiple other virulence-associated, mobile genetic elements (423, 424). Among these, the chromosomally located Shigella pathogenicity islands (SHIs) SHI-1 and SHI-2 (3), SHI-O, and SRL (Shigella resistance locus) contribute a plethora of additional virulence factors and resistance to several antibiotics, which played a critical role during pathogen evolution (424). The highly dynamic Shigella genome harbors 300 to 650 additional insertion sequence elements compared to E. coli K-12 and thus affords the pathogen its pathogenic flexibility (425). In addition, the Shigella genome comprises numerous deletions (black holes) and more than 200 pseudogenes (425–427). Many metabolic pathways became dispensable due to the intracellular lifestyle of Shigella (424), especially the inactivation and depletion of genes that encode bacterial antivirulence factors that otherwise would oppress Shigella/EIEC pathogenicity, and are regarded as crucial elements of pathoadaptation; these were attained by convergent evolution and were comprehensively reviewed recently (38). In contrast to the acquisition of novel virulence factors, the identification of antivirulence genes that have been silenced by deletion, insertion, or point mutations is more challenging. Nevertheless, several inactivated antivirulence genes have been identified in EIEC and Shigella. Among these, nadA and nadB encode enzymes of the NAD synthesis pathway that catalyze the synthesis of quinolinic acid, an inhibitor of virulence. In contrast, ompT encodes a protease that directly triggers proteolytic degradation of VirG, a crucial effector for intracellular motility. Moreover, silencing of the speG gene results in the beneficial accumulation of spermidine, which increases resistance to oxidative stress encountered during infection. ArgT was observed to attenuate Shigella virulence and consequently was found to be downregulated at 37°C, the temperature of the host environment. Finally, lysine decarboxylase (LDC) is an important virulence-attenuating factor that is absent in both EIEC and Shigella. For Shigella, it has been shown that a product of lysine decarboxylation, cadaverine, attenuates the activity of bacterial enterotoxins, inhibits the migration of polymorphonuclear leukocytes across the epithelial barrier, and blocks phagosomal escape (426, 428, 429). All of these are important elements of EIEC and Shigella virulence. LDC is encoded by the gene cadA, which is part of the cadBA operon and under positive control of the regulator CadC. EIEC and Shigella reveal different strategies to block LDC synthesis. In most Shigella strains, the cadA gene is either deleted or inactivated, whereas most EIEC strains still possess cadA but have an inactivated cadC (426, 430).
Table 6.
Shigella model of elements acquired or disposed of during pathoadaptationa
| Adaptation | Category and element(s) | Role in pathogenesisb |
|---|---|---|
| Acquisition | Invasion plasmid elements | |
| T3SS | Translocation of T3S effectors into host | |
| T3SS substrates/translocators | Subversion of host cell processes for bacterial invasion, intracellular survival, and intra- and intercellular motility | |
| Transcriptional regulators | Coordination of T3SS-associated gene transcription, stabilization of T3S effectors in bacterial cytosol | |
| Chaperones | Watery diarrhea, inflammation | |
| Enterotoxin ShET2 | ||
| Chromosomal Shigella PAIs | ||
| SHI-1(including enterotoxin ShET1 and SPATEs Pic and SigA) | Watery diarrhea, mucus permeabilization, serum resistance and hemagglutination | |
| SHI-2/3 | Iron acquisition, regulation of inflammation | |
| SHI-O | Host immune evasion | |
| SRL | Iron acquisition, multidrug resistance | |
| Toxin | ||
| Shiga toxin (primarily in S. dysenteriae 1) | Inhibition of host protein synthesis, induction of apoptosis | |
| Loss or inactivation | Antivirulence genes | |
| ompT | Inhibition of intra- and intercellular motility | |
| cadA (for EIEC often cadC) | Attenuation of PMN transepithelial migration, enterotoxin activity, and phagosomal escape | |
| nadA, nadB | Inhibition of PMN transepithelial migration, cell-to-cell spread, and T3SS | |
| speG | Decrease of oxidative stress tolerance | |
| argT | Inhibition of cell invasion | |
| Surface structures | ||
| Flagella, fimbriae | Activators of host immune system; flagella negligible due to acquired T3S effector-mediated, actin-based motility | |
| Diverse catabolic pathways | Redundant due to pathogenic lifestyle |
Epidemiology
Transmission and reservoirs.
Conventional host-to-host transmission of EIEC and Shigella is mediated via the fecal-oral route mainly through contaminated water and food or direct person-to-person spread (431, 432) (Fig. 5). In addition, flies and living amoebae have been reported to play a role in Shigella transmission (433, 434). Disease is caused in humans and higher nonhuman primates with an infectious dose of as little as 10 to 100 CFU of Shigella, as observed in volunteer studies (435). EIEC infection tends to be less effective, and higher infectious doses have been reported from volunteer studies (436).
Incidence and outbreaks.
EIEC is underrepresented in epidemiological surveys due to its less severe clinical manifestations. In addition, EIEC strains with very close biochemical, genetic and pathogenic similarity to Shigella might be misclassified, whereas EIEC strains with commensal E. coli characteristics, such as lactose fermentation, might remain undetected in respective surveys. Hence, no specific geographical pattern can be associated with EIEC incidences, as the epidemiology of EIEC can be properly addressed only by studies that are more specifically designed for EIEC, which would include EIEC-specific genetic markers or laborious EIEC typing.
No EIEC episodes have been reported in recent health-related government surveillance programs led in the United States, Canada, Europe, or Australia (193, 195, 437, 438). However, rare cases do occur, including a 1985 food-borne EIEC outbreak which affected 370 people in Texas, United States (439). In the last decade only a few disease-associated EIEC episodes with more than 10 cases have been reported from studies in Central and South America, Africa, and Asia (65, 440–443).
In contrast, the epidemiology of Shigella is well documented. Shigella causes diarrhea and is associated with about 30 to 50% of bacillary dysentery cases worldwide (444). A study in 1999 estimated 164.7 million Shigella episodes annually, with 99% affecting developing countries, including 1.1 million deaths per year. Approximately two-thirds of episodes and deaths involved children under 5 years old (445). More recent studies have reported a declining trend of the Shigella disease burden, a common trend for diarrheal diseases in general (5, 446, 447). This is more likely a consequence of the dramatic drop in case-fatality rates, which is also reflected by the last WHO figures in 2009 estimating 90 million Shigella episodes and 108,000 deaths per year worldwide (http://www.who.int/vaccine_research/diseases/diarrhoeal/en/index6.html), than of changes in overall morbidity. Improved primary health care, education, and hygienic standards in areas of endemicity and the absence of devastating S. dysenteriae 1 epidemics are plausible reasons for this recent development (448). Nevertheless, Shigella still remains a major public health concern, with a high mortality rate, evolving epidemiology, increasing multidrug resistance, and a high incidence of unreported cases. GEMS reported that Shigella was among the four major pathogens associated with moderate to severe pediatric diarrhea, being the fourth most common isolate among infants of 0 to 11 months, second among toddlers of 12 to 23 months, and first among children of 24 to 59 months (7).
All Shigella species are covered by most epidemiological studies. The Shiga toxin-producing S. dysenteriae 1 was responsible for the high mortality rate during devastating pandemics in Central America in the 1960s, South Asia in the 1970s, Central Africa in the 1980s, and East Africa in the 1990s (449–452). The last major S. dysenteriae 1 epidemic was recorded in Sierra Leone, West Africa, in 1999 (453). Shigellosis due to S. dysenteriae serotypes other than type 1 is uncommon. S. boydii-mediated disease also is rare and mainly restricted to the Indian subcontinent (447). In contrast, little geographical restriction applies for S. flexneri, which is the most frequently isolated Shigella sp. worldwide and is endemic in developing countries. A literature-based study in 2007 incorporating countries in Asia, Africa, and South America demonstrated that in most places, S. flexneri was responsible for about two-thirds of shigellosis cases, with S. flexneri 2a being the single most prevalent subserotype (454). More recent reports from India, Pakistan, Bangladesh, Argentina, and China confirm S. flexneri as the most prominent Shigella sp. being detected (455–458). However, a major shift from S. flexneri to S. sonnei is becoming more evident in transition countries and areas with improving socioeconomic conditions (454, 459, 460). In developed countries, S. sonnei is the most prevalent Shigella species and causes sporadic disease and outbreaks in epidemiological niches such as day care centers (461). In general, the average Shigella incidence rate in developed countries ranges from 1.6 to 4 cases per 100,000 population according to recent estimations by surveillance programs in the United States, Canada, Europe, and Australia (193, 195, 437, 438). Shigella infections that affect travelers and workers from industrialized countries who entered areas of endemicity account for 2 to 8% of traveler's disease (1 to 2% for EIEC) and usually involve S. flexneri and S. sonnei (462). Changing traveling habits in a rapidly globalizing world are an important aspect affecting the propagation of pathogenic agents. Holt et al. recently performed a phylogeographic analysis based on results from whole-genome sequencing of 132 globally distributed S. sonnei isolates that have been collected over the course of 60 years. They showed that most current S. sonnei infections are caused by only a small number of successful, often antibiotic-resistant strains that originally disseminated from Europe and spread globally (463). This rapid global dispersion of robust and infectious strains illustrates that Shigella is still a massive global health threat with rapidly evolving multidrug-resistant strains.
Pathogenesis
EIEC and Shigella are highly invasive pathogens that use the intracellular milieu of intestinal epithelial cells (IECs) in the large intestine as their replicative niche. These pathogens are readily adaptable to the various environmental challenges they face during the course of infection, including low gastric pH, temperature changes, oxygen availability, and oxidative stress, as well as osmolarity (464). Advances in S. flexneri research provide the basis for our current understanding of both EIEC and Shigella pathogenesis at the cellular and molecular levels, which has been comprehensively summarized in recent reviews (424, 465). Successful infection is dependent on essential virulence determinants that are encoded by both chromosomal and plasmid loci. Key plasmid-encoded virulence factors include components of the T3SS needle complex (Mxi-Spa proteins), chaperones (IpgA, IpgC, IpgE, and Spa15), transcriptional regulators (VirF, VirB, and MxiE), translocators (IpaB, IpaC, and IpaD), and approximately 25 effector proteins. An additional 5 to 7 T3SS substrates of the IpaH family and further T3SS-independent virulence factors are chromosome encoded (424). Shigella infection is a multistep process involving penetration of the epithelial barrier, induction of macrophage cell death, IEC invasion, suppression of the immune response, intra- and intercellular movement, and modulation of epithelial integrity (Fig. 6).
Penetration of epithelial barrier and macrophage cell death.
In order to gain access to the basolateral surface of IECs, bacteria first penetrate the epithelial barrier through M cells by transcytosis. In the underlying submucosa, Shigella is phagocytosed by resident macrophages and evades its own degradation by effector-dependent phagosomal escape and induction of caspase I-dependent pyroptosis-like macrophage cell death (466–468).
Invasion.
Shigella's subsequent basolateral invasion of IECs, its intracellular survival and replication, and cell-to-cell spread are governed by the ability of its effector repertoire to subvert host cell signaling pathways (424). Bacterial adhesion to the host cell is mediated by IpaB and an IpaBCD complex, which bind to the host hyaluronan receptor CD44 and α5β1 integrin, respectively (469, 470). IpaC, IpgB1, IpgD, IpaA, and VirA have been implicated in extensive host cytoskeleton reorganization and membrane ruffling to promote pathogen uptake into the phagosome (471–476). Subsequently, Shigella escapes the phagosome by utilizing the effectors IpaB, IpaC, IpaD, and IpaH7.8 (477–480).
Suppression of host immune response.
Multiple effectors counteract the host immune defense (481). Host inflammatory responses, such as the mitogen-activated protein kinase (MAPK) and NF-κB pathways, as well as cytokine production, are targeted and dampened by multiple effectors (481), including OspG, OspF, OspB, OspZ, OspI, IpaH4.5, and IpaH9.8 (126, 482–488). Additionally, Shigella employs strategies to manipulate and escape the host immune response, such as suppression of antimicrobial peptide expression, induction of apoptosis in dentritic cells, and impairment of T-cell function (489–491). Moreover, the T3S effectors VirA and IcsB help Shigella to circumvent its autophagy-mediated degradation (492, 493).
Intra- and intercellular movement.
T3S effectors are responsible for the intra- and intercellular movement of otherwise nonmotile Shigella. VirG induces the acquisition of N-WASP, which then leads to the formation of an ARP2/3 complex-mediated unipolar actin tail on the bacterial surface. This tail provides the propulsive force required for directed motility of the pathogen (494, 495). Furthermore, it has been suggested that VirA, with its capability to stimulate microtubule destabilization, is another essential effector that allows Shigella to efficiently spread through the dense intracellular cytoskeletal network (496). The acquired motility also helps the pathogen to disseminate into neighboring cells, which was recently found to occur primarily at tricellular tight junctions (497).
Epithelial integrity.
Epithelial integrity plays a fundamental role during Shigella infection (498). Early in infection, Shigella promotes host cell survival and integrity. Inhibition of IEC turnover and detachment by IpaB and OspE, respectively, and IpgD- or VirA-mediated blockage of host cell death are known mechanisms employed by the pathogen to conserve its replicative niche and promote colonization (499–502). However, Shigella has also evolved strategies to breach the epithelium to facilitate access to the basolateral surface of IECs and ultimately disembarkation from its replicative niche. This includes processes such as cell surface manipulation by IpaB-mediated disruption of Golgi network-driven secretion, destabilization of epithelial tight junction sealing, and induction of IEC death (503–505). The massive inflammatory response associated with apoptotic macrophages and IEC invasion, including infiltrating polymorphonuclear leukocytes, further perforates the epithelial barrier and ultimately leads to tissue lesions, which are characteristic for the pathology of shigellosis (424, 506).
Toxins.
Additional virulence factors are known to contribute to the clinical manifestations of EIEC/Shigella infections. The common observation of watery diarrhea has been attributed to Shigella enterotoxins 1 and 2 (ShET1 and ShET2) (507–509). The chromosome-encoded ShET1 is limited to S. flexneri, whereas the pINV-encoded and T3S ShET2 has been found in Shigella strains of different serotypes and EIEC (510–512). In addition to its contribution to secretory intestinal activity, ShET2 has also been implicated in Shigella-induced inflammation in IECs (510). Besides ShET1 and ShET2, two additional enterotoxigenic factors, namely, Pic, which is encoded by a chromosomally located gene that comprises the open reading frame of ShET1 on its opposite strand, and the pINV-encoded SepA, have been recently described to account for intestinal secretion in a mouse tissue model (513, 514). Both Pic and SepA belong to the family of serine protease autotransporters of Enterobacteriaceae (SPATEs). SPATEs are serine proteases that engage the autotransporter pathway and self-cleavage for their secretion. Shigella encodes two more members of the SPATE family, the cytotoxins Sat and SigA. SigA was found to contribute to intestinal fluid accumulation in the rabbit ileal loop model (515). SPATEs are not limited to EIEC and Shigella but are commonly found in a multiplicity of pathogens, including pathotypes described here, and are summarized in a recent review (516). Lastly, the often severe and lethal complications of S. dysenteriae 1 infections are associated mainly with Stx. Stx is mostly only present in S. dysenteriae 1 and is almost identical to Stx1 produced by other STEC strains. Besides its inhibitory impact on host protein synthesis by catalytic inactivation of eukaryotic ribosomes, Stx has been described to induce apoptosis in various cell types. As such, it was found to be responsible for the development of vascular lesions in the colon, kidneys, and central nervous system (517–519). In contrast to the case for STEC, the original Stx-encoding bacteriophage of S. dysenteriae 1 lost its functionality and thus transducibility (520). An S. sonnei isolate that carried a phage-encoded S. dysenteriae 1-like Stx was reported, revealing that other Shigella serotypes may have the potential to acquire Stx via horizontal gene transfer and thus increase their virulence (521).
EIEC versus Shigella.
In general, EIEC and Shigella employ the same strategies to invade host cells. Nevertheless, EIEC exhibits reduced virulence compared to that of Shigella, including reduced expression of virulence genes, less efficient macrophage killing, reduced cell-to-cell spread, and decreased induction of a proinflammatory host response (522, 523), which correlates with the less severe disease induced by EIEC.
Clinical Considerations
Symptoms.
The clinical presentation, progression, and complications of bacillary dysentery, or shigellosis, vary depending on the infectious agent, the immunological background of the host, and available medical infrastructure. The reader is referred to earlier literature for a more detailed overview of clinical symptoms (524, 525). Briefly, mild watery diarrhea, fatigue, malaise, fever, and anorexia develop in the early stages of disease. This is typically accompanied by abdominal cramps, tenesmus, scanty stools with blood and mucus, and dehydration. In most cases, shigellosis is self-limiting. However, severe and life-threatening complications can occur, especially in areas of the developing world where the disease is epidemic or endemic, where mainly young, often malnourished and strongly immunosuppressed children are infected and lack access to adequate treatment. Severe shigellosis complications are often associated with the Shiga toxin-producing serotype S. dysenteriae 1 and can range from local intestinal disorders to systemic manifestations. Shigellosis complications include toxic megacolon, intestinal perforation, peritonitis, hyponatremia, hypoglycemia, pneumonia, and HUS. HUS was estimated to occur in up to 13% of S. dysenteriae 1-infected, dysenteric patients, with a case-fatality rate of about 36%, in Africa and Asia (526). S. dysenteriae 1-caused HUS is very similar to HUS caused by STEC (see discussion in the section on Shiga toxin-producing E. coli above). However, disseminated intravascular coagulation with consumption of coagulation factors can be present in S. dysenteriae 1-associated HUS cases but is rare with STEC (527). Further shigellosis complications include septicemia and neurological disorders, such as encephalopathy and seizures (528, 529). Incidences of postinfectious irritable bowel syndrome after Shigella infection have also been reported (530).
EIEC infection often leads only to self-limiting, mild watery diarrhea. However, in rare situations it can cause shigellosis-like symptoms (8). Complications have been reported, but are very uncommon (531).
Detection.
A classical approach to identify EIEC and Shigella in a stool sample begins with the determination of invasiveness of the bacterial isolate. For this purpose, the guinea pig keratoconjunctivitis (Sereny) test, which requires prior isolation of the bacterial species, and the microscopic detection of numerous polymorphonuclear leukocytes in blood-containing stool samples are two possible phenotypic assays (525, 532, 533). In addition, a tissue culture plaque assay that exploits the ability of Shigella to form plaques in epithelial cell monolayers can be used to test invasiveness (534). However, the Sereny test is laborious and involves animal work, the plaque assay requires appropriate infrastructure, and both assays do not result in an unambiguous identification of either EIEC or Shigella. Therefore, alternative diagnostic tools are necessary and are often based on the biochemical distinction of EIEC and Shigella from other bacteria. For efficient diagnosis, the bacterial isolates need to be obtained from either fresh stool samples or stool specimens that were stored in adequate transport medium, such as buffered glycerol saline or Cary-Blair transport medium (524). Biochemical identification of Shigella, which can often be employed for EIEC, uses differential/selective media and focuses mostly on the following characteristics of almost all Shigella strains: inability to ferment lactose and utilize citrate; absence of motility, lysine decarboxylase, and urease activity; and acid but no gas and H2S production upon sugar fermentation (447, 524, 535). If adequate equipment and scientific training are available, then PCR-based detection of pathotype-specific genetic markers, such as the invasion plasmid antigen H gene (ipaH) or the invasion-associated locus gene (ial), is a common diagnostic tool for both EIEC and Shigella detection (536). The latter is frequently performed as a multiplex PCR to simultaneously identify EIEC/Shigella and other pathogenic agents (537, 538). The recent development of a TaqMan Array Card platform, which is potentially able to multiplex up to 384 targets and was used to detect 19 pathogens, including 5 viruses, 9 bacteria (including EIEC/Shigella), 3 protozoa, and 2 helminths, represents a prospective tool for fast and comprehensive surveillance data processing on the next level of multiplexing technologies (539). Once the bacterial isolate is identified as EIEC or Shigella, the serogroup and serotype, respectively, can be confirmed by slide agglutination tests using commercially available antisera (447, 524, 535). Alternative approaches to antiserum-based serotyping have also been developed. Coimbra et al. designed a software-supported and restriction fragment length polymorphism-based technique which is able to distinctly identify most but not all Shigella serotypes (540, 541). In another study, molecular serotyping of S. flexneri was achieved by a multiplex PCR assay targeting several specific O-antigen synthesis and modification genes (542). The analysis of single nucleotide polymorphisms also has been employed to successfully determine Shigella serogroups (543). The increasing availability of genomic sequencing data facilitates the search for novel genetic markers that can be used for unambiguous Shigella and EIEC serotyping. In locations where the appropriate technological infrastructure is available, additional genome-based typing methods for precise assessment of clinical isolates and are on the rise thus provide a major contribution to identification, surveillance, and risk assessment of EIEC and Shigella. MLST constitutes a valuable tool to identify clinical isolates and characterize their degree of genomic relationship (13). By June 2013, the sequence types of 50 EIEC and more than 140 Shigella strains were listed and available in an open-access database (20; http://www.mlst.net). Comparative genomics represents an effective measure in the proper assessment of pathogenic isolates. For example, the high discriminatory power of a retrospective whole-genome analysis of an S. sonnei outbreak allowed the determination of various phylogenetic lineages and proved to be superior to conventional typing techniques in defining the outbreak (544). However, DNA sequence-based typing approaches are often difficult to implement in the field, where technological restrictions apply. The introduction of dipsticks has proven to be an applicable approach for rapid diagnosis of S. flexneri 2a and S. dysenteriae 1 in the field (545, 546). This format detects serotype-specific lipopolysaccharide (LPS) in less than 15 min by using serotype-specific monoclonal antibodies that are coupled to gold particles and displayed on a one-step immunochromatographic dipstick.
Due to their phenotypic, biochemical, and genetic relatedness it is difficult to distinguish between EIEC and Shigella. Generally, Shigella tests negative on motility, lactose, and mucate fermentation and l-serine, d-xylose, and/or sodium acetate utilization. Some EIEC strains test positive in one or more of these categories and can thus be distinguished from Shigella (417). However, these are not very reliable methods, since most EIEC strains would also show a negative Shigella-like phenotype. Recently, van den Beld and Reubsaet proposed a key for EIEC and Shigella identification combining IpaH gene PCR with several biochemical assays (547). In this detection strategy scheme, the authors suggested that the use of agglutination tests using EIEC- or Shigella-specific antisera is often unavoidable in order to clearly discriminate between EIEC and Shigella.
Some antisera cross-react with O antigens of both EIEC and Shigella (412, 413) (Table 5). In these cases the PCR-based detection of the lysine decarboxylase cadA gene, which is mostly absent in Shigella but often present in EIEC, may provide better distinction (413, 430). In general, genotypic analyses, such as MLST, provide higher resolution, which allows discrimination between Shigella and EIEC.
Treatment.
The essential steps for effective treatment of shigellosis have been extensively reviewed and slightly adapted in the last decade (444, 524, 525, 548). Generally, mild to moderate shigellosis is considered to be self-limiting provided that proper rehydration is guaranteed. This essential first step in diarrhea therapy is manifested in a formula for effective oral rehydration therapy, which has been developed by the WHO (525). As a second step, antimicrobial treatment has proven to effectively shorten the duration of symptoms and to reduce the risk of serious complications and death (524, 549). In contrast to the case for STEC, early antimicrobial treatment was also reported to be efficient in reducing the risk of S. dysenteriae 1-associated HUS (550). In general, the severity of disease, age of the patient, and local antibiotic susceptibility pattern should be taken into account for an adequate antibiotic-based strategy. The WHO guidelines for antibiotic treatment of shigellosis from 2005 were slightly revised recently (444, 525, 548). Recommended antibiotics for pediatric treatment include the macrolide azithromycin, the third-generation cephalosporin ceftriaxone, and the fluoroquinolone ciprofloxacin. Fluoroquinolones and azithromycin are recommended for adult treatment. The milder clinical manifestations of EIEC infection often do not require antibiotic treatment and are self-limiting (551). However, in the rare case of severe symptoms, an antibiotic strategy similar to that for shigellosis is recommended (548). In addition, zinc supplementation has been recommended due to its beneficial effect in reducing the severity and duration of diarrheal diseases in general (525, 552). Also, nutritional therapy with green plantains has been reported to be health supportive by increasing the weight gain of the diarrheal patient while decreasing the duration of the disease (553, 554).
Antibiotic resistance.
Unrestricted use of antibiotics as a therapeutic agent has caused a dramatic increase in multidrug resistance among Shigella, an alarming fact with regard to the future treatment of dysentery (447, 555). High incidences of antibiotic resistance among Shigella were reported as early as the 1950s in a case of dysentery due to sulfonamide-resistant S. sonnei (556). Since then, Shigella has acquired resistance to all first-line antibiotics, including sulfonamides, ampicillin, tetracycline, chloramphenicol, sulfamethoxazole/trimethoprim, and nalidixic acid (557–560). Although less reported due to its lower incidence rate and morbidity, EIEC seems to follow a similar trend. A comprehensive analysis of both Shigella and EIEC strains that were collected from North and South America, Africa, and Asia between 1970 and 2000 showed tetracycline resistance within 70 to 86% of Shigella isolates and 48% of EIEC isolates (561). Also, resistance to all first-line antibiotics was reported for an EIEC isolate in Japan (562). Even more alarming is the increasing occurrence of fluoroquinolone resistance, especially among S. flexneri but also within other Shigella subgroups (563). Up to 6% of ciprofloxacin-resistant Shigella strains were isolated from diarrhea episodes in Asia between 2000 and 2004 (447). More recent studies revealed an occasional ciprofloxacin resistance rate of up to 10% in Asia (457, 564). In addition, one study in China reported a decrease in susceptibility to ciprofloxacin in approximately 80% of isolates (564). In contrast, resistance against the two other current therapeutics of choice, azithromycin and ceftriaxone, is still rare, but reports of sporadic incidences are on the rise (457, 565, 566). Antibiotic resistance in EIEC/Shigella is most commonly plasmid and/or chromosome borne (424, 567). The acquisition of mobile elements, such as the S. flexneri chromosomal pathogenicity island “Shigella resistance locus,” which encodes resistance to streptomycin, ampicillin, chloramphenicol, and tetracycline, plays a crucial role in the fast evolution of drug-resistant strains (568). Alternatively, plant-based therapeutic approaches such as use of the Aegle marmelos fruit lectin or the essential oil from the Brazilian medicinal plant Cymbopogon martinii have shown promising antimicrobial activity in vitro (569, 570).
Despite some promising alternatives being in development, antibiotic treatment still remains the most effective treatment against EIEC and Shigella. Controlled and considerate use of antibiotics that also takes into account the regional antibiotic susceptibility patterns of EIEC and Shigella will slow down the emergence of multidrug-resistant strains. The improvement of health education, sanitary conditions, and access to clean drinking water, especially in impoverished areas of endemic diarrhea, is an important step toward reduction of Shigella- and EIEC-mediated disease.
Vaccines.
An effective vaccine would constitute another preventive and sustainable approach to eliminate the disease burden of bacillary dysentery. However, no vaccine is currently available for either EIEC or Shigella. The development of an effective Shigella vaccine has therefore been the focus of many laboratories over the last decade. The progress made and the hurdles encountered in the development of a Shigella vaccine have been well documented in recent reviews (454, 571, 572). Briefly summarized, recent research pursues the design of a multivalent vaccine protecting against the most prevalent serotypes and subserotypes, including S. dysenteriae 1, S. sonnei, and all 14 types of S. flexneri. Multiple strategies were implemented to engineer both parenteral and mucosal candidate vaccines that have shown various levels of efficacy in clinical trials. The most promising candidates include live attenuated strains of S. flexneri 2a, S. sonnei, and S. dysenteriae 1, as well as inactivated whole-cell vaccines derived from inactivated S. sonnei and S. flexneri 2a strains. Subunit-based approaches involve covalent and noncovalent O-polysaccharide–protein conjugates targeting S. flexneri, S. sonnei, and S. dysenteriae, LPS-Ipa-protein complexes protecting against S. flexneri 2a and S. sonnei, and S. flexneri 2a-directed outer membrane vesicles. These candidates, together with major efforts to increase the immunogenicity of mucosal vaccines as well as the selection and design of potent adjuvants and antigen carriers, promise fast progress toward a long-awaited safe and powerful vaccine against Shigella (571, 573). Unfortunately, various factors have hindered a rapid solution thus far (454). The diversity of epidemiologically relevant Shigella serotypes, the fragile nature of its pediatric target group in developing countries, and the geographic divergence of shigellosis incidence, including different diets, sanitary standards, and target groups with various immunological backgrounds, are just some of the hurdles that complicate vaccine development process.
ENTEROAGGREGATIVE E. COLI
Enteroaggregative E. coli (EAEC or EAggEC) was identified by comparing adherence patterns of over 500 isolates from a case-control study in 1987 (574). Since its initial identification, EAEC has been identified in worldwide endemic and epidemic diarrheal diseases (575), and in several large-scale studies it has even been found to be the most common bacterial pathogen identified in diarrheal stool samples (576, 577). It causes persistent diarrhea in children in areas where EAEC is endemic (578) and persistent diarrhea in human immunodeficiency virus (HIV)-infected patients (579), and it is a causative agent for traveler's diarrhea (580).
Classification
After EAEC was first described, a method to identify the pathotype was developed using a probe that was later found to hybridize with an ATP-binding cassette transporter apparatus that translocates dispersin across the bacterial cell membrane (577, 581, 582). The majority of these probe-positive samples were also found to carry aggR, but not all diarrheagenic strains were positive for aggR, which led to a general classification of EAEC into typical (containing aggR) and atypical (lacking aggR) groups (583). Further classification of EAEC is possible based upon adherence patterns, as some strains preferentially infect the small bowel, while others infect both the small bowel and colon (580).
In a study of 143 EAEC isolates gathered from outbreaks, sporadic cases of diarrhea, and hospital-based studies from 1985 to 2006 in the United Kingdom, 43 could not be serotyped (584). Of those that could, the most common serotypes isolated were O126:H27 (5 isolates), O44:H18 (6 isolates), O111ab:H21 (8 isolates), O73:H18 (2 isolates), O92:H33 (2 isolates) O126:H27 (2 isolates), and O136:H2 (2 isolates) (584). However, O serotyping of EAEC for the purposes of classification is problematic, as many strains autoagglutinate, there are a variety of serotypes, and even EAEC strains that share serotypes differentially adhere to HEp-2 cells, which is a common method used for detection and classification of different E. coli pathovars (584–586). Using MLST on 126 E. coli isolates from a Nigerian case-control study, researchers recently concluded that the sequence type complexes present in the Nigerian study were shared with well-studied EAEC strains from other parts of the world (16). Additionally, they found that the category of EAEC encompasses multiple pathogenic lineages, emphasizing the global heterogeneity of EAEC (16). Importantly, they concluded that no single strain can be considered representative of EAEC and suggested that to properly study EAEC, researchers instead must first identify specific virulence genes and mechanisms within these separate lineages.
Epidemiology
Incidence and outbreaks.
As global surveillance for all strains of diarrheagenic E. coli was limited in the past, with much of the historical data regarding outbreaks of EAEC limited to North America, Europe, and South America, a comprehensive picture of EAEC incidence and outbreaks is still an ongoing research goal. However, EAEC was found to be the most common bacterial cause of diarrhea in the emergency departments and outpatient clinics of two large academic hospitals in Maryland and Connecticut in a large-scale study (577). EAEC has caused many significant outbreaks, including a 1997 school lunch outbreak in Japan that sickened 2,697 children. In 1997 an EAEC diarrheal epidemic occurred in a village in India and sickened approximately 15% of the population (587). More recent surveillance for EAEC has detected diarrheagenic EAEC strains in Mali (588), Libya (589), sub-Saharan Africa (7), and Nigeria (16), establishing EAEC's presence on the African continent.
Hybrid strains of EAEC and STEC are also becoming recognized. A notable 2011 outbreak in Germany sickened 4,321 previously healthy individuals from 16 countries, where over 900 patients developed HUS and there were more than 50 deaths (590). This Stx-encoding strain was identified as E. coli O104:H4, a hybrid pathogen that carried virulence genes found in both typical EAEC strains (aggA, aggR, set1, pic, and aap) and STEC (stx2) (45, 46, 330, 585, 591). The stx-carrying phage was shown to be most closely related to the phage of an STEC O111:NM strain (592) and was probably acquired quite recently in the phylogenetic history of the outbreak strain. EAEC has an established pattern of acquiring Shiga toxins, as reports from Japan (42), France (43), Northern Ireland (593), and Central African Republic (594) describe stx-positive EAEC isolates from patients with HUS. These cases indicate the possibility of outbreaks in the future.
Transmission and reservoirs.
Transmission of traveler's diarrhea, which is often caused by EAEC, occurs mostly through contaminated water and food, such as salads (595) (Fig. 5), with desserts and salsa being common contaminated food sources of EAEC in Mexico (596–598). Additionally, food handlers may be carriers of EAEC, emphasizing how important sanitary food handling practices are in prevention of EAEC transmission (599, 600). Remarkably, individuals with an AA rather than an AT or TT genotype at the −251 position of the interleukin-8 (IL-8) gene generated greater fecal IL-8 in response to EAEC and were associated with increased infection with EAEC (601). While atypical EAEC has also been identified in calves, piglets, and horses, animals are not an important reservoir of human-pathogenic typical EAEC (602). However, because EAEC isolates are so heterogeneous, it is premature to rule out animal reservoirs for undercharacterized lineages of EAEC.
Stx-containing strains.
During the 2011 European outbreak of 4,321 cases, fenugreek sprouts were identified as the most likely source of infection (603). The seeds were imported as a lot in late 2009 from Egypt, and it is still unknown if the point of contamination occurred at the site where seeds were produced, during transportation, or at the importer (603). By PCR screening of 1,468 French cattle, it was determined that cattle were not a reservoir for the 2011 European outbreak of STEC O104:H4, despite the observation that domestic ruminants are natural reservoirs of STEC (604). Finally, delayed human-to-human transmission did occasionally occur during the STEC O104:H4 infection in Poland, France, Germany, and The Netherlands during the 2011 European outbreak, indicating that while it is rare, humans can act as reservoirs of STEC O104:H4 (44, 605–608).
Pathogenesis
For non-Stx variants, EAEC strain 042 has been used as a prototypical strain to study virulence factors and pathogenicity of diarrheagenic EAEC, as it causes diarrhea in the majority of volunteers (609). However, the encoding genes for numerous adhesins, toxins, and proteins associated with virulence are highly variable among strains (3, 8, 16, 583, 585, 610). Even the site of infection in the gastrointestinal tract is not uniform. For example, the EAEC 042 strain has been isolated from the jejunum in infected volunteers, and in tissue culture it adheres strongly to samples of jejunal, ileal, and colonic mucosa (609, 611). In a controlled study looking at five different non-Stx EAEC isolates from children, each strain had a different affinity for the jejunal, ileal, and colonic mucosae (612). Despite the heterogeneity among the different non-Stx EAEC strains, a general three-part model has emerged for non-Stx EAEC pathogenesis: (i) adherence to the intestinal mucosa, (ii) production of enterotoxins and cytotoxins, and (iii) mucosal inflammation (585).
Adherence.
During the first stage of its pathogenesis, adherence to the intestinal mucosa, EAEC aggregative adherence is associated with both fimbrial (aggregative adhesion fimbria [AAF]) and afimbrial adhesins. As reviewed by Estrada-Garcia and Navarro-Garcia in 2012 (585), several different EAEC strains contain different afimbrial adhesins, otherwise known as outer membrane proteins associated with aggregative adherence (585, 613–617) (Fig. 6). Along with the diversity in afimbiral adhesins found among different EAEC strains, there have also been several different AAFs observed by electron microscopy (618–620). Depending upon the EAEC strain, the major structural subunit of AAF has four different variants: AggA (AAF/I), AafA (AAF/II), Agg3A (AAF/III), and Agg4A (AAF/IV) (621–624). These variants are all encoded on the pAA virulence plasmid, which also encodes AggR, a transcription factor that regulates AAF biogenesis (625, 626). In addition to these AAF variants, other EAEC strains can encode alternative fimbrial structures, such as a type IV pili in EAEC strain C1096 (627, 628). For strains that lack AAFs, aggregative adherence has been linked to the hraI gene on the genome (also known as hek) or to possession of alternate adhesins such as Hda (629).
Toxin production.
For the second stage of EAEC pathogenesis, many different putative virulence factors have been described and reviewed (585, 588, 630–632). The effects of these toxins include microvillus vesiculation, enlarged crypt openings, and increased epithelial cell extrusion (583). These putative toxins include Pet, a cytoskeleton-altering toxin (633), EAST-1, a heat-stable enterotoxin encoded by many other pathogens besides EAEC (634), ShET1, a subunit toxin that appears to induce intestinal cyclic AMP (cAMP)- and cGMP-mediated secretion and is also encoded by Shigella flexneri (508), and Pic, a mucinase shared among many different pathogenic E. coli and Shigella strains (635). Interestingly, the Pet toxin of EAEC belongs to the same protein family as the EspC toxin of EPEC, where the two toxins cleave the actin-binding protein fodrin but have dramatically different mechanisms of entering the host cell (636). Other, uncharacterized toxins identified in genome sequencing and metabolic profiling screens include a potential hemolysin encoded by the hlyE gene (27, 577), as well as dispersin, encoded by the aap gene (antiaggregation protein, previously named aspU [587, 637, 638]). Finally, the EAEC 042 strain also carries the shF gene, which is predicted to be similar to IcaB, a mediator of biofilm formation in Staphylococcus epidermidis (590, 637, 639). It is known that the AAFs also mediate biofilm formation in EAEC (585, 622, 640). Characterization of toxins encoded by different EAEC strains remains an ongoing and active field of research. AggR is involved in the regulation of many of the virulence genes involved in both the aggregation and toxin production stages of EAEC pathogenesis (45, 46, 330, 625). AggR is a positive regulator and belongs to the AraC family of regulators. It is negatively regulated by the global regulator H-NS and positively regulated by itself and the DNA-binding protein FIS (43, 641).
Inflammatory diarrhea.
During the final stage of the EAEC pathogenesis model, there are multiple factors that influence the scale of inflammation, including the host innate immune system and the strain of EAEC (585, 597). During infection of the gastrointestinal tract, EAEC adherence to intestinal epithelial cells stimulates the release of IL-8 and CCL20, which recruits neutrophils and stimulates inflammatory diarrhea (599, 600, 642, 643). Proteomics studies with the EAEC-T8 strain (644) have shown that it interacts with Gp96, a cell surface protein that has significant homology with Hsp90, a protein which triggers signaling pathways in response to binding by other pathogens and leads to activation of inflammatory/immune responses through NF-κB and MAPK (601, 644, 645). In the same proteomics study (644), it was also found that the EAEC-T8 strain interacts with TSP1, an ECM glycoprotein that is rapidly secreted at high levels in inflamed and damaged tissues and has been implicated as a molecular bridge between Gram-positive pathogens and host tissue cells (603, 644). Additionally, AAF/II can elicit a basolateral release of IL-8 from polarized monolayers of T84 human colonic epithelial cells (603, 646). Furthermore, AAFs mediate EAEC-induced polymorphonuclear neutrophil (PMN) infiltration both in vitro and in vivo by triggering host cells to release an eicosanoid-based PMN chemoattractant generated through 12/15 lipoxygenase (12/15-LOX) activity (647). The released eicosanoid guides PMNs across the intestinal epithelium to the luminal surface by forming a chemotactic gradient (647–649). Finally, the presence of many of the putative virulence factors outlined above correlates with elevated levels of fecal cytokines, interleukins (IL-1ra, IL-1β, and IL-8), alpha interferon (IFN-α), lactoferrin, fecal leukocytes, and occult blood (as reviewed in references 585 and 609).
Clinical Considerations
Symptoms.
