Abstract
Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae strains have spread worldwide and become a significant public health threat. blaKPC, the plasmid-borne KPC gene, was frequently identified on numerous transferable plasmids in different incompatibility replicon groups. Here we report the complete nucleotide sequence of a novel blaKPC-3-harboring IncI2 plasmid, pBK15692, isolated from a multidrug-resistant K. pneumoniae ST258 strain isolated from a New Jersey hospital in 2005. pBK15692 is 78 kb in length and carries a backbone that is similar to those of other IncI2 plasmids (pR721, pChi7122-3, pHN1122-1, and pSH146-65), including the genes encoding type IV pili and shufflon regions. Comparative genomics analysis of IncI2 plasmids reveals that they possess a conserved plasmid backbone but are divergent with respect to the integration sites of resistance genes. In pBK15692, the blaKPC-3-harboring Tn4401 was inserted into a Tn1331 element and formed a nested transposon. A PCR scheme was designed to detect the prevalence of IncI2 and pBK15692-like plasmids from a collection of clinical strains from six New Jersey and New York hospitals isolated between 2007 and 2011. IncI2 plasmids were found in 46.2% isolates from 318 clinical K. pneumoniae strains. Notably, 59 pBK15692-like plasmids (23%) have been identified in 256 KPC-bearing K. pneumoniae strains, and all carried KPC-3 and belong to the epidemic ST258 clone. Our study revealed that the prevalence of IncI2 plasmids has been considerably underestimated. Further studies are needed to understand the distribution of this plasmid group in other health care regions and decipher the association between IncI2 plasmids and blaKPC-3-bearing ST258 strains.
INTRODUCTION
Since the initial report in 2001, Klebsiella pneumoniae strains producing K. pneumoniae carbapenemase (KPC) have spread worldwide and emerged as a significant public health threat. The KPC gene, blaKPC, is located on Tn3-like transposon Tn4401 and is commonly carried on numerous transferable plasmids, thereby facilitating its inter- and intraspecies dissemination (1–3). Presently, blaKPC is identified on plasmids that are distinguished on the basis of their different incompatibility (Inc) replicon groups, including IncFII, IncL/M, IncN, IncA/C, IncR, IncX, and ColE1 (2, 4, 5). Nevertheless, Tn4401 and the blaKPC gene have not been found on IncI plasmids, although IncI plasmids are associated with multidrug-resistant Enterobacteriaceae worldwide and known to harbor numerous β-lactmase genes, including blaCMY, blaCTX-M, blaSHV, blaTEM, and blaVIM (6, 7).
Originally, IncI plasmids were identified on the basis of their susceptibility to the similar filamentous bacteriophages If1 (8) and PR64FS (9). The phages recognize type IV pili, which are expressed from the pil gene cluster harbored on IncI plasmids. In addition to the pil genes, IncI plasmids are also characterized by containing the shufflon region that is involved in changing the C-terminal segment of the PilV protein, and in determining the recipient specificity in liquid mating (10). IncI plasmids, mostly IncI1 but also IncI2, have been associated with the spread of several antimicrobial resistance genes in humans, livestock, and wild animals (6, 11–14).
Currently, four IncI2 plasmids, from different bacterial species and hosts, have been completely sequenced and their sequences have been deposited in GenBank, including the plasmid pR721, from a trimethoprim-resistant clinical Escherichia coli strain identified in the early 1970s (15); pChi7211-3, from an avian pathogenic E. coli (APEC) strain, χ7122, isolated from a diseased turkey (16); pSH146-65, from a Salmonella enterica serovar Heidelberg strain isolated from porcine diagnostic specimen in 2002 (17); and pHN1122-1, from an E. coli strain isolated from dog feces (GenBank accession no. JN797501). Here we report the complete sequence of the first blaKPC-harboring IncI2 plasmid (pBK15692) isolated from a strain of the epidemic K. pneumoniae clone ST258 in New Jersey. A survey of this plasmid among a collection of KPC-positive and -negative K. pneumoniae strains from New York and New Jersey hospitals surprisingly uncovered its significant prevalence and its association with blaKPC-3-harboring ST258 strains.
MATERIALS AND METHODS
Bacterial strains.
A multidrug-resistant K. pneumoniae strain, BK15692, was identified from a retrospective study of carbapenem-resistant K. pneumoniae from New Jersey and New York hospitals. BK15692 was isolated from a northern New Jersey hospital in 2005, but the patient's demographic information, underlying disease, and site of isolation were not recorded at that time. Three hundred eighteen K. pneumoniae unique clinical isolates collected from six hospitals in the New Jersey and New York area were included to check the prevalence of IncI2 and pBK15692-like plasmids, using a PCR approach (described below). An additional 19 non-K. pneumoniae Enterobacteriaceae KPC-positive isolates, including 11 Enterobacter species, 6 E. coli, and 2 Citrobacter freundii isolates collected from two of the six hospitals between 2009 and 2011, were also subject to the PCR screening.