Diarrheagenic EAEC presents with watery secretory diarrhea, often with mucus, and can be accompanied by a low-grade fever, nausea, vomiting, abdominal pain, and occasionally bloody stool (as reviewed in references 3, 8, 583, 585, 597, 610, 650, 651, and 652). In both developing and developed countries, EAEC affects both adults and children (609, 611, 632, 653–655). It also causes acute and persistent diarrhea in HIV-infected patients (612, 656, 657) and persistent diarrhea in children, which leads to growth and intellectual shortfalls (as reviewed in references 585 and 658). The most significant public health concern stemming from EAEC infections is malnourishment in children in developing countries, as persistent EAEC infections lead to chronic inflammation, which damages the intestinal epithelium and reduces its ability to absorb nutrients (585, 618–620, 658–660). Additionally, the odds of developing postinfectious irritable bowel syndrome are dramatically increased after acute infectious gastroenteritis (661), with some studies reporting EAEC, among other enteropathogens, as a causative agent (662).
Detection.
The gold standard method to identify EAEC is to culture five colonies per patient in static Luria broth at 37°C and then infect semiconfluent HEp-2 cells for 3 h and look for the aggregative adherence (AA) pattern (Fig. 7). When treated this way, EAEC aggregates and produces a hallmark “stacked-brick” appearance, where the bacilli are elongated and sometimes line up in a single layer on the surface of the cell (574, 627, 628). However, this method does not distinguish between pathogenic and nonpathogenic strains and is unsuitable for surveillance for EAEC outbreaks, as it requires specialized equipment and is labor-intensive. In a meta-analysis of four different studies examining the distribution of EAEC genes in children who were excreting EAEC, the aafA, astA, and 4A18 genes were all associated with acute diarrheal illness among children from developing regions. However, the EAEC strains in all of these 4 studies were heterogeneous, with the detection of these genes in each study quite different (632). Additionally, in the same meta-analysis, a DNA probe commonly used for EAEC, which is based upon a 1-kb DNA fragment from the 60-MDa plasmid of EAEC strain 17-2, showed a wide range in sensitivity (15% to 89%), reflective of the heterogeneity among EAEC strains and the lack of defined virulence properties in many EAEC strains (632). Using molecular biology assays such as PCR to identify hallmark virulence genes is problematic, as not all EAEC strains that have the AA binding pattern are diarrheal, and in those that are, there is no single gene or set of genes that distinguishes between pathogenic and nonpathogenic EAEC strains (585).
Historically, the aggR gene was thought to be a promising candidate for molecular detection, as it encodes a positive regulator for many virulence factors involved in adherence and toxin production by typical EAEC strains and is carried by more than 70% of EAEC isolates from human patients (583, 663–665). However, nondiarrheal EAEC strains carrying the aggR gene have also been isolated from healthy patients, and detection of this gene in a bacterial isolate from a diseased patient cannot be considered conclusive that it is the causative agent of diarrhea (586, 633, 663–665). Nonetheless, multiple PCR-based assays have been developed to identify the aggR gene, and detection of additional virulence genes, such as aap, astA, and set1A, significantly increases the detection of strains associated with causing diarrhea in U.S. and European patients (634, 666, 667). However, strains that do not carry the aggR gene have also been isolated from gastrointestinal outbreaks (627).
Currently, amplification by multiplex PCR of either the plasmid-carried gene aatA or the chromosomally carried aaiC locus is considered sufficient to confirm EAEC in the recently released GEMS, an initiative to comprehensively identify major enteric pathogens rapidly at sites where the diarrheal burden is high (535). aatA encodes an outer membrane protein that is part of a protein transporter system (581, 668). In a 2009 study of 252 E. coli isolates from children with diarrhea, aatA was amplified only in EAEC strains and not in other diarrheagenic E. coli strains (669). The other gene used in GEMS, aaiC, encodes a protein secreted by the prototypical EAEC strain 042, with AaiC secretion regulated by AggR. aaiC was detected in 26 EAEC strains in a study of 35 EAEC strains from geographically distinct regions, indicating that it may be found in high prevalence in EAEC (625). However, amplification of either of these genes does not conclusively distinguish diarrheagenic EAEC from nondiarrheagenic strains (631). In a subsequent study of all EAEC strains that were positive for either aaiC or aatA, one set of genes in combination, aaiC and agg3/4C but lacking agg4A and orf61, was associated with diarrhea cases, and another one, orf61 in the absence of pet and aafA, was correlated with control children (631). Additionally, as reviewed in reference 585, in Brazilian children with diarrhea compared to those without, the presence of the kps (capsule), hly, aero (aerobactin), and aggA genes was strongly associated with disease (586), suggesting that development of an assay to detect these genes may be a promising method for detection of EAEC strains that cause disease in children in non-European and non-U.S. locations. While current studies to identify conclusive markers for diarrheagenic EAEC strains are promising, identification of molecular markers for rapid detection will remain a challenge due to the heterogeneity of EAEC (585).
Treatment.
(i) Treatment of traveler's diarrhea.
Antibiotics are generally recommended for treatment of traveler's diarrhea, but because EAEC is increasingly resistant to various antibiotics, selection of the appropriate antibiotic should take into account the region of the world where the infection was acquired, as there are different antimicrobial susceptibility patterns for each geographical region (670–672). Like Shigella infections, EAEC infections are often successfully treated with ciprofloxacin and other fluoroquinolones, but there are multiple antibiotic-resistant strains (597, 673). For example, in southern India, EAEC is increasingly resistant to quinolones (672). In adult patients in the United States, EAEC is susceptible to rifaximin or a single dose of azithromycin with or without loperamide (670, 674, 675). Elimination of persistent diarrhea may be problematic, as internalized EAEC was observed in intestinal epithelial cells in vitro, suggesting that EAEC may protect against host clearance by intracellular persistence (676).
(ii) Treatment of infections caused by Stx-containing EAEC strains.
Prior to the 2011 outbreak in northern Germany, there was no standardized treatment for Stx-containing EAEC. During the 2011 German outbreak, 3 children with Stx-associated HUS showed rapid clinical improvement with eculizumab, but results from a subsequent nonrandomized trial with 298 patients were unclear (677–679). Patients who had no clinical improvement during plasmapheresis and/or were suffering from severe neurological complications were preferentially selected for the trial, leading to a selection bias that complicates the results (677, 679). As eculizumab disrupts the complement cascade, clinicians at the time were required to treat with a prophylactic antibiotic to prevent meningitis. In general, antibiotics are normally not recommended for STEC, as they increases the risk for development of HUS by stimulating Stx production. Because of this risk, clinicians treating stx-expressing EAEC strain O104:H4 selected azithromycin, which in vitro represses stx expression (677, 679, 680). Monitoring of STEC shedding in patients receiving azithromycin showed that these patients were rapidly decolonized (677, 681). Because of this, long-term (>28 days) carriers of STEC O104:H4 were treated with azithromycin, and after a 3-day course of treatment, all 15 were negative for shedding as well as HUS-related symptoms (677, 681). Further studies have since shown that subinhibitory concentrations of ciprofloxacin increase Stx production in STEC O104:H4 but that meropenem, rifaximin, tigecycline, and azithromycin do not (677, 680). Stx production by STEC O157:H7 responds differently to these same antibiotics (677, 680). Nonetheless, the use of azithromycin to eliminate Shiga toxin-containing strains such as O104:H4 from patients is still considered a controversial treatment; if used early in treatment, it is still unclear if it plays a role in the development of HUS (682), and if used later in treatment, it may actually increase the risk of sudden cardiac death (683).
(iii) Treatment of immunocompromised patients.
In a study of HIV-infected children in Peru, 74% of the diarrheagenic E. coli strains detected were highly resistant to ampicillin (74%) and co-trimoxazole (70%) (684). The current Infectious Disease Society of America practice guidelines for the management of infectious diarrhea recommend fluoroquinolones for the treatment of immunocompromised patients (685). AIDS patients with EAEC-induced diarrhea have been successfully treated with ciprofloxacin, with fecal shedding eliminated and 79% of total symptoms improved or eliminated (686).
Vaccines and other preventative treatments.
As EAEC proteins are antigenic, it remains possible that a vaccine could be developed, but as of yet, there is none. As reviewed in reference 670, in a vaccine study using the ETEC heat-labile toxin, the rate of infection and severity of disease caused by ETEC was decreased, and despite the presence of EAEC in the placebo groups, the vaccine-treated group had no EAEC detected, suggesting that the vaccine may also exert protection against EAEC (687). In vitro treatment with lactoferrin inhibits EAEC enteroadhesion and biofilm formation, making it a potential but untested nonantibiotic treatment for the prevention of EAEC (670, 688).
ENTEROTOXIGENIC E. COLI
ETEC is a diverse pathotype that is a major cause of traveler's diarrhea and endemic in most underdeveloped countries (30). In these developing countries, ETEC can be isolated from both symptomatic and asymptomatic carriers, with significant mortality rates in children (30, 689). ETEC is defined by its ability to produce either a heat-labile (LT) or a heat-stable (ST) enterotoxin, and it carries a diverse set of colonization factors (CFs) for adherence to the intestinal epithelium. In addition to the impact of ETEC on humans, the swine industry is also adversely affected, as ETEC-induced diarrhea in neonatal and postweaning piglets causes much morbidity and mortality. This review of ETEC will focus on its human-related illnesses, building on recent knowledge from the excellent reviews by Nataro and Kaper and by Qadri et al. (8, 30).
Classification
Common serotypes.
The distribution and occurrence of O and H antigens across various regions were thoroughly reviewed by Wolf in 1997 (690). There are at least 78 detectable O antigens and 34 H antigens associated with ETEC. Among the most common serotypes were O6:H16 (LT/ST), O8:H9 (ST only), O25:NM (LT only), O78:H12 (ST only), O148:H28 (ST only), and O153:H45 (ST only); however, their prevalence and phenotypes (e.g., which toxins they harbor or CFs they express) can vary depending on the location (690). Many of these serotypes were also commonly isolated from surface water in Bangladesh (691) and from outbreaks in the United States (692) and Japan (693). More recently, O169:H41 (ST only) has become an emerging serotype that has been involved in several outbreaks in the United States, where it was rarely seen before 1996 (694, 695).
Lineages.
MLST of ETEC isolates groups them across all phylogroups (696). Further studies on a set of 1,019 human isolates found that they could be grouped into 42 distinct clonal groups, with diverse toxin and CF profiles spread across all lineages (18).
Epidemiology
While ETEC is a major cause of traveler's diarrhea, there is significant morbidity and mortality associated with ETEC-induced diarrhea in children in developing countries. Cohort studies following children in Argentina, Bangladesh, Egypt, and Guinea-Bissau have looked at the distribution of toxins and CFs and their frequency and contribution in diarrheal disease (697–700). In these studies, ST-positive ETEC strains were commonly associated with diarrhea. The GEMS case-control study also reported ST-positive ETEC, but not LT-only-positive ETEC, as a major contributor to infantile diarrhea (7). ST-positive ETEC strains were also found to be associated with an increased risk of death in children.
Incidence.
A review of population-based studies estimates that there are nearly 840 million annual cases of ETEC in developing countries, with approximately 280 million of these in children aged 0 to 4 (701). Children under the age of 4 can experience up to 0.47 to 0.54 ETEC episode/person/year, which drops to 0.11 to 0.15 ETEC episode/person/year after the age of 5 (701), possibly due to immunity acquired from prior infections (702). A review of population studies in developing countries showed that ETEC was the etiological agent isolated in a median of 13% of diarrheal cases in children (689). The authors of that review also estimated that 325,000 children under the age of 5 die each year due to ETEC-associated illness (689). An additional 46.6 million children under the age of 4 are thought to be asymptomatic carriers of ETEC (701). In rural Egypt, a study showed that 84% of children under the age of 3 will have at least one ETEC-related episode of diarrhea (703). The authors estimated that there were approximately 1.5 episodes/child/year related to ETEC.
In developed countries, ETEC is not routinely tested for in patients presenting with diarrhea (704); therefore, incidences of ETEC illness and epidemiological studies in these areas are lacking. However, travelers to high-risk, developing regions such as Latin America, Africa, and certain regions of Asia are at risk of developing traveler's diarrhea. Despite a decrease in overall ETEC-related traveler's diarrhea cases in Latin America and Africa, ETEC is still the most common etiological agent detected and makes up approximately 30% of these cases (462).
Adhesin and toxin profiles of ETEC isolates are also variable and differ among different geographical regions and populations. A systematic review of the literature across 35 countries found overall that approximately 45% of ETEC isolates expressed ST only, 27% expressed LT only, and 33% expressed both ST and LT (705). Furthermore, the authors also determined the regional prevalence of colonization factors and showed that CFs such as CFA/I, CS6, and CS21, were among the most common CFs detected in ETEC isolates worldwide (705).
Host factors.
A study in Bangladesh, following ETEC diarrhea of children during their first 2 years of life, noted that ST-expressing ETEC more frequently infected children with blood type A or AB than children with blood group O (698). Further studies determined that children with Lewis antigen Le(a+b−), but not Le(a−b+), were more susceptible to illness caused by ETEC that expresses the CFA/I group of CFs (706). This association was suggested to be because of the ability of CFA/I to adhere to Lea-terminated glycosphingolipids but not Leb-terminated glycosphingolipids on host cells (706, 707).
Transmission, reservoirs, and sources.
ETEC infections are transmitted through the fecal-oral route. Exposure to ETEC is usually from contaminated food and drinking water (Fig. 5). Some examples of high-risk foods contaminated with etiological agents for traveler's diarrhea include food that is left at room temperature, table-top sauces, certain fruits, and food from street vendors (reviewed in references 708 and 709). Additionally, surface water in developing regions such as Bangladesh can also contain these organisms and may serve as an important source of infection during contact with this water (691). Foods can be contaminated by infected food handlers (710), by asymptomatic carriers (711), or when vegetable crops are irrigated with untreated water (712). The persistence and ability of ETEC to survive in these environments are mostly unknown. One study showed that ETEC was able to survive for up to 3 months in freshwater (713) and was able to form biofilms in drinking water sources (714). The infectious dose, compared to that of other E. coli pathotypes, is relatively high and is thought to be between 106 and 108 organisms (715). However, this infectious dose is based on laboratory-grown cultures and may differ from the actual dose from natural transmission. One investigation of an outbreak in a Japanese prison estimated that the infectious dose in contaminated pickles was between 25 and 1,000 organisms (716).
Outbreaks.
Outbreaks in developed countries are sporadic and can consist of more than one serotype (692). These outbreaks are likely underdetected, as ETEC is rarely sought in diarrheal cases (704). A study of the CDC's ETEC outbreak investigations from 1996 to 2003 identified 3 outbreaks related to international cruise ships, mainly linked to drinking water, and 13 domestic outbreaks (694). This study identified serotype O169:H41 (ST only) as the most common isolate, which is considered an emerging strain in the United States (694) and which affected over 30 people in an outbreak in Tennessee (695). The largest ETEC outbreak reported in the United States was from the state of Illinois in 1998, where serotype O6:H16 (LT/ST) was isolated from several patients. Although only 120 cases were documented, it was estimated that 3,300 people may have become ill due to food prepared by a delicatessen that catered several events over the span of a few days (717). Further outbreaks were reported from a sushi restaurant in Nevada (710) and a buffet-style lunch in Illinois (718).
Outbreaks have occurred in other developed countries outside the United States. In 2006 a mixed outbreak of ETEC and Salmonella, possibly linked to pesto, caused 217 illnesses in a Danish school (719). Denmark had an additional 5 outbreaks of ETEC mixed with norovirus in early 2010, which were linked to contaminated lettuce (720). Additionally, Japan has had 131 ETEC-related outbreaks from 1966 to 2009 that originated from various contaminated food and water sources (693), while Israel had a large outbreak of ETEC, likely due to improperly sanitized water, that affected 175 military personnel and at least 54 civilians (721).
Although ETEC is endemic in most developing countries, epidemic outbreaks have occurred during major floods in Bangladesh, where ETEC was identified in 18% of diarrheal cases studied, second only to Vibrio cholerae (722). In hospitals, outbreaks have been reported in neonatal intensive care units, likely due to contaminated milk prepared by an asymptomatic carrier (711).
Pathogenesis
Analysis of ETEC genomes has shown that there may be a core set of genes shared between ETEC isolates; however, no virulence factors were attributed to these core sequences (24). Indeed, it has been suggested that the acquisition of plasmid-borne toxins and virulence factors may be the major driving force that makes ETEC a pathogen (696, 723). ETEC causes disease by colonizing the small bowel through attachment to the host epithelial lining by surface proteins called CFs and possibly other surface structures (Fig. 6). Adherent ETEC then elaborates enterotoxins that cause the clinical manifestations typical of ETEC-induced diarrhea. Unlike for most other pathotypes, almost all known virulence factors are encoded exclusively on plasmids. With more whole genomes being sequenced, it is possible that chromosomal virulence factors will be identified in the future.
Adhesion.
ETEC CFs are named coli surface antigens (CS), suffixed with a number, except for CFA/I. They are surface structures that are required for colonization and disease. Currently, at least 22 CFs have been characterized; however, between 30 and 50% of ETEC isolates have undetectable CFs (30). This suggests that there are still unknown CFs or that they are undetectable under typical laboratory conditions. The advancement of whole-genome sequencing could identify potential new CFs. The most recently identified CF, CS23, is an afimbrial adhesin (724).
These surface structures are either fimbrial, afimbrial, helical, or fibrillar and are encoded mostly on virulence plasmids (30, 725). The molecular mechanisms of their assembly and structure have been reviewed elsewhere (726). CFs are subdivided into different families, with CFA/I, CFA/II, and CFA/IV being the most common, depending on the region (30, 725).
CFs bind to different receptors on host cells. For example, CS6 was found to bind fibronectin (727) and sulfatide (SO3-3Galβ1 ceramide), a major glycosphingolipid (728), while sequence variation changes binding affinity to host cells (729). CFA/I was also found to bind to glycoproteins and glycosphingolipids found on the surface of host cells in the small intestine (707). CFs can also function beyond host cell adherence; longus (CS21) has been shown to be involved in twitching motility (730) and self-aggregation, which was shown to be protective against antimicrobials, in vitro (731).
Other adhesive proteins found in some ETEC isolates are encoded in the tia and tib loci. Based on a Chilean study, tia and tib were found in 9% and 11% of isolates, respectively (732). Genes from the tia locus (also found in strains of EPEC, EAEC, and S. sonnei) have been shown to be involved in invasion of and adherence to epithelial cells (733, 734) and interact with heparan sulfate proteoglycans (735). The tib locus is also involved in adherence and invasion (736, 737). More recent studies have demonstrated the involvement of TibA in autoaggregation and biofilm formation (738, 739). Finally, a protein, EtpA, was found to contribute to adherence to epithelial cells and is thought to act by being transiently exposed at the tip of the flagellum to mediate attachment (740, 741). EtpA is found in approximately 75% of ETEC isolates, based on work by Del Canto et al. (732).
ST.
Harboring a heat-stable enterotoxin (ST) is a characteristic that can define an ETEC strain. Two STa (or ST-I) variants have been found in human disease, STp and STh (8), and will be referred to here as ST. STb (or ST-II) can be found in strains that colonize animals. These toxins can be found as the only enterotoxin (ST only) or in combination with LT (LT/ST). Overall, ST is found in about 75 to 80% of ETEC isolates (approximately 45% ST only and 33% LT/ST) (705) and is found more frequently in severe human disease than LT-only isolates (30). STs are small enterotoxins in comparison to LT and are generally 18 or 19 amino acids in length, following processing and secretion from the bacterial cell.
The molecular mechanisms of ST are well described in the literature (for a review, see reference 742), but the dogma on secretion has been challenged (reviewed in reference 743). Generally, these small peptides mimic the hormone guanylin and thus bind guanylyl cyclase C (GC-C) receptors on the intestinal epithelium. This overactivation of GC-C, caused by ST binding, leads to an accumulation of cGMP, which indirectly promotes cystic fibrosis transmembrane receptor (CFTR) to secrete chloride into the intestinal lumen and also indirectly inhibits the sodium-hydrogen exchanger, preventing absorption. Although ST-induced chloride and fluid secretion into the lumen is widely regarded as the cause of watery diarrhea, an in vivo study using volume recovery in the rat jejunum suggested that diarrhea caused by ST may instead be due to impaired fluid absorption (744; recently discussed in reference 743).
LT.
Heat-labile enterotoxin (LT) is the second enterotoxin that can define ETEC. There are two classes of LTs, the prototypical LT-I (usually referred to as LT, which will be followed in this review) and LT-II, which has differences in its B subunit receptor affinity and immune properties (reviewed in reference 745). LT-II can also be subcategorized into LT-IIa, LT-IIb, and the more recently discovered LT-IIc (746). Generally, LT-I is more consequential in human illness, but LT-II has been isolated from human cases of ETEC diarrhea (747). While LT is found encoded on virulence plasmids, all LT-II variants have been found encoded in the chromosome and may have been acquired by a phage (747).
Assembled as an AB5 toxin, LT is a large toxin that is about 80% identical to the cholera toxin from V. cholerae. As with ST, the molecular mechanisms of LT are well studied and extensively reviewed (748). Briefly, the pentameric B subunits bind to GM1 gangliosides at lipid rafts to deliver the catalytic A subunit inside the cell. Once inside the cell, the A subunit is trafficked through the endoplasmic reticulum (retrograde) and delivered to the host cytoplasm, where it ADP-ribosylates Gsα, inhibiting its GTPase activity and increasing cyclic AMP levels. Increased cAMP opens the CFTR, resulting in electrolytes and fluid loss into the intestinal lumen. Additionally, fluid loss into the intestinal lumen may also be a consequence of intestinal barrier disruption caused by LT (749). As discussed in reference 748, LT has also been shown to interact with lipid A of LPS (association with OMVs), blood type A sugars, and non-GM1 gangliosides, albeit at lower affinities.
Additional roles have been characterized for LT, including contributing to adherence to host cells through activation of host signaling pathways (750, 751), as well as suppression of antimicrobial peptide expression (752).
EAST1.
Very little is known about the involvement of EAST1 in disease, but it is carried by many pathogenic and commensal E. coli strains (148), leading to uncertainty about EAST1 involvement in diarrhea. A recent study showed that EAST1 in ETEC may not be involved in diarrhea (753).
Other virulence factors.
The SPATE EatA is a 104-kDa plasmid-encoded protein that contributes to increased virulence in a rabbit ileal loop model (754). More recent work has shown that EatA degrades EtpA and contributes to LT delivery, as an eatA mutant was hyperadherent to epithelial cells but was unable to efficiently deliver LT (755). Del Canto and colleagues identified EatA in approximately 70% of Chilean ETEC isolates and suggested that it could be suitable as a marker for epidemiological studies (732).
A small, 12.6-kDa secreted protein, called CexE, was identified to be regulated by a known virulence regulator (756) and has been suggested to function similarly to dispersin from EAEC (723).
Clinical Considerations
Symptoms and onset of disease.
ETEC causes mild to severe watery diarrhea (usually nonbloody) that has a clinical presentation very similar to that of cholera and can rapidly lead to dehydration (30). Illness is sometimes accompanied by headaches, fever, abdominal cramping, nausea, and vomiting (30, 692, 694). The onset of ETEC-related diarrhea is generally quick, with an incubation period that can be as short as 5 h but generally averages between 1 and 2 days following ingestion (692, 694). The duration of diarrhea is usually about 3 to 5 days (692, 694) but can be prolonged for more than a week (718). Mortality rates with proper treatment are very low (<1%) (30).
Complications.
ETEC diarrheal illness in children in developing countries under the age of 2 has implications for growth. A study following children in Bangladesh found that children who experienced an episode of ETEC diarrhea were more malnourished and growth stunted than those who had non-ETEC diarrhea (698). There is also a suggestion in the literature that people who have experienced traveler's diarrhea have a 5-fold-increased risk of developing postinfectious irritable bowel syndrome (757). Although ETEC is commonly implicated in traveler's diarrhea, the etiological agent responsible for this increased risk of irritable bowel syndrome is unclear (662).
Detection.
ETEC is defined by the presence of either enterotoxin, and thus identification is made by detection of LT, ST, or both. As with most other E. coli pathotypes, ETEC cannot be readily distinguished on typical MacConkey media from commensal E. coli, and thus molecular techniques must be used. The history of ETEC detection with methods such as monosialoganglioside GM1 enzyme-linked immunosorbent assay (ELISA) for LT and ST, multiplex PCR for toxins and CFs, and monoclonal anti-CF antibodies has been reviewed by Qadri and colleagues (30). A recent paper highlighted the importance of screening multiple colonies for ETEC, as testing only 5 colonies missed approximately 50% of ETEC-positive stools identified by screening 20 colonies (758). Additionally, toxin-encoding genes may be lost during handling, as one study showed that toxin genes became undetectable in 119 of 1,197 isolates (702).
A comparison of the phenotypic and genotypic ETEC detection methods suggested that PCR methods for enterotoxin detection were highly specific and sensitive, and CFs could also be detected by PCR or dot blots using monoclonal antibodies (759). PCR-based approaches are particularly useful in studies where monoclonal antibodies to enterotoxins or CFs are unavailable. A multiplex assay that was able to detect 19 different ETEC virulence factors was developed (760). Additional studies have shown that fecal smears on hemocult cards allowed for long-term storage and isolation of ETEC DNA, which could be useful in epidemiological studies when standard methods for culture are not readily available (761).
Treatment.
ETEC-associated diarrhea is self-limiting, and in general, oral rehydration and maintaining fluid and electrolyte balance through diet are very effective for both children and adults (8, 30). In severe cases of dehydration, intravenous rehydration may be necessary (691). Antisecretory drugs, such as loperamide, may be beneficial in reducing the number of stools in adults and are usually safe to use in combination with antimicrobials (reviewed in reference 762). Antibiotics such as fluoroquinolones, azithromycin, and rifamixin can lessen the duration of infection and are commonly used in self-treatment of traveler's diarrhea. However, the decision to use antibiotics has to be weighed against possible adverse effects, such as the selection of antibiotic-resistant bacteria and the possibility of increasing the predisposition to developing other intestinal infections (762). Recommendations for preventative measures for traveler's diarrhea, such as chemoprophylaxis, immunoprophylaxis, and food safety, have been discussed elsewhere (763).
Antimicrobial resistance.
A study looking at the changing antibiotic resistance profiles of ETEC showed that nearly 60% of isolates were resistant to trimethoprim-sulfamethoxazole and tetracycline and approximately 50% to ampicillin for isolates from 2001 to 2004 (764). Of concern is the noted increase of ciprofloxacin resistance from 1% in the period of 1994 to 1997 to 8% in 2001 to 2004, where most of the ciprofloxacin resistant ETEC strains were isolated from patients who traveled to India. Ciprofloxacin-resistant isolates were also recovered from travelers from North Africa and Southeast Asia. This trend of emerging antibiotic resistance in ETEC was reviewed by Qadri et al. (30). In addition, it has been shown that ETEC rifaximin-resistant mutants can be selected in vitro and might occur more frequently than ciprofloxacin-resistant mutants (765).
Vaccines.
Immunity against ETEC reinfections has been observed in adult volunteers and naturally in children (702, 715) and supports the concept that a successful ETEC vaccine may be possible. Unfortunately, the diversity of ETEC strains has made vaccine development challenging; there are too many serotypes to target the O antigen, and ETEC has a virulence profile that differs between isolates, depending on geography (i.e., CF and toxin distribution).
Vaccines that are currently being evaluated can be grouped into different types, such as toxin-based, live attenuated, inactivated whole-cell, hybrid, and fimbrial antigen vaccines (766). LT is strongly immunogenic and cross-reacts with the B subunit of cholera toxin (CTB). The recombinant CTB (rCTB) has been used in an inactivated whole-cell oral vaccine (Dukoral), where it provides short-term protection against ETEC in travelers (as discussed in reference 709), as well as in the live attenuated cholera vaccine Peru-15pCTB, where sera from mice and rabbits could neutralize LT toxins (767). A transdermal patch made up of LT was shown to have few adverse effects and to elicit an immune response to the toxin (768, 769). A phase II study showed that the transdermal patch reduced the occurrences of traveler's diarrhea in travelers to Mexico and Guatemala compared to that in placebo-controlled travelers (687). Expression of LTB in the Salmonella enterica serovar Typhi live attenuated vaccine has been shown to elicit antibodies to LT in mice and in approximately 70% of inoculated human volunteers (770, 771). Additionally, LTB expressed in potatoes and corn and then ingested by human volunteers was able to cause development of IgG antibodies to LT in most volunteers (772), while rice expressing CTB (MucoRice-CTB) was protective against V. cholerae and ETEC challenge in mice, through mucosal IgA (773). Work is also being done to improve the safety of LT. A recent study characterized a new mutant LT called dmLT that is quickly degraded, has reduced toxicity, and may be useful in oral vaccines (774).
LT is present in only approximately 60% of isolates (LT only and ST/LT), while the poorly immunogenic ST is found in about 75 to 80% (705). Despite the poor immune properties, there is still interest in developing ST as a vaccine. In a recent study a hybrid toxoid with ST fused to LT was made, where both proteins were mutated to abate any toxicity, and showed an antibody response to both LT and ST (775, 776). A green fluorescent protein (GFP)-ST-LTB fusion that was expressed in Lactobacillus reuteri was given as a live oral vaccine to mice and was able to elicit antibodies to the protein fusion and protect mice against ETEC challenge (777). However, since ST is similar to guanylin, there is concern that there may be cross-reactivity issues with vaccines that target ST and its GCC-C receptor (reviewed in reference 778).
An oral, inactivated vaccine that expressed a mix of CFs and is supplemented with rCTB (rCTB-CF ETEC) has been extensively studied (discussed and reviewed in reference 779). This vaccine was studied in individuals traveling to Mexico and Guatemala, where the severity of diarrhea was milder in vaccinated travelers but the number of diarrheal incidences was not significantly different than that in placebo controls (780). This was further extended to a study in Egyptian children, aged 6 to 18 months, where the vaccine was not protective against diarrhea (779). To improve protection of ETEC vaccines, studies have started to optimize expression of CFs in nonpathogenic E. coli hosts. Overexpression of various CFs and hybrid CFs in a murine model showed an immune response to the CFs that was higher than that for reference ETEC strains (781–784). A recent publication described a phase 1 trial in volunteers using an E. coli strain overexpressing CFA/I coupled with a hybrid LT and CT (LCTBA) and demonstrated that it was safe and elicited an antibody response to both LTB and CFA/I (785).
Surface antigens may provide an alternate strategy for vaccine development. Recently, a study of chromosomally expressed autotransporter genes in ETEC that are not found in commensal K-12 E. coli identified two proteins, pAT and Ag43, that are detected using sera from ETEC-infected patients (786). The passenger domains of these proteins were found to be protective in a murine model. Other studies have shown that surface proteins, such as LT, flagellin, EatA, EtpA, CexE, and TibA, could be detected by sera from ETEC-infected mice or humans (787), where it was previously demonstrated that recombinant flagella and EtpA were protective in mice (788). Follow-up studies demonstrated that many of these proteins could be found in OMVs that provide protection against ETEC colonization in mice (789).
Live attenuated vaccines expressing either CFA/I (ACAM2010), CFA/II (PTL-003), or a combination of CS1, CS2, and CS3 (ACAM2017) were first evaluated in phase 1 trials and resulted in an antibody response to their respective CFs (790–792). Follow-up studies with PTL-003-vaccinated human volunteers did not show adequate protection against ETEC-induced diarrhea (793). This idea of a live attenuated vaccine was further developed by engineering three strains that, collectively, covered 6 CFs as well as LTB when mixed together to form ACE527 (794). Phase 1 studies with ACE527 demonstrated safe use in human volunteers and immune responses to all CFs and LT (795). A recent phase 2 study showed a 27% reduction in the incidence of moderate to severe diarrhea compared to that in placebo controls following ETEC challenge, but it was not significant. However, there were improved outcomes noted from secondary endpoints (796).
The expression and delivery of ETEC antigens has been attempted in other bacterial hosts. For example, Bifidobacterium infantis strains were recently used to express CFA/I or LTB, and these strains together conferred the most protection to rats following challenge with ETEC (797). Other bacteria, such as Shigella spp. (798, 799), V. cholerae (781), and STEC O157:H7 (800) have also been used to deliver ETEC antigens to experimental animals.
DIFFUSELY ADHERENT E. COLI
The diffusely adherent E. coli (DAEC) pathotype describes diarrheagenic E. coli strains that attach to cells but do not fall into classical patterns of adherence, such as localized or A/E (8). They have now emerged as a unique group and are considered distinct from other pathotypes, but because of difficulties in classification and identification, the designation of DAEC as a distinct enteric E. coli pathotype (801) requires further epidemiological studies.
Classification
DAEC has been classically defined by its diffuse adherence (DA) to cultured epithelial HEp-2 cells, where bacterial adherence occurs over the entire surface of the cell in a scattered pattern (Fig. 7) (86). In a 2005 study of 112 DAEC isolates from diarrheagenic Brazilian patients and controls, 45 total serotypes were found, with 19 of them unique to patients with diarrhea (802).
Epidemiology
Incidence, outbreaks, transmission, and reservoirs.
As mentioned above, detection methods for diarrheagenic strains of DAEC are still being developed, and currently there is no universal method to detect strains in a clinical setting that are responsible for diarrhea. Thus, the epidemiology of diarrheagenic DAEC remains unclear. DAEC isolates have been identified from children with diarrhea in Chile (75), Mexico (803), Australia (804), the United Kingdom (805), Brazil (802), Peru (806), Columbia (807), and many other countries, including the United States. E. coli isolates from 200 children in Brazil who had diarrhea revealed DAEC in 23% of 1,801 isolates, further suggesting it is a widespread pathotype that causes diarrhea (808). However, in some of these studies DAEC was also detected in healthy age-matched controls who did not present with diarrhea at rates similar to those for DAEC detection in ill patients, underlining the importance of developing a method to rapidly and conclusively identify DAEC strains that are causative for diarrhea. It is unknown how DAEC is transmitted or its reservoir.
Pathogenesis
As extensively reviewed by Le Bouguénec and Servin in 2006 and by Servin in 2005, the prototypical strain C1845 is a DAEC strain that encodes Afa/Dr adhesins. The Afa/Dr adhesins are a class of adhesins that includes the AfaE-I, AfaE-II, AfaE-III, AfaE-V, Dr, Dr-II, F1845, and NFA-I adhesins (809, 810). In Afa/Dr DAEC, the Dr and the F1845 adhesins bind to brush border-associated decay-accelerating factor (DAF), a molecule that is highly expressed on the apical surface of polarized epithelial cells. After binding, cytoskeleton rearrangement is induced, destroying or partially rearranging microvilli (809–814) (Fig. 6). Some Afa/Dr DAEC strains also bind the human carcinoembryonic antigen-related cell adhesion molecule (CAECAM) family of receptors in a process that leads to internalization into undifferentiated epithelial cells (815). The internalization mechanism is microfilament independent, microtubule dependent, and lipid raft dependent, and DAEC survival is better for bacteria internalized in the vacuoles of cells expressing DAF (809, 816). In addition to binding DAF and the CAECAM family of receptors, Afa/Dr DAEC has also been shown to bind type IV collagen through the Dr adhesin, an interaction that is important for urinary tract infections caused by Afa/Dr DAEC. However, it remains uncertain if this interaction plays a role in diarrheal disease, as type IV collagen is never present at the apical domain of polarized epithelial cells, the site of Afa/Dr DAEC colonization (810).
After binding DAF, disassembly of F actin and villin results in brush border lesions. This eventually leads to a loss of microvilli due to defective expression of brush border-associated functional intestinal proteins (811, 817). Rearrangement of the tight-junction-associated proteins ZO-1 and occludin after infection by Afa/Dr DAEC strains leads to increased paracellular permeability but does not affect transepithelial electrical resistance (813).
After binding of Afa/Dr DAEC strains with DAF, the proinflammatory cytokine IL-8 is produced through flagellar stimulation of Toll-like receptor 5 (TLR5), resulting in activation of mitogen-activated protein kinase, extracellular signal-regulated kinases 1 and 2 (ERK1/2), P38, and Jun-C kinase (818–822). After IL-8 activation, PMNs migrate across the epithelial barrier, promoting TNF-α and IL-1β production, which results in upregulation of DAF (820). Some have speculated that because DAF is present at increased levels in Crohn's disease (CD) patients, persistent carriage of Afa/Dr DAEC strains by these patients may contribute to inflammation (809, 822, 823). From the use of probes to detect Afa/Dr DAEC, it is known that this pathotype is present in ileal mucosa isolated from patients with CD (809, 824).
While the pathogenesis of typical Afa/Dr DAEC is beginning to be characterized, much remains to be discovered for atypical diarrheagenic DAEC strains. There are two different subclasses of atypical DAEC. One subclass contains all the adhesins typical of the Afa/Dr family of adhesins in another E. coli background, such as diffusely adherent EPEC (810). In the other subclass of atypical DAEC, the bacterium does not bind DAF and expresses a different array of adhesins on its surface, including AfaE-VII, AfaE-VIII, AAF-I, AAF-II, and AAF-III (810). In this subclass of atypical DAEC, IL-8 is still stimulated by DAEC strains that have been internalized by an uncharacterized mechanism, suggesting that it may elicit pathogenesis mechanisms similar to those of typical Afa/Dr DAEC (825).
Clinical Considerations
Symptoms and detection.
DAEC is associated with watery diarrhea that can become persistent in young children, with an increase in severity of disease from the age of 18 months to 5 years (3, 810, 826). It is thought that older adults become asymptomatic carriers, and it has been speculated that DAEC carriage may lead to chronic inflammatory intestinal diseases such as CD (809).
DAEC strain C1845 has surface fimbriae (F1845) that are homologous with members of the Afa/Dr family of adhesins. Therefore, in classical hybridization and subsequent PCR-based screens for detection of DAEC strains, probes and PCR primers were developed to detect daaC, daaE, and afaB and -C, which are common genes in operons that encode the Afa/Dr family of adhesins (827–830). However, in a study of 40 infants under 1 year old who had diarrhea in Brazil, 10% of the adherent E. coli strains detected were diffusely adherent in tissue culture but were not detected with the daaC probe (831), indicating that the absence or presence of these genes cannot be considered conclusive in the identification of all DAEC in clinical screens. Additionally, the daaC probe cross-reacts with a subset of EAEC strains that cause diarrhea, further confounding the conclusive identification of diarrheagenic DAEC in the field (801). More recent studies have shown that in 61 diarrheagenic patients, only 9.8% of the DAEC isolates hybridized with daaE, while 17.6% of healthy controls also contained DAEC strains that hybridized with daaE (802). This study revealed that for the DAEC isolates from patients there was wide array of probes that corresponded with virulence, but no single overall pattern was immediately obvious that could be used to universally detect pathogenic DAEC isolates (802). These probes included sequences for hemolysin (hly), aerobactin siderophore (iucA), yersiniabactin operon (irp2), EAST1 toxin (astA), shET1, cytotoxic necrotizing factor 1 (cnF1), outer membrane protein AIDA-I (a 450-bp EcoRI fragment of pIB6), afimbrial adhesion (afa), adhesion subunit type 1 fimbriae (fimH), aggregative adhesion fimbria type III (agg-3A), P fimbria (pap), and S fimbria (sfa) (802).
Treatment, antibiotic resistance, and vaccines.
Rehydration is currently the only treatment recommended for the watery diarrhea caused by DAEC. Among 112 DAEC strains isolated in Brazilian children, 70% of the strains were resistant to multiple antibiotics, and over 50% were resistant to either ampicillin, cephalothin, co-trimoxazole, streptomycin, sulfonamide, or tetracycline (802). Only 20% of the strains were resistant to chloramphenicol, and all DAEC isolates were susceptible to ceftazidime, gentamicin, lomefloxacin, ofloxacin, and nalidixic acid (802). At the date of this publication, no vaccines for any strain of DAEC exist.
ADHERENT INVASIVE E. COLI
Adherent invasive E. coli (AIEC) has been implicated as one of the causative agents for CD, which is a cause of inflammatory bowel disease (IBD) affecting mainly the small bowel. There is no single causative agent of CD identified, and the current hypothesis is that disease is caused by a combination of factors, including genetics, the intestinal microbiota, environmental factors, and enteric pathogens. A German nationwide study compared CD concordance in twin pairs and showed that 35% of monozygotic pairs were concordant for CD, compared to only 3% of dizygotic pairs (832). Concordance of CD in monozygotic twin pairs provides strong evidence for a genetic predisposition driving disease, while the 65% discordance of disease points to the importance of other environmental factors leading to disease. Current research supports the hypothesis that a breach in the intestinal epithelium, driven by genetic predisposition or largely by as-yet-unidentified environmental factors, occurs, allowing luminal contents and the microbiota to interact with the underlying immune cells of the lamina propria (833). In genetically susceptible individuals (834, 835), this breach is followed by a weak inflammatory response and impaired clearance of the luminal matter, leading to chronic inflammation and development of CD lesions (835).
In addition to microbial dysbiosis and exposure to adverse environmental factors, several enteric pathogens have been implicated as the causative agent of the inflammation in CD patients (836). Pathogens of interest include Mycobacterium avium subsp. paratuberculosis, Campylobacter species, cytomegalovirus, and adherent invasive E. coli (AIEC) (836).
Classification
The AIEC pathotype does not express common virulence factors found in various other pathogenic E. coli strains and the genetic basis for its proinflammatory and invasive phenotype is not fully understood (837). Currently the virulence-like features associated with AIEC are detectable only phenotypically, but recent genomic studies are beginning to define some unique genetic determinants that could be used for pathotype identification in the future (837). AIEC strains have a high variability of O:H serotypes, with the O6 and O22 serogroups being most prevalent (838). AIEC strains cluster within the B2 phylogenetic group and are most closely related to extraintestinal pathogenic E. coli (ExPEC), which is associated with urinary tract infections (referred to as uropathogenic E. coli [UPEC]) and neonatal meningitis (838). However, ExPEC strains rarely share the adhesion, invasion, and intracellular replication traits that identify the AIEC pathotype (839).
Pathoadaptation.
Whole-genome sequencing of LF82, the prototypical strain of the AIEC pathotype, and other clinically isolated AIEC strains has aided in the characterization of a number of acquired genes, gene clusters, and pathoadaptive mutations responsible for the AIEC phenotype (837, 840, 841). The organization of the AIEC genome is similar to that of other pathogenic E. coli strains, with large regions of the core genome interrupted with genomic islands probably acquired by horizontal transfer (841). The presence of these 35 unique genomic islands likely represents the genetic determinants accounting for the AIEC pathotype contributing to bacterial adherence, invasion, and intracellular survival (837, 841). Genome sequencing has aided the identification of four potential virulence determinants: first, a type VI secretion system, which is utilized by Gram-negative bacteria to export proteins across the cell envelope and is thought to support intracellular survival (837, 841); second, several genes encoding adhesins, specifically type I pili, the associated FimH adhesion tip protein, which is important for adhesion to the intestinal epithelium, and long polar fimbriae that play a crucial role in invasion; third, various transcriptional regulators of virulence genes, which require further study by functional genomics to understand their roles in AIEC survival and growth (837); and finally, genes involved in iron acquisition, which has been shown to be an essential virulence trait in ExPEC strains (837). Although the LF82 strain is considered prototypic for the AIEC pathotype, genome sequencing revealed the presence of an additional plasmid resembling the Yersinia pestis pMT1, which is not present in other clinical AIEC isolates (841).
Epidemiology
Several clinical studies have found adhesive and invasive E. coli in over 30% of patients with CD (842–845). In a study including 63 CD patients, AIEC was isolated from approximated 36 to 38% of ileal lesions, whereas it was only found in 6% of 102 healthy individuals (843). Further CD patients have increased serum antibody titers raised against E. coli (846), especially to outer membrane protein C (OmpC) (847). Increased serum reactivity to OmpC correlates with increased disease severity and can be used as an indicator of CD severity and progression in patients (847). With respect to CD, recent epidemiological studies have shown that incidence and prevalence is rising in all ethnic groups of the developed world; however, the highest incidence is seen in North America, northern Europe, and the United Kingdom. It is estimated that 1.4 million persons in the United States and 2.2 million persons in Europe suffer from this debilitating disease (848). Although epidemiological data on AIEC in connection with CD are limited, the available data suggest a correlation between CD occurrence and the presence of AIEC, making this pathotype a relevant health threat and therapeutic target.
Pathogenesis
The AIEC pathotype is defined by the ability to adhere to and invade epithelial cells and replicate within epithelial cells and macrophages (Fig. 6). Interestingly, many of the virulence strategies used by AIEC are facilitated by gene mutations associated with susceptibility to CD. The initial stage of AIEC pathogenesis in the ileum requires adhesion to host cells. AIEC requires flagellum-mediated motility and utilizes surface expression of type 1 pili to adhere to CAECAM6 expressed on host epithelial cells (849, 850). CAECAM6 expression is increased in CD patients, and this could be due to genetic predisposition or through stimulation of TNF-α and IFN-γ production by AIEC colonization, perpetuating disease pathology (850). After adhesion to epithelial cells, AIEC utilizes several virulence factors to facilitate invasion. One such virulence strategy is the production of OMVs, which deliver bacterial effectors to host epithelial cells (851). OMVs deliver effectors by fusing with epithelial cells through the expression of OmpA, a major membrane-bound protein, which binds the host ER stress response chaperone, Gp96 (852). A subset of CD patients have an SNP in the XBP1 gene locus, causing abnormalities in the ER stress response pathway and increased expression of Gp96 in the ileum (852, 853). The increased expression of Gp96 in CD patients promotes AIEC virulence by enhancing Gp96-mediated invasion. In addition to invasion of intestinal epithelial cells, AIEC is also capable of translocating through the epithelium to gain access to the underlying lymphoid tissue, the lamina propria. AIEC expresses long polar fimbriae that interact with M cells, a monolayer of specialized epithelial cells at the surface of Peyer's patches, allowing AIEC to access the lamina propria (305). Once AIEC has gained access to the lamina propria, it can infect and replicate within the phagolysosomes of macrophages without inducing macrophage cell death (854, 855). Continuous replication of AIEC in infected macrophages results in secretion of high levels of TNF-α, causing intestinal inflammation and granuloma formation in CD patients (855). CD can be associated with genetic polymorphisms in autophagy-related genes, such as those for ATG16L1 (autophagy-related like 1) and IRGM (immunity-related GTPase family M). Autophagy plays a major role in the prevention of intracellular bacterial replication. Thus, respective genetic polymorphisms can enhance intracellular replication of AIEC in macrophages and epithelial cells, causing heightened inflammation and pathology (856, 857).
Clinical Considerations
Symptoms.
AIEC has been implicated in inflammatory bowel diseases in humans and animals, including CD in humans and granulomatous colitis in boxer dogs, chickens, and turkeys (858, 859). Human studies have found that AIEC colonizes the intestinal mucosae of CD patients and is associated with both early and chronic lesions, suggesting that AIEC may initiate disease as well as contribute to chronic inflammation (17, 843, 860). CD presents with abdominal pain, fever, and bowel obstruction or diarrhea with the presence of blood and/or mucus (861).
Detection.
In order to identify AIEC strains in intestinal specimens, E. coli strains are screened for their ability to invade epithelial cells and to survive and replicate within macrophages using a standard gentamicin protection assay (843).
Treatment.
AIEC-mediated CD could be treated utilizing strategies involved in the prevention of AIEC replication, adhesion, and invasion. It has been documented that clinical symptoms of CD improve when luminal bacteria concentrations are reduced via intestinal washes or antibacterial drugs, such as antibiotics (862). Although antibiotics have been used extensively in the clinic to treat CD and other IBDs, continued use should be carefully considered due to their drastic effects on the microbial composition and immune status of the intestine (863). Future therapeutic strategies could involve blocking AIEC adhesion. Based on the recent crystal structure of the interaction between type 1 pili and CAECAM6, it would be feasible to utilize natural or engineered mannose antagonists to block the interaction between FimH and CAECAM6, thereby inhibiting colonization (864, 865). Adhesin-based vaccines could also be an effective strategy to block AIEC colonization in the gut, as these vaccines have shown success in blocking UPEC colonization in the mouse bladder (866). The function of type 1 pili, and hence ability of AIEC to colonize the ileum, could also be inhibited by preventing pilus assembly using currently available pilicides (867). Adhesion of AIEC could be competitively inhibited through administration of yeast-based probiotics, as they express cell walls that are rich in free mannose residues, which would act as decoy ligands for type 1 pili (865). Lastly, therapeutic strategies could be developed that target the host ER stress response chaperone, Gp96, or OMV surface-expressed proteins, specifically OmpA, in order to block AIEC invasion of intestinal epithelial cells (852).
CONCLUSIONS
The diversity of E. coli strains is remarkable, ranging from harmless inhabitants of the gastrointestinal tract to diverse pathogens capable of causing intestinal or extraintestinal diseases. While some clinical outcomes are more severe than others, E. coli still remains a major public health concern. It is clear that pathogenic E. coli continues to evolve, as exemplified by the EAEC and STEC hybrid strain that caused a large outbreak throughout Europe in 2011. This also points to the ease of transmissibility of pathogenic E. coli; the low infectious doses of many of the pathotypes and the potential to disseminate among a variety of sources are extraordinary. Food, water, companion pets, animals, and other people are all potential points of contamination and transmission.
New technologies are needed to rapidly identify, characterize, and monitor pathogenic E. coli. It is hoped that the development of rapid pathogen identification strategies will provide clinicians faster diagnosis for quick and appropriate illness management for patients. The depth and wealth of information that can be acquired through high-throughput sequencing will help inform clinical needs (e.g., potential antibiotic resistance and identification of pathogen-specific virulence factors) and epidemiology and help monitor spread of pathogens, as has been nicely reviewed elsewhere (868). An excellent example of the possibility and utility of rapid sequencing occurred during the German STEC O104:H4 outbreak (396), and this was further exemplified by the assembly of a draft genome of STEC O104:H4 using high-throughput sequencing during a retrospective study of stool samples (869). Whole-genome sequences can also provide high-resolution detail for typing and outbreak investigations, as it has been shown that analysis of genome sequences can provide high-resolution typing of outbreak strains (870). The limiting factor for all this information, however, is still meaningful and reliable bioinformatics. While it is very possible to sequence and generate a draft genome of a given pathogen in a day or two, quality assurance is needed for the nucleic acid information. In addition, there are still too many hypothetical genes and genes of unknown function, which hinders our ability to fully understand these pathogens. By generating more genomic data sets, we will undoubtedly uncover more genes (i.e., contributing to the pangenome). The amount of information derived from whole-genome sequences will provide a deeper understanding of the relationships between pathogens and their hosts, how they evolve, and how this ultimately affects human health.
ACKNOWLEDGMENTS
Work in the laboratory of B.B.F. is supported by the Canadian Institutes of Health Research (CIHR) (funding reference no. MOP-13452). B.B.F. is the University of British Columbia Peter Wall Distinguished Professor. R.J.L is supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) postgraduate scholarship. K.M.K. is supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award FAIO85761A. M.W. is supported by a CIHR Frederick Banting and Charles Best Canada graduate scholarship.
Biographies
Matthew Croxen finished his doctoral degree at Dalhousie University, Halifax, Nova Scotia, Canada, in the Department of Microbiology and Immunology under Dr. Paul Hoffman. He then moved to Vancouver, British Columbia, Canada, for postdoctoral training with Professor B. Brett Finlay at the University of British Columbia. With almost 12 years of microbiology and molecular biology experience, he is currently a research associate with the Department of Pathology and Laboratory Medicine at the University of British Columbia and is interested in enteric pathogens.
Robyn Law is a Ph.D. candidate in the lab of Professor B. Brett Finlay at the University of British Columbia in Vancouver, Canada. She obtained a B.Sc. in microbiology at the University of Manitoba and an M.Sc. in the laboratory of Dr. Silvia T. Cardona at the University of Manitoba, Winnipeg, Canada. She has had a long-standing interest in microbial pathogenesis and bacterium-host cell interactions. Her current research is focused on identifying molecular functions of enteropathogenic Escherichia coli type III secreted effectors during host cell infection. She has been awarded a Natural Sciences and Engineering Research Council of Canada Scholarship at both the master's and doctoral levels and currently holds a Four Year Fellowship from the University of British Columbia.
Roland Scholz received his training in biochemistry and microbiology at the University of Regensburg, Regensburg, Germany, the University of Colorado at Boulder, Boulder, CO, USA, and the Max Planck Institute of Biochemistry, Martinsried, Germany. He obtained his Ph.D. training at the Institute of Cell Biology, ETH Zurich, Zurich, Switzerland. Currently he is a postdoctoral trainee in the laboratory of Professor B. Brett Finlay at the University of British Columbia, Vancouver, Canada. With more than 12 years of experience in the fields of biochemistry, cell biology, and microbiology, Dr. Scholz currently studies the molecular mechanisms of host-pathogen interactions, with a focus on the human pathogens enteropathogenic and enterohemorrhagic Escherichia coli. By unraveling the complex molecular interplay between host and pathogen, he aims to provide the basis for novel therapeutic approaches. Dr. Scholz is an author of 13 peer-reviewed scientific publications.
Kristie Keeney has more than 12 years of experience in microbiology and immunology and has eight peer-reviewed scientific publications. Dr. Keeney is a senior postdoctoral trainee in the laboratory of Professor B. Brett Finlay, a world leader in the field of microbiology and bacterial pathogenesis, at the University of British Columbia, Vancouver, Canada. She obtained her B.A. in biochemistry and molecular biology at Reed College in Portland, OR, and her Ph.D. in microbiology and immunology at the University of Michigan, Ann Arbor. In her postdoctoral study, Dr. Keeney is studying the human pathogen enteropathogenic Escherichia coli by using Citrobacter rodentium in a mouse model of infection and is identifying new targets for drug design and probiotic treatment for the prevention of pathogenic colonization. Dr. Keeney has won several awards and is currently a Ruth L. Kirschstein National Research Service Award (NRSA) postdoctoral fellow.
Marta Wlodarska is currently a senior Ph.D. candidate in the laboratory of Professor B. Brett Finlay. She has received several prestigious awards for her doctoral work, including the Canada Graduate Scholarship from the Canadian Institutes of Health Research, a Michael Smith Foundation for Health Research and Crohns & Colitis Foundation Trainee Award, and the Pacific Century Graduate Award and Four Year Fellowship Award from the University of British Columbia. She obtained her B.Sc. also at the University of British Columbia with honors in microbiology and immunology. Currently her research focuses on understanding intestinal mucin dynamics and their implications for gastrointestinal health.
B. Brett Finlay is a Professor in the Michael Smith Laboratories and the Departments of Biochemistry and Molecular Biology and of Microbiology and Immunology at the University of British Columbia. He obtained a B.Sc. (Honors) in biochemistry at the University of Alberta, where he also did his Ph.D. in biochemistry under Dr. William Paranchych. His postdoctoral studies were performed with Dr. Stanley Falkow at the Department of Medical Microbiology and Immunology at Stanford University School of Medicine. In 1989, he joined the University of British Columbia as an Assistant Professor in the Biotechnology Laboratory. Dr. Finlay's research interests are focused on host-pathogen interactions at the molecular level. His laboratory studies several pathogenic bacteria, where interactions of Salmonella and pathogenic E. coli with host cells are the primary focus. Additionally, he is interested in the role of microbiota in enteric and allergic diseases.
REFERENCES
- 1.Escherich T, Bettelheim KS. 1988. The intestinal bacteria of the neonate and breast-fed infant. Rev. Infect. Dis. 10:1220–1225 [DOI] [PubMed] [Google Scholar]
- 2.Cowan ST. 1954. A review of names for coliform organisms. Int. Bull. Bacteriol. Nomencl. Taxon. 4:119–124 [Google Scholar]
- 3.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 4.Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8:26–38 [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization 2012. World health statistics 2012 WHO Press, Geneva, Switzerland [Google Scholar]
- 6.Levine MM, Kotloff KL, Nataro JP, Muhsen K. 2012. The Global Enteric Multicenter Study (GEMS): impetus, rationale, and genesis. Clin. Infect. Dis. 55(Suppl 4):S215–S224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotloff KL, Nataro JP, Blackwelder W, Nasrin D, Farag T. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicentre Study, GEMS): a prospective, case-control study. Lancet 382(9888):209–222 [DOI] [PubMed] [Google Scholar]
- 8.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Rothemund D, Curd H, Reeves PR. 2003. Species-wide variation in the Escherichia coli flagellin (H-antigen) gene. J. Bacteriol. 185:2936–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DebRoy C, Roberts E, Fratamico PM. 2011. Detection of O antigens in Escherichia coli. Anim. Health Res. Rev. 12:169–185 [DOI] [PubMed] [Google Scholar]
- 11.Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noller AC, McEllistrem MC, Pacheco AGF, Boxrud DJ, Harrison LH. 2003. Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 41:5389–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi SY, Jeon Y-S, Lee JH, Choi B, Moon SH, von Seidlein L, Clemens JD, Dougan G, Wain J, Yu J, Lee JC, Seol SY, Lee BK, Song J-H, Song M, Czerkinsky C, Chun J, Kim DW. 2007. Multilocus sequence typing analysis of Shigella flexneri isolates collected in Asian countries. J. Med. Microbiol. 56:1460–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenke C, Lindstedt BA, Harmsen D, Karch H, Brandal LT, Mellmann A. 2011. Comparison of multilocus variable-number tandem-repeat analysis and multilocus sequence typing for differentiation of hemolytic-uremic syndrome-associated Escherichia coli (HUSEC) collection strains. J. Clin. Microbiol. 49:3644–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacher DW, Steinsland H, Blank TE, Donnenberg MS, Whittam TS. 2007. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J. Bacteriol. 189:342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okeke IN, Wallace-Gadsden F, Simons HR, Matthews N, Labar AS, Hwang J, Wain J. 2010. Multi-locus sequence typing of enteroaggregative Escherichia coli isolates from Nigerian children uncovers multiple lineages. PLoS One 5:e14093. 10.1371/journal.pone.0014093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sepehri S, Khafipour E, Bernstein CN, Coombes BK, Pilar AV, Karmali M, Ziebell K, Krause DO. 2011. Characterization of Escherichia coli isolated from gut biopsies of newly diagnosed patients with inflammatory bowel disease. Inflamm. Bowel Dis. 17:1451–1463 [DOI] [PubMed] [Google Scholar]
- 18.Steinsland H, Lacher DW, Sommerfelt H, Whittam TS. 2010. Ancestral lineages of human enterotoxigenic Escherichia coli. J. Clin. Microbiol. 48:2916–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao W, Allen VG, Jamieson FB, Low DE, Alexander DC. 2012. Phylogenetic incongruence in E. coli O104: understanding the evolutionary relationships of emerging pathogens in the face of homologous recombination. PLoS One 7:e33971. 10.1371/journal.pone.0033971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50:1355–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérez-Losada M, Cabezas P, Castro-Nallar E, Crandall KA. 2013. Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 16C:38–53 [DOI] [PubMed] [Google Scholar]
- 23.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne Matic VI, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EPC, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahl JW, Steinsland H, Redman JC, Angiuoli SV, Nataro JP, Sommerfelt H, Rasko DA. 2011. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect. Immun. 79:950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leimbach A, Hacker J, Dobrindt U. 2013. E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr. Top. Microbiol. Immunol. 358:3–32 [DOI] [PubMed] [Google Scholar]
- 26.Kaas RS, Friis C, Ussery DW, Aarestrup FM. 2012. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 13:577. 10.1186/1471-2164-13-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhuri RR, Sebaihia M, Hobman JL, Webber MA, Leyton DL, Goldberg MD, Cunningham AF, Scott-Tucker A, Ferguson PR, Thomas CM, Frankel G, Tang CM, Dudley EG, Roberts IS, Rasko DA, Pallen MJ, Parkhill J, Nataro JP, Thomson NR, Henderson IR. 2010. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS One 5:e8801. 10.1371/journal.pone.0008801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 29.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722–732 [DOI] [PubMed] [Google Scholar]
- 30.Qadri F, Svennerholm A-M, Faruque ASG, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:1664–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207–217 [DOI] [PubMed] [Google Scholar]
- 33.Asadulghani M, Ogura Y, Ooka T, Itoh T, Sawaguchi A, Iguchi A, Nakayama K, Hayashi T. 2009. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5:e1000408. 10.1371/journal.ppat.1000408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11–22 [DOI] [PubMed] [Google Scholar]
- 35.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, Tobe T, Hattori M, Hayashi T. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:17939–17944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hershberg R, Tang H, Petrov DA. 2007. Reduced selection leads to accelerated gene loss in Shigella. Genome Biol. 8:R164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Y, Chen Z, Liu S-L. 2011. Gene decay in Shigella as an incipient stage of host-adaptation. PLoS One 6:e27754. 10.1371/journal.pone.0027754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prosseda G, Di Martino ML, Campilongo R, Fioravanti R, Micheli G, Casalino M, Colonna B. 2012. Shedding of genes that interfere with the pathogenic lifestyle: the Shigella model. Res. Microbiol. 163:399–406 [DOI] [PubMed] [Google Scholar]
- 39.Hwang J, Mattei LM, VanArendonk LG, Meneely PM, Okeke IN. 2010. A pathoadaptive deletion in an enteroaggregative Escherichia coli outbreak strain enhances virulence in a Caenorhabditis elegans model. Infect. Immun. 78:4068–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jores J, Torres AG, Wagner S, Tutt CB, Kaper JB, Wieler LH. 2006. Identification and characterization of “pathoadaptive mutations” of the cadBA operon in several intestinal Escherichia coli. Int. J. Med. Microbiol. 296:547–552 [DOI] [PubMed] [Google Scholar]
- 41.Vazquez-Juarez RC, Kuriakose JA, Rasko DA, Ritchie JM, Kendall MM, Slater TM, Sinha M, Luxon BA, Popov VL, Waldor MK, Sperandio V, Torres AG. 2008. CadA negatively regulates Escherichia coli O157:H7 adherence and intestinal colonization. Infect. Immun. 76:5072–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyoda S, Tamura K, Itoh K, Izumiya H, Ueno N, Nagata K, Togo M, Terajima J, Watanabe H. 2000. Inducible stx2 phages are lysogenized in the enteroaggregative and other phenotypic Escherichia coli O86:HNM isolated from patients. FEMS Microbiol. Lett. 191:7–10 [DOI] [PubMed] [Google Scholar]
- 43.Morabito S, Karch H, Mariani-Kurkdjian P, Schmidt H, Minelli F, Bingen E, Caprioli A. 1998. Enteroaggregative, Shiga toxin-producing Escherichia coli O111:H2 associated with an outbreak of hemolytic-uremic syndrome. J. Clin. Microbiol. 36:840–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Müller L, King LA, Rosner B, Buchholz U, Stark K, Krause G. 2011. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 365:1771–1780 [DOI] [PubMed] [Google Scholar]
- 45.Bielaszewska M, Mellmann A, Zhang W, Köck R, Fruth A, Bauwens A, Peters G, Karch H. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11:671–676 [DOI] [PubMed] [Google Scholar]
- 46.Brzuszkiewicz E, Thürmer A, Schuldes J, Leimbach A, Liesegang H, Meyer F-D, Boelter J, Petersen H, Gottschalk G, Daniel R. 2011. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: entero-aggregative-haemorrhagic Escherichia coli (EAHEC). Arch. Microbiol. 193:883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steyert SR, Sahl JW, Fraser CM, Teel LD, Scheutz F, Rasko DA. 2012. Comparative genomics and stx phage characterization of LEE-negative Shiga toxin-producing Escherichia coli. Front. Cell. Infect. Microbiol. 2:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neter E, Westphal O, Luderitz O, Gino RM, Gorzynski EA. 1955. Demonstration of antibodies against enteropathogenic Escherichia coli in sera of children of various ages. Pediatrics 16:801–808 [PubMed] [Google Scholar]
- 49.Robins-Browne RM. 1987. Traditional enteropathogenic Escherichia coli of infantile diarrhea. Rev. Infect. Dis. 9:28–53 [DOI] [PubMed] [Google Scholar]
- 50.Bray J. 1945. Isolation of antigenically homogenous strains of Bact. coli from summer diarrhoea of infants. J. Pathol. Bacteriol. 57:239–247 [Google Scholar]
- 51.Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64–67 [DOI] [PubMed] [Google Scholar]
- 52.Scaletsky IC, Souza TB, Aranda KRS, Okeke IN. 2010. Genetic elements associated with antimicrobial resistance in enteropathogenic Escherichia coli (EPEC) from Brazil. BMC Microbiol. 10:25. 10.1186/1471-2180-10-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trabulsi LR, Keller R, Tardelli Gomes TA. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ochoa TJ, Contreras CA. 2011. Enteropathogenic Escherichia coli infection in children. Curr. Opin. Infect. Dis. 24:478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whittam TS, Wolfe ML, Wachsmuth IK, Orskov F, Orskov I, Wilson RA. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng P, Lampel KA, Karch H, Whittam TS. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750–1753 [DOI] [PubMed] [Google Scholar]
- 57.Zhou Z, Li X, Liu B, Beutin L, Xu J, Ren Y, Feng L, Lan R, Reeves PR, Wang L. 2010. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLoS One 5:e8700. 10.1371/journal.pone.0008700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hernandes RT, Elias WP, Vieira MAM, Gomes TAT. 2009. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol. Lett. 297:137–149 [DOI] [PubMed] [Google Scholar]
- 59.Bortolini MR, Trabulsi LR, Keller R, Frankel G, Sperandio V. 1999. Lack of expression of bundle-forming pili in some clinical isolates of enteropathogenic Escherichia coli (EPEC) is due to a conserved large deletion in the bfp operon. FEMS Microbiol. Lett. 179:169–174 [DOI] [PubMed] [Google Scholar]
- 60.Okeke IN, Borneman JA, Shin S, Mellies JL, Quinn LE, Kaper JB. 2001. Comparative sequence analysis of the plasmid-encoded regulator of enteropathogenic Escherichia coli strains. Infect. Immun. 69:5553–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ochoa TJ, Barletta F, Contreras C, Mercado E. 2008. New insights into the epidemiology of enteropathogenic Escherichia coli infection. Trans. R. Soc. Trop. Med. Hyg. 102:852–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maranhão HS, Medeiros MCC, Scaletsky ICA, Fagundes-Neto U, Morais MB. 2008. The epidemiological and clinical characteristics and nutritional development of infants with acute diarrhoea, in north-eastern Brazil. Ann. Trop. Med. Parasitol. 102:357–365 [DOI] [PubMed] [Google Scholar]
- 63.Centers for Disease Control and Prevention (CDC) 2013. Surveillance for foodborne disease outbreaks—United States, 2009-2010. MMWR Morb. Mortal. Wkly. Rep. 62:41–47 [PMC free article] [PubMed] [Google Scholar]
- 64.Afset JE, Bevanger L, Romundstad P, Bergh K. 2004. Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J. Med. Microbiol. 53:1137–1144 [DOI] [PubMed] [Google Scholar]
- 65.Ratchtrachenchai O-A, Subpasu S, Hayashi H, Ba-Thein W. 2004. Prevalence of childhood diarrhoea-associated Escherichia coli in Thailand. J. Med. Microbiol. 53:237–243 [DOI] [PubMed] [Google Scholar]
- 66.Franzolin MR, Alves RCB, Keller R, Gomes TAT, Beutin L, Barreto ML, Milroy C, Strina A, Ribeiro H, Trabulsi LR. 2005. Prevalence of diarrheagenic Escherichia coli in children with diarrhea in Salvador, Bahia, Brazil. Mem. Inst. Oswaldo Cruz 100:359–363 [DOI] [PubMed] [Google Scholar]
- 67.Bakhshi B, Fallahzad S, Pourshafie MR. 2013. The occurrence of atypical enteropathogenic Escherichia coli strains among children with diarrhea in Iran. J. Infect. Chemother 19:615–620 [DOI] [PubMed] [Google Scholar]
- 68.Afset JE, Bergh K, Bevanger L. 2003. High prevalence of atypical enteropathogenic Escherichia coli (EPEC) in Norwegian children with diarrhoea. J. Med. Microbiol. 52:1015–1019 [DOI] [PubMed] [Google Scholar]
- 69.Nguyen RN, Taylor LS, Tauschek M, Robins-Browne RM. 2006. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg. Infect. Dis. 12:597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alikhani MY, Mirsalehian A, Aslani MM. 2006. Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J. Med. Microbiol. 55:1159–1163 [DOI] [PubMed] [Google Scholar]
- 71.Levine MM, Edelman R. 1984. Enteropathogenic Escherichia coli of classic serotypes associated with infant diarrhea: epidemiology and pathogenesis. Epidemiol. Rev. 6:31–51 [DOI] [PubMed] [Google Scholar]
- 72.Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, Schoolnik GK. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114–2118 [DOI] [PubMed] [Google Scholar]
- 73.Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119–1122 [DOI] [PubMed] [Google Scholar]
- 74.Behiry IK, Abada EA, Ahmed EA, Labeeb RS. 2011. Enteropathogenic Escherichia coli associated with diarrhea in children in Cairo, Egypt. Sci. World J. 11:2613–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levine MM, Ferreccio C, Prado V, Cayazzo M, Abrego P, Martinez J, Maggi L, Baldini MM, Martin W, Maneval D. 1993. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am. J. Epidemiol. 138:849–869 [DOI] [PubMed] [Google Scholar]
- 76.Sekse C, Sunde M, Lindstedt B-A, Hopp P, Bruheim T, Cudjoe KS, Kvitle B, Urdahl AM. 2011. Potentially human-pathogenic Escherichia coli O26 in Norwegian sheep flocks. Appl. Environ. Microbiol. 77:4949–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moura RA, Sircili MP, Leomil L, Matté MH, Trabulsi LR, Elias WP, Irino K, Pestana de Castro AF. 2009. Clonal relationship among atypical enteropathogenic Escherichia coli strains isolated from different animal species and humans. Appl. Environ. Microbiol. 75:7399–7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Swennes AG, Buckley EM, Parry NMA, Madden CM, García A, Morgan PB, Astrofsky KM, Fox JG. 2012. Enzootic enteropathogenic Escherichia coli infection in laboratory rabbits. J. Clin. Microbiol. 50:2353–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goosney DL, Gruenheid S, Finlay BB. 2000. Gut feelings: enteropathogenic E. coli (EPEC) interactions with the host. Annu. Rev. Cell Dev. Biol. 16:173–189 [DOI] [PubMed] [Google Scholar]
- 80.Moon HW, Whipp SC, Argenzio RA, Levine MM, Giannella RA. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rosenshine I, Ruschkowski S, Stein M, Reinscheid DJ, Mills SD, Finlay BB. 1996. A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 15:2613–2624 [PMC free article] [PubMed] [Google Scholar]
- 82.Kenny B. 2002. Mechanism of action of EPEC type III effector molecules. Int. J. Med. Microbiol. 291:469–477 [DOI] [PubMed] [Google Scholar]
- 83.Donnenberg MS, Kaper JB. 1992. Enteropathogenic Escherichia coli. Infect. Immun. 60:3953–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dean P, Kenny B. 2009. The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr. Opin. Microbiol. 12:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Girón JA, Ho AS, Schoolnik GK. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710–713 [DOI] [PubMed] [Google Scholar]
- 86.Scaletsky IC, Silva ML, Trabulsi LR. 1984. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 45:534–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baldini MM, Kaper JB, Levine MM, Candy DC, Moon HW. 1983. Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J. Pediatr. Gastroenterol. Nutr. 2:534–538 [DOI] [PubMed] [Google Scholar]
- 88.Brinkley C, Burland V, Keller R, Rose DJ, Boutin AT, Klink SA, Blattner FR, Kaper JB. 2006. Nucleotide sequence analysis of the enteropathogenic Escherichia coli adherence factor plasmid pMAR7. Infect. Immun. 74:5408–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tobe T, Hayashi T, Han CG, Schoolnik GK, Ohtsubo E, Sasakawa C. 1999. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect. Immun. 67:5455–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mellies JL, Barron AMS, Carmona AM. 2007. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect. Immun. 75:4199–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rodrigues J, Scaletsky IC, Campos LC, Gomes TA, Whittam TS, Trabulsi LR. 1996. Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect. Immun. 64:2680–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Badea L, Doughty S, Nicholls L, Sloan J, Robins-Browne RM, Hartland EL. 2003. Contribution of Efa1/LifA to the adherence of enteropathogenic Escherichia coli to epithelial cells. Microb. Pathog. 34:205–215 [DOI] [PubMed] [Google Scholar]
- 93.Klapproth J-MA, Sasaki M, Sherman M, Babbin B, Donnenberg MS, Fernandes PJ, Scaletsky ICA, Kalman D, Nusrat A, Williams IR. 2005. Citrobacter rodentium lifA/efa1 is essential for colonic colonization and crypt cell hyperplasia in vivo. Infect. Immun. 73:1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Deng W, Yu HB, de Hoog CL, Stoynov N, Li Y, Foster LJ, Finlay BB. 2012. Quantitative proteomic analysis of type III secretome of enteropathogenic Escherichia coli reveals an expanded effector repertoire for attaching/effacing bacterial pathogens. Mol. Cell. Proteomics 11:692–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vieira MAM, Salvador FA, Silva RM, Irino K, Vaz TMI, Rockstroh AC, Guth BEC, Gomes TAT. 2010. Prevalence and characteristics of the O122 pathogenicity island in typical and atypical enteropathogenic Escherichia coli strains. J. Clin. Microbiol. 48:1452–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Afset JE, Bruant G, Brousseau R, Harel J, Anderssen E, Bevanger L, Bergh K. 2006. Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. J. Clin. Microbiol. 44:3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rendón MA, Saldaña Z, Erdem AL, Monteiro-Neto V, Vázquez A, Kaper JB, Puente JL, Girón JA. 2007. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. U. S. A. 104:10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saldaña Z, Erdem AL, Schüller S, Okeke IN, Lucas M, Sivananthan A, Phillips AD, Kaper JB, Puente JL, Girón JA. 2009. The Escherichia coli common pilus and the bundle-forming pilus act in concert during the formation of localized adherence by enteropathogenic E. coli. J. Bacteriol. 191:3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cleary J, Lai L-C, Shaw RK, Straatman-Iwanowska A, Donnenberg MS, Frankel G, Knutton S. 2004. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology. 150:527–538 [DOI] [PubMed] [Google Scholar]
- 100.Gruenheid S, DeVinney R, Bladt F, Goosney D, Gelkop S, Gish GD, Pawson T, Finlay BB. 2001. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3:856–859 [DOI] [PubMed] [Google Scholar]