Characterization of strain BK15692 and manipulation of plasmids.
Strain BK15692 was initially screened by a multiplex real-time PCR for K. pneumoniae ST258 clone identification and the presence of blaKPC (18), and the sequence type (ST) was further confirmed by multilocus sequence typing (MLST) (19). β-Lactam (blaCTX-M, blaSHV, blaTEM, blaGES, blaNDM, blaVIM, blaIMP, blaOXA-48, blaACT-1, blaACC, blaBIL-1, blaCMY, blaDHA, blaFOX, blaLAT, blaMIR-1, and blaMOX), aminoglycoside (aadA1, aadA2, aadB, aadA5, strA, strB, aphA1, aphA2, aphA6, aacC1, aacC2, aacC4, aacA4, armA, rmtC, and rmtB), and fluoroquinolone (qnrA, qnrB, qnrC, qnrD, qnrS, oqxA, and oqxB) resistance genes were investigated by PCR using methods described elsewhere (20–24). Plasmid incompatibility groups were determined using the PCR methods described previously (25, 26). Specifically, 18 replicon types (FIA, FIB, FIC, HI1, HI2, I1-Iγ, L/M, N, P, W, T, A/C, K, B/O, X, Y, F, and FIIA) were screened by the methods of Carattoli et al. (25), and an additional 3 types (ColE, R, and U) were investigated by the method of García-Fernández et al. (26).
Plasmid DNA in strain BK15692 was extracted using a Qiagen plasmid maxikit (Qiagen, Valencia, CA), followed by electroporation into E. coli DH10B (Invitrogen) using a Gene Pulser II instrument (Bio-Rad Laboratories). E. coli DH10B transformants were selected on Luria-Bertani (LB) agar plates containing 0.5 μg/ml of imipenem and then confirmed by multiplex real-time PCR for the presence of blaKPC genes (27). Transferability of blaKPC-bearing plasmid was examined by conjugation experiments using BK15692 as the donor and E. coli J53 Azr as the recipient as described previously (28). MICs of isolate BK15692 and its E. coli DH10B transformant were determined by broth microdilution in cation-adjusted Mueller-Hinton broth (MHB) using Sensititre GNX2F panels (Thermo Fisher Scientific, Waltham, MA) according to Clinical and Laboratory Standards Institute methods and interpretations (29, 30).
Plasmid sequencing and bioinformatics.
Plasmid DNA from the E. coli DH10B transformant was extracted as described above using a Qiagen plasmid maxikit. The plasmid DNA was sequenced using a Roche 454 GS-FLX system. Sequencing reads were de novo assembled into contigs using the Roche Genome Sequencer FLX software GSA assembler, version 2.5.3. Gaps between contigs were closed by PCR with standard Sanger sequencing. Open reading frames (ORFs) were predicted and annotated using the RAST (http://rast.nmpdr.org) server (31).
PCR screening for pBK15692-like plasmids.
Based on the complete sequence of pBK15692, a PCR scheme, including three individual reactions, was designed to detect pBK15692-like plasmids (Fig. 1). PCR-1 was designed to target the IncI2 plasmid-specific replication genes repA and repR. PCR-2 and PCR-3 were designed to target the junction between Tn4401 (ISKpn6) and the upstream IncI2 plasmid backbone and the junction between Tn1331 (blaTEM-1) and the downstream IncI2 plasmid backbone, respectively. PCR-1 was able to identify the presence of IncI2 plasmids, and a combination of PCR-1 to -3 was able to detect pBK15692-like plasmids. The primer sequences and locations are illustrated in Fig. 1. The PCR cycling conditions were as follows: an initial denaturation step of 95°C for 4 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 2 min (PCR-2) or 45 s (PCR-1 and PCR-3) and a final extension step of 72°C for 7 min. The K. pneumoniae ST258 sequence type and blaKPC variants of all strains were characterized by two multiplex real-time PCR methods developed by our lab (18, 27). DNAs from strain BK15692 and E. coli DH10B were used as positive and negative controls in each PCR run.
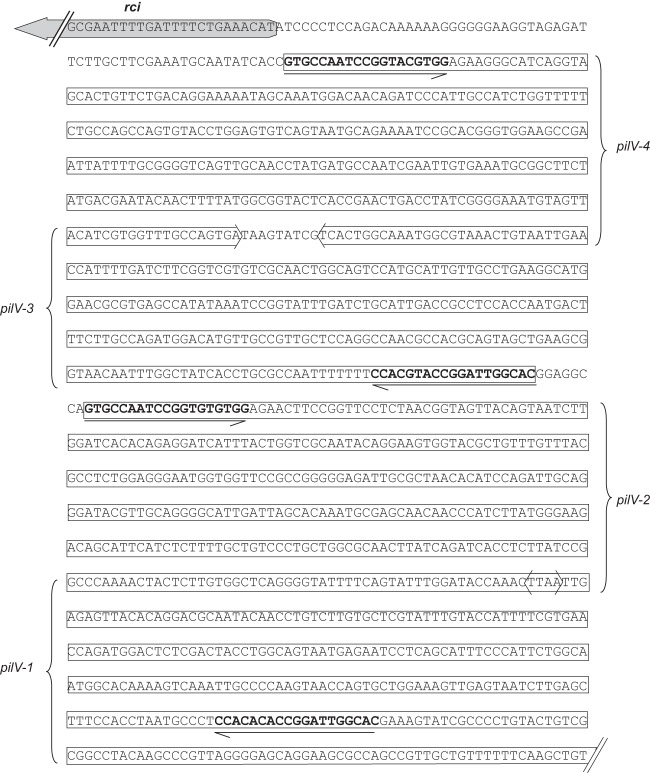
Fig 1.
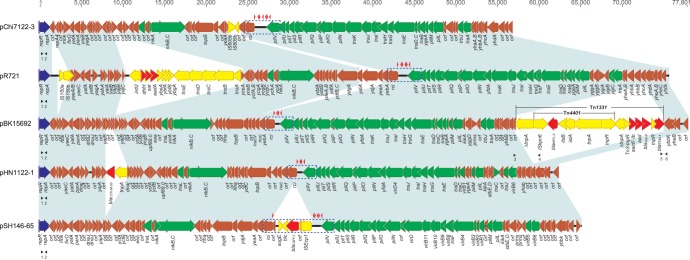
Comparative analysis of IncI2 plasmids pChi7122-3 (FR851304), pR721 (AP002527), pBK15692 (KC845573), pHN1122-1 (JN797501), and pSH146-65 (JN983044). Light blue shading denotes shared regions of homology. ORFs are represented by arrows and colored based on predicted gene function. Orange arrows indicate plasmid scaffold regions. The genes associated with plasmid transfer, including tra, pil, and nikABC loci, are indicated by green arrows, and replication-associated genes are represented as dark blue arrows. Antimicrobial resistance genes are indicated by red arrows, while other genes in the accessory region are indicated by yellow arrows. The shufflon regions, including their associated rci and pilV genes, are displayed by blue dashed box, while the red arrowheads above the shufflon regions indicate the locations and directions of the shufflon-associated 19-bp repeats. Small black arrowheads beneath the plasmids indicate the locations of primers used for PCR screening of pBK15692-like plasmids. Primers used for PCRs were as follows: 1 (I2-repR-F, TTACAGTGCAAGCTAAGTGCAG), 2 (I2-repA-R, GATTCACGGTCCCATATCGT) (615 bp), 3 (15692-F1, TTTAATGATTTGCTCATTCGTGA), 4 (15692-R1, GCCTCAGATAGATGCGGTAGC) (2,337 bp), 5 (15692-F2, AGCCCTCCCGTATCGTAGTT), and 6 (15692-R2, GAAGGCAGAAGGGGAGAAAC) (611 bp).
Statistical analysis.
The statistical significance of differences in the distributions of IncI2 and pBK15692-like plasmids were determined using the chi-square test (Prism, GraphPad, San Diego, CA). Differences were considered statistically significant at a P value of ≤0.05.
Nucleotide sequence accession number.
The complete nucleotide sequence of pBK15692 was deposited in GenBank under accession no. KC845573.
RESULTS
Microbiological and genetic characterization of strain BK15692.
Susceptibility testing showed that BK15692 was resistant to imipenem (MIC = 8 μg/ml), ertapenem (≥4 μg/ml), meropenem (≥8 μg/ml), doripenem (≥4 μg/ml), cefepime (≥32 μg/ml), cefotaxime (≥32 μg/ml), ceftazidime (≥16 μg/ml), aztreonam (≥16 μg/ml), levofloxacin (≥8 μg/ml), ciprofloxacin (≥4 μg/ml), and co-trimoxazole (≥4/76 μg/ml) and was intermediate to amikacin (32 μg/ml) and gentamicin (8 μg/ml)). In contrast, BK15692 was susceptible to tetracyclines (doxycycline and minocycline), colistin (1 μg/ml), tigecycline (1 μg/ml), and polymyxin B (1 μg/ml). MLST PCR amplification and sequencing of antimicrobial resistance genes revealed that this clinical isolate belonged to the epidemic K. pneumoniae ST258 clone and harbored the β-lactamase genes blaKPC-3, blaCTX-M-15, blaOXA-9, blaTEM-1, and blaSHV-11; the aminoglycoside-modifying enzyme genes aadA1, aadA2, aphA1, and aac(6′)-Ib (aacA4); and quinolone resistance genes oqxA and oqxB. PCR-based replicon typing (PBRT) revealed that BK15692 contained IncFII and colE1 plasmids.