- 101.Kalman D, Weiner OD, Goosney DL, Sedat JW, Finlay BB, Abo A, Bishop JM. 1999. Enteropathogenic E. coli acts through WASP and Arp2/3 complex to form actin pedestals. Nat. Cell Biol. 1:389–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520 [DOI] [PubMed] [Google Scholar]
- 103.Frankel G, Candy DC, Everest P, Dougan G. 1994. Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii, and Hafnia alvei. Infect. Immun. 62:1835–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tardelli Gomes TA, Gonzalez-Pedrajo B. 2010. Enteropathogenic Escherichia coli (EPEC), p 66–126 In Torres AG. (ed), Pathogenic Escherichia coli in Latin America. Benthem Science Publishers, Oak Park, IL [Google Scholar]
- 105.Schmidt MA. 2010. LEEways: tales of EPEC, ATEC and EHEC. Cell. Microbiol. 12:1544–1552 [DOI] [PubMed] [Google Scholar]
- 106.Müller D, Benz I, Liebchen A, Gallitz I, Karch H, Schmidt MA. 2009. Comparative analysis of the locus of enterocyte effacement and its flanking regions. Infect. Immun. 77:3501–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vázquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 101:3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Frankel G, Phillips AD, Rosenshine I, Dougan G, Kaper JB, Knutton S. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911–921 [DOI] [PubMed] [Google Scholar]
- 109.Iguchi A, Thomson NR, Ogura Y, Saunders D, Ooka T, Henderson IR, Harris D, Asadulghani M, Kurokawa K, Dean P, Kenny B, Quail MA, Thurston S, Dougan G, Hayashi T, Parkhill J, Frankel G. 2009. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191:347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Petty NK, Bulgin R, Crepin VF, Cerdeño-Tárraga AM, Schroeder GN, Quail MA, Lennard N, Corton C, Barron A, Clark L, Toribio AL, Parkhill J, Dougan G, Frankel G, Thomson NR. 2010. The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J. Bacteriol. 192:525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ruchaud-Sparagano M-H, Mühlen S, Dean P, Kenny B. 2011. The enteropathogenic E. coli (EPEC) Tir effector inhibits NF-κB activity by targeting TNFα receptor-associated factors. PLoS Pathog. 7:e1002414. 10.1371/journal.ppat.1002414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ma C, Wickham ME, Guttman JA, Deng W, Walker J, Madsen KL, Jacobson K, Vogl WA, Finlay BB, Vallance BA. 2006. Citrobacter rodentium infection causes both mitochondrial dysfunction and intestinal epithelial barrier disruption in vivo: role of mitochondrial associated protein (Map). Cell. Microbiol. 8:1669–1686 [DOI] [PubMed] [Google Scholar]
- 113.Guttman JA, Samji FN, Li Y, Deng W, Lin A, Finlay BB. 2007. Aquaporins contribute to diarrhoea caused by attaching and effacing bacterial pathogens. Cell. Microbiol. 9:131–141 [DOI] [PubMed] [Google Scholar]
- 114.Nougayrède J-P, Donnenberg MS. 2004. Enteropathogenic Escherichia coli EspF is targeted to mitochondria and is required to initiate the mitochondrial death pathway. Cell. Microbiol. 6:1097–1111 [DOI] [PubMed] [Google Scholar]
- 115.Guttman JA, Li Y, Wickham ME, Deng W, Vogl AW, Finlay BB. 2006. Attaching and effacing pathogen-induced tight junction disruption in vivo. Cell. Microbiol. 8:634–645 [DOI] [PubMed] [Google Scholar]
- 116.Hardwidge PR, Deng W, Vallance BA, Rodriguez-Escudero I, Cid VJ, Molina M, Finlay BB. 2005. Modulation of host cytoskeleton function by the enteropathogenic Escherichia coli and Citrobacter rodentium effector protein EspG. Infect. Immun. 73:2586–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shames SR, Deng W, Guttman JA, de Hoog CL, Li Y, Hardwidge PR, Sham HP, Vallance BA, Foster LJ, Finlay BB. 2010. The pathogenic E. coli type III effector EspZ interacts with host CD98 and facilitates host cell prosurvival signalling. Cell. Microbiol. 12:1322–1339 [DOI] [PubMed] [Google Scholar]
- 118.Wong ARC, Raymond B, Collins JW, Crepin VF, Frankel G. 2012. The enteropathogenic E. coli effector EspH promotes actin pedestal formation and elongation via WASP-interacting protein (WIP). Cell. Microbiol. 14:1051–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wong ARC, Clements A, Raymond B, Crepin VF, Frankel G. 2012. The interplay between the Escherichia coli Rho guanine nucleotide exchange factor effectors and the mammalian RhoGEF inhibitor EspH. mBio 3(1):e00250–11. 10.1128/mBio.00250-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dong N, Liu L, Shao F. 2010. A bacterial effector targets host DH-PH domain RhoGEFs and antagonizes macrophage phagocytosis. EMBO J. 29:1363–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Iizumi Y, Sagara H, Kabe Y, Azuma M, Kume K, Ogawa M, Nagai T, Gillespie PG, Sasakawa C, Handa H. 2007. The enteropathogenic E. coli effector EspB facilitates microvillus effacing and antiphagocytosis by inhibiting myosin function. Cell Host Microbe 2:383–392 [DOI] [PubMed] [Google Scholar]
- 122.Baruch K, Gur-Arie L, Nadler C, Koby S, Yerushalmi G, Ben-Neriah Y, Yogev O, Shaulian E, Guttman C, Zarivach R, Rosenshine I. 2011. Metalloprotease type III effectors that specifically cleave JNK and NF-κB. EMBO J. 30:221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gao X, Wan F, Mateo K, Callegari E, Wang D, Deng W, Puente J, Li F, Chaussee MS, Finlay BB, Lenardo MJ, Hardwidge PR. 2009. Bacterial effector binding to ribosomal protein s3 subverts NF-κB function. PLoS Pathog. 5:e1000708. 10.1371/journal.ppat.1000708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gao X, Wang X, Pham TH, Feuerbacher LA, Lubos M-L, Huang M, Olsen R, Mushegian A, Slawson C, Hardwidge PR. 2013. NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-κB activation. Cell Host Microbe 13:87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nadler C, Baruch K, Kobi S, Mills E, Haviv G, Farago M, Alkalay I, Bartfeld S, Meyer TF, Ben-Neriah Y, Rosenshine I. 2010. The type III secretion effector NleE inhibits NF-κB activation. PLoS Pathog. 6:e1000743. 10.1371/journal.ppat.1000743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Newton HJ, Pearson JS, Badea L, Kelly M, Lucas M, Holloway G, Wagstaff KM, Dunstone MA, Sloan J, Whisstock JC, Kaper JB, Robins-Browne RM, Jans DA, Frankel G, Phillips AD, Coulson BS, Hartland EL. 2010. The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-κB p65. PLoS Pathog. 6:e1000898. 10.1371/journal.ppat.1000898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pearson JS, Riedmaier P, Marchès O, Frankel G, Hartland EL. 2011. A type III effector protease NleC from enteropathogenic Escherichia coli targets NF-κB for degradation. Mol. Microbiol. 80:219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sham HP, Shames SR, Croxen MA, Ma C, Chan JM, Khan MA, Wickham ME, Deng W, Finlay BB, Vallance BA. 2011. Attaching and effacing bacterial effector NleC suppresses epithelial inflammatory responses by inhibiting NF-κB and p38 mitogen-activated protein kinase activation. Infect. Immun. 79:3552–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yen H, Ooka T, Iguchi A, Hayashi T, Sugimoto N, Tobe T. 2010. NleC, a type III secretion protease, compromises NF-κB activation by targeting p65/RelA. PLoS Pathog. 6:e1001231. 10.1371/journal.ppat.1001231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marchès O, Covarelli V, Dahan S, Cougoule C, Bhatta P, Frankel G, Caron E. 2008. EspJ of enteropathogenic and enterohaemorrhagic Escherichia coli inhibits opsono-phagocytosis. Cell. Microbiol. 10:1104–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim J, Thanabalasuriar A, Chaworth-Musters T, Fromme JC, Frey EA, Lario PI, Metalnikov P, Rizg K, Thomas NA, Lee SF, Hartland EL, Hardwidge PR, Pawson T, Strynadka NC, Finlay BB, Schekman R, Gruenheid S. 2007. The bacterial virulence factor NleA inhibits cellular protein secretion by disrupting mammalian COPII function. Cell Host Microbe 2:160–171 [DOI] [PubMed] [Google Scholar]
- 132.Thanabalasuriar A, Koutsouris A, Weflen A, Mimee M, Hecht G, Gruenheid S. 2010. The bacterial virulence factor NleA is required for the disruption of intestinal tight junctions by enteropathogenic Escherichia coli. Cell. Microbiol. 12:31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thanabalasuriar A, Bergeron J, Gillingham A, Mimee M, Thomassin J-L, Strynadka N, Kim J, Gruenheid S. 2012. Sec24 interaction is essential for localization and virulence-associated function of the bacterial effector protein NleA. Cell. Microbiol. 14:1206–1218 [DOI] [PubMed] [Google Scholar]
- 134.Hemrajani C, Berger CN, Robinson KS, Marchès O, Mousnier A, Frankel G. 2010. NleH effectors interact with Bax inhibitor-1 to block apoptosis during enteropathogenic Escherichia coli infection. Proc. Natl. Acad. Sci. U. S. A. 107:3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wong ARC, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. 2011. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80:1420–1438 [DOI] [PubMed] [Google Scholar]
- 136.Navarro-Garcia F, Canizalez-Roman A, Sui BQ, Nataro JP, Azamar Y. 2004. The serine protease motif of EspC from enteropathogenic Escherichia coli produces epithelial damage by a mechanism different from that of Pet toxin from enteroaggregative E. coli. Infect. Immun. 72:3609–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mellies JL, Navarro-Garcia F, Okeke I, Frederickson J, Nataro JP, Kaper JB. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vidal JE, Navarro-García F. 2008. EspC translocation into epithelial cells by enteropathogenic Escherichia coli requires a concerted participation of type V and III secretion systems. Cell. Microbiol. 10:1975–1986 [DOI] [PubMed] [Google Scholar]
- 139.Vidal JE, Navarro-García F. 2006. Efficient translocation of EspC into epithelial cells depends on enteropathogenic Escherichia coli and host cell contact. Infect. Immun. 74:2293–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Salinger N, Kokona B, Fairman R, Okeke IN. 2009. The plasmid-encoded regulator activates factors conferring lysozyme resistance on enteropathogenic Escherichia coli strains. Appl. Environ. Microbiol. 75:275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Drago-Serrano ME, Parra SG, Manjarrez-Hernández HA. 2006. EspC, an autotransporter protein secreted by enteropathogenic Escherichia coli (EPEC), displays protease activity on human hemoglobin. FEMS Microbiol. Lett. 265:35–40 [DOI] [PubMed] [Google Scholar]
- 142.Dutta PR, Cappello R, Navarro-Garcia F, Nataro JP. 2002. Functional comparison of serine protease autotransporters of enterobacteriaceae. Infect. Immun. 70:7105–7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xicohtencatl-Cortes J, Saldaña Z, Deng W, Castañeda E, Freer E, Tarr PI, Finlay BB, Puente JL, Girón JA. 2010. Bacterial macroscopic rope-like fibers with cytopathic and adhesive properties. J. Biol. Chem. 285:32336–32342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dulguer MV, Fabbricotti SH, Bando SY, Moreira-Filho CA, Fagundes-Neto U, Scaletsky ICA. 2003. Atypical enteropathogenic Escherichia coli strains: phenotypic and genetic profiling reveals a strong association between enteroaggregative E. coli heat-stable enterotoxin and diarrhea. J. Infect. Dis. 188:1685–1694 [DOI] [PubMed] [Google Scholar]
- 145.Hedberg CW, Savarino SJ, Besser JM, Paulus CJ, Thelen VM, Myers LJ, Cameron DN, Barrett TJ, Kaper JB, Osterholm MT. 1997. An outbreak of foodborne illness caused by Escherichia coli O39:NM, an agent not fitting into the existing scheme for classifying diarrheogenic E. coli. J. Infect. Dis. 176:1625–1628 [DOI] [PubMed] [Google Scholar]
- 146.Viljanen MK, Peltola T, Junnila SY, Olkkonen L, Järvinen Kuistila HM, Huovinen P. 1990. Outbreak of diarrhoea due to Escherichia coli O111:B4 in schoolchildren and adults: association of Vi antigen-like reactivity. Lancet 336:831–834 [DOI] [PubMed] [Google Scholar]
- 147.Yatsuyanagi J, Saito S, Miyajima Y, Amano K-I, Enomoto K. 2003. Characterization of atypical enteropathogenic Escherichia coli strains harboring the astA gene that were associated with a waterborne outbreak of diarrhea in Japan. J. Clin. Microbiol. 41:2033–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Savarino SJ, McVeigh A, Watson J, Cravioto A, Molina J, Echeverria P, Bhan MK, Levine MM, Fasano A. 1996. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J. Infect. Dis. 173:1019–1022 [DOI] [PubMed] [Google Scholar]
- 149.Celli J, Olivier M, Finlay BB. 2001. Enteropathogenic Escherichia coli mediates antiphagocytosis through the inhibition of PI 3-kinase-dependent pathways. EMBO J. 20:1245–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ruchaud-Sparagano M-H, Maresca M, Kenny B. 2007. Enteropathogenic Escherichia coli (EPEC) inactivate innate immune responses prior to compromising epithelial barrier function. Cell. Microbiol. 9:1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dean P, Maresca M, Schüller S, Phillips AD, Kenny B. 2006. Potent diarrheagenic mechanism mediated by the cooperative action of three enteropathogenic Escherichia coli-injected effector proteins. Proc. Natl. Acad. Sci. U. S. A. 103:1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gill RK, Borthakur A, Hodges K, Turner JR, Clayburgh DR, Saksena S, Zaheer A, Ramaswamy K, Hecht G, Dudeja PK. 2007. Mechanism underlying inhibition of intestinal apical Cl/OH exchange following infection with enteropathogenic E. coli. J. Clin. Invest. 117:428–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Viswanathan VK, Koutsouris A, Lukic S, Pilkinton M, Simonovic I, Simonovic M, Hecht G. 2004. Comparative analysis of EspF from enteropathogenic and enterohemorrhagic Escherichia coli in alteration of epithelial barrier function. Infect. Immun. 72:3218–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Matsuzawa T, Kuwae A, Abe A. 2005. Enteropathogenic Escherichia coli type III effectors EspG and EspG2 alter epithelial paracellular permeability. Infect. Immun. 73:6283–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dean P, Kenny B. 2004. Intestinal barrier dysfunction by enteropathogenic Escherichia coli is mediated by two effector molecules and a bacterial surface protein. Mol. Microbiol. 54:665–675 [DOI] [PubMed] [Google Scholar]
- 156.Donnenberg MS, Tacket CO, James SP, Losonsky G, Nataro JP, Wasserman SS, Kaper JB, Levine MM. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J. Clin. Invest. 92:1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Knutton S, Baldwin T, Williams PH, McNeish AS. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Knutton S, Phillips AD, Smith HR, Gross RJ, Shaw R, Watson P, Price E. 1991. Screening for enteropathogenic Escherichia coli in infants with diarrhea by the fluorescent-actin staining test. Infect. Immun. 59:365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shariff M, Bhan MK, Knutton S, Das BK, Saini S, Kumar R. 1993. Evaluation of the fluorescence actin staining test for detection of enteropathogenic Escherichia coli. J. Clin. Microbiol. 31:386–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cravioto A, Gross RJ, Scotland SM, Rowe B. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr. Microbiol. 3:95–99 [Google Scholar]
- 161.Donnenberg MS, Nataro JP. 1995. Methods for studying adhesion of diarrheagenic Escherichia coli. Methods Enzymol. 253:324–336 [DOI] [PubMed] [Google Scholar]
- 162.Subramanian K, Selvakkumar C, Vinaykumar KS, Goswami N, Meenakshisundaram S, Balakrishnan A, Lakshmi BS. 2009. Tackling multiple antibiotic resistance in enteropathogenic Escherichia coli (EPEC) clinical isolates: a diarylheptanoid from Alpinia officinarum shows promising antibacterial and immunomodulatory activity against EPEC and its lipopolysaccharide-induced inflammation. Int. J. Antimicrob. Agents 33:244–250 [DOI] [PubMed] [Google Scholar]
- 163.Manning SD. 2010. Escherichia coli infections, 2nd ed, p 72–84 Infobase Publishing, New York, NY [Google Scholar]
- 164.Moyenuddin M, Wachsmuth IK, Moseley SL, Bopp CA, Blake PA. 1989. Serotype, antimicrobial resistance, and adherence properties of Escherichia coli strains associated with outbreaks of diarrheal illness in children in the United States. J. Clin. Microbiol. 27:2234–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gross RJ, Ward LR, Threlfall EJ, King H, Rowe B. 1982. Drug resistance among infantile enteropathogenic Escherichia coli strains isolated in the United Kingdom. Br. Med. J. Clin. Res. Ed. 285:472–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gross RJ, Rowe B, Threlfall EJ. 1985. Escherichia coli 0142.H6; a drug-resistant enteropathogenic clone? J. Hyg. (Lond.) 94:181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jenkins C, Smith HR, Lawson AJ, Willshaw GA, Cheasty T, Wheeler JG, Tompkins DS. 2006. Serotypes, intimin subtypes, and antimicrobial resistance patterns of atypical enteropathogenic Escherichia coli isolated in England from 1993 to 1996. Eur. J. Clin. Microbiol. Infect. Dis. 25:19–24 [DOI] [PubMed] [Google Scholar]
- 168.Mitra M, Ahmad P, Mehdi R, Hosein A, Ahmad K. 2011. Multiple drug resistance of enteropathogenic Escherichia coli isolated from children with diarrhea in Kashan, Iran. Afr. J. Microbiol. Res. 5:3305–3309 [Google Scholar]
- 169.Lim YS, Ngan CC, Tay L. 1992. Enteropathogenic Escherichia coli as a cause of diarrhoea among children in Singapore. J. Trop. Med. Hyg. 95:339–342 [PubMed] [Google Scholar]
- 170.Costa-Carvalho BT, Bertipaglia A, Solé D, Naspitz CK, Scaletsky IC. 1994. Detection of immunoglobulin (IgG and IgA) anti-outer-membrane proteins of enteropathogenic Escherichia coli (EPEC) in saliva, colostrum, breast milk, serum, cord blood and amniotic fluid. Study of inhibition of localized adherence of EPEC to HeLa cells. Acta Paediatr. 83:870–873 [DOI] [PubMed] [Google Scholar]
- 171.Cravioto A, Tello A, Villafán H, Ruiz J, del Vedovo S, Neeser JR. 1991. Inhibition of localized adhesion of enteropathogenic Escherichia coli to HEp-2 cells by immunoglobulin and oligosaccharide fractions of human colostrum and breast milk. J. Infect. Dis. 163:1247–1255 [DOI] [PubMed] [Google Scholar]
- 172.Carbonare SB, Silva ML, Palmeira P, Carneiro-Sampaio MM. 1997. Human colostrum IgA antibodies reacting to enteropathogenic Escherichia coli antigens and their persistence in the faeces of a breastfed infant. J. Diarrhoeal Dis. Res. 15:53–58 [PubMed] [Google Scholar]
- 173.Loureiro I, Frankel G, Adu-Bobie J, Dougan G, Trabulsi LR, Carneiro-Sampaio MM. 1998. Human colostrum contains IgA antibodies reactive to enteropathogenic Escherichia coli virulence-associated proteins: intimin, BfpA, EspA, and EspB. J. Pediatr. Gastroenterol. Nutr. 27:166–171 [DOI] [PubMed] [Google Scholar]
- 174.Parissi-Crivelli A, Parissi-Crivelli JM, Girón JA. 2000. Recognition of enteropathogenic Escherichia coli virulence determinants by human colostrum and serum antibodies. J. Clin. Microbiol. 38:2696–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Quintana Flores VM, Campos de Souza Fernandes RC, Sousa de Macedo Z, Medina-Acosta E. 2002. Expression and purification of the recombinant enteropathogenic Escherichia coli vaccine candidates BfpA and EspB. Protein Expr. Purif. 25:16–22 [DOI] [PubMed] [Google Scholar]
- 176.De Souza Campos Fernandes RC, Quintana Flores VM, Sousa de Macedo Z, Medina-Acosta E. 2003. Coproantibodies to the enteropathogenic Escherichia coli vaccine candidates BfpA and EspB in breastfed and artificially fed children. Vaccine 21:1725–1731 [DOI] [PubMed] [Google Scholar]
- 177.Schriefer A, Maltez JR, Silva N, Stoeckle MY, Barral-Netto M, Riley LW. 1999. Expression of a pilin subunit BfpA of the bundle-forming pilus of enteropathogenic Escherichia coli in an aroA live Salmonella vaccine strain. Vaccine 17:770–778 [DOI] [PubMed] [Google Scholar]
- 178.Stakenborg T, Vandekerchove D, Mariën J, Laevens H, Imberechts H, Peeters J. 2006. Protection of rabbits against enteropathogenic Escherichia coli (EPEC) using an intimin null mutant. BMC Vet. Res. 2:22. 10.1186/1746-6148-2-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tacket CO, Sztein MB, Losonsky G, Abe A, Finlay BB, McNamara BP, Fantry GT, James SP, Nataro JP, Levine MM, Donnenberg MS. 2000. Role of EspB in experimental human enteropathogenic Escherichia coli infection. Infect. Immun. 68:3689–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Liu J, Wang WD, Liu YJ, Liu S, Zhou B, Zhu LW, Ji X, Sun Y, Feng SZ. 2012. Mice vaccinated with enteropathogenic Escherichia coli ghosts show significant protection against lethal challenges. Lett. Appl. Microbiol. 54:255–262 [DOI] [PubMed] [Google Scholar]
- 181.Donnenberg MS, Tacket CO, Losonsky G, Frankel G, Nataro JP, Dougan G, Levine MM. 1998. Effect of prior experimental human enteropathogenic Escherichia coli infection on illness following homologous and heterologous rechallenge. Infect. Immun. 66:52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Blanco M, Blanco JE, Mora A, Dahbi G, Alonso MP, González EA, Bernárdez MI, Blanco J. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-ξ). J. Clin. Microbiol. 42:645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Levine MM. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J. Infect. Dis. 155:377–389 [DOI] [PubMed] [Google Scholar]
- 184.Anonymous 2012. Shiga-toxn producing Escherichia coli in certain raw beef products. Fed. Regist. 77:31975–31981 [Google Scholar]
- 185.Johnson KE, Thorpe CM, Sears CL. 2006. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 43:1587–1595 [DOI] [PubMed] [Google Scholar]
- 186.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 192:1422–1429 [DOI] [PubMed] [Google Scholar]
- 187.Vally H, Hall G, Dyda A, Raupach J, Knope K, Combs B, Desmarchelier P. 2012. Epidemiology of Shiga toxin producing Escherichia coli in Australia, 2000-2010. BMC Public Health 12:63. 10.1186/1471-2458-12-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bielaszewska M, Köck R, Friedrich AW, von Eiff C, Zimmerhackl LB, Karch H, Mellmann A. 2007. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS One 2:e1024. 10.1371/journal.pone.0001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yang Z, Kovar J, Kim J, Nietfeldt J, Smith DR, Moxley RA, Olson ME, Fey PD, Benson AK. 2004. Identification of common subpopulations of non-sorbitol-fermenting, β-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70:6846–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, Mladonicky JM, Somsel P, Rudrik JT, Dietrich SE, Zhang W, Swaminathan B, Alland D, Whittam TS. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Allos BM, Moore MR, Griffin PM, Tauxe RV. 2004. Surveillance for sporadic foodborne disease in the 21st century: the FoodNet perspective. Clin. Infect. Dis. 38(Suppl 3):S115–S120 [DOI] [PubMed] [Google Scholar]
- 192.Scallan E. 2007. Activities, achievements, and lessons learned during the first 10 years of the Foodborne Diseases Active Surveillance Network: 1996-2005. Clin. Infect. Dis. 44:718–725 [DOI] [PubMed] [Google Scholar]
- 193.CDC 2012. Foodborne disease active surveillance network (FoodNet): FoodNet surveillance report for 2011 (final report). CDC, Atlanta, GA [Google Scholar]
- 194.Stigi KA, Macdonald JK, Tellez-Marfin AA, Lofy KH. 2012. Laboratory practices and incidence of non-O157 Shiga toxin-producing Escherichia coli infections. Emerg. Infect. Dis. 18:477–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.National Enteric Surveillance Program 2010. National Enteric Surveillance Program (NESP) annual summary 2010. The National Microbiology Laboratory (NML) and Centre for Food-borne, Environmental and Zoonotic Infectious Diseases (CFEZID), Public Health Agency of Canada and Provincial Public Health Microbiology Laboratories, Winnipeg, Canada [Google Scholar]
- 196.European Centre for Disease Prevention and Control and European Food Safety Authority 2011. Shiga toxin/verotoxin-producing Escherichia coli in humans, food and animals in the EU/EEA, with special reference to the German outbreak strain STEC O104. European Centre for Disease Prevention and Control and European Food Safety Authority, Stockholm, Sweden [Google Scholar]
- 197.Rivero MA, Passucci JA, Rodriguez EM, Parma AE. 2010. Role and clinical course of verotoxigenic Escherichia coli infections in childhood acute diarrhoea in Argentina. J. Med. Microbiol. 59:345–352 [DOI] [PubMed] [Google Scholar]
- 198.Bentancor AB, Ameal LA, Calviño MF, Martinez MC, Miccio L, Degregorio OJ. 2012. Risk factors for Shiga toxin-producing Escherichia coli infections in preadolescent schoolchildren in Buenos Aires, Argentina. J. Infect. Dev. Ctries. 6:378–386 [DOI] [PubMed] [Google Scholar]
- 199.De Souza RL, Abreu Carvalhaes JT, Sanae Nishimura L, de Andrade MC, Cabilio Guth BE. 2011. Hemolytic uremic syndrome in pediatric intensive care units in São Paulo, Brazil. Open Microbiol. J. 5:76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Irino K, Vaz TMI, Kato MAMF, Naves ZVF, Lara RR, Marco MEC, Rocha MMM, Moreira TP, Gomes TAT, Guth BEC. 2002. O157:H7 Shiga toxin-producing Escherichia coli strains associated with sporadic cases of diarrhea in São Paulo, Brazil. Emerg. Infect. Dis. 8:446–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Effler E, Isaäcson M, Arntzen L, Heenan R, Canter P, Barrett T, Lee L, Mambo C, Levine W, Zaidi A, Griffin PM. 2001. Factors contributing to the emergence of Escherichia coli O157 in Africa. Emerg. Infect. Dis. 7:812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sehgal R, Kumar Y, Kumar S. 2008. Prevalence and geographical distribution of Escherichia coli O157 in India: a 10-year survey. Trans. R. Soc. Trop. Med. Hyg. 102:380–383 [DOI] [PubMed] [Google Scholar]
- 203.Tuttle J, Gomez T, Doyle MP, Wells JG, Zhao T, Tauxe RV, Griffin PM. 1999. Lessons from a large outbreak of Escherichia coli O157:H7 infections: insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol. Infect. 122:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Paton AW, Ratcliff RM, Doyle RM, Seymour-Murray J, Davos D, Lanser JA, Paton JC. 1996. Molecular microbiological investigation of an outbreak of hemolytic-uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J. Clin. Microbiol. 34:1622–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Tilden J, Jr, Young W, McNamara AM, Custer C, Boesel B, Lambert-Fair MA, Majkowski J, Vugia D, Werner SB, Hollingsworth J, Morris JG., Jr 1996. A new route of transmission for Escherichia coli: infection from dry fermented salami. Am. J. Public Health 86:1142–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dinu L-D, Bach S. 2011. Induction of viable but nonculturable Escherichia coli O157:H7 in the phyllosphere of lettuce: a food safety risk factor. Appl. Environ. Microbiol. 77:8295–8302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gyles CL. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85:E45–E62 [DOI] [PubMed] [Google Scholar]
- 208.Bosilevac JM, Koohmaraie M. 2011. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl. Environ. Microbiol. 77:2103–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ferens WA, Hovde CJ. 2011. Escherichia coli O157:H7: animal reservoir and sources of human infection. Foodborne Pathog. Dis. 8:465–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Karch H, Bielaszewska M. 2001. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J. Clin. Microbiol. 39:2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wieler LH, Semmler T, Eichhorn I, Antao EM, Kinnemann B, Geue L, Karch H, Guenther S, Bethe A. 2011. No evidence of the Shiga toxin-producing E. coli O104:H4 outbreak strain or enteroaggregative E. coli (EAEC) found in cattle faeces in northern Germany, the hotspot of the 2011 HUS outbreak area. Gut Pathog. 3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hale CR, Scallan E, Cronquist AB, Dunn J, Smith K, Robinson T, Lathrop S, Tobin-D'Angelo M, Clogher P. 2012. Estimates of enteric illness attributable to contact with animals and their environments in the United States. Clin. Infect. Dis. 54(Suppl 5):S472–S479 [DOI] [PubMed] [Google Scholar]
- 213.Chase-Topping ME, McKendrick IJ, Pearce MC, MacDonald P, Matthews L, Halliday J, Allison L, Fenlon D, Low JC, Gunn G, Woolhouse MEJ. 2007. Risk factors for the presence of high-level shedders of Escherichia coli O157 on Scottish farms. J. Clin. Microbiol. 45:1594–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cobbold RN, Hancock DD, Rice DH, Berg J, Stilborn R, Hovde CJ, Besser TE. 2007. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl. Environ. Microbiol. 73:1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chase-Topping M, Gally D, Low C, Matthews L, Woolhouse M. 2008. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat. Rev. Microbiol. 6:904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Strachan NJC, Dunn GM, Locking ME, Reid TMS, Ogden ID. 2006. Escherichia coli O157: burger bug or environmental pathogen? Int. J. Food Microbiol. 112:129–137 [DOI] [PubMed] [Google Scholar]
- 217.Ogden ID, Hepburn NF, MacRae M, Strachan NJC, Fenlon DR, Rusbridge SM, Pennington TH. 2002. Long-term survival of Escherichia coli O157 on pasture following an outbreak associated with sheep at a scout camp. Lett. Appl. Microbiol. 34:100–104 [DOI] [PubMed] [Google Scholar]
- 218.Bolton DJ, Monaghan A, Byrne B, Fanning S, Sweeney T, McDowell DA. 2011. Incidence and survival of non-O157 verocytotoxigenic Escherichia coli in soil. J. Appl. Microbiol. 111:484–490 [DOI] [PubMed] [Google Scholar]
- 219.Kudva IT, Blanch K, Hovde CJ. 1998. Analysis of Escherichia coli O157:H7 survival in ovine or bovine manure and manure slurry. Appl. Environ. Microbiol. 64:3166–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Williams AP, McGregor KA, Killham K, Jones DL. 2008. Persistence and metabolic activity of Escherichia coli O157:H7 in farm animal faeces. FEMS Microbiol. Lett. 287:168–173 [DOI] [PubMed] [Google Scholar]
- 221.Avery LM, Williams AP, Killham K, Jones DL. 2008. Survival of Escherichia coli O157:H7 in waters from lakes, rivers, puddles and animal-drinking troughs. Sci. Total Environ. 389:378–385 [DOI] [PubMed] [Google Scholar]
- 222.Van Elsas JD, Semenov AV, Costa R, Trevors JT. 2011. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 5:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Chekabab SM, Paquin-Veillette J, Dozois CM, Harel J. 2013. The ecological habitat and transmission of Escherichia coli O157:H7. FEMS Microbiol. Lett. 341:1–12 [DOI] [PubMed] [Google Scholar]
- 224.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Solomon EB, Yaron S, Matthews KR. 2002. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68:397–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chauret C. 2011. Survival and control of Escherichia coli O157:H7 in foods, beverages, soil and water. Virulence 2:593–601 [DOI] [PubMed] [Google Scholar]
- 227.Shaw RK, Berger CN, Feys B, Knutton S, Pallen MJ, Frankel G. 2008. Enterohemorrhagic Escherichia coli exploits EspA filaments for attachment to salad leaves. Appl. Environ. Microbiol. 74:2908–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Saldaña Z, Sánchez E, Xicohtencatl-Cortes J, Puente JL, Girón JA. 2011. Surface structures involved in plant stomata and leaf colonization by Shiga-toxigenic Escherichia coli O157:H7. Front. Microbiol. 2:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Brandl MT. 2008. Plant lesions promote the rapid multiplication of Escherichia coli O157:H7 on postharvest lettuce. Appl. Environ. Microbiol. 74:5285–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Snedeker KG, Shaw DJ, Locking ME, Prescott RJ. 2009. Primary and secondary cases in Escherichia coli O157 outbreaks: a statistical analysis. BMC Infect. Dis. 9:144. 10.1186/1471-2334-9-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Stritt A, Tschumi S, Kottanattu L, Bucher BS, Steinmann M, von Steiger N, Stephan R, Hächler H, Simonetti GD. 2013. Neonatal hemolytic uremic syndrome after mother-to-child transmission of a low-pathogenic stx2b harboring Shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 56:114–116 [DOI] [PubMed] [Google Scholar]
- 232.Charatan F. 1999. New York outbreak of E coli poisoning affects 1000 and kills two. BMJ 319:873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Anonymous 2000. Waterborne outbreak of gastroenteritis associated with a contaminated municipal water supply, Walkerton, Ontario, May-June 2000. Can. Commun. Dis. Rep. 26:170–173 [PubMed] [Google Scholar]
- 234.Germani Y, Cunin P, Tedjouka E, Ncharre CB, Morvan J, Martin P. 1998. Enterohaemorrhagic Escherichia coli in Ngoïla (Cameroon) during an outbreak of bloody diarrhoea. Lancet 352:625–626 [DOI] [PubMed] [Google Scholar]
- 235.Koyange L, Ollivier G, Muyembe J-J, Kebela B, Gouali M, Germani Y. 2004. Enterohemorrhagic Escherichia coli O157, Kinshasa. Emerg. Infect. Dis. 10:968–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bradley KK, Williams JM, Burnsed LJ, Lytle MB, McDermott MD, Mody RK, Bhattarai A, Mallonee S, Piercefield EW, McDonald-Hamm CK, Smithee LK. 2012. Epidemiology of a large restaurant-associated outbreak of Shiga toxin-producing Escherichia coli O111:NM. Epidemiol. Infect. 140:1644–1654 [DOI] [PubMed] [Google Scholar]
- 237.Piercefield EW, Bradley KK, Coffman RL, Mallonee SM. 2010. Hemolytic uremic syndrome after an Escherichia coli O111 outbreak. Arch. Intern. Med. 170:1656–1663 [DOI] [PubMed] [Google Scholar]
- 238.Schimmer B, Nygard K, Eriksen H-M, Lassen J, Lindstedt B-A, Brandal LT, Kapperud G, Aavitsland P. 2008. Outbreak of haemolytic uraemic syndrome in Norway caused by stx2-positive Escherichia coli O103:H25 traced to cured mutton sausages. BMC Infect. Dis. 8:41. 10.1186/1471-2334-8-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Brown JA, Hite DS, Gillim-Ross LA, Maguire HF, Bennett JK, Patterson JJ, Comstock NA, Watkins AK, Ghosh TS, Vogt RL. 2012. Outbreak of Shiga toxin-producing Escherichia coli serotype O26:H11 infection at a child care center in Colorado. Pediatr. Infect. Dis. J. 31:379–383 [DOI] [PubMed] [Google Scholar]
- 240.Alpers K, Werber D, Frank C, Koch J, Friedrich AW, Karch H, An Heiden DER M, Prager R, Fruth A, Bielaszewska M, Morlock G, Heissenhuber A, Diedler A, Gerber A, Ammon A. 2009. Sorbitol-fermenting enterohaemorrhagic Escherichia coli O157:H− causes another outbreak of haemolytic uraemic syndrome in children. Epidemiol. Infect. 137:389–395 [DOI] [PubMed] [Google Scholar]
- 241.Pollock KGJ, Locking ME, Beattie TJ, Maxwell H, Ramage I, Hughes D, Cowieson J, Allison L, Hanson M, Cowden JM. 2010. Sorbitol-fermenting Escherichia coli O157, Scotland. Emerg. Infect. Dis. 16:881–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, Blattner FR. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo CH, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, Honda T, Han CG, Ohtsubo E, Kasamatsu M, Hayashi T, Kuhara S, Shinagawa H. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 5:1–9 [DOI] [PubMed] [Google Scholar]
- 244.Brunder W, Karch H, Schmidt H. 2006. Complete sequence of the large virulence plasmid pSFO157 of the sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H- strain 3072/96. Int. J. Med. Microbiol. Ijmm. 296:467–474 [DOI] [PubMed] [Google Scholar]
- 245.Fratamico PM, Yan X, Caprioli A, Esposito G, Needleman DS, Pepe T, Tozzoli R, Cortesi ML, Morabito S. 2011. The complete DNA sequence and analysis of the virulence plasmid and of five additional plasmids carried by Shiga toxin-producing Escherichia coli O26:H11 strain H30. Int. J. Med. Microbiol. 301:192–203 [DOI] [PubMed] [Google Scholar]
- 246.Yan X, Fratamico PM, Needleman DS, Bayles DO. 2012. DNA sequence and analysis of a 90.1-kb plasmid in Shiga toxin-producing Escherichia coli (STEC) O145:NM 83-75. Plasmid 68:25–32 [DOI] [PubMed] [Google Scholar]
- 247.Newton HJ, Sloan J, Bulach DM, Seemann T, Allison CC, Tauschek M, Robins-Browne RM, Paton JC, Whittam TS, Paton AW, Hartland EL. 2009. Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg. Infect. Dis. 15:372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, Reid-Smith R, Isaac-Renton J, Clark C, Rahn K, Kaper JB. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Bugarel M, Beutin L, Fach P. 2010. Low-density macroarray targeting non-locus of enterocyte effacement effectors (nle genes) and major virulence factors of Shiga toxin-producing Escherichia coli (STEC): a new approach for molecular risk assessment of STEC isolates. Appl. Environ. Microbiol. 76:203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Coombes BK, Wickham ME, Mascarenhas M, Gruenheid S, Finlay BB, Karmali MA. 2008. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 74:2153–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.European Food Safety Authority Panel on Biological Hazards (BIOHAZ) 2013. Scientific opinion on VTEC-seropathotype and scientific criteria regarding pathogenicity assessment. EFSA J. 11:3138 [Google Scholar]
- 252.Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnson RP, Gyles CL. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Friedrich AW, Bielaszewska M, Zhang W-L, Pulz M, Kuczius T, Ammon A, Karch H. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84 [DOI] [PubMed] [Google Scholar]
- 254.Bielaszewska M, Mellmann A, Bletz S, Zhang W, Köck R, Kossow A, Prager R, Fruth A, Orth-Höller D, Marejková M, Morabito S, Caprioli A, Piérard D, Smith G, Jenkins C, Curová K, Karch H. 2013. Enterohemorrhagic Escherichia coli O26:H11/H−: a new virulent clone emerges in Europe. Clin. Infect. Dis. 56:1373–1381 [DOI] [PubMed] [Google Scholar]
- 255.Wickham ME, Lupp C, Mascarenhas M, Vazquez A, Coombes BK, Brown NF, Coburn BA, Deng W, Puente JL, Karmali MA, Finlay BB. 2006. Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J. Infect. Dis. 194:819–827 [DOI] [PubMed] [Google Scholar]