With selection on imipenen-containing agar (0.5 μg/ml), we were successful in transferring carbapenem resistance from K. pneumoniae BK15692 to E. coli J53 by conjugation and into E. coli DH10B by electroporation. A representative E. coli DH10B transformant displayed antimicrobial susceptibility profiles similar to those of the parent strain but was susceptible to ciprofloxacin and levofloxacin and was less resistant to ertapenem (2 μg/ml), imipenem (2 μg/ml), meropenem (1 μg/ml), and doripenem (1 μg/ml). Surprisingly, PBRT of the transformant was negative, raising the possibility that the blaKPC-3 gene was on a plasmid with an uncommon incompatibility type. Consequently, plasmid pBK15692 from the E. coli DH10B transformant was sequenced.
Structure of blaKPC-3-harboring IncI2 plasmid pBK15692.
pBK15692 is an IncI2 plasmid 77,801 bp in length with an average G+C content of 45.4%, which is lower than the G+C content of K. pneumoniae genomes (∼57.5%), and harbored 105 predicted ORFs (Fig. 1). Three additional PilV proteins with different C-terminal segments could be created by shufflon multiple inversions as demonstrated in other IncI1 or I2 plasmids (10) (Fig. 2). Thus, 108 different proteins are encoded on plasmid pBK15692. The overall structure of pBK15692 is highly similar to that of the other four completely sequenced IncI2 plasmids, with 73%, 68%, 65%, and 64% query coverage and a maximum of 100% nucleotide identity to plasmids pHN1122-1, pSH146-65, pR721, and pChi7211-3 by Blast. All IncI2 plasmids shared a common backbone set of sequences that are responsible for plasmid replication, maintenance, and transfer (Fig. 1).
Fig 2.
DNA sequences of shufflon region in pBK15692. Four 19-bp repeat sequences are in bold, and the directions are shown by the arrow beneath the repeats. The upstream rci gene and four forms of pilV genes (from the 19-bp repeat to the stop codon) are shown in boxed arrows. The predicted pilV-1 to -4 genes are 1,245, 1,377, 1,287, and 1,317 bp, respectively. Corresponding PilV-1 to -4 proteins are predicted to be 414, 459, 428, and 438 amino acids, sharing identical N-terminal 345 amino acids but distinct C-terminal sequences.
The replication region (1,066 bp) of pBK15692, including the replication initiation protein gene repA and replication regulatory protein gene repR, share >96.5% nucleotide similarities with other four IncI2 plasmids (data not shown). We tentatively assigned the downstream locus of the replication region as the stability region because of the presence of several genes presumably required for plasmid maintenance and stability. For example, the gene yaeC encodes a protein that belongs to the FinO protein family and may function as a conjugal transfer repressor; yafA encodes a ribbon-helix-helix CopG family protein, presumably involving control of plasmid copy number (32); parA encodes the protein for plasmid partition; and topB encodes DNA topoisomerase III, which regulates the overwinding or underwinding of DNA. Juxtaposed to the stability region is the transfer region, which is organized into different gene clusters, including the nikABC gene operon for conjugative DNA processing, tra operon for general conjugation and regulation, and pil gene cluster for type IV pilus biogenesis. The replication, stability, and transfer gene clusters among the five characterized IncI2 plasmids have the same syntenic order (Fig. 1).
The characteristic shufflon region of IncI plasmids is also conserved in all IncI2 plasmids. The shufflons are located between the shufflon-specific recombinase gene, rci, and one of the pilus genes, pilV (Fig. 1). Shufflons are a multiple inversion system, originally described for IncI1 plasmid R64 (33) and later also found in IncI2 plasmids (34). The R64 shufflon consists of four DNA segments designated A, B, C, and D (33). They are flanked and separated by seven 19-bp repeat sequences oriented in either direction. Site-specific recombination (mediated by the above-described rci gene product) between any of two inverted repeats results in the inversion of DNA segments either independently or in groups. In plasmid pBK15692, four repeat sequences have been identified; therefore, four different PilV proteins can be presumably created, with the same PilV N-terminal sequences but with variable C-terminal sequences (Fig. 2). Similar to pBK15692, pHN1122-1 also harbors four repeats, and the other three IncI2 plasmids carry either six (pR721 and pSH146-65) or seven (pChi7122-3) repeats (Fig. 1). Interestingly, the shufflon region in pSH146-65 was disrupted by the insertion of a 4.7-kb transposon-like element (ISEcp1-blaCMY-2-blc-sugE), which may affect the function of shufflon, as the recombinase gene rci is distant from the gene pilV.
The major differences between pBK15692 and other IncI2 plasmids are found in the number of acquired genes. An 18-kb acquired resistance gene region is identified downstream of tra and pil operon in pBK15692, located between an unknown orf and the gene ybbk (Fig. 1), and includes β-lactamase genes blaKPC-3, blaOXA-9, and blaTEM-1 and aminoglycoside resistance genes aac(6′)-Ib and aadA1. The G+C content of this region is 56.6%, significantly higher than that of the rest of the plasmid (42.1%), suggesting that this region was acquired recently. Inspection of this region reveals that the blaKPC-3-harboring Tn4401b element is inserted into the transposase gene (tnpA) of Tn1331, generating a 5-bp duplication of the target sequence (AGAAC) (Fig. 1) and forming a nested transposon.
A total of six unique integration sites for acquired regions were identified among the five IncI2 plasmids, including the insertion of the ISEcp1-blaCMY-2-blc-sugE element within the shufflon region in pSH146-65 (described above) (Fig. 1). Except for pChi7122-3, the remaining four completely sequenced IncI2 plasmids harbor at least one resistance gene: pBK15692 contains blaKPC-3, blaTEM-1, blaOXA-9, aadA1, and aac(6′)-Ib; pR721 harbors dfrA1 (trimethoprim resistance), sat1 (streptothricin resistance), and aadA1; blaCTX-M-55 (β-lactam resistance) is identified on pHN1122-1; and blaCMY-2 (β-lactam resistance) is found on pSH146-65.
Prevalence and dissemination of IncI2 and pBK15692-like plasmids.
As part of an ongoing surveillance project, hospitals in New York and New Jersey routinely submit carbapenem-resistant and -susceptible Enterobacteriaceae to our laboratory for genotyping. A total of 318 clinical K. pneumoniae isolates from 6 hospitals were evaluated by PCR for the presence of IncI2 and pBK15692-like plasmid markers. Among them, 256 were blaKPC positive (96 blaKPC-2 and 160 blaKPC-3), while 62 were blaKPC negative; 261 (82%) belonged to the ST258 clone (Table 1).
Table 1.
Distributions of IncI2 and pBK15692-like plasmids in different groups
| Enzyme or clone | IncI2 plasmids, n (%) | pBK15692-like plasmids, n (%) | Total (n) |
|---|---|---|---|
| KPC vs non-KPC | |||
| non-KPC | 19 (30.6) | 0 (0) | 62 |
| KPC | 128 (50.0)a | 59 (23.0)a | 256 |
| KPC-2 vs KPC-3 | |||
| KPC-2 | 3 (3.1) | 0 (0) | 96 |
| KPC-3 | 125 (78.1)b | 59 (36.9)b | 160 |
| ST258 vs non-ST258 | |||
| non-ST258 | 17 (29.8) | 0 (0) | 57 |
| ST258 | 130 (49.8)c | 59 (22.6)c | 261 |
P < 0.01 compared with non-KPC strains.
P < 0.01 compared with KPC-2-bearing strains.
P < 0.01 compared with non-ST258 strains.
Surprisingly, this uncommon plasmid was found to be highly prevalent and a major vector for Tn4401. Among these 318 isolates, 46.2% (n = 147) were positive for IncI2 rep PCR (PCR-1), including both blaKPC-positive and -negative strains. However, the prevalence of IncI2 plasmids in KPC-bearing isolates (50.0%) was significantly higher than that in non-KPC isolates (30.6%) (P < 0.01). Interestingly, IncI2 plasmids were found to be substantially associated with KPC-3-positive isolates: 78.1% of KPC-3 isolates carried IncI2 plasmids, in contrast to 3.1% for KPC-2 isolates. Among 256 blaKPC-positive isolates, 59 (23.0%) pBK15692-like plasmids were identified (positive for PCR-1 to -3), covering isolates from all six hospitals. All 59 isolates are exclusively ST258 and carry blaKPC-3, accounting for 36.9% of the total 160 blaKPC-3-positive isolates in this study.
Among 19 non-K. pneumoniae KPC-carrying Enterobacteriaceae isolates, 12 were found to carry IncI2 plasmids (9 Enterobacter spp. and 3 E. coli isolates). pBK15692-like plasmids were identified in all nine Enterobacter isolates (positive for PCR-1 to -3), and they all harbor blaKPC-3. The three E. coli strains were negative for pBK15692-like plasmids.