- 256.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Abu-Ali GS, Ouellette LM, Henderson ST, Lacher DW, Riordan JT, Whittam TS, Manning SD. 2010. Increased adherence and expression of virulence genes in a lineage of Escherichia coli O157:H7 commonly associated with human infections. PLoS One 5:e10167. 10.1371/journal.pone.0010167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Abu-Ali GS, Ouellette LM, Henderson ST, Whittam TS, Manning SD. 2010. Differences in adherence and virulence gene expression between two outbreak strains of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 156:408–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Neupane M, Abu-Ali GS, Mitra A, Lacher DW, Manning SD, Riordan JT. 2011. Shiga toxin 2 overexpression in Escherichia coli O157:H7 strains associated with severe human disease. Microb. Pathog. 51:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kulasekara BR, Jacobs M, Zhou Y, Wu Z, Sims E, Saenphimmachak C, Rohmer L, Ritchie JM, Radey M, McKevitt M, Freeman TL, Hayden H, Haugen E, Gillett W, Fong C, Chang J, Beskhlebnaya V, Waldor MK, Samadpour M, Whittam TS, Kaul R, Brittnacher M, Miller SI. 2009. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 77:3713–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Hong W, Wu YE, Fu X, Chang Z. 2012. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol. 20:328–335 [DOI] [PubMed] [Google Scholar]
- 262.Farfan MJ, Torres AG. 2012. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect. Immun. 80:903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Obrig TG. 2010. Escherichia coli Shiga toxin mechanisms of action in renal disease. Toxins 2:2769–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Schüller S. 2011. Shiga toxin interaction with human intestinal epithelium. Toxins 3:626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Bauwens A, Betz J, Meisen I, Kemper B, Karch H, Müthing J. 2013. Facing glycosphingolipid-Shiga toxin interaction: dire straits for endothelial cells of the human vasculature. Cell. Mol. Life Sci. 70:425–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Mayer CL, Leibowitz CS, Kurosawa S, Stearns-Kurosawa DJ. 2012. Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins 4:1261–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Scheutz F, Teel LD, Beutin L, Piérard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 50:2951–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Neely MN, Friedman DI. 1998. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 28:1255–1267 [DOI] [PubMed] [Google Scholar]
- 269.Zhang X, McDaniel AD, Wolf LE, Keusch GT, Waldor MK, Acheson DW. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664–670 [DOI] [PubMed] [Google Scholar]
- 270.McGannon CM, Fuller CA, Weiss AA. 2010. Different classes of antibiotics differentially influence Shiga toxin production. Antimicrob. Agents Chemother. 54:3790–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Friedrich AW, Zhang W, Bielaszewska M, Mellmann A, Köck R, Fruth A, Tschäpe H, Karch H. 2007. Prevalence, virulence profiles, and clinical significance of Shiga toxin-negative variants of enterohemorrhagic Escherichia coli O157 infection in humans. Clin. Infect. Dis. 45:39–45 [DOI] [PubMed] [Google Scholar]
- 272.Mellmann A, Bielaszewska M, Zimmerhackl LB, Prager R, Harmsen D, Tschäpe H, Karch H. 2005. Enterohemorrhagic Escherichia coli in human infection: in vivo evolution of a bacterial pathogen. Clin. Infect. Dis. 41:785–792 [DOI] [PubMed] [Google Scholar]
- 273.Bielaszewska M, Middendorf B, Köck R, Friedrich AW, Fruth A, Karch H, Schmidt MA, Mellmann A. 2008. Shiga toxin-negative attaching and effacing Escherichia coli: distinct clinical associations with bacterial phylogeny and virulence traits and inferred in-host pathogen evolution. Clin. Infect. Dis. 47:208–217 [DOI] [PubMed] [Google Scholar]
- 274.Mellmann A, Lu S, Karch H, Xu J, Harmsen D, Schmidt MA, Bielaszewska M. 2008. Recycling of Shiga toxin 2 genes in sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl. Environ. Microbiol. 74:67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lee M-S, Cherla RP, Tesh VL. 2010. Shiga toxins: intracellular trafficking to the ER leading to activation of host cell stress responses. Toxins 2:1515–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Betz J, Bielaszewska M, Thies A, Humpf H-U, Dreisewerd K, Karch H, Kim KS, Friedrich AW, Müthing J. 2011. Shiga toxin glycosphingolipid receptors in microvascular and macrovascular endothelial cells: differential association with membrane lipid raft microdomains. J. Lipid Res. 52:618–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Betz J, Bauwens A, Kunsmann L, Bielaszewska M, Mormann M, Humpf H-U, Karch H, Friedrich AW, Müthing J. 2012. Uncommon membrane distribution of Shiga toxin glycosphingolipid receptors in toxin-sensitive human glomerular microvascular endothelial cells. Biol. Chem. 393:133–147 [DOI] [PubMed] [Google Scholar]
- 278.Tesh VL. 2012. Activation of cell stress response pathways by Shiga toxins. Cell. Microbiol. 14:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Etienne-Mesmin L, Chassaing B, Sauvanet P, Denizot J, Blanquet-Diot S, Darfeuille-Michaud A, Pradel N, Livrelli V. 2011. Interactions with M cells and macrophages as key steps in the pathogenesis of enterohemorrhagic Escherichia coli infections. PLoS One 6:e23594. 10.1371/journal.pone.0023594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Janka A, Bielaszewska M, Dobrindt U, Greune L, Schmidt MA, Karch H. 2003. Cytolethal distending toxin gene cluster in enterohemorrhagic Escherichia coli O157:H− and O157:H7: characterization and evolutionary considerations. Infect. Immun. 71:3634–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Friedrich AW, Lu S, Bielaszewska M, Prager R, Bruns P, Xu J-G, Tschäpe H, Karch H. 2006. Cytolethal distending toxin in Escherichia coli O157:H7: spectrum of conservation, structure, and endothelial toxicity. J. Clin. Microbiol. 44:1844–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Bielaszewska M, Fell M, Greune L, Prager R, Fruth A, Tschäpe H, Schmidt MA, Karch H. 2004. Characterization of cytolethal distending toxin genes and expression in Shiga toxin-producing Escherichia coli strains of non-O157 serogroups. Infect. Immun. 72:1812–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Elwell CA, Dreyfus LA. 2000. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol. Microbiol. 37:952–963 [DOI] [PubMed] [Google Scholar]
- 284.Bielaszewska M, Sinha B, Kuczius T, Karch H. 2005. Cytolethal distending toxin from Shiga toxin-producing Escherichia coli O157 causes irreversible G2/M arrest, inhibition of proliferation, and death of human endothelial cells. Infect. Immun. 73:552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Schmidt H, Beutin L, Karch H. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Schmidt H, Maier E, Karch H, Benz R. 1996. Pore-forming properties of the plasmid-encoded hemolysin of enterohemorrhagic Escherichia coli O157:H7. Eur. J. Biochem. 241:594–601 [DOI] [PubMed] [Google Scholar]
- 287.Aldick T, Bielaszewska M, Uhlin BE, Humpf H-U, Wai SN, Karch H. 2009. Vesicular stabilization and activity augmentation of enterohaemorrhagic Escherichia coli haemolysin. Mol. Microbiol. 71:1496–1508 [DOI] [PubMed] [Google Scholar]
- 288.Aldick T, Bielaszewska M, Zhang W, Brockmeyer J, Schmidt H, Friedrich AW, Kim KS, Schmidt MA, Karch H. 2007. Hemolysin from Shiga toxin-negative Escherichia coli O26 strains injures microvascular endothelium. Microbes Infect. 9:282–290 [DOI] [PubMed] [Google Scholar]
- 289.Brockmeyer J, Aldick T, Soltwisch J, Zhang W, Tarr PI, Weiss A, Dreisewerd K, Müthing J, Bielaszewska M, Karch H. 2011. Enterohaemorrhagic Escherichia coli haemolysin is cleaved and inactivated by serine protease EspPα. Environ. Microbiol. 13:1327–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Brockmeyer J, Bielaszewska M, Fruth A, Bonn ML, Mellmann A, Humpf H-U, Karch H. 2007. Subtypes of the plasmid-encoded serine protease EspP in Shiga toxin-producing Escherichia coli: distribution, secretion, and proteolytic activity. Appl. Environ. Microbiol. 73:6351–6359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Brunder W, Schmidt H, Karch H. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767–778 [DOI] [PubMed] [Google Scholar]
- 292.Orth D, Ehrlenbach S, Brockmeyer J, Khan AB, Huber G, Karch H, Sarg B, Lindner H, Würzner R. 2010. EspP, a serine protease of enterohemorrhagic Escherichia coli, impairs complement activation by cleaving complement factors C3/C3b and C5. Infect. Immun. 78:4294–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Tobe T, Beatson SA, Taniguchi H, Abe H, Bailey CM, Fivian A, Younis R, Matthews S, Marches O, Frankel G, Hayashi T, Pallen MJ. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. U. S. A. 103:14941–14946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Curtis MM, Sperandio V. 2011. A complex relationship: the interaction among symbiotic microbes, invading pathogens, and their mammalian host. Mucosal Immunol. 4:133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sperandio V. 2010. SdiA sensing of acyl-homoserine lactones by enterohemorrhagic E. coli (EHEC) serotype O157:H7 in the bovine rumen. Gut Microbes 1:432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kendall MM, Gruber CC, Parker CT, Sperandio V. 2012. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3(3):e00050–12. 10.1128/mBio.00050-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Nakanishi N, Tashiro K, Kuhara S, Hayashi T, Sugimoto N, Tobe T. 2009. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology 155:521–530 [DOI] [PubMed] [Google Scholar]
- 298.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492:113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Farfan MJ, Cantero L, Vidal R, Botkin DJ, Torres AG. 2011. Long polar fimbriae of enterohemorrhagic Escherichia coli O157:H7 bind to extracellular matrix proteins. Infect. Immun. 79:3744–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Samadder P, Xicohtencatl-Cortes J, Saldaña Z, Jordan D, Tarr PI, Kaper JB, Girón JA. 2009. The Escherichia coli ycbQRST operon encodes fimbriae with laminin-binding and epithelial cell adherence properties in Shiga-toxigenic E. COLI O157:H7. Environ. Microbiol. 11:1815–1826 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 301.Torres AG, Giron JA, Perna NT, Burland V, Blattner FR, Avelino-Flores F, Kaper JB. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Torres AG, Kanack KJ, Tutt CB, Popov V, Kaper JB. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol. Lett. 238:333–344 [DOI] [PubMed] [Google Scholar]
- 303.Fitzhenry R, Dahan S, Torres AG, Chong Y, Heuschkel R, Murch SH, Thomson M, Kaper JB, Frankel G, Phillips AD. 2006. Long polar fimbriae and tissue tropism in Escherichia coli O157:H7. Microbes Infect. 8:1741–1749 [DOI] [PubMed] [Google Scholar]
- 304.Toma C, Higa N, Iyoda S, Rivas M, Iwanaga M. 2006. The long polar fimbriae genes identified in Shiga toxin-producing Escherichia coli are present in other diarrheagenic E. coli and in the standard E. coli Collection of Reference (ECOR) strains. Res. Microbiol. 157:153–161 [DOI] [PubMed] [Google Scholar]
- 305.Chassaing B, Rolhion N, de Vallée A, Salim SY, Prorok-Hamon M, Neut C, Campbell BJ, Söderholm JD, Hugot J-P, Colombel J-F, Darfeuille-Michaud A. 2011. Crohn disease-associated adherent-invasive E. coli bacteria target mouse and human Peyer's patches via long polar fimbriae. J. Clin. Invest. 121:966–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Torres AG, Blanco M, Valenzuela P, Slater TM, Patel SD, Dahbi G, López C, Barriga XF, Blanco JE, Gomes TAT, Vidal R, Blanco J. 2009. Genes related to long polar fimbriae of pathogenic Escherichia coli strains as reliable markers to identify virulent isolates. J. Clin. Microbiol. 47:2442–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Chingcuanco F, Yu Y, Kus JV, Que L, Lackraj T, Lévesque CM, Barnett Foster D. 2012. Identification of a novel adhesin involved in acid-induced adhesion of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 158:2399–2407 [DOI] [PubMed] [Google Scholar]
- 308.Xicohtencatl-Cortes J, Monteiro-Neto V, Ledesma MA, Jordan DM, Francetic O, Kaper JB, Puente JL, Girón JA. 2007. Intestinal adherence associated with type IV pili of enterohemorrhagic Escherichia coli O157:H7. J. Clin. Invest. 117:3519–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Xicohtencatl-Cortes J, Monteiro-Neto V, Saldaña Z, Ledesma MA, Puente JL, Girón JA. 2009. The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J. Bacteriol. 191:411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Ledesma MA, Ochoa SA, Cruz A, Rocha-Ramírez LM, Mas-Oliva J, Eslava CA, Girón JA, Xicohtencatl-Cortes J. 2010. The hemorrhagic coli pilus (HCP) of Escherichia coli O157:H7 is an inducer of proinflammatory cytokine secretion in intestinal epithelial cells. PLoS One 5:e12127. 10.1371/journal.pone.0012127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Brunder W, Khan AS, Hacker J, Karch H. 2001. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H−. Infect. Immun. 69:4447–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Müsken A, Bielaszewska M, Greune L, Schweppe CH, Müthing J, Schmidt H, Schmidt MA, Karch H, Zhang W. 2008. Anaerobic conditions promote expression of Sfp fimbriae and adherence of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM to human intestinal epithelial cells. Appl. Environ. Microbiol. 74:1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Bielaszewska M, Prager R, Vandivinit L, Müsken A, Mellmann A, Holt NJ, Tarr PI, Karch H, Zhang W. 2009. Detection and characterization of the fimbrial sfp cluster in enterohemorrhagic Escherichia coli O165:H25/NM isolates from humans and cattle. Appl. Environ. Microbiol. 75:64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Stevens MP, Roe AJ, Vlisidou I, van Diemen PM, La Ragione RM, Best A, Woodward MJ, Gally DL, Wallis TS. 2004. Mutation of toxB and a truncated version of the efa-1 gene in Escherichia coli O157:H7 influences the expression and secretion of locus of enterocyte effacement-encoded proteins but not intestinal colonization in calves or sheep. Infect. Immun. 72:5402–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Tatsuno I, Horie M, Abe H, Miki T, Makino K, Shinagawa H, Taguchi H, Kamiya S, Hayashi T, Sasakawa C. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69:6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Abu-Median A-B, van Diemen PM, Dziva F, Vlisidou I, Wallis TS, Stevens MP. 2006. Functional analysis of lymphostatin homologues in enterohaemorrhagic Escherichia coli. FEMS Microbiol. Lett. 258:43–49 [DOI] [PubMed] [Google Scholar]
- 317.Tarr PI, Bilge SS, Vary JC, Jr, Jelacic S, Habeeb RL, Ward TR, Baylor MR, Besser TE. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Torres AG, Perna NT, Burland V, Ruknudin A, Blattner FR, Kaper JB. 2002. Characterization of Cah, a calcium-binding and heat-extractable autotransporter protein of enterohaemorrhagic Escherichia coli. Mol. Microbiol. 45:951–966 [DOI] [PubMed] [Google Scholar]
- 319.Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, Webb RI, Allsopp LP, Gobius KS, Gally DL, Schembri MA. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ. Microbiol. 10:589–604 [DOI] [PubMed] [Google Scholar]
- 320.Wells TJ, McNeilly TN, Totsika M, Mahajan A, Gally DL, Schembri MA. 2009. The Escherichia coli O157:H7 EhaB autotransporter protein binds to laminin and collagen I and induces a serum IgA response in O157:H7 challenged cattle. Environ. Microbiol. 11:1803–1814 [DOI] [PubMed] [Google Scholar]
- 321.Campellone KG. 2010. Cytoskeleton-modulating effectors of enteropathogenic and enterohaemorrhagic Escherichia coli: Tir, EspFU and actin pedestal assembly. FEBS J. 277:2390–2402 [DOI] [PubMed] [Google Scholar]
- 322.Lathem WW, Grys TE, Witowski SE, Torres AG, Kaper JB, Tarr PI, Welch RA. 2002. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 45:277–288 [DOI] [PubMed] [Google Scholar]
- 323.Lathem WW, Bergsbaken T, Welch RA. 2004. Potentiation of C1 esterase inhibitor by StcE, a metalloprotease secreted by Escherichia coli O157:H7. J. Exp. Med. 199:1077–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Grys TE, Siegel MB, Lathem WW, Welch RA. 2005. The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect. Immun. 73:1295–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Szabady RL, Lokuta MA, Walters KB, Huttenlocher A, Welch RA. 2009. Modulation of neutrophil function by a secreted mucinase of Escherichia coli O157:H7. PLoS Pathog. 5:e1000320. 10.1371/journal.ppat.1000320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Karch H, Schubert S, Zhang D, Zhang W, Schmidt H, Olschläger T, Hacker J. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67:5994–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66:480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Ito H, Terai A, Kurazono H, Takeda Y, Nishibuchi M. 1990. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8:47–60 [DOI] [PubMed] [Google Scholar]
- 329.Paton AW, Woodrow MC, Doyle RM, Lanser JA, Paton JC. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin C-S, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Møller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Doughty S, Sloan J, Bennett-Wood V, Robertson M, Robins-Browne RM, Hartland EL. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70:6761–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Paton AW, Srimanote P, Woodrow MC, Paton JC. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Jenkins C, Perry NT, Cheasty T, Shaw DJ, Frankel G, Dougan G, Gunn GJ, Smith HR, Paton AW, Paton JC. 2003. Distribution of the saa gene in strains of Shiga toxin-producing Escherichia coli of human and bovine origins. J. Clin. Microbiol. 41:1775–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Herold S, Paton JC, Paton AW. 2009. Sab, a novel autotransporter of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli O113:H21, contributes to adherence and biofilm formation. Infect. Immun. 77:3234–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Lu Y, Iyoda S, Satou H, Satou H, Itoh K, Saitoh T, Watanabe H. 2006. A new immunoglobulin-binding protein, EibG, is responsible for the chain-like adhesion phenotype of locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 74:5747–5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Merkel V, Ohder B, Bielaszewska M, Zhang W, Fruth A, Menge C, Borrmann E, Middendorf B, Müthing J, Karch H, Mellmann A. 2010. Distribution and phylogeny of immunoglobulin-binding protein G in Shiga toxin-producing Escherichia coli and its association with adherence phenotypes. Infect. Immun. 78:3625–3636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Luck SN, Bennett-Wood V, Poon R, Robins-Browne RM, Hartland EL. 2005. Invasion of epithelial cells by locus of enterocyte effacement-negative enterohemorrhagic Escherichia coli. Infect. Immun. 73:3063–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Luck SN, Badea L, Bennett-Wood V, Robins-Browne R, Hartland EL. 2006. Contribution of FliC to epithelial cell invasion by enterohemorrhagic Escherichia coli O113:H21. Infect. Immun. 74:6999–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Rogers TJ, Thorpe CM, Paton AW, Paton JC. 2012. Role of lipid rafts and flagellin in invasion of colonic epithelial cells by Shiga-toxigenic Escherichia coli O113:H21. Infect. Immun. 80:2858–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Melton-Celsa AR, Darnell SC, O'Brien AD. 1996. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect. Immun. 64:1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Fuller CA, Pellino CA, Flagler MJ, Strasser JE, Weiss AA. 2011. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 79:1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Bielaszewska M, Friedrich AW, Aldick T, Schürk-Bulgrin R, Karch H. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160–1167 [DOI] [PubMed] [Google Scholar]
- 343.Paton AW, Srimanote P, Talbot UM, Wang H, Paton JC. 2004. A new family of potent AB5 cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200:35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Michelacci V, Tozzoli R, Caprioli A, Martínez R, Scheutz F, Grande L, Sánchez S, Morabito S. 2013. A new pathogenicity island carrying an allelic variant of the Subtilase cytotoxin is common among Shiga toxin producing Escherichia coli of human and ovine origin. Clin. Microbiol. Infect. 19:E149–E156 [DOI] [PubMed] [Google Scholar]
- 345.Tozzoli R, Caprioli A, Cappannella S, Michelacci V, Marziano ML, Morabito S. 2010. Production of the subtilase AB5 cytotoxin by Shiga toxin-negative Escherichia coli. J. Clin. Microbiol. 48:178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Furukawa T, Yahiro K, Tsuji AB, Terasaki Y, Morinaga N, Miyazaki M, Fukuda Y, Saga T, Moss J, Noda M. 2011. Fatal hemorrhage induced by subtilase cytotoxin from Shiga-toxigenic Escherichia coli. Microb. Pathog. 50:159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Wang H, Paton JC, Paton AW. 2007. Pathologic changes in mice induced by subtilase cytotoxin, a potent new Escherichia coli AB5 toxin that targets the endoplasmic reticulum. J. Infect. Dis. 196:1093–1101 [DOI] [PubMed] [Google Scholar]
- 348.Wang H, Paton AW, McColl SR, Paton JC. 2011. In vivo leukocyte changes induced by Escherichia coli subtilase cytotoxin. Infect. Immun. 79:1671–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Paton AW, Paton JC. 2010. Escherichia coli subtilase cytotoxin. Toxins 2:215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Hu C-CA, Dougan SK, Winter SV, Paton AW, Paton JC, Ploegh HL. 2009. Subtilase cytotoxin cleaves newly synthesized BiP and blocks antibody secretion in B lymphocytes. J. Exp. Med. 206:2429–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Tsutsuki H, Yahiro K, Suzuki K, Suto A, Ogura K, Nagasawa S, Ihara H, Shimizu T, Nakajima H, Moss J, Noda M. 2012. Subtilase cytotoxin enhances Escherichia coli survival in macrophages by suppression of nitric oxide production through the inhibition of NF-κB activation. Infect. Immun. 80:3939–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Bell BP, Goldoft M, Griffin PM, Davis MA, Gordon DC, Tarr PI, Bartleson CA, Lewis JH, Barrett TJ, Wells JG. 1994. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA 272:1349–1353 [PubMed] [Google Scholar]
- 353.Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086 [DOI] [PubMed] [Google Scholar]
- 354.Jelacic JK, Damrow T, Chen GS, Jelacic S, Bielaszewska M, Ciol M, Carvalho HM, Melton-Celsa AR, O'Brien AD, Tarr PI. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719–729 [DOI] [PubMed] [Google Scholar]
- 355.Pai CH, Ahmed N, Lior H, Johnson WM, Sims HV, Woods DE. 1988. Epidemiology of sporadic diarrhea due to verocytotoxin-producing Escherichia coli: a two-year prospective study. J. Infect. Dis. 157:1054–1057 [DOI] [PubMed] [Google Scholar]
- 356.Vonberg RP, Höhle M, Aepfelbacher M, Bange FC, Belmar Campos C, Claussen K, Christner M, Cramer JP, Haller H, Hornef M, Fickenscher H, Fraedrich K, Knobloch JK, Kühbacher T, Manns MP, Nitschke M, Peters G, Pulz M, Rohde H, Roseland RT, Sayk F, Schaumburg F, Schöcklmann HO, Schubert S, Solbach W, Karch H, Suerbaum S. 2013. Duration of fecal shedding of Shiga toxin-producing Escherichia coli O104:H4 in patients infected during the 2011 outbreak in Germany: a multicenter study. Clin. Infect. Dis. 56:1132–1140 [DOI] [PubMed] [Google Scholar]
- 357.Karmali MA, Steele BT, Petric M, Lim C. 1983. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet i:619–620 [DOI] [PubMed] [Google Scholar]
- 358.Wong CS, Mooney JC, Brandt JR, Staples AO, Jelacic S, Boster DR, Watkins SL, Tarr PI. 2012. Risk factors for the hemolytic uremic syndrome in children infected with Escherichia coli O157:H7: a multivariable analysis. Clin. Infect. Dis. 55:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Spinale JM, Ruebner RL, Copelovitch L, Kaplan BS. 4 January 2013. Long-term outcomes of Shiga toxin hemolytic uremic syndrome. Pediatr. Nephrol. 10.1007/s00467-012-2383-6 [DOI] [PubMed] [Google Scholar]
- 360.Rosales A, Hofer J, Zimmerhackl L-B, Jungraithmayr TC, Riedl M, Giner T, Strasak A, Orth-Höller D, Würzner R, Karch H. 2012. Need for long-term follow-up in enterohemorrhagic Escherichia coli-associated hemolytic uremic syndrome due to late-emerging sequelae. Clin. Infect. Dis. 54:1413–1421 [DOI] [PubMed] [Google Scholar]
- 361.Paton JC, Paton AW. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Gould LH. 2012. Update: recommendations for diagnosis of Shiga toxin-producing Escherichia coli infections by clinical laboratories. Clin. Microbiol. Newsl. 34:75–83 [Google Scholar]
- 363.Gould LH, Bopp C, Strockbine N, Atkinson R, Baselski V, Body B, Carey R, Crandall C, Hurd S, Kaplan R, Neill M, Shea S, Somsel P, Tobin-D'Angelo M, Griffin PM, Gerner-Smidt P. 2009. Recommendations for diagnosis of Shiga toxin-producing Escherichia coli infections by clinical laboratories. MMWR Recommend. Rep. 58:1–14 [PubMed] [Google Scholar]
- 364.March SB, Ratnam S. 1986. Sorbitol-MacConkey medium for detection of Escherichia coli O157:H7 associated with hemorrhagic colitis. J. Clin. Microbiol. 23:869–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Church DL, Emshey D, Semeniuk H, Lloyd T, Pitout JD. 2007. Evaluation of BBL CHROMagar O157 versus sorbitol-MacConkey medium for routine detection of Escherichia coli O157 in a centralized regional clinical microbiology laboratory. J. Clin. Microbiol. 45:3098–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.March SB, Ratnam S. 1989. Latex agglutination test for detection of Escherichia coli serotype O157. J. Clin. Microbiol. 27:1675–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Bielaszewska M, Tarr PI, Karch H, Zhang W, Mathys W. 2005. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J. Clin. Microbiol. 43:452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Bielaszewska M, Middendorf B, Tarr PI, Zhang W, Prager R, Aldick T, Dobrindt U, Karch H, Mellmann A. 2011. Chromosomal instability in enterohaemorrhagic Escherichia coli O157:H7: impact on adherence, tellurite resistance and colony phenotype. Mol. Microbiol. 79:1024–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Bielaszewska M, Prager R, Köck R, Mellmann A, Zhang W, Tschäpe H, Tarr PI, Karch H. 2007. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl. Environ. Microbiol. 73:3144–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Hermos CR, Janineh M, Han LL, McAdam AJ. 2011. Shiga toxin-producing Escherichia coli in children: diagnosis and clinical manifestations of O157:H7 and non-O157:H7 infection. J. Clin. Microbiol. 49:955–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Mackenzie A, Orrbine E, Hyde L, Benoit M, Chan F, Park C, Alverson J, Lembke A, Hoban D, Kennedy W. 2000. Performance of the ImmunoCard STAT! E. coli O157:H7 test for detection of Escherichia coli O157:H7 in stools. J. Clin. Microbiol. 38:1866–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Lin A, Nguyen L, Lee T, Clotilde LM, Kase JA, Son I, Carter JM, Lauzon CR. 2011. Rapid O serogroup identification of the ten most clinically relevant STECs by Luminex microbead-based suspension array. J. Microbiol. Methods 87:105–110 [DOI] [PubMed] [Google Scholar]
- 373.Liu Y, Fratamico P. 2006. Escherichia coli O antigen typing using DNA microarrays. Mol. Cell. Probes 20:239–244 [DOI] [PubMed] [Google Scholar]
- 374.Norman KN, Strockbine NA, Bono JL. 2012. Association of nucleotide polymorphisms within the O-antigen gene cluster of Escherichia coli O26, O45, O103, O111, O121, and O145 with serogroups and genetic subtypes. Appl. Environ. Microbiol. 78:6689–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Gonzales TK, Kulow M, Park D-J, Kaspar CW, Anklam KS, Pertzborn KM, Kerrish KD, Ivanek R, Döpfer D. 2011. A high-throughput open-array qPCR gene panel to identify, virulotype, and subtype O157 and non-O157 enterohemorrhagic Escherichia coli. Mol. Cell. Probes 25:222–230 [DOI] [PubMed] [Google Scholar]
- 376.Marcon MJ, Kiska DL, Riddell SW. 2011. Should all stools be screened for Shiga toxin-producing Escherichia coli? J. Clin. Microbiol. 49:2390–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Ahn CK, Holt NJ, Tarr PI. 2009. Shiga-toxin producing Escherichia coli and the hemolytic uremic syndrome: what have we learned in the past 25 years? Adv. Exp. Med. Biol. 634:1–17 [DOI] [PubMed] [Google Scholar]
- 378.Hickey CA, Beattie TJ, Cowieson J, Miyashita Y, Strife CF, Frem JC, Peterson JM, Butani L, Jones DP, Havens PL, Patel HP, Wong CS, Andreoli SP, Rothbaum RJ, Beck AM, Tarr PI. 2011. Early volume expansion during diarrhea and relative nephroprotection during subsequent hemolytic uremic syndrome. Arch. Pediatr. Adolesc. Med. 165:884–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Holtz LR, Neill MA, Tarr PI. 2009. Acute bloody diarrhea: a medical emergency for patients of all ages. Gastroenterology 136:1887–1898 [DOI] [PubMed] [Google Scholar]
- 380.Goldwater PN, Bettelheim KA. 2012. Treatment of enterohemorrhagic Escherichia coli (EHEC) infection and hemolytic uremic syndrome (HUS). BMC Med. 10:12. 10.1186/1741-7015-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Rocha LB, Luz DE, Moraes CTP, Caravelli A, Fernandes I, Guth BEC, Horton DSPQ, Piazza RMF. 2012. Interaction between Shiga toxin and monoclonal antibodies: binding characteristics and in vitro neutralizing abilities. Toxins 4:729–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.López EL, Contrini MM, Glatstein E, González Ayala S, Santoro R, Allende D, Ezcurra G, Teplitz E, Koyama T, Matsumoto Y, Sato H, Sakai K, Hoshide S, Komoriya K, Morita T, Harning R, Brookman S. 2010. Safety and pharmacokinetics of urtoxazumab, a humanized monoclonal antibody, against Shiga-like toxin 2 in healthy adults and in pediatric patients infected with Shiga-like toxin-producing Escherichia coli. Antimicrob. Agents Chemother. 54:239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Bitzan M, Poole R, Mehran M, Sicard E, Brockus C, Thuning-Roberson C, Rivière M. 2009. Safety and pharmacokinetics of chimeric anti-Shiga toxin 1 and anti-Shiga toxin 2 monoclonal antibodies in healthy volunteers. Antimicrob. Agents Chemother. 53:3081–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Dowling TC, Chavaillaz PA, Young DG, Melton-Celsa A, O'Brien A, Thuning-Roberson C, Edelman R, Tacket CO. 2005. Phase 1 safety and pharmacokinetic study of chimeric murine-human monoclonal antibody cαStx2 administered intravenously to healthy adult volunteers. Antimicrob. Agents Chemother. 49:1808–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Watanabe-Takahashi M, Sato T, Dohi T, Noguchi N, Kano F, Murata M, Hamabata T, Natori Y, Nishikawa K. 2010. An orally applicable Shiga toxin neutralizer functions in the intestine to inhibit the intracellular transport of the toxin. Infect. Immun. 78:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Stearns-Kurosawa DJ, Collins V, Freeman S, Debord D, Nishikawa K, Oh S-Y, Leibowitz CS, Kurosawa S. 2011. Rescue from lethal Shiga toxin 2-induced renal failure with a cell-permeable peptide. Pediatr. Nephrol. Berl. Ger. 26:2031–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Li X, Wu P, Cheng S, Lv X. 2012. Synthesis and assessment of globotriose-chitosan conjugate, a novel inhibitor of Shiga toxins produced by Escherichia coli. J. Med. Chem. 55:2702–2710 [DOI] [PubMed] [Google Scholar]
- 388.Trachtman H, Cnaan A, Christen E, Gibbs K, Zhao S, Acheson DWK, Weiss R, Kaskel FJ, Spitzer A, Hirschman GH. 2003. Effect of an oral Shiga toxin-binding agent on diarrhea-associated hemolytic uremic syndrome in children: a randomized controlled trial. JAMA 290:1337–1344 [DOI] [PubMed] [Google Scholar]
- 389.Medellin-Peña MJ, Griffiths MW. 2009. Effect of molecules secreted by Lactobacillus acidophilus strain La-5 on Escherichia coli O157:H7 colonization. Appl. Environ. Microbiol. 75:1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Mogna L, Del Piano M, Deidda F, Nicola S, Soattini L, Debiaggi R, Sforza F, Strozzi G, Mogna G. 2012. Assessment of the in vitro inhibitory activity of specific probiotic bacteria against different Escherichia coli strains. J. Clin. Gastroenterol. 46(Suppl):S29–S32 [DOI] [PubMed] [Google Scholar]
- 391.Mukhopadhyay S, Linstedt AD. 2012. Manganese blocks intracellular trafficking of Shiga toxin and protects against Shiga toxicosis. Science 335:332–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Gaston MA, Pellino CA, Weiss AA. 2013. Failure of manganese to protect from Shiga toxin. PLoS One 8:e69823. 10.1371/journal.pone.0069823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Buvens G, Bogaerts P, Glupczynski Y, Lauwers S, Piérard D. 2010. Antimicrobial resistance testing of verocytotoxin-producing Escherichia coli and first description of TEM-52 extended-spectrum β-lactamase in serogroup O26. Antimicrob. Agents Chemother. 54:4907–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Venturini C, Beatson SA, Djordjevic SP, Walker MJ. 2010. Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J. 24:1160–1166 [DOI] [PubMed] [Google Scholar]
- 395.Torpdahl M, Nielsen EM, Scheutz F, Olesen B, Hansen DS, Hasman H. 2013. Detection of a Shiga toxin- and extended-spectrum-β-lactamase-producing Escherichia coli O157:H7 human clinical isolate. J. Antimicrob. Chemother. 68:1203–1204 [DOI] [PubMed] [Google Scholar]
- 396.Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, Chen W, Pu F, Peng Y, Li J, Xi F, Li S, Li Y, Zhang Z, Yang X, Zhao M, Wang P, Guan Y, Cen Z, Zhao X, Christner M, Kobbe R, Loos S, Oh J, Yang L, Danchin A, Gao GF, Song Y, Li Y, Yang H, Wang J, Xu J, Pallen MJ, Wang J, Aepfelbacher M, Yang R. 2011. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. Engl. J. Med. 365:718–724 [DOI] [PubMed] [Google Scholar]
- 397.Orth D, Grif K, Dierich MP, Würzner R. 2007. Variability in tellurite resistance and the ter gene cluster among Shiga toxin-producing Escherichia coli isolated from humans, animals and food. Res. Microbiol. 158:105–111 [DOI] [PubMed] [Google Scholar]
- 398.Potter AA, Klashinsky S, Li Y, Frey E, Townsend H, Rogan D, Erickson G, Hinkley S, Klopfenstein T, Moxley RA, Smith DR, Finlay BB. 2004. Decreased shedding of Escherichia coli O157:H7 by cattle following vaccination with type III secreted proteins. Vaccine 22:362–369 [DOI] [PubMed] [Google Scholar]
- 399.Cai K, Gao X, Li T, Wang Q, Hou X, Tu W, Xiao L, Tian M, Liu Y, Wang H. 2011. Enhanced immunogenicity of a novel Stx2Am-Stx1B fusion protein in a mice model of enterohemorrhagic Escherichia coli O157:H7 infection. Vaccine 29:946–952 [DOI] [PubMed] [Google Scholar]
- 400.Gao X, Cai K, Li T, Wang Q, Hou X, Tian R, Liu H, Tu W, Xiao L, Fang L, Luo S, Liu Y, Wang H. 2011. Novel fusion protein protects against adherence and toxicity of enterohemorrhagic Escherichia coli O157:H7 in mice. Vaccine 29:6656–6663 [DOI] [PubMed] [Google Scholar]
- 401.Gu J, Ning Y, Wang H, Xiao D, Tang B, Luo P, Cheng Y, Jiang M, Li N, Zou Q, Mao X. 2011. Vaccination of attenuated EIS-producing Salmonella induces protective immunity against enterohemorrhagic Escherichia coli in mice. Vaccine 29:7395–7403 [DOI] [PubMed] [Google Scholar]
- 402.Zhang X-H, He K-W, Zhang S-X, Lu W-C, Zhao P-D, Luan X-T, Ye Q, Wen L-B, Li B, Guo R-L, Wang X-M, Lv L-X, Zhou J-M, Yu Z-Y, Mao A-H. 2011. Subcutaneous and intranasal immunization with Stx2B-Tir-Stx1B-Zot reduces colonization and shedding of Escherichia coli O157:H7 in mice. Vaccine 29:3923–3929 [DOI] [PubMed] [Google Scholar]
- 403.Amani J, Mousavi SL, Rafati S, Salmanian AH. 2011. Immunogenicity of a plant-derived edible chimeric EspA, Intimin and Tir of Escherichia coli O157:H7 in mice. Plant Sci. Int. J. Exp. Plant Biol. 180:620–627 [DOI] [PubMed] [Google Scholar]
- 404.Fujii J, Naito M, Yutsudo T, Matsumoto S, Heatherly DP, Yamada T, Kobayashi H, Yoshida S-I, Obrig T. 2012. Protection by a recombinant Mycobacterium bovis Bacillus Calmette-Guerin vaccine expressing Shiga toxin 2 B subunit against Shiga toxin-producing Escherichia coli in mice. Clin. Vaccine Immunol. 19:1932–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Oliveira AF, Cardoso SA, Almeida FB dos, de Oliveira RLL, Pitondo-Silva A, Soares SG, Hanna ES. 2012. Oral immunization with attenuated Salmonella vaccine expressing Escherichia coli O157:H7 intimin γ triggers both systemic and mucosal humoral immunity in mice. Microbiol. Immunol. 56:513–522 [DOI] [PubMed] [Google Scholar]
- 406.Bentancor LV, Bilen M, Brando RJF, Ramos MV, Ferreira LCS, Ghiringhelli PD, Palermo MS. 2009. A DNA vaccine encoding the enterohemorragic Escherichia coli Shiga-like toxin 2 A2 and B subunits confers protective immunity to Shiga toxin challenge in the murine model. Clin. Vaccine Immunol. 16:712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Shiga K. 1898. Ueber den erreger der dysenterie in Japan (vorläufige mitteilung). Zentralbl. Bakteriol. Mikrobiol. Hyg. 599–600. [Google Scholar]
- 408.Trofa AF, Ueno-Olsen H, Oiwa R, Yoshikawa M. 1999. Dr. Kiyoshi Shiga: discoverer of the dysentery bacillus. Clin. Infect. Dis. 29:1303–1306 [DOI] [PubMed] [Google Scholar]
- 409.Lan R, Alles MC, Donohoe K, Martinez MB, Reeves PR. 2004. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 72:5080–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Scheutz F, Strockbine NA. 2005. Genus I. Escherichia, p 607–624 In Garrity GM, et al. (ed), Bergey's manual of systematic bacteriology. Springer Publishing Company, New York, NY [Google Scholar]
- 411.Strockbine NA, Maurelli AT. 2005. Genus XXXV. Shigella, p 811–823 In Garrity GM, et al. (ed), Bergey's manual of systematic bacteriology. Springer Publishing Company, New York, NY [Google Scholar]
- 412.Cheasty T, Rowe B. 1983. Antigenic relationships between the enteroinvasive Escherichia coli O antigens O28ac, O112ac, O124, O136, O143, O144, O152, and O164 and Shigella O antigens. J. Clin. Microbiol. 17:681–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Li Y, Cao B, Liu B, Liu D, Gao Q, Peng X, Wu J, Bastin DA, Feng L, Wang L. 2009. Molecular detection of all 34 distinct O-antigen forms of Shigella. J. Med. Microbiol. 58:69–81 [DOI] [PubMed] [Google Scholar]
- 414.Ewing WH. 1949. Shigella nomenclature. J. Bacteriol. 57:633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Kalluri P, Cummings KC, Abbott S, Malcolm GB, Hutcheson K, Beall A, Joyce K, Polyak C, Woodward D, Caldeira R, Rodgers F, Mintz ED, Strockbine N. 2004. Epidemiological features of a newly described serotype of Shigella boydii. Epidemiol. Infect. 132:579–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Hyma KE, Lacher DW, Nelson AM, Bumbaugh AC, Janda JM, Strockbine NA, Young VB, Whittam TS. 2005. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 187:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Lan R, Reeves PR. 2002. Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 4:1125–1132 [DOI] [PubMed] [Google Scholar]