DISCUSSION
The rapid global dissemination of KPC-producing K. pneumoniae strains has been largely associated with the epidemic ST258 clone, although KPC enzymes have been detected in a number of other K. pneumoniae sequence types (18, 35–37). One possible explanation for the “epidemiological success of ST258” could be attributed to chromosomal and/or specific plasmids factors that contribute to the compatibility or fitness of this strain (38). Clearly, unraveling these factors will play a key role in understanding the current epidemiology associated with carbapenem-resistant ST258 K. pneumoniae and potentially lead to measures that may assist with infection control and prevention. A number of ST258-associated KPC-producing plasmids have been characterized. One example is the IncFIIK2 plasmid pKpQIL, which was originally reported from ST258 strains from Israel and has now been detected in the United States, Poland, Italy, and other regions (39–43). Interspecies transfer of pKpQIL has also been documented (44). Other completely characterized ST258 associated blaKPC-harboring plasmids include IncX3 plasmid pKpS90 and IncFIIK1 plasmid pBK32179 (45, 46).
To our knowledge, IncI2 plasmids have never been reported for ST258 strains or been associated with carbapenem-resistant genes. This study yielded the first description of the presence and complete sequence of blaKPC-bearing IncI2 plasmids in K. pneumoniae and the first observation that this resistant plasmid is relatively common in our region. Until now, the prevalence of IncI2 plasmids in ST258 was unappreciated, primarily because this plasmid is not included in the PCR-based replicon typing (PBRT) panel (25, 26). This limitation of the current replicon typing method needs to be expanded as more plasmids are described and more replication proteins are identified.
Our results show that among 318 clinical isolates of K. pneumoniae tested, IncI2 plasmids were found in 147 (46%) of them (Table 1). Having uncovered the successful spread of IncI2 plasmids raises the question of whether their unique plasmid structure could contribute to their dissemination. Genetically, IncI plasmids have the pil gene cluster, encoding type IV pili, and they encode a specific shufflon (clustered inversion) region that functions as a biological switch to select the C-terminal segment of the PilV protein (encoded by one of the pil genes, pilV). As an example, the IncI1 plasmid pR64 shufflon determines the recipient specificity in liquid mating experiments by selecting the C-terminal segments of PilV proteins, and DNA rearrangement of the shufflon in the pilV gene can alter this specificity to mate with different recipients (10). The type IV pili are also a potential virulence factor, and the association of virulence and resistance determinants may favor the positive selection of plasmids belonging to the IncI family (6).
IncI2 plasmids maintain both the pil gene cluster and shufflon regulation, and presumably they contribute to their ability to transfer into different strains and species. Our finding that IncI2 plasmids are found in different K. pneumoniae sequence types and in other Enterobacteriaceae species suggests that this plasmid is widely disseminated. In addition, IncI2 plasmid pChi7122-3 showed the properties of acid resistance and enhanced biofilm production, which could be related not only to tra operon expression but also to type IV fimbriae (47). Acid resistance and plasmid-driven biofilms in IncI2 plasmids could be essential for bacterial survival by enhancing persistence in acidic environments (e.g., human stomach) (47).
IncI2 plasmids share similar core genes essential for their replication, stability, and transferability, but their acquired regions are otherwise plastic. This is in contrast to IncN, IncX, and IncI1 plasmids, in which acquired genes share similar integration sites (4, 7, 48). As shown in Fig. 1, IncI2 plasmids have variable integration sites, which may facilitate their ability to rapidly adapt and acquire additional resistance genes to survive in an antibiotic-rich environment.
In pBK15692, the blaKPC-bearing Tn4401b transposon was inserted into another transposon, Tn1331. Tn1331 carries Tn3-like transposase and resolvase genes (tnpA and tnpR), aminoglycoside-modifying enzyme genes aac(6′)-Ib and aadA1, and β-lactamase genes blaOXA-9 and blaTEM-1 (49). The insertion of Tn4401 within Tn1331 disrupts the transposase gene, which presumably deactivates its mobility. In this case, it is most likely that Tn4401 was inserted after the integration of Tn1331 in pBK15692, instead of transferring as a nested transposon. Interestingly, a similar Tn4401/Tn1331 nested transposon was previously described for plasmid pLRM24 from K. pneumoniae strain VA367, but this plasmid is different from pBK15692 in that it also harbors a qnrB19-harboring Tn5387 element that inserted in the blaOXA-9 gene (50). However, it is not clear if pLRM24 was the same plasmid as pBK15692 or if the same insertion of Tn4401 into Tn1331 happened on multiple occasions.