- 418.Pupo GM, Karaolis DK, Lan R, Reeves PR. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Pupo GM, Lan R, Reeves PR. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. U. S. A. 97:10567–10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Wang RF, Cao WW, Cerniglia CE. 1997. Phylogenetic analysis and identification of Shigella spp. by molecular probes. Mol. Cell. Probes 11:427–432 [DOI] [PubMed] [Google Scholar]
- 421.Peng J, Yang J, Jin Q. 2009. The molecular evolutionary history of Shigella spp. and enteroinvasive Escherichia coli. Infect. Genet. Evol. 9:147–152 [DOI] [PubMed] [Google Scholar]
- 422.Yang J, Nie H, Chen L, Zhang X, Yang F, Xu X, Zhu Y, Yu J, Jin Q. 2007. Revisiting the molecular evolutionary history of Shigella spp. J. Mol. Evol. 64:71–79 [DOI] [PubMed] [Google Scholar]
- 423.Johnson TJ, Nolan LK. 2009. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73:750–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Schroeder GN, Hilbi H. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin. Microbiol. Rev. 21:134–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Yang F, Yang J, Zhang X, Chen L, Jiang Y, Yan Y, Tang X, Wang J, Xiong Z, Dong J, Xue Y, Zhu Y, Xu X, Sun L, Chen S, Nie H, Peng J, Xu J, Wang Y, Yuan Z, Wen Y, Yao Z, Shen Y, Qiang B, Hou Y, Yu J, Jin Q. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 33:6445–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Maurelli AT, Fernández RE, Bloch CA, Rode CK, Fasano A. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:3943–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, Mayhew GF, Plunkett G, III, Rose DJ, Darling A, Mau B, Perna NT, Payne SM, Runyen-Janecky LJ, Zhou S, Schwartz DC, Blattner FR. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71:2775–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Fernandez IM, Silva M, Schuch R, Walker WA, Siber AM, Maurelli AT, McCormick BA. 2001. Cadaverine prevents the escape of Shigella flexneri from the phagolysosome: a connection between bacterial dissemination and neutrophil transepithelial signaling. J. Infect. Dis. 184:743–753 [DOI] [PubMed] [Google Scholar]
- 429.McCormick BA, Fernandez MI, Siber AM, Maurelli AT. 1999. Inhibition of Shigella flexneri-induced transepithelial migration of polymorphonuclear leucocytes by cadaverine. Cell. Microbiol. 1:143–155 [DOI] [PubMed] [Google Scholar]
- 430.Casalino M, Latella MC, Prosseda G, Colonna B. 2003. CadC is the preferential target of a convergent evolution driving enteroinvasive Escherichia coli toward a lysine decarboxylase-defective phenotype. Infect. Immun. 71:5472–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Harris JR, Mariano J, Wells JG, Payne BJ, Donnell HD, Cohen ML. 1985. Person-to-person transmission in an outbreak of enteroinvasive Escherichia coli. Am. J. Epidemiol. 122:245–252 [DOI] [PubMed] [Google Scholar]
- 432.Lampel KA. 2012. Shigella species, p 25–28 In Bad bug book, 2nd ed. Center for Food Safety and Applied Nutrition, Food and Drug Administration, Washington, DC [Google Scholar]
- 433.Levine OS, Levine MM. 1991. Houseflies (Musca domestica) as mechanical vectors of shigellosis. Rev. Infect. Dis. 13:688–696 [DOI] [PubMed] [Google Scholar]
- 434.Saeed A, Abd H, Edvinsson B, Sandström G. 2009. Acanthamoeba castellanii an environmental host for Shigella dysenteriae and Shigella sonnei. Arch. Microbiol. 191:83–88 [DOI] [PubMed] [Google Scholar]
- 435.DuPont HL, Levine MM, Hornick RB, Formal SB. 1989. Inoculum size in shigellosis and implications for expected mode of transmission. J. Infect. Dis. 159:1126–1128 [DOI] [PubMed] [Google Scholar]
- 436.Hsia RC, Small PL, Bavoil PM. 1993. Characterization of virulence genes of enteroinvasive Escherichia coli by TnphoA mutagenesis: identification of invX, a gene required for entry into HEp-2 cells. J. Bacteriol. 175:4817–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.European Centre for Disease Prevention and Control 2011. Annual epidemiological report 2011. Reporting on 2009 surveillance data and 2010 epidemic intelligence data. European Centre for Disease Prevention and Control, Stockholm, Sweden [Google Scholar]
- 438.OzFoodNet Working Group 2012. Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet network, 2010. Commun. Dis. Intell. 36:E213–E241 [DOI] [PubMed] [Google Scholar]
- 439.Gordillo ME, Reeve GR, Pappas J, Mathewson JJ, DuPont HL, Murray BE. 1992. Molecular characterization of strains of enteroinvasive Escherichia coli O143, including isolates from a large outbreak in Houston, Texas. J. Clin. Microbiol. 30:889–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Gassama-Sow A, Sow PS, Guèye M, Guèye-N′diaye A, Perret JL, M'boup S, Aïdara-Kane A. 2004. Characterization of pathogenic Escherichia coli in human immunodeficiency virus-related diarrhea in Senegal. J. Infect. Dis. 189:75–78 [DOI] [PubMed] [Google Scholar]
- 441.Okeke IN. 2009. Diarrheagenic Escherichia coli in sub-Saharan Africa: status, uncertainties and necessities. J. Infect. Dev. Ctries. 3:817–842 [DOI] [PubMed] [Google Scholar]
- 442.Pérez C, Gómez-Duarte OG, Arias ML. 2010. Diarrheagenic Escherichia coli in children from Costa Rica. Am. J. Trop. Med. Hyg. 83:292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Vieira N, Bates SJ, Solberg OD, Ponce K, Howsmon R, Cevallos W, Trueba G, Riley L, Eisenberg JNS. 2007. High prevalence of enteroinvasive Escherichia coli isolated in a remote region of northern coastal Ecuador. Am. J. Trop. Med. Hyg. 76:528–533 [PMC free article] [PubMed] [Google Scholar]
- 444.Pfeiffer ML, DuPont HL, Ochoa TJ. 2012. The patient presenting with acute dysentery—a systematic review. J. Infect. 64:374–386 [DOI] [PubMed] [Google Scholar]
- 445.Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. World Health Organ. 77:651–666 [PMC free article] [PubMed] [Google Scholar]
- 446.Bardhan P, Faruque ASG, Naheed A, Sack DA. 2010. Decrease in shigellosis-related deaths without Shigella spp.-specific interventions, Asia. Emerg. Infect. Dis. 16:1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Von Seidlein L, Kim DR, Ali M, Lee H, Wang X, Thiem VD, Canh DG, Chaicumpa W, Agtini MD, Hossain A, Bhutta ZA, Mason C, Sethabutr O, Talukder K, Nair GB, Deen JL, Kotloff K, Clemens J. 2006. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 3:e353. 10.1371/journal.pmed.0030353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Sansonetti PJ. 2006. Shigellosis: an old disease in new clothes? PLoS Med. 3:e354. 10.1371/journal.pmed.0030354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Aragón M, Barreto A, Chambule J, Noya A, Tallarico M. 1995. Shigellosis in Mozambique: the 1993 outbreak rehabilitation—a follow-up study. Trop. Doct. 25:159–162 [DOI] [PubMed] [Google Scholar]
- 450.Ebright JR, Moore EC, Sanborn WR, Schaberg D, Kyle J, Ishida K. 1984. Epidemic Shiga bacillus dysentery in Central Africa. Am. J. Trop. Med. Hyg. 33:1192–1197 [DOI] [PubMed] [Google Scholar]
- 451.Gangarosa EJ, Perera DR, Mata LJ, Mendizábal-Morris C, Guzmán G, Reller LB. 1970. Epidemic Shiga bacillus dysentery in Central America. II. Epidemiologic studies in 1969. J. Infect. Dis. 122:181–190 [DOI] [PubMed] [Google Scholar]
- 452.Rahaman MM, Khan MM, Aziz KM, Islam MS, Kibriya AK. 1975. An outbreak of dysentery caused by Shigella dysenteriae type 1 on a coral island in the Bay of Bengal. J. Infect. Dis. 132:15–19 [DOI] [PubMed] [Google Scholar]
- 453.Guerin PJ, Brasher C, Baron E, Mic D, Grimont F, Ryan M, Aavitsland P, Legros D. 2003. Shigella dysenteriae serotype 1 in west Africa: intervention strategy for an outbreak in Sierra Leone. Lancet 362:705–706 [DOI] [PubMed] [Google Scholar]
- 454.Levine MM, Kotloff KL, Barry EM, Pasetti MF, Sztein MB. 2007. Clinical trials of Shigella vaccines: two steps forward and one step back on a long, hard road. Nat. Rev. Microbiol. 5:540–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Mandal J, Emelda V GJ, Parija S MSC. 2012. The recent trends of shigellosis: a JIPMER perspective. J. Clin. Diagn. Res. 6:1474–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Rolfo F, Marin GH, Silberman M, Pattin J, Giugnio S, Gatti B, Bettiol M, Rigoni A. 2012. Epidemiological study of shigellosis in an urban area of Argentina. J. Infect. Dev. Ctries. 6:324–328 [DOI] [PubMed] [Google Scholar]
- 457.Tariq A, Haque A, Ali A, Bashir S, Habeeb MA, Salman M, Sarwar Y. 2012. Molecular profiling of antimicrobial resistance and integron association of multidrug-resistant clinical isolates of Shigella species from Faisalabad, Pakistan. Can. J. Microbiol. 58:1047–1054 [DOI] [PubMed] [Google Scholar]
- 458.Xia S, Xu B, Huang L, Zhao J-Y, Ran L, Zhang J, Chen H, Pulsrikarn C, Pornruangwong S, Aarestrup FM, Hendriksen RS. 2011. Prevalence and characterization of human Shigella infections in Henan Province, China, in 2006. J. Clin. Microbiol. 49:232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Koh XP, Chiou CS, Ajam N, Watanabe H, Ahmad N, Thong KL. 2012. Characterization of Shigella sonnei in Malaysia, an increasingly prevalent etiologic agent of local shigellosis cases. BMC Infect. Dis. 12:122. 10.1186/1471-2334-12-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Qu F, Bao C, Chen S, Cui E, Guo T, Wang H, Zhang J, Wang H, Tang Y-W, Mao Y. 2012. Genotypes and antimicrobial profiles of Shigella sonnei isolates from diarrheal patients circulating in Beijing between 2002 and 2007. Diagn. Microbiol. Infect. Dis. 74:166–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Arvelo W, Hinkle CJ, Nguyen TA, Weiser T, Steinmuller N, Khan F, Gladbach S, Parsons M, Jennings D, Zhu BP, Mintz E, Bowen A. 2009. Transmission risk factors and treatment of pediatric shigellosis during a large daycare center-associated outbreak of multidrug resistant Shigella sonnei: implications for the management of shigellosis outbreaks among children. Pediatr. Infect. Dis. J. 28:976–980 [DOI] [PubMed] [Google Scholar]
- 462.Shah N, DuPont HL, Ramsey DJ. 2009. Global etiology of travelers' diarrhea: systematic review from 1973 to the present. Am. J. Trop. Med. Hyg. 80:609–614 [PubMed] [Google Scholar]
- 463.Holt KE, Baker S, Weill F-X, Holmes EC, Kitchen A, Yu J, Sangal V, Brown DJ, Coia JE, Kim DW, Choi SY, Kim SH, da Silveira WD, Pickard DJ, Farrar JJ, Parkhill J, Dougan G, Thomson NR. 2012. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat. Genet. 44:1056–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Marteyn B, Gazi A, Sansonetti P. 2012. Shigella: a model of virulence regulation in vivo. Gut Microbes 3:104–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Carayol N, Tran Van Nhieu G. 2013. Tips and tricks about Shigella invasion of epithelial cells. Curr. Opin. Microbiol 16:32–37 [DOI] [PubMed] [Google Scholar]
- 466.Hilbi H, Moss JE, Hersh D, Chen Y, Arondel J, Banerjee S, Flavell RA, Yuan J, Sansonetti PJ, Zychlinsky A. 1998. Shigella-induced apoptosis is dependent on caspase-1 which binds to IpaB. J. Biol. Chem. 273:32895–32900 [DOI] [PubMed] [Google Scholar]
- 467.Senerovic L, Tsunoda SP, Goosmann C, Brinkmann V, Zychlinsky A, Meissner F, Kolbe M. 2012. Spontaneous formation of IpaB ion channels in host cell membranes reveals how Shigella induces pyroptosis in macrophages. Cell Death Dis. 3:e384. 10.1038/cddis.2012.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Zychlinsky A, Prevost MC, Sansonetti PJ. 1992. Shigella flexneri induces apoptosis in infected macrophages. Nature 358:167–169 [DOI] [PubMed] [Google Scholar]
- 469.Skoudy A, Mounier J, Aruffo A, Ohayon H, Gounon P, Sansonetti P, Tran Van Nhieu G. 2000. CD44 binds to the Shigella IpaB protein and participates in bacterial invasion of epithelial cells. Cell. Microbiol. 2:19–33 [DOI] [PubMed] [Google Scholar]
- 470.Watarai M, Funato S, Sasakawa C. 1996. Interaction of Ipa proteins of Shigella flexneri with α5β1 integrin promotes entry of the bacteria into mammalian cells. J. Exp. Med. 183:991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Handa Y, Suzuki M, Ohya K, Iwai H, Ishijima N, Koleske AJ, Fukui Y, Sasakawa C. 2007. Shigella IpgB1 promotes bacterial entry through the ELMO-Dock180 machinery. Nat. Cell Biol. 9:121–128 [DOI] [PubMed] [Google Scholar]
- 472.Huang Z, Sutton SE, Wallenfang AJ, Orchard RC, Wu X, Feng Y, Chai J, Alto NM. 2009. Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat. Struct. Mol. Biol. 16:853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Mounier J, Popoff MR, Enninga J, Frame MC, Sansonetti PJ, Van Nhieu GT. 2009. The IpaC carboxyterminal effector domain mediates Src-dependent actin polymerization during Shigella invasion of epithelial cells. PLoS Pathog. 5:e1000271. 10.1371/journal.ppat.1000271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Niebuhr K, Giuriato S, Pedron T, Philpott DJ, Gaits F, Sable J, Sheetz MP, Parsot C, Sansonetti PJ, Payrastre B. 2002. Conversion of PtdIns4,5P2 into PtdIns5P by the S. flexneri effector IpgD reorganizes host cell morphology. EMBO J. 21:5069–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Park H, Valencia-Gallardo C, Sharff A, Tran Van Nhieu G, Izard T. 2011. Novel vinculin binding site of the IpaA invasin of Shigella. J. Biol. Chem. 286:23214–23221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Yoshida S, Katayama E, Kuwae A, Mimuro H, Suzuki T, Sasakawa C. 2002. Shigella deliver an effector protein to trigger host microtubule destabilization, which promotes Rac1 activity and efficient bacterial internalization. EMBO J. 21:2923–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Fernandez-Prada CM, Hoover DL, Tall BD, Hartman AB, Kopelowitz J, Venkatesan MM. 2000. Shigella flexneri IpaH7.8 facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect. Immun. 68:3608–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Harrington A, Darboe N, Kenjale R, Picking WL, Middaugh CR, Birket S, Picking WD. 2006. Characterization of the interaction of single tryptophan containing mutants of IpaC from Shigella flexneri with phospholipid membranes. Biochemistry 45:626–636 [DOI] [PubMed] [Google Scholar]
- 479.High N, Mounier J, Prévost MC, Sansonetti PJ. 1992. IpaB of Shigella flexneri causes entry into epithelial cells and escape from the phagocytic vacuole. EMBO J. 11:1991–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Picking WL, Nishioka H, Hearn PD, Baxter MA, Harrington AT, Blocker A, Picking WD. 2005. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect. Immun. 73:1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Ashida H, Ogawa M, Kim M, Suzuki S, Sanada T, Punginelli C, Mimuro H, Sasakawa C. 2011. Shigella deploy multiple countermeasures against host innate immune responses. Curr. Opin. Microbiol. 14:16–23 [DOI] [PubMed] [Google Scholar]
- 482.Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, Parsot C, Sansonetti PJ. 2007. An injected bacterial effector targets chromatin access for transcription factor NF-κB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8:47–56 [DOI] [PubMed] [Google Scholar]
- 483.Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C. 2010. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKγ to dampen the host NF-κB-mediated inflammatory response. Nat. Cell Biol. 12:66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Kim DW, Lenzen G, Page A-L, Legrain P, Sansonetti PJ, Parsot C. 2005. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. U. S. A. 102:14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou J-M, Shao F. 2007. The phosphothreonine lyase activity of a bacterial type III effector family. Science 315:1000–1003 [DOI] [PubMed] [Google Scholar]
- 486.Sanada T, Kim M, Mimuro H, Suzuki M, Ogawa M, Oyama A, Ashida H, Kobayashi T, Koyama T, Nagai S, Shibata Y, Gohda J, Inoue J, Mizushima T, Sasakawa C. 2012. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483:623–626 [DOI] [PubMed] [Google Scholar]
- 487.Wang F, Jiang Z, Li Y, He X, Zhao J, Yang X, Zhu L, Yin Z, Li X, Wang X, Liu W, Shang W, Yang Z, Wang S, Zhen Q, Zhang Z, Yu Y, Zhong H, Ye Q, Huang L, Yuan J. 2013. Shigella flexneri T3SS effector IpaH4.5 modulates the host inflammatory response via interaction with NF-κB p65 protein. Cell. Microbiol. 15:474–485 [DOI] [PubMed] [Google Scholar]
- 488.Zurawski DV, Mumy KL, Faherty CS, McCormick BA, Maurelli AT. 2009. Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol. Microbiol. 71:350–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Kim DW, Chu H, Joo DH, Jang MS, Choi JH, Park S-M, Choi Y-J, Han SH, Yun C-H. 2008. OspF directly attenuates the activity of extracellular signal-regulated kinase during invasion by Shigella flexneri in human dendritic cells. Mol. Immunol. 45:3295–3301 [DOI] [PubMed] [Google Scholar]
- 490.Konradt C, Frigimelica E, Nothelfer K, Puhar A, Salgado-Pabon W, di Bartolo V, Scott-Algara D, Rodrigues CD, Sansonetti PJ, Phalipon A. 2011. The Shigella flexneri type three secretion system effector IpgD inhibits T cell migration by manipulating host phosphoinositide metabolism. Cell Host Microbe 9:263–272 [DOI] [PubMed] [Google Scholar]
- 491.Sperandio B, Regnault B, Guo J, Zhang Z, Stanley SL, Jr, Sansonetti PJ, Pédron T. 2008. Virulent Shigella flexneri subverts the host innate immune response through manipulation of antimicrobial peptide gene expression. J. Exp. Med. 205:1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Dong N, Zhu Y, Lu Q, Hu L, Zheng Y, Shao F. 2012. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150:1029–1041 [DOI] [PubMed] [Google Scholar]
- 493.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. 2005. Escape of intracellular Shigella from autophagy. Science 307:727–731 [DOI] [PubMed] [Google Scholar]
- 494.Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier MF. 1999. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 146:1319–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Goldberg MB, Theriot JA. 1995. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc. Natl. Acad. Sci. U. S. A. 92:6572–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Yoshida S, Handa Y, Suzuki T, Ogawa M, Suzuki M, Tamai A, Abe A, Katayama E, Sasakawa C. 2006. Microtubule-severing activity of Shigella is pivotal for intercellular spreading. Science 314:985–989 [DOI] [PubMed] [Google Scholar]
- 497.Fukumatsu M, Ogawa M, Arakawa S, Suzuki M, Nakayama K, Shimizu S, Kim M, Mimuro H, Sasakawa C. 2012. Shigella targets epithelial tricellular junctions and uses a noncanonical clathrin-dependent endocytic pathway to spread between cells. Cell Host Microbe 11:325–336 [DOI] [PubMed] [Google Scholar]
- 498.Kim M, Ashida H, Ogawa M, Yoshikawa Y, Mimuro H, Sasakawa C. 2010. Bacterial interactions with the host epithelium. Cell Host Microbe 8:20–35 [DOI] [PubMed] [Google Scholar]
- 499.Bergounioux J, Elisee R, Prunier A-L, Donnadieu F, Sperandio B, Sansonetti P, Arbibe L. 2012. Calpain activation by the Shigella flexneri effector VirA regulates key steps in the formation and life of the bacterium's epithelial niche. Cell Host Microbe 11:240–252 [DOI] [PubMed] [Google Scholar]
- 500.Iwai H, Kim M, Yoshikawa Y, Ashida H, Ogawa M, Fujita Y, Muller D, Kirikae T, Jackson PK, Kotani S, Sasakawa C. 2007. A bacterial effector targets Mad2L2, an APC inhibitor, to modulate host cell cycling. Cell 130:611–623 [DOI] [PubMed] [Google Scholar]
- 501.Kim M, Ogawa M, Fujita Y, Yoshikawa Y, Nagai T, Koyama T, Nagai S, Lange A, Fässler R, Sasakawa C. 2009. Bacteria hijack integrin-linked kinase to stabilize focal adhesions and block cell detachment. Nature 459:578–582 [DOI] [PubMed] [Google Scholar]
- 502.Pendaries C, Tronchère H, Arbibe L, Mounier J, Gozani O, Cantley L, Fry MJ, Gaits-Iacovoni F, Sansonetti PJ, Payrastre B. 2006. PtdIns5P activates the host cell PI3-kinase/Akt pathway during Shigella flexneri infection. EMBO J. 25:1024–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Carneiro LAM, Travassos LH, Soares F, Tattoli I, Magalhaes JG, Bozza MT, Plotkowski MC, Sansonetti PJ, Molkentin JD, Philpott DJ, Girardin SE. 2009. Shigella induces mitochondrial dysfunction and cell death in nonmyleoid cells. Cell Host Microbe 5:123–136 [DOI] [PubMed] [Google Scholar]
- 504.Mounier J, Boncompain G, Senerovic L, Lagache T, Chrétien F, Perez F, Kolbe M, Olivo-Marin J-C, Sansonetti PJ, Sauvonnet N. 2012. Shigella effector IpaB-induced cholesterol relocation disrupts the Golgi complex and recycling network to inhibit host cell secretion. Cell Host Microbe 12:381–389 [DOI] [PubMed] [Google Scholar]
- 505.Sakaguchi T, Köhler H, Gu X, McCormick BA, Reinecker H-C. 2002. Shigella flexneri regulates tight junction-associated proteins in human intestinal epithelial cells. Cell. Microbiol. 4:367–381 [DOI] [PubMed] [Google Scholar]
- 506.Perdomo OJ, Cavaillon JM, Huerre M, Ohayon H, Gounon P, Sansonetti PJ. 1994. Acute inflammation causes epithelial invasion and mucosal destruction in experimental shigellosis. J. Exp. Med. 180:1307–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Fasano A, Noriega FR, Maneval DR, Jr, Chanasongcram S, Russell R, Guandalini S, Levine MM. 1995. Shigella enterotoxin 1: an enterotoxin of Shigella flexneri 2a active in rabbit small intestine in vivo and in vitro. J. Clin. Invest. 95:2853–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Fasano A, Noriega FR, Liao FM, Wang W, Levine MM. 1997. Effect of Shigella enterotoxin 1 (ShET1) on rabbit intestine in vitro and in vivo. Gut 40:505–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Nataro JP, Seriwatana J, Fasano A, Maneval DR, Guers LD, Noriega F, Dubovsky F, Levine MM, Morris JG., Jr 1995. Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infect. Immun. 63:4721–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Farfán MJ, Toro CS, Barry EM, Nataro JP. 2011. Shigella enterotoxin-2 is a type III effector that participates in Shigella-induced interleukin 8 secretion by epithelial cells. FEMS Immunol. Med. Microbiol. 61:332–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Vargas M, Gascon J, Jimenez De Anta MT, Vila J. 1999. Prevalence of Shigella enterotoxins 1 and 2 among Shigella strains isolated from patients with traveler's diarrhea. J. Clin. Microbiol. 37:3608–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Yavzori M, Cohen D, Orr N. 2002. Prevalence of the genes for Shigella enterotoxins 1 and 2 among clinical isolates of Shigella in Israel. Epidemiol. Infect. 128:533–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Behrens M, Sheikh J, Nataro JP. 2002. Regulation of the overlapping pic/set locus in Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 70:2915–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Faherty C, Harper JM, Shea-Donohue T, Barry EM, Kaper JB, Fasano A, Nataro JP. 2012. Chromosomal and plasmid-encoded factors of Shigella flexneri induce secretogenic activity ex vivo. PLoS One 7:e49980. 10.1371/journal.pone.0049980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Al-Hasani K, Henderson IR, Sakellaris H, Rajakumar K, Grant T, Nataro JP, Robins-Browne R, Adler B. 2000. The sigA gene which is borne on the she pathogenicity island of Shigella flexneri 2a encodes an exported cytopathic protease involved in intestinal fluid accumulation. Infect. Immun. 68:2457–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Ruiz-Perez F, Nataro JP. 25 April 2013. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell. Mol. Life Sci. 10.1007/s00018-013-1355-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Cherla RP, Lee S-Y, Tesh VL. 2003. Shiga toxins and apoptosis. FEMS Microbiol. Lett. 228:159–166 [DOI] [PubMed] [Google Scholar]
- 518.Johannes L, Römer W. 2010. Shiga toxins—from cell biology to biomedical applications. Nat. Rev. Microbiol. 8:105–116 [DOI] [PubMed] [Google Scholar]
- 519.Tesh VL. 2010. Induction of apoptosis by Shiga toxins. Future Microbiol. 5:431–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.McDonough MA, Butterton JR. 1999. Spontaneous tandem amplification and deletion of the Shiga toxin operon in Shigella dysenteriae 1. Mol. Microbiol. 34:1058–1069 [DOI] [PubMed] [Google Scholar]
- 521.Beutin L, Strauch E, Fischer I. 1999. Isolation of Shigella sonnei lysogenic for a bacteriophage encoding gene for production of Shiga toxin. Lancet 353:1498. [DOI] [PubMed] [Google Scholar]
- 522.Bando SY, Moreno ACR, Albuquerque JAT, Amhaz JMK, Moreira-Filho CA, Martinez MB. 2010. Expression of bacterial virulence factors and cytokines during in vitro macrophage infection by enteroinvasive Escherichia coli and Shigella flexneri: a comparative study. Mem. Inst. Oswaldo Cruz 105:786–791 [DOI] [PubMed] [Google Scholar]
- 523.Moreno ACR, Ferreira LG, Martinez MB. 2009. Enteroinvasive Escherichia coli vs. Shigella flexneri: how different patterns of gene expression affect virulence. FEMS Microbiol. Lett. 301:156–163 [DOI] [PubMed] [Google Scholar]
- 524.Niyogi SK. 2005. Shigellosis. J. Microbiol. 43:133–143 [PubMed] [Google Scholar]
- 525.World Health Organization 2005. Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1. WHO Press, Geneva, Switzerland [Google Scholar]
- 526.Butler T. 2012. Haemolytic uraemic syndrome during shigellosis. Trans. R. Soc. Trop. Med. Hyg. 106:395–399 [DOI] [PubMed] [Google Scholar]
- 527.Mark Taylor C. 2008. Enterohaemorrhagic Escherichia coli and Shigella dysenteriae type 1-induced haemolytic uraemic syndrome. Pediatr. Nephrol. 23:1425–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Khan WA, Dhar U, Salam MA, Griffiths JK, Rand W, Bennish ML. 1999. Central nervous system manifestations of childhood shigellosis: prevalence, risk factors, and outcome. Pediatrics 103:E18. [DOI] [PubMed] [Google Scholar]
- 529.Struelens MJ, Patte D, Kabir I, Salam A, Nath SK, Butler T. 1985. Shigella septicemia: prevalence, presentation, risk factors, and outcome. J. Infect. Dis. 152:784–790 [DOI] [PubMed] [Google Scholar]
- 530.Ji S, Park H, Lee D, Song YK, Choi JP, Lee S-I. 2005. Post-infectious irritable bowel syndrome in patients with Shigella infection. J. Gastroenterol. Hepatol. 20:381–386 [DOI] [PubMed] [Google Scholar]
- 531.Ephros M, Cohen D, Yavzori M, Rotman N, Novic B, Ashkenazi S. 1996. Encephalopathy associated with enteroinvasive Escherichia coli 0144:NM infection. J. Clin. Microbiol. 34:2432–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Kopecko DJ. 1994. Experimental keratoconjunctivitis (Sereny) assay. Methods Enzymol. 235:39–47 [DOI] [PubMed] [Google Scholar]
- 533.Wood PK, Morris JG, Jr, Small PL, Sethabutr O, Toledo MR, Trabulsi L, Kaper JB. 1986. Comparison of DNA probes and the Sereny test for identification of invasive Shigella and Escherichia coli strains. J. Clin. Microbiol. 24:498–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Oaks EV, Wingfield ME, Formal SB. 1985. Plaque formation by virulent Shigella flexneri. Infect. Immun. 48:124–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Panchalingam S, Antonio M, Hossain A, Mandomando I, Ochieng B, Oundo J, Ramamurthy T, Tamboura B, Zaidi AKM, Petri W, Houpt E, Murray P, Prado V, Vidal R, Steele D, Strockbine N, Sansonetti P, Glass RI, Robins-Browne RM, Tauschek M, Svennerholm A-M, Kotloff K, Levine MM, Nataro JP. 2012. Diagnostic microbiologic methods in the GEMS-1 case/control study. Clin. Infect. Dis. 55(Suppl 4):S294–S302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Venkatesan MM, Buysse JM, Kopecko DJ. 1989. Use of Shigella flexneri ipaC and ipaH gene sequences for the general identification of Shigella spp. and enteroinvasive Escherichia coli. J. Clin. Microbiol. 27:2687–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Barletta F, Ochoa TJ, Cleary TG. 2013. Multiplex real-time PCR (MRT-PCR) for diarrheagenic. Methods Mol. Biol. 943:307–314 [DOI] [PubMed] [Google Scholar]
- 538.Cunningham SA, Sloan LM, Nyre LM, Vetter EA, Mandrekar J, Patel R. 2010. Three-hour molecular detection of Campylobacter, Salmonella, Yersinia, and Shigella species in feces with accuracy as high as that of culture. J. Clin. Microbiol. 48:2929–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, Verweij JJ, Taniuchi M, Sobuz SU, Haque R, Haverstick DM, Houpt ER. 2013. A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J. Clin. Microbiol. 51:472–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Coimbra RS, Grimont F, Grimont PA. 1999. Identification of Shigella serotypes by restriction of amplified O-antigen gene cluster. Res. Microbiol. 150:543–553 [DOI] [PubMed] [Google Scholar]
- 541.Coimbra RS, Artiguenave F, Jacques LSRZ, Oliveira GC. 2010. MST (molecular serotyping tool): a program for computer-assisted molecular identification of Escherichia coli and Shigella O antigens. J. Clin. Microbiol. 48:1921–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Sun Q, Lan R, Wang Y, Zhao A, Zhang S, Wang J, Wang Y, Xia S, Jin D, Cui Z, Zhao H, Li Z, Ye C, Zhang S, Jing H, Xu J. 2011. Development of a multiplex PCR assay targeting O-antigen modification genes for molecular serotyping of Shigella flexneri. J. Clin. Microbiol. 49:3766–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Hayford AE, Mammel MK, Lacher DW, Brown EW. 2011. Single nucleotide polymorphism (SNP)-based differentiation of Shigella isolates by pyrosequencing. Infect. Genet. Evol. 11:1761–1768 [DOI] [PubMed] [Google Scholar]
- 544.McDonnell J, Dallman T, Atkin S, Turbitt DA, Connor TR, Grant KA, Thomson NR, Jenkins C. 2013. Retrospective analysis of whole genome sequencing compared to prospective typing data in further informing the epidemiological investigation of an outbreak of Shigella sonnei in the UK. Epidemiol. Infect. 21:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Nato F, Phalipon A, Nguyen TLP, Diep TT, Sansonetti P, Germani Y. 2007. Dipstick for rapid diagnosis of Shigella flexneri 2a in stool. PLoS One 2:e361. 10.1371/journal.pone.0000361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Taneja N, Nato F, Dartevelle S, Sire JM, Garin B, Thi Phuong LN, Diep TT, Shako JC, Bimet F, Filliol I, Muyembe J-J, Ungeheuer MN, Ottone C, Sansonetti P, Germani Y. 2011. Dipstick test for rapid diagnosis of Shigella dysenteriae 1 in bacterial cultures and its potential use on stool samples. PLoS One 6:e24830. 10.1371/journal.pone.0024830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Van den Beld MJC, Reubsaet FAG. 2012. Differentiation between Shigella, enteroinvasive Escherichia coli (EIEC) and noninvasive Escherichia coli. Eur. J. Clin. Microbiol. Infect. Dis. 31:899–904 [DOI] [PubMed] [Google Scholar]
- 548.DuPont HL. 2009. Clinical practice. Bacterial diarrhea. N. Engl. J. Med. 361:1560–1569 [DOI] [PubMed] [Google Scholar]
- 549.Salam MA, Bennish ML. 1991. Antimicrobial therapy for shigellosis. Rev. Infect. Dis. 13(Suppl 4):S332–S341 [DOI] [PubMed] [Google Scholar]
- 550.Bennish ML, Khan WA, Begum M, Bridges EA, Ahmed S, Saha D, Salam MA, Acheson D, Ryan ET. 2006. Low risk of hemolytic uremic syndrome after early effective antimicrobial therapy for Shigella dysenteriae type 1 infection in Bangladesh. Clin. Infect. Dis. 42:356–362 [DOI] [PubMed] [Google Scholar]
- 551.Feng P. 2012. Pathogenic E. coli group, p 69–81 In Bad bug book, 2nd ed. Center for Food Safety and Applied Nutrition, Food and Drug Administration, Washington, DC [Google Scholar]
- 552.Bhandari N, Mazumder S, Taneja S, Dube B, Agarwal RC, Mahalanabis D, Fontaine O, Black RE, Bhan MK. 2008. Effectiveness of zinc supplementation plus oral rehydration salts compared with oral rehydration salts alone as a treatment for acute diarrhea in a primary care setting: a cluster randomized trial. Pediatrics 121:e1279–e1285 [DOI] [PubMed] [Google Scholar]
- 553.Alvarez-Acosta T, León C, Acosta-González S, Parra-Soto H, Cluet-Rodriguez I, Rossell MR, Colina-Chourio JA. 2009. Beneficial role of green plantain [Musa paradisiaca] in the management of persistent diarrhea: a prospective randomized trial. J. Am. Coll. Nutr. 28:169–176 [DOI] [PubMed] [Google Scholar]
- 554.Rabbani GH, Ahmed S, Hossain I, Islam R, Marni F, Akhtar M, Majid N. 2009. Green banana reduces clinical severity of childhood shigellosis: a double-blind, randomized, controlled clinical trial. Pediatr. Infect. Dis. J. 28:420–425 [DOI] [PubMed] [Google Scholar]
- 555.Niyogi SK. 2007. Increasing antimicrobial resistance–an emerging problem in the treatment of shigellosis. Clin. Microbiol. Infect. 13:1141–1143 [DOI] [PubMed] [Google Scholar]
- 556.Cooper ML, Keller HM. 1950. Dysentery due to sulfonamide-resistant Shigella sonnei controlled with chloromycetin. Ama Am. J. Dis. Child. 80:911–920 [DOI] [PubMed] [Google Scholar]
- 557.Griffin PM, Tauxe RV, Redd SC, Puhr ND, Hargrett-Bean N, Blake PA. 1989. Emergence of highly trimethoprim-sulfamethoxazole-resistant Shigella in a native American population: an epidemiologic study. Am. J. Epidemiol. 129:1042–1051 [DOI] [PubMed] [Google Scholar]
- 558.Gross RJ, Rowe B, Cheasty T, Thomas LV. 1981. Increase in drug resistance among Shigella dysenteriae, Sh flexneri, and Sh boydii. Br. Med. J. Clin. Res. Ed. 283:575–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Panhotra BR, Desai B, Sharma PL. 1985. Nalidixic-acid-resistant Shigella dysenteriae I. Lancet i:763. [DOI] [PubMed] [Google Scholar]
- 560.Ross S, Controni G, Khan W. 1972. Resistance of Shigellae to ampicillin and other antibiotics. Its clinical and epidemiological implications. JAMA 221:45–47 [PubMed] [Google Scholar]
- 561.Hartman AB, Essiet II, Isenbarger DW, Lindler LE. 2003. Epidemiology of tetracycline resistance determinants in Shigella spp. and enteroinvasive Escherichia coli: characterization and dissemination of tet(A)-1. J. Clin. Microbiol. 41:1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Ahmed AM, Miyoshi S, Shinoda S, Shimamoto T. 2005. Molecular characterization of a multidrug-resistant strain of enteroinvasive Escherichia coli O164 isolated in Japan. J. Med. Microbiol. 54:273–278 [DOI] [PubMed] [Google Scholar]
- 563.Gu B, Cao Y, Pan S, Zhuang L, Yu R, Peng Z, Qian H, Wei Y, Zhao L, Liu G, Tong M. 2012. Comparison of the prevalence and changing resistance to nalidixic acid and ciprofloxacin of Shigella between Europe-America and Asia-Africa from 1998 to 2009. Int. J. Antimicrob. Agents 40:9–17 [DOI] [PubMed] [Google Scholar]
- 564.Zhang W, Luo Y, Li J, Lin L, Ma Y, Hu C, Jin S, Ran L, Cui S. 2011. Wide dissemination of multidrug-resistant Shigella isolates in China. J. Antimicrob. Chemother. 66:2527–2535 [DOI] [PubMed] [Google Scholar]
- 565.Goel N, Wattal C, Kaul D, Khanna VK. 2013. Emergence of ceftriaxone resistant Shigella. Indian J. Pediatr. 80:70–71 [DOI] [PubMed] [Google Scholar]
- 566.Sjölund Karlsson M, Bowen A, Reporter R, Folster JP, Grass JE, Howie RL, Taylor J, Whichard JM. 2013. Outbreak of infections caused by Shigella sonnei with reduced susceptibility to azithromycin in the United States. Antimicrob. Agents Chemother. 57:1559–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Pazhani GP, Niyogi SK, Singh AK, Sen B, Taneja N, Kundu M, Yamasaki S, Ramamurthy T. 2008. Molecular characterization of multidrug-resistant Shigella species isolated from epidemic and endemic cases of shigellosis in India. J. Med. Microbiol. 57:856–863 [DOI] [PubMed] [Google Scholar]
- 568.Luck SN, Turner SA, Rajakumar K, Sakellaris H, Adler B. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69:6012–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Duarte MCT, Leme EE, Delarmelina C, Soares AA, Figueira GM, Sartoratto A. 2007. Activity of essential oils from Brazilian medicinal plants on Escherichia coli. J. Ethnopharmacol. 111:197–201 [DOI] [PubMed] [Google Scholar]
- 570.Raja SB, Murali MR, Kumar NK, Devaraj SN. 2011. Isolation and partial characterisation of a novel lectin from Aegle marmelos fruit and its effect on adherence and invasion of Shigellae to HT29 cells. PLoS One 6:e16231. 10.1371/journal.pone.0016231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Camacho AI, Irache JM, Gamazo C. 2013. Recent progress towards development of a Shigella vaccine. Expert Rev. Vaccines 12:43–55 [DOI] [PubMed] [Google Scholar]
- 572.Kweon M-N. 2008. Shigellosis: the current status of vaccine development. Curr. Opin. Infect. Dis. 21:313–318 [DOI] [PubMed] [Google Scholar]
- 573.Pavot V, Rochereau N, Genin C, Verrier B, Paul S. 2012. New insights in mucosal vaccine development. Vaccine 30:142–154 [DOI] [PubMed] [Google Scholar]
- 574.Nataro JP, Kaper JB, Robins-Browne R, Prado V, Vial P, Levine MM. 1987. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr. Infect. Dis. J. 6:829–831 [DOI] [PubMed] [Google Scholar]
- 575.Bhan MK, Raj P, Levine MM, Kaper JB, Bhandari N, Srivastava R, Kumar R, Sazawal S. 1989. Enteroaggregative Escherichia coli associated with persistent diarrhea in a cohort of rural children in India. J. Infect. Dis. 159:1061–1064 [DOI] [PubMed] [Google Scholar]
- 576.Cohen MB, Nataro JP, Bernstein DI, Hawkins J, Roberts N, Staat MA. 2005. Prevalence of diarrheagenic Escherichia coli in acute childhood enteritis: a prospective controlled study. J. Pediatr. 146:54–61 [DOI] [PubMed] [Google Scholar]
- 577.Nataro JP, Mai V, Johnson J, Blackwelder WC, Heimer R, Tirrell S, Edberg SC, Braden CR, Glenn Morris J, Jr, Hirshon JM. 2006. Diarrheagenic Escherichia coli infection in Baltimore, Maryland, and New Haven, Connecticut. Clin. Infect. Dis. 43:402–407 [DOI] [PubMed] [Google Scholar]
- 578.Wanke CA, Schorling JB, Barrett LJ, Desouza MA, Guerrant RL. 1991. Potential role of adherence traits of Escherichia coli in persistent diarrhea in an urban Brazilian slum. Pediatr. Infect. Dis. J. 10:746–751 [DOI] [PubMed] [Google Scholar]
- 579.Mathewson JJ, Salameh BM, DuPont HL, Jiang ZD, Nelson AC, Arduino R, Smith MA, Masozera N. 1998. HEp-2 cell-adherent Escherichia coli and intestinal secretory immune response to human immunodeficiency virus (HIV) in outpatients with HIV-associated diarrhea. Clin. Diagn. Lab. Immunol. 5:87–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Okhuysen PC, Dupont HL. 2010. Enteroaggregative Escherichia coli (EAEC): a cause of acute and persistent diarrhea of worldwide importance. J. Infect. Dis. 202:503–505 [DOI] [PubMed] [Google Scholar]