In this study, pBK15692-like plasmids were identified in 23% of KPC-positive K. pneumoniae isolates from all six hospitals, suggesting the widespread nature of this single plasmid in our region. Moreover, pBK15692 was exclusively associated with KPC-3-bearing ST258 isolates in K. pneumoniae, indicating clonal dissemination with ST258 isolates. Remarkably, the finding of pBK15692-like plasmids in Enterobacter spp. from the same hospitals also suggested that the interspecies transfer could contribute to the spread of this plasmid.
In conclusion, this report presents the first complete sequence of a blaKPC-harboring IncI2 plasmid, pBK15692, and the finding that Tn4401 inserted into Tn1331, similar to the structure found in plasmid pLRM24. Comparative genomic analysis of IncI2 plasmids reveals that they possess highly conserved plasmid backbones but are quite divergent with respect to the integration sites of resistance genes. A screening study from six New Jersey and New York hospitals reveals that IncI2 and pBK15692-like plasmids are widely spread in the New York-New Jersey region and are significantly associated with KPC-3-harboring ST258 strains. Further studies are required to determine the distributions of this plasmid in other geographic areas and to understand its contribution to KPC epidemiology and virulence.
ACKNOWLEDGMENTS
This study was supported by a grant (to B.N.K.) from the National Institutes of Health (1R01AI090155). This work was also supported by Public Health Service grant R01AI072219 and R01AI063517 (to R.A.B.) from the National Institutes of Health and funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program, and the Geriatric Research Education and Clinical Center VISN 10 to R.A.B.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print 29 July 2013
REFERENCES
- 1.Naas T, Cuzon G, Villegas M-V, Lartigue M-F, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 53:1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53:4333–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob. Agents Chemother. 57:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob. Agents Chemother. 54:4493–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson TJ, Shepard SM, Rivet B, Danzeisen JL, Carattoli A. 2011. Comparative genomics and phylogeny of the IncI1 plasmids: a common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid 66:144–151 [DOI] [PubMed] [Google Scholar]
- 8.Meynell GG, Lawn AM. 1968. Filamentous phages specific for the I sex factor. Nature 217:1184–1186 [DOI] [PubMed] [Google Scholar]
- 9.Coetzee JN, Sirgel FA, Lecatsas G. 1980. Properties of a filamentous phage which adsorbs to pili coded by plasmids of the IncI complex. J. Gen. Microbiol. 117:547–551 [DOI] [PubMed] [Google Scholar]
- 10.Komano T, Kim SR, Nisioka T. 1987. Distribution of shufflon among IncI plasmids. J. Bacteriol. 169:5317–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaheen BW, Nayak R, Foley SL, Kweon O, Deck J, Park M, Rafii F, Boothe DM. 2011. Molecular characterization of resistance to extended-spectrum cephalosporins in clinical Escherichia coli isolates from companion animals in the United States. Antimicrob. Agents Chemother. 55:5666–5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Börjesson S, Bengtsson B, Jernberg C, Englund S. 2013. Spread of extended-spectrum beta-lactamase producing Escherichia coli isolates in Swedish broilers mediated by an IncI plasmid carrying blaCTX-M-1. Acta Vet. Scand. 55:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antunes P, Coque TM, Peixe L. 2010. Emergence of an IncIg plasmid encoding CMY-2 b-lactamase associated with the international ST19 OXA-30-producing b-lactamase Salmonella Typhimurium multidrug-resistant clone. J. Antimicrob. Chemother. 65:2097–2100 [DOI] [PubMed] [Google Scholar]
- 14.Wise PJ, Towner KJ, Webster CA, Slack RC, Jones TO. 1985. Trimethoprim resistance plasmids in Escherichia coli isolated from cases of diarrhoea in cattle, pigs and sheep. J. Appl. Bacteriol. 58:555–561 [DOI] [PubMed] [Google Scholar]
- 15.Jobanputra RS, Datta N. 1974. Trimethoprim R factors in enterobacteria from clinical specimens. J. Med. Microbiol. 7:169–177 [DOI] [PubMed] [Google Scholar]
- 16.Provence DL, Curtiss R., III 1992. Role of crl in avian pathogenic Escherichia coli: a knockout mutation of crl does not affect hemagglutination activity, fibronectin binding, or curli production. Infect. Immun. 60:4460–4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han J, Lynne AM, David DE, Tang H, Xu J, Nayak R, Kaldhone P, Logue CM, Foley SL. 2012. DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype Heidelberg isolates. PLoS One 7:e51160. 10.1371/journal.pone.0051160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Chavda KD, Mediavilla JR, Zhao Y, Fraimow HS, Jenkins SG, Levi MH, Hong T, Rojtman AD, Ginocchio CC, Bonomo RA, Kreiswirth BN. 2012. Multiplex real-time PCR for detection of an epidemic KPC-producing Klebsiella pneumoniae ST258 clone. Antimicrob. Agents Chemother. 56:3444–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 21.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, Chen Z, Chen S, Deng Y, Liu Y, Tian W, Huang X, Wu C, Sun Y, Zeng Z, Liu JH. 2010. Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob. Agents Chemother. 54:4219–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho YJ, Moon DC, Jin JS, Choi CH, Lee YC, Lee JC. 2009. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn. Microbiol. Infect. Dis. 64:185–190 [DOI] [PubMed] [Google Scholar]