- 581.Baudry B, Savarino SJ, Vial P, Kaper JB, Levine MM. 1990. A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli, a recently discovered diarrheal pathogen. J. Infect. Dis. 161:1249–1251 [DOI] [PubMed] [Google Scholar]
- 582.Schmidt H, Knop C, Franke S, Aleksic S, Heesemann J, Karch H. 1995. Development of PCR for screening of enteroaggregative Escherichia coli. J. Clin. Microbiol. 33:701–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Harrington SM, Dudley EG, Nataro JP. 2006. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol. Lett. 254:12–18 [DOI] [PubMed] [Google Scholar]
- 584.Jenkins C, Chart H, Willshaw GA, Cheasty T, Smith HR. 2006. Genotyping of enteroaggregative Escherichia coli and identification of target genes for the detection of both typical and atypical strains. Diagn. Microbiol. Infect. Dis. 55:13–19 [DOI] [PubMed] [Google Scholar]
- 585.Estrada-Garcia T, Navarro-Garcia F. 2012. Enteroaggregative Escherichia coli pathotype: a genetically heterogeneous emerging foodborne enteropathogen. FEMS Immunol. Med. Microbiol. 66:281–298 [DOI] [PubMed] [Google Scholar]
- 586.Regua-Mangia AH, Gomes TAT, Vieira MAM, Andrade JRC, Irino K, Teixeira LM. 2004. Frequency and characteristics of diarrhoeagenic Escherichia coli strains isolated from children with and without diarrhoea in Rio de Janeiro, Brazil. J. Infect. 48:161–167 [DOI] [PubMed] [Google Scholar]
- 587.Pai M, Kang G, Ramakrishna BS, Venkataraman A, Muliyil J. 1997. An epidemic of diarrhoea in south India caused by enteroaggregative Escherichia coli. Indian J. Med. Res. 106:7–12 [PubMed] [Google Scholar]
- 588.Boisen N, Scheutz F, Rasko DA, Redman JC, Persson S, Simon J, Kotloff KL, Levine MM, Sow S, Tamboura B, Toure A, Malle D, Panchalingam S, Krogfelt KA, Nataro JP. 2012. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J. Infect. Dis. 205:431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Dow MA, Tóth I, Malik A, Herpay M, Nógrády N, Ghenghesh KS, Nagy B. 2006. Phenotypic and genetic characterization of enteropathogenic Escherichia coli (EPEC) and entero-aggregative E. coli (EAEC) from diarrhoeal and non-diarrhoeal children in Libya. Comp. Immunol. Microbiol. Infect. Dis. 29:100–113 [DOI] [PubMed] [Google Scholar]
- 590.Grad YH, Lipsitch M, Feldgarden M, Arachchi HM, Cerqueira GC, Fitzgerald M, Godfrey P, Haas BJ, Murphy CI, Russ C, Sykes S, Walker BJ, Wortman JR, Young S, Zeng Q, Abouelleil A, Bochicchio J, Chauvin S, Desmet T, Gujja S, McCowan C, Montmayeur A, Steelman S, Frimodt-Møller J, Petersen AM, Struve C, Krogfelt KA, Bingen E, Weill F-X, Lander ES, Nusbaum C, Birren BW, Hung DT, Hanage WP. 2012. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc. Natl. Acad. Sci. U. S. A. 109:3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. 10.1371/journal.pone.0022751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Laing CR, Zhang Y, Gilmour MW, Allen V, Johnson R, Thomas JE, Gannon VPJ. 2012. A comparison of Shiga-toxin 2 bacteriophage from classical enterohemorrhagic Escherichia coli serotypes and the German E. coli O104:H4 outbreak strain. PLoS One 7:e37362. 10.1371/journal.pone.0037362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Dallman T, Smith GP, O'Brien B, Chattaway MA, Finlay D, Grant KA, Jenkins C. 2012. Characterization of a verocytotoxin-producing enteroaggregative Escherichia coli serogroup O111:H21 strain associated with a household outbreak in Northern Ireland. J. Clin. Microbiol. 50:4116–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Mossoro C, Glaziou P, Yassibanda S, Lan NTP, Bekondi C, Minssart P, Bernier C, Le Bouguénec C, Germani Y. 2002. Chronic diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome associated with HEp-2 adherent Escherichia coli in adults infected with human immunodeficiency virus in Bangui, Central African Republic. J. Clin. Microbiol. 40:3086–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Tompkins DS, Hudson MJ, Smith HR, Eglin RP, Wheeler JG, Brett MM, Owen RJ, Brazier JS, Cumberland P, King V, Cook PE. 1999. A study of infectious intestinal disease in England: microbiological findings in cases and controls. Commun. Dis. Public Health 2:108–113 [PubMed] [Google Scholar]
- 596.Koo HL, Jiang Z-D, Brown E, Garcia C, Qi H, Dupont HL. 2008. Coliform contamination of vegetables obtained from popular restaurants in Guadalajara, Mexico, and Houston, Texas. Clin. Infect. Dis. 47:218–221 [DOI] [PubMed] [Google Scholar]
- 597.Okeke IN, Nataro JP. 2001. Enteroaggregative Escherichia coli. Lancet Infect. Dis. 1:304–313 [DOI] [PubMed] [Google Scholar]
- 598.Vigil KJ, Jiang Z-D, Chen JJ, Palumbo KL, Galbadage T, Brown EL, Yiang J, Koo H, DuPont MW, Ericsson C, Adachi JA, DuPont HL. 2009. Coliform and Escherichia coli contamination of desserts served in public restaurants from Guadalajara, Mexico, and Houston, Texas. Am. J. Trop. Med. Hyg. 80:606–608 [PubMed] [Google Scholar]
- 599.Oundo JO, Kariuki SM, Boga HI, Muli FW, Iijima Y. 2008. High incidence of enteroaggregative Escherichia coli among food handlers in three areas of Kenya: a possible transmission route of travelers' diarrhea. J. Travel Med. 15:31–38 [DOI] [PubMed] [Google Scholar]
- 600.Smith HR, Cheasty T, Rowe B. 1997. Enteroaggregative Escherichia coli and outbreaks of gastroenteritis in UK. Lancet 350:814–815 [DOI] [PubMed] [Google Scholar]
- 601.Jiang Z-D, Okhuysen PC, Guo D-C, He R, King TM, DuPont HL, Milewicz DM. 2003. Genetic susceptibility to enteroaggregative Escherichia coli diarrhea: polymorphism in the interleukin-8 promotor region. J. Infect. Dis. 188:506–511 [DOI] [PubMed] [Google Scholar]
- 602.Uber AP, Trabulsi LR, Irino K, Beutin L, Ghilardi ACR, Gomes TAT, Liberatore AMA, de Castro AFP, Elias WP. 2006. Enteroaggregative Escherichia coli from humans and animals differ in major phenotypical traits and virulence genes. FEMS Microbiol. Lett. 256:251–257 [DOI] [PubMed] [Google Scholar]
- 603.Buchholz U, Bernard H, Werber D, Böhmer MM, Remschmidt C, Wilking H, Deleré Y, an der Heiden M, Adlhoch C, Dreesman J, Ehlers J, Ethelberg S, Faber M, Frank C, Fricke G, Greiner M, Höhle M, Ivarsson S, Jark U, Kirchner M, Koch J, Krause G, Luber P, Rosner B, Stark K, Kühne M. 2011. German outbreak of Escherichia coli O104:H4 associated with sprouts. N. Engl. J. Med. 365:1763–1770 [DOI] [PubMed] [Google Scholar]
- 604.Auvray F, Dilasser F, Bibbal D, Kérourédan M, Oswald E, Brugère H. 2012. French cattle is not a reservoir of the highly virulent enteroaggregative Shiga toxin-producing Escherichia coli of serotype O104:H4. Vet. Microbiol. 158:443–445 [DOI] [PubMed] [Google Scholar]
- 605.Aldabe B, Delmas Y, Gault G, Vendrely B, Llanas B, Charron M, Castor C, Ong N, Weill F, Mariani-Kurkdjian P, Terrier F, Desjardin M, Simoes J, Le Bihan B, Combe C, Rolland P. 2011. Household transmission of haemolytic uraemic syndrome associated with Escherichia coli O104:H4, south-western France, June 2011. Euro Surveill. 16(31):pii=19934 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19934 [PubMed] [Google Scholar]
- 606.Januszkiewicz A, Szych J, Rastawicki W, Wołkowicz T, Chróst A, Leszczyńska B, KuŸma E, Roszkowska-Blaim M, Gierczyński R. 2012. Molecular epidemiology of a household outbreak of Shiga-toxin-producing Escherichia coli in Poland due to secondary transmission of STEC O104:H4 from Germany. J. Med. Microbiol. 61:552–558 [DOI] [PubMed] [Google Scholar]
- 607.Kuijper EJ, Soonawala D, Vermont C, van Dissel JT. 2011. Household transmission of haemolytic uraemic syndrome associated with Escherichia coli O104:H4 in the Netherlands, May 2011. Euro Surveill. 16(25):pii=19897 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19897 [PubMed] [Google Scholar]
- 608.Sin MA, Takla A, Flieger A, Prager R, Fruth A, Tietze E, Fink E, Korte J, Schink S, Höhle M, Eckmanns T. 2013. Carrier prevalence, secondary household transmission, and long-term shedding in 2 districts during the Escherichia coli O104:H4 outbreak in Germany, 2011. J. Infect. Dis. 207:432–438 [DOI] [PubMed] [Google Scholar]
- 609.Nataro JP, Deng Y, Cookson S, Cravioto A, Savarino SJ, Guers LD, Levine MM, Tacket CO. 1995. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J. Infect. Dis. 171:465–468 [DOI] [PubMed] [Google Scholar]
- 610.Weintraub A. 2007. Enteroaggregative Escherichia coli: epidemiology, virulence and detection. J. Med. Microbiol. 56:4–8 [DOI] [PubMed] [Google Scholar]
- 611.Nataro JP, Hicks S, Phillips AD, Vial PA, Sears CL. 1996. T84 cells in culture as a model for enteroaggregative Escherichia coli pathogenesis. Infect. Immun. 64:4761–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Hicks S, Candy DC, Phillips AD. 1996. Adhesion of enteroaggregative Escherichia coli to pediatric intestinal mucosa in vitro. Infect. Immun. 64:4751–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Chart H, Smith HR, Rowe B. 1995. Enteroaggregative strains of Escherichia coli belonging to serotypes O126:H27 and O44:H18 express antigenically similar 18 kDa outer membrane-associated proteins. FEMS Microbiol. Lett. 132:17–22 [DOI] [PubMed] [Google Scholar]
- 614.Debroy C, Yealy J, Wilson RA, Bhan MK, Kumar R. 1995. Antibodies raised against the outer membrane protein interrupt adherence of enteroaggregative Escherichia coli. Infect. Immun. 63:2873–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Mancini J, Weckselblatt B, Chung YK, Durante JC, Andelman S, Glaubman J, Dorff JD, Bhargava S, Lijek RS, Unger KP, Okeke IN. 2011. The heat-resistant agglutinin family includes a novel adhesin from enteroaggregative Escherichia coli strain 60A. J. Bacteriol. 193:4813–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Monteiro-Neto V, Bando SY, Moreira-Filho CA, Girón JA. 2003. Characterization of an outer membrane protein associated with haemagglutination and adhesive properties of enteroaggregative Escherichia coli O111:H12. Cell. Microbiol. 5:533–547 [DOI] [PubMed] [Google Scholar]
- 617.Wai SN, Takade A, Amako K. 1996. The hydrophobic surface protein layer of enteroaggregative Escherichia coli strains. FEMS Microbiol. Lett. 135:17–22 [DOI] [PubMed] [Google Scholar]
- 618.Knutton S, Shaw RK, Bhan MK, Smith HR, McConnell MM, Cheasty T, Williams PH, Baldwin TJ. 1992. Ability of enteroaggregative Escherichia coli strains to adhere in vitro to human intestinal mucosa. Infect. Immun. 60:2083–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Suzart S, Guth BE, Pedroso MZ, Okafor UM, Gomes TA. 2001. Diversity of surface structures and virulence genetic markers among enteroaggregative Escherichia coli (EAEC) strains with and without the EAEC DNA probe sequence. FEMS Microbiol. Lett. 201:163–168 [DOI] [PubMed] [Google Scholar]
- 620.Vial PA, Robins-Browne R, Lior H, Prado V, Kaper JB, Nataro JP, Maneval D, Elsayed A, Levine MM. 1988. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J. Infect. Dis. 158:70–79 [DOI] [PubMed] [Google Scholar]
- 621.Bernier C, Gounon P, Le Bouguénec C. 2002. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect. Immun. 70:4302–4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Boisen N, Struve C, Scheutz F, Krogfelt KA, Nataro JP. 2008. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect. Immun. 76:3281–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Czeczulin JR, Balepur S, Hicks S, Phillips A, Hall R, Kothary MH, Navarro-Garcia F, Nataro JP. 1997. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect. Immun. 65:4135–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Nataro JP, Deng Y, Maneval DR, German AL, Martin WC, Levine MM. 1992. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect. Immun. 60:2297–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 61:1267–1282 [DOI] [PubMed] [Google Scholar]
- 626.Nataro JP, Yikang D, Yingkang D, Walker K. 1994. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J. Bacteriol. 176:4691–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Cobeljić M, Miljković-Selimović B, Paunović-Todosijević D, Velicković Z, Lepsanović Z, Zec N, Savić D, Ilić R, Konstantinović S, Jovanović B, Kostić V. 1996. Enteroaggregative Escherichia coli associated with an outbreak of diarrhoea in a neonatal nursery ward. Epidemiol. Infect. 117:11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.Dudley EG, Abe C, Ghigo J-M, Latour-Lambert P, Hormazabal JC, Nataro JP. 2006. An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli strain C1096 to cultured cells and abiotic surfaces. Infect. Immun. 74:2102–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.Bhargava S, Johnson BB, Hwang J, Harris TA, George AS, Muir A, Dorff J, Okeke IN. 2009. Heat-resistant agglutinin 1 is an accessory enteroaggregative Escherichia coli colonization factor. J. Bacteriol. 191:4934–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Okeke IN, Lamikanra A, Czeczulin J, Dubovsky F, Kaper JB, Nataro JP. 2000. Heterogeneous virulence of enteroaggregative Escherichia coli strains isolated from children in Southwest Nigeria. J. Infect. Dis. 181:252–260 [DOI] [PubMed] [Google Scholar]
- 631.Lima IFN, Boisen N, Quetz Jda Havt SA, de Carvalho EB, Soares AM, Lima NL, Mota RMS, Nataro JP, Guerrant RL, Lima  AM. 2013. Prevalence of enteroaggregative Escherichia coli and its virulence-related genes in a case-control study among children from north-eastern Brazil. J. Med. Microbiol. 62:683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Huang DB, Nataro JP, DuPont HL, Kamat PP, Mhatre AD, Okhuysen PC, Chiang T. 2006. Enteroaggregative Escherichia coli is a cause of acute diarrheal illness: a meta-analysis. Clin. Infect. Dis. 43:556–563 [DOI] [PubMed] [Google Scholar]
- 633.Navarro-Garcia F, Elias WP.2011. Autotransporters and virulence of enteroaggregative E. coli. Gut Microbes 2:13–24 [DOI] [PubMed] [Google Scholar]
- 634.Vila J, Gene A, Vargas M, Gascon J, Latorre C, Jimenez de Anta MT. 1998. A case-control study of diarrhoea in children caused by Escherichia coli producing heat-stable enterotoxin (EAST-1). J. Med. Microbiol. 47:889–891 [DOI] [PubMed] [Google Scholar]
- 635.Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. 1999. Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67:5587–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Navarro-Garcia F, Sonnested M, Teter K. 2010. Host-toxin interactions involving EspC and Pet, two serine protease autotransporters of the Enterobacteriaceae. Toxins 2:1134–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Czeczulin JR, Whittam TS, Henderson IR, Navarro-Garcia F, Nataro JP. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67:2692–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Sheikh J, Czeczulin JR, Harrington S, Hicks S, Henderson IR, Le Bouguénec C, Gounon P, Phillips A, Nataro JP. 2002. A novel dispersin protein in enteroaggregative Escherichia coli. J. Clin. Invest. 110:1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083–1091 [DOI] [PubMed] [Google Scholar]
- 640.Sheikh J, Hicks S, Dall'Agnol M, Phillips AD, Nataro JP. 2001. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 41:983–997 [DOI] [PubMed] [Google Scholar]
- 641.Morin N, Tirling C, Ivison SM, Kaur AP, Nataro JP, Steiner TS. 2010. Autoactivation of the AggR regulator of enteroaggregative Escherichia coli in vitro and in vivo. FEMS Immunol. Med. Microbiol. 58:344–355 [DOI] [PubMed] [Google Scholar]
- 642.Edwards LA, Bajaj-Elliott M, Klein NJ, Murch SH, Phillips AD. 2011. Bacterial-epithelial contact is a key determinant of host innate immune responses to enteropathogenic and enteroaggregative Escherichia coli. PLoS One 6:e27030. 10.1371/journal.pone.0027030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Khan K, Konar M, Goyal A, Ghosh S. 2010. Enteroaggregative Escherichia coli infection induces IL-8 production via activation of mitogen-activated protein kinases and the transcription factors NF-κB and AP-1 in INT-407 cells. Mol. Cell. Biochem. 337:17–24 [DOI] [PubMed] [Google Scholar]
- 644.Konar M, Sachin O, Priya A, Ghosh S. 2012. Identification of key proteins of cultured human intestinal cells involved in interaction with enteroaggregative Escherichia coli. FEMS Immunol. Med. Microbiol. 66:177–190 [DOI] [PubMed] [Google Scholar]
- 645.Jin S, Song YC, Emili A, Sherman PM, Chan VL. 2003. JlpA of Campylobacter jejuni interacts with surface-exposed heat shock protein 90α and triggers signalling pathways leading to the activation of NF-κB and p38 MAP kinase in epithelial cells. Cell. Microbiol. 5:165–174 [DOI] [PubMed] [Google Scholar]
- 646.Harrington SM, Strauman MC, Abe CM, Nataro JP. 2005. Aggregative adherence fimbriae contribute to the inflammatory response of epithelial cells infected with enteroaggregative Escherichia coli. Cell. Microbiol. 7:1565–1578 [DOI] [PubMed] [Google Scholar]
- 647.Boll EJ, Struve C, Sander A, Demma Z, Krogfelt KA, McCormick BA. 2012. Enteroaggregative Escherichia coli promotes transepithelial migration of neutrophils through a conserved 12-lipoxygenase pathway. Cell. Microbiol. 14:120–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Mumy KL, Bien JD, Pazos MA, Gronert K, Hurley BP, McCormick BA. 2008. Distinct isoforms of phospholipase A2 mediate the ability of Salmonella enterica serotype Typhimurium and Shigella flexneri to induce the transepithelial migration of neutrophils. Infect. Immun. 76:3614–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Pazos M, Siccardi D, Mumy KL, Bien JD, Louie S, Shi HN, Gronert K, Mrsny RJ, McCormick BA. 2008. Multidrug resistance-associated transporter 2 regulates mucosal inflammation by facilitating the synthesis of hepoxilin A3. J. Immunol. 181:8044–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Adachi JA, Ericsson CD, Jiang Z-D, DuPont MW, Pallegar SR, DuPont HL. 2002. Natural history of enteroaggregative and enterotoxigenic Escherichia coli infection among US travelers to Guadalajara, Mexico. J. Infect. Dis. 185:1681–1683 [DOI] [PubMed] [Google Scholar]
- 651.Nataro JP. 2005. Enteroaggregative Escherichia coli pathogenesis. Curr. Opin. Gastroenterol. 21:4–8 [PubMed] [Google Scholar]
- 652.Paul M, Tsukamoto T, Ghosh AR, Bhattacharya SK, Manna B, Chakrabarti S, Nair GB, Sack DA, Sen D, Takeda Y. 1994. The significance of enteroaggregative Escherichia coli in the etiology of hospitalized diarrhoea in Calcutta, India and the demonstration of a new honey-combed pattern of aggregative adherence. FEMS Microbiol. Lett. 117:319–325 [DOI] [PubMed] [Google Scholar]
- 653.Al-Gallas N, Bahri O, Bouratbeen A, Ben Haasen A, Ben Aissa R. 2007. Etiology of acute diarrhea in children and adults in Tunis, Tunisia, with emphasis on diarrheagenic Escherichia coli: prevalence, phenotyping, and molecular epidemiology. Am. J. Trop. Med. Hyg. 77:571–582 [PubMed] [Google Scholar]
- 654.Denno DM, Shaikh N, Stapp JR, Qin X, Hutter CM, Hoffman V, Mooney JC, Wood KM, Stevens HJ, Jones R, Tarr PI, Klein EJ. 2012. Diarrhea etiology in a pediatric emergency department: a case control study. Clin. Infect. Dis. 55:897–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Okeke IN, Ojo O, Lamikanra A, Kaper JB. 2003. Etiology of acute diarrhea in adults in southwestern Nigeria. J. Clin. Microbiol. 41:4525–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Cárcamo C, Hooton T, Wener MH, Weiss NS, Gilman R, Arevalo J, Carrasco J, Seas C, Caballero M, Holmes KK. 2005. Etiologies and manifestations of persistent diarrhea in adults with HIV-1 infection: a case-control study in Lima, Peru. J. Infect. Dis. 191:11–19 [DOI] [PubMed] [Google Scholar]
- 657.Johnson S, Hendson W, Crewe-Brown H, Dini L, Frean J, Perovic O, Vardas E. 2000. Effect of human immunodeficiency virus infection on episodes of diarrhea among children in South Africa. Pediatr. Infect. Dis. J. 19:972–979 [DOI] [PubMed] [Google Scholar]
- 658.Lima AA, Guerrant RL. 1992. Persistent diarrhea in children: epidemiology, risk factors, pathophysiology, nutritional impact, and management. Epidemiol. Rev. 14:222–242 [DOI] [PubMed] [Google Scholar]
- 659.Guerrant RL, Oriá RB, Moore SR, Oriá MOB, Lima AAM. 2008. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr. Rev. 66:487–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Steiner TS, Lima AA, Nataro JP, Guerrant RL. 1998. Enteroaggregative Escherichia coli produce intestinal inflammation and growth impairment and cause interleukin-8 release from intestinal epithelial cells. J. Infect. Dis. 177:88–96 [DOI] [PubMed] [Google Scholar]
- 661.Thabane M, Kottachchi DT, Marshall JK. 2007. Systematic review and meta-analysis: the incidence and prognosis of post-infectious irritable bowel syndrome. Aliment. Pharmacol. Ther. 26:535–544 [DOI] [PubMed] [Google Scholar]
- 662.Okhuysen PC, Jiang ZD, Carlin L, Forbes C, DuPont HL. 2004. Post-diarrhea chronic intestinal symptoms and irritable bowel syndrome in North American travelers to Mexico. Am. J. Gastroenterol. 99:1774–1778 [DOI] [PubMed] [Google Scholar]
- 663.Aslani MM, Alikhani MY, Zavari A, Yousefi R, Zamani AR. 2011. Characterization of enteroaggregative Escherichia coli (EAEC) clinical isolates and their antibiotic resistance pattern. Int. J. Infect. Dis. 15:e136–e139 [DOI] [PubMed] [Google Scholar]
- 664.Cerna JF, Nataro JP, Estrada-Garcia T. 2003. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. J. Clin. Microbiol. 41:2138–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Jenkins C, Tembo M, Chart H, Cheasty T, Willshaw GA, Phillips AD, Tompkins D, Smith H. 2006. Detection of enteroaggregative Escherichia coli in faecal samples from patients in the community with diarrhoea. J. Med. Microbiol. 55:1493–1497 [DOI] [PubMed] [Google Scholar]
- 666.Cennimo D, Abbas A, Huang DB, Chiang T. 2009. The prevalence and virulence characteristics of enteroaggregative Escherichia coli at an urgent-care clinic in the U. S. A.: a case-control study. J. Med. Microbiol. 58:403–407 [DOI] [PubMed] [Google Scholar]
- 667.Huang DB, Mohamed JA, Nataro JP, DuPont HL, Jiang Z-D, Okhuysen PC. 2007. Virulence characteristics and the molecular epidemiology of enteroaggregative Escherichia coli isolates from travellers to developing countries. J. Med. Microbiol. 56:1386–1392 [DOI] [PubMed] [Google Scholar]
- 668.Nishi J, Sheikh J, Mizuguchi K, Luisi B, Burland V, Boutin A, Rose DJ, Blattner FR, Nataro JP. 2003. The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J. Biol. Chem. 278:45680–45689 [DOI] [PubMed] [Google Scholar]
- 669.Monteiro BT, Campos LC, Sircili MP, Franzolin MR, Bevilacqua LF, Nataro JP, Elias WP. 2009. The dispersin-encoding gene (aap) is not restricted to enteroaggregative Escherichia coli. Diagn. Microbiol. Infect. Dis. 65:81–84 [DOI] [PubMed] [Google Scholar]
- 670.Flores J, Okhuysen PC. 2009. Enteroaggregative Escherichia coli infection. Curr. Opin. Gastroenterol. 25:8–11 [DOI] [PubMed] [Google Scholar]
- 671.Ouyang-Latimer J, Jafri S, VanTassel A, Jiang Z-D, Gurleen K, Rodriguez S, Nandy RK, Ramamurthy T, Chatterjee S, McKenzie R, Steffen R, DuPont HL. 2011. In vitro antimicrobial susceptibility of bacterial enteropathogens isolated from international travelers to Mexico, Guatemala, and India from 2006 to 2008. Antimicrob. Agents Chemother. 55:874–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 672.Raju B, Ballal M. 2009. Multidrug resistant enteroaggregative Escherichia coli diarrhoea in rural southern Indian population. Scand. J. Infect. Dis. 41:105–108 [DOI] [PubMed] [Google Scholar]
- 673.Mortensen NP, Fowlkes JD, Maggart M, Doktycz MJ, Nataro JP, Drusano G, Allison DP. 2011. Effects of sub-minimum inhibitory concentrations of ciprofloxacin on enteroaggregative Escherichia coli and the role of the surface protein dispersin. Int. J. Antimicrob. Agents 38:27–34 [DOI] [PubMed] [Google Scholar]
- 674.Dupont HL, Jiang Z-D, Belkind-Gerson J, Okhuysen PC, Ericsson CD, Ke S, Huang DB, Dupont MW, Adachi JA, De La Cabada FJ, Taylor DN, Jaini S, Martinez Sandoval F. 2007. Treatment of travelers' diarrhea: randomized trial comparing rifaximin, rifaximin plus loperamide, and loperamide alone. Clin. Gastroenterol. Hepatol. 5:451–456 [DOI] [PubMed] [Google Scholar]
- 675.Ericsson CD, DuPont HL, Okhuysen PC, Jiang Z-D, DuPont MW. 2007. Loperamide plus azithromycin more effectively treats travelers' diarrhea in Mexico than azithromycin alone. J. Travel Med. 14:312–319 [DOI] [PubMed] [Google Scholar]
- 676.Pereira AC, Britto-Filho JD, José de Carvalho J, de Luna M, Rosa AC. 2008. Enteroaggregative Escherichia coli (EAEC) strains enter and survive within cultured intestinal epithelial cells. Microb. Pathog. 45:310–314 [DOI] [PubMed] [Google Scholar]
- 677.Hauswaldt S, Nitschke M, Sayk F, Solbach W, Knobloch JK-M. 2013. Lessons learned from outbreaks of Shiga toxin producing Escherichia coli. Curr. Infect. Dis. Rep. 15:4–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678.Lapeyraque A-L, Malina M, Fremeaux-Bacchi V, Boppel T, Kirschfink M, Oualha M, Proulx F, Clermont M-J, Le Deist F, Niaudet P, Schaefer F. 2011. Eculizumab in severe Shiga-toxin-associated HUS. N. Engl. J. Med. 364:2561–2563 [DOI] [PubMed] [Google Scholar]
- 679.Menne J, Nitschke M, Stingele R, Abu-Tair M, Beneke J, Bramstedt J, Bremer JP, Brunkhorst R, Busch V, Dengler R, Deuschl G, Fellermann K, Fickenscher H, Gerigk C, Goettsche A, Greeve J, Hafer C, Hagenmüller F, Haller H, Herget-Rosenthal S, Hertenstein B, Hofmann C, Lang M, Kielstein JT, Klostermeier UC, Knobloch J, Kuehbacher M, Kunzendorf U, Lehnert H, Manns MP, Menne TF, Meyer TN, Michael C, Münte T, Neumann-Grutzeck C, Nuernberger J, Pavenstaedt H, Ramazan L, Renders L, Repenthin J, Ries W, Rohr A, Rump LC, Samuelsson O, Sayk F, Schmidt BMW, Schnatter S, Schöcklmann H, Schreiber S, von Seydewitz CU, Steinhoff J, Stracke S, Suerbaum S, van de Loo A, Vischedyk M, Weissenborn K, Wellhöner P, Wiesner M, Zeissig S, Büning J, Schiffer M, Kuehbacher T. 2012. Validation of treatment strategies for enterohaemorrhagic Escherichia coli O104:H4 induced haemolytic uraemic syndrome: case-control study. BMJ 345:e4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 680.Bielaszewska M, Idelevich EA, Zhang W, Bauwens A, Schaumburg F, Mellmann A, Peters G, Karch H. 2012. Effects of antibiotics on Shiga toxin 2 production and bacteriophage induction by epidemic Escherichia coli O104:H4 strain. Antimicrob. Agents Chemother. 56:3277–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Nitschke M, Sayk F, Härtel C, Roseland RT, Hauswaldt S, Steinhoff J, Fellermann K, Derad I, Wellhöner P, Büning J, Tiemer B, Katalinic A, Rupp J, Lehnert H, Solbach W, Knobloch JK-M. 2012. Association between azithromycin therapy and duration of bacterial shedding among patients with Shiga toxin-producing enteroaggregative Escherichia coli O104:H4. JAMA 307:1046–1052 [DOI] [PubMed] [Google Scholar]
- 682.Seifert ME, Tarr PI. 2012. Therapy: azithromycin and decolonization after HUS. Nat. Rev. Nephrol. 8:317–318 [DOI] [PubMed] [Google Scholar]
- 683.Seifert ME, Tarr PI. 2012. Azithromycin decolonization of STEC—a new risk emerges. Nat. Rev. Nephrol. 8:429. [DOI] [PubMed] [Google Scholar]
- 684.Medina AM, Rivera FP, Romero LM, Kolevic LA, Castillo ME, Verne E, Hernandez R, Mayor YE, Barletta F, Mercado E, Ochoa TJ. 2010. Diarrheagenic Escherichia coli in human immunodeficiency virus (HIV) pediatric patients in Lima, Peru. Am. J. Trop. Med. Hyg. 83:158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 685.Guerrant RL, Van Gilder T, Steiner TS, Thielman NM, Slutsker L, Tauxe RV, Hennessy T, Griffin PM, DuPont H, Sack RB, Tarr P, Neill M, Nachamkin I, Reller LB, Osterholm MT, Bennish ML, Pickering LK. 2001. Practice guidelines for the management of infectious diarrhea. Clin. Infect. Dis. 32:331–351 [DOI] [PubMed] [Google Scholar]
- 686.Wanke CA, Gerrior J, Blais V, Mayer H, Acheson D. 1998. Successful treatment of diarrheal disease associated with enteroaggregative Escherichia coli in adults infected with human immunodeficiency virus. J. Infect. Dis. 178:1369–1372 [DOI] [PubMed] [Google Scholar]
- 687.Frech SA, Dupont HL, Bourgeois AL, McKenzie R, Belkind-Gerson J, Figueroa JF, Okhuysen PC, Guerrero NH, Martinez-Sandoval FG, Meléndez-Romero JH, Jiang Z-D, Asturias EJ, Halpern J, Torres OR, Hoffman AS, Villar CP, Kassem RN, Flyer DC, Andersen BH, Kazempour K, Breisch SA, Glenn GM. 2008. Use of a patch containing heat-labile toxin from Escherichia coli against travellers' diarrhoea: a phase II, randomised, double-blind, placebo-controlled field trial. Lancet 371:2019–2025 [DOI] [PubMed] [Google Scholar]
- 688.Ochoa TJ, Cleary TG. 2009. Effect of lactoferrin on enteric pathogens. Biochimie 91:30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689.Gupta SK, Keck J, Ram PK, Crump JA, Miller MA, Mintz ED. 2008. Analysis of data gaps pertaining to enterotoxigenic Escherichia coli infections in low and medium human development index countries, 1984-2005. Epidemiol. Infect. 136:721–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 690.Wolf MK. 1997. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin. Microbiol. Rev. 10:569–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691.Begum YA, Talukder KA, Nair GB, Qadri F, Sack RB, Svennerholm A-M. 2005. Enterotoxigenic Escherichia coli isolated from surface water in urban and rural areas of Bangladesh. J. Clin. Microbiol. 43:3582–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.Dalton CB, Mintz ED, Wells JG, Bopp CA, Tauxe RV. 1999. Outbreaks of enterotoxigenic Escherichia coli infection in American adults: a clinical and epidemiologic profile. Epidemiol. Infect. 123:9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 693.Konishi N, Obata H, Monma C, Nakama A, Kai A, Tsuji T. 2011. Bacteriological and epidemiological characteristics of enterotoxigenic Escherichia coli isolated in Tokyo, Japan, between 1966 and 2009. J. Clin. Microbiol. 49:3348–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694.Beatty ME, Bopp CA, Wells JG, Greene KD, Puhr ND, Mintz ED. 2004. Enterotoxin-producing Escherichia coli O169:H41, United States. Emerg. Infect. Dis. 10:518–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 695.Devasia RA, Jones TF, Ward J, Stafford L, Hardin H, Bopp C, Beatty M, Mintz E, Schaffner W. 2006. Endemically acquired foodborne outbreak of enterotoxin-producing Escherichia coli serotype O169:H41. Am. J. Med. 119:168.e7–e10. [DOI] [PubMed] [Google Scholar]
- 696.Turner SM, Chaudhuri RR, Jiang Z-D, DuPont H, Gyles C, Penn CW, Pallen MJ, Henderson IR. 2006. Phylogenetic comparisons reveal multiple acquisitions of the toxin genes by enterotoxigenic Escherichia coli strains of different evolutionary lineages. J. Clin. Microbiol. 44:4528–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697.Abu-Elyazeed R, Wierzba TF, Mourad AS, Peruski LF, Kay BA, Rao M, Churilla AM, Bourgeois AL, Mortagy AK, Kamal SM, Savarino SJ, Campbell JR, Murphy JR, Naficy A, Clemens JD. 1999. Epidemiology of enterotoxigenic Escherichia coli diarrhea in a pediatric cohort in a periurban area of lower Egypt. J. Infect. Dis. 179:382–389 [DOI] [PubMed] [Google Scholar]
- 698.Qadri F, Saha A, Ahmed T, Al Tarique A, Begum YA, Svennerholm A-M. 2007. Disease burden due to enterotoxigenic Escherichia coli in the first 2 years of life in an urban community in Bangladesh. Infect. Immun. 75:3961–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 699.Steinsland H, Valentiner-Branth P, Perch M, Dias F, Fischer TK, Aaby P, Mølbak K, Sommerfelt H. 2002. Enterotoxigenic Escherichia coli infections and diarrhea in a cohort of young children in Guinea-Bissau. J. Infect. Dis. 186:1740–1747 [DOI] [PubMed] [Google Scholar]
- 700.Viboud GI, Jouve MJ, Binsztein N, Vergara M, Rivas M, Quiroga M, Svennerholm AM. 1999. Prospective cohort study of enterotoxigenic Escherichia coli infections in Argentinean children. J. Clin. Microbiol. 37:2829–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 701.Wennerås C, Erling V. 2004. Prevalence of enterotoxigenic Escherichia coli-associated diarrhoea and carrier state in the developing world. J. Health Popul. Nutr. 22:370–382 [PubMed] [Google Scholar]
- 702.Steinsland H, Valentiner-Branth P, Gjessing HK, Aaby P, Mølbak K, Sommerfelt H. 2003. Protection from natural infections with enterotoxigenic Escherichia coli: longitudinal study. Lancet 362:286–291 [DOI] [PubMed] [Google Scholar]
- 703.Rao MR, Abu-Elyazeed R, Savarino SJ, Naficy AB, Wierzba TF, Abdel-Messih I, Shaheen H, Frenck RW, Jr, Svennerholm A-M, Clemens JD. 2003. High disease burden of diarrhea due to enterotoxigenic Escherichia coli among rural Egyptian infants and young children. J. Clin. Microbiol. 41:4862–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.Lynch M, Painter J, Woodruff R, Braden C. 2006. Surveillance for foodborne-disease outbreaks—United States, 1998–2002. MMWR Surveill. Summ. 55:1–42 [PubMed] [Google Scholar]
- 705.Isidean SD, Riddle MS, Savarino SJ, Porter CK. 2011. A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 29:6167–6178 [DOI] [PubMed] [Google Scholar]
- 706.Ahmed T, Lundgren A, Arifuzzaman M, Qadri F, Teneberg S, Svennerholm A-M. 2009. Children with the Le(a+b−) blood group have increased susceptibility to diarrhea caused by enterotoxigenic Escherichia coli expressing colonization factor I group fimbriae. Infect. Immun. 77:2059–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 707.Jansson L, Tobias J, Lebens M, Svennerholm A-M, Teneberg S. 2006. The major subunit, CfaB, of colonization factor antigen i from enterotoxigenic Escherichia coli is a glycosphingolipid binding protein. Infect. Immun. 74:3488–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.DuPont HL. 2006. Travellers' diarrhoea: contemporary approaches to therapy and prevention. Drugs 66:303–314 [DOI] [PubMed] [Google Scholar]
- 709.Hill DR, Beeching NJ. 2010. Travelers' diarrhea. Curr. Opin. Infect. Dis. 23:481–487 [DOI] [PubMed] [Google Scholar]
- 710.Jain S, Chen L, Dechet A, Hertz AT, Brus DL, Hanley K, Wilson B, Frank J, Greene KD, Parsons M, Bopp CA, Todd R, Hoekstra M, Mintz ED, Ram PK. 2008. An outbreak of enterotoxigenic Escherichia coli associated with sushi restaurants in Nevada, 2004. Clin. Infect. Dis. 47:1–7 [DOI] [PubMed] [Google Scholar]
- 711.Taneja N, Das A, Raman Rao DSV, Jain N, Singh M, Sharma M. 2003. Nosocomial outbreak of diarrhoea by enterotoxigenic Escherichia coli among preterm neonates in a tertiary care hospital in India: pitfalls in healthcare. J. Hosp. Infect. 53:193–197 [DOI] [PubMed] [Google Scholar]
- 712.Castro-Rosas J, Cerna-Cortés JF, Méndez-Reyes E, Lopez-Hernandez D, Gómez-Aldapa CA, Estrada-Garcia T. 2012. Presence of faecal coliforms, Escherichia coli and diarrheagenic E. coli pathotypes in ready-to-eat salads, from an area where crops are irrigated with untreated sewage water. Int. J. Food Microbiol. 156:176–180 [DOI] [PubMed] [Google Scholar]
- 713.Lothigius A, Sjöling A, Svennerholm A-M, Bölin I. 2010. Survival and gene expression of enterotoxigenic Escherichia coli during long-term incubation in sea water and freshwater. J. Appl. Microbiol. 108:1441–1449 [DOI] [PubMed] [Google Scholar]
- 714.Ahmed D, Islam MS, Begum YA, Janzon A, Qadri F, Sjöling A. 2013. Presence of enterotoxigenic Escherichia coli in biofilms formed in water containers in poor households coincides with epidemic seasons in Dhaka. J. Appl. Microbiol. 114:1223–1229 [DOI] [PubMed] [Google Scholar]
- 715.Levine MM, Nalin DR, Hoover DL, Bergquist EJ, Hornick RB, Young CR. 1979. Immunity to enterotoxigenic Escherichia coli. Infect. Immun. 23:729–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716.Hara-Kudo Y, Takatori K. 2011. Contamination level and ingestion dose of foodborne pathogens associated with infections. Epidemiol. Infect. 139:1505–1510 [DOI] [PubMed] [Google Scholar]
- 717.Beatty ME, Adcock PM, Smith SW, Quinlan K, Kamimoto LA, Rowe SY, Scott K, Conover C, Varchmin T, Bopp CA, Greene KD, Bibb B, Slutsker L, Mintz ED. 2006. Epidemic diarrhea due to enterotoxigenic Escherichia coli. Clin. Infect. Dis. 42:329–334 [DOI] [PubMed] [Google Scholar]
- 718.Yoder JS, Cesario S, Plotkin V, Ma X, Kelly-Shannon K, Dworkin MS. 2006. Outbreak of enterotoxigenic Escherichia coli infection with an unusually long duration of illness. Clin. Infect. Dis. 42:1513–1517 [DOI] [PubMed] [Google Scholar]
- 719.Pakalniskiene J, Falkenhorst G, Lisby M, Madsen SB, Olsen KEP, Nielsen EM, Mygh A, Boel J, Mølbak K. 2009. A foodborne outbreak of enterotoxigenic E. coli and Salmonella Anatum infection after a high-school dinner in Denmark, November 2006. Epidemiol. Infect. 137:396–401 [DOI] [PubMed] [Google Scholar]
- 720.Ethelberg S, Lisby M, Bottiger B, Schultz AC, Villif A, Jensen T, Olsen KE, Scheutz F, Kjelso C, Muller L. 2010. Outbreaks of gastroenteritis linked to lettuce, Denmark, January 2010. Euro Surveill. 15(6):pii-1948 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19484 [PubMed] [Google Scholar]
- 721.Huerta M, Grotto I, Gdalevich M, Mimouni D, Gavrieli B, Yavzori M, Cohen D, Shpilberg O. 2000. A waterborne outbreak of gastroenteritis in the Golan Heights due to enterotoxigenic Escherichia coli. Infection 28:267–271 [DOI] [PubMed] [Google Scholar]
- 722.Qadri F, Khan AI, Faruque ASG, Begum YA, Chowdhury F, Nair GB, Salam MA, Sack DA, Svennerholm A-M. 2005. Enterotoxigenic Escherichia coli and Vibrio cholerae diarrhea, Bangladesh, 2004. Emerg. Infect. Dis. 11:1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 723.Crossman LC, Chaudhuri RR, Beatson SA, Wells TJ, Desvaux M, Cunningham AF, Petty NK, Mahon V, Brinkley C, Hobman JL, Savarino SJ, Turner SM, Pallen MJ, Penn CW, Parkhill J, Turner AK, Johnson TJ, Thomson NR, Smith SGJ, Henderson IR. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192:5822–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 724.Del Canto F, Botkin DJ, Valenzuela P, Popov V, Ruiz-Perez F, Nataro JP, Levine MM, Stine OC, Pop M, Torres AG, Vidal R. 2012. Identification of coli surface antigen 23, a novel adhesin of enterotoxigenic Escherichia coli. Infect. Immun. 80:2791–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.Gaastra W, Svennerholm AM. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444–452 [DOI] [PubMed] [Google Scholar]
- 726.Turner SM, Scott-Tucker A, Cooper LM, Henderson IR. 2006. Weapons of mass destruction: virulence factors of the global killer enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 263:10–20 [DOI] [PubMed] [Google Scholar]
- 727.Ghosal A, Bhowmick R, Banerjee R, Ganguly S, Yamasaki S, Ramamurthy T, Hamabata T, Chatterjee NS. 2009. Characterization and studies of the cellular interaction of native colonization factor CS6 purified from a clinical isolate of enterotoxigenic Escherichia coli. Infect. Immun. 77:2125–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Jansson L, Tobias J, Jarefjäll C, Lebens M, Svennerholm A-M, Teneberg S. 2009. Sulfatide recognition by colonization factor antigen CS6 from enterotoxigenic Escherichia coli. PLoS One 4:e4487. 10.1371/journal.pone.0004487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729.Sabui S, Ghosal A, Dutta S, Ghosh A, Ramamurthy T, Nataro JP, Hamabata T, Chatterjee NS. 2010. Allelic variation in colonization factor CS6 of enterotoxigenic Escherichia coli isolated from patients with acute diarrhoea and controls. J. Med. Microbiol. 59:770–779 [DOI] [PubMed] [Google Scholar]
- 730.Mazariego-Espinosa K, Cruz A, Ledesma MA, Ochoa SA, Xicohtencatl-Cortes J. 2010. Longus, a type IV pilus of enterotoxigenic Escherichia coli, is involved in adherence to intestinal epithelial cells. J. Bacteriol. 192:2791–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 731.Clavijo AP, Bai J, Gómez-Duarte OG. 2010. The Longus type IV pilus of enterotoxigenic Escherichia coli (ETEC) mediates bacterial self-aggregation and protection from antimicrobial agents. Microb. Pathog. 48:230–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732.Del Canto F, Valenzuela P, Cantero L, Bronstein J, Blanco JE, Blanco J, Prado V, Levine M, Nataro J, Sommerfelt H, Vidal R. 2011. Distribution of classical and nonclassical virulence genes in enterotoxigenic Escherichia coli isolates from Chilean children and tRNA gene screening for putative insertion sites for genomic islands. J. Clin. Microbiol. 49:3198–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Fleckenstein JM, Kopecko DJ, Warren RL, Elsinghorst EA. 1996. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64:2256–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 734.Mammarappallil JG, Elsinghorst EA. 2000. Epithelial cell adherence mediated by the enterotoxigenic Escherichia coli tia protein. Infect. Immun. 68:6595–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 735.Fleckenstein JM, Holland JT, Hasty DL. 2002. Interaction of an outer membrane protein of enterotoxigenic Escherichia coli with cell surface heparan sulfate proteoglycans. Infect. Immun. 70:1530–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736.Elsinghorst EA, Weitz JA. 1994. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62:3463–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 737.Lindenthal C, Elsinghorst EA. 2001. Enterotoxigenic Escherichia coli TibA glycoprotein adheres to human intestine epithelial cells. Infect. Immun. 69:52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 738.Côté J-P, Mourez M. 2011. Structure-function analysis of the TibA self-associating autotransporter reveals a modular organization. Infect. Immun. 79:1826–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 739.Sherlock O, Vejborg RM, Klemm P. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 73:1954–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740.Fleckenstein JM, Roy K, Fischer JF, Burkitt M. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 74:2245–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 741.Roy K, Hilliard GM, Hamilton DJ, Luo J, Ostmann MM, Fleckenstein JM. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457:594–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 742.Weiglmeier PR, Rösch P, Berkner H. 2010. Cure and curse: E. coli heat-stable enterotoxin and its receptor guanylyl cyclase C. Toxins 2:2213–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 743.Lucas ML. 2010. Diarrhoeal disease through enterocyte secretion: a doctrine untroubled by proof. Exp. Physiol. 95:479–484 [DOI] [PubMed] [Google Scholar]
- 744.Lucas ML, Thom MMM, Bradley JM, O'Reilly NF, McIlvenny TJ, Nelson YB. 2005. Escherichia coli heat stable (STa) enterotoxin and the upper small intestine: lack of evidence in vivo for net fluid secretion. J. Membr. Biol. 206:29–42 [DOI] [PubMed] [Google Scholar]
- 745.Liang S, Hajishengallis G. 2010. Heat-labile enterotoxins as adjuvants or anti-inflammatory agents. Immunol. Invest. 39:449–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 746.Nawar HF, King-Lyons ND, Hu JC, Pasek RC, Connell TD. 2010. LT-IIc, a new member of the type II heat-labile enterotoxin family encoded by an Escherichia coli strain obtained from a nonmammalian host. Infect. Immun. 78:4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 747.Jobling MG, Holmes RK. 2012. Type II heat-labile enterotoxins from 50 diverse Escherichia coli isolates belong almost exclusively to the LT-IIc family and may be prophage encoded. PLoS One 7:e29898. 10.1371/journal.pone.0029898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 748.Mudrak B, Kuehn MJ. 2010. Heat-labile enterotoxin: beyond Gm1 binding. Toxins 2:1445–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 749.Kreisberg RB, Harper J, Strauman MC, Marohn M, Clements JD, Nataro JP. 2011. Induction of increased permeability of polarized enterocyte monolayers by enterotoxigenic Escherichia coli heat-labile enterotoxin. Am. J. Trop. Med. Hyg. 84:451–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 750.Johnson AM, Kaushik RS, Francis DH, Fleckenstein JM, Hardwidge PR. 2009. Heat-labile enterotoxin promotes Escherichia coli adherence to intestinal epithelial cells. J. Bacteriol. 191:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Wang X, Gao X, Hardwidge PR. 2012. Heat-labile enterotoxin-induced activation of NF-κB and MAPK pathways in intestinal epithelial cells impacts enterotoxigenic Escherichia coli (ETEC) adherence. Cell. Microbiol. 14:1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 752.Chakraborty K, Ghosh S, Koley H, Mukhopadhyay AK, Ramamurthy T, Saha DR, Mukhopadhyay D, Roychowdhury S, Hamabata T, Takeda Y, Das S. 2008. Bacterial exotoxins downregulate cathelicidin (hCAP-18/LL-37) and human β-defensin 1 (HBD-1) expression in the intestinal epithelial cells. Cell. Microbiol. 10:2520–2537 [DOI] [PubMed] [Google Scholar]
- 753.Ruan X, Crupper SS, Schultz BD, Robertson DC, Zhang W. 2012. Escherichia coli expressing EAST1 toxin did not cause an increase of cAMP or cGMP levels in cells, and no diarrhea in 5-day old gnotobiotic pigs. PLoS One 7:e43203. 10.1371/journal.pone.0043203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 754.Patel SK, Dotson J, Allen KP, Fleckenstein JM. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72:1786–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 755.Roy K, Kansal R, Bartels SR, Hamilton DJ, Shaaban S, Fleckenstein JM. 2011. Adhesin degradation accelerates delivery of heat-labile toxin by enterotoxigenic Escherichia coli. J. Biol. Chem. 286:29771–29779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 756.Pilonieta MC, Bodero MD, Munson GP. 2007. CfaD-dependent expression of a novel extracytoplasmic protein from enterotoxigenic Escherichia coli. J. Bacteriol. 189:5060–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 757.Stermer E, Lubezky A, Potasman I, Paster E, Lavy A. 2006. Is traveler's diarrhea a significant risk factor for the development of irritable bowel syndrome? A prospective study. Clin. Infect. Dis. 43:898–901 [DOI] [PubMed] [Google Scholar]
- 758.Galbadage T, Jiang Z-D, DuPont HL. 2009. Improvement in detection of enterotoxigenic Escherichia coli in patients with travelers' diarrhea by increasing the number of E. coli colonies tested. Am. J. Trop. Med. Hyg. 80:20–23 [PubMed] [Google Scholar]
- 759.Sjöling A, Wiklund G, Savarino SJ, Cohen DI, Svennerholm A-M. 2007. Comparative analyses of phenotypic and genotypic methods for detection of enterotoxigenic Escherichia coli toxins and colonization factors. J. Clin. Microbiol. 45:3295–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 760.Rodas C, Iniguez V, Qadri F, Wiklund G, Svennerholm A-M, Sjöling A. 2009. Development of multiplex PCR assays for detection of enterotoxigenic Escherichia coli colonization factors and toxins. J. Clin. Microbiol. 47:1218–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 761.Grimes KA, Mohamed JA, Dupont HL, Padda RS, Jiang Z-D, Flores J, Belkind-Gerson J, Martinez-Sandoval FG, Okhuysen PC. 2008. PCR-based assay using occult blood detection cards for detection of diarrheagenic Escherichia coli in specimens from U.S. travelers to Mexico with acute diarrhea. J. Clin. Microbiol. 46:2227–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 762.DuPont HL, Ericsson CD, Farthing MJG, Gorbach S, Pickering LK, Rombo L, Steffen R, Weinke T. 2009. Expert review of the evidence base for self-therapy of travelers' diarrhea. J. Travel Med. 16:161–171 [DOI] [PubMed] [Google Scholar]
- 763.DuPont HL, Ericsson CD, Farthing MJG, Gorbach S, Pickering LK, Rombo L, Steffen R, Weinke T. 2009. Expert review of the evidence base for prevention of travelers' diarrhea. J. Travel Med. 16:149–160 [DOI] [PubMed] [Google Scholar]
- 764.Mendez Arancibia E, Pitart C, Ruiz J, Marco F, Gascón J, Vila J. 2009. Evolution of antimicrobial resistance in enteroaggregative Escherichia coli and enterotoxigenic Escherichia coli causing traveller's diarrhoea. J. Antimicrob. Chemother. 64:343–347 [DOI] [PubMed] [Google Scholar]
- 765.Ruiz J, Mensa L, Pons MJ, Vila J, Gascon J. 2008. Development of Escherichia coli rifaximin-resistant mutants: frequency of selection and stability. J. Antimicrob. Chemother. 61:1016–1019 [DOI] [PubMed] [Google Scholar]
- 766.Walker RI, Steele D, Aguado T. 2007. Analysis of strategies to successfully vaccinate infants in developing countries against enterotoxigenic E. coli (ETEC) disease. Vaccine 25:2545–2566 [DOI] [PubMed] [Google Scholar]
- 767.Roland KL, Cloninger C, Kochi SK, Thomas LJ, Tinge SA, Rouskey C, Killeen KP. 2007. Construction and preclinical evaluation of recombinant Peru-15 expressing high levels of the cholera toxin B subunit as a vaccine against enterotoxigenic Escherichia coli. Vaccine 25:8574–8584 [DOI] [PubMed] [Google Scholar]
- 768.Glenn GM, Villar CP, Flyer DC, Bourgeois AL, McKenzie R, Lavker RM, Frech SA. 2007. Safety and immunogenicity of an enterotoxigenic Escherichia coli vaccine patch containing heat-labile toxin: use of skin pretreatment to disrupt the stratum corneum. Infect. Immun. 75:2163–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.McKenzie R, Bourgeois AL, Frech SA, Flyer DC, Bloom A, Kazempour K, Glenn GM. 2007. Transcutaneous immunization with the heat-labile toxin (LT) of enterotoxigenic Escherichia coli (ETEC): protective efficacy in a double-blind, placebo-controlled challenge study. Vaccine 25:3684–3691 [DOI] [PubMed] [Google Scholar]
- 770.Khan S, Chatfield S, Stratford R, Bedwell J, Bentley M, Sulsh S, Giemza R, Smith S, Bongard E, Cosgrove CA, Johnson J, Dougan G, Griffin GE, Makin J, Lewis DJM. 2007. Ability of SPI2 mutant of S. typhi to effectively induce antibody responses to the mucosal antigen enterotoxigenic E. coli heat labile toxin B subunit after oral delivery to humans. Vaccine 25:4175–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 771.Stratford R, McKelvie ND, Hughes NJ, Aldred E, Wiseman C, Curtis J, Bellaby T, Bentley M, Hindle Z, Brennan FR, Chatfield SN, Dougan G, Khan SA. 2005. Optimization of Salmonella enterica serovar Typhi ΔaroC ΔssaV derivatives as vehicles for delivering heterologous antigens by chromosomal integration and in vivo inducible promoters. Infect. Immun. 73:362–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 772.Tacket CO. 2007. Plant-based vaccines against diarrheal diseases. Trans. Am. Clin. Climatol. Assoc. 118:79–87 [PMC free article] [PubMed] [Google Scholar]
- 773.Tokuhara D, Yuki Y, Nochi T, Kodama T, Mejima M, Kurokawa S, Takahashi Y, Nanno M, Nakanishi U, Takaiwa F, Honda T, Kiyono H. 2010. Secretory IgA-mediated protection against V. cholerae and heat-labile enterotoxin-producing enterotoxigenic Escherichia coli by rice-based vaccine. Proc. Natl. Acad. Sci. U. S. A. 107:8794–8799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 774.Norton EB, Lawson LB, Freytag LC, Clements JD. 2011. Characterization of a mutant Escherichia coli heat-labile toxin, LT(R192G/L211A), as a safe and effective oral adjuvant. Clin. Vaccine Immunol. 18:546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 775.Liu M, Ruan X, Zhang C, Lawson SR, Knudsen DE, Nataro JP, Robertson DC, Zhang W. 2011. Heat-labile- and heat-stable-toxoid fusions (LTR192G-STaP13F) of human enterotoxigenic Escherichia coli elicit neutralizing antitoxin antibodies. Infect. Immun. 79:4002–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 776.Liu M, Zhang C, Mateo K, Nataro JP, Robertson DC, Zhang W. 2011. Modified heat-stable toxins (hSTa) of enterotoxigenic Escherichia coli lose toxicity but display antigenicity after being genetically fused to heat-labile toxoid LT(R192G). Toxins 3:1146–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 777.Wu C-M, Chung T-C. 2007. Mice protected by oral immunization with Lactobacillus reuteri secreting fusion protein of Escherichia coli enterotoxin subunit protein. FEMS Immunol. Med. Microbiol. 50:354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Taxt A, Aasland R, Sommerfelt H, Nataro J, Puntervoll P. 2010. Heat-stable enterotoxin of enterotoxigenic Escherichia coli as a vaccine target. Infect. Immun. 78:1824–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 779.Svennerholm A-M. 2011. From cholera to enterotoxigenic Escherichia coli (ETEC) vaccine development. Indian J. Med. Res. 133:188–196 [PMC free article] [PubMed] [Google Scholar]
- 780.Sack DA, Shimko J, Torres O, Bourgeois AL, Francia DS, Gustafsson B, Kärnell A, Nyquist I, Svennerholm A-M. 2007. Randomised, double-blind, safety and efficacy of a killed oral vaccine for enterotoxigenic E. coli diarrhoea of travellers to Guatemala and Mexico. Vaccine 25:4392–4400 [DOI] [PubMed] [Google Scholar]
- 781.Tobias J, Lebens M, Bölin I, Wiklund G, Svennerholm A-M. 2008. Construction of non-toxic Escherichia coli and Vibrio cholerae strains expressing high and immunogenic levels of enterotoxigenic E. coli colonization factor I fimbriae. Vaccine 26:743–752 [DOI] [PubMed] [Google Scholar]
- 782.Tobias J, Holmgren J, Hellman M, Nygren E, Lebens M, Svennerholm A-M. 2010. Over-expression of major colonization factors of enterotoxigenic Escherichia coli, alone or together, on non-toxigenic E. coli bacteria. Vaccine 28:6977–6984 [DOI] [PubMed] [Google Scholar]
- 783.Tobias J, Svennerholm A-M, Holmgren J, Lebens M. 2010. Construction and expression of immunogenic hybrid enterotoxigenic Escherichia coli CFA/I and CS2 colonization fimbriae for use in vaccines. Appl. Microbiol. Biotechnol. 87:1355–1365 [DOI] [PubMed] [Google Scholar]
- 784.Tobias J, Svennerholm A-M, Carlin NIA, Lebens M, Holmgren J. 2011. Construction of a non-toxigenic Escherichia coli oral vaccine strain expressing large amounts of CS6 and inducing strong intestinal and serum anti-CS6 antibody responses in mice. Vaccine 29:8863–8869 [DOI] [PubMed] [Google Scholar]
- 785.Lundgren A, Leach S, Tobias J, Carlin N, Gustafsson B, Jertborn M, Bourgeois L, Walker R, Holmgren J, Svennerholm A-M. 2013. Clinical trial to evaluate safety and immunogenicity of an oral inactivated enterotoxigenic Escherichia coli prototype vaccine containing CFA/I overexpressing bacteria and recombinantly produced LTB/CTB hybrid protein. Vaccine 31:1163–1170 [DOI] [PubMed] [Google Scholar]
- 786.Harris JA, Roy K, Woo-Rasberry V, Hamilton DJ, Kansal R, Qadri F, Fleckenstein JM. 2011. Directed evaluation of enterotoxigenic Escherichia coli autotransporter proteins as putative vaccine candidates. PLoS Negl. Trop. Dis. 5:e1428. 10.1371/journal.pntd.0001428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 787.Roy K, Bartels S, Qadri F, Fleckenstein JM. 2010. Enterotoxigenic Escherichia coli elicits immune responses to multiple surface proteins. Infect. Immun. 78:3027–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 788.Roy K, Hamilton D, Ostmann MM, Fleckenstein JM. 2009. Vaccination with EtpA glycoprotein or flagellin protects against colonization with enterotoxigenic Escherichia coli in a murine model. Vaccine 27:4601–4608 [DOI] [PubMed] [Google Scholar]
- 789.Roy K, Hamilton DJ, Munson GP, Fleckenstein JM. 2011. Outer membrane vesicles induce immune responses to virulence proteins and protect against colonization by enterotoxigenic Escherichia coli. Clin. Vaccine Immunol. 18:1803–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 790.Daley A, Randall R, Darsley M, Choudhry N, Thomas N, Sanderson IR, Croft NM, Kelly P. 2007. Genetically modified enterotoxigenic Escherichia coli vaccines induce mucosal immune responses without inflammation. Gut 56:1550–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 791.McKenzie R, Bourgeois AL, Engstrom F, Hall E, Chang HS, Gomes JG, Kyle JL, Cassels F, Turner AK, Randall R, Darsley M, Lee C, Bedford P, Shimko J, Sack DA. 2006. Comparative safety and immunogenicity of two attenuated enterotoxigenic Escherichia coli vaccine strains in healthy adults. Infect. Immun. 74:994–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 792.Turner AK, Beavis JC, Stephens JC, Greenwood J, Gewert C, Thomas N, Deary A, Casula G, Daley A, Kelly P, Randall R, Darsley MJ. 2006. Construction and phase I clinical evaluation of the safety and immunogenicity of a candidate enterotoxigenic Escherichia coli vaccine strain expressing colonization factor antigen CFA/I. Infect. Immun. 74:1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 793.McKenzie R, Darsley M, Thomas N, Randall R, Carpenter C, Forbes E, Finucane M, Sack RB, Hall E, Bourgeois AL. 2008. A double-blind, placebo-controlled trial to evaluate the efficacy of PTL-003, an attenuated enterotoxigenic E. coli (ETEC) vaccine strain, in protecting against challenge with virulent ETEC. Vaccine 26:4731–4739 [DOI] [PubMed] [Google Scholar]
- 794.Turner AK, Stephens JC, Beavis JC, Greenwood J, Gewert C, Randall R, Freeman D, Darsley MJ. 2011. Generation and characterization of a live attenuated enterotoxigenic Escherichia coli combination vaccine expressing six colonization factors and heat-labile toxin subunit B. Clin. Vaccine Immunol. 18:2128–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 795.Harro C, Sack D, Bourgeois AL, Walker R, DeNearing B, Feller A, Chakraborty S, Buchwaldt C, Darsley MJ. 2011. A combination vaccine consisting of three live attenuated enterotoxigenic Escherichia coli strains expressing a range of colonization factors and heat-labile toxin subunit B is well tolerated and immunogenic in a placebo-controlled double-blind phase I trial in healthy adults. Clin. Vaccine Immunol. 18:2118–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 796.Darsley MJ, Chakraborty S, DeNearing B, Sack DA, Feller A, Buchwaldt C, Bourgeois AL, Walker R, Harro CD. 2012. The oral, live attenuated enterotoxigenic Escherichia coli vaccine ACE527 reduces the incidence and severity of diarrhea in a human challenge model of diarrheal disease. Clin. Vaccine Immunol. 19:1921–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 797.Ma Y, Luo Y, Huang X, Song F, Liu G. 2012. Construction of Bifidobacterium infantis as a live oral vaccine that expresses antigens of the major fimbrial subunit (CfaB) and the B subunit of heat-labile enterotoxin (LTB) from enterotoxigenic Escherichia coli. Microbiology 158:498–504 [DOI] [PubMed] [Google Scholar]
- 798.Barry EM, Wang J, Wu T, Davis T, Levine MM. 2006. Immunogenicity of multivalent Shigella-ETEC candidate vaccine strains in a guinea pig model. Vaccine 24:3727–3734 [DOI] [PubMed] [Google Scholar]
- 799.Ranallo RT, Fonseka CP, Cassels F, Srinivasan J, Venkatesan MM. 2005. Construction and characterization of bivalent Shigella flexneri 2a vaccine strains SC608(pCFAI) and SC608(pCFAI/LTB) that express antigens from enterotoxigenic Escherichia coli. Infect. Immun. 73:258–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 800.Byrd W, Boedeker EC. 2013. Attenuated Escherichia coli strains expressing the colonization factor antigen I (CFA/I) and a detoxified heat-labile enterotoxin (LThK63) enhance clearance of ETEC from the lungs of mice and protect mice from intestinal ETEC colonization and LT-induced fluid accumulation. Vet. Immunol. Immunopathol. 152:57–67 [DOI] [PubMed] [Google Scholar]
- 801.Snelling AM, Macfarlane-Smith LR, Fletcher JN, Okeke IN. 2009. The commonly-used DNA probe for diffusely-adherent Escherichia coli cross-reacts with a subset of enteroaggregative E. coli. BMC Microbiol. 9:269. 10.1186/1471-2180-9-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 802.Lopes LM, Fabbricotti SH, Ferreira AJP, Kato MAMF, Michalski J, Scaletsky ICA. 2005. Heterogeneity among strains of diffusely adherent Escherichia coli isolated in Brazil. J. Clin. Microbiol. 43:1968–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 803.Girón JA, Jones T, Millán-Velasco F, Castro-Muñoz E, Zárate L, Fry J, Frankel G, Moseley SL, Baudry B, Kaper JB. 1991. Diffuse-adhering Escherichia coli (DAEC) as a putative cause of diarrhea in Mayan children in Mexico. J. Infect. Dis. 163:507–513 [DOI] [PubMed] [Google Scholar]
- 804.Gunzburg ST, Chang BJ, Elliott SJ, Burke V, Gracey M. 1993. Diffuse and enteroaggregative patterns of adherence of enteric Escherichia coli isolated from aboriginal children from the Kimberley region of Western Australia. J. Infect. Dis. 167:755–758 [DOI] [PubMed] [Google Scholar]
- 805.Knutton S, Shaw R, Phillips AD, Smith HR, Willshaw GA, Watson P, Price E. 2001. Phenotypic and genetic analysis of diarrhea-associated Escherichia coli isolated from children in the United Kingdom. J. Pediatr. Gastroenterol. Nutr. 33:32–40 [DOI] [PubMed] [Google Scholar]
- 806.Ochoa TJ, Rivera FP, Bernal M, Meza R, Ecker L, Gil AI, Cepeda D, Mosquito S, Mercado E, Maves RC, Hall ER, Svennerholm A-M, McVeigh A, Savarino S, Lanata CF. 2010. Detection of the CS20 colonization factor antigen in diffuse-adhering Escherichia coli strains. FEMS Immunol. Med. Microbiol. 60:186–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 807.Gómez-Duarte OG, Arzuza O, Urbina D, Bai J, Guerra J, Montes O, Puello M, Mendoza K, Castro GY. 2010. Detection of Escherichia coli enteropathogens by multiplex polymerase chain reaction from children's diarrheal stools in two Caribbean-Colombian cities. Foodborne Pathog. Dis. 7:199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 808.Gomes TA, Vieira MA, Abe CM, Rodrigues D, Griffin PM, Ramos SR. 1998. Adherence patterns and adherence-related DNA sequences in Escherichia coli isolates from children with and without diarrhea in São Paulo city, Brazil. J. Clin. Microbiol. 36:3609–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 809.Le Bouguénec C, Servin AL. 2006. Diffusely adherent Escherichia coli strains expressing Afa/Dr adhesins (Afa/Dr DAEC): hitherto unrecognized pathogens. FEMS Microbiol. Lett. 256:185–194 [DOI] [PubMed] [Google Scholar]
- 810.Servin AL. 2005. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin. Microbiol. Rev. 18:264–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 811.Bernet-Camard MF, Coconnier MH, Hudault S, Servin AL. 1996. Pathogenicity of the diffusely adhering strain Escherichia coli C1845: F1845 adhesin-decay accelerating factor interaction, brush border microvillus injury, and actin disassembly in cultured human intestinal epithelial cells. Infect. Immun. 64:1918–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 812.Goluszko P, Selvarangan R, Popov V, Pham T, Wen JW, Singhal J. 1999. Decay-accelerating factor and cytoskeleton redistribution pattern in HeLa cells infected with recombinant Escherichia coli strains expressing Dr family of adhesins. Infect. Immun. 67:3989–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 813.Peiffer I, Blanc-Potard AB, Bernet-Camard MF, Guignot J, Barbat A, Servin AL. 2000. Afa/Dr diffusely adhering Escherichia coli C1845 infection promotes selective injuries in the junctional domain of polarized human intestinal Caco-2/TC7 cells. Infect. Immun. 68:3431–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 814.Peiffer I, Servin AL, Bernet-Camard MF. 1998. Piracy of decay-accelerating factor (CD55) signal transduction by the diffusely adhering strain Escherichia coli C1845 promotes cytoskeletal F-actin rearrangements in cultured human intestinal INT407 cells. Infect. Immun. 66:4036–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 815.Guignot J, Peiffer I, Bernet-Camard MF, Lublin DM, Carnoy C, Moseley SL, Servin AL. 2000. Recruitment of CD55 and CD66e brush border-associated glycosylphosphatidylinositol-anchored proteins by members of the Afa/Dr diffusely adhering family of Escherichia coli that infect the human polarized intestinal Caco-2/TC7 cells. Infect. Immun. 68:3554–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 816.Guignot J, Hudault S, Kansau I, Chau I, Servin AL. 2009. Human decay-accelerating factor and CEACAM receptor-mediated internalization and intracellular lifestyle of Afa/Dr diffusely adhering Escherichia coli in epithelial cells. Infect. Immun. 77:517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 817.Peiffer I, Guignot J, Barbat A, Carnoy C, Moseley SL, Nowicki BJ, Servin AL, Bernet-Camard MF. 2000. Structural and functional lesions in brush border of human polarized intestinal Caco-2/TC7 cells infected by members of the Afa/Dr diffusely adhering family of Escherichia coli. Infect. Immun. 68:5979–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 818.Arikawa K, Nishikawa Y. 2010. Interleukin-8 induction due to diffusely adherent Escherichia coli possessing Afa/Dr genes depends on flagella and epithelial Toll-like receptor 5. Microbiol. Immunol. 54:491–501 [DOI] [PubMed] [Google Scholar]
- 819.Arikawa K, Meraz IM, Nishikawa Y, Ogasawara J, Hase A. 2005. Interleukin-8 secretion by epithelial cells infected with diffusely adherent Escherichia coli possessing Afa adhesin-coding genes. Microbiol. Immunol. 49:493–503 [DOI] [PubMed] [Google Scholar]
- 820.Bétis F, Brest P, Hofman V, Guignot J, Kansau I, Rossi B, Servin A, Hofman P. 2003. Afa/Dr diffusely adhering Escherichia coli infection in T84 cell monolayers induces increased neutrophil transepithelial migration, which in turn promotes cytokine-dependent upregulation of decay-accelerating factor (CD55), the receptor for Afa/Dr adhesins. Infect. Immun. 71:1774–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 821.Bétis F, Brest P, Hofman V, Guignot J, Bernet-Camard M-F, Rossi B, Servin A, Hofman P. 2003. The Afa/Dr adhesins of diffusely adhering Escherichia coli stimulate interleukin-8 secretion, activate mitogen-activated protein kinases, and promote polymorphonuclear transepithelial migration in T84 polarized epithelial cells. Infect. Immun. 71:1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 822.Tieng V, Le Bouguénec C, du Merle L, Bertheau P, Desreumaux P, Janin A, Charron D, Toubert A. 2002. Binding of Escherichia coli adhesin AfaE to CD55 triggers cell-surface expression of the MHC class I-related molecule MICA. Proc. Natl. Acad. Sci. U. S. A. 99:2977–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 823.Berstad AE, Brandtzaeg P. 1998. Expression of cell membrane complement regulatory glycoproteins along the normal and diseased human gastrointestinal tract. Gut 42:522–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 824.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel JF. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115:1405–1413 [DOI] [PubMed] [Google Scholar]
- 825.Meraz IM, Arikawa K, Ogasawara J, Hase A, Nishikawa Y. 2006. Epithelial cells secrete interleukin-8 in response to adhesion and invasion of diffusely adhering Escherichia coli lacking Afa/Dr genes. Microbiol. Immunol. 50:159–169 [DOI] [PubMed] [Google Scholar]
- 826.Le Bouguénec C. 1999. Diarrhea-associated diffusely adherent Escherichia coli. Clin. Microbiol. Rev. 12:180–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 827.Bilge SS, Clausen CR, Lau W, Moseley SL. 1989. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J. Bacteriol. 171:4281–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 828.Le Bouguenec C, Archambaud M, Labigne A. 1992. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microbiol. 30:1189–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 829.Le Bouguénec C, Lalioui L, du Merle L, Jouve M, Courcoux P, Bouzari S, Selvarangan R, Nowicki BJ, Germani Y, Andremont A, Gounon P, Garcia MI. 2001. Characterization of AfaE adhesins produced by extraintestinal and intestinal human Escherichia coli isolates: PCR assays for detection of Afa adhesins that do or do not recognize Dr blood group antigens. J. Clin. Microbiol. 39:1738–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 830.Nowicki B, Svanborg-edén , Hull CR, Hull S. 1989. Molecular analysis and epidemiology of the Dr hemagglutinin of uropathogenic Escherichia coli. Infect. Immun. 57:446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 831.Scaletsky IC, Pedroso MZ, Oliva CA, Carvalho RL, Morais MB, Fagundes-Neto U. 1999. A localized adherence-like pattern as a second pattern of adherence of classic enteropathogenic Escherichia coli to HEp-2 cells that is associated with infantile diarrhea. Infect. Immun. 67:3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 832.Spehlmann ME, Begun AZ, Burghardt J, Lepage P, Raedler A, Schreiber S. 2008. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm. Bowel Dis. 14:968–976 [DOI] [PubMed] [Google Scholar]
- 833.Marks DJB. 2011. Defective innate immunity in inflammatory bowel disease: a Crohn's disease exclusivity? Curr. Opin. Gastroenterol. 27:328–334 [DOI] [PubMed] [Google Scholar]
- 834.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot J-P, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ. 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40:955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 835.Tsianos EV, Katsanos KH, Tsianos VE. 2012. Role of genetics in the diagnosis and prognosis of Crohn's disease. World J. Gastroenterol. 18:105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 836.Mann EA, Saeed SA. 2012. Gastrointestinal infection as a trigger for inflammatory bowel disease. Curr. Opin. Gastroenterol. 28:24–29 [DOI] [PubMed] [Google Scholar]
- 837.Nash JH, Villegas A, Kropinski AM, Aguilar-Valenzuela R, Konczy P, Mascarenhas M, Ziebell K, Torres AG, Karmali MA, Coombes BK. 2010. Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC Genomics 11:667. 10.1186/1471-2164-11-667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 838.Martinez-Medina M, Aldeguer X, Lopez-Siles M, González-Huix F, López-Oliu C, Dahbi G, Blanco JE, Blanco J, Garcia-Gil LJ, Darfeuille-Michaud A. 2009. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm. Bowel Dis. 15:872–882 [DOI] [PubMed] [Google Scholar]
- 839.Martinez-Medina M, Mora A, Blanco M, López C, Alonso MP, Bonacorsi S, Nicolas-Chanoine M-H, Darfeuille-Michaud A, Garcia-Gil J, Blanco J. 2009. Similarity and divergence among adherent-invasive Escherichia coli and extraintestinal pathogenic E. coli strains. J. Clin. Microbiol. 47:3968–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 840.Krause DO, Little AC, Dowd SE, Bernstein CN. 2011. Complete genome sequence of adherent invasive Escherichia coli UM146 isolated from Ileal Crohn's disease biopsy tissue. J. Bacteriol. 193:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 841.Miquel S, Peyretaillade E, Claret L, de Vallée A, Dossat C, Vacherie B, Zineb EH, Segurens B, Barbe V, Sauvanet P, Neut C, Colombel J-F, Medigue C, Mojica FJM, Peyret P, Bonnet R, Darfeuille-Michaud A. 2010. Complete genome sequence of Crohn's disease-associated adherent-invasive E. coli strain LF82. PLoS One 5(9):e12714. 10.1371/journal.pone.0012714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 842.Ambrose NS, Johnson M, Burdon DW, Keighley MR. 1984. Incidence of pathogenic bacteria from mesenteric lymph nodes and ileal serosa during Crohn's disease surgery. Br. J. Surg. 71:623–625 [DOI] [PubMed] [Google Scholar]
- 843.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser A-L, Barnich N, Bringer M-A, Swidsinski A, Beaugerie L, Colombel J-F. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127:412–421 [DOI] [PubMed] [Google Scholar]
- 844.Giaffer MH, Holdsworth CD, Duerden BI. 1992. Virulence properties of Escherichia coli strains isolated from patients with inflammatory bowel disease. Gut 33:646–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 845.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. 2004. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology 127:80–93 [DOI] [PubMed] [Google Scholar]
- 846.Tabaqchali S, O'Donoghue DP, Bettelheim KA. 1978. Escherichia coli antibodies in patients with inflammatory bowel disease. Gut 19:108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 847.Arnott IDR, Landers CJ, Nimmo EJ, Drummond HE, Smith BKR, Targan SR, Satsangi J. 2004. Sero-reactivity to microbial components in Crohn's disease is associated with disease severity and progression, but not NOD2/CARD15 genotype. Am. J. Gastroenterol. 99:2376–2384 [DOI] [PubMed] [Google Scholar]
- 848.Loftus EV., Jr 2004. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology 126:1504–1517 [DOI] [PubMed] [Google Scholar]
- 849.Barnich N, Boudeau J, Claret L, Darfeuille-Michaud A. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease. Mol. Microbiol. 48:781–794 [DOI] [PubMed] [Google Scholar]
- 850.Barnich N, Carvalho FA, Glasser A-L, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel J-F, Darfeuille-Michaud A. 2007. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 117:1566–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 851.Rolhion N, Barnich N, Claret L, Darfeuille-Michaud A. 2005. Strong decrease in invasive ability and outer membrane vesicle release in Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. J. Bacteriol. 187:2286–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 852.Rolhion N, Barnich N, Bringer M-A, Glasser A-L, Ranc J, Hébuterne X, Hofman P, Darfeuille-Michaud A. 2010. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut 59:1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 853.Kaser A, Lee A-H, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EES, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. 2008. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134:743–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 854.Bringer M-A, Barnich N, Glasser A-L, Bardot O, Darfeuille-Michaud A. 2005. HtrA stress protein is involved in intramacrophagic replication of adherent and invasive Escherichia coli strain LF82 isolated from a patient with Crohn's disease. Infect. Immun. 73:712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 855.Glasser AL, Boudeau J, Barnich N, Perruchot MH, Colombel JF, Darfeuille-Michaud A. 2001. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect. Immun. 69:5529–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 856.Lapaquette P, Glasser A-L, Huett A, Xavier RJ, Darfeuille-Michaud A. 2010. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell. Microbiol. 12:99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 857.Lapaquette P, Bringer M-A, Darfeuille-Michaud A. 2012. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell. Microbiol. 14:791–807 [DOI] [PubMed] [Google Scholar]
- 858.Schofield FW. 1947. Hjarre and Wramby disease in turkeys (coli-granuloma). Can. J. Comp. Med. Vet. Sci. 11:141–143 [PubMed] [Google Scholar]
- 859.Simpson KW, Dogan B, Rishniw M, Goldstein RE, Klaessig S, McDonough PL, German AJ, Yates RM, Russell DG, Johnson SE, Berg DE, Harel J, Bruant G, McDonough SP, Schukken YH. 2006. Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect. Immun. 74:4778–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 860.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect. Immun. 67:4499–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 861.Baumgart DC, Sandborn WJ. 2012. Crohn's disease. Lancet 380:1590–1605 [DOI] [PubMed] [Google Scholar]
- 862.Prantera C, Scribano ML. 2009. Antibiotics and probiotics in inflammatory bowel disease: why, when, and how. Curr. Opin. Gastroenterol. 25:329–333 [DOI] [PubMed] [Google Scholar]
- 863.Wlodarska M, Finlay BB. 2010. Host immune response to antibiotic perturbation of the microbiota. Mucosal Immunol. 3:100–103 [DOI] [PubMed] [Google Scholar]
- 864.Wellens A, Garofalo C, Nguyen H, Van Gerven N, Slättegård R, Hernalsteens J-P, Wyns L, Oscarson S, De Greve H, Hultgren S, Bouckaert J. 2008. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS One 3:e2040. 10.1371/journal.pone.0002040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 865.Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CHF, Stanners CP, Darfeuille-Michaud A. 2009. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J. Exp. Med. 206:2179–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 866.Poggio TV, La Torre JL, Scodeller EA. 2006. Intranasal immunization with a recombinant truncated FimH adhesin adjuvanted with CpG oligodeoxynucleotides protects mice against uropathogenic Escherichia coli challenge. Can. J. Microbiol. 52:1093–1102 [DOI] [PubMed] [Google Scholar]
- 867.Pinkner JS, Remaut H, Buelens F, Miller E, Aberg V, Pemberton N, Hedenström M, Larsson A, Seed P, Waksman G, Hultgren SJ, Almqvist F. 2006. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:17897–17902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 868.Chan JZ-M, Pallen MJ, Oppenheim B, Constantinidou C. 2012. Genome sequencing in clinical microbiology. Nat. Biotechnol. 30:1068–1071 [DOI] [PubMed] [Google Scholar]
- 869.Loman NJ, Constantinidou C, Christner M, Rohde H, Chan JZ-M, Quick J, Weir JC, Quince C, Smith GP, Betley JR, Aepfelbacher M, Pallen MJ. 2013. A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA 309:1502–1510 [DOI] [PubMed] [Google Scholar]
- 870.Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc. Natl. Acad. Sci. U. S. A. 108:20142–20147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 871.Mellmann A, Bielaszewska M, Köck R, Friedrich AW, Fruth A, Middendorf B, Harmsen D, Schmidt MA, Karch H. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 14:1287–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]