- 24.Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60:394–397 [DOI] [PubMed] [Google Scholar]
- 25.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 26.García-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 63:274–281 [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J. Clin. Microbiol. 49:579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.CLSI 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—ninth edition (M07-A9) CLSI, Wayne, PA [Google Scholar]
- 30.CLSI 2013. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement, M100-S23 CLSI, Wayne, PA [Google Scholar]
- 31.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomis-Rüth FX, Sola M, Acebo P, Parraga A, Guasch A, Eritja R, Gonzalez A, Espinosa M, del Solar G, Coll M. 1998. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J. 17:7404–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komano T, Kubo A, Kayanuma T, Furuichi T, Nisioka T. 1986. Highly mobile DNA segment of IncI alpha plasmid R64: a clustered inversion region. J. Bacteriol. 165:94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komano T, Fujitani S, Funayama N, Kanno A, Sakuma K. 1990. Physical and genetic analyses of IncI2 plasmid R721: evidence for the presence of shufflon. Plasmid 23:248–251 [DOI] [PubMed] [Google Scholar]
- 35.Giakkoupi P, Papagiannitsis CC, Miriagou V, Pappa O, Polemis M, Tryfinopoulou K, Tzouvelekis LS, Vatopoulos AC. 2011. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009–10). J. Antimicrob. Chemother. 66:1510–1513 [DOI] [PubMed] [Google Scholar]
- 36.Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, Gales AC, Venezia SN, Quinn JP, Nordmann P. 2010. Worldwide diversity of Klebsiella pneumoniae that produce b-lactamase blaKPC-2 gene. Emerg. Infect. Dis. 16:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chmelnitsky I, Shklyar M, Hermesh O, Navon-Venezia S, Edgar R, Carmeli Y. 2013. Unique genes identified in the epidemic extremely drug-resistant KPC-producing Klebsiella pneumoniae sequence type 258. J. Antimicrob. Chemother. 68:74–83 [DOI] [PubMed] [Google Scholar]
- 39.Warburg G, Hidalgo-Grass C, Partridge SR, Tolmasky ME, Temper V, Moses AE, Block C, Strahilevitz J. 2012. A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6′)-Ib. J. Antimicrob. Chemother. 67:898–901 [DOI] [PubMed] [Google Scholar]
- 40.Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. 2010. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J. Antimicrob. Chemother. 65:243–248 [DOI] [PubMed] [Google Scholar]
- 41.García-Fernández A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56:2143–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leavitt A, Carmeli Y, Chmelnitsky I, Goren MG, Ofek I, Navon-Venezia S. 2010. Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob. Agents Chemother. 54:3002–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, Bojarska K, Zabicka D, Kania-Pudlo M, Mlynarczyk G, Zak-Pulawska Z, Hryniewicz W, Gniadkowski M. 2011. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. Antimicrob. Agents Chemother. 55:5493–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goren MG, Carmeli Y, Schwaber MJ, Chmelnitsky I, Schechner V, Navon-Venezia S. 2010. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg. Infect. Dis. 16:1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kassis-Chikhani N, Frangeul L, Drieux L, Sengelin C, Jarlier V, Brisse S, Arlet G, Decre D. 2013. Complete nucleotide sequence of the first KPC-2- and SHV-12-encoding IncX plasmid, pKpS90, from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57:618–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Chavda KD, Melano RG, Jacobs MR, Levi MH, Bonomo RA, Kreiswirth BN. 2013. Complete sequence of a blaKPC-2-harboring IncFIIK1 plasmid from a Klebsiella pneumoniae sequence type 258 strain. Antimicrob. Agents Chemother. 57:1542–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellata M, Maddux JT, Nam T, Thomson N, Hauser H, Stevens MP, Mukhopadhyay S, Sarker S, Crabbe A, Nickerson CA, Santander J, Curtiss R., III 2012. New insights into the bacterial fitness-associated mechanisms revealed by the characterization of large plasmids of an avian pathogenic E. coli. PLoS One 7:e29481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50 [DOI] [PubMed] [Google Scholar]
- 49.Sarno R, McGillivary G, Sherratt DJ, Actis LA, Tolmasky ME. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 46:3422–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice LB, Carias LL, Hutton RA, Rudin SD, Endimiani A, Bonomo RA. 2008. The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52:3427–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